
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 April 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.785441
In plants, the key enzyme in ethylene biosynthesis is 1-aminocyclopropane-1 carboxylic acid (ACC) synthase (ACS), which catalyzes S-adenosyl-L-methionine (SAM) to ACC, the precursor of ethylene. Ethylene binds to its receptors, such as ethylene response 1 (ETR1), to switch on ethylene signal transduction. To understand the function of ACS and ETR1 in orchids, Oncidium ACC synthase 12 (OnACS12) and Oncidium ETR1 (OnETR1) from Oncidium Gower Ramsey were functionally analyzed in Arabidopsis. 35S::OnACS12 caused late flowering and anther indehiscence phenotypes due to its effect on GA–DELLA signaling pathways. 35S::OnACS12 repressed GA biosynthesis genes (CPS, KS, and GA3ox1), which caused the upregulation of DELLA [GA-INSENSITIVE (GAI), RGA-LIKE1 (RGL1), and RGL2] expression. The increase in DELLAs not only suppressed LEAFY (LFY) expression and caused late flowering but also repressed the jasmonic acid (JA) biosynthesis gene DAD1 and caused anther indehiscence by downregulating the endothecium-thickening-related genes MYB26, NST1, and NST2. The ectopic expression of an OnETR1 dominant-negative mutation (OnETR1-C65Y) caused both ethylene and JA insensitivity in Arabidopsis. 35S::OnETR1-C65Y delayed flower/leaf senescence by suppressing downstream genes in ethylene signaling, including EDF1-4 and ERF1, and in JA signaling, including MYC2 and WRKY33. JA signaling repression also resulted in indehiscent anthers via the downregulation of MYB26, NST1, NST2, and MYB85. These results not only provide new insight into the functions of ACS and ETR1 orthologs but also uncover their functional interactions with other hormone signaling pathways, such as GA–DELLA and JA, in plants.
The gaseous phytohormone ethylene affects many plant developmental processes, including seed germination, seed dormancy, root nodulation, flowering time, fruit ripening, flower/leaf senescence, biotic/abiotic stress responses, and sex determination (Bleecker and Kende, 2000; Guo and Ecker, 2004; Young et al., 2004; Achard et al., 2006, 2007; Corbineau et al., 2014; Li et al., 2019; Feng et al., 2020; Perata, 2020; Chen et al., 2021; Dar et al., 2021).
There are two main steps in the ethylene biosynthetic pathway. The first step is the conversion of S-adenosyl-L-methionine (SAM) to 1-aminocyclopropane-1 carboxylic acid (ACC), the precursor of ethylene, by ACC synthase (ACS), and the second step is the further conversion of ACC to ethylene by ACC oxidase (ACO) (Adams and Yang, 1979; Hamilton et al., 1991; Van Der Straeten et al., 1993; Pattyn et al., 2021). In addition to functioning in the ethylene pathway, ACC has been reported to function independently of ethylene as a signaling molecule to regulate several developmental processes, such as pollen tube attraction, guard mother cell terminal division, and early vegetative development (Vanderstraeten et al., 2019; Yin et al., 2019; Mou et al., 2020). ACS has been thought to be a rate-limiting enzyme in ethylene biosynthesis (Adams and Yang, 1979). In plants, the biochemical characterization of ACS and ACO indicates that both are encoded by a divergent gene family (Alexander and Grierson, 2002; Tsuchisaka et al., 2009; Booker and DeLong, 2015; Liu et al., 2015; Houben and Van de Poel, 2019) and are differentially regulated in a tissue-specific manner at the transcriptional, posttranscriptional, and posttranslational levels (Woeste et al., 1999; Yamagami et al., 2003; Rodrigues et al., 2014; Datta et al., 2015; Park et al., 2018; Pattyn et al., 2021). It has been reported that a mutation in maize ACS (ZmACS6) alters ethylene synthesis and results in delayed leaf senescence (Young et al., 2004). In tomato, an acs2-1 mutant showed an increased ACS activity and ethylene overproduction, thereby promoting leaf senescence and accelerating fruit ripening (Sharma et al., 2021). In contrast, decreased ACS activity and ethylene production were observed in an acs2-2 mutant, which showed delayed leaf senescence and fruit ripening (Sharma et al., 2021).
Ethylene triggers ethylene responses by first binding to the endoplasmic reticulum-localized receptors ethylene response 1 (ETR1), ethylene response sensor 1 (ERS1), ETR2, ERS2, and ethylene insensitive 4 (EIN4) to inhibit these receptors from interacting with and activating the negative ethylene signaling regulator constitutive triple response 1 (CTR1; Bleecker, 1999; Chang and Shockey, 1999; Alonso and Stepanova, 2004; Merchante et al., 2013; Ju and Chang, 2015; Yang et al., 2015; Azhar et al., 2019; Binder, 2020; Zhao et al., 2020; Liu and Chen, 2021). The inhibition of the interaction for receptors to CTR1 causes the activation of several positive regulators of the ethylene response downstream of CTR1, including EIN2, EIN3, ethylene response DNA-binding factor 1-4 (EDF1-4), and ethylene response factors (ERFs), resulting in downstream ethylene responses. EIN2 is a membrane protein that encodes an Nramp family protein (Alonso et al., 1999). EIN3 encodes a nuclear protein and works downstream of EIN2 (Chao et al., 1997). The ethylene response DNA-binding factor (EDF) family and ERF are transcription factors involved in ethylene signal transduction (Stepanova and Alonso, 2005; Chen et al., 2021). In the absence of ethylene, these ethylene receptors (ETR1, ERS1, ETR2, ERS2, and EIN4) can activate CTR1 and suppress ethylene signaling (Binder, 2020; Zhao et al., 2020; Liu and Chen, 2021).
In Arabidopsis, etr1 and ein2 mutants are insensitive to ethylene and cause delayed leaf/flower senescence (Grbic and Bleecker, 1995; Patterson and Bleecker, 2004; Graham et al., 2012). Interestingly, the ectopic expression of a mutant Arabidopsis ETR1-1 with cysteine 65 substituted with tyrosine (Cys65Tyr) causes the loss of copper cofactor binding activity to its transmembrane domain and prevents ethylene from binding to itself (Schaller and Bleecker, 1995; Wilkinson et al., 1997; Rodriguez et al., 1999), not only causing an ethylene insensitivity phenotype but also delaying flower and leaf senescence in transgenic tomato, petunia (Wilkinson et al., 1997) and tobacco (Yang et al., 2008). In contrast, mutations in the CTR1 gene result in a constitutive ethylene response, including early senescence and flower abscission (Kieber et al., 1993; Huang et al., 2003).
Ethylene has been thought to perform its functions by interacting with other plant hormones (Yoo et al., 2009; Van de Poel et al., 2015; Zhou et al., 2019). Ethylene can act synergistically with GAs but antagonistically with ABA in regulating seed germination (Arc et al., 2013; Corbineau et al., 2014; Van de Poel et al., 2015). It has been reported that the downstream gene of ethylene signaling ERF1 could regulate flowering time in Arabidopsis through direct suppression of FLOWERING LOCUS T (FT) expression (Chen et al., 2021). Ethylene can also regulate gibberellin–DELLA signaling pathways in controlling flowering time (Achard et al., 2007; Yoo et al., 2009). In this case, DELLA proteins could delay the timing of the floral transition, possibly through the suppression of LEAFY (LFY; Wen and Chang, 2002; Harberd, 2003; Achard et al., 2006) and WRKY75 (Zhang et al., 2018). Crosstalk between ethylene and auxin in the regulation of root development has also been reported (Ruzicka et al., 2007; Swarup et al., 2007; Mao et al., 2016). Since jasmonic acid (JA) can also promote senescence in Arabidopsis (He et al., 2002) and guanine-induced plant immunity to pathogens depends on not only ethylene but also JA signaling (Wang et al., 2022), JA it may also engage in crosstalk with ethylene in regulating these processes.
Genes involved in ethylene biosynthesis and signaling have been characterized and reported in representative dicot/monocot species (Yang et al., 2015; Liu and Chen, 2021; Pattyn et al., 2021), as well as in the charophyte alga Spirogyra pratensis (Ju et al., 2015). The possible functional diversity of these genes among different plant species has been reported (Kim et al., 2012; Yang et al., 2015; Liu and Chen, 2021) and remains under investigation. Monocot orchids comprise more than 25,000 species in a large plant family, the Orchidaceae. Surprisingly, relatively few molecular investigations of genes involved in ethylene biosynthesis and signaling have been reported (Huang et al., 2007; Lerslerwong and Ketsa, 2008; Almasi and Mohamed, 2020). In this study, we isolated and functionally analyzed Oncidium ACC synthase 12 (OnACS12) and Oncidium ETR1 (OnETR1) from Oncidium orchids. We ectopically expressed OnACS12 and a dominant-negative OnETR1 mutation (OnETR1-C65Y) in Arabidopsis. We found that 35S::OnACS12 could regulate the GA–DELLA signaling pathways controlling flowering time by suppressing LFY expression and could regulate anther dehiscence by downregulating the JA synthesis gene DAD1. Furthermore, 35S::OnETR1-C65Y caused a delay in flower/leaf senescence due to insensitivity to ethylene and JA signaling, which also resulted in anther indehiscence. Thus, our study provides new insight into the functions of ACS and ETR1 orthologs, as well as their interactions with other plant hormones, specifically GA and JA.
Plants of Oncidium Gower Ramsey, a hybrid (Oncidium Goldiana × Oncidium Guinea Gold; Chang et al., 2010) orchid, were grown at day/night temperature of 26/23°C under long-day conditions in the greenhouse of National Chung-Hsing University, Taichung, Taiwan. Arabidopsis thaliana ecotype Columbia was used for all experiments. Arabidopsis etr1-1 mutants in the Columbia background were obtained from Arabidopsis Biological Resource Center, Ohio State University, Columbia, OH, United States. Seeds of Arabidopsis thaliana were sterilized by 75% ethanol and placed on agar plates medium (1/2 Murashige-Skoog salt, 1% sucrose, 0.5% agar) with selected antibiotics (Murashige and Skoog, 1962) at 4°C for 2 days. The seedlings were moved into growth chambers under long-day conditions (16 h light/8 h dark) at 22°C for 10–14 days before being transplanted into soil. The light intensity of the growth chambers was 150 μE m−2s−1.
Total RNA extracted from approximately 0.1 g of flower buds (fb) from Oncidium was used for cDNA synthesis. The full-length cDNAs of OnACS12 and OnETR1 were amplified by PCR using the specific 5′ and 3′ primers listed in Supplementary Table 2. Full-length cDNA fragments of OnACS12 and OnETR1 were cloned into the linker region in the binary vector pEpyon-12K (CHY Lab, Taichung, Taiwan) under control of the cauliflower mosaic virus (CaMV) 35S promoter for further use in plant transformation.
To generate OnETR1-C65Y fragment, two fragments amplified by PCR using the specific 5′ and 3′ primers listed in Supplementary Table 2 were performed. The front fragment (206 bp) of OnETR1-C65Y was PCR-amplified from start codon to the point mutation. The end fragment (1717 bp) of OnETR1-C65Y was PCR-amplified from the point mutation to the stop codon. The full-length cDNAs of OnETR1-C65Y were then PCR-amplified using the specific 5′ and 3′ primers listed in Supplementary Table 1 and were cloned into the linker region in the binary vector pEpyon-22K (CHY Lab, Taichung, Taiwan) under control of the CaMV 35S promoter for further use in plant transformation.
The generated 35S::OnACS12, 35S::OnETR1, and 35S::OnETR1-C65Y constructs were transformed into the Agrobacterium tumefaciens strain GV3101. These A. tumefaciens were then infiltrated into Arabidopsis plants through the floral dip method as described previously (Clough and Bent, 1998). Transformants were selected in a medium containing 50 μg ml−1 kanamycin and were further verified by PCR and real-time PCR (RT-PCR.)
Real-time quantitative PCR was performed on Mini Opticon (Bio-Rad Laboratories, Hercules, CA, United States) and SYBR Green Master Mix (KAPA, Biosystem Wilmington, MA, United States). The amplification condition was 95°C for 3 min, followed by 40 cycles of amplification (95°C for 3 s, 60°C for 30 s, and 72°C for 1 s) and melted (65–95°C with readings every 1°C). Sequences for the primers used for real-time quantitative RT-PCR for Oncidium and Arabidopsis genes are listed in Supplementary Table 2. The housekeeping gene UBQ10 was used as normalization control for Arabidopsis, and OnUBQ gene was used as normalization control for Oncidium. Data were analyzed using CFX Manager TM Software (version 1.5, Bio-Rad).1
Anthers were imaged by an Olympus FV1000 confocal microscope (Olympus FV1000, Tokyo, Japan). The plant cell cellulose was stained with calcofluor white excited by 405 nm helium/neon laser line, and emission was collected at 415 ~ 470 nm. The lignin was stained with auramine O excited by 488 nm helium/neon laser line, and emission was collected at 470 nm.
Ethylene was measured from seedlings. Sixty 12-day-old seedlings were collected, and fresh weight was measured. The sample was placed in an airtight 30-ml flask and incubated for 3 h in growth chamber. By using a gas-tight hypodermic syringe, 1 ml of headspace was sampled from each flask and injected into a gas chromatograph. The gas chromatograph is equipped with an alumina-based capillary column and a flame ionization detector. Three replicates were conducted and the average and SD reported.
For ethylene responses, mature wild-type (WT) and transgenic Arabidopsis plants were sealed in plastic chambers and gassed with air or air containing 6 ppm ethylene for 3 days in a 16 h light/8 h dark cycle as described previously (Chen and Bleecker, 1995; Chen et al., 2011, 2015).
For triple responses, WT and transgenic Arabidopsis seedlings were grown on MS plates in the presence or absence of 6 mM ethylene precursor ACC for 7 days in the dark (4 days in 4°C, 3 days in 22°C) and phenotype analyzed.
The dark- and hormone-induced leaf senescence assay was modified as described previously (Li et al., 2013). Detached rosette leaves from mature wild-type and transgenic Arabidopsis plants were excised and floated in sealed Petri dishes with 10 ml distilled water in the presence of 75 μM ACC or 50 μM MeJA in the dark for 4 days at 23°C.
For chlorophyll extraction, leaves from mature wild-type and transgenic Arabidopsis plants were incubated in 80% acetone (v/v) for 24 h in darkness as described previously (Li et al., 2013). Absorbance was measured at 645 and 657 nm, and chlorophyll content was calculated based on (20.2 *A645 + 8.02* A657)/g fresh weight.
The Oncidium ACC synthase 12 (OnACS12) gene was identified through a search in our Oncidium EST database using BLAST with published Arabidopsis sequences as templates. The full-length OnACS12 cDNA encodes 508 amino acids (Supplementary Figure 1) that share 55% similarity with Arabidopsis ACS12 (AT5G51690.1). Oncidium OnACS12 had seven known conserved boxes identified in the ACS orthologs (Supplementary Figure 1). In addition, OnACS12 has three vital characteristics of ACC synthase: substrate AdoMet specificity (Mccarthy et al., 2001), a pyridoxal phosphate (PLP) binding site (Capitani et al., 1999), and a PLP cofactor active site (Supplementary Figure 1). These results suggested that OnACS12 functions as an active synthase to produce ACC, the precursor of ethylene. A phylogenetic tree was constructed based on an amino acid sequence alignment and showed that OnACS12 is closely related to ACS in monocots (Supplementary Figure 2).
To determine the expression profile of OnACS12, OnACS12 expression was detected in different flower organs (lips/sepals/petals) of O. Gower Ramsey flowers at different developmental stages (Figures 1A,B). OnACS12 was expressed in all flower organs in young fb, mature flower buds (mfb), and mature flowers (mf; Figure 1C). OnACS12 expression was relatively low in the floral organs of young flower buds and increased in mfb and in mf (Figure 1C). This result indicated that having the lowest expression of OnACS12 in young flower buds ensured that senescence will not occur during early Oncidium flower development.
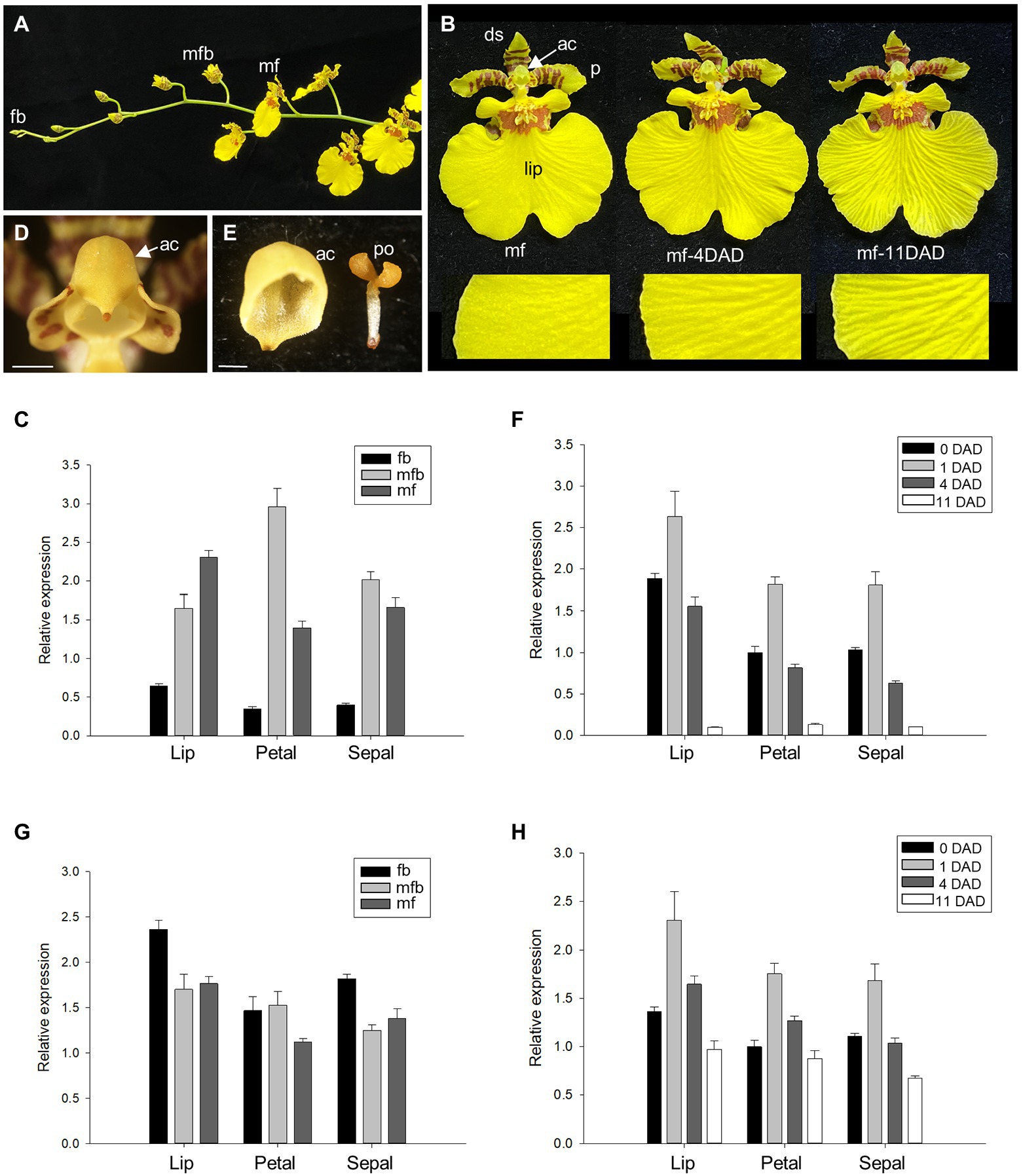
Figure 1. Detection of the expression of Oncidium ACC synthase 12 (OnACS12) and Oncidium ETR1 (OnETR1) during Oncidium flower development. (A) Flower buds (fb), mature flower buds (mfb), and mature flowers (mf) along an Oncidium inflorescence. (B) Mature Oncidium flower (mf) with the anther cap (ac) attached (left), and 4 days [middle, mf-4 days after anther cap detachment (DAD)] or 11 days (right, mf-11 DAD) after the anther cap detached. A close-up of the lips (lip) is shown at the bottom. p; petals, ds: dorsal sepals. (C,G) Detection of the expression of OnACS12 (C) and OnETR1 (G) in the lips, petals, and sepals of Oncidium fb, mfb, and mf by real-time quantitative PCR. (D) Close up of the anther cap (ac) from (B, left). Bar = 0.8 mm. (E) Close up of the detached anther cap (ac) and pollinium (po). Bar = 0.6 mm. (F,H) Expression of OnACS12 (F) and OnETR1 (H) in the lips, petals, and sepals of flowers 0, 1, 4, and 11 DAD as detected via real-time quantitative PCR. In (C,F,G,H), the transcript levels of the genes were determined using two to three replicates and were normalized to the OnUBQ gene. The error bars represent SD.
It has been reported that Oncidium and Cymbidium flower senescence is induced by the detachment of the anther cap (ac; Figures 1B,D,E; Woltering, 1990; Huang et al., 2007). To further analyze the relationship between flower senescence and the expression levels of OnACS12, mRNA from flower organs on different days after ac detachment (DAD; Figure 1B) was used to detect the expression of OnACS12. The results showed that OnACS12 expression was clearly upregulated at one DAD (Figure 1F), at which time no sign of senescence in the perianth was observed. OnACS12 expression was decreased at 4 DAD (Figure 1F), when slight senescence occurred in the perianth (Figure 1B, middle), and it was significantly downregulated at 11 DAD (Figure 1F), when flower organs were severely senescent (Figure 1B, right). This result indicated that anther cap detachment could induce the expression of OnACS12 and start the senescence process very quickly. Once the process was initiated, OnACS12 expression gradually decreased to its lowest level when the flower was severely senescent.
To further explore its role, OnACS12 cDNA driven by the CaMV 35S promoter was constructed and transformed into Arabidopsis. To verify ethylene synthesis in 35S::OnACS12 Arabidopsis, we measured ethylene production from wild-type and 35S::OnACS12 transgenic Arabidopsis. The results indicated that ethylene production was 28.20 μl C2H4/kg*Hr on average for 35S::OnACS12 Arabidopsis, which was much higher than that of wild-type Arabidopsis (8.91 μl C2H4/kg*Hr on average; Figure 2A). This result indicated that the ectopic expression of OnACS12 could increase ethylene production in Arabidopsis.
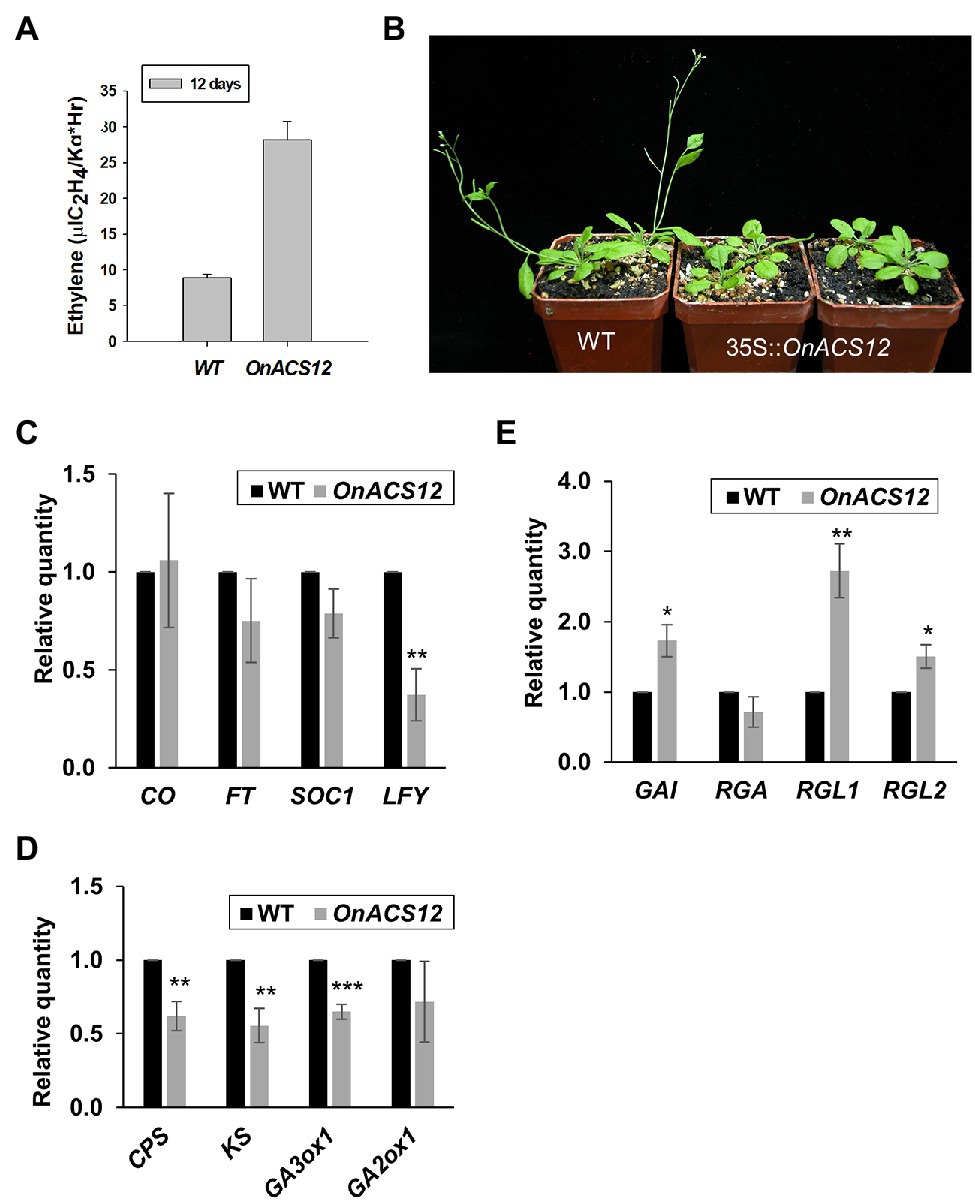
Figure 2. Phenotypic analysis and detection of gene expression in Arabidopsis ectopically expressing OnACS12. (A) Detection of ethylene production in 12-day-old wild-type (WT) and 35S::OnACS12 seedlings. (B) Four 38-day-old 35S::OnACS12 plants (middle and right) flowered at 43 days, much later than wild-type plants (left) when grown under LD conditions. (C–E) Detection of the expression of flowering time genes, GA biosynthesis genes and DELLAs via real-time quantitative PCR. Total RNA isolated from 35S::OnACS12 transgenic Arabidopsis and WT Columbia plants was used as a template to detect the expression of the flowering time genes CONSTANS (CO), FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1), and LFY (C), the GA biosynthetic genes CPS, KS, GA3ox1, and GA2ox1 (D) and the DELLAs GA-INSENSITIVE (GAI), REPRESSOR OF GA1-3 (RGA), RGA-LIKE1 (RGL1), and RGL2 (E). The transcript levels of these genes were determined using two to three replicates and were normalized to UBQ10. The expression of each gene in the transgenic plants is given relative to that of the wild-type plant, which was set at 1. The error bars represent SD. The asterisks indicate a significant difference from the WT value (*p < 0.05; **p < 0.01; and ***p < 0.001). Statistical analysis was conducting using two-tailed Student’s t-tests.
In this study, 80 independent OnACS12 transgenic Arabidopsis T1 plants were obtained. Fifty-four plants were phenotypically indistinguishable from wild-type plants, whereas the other 26 plants showed a late flowering phenotype (Figure 2B). 35S::OnACS12 Arabidopsis flowered 39 days after germination on average, which was obviously later than that of wild-type plants, which flowered at 31 days (Supplementary Table 3). To explore the relationship between OnACS12 and flowering time genes, the expression of CONSTANS (CO), FT, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1), and LFY was analyzed using quantitative RT–PCR. As shown in Figure 2C, the expression of LFY was significantly downregulated, whereas the expression of FT and SOC1 was slightly suppressed in transgenic 35S::OnACS12 plants. This result indicated that the late-flowering phenotype of 35S::OnACS12 was mainly correlated with the downregulation of LFY.
It has been thought that ethylene acts as a plant stress hormone and delays flowering by regulating gibberellin–DELLA signaling (Achard et al., 2007; Yoo et al., 2009). GA has been reported to promote the development and elongation of flower organs by suppressing the functions of DELLA proteins (Cheng et al., 2004; Tyler et al., 2004), which contain an N-terminal DELLA domain responsible for the activity of the DELLA proteins in response to GA (Peng et al., 1997; Silverstone et al., 1998; Dill and Sun, 2001). DELLA proteins act as repressors of GA signaling and delay the timing of the floral transition, possibly through the downregulation of LFY (Wen and Chang, 2002; Harberd, 2003; Achard et al., 2006). Since 35S::OnACS12 increased ethylene production and delayed flowering time, exploring the relationship between OnACS12 and gibberellin–DELLA signaling pathways was necessary. When the expression of three GA biosynthesis genes, CPS, KS, GA3ox1, and one GA deactivation gene, GA2ox1, was analyzed in 35S::OnACS12 Arabidopsis, a clear downregulation of CPS, KS, and GA3ox1 was observed (Figure 2D). The expression of GA2ox1 was not affected (Figure 2D). When the expression of the DELLA genes GA-INSENSITIVE (GAI), REPRESSOR OF GA1-3 (RGA), and RGA-LIKE1 (RGL1) and RGL2 was further analyzed, the expression levels of GAI, RGL1, and RGL2 were clearly upregulated in 35S::OnACS12 plants (Figure 2E). These results implied that OnACS12 reduced the levels of bioactive GA and promoted DELLA gene expression to further suppress LFY expression and delay flowering.
In addition to delaying the flowering time, 35S::OnACS12 Arabidopsis also showed a sterility phenotype by producing unelongated siliques during late development (Figures 3A,C; Supplementary Figures 3A–D); these siliques were significantly different from the elongated and fully developed siliques of the wild-type plants (Figures 3A,B). 35S::OnACS12 flowers produced normal sepals, petals, and carpels (Figure 3D). However, the anthers on the stamens were indehiscent at all stages of flower development (Figures 3D,E). Thus, the 35S::OnACS12 flowers were sterile and unable to set seeds due to the inability to successfully pollinate. Alexander’s staining indicated that the indehiscent anthers of 35S::OnACS12 Arabidopsis contained viable pollen grains (Figures 3F,G) that closely resembled wild-type pollen (Figures 3H,I). To further examine pistil activity, wild-type pollen was crossed with stigmas of sterile 35S::OnACS12 Arabidopsis. The siliques exhibited normal elongation and produced normal seeds after cross-pollination (Figures 3J,K; Supplementary Figures 3E,F). These results indicated that the sterility of 35S::OnACS12 plants was associated with indehiscent anthers.
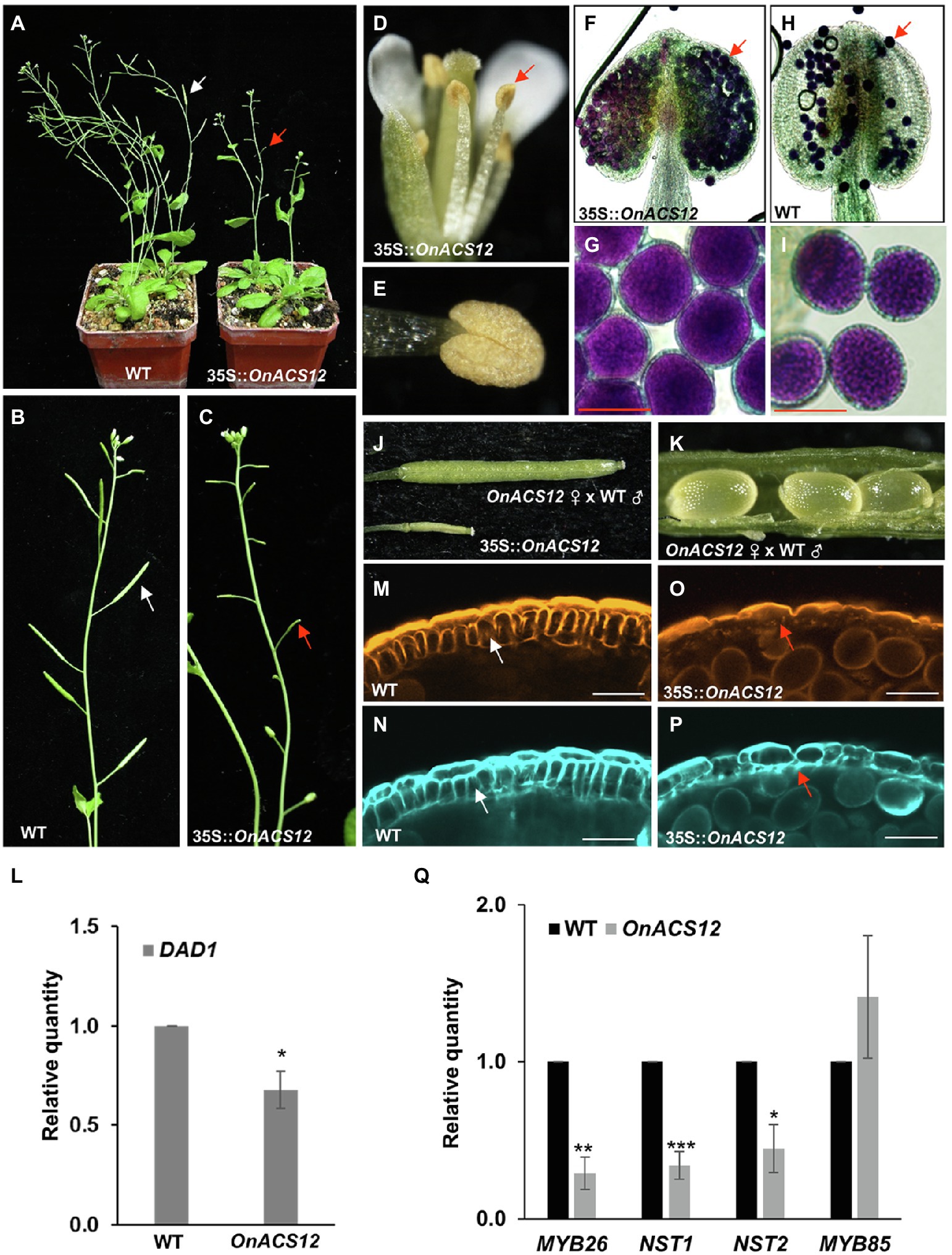
Figure 3. Analysis of the indehiscent anther phenotype and detection of gene expression in 35S::OnACS12 Arabidopsis. (A) A 52-day-old 35S::OnACS12 plant produced short siliques (red arrow on the right), whereas the wild-type plant produced normal elongated siliques (white arrow on the left). (B,C) Wild-type inflorescence showing elongated siliques (white arrow in B) and a 35S::OnACS12 plant with short siliques (red arrow in C). (D) A 35S::OnACS12 flower with an indehiscent anther (arrow). (E) Close-up view of an indehiscent anther from (D). (F–I) Pollen grain viability analysis of 35S::OnACS12 male-sterile anthers via Alexander staining. Anthers of 35S::OnACS12 (F) and wild-type (H) Arabidopsis had similar viable pollen grains (arrows). Close-up of pollen grains with similar shapes and normal viability from 35S::OnACS12 (G) and wild-type (I) plants. Bar = 20 μm. (J) The normal elongated silique of a 35S::OnACS12 plant after cross-pollination with wild-type pollen (top) along with a short 35S::OnACS12 silique without pollination (bottom). (K) Normal seed development was observed in a 35S::OnACS12 silique after cross-pollination. (L,Q) The mRNA levels were determined via real-time quantitative PCR. Total RNA isolated from 35S::OnACS12 transgenic plants and wild-type Columbia plants was used as a template to detect the expression of DAD1 (L), MYB26, NST1/2, and MYB85 (Q). The transcript levels of these genes were determined using two to three replicates and were normalized to UBQ10. The expression of each gene in the transgenic plants is given relative to that of the wild-type plant, which was set at 1. The error bars represent SDs. The asterisks indicate a significant difference from the WT value (*p < 0.05; **p < 0.01; and ***p < 0.001). Statistical analysis was conducting using two-tailed Student’s t-tests. (M–P) Confocal laser-scanning microscopy (CLSM) of anther cell walls in 35S::OnACS12 (O,P) and wild-type (M,N) plants. The anthers were double stained with auramine O for lignin, indicated in orange (M,O), and calcofluor white for cellulose, indicated in cyan (N,P). Secondary thickening is visible in the endothecium (arrow) of the wild-type anthers (M,N) and is absent in 35S::OnACS12 anthers (arrow; O,P). Bar = 25 μm.
The anther indehiscence phenotype in 35S::OnACS12 plants was similar to that observed in mutants of the Arabidopsis JA biosynthesis gene DAD1 (Ishiguro et al., 2001); GA has been reported to be able to regulate anther dehiscence by repressing the DELLA proteins and sequentially activating the JA biosynthesis gene DAD1 (Cheng et al., 2009; Peng, 2009; Marciniak and Przedniczek, 2019); and GA was suppressed, whereas DELLA genes were upregulated in 35S::OnACS12 plants. Thus, the relationship between OnACS12 and DAD1 expression was analyzed in 35S::OnACS12 flowers. As shown in Figure 3L, the level of DAD1 transcripts was clearly downregulated in 35S::OnACS12 flowers. This result indicated that the anther indehiscence in 35S::OnACS12 plants was likely due to the suppressed expression of the JA biosynthesis gene DAD1, which was caused by the decrease in GA levels and increased DELLA activity in 35S::OnACS12 plants.
Anther dehiscence is the final action of the anther and requires the lignification and secondary cell wall thickening of the endothecium to generate forces to rupture the anther cell wall and permit pollen release (Bonner and Dickinson, 1989; Yang et al., 2007; Cecchetti et al., 2013). To further examine the secondary wall thickening and analyze the cellular basis for anther dehiscence, lignin staining with auramine O and cellulose staining with calcofluor white were performed in the endothecium of developing anthers in 35S::OnACS12 and wild-type plants. The results indicated that in wild-type plants, secondary thickening occurred in the endothecium, with striated lignin formation before anther dehiscence, and the surrounding cell layers of the anther did not undergo secondary thickening (Figures 3M,N). In contrast, the secondary cell wall could not thicken and lignin formation was absent in the endothecium of 35S::OnACS12 plants (Figures 3O,P).
MYB26, MYB85, NAC SECONDARY WALL PROMOTING FACTOR1 (NST1), and NST2 have been reported to be involved in regulating secondary thickness in the anther endothecium (Mitsuda et al., 2005; Zhong et al., 2008). nst1nst2 double mutants exhibited male sterility, with a similar anther-indehiscent phenotype as that of 35S::OnACS12 plants, due to the loss of secondary wall thickening in the anther endothecium (Mitsuda et al., 2005). To confirm whether the failure of secondary thickening in the anther endothecium of 35S::OnACS12 plants was associated with the altered expression of the MYB26, MYB85, NST1, and NST2 genes, the expression of these genes was analyzed in 35S::OnACS12 flowers. The results indicated that the expression of MYB26, NST1, and NST2 was downregulated in 35S::OnACS12 plants, whereas the expression of MYB85 was unaffected (Figure 3Q). Thus, OnACS12 also caused indehiscent anthers due to the failure of secondary cell wall thickening through the downregulation of MYB26, NST1, and NST2 in Arabidopsis.
The Oncidium ETR1 (OnETR1) gene was identified through a search in our Oncidium EST database using BLAST with published Arabidopsis sequences as templates. The full-length OnETR1 cDNA encodes 631 amino acids (Supplementary Figure 4) that share 71% similarity with Arabidopsis ETR1 (AT1G66340.1). Oncidium OnETR1 had four conserved domains, including an ethylene binding site, a GAF domain, a histidine kinase domain, and a cyan box, which were identified as putative receiver domains in the ETR1 orthologs (Supplementary Figure 4; Parkinson and Kofoid, 1992). Sequence and domain conservation suggested that OnETR1 functions as an ETR1 ortholog in Oncidium. A phylogenetic tree was constructed based on an amino acid sequence alignment and showed that OnETR1 is closely related to the ETR1 orthologs from the orchids Dendrobium (DeETR) and Phalaenopsis (PhETR; Supplementary Figure 5).
To determine the expression profile of OnETR1, OnETR1 expression was detected in different flower organs at different developmental stages in O. Gower Ramsey flowers (Figures 1A,B). In contrast to OnACS12, OnETR1 was expressed in all flower organs (lips/sepals/petals) at a similar level in young flower buds, mature flower buds, and mature flowers (Figure 1G). When the expression level of OnETR1 from flower organs on different DAD (Figure 1B) was examined, OnETR1 expression was upregulated in all flower organs at one DAD (Figure 1H), similar to OnACS12 expression (Figure 1F). This result may due to the effect of a negative feedback loop for ethylene signaling (Azhar et al., 2019). It has been reported that this negative feedback loop could desensitize plants to ethylene and functioned at the level of the ethylene receptors (Azhar et al., 2019). Thus, early after the removing of the anther cap may increase the ethylene level and sequentially cause the upregulation of ethylene receptor OnETR1 in Oncidium through the negative feedback loop mechanism. However, at 4 and 11 DAD, OnETR1 expression decreased in the flowers to a level similar to that observed in control flowers at 0 DAD (Figure 1H). These results indicated that as a receptor, OnETR1 was constitutively expressed at a certain level during different stages of Oncidium flower development, as well as during the entire process of flower senescence.
To further explore the role of Oncidium OnETR1, a dominant-negative mutation, OnETR1-C65Y, in which cysteine 65 was substituted with tyrosine, was generated and ectopically expressed in Arabidopsis; this residue mediates ethylene binding to ethylene receptors, is a candidate ligand for the Cu(I) cofactor, causes the loss of ethylene binding and suppresses the sequential ethylene response (Schaller et al., 1995; Wilkinson et al., 1997; Rodriguez et al., 1999). Eighty independent 35S::OnETR1-C65Y transgenic Arabidopsis plants were obtained. Sixty-four OnETR1-C65Y plants were phenotypically indistinguishable from wild-type plants, whereas the other 16 plants showed a delayed flower senescence phenotype (Figures 4A,B). Flower organs were senescent and abscised at positions 3–4 in wild-type plants (Figures 4E,F) and in 35S::OnETR1 Arabidopsis (Figures 4D,F). In etr1-1 mutant plants, senescence was delayed until position 12 (Figures 4A,C,F). In 35S::OnETR1-C65Y transgenic Arabidopsis, senescence was delayed until position 8 (Figures 4A,B,F). To examine whether the ethylene response was affected by 35S::OnETR1-C65Y, the expression of downstream genes in the ethylene response, such as ethylene response DNA-binding factor 1-4 (EDF1-4) and ethylene response factor 1 (ERF1; Stepanova and Alonso, 2005; Chen et al., 2021), was analyzed in 35S::OnETR1-C65Y Arabidopsis that showed high expression of OnETR1-C65Y (Figure 4G). The results indicated that the expression of EDF1-4 and ERF1 was significantly downregulated in 35S::OnETR1-C65Y Arabidopsis (Figure 4H). In the etr1-1 mutant, only EDF1, EDF4, and ERF1 were downregulated (Figure 4H). These results indicated that the delayed flower senescence in 35S::OnETR1-C65Y Arabidopsis was mainly due to the inhibition of the ethylene signaling pathway.
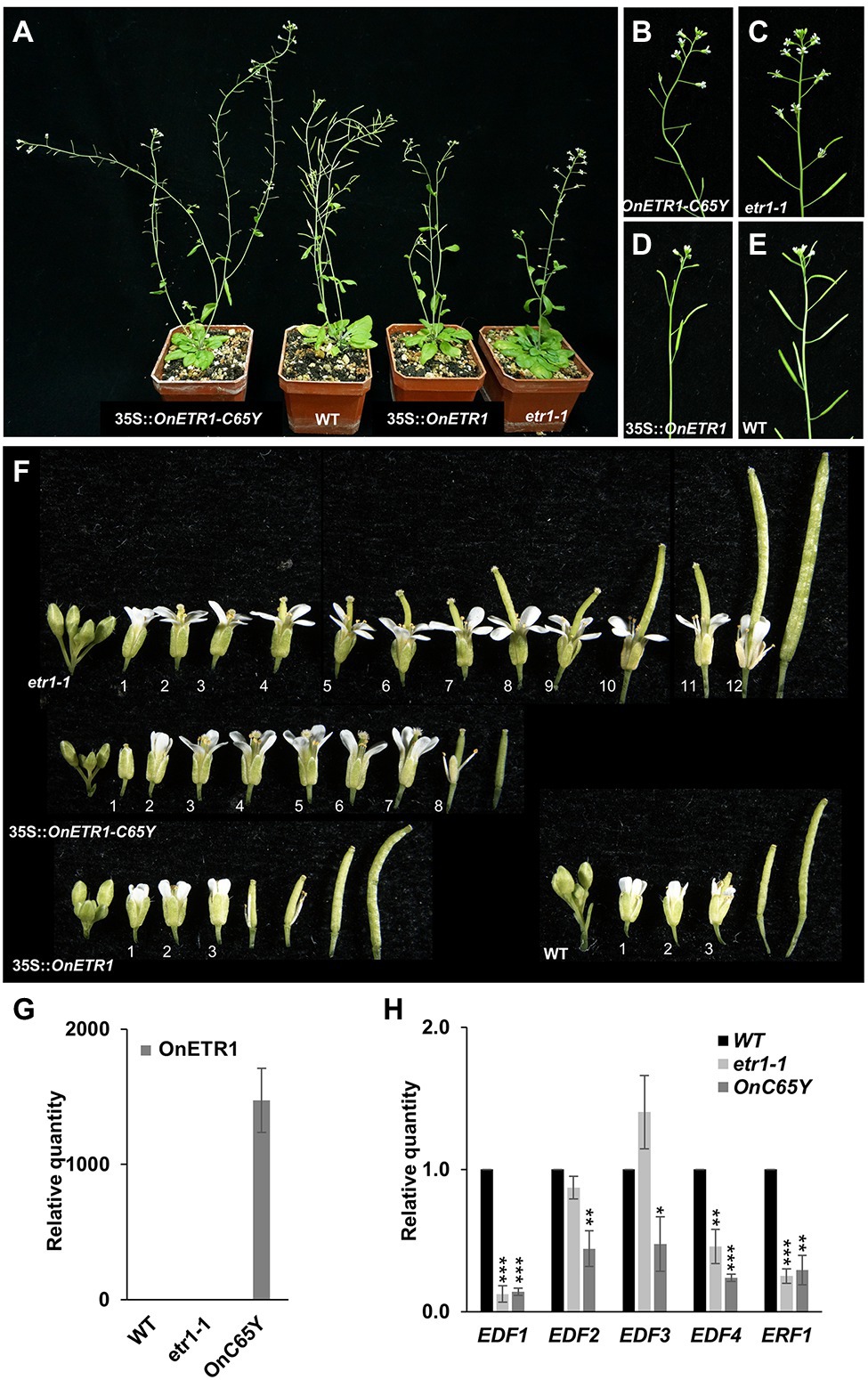
Figure 4. Phenotypic analysis and detection of gene expression in Arabidopsis ectopically expressing OnETR1-C65Y. (A) Mature 35S::OnETR1-C65Y, WT, 35S::OnETR1, and etr1-1 plants (from left to right). (B–E) Close-up of the inflorescences of the 35S::OnETR1-C65Y (B), etr1-1 (C), 35S::OnETR1 (D), and wild-type (E) plants from (A). (F) Flowers along inflorescences from etr1-1, 35S::OnETR1-C65Y, 35S::OnETR1 and WT Arabidopsis. The senescence and abscission of flower organs in etr1-1 and 35S::OnETR1-C65Y flowers were significantly delayed. The numbers indicate the positions of the flowers. (G) Relative expression of OnETR1-C65Y in WT, etr1-1 and 35S::OnETR1-C65Y transgenic Arabidopsis based on real-time quantitative PCR. (H) Relative expression of the downstream ethylene signaling genes EDF1-4 and ERF1 in WT, etr1-1 and 35S::OnETR1-C65Y transgenic Arabidopsis based on real-time quantitative PCR. The transcript levels of these genes were determined using two to three replicates and were normalized to UBQ10. The expression of each gene in the transgenic plants is given relative to that of the wild-type plant, which was set at 1. The error bars represent SDs. The asterisks indicate a significant difference from the WT value (*p < 0.05; **p < 0.01; and ***p < 0.001). Statistical analysis was conducting using two-tailed Student’s t-tests.
To further examine the ethylene response in OnETR1-C65Y Arabidopsis, 35S::OnETR1-C65Y, etr1-1, and wild-type Arabidopsis were exposed to air containing 6 ppm ethylene for 3 days. In the presence of ethylene, the perianth organs senesced and abscised early (at position 1) in wild-type Arabidopsis (Figure 5A, bottom). In contrast, in the presence of ethylene, the perianth organs of the 35S::OnETR1-C65Y and etr1-1 flowers were turgid and not senescent (Figure 5A, middle and top). These results indicated that the delayed senescence/abscission of the flower organs in 35S::OnETR1-C65Y Arabidopsis was due to ethylene insensitivity, similar to that in etr1-1 mutants.
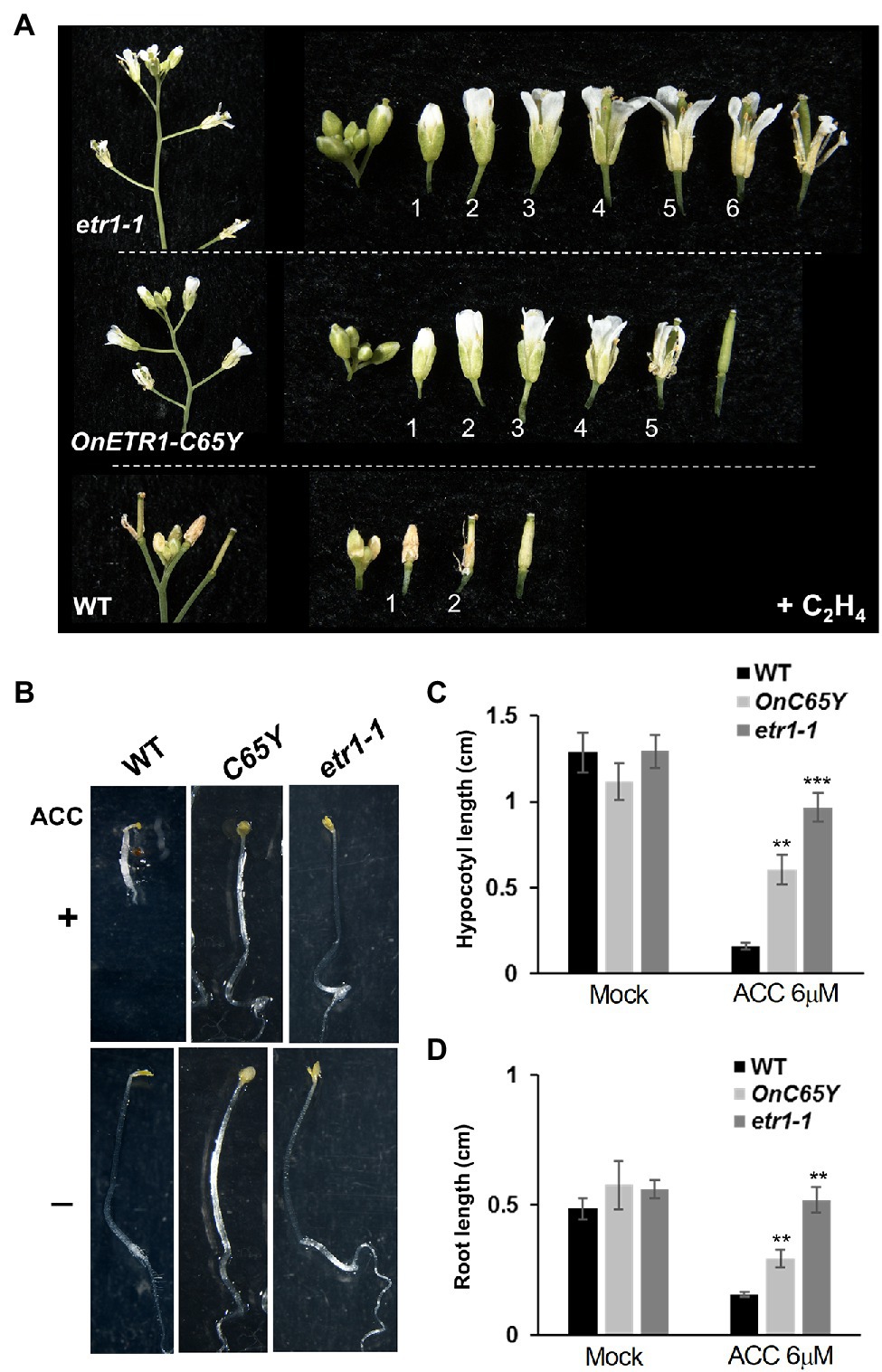
Figure 5. Ethylene treatment and triple response assay in 35S::OnETR1-C65Y and etr1-1 Arabidopsis. (A) A comparison of etr1-1 (top), 35S::OnETR1-C65Y (middle), and wild-type (bottom) flowers exposed to ethylene (+C2H4). In the wild type, the flowers were senescent at position 1, whereas etr1-1 and 35S::OnETR1-C65Y flowers were not senescent at positions 5–6 after ethylene treatment. (B) WT, 35S::OnETR1-C65Y (C65Y), and etr1-1 Arabidopsis seedlings were grown on MS plates in the presence (+, top row) or absence (−, bottom row) of 6 μM 1-aminocyclopropane-1 carboxylic acid (ACC), the ethylene precursor, for 7 days in the dark. Wild-type seedlings showed apical hooks and short hypocotyls and roots after ACC treatment. Triple response phenotypes were not observed in the etr1-1 or 35S::OnETR1-C65Y seedlings after ACC treatment. (C,D) Measurement of hypocotyl (C) and root (D) length in wild-type, 35S::OnETR1-C65Y (C65Y) and etr1-1 Arabidopsis seedlings in the presence (ACC 6 μM) or absence (Mock) of ACC. Values are the average lengths (mean ± SD) of >15 hypocotyls or roots. The error bars represent SD. The asterisks indicate a significant difference from the WT value (**p < 0.01 and ***p < 0.001). Statistical analyses were conducted using two-tailed Student’s t-tests.
The application of ethylene to etiolated seedlings results in the triple response, including a curvature of the apical hook and a shortening and thickening of the hypocotyls and roots (Guzman and Ecker, 1990; Bakshi et al., 2015). To confirm the ethylene insensitivity of 35S::OnETR1-C65Y plants, the triple response was analyzed in 35S::OnETR1-C65Y, etr1-1, and wild-type seedlings by growing seedlings in the presence or absence of 6 M ACC, the ethylene precursor. Wild-type seedlings showed apical hooks and extremely short hypocotyls and roots upon ACC treatment (Figures 5B–D). In contrast, 35S::OnETR1-C65Y and etr1-1 seedlings showed no obvious apical hooks and produced longer hypocotyls and roots than did wild-type seedlings after ACC treatment (Figures 5B–D). The lack of a triple response in 35S::OnETR1-C65Y seedlings, similar to that in etr1-1 seedlings, further confirmed that OnETR1-C65Y could cause ethylene insensitivity and thus repress ethylene signal transduction in Arabidopsis.
Similar to ethylene, JA has also been demonstrated to promote senescence in Arabidopsis (He et al., 2002). To further analyze the effect of ethylene and JA on leaf senescence in 35S::OnETR1-C65Y plants, detached leaves from 35S::OnETR1-C65Y, etr1-1 and wild-type plants were treated with ACC and JA in the dark, and the phenotypes were analyzed. The results indicated that the detached leaves of wild-type Arabidopsis were yellow and withered in the dark after 4 days (Figure 6A, second row, left) compared to the control (Figure 6A, first row, left). In addition, the wild-type leaves showed ACC- and JA-induced senescence after treatment with ACC or JA in the dark for 4 days (Figure 6A, third and fourth rows, left). In contrast, the detached leaves from 35S::OnETR1-C65Y and etr1-1 stayed green without signs of senescence upon treatment with ACC or JA in the dark for 4 days (Figure 6A, second to fourth rows, middle and right). Furthermore, chlorophyll remained significantly higher in the detached leaves from 35S::OnETR1-C65Y and etr1-1 plants than in wild-type plants after dark, ACC or JA treatments (Figure 6B). These results indicated that 35S::OnETR1-C65Y could inhibit leaf senescence by repressing the signal transduction of not only ethylene but also JA.
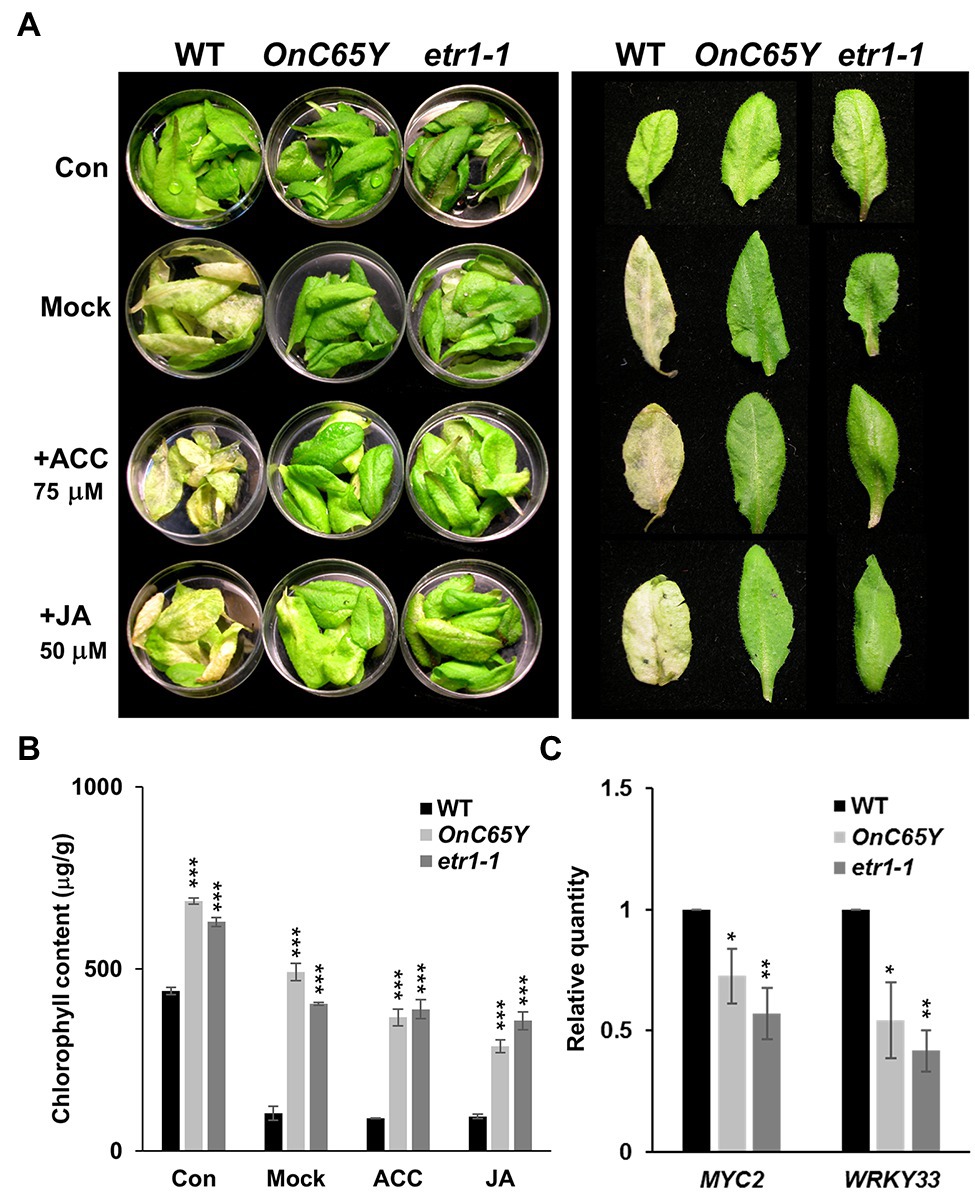
Figure 6. Assay of the effect ACC and jasmonic acid (JA) treatments on detached leaf senescence in 35S::OnETR1-C65Y and etr1-1 Arabidopsis. (A) Phenotypes of detached leaves from WT, 35S::OnETR1-C65Y (OnC65Y) and etr1-1 Arabidopsis treated with water (Mock), ACC or JA in the dark for 4 days. The control (con) did not receive a dark or ACC/JA treatment. The left panel shows the detached leaves in Petri dishes, whereas the right panel shows a single detached leaf. Detached wild-type leaves were clearly senescent, whereas detached 35S::OnETR1-C65Y and etr1-1 leaves were still green and showed no sign of senescence after ACC or JA treatment. (B) Measurement of the chlorophyll contents of the detached leaves from (A). The error bars represent SD. The asterisks indicate a significant difference from the WT value (***p < 0.001). Statistical analyses were conducted using two-tailed Student’s t-tests. (C) Detections of MYC2 and WRKY33 gene expression via real-time quantitative PCR. Total RNA isolated from 35S::OnETR1-C65Y, etr1-1 and wild-type Arabidopsis was used as a template to detect the expression of MYC2 and WRKY33. The transcript levels of these genes were determined using two to three replicates and were normalized to UBQ10. The expression of each gene in the transgenic plants is given relative to that of the wild-type plant, which was set at 1. The error bars represent SDs. The asterisks indicate a significant difference from the WT value (*p < 0.05 and **p < 0.01). Statistical analysis was conducting using two-tailed Student’s t-tests.
MYC2 and WRKY33 act as downstream genes in JA signaling and activate JA responses (Gfeller et al., 2010). Since 35S::OnETR1-C65Y and etr1-1 displayed inhibited senescence and JA insensitivity, an analysis of the expression of MYC2 and WRKY33 in 35S::OnETR1-C65Y and etr1-1 plants was performed. The results clearly indicated that MYC2 and WRKY33 expression was significantly downregulated in 35S::OnETR1-C65Y and etr1-1 plants (Figure 6C). This result supported the idea that delayed leaf and flower senescence by 35S::OnETR1-C65Y was also due to the suppression of JA signaling through the downregulation of MYC2 and WRKY33 expression.
In addition to the delay in flower senescence, 35S::OnETR1-C65Y also caused male sterility, with the production of indehiscent anthers and unelongated siliques (Figures 7A–C). Interestingly, this indehiscent anther phenotype was not observed in etr1-1 mutants, which produced dehiscent anthers and elongated siliques (Figures 7D–F). When Alexander’s staining was used, the anthers of 35S::OnETR1-C65Y Arabidopsis contained viable pollen grains (Figure 7H) that closely resembled those in wild-type (Figure 7G) and etr1-1 (Figure 7I) plants. To further examine pistil activity, wild-type pollen was crossed with stigmas of sterile 35S::OnETR1-C65Y Arabidopsis. The siliques exhibited normal elongation and produced normal seeds after cross-pollination (Figures 7J–L). The results indicated that the sterility observed in 35S::OnETR1-C65Y plants was associated with the production of indehiscent anthers.
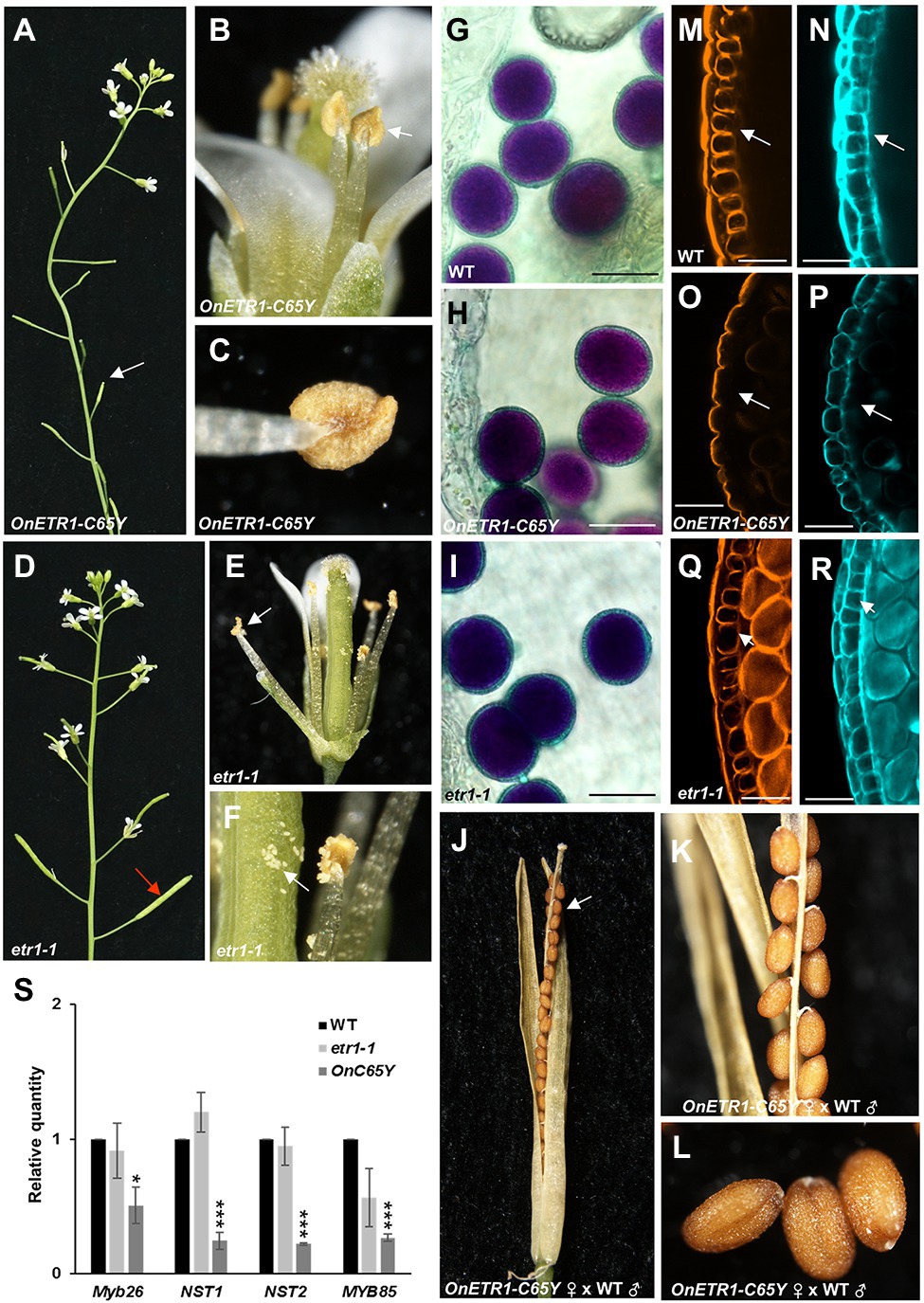
Figure 7. Analysis of the indehiscent anther phenotype and the detection of gene expression in 35S::OnETR1-C65Y Arabidopsis. (A) Inflorescence of a 35S::OnETR1-C65Y plant showing short siliques (arrow). (B) A 35S::OnETR1-C65Y flower with an indehiscent anther (arrow). (C) Close-up view of an indehiscent anther from (B). (D) Inflorescence of an etr1-1 mutant with elongated siliques (arrow). (E) An etr1-1 flower with a dehiscent anther (arrow). (F) Close-up view of a dehiscent anther and released pollen (arrow) from (E). (G–I) Pollen grain viability analysis of 35S::OnETR1-C65Y male-sterile anthers via Alexander staining. Wild-type (G), 35S::OnETR1-C65Y (H), and etr1-1 (I) Arabidopsis had similar viable pollen grains with similar shapes. Bar = 20 μm. (J) The normal elongated silique and normal seeds (arrow) of a 35S::OnETR1-C65Y plant after pollination with wild-type pollen. (K,L) Close up of the normal seeds in the silique from (J). (M–R) CLSM of anther cell walls in wild-type (M,N), 35S::OnETR1-C65Y (O,P), and etr1-1 (Q,R) Arabidopsis. The anthers were double stained with auramine O for lignin, indicated in orange (M,O,Q), and with calcofluor white for cellulose, indicated cyan (N,P,R). Secondary thickening is visible in the endothecium (arrowed) of the wild-type (M,N) and etr1-1 (Q,R) anthers but is absent in 35S::OnETR1-C65Y (O,P) anthers (arrowed). Bar = 25 μm. (S) mRNA levels as determined via real-time quantitative PCR. Total RNA isolated from wild-type, etr1-1 and 35S::OnETR1-C65Y plants was used as a template to detect the expression of MYB26, NST1/2, and MYB85. The transcript levels of these genes were determined using two to three replicates and were normalized to UBQ10. The expression of each gene in the transgenic plants is given relative to that of the wild-type plant, which was set at 1. The error bars represent SDs. The asterisks indicate a significant difference from the WT value (*p < 0.05 and ***p < 0.001). Statistical analysis was conducting using two-tailed Student’s t-tests.
To analyze the anther indehiscence phenotype in 35S::OnETR1-C65Y plants, the secondary cell wall thickness of the endothecium was further examined by double staining with auramine O for lignin, which is indicated in orange (Figures 7M,O,Q), and with calcofluor white for cellulose, which is indicated in cyan (Figures 7N,P,R). The results indicated that the secondary cell wall thickened normally in wild-type anthers, with striated lignin formation in the endothecium (Figures 7M,N). In contrast, 35S::OnETR1-C65Y plants could not develop secondary cell wall thickening, and lignin formation was absent in the endothecium (Figures 7O,P). In contrast to 35S::OnETR1-C65Y plants, in etr1-1 anthers, normal secondary cell wall thickening was also observed, with striated lignin formation in the endothecium (Figures 7Q,R). When the expression of MYB26, MYB85, and NST1/2, which have been thought to regulate secondary thickening in anther endothecium (Mitsuda et al., 2005; Zhong et al., 2008), was analyzed, all four genes were clearly downregulated in 35S::OnETR1-C65Y plants and were not affected in etr1-1 mutants (Figure 7S). These results indicated that 35S::OnETR1-C65Y caused a failure in secondary cell wall thickening due to the downregulation of MYB26/MYB85/NST1/2, which led to the production of indehiscent anthers.
The regulatory role of ethylene in plant senescence has been well characterized, and ethylene-induced senescence is a considerable physiological problem in cut Oncidium flowers. Removing the pollinia caps of Oncidium increases ethylene production and leads to flower senescence during harvest and transport (Huang et al., 2007). Thus, the control of ethylene synthesis and signal transduction genes in Oncidium is important. In this study, we identified OnACS12, an ethylene synthesis gene, and OnETR1, an ethylene signal transduction gene, from Oncidium and functionally characterized them in transgenic Arabidopsis.
The ectopic expression of OnACS12 in Arabidopsis can cause increased ethylene production and a late flowering phenotype. Ethylene regulates flowering time via GA–DELLA signaling (Achard et al., 2007). GA is perceived by GA INSENSITIVE DWARF (GID) 1 and 2, which are soluble nuclear receptors. GIDs can bind to all bioactive gibberellins and interact with DELLAs to enhance the binding of the ubiquitin E3 ligase complex SCF to DELLAs, thereby promoting DELLA degradation via the 26S proteasome pathway (Sasaki et al., 2003; Griffiths et al., 2006). The DELLA proteins include five members, GAI, RGA, and RGA-LIKE (RGL1, RGL2, and RGL3); they act as repressors of GA signaling and delay the timing of the floral transition, possibly through the downregulation of LFY (Wen and Chang, 2002; Harberd, 2003; Achard et al., 2006; Figure 8). We verified that the late flowering phenotype was correlated with the alteration of GA–DELLA signaling pathways in 35S::OnACS12 Arabidopsis. Three GA biosynthetic genes, CPS, KS, and GA3ox1, were downregulated; three DELLA genes, GAI, RGL1, and RGL2, were upregulated; and LFY expression was suppressed in 35S::OnACS12 plants. These results implied that bioactive GA levels were downregulated by decreasing gene transcripts encoding GA biosynthetic enzymes, which caused the accumulation of DELLAs, thus enhancing DELLA activity to suppress the expression of LFY and cause a delay in flowering. Thus, the ectopic expression of orchid OnACS12 extended the vegetative phase of Arabidopsis by increasing ethylene production, which sequentially influenced GA–DELLA signaling and the reduction of LFY expression.
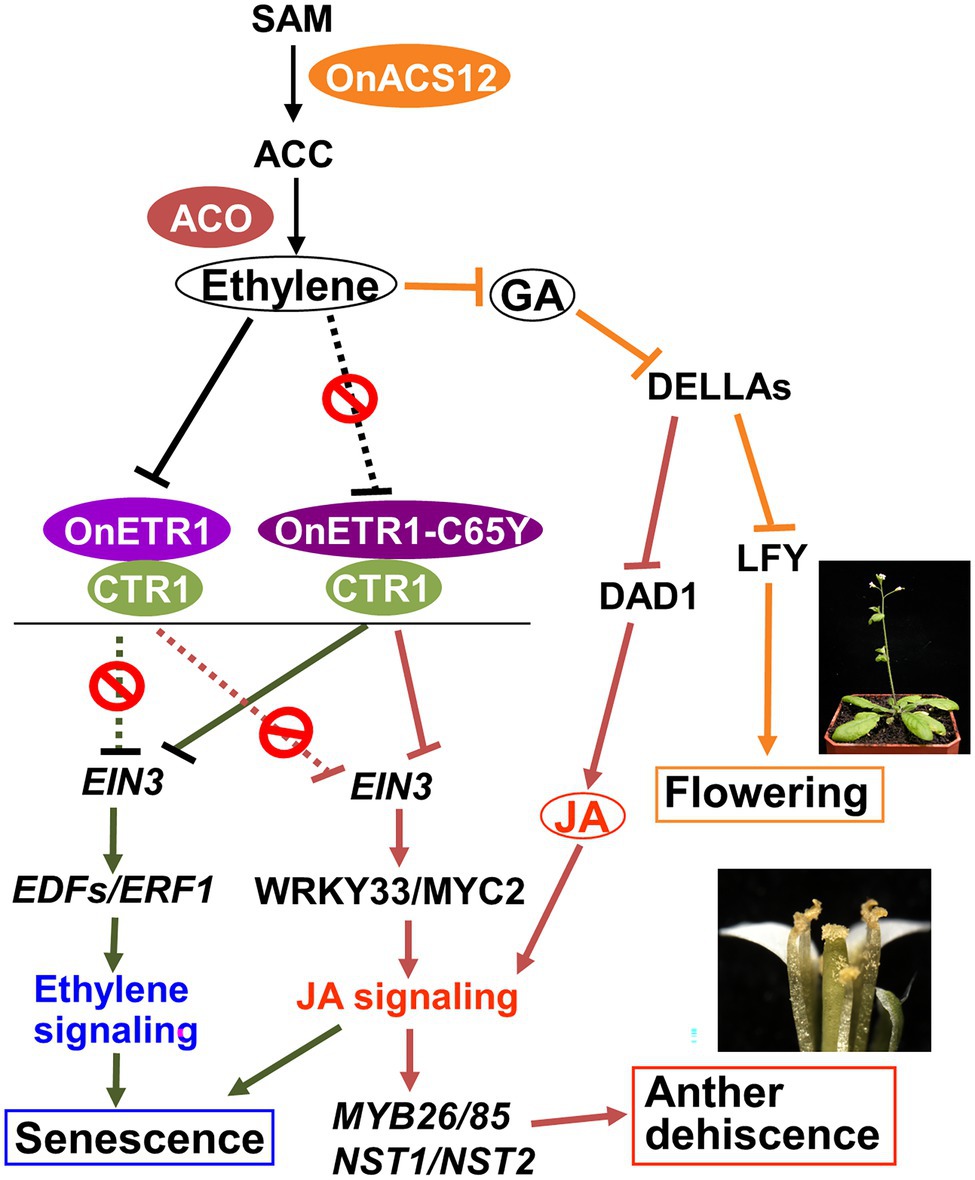
Figure 8. Model for the function of OnACS12 and OnETR1-C65Y in regulating ethylene, GA-DELLA and JA signaling in Arabidopsis. The ectopic expression of Oncidium OnACS12 delayed flowering and produced indehiscent anthers in Arabidopsis through an increase ( ) in ACC and ethylene production, which influenced GA–DELLA signaling by suppressing (
) in ACC and ethylene production, which influenced GA–DELLA signaling by suppressing ( ) GA production and activating DELLA activity. The increased DELLA activity sequentially suppressed (
) GA production and activating DELLA activity. The increased DELLA activity sequentially suppressed ( ) the expression of the flowering time gene LFY and caused late flowering phenotypes. The increased DELLA activity also suppressed (
) the expression of the flowering time gene LFY and caused late flowering phenotypes. The increased DELLA activity also suppressed ( ) the expression of the JA biosynthesis gene DAD1, which caused a decrease in JA activity, the suppression of MYB26, NST1/2, and MYB85 expression and the failure of secondary thickening in the endothecium, resulting in the production of indehiscent anthers. In wild-type plants, Oncidium OnETR1 can bind to and be suppressed (
) the expression of the JA biosynthesis gene DAD1, which caused a decrease in JA activity, the suppression of MYB26, NST1/2, and MYB85 expression and the failure of secondary thickening in the endothecium, resulting in the production of indehiscent anthers. In wild-type plants, Oncidium OnETR1 can bind to and be suppressed (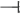 ) by ethylene, resulting in the inability (
) by ethylene, resulting in the inability (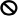 ) to activate CTR1 to (
) to activate CTR1 to (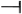 ) suppress positive regulators (i.e., EIN3) and downstream genes (EDFs and ERF1) of the ethylene response, as well as the downstream JA signaling genes MYC2 and WRKY33. This caused a normal ethylene and JA response and led to leaf/flower senescence and anther dehiscence, respectively. The ectopic expression of OnETR1-C65Y blocked (
) suppress positive regulators (i.e., EIN3) and downstream genes (EDFs and ERF1) of the ethylene response, as well as the downstream JA signaling genes MYC2 and WRKY33. This caused a normal ethylene and JA response and led to leaf/flower senescence and anther dehiscence, respectively. The ectopic expression of OnETR1-C65Y blocked (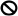 ) normal ethylene binding and sequentially activated CTR1 to suppress (
) normal ethylene binding and sequentially activated CTR1 to suppress ( ) the positive regulators (i.e., EIN3) and downstream genes (EDFs and ERF1) of the ethylene response, as well as the downstream JA signaling genes MYC2 and WRKY33, resulting in delayed leaf/flower senescence and indehiscent anthers in Arabidopsis.
) the positive regulators (i.e., EIN3) and downstream genes (EDFs and ERF1) of the ethylene response, as well as the downstream JA signaling genes MYC2 and WRKY33, resulting in delayed leaf/flower senescence and indehiscent anthers in Arabidopsis.
In addition to delaying flowering, 35S::OnACS12 also caused male sterility due to anther indehiscence in transgenic Arabidopsis. It has been reported that GA can regulate anther dehiscence by repressing DELLA proteins and sequentially activating JA biosynthetic genes such as DAD1 (Cheng et al., 2009; Peng, 2009; Marciniak and Przedniczek, 2019). JA is thought to play an important role in regulating anther dehiscence (Sanders et al., 2000; Zhao and Ma, 2000; Ishiguro et al., 2001; Scott et al., 2004), and mutations in genes that participate in JA biosynthesis, such as DAD1 and OPR3, cause a similar delay in anther dehiscence (Sanders et al., 1999, 2000; Stintzi and Browse, 2000; Scott et al., 2004). GA also regulates anther dehiscence in yellow lupine (Lupinus luteus L.) by regulating genes involved in controlling anther structure such as secondary thickening in the endothecium (Marciniak and Przedniczek, 2021). Since we have shown that 35S::OnACS12 can suppress GA levels and upregulate the accumulation of DELLA proteins, the exploration of the relationship between 35S::OnACS12 and JA biosynthesis was necessary. Unsurprisingly, the expression of the key JA biosynthetic gene DAD1 was downregulated in 35S::OnACS12 Arabidopsis. Thus, the enhancement of DELLA activity through the suppression of GA activity not only caused late flowering by suppressing LFY expression but also caused anther indehiscence by suppressing DAD1 expression and JA activity in 35S::OnACS12 Arabidopsis (Figure 8). In addition to the downregulation of the JA biosynthetic gene DAD1, the expression of MYB26 and NST1/NST2, which are thought to regulate secondary thickening in the anther endothecium (Mitsuda et al., 2005; Zhong et al., 2008), was clearly downregulated in 35S::OnACS12 Arabidopsis. The Arabidopsis myb26 mutant exhibits anther indehiscence, and secondary thickening does not occur in the endothecium (Yang et al., 2007). NST1 and NST2, which are thought to act downstream of MYB26, are associated with secondary cell wall thickening and endothecium lignification. Thus, once ectopically expressed in Arabidopsis, orchid OnACS12 can increase ethylene production, which affects GA–DELLA signaling and lowers JA activity, resulting in the failure of secondary thickening in the anther endothecium and the production of an anther indehiscence phenotype.
In this study of the Oncidium OnETR1 gene, the dominant-negative mutation OnETR1-C65Y, which could potentially prevent ethylene binding and block ethylene signaling (Wilkinson et al., 1997; Rodriguez et al., 1999), was ectopically expressed in Arabidopsis. Unsurprisingly, a significant delay in flower senescence was observed in 35S::OnETR1-C65Y Arabidopsis, which was similar to the phenotype observed in the etr1-1 mutant and in 35S::AtETR1-C65Y transgenic tomato, petunia (Wilkinson et al., 1997) and tobacco (Yang et al., 2008). Similar to the etr1-1 mutant, 35S::OnETR1-C65Y flowers also showed insensitivity to ethylene treatment, and seedlings lacked the triple response phenotype after exposure to an external supply of ACC. In addition, detached leaves from 35S::OnETR1-C65Y and etr1-1 did not show senescence after ACC treatment. Thus, our results demonstrated that orchid OnETR1 has the same ability as Arabidopsis AtETR1 to act as an ethylene receptor in transgenic Arabidopsis. The ectopic expression of OnETR1-C65Y in Arabidopsis altered ethylene binding and suppressed ethylene signaling, which downregulated the expression of the ethylene signal transduction genes EDFs and ERF1 (Stepanova and Alonso, 2005; Chen et al., 2021) and resulted in an ethylene insensitive dominant-negative mutant phenotype, such as a delay in flower/leaf senescence (Figure 8).
One interesting result is that detached leaves from 35S::OnETR1-C65Y and the etr1-1 mutant also showed JA insensitivity since the detached leaves did not senesce after JA treatment. JA plays a role in leaf senescence (Ueda and Kato, 1980), and EIN3 in ethylene signaling is required for JA-induced leaf senescence (Li et al., 2013). Thus, 35S::OnETR1-C65Y should also be able to suppress JA signaling due to the suppression of ethylene signaling and result in ethylene/JA insensitivity. This assumption was further supported by the downregulation of the downstream JA signaling genes MYC2 and WRKY33 (Gfeller et al., 2010) in 35S::OnETR1-C65Y Arabidopsis. Thus, our results demonstrated that ethylene signaling can regulate some ethylene responses, such as senescence, through the activation of JA signal transduction in plants (Figure 8). This finding could be used to explain the additional indehiscent anther phenotype observed in transgenic 35S::OnETR1-C65Y Arabidopsis. As described above, JA can regulate anther dehiscence (Sanders et al., 2000; Zhao and Ma, 2000; Ishiguro et al., 2001; Scott et al., 2004) since mutations in genes that participate not only in JA biosynthesis (Sanders et al., 1999, 2000; Stintzi and Browse, 2000; Scott et al., 2004) but also in JA signaling, such as coronatine insensitive 1 (coi1), cause similar anther indehiscence phenotypes (Feys et al., 1994; Xie et al., 1998). It is thus reasonable to propose that 35S::OnETR1-C65Y suppressed ethylene signaling, which downregulated the JA signaling genes MYC2 and WRKY33 and resulted in the suppression of MYB26/MYB85/NST1/2 expression and the anther indehiscence phenotype, as seen in our results (Figure 8). In contrast, in wild-type plants, the normal binding of ethylene to AtETR1/OnETR1 activated ethylene signaling, which activated the expression of the JA signal transduction genes MYC2/WRKY33 and MYB26/MYB85/NST1/2 and resulted in normal anther dehiscence. This result further confirmed the crosstalk between ethylene and JA in regulating both flower/leaf senescence and anther dehiscence in plants (Figure 8).
Notably, the anther indehiscence phenotype was not seen in etr1-1 mutants. Despite a similar delayed flower senescence phenotype as that of 35S::OnETR1-C65Y plants, etr1-1 mutants exhibited normal anther dehiscence and pollen release. This phenotype was confirmed based on the normal expression of MYB26/MYB85/NST1/2 in etr1-1 mutant flowers. One possible explanation is that ETR1 does not normally function or has a smaller role among ethylene receptors during anther development. In etr1-1 mutants, other ethylene receptors (ERS1, ETR2, ERS2, and EIN4) may still interact with ethylene to sequentially activate ethylene signaling and responses, resulting in the activation of JA signaling and normal anther dehiscence. In 35S::OnETR1-C65Y plants, the ectopic expression of OnETR1-C65Y during anther development generated dominant-negative mutations for all ethylene receptors, which blocked the binding of ethylene and caused the sequential suppression of ethylene signaling/response and JA signaling, resulting in the production of indehiscent anthers.
In summary, the Oncidium ethylene synthesis gene OnACS12 and the ethylene signal transduction gene OnETR1 were functionally characterized in transgenic Arabidopsis. We found that OnACS12 and OnETR1 could perform their functions in the ethylene response by interacting with the plant hormones GA and JA. The ectopic expression of OnACS12 could delay flowering via the regulation of GA–DELLA signaling pathways and LFY expression. OnETR1-C65Y could regulate flower/leaf senescence through the suppression of ethylene and JA signaling. Most interestingly, both OnACS12 and OnETR1-C65Y could regulate anther dehiscence by downregulating the JA biosynthetic gene DAD1 or by suppressing JA signaling, respectively. The characterization of the OnACS12 and OnETR1 genes in this study not only provides useful data for understanding the functions of ACS and ETR1 orthologs in regulating ethylene/GA/JA responses during various plant developmental processes but also provides a feasible future strategy to control flower senescence in Oncidium orchids by introducing a dominant-negative OnETR1-C65Y mutation or to generate male sterility for valuable crops through the ectopic expression of either OnACS12 or OnETR1-C65Y.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
C-HY developed the overall strategy, designed experiments, coordinated the project, and prepared and revised the manuscript. T-HH performed gene cloning, transgenic plants, and gene expression experiments. W-HH performed Alexander, Calcofluor White, and Auramine O staining. W-TM performed orchid gene expression analyses. All authors contributed to the article and approved the submitted version.
This work was supported by grants to C-HY from the Ministry of Science and Technology, Taiwan, grant number: MOST 103-2313-B-005-001-MY3 and MOST 106-2321-B-005-010. This work was also financially supported (in part) by the Advanced Plant Biotechnology Center from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.785441/full#supplementary-material
Achard, P., Baghour, M., Chapple, A., Hedden, P., Van Der Straeten, D., Genschik, P., et al. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. U. S. A. 104, 6484–6489. doi: 10.1073/pnas.0610717104
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. doi: 10.1126/science.1118642
Adams, D. O., and Yang, S. F. (1979). Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. U. S. A. 76, 170–174. doi: 10.1073/pnas.76.1.170
Alexander, L., and Grierson, D. (2002). Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 53, 2039–2055. doi: 10.1093/jxb/erf072
Almasi, P., and Mohamed, M. T. M. (2020). Ethylene sensitivity in orchid flowers and its management using 1-MCP: a review. Fund. Appl. Agricult. 5, 10–20. doi: 10.5455/faa.80674
Alonso, J. M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J. R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. doi: 10.1126/science.284.5423.2148
Alonso, J. M., and Stepanova, A. N. (2004). The ethylene signaling pathway. Science 306, 1513–1515. doi: 10.1126/science.1104812
Arc, E., Sechet, J., Corbineau, F., Rajjou, L., and Marion-Poll, A. (2013). ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 4:63. doi: 10.3389/fpls.2013.00063
Azhar, B. J., Zulfiqar, A., Shakeel, S. N., and Schaller, G. E. (2019). Amplification and adaptation in the ethylene signaling pathway. Small Methods 4:1900452. doi: 10.1002/smtd.201900452
Bakshi, A., Shemansky, J. M., Chang, C. R., and Binder, B. M. (2015). History of research on the plant hormone ethylene. J. Plant Growth Regul. 34, 809–827. doi: 10.1007/s00344-015-9522-9
Binder, B. M. (2020). Ethylene signaling in plants. J. Biol. Chem. 295, 7710–7725. doi: 10.1074/jbc.REV120.010854
Bleecker, A. B. (1999). Ethylene perception and signaling: an evolutionary perspective. Trends Plant Sci. 4, 269–274. doi: 10.1016/S1360-1385(99)01427-2
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Bonner, L., and Dickinson, H. (1989). Anther dehiscence in Lycopersicon esculentum mill. New Phytol. 113, 97–115. doi: 10.1111/j.1469-8137.1989.tb02399.x
Booker, M. A., and DeLong, A. (2015). Producing the ethylene signal: regulation and diversification of ethylene biosynthetic enzymes. Plant Physiol. 169, 42–50. doi: 10.1104/pp.15.00672
Capitani, G., Hohenester, E., Feng, L., Storici, P., Kirsch, J. F., and Jansonius, J. N. (1999). Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene. J. Mol. Biol. 294, 745–756. doi: 10.1006/jmbi.1999.3255
Cecchetti, V., Altamura, M. M., Brunetti, P., Petrocelli, V., Falasca, G., Ljung, K., et al. (2013). Auxin controls Arabidopsis anther dehiscence by regulating endothecium lignification and jasmonic acid biosynthesis. Plant J. 74, 411–422. doi: 10.1111/tpj.12130
Chang, Y. Y., Kao, N. H., Li, J. Y., Hsu, W. H., Liang, Y. L., Wu, J. W., et al. (2010). Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol. 152, 837–853. doi: 10.1104/pp.109.147116
Chang, C., and Shockey, J. A. (1999). The ethylene-response pathway: signal perception to gene regulation. Curr. Opin. Plant Biol. 2, 352–358. doi: 10.1016/S1369-5266(99)00004-7
Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J. R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144. doi: 10.1016/S0092-8674(00)80300-1
Chen, Q. G., and Bleecker, A. B. (1995). Analysis of ethylene signal-transduction kinetics associated with seedling-growth response and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiol. 108, 597–607. doi: 10.1104/pp.108.2.597
Chen, M. K., Hsu, W. H., Lee, P. F., Thiruvengadam, M., Chen, H. I., and Yang, C. H. (2011). The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J. 68, 168–185. doi: 10.1111/j.1365-313X.2011.04677.x
Chen, W. H., Li, P. F., Chen, M. K., Lee, Y. I., and Yang, C. H. (2015). FOREVER YOUNG FLOWER negatively regulates ethylene response DNA-binding factors by activating an ethylene-responsive factor to control Arabidopsis floral organ senescence and abscission. Plant Physiol. 168, 1666–1683. doi: 10.1104/pp.15.00433
Chen, Y., Zhang, L., Zhang, H., Chen, L., and Yu, D. (2021). ERF1 delays flowering through direct inhibition of FLOWERING LOCUS T expression in Arabidopsis. J. Integr. Plant Biol. 63, 1712–1723. doi: 10.1111/jipb.13144
Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D. E., Cao, D., et al. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064. doi: 10.1242/dev.00992
Cheng, H., Song, S., Xiao, L., Soo, H. M., Cheng, Z., Xie, D., et al. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5:e1000440. doi: 10.1371/journal.pgen.1000440
Clough, S., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Corbineau, F., Xia, Q., Bailly, C., and El-Maarouf-Bouteau, H. (2014). Ethylene, a key factor in the regulation of seed dormancy. Front. Plant Sci. 5:539. doi: 10.3389/fpls.2014.00539
Dar, R. A., Nisar, S., and Tahir, I. (2021). Ethylene: a key player in ethylene sensitive flower senescence: a review. Sci. Hortic. 290:110491. doi: 10.1016/j.scienta.2021.110491
Datta, R., Kumar, D., Sultana, A., Hazra, S., Bhattacharyya, D., and Chattopadhyay, S. (2015). Glutathione regulates 1-aminocyclopropane-1-carboxylate synthase transcription via WRKY33 and 1-aminocyclopropane-1-carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress. Plant Physiol. 169, 2963–2981. doi: 10.1104/pp.15.01543
Dill, A., and Sun, T. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159, 777–785. doi: 10.1093/genetics/159.2.777
Feng, G., Sanderson, B. J., Keefover-Ring, K., Liu, J., Ma, T., Yin, T., et al. (2020). Pathways to sex determination in plants: how many roads lead to Rome? Curr. Opin. Plant Biol. 54, 61–68. doi: 10.1016/j.pbi.2020.01.004
Feys, B. J., Benedetti, C. E., Penfold, C. N., and Turner, J. G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. doi: 10.2307/3869877
Gfeller, A., Liechti, R., and Farmer, E. E. (2010). Arabidopsis jasmonate signaling pathway. Sci. Signal. 3:cm4. doi: 10.1126/scisignal.3109cm4
Graham, L. E., Schippers, J. H. M., Dijkwel, P. P., and Wagstaff, C. (2012). Ethylene and senescence processes. Annu. Plant Rev. 44, 305–341. doi: 10.1002/9781118223086.ch12
Grbic, V., and Bleecker, A. B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8, 595–602. doi: 10.1046/j.1365-313X.1995.8040595.x
Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z. L., Powers, S. J., et al. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18, 3399–3414. doi: 10.1105/tpc.106.047415
Guo, H., and Ecker, J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7, 40–49. doi: 10.1016/j.pbi.2003.11.011
Guzman, P., and Ecker, J. R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523. doi: 10.1105/tpc.2.6.513
Hamilton, A. J., Bouzayen, M., and Grierson, D. (1991). Identification of a tomato gene for the ethylene-forming enzyme by expression in yeast. Proc. Natl. Acad. Sci. U. S. A. 88, 7434–7437. doi: 10.1073/pnas.88.16.7434
Harberd, N. P. (2003). Relieving DELLA restraint. Science 299, 1853–1854. doi: 10.1126/science.1083217
He, Y., Fukushige, H., Hildebrand, D. F., and Gan, S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128, 876–884. doi: 10.1104/pp.010843
Houben, M., and Van de Poel, B. (2019). 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO): the enzyme that makes the plant hormone ethylene. Front. Plant Sci. 10:695. doi: 10.3389/fpls.2019.00695
Huang, W.-F., Huang, P.-L., and Do, Y.-Y. (2007). Ethylene receptor transcript accumulation patterns during flower senescence in Oncidium ‘Gower Ramsey’ as affected by exogenous ethylene and pollinia cap dislodgment. Postharvest Biol. Technol. 44, 87–94. doi: 10.1016/j.postharvbio.2006.12.012
Huang, Y., Li, H., Hutchison, C. E., Laskey, J., and Kieber, J. J. (2003). Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 33, 221–233. doi: 10.1046/j.1365-313X.2003.01620.x
Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchoronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209. doi: 10.1105/tpc.010192
Ju, C., and Chang, C. (2015). Mechanistic insights in ethylene perception and signal transduction 1. Plant Physiol. 169, 85–95. doi: 10.1104/pp.15.00845
Ju, C., Van de Poel, B., Cooper, E. D., Thierer, J. H., Gibbons, T. R., Delwiche, C. F., et al. (2015). Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants 1:14004. doi: 10.1038/nplants.2014.4
Kieber, J. J., Rothenberg, M., Roman, G., Feldmann, K. A., and Ecker, J. R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441. doi: 10.1016/0092-8674(93)90119-B
Kim, J., Wilson, R. L., Case, J. B., and Binder, B. M. (2012). A comparative study of ethylene growth response kinetics in eudicots and monocots reveals a role for gibberellin in growth inhibition and recovery. Plant Physiol. 160, 1567–1580. doi: 10.1104/pp.112.205799
Lerslerwong, L., and Ketsa, S. (2008). Autocatalytic ethylene production by Dendrobium flowers during senescence induced by exogenous ethylene. Thai J. Agricul. Sci. 41, 91–99.
Li, X., Chen, T., Li, Y., Wang, Z., Cao, H., Chen, F., et al. (2019). ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression. Plant Cell 31, 832–847. doi: 10.1105/tpc.18.00449
Li, Z., Peng, J., Wen, X., and Guo, H. (2013). ETHYLENE-INSENSITIVE3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25, 3311–3328. doi: 10.1105/tpc.113.113340
Liu, S., and Chen, H. (2021). Ethylene signaling facilitates plant adaption to physical barriers. Front. Plant Sci. 12:697988. doi: 10.3389/fpls.2021.697988
Liu, M., Pirrello, J., Chervin, C., Roustan, J. P., and Bouzayen, M. (2015). Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol. 169, 2380–2390. doi: 10.1104/pp.15.01361
Mao, J. L., Miao, Z. Q., Wang, Z., Yu, L. H., Cai, X. T., and Xiang, C. B. (2016). Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet. 12:e1005760. doi: 10.1371/journal.pgen.1005760
Marciniak, K., and Przedniczek, K. (2019). Comprehensive insight into gibberellin- and jasmonate-mediated stamen development. Gene 10:811. doi: 10.3390/genes10100811
Marciniak, K., and Przedniczek, K. (2021). Anther dehiscence is regulated by gibberellic acid in yellow lupine (Lupinus luteus L.). BMC Plant Biol. 21:314. doi: 10.1186/s12870-021-03085-4
Mccarthy, D. L., Capitani, G., Feng, L., Gruetter, M. G., and Kirsch, J. F. (2001). Glutamate 47 in 1-aminocyclopropane-1-carboxylate synthase is a major specificity determinant. Biochemistry 40, 12276–12284. doi: 10.1021/bi011050z
Merchante, C., Alonso, J. M., and Stepanova, A. N. (2013). Ethylene signaling: simple ligand, complex regulation. Curr. Opin. Plant Biol. 16, 554–560. doi: 10.1016/j.pbi.2013.08.001
Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. doi: 10.1105/tpc.105.036004
Mou, W., Kao, Y.-T., Michard, E., Simon, A. A., Li, D., Wudick, M. M., et al. (2020). Ethylene-independent signaling by the ethylene precursor ACC in Arabidopsis ovular pollen tube attraction. Nat. Commun. 11:4082. doi: 10.1038/s41467-020-17819-9
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–476. doi: 10.1111/j.1399-3054.1962.tb08052.x
Park, C. H., Roh, J., Youn, J. H., Son, S.-H., Park, J. H., Kim, S. Y., et al. (2018). Arabidopsis ACC oxidase 1 coordinated by multiple signals mediates ethylene biosynthesis and is involved in root development. Mol. Cell 41, 923–932. doi: 10.14348/molcells.2018.0092
Parkinson, J. S., and Kofoid, E. C. (1992). Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26, 71–112. doi: 10.1146/annurev.ge.26.120192.000443
Patterson, S. E., and Bleecker, A. B. (2004). Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol. 134, 194–203. doi: 10.1104/pp.103.028027
Pattyn, J., Vaughan-Hirsch, J., and Van de Poel, B. (2021). The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 229, 770–782. doi: 10.1111/nph.16873
Peng, J. (2009). Gibberellin and jasmonate crosstalk during stamen development. J. Integr. Plant Biol. 51, 1064–1070. doi: 10.1111/j.1744-7909.2009.00881.x
Peng, J., Carol, P., Richards, D. E., King, K. E., Cowling, R. J., Murphy, G. P., et al. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205. doi: 10.1101/gad.11.23.3194
Perata, P. (2020). Ethylene signaling controls fast oxygen sensing in plants. Trends Plant Sci. 25, 3–6. doi: 10.1016/j.tplants.2019.10.010
Rodrigues, M. A., Bianchetti, R. E., and Freschi, L. (2014). Shedding light on ethylene metabolism in higher plants. Front. Plant Sci. 5:665. doi: 10.3389/fpls.2014.00665
Rodriguez, F. I., Esch, J. J., Hall, A. E., Binder, B. M., Schaller, G. E., and Bleecker, A. B. (1999). A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283, 996–998. doi: 10.1126/science.283.5404.996
Ruzicka, K., Ljung, K., Vanneste, S., Podhorská, R., Beeckman, T., Friml, J., et al. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212. doi: 10.1105/tpc.107.052126
Sanders, P. M., Bui, A. Q., Weterings, K., McIntire, K. N., Hsu, Y. C., Lee, P. Y., et al. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. doi: 10.1007/s004970050158
Sanders, P. M., Lee, P. Y., Biesgen, C., Boone, J. D., Beals, T. P., Weiler, E. W., et al. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12, 1041–1061. doi: 10.1105/tpc.12.7.1041
Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., et al. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898. doi: 10.1126/science.1081077
Schaller, G. E., and Bleecker, A. B. (1995). Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270, 1809–1811. doi: 10.1126/science.270.5243.1809
Schaller, G. E., Ladd, A. N., Lanahan, M. B., Spanbauer, J. M., and Bleecker, A. B. (1995). The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J. Biol. Chem. 270, 12526–12530. doi: 10.1074/jbc.270.21.12526
Scott, J. R., Spielman, M., and Dickinson, H. G. (2004). Stamen structure and function. Plant Cell 16, S46–S60. doi: 10.1105/tpc.017012
Sharma, K., Gupta, S., Sarma, S., Rai, M., Sreelakshmi, Y., and Sharma, R. (2021). Mutations in tomato 1-aminocyclopropane carboxylic acid synthase2 uncover its role in development beside fruit ripening. Plant J. 106, 95–112. doi: 10.1111/tpj.15148
Silverstone, A. L., Ciampaglio, C. N., and Sun, T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169. doi: 10.1105/tpc.10.2.155
Stepanova, A. N., and Alonso, J. M. (2005). Arabidopsis ethylene signaling pathway. Sci. STKE 276:cm4. doi: 10.1126/stke.2762005cm4
Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. U. S. A. 97, 10625–10630. doi: 10.1073/pnas.190264497
Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G. T. S., Sandberg, G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196. doi: 10.1105/tpc.107.052100
Tsuchisaka, A., Yu, G., Jin, H., Alonso, J. M., Ecker, J. R., Zhang, X., et al. (2009). A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 183, 979–1003. doi: 10.1534/genetics.109.107102
Tyler, L., Thomas, S. G., Hu, J., Dill, A., Alonso, J. M., Ecker, J. R., et al. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135, 1008–1019. doi: 10.1104/pp.104.039578
Ueda, J., and Kato, J. (1980). Identification of a senescence-promoting substance from wormwood (Artemisia absinthum L.). Plant Physiol. 66, 246–249. doi: 10.1104/pp.66.2.246
Van de Poel, B., Smet, D., and Van Der Straeten, D. (2015). Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol. 169, 61–72. doi: 10.1104/pp.15.00724
Van Der Straeten, D., Djudzman, A., Van Caeneghem, W., Smalle, J., and Van Montagu, M. (1993). Genetic and physiological analysis of a new locus in Arabidopsis that confers resistance to 1-aminocyclopropane-1-carboxylic acid and ethylene and specifically affects the ethylene signal transduction pathway. Plant Physiol. 102, 401–408. doi: 10.1104/pp.102.2.401
Vanderstraeten, L., Depaepe, T., Bertrand, S., and Van Der Straeten, D. (2019). The ethylene precursor ACC affects early vegetative development independently of ethylene signaling. Front. Plant Sci. 10:1591. doi: 10.3389/fpls.2019.01591
Wang, L., Liu, H., Yin, Y., Li, Y., Lu, C., and Wang, Q. (2022). A novel guanine elicitor stimulates immunity in Arabidopsis and rice by ethylene and jasmonic acid signaling pathways. Front. Plant Sci. 13:841228. doi: 10.3389/fpls.2022.841228
Wen, C. K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14, 87–100. doi: 10.1105/tpc.010325
Wilkinson, J. Q., Lanahan, M. B., Clark, D. G., Bleeker, A. B., and Chang, C. (1997). A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat. Biotechnol. 15, 444–447. doi: 10.1038/nbt0597-444
Woeste, K., Ye, C., and Kieber, J. (1999). Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Physiol. 119, 521–530. doi: 10.1104/pp.119.2.521
Woltering, E. J. (1990). Interrelationship between the different flower parts during emasculation-induced senescence in cymbidium flowers. J. Exp. Bot. 41, 1021–1029. doi: 10.1093/jxb/41.8.1021
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Yamagami, T., Tsuchisaka, A., Yamada, K., Haddon, W. F., Harden, L. A., and Theologis, A. (2003). Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 278, 49102–49112. doi: 10.1074/jbc.M308297200
Yang, T. F., Gonzalez-Carranza, Z. H., Maunders, M. J., and Roberts, J. A. (2008). Ethylene and the regulation of senescence processes in transgenic Nicotiana sylvestris plants. Ann. Bot. 101, 301–310. doi: 10.1093/aob/mcm229
Yang, C., Lu, X., Ma, B., Chen, S. Y., and Zhang, J. S. (2015). Ethylene signaling in rice and Arabidopsis: conserved and diverged aspects. Mol. Plant 8, 495–505. doi: 10.1016/j.molp.2015.01.003
Yang, C., Xu, Z., Song, J., Conner, K., Barrena, G. V., and Wilson, Z. A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19, 534–548. doi: 10.1105/tpc.106.046391
Yin, J., Zhang, X., Zhang, G., Wen, Y., Liang, G., and Chen, X. (2019). Aminocyclopropane-1-carboxylic acid is a key regulator of guard mother cell terminal division in Arabidopsis thaliana. J. Exp. Bot. 70, 897–908. doi: 10.1093/jxb/ery413
Yoo, S. D., Cho, Y., and Sheen, J. (2009). Emerging connections in the ethylene signaling network. Trends Plant Sci. 14, 270–279. doi: 10.1016/j.tplants.2009.02.007
Young, T. E., Meeley, R. B., and Gallie, D. R. (2004). ACC synthase expression regulates leaf performance and drought tolerance in maize. Plant J. 40, 813–825. doi: 10.1111/j.1365-313X.2004.02255.x
Zhang, L., Chen, L., and Yu, D. (2018). Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 176, 790–803. doi: 10.1104/pp.17.00657
Zhao, D., and Ma, H. (2000). Male fertility: a case of enzyme identity. Curr. Biol. 10, R904–R907. doi: 10.1016/S0960-9822(00)00848-4
Zhao, H., Yin, C.-C., Ma, B., Chen, S.-Y., and Zhang, J.-S. (2020). Ethylene signaling in rice and Arabidopsis: new regulators and mechanisms. J. Integr. Plant Biol. 63, 102–125. doi: 10.1111/jipb.13028
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L., and Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Keywords: anther dehiscence, ACC synthase, ethylene response 1, ethylene signaling, Oncidium orchids, senescence
Citation: Huang T-H, Hsu W-H, Mao W-T and Yang C-H (2022) The Oncidium Ethylene Synthesis Gene Oncidium 1-Aminocyclopropane-1 Carboxylic Acid Synthase 12 and Ethylene Receptor Gene Oncidium ETR1 Affect GA–DELLA and Jasmonic Acid Signaling in Regulating Flowering Time, Anther Dehiscence, and Flower Senescence in Arabidopsis. Front. Plant Sci. 13:785441. doi: 10.3389/fpls.2022.785441
Received: 29 September 2021; Accepted: 07 March 2022;
Published: 01 April 2022.
Edited by:
Chris Helliwell, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaReviewed by:
Xin Zhou, Shanghai Normal University, ChinaCopyright © 2022 Huang, Hsu, Mao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang-Hsien Yang, Y2h5YW5nQGRyYWdvbi5uY2h1LmVkdS50dw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.