
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 03 June 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.777332
Plant Cellulose synthase genes constitute a supergene family that includes the Cellulose synthase (CesA) family and nine Cellulose synthase-like (Csl) families, the members of which are widely involved in the biosynthesis of cellulose and hemicellulose. However, little is known about the Cellulose synthase superfamily in the family Orchidaceae, one of the largest families of angiosperms. In the present study, we identified and systematically analyzed the CesA/Csl family members in three fully sequenced Orchidaceae species, i.e., Dendrobium officinale, Phalaenopsis equestris, and Apostasia shenzhenica. A total of 125 Cellulose synthase superfamily genes were identified in the three orchid species and classified into one CesA family and six Csl families: CslA, CslC, CslD, CslE, CslG, and CslH according to phylogenetic analysis involving nine representative plant species. We found species-specific expansion of certain gene families, such as the CslAs in D. officinale (19 members). The CesA/Csl families exhibited sequence divergence and conservation in terms of gene structure, phylogeny, and deduced protein sequence, indicating multiple origins via different evolutionary processes. The distribution of the DofCesA/DofCsl genes was investigated, and 14 tandemly duplicated genes were detected, implying that the expansion of DofCesA/DofCsl genes may have originated via gene duplication. Furthermore, the expression profiles of the DofCesA/DofCsl genes were investigated using transcriptome sequencing and quantitative Real-time PCR (qRT-PCR) analysis, which revealed functional divergence in different tissues and during different developmental stages of D. officinale. Three DofCesAs were highly expressed in the flower, whereas DofCslD and DofCslC family genes exhibited low expression levels in all tissues and at all developmental stages. The 19 DofCslAs were differentially expressed in the D. officinale stems at different developmental stages, among which six DofCslAs were expressed at low levels or not at all. Notably, two DofCslAs (DofCslA14 and DofCslA15) showed significantly high expression in the stems of D. officinale, indicating a vital role in mannan synthesis. These results indicate the functional redundancy and specialization of DofCslAs with respect to polysaccharide accumulation. In conclusion, our results provide insights into the evolution, structure, and expression patterns of CesA/Csl genes and provide a foundation for further gene functional analysis in Orchidaceae and other plant species.
cellulose, hemicellulose, and pectin are three major types of polysaccharides in plant cell walls and play vital roles in controlling cell shape, expansion, overall development, and interactions with the environment (McFarlane et al., 2014). cellulose is a paracrystalline polymer of β-1,4 glucan chains and is the major determinant of the load-bearing capacity of cell walls constituting approximately one-third of the total plant mass (Nishiyama, 2009; McFarlane et al., 2014). Hemicelluloses are polysaccharides that have equatorial β-(1→4)-linked backbones, including xyloglucans, xylans, mannans, glucomannans, and β-(1→3,1→4)-glucans (Scheller and Ulvskov, 2010). Hemicellulose and pectin polysaccharide matrix fill in the gaps between cellulose microfibrils.
The Cellulose synthase (CesA) superfamily comprises the CesA family and nine CesA-like (Csl) families, which all belong to the glycosyltransferase-2 (GT2) superfamily, typically with a catalytic domain containing a DDDQXXRW motif (Pear et al., 1996; Richmond and Somerville, 2000). The number of CesA genes is rather stable among plant species, usually around ten members; however, these genes underwent expansion, which then had 18 genes (Yin et al., 2009). Csl family members also underwent large gene expansion, from the presence of a single CslA/C-like gene in green algae to up to 50 genes in terrestrial plant species. The expansion of plant Cellulose synthase genes is closely linked to major events in the evolution of plant and algal lineages, including multicellularity, terrestrialization, and vascularization (Popper et al., 2011). cellulose is synthesized by plasma membrane-localized Cellulose synthase complexes (CSCs) with access to the GDP-glucose pool in the cytosol. In plants, the first CesA gene in cotton (Gossypium hirsutum) fibers was identified based on the sequence similarity to a bacterial CesA gene (Pear et al., 1996). To date, CesA/Csl family genes have been identified in many plant species, including Arabidopsis (Persson et al., 2007), tomato (Song et al., 2018), rice (Shu et al., 2007), sorghum (Paterson et al., 2009), Populus trichocarpa (Takata and Taniguchi, 2015), Pyrus bretschneideri (Li et al., 2020), and cotton (Li et al., 2017; Cui et al., 2020). Ten CesA genes are present in the Arabidopsis genome and are required for primary (CesA1−3, −5, −6, and −9) and secondary (CesA4, −7, and −8) cell wall synthesis (Taylor et al., 2003; Takata and Taniguchi, 2015). Mutations of cesa1 and cesa3 are gamete lethal, whereas single-knockout mutants of the other primary cell wall-related CesA genes exhibit more moderate phenotypes (Desprez et al., 2007; Persson et al., 2007).
Hemicellulose is synthesized in the Golgi apparatus by membrane-localized enzymes encoded by Cellulose synthase-like (Csl) genes, which share relatively high sequence similarity to those of CesA family genes. In plants, the Csl families usually comprise 30–50 members, which are classified into nine subgroups: CslA-CslH and CslJ (Keegstra and Walton, 2006; Fincher, 2009; Yin et al., 2014). Of these, CslA, CslC, and CslD are commonly found in all land plants, and CslE and CslJ are common to angiosperms (Yin et al., 2009, 2014; Popper et al., 2011). CslH and CslF are specific to grasses, whereas CslB and CslG are found in non-grass angiosperms (Keegstra and Walton, 2006; Suzuki et al., 2006; Popper et al., 2011; Yin et al., 2014). Csl family genes are reported to be involved in the synthesis of various cell wall polysaccharides (Kaur et al., 2017). CslAs encode proteins with both mannan and glucomannan synthase activity (Liepman et al., 2010). In Arabidopsis, the stems of csla-knockout mutants have no glucomannan, indicating that CslA family genes have an exclusive role in mannan biosynthesis (Goubet et al., 2009). The members of the CslC family are involved in the synthesis of the β-1,4-linked glucan backbone of xyloglucan in Tropaeolum majus (Cocuron et al., 2007). CslDs in Arabidopsis are involved in mannan and Cellulose synthesis, especially in tip-growing root hairs and pollen tubes (Park et al., 2011). In tobacco, CslD-overexpressing plants have high mannan synthase activity (Verhertbruggen et al., 2011). The rice OsCSLD1 gene is required for root hair morphogenesis (Kim et al., 2007). The CslF, CslH, and CslJ genes are involved in the synthesis of mixed-linkage glucan polymers in grasses such as rice and barley (Burton et al., 2006; Doblin et al., 2009; Fincher, 2009). However, the roles of the remaining Csl family members (CslB, CslD, CslE, CslG) remain unclear.
Orchidaceae is one of the largest families of angiosperms whose members exhibit vastly different morphotypes, lifestyles, and remarkable adaptations to environmental conditions. Dendrobium is one of the largest genera of the Orchidaceae family, containing approximately 1,450 species (Zhang et al., 2016); these species are characterized by bioactive ingredients with immunomodulatory hepatoprotective activities, such as dendrobine and polysaccharides (Ng et al., 2012). Dendrobium officinale contains abundant polysaccharides in flesh stems, primarily glucomannan (GM) and galactoglucomannan (GGM) (Xing et al., 2014). Previous studies have shown that Csl genes, especially CslAs, play important roles in the synthesis of GM and GGM. However, little is known about the evolutionary pattern and functional diversification of the Csl genes involved in polysaccharide content accumulation. In this study, we performed a global analysis of the identification and characterization of the CesA/Csl family in three fully sequenced orchid species: D. officinale (Zhang et al., 2016), P. equestris (Cai et al., 2014), and A. shenzhenica (Zhang et al., 2017). Moreover, the contents of water-soluble polysaccharides and constituting monosaccharides were also measured to elucidate the potential functional divergence of the Csl genes in D. officinale. The results provide insights into the evolutionary patterns and functional divergence of CesA/Csls in Orchidaceae species.
The genome sequences of three Orchidaceae species, D. officinale (Zhang et al., 2016), P. equestris (Cai et al., 2014), and A. shenzhenica (Zhang et al., 2017) were downloaded from the NCBI database1. The Cellulose synthase family proteins contain two different pfam domains: PF03552 for CesAs and PF00535 for Csls. The seed sequences of the two domains were then downloaded from the TIGRFAMs database2. HMMsearch from the HMMER suite (version 3.1; Finn et al., 2011) was used to search for Cellulose synthase proteins in D. officinale, P. equestris, and A. shenzhenica with a cutoff E-value of 1e––4. The molecular weight and theoretical isoelectric point (pI) of the proteins were predicted using the ExPASy website3. Transmembrane domains and subcellular locations were predicted using TMHMM4 and WoLF PSORT5, respectively.
The amino acid sequences of CesA/Csls were retrieved for nine species, including algae, mosses, lycophytes, monocots, and dicots: Chlamydomonas reinhardtii (Cre), Volvox carteri (Vca), Physcomitrella patens (Ppa), Selaginella moellendorffii (Smo), Oryza sativa (Osa), A. shenzhenica (As), D. officinale (Dof), P. equestris (Peq), and Arabidopsis thaliana (AT). The protein sequences were downloaded from the Pfam database6. MEGA 6 (Tokyo Metropolitan University, Tokyo, Japan; Tamura et al., 2013) was used to analyze the protein sequences and construct phylogenetic trees. First, the MUSCLE program of MEGA 6 was used for multiple sequence alignment, and then the maximum likelihood method (ML) method with the Jones--Taylor--Thornton (JTT) model was used to construct a phylogenetic tree with 1,000 bootstrap replicates. A partial deletion with a site coverage cutoff of 70% was used for gap treatment. The phylogenetic trees were visualized and modified using MEGA 6 and iTOL7.
The Gene Structure Display Server tool8 (v2.0; Hu et al., 2015) was used to analyze the gene structure of all the CesA/Csls identified in the three Orchidaceae species. Gene distribution in the genome of D. officinale was visualized using the TBtools program (Chen et al., 2020), and conserved domains were identified with the NCBI Web CD-Search tool9. MEME software10 (v4.11.0) was used to search for sequences of motifs of CesA/Csl proteins, with a motif window length from 10 to 100 bp (Bailey et al., 2009), with the maximum number of motifs set at 10, and where motifs present in at least three proteins were considered true motifs. Multiple alignments of CesA/Csls in the three orchid species were conducted using DNAMAN software (version 9; Lynnon Biosoft Company, Quebec, QC, Canada).
Two transcriptome sequencing analyses were performed using four different organs of a 3-year-old D. officinale and the stems of three developmental stages of D. officinale. The four different organs of the 3-year-old D. officinale used in the present study include flower, leaf, flower, and root. Three different D. officinale developmental stages were investigated: 1-year old (Y1), 2 years old (Y2), and 3 years old (Y3). The plants were grown in glasshouses at the Mulberry Field Station of Zhejiang Academy of Agriculture Science (Hangzhou, China). The tissues were collected and frozen in liquid nitrogen and stored at –80°C until use. For each tissue, five plants were treated as an independent biological replicate, and three biological replicates were performed. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions and sequenced on an Illumina HiSeq 2000 platform. Subsequently, the expression profiles of all D. officinale genes were obtained with FPKM (fragments per kilobase of exon per million fragments mapped) values using Cufflinks software11 (v2.2.1) according to annotated gene models with a GFF file. The expression profiles of the CesA/Csl genes from each sample were analyzed using the HemI program12 with the average hierarchical clustering method.
Stems of D. officinale plants at five different growth stages, i.e., 3 months (3M), 9 months (9M), 1 year (Y1), 2 years (Y2), and 3 years (Y3), were collected (three replicates for each sample) and dried in an oven at 105°C until constant weight. The 3-M and 9-M D. officinale seedlings were cultured on half-strength Murashige and Skoog (MS) (Murashige and Skoog, 1962) media containing 0.1% activated carbon, 2% sucrose, and 0.6% agar (pH 5.4) in a growth chamber (25.5 ± 1°C, 45 μmol m-2 s-1 irradiance, 12-h light/dark photoperiod, 60% relative humidity). Y1, Y2, and Y3 D. officinale plants were grown in glasshouses at the Mulberry Field Station of Zhejiang Academy of Agriculture Science (Hangzhou, China). The samples were shattered into fine powder independently by a mixing mill (MM 400, Retsch). The total polysaccharide was extracted using the water extraction and alcohol precipitation method, and the contents of total polysaccharides were measured using the phenol–sulfuric acid method as described by Wang et al. (2021). Glucose was used as a reference for subsequent calculations in which total polysaccharides (μg/g dry weight) = (A + 0.0037)÷7.981 × V1÷V2 × V3÷W × 1000 = 626. 49 × (A + 0.0037)÷W, where V1 represents the redissolved volume after alcohol precipitation (1 mL), V2 represents the volume of alcohol precipitation (0.2 ml), V3 represents the volume of water added during extraction (1 mL), and W represents sample weight in grams (1,000 g), serving as a coefficient converting milligrams to micrograms.
After drying, the samples from the D. officinale stems of the five growth stages were shattered into fine powder independently by a mixing mill (MM 400, Retsch). The constituting monosaccharides, including mannose, glucose, and galactose, were extracted using the GC–MS/MS method (Sun et al., 2016). Briefly, 20 mg of powder was diluted in a 500 μL solution and the extracts were centrifuged at 14,000 rpm at 4°C for 3 min. Then, the supernatants were mixed, evaporated, and freeze-dried. The residue was used for further derivatization. The small-molecule carbohydrates and 100 μL of a solution of methoxyamine hydrochloride in pyridine (15 mg/mL) were mixed together and incubated at 37°C for 2 h. Then, 100 μL of BSTFA was added and the mixture was maintained at 37°C for 30 min after being vortexed. The mixture was subsequently diluted and analyzed by GC–MS/MS according to the methods of Gómez-González et al. (2010) and Sun et al. (2016) with modifications. Agilent 7890B gas chromatograph coupled to a 7000D mass spectrometer equipped with a DB-5MS column (30 m length × 0.25 mm i.d. × 0.25 μm film thickness, J&W Scientific, Santa Clara, CA, United States) was employed for GC–MS/MS analysis of the monosaccharides.
Total RNA was extracted from all the samples of D. officinale stems at the five growth stages (3M, 9M, Y1, Y2, and Y3) using TRIzol reagent (Invitrogen, Carlsbad, CA, United States). The potential contaminating genomic DNA was eliminated with DNase I. The quality of the RNA samples was checked with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Beijing, China) and 1% denaturing agarose gels. The RNA was used as a template for first-strand cDNA synthesis using PrimeScript reverse transcriptase (TaKaRa Biotechnology, Dalian, China). Gene-specific primers were designed with the Primer Premier 5.0 program. The DnActin (comp205612_c0) gene was used as an internal standard for normalizing the gene expression data. The DofCesA/DofCsl expression levels were analyzed using a quantitative Real-time PCR (qRT–PCR) assay, which was performed by using a SYBR Green qPCR Kit (TaKaRa Biotechnology, Dalian, China) and a Stratagene Mx3000P thermocycler (Agilent, Santa Clara, CA, United States). The PCR program was as follows: 95°C for 5 min followed by 40 cycles at 95°C for 15 s and then 60°C for 30 s. The relative gene expression levels were calculated with the 2–ΔΔCt method (Livak and Schmittgen, 2001). Three biological replicates were subjected to the analysis, each with three technical replicates. The expression levels in the different tissues were visualized using GraphPad Prism (v. 8.4.3).
Statistical analysis was performed, and the average values and standard errors of the three replicates were calculated. SPSS software (v. 16.0) was used to determine the significant differences in polysaccharide and monosaccharide contents between the different developmental stages using a one-way ANOVA procedure and post hoc analysis. The value p < 0.05 indicates a significant difference and is represented by an asterisk (*) in the figures, and p < 0.01 indicates a very significant difference and is represented by two asterisks (**) in the figures. (***) indicates p value < 0.001 and (****) indicates p value < 0.0001.
To investigate the potential roles of Cellulose synthase superfamily genes in orchids, we performed a genome-wide identification and characterization of CesA/Csl genes in three sequenced Orchidaceae species: D. officinale (Zhang et al., 2016), P. equestris (Cai et al., 2014), and A. shenzhenica (Zhang et al., 2017). A total of 125 Cellulose synthase superfamily genes were identified in the three orchid species, which were designated ‘DofCesA/DofCsl’ for D. officinale, ‘PeqCesA/PeqCsl’ for P. equestris, and ‘AsCesA/AsCsl’ for A. shenzhenica (Supplementary Table S1). The CesA superfamily members in the Orchidaceae family were classified into seven families: CesA, CslA, CslC, CslD, CslE, CslG, and CslH according to the sequence similarities with that of A. thaliana and O. sativa (Figure 1, Supplementary Figure S1, and Supplementary Table S1).
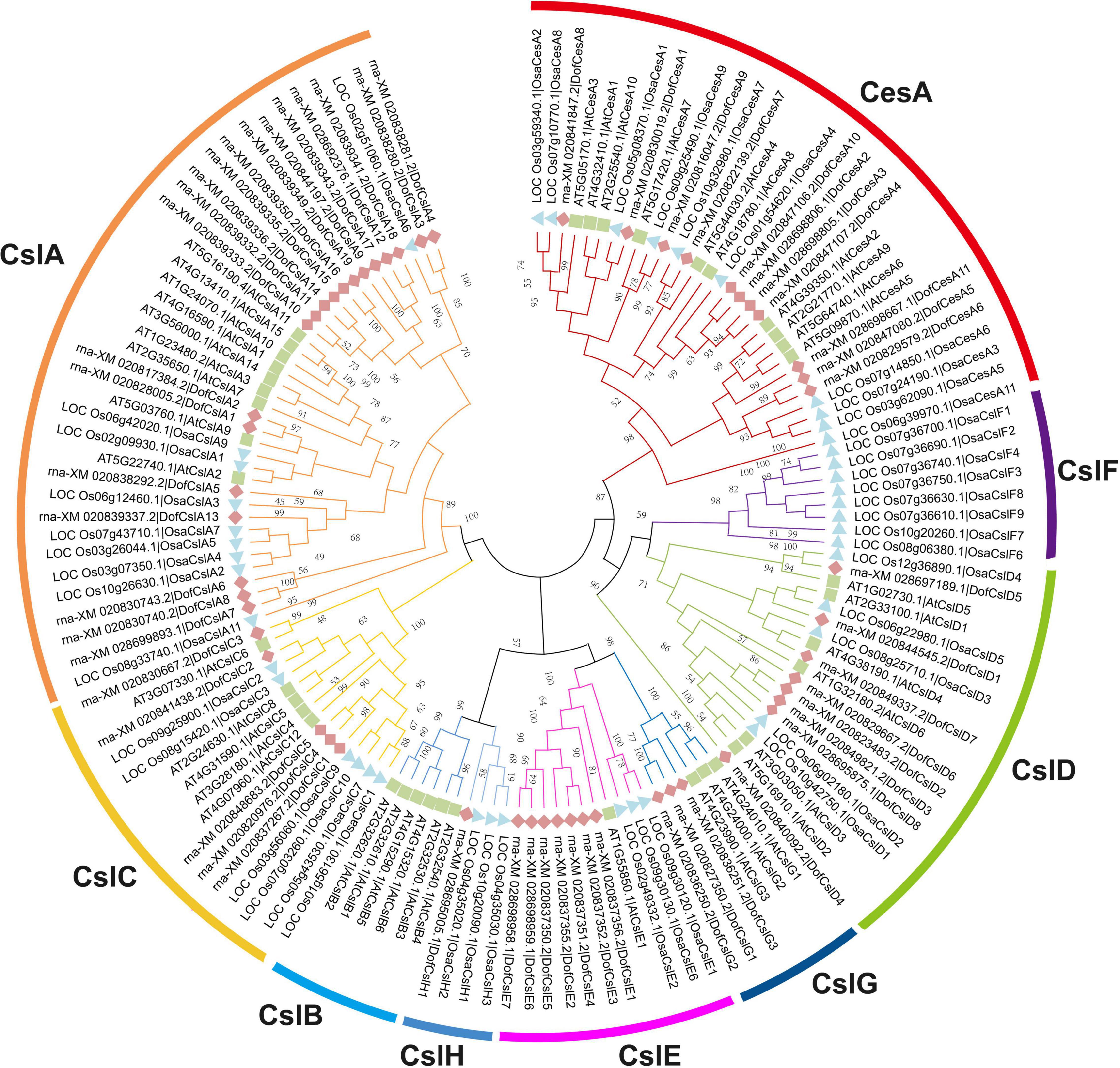
Figure 1. Phylogenetic tree of the Cellulose synthase superfamily in D. officinale (Dof), rice (Osa) and Arabidopsis (At). The phylogenetic tree was constructed using MEGA 6.0 with the neighbor-joining (NJ) method and 1,000 bootstrap replicates. The CesA/Csl proteins were grouped into one CesA family and eight Csl families: CesA, CslA, CslB, CslC, CslD, CslE, CslF, CslG, and CslH. The families are marked by different arc lines and branch colors, and individual species are distinguished by triangles, squares, or rhombuses in different colors.
We identified 54 CesA/Csl proteins from the D. officinale genome, which comprised 11 CesAs and 43 Csls (Table 1). The 54 proteins were further classified into DofCesA (11 members) and six different Csl families, i.e., DofCslA (19 proteins), DofCslC (5 proteins), DofCslD (8 proteins), DofCslE (7 proteins), DofCslG (3 proteins), and DofCslH1, according to the genome annotation information and sequence similarity with those of Arabidopsis and rice (Figure 1 and Table 1). P. equestris was identified with 37 CesA/Csl proteins, which were classified into PeqCesA (8 proteins) and six Csl families: PeqCslA (10 proteins), PeqCslD (10 proteins), PeqCslH (3 proteins), PeqCslE (4 proteins), PeqCslC1, and PeqCslG1. The A. shenzhenica genome was identified with 34 CesA/Csl proteins, including AsCesA (9 proteins), AsCslA (10 proteins), AsCslC (4 proteins), AsCslD (7 proteins), AsCslG (2 proteins), AsCslE1, and AsCslH1. These results indicated that the Cellulose synthase superfamily in D. officinale had greatly expanded, whereas that in the other two Orchidaceae species was similar to Arabidopsis (Persson et al., 2007), tomato (Song et al., 2018), and rice (Wang et al., 2010). Notably, although the number of DofCesAs was similar to that in other species, the DofCsl family had significantly expanded, especially DofCslA.
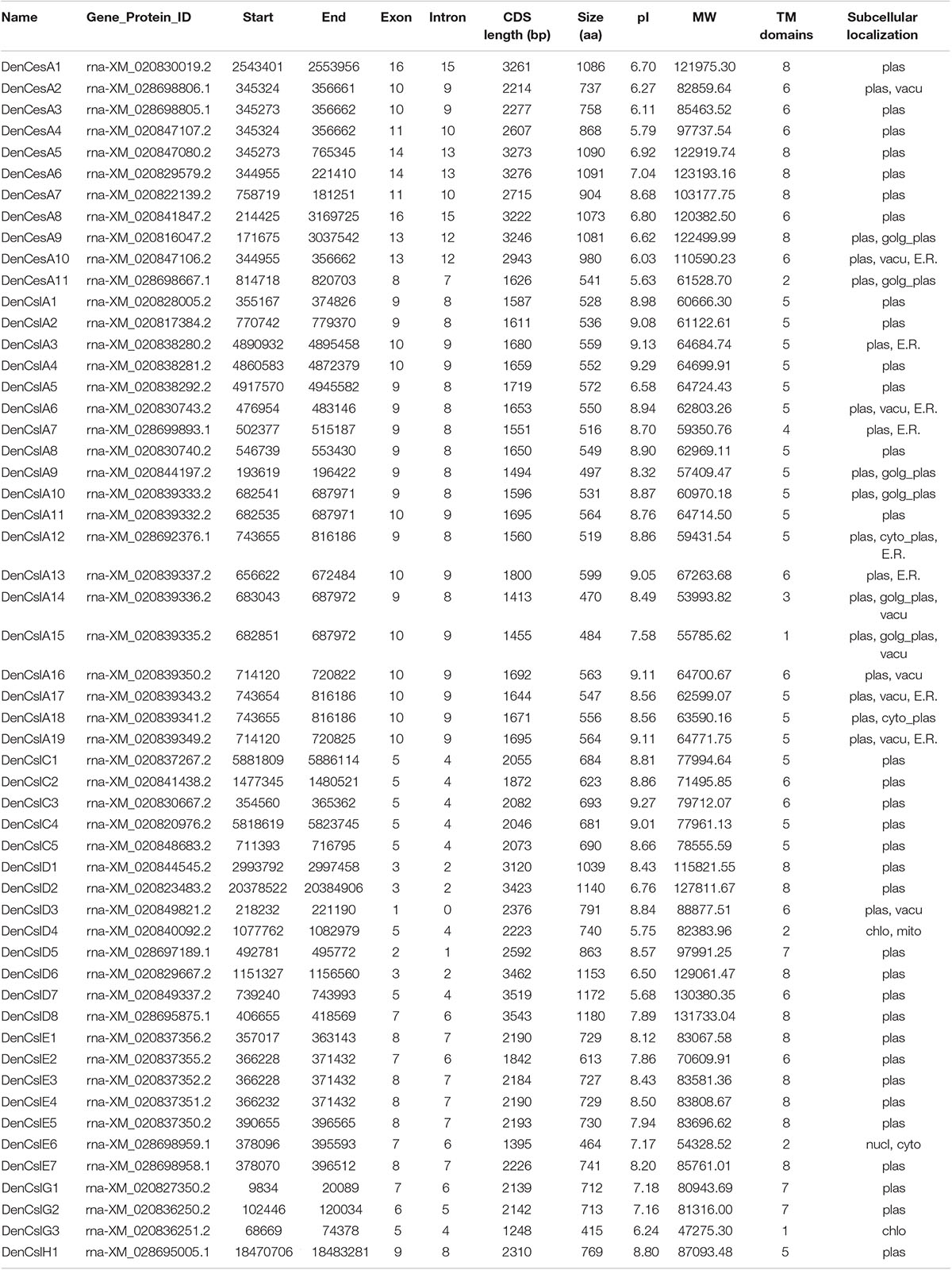
Table 1. Physical and molecular characteristics of Cellulose synthase superfamily genes in D. officinale.
Three of the 125 CesA/Csl proteins (AsCslA2, PeqCslE4, and PeqCslH3) had extremely short amino acid sequences (less than 300 aa), even after reannotation via Softberry13. This may be due to genome sequencing errors or improper assembly. Thus, the three proteins were excluded from the subsequent statistical analysis. The molecular weights of the remaining 122 CesA/Csls ranged from 42.84 kDa (PeqCslA9) to 146.86 kDa (AsCslD5), with pI values ranging from 5.63 (DofCesA11) to 9.29 (AsCslA3 and DofCslA4) (Supplementary Table S1). The longest protein was AsCslD5 (1392 aa), whereas the shortest was PeqCslA9/PeqCslA10 (374 aa). The amino acid sequences of the CesA/Csls ranged from 415 aa to 1,180 aa in D. officinale, 515 aa to 1157 aa in P. equestris, and 438 aa to 1329 aa in A. shenzhenica. Except for four proteins, most of the CesAs and CslDs in the three orchid species were approximately 800–1,000 aa in length with 6–8 transmembrane (TMs) domains (Supplementary Table S1). The DofCslAs and DofCslCs were generally ∼500 aa and ∼700 aa in length, respectively, with 5 or 6 TM domains. Moreover, subcellular localization prediction showed that 94% of the CesA/Csls were localized on the plasma membrane. Nonetheless, two AsCslAs and four DofCslAs were predicted to localize on both the plasma membrane and the Golgi apparatus plasma. Detailed information on the CesA/Csl proteins in the three orchid species, including their gene/protein ID, CDS length, subcellular localization, TM domains, molecular weight, and pI value are shown in Table 1 and Supplementary Table S1.
To investigate the evolution of the Cellulose synthase gene superfamily, phylogenetic analysis was performed for nine representative plant and algal species, i.e., two algal species (Volvox carteri and Chlamydomonas reinhardtii), Physcomitrella patens, Selaginella moellendorffii, Oryza sativa, Arabidopsis, and three orchid species (D. officinale, P. equestris, and A. shenzhenica). A phylogenetic tree was constructed based on 273 amino acid sequences using MEGA 6 software and the maximum likelihood (ML) method with 1,000 bootstraps. The 273 CesA/Csls were classified into CesA and eight Csl families: CslA, CslB, CslC, CslD, CslE, CslF, CslG, and CslH (Figure 2A and Supplementary Figure S2). The lower land plants, i.e., the bryophyte P. patens and the lycophyte S. moellendorffii, are identified with representatives of the CesA, CslA, CslC, and CslD families, which is generally consistent with previous findings (Roberts and Bushoven, 2007; Yin et al., 2014). Each of the two green algae was identified as containing a single copy of the CslA/C-like gene (VcaCslC for V. carteri and CreCslA for C. reinhardtii), implying that the possible duplication of the common ancestor of CslA/CslC occurred in land plants after they diverged from green algae (Yin et al., 2009; Popper et al., 2011). Among the eight Csl families, CslA and CslC are distantly related to the other Csl families, supporting previous suggestions that the CslA and CslC family members evolved from a separate cyanobacterial endosymbiotic event (Yin et al., 2009). CslB was found only in the dicot species Arabidopsis, whereas CslH was specific to the four monocots (Figure 2C). Notably, CslG family members were found in all three Orchidaceae species.
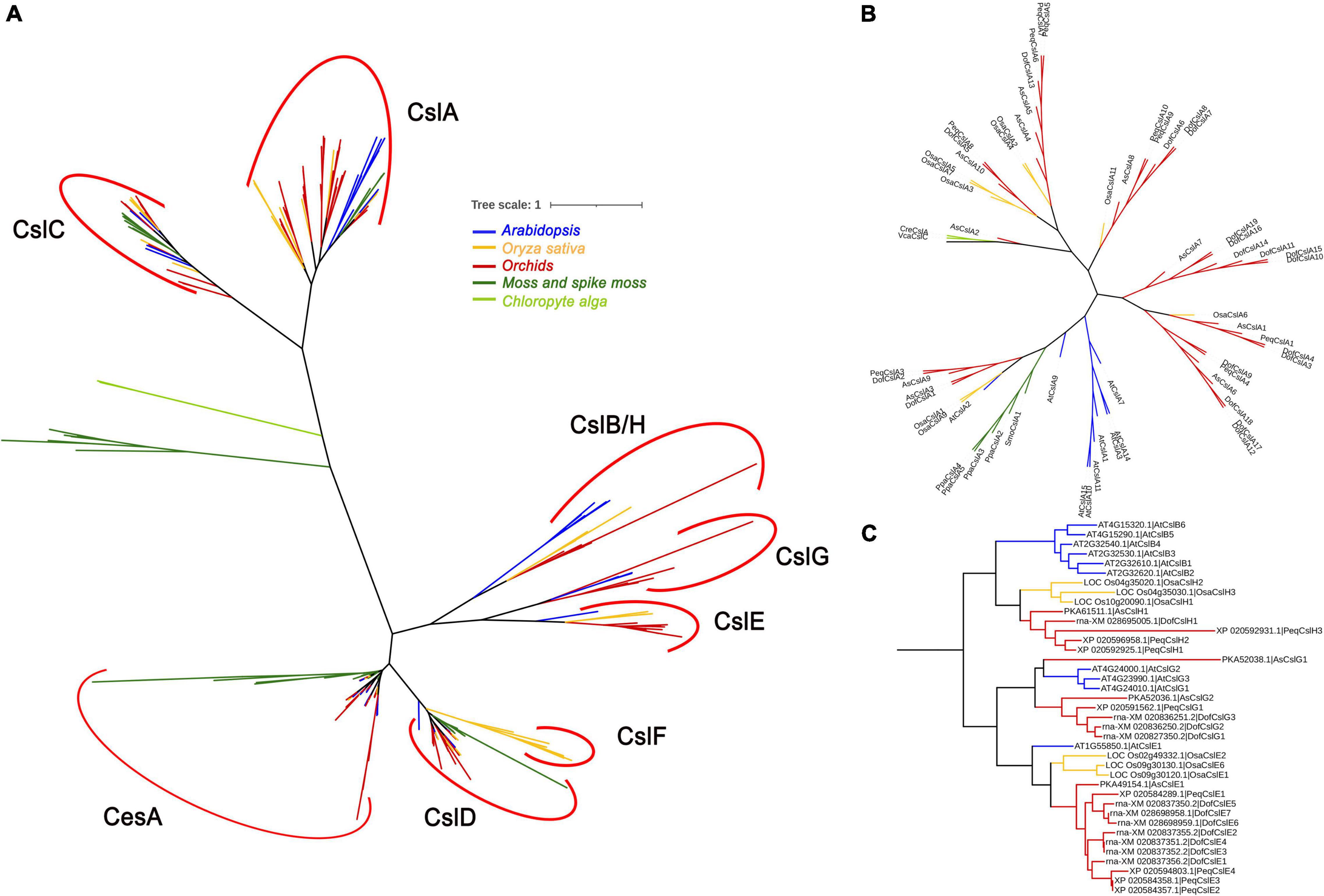
Figure 2. Phylogenetic relationships between Cellulose synthase proteins from nine representative species: two chlorophyte algae (light green), a moss and spike moss (dark green), three orchid species (red), rice (yellow), and Arabidopsis (blue). The CesA/Csl proteins were grouped into nine families: CesA, CslA, CslB, CslC, CslD, CslE, CslF, CslG, and CslH. (A) The phylogenetic tree contains all 273 CesA/Csl protein sequences; (B) phylogenetic tree showing only the CslA cluster; (C) phylogenetic tree showing only the CslB/H/E/G cluster. The sequences in (B,C) were extracted from (A) and realigned.
The number of CesAs was rather conserved among angiosperms, with each generally having 8∼13 members (except for S. moellendorffii), whereas the members in the other families greatly varied. P. patens had the highest number of CesAs (13 members) and CslDs (10 members), whereas S. moellendorffii had the lowest (five SmoCesAs and three SmoCslDs). The CslCs were most abundant in S. moellendorffii (10 SmoCslCs); however, the family had greatly contracted in other species, especially in P. equestris (containing only PeqCslC1). Notably, the CslA family had significantly expanded in D. officinale (with 19 DofCslAs) compared with the remaining species (Figure 2B), which may have resulted in functional redundancy or innovation.
The sequences of conserved domains of the 125 CesA/Csl proteins in three Orchidaceae species were searched and analyzed. Five conserved domains: Cellulose synthase domain (cellulose_synt), zf-UDP, zf-RING_4, and two glycosyltransferase family domains (Glyco_transf_2, Glyco_trans_2_3) were found (Figure 3 and Supplementary Figures S3, S4). We found that the Cellulose synthase domain was present in CesA and CslD/E/G/H, whereas all the CslA/CslC proteins were found to contain two glycosyltransferase domains (Table 2), which support the CslA/CslC families evolved from an independent cyanobacterial endosymbiotic event (Yin et al., 2009). The number of CesA and CslD proteins that contain zf-UDP/zf-RING_4 domains varied among the three Orchidaceae species. For example, the zf-UDP domain was found in all nine AsCesA proteins but was absent from three DofCesAs and one PeqCesA. The motif patterns also varied among different species and families; however, we found species-specific motifs and similar conserved motif patterns within the same CesA/Csl family (Table 2, Figure 3 and Supplementary Figures S3, S4). These results suggested that CesA/Csl proteins among different families in Orchidaceae species might have different functional properties. Furthermore, multiple alignments of the predicted Cellulose synthase amino acid sequences showed that 18 of the 125 CesA/Csl proteins had no “D,D,D,QxxRW” integrated active site amino acid sequence (Supplementary Figures S5–S7), implying possible functional redundancy.
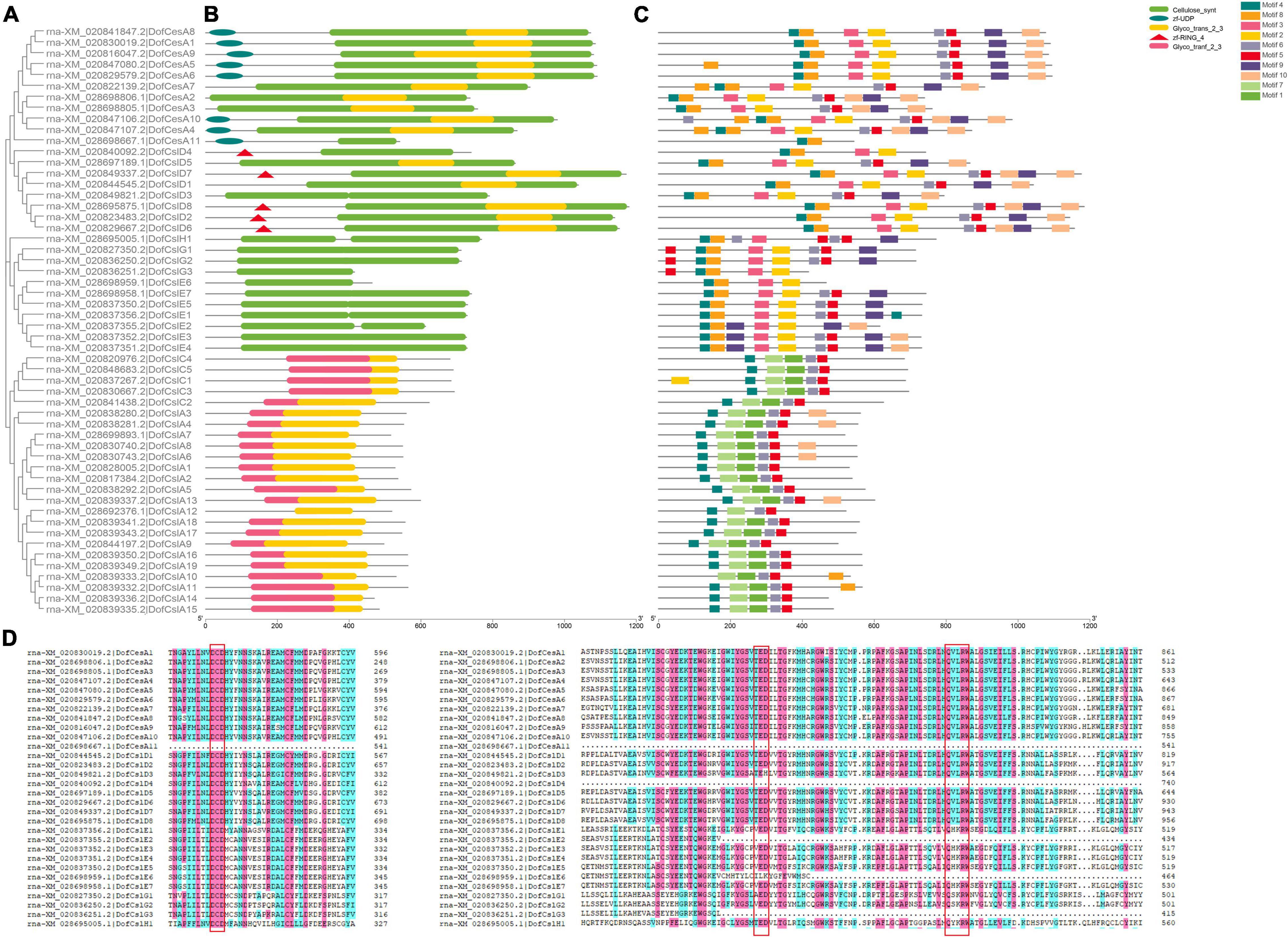
Figure 3. Phylogenetic, conserved domain and motif analyses of Cellulose synthase proteins in D. officinale (Dof). (A) Phylogenetic tree of the DofCesA/DofCsl proteins; (B) Schematic presentation of the domains of the DofCesA/DofCsl proteins; (C) Schematic presentation of the conserved motifs in the DofCesA/DofCsl proteins; (D) Comparison of the deduced amino acid sequences of Cellulose synthase in D. officinale. The conserved cysteine residues are marked by red frames. The full versions of multiple alignments of the DofCesA/DofCsl protein sequences are shown in Supplementary Figure S5. The scale bar (aa) indicates the amino acid position within the corresponding conserved domain/motif.
To investigate the sequence diversity of the CesA/Csl proteins in the three orchid species, putative motifs were identified using MEME Suite 5.0.2; a total of 10 conserved motifs were found (Figure 3C and Supplementary Figures S3, S4). Although the motifs varied among the different species and families, the CesA/Csl proteins in each family in the same species shared several unique motifs. For example, in D. officinale, motif4, motif8, and motif2 were all observed in the DofCesA, DofCslD, DofCslG, and DofCslE families but not in the DofCesA11 family (Figure 3C and Supplementary Figure S5A). The DofCslA and DofCslC family proteins shared five common motifs: motif4, motif7, motif1, motif6, and motif5; however, DofCslA10 lacked motif 4. Among these motifs, motif7 and motif1 were only observed in DofCslA and DofCslC families. In P. equestris, the conserved motifs in PeqCesA, PeqCslD, PeqCslE, PeqCslH, and PeqCslG were similar; except for four proteins, all contained motif1–motif7 and motif10 (Supplementary Figure S3). Moreover, motif9 was specific to members of the PeqCslA and PeqCslC families. In A. shenzhenica, motif1, motif5, and motif2 were uniformly observed in all CesA/Csl proteins, with the exceptions of AsCesA9 and AsCslA2. Members of AsCesA, AsCslD, AsCslE, AsCslH, and AsCslG also shared similar motifs (Supplementary Figure S4), and motif 10 was specifically present in the AsCslA and AsCslC proteins. Taken together, these results revealed species-specific motifs and similar motifs shared among certain families. Thus, CesA/Csl proteins among different families and species might have different functional properties.
To gain more information on the evolutionary patterns of Cellulose synthase genes, the gene structures of the CesA/Csls in D. officinale, P. equestris, and A. shenzhenica were analyzed (Supplementary Table S1). The intron–exon structure of the CesA/Csls was highly diverse among the different families within the same species (Figures 4A,B). Nonetheless, the same family members have similar exon/intron numbers among the three Orchidaceae species. For example, except for four genes, AsCesAs and PeqCesAs usually have 13 or 14 exons. The exon number in DenCesAs varied more significantly compared to the other two orchids and ranged from 8 to 16 exons. CslAs generally contain 5–10 exons, and most have 9 or 10 exons. The exon–intron structures and intron phases of the genes were relatively conserved within the same family (Figure 4B and Supplementary Figures S3, S4). In D. officinale, all of the DofCslCs had 5 exons and 4 introns. The gene structure varied the most within the CslD genes, from 0–7 or 8 introns/exons. These results suggested that possible functional diversity occurred within the same family and evolutionary conservation in Orchidaceae species.
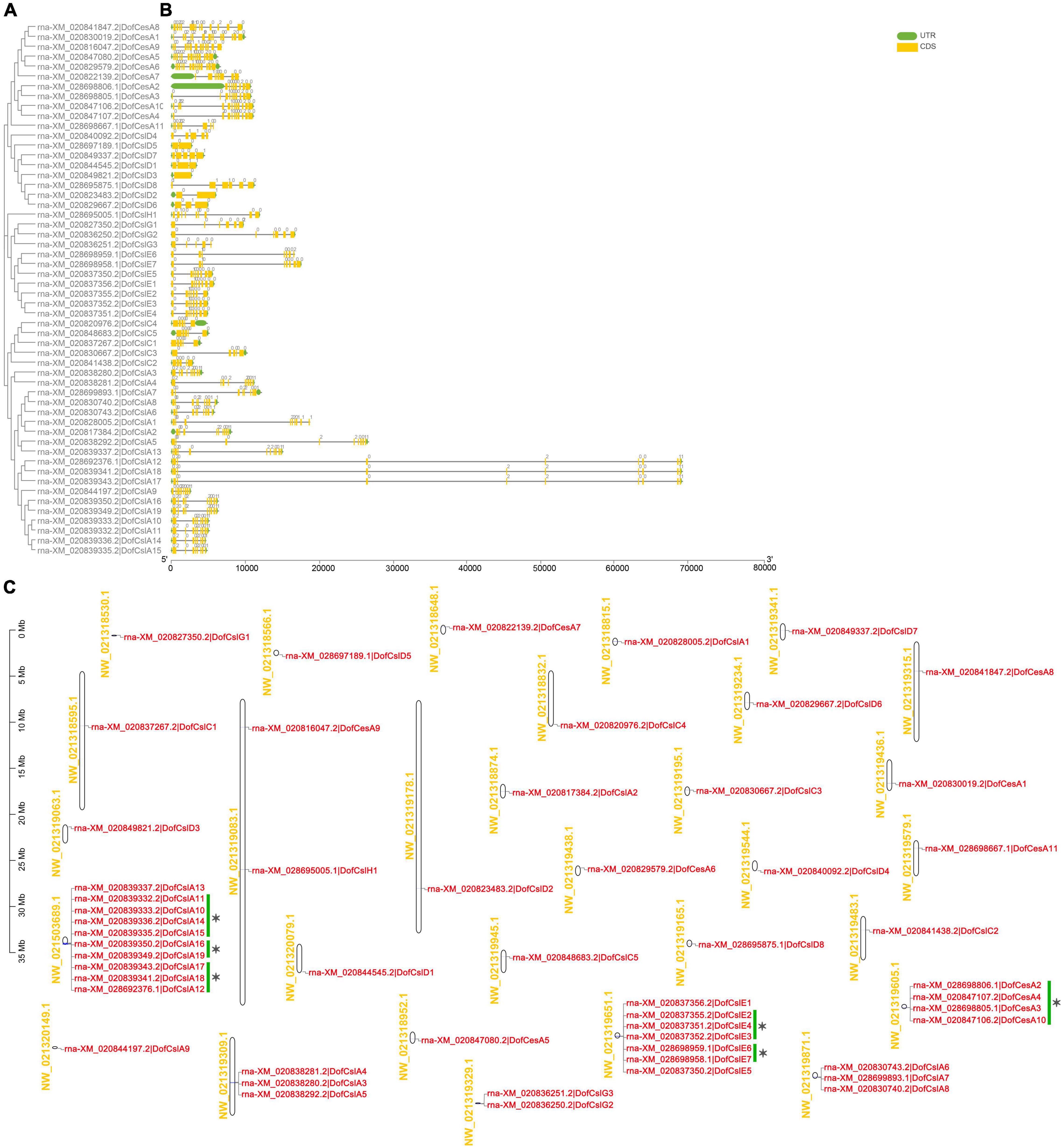
Figure 4. Phylogenetic, gene structure, and distribution analyses of Cellulose synthase superfamily genes in D. officinale (Dof). (A) Phylogenetic relationships of the DofCesA/DofCsl genes; (B) Sequence analysis of exon–intron structures of the Cellulose synthase genes. UTRs and exons are indicated using green and yellow rectangles, respectively. The solid lines indicate introns. The numbers above the solid lines represent the intron phase. The scale bar (bp) indicates the length of the corresponding genes; (C) distribution of theCellulose synthasegenes in the D. officinale genome. The scaffold names are shown on the left, and the gene names are on the right. The scale bar (Mb) indicates the length of the corresponding scaffolds. A total of 14 tandemly duplicated genes (with two or more homologous genes ≤ 100 kb apart) were found. Green scale bars and asterisks indicated alternative splicing genes.
Based on the physical positions of the Cellulose synthase genes in the D. officinale genome (Zhang et al., 2016), the distribution of 54 DofCesA/DofCsl genes was mapped to 30 scaffolds (Figure 4C and Table 1). Among them, 23 scaffolds had only one Cellulose synthase gene and two scaffolds each had two genes. Apart from the alternative splicing genes (indicated with green scale bars and asterisks), 14 tandemly duplicated genes (with two or more homologous genes ≤ 100 kb apart) were detected on six scaffolds (Figure 4C). A maximum number of 10 genes were detected on scaffold NW_021503689.1, all of which were DofCslA members (DofCslA10-19); after merging alternative splicing genes, there were four tandemly duplicated genes. DofCslA3, -4, -5 and DofCslA6, -7, -8 were also tandem duplicated genes located on scaffold NW_021319309.1 and NW_021319871.1, respectively. In addition, the DofCslEs also contain tandemly duplicated members that clustered on scaffold NW_02131965.1 (Figure 4C). Moreover, we found that the structures of alternative splicing genes, such as the number of exons and the length of introns, varied in different degrees among the splicing variants of the same gene (Supplementary Figure S8 and Supplementary Table S2). These results revealed that the expansion of the DofCesA/DofCsl genes mainly originated through gene duplication.
To analyze the roles of the CesA/Csl genes in D. officinale, their expression patterns were investigated in different organs in D. officinale at different developmental stages. Transcriptome sequencing was performed on four organs of D. officinale (i.e., the leave, stem, flower, and root) and the stems of three different stages (Y1, Y2, and Y3). The expression levels of the DofCesA/DofCsl genes are presented in heatmaps in Figure 5 and Supplementary Table S3). The results revealed diverse expression patterns of the DofCesA/DofCsl genes, suggesting functional divergence had occurred after gene expansion.
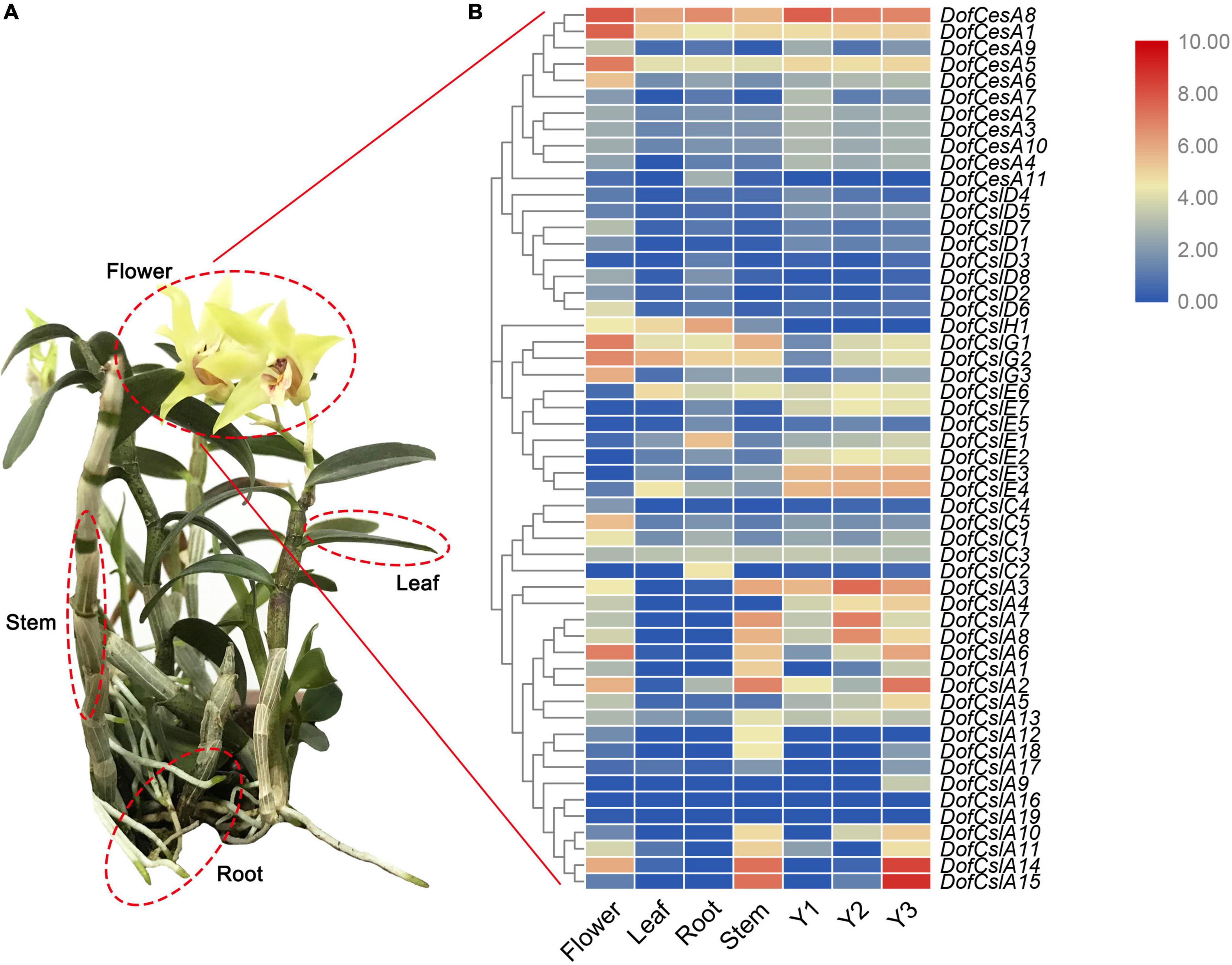
Figure 5. Hierarchical clustering of the expression profiles of Cellulose synthase superfamily genes in different organs (flowers, roots, stems, and leaves) of D. officinale and at different growth stages (Y1, Y2, and Y3). (A) Image of a 3-year-old D. officinale plant; (B) FPKM values visualized as the heatmap.
In the DofCesA family, three genes (DofCesA1, DofCesA5, and DofCesA8) were consistently expressed across different organs and developmental stages and were all significantly expressed in the flowers. Among them, DofCesA8 also had high expression levels in the roots and low expression in the leaves and stems; both DofCesA1 and DofCesA5 were expressed at low levels in the leaves, roots, and stems. Notably, DofCesA8 was also highly expressed in the Y1, Y2, and Y3 stems of D. officinale. The DofCslD and DofCslC family genes exhibited similar expression patterns, with low expression across the four organs and three developmental stages. The DofCslEs showed organ-specific expression patterns; for example, DofCslE1 and DofCslE4 exhibited moderate expression in the roots and leaves, respectively, whereas DofCslE6 was expressed in the leaves, roots, and stems and across all three growth stages. DofCslH1 was expressed in three organs (flowers, leaves, and roots), with the highest expression level in the roots. Moreover, the DofCslG family genes showed similar expression patterns; all exhibited higher expression levels in the flowers than in the roots, leaves, and stems. DofCslG1 and DofCslG2 also exhibited moderate expression in Y2 and Y3 stems.
CslAs are known for their participation in the synthesis of polysaccharides, especially mannans and glucomannans. Interestingly, the DofCslAs exhibited apparent organ- and development-specific expression in D. officinale. Most of these genes were predominantly expressed in the stems and/or flowers, with very low levels in the roots and leaves. For example, DofCslA2, DofCslA14, and DofCslA15 had significantly higher expression levels in the stems than in the other organs (especially DofCslA15); DofCslA2 and DofCslA14 were also expressed in the flowers. However, several genes had almost no expression in any of the four organs. In addition, DofCslA6 was specifically expressed in the flowers. Among the three growth stages, some genes showed similar expression patterns, such as DofCslA7 and DofCslA8, both of which were expressed at higher levels in the Y2 stems. Notably, DofCslA14 and DofCslA15 had significantly high expression in Y3 stems. However, five of the 19 DofCslAs showed low levels of or no expression in the Y1, Y2, and Y3 stems. DofCslA2 was expressed at moderate levels in the Y3 stems and at low levels in the Y1 stems, whereas DofCslA3 showed a consistent expression pattern across all three stages, with the highest expression levels occurring in Y2 stems. These results revealed the possible functional specialization of DofCslAs in the polysaccharide synthesis in D. officinale.
A previous study showed that polysaccharide accumulation in D. officinale occurred predominantly in the stems (He et al., 2015). To further investigate the polysaccharide synthesis and functions of Cellulose synthase superfamily genes in Orchidaceae species, we measured the contents of total water-soluble polysaccharides, and their component monosaccharides (glucose, mannose, and galactose) in the stems of D. officinale at five different growth stages, i.e., 3 months (3M), 9 months (9M), 1 year (Y1), 2 years (Y2), and 3 years (Y3) (Figure 6). The results showed that the total polysaccharide content varied significantly among the different stages (Figure 6D and Table 3); it was lowest in the 3M stems (∼18.06 mg/g) and highest in the Y3 stems (∼93.02 mg/g). The total polysaccharide contents varied slightly between the 3M and 9M stems and between Y2 and Y3 stems, but increased significantly from 3M/9M to Y2/Y3.
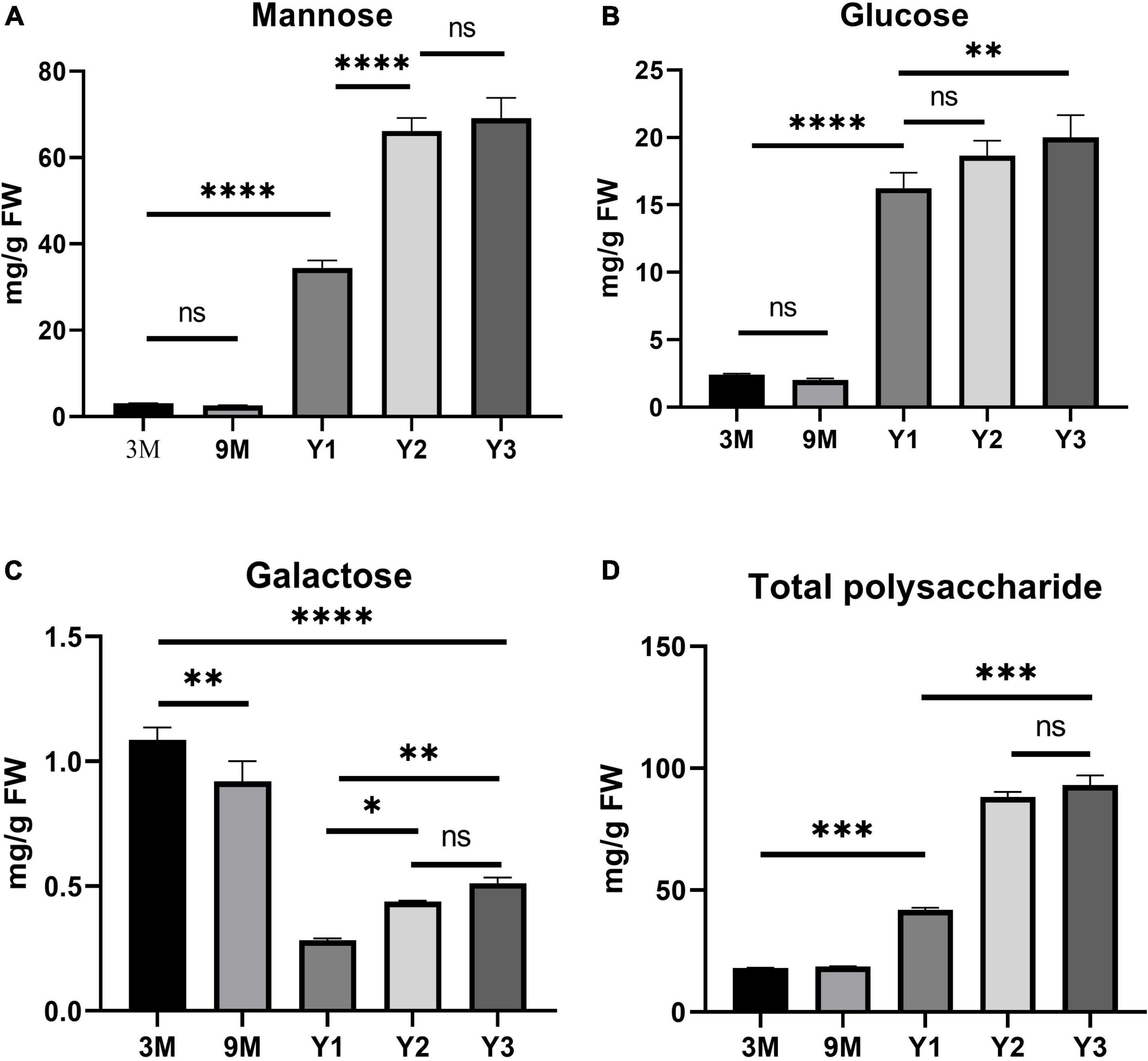
Figure 6. Histogram of the contents of water-soluble polysaccharides and monosaccharide components (mg/g) in the stems of D. officinale in different growth stages, including 3M, 9M, Y1, Y2, and Y3. (A) Mannose content; (B) glucose content; (C) galactose content; (D) total polysaccharide content. *p value < 0.05, **p value < 0.01, ***p value < 0.001, ****p value < 0.0001.
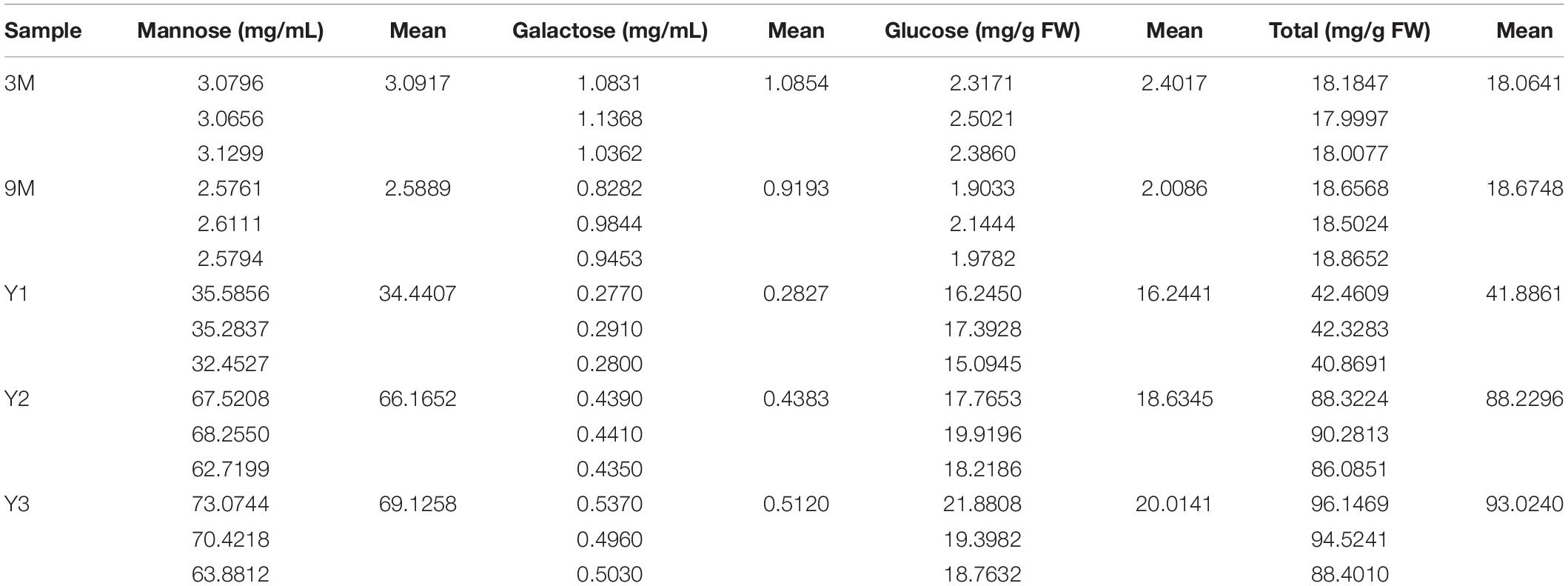
Table 3. Contents of water-soluble polysaccharides and constituting monosaccharide (mg/g) in the stems of D. officinale at different growth stages, including 3M, 9M, Y1, Y2, and Y3.
To determine the composition and contents of monosaccharides in the stems, the neutral monosaccharide composition was analyzed at the five growth stages. The results revealed that mannose was the most abundant neutral monosaccharide in the stems of D. officinale across all five growth stages, followed by glucose and galactose (Figure 6). The contents of glucose and mannose ranged from 2.40 to 20.01 mg/g and from 3.09 to 69.13 mg/g, respectively, with the highest detected in the Y3 stems and the lowest in the 3M stems. Mannose and glucose in the stems increased remarkably during D. officinale development, corresponding to the increases in total polysaccharides. Interestingly, the galactose content was highest in the 3M stems; however, this content decreased as D. officinale developed, lowest in the Y1 stems, and then increased slightly from Y2 to Y3.
To better understand the roles of CslAs in polysaccharide synthesis in D. officinale, qPCR analyses of the 19 DofCslAs were performed on D. officinale stems at five different growth stages: 3M, 9M, Y1, Y2, and Y3. The results for Y1, Y2, and Y3 were generally consistent with the transcriptome sequencing data. In general, most of the DofCslAs exhibited high expression levels in Y2 and/or Y3 stems (Figure 7 and Supplementary Table S4), which was in agreement with the accumulation of water-soluble polysaccharides and monosaccharides. Among them, five genes, i.e., DofCslA2, DofCslA6, DofCslA11, DofCslA14, and DofCslA15, exhibited significantly high expression levels in the Y3 stems (especially DofCslA15). Four genes, i.e., DofCslA3, DofCslA4, DofCslA7, and DofCslA8, were highly expressed in the Y2 stems compared with the Y3 stems. Notably, several genes, such as DofCslA2, DofCslA3, and DofCslA7, were also expressed in the 9M stems; DofCslA8 also exhibited moderate expression in the 3M stems. In addition, six DofCslAs, such as DofCslA16-19, showed low expression levels. The primer sequences for qPCR analysis are provided in Supplementary Table S5. Taken together, these results suggest that DofCslA genes may exhibit functional redundancy and may be specialized for activity at different growth stages.
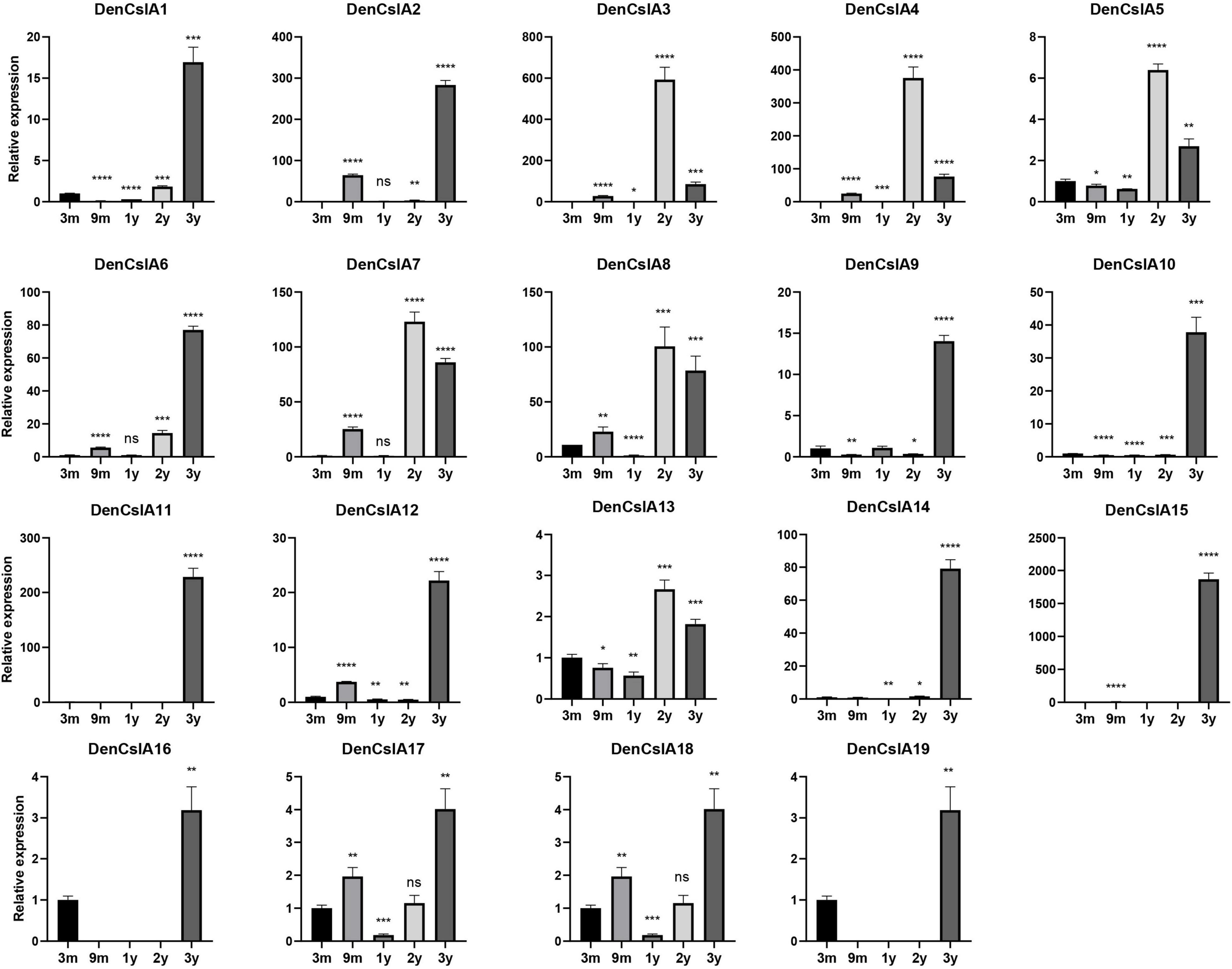
Figure 7. Expression levels of DofCslA genes in the stems of D. officinale in different growth stages (including 3M, 9M, Y1, Y2, and Y3), as determined by qRT–PCR analysis. The results are shown as the means ± SDs of three independent experiments. The presented expression levels are relative to the expression of the reference gene. *p value < 0.05, **p value < 0.01, ***p value < 0.001, ****p value < 0.0001.
Owing to its important roles in the biosynthesis of plant cell wall polysaccharides, primarily cellulose and hemicellulose, the Cellulose synthase superfamily has been the subject of intense studies. Much effort has been devoted to the identification of the CesA/Csl family in plants, including mosses, lycophytes, monocots, and dicots, as well as in green algae (Persson et al., 2007; Yin et al., 2014; Takata and Taniguchi, 2015; Li et al., 2017, 2020; Song et al., 2018; Cui et al., 2020). However, knowledge of the Cellulose synthase superfamily is still lacking in the family Orchidaceae. In the present study, we performed genome-wide characterizations of CesA/Csl family members in three fully sequenced Orchidaceae species (Cai et al., 2014; Zhang et al., 2016, 2017), which revealed the evolution, structural divergence, and expression profiles of CesA/Csls.
The Cellulose synthase superfamily belongs to the glycosyltransferase-2 superfamily, which is widely distributed throughout the plant kingdom (Kim et al., 2007; Cantarel et al., 2009). In the present study, we identified a total of 125 Cellulose synthase superfamily members from the three sequenced orchids using two Pfam domains (PF03552/PF00535; Table 1 and Supplementary Table S1). Among them, 54 DofCesA/DofCsls, 37 PeqCesA/PeqCsls, and 34 AsCesA/AsCsls were identified in the genomes of D. officinale, P. equestris, and A. shenzhenica, respectively. The results revealed species-specific expansion of the Cellulose synthase superfamily in D. officinale.
In land plants, the Cellulose synthase superfamily can be further classified into a CesA family and nine Csl families: CslA-CslH and CslJ (Fincher, 2009; Yin et al., 2014; Schwerdt et al., 2015). Previous studies indicate that PF03552 encompasses the CesA family and seven Csl families (CslB/D/E/F/G/H/J), whereas PF03552 is absent from the CslA/CslC families (Yin et al., 2009). In the present study, the CesA superfamily genes in Orchidaceae were grouped into one CesA family and six Csl families: CslA, CslC, CslD, CslE, CslG, and CslH (Figure 1 and Supplementary Figure S1). The number of CesAs in the three orchid species was largely the same, while the number of Csls greatly varied, from 25 AsCsls to 43 DofCsls. Interestingly, the Csl proteins exhibited family-specific expansion among the three orchid species; for example, there were 19 DofCslAs, almost twice that in the other two orchids. The results of similar expansion were also observed in Csl families such as PeqCslDs and PeqCslHs. The expansion of CesA/Csl families likely resulted in functional redundancy or innovation. To further investigate the evolution of CesA/Csls, a phylogenetic tree based on 273 amino acid sequences from nine representative plant species was constructed (Figure 2). CesA/Csls were classified into one CesA family and eight Csl families: CslA, CslB, CslC, CslD, CslE, CslF, CslG, and CslH. These results are consistent with those of previous findings in which CslA, CslC, and CslD were found to be conserved in all land plants (Farrokhi et al., 2006; Yin et al., 2014). CslF and CslH may be restricted to monocots, and CslFs were absent from the three Orchidaceae species. CslG members were previously thought to be confined to eudicots (Fincher, 2009). However, CslG family members were found in all three Orchidaceae species, supporting the proposal that CslG should no longer be considered a dicot-specific family (Yin et al., 2014). In addition, our results strongly supported previous findings that the numbers of CesA/Csls vary greatly among different plant species (Paterson et al., 2009; Takata and Taniguchi, 2015; Song et al., 2018; Zou et al., 2018; Li et al., 2020). The number of CesAs is largely conserved among angiosperms, most of which have 8–13 members (except for S. moellendorffii), whereas the members in other families greatly varied among different species, indicating that extensive expansion and diversification have occurred.
We identified three proteins in P. equestris and A. shenzhenica that had extremely short amino acid sequences (less than 300 aa). The molecular weights of the remaining 122 CesA/Csls in the three orchid species ranged from 42.84 to 146.86 kDa. The longest protein was AsCslD5-1392 aa, which is shorter than the 2038 aa sequence in tomato (Song et al., 2018). Previous studies have indicated that cellulose is synthesized by plasma membrane-localized Cellulose synthase complexes with access to the cytosolic GDP-glucose pool (Taylor, 2008; Endler and Persson, 2011; McFarlane et al., 2014; Schneider et al., 2016). In the present study, subcellular localization predictions showed that most of the CesA/Csls were located on the plasma membrane.
The GT2 superfamily is characterized by conserved cytosolic substrate binding and catalytic residues. These residues are positioned in a loop between TM domains 2 and 3 and contain a D,D,D,QXXRW motif (Pear et al., 1996). In addition, plant CesA proteins also contain an extended N-terminal zinc-finger domain and two insertions in the catalytic loop, which are presumably involved in specific functions in higher plants, such as multimerization (Pear et al., 1996; Sethaphong et al., 2013). In the present study, five conserved domains were identified from the 125 orchid CesA/Csls (Figure 3 and Supplementary Figures S3, S4). We found that the Cellulose synthase domain was present in CesA and CslD/E/G/H, whereas all the CslA/CslC proteins were found to contain two glycosyltransferase domains, which support the CslA/CslC families evolved from an independent cyanobacterial endosymbiotic event (Yin et al., 2009). The number of CesA and CslD proteins that contain zf-UDP/zf-RING_4 domains varied among the three Orchidaceae species. For example, the zf-UDP domain was found in all nine AsCesA proteins but was absent from three DofCesAs and one PeqCesA. The motif patterns also varied among different species and families; however, we found species-specific motifs and similar conserved motif patterns within the same CesA/Csl family (Figure 3 and Supplementary Figures S3, S4). These results suggested that CesA/Csl proteins among different families in Orchidaceae species might have different functional properties. Furthermore, multiple alignments of the predicted Cellulose synthase amino acid sequences showed that 18 of the 125 CesA/Csl proteins had no “D,D,D,QxxRW” integrated active site amino acid sequence (Supplementary Figures S5–S7), implying possible functional redundancy.
Gene structure and duplication analysis can facilitate the understanding of gene evolution, structural divergence, and functional conservation within a gene family (Worberg et al., 2008; Xu et al., 2012; Nawaz et al., 2017). We found highly diverse structures among the different families but similar exon/intron numbers in the same family among the three orchid species (Figure 4B). The results revealed that there was functional diversity within the same family and that evolutionary conservation occurred among different orchid species. Moreover, a total of 14 tandemly duplicated genes were detected (Figure 4C and Table 1), indicating that the expansion of DofCesA/DofCsl genes might result from tandem duplication. These results provide insight into the evolutionary divergence and origins of CesA/Csl in Orchidaceae species.
Cellulose synthase superfamily genes are involved in the synthesis of the subunits of cellulose and hemicellulose (Burton et al., 2004; Scheller and Ulvskov, 2010; Takata and Taniguchi, 2015). The expansion and diversification of the plant CesA superfamily are linked tightly with major events in the evolution of plant and algal lineages, including multicellularity, terrestrialization, and vascularization (Popper et al., 2011). To understand the functions of the CesA/Csl genes in orchids, we investigated gene expression profiles in different organs of D. officinale and at different developmental stages (Figure 5). The results revealed diverse expression patterns of the DofCesA/DofCsl genes, suggesting functional divergence had occurred after gene expansion.
Three DofCesAs (DofCesA1, DofCesA5, and DofCesA8) were significantly expressed in the flowers of D. officinale. DofCesA8 also exhibited high expression in the roots and moderate expression in the leaves and stems. Except for DofCesA6, which exhibited low expression in the flowers, the expression levels of the remaining DofCesAs were minimal in all the tested organs. In addition, DofCesA8 was also highly expressed in Y1 stems and moderately expressed in the Y2 and Y3 stems. In the Arabidopsis genome, ten AtCesA genes were identified and found to be responsible for primary (CesA1-3, -5, -6, and -9) and secondary (AtCesA4, -7, and -8) cell wall synthesis (Taylor et al., 2003; Burton et al., 2004; Takata and Taniguchi, 2015). Mutations in secondary cell wall AtCesAs result in collapsed vasculature or irregular xylem phenotypes (Turner and Somerville, 1997; Taylor et al., 2003). Moreover, AtCesA1 and AtCesA3 also play roles in flowering; mutations in atcesa1 and atcesa3 result in gamete lethal phenotypes, whereas single-knockout AtCesA2, -5, -6, and -9 mutants exhibit more moderate phenotypes (Scheible et al., 2001; Caño-Delgado et al., 2003). The AtCesA1 was clustered in the same subgroup with DofCesA1, and AtCesA3 was clustered with DofCesA8. Therefore, the results, combined with the expression patterns, suggested that the three DofCesAs likely play roles in flower organ development, and that DofCesA8 may play additional roles in cellulose deposition in the stems of D. officinale.
In plants, the synthesis of hemicellulose occurs in the Golgi membrane by proteins encoded by Csl genes (Kaur et al., 2017). In land plants, CslAs and CslCs encode proteins that are involved in the synthesis of mannan and the β-1,4-linked glucan backbone of xyloglucan, respectively (Liepman et al., 2005, 2010; Cocuron et al., 2007). Moreover, CslD members have been reported to have mannan and Cellulose synthase activities, especially in tip-growing root hairs and pollen tubes of plant species such as rice and Arabidopsis. In rice, the OsCSLD1 gene is required for root hair morphogenesis (Kim et al., 2007). AtCslD3 also plays a role in Cellulose synthesis, as atcsld3 mutants exhibit defects in polarized growth of root hairs and abnormal distribution of cellulose and xyloglucan (Pauly et al., 2001; Park et al., 2011). CslCs are involved in the biosynthesis of the β-1,4-linked glucan backbone of xyloglucan (Cocuron et al., 2007). However, the expression of the DofCslD and DofCslC family genes was either low or absent from the four organs and across the three developmental stages, indicating minimal effects on polysaccharide synthesis in D. officinale. Interestingly, most of the DofCslAs were predominantly expressed in the stems and/or flowers. DofCslA2, -14, and -15 had significantly high expression levels in the stem; DofCslA6 was specifically expressed in the flowers. Moreover, DofCslAs were differentially expressed among the three growth stages. For example, DofCslA14 and DofCslA15 exhibited significantly high expression in Y3 stems, while DofCslA7 and DofCslA8 were expressed at higher levels in Y2 stems. Five of the 19 DofCslAs exhibited low expression or none in the stems of plants at all three growth stages. These results revealed functional divergence and possible redundancy of the DofCslAs involved in polysaccharide synthesis in D. officinale. The functional role of the CslB/E/G families remains unclear, but CslF/H genes have been suggested to be involved in the synthesis of mixed-linkage glucans (MLGs) (Prasad et al., 2005; Burton et al., 2006). We found that the DofCslGs all presented higher expression levels in the flowers than in the rest of the organs. DofCslG1 and DofCslG2 also had moderate expression in Y2 and Y3 stems. Our phylogenetic study showed that DofCslE had significantly expanded in D. officinale compared with other species (Figure 2). However, the expression of most DofCslEs was very low in different organs and growth stages. Thus, further investigations are needed to elucidate the exact roles of DofCslE/G/H family genes.
Containing approximately 1,450 species, Dendrobium is one of the largest genera of the Orchidaceae family (Zhang et al., 2016). Dendrobium species are characterized by their diverse growth habits and bioactive constituents with immunomodulatory hepatoprotective activities, such as dendrobine and polysaccharides (Ng et al., 2012). D. officinale stems contain abundant polysaccharides, primarily those consisting of GM and GGM (Xing et al., 2014). The high amount of polysaccharides in D. officinale stems supposedly helps maintain osmotic pressure and improve drought tolerance, as this species is epiphytic in natural habitats (Zotz and Tyree, 1996; Wu et al., 2009). CslAs encode proteins with both mannan and glucomannan synthase activity (Liepman et al., 2005, 2010). In Arabidopsis, the stems of csla knockout mutants have no glucomannan, indicating an exclusive role in mannan biosynthesis of the CslA family genes (Goubet et al., 2009). To determine the roles of the 19 DofCslAs in polysaccharide accumulation in D. officinale stems across different growth stages, we performed qRT–PCR and measured the content of total polysaccharides and their monosaccharide components in the stems of 3M, 9M, Y1, Y2, and Y3 plants. The total polysaccharide content was lowest in the 3M stems and highest in the Y3 stems (Figure 6 and Table 3). The content varied slightly between 3M and 9M stems but increased significantly in the mature stems. Monosaccharide composition analysis showed that mannose was the most abundant component across all development stages, supporting previous findings in Dendrobium species (Meng et al., 2013). The amounts of glucose and mannose in the stems also experienced remarkable increases from 3M to Y3. In agreement with the changes in total polysaccharide and monosaccharide component contents, most of the DofCslAs exhibited high expression levels in the Y2 and/or Y3 stems (Figure 7). Among them, five genes exhibited significantly high expression levels in Y3 stems (especially DofCslA15); four genes were highly expressed in Y2 stems. Thus, these genes likely play roles in the biosynthesis of mannan and glucomannan in the stems of D. officinale. In addition, six genes showed low expression levels, suggesting that possible redundancy occurred after gene expansion. These results imply that the DofCslAs may experience functional specialization with respect to polysaccharide accumulation in different growth stages.
The raw data of RNA-seq experiment is deposited in Sequence Read Archive (NCBI): PRJNA680456 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA680456) and PRJNA762115 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA762115). All data and material used in this study are available from the corresponding author upon reasonable request.
YW and CS conceived and designed the experiments, performed the experiments, analyzed the data, prepared the figures and/or tables, authored or reviewed the drafts of the study, and approved the final draft. KZ and YC analyzed the data, authored or reviewed the drafts of the study, and approved the final draft. XC, QW, and HW analyzed the data, contributed reagents, materials, and analysis tools, authored or reviewed the drafts of the study, and approved the final draft. CS conceived the experiments, authored or reviewed the drafts of the study, and approved the final draft. All authors contributed to the article and approved the submitted version.
This study was funded by the Zhejiang Provincial Natural Science Foundation (grant number LQ20C150002), Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding (grant number 2021C02071-5).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.777332/full#supplementary-material
Supplementary Figure S1 | Phylogenetic tree of the Cellulose synthase superfamily members in D. officinale (Dof), A. shenzhenica (Apo), and P. equestris (Peq). The phylogenetic tree was constructed using MEGA 6.0 with the neighbor-joining (NJ) method and 1000 bootstrap replicates. The CesA/Csl proteins were grouped into one CesA family and six Csl families: CslA, CslC, CslD, CslE, CslG, and CslH. The families are marked by different arc lines and branch colors. The D. officinale, A. shenzhenica, and P. equestris are distinguished by red, green and blue rhombuses, respectively.
Supplementary Figure S2 | Phylogenetic tree of Cellulose synthase superfamily members from nine representative plant species. The phylogenetic tree was constructed using MEGA 6.0 with the neighbor-joining (NJ) method and 1000 bootstrap replicates. The CesA/Csl proteins were grouped into nine families: CesA, CslA, CslB, CslC, CslD, CslE, CslF, CslG, and CslH. The species included (with the gene code prefixes shown in parentheses) are as follows: Chlamydomonas reinhardtii (Cre), Volvox carteri (Vca), Physcomitrella patens (Ppa), Selaginella moellendorffii (Smo), A. shenzhenica (Apo), D. officinale (Den), P. equestris (Peq), Oryza sativa (Osa), and Arabidopsis thaliana (AT). The families are marked by different arc lines and branch colors.
Supplementary Figure S3 | Phylogenetic, conserved domain and motif and gene structure analyses of Cellulose synthase proteins in P. equestris (Peq). (A) Phylogenetic relationships and conserved domains of the PeqCesA/PeqCsl proteins. The scale bar (aa) indicates the amino acid position in the corresponding conserved domain. (B) Phylogenetic relationships, conserved motifs and gene structure of PeqCesA/PeqCsl genes. UTRs and exons are indicated using green and yellow rectangles, respectively. The solid lines indicate introns. The numbers above the solid lines represent the intron phase. Scale bars (bp/aa) indicate the length/amino acid position of corresponding genes/proteins; (C) Sequence logos of the 10 motifs.
Supplementary Figure S4 | Phylogenetic, conserved domain and motif and gene structure analyses of Cellulose synthase proteins in A. shenzhenica (Apo). (A) Phylogenetic relationships and conserved domains of the AsCesA/AsCsl proteins. The scale bar (aa) indicates the amino acid position in the corresponding conserved domains. (B) Phylogenetic relationships, conserved motifs and gene structure of the AsCesA/AsCsl genes. Exons are indicated in yellow rectangles. The solid lines indicate introns. The numbers above the solid lines represent the intron phase. The scale bars (bp/aa) indicate the length/amino acid position of corresponding genes/proteins; (C) sequence logos of the 10 motifs.
Supplementary Figure S5 | Multiple alignment and motif logos of D. officinale. (A) Multiple comparisons of the deduced amino acid sequences of Cellulose synthase in D. officinale. Conserved cysteine residues are marked by red frames. (B) Sequence logos of the 10 motifs.
Supplementary Figure S6 | Multiple alignment of the deduced amino acid sequences of Cellulose synthases in P. equestris. Conserved cysteine residues are marked by red frames.
Supplementary Figure S7 | Multiple alignment of the deduced amino acid sequences of Cellulose synthase in A. shenzhenica. Conserved cysteine residues are marked by red frames.
Supplementary Figure S8 | Sequence structures of the alternative splicing transcripts of the D. officinale genes. UTRs and exons are indicated using blue and yellow rectangles, respectively. The solid lines indicate introns. The scale bar (bp) indicates the length of the corresponding transcripts.
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic. Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Burton, R. A., Shirley, N. J., Harvey, A. J., and Fincher, G. B. (2004). The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 134, 224–236. doi: 10.1104/pp.103.032904
Burton, R. A., Wilson, S. M., Hrmova, M., Harvey, A. J., Shirley, N. J., Medhurst, A., et al. (2006). Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science 311, 1940–1942. doi: 10.1126/science.1122975
Cai, J., Liu, X., Vanneste, K., Proost, S., Tsai, W. C., Liu, K. W., et al. (2014). The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47, 65–72.
Caño-Delgado, A., Penfield, S., Smith, C., Catley, M., and Bevan, M. (2003). Reduced Cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34, 351–362. doi: 10.1046/j.1365-313x.2003.01729.x
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic. Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., and Xia, R. (2020). TBtools: an integrative Toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cocuron, J. C., Lerouxel, O., Drakakaki, G., Alonso, A. P., Liepman, A. H., Keegstra, K., et al. (2007). A gene from the Cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. U. S. A. 104, 8550–8555. doi: 10.1073/pnas.0703133104
Cui, Z., Sun, G., and Zhao, Q. (2020). Phylogenetic and functional analysis of CesA genes in cotton. Res. Square [Preprint]. doi: 10.21203/rs.3.rs-101509/v1
Desprez, T. T., Juraniec, M. M., Crowell, E. F. E., Jouy, H. H., Pochylova, Z., Parcy, F., et al. (2007). Organization of Cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 104, 15572–15577. doi: 10.1073/pnas.0706569104
Doblin, M. S., Pettolino, F. A., Wilson, S. M., Campbell, R., Burton, R. A., and Fincher, G. B. (2009). A barley Cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 14, 5996–6001. doi: 10.1073/pnas.0902019106
Endler, A., and Persson, S. (2011). Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 4, 199–211.
Farrokhi, N., Burton, R. A., Brownfield, L., Hrmova, M., Wilson, S. M., Bacic, A., et al. (2006). Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol. J. 4, 145–167. doi: 10.1111/j.1467-7652.2005.00169.x
Fincher, G. B. (2009). Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol. 149, 27–37. doi: 10.1104/pp.108.130096
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic. Acids Res. 39, 29–37. doi: 10.1093/nar/gkr367
Gómez-González, S., Ruiz-Jiménez, J., Priego-Capote, F., and Luque de Castro, M. D. (2010). Qualitative and quantitative sugar profiling in olive fruits, leaves, and stems by Gas Chromatography-Tandem Mass Spectrometry (GC-MS/MS) after ultrasound-assisted Leaching. J. Agric. Food Chem. 58, 12292–12299. doi: 10.1021/jf102350s
Goubet, F., Barton, C. J., Mortimer, J. C., Yu, X., Zhang, Z., Miles, G. P., et al. (2009). Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 60, 527–538. doi: 10.1111/j.1365-313X.2009.03977.x
He, C., Zhang, J., Liu, X., Zeng, S., Wu, K., Yu, Z., et al. (2015). Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol. Biol. 88, 219–231.
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Kaur, S., Dhugga, K. S., Beech, R., and Singh, J. (2017). Genome-wide analysis of the Cellulose synthase-like (Csl) gene family in bread wheat (Triticum aestivum L.). BMC Plant Biol. 17:193. doi: 10.1186/s12870-017-1142-z
Keegstra, K., and Walton, J. (2006). Plant science. Beta-glucans-brewer’s bane, dietician’s delight. Science 311, 1872–1873. doi: 10.1126/science.1125938
Kim, C. M., Park, S. H., Je, B. I., Park, S. H., Park, S. J., Piao, H. L., et al. (2007). OsCSLD1, a Cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol. 143, 1220–1230. doi: 10.1104/pp.106.091546
Li, G., Liu, X., Liang, Y., Zhang, Y., and Cai, Y. (2020). Genome-wide characterization of the Cellulose synthase gene superfamily in Pyrus bretschneideri and reveal its potential role in stone cell formation. Funct. Integr. Genomic. 20, 857–858. doi: 10.1007/s10142-020-00747-8
Li, Y., Yang, T., Dai, D., Hu, Y., Guo, X., and Guo, H. (2017). Evolution, gene expression profiling and 3D modeling of CSLD proteins in cotton. BMC Plant Biol. 17:119. doi: 10.1186/s12870-017-1063-x
Liepman, A. H., Wightman, R., Geshi, N., Turner, S. R., and Scheller, H. V. (2010). Arabidopsis - a powerful model system for plant cell wall research. Plant J. 61, 1107–1121. doi: 10.1111/j.1365-313X.2010.04161.x
Liepman, A. H., Wilkerson, C. G., and Keegstra, K. (2005). Expression of Cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. U. S. A. 102, 2221–2226. doi: 10.1073/pnas.0409179102
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
McFarlane, H. E., Döring, A., and Persson, S. (2014). The Cell Biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94. doi: 10.1146/annurev-arplant-050213-040240
Meng, L. Z., Lv, G. P., Hu, D. J., Cheong, K. L., Xie, J., Zhao, J., et al. (2013). Effects of polysaccharides from different species of Dendrobium (Shihu) on macrophage function. Molecules 18, 5779–5791. doi: 10.3390/molecules18055779
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497.
Nawaz, M. A., Rehman, H. M., Baloch, F. S., Ijaz, B., Ali, M. A., Khan, I. A., et al. (2017). Genome and transcriptome-wide analyses of Cellulose synthase gene superfamily in soybean. J. Plant Physiol. 215, 163–175. doi: 10.1016/j.jplph.2017.04.009
Ng, T. B., Liu, J., Wong, J. H., Ye, X., Sze, S. C. W., Tong, Y., et al. (2012). Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 93, 1795–1803.
Nishiyama, Y. (2009). Structure and properties of the cellulose microfibril. J. Wood Sci. 55, 241–249.
Park, S., Szumlanski, A. L., Gu, F., Guo, F., and Nielsen, E. (2011). A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 13, 973–980. doi: 10.1038/ncb2294
Paterson, A. H., Bowers, J. E., Bruggmann, R., Dubchak, I., Grimwood, J., Gundlach, H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556. doi: 10.1038/nature07723
Pauly, M., Qin, Q., Greene, H., Albersheim, P., Darvill, A., and York, W. S. (2001). Changes in the structure of xyloglucan during cell elongation. Planta 212, 842–850. doi: 10.1007/s004250000448
Pear, J. R., Kawagoe, Y., Schreckengost, W. E., Delmer, D. P., and Stalker, D. M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of Cellulose synthase. Proc. Natl. Acad. Sci. U. S. A. 93, 12637–12642. doi: 10.1073/pnas.93.22.12637
Persson, S., Paredez, A., Carroll, A., Palsdottir, H., Doblin, M., Poindexter, P., et al. (2007). Genetic evidence for three unique components in primary cell-wall Cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 104, 15566–15571. doi: 10.1073/pnas.0706592104
Popper, Z. A., Michel, G., Hervé, C., Domozych, D. S., and Stengel, D. B. (2011). Evolution and diversity of plant cell walls: from algae to flowering plants. Annu. Rev. Plant Biol. 62, 567–590. doi: 10.1146/annurev-arplant-042110-103809
Prasad, V., Stromberg, C. A. E., Alimohammadian, H., and Sahni, A. (2005). Dinosaur coprolites and the early evolution of grasses and grazers. Science 310, 1177–1180. doi: 10.1126/science.1118806
Richmond, T. A., and Somerville, C. R. (2000). The Cellulose synthase superfamily. Plant Physiol. 124, 495–498.
Roberts, A. W., and Bushoven, J. T. (2007). The Cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol. Biol. 63, 207–219. doi: 10.1007/s11103-006-9083-1
Scheible, W. R., Eshed, R., Richmond, T., Delmer, D., and Somerville, C. (2001). Modifications of Cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis ixr1 mutants. Proc. Natl. Acad. Sci. U. S. A. 98, 10079–10084. doi: 10.1073/pnas.191361598
Schneider, R., Hanak, T., Persson, S., and Voigt, C. A. (2016). cellulose and callose synthesis and organization in focus, what’s new? Curr. Opin. Plant Biol. 34, 9–16. doi: 10.1016/j.pbi.2016.07.007
Schwerdt, J. G., MacKenzie, K., Wright, F., Oehme, D., Wagner, J. M., Harvey, A. J., et al. (2015). Evolutionary dynamics of the Cellulose synthase gene superfamily in grasses. Plant Physiol. 168, 968–983. doi: 10.1104/pp.15.00140
Sethaphong, L., Haigler, C. H., Kubicki, J. D., Zimmer, J., Bonetta, D., DeBolt, S., et al. (2013). Tertiary model of a plant Cellulose synthase. Proc. Natl. Acad. Sci. U. S. A. 110, 7512–7517. doi: 10.1073/pnas.1301027110
Shu, O., Wei, Z., John, H., Lin, H., Matthew, C., Kevin, C., et al. (2007). The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic. Acids Res. 35, D883–D887. doi: 10.1093/nar/gkl976
Song, X. M., Li, X., Yu, J. W., Tian, P., Hu, X., Wang, Q. J., et al. (2018). Genome-wide characterization of the Cellulose synthase gene superfamily in Solanum lycopersicum. Gene 688, 71–83.
Sun, S. H., Wang, H., Xie, J. P., and Sun, Y. (2016). Simultaneous determination of rhamnose, xylitol, arabitol, fructose, glucose, inositol, sucrose, maltose in jujube (Zizyphus jujube Mill.) extract: comparison of HPLC-ELSD, LC-ESI-MS/MS and GC-MS. Chem. Central J. 10:25. doi: 10.1186/s13065-016-0171-2
Suzuki, S., Li, L., Sun, Y. H., and Chiang, V. L. (2006). The Cellulose synthase gene superfamily and biochemical functions of xylem-specific Cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 142, 1233–1245. doi: 10.1104/pp.106.086678
Takata, N., and Taniguchi, T. (2015). Expression divergence of Cellulose synthase (CesA) genes after a recent whole genome duplication event in Populus. Planta 241, 29–42. doi: 10.1007/s00425-014-2217-9
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). Mega6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729.
Taylor, N. G. (2008). cellulose biosynthesis and deposition in higher plants. New Phytol. 178, 239–252. doi: 10.1111/j.1469-8137.2008.02385.x
Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K., and Turner, S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U. S. A. 100, 1450–1455. doi: 10.1073/pnas.0337628100
Turner, S. R., and Somerville, C. R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. doi: 10.1105/tpc.9.5.689
Verhertbruggen, Y., Yin, L., Oikawa, A., and Scheller, H. V. (2011). Mannan synthase activity in the CSLD family. Plant Signal. Behav. 6:1620. doi: 10.4161/psb.6.10.17989
Wang, Y., Chen, Y., Wei, Q., Wan, H., and Sun, C. (2021). Phylogenetic relationships of sucrose transporters (SUTs) in plants and genome-wide characterization of SUT genes in Orchidaceae. PeerJ 9:e11961.
Wang, L. Q., Guo, K., Li, Y., Tu, Y. Y., Hu, H. Z., Wang, B. R., et al. (2010). Expression profiling and integrative analysis of the CesA/Csl superfamily in rice. BMC Plant Biol. 10:282. doi: 10.1186/1471-2229-10-282
Worberg, A., Quandt, D., Barniske, A. M., Löhne, C., Hilu, K. W., and Borsch, T. (2008). Phylogeny of basal eudicots: insights from non-coding and rapidly evolving DNA. Org. Divers. Evol. 7, 55–77.
Wu, Z. Y., Raven, P. H., and Hong, D. Y. (2009). Flora of China (Orchidaceae). Beijing: Science Press.
Xing, X., Cui, S. W., Nie, S., Phillips, G. O., Goff, H. D., and Wang, Q. (2014). Study on Dendrobium officinale O-acetyl-glucomannan (Dendronan R): part I. Extraction, purification, and partial structural characterization. Bioact. Carbohydr. Diet. Fibre 4, 74–83.
Xu, G., Guo, C., Shan, H., and Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U. S. A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Yin, Y., Huang, J., and Xu, Y. (2009). The Cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 9:99. doi: 10.1186/1471-2229-9-99
Yin, Y., Johns, M. A., Cao, H., and Rupani, M. (2014). A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the Cellulose synthase superfamily. BMC Genom. 15:260. doi: 10.1186/1471-2164-15-260
Zhang, G. Q., Liu, K. W., Li, Z., Lohaus, R., Hsiao, Y. Y., Niu, S. C., et al. (2017). The Apostasia genome and the evolution of orchids. Nature 549, 379.
Zhang, G. Q., Xu, Q., Bian, C., Tsai, W. C., Yeh, C. M., Liu, K. W., et al. (2016). The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 6:19029. doi: 10.1038/srep19029
Zou, X., Zhen, Z., Ge, Q., Fan, S., Liu, A., Gong, W., et al. (2018). Genome-wide identification and analysis of the evolution and expression patterns of the cellulose synthase gene superfamily in Gossypium species. Gene 646, 28–38.
Keywords: Cellulose synthase, CesA/Csl, Orchidaceae, gene expansion, polysaccharide synthesis
Citation: Wang Y, Zhao K, Chen Y, Wei Q, Chen X, Wan H and Sun C (2022) Species-Specific Gene Expansion of the Cellulose synthase Gene Superfamily in the Orchidaceae Family and Functional Divergence of Mannan Synthesis-Related Genes in Dendrobium officinale. Front. Plant Sci. 13:777332. doi: 10.3389/fpls.2022.777332
Received: 15 September 2021; Accepted: 02 May 2022;
Published: 03 June 2022.
Edited by:
Ming Chen, Zhejiang University, ChinaReviewed by:
Jia-Yu Xue, Nanjing Agricultural University, ChinaCopyright © 2022 Wang, Zhao, Chen, Wei, Chen, Wan and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chongbo Sun, Y2hvbmdwbzEyMzBAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.