- 1Key Laboratory of Plant-Soil Interaction (MOE), Centre for Resources, Environment and Food Security, College of Resources and Environmental Sciences, China Agricultural University, Beijing, China
- 2Key Lab of Crop Heterosis and Utilization of Ministry of Education, Beijing Key Lab of Crop Genetic Improvement, China Agricultural University, Beijing, China
Iron (Fe) is a limiting factor in crop growth and nutritional quality because of its low solubility. However, the current understanding of how major crops respond to Fe deficiency and the genetic basis remains limited. In the present study, Fe-efficient inbred line Ye478 and Fe-inefficient inbred line Wu312 and their recombinant inbred line (RIL) population were utilized to reveal the physiological and genetic responses of maize to low Fe stress. Compared with the Fe-sufficient conditions (+Fe: 200 μM), Fe-deficient supply (−Fe: 30 μM) significantly reduced shoot and root dry weights, leaf SPAD of Fe-efficient inbred line Ye478 by 31.4, 31.8, and 46.0%, respectively; decreased Fe-inefficient inbred line Wu312 by 72.0, 45.1, and 84.1%, respectively. Under Fe deficiency, compared with the supply of calcium nitrate (N1), supplying ammonium nitrate (N2) significantly increased the shoot and root dry weights of Wu312 by 37.5 and 51.6%, respectively; and enhanced Ye478 by 23.9 and 45.1%, respectively. Compared with N1, N2 resulted in a 70.0% decrease of the root Fe concentration for Wu312 in the −Fe treatment, N2 treatment reduced the root Fe concentration of Ye478 by 55.8% in the −Fe treatment. These findings indicated that, compared with only supplying nitrate nitrogen, combined supply of ammonium nitrogen and nitrate nitrogen not only contributed to better growth in maize but also significantly reduced Fe concentration in roots. In linkage analysis, ten quantitative trait loci (QTLs) associated with Fe deficiency tolerance were detected, explaining 6.2–12.0% of phenotypic variation. Candidate genes considered to be associated with the mechanisms underlying Fe deficiency tolerance were identified within a single locus or QTL co-localization, including ZmYS3, ZmPYE, ZmEIL3, ZmMYB153, ZmILR3 and ZmNAS4, which may form a sophisticated network to regulate the uptake, transport and redistribution of Fe. Furthermore, ZmYS3 was highly induced by Fe deficiency in the roots; ZmPYE and ZmEIL3, which may be involved in Fe homeostasis in strategy I plants, were significantly upregulated in the shoots and roots under low Fe stress; ZmMYB153 was Fe-deficiency inducible in the shoots. Our findings will provide a comprehensive insight into the physiological and genetic basis of Fe deficiency tolerance.
Introduction
Iron (Fe) is an essential micronutrient with numerous cellular functions in animals and plants (Zhou et al., 2013). About 80% of the cellular Fe in plant leaves is located in the chloroplasts, which is the vital pigment required for photosynthesis (Zhang et al., 2017; Long et al., 2020). Also, the evolutionary ability of Fe to change oxidation states between Fe(III) and Fe(II) renders it irreplaceably important in many essential processes associated with basic redox reactions (Li and Lan, 2015). However, Fe is found in nature mostly as insoluble Fe(III) oxyhydroxides that are sparingly soluble in aerobic, neutral pH soils because of many adverse soil properties (Tognetti et al., 2007; Moreno-Jiménez et al., 2019; Housh et al., 2021). Fe deficiency can significantly reduce crop yield and quality, which, in turn, affects the immune and nervous systems, the mental and intellectual development of humans who consume these crops (Li et al., 2019). Fe cannot be synthesized by humans and must be acquired through the diet (Hindu et al., 2018). In developing countries that rely on cereal diets, for example, in China, more than 300 million people are at risk of Fe and zinc (Zn) deficiencies (Zhao et al., 2020). Additionally, inadequate intake of Fe results in Fe-deficiency anemia (IDA), affecting more than half of the world’s population (Wang et al., 2019). In the worst-case scenario, Fe deficiency may cause human death (Prasad, 2013; WHO, 2013).
Maize is the world’s most-produced food crop and accounts for 41% of the world’s total grain production (Tian et al., 2019). And maize provides 15 and 20% of the total proteins and calories, respectively, consumed by the world’s population (Cheah et al., 2019; Guo et al., 2020). And maize ranks as the top cereal crop in terms of total production and planting area in China (Food and Agriculture Organization of the United Nations [FAO], 2018). Fe deficiency considerably restricts maize production, hence threatening food security. It supplies not only energy in the form of carbohydrates, fat, and proteins but also vitamins and many minerals, including Fe (Wang et al., 2019). Meanwhile, maize is not only a major crop plant for the feed industry and food, but also a model plant for genetics and evolutionary study (Zhou et al., 2013). Maize is considered a graminaceous species with low phytosiderophore release, and little is known about other determinants contributing to its tolerance to Fe deficiency-induced chlorosis (Shi et al., 2018). Investigating the mechanisms of Fe acquisition, translocation, and homeostasis in maize may support a model for understanding that in other crops and provide gene resources for further breeding maize varieties with enhanced Fe content.
Moreover, Fe deficiency in humans is a global health issue, affecting about 25% of the world’s population (1.62 billion) (Chan-Rodriguez and Walker, 2018). At present, information on the molecular mechanisms of Fe absorption, translocation, and accumulation in maize under low Fe stress is still limited. The main reason is that there are few works to identify related genes by using methods such as population genetics approach. These works are of great significance for maintaining Fe homeostasis and bio-enhancement in plants under Fe starvation. Under low Fe stress, plants have evolved two distinct strategies to solubilize and transport Fe, such as the reduction-based strategy (Strategy I) in dicotyledonous and non-graminaceous monocotyledonous plants, and the chelation-based strategy (Strategy II) for graminaceous monocots (Li et al., 2014; Kurt et al., 2019). The characteristic of strategy I is the release of a proton (H+) from the roots (mediated by AHA2), which facilitates the mobilization and subsequent reduction of Fe(III) to Fe(II) via the actions of plasma membrane-bound Fe(III), chelate reductase (FRO2) (Guerinot, 2007; Benke et al., 2015; Li and Lan, 2015). The soluble Fe(II) is finally transported into the root epidermis by the divalent metal transporter (iron-regulated Transporter 1, IRT1) (Vert et al., 2002; Mallikarjuna et al., 2020). Strategy II plants, like maize, secrete mugineic acids (MAs; also known as phytosiderophores) into the rhizosphere using efflux transporters (TOM1 and TOM2) (Nozoye et al., 2011). The MAs chelate inorganic Fe(III) in the soil, producing Fe(III)-MAs complexes that are subsequently absorbed through the specific transporter yellow stripe 1 (YS1) (Benke et al., 2015).
Transcriptomic analysis of Fe deficiency response reveals that a strong modulation of genes involved in regulatory aspects, Fe translocation, root morphological modification, primary metabolic pathways, and hormonal metabolism are induced by the Fe nutritional stress (Zanin et al., 2017). Fe-deficiency stress is signaled by many plant hormones, including indole acetic acid (IAA), ethylene, abscisic acid (ABA), gibberellins, cytokinins, and brassinosteroids (Ivanov et al., 2012; Wang et al., 2012). The interaction between the auxin and NO modulates the root growth under Fe deficiency in rice (Sun et al., 2017). Also, ethylene regulates FRO2 and IRT1 gene expression through the modulation of the major transcription factor FER or FIT (FER-like iron deficiency-induced transportation factor) (Ye et al., 2015). The FIT forms heterodimers with bHLH38 and bHLH39, and positively regulates a subset of Fe-responsive genes (Yuan et al., 2008). In addition, bHLH115 proteins have bZIP, bHLH, and MYB transcription factor-binding sites, which strengthen their engagement in various metabolic ways (Li et al., 2016). The POPEYE (PYE) plays a negative role in the Fe deficiency response in Arabidopsis by repressing the expression of nicotianamine synthase 4 (NAS4), FRO3, and zinc-induced facilitator1 (ZIF1) (Kurt et al., 2019). And, in Arabidopsis, the mitogen-activated protein kinase 3 and 6 (MPK3/MPK6)-regulated ACS2/6 activation is part of the Fe deficiency-induced ethylene production-signaling pathway (Ye et al., 2015). These Fe deficiency-inducible genes are regulated at transcriptional, translational, and post-translational levels. However, the functions of these genes are mainly verified in the strategy I plants.
Few genes related to Fe homeostasis have been studied in maize. ZmYS1 expression at both the messenger RNA (mRNA) and protein levels responds rapidly to changes in Fe availability (Chorianopoulou et al., 2015). ZmNAS proteins of Class I are mostly responsible for the long-distance translocation of Fe in stems, and their expression can be expanded in the epidermis, shoot apices as a response to Fe starvation (Zhou et al., 2013). The ZmZIP5 may play a role in Zn and Fe uptake and root-to-shoot translocation (Li et al., 2019). Fe concentrations are elevated in the shoots and roots of ectopic ZmZIP7-overexpressing plants, while the endogenic Fe deficiency-inducible genes are stimulated in transgenic Arabidopsis (Li et al., 2016). The ZmFDR4 functions as a Fe transporter in monocot plastids, which subsequently increases chlorophyll content and enhances photosynthetic efficiency, thus, affects plastid development (Zhang et al., 2017). Genes related to Fe homeostasis could be employed to fortify the Fe contents in plants, thus boosting micronutrient availability for human populations, depending on staple crops (Ricachenevsky et al., 2018; Li et al., 2019). Therefore, identifying and analyzing more genes related to low Fe stress tolerance in maize may play an important role in the growth and development of maize and human health.
In the current study, several supply levels of Fe(III)-EDTA were designed to evaluate more reasonable and effective indicators to characterize Fe deficiency tolerance between the Fe-inefficient (Wu312) and Fe-efficient (Ye478) inbred lines at the seedling stage in Experiment 1, which allowed for linkage analysis in the RIL population in Experiment 3. Under low Fe stress, there is still very limited information on the differences in the response of Fe-inefficient and Fe-efficient maize inbred lines to different nitrogen supply forms. Also, so far, mainly supplying nitrate nitrogen, which is recommended to use in the Hoagland nutrient solution, has significantly affected the accuracy for measuring Fe concentration in roots. According to the results in Experiment 1, Experiment 2 was designed to analyze the effects of different nitrogen supply forms on the plant growth, root Fe concentration under Fe-deficient and Fe-sufficient conditions. Furthermore, the recombinant inbred line (RIL) population derived from the Fe-inefficient (Wu312) and Fe-efficient (Ye478) inbred lines were utilized to identify the quantitative trait loci and candidate genes associated with Fe efficiency in maize. The expression of candidate genes probably involved in the mechanisms underlying Fe deficiency tolerance in maize was assessed in the shoots and roots of Fe-inefficient (Wu312) and Fe-efficient (Ye478) parents under different Fe nutritional statuses.
Materials and Methods
Plant Material and Experimental Design
In Experiment 1, eight different Fe treatments were designed, containing 0, 10, 30, 50, 70, 100, 200, and 300 μM Fe(III)-EDTA. Two seedings of Wu312 and Ye478 were grown in a 3.3-L container under different Fe nutritional statuses in the same growth chamber (Figure 1A) as a replicate. In Experiment 2, two seedlings for each inbred line were grown in a 3.3-L container supplied with Ca (NO3)2 [4 mM NO3–-N] or NH4NO3 [2 mM NO3–-N, 2 mM NH4+-N] under Fe-deficient [−Fe: 30 μM Fe(III)-EDTA] and Fe-sufficient conditions [+Fe: 200 μM Fe(III)-EDTA] as a replicate (Figure 1B). Each treatment contained three replicates, and a total of six seedlings for each inbred line were used for each treatment in Experiment 2.
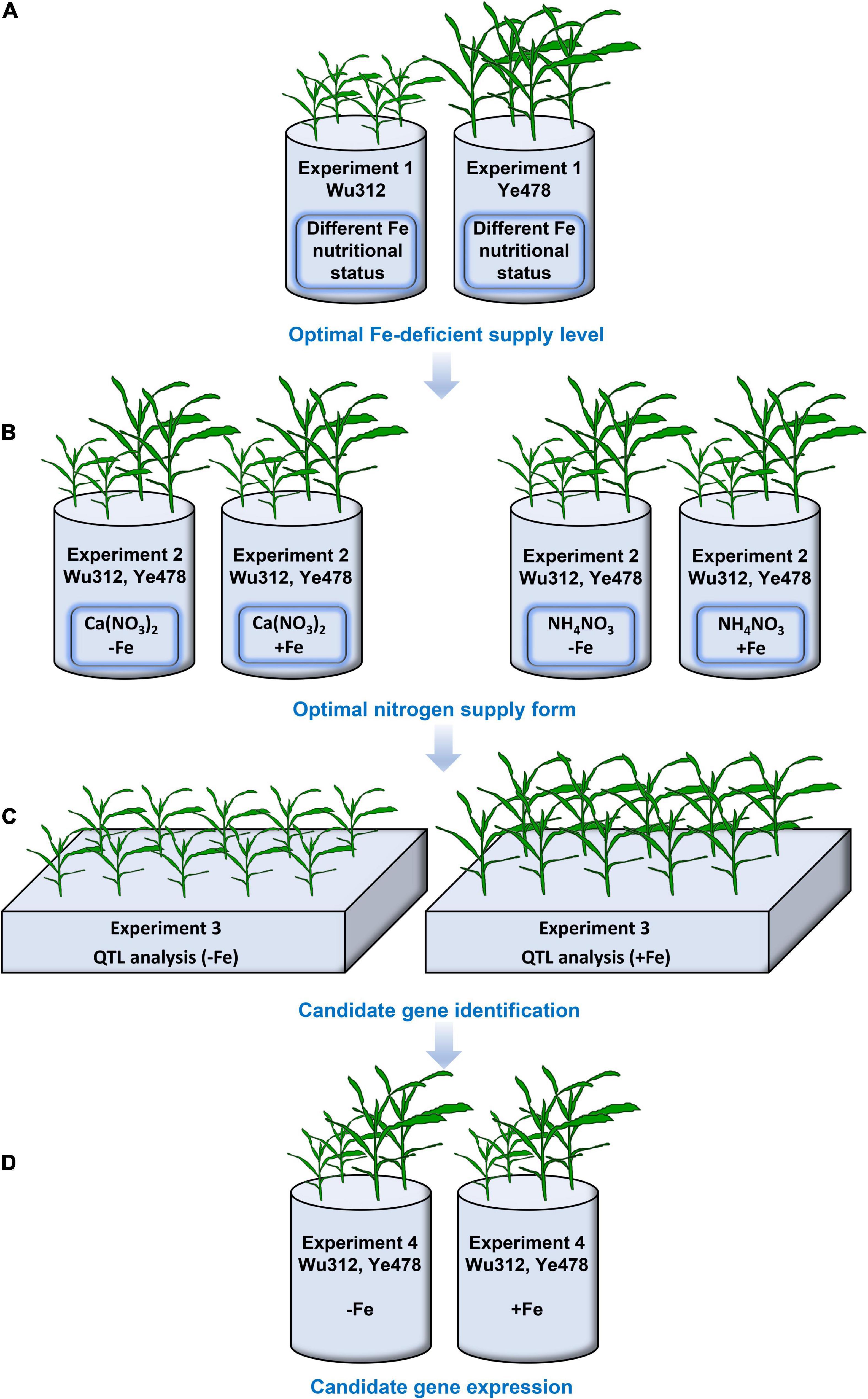
Figure 1. An experimental setup in this study. (A) In Experiment 1, eight different Fe(III)-EDTA levels (0, 10, 30, 50, 70, 100, 200, and 300 μM) were designed. And four seedlings of inbred lines Wu312 or Ye478 were hydroponically grown in a 3.3-L container as a replicate. Each treatment contained three replicates. (B) In Experiment 2, two seedlings of each inbred line were grown in a 3.3-L container supplied with Ca (NO3)2 [4 mM NO3–-N] or NH4NO3 [2-mM NO3–-N and 2-mM NH4+-N] under Fe-deficient [–Fe: 30-μM Fe(III)-EDTA] and Fe-sufficient conditions [+Fe: 200-μM Fe(III)-EDTA] as a replicate. Each treatment contained three replicates. (C) In Experiment 3, a recombinant inbred line (RIL) population derived from Fe-inefficient inbred line Wu312 and Fe-efficient inbred line Ye478 was grown under the –Fe and +Fe conditions to identify the quantitative trait loci associated with Fe deficiency tolerance. Each treatment contained three replicates. (D) In Experiment 4, the expression of candidate genes identified from Experiment 3 was analyzed in the shoots and roots of Wu312 and Ye478 under the –Fe and +Fe conditions. Each treatment contained three replicates.
In Experiment 3, the Ye478 × Wu312 RIL population, which consists of 218 lines, was derived from Ye478 (a female parent) and Wu312 (a male parent) as described by Liu et al. (2011), and their parents were hydroponically grown under Fe-deficient [−Fe: 30 μM Fe(III)-EDTA] and Fe-sufficient conditions [+Fe: 200 μM Fe(III)-EDTA] (Figure 1C). Each treatment contained three replicates.
In Experiment 4, the expression of candidate genes associated with Fe deficiency tolerance identified from Experiment 3 was analyzed in the shoots and roots of Fe-inefficient (Wu312), and Fe-efficient (Ye478) parents under Fe-deficient [−Fe: 30 μM Fe(III)-EDTA] and Fe-sufficient [+Fe: 200 μM Fe(III)-EDTA] conditions (Figure 1D).
Plant Growth and Hydroponic Culture
Maize seeds were sterilized for 30 min in a 10% solution of H2O2, washed with distilled water, soaked in saturated CaSO4 for 10 h, and then germinated on moist filter paper in the dark at room temperature. Two days later, the germinated seeds were wrapped in a moist filter paper roll and grown. At the stage of two visible leaves, the seedlings were selected and transferred into a 3.3-L container (Experiments 1, 2 and 4) or a 40-L container (Experiment 3). The adjusted Hoagland nutrient solution contained (mM) 0.75 K2SO4, 0.65 MgSO4, 0.1 KCl, 0.25 KH2PO4, 1.0 × 10–3 H3BO3, 1 × 10–2 ZnSO4, 1 × 10–4 CuSO4, 1 × 10–3 MnSO4, 5 × 10–6 (NH4) Mo7O24, 1 × 10–4 NiCl. Solution pH was set at 5.5–6. The nutrient solution was renewed every 3 days and aerated by a pump. The growth chamber condition was set as a 14-h light period from 8:00 to 22:00 with 28°C and a 10-h-dark period with 22°C. The average light intensity measured at a canopy was 350 μmol m–2 s–1.
Data Collection
During the period from 9:00 to 11:00 a.m. on the 14th day after transfer; the SPAD value of the fourth leaf was measured on the 1/3 parts from the leaf base three times using a SPAD-502 leaf chlorophyll meter. The average of three observed values was recorded for each plant. Plants of all experiments were harvested on the 14th day after transfer. Plant height and root length for each plant were recorded. In Experiment 1, active Fe concentration was determined in young leaves and old leaves for each plant. About 2 g fresh samples were soaked in 20 ml 1 M HCl solution and shaken for 5 h at 25°C. Then, the solution was filtered, and Fe concentration was measured by ICP-AES (Inductively Coupled Plasma-Atomic Emission Spectroscopy). Active Fe concentration was expressed as μg g–1 fresh weight (FW). The roots in Experiment 1 were only washed by distilled water. The roots in Experiment 2 were washed using a mixed solution of EDTA-Na and CaSO4, subsequently washed by 1 M HCl solution, and finally washed using distilled water. Then, the shoot and root for each plant were separately collected in an envelope. All samples were heat-treated at 105°C for 30 min and dried at 75°C until constant weight. Subsequently, Fe concentrations in shoots and roots were measured by ICP-AES. Fe efficiency based on shoot (root) dry weight, Fe content, Fe uptake efficiency, and Fe transport efficiency was estimated using the following equations from (1) to (4), respectively.
(1) Fe efficiency based on shoot (root) dry weight (%) = shoot (root) dry weight under Fe-deficient conditions × 100%/shoot (root) dry weight under Fe-sufficient conditions.
(2) Fe content (μg plant–1) = concentration × dry weight.
(3) Fe uptake efficiency (μg root dry weight g–1) = (shoot Fe content + root Fe content)/root dry weight.
(4) Fe transport efficiency (%) = shoot Fe content × 100%/(shoot Fe content + root Fe content).
Statistical Analysis
The mean for each trait was compared using one-way ANOVA at a 0.05 level of probability followed by the least significant difference (LSD) test. Means between inbred lines Wu312 and Ye478 in each experiment and the means of each parent between the −Fe and +Fe treatments in Experiment 3 were compared using Student’s t-test. In Experiment 3, the linear mixed effect function lmer from the lme4 package in R was fitted to each RIL to obtain the best linear unbiased prediction (BLUP) value for each trait. Broad-sense heritability for each trait was calculated using H2 = σg2/(σg2 + σge2/e + σε2/re), where σg2 is genetic variance, σge2 is the interaction of genotype and treatment, σε2 is a residual error, while e and r are the number of environments and replicates, respectively. The Kolmogorov-Smirnova test was performed as the statistical method to verify the normality in the distribution for each trait in the RIL population.
Quantitative Trait Locus Analysis
Quantitative trait locus (QTL) analysis was performed using composite interval mapping (CIM) in the Windows QTL Cartographer version 2.5. Model 6 was selected for detecting QTL and estimating their effects. The threshold logarithm of odds (LOD) values to declare the putative QTL was estimated by permutation tests with a minimum of 1,000 replicates at a significant level of p < 0.05 (LOD = 2.9). The confidence interval for each QTL was determined using the 1-LOD interval method, which was calculated and defined by left and right markers. The QTLs for the same trait having the same confidence intervals were defined as identical QTL.
Annotation of Candidate Genes
According to the physical distance, genes within the refined interval and their functional descriptions were identified using the maize B73 reference genome assembly available on the MaizeGDB database1 and Gramene Database2.
RNA Extraction and Gene Expression Quantification
In Experiment 4, the total RNA was extracted from shoots and roots of plants using the Total RNA Extraction Kit (TIANGEN, China). The complementary DNA (cDNA) was synthesized in accordance with Fast Quant RT Super MixReverse Transcription Kit instructions (TransGene, Beijing, China). Quantitative real-time PCR was performed using SYBR Green Real-time RT-PCR (Takara) and an ABI7500 Fast Real-Time PCR System (Applied Biosystems). The primers used for real-time PCR are shown in Supplementary Table 1. The relative gene expression level was calculated using the 2–ΔΔCt method. Each real-time PCR experiment contained three technical replicates.
Results
Effect of Different Fe(III)-EDTA Supply Levels on the Physiological Characteristics of Maize With Different Fe Efficiencies
The shoot dry weight of Wu312 and Ye478 gradually increased with the increasing supply of Fe(III)-EDTA (Figure 2A). When the Fe supply level was 30 μM, Ye478 produced shoot dry weight like that on the supply of 100 μM. The shoot dry weight of two inbred lines reached the highest value when the Fe supply level was 200 μM. Nevertheless, when the Fe supply concentration was 300 μM, their plant growth was inhibited. Especially, the plant growth and development of Wu312 were significantly suppressed. Leaf SPAD of Ye478 and Wu312 increased with increasing Fe supply levels except for the leaf SPAD on the supply exceeding 100 μM (Figure 2D). In all treatments, leaf SPAD of Ye478 was significantly higher than that of Wu312 (Figure 2D). The leaf SPAD of Wu312 and Ye478 on the supply of 200 μM Fe(III)-EDTA was 5.8 and 1.8-fold higher than those on the supply of 30 μM, respectively.
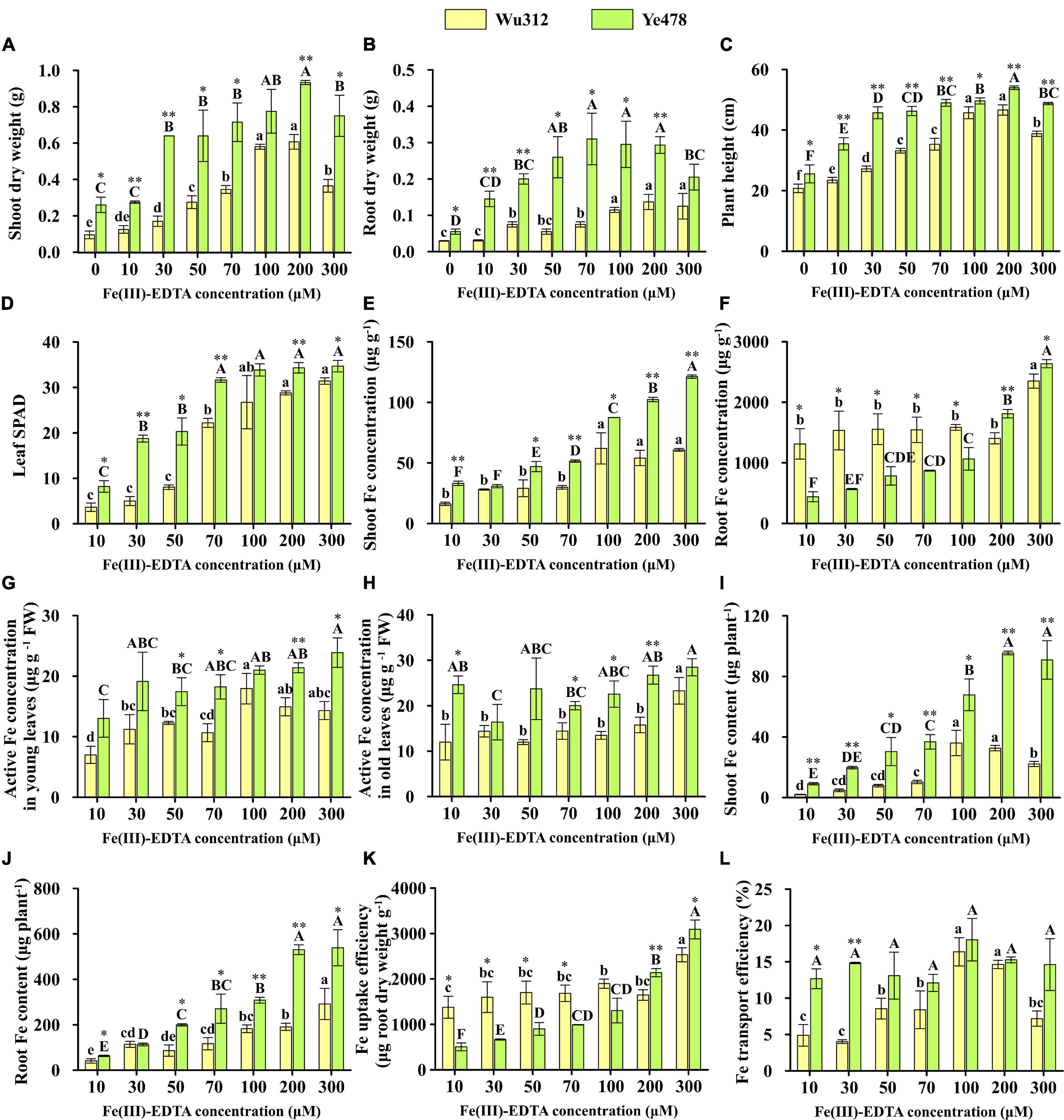
Figure 2. Shoot (A) and root dry weights (B), Plant height (C), SPAD value of the fourth leaf (D), shoot (E) and root Fe concentrations (F), active Fe concentration in young leaves (G) and old leaves (H), shoot (I) and root Fe contents (J), Fe uptake (K), and transport (L) efficiencies of Fe-inefficient inbred line Wu312 and Fe-efficient inbred line Ye478 under different Fe(III)-EDTA levels in Experiment 1. Different lowercase and upper letters indicate significant difference (p < 0.05) of Wu312 and Ye478 among treatments, respectively. * and ** indicate significant difference between Wu312 and Ye478 at p < 0.05 and p < 0.01, respectively.
Considering the dry matter weight, plant height, and leaf SPAD, as well as the observed plant growth in Experiment 1, this study determined the Fe supply level 200 μM as Fe-sufficient supply for normal growth. When the Fe supply level was 30 μM, the phenotypic difference between two inbred lines was the largest (Figures 2A–D). The shoot and root dry weights, plant height, and leaf SPAD of Ye478 were 3.8-, 2.7-, 1.7-, and 3.8-fold higher than those of Wu312 when the iron supply level was 30 μM Fe(III)-EDTA, respectively. Compared with the Fe-sufficient treatment, the shoot and root dry weights, plant height, and leaf SPAD of Ye478 on the supply of 30 μM Fe(III)-EDTA were decreased by 31.4, 31.8, 15.3 and 46.0%, respectively; while Wu312 was decreased by 72.0, 45.1, 41.6 and 84.1%, respectively. The R/S ratio of Ye478 showed no significant difference between the Fe-deficient treatment [30 μM Fe(III)-EDTA] and the Fe-sufficient treatment; while the R/S ratio of Wu312 on the supply of 30 μM Fe(III)-EDTA was the highest among all treatments (Supplementary Figure 1). In this study, Fe efficiency was calculated using the ratio of the value on the Fe-deficient supply of 30 μM to the value on the Fe-sufficient supply of 200 μM. As shown in Figure 3, the Fe efficiency based on the shoot and root dry weight of Ye478 was 68.8 and 66.7%, which was 2.5-fold and 1.1-fold significantly higher than those of Wu312, respectively.
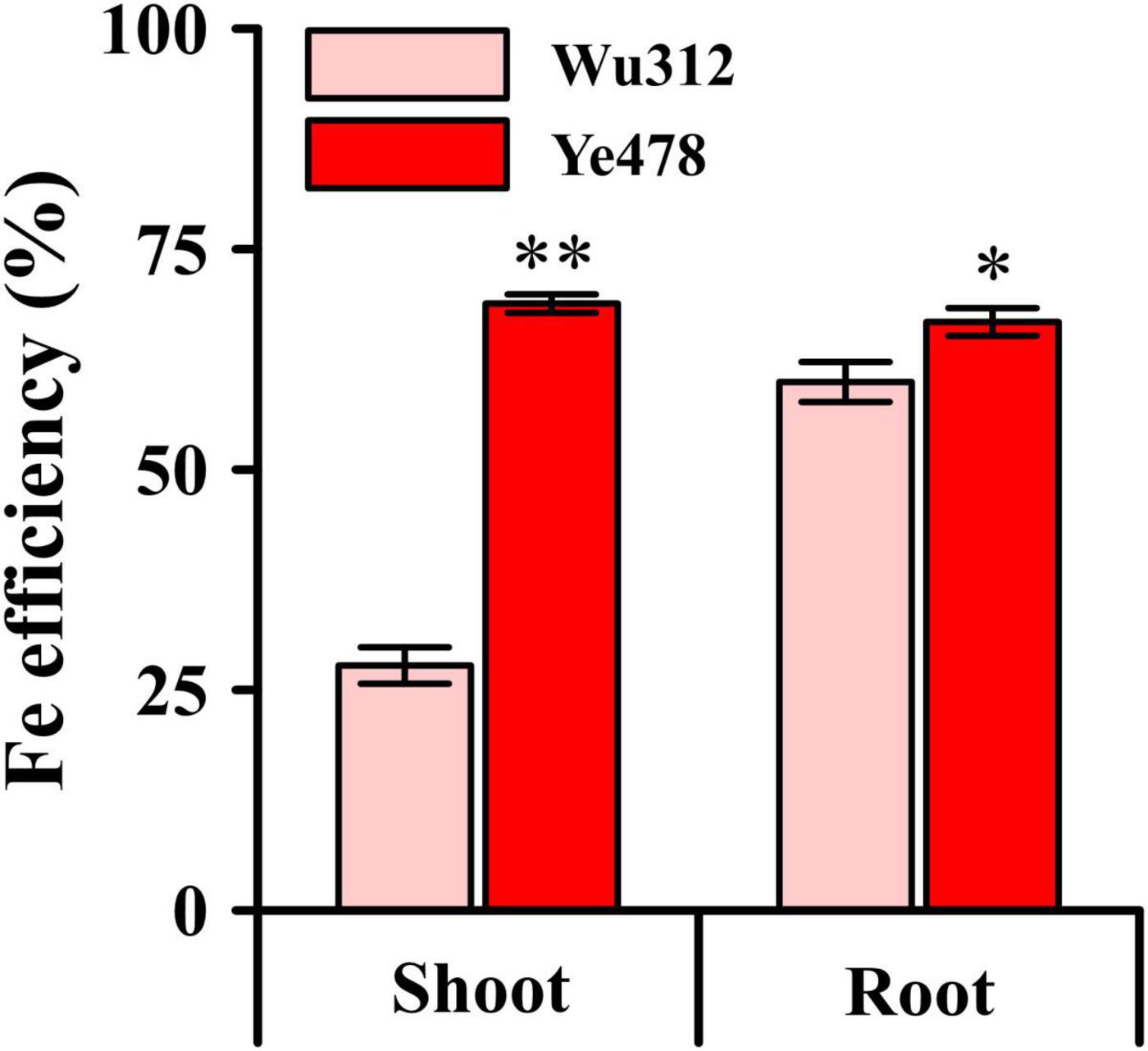
Figure 3. Iron (Fe) efficiency based on shoot and root dry weights. * and ** indicate significant difference between the Fe-inefficient (Wu312) and Fe-efficient (Ye478) inbred lines at p < 0.05 and p < 0.01, respectively.
The shoot Fe concentration of the two inbred lines showed an increasing trend as the Fe supply level increased (Figure 2E). Furthermore, the shoot Fe concentration of Ye478 was higher than that of Wu312. However, when the Fe supply levels ranged from 10 to 100 μM, the Fe concentrations in the roots of Wu312 were significantly higher than those of Ye478 (Figure 2F). In addition, a large amount of Fe might have accumulated in the root apoplast and root surface when only nitrate nitrogen was supplied, which may cause the root Fe concentration of two inbred lines to be too high. In both young and old leaves, the active Fe concentration of Ye478 was higher than that of Wu312 in most treatments (Figures 2G,H). However, there were no significant differences in active Fe concentration in young and old leaves of two inbred lines between the Fe-deficient and Fe-sufficient treatments. In the old leaves, the average active Fe concentrations of Ye478 and Wu312 were 23.2 and 15 μg g–1 FW, respectively. In the new leaves, the average active Fe concentration of Ye478 and Wu312 was 19.1 and 12.6 μg g–1 FW, respectively.
Fe contents in the shoots and roots of Wu312 and Ye478 displayed an increasing trend with the increase of Fe(III)-EDTA supply (Figures 2I,J). The shoot Fe contents of Wu312 and Ye478 on the supply of 200 μM were 7.5-fold and 3.4-fold higher than those under Fe-deficient supply (30 μM), respectively. Fe uptake efficiency of Ye478 increased with increasing Fe supply levels, and the uptake efficiency of Wu312 displayed no significant difference among the treatments except for 300 μM (Figure 2K). Moreover, the uptake efficiency of Wu312 on the supply of 30 μM was significantly higher than that of Ye478 under Fe-deficient supply (30 μM) and was markedly lower than that of Ye478 under Fe-sufficient supply (200 μM). The Fe transport efficiency of Wu312 displayed a significant reduction under the Fe-deficient condition when compared with the Fe-sufficient condition, and Ye478 displayed no significant difference between the two treatments (Figure 2L).
Impact of Nitrogen Form on Maize With Different Iron Efficiencies Under Low Fe Stress
Under Fe deficiency, the SPAD for Ye478 in the N2 was 1.2-fold higher than that in the N1, whereas there was no significant difference in the SPAD of Wu312 between the N1 and N2 treatments (Figure 4A). Except for Wu312 under Fe-sufficient condition, shoot and root dry weights for two inbred lines supplied with nitrate-nitrogen (N1) were significantly lower than those simultaneously supplied with equivalent NH4+-N and NO3–-N(N2) (Figures 4C,D). Under low Fe stress, compared with N1, N2 increased the shoot and root dry weights of Wu312 by 37.5 and 51.6% and increased the shoot and root dry weights of Ye478 by 23.9 and 45.1%, respectively. Under Fe-sufficient condition, compared with N1, the shoot and root dry weights of Ye478 in the N2 treatment were increased by 32.8 and 27.6%, respectively. N2 treatment also increased the plant heights of two inbred lines except for the plant height of Wu312 under Fe-sufficient condition (Figure 4B). Under Fe-deficient condition, compared with N1, N2 reduced the shoot Fe concentration of Ye478 by 35.4% (Figure 4E).
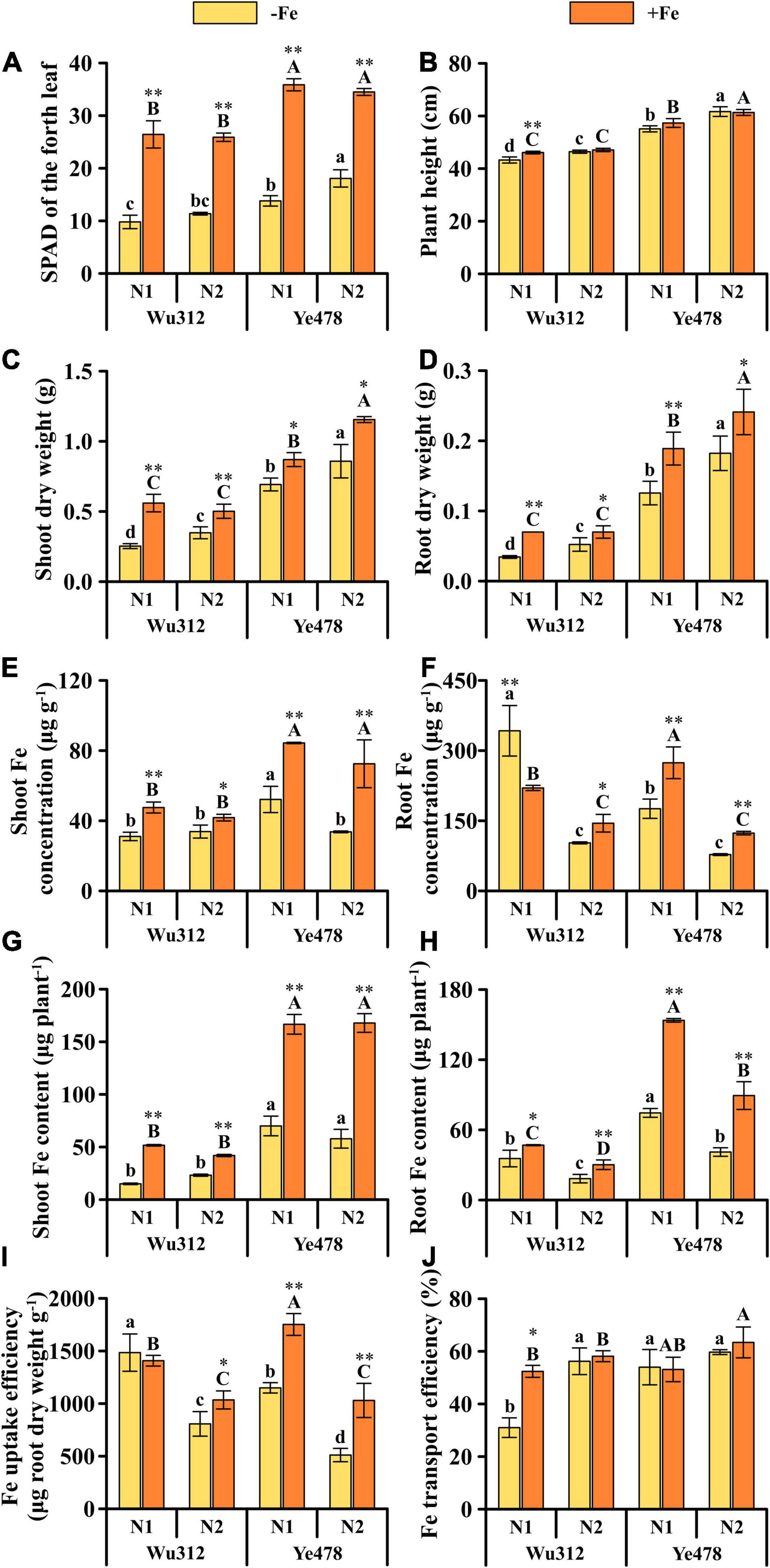
Figure 4. SPAD value of the fourth leaf (A), plant height (B), shoot (C) and root dry weights (D), shoot (E) and root Fe concentrations (F), shoot (G) and root Fe contents (H), and Fe uptake (I) and transport efficiencies (J) of Wu312 and Ye478 under Fe-deficient [–Fe: 30 μM Fe(III)-EDTA] and Fe-sufficient conditions [+Fe: 200 μM Fe(III)-EDTA] supplied with N1 [Ca(NO3)2: 2 mM NO3–-N] or N2 [NH4NO3: 2 mM NO3–-N, 2 mM NH4+-N]. Different lowercase and upper letters indicate significant difference (p < 0.05) of inbred lines under –Fe and +Fe conditions, respectively. * and ** indicate significant difference between the –Fe and +Fe treatments at p < 0.05 and p < 0.01, respectively.
The use of hydrochloric acid combined with a mixed cleaning solution of EDTA-Na and CaSO4 could significantly reduce the precipitation and fixation of Fe in the roots (Figures 2F, 4F). Compared with N1, N2 significantly reduced the root Fe concentration (Figure 4F). N2 decreased root Fe concentration of Wu312 under Fe-deficient and Fe-sufficient conditions by 70.0 and 34.3%, respectively; and reduced Ye478 by 55.8 and 54.9%, respectively. There were no significant differences in the shoot Fe contents of two inbred lines between the N1 and N2 treatments (Figure 4G). However, the root Fe contents of two inbred lines in the N1 treatment were higher than those in the N2 treatment (Figure 4H). Compared with the N1 treatment, N2 treatment decreased root Fe content of Wu312 in the −Fe and +Fe treatments by 48.5 and 35.6% and decreased Ye478 by 44.9 and 41.9%, respectively. Compared with N1, the Fe uptake efficiency of the two inbred lines also showed a significant reduction in the N2 treatment (Figure 4I). Under Fe-deficient conditions, the Fe uptake efficiencies of Wu312 and Ye478 were decreased by 45.6 and 55.5%, respectively. Also, compared with N1, the supply of equivalent NH4+-N and NO3–-N improved the Fe transport efficiency of Wu312 under Fe starvation (Figure 4J).
Quantitative Trait Locus Analysis of Fe Deficiency Tolerance in Maize
Phenotypic Data Analysis
The Ye478 × Wu312 RIL population consisting of 218 lines was used to identify the loci associated with Fe deficiency tolerance in maize. In this study, phenotypic data of each trait under Fe-deficient condition (−Fe) and the ratios of the values under Fe-deficient condition to the values under Fe-sufficient condition (−Fe/+Fe) were used for QTL mapping. Here, the ratio of −Fe to +Fe for each trait was calculated using the BLUPs for each trait in the −Fe and +Fe treatment. The means and range of each trait for the parents and their RIL population are shown in Table 1. Our results indicated that Ye478 exhibited more tolerant to Fe deficiency when compared with Wu312. Fe-efficient inbred line Ye478 obtained higher leaf SPAD, plant height, shoot and root dry weights. Fe deficiency resulted in 64.5 and 74.2% decreases in leaf SPAD of Ye478 and Wu312, respectively. Fe deficiency led to a 66 and 75% reduction in leaf SPAD of Ye478 and Wu312, respectively. Plant heights of Ye478 and Wu312 under Fe deficiency were significantly decreased by 9 and 13%, respectively. Furthermore, Fe deficiency led to a 22 and 59% reduction in shoot dry weights of Ye478 and Wu312, respectively. Compared with Fe-sufficient condition, root dry weight of Ye478 under Fe-deficient condition was markedly decreased by 15%, and root dry weight of Wu312 was significantly reduced by 36%. Under Fe deficiency, the R/S ratio for Wu312 was markedly increased by 57%, whereas the R/S ratio of Ye478 displayed no significant difference between different treatments. Interestingly, the root length of Wu312 in the −Fe treatment was significantly higher than that of Ye478.
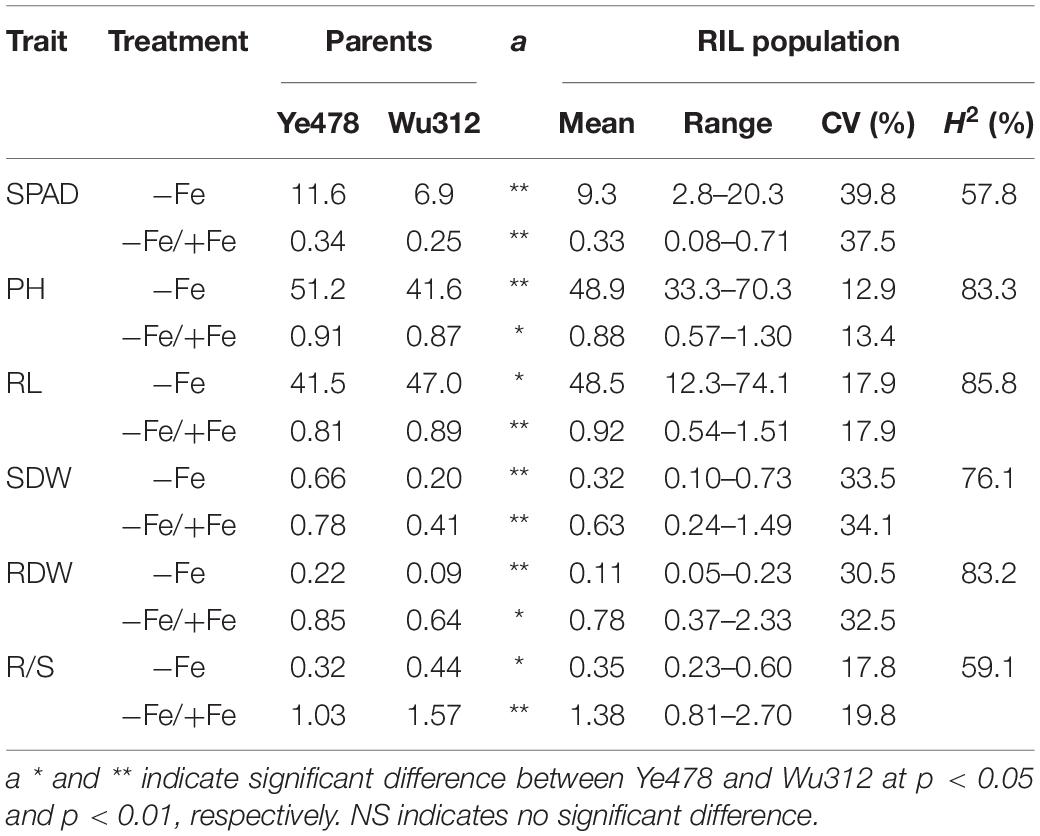
Table 1. Statistical analysis of phenotypic variation in parents and their recombinant inbred line (RIL) population.
In the −Fe and −Fe/+Fe treatments, the means for leaf SPAD, plant height, shoot and root dry weights, R/S ratio were between the values of Fe-efficient inbred line Ye478 and Fe-inefficient inbred line Wu312 (Table 1 and Figure 5). The coefficient of variation (CV) for each trait under Fe-deficient nutritional status ranged from 12.9 to 39.8%, indicating large phenotypic variation among 218 RILs. Most traits displayed normal distributions based on the BLUPs for each trait among the RILs (Figure 5 and Supplementary Table 2).
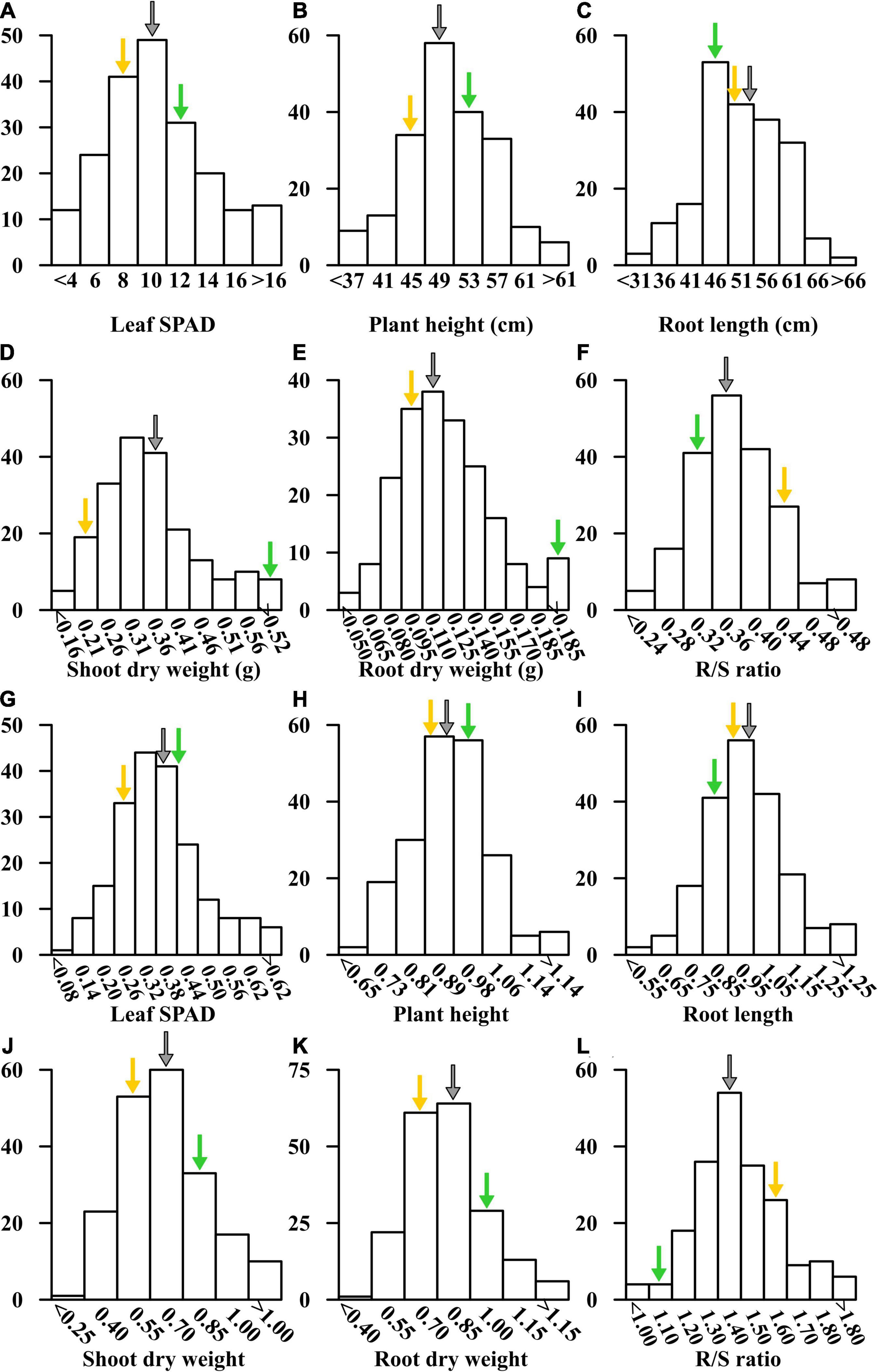
Figure 5. Distribution of SPAD value of the fourth leaf (A,G), plant height (B,H), root length (C,I), shoot (D,J) and root dry weights (E,K), and R/S ratio (F,L) in the –Fe and –Fe/+Fe treatments are shown in (A–L), respectively. Yellow and green arrows represent the mean values for each trait of Fe-inefficient parent Wu312 and Fe-efficient parent Ye478, respectively. Gray arrows indicate the mean values of each trait in the Ye478 × Wu312 RIL population.
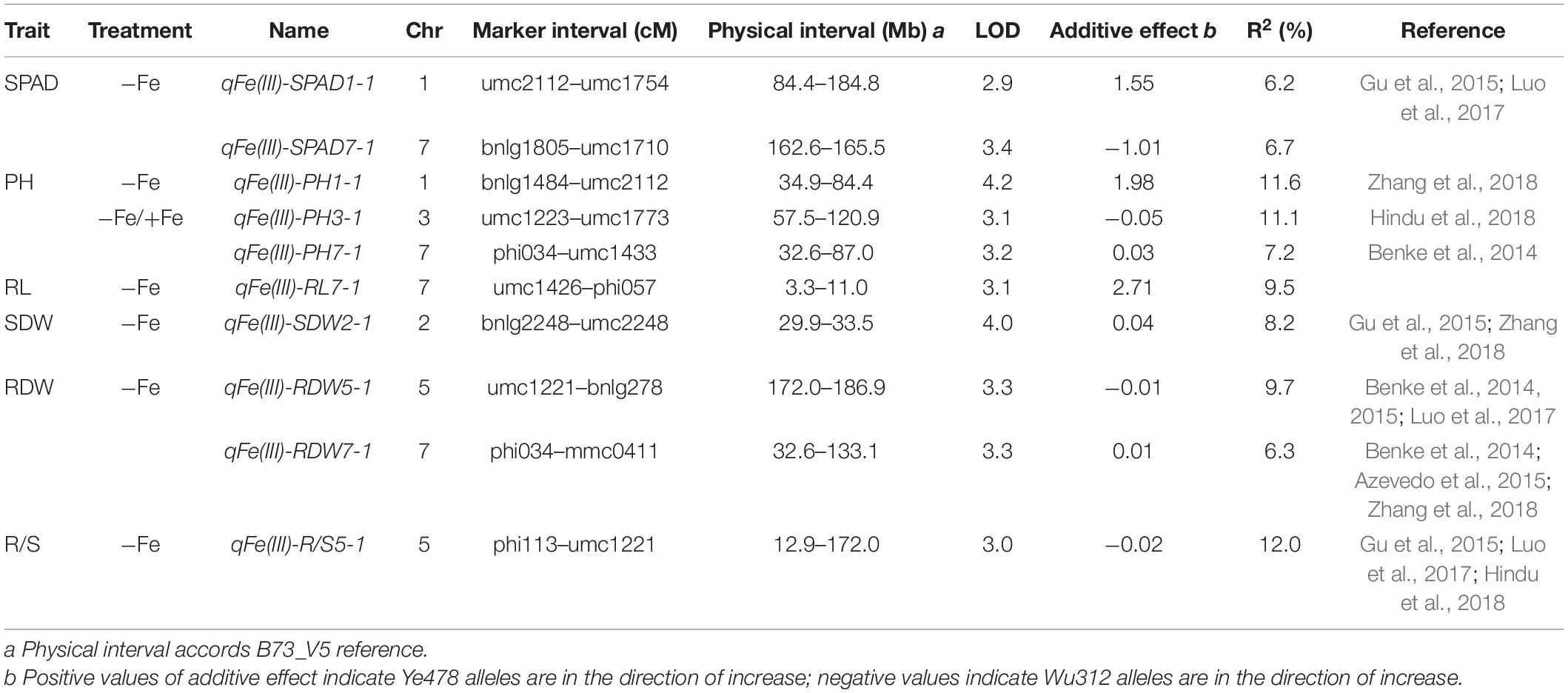
Pearson’s correlation analysis among different traits in the −Fe treatment was performed, and the coefficients of correlation between each trait pair are shown in Table 2. There were positive significant correlations among plant height, root length, shoot and root dry weights under Fe-deficient nutritional status (r = 0.163–0.859, p < 0.05) (Table 2), indicating that inbred lines, which had higher plant height, may also have higher root length as well as more biomass accumulation.
Quantitative Trait Locus Detection
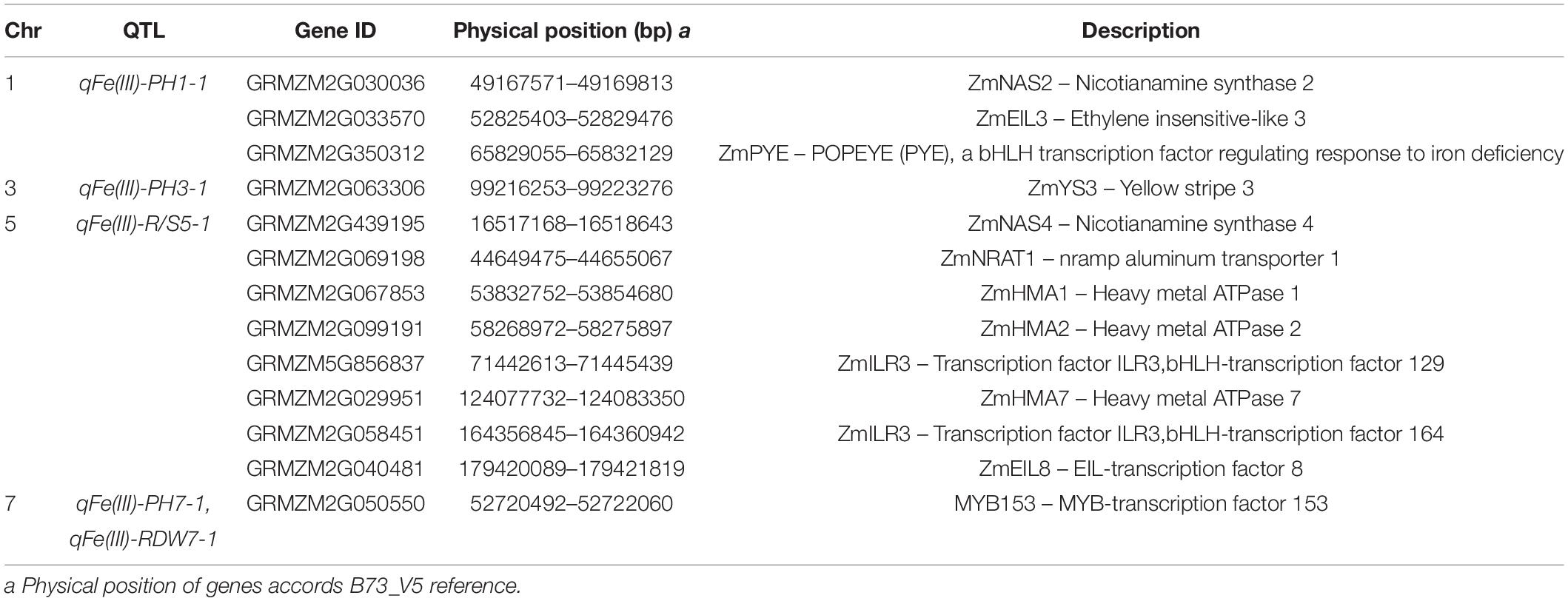
In this study, the BLUPs under Fe deficiency and the ratios of the BLUPs in the −Fe treatment to the values in the + Fe treatment for each trait were used to perform QTL analysis. Physical positions for each locus distributed detected in this study are shown in Figure 6. Details of each detected QTL, including QTL name, chromosomal position, 1-LOD marker interval, and physical interval, LOD value, additive effect, and percentages of total phenotypic variance explained (R2), are shown in Table 3. A total of ten quantitative trait loci controlling leaf SPAD, plant height, root length, shoot and root dry weights, and R/S ratio were identified on Chromosomes 1, 2, 3, 5, and 7, explaining 6.2–12.0% of phenotypic variation.
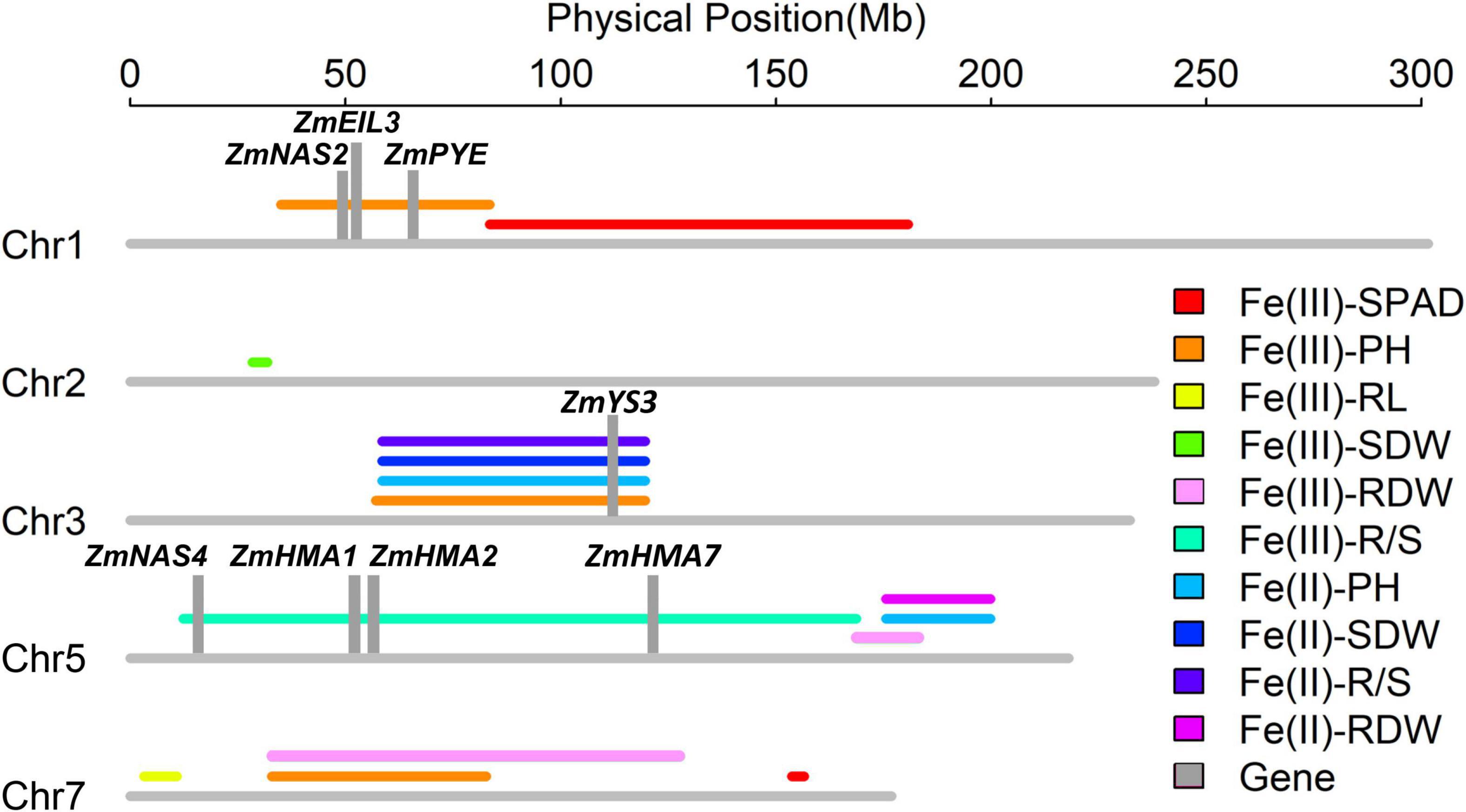
Figure 6. Ten quantitative trait loci associated with Fe deficiency tolerance detected on Chromosomes 1, 2, 3, 5, and 7 in the Ye478 × Wu312 recombinant inbred line (RIL) population and the co-localizations identified in the same population on the supply of Fe(III)-EDTA (Xu et al., 2022). Important candidate genes (ZmYS3, ZmHMA1, ZmHMA2, ZmHMA7, ZmNAS2, ZmNAS4, ZmPYE, and ZmEIL3) are depicted in gray columns. The lines of different colors indicate the QTLs controlling different traits, including leaf SPAD (SPAD), plant height (PH), root length (RL), shoot (SDW), and root (RDW) dry weights, and R/S ratio (R/S).
Two minor-effect QTLs controlling leaf SPAD were identified under Fe deficiency (Table 3). On Chromosome 1, qFe(III)-SPAD1-1 was located in the interval of umc2112-umc1754, explaining 6.2% of phenotypic variation. Alleles from Ye478 increased leaf SPAD by 1.55 at this locus. qFe(III)-SPAD7-1 was flanked by bnlg1805-umc1710 on Chromosome 7, accounting for 6.7% of phenotypic variation. Alleles from Wu312 contributed to increased leaf SPAD.
Three QTLs controlling plant height in the −Fe and −Fe/+Fe treatments and a single locus controlling root length under Fe-deficient condition were detected on Chromosomes 1, 3, and 7 (Table 3). The major-effect QTL qFe(III)-PH1-1 detected under Fe deficiency was located in the interval of bnlg1484-umc2112 on Chromosome 1, explaining 11.6% of phenotypic variation. Alleles from Ye478 at this QTL increased plant height by 1.98 cm. Major-effect QTL qFe(III)-PH3-1 and minor-effect QTL qFe(III)-PH7-1 controlling plant height were identified on Chromosomes 3 and 7 in the −Fe/+Fe treatment, and accounted for 11.1 and 7.2% of phenotypic variation, respectively. A minor-effect QTL qFe(III)-RL7-1 controlling root length was flanked by umc1426-phi057 on Chromosome 7. Alleles from Wu312 contributed to increased root length.
Four QTLs controlling shoot and root dry weights and the R/S ratio under Fe deficiency were identified on Chromosomes 2, 5, and 7, accounting for 6.3–12.0% of phenotypic variation. qFe(III)-SDW2-1 controlling shoot dry weight was flanked by bnlg2248-umc2248 on Chromosome 2 and explained 8.2% of phenotypic variation. A favorable allele at this locus came from Fe-efficient parent Ye478. Two minor-effect QTLs [qFe(III)-RDW5-1, qFe(III)-RDW7-1] controlling root dry weight were identified on Chromosomes 5 and 7, explaining 9.7 and 6.3% of phenotypic variation, respectively. Major-effect QTL qFe(III)-R/S5-1 was located in the interval of phi113-umc1221 on Chromosome 5, accounting for 12.0% of phenotypic variation.
Quantitative Trait Locus Co-localization and Candidate Gene Identification
As shown in Figure 6, three QTL co-localization regions were identified on Chromosomes 3, 5, and 7. In this study, qFe(III)-PH7-1 controlling plant height was co-localized with qFe(III)-RDW7-1 for root dry weight on Chromosome 7. Besides, another two loci in this study were co-localized with the loci identified in the Ye478 × Wu312 RIL population on the supply of Fe(III)-EDTA (Xu et al., 2022). On Chromosome 3, qFe(III)-PH3-1 detected on deficient supply of Fe(III)-EDTA in this study was co-localized with three loci [qFe(II)-PH3-1, qFe(II)-SDW3-1, and qFe(II)-R/S3-1] detected on the supply of ferrous iron. On Chromosome 5, qFe(III)-RDW5-1 controlling root dry weight was co-localized with two QTLs [qFe(II)-PH5-1 and qFe(II)-RDW5-1] mapped on the supply of ferrous iron.
Based on physical position, three QTL co-localization identified by nine QTLs for different traits on Chromosomes 3, 5 and 7 were selected for the identification of candidate genes. Three intervals were identified at 32.6–87.0 Mb on Chromosomes 7, 59.0–120.9 Mb on Chromosome 3, and 178.9–186.9 Mb on Chromosome 5. A total of 426 candidate genes were identified: 149 genes on Chromosome 7 (Supplementary Table 3), 165 genes on Chromosome 3 (Supplementary Table 4), and 112 genes on Chromosome 5 (Supplementary Table 5).
Based on the functional descriptions of 426 candidate genes in Arabidopsis and rice on MaizeGDB Database (see text footnote 1) and Gramene Database (see text footnote 2), three candidate genes were considered to be associated with Fe deficiency tolerance, including ZmYS3 (GRMZM2G063306), ZmEIL8 (GRMZM2G040481), and ZmMYB153 (GRMZM2G050550) (Table 4). Beyond that, according to previous reports on Fe deficiency, ten genes associated with the tolerance to Fe deficiency were identified within three loci [qFe(III)-PH1-1, qFe(III)-PH3-1, and qFe(III)-R/S5-1], including ZmNAS2 (GRMZM2G030036), ZmNAS4 (GRMZM2G439195), ZmEIL3 (GRMZM2G033570), ZmPYE (GRMZM2G350312), ZmHMA1 (GRMZM2G067853), ZmHMA2 (GRMZM2G099191), ZmHMA7 (GRMZM2G029951), and ZmILRs (GRMZM5G 856837, GRMZM2G058451) (Table 4).
Expression of Candidates Associated With Fe Deficiency Tolerance in Maize
According to the candidate genes identified in Experiment 3, the expression of ZmYS3 (GRMZM2G063306), ZmPYE (GRMZM2G350312), ZmEIL3 (GRMZM2G033570), and ZmMYB153 (GRMZM2G050550) was assessed in Fe-deficient and Fe-sufficient shoots and roots of Fe-inefficient parent Wu312 and Fe-efficient parent Ye478 (Figure 7).
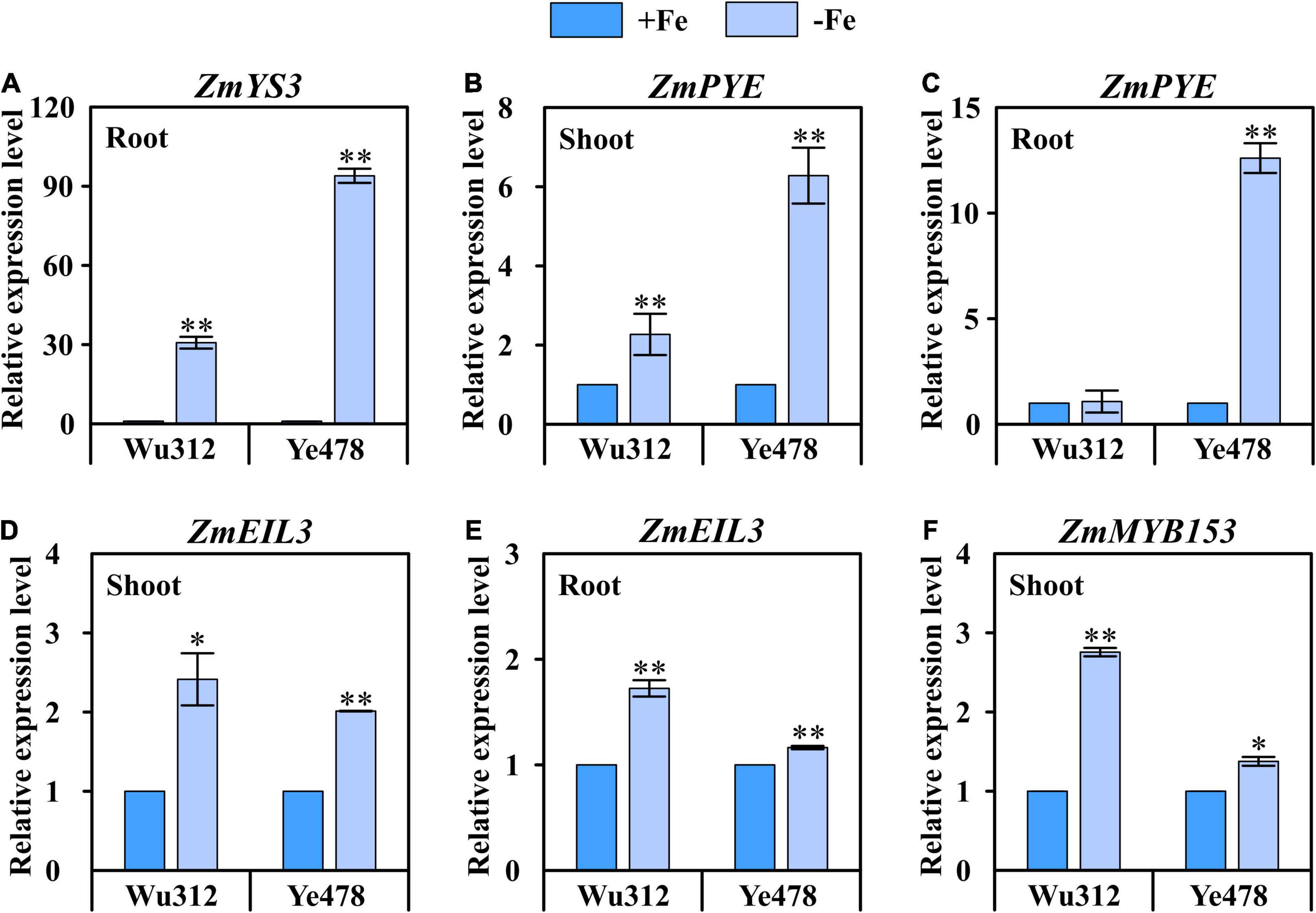
Figure 7. Expression of four candidate genes in the shoots and roots of Fe-inefficient (Wu312) and Fe-efficient (Ye478) parents under Fe-deficient [–Fe: 30 μM Fe(III)-EDTA] and Fe-sufficient [+Fe: 200 μM Fe(III)-EDTA] conditions. (A) ZmYS3 expression in the roots, (B,C) ZmPYE and (D,E) ZmEIL3 expression in the shoots and roots, (F) ZmMYB153 expression in the shoots. * and ** indicate significant difference between the –Fe and +Fe treatments at p < 0.05 and p < 0.01, respectively.
It should be noted that ZmYS3, which encodes a yellow-stripe protein, was only expressed in the roots of Wu312 and Ye478. For Fe-inefficient parent Wu312, ZmYS3 was significantly (p < 0.01, 31-fold) upregulated under Fe deficiency. A 94-fold upregulation was observed in Fe-deficient roots of Fe-efficient parent Ye478, which was more than three times higher than that for Wu312 (Figure 7A). Interestingly, ZmPYE, a homolog of POPEYE, which plays a vital role in Fe homeostasis in strategy I plants in rice and non-gramineous plants, was significantly highly induced by Fe deficiency in the shoots and roots of two inbred lines. In the shoots, ZmPYE was 2-fold and 6-fold upregulated by Fe deficiency in Wu312 and Ye478, respectively (Figure 7B). In the roots, 13-fold upregulation induced by Fe deficiency was found in Ye478; however, no change in expression was observed in Wu312 (Figure 7C). Similar to ZmYS3, the upregulation of ZmPYE induced by low Fe stress was greater in Ye478 than in Wu312. These findings implicated that candidate genes ZmYS1 and ZmPYE may be responsible for untangling genotypic differences in tolerance to Fe deficiency between the Fe-inefficient parent Wu312 and Fe-efficient parent Ye478.
The ZmEIL3, a homolog of ETHYLENE INTENSITIVE3, which is the key for ethylene regulation and essential for Fe homeostasis in Arabidopsis, was induced by low Fe stress in both shoots and roots of two inbred lines (Figures 7D,E). Moreover, ZmMYB153 was only significantly upregulated in the shoots of two parents (Figure 7F). These results indicated that ZmEIL3 and ZmMYB153 may be involved in the mechanisms of Fe deficiency tolerance in maize.
Discussion
Physiological Evaluation of Fe Efficiency in Maize Seedlings Adapted to Low-Fe Stress
Previous studies have shown that Fe-efficient genotypes should not only be able to absorb more Fe from deficient soils but should also produce more dry matter and grain yield (Krishnasamy et al., 2005; Long et al., 2020), which was consistent with the results in the current study. On the supply of 30 μM Fe(III)-EDTA in Experiment 1, shoot and root dry weights of Fe-efficient Ye478 were 3.8-fold and 2.7-fold higher than those of Fe-inefficient Wu312, respectively (Figures 2A,B). Also, the Fe efficiencies based on the shoot and root dry weights of Ye478 were 68.8 and 66.7%, respectively, which were significantly higher than those of Wu312 (Figure 3). Fe is an essential component of several Fe-S proteins involved in the chloroplast electron transfer chain (Yousfi et al., 2009). The reason why the dry weight of Wu312 decreased significantly may be related to the alteration in chloroplasts, the corresponding inhibition of cell division, and the activation of several glycolytic enzymes (Rabotti et al., 1995; Espen et al., 2000). Rengel and Römheld (2000) demonstrated that the shoot dry weight and relative shoot growth were the most suitable parameters for distinguishing the ability of plants to tolerate Fe deficiency. Moreover, Fe deficiency response requires an overall increase in carbon import to the roots, favoring root expansion (Jin et al., 2008). Fe deficiency stress leads to an increase in the R/S ratio, indicating a less reduction of shoot growth when compared with root growth (Cianzio and Voss, 1994). In this study, compared with Fe-sufficient supply, we observed a significant increase of the R/S ratio in Fe-inefficient inbred line Wu312 and no significant difference of the R/S ratio in Fe-efficient inbred line Ye478 on the Fe supply of 30 μM (Figure 2C). This indicated that the R/S ratio displayed a markedly genotypic difference of the allocation of biomass in different organs between Fe-efficient and Fe-inefficient genotypes in response to low-Fe nutrient availability. Crowley et al. (2002) reported that Fe starvation altered R/S ratios in different barley cultivars and suggested this parameter as a criterion for selecting Fe-efficient plants. Phytosiderophores (PS) not only promote the root uptake of Fe but also regulate the translocation of Fe to the shoot as the Fe-PS complex (Nogiya et al., 2016). However, Zhang et al. (1991) reported a relatively poor relationship between PS exudation and Fe deficiency tolerance of barley genotypes. And, in several maize cultivars, Fe deficiency tolerance was reported not to be related to the total amounts of secreted PS (von Wirén et al., 1994; Yousfi et al., 2009).
Shoot Fe concentration of Ye478 was higher than that of Wu312 (Figure 2E). However, when the Fe supply level changed from 10 to 100 μM, the root Fe concentration of Wu312 was significantly higher than that of Ye478 (Figure 2F). The confirmed results from some cereal species indicated that the concentration of Fe is not a reliable indicator for distinguishing sensitivity to Fe deficiency among genotypes (Mohamed et al., 2003; Essa et al., 2015). Moreover, it has been established that, when plants are grown under Fe deficiency in field conditions, no significant correlation is found between Fe concentration and crop tolerance to Fe deficiency (Eshghizadeh et al., 2015). Iron chlorosis is due to chlorophyll dilution when leaves continue to grow at a normal rate under Fe deficiency (Abadía et al., 1999). The difference in the pigment dilution rates was directly related to the dissimilarity in growth rates (leaf expansion rate) (Yousfi et al., 2009). Combined with the results of visual observation and shoot dry weight, the growth rate of Ye478 plants was significantly higher than that of Wu312 (Figure 2A). Simultaneously, the SPAD value of Ye478 was also higher than that of Wu312 (Figure 2D). Therefore, the SPAD value is also a suitable index for evaluating Fe efficiency. An efficient root-to-shoot translocation of Fe, under Fe-deficient condition, has been causally related to the Fe deficiency tolerance in rice and wheat (Nogiya et al., 2016). However, our results showed that the Fe transport efficiency of two inbred lines did not display a significant difference in most treatments (Figure 2L). Taken together, shoot and root dry weights, R/S ratio, leaf SPAD, and plant height are suitable for evaluating the tolerance to Fe deficiency in maize.
Effects of Different Nitrogen Forms on Maize Growth and Fe Nutritional Status
In this study, the nitrogen (N) supply form of Experiment 1 was to supply only nitrate-nitrogen recommended by Hoagland nutrient solution. The results showed that, when the Fe supply concentration of Wu312 was as high as 300 μM, it had an inhibitory effect on plant growth. There was still obvious inter-vein chlorosis. The roots of two inbred lines precipitated and fixed a lot of Fe. Because the Fe nutrition of plants can be significantly affected by the form of N supply through altering the rhizosphere pH, apoplastic pH, and the uptake ratio of cations and anions (Zou et al., 2001). For example, in sunflower, supply of NO3–-N increased leaf apoplastic pH and depressed Fe(III) reductase activity and hence induced Fe chlorosis (Kosegarten et al., 1999). However, in ryegrass, switching the root N source from nitrate to ammonium causes a concomitant decrease in apoplastic pH (Schjoerring et al., 2002). The Fe availability to plants could increase 1,000-fold with a decrease in pH, such as from pH 8 to pH 7 (Guo et al., 2014). This study used different nitrogen supply forms to improve the growth of plants under Fe starvation and, at the same time, reduce the accumulation of Fe in roots.
It has been reported that ammonium as the sole N source can inhibit plant growth, including maize seedlings (Wang et al., 2015; Esteban et al., 2016). However, ammonium nitrogen can improve plant tolerance to a variety of abiotic stresses and enhance resistance to pathogen infection (Hessini et al., 2013; Vega-Mas et al., 2017; Xun et al., 2020). In addition, nitrate may stimulate the xylem loading of ammonium and/or its assimilation through the enhanced biosynthesis of amino acids (Kiyomiya et al., 2001). However, co-provision of nitrate and ammonium is beneficial for plant growth beyond that observed with either N source provided alone (Carlisle et al., 2012; Luo et al., 2013). Like other major crops, maize plants achieve optimal growth and yield under mixed N nutrition of nitrate and ammonium (Gu et al., 2013; Hessini et al., 2017; Ravazzolo et al., 2020), which were consistent with our results. In Experiment 2 of this study, N2 significantly increased the shoot and root dry weights of Wu312 by 37.5 and 51.6% and enhanced the shoot and root dry weights of Ye478 by 23.9 and 45.1% under Fe-deficient condition, respectively (Figures 4C,D). Beyond that, under Fe-sufficient condition, the shoot and root dry weights of Ye478 were increased by 32.8 and 27.6%, respectively (Figures 4C,D). The application of ammonium stimulates lateral roots (LRs) branching, whereas nitrate stimulates LR elongation, suggesting that the application of nitrate and ammonium has local and complementary effects on LR development (Hachiya and Sakakibara, 2017). This may also improve the tolerance to low-Fe stress of the two inbred lines. The synergistic beneficial effects of co-provision mainly arise from the reciprocal influences between NH4+ and NO3– on their uptake, root morphology, transport of N compounds from roots to shoots, and plant C metabolism (Prinsi and Espen, 2018).
In this study, compared with Experiment 1, root Fe concentration in Experiment 2 was substantially decreased, which may be mainly due to the cleaning of roots using a mixed solution of EDTA-Na and CaSO4 and the solution of HCl. In this research, the supply of ammonium nitrate (N2) markedly reduced the root Fe concentrations of Wu312 and Ye478 in the −Fe and +Fe treatments (Figure 4F). This could be that external Fe was immobilized in the root apoplast or onto the root surface due to high pH and typical nucleation of precipitated Fe (Strasser et al., 1999). However, co-provision of nitrate and ammonium could have caused dissolution or remobilization dissolution of the Fe by lowering pH, resulting in a relatively low concentration of Fe in the roots (Zou et al., 2001). Besides, N2 also significantly reduced the root Fe contents of these two inbred lines under Fe-deficient and Fe-sufficient conditions; however, such differences were not shown in shoot Fe contents (Figures 4G,H). This may be attributed to the reduction of Fe(III)-citrate that might be depressed by the high root apoplastic pH, disrupting Fe translocation to the shoots (Mengel et al., 1994).
Comparison of Detected Loci With Previous Studies
Numerous studies have been related to the concentrations of Fe and other heavy metals in grains and seedling leaves (Blair et al., 2010a; Mamo et al., 2014; Zdunić et al., 2014; Gu et al., 2015; Hindu et al., 2018; Izquierdo et al., 2018; Jeong et al., 2020). Nevertheless, there are very few previous studies focusing on Fe deficiency tolerance in crops (Blair et al., 2010b; Peiffer et al., 2012), and only a few researchers have reported the loci associated with the performance under Fe deficiency in maize (Benke et al., 2014, 2015).
In total, eight of ten QTLs detected in this study were co-localized with the loci controlling different traits reported in other pieces of research (Table 3). The qFe(III)SPAD1-1, qFe(III)-SDW2-1, qFe(III)PH3-1, and qFe(III)-RDW5-1 identified in this study were co-localized with the loci controlling Zn and manganese (Mn) concentration in grains (Gu et al., 2015; Hindu et al., 2018), indicating that these QTL regions may have pleiotropic effects on mineral concentration of grains in maize. Intervals of qFe(III)SPAD1-1, qFe(III)-R/S5-1, and qFe(III)-RDW5-1 were overlapped by the genomic regions identified by the loci controlling plant height under salt stress detected by Luo et al. (2017), which suggested that these QTL regions may have pleiotropic effects on plant height under abiotic stress. Two QTLs [qFe(III)-R/S5-1 and qFe(III)-RDW5-1] controlling root dry weight under Fe deficiency in this work were co-localized with the loci controlling the same trait under Fe-deficient condition identified by linkage and association analysis (Benke et al., 2014, 2015). Besides, intervals of these two loci were also overlapped by the genomic regions of the QTLs controlling plant height under normal and external ethylene conditions, root diameter under low-P stress, and SPAD under low-Fe stress (Benke et al., 2014; Azevedo et al., 2015; Luo et al., 2017; Zhang et al., 2018). This implicated that these gnomic regions may harbor several genes with pleiotropic effect not only on root biomass accumulation but also on plant height, root morphological traits, and leaf SPAD at the seedling stage.
Candidate Genes Associated With Fe Deficiency Tolerance
The GRMZM2G063306, also known as ZmYS3, was detected in the QTL co-localization of qFe(III)-PH3-1, qFe(II)-PH3-1, qFe(II)-SDW3-1 and qFe(II)-R/S3-1, which were identified on the supply of Fe(II) and Fe(III). It is reported that ZmYS3 was substantially induced by Fe deficiency in maize roots (Nozoye et al., 2013). Fe deficiency-inducible genes, such as ZmTOM1, ZmDMAS1 and ZmIRT1, are upregulated in the roots of the ys3 mutant, even under Fe-sufficient condition, suggesting that ys1 and ys3 plants are Fe deficient during growth in presence of Fe (Nozoye et al., 2013, 2015). Unspliced introns of ZmTOM1 were detected only in ys3, rather than in YS1YS3 or ys1, suggesting that ZmTOM1 may be involved in the ys3 phenotype (Nozoye et al., 2013, 2015). Furthermore, our results indicated that ZmYS3 was remarkably upregulated in the roots of Wu312 and Ye478 under Fe deficiency, and the upregulation was greater in the Fe-efficient genotype than in the Fe-inefficient genotype (Figure 7A). These findings indicated that ZmYS3 is involved in strategy II Fe acquisition and may play important roles in differing the ability to tolerate low-Fe stress in maize.
Apart from ZmYS3, other candidate genes may be involved in Fe homeostasis in strategy I plants and heavy metals transport in plants, which may form a sophisticated network underlying Fe deficiency tolerance. The GRMZM2G350312 identified within qFe(III)-PH1-1 under deficient supply of Fe(III) in this study, encodes a bHLH transcription factor regulating the response to Fe deficiency and is identified to be POPEYE (PYE) in maize. The POPEYE (PYE belongs to IVb subgroup bHLHs) functions as an important repressor in the Fe deficiency regulatory network (Wang et al., 2018; Cheng et al., 2020). The PYE targets key genes involved in metal ion homeostasis, including a Zn transport facilitator (ZIF1), a plasma membrane-localized ferric chelate reductase from the same family as FRO2 (FRO3), and a nicotianamine synthase (NAS4) (Dinneny et al., 2008; Jeong et al., 2008; Curie et al., 2009). The PYE protein is localized in the nuclei of all cells within Fe-deficient Arabidopsis roots and is most highly expressed in the pericycle within the maturation zone (Long et al., 2010). The pye-1 mutant exhibits chlorosis and root growth inhibition under Fe deficiency, which is associated with decreased elongation and swelling of root cells (Hindt and Guerinot, 2012; Kobayashi and Nishizawa, 2012). Additionally, in pye-1, several other Fe homeostasis genes are upregulated, including OPT3, FRD3, NRAMP4, bHLH39 and bHLH101 (Green and Rogers, 2004; Stacey et al., 2008; Long et al., 2010). In this research, we found that ZmPYE was not only significantly upregulated in the roots but also in shoots under Fe deficiency, and a greater upregulation was observed in Fe-efficient maize inbred line Ye478 (Figures 7B,C). This indicates that ZmPYE, which is involved in Fe homeostasis in strategy I plants, may play a vital role in Fe efficiency in maize.
The GRMZM2G033570, also known as EIL3 encoding ETHYLENE INSENSITIVE3-LIKE1 protein in maize, was identified within qFe(III)-PH1-1 under Fe-deficient condition. Besides a potential role in ethylene signaling, EIL3 (also called SLIM1 for SULFUR LIMITATION1), which participates in the modulation of S-deficiency response, is the only identified transcription factor in plant S-metabolism (Van der Ent et al., 2008; Filiz et al., 2017). The EIL3 function as a central transcriptional regulator, which control both the activation of sulfate acquisition and degradation of glucosinolates under –S conditions (Maruyama-Nakashita et al., 2006). It is reported that MYB72 physically interacts in vitro with the EIL3, which is induced under Fe deficiency (Van der Ent et al., 2008; García et al., 2011). This tandem MYB72-EIL3 may play a role in the regulation of Fe-deficiency responses (García et al., 2010).
In addition, GRMZM2G050550 that encodes MYB-transcription factor 153 was detected within qFe(III)-PH7-1 in the −Fe/+Fe treatment. MYB transcription factors play pivotal roles in hormone conduction signaling and abiotic stress response (Zhang et al., 2021). Some members of the MYB family (members of the myeloblastosis family), which may be implicated in the PYE-mediated Fe deficiency response, have been implicated in Fe redistribution through the regulation of NAS4 (Palmer et al., 2013; Brumbarova et al., 2015). The TPL/TPR transcriptional co-repressors play very important roles in various phytohormone signaling pathways (including jasmonic acid and strigolactone). Some studies speculate that ZmMYB153 recruits the LisH or CTLH domains of some proteins in the TPL/TPRs family through its LSLSL-type EAR motif to form a complex, thereby inhibiting the expression of the relevant genes (Bai et al., 2020). This affects how maize responds to biotic and abiotic stresses.
Our results indicated that ZmEIL3 was significantly upregulated in the Fe-deficient shoots and roots of maize inbred lines (Figures 7D,E), and ZmMYB153 was Fe-deficiency inducible in maize roots (Figure 7F), further indicating that ZmEIL3 and ZmMYB153 may be involved in Fe deficiency tolerance in maize, which is considered as a strategy II plant.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
JX, FYa, and XZhu performed the experiments. JX and XZhu analyzed the data. JX, XZhu, FYa, and HZ wrote the manuscript. FYu designed the study and modified the manuscript. All authors contributed to the article and approved the submitted version.
Funding
National Key Research and Development Program of China (2016YFD0200405).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Professor Fanjun Chen (China Agricultural University) for providing inbred lines Wu312, Ye478, and the Ye478 × Wu312 RIL population.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.805247/full#supplementary-material
Footnotes
References
Abadía, J., Morales, F., and Abadía, A. (1999). Photosystem II efficiency in low chlorophyll, iron-deficient leaves. Plant Soil 215, 183–192. doi: 10.1023/A:1004451728237
Azevedo, G. C., Cheavegatti-Gianotto, A., Negri, B. F., Hufnagel, B., E Silva, L. C., Magalhaes, J. V., et al. (2015). Multiple interval QTL mapping and searching for PSTOL1 homologs associated with root morphology, biomass accumulation and phosphorus content in maize seedlings under low-P. BMC Plant Biol. 15:172. doi: 10.1186/s12870-015-0561-y
Bai, H., Cao, H., Liu, P., Zhou, F., Zang, J., Dong, J., et al. (2020). Study of interaction between transcription factor ZmMYB153 and TPL/TPRs proteins. J. Hebei Agric. Univ. 43, 61–67. doi: 10.13320/j.cnki.jauh.2020.0053
Benke, A., Urbany, C., Marsian, J., Shi, R., Wirén, N. V., and Stich, B. (2014). The genetic basis of natural variation for iron homeostasis in the maize IBM population. BMC Plant Biol. 14:12. doi: 10.1186/1471-2229-14-12
Benke, A., Urbany, C., and Stich, B. (2015). Genome-wide association mapping of iron homeostasis in the maize association population. BMC Genet. 16:1. doi: 10.1186/s12863-014-0153-0
Blair, M. W., Medina, J. I., Astudillo, C., Rengifo, J., Beebe, S. E., Machado, G., et al. (2010a). QTL for seed iron and zinc concentration and content in a Mesoamerican common bean (Phaseolus vulgaris L.) population. Theor. Appl. Genet. 121, 1059–1070. doi: 10.1007/s00122-010-1371-0
Blair, M. W., Knewtson, S. J., Astudillo, C., Li, C. M., Fernandez, A. C., and Grusak, M. A. (2010b). Variation and inheritance of iron reductase activity in the roots of common bean (Phaseolus vulgaris L.) and association with seed iron accumulation QTL. BMC Plant Biol. 10:215. doi: 10.1186/1471-2229-10-215
Brumbarova, T., Bauer, P., and Ivanov, R. (2015). Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 20, 124–133. doi: 10.1016/j.tplants.2014.11.004
Carlisle, E., Myers, S., Raboy, V., and Bloom, A. (2012). The effects of inorganic nitrogen form and CO2 concentration on wheat yield and nutrient accumulation and distribution. Front. Plant Sci. 3:195. doi: 10.3389/fpls.2012.00195
Chan-Rodriguez, D., and Walker, E. L. (2018). Analysis of Yellow Striped mutants of Zea mays reveals novel loci contributing to iron deficiency chlorosis. Front. Plant Sci. 9:157. doi: 10.3389/fpls.2018.00157
Cheah, Z. X., Kopittke, P. M., Harper, S. M., O’Hare, T. J., Wang, P., Paterson, D. J., et al. (2019). In situ analyses of inorganic nutrient distribution in sweetcorn and maize kernels using synchrotron-based X-ray fluorescence microscopy. Ann. Bot. 123, 543–556. doi: 10.1093/aob/mcy189
Cheng, N., Yu, H., Rao, X., Park, S., Connolly, E. L., Hirschi, K. D., et al. (2020). Alteration of iron responsive gene expression in Arabidopsisglutaredoxin S17 loss of function plants with or without iron stress. Plant Signal Behav. 15:1758455. doi: 10.1080/15592324.2020.1758455
Chorianopoulou, S. N., Saridis, Y. I., Dimou, M., Katinakis, P., and Bouranis, D. L. (2015). Arbuscular mycorrhizal symbiosis alters the expression patterns of three key iron homeostasis genes, ZmNAS1, ZmNAS3, and ZmYS1, in S deprived maize plants. Front. Plant Sci. 6:257. doi: 10.3389/fpls.2015.00257
Cianzio, S. R., and Voss, B. K. (1994). Three strategies for population development in breeding high-yielding soybean cultivars with improved iron efficiency. Crop Sci. 34, 355–359. doi: 10.2135/cropsci1994.0011183X003400020009x
Crowley, D. E., Wu, C. L., Gries, D., Brünn, S., and Parker, D. R. (2002). Quantitative traits associated with adaptation of three barley (Hordeum vulgare L.) cultivars to suboptimal iron supply. Plant Soil 241, 57–65. doi: 10.1023/A:1016047101677
Curie, C., Cassin, G., Couch, D., Divol, F., and Higuchi, K. (2009). Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. 103, 1–11. doi: 10.1093/aob/mcn207
Dinneny, J. R., Long, T. A., Wang, J. Y., Jung, J. W., Mace, D., Pointer, S., et al. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945. doi: 10.1126/science.1153795
Eshghizadeh, H. R., Khoshgoftarmanesh, A. H., and Zahedi, M. (2015). Iron efficiency of different corn hybrids grown in nutrient solution culture. Arch. Agron. Soil Sci. 61, 877–887. doi: 10.1080/03650340.2014.957192
Espen, L., Dell’Orto, M., De Nisi, P., and Zocchi, G. (2000). Metabolic responses in cucumber (Cucumis sativus L.) roots under Fe-deficiency: a 31P-nuclear magnetic resonance in-vivo study. Planta 210, 985–992. doi: 10.1007/s004250050707
Essa, E. M., Zaghloul, S. M., Salama, Z. A., Rakha, F. A., Mabrouk, Y., and El-Bendary, A. A. (2015). Physiological and biochemical evaluation of Fe-efficiency in Fe-deficient maize genotypes. Am. J Agri. Biolog. Sci. 10, 55–62. doi: 10.3844/ajabssp.2015.55.62
Esteban, R., Ariz, I., Cruz, C., and Moran, J. F. (2016). Review: mechanisms of ammonium toxicity and the quest for tolerance. Plant Sci. 248, 92–101. doi: 10.1016/j.plantsci.2016.04.008
Filiz, E., Vatansever, R., Ozyigit, I. I., Uras, M. E., Sen, U., Anjum, N. A., et al. (2017). Genome-wide identification and expression profiling of EIL gene family in woody plant representative poplar (Populustrichocarpa). Arch. Biochem. Biophys. 627, 30–45. doi: 10.1016/j.abb.2017.06.012
García, M. J., Lucena, C., Romera, F. J., Alcántara, E., and Pérez-Vicente, R. (2010). Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J. Exp. Bot. 61, 3885–3899. doi: 10.1093/jxb/erq203
García, M. J., Suárez, V., Romera, F. J., Alcántara, E., and Pérez-Vicente, R. (2011). A new model involving ethylene, nitric oxide and Fe to explain the regulation of Fe-acquisition genes in strategy I plants. Plant Physiol. Biochem. 49, 537–544. doi: 10.1016/j.plaphy.2011.01.019
Green, L. S., and Rogers, E. E. (2004). FRD3 controls iron localization in Arabidopsis. Plant Physiol. 136, 2523–2531. doi: 10.1104/pp.104.045633
Gu, R., Chen, F., Liu, B., Wang, X., Liu, J., Li, P., et al. (2015). Comprehensive phenotypic analysis and quantitative trait locus identification for grain mineral concentration, content, and yield in maize (Zea mays L.). Theor. Appl. Genet. 128, 1777–1789. doi: 10.1007/s00122-015-2546-5
Gu, R., Duan, F., An, X., Zhang, F., von Wirén, N., and Yuan, L. (2013). Characterization of AMT-mediated high-affinity ammonium uptake in roots of maize (Zea mays L.). Plant Cell Physiol. 54, 1515–1524. doi: 10.1093/pcp/pct099
Guerinot, M. L. (2007). It’s elementary: enhancing Fe3+ reduction improves rice yields. Proc. Natl. Acad. Sci. U.S.A. 104, 7311–7312. doi: 10.1073/pnas.0701954104
Guo, S., Chen, Y., Chen, X., Chen, Y., Yang, L., Wang, L., et al. (2020). Grain mineral accumulation changes in Chinese maize cultivars released in different decades and the responses to nitrogen fertilizer. Front. Plant Sci. 10:1662. doi: 10.3389/fpls.2019.01662
Guo, X., Xiong, H., Shen, H., Qiu, W., Ji, C., Zhang, Z., et al. (2014). Dynamics in the rhizosphere and iron-uptake gene expression in peanut induced by intercropping with maize: role in improving iron nutrition in peanut. Plant Physiol. Biochem. 76, 36–43. doi: 10.1016/j.plaphy.2013.12.019
Hachiya, T., and Sakakibara, H. (2017). Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J. Exp. Bot. 68, 2501–2512. doi: 10.1093/jxb/erw449
Hessini, K., Hamed, K. B., Gandour, M., Mejri, M., Abdelly, C., and Cruz, C. (2013). Ammonium nutrition in the halophyte Spartina alterniflora under salt stress: evidence for a priming effectof ammonium? Plant Soil 370, 163–173. doi: 10.1007/s11104-013-1616-1
Hessini, K., Kronzucker, H. J., Abdelly, C., and Cruz, C. (2017). Drought stress obliterates the preference for ammonium as an N source in the C4 plant Spartina alterniflora. J. Plant Physiol. 213, 98–107.
Hindt, M. N., and Guerinot, M. L. (2012). Getting a sense for signals: regulation of the plant iron deficiency response. Biochim. Biophys. Acta. 1823, 1521–1530. doi: 10.1016/j.bbamcr.2012.03.010
Hindu, V., Palacios-Rojas, N., Babu, R., Suwarno, W. B., Rashid, Z., Usha, R., et al. (2018). Identification and validation of genomic regions influencing kernel zinc and iron in maize. Theor. Appl. Genet. 131, 1443–1457. doi: 10.1007/s00122-018-3089-3
Housh, A. B., Powell, G., Scott, S., Anstaett, A., Gerheart, A., Benoit, M., et al. (2021). Functional mutants of Azospirillum brasilense elicit beneficial physiological and metabolic responses in Zea mays contributing to increased host iron assimilation. ISME J. 15, 1505–1522.
Ivanov, R., Brumbarova, T., and Bauer, P. (2012). Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 5, 27–42. doi: 10.1093/mp/ssr065
Izquierdo, P., Astudillo, C., Blair, M. W., Iqbal, A. M., Raatz, B., and Cichy, K. A. (2018). Meta-QTL analysis of seed iron and zinc concentration and content in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 131, 1645–1658. doi: 10.1007/s00122-018-3104-8
Jeong, J., Cohu, C., Kerkeb, L., Piln, M., Connolly, E. L., and Guerinot, M. L. (2008). Chloroplast Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proc. Natl. Acad. Sci. 105, 10619–10624. doi: 10.1073/pnas.0708367105
Jeong, O. Y., Lee, J. H., Jeong, E. G., Chun, A., Bombay, M., Ancheta, M. B., et al. (2020). Analysis of QTL responsible for grain iron and zinc contentin doubled haploid lines of rice (Oryza sativa) derived from anintra-japonica cross. Plant Breed. 139, 344–355. doi: 10.1111/pbr.12787
Jin, C. W., Chen, W. W., Meng, Z. B., and Zheng, S. J. (2008). Iron deficiency-induced increase of root branching contributes to the enhanced root ferric chelate reductase activity. J. Integr. Plant Biol. 50, 1557–1562. doi: 10.1111/j.1744-7909.2008.00654.x
Kiyomiya, S., Nakanishi, H., Uchida, H., Tsuji, A., Nishiyama, S., Futatsubashi, M., et al. (2001). Real time visualization of 13N-translocation in rice under different environmental conditions using positron emitting Ttacer imaging system. Plant Physiol. 125, 1743–1753. doi: 10.1104/pp.125.4.1743
Kobayashi, T., and Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. doi: 10.1146/annurev-arplant-042811-105522
Kosegarten, H. U., Hoffmann, B., and Mengel, K. (1999). Apoplastic pH and Fe3+ reduction in intact sunflower leaves. Plant Physiol. 121, 1069–1079. doi: 10.1104/pp.121.4.1069
Krishnasamy, R., Jegadeeswari, D., Surendran, U., and Sudhalakshmi, C. (2005). Screening of sorghum (Sorghum bicolor) genotypes for their iron efficiency. World J. Agric. Sci. 1, 98–100.
Kurt, F., Filiz, E., and Kurt, B. (2019). A key gene bHLH115 in iron homeostasis: comprehensive bioinformatics analyses in Arabidopsis, tomato, rice, and maize. Biometals 32, 641–656. doi: 10.1007/s10534-019-00199-z
Li, S., Liu, X., Zhou, X., Li, Y., Yang, W., and Chen, R. (2019). Improving zinc and iron accumulation in maize grains using the zinc and iron transporter ZmZIP5. Plant Cell Physiol. 60, 2077–2085. doi: 10.1093/pcp/pcz104
Li, W., and Lan, P. (2015). Genome-wide analysis of overlapping genes regulated by iron deficiency and phosphate starvation reveals new interactions in Arabidopsis roots. BMC Res. Notes 8:555. doi: 10.1186/s13104-015-1524-y
Li, X., Zhang, H., Ai, Q., Liang, G., and Yu, D. (2016). Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 170, 2478–2493. doi: 10.1104/pp.15.01827
Li, Y., Wang, N., Zhao, F., Song, X., Yin, Z., Huang, R., et al. (2014). Changes in the transcriptomic profiles of maize roots in response to iron-deficiency stress. Plant Mol. Biol. 85, 349–363. doi: 10.1007/s11103-014-0189-6
Liu, J., Cai, H., Chu, Q., Chen, X., Chen, F., Yuan, L., et al. (2011). Genetic analysis of vertical root pulling resistance (VRPR) in maize using two genetic populations. Mol. Breeding 28, 463–474. doi: 10.1007/s11032-010-9496-z
Long, T. A., Tsukagoshi, H., Busch, W., Lahner, B., Salt, D. E., and Benfey, P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22, 2219–2236. doi: 10.1105/tpc.110.074096
Long, W., Li, Q., Wan, N., Feng, D., Kong, F., Zhou, Y., et al. (2020). Root morphological and physiological characteristics in maize seedlings adapted to low iron stress. PLoS One 15:e0239075. doi: 10.1371/journal.pone.0239075
Luo, J., Qin, J., He, F., Li, H., Liu, T., Polle, A., et al. (2013). Net fluxes of ammonium and nitrate in association with H+ fluxes in fine roots of Populus popularis. Planta 237, 919–931. doi: 10.1007/s00425-012-1807-7
Luo, M., Zhao, Y., Zhang, R., Xing, J., Duan, M., Li, J., et al. (2017). Mapping of a major QTL for salt tolerance of mature field-grown maize plants based on SNP markers. BMC Plant Biol. 17:140. doi: 10.1186/s12870-017-1090-7
Mallikarjuna, M. G., Thirunavukkarasu, N., Sharma, R., Shiriga, K., Hossain, F., Bhat, J. S., et al. (2020). Comparative transcriptome analysis of iron and zinc deficiency in maize (Zea mays L.). Plants 9:1812. doi: 10.3390/plants9121812
Mamo, B. E., Barber, B. L., and Steffenson, B. J. (2014). Genome-wide association mapping of zinc and iron concentration inbarley landraces from Ethiopia and Eritrea. J. Cereal Sci. 60, 497-506. doi: 10.1016/j.jcs.2014.08.007
Maruyama-Nakashita, A., Nakamura, Y., Tohge, T., Saito, K., and Takahashi, H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18, 3235–3251. doi: 10.1105/tpc.106.046458
Mengel, K., Planker, R., and Hoffman, B. (1994). Relationship between leaf apoplast pH and iron chlorosis of sunflower (Helianthus annuus L.). J. Plant Nutr. 17, 1053–1065. doi: 10.1080/01904169409364787
Mohamed, A. A., El-Baz, F. K., and Khalifa, R. K. H. M. (2003). Genotypic differences of tow wheat cultivars for the enzymes activity, amino acids and protein profile under Fe deficiency. J. Biol. Sci. 3, 864–874. doi: 10.3923/jbs.2003.864.874
Moreno-Jiménez, E., Plaza, C., Saiz, H., Manzano, R., Flagmeier, M., and Maestre, F. T. (2019). Aridity and reduced soil micronutrient availability in global drylands. Nat. Sustain. 2, 371–377. doi: 10.1038/s41893-019-0262-x
Nogiya, M., Pandey, R. N., and Singh, B. (2016). Physiological basis of iron chlorosis tolerance in rice (Oryza sativa) in relation to the root exudation capacity. J. Plant Nutr. 39, 1536–1546. doi: 10.1080/01904167.2016.1161786
Nozoye, T., Nagasaka, S., Kobayashi, T., Takahashi, M., Sato, Y., Sato, Y., et al. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286, 5446–5454. doi: 10.1074/jbc.M110.180026
Nozoye, T., Nakanishi, H., and Nishizawa, N. K. (2013). Characterizing the crucial components of iron homeostasis in the maize mutants ys1 and ys3. PLoS One 8:e62567. doi: 10.1371/journal.pone.0062567
Nozoye, T., Nakanishi, H., and Nishizawa, N. K. (2015). Transcriptomic analyses of maize ys1 and ys3 mutants reveal maize iron homeostasis. Geno. Data 5, 97–99. doi: 10.1016/j.gdata.2015.05.021
Palmer, C. M., Hindt, M. N., Schmidt, H., Clemens, S., and Guerinot, M. L. (2013). MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 9:e1003953. doi: 10.1371/journal.pgen.1003953
Peiffer, G. A., King, K. E., Severin, A. J., May, G. D., Cianzio, S. R., Lin, S. F., et al. (2012). Identification of candidate genes underlying an iron efficiency quantitative trait locus in soybean. Plant Physiol. 158, 1745–1754.
Prasad, A. S. (2013). Discovery of human zinc deficiency: its impact on human health and disease. Adv. Nutr. 4, 176–190. doi: 10.3945/an.112.003210
Prinsi, B., and Espen, L. (2018). Time-course of metabolic and proteomic responses to different nitrate/ammonium availabilities in roots and leaves of maize. Int. J. Mol. Sci. 19:2202. doi: 10.3390/ijms19082202
Rabotti, G., De Nisi, P., and Zocchi, G. (1995). Metabolic implications in the biochemical responses to iron deficiency in cucumber (Cucumis sativus L.) roots. Plant Physiol. 107, 1195–1199. doi: 10.1104/pp.107.4.1195
Ravazzolo, L., Trevisan, S., Forestan, C., Varotto, S., Sut, S., Dall’Acqua, S., et al. (2020). Nitrate and ammonium affect the overall maize response to nitrogen availability by triggering specific and common transcriptional signatures in roots. Int. J. Mol. Sci. 21:686. doi: 10.3390/ijms21020686
Rengel, Z., and Römheld, V. (2000). Differential tolerance to Fe and Zn deficiencies in wheat germplasm. Euphytica 113, 219–225. doi: 10.1023/A:100396500730
Ricachenevsky, F. K., Punshon, T., Lee, S., Oliveira, B. H. N., Trenz, T. S., Maraschin, F. D. S., et al. (2018). Elemental profiling of rice FOX lines leads to characterization of a new Zn plasma membrane transporter, OsZIP7. Front. Plant Sci. 9:865. doi: 10.3389/fpls.2018.00865
Schjoerring, J. K., Husted, S., Mäck, G., and Mattsson, M. (2002). The regulation of ammonium translocation in plants. J. Exp. Bot. 53, 883–890. doi: 10.1093/jexbot/53.370.883
Shi, R., Melzer, M., Zheng, S., Benke, A., Stich, B., and von Wirén, N. (2018). Iron retention in root hemicelluloses causes genotypic variability in the tolerance to iron deficiency-induced chlorosis in maize. Front. Plant Sci. 9:557. doi: 10.3389/fpls.2018.00557
Stacey, M. G., Patel, A., McClain, W. E., Mathieu, M., Remley, M., and Rogers, E. E. (2008). The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 146, 589–601. doi: 10.1104/pp.107.108183
Strasser, O., Köhl, K., and Römheld, V. (1999). Overestimation of apoplastic Fe in roots of soil grown plants. Plant Soil 210, 179–187. doi: 10.1023/A:1004650506592
Sun, H., Feng, F., Liu, J., and Zhao, Q. (2017). The interaction between auxin and nitric oxide regulates root growth in response to iron deficiency in rice. Front. Plant Sci. 8:2169. doi: 10.3389/fpls.2017.02169
Tian, J., Wang, C., Xia, J., Wu, L., Xu, G., Wu, W., et al. (2019). Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365, 658–664. doi: 10.1126/science.aax5482
Tognetti, V. B., Zurbriggen, M. D., Morandi, E. N., Fillat, M. F., Valle, E. M., Hajirezaei, M. R., et al. (2007). Enhanced plant tolerance to iron starvation by functional substitution of chloroplast ferredoxin with a bacterial flavodoxin. Proc. Natl. Acad. Sci. U.S.A. 104, 11495–11500. doi: 10.1073/pnas.0704553104
Van der Ent, S., Verhagen, B. W., Van Doorn, R., Bakker, D., Verlaan, M. G., Pel, M. J., et al. (2008). MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 146, 1293–1304. doi: 10.1104/pp.107.113829
Vega-Mas, I., Pérez-Delgado, C. M., Marino, D., Fuertes-Mendizábal, T., González-Murua, C., Márquez, A. J., et al. (2017). Elevated CO2 induces root defensive mechanisms in tomato plants when dealing with ammonium toxicity. Plant Cell Physiol. 58, 2112–2125. doi: 10.1093/pcp/pcx146
Vert, G., Grotz, N., Dédaldéchamp, F., Gaymard, F., Guerinot, M. L., Briat, J. F., et al. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 14, 1223–1233. doi: 10.1105/tpc.001388
von Wirén, N., Marschner, H., and Römheld, V. (1994). Uptake kinetics of iron-phytosiderophores in two maize genotypes differing in iron efficiency. Physiol. Plant 93, 611–616. doi: 10.1111/j.1399-3054.1995.tb05107.x
Wang, B., Li, Y., and Zhang, W. H. (2012). Brassinosteroids are involved in response of cucumber (Cucumis sativus) to iron deficiency. Ann. Bot. 110, 681–688. doi: 10.1093/aob/mcs126
Wang, F. P., Wang, X. F., Zhang, J., Ma, F., and Hao, Y. J. (2018). MdMYB58 modulates Fe homeostasis by directly binding to the MdMATE43 promoter in plants. Plant Cell Physiol. 59, 2476–2489. doi: 10.1093/pcp/pcy168
Wang, S., Ouerdane, L., Hoekenga, O., Szpunar, J., and Lobinski, R. (2019). Mass spectrometry-based analytical developments to link iron speciation to iron bioavailability in maize. Food Chem. 294, 414–422. doi: 10.1016/j.foodchem.2019.05.014
Wang, Z., Miao, Y., and Li, S. (2015). Effect of ammonium and nitrate nitrogen fertilizers on wheat yield in relation to accumulated nitrate at different depths of soil in drylands of China. Field Crop Res. 183, 211–224. doi: 10.1016/j.fcr.2015.07.019
WHO (2013). Research for Universal Health Coverage: World Health Report 2013. Geneva: World Health Organization (WHO).
Xu, J., Qin, X., Zhu, H., Chen, F., Fu, X., and Yu, F. (2022). Mapping of the quantitative trait loci and candidate genes associated with iron efficiency in maize. Front. Plant Sci. 13:855572. doi: 10.3389/fpls.2022.855572
Xun, Z., Guo, X., Li, Y., Wen, X., Wang, C., and Wang, Y. (2020). Quantitative proteomics analysis of tomato growth inhibition by ammonium nitrogen. Plant Physiol. Biochem. 154, 129–141. doi: 10.1016/j.plaphy.2020.05.036
Ye, L., Li, L., Wang, L., Wang, S., Li, S., Du, J., et al. (2015). MPK3/MPK6 are involved in iron deficiency-induced ethylene production in Arabidopsis. Front Plant Sci. 6:953. doi: 10.3389/fpls.2015.00953
Yousfi, S., Rabhi, M., Abdelly, C., and Gharsalli, M. (2009). Iron deficiency tolerance traits in wild (Hordeummaritimum) and cultivated barley (Hordeum vulgare). C. R. Biol. 332, 523–533. doi: 10.1016/j.crvi.2009.03.006
Yuan, Y., Wu, H., Wang, N., Li, J., Zhao, W., Du, J., et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18, 385–397. doi: 10.1038/cr.2008.26
Zanin, L., Venuti, S., Zamboni, A., Varanini, Z., Tomasi, N., and Pinton, R. (2017). Transcriptional and physiological analyses of Fe deficiency response in maize reveal the presence of strategy I components and Fe/P interactions. BMC Genomics 18:154. doi: 10.1186/s12864-016-3478-4
Zdunić, Z., Grljušić, S., Ledenčan, T., Duvnjak, T., and Simić, D. (2014). Quantitative trait loci mapping of metal concentrations in leaves of the maize IBM population. Hereditas 151, 55–60. doi: 10.1111/hrd2.00048
Zhang, F., Römheld, V., and Marschner, H. (1991). Diurnal rhythm of release of phytosiderophores and uptake rate of zinc in iron-deficient wheat. Soil Sci. Plant Nutr. 37, 671–678. doi: 10.1080/00380768.1991.10416935
Zhang, P. Y., Qiu, X., Fu, J. X., Wang, G. R., Wei, L., and Wang, T. C. (2021). Systematic analysis of differentially expressed ZmMYB genes related to drought stress in maize. Physiol. Mol. Biol. Plants 27, 1295–1309. doi: 10.1007/s12298-021-01013-2
Zhang, W., Li, Z., Fang, H., Zhang, M., and Duan, L. (2018). Analysis of the genetic basis of plant height-related traits in response to ethylene by QTL mapping in maize (Zea mays L.). PLoS One 13:e0193072. doi: 10.1371/journal.pone.0193072
Zhang, X. Y., Zhang, X., Zhang, Q., Pan, X. X. K., Yan, L. C., Ma, X. J., et al. (2017). Zea mays Fe deficiency-related 4 (ZmFDR4) functions as an iron transporter in the plastids of monocots. Plant J. 90, 147–163. doi: 10.1111/tpj.13482
Zhao, Q. Y., Xu, S. J., Zhang, W. S., Zhang, Z., Yao, Z., Chen, X. P., et al. (2020). Identifying key drivers for geospatial variation of grain micronutrient concentrations in major maize production regions of China. Environ. Pollut. 266:115114. doi: 10.1016/j.envpol.2020.115114
Zhou, X., Li, S., Zhao, Q., Liu, X., Zhang, S., Sun, C., et al. (2013). Genome-wide identification, classification and expression profiling of nicotianamine synthase (NAS) gene family in maize. BMC Genomics 14:238. doi: 10.1186/1471-2164-14-238
Keywords: maize (Zea mays L.), iron (Fe) deficiency tolerance, iron efficiency, nitrogen form, quantitative trait locus (QTL)
Citation: Xu J, Zhu X, Yan F, Zhu H, Zhou X and Yu F (2022) Identification of Quantitative Trait Loci Associated With Iron Deficiency Tolerance in Maize. Front. Plant Sci. 13:805247. doi: 10.3389/fpls.2022.805247
Received: 30 October 2021; Accepted: 07 March 2022;
Published: 14 April 2022.
Edited by:
Ricardo Fabiano Hettwer Giehl, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), GermanyReviewed by:
Dhriti Singh, National Institute of Plant Genome Research (NIPGR), IndiaZi Shi, Maize Research Center, Beijing Academy of Agriculture and Forestry Sciences, China
Copyright © 2022 Xu, Zhu, Yan, Zhu, Zhou and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Futong Yu, ZnV0b25neXVAY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Jianqin Xu
Jianqin Xu Xiaoyang Zhu2†
Xiaoyang Zhu2† Futong Yu
Futong Yu