
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 May 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.729128
This article is part of the Research Topic Targeted Genome Editing for Crop Improvement View all 20 articles
Polygalacturonase (PG) gene has been documented as a key candidate for the improvement of fruit firmness, which is a target trait for tomato production because it facilitates transportation and storage. To reduce the expression of the PG gene, most of the elite commercial tomato varieties were obtained by RNA interference technology. However, this approach of producing commercialized tomatoes by integration of the exogenous gene is controversial. In this work, CRISPR/Cas9 technology was used to induce the targeted mutagenesis of the SlPG gene to delay the softening of tomato fruit. Results showed that the SlPG gene was frameshift mutated by 4 bp deletion, 10 bp deletion, and 1 bp insertion, which generated premature translation termination codons. Compared with wild-type (WT), homozygous T1-generation tomato plants exhibited late fruit softening under natural conditions. Consistent with this phenomenon, the firmness value of WT fruit was lower in slpg mutant fruit, and the physiological loss of water was higher. Collectively, these data demonstrate that the mutation of the SlPG gene delays tomato fruit softening. More importantly, 8 out of 20 transgene-free tomato plants, which were homozygous for null alleles of SlPG, were separated in the T3-generation of line slpgT2-#2. This transgene-free slpg may provide materials for more in-depth research of SlPG functions and the molecular mechanism of fruit softening in tomatoes.
Tomato (Solanum lycopersicum), one of the most significant commercial vegetable crops grown in the world, has abundant nutritive compounds and elements (e.g., lycopene, zeaxanthin, vitamin C, and potassium) for human health (Vincent et al., 2013). Fruit softening is the most common defect consideration of commercial importance and has tremendous associated costs in terms of fruit shelf life, frequency of harvest, transportation, and storage (Garcia-Gago et al., 2009). During fruit ripening, the biochemical and physiological processes are sophisticated, and the solubilization of the cell wall is caused by a group of enzymes (Payasi et al., 2009). One of the key enzymes is PG (EC 3.2.1.15), which functions in the breakdown of pectin, a polymer of galacturonic acid that forms part of the structural support of cell wall (Bird et al., 1988). In 1965, the PG enzyme was first reported in relation to the firmness of the tomato fruit (Hobson, 1965), and the PG gene sequence was first cloned from tomatoes in 1986 (Grierson et al., 1986). In 1988, with the application of molecular genetics, PG gene expression was first manipulated with transgenic approaches in tomatoes, where the suppression of PG gene expression resulted in significantly firmer fruit (Bird et al., 1988; Sheehy et al., 1988; Smith et al., 1988). Since then, the suppression of PG gene expression in other fruits, including strawberries (Quesada et al., 2009; Pose et al., 2013), pear (Zhang S. et al., 2019), and kiwifruit (Huang et al., 2020), have been reported and the desired traits were also obtained. Mostly, these fruits are reformed using antisense RNA (Smith et al., 1990; Ju et al., 1994; Kramer and Redenbaugh, 1994) or site-selected insertion (Cooley and Yoder, 1998) through transgene in elite commercial background varieties. Similar to other transgenic plants, the random integration of exogenous transgenes into the tomato genome may cause unstable and/or off-target effects, thereby causing public concern for human consumption and the commercialization of these tomatoes is restricted by complicated safety evaluation. Therefore, delayed tomato fruit softening that does not compromise exogenous genes is a key characteristic in modern tomato breeding.
In 2014, a developed technology, clustered regularly interspaced short palindromic repeat (CRISPR)/Cas (CRISPR-associated) (Doudna and Charpentier, 2014), as a robust and effective tool for the targeted gene of interest, has rapidly emerged in plant breeding with simple design and flexibility of operation (Xie et al., 2014; Zhang Y. et al., 2019). Since then, this system has been applied to generate and estimate targeted mutations in many representative plants, such as Arabidopsis (Li et al., 2013), tobacco (Nekrasov et al., 2013), rice (Shan et al., 2013), soybean (Michno et al., 2015), wheat (Upadhyay et al., 2013), and maize (Svitashev et al., 2015). CRISPR/Cas9-mediated genome editing in tomatoes was first successfully achieved in 2014 (Brooks et al., 2014) by designing two single guide RNAs (sgRNAs) to create mutagenesis in the SlAGO7 gene. In 2017, the high rates of homozygous and biallelic mutants of SlPIF4 were generated in the first generation by applying CRISPR/Cas9 technology, and the gene modifications were stably transmitted to the next generation (Pan et al., 2017). In 2018, the promoter of sgRNA, which provides a strong technical basis for creating mutagenesis in tomato, was optimized for this technology (Yan et al., 2018). In 2019, cytidine base editors, CRISPR/Cas9 derived tools, were applied in tomato successfully (Veillet et al., 2019), and the acetolactate synthase (ALS) gene was edited with the rate of 12.9%. These works have shown that CRISPR/Cas9 is a simple, efficient, and powerful gene editing tool in tomato molecular breeding.
In this study, CRISPR/Cas9 technology was used to induce the targeted mutagenesis of the SlPG gene in tomatoes. The use of the transient assay to select the high editing rate of sgRNA has not been reported previously. Out of 10 stable transgenic tomato events, 7 slpg mutants were obtained via Agrobacterium-mediated transformation. In T1-generation tomato plants, three homozygous for null alleles of SlPG frameshift mutated by short deletions (4 and 10 bp) and insertion (1 bp) exhibited late fruit softening under greenhouse conditions. The homozygous T2 slpg mutants, which have mutation types that were consistent with T1 generation, also exhibited late fruit softening. More importantly, in T3-generation tomato plants, “transgene-free” slpg mutants without any transgenic element were separated successfully. Such an approach is expected to reduce the deleterious effects caused by the random integration of the transgene into the tomato genome. These slpg mutants will provide materials for more in-depth research on improving tomato quality.
The tomato cultivar Micro-Tom was used for transformation in this study. Micro-Tom refers to a miniature dwarf tomato that can grow at a high density, has a short life cycle, and can be transformed efficiently. It became a new model plant for functional genomic study. The origin, background, and biological characteristics of Micro-Tom were clear (Liu et al., 2008). WT Micro-tom (as a control) and all seeds collected from T0 mutant plants were sown under greenhouse conditions (16 h light/8 h dark and 22°C/18°C of day/night temperature, 80% humidity, and 10,000 lux light intensity).
The tomato gene SlPG sequence (GenBank Accession No.: NM_001247092) was first cloned and published in 1986 (Grierson et al., 1986) and its sequence information (solyc10g080210) was downloaded from Solanum lycopersicum ITAG3.2 of the Phytozome website1 (Goodstein et al., 2011). For the CRISPR/Cas9 vector construction, the sgRNAs used for genome editing were designed by using the website tool CRISPOR2 (Concordet and Haeussler, 2018). In this study, five sgRNAs that target the SlPG gene were selected and named SP1, SP2, SP3, SP4, and SP5. All the sgRNA sites were located at the front exons of the SlPG gene (Figure 1A). The sgRNA sequences were synthesized by Genewiz (Suzhou). For each sgRNA, a pair of DNA oligonucleotides was annealed to generate dimers, which were subsequently integrated upstream of the sgRNA scaffolds in the plasmid vector while expressing Cas9 and sgRNA. The sequence of Cas9 was codon-optimized for maize and assembled downstream of the double cauliflower mosaic virus (CaMV) 35S promoter together with a customized sgRNA driven by the AtU6-26 promoter. The kanamycin resistance gene, neomycin phosphotransferase II (nptII), driven by a CaMV 35S promoter, was selected as a plant screening marker. The schematic of the SlPG-CRISPR/Cas9 vector is shown in Figure 1B. In this study, SlPG-CRISPR/Cas9 vector containing sgRNA SP1, SP2, SP3, SP4, and SP5 was named pg1, pg2, pg3, pg4, and pg5, respectively. The schematic of pg1 vector is shown in Figure 1B.
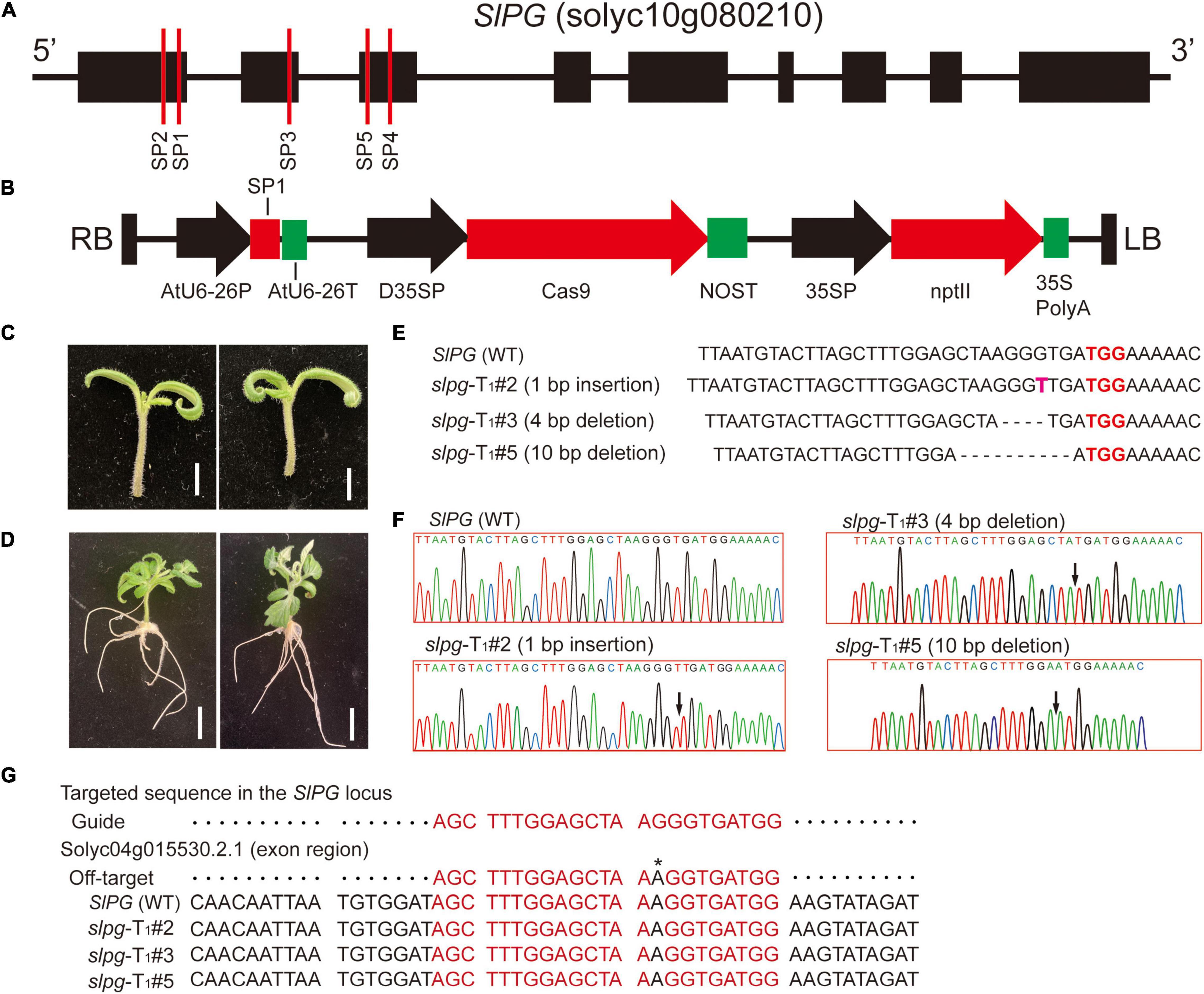
Figure 1. CRISPR/Cas9 technology was used to induce the targeted mutagenesis of the SlPG gene in tomatoes. (A) Schematic representation of tomato SlPG gene and target sites used in this study. Black boxes and lines represent exons and non-coding regions, respectively. Red vertical lines indicate the target site position and name of designed sgRNAs. (B) Schematic illustrating the pg1 vector. RB/LB, right/left border of T-DNA; AtU6-26P, Arabidopsis U6-26 promoter; SP1, sgRNA-SP1; AtU6-26T, Arabidopsis U6-26 terminator; D35SP, double cauliflower mosaic virus (CaMV) 35S promoter; Cas9, coding region of Cas9; NOST, Nos terminator; 35SP, 35S promoter; nptII, kanamycin resistance gene; 35S PolyA, 35S PolyA terminator. (C) Tomato seedlings were selected as explants used for transient transformation. Scale bar, 1 cm. (D) Well-developed hairy roots. Scale bar, 1 cm. (E) Sequences of WT plant and T1 homozygous slpg mutants (slpg-T1#2, slpg-T1#3, and slpg-T1#5) and representative mutation types induced at target site SP1 are presented, respectively. Nucleotides in red represent PAM sequences. Nucleotide in pink represents insertions and dashes between nucleotides represent deletions. (F) Sequence peaks of WT plant and T1 homozygous slpg mutants (slpg-T1#2, slpg-T1#3, and slpg-T1#5). The black arrowheads indicate the location of mutations. (G) Representative putative off-target site recognized by the gRNA designed in this study. One representative site was predicted by the website tool CRISOR. The sequence at the top of the alignment is predicted off-targeted sequence. Red sequences, nucleotides corresponding to the site recognized by the gRNA designed in this study; Black nucleotide with * on top, nucleotide mismatch in the sgRNA region.
To test sgRNA editing activity, the constructed vectors (pg1, pg2, pg3, pg4, and pg5) were mobilized into A. rhizogenes K599 via electroporation. Hairy root transformation was performed following the reference (Pavli and Skaracis, 2010) with minor revision. The healthy and well-developed seedlings were selected as explants for root transformation. The hypocotyl was cut with the sterile blade at the bottom of the seedling (Figure 1C) and immersed the whole seedling into the K599 suspension for 10 min. Inoculated seedlings (5–7 seedlings per petri dish) were placed on square dishes that contained a cocultivation medium used in stable transformation and placed vertically in the greenhouse for 2 days. The seedlings were transferred into growth boxes that contained seedling germination medium, supplemented with appropriate concentrations of cefotaxime and antibiotics to eliminate bacteria and select transformed roots. After 15 days, seedlings with well-developed hairy roots emerged (Figure 1D) and could be directly used for the next analysis. To evaluate the ratio of mutagenesis of SlPG, genomic DNA was extracted from the hairy roots of each plant, and then the regions spanning the target sites were amplified by PCR. The PCR products were purified and sequenced. Target mutagenesis can be viewed via sequence peaks overlapping at the target site.
CRISPR/Cas9 expression vectors were individually transformed into A. tumefaciens EHA105 via electroporation. The tomato cotyledon was used for tissue culture and transformation explant according to the protocol previously reported (Mallikarjunaiah and Moudgalya, 2017). Genomic DNA was extracted from the leaves of each plant in the T0 generation, and transgenic plants were identified by PCR primer Cas9-F/R and then used for subsequent gene editing analysis using primer SP1-F/R (Supplementary Table 1). Different types of gene editing can be identified via sequence peaks. Subsequently, the homozygous mutant types were identified by sequence alignment with the WT plant sequence. This method was also used in the T1 and T2 generations.
Genomic DNA of three T1 homozygous slpg mutants and WT tomato plants was used to evaluate the off-target effect of sgRNA SP1 used in this study. Off-target sites were estimated by the web tool CRISOR (see footnote 2) (Concordet and Haeussler, 2018). PCR analysis was performed using primers specific to the targeted gene (Supplementary Table 1). The amplified products were sequenced and evaluated the off-target effect of the sgRNA SP1.
The WT plants and T2 homozygous slpg mutants grown in a greenhouse were used to compare the expression levels of SlPG. Total RNAs were extracted from frozen tissue using TransZol by following the protocol provided by the product supplier (TransGen Biotech, Beijing, China). For reverse transcription, 2 μg of total RNA was used to synthesize first-strand cDNA using TransScript® II One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). For qRT-PCR, the reactions were performed using TransStart® Green qPCR SuperMix (TransGen Biotech, Beijing, China). The PCR amplification conditions begin with a denaturing step for 20 s at 95°C, followed by 40 cycles of 95°C for 5 s and a primer extension reaction at 60°C for 30 s. PCR products were monitored using Bio-Rad CFX connect (Bio-Rad). All PCR reactions were run with three biological replicates each. Data were analyzed using the 2–ΔΔCt method. SlActin (solyc03g078400) was selected as the internal reference.
Fruit firmness was measured using a GY-4 digital fruit hardness tester fruit sclerometer (ALIYIQI, China) according to a previously reported method (Yang et al., 2017). Fresh intact fruits from different developmental stages, including breaker stage (BR), pink stage (PK), light red stage (LR), and red ripe stage (RR), were collected. A total of 20 fruits from different plants were taken at each stage, and each stage fruit was tested twice at equidistant points along the equatorial plane of the fruit. The fresh weight and size of each fruit were also measured to calculate the physiological loss of water. For the fruit softening analysis, more than 20 fruits at the RR stage were collected, the surface was sterilized first, and then stored at room temperature (23–25°C and 55–60% relative humidity). Every 10 days, fruit firmness was measured, and visual softening and collapse of fruit were assessed (Nambeesan et al., 2010). A total of 20 fruits were taken from different plants at each stage for analysis.
All experiments were repeated three times independently, and all results are reproducible. Statistical analyses were performed using SPSS 19.0 software. Two-tailed Student’s t-test was applied to compare the significance of differences between different groups. The P-values less than 0.05 were recognized as significant. OriginPro8 was used for drawing box plots and histograms. Values represent mean ± standard error (SE; n = 20).
The primer sequences used for screening the transgenic positive transformants in transient and stable transformation, detecting the mutation induced by CRISPR/Cas9, potential off-target analysis, identifying transgene-free slpg mutant lines, and qRT-PCR analysis are listed in Supplementary Table 1.
Hairy roots that contain different sgRNAs (SP1, SP2, SP3, SP4, and SP5) grown to a length of 5–6 cm (Figure 1D) were harvested individually and mutations at the targeted sites were examined. According to the results (Table 1), 15 out of 20 hairy roots show gene editing by the sgRNA SP1, which was predicted to target the first exon of the SlPG gene. In terms of editing efficiency, the ratio of SP1 gene editing is relatively high and reached 75%. Subsequently, the vector pg1 was chosen for further stable transformation.
The mutants of SlPG at the target sites were named slpg. The T0 transgenic line was identified, and 7 out of 10 had heterozygous-targeted mutations in SlPG. The site-directed mutagenesis of SlPG was also observed at the target site in the T1 generation. A total of five T1 plants that were homozygous of SlPG induced by CRISPR/Cas9 were identified and three types of mutations for null alleles of SlPG were founded at target site SP1 [1 bp insertion (slpg-T1#2), 4 bp deletion (slpg-T1#3) and 10 bp deletion (slpg-T1#5)] (Figure 1E). DNA sequencing peaks showing successful gene editing at the target site are shown in Figure 1F. All types of frameshift mutations induced by CRISPR/Cas9 at the target site generated premature translation termination codons (PTCs) (Supplementary Figure 1). These results demonstrated that CRISRP/Cas9 generates the targeted mutations and loss of function in the tomato SlPG gene.
According to the predicted result, the one most likely to be the off-target site of SP1 target sites of SlPG was found in an exon of Solyc04g015530.2.1 (Figure 1G). They examined potential off-target site only possessed mismatches of 1 bp compared with the on-target guide sequences (Figure 1G). Sequencing analysis demonstrated that sequences of the off-target site in all three homozygous slpg lines completely matched the WT sequence in tomato. This result indicates that no mutations were induced at the off-target site.
To determine whether homozygous slpg can transmit the induced mutations and phenotypes to their progenies, a total of 20 T2-generation plants of each mutation type deriving from three individuals of corresponding T1 slpg lines were selected randomly, respectively. The result demonstrated that the targeted mutagenesis of SlPG was stably inherited and had maintained consistent mutation types from the T1 generation. To assess the consequences of the site-directed mutagenesis in the targeted loci, we examined the morphological characteristics of the plant body and fruits in all three slpg mutants and WT plants and detected no morphological differences (Supplementary Figure 2). In addition, some of the agronomic characteristics of all slpg mutants and WT plants were also evaluated (Supplementary Table 2). Results indicate that the growth and development of all slpg mutants and WT plants are consistent and are not affected by gene editing.
To assess the potential roles of SlPG throughout the development of the tomato fruit, we conducted detailed quantitative real-time PCR (qRT-PCR) to examine its transcription in five different fruit development sections [e.g., mature green (MG), BR, PK, LR, and RR]. In WT plants, the expression of SlPG was relatively low and negligible in MG fruit. However, the expression level was drastically enhanced from the PK section and reached a maximum in the LR section (Figure 2A). The result showed that the SlPG expression level was relatively high in the PK and LR sections while relatively low in the RR section, indicating that SlPG functioned mainly in the fruit ripening section and secondary in the fruit softening. In slpg mutants, real-time PCR results showed that SlPG transcripts were significantly decreased in the PK section when compared with WT plants (Figure 2B).
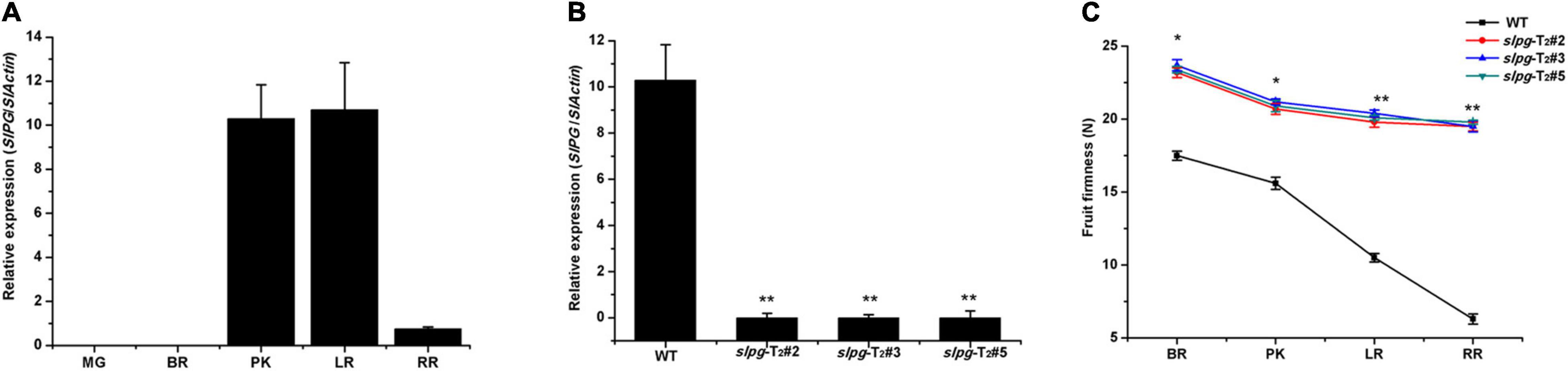
Figure 2. Expression patterns of SlPG and phenotype identification of tomato slpg mutant. (A) Expression of SlPG in fruit at different developmental stages: MG, BR, PK, LR, and RR; (B) Expression of SlPG in WT plants and T2 homozygous slpg mutants (slpg-T2#2, slpg-T2#3, and slpg-T2#5). Relative transcript levels of SlPG were analyzed by qRT-PCR and normalized to SlActin. Quantitative PCR data represent means values for three independent biological replicates (n = 3); (C) Phenotype of enhanced fruit firmness at different fruit development stages in slpg mutants. The fruit firmness values represent the means ± standard error (SE) of 20 individual fruit per line at each stage. * and ** indicate significant differences between WT plants and T2 homozygous slpg mutants (slpg-T2#2, slpg-T2#3, and slpg-T2#5) with p < 0.05 and p < 0.01, respectively, as determined by t-test.
The suitable values for the weight and size of fresh tomato fruit were 4.5 ± 0.2 g and 1.2 ± 0.1 cm, respectively. We next measured the fruit firmness at four different ripening stages (e.g., BR, PK, LR, and RR). The firmness in the WT fruit decreased gradually with fruit ripening, with values of 17.5 ± 0.31, 15.6 ± 0.42, 10.5 ± 0.29, and 6.3 ± 0.34 newton (N) for each stage. In line slpg-T2#2, the values of fruit firmness for each stage were 23.2 ± 0.34, 20.7 ± 0.38, 19.8 ± 0.29, and 19.5 ± 0.27 N. In line slpg-T2#3, the values of fruit firmness for each stage were 23.7 ± 0.39, 21.2 ± 0.21, 20.4 ± 0.23, and 19.5 ± 0.38 N. In line slpg-T2#5, the values of fruit firmness for each stage were 23.4 ± 0.22, 20.9 ± 0.38, 20.1 ± 0.17, and 19.8 ± 0.14 N. All slpg mutant lines showed similar symptoms of firmness, implying that the targeted mutagenesis of SlPG-enhanced fruit firmness (Figure 2C). The WT and slpg mutant fruits were harvested at the RR stage and stored at the greenhouse. WT fruits were wrinkled after 10 days of storage, whereas slpg mutant fruits showed similar symptoms of senescence after 20 days of storage. Comparative analysis of images of fruit after storage of 0, 10, and 20 days showed fewer wrinkles for slpg mutant than WT fruit (Figure 3A). During storage, fruit firmness was much higher in slpg mutant than in WT fruits (Figure 3B). In addition, the slpg mutant fruits exhibited lower physiological loss of water than WT fruits (Figure 3C). We compared the fruit softening time between all slpg mutants with WT plants and found that slpg mutants delayed fruit softening.
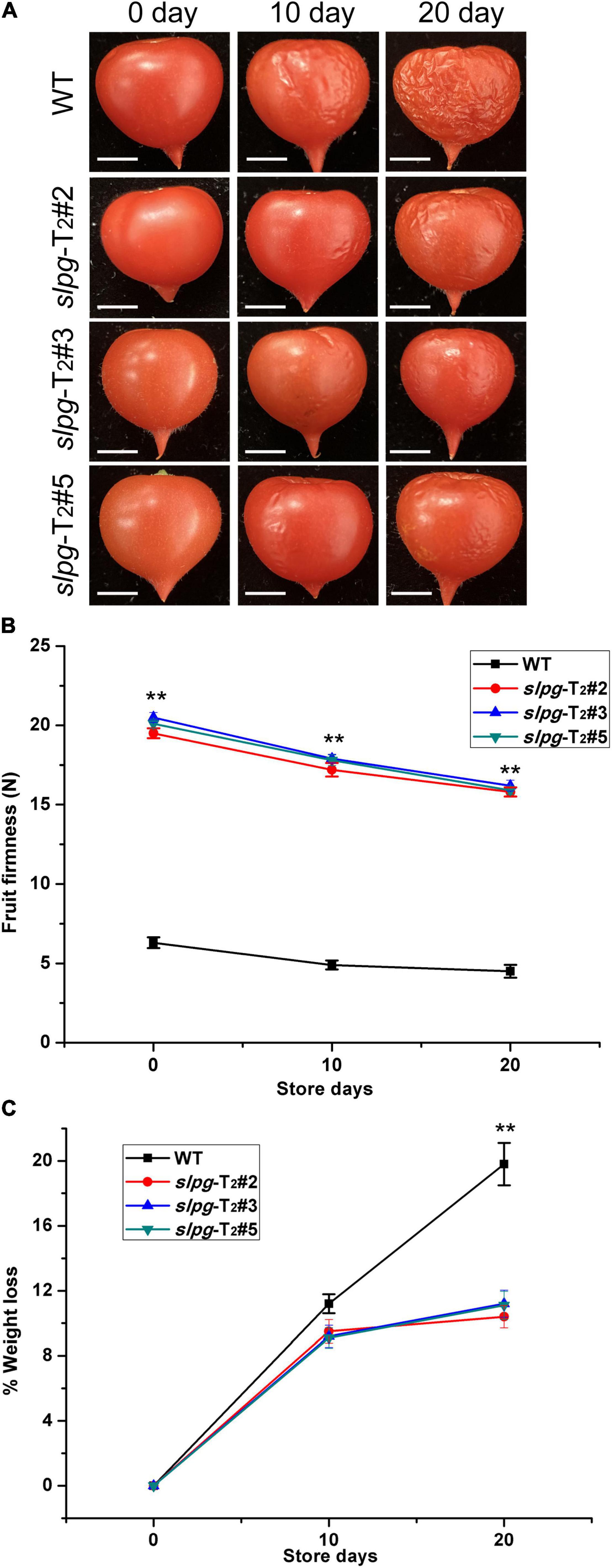
Figure 3. Three slpg mutants (slpg-T2#2, slpg-T2#3, and slpg-T2#5) show extended softening time. (A) RR fruit from three slpg mutants and WT plants after storage at room temperature for 0, 10, and 20 days. Scale bar, 0.5 cm. (B) Change of fruit firmness in three slpg mutants and WT plants. (C) Physiological loss of water (PLW) in three slpg mutants and WT plants. The fruit firmness per fruit and PLW were measured after storage of 0, 10, and 20 days. Average and SE (standard error) values were shown for each data point. ** Indicate significant differences between WT plants and three slpg mutants with p < 0.01, as determined by the t-test.
To obtain tomato homozygous lines for endogenous SlPG mutations without any transgenic element, “transgene-clean” plants were sought via a PCR strategy that used four sets of primer pairs spanning four distinct regions in the T-DNA of sgRNA/Cas9 vectors and one region in the vector backbone. In this assay, we found that 8, 2, and 2 out of 20 T3 slpg mutants from slpg-T2#2 (Figure 4A), slpg-T2#3 (Figure 4B), and slpg-T2#5 (Figure 4C) and their offspring plants were transgene-clean homozygous slpg mutants, respectively.
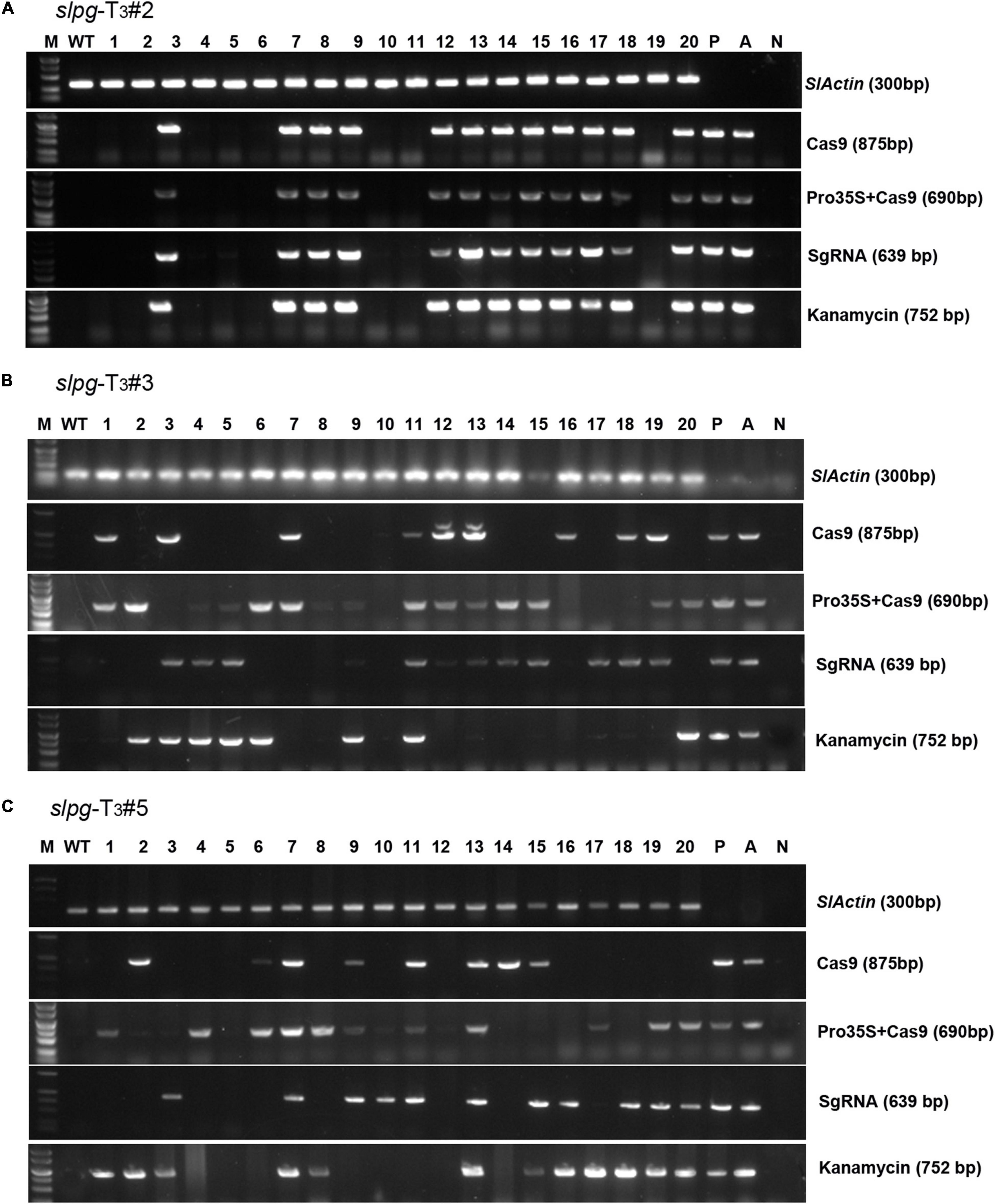
Figure 4. Identifying transgene-free homozygous slpg mutants [(A) slpg-T3#2, (B) slpg-T3#3, and (C) slpg-T3#5]. Gel image of PCR products obtained with primer sets for four T-DNA regions of sgRNA/Cas9 vector. SlActin (300 bp), part of the tomato Actin coding sequence, was used as a normalization control; Cas9 (875 bp), part of the Cas9 coding sequence; Pro35S + Cas9 (690 bp), region from the downstream of the 35S promoter to the upstream of the Cas9 coding sequence; SgRNA (639 bp), region from the AtU6 promoter to the downstream vector sequence spanning the sgRNA; Kanamycin (752 bp), part of the nptII coding sequence; M, DL5000 ladder DNA marker; WT, DNA of WT plant as a template; Lane 1-20, individual mutant lines; P, positive control (sgRNA/Cas9 vector plasmid as template); A, positive control (Agrobacterium containing sgRNA/Cas9 vector as template); N, negative control (water as template).
The CRISPR/Cas9 system has emerged as a robust technology for efficient genome editing and has been successfully applied to many crops (Shan et al., 2013; Upadhyay et al., 2013; Michno et al., 2015; Svitashev et al., 2015). However, some genome editing events do not generate desired specific mutation because of unknown reasons. To overcome this problem, A. rhizogenes-mediated transient transformation is an attractive system for assessing the efficiency of sgRNAs in generating InDels at target sites. Different from A. tumefaciens, A. rhizogenes promote the rapid proliferation of adventitious roots that emerge at the wounding site, which are called hairy roots. The gene-editing efficacy of the sgRNAs in generating mutation at target sites can be evaluated in 15 days by sequencing PCR products amplified from the genomic DNA of pooled hairy roots. Before the whole plant’s stable transform, this transient system can be time-saving, less labor-intensive, and more effective, and many researchers have already applied this system in their works (Pavli and Skaracis, 2010; Li et al., 2019). In this work, we designed five sgRNAs and evaluated the gene-editing efficacy of each sgRNA in the tomato SlPG gene. Our data demonstrated that the editing efficacy of sgRNA SP1 is relatively high (75%, in Table 1), and SP1 was chosen and applied in the next stable transformation.
Tomato genomes are ca. 900 Mb arranged in 12 chromosomes with 35,768 loci containing protein-coding transcripts3. It offers many possible binding sites for CRISPR/Cas9 to generate some additional unwanted mutations. Therefore, a certain risk of off-target activity always remains in the application of CRISPR/Cas9. Previously, several measures, such as choice of suitable nuclease (Fauser et al., 2014), target site choice with CRISPOR (Concordet and Haeussler, 2018), and nuclease delivery method (Hendel et al., 2015), should be considered to minimize the chances for an off-target cleavage to occur. A suitably engineered nucleases can increase the efficiency of targeted mutagenesis. Different types of codon-optimized Cas9 nucleases, including maize codon-optimized (Svitashev et al., 2015), were expressed in our transient assay and the results showed that maize codon-optimized nucleases possess the highest rate of targeted mutagenesis in monocot and dicot plants (paper unpublished). In this study, to avoid potential off-target mutations, a zCas9 nuclease (maize codon-optimized deactivated Cas9), a high specificity score (94) target site SP1 from CRISPOR, and Agrobacterium-mediated transformation were applied.
CRISPR/Cas9-induced mutations were determined from the T0 to T1 generation. All T0 heterozygous biallelic and homozygous slpg edited lines could transmit the targeted mutations to the T1 generation. We still obtained some novel targeted mutagenesis in T1 generation from T0 heterozygous monoallelic slpg lines. In these lines, CRISPR/Cas9-induced mutation exists only in one allele and the other allele without any mutation, and the targeted mutations of the SlPG allele induced by CRISPR/Cas9 were stably inherited and maintained consistent mutation types from T0 to T1 generation. At the same time, the other allele without any mutations can be induced by CRISPR/Cas9 and generate novel targeted mutagenesis in the T1 generation. These results demonstrated that CRISPR/Cas9 can induce targeted mutagenesis in the next generation if monoallelic edits are in the previous generation. This phenomenon is consistent with the previous research on soybean (Bai et al., 2020).
Fruit softening is one of the most characteristic physiological processes that occur during the ripening of fleshy fruits (Garcia-Gago et al., 2009). A softening process results from the degradation of cell wall polymers by the action of the cell wall-associated hydrolytic enzymes. In the past few decades, many enzymes, such as pectate lyase (PL) (Uluisik et al., 2016; Yang et al., 2017), β-galactosidase (Smith et al., 2002), expansin (Brummell et al., 1999), and GA2-oxidase (Li et al., 2020), have been studied as candidates for delaying fruit softening, and some changes were observed in fruit softening. For example, the PL gene has already been chosen to manipulate fruit softening in tomatoes (Marín-Rodríguez et al., 2002; Yang et al., 2017), bananas (Payasi and Sanwal, 2003), and mangos (Chourasia et al., 2006). Among the cell hydrolytic enzymes involved in fruit softening, PG is the best and the earliest characterized (Brady et al., 1982). In 1986, the PG gene was first cloned and sequenced (Grierson et al., 1986). In 1994, The FLAVR SAVR™ tomato is the first genetically engineered commodity to be sold in commerce (Kramer and Redenbaugh, 1994). By 2000, the members of the homologs of SlPG have reached 7 (Hong and Tucker, 2000). The last cloned PG gene, TPG7, is highly expressed in pistils and shares 39% sequence identity with the tomato fruit PG genes. The PG gene locus, solyc10g080210, was proved to be highly expressed in tomato fruit, especially in fruit PK and LR stage (Figure 2A). These results were also basically consistent with the data obtained from the TomExpress platform4, thereby further implying a possible crucial role of SlPG during fruit ripening and softening. The FLAVR SAVR™ tomato was developed by using antisense RNA to suppress the expression of PG in ripening tomato fruit. Subsequently, this antisense RNA strategy was applied to other fruits to produce firmer fruits (Garcia-Gago et al., 2009; Zhang S. et al., 2019; Huang et al., 2020). However, these commodities were developed through a transgenic approach, which may generate negative results, such as the disruption of plant endogenous genes or exogenous gene silencing (Napoli et al., 1990). Although no scientific evidence suggests that genetically modified organism (GMO) products are harmful to human health, debates about GMO safety continue. Previously, many transgene-free targeted mutagenesis plants, such as soybean (Cai et al., 2018), rice (Li et al., 2016), wheat (Zhang et al., 2016), and tomato (Soyk et al., 2017), have been generated by CRISPR/Cas9. In this study, CRISPR/Cas9 was applied to induce targeted SlPG mutagenesis, which could delay fruit softening without any exogenous gene fragment. Moreover, CRISPR/Cas9 was proven to enhance fruit firmness without compromising other fruit qualities (Li et al., 2020), indicating that the targeted mutagenesis of the SlPG gene in tomatoes could delay fruit softening without impacting the tomato flavor, aroma, taste, and nutritional value. This conclusion was supported by Kramer and Redenbaugh’s research (Kramer and Redenbaugh, 1994). This is a key target characteristic in modern tomato breeding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
HN and GX designed the project. HN and YS performed the experiment. HN and XG analyzed the data. HN wrote the manuscript. GX made suggestions for improving the procedure and proofread the manuscript. All authors read and approved the final manuscript.
This work was supported by the Shanxi Province Key R&D Plan (201903D211011), the National Natural Science Foundation of China (31501750), and the Shanghai International Scientific and Technological Cooperation Foundation (19390743400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.729128/full#supplementary-material
Bai, M., Yuan, J., Kuang, H., Gong, P., Li, S., Zhang, Z., et al. (2020). Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol. J. 18, 721–731. doi: 10.1111/pbi.13239
Bird, C. R., Smith, C. J., Ray, J. A., Moureau, P., Bevan, M. W., Bird, A. S., et al. (1988). The tomato polygalacturonase gene and ripening-specific expression in transgenic plants. Plant Mol. Biol. 11, 651–662. doi: 10.1007/BF00017465
Brady, C. J., Macalpine, G., Mcglasson, W. B., and Veda, Y. (1982). Polygalacturonase in tomato fruits and the induction of ripening. J. Funct. Plant Biol. 9, 171–178.
Brooks, C., Nekrasov, V., Lippman, Z., and Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-Associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Brummell, D. A., Harpster, M. H., Civello, P. M., Palys, J. M., Bennett, A. B., and Dunsmuir, P. (1999). Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant cell 11, 2203–2216. doi: 10.1105/tpc.11.11.2203
Cai, Y., Chen, L., Liu, X., Guo, C., Sun, S., Wu, C., et al. (2018). CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 16, 176–185. doi: 10.1111/pbi.12758
Chourasia, A., Sane, V. A., and Nath, P. (2006). Differential expression of pectate lyase during ethylene-induced postharvest softening of mango (Mangifera indica var. Dashehari). Physiol. Plantarum 128, 546–555. doi: 10.1111/j.1399-3054.2006.00752.x
Concordet, J.-P., and Haeussler, M. (2018). CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245. doi: 10.1093/nar/gky354
Cooley, M. B., and Yoder, J. I. (1998). Insertional inactivation of the tomato polygalacturonase gene. Plant Mol. Biol. 38, 521–530. doi: 10.1023/a:1006086004262
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096
Fauser, F., Schiml, S., and Puchta, H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359. doi: 10.1111/tpj.12554
Garcia-Gago, J. A., Pose, S., Munoz-Blanco, J., Quesada, M. A., and Mercado, J. A. (2009). The polygalacturonase FaPG1 gene plays a key role in strawberry fruit softening. Plant Signal Behav. 4, 766–768. doi: 10.1104/pp.109.138297
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2011). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Grierson, D., Tucker, G. A., Keen, J., Ray, J., Bird, C. R., and Schuch, W. (1986). Sequencing and identification of a cDNA clone for tomato polygalacturonase. Nucleic Acids Res. 14, 8595–8603. doi: 10.1093/nar/14.21.8595
Hendel, A., Fine, E. J., Bao, G., and Porteus, M. H. (2015). Quantifying on- and off-target genome editing. Trends Biotechnol. 33, 132–140. doi: 10.1016/j.tibtech.2014.12.001
Hobson, G. E. (1965). The firmness of tomato fruit in relation to polygalacturonase activity. J. Horticult. Sci. 40, 66–72. doi: 10.1080/00221589.1965.11514121
Hong, S. B., and Tucker, M. L. (2000). Molecular characterization of a tomato polygalacturonase gene abundantly expressed in the upper third of pistils from opened and unopened flowers. Plant Cell Rep. 19, 680–683. doi: 10.1007/s002999900175
Huang, W., Chen, M., Zhao, T., Han, F., Zhang, Q., Liu, X., et al. (2020). Genome-wide identification and expression analysis of polygalacturonase gene family in Kiwifruit (Actinidia chinensis) during fruit softening. Plants 9, 1–16. doi: 10.3390/plants9030327
Ju, R., Tian, Y., Shen, Q., Liu, C., and Mang, K. (1994). Cloning of polygalacturonase (PG) cDNA and inhibition effects of its antisense RNA on the expression of PG gene in transgenic tomato plants. Chin. J. Biotechnol. 10, 67–74.
Kramer, M. G., and Redenbaugh, K. (1994). Commercialization of a tomato with an antisense polygalacturonase gene: the FLAVR SAVR™ tomato story. Euphytica 79, 293–297. doi: 10.1007/BF00022530
Li, C., Nguyen, V., Liu, J., Fu, W., Chen, C., Yu, K., et al. (2019). Mutagenesis of seed storage protein genes in Soybean using CRISPR/Cas9. BMC Res. Notes 12:176. doi: 10.1186/s13104-019-4207-2
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Li, M., Li, X., Zhou, Z., Wu, P., Fang, M., Pan, X., et al. (2016). Reassessment of the Four Yield-related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front. Plant Sci. 7:377. doi: 10.3389/fpls.2016.00377
Li, R., Sun, S., Wang, H., Wang, K., Yu, H., Zhou, Z., et al. (2020). FIS1 encodes a GA2-oxidase that regulates fruit firmness in tomato. Nat. Commun. 11:5844. doi: 10.1038/s41467-020-19705-w
Liu, X. H., Zhang, L. L., Zhu, C. Q., Chen, K. S., and Xu, C. J. (2008). Mechanisms for miniature dwarf characteristics of Micro-Tom tomato and its application in plant functional genomics studies. Yi Chuan 30, 1257–1264. doi: 10.3724/sp.j.1005.2008.01257
Mallikarjunaiah, S., and Moudgalya, S. (2017). Agrobacterium tumefaciens-mediated transformation of tomato (Solanum lycopersicum). IJARS 6:6. doi: 10.20908/ijars.v6i6.8004
Marín-Rodríguez, M. C., Orchard, J., and Seymour, G. B. (2002). Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 53, 2115–2119. doi: 10.1093/jxb/erf089
Michno, J. M., Wang, X., Liu, J., Curtin, S. J., Kono, T. J., and Stupar, R. M. (2015). CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6, 243–252. doi: 10.1080/21645698.2015.1106063
Nambeesan, S., Datsenka, T., Ferruzzi, M., Malladi, A., Mattoo, A., and Handa, A. (2010). Overexpression of yeast spermidine synthase impacts ripening, senescence and decay symptoms in tomato. Plant J. 63, 836–847. doi: 10.1111/j.1365-313X.2010.04286.x
Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2, 279–289. doi: 10.1105/tpc.2.4.279
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2017). Corrigendum: CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep-UK 7:46916. doi: 10.1038/srep46916
Pavli, O., and Skaracis, G. (2010). Fast and efficient genetic transformation of sugar beet by Agrobacterium rhizogenes. Nat. Protoc. 1–6. doi: 10.1038/nprot.2010.98
Payasi, A., Mishra, N. N., Singh, C. R. J. P., and Plants, M.B.o (2009). Biochemistry of fruit softening: an overview. Physiol. Mol. Biol. Pla 15, 103–113. doi: 10.1007/s12298-009-0012-z
Payasi, A., and Sanwal, G. G. (2003). Pectate lyase activity during ripening of banana fruit. Phytochemistry 63, 243–248. doi: 10.1016/S0031-9422(03)00027-X
Pose, S., Paniagua, C., Cifuentes, M., Blanco-Portales, R., Quesada, M. A., and Mercado, J. A. (2013). Insights into the effects of polygalacturonase FaPG1 gene silencing on pectin matrix disassembly, enhanced tissue integrity, and firmness in ripe strawberry fruits. J. Exp. Bot. 64, 3803–3815. doi: 10.1093/jxb/ert210
Quesada, M. A., Blanco-Portales, R., Pose, S., Garcia-Gago, J. A., Jimenez-Bermudez, S., Munoz-Serrano, A., et al. (2009). Antisense down-regulation of the FaPG1 gene reveals an unexpected central role for polygalacturonase in strawberry fruit softening. Plant Physiol. 150, 1022–1032. doi: 10.1104/pp.109.138297
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Sheehy, R. E., Kramer, M., and Hiatt, W. R. (1988). Reduction of polygalacturonase activity in tomato fruit by antisense RNA. PNAS 85, 8805–8809. doi: 10.1073/pnas.85.23.8805
Smith, C. J., Watson, C. F., Bird, C. R., Ray, J., Schuch, W., and Grierson, D. (1990). Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Mol. Gen. Genet. 224, 477–481. doi: 10.1007/BF00262443
Smith, C. J. S., Watson, C. F., Ray, J., Bird, C. R., Morris, P. C., Schuch, W., et al. (1988). Antisense RNA inhibition of polygalacturonase gene expression in transgenic tomatoes. Nature 334, 724–726. doi: 10.1038/334724a0
Smith, D. L., Abbott, J. A., and Gross, K. C. (2002). Down-regulation of tomato beta-galactosidase 4 results in decreased fruit softening. Plant Physiol. 129, 1755–1762. doi: 10.1104/pp.011025
Soyk, S., Müller, N. A., Park, S. J., Schmalenbach, I., Jiang, K., Hayama, R., et al. (2017). Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 49, 162–168. doi: 10.1038/ng.3733
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., and Cigan, A. M. (2015). Targeted mutagenesis, Precise gene editing, and site-specific gene insertion in maize using Cas9 and Guide RNA. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Uluisik, S., Chapman, N. H., Smith, R., Poole, M., Adams, G., Gillis, R. B., et al. (2016). Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 34, 950–952. doi: 10.1038/nbt.3602
Upadhyay, S. K., Kumar, J., Alok, A., and Tuli, R. (2013). RNA-guided genome editing for target gene mutations in wheat. G3 3, 2233–2238. doi: 10.1534/g3.113.008847
Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M.-P., Guyon-Debast, A., Chauvin, J.-E., et al. (2019). Transgene-free genome editing in tomato and potato plants using Agrobacterium-Mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 20:402. doi: 10.3390/ijms20020402
Vincent, H., Wiersema, J., Kell, S., Fielder, H., Dobbie, S., Castañeda-Álvarez, N. P., et al. (2013). A prioritized crop wild relative inventory to help underpin global food security. Biol. Conserv. 167, 265–275. doi: 10.1016/j.biocon.2013.08.011
Xie, K., Zhang, J., and Yang, Y. (2014). Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol. Plant 7, 923–926. doi: 10.1093/mp/ssu009
Yan, P. U., Liu, C., Ji-Yang, L. I., Gulitashi, A., Yan, H. U., and Liu, X. D. (2018). Different SlU6 promoters cloning and establishment of CRISPR/Cas9 mediated gene editing system in tomato. J.S.A.S. 51, 315–326. doi: 10.3864/j.issn.0578-1752.2018.02.011
Yang, L., Huang, W., Xiong, F., Xian, Z., Su, D., Ren, M., et al. (2017). Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 15, 1544–1555. doi: 10.1111/pbi.12737
Zhang, S., Ma, M., Zhang, H., Zhang, S., Qian, M., Zhang, Z., et al. (2019). Genome-wide analysis of polygalacturonase gene family from pear genome and identification of the member involved in pear softening. BMC Plant Biol. 19:587. doi: 10.1186/s12870-019-2168-1
Zhang, Y., Malzahn, A. A., Sretenovic, S., and Qi, Y. (2019). The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 5, 778–794. doi: 10.1038/s41477-019-0461-5
Keywords: tomato, polygalacturonase, CRISPR/Cas9, genome editing, fruit softening
Citation: Nie H, Shi Y, Geng X and Xing G (2022) CRISRP/Cas9-Mediated Targeted Mutagenesis of Tomato Polygalacturonase Gene (SlPG) Delays Fruit Softening. Front. Plant Sci. 13:729128. doi: 10.3389/fpls.2022.729128
Received: 24 June 2021; Accepted: 21 April 2022;
Published: 19 May 2022.
Edited by:
Kejian Wang, China National Rice Research Institute (CAAS), ChinaReviewed by:
Prasenjit Saha, University of Illinois Urbana-Champaign, United StatesCopyright © 2022 Nie, Shi, Geng and Xing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoming Xing, a2FpYW5kZXJAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.