
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 01 February 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.790140
Cassava bacterial blight (CBB) caused by Xanthomonas axonopodis pv. manihotis (Xam) seriously affects cassava yield. Genes encoding nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains are among the most important disease resistance genes in plants that are specifically involved in the response to diverse pathogens. However, the in vivo roles of NBS-LRR remain unclear in cassava (Manihot esculenta). In this study, we isolated four MeLRR genes and assessed their expression under salicylic acid (SA) treatment and Xam inoculation. Four MeLRR genes positively regulate cassava disease general resistance against Xam via virus-induced gene silencing (VIGS) and transient overexpression. During cassava-Xam interaction, MeLRRs positively regulated endogenous SA and reactive oxygen species (ROS) accumulation and pathogenesis-related gene 1 (PR1) transcripts. Additionally, we revealed that MeLRRs positively regulated disease resistance in Arabidopsis. These pathogenic microorganisms include Pseudomonas syringae pv. tomato, Alternaria brassicicola, and Botrytis cinerea. Our findings shed light on the molecular mechanism underlying the regulation of cassava resistance against Xam inoculation.
Disease resistance genes (R genes) usually act as receptors of pathogen-encoded effector proteins, which are often secreted by pathogens directly into host cells (Urbach and Ausubel, 2017). R genes are specifically involved in the response to diverse pathogens, including fungi, bacteria, viruses, nematodes, insects, and oomycetes (Dalio et al., 2017). In the past 30 years, more than 300 R genes have been cloned from many plant species (Kourelis and van der Hoorn, 2018). Among them, genes encoding nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains are important R genes in plants (van der Hoorn and Kamoun, 2008; Pandolfi et al., 2017). The amino terminal (N-terminal) of NBS-LRR proteins usually contain the Toll/interleukin-1 receptor-like (TIR) domain, coiled-coil (CC) domain, or resistance to powdery mildew 8 (RPW8) domain, and the carboxyl terminus (C-terminus) contain a zinc-finger transcription factor-related domain containing the WRKY sequence (WRKY domain) (Shao et al., 2006). Based on the N-terminal domains, NBS-LRR was usually divided into three subclasses, namely TIR-NBS-LRR (TNL), CC-NBS-LRR (CNL), and RPW8-NBS-LRR (RNL) proteins (Shao et al., 2006).
In plant genome, about 0.2–1.6% of genes are predicted as NBS-LRR-coding genes (Jia et al., 2015). For instance, there are 150–175 NBS-LRR genes in Arabidopsis thaliana genome (Meyers et al., 2003; Joshi et al., 2011), constituting about 0.6% of its 25,000 genes, and there are approximately 600 NBS-LRR genes in rice (Oryza sativa ssp. japonica) genome (Goff et al., 2002; Chen et al., 2015), constituting about 1.5% of its 40,000 genes (Goff et al., 2002). In the past few years, NBS-LRR genes in several plant species have been isolated via genome-wide analysis, including mango (Mangifera indica) (Lei et al., 2014), cassava (Manihot esculenta) (Lozano et al., 2015; Utsumi et al., 2016), sorghum (Sorghum bicolor) (Yang and Wang, 2016), wheat (Triticum aestivum) (Li et al., 2017), cotton (Gossypium hirsutum) (Deng et al., 2019), maize (Zea mays) (Xu et al., 2018), soybean (Glycine max) (Zhao et al., 2018), grapevine (Vitis vinifera) (Goyal et al., 2020), and yam (Dioscorea rotundata) (Zhang et al., 2020). In recent years, accumulated evidence has confirmed that NBS-LRR protein is widely involved in plant development and stress response (Li et al., 2017, 2018a,2018b; Xu et al., 2018; Zhao et al., 2018; Deng et al., 2019). Activated by effector proteins, NBS-LRR proteins could elicit robust defense responses, inducing the biosynthesis and accumulation of SA and increasing expression of pathogenesis-related (PR) genes (Wu et al., 2014; Palmer et al., 2019). In cotton, silencing of the NB-ARC domain-containing (GbaNA1) gene impaired cotton resistance to Verticillium dahliae Vd991 (Li et al., 2018a). Similarly, heterologous expression of the maize NBS-LRR gene ZmNBS25 enhanced resistance to P. syringae pv. tomato DC3000 in rice and Arabidopsis by induced the defense-related gene expression, but grain yield was not affected (Xu et al., 2018). NBS-LRR proteins and SA are involved in pathogen-host interactions (Bonardi et al., 2011; Zhao et al., 2018). Yoodee et al. (2018) found that exogenous application of SA could elevate the defense resistance of cassava to Xam inoculation. Although 228 NBS-LRR genes have been identified in cassava (Lozano et al., 2015), their role remains unknown in vivo.
Cassava is a widely grown drought-tolerant crop that can be cultivated as an annual crop in marginal soils in tropical and subtropical regions of the world (Lozano et al., 2015; Bredeson et al., 2016). However, as a clonally propagated crop, cassava is especially vulnerable to pathogens, especially cassava bacterial blight (X. axonopodis pv. manihotis = X. phaseoli pv. manihotis) (Bredeson et al., 2016; Constantin et al., 2016; Zárate-Chaves et al., 2021), cassava brown streak disease (Cassava brown streak virus, CBSV) and anthracnose disease (Colletotrichum gloeosporioides) (Utsumi et al., 2016). Therefore, it is best to identify the NBS-LRR proteins in cassava. Results presented by Utsumi et al. (2016) indicated that the transcript level of NBS-LRRs was inducted by C. gloeosporioides infection. Similar results were obtained under plants infected by viruses (Louis and Rey, 2015; Lozano et al., 2015; Amuge et al., 2017; Masumba et al., 2017). A cluster of NBS-LRR genes on chromosome 11 of cassava genome was associated with resistance to cassava brown streak disease via genome-wide associated mapping and genomic selection (Kayondo et al., 2018). However, the mechanisms remain unclear, particularly in experimental investigation and verification.
In this study, we analyzed the published transcriptome databases of cassava-pathogens interaction (Lozano et al., 2015; Utsumi et al., 2016). Within the database, four NBS-LRR genes that showed high transcription level after pathogen infection attracted our attention. The expression levels of four chosen MeLRRs were significantly induced by exogenous application of SA treatment and Xam inoculation. Moreover, these genes positively regulated cassava resistance to Xam inoculation. The functional analysis of MeLRR genes will offer potential roles in genetic breeding for disease-resistant cassava.
There are 228 NBS-LRRs in cassava, including both TIR-NBS-LRR and CC-NBS-LRR. Their transcript levels were analyzed through RNA-seq in response to CBSV and C. gloeosporioides infection (Lozano et al., 2015; Utsumi et al., 2016). Among these, four MeLRRs (MeLRR1, MeLRR2, MeLRR3, and MeLRR4) were both induced under CBSV and C. gloeosporioides infection and selected for further analysis. The four MeLRR proteins have typical leucine-rich repeats, which are named MeLRR1 (Manes. 11G053000.1), MeLRR2 (Manes. 03G071700.1), MeLRR3 (Manes. 13G036800.1), and MeLRR4 (Manes. 07G107800.1), located on chromosomes 11, 3, 13, and 7, respectively. MeLRR1, MeLRR3, and MeLRR4 belong to CC-NBS-LRR protein, while MeLRR2 is one of the TIR-NBS-LRR protein. Bioinformatics predicted that the MeLRR proteins were unstable and hydrophilic (Supplementary Table 1). The phylogenetic analysis showed that MeLRR1 clustered with XP_012073222.1 of Jatropha curcas, MeLRR2 clustered with XP_021684995.1 of Hevea brasiliensis, MeLRR3 clustered with XP_020535356.1 of J. curcas, and MeLRR4 clustered with KAF2295929.1 of H. brasiliensis based on whole protein sequences (Supplementary Figure 1).
To investigate the subcellular localization of the MeLRR proteins, the coding sequences (CDSs) of MeLRRs were cloned and inserted into the poly-cloning sites of the fusion expression vector pEGAD and fused upstream to a green florescence protein (GFP) fusion partner by the constitutive CaMV35S promoter. The Agrobacterium tumefaciens strain GV3101 cell culture harboring the pEGAD empty vector containing 35S:GFP was used as a control, and tobacco (Nicotiana benthamiana) leaves were infected with 35S:GFP or 35S:GFP-MeLRR1,−2,−3,−4 plasmid as described by Sparkes et al. (2006). The fluorescence of transiently expressing MeLRR proteins in tobacco leaf epidermal cells was detected in the nucleus, cytoplasm and cytomembrane, similar to that of 35S:GFP (Figure 1).
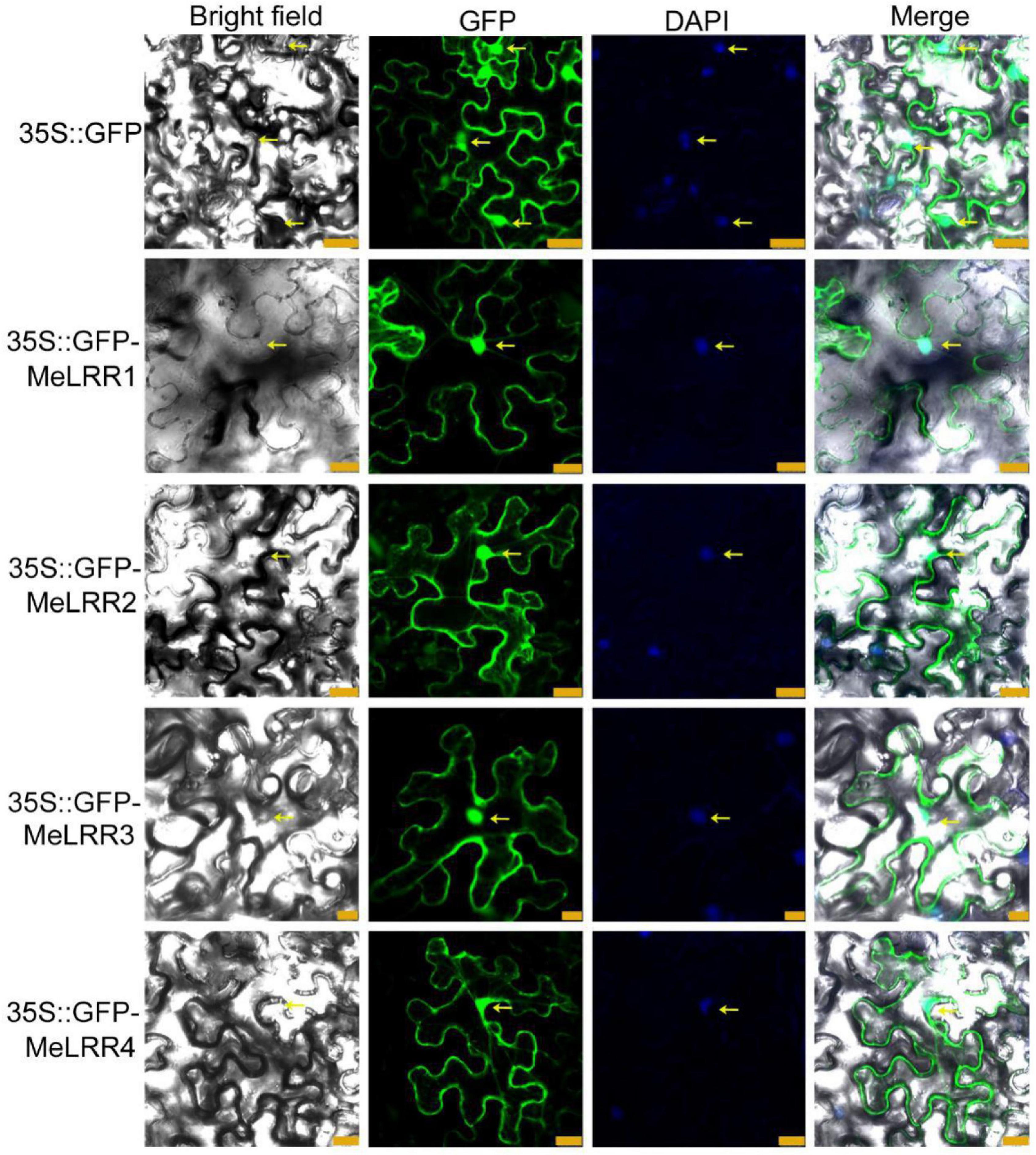
Figure 1. Subcellular localization of MeLRR proteins in N. benthamiana leaves. Transient expression of Agrobacterium GV3101 with 35S:GFP and 35S:GFP-MeLRRs plasmids in N. benthamiana leaves. After 2 dpi, the fluorescence was scanned by a Leica confocal microscopy system (Leica TCS SP8, Solms, Germany) with an excitation wavelength of 488 nm and a 505–530 nm bandpass emission filter. The empty vector 35S:GFP was used as a control. Nuclei were stained using DAPI (4’,6-diamidino-2-phenylindole). Scale bar = 25 μm.
The expression profile of MeLRRs in response to SA treatment and Xam inoculation were analyzed by qRT-PCR (real-time quantitative reverse transcription PCR). Under SA treatment, the expressions of MeLRR1, MeLRR3, and MeLRR4 were induced and peaked at 1 h post treatment (hpt), while MeLRR2 showed the highest level at 3 hpt (Figure 2). Following infection by Xam, the expression level of MeLRRs at 1–24 hpt hpi was significantly higher than that at 0 hpi (Figure 2). Moreover, the expression of MeLRR1, MeLRR2, and MeLRR3 were induced and peaked at 3 hpt, while the expression of MeLRR4 reached the peak at 12 hpt (Figure 2).
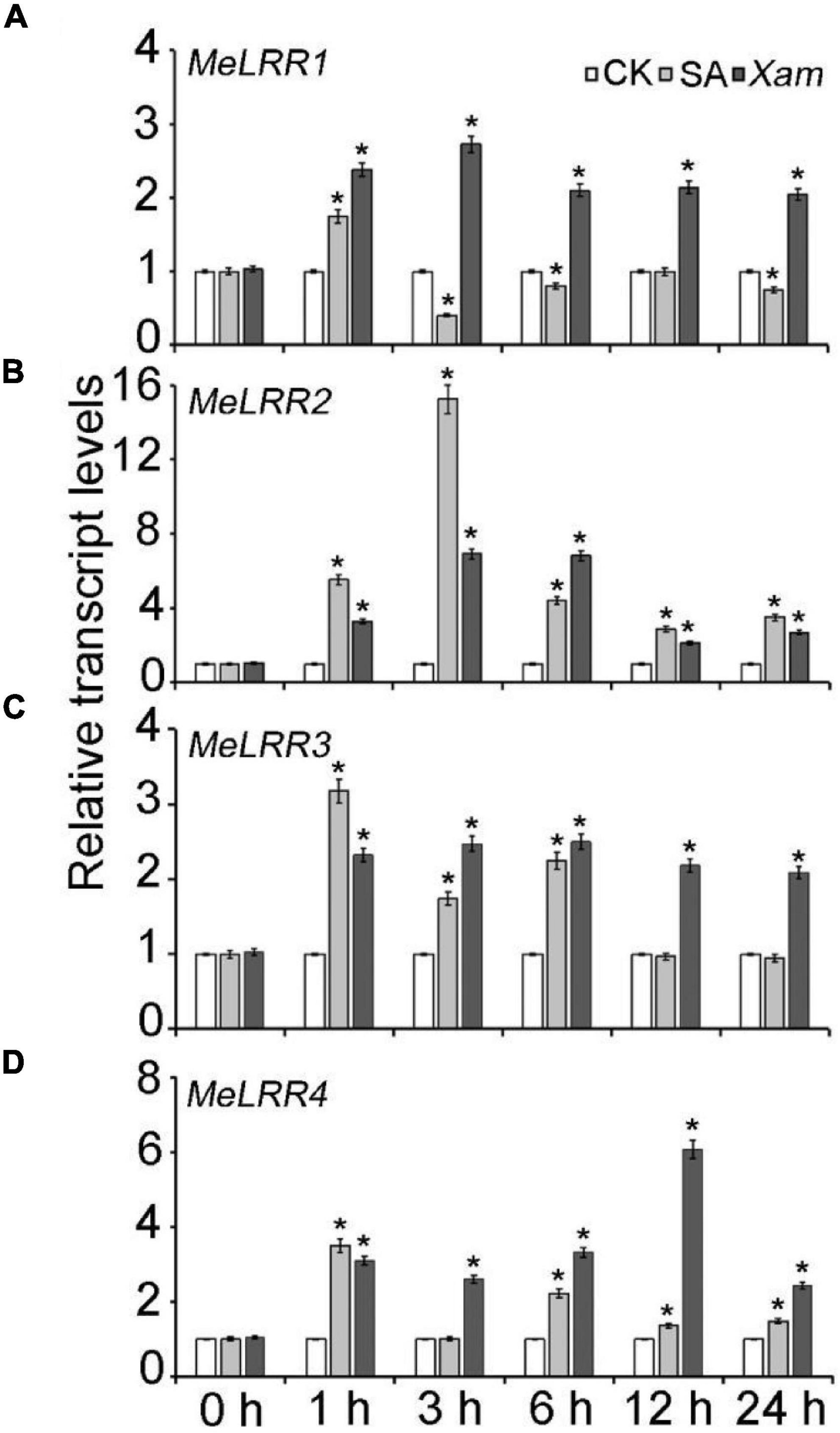
Figure 2. The expression profiles of MeLRR genes in cassava in response to SA treatment and Xam inoculation. For the assays, 4-week-old cutting seedlings of cassava leaves were sprayed with water, 5 mmol/L salicylic acid, or 4 × 108 cfu/mL Xam suspension for 0, 1, 3, 6, 12, or 24 h. The transcript levels of MeLRR1 (A), MeLRR2 (B), MeLRR3 (C), and MeLRR4 (D) at 0 h of the mock treatment was normalized to 1. Asterisks (*) indicate significant differences at p < 0.05.
To analyze the function of MeLRRs, we constructed MeLRR-silenced cassava plants by virus-induced gene silencing (VIGS). The partial sequences of MeLRR1 (453 bp), MeLRR2 (441 bp), MeLRR3 (433 bp) and MeLRR4 (423 bp) were individually inserted into pTRV2 plasmid to construct VIGS vector. At 14 days post-infection (dpi) in cassava infected with Agrobacterium GV3101 carrying the pTRV-MeLRR plasmids, qRT-PCR was performed to detect the target gene transcript level. The transcript level of the target MeLRR-1,-2,-3,-4 genes were significantly decreased in the MeLRR-silenced cassava leaves compared to the pTRV empty vector. The silencing efficiency of MeLRR-1,-2,-3,-4 was 46.33 (± 2.31)%, 15.28 (± 0.49)%, 30.22 (± 2.28)%, and 17.45 (± 0.87)% (Mean ± SD, n = 3), respectively (Figure 3A). It was noteworthy that the silenced of MeLRR1 did not affected the transcription of MeLRR2,-3,-4. Similar results were verified in MeLRR2-, MeLRR3-, and MeLRR4-silenced plants (Supplementary Figure 2). When co-silenced four target genes (MeLRR-1,-2,-3,-4) in one VIGS line, the transcript levels of all four target genes were significantly decreased (Supplementary Figure 3). On the contrary, the bacteria number was significantly higher than that in the pTRV empty vector-infected cassava leaves (Figure 3B). Moreover, the cassava MePR1 transcript level was significantly decreased (Figure 3C). And the transcript level of MePR1 in pTRV-MeLRR1,−2,−3,−4 cassava was reduced on average to 84, 7, 58, and 69%, respectively, of the transcript level in the pTRV control at 14 dpi (Figure 3C). Additionally, the transcript level of MePR1 in MeLRR-1,-2,-3,-4-silenced plant was significantly reduced to 66% (Supplementary Figure 3). Silencing of MeLRRs conferred increased disease susceptibility in cassava leaves (Figure 3D). Moreover, MeLRRs-silenced cassava leaves showed significantly lower ROS burst measurements than the empty vector (Figures 3E,F). These results indicate that silencing of MeLRRs impairs cassava resistance to Xam.
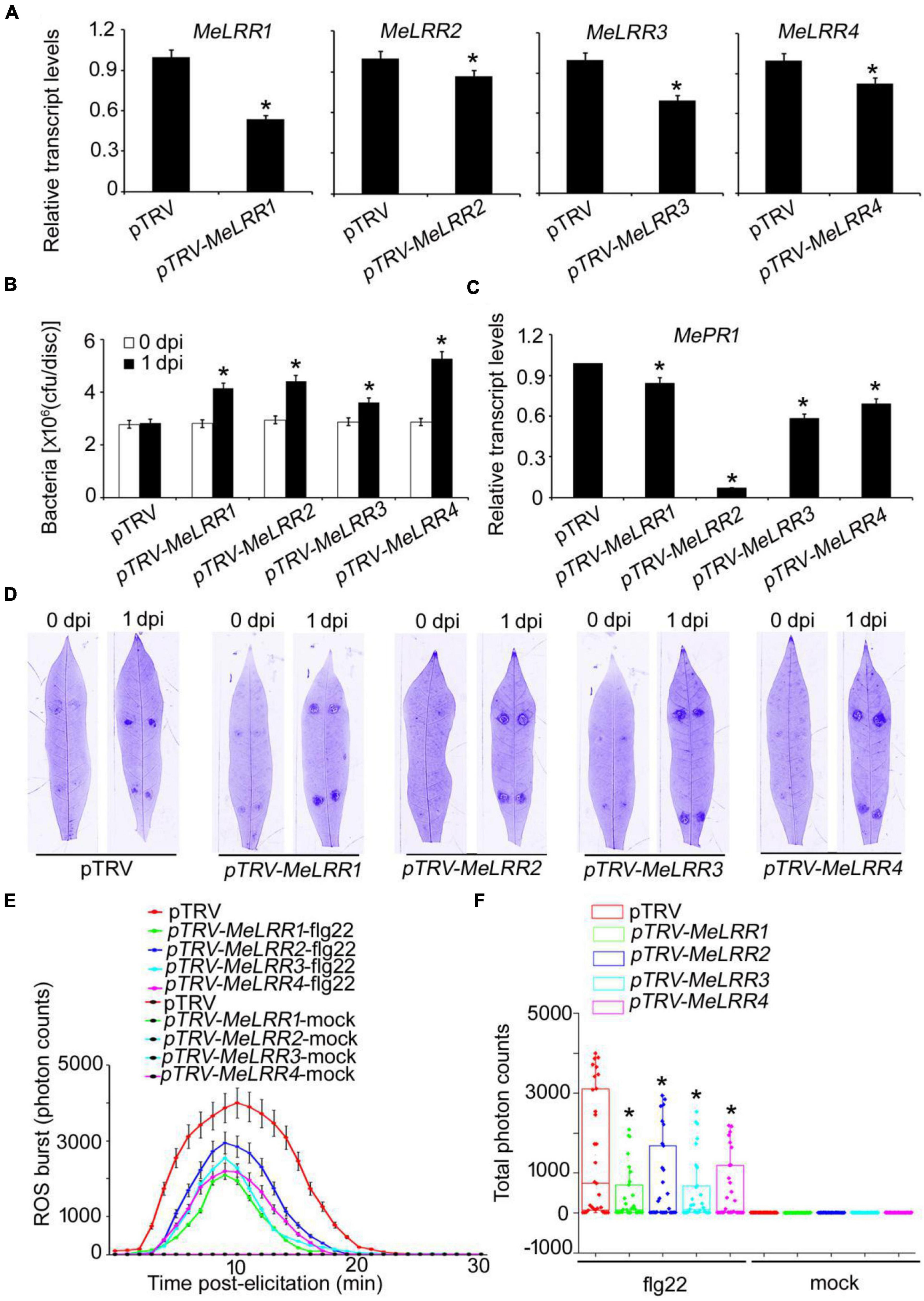
Figure 3. The VIGS of MeLRRs reduced disease resistance against cassava bacterial blight. (A) At 14 dpi, the new leaves were used for relative transcript levels of MeLRRs in MeLRR-silenced leaves and the pTRV control leaves. Then, the new leaves were syringe infiltrated with 4 × 108 cfu/mL of pathogenic bacteria Xam used for disease resistance assay. (B) The number of Xam populations in MeLRR-silenced cassava and the pTRV control leaves at 0 and 1 dpi, respectively. (C) The pathogenesis-related gene (MePR1) transcript level was quantitatively analyzed by qRT-PCR at 1 dpi. The relative transcript level of MePR1 in the pTRV control leaves was normalized to 1.0. (D) Cassava leaves were observed using a Coomassie brilliant blue imaging system Fusion FX7-826 apparatus (Vilber Lourmat, France). (E) Dynamic of ROS accumulation in response to flg22 elicitation in MeLRR-silenced cassava and the pTRV control leaves. The flg22-triggered ROS burst were measured using luminol-based assay by a GloMax 96 Microplate Luminometer. (F) Total photon of MeLRR-silenced cassava and the pTRV control leaves. Multiple comparisons of total photon were calculated by Student’s t-test. Asterisks (*) indicate significant differences at p < 0.05. dpi is days post-infection.
To further verify the function of MeLRRs, 35S:GFP-MeLRR recombinant plasmids were constructed and introduced into Agrobacterium strain GV3101. Cassava leaves were infected with Agrobacterium containing the recombinant plasmids or empty vector for 3 days. The transcript level of the target MeLRR-1,-2,-3,-4 genes were significantly higher than that in the 35S:GFP empty vector (Figure 4A). It is similar in silenced plant, overexpressing MeLRR1 plant did not affected the transcription of MeLRR2,-3,-4, and the same as in MeLRR2-, MeLRR3-, and MeLRR4-overexpressed plants (Supplementary Figure 4). However, the transcript levels of the four target genes were significantly enhanced in co-overexpression MeLRR-1,-2,-3,-4 plants (Supplementary Figure 5). On the contrary, the bacteria number was significantly lower than that in the control (Figure 4B). However, the transcript levels of MePR1 in 35S:GFP-MeLRR1,−2,−3,−4 cassava were increased by 3. 77-, 23. 73-, 10. 70-, and 1.39-fold, respectively, compared to those in the control at 3 dpi (Figure 4C). Similarly, the transcript level of MePR1 in co-overexpression MeLRR-1,−2,−3,−4 lines was significantly increased by 24.03-fold (Supplementary Figure 5). Interestingly, overexpression of MeLRRs conferred improved disease resistance in cassava leaves (Figure 4D). Moreover, cassava leaves that overexpressed MeLRRs exhibited significantly higher ROS burst than 35S:GFP control during flg22 treatment (Figures 4E,F). These results suggest that MeLRRs positively regulated cassava resistance to Xam. In addition, trypan blue staining showed no cell death phenotype at 2 dpi at transient expression of MeLRRs in cassava and N. benthamiana leaves (Supplementary Figure 6).
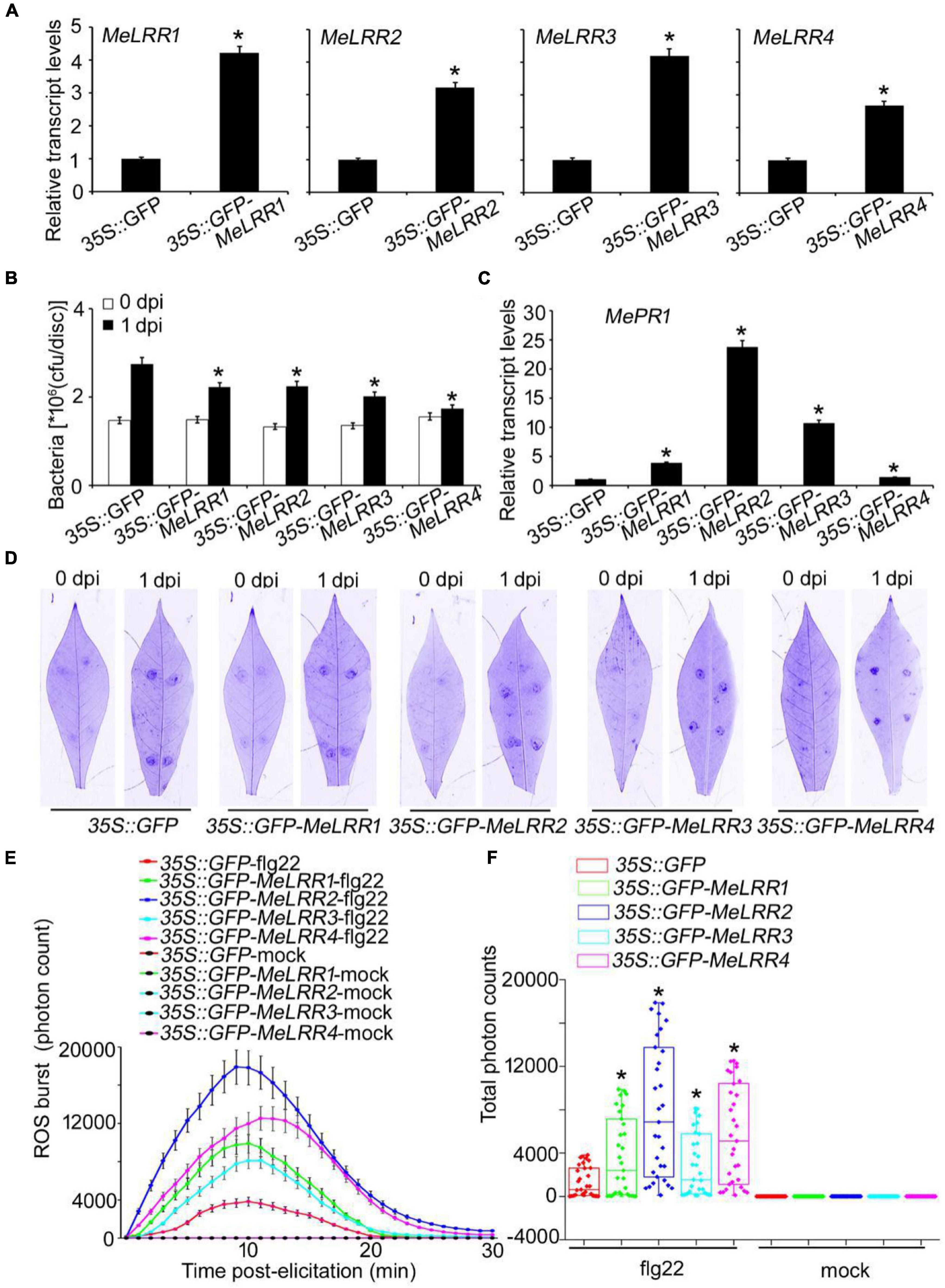
Figure 4. Transient overexpression of MeLRRs improved disease resistance against cassava bacterial blight. Cassava leaves inject with recombinant pEGAD plasmids and empty vector of Agrobacterium GV3101, respectively. (A) At 3 days later, the relative transcript levels of MeLRRs in MeLRR-overexpression cassava and the pEGAD control leaves. The relative transcript levels of MeLRRs in the pEGAD control leaves was normalized to 1.0. Then, the cassava leaves were syringe infiltrated with 4 × 108 cfu/mL of pathogenic bacteria Xam used for disease resistance assay. (B) The number of Xam populations in MeLRR-overexpression cassava and the pEGAD control leaves at 0 and 1 dpi, respectively. (C) The pathogenesis-related gene (MePR1) transcript level was quantitatively analyzed by qRT-PCR at 1 dpi. The relative transcript level of MePR1 in the pEGAD control leaves was normalized to 1.0. (D) Cassava leaves were observed using a Coomassie brilliant blue imaging system Fusion FX7-826 apparatus (Vilber Lourmat, France). (E) Dynamics of ROS accumulation in response to flg22 elicitation in MeLRR-overexpression cassava and the pEGAD control leaves. The flg22-triggered ROS burst were measured using luminol-based assay using a GloMax 96 Microplate Luminometer. (F) Total photon of MeLRR-overexpression cassava and the pEGAD control leaves. Multiple comparisons of total photon were calculated using Student’s t-test. Asterisks (*) indicate significant differences at p < 0.05. dpi is days post-infection.
To further analyze the mechanism of MeLRRs in response to Xam inoculation, the SA content was measured. As shown in Figure 5, the SA level in MeLRR1,−2,−3,−4-silencing was significantly decreased compared with that in pTRV control cassava leaves (Figure 5A). By contrast, the SA level in MeLRRs overexpression was significantly increased compared with the control cassava leaves (Figure 5B). These results suggested that MeLRR1,−2,−3,−4 positively participated in cassava immune responses via SA accumulation.
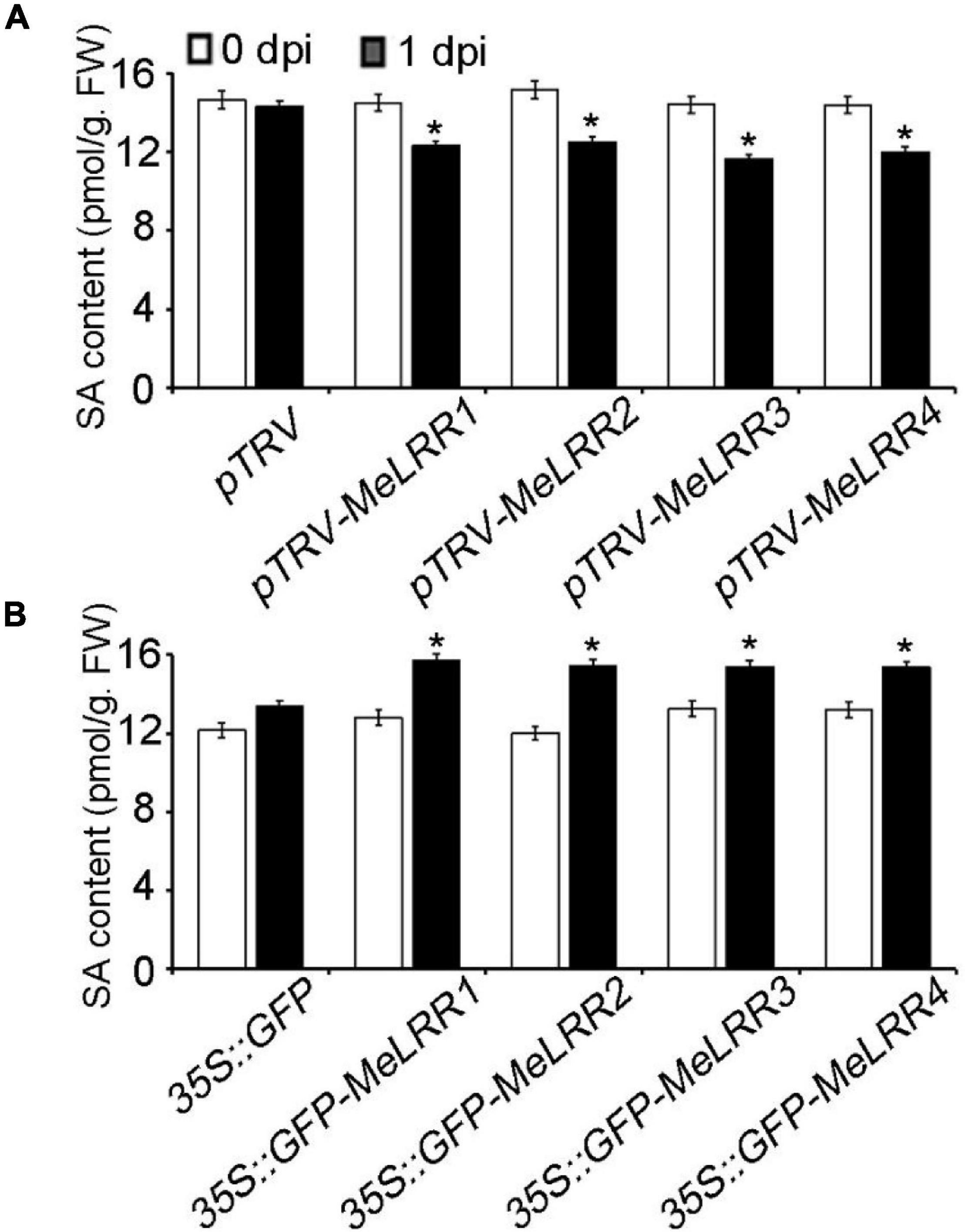
Figure 5. The salicylic acid content in cassava. The SA content in MeLRR-silenced (A) and MeLRR-overexpression (B) cassava leaves. Transient expression of MeLRRs conferred immunity. At 0 and 1 d infection with Xam, the cassava leaves were used for analyzing the content of SA. Asterisks (*) indicate significant differences at p < 0.05. dpi is days post-infection.
To further confirm the MeLRR function, MeLRRs were overexpressed in Arabidopsis. Quantification of endogenous SA levels indicated that MeLRR-overexpressing lines accumulated significantly higher levels than WT leaves (Supplementary Figure 7). The MeLRRs overexpression plants displayed slight symptoms of wilting in response to P. syringae pv. tomato, A. brassicicola, and B. cinerea infection support the hypothesis that MeLRRs functions in a pathogen response pathway. A difference was already observed in the WT, suggesting that restricted bacterial entry into the leaves may underlie part of the apparent resistance (Figure 6A). Unlike P. syringae pv. tomato, A. brassicicola, and B. cinerea can enter hosts by penetrating the cuticle. Consistently, there was less fungal growth in leaves overexpressing these factors than WT plants by analyzing the transcript levels of the A. brassicicola AbAct (JQ671669.1) gene and B. cinerea BcActA (XM_024697950.1) gene (Liao et al., 2016) with the Arabidopsis AtAct2 gene as an internal control at 2 and 4 dpi, respectively (Figures 6B,C).
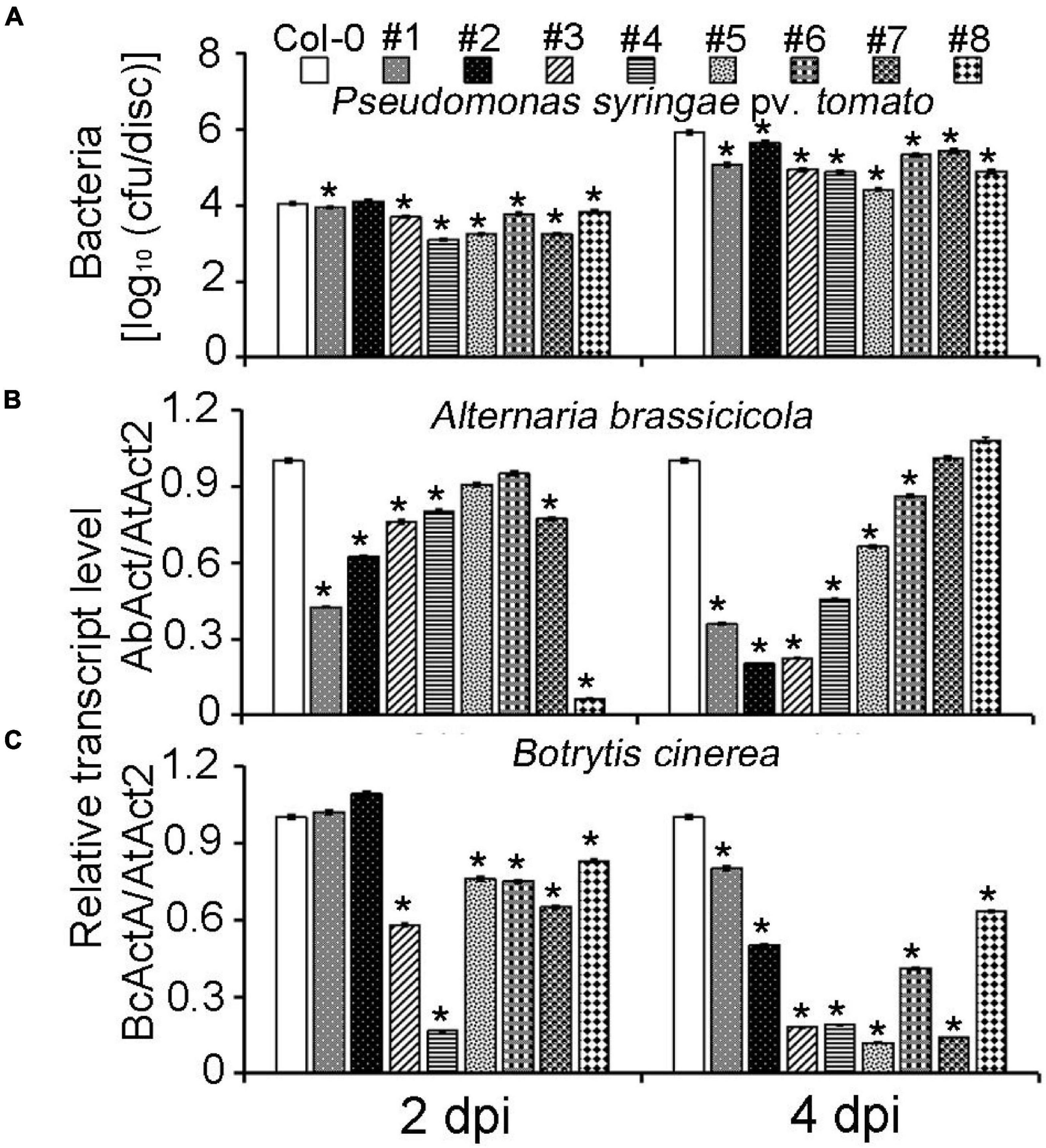
Figure 6. Overexpression of MeLRRs in Arabidopsis enhances resistance to plant pathogens. (A) The number of P. syringae pv. tomato populations in overexpression Arabidopsis leaves and the wild type. The relative transcript levels of AbAct/AtAct2 (B) and BcActA/AtAct2 (C) in overexpression Arabidopsis leaves and the wild type after infection with A. brassicicola and B. cinerea, respectively. Asterisks (*) indicate significant differences at p < 0.05. Col-0 is A. thaliana ecotype Columbia-0. #1 and #2, #3 and #4, #5 and #6, and #7 and #8 are overexpression of MeLRR3 in A. thaliana Col-0 lines, respectively. dpi is days post-infection.
To determine whether the enhanced resistance to plant pathogens was related to changing the defense response genes expression level, we used qRT-PCR to analyze the expression levels of AtICS1, AtPDF1.2, AtPR1, AtPR2, AtPR5, and AtTGA3 in WT and MeLRR overexpression lines upon A. brassicicola, B. cinerea, and P. syringae pv. tomato DC3000 infection (Supplementary Figure 8). Particularly, the relative expression levels of genes involved in the SA synthesis pathway and pathogen resistance showed higher level in overexpression MeLRR1 and MeLRR2 in Arabidopsis plants than in control plants without A. brassicicola, B. cinerea, and P. syringae pv. tomato DC3000 infection. Similar results were observed in plant pathogen-infected overexpression of MeLRR3 in Arabidopsis plants compared with control plants. However, AtPDF1.2 and AtPR1 were significantly down-regulated in overexpression of MeLRR3 in Arabidopsis plants than in control plants without P. syringae pv. tomato DC3000 infection. On the other hand, the expression levels of AtPR2 and AtTGA3 were significantly up-regulated in overexpression of MeLRR4 in Arabidopsis plants than in control plants. AtICS1, AtPDF1.2, AtPR1, and AtPR5 genes were up-regulated or down-regulated under different plant pathogen infections. These results indicate that overexpression of MeLRRs resulted in enhanced resistance simultaneously against pathogenic bacteria and pathogenic fungi, demonstrating the requirement of MeLRRs for resistance to plant pathogens.
NBS-LRR proteins play important roles in pathogen recognition and defense response signal transduction (Urbach and Ausubel, 2017). An increasing number of NBS-LRR proteins that conferred resistance to pathogens have been cloned from higher plants (Liu et al., 2017), such as TaRCR1 (Zhu et al., 2017), ZmNBS25 (Xu et al., 2018), GbaNA1 (Li et al., 2018a,b), GhDSC1 (Li et al., 2019), and OsRLR1 (Du et al., 2021). In this study, we found that MeLRR1,-2,-3,-4 expression could be induced by Xam inoculation. Similar expression patterns have been observed in other plant NBS-LRR genes, such as AhRRS5 (Zhang et al., 2017) and SacMi (Zhou et al., 2018). NBS-LRRs mainly participate in plant resistance against pathogen infection, and we speculated that the up-regulation of MeLRRs could help cassava successfully evade Xam inoculation.
SA is a secondary messenger for systemic acquired resistance (SAR), and its production in plants represents the successful recognition of pathogen infection and pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Divi et al., 2010; Peng et al., 2021). In cassava, SA also plays an important role in the regulation of cassava resistance to CBB (Liu C. et al., 2018; Chang et al., 2020; Wei et al., 2021a,b) and to whitefly (Irigoyen et al., 2020). Wei et al. (2018) found that MeHsf3 regulates cassava resistance to cassava bacterial blight through modulation of SA accumulation. Cassava co-chaperones MeHSP90.9 interacts with MeSRS1 and MeWRKY20 to activate SA biosynthesis, accumulation of SA, and thus improve resistance to CBB (Wei et al., 2021b). Therefore, endogenous SA accumulation levels are an indicator of resistance to CBB. We found that the expression levels of MeLRR were significantly increased by SA treatment, which showed the similar expression pattern of NPR1 in Arabidopsis, ZmNBS25 in maize, and GhDSC1 in cotton. In response to pathogen infection, plant endogenous SA is quickly and strongly induced.
Moreover, multiple transcription activator-like (TAL) effectors and type III effectors (T3Es) of Xam regulate plant immune (Castiblanco et al., 2013; Medina et al., 2018). Such as, TALE1Xam (Castiblanco et al., 2013), Xop (Arrieta-Ortiz et al., 2013), avrBS2, xopQ, XopR, XopAO1, and similar factors (Bart et al., 2012; Cohn et al., 2016; Medina et al., 2018; Mondal et al., 2020). Flagellin peptide (flg22) treatment regulates the expression of MebZIP3, -5 (Li et al., 2017), MeBIK1 (Li et al., 2018c), MeDELLAs (Li et al., 2018d), MeWHYs (Liu W. et al., 2018), and MeASMT2 (Wei et al., 2017). Moreover, these genes mediated cassava resistance to CBB. Flg22 is a bacterial PAMP. In Arabidopsis and tomato, flg22 was used to instead of P. syringae and Xanthomonas to measure the ROS burst, respectively (de Torres Zabala et al., 2015; Bhattarai et al., 2016). Interestingly, MeLRRs regulated ROS burst was induced by flg22 (Zipfel et al., 2004). As a homolog protein of MeLRR3, AtLRRAC1 is induced by flg22 treatment and leads to production ROS and induction of pathogen-responsive genes (Bigeard et al., 2015; Bianchet et al., 2019). Therefore, we hypothesized that MeLRRs and effectors of Xam conform to the gene for gene theory.
AtPDF1.2, AtPR1, AtPR2, and AtPR5 are widely known as marker genes for innate immune response (Wang et al., 2017; Xu et al., 2018). AtICS1 is a key enzyme for SA biosynthesis (Macaulay et al., 2017). AtTGA3 showed strong affinity for the NPR1 protein (Zhou et al., 2000; Yuan et al., 2009). In pathogenic microorganism infection, the SAR defense response is triggered by elevated SA through an SA-NPR1-TGA-PR1 signaling pathway (Zhang, 2003). Further analysis of gene expression in overexpression of MeLRR1,−2,−3,−4 at Arabidopsis leaves suggested that these genes might exert their function through SA biosynthesis and immune responses. This is similar to the function of MeHsf3 (Wei et al., 2018), and MebZIP3, -5 (Li et al., 2017), which were confirmed to regulate cassava resistance against cassava bacterial blight. Hence, we conclude that MeLRR1,−2,−3,−4 may regulate the plant immune response through SA and ROS accumulation, and the transcription of disease resistance genes. Taken together, the MeLRR genes encode a class of NBS-LRR proteins, which controls immunity to Xanthomonas axonopodis pv. manihotis in cassava. Further investigation of the role of the MeLRRs will build an important foundation for future development of resistant cultivars, which may be the most effective means of controlling this devastating disease.
Cassava (M. esculenta), variety South China 124 (SC124), and N. benthamiana were cultivated in mixed soil (vermiculite/nutritional soil = 2:1, v.v.) in a greenhouse with 16/8 h light/dark at 28/22°C, 60–70% relative humidity with irradiance of 130–150 uE.m–2.s–1. A. thaliana ecotype Col-0 (Columbia-0) seedlings were cultivated in the mixed soil under fluorescent light (130–150 uE.m–2.s–1) and were grown under 16/8 h light/dark at 22°C. For axenic growth, N. benthamiana, and A. thaliana seeds were sterilized (10% NaClO for 1 min, washed five times with sterile water) and sown on half-strength MS (Murashige and Skoog) medium (PhytoTechnology Laboratories, Kansas, United States) with 0.4% agar powder and 2% (w/v) sucrose. The seeds were grown in chambers under 16/8 h light/dark at 22°C and 130–150 uE.m–2.s–1. For expression analysis, 4-week-old cutting seedlings of cassava leaves were sprayed with 5 mmol/L salicylic acid or Xam suspension for 0, 1, 3, 6, 12, or 24 h, and the bacterial solution was diluted to 4 × 108 colony-forming units/mL (cfu/mL) using 10 mmol/L MgCl2 with 0.05% Silwet L-77.
The sequences of MeLRR genes were searched and obtained from the cassava genome database, M. esculenta v6.1 (Phytozome v131) (Muñoz-Bodnar et al., 2014; Lozano et al., 2015; Bredeson et al., 2016). The ProtParam tool2 was used to predict the number of amino acids, relative molecular mass of protein, isoelectric point, total average hydrophilicity stability index, fat coefficient, and instability index (Gasteiger et al., 2003). Alignments between MeLRRs and other NBS-LRR proteins were performed used DNAMAN 6.0, and the phylogenetic tree was constructed by the neighbor-joining method based on the whole protein sequences and considering 1,000 bootstrap replicates using ClustalW tool and MEGA 7 (Kumar et al., 2016). The 24 NBS-LRR protein amino acid sequences in 13 species were screened based on the principles of encoding nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains, and were validated through comparisons of the protein basic local alignment search tool (BLASTP) with the National Center for Biotechnology Information (NCBI). The 24 NBS-LRR proteins were derived from A. thaliana (CAA0374684.1, CAD5320387.1, CAE6029947.1, NP_181039.1, OAP10808.1, VYS54481.1), Durio zibethinus (XP_022746274.1), H. brasiliensis (XP_021646775.1, XP_021652057.1, XP_021646749.1, XP_021684995.1, KAF229 5929.1), J. curcas (XP_012073222.1, KDP37136. 1. XP_02053 5356.1), Populus alba (XP_034892116.1, XP_034896332.1), P. euphratica (XP_011001622.1), P. trichocarpa (RQO87881.1), Ricinus communis (EEF44774.1), Theobroma cacao (XP_01796 9995.1), Vernicia montana (AMM43068.1), V. vinifera (XP_010657.1), and Ziziphus jujuba (XP_024924720.1), respectively.
Total RNA was extracted from three independent pools, and DNA contamination was removed using the Tiangen RNA prep pure plant plus kit (Tiangen Biotech, Beijing, China, Cat# DP441). cDNA synthesis was performed using the Tiangen FastQuant RT kit (Tiangen Biotech, Beijing, China, Cat# KR116) with 20-μl reaction mixture. qRT-PCR analysis was performed using UltraSYBR Mixture (low ROX) (CoWin Biosciences, Beijing, China, Cat# CW0956) in an ABI QuantStudio™ 6 flex Real-Time PCR System (ABI, CA, United States). The PCR cycling conditions were 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The Arabidopsis and cassava gene transcripts were normalized to the AtAct2 gene (AT3G18780) and elongation factor 1α (EF1α, Me.15G054800) using the comparative 2–Δ Δ Ct method, respectively (Livak and Schmittgen, 2001). The qRT-PCR primers of MeEF1a, MePR1 were obtained from Wei et al. (2018), AtPR1, AtPR2, AtPR5, AtPDF1.2, AtICS1, AtAct2, and BcActA were obtained from Mhamdi and Noctor (2016), and AtTGA3 was obtained from Ndamukong et al. (2017), respectively. The qRT-PCR primers of MeLRRs, and AbAct (JQ671669.1) of A. brassicicola were designed by Primer3Plus3 to find optimal primers (Untergasser et al., 2007), and then the specificity of the melt curve analyzed performed to determine. In addtion, the qRT-PCR fragments and VIGS fragments are different CDS regions of MeLRRs. The primers used are listed in Supplementary Table 2.
For overexpression, the full-length coding regions of MeLRR1,-2,-3,-4 were amplified and cloned into the pEGAD vector (Promoter CaMV35S:GFP) via appropriate restriction enzyme digestion and T4 DNA ligase. The recombinant plasmids and empty vector were transformed into Agrobacterium GV3101. Then, the A. tumefaciens suspension was used to infect the leaves of cassava or tobacco as described by Sparkes et al. (2006) and Zeng et al. (2019). Tobacco leaves injected with Agrobacterium GV3101 for 2 days, the GFP fluorescence and DAPI (4’,6-diamidino-2-phenylindole, Thermo Fisher Scientific, Shanghai, China)-stained cell nuclei were imaged under a fluorescence microscope (Leica TCS SP8, Solms, Germany), with an excitation wavelength of 488 nm and a 505–530-nm bandpass emission filter. Cassava leaves inject with recombinant pEGAD plasmids or empty vector of Agrobacterium GV3101. Then, 3 days later, the cassava leaves were syringe infiltrated with 4 × 108 cfu/mL of pathogenic bacteria Xam used for disease resistance assay, include number of Xam populations, MePR1 transcript level, and symptoms of cassava bacterial blight at 0 and 1 dpi, respectively.
VIGS constructs are usually prepared using 300--500 bp partial CDS regions of MeLRRs and the online siDirect 2.04 tools (Naito et al., 2009) are available for predicting regions with high siRNA generating capability (Naito et al., 2009; Ui-Tei and Naito, 2013). Zeng et al. (2019) constructs the method about Agrobacterium-mediated Tobacco Rattle Virus (TRV)-based gene silencing in cassava. For VIGS in cassava, the specific CDS fragments of MeLRR1,−2,−3,−4 were amplified and cloned into the pTRV2 vector through appropriate restriction enzyme digestion and T4 DNA ligase. The recombinant plasmids and empty vectors were transformed into Agrobacterium GV3101. Then, the Agrobacterium suspension, as well as pTRV1, was used to infect the leaves of cassava as previously described (Zeng et al., 2019). At 14 dpi, the new leaves were syringe infiltrated with 4 × 108 cfu/mL of pathogenic bacteria Xam used for disease resistance assay. The sequences of primers used for vector construction in this study are listed in Supplementary Table 2.
Arabidopsis thaliana ecotype Col-0 was used as wild-type. Overexpressing lines were transformed by floral dip transformation method of 35S:GFP-MeLRR recombinant plasmids constructs with Agrobacterium GV3101 (Bechtold and Pelletier, 1998). The overexpressing lines were selected by 100 mg/L kanamycin and 20 mg/L glufosinate (Basta; Sangon Biotech. Shanghai, China) resistance and further confirmed by PCR. Single insertion transgenic lines were chosen for further analysis in transgenic third generations (T3).
The endogenous SA content in leaves was determined as previously described (Wei et al., 2018). Briefly, leaves were flash-frozen in liquid nitrogen and ground to a very fine powder. SA was extracted from 0.1 g powder using phosphate-buffered solution (PBS, pH 7.4, 0.15 M) on ice. Then, the supernatant was used for SA quantification using a plant SA ELISA (enzyme-linked immunosorbent assay) kit (Jiangsu Meimian Industrial, Jiangsu, China, Cat#HLE01901) according to the manufacturer’s instructions.
The ROS burst in leaves was determined as described previously (de Torres Zabala et al., 2015; Chang et al., 2020; Yan et al., 2021). In tomato, flg22 was used to instead of Xanthomonas to measure the ROS burst (Bhattarai et al., 2016). Similar methods were applied to study the cassava resistance to Xam, such as MeCAMTA3 (Chang et al., 2020), MeRAV5 (Yan et al., 2021). Herein, to measure the ROS burst, 48 leaf discs (5 mm in diameter) of cassava were placed in 48 single wells of 96-well black plates and placed in the dark for 12 h in 100 μL double-distilled water. After 12 h, the 48 leaf discs were divided into two groups. In one group, the water was replaced with 100 μL incubation solution containing 0.2 μmol/L luminol (AppliChem, Darmstadt, Germany) and 10 μg/mL horseradish peroxidase (AppliChem, Darmstadt, Germany). In the other group, the water was then replaced with 100 μL incubation solution containing 0.2 μmol/L luminol, 10 μg/mL horseradish peroxidase and 1 μmol/L flg22 (Phyto Technology Laboratories, Lenexa, KS, United States). Luminescence was measured immediately for 30 min using a GloMax 96 Microplate Luminometer (Promega, Madison, WI, United States). Luminescence readout is given in relative light emitting units (RLU).
The cassava or N. benthamiana leaves were boiled for 1 min in the trypan blue working solution (100 mL lactic acid, 100 mL glycerol, 100 g phenol, and 0.2 g trypan blue, dissolved in 100 mL distilled water) for 24 h at room temperature (Luo et al., 2017). The leaves were transferred into a chloral hydrate solution (2.5 g/mL) and repeatedly reduced until the background was gone (Luo et al., 2017).
The pathogenic bacterium P. syringae pv. tomato (Pst) DC3000 was streaked on LB medium with 50 mg/L of rifampicin at 28°C and shaken to OD600 reached 0.6. Thereafter, a fresh bacterial culture of Pst DC3000 was diluted to 4 × 108 cfu/mL in 10 mmol/L MgCl2 and 0.05% Silwet L-77 and then sprayed on 24-day-old Arabidopsis leaves. The A. brassicicola and B. cinerea strains were cultured on potato dextrose agar (PDA) medium with 2% (w/v) sucrose at 28°C. Conidia were suspended in distilled water for plant infection. Spore suspensions (about 4 × 106 spores/mL) of A. brassicicola and B. cinerea were sprayed on Arabidopsis leaves. The infected plants were grown in an incubator at 90% RH and 22°C. At 0, 2, and 4 dpi, the number of Pst DC3000 bacteria was determined, as well as the fungal actin gene transcript in leaves of Col-0 and mutants infected with B. cinerea and A. brassicicola (Veronese et al., 2006; Mhamdi and Noctor, 2016).
Mean and standard deviations are displayed as representative values for data in the figures. Analysis of variance (ANOVA) with Duncan’s test and Student’s t-test were applied to the obtained data with the help of IBM SPSS v20. Statistical significance (*) was set at p < 0.05. Each assay contained three independent replicates.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
YW and HZ designed the research. HZ did most experimental works and wrote the manuscript. ZY, ZL, and YS did experimental works and database analysis. XL, JW, and GZ did experimental works. YW supervised this project. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Key R&D Program of China (2019YFD1000500), National Natural Science Foundation of China (31671489), Hainan Provincial Natural Science Foundation of China (2019RCI55), and Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Yule Liu from Tsinghua University and Haitao Shi from Hainan University for kindly providing the pTRV1 and pTRV2 vectors. We are also grateful to Haitao Shi from Hainan University for kindly providing the pEGAD vector, Pseudomonas syringae pv. tomato DC3000, Alternaria brassicicola, and Botrytis cinerea strains. We would like to thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.790140/full#supplementary-material
Amuge, T., Berger, D. K., Katari, M. S., Myburg, A. A., Goldman, S. L., and Ferguson, M. E. (2017). A time series transcriptome analysis of cassava (Manihot esculenta Crantz) varieties challenged with Ugandan cassava brown streak virus. Sci. Rep. 7:9747. doi: 10.1038/s41598-017-09617-z
Arrieta-Ortiz, M. L., Rodríguez-R, L. M., Pérez-Quintero, ÁL., Poulin, L., Díaz, A. C., Arias, R. N., et al. (2013). Genomic survey of pathogenicity determinants and VNTR markers in the cassava bacterial pathogen Xanthomonas axonopodis pv. manihotis strain CIO151. PLoS One 8:e79704. doi: 10.1371/journal.pone.0079704
Bart, R., Cohn, M., Kassen, A., McCallum, E. J., Shybut, M., Petriello, A., et al. (2012). High-throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance. Proc. Natl. Acad. Sci. U.S.A. 109, E1972–E1979. doi: 10.1073/pnas.1208003109
Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. doi: 10.1385/0-89603-391-0:259
Bhattarai, K., Louws, F. J., Williamson, J. D., and Panthee, D. P. (2016). Differential response of tomato genotypes to Xanthomonas-specific pathogen-associated molecular patterns and correlation with bacterial spot (Xanthomonas perforans) resistance. Hortic. Res. 3:16035. doi: 10.1038/hortres.2016.35
Bianchet, C., Wong, A., Quaglia, M., Alqurashi, M., Gehring, C., Ntoukakis, V., et al. (2019). An Arabidopsis thaliana leucine-rich repeat protein harbors an adenylyl cyclase catalytic center and affects responses to pathogens. J. Plant Physiol. 232, 12–22. doi: 10.1016/j.jplph.2018.10.025
Bigeard, J., Colcombet, J., and Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Bonardi, V., Tang, S., Stallmann, A., Roberts, M., Cherkis, K., and Dangl, J. L. (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. U.S.A. 108, 16463–16468. doi: 10.1073/pnas.1113726108
Bredeson, J., Lyons, J., Prochnik, S. E., Wu, G., Ha, C., Edsinger-Gonzales, E., et al. (2016). Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 34, 562–570. doi: 10.1038/nbt.3535
Castiblanco, L. F., Gil, J., Rojas, A., Osorio, D., Gutiérrez, S., Muñoz-Bodnar, A., et al. (2013). TALE1 from Xanthomonas axonopodis pv. manihotis acts as a transcriptional activator in plant cells and is important for pathogenicity in cassava plants. Mol. Plant Pathol. 14, 84–95. doi: 10.1111/j.1364-3703.2012.00830.x
Chang, Y., Bai, Y., Wei, Y., and Shi, H. (2020). CAMTA3 negatively regulates disease resistance through modulating immune response and extensive transcriptional reprogramming in cassava. Tree Physiol. 40, 1520–1533. doi: 10.1093/treephys/tpaa093
Chen, J., Peng, P., Tian, J., He, Y., Zhang, L., Liu, Z., et al. (2015). Pike, a rice blast resistance allele consisting of two adjacent NBS–LRR genes, was identified as a novel allele at the Pik locus. Mol. Breed. 35:117. doi: 10.1007/s11032-015-0305-6
Cohn, M., Morbitzer, R., Lahaye, T., and Staskawicz, B. J. (2016). Comparison of gene activation by two TAL effectors from Xanthomonas axonopodis pv. manihotis reveals candidate host susceptibility genes in cassava. Mol. Plant Pathol. 17, 875–889. doi: 10.1111/mpp.12337
Constantin, E. C., Cleenwerck, I., Maes, M., Baeyen, S., Malderghem, C. V., Vos, P. D., et al. (2016). Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol. 65, 792–806. doi: 10.1111/ppa.12461
Dalio, R. J. D., Magalhães, D. M., Rodrigues, C. M., Arena, G. D., Oliveira, T. S., Souza-Neto, R. R., et al. (2017). PAMPs, PRRs, effectors and R-genes associated with citrus-pathogen interactions. Ann. Bot. 119, 749–774. doi: 10.1093/aob/mcw238
de Torres Zabala, M., Littlejohn, G., Jayaraman, S., Studholme, D., Bailey, T., Lawson, T., et al. (2015). Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat Plants 1:15074. doi: 10.1038/nplants.2015.74
Deng, J., Fang, L., Zhu, X., Zhou, B., and Zhang, T. (2019). A CC-NBS-LRR gene induces hybrid lethality in cotton. J. Exp. Bot. 70, 5145–5156. doi: 10.1093/jxb/erz312
Divi, U., Rahman, T., and Krishna, P. (2010). Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 10:151. doi: 10.1186/1471-2229-10-151
Du, D., Zhang, C., Xing, Y., Lu, X., Cai, L., Yun, H., et al. (2021). The CC-NB-LRR OsRLR1 mediates rice disease resistance through interaction with OsWRKY19. Plant Biotechnol. J. 19, 1052–1064. doi: 10.1111/pbi.13530
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Goff, S. A., Ricke, D., Lan, T., Presting, G., Wang, R., Dunn, M., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 79–92. doi: 10.1126/science.1068275
Goyal, N., Bhatia, G., Sharma, S., Garewal, N., Upadhyay, A., Upadhyay, S., et al. (2020). Genome-wide characterization revealed role of NBS-LRR genes during powdery mildew infection in Vitis vinifera. Genomics 112, 312–322. doi: 10.1016/j.ygeno.2019.02.011
Irigoyen, M. L., Garceau, D. C., Bohorquez-Chaux, A., Lopez-Lavalle, L. A. B., Perez-Fons, L., Fraser, P. D., et al. (2020). Genome-wide analyses of cassava pathogenesis-related (PR) gene families reveal core transcriptome responses to whitefly infestation, salicylic acid and jasmonic acid. BMC Genomics 21:93. doi: 10.1186/s12864-019-6443-1
Jia, Y., Yuan, Y., Zhang, Y., Yang, S., and Zhang, X. (2015). Extreme expansion of NBS-encoding genes in Rosaceae. BMC Genet. 16:48. doi: 10.1186/s12863-015-0208-x
Joshi, R. K., Kar, B., and Nayak, S. (2011). Survey and characterization of NBS-LRR (R) genes in Curcuma longa transcriptome. Bioinformation 6, 360–363. doi: 10.6026/97320630006360
Kayondo, S. I., Pino Del, C. D., Lozano, R., Ozimati, A., Wolfe, M., Baguma, Y., et al. (2018). Genome-wide association mapping and genomic prediction for CBSD resistance in Manihot esculenta. Sci. Rep. 8:1549. doi: 10.1038/s41598-018-19696-1
Kourelis, J., and van der Hoorn, R. A. L. (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299. doi: 10.1105/tpc.17.00579
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lei, X., Yao, Q., Xu, X., and Liu, Y. (2014). Isolation and characterization of NBS-LRR resistance gene analogues from mango. Biotechnol. Biotechnol. Equip. 28, 417–424. doi: 10.1080/13102818.2014.931706
Li, N., Ma, X., Short Dylan, P. G., Li, T., Zhou, L., Gui, Y., et al. (2018a). The island cotton NBS-LRR gene GbaNA1 confers resistance to the non-race 1 Verticillium dahliae isolate Vd991. Mol. Plant Pathol. 19, 1466–1479. doi: 10.1111/mpp.12630
Li, N., Zhou, L., Zhang, D., Klosterman Steven, J., Li, T., Gui, Y., et al. (2018b). GbaNA1 heterologous expression of the cotton NBS-LRR gene enhances Verticillium wilt resistance in Arabidopsis. Front. Plant Sci. 9:119. doi: 10.3389/fpls.2018.00119
Li, K., Xiong, X., Zhu, S., Liao, H., Xiao, X., Tang, Z., et al. (2018c). MeBIK1, a novel cassava receptor-like cytoplasmic kinase, regulates PTI response of transgenic Arabidopsis. Funct. Plant Biol. 45, 658–667. doi: 10.1071/FP17192
Li, X., Liu, W., Li, B., Liu, G., Wei, Y., He, C., et al. (2018d). Identification and functional analysis of cassava DELLA proteins in plant disease resistance against cassava bacterial blight. Plant Physiol. Biochem. 124, 70–76. doi: 10.1016/j.plaphy.2017.12.022
Li, T., Wang, B., Yin, C., Zhang, D., Wang, D., Song, J., et al. (2019). The Gossypium hirsutum TIR-NBS-LRR gene GhDSC1 mediates resistance against Verticillium wilt. Mol. Plant Pathol. 20, 857–876. doi: 10.1111/mpp.12797
Li, X., Fan, S., Hu, W., Liu, G., Wei, Y., He, C., et al. (2017). Two cassava basic leucine zipper (bZIP) transcription factors (MebZIP3 and MebZIP5) confer disease resistance against cassava bacterial blight. Front. Plant Sci. 8:2110. doi: 10.3389/fpls.2017.02110
Liao, C., Lai, Z., Lee, S., Yun, D., and Mengiste, T. (2016). Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell 28, 1662–1681. doi: 10.1105/tpc.16.00105
Liu, C., Chen, X., Ma, P., Zhang, S., Zeng, C., Jiang, X., et al. (2018). Ethylene responsive factor MeERF72 negatively regulates sucrose synthase 1 gene in cassava. Int. J. Mol. Sci. 19:1281. doi: 10.3390/ijms19051281
Liu, H., Dong, S., Gu, F., Liu, W., Yang, G., Huang, M., et al. (2017). NBS-LRR protein Pik-H4 interacts with OsBIHD1 to balance rice blast resistance and growth by coordinating ethylene-brassinosteroid pathway. Front. Plant Sci. 8:127. doi: 10.3389/fpls.2017.00127
Liu, W., Yan, Y., Zeng, H., Li, X., Wei, Y., Liu, G., et al. (2018). Functional characterization of WHY-WRKY75 transcriptional module in plant response to cassava bacterial blight. Tree Physiol. 38, 1502–1512. doi: 10.1093/treephys/tpy053
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Louis, B., and Rey, C. (2015). Resistance gene analogs involved in tolerant cassava-geminivirus interaction that shows a recovery phenotype. Virus Genes 51, 393–407. doi: 10.1007/s11262-015-1246-1
Lozano, R., Hamblin, M. T., Prochnik, S., and Jannink, J. L. (2015). Identification and distribution of the NBS-LRR gene family in the cassava genome. BMC Genomics 16:360. doi: 10.1186/s12864-015-1554-9
Luo, X. M., Xu, N., Huang, J. K., Gao, F., Zou, H. S., Boudsocq, M., et al. (2017). A lectin receptor-like kinase mediates pattern-triggered salicylic acid signaling. Plant Physiol. 174, 2501–2514. doi: 10.1104/pp.17.00404
Macaulay, K. M., Heath Geraldine, A., Ciulli, A., Murphy Alex, M., Abell, C., Carr John, P., et al. (2017). The biochemical properties of the two Arabidopsis thaliana isochorismate synthases. Biochem. J. 474, 1579–1590. doi: 10.1042/BCJ20161069
Masumba, E. A., Kapinga, F., Mkamilo, G., Salum, K., Kulembeka, H., Rounsley, S., et al. (2017). QTL associated with resistance to cassava brown streak and cassava mosaic diseases in a bi-parental cross of two Tanzanian farmer varieties, Namikonga and Albert. Theor. Appl. Genet. 130, 2069–2090. doi: 10.1007/s00122-017-2943-z
Medina, C. A., Reyes, P. A., Trujillo, C. A., Gonzalez, J. L., Bejarano, D. A., Montenegro, N. A., et al. (2018). The role of type III effectors from Xanthomonas axonopodis pv. manihotis in virulence and suppression of plant immunity. Mol. Plant Pathol. 19, 593–606. doi: 10.1111/mpp.12545
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R. W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308
Mhamdi, A., and Noctor, G. (2016). High CO2 primes plant biotic stress defences through redox-linked pathways. Plant Physiol. 172, 929–942. doi: 10.1104/pp.16.01129
Mondal, K. K., Soni, M., Verma, G., Kulshreshtha, A., Mrutyunjaya, S., and Kumar, R. (2020). Xanthomonas axonopodis pv. punicae depends on multiple non-TAL (Xop) T3SS effectors for its coveted growth inside the pomegranate plant through repressing the immune responses during bacterial blight development. Microbiol. Res. 240:126560. doi: 10.1016/j.micres.2020.126560
Muñoz-Bodnar, A., Perez-Quintero Alvaro, L., Gomez-Cano, F., Gil, J., Michelmore, R., Bernal, A., et al. (2014). RNAseq analysis of cassava reveals similar plant responses upon infection with pathogenic and non-pathogenic strains of Xanthomonas axonopodis pv. manihotis. Plant Cell Rep. 33, 1901–1912. doi: 10.1007/s00299-014-1667-7
Naito, Y., Yoshimura, J., Morishita, S., and Ui-Tei, K. (2009). siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics 10:392. doi: 10.1186/1471-2105-10-392
Ndamukong, I., Abdallat, A. A., Thurow, C., Fode, B., Zander, M., Weigel, R., et al. (2017). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50, 128–139. doi: 10.1111/j.1365-313X.2007.03039.x
Palmer, I. A., Chen, H., Chen, J., Chang, M., Li, M., Liu, F., et al. (2019). Novel salicylic acid analogs induce a potent defense response in Arabidopsis. Int. J. Mol. Sci. 20:3356. doi: 10.3390/ijms20133356
Pandolfi, V., Neto, J. R. C. F., da Silva, M. D., Amorim, L. L. B., Wanderley-Nogueira, A. C., de Oliveira Silva, R. L., et al. (2017). Resistance (R) genes: applications and prospects for plant biotechnology and breeding. Curr. Protein Pept. Sci. 18, 323–334. doi: 10.2174/1389203717666160724195248
Peng, Y., Yang, J., Li, X., and Zhang, Y. (2021). Salicylic acid: biosynthesis and signaling. Annu. Rev. Plant Biol. 72, 761–791. doi: 10.1146/annurev-arplant-081320-092855
Shao, Z., Zhang, Y., Hang, Y., Xue, J., Zhou, G., Wu, P., et al. (2006). Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: understanding gained from and beyond the legume family. Plant Physiol. 166, 217–234. doi: 10.1104/pp.114.243626
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Ui-Tei, K., and Naito, Y. (2013). Designing functional siRNA with reduced off-target effects. Methods Mol. Biol. 942, 57–68. doi: 10.1007/978-1-62703-119-6_3
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., and Leunissen, J. A. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74. doi: 10.1093/nar/gkm306
Urbach, J. M., and Ausubel, F. M. (2017). The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events. Proc. Natl. Acad. Sci. U.S.A. 114, 1063–1068. doi: 10.1073/pnas.1619730114
Utsumi, Y., Tanaka, M., Kurotani, A., Yoshida, T., Mochida, K., Matsui, A., et al. (2016). Cassava (Manihot esculenta) transcriptome analysis in response to infection by the fungus Colletotrichum gloeosporioides using an oligonucleotide-DNA microarray. J. Plant Res. 129, 711–726. doi: 10.1007/s10265-016-0828-x
van der Hoorn, R. A., and Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017. doi: 10.1105/tpc.108.060194
Veronese, P., Nakagami, H., Bluhm, B., Abuqamar, S., Chen, X., Salmeron, J., et al. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273. doi: 10.1105/tpc.105.035576
Wang, X., Guo, R., Tu, M., Wang, D., Guo, C., Wan, R., et al. (2017). Arabidopsis thaliana ectopic expression of the wild grape WRKY transcription factor VqWRKY52 in enhances resistance to the biotrophic pathogen powdery mildew but not to the necrotrophic pathogen. Front. Plant Sci. 8:97. doi: 10.3389/fpls.2017.00097
Wei, Y., Liu, G., Bai, Y., Xia, F., He, C., Shi, H., et al. (2017). Two transcriptional activators of N-acetylserotonin O-methyltransferase 2 and melatonin biosynthesis in cassava. J. Exp. Bot. 68, 4997–5006. doi: 10.1093/jxb/erx305
Wei, Y., Liu, G., Chang, Y., He, C., and Shi, H. (2018). Heat shock transcription factor 3 regulates plant immune response through modulation of salicylic acid accumulation and signalling in cassava. Mol. Plant Pathol. 19, 2209–2220. doi: 10.1111/mpp.12691
Wei, Y., Zeng, H., Liu, W., Cheng, X., Zhu, B., Guo, J., et al. (2021a). Autophagy-related genes serve as heat shock protein 90 co-chaperones in disease resistance against cassava bacterial blight. Plant J. 107, 925–937. doi: 10.1111/tpj.15355
Wei, Y., Zhu, B., Liu, W., Cheng, X., Lin, D., He, C., et al. (2021b). Heat shock protein 90 co-chaperone modules fine-tune the antagonistic interaction between salicylic acid and auxin biosynthesis in cassava. Cell Rep. 34:108717. doi: 10.1016/j.celrep.2021.108717
Wu, L., Chen, H., Curtis, C., and Fu, Z. Q. (2014). Go in for the kill: how plants deploy effector-triggered immunity to combat pathogens. Virulence 5, 710–721. doi: 10.4161/viru.29755
Xu, Y., Liu, F., Zhu, S., and Li, X. (2018). ZmNBS25, the maize NBS-LRR gene enhances disease resistance in rice and Arabidopsis. Front. Plant Sci. 9:1033. doi: 10.3389/fpls.2018.01033
Yan, Y., Wang, P., Wei, Y., Bai, Y., Lu, Y., Zeng, H., et al. (2021). The dual interplay of RAV5 in activating nitrate reductases and repressing catalase activity to improve disease resistance in cassava. Plant Biotechnol. J. 19, 785–800. doi: 10.1111/pbi.13505
Yang, X., and Wang, J. (2016). Genome-wide analysis of NBS-LRR genes in sorghum genome revealed several events contributing to NBS-LRR gene evolution in grass species. Evol. Bioinform. Online 12, 9–21. doi: 10.4137/EBO.S36433
Yoodee, S., Kobayashi, Y., Songnuan, W., Boonchird, C., Thitamadee, S., Kobayashi, I., et al. (2018). Phytohormone priming elevates the accumulation of defense-related gene transcripts and enhances bacterial blight disease resistance in cassava. Plant Physiol. Biochem. 122, 65–77. doi: 10.1016/j.plaphy.2017.11.016
Yuan, Y., Chung, J. D., Fu, X., Johnson Virgil, E., Ranjan, P., Booth, S. L., et al. (2009). Alternative splicing and gene duplication differentially shaped the regulation of isochorismate synthase in Populus and Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22020–22025. doi: 10.1073/pnas.0906869106
Zárate-Chaves, C. A., de la Cruz, D. G., Verdier, V., López, C. E., Bernal, A., and Szurek, B. (2021). Cassava diseases caused by Xanthomonas phaseoli pv. manihotis and Xanthomonas cassavae. Mol. Plant Pathol. 22, 1520–1537. doi: 10.1111/mpp.13094
Zeng, H., Xie, Y., Liu, G., Wei, Y., Hu, W., and Shi, H. (2019). Agrobacterium-mediated gene transient overexpression and Tobacco rattle virus (TRV)-based gene silencing in cassava. Int. J. Mol. Sci. 20:3976. doi: 10.3390/ijms20163976
Zhang, C., Chen, H., Cai, T., Deng, Y., Zhuang, R., Zhang, N., et al. (2017). Overexpression of a novel peanut NBS-LRR gene AhRRS5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol. J. 15, 39–55. doi: 10.1111/pbi.12589
Zhang, J. Z. (2003). Overexpression analysis of plant transcription factors. Curr. Opin. Plant Biol. 6, 430–440. doi: 10.1016/s1369-5266(03)00081-5
Zhang, Y., Chen, M., Sun, L., Wang, Y., Yin, J., Liu, J., et al. (2020). Genome-wide identification and evolutionary analysis of NBS-LRR genes from Dioscorea rotundata. Front. Genet. 11:484. doi: 10.3389/fgene.2020.00484
Zhao, Q., Li, H., Sun, H., Li, A., Liu, S., Yu, R., et al. (2018). Salicylic acid and broad spectrum of NBS-LRR family genes are involved in SMV-soybean interactions. Plant Physiol. Biochem. 123, 132–140. doi: 10.1016/j.plaphy.2017.12.011
Zhou, J., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., et al. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13, 191–202. doi: 10.1094/MPMI.2000.13.2.191
Zhou, X., Liu, J., Bao, S., Yang, Y., and Zhuang, Y. (2018). Molecular cloning and characterization of a wild eggplant Solanum aculeatissimum NBS-LRR gene, involved in plant resistance to Meloidogyne incognita. Int. J. Mol. Sci. 19:583. doi: 10.3390/ijms19020583
Zhu, X., Lu, C., Du, L., Ye, X., Liu, X., Coules, A., et al. (2017). The wheat NB-LRR gene TaRCR1 is required for host defence response to the necrotrophic fungal pathogen Rhizoctonia cerealis. Plant Biotechnol. J. 15, 674–687. doi: 10.1111/pbi.12665
Keywords: cassava, cassava bacterial blight, resistance genes, salicylic acid, ROS, NBS-LRR
Citation: Zhang H, Ye Z, Liu Z, Sun Y, Li X, Wu J, Zhou G and Wan Y (2022) The Cassava NBS-LRR Genes Confer Resistance to Cassava Bacterial Blight. Front. Plant Sci. 13:790140. doi: 10.3389/fpls.2022.790140
Received: 06 October 2021; Accepted: 07 January 2022;
Published: 01 February 2022.
Edited by:
Daguang Cai, University of Kiel, GermanyReviewed by:
Divya Chandran, Regional Centre for Biotechnology (RCB), IndiaCopyright © 2022 Zhang, Ye, Liu, Sun, Li, Wu, Zhou and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinglang Wan, eWx3YW5AaGFpbmFudS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.