- 1College of Life Science and Agronomy, Zhoukou Normal University, Zhoukou, China
- 2State Key Laboratory of Crop Stress Biology in Arid Areas, College of Life Sciences, Northwest A&F University, Yangling, Shaanxi, China
- 3Key Laboratory of Plant Genetics and Molecular Breeding, Zhoukou Normal University, Zhoukou, China
As the critical sensors and decoders of calcium signal, calcium-dependent protein kinase (CDPK) has become the focus of current research, especially in plants. However, few resources are available on the properties and functions of CDPK gene family in Triticum aestivum (TaCDPK). Here, a total of 79 CDPK genes were identified in the wheat genome. These TaCDPKs could be classified into four subgroups on phylogenesis, while they may be classified into two subgroups based on their tissue and organ-spatiotemporal expression profiles or three subgroups according to their induced expression patterns. The analysis on the signal network relationships and interactions of TaCDPKs and NADPH (reduced nicotinamide adenine dinucleotide phosphate oxidases, NOXs), the key producers for reactive oxygen species (ROS), showed that there are complicated cross-talks between these two family proteins. Further experiments demonstrate that, two members of TaCDPKs, TaCDPK2/4, can interact with TaNOX7, an important member of wheat NOXs, and enhanced the TaNOX7-mediated ROS production. All the results suggest that TaCDPKs are highly expressed in wheat with distinct tissue or organ-specificity and stress-inducible diversity, and play vital roles in plant development and response to biotic and abiotic stresses by directly interacting with TaNOXs for ROS production.
1 Introduction
In multicellular organisms, including plants calcium ion (Ca2+) is recognized as a vital and conserved secondary messenger that is necessary for signaling transduction. As the Ca2+ sensors and responders, calcium-dependent protein kinase CDPKs/CPKs universally present in green algae, oomycetes, protists, especially in higher plants, but absent in animals and fungi (Valmonte et al., 2014), while CDPK-related receptor-like kinases CRKs, that share some conserved homology from the parent CDPKs, are only observed in plants (Hrabak et al., 2003). For example, 34 AtCDPKs and 8 CRKs in Arabidopsis (Arabidopsis Thaliana) (Yip Delormel and Boudsocq, 2019), 29 OsCDPKs in rice (Oryza sativa L.) (Asano et al., 2005), 42 ZmCPKs in maize (Zea mays) (Li et al., 2018), 44 BaCDPKs in banana (Musa paradisiaca) (Li et al., 2020), 30 PtCDPKs in black cottonwood (Populus trichocarpa) (Zuo et al., 2013), 128 WmCDPKs and WmCRKs in watermelon (Citrullus lanatus) (Wei et al., 2019), and so on, were all identified and described in plants. CDPKs play important roles in many biological processes, such as growth and development, physiological regulation and control, and response to biotic and abiotic stresses in plants. For example, in Arabidopsis, AtCPK1 was found to be involved in the regulation of cell death by phosphorylating the senescence master regulator ORE1, a NAC transcription factor also called AtNAC2/ANAC092 (Durian et al., 2020); AtCPK12 performed a negative ABA-signaling regulator functions in seed germination and post-germination growth (Zhao et al., 2011); AtCPK33 plays an important role in strigolactones (SLs) induced stomatal closure (Wang et al., 2019a). In rice, OsCDPK5/13, as the negative regulators, participate in aerenchyma formation of roots (Yamauchi et al., 2017). While, OsCPK12 performs the positive effect on delaying leaf senescence and providing the potential productivity in plants (Wang et al., 2019b). In okra (Abelmoschus esculentus L.), AeCDPK6 can prolong full-blooming period by regulating hyperoside biosynthesis indirectly (Yang et al., 2020a). Moreover, AtCPK28 is not only involved in the regulation of stem elongation and secondary growth (Matschi et al., 2013), but also acts as a negative regulator and plays a crucial role in immune signaling (Monaghan et al., 2014). Similarly, GmCDPK38 also plays a dual role in coordinating flowering time regulation and insect resistance of soybean (Glycine max) (Li et al., 2022a). In addition, AtCPK5 directly phosphorylates AtLYK5, a lysin motif receptor-like kinases, and regulates chitin-induced defense responses in Arabidopsis (Huang et al., 2020). In terms of abiotic stress, AtCPK12 is involved in plant adaptation to salt stress by regulating Na+ and H2O2 homeostasis (Zhang et al., 2018); StCDPK32 positively modulates physiological properties and photosynthesis in response to salinity stress in potato (Solanum tuberosum) (Zhu et al., 2021); overexpression of GmCDPK3 improved soybean tolerance to drought and salt stresses (Wang et al., 2019c). Whereas, PheCDPK22 functions as a negative regulator of drought stress in moso bamboo (Phyllostachys edulis) (Wu et al., 2020).
CDPKs also participate in many biological signaling networks in plants, especially by interacting with NOX (also called respiratory burst oxidase homolog, RBOH/Rboh) family proteins, the key producers of reactive oxygen species (ROS) of plants. For example, StCDPK5 directly activates and phosphorylates StRbohB in a calcium-dependent manner to regulate the oxidative burst for defense responses to pathogens (Kobayashi et al., 2007); AtCPK5 phosphorylates AtRbohD and thereby enhances ROS production for defense responses and bacterial resistance (Dubiella et al., 2013). In addition, BnaCPK6L was reported to play an important role in ROS accumulation and hypersensitive response (HR)-like cell death by interacting with and phosphorylating BnaRbohD (Pan et al., 2019). Additionally, StCDPK23 may participate in the wound healing of potato tubers by regulating StRbohs for H2O2 production (Ma et al., 2022). Intriguingly, OsCPK12 promotes the tolerance of rice to salt stress by repressing the expression level of OsRbohI and reducing the accumulation of ROS (Asano et al., 2012; Boudsocq and Sheen, 2013). More importantly, a MtCDPK5 can directly phosphorylate three Rbohs MtRbohB, MtRbohC, and MtRbohD respectively, which can trigger immune responses to regulating rhizobial colonization in symbiotic cells of barrel medic (Medicago Truncatula) (Yu et al., 2018a). Conversely, an OsRbohH can be stimulated by two CDPKs CDPK5 and CDPK13 for ROS production, which is essential for aerenchyma formation in rice roots (Yamauchi et al., 2017). In wheat (Triticum aestivum), our previous studies showed that TaCDPK13 directly interacts with and activates TaNOX7 for ROS production for plant fertility regulation and drought tolerance (Hu et al., 2020a). Besides these, NADPH oxidases TaNOXs, as the key producers of ROS, play crucial roles in various biological processes in plants (Hu et al., 2018). All the results mentioned above prompt us to speculate that there may also be complicated interactions between TaCDPKs and TaNOXs family members in wheat.
However, as of today, only a few of CDPKs in wheat were characterized according to their gene evolutionary (Geng et al., 2011) and expressional characteristics (Martínez-Noël et al., 2007), the functions and signal network relationships of wheat CDPK family genes involved in plant growth regulation and environmental stress response are still largely unknown. In the present study, comprehensive analyses based on bioinformatics approaches and experimental methods were performed to identify the wheat CDPK family genes, their functions and signal network relationships during the plant development and stress response. Based on the results, the interactions between TaCDPK2/4/14/16/20/21 and TaNOX7 were further studied and verified that TaCDPK2/4 can interact with TaNOX7 and co-expression of TaCDPK2/4 with TaNOX7 enhanced ROS production in plants. The results obtained here will largely broaden our understanding of the roles of TaCDPKs and the signal network relationships between TaNOXs and TaCDPKs in wheat.
2 Materials and methods
2.1 Sequence retrieval and identification of the CDPK gene family in wheat
We retrieved the potential sequences of CDPK members in wheat from IWGSC (http://www.wheatgenome.org/, last accessed May 25, 2021), NCBI (https://www.ncbi.nlm.nih.gov/, last accessed May 20, 2021), and Ensembl Plants (http://plants.ensembl.org/Triticum_aestivum/Info/Index, last accessed May 20, 2021) websites, with the well-known CDPK sequences as queries. We identified each CDPK member by predicting the conserved domains. For further information, we analyzed some physicochemical parameters, predicted the subcellular localization and the numbers of transmembrane helix, and performed amino acid sequence alignment (the detailed information in Table S1 and Figure 1).
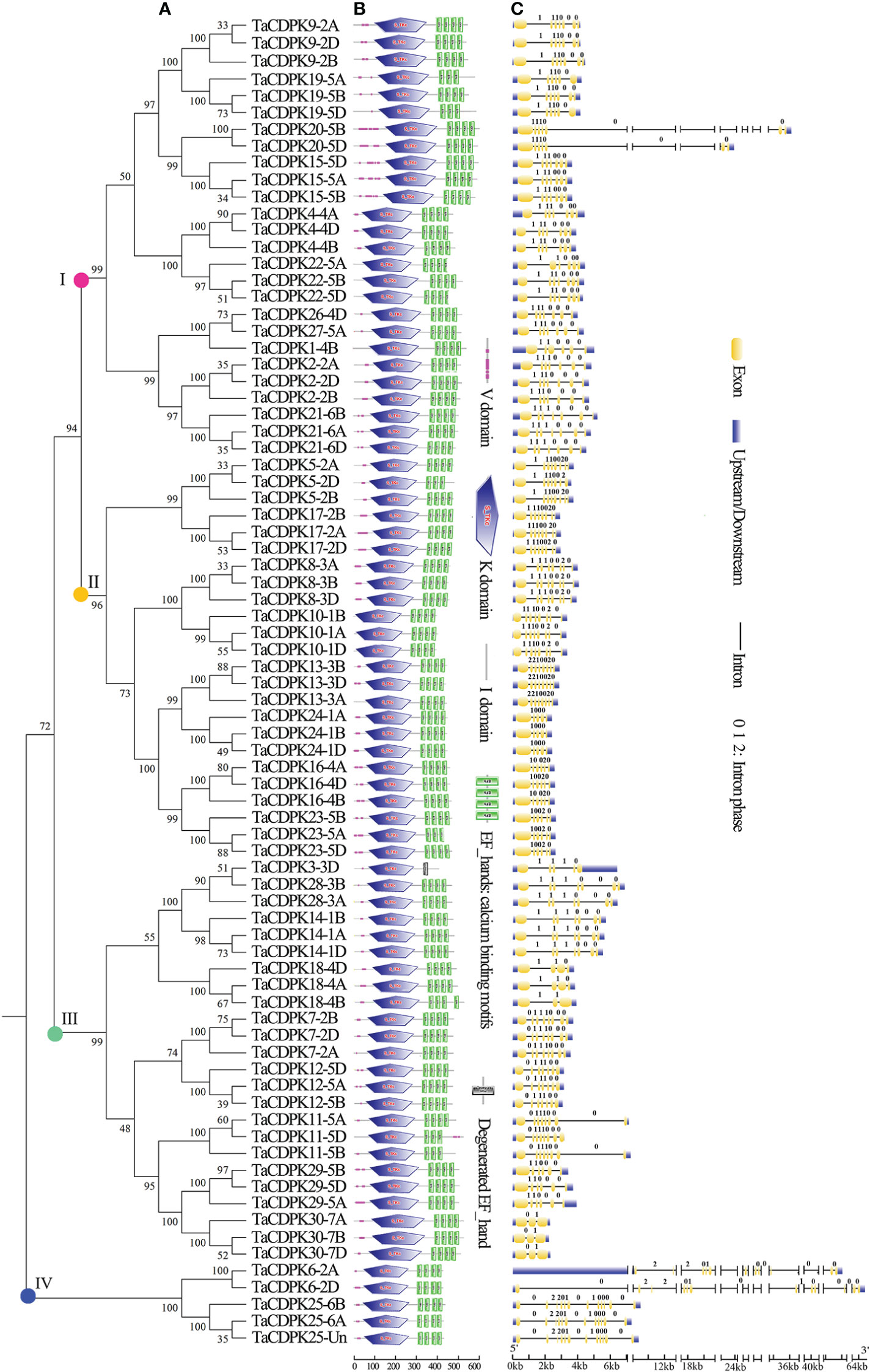
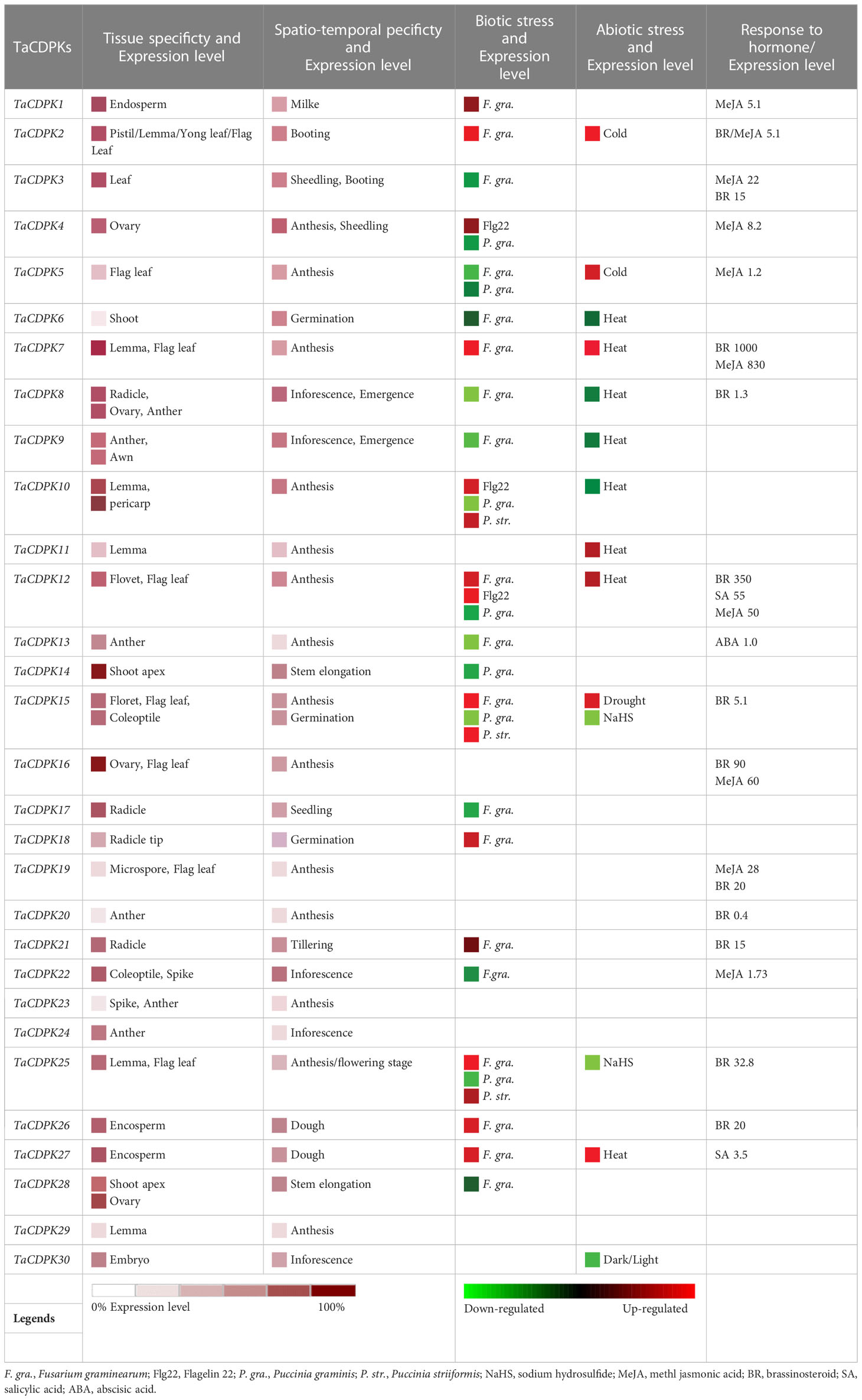
Figure 1 Phylogenetic relationship, domain organization, and exon/intron structure analysis of CDPK family members in wheat. (A) The unrooted maximum-likelihood phylogenetic tree of TaCDPK family members were made with MEGA 6.06. Numbers above the nodes represent bootstrap values from 1,000 replications. (B) Domain organization of the TaCDPKs. The logos of domain organization were obtained from EMBL-EBI and SMART websites and were amended with Adobe_Photoshop_CS6. The domains: V represents variable domain; K represents catalytic domain; I represents auto-inhibitory domain; C represents the region of calcium binding motifs: EF_hands. (C) The exon/intron structures of CDPK family genes in wheat. The numbers 0, 1, and 2 represent the phase of each intron in the sequence.
2.2 Sequence alignment and protein structure analysis
The phylogenetic tree of wheat CDPK family members was constructed with MEGA 6.06 (Figure 1A). The logos of domain organization were obtained from EMBL-EBI (http://pfam.xfam.org/search#tabview=tab1) or SMART (http://smart.embl-heidelberg.de/) websites and were amended with Adobe_Photoshop_CS6. The four conserved domains: N-terminal variable domain (V), kinase active region (K), Auto-inhibitory domain (I), and calcium binding motif (C) in each CDPK protein sequence, were generated by MEME suite (http://meme-suite.org/) (Figure 1B).
2.3 Exon/intron structure analysis and chromosomal location
The exon/intron logos of individual CDPK genes were obtained from the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn) by aligning the coding or cDNA sequences with their corresponding genomic DNA sequences (for the detailed information in Figure 1C). The chromosomal distributions of 79 candidate genes TaCDPKs were displayed using TBtools software (https://www.yuque.com/cjchen/hirv8i/ra35nv). MCScanX and BLASTP were used to analyze gene duplication events of TaCDPK genes in the Triticum aestivum genome (Figure 2). (See the detailed information about the synteny between homologous genes in Table S2).
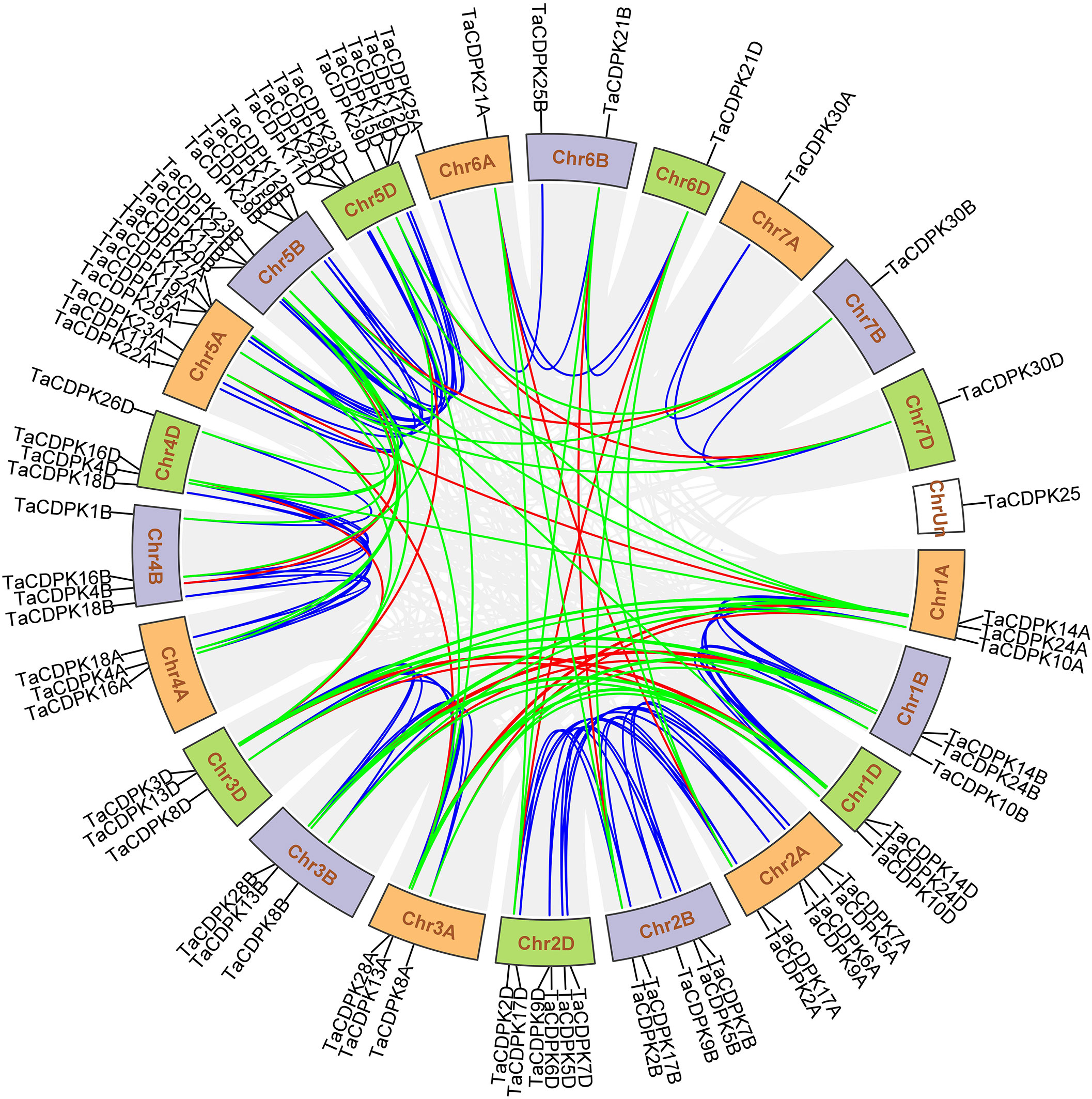
Figure 2 Chromosomal locations of TaCDPK genes and their synteny in wheat. Chromosomal locations of TaCDPK genes and their synteny are illustrated by the circos diagram. Colored lines indicate similarity. The blue line shows the synteny between homologous genes on homologous chromosomes from different subgenome, such as Chr1A, 1B and 1D; The green lines represent the synteny between homologous genes on non-homologous chromosomes from the same subgenome, such as Chr1A-3A, 1A-5A; The red line shows the synteny of homologous genes between non-homologous chromosomes from different subgenome, such as Chr1A-3B and Chr1A-4D.
2.4 Prediction and functional analysis of cis-regulatory elements
We selected 2,000-bp genomic DNA sequences upstream of the transcriptional start sites of TaCDPKs as the promoter sequences to analyze the cis-acting elements using the databases: PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) according to the method we previously used (Hu et al., 2018).
2.5 Signal network relationships analysis between the members of CDPK and NOX family
The signal network relationships between the members of CDPK family, NOX family were drawn by using Cytoscape software and Adobe Photoshop, based on the information from STRING (http://string-db.org/cgi/input.pl?sessionId=bdYxf9Fv5NiI&input_page_show_search=on). (See the detailed information in Figure 3 and Table S3).
Figure 3 The signal network relationships between the members of CDPK and NOX family. The network signal relationships between the members of CDPK family and NOX family were preliminarily predicted on STRING (http://string-db.org/cgi/input.pl?sessionId=bdYxf9Fv5NiI&input_page_show_search=on), and then the network signal diagram between them were also drawn by softwares of TBtools Cytoscape and Adobe Photoshop. The edge(line) width is positively correlated with the combined score in STRING (Min 1- Max 3).
2.6 Firefly luciferase complementation imaging (LCI) assay
To verify the interaction between TaNOX7 and TaCDPK2/4/14/16/20/21, the firefly luciferase complementation imaging (LCI) assay was performed according to a previously described method (Hu et al., 2020a), with TaCDPK13 as the positive control group. First, we constructed the expression vectors TaCDPK2/4/14/16/20/21-cLUC (the C-terminal gene fragment of the Luciferase) and TaNOX7-nLUC (the N-terminal gene fragment of the Luciferase), then transformed them into wild tobacco (Nicotiana benthamiana) leaves by an Agrobacterium-mediated transient transform method mentioned above. After inoculating for 2 or 3 days, the chemiluminescence images and the fluorescence intensity profiles were all taken by a plant living imaging system (Lumazone Pylon2048B, Princeton). The primers used for the vector construction are listed in the Supporting Information (Table S4).
2.7 Bimolecular fluorescence complementation (BiFC) assay
The bimolecular fluorescence complementation (BiFC) assay was performed with TaCDPK13-TaNOX7 interaction as the positive control group (Hu et al., 2020a), according to the method described by Walter and others (Walter et al., 2004). The coding regions of TaCDPK2/4/16 genes were cloned into the pSPYNE vector with the N-terminal gene fragment of the yellow fluorescent protein (nYFP), and TaNOX7 was cloned into the pSPYCE vector with the C-terminal gene fragment of the yellow fluorescent protein (cYFP). Then, the Agrobacterium-mediated transient transform method was used to transiently coexpress TaNOX7-cYFP and TaCDPK2/4/16-nYFP in N. benthamiana leaves. The fluorescence visualization in leaves was ultimately observed with a confocal microscope (A1R, Nikon, Tokyo, Japan). The primers used for the vector construction are listed in the Supporting Information (Table S4).
2.8 Co-immunoprecipitation (Co-IP) assays
The total proteins were extracted from N. benthamiana leaves using a membrane protein extraction method with some modification (Liu et al., 2016). The protein extracts were denatured and separated by SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) and then the gel was stained with Coomassie Brilliant Blue. For the co-immunoprecipitation (Co-IP) assay, TaNOX7(1047 bp)-GFP-tagged and TaCDPK2/4/13-6*Myc-tagged proteins were detected by monoclonal anti-GFP antibody and anti-Myc antibody (SA003; ABclonal, Wuhan, China), respectively. HRP (horseradish peroxidase) goat anti-mouse IgG antibody (SA003; ABclonal) and antigen-protein complex were detected using the ECL protein gel blot detection kit (GE Healthcare Life Sciences, Beijing, China) and Light-Capture equipped with a CCD camera (ATTO, Shanghai, China) as described by Kobayashi and others (Kobayashi et al., 2007). The primers used for the vector construction are listed in Supplementary Table S4. TaNOX7(1047 bp) represents the truncated gene sequence from the start codon ATG to the 1047th base of TaNOX7, and TaNOX7(1047 bp) includes the conserved functional domain NADPH_Ox and CDPK binding sites (Hu et al., 2018).
2.9 Detection of ROS production
The histochemical analyses of H2O2 accumulation in plant tissues were conducted by 3, 3′- diaminobenzidine (DAB) using an Agrobacterium-mediated instantaneous transformation system according to a previously described method (Kumar et al., 2014). The leaves of N. benthamiana after agroinfiltrated for 2-3 days were separated from plants and put into DAB staining solutions in darkness at room temperature for several hours. After exposed in light for 2~3 h, the samples were then immersed into bleaching solution (ethanol: acetic acid: glycero = 3:1:1) and boiling in water bath for 10~15 min. The bleaching process was repeated 2~3 times for the clearer photographs.
2.10 Subcellular localization analysis
The subcellular location of TaCDPK2/4 and TaNOX7 were examined with N. benthamiana as the materials using an Agrobacterium-mediated instantaneous transformation system according to the method with some modifications (Chen et al., 2009). The full-length open reading frame of TaNOX7 and TaCDPK2/4 gene sequences were used to construct fusion expression vectors containing the gene sequences of GFP (green fluorescent protein) or mCherry (red fluorescent protein): pCAMBIA1301-2*35S-TaNOX7-eGFP, pCAMBIA131-2*35S-TaCDPK2/4-mCherry. At the same time, the membrane protein AtCBL1n-eGFP was used as a positive control, and the constructed expression vector was transformed into tobacco mesophyll cells by transient transformation mediated by Agrobacterium tumefaciens. After 60-84 h of co-culture, the leaves were isolated, and the subcellular localization of proteins were observed by using laser confocal (A1R, Nikon, Tokyo, Japan) at 488 nm (eGFP), 561 nm (mCherry) and 637 nm (Chlorophyll) emission wavelengths. The primers used for the vector construction are listed in the Supporting Information (Table S4).
2.11 Plant materials, treatments, expression profile analysis
Wheat (T. aestivum cv. Chinese Spring) seedlings growing in field were harvested from different developmental stages and used for gene cloning and expression profile analysis. For analysis of the inducible expression profiles of the CDPK genes, the spikelet at the early stage of wheat flowering infected by Fusarium graminae spore with the method of single flower infusion, and the 10 d old hydroponic seedlings treated with 4°C, 200 mM NaCl, 20% polyethylene glycol 6000 (PEG6000), 100 μM methl jasmonic acid (MeJA), 100 μM abscisic acid (ABA), 500 μM salicylic acid (SA), and 50 μM brassinosteroids (BR), respectively, for 0 h, 24 h and/or 48 h, and with 40°C for 0 h, 12 h and/or 24 h, were all used as the materials for RNA extraction using RNAiso TM Plus (Takara, Dalian, China) performance. In addition, tissue-specific expression profiles, inducible expression profiles of TaCDPK genes in wheat were performed using bioinformatics methods based on the online database Genevestigator (https://genevestigator.com/gv/) and/or by quantitative real-time PCR (qRT-PCR) with TaActin (AB181991.1) and TaGAPDH (ABS59297.1) as the internal transcript level controls. All the results mentioned above were presented as heat maps or histograms. All the expression levels represent the mean ± SD of data collected from three independent experiments with each having three or four replicates. The primers used for qRT-PCR are listed in the Supplementary data (Table S4).
3 Results
3.1 Identification of CDPK family genes in wheat genome
A Hidden Markov Model (HMM) search was performed to investigate and characterize the CDPK gene family in wheat genome, and a total of 79 candidates were identified (Table S1). The homologous genes from different subgenomes (A, B, and D) were assigned the same number in gene denomination due to their similarity in gene structure and protein size (Figure 1). Intriguingly, the CDPK family genes in wheat genome are regularly distributed across the chromosomes (Figure 2). It seems that all the predicted CDPK candidates are mainly distributed on Chr 5, followed by Chr 2, 4, 1, 3, 6, and 7 in turn. Moreover, the location information of CDPK25 is unclear, which is referred to as Chr Un. Considering the objective fact that the distributions of homologous genes (such as TaCDPK2A/B/C on Chr 2) are symmetrical on subchromosomes. Therefore, we speculate that CDPK25 on Chr Un actually distributes on Chr 6D. This prediction was further confirmed by the cluster analysis using protein sequences of CDPK family as reference (Figure 1). In addition, the asymmetrical distribution of CDPK4/16/18 between Chr 4B/D and A in Figure 2, implying that there are orientation errors on chromosome localization. All these anomalies may provide references for the precise localization of CDPK4/16/18-colinked genes.
3.2 Gene structure and domain composition
As can be seen in Figure 1C, the gene structures are quite diverse between the TaCDPKs with different intron numbers and length except for the certain homologous genes from different subgenomes (A, B, and D). Except for the members TaCDPK6/20 (64 Kb and 36 Kb, respectively), the length of different TaCDPK genes varies from 2 Kb to 8 Kb. As shown in Figure 1B, almost all the members of CDPK family have four conserved domains, namely N-terminal variable domain (V), Catalytic domain (S_TKc)(also known as kinase active region; K), auto-inhibitory domain (I), and calcium binding domain (C) conceived with three or four EF_hand motifs. Based on their sequence homology, all the 79 members of TaCDPKs could be divided into four subgroups I, II, III, and IV. The domain composition of the proteins is also different between the subgroups. For example, the members of TaCDPK22/1/10/13 in subgroup I and II have no variable domains (V); TaCDPK3 in the subgroup III has only one degenerated EF_hand motif, and the variable domain (V) of TaCDPK11-5D is also abnormally present on the C-side. As speculated above, the member TaCDPK25 mapped on Chr Un were clustered together with their homologue TaCDPK25 from Chr 6A/6B, indicating that gene TaCDPK25-Un may be objectively present on Chr 6D. Surprisingly, the members come from different Chr with different serial numbers are also clustered together, such as TaCDPK26-4D and TaCDPK27-5A. In addition, different numbered members with different structures are also firstly grouped together, such as TaCDPK28-3A/B and TaCDPK3-3D (Figure 1A). Taken together, the complexity of genes and protein structures and the confusing clustering relationships imply some complex evolutionary relationships and functional diversity between CDPK family members.
3.3 Tissue and spatio-temporal specific expression of CDPK family genes in wheat
To clarify the tissue and spatio-temporal expression profiles of CDPK family genes during the development of wheat, a set of microarray data for the gene expression was obtained from Genevestigator v3 (Figures S1, S2). To simplify the phraseology in the following experiments and optimize the graphs in this paper, the homologous genes located on different subchromosomes (Chr A, B, and C) that are referred to as TaCDPKx; for example, TaCDPK5-2A, -2B, and -2D were all named as TaCDPK5.
The expression levels of TaCDPKs at 10 developmental stages and 43 tissues were presented different expression patterns with some genes dominantly expressed at a certain stage or tissue (Figures S1, S2). Comparison between Figures S1, S2 showed that the tissue and spatio-temporal specific expression profiles of 79 genes echo confirm and complement each other. For example, almost all the members in Figures S1, S2 were all divided into two identical groups I and II except for TaCDPK17. The members in group I are all widely expressed in whole developmental stages and most of tissues with the highest level of TaCDPK26/27 in endosperm at dough development stage, TaCDPK16 in ovary at inflorescence emergence stage (Heading stage) and anthesis stage. Furthermore, TaCDPK9 in group I expressed with the peaking level in awn, anther, and glum at florescence emergence stages. TaCDPK4/10/12 are, in turn, expressed at the highest level with TaCDPK4 in pistio, spikelet, ovary, TaCDPK10 in pericarp, TaCDPK12 in coleoptile, floret; while they all expressed with the highest level at anthesis stage. The members in group II were expressed with low level or expressed restrictively in a certain tissue or at a certain stage. TaCDPK24 is expressed exclusively in anther at inflorescence emergence stage (Heading stage) as well as TaCDPK30 in embryo at inflorescence emergence stage. Besides these, there are also some contradictions between Figures S1, S2. For example, in Figure S1, the expression peak of TaCDPK5 is at germinating seeds, TaCDPK11 is at anthesis stage, TaCDPK18 is at seeding stage, TaCDPK17 is at tillering stage, and TaCDPK3/14/28 are at stem elongation stage; while, in Figure S2, the expression peak of them, in turn, is at seedling, radicle, shoot apex/ovary. Unsurprisingly, the members with high homology at protein sequence, such as TaCDPK26 and TaCDPK27, also have the similar expression patterns, as well as TaCDPK3 and TaCDPK28 in Figures S1, S2. Obviously, the different results in Figures S1, S2 may be attributed to the different experimental materials, which indicating that the results in Figures S1, S2 are complementary as well as confirming each other.
In order to further study the expression specificity of TaCDPKs, the tissue and spatio-temporal expression profiles were also performed in 21 tissues from 8 different development stages by qRT-PCR (Figure 4). Due to the low expression level of TaCDPK6/11/18/23/24 as shown in Figure S1/S2, or the nonspecific amplification of TaCDPK9/10/14/17/28/29/30, the expression profiles of them could not be obtained here. From Figure 4, we can see that every member of TaCDPKs has its special expression pattern. For example, TaCDPK2, 4, 7 are expressed with peak in leaf at seedling stage, in sheath at seedling stage, and in flag leaf 1 at heading stage, respectively, though they are all highly expressed in the whole plant. Significantly, the expressive peaks of 9 TaCDPK members TaCDPK3/5/12/16/19/20/21/25/26, are all in the flag leaves at flowering stage. TaCDPK13 is mainly expressed in spikes at milk and heading stages; TaCDPK8 is expressed in all the tissues with no specialty. In addition, compared with Figures S1, S2, the expression patterns of TaCDPKs in Figure 4, are not always consistent. These different results are undoubtedly due to the different experimental methods, sampling period, or growth environment conditions. Therefore, based on previous studies and comprehensive analysis of Figure S1/S2 and Figure 4, we systematically illuminate the unique tissue and developmental expression profiles of TaCDPK family members (Shown in Table 1).
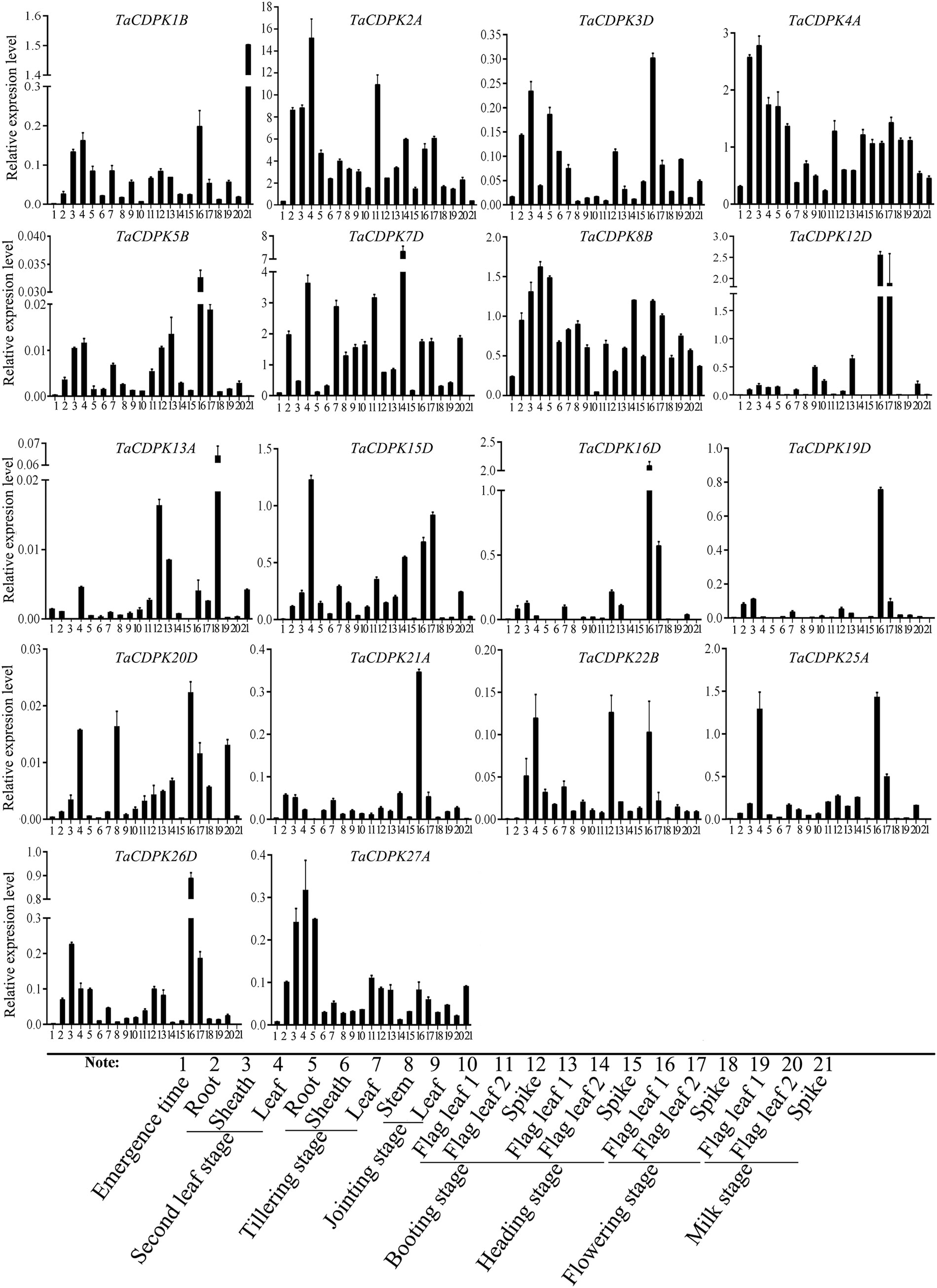
Figure 4 The tissue-specific and spatio-temporal expression profiles of TaCDPKs in wheat by qRT-PCR. Expression analysis of TaCDPKs in 21 tissues from 8 different development stages of wheat by qRT-PCR. All the expression levels represent the mean ± SD of data collected from the experiments with each having three or four replicates.
3.4 Inducible expression profiles of TaCDPK family genes
To further study the expression characteristics of wheat CDPK family genes under suboptimal conditions, we carried out a comprehensive analysis using both the wheat microarray data in Genevestigator v3 (Figure 5) and qRT-PCR experiment (Figure 6). As can be seen in Figure 5, different inducible expression patterns of CDPK genes could be seen in responding to different biotic and/or abiotic stresses. According to the expression patterns in Figure 5, all the members can be simply classified into three groups: Group I including TaCDPK1/2/4/7/12/14/15/18/21/25/26, and most of them were upregulated under the biotic stresses except down-regulated under P. graminis (Puccinia graminis) stress; Group II including TaCDPK3/5/6/8/9/13/16/17/22/28 were down-regulated under F. graminearum (Fusarium graminearum); Group III including TaCDPK11/19/20/23/24/29/30 had no obvious changes under all the biotic or abiotic stresses, but had high expression in another development. Interestingly, these members of Group III also had lower tissue and spatiotemporal expression level comparing with other members in Figures S1, S2. Furthermore, the high expression level of TaCDPK2/7/12/15/25/26 in group I, was further verified under Fusarium Head Blight (FHB) stress in Figure 6B. In addition, the expression of most TaCDPK genes were significantly up-regulated under the treatment of hormone MeJA and BR (Figure 6A), which are consistent with the results that hormone responsive element JARE and ABRE are distributed on almost all the promoters of TaCDPK genes (Figure S3). Only a few members, such as TaCDPK27 had obvious response to SA and heat (Figure 6A). In order to more intuitively dissect the possible functions of TaCDPKs, we listed the specific expressions of each member in plant development or under stress treatment in Table 1.
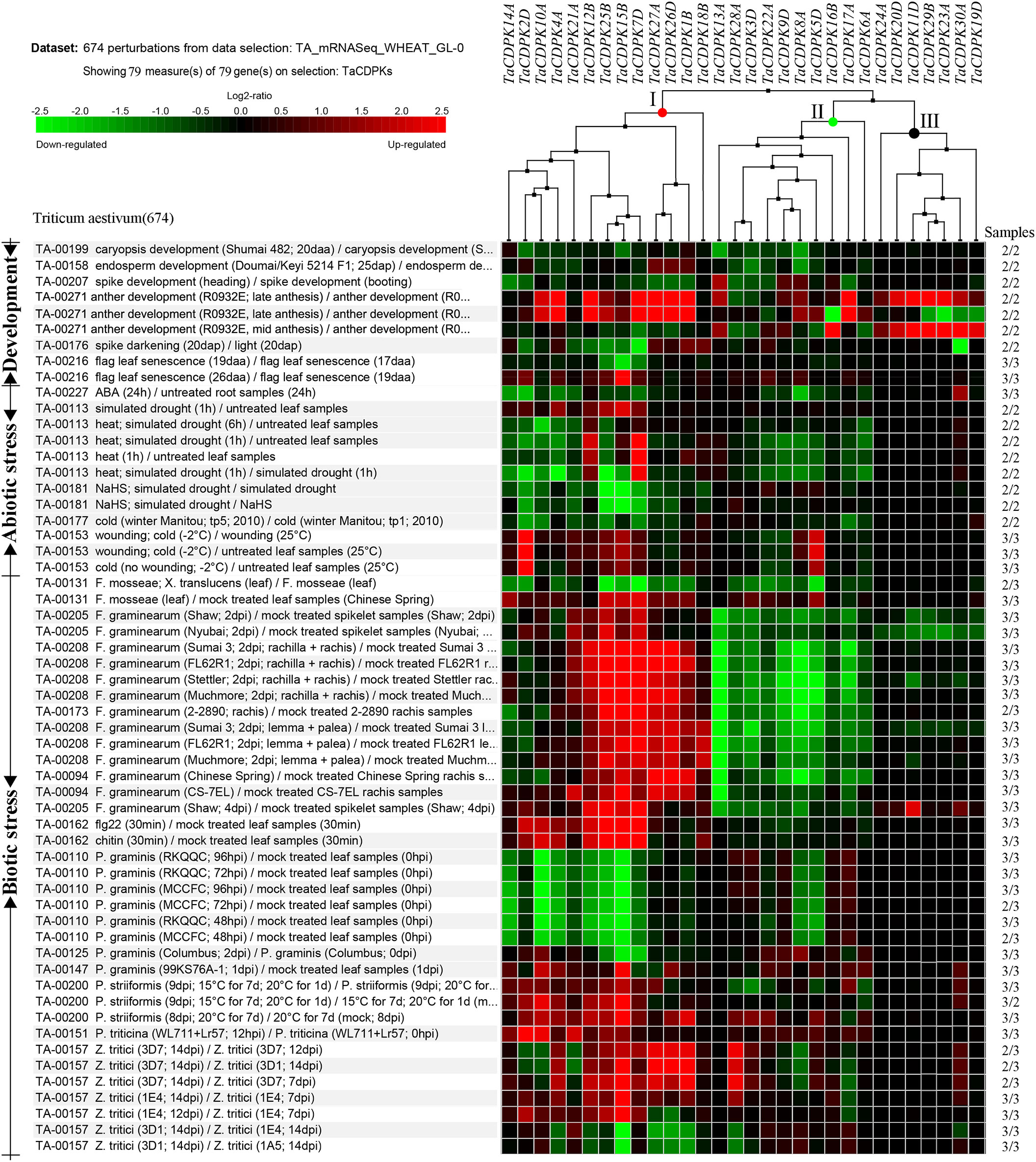
Figure 5 Inducible expression profiles of TaCDPKs in wheat. Inducible expression profiles of TaCDPKs under a wide of environmental stresses and hormone treatments were selected from the Ta_mRNASeq_WHEAT_GL-0 database in Genevestigator v3. For clear and beautiful presentation of the profiles, each of the 30 members shown in the Figure 5 contains its homologues (which had been deleted), so there are 79 members in total.
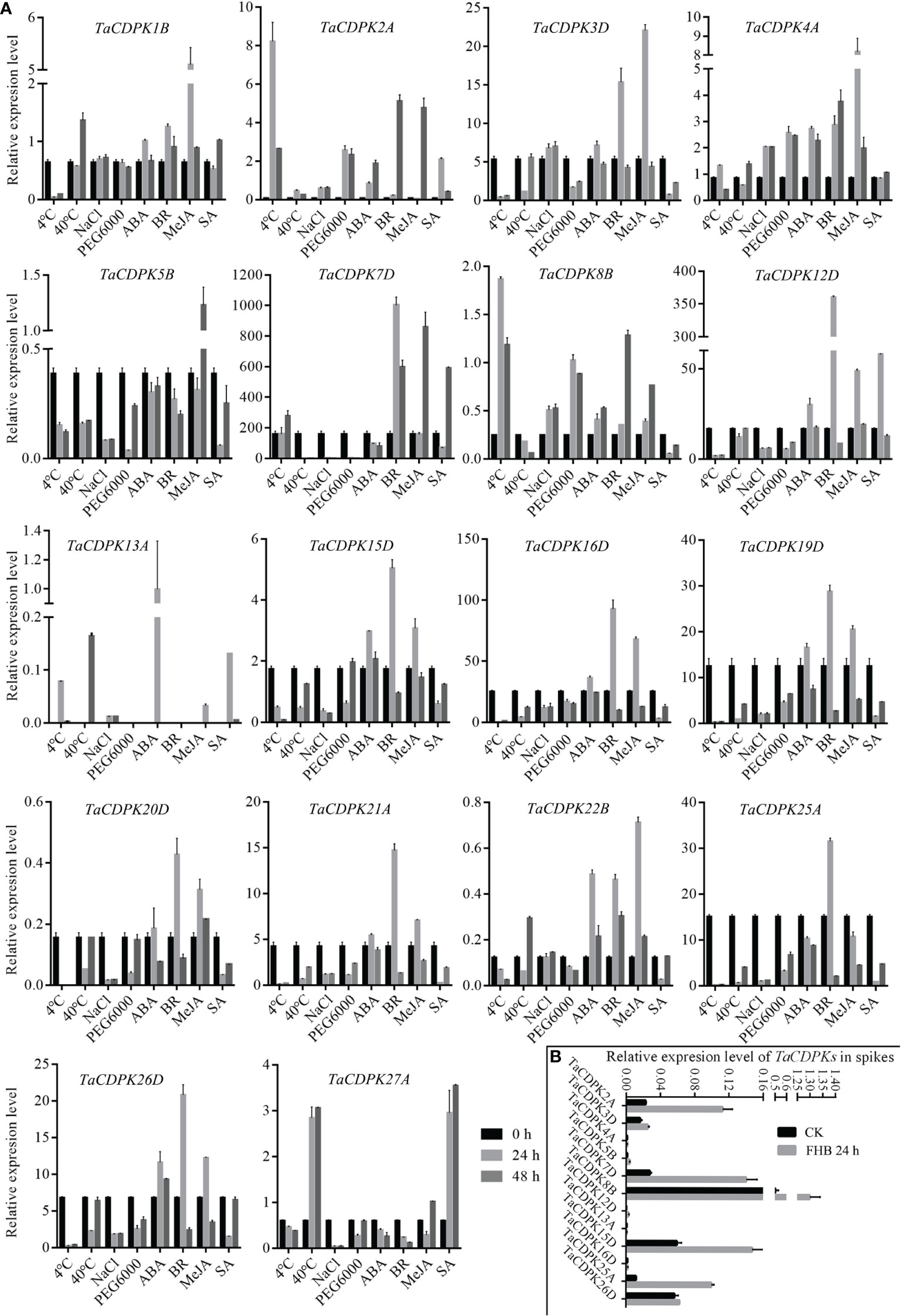
Figure 6 Inducible expression profiles of TaCDPKs in wheat by qRT-PCR. (A) The inducible expression patterns performed by qRT-PCR under cold (4°C), heat (40°C), 20 % PEG6000, salt (200 mM NaCl), ABA (100 μM), SA (500 μM), MeJA (100 μM) hormone treatments. In all treatments, the heat (40°C) treatment lasted for 12 h and 24 h, respectively, instead of 24 h and 48 h as indicated in the (A). The 10-day old hydroponic seedlings were used for the analysis. (B) Inducible expression profiles of TaCDPKs in wheat spike treated with Fusarium graminae spore suspension for 24 h. The expression level of every gene is the mean of results from three independent experiments, each having three or four replicates. CK: the control group; FHB: the experimental group treated with Fusarium graminae spore suspension, which are associated with Fusarium Head Blight (FHB).
3.5 Interaction and co-localization relationships between the members of TaCDPKs and TaNOX7
Lots of studies have addressed that the roles of CDPKs in plant growth regulation and various stress responses are closely associated with NOX-/RBOH mediated ROS production in a Ca2+-dependent manner (Potocký et al., 2007; Potocký et al., 2012; Boisson-Dernier et al., 2013; Yamauchi et al., 2017). In addition, numerous literatures confirmed that CDPKs can directly interact with NOXs/RBOHs, and both of which synergistically involved in plant development and response to environmental stress (Kobayashi et al., 2007; Dubiella et al., 2013; Majumdar and Kar, 2018). Therefore, in order to obtain more insights into the function of TaCDPKs, the network signal relationships between 26 members of CDPK family and 9 members of NOX family from the network of STRING had been obtained and drawn with the software of Cytoscape and Adobe Photoshop (Figure 3).
As expected, there are indeed complicated signal relationships between CDPK and NOX family members (Figure 3). In addition, our previous research showed that TaCDPK13 could directly interact with and activate TaNOX7 for ROS production, which perform a crucial role during plant development and stress tolerance (Hu et al., 2020a). Therefore, based on the results mentioned above, coupled with the information associated with subcellular localization of TaCDPKs (Table S1), we selected TaCDPK2/4/14/16/20/21 as the represents from plasma membrane, cytoplasm, whole cell, chloroplast, and mitochondrion localized members as shown in Table S1, and analyzed the relationships between them and TaNOX7 with the physical interaction between TaNOX7 and TaCDPK13 as a positive control. LCI assay showed that there are different intensity of fluorescence signals, presenting TaNOX7 has different interaction with TaCDPK2/4/14/16. The signals from TaCDPK4-TaNOX7 were the strongest, followed by TaCDPK2-TaNOX7, TaCDPK16-TaNOX7, and there are no obvious signals between TaCDPK14/20/21 and TaNOX7 (Figure 7A). These indicated that TaNOX7 may interact with TaCDPK2/4/16, respectively, but not with TaCDRK14. Furthermore, as shown in Figure 7B, BiFC experiment further verified that TaNOX7 can interact with TaCDPK2/4, but not with TaCDPK16. In addition, Co-IP assays confirmed the conclusion once again (Figure 7C). Moreover, the results of subcellular localization indicated that TaCDPK2/4 were all co-located on the cell membrane with TaNOX7 (Figure S4), which further provide theoretical support for the TaCDPK2/4-TaNOX7 interaction.
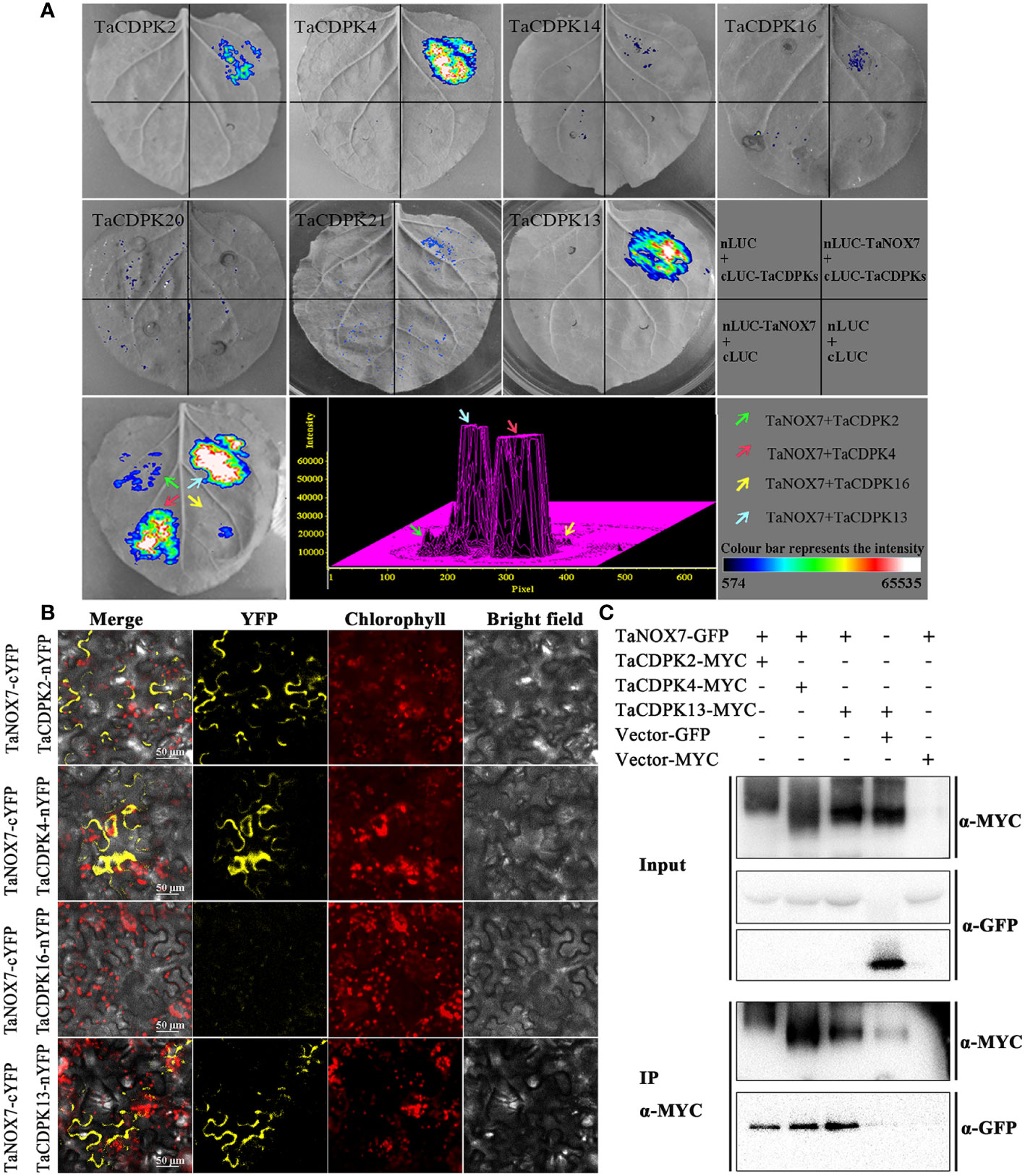
Figure 7 Protein interactions between TaCDPKs and TaNOX7. (A) Verification of protein interactions between TaCDPK2/4/14/16/20/21 and TaNOX7 were performed using the method of firefly luciferase complementation imaging (LCI) assay; (B) Bimolecular fluorescence complementation (BiFC) assay showing the interactions between TaCDPK2/4/16 and TaNOX7. (C) The interactions between TaCDPK2/4 and TaNOX7 were confirmed using the assay of co-immunoprecipitation (Co-IP), in which input and immunoprecipitates were analyzed by immunoblotting using anti-GFP and anti-MYC antibodies.
3.6 Coexpression of TaNOX7 and TaCDPK2/4 promoted ROS production
Increasing literatures have reported that CDPK-mediated NOX activation promotes the production of ROS, which plays important role in plants. Consistent with these mentioned above, the results in Figure 8 showed the red-brown precipitates in the regions that co-expressing of cLUC-TaCDPK2/4 and nLUC-TaNOX7 were significantly higher than those in the control group, which implying that coexpression of TaNOX7 and TaCDPK2/4 promotes ROS accumulation in plant leaves. Hence, what is the biological significance of the interaction between TaCDPK2/4 and TaNOX7? Therefore, we further constructed the sophisticated tissue expression profiles of TaNOX7 and TaCDPK2/4 at wheat spikes from 12 different developmental stages (Figure 9A). As shown in Figure 9A, compared with flag leaves, TaNOX7 was expressed at an absolutely higher level in the young panicles at all the examined stages. Unexpectedly, the expression level of TaCDPK2/4 was much lower in each stage of young panicles than that of flag leaves, which suggesting that there are no co-expression relationships between TaNOX7 and TaCDPK2/4 during wheat panicle development. Therefore, we further constructed tissue expression profiles of TaNOX7 and TaCDPK2/4 in six flower organs at heading stage (Figure 9B). As shown in Figure 9B, the expression level of TaCDPK2 was still lower than that of flag leaves. Intriguingly, TaCDPK4 is expressed in the pistils with peak level. These results, coupled with the previous tissue expressions and protein interactions, we concluded that TaCDPK2 was mainly expressed in young leaves and flag leaves, and the interaction with NOX7 might be involved in the vegetative and reproductive growth of plants; TaCDPK4 was mainly expressed in the pistils, and its interaction with NOX7 might contribute to seed development.
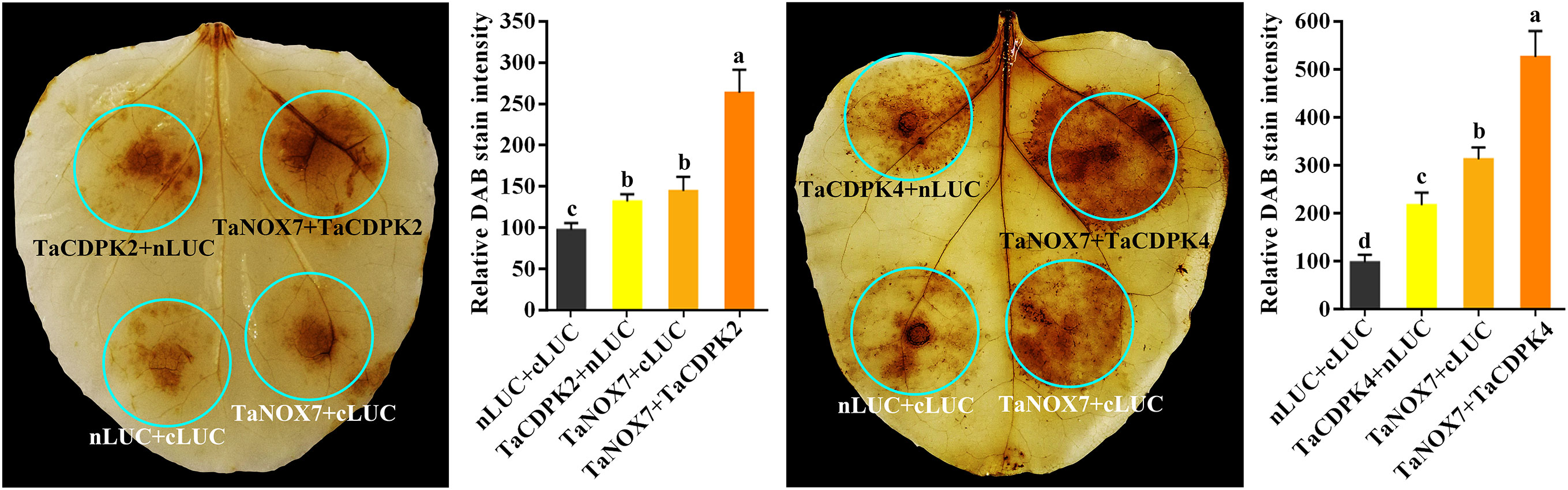
Figure 8 TaCDPK2/4-TaNOX7 interactions enhanced ROS production in plants. Transient coexpression of TaNOX7 with TaCDPK2 or TaCDPK4 all enhanced ROS production in the leaves of N. benthamiana. The level of ROS accumulation was detected by the method of DAB (3, 3´- diaminobenzidine) staining method. The DAB staining intensity in in situ ROS levels of the agroinfiltrated tobacco leaves was calculated based on the stain intensity of the control “cLUC + nLUC”. Data are means ± SD (n = 10~15 leaves) from more than three independent experiments. Bars annotated with different letters represent values that are significantly different at P ≤ 0.05 according to one-way analysis of variance (ANOVA) analysis.
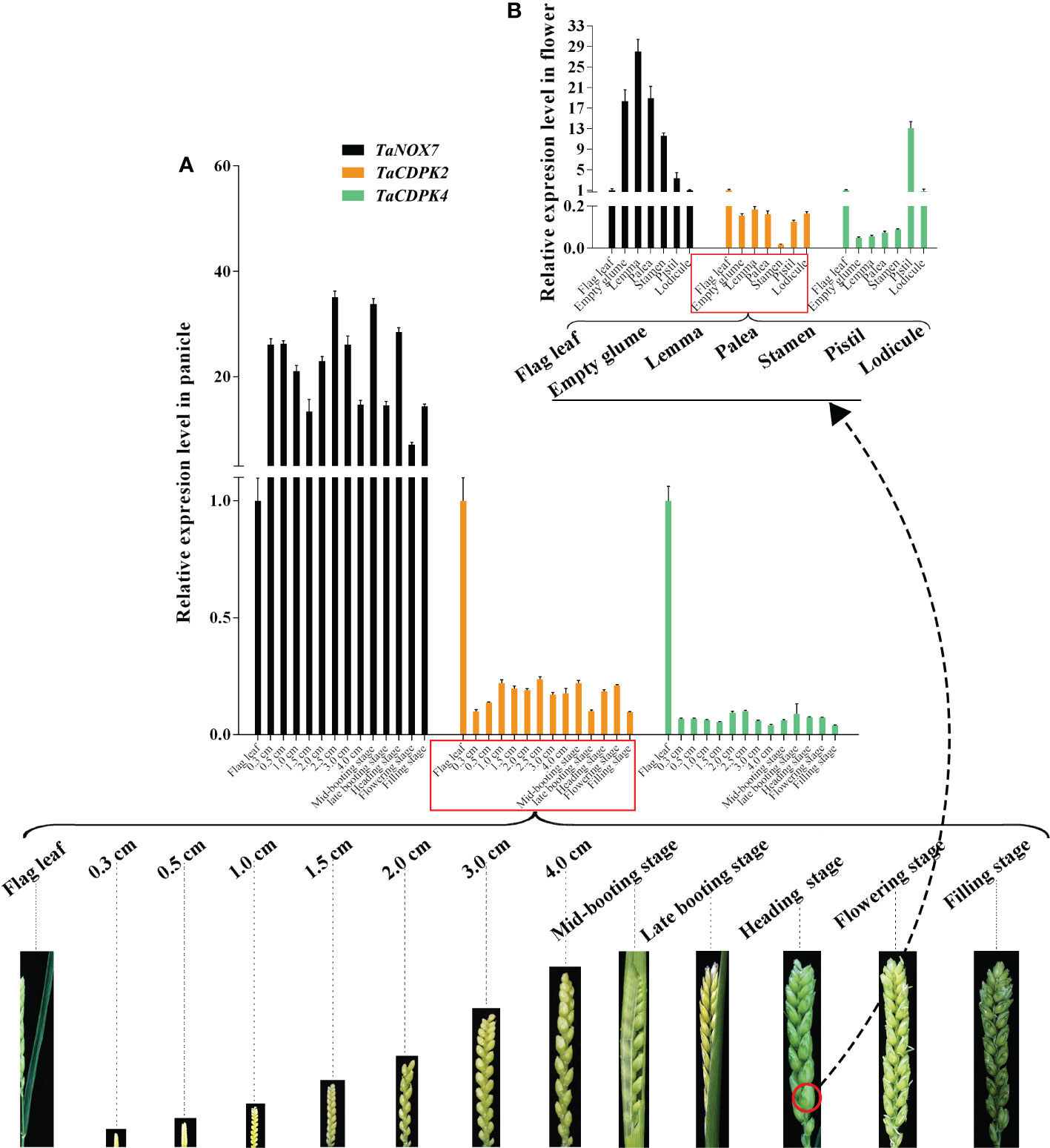
Figure 9 Co-expression interactions between TaNOX7 and TaCDPK2/4 in wheat. (A) Co-expression of TaCDPK2/4 and TaNOX7 in panicles from 12 different developmental stages; (B) Co-expression of TaCDPK2/4 and TaNOX7 in six flower organs at heading stage.
4 Discussion
4.1 Wheat CDPKs are diverse in members and structures with a complicated evolution history
In this study, a total of 79 CDPK family genes, which encode 30 TaCDPKs, were identified according to the sequence analysis and domain composition (Table S1). Interestingly, not every protein has three homologous genes distributed on the subchromosomes A, B, and D. For example, TaCDPK6 includes two homologous genes (TaCDPK6-6A and TaCDPK6-6D) but TaCDPK1 has only one (TaCDPK1-4B) (Figure 1A). These means that their homologs on a certain chromosome might be lost during the long-term evolution and natural selection. In addition, the homologous genes from different subgenomes, such as TaCDPK27-4D and TaCDPK28-5A in Figure 1A, clustered together firstly, which may be attributed to the gene duplication, and/or exon shuffling. Moreover, the phenomenon of motif EF_hand loss is also common in TaCDPK protein sequences. In fact, these anomalies probably belong to genovariation including gene structural variation, rearrangement, DNA sequence loss, and transposon activation occurred frequently during the polyploidization of genome in plants (Jackson and Chen, 2010). Furthermore, degeneration and/or lose of the motif EF_hand in TaCDPKs (such as TaCDPK3) supports the viewpoint that the evolutionary process from TaCDPKs to CDPK-related receptor-like kinases TaCRKs (TaCRKs possess the conserved domains that the typical TaCDPKs have, but lack the EF_hand motifs). In summary, all the information mentioned above, along with the non-random distribution of TaCDPKs on 21 chromosomes (Figure 2), suggesting that wheat CDPKs underwent a complicated evolutionary history, which might endow TaCDPK family with the gene expansion, gene variation, and functional divergence, though further researches are needed to confirm these. In addition, the analyses of gene structure (Figure 1C), protein cluster (Figure 1A), and gene karyotype (Figure 2) indicated that the map of TaCDPK25 may be mistake, but the corresponding location of TaCDPK25 on 6D may be more reasonable, which will provide references for the precise mapping of them and their linked genes.
4.2 Wheat CDPKs exhibit a great specificity in expression and play vital roles in both the plant growth regulation and stress response
Specific expression is a common characteristic of the genes of a certain protein family in plants, which often reflects the cross-talk and/or difference in the functions of the family members (Hu et al., 2018). In this study, we found that the expression patterns of CDPK family members in wheat showed overall regularity and individual specificity, which indicating their functional synergy and specificity.
Firstly, in terms of organization and spatio-temporal expression, 67.7% of the family members (TaCDPK2-5/7-13/15/16/19/20/22-27/29/30) all expressed with peak level in different reproductive organs, suggesting that they probably all involved in the regulation of plant reproductive growth but in different reproductive organs (Figures S1-2, 4 and Table 1). For instance, the expression profile that TaCDPK13 expressed in anther at the anthesis stage with peak level, which was consistent with our previous results that TaCDPK13 functioned in plant fertility (Hu et al., 2020a). Moreover, increasing reports found that ZmCPK32, AtCPK32, and GmCDPK38 as the homologue of TaCDPK23, played important roles in modulating flowering time and pollen tube growth (Zhou et al., 2014; Li et al., 2018; Li et al., 2022a; Li et al., 2022b), implying that TaCDPK23 probably plays a crucial role in regulating the development of spike/anther at the anthesis stage. OsCDPK1 was proved that it played a functional role in rice seed development (Jiang et al., 2018). As shown in Table S5, TaCDPK8 and OsCDPK1 are also the homologues with the highest identification 94.2%, which tempt us to speculate that TaCDPK8 perhaps performs the similar functions to OsCDPK1 by regulating the development of ovary/anther at the inforescence stage in wheat.
Secondly, under abiotic stresses, the expression of TaCDPK6-12 were all sensitive to heat and obviously upregulated under heat stress, as well as TaCDPK2/5 to cold stress, TaCDPK15 to drought stress, and TaCDPK15/25 to sodium hydrosulfide (NaHS) stress (Figure 5 and Table 1). Previous studies showed that the homologues of CDPKs played versatile functions in plants in response to different abiotic stresses. For instance, StCDPK32 (Zhu et al., 2021), ZmCPK11 (Borkiewicz et al., 2020), AtCPK12 (Zhang et al., 2018), OsCDPK21 (Asano et al., 2011), and AtCPK3 (Mehlmer et al., 2010) were all required for plant adaptation or response to salinity stress. In rice, the expression level of OsCDPK13 was also increased in leaf sheath segments upon subjected to cold stress (Yang et al., 2003). On the contrary, ZmCPK1 was identified as a negative regulator in cold stress signaling (Weckwerth et al., 2015). CsCDPK20 and CsCDPK26 might act as a positive regulator in tea plant (Camellia sinensis) response to heat stress (Wang et al., 2018). In foxtail millet (Setaria italica), overexpression of SiCDPK24 in plants enhanced drought resistance and improved the survival rate under drought stress (Yu et al., 2018b). Moreover, overexpression of GmCDPK3 also improved plant tolerance to drought as well as salt stresses (Wang et al., 2019b). Besides these, OsCDPK1 also conferred drought tolerance in rice seedlings besides of its functions in seed development (Ho et al., 2013; Jiang et al., 2018). Based on the inducible expression profile of TaCDPK2, the high identification between TaCDPK2 and OsCDPK13 with 95.9%, we speculate that TaCDPK2 perhaps play the potential role in plant response to cold stress. In addition, although the induced expression profile (Figures 3, 7) did not give a clear picture of the response of TaCDPK27 to cold stress, the more cold response-elements in the promoter of TaCDPK27 (Figure S3) and its high identification with ZmCPK1 (83.0% in Table S5) also suggesting its potential role in plant response to cold stress.
Thirdly, under biological stresses, the expression levels of different groups showed different responses (in Figures 5, 6B and Table 1). For example, the members from the group I (TaCDPK2/7/12/15/25/26) were upregulated, but downregulated in the group II (TaCDPK5/6/8/9/13/17/22/28), and barely responded in the group III (TaCDPK11/19/20/23/24/29/30), under the treatment of F. graminearum, which is the main pathogen of FHB. While, TaCDPK4/10/12/14/15/25 were all downregulated under P. graminis infection, which is the main pathogen of wheat stem rust. TaCDPK5/10/15/25 were susceptive to P. striiformis (Puccinia striiformis, wheat stripe rust pathogen), as well as TaCDPK4/10/12 to flg22 (pattern pathogenic elicitors from bacteria). These results suggested that different members with different genes and protein structures may also have the similar biological functions, such as TaCDPK15 and TaCDPK25 to F. graminearum as well as to P. graminis and P. striiformis in plants. On the other hand, the same members of TaCDPKs may show different biological functions in response to different biological stresses. For example, the expression level of TaCDPK12 was upregulated under F. graminearum or flg22 stresses, but downregulated under P. graminis stress. Up to now, lots of CDPK homologs have been reported to be involved in plant immune processes. In Arabidopsis, AtCPK28, the homologs of TaCDPK6 (with the identification of 81.2%), as a negative regulator that continually buffers immune signaling by controlling the turnover of the plasma-membrane-associated cytoplasmic kinase BIK1, which is a rate limiting in pathogen-associated molecular patterns (PAMP)-triggered immunity (PTI) signaling (Monaghan et al., 2014). In addition, AtCPK5/CPK6 signaling pathways contribute to defense against Botrytis cinerea by promoting the biosynthesis of 4-methoxyindole-3-ylmethylglucosinolate and camalexin in plants (Yang et al., 2020b).
Finally, the induced expression profiles in Figure 6A indicated that TaCDPK1-5/7/12/16/19/22 were all sensitive to hormone MeJA, which are consistent with the results that most members of TaCDPKs harboring MeJA-responsive element (JARE) in their promoters (Figure S3). The results mentioned above suggesting that these TaCDPK family members perhaps widely involved in MeJA-mediated signaling pathways, which play important roles in plant growth, development, senescence, and in response to biotic and abiotic stresses (Shu et al., 2020; Ma et al., 2021; Raza et al., 2021; Wei et al., 2021). Whereas, TaCDPK2/3/7/8/12/15/16/19-21/25/26 were all sensitive to hormone BR as well as TaCDPK12/27 to SA and TaCDPK13 to ABA. Moreover, increasingly compelling evidences indicated that CDPKs involved in many hormone-mediated signaling in plants. In Arabidopsis, AtCPK6 the homologue of TaCDPK1 (identification 70.3%) was demonstrated that it functioned as a positive regulator in MeJA signaling in Arabidopsis guard cells as well as ABA-induced stomatal closure (Munemasa et al., 2011; Brandt et al., 2012). Another report showed that AtCPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth (Zhao et al., 2011). In addition, AtCPK29 was involved in the process of auxin efflux transport, polarity and auxin responses by specifically phosphorylating the target residues on the auxin efflux transporter (PIN) (Lee et al., 2021). In this paper, the expression level of TaCDPK13 in anther/flowering stage and its sensitivity to ABA (Table 1) were all consistent with our previous study, that TaCDPK13 played crucial roles in plant fertility, and drought tolerance (Hu et al., 2020a). Such results strongly support the idea that TaCDPK13 may be involved in drought response via ABA-dependent signal pathways.
4.3 The complicated interactions between TaCDPKs and TaNOXs perhaps play vital roles in plant development and response to stresses by regulating ROS production
It is well known that ROS and Ca2+ are universal and important intracellular signaling molecules, and their homeostasis play important roles in plant growth and development as well as response to biotic and abiotic stresses. More importantly, ROS and Ca2+, both as the signaling messengers, have complex cross-talks during signaling. For instance, Ca2+ binding can activate CDPKs to phosphorylate NADPH oxidases OsRbohB for ROS production (Kobayashi et al., 2007), which is necessary for Ca2+ influx, and then the induced ROS in turn may trigger Ca2+ efflux from intracellular Ca2+ stores in vivo (McAinsh MR et al., 1996; Pei et al., 2000). It should be added here that NADPH oxidases (NOXs), mostly known as respiratory burst oxidase homologs (RBOHs), are the key producers of ROS in plants (Hu et al., 2020b). Moreover, both TaCDPKs and TaNOXs all contain the EF_Hand, which are capable of calcium binding domains and thereby activate the protease activity of CDPKs and NOXs by cooperating with other particles (Hu et al., 2020b). Therefore, there must be complicated interactions between the members of CDPKs and NOXs in plants. Previously, we found that TaCDPK13 can interact with TaNOX7 for the plant fertility and drought tolerance (Hu et al., 2020a). Here, we found that the two others, TaCDPK2/4, can also directly interact with TaNOX7 and coexpression of the CDPKs with TaNOX7 enhanced ROS production (Figures 7, 8). Intriguingly, increasing literatures reported that CDPKs-NOXs/RBOHs interactions played important roles in plants by regulating ROS homeostasis. For example, OsRboh mediated ROS production, which is induced by OsCDPK5/OsCDPK13, is essential for aerenchyma formation in rice roots (Yamauchi et al., 2017). In addition, StCDPK23 may participate in the wound healing of potato tubers by regulating StRBOHs for H2O2 production (Ma et al., 2022). AtCPK5 phosphorylates AtRbohD and enhances ROS production for defense responses and bacterial resistance (Dubiella et al., 2013); BnaCPK6L phosphorylates BnaRBOHD and increases the accumulation of ROS and HR-like cell death (Pan et al., 2019). More intriguingly, OsCPK12 promotes the tolerance of rice to salt stress by repressing the expression level of OsRbohI and reducing the accumulation of ROS (Asano et al., 2012; Boudsocq and Sheen, 2013). Therefore, CDPKs can regulate the activity of NOXs/RBOHs for regulating ROS homeostasis in plants, which plays diverse and vital functions in plants. Then, what are the biological significances of TaCDPK2/4-TaNOX7 interactions? All the expression patterns and analyses from Figures S1/S2, 4–6, 9 and Table 1 showed that TaCDPK2 was mainly expressed in young leaves, flag leaves and has significant response to F.graminis and cold stress; TaCDPK4 was mainly expressed in the pistils and has obvious response to Flagelin 22 (Flg22). In addition, our previous results showed that TaNOX7 was expressed in almost all the tissues of wheat and had high sensitivity to many stresses (Hu et al., 2018). TaCDPK13 can also interact with and activate TaNOX7 for ROS production, which can enhance plant fertility and drought tolerance (Hu et al., 2020a). Based on all the results, we concluded that, as shown in the model in Figure 10, TaCDPK2/4-TaNOX7 interactions mediated ROS homeostasis perhaps also plays crucial roles during the progress of vegetative and reproductive growth, seed development, and fertility in plants, respectively. In addition, they perhaps also play essential roles in plant response to biotic and abiotic stresses, such as cold, F.graminis stresses and so on.
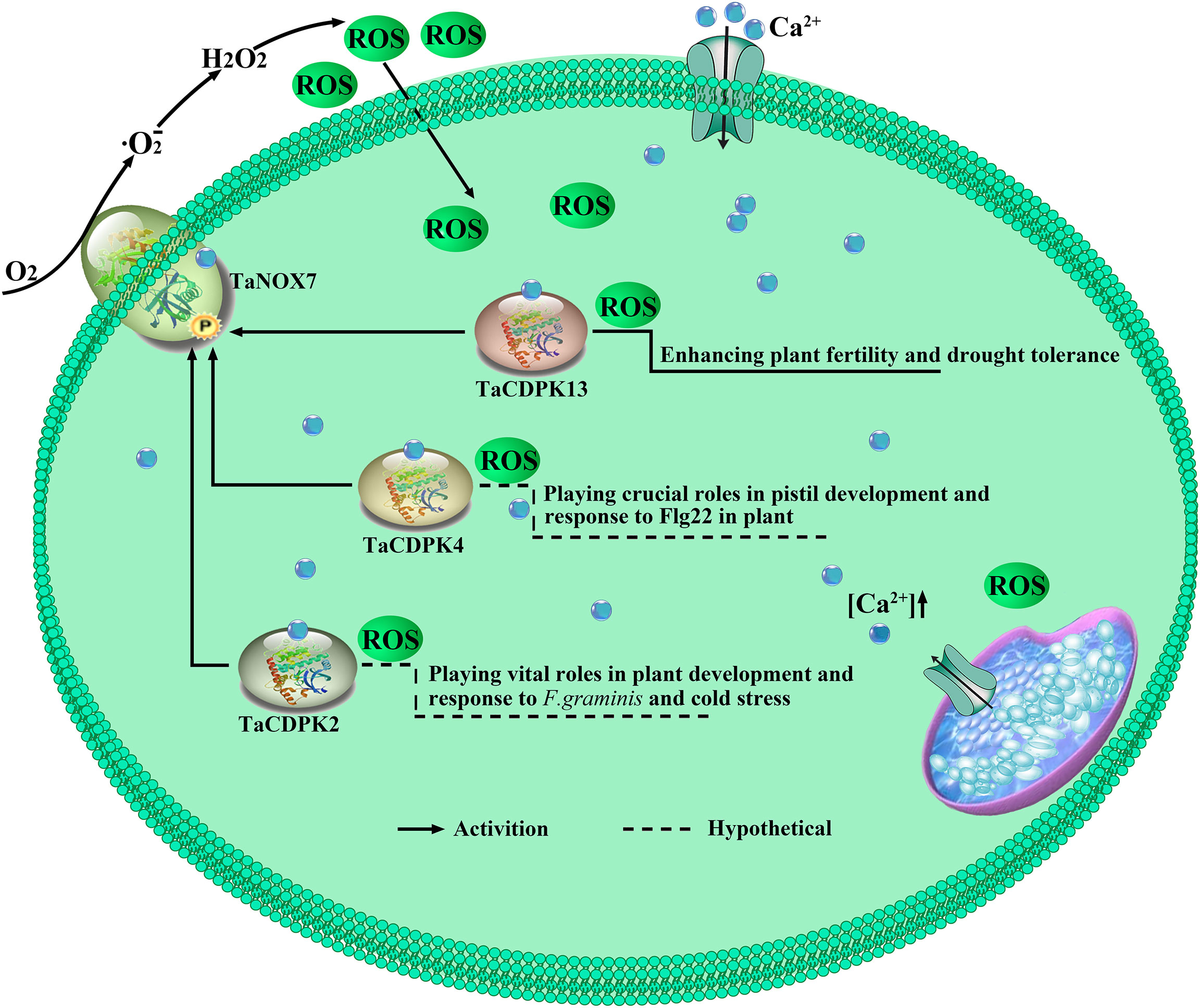
Figure 10 TaCDPK2/4/13-TaNOX7 interactions play crucial roles in plant by regulating ROS production. TaCDPK2/4 and TaCDPK13 (Hu et al., 2020a) can interact with and activate TaNOX7 for ROS production, which plays crucial roles in plant development.
In summary, wheat has multiple members of CDPKs with diverse but vital functions in plant growth, development regulation and stress responses. Every member of TaCDPKs has its specific expression pattern and function. Moreover, the synergistic or antagonistic interactions between TaCDPKs and TaNOXs are complicated and play important roles by regulating ROS level in plant, though the regulatory mechanism and biological significance of them are still under investigation. Therefore, the results obtained here have provided a valuable foundation for further exploring the functions and the signal pathways of CDPK superfamily members, especially the interactions between TaCDPKs and TaNOXs in wheat.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
K-M C, L-L L, and K-S M proposed the concept and content. C-H H, and B-B L wrote the manuscript. P C, H-Y S, and W-G X revised the manuscript. Y Z, Z-H Y and H-X W helped in the sample collection and experiment. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 31270299 and 32102487), the Program for New Century Excellent Talents in University of China (NCET-11-0440), the Key Scientific Research Project of Colleges and Universities in Henan Province (23A210030), and the Foundation of Zhoukou Normal University College Student Scientific Research Innovation Fund Project (ZKNUD2022015 and ZKNUD2022062).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1108622/full#supplementary-material
References
Asano, T., Hakata, M., Nakamura, H., Aoki, N., Komatsu, S., Ichikawa, H., et al. (2011). Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 75, 179–191. doi: 10.1007/s11103-010-9717-1
Asano, T., Hayashi, N., Kobayashi, M., Aoki, N., Miyao, A., Mitsuhara, I., et al. (2012). A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 69, 26–36. doi: 10.1111/j.1365-313X.2011.04766.x
Asano, T., Tanaka, N., Yang, G., Hayashi, N., Komatsu, S. (2005). Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 46, 356–366. doi: 10.1093/pcp/pci035
Boisson-Dernier, A., Lituiev, D. S., Nestorova, A., Franck, C. M., Thirugnanarajah, S., Grossniklaus, U. (2013). ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PloS Biol. 11, e1001719. doi: 10.1371/journal.pbio.1001719
Borkiewicz, L., Polkowska-Kowalczyk, L., Ciesla, J., Sowinski, P., Jonczyk, M., Rymaszewski, W., et al. (2020). Expression of maize calcium-dependent protein kinase (ZmCPK11) improves salt tolerance in transgenic Arabidopsis plants by regulating sodium and potassium homeostasis and stabilizing photosystem II. Physiol. Plant 168, 38–57. doi: 10.1111/ppl.12938
Boudsocq, M., Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Brandt, B., Brodsky, D. E., Xue, S. W., Negi, J., Iba, J., Kangasjärvi, J., et al (2012). Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. U.S.A. 109, 10593–10598. doi: 10.1073/pnas.1116590109
Chen, Y. F., Li, L. Q., Xu, Q., Kong, Y. H., Wang, H., Wu, W. H. (2009). The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low pi stress in Arabidopsis. Plant Cell 21, 3554–3566. doi: 10.1105/tpc.108.064980
Dubiella, U., Seybold, H., Durian, G., Komander, E., Lassig, R., Witte, C. P., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U.S.A. 110, 8744–8749. doi: 10.1073/pnas.1221294110
Durian, G., Sedaghatmehr, M., Matallana-Ramirez, L. P., Schilling, S. M., Schaepe, S., Guerra, T., et al. (2020). Calcium-dependent protein kinase CPK1 controls cell death by In vivo phosphorylation of senescence master regulator ORE1. Plant Cell 32, 1610–1625. doi: 10.1105/tpc.19.00810
Geng, S., Zhao, Y., Tang, L., Zhang, R., Sun, M., Guo, H., et al. (2011). Molecular evolution of two duplicated CDPK genes CPK7 and CPK12 in grass species: a case study in wheat (Triticum aestivum l.). Gene 475, 94–103. doi: 10.1016/j.gene.2010.12.015
Ho, S. L., Huang, L. F., Lu, C. A., He, S. L., Wang, C. C., Yu, S. P., et al. (2013). Sugar starvation- and GA-inducible calcium-dependent protein kinase 1 feedback regulates GA biosynthesis and activates a 14-3-3 protein to confer drought tolerance in rice seedlings. Plant Mol. Biol. 81, 347–361. doi: 10.1007/s11103-012-0006-z
Hrabak, E. M., Chan, C. W., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. doi: 10.1104/pp.102.011999
Huang, C., Yan, Y., Zhao, H., Ye, Y., Cao, Y. (2020). Arabidopsis CPK5 phosphorylates the chitin receptor LYK5 to regulate plant innate immunity. Front. Plant Sci. 11, 702. doi: 10.3389/fpls.2020.00702
Hu, C. H., Wang, P. Q., Zhang, P. P., Nie, X. M., Li, B. B., Tai, L., et al. (2020b). NADPH oxidases: The vital performers and center hubs during plant growth and signaling. Cells 9, 437. doi: 10.3390/cells9020437
Hu, C. H., Wei, X. Y., Yuan, B., Yao, L. B., Ma, T. T., Zhang, P. P., et al. (2018). Genome-wide identification and functional analysis of NADPH oxidase family genes in wheat during development and environmental stress responses. Front. Plant Sci. 9, 906. doi: 10.3389/fpls.2018.00906
Hu, C. H., Zeng, Q. D., Tai, L., Li, B. B., Zhang, P. P., Nie, X. M., et al. (2020a). Interaction between TaNOX7 and TaCDPK13 contributes to plant fertility and drought tolerance by regulating ROS production. J. Agric. Food Chem. 68, 7333–7347. doi: 10.1021/acs.jafc.0c02146
Jackson, S., Chen, Z. J. (2010). Genomic and expression plasticity of polyploidy. Curr. Opin. Plant Biol. 13, 153–159. doi: 10.1016/j.pbi.2009.11.004
Jiang, J. Z., Kuo, C. H., Chen, B. H., Chen, M. K., Lin, C. S., Ho, S. L. (2018). Effects of OsCDPK1 on the structure and physicochemical properties of starch in developing rice seeds. Int. J. Mol. Sci. 19, 3247. doi: 10.3390/ijms19103247
Kobayashi, M., Ohura, I., Kawakita, K., Yokota, N., Fujiwara, M., Shimamoto, K., et al. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19, 1065–1080. doi: 10.1105/tpc.106.048884
Kumar, D., Yusuf, M., Singh, P., Sardar, M., Sarin, N. (2014). Histochemical detection of superoxide and H2O2 accumulation in brassica juncea seedlings. Bio-Protocol 4, 1108. doi: 10.21769/BioProtoc.1108
Lee, H., Ganguly, A., Baik, S., Cho, H. T. (2021). Calcium-dependent protein kinase 29 modulates PIN-FORMED polarity and Arabidopsis development via its own phosphorylation code. Plant Cell 33, 3513–3531. doi: 10.1093/plcell/koab207
Li, X., Chen, L., Yao, L., Zou, J., Hao, J., Wu, W. (2022b). Calcium-dependent protein kinase CPK32 mediates calcium signaling in regulating Arabidopsis flowering time. Natl. Sci. Rev. 9, nwab180. doi: 10.1093/nsr/nwab180
Li, X., Hu, D., Cai, L., Wang, H., Liu, X., Du, H., et al. (2022a). CALCIUM-DEPENDENT PROTEIN KINASE38 regulates flowering time and common cutworm resistance in soybean. Plant Physiol. 190, 480–499. doi: 10.1093/plphys/kiac260
Li, M., Hu, W., Ren, L., Jia, C., Liu, J., Miao, H., et al. (2020). Identification, expression, and interaction network analyses of the CDPK gene family reveal their involvement in the development, ripening, and abiotic stress response in banana. Biochem. Genet. 58, 40–62. doi: 10.1007/s10528-019-09916-2
Li, J., Li, Y., Deng, Y., Chen, P., Feng, F., Chen, W., et al. (2018). A calcium-dependent protein kinase, ZmCPK32, specifically expressed in maize pollen to regulate pollen tube growth. PloS One 13, e0195787. doi: 10.1371/journal.pone.0195787
Liu, L. J., Sonbol, F. M., Huot, B., Gu, Y. N., Withers, J., Mwimba, M., et al. (2016). Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7, 13099. doi: 10.1038/ncomms13099
Ma, L., Jiang, H., Ren, Y. Y., Yang, J. W., Han, Y., Si, H. J., et al. (2022). Overexpression of StCDPK23 promotes wound healing of potato tubers by regulating StRbohs. Plant Physiol. Biochem. 185, 279–289. doi: 10.1016/j.plaphy.2022.06.010
Majumdar, A., Kar, R. K. (2018). Congruence between PM h(+)-ATPase and NADPH oxidase during root growth: a necessary probability. Protoplasma 255, 1129–1137. doi: 10.1007/s00709-018-1217-1
Ma, B., Liu, X., Guo, S., Xie, X., Zhang, J., Wang, J., et al. (2021). RtNAC100 involved in the regulation of ROS, na(+) accumulation and induced salt-related PCD through MeJA signal pathways in recretohalophyte reaumuria trigyna. Plant Sci. 310, 110976. doi: 10.1016/j.plantsci.2021.110976
Martínez-Noël, G., Nagaraj, V. J., Caló, G., Wiemken, A., Pontis, H. G. (2007). Sucrose regulated expression of a Ca2+-dependent protein kinase (TaCDPK1) gene in excised leaves of wheat. Plant Physiol. Biochem. 45, 410–419. doi: 10.1016/j.plaphy.2007.03.004
Matschi, S., Werner, S., Schulze, W. X., Legen, J., Hilger, H. H., Romeis, T. (2013). Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 73, 883–896. doi: 10.1111/tpj.12090
McAinsh MR, C. H., Mansfield, T. A., Hetherington, A. M. (1996). Changes in stomatal behavior and guard cell cytosolic free calcium in response to oxidative stress. Plant Physiol. 111, 1031–1042. doi: 10.1104/pp.111.4.1031
Mehlmer, N., Wurzinger, B., Stael, S., Hofmann-Rodrigues, D., Csaszar, E., Pfister, B., et al. (2010). The Ca(2+) -dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 63, 484–498. doi: 10.1111/j.1365-313X.2010.04257.x
Monaghan, J., Matschi, S., Shorinola, O., Rovenich, H., Matei, A., Segonzac, C., et al. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe 16, 605–615. doi: 10.1016/j.chom.2014.10.007
Munemasa, S., Hossain, M. A., Nakamura, Y., Mori, I. C., Murata, Y. (2011). The Arabidopsis calcium-dependent protein kinase, CPK6, functions as a positive regulator of methyl jasmonate signaling in guard cells. Plant Physiol. 155, 553–561. doi: 10.1104/pp.110.162750
Pan, G., Zhang, H., Chen, B., Gao, S., Yang, B., Jiang, Y. Q. (2019). Rapeseed calcium-dependent protein kinase CPK6L modulates reactive oxygen species and cell death through interacting and phosphorylating RBOHD. Biochem. Biophys. Res. Commun. 518, 719–725. doi: 10.1016/j.bbrc.2019.08.118
Pei, Z. M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734. doi: 10.1038/35021067
Potocký, M., Jones, M., Bezvoda, R., Smirnoff, N., Žárský, V. (2007). Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 146, S269–S270. doi: 10.1016/j.cbpa.2007.01.605
Potocký, M., Pejchar, P., Gutkowska, M., Jimeínez-Quesada, M. J., Potockaí, A., Alche Jde, D., et al. (2012). NADPH oxidase activity in pollen tubes is affected by calcium ions, signaling phospholipids and Rac/Rop GTPases. J. Plant Physiol. 169, 1654–1663. doi: 10.1016/j.jplph.2012.05.014
Raza, A., Charagh, S., Zahid, Z., Mubarik, M. S., Javed, R., Siddiqui, M. H., et al. (2021). Jasmonic acid: a key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 40, 1513–1541. doi: 10.1007/s00299-020-02614-z
Shu, P., Li, Z., Min, D., Zhang, X., Ai, W., Li, J., et al. (2020). CRISPR/Cas9-mediated SlMYC2 mutagenesis adverse to tomato plant growth and MeJA-induced fruit resistance to botrytis cinerea. J. Agric. Food Chem. 68, 5529–5538. doi: 10.1021/acs.jafc.9b08069
Valmonte, G. R., Arthur, K., Higgins, C. M., MacDiarmid, R. M. (2014). Calcium-dependent protein kinases in plants: evolution, expression and function. Plant Cell Physiol. 55, 551–569. doi: 10.1093/pcp/pct200
Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. doi: 10.1111/J.1365-313X.2004.02219.X
Wang, M., Li, Q., Sun, K., Chen, X., Zhou, Q., Li, H., et al. (2018). Involvement of CsCDPK20 and CsCDPK26 in regulation of thermotolerance in tea plant (Camellia sinensis). Plant Mol. Biol. Rep. 36, 176–187. doi: 10.1007/s11105-018-1068-0
Wang, D., Liu, Y. X., Yu, Q., Zhao, S. P., Zhao, J. Y., Ru, J. N., et al. (2019c). Functional analysis of the soybean GmCDPK3 gene responding to drought and salt stresses. Int. J. Mol. Sci. 20, 5909. doi: 10.3390/ijms20235909
Wang, X., Lv, S., Han, X., Guan, X., Shi, X., Kang, J., et al. (2019a). The calcium-dependent protein kinase CPK33 mediates strigolactone-induced stomatal closure in Arabidopsis thaliana. Front. Plant Sci. 10, 1630. doi: 10.3389/fpls.2019.01630
Wang, B., Zhang, Y., Bi, Z., Liu, Q., Xu, T., Yu, N., et al. (2019b). Impaired function of the calcium-dependent protein kinase, OsCPK12, leads to early senescence in rice (Oryza sativa l.). Front. Plant Sci. 10, 52. doi: 10.3389/fpls.2019.00052
Weckwerth, P., Ehlert, B., Romeis, T. (2015). ZmCPK1, a calcium-independent kinase member of the zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 38, 544–558. doi: 10.1111/pce.12414
Wei, X., Guan, W., Yang, Y., Shao, Y., Mao, L. (2021). Methyl jasmonate promotes wound healing by activation of phenylpropanoid metabolism in harvested kiwifruit. Postharvest Biol. Technol. 175, 111472. doi: 10.1016/j.postharvbio.2021.111472
Wei, C., Zhang, R., Yang, X., Zhu, C., Li, H., Zhang, Y., et al. (2019). Comparative analysis of calcium-dependent protein kinase in cucurbitaceae and expression studies in watermelon. Int. J. Mol. Sci. 20, 2527. doi: 10.3390/ijms20102527
Wu, M., Liu, H., Gao, Y., Shi, Y., Pan, F., Xiang, Y. (2020). The moso bamboo drought-induced 19 protein PheDi19-8 functions oppositely to its interacting partner, PheCDPK22, to modulate drought stress tolerance. Plant Sci. 299, 110605. doi: 10.1016/j.plantsci.2020.110605
Yamauchi, T., Yoshioka, M., Fukazawa, A., Mori, H., Nishizawa, N. K., Tsutsumi, N., et al. (2017). An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 29, 775–790. doi: 10.1105/tpc.16.00976
Yang, Q., Dong, B., Wang, L., Song, Z., Niu, L., Li, H., et al. (2020a). CDPK6 phosphorylates and stabilizes MYB30 to promote hyperoside biosynthesis that prolongs the duration of full-blooming in okra. J. Exp. Bot. 71, 4042–4056. doi: 10.1093/jxb/eraa174
Yang, G., Shen, S., Yang, S., Komatsu, S. (2003). OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced in response to cold and gibberellin. Plant Physiol. Biochem. 41, 369–374. doi: 10.1016/S0981-9428(03)00032-9
Yang, L., Zhang, Y., Guan, R., Li, S., Xu, X., Zhang, S., et al. (2020b). Co-Regulation of indole glucosinolates and camalexin biosynthesis by CPK5/CPK6 and MPK3/MPK6 signaling pathways. J. Integr. Plant Biol. 62, 1780–1796. doi: 10.1111/jipb.12973
Yip Delormel, T., Boudsocq, M. (2019). Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 224, 585–604. doi: 10.1111/nph.16088
Yu, H., Xiao, A., Dong, R., Fan, Y., Zhang, X., Liu, C., et al. (2018a). Suppression of innate immunity mediated by the CDPK-rboh complex is required for rhizobial colonization in medicago truncatula nodules. New Phytol. 220, 425–434. doi: 10.1111/nph.15410
Yu, T. F., Zhao, W. Y., Fu, J. D., Liu, Y. W., Chen, M., Zhou, Y. B., et al. (2018b). Genome-wide analysis of CDPK family in foxtail millet and determination of SiCDPK24 functions in drought stress. Front. Plant Sci. 9, 651. doi: 10.3389/fpls.2018.00651
Zhang, H., Zhang, Y., Deng, C., Deng, S., Li, N., Zhao, C., et al. (2018). The Arabidopsis Ca(2+)-dependent protein kinase CPK12 is involved in plant response to salt stress. Int. J. Mol. Sci. 19, 4062. doi: 10.3390/ijms19124062
Zhao, R., Sun, H. L., Mei, C., Wang, X. J., Yan, L., Liu, R., et al. (2011). The Arabidopsis Ca(2+) -dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth. New Phytol. 192, 61–73. doi: 10.1111/j.1469-8137.2011.03793.x
Zhou, L., Lan, W., Jiang, Y., Fang, W., Luan, S. (2014). A calcium-dependent protein kinase interacts with and activates a calcium channel to regulate pollen tube growth. Mol. Plant 7, 369–376. doi: 10.1093/mp/sst125
Zhu, X., Hong, X., Liu, X., Li, S., Yang, J., Wang, F., et al. (2021). Calcium-dependent protein kinase 32 gene maintains photosynthesis and tolerance of potato in response to salt stress. Scientia Hortic. 285, 123. doi: 10.1016/j.scienta.2021.110179
Keywords: wheat (Triticum aestivum), TaCDPK protein, identification, expression analysis, TaCDPK2/4-TaNOX7 interactions; ROS production
Citation: Hu C-H, Li B-B, Chen P, Shen H-Y, Xi W-G, Zhang Y, Yue Z-H, Wang H-X, Ma K-S, Li L-L and Chen K-M (2023) Identification of CDPKs involved in TaNOX7 mediated ROS production in wheat. Front. Plant Sci. 13:1108622. doi: 10.3389/fpls.2022.1108622
Received: 26 November 2022; Accepted: 22 December 2022;
Published: 23 January 2023.
Edited by:
Rupesh Tayade, Kyungpook National University, Republic of KoreaReviewed by:
Wen-Cheng Liu, Henan University, ChinaAbu Imran Baba, Swedish University of Agricultural Sciences, Sweden
Copyright © 2023 Hu, Li, Chen, Shen, Xi, Zhang, Yue, Wang, Ma, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun-Ming Chen, a3VubWluZ2NoZW5AbndzdWFmLmVkdS5jbg==; Li-Li Li, bGlsaWxpQHprbnUuZWR1LmNu; Ke-Shi Ma, emtudW1rc0AxMjYuY29t
†These authors have contributed equally to this work
 Chun-Hong Hu
Chun-Hong Hu Bin-Bin Li
Bin-Bin Li Peng Chen
Peng Chen Hai-Yan Shen
Hai-Yan Shen Wei-Gang Xi
Wei-Gang Xi Yi Zhang
Yi Zhang Zong-Hao Yue
Zong-Hao Yue Hong-Xing Wang
Hong-Xing Wang Ke-Shi Ma
Ke-Shi Ma Li-Li Li
Li-Li Li Kun-Ming Chen
Kun-Ming Chen