
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 24 February 2022
Sec. Plant Systematics and Evolution
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.788911
This article is part of the Research TopicMolecular Organization, Evolution, and Function of Ribosomal DNAView all 15 articles
The ubiquitous presence of rRNA genes in nuclear, plastid, and mitochondrial genomes has provided an opportunity to use genomic markers to infer patterns of molecular and organismic evolution as well as to assess systematic issues throughout the tree of life. The number, size, location, and activity of the 35S rDNA cistrons in plant karyotypes have been used as conventional cytogenetic landmarks. Their scrutiny has been useful to infer patterns of chromosomal evolution and the data have been used as a proxy for assessing species discrimination, population differentiation and evolutionary relationships. The correct interpretation of rDNA markers in plant taxonomy and evolution is not free of drawbacks given the complexities derived from the lability of the genetic architecture, the diverse patterns of molecular change, and the fate and evolutionary dynamics of the rDNA units in hybrids and polyploid species. In addition, the terminology used by independent authors is somewhat vague, which often complicates comparisons. To date, no efforts have been reported addressing the potential problems and limitations involved in generating, utilizing, and interpreting the data from the 35S rDNA in cytogenetics. This review discusses the main technical and conceptual limitations of these rDNA markers obtained by cytological and karyological experimental work, in order to clarify biological and evolutionary inferences postulated in a systematic and phylogenetic context. Also, we provide clarification for some ambiguity and misconceptions in terminology usually found in published work that may help to improve the usage of the 35S ribosomal world in plant evolution.
Ribosomes are universal cellular components found across all domains of life. Research suggests they represent the most critical macromolecular machine in living organisms, as they are trusted with carrying out protein synthesis in cells by converting information encoded within mRNA into peptides (Ojha et al., 2020). Ribosome biogenesis in eukaryotes is a process of extraordinary complexity (Thomson et al., 2013). Four rRNA species are transcribed by two RNA polymerases, RNA Pol I (18S, 5.8S, 26S rRNA) and RNA Pol III (5S rRNA) being extensively modified during their subsequent maturation in the macromolecular complex of the nucleolus, the nucleus and the cytoplasm (Sloan et al., 2017).
The ubiquitous presence of rDNA genes in nuclear, plastid and mitochondrial genomes has provided an opportunity to use ribosomal sequences as homologous markers to infer evolutionary processes and to assess systematic issues at the three basic domains of life, Bacteria, Archaea, Eukarya, including plants (Álvarez and Wendel, 2003; Nieto Feliner and Rosselló, 2007). In fact, the 5S intergenic spacers and the internal transcribed spacers (ITS) from the nuclear ribosomal 35S repeat have been proposed as nuclear standards for species identification (Gemeinholzer et al., 2006; Doveri and Lee, 2007; Gao et al., 2010), species delimitation (Müller et al., 2007) and DNA barcoding (Kress et al., 2005; Chase et al., 2007; CBOL Plant Working Group, 2009) in land plant lineages.
Ribosomal markers are not only limited to the use of rDNA sequences. Since the pioneering studies on the topic (Heitz, 1931), there has been extensive karyological work characterizing rDNA (García et al., 2017). The study of the development of the nucleolus, the assessment of karyological landmarks related to the linear differentiation of chromosomes, like non-centromeric constrictions and associated (satellite) regions, and the physical mapping and linkage between 35S rDNA and 5S rDNA families, have provided anchor points for comparative studies (Weiss-Schneeweiss and Schneeweiss, 2013). These include assessing the molecular evolution of rDNA units, gene silencing, evolutionary trends on karyotype differentiation, producing genetic maps and identifying ancestors in hybrids and polyploid species (Weiss-Schneeweiss et al., 2013). These karyo-evolutionary trends have complemented the knowledge on nuclear rDNA using DNA sequencing and are clearly relevant for providing cytogenetic markers in species that are routinely used for plant systematics and evolution purposes, and to postulate phylogenetic hypotheses (Totta et al., 2017).
The number, size, location and transcriptional activity of the 35S rDNA cistrons in plant karyotypes are some of the traditional cytogenetic landmarks reported in conventional karyotype descriptions (Battaglia, 1955). Their scrutiny has been useful to infer patterns of chromosomal evolution and the data have been used as a proxy for species discrimination, population differentiation and relationships at several evolutionary levels (Guerra, 2012). In agreement with their continued use in the field of cytotaxonomy, no major concerns have been reported for their application as cytogenetic markers in plant taxonomy and evolution (García et al., 2017).
The correct interpretation of 35S rDNA markers in plant taxonomy and phylogenetics is not free of drawbacks. This is not surprising given the complexities derived from the lability of the genetic architecture, the patterns of molecular evolution, and the evolutionary dynamics of the rDNA units, including the number and position of rDNA sites in the genome, sequence homogenization, structure and organization of rRNA genes (Feliner and Rosselló, 2012). In addition, the terminology used by independent authors is somewhat vague, which often complicates comparisons.
This complexity does not appear to be fully appreciated by some systematic plant studies that use chromosomal, or cytological-based rDNA markers, alone or in combination with genomic data. Unfortunately, misconceptions and misuses may lead to far-reaching conclusions based on risky assumptions that do not have general value, or on the use of confusing terminology (Tables 1#x2013;4). To date, no efforts acknowledging the potential problems and limitations involved in generating, utilizing, and interpreting the raw ribosomal data have been reported.

Table 1. The nuclear 35S rDNA landmarks and significant terminology associated to its activity, detection and morphology.
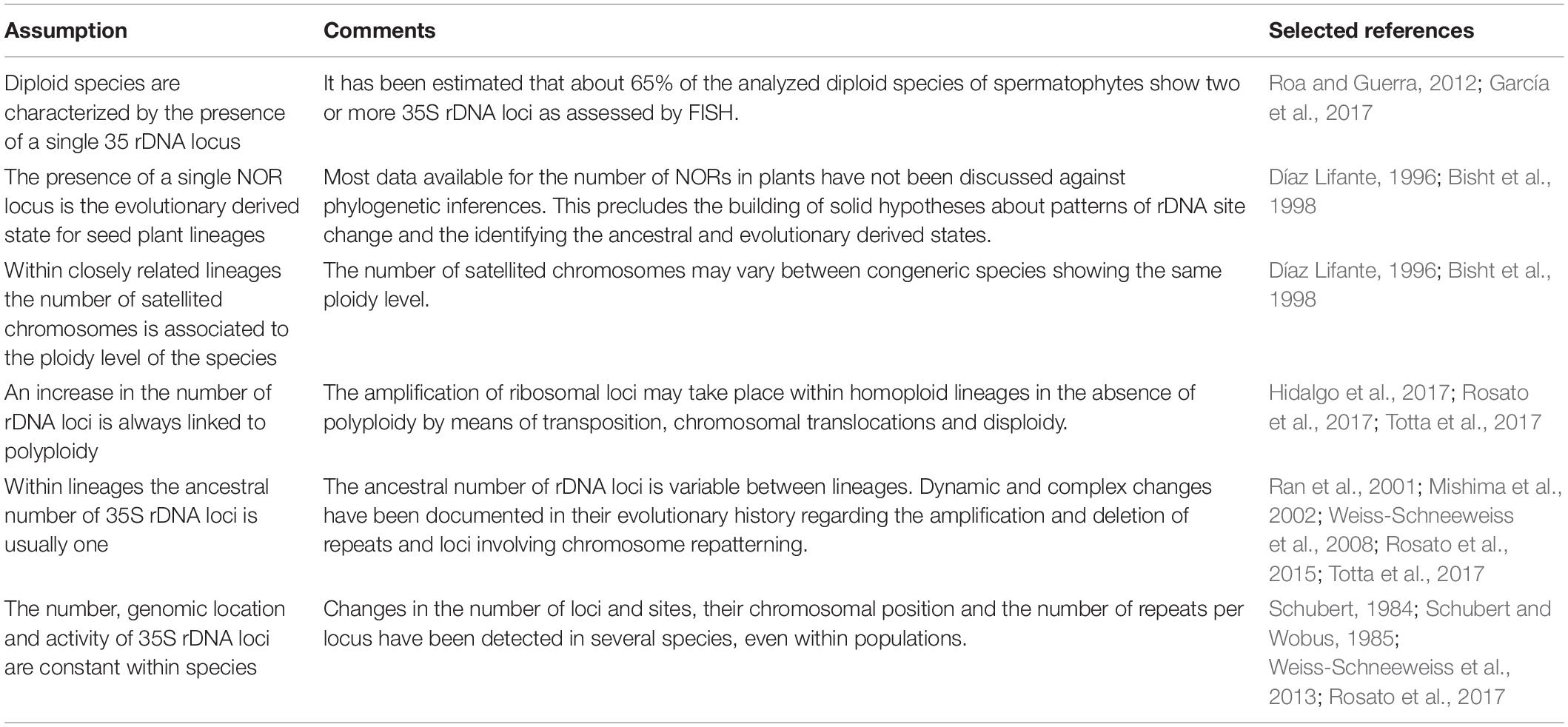
Table 3. Evolutionary trends in the number and activity of rDNA and NOR loci: cautions and limitations.
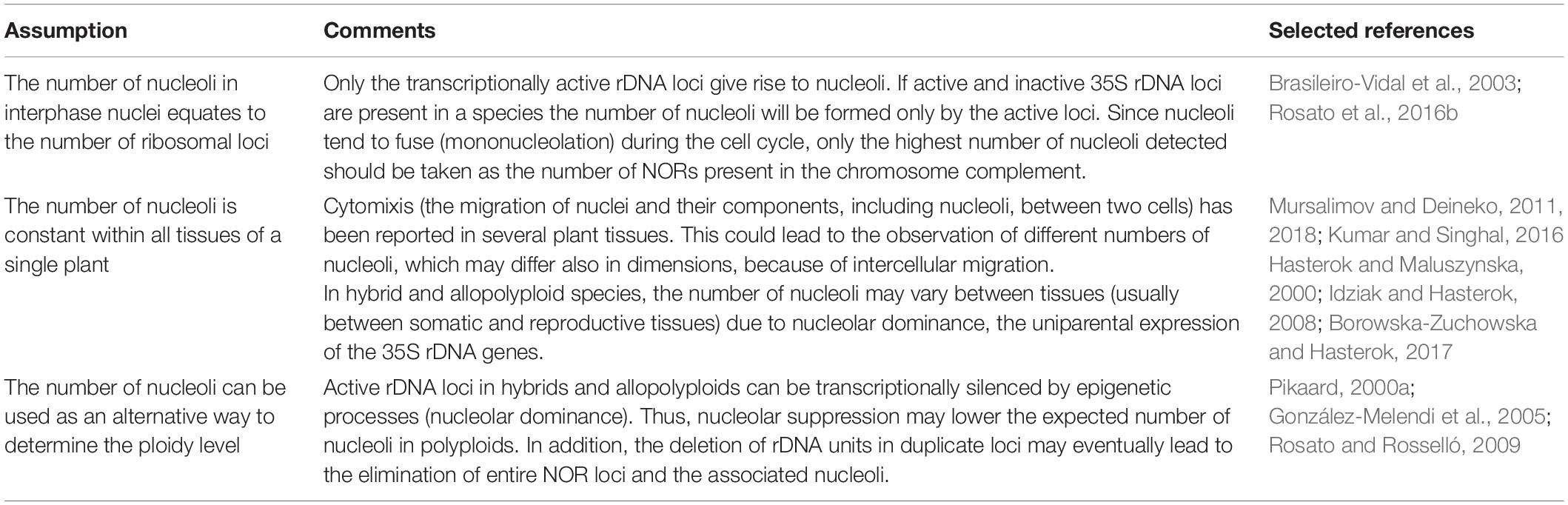
Table 4. Intra- and interindividual variation in nucleoli number: assumptions about their use in plant evolution.
This paper aims to discuss the limitations of nuclear 35S rDNA markers (i.e., site number, transcriptional activity, nucleolar dominance) based on cytological and karyological experimental work to draw sound biological and evolutionary conclusions in a systematic and phylogenetic context. Also, we provide clarification for some conceptual misconceptions usually found in published work that could help lead to an insightful utilization of the ribosomal world in plant evolution.
Most of the following terms are often used interchangeably in scientific literature: secondary constriction, nucleolar constriction, satellite, sputnik, intercalary satellite, nucleolar organizing regions (NORs), satellite(d) chromosome, SAT-chromosome, nucleolar chromosome, NOR chromosome, and 35S rDNA locus (Figure 1). However, in some cases there are significant differences that might not be rightly appreciated. Thus, inappropriate or confusing denominations may cause incorrect judgment on technically accurate data (Table 1).
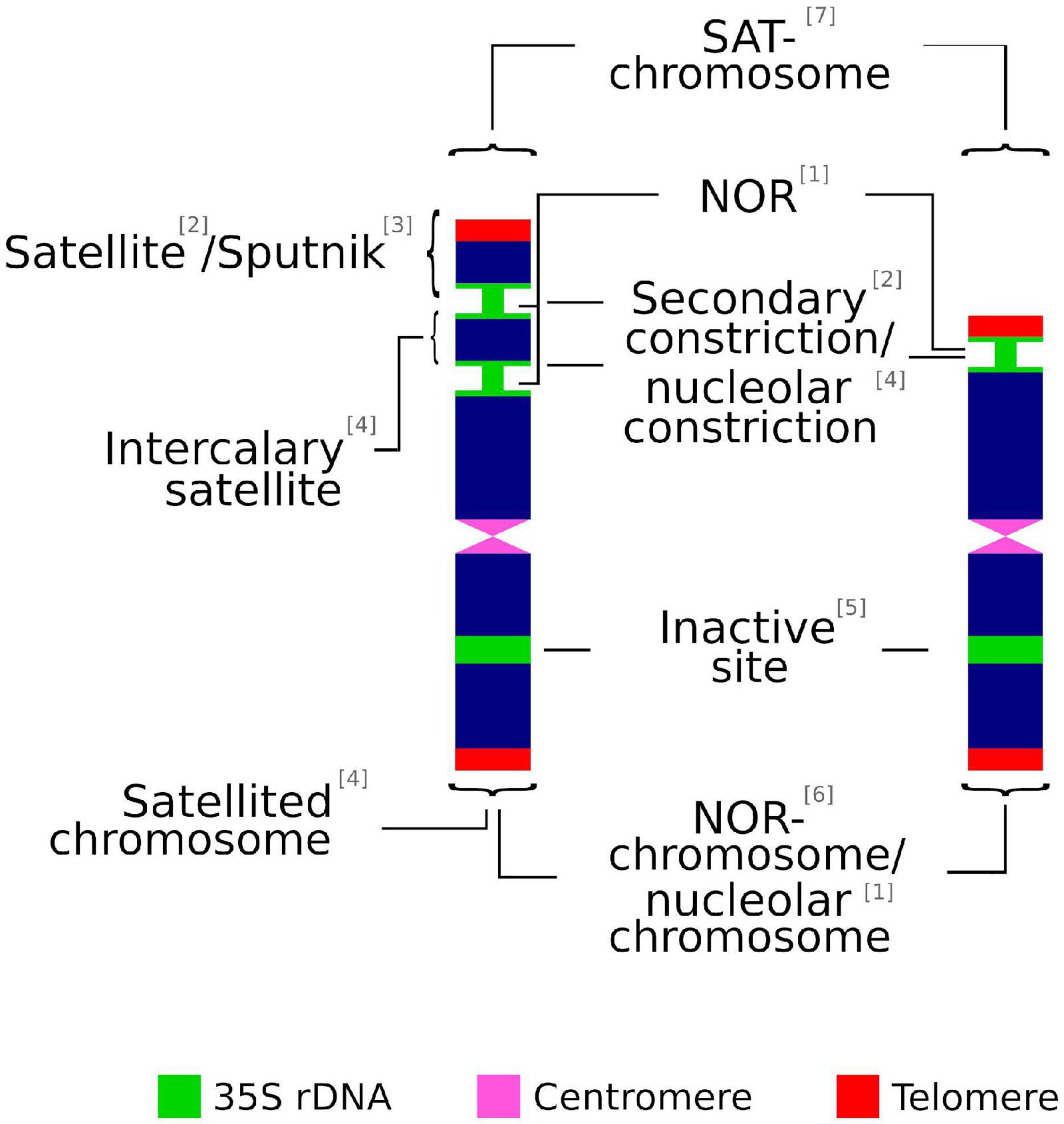
Figure 1. Graphical summary of terms used to describe the chromosomic domains associated to the 35S rDNA locus. (1) McClintock, 1934; (2) Darlington, 1926; (3) Battaglia, 1999; (4) Battaglia, 1955; (5) Leitch and Heslop-Harrison, 1993; (6) Schwarzacher and Wachtler, 1983; (7) Heitz, 1931.
The association of non-centromeric (i.e., secondary) constrictions (Darlington, 1926) and nucleolus formation during the interphase was noted early on by Heitz (1931) and McClintock (1934). The latter is credited for coining the term nucleolar organizer (later changed to NOR) to describe the chromosome region in Zea mays that was involved in the formation of the nucleolus (Pikaard, 2000a). In addition, a heterochromatic knob of the chromosome, the satellite, is present at the telomeric-proximal site of the secondary constriction, but is not involved in the formation of the nucleolus (Chen et al., 2000).
The term satellite chromosome is commonly used in the same way as SAT-chromosome. However, this is a misinterpretation recognized early on by Berger (1940), whose efforts to distinguish both technical terms have been repeatedly ignored in cytological literature (Battaglia, 1955). SAT was coined by Heitz (1931) as an abbreviation of Sine Acido Thymonucleinico (Without Thymonucleic Acid, the early denomination of DNA) and it refers to secondary constrictions. The decondensed chromatin at the NOR observed by Heitz (1931) showed less stained intensity (achromatic) than other chromosomal regions and was wrongly interpreted as lacking DNA. Accordingly, and following the original meaning, SAT-chromosome is not a synonym for satellited-chromosome but implies either a satellited chromosome or a chromosome with a secondary constriction that is associated with the formation of the nucleolus but does not have a satellite (Berger, 1940). In addition, and in contrast to the original meaning coined by McClintock (1934), some authors broadly define NOR sites as the chromosomal segments that contain ribosomal genes, irrespective of their transcriptional activity (Trerè, 2000).
In maize, there is a single pair of chromosomes showing secondary constrictions (i.e., two nucleolar chromosomes) that are also satellited chromosomes, a single active 35S rDNA locus or NOR, and a single associated nucleolus (Chen et al., 2000). This chromosomal rDNA pattern shows a complete agreement between chromatin decondensation sites (secondary constrictions), transcriptional active regions (NOR), the physical location of 35S rDNA units and the number of nucleoli formed (McClintock, 1934; Khuong and Schubert, 1985; Sadder and Weber, 2001).
Complexity may arise in lineages, however, when mechanisms involved in altering the number of rDNA loci have occurred along their evolutionary history. These bursts of rDNA amplification include the dispersion of loci by structural chromosome rearrangements at homoploid levels as well as the transposition and amplification of rDNA copy numbers (Datson and Murray, 2006; Rosato et al., 2017), and have involved both diploid lineages and complex scenarios of ancestral and more recent polyploid events (Rosato et al., 2015). These events may have resulted in karyotypes that eventually contain more 35S rDNA sites than expected when compared to the ancestral or parental lineages. When karyotypes contain more than one 35S rDNA locus, it clearly shows that care should be exercised to describe the chromosomal rDNA pattern, since not all the commonly used terms are equivalent and should not be broadly used as mere alternatives with similar or identical meanings (Figure 2).
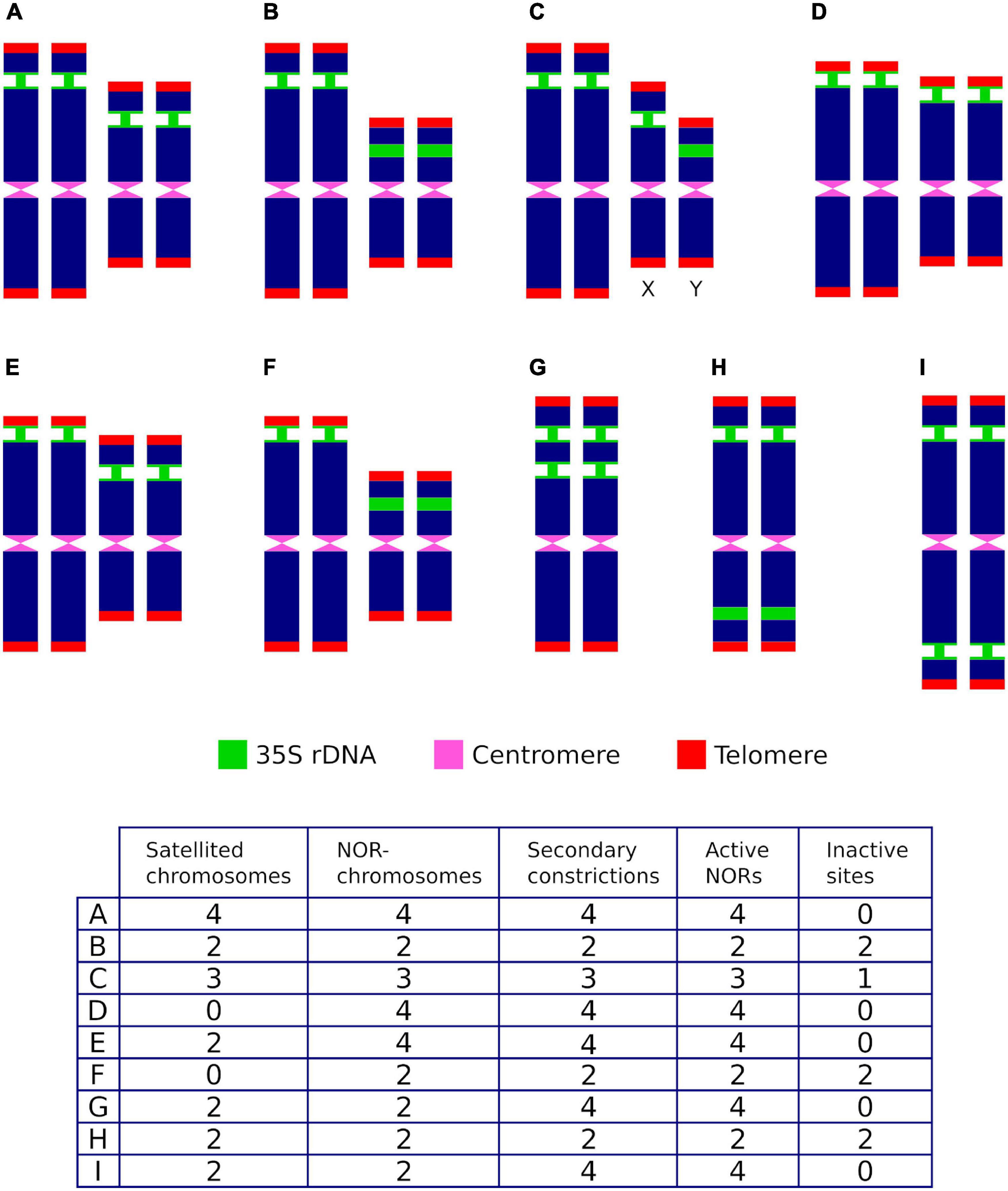
Figure 2. Hypothetical ribosomal phenotypes (A–I) and their associated ribosomal descriptors for individuals showing two rDNA loci and differing in transcriptional activity, the number of chromosome pairs where they are located, the position of the loci along the chromosome arms, the presence of sites in sexual chromosomes (X,Y), the number of secondary constrictions and the presence of satellites. Active sites are represented with narrow green stalks.
As an example that illustrates this issue, let us consider that the chromosomal complement of a hypothetical diploid species contains two 35S rDNA loci. After taking into account all heretofore reported relevant genomic locations and transcriptional activity alternatives that occur in the karyograms of seed plants (i.e., the number of active and silenced sites, the number of chromosome pairs where they are located, the position of the loci along the chromosome’s arms, the presence of sites in sexual chromosomes, the number of secondary constrictions and the presence of satellites), up to eight potential ribosomal phenotypes may be described (Figure 2) by common descriptors frequently used in the literature (the number of SAT-chromosomes and satellited chromosomes, secondary constrictions and NOR sites). Thus, for a given karyotype (1) the number of SAT-chromosomes, satellite(d) chromosomes and nucleolar chromosomes may differ, (2) the counting of secondary constrictions may not be predictive of the overall number of rDNA loci and satellite(d) chromosomes, (3) some 35S rDNA loci may not be transcriptionally active and are not, accordingly, NOR sites, and (4) an odd number of NOR sites does not point to positional hemizygosity and may be attained instead by epigenetic silencing.
Several methods are currently used to physically map the 35S rDNA units in plant cells. These include (1) the observation of chromosomes with conventional stains (e.g., acetic orcein, hematoxylin, Feulgen reagent, carmin acetic acid, Giemsa C-banding) or fluorescent dyes binding the DNA, e.g., DAPI (4’,6-diamidino-2-phenylindole) and CMA (Chromomycin A3) (Figures 3A,B), looking for the presence of secondary constrictions that are associated with the nucleolus (Maluszynska et al., 1998), (2) the observation in secondary constrictions of acidic, non-histone proteins that bind silver ions (argyrophilic) and are differentially stained by silver impregnation (Goodpasture and Bloom, 1975; Tucker et al., 2010; Figure 3C), (3) the distribution of epigenetic marks at the rDNA loci such as methylation and deacetylation patterns of histone H3 and the pattern of DNA methylation (5-methylcytosine sites) (Marques et al., 2011; Borowska-Zuchowska and Hasterok, 2017), and (4) the detection of tandemly repeated rDNA copies using in situ hybridization techniques (Heslop-Harrison and Schwarzacher, 2011; Jiang, 2019; Figure 3D).
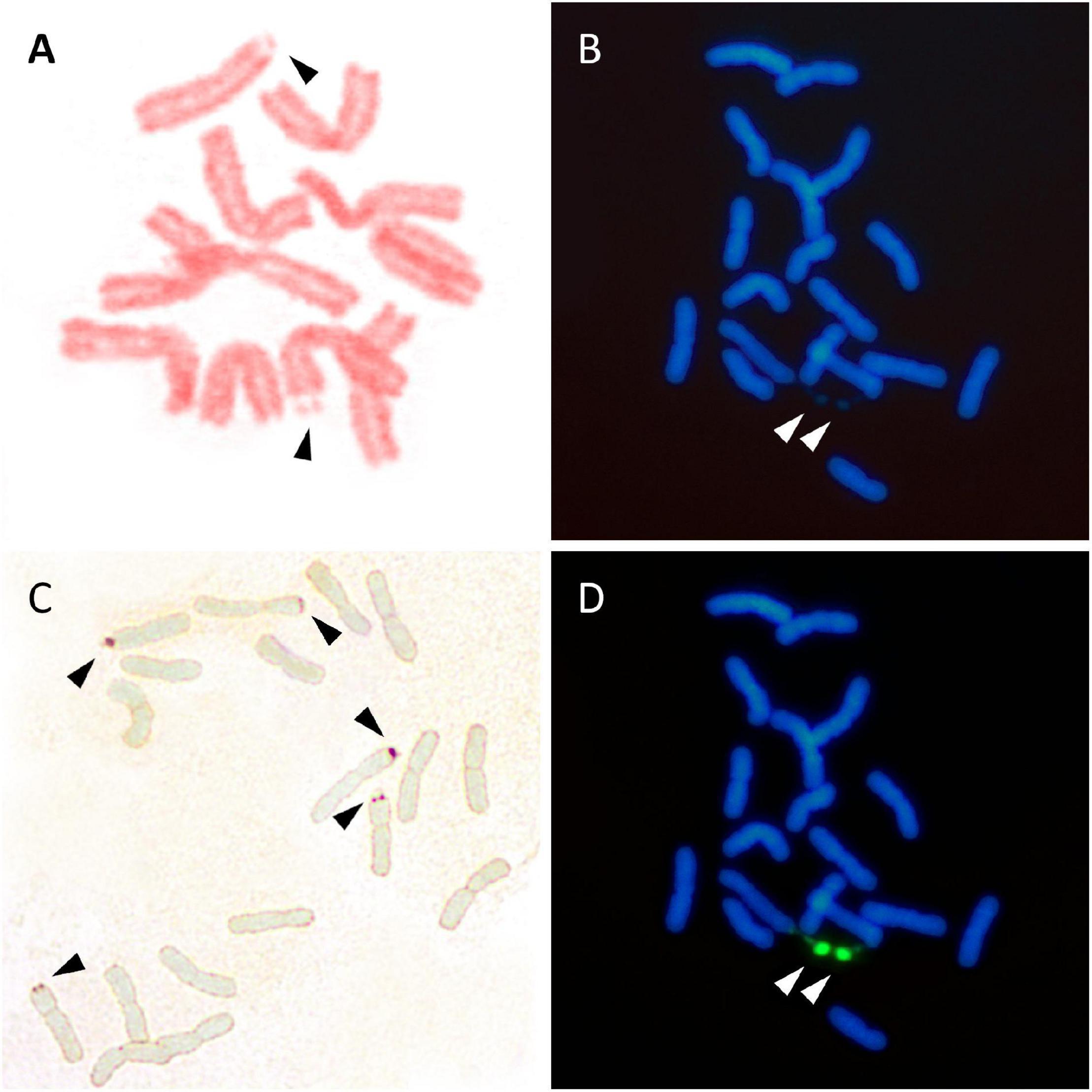
Figure 3. Visualization of 35S rDNA locus using conventional staining (A,B), silver impregnation (C), and FISH (D). (A) Lathyrus pisiformis. A single chromosome pair with secondary constrictions after Feulgen staining. (B) Achillea maritima. The presence of two secondary constrictions (arrows) are observed using DAPI staining. (C) Anacyclus homogamos. Five NORs are present after Ag-NOR staining (arrows). (D) Achillea maritima. A single 35SrDNA locus is located on the secondary constrictions after FISH (green signals).
The above procedures differ in the level of specificity, sensitivity, and reproducibility in the required quality of the target cells, as well as in the time and complexity of the experimental work. Most importantly, the techniques differ in the type of information retrieved. The first three approaches (secondary constrictions, silver staining, and epigenetic patterns) basically identify the 35S rDNA sites which are transcriptionally active in the cell, and which may be cytologically visible from interphase to metaphase of mitosis and prophase I of meiosis. In contrast, in situ hybridization techniques on nuclei and chromosomes using DNA labeled probes identify both the active and inactive 35S rDNA sites. None of the available techniques are free of experimental drawbacks and limitations and the best results are usually obtained with a combination of methods.
The visualization of NORs by observing secondary constrictions using conventional and fluorochrome staining is a fast but rather crude technique which is still in use. It has been reported that the decondensation of the rDNA chromatin occurs in different ways depending on the cell type and the analyzed species (Leitch, 2000). It has even been suggested that the activity of rDNA at different loci may vary over the course of the cell cycle, with their transcription being determined in a time and region-specific manner (Li et al., 2006; Chandrasekhara et al., 2016). In addition, the length of the decondensed chromatin is connected to the number of rDNA units being transcribed. Thus, the observation of secondary constrictions on mitotic chromosomes may be difficult to obtain unless other confirmatory, more powerful methods are used. The highest level of chromatin decondensation displayed in meiosis, in contrast to mitotic chromosomes, makes prophase I (generally, from pachytene to diakinesis) an excellent stage for a highly accurate physical mapping of chromosomal landmarks (De Jong et al., 1999; Sepsi et al., 2018), including NORs. However, the association of the homologous chromosomes bearing NORs (nucleolar bivalent or NOR-bivalent) with the nucleolus may appear disconnected and is not always easily observed (McClintock, 1934) in a similar way as has been reported for somatic chromosomes. These explanations alone or in combination may be related to the unsuccessful cytological detection of secondary constrictions in plant chromosomes and should considerably mitigate their use when unorthodox observations are obtained. For instance, data on the presence of secondary constrictions in the karyotypes of the endemic flora of the Balearic Islands using conventional karyological techniques is known for only 13% of the species (Rosselló and Castro, 2008).
Nucleolar organizing regions contain argyrophilic proteins that are selectively stained by silver methods allowing their identification throughout the nucleolar area (Ag-NORs). Unfortunately, the use of such a sensitive stain is difficult to standardize and many technical improvements have been reported for plant and animal organisms from time to time (e.g., Biliński and Bilińska, 1996; Trerè, 2000). However, non-optimal silver impregnation resulting in a bright background of nuclear DNA (Goodpasture and Bloom, 1975) or unspecific results (for instance, the staining of centromeric regions; Báez et al., 2020) may compromise the reliability of the technique. Since the presence of NORs in a decondensed state is one of the prerequisites for positive silver staining (Jiménez et al., 1988), the detection of Ag-NORs is not always reliable. Research has revealed the presence of active rDNA sites which are barely visible or not reflected by secondary constrictions on metaphase chromosomes, even after Ag-NOR staining, in species from lineages that are not closely related (Berg and Greilhuber, 1993). These observations may have various origins, like a small number of active ribosomal genes, a low transcriptional activity in some tissues or a different condensation level of chromatin in the NORs.
Fluorescence in situ hybridization (FISH) is by far the preferred karyological technique to assess 35S rDNA loci in plant chromosomes and very thorough compilations are available online (García et al., 2017). However, this approach alone does not differentiate the active and transcriptionally silent loci, and sequential methods (previous Ag-staining or the immunolocalization of epigenetic markers followed by FISH) are needed to obtain the most comprehensive results related to the number and functionality of 35S rDNA. FISH can reveal the major 35S rDNA loci of the genome, but it also has limitations or a lack of repeatability to detect minor loci characterized by a small number of rDNA units.
Assessments have reported that the number of secondary constrictions is gender dependent in several gymnosperm species, as Ephedra foliata (Mehra and Khitha, 1981), Cycas sp. pl. (Abraham and Mathew, 1962; Sangduen et al., 2007, 2009), Ginkgo biloba (Nakao et al., 2005), and Stangeria eriopus (Kokubugata et al., 2002). Observations in Ephedra and Cycas were based on conventional cytological techniques suggesting that faint satellites located at the end of the postulated sexual chromosomes (XY system), differentiate between X (with a satellite at each arm end) and Y chromosomes (a satellite at one end only, or none). Sangduen et al. (2009) concluded that the karyotype of male and female Cycas species could be clearly distinguished by the number of homomorphic and heteromorphic chromosome pairs possessing secondary constrictions. Although uncertainties about the accuracy of these cytological observations have been expressed by some authors dealing with sex chromosomes in gymnosperms (Ming et al., 2011), no available experimental work has been produced to dispute these findings, some of which were corroborated in somatic and generative tissues (Abraham and Mathew, 1962). Two heteromorphic chromosome pairs connected to rDNA sites were detected in Stangeria using a homologous probe (Kokubugata et al., 2002). The failure to associate this finding with sex chromosomes was due to the fact that the sex of the individual investigated was unknown.
Similar observations were reported in G. biloba chromosomes, where male individuals showed three chromosomes with secondary constrictions whereas four were present in female plants (Nakao et al., 2005). Additional studies by Lan et al. (2008) reported that in male individuals, the satellites of chromosome 1 (the biggest of the complement) were homomorphic, while in females they were heteromorphic, and one appeared to be bigger than the other. Both male and female plants showed the same number of rDNA sites (four) when performing FISH using a homologous rDNA probe (Nakao et al., 2005). Therefore, the differential epigenetic silencing of a whole rDNA site associated to the presence of a different rDNA copy number in homologous chromosomes may explain these cytological singularities in Ginkgo.
Accessory or supernumerary B chromosomes (Bs) are one of the most captivating topics of the evolution of the nuclear eukaryote genome. Bs are additional dispensable genetic components that contribute as part of the genome of a great diversity of organisms including plants (D’Ambrosio et al., 2017; Houben et al., 2019; Pokorná and Reifová, 2021). Several studies have revealed the lack of essential genes in their composition, except for the eventual presence of 35S and 5S rDNA families (Sýkorová et al., 2003; Jones et al., 2008; Houben et al., 2013). The activity of 35S rDNA sites located on Bs have been analyzed, and diversity in the transcriptional activity has been revealed in a similar way as is known for the chromosomes of the regular chromosomal complement (A chromosomes).
In Brachyscome dichromosomatica the 35S rDNA sites located at the large and micro Bs are silenced and are not associated to the nucleolus (Marschner et al., 2007). In contrast, in Plantago lagopus and Crepis capillaris the Bs are transcriptionally active (NOR) and contribute to the genesis of the nucleolus (Jones, 1995; Dhar et al., 2002). In Nierembergia aristata, Bs possess not only strong nucleolar activity, but also show nucleolar competition with the A chromosomes (Acosta and Moscone, 2011). This phenomenon could be analogous to the nucleolar dominance that occurs in interspecific hybrids (see below). Moreover, Secale cereale show the presence of B chromosomes without 35S rDNA sites. Interestingly, it has been suggested that Bs changes the rDNA organization pattern in interphase nuclei as detected by a drastic increase of rDNA condensed blocks inside the nucleolus (Delgado et al., 2004). Available evidence suggests that the rDNA alteration is caused by the presence of the B chromosomes themselves rather than by an obvious dosage effect (Delgado et al., 2004). The singular nature of B chromosomes exhibiting specific genomic features illustrates the need for caution when analyzing the pattern of nucleolar activity (Table 2).
Differential amphiplasty, also known as nucleolar dominance or reversible NOR silencing, is a conspicuous cytological and complex molecular phenomenon which was known to early plant cytogeneticists (Navashin, 1934). When the nuclear genomes of related species are merged by hybridization processes their NORs may differ in their competitive ability to transcribe the ribosomal genes and form the nucleolus. The net result of this is that a set of NORs from one of the parental species is epigenetically suppressed and fails to form secondary constrictions, thus leading to a decrease in the number of nucleoli. Accordingly, the NORs of hybrid and allopolyploids may not be added together and thus fail to reveal all the active rDNA sites inherited from the two parents (Tucker et al., 2010).
Interestingly, in individuals of hybrid origin, the presence of nucleolar dominance may be tissue-specific and the rDNA sites could be differentially expressed (Chen and Pikaard, 1997; Borowska-Zuchowska et al., 2021). These combined uniparental and biparental patterns of NOR silencing showing a contrasting number of NORs has detected in vegetative and reproductive tissues. In allotetraploid Brassica species (B. carinata, B. juncea, B. napus), rRNA genes silenced in leaves were found to be transcribed in all floral organs where biparental gene expression was maintained (Chen and Pikaard, 1997; Sochorová et al., 2017). However, Hasterok and Maluszynska (2000) reported that nucleolar dominance did not occur in root tip cells from these polyploid species. Biparental rDNA expression was found in roots, flowers and callus in the allotetraploid Tragopogon mirus (Dobešová et al., 2015). However, uniparental dominance was maintained in its leaves. These observations clearly indicate that silenced and derepressed rRNA genes may occur not only during developmental stages, but within vegetative and reproductive tissues. Since cytogenetic observations are preferentially limited to favorable cell types such as root tips and pollen mother cells, there may be contrasting reports on the number of active NOR sites (Table 2).
Detailed knowledge of the number of rDNA loci, their genomic location and the linkage of the 35S and 5S rDNA units has been assessed for a substantial number of species (García et al., 2012, 2014; Roa and Guerra, 2012; Vitales et al., 2017). In contrast, compilations of NOR activity are very few and, unfortunately, this knowledge has not been updated since the work of Lima-de-Faria (1976). In addition, most information on the dynamics of rDNA loci variation has not been thoroughly analyzed under explicit phylogenetic frameworks, and the data has been subject to speculative interpretations. In sharp contrast with what is known in selected polyploid species, the evolutionary patterns of rDNA loci number in predominantly diploid lineages are insufficiently understood and have received less attention (e.g., Datson and Murray, 2006; Totta et al., 2017). Moreover, most previous studies on rDNA loci changes lack explicit temporal frames, and as a result, their dynamics could not be assessed with certainty. The fact that surprisingly few studies have addressed intrapopulation, interpopulation and interspecific levels of rDNA variability in non-model wild plants, may question the assumptions of generalized evolutionary trends that are usually based on very few case studies.
Thus, the pervading perceptions that (1) the number, genomic location, and activity of 35S rDNA loci are constant within species, (2) diploid species are usually characterized by a single rDNA locus, (3) the ancestral number of 35S rDNA loci is usually one, (4) the presence of a single NOR locus is the derived state for seed plant lineages and (5) the increase in the number of rDNA loci is mostly linked to polyploidy, should all be checked on a case-by-case basis (Table 3).
The cytogenetic research conducted on the Mediterranean Anacyclus (Asteraceae), a diploid genus comprising nine species of weedy annuals and a few perennials, has provided relevant results illustrating how the above statements are not generally applicable in plants. Available karyological rDNA data was first obtained by Schweizer and Ehrendorfer (1976) and Ehrendorfer et al. (1977), who determined the number of active rDNA loci for all species based on a small number of accessions. Later, Rosato et al. (2017) determined the number and chromosomal position of 35S rDNA sites in 196 individuals from 47 populations in all Anacyclus species using FISH. The following conclusions could be firmly established from the results obtained by both research teams. First, the level of rDNA site-number variation detected within most Anacyclus species was outstanding and included both intra-specific and intra-population polymorphisms that encompassed a large part of the range of variation found in all angiosperms. Second, no clear association could be established between the phylogenetic position of the species and the number of rDNA sites. Third, the cytogenetic changes underlying the inferred rDNA dynamism were not related to polyploidy and were likely triggered by genomic rearrangements resulting from contemporary hybridization. Finally, the number of NORs in the genus was not associated to the phylogenetic ancestry of the species; the perennial clade showed two loci whereas the most derived annual species presented three loci.
Inferring polyploidy based on the number of NOR and rDNA sites may be misleading. An increase of NORs, and thus of nucleoli, is accomplished not only by genome duplication (Table 3), as had been earlier postulated (Gates, 1942; Fankhauser and Humphrey, 1943). Additional processes including structural rearrangements, ectopic recombination and rDNA transposition have been proposed as alternative mechanisms to explain NOR amplification within genomes (Pikaard, 2000b; Cabrero and Camacho, 2008, and references therein). Specifically, the intragenomic mobility of rRNA genes because of transposon activity, which can produce a translocation of rDNA copies to new genomic sites has been substantiated in seed plants, and it has been hypothesized that it is one of the major forces driving rDNA locus evolution in connection with the origin of new 35S rDNA sites (Dubcovsky and Dvorák, 1995; Raskina et al., 2004; Datson and Murray, 2006).
The basal chromosome number for each major lineage of early land plants (liverworts, mosses, and hornworts) is not known with certainty, and the topic has been greatly debated. The fact that two NOR loci were reported in some liverwort species (Riccardia pinguis) led Berrie (1958a,b) to hypothesize that species with n = 10 were polyploids derived from n = 5 hornwort ancestors. He further suggested that liverworts with gametophytic numbers of chromosomes of n = 8, 9 or 10 are basically polyploids that evolved from n = 10 ancestors through disploidy. The hypothesis of Berrie (1958a,b) was based on the misconception that the presence of two nucleolar chromosomes in the haploid complement of a plant is a marker of polyploidy. Recent work has shown that in bryophytes the number of 35S rDNA loci and copies are not correlated to ploidy level (Rosato et al., 2016a). Also, a lack of association between the number of 35S rDNA sites and polyploidy is known in Medicago (Fabaceae) where some tetraploid species (M. arborea, M. strasseri) show a single rDNA site, the same number usually present in diploid lineages (Rosato et al., 2008; Figure 4).
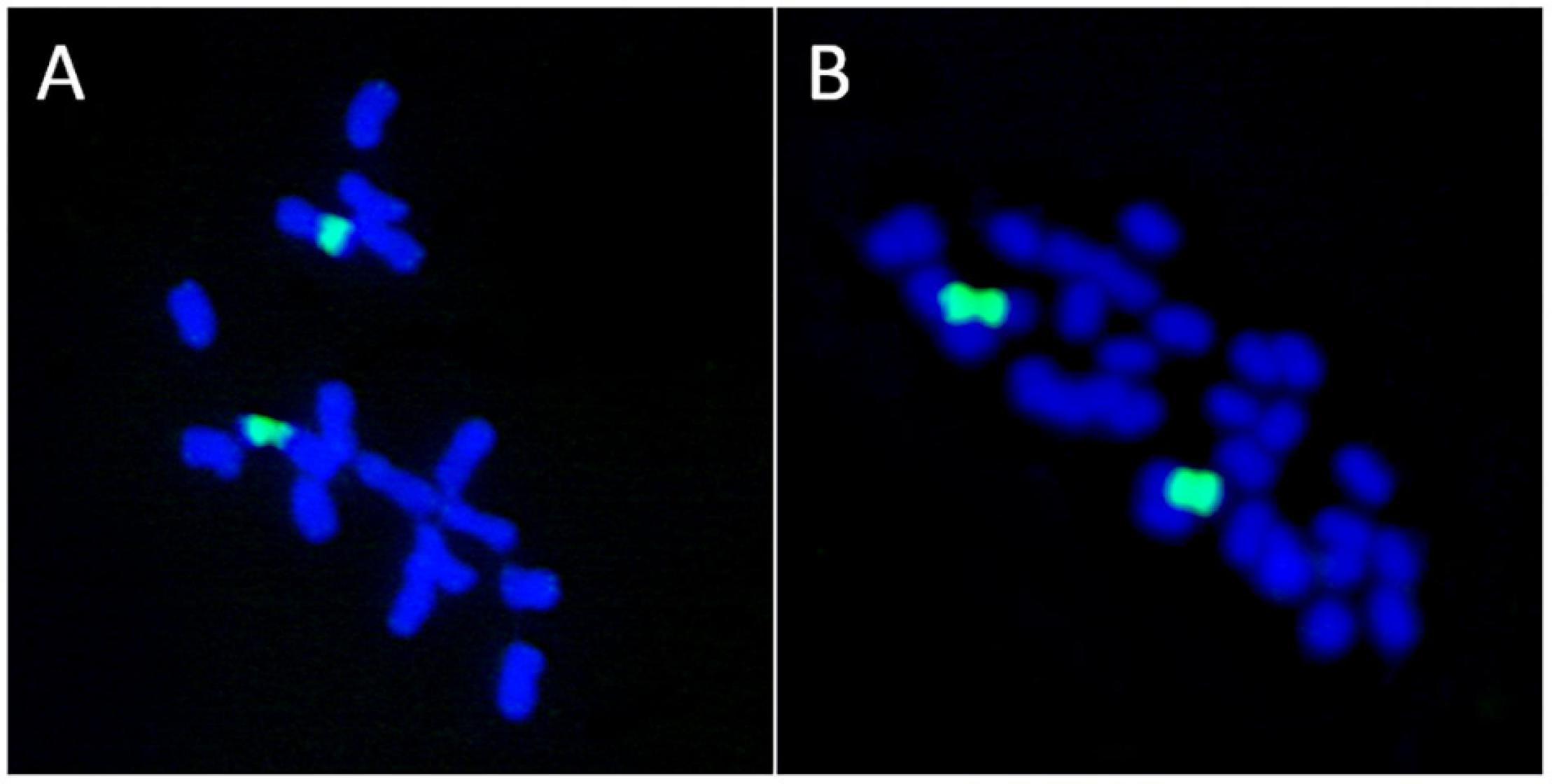
Figure 4. Lack of association between the number of ribosomal loci and polyploidy. (A) Medicago marina, 2n = 2x = 16. (B) M. arborea, 2n = 4x = 32. Both species possess a single 35S rDNA locus as revealed by FISH (green signals).
It is known that several independent events of entire genome duplications are characteristic of almost all fundamental lineages of land plants, and they are considered a major driving force in species diversification (Clark and Donoghue, 2018; Ren et al., 2018). Hence, attempts made to associate species with low chromosome numbers to diploid entities are confirmed to be incorrect as more and more events of paleoploidization are discovered (Clark and Donoghue, 2018). This suggests that overall assumptions connecting chromosome number and the ancestral or derived state to the number of 35S rDNA loci and NOR loci should be viewed with utmost caution. Despite these caveats, it has been estimated that 64.9–66% of the analyzed diploid species of seed plants show more than a single 35S rDNA locus as assessed by FISH (Roa and Guerra, 2012; García et al., 2017). These figures clearly disagree with the assumptions that the presence of a single rDNA in the genome supports the diploid status of a species.
One of the flowering plant genera that has received many efforts to infer the patterns and evolutionary significance of satellite chromosomes (referred to as SAT-chromosomes in the literature) is Taraxacum (dandelions), a complex genus where sexual (diploid) and, mostly, apomitic (polyploid) lineages occur. Mogie and Richards (1983) noticed the presence of two markedly divergent satellited chromosomes in the genus differing in the location and visibility of the secondary constrictions as observed after conventional Feulgen staining. The Taraxacum-type patterns were characterized by the presence of a conspicuous and intercalary secondary constriction, whereas the identified so-called conventional type satellite had a subterminal location and an extremely small distal euchromatic region Mogie and Richards (1983). The karyological inspection of 123 dandelion species by their SAT-chromosomes showed that the species belonging to the 10 sections hypothesized to be most primitive in the genus lack satellited chromosomes. Surprisingly, most analyzed species lacking SAT-chromosomes were reported to be sexual diploids, whereas a variable and usually unstable number of Taraxacum-type or conventional-type satellites was found in polyploids. Additional observations revealed a most inconsistent pattern of satellites and secondary constrictions in a single species, involving their number, size and chromosomal location, even within individuals (Richards, 1989). The variation in these cytological rDNA markers did not appear to be associated with the geographic origin of the populations sampled or the taxonomic adscription of the species, and accordingly, a constant number of satellites was rarely recorded for any species (Den Nijs et al., 1978; Krahulcová, 1993). It was suggested that such uncertainties regarding secondary constrictions might partially be explained by the experimental vagueness associated to the lack of strict uniformity of the pretreatment procedures (Den Nijs et al., 1978). Despite these concerns, however, there has been an increase in the number of reports on the number, size and morphology of satellited chromosomes for taxonomic and evolutionary purposes in Taraxacum (e.g., Gürdal and Özhatay, 2018; Watanabe et al., 2021). Again, the apparent lack of secondary constrictions continues to be reported in some species (T. scaturiginosum) (Gürdal and Özhatay, 2018). A recent study investigated the position and number of 35S rDNA loci in 38 Taraxacum species covering different reproduction modes, geographical regions and putative phylogenetic groups using FISH (Macháčková et al., 2021). Interestingly, these authors do not support the presence of the previously differentiated Taraxacum-type and conventional-type satellites by Mogie and Richards (1983). Most importantly, all analyzed species showed at least one secondary constriction, refuting the view that the lack of SAT-chromosomes is a reliable karyological marker in dandelions, as previously indicated (Mogie and Richards, 1983; Gürdal and Özhatay, 2018). However, the work of Macháčková et al. (2021) raises additional questions. These authors reported and provided images of the presence of conspicuous secondary constrictions in chromosomes lacking FISH signals for 35S rDNA. These findings are surprising and require additional verification to use nucleolar chromosomes as a meaningful karyological and evolutionary marker in Taraxacum.
The location and number of secondary constrictions have also been used as relevant cytogenetic features to investigate the karyotype evolution in the Lilieae tribe, as assessed by conventional staining (Gao et al., 2012). These authors characterized the Notholirion genus by the absence of nucleolar constrictions in the three analyzed species, in contrast with the recognized presence in all species of the related Cardiocrinum, Fritillaria, Lilium, and Nomocharis genera (Gao et al., 2012). Since Notholirion showed a basal position in the inferred phylogenetic tree based on nuclear ribosomal Internal Transcribed Spacer sequences, Gao et al. (2012) hypothesized that the lack of secondary constrictions was the ancestral state in Lilieae and that they emerged and evolved as the apparition of genera took place over time. Since the secondary constrictions on chromosomes represent the expression of rRNA genes which were transcribed during the preceding interphase (Tucker et al., 2010), species in the Lilieae tribe for which no nucleolar constrictions were recorded by Gao et al. (2012) should obviously present at least one chromosome pair showing active rDNA units. Clearly, the evolutionary interpretations drawn from the data appear unreliable, and new research using FISH is imperative to assess the pattern of NOR evolution in the group.
The nucleolus is a conspicuous domain delimiting the nuclear territory of transcriptionally active and mostly de-condensed ribosomal 35S rRNA genes, where the ribosomal units are assembled (Bersaglieri and Santoro, 2019). Nucleoli can be easily observed and analyzed in interphase nuclei (also in meiotic prophase I) by the same conventional silver nitrate staining and protocols that are applied to NOR loci (Lacadena et al., 1984; Xu and Earle, 1996). In recent years, several in situ markers of plant nucleoli have been implemented which involve methods for tagging specific nucleolar proteins with fluorescent tags, by raising antibodies or through nascent DNA and RNA using the so-called click iT chemistry (Dvořáčková and Fajkus, 2018). Nevertheless, silver staining is a relatively easy and fast approach that is still in use and advantageous over many more demanding, expensive and time-consuming protocols. In addition, silver staining allows sequential FISH to be done in the same slides, which reveals the number, location and transcriptional activity of the 35S rDNA loci (e.g., Castro et al., 2018). Observing the nucleoli in interphase nuclei has its own value and may complement the information gained by analyzing the 35S rDNA loci in metaphase. However, care should be exercised when nucleolus descriptors (usually the number) are used as evolutionary markers in plants (Table 4) since nucleoli are dynamic substructures regarding their activity, size, position, and number (Leitch, 2000) and their morphology can be modified when several types of stress occur (Hayashi and Matsunaga, 2019).
The number of nucleoli has been correlated with the number of NORs present in the chromosome complement and in the ploidy level (Venkatesh et al., 2019). However, earlier claims suggesting a close connection (i.e., linearity) between the number of nucleoli and polyploidy (Gates, 1942; Fankhauser and Humphrey, 1943) are certainly not universally used. Thus, ploidy is not related to the nucleolar number but to the nucleolar size in the fruit cells of Solanum lycopersicum (Bourdon et al., 2012).
Counting nucleoli is one of the techniques used to estimate the ploidy level of individuals without having dividing cells, which may be useful when using tissues with a low mitotic index (Ochatt and Seguí-Simarro, 2021). Since nucleolar fusion frequently occurs during the cell cycle (mononucleation), only the highest number of nucleoli detected should be taken as evidence of the number of active rDNA loci (Ochatt and Seguí-Simarro, 2021; Figures 5A–C). A confident determination of the number of nucleoli should be estimated in a large sample size on this basis. Nucleoli have been detected in fossilized stems of a royal fern (Osmundaceae) dating back 180 million years (Bomfleur et al., 2014) and in herbarium specimens (Laane and Höiland, 1986), paving the way to assess the estimation of ploidy levels in extinct species and populations of well-preserved museum specimens.
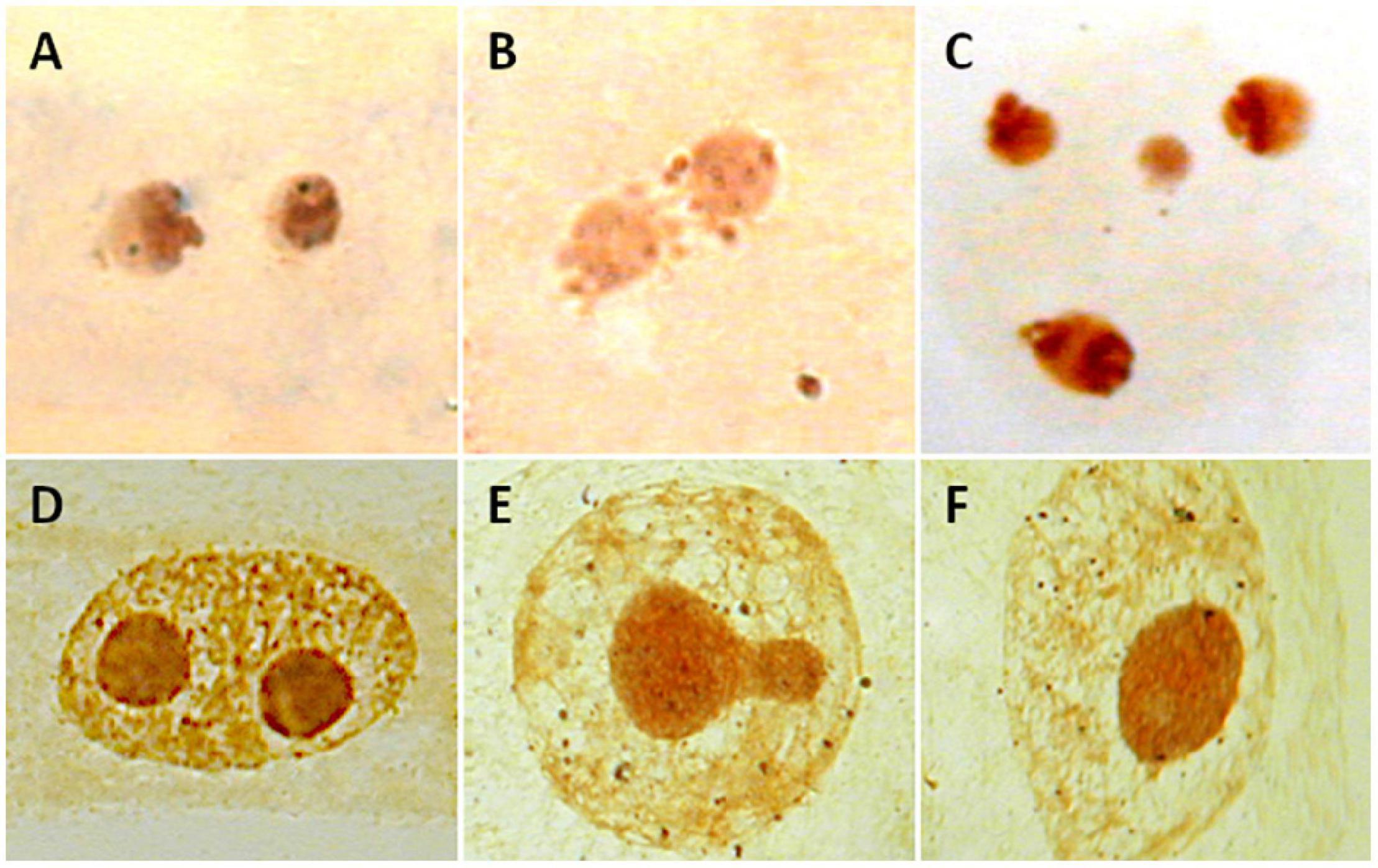
Figure 5. Nucleoli in interphase nuclei of Medicago species (A–C) and Cistus heterophyllus (D–F) after Ag-NOR staining. (A) Two nucleoli are observed in diploid M. marina (2n = 2x = 16). (B) A single nucleoli is observed in M. arborea (2n = 4x = 32). (C) Four nucleoli are shown in hexaploid M. citrina (2n = 6x = 48). (D–F) Process of mononucleation (nucleoli fusion) in C. heterophyllus.
The assumption that the number of nucleoli is constant within individuals and species is risky and should be validated on a case-by-case basis. The variability of NOR activity, expressed as the number and size of silver-stained active sites, has been detected between cells of single individuals, and also between individuals of the same population in cock’s-foot (Besendorfer et al., 2002). In addition, atypical nucleolar behaviors across the cell cycle, including the presence of numerous small silver-positive bodies in nuclei and cytoplasm, have been reported in some species and may also compromise the interpretation of the results (Dagne and Heneen, 1992). Moreover, the migration of nuclei, including nucleoli, between plant cells (cytomixis) is a cellular phenomenon frequently observable in the male meiosis of higher plants, but the causes and consequences of cytomixis are still not entirely understood (Mursalimov and Deineko, 2018). This intercellular migration could lead to the detection of a different number of nucleoli in cells associated with the production of pollen. Finally, in hybrid and allopolyploid species, the number of nucleoli may vary between tissues (usually between vegetative and reproductive tissues and at several developmental stages) due to nucleolar dominance, the uniparental expression of the NORs (Chen and Pikaard, 1997; Hasterok and Maluszynska, 2000; Dobešová et al., 2015) as indicated above.
Additional inconsistencies related to a lack of full correspondence between the number of NORs present in the chromosome complement and the number of nucleoli have been noted. For several species, there have been reports indicating that the number of Ag-NORs in metaphase may disagree with the maximum number of nucleoli recorded in interphase nuclei (e.g., Scaldaferro et al., 2016). In Solanum lycopersicum meiosis, pachytene chromosomes show five 35S rDNA loci as revealed by FISH (Xu and Earle, 1996). However, only the major rDNA locus is associated to the nucleolus by Ag-NOR staining, whereas the remaining four minor loci, which are also active after silver impregnation, are not. Thus, in this species there is a correspondence between the number of Ag-NOR pachytene chromosomes and the maximum number of nucleoli observed (five), but not all NOR-bivalents could be associated to a nucleolus (Xu and Earle, 1996).
Several hypotheses attempt to explain this fact, including nucleolar association, the merging of the nucleolus during the interphase (Lacadena et al., 1984), or the occurrence of interchromosomal nucleolar dominance, where NORs from different pairs of chromosomes compete to make up the nucleoli (Tapia-Pastrana, 2020). The presence of cryptic NORs, chromosome regions apparently lacking rDNA loci, as revealed by FISH and silver staining, but which give rise to small nucleoli in interphase has been reported in plant and animal species (Sato et al., 1981; Cabrero and Camacho, 2008).
The biological reasons underlying the unsteadiness in the number of chromosomes with NOR sites and the number of nucleoli may be varied and are not fully understood. However, past and ongoing interspecific hybridization has been suggested as one of the most outstanding causes involved in the generation of cytological abnormalities that modify the regulatory system of the cell and contribute to decreasing the connection between the number of NORs and nucleoli (Levin, 1973; Tapia-Pastrana, 2020). For instance, in Phlox hybrida a high and significant correlation between the population hybridity and incidence of accessory nucleoli was detected (Levin, 1973). Supernumerary nucleoli were observed in Crotalaria agatiflora and their presence was attributed to the hybrid origin of this species (Verma and Raina, 1981). However, hybridization was rejected as the driving factor involved in the presence of accessory nucleoli in the diploid species Trigonella foenum-graecum although no alternative hypothesis was substantiated (Lakshmi and Raghavaiah, 1984).
Lack of association between number of nucleoli and polyploidy was detected in Medicago species (Rosato and Rosselló, 2009). A single nucleolus was present both in diploid (M. marina; 2n = 16) and tetraploid species (M. arborea, M. strasseri; 2n = 32). In addition, hexaploid M. citrina (2n = 48), for which the presence of three nucleoli should be theoretically expected, showed only two nucleoli (Rosato and Rosselló, 2009; Figures 5D–F). Inconsistencies also apply to non-flowering plants like bryophytes. Populations showing one or two nucleoli were registered in the gametophytic haploid (n = 9) Pellia endiviifolia noted (Newton, 1988). In contrast, a single nucleolus was present in diploid (n = 18) P. borealis (Newton, 1986).
JR: conceptualization, writing—original draft preparation, and funding acquisition. AM, MR, and JR: writing–review and editing. MR: supervision. All authors have equally contributed, read and agreed to the published version of the manuscript.
This study was supported by a grant from the Spanish Ministry of Economy and Competitivity through the project CGL2017-88500-P.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abd El-Twab, M. H., and Kondo, K. (2010). Characterization of chromosome complement in Tridactylina kirilowii (Turcz. ex DC.) Schultz-Bip. by aceto-orcein, CMA, DAPI and FISH. Chromosom. Bot. 5, 15–21. doi: 10.3199/iscb.5.15
Abraham, A., and Mathew, P. (1962). Cytological studies in the Cycads: sex chromosomes in Cycas. Ann. Bot. 26, 261–266. doi: 10.1093/oxfordjournals.aob.a083792
Acosta, M. C., and Moscone, E. A. (2011). B Chromosomes in Nierembergia aristata (Solanaceae): nucleolar activity and competition with the A chromosomes. Cytogenet. Genome Res. 132, 105–112. doi: 10.1159/000320705
Álvarez, I., and Wendel, J. F. (2003). Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29, 417–434. doi: 10.1016/S1055-7903(03)00208-2
Báez, M., Souza, G., and Guerra, M. (2020). Does the chromosomal position of 35S rDNA sites influence their transcription? A survey on Nothoscordum species (Amaryllidaceae). Genet. Mol. Biol. 43, e20180194. doi: 10.1590/1678-4685-GMB-2018-0194
Bairu, M. W., Aremu, A. O., and Van Staden, J. (2011). Somaclonal variation in plants: causes and detection methods. Plant Growth Regul. 63, 147–173. doi: 10.1007/s10725-010-9554-x
Battaglia, E. (1955). Chromosome morphology and terminology. Caryologia 8, 179–187. doi: 10.1080/00087114.1955.10797556
Battaglia, E. (1999). The chromosome satellite [Navashin’s sputnik or satelles]: a terminological comment. Acta Biol. Crac. Ser. Bot. 41, 15–18.
Berg, C., and Greilhuber, J. (1993). Cold-sensitive chromosome regions and heterochromatin in Cestrum aurantiacum (Solanaceae). Plant Syst. Evol. 185, 259–273. doi: 10.1007/BF00937662
Berrie, G. K. (1958a). The nucleolar chromosome in hepatics. I. Trans. Br. Bryol. Soc. 3, 422–426. doi: 10.1179/006813858804829451
Berrie, G. K. (1958b). The nucleolar chromosome in hepatics, II. A phylogenetic speculation. Trans. Br. Bryol. Soc. 3, 427–429. doi: 10.1179/006813858804829334
Bersaglieri, C., and Santoro, R. (2019). Genome organization in and around the nucleolus. Cells 8:579. doi: 10.3390/cells8060579
Besendorfer, V., Samardzija, M., Zoldos, V., Solic, M. E., and Papes, D. (2002). Chromosomal organization of ribosomal genes and NOR-associated heterochromatin, and NOR activity in some populations of Allium commutatum Guss. Bot. J. Linn. Soc. 139, 99–108. doi: 10.1046/j.1095-8339.2002.00047.x
Biliński, S. M., and Bilińska, B. (1996). A new version of the Ag-NOR technique. A combination with DAPI staining. Histochem. J. 28, 651–656. doi: 10.1007/BF02331386
Bisht, M. S., Kesavacharyulu, K., and Raina, S. N. (1998). Nucleolar chromosome variation and evolution in the genus Vicia. Caryologia 51, 133–147. doi: 10.1080/00087114.1998.10589128
Bomfleur, B., McLoughlin, S., and Vajda, V. (2014). Fossilized nuclei and chromosomes reveal 180 million years of genomic stasis in royal ferns. Science 343, 1376–1377. doi: 10.1126/science.1249884
Borowska-Zuchowska, N., and Hasterok, R. (2017). Epigenetics of the preferential silencing of Brachypodium stacei-originated 35S rDNA loci in the allotetraploid grass Brachypodium hybridum. Sci. Rep. 7:5260. doi: 10.1038/s41598-017-05413-x
Borowska-Zuchowska, N., Robaszkiewicz, E., Mykhailyk, S., Wartini, J., Piński, A., Kovarik, A., et al. (2021). To be or not to be Expressed: the first evidence of a nucleolar dominance tissue-specificity in Brachypodium hybridum. Front. Plant Sci. 12:768347. doi: 10.3389/fpls.2021.768347
Bourdon, M., Pirrello, J., Cheniclet, C., Coriton, O., Bourge, M., Brown, S., et al. (2012). Evidence for karyoplasmic homeostasis during endoreduplication and a ploidy-dependent increase in gene transcription during tomato fruit growth. Development 139, 3817–3826. doi: 10.1242/dev.084053
Brasileiro-Vidal, A. C., Cuadrado, A., Brammer, S., Zanatta, A. C., Prestes, A., Moraes-Fernandes, M. I. B., et al. (2003). Chromosome characterization in Thinopyrum ponticum (Triticeae, Poaceae) using in situ hybridization with different DNA sequences. Genet. Mol. Biol. 26, 505–510. doi: 10.1590/S1415-47572003000400014
Cabrero, J., and Camacho, J. P. M. (2008). Location and expression of ribosomal RNA genes in grasshoppers: abundance of silent and cryptic loci. Chromosome Res. 16, 595–607. doi: 10.1007/s10577-008-1214-x
Castro, C., Carvalho, A., Pavia, I., Leal, F., Moutinho-Pereira, J., and Lima-Brito, J. (2018). Nucleolar activity and physical location of ribosomal DNA loci in Vitis vinifera L. by silver staining and sequential FISH. Sci. Hortic. 232, 57–62. doi: 10.1016/j.scienta.2017.12.064
CBOL Plant Working Group (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. U. S. A. 106, 12794–12797. doi: 10.1073/PNAS.0905845106
Chandrasekhara, C., Mohannath, G., Blevins, T., Pontvianne, F., and Pikaard, C. S. (2016). Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes Dev. 30, 177–190. doi: 10.1101/gad.273755.115
Chase, M. W., Cowan, R. S., Hollingsworth, P. M., Van Den Berg, C., Madriñán, S., Petersen, G., et al. (2007). A proposal for a standardised protocol to barcode all land plants. Taxon 56, 295–299. doi: 10.1002/tax.562004
Chen, C. C., Chen, C. M., Hsu, F. C., Wang, C. J., Yang, J. T., and Kao, Y. Y. (2000). The pachytene chromosomes of maize as revealed by fluorescence in situ hybridization with repetitive DNA sequences. Theor. Appl. Genet. 101, 30–36. doi: 10.1007/s001220051445
Chen, Z. J., and Pikaard, C. S. (1997). Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc. Natl. Acad. Sci. U. S. A. 94, 3442–3447. doi: 10.1073/pnas.94.7.3442
Chiavarino, A. M., Rosato, M., Manzanero, S., Jiménez, G., González-Sánchez, M., and Puertas, M. J. (2000). Chromosome nondisjunction and instabilities in tapetal cells are affected by B chromosomes in maize. Genetics 155, 889–897. doi: 10.1093/genetics/155.2.889
Clark, J. W., and Donoghue, P. C. (2018). Whole-genome duplication and plant macroevolution. Trends Plant Sci. 23, 933–945. doi: 10.1016/j.tplants.2018.07.006
Cuñado, N., De la Herrán, R., Santos, J. L., Ruiz Rejón, C., Garrido-Ramos, M. A., and Ruiz Rejón, M. (2000). The evolution of the ribosomal loci in the subgenus Leopoldia of the genus Muscari (Hyacinthaceae). Plant Syst. Evol. 221, 245–252. doi: 10.1007/BF01089296
Dagne, K., and Heneen, W. K. (1992). The karyotype and nucleoli of Guizotia abyssinica (Compositae). Hereditas 117, 73–83. doi: 10.1111/j.1601-5223.1992.tb00010.x
D’Ambrosio, U., Alonso-Lifante, M. P., Barros, K., Kovaøík, A., de Xaxars, G. M., and García, S. (2017). B-chrom: a database on B-chromosomes of plants, animals and fungi. New Phytol. 216, 635–642. doi: 10.1111/nph.14723
Darlington, C. D. (1926). Chromosome studies in the Scilleae. J. Genet. 16, 237–251. doi: 10.1007/BF02983000
Datson, P. M., and Murray, B. G. (2006). Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosome Res. 14, 845–857. doi: 10.1007/s10577-006-1092-z
De Jong, J. H., Fransz, P., and Zabel, P. (1999). High resolution FISH in plants-techniques and applications. Trends Plant Sci. 4, 258–263. doi: 10.1016/S1360-1385(99)01436-3
Delgado, M., Caperta, A., Ribeiro, T., Viegas, W., Jones, R. N., and Morais-Cecilio, L. (2004). Different numbers of rye B chromosomes induce identical compaction changes in distinct A chromosome domains. Cytogenet. Genome Res. 106, 320–324. doi: 10.1159/000079306
Den Nijs, J. C. M., Sterk, A. A., and Van der Hammen, H. (1978). Cytological and ecological notes on the Taraxacum sections Erythrosperma and Obliqua of the coastal area of the Netherlands. Acta Bot. Neerl. 27, 287–305. doi: 10.1111/j.1438-8677.1978.tb00303.x
Deng, C. L., Qin, R. Y., Wang, N. N., Cao, Y., Gao, J., Gao, W. J., et al. (2012). Karyotype of asparagus by physical mapping of 45S and 5S rDNA by FISH. J. Genet. 91, 209–212. doi: 10.1007/s12041-012-0159-1
Dhar, M. K., Friebe, B., Koul, A. K., and Gill, B. S. (2002). Origin of an apparent B chromosome by mutation, chromosome fragmentation and specific DNA sequence amplification. Chromosoma 111, 332–340. doi: 10.1007/s00412-002-0214-4
Díaz Lifante, Z. (1996). A karyological study of Asphodelus L. (Asphodelaceae) from the Western Mediterranean. Bot. J. Linn. Soc. 121, 285–344. doi: 10.1111/j.1095-8339.1996.tb00760.x
Dobešová, E., Malinská, H., Matyášek, R., Leitch, A. R., Soltis, D. E., Soltis, P. S., et al. (2015). Silenced rRNA genes are activated and substitute for partially eliminated active homeologs in the recently formed allotetraploid, Tragopogon mirus (Asteraceae). Heredity 114, 356–365. doi: 10.1038/hdy.2014.111
Doveri, S., and Lee, D. (2007). Development of sensitive crop-specific polymerase chain reaction assays using 5S DNA: applications in food traceability. J. Agric. Food Chem. 55, 4640–4644. doi: 10.1021/jf063259v
Dubcovsky, J., and Dvorák, J. (1995). Ribosomal RNA multigene loci: nomads of the Triticeae genomes. Genetics 140, 1367–1377. doi: 10.1093/genetics/140.4.1367
Dvořáčková, M., and Fajkus, J. (2018). Visualization of the nucleolus using ethynyl uridine. Front. Plant Sci. 9:177. doi: 10.3389/fpls.2018.00177
Ehrendorfer, F., Schweizer, D., Greger, H., and Humphries, C. (1977). Chromosome banding and synthetic systematics in Anacyclus (Asteraceae Anthemideae). Taxon 26, 387–394. doi: 10.2307/1220037
Fankhauser, G., and Humphrey, R. R. (1943). The relation between number of nucleoli and number of chromosome sets in animal cells. Proc. Natl. Acad. Sci. U. S. A. 29, 344–350. doi: 10.2307/87559
Fehrer, J., Slavíková, R., Paštová, L., Josefiová, J., Mráz, P., Chrtek, J., et al. (2021). Molecular evolution and organization of ribosomal DNA in the hawkweed tribe Hieraciinae (Cichorieae, Asteraceae). Front. Plant Sci. 12:395. doi: 10.3389/fpls.2021.647375
Feliner, G. N., and Rosselló, J. A. (2012). “Concerted evolution of multigene families and homoeologous recombination,” in Plant Genome Diversity Volume 1, ed. J. F. Wendel (Vienna: Springer), 171–193. doi: 10.1007/978-3-7091-1130-7_12
Fujisawa, M., Nakayama, S., Nishio, T., Fujishita, M., Hayashi, K., Ishizaki, K., et al. (2003). Evolution of ribosomal DNA unit on the X chromosome independent of autosomal units in the liverwort Marchantia polymorpha. Chromosome Res. 11, 695–703. doi: 10.1023/A:1025941206391
Galián, J. A., Rosato, M., and Rosselló, J. A. (2012). Early evolutionary colocalization of the nuclear ribosomal 5S and 45S gene families in seed plants: evidence from the living fossil gymnosperm Ginkgo biloba. Heredity 108, 640–646. doi: 10.1038/hdy.2012.2
Gao, T., Yao, H., Song, J., Liu, C., Zhu, Y., Ma, X., et al. (2010). Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J. Ethnopharmacol. 130, 116–121. doi: 10.1016/j.jep.2010.04.026
Gao, Y. D., Zhou, S. D., He, X. J., and Wan, J. (2012). Chromosome diversity and evolution in tribe Lilieae (Liliaceae) with emphasis on Chinese species. J. Plant Res. 125, 55–69. doi: 10.1007/s10265-011-0422-1
García, S., Gálvez, F., Gras, A., Kovaøík, A., and Garnatje, T. (2014). Plant rDNA database: update and new features. Database 2014:bau063. doi: 10.1093/database/bau063
García, S., Garnatje, T., and Kovaøík, A. (2012). Plant rDNA database: ribosomal DNA loci information goes online. Chromosoma 121, 389–394. doi: 10.1007/s00412-012-0368-7
García, S., Kovaøík, A., Leitch, A. R., and Garnatje, T. (2017). Cytogenetic features of rRNA genes across land plants: analysis of the Plant rDNA database. Plant J. 89, 1020–1030. doi: 10.1111/tpj.13442
Gates, R. R. (1942). Nucleoli and related nuclear structures. Bot. Rev. 8, 337–409. doi: 10.1007/BF02882158
Gemeinholzer, B., Oberprieler, C., and Bachmann, K. (2006). Using GenBank data for plant identification: possibilities and limitations using the ITS 1 of Asteraceae species belonging to the tribes Lactuceae and Anthemideae. Taxon 55, 173–187. doi: 10.2307/25065539
González-Melendi, P., Ramírez, C., Testillano, P. S., Kumlehn, J., and Risueño, M. C. (2005). Three dimensional confocal and electron microscopy imaging define the dynamics and mechanisms of diploidisation at early stages of barley microspore-derived embryogenesis. Planta 222, 47–57. doi: 10.1007/s00425-005-1515-7
Goodpasture, C., and Bloom, S. E. (1975). Visualization of nucleolar organizer regions in mammalian chromosomes using silver staining. Chromosoma 53, 37–50. doi: 10.1007/bf00329389
Guerra, M. (2012). Cytotaxonomy: the end of childhood. Plant Biosyst. 146, 703–710. doi: 10.1080/11263504.2012.717973
Guimond, A., and Moss, T. (1999). A ribosomal orphon sequence from Xenopus laevis flanked by novel low copy number repetitive elements. Biol. Chem. 380, 167–174. doi: 10.1515/bc.1999.025
Gürdal, B., and Özhatay, N. (2018). Karyological study on 12 species of the genus Taraxacum (Asteraceae) grown in Turkey. Flora 28, 429–439. doi: 10.7320/FlMedit28.429
Hasterok, R., and Maluszynska, J. (2000). Nucleolar dominance does not occur in root tip cells of allotetraploid Brassica species. Genome 43, 574–579. doi: 10.1139/g00-005
Hayashi, K., and Matsunaga, S. (2019). Heat and chilling stress induce nucleolus morphological changes. J. Plant Res. 132, 395–403. doi: 10.1007/s10265-019-01096-9
Heitz, E. (1931). Die ursache der gesetzmässigen zahl, lage, form und grösse pflanzlicher nukleolen. Planta 12, 775–844. doi: 10.1007/BF01912443
Heslop-Harrison, J. S., and Schwarzacher, T. (2011). Organisation of the plant genome in chromosomes. Plant J. 66, 18–33. doi: 10.1111/j.1365-313X.2011.04544.x
Hidalgo, O., Vitales, D., Vallès, J., Garnatje, T., Siljak-Yakovlev, S., Leitch, I. J., et al. (2017). Cytogenetic insights into an oceanic island radiation: the dramatic evolution of pre-existing traits in Cheirolophus (Asteraceae: Cardueae: Centaureinae). Taxon 66, 146–157. doi: 10.12705/661.8
Houben, A., Banaei-Moghaddam, A. M., and Klemme, S. (2013). “Biology and evolution of B chromosomes,” in Plant Genome Diversity Volume 2, ed. I. J. Leitch (Vienna: Springer), 149–165. doi: 10.1007/978-3-7091-1160-4
Houben, A., Jones, N., Martins, C., and Trifonov, V. (2019). Evolution, composition and regulation of supernumerary B chromosomes. Genes 10:161. doi: 10.3390/genes10020161
Idziak, D., and Hasterok, R. (2008). Cytogenetic evidence of nucleolar dominance in allotetraploid species of Brachypodium. Genome 51, 387–391. doi: 10.1139/G08-017
Jiang, J. (2019). Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosome Res. 27, 153–165. doi: 10.1007/s00425-00018-03033-00424
Jiménez, R., Burgos, M., and de la Guardia, R. (1988). A study of the Ag-staining significance in mitotic NOR’s. Heredity 60, 125–127. doi: 10.1038/hdy.1988.18
Jones, R. N. (1995). B chromosomes in plants. New Phytol. 131, 411–434. doi: 10.1111/j.1469-8137.1995.tb03079.x
Jones, R. N., Gonzalez-Sanchez, M., Gonzalez-García, M., Vega, J. M., and Puertas, M. J. (2008). Chromosomes with a life of their own. Cytogenet. Genome Res. 120, 265–280. doi: 10.1159/000121076
Khuong, N. T., and Schubert, I. (1985). Silver staining of nucleolus organizing regions in Zea mays. Caryologia 38, 331–334. doi: 10.1080/00087114.1985.10797757
Kokubugata, G., Hill, K. D., and Kondo, K. (2002). Ribosomal DNA distribution in somatic chromosomes of Stangeria eriopus (Stangeriaceae, Cycadales) and molecular-cytotaxonomic relationships to some other cycad genera. Brittonia 54, 1–5.
Krahulcová, A. (1993). New chromosome numbers in Taraxacum with reference to SAT-chromosomes. Geobot. Phytotax. 28, 289–294. doi: 10.1007/BF02853516
Kress, W. J., Wurdack, K. J., Zimmer, E. A., Weigt, L. A., and Janzen, D. H. (2005). Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. U. S. A. 102, 8369–8374. doi: 10.1073/pnas.0503123102
Kumar, P., and Singhal, V. K. (2016). Nucleoli migration coupled with cytomixis. Biologia 71, 651–659. doi: 10.1515/biolog-2016-0076
Laane, M. M., and Höiland, K. (1986). Chromosome number and meiosis in herbarium specimens from the extinct Scandinavian population of Crepis multicaulis. Hereditas 105, 187–192. doi: 10.1111/j.1601-5223.1986.tb00660.x
Lacadena, J. R., Cermeno, M., Orellana, J., and Santos, J. L. (1984). Evidence for wheat-rye nucleolar competition (amphiplasty) in triticale by silver-staining procedure. Theor. Appl. Genet. 67, 207–213. doi: 10.1007/BF00317037
Lakshmi, N., and Raghavaiah, P. V. (1984). Accessory nucleoli in Trigonella foenum-graecum L. Cytologia 49, 401–405. doi: 10.1508/cytologia.49.401
Lan, T., Chen, R. Y., Li, X. L., Dong, F. P., Qi, Y. C., and Song, W. Q. (2008). Microdissection and painting of the W chromosome in Ginkgo biloba showed different labelling patterns. Bot. Stud. 49, 33–37.
Leitch, A. R. (2000). Higher levels of organization in the interphase nucleus of cycling and differentiated cells. Microbiol. Mol. Biol. Rev. 64, 138–152. doi: 10.1128/MMBR.64.1.138-152.2000
Leitch, A. R., and Heslop-Harrison, J. S. (1993). “Ribosomal RNA gene expression and localization in cereals,” in Chromosomes today Volume 11, eds A. T. Sumner and A. C. Chandley (Dordrecht: Springer), 91–100. doi: 10.1007/978-94-011-1510-0
Levin, D. A. (1973). Accessory nucleoli in microsporocytes of hybrid Phlox. Chromosoma 41, 413–420. doi: 10.1007/BF00396499
Li, Z. Y., Fu, M. L., Hu, F. F., Huang, S. F., and Song, Y. C. (2006). Visualization of the ribosomal DNA (45S rDNA) of indica rice with FISH on some phases of cell cycle and extended DNA fibers. Biocell 30, 27–32. doi: 10.32604/biocell.2006.30.027
Lima-de-Faria, A. (1976). The chromosome field: I. Prediction of the location of ribosomal cistrons. Hereditas 83, 1–22. doi: 10.1111/j.1601-5223.1976.tb01565.x
Macas, J., Navrátilová, A., and Mészáros, T. (2003). Sequence subfamilies of satellite repeats related to rDNA intergenic spacer are differentially amplified on Vicia sativa chromosomes. Chromosoma 112, 152–158. doi: 10.1007/s00412-003-0255-3
Macháčková, P., Majeský, L’, Hroneš, M., Bílková, L., Hřibová, E., and Vašut, R. J. (2021). New insights into ribosomal DNA variation in apomictic and sexual Taraxacum (Asteraceae). Bot. J. Linn. 20, 1–26. doi: 10.1093/botlinnean/boab094
Maggini, F., Cremonini, R., Zolfino, C., Tucci, G. F., D’ovidio, R., Delre, V., et al. (1991). Structure and chromosomal localization of DNA sequences related to ribosomal subrepeats in Vicia faba. Chromosoma 100, 229–234. doi: 10.1007/BF00344156
Maluszynska, J., Hasterok, R., and Weiss, H. (1998). “rRNA genes-their distribution and activity in plants,” in Prace Naukowe Uniwersytetu Śląskiego w Katowicach, ed. J. Małuszynska (Katowice: University of Silesia), 75–95.
Marques, A., Fuchs, J., Ma, L., Heckmann, S., Guerra, M., and Houben, A. (2011). Characterization of eu-and heterochromatin of Citrus with a focus on the condensation behavior of 45S rDNA chromatin. Cytogenet. Genome Res. 134, 72–82. doi: 10.1159/000323971
Marschner, S., Meister, A., Blattner, F. R., and Houben, A. (2007). Evolution and function of B chromosome 45S rDNA sequences in Brachycome dichromosomatica. Genome 50, 638–644. doi: 10.1139/G07-048
McClintock, B. (1934). The relationship of a particular chromosomal element to the development of the nucleoli in Zea mays. Zeit. Zellforsch. Mik. Anat. 21, 294–328. doi: 10.1007/BF00374060
Mehra, P. N., and Khitha, S. C. (1981). Karyotype and mechanism of sex determination in Ephedra foliata Boiss—a dioecious gymnosperm. Cytologia 46, 173–181. doi: 10.1508/cytologia.46.173
Milioto, V., Vlah, S., Mazzoleni, S., Rovatsos, M., and Dumas, F. (2019). Chromosomal localization of 18S-28S rDNA and (TTAGGG)n sequences in two South African dormice of the genus Graphiurus (Rodentia: Gliridae). Cytogenet. Genome Res. 158, 145–151. doi: 10.1159/000500985
Ming, R., Bendahmane, A., and Renner, S. S. (2011). Sex chromosomes in land plants. Annu. Rev. Plant Biol. 62, 485–514. doi: 10.1146/annurev-arplant-042110-103914
Mishima, M., Ohmido, N., Fukui, K., and Yahara, T. (2002). Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae). Chromosoma 110, 550–558. doi: 10.1007/s00412-001-0175-z
Mogie, M., and Richards, A. J. (1983). Satellited chromosomes, systematics and phylogeny in Taraxacum (Asteraceae). Plant Syst. Evol. 141, 219–229. doi: 10.1007/BF00989003
Müller, T., Philippi, N., Dandekar, T., Schultz, J., and Wolf, M. (2007). Distinguishing species. RNA 13, 1469–1472. doi: 10.1261/rna.617107
Mursalimov, S., and Deineko, E. (2018). Cytomixis in plants: facts and doubts. Protoplasma 255, 719–731. doi: 10.1007/s00709-017-1188-7
Mursalimov, S. R., and Deineko, E. V. (2011). An ultrastructural study of cytomixis in tobacco pollen mother cells. Protoplasma 248, 717–724. doi: 10.1007/s00709-010-0234-5
Nakao, Y., Taira, T., Horiuchi, S., Kawase, K., and Mukai, Y. (2005). Chromosomal difference between male and female trees of Ginkgo biloba examined by karyotype analysis and mapping of rDNA on the chromosomes by Fluorescence in situ Hybridization. J. Jpn. Soc. Hortic. Sci. 74, 275–280. doi: 10.2503/jjshs.74.275
Nakayama, S., Fujishita, M., Sone, T., and Ohyama, K. (2001). Additional locus of rDNA sequence specific to the X chromosome of the liverwort, Marchantia polymorpha. Chromosome Res. 9, 469–473. doi: 10.1023/A:1011676328165
Navashin, M. (1934). Chromosome alterations caused by hybridisation and their bearing upon certain general genetic problems. Cytologia 5, 169–203. doi: 10.1508/cytologia.5.169
Newton, M. E. (1986). Pellia borealis Lorbeer: its cytological status and discovery in Britain. J. Bryol. 14, 215–230. doi: 10.1179/jbr.1986.14.2.215
Newton, M. E. (1988). Cytological diversity in Pellia endiviifolia (Dicks.) Dum. J. Bryol. 15, 303–314. doi: 10.1179/jbr.1988.15.2.303
Nieto Feliner, G., and Rosselló, J. A. (2007). Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol. Phylogenet. Evol. 44, 911–919. doi: 10.1016/j.ympev.2007.01.013
Ochatt, S. J., and Seguí-Simarro, J. M. (2021). “Analysis of ploidy in haploids and doubled haploids,” in Doubled Haploid Technology, ed. J. M. Seguí-Simarro (New York: Humana Press), 105–125. doi: 10.1007/978-1-0716-1315-3_4
Ojha, S., Malla, S., and Lyons, S. M. (2020). snoRNPs: functions in ribosome biogenesis. Biomolecules 10:783. doi: 10.3390/biom10050783
Pikaard, C. S. (2000a). The epigenetics of nucleolar dominance. Trends Genet. 16, 495–500. doi: 10.1016/S0168-9525(00)02113-2
Pikaard, C. S. (2000b). Nucleolar dominance: uniparental gene silencing on a multi-megabase scale in genetic hybrids. Plant Mol. Biol. 43, 163–177. doi: 10.1023/A:1006471009225
Poczai, P., and Hyvönen, J. (2010). Nuclear ribosomal spacer regions in plant phylogenetics: problems and prospects. Mol. Biol. Rep. 37, 1897–1912. doi: 10.1007/s11033-009-9630-3
Pokorná, M. J., and Reifová, R. (2021). Evolution of B Chromosomes: from dispensable parasitic chromosomes to essential genomic players. Front. Genet. 12:727570. doi: 10.3389/fgene.2021.727570
Ran, Y., Hammett, K. R., and Murray, B. G. (2001). Phylogenetic analysis and karyotype evolution in the genus Clivia (Amaryllidaceae). Ann. Bot. 87, 823–830. doi: 10.1006/anbo.2001.1422
Raskina, O., Belyayev, A., and Nevo, E. (2004). Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosome Res. 12, 153–161. doi: 10.1023/B:CHRO.0000013168.61359.43
Ren, R., Wang, H., Guo, C., Zhang, N., Zeng, L., Chen, Y., et al. (2018). Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol. Plant 11, 414–428. doi: 10.1016/j.molp.2018.01.002
Richards, A. J. (1989). A comparison of within-plant karyological heterogeneity between agamospermous and sexual Taraxacum (Compositae) as assessed by the nucleolar organiser chromosome. Plant Syst. Evol. 163, 177–185. doi: 10.1007/BF00936513
Roa, F., and Guerra, M. (2012). Distribution of 45S rDNA sites in chromosomes of plants: structural and evolutionary implications. BMC Evol. Biol. 12:225. doi: 10.1186/1471-2148-12-225
Rosato, M., Álvarez, I., Nieto Feliner, G., and Rosselló, J. A. (2017). High and uneven levels of 45S rDNA site-number variation across wild populations of a diploid plant genus (Anacyclus, Asteraceae). PLoS One 12:e0187131. doi: 10.1371/journal.pone.0187131
Rosato, M., Castro, M., and Rosselló, J. A. (2008). Relationships of the woody Medicago species (section Dendrotelis) assessed by molecular cytogenetic analyses. Ann. Bot. 102, 15–22. doi: 10.1093/aob/mcn055
Rosato, M., Kovaøík, A., Garilleti, R., and Rosselló, J. A. (2016a). Conserved organisation of 45S rDNA sites and rDNA gene copy number among major clades of early land plants. PLoS One 11:e0162544. doi: 10.1371/journal.pone.0162544
Rosato, M., Ferrer-Gallego, P., Totta, C., Laguna, E., and Rosselló, J. A. (2016b). Nuclear rDNA instability in in vitro-generated plants is amplified after sexual reproduction with conspecific wild individuals. Bot. J. Linn. Soc. 181, 127–137. doi: 10.1111/boj.12392
Rosato, M., Moreno-Saiz, J. C., Galián, J. A., and Rossello, J. A. (2015). Evolutionary site-number changes of ribosomal DNA loci during speciation: complex scenarios of ancestral and more recent polyploid events. AoB Plants 7:lv135. doi: 10.1093/aobpla/plv135
Rosato, M., and Rosselló, J. A. (2009). Karyological observations in Medicago section Dendrotelis (Fabaceae). Folia Geobot. 44, 423–433. doi: 10.1007/s12224-009-9048-7
Rosselló, J. A., and Castro, M. (2008). Karyological evolution of the angiosperm endemic flora of the Balearic Islands. Taxon 57, 259–273. doi: 10.2307/25065967
Sadder, M. T., and Weber, G. (2001). Karyotype of maize (Zea mays L.) mitotic metaphase chromosomes as revealed by fluorescence in situ hybridization (FISH) with cytogenetic DNA markers. Plant Mol. Biol. Rep. 19, 117–123. doi: 10.1007/BF02772153
Sangduen, N., Toahsakul, M., and Hongtrakul, V. (2007). Karyomorphological study of some selected Cycads. Au J. T. 11, 1–6.
Sangduen, N., Toahsakul, M., and Hongtrakul, V. (2009). Comparative karyomorphological study between male and female plants of some Cycas and Zamia species. Nat. Resour. J. 43, 476–485.
Sato, S., Matsumoto, E., and Kuroki, Y. (1981). Satellite association of the nucleolar chromosomes in a plant. Protoplasma 108, 139–147. doi: 10.1007/BF01276888
Scaldaferro, M. A., da Cruz, M. V. R., Cecchini, N. M., and Moscone, E. A. (2016). FISH and AgNor mapping of the 45S and 5S rRNA genes in wild and cultivated species of Capsicum (Solananceae). Genome 59, 95–113. doi: 10.1139/gen-2015-0099
Schubert, I. (1984). Mobile nucleolus organizing regions (NORs) in Allium (Liliaceae s. lat.)? —Inferences from the specifity of silver staining. Plant Syst. Evol. 144, 291–305. doi: 10.1007/BF00984139
Schubert, I., and Wobus, U. (1985). In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92, 143–148. doi: 10.1007/BF00328466
Schwarzacher, H. G., and Wachtler, F. (1983). Nucleolus organizer regions and nucleoli. Hum. Genet. 63, 89–99. doi: 10.1007/BF00291525
Schweizer, D., and Ehrendorfer, F. (1976). Giemsa banded karyotypes, systematics, and evolution in Anacyclus (Asteraceae-Anthemideae). Plant Syst. Evol. 126, 107–148. doi: 10.1007/BF00981668
Sepsi, A., Fábián, A., Jäger, K., Heslop-Harrison, J. S., and Schwarzacher, T. (2018). ImmunoFISH: simultaneous visualisation of proteins and DNA sequences gives insight into meiotic processes in nuclei of grasses. Front. Plant Sci. 9:1193. doi: 10.3389/fpls.2018.01193
Sloan, K. E., Warda, A. S., Sharma, S., Entian, K. D., Lafontaine, D. L., and Bohnsack, M. T. (2017). Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152. doi: 10.1080/15476286.2016.1259781
Sochorová, J., Coriton, O., Kuderová, A., Lunerová, J., Chevre, A. M., and Kovaøík, A. (2017). Gene conversion events and variable degree of homogenization of rDNA loci in cultivars of Brassica napus. Ann. Bot. 119, 13–26. doi: 10.1093/aob/mcw187
Sýkorová, E., Lim, K. Y., Fajkus, J., and Leitch, A. R. (2003). The signature of the Cestrum genome suggests an evolutionary response to the loss of (TTTAGGG)n telomeres. Chromosoma 112, 164–172. doi: 10.1007/s00412-003-0256-2
Tapia-Pastrana, F. (2020). Differential amphiplasty and nucleolar dominance in somatic metaphase cells as evidence of hybridization in Prosopis juliflora (Leguminosae, Mimosoideae). Cytologia 85, 295–299. doi: 10.1508/cytologia.85.295
Terasaka, O., and Tanaka, R. (1974). Absence of the nucleolar constriction in the division of generative nucleus in the pollen of some angiosperms. Cytologia 39, 97–106. doi: 10.1508/cytologia.39.97
Thomson, E., Ferreira-Cerca, S., and Hurt, E. (2013). Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 126, 4815–4821. doi: 10.1242/jcs.111948
Totta, C., Rosato, M., Ferrer-Gallego, P., Lucchese, F., and Rosselló, J. A. (2017). Temporal frames of 45S rDNA site-number variation in diploid plant lineages: lessons from the rock rose genus Cistus (Cistaceae). Biol. J. Linn. Soc. 120, 626–636. doi: 10.1111/bij.12909
Trerè, D. (2000). AgNOR staining and quantification. Micron 31, 127–131. doi: 10.1016/S0968-4328(99)00069-4
Tucker, S., Vitins, A., and Pikaard, C. S. (2010). Nucleolar dominance and ribosomal RNA gene silencing. Curr. Opin. Cell Biol. 22, 351–356. doi: 10.1016/j.ceb.2010.03.009
Venkatesh, K. H., Pragathi, G. S., and Shivashankar, M. (2019). Meiotic behavior and chromosomal association of two diploid mulberry varieties, (Moraceae). Chromosome Bot. 13, 68–70. doi: 10.3199/iscb.13.68
Verma, R. C., and Raina, S. N. (1981). Cytogenetics of Crotalaria V. Supernumerary nucleoli in C. agatiflora (Leguminosae). Genetica 56, 75–80. doi: 10.1007/BF00126933
Vitales, D., D’Ambrosio, U., Gálvez, F., Kovaøík, A., and García, S. (2017). Third release of the plant rDNA database with updated content and information on telomere composition and sequenced plant genomes. Plant Syst. Evol. 303, 1115–1121. doi: 10.1007/s00606-017-1440-9
Watanabe, K., Shibaike, H., Suzuki, T., Ito, M., and Hoya, A. (2021). DNA Contents and karyotypes of the natural hybrids in Taraxacum (Asteraceae) in Japan. Acta Phytotaxon. Geobot. 72, 135–144. doi: 10.18942/apg.202013
Weiss-Schneeweiss, H., Emadzade, K., Jang, T. S., and Schneeweiss, G. M. (2013). Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenet. Genome Res. 140, 137–150. doi: 10.1159/000351727
Weiss-Schneeweiss, H., and Schneeweiss, G. M. (2013). “Karyotype diversity and evolutionary trends in angiosperms,” in Plant Genome Diversity Volume 2, ed. I. J. Leitch (London: Springer), 209–230. doi: 10.1007/978-3-7091-1160-4_13
Weiss-Schneeweiss, H., Tremetsberger, K., Schneeweiss, G. M., Parker, J. S., and Stuessy, T. F. (2008). Karyotype diversification and evolution in diploid and polyploid South American Hypochaeris (Asteraceae) inferred from rDNA localization and genetic fingerprint data. Ann. Bot. 101, 909–918. doi: 10.1093/aob/mcn023
Keywords: 35S rDNA, secondary constriction, satellite chromosome, NOR, nucleolus, amphiplasty, cytogenetic markers, rRNA genes
Citation: Rosselló JA, Maravilla AJ and Rosato M (2022) The Nuclear 35S rDNA World in Plant Systematics and Evolution: A Primer of Cautions and Common Misconceptions in Cytogenetic Studies. Front. Plant Sci. 13:788911. doi: 10.3389/fpls.2022.788911
Received: 03 October 2021; Accepted: 27 January 2022;
Published: 24 February 2022.
Edited by:
Sonia Garcia, Botanical Institute of Barcelona (CSIC), SpainReviewed by:
Teresa Garnatje, Spanish National Research Council (CSIC), SpainCopyright © 2022 Rosselló, Maravilla and Rosato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcela Rosato, bWFyY2VsYS5yb3NhdG9AdXYuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.