
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 March 2022
Sec. Aquatic Photosynthetic Organisms
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.749982
 Yichao Wang1,2,3,4
Yichao Wang1,2,3,4 Shuya Liu1,2,4
Shuya Liu1,2,4 Jing Wang1,2,4
Jing Wang1,2,4 Yanxin Yao1,2,3,4
Yanxin Yao1,2,3,4 Yang Chen1,2,3,4
Yang Chen1,2,3,4 Qing Xu1,2,4,5
Qing Xu1,2,4,5 Zengxia Zhao2,6
Zengxia Zhao2,6 Nansheng Chen1,2,4,7*
Nansheng Chen1,2,4,7*Diatoms (Bacillariophyta) constitute one of the most diverse and ecologically significant groups of phytoplankton, comprising 100,000–200,000 species in three classes Bacillariophyceae, Mediophyceae, and Coscinodiscophyceae. However, due to the limited resolution of common molecular markers including 18S rDNA, 28S rDNA, ITS, rbcL, and cox1, diatom biodiversity has not been adequately ascertained. Organelle genomes including mitochondrial genomes (mtDNAs) have been proposed to be “super barcodes” for distinguishing diatom species because of their rich genomic content, and the rapid progress of DNA sequencing technologies that has made it possible to construct mtDNAs with increasing throughout and decreasing cost. Here, we constructed complete mtDNAs of 15 diatom species including five Coscinodiscophyceae species (Guinardia delicatula, Guinardia striata, Stephanopyxis turris, Paralia sulcata, and Actinocyclus sp.), four Mediophyceae species (Hemiaulus sinensis, Odontella aurita var. minima, Lithodesmioides sp., and Helicotheca tamesis), and six Bacillariophyceae species (Nitzschia ovalis, Nitzschia sp., Nitzschia traheaformis, Cylindrotheca closterium, Haslea tsukamotoi, and Pleurosigma sp.) to test the practicality of using mtDNAs as super barcodes. We found that mtDNAs have much higher resolution compared to common molecular markers as expected. Comparative analysis of mtDNAs also suggested that mtDNAs are valuable in evolutionary studies by revealing extensive genome rearrangement events with gene duplications, gene losses, and gains and losses of introns. Synteny analyses of mtDNAs uncovered high conservation among species within an order, but extensive rearrangements including translocations and/or inversions between species of different orders within a class. Duplication of cox1 was discovered for the first time in diatoms in Nitzschia traheaformis and Haslea tsukamotoi. Molecular dating analysis revealed that the three diatom classes split 100 Mya and many diatom species appeared since 50 Mya. In conclusion, more diatom mtDNAs representing different orders will play great dividends to explore biodiversity and speciation of diatoms in different ecological regions.
Diatoms (Bacillariophyta) constitute a major group of phytoplankton as free living organisms in both marine and freshwater environments or as endosymbionts in dinoflagellates and foraminifers (Field et al., 1998; Pierella Karlusich et al., 2020). Diatoms are unicellular photosynthetic microalgae deemed to be of crucial significance in biogeochemical cycles and the functioning of aquatic food webs (Smetacek, 1998; Armbrust, 2009; Falciatore et al., 2020). Diatoms have the most effective RuBisCO enzyme within autotrophs and work as an essential part in the cycling of CO2, which is predicted to be comparable to that of all terrestrial rainforests combined in global carbon cycling (Field et al., 1998; Giordano et al., 2005) and constitute a large part of aquatic biomass contributing approximately 20% of the total primary production on Earth (Nelson et al., 1995; Falkowski et al., 1998). Diatoms also have great value in commercial applications. Combination of different diatom species provides better balanced nutrition and improves aquaculture animal growth better such as Chaetoceros, Skeletonema, and Thalassiosira (Spolaore et al., 2006). Odontella aurita is a marine diatom with rich eicosapentaenoic acid (EPA) content and displays beneficial effect in reducing risk factors for high-fat induced metabolic syndrome (Haimeur et al., 2012). The high neutral lipid content and growth rate of Fistulifera solaris are beneficial for biodiesel production (Tanaka et al., 2015). Diatom species can also pose substantial negative impact on environment. Extreme successions of some diatom species under certain situation can cause harmful algal blooms (HABs) with serious negative consequences on human health, as well as economic loss and social disruption (Jin et al., 2008; Dyson and Huppert, 2010; Lelong et al., 2012; Crosman et al., 2019; Moore et al., 2020). For example, Pseudo-nitzschia pungens has been found to cause HABs in coastal regions of New Zealand and the west coastal regions of America (Patrick et al., 2016).
The critical importance of diatoms is to some degree dictated by their biodiversity. Diatoms are divided into three classes Bacillariophyceae, Mediophyceae, and Coscinodiscophyceae (Medlin and Kaczmarska, 2019). Diatom species show high variations in cell sizes, ranging from a few micrometers to a few millimeters, with single cells forming chains of connected cells for many species (Foster et al., 2011). While morphological features have been effectively used as defining criteria for taxonomy of the diatom species, taxonomy for diatom species with small-cell sizes, and few discrete morphological characters under light microscope are challenging (Mann et al., 2021). Many molecular markers including 18S rDNA, 28S rDNA, ITS, rbcL, and cox1 have been successfully applied alone or in combination with microscopy to study diatom species (Evans et al., 2007; Hamsher et al., 2013; Park et al., 2017). While these common molecular markers are normally effective for distinguishing higher taxa, they are often inadequate for distinguishing congener species, let alone intra-species genetic diversity (Wilcox, 1998; Hollingsworth et al., 2011; Song et al., 2020). As such, although between 100,000 and 200,000 diatom species have been estimated to exist (Malviya et al., 2016), currently only about 12,000 species have been identified (Mann and Vanormelingen, 2013). Lack of molecular markers with sufficient resolution impedes in-depth analysis of diatom biodiversity and speciation, and particularly their differential contribution to ecological systems.
With the development of DNA sequencing technologies and bioinformatics analysis algorithms, mitochondrial genomes (mtDNAs) of thousands species have been fully constructed, which helped to uncover valuable knowledge about phylogenetic relationships and evolutionary trajectories (Smith, 2016). The research of diatom mtDNAs began with the construction of the mtDNA (a circular DNA of 43,827 bp) of Thalassiosira pseudonana (Armbrust et al., 2004). Subsequent construction of the Phaeodactylum tricornutum mtDNA revealed a 35,454-bp repeat sequence, providing a first glimpse of rich divergence among diatoms mtDNAs (Secq and Green, 2011). Further analysis of mtDNAs uncovered the existence of group I intron, encoding a LAGLIDADG endonuclease in the mtDNA of the endosymbiont diatom Kryptoperidinium foliaceum (Imanian et al., 2012). Comparative analysis of five mtDNAs (Toxarium undulatum, Psammoneis japonica, Eunotia naegelii, Cylindrotheca closterium, and Nitzschia sp.) with eleven publicly available diatom mtDNAs suggested that diatom mtDNAs shared strongly conserved genes and highly variable intron content caused by intron loss and horizontal transfer (Guillory et al., 2018). The mtDNAs have also been used as “super barcodes” with higher resolution than common molecular marker genes for comparative genomics analysis (Lv et al., 2020). Mitochondrial genomes have been used to track the genetic diversity of the harmful algal species Eucampia zodiacus (Zhang W. et al., 2021). Despite their critical importance, mtDNAs of only 53 diatom species of seventeen orders have been successfully constructed, with many orders underrepresented. In particular, only four mtDNAs have been constructed for species in the class Coscinodiscophyceae. Such limited availability of mtDNAs hinders in-depth research on diatom biodiversity and the speciation of diatoms.
The Jiaozhou Bay, which is connected to the Yellow Sea with a small opening, is an epitome of China’s offshore ecosystem (Liu et al., 2004). Previous studies revealed rich diatom composition in the Jiaozhou Bay, which makes it suitable for studying the biodiversity and speciation of diatom (Liu and Chen, 2021). In this study, we constructed complete mtDNAs of 15 diatom species, which was the first step in constructing mtDNAs for all diatom species in the Jiaozhou Bay. All of these 15 strains were isolated from the Jiaozhou Bay, consisting of strains from 10 diatom orders including two orders Bacillariales (four species) and Naviculales (two species) of the class Bacillariophyceae, four orders Eupodiscales (one species), Hemiaulales (one species), Briggerales (one species) and Lithodesmiales (one species) of the class Mediophyceae, and four orders from Paraliales (one species), Stephanopyxales (one species), Rhizosoleniales (two species), and Coscinodiscales (one species) of the class Coscinodiscophyceae. For accurate comparison, we re-annotated 53 published mtDNAs publicly available at GenBank at NCBI and made several annotation corrections. This work enriched diatoms’ mtDNA resources for comparative analysis of biodiversity and speciation, and represents the first large-scale construction of mtDNAs for diatom species from the Jiaozhou Bay.
Fifteen diatom strains (CNS00113, CNS00114, CNS00354, CNS00378, CNS00381, CNS00407, CNS00413, CNS00418, CNS00428, CNS00432, CNS00433, CNS00513, CNS00514, CNS00558, and CNS00559) were isolated from water samples collected in the Jiaozhou Bay (Figure 1), onboard the R/V Chuangxin, which was operated by The Jiaozhou Bay Marine Ecosystem Research Station using single-cell capillary methods. Cells were cultured in L1 medium with 1‰ volume fraction Na2SiO3 with H2O added. The culture temperature was 18–20°C, and the illuminance was from 2,000 to 3,000 Lx at the photoperiod of 12 h light–12 h dark.
Total DNA was extracted with DNAsecure Plant Kit (Tiangen Biotech, Beijing, China). The genomic DNA samples were fragmented by sonication (Covaris S220, Covaris, WBN, United States) to a size of 350 bp. DNA fragments were then end polished, A-tailed, and ligated with the full-length adapters for Illumina sequencing, followed by PCR (MiniAmp Thermal Cycler, ThermoFisher, MA, United States) enrichment using generic adapter P5 and P7 oligos. The DNA libraries were sequenced using NovaSeq PE150 (Illumina, San Diego, CA, United States) and paired-end reads in size of 150 bp were generated.
All diatom strains were observed using microscope (Zeiss Axio Imager Z2) (Figure 2A). Common molecular marker 18S rDNA for each strain were constructed using whole genome sequencing results. The phylogenetic trees of 18S rDNA were constructed using MEGA7 (Kumar et al., 2016). Phylogenetic relationships were inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) was shown next to the branches (Felsenstein, 1985). The analyses involved 50 nucleotide sequences in 18S rDNA gene. All positions containing gaps and missing data were eliminated. There were a total of 1,518 positions in the final dataset of 18S rDNA gene.

Figure 2. Morphological and molecular anlaysis of diatom species analyzed in this study. (A) Representative micrographs of 15 diatom species studied in this project. (B) Phylogenetic analysis based on 18S ribosomal DNA (18S rDNA) gene. Numbers at the branches represent bootstrap values. Branch lengths are proportional to the genetic distances, which are indicated by the scale bar.
Raw data was trimmed using Trimmomatic-0.39 with the parameters: LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:75 (Bolger et al., 2014). Clean reads were assembled to get complete mtDNAs using GetOrganelle-1.7.4.1 (Jin et al., 2020). The mtDNA sequences were validated using BWA-0.7.17 (Li and Durbin, 2009), SAMtools-1.10 (Li et al., 2009), and IGV-2.7.2 (Thorvaldsdottir et al., 2013). Annotations were conducted with MFannot1 and NCBI’s ORF Finder,2 completed in NCBI’s Sequin15.103 with genetic code of Mold, Protozoan, and Coelenterate Mitochondrial; Mycoplasma/Spiroplasma. For the accuracy of comparative analysis, we had inspected and re-annotated the 53 mtDNAs which were downloaded from the NCBI.
The amino acid sequences encoded by 27 mitochondrial protein-coding genes (atp6, 8, 9; cob; cox1, 2, 3; nad1-7, 4L, 9, 11; rpl2, 6, 14, 16; rps3, 4, 8, 11, 14; and tatC) from each diatom species were extracted and concatenated for phylogenetic analysis and Phytophthora ramorum in Oomycota was used as outgroup according to previous studies (Martin, 2008; An et al., 2017; Liu et al., 2019a). The amino acid sequences encoded by the 27 genes were individually aligned using MAFFT (Katoh and Standley, 2013). Ambiguously aligned regions were trimmed with using trimAl 1.2rev59 (Capella-Gutierrez et al., 2009), and then concatenated using Phyutility (Smith and Dunn, 2008). The best-fit model was found by ModelFinder (Kalyaanamoorthy et al., 2017). The incongruence length difference test (ILD, also called the partition homogeneity test) was conducted via PAUP4.0 with the following parameters: number of replicates = 100; optimality criterion = maximum parsimony (Swofford, 2002). The p-value (0.01) indicated that combining data would not affect the phylogenetic accuracy. When the ILD detects p values lower than 0.001, the combined data performs less well than the individual partitions (Cunningham, 1997). The phylogenetic tree was constructed with IQ-TREE (Trifinopoulos et al., 2016). The ultrafast bootstrap analysis with 1,000 replicates of the dataset and approximate Bayes test were conducted to estimate statistical reliability (Anisimova et al., 2011; Bui Quang et al., 2013). The datasets used for phylogenetic inference are provided in Supplementary Material.
Synteny analysis of mtDNA sequences was carried out with the program Mauve v2.3.1 using progressiveMauve (Darling et al., 2010). The comparative illustration of mtDNAs was performed using circos-0.69 (Krzywinski et al., 2009).
Phylogenetic analysis and molecular dating were analyzed by calculating the codon evolution rate of 27 mitochondrial protein-coding gene nucleotide sequences (atp6, 8, 9; cob; cox1, 2, 3; nad1-7, 4L, 9, 11; rpl2, 6, 14, 16; rps3, 4, 8, 11, 14; and tatC). The Ectocarpus siliculosus was used as outgroup with its fossil age (Supplementary Table 1). The nucleotide sequence of 27 protein-coding genes (PCGs) were aligned using MAFFT with Codon and concatenated in the software PhyloSuite (Zhang et al., 2020). The phylogenetic tree was performed with IQ-TREE. Molecular dating was conducted using PAML package (Yang, 2007). Briefly, rough estimation of substitution rate was conducted using baseml, and estimation of divergence times with the approximate likelihood method was performed using mcmctree. Five calibration points were used in this analysis (Supplementary Table 1). The phylogenetic tree was displayed in the Figtree and visualized with 95% highest posterior density interval (HPD) for each node.
Fifteen diatom strains analyzed in this study were all isolated from the Jiaozhou Bay, China (Figure 1). These strains were annotated based on their morphological features (Figure 2A) and the similarity of the molecular marker 18S rDNA sequence of each strain to those of known species (Figure 2B), with percentage identity (PID) ≥ 99% to corresponding reference sequences. Three strains CNS00433, CNS00113, and CNS00418, which were unicellular cells that were long, straight, and ovoid, shared morphologies that resembled Nitzschia species (Trobajo et al., 2013). The strain CNS00433 was annotated as Nitzschia traheaformis because its full-length 18S rDNA clustered closely with that of Nitzschia traheaformis in the phylogenetic tree (Figure 2B) with high PID (99.69% with KT943644) (Table 1) (Witkowski et al., 2016). The strain CNS00113 was annotated as Nitzschia ovalis (PID = 99.82% with FR865500) (Gachon et al., 2013), while the strain CNS00418 was annotated as Nitzschia sp. (PID = 98.70% with KY320394). The strain CNS00381, whose cells were unicellular and long with cylindrical central part and elongated ends, was annotated as Cylindrotheca closterium (99.71% with DQ019446) (Williams, 2007). The cells of strains CNS00413 and CNS00432 were all spindle-like. The strain CNS00413 was annotated as Pleurosigma sp. (98.31% PID with AY485514) (Damste et al., 2004), while the strain CNS00432, which had a “sandwich-type” valve structure, was annotated as Haslea tsukamotoi (99.61% PID with KY937691) (Li et al., 2017). The strain CNS00514, whose cells had protrusions at the ends that connected the cells into a curved chain, was annotated as Hemiaulus sinensis (100% PID with HQ912624) (Theriot et al., 2010). The strain CNS00407, whose flat cells were tightly connected and twisted into a chain, was annotated as Helicotheca tamesis (99.64% PID with EF192982) (Rampen et al., 2007). The strain CNS00559, whose cells were irregular polygon and possessed numerous centrally located plastids, was annotated as Lithodesmioides sp. (98.80% PID with HQ912655) (Theriot et al., 2010). The strain CNS00354, whose cells had long thorns in the middle was annotated as Odontella aurita var. minima (99.63% PID with KC309511) (Ashworth et al., 2013). The strains CNS00378, CNS00428, CNS00558, CNS00513, and CNS00114 were annotated as species in the class Coscinodiscophyceae based on their morphological features (Figure 2A). The strains CNS00378, CNS00558, and CNS00513 were annotated as Stephanopyxis turris (99.94% PID with HQ912657), Guinardia delicatula (99.94% PID with AY485487), and Guinardia striata (99.94% PID with KT861015), respectively (Damste et al., 2004; Theriot et al., 2010). The strain CNS00428 was annotated as Paralia sulcata (99.71% PID with HQ912573) (Theriot et al., 2010). The strain CNS00114, whose cells were round did not form chain, was annotated as Actinocyclus sp. (98.86% PID with X85401) (Kooistra and Medlin, 1996).
We successfully constructed the complete mtDNA for all 15 diatom strains, among which 14 mtDNAs representing 14 species were constructed for the first time (Figure 3). The mtDNA of Cylindrotheca closterium, whose mtDNA was previously published, was constructed for a second strain collected from Jiaozhou Bay for comparative analysis. The sizes of these 15 mtDNAs varied substantially, ranging from 34,323 bp (Guinardia delicatula, order Rhizosoleniales, class Coscinodiscophyceae) to 57,953 bp (Nitzschia ovalis, order Bacillariales, class Bacillariophyceae) (Table 2). The AT contents also varied substantially, ranging from 68.0% (Nitzschia ovalis) to 77.8% (Actinocyclus sp.). Interestingly, of these mtDNAs, genome sizes and AT contents showed a significant negative correlation (Pearson correlation coefficient = −0.59, p < 0.05), suggesting that mtDNAs with large sizes tend to have relatively lower AT contents. Further study on more mtDNAs is needed to ascertain the relationship between AT contents and mtDNA genome sizes.
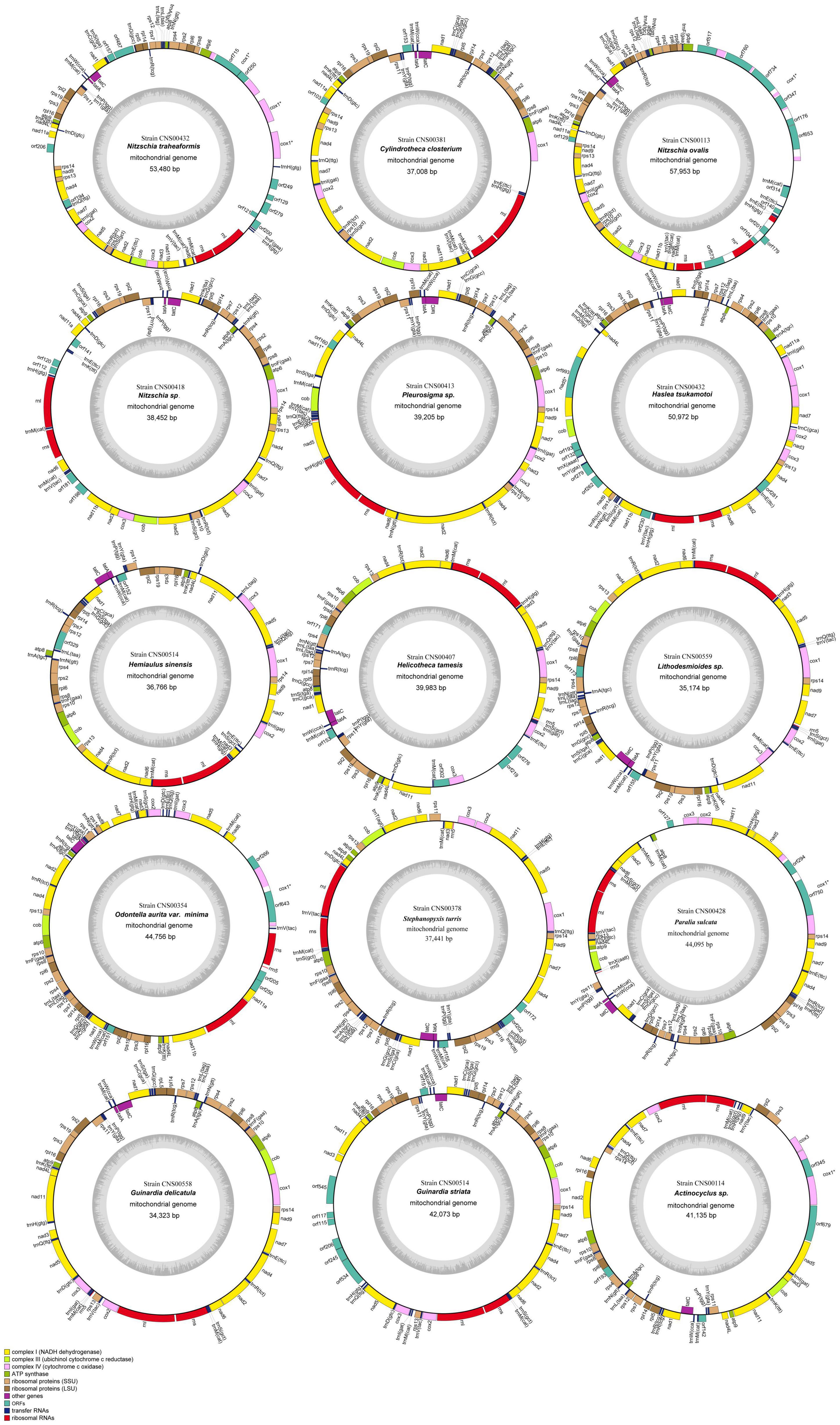
Figure 3. Circular maps of 15 complete mtDNAs. Genes shown on the inside of the map are transcribed in a clockwise direction, whereas those on the outside of the map are transcribed counterclockwise. The assignment of genes into different functional groups is indicated by colors. The ring of bar graphs on the inner circle shows the AT content in light gray.
With the 15 mtDNAs constructed in this study, there are altogether 68 mtDNA constructed for all Bacillariophyta species. Of these 68 mtDNAs, 39 mtDNAs were for species in Bacillariophyceae, 20 mtDNAs were for species in Mediophyceae, while only nine mtDNAs were for species in Coscinodiscophyceae (Table 3). Most mtDNAs contained 35 PCGs and two non-coding rRNA genes (rns and rnl). The mtDNAs PCGs of Nitzschia traheaformis and Haslea tsukamotoi were exceptional because they possessed three and two copies of cox1, respectively (Figure 4). Different copies of cox1 genes had different lengths, suggesting prolonged evolution after the duplication events. The numbers of tRNA genes varied from 20 (Actinocyclus sp.) to 28 (Psammoneis japonica). Most instances of introns were found in cox1 and rnl (Table 3). However, introns were also found in other genes including nad5 and nad11 in the mtDNAs of Haslea tsukamotoi and Pleurosigma sp., in cox2, cox3, cob, nad7, and rns in Halamphora calidilacuna mtDNA, and nad2 in Eunotia naegelii mtDNA. Among these introns, only four group I introns were found, including one found in cox1 in the mtDNAs of Nitzschia ovalis, one found in rns in the mtDNA of Halamphora calidilacuna, and two found in cox1 and rnl in the mtDNA of Endosymbiont of Kryptoperidinium foliaceum (Table 3).
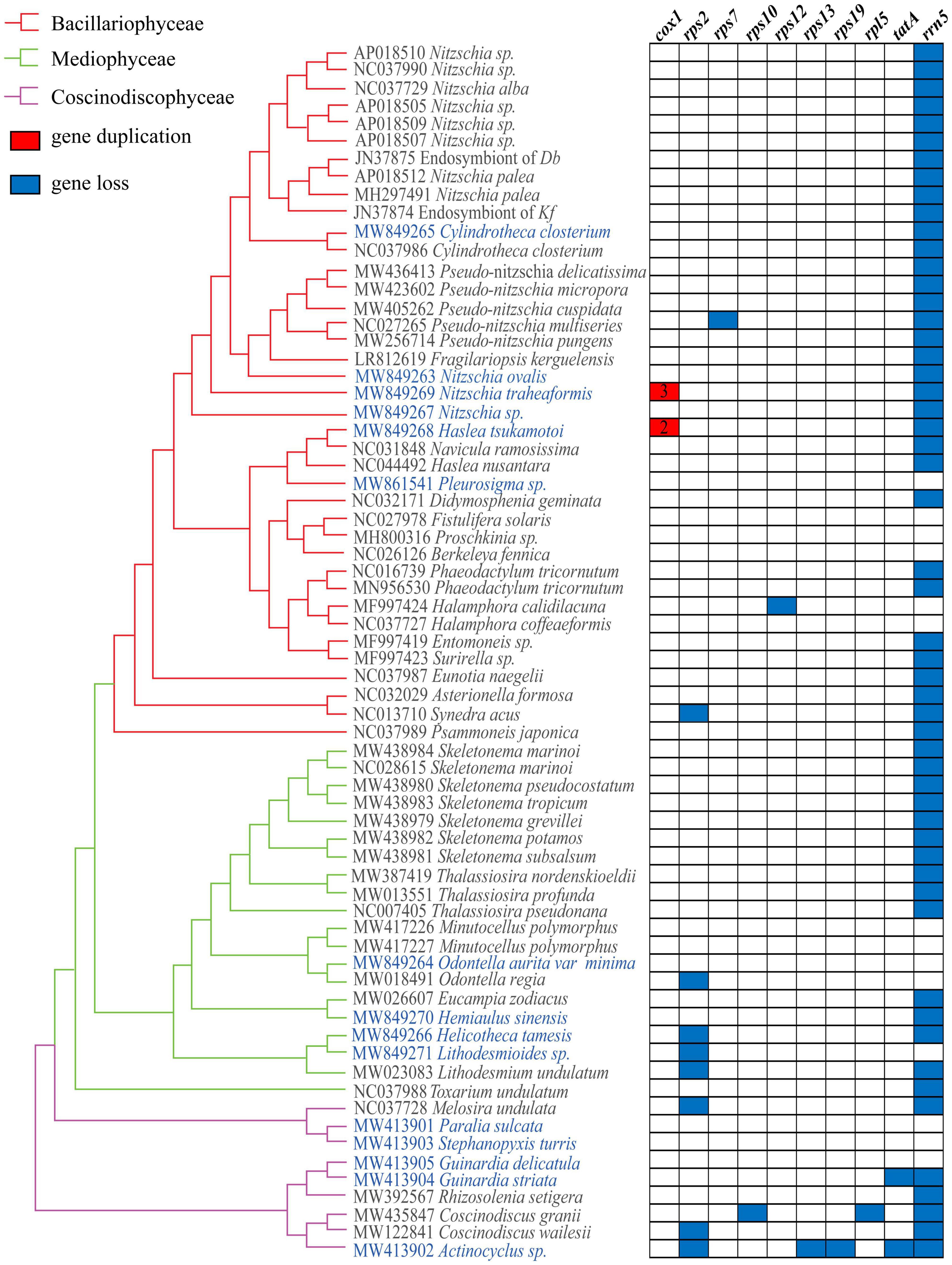
Figure 4. Gene losses and gains in diatom mtDNAs. The matrix shows 10 genes variably present among 68 mtDNAs.
Independent gene loss events have been identified in these mtDNAs. While the 5S rRNA gene (rrn5) was missing in the 52 of 68 mtDNAs, most other genes were missing in mtDNAs of only a few species. The gene rps2 was found missing in eight mtDNAs, including four mtDNAs of species in Mediophyceae and three mtDNAs of species in Coscinodiscophyceae (Figure 4). The gene tatA was found missing in two mtDNAs, both of which were species in Coscinodiscophyceae. Other genes, including rps7, rps10, rps12, rps13, rps19, and rpl5, were found to be missing in single instances of these 68 mtDNAs, suggesting that these genes were likely disposable from the mtDNAs. Indeed, similarity searches of genes rps2, rps13, rps19, and tatA that were lost in mtDNAs in their corresponding nuclear genomes did not identify close homologs. In the class Mediophyceae, mtDNAs of Lithodesmioides sp., Odontella regia, and Lithodesmium undulatum each lost a single PCG rps2. In the class Coscinodiscophyceae, Guinardia striata mtDNA lost a single PCG tatA, while Actinocyclus sp. mtDNA lost four PCGs rps2, rps13, rps19, and tatA. In addition, Coscinodiscus grani mtDNA lost two PCGs rps10 and rpl5, mtDNAs of Coscinodiscus wailesi and Melosira undulata each lost a single PCG rps2. In the class Bacillariophyceae, mtDNAs of three species each lost a single PCGs. Synedra acus mtDNA lost rps2, Pseudo-nitzschia multiseries mtDNA lost rps7, while Halamphora calidilacuna mtDNA lost rps12.
To explore the evolutionary relationships between mtDNAs of 15 species constructed in this project and those of other diatom species reported previously, we constructed a phylogenetic tree using amino acid sequences of PCGs shared by these mtDNAs using maximum likelihood method. As expected, all diatom species generally fell into three clades, corresponding to three distinct classes Bacillariophyceae, Mediophyceae, and Coscinodiscophyceae (Figure 5). Mitochondrial genomes representing 39 species in the class Bacillariophyceae were classified into nine orders. Of the 15 mtDNAs constructed in this study, six mtDNAs belonged to the orders Bacillariales and Naviculales in this class. In Bacillariales, Cylindrotheca closterium (MW849265 of the strain CNS00381) clustered with another strain of Cylindrotheca closterium (NC_037986) as expected (Guillory et al., 2018). The mtDNAs of Nitzschia ovalis, Nitzschia traheaformis, and Nitzschia sp. did not cluster with mtDNAs of other species in the genus Nitzschia, suggesting that that the genus Nitzschia may actually represent multiple genera. In the order Naviculales, Haslea tsukamotoi was found to group more closely with Navicula ramosissima than with Haslea nusantara suggesting that that the genus Navicula and Haslea had more complex relationships (An et al., 2016; Prasetiya et al., 2019). And the mtDNA of Pleurosigma sp. was found to form an independent clade (Figure 5).
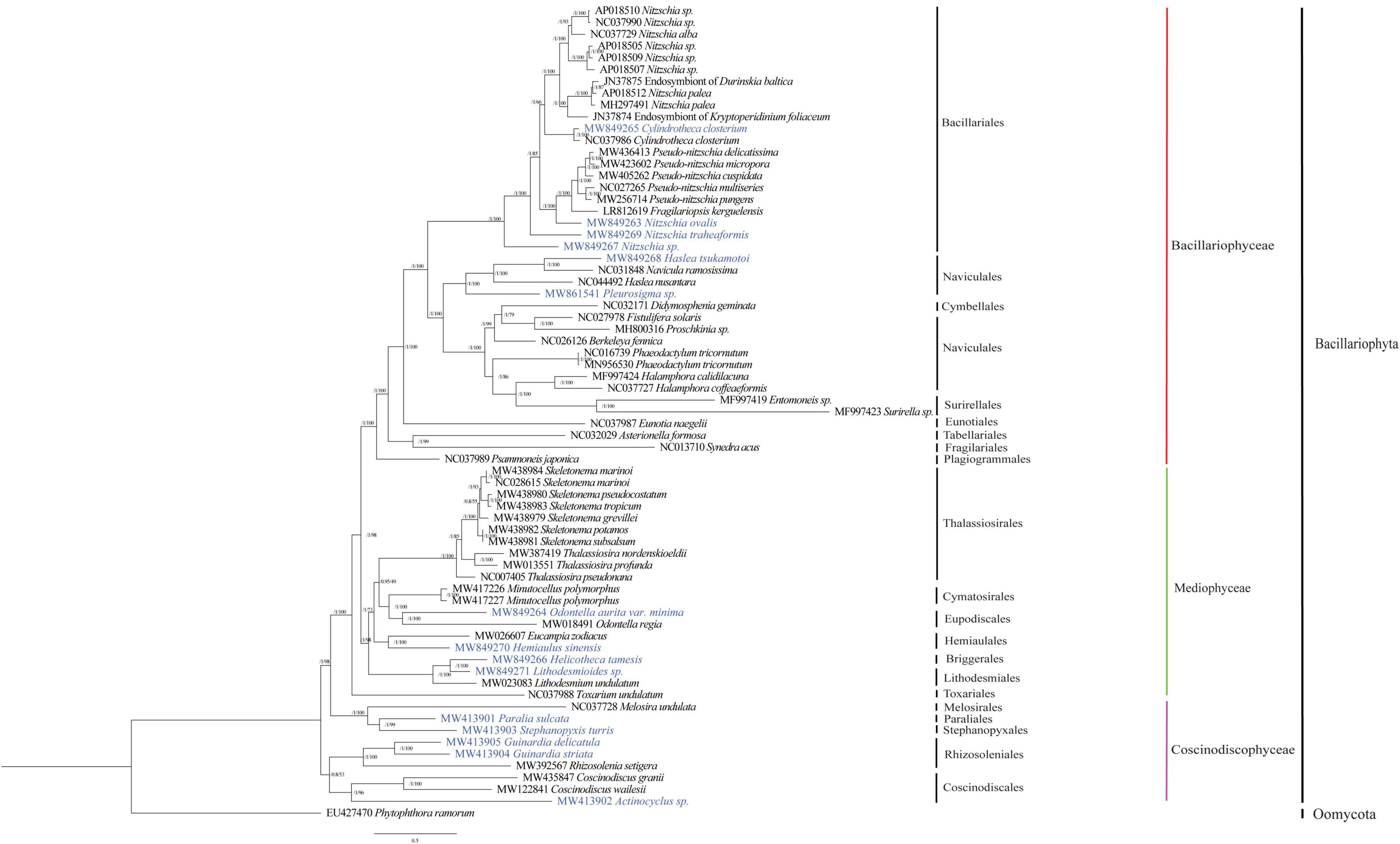
Figure 5. Maximum likelihood (ML) phylogenetic tree based on concatenated amino acid sequences encoded by 27 mitochondrial protein-coding genes (atp6, 8, 9; cob; cox1, 2, 3; nad1-7, 4L, 9, 11; rpl2, 6, 14, 16; rps3, 4, 8, 11, 14; and tatC). Mitochondrial protein-coding genes of Phytophthora ramorum were used as outgroup taxa. Numbers on the left and right side at the branches represent Bayesian posterior probabilities and bootstrap values. Branch lengths were proportional to the amount of sequence change, which are indicated by the scale bar below the trees.
Twenty mtDNAs (including four mtDNAs constructed in this study) were found in the Mediophyceae clade, including species representing seven orders. Odontella aurita var. minima mtDNA was grouped with that of Odontella regia in the order Eupodiscales as expected (Wang et al., 2021). Hemiaulus sinensis mtDNA was clustered with that of Eucampia zodiacus (Zhang M. et al., 2021) in the order Hemiaulales. Lithodesmioides sp. mtDNA, which belonged to the order Lithodesmiales, was clustered more closely with the mtDNA of Helicotheca tamesis from another order Briggerales than with the mtDNA of Lithodesmium undulatum that belongs to the same order Lithodesmiales (Figure 5).
In the class Coscinodiscophyceae, nine mtDNAs (including five mtDNAs constructed in this study) representing five orders have been constructed. Mitochondrial genome of Paralia sulcata, which represented the first species of the order Paraliales, was clustered with the mtDNA of Stephanopyxis turris, which also represented the first species of the order Stephanopyxales. In the order Rhizosoleniales, mtDNAs of Guinardia delicatula and Guinardia striata from the same genus Guinardia formed a clade within the same order that also included Rhizosolenia setigera. Finally, mtDNA of Actinocyclus sp. was clustered with that of Coscinodiscus granii and Coscinodiscus wailesii (Figure 5).
Taken together, the 15 mtDNAs constructed in this study were grouped into 14 orders of diatom species. Notably, the mtDNAs of Helicotheca tamesis, Paralia sulcata, and Stephanopyxis turris, represented the first mtDNAs of the orders Briggerales, Paraliales, and Stephanopyxales, respectively, providing first reference mtDNAs for these orders.
We further explored the sizes of intergenic regions of mtDNAs in the context of the phylogenetic relationships (Figure 6). As expected, the intergenic regions of diatom mtDNAs were generally small, with only minor exceptions. Coding sequence of the gene nad11 was split into two segments in the mtDNAs of many diatom species. The availability of 15 additional mtDNAs allowed us to systematically examine the evolutionary features of nad11 (Figure 6). Among 39 mtDNAs of class Bacillariophyceae, nad11 showed split model in 33 mtDNAs (Figure 6). The gene nad11 was not split in the mtDNAs of six species including Pleurosigma sp., Fistulifera solaris, Proschkinia sp., Psammoneis japonica, Asterionella Formosa, and Synedra acus. In contrast, the gene nad11 was not split in all mtDNAs of class Mediophyceae except Odontella aurita var.minima. In the class Coscinodiscophyceae, none of the nad11 genes were split in the mtDNAs of all species, suggesting that the split of nad11 was occurred multiple times in evolution.
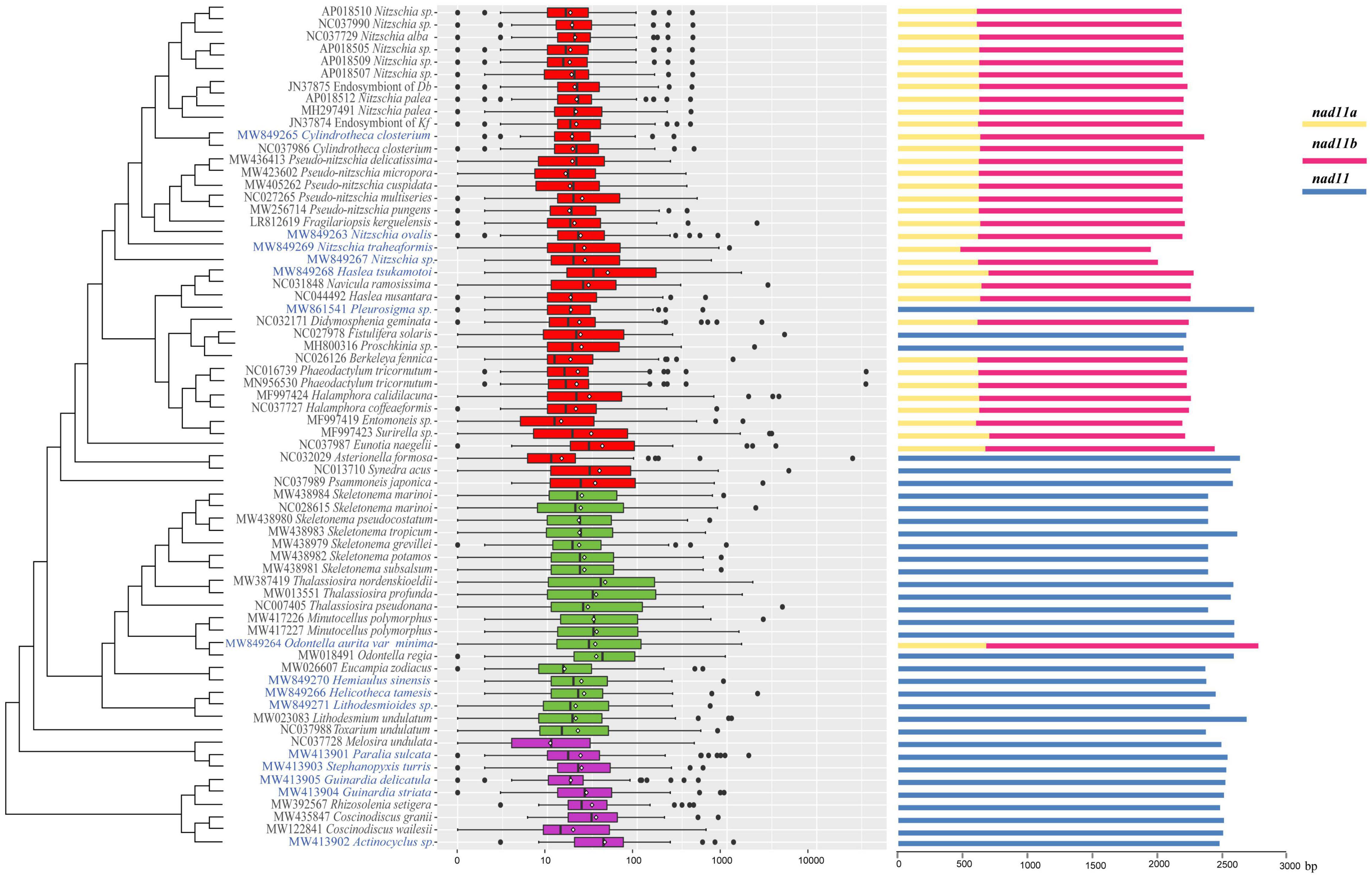
Figure 6. Boxplot of intergenic region length and the structure of nad11gene of 68 mtDNAs. On the left part, red boxes, green boxes, and purple boxes represented the three classes Bacillariophyceae, Mediophyceae, and Coscinodiscophyceae, respectively. The white point and black vertical line of every box were average and median of intergenic region length, respectively. The scale of intergenic region length is in a log10 units. On the right part, nad11a and nad11b were two unit caused by the break of the nad11 gene.
In the class Bacillariophyceae, six mtDNAs constructed in this study exhibited highly diverse gene arrangements (Figure 7A). Similar situation was also found in the classes Mediophyceae and Coscinodiscophyceae. Four mtDNAs constructed in this study that belonged to the class Mediophyceae and five mtDNAs constructed in this study that belonged to the class Coscinodiscophyceae also had a variety of rearrangements, respectively (Figures 7B,C).

Figure 7. Synteny analysis of diatom mtDNAs constructed in this study. (A) Synteny relationships among six newly constructed mtDNAs of Bacillariophyceae based Mauve analysis. (B) Synteny relationships among four newly constructed mtDNAs of Mediophyceae. (C) Synteny relationships among five newly constructed mtDNAs of Coscinodiscophyceae. Each colored block indicates a synteny block among different mitochondrial genomes.
For exploring the degree of conservation of the mtDNAs at the order level, we conducted a series of synteny analysis in six orders that contained mtDNAs constructed in this study (Figure 8). The mtDNAs of the same order showed more conservation with fewer genome rearrangement events than that of different orders. For example, the mtDNAs of two species Lithodesmioides sp. and Lithodesmium undulatum of the order Lithodesmiales showed nearly identical genome organization. Gene-level analysis revealed that gene translocations and/or inversion in the block of cox2-trnE-trnM-3 appeared in Lithodesmioides sp. and Lithodesmium undulatum, Helicotheca tamesis, and Lithodesmioides sp. (Figures 9D,E). The mtDNAs of three species Guinardia delicatula, Guinardia striata, and Rhizosolenia setigera of the order Rhizosoleniales were also highly syntonic with no rearrangements. Gene-level analysis of Guinardia striata and Guinardia delicatula mtDNAs showed that most genes arrangements were conserved except for the translocations of trnM-1, trnH, and nad3 (Figure 9F).
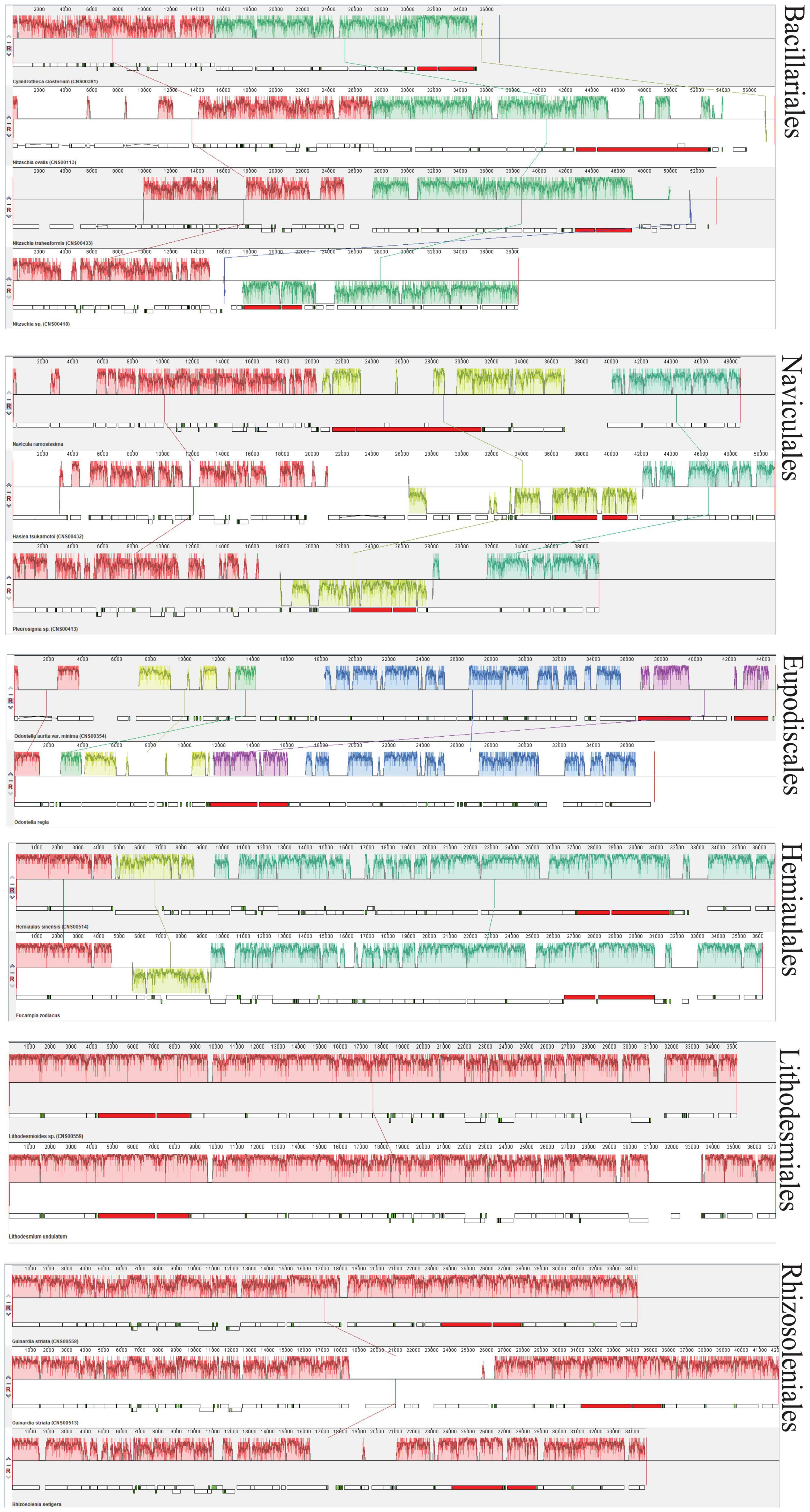
Figure 8. Synteny relationships using Mauve analysis for six orders of diatoms (Bacillariales, Naviculales, Eupodiscales, Hemiaulales, Lithodesmiales, and Rhizosoleniales) that included mtDNAs constructed in this study. Each colored block indicates a synteny block among different mitochondrial genomes.
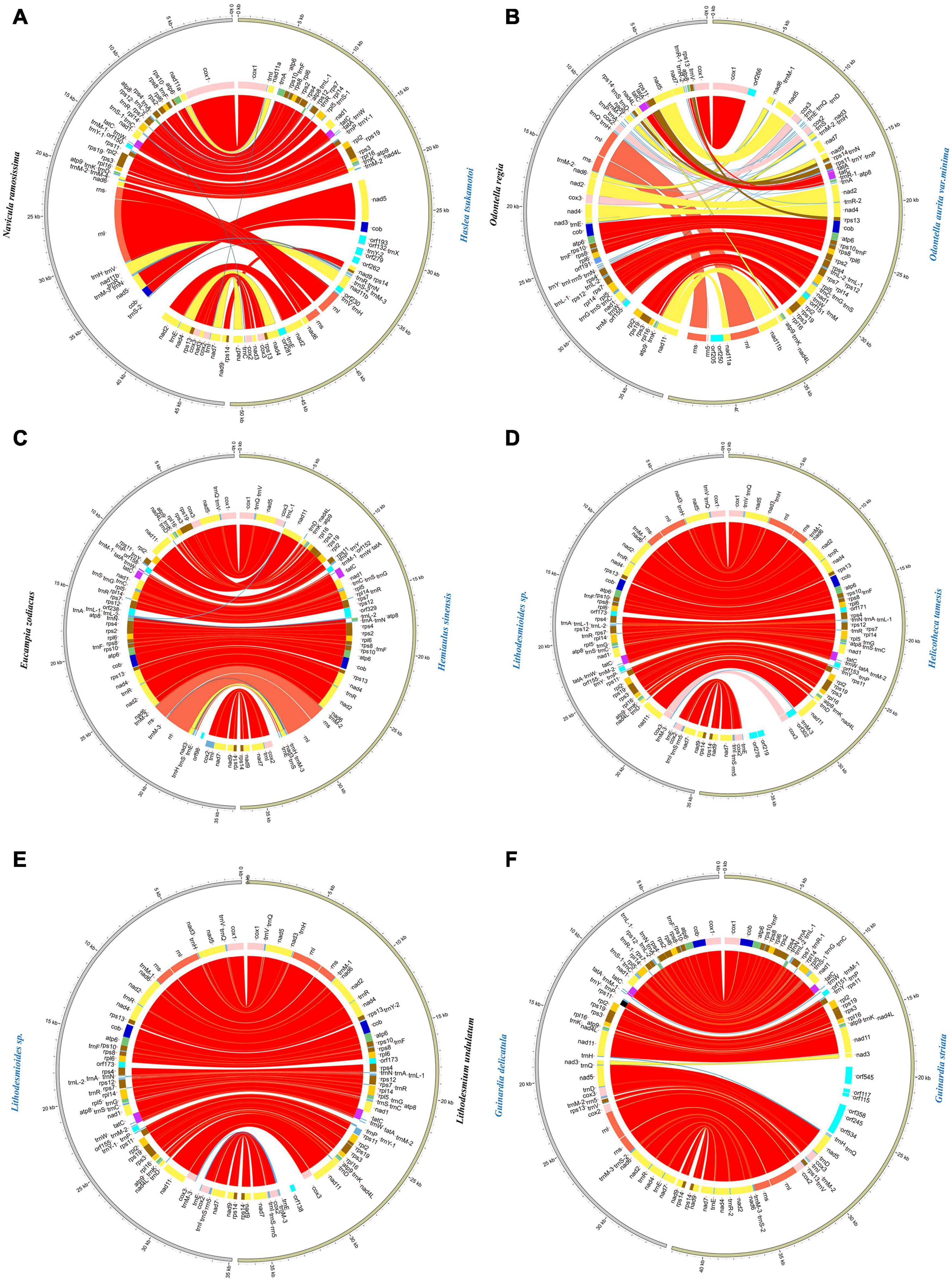
Figure 9. Pair-wise comparison of representative mtDNAs. (A) The comparative analysis of Haslea tsukamotoi and Navicula ramosissima mtDNAs. (B) The comparative analysis of Odontella aurita var. minima and Odontella regia mtDNAs. (C) The comparative analysis of Hemiaulus sinensis and Eucampia zodiacus mtDNAs. (D) The comparative analysis of Helicotheca tamesis and Lithodesmioides sp. mtDNAs. (E) The comparative analysis of Lithodesmium undulatum and Lithodesmioides sp. mtDNAs. (F) The comparative analysis of Guinardia striata and Guinardia delicatula mtDNAs. The assignment of genes into different functional groups is indicated by colors. The conserved back-to-back arrangement of genes was all shown with same color.
In other orders, some genome rearrangements were identified. In the order Bacillariales, a major genome arrangement event (i.e., an inversion) was identified between the mtDNA of Nitzschia sp. and the mtDNAs of three other species in this order (Cylindrotheca closterium, Nitzschia ovalis, and Nitzschia traheaformis) (Figure 8). In the order Naviculales, a major genome rearrangement event (i.e., an inversion) was also identified between the mtDNA of Navicula ramosissima and mtDNAs of two other species (Haslea tsukamotoi and Pleurosigma sp.) (Figure 8). Further gene level analysis of Navicula ramosissima and Haslea tsukamotoi mtDNAs revealed multiple translocation events including trnI, nad11a, trnA, trnR, trnN, trnS, trnM-3, nad11b, trnC, nad4, and nad7 and the inversion of a gene block containing rnl, rns, and nad6 (Figure 9A). In the order Eupodiscales, multiple translocation events were identified between the mtDNAs of two species of a same genus, Odontella aurita var.minima and Odontella regia, breaking the genes in these two genomes into five gene blocks (Figures 8, 9B). Notably, all genes on the mtDNAs of both Odontella aurita var. minima and Odontella regia were located on a single strand (Figure 8). In the order Hemiaulales, a major inversion event was identified between two species, Hemiaulus sinensis and Eucampia zodiacus. The gene arrangements of Hemiaulus sinensis and Eucampia zodiacus mtDNAs were almost consistent with seven gene rearrangements including trnL-1, trnL-2, rns, rnl, trnM-3, trnH, and nad3 (Figure 9C).
The availability of mtDNAs allowed us to explore the divergence time between diatom species, the speciation of which originated in Jurassic period (95% HPD: 173–192 Mya) (Lewitus et al., 2018) (Figure 10). The divergence between the classes Coscinodiscophyceae and Mediophyceae occurred 141 million years ago (Mya, 95% HPD: 159–130 Mya), while the divergence between the classes Mediophyceae and Bacillariophyceae occurred in 131 Mya (95% HPD: 146–120 Mya). The class Bacillariophyceae were represented by two major clades, including Clade A containing a single order Bacillariales, and Clade B containing three orders (e.g., Surirellale, Naviculales, and Cymbellales). The divergence between of the Clade A and Clade B occurred 109 Mya (95% HPD: 124–99 Mya). In Clade B, the order Naviculales was divided into two subclades, suggesting that this order should be divided into two independent orders.
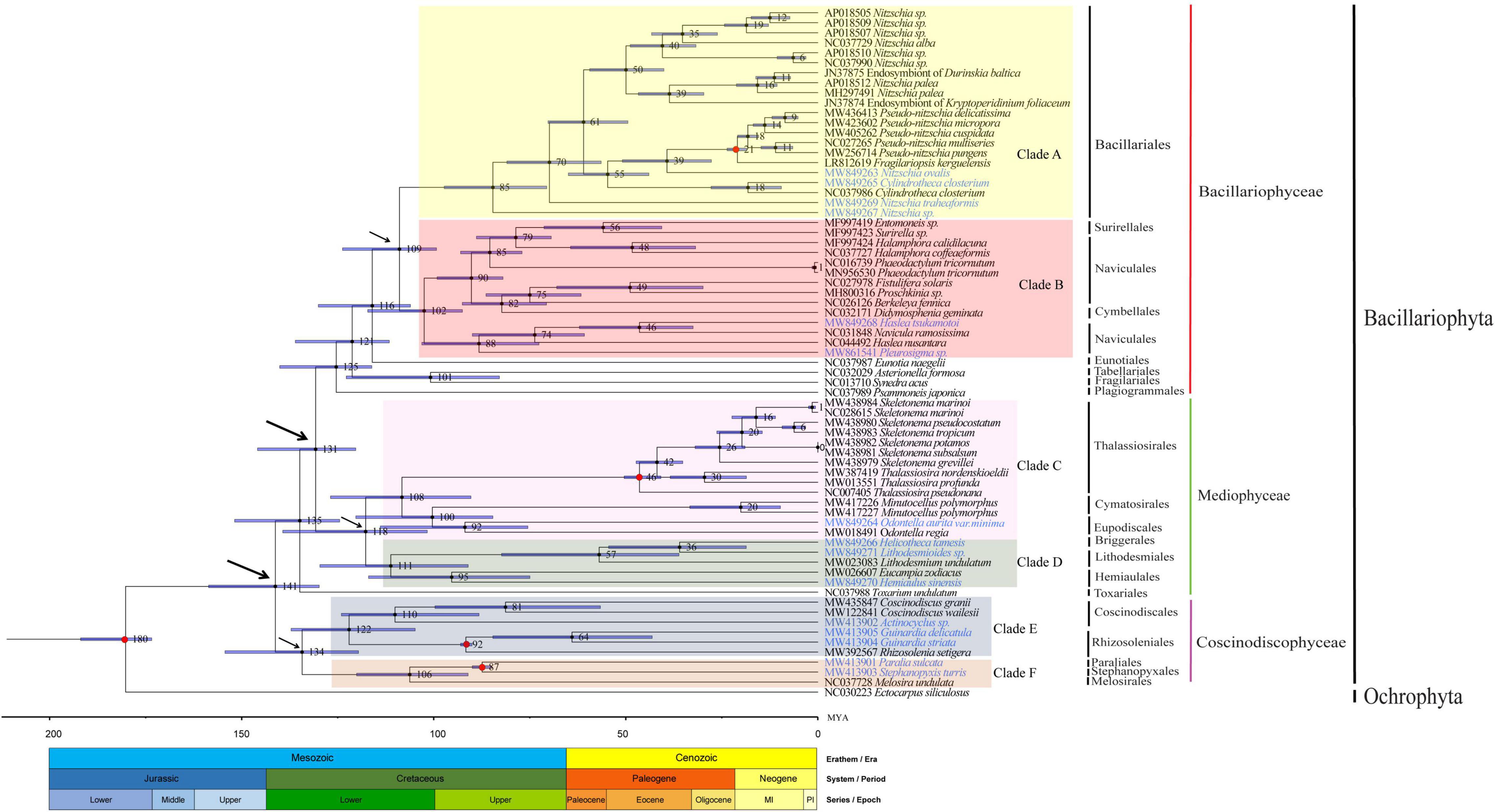
Figure 10. Time-calibrated phylogeny of 68 mtDNAs based on 27 shared PCGs in the mtDNAs of three classes of Bacillariophyta and the outgroup (Ectocarpus siliculosus). The red dots represent calibration point and the 95% highest posterior density interval for node ages are shown with translucent blue bars.
The class Mediophyceae contained Clade C that consisted of the orders Thalassiosirales, Cymatosirales, and Eupodiscales, and Clade D that consisted of the orders Hemiaulales, Lithodesmiales, and Briggerales. In Clade C, the divergence between the order Thalassiosirales and the orders Cymatosirales and Eupodiscales occurred 108 Mya (95% HPD: 127–90 Mya), followed by the divergence of the orders Cymatosirales and Eupodiscales at 100 Mya (95% HPD: 120–85 Mya). In Clade D, the divergence between the orders Briggerales, Lithodesmiales, and Hemiaulales occurred at 111 Mya (95% HPD: 130–91 Mya), (95% HPD: 130–91 Mya), followed by the divergence of the order Briggerales from the order Lithodesmiales at 36 Mya (95% HPD: 55–19 Mya).
The class Coscinodiscophyceae contained Clade E that consisted of the orders Coscinodiscales and Rhizosoleniales, and Clade F that consisted of the orders Paraliales, Stephanopyxales, and Melosirales. These two clades diverged at 134 Mya (95% HPD: 154–120 Mya). In Clade E, the divergence between the orders Coscinodiscales and Rhizosoleniales occurred 122 Mya (95% HPD: 137–105 Mya). In clade F, the divergence between the orders Paraliales and Stephanopyxales from Melosirales occurred 106 Mya (95% HPD: 120–91 Mya).
Although diatoms have been regarded to be enormously species-rich with 100,000–200,000 species, only 12,000 species have been described based on their morphological features with some molecular support (Mann and Vanormelingen, 2013). As a consequence, the quantitative understanding of diatom biodiversity has been limited. The recent development of remarkable genomics technologies and bioinformatics methods have made it possible to systematically construct mtDNAs with high throughput and low cost. Thus, organelle genomes including mtDNAs have been proposed to be used as “super barcodes” that provide higher resolution for distinguishing. As a demonstration of this idea, we have constructed 15 mtDNAs for diatom species isolated from the Jiaozhou Bay, which is an epitome of China’s offshore ecosystem (Liu et al., 2004). This research has demonstrated that systematic construction of diatom mtDNAs is an effective and fruitful approach for exploring diatom biodiversity and speciation.
Ribosomal PCGs of the mtDNAs have been lost more often than other genes that were related to respiration including cytochrome b (cob) and cytochrome oxidase subunit 1 (cox1) (Adamsa and Palmer, 2003). Among 68 diatom mtDNAs, the absence of specific ribosomal PCGs occurred sporadically and independently in only a few mtDNAs instead of widespread loss, which indicated that the function of the missing genes was still needed and may be directly replaced by some preexisting genes just like the gene substitution in yeast and plant mtDNAs (Karlberg et al., 2000; Adams et al., 2002). The loss of tatA from mtDNAs was first discovered in the diatom species Guinardia striata and Actinocyclus sp., and rrn5 was found to be absent from most diatom mtDNAs (Oudot-Le Secq et al., 2001).
Gene duplication and subsequent divergence played a vital role in evolution by providing genetic material for novel functions (Hoshino et al., 2001; Taylor et al., 2001). In this study, the duplication and subsequent divergence of cox1 gene in the mtDNAs of Nitzschia traheaformis and Haslea tsukamotoi was found for the first time in diatom mtDNAs. Two cox2 genes were previously found in mtDNA of rapeseed (Brassica napus L.) (Handa, 2003). Further study of more mtDNAs will help us to learn the origin of gene duplication and explore the role gene duplication plays in the evolution of diatom mtDNAs, which will in turn facilitate research on diatom biodiversity and speciation.
Many studies had been conducted to explore the evolutionary relationships among diatom mtDNAs. For instance, the class Coscinodiscophyceae was found to be non-monophyletic according to the comparative analysis of 17 mtDNAs of diatoms (Pogoda et al., 2019), while the class Mediophyceae was found to be non-monophyletic and was a sister group of the class Bacilariophyceae according to the analysis of 35 mtDNAs of diatoms (Wang et al., 2021). In this study, the phylogenetic analysis of 68 mtDNAs covering 20 orders indicated that Mediophyceae and Coscinodiscophyceae were non-monophyletic. In the order Bacillariales, the taxonomy of species-rich Nitzschia was found to be unstable (Trobajo et al., 2013), which may contribute to the complicated genetic evolution relationships in genus Nitzschia. The published study had proposed transferring Haslea tsukamotoi to Navicula according to morphological data and molecular marker analysis (Li et al., 2017). That Haslea tsukamotoi was closer to Navicula ramosissima than Haslea nusantara in our tree also underscored this point. Similar situation was found in the order Thalassiosirales in which Skeletonema species was found to be closer to species of the genus Thalassiosira rather than Thalassiosira pseudonana. T. pseudonana has been proposed to be more appropriately classified as Cyclotella nana on the basis of the phylogenetic analyses of morphological and molecular datasets (Alverson et al., 2011). More mtDNAs from different orders are needed to facilitate understanding the complex evolutionary relationships between pairs of orders including Naviculales and Cymbellales, Briggerales and Lithodesmiales, and Paraliales and Stephanopyxales.
The sizes of intergenic regions usually correlated linearly with the genome size (Watzlowik et al., 2021). The largest differences were due to repeat sequences. The longest intergenic region was found in Phaeodactylum tricornutum and Asterionella formosa which contained 35 and 25 kb-long repeats, respectively (Secq and Green, 2011; Villain et al., 2017). The nad11 split coding region made nad11 gene undergo fission into two separate submits with its own start and stop codons, respectively and corresponds to the iron-sulfur binding (nad11a) and molybdopterin-binding domains (nad11b) (Guillory et al., 2018). In distinct contrast to the balance of intergenic region average length, the presence of nad11 split coding region was obviously different in the three classes of diatom, only existing in the class of Bacillariophyceae and Mediophyceae, which pointed to that the break of nad11 gene was produced during the evolution of diatoms diversity instead of generating randomly. In the class Coscinodiscophyceae, none of the nad11 genes were split in the mtDNAs of all species suggesting that the split model of nad11 was the derived version, while the non-split model of nad11 was the original version.
There were a variety of rearrangements in three classes, which pointed to that the mtDNA structure was diverse between species in the phylum of Bacillariophyta. Other comparative analyses of mtDNAs had also shown a number of gene rearrangements in the class Eustigmatophyceae and class Phaeophyceae of algae, respectively (Sevcikova et al., 2016; Liu et al., 2019b). Compared to mtDNAs of different orders in the phylum of Bacillariophyta, the mtDNAs within same orders were more conserved. Interestingly, the transcriptional direction of genes on mtDNAs showed substantial variations. While genes are found on both strands of most mtDNAs, genes of mtDNAs of the genus Odontella (Eupodiscales) were found to be located on the same strand. The functional implication of such genome arrangement was yet unknown, but may represent a more economic transcription mode, as suggested previously in Ulva pertusa (Liu et al., 2017). Obviously further studies are needed to explore how such genome arrangement was acquired by Odontella species during evolution, and the functional implication it has in the formation of diversity in Odontella. Aside from the structural similarity, mtDNAs of Lithodesmioides sp. was closer to Helicotheca tamesis than Lithodesmium undulatum in the phylogenetic tree, suggesting that the mtDNA sequences evolved faster than their structures within order Lithodesmiales.
Fossil evidence suggested that diatoms originated in the late Jurassic period and became more common in the mid-Cretaceous (Round et al., 1990; Sims et al., 2006; Lewitus et al., 2018; Nakov et al., 2018). Despite the reliability of fossil records, many diatom deposits have undoubtedly been lost through diagenesis, making it difficult to track evolutionary trajectory of diatoms. Comparative analysis of diatom mtDNAs could provide valuable complementary genetic insights into diatom evolution. In this study, we first used diatom mtDNAs as a “super barcode” to infer the time frame within which the diatoms originated and diversified. Analysis of fossil records revealed that Mediophyceae and Coscinodiscophyceae diatoms evolved before Bacillariophyceae diatoms (Round et al., 1990). In our study, diatom species of the class Coscinodiscophyceae was first established, followed by diatom species of the class Mediophyceae, and diatom species of the class Bacillariophyceae were the last to appear, which was also generally consistent with the molecular phylogenetic dating analysis using nuclear-encoded small-subunit ribosomal RNA (Sorhannus, 2007). Our study indicated that all three classes of diatom emerged 100 Mya during the Cretaceous period (Figure 10). During this time frame, diatoms played dominant role in the carbon cycle, when CO2 levels of atmosphere were about five times higher than they are today and O2 levels were rising (Falkowski et al., 2005; Armbrust, 2009). The evolution of the habitat of the class Bacillariophyceae had happened in approximately 120 Mya which reflected the propensity of diatoms to occupy and adapt to a variety of habitats, from marine to the freshwater (Falciatore et al., 2020). Our study also suggested that freshwater species of Bacillariophyceae had appeared in 121 Mya (95% HPD: 112–136 Mya) such as Asterionella formosa. Many species began to appear after the end of Cretaceous, especially those of the order Bacillariales and Thalassiosirales. The mass extinction about 65 Mya which caused loss of about 85% of all species except diatoms and provided ecological opportunities for the establishment of new species (Falkowski et al., 2004). Throughout the Cenozoic, the rising diversity of diatoms had been attributed to an expanded bioavailability of silica from increased weathering of silicate rock (Cermeno et al., 2015) and an influx of nutrient-rich seawater into the South Atlantic brought by the Antarctic Circumpolar Current (Barker, 2001; Scher and Martin, 2006).
The availability of 15 mtDNAs grouped in ten orders of diatom species provided valuable reference sequences for further evolutionary studies. The 15 newly constructed mtDNAs display several new features of mtDNAs including the identification of first duplication events of the cox1 gene. Further study could help understand the role they play in the evolution and diversity formation of diatoms. The three classes of diatom are separate from each other. The evolutionary relationships for diatom species from different orders were complex, with mtDNAs within orders showing higher similarity. Diatoms of the class Coscinodiscophyceae appeared first in evolution. All three classes emerged 100 Mya during the Cretaceous period, the diversity of diatom began to rise obviously after the mass extinction about 65 Mya and many diatom species were generated 50 Mya. With the construction of more mtDNAs of more diatom species, more insight will be gained into the diatom biodiversity and speciation.
The datasets presented in this study can be found in online repositories. The names of the repositories and accession numbers can be found below: https://www.ncbi.nlm.nih.gov/, MW413901; https://www.ncbi.nlm.nih.gov/, MW413902; https://www.ncbi.nlm.nih.gov/, MW413903; https://www.ncbi.nlm.nih.gov/, MW413904; https://www.ncbi.nlm.nih.gov/, MW413905; https://www.ncbi.nlm.nih.gov/, MW849263; https://www.ncbi.nlm.nih.gov/, MW849264; https://www.ncbi.nlm.nih.gov/, MW849265; https://www.ncbi.nlm.nih.gov/, MW849266; https://www.ncbi.nlm.nih.gov/, MW849267; https://www.ncbi.nlm.nih.gov/, MW849268; https://www.ncbi.nlm.nih.gov/, MW849269; https://www.ncbi.nlm.nih.gov/, MW849270; https://www.ncbi.nlm.nih.gov/, MW849271; https://www.ncbi.nlm.nih.gov/, MW861541; and https://www.ncbi.nlm.nih.gov/, PRJNA746723.
NC conceived of the project. YW, SL, JW, YY, YC, and ZZ organized the strain selection, cultivation, and genome sequencing. YW and QX organized the assembly, annotation, quality control, and analyzed the data with suggestions from others. YW and NC wrote the manuscript with inputs from others. All authors read and approved the final version of the manuscript.
This research was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB42000000), the Chinese Academy of Sciences Pioneer Hundred Talents Program (to NC), the Taishan Scholar Project Special Fund (to NC), the Qingdao Innovation and Creation Plan (Talent Development Program – 5th Annual Pioneer and Innovator Leadership Award to NC, 19-3-2-16-zhc), and the National Key Research and Development Program of China (2017YFC1404300).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.749982/full#supplementary-material
Adams, K. L., Daley, D. O., Whelan, J., and Palmer, J. D. (2002). Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell 14, 931–943. doi: 10.1105/tpc.010483
Adamsa, K. L., and Palmer, J. D. (2003). Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 29, 380–395. doi: 10.1016/s1055-7903(03)00194-5
Alverson, A. J., Beszteri, B., Julius, M. L., and Theriot, E. C. (2011). The model marine diatom Thalassiosira pseudonana likely descended from a freshwater ancestor in the genus Cyclotella. BMC Evol. Biol. 11:125. doi: 10.1186/1471-2148-11-125
An, S. M., Kim, S. Y., Noh, J. H., and Yang, E. C. (2017). Complete mitochondrial genome of Skeletonema marinoi (Mediophyceae, Bacillariophyta), a clonal chain forming diatom in the west coast of Korea. Mitochondrial DNA A DNA Mapp. Seq. Anal. 28, 19–20. doi: 10.3109/19401736.2015.1106523
An, S. M., Noh, J. H., Lee, H. R., Choi, D. H., Lee, J. H., and Yang, E. C. (2016). Complete mitochondrial genome of biraphid benthic diatom, Navicula ramosissima (Naviculales, Bacillariophyceae). Mitochondrial DNA Part B Resour. 1, 549–550. doi: 10.1080/23802359.2016.1198997
Anisimova, M., Gil, M., Dufayard, J.-F., Dessimoz, C., and Gascuel, O. (2011). Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699. doi: 10.1093/sysbio/syr041
Armbrust, E. V. (2009). The life of diatoms in the world’s oceans. Nature 459, 185–192. doi: 10.1038/nature08057
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Ashworth, M. P., Nakov, T., and Theriot, E. C. (2013). Revisiting Ross and Sims (1971): toward a molecular phylogeny of the Biddulphiaceae and Eupodiscaceae (Bacillariophyceae). J. Phycol. 49, 1207–1222. doi: 10.1111/jpy.12131
Barker, P. F. (2001). Scotia Sea regional tectonic evolution: implications for mantle flow and palaeocirculation. Earth Sci. Rev. 55, 1–39. doi: 10.1016/s0012-8252(01)00055-1
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bui Quang, M., Nguyen, M. A., and von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Cermeno, P., Falkowski, P. G., Romero, O. E., Schaller, M. F., and Vallina, S. M. (2015). Continental erosion and the Cenozoic rise of marine diatoms. Proc. Natl. Acad. Sci. U.S.A. 112, 4239–4244. doi: 10.1073/pnas.1412883112
Crosman, K. M., Petrou, E. L., Rudd, M. B., and Tillotson, M. D. (2019). Clam hunger and the changing ocean: characterizing social and ecological risks to the Quinault razor clam fishery using participatory modeling. Ecol. Soc. 24:16. doi: 10.5751/es-10928-240216
Cunningham, C. W. (1997). Can three incongruence tests predict when data should be combined? Mol. Biol. Evol. 14, 733–740.
Damste, J. S. S., Muyzer, G., Abbas, B., Rampen, S. W., Masse, G., Allard, W. G., et al. (2004). The rise of the rhizosolenid diatoms. Science 304, 584–587. doi: 10.1126/science.1096806
Darling, A. E., Mau, B., and Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e0011147. doi: 10.1371/journal.pone.0011147
Dyson, K., and Huppert, D. D. (2010). Regional economic impacts of razor clam beach closures due to harmful algal blooms (HABs) on the Pacific coast of Washington. Harmful Algae 9, 264–271. doi: 10.1016/j.hal.2009.11.003
Evans, K. M., Wortley, A. H., and Mann, D. G. (2007). An assessment of potential diatom “barcode” genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist 158, 349–364. doi: 10.1016/j.protis.2007.04.001
Falciatore, A., Jaubert, M., Bouly, J. P., Bailleul, B., and Mock, T. (2020). Diatom molecular research comes of age: model species for studying phytoplankton biology and diversity(OPEN). Plant Cell 32, 547–572. doi: 10.1105/tpc.19.00158
Falkowski, P. G., Barber, R. T., and Smetacek, V. (1998). Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–206. doi: 10.1126/science.281.5374.200
Falkowski, P. G., Katz, M. E., Knoll, A. H., Quigg, A., Raven, J. A., Schofield, O., et al. (2004). The evolution of modern eukaryotic phytoplankton. Science 305, 354–360. doi: 10.1126/science.1095964
Falkowski, P. G., Katz, M. E., Milligan, A. J., Fennel, K., Cramer, B. S., Aubry, M. P., et al. (2005). The rise of oxygen over the past 205 million years and the evolution of large placental mammals. Science 309, 2202–2204. doi: 10.1126/science.1116047
Felsenstein, J. (1985). Confidence-limits on phylogenies - an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Field, C. B., Behrenfeld, M. J., Randerson, J. T., and Falkowski, P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. doi: 10.1126/science.281.5374.237
Foster, R. A., Kuypers, M. M. M., Vagner, T., Paerl, R. W., Musat, N., and Zehr, J. P. (2011). Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. ISME J. 5, 1484–1493. doi: 10.1038/ismej.2011.26
Gachon, C. M. M., Heesch, S., Küpper, F. C., Achilles-Day, U. E. M., Brennan, D., Campbell, C. N., et al. (2013). The CCAP KnowledgeBase: linking protistan and cyanobacterial biological resources with taxonomic and molecular data. Syst. Biodivers. 11, 407–413. doi: 10.1080/14772000.2013.859641
Giordano, M., Beardall, J., and Raven, J. A. (2005). CO2 concentrating mechanisms in algae: mechanisms, environmental modulation, and evolution. Annu. Rev. Plant Biol. 56, 99–131. doi: 10.1146/annurev.arplant.56.032604.144052
Guillory, W. X., Onyshchenko, A., Ruck, E. C., Parks, M., Nakov, T., Wickett, N. J., et al. (2018). Recurrent loss, horizontal transfer, and the obscure origins of mitochondrial introns in Diatoms (Bacillariophyta). Genome Biol. Evol. 10, 1504–1515. doi: 10.1093/gbe/evy103
Haimeur, A., Ulmann, L., Mimouni, V., Gueno, F., Pineau-Vincent, F., Meskini, N., et al. (2012). The role of Odontella aurita, a marine diatom rich in EPA, as a dietary supplement in dyslipidemia, platelet function and oxidative stress in high-fat fed rats. Lipids Health Dis. 11:147. doi: 10.1186/1476-511x-11-147
Hamsher, S. E., LeGresley, M. M., Martin, J. L., and Saunders, G. W. (2013). A comparison of morphological and molecular-based surveys to estimate the species richness of chaetoceros and Thalassiosira (Bacillariophyta), in the bay of fundy. PLoS One 8:e0073521. doi: 10.1371/journal.pone.0073521
Handa, H. (2003). The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 31, 5907–5916. doi: 10.1093/nar/gkg795
Hollingsworth, P. M., Graham, S. W., and Little, D. P. (2011). Choosing and using a plant DNA barcode. PLoS One 6:e0019254. doi: 10.1371/journal.pone.0019254
Hoshino, A., Johzuka-Hisatomi, Y., and Iida, S. (2001). Gene duplication and mobile genetic elements in the morning glories. Gene 265, 1–10. doi: 10.1016/s0378-1119(01)00357-2
Imanian, B., Pombert, J.-F., Dorrell, R. G., Burki, F., and Keeling, P. J. (2012). Tertiary endosymbiosis in two dinotoms has generated little change in the mitochondrial genomes of their dinoflagellate hosts and diatom endosymbionts. PLoS One 7:e0043763. doi: 10.1371/journal.pone.0043763
Jin, D., Thunberg, E., and Hoagland, P. (2008). Economic impact of the 2005 red tide event on commercial shellfish fisheries in New England. Ocean Coast. Manag. 51, 420–429. doi: 10.1016/j.ocecoaman.2008.01.004
Jin, J.-J., Yu, W.-B., Yang, J.-B., Song, Y., dePamphilis, C. W., Yi, T.-S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241. doi: 10.1186/s13059-020-02154-5
Kalyaanamoorthy, S., Bui Quang, M., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Karlberg, O., Canback, B., Kurland, C. G., and Andersson, S. G. E. (2000). The dual origin of the yeast mitochondrial proteome. Yeast 17, 170–187.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kooistra, W. H. C. F., and Medlin, L. K. (1996). Evolution of the Diatoms (Bacillariophyta): IV. A reconstruction of their age from small subunit rRNA coding regions and the fossil record. Mol. Phylogenet. Evol. 6, 391–407. doi: 10.1006/mpev.1996.0088
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lelong, A., Hegaret, H., Soudant, P., and Bates, S. S. (2012). Pseudo-nitzschia (Bacillariophyceae) species, domoic acid and amnesic shellfish poisoning: revisiting previous paradigms. Phycologia 51, 168–216. doi: 10.2216/11-37
Lewitus, E., Bittner, L., Malviya, S., Bowler, C., and Morlon, H. (2018). Clade-specific diversification dynamics of marine diatoms since the Jurassic. Nat. Ecol. Evol. 2, 1715–1723. doi: 10.1038/s41559-018-0691-3
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, Y., Chen, X., Sun, Z., and Xu, K. (2017). Taxonomy and molecular phylogeny of three marine benthic species of Haslea (Bacillariophyceae), with transfer of two species to Navicula. Diatom Res. 32, 451–463. doi: 10.1080/0269249x.2017.1401008
Liu, D. Y., Sun, J., Liu, Z., Chen, H. T., Wei, H., and Zhang, J. (2004). The effects of spring-neap tide on the phytoplankton community development in the Jiaozhou Bay. China Acta Oceanol. Sin. 23, 687–697.
Liu, F., Liu, S., Huang, T., and Chen, N. (2019a). Construction and comparative analysis of mitochondrial genome in the brown tide forming alga Aureococcus anophagefferens (Pelagophyceae. Ochrophyta). J. Appl. Phycol. 32, 441–450. doi: 10.1007/s10811-019-01952-0
Liu, F., Melton, J. T., and Bi, Y. P. (2017). Mitochondrial genomes of the green macroalga Ulva pertusa (Ulvophyceae, Chlorophyta): novel insights into the evolution of mitogenomes in the Ulvophyceae. J. Phycol. 53, 1010–1019. doi: 10.1111/jpy.12561
Liu, F., Zhang, Y., Bi, Y., Chen, W., and Moejes, F. W. (2019b). Understanding the evolution of mitochondrial genomes in phaeophyceae inferred from mitogenomes of ishige okamurae (Ishigeales) and Dictyopteris divaricata (Dictyotales). J. Mol. Evol. 87, 16–26. doi: 10.1007/s00239-018-9881-5
Liu, S., and Chen, N. (2021). Advances in biodiversity analysis of phytoplankton and harmful algal bloom species in the Jiaozhou Bay. Mar. Sci. 45, 170–188.
Lv, W. Q., Jiang, H. F., Bo, J., Wang, C., Yang, L. D., and He, S. P. (2020). Comparative mitochondrial genome analysis of Neodontobutis hainanensis and Perccottus glenii reveals conserved genome organization and phylogeny. Genomics 112, 3862–3870. doi: 10.1016/j.ygeno.2020.06.039
Malviya, S., Scalco, E., Audic, S., Vincenta, F., Veluchamy, A., Poulain, J., et al. (2016). Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. U.S.A. 113, E1516–E1525. doi: 10.1073/pnas.1509523113
Mann, D. G., Trobajo, R., Sato, S., Li, C., Witkowski, A., Rimet, F., et al. (2021). Ripe for reassessment: a synthesis of available molecular data for the speciose diatom family Bacillariaceae. Mol. Phylogenet. Evol. 158:106985. doi: 10.1016/j.ympev.2020.106985
Mann, D. G., and Vanormelingen, P. (2013). An inordinate fondness? The number, distributions, and origins of diatom species. J. Eukaryot. Microbiol. 60, 414–420. doi: 10.1111/jeu.12047
Martin, F. N. (2008). Mitochondrial haplotype determination in the oomycete plant pathogen Phytophthora ramorum. Curr. Genet. 54, 23–34. doi: 10.1007/s00294-008-0196-8
Medlin, L. K., and Kaczmarska, I. (2019). Evolution of the diatoms: V. Morphological and cytological support for the major clades and a taxonomic revision. Phycologia 43, 245–270. doi: 10.2216/i0031-8884-43-3-245.1
Moore, S. K., Dreyer, S. J., Ekstrom, J. A., Moore, K., Norman, K., Klinger, T., et al. (2020). Harmful algal blooms and coastal communities: socioeconomic impacts and actions taken to cope with the 2015 US West Coast domoic acid event. Harmful Algae 96:101799. doi: 10.1016/j.hal.2020.101799
Nakov, T., Beaulieu, J. M., and Alverson, A. J. (2018). Insights into global planktonic diatom diversity: the importance of comparisons between phylogenetically equivalent units that account for time. ISME J. 12, 2807–2810. doi: 10.1038/s41396-018-0221-y
Nelson, D. M., Treguer, P., Brzezinski, M. A., Leynaert, A., and Queguiner, B. (1995). Production and dissolution of biogenic silica in the ocean - revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cycles 9, 359–372. doi: 10.1029/95gb01070
Oudot-Le Secq, M. P., Fontaine, J. M., Rousvoal, S., Kloareg, B., and Loiseaux-de Goer, S. (2001). The complete sequence of a brown algal mitochondrial genome, the ectocarpale Pylaiella littoralis (L.) Kjellm. J. Mol. Evol. 53, 80–88. doi: 10.1007/s002390010196
Park, J. S., Jung, S. W., Ki, J. S., Guo, R., Kim, H. J., Lee, K. W., et al. (2017). Transfer of the small diatoms Thalassiosira proschkinae and T. spinulata to the genus Minidiscus and their taxonomic re-description. PLoS One 12:e0181980. doi: 10.1371/journal.pone.0181980
Patrick, L., Nicolas, C., Philipp, H., and Elisabeth, N. (2016). Toxic and Harmful Microalgae of the World Ocean. Paris: UNESCO.
Pierella Karlusich, J. J., Ibarbalz, F. M., and Bowler, C. (2020). Phytoplankton in the Tara Ocean. Ann. Rev. Mar. Sci. 12, 233–265. doi: 10.1146/annurev-marine-010419-010706
Pogoda, C. S., Keepers, K. G., Hamsher, S. E., Stepanek, J. G., Kane, N. C., and Kociolek, J. P. (2019). Comparative analysis of the mitochondrial genomes of six newly sequenced diatoms reveals group II introns in the barcoding region of cox1. Mitochondrial DNA A DNA Mapp. Seq. Anal. 30, 43–51. doi: 10.1080/24701394.2018.1450397
Prasetiya, F. S., Gastineau, R., Poulin, M., Lemieux, C., Turmel, M., Syakti, A. D., et al. (2019). Haslea nusantara (Bacillariophyceae), a new blue diatom from the Java Sea, Indonesia: morphology, biometry and molecular characterization. Plant Ecol. Evol. 152, 188–202. doi: 10.5091/plecevo.2019.1623
Rampen, S. W., Schouten, S., Abbas, B., Panoto, F. E., Muyzer, G., Campbell, C. N., et al. (2007). On the origin of 24-norcholestanes and their use as age-diagnostic biomarkers. Geology 35, 419–422. doi: 10.1130/g23358a.1
Round, F. E., Crawford, R. M., and Mann, D. G. (1990). The Diatoms : Biology and Morphology of the Genera. Cambridge: Cambridge University Press.
Saitou, N., and Nei, M. (1987). The neighbor-joining method - a new method for reconstructing phylogenetic trees. Mol. Bio. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Scher, H. D., and Martin, E. E. (2006). Timing and climatic consequences of the opening of Drake Passage. Science 312, 428–430. doi: 10.1126/science.1120044
Secq, M.-P. O.-L., and Green, B. R. (2011). Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene 476, 20–26. doi: 10.1016/j.gene.2011.02.001
Sevcikova, T., Klimes, V., Zbrankova, V., Strnad, H., Hroudova, M., Vlcek, C., et al. (2016). A Comparative analysis of mitochondrial genomes in eustigmatophyte algae. Genome Biol. Evol. 8, 705–722. doi: 10.1093/gbe/evw027
Sims, P. A., Mann, D. G., and Medlin, L. K. (2006). Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402. doi: 10.2216/05-22.1
Smetacek, V. (1998). Biological oceanography – Diatoms and the silicate factor. Nature 391, 224–225. doi: 10.1038/34528
Smith, D. R. (2016). The past, present and future of mitochondrial genomics: have we sequenced enough mtDNAs? Brief. Funct. Genomics 15, 47–54. doi: 10.1093/bfgp/elv027
Smith, S. A., and Dunn, C. W. (2008). Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24, 715–716. doi: 10.1093/bioinformatics/btm619
Song, H. Y., Liu, F., Li, Z. L., Xu, Q., Chen, Y., Yu, Z. M., et al. (2020). Development of a high-resolution molecular marker for tracking Phaeocystis globosa genetic diversity through comparative analysis of chloroplast genomes. Harmful Algae 99:101911. doi: 10.1016/j.hal.2020.101911
Sorhannus, U. (2007). A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Mar. Micropaleontol. 65, 1–12. doi: 10.1016/j.marmicro.2007.05.002
Spolaore, P., Joannis-Cassan, C., Duran, E., and Isambert, A. (2006). Commercial applications of microalgae. J. Biosci. Bioeng. 101, 87–96. doi: 10.1263/jbb.101.87
Swofford, D. L. (2002). PAUP*: Phylogenetic Analysis Using Parsimony. Version 4.0b10. Sunderland, MA: Sinauer Associates, Inc.
Tanaka, T., Maeda, Y., Veluchamy, A., Tanaka, M., Abida, H., Marechal, E., et al. (2015). Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. Plant Cell 27, 162–176. doi: 10.1105/tpc.114.135194
Taylor, J. S., Van de Peer, Y., and Meyer, A. (2001). Genome duplication, divergent resolution and speciation. Trends Genet. 17, 299–301. doi: 10.1016/s0168-9525(01)02318-6
Theriot, E. C., Ashworth, M., Ruck, E., Nakov, T., and Jansen, R. K. (2010). A preliminary multigene phylogeny of the diatoms (Bacillariophyta): challenges for future research. Plant Ecol. Evol. 143, 278–296. doi: 10.5091/plecevo.2010.418
Thorvaldsdottir, H., Robinson, J. T., and Mesirov, J. P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. doi: 10.1093/bib/bbs017
Trifinopoulos, J., Lam-Tung, N., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Trobajo, R., Rovira, L., Ector, L., Wetzel, C. E., Kelly, M., and Mann, D. G. (2013). Morphology and identity of some ecologically important small Nitzschia species. Diatom Res. 28, 37–59. doi: 10.1080/0269249x.2012.734531
Villain, A., Kojadinovic, M., Puppo, C., Prioretti, L., Hubert, P., Zhang, Y., et al. (2017). Complete mitochondrial genome sequence of the freshwater diatom Asterionella formosa. Mitochondrial DNA Part B Resour. 2, 97–98. doi: 10.1080/23802359.2017.1285210
Wang, Y., Chen, Y., Wang, J., Liu, F., and Chen, N. (2021). Mitochondrial genome of the harmful algal bloom species Odontella regia (Mediophyceae, Bacillariophyta). J. Appl. Phycol. 33, 855–868. doi: 10.1007/s10811-020-02364-1
Watzlowik, M. T., Das, S., Meissner, M., and Längst, G. (2021). Peculiarities of Plasmodium falciparum gene regulation and chromatin structure. Int. J. Mol. Sci. 22:5168. doi: 10.3390/ijms22105168
Wilcox, T. P. (1998). Large-subunit ribosomal RNA systematics of symbiotic dinoflagellates: morphology does not recapitulate phylogeny. Mol. Phylogenet. Evol. 10, 436–448. doi: 10.1006/mpev.1998.0546
Williams, D. M. (2007). Diatom phylogeny: fossils, molecules and the extinction of evidence. Comptes Rendus Palevol. 6, 505–514. doi: 10.1016/j.crpv.2007.09.016
Witkowski, A., Li, C., Zglobicka, I., Yu, S.-X., Ashworth, M., Bek, P. D., et al. (2016). multigene assessment of biodiversity of diatom(Bacillariophyceae) assemblages from the littoral zone of the bohai and yellow seas in yantai region of northeast china with some remarks on ubiquitous taxa. J. Coast. Res. 74, 166–195. doi: 10.2112/si74-016.1
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Li, W. X., et al. (2020). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, M., Cui, Z., Liu, F., and Chen, N. (2021). Definition of a high-resolution molecular marker for tracking the genetic diversity of the harmful algal species Eucampia zodiacus Through comparative analysis of mitochondrial genomes. Front. Microbiol. 12:631144.
Keywords: diatom, mitochondrial genome, comparative genomics, evolution, diversity, divergence time
Citation: Wang Y, Liu S, Wang J, Yao Y, Chen Y, Xu Q, Zhao Z and Chen N (2022) Diatom Biodiversity and Speciation Revealed by Comparative Analysis of Mitochondrial Genomes. Front. Plant Sci. 13:749982. doi: 10.3389/fpls.2022.749982
Received: 05 August 2021; Accepted: 23 February 2022;
Published: 24 March 2022.
Edited by:
Gwenael Piganeau, UMR 7232 Biologie Intégrative des Organismes Marins (BIOM), FranceReviewed by:
Denis Baurain, University of Liège, BelgiumCopyright © 2022 Wang, Liu, Wang, Yao, Chen, Xu, Zhao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nansheng Chen, Y2hlbm5AcWRpby5hYy5jbg==, Y2hlbm5Ac2Z1LmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.