
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 January 2023
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1118398
This article is part of the Research TopicThe Role of ROS and Phytohormones in Crops Under Environmental StressView all 12 articles
 Bin Deng1,2†
Bin Deng1,2† Xiaoyan Gu1,2†
Xiaoyan Gu1,2† Sen Chen1,2
Sen Chen1,2 Meng Zhang1,2
Meng Zhang1,2 Suwei Hao1,2
Suwei Hao1,2 Lixian Wei1,2
Lixian Wei1,2 Ying Cao1,2*
Ying Cao1,2* Shanglian Hu1,2*
Shanglian Hu1,2*Sucrose is the main transported form of photosynthetic products. Sucrose transporter (SUT) participates in the translocation of sucrose from source to sink, which is important for the growth and development of plants. Dendrocalamus farinosus is an important economic crop in southwestern China because of its high growth rate, high fiber content, and dual usage for food and timber, but the mechanism of sucrose transportation in D. farinosus is unclear. In this study, a total of 12 SUT transporter genes were determined in D. farinosus by whole-genome identification. DfSUT2, DfSUT7, and DfSUT11 were homologs of rice OsSUT2, while DfSUT4 was a homolog of OsSUT4, and these four DfSUT genes were expressed in the leaf, internode, node, and bamboo shoots of D. farinosus. In addition, DfSUT family genes were involved in photosynthetic product distribution, ABA/MeJA responses, and drought resistance, especially DfSUT4. The function of DfSUT4 was then verified in Nicotiana tabacum. DfSUT4 was localized mainly in the leaf mesophyll and stem phloem of pDfSUT4::GUS transgenic plant. The overexpression of DfSUT4 gene in transgenic plant showed increases of photosynthetic rate, above-ground biomass, thousand grain weight, and cellulose content. Our findings altogether indicate that DfSUT4 can be a candidate gene that can be involved in phloem sucrose transportation from the source leaves to the sink organs, phytohormone responses, abiotic stress, and fiber formation in plants, which is very important in the genetic improvement of D. farinosus and other crops.
Sucrose is the major long-distance-transported form of carbohydrate metabolites in plants (Deol et al., 2013; Wang et al., 2021). After having been synthesized by photosynthesis in mature leaves (source), sucrose is further transported to a variety of heterotrophic organs (sink) such as roots, developing leaves, flowers, and seeds (Barker et al., 2000; Sun et al., 2010). Sucrose transporters (SUTs; also called SUCs) are sucrose–H+ symporters that are localized in the vacuole membrane or plasma membrane (Zhu et al., 2015), and they are members of the major facilitator superfamily that contains 12 transmembrane-spanning helices (Aoki et al., 2012). In addition, the SUT family genes play key roles in sucrose loading and unloading from t source tissue to sink cells through the phloem to effect multiple processes of plant growth and development, such as phytohormone biosynthesis, abiotic stresses, photosynthesis, flower development, and fiber synthesis in many species (Srivastava et al., 2009; Slewinski and Braun, 2010; Payyavula et al., 2011; Wang et al., 2021). According to the direction and type of transmembrane transport of sucrose, SUT transporters in plants can be divided into three types: plasma membrane efflux carriers are active in the efflux of sucrose from the mesophyll cells to the apoplast, plasma membrane influx carriers function in the entry of sucrose from the apoplast into phloem cells, and tonoplast carriers are involved in the transport of sucrose between the vacuole and the cytoplasm (Ward et al., 1998; Lemoine, 2000).
To date, there are numerous studies that have shown SUTs as crucial not only for carbohydrate partitioning but also for responses to various phytohormones, abiotic stresses, and environmental factors in plants—for example, Arabidopsis suc2 seedlings were smaller than the wild type in the absence of sucrose, and ABA treatment on Arabidopsis suc2 mutant could increase the sucrose content in shoots but decrease the sucrose content in roots (Gottwald et al., 2000), suggesting that AtSUC2 is crucial for plant development and sucrose loading in Arabidopsis root (Gong et al., 2015). In contrast to other AtSUC transporters, AtSUC4 is localized in the tonoplast and participate in the translocation of sucrose from the vacuole into the cytoplasm (Weise et al., 2000). Consistently with SUC4 from Arabidopsis, PtaSUT4 from poplar and OsSUT2 from rice are also localized to the tonoplast and mediate sucrose efflux from the source leaves (Eom et al., 2011). Moreover, PtaSUT4-repressed poplar showed increased leaf-to-stem biomass ratio and sucrose content in the source leaves because of the translocation of sucrose to the sink organs which was inhibited in transgenic plants (Payyavula et al., 2011). The plasma membrane-localized OsSUT4 functions in the apoplastic phloem loading of sucrose. The rice sut4 mutant showed a dwarf phenotype, and the yield of rice was decreased (Chung et al., 2014).
In addition, AtSUC2, AtSUC4, and AtSUC9 are all associated with ABA treatment and drought/cold stress (Gong et al., 2015; Jia et al., 2015). OsSUT2 promotes photosynthesis and sucrose distribution in plants under drought and salt stress (Ibraheem et al., 2011). PtaSUT4 transporters also function in photosynthesis (Frost et al., 2012) and drought response (Harding et al., 2020). Furthermore, PaSUT and OsSUT1 transporters are associated with phytohormone (auxin and cytokinin) signaling pathways in the translocation of sucrose in Petunia axillaris flower (Rolland et al., 2002; Iftikhar et al., 2020) and rice seeds (Zhao et al., 2022), respectively. More importantly, PttSUT3 transporters are responsible for carbon delivery and formation of wood fibers in a hybrid aspen (Mahboubi et al., 2013).
Bamboo is widely scattered in China, and products made of bamboo have been utilized by humans since the ancient times (Qi et al., 2015). D. farinosus is one of the essential economic bamboo species in southwestern China, which has many advantages including high growth rate and disease resistance, cold and drought tolerance, and dual usage for food and timber (Zakikhani et al., 2014; Chen et al., 2021; Pei et al., 2022). In particular, D. farinosus is also an excellent paper pulp bamboo species because of its high fiber content, stiffness, and low density (Xiong et al., 2012; Imran et al., 2022). Sucrose is the predominant form of transported carbon, which is necessary to synthesize fiber (Rennie and Turgeon, 2009), and SUT transporter genes are crucial to carbon delivery and fiber formation in some plants (Zhang et al., 2017; Yadav et al., 2022). However, the mechanisms of sucrose translocation between the source leaves and the sink organs of D. farinosus are unclear. In this study, we reported the phylogenetic analysis and the whole-genome identification of SUT genes as well as their chromosomal localization and expression pattern in D. farinosus. Most DfSUT genes were involved in ABA/MeJA signaling pathways and drought resistance. We then verified the transport function of D. farinosus DfSUT4 as a rice OsSUT4 homolog in transgenic Nicotiana tabacum. Our results indicated that DfSUT4 is a candidate gene in carbohydrate transport and improving the photosynthetic rate as well as involved in the phloem transportation of sucrose from the source leaves to the sink organs, phytohormone responses, abiotic stress, and fiber formation in D. farinosus and other crops.
The identified SUT sequences in rice were searched for the corresponding SUT homologous gene in D. farinosus. According to the chromosomal location, we identified 12 genes of the DfSUT transporter family (Table 1), namely DfSUT1–DfSUT12. Detailed information about the 12 DfSUT genes, including the predicted length of CDSs, encoded proteins, and physicochemical parameters, are presented in Table 1. The length of DfSUT CDSs ranged from 291 to 1,803 bp, with an average length of 1,481 bp. The protein size of DfSUT was an average of 493 aa, which ranged from 96 to 600 aa. The molecular weight ranged from 10.54 kDa (DfSUT12) to 64.06 kDa (DfSUT9). The pI for all DfSUT genes was below 10.0, with an average value of 8.1; only one gene product (DfSUT1) was below 5.0. DfSUT genes were predicted to localize to the plasma membrane or tonoplast. Among them, DfSUT1 was not assembled on the chromosome of Liang Shan Cichlid, so it was not discussed in the following analysis.
To comprehensively analyze the evolutionary relationships between SUT families in D. farinosus, Phyllostachys edulis, D. latiflorus munro, Arabidopsis thaliana, rice, and Populus L., we constructed a neighbor-joining (NJ) phylogenetic tree of SUTs based on 55 full-length SUT protein sequences, including 12 sequences from D. farinosus, 10 sequences from Phyllostachys edulis, 13 sequences from D. latiflorus munro, nine sequences from Arabidopsis thaliana, five sequences from rice, and six sequences from Populus L. (Figure 1A). All three woody bamboos contain more SUT genes than Arabidopsis thaliana, rice, and Populus L. According to the phylogenetic analysis, plant SUTs are divided into three types. Type I and type II SUTs are localized to the plasma membrane, and type III SUTs are localized to the vacuolar membrane (Reinders et al., 2012). The phylogenetic tree showed that the 55 SUT proteins could be divided into three distinct groups (clade I–clade III; Figure 1A). All 12 SUT genes in D. farinosus could be divided into two clades (clade II and clade III), such as three DfSUTs (DfSUT2, DfSUT7, and DfSUT11) that belonged to clade II. The other nine DfSUTs belonged to clade III. However, none of the SUTs in all three woody bamboos and rice belonged to clade I. DfSUT2, DfSUT7, and DfSUT11 were homologs of rice OsSUT2, Arabidopsis AtSUC4, and Populus L. PtaSUT4. While DfSUT4, DfSUT9, and DfSUT12 were homologs of AtSUC3, OsSUT4, PtaSUT5, and PtaSUT6, suggesting that they might have a similar function. Interestingly, we found that all three woody bamboos contained almost an equal number of SUT genes, and they all had more SUT genes than the other species.
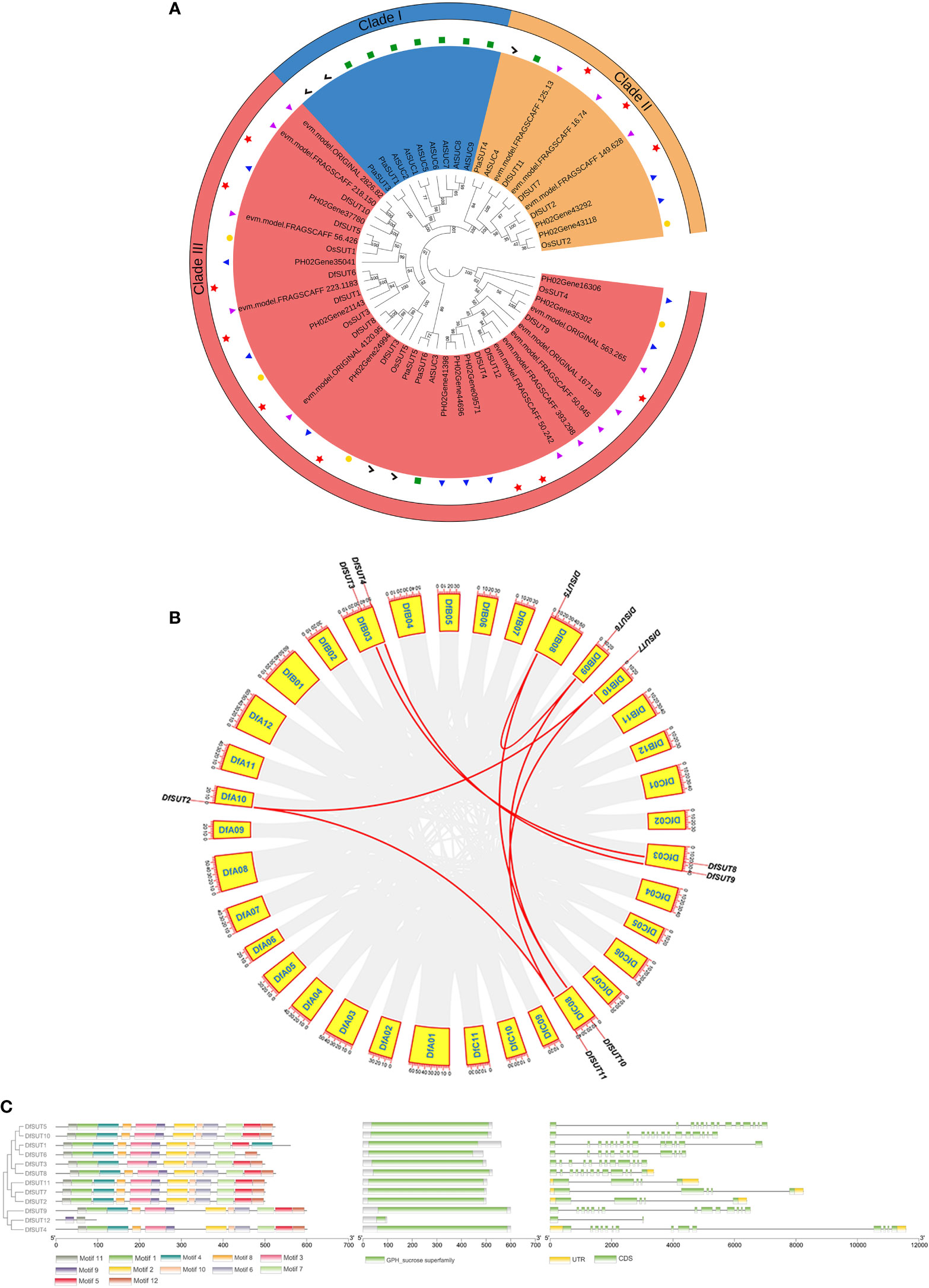
Figure 1 Analysis of the phylogenetic evolutionary, chromosomal localization and gene structure of the SUT family in D. farinosus. (A) Phylogenetic tree of the SUT family in D. farinosus. Based on the amino acid sequences of D. farinosus, Phyllostachys edulis, D. latiflorus munro, Arabidopsis, rice, and Populus L., phylogenetic trees were generated by the neighbor joining method using MEGA 7.0.21. The SUT members are divided into three branches: clades I, II, and III. The species are labeled with different colors: Df, D. farinosus (red); Phyllostachys edulis (blue); D. latiflorus munro (purple); At, Arabidopsis (green); Os, rice (yellow); Pta, Populus L. (black). (B) Chromosomal localization and gene duplication analysis of DfSUT genes in the genome of D. farinosus. The DfSUT genes are localized on different chromosomes. Chromosome numbers are indicated in the yellow box. The numbers on the chromosome boxes represent the sequence length in megabases. Gene pairs with sibling relationships are connected by a red line. (C) Conserved motif and gene structure analysis of the SUT family in D. farinosus. The three maps of conserved motif analysis, conserved structural domains, and exon/intron structure of SUT genes were merged in the order of phylogenetic tree by the Gene Structure View online software analysis of TBtools software.
The chromosomal distribution showed that 10 of the 12 DfSUT genes were mapped on seven chromosomes in D. farinosus (Figure 1B) (Supplementary Table S4). The result showed that four chromosomes (DfA10, DfB08, DfB09, and DfB10) harbored only one gene, and they were DfSUT2, DfSUT5, DfSUT6, and DfSUT7, respectively. Three chromosomes (DfB03, DfC03, and DfC08) contained two genes, such that chromosome DfB03 contained DfSUT3 and DfSUT4, chromosome DfC03 contained DfSUT8 and DfSUT9, and chromosome DfC08 contained DfSUT10 and DfSUT11. In addition, among the 12 DfSUT family genes, there were 10 genes with existing gene duplication. Seven gene pairs connected with each other, representing fragment duplication, such as DfSUT2 and DfSUT7, DfSUT2 and DfSUT11, DfSUT7 and DfSUT11, DfSUT3 and DfSUT8, DfSUT4 and DfSUT9, DfSUT5 and DfSUT6, DfSUT5 and DfSUT10 (Figure 1B). In addition, we estimated the approximate dates of DfSUT gene duplication events and the gene evolutionary selection pressure by measuring the Ka and Ks values and the Ka/Ks ratios (Supplementary Table S5). The Ka/Ks ratios of the DfSUT genes were all less than 1, indicating that they were all in a purifying selection condition.
We then analyzed the conserved motif and gene structure of the SUT family in D. farinosus (Figure 1C). The amino acid sequences of DfSUT in Table 1 were used to construct the phylogenetic tree separately. The conserved motifs and structural domains of the family were identified using MEME and GSDS online tools, respectively. The structural analysis maps of the above-mentioned three sequences were combined and plotted using TBtools (Figure 1C). The result showed that all DfSUT proteins contain 11 or 12 motifs and have similar conserved structural domains, except for DfSUT12 which has only two motifs. In the SUT family, eight SUT genes contain 14 exons and 13 introns, while three SUT genes contain five exons and four introns. Interestingly, we found that DfSUT12 only has two exons and one intron, and seven SUT genes including DfSUT12 have no upstream and downstream untranslated regions (Figure 1C).
To determine the dynamic gene expression of SUT genes in D. farinosus, we performed RNA-Seq analysis of 12 SUT protein-coding genes in 14 organs and developmental stages of D. farinosus, including branch, culm sheath (CSh), internode (Inod), lateral bud (Labud), mature leaf (MLeaf), node, root, sheath of bamboo shoot (ShB), young leaf (Yleaf), 10-cm shoot (C10), 50-cm shoot (C50), 200-cm shoot (C200), 400-cm shoot (C400), and 800-cm shoot (C800). According to the transcriptome heat map, DfSUT2, DfSUT4, DfSUT7, and DfSUT11 showed a wide range of expression levels and distinct regulation during D. farinosus development (Figure 2A). These four SUT-encoding genes (DfSUT2, DfSUT4, DfSUT7, and DfSUT11) were abundantly expressed in the branch, leaf, and root (Figure 2A), suggesting that they were associated with the transportation of sucrose from the source leaf to the sink organs. They were all highly expressed in bamboo shoots (Figure 2A), such as DfSUT2 and DfSUT11 in 400-cm-tall shoot, DfSUT4 in 200-cm- and 800-cm-tall shoots, and DfSUT7 in 200-cm-tall shoot, suggesting that they were all associated with the rapid growth of shoots. Interestingly, DfSUT4, DfSUT7, and DfSUT11 were abundantly expressed in internode and node, suggesting that they were involved in sucrose accumulation in the node and internode of D. farinosus. DfSUT8 was only highly expressed in the lateral bud and 10-cm shoot, suggesting that it is important for the development of buds and shoots.
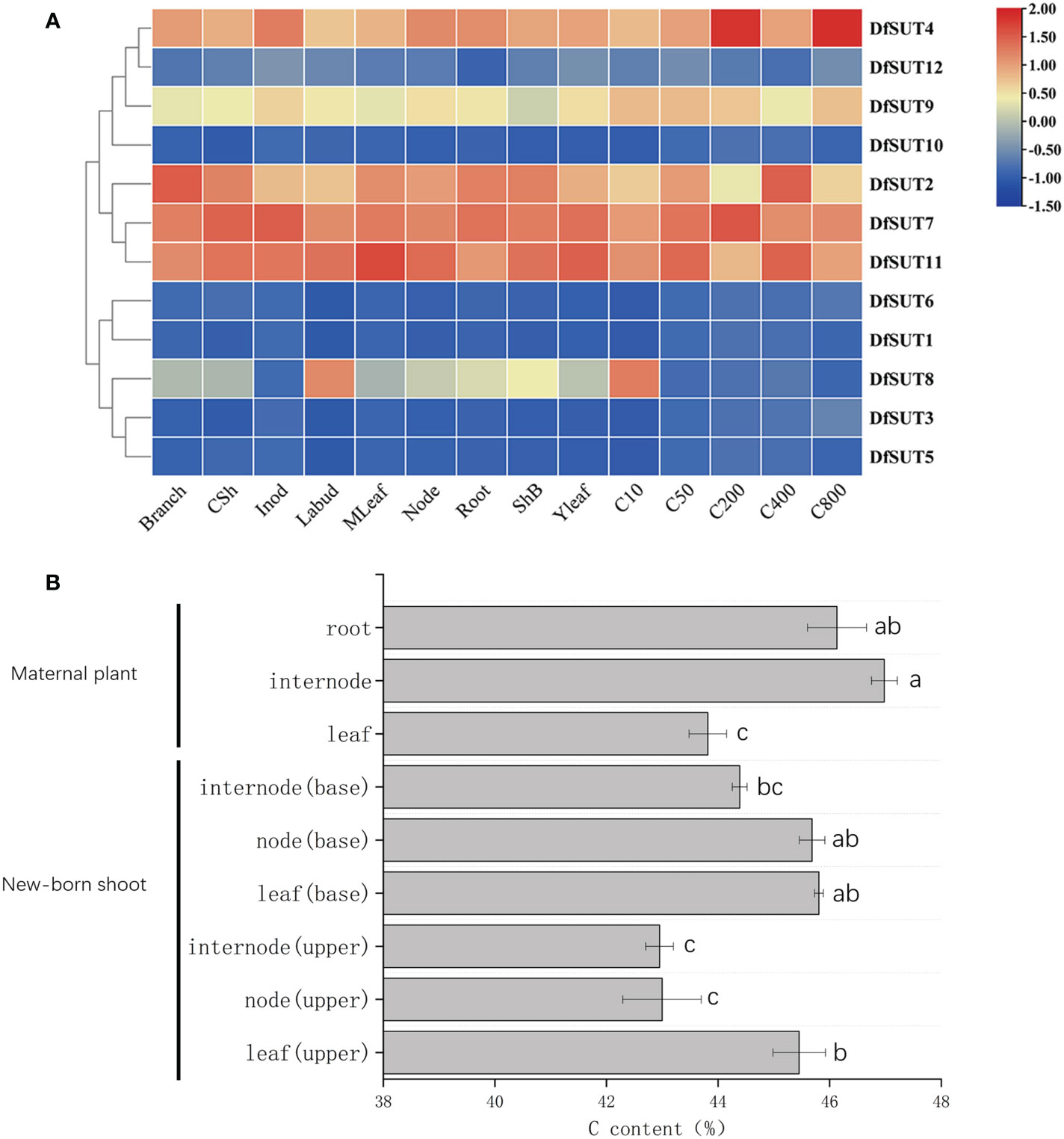
Figure 2 Analysis of transcriptome expression profiles of SUT and C content in different developmental phases/tissues of D. farinosus. (A) Transcriptome expression profiles of SUT at different developmental stages and various parts in D. farinosus. The red and blue colors correspond to the strong and weak expression of the genes, respectively. (B) C content in different developmental phases and tissues in D. farinosus. Data are detected as the mean ± SD from three independent experiments. The P-values of Student’s test for different developmental phases and tissues in D. farinosus are denoted with letters (P < 0.05).
We then measured the C content in different developmental phases and tissues in D. farinosus (Figure 2B). Consistent with the expression patterns of SUT genes, the high C content in the mature internode suggested that carbohydrate transportation by DfSUT proteins was necessary to synthesize cellulose in the internode of D. farinosus. More importantly, the new leaf (base), new node (base), and mature root of D. farinosus contained the same high C content (Figure 2B). These results showed that, except the leaf source and the root sink, the bamboo node that consists of complex vascular bundles in D. farinosus might be a temporary sink for the accumulation of sucrose for some specific functions.
To investigate the responses to various factors by DfSUT members, the promoter (2 kb upstream) of these genes was submitted to the PlantCARE server to predict its promoter cis-regulatory elements. We identified 20 cis-regulatory elements in DfSUT gene promoter, such as phytohormone response elements (abscisic acid, auxin, methyl jasmonate, gibberellin, and salicylic acid), abiotic stress response elements (anaerobic, low-temperature, drought, defense, wound, and light), and growth and development response elements (meristem expression, circadian control, mesophyll cells differentiation, cell cycle regulation, and seed-specific regulation) (Figure 3). The result showed that most DfSUT genes were associated with plant hormones, stress, growth, and development, suggesting that DfSUT genes are involved in multiple physiological processes through a variety of environmental adaptations. In addition, ABRE-motif, CGTCA-motif, TGACG-motif, MBS-motif, and G-Box-motif were most enriched in the DfSUT promoter regions (Figure 3), which indicated that most DfSUT genes might be involved in the responses of ABA, MeJA, drought, and light—for example, six DfSUT genes (DfSUT2, DfSUT4, DfSUT5, DfSUT6, DfSUT8, and DfSUT10) were related to the responses of ABA, and five DfSUT genes (DfSUT2, DfSUT4, DfSUT5, DfSUT7, and DfSUT11) were related to the responses of MeJA. Moreover, DfSUT2 and DfSUT5 included more than 10 motifs in the promoter that were associated with ABA and light, respectively. DfSUT4 contained about five motifs in the promoter that were associated with ABA, MeJA, and light, respectively.
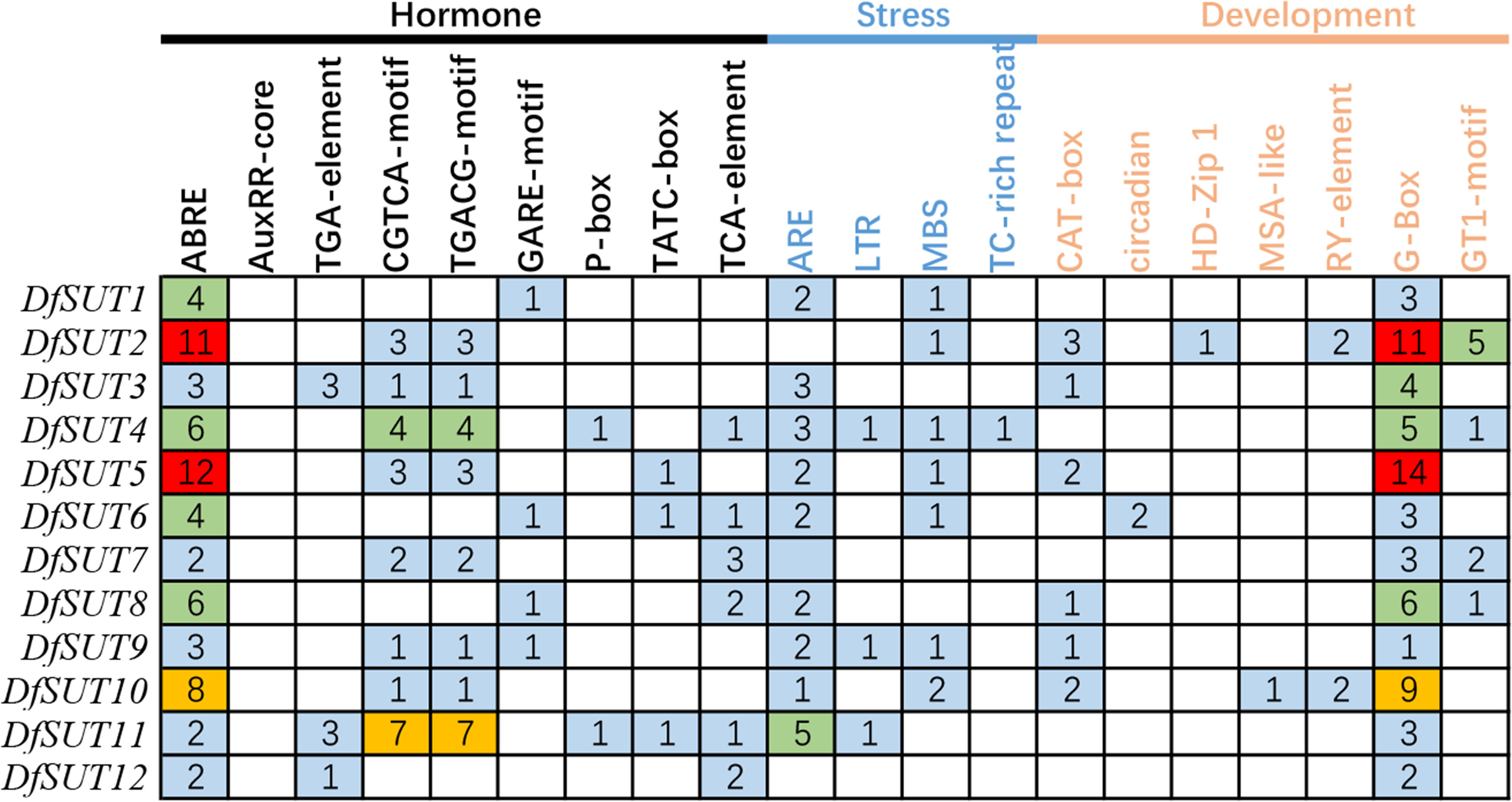
Figure 3 The cis-regulatory elements involved in phytohormone, development, and stress responses in the upstream regions of DfSUT gene promoters. ABRE, abscisic acid-responsive element; AuxRR core and TGA element, auxin-responsive element; CGTCA-motif and TGACG-motif, MeJA-responsive elements; GARE-motif, P-box and TATC-box, gibberellin-responsive elements; TCA element, salicylic acid-responsive elements; ARE, involved in anaerobic induction; LTR, low temperature-responsive element; MBS, TC-rich repeats, involved in defense and stress response; CAT box, circadian, HD-Zip I, MSA-like, and RY-element involved in meristem expression, circadian control differentiation of the palisade mesophyll, cell cycle regulation, and seed-specific regulation, respectively; G-box, GT1-motif, light-responsive elements.
To determine the transport of photosynthetic products by SUT proteins, we measured the transcript levels and net photosynthetic reaction rate in D. farinosus leaves every 3 h (from 6 a.m. to 9 p.m.). The results showed that the expression of DfSUT remained basically consistent with the change in the net photosynthetic reaction rate, except DfSUT7 (Figure 4A). The transcript levels of five DfSUT genes (DfSUT2, DfSUT4, DfSUT8, DfSUT9, and DfSUT11) increased with the increase of the net photosynthetic reaction rate in D. farinosus, and both of them reached the highest levels at 9 a.m. These results indicated that the expression of these DfSUT genes was involved in the transport of photosynthetic products.
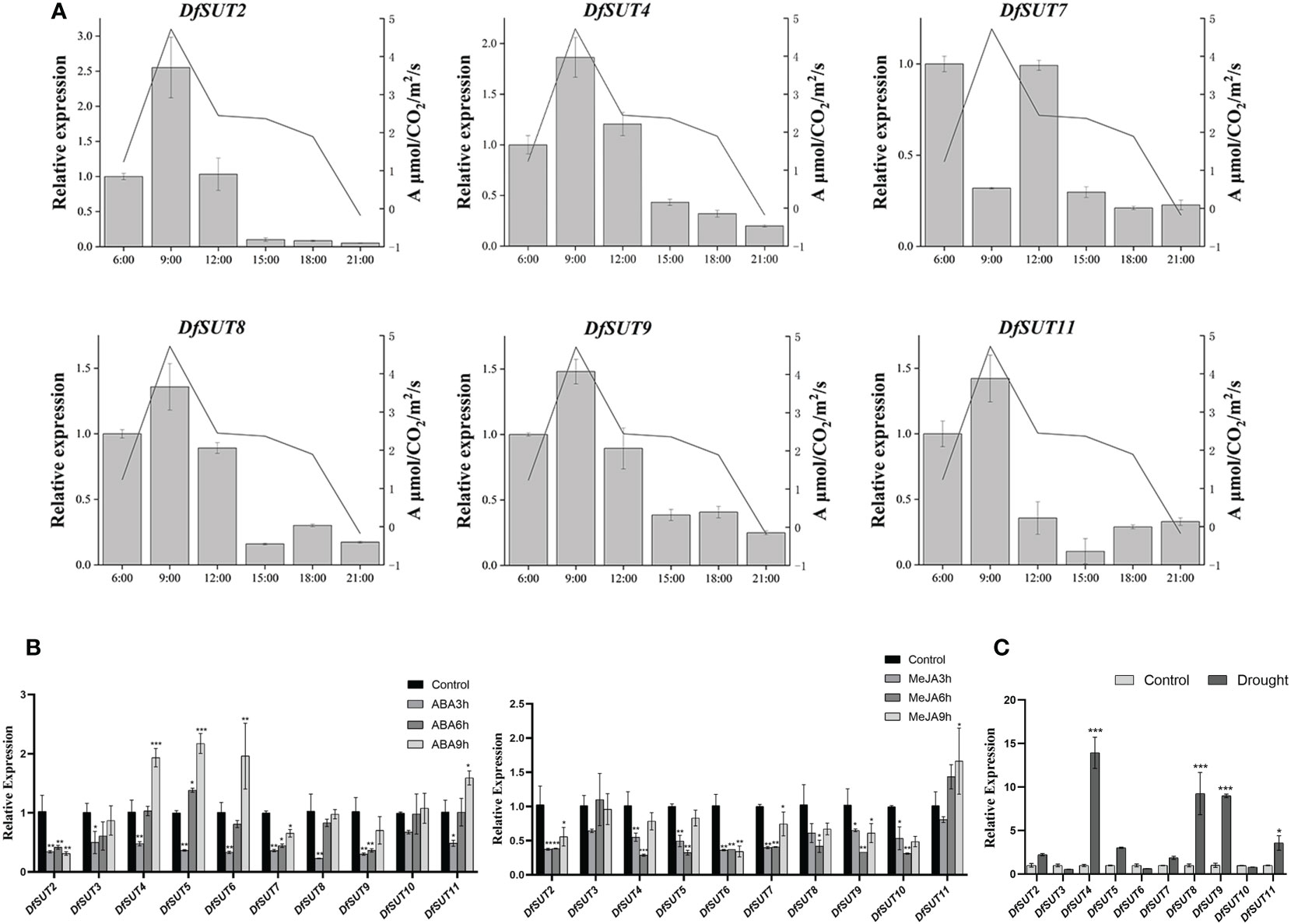
Figure 4 Analysis of photosynthetic rate and response to different treatments of the SUT family in D. farinosus. (A) Relative expression of DfSUT at different times of the day and the net photosynthetic reaction rate at the corresponding time periods. Total RNA was extracted from D. farinosus leaves every 3 h (from 6 a.m. to 9 p.m.) during the day. (B) Relative expression of DfSUT in response to 100 mM ABA treatment and 100 mM MeJA treatment for 3, 6, and 9 h, respectively. (C) Relative expression of DfSUT under drought treatment. *, P < 0.05, **, P < 0.01, ***, P < 0.001, Student's t-test.
More importantly, we determined the response of DfSUT genes to phytohormone treatment (ABA and MeJA) and drought stress. Consistent with the analysis of the cis-regulatory elements of the DfSUT gene promoter, the results showed that the expression of nine DfSUT genes decreased within 3 h and then increased within 9 h under the treatment of exogenous ABA, except DfSUT2. Compared with the control that was untreated, the expression of DfSUT4, DfSUT5, and DfSUT6 was much higher in response to ABA within 9 h, but the transcript levels of DfSUT2 and DfSUT7 showed significant declines within 9 h in response to ABA (Figure 4B). However, the transcript levels of six DfSUT genes (DfSUT2, DfSUT3, DfSUT4, DfSUT5, DfSUT7, and DfSUT11) decreased within 3 h and then showed a rebound within 9 h in response to MeJA. Compared with the control that was untreated, the transcript levels of DfSUT2, DfSUT6, and DfSUT10 showed significant declines, but the expression of DfSUT11 showed an increase within 9 h in response to MeJA (Figure 4B). Moreover, the transcript levels of DfSUT4, DfSUT8, DfSUT9, and DfSUT11 showed significant increases in response to the drought treatment (Figure 4C).
Since we have found that the DfSUT4 gene was involved in the photosynthetic product transport and the responses to various phytohormone treatment (ABA and MeJA) and drought stress, we then verified the transport function of DfSUT4 in transgenic N. tabacum lines. The result showed that GUS activity was mainly detected in the mesophyll of leaves, primary roots, lateral roots, and root hair in pDfSUT4::GUS transgenic plant, but not in flower organ (Figures 5A–C). The DfSUT4 gene was also found to be expressed in the phloem of the stem section in N. tabacum (Figure 5D).

Figure 5 Expression of pDfSUT4::GUS in N. tabacum. (A) Intact plant. (B) Leaf. (C) Flower. (D) Stem section. Xy, xylem, Ph, phloem.
Moreover, the result showed that, compared with CK, DfSUT4 transgenic N. tabacum showed phenotypes of taller stem and noticeably larger leaves, flowers, and fruits (Figures 6A–D, F–I). The net photosynthetic reaction rate increased nearly twofold in transgenic N. tabacum (Figure 6E), suggesting that DfSUT4 genes might be involved in the distribution of carbohydrate from the source leaves. Carbohydrates can be synthesized by photosynthesis in mature leaves and then transported to other sink tissues (Barker et al., 2000; Sun et al., 2010). To explore the distribution mechanism of the increased photosynthetic products, we measured the above-ground biomass and cellulose content of stem in three transgenic lines. The results showed that, compared with the control line, the above-ground biomass in transgenic N. tabacum line 3 has increased nearly twofold compared with the control lines (Figure 6J), and the cellulose content of the stem in line 3 was increased by more than threefold (Figure 6K). Therefore, these results suggested that DfSUT4 is important for the translocation of sucrose from the leaf to the sink organs through the phloem and promotes cellulose synthesis in plants.
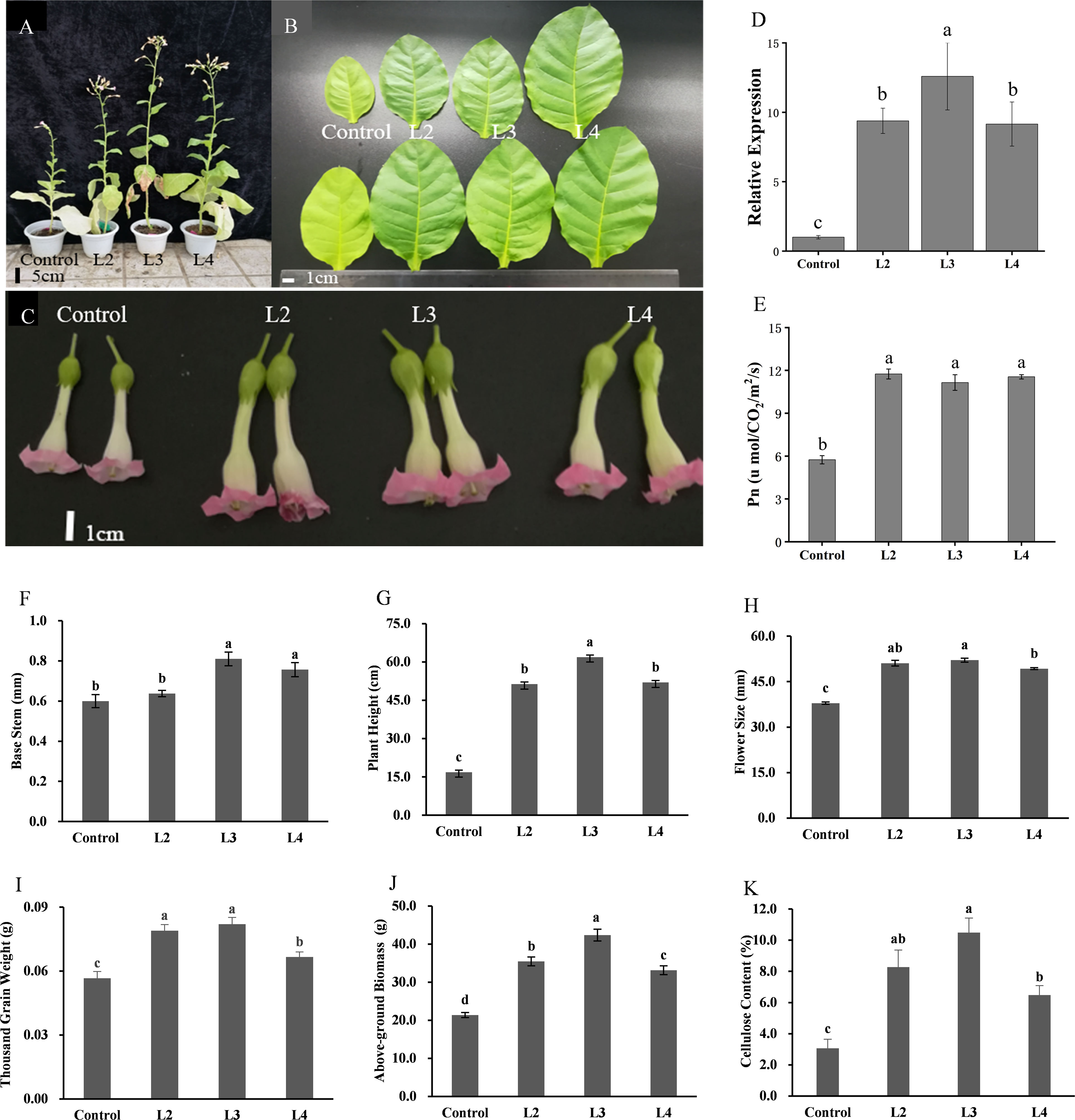
Figure 6 Morphological phenotypes, statistics, and above-ground biomass/cellulose content of control and three transgenic N. tabacum lines. (A) 3-month-old plants. (B) Leaf blades of 2-month-old plants. (C) Phenotype of the flower. (D) Relative expression of DfSUT4 in transgenic N. tabacum. (E) Net photosynthetic reaction rate of control and transgenic lines. (F–I) Morphologic index of control and transgenic lines. (J, K) Above-ground biomass and cellulose content of control and transgenic lines. Data are expressed as the mean ± SD from six independent experiments. The P-values of Student’s test for control and transgenic lines are denoted with letters (P < 0. 05).
In higher plants, sucrose is produced by leaf photosynthesis and then metabolized into hexoses (glucose and fructose), which are necessary to synthesize cellulose, proteins, and starch and generate energy in the sink tissues (Ruan, 2014). The SUT proteins play key roles in the translocation of sucrose from the source to the sink through the phloem to support plant growth and development (Slewinski and Braun, 2010; Deol et al., 2013). D. farinosus is an ideal economic crop in southwestern China because of its high fiber content and dual usage for food and paper pulp (Chen et al., 2021; Imran et al., 2022; Pei et al., 2022). Sucrose is the main substrate of fiber synthesis, and studying the function of SUT genes in regulating sucrose transportation helps to illustrate the synthesis mechanism of fiber in plants (Yadav et al., 2022)—for example, GhSUT1 and GhSUT3 are crucial to regulate sucrose accumulation in cotton fibers (Zhang et al., 2017; Yadav et al., 2022). PttSUT3 transporters are responsible for the synthesis of wood fibers in a hybrid aspen (Mahboubi et al., 2013).
Herein we identified 12 main SUT transporter genes in D. farinosus, which were distributed in eight chromosomes in groups A, B, and C of D. farinosus, respectively (Table 1; Figure 1B). We analyzed the phylogenetic relationships of SUT genes from three woody bamboos including D. farinosus, Phyllostachys edulis, and D. latiflorus munro and found that some SUT genes from Phyllostachys edulis and D. latiflorus munro clustered together, respectively. However, the SUT genes from D. farinosus were relatively scattered (Figure 1A). All three woody bamboos contained more SUT genes than Arabidopsis and rice, suggesting that SUT genes were vital to synthesize fiber with high quality and content in woody bamboos. The phylogenetic analysis also showed that DfSUT2, DfSUT7, and DfSUT11 were homologs of OsSUT2 and PtaSUT4, and DfSUT4 is a homolog of OsSUT4 (Figure 1A); all of these four DfSUT genes were mainly expressed in the leaves of D. farinosus (Figure 2A). These results suggested that DfSUT2, DfSUT7, and DfSUT11 might be important for sucrose efflux from the vacuole to the apoplast in the source leaves of D. farinosus (Eom et al., 2011), and DfSUT4 might function in the apoplastic loading of sucrose from the leaf mesophyll to the phloem (Chung et al., 2014). Chen et al. (2022) revealed that the bamboo node consists of sophisticated porous vascular bundles, microfibers, and twist-aligned nanofibers, and they are essential to the structural stability maintenance and body growth of bamboo. We found that DfSUT4, DfSUT7, and DfSUT11 were highly expressed in both the internode and node of D. farinosus (Figure 2A). The same high C content was detected in the new leaf (base), new node (base), and mature root in D. farinosus (Figure 2B), suggesting that the directional channels, including multiscale sieve tubes and vessels in the node, might serve as a temporary sink for the accumulation of sucrose for some specific functions, such as supply of nutrients for the rapid growth of bamboo at night, and these DfSUT genes were also involved in fiber formation in the internode of D. farinosus.
The involvement of sucrose transporters in phytohormone signaling and abiotic responses was already described in other species—for example, SlSUT2 and StSUT4 proteins were found to interact with brassinosteroid signaling, ethylene sensing, gibberellic acid responses, and auxin transport in tomato and potato (Bitterlich et al., 2014; Garg et al., 2022a). PaSUT and OsSUT1 were found to interact with hormone (auxin and cytokinin) signaling pathways to regulate the carbohydrate partitioning in Petunia axillaris flower (Rolland et al., 2002; Iftikhar et al., 2020) and the reproductive organ of rice (Zhao et al., 2022), respectively. Consistent with these conclusions, we found that DfSUT4, DfSUT5, and DfSUT6 showed significant increases in expression in response to ABA treatment within 9 h (Figure 4B). DfSUT2, DfSUT6, and DfSUT10 showed significant decreases in response to MeJA treatment within 9 h (Figure 4B). The drought treatment also increased the transcript levels of DfSUT4, DfSUT8, DfSUT9, and DfSUT11 (Figure 4C), so these results showed that the DfSUT family genes interacted with ABA and MeJA signaling pathways and drought resistance, which helped the plants to be more adaptable to environmental changes, especially the DfSUT4 gene (Figure 4). We also found that the DfSUT genes were resistant to Puccinia striiformis (not shown).
Then, we verified the transport function of DfSUT4 in N. tabacum. The OsSUT4 transporters were localized in the plasma membrane (PM), functioned in apoplastic phloem loading of sucrose, and increased the rice yield (Chung et al., 2014). Consistently, we found that DfSUT4 was localized in the plasma membrane (not shown), and the GUS activity was expressed in the mesophyll, roots, and stem phloem of pDfSUT4::GUS transgenic plant (Figure 5). The overexpression of the DfSUT4 gene in transgenic plant showed increases of photosynthesis capacity, above-ground biomass, thousand grain weight, and cellulose content and promoted the development of flower and seed in transgenic plant (Figure 6). These results suggested that DfSUT4 can be a candidate gene to regulate the phloem transportation of sucrose from the source leaves to the sink organs, phytohormone responses, abiotic stress, and fiber formation in the internode of D. farinosus.
Partially functional redundancy existed in nine different Arabidopsis AtSUC genes (Garg et al., 2022b), such as the PM-localized AtSUC1 protein which interacted with the vacuolar AtSUC4 protein to mediate sucrose allocation in plants (Schulze et al., 2003; Kruegel et al., 2012). Recent studies revealed that SUT proteins are dynamic within cells, and SNARE protein is important to the subcellular movement of SUT transporters (Garg et al., 2020). At present, the interacting proteins of DfSUT, the functional redundancy between different DfSUT proteins, and the mechanisms of the DfSUT4 genes involved in phytohormone responses, abiotic stress, and fiber formation in D. farinosus need to be analyzed further.
The SUT family genes of rice were queried using the Uniprot database (https://www.uniprot.org/), and the candidate SUT sequences were obtained from the whole genomic data of D. farinosus (https://www.ncbi.nlm.nih.gov/, PRJNA923443) using HMMER 3.0 (E ≤ 10–20). The Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) online software in NCBl was used to remove the sequences that did not include the GPH sucrose superfamily domain. There were 12 SUT family members annotated in D. farinosus for further study (Table 1).
The SUT family protein sequences of Arabidopsis thaliana, rice, Populus L., Phyllostachys edulis, and Dendrocalamus latiflorus munro were downloaded from UniProt (http://www.uniprot.org) (Supplementary Table S1). By using the Clustal W program in MEGA 6.06 software, we performed a multiple sequence alignment analysis of the SUT protein between D. farinosus and other species and constructed a phylogenetic tree by using the NJ method using MEGA 7.0.21 with bootstrap option n = 1,000 (Chen et al., 2020).
The Multiple Collinearity Scan toolkit (MCScanX) was used to analyze the internal synteny relationship and obtain syntenic gene pairs in the SUT family genes of D. farinosus (Wang et al., 2012). The syntenic analysis maps were then constructed using the Dual Systeny Plotter software (https://github.com/CJ-Chen/TBtools) (Liu et al., 2017).
The conserved motifs of DfSUTs were analyzed using the MEME online tool (version 5.3.0, http://meme-suite.org/tools/meme) (Bailey et al., 2015), and the maximum motif search value was set at 10. Motif analysis and gene structure visualization were performed via TBtools. The Gene Structure Display Server 2.0 online tool (http://gsds.gao-lab.org/index.php) (Hu et al., 2015) was used to display the structure of DfSUT genes and the genomic length and organization of introns/exons. The 2,000-bp upstream promoter sequences of DfSUT genes were uploaded to the PlantCARE database (Lescot et al., 2002) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), and the cis-elements were subsequently screened manually.
One-year-old D. farinosus materials were potted in a growing chamber wherein the condition was kept at 16-h light/8-h dark, and the temperature was 23–25°C. The samples from maternal plants and newborn shoots which included root, internode, leaf, and node were collected for the analysis of C content. All samples were de-enzymed at 100°C, dried at 85°C to a constant weight, and then ground. The total C content was measured using the potassium dichromate–sulfuric acid oxidation method (Cai et al., 2011). The experiment was repeated three times.
All materials used in this study were grown in a greenhouse at the Institute of Bamboo Research, Southwest University of Science and Technology. The leaves of 1-year-old D. farinosus were collected every 3 h (from 6:00 a.m. to 9:00 p.m.) during the day for the measurement of net photosynthetic reaction rate by using LCpro-SD photosynthesis equipment, and six independent biological replicates were performed. In addition, the young leaves of 1-year-old D. farinosus were sprayed with 100 mM ABA and MeJA, respectively, and the leaf samples were taken before treatment and after 3, 6, and 9 h of treatment.
Total RNA was extracted from D. farinosus leaves by using TRIzol reagent (Tiangen, Beijing, China), and DNase I was used to purify potentially genomic DNA. The quality of total RNA was checked by 1% denaturing agarose gel and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Beijing, China). First-strand cDNA synthesis was performed by using the FastKing cDNA First Strand Synthesis Kit (Tiangen, Beijing, China). Specific primers were designed by using Primer Premier 5.0 (Supplementary Table S2). The internal reference gene was TUBLIN. The transcript levels of DfSUT were analyzed by real-time quantitative PCR assay using SYBR qPCR Master MIX kit (Vazyme, Nanjing, China) and CFX96TM Real-Time System thermal cycler (Bio-Rad, CA, USA). The relative expression levels of the SUT gene were calculated by using the 2-ΔΔCt method (Livak and Schmittgen, 2001). The analysis included three biological replicates, and each had three technical replicates. The expression levels under different treatments were shown by histograms using the mean values.
One-year-old D. farinosus materials potted in the growing chamber were used for RNA sequencing. The samples included branch, culm sheath, internode, lateral bud, young and mature leaves, node, root, sheath of bamboo shoot, and shoots of different height, which were collected for RNA sequencing to be performed. Each sample had three biological replicates. Total RNA was extracted by using TRIzol reagent (Tiangen, Beijing, China), and DNase I was used to purify potentially genomic DNA. The quantity and the quality of total RNA were checked by using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Beijing, China). All 14 samples were subjected to the HiSeq 2500 platform (Illumina, Beijing, China), and the data of RNA sequencing (Supplementary Table S3) was used to construct the heat map of the DfSUT genes.
The recombinant plasmid of pCAMBIA1303-N-DfSUT4 with CaMV 35S promoter was transferred into Agrobacterium tumefaciens EHA105. Transgenic tobacco overexpression lines were obtained by the leaf disc method (Zhang et al., 2019). Infected tobacco leaf discs were cultivated in the dark on MS medium containing 9 mg/L Hyg, 400 mg/L cephalexin, 0.5 mg/L 6-BA, and 0.1 mg/L NAA at 27 ± 1°C for 2 days. The regenerated shoots were transferred to 1/2 MS medium with 9 mg/L Hyg, 400 mg/L cephalexin, and 0.1 mg/L NAA for the formation of whole plants. Genomic DNA was extracted from the leaves of transgenic plants and wild-type and amplified under the following conditions: preheating at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 2 min, and finally extension at 72°C for 10 min. The PCR products were detected by 1% agarose gel electrophoresis to confirm the insertion of DfSUT4 into the transgenic plants. Then, qRT-PCR was used to detect the expression of DfSUT4 in transgenic plants. The internal reference gene was N. tabacum NtActin (Wang et al., 2022).
Fresh plant organs of transgenic tobacco were fixed by 90% acetone on ice for 20 min and then placed in GUS staining solution containing 2 mM X-Gluc in GUS assay buffer (200 mM phosphate buffer, pH 7.0, 0.1% Triton X-100, 20 mM EDTA, and 0.5 mM potassium ferricyanide and potassium ferrocyanide), vacuum infiltrated, and incubated at 37°C overnight. The stained samples were then dehydrated in an ethanol series. Photographs were taken with a stereomicroscope.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA923443.
SHu and YC planned and designed the research. BD, MZ, SC, SHa, and LW performed all experiments and analyzed the data. XG and BD wrote the original manuscript. SHu and YC proofread the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Key R&D Program of China (2021YFD2200504_1 and 2021YFD2200505_2), Science and Technology Project of Sichuan Province, China (2021YFYZ0006 and 2022NSFSC0093), and Ph.D. Foundation (no. 22zx7147) from Southwest University of Science and Technology.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1118398/full#supplementary-material
Aoki, N., Hirose, T., Furbank, R. T. (2012). Sucrose transport in higher plants: From source to sink. In Photosynthesis Springer 34, 703–729. doi: 10.1007/978-94-007-1579-0_28
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43 (W1), W39–W49. doi: 10.1093/nar/gkv416
Barker, L., Kuhn, C., Weise, A., Schulz, A., Gebhardt, C., Hirner, B., et al. (2000). SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12 (7), 1153–1164. doi: 10.1105/tpc.12.7.1153
Bitterlich, M., Kruegel, U., Boldt-Burisch, K., Franken, P., Kuehn, C. (2014). The sucrose transporter SlSUT2 from tomato interacts with brassinosteroid functioning and affects arbuscular mycorrhiza formation. Plant J. 78 (5), 877–889. doi: 10.1111/tpj.12515
Cai, Y., Peng, C., Qiu, S., Li, Y., Gao, Y. (2011). Dichromate digestion-spectrophotometric procedure for determination of soil microbial biomass carbon in association with fumigation-extraction. Commun. Soil Sci. Plant Anal. 42 (22), 2824–2834. doi: 10.1080/00103624.2011.623027
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, S. M., Zhang, S. C., Gao, H. L., Wang, Q., Zhou, L., Zhao, H. Y., et al. (2022). Mechanically robust bamboo node and its hierarchically fibrous structural design. Natl. Sci. Rev. nwac195. doi: 10.1093/nsr/nwac195
Chen, P., Zheng, R. H., Wang, Y., Hu, Y. B. (2021). Study on clonal selection of dendrocalamus farinosus in south sichuan. J. Sichuan Forestry Sci. Technol. 42 (1), 109–113. doi: 10.12172/202007270002
Chung, P., Hsiao, H. H., Chen, H. J., Chang, C. W., Wang, S. J. (2014). Influence of temperature on the expression of the rice sucrose transporter 4 gene, OsSUT4, in germinating embryos and maturing pollen. Acta Physiologiae Plantarum 36 (1), 217–229. doi: 10.1007/s11738-013-1403-x
Deol, K. K., Mukherjee, S., Gao, F., Brule-Babel, A., Stasolla, C., Ayele, B. T. (2013). Identification and characterization of the three homeologues of a new sucrose transporter in hexaploid wheat (Triticum aestivum l.). BMC Plant Biol. 13, 181. doi: 10.1186/1471-2229-13-181
Eom, J. S., Cho, J. I., Reinders, A., Lee, S. W., Yoo, Y., Pham Quoc, T., et al. (2011). Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 157 (1), 109–119. doi: 10.1104/pp.111.176982
Frost, C. J., Nyamdari, B., Tsai, C. J., Harding, S. A. (2012). The tonoplast-localized sucrose transporter in populus (PtaSUT4) regulates whole-plant water relations, responses to water stress, and photosynthesis. PloS One 7 (8), e44467. doi: 10.1371/journal.pone.0044467
Garg, V., Hackel, A., Kuehn, C. (2020). Subcellular targeting of plant sucrose transporters is affected by their oligomeric state. Plants 9 (2), 158. doi: 10.3390/plants9020158
Garg, V., Kuhn, C. (2022b). Subcellular dynamics and protein-protein interactions of plant sucrose transporters. J. Plant Physiol. 273, 1–8. doi: 10.1016/j.jplph.2022.153696
Garg, V., Reins, J., Hackel, A., Kuhn, C. (2022a). Elucidation of the interactome of the sucrose transporter StSUT4: sucrose transport is connected to ethylene and calcium signaling. J. Exp. Bot. 73 (22), 7401–7416. doi: 10.1093/jxb/erac378
Gong, X., Liu, M., Zhang, L., Ruan, Y., Ding, R., Ji, Y., et al. (2015). Arabidopsis AtSUC2 and AtSUC4, encoding sucrose transporters, are required for abiotic stress tolerance in an ABA-dependent pathway. Physiologia Plantarum 153 (1), 119–136. doi: 10.1111/ppl.12225
Gottwald, J. R., Krysan, P. J., Young, J. C., Evert, R. F., Sussman, M. R. (2000). Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. U. S. A. 97 (25), 13979–13984. doi: 10.1073/pnas.250473797
Harding, S. A., Frost, C. J., Tsai, C. J. (2020). Defoliation-induced compensatory transpiration is compromised in SUT4-RNAi populus. Plant Direct 4 (9), e00268. doi: 10.1002/pld3.268
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, Z., Tang, Z., Zhang, Y., Niu, L., Yang, F., Zhang, D., et al. (2021). Rice SUT and SWEET transporters. Int. J. Mol. Sci. 22 (20), 11198. doi: 10.3390/ijms222011198
Ibraheem, O., Dealtry, G., Roux, S., Bradley, G. (2011). The effect of drought and salinity on the expressional levels of sucrose transporters in rice (Oryza sativa nipponbare) cultivar plants. Plant Omics 4 (2), 68–74. doi: 10.1105/tpc.110.081794
Iftikhar, J., Lyu, M., Liu, Z., Mehmood, N., Munir, N., Ahmed, M. A. A., et al. (2020). Sugar and hormone dynamics and the expression profiles of SUT/SUC and SWEET sugar transporters during flower development in petunia axillaris. Plants-Basel 9 (12) 1770. doi: 10.3390/plants9121770
Imran, M., Luo, X., Hu, S., Cao, Y., Long, Z. (2022). Epigenetic and somaclonal divergence in dendrocalamus farinosus for physiological augmentation and lignin degradation. Biotechnol. Appl. Biochem. 69 (4), 1545–1556. doi: 10.1002/bab.2226
Jia, W., Zhang, L., Wu, D., Liu, S., Gong, X., Cui, Z., et al. (2015). Sucrose transporter AtSUC9 mediated by a low sucrose level is involved in arabidopsis abiotic stress resistance by regulating sucrose distribution and ABA accumulation. Plant Cell Physiol. 56 (8), 1574–1587. doi: 10.1093/pcp/pcv082
Kruegel, U., He, H.-X., Gier, K., Reins, J., Chincinska, I., Grimm, B., et al. (2012). The potato sucrose transporter StSUT1 interacts with a DRM-associated protein disulfide isomerase. Mol. Plant 5 (1), 43–62. doi: 10.1093/mp/ssr048
Lemoine, R. (2000). Sucrose transporters in plants: update on function and structure. Biochim. Et Biophys. Acta-Biomembranes 1465 (1-2), 246–262. doi: 10.1016/s0005-2736(00)00142-5
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Liu, C., Xie, T., Chen, C., Luan, A., Long, J., Li, C., et al. (2017). Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genomics 18 (1), 503. doi: 10.1186/s12864-017-3896-y
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Mahboubi, A., Ratke, C., Gorzsas, A., Kumar, M., Mellerowicz, E. J., Niittyla, T. (2013). Aspen SUCROSE TRANSPORTER3 allocates carbon into wood fibers. Plant Physiol. 163 (4), 1729–1740. doi: 10.1104/pp.113.227603
Payyavula, R. S., Tay, K. H. C., Tsai, C. J., Harding, S. A. (2011). The sucrose transporter family in populus: the importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant J. 65 (5), 757–770. doi: 10.1111/j.1365-313X.2010.04463.x
Pei, J., Wang, Y., Zhuo, J., Gao, H., Vasupalli, N., Hou, D., et al. (2022). Complete chloroplast genome features of dendrocalamus farinosus and its comparison and evolutionary analysis with other bambusoideae species. Genes 13 (9) 1519. doi: 10.3390/genes13091519
Qi, J., Xie, J., Yu, W., Chen, S. (2015). Effects of characteristic inhomogeneity of bamboo culm nodes on mechanical properties of bamboo fiber reinforced composite. J. Forestry Res. 26 (4), 1057–1060. doi: 10.1007/s11676-015-0106-0
Reinders, A., Sivitz, A. B., Ward, J. M. (2012). Evolution of plant sucrose uptake transporters. Front. Plant Sci. 3 (22). doi: 10.3389/fpls.2012.00022
Rennie, E. A., Turgeon, R. (2009). A comprehensive picture of phloem loading strategies. Proc. Natl. Acad. Sci. U. S. A. 106 (33), 14162–14167. doi: 10.1073/pnas.0902279106
Rolland, F., Moore, B., Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14, 185–205. doi: 10.1105/tpc.010455
Ruan, Y. L. (2014). Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. doi: 10.1146/annurev-arplant-050213-040251
Schulze, W. X., Reinders, A., Ward, J., Lalonde, S., Frommer, W. B. (2003). Interactions between co-expressed arabidopsis sucrose transporters in the split-ubiquitin system. BMC Biochem. 4, 3. doi: 10.1186/1471-2091-4-3
Slewinski, T. L., Braun, D. M. (2010). Current perspectives on the regulation of whole-plant carbohydrate partitioning. Plant Sci. 178 (4), 341–349. doi: 10.1016/j.plantsci.2010.01.010
Srivastava, A. C., Ganesan, S., Ismail, I. O., Ayre, B. G. (2009). Effective carbon partitioning driven by exotic phloem-specific regulatory elements fused to the arabidopsis thaliana AtSUC2 sucrose-proton symporter gene. BMC Plant Biol. 9, 7. doi: 10.1186/1471-2229-9-7
Sun, Y., Reinders, A., LaFleur, K. R., Mori, T., Ward, J. M. (2010). Transport activity of rice sucrose transporters OsSUT1 and OsSUT5. Plant Cell Physiol. 51 (1), 114–122. doi: 10.1093/pcp/pcp172
Wang, Y., Chen, Y., Wei, Q., Wan, H., Sun, C. (2021). Phylogenetic relationships of sucrose transporters (SUTs) in plants and genome-wide characterization of SUT genes in orchidaceae reveal roles in floral organ development. Peer J. 9, e11961. doi: 10.7717/peerj.11961
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi: 10.1093/nar/gkr1293
Wang, C., Wang, L., Lei, J., Chai, S., Jin, X., Zou, Y., et al. (2022). IbMYB308, a sweet potato R2R3-MYB gene, improves salt stress tolerance in transgenic tobacco. Genes 13 (8), 1476. doi: 10.3390/genes13081476
Ward, J. M., Kuhn, C., Tegeder, M., Frommer, W. B. (1998). Sucrose transport in higher plants. Int. Rev. Cytology 178, 41–71. doi: 10.1016/s0074-7696(08)62135-x
Weise, A., Barker, L., Kuhn, C., Lalonde, S., Buschmann, H., Frommer, W. B., et al. (2000). A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12 (8), 1345–1355. doi: 10.1105/tpc.12.8.1345
Xiong, Z., Fu., Z. H., Yan, W. X., Gan, S. X., Cao, X. J., Feng, S. J. (2012). The preliminary studies on production status and cultivation techniques of dendrocalamus farinosus. World Bamboo Rattan 10 (5), 7–11. doi: 10.13640/j.cnki.wbr.2012.05.008
Yadav, U. P., Evers, J. F., Shaikh, M. A., Ayre, B. G., Hancock, R. (2022). Cotton phloem loads from the apoplast using a single member of its nine-member sucrose transporter gene family. J. Exp. Bot. 73 (3), 848–859. doi: 10.1093/jxb/erab461
Zakikhani, P., Zahari, R., Sultan, M. T. H., Majid, D. L. (2014). Extraction and preparation of bamboo fibre-reinforced composites. Materials Design 63, 820–828. doi: 10.1016/j.matdes.2014.06.058
Zhang, H., Gao, X., Zhi, Y., Li, X., Zhang, Q., Niu, J., et al. (2019). A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol. 223 (4), 1918–1936. doi: 10.1111/nph.15925
Zhang, Z., Ruan, Y. L., Zhou, N., Wang, F., Guan, X., Fang, L., et al. (2017). Suppressing a putative sterol carrier gene reduces plasmodesmal permeability and activates sucrose transporter genes during cotton fiber elongation. Plant Cell 29 (8), 2027–2046. doi: 10.1105/tpc.17.00358
Zhao, Z., Wang, C., Yu, X., Tian, Y., Wang, W., Zhang, Y., et al. (2022). Auxin regulates source-sink carbohydrate partitioning and reproductive organ development in rice. Proc. Natl. Acad. Sci. U. S. A. 119 (36), 1–11. doi: 10.1073/pnas.2121671119
Keywords: Dendrocalamus farinosus, SUT gene, phytohormone responses, abiotic stress, genome-wide analysis, sucrose transportation, fiber formation
Citation: Deng B, Gu X, Chen S, Zhang M, Hao S, Wei L, Cao Y and Hu S (2023) Genome-wide analysis and characterization of Dendrocalamus farinosus SUT gene family reveal DfSUT4 involvement in sucrose transportation in plants. Front. Plant Sci. 13:1118398. doi: 10.3389/fpls.2022.1118398
Received: 07 December 2022; Accepted: 23 December 2022;
Published: 20 January 2023.
Edited by:
Ben Zhang, Shanxi University, ChinaReviewed by:
Yunming Zhang, Huazhong Agricultural University, ChinaCopyright © 2023 Deng, Gu, Chen, Zhang, Hao, Wei, Cao and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanglian Hu, aHVzaGFuZ2xpYW5Ac3d1c3QuZWR1LmNu; Ying Cao, Y2FveWluZ0Bzd3VzdC5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.