- 1Institute of Vegetables, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2College of Horticulture Science, Zhejiang Agriculture and Forestry (A&F) University, Hangzhou, China
- 3State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
The basic leucine zipper (bZIP) as a well-known transcription factor family, figures prominently in diverse biological and developmental processes and response to abiotic/biotic stresses. However, no knowledge of the bZIP family is available for the important edible Cucurbitaceae crop bottle gourd. Herein, we identified 65 putative LsbZIP genes and characterized their gene structure, phylogenetic and orthologous relationships, gene expression profiles in different tissues and cultivars, and responsive genes under cold stress. The phylogenetic tree of 16 released Cucurbitaceae plant genomes revealed the evolutionary convergence and divergence of bZIP family. Based on the specific domains, LsbZIP family were classified into 12 clades (A–K, S) with similar motifs and exon-intron distribution. 65 LsbZIP genes have undergone 19 segmental and two tandem duplication events with purifying selection. The expression profiling of LsbZIP genes showed tissue-specific but no cultivar-specific pattern. The cold stress-responsive candidate LsbZIP genes were analyzed and validated by RNA-Seq and RT-PCR, providing new insights of transcriptional regulation of bZIP family genes in bottle gourd and their potential functions in cold-tolerant variety breeding.
Introduction
Plants are subjected to various abiotic stresses such as drought, chilling, salinity and high temperature with the increasing environmental degradation and changing weather, which extremely limits the growth and production of crops. Therefore, plants have evolved complicated stress-responsive defense tactics involving in a number of proteins, containing phosphatase, protein kinase and transcription factors (TFs), to adapt, survive and reproduce in harsh environment (Sornaraj et al., 2016). TFs, as the primary modulators in stress response, bind specific promoter regions of downstream target genes to alter transcription activity and stimulate or suppress gene expressions (Kong et al., 2015). Among more than 80 TF families, the basic leucine zipper (bZIP) family is one of the largest and diverse groups (Llorca et al., 2014; Yin et al., 2017). The bZIP domain is consist of two functional regions, the basic area for DNA binding and the leucine zipper for protein dimerization (Li et al., 2019). The basic region is highly conserved with 16 amino acid residues, including a specific motif of N-X7-R/K for nuclear localization signal and sequence-specific binding, while the leucine zipper motif is predisposed to form an alpha helical loop with dimerization specificity (Wang et al., 2011).
The bZIP TFs with plenty of diverse members have been found to be involved in the regulation of multiple biological processes, namely flower and seed development (Zhang et al., 2016; Jain et al., 2017), hormone signal transduction (Sirichandra et al., 2010), energy metabolism (Weiste et al., 2017), embryogenesis (Huang et al., 2012), photomorphogenesis (Lindemose et al., 2013), lateral root development and leaf senescence (Dankov et al., 2009). Additionally, bZIP TFs play vital roles in response to abiotic/biotic stresses including extreme temperatures, water shortage, high salinity and hormone-induced defense from pathogens (Zander et al., 2010; Lindemose et al., 2013; Sornaraj et al., 2016; Wang et al., 2020). The regulatory network of downstream genes can be controlled by the combination of bZIPs and corresponding cis-elements such as A-box (TACGTA), C-box (GACGTC), G-box (CACGTG) and abscisic acid (ABA)-responsive elements (ABRE) (CCACGTGG) (Foster et al., 1994; Zhang et al., 2008; Tang et al., 2015). In Arabidopsis, four bZIP factors ABF1 (AtbZIP35), ABF2 (AtbZIP36), ABF3 (AtbZIP37) and ABF4 (AtbZIP38) are predominant in regulating downstream gene expression of ABA signaling in response to osmotic stresses like drought and salt (Jakoby et al., 2002; Yoshida et al., 2015). And TGA2 (AtbZIP20), TGA5 (AtbZIP26) and TGA6 (AtbZIP45) mediate salicylic acid (SA)-dependent pathway and meanwhile trigger ethylene and jasmonic acid (JA) pathways under abiotic stress (Zander et al., 2010). OsbZIP1 increases pathogen resistance to Magnaporthe grisea in rice through SA, JA and ABA signal transduction pathway (Meng et al., 2005). ZmbZIP17 is capable of binding ABREs and transduces stress signals through ABA pathway during germination and post-germination seedling establishment (Yang et al., 2013). Likewise, LIP19 acts as a molecular switch for low-temperature signal transduction in rice (Shimizu et al., 2005), while OsBZ8 positively regulates salt tolerance with ABA pathway (Mukherjee et al., 2006). With respect to number of bZIPs, 78 bZIP genes have been identified in Arabidopsis and classified into 13 groups (Jakoby et al., 2002; Dröge-Laser et al., 2018). So far, the genome-wide characterization of bZIP genes from many plant species have been reported, including 89 in rice (Nijhawan et al., 2008), 125 in maize (Wei et al., 2012), 247 in rapeseed (Zhou et al., 2017), 89 in barely (Pourabed et al., 2015), 92 in sorghum (Wang et al., 2011), 131 in soybean (Liao et al., 2008), 99 in poplar (Zhao et al., 2021), 56 in potato (Mirzaei et al., 2020) and 227 in wheat (Liang et al., 2022). The diversity and expansion of bZIP gene members is attributed to the whole genome, tandem and segmental duplication events (Panchy et al., 2016; Qiao et al., 2019). It’s one of the evolutionary strategies to adapt various stresses by expanding TFs family members like bZIP to achieve targeted regulation and control of complex gene expression.
Cucurbitaceae is one of the most important edible plant family in the world and encompasses nearly 1000 species from 115 genera (Schaefer et al., 2009). Bottle gourd [Lagenaria siceraria (Molina) Standl.], known as calabash or white-flowered gourd, is an annual Cucurbitaceae plant grown for its fruit. It is one of the world’s first cultivated plants not primarily for food, but also for use as containers, decorations, musical instruments and so on (Schlumbaum and Vandorpe, 2012). Bottle gourd is planted all over China and widely cultivated in tropical to temperate regions of the world. It is suitable to grow in conditions with moderate temperature and sufficient sunlight. However, cold stress owing to early-spring low temperature is a major abiotic stress and limiting factor for its industry (Chimonyo and Modi, 2013). Low temperature can influence many physiological and biochemical metabolisms like photosynthesis, osmotic regulation, the activity of antioxidants and so on. Breeding new bottle gourd varieties with low temperature resistance is necessary to elevate its production and quality. To accelerate the breeding process, the landscape of gene families which influence growth, development and stress response should also be comprehensively studied. As the release of our recently assembly high quality genome of bottle gourd variety “HZCG” (Xu et al., 2021), it allows us to carry out a genome-wide identification and analysis of LsbZIP gene family and its expression pattern under cold stress. The comparison of all bZIPs from present available Cucurbitaceae genomes in this research aims to reveal the evolution and selection in Cucurbitaceae species differentiation. This study provides the full-scale description of LsbZIP genes involved in response to cold stress, and lays a foundation for further molecular breeding in bottle gourd.
Materials and methods
Sequence retrieval and domain identification
The reference genome of bottle gourd (genome assembly ZAAS_Lsic_2.0) was obtained from our previous study (Xu et al., 2021), the query sequences 78 AtbZIPs (genome assembly TAIR10) from the Arabidopsis information resource (TAIR) (https://www.arabidopsis.org/index.jsp) were retrieved (Jakoby et al., 2002; Dröge-Laser et al., 2018), and the other genomes of selected 15 Cucurbitaceae species were downloaded from Cucurbit Genomics Database (CuGenDBv2, http://cucurbitgenomics.org/v2/) (Wang et al., 2020). A local protein database was created by TBtools v1.098769 to blast the underlying Cucurbitaceae bZIP sequences using an e value cut-off of 1e-5 (Chen et al., 2020). To avoid missing potential bZIPs, a hidden Markov model (HMM) file of the bZIP domain (PF00170 and PF07716) was downloaded (https://pfam.xfam.org/) and used as the template to identify bZIP sequences in the respective genome. The primary candidate sequences were further submitted in the Plant Transcription Factor Database (http://planttfdb.gao-lab.org/), SMART database (http://smart.embl.de/), NCBI CDD (https://www.ncbi.nlm.nih.gov/cdd/) for confirmation. The physicochemical properties were calculated based on the high confidence bZIPs by ProtParam (https://web.expasy.org/protparam/) (Gasteiger et al., 2003).
Multiple alignments and phylogenetic tree construction
All bZIP sequences from Arabidopsis and 15 Cucurbitaceae were screened and aligned by Muscle algorithm (Edgar, 2004). TrimAl v1.2 was invoked to remove the unambiguous aligments using gappyout trimming mode (Capella-Gutiérrez et al., 2009). The construction of phylogenetic trees was executed with IQ-tree software, using a maximal likelihood (ML) method with 1,000 bootstrap replications (Nguyen et al., 2015). Amino acid sequences from bottle gourd, as well as 78 AtbZIPs were completely aligned by Clustal X2.1 and converted ALN file into MEGA 7.0.14 to build a neighbor-joining (NJ) phylogenetic tree with following parameters: poission model, and pairwise deletion for the reliability of interior branches.
Gene distribution and duplication visualization
The chromosomal locations of LsbZIP genes and their tandem duplications were represented according to previous studies (Wang et al., 2022). Gene duplication events were predicted by multiple collinearity scan toolkit (MCScanX) (Wang et al., 2012). The Ka and Ks were calculated using MCScanX package to assess the selection history and divergence time of LsbZIP gene families. The distribution and collinearity relationships were displayed with Advanced Circos function in Tbtools.
Prediction of conserved motifs and exon/intron structure
To better decipher the structure diversity of LsbZIP genes, motifs were detected using the Multiple Expectation Maximization for Motif Elicitation (MEME)/Motif Alignment and Search Tool (MAST) system (http://meme-suite.org/) with default set (Bailey et al., 2009). The exon-intron structure and the intron phases of bZIP gene family were visualized by the online program Gene Structure Display Server 2.0(GSDS, http://gsds.gao-lab.org/) (Hu et al., 2015).
Orthologous gene clusters analysis of eight selected species
Eight common and important species genomes namely Arabidopsis thaliana, Brassica napus, Cucumis sativus, Glycine max, Oryza sativa, Solanum lycopersicum, Vitis vinifera and Lagenaria siceraria were blasted through Ortho Venn 2.0 web server (https://orthovenn2.bioinfotoolkits.net/home). 65 LsbZIP protein sequences were uploaded and analyzed with e-value 1e-5 and inflation value of 1.5 (Wang et al., 2022).
Ab initio prediction of the core promoter in LsbZIPs
The putative promoter sequences were extracted from 2000 base pair (bp) upstream of the transcriptional starting site (ATG) of genomic DNA sequences. The cis-regulatory elements (CREs) were obtained by introducing promoter sequences into the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
The expression patterns analysis of LsbZIP genes in different tissues and varieties
To determine the gene expression profiles of bZIP family, RNA-Seq data of various tissues including leaf, root, stem, flower and fruit were downloaded from NCBI sequence read archive (SRA) with accession SRP107894 (Wu et al., 2017). A collection of re-sequencing data of 5 bottle gourd accessions were also downloaded from SRP095913 (Xu et al., 2021). The expression of each LsbZIP gene was quantified with transcripts per million (TPM). The transcript abundances were output by kallisto in TPM units. Normalization (log2) and heat-maps were generated using Tbtools software.
Plant materials and treatments
‘YZ’, a bottle gourd variety of rootstock type tolerance to low temperature, was used in this experiment. Healthy and sterilized seeds were germinated and sown in plastic pots. The seedings were placed in growth chamber at 28°C/22°C (16 h/8 h) day/night temperatures with a relative humidity of 75%. At 2~3 leaf stage, uniform seedlings were divided into two groups for room temperature (24°C) or cold treatment (4°C). The leaf samples were harvested for gene expression validation at 0, 1, 6, 12, 24 h after cold treatment. And three biological replications were used in this study.
Transcriptome and qRT-PCR analysis
The transcriptional profile of bZIP genes of bottle gourd under cold stress were downloaded under accession PRJNA553072 from our previous study (Wang et al., 2020). The differentially expressed genes (DEGs) of LsbZIPs were quantified with fragments per kilobase of transcript per million mapped fragments (FPKM) method between control and cold treatment. DEGs were screened according the criteria (false discovery rate <0.5 and absolute fold change ≥2). For cold-induced LsbZIPs expression, total RNA was extracted from RNAprep Pure Plant Kit (Tiangen, China). TuB-α gene (BG_GLEAN_10019523) was used as the internal control gene in bottle gourd (Li et al., 2021). qRT-PCR was performed on a Bio-Rad CFX96 (BIO RAD, USA) with SuperReal PreMix Plus/SYBR Green (Tiangen, China). Triplicate replications were performed for analysis. The gene relative expression level was calculated by the 2–ΔΔCt method. All primers used for qRT-PCR are listed in Supplementary Table S1.
Results and discussions
Systematic identification and classification of bZIPs
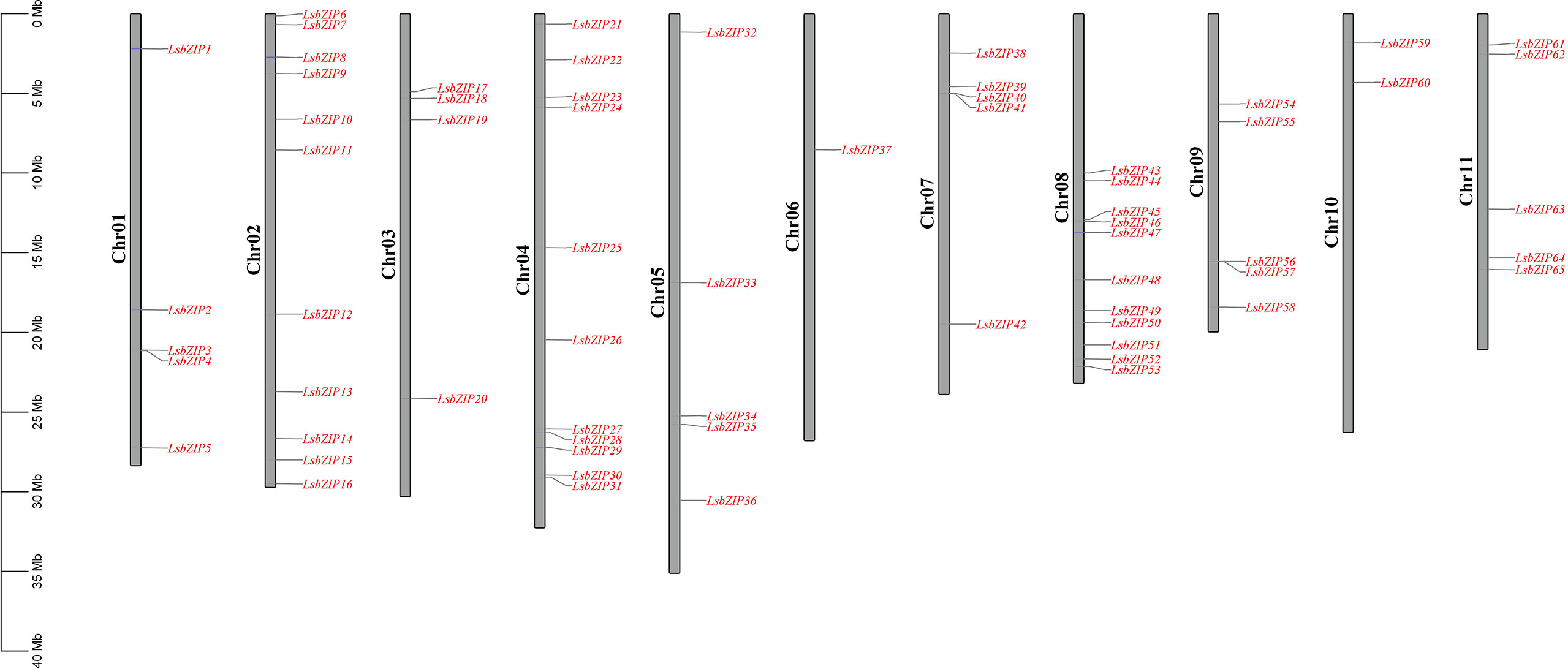
So far two genomes of bottle gourd inbred lines, HZCG (food-type, China) assembled with Illumina paired-end and USVL1VR-Ls (rootstock-type, India) assembled with PacBio and BioNano, have been released (Wu et al., 2017; Xu et al., 2021). Based on the specificity of bZIP family domain, 65 and 60 bZIP genes were identified in HZCG and USVL1VR-Ls, respectively (Table S2, Figure S1). The comparison of bZIP genes in two bottle gourd reference genomes showed that each bZIP in USVL1VR-Ls was able to find its homolog in HZCG (Supplementary Figure S1). Thus, we selected 65 bZIPs in HZCG genome for our subsequent analysis and named them as LsbZIP1~LsbZIP65 according to their location (Table S2). Generally, 65 LsbZIPs were roughly spread equally across 11 all chromosomes (Chr) while only Chr 6 contained one bZIP gene and Chr 10 contained two bZIP genes (Figure 1). The physicochemical parameters showed that the length of amino acid (AA) residues ranged from 129 AA (LsbZIP36) to 1225 AA (LsbZIP8), with isoelectric point (pI) ranging from 4.55 (LsbZIP6) to 9.89 (LsbZIP33) (Supplementary Table S2). The molecular weight (MW) ranged from 15.14 kDa (LsbZIP36) to 135.15 kDa (LsbZIP8) with average 39.74 kDa. In addition, the prediction of subcellular localization of all LsbZIP proteins were distributed in nucleus except LsbZIP8 locating on plasma membrane and LsbZIP34 locating in cytoplasmic zone.
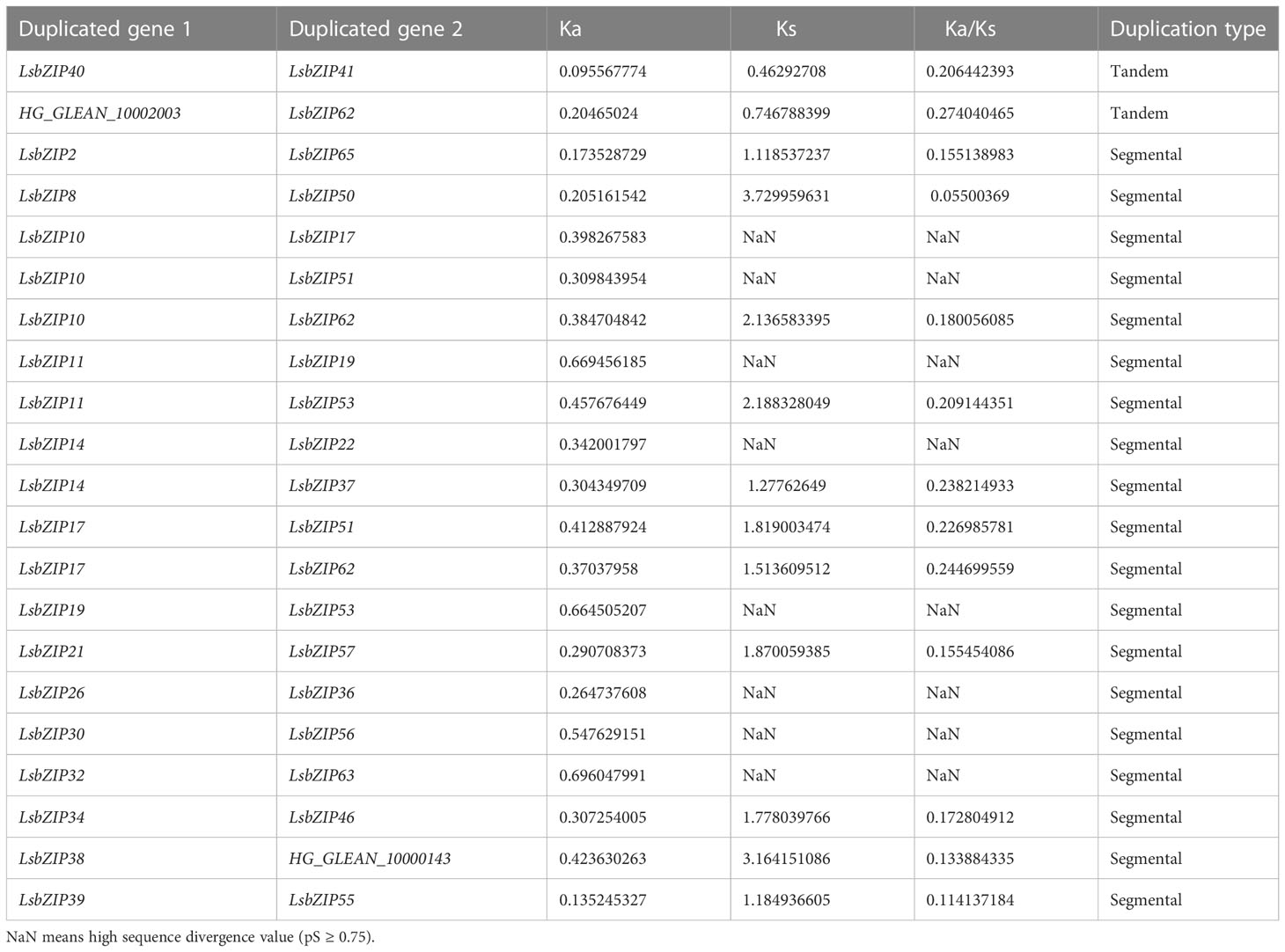
For the gene duplication events, 19 gene pairs were segmentally duplicated on all chromosomes except Chr 10 (Figure 2; Table 1). There were only two tandem duplication events with gene pairs (LsbZIP40/LsbZIP41, HG_GLEAN_10002003/LsbZIP62) in bottle gourd. To estimate the selective pattern, Ka/Ks is calculated to uncover the diversity of duplicated gene pairs in the evolution. In this study, all duplicated gene pairs were subjected to strong purifying selection for the purpose of eliminating detrimental mutations as Ka/Ks<1 while 8 duplicated gene pairs showed NaN caused by high sequence divergence value (Table 1).
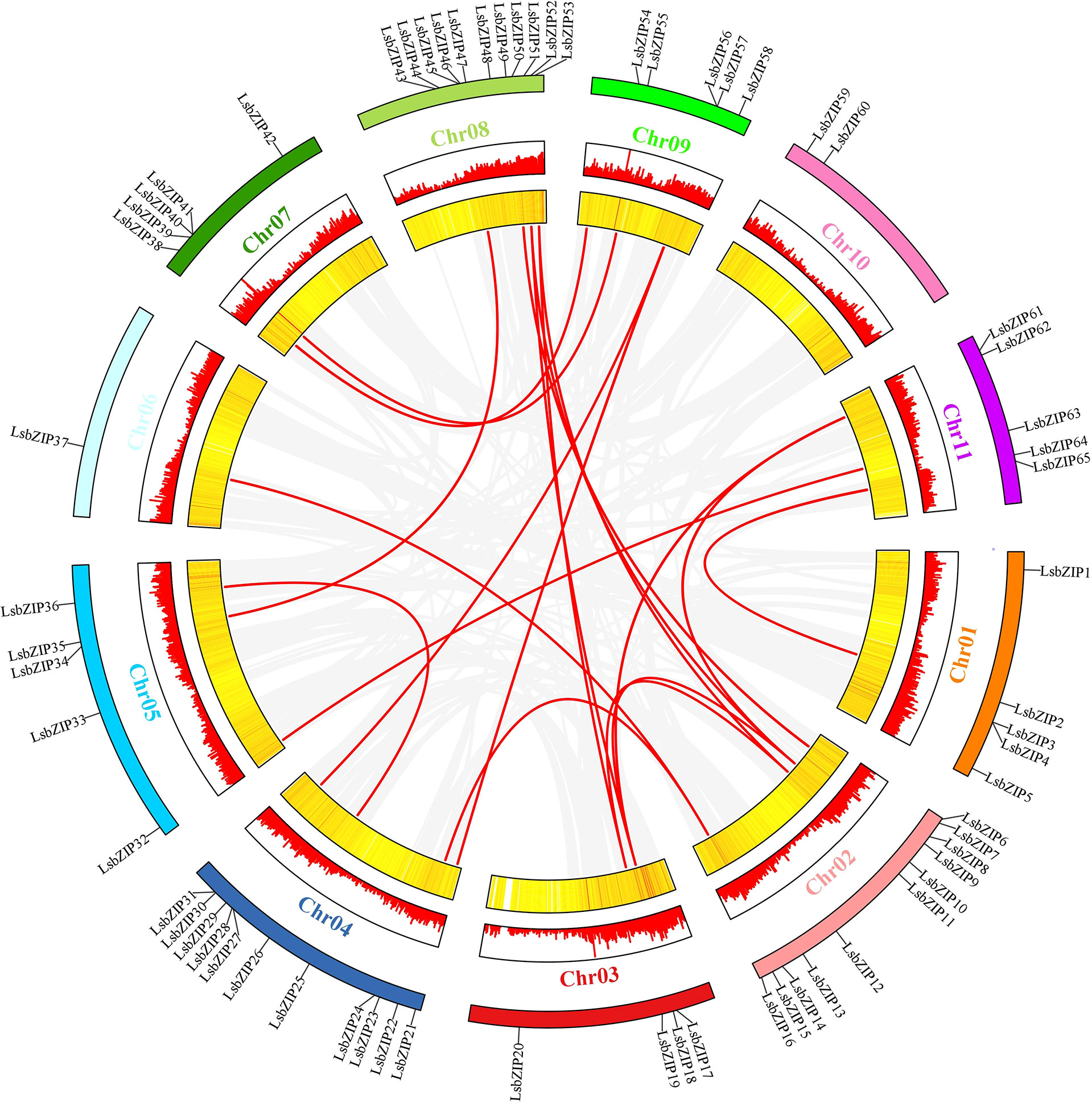
Figure 2 Interchromosomal synteny of linked LsbZIP genes. The red bars and yellow color strips indicated the gene density in the chromosomes.
Phylogenetic analysis of Cucurbitaceae species
Based on the specific domains bZIP_1 (PF00170) and bZIP_2 (PF07716), we finally identified and confirmed 1510 sequences from 17 Cucurbitaceae species including two different bottle gourd genomes (Tables 2, S3, S4; Figure 3). Apart from the splicing variants found in Sechium edule and Trichosanthes anguina, only 1235 unique bZIP sequences were retrieved and grouped into 13 subfamilies in Cucurbit genomes. Generally, clade A, D, E, I and S occupied the numerous bZIP members exceeding 100 while other clades B, J, K contained limited numbers of bZIP homologs less than 20 (Table 2). 11 of 13 clades were existed in all selected cucurbit crops except clades B, K and M. Clade B and K usually owned 1~3 and only one bZIP gene in 15 species (not found in cushaw pumpkin and ridge gourd) respectively, and clade M was consist of 1~ 3 bZIP members in 12 species (not identified in wax, cucumber, bottle gourd and bitter gourd). The number showed diverse distribution not only in different clades, but also in different species. The total number of bZIPs in 16 species could be divided into two categories. One type mostly belonged to Cucurbita genera including C. argyosperma, C. maxima, C. moschata and C. pepo besides S. edule. The gene family number in this class was approximately two-fold than it in another class reaching the amount of average 65 members (Table 2). The majority of all clades were quite distinct from each other, but clade F was sandwiched between two separated clades S.
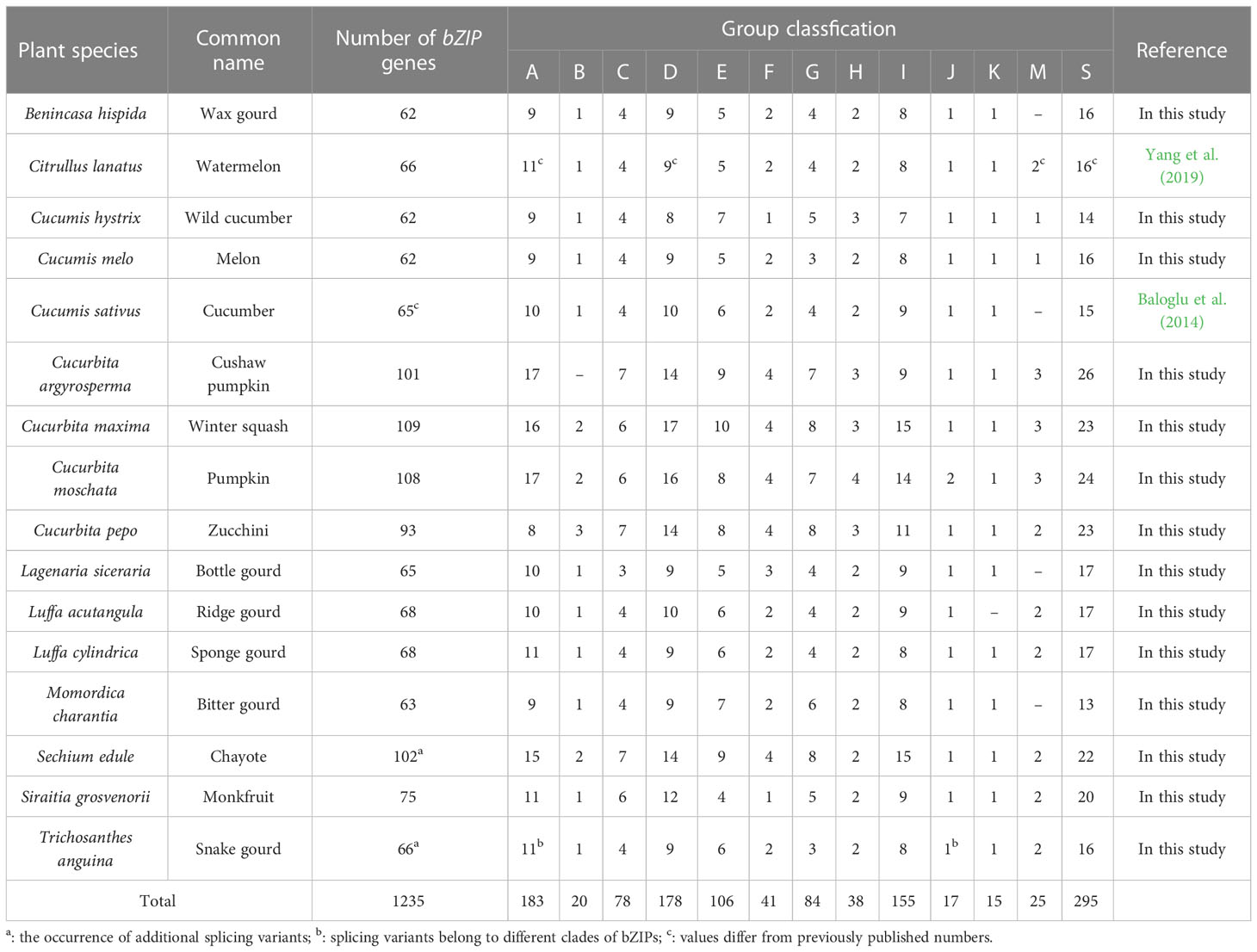
Table 2 bZIP family members of 16 Cucurbitaceae plant species and their phylogenetic classifications.
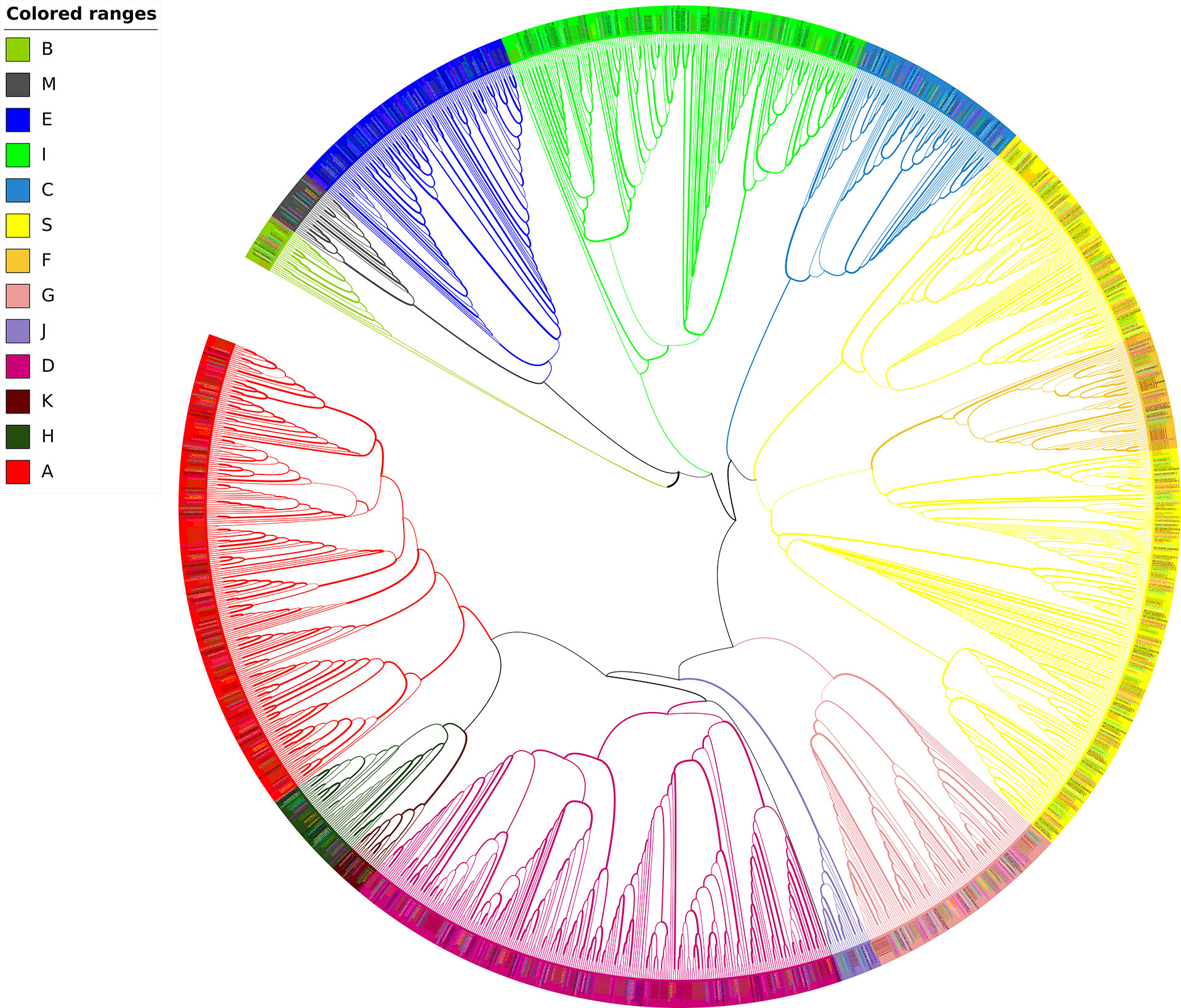
Figure 3 Unrooted Maximum likelihood phylogenetic tree of bZIP genes in 16 Cucurbitaceae species. Different colors represent bZIPs in different clades according to Dröge-Laser et al. (2018).
Gene structure and motif analysis
According to the classification criterion of AtbZIPs by Dröge-Laser et al. (2018), 65 LsbZIPs were divided into 12 clades (LsbZIP-A, LsbZIP-B, LsbZIP-C, LsbZIP-D, LsbZIP-E, LsbZIP-F, LsbZIP-G, LsbZIP-H, LsbZIP-I, LsbZIP-J, LsbZIP-K and LsbZIP-S) based on their characterized domains (Figure 4). To recapitulate briefly, different clades had diverse phylogenetic clustering. For the relatively small clade like clade J, H and K, bZIPs from two species showed highly homologous with nearly 100 bootstrap value. In some clades like clade B, I and S, the whole clade can be further separated into two or three subfamilies because of the dissimilar divergences. On the other hand, LsbZIPs didn’t present one to one mapping relationship with AtbZIPs, indicating the selective evolution from gene duplication.
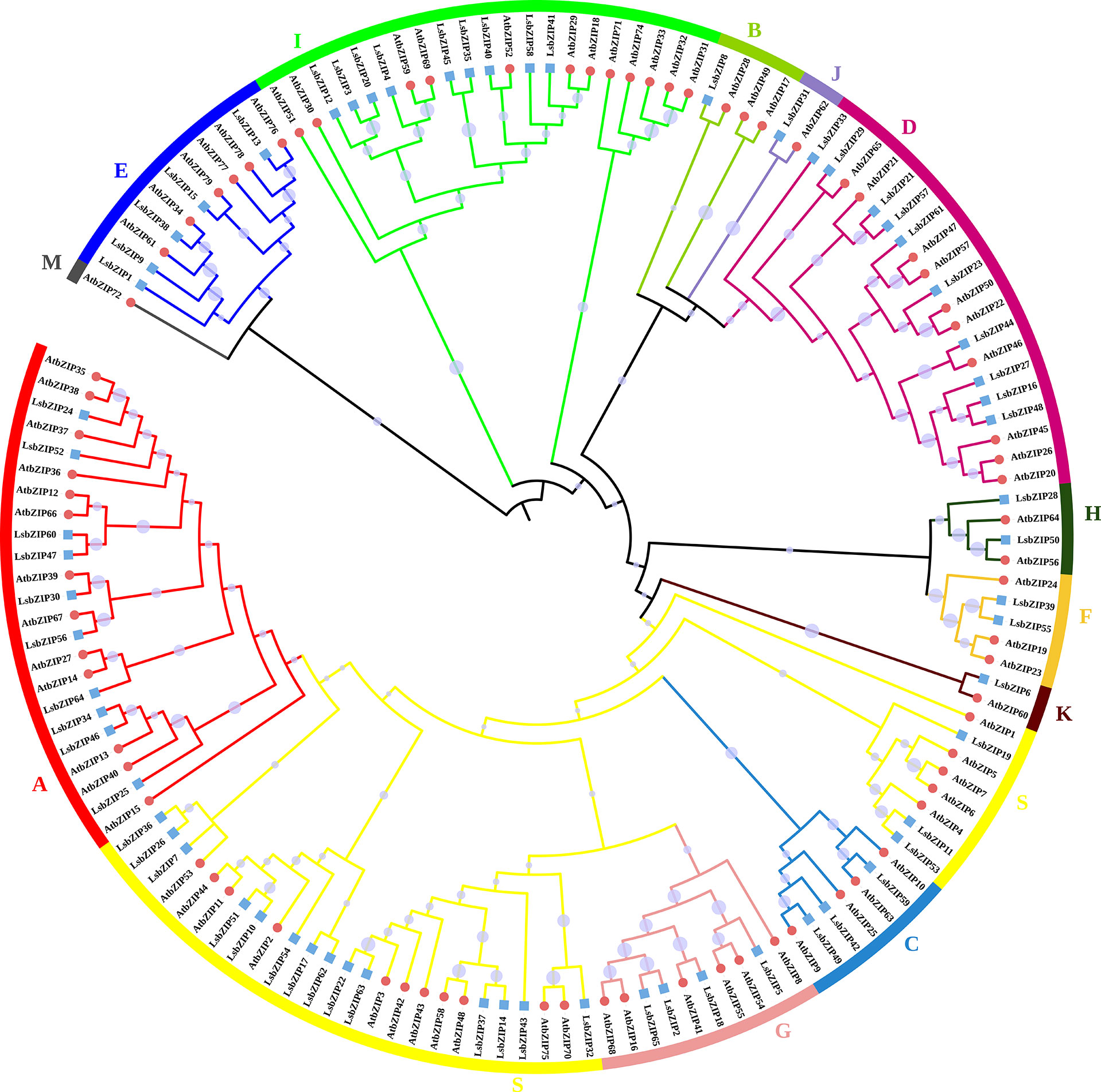
Figure 4 Phylogenetic tree of bZIP genes from Arabidopsis and bottle gourd. The tree is constructed by Neighbor-Joining method with bootstrap values of 1000 replicates. Different colored branches and stripes indicate different subgroups. The red circles and blue squares represent bZIP genes from Arabidopsis and bottle gourd, respectively.
To better depict the specific domain, the phylogeny, exon/intron structure and conserved motifs distribution were integrated in Figure 5. The phylogenetic classification was consistent with previous results. According to MEME analysis, 10 highly conserved motifs were identified in LsbZIPs. And all 65 bZIPs roughly split into 7 types in consideration of the number and order of motifs. Four clades (B, J, H and K) shared the same distribution pattern with motif 1 and motif 3. LsbZIPs containing motif 9 at the C-terminal end were found in type C, S, F while clade A had motif 10 at the N-terminal end. Motifs 5 and 8 can be detected only in clade I, and then 4 motifs (7, 2, 6, 4) can be identified only in clade D. In addition, Clade G owned a replication with motif 3. The results indicated that members classified into the same clade mostly shared the same or similar conserved motifs (Figure 5 and Table S5). The distributed positions of exons/introns on LsbZIP genes were investigated by GSDS, to gain further insight into the structure diversity in bottle gourd. The number of exons ranged from 1 to 22, and clade G and D had relative more exons than other clades. The phylogenetic classification of LsbZIPs was not obviously consistent with exon/intron distribution. However, some genes in the same type shared similar structures, such clade C possessed six exons and five introns.
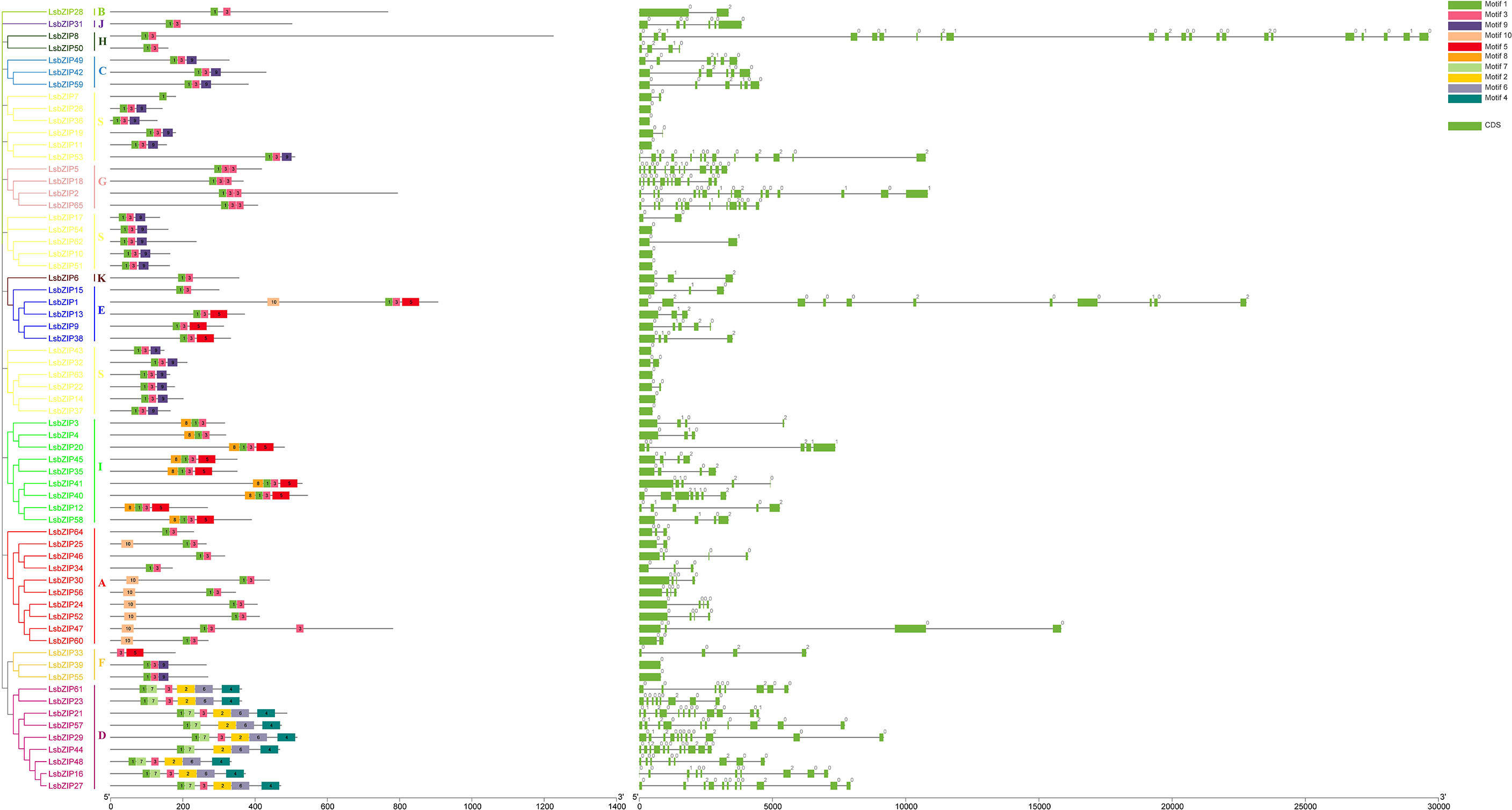
Figure 5 Conserved motifs and gene structures of LsbZIP genes in the bottle gourd. Left: 10 Motifs are indicated by different colored boxes. Right: Green boxes indicate the exons, and grey lines indicate the introns. Different numbers indicate the phase of introns.
Orthologous clusters comparisons in different species
The comparison and annotation of orthologous gene clusters among multiple species were displayed in Figure 6A (https://orthovenn2.bioinfotoolkits.net/task/result/3515a11fe06d38df8aca0e62cc1739d9). The number of orthologous proteins in each cluster varied from 4 to 352. In summary, a total of 60 orthologous proteins in C. sativus were predicted to have similar conserved domain with LsbZIPs. O. sativa had the lowest homology with 30 proteins while the orthologous proteins in the rest species varied from 37 (S. lycopersicum) to 42 (G. max). 8 LsbZIPs were only orthologous in C. sativus, LsbZIP33 and LsbZIP34 was only orthologous in G. max and B. napus, respectively (Figure 6B, Table S6). 20 LsbZIP proteins were able to find their orthologous proteins in each species. 26 clusters including 352 proteins were overlapped in all 8 species and 26 proteins belonged to L. siceraria were corresponding to orthologous proteins, respectively (Figure 6A). 3 singleton proteins (LsbZIP43, LsbZIP20, LsbZIP40) were identified to have no orthologs in any of these species.
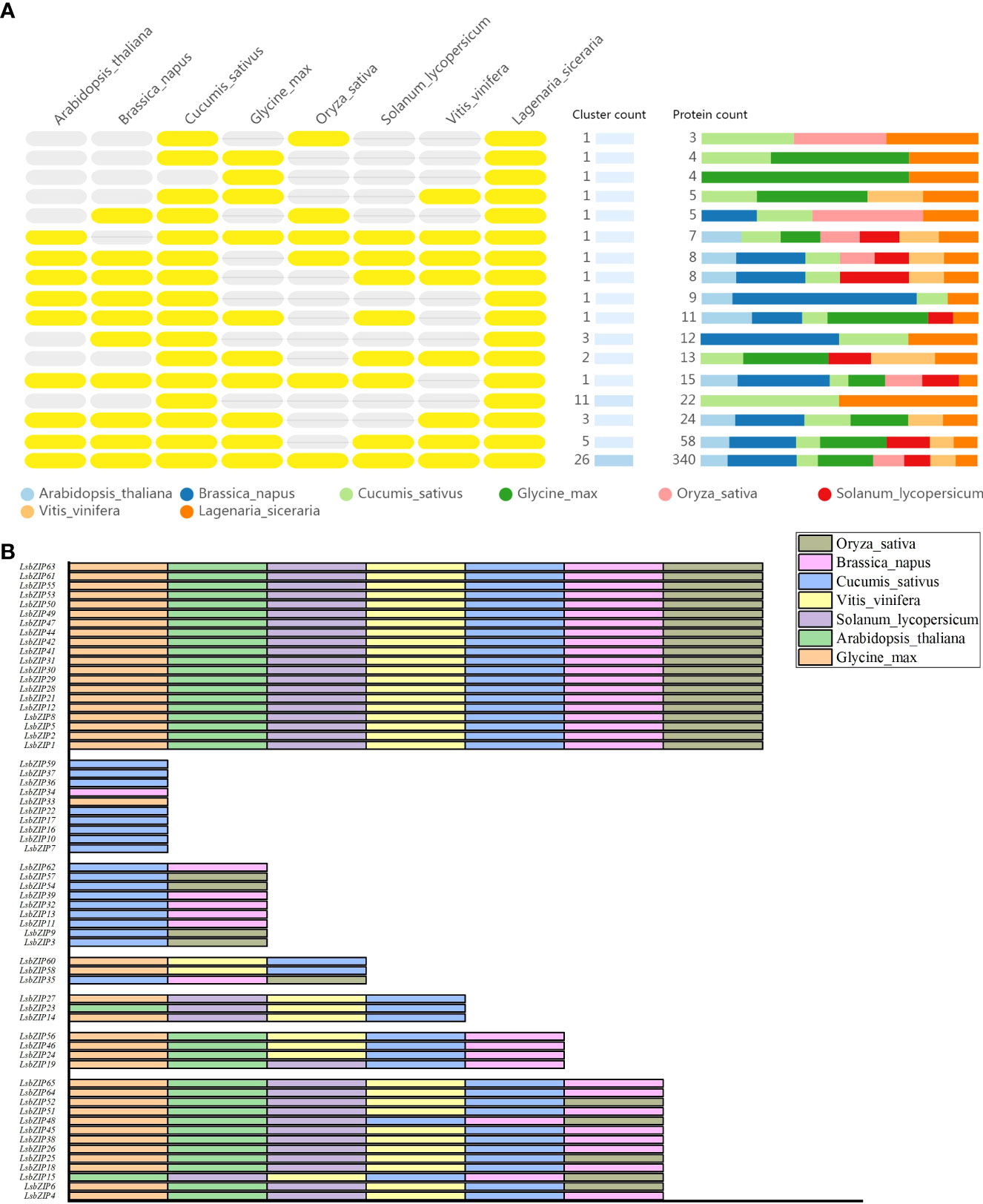
Figure 6 Comparative analysis of orthologous gene clusters among multiple species. (A) Yellow ovals on the left side represent the presence of orthologous genes while grey ovals represent the absence of orthologous genes. Different colors bars on the right side indicate the number and ratio of orthologous proteins in 8 plant species. (B) Layout of the different orthologous gene pairs. Each species was marked with different color.
Cis-regulatory elements analysis of LsbZIP genes
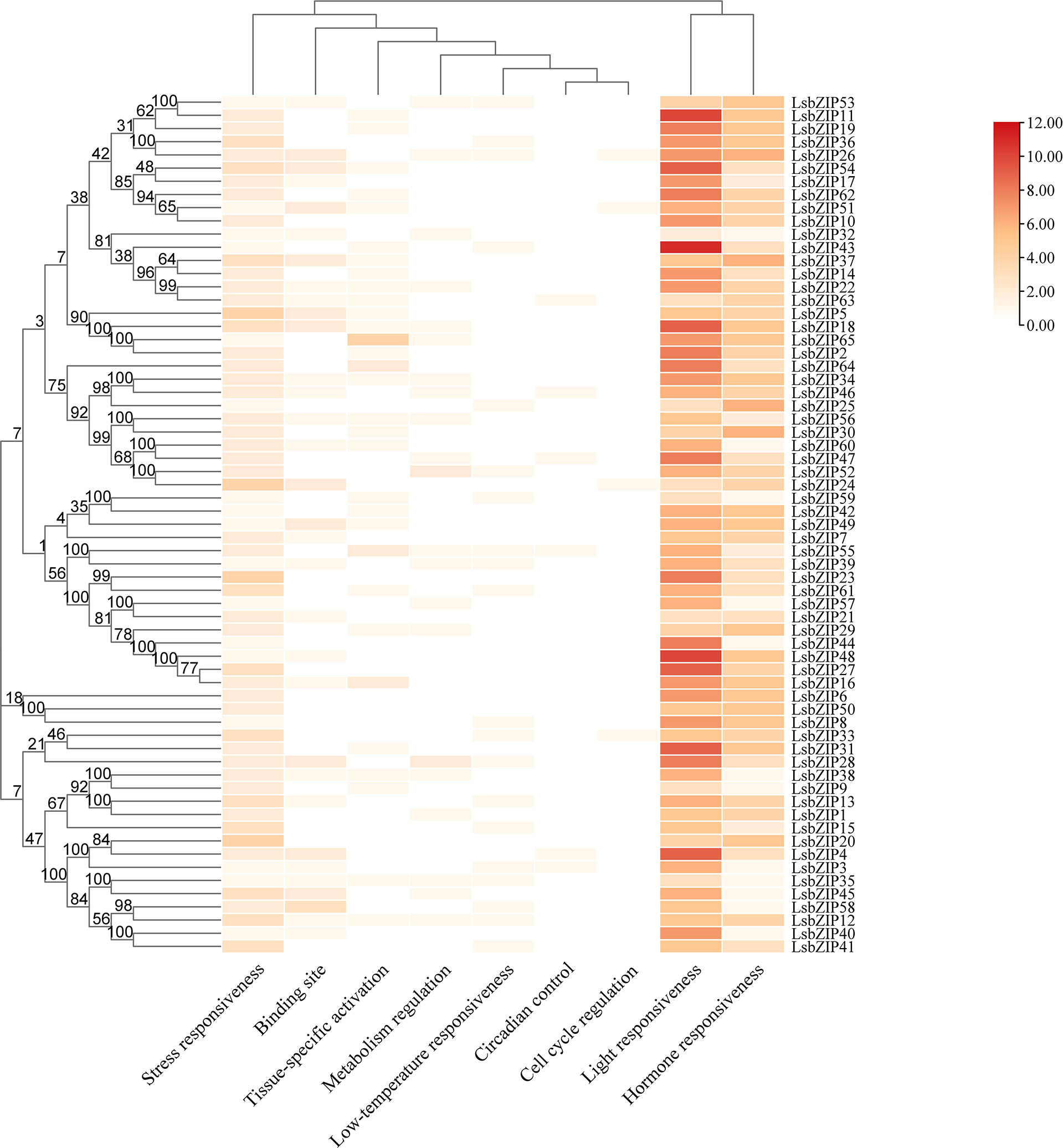
To further understand the transcriptional regulation of bZIP genes in bottle gourd, 2000 bp sequences upstream of the translation initiation site (ATG) were extracted to predict cis-elements by PlantCARE database. In a summary, we identified 64 cis-acting elements which can be grouped into 9 different responsive processes based on the diverse functions (Table S7, Figure 7). Besides the core promoter elements (CAAT-box and TATA-box) found in all 65 bZIP genes, the column clustering had a certain degree of preferences. CREs involved in light responsiveness, hormone responsiveness and stress responsive were ubiquitous in each LsbZIP, accounting for the majority of putative promoters. Three genes (LsbZIP11, LsbZIP43, LsbZIP48) had more than 10 different light-responsive CREs. The numbers of concerned CREs in stress responsiveness contained low-temperature responsive elements (LTR) in 20 LsbZIPs, anaerobic-responsive element (ARE) in 57 LsbZIPs, wound-responsive element (WUN-motif) in 7 LsbZIPs. Twelve hormone responsive elements were found, in which included abscisic acid responsiveness element (ABRE) in 41 LsbZIPs, auxin-responsive element (AuxRR-core, TGA-element) in 6 LsbZIPs, gibberellin-responsive element (GARE-motif, P-box, and TATC-box) in 31 LsbZIPs, salicylic acid responsiveness (SARE, TCA-element) and MeJA-responsiveness (CGTCA-motif and TGACG-motif) in 35 and 41 LsbZIPs, respectively (Table S7). Moreover, circadian control was found in 6 LsbZIPs as well as zein metabolism regulation (O2-site) and flavonoid biosynthetic gene regulation (MBSI) were found in 15 and 4 LsbZIPs, respectively. The results demonstrated that bZIP TFs acted as an important role not only in the regulation of multiple biological processes, but also in response to many abiotic stresses.
Expression patterns of LsbZIPs in various tissues and cultivars
To dissect the tissue-specific expression of LsbZIP genes, the expression of 65 LsbZIPs were determined in five tissues (root, leaf, flower, fruit and stem) from RNA-Seq data SRP107894 (Wu et al., 2017). As shown in Figure 8A, four genes (LsbZIP29, LsbZIP56, LsbZIP7, LsbZIP33) were found not totally expressed in all tissues under normal conditions, indicating that they may be pseudogenes or act in response to abiotic/biotic stresses. And 17 genes were expressed at a low level (log2(TPM+1)<3), especially in roots and leaves. Meanwhile, 6 genes (LsbZIP10, LsbZIP51, LsbZIP26, LsbZIP39, LsbZIP50, LsbZIP54) were highly expressed in leaves, flowers, fruits and stems except in roots. These genes may be involved in the development process of various organs. In roots, 44 of 65 identified LsbZIP genes were regarded as silent while 11 LsbZIP genes exhibited high transcript expression level (Table S8). Three genes (LsbZIP17, LsbZIP43, LsbZIP63) were root-specific expressed while four genes (LsbZIP36, LsbZIP45, LsbZIP14 and LsbZIP37) were relatively high induced in flowers and stems. To sum it up, tissue-specific genes with high expressions might be involved in corresponding tissue defense system. And the illustrated heatmap showed that LsbZIPs were most active in flower and not relatively active in leaves. To further discuss the expression patterns in different varieties, our previous RNA-Seq data with five bottle gourd accession were used in this study (Xu et al., 2021). The results showed that 5 genes (LsbZIP21, LsbZIP56, LsbZIP29, LsbZIP33, LsbZIP43) were rarely expressed in all cultivars, similar to the tendency in Figure 8B. The genes with high expression level in different cultivars were exactly same in different tissues, demonstrating these genes played important roles in manipulating normal growth and development in plants. However, three genes (LsbZIP36, LsbZIP50, LsbZIP63) were highly expressed in variety HZ and YD-4 whilst low expression in G22, J104 and J129. And LsbZIP37 had relative high transcript abundance in J104 and J129 than in other 3 cultivars (Table S9). It revealed that a few LsbZIP genes had cultivar-specific expression, to a certain extent.
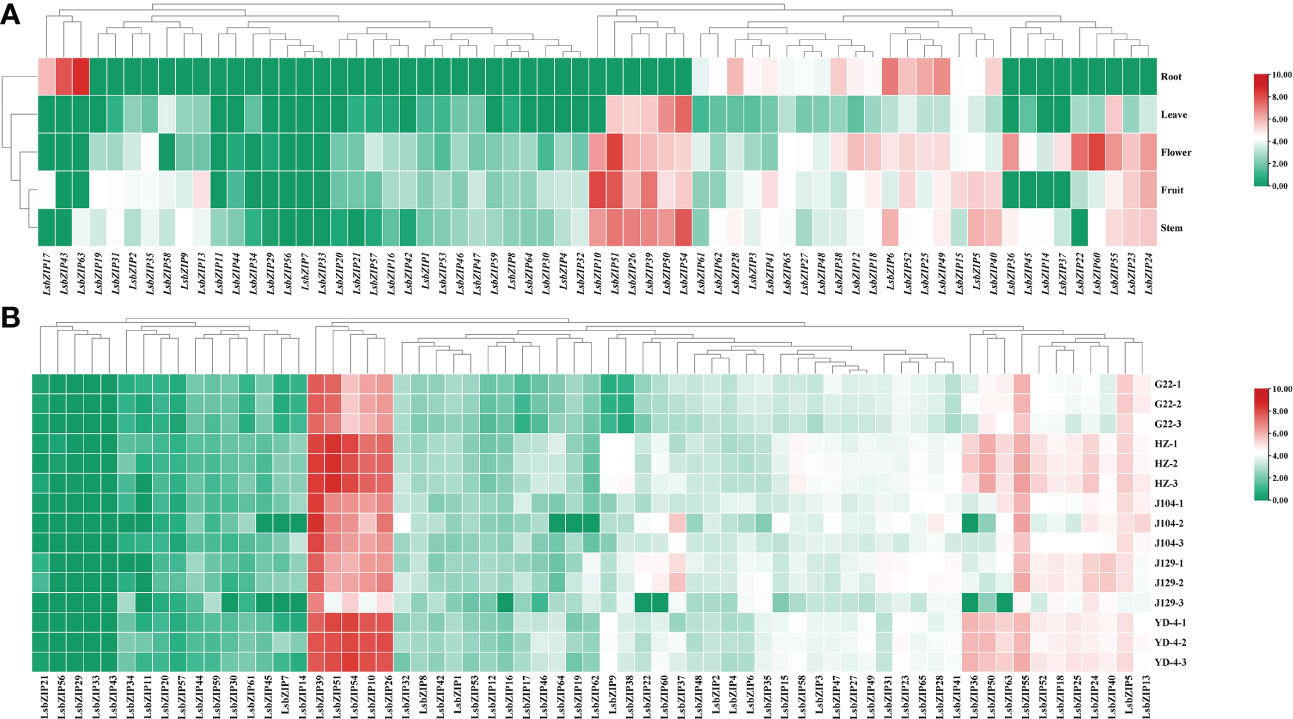
Figure 8 Heatmaps of LsbZIP genes displaying tissue-specific and cultivar-specific expression. (A) Hierarchical clustering of expression profile of bZIP genes in different tissues (root, leaf, flower, fruit, stem) of bottle gourd under normal condition. (B) The expression profile of bZIP genes in 5 bottle gourd varieties (G22, HZ, J104, J129, YD-4) under normal condition. Data were transformed with a log2(TPM+1) transformation.
Expression of LsbZIP genes under cold stress
To investigate early changes in gene expression, we retrieved the transcriptional data under the accession number PRJNA553072 in the bottle gourd cultivar “YZ”, which is a rootstock type showing high tolerance to low temperature (Wang et al., 2020). The differentially expressed genes (DEGs) in accordance with false discovery rate <0.5 and absolute fold change ≥2 were selected and shown in Figure 9A. In general, 19 LsbZIPs were detected in YZ under low temperature treatment for 2 days. 7 genes were significantly down-regulated and 10 genes were up-regulated remarkably (Figure 9A and Table S10). Intriguingly, two genes (LsbZIP7 and LsbZIP62) were not expressed at room temperature similar to the results found in Figure 8A. And LsbZIP7 was highly induced after low temperature treatment while LsbZIP62 was slightly induced, as well. Three genes LsbZIP55, LsbZIP37, and LsbZIP24 which kept relative high gene expression exhibited opposite expression patterns. The expression of LsbZIP55 cut in half, but the expression level of LsbZIP37 and LsbZIP24 increased 2-3 folds compared to respective control (Table S10). In addition, another three genes namely LsbZIP28, LsbZIP61, LsbZIP6 increased their expression up to 5, 7, 30 times respectively in response to cold stress. Five genes (LsbZIP56, LsbZIP40, LsbZIP16, LsbZIP31 and LsbZIP22) were dramatically down-regulated by low temperature. To gain insight into the changing trends in response to cold stress, 10 selected LsbZIP genes were determined by qRT-PCR to validate the RNA-Seq data (Figure 9B). The leaves were harvested at 5 different time-points (0h, 1h, 6h, 12h, 24h) after cold treatment at 4°C. The plants without 0 h cold treatment were considered as control. The relative expression level of selected LsbZIP genes was evaluated by the 2–ΔΔCt method. Statistical significance was detected by LSD test using SPSS v16.0 (SPSS, Inc., Chicago, IL, USA). 8 genes were activated fast within one hour cold stress, indicating the responsive rate varied in different gene expression. LsbZIP6 was quickly induced to be remarkably expressed at 1 hour after treatment (HAT) and continuously rise up to 20 folds than 0 HAT. The parallel increasing pattern was observed in LsbZIP61 and LsbZIP28, but the expression of these two genes were initiated to ascend 12 HAT. Three genes (LsbZIP16, LsbZIP22 and LsbZIP31) presented earlier increase and later decrease trend. On the other hand, three genes including LsbZIP40, LsbZIP56, LsbZIP65 were down-regulated within 1 HAT and subsequently inhibited all the time till 24 HAT. In addition, only one gene LsbZIP44 showed a distinct expression pattern. It was highly expressed within 1 HAT, then suppressed after 6 h cold treatment. Interestingly, LsbZIP44 was again induced, followed by gradual increase at 12 and 24 h.
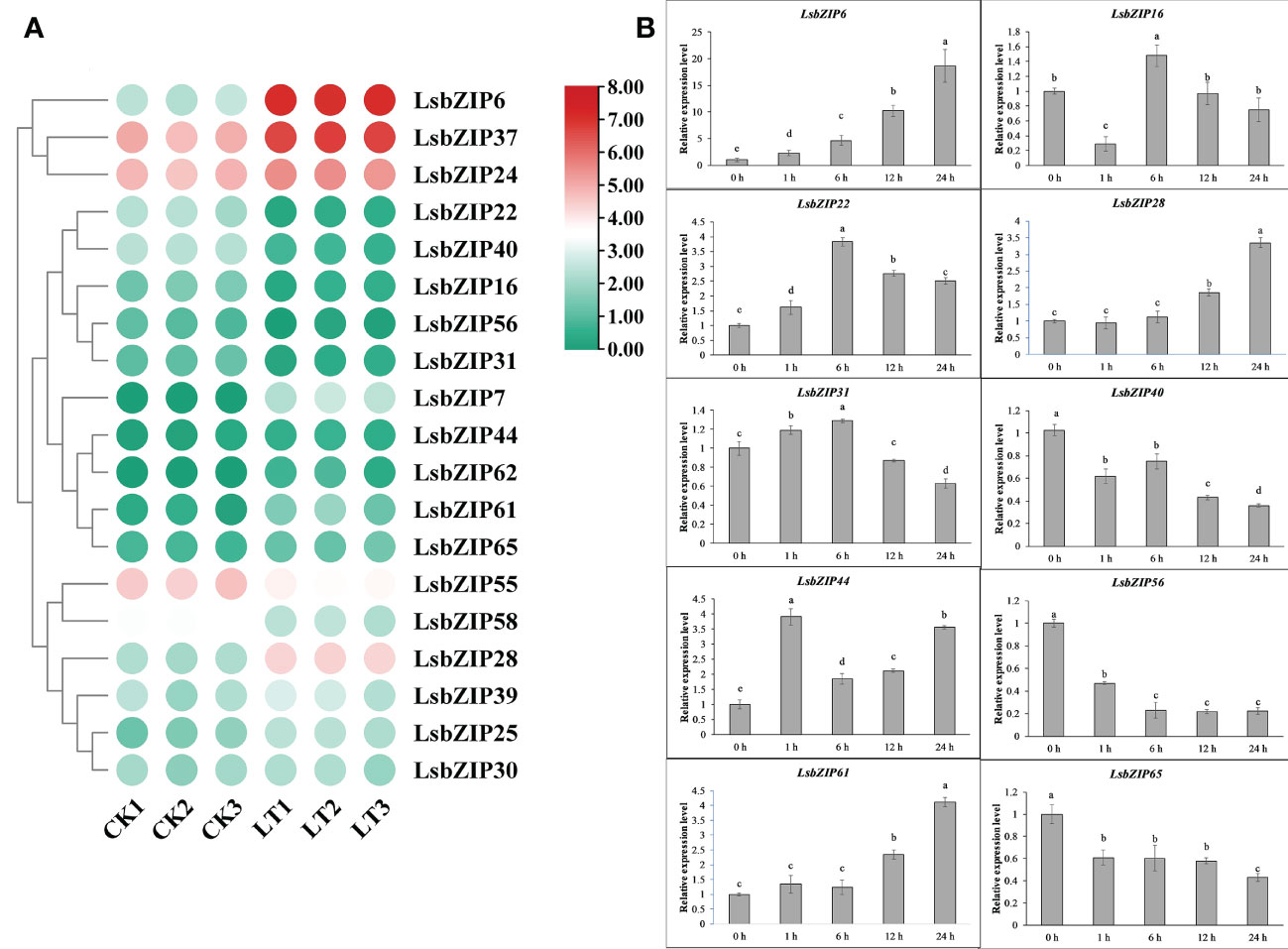
Figure 9 Expression profile of LsbZIP genes in leaves under cold stress. (A) The expression levels of DEGs in leaves under cold stress. (B) Relative expression of selected 10 LsbZIP genes in response to cold stress at 5 time points. Mean ± SE of three replicates represent the significant difference with LSD test. The data followed by the different letter in the graph are significant at P≥0.05.
Discussion
Increasing evidences have suggested that bZIP TFs play vital roles in many biological and physiological processes and regulating signal transductions in response to various abiotic and biotic stresses, such as seed germination (Toh et al., 2012), flower development (Strathmann et al., 2001), cold tolerance (Ritonga and Chen, 2020), salt stress (Wang et al., 2010) and pathogen defense (Thurow et al., 2005), etc. So far, a number of plant species have systematically analyzed bZIP family genes in the genome-wide scale, including some crucial crops like A. thaliana (78) (Dröge-Laser et al., 2018), O. sativa (89) (Nijhawan et al., 2008), Z. mays (125) (Wei et al., 2012) and a few cucurbit vegetables cucumber (64) (Baloglu et al., 2014) and watermelon (62) (Yang et al., 2019). However, the genome-wide characterization of bZIP family genes in Lagenaria siceraria has not been reported. In this study, we compared the former published bottle gourd genome to our newly assembled genome and finally identified 65 unique LsbZIP sequences.
Cucurbitaceous plants, also known as the gourd family, are one of the most important edible plant families and have great economic value. Currently, at least 16 genomes of Cucurbitaceae species including diverse varieties in each species have been released (Wang et al., 2020). To dig deeply into the evolution and divergence in Cucurbit, the available known cucurbit genomes were collected and exhaustively searched for the putative bZIP genes. A total number of 1235 non-redundant protein sequences from 16 Cucurbitaceae species were subdivided into 13 clades with well-supported bootstrap values (Figure 3, Table S4). In clade B, three AtbZIPs (AtbZIP17, AtbZIP49, AtbZIP28) formed a small unique cluster independent from other Cucurbitaceae bZIPs. This scenario also emerged in clade A, F, I, suggesting that these group-specific sequence characteristics of the bZIP members may form before the divergence of Arabidopsis and Cucurbitaceae since conserved domains were homologous in the same clade containing both Arabidopsis and Cucurbitaceae bZIP genes. However, it seemed that intraspecific duplication and parallel evolution of the bZIP family in each Cucurbitaceae has occurred afterward and led to the member variation in each clade. The majority of bZIPs in other clades originated from the whole genome duplication to evolve both generically and specifically in Cucurbitaceae. The results were in consistent with the conclusion in legumes (Wang et al., 2020).
In bottle gourd, 65 LsbZIP genes were undoubtably grouped into 12 subfamilies except clade M (Table S2, Figure 4). The general idea is that bZIP proteins in plant had extra functional conserved domains, in addition to specific bZIP1 (PF00170) and bZIP2 (PF07716) domains, participating in various biological processes (Jin et al., 2014). The domain basic leucine-zipper C terminal (bZIP_C) was identified only in clade C. Similar phenomena happened in clade G with G-box binding protein MFMR, and in clade D with seed dormancy control protein DOG1 as well (Table S2). The structure of motifs and exon-intron composition followed the classification pattern of phylogeny (Figure 5). Motif 2 and 6 were characteristic in clade D, composing DOG1 domain found in Arabidopsis (Dröge-Laser et al., 2018). The number of introns in LsbZIPs varied from 0 to 14, besides LsbZIP8 owning incredible 21 introns (Figure 5). It was basically coincident with the results in V. vinifera, Brachypodium distachyon, S. lycopersicum, cassava and potato (Li et al., 2015a; Liu and Chu, 2015; Hu et al., 2016b; Mirzaei et al., 2020; Wang et al., 2020). Furthermore, most bZIP genes in clade F and S were prevalently intronless (Figure 5). It has been demonstrated that lack of introns contribute to accelerating the post-transcription in response to abiotic stresses (Zhou et al., 2018). And similar findings were observed in soybean and watermelon, revealing evolutionary imprint and functional divergence (Li et al., 2015b; Liu et al., 2017; Zhang et al., 2018; Yang et al., 2019). Compared to the intronless phenomenon, majority of clade D and G obviously possessed more introns (Figure 5). Previous researches suggested that the rate of evolution in genes without introns would increase after gene duplication (Lecharny et al., 2003; Lurin, 2004; Jain et al., 2006). It is deduced that bZIPs in clade D and G might derive earliest than other clades (Hu et al., 2016b). The orthologous genes in clade D and G shared overlaps in seven or eight species while genes in clade F shared overlaps in one or two species (Figure 6A, Table S6). It verified the speculation on the other side. Gene duplication is the main reason of genome evolution and expansion in multigene family (Si et al., 2019). In this study, 19 segmental duplication events in bottle gourd genome led to the expansion of bZIP family genes as in rice (Nijhawan et al., 2008), sorghum (Wang et al., 2011), maize (Wei et al., 2012), tomato (Li et al., 2015a) and peanut (Wang et al., 2019).
The genome-wide expression pattern of LsbZIPs exhibited tissue-specific trait in various organs in Figure 8A. For example, three genes including LsbZIP17, LsbZIP43 and LsbZIP63 were highly expressed and preferentially enriched in roots, indicating that they might play roles in root development. Similar tissue-specific pattern were obtained in many species such as apple (Li et al., 2016), grape (Liu et al., 2014), banana (Hu et al., 2016a), watermelon (Yang et al., 2019) and Prunus mume (Li et al., 2022). LsbZIP50 and LsbZIP24, which were found to be highly expressed in all tissues except root, were orthologous of HY5 (At5g11260) and abscisic acid responsive elements-binding factor (ABF3, At4g34000), respectively (Figure 7; Table S6). The function of HY5 in numerous developmental processes has been well-established, like photomorphogenesis generation, chloroplast development and pigment accumulation (Gangappa and Botto, 2016; Yang et al., 2018). Likewise, LsbZIPs of clade A (52, 30), clade D (48, 61) and clade G (18, 5, 65) with high expression in all tissues were homologous to ABFs, TGA factors (TGAs) and G-box-binding factors (GBFs) in Arabidopsis. The ABFs have been found to act as the core of ABA signaling which counteract abiotic stresses (Banerjee and Roychoudhury, 2017). TGAs redundantly are regarded as vital transcriptional regulators by triggering systemic acquired resistance to interact with nonexpressor of pathogenesis-related genes 1 (NPR1), which is key gene in plant pathogen response (Fu and Dong, 2013). In addition, LsbZIP37 showed cultivar-specific expression pattern in J104 and J129 which are relative resistant to cold stress, implying that LsbZIP37 might participate in the regulation of cold stress response (Figure 8B).
Although bottle gourd is routinely used as a rootstock of other Cucurbitaceae crops due to its climate resiliency and high resistance to biotic stresses (Wang et al., 2020), the mechanism of which bottle gourd respond to cold stress has not been clearly understood. And plants have been found to evolve different strategies to adapt environmental stresses like cold (Xi et al; Wu et al., 2016; Xu et al., 2022). In the previous reports, a number of bZIPs including OsbZIP73, OsbZIP52/RISBZ5, MdHY5 acts as positive or negative roles in response to cold stress (Liu et al., 2012; An et al., 2017; Liu et al., 2018). Based on the previous transcriptome profile, 19 LsbZIP genes were differentially expressed (12 up-regulated, 7 down-regulated) under cold stress (Figure 9A). Subsequently, 10 selected LsbZIPs were validated through RT-PCR at different time points after cold treatment (Figure 9B). LsbZIP6 (clade K), LsbZIP28 (clade B) and LsbZIP61 (clade D) were quickly activated in response to cold stress. Their homologous genes in Arabidopsis contain three members of group B (AtbZIP17, AtbZIP28, AtbZIP49) and one member of group K (AtbZIP60), and have important effects on evolutionary conserved endoplasmic reticulum (ER) stress response (Howell, 2013). Transgenic plants overexpressed AtbZIP17 showed salt tolerance and truncated AtbZIP60 actively moved to the nucleus to regulate ER stress-genes (Iwata and Koizumi, 2005; Iwata et al., 2008; Liu et al., 2010). Therefore, B-bZIP LsbZIP6 and K-bZIP LsbZIP28 might converge on ER-stress pathways targeting specific gene sets like AtbZIP60 and AtbZIP28/bZIP17 were existed in heterodimer (Kim et al., 2018). LsbZIP61 had orthologous relationship with TGA1, which interacts with NPR1 and induces apoplastic defense and ER stress response (Shearer et al., 2012; Wang and Fobert, 2013). G-bZIP LsbZIP65, which was orthologous to GBF1, exhibited a decline expression trend after cold treatment (Figure 9B). GBF1 is a well-known negative regulator of hypocotyl expansion (Gangappa et al., 2013), and can bind to the catalase2 promoter to inhibit the H2O2-scavenging activity, leading to ROS accumulation (Giri et al., 2017). The down-regulated expression of LsbZIP65 might decrease the accumulation of H2O2 through less binding catalase2 promoter in order to endure cold stress in bottle gourd.
Conclusion
In this study, we firstly identified and characterized 65 non-duplicated bZIP genes in L. siceraria and divided into 12 subfamilies at a genome-wide scale. The phylogenetic analysis based on 1235 bZIP sequences from 16 Cucurbitaceae revealed evolutionary convergence and divergence of bZIP gene family. The analysis of LsbZIPs domain, structure and orthologous relationships were consistent with the classification. The tissue-specific expression pattern of LsbZIPs indicated the pivotal roles in various physiological and biological processes. We elaborated the expression profiles of LsbZIPs in response to low-temperature stress, which provide valuable information of bZIPs under cold condition and strategies to breed cultivars conferred cold tolerance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JW analyzed the data. YW and XinW conducted the qPCR analysis. JW drew the figures and edited the manuscript. BW, ZL and LZ contributed regents/materials/analysis tool. JW, GL and XiaW conceived and designed the study, obtained funds, and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was mainly supported by Key Research and Development Programs of Zhejiang Province (Grant No. 2021C02052 and 2021C02042), the Key Science Project of Vegetable Breeding in Zhejiang (14th Five-Year Plan) (2021C02065).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1128007/full#supplementary-material
Supplementary Figure 1 | Phylogenetic analysis of bZIPs between two bottle gourd genomes (‘USVL1VR-Ls’ and ‘HZCG’).
References
An, J. P., Qu, F. J., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., et al. (2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Horticult. Res. 4, 17023. doi: 10.1038/hortres.2017.23
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Baloglu, M. C., Eldem, V., Hajyzadeh, M., Unver, T. (2014). Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS One 9, e96014. doi: 10.1371/journal.pone.0096014
Banerjee, A., Roychoudhury, A. (2017). Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254, 3–16. doi: 10.1007/s00709-015-0920-4
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chimonyo, V. G. P., Modi, A. T. (2013). Seed performance of selected bottle gourd (Lagenaria siceraria (Molina) standl.). Am. J. Exp. Agric. 3, 740. doi: 10.9734/ajea/2013/4114
Dankov, K., Dobrikova, A., Bogos, B., Gombos, Z., Apostolova, E. (2009). The role of anionic lipids in LHCII organization and in photoinhibition of photosynthetic apparatus. Comptes rendus l’Académie bulgare Des. Sci. 62, 941–948.
Dröge-Laser, W., Snoek, B. L., Snel, B., Weiste, C. (2018). The arabidopsis bZIP transcription factor family–an update. Curr. Opin. Plant Biol. 45, 36–49. doi: 10.1016/j.pbi.2018.05.001
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Foster, R., Izawa, T., Chua, N.-H. (1994). Plant bZIP proteins gather at ACGT elements. FASEB J. 8, 192–200. doi: 10.1096/fasebj.8.2.8119490
Fu, Z. Q., Dong, X. (2013). Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gangappa, S. N., Botto, J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant. 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Gangappa, S. N., Srivastava, A. K., Maurya, J. P., Ram, H., Chattopadhyay, S. (2013). Z-box binding transcription factors (ZBFs): A new class of transcription factors in arabidopsis seedling development. Mol. Plant 6, 1758–1768. doi: 10.1093/mp/sst140
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Giri, M. K., Singh, N., Banday, Z. Z., Singh, V., Ram, H., Singh, D., et al. (2017). GBF1 differentially regulates CAT2 and PAD4 transcription to promote pathogen defense in arabidopsis thaliana. Plant J. 91, 802–815. doi: 10.1111/tpj.13608
Howell, S. H. (2013). Endoplasmic reticulum stress responses in plants Annu. Rev. Plant Biol. 64, 477–499. doi: 10.1146/annurev-arplant-050312-120053
Huang, X., Ouyang, X., Yang, P., Lau, O. S., Li, G., Li, J., et al. (2012). Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-b light. Plant Cell 24, 4590–4606. doi: 10.1105/tpc.112.103994
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, W., Wang, L., Tie, W., Yan, Y., Ding, Z., Liu, J., et al. (2016a). Genome-wide analyses of the bZIP family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci. Rep. 6, 30203. doi: 10.1038/srep30203
Hu, W., Yang, H., Yan, Y., Wei, Y., Tie, W., Ding, Z., et al. (2016b). Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 6, 1–12. doi: 10.1038/srep22783
Iwata, Y., F., N. V., Koizumi, N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20, 3107–3121. doi: 10.1105/tpc.108.061002
Iwata, Y., Koizumi, N. (2005). An arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. United States America 102, 5280–5285. doi: 10.1073/pnas.0408941102
Jain, P., Shah, K., Sharma, N., Kaur, R., Singh, J., Vinson, C., et al. (2017). A-ZIP53, a dominant negative reveals the molecular mechanism of heterodimerization between bZIP53, bZIP10 and bZIP25 involved in arabidopsis seed maturation. Sci. Rep. 7, 1–17. doi: 10.1038/s41598-017-14167-5
Jain, M., Tyagi, A. K., Khurana, J. P. (2006). Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88, 360–371. doi: 10.1016/j.ygeno.2006.04.008
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jin, Z., Xu, W., Liu, A. (2014). Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis l.). Planta 239, 299–312. doi: 10.1007/s00425-013-1979-9
Kim, J.-S., Yamaguchi-Shinozaki, K., Shinozaki, K. (2018). ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol. 176, 2221–2230. doi: 10.1104/pp.17.01414
Kong, S., Park, S.-Y., Lee, Y.-H. (2015). Systematic characterization of the bZIP transcription factor gene family in the rice blast fungus, m agnaporthe oryzae. Environ. Microbiol. 17, 1425–1443. doi: 10.1111/1462-2920.12633
Lecharny, A., Boudet, N., Gy, I., Aubourg, S., Kreis, M. (2003). Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. J. Struct. Funct. Genomics 3, 111–116. doi: 10.1023/A:1022614001371
Liang, Y., Xia, J., Jiang, Y., Bao, Y., Chen, H., Wang, D., et al. (2022). Genome-wide identification and analysis of bZIP gene family and resistance of TaABI5 (TabZIP96) under freezing stress in wheat (Triticum aestivum). Int. J. Mol. Sci. 23, 2351. doi: 10.3390/ijms23042351
Liao, Y., Zou, H.-F., Wei, W., Hao, Y.-J., Tian, A.-G., Huang, J., et al. (2008). Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic arabidopsis. Planta 228, 225–240. doi: 10.1007/s00425-008-0731-3
Li, D., Fu, F., Zhang, H., Song, F. (2015a). Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum l.). BMC Genomics 16, 771. doi: 10.1186/s12864-015-1990-6
Li, X., Gao, S., Tang, Y., Li, L., Zhang, F., Feng, B., et al. (2015b). Genome-wide identification and evolutionary analyses of bZIP transcription factors in wheat and its relatives and expression profiles of anther development related TabZIP genes. BMC Genomics. 16, 976. doi: 10.1186/s12864-015-2196-7
Li, Y.-Y., Meng, D., Li, M., Cheng, L. (2016). Genome-wide identification and expression analysis of the bZIP gene family in apple (Malus domestica). Tree Genet. Genomes 12, 1–17. doi: 10.1007/s11295-016-1043-6
Lindemose, S., O’Shea, C., Jensen, M. K., Skriver, K. (2013). Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 14, 5842–5878. doi: 10.3390/ijms14035842
Liu, J., Chen, N., Chen, F., Cai, B., Dal Santo, S., Tornielli, G. B., et al. (2014). Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genomics 15, 281. doi: 10.1186/1471-2164-15-281
Liu, X., Chu, Z. (2015). Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in brachypodium distachyon. BMC Genomics 16, 227. doi: 10.1186/s12864-015-1457-9
Liu, C., Ou, S., Mao, B., Tang, J., Wang, W., Wang, H., et al. (2018). Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat. Commun. 9, 3302. doi: 10.1038/s41467-018-05753-w
Liu, J. X., Srivastava, R., Howell, S. H. (2010). Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in arabidopsis. Plant Cell Environ. 31, 1735–1743. doi: 10.1111/j.1365-3040.2008.01873.x
Liu, C., Wu, Y., Wang, X. (2012). bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta 235, 1157–1169. doi: 10.1007/s00425-011-1564-z
Liu, H., Zhong, Y., Guo, C., Wang, X. L., Xiong, J., Cheng, Q., et al. (2017). Genome-wide analysis and evolution of the bZIP transcription factor gene family in six fragaria species. Plant Systematics Evol. 303, 1225–1237. doi: 10.1007/s00606-017-1433-8
Li, Y., Wang, Y., Wu, X., Wang, J., Wu, X., Wang, B., et al. (2021). Novel genomic regions of fusarium wilt resistance in bottle gourd [Lagenaria siceraria (Mol.) standl.] discovered in genome-wide association study. Front. Plant Sci. 12, 715. doi: 10.3389/fpls.2021.650157
Li, Q., Xiong, L., Gao, J., Zhang, H.-Y. (2019). Molecular mechanisms of the protein-protein interaction–regulated binding specificity of basic-region leucine zipper transcription factors. J. Mol. Modeling 25, 1–11. doi: 10.1007/s00894-019-4138-9
Li, P., Zheng, T., Li, L., Wang, J., Cheng, T., Zhang, Q. (2022). Genome-wide investigation of the bZIP transcription factor gene family in prunus mume: classification, evolution, expression profile and low-temperature stress responses. Hortic. Plant J. 8, 230–242. doi: 10.1016/j.hpj.2021.01.009
Llorca, C. M., Potschin, M., Zentgraf, U. (2014). bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00169
Lurin, C. (2004). Genome-wide analysis of arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. doi: 10.1105/tpc.104.022236
Meng, X.-B., Zhao, W.-S., Lin, R.-M., Wang, M., Peng, Y.-L. (2005). Identification of a novel rice bzip-type transcription factor gene, osbzip1, involved in response to infection ofmagnaporthe grisea. Plant Mol. Biol. Rep. 23, 301–302. doi: 10.1007/BF02772762
Mirzaei, K., Bahramnejad, B., Fatemi, S. (2020). Genome-wide identification and characterization of the bZIP gene family in potato (Solanum tuberosum). Plant Gene 24, 100257. doi: 10.1016/j.plgene.2020.100257
Mukherjee, K., Choudhury, A. R., Gupta, B., Gupta, S., Sengupta, D. N. (2006). An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of indica rice. BMC Plant Biol. 6, 1–14. doi: 10.1186/1471-2229-6-18
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nijhawan, A., Jain, M., Tyagi, A. K., Khurana, J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 146, 333. doi: 10.1104/pp.107.112821
Panchy, N., Lehti-Shiu, M., Shiu, S.-H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. doi: 10.1104/pp.16.00523
Pourabed, E., Ghane Golmohamadi, F., Soleymani Monfared, P., Razavi, S. M., Shobbar, Z.-S. (2015). Basic leucine zipper family in barley: genome-wide characterization of members and expression analysis. Mol. Biotechnol. 57, 12–26. doi: 10.1007/s12033-014-9797-2
Qiao, X., Li, Q., Yin, H., Qi, K., Li, L., Wang, R., et al. (2019). Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 20, 1–23. doi: 10.1186/s13059-019-1650-2
Ritonga, F. N., Chen, S. (2020). Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants 9, 560. doi: 10.3390/plants9050560
Schaefer, H., Heibl, C., Renner, S. S. (2009). Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc. R. Soc. B: Biol. Sci. 276, 843–851. doi: 10.1098/rspb.2008.1447
Schlumbaum, A., Vandorpe, P. (2012). A short history of lagenaria siceraria (bottle gourd) in the Roman provinces: morphotypes and archaeogenetics. Vegetation history archaeobotany 21, 499–509. doi: 10.1007/s00334-011-0343-x
Shearer, H. L., Cheng, Y. T., Wang, L., Liu, J., Boyle, P., Després, C., et al. (2012). Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion. Mol. Plant Microbe Interact. 25, 1459–1468. doi: 10.1094/MPMI-09-11-0256
Shimizu, H., Sato, K., Berberich, T., Miyazaki, A., Ozaki, R., Imai, R., et al. (2005). LIP19, a basic region leucine zipper protein, is a fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol. 46, 1623–1634. doi: 10.1093/pcp/pci178
Si, W., Hang, T., Guo, M., Chen, Z., Ding, T. (2019). Whole-genome and transposed duplication contributes to the expansion and diversification of TLC genes in maize. Int. J. Mol. Sci. 20, 5484. doi: 10.3390/ijms20215484
Sirichandra, C., Davanture, M., Turk, B. E., Zivy, M., Valot, B., Leung, J., et al. (2010). The arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PloS One 5, e13935. doi: 10.1371/journal.pone.0013935
Sornaraj, P., Luang, S., Lopato, S., Hrmova, M. (2016). Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta (BBA)-General Subj. 1860, 46–56. doi: 10.1016/j.bbagen.2015.10.014
Strathmann, A., Kuhlmann, M., Heinekamp, T., Dröge-Laser, W. (2001). BZI-1 specifically heterodimerises with the tobacco bZIP transcription factors BZI-2, BZI-3/TBZF and BZI-4, and is functionally involved in flower development. Plant J. 28, 397–408. doi: 10.1046/j.1365-313X.2001.01164.x
Tang, W., Ru, Y., Hong, L., Zhu, Q., Zuo, R., Guo, X., et al. (2015). System-wide characterization of bZIP transcription factor proteins involved in infection-related morphogenesis of m agnaporthe oryzae. Environ. Microbiol. 17, 1377–1396. doi: 10.1111/1462-2920.12618
Thurow, C., Schiermeyer, A., Krawczyk, S., Butterbrodt, T., Nickolov, K., Gatz, C. (2005). Tobacco bZIP transcription factor TGA2. 2 and related factor TGA2. 1 have distinct roles in plant defense responses and plant development. Plant J. 44, 100–113. doi: 10.1111/j.1365-313X.2005.02513.x
Toh, S., McCourt, P., Tsuchiya, Y. (2012). HY5 is involved in strigolactone-dependent seed germination in arabidopsis. Plant Signaling Behav. 7, 556–558. doi: 10.4161/psb.19839
Wang, L., Fobert, P. R. (2013). Arabidopsis clade I TGA factors regulate apoplastic defences against the bacterial pathogen pseudomonas syringae through endoplasmic reticulum-based processes. PLoS One 8, e77378. doi: 10.1371/journal.pone.0077378
Wang, Y., Gao, C., Liang, Y., Wang, C., Yang, C., Liu, G. (2010). A novel bZIP gene from tamarix hispida mediates physiological responses to salt stress in tobacco plants. J. Plant Physiol. 167, 222–230. doi: 10.1016/j.jplph.2009.09.008
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49. doi: 10.1093/nar/gkr1293
Wang, Y., Wang, L., Xing, N., Wu, X., Wu, X., Wang, B., et al. (2020). A universal pipeline for mobile mRNA detection and insights into heterografting advantages under chilling stress. Horticult. Res. 7, 13. doi: 10.1038/s41438-019-0236-1.
Wang, J., Yang, C., Wu, X., Wang, Y., Wang, B., Wu, X., et al. (2022). Genome-wide characterization of NBS-LRR family genes and expression analysis under powdery mildew stress in lagenaria siceraria. Physiol. Mol. Plant Pathol. 118, 101798. doi: 10.1016/j.pmpp.2022.101798
Wang, Z., Yan, L., Wan, L., Huai, D., Kang, Y., Shi, L., et al. (2019). Genome-wide systematic characterization of bZIP transcription factors and their expression profiles during seed development and in response to salt stress in peanut. BMC Genomics 20, 51. doi: 10.1186/s12864-019-5434-6
Wang, J., Zhou, J., Zhang, B., Vanitha, J., Ramachandran, S., Jiang, S.-Y. (2011). Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum f. J. Integr. Plant Biol. 53, 212–231. doi: 10.1111/j.1744-7909.2010.01017.x
Wei, K., Chen, J., Wang, Y., Chen, Y., Chen, S., Lin, Y., et al. (2012). Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 19, 463–476. doi: 10.1093/dnares/dss026
Weiste, C., Pedrotti, L., Selvanayagam, J., Muralidhara, P., Fröschel, C., Novák, O., et al. (2017). The arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PloS Genet. 13, e1006607. doi: 10.1371/journal.pgen.1006607
Wu, S., Shamimuzzaman, M., Sun, H., Salse, J., Sui, X., Wilder, A., et al. (2017). The bottle gourd genome provides insights into cucurbitaceae evolution and facilitates mapping of a papaya ring-spot virus resistance locus. Plant J. 92, 963–975. doi: 10.1111/tpj.13722
Wu, L., Wu, J., Liu, Y., Gong, X., Xu, J., Lin, D., et al. (2016). The rice pentatricopeptide repeat gene TCD10 is needed for chloroplast development under cold stress. Rice 9, 67. doi: 10.1186/s12284-016-0134-1
Xi, L., Jie, L., Yunshuai, H., Penghui, C., Chunlei, Z., Yaken, R., et al. (2018). WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J. Exp. Bot. 69, 3949–3961. doi: 10.1093/jxb/ery214
Xu, L., Cao, M., Wang, Q., Xu, J., Liu, C., Ullah, N., et al. (2022). Insights into the plateau adaptation of salvia castanea by comparative genomic and WGCNA analyses. J. Advanced Res. 42, 221–235. doi: 10.1016/j.jare.2022.02.004
Xu, P., Wang, Y., Sun, F., Wu, R., Du, H., Wang, Y., et al. (2021). Long-read genome assembly and genetic architecture of fruit shape in the bottle gourd. Plant J. 107, 956–968. doi: 10.1111/tpj.15358
Yang, Y., Liang, T., Zhang, L., Shao, K., Gu, X., Shang, R., et al. (2018). UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in arabidopsis. Nat. Plants 4, 98–107. doi: 10.1038/s41477-017-0099-0
Yang, Y., Li, J., Li, H., Yang, Y., Guang, Y., Zhou, Y. (2019). The bZIP gene family in watermelon: genome-wide identification and expression analysis under cold stress and root-knot nematode infection. PeerJ 7, e7878. doi: 10.7717/peerj.7878
Yang, Y.-G., Lv, W.-T., Li, M.-J., Wang, B., Sun, D.-M., Deng, X. (2013). Maize membrane-bound transcription factor Zmbzip17 is a key regulator in the cross-talk of ER quality control and ABA signaling. Plant Cell Physiol. 54, 2020–2033. doi: 10.1093/pcp/pct142
Yin, W., Cui, P., Wei, W., Lin, Y., Luo, C. (2017). Genome-wide identification and analysis of the basic leucine zipper (bZIP) transcription factor gene family in ustilaginoidea virens. Genome 60, 1051–1059. doi: 10.1139/gen-2017-0089
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2015). Four a rabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi: 10.1111/pce.12351
Zander, M., La Camera, S., Lamotte, O., Métraux, J.-P., Gatz, C. (2010). Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 61, 200–210. doi: 10.1111/j.1365-313X.2009.04044.x
Zhang, M., Liu, Y. H., Shi, Y. H., Guo, M. L., Chai, M. N., He, Q., et al. (2018). Evolutionary and expression analyses of soybean basic leucine zipper transcription factor family. BMC Genomics. 19, 159. doi: 10.1186/s12864-018-4511-6
Zhang, L., Yu, H., Lin, S., Gao, Y. (2016). Molecular characterization of FT and FD homologs from eriobotrya deflexa nakai forma koshunensis. Front. Plant Sci. 7, 8. doi: 10.3389/fpls.2016.00008
Zhang, Y., Zhang, G., Xia, N., Wang, X.-J., Huang, L.-L., Kang, Z.-S. (2008). Cloning and characterization of a bZIP transcription factor gene in wheat and its expression in response to stripe rust pathogen infection and abiotic stresses. Physiol. Mol. Plant Pathol. 73, 88–94. doi: 10.1016/j.pmpp.2009.02.002
Zhao, K., Chen, S., Yao, W., Cheng, Z., Zhou, B., Jiang, T. (2021). Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 21, 1–16. doi: 10.1186/s12870-021-02879-w
Zhou, Y., Xu, D., Jia, L., Huang, X., Ma, G., Wang, S., et al. (2017). Genome-wide identification and structural analysis of bZIP transcription factor genes in brassica napus. Genes 8, 288. doi: 10.3390/genes8100288
Keywords: Lagenaria siceraria, bZIP transcription factor, Cucurbitaceae evolution, expression pattern, cold stress
Citation: Wang J, Wang Y, Wu X, Wang B, Lu Z, Zhong L, Li G and Wu X (2023) Insight into the bZIP gene family in Lagenaria siceraria: Genome and transcriptome analysis to understand gene diversification in Cucurbitaceae and the roles of LsbZIP gene expression and function under cold stress. Front. Plant Sci. 13:1128007. doi: 10.3389/fpls.2022.1128007
Received: 20 December 2022; Accepted: 30 December 2022;
Published: 17 February 2023.
Edited by:
Meng Jiang, Zhejiang University, ChinaReviewed by:
Chong Yang, Guangdong Academy of Science (CAS), ChinaLing Xu, Zhejiang Sci-Tech University, China
Muhammad Awais Ghani, University of Agriculture, Faisalabad, Pakistan
Copyright © 2023 Wang, Wang, Wu, Wang, Lu, Zhong, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohua Wu, d3V4aWFvaHVhMjAwMUAxMjYuY29t
 Jian Wang
Jian Wang Ying Wang1
Ying Wang1 Xinyi Wu
Xinyi Wu Guojing Li
Guojing Li