- 1College of Horticulture, Qingdao Agricultural University, Qingdao, China
- 2Haidu College, Qingdao Agricultural University, Laiyang, China
KNOTTED1-like homeobox (KNOX) transcription factors (TFs) belonging to the homeobox TF family play important roles in plant growth, development, and responses to abiotic and biotic stress. However, little information is available on KNOX TF in pear (Pyrus). In this study, 19 PbKNOXs TFs were re-identified in pear (Pyrus bretschneideri Rehd.). Phylogenetic analysis revealed that the TFs were clustered into three groups with 10 conserved motifs, some of which were group- or subgroup-specific, implying that they are important for the functions of the KNOX in these clades. PbKNM1 and PbKNM2 are KNM (encodes a MEINOX domain but not a homeodomain) genes identified in pear for the first time. KNOX genes in Pyrus and Malus were closely related, and a collinear relationship among PbKNOX genes in Pyrus and Malus was observed. Analysis of the expression patterns of PbKNOX genes in different tissues, at various growth stages, and in response to abiotic and biotic stress revealed that PbKNOXs are involved in plant growth and development. Our comparative transcriptional analysis of dwarf mutant varieties revealed that genes belonging to class I are highly expressed compared with genes in other classes. Analysis of the expression of PbKNOX genes in the hybrid offspring of vigorous and dwarf varieties revealed that PbKNOX genes were highly expressed in the vigorous offspring and weakly expressed in the dwarf offspring. These findings provide new insight into the function of KNOX TFs in pear and will aid future studies of dwarf fruit trees.
Introduction
Pear is one of the world’s most important fruits. In 2019, the world planting area of pear reached 1,379,387 ha, and the total output reached 23,919,075 tons. China’s output of pear accounts for 71.4% of the world’s output (FAO, 2021; http://www.fao.org/faostat/zh/#data/QCL/visualize). Labor-saving cultivation modes, such as dwarf close planting, have become important cultivation modes for fruit trees both within and outside of China for their various advantages, including early fruiting, high yield, high quality, ease of management, low costs, and high profit. However, the shortage of dwarf rootstocks or dwarf varieties has greatly impeded the development of the pear industry. The mechanism of dwarfism has become a major focus of research because of the development of biotechnology breeding.
Dwarfism is typically a result of reduced cell division and/or cell elongation. These processes are generally regulated by regulatory genes. Plant transcription factors (TFs) are important regulators that activate or inhibit the transcription of downstream genes during plant development and in response to environmental stimuli. Multiple TFs can play a role in dwarfism, including WRKY, BHLH, ARF, NAC, and homeobox TFs (Kato et al., 2010; Ou et al., 2015; Zhang et al., 2015). Previous studies have shown that there is an important relationship between internode development and shoot tip meristem and that homeobox family genes play an important role in shoot tip development. The homeobox is a conserved 60-amino acid motif called the homeodomain (HD), which can bind to specific DNA sequences. The HD has a three-helix structure and plays a role in development in all eukaryotic organisms. HD-containing proteins can be classified into six groups based on differences in the sequence, size, and location of the HD: homeodomain-leucine zipper (HD-Zip), Wuschel-related homeobox (WOX), plant homeodomain associated to a finger domain (PHD-finger), bell domain (BELL), knotted related homeobox (KNOX), and zinc finger-homeodomain (ZF-HD; Moens and Selleri, 2006; Ariel et al., 2010).
KNOX family members have been shown to be involved in plant dwarfism and internode development. The first KNOTTED1 gene to be identified in plants was KNOTTED1 (kn1) in maize (Vollbrecht et al., 1991). Following this discovery, several studies of KNOTTED1 proteins have been carried out in model and non-model plants, named KNOTTED1-like. The functions of KNOX proteins have been studied extensively in Arabidopsis, rice, and cotton (Bhattacharjee et al., 2016; Yang et al., 2017; Wang et al., 2019). Based on sequence similarity, intron position, expression patterns, and phylogenetic analysis, the KNOX genes can be divided into two subclasses: class I and class II (Furumizu et al., 2015). The functions of class I KNOX genes have been extensively studied. In Arabidopsis, there are four class I genes: SHOOTMERISTMELESS (STM), KNAT1(BP), KNAT2, and KNAT6. The gene STM is essential for the formation and maintenance of the shoot apical meristem (SAM; Aguilar-Martinez et al., 2015). KNOX1 proteins form heterodimers with other HDs (e.g., BEL-like HD) in the TALE superclass and regulate the activity of downstream genes with different combinations of KNOX/BLH TFs (Arnaud and Pautot, 2014). BELL1-like homeobox (BLH) TFs are known to heterodimerize with KNOTTED1-like homeobox TFs and play important roles in shoot meristem maintenance, maintain intercalary meristems that accumulate KN1, and promote internode elongation (Tsuda et al., 2017). Other class I proteins can regulate flower patterning, promote the outgrowth of primordia, create leaf tips, and create the basipetal streams of auxin from the tip through the internal tissue to induce the differentiation of the vascular strand (Thomas et al., 2006; Wenzel et al., 2007; Li et al., 2012a).
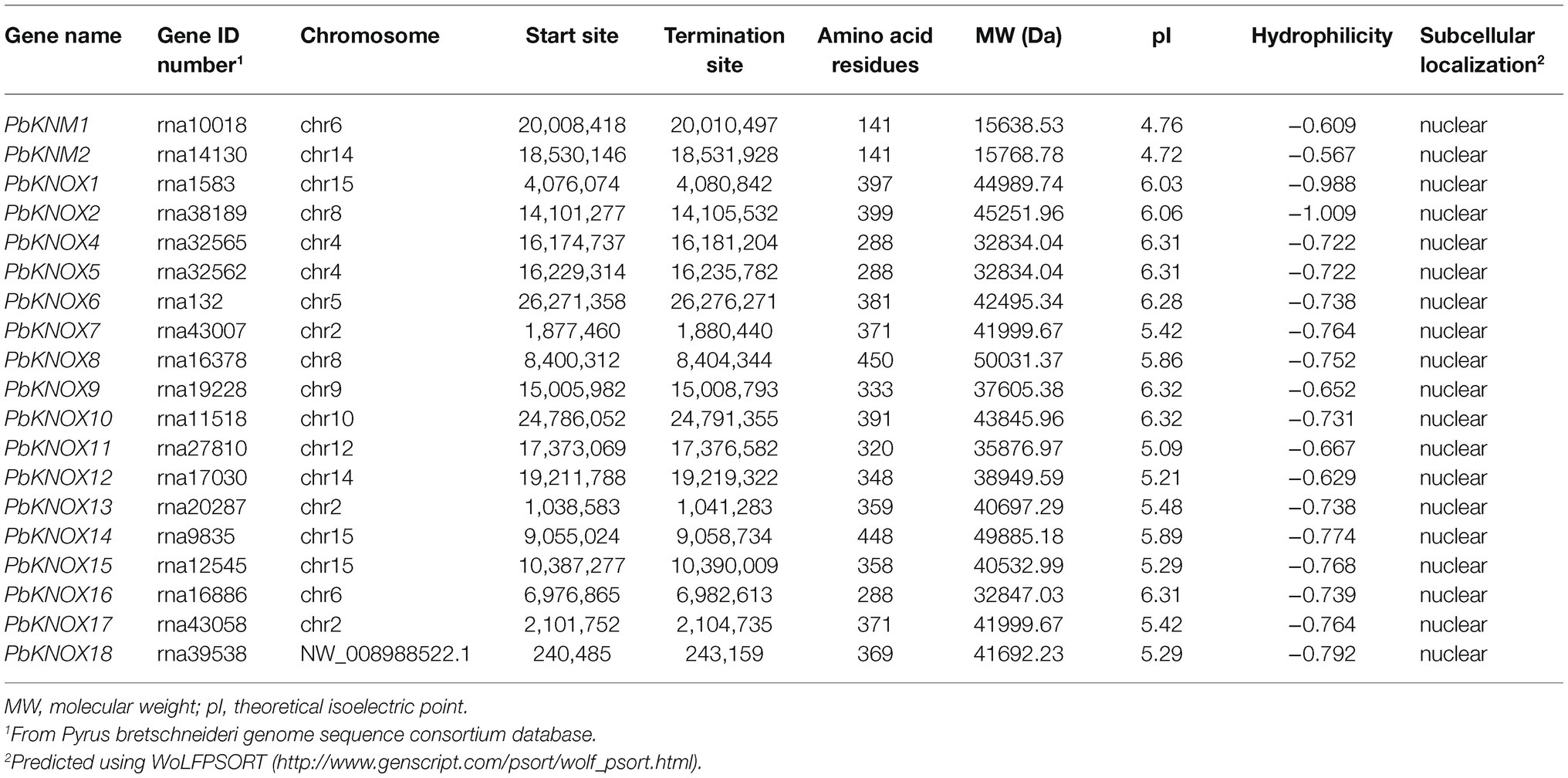
Among class II genes, MdKNOX19 in apple is a positive regulator of ABI5 expression, and the conserved module MdKNOX19-MdABI5-ABA may contribute to organ development. However, functional studies examining class II genes are fewer than those examining class I genes (Jia et al., 2021). Class I and class II genes perform non-redundant functions to control the development of all the aboveground organs of plants (Furumizu et al., 2015). With the continually increasing amount of plant genome data published, genome-wide identification and analysis have become effective methods for rapidly predicting gene functions from a large family of genes. Several gene families in plants have been studied. Eighteen PbKNOXs family genes were identified in pear. Based on the phylogenetic tree and chromosomal localization, the 18 PbKNOX genes were divided into five subfamilies [SHOOT MERISTEMLESS (STM)-like, BREVIPEDICELLUS (BP)-like, KNOTTED ARABIDOPSIS THALIANA 2/6 (KNAT2/6)-like, KNAT7-like, and KNAT3-5-like] and were distributed among 10 chromosomes (Cheng et al., 2019). With the improvement of genomic data, the bioinformatics of these PbKNOXs have changed, such as the characteristics of protein, sequence information, and chromosome location. Therefore, in this study, we re-identified the relevant information to improve the accuracy of sequence information and bioinformatics information of related proteins.
According to previous studies, we speculate that KNOXs family genes are important dwarfing-related genes. Dwarf negative regulatory genes are important for genetic engineering breeding by gene editing. However, dwarfism is often accompanied by weak plant growth and survival challenges for plants. “Nain Vert,” a French pear cultivar with short internodes and stature, originated from a chance seedling (Bagnara and Rivalta, 1989). This germplasm resource can be used in crossbreeding to create new dwarf scion cultivars with high quality. We introduced seeds of “Nain Vert” from the East Malling Research Station, West Malling, United Kingdom, in 1991. Several seedlings were then raised, and one dwarf seedling showing similar features to “Nain Vert” was identified and named “Aihuali.” “Chili,” which belongs to Pyrus bretschneideri Rehd., is famous for its fruit quality and is mainly cultivated in Shandong Province, China. It has higher photosynthetic capacity, short internodes, and thick branches; it is thus an excellent material for studying the mechanism underlying dwarfism.
Here, we re-conducted a genome-wide analysis of Chinese white pear (Pyrus bretschneideri Rehd.) PbKNOXs investigated their potential functions in plant development and responses to stress. We used dwarf pear materials to identify the expression pattern of PbKNOXs in dwarfism.
Materials and Methods
Database Search and Sequence Retrieval
The complete genome assembly of pear (Pyrus bretschneideri Rehd.) along with the complete proteome sequence file was downloaded from the Genome Database for Rosaceae.1 Conserved amino acid sequences were used as a query to identify PbKNOXs TFs in the Pyrus proteome sequence file using CLC Sequence Viewer v7.6.1 (Jin et al., 2017).
Putative PbKNOX TFs were confirmed by BLASTP searches of the NCBI database. This database was also used to obtain gene accession numbers, chromosome number, genomic information, and the protein size of KNOX TFs. The nucleotide sequences of all identified PbKNOX were also retrieved from NCBI. The ExPASY online tool2 was used to calculate molecular weight, pI, and GRAVY of all PbKNOX proteins. WoLF PSORT3 was used to predict the subcellular localization of PbKNOX proteins.
Chromosomal Mapping, Intron/Exon Distribution, Conserved Domain, and cis-Elements Analysis
The NCBI database was used to record the positions of the identified PbKNOX genes on chromosomes, and Map Chart v2.32 was used to draw a map of chromosomal locations of PbKNOX genes to scale (Voorrips, 2002). Gene Structure Display Server (v2.0 http://gsds.cbi.pku.edu.cn/) was used to draw and visualize the intron/exon organization of PbKNOX genes (Hu et al., 2014). The whole genome sequence and the coding sequences of all PbKNOX genes were used to construct the gene structure map containing introns. In order to identify domains conserved among all PbKNOXs, their protein sequences were analyzed with MEME v5.0.3 (Bailey et al., 2015) using default parameters, except that the minimum number of motif sites was set to 10. The 2,000-bp sequence upstream from each PbKNOX initiation codon was analyzed, and the online tool PlantCARE4 was used for the prediction of cis-elements (Lescot et al., 2002).
Phylogenetic Comparison of Homoebox and KNOX Proteins in Pyrus bretschneideri, Malus domestica, and Arabidopsis thaliana
A phylogenetic tree was constructed using the protein sequences of putative Homeobox TFs from Pyrus bretschneideri. Another phylogenetic tree was constructed using the protein sequences of putative KNOX TFs from Pyrus bretschneideri and Malus domestica, with Arabidopsis thaliana sequences used as a reference (Supplementary Table S1). Multiple sequence alignment of the proteins was performed using ClustalW v1.83. The tree was used to infer the evolutionary history and functions of PbKNOX TFs. Their coding sequences were used for pairwise alignment with the built-in ClustalW and PAM protein weight matrix of MEGA7 (Kumar et al., 2016). The resultant alignments were analyzed for DNA sequence polymorphisms and DnaSP v5.10.01 (Librado and Rozas, 2009) was used to compute the synonymous substitution rate (Ks) and nonsynonymous substitution rate (Ka). Ks/Ka was also calculated to evaluate codon selection during evolution.
Expression Patterns of PbKNOXs in Various Tissues of Pear at Different Developmental Stages
The expression patterns of PbKNOX genes in various tissues at different developmental stages were obtained from the pear gene expression atlas. Bud was collected from flower bud differentiation stage. Leaf, petal, sepal, oval, and stem were collected from full flowering stage. Stem was the stem tissue of annual branches, and fruit was collected from commercial maturity stage. The raw sequence reads from seven pear tissues were analyzed using the NCBI web browser (https://www.ncbi.nlm.nih.gov/bioproject/; accession no. PRJNA498777; Li et al., 2019a). The expression patterns of PbKNOX genes at three different stages of ovary development and under three treatments [unpollinated (control), hand-pollinated, and unpollinated overy with GA treatment] were analyzed (Liu et al., 2018a, 2021). For the analysis of expression patterns of PbKNOX genes in leaf samples at different developmental stages of pear, leaves were collected at 30, 45, 60, 75, and 90 DAB; the RNA sequencing (RNA-seq) data were downloaded from the Sequence Read Archive (SRA; accession nos. SRR10997902–SRR10997912; Zhao et al., 2020). Finally, the expression profiles of PbKNOX genes based on RNA-seq data from four pear varieties at seven fruit development stages were downloaded from the SRA (accession no. SRP070620). The following fruit development stages were analyzed as: fruit setting (period 1), physiologic fruit dropping (period 2), rapid fruit enlargement (period 3), 1 month after fruit enlargement (period 4), pre-maturity (period 5), and maturity (period 6); additionally, one fruit senescence stage after harvest (period 7) was included in the analysis (Zhang et al., 2016).
Expression Patterns of PbKNOXs Under Different Stress Conditions
Expression patterns of PbKNOXs in response to abiotic and biotic stress were based on microarray data downloaded from the SRA (series matrix accession nos. SRP051914 and SRP148620; Yang et al., 2015; Li et al., 2016) and data from the present study.
Salt Stress
Two varieties of pear, “QAUP-1” (Pyrus ussuriensis Maxim.) and “Qingzhen D1” (Pyrus communis L. × Pyrus bretschneideri Rehd.), were used. Salt stress treatment was applied as: half-strength nutrient solution and 100 mmol/L NaCl. Leaf and root samples were collected from the plants 0, 12, and 24 h after treatment along with control samples. Each treatment contained three replicates of 50 plants (Liu et al., 2021).
Drought Stress
Pyrus betulaefolia plants used in this experiment were 3 months old.
Seedlings were transferred to clean filter paper (90 mm × 90 mm) and allowed to dry for 0, 1, 3, and 6 h at 26°C, followed by recovery in water at 26°C for 24 h. Each treatment contained three biological repeats (Li et al., 2016; Liu et al., 2021).
Cork Spots
The superior pear line “1–43” is the hybrid from “Xinli No. 7” (Pyrus bretschneideri Rehd.) × “Zhongxiang Pear” (Pyrus bretschneideri Rehd.). Its fruit showed cork spots at 180 days after anthesis. We took normal pear flesh without disease, pear flesh with moderate disease, and pear flesh with serious cork spots 180 days after flowering as materials, including three biological repeats (Liu et al., 2021).
Pear Black Spot Disease
The spore suspension was sprayed into detached young leaves of “Jinjing” and “Hongfen” pear varieties with a glass atomizer. Control leaves were sprayed with distilled water. Samples H-CK, H-P, J-CK, and J-P were used in the experimental design and data analysis. All of the samples were tested in triplicate, and the experiments were performed on three biological replicates (Yang et al., 2015; Liu et al., 2021).
Expression Patterns of PbKNOXs in Pear Varieties With Different Growth Potential
The experiment was carried out in the Jiaozhou Experiment and Demonstration Station of Qingdao Agricultural University (36.44°N, 120.09°E), which features a warm temperate monsoon climate. The branches of dwarfing pear “601D” were treated with Co-r (60; 5,000 rad) radiation and then grafted on the rootstocks of one-year-old Pyrus betulifolia Bunge. One vigorous mutant was observed and referred to as “601V.” “601D” and “601V” were then grafted on the five-year-old rootstocks of Pyrus betulifolia Bunge. “Laiyang chili” pear and dwarf pear were used for hybridization. There were two phenotypes in the hybrid offspring population: non-dwarf varying in height and dwarf. Fifteen individual trees of arboreal and dwarf progenies were selected for the expression pattern analysis of PbKNOX genes. The samples were collected from the shoot tips of three-year-old branches. All samples were immediately placed into liquid nitrogen and then stored at −80°C until further use.
Plant Growth Conditions and Quantitative Real-Time PCR Analysis
Total RNA of pear peel samples at the three different stages was extracted using the RNAprep Pure Plant Plus Kit (Tiangen, Beijing, China) and treated with DNase I (RNase-free; Takara Bio, Dalian, China) to eliminate residual contaminating genomic DNA. For qRT-PCR, 1.5 μg total RNA was used to synthesize first-strand cDNA with the PrimeScript II First-Strand cDNA Synthesis kit (Takara Bio) according to the manufacturer’s instructions. qRT-PCR amplification was carried out as follows: 95°C for 5 min, 45 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s using the Roche 480 real-time PCR system (Basil, Switzerland) in standard mode with the FastStart Essential DNA Green Master kit. All reactions were performed in triplicate at a volume of 20 μl, containing 2 μl of 10-fold diluted cDNA. The Pyrus ACTIN gene was used as the reference to normalize the qRT-PCR data, and relative gene expression levels were determined via the 2−ΔΔCT method (Livak and Schmittgen, 2001). All experiments were performed with three biological replicates and all primer sequences are shown in Supplementary Table S2.
Statistical Analysis
Statistical analyses were conducted using SPSS 23.0 (SPSS, Chicago, IL, United States). Values are represented as the mean ± SD of three independent biological replicates. Data were analyzed with Duncan’s test, and p ≤ 0.05 was considered significant.
Results
Identification and Phylogenetic Analysis of PbKNOX TFs in Pear
Putative pear PbKNOX sequences were identified by searching the Rosaceae genome using the Basic Local Alignment Search Tool for protein sequences (BLASTP). A total of 19 KNOX protein sequences were screened based on domain and named according to Cheng’s research (Table 1; Cheng et al., 2019). However, the PbKNOXs protein sequence has been modified in the latest reference genome. In previous studies, PbKNOX3 was not located on the chromosome, and no similar sequence was found in the latest version, which was deleted (Supplementary Table S1). Two PbKNM genes were found according to the conserved domain. All of the identified family members contained the KNOX I and/or KNOX II domains. The polypeptide lengths of the predicted pear PbKNOXs ranged from 141 to 450 amino acids. The predicted subcellular localization was the nucleus for all PbKNOXs. The molecular weight, isoelectric point (pI), and grand average of hydropathicity (GRAVY) are summarized in Table 1. We examined the phylogenetic relationships among pear homeobox and PbKNOX genes by generating a phylogenetic tree using the neighbor-joining method. A total of 72 homeobox genes were identified in pear, and the family was divided into five groups. Among them, 17 genes in KNOX group are PbKNOX genes. Therefore, we analyzed the evolutionary relationships among these 19 genes with 21 Malus domestica and eight Arabidopsis KNOX genes (Figure 1). The family was divided into three groups (Figure 1). PbKNOX6, PbKNOX2, PbKNOX9, PbKNOX10, PbKNOX11, PbKNOX12, and PbKNOX1 were more closely related to STM, KNAT1, KNAT2, and KNAT6, which belong to the class I family. KNATM and three KNMs of apple were closely related to PbKNM1 and PbKNM2. The remaining PbKNOXs are homologous to class II genes.
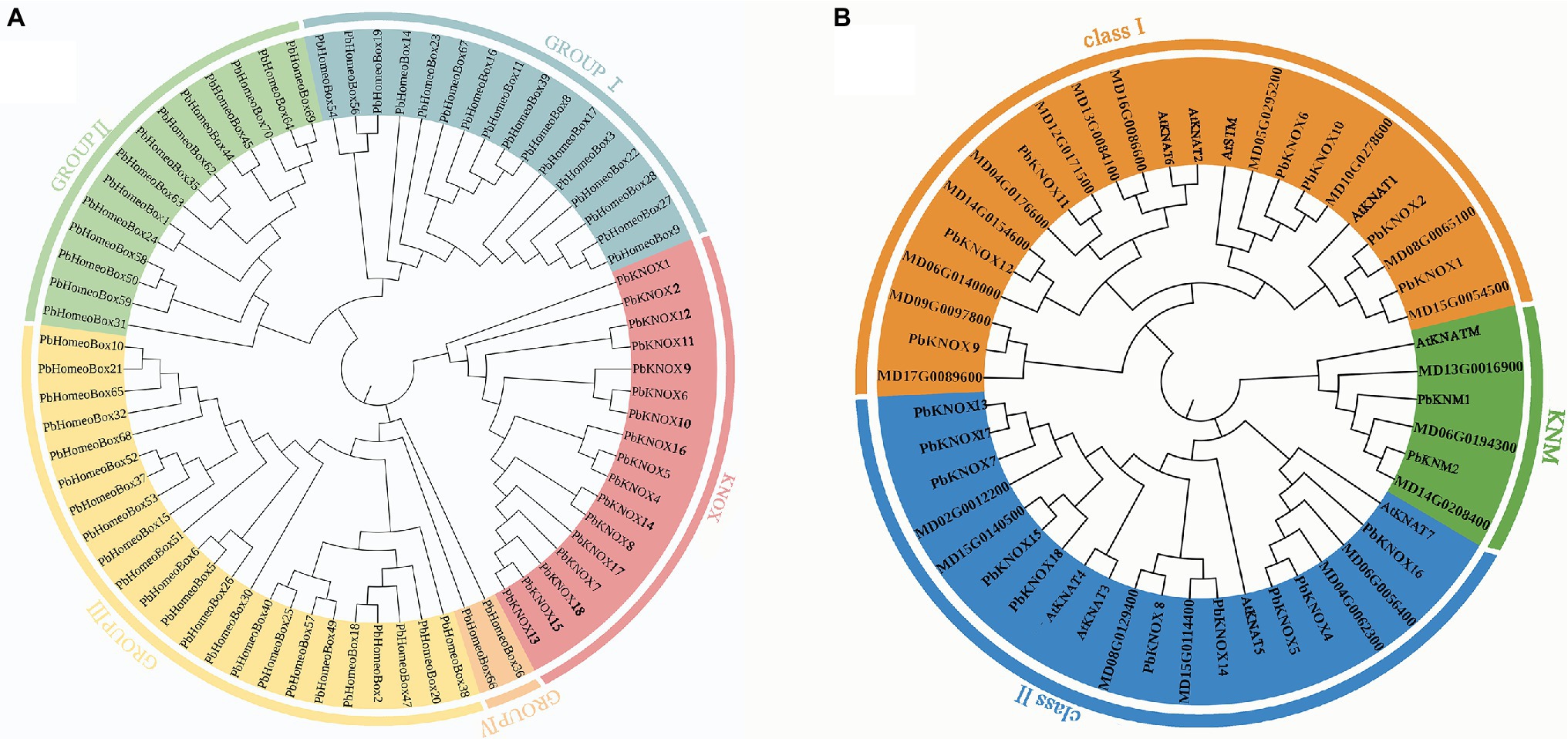
Figure 1. Phylogenetic tree constructed with the neighbor-joining method using homeobox TF domains in Pyrus bretschneideri (A) and KNOX TF domains in Pyrus bretschneideri, Malus domestica, and Arabidopsis (B). The tree was divided into three groups (class I, class II, and KNM).
Chromosomal Distribution and Orthologous Relationships of PbKNOX Genes in Pyrus bretschneideri, Malus domestica, and Arabidopsis thaliana
Physical mapping of PbKNOXs on the 17 chromosomes of P. bretschneideri revealed that the distribution of PbKNOXs was uneven (Figure 2); the genes were distributed on 10 chromosomes, with one or two genes per chromosome. The scaffold of PbKNOX18 genome database is not attached to the chromosome, which may be a problem of gene assembly. The exact position (in bp) of each PbKNOX is shown in Table 1. Two PbKNOX genes were tandem repeats, and 12 (six pairs) were segmentally duplicated (Figure 2B). The Ka/Ks ratios of the gene pairs displaying segmental duplication were less than 1, indicating that they might have undergone purification selection (Supplementary Table S4). To establish the orthologous relationships of PbKNOXs, we compared the physical location of PbKNOXs in the genomes of Pyrus bretschneideri, Malus domestica, and Arabidopsis thaliana (Figure 2C). PbKNOX6, PbKNOX10, and PbKNOX1 were closely related to KNOX in Arabidopsis. There was a collinear relationship among KNOX in Malus and Pyrus.
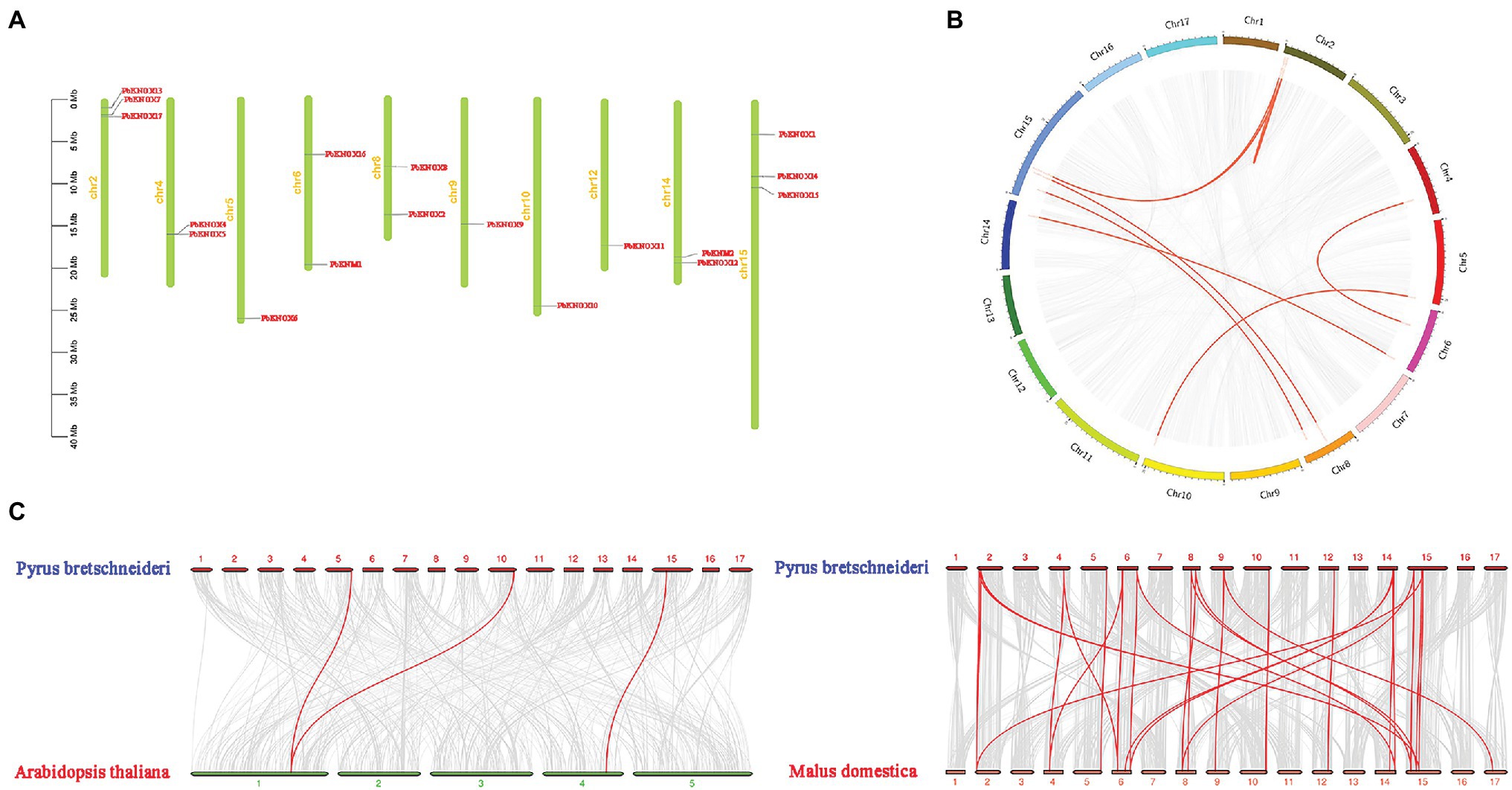
Figure 2. Chromosomal distribution of PbKNOX genes drawn using MapChart v2.2. The scale represents chromosome length: 0–40 Mb (A). Collinearity of KNOXs (B). Distribution of segmentally duplicated PbKNOX genes on Pyrus bretschneideri chromosomes. Gray lines indicate collinear blocks in the whole Pyrus bretschneideri genome, and red lines indicate duplicated PbKNOX gene pairs. (C) Collinearity of Pyrus bretschneideri, Arabidopsis, and Malus domestica KNOX genes. Red lines indicate syntenic gene pairs.
Cis-Elements, Conserved Motifs, and Gene Structure of PbKNOX TFs
A total of 19 PbKNOX promoter regions (2,000 bp) were analyzed. There was a large number of cis-acting elements in the promoter region, which mainly included hormone signals and stress regulatory networks, such as the GARE-motif, P-box (related to gibberellins), CGTCA-motif, and TGACG-motif (related to JA; Supplementary Table S3).
The conserved motifs of PbKNOXs were analyzed using Multiple Em for Motif Elicitation (MEME). Ten conserved motifs were predicted in PbKNOXs (Figure 3; Supplementary Figure S1); their sizes ranged from 11 to 80 amino acids. Motif 3 was present in all PbKNOXs; motif 2 was detected in class II members; motif 1 was present in class I and class II group members; motif 9 was present in KNM and class I group members; the other motifs were observed in class II members; and motif 5 and motif 10 were detected in PbKNOX10. Most PbKNOX genes had 4 ~ 7 exons and 3 ~ 6 introns (Figure 3).
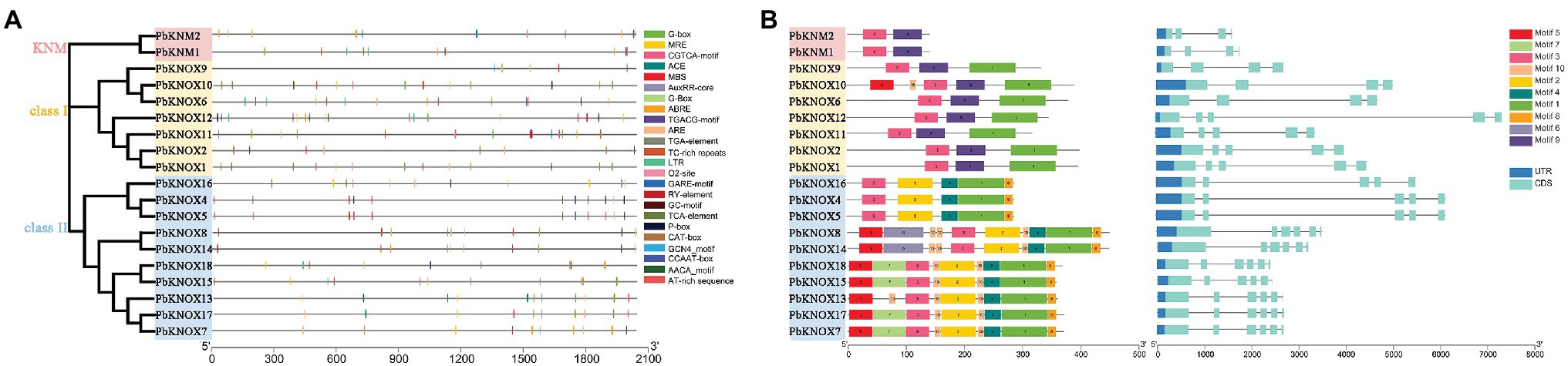
Figure 3. A list of motifs detected in the promoter regions of PbKNOX genes (A). Schematic representation of protein and gene structures of Pyrus bretschneideri PbKNOX genes (B). Motifs 1–10 identified using the MEME search tool are marked on the protein sequences in each clade (1–3). The length and order of each motif correspond to the actual length and position of each motif in the protein sequences, respectively. The coding sequence and untranslated regions are represented by filled dark blue and orange boxes, respectively. Evolutionary analysis shows the homology of PbKNOXs in pear.
Expression Pattern of PbKNOX Genes During Pear Development
We analyzed the spatiotemporal expression patterns of PbKNOX genes in seven different tissues and at different developmental stages using publicly available gene expression data. Of the examined PbKNOX genes, one was not expressed in any of the tissue types; most of the genes were expressed in all seven tissue types, and their expression was constitutive (fragments per kilobase of transcript per million mapped reads (FPKM) > 10). Among class I genes, PbKNOX6, PbKNOX2, PbKNOX9, PbKNOX10, and PbKNOX1 were highly expressed in buds and stems, especially stems (Figure 4). During leaf development, the expression of PbKNOX5 decreased gradually with leaf development time in class II, and the expression of PbKNOX16 and PbKNOX14 increased gradually with leaf development time. However, the expression of genes was low in class I and KNM group, and there was no significant difference in the expression of these genes between these two groups (Figure 4A). During ovary development, the expression of genes in class I and class II was significantly different. In class I, the expression of PbKNOX11/9/10 became downregulated as development progressed, and in class II, the expression of most genes became upregulated as development progressed (PbKNOX13/15/18/14/8/16; Figure 4B). PbKNOXs are involved in fruit development, and PbKNOX14/8/12 were highly expressed (fragments per kilobase of transcript per million mapped reads (FPKM) > 10) in four pear varieties.
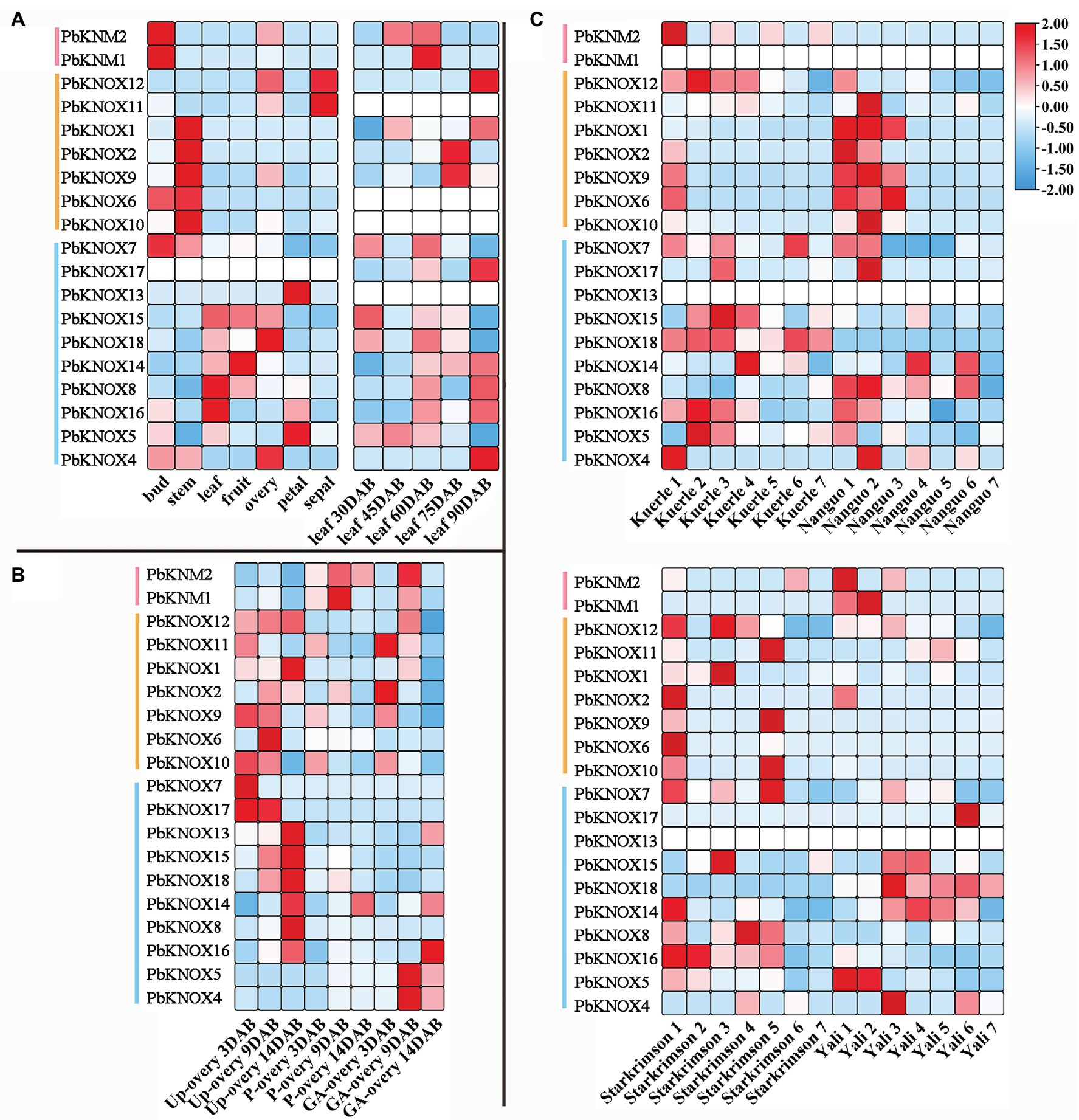
Figure 4. Expression profiles of PbKNOXs in pear. (A,B) Expression profile of PbKNOXs in leaf (A) and ovary at different development stages (B) of Pyrus bretschneideri. (C) Expression profile of PbKNOXs at different fruit development stages in various Pyrus bretschneideri varieties. P-ovary, pollinated ovary; Up-ovary, unpollinated ovary.
Expression of PbKNOX Genes in Response to Abiotic and Biotic Stress
To determine whether PbKNOXs expression is affected by different types of stress, we analyzed the expression patterns of genes following exposure to abiotic and biotic stress (Figure 5). Under salt stress, the expression of class I and class II group genes significantly differed among tissues. Most genes in class II group were highly responsive to salt stress in leaves, whereas genes in class I group were highly expressed in roots. Comparison of expression under drought stress and black spot infection revealed that the expression of class II group genes was high under different types of stress. The expression of class I and KNM group genes was lower across all treatments. The expression of PbKNOX7/13 increased under drought stress and decreased after plants were covered with water, whereas the expression of PbKNOX5/16 was inhibited under drought stress and increased after plants were covered with water.
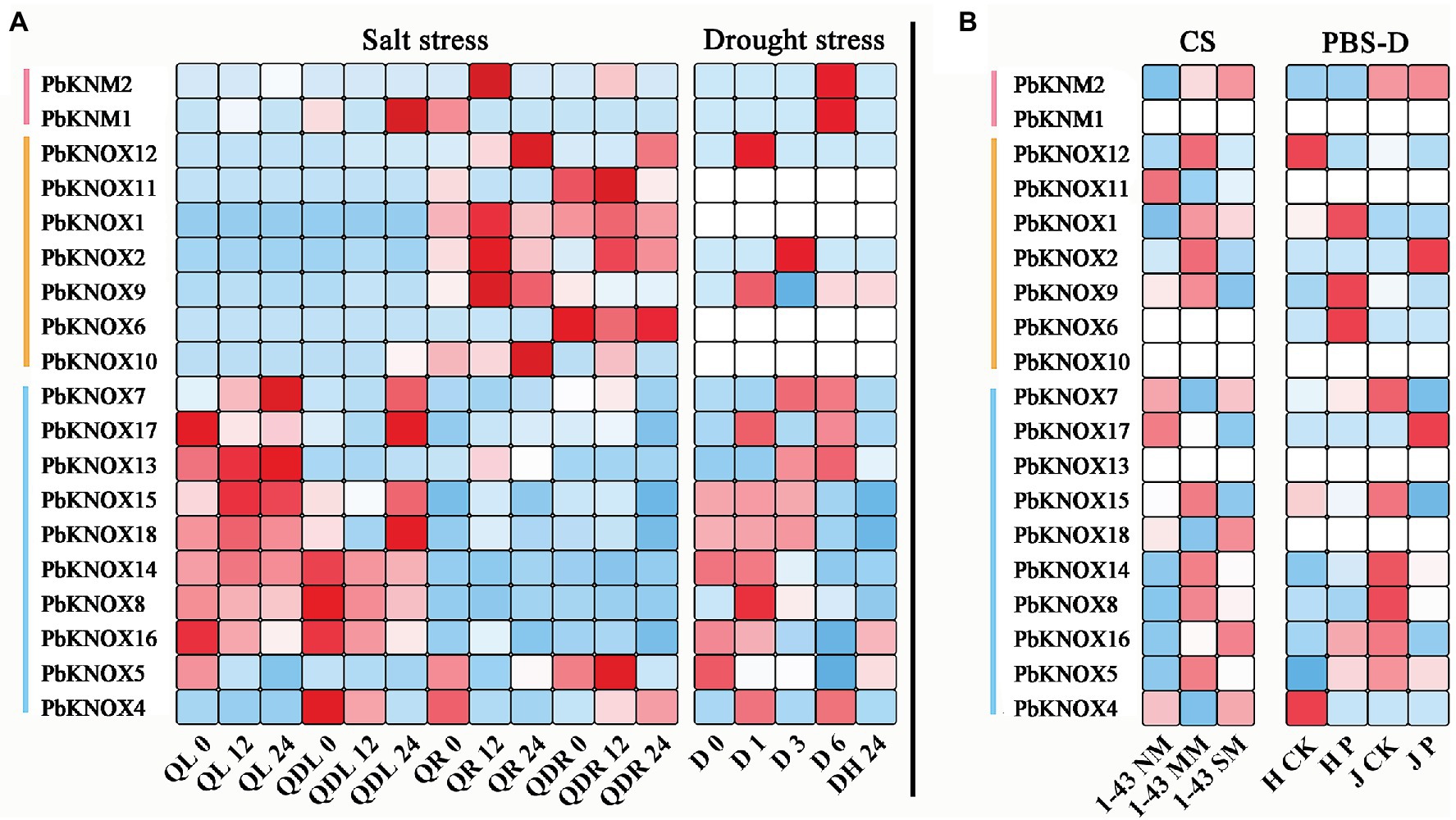
Figure 5. Expression profiles of PbKNOX genes under abiotic and biotic stress. (A) Abiotic stress included salt and drought stress. (B) Biotic stress included cork spot and pear black spot disease. CS, cork spot; MM, moderate disease mesocarp; NM, normal mesocarp; PBS-D, pear black spot disease; QL, “Qaup-1” leaf, QR, “Qaup-1” root; QZL, “Qingzhen D1” leaf; QZR, “Qingzhen D1” root; and SM, severe disease mesocarp.
Expression of PbKNOX Genes in Dwarf Plants
We used the dwarf pear variety “601D” and its vigorous mutant “601V” to study the role of PbKNOXs in dwarf plants. The internodes of “601D” were shorter than those of “601V”; the tree body was dwarf, the tree was sturdy, and the branches were significantly higher than those of “601V” (Figure 6A). A total of 2,741 differentially expressed genes were identified, including 1,636 upregulated and 1,105 downregulated genes. PbKNOXs in class I were all differentially expressed (log2fold change >5; Figure 6B). The expression of PbKNOX genes was much higher in “601V” than in “601D” (Figure 6C).
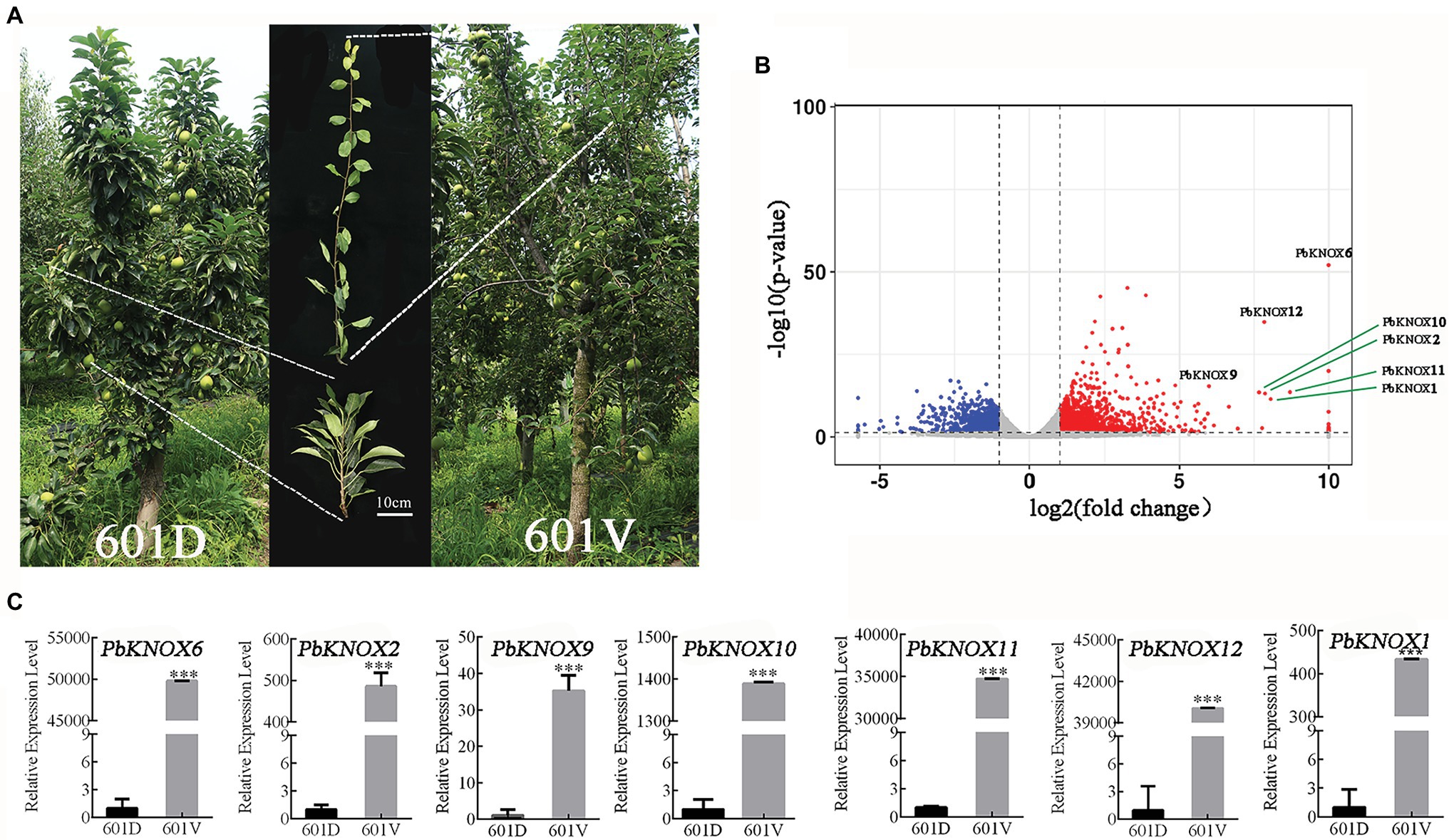
Figure 6. Expression profile of PbKNOXs in “601D” and the new vigorous mutant “601V.” (A) Photographs of “601D” and “601V.” (B) Volcano map of differentially expressed genes in the “601D” and “601V” transcriptome. (C) Expression profile of PbKNOXs in “601D” and “601V” pear. ***p < 0.001 (Student’s t-test). The expression levels of genes in “601D” pear under control conditions were normalized to 1.0.
We used the hybrid progeny population of “Laiyang Chili” pear and dwarf pear to screen 30 plants: 15 non-dwarf plants and 15 dwarf plants. The dwarf population had the dwarf phenotype, which is characterized by short internodes and thick branches. The expression of PbKNOX genes in class I group was significantly higher in vigorous plants than in dwarf plants (Figure 7).
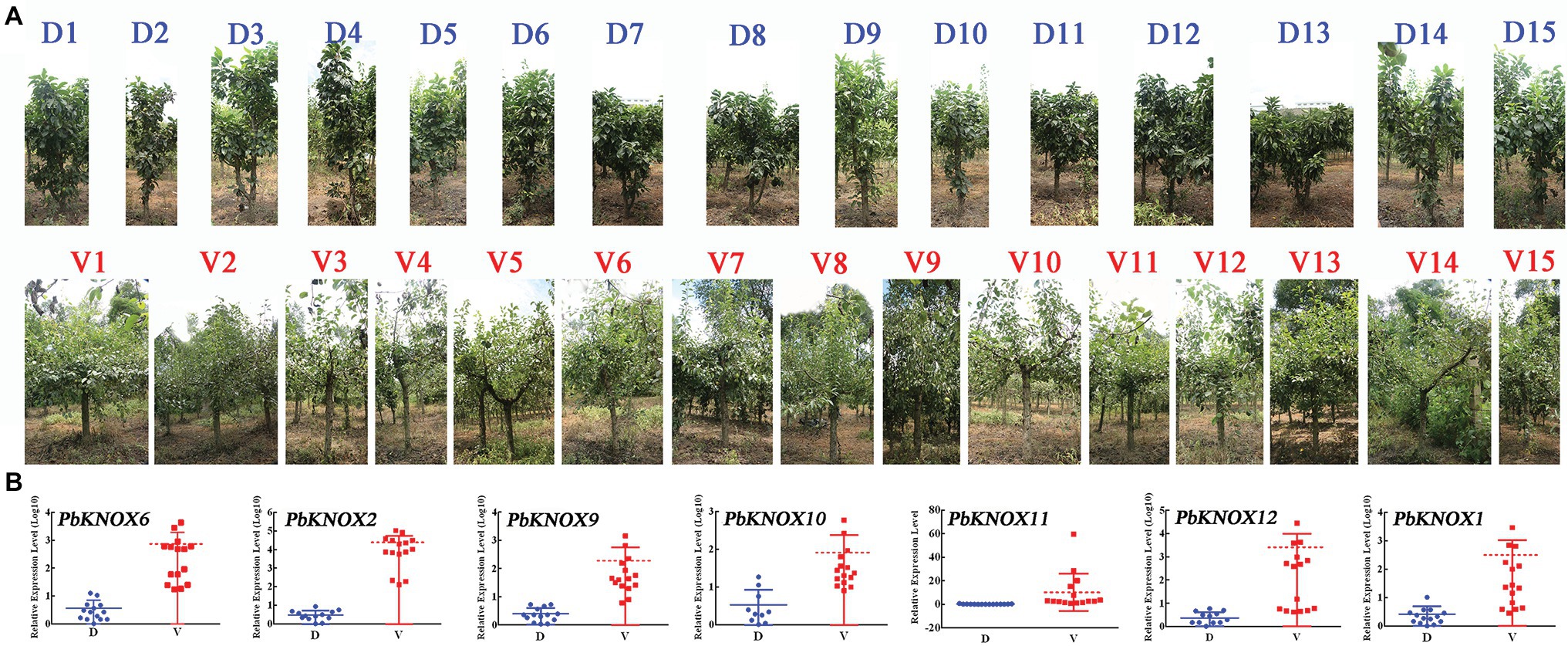
Figure 7. Expression profile of PbKNOXs of hybrid progeny. (A) Representative types of 15 dwarf and 15 vigorous pear trees. (B) Expression pattern of PbKNOX class I genes in offspring.
Discussion
Homeobox genes, including HD-Zip, WOX, PHD-finger, BELL, KNOX, and ZF-HD genes, have been studied in other species. These genes are involved in early leaf development and lateral branch growth. They also respond to different types of stress (e.g., aluminum stress and drought stress) and play a role in the signal transduction of multiple hormones (Nakata et al., 2012; Alvarez et al., 2015; Lim et al., 2018; Li et al., 2019b; Feng et al., 2020). Previous studies have conducted bioinformatics analysis on the PbKNOX gene family, and the research shows that there are 18 PbKNOX genes in pear. In our study, we named it according to sequence, chromosome location, and previous naming (Cheng et al., 2019). However, PbKNOX3 is not displayed in the current study, because there is no chromosome location and there is no consistent sequence in sequence alignment. Meanwhile, two PbKNM genes were identified. Compared with previous studies, the sequence information of nine genes was corrected, of which seven had amino acid deletion. Due to incomplete chromosome assembly, the length of some chromosomes is not enough, and gene chromosome mapping is inaccurate. So we used the latest chromosome information to relocate all PbKNOXs. In this study, a total of 72 homeobox proteins were identified in Pyrus bretschneideri. There were 17 proteins in KNOX group. Evolutionary analysis of the homologs of pear PbKNOXs in Arabidopsis and Malus was conducted. PbKNOX6/2/9/10/11/12/1 are closely related to STM, KNAT1, KNAT2, and KNAT6 in Arabidopsis, indicating that these are class I genes. We identified two PbKNM paralogs in Pyrus and three paralogs in malus; PbKNM1 and PbKNM2 have high homology with MdKNM in apple. Conservative domain analysis of these two proteins revealed the loss of the homeobox domain. Previous studies have shown that KNATM family KNOX proteins (i.e., proteins with only KNOX1 and KNOX2 domains) occur exclusively in eudicots, and some species contain more than one homolog of the KNM protein (Gao et al., 2015). PbKNM1 and PbKNM2 are KNM genes identified in pear for the first time in this study.
The number of PbKNOX genes in pear (19) is greater than the number of PbKNOX genes in Arabidopsis (8); this might stem from gene duplication in plants, which is considered a fundamental driver of the evolution of genomes (Kong et al., 2007), as it provides raw material for new genes that can then lead to the emergence of new functions. Segmental duplication, tandem duplication, and transposition events, such as retro- and replicative transposition (Moore and Purugganan, 2003), are the three main mechanisms of gene evolution; the first two are thought to underlie gene family expansion in plants (Cannon et al., 2004). Tandem duplications can be identified based on the presence of multiple members of a single gene family within the same or in neighboring intergenic regions (Ramamoorthy et al., 2008). Among the 19 PbKNOX genes in pear, there were one groups of tandem repeats (PbKNOX4 and PbKNOX5). We also detected six segmental duplication pairs involving 12 PbKNOX genes, suggesting that segmental duplication was the main driver of the expansion of PbKNOX genes in Pyrus. We also found that tandem repeats and segmentally duplicated pairs were concentrated in class II subfamily, which might indicate that the genes in the class I family are more conservative. Variation in the class II family contributes to the increase in the number of PbKNOX genes in pears. A collinear relationship of PbKNOX6 and PbKNOX10 with STM, KNAT2, KNAT4, and KNAT5 in Arabidopsis was detected. A total of 17 PbKNOX genes in pear were collinear with KNOX genes in Malus. Syntenic analysis of KNOX genes in Pyrus, Arabidopsis, and Malus showed that PbKNOX genes had higher homology with KNOX genes in Malus and lower homology with those in Arabidopsis.
In plants, gene expression is regulated by various physical and chemical factors. Regulation of expression at the transcriptional level is the most important mode of gene expression regulation. Cis-acting elements in the promoter, terminator, and UTR sequences play an important role in regulating expression. Several cis-elements responsive to plant growth regulation, auxin, GA, SA, biotic stress, and abiotic stress are present in the promoter of PbKNOX genes. This indicates that PbKNOX genes are induced by different factors, which is consistent with the observed expression patterns related to growth, development, and stress. Conserved domains or amino acid motifs in TFs are frequently involved in DNA binding, nuclear localization, protein–protein interactions, and transcriptional activity (Du et al., 2013), and TFs with similar domains or motifs likely have similar functions. KNM is a subfamily gene with the least conserved domain. The expression of PbKNM1 and PbKNM2 was generally low. Motif 2 and motif 4 conserved domains in class II group are unique, and motif 9 is unique in class I group. The distribution of these motifs may account for the variable responses of PbKNOXs to different conditions.
Based on the above analysis, we used transcriptome data to analyze the expression patterns of 19 PbKNOX genes in the process of growth and development and under biotic and abiotic stress to explore the possible functions of PbKNOXs. Most PbKNOX genes identified in our study are involved in these processes. Moreover, PbKNOX genes show significant tissue-specific expression, in which genes in class I group are highly expressed in growth point areas, such as the stem and bud, which indicates that class I genes play an important role in the function of the meristem and growth and development. Class I group genes have more cis-elements of P-box and CAT-box, which may have an important relationship with its functions of regulating growth and promoting meristem development. In other species, class I group genes have been shown to promote the development of lateral branches and internodes (Aguilar-Martinez et al., 2015; Tsuda et al., 2017). In class II group, the expression of PbKNOX5 decreased gradually with leaf development time, whereas the expression of PbKNOX14 and PbKNOX16 increased gradually with leaf development time. Meanwhile, PbKNOXs is also highly expressed during ovary development and fruit development, indicating that it plays an important role in these processes.
Under salt stress treatment, the expression of most genes in class II group was highly responsive in leaves, whereas class I group genes were highly expressed in roots. This indicates that PbKNOX genes in class I and class II differ in their functions, especially in growth and development and the response to salt stress. This difference in expression might be related to differences in their conserved domains. Some members of the family also showed responses to drought stress, such as the increased expression of PbKNOX7/13 under drought stress and after plants were covered with water. The expression of PbKNOX5/16 was inhibited under drought stress and increased after plants were covered with water. PbKNOX genes in pear have important regulatory functions in growth and development and responses to stress.
PbKNOXs play an important role in the regulation of dwarf plants. We examined the specific expression pattern of PbKNOX genes in internode development in the pear varieties “601D” and “601V.” “601D” had an obvious dwarf phenotype compared with “601V,” and its internodes were significantly shorter than those of “601V.” Transcriptomic data showed that class I genes were significantly overexpressed in “601V,” and their log2fold change was 5-fold higher compared with “601D.” Class I genes play an important role in SAM. Loss of STM function prevents the formation of SAM and thus the continuous generation of lateral organs; this inhibition of growth leads to dwarf but strong plants (Liu et al., 2018b), which is consistent with our findings. In order to further verify the expression pattern of class I gene in dwarf plants, we used the dwarfing hybrid population to screen 30 seedlings showing significant variation in height in the offspring population, including 15 vigorous and 15 dwarf trees. The expression of PbKNOX genes was higher in offspring plants than in dwarf plants. This indicates that class I genes contribute to dwarfism in plants. Previous studies have reported that the member genes of class I have the function of regulating plant dwarf. The rice homeobox gene OSH15 (Oryza sativa homeobox) is a member of the knotted1-type homeobox gene family. Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants (Sato et al., 1999). The bp mutant of Arabidopsis which exhibits bends in inflorescence stems and pedicels as well as reductions in their lengths (Douglas et al., 2002). These data demonstrate a previously unknown link between KNOX gene activity and dicot stem development, whether PbKNOXs can cause plant dwarfing in pear is also unknown, which will be the focus of our next research.
Conclusion
In this study, we identified 19 PbKNOX TFs from pear based on genome sequences. PbKNM1 and PbKNM2 are KNM genes identified in pear for the first time. Class I genes play an important role in the function of the meristem and growth and development. Class II genes play an important role in the response to salt stress and drought stress. Analysis of PbKNOXs expression in vigorous and dwarf pear trees and hybrid progeny revealed that the expression of class I genes was significantly correlated with height. The results of our study provide candidate genes that could be used to modify the height of pear. Our study also provides new insights that could be used by future studies to clarify the function of PbKNOX genes.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author.
Author Contributions
JL, JS, and CL contributed to the conception and design of the study. JL, CZ, JH, XF, and HX organized the database. JL performed the statistical analysis and wrote the first draft of the manuscript. RW, DL, YY, and ZC provided the resources. JL and JS contributed to project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFD1001404-3), the Agricultural Improved Variety Project Program of Shandong Province (2019LZGC008), the Natural Science Foundation of Shandong Province (ZR2020QC150), and Open Funds of the State Key Laboratory of Crop Genetics and Germplasm Enhancement (no. ZW202004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.806765/full#supplementary-material
Footnotes
1. ^https://www.rosaceae.org/species/pyrus/all
2. ^https://web.expasy.org/protparam
3. ^http://www.genscript.com/psort/wolf_psort.html
4. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Aguilar-Martinez, J. A., Uchida, N., Townsley, B., West, D. A., Yanez, A., Lynn, N., et al. (2015). Transcriptional, posttranscriptional, and posttranslational regulation of SHOOT MERISTEMLESS gene expression in Arabidopsis determines gene function in the shoot apex. Plant Physiol. 2, 424–442. doi: 10.1104/pp.114.248625
Alvarez, J. M., Sohlberg, J., Engstrom, P., Zhu, T. Q., Englund, M., Moschou, P. N., et al. (2015). The WUSCHEL-RELATED HOMEOBOX 3 gene PaWOX3 regulates lateral organ formation in Norway spruce. New Phytol. 4, 1078–1088. doi: 10.1111/nph.13536
Ariel, F., Diet, A., Verdenaud, M., Gruber, V., Frugier, F., Chan, R., et al. (2010). Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-zip I transcription factor HB1. Plant Cell 22, 2171–2183. doi: 10.1105/tpc.110.074823
Arnaud, N., and Pautot, V. (2014). Ring the BELL and tie the KNOX: roles for TALEs in gynoeciurn development. Front. Plant Sci. 5:93. doi: 10.3389/fpls.2014.00093
Bagnara, G. L., and Rivalta, L. (1989). Dwarfing in pear. Acta Hortic. 256, 103–110. doi: 10.17660/ActaHortic.1989.256.11
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. W1, 39–49. doi: 10.1093/nar/gkv416
Bhattacharjee, A., Khurana, J. P., and Jain, M. (2016). Characterization of rice homeobox genes, OsHOX22 and OsHOX24, and over-expression of OsHOX24 in transgenic Arabidopsis suggest their role in abiotic stress response. Front. Plant Sci. 7:627. doi: 10.3389/fpls.2016.00627
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Cheng, X., Li, M. L., Abdullah, M., Li, G. H., Zhang, J. Y., Manzoor, M. A., et al. (2019). In Silico genome-wide analysis of the pear (Pyrus bretschneideri) KNOX family and the functional characterization of PbKNOX1, an Arabidopsis BREVIPEDICELLUS orthologue gene, involved in cell wall and lignin biosynthesis. Front. Genet. 10:632. doi: 10.3389/fgene.2019.00632
Douglas, S. J., Chuck, G., Dengler, R. E., Pelecanda, L., and Riggs, C. D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14, 547–558. doi: 10.1105/tpc.010391
Du, D., Hao, R., Cheng, T., Pan, H., Yang, W., Wang, J., et al. (2013). Genome-wide analysis of the AP2/ERF gene family in Prunus mume. Plant Mol. Biol. Report. 31, 741–750. doi: 10.1007/s11105-012-0531-6
FAO (2021). FAOSTAT, FAO statistical databases (Food and Agriculture Organization of the United Nations). Available at: http://www.fao.org/faostat/en/#data/QC (Accessed December 31, 2021).
Feng, X., Liu, W. X., Dai, H. X., Qiu, Y., Zhang, G. P., Chen, Z. H., et al. (2020). HvHOX9, a novel homeobox leucine zipper transcription factor revealed by root miRNA and RNA sequencing in tibetan wild barley, positively regulates Al tolerance. J. Exp. Bot. 19, 6057–6073. doi: 10.1093/jxb/eraa290
Furumizu, C., Alvarez, J. P., Sakakibara, K., and Bowman, J. L. (2015). Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet. 11:e1004980. doi: 10.1371/journal.pgen.1004980
Gao, J., Yang, X., Zhao, W., Lang, T., and Samuelsson, T. (2015). Evolution, diversification, and expression of KNOX proteins in plants. Front. Plant Sci. 6:882. doi: 10.3389/fpls.2015.00882
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jia, P., Xing, L. B., Zhang, C. G., Zhang, D., Ma, J. J., Zhao, C. P., et al. (2021). MdKNOX19, a class II knotted-like transcription factor of apple, plays roles in ABA signalling/sensitivity by targeting ABI5 during organ development. Plant Sci. 302:110701. doi: 10.1016/j.plantsci.2020.110701
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). Plant TFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Kato, H., Motomura, T., Komeda, Y., Saito, K., and Kato, A. (2010). Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 167, 571–577. doi: 10.1016/j.jplph.2009.11.004
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Ma, H., and DePamphilis, C. W. (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885. doi: 10.1111/j.1365-313X.2007.03097.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, Y., Pi, L., Huang, H., and Xu, L. (2012a). ATH1 and KNAT2 proteins act together in regulation of plant inflorescence architecture. J. Exp. Bot. 63, 1423–1433. doi: 10.1093/jxb/err376
Li, Q., Qiao, X., Yin, H., Zhou, Y., Dong, H., Qi, K., et al. (2019a). Unbiased subgenome evolution following a recent wholegenome duplication in pear (Pyrus bretschneideri Rehd.). Hortic. Res. 6:34. doi: 10.1038/s41438-018-0110-6
Li, K. Q., Xu, X. Y., and Huang, X. S. (2016). Identification of differentially expressed genes related to dehydration resistance in a highly drought-tolerant pear, Pyrus betulaefolia, as through RNA-Seq. PLoS One 11:e0149352. doi: 10.1371/journal.pone.0149352
Li, Y. D., Zhang, S. L., Dong, R. Z., Wang, L., Yao, J., van Nocker, S., et al. (2019b). The grapevine homeobox gene VvHB58 influences seed and fruit development through multiple hormonal signaling pathways. BMC Plant Biol. 1:523. doi: 10.1186/s12870-019-2144-9
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lim, C. W., Hong, E., Bae, Y., and Lee, S. C. (2018). The pepper dehydration-responsive homeobox 1, CaDRHB1, plays a positive role in the dehydration response. Environ. Exp. Bot. 147, 104–115. doi: 10.1016/j.envexpbot.2017.11.015
Liu, J. L., Deng, Z. W., Liang, C. L., Sun, H. W., Li, D. L., Song, J. K., et al. (2021). Genome-wide analysis of RAV transcription factors and functional characterization of anthocyanin-biosynthesis-related RAV genes in pear. Int. J. Mol. Sci. 22:5567. doi: 10.3390/ijms22115567
Liu, L., Li, C. Y., Song, S. Y., Teo, Z. W. N., Shen, L. S., Wang, Y. W., et al. (2018a). FTIP-dependent STM trafficking regulates shoot meristem development in Arabidopsis. Cell Rep. 23:1879. doi: 10.1016/j.celrep.2018.04.033
Liu, L., Wang, Z., Liu, J., Liu, F., Zhai, R., Zhu, C., et al. (2018b). Histological, hormonal and transcriptomic reveal the changes upon gibberellininduced parthenocarpy in pear fruit. Hortic. Res. 5:1. doi: 10.1038/s41438-017-0012-z
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Moens, C. B., and Selleri, L. (2006). Hox cofactors in vertebrate development. Dev. Biol. 291, 193–206. doi: 10.1016/j.ydbio.2005.10.032
Moore, R. C., and Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. 100, 15682–15687. doi: 10.1073/pnas.2535513100
Nakata, M., Matsumoto, N., Tsugeki, R., Rikirsch, E., Laux, T., and Okada, K. (2012). Roles of the middle domain–specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 2, 519–535. doi: 10.1105/tpc.111.092858
Ou, C., Jiang, S., Wang, F., Tang, C., and Hao, N. (2015). An RNA–Seq analysis of the pear (Pyrus communis L.) transcriptome, with a focus on genes associated with dwarf. Plant Gene. 4, 69–77. doi: 10.1016/j.plgene.2015.08.003
Ramamoorthy, R., Jiang, S. Y., Kumar, N., Venkatesh, P. N., and Ramachandran, S. (2008). A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 49, 865–879. doi: 10.1093/pcp/pcn061
Sato, Y., Sentoku, N., Miura, Y., Hirochika, H., Kitano, H., and Matsuoka, M. (1999). Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 18, 992–1002. doi: 10.1093/emboj/18.4.992
Thomas, B., Enrico, S., Jiri, F., and Danielle, M. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015–1027. doi: 10.1101/gad.1402406
Tsuda, K., Abraham-Juarez, M. J., Maeno, A., Dong, Z., Aromdee, D., Meeley, R., et al. (2017). KNOTTED1 cofactors, BLH12 and BLH14, regulate internode patterning and vein anastomosis in maize. Plant Cell 5, 1105–1118. doi: 10.1105/tpc.16.00967
Vollbrecht, E., Veit, B., Sinha, N., and Hake, S. (1991). The developmental gene Knotted-1 is a member of a maize homebox gene family. Nature 350, 241–243. doi: 10.1038/350241a0
Voorrips, R. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, S. M., Yamaguchi, M., Grienenberger, E., Martone, P. T., Samuels, A. L., and Mansfield, S. D. (2019). The class II KNOX genes KNAT3 and KNAT7 work cooperatively to influence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J. 2, 293–309. doi: 10.1111/tpj.14541
Wenzel, C. L., Schuetz, M., Yu, Q., and Mattsson, J. (2007). Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J. 49, 387–398. doi: 10.1111/j.1365-313X.2006.02977.x
Yang, Z. E., Gong, Q., Qin, W. Q., Yang, Z. R., Cheng, Y., Lu, L. L., et al. (2017). Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 17:113. doi: 10.1186/s12870-017-1065-8
Yang, X., Hu, H., Yu, D., Sun, Z., He, X., Zhang, J., et al. (2015). Candidate resistant genes of sand pear (Pyrus pyrifolia Nakai) to Alternaria alternate revealed by transcriptome sequencing. PLoS One 10:e0135046. doi: 10.1371/journal.pone.0135046
Zhang, D., Ren, L., Yue, J. H., Shi, Y. B., Zhuo, L. H., Wang, L., et al. (2015). RNA–Seq–based transcriptome analysis of stem development and dwarfing regulation in Agapanthus praecox ssp. orientalis (Leighton) Leighton. Gene 565, 252–267. doi: 10.1016/j.gene.2015.04.013
Zhang, M. Y., Xue, C., Xu, L., Sun, H., Qin, M. F., Zhang, S., et al. (2016). Distinct transcriptome profiles reveal gene expression patterns during fruit development and maturation in five main cultivated species of pear (Pyrus L.). Sci. Rep. 6:28130. doi: 10.1038/srep28130
Keywords: pear, KNOX, genome-wide analysis, functional characterization, dwarf mechanism
Citation: Liu J, Zhang C, Han J, Fang X, Xu H, Liang C, Li D, Yang Y, Cui Z, Wang R and Song J (2022) Genome-Wide Analysis of KNOX Transcription Factors and Expression Pattern of Dwarf-Related KNOX Genes in Pear. Front. Plant Sci. 13:806765. doi: 10.3389/fpls.2022.806765
Edited by:
Guillaume Pilot, Virginia Tech, United StatesReviewed by:
Jose Manuel Alvarez, University of Oviedo, SpainKatsutoshi Tsuda, National Institute of Genetics, Japan
Copyright © 2022 Liu, Zhang, Han, Fang, Xu, Liang, Li, Yang, Cui, Wang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiankun Song, cWF1c2prQDEyNi5jb20=
 Jianlong Liu
Jianlong Liu Chenxiao Zhang1
Chenxiao Zhang1 Jingyue Han
Jingyue Han Zhenhua Cui
Zhenhua Cui