- Key Laboratory of Forest Genetics & Biotechnology of Ministry of Education, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
Introduction: Wood formation is closely related to lignin biosynthesis. Cinnamoyl-CoA reductase (CCR) catalyzes the conversion of cinnamoyl-CoA to cinnamaldehydes, which is the initiation of the lignin biosynthesis pathway and a crucial point in the manipulation of associated traits. Liriodendron chinense is an economically significant timber tree. Nevertheless, the underlying mechanism of wood formation in it remains unknown; even the number of LcCCR family members in this species is unclear.
Materials and Results: This study aimed to perform a genome-wide identification of genes(s) involved in lignin biosynthesis in L. chinense via RT-qPCR assays and functional verification. Altogether, 13 LcCCR genes were identified that were divided into four major groups based on structural and phylogenetic features. The gene structures and motif compositions were strongly conserved between members of the same groups. Subsequently, the expression patterns analysis based on RNA-seq data indicated that LcCCR5/7/10/12/13 had high expression in the developing xylem at the stem (DXS). Furthermore, the RT-qPCR assays showed that LcCCR13 had the highest expression in the stem as compared to other tissues. Moreover, the overexpression of the LcCCR13 in transgenic tobacco plants caused an improvement in the CCR activity and lignin content, indicating that it plays a key role in lignin biosynthesis in the stems.
Discussion: Our research lays a foundation for deeper investigation of the lignin synthesis and uncovers the genetic basis of wood formation in L. chinense.
Introduction
Lignin is an aromatic phenolic compound formed by the polymerization of three monolignols (p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol, also called the H, G, and S subunits) in plants. It is extremely abundant in nature and is only second to cellulose in the constituents of natural biomass (Li et al., 2009). Lignin plays a pivotal role in maintaining the cell structure and imparting resistance to biotic and abiotic stresses (Bhuiyan et al., 2009; Weng et al., 2010; Srivastava et al., 2015). Lignin content is also a critical factor in determining the specific applications of different types of wood. Wood with high lignin content is more rigid and usually used in furniture manufacture, while wood with low lignin content is easier to degrade and is often used in the pulp and paper industries (Boerjan et al., 2003).
Due to its agricultural and economic importance, the lignin biosynthesis pathway has been comprehensively investigated in the past. The phenylpropanoid pathway, an important pathway in lignin synthesis, requires the participation of a series of enzymes. Enzymes in this process are associated with 4-coumarate: CoA ligase (4CL), cinnamate-4-hydroxylase (C4H), phenylalanine ammonia-lyase (PAL), caffeic acid O-methyltransferase (COMT), hydroxycinnamoyl-CoA shikimate/quinate hydroxy-cinnamoyl transferase (HCT), cinnamate-3-hydroxylase (C3H), caffeoyl shikimate esterase (CSE), caffeoyl-CoA 3-O-methyl-transferase (CCoAOMT), cinnamoyl-CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), and ferulate-5-hydroxy-lase (F5H). Briefly, phenylalanine is converted to p-coumaroyl-CoA via PAL, C4H, and 4CL, where p-coumaroyl-CoA is the branching point in the flavonoid pathway (Vogt, 2010). F5H and COMT convert coniferaldehyde to sinapaldehyde, and CCR triggers the reduction of feruloyl-CoA to coniferaldehyde (Li et al., 2005; Leple et al., 2007; Vanholme et al., 2008; Yan et al., 2019). And CAD is involved in the last enzymatic step of monolignol biosynthesis by reducing the substrates, coniferaldehyde, and sinapaldehyde, to the G and S monolignols (Yan et al., 2019). Moreover, it has been reported that most CAD enzyme-coding genes can influence plant growth by participating in lignin biosynthesis. In Gossypium hirsutum, Gh4CL-silencing and -overexpressing plants have a ~ 20% reduction and a ~ 10% increase in lignin content (Sun et al., 2020). In Populus trichocarpa, monolignol biosynthesis is influenced by the PtrCAD1-PtrCCR2 protein complex (Yan et al., 2019).
The first step of the lignin reduction reaction requires cinnamyl-CoA reductase, which employs five hydroxyl cinnamic acid coenzyme CoA esters (p-coumaryl-CoA, caffeoyl-CoA, feruryl-CoA, 5-hydroxyeruloyl-CoA, and sinapyl-CoA) as substrates to catalyze the production of cinnamaldehyde, leading to the generation of lignin (Lewis and Yamamoto, 1990; Goujon et al., 2003). Therefore, the cinnamyl-CoA reductase encoded by the CCR gene is the point of initiation for the lignin synthesis pathway (Lacombe et al., 1997; Humphreys and Chapple, 2002; Larsen, 2004). The carbon flux toward lignin is regulated by the CCR-coding enzyme, thus, it is a suitable target to alter lignin levels (Lacombe et al., 1997). Due to their key roles in monolignol biosynthesis, CCRs have been cloned and characterized in many plants, including the monocotyledons, Zea mays (Pichon et al., 1998), Triticum aestivum (Ma, 2007), Lolium perenne (Tu et al., 2010), and Lilium Oriental Hybrids (Li et al., 2009), as well as the dicotyledons, Caragana korshinskii (Li et al., 2014), Salvia miltiorrhiza (Wang et al., 2012), Arabidopsis thaliana (Lauvergeat et al., 2001), Populus tomentosa (Chao et al., 2017), and Betula platyphylla (Wei, 2012), etc.
CCR genes are preferentially expressed in the stems or roots of plants (Pichon et al., 1998; Lauvergeat et al., 2001; Larsen, 2004), and have thus been thought to be involved in lignification. Moreover, manipulation of the CCR expression typically results in a large variation in the lignin content and composition. Plants with heavily down-regulated CCR expression often exhibit stunted growth and delayed development, along with altered carbon fluxes across lignin and other metabolic pathways (Yin et al., 2021). The overexpression of BnCCR1 and BnCCR2 has been reported to increase lignin content in stems and roots of Brassica napus, which improved the lodging resistance in transgenic BnCCRox lines (Yin et al., 2021). Su et al. reported that PbCCR1/2 are related to lignin biosynthesis in overexpression transgenic plants (Su et al., 2019). A similar phenotype was observed in birch; Zhang et al. reported that the overexpression of BpCCR1 increases lignin content in transgenic plants (Zhang et al., 2015). Giordano et al. demonstrated that the spatiotemporal expression pattern of CCR1 cDNAs from Paspalum dilatatum correlates with the developmental profile of lignin deposition (Giordano et al., 2014). Compared to the wildtype (WT) plants, the lignin content in A. thaliana mutant irx4 plants is established to reduce significantly (50% of that in the WT plants), leading to abnormal growth of the irx4 plants (Smith et al., 2017). Based on previous studies, regulating the expression of the CCR gene may be an effective way to change the lignin content in plants. As more plant genome resources become available, genome-wide surveys will enable systematic characterizations of key enzymes and their corresponding family members. Identification and functional analysis of lignin biosynthesis enzymes and their associated genes will lay a foundation for the systematic analysis of carbon flux through lignin metabolism (Wang et al., 2019).
Liriodendron, a tertiary relic genus, belongs to the Magnoliaceae family. At present, there are only two natural species in this genus, L. tulipifera L and L. chinense (Hemsl.) Sarg. L. tulipifera is distributed throughout eastern North America, while L. chinense is scattered in southern China and northern Vietnam (Yang et al., 2021). Due to their fast growth rate, strong stress resistance, and good wood quality, Liriodendron trees are widely cultivated for use in timber, furniture, and paper-making industries. Lignin is closely related to wood quality, and CCR is a key enzyme-encoding gene in lignin synthesis (Chao et al., 2017). Nevertheless, little is known about the key genes involved in lignin biosynthesis in L. chinense. Therefore, this study aims to identify LcCCR gene family members in L. chinense and analyze their potential roles in lignin synthesis. Our results revealed that the LcCCR13 gene participates in lignin synthesis and is useful for the elucidation of the mechanism of wood formation in L. chinense.
Materials and methods
Plant materials
All plant materials were obtained from a provenance test plantation of L. chinense in Xiashu Forest Farm, Jurong City, Jiangsu Province (119°13′E, 32°7′N), China. The provenance of the sample tree was the Lushan Natural Reserve, Jiangxi Province (116°0′E, 29°32′N). From March to July, 2021, we collected leaves, shoots, roots, petals, and stems, which were quickly frozen in liquid nitrogen and stored in the refrigerator at –80°C before RNA extraction.
Nicotiana alata seedlings were sterilized with 10% NaClO for 15 min and then sown in 1/2 MS medium. After 2 days of vernalization (4°C dark), the seedlings were placed in an incubator (22°C with 16-h light and 8-h dark photoperiod) for a week. They were then transplanted into a medium for culturing.
Identification and characterization of LcCCRs
We downloaded the Liriodendron Genome resources from the NCBI database (https://www.ncbi.nlm.nih.gov/; PRJNA418360; Chen et al., 2019). Thirteen AtCCRs that were identified to be involved in the biosynthesis of lignin precursors in A. thaliana were obtained from the TAIR databases (https://www.arabidopsis.org/). Thirteen AtCCR proteins (Table S1) were used as alignment sequences to perform BLAT alignment with the protein database of L. chinense. The E-value was set to 0.001 to obtain the candidate CCR sequences of L. chinense. All candidate CCR sequences were assessed based on the presence of the conserved domain with InterPro (http://www.ebi.ac.uk/interpro/search/sequence/) and CDD search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) procedures. Finally, sequences with complete CCR domains were selected, and the sequences with more than 97% similarity between the different databases were deleted. The physical and chemical characteristics of LcCCR proteins identified from the L. chinense genome were predicted by ProtParam online tool (https://web.expasy.org/protparam/; Wilkins et al., 1999). The WoLF PSORT (https://wolfpsort.hgc.jp/) and TargetP-2.0 Server (http://www.cbs.dtu.dk/services/TargetP/) were used to predict the subcellular localization of the LcCCR proteins.
Multiple alignment and phylogenetic analysis
To group the CCR proteins in L. chinense, we analyzed the phylogenetic relationships between L. chinense and other plants. We constructed a phylogenetic tree using the CCR protein sequences from A. thaliana, P. tomentosa, L. perenne, Oryza sativa, Z. mays, and L. chinense (Table S1). DNAMAN (version 6.0) software was used to perform multiple comparisons among these protein sequences, and a phylogenetic tree was constructed by the maximum likelihood method via MEGA (version 5.1) software. Substitution and site change rates were calculated using the Jones–Taylor–Thornton (JTT) model and the Gamma distributed with Invariant sites (G+I) model. Bootstrap analysis was performed with 1000 replicates to calculate the reliability of the phylogenetic tree. Finally, the network profile of the phylogenetic tree was visualized by Evoview (https://www.evolgenius.info/evolview/) (He et al., 2016). Last, we used DNAMAN (version 6.0) to compare the 13 candidate LcCCR genes with other bona fide CCR genes.
Conserved motif and gene structure analysis
To analyze the exon-intron structure of CCR genes, the annotation profile was retrieved from the L. chinense genome (Chen et al., 2019). Information on the introns and exons of the LcCCR gene family was visualized by GSDS (version 2.0) (http://gsds.cbi.pku.edu.cn/) online tool (Hu et al., 2014). The conserved motifs in LcCCRs were visualized by MEME (version 5.3, https://meme-suite.org/meme/tools/meme) online tool (Bailey et al., 2006). The parameters were set as follows: an optimum motif could contain no less than 6 and no greater than 200 residues; the maximum number of motifs allowed was 10 (Chen et al., 2020b). The results of these motifs were then visualized via the TBtools software (Chen et al., 2020a).
Prediction of cis-acting elements in the promoters of LcCCRs
To identify the cis-acting elements in the promoter sequences of the 13 CCRs in L. chinense, the 2000-bp upstream sequence of the start codon (ATG) in CCRs was analyzed. The types and numbers of cis-elements in LcCCRs were evaluated via the Plant CARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) online tool (Lescot et al., 2002).
Expression profile analysis of CCR genes in different tissues
To quantify the expression levels of LcCCR genes in the different tissues (leaf, shoot apex, and developing xylem at the stem (DXS)), we analyzed the expression profiles based on the transcriptome data (Unpublished data from our laboratory). The RPKM (reads per kilobase per million mapped reads) approach was used to represent the expression abundance of each LcCCRs. The TBtools (version 1.0) software was used to draw the gene expression heatmap (Chen et al., 2020a).
RNA extraction and RT-qPCR analysis of CCR genes
To identify genes involved in lignin biosynthesis, we conducted the RT-qPCR analysis of CCR genes in five tissues (leaves, shoots, roots, petals, and stems). The detailed protocols are as follows:
Total RNA was isolated from each sample using a total RNA isolation kit (TIANGEN, China) following the manufacturer’s protocol. RNA-free deoxyribonuclease (DNase I) was used to remove trace DNA from the extracted RNA. The integrity of the total RNA was detected via 1.0% agarose gel electrophoresis. The concentration and purity of the total RNA were analyzed by a NanoDrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Reverse transcription was performed using the Evo M-MLV RT Premix for qPCR (AG11706, ACCURATE BIOTECHNOLOGY, HUNAN, Co., Ltd). Quantitative primers for the CCR genes were designed using Oligo 7.0 software according to strict requirements (Table S2). The RT-qPCR was run in a StepOnePlusTM System (Applied Biosystems) as a 10-μL reaction mixture containing 5 μL of 2× SYBR Premix Ex Taq, 0.2 μL of 50× ROX Reference Dye (SYBR Green Premix Pro Taq HS qPCR Kit (AG11701, ACCURATE BIOTECHNOLOGY, HUNAN, Co., Ltd)), primers, and cDNA. The LcActin97 (Tu et al., 2019) and NtActin (AJ421411) primers were used for the amplification of the internal reference gene. To ensure the accuracy of the results, three biological replicates (where each biological replicate had three technical replicates) were conducted, and the data were examined using the 2−ΔΔCT method. Significance was determined by the t-test using the SPSS statistical software (version 20, IBM, New York, NY, USA; *p < 0.05, **p < 0.01).
Function validation of LcCCR13 using genetic transformation
LcCCR13 was retrieved from the existing L. chinense genome data (Chen et al., 2019). We designed the intermediate fragment-specific PCR primers via the Oligo (version 7.0) software (Table S2). Reverse-transcribed cDNA was used as a template for PCR to synthesize intermediate fragments. The PCR products were cloned into the pEASY-Blunt Zero Cloning Kit (Transgen Biotech, Beijing, China) and transformed into E. coli (DH5α). Finally, the amplicons were sequenced by Jie Li Biology (Shanghai, China).
The sequenced ORF of the LcCCR13 gene was inserted into the overexpression vector, which was digested with Xba I and BamH I QuickCut enzymes (Takara Biomedical Technology, Dalian, China). The construct was then transformed into Agrobacterium tumefaciens strain EHA105 by the freeze-thaw method (Holsters et al., 1978, Zhang et al., 2012). The target gene was transferred into WT tobacco by the leaf-disc conversion method. Six transgenic tobacco lines featuring independent recombination events were identified by molecular detection; they were all heterozygous with similar phenotypes. To detect the relative expression levels of LcCCR13 in the different lines, three transgenic lines together with the WT strain were subjected to a semi-quantitative PCR assay. The relative expression level of LcCCR13 was quantified from three biological replicates, where each replicate had three technical replicates.
Meanwhile, the histochemical staining of stem segments from transgenic tobacco was performed via the Phloroglucinol-HCL and safranin staining methods (Yang et al., 2022). The stained sections were observed under a Zeiss Axio microscope. The ImageJ software (version 2.3.0) was then used to calculate the size of the thickened and lignified cells (μm).
All the transgenic and WT plants were acclimated and grown in a greenhouse at 18–23°C with 60% humidity under a 16-h light and 8-h dark photoperiod at NJFU (Nanjing Forestry University, Nanjing, China).
Determination of plant height, CCR activity, and lignin content
The height and the diameter were measured in 2-month-old transgenic and WT plants. The measurements for each plant line included at least 5 replicates.
The CCR activity was determined following the method of Hamedan et al. (Hamedan et al., 2019).
According to the method of Bhaskara et al. (1999), a 10 mg powdered sample was weighed and ground into a homogenate in a mortar with 95% ethanol. The homogenate was then transferred to a 50-mL centrifuge tube for centrifugal separation at 4000 rpm for 5 min. The pellet thus obtained was washed twice with 5 mL of 95% ethanol and 6 mL of ethanol: n-hexane (1:2 (v/v)). The pellet was dried naturally. Six milliliters of 25% acetyl bromide solution in glacial acetic acid were used to dissolve the precipitate, followed by being covered and sealed in a water bath at 70°C for 30 min. Subsequently, 0.9 mL of 2 mol·L−1 NaOH, 5 mL of glacial acetic acid, 0.1 mL of 7.5 mol·L−1 hydroxylamine hydrochloride, and 15 mL of glacial acetic acid were added to the centrifuge tube. Instead of the substrate, distilled water was used for the same reaction as a control. The absorbance of the supernatant was measured at 280 nm using the GeneQuant pro ultraviolet spectrophotometer (Biochrom Ltd, Cambridge, UK), and the absorbance per gram of the dry sample at 280 nm represented the lignin content per gram (OD g−1 DW) (OD: Optical Density, DW: dry weight). The statistical significance of the data was estimated by analysis of variance (ANOVA); the Pearson coefficient was calculated using the SPSS statistical software (version 20, IBM, New York, NY, USA; *p < 0.05, **p < 0.01).
Data processing and statistical analysis
The statistical significance of the data using ANOVA was assessed using the SPSS statistical software (version 20, IBM, New York, NY, USA). Multiple comparative analyses were performed using the t-test (*p < 0.05, **p < 0.01). To identify differences among the samples or plant lines. One-way ANOVA followed by Duncan’s multiple comparisons test was used.
Results
Genome-wide identification of the LcCCRs in L. chinense
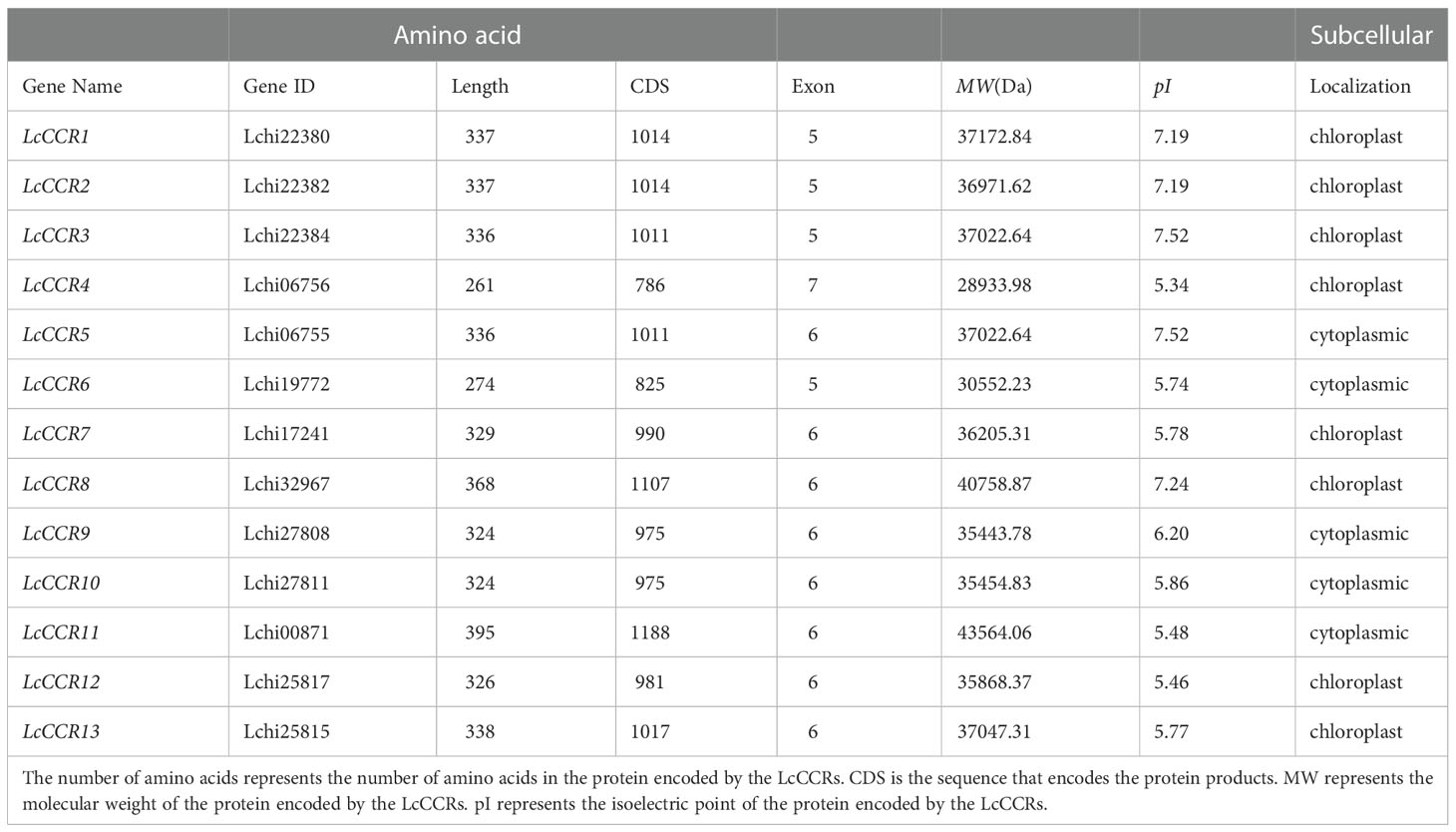
Thirteen LcCCRs were identified in this study containing the complete “X-W-Y-X-X” functional domain. Detailed characteristics of the 13 LcCCRs are presented in Table 1. These CCR genes were named LcCCR1-LcCCR13. Briefly, the length of the coding DNA sequences (CDS) ranged from 786 to 1188 bp with 5–7 exons in each, and the amino acid length ranged from 261 (LcCCR4) to 368 (LcCCR48) amino acids, with the number of amino acids being greater than 300, except for in two proteins (LcCCR4/6). In addition, the molecular weight (MW) of the proteins ranged from 28933.98 (LcCCR4) to 43564.06 (LcCCR11) Da, where most protein’s MW was greater than 30.00 kDa. The isoelectric points (pI) varied from 5.34 (LcCCR4) to 7.52 (LcCCR3/5). The average amino acid number, MW, and pI of these protein sequences were 330, 36309.11 Da, and 6.33, respectively. We used WoLF POST and online analysis with the TargetP-2.0 Server to predict the subcellular localization of the LcCCR proteins. Both online tools indicated that LcCCR proteins are localized in the cytoplasm or the chloroplast (Table 1).
The subcellular localization was predicted by online tools: WoLF PSORT (https://wolfpsort.hgc.jp/) and TargetP-2.0 Server (http://www.cbs.dtu.dk/services/TargetP/). The physical and chemical characteristics of LcCCR proteins identified in the L. chinense genome were predicted using the online tool, ProtParam (https://web.expasy.org/protparam/).
Multiple sequence alignment and phylogenetic analysis of the LcCCR gene family
To understand the sequence characteristics, a multiple sequence alignment analysis of the 13 LcCCR proteins was performed by DNAMAN software with default parameters. Two AtCCR proteins (AtCCR5/9) and three PtoCCR proteins (PtoCCR1/4/7) were selected as representatives for further comparison. The conserved domain structures in LcCCRs are displayed in Figure 1. It was predicted that the amino acid sequences in each member of the LcCCRs were significantly different from other CCR proteins, but the C-terminal and the middle sequences were highly conserved. Almost all of the above sequences contained the conserved domain, “X-W-Y-X-X,” from the cinnamyl-CoA reductase family of proteins and the recognition domain of NAD(P)-dependent short-chain reductase (SDR). The structure also included the typical “Rossmann fold” and the conserved catalytic center, the Ser-Tyr-Lyr triad (Kallberg et al., 2002).
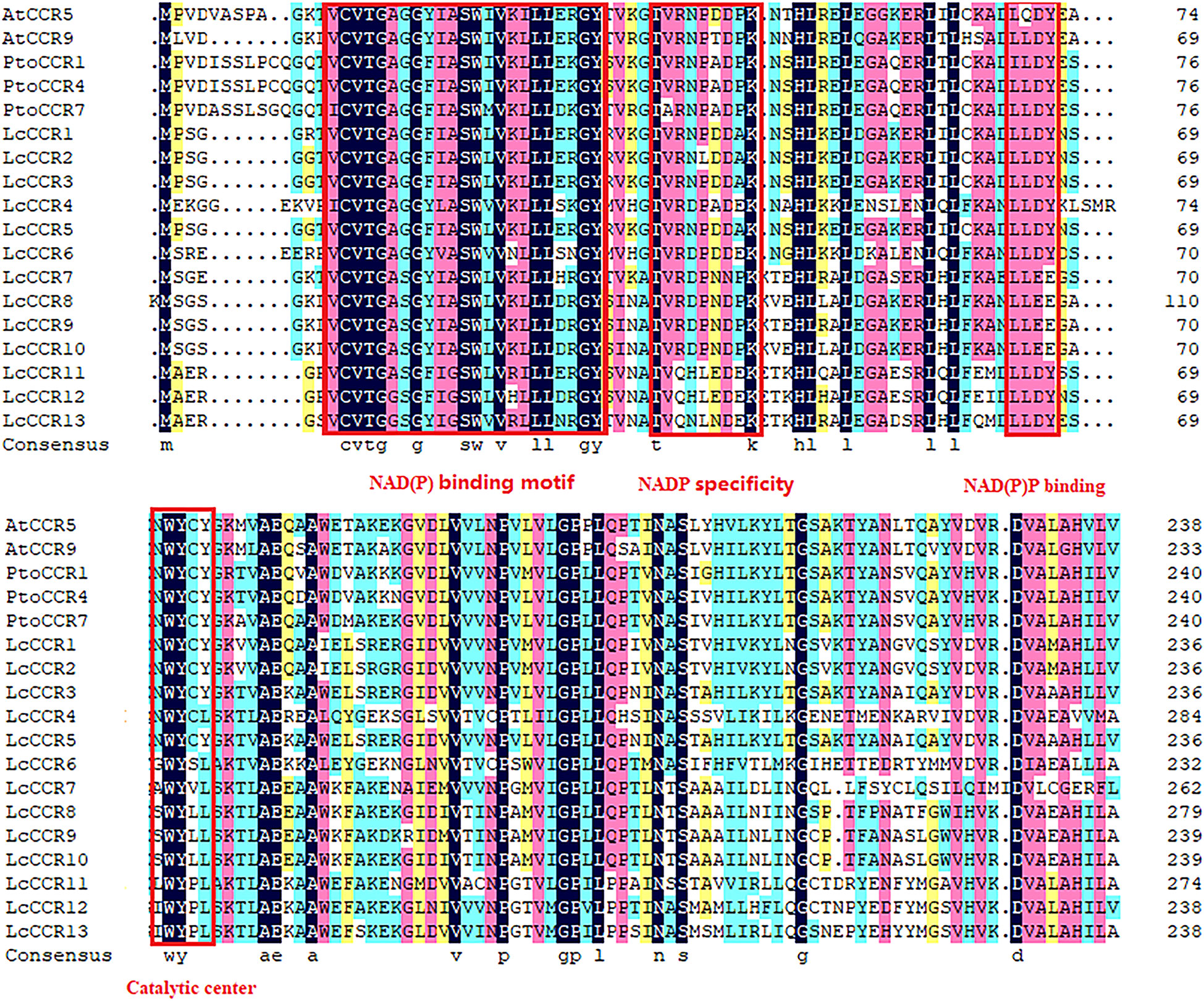
Figure 1 Characteristics of the amino acid sequences of LcCCRs (LcCCR1-13) and other CCR proteins (AtCCR5: AT1G15950; AtCCR9: AT1G80820; PtoCCR1: KP281597; PtoCCR4: KP281599; PtoCCR7: KF145198). Multiple sequence alignment was performed by DNAMAN (version 6.0). The conservative functional domains: NAD(P) binding motif (VCVTGAGGXIASWXVKXLLXXGY), NAD(P) specific site (TVRNPDDPK), NAD(P) binding (LLLDY), and catalytic center were surrounded by red boxes (NWYCY).
To clarify the phylogenetic relationship between the LcCCRs and other CCR proteins, an unrooted phylogenetic tree was constructed with 59 CCR proteins (13 LcCCR, 13 AtCCR, 11 PtoCCR, 10 LpCCR, 10 OsCCR, and 2 ZmCCR proteins) based on the maximum likelihood method. In the phylogenetic tree, all 59 CCRs mentioned above could be roughly divided into four groups (Groups I to IV; Figure 2). Our results showed that Group I was divided into three subgroups: subgroups Ia, Ib, and Ic. Subgroup Ia contained 14 CCR members (3 LcCCRs, 2 AtCCRs, 3 LpCCRs, 3 PtCCRs, 2 OsCCRs, and 1 ZmCCRs), which were related to lignin biosynthesis [AtCCR5/9 (Goujon et al., 2003), ZmCCR1 (Pichon et al., 1998), PtCCR1/4/7 (Chao et al., 2017)]. The entire Group I comprised CCRs from dicotyledons and monocotyledons. Subgroup Ib contained 8 CCR members (4 LpCCRs, 3 OsCCRs, and 1 ZmCCR). Subgroup Ic contained 8 CCR members (3 LpCCRs and 5 OsCCRs). Subgroups Ib and Ic consisted only of CCR proteins from the monocotyledons. Group II, shown in blue color, contained 5 CCR members (3 LcCCRs and 2 PtoCCRs). Group III in green contained 8 CCR members (3 LcCCRs, 3 PtoCCRs, and 2 AtCCRs). Group IV in pink contained 16 CCR members (4 LcCCRs, 4 PtoCCRs, and 8 AtCCRs) (Figure 2). Notably, most CCRs were evenly distributed in four groups (Barakat et al., 2011). Moreover, it was found that the LcCCRs had close phylogenetic relations with their ancestors’ species in each group. The distribution of CCR proteins in the phylogenetic tree suggested that CCR proteins were duplicated multiple times before becoming specific to the monocotyledon and dicotyledon species in which they were observed.
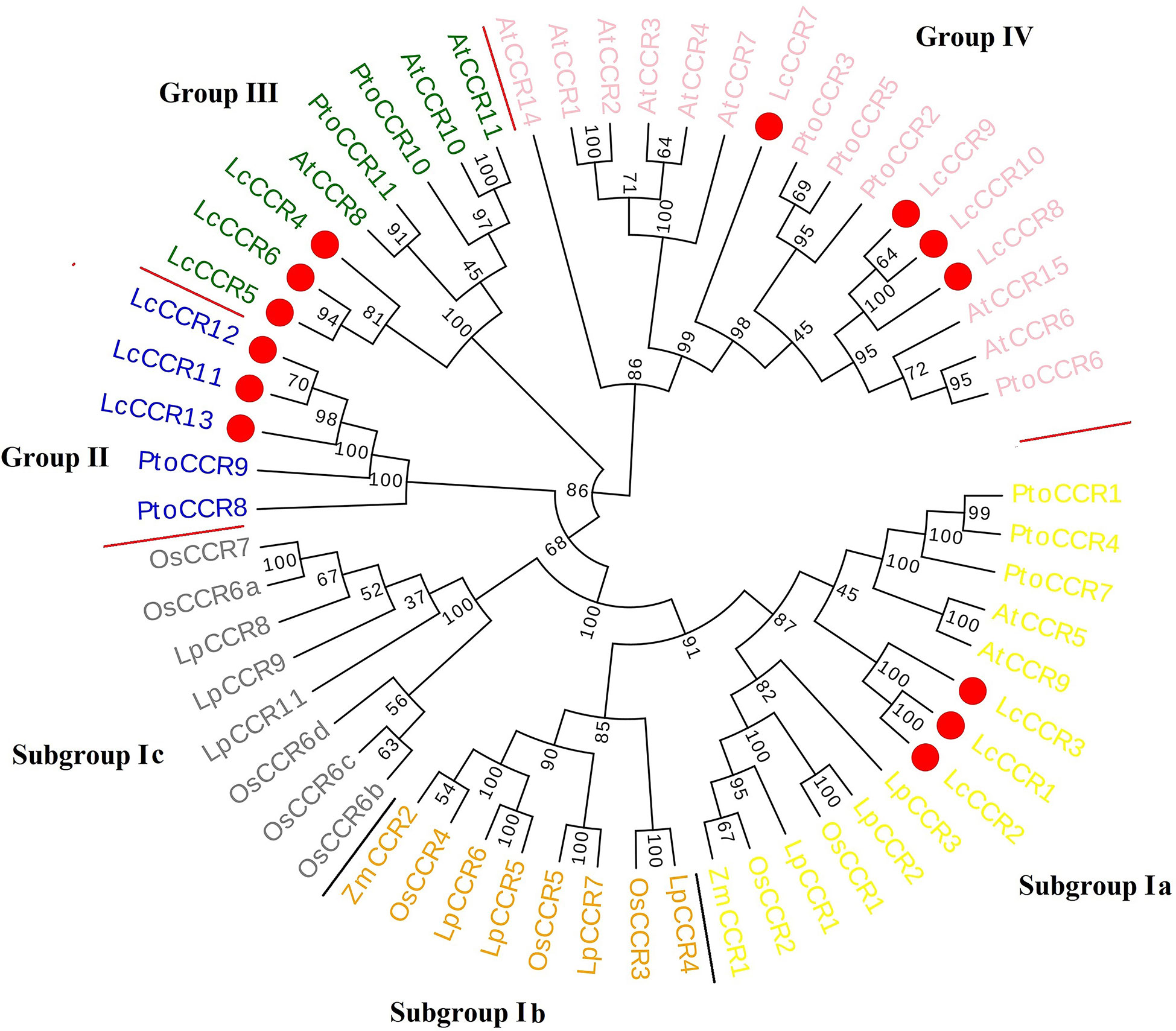
Figure 2 Phylogenetic tree of CCR protein sequences from various plant species. Circular phylogenetic tree of CCR protein sequences of various plants. The phylogenetic tree consisted of 59 CCR proteins (13 LcCCRs, 13 AtCCRs – A. thaliana, 11 PtoCCRs – P. tomentosa,10 LpCCRs – L. perenne, 10 OsCCRs – O. sativa, 2 ZmCCRs – Z. mays). Protein sequences were aligned and a phylogenetic tree was constructed using MEGA5.1 with the maximum likelihood method and visualized by Evoview software. Substitution and site change rates were calculated using the Jones–Taylor–Thornton (JTT) model and the Gamma distributed with Invariant sites (G+I) model. Bootstrap analysis was performed on 1000 replicates to calculate the reliability of the phylogenetic tree. Bootstrap values are shown at each branch as percentages. The red circle indicated LcCCR protein sequences; different color backgrounds and red lines distinguish the different groups. The phylogenetic tree consists of four groups: Group I (Subgroup Ia – yellow, Subgroup Ib – orange, Subgroup Ic – grey), Group II – blue, Group III – green, and Group IV – pink. The higher value on the branch of the evolutionary tree, the higher reliability of this branch.
Gene structures and conserved motif composition in the LcCCR gene family
The exon-intron configurations in the LcCCR genes were examined to further explore the probable structural evolution of this family of genes (Figure 3A). Similar structures were usually observed in the same group. e.g., the members of Group II contained 5 exons, and the members of Group III/IV contained 6 exons. The length and number of introns were generally similar in each group, which was consistent with the clusters of LcCCRs. However, there were some exceptions. For example, the members of Group I did not have a similar exon-intron pattern.
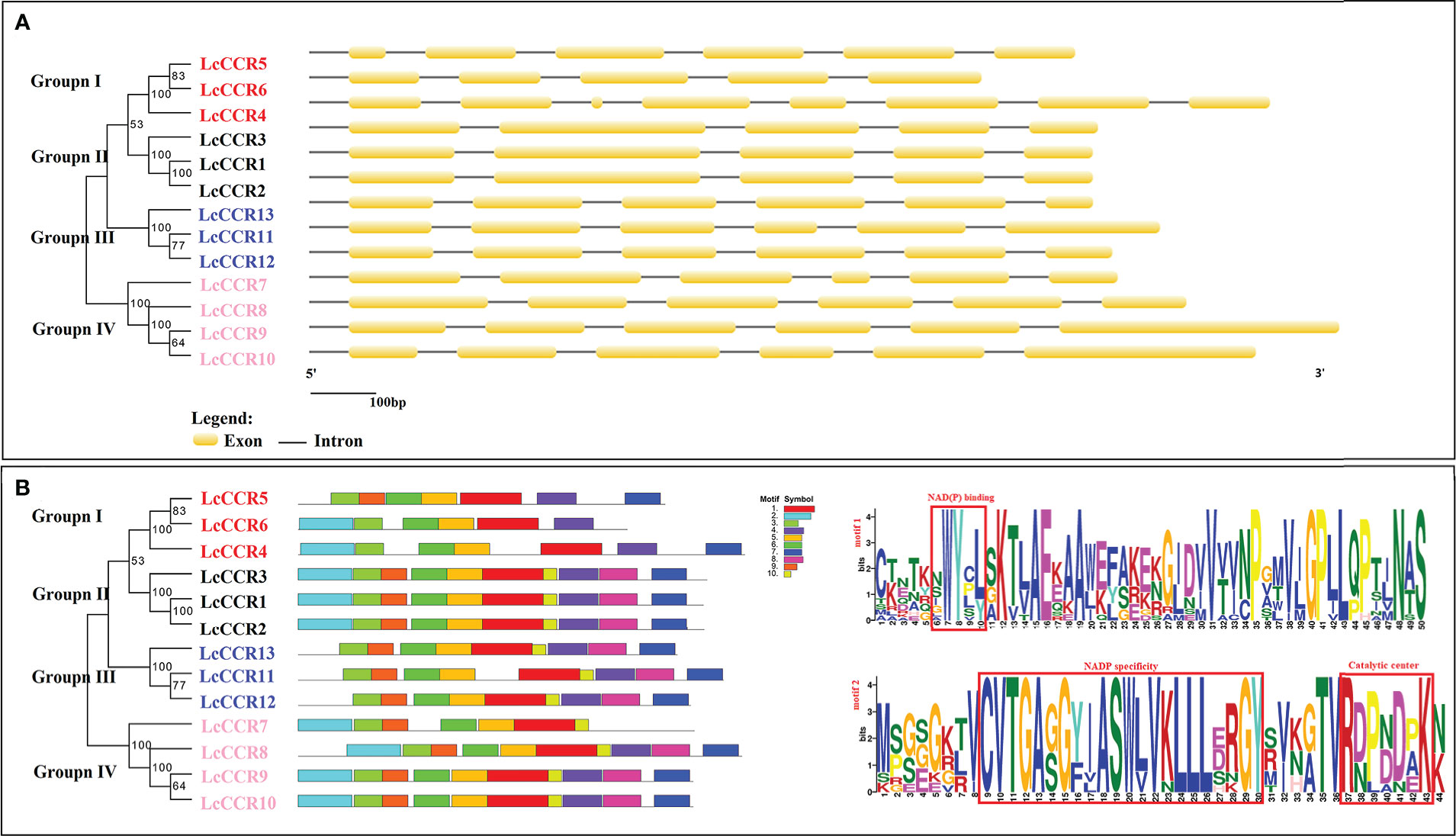
Figure 3 Map of the gene structures and conserved motifs patterns in L. chinense CCR proteins. The phylogenetic relationship was based on LcCCRs. According to phylogenetic relationships, 13 LcCCRs were divided into four distinct groups (I–IV). Different subfamilies were represented by different colors and names. The red background denotes Group I; The grey color background denotes group II; The blue background denotes Group III; The orange color background denotes Group IV. (A) The exon–intron structure (visualized by the GSDS online tool) of LcCCR proteins. The yellow squares indicate exons, and the black lines indicate introns. (B) Distribution of 10 conserved motifs (visualized by MEME online tool) in the LcCCR proteins. Differently colored squares indicate different motifs. The bottom scale shows the protein length, and the details of two crucial conserved motifs in the LcCCR proteins are shown at the back of the scale. Motif 1: CKETKNWYCLSKTLAEKAAWEFAKEKGJDVVTVNPGMVJGPLLQPTJNAS, contains the NAD(P) binding; Motif 2: MPGEGKTVCVTGAGGYIASWLVKLLLERGYSVKGTVRDPBDPKN, contains the NAD(P) specific site and the catalytic center. More detail of the motif sequences is presented in Table S3.
Furthermore, we investigated the full-length protein sequences of the 13 LcCCRs to identify their conserved motifs. 10 conserved motifs were predicted to be present in 13 LcCCRs. The number of amino acids in the 10 motifs ranged from 11 to 50 (Table S3). The patterns of the conserved motifs are shown in Figure 3B. As motif 1 constituted the main structure of the CCR domain (“NWYCY”), it was identified in all LcCCR proteins. LcCCR proteins in the same group tended to have similar motif compositions, suggesting that protein structures were conserved in a specific subfamily (Song and Peng, 2019). Furthermore, only the members of Group I of LcCCR proteins and LcCCR7 did not contain Motif 8. We speculate that LcCCR7 may have a deletion, leading to the lack of this discrepancy. Overall, members within each group shared the same genetic structure, which was consistent with their phylogenetic relationships. The stability of group classifications was maintained as per our findings from surveys on the conserved motif compositions, gene structures, and phylogenetic relationships.
Cis-acting element prediction in the promoters of 13 LcCCR genes
To explore gene function and regulation patterns, we surveyed the cis-elements in a region 2000 bp upstream of the initiation codon in each LcCCR gene. The cis-acting elements of the LcCCR genes were then predicted by searching the promoter sequences from the PlantCARE database (Song and Peng, 2019). As shown in Figure 4, almost all LcCCR gene promoters contained an AC element (Raes et al., 2003), along with the methyl jasmonate (MejA), salicylic acid (SA), and abscisic acid (ABA) elements. Some promoters contained light response seed-specific regulation (Chen et al., 2020b) or low-temperature response elements. The number of cis-acting elements for each of the 13 LcCCR genes ranged from 4 to 22. Promoters of different genes in the same group contained similar elements with only minor differences.
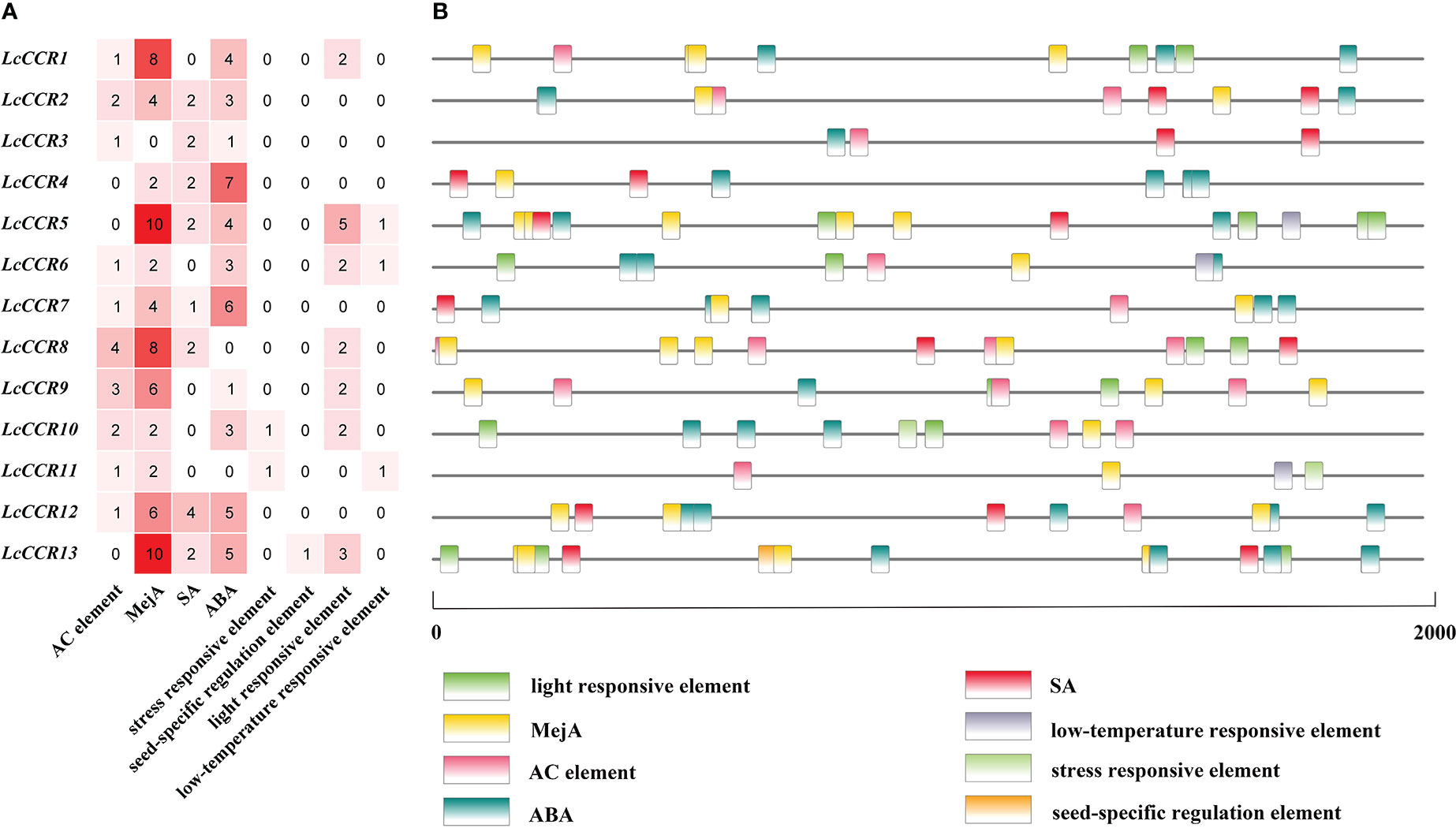
Figure 4 Analysis of cis-elements in LcCCR promoters (2000 bp upstream of the translation start site). (A) Number of distinct cis-elements in each LcCCR promoters. (B) location of distinct cis-elements in each LcCCR promoters. The figure shows the cis-elements that were related to different stress and hormone responses in the putative promoters of LcCCRs. All LcCCR gene promoters mainly contained the following eight elements: AC element, MejA (cis-acting regulatory element involved in the MeJA-responsiveness), SA (cis-acting element involved in salicylic acid responsiveness), ABA (cis-acting element involved in the abscisic acid responsiveness), stress responsive element, seed-specific regulation element, light-responsive element, and low-temperature element. Detailed information on cis-acting elements is given in Table S4.
The promoter sequences of most genes (70%) had AC elements, such as LcCCR1/2/3/6/7/8/8/10/11/12 (Figure 4A), suggesting that these genes may be involved in the regulation of phenylpropanol metabolism and other unknown functions. In addition, these cis-elements are evenly distributed in the promoter region of LcCCRs (Figure 4B).
Cis-acting elements, such as ABRE, TCA-element, and TGACG-motif, are related to the signaling pathways of ABA, SA, and MejA, respectively. All of these metabolites are related to stress resistance in plants. Almost all promoters contain these three cis-acting elements, therefore, it can be reasonably inferred that some LcCCRs might be involved in plants’ responses to biotic and abiotic stresses.
Other promoters contained cis-elements for seed-specific regulation, light response, and low-temperature response, indicating that they may have a certain influence on the seed. The low-temperature response was related to the light response. Detailed information is shown in Table S4.
Tissue expression patterns of LcCCRs revealed by RNA-seq data
We investigated the expression patterns of LcCCRs in different tissues (leaf, shoot apex, and DXS) with Illumina RNA-seq data (Unpublished data from our laboratory). The results showed that the 13 LcCCR genes were expressed in all three tissues but some genes exhibited tissue-specific expression. For instance, LcCCR1/3/4/6/9 were highly expressed in the leaf. While in the shoot apex, the expression levels of LcCCR2/11 were the highest. Meanwhile, LcCCR8 displayed high expression in the leaf and DXS. Furthermore, it was found that LcCCR5/7/10/12/13 had the highest expression in the DXS (Figure 5). Collectively, the different expression patterns of LcCCRs suggest that they may play different roles in the various developmental stages/tissues in L. chinense.
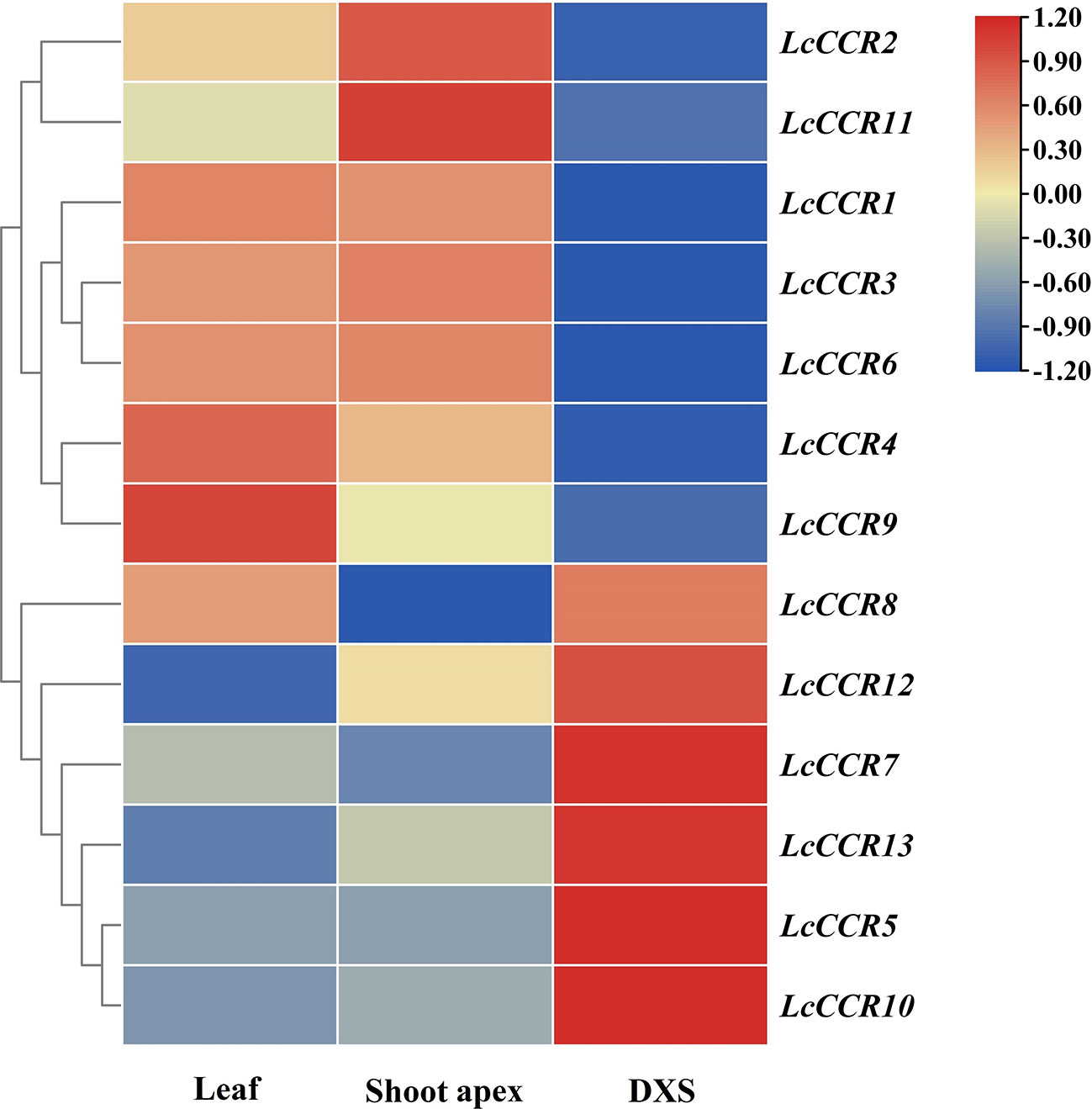
Figure 5 Tissue expression patterns of 13 CCR genes across various tissues in L. chinense based on FPKM values from transcriptomic data. Heatmap showing the relative expression levels of each LcCCRs in three tissues, including leaf, shoot apex, and DXS (developing xylem at stem) (based on RNA-seq data which will be released in another paper). FPKM (fragments per kilobase of transcript per million fragments mapped) values shown were standardized by the Z-score, and the heatmap was constructed by TBtools (version 1.0) software. The x-axis shows the different tissues, and the y-axis shows the different genes. The red box indicates a relatively high expression level, while the blue box indicates a relatively low expression level.
Tissue expression specificity analysis of LcCCR5/7/10/12/13
To obtain insights into the potential roles of LcCCR5/7/10/12/13, which were predicted to be highly expressed in DXS (Figure 5), the expression level of LcCCR genes was further determined by RT-qPCR in five tissues (root, stem, leaf, shoot, and petal). The results showed that the LcCCR5/7/10/12/13 were expressed in all the tested tissues, but their expression levels were different (Figure 6). The relative expressions of LcCCR5/10/12 were similar in all tissues with the highest expression level in the root. Moreover, the relative expression level of LcCCR7 in the petal was the highest. Furthermore, LcCCR13 had the highest expression level in the stem, too, which was extremely significantly different from the expression in the root, leaf, petal, and shoot. Although the expression of the LcCCR13 gene was not xylem-specific, it was mainly expressed in lignin-forming tissues, such as stem, suggesting that LcCCR13 may play an important role in lignin biosynthesis in L. chinense.
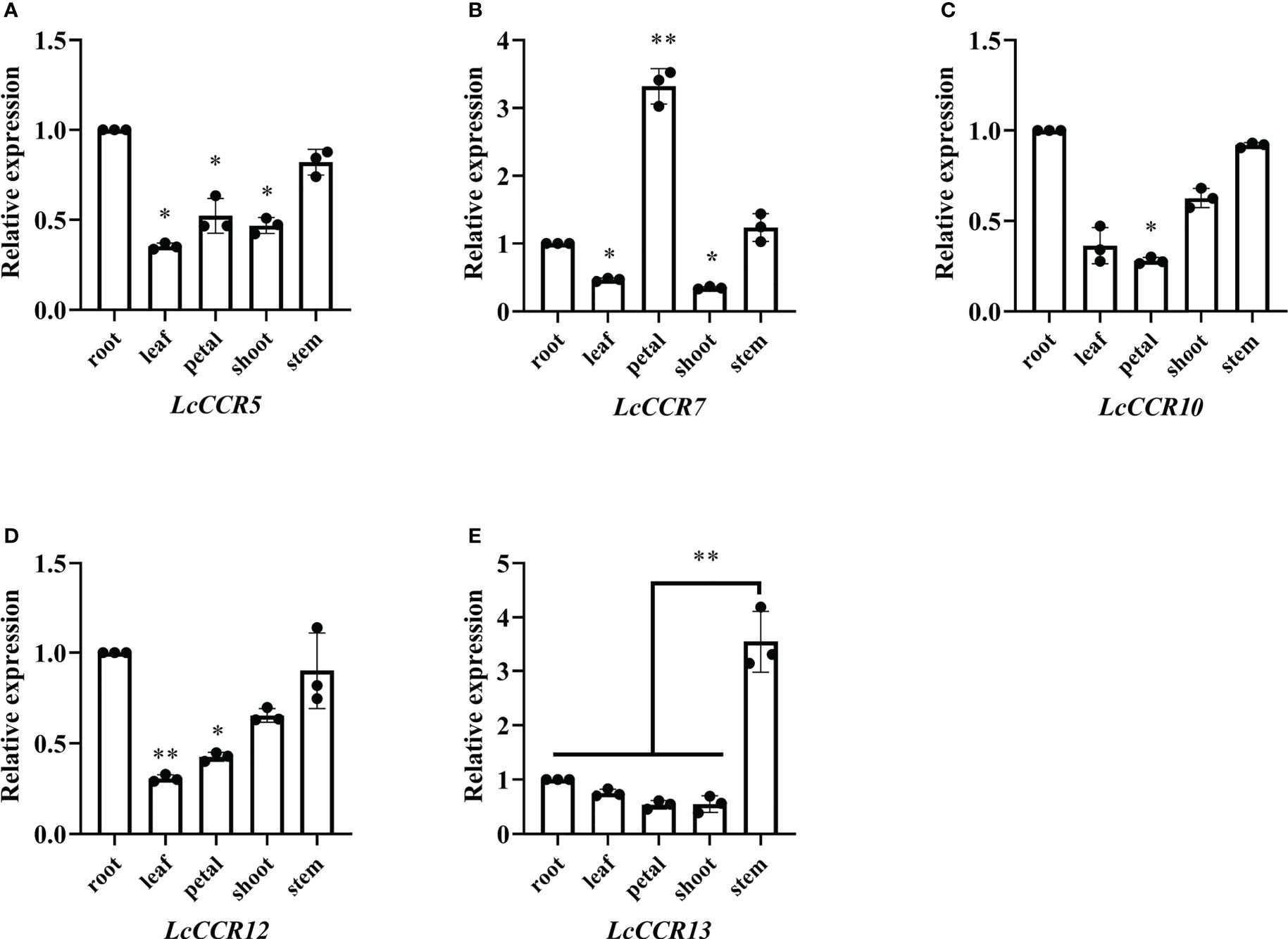
Figure 6 Tissue expression specificity analysis of LcCCR5/7/10/12/13. (A–E) The x-axis represents different tissues (root, stem, leaf, petal, and shoot). The value of gene expression in root was set to 1. Tubulin was used as the internal control. Tissue-specific expression analyses were performed using real-time qPCR; the values are presented as means ± SD from three biologically independent replicates (each biological replicate had three technical replicates). Gene expression profiles were evaluated by the 2−ΔΔCT method. The y-axis represents the relative expression level. Asterisks above the bars in figures indicate significant differences (Student’s t-test; *p < 0.05, **p < 0.01) as compared to the roots (LcCCR5/7/10/12)/stems (LcCCR13). The raw data are marked as a point on the histogram. The error bars in the charts indicate the standard deviation from the mean of the triplicate treatments.
Growth and morphological characteristics of LcCCR13 transgenic tobacco
To further investigate the function of the LcCCR13 gene in lignification, a transgenic assay was used. Tobacco leaves were transformed with the 35s::LcCCR13 vector to induce high expression of the gene (Figures 7A, B); six LcCCR13 positive lines were thus obtained (Figure 7C). To investigate the influence of the LcCCR13 gene on plant growth, the WT controls and the three transgenic tobacco lines were monitored. Significant differences were observed in the growth phenotypes of the transgenic lines and the WT plants (Figures 8A, B). For example, there was a significant height difference between the L4 lines and the WT plants (0.7 fold; Figure 8C). We found that the transgenic lines had a significantly greater stem diameter than the WT (1.6 fold; Figure 8D). All in all, transgenic lines exhibited altered growth characteristics as compared to the WT plants.
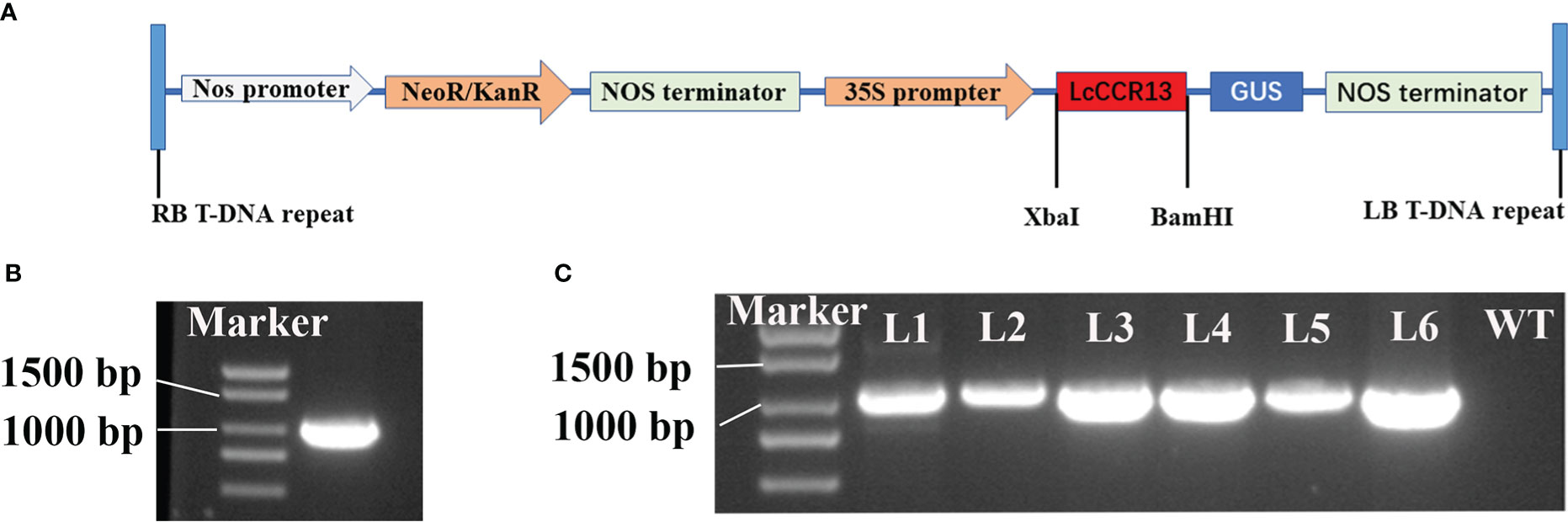
Figure 7 The genetic transformation of the LcCCR13 gene. (A) Schematic diagram of the overexpression vector construction. 35S: cauliflower mosaic virus 35S promoter. GUS: β-glucuronidase. Nos terminator: nopaline synthase gene terminator. KanR: Kana resistance gene. RB and LB: the right and left borders, respectively. (B) ORF cloning of the LcCCR13 gene, and the length of LcCCR13 was 1017 bp. (C) Verification of LcCCR13 tobacco transgenic lines by PCR amplification. DNA was extracted from the leaves of 2-month-old tobacco plants. The marker length was 2000 bp.
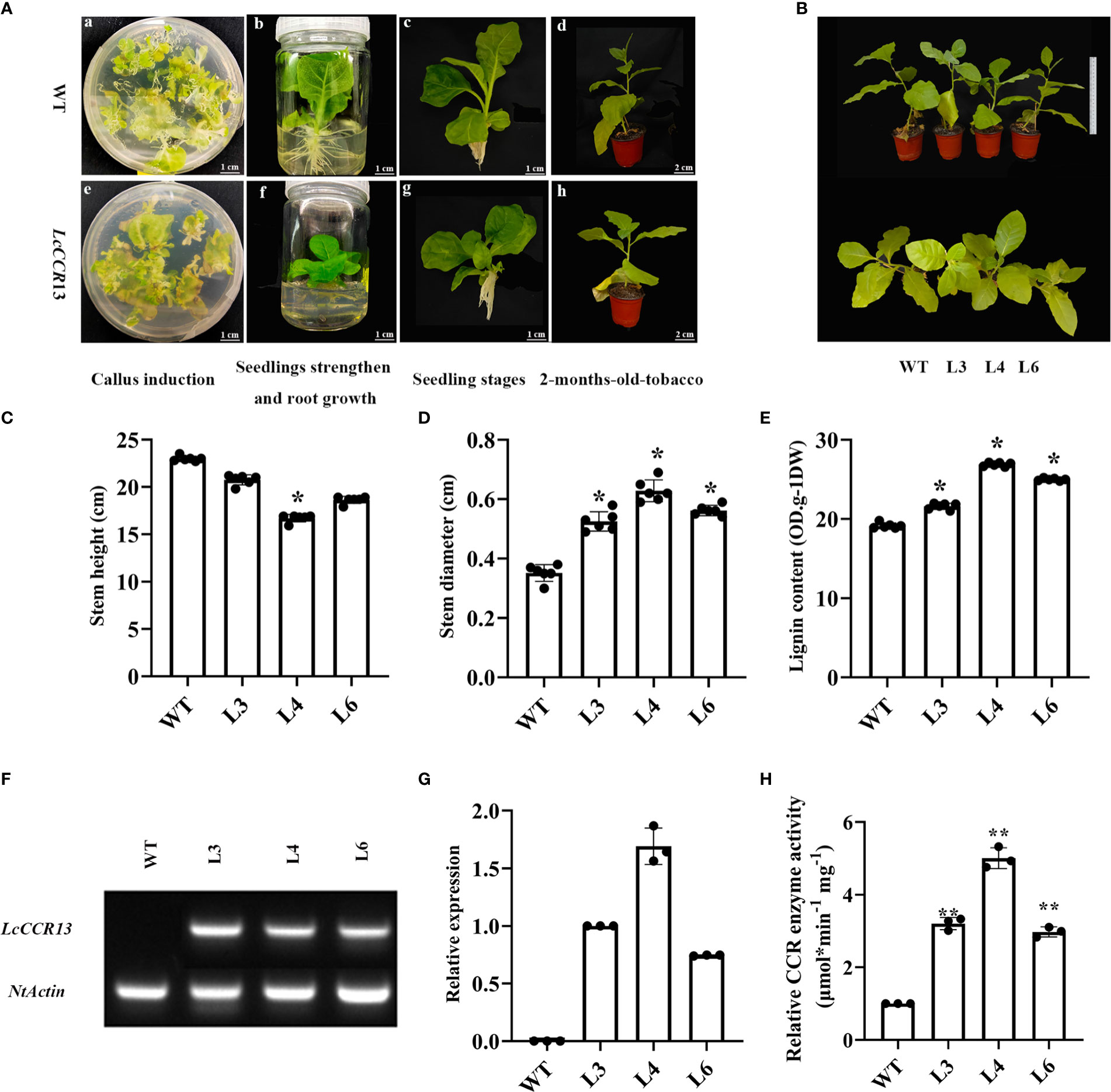
Figure 8 The phenotypic and physiological traits of transgenic tobacco. (A) The different growth stages of the WT and transgenic tobaccos. The a–d figures show the representative images of wild-type (WT), and the e–f figures show the representative images of LcCCR13 transgenic tobaccos. The a and e figures show the periods of callus induction. The b and f figures show the periods of seedlings’ strengthening and root growth. The c and g figures show the seedling stage. The d and h figures show the periods of 2-month-old tobacco. Four diverse mediums included: differentiation medium (MS + 0.05 mg/L NAA + 2.0 mg/L 6-BA + 40 mg/L Kan + 150 mg/L TMT), selective medium (MS + 0.05 mg/L NAA + 2.0 mg/L 6-BA + 40 mg/L Kan + 200 mg/L TMT), seedling strengthened medium (MS + 0.05 mg/L NAA + 2.0 mg/L 6-BA + 40 mg/L Kan + 250 mg/L TMT), and root growth medium (1/2 MS + 250 mg/L TMT), respectively. Bar = 1 cm (a-c, e-g); Bar = 2 cm (d and h). (B) The growth conditions of the WT and the transgenic tobacco lines (L3, L4, L6) for 3 months. Bar = 20 cm. (C) Stem height, (D) Stem diameter, and (E) lignin content of the WT and 2-month-old transgenic tobacco lines from six biological repeats. (F) A semi-quantitative (NtActin was detected in all the transgenic lines and WT; LcCCR13 was detected in all the transgenic lines except WT) (G) RT-qPCR assay of LcCCR13 with WT, L3, L4, and L6 lines (the expression level of LcCCR13 in L3 was set to 1). (H) The relative activity of CCR enzyme in WT and transgenic plants (The activity of CCR of WT was set to 1, and the value of L3/4/6 CCR activity were calculated when compared to the value of WT). There were three biologically independent replicates, where each biological replicate had three technical replicates. Error bars in the charts indicate the standard deviation from the mean of each repeat. Asterisks indicate significant differences between the transgenic tobacco and WT plants (Student’s t-test; *p < 0.05, **p < 0.01). The data were the Mean ± standard error.
LcCCR13 overexpression positively regulates lignin synthesis
A significant increase was noted in the lignin content of the transgenic lines (1.3 fold; Figure 8E), and the L4 lines were selected for histochemical staining due to the highest LcCCR13 gene relative expression in them (Figures 8F, G). The difference in the CCR activity of transgenic lines was measured. We found that the relative activity of CCR in transgenic plants was significantly higher than that in the WT plants, and the L4 lines had the highest CCR activity compared to other transgenic lines (Figure 8H).
When observed under higher magnification, the tissue sections of the L4 lines showed a more highly developed xylem with wider and deeper brown color than in the WT plants (Figures 9A–F). Furthermore, the size of the thickened and lignified cells in transgenic tobacco was 306.20 μm, and they were ~1.75× (p < 0.01) as thick as the WT cells (174.52 μm; Figure 9G). Therefore, higher lignin deposition occurred in the L4 lines than in the WT plants. These results suggest that the overexpression of LcCCR13 might positively regulate lignin synthesis.
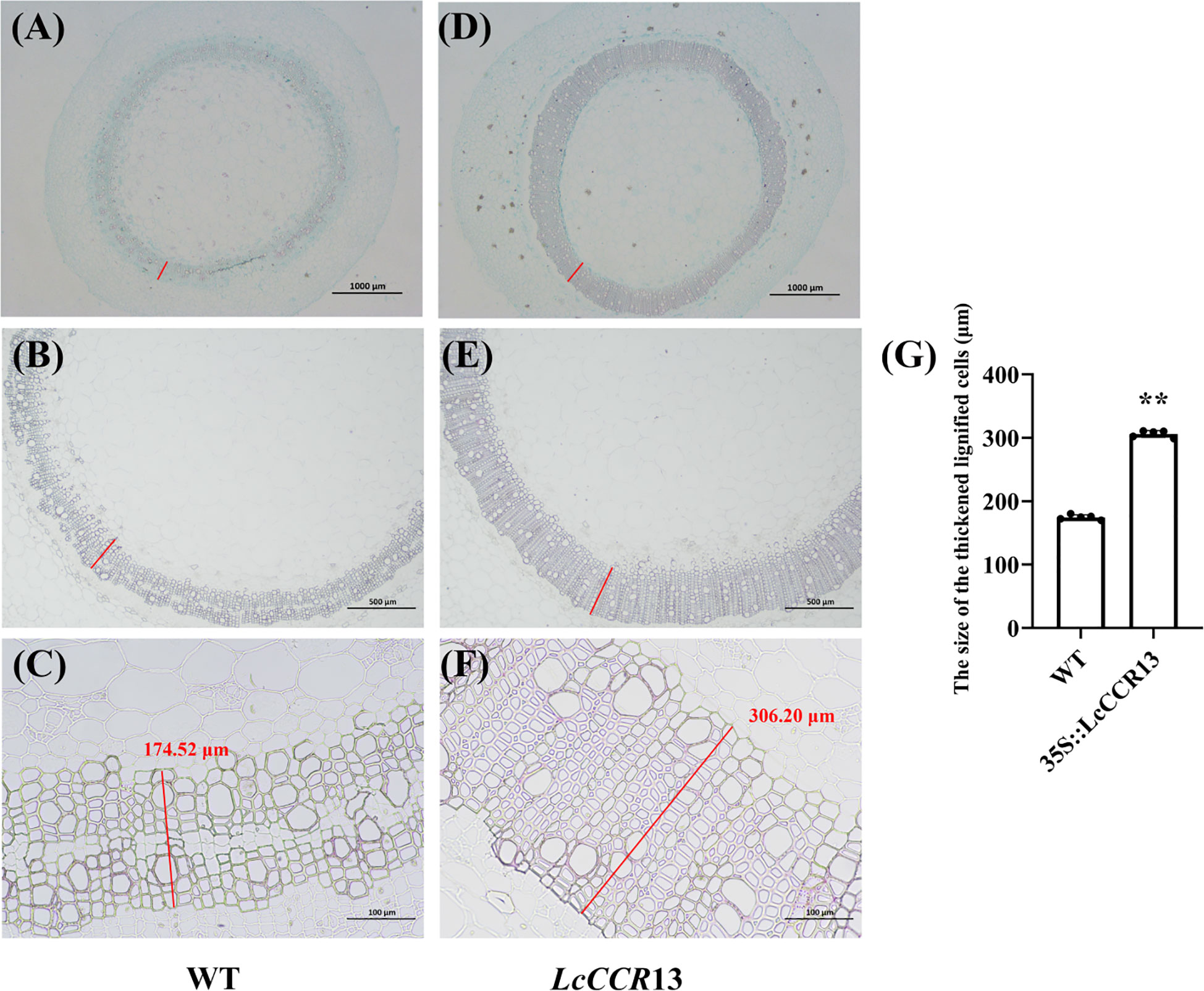
Figure 9 Sectioned and microscopic observation of the wildtype (WT) and transgenic tobacco stem. Sections stained with phloroglucinol-HCl and safranin of transgenic tobacco stems. (A-C) WT tobacco; (D-F) Transgenic tobacco lines (L4). The brown region represents the site of lignin deposition. (A, D) Cross sections of 2-month-old L4 and WT plants with the 2nd internode (IN). The cross sections were stained with safranin O and Fast Green. Bar, 1000 μm. (B, C, E, F) Cross sections of 2-month-old L4 and WT plants with the 2nd internode (IN). The cross sections were stained with phloroglucinol-HCl. Bar, 1000 μm (B, E) and 100 μm (C, F). The red lines were the size of the thickened and lignified cells, and the number in the figure denotes the length (μm, measured by ImageJ, version 2.3.0). (G) The size of the thickened and lignified cells (μm) in WT and transgenic tobacco. Error bars in the charts indicate the standard deviation from the mean for the three repeats. Asterisks indicate significant differences between the transgenic tobacco and WT plants (Student’s t-test; *p < 0.05, **p < 0.01). The data are the mean ± standard error.
Discussion
Sequence characteristics and differential analysis of LcCCRs in L. chinense revealed functional differentiation
Lignin synthesis starts from the common phenylpropanoid pathway and is regulated via a variety of enzymes, but the cinnamyl-CoA reductase encoded by the CCR is one of the key enzymes in lignin biosynthesis (Lewis and Yamamoto, 1990; Raes et al., 2003). As for most forest tree species and associated metabolic pathways, genome-wide analysis is a crucial method to elucidate the biological functions of the CCR family in plants. Here, we report the phylogeny and genome structure of the CCR genes in basal angiosperms for the first time. After further characterization, 13 LcCCR genes were identified in L. chinense.
The expansion of the CCR family members is important to plant evolution. After years of continuous research, studies have identified and cloned the full-length or partial coding sequence (CDS) of CCR genes from the xylem or other tissues in many plant species. For example, seven CCR genes involved in lignin biosynthesis have been identified in P. tomentosa (Chao et al., 2017), 11 in Populus tremuloides (Li et al., 2006), 4 in Boehmeria nivea (Tang et al., 2019), and 11 in A. thaliana (Raes et al., 2003). In general, there are more CCR genes in woody species. We hypothesize that the number of CCR family members in different species may be affected by genomic differences, the redundancy of gene functions, or species differences.
In this study, 13 LcCCR genes were screened by proteomic mining of the complete sequence genome of L. chinense. The molecular weight of these LcCCR proteins was greater than 35 kDa each, which is the same as most CCR proteins reported in plants (Prasad et al., 2010). Their isoelectric points were between 5.5 and 7.5, which is consistent with those for the BnCCR proteins (Tang et al., 2019). Lauvergeat et al. examined the protein structures of the bona fide Arabidopsis CCR proteins, AtCCR5 (AT1G15950) and AtCCR9 (AT1G80820) (Lauvergeat et al., 2001), to reveal that the region of the N-terminal portion of AtCCR5/9 involved in the NADP(H) cofactor binding site was conserved. A very well-conserved motif, KNWYCY, which is thought to be involved in the catalytic site of this enzyme, also exhibits the signature of CCRs (Lacombe et al., 1997). Previous studies have shown that the second and third amino acids (“W” and “Y”) are crucial for the binding of the enzyme to the substrate, and thus, they are rarely replaced (Barakat et al., 2011). The conserved domain, “X-W-Y-X-X”, in the cinnamyl-CoA reductase family of proteins was found in all 13 LcCCR protein sequences here. A similar phenomenon was observed in Pyrus bretschneideri (Chen et al., 2020b) indicating that LcCCRs remained highly conserved during the evolution of L. chinense.
Based on the results of MEME analysis with default parameters, the same group of LcCCR proteins shared common motifs in the phylogenetic tree, suggesting that these LcCCRs are highly conserved, strongly supporting the reliability of the group classifications (Bailey et al., 2006). In addition, gene structure and motif analysis methods were employed to discover the potential features of genes (Barakat et al., 2011). Phylogenetic analysis shows that LcCCR genes could be divided into four groups. The structures of exon-intron regions in the genes from the same group were similar. The change in the number of CCR genes and the diversification of characteristic motifs provides new insights to understand the evolution and gene function of the LcCCR gene family.
Role of the LcCCR13 gene in the lignin biosynthesis
The phenylpropanoid pathway, the main pathway in lignin biosynthesis that is regulated by a variety of enzymes, is well understood. Cinnamoyl-CoA reductase (CCR) is considered to be the first committed enzyme in the lignin-specific branch because it can catalyze the conversion of cinnamoyl-CoA to cinnamaldehyde in the monolignol biosynthetic pathway, which can further be transferred into three lignin monomers (G, S, and H) (Gayoso et al., 2010). Besides, cinnamoyl-CoA can be synthesized into phenolic substances (such as anthocyanins and flavonoids) when the function of CCR has been lost. For this reason, the key role of the CCR gene in lignin synthesis has been confirmed in many plant species, and changing its expression levels might significantly affect the lignin content and growth of plants (Liu et al., 2021). In our study, we measured the plant height and the lignin content in transgenic lines and observed that the overexpression of the LcCCR13 gene significantly increased lignin accumulation in the stems and decreased plant height. These results corroborate that the LcCCR13 is a key gene for lignin synthesis in the stems of L. chinense.
The synthesis of lignin is tissue specific. In general, lignin synthesis occurs in tissues with higher lignification, and the CCR genes are strongly expressed in such tissues. For example, AtCCR5 is mainly concentrated in the tissues that are being lignified and participate in the process of tissue lignification (Lauvergeat et al., 2001; Goujon et al., 2003); the activity of PtoCCR1/4/7 affects the accumulation of lignin (Chao et al., 2017). The relative expression levels of the EgCCR gene are also the highest in the stems, followed by their expression in the roots and other tissues (Lacombe et al., 1997). TaCCR2 is mainly expressed in the roots but also participates in the lignin synthesis in stems, indicating that it plays an important role in lignin synthesis in T. aestivum (Lin et al., 2001). The expression analysis of 10 PoptrCCRs showed that all of them were expressed in the bark, leaves, and xylem, but only the bona fide PoptrCCR12/14 had the highest expression levels in the xylem, where they showed a significant difference from the expression levels in the leaf/bark (Barakat et al., 2011). Our transcriptome data indicated that only LcCCR5/7/10/12/13 genes were predominantly expressed in the DXS (stem developmental xylem). RT-qPCR assays showed that the LcCCR13 had the highest expression level in the stem, which was significantly different from the expression level in other tissues. These observations were largely consistent with previous studies; For instance, the bona fide CCR genes are highly expressed in the tissues/organs with high lignification (Zhang et al., 2015). In this study, only LcCCR13 was highly expressed in the stem, indicating that it might play potential roles in stem development. Therefore, we overexpressed LcCCR13 in tobacco to determine whether it was associated with lignin synthesis.
Transgenic technologies have been widely used to control the lignin content and composition in various plants. Up-regulation or down-regulation of AtCCR expression in A. thaliana significantly affected the lignin content and other characteristics related to plant growth and development (Goujon et al., 2003). As compared to the WT plants, the transgenic lines of B. napus over-expressing BnC.CCR2b had significantly higher lignin content in the stems (Prasad et al., 2010). When BnCCR1 activity was increased in B. platyphylla, the lignin content also increased, and the height of the transgenic plants was reduced (Zhang et al., 2015). The lignin content increased and the plant height decreased in the antisense transgenic tobacco plants, and their plant phenotypes (such as plant height, seed quality, and length of the leaves) was significantly altered (Chabannes et al., 2001). These findings indicate that the manipulation of CCR gene expression affects lignin content, and the changes in lignin content in the transgenic plants is incompatible with normal phenotypes.
In this study, we found that the over-expression of the LcCCR13 gene affects the growth and development of transgenic tobacco. As compared to other transgenic lines, transgenic plants with the highest CCR activity showed the highest lignin content, indicating that CCR might be the key gene involved in lignin biosynthesis. Hamedan et al. also reported that increased lignin content coincides with CCR activities in Gerbera jamesonii (Hamedan et al., 2019). Moreover, according to the measurement of lignin content and its deposition sites in transgenic plants, we conclude that the LcCCR13 gene has a significant effect on lignin synthesis in the stem. Based on the fact that increasing the expression of LcCCR13 reduced the height growth and increased the lignin content in the stem, we assume that LcCCR13 is involved in the thickening of the cell wall by increasing the levels of all three types of subunits. It further regulates the ligin content in plants. Tu et al. found that the downregulation of CCR1 expression in transgenic L. perenne plants reduced the lignin content by lowering the levels of all three lignin monomers (Tu et al., 2010). Interestingly, Zhang et al. found that BpCCR1 could control the height growth and lignin content by lignifying the cell wall (Zhang et al., 2015), which was consistent with our results. All in all, these findings might be significant for understanding the roles of the LcCCR13 gene in lignin biosynthesis in the stems of L. chinense.
Conclusions
In this study, 13 LcCCR genes were identified in the L. chinense genome, among which the LcCCR13 is speculated to potentially play a role in lignin synthesis in the stem as per the results of phylogenetic and bioinformatics analysis, gene expression profiling via RT-qPCR assays, and function verification via gene transformation in tobacco. In conclusion, this study lays a foundation to uncover the mechanism of wood formation in L. chinense.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
WL designed the experiments, performed the experiments, analyzed the experimental data, and wrote the paper. ZH, LY and JW collected plant materials and formally analyzed the experimental data. HX and ZT performed the experiments and formally analyzed the experimental data. ZC collected plant materials and analyzed the experimental data. HL conceived and designed the experiments, gave comments on the data analysis, and revised the paper. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by funds from the National Natural Science Foundation of China (No. 31770718 and No. 31470660) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1110639/full#supplementary-material
References
Bailey, T. L., Nadya, W., Chris, M., Li, W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic. Acids Res. 34, 369–W373. doi: 10.1093/nar/gkl198
Barakat, A., Yassin, N. B. M., Park, J. S., Choi, A., Herr, J., Carlson, J. E. (2011). Comparative and phylogenomic analyses of cinnamoyl-CoA reductase and cinnamoyl-CoA-reductase-like gene family in land plants. Plant Sci. 181, 249–257. doi: 10.1016/j.plantsci.2011.05.012
Bhaskara, R. M. V., Arul, J., Angers, P., Couture, L. (1999). Treatment of wheat seeds induces resistance to fusarium graminearum and improves seed quality. J. Agr. Food. Chem. 47, 1208–1216. doi: 10.1021/jf981225k
Bhuiyan, N. H., Selvaraj, G., Wei, Y., King, J. (2009). Role of lignification in plant defense. Plant Signal. Behav. 4, 158–159. doi: 10.4161/psb.4.2.7688
Boerjan, W., Ralph, J., Baucher, M. (2003). Lignin biosynthesis. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Chabannes, M., Barakate, A., Lapierre, C., Marita, J. M., Ralph, J., Pean, M., et al. (2001). Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 28, 257–270. doi: 10.1046/j.1365-313x.200101140.x
Chao, N., Li, N., Qi, Q., Li, S., Lv, T., Jiang, X. N., et al. (2017). Characterization of the cinnamoyl-CoA reductase (CCR) gene family in populus tomentosa reveals the enzymatic active sites and evolution of CCR. Planta 245, 61–75. doi: 10.1007/s00425-016-2591-6
Chen, J., Hao, Z., Guang, X., Zhao, C., Wang, P., Xue, L., et al. (2019). Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat. Plants 5, 18–25. doi: 10.1038/s41477-019-0368-1
Chen, C., Chen, H., Zhang., Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Li, M., Li, D., Zhang, J., Jin, Q., Sheng, L., et al. (2020b). Correction: Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biol. Open 6, 1602–1613. doi: 10.1242/bio.052985
Gayoso, C., Pomar, F., Novo-Uzal, E., Merino, F., de Ilárduya, O. M. (2010). The ve-mediated resistance response of the tomato to verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 10, 232. doi: 10.1186/1471-2229-10-232
Giordano, A., Liu, Z., N.Panter, S., M.Dimech, A., Shang, Y., Wijesinghe, H., et al. (2014). Reduced lignin content and altered lignin composition in the warm season forage grass paspalum dilatatum by down-regulation of a cinnamoyl CoA reductase gene. Transgenic Res. 23, 503–517. doi: 10.1007/s11248-014-9784-1
Goujon, T., Ferret, V., Mila, I., Pollet, B., Ruel, K., Burlat, V., et al. (2003). Down-regulation of the AtCCR1 gene in arabidopsis thaliana: effects on phenotype, lignins and cell wall degradability. Planta 217, 218–228. doi: 10.1007/s00425-003-0987-6
Hamedan, J. H., Sohani, M. M., Aalami, A., Nazarideljou, M. J. (2019). Genetic engineering of lignin biosynthesis pathway improved stem bending disorder in cut gerbera (Gerbera jamesonii) flowers. Sci. Hortic. 245, 274–279. doi: 10.1016/j.scienta.2018.10.013
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W., Hu, S. N. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic. Acids Res. 44, 236–241. doi: 10.1093/nar/gkw370
Holsters, M., Waele, D. D., Depicker, A., Messens, E., Montagu, M. V., Schell, J. (1978). Transfection and transformation of agrobacterium tumefaciens. Mol. Genet. Genet. 163, 181–187. doi: 10.1007/bf00267408
Hu, B., Jin, J., Guo, A., Zhang, H., Luo, J., Gao, G. (2014). GSDS 2.0: an upgraded gene features visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Humphreys, J. M., Chapple, C. (2002). Rewriting the lignin road map. Plant Biol. 5, 224–229. doi: 10.1016/s1369-5266(02)00257-1
Kallberg, Y., Oppermann, U., Jörnvall, H., Person, B. (2002). Short-chain dehydrogenase/reductase (SDR) relationships: A large family with eight clusters common to human, animal, and plant genomes. Protein Sci. 11, 636–641. doi: 10.1110/ps.26902
Lacombe, E., Hawkins, S., Van, D. J., Piquemal, J., Goffner, D., Poeydomenge, O., et al. (1997). Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant J. 11, 429–441. doi: 10.1046/j.1365-313x.199711030429.x
Larsen, K. (2004). Cloning and characterization of a ryegrass (Lolium perenne) gene encoding cinnamoyl-CoA reductase (CCR). Plant sSci. 166, 569–581. doi: 10.1016/j.plantsci.2003.09.026
Lauvergeat, V., Lacomme, C., Lacombe, E., Lasserre, E., Roby, D., Grima, P. J. (2001). Two cinnamoyl-CoA reductase (CCR) genes from arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57, 1187–1195. doi: 10.1016/s0031-9422(01)00053-x
Leple, J. C., Dauwe, R., Morreel, K., Storme, V., Lapierre, C., Pollet, B., et al. (2007). Downregulation of cinnamoyl-coenzyme a reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19, 3669–3691. doi: 10.1105/tpc.107.054148
Lescot, M. (2002). Plant CARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Lewis, N. G., Yamamoto, E. (1990). Lignin: Occurrence, biogenesis and biodegradation. Plant Biol. 41, 455–496. doi: 10.1146/annurev.pp.41.060190.002323
Li, X. R. (2009). Cloning and functional analysis of genes related to lignin synthesis in lilium "Oriental hybrids".
Li, L., Cheng, X., Lu, S., Nakatsubo, T., Umezawa, T., Chiang, V. L. (2005). Clarification of cinnamoyl co-enzyme a reductase catalysis in monolignol biosynthesis of aspen. Plant Cell Physiol. 46, 1073–1082. doi: 10.1093/pcp/pci120
Li, B., Liang, Y., Chai, Y. (2006). Achievements in research on plant cinnamoyl-CoA reductase (CCR) genes. Mol. Plant Breed. S1, 55–65. doi: CNKISUNFZZW.0.2006-S1-016
Lin, Z., Ma, Q., Ma, M. (2001). Isolation and functional analysis of cinnamyl coenzyme a reductase (CCR) gene from wheat. Acta Botanica Sin. 43 (10), 1043–1046. doi: 10.3321/j.issn:1672-9072.2001.10.008
Li, W., Tan, X., Chen, H. (2009). Structure, function and application potential of cinnamyl-CoA reductase gene in plant. Economic For. Res. 27, 7–12. doi: 10.3969/j.issn.1003-8981.2009.01.002
Liu, D., Wu, J., Lin, L., Li, P., Li, S., Wang, Y., et al. (2021). Overexpression of cinnamoyl-CoA reductase 2 in brassica napus increases resistance to sclerotinia sclerotiorum by affecting lignin biosynthesis. Front. Plant Sci. 9. doi: 10.3389/fpls.2021.732733
Li, G., Yang, Q., Zhang, Y., Wang, Y., Zhang, T., Li, G. (2014). Cloning and analysis of cinnamyl coenzyme a reductase gene from caragana korshinskii. China Biotechnol. 34, 50–56. doi: 10.13523/j.cb.20140108
Ma, Q. (2007). Characterization of a cinnamoyl-CoA reductase that is associated with stem development in wheat. J. Exp. Bot. 58, 2011–2021. doi: 10.1093/jxb/erm064
Pichon, M., Courbou, I., Beckert, M., Boudet, A., Grima-Pettenati, J. (1998). Cloning and characterization of two maize cDNAs encoding cinnamoyl-CoA reductase (CCR) and differential expression of the corresponding genes. Plant Mol. Biol. 38, 671–676. doi: 10.1023/a:1006060101866
Prasad, N. K., Vindal, V., Kumar, V., Kabra, A., Phogat, N., Kumar, M. (2010). Structural and docking studies of leucaena leucocephala cinnamoyl CoA reductase. J. Mol. Model. 17, 533–541. doi: 10.1007/s00894-010-0744-2
Raes, J., Rohde, A., Christensen, J. H., Van, D., Peer, Y., Boerjan, W. (2003). Genome-wide characterization of the lignification toolbox in arabidopsis. Plant Physiol. 133, 1051–1071. doi: 10.1104/pp.103.026484
Smith, R. A., Cass, C. L., Mazaheri, M., Sekhon, R. S., Heckwolf, M., Kaeppler, H. (2017). Suppression of CINNAMOYL-CoA REDUCTASE increases the level of monolignol ferulates incorporated into maize lignins. Biotechnol. Biofuels. 10, 109. doi: 10.1186/s13068-017-0793-1
Song, M., Peng, X. (2019). Genome-wide identification and characterization of DIR genes in medicago truncatula. Bio. Genet. 57, 487–506. doi: 10.1007/s10528-019-09903-7
Srivastava, S., Vishwakarma, R. K., Arafat, Y. A., Gupta, S. K., Khan, B. M. (2015). Abiotic stress induces change in cinnamoyl CoA reductase (CCR) protein abundance and lignin deposition in developing seedlings of leucaena leucocephala. Physiol. Mol. Biol. Plants. 21, 197–205. doi: 10.1007/s12298-015-0289-z
Sun, S., Xiong, X., Zhang, X., Feng, H., Zhu, Q., Sun, J., et al. (2020). Characterization of the Gh4CL gene family reveals a role of Gh4CL7 in drought tolerance. BMC Plant Biol. 20, 125. doi: 10.1186/s12870-020-2329-2
Su, X., Zhao, Y., Wang, H., Li, G., Cheng, X., Jin, Q., et al. (2019). Transcriptomic analysis of early fruit development in Chinese white pear (Pyrus bretschneideri rehd.) and functional identification of PbCCR1 in lignin biosynthesis. BMC Plant Biol. 19, 417. doi: 10.1186/s12870-019-2046-x
Tang, Y., Liu, F., Xing, H., Mao, K., Chen, G., Guo, Q., et al. (2019). Correlation analysis of lignin accumulation and expression of key genes involved in lignin biosynthesis of ramie (Boehmeria nivea). Genes 10, 389. doi: 10.3390/genes10050389
Tu, Z., Hao, Z., Zhong, W., Li, H. (2019). Identification of suitable reference genes for RT-qPCR assays in liriodendron chinense (Hemsl.) sarg. Forests 10, 441. doi: 10.3390/f10050441
Tu, Y., Rochfort, S., Liu, Z., Ran, Y., Griffith, M., Badenhorst, P., et al. (2010). Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-Reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 22, 3357–3373. doi: 10.1105/tpc.109.072827
Vanholme, R., Morreel, K., Ralph, J., Boerjan, W. (2008). Lignin engineering. Curr. Opin. Plant Biol. 11, 278–285. doi: 10.1016/j.pbi.2008.03.005
Wang, Z., Cui, L., Chen, C., Liu, X., Yan, Y., Wang, Z. (2012). Downregulation of cinnamoyl CoA reductase affects lignin and phenolic acids biosynthesis in salvia miltiorrhiza bunge. Plant Mol. Biol. 30, 1229–1236. doi: 10.1007/s11105-012-0444-4
Wang, J., Liu, B., Sun, Y., Chiang, V., Sederoff, R. (2019). Enzyme-enzyme interactions in monolignol biosynthesis. Front. Plant Sci. 11. doi: 10.3389/fpls.2018.01942
Weng, J., Akiyama, T., Bonawitz, N. D., Li, X., Ralph, J., Chapple, C. (2010). Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte selaginella and flowering plants. Plant Cell 22, 1033–1045. doi: 10.1105/tpc.109.073528
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Yang, L., Liu, H., Hao, Z., Zong, Y., Xia, H., Shen, Y., et al. (2021). Genome-wide identification and expression analysis of R2R3-MYB family genes associated with petal pigment synthesis in liriodendron. Int. J. Mol. Sci. 22, 11291. doi: 10.3390/ijms222011291
Yang, J., Xu, J., Zhang, Y., Cui, J., Hu, H. (2022). Transcriptome-wide identification, characterization, and expression analysis of R2R3-MYB gene family during lignin biosynthesis in Chinese cedar (Cryptomeria fortunei hooibrenk). Ind. Crop Prod. 182, 114883. doi: 10.1016/j.indcrop.2022.114883
Yan, X., Liu, J., Kim, H., Liu, B., Huang, X., Yang, Z., et al. (2019). CAD1 and CCR2 protein complex formation in monolignol biosynthesis in populus trichocarpa. New Phytol. 222, 244–260. doi: 10.1111/nph.15505
Yin, N., Li, B., Liu, X., Liang, Y., Lian, J., Xue, Y., et al. (2021). Two types of cinnamoyl-CoA reductase function divergently in accumulation of lignins, flavonoids and glucosinolates and enhance lodging resistance in brassica napus. Crop J. 3, 647–660. doi: 10.1016/j.cj.2021.10.002
Zhang, Y., Wang, Y., Wang, C. (2012). Gene overexpression and gene silencing in birch using an agrobacterium-mediated transient expression system. Mol. Biol. Rep. 39, 5537–5541. doi: 10.1007/s11033-011-1357-2
Keywords: Liriodendron chinense, lignin biosynthesis, cinnamoyl-CoA reductase, CCR gene family, expression analysis
Citation: Li W, Hao Z, Yang L, Xia H, Tu Z, Cui Z, Wu J and Li H (2023) Genome-wide identification and characterization of LcCCR13 reveals its potential role in lignin biosynthesis in Liriodendron chinense. Front. Plant Sci. 13:1110639. doi: 10.3389/fpls.2022.1110639
Received: 29 November 2022; Accepted: 23 December 2022;
Published: 16 January 2023.
Edited by:
Qingzhang Du, Beijing Forestry University, ChinaReviewed by:
Aimin Wu, South China Agricultural University, ChinaSian Liu, Yangzhou University, China
Copyright © 2023 Li, Hao, Yang, Xia, Tu, Cui, Wu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huogen Li, hgli@njfu.edu.cn
 Wei Li
Wei Li Ziyuan Hao
Ziyuan Hao Lichun Yang
Lichun Yang Zhonghua Tu
Zhonghua Tu Junpeng Wu
Junpeng Wu Huogen Li
Huogen Li