
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 16 December 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1102908
This article is part of the Research Topic Plant Defense Mechanisms in Plant-pathogen Interactions View all 20 articles
Wheat powdery mildew caused by a biotrophic fungus Blumeria graminis f. sp. tritici (Bgt), is a widespread airborne disease which continues to threaten global wheat production. One of the most chemical-free and cost-effective approaches for the management of wheat powdery mildew is the exploitation of resistant cultivars. Accumulating evidence has reported that more than 100 powdery mildew resistance genes or alleles mapping to 63 different loci (Pm1-Pm68) have been identified from common wheat and its wild relatives, and only a few of them have been cloned so far. However, continuous emergence of new pathogen races with novel degrees of virulence renders wheat resistance genes ineffective. An essential breeding strategy for achieving more durable resistance is the pyramiding of resistance genes into a single genotype. The genetics of host-pathogen interactions integrated with temperature conditions and the interaction between resistance genes and their corresponding pathogen a virulence genes or other resistance genes within the wheat genome determine the expression of resistance genes. Considerable progress has been made in revealing Bgt pathogenesis mechanisms, identification of resistance genes and breeding of wheat powdery mildew resistant cultivars. A detailed understanding of the molecular interactions between wheat and Bgt will facilitate the development of novel and effective approaches for controlling powdery mildew. This review gives a succinct overview of the molecular basis of interactions between wheat and Bgt, and wheat defense mechanisms against Bgt infection. It will also unleash the unsung roles of epigenetic processes, autophagy and silicon in wheat resistance to Bgt.
Wheat (Triticum aestivum, 2n=42; AABBDD) significantly contributes to human calorie consumption, food security and global agricultural sustainability (Godfray et al., 2010). Wheat production demand is driven by the increasing human population; however, it is continuously threatened by various fungal pathogens. Wheat powdery mildew caused by an obligate biotrophic fungal pathogen Blumeria graminis f. sp. tritici (Bgt), is one of the most destructive wheat diseases, and may cause yield losses of 10–40% and, in extreme cases, even up to 50% (Parlange et al., 2015; Zhu et al., 2015; Zhang et al., 2017). The rapid spread and adaptation of the pathogen is enhanced by its short life cycle, the ease with which airborne spores may be spread over long distances, and the possibility of sexual recombination leading to the generation of new virulent races (Jankovics et al., 2015). Powdery mildew infection induces immense transcriptional reprogramming which is strictly regulated by various mechanisms (Buscaill and Rivas, 2014; Jones et al., 2016; Adachi and Tsuda, 2019). Accumulating evidence has reported that histone post-translational modifications and other epigenetic processes, which typically affect the accessibility of DNA to the transcriptional machinery, fine-tune the expression of genes involved in plant defense responses to pathogen infection (Ding and Wang, 2015; Ramirez-Prado et al., 2018; Kong et al., 2020a; Kong et al., 2020b).
Wheat resistance against powdery mildew can either be race-specific or non-race specific resistance. Race-specific resistance imparts full resistance to some particular pathogens, but not others, and it is conferred by single resistance (R) (major effect) genes and is relatively heritable. Non-race specific resistance imparts partial resistance which does not rely on specific pathogen avirulence genes and thus allows for infection but reduces pathogen proliferation (Maurya et al., 2021; Mapuranga et al., 2022c). However, pathogens are endowed with exceptional adaptability by long-term survival pressure and environmental cues (Turrà et al., 2014), leading to high pathogen variations (Lei et al., 2017). Since the wheat-Bgt interaction is host- or race-specific, new, more virulent races might swiftly evolve and arise, rendering the numerous race-specific resistance genes added in wheat cultivars ineffective in preventing the development of powdery mildew (Kang et al., 2020). Therefore, it is critical to explore new wheat powdery mildew (Pm) resistance genes to develop durable prevention and management approaches of this disease. Furthermore, uncovering the underlying regulatory mechanisms of bread wheat defense responses to biotic stresses is crucial for increasing resistance to powdery mildew (Liu et al., 2016). This review gives a critical overview of the molecular basis of interactions between wheat and Bgt, and wheat defense response mechanisms to Bgt infection, including the role of autophagy and silicon in conferring resistance to powdery mildew. It will also discuss the recent developments in understanding the epigenetic processes and elements such as DNA methylation, and histone modification involved in wheat responses to Bgt infection.
The infection strategies and pathogenicity mechanisms of biotrophic fungal plant pathogens including Bgt have been extensively reviewed (Chaudhari et al., 2014; Bourras et al., 2015; Bindschedler et al., 2016; Bourras et al., 2016; Bourras et al., 2018; Liang et al., 2018; Sharma and Gautam, 2019; Jaswal et al., 2020; Mapuranga et al., 2022a; Mapuranga et al., 2022b). A prerequisite for compatibility establishment between a pathogen and its hosts is successful penetration. The conidia and ascospores produced by Bgt in cleistothecia, are primarily responsible for its propagation, and the disease most often affects a large area after being spread by wind (Jankovics et al., 2015). Following the attachment of conidia or ascospores to the surface of a photosynthetically active wheat leaf, a sophisticated germ tube is produced which then elongates into a thread-like hypha with appressoria (Acevedo-Garcia et al., 2017). Appressoria formation is triggered by the host’s chemical or physical signals, topographic signals, substratum hydrophobicity, and cutin monomers (Jones et al., 2001; Kamakura et al., 2002; Sharma and Gautam, 2019; Chethana et al., 2021). Subsequently, the digitate hypha develops into a penetration peg and ramifies a haustorium in order to facilitate the direct breaching of the host epidermal cell via turgor pressure and dynamic enzymes (Glawe, 2008). Subsequently, the fungus will only colonize the cell walls of the epidermal cell layer of leaf tissues. It will then form haustoria, which are the only fungal structures that will grow inside the host tissues, but it will do so without shattering the cell membrane. The haustoria are at the hub of Bgt biotrophic interaction through nutrient acquisition from the host and delivering effector proteins into the infected host cells to suppress host immunity (Jankovics et al., 2015). Only a few Bgt effectors including AvrPm2, AvrPm3a2/f2, AvrPm3b2/c2, and SvrPm3a1/f1 have been identified and cloned so far (Bourras et al., 2015; Praz et al., 2017; Bourras et al., 2019). These effectors belong to variable effector families and they are all highly induced upon Bgt infection to enhance pathogenesis. SvrPm3a1/f1 is a ribonucleic effector which suppresses the recognition of AvrPm2, AvrPm3a2/f2, AvrPm3b2/c2 by their cognate host resistance proteins (Bourras et al., 2019). Bgt infection process is also enhanced by mild temperatures (10-22°C). Furthermore, development of conidia that are produced asexually and secondary infections is enhanced by an increase in relative humidity and this will result in polycyclic disease development and many epidemics (Te Beest et al., 2008). Accumulating evidence has reported that wheat surface signals from the cuticle are responsible for inducing the pre-penetration growth of fungal pathogens (Wang et al., 2020; Arya et al., 2021). For instance, knockdown of a cuticle biosynthesis regulator gene, TaWIN1 reduced cuticle biosynthesis and conidial germination of Bgt (Kong and Chang, 2018). Intriguingly, the exogenous application of wax very-long-chain aldehydes, which are absent from the cuticle wax of TaWIN1-silenced plants, was able to fully rescue the Bgt germination penalty. This suggests that Bgt exploits the wax very-long-chain aldehydes biosynthesis which is positively regulated by TaWIN1 to trigger conidial germination (Kong and Chang, 2018). Consistent with this, the silencing of TaKCS6 and TaECR in bread wheat led to reduced Bgt germination and biosynthesis of wax (Wang et al., 2019; Kong et al., 2020b). The use of cuticular wax derived from wild-type wheat plants restored the Bgt germination penalty on TaKCS6- or TaECR-silenced plants (Wang et al., 2019; Kong et al., 2020b). Findings from these studies support the hypothesis that Bgt exploits the cuticle biosynthesis genes TaECR, TaWIN1, and TaKCS6 as susceptibility genes to enhance its pre-penetration growth.
The ability of plants to defend themselves against pathogen infection varies widely, and the interaction outcome is determined by the genetic status of both the pathogen and its host.
In order to ward against invading pathogens, plants have developed a multifaceted innate immune system (Dodds et al., 2006). The recognition of pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs) by membrane localized pattern recognition receptors (PRRs) triggers the first layer of innate immunity known at the PAMP-triggered immunity (PTI), which curtails pathogen invasion (Jones and Dangl, 2006; Jones et al., 2016). Downstream effects including mitogen-activated protein kinase (MAPK) pathways activation, defense-responsive genes expression, expression of pathogenesis-related genes, stomatal closure, reactive oxygen species (ROS) burst and callose deposition are triggered by PTI activation (Stael et al., 2015; Jaswal et al., 2020). By secreting effector molecules, pathogens may circumvent PTI and activate effector-triggered susceptibility (ETS), which is then countered by the host’s effector-triggered immunity (ETI), the second layer of innate immunity (Figure 1). ETI activation causes a dynamic reprogramming of several defense-responsive genes like MAPK, ROS burst, increase of Ca2+ levels, and hormonal change, and is usually accompanied by hypersensitive response and programmed cell death at the infection sites to curb pathogen invasive growth and proliferation (Jones and Dangl, 2006; Dangl et al., 2013; Jones et al., 2016). As proposed initially by Jones and Dangl, the PTI-ETS-ETI cycle continues in a zig-zag model (Jones and Dangl, 2006). Wheat calcium-dependent protein kinases (CDPKs) are critical sensors of calcium concentration changes that occur in response to a variety of biotic and abiotic stresses (Li et al., 2008). CDPK genes in wheat respond to a variety of biotic and abiotic stresses, including Bgt infection, low temperatures, salt, drought, and abscisic acid. It was found that each CDPK gene responded well to several treatments, which lends credence to the notion that wheat CDPKs serve as convergence points for many signal transduction pathways (Li et al., 2008). During wheat-Bgt interaction, the expression of CDPK2 was significantly upregulated, hence it was speculated that CDPK2 plays an essential role in wheat defense responses to Bgt infection (Li et al., 2008). Receptor-like kinases (RLKs) which recognize microbial presence in the apoplast, have an extracellular domain, a transmembrane domain and a cytoplasmic kinase domain (Kanyuka and Rudd, 2019). Wheat resistance to powdery mildew was recently found to be positively regulated by TaRLK1 and TaRLK2 (Chen et al., 2016), RLK-V (Hu et al., 2018) and LecRK-V from Haynaldia villosa, a diploid wheat relative highly resistant to powdery mildew (Figure 1) (Wang et al., 2018). TaRLK1 and TaRLK2 are leucine-rich repeat receptor-like kinases (LRR-RLKs), and their ectopic overexpression in wheat resulted in increased ROS accumulation in fungal penetration sites, leading to Bgt resistance (Chen et al., 2016). Overexpression of the lectin type RLK gene LecRK-V enhanced wheat resistance to Bgt (Wang et al., 2018). Wheat basal defense and Pm21-mediated resistance to Bgt were both found to be regulated by the malectin-like/LRR-RLK gene LRK-V (Hu et al., 2018).
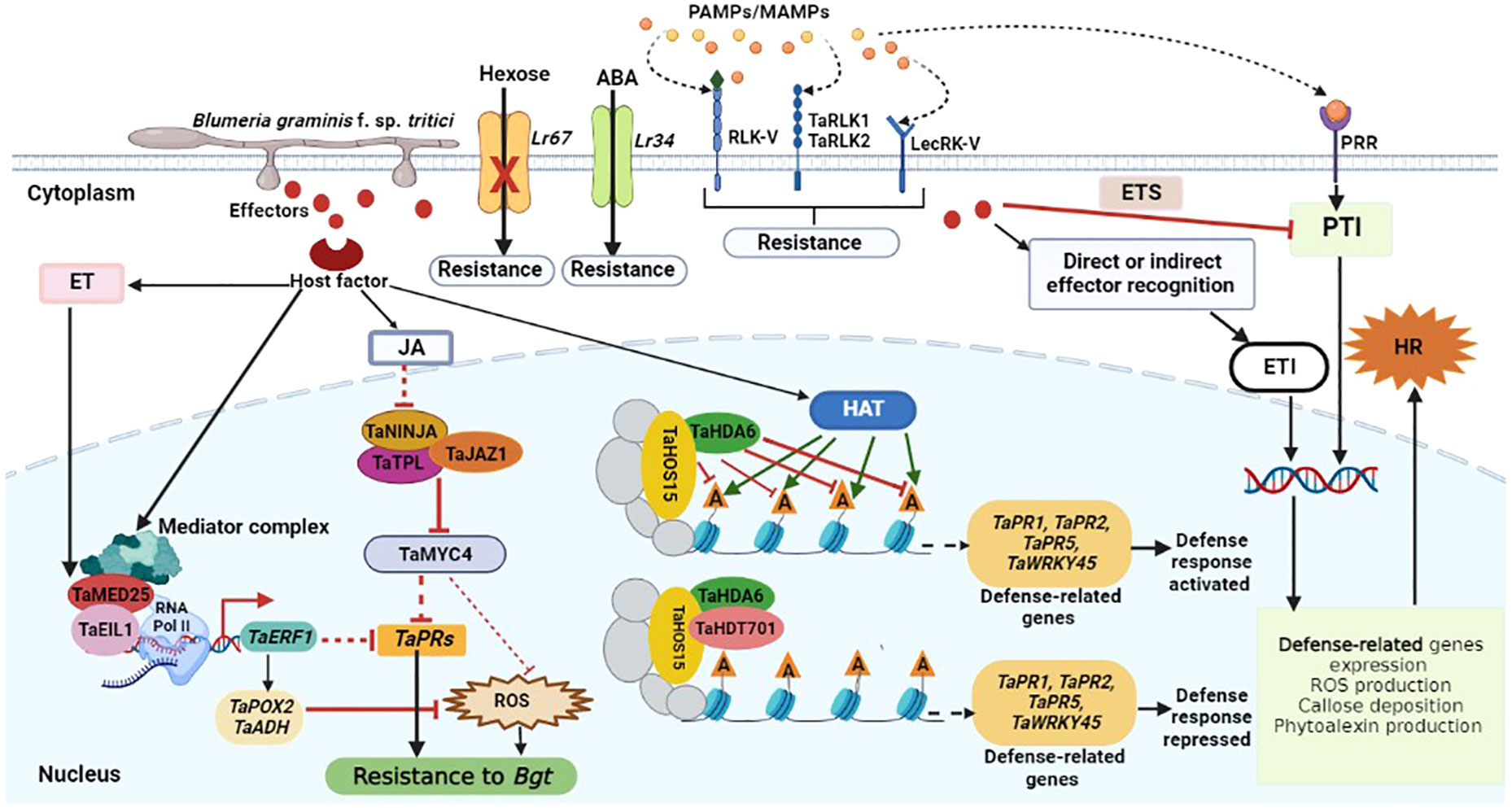
Figure 1 Schematic representation of wheat defense responses to Bgt infection. Pathogen-derived conserved molecules (PAMPs/MAMPs) are recognized by pattern recognition receptors (PRRs), and this activates PAMP-triggered immunity (PTI). Pathogens induce susceptibility by interfering with the immune signaling network through the secretion of effectors, resulting in effector-triggered susceptibility (ETS). Following direct or indirect effector recognition, plant R proteins activate host defense responses to inhibit pathogen growth, and this is called effector-triggered immunity (ETI). The receptor-like kinases (TaRLK1, TaRLK2), malectin-like/LRR-RLK (RLK-V), and lectin type RLK (LecRK-V) resistance genes, encode proteins that are involved in the perception of fungal determinants and transmission of defense signals to the cell interior. Upon Bgt, reactive oxygen species (ROS) and endogenous pathogenesis-related (PR) genes are highly induced to enhance defense response to infection. Meanwhile, the physical interaction between TaMED25 and TaEIL1 may lead to the recruitment of RNA Pol II to enhance the transcription of TaERF1. TaERF1 may further suppress ROS accumulation and the expression of PR genes to partially attenuate wheat basal resistance to Bgt infection. TaJAZ1, TaNINJA and TaTPL form a co-repressor complex that suppresses TaMYC4 transcriptional activity under normal conditions. Jasmonate (JA) accumulation in response to Bgt infection results in the degradation of TaJAZ1 to release TaMYC4 and other transcriptional factors, consequently leading to the suppression of TaPRs expression and ROS accumulation in wheat. Ethylene (ET) also inhibits ROS accumulation and the expression of TaPRs to negatively regulate wheat resistance against Bgt infection. Furthermore, Bgt infection also results in the recruitment of more histone acetyltransferases (HATs) and thus increase histone acetylation at the defense related genes (represented by green arrows), thus activating the expression of these genes. The TaHDT701-TaHDA6-TaHOS15 histone deacetylase complex mediates histone deacetylation at the wheat defense-related genes like TaPR1, TaPR2, TaPR5 and TaWRKY45, which results in the suppression of defense-related genes transcription and defense responses to Bgt.
COI1 serves as a key regulator in the jasmonate (JA) signaling pathway. During both compatible and incompatible interactions between wheat and Bgt, TaCOI1 expression was induced. The rate at which Bgt was able to successfully penetrate all TaCOI1-silenced Brock was greater, but the proportion of macrocephalic appressoria and resistance responses was lower than in the control group. Based on these results, TaCOI1 gene positively regulates wheat resistance to Bgt infection (Liu et al., 2018). Copines are calcium-dependent phospholipid-binding proteins that are made up of two highly conserved C2 domains at their N-termini, and a von Willebrand factor A (vWA) domain at their C-termini (Creutz et al., 1998). It was found that wheat resistance to Bgt is negatively regulated by the wheat copine genes TaBON1 and TaBON3. Both variations in temperature and the presence of pathogens affected the expression of TaBON1 and TaBON3. These findings demonstrated a conserved role of plant copine genes in the plant immunity and revealed new genetic resources for the development of wheat varieties with durable resistance to powdery mildew (Zou et al., 2018a).
The glucan synthase-like (GSL) gene family is found in many plants where it produces callose which is not only present at areas of fungal penetration, but also in callus plugs that form in wounds and in the cells wall of pollen (Voigt et al., 2006). In wheat, a total of eight distinct TaGSL genes were found and confirmed to mediate the synthesis and regulation of callose in diverse tissues under distinct environmental cues (Voigt et al., 2006). Recently, Cheng and colleagues showed that expression level of TaGSL22 was substantially upregulated by Bgt infection. Callose deposition was reduced by the silencing of TaGSL22 which led to increased susceptibility of wheat to Bgt (Wang et al., 2022b). Therefore, TaGSL22 positively regulates wheat resistance to Bgt. Genes known as Recognition of Peronospora parasitica 13-like, (RPP13-like) genes which are members of the NBS-LRR superfamily serve important roles in the resistance of various plant species to many pathogens. It was recently established that wheat resistance to powdery mildew is positively regulated by TaRPP13L1-3D gene via a RanGAP-WPP complex-mediated nucleocytoplasmic pathway (Zhang et al., 2022). These fresh insights into TaRPP13L1-3D will be beneficial in dissecting the methods by which wheat develops its resistance to biotrophic fungal pathogens and understanding the link between R gene expression and pathogen defense. Generally, different degrees of plant resistance to a particular pathogen species are dependent on how the host interacts with the pathogen, either via broad-spectrum (non-race specific resistance) or race-specific (specific resistance to a single race) resistance.
Race-specific resistance is also called qualitative resistance, all stage resistance or monogenic resistance, and is generally mediated by a single major R gene, which becomes active upon detection of the pathogen’s effectors in a gene-for-gene paradigm (Maurya et al., 2021; Mapuranga et al., 2022c). Although wheat powdery mildew resistance genes have been shown to activate the PTI and ETI pathways, the nature of this activation has remained unclear. Due to the rapid evolution of new pathogen races, major R genes are known to deteriorate easily, which results in a short lifetime for resistance. This results in a restricted window of opportunity for protection (McDonald and Linde, 2002). To determine which resistance genes are ineffective, it is necessary to know the identification of the race-specific resistance genes present in wheat cultivars. Molecular marker assisted selective breeding is an efficient and rapid approach for identifying resistance genes. Wheat disease resistance genes have been mapped with the use of molecular markers. Recent research into Pm genes has made extensive use of simple sequence repeat (SSR) and single-nucleotide polymorphism (SNP) markers (Wu et al., 2019).
The longevity of race-specific resistance genes in the field may be extended by the use of gene combinations, strategic gene deployment, and multiline cultivars (Burdon et al., 2014; Li et al., 2014; Mundt, 2018). However, there is a tendency of many pathogen isolates to coexist in their respective ecological habitats. As a consequence of genetic variety in the pathogen population, this may increase the probability of a disease epidemic. Because of this, it’s possible that gene stacking won’t be enough to halt the spread of disease for very long, and the cultivars will lose their efficacy. In order to increase the efficacy of gene stacking, allelic mining was proposed for the improvement of agriculture (Bhullar et al., 2010). Accumulating evidence has reported that multi-allelic race specific resistance genes evolve under intense diversifying selection to detect specific pathogen avirulence alleles (Bhuiyan et al., 2009; Kanzaki et al., 2012). For example, at the Pm3 locus, race-specific resistance is conferred by 17 functional alleles (Brunner et al., 2010). Pm3 encodes a CC-NBS-LRR protein, with complete sequence conservation in the CC domain across these alleles, but sequence exchange is shown in the NBS and LRR sections, suggesting gene conversion or intragenic recombination between the alleles (Bourras et al., 2015). Using map-based cloning of haploid F1 populations, some studies identified Bgt effectors AvrPm2, AvrPm3a2/f2, AvrPm3b2/c2, and SvrPm3a1/f1 corresponding to some NLR resistance genes in wheat (Bourras et al., 2015; Praz et al., 2017; Bourras et al., 2019). Bourras and colleagues characterized the corresponding Avr loci of Pm3 alleles and proposed the involvement of a pathogen-encoded suppressor of avirulence (Svr) in race specificity (Bourras et al., 2015). It was found that the function of detected avirulence alleles is blocked by an active suppressor, resulting in a disease. The presence of Svr in the powdery mildew will, in combination with the avirulence allele and the R allele, regulate plant resistance in the wheat-powdery mildew pathosystem. This suggests that there are additional layers of a simple gene for the gene model controlling Avr-R interactions in the wheat-powdery mildew system. A recent study on the AvrPm3-Pm3 model, further demonstrated the importance of Pm3 NLR receptors in concurrently determining host-specificity for grass mildews and race-specific resistance to wheat powdery mildew (Bourras et al., 2019). Pm12 and Pm21 have recently been found to be orthologous genes (Zhu et al., 2022). Distinct intramolecular interaction patterns and race specificities in the two orthologous resistance genes Pm12 and Pm21 were clearly demonstrated (Zhu et al., 2022). Multiple alleles at the Pm2 locus have been discovered across a wide range of genotypes (Jin et al., 2018). Powdery mildew resistance genes Pm2a, Pm2b and PmCH1357 were mapped to the same genomic level, and cloning of these genes using different approaches revealed that they genes confer different levels of resistance, suggesting that it might be imparted by distinct resistance genes or alleles (Sánchez-Martín et al., 2016; Chen et al., 2019a; Jin et al., 2022).
Non-race specific resistance is also called quantitative resistance, minor gene resistance or adult plant resistance. This type of resistance often manifests as a partial resistance phenotype, in which pathogen invasive growth and proliferation is suppressed without any immune response overtly shown (Maurya et al., 2021; Mapuranga et al., 2022c). It is also called minor gene resistance because just a few genes are responsible for imparting the resistance (Maurya et al., 2021). Because it provides partial levels of resistance and field tolerance, it is also known as standard/partial/field resistance. Adult stages of wheat development is associated with the manifestation of this resistance, and is hence regarded as adult plant resistance (Li et al., 2014). Most of the identified Pm genes are race specific all-stage resistance genes, and only a few pleiotropic partial resistance genes, such as Pm38/Yr18/Lr34/Sr57/Lt1 (Singh et al., 2012), Lr46/Yr29/Pm39/Sr58/Ltn2 (Singh et al., 2013) and Pm46/Yr46/Lr67/Sr55/Ltn3 (Herrera-Foessel et al., 2014), which confer partial resistance to powdery mildew and other rust pathogens at the adult plant stage, hence they are adult plant resistance genes. Isolation of adult plant resistance genes Pm38 and Pm46 found a single gene encoding an ATP-Binding Cassette (ABC) transporter (Krattinger et al., 2009) and a hexose transporter (Moore et al., 2015) at the multi-pathogen resistance locus, respectively, which confers a dual resistance for wheat leaf rust and stripe rust in addition to resistance to powdery mildew (Figure 1). It was suggested that these two genes would go through a defense activation process associated with a coordinated interaction between abscisic acid and sugar signaling (Moore et al., 2015; Krattinger et al., 2019). These sugar signals may operate as priming molecules that lead to PTI and ETI (Bolouri Moghaddam and Van den Ende, 2012), modulating plant metabolism that is induced by the fungal pathogen. The activation of sugar transporters is necessary for upsetting the carbohydrates equilibrium between sources and sinks, which is often seen in response to biotic stresses (Valluru and Van den Ende, 2011). Accumulating evidence has established that non-race specific resistance is more robust and durable than race-specific resistance in combating the new emerging virulent pathogen races (Liu et al., 2001; Li et al., 2014). For example, after 20 years of cultivation, the cultivar Knox has shown consistent adult plant resistance effectiveness to powdery mildew (Liu et al., 2001). In particular, breeders are interested in powdery mildew adult plant resistance genes that confer resistance to a wide range of pathogens.
The first powdery mildew resistance gene in wheat was found in wheat variety ‘Thew’ by an Australian researcher Waterhouse in 1930 (Zeller, 1973). Since then, new Pm genes have been identified from common wheat and wheat relatives. Although more than 100 powdery mildew resistance genes/alleles in 63 loci (Pm1-Pm66) have been documented (McIntosh et al., 2019; Zhang et al., 2019), only a few of them have been cloned and characterized so far (Table 1) including Pm1a (Hewitt et al., 2021), Pm2a (Sánchez-Martín et al., 2016), Pm2b (Jin et al., 2022), Pm3b (Yahiaoui et al., 2004), Pm5e (Xie et al., 2020), Pm8 (Hurni et al., 2013), Pm17 (Singh et al., 2018), Pm21 (Xing et al., 2018; He et al., 2018), Pm24 (Lu et al., 2020), Pm38 (Krattinger et al., 2009), Pm41 (Li et al., 2020), Pm46 (Moore et al., 2015), and Pm60 (Zou et al., 2018b). The integration of the high-quality wheat genome sequence (IWGSC Ref-Seq v1.0) (Avni et al., 2017; Appels et al., 2018; Maccaferri et al., 2019), and various advanced molecular cloning approaches for reducing the complexity of the genome to allow the performance of targeted resequencing analyses has facilitated the cloning of wheat Pm genes. Some of these genes including Pm1a (Hewitt et al., 2021), Pm2a (Sánchez-Martín et al., 2016; Manser et al., 2021), and Pm4b (Sánchez-Martín et al., 2021), were cloned using the Target-sequence Enrichment Sequencing (TEnSeq) pipelines. TEnSeq pipelines include various approaches such as Mutagenesis and Resistance gene enrichment Sequencing (MutRenSeq), Mutagenesis Chromosome flow sorting and short-read Sequencing (MutChromSeq), Association genetics with Resistance gene enrichment Sequencing (AgRenSeq) and Targeted Chromosome-based Cloning via long-range Assembly (TACCA). AgRenSeq and MutRenSeq are based on NLR-targeted DNA capture and hybridization, while MutChromSeq and TACCA are based on purification of individual chromosomes from wheat lines (Zhang et al., 2020). The majority of the powdery mildew resistance genes were cloned using the map-based cloning approach (Yahiaoui et al., 2004; Krattinger et al., 2009; Yahiaoui et al., 2009; Moore et al., 2015; Sánchez-Martín et al., 2016; Zou et al., 2018b; Li et al., 2020; Lu et al., 2020; Hewitt et al., 2021), with a few having been cloned using the homology-based cloning approach (Hurni et al., 2013; Singh et al., 2018).
Except for Pm24, Pm38, and Pm46, which encode tandem kinase gene WTK3, an ABC transporter, and a hexose transporter, respectively, most of the cloned powdery mildew resistance genes in wheat encode NLR proteins which are race-specific (Table 1). In recent years, tandem kinase protein (TKP) has emerged as a novel kinase protein family in plant innate immunity. A serine/threonine non-arginine-aspartate (non-RD) receptor-like protein kinase with two putative tandem kinase domains (Kin I and Kin II) and lacking an extracellular domain is encoded by the Chinese wheat landrace Hulutou’s Pm24 (WTK3) gene. A rare 6-bp natural deletion of lysine-glycine codons was found to be endemic to wheat landraces in Shaanxi Province, China, in the kinase I domain (Kin I) of WTK3, which is essential for the gain of function of powdery mildew disease resistance (Lu et al., 2020). The isolation of WTK3 with the tandem kinase domains laid a foundation for comprehension of the signal transduction function of TKP in wheat powdery mildew resistance and plant innate immunity. Understanding the molecular mechanisms of this protein family’s role in plant innate immunity will be enhanced by the characterization of the TKP kinase functions, related proteins, and phosphorylation processes. The cloning of powdery mildew resistance genes paves a way for the development of high-throughput diagnostic functional markers that can be utilized in markers-assisted selection for fungal-resistance breeding programmes.
The network of resistance can be analyzed using transcriptomics (gene regulation and expression profiling), proteomics (protein identification and effects), and metabolomics (metabolite profiling, regulation, pathway and intermediates). However, there are no metabolic profiling studies that have been reported so far during wheat-Bgt interactions, hence this section will specifically focus on transcriptomics and proteomics.
Expression and regulation of genes, as well as the identification of important genes involved in the stress response pathway, may all be understood by transcriptome profiling. Depending on the amount of available genomic resources generated and the type of plant being studied, different methods including RNA-sequencing, Affymetrix GeneChips, and spotted micro arrays, expressed sequence tags, sequencing in conjunction with suppression subtractive hybridization, are employed to study the transcriptome. RNA-Seq surpassed other transcriptomics methods because it is the most high-throughput, cost-effective and efficient method due to recent developments in next-generation sequencing technology. Transcriptomics research has so far been extensively conducted in wheat. Transcriptional and translational alterations of plants are also triggered by pathogen attack and this results in the activations of several genes and metabolic pathways as a defense mechanism. Epidermal Bgt infection in diploid wheat (Triticum monococcum) triggered the expression of 12 genes involved in methyl unit biosynthesis and supply, showing that genes involved in methyl unit production pathways are also important for the host defense response (Bhuiyan et al., 2007). An mRNA tag analysis was performed to explore the cellular metabolic processes that are activated by H2O2 treatment in wheat. It was found that 260 transcripts had conserved expression across all three wheat lines and these genes might represent a subset of basal H2O2 sensitive genes involved in signal transduction, cell defense, redox homeostasis, photosynthesis, lipid metabolism, glucose metabolism, and transport (Li et al., 2011).
Another comparative transcriptome analysis of wheat in response to Bgt infection established an approximately equal gene expression level in both resistant and susceptible cultivars before Bgt inoculation (Xin et al., 2012). Various pathways such as flavonoid biosynthesis, cell wall fortification and metabolic processes were found to be involved in wheat defense responses to powdery mildew (Xin et al., 2012). SGT1 is responsible for the positive regulation of many plant R genes that confer race-specific resistance (Muskett and Parker, 2003; Azevedo et al., 2006). The expression of SGT1 was shown to have a very close relationship with the expression of R genes, the hypersensitive response, and the activation of specific resistance proteins (Bhaskar et al., 2008; Meldau et al., 2011). Haynaldia villosa Hv-SGT1 transcript levels were substantially increased by Bgt infection. Transcript levels of Hv-SGT1 were also substantially upregulated by H2O2 and methyl jasmonate treatments (MeJA). However, these levels were only minimally stimulated following exposure to ethephon or abscisic acid; but were not changed by salicylic acid (SA) treatment (Xing et al., 2013). As a result, the involvement of Hv-SGT1 in the production of H2O2 correlates with the hypersensitive response as well as jasmonic acid signaling. This innovative demonstration that wheat with overexpressed Hv-SGT1 had higher resistance to powdery mildew, implies that it might serve as a transgenic genetic resource in wheat breeding for the purpose of producing wheat cultivars with durable resistance to multiple pathogens (Xing et al., 2013).
Powdery mildew infection also resulted in differential expression of wheat genes in which 289 transcripts were upregulated and 154 transcripts were downregulated during R-gene mediated incompatible interactions between wheat and Bgt (Li et al., 2013). Another comparative transcriptome analysis also revealed different activations in wheat’s response to powdery mildew and stripe rust infections (Zhang et al., 2014). In Bgt-infected leaves, the differentially expressed genes were grouped into seven pathways, whereas in Pst-infected leaves they were grouped into four enriched Kyto Encyclopedia of Genes and Genomes (KEGG) pathways. Many genes and pathways were activated in the wheat-pathogen interaction pathways in response to Bgt infection than in response to Pst infection (Zhang et al., 2014). Liu et al. analyzed the transcriptomic changes in wheat response to Bgt infection in a continuous time period and their findings confirmed involvement of CMPG1-V, which encodes the U-box E3 ubiquitin ligase, in augmentation of broad-spectrum resistance against powdery mildew (Liu et al., 2020). A weighted gene co-expression network analysis revealed important CMPG1-V regulatory candidates including glucosyltransferase in flavonoid biosynthesis, serine/threonine-protein kinase in phosphorylation, peroxidase in oxidative stress, and defense factor WRKYs (Liu et al., 2020). This study laid a foundation for the elucidation of CMPG1-V resistant regulatory network and the identification and molecular characterization of the key candidates that might be exploited in resistant breeding programs.
Another recent transcriptomics study demonstrated that the gene(s) located at 2Mb in the Chinese Spring (CS)-Aegilops biuncialis 2Mb disomic addition line TA7733 imparted a high degree of resistance Bgt. When compared to CS, there were 7 278 identified unigenes that displayed unique expression in TA7733 both pre-and post-Bgt infection. It was found that 53 out of the 295 unigenes genes encoded NLR proteins and were unique to 2Mb and had the potential to be involved in powdery mildew resistance. The establishment of TA7733 broad-spectrum resistance to Bgt and availability of putative candidate R-gene specific molecular markers derived from 2Mb laid a foundation for transferring powdery mildew resistance from 2Mb to common wheat by inducing CS-Ae. biuncialis homoeologous recombination (Li et al., 2019). Using bulked segregant RNA-Seq, Ma and colleagues analyzed 2 973 differentially expressed genes to study resistance gene regulatory genes. They found that after Bgt invasion, six disease-related genes had unique expression patterns, making them prime candidates for elucidating resistance mechanisms and enhancing long-term resistance in wheat to powdery mildew (Ma et al., 2021). Glutathione S-transferase (GST), epitomizes a group of ubiquitous proteins found in plants that comprise several functional proteins. A lot of studies indicated that GSTs are involved in secondary metabolism (Mueller et al., 2000), growth and development (Gong et al., 2005), and biotic and abiotic stress responses in plants (Bianchi et al., 2002). The presence of GST in the leaves of emmer wheat leads not only to a reduction in oxidative stress but also to an enhancement in the plant’s resistance to herbicides (Karpenko et al., 2019). In common wheat, a comprehensive genome-wide analysis uncovered 346 GST genes and 87 TaGSTU family members. During wheat-Bgt incompatible interaction, TaGSTU6, TaGSTU4, and TaGSTU7 genes were are highly upregulated (Hao et al., 2021).
Proteomics is the study of the structural and functional features of all of the proteins present in a live organism at the same time. It can be performed using various approaches including, two-dimensional (2-D) gel electrophoresis, western blotting, matrix-assisted laser desorption ionization-time of flight (MALDI-TOF), mass spectrometry, and enzyme-linked immunosorbent assay (ELISA), together with several bioinformatic tools (Baggerman et al., 2005; Gevaert et al., 2011; Chaudhary et al., 2019). Recent advances in the field of proteomics have made it possible to conduct high-throughput proteome analyses. The majority of proteomic studies has been done on crops including wheat, rice, barley, maize, potato, and soybeans since these crops are the only ones whose whole genome sequences are accessible in the public domain (Yadav et al., 2022). A comparative proteomic analysis of wheat’s response to Bgt infection in wheat Pm30 near-isogenic found 97 differentially expressed proteins and 26 of them belonged to a variety of functional categories, including energy and metabolism, disease and defense, transcription and translation, and signal transduction (Wang et al., 2012). Another proteomic analysis of wheat defense response to Bgt also established that 44 differentially expressed proteins were related to the defensive response, photosynthesis, metabolism, and other cellular activities in wheat (Mandal et al., 2014). Quantitative proteomic analysis of N9134, a resistant wheat line, identified 394 protein-species that showed differential accumulation (Fu et al., 2016). These differentially accumulated protein-species predominantly consisted of oxidative stress responsive proteins, pathogenesis-related polypeptides, and primary metabolic pathway components (Fu et al., 2016). MAPKs are involved in the regulation of a variety of plant processes, such as metabolism and the signaling processes involved in abiotic and biotic responses (Gawroński et al., 2014). During incompatible wheat-Bgt interaction, MAPK5 protein level was substantially upregulated, suggesting that it serves an essential role in wheat resistance to Bgt (Li et al., 2017b).
Liang and colleagues analyzed the proteome of three distinct wheat cultivars at three different time intervals and found that various proteins were upregulated and downregulated in response to Bgt infection in wheat cultivar with Pm40 gene (Liang et al., 2019). It was inferred that the proteins have the capacity to mediate the immune responses and coordinate various physiological and cellular processes that take place during wheat defense response to Bgt infection (Liang et al., 2019). Recently a quantitative proteomic analysis revealed that TaGSTU6/TaCBSX3 interaction serves a crucial role in wheat resistance to Bgt infection (Wang et al., 2022a). Seven hundred and forty-one differentially accumulated proteins were identified and intriguingly, 42 of them responded to both Bgt and Pst infection. These findings imply that wheat has distinct mechanisms of responding to Pst and Bgt infections, since the interaction of TaGSTU6 with TaCBSX3 only imparts resistance to Bgt (Wang et al., 2022a). Altogether, these proteomics studies provide a new basis for broadening our understanding of the molecular mechanism of the wheat-Bgt interactions at the protein level, as well as a theoretical framework for resistance breeding and the long-term management of powdery mildew.
Although accumulating evidence has reported the transcriptional regulation of wheat’s defense response to biotic stresses; the transcriptional regulation of wheat resistance to Bgt infection remains to be explored. The molecular and functional characterization of transcription factors can be exploited in wheat breeding programs for the development of durable broad-spectrum resistance in wheat cultivars. Three transcription factors namely, T16.17353, T4.32876, and T19.62870, belonging to the TGA family of bZIP were found to be involved in wheat resistance to Bgt infection (Zhang et al., 2014). Furthermore, there was significant upregulation of WRKY33 gene orthologs T13.35253 and T16.5876 during wheat-Bgt interactions and they were enriched in the plant-pathogen interaction pathway (Zhang et al., 2014). This clearly demonstrated the involvement of WRKY33 transcription factor in the defense response of wheat to Bgt infection through the PTI (Mapuranga et al., 2022c). Eukaryotes have a conserved multisubunit complex called mediator that enhances transcription by bridging certain transcription factors with RNA polymerase II (Chadick and Asturias, 2005). The MED25 Mediator component has been studied in Arabidopsis for its role in regulating plant defensive responses through interactions with particular transcription factors involved in plant hormone signaling (Kidd et al., 2009; Chen et al., 2012). Wheat’s resistance to powdery mildew was found to be negatively regulated by the TaMED25-TaEIL1-TaERF1 signaling module (Liu et al., 2016). It was demonstrated that TaMED25 promotes TaEIL1-mediated activation of the powdery mildew-responsive gene TaERF1. It was also shown that the TaMED25-TaEIL1-TaERF1 signaling module represses the expression of particular pathogenesis-related genes (TaPRs) and prevents ROS accumulation in bread wheat leaf cells to fine-tune resistance to powdery mildew. Both TaMED25 and TaEIL1 negatively regulated wheat response to Bgt. Pol II can be recruited to the promoter of TaEIL1 and target gene TaERF1 by the interaction between TaMED25 and TaEIL1, which further suppresses ROS accumulation by upregulating the expression of ROS-scavenging genes (Figure 1) (Liu et al., 2016). Zhou and colleagues established that, overexpression of wheat NAC transcription factor TaNAC6-A enhances resistance to Bgt, but resistance against Bgt was compromised by the silencing of the TaNAC6s (Zhou et al., 2018). This implies that wheat TaNAC6s are essential for the positive regulation of wheat broad-spectrum resistance to Bgt. Moreover, it was also suggested that wheat TaNAC6s might be regulated by JA, and this feedback regulation of the JA pathway increases resistance against Bgt infection. Therefore, wheat TaNAC6s share a common signaling pathway with their orthologs HvNAC6 and ATAF1 to confer resistance to powdery mildew (Zhou et al., 2018).
Accumulating evidence has reported the involvement of JA in the regulation of wheat responses to abiotic and biotic stresses (Cao et al., 2014; Zhao et al., 2014; Xu et al., 2016; Li et al., 2017a). Based on wheat genome data, Wang and colleagues found 14 JAZ (Jasmonate-ZIM domain) genes and demonstrated that the transcription of some of these JAZ genes was affected by a wide range of abiotic stress treatments and phytohormones (Wang et al., 2017). Overexpression of a truncated TaJAZ1 without the Jas motif was demonstrated to enhance the expression of TaPR1/2 gene and accumulation of ROS in transgenic bread wheat lines, thereby increasing resistance to powdery mildew. Based on these observations, it seems that there is a synergistic action of the JA and ET signaling pathways to regulate the resistance of wheat against powdery mildew, most likely via the modification of the common downstream processes of ROS accumulation and expression of pathogenesis-related genes (Jing et al., 2019). The transcriptional activity of TaMYC4, a JA-induced bHLH transcription factor was repressed by TaJAZ1 via direct interaction (Figure 1). While the N-terminal EAR motif of TaNINJA was shown to interact with the transcriptional co-repressor TaTPL, the ZIM domain interacted with the C-terminus of TANINJA. These results collectively demonstrated that TaJAZ1 positively regulates wheat resistance to Bgt infection (Jing et al., 2019). To increase bread wheat’s resistance to powdery mildew, scientists may utilize genome-editing tools like CRISPR/Cas9 to generate gain-of-function mutations by modifying the Jas motif of specific TaJAZ genes like TaJAZ1. The TaJAZ genes have the potential to be used in molecular breeding, but how they affect the susceptibility/resistance of wheat against other biotrophic fungal pathogens other than Bgt remains to be figured out. The expression of Pm2b was highly upregulated by Bgt infection isolate E09. Self-association of Pm2b through its NB domain was also detected. Intriguingly, it was found that Pm2b interacts with TaWRKY76-D and TaWRKY76-D silencing showed negatively regulated wheat resistance to powdery mildew (Jin et al., 2022). Pm2b probably interacted with TaWRKY76-D to alleviate TaWRKY76D’s suppression of downstream genes. Pm2 has been shown to either directly or indirectly interact with its avirulence gene AVRPM2 when infected with Bgt isolates, resulting in strong hypersensitive response (Praz et al., 2017). Therefore, it was suggested that the interaction between Pm2 and TaWRKY76-D was enhanced by AVRPM2, leading to improved powdery mildew resistance in wheat. Likewise, the recognition of PM2 by AVRPM2 could be facilitated by WRKY76-D, via interaction with Pm2 and this will initiate a defense response to infection. Nevertheless, TaWRKY76-D silencing failed to confer complete resistance to Bgt infection, suggesting that TaWRKY76-D may serve a crucial role in the PM2-mediated powdery mildew resistance pathway (Jin et al., 2022).
Epigenetics refers to heritable gene expression alterations that occur between DNA and its surrounding chromatin without changing its DNA sequence and resulting in substantial changes in any organisms’ genome (Kong et al., 2020a). Several studies reported the direct involvement of epigenetic processes and elements in the transcriptional reprogramming and regulation of defense response genes (Deleris et al., 2016; Kim et al., 2017; Chen et al., 2019b), and in establishing memory to environmental cues (He and Li, 2018; Ashapkin et al., 2020). Therefore, such responses are influenced by both the host’s epigenetic configuration and the effects of biotrophic interactions (Ramos-Cruz et al., 2021). DNA methylation, histone modifications, and chromatin remodeling are the major components of the epigenetic make up of plants. Local chromatin accessibility is dictated by single or combined epigenetic marks, which in turn regulates gene expression and, thus may play a role in plant defense, and other processes (Ramos-Cruz et al., 2021).
DNA methylation is a type of DNA chemical modification that regulates DNA stability, chromatin structure and even gene expression without altering the nucleotide sequence (Kong et al., 2020a). In plants, cytosine methylation is detected in the context of CG, CHG, and CHH (where H is any nucleotide except G) (Henderson and Jacobsen, 2007; Law and Jacobsen, 2010), in which CG is the most plentiful and common methylation site (Kong et al., 2020a). DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) is the enzyme that catalyzes plant de novo methylation via the RNA-directed DNA methylation (RdDM) pathway which is a plant-specific pathway (Matzke and Mosher, 2014; Kong et al., 2020a). DNA methylation is important for both transcriptional gene silencing and preservation of genomic integrity by silencing transposable elements. Several studies have recently reported the significance of DNA methylation in plant defense responses to abiotic and biotic stresses (Dowen et al., 2012; Yu et al., 2013; Le et al., 2014; Deng et al., 2017). DNA methylation, and CHH methylation to be specific, has recently been linked to the regulation of defense responses to Bgt infection in Aegilops tauschii (Geng et al., 2019). There was a significant downregulation of ARGONAUTE4a (AGO4a) in A. tauschii following Bgt infection. This was accompanied by a significant reduction in the amounts of AGO4a-sorted 24-nt siRNA, particularly for genes near transposable elements (TGAs). The downregulation of AGO4a and 24-nt siRNAs suggests the presence of an active defense mechanism in A. tauschii that coordinately downregulates DNA methylation, thereby upregulating the expression of defense-related genes. The differential expression of 24-nt siRNAs between compatible and incompatible interactions with Bgt supports the hypothesis that siRNAs are key players in basal defense responses by directing DNA methylation. The presence of many differentially methylated regions (DMRs) was associated with hypomethylation (Geng et al., 2019). Furthermore, it was established that TGAs with CHH-hypomethylated DMRs were enriched in genes for stress response functions including the receptor kinases, pathogenesis-related genes and peroxidases, indicating the involvement of DNA methylation in the regulation of wheat responses to powdery mildew infection (Geng et al., 2019). The upregulation of a pathogenesis-related β-1,3-glucanase gene in response to Bgt infection exemplified the effect of CHH-hypomethylation. These results support the hypothesis that dynamic DNA methylation constitutes a regulatory layer within the intricate mechanisms of plant immunity, that may be targeted in an effort to enhance wheat resistance against powdery mildew. However, more research is required to determine whether Bgt-derived sRNAs downregulate AGO4a and decrease the total siRNA or CHH methylation levels during wheat-Bgt interactions (Geng et al., 2019).
Chromatin modifications and remodeling have been reported to be involved in the transcriptional reprogramming and regulation of defense related genes in plants (Ding and Wang, 2015; Espinas et al., 2016). Histones in chromatin can be subjected to a variety of covalent modifications, including methylation, acetylation, ubiquitination, and phosphorylation. These modifications can change the structure of the chromatin so that it is either permissive or repressive to transcription by modifying interactions between DNA and the histones (Jenuwein and Allis, 2001). The acetylation of histone Lys residues catalyzed by histone acetyltransferases (HATs), can relax the chromatin structure and promote transcription, whereas the process of chromatin compaction involving removal of the acetyl group, which is induced by histone deacetylases (HDACs), is associated with transcriptional repression (Kurdistani and Grunstein, 2003). Recently, various HDACs have been reported to serve as positive and negative regulators of plant defense response to pathogen infections (Ding and Wang, 2015; Ramirez-Prado et al., 2018; Kong et al., 2020a). Scaffolding proteins, also known as proteins with WD repeats (WDRs), often act as platforms onto which protein complexes may be assembled, indicating that they are involved in various cellular processes including secondary metabolism, defense responses, and abiotic stress tolerance (Guerriero et al., 2015).
Transcription factors have been demonstrated as key players in the recruitment of chromatin-remodeling and histone modification machinery to key gene promoters (Luo et al., 2012; Jing et al., 2013). TaHDA6, a wheat RPD3 type deacetylase interacted with TaHOS15, a WDR protein, to inhibit expression of defense-related genes via histone deacetylation, thus negatively regulating wheat response to Bgt (Figure 1) (Liu et al., 2019). It was found that both TaHDA6 and TaHOS15 can bind to promoters of defense-related genes. Nevertheless, TaHDA6 binding ability to these promoters was eliminated when TaHOS15 was silenced, suggesting that TaHOS15 directs TAHDA6 to these promoters for histone deacetylation. These findings laid a foundation for future studies to further identify the transcription factors associated with TaHOS15 and characterize their roles in the TaHOS15-TaHDA6 HDAC complex recruitment to actual promoters (Liu et al., 2019). The wheat histone deacetylase TaHDT701, a member of the histone deacetylase 2 (HD2), is involved in the regulation of wheat resistance to Bgt infection. Recently, it was found that TaHDT701 interacts with TaHDA6 and TaHOS15 to form a histone deacetylase complex. It was shown that the deacetylase complex TaHDT701-TaHDA6-TaHOS15 negatively regulates wheat resistance against powdery mildew by altering the chromatin state of defense-related genes such as TaWRKY45, TaPR1, TaPR2, and TaPR5 (Figure 1). The expression of WRRKY45, TaPR1, TaPR2 and TaPR5 was significantly upregulated by the knockdown of TaHDT701, TaHDA6 and TaHOS15. This was accompanied by increased histone acetylation, methylation and reduced nucleosome occupancy at the promoters of these genes, suggesting that the histone deacetylase complex TaDHT701-TaDHA6-TaHOS15 negatively regulates wheat resistance to Bgt by modifying altering chromatin state of defense related genes (Figure 1) (Zhi et al., 2020). This was the first study to show that the histone deacetylase complex HD2-RPD3-WD40 has a role in controlling plant defense responses to pathogen infection.
The process of chromatin remodeling, which alters the accessibility of particular DNA sections to transcription machinery may also play a role in regulating chromatin structure and gene expression (Narlikar et al., 2013; Chen et al., 2017). It was recently demonstrated that wheat CHD-type chromatin remodeling factor TaCHR729 interacts with the TaKPAB1, a bHLH transcription factor which recognizes the E-box cis element in the TaKCS6 promoter, resulting in the transcriptional activation of TaKCS6, a wax biosynthesis gene (Wang et al., 2019). Intriguingly, it was found that TaCHR729 enhances H3K4me at TaKCS6 promoter region, thereby promoting TaKCS6 transcription, and positively regulates the biosynthesis of wheat cuticular wax. Silencing of the expression of TaKCS6 and TaKPAB1 downregulated wax accumulation, demonstrating that wheat cuticular accumulation might be positively regulated by the transactivation of TaKCS6 by TaKPAB1 (Wang et al., 2019). The TaGCN5-TaADA2 module, in association with TaEPBM1, mediates histone acetylation in the TaECR promoter region (Kong et al., 2020b). TaGCN5 and TaADA2 knockdown downregulated the expression of TaECR and significantly reduced wax accumulation, signifying that wheat wax biosynthesis is triggered by the epigenetic activation of TaECR caused by the TaGCN5-TaADA2 histone acetyltransferase complex (Kong et al., 2020b). Similarly, it was also demonstrated that TaCHR729 knockdown reduced cuticular wax biosynthesis and Bgt conidia germination, suggesting that wheat chromatin remodeling factor TaCHR729 mediates histone methylation and fine-tunes cuticular wax biosynthesis to regulate the interaction between wheat and Bgt (Wang et al., 2019). The findings from these studies clearly demonstrate that various epigenetic regulators like the histone modifying enzymes and chromatin remodeling elements epigenetically regulate wheat cuticle biosynthesis gene expression.
Autophagy, also known as self-eating, is a process whereby cell organelles and cytosolic macromolecules are digested in lysosomes (or vacuoles in plants and yeast) in order to recycle nutrients (Liu and Bassham, 2012; Li and Vierstra, 2012). In plants, the process of autophagy is responsible for both “pro-death” and “pro-survival” functions in regulating programmed cell death associated with ETI. Autophagy contributes to defense responses during plant-biotrophic pathogen interactions (Hofius et al., 2009; Minina et al., 2014). Autophagy-related proteins ATG4 and ATG8 are essential for the biogenesis of autophagy. In the early stages of the Pm21- and Pm3f-triggered wheat incompatible responses to Bgt, two times of transcript accumulation of ATG4s/ATG8s were detected. The first transcript accumulation was detected during early stages of infection and the second one was detected during late stages of infection. Fluorescence microscopy also revealed a Bgt-induced upregulated wheat autophagy level in the Pm21-triggered incompatible interaction. Bgt-induced transcript accumulation of ATG4s/ATG8s was also observed in the late stages of infection of a susceptible line isogenic to the Pm21 resistance line although it was not much higher than that in incompatible interactions. These findings showed that ATG4/ATG8-associated autophagy in the early stage and late stage positively and negatively regulates wheat immune responses to Bgt, respectively (Pei et al., 2014). Furthermore, the expression of wheat ATG4s/ATG8s was shown to be elevated by abiotic stress factors and typically regulated by various phytohormones (Pei et al., 2014). ATG6 proteins are pleiotropic proteins that serve in autophagy as well as the phosphatidylinositol 3-phosphate signaling pathways. Wheat ATG6s are involved in powdery mildew immunity, in which they positively regulate Pm21-triggered resistance response and negatively regulate basal resistance in susceptible plants. Silencing of the ATG-heterologous protein in wheat inhibited the broad-spectrum resisted conferred by Pm21, demonstrating that autophagy positively regulates wheat resistance to Bgt (Yue et al., 2015). The inevitable arrest of successful conidia penetration on knockdown plants before developing into large mycelia suggests that TaATG6 serves a weak positive function in the Pm21-triggered resistance response, most likely in the early stages of Bgt invasion. This implies that, the dynamics of host-pathogen interactions are influenced by autophagy-mediated defense responses. Bgt targets the autophagy process in its host cells because it remobilizes cellular nutrients which it may exploit to accomplish the nutritional supply for its invasive growth and proliferation.
Silicon (Si) is the second most common element in the Earth’s crust, yet its importance in biota is still up for debate (Exley, 1998). It is considered as a quasi-essential bioactive element not important for the growth and development of plants (Epstein, 1972), but accumulating evidence has reported its significant involvement in plant defense responses to abiotic and biotic stresses. Initial speculation about Si’s protective role focused on its ability to strengthen the cell wall, for example, against the invasion of fungal hyphae, but subsequent research has shown that Si’s action on plants is far more complex, involving a cross-talk with the cell interior and an effect on the metabolic processes of the plant (Luyckx et al., 2017). Physical barriers, such as wax, cuticle, and cell-wall protection, and post-formed defensive barriers, such as cell-wall reinforcement and papillae deposition at infection sites, are the basis for the resistance induced by Si against a broad array of pathogens. During plant-pathogens interactions, Si induces or reinforces biochemical or molecular mechanisms to enhance plant resistance against biotic stresses. This includes the activation of defense-related compounds like phytoalexins, phenolics and momilactones and defense-related enzymes like polyphenol oxidase, peroxidase, phenylalanine ammonia lyase and lipoxygenase (Rémus-Borel et al., 2005; Rahman et al., 2015). The unleashing of the unsung roles of silicon in wheat resistance to powdery mildew will benefit many disciplines including agriculture, and ecology (Figure 2).
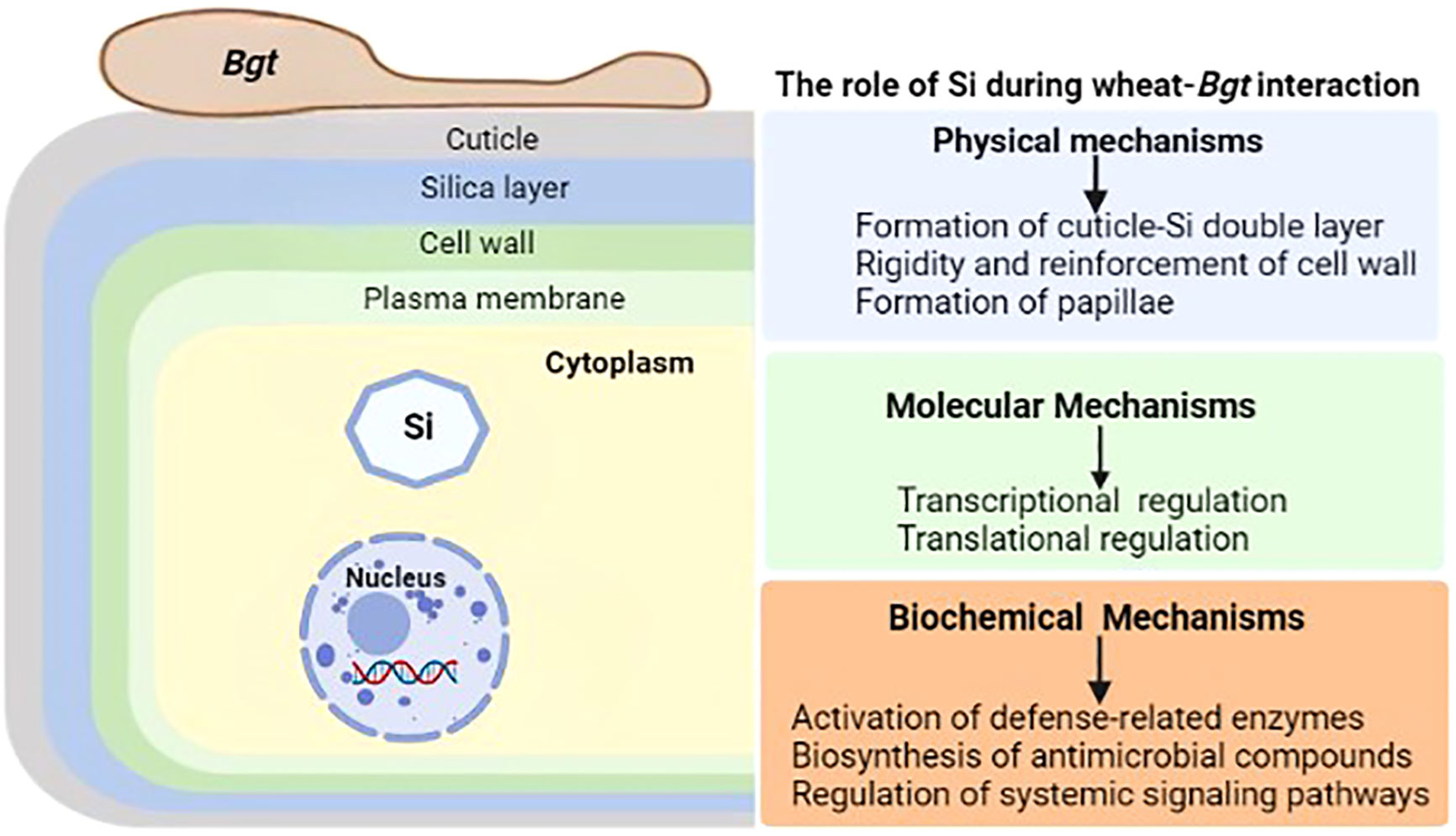
Figure 2 General illustration of the role of silicon (Si) during wheat-Bgt interactions. Wheat defense responses mediated by Si are generally classified into physical, molecular and biochemical mechanisms. Physical mechanisms involve the formation of cuticle-Si double layer, reinforcement of cell wall and papillae formation. Molecular mechanisms involve the transcriptional and translational regulation of key defense-related genes and proteins. Biochemical mechanisms are attributed to the activation of defense-related enzymes, stimulation of antimicrobial compounds biosynthesis and regulation of the complex systemic network of signaling pathways.
Plants exposed to Si form a silica layer in between cuticle and cell wall, which serves as a physical barrier and promotes plant resistance to pathogenic fungal infection (Islam et al., 2020). It was established that Si can mitigate the effects of various wheat fungal diseases including powdery mildew by modifying their monocyclic components like infection efficiency, incubation time, quantity of lesions per unit area of infected leaf, lesion size, and the rate at which the lesion expands (Pazdiora et al., 2018). Both physical barriers and metabolic defenses act synergistically to reduce disease severity (Debona et al., 2017). Si deposition below the cuticle and an increase in papillae formation at infection sites have been attributed to the physical barrier in wheat (Bélanger et al., 2003; Filha et al., 2011). Si also increased defense enzyme activity, biosynthesis of phytoalexins and phenolic compounds, and H2O2 accumulation at the infection sites, thus priming the biochemical defense responses (Pazdiora et al., 2021). The presence of a phenolic-like material associated with degraded powdery mildew haustoria was observed in epidermal cells of Si-treated wheat leaves infected with Bgt (Bélanger et al., 2003). Fungitoxic aglycones were found in different concentrations in plants that were treated with Si compared to the control. In wheat plants that were not treated with Si, Bgt formed a haustorium that was well developed, whereas in Si-treated plants osmiophilic deposits remained behind (Bélanger et al., 2003). Furthermore, collapsed conidial chains and intense fluorescence was observed in Bgt-infected wheat leaves at the sections that were treated with Si. These findings strongly suggest that wheat plants treated with Si are able to produce phytoalexins in response to Bgt infection, that is, active localized defense responses to Bgt infection are mediated by Si (Bélanger et al., 2003; Rémus-Borel et al., 2005). Si can bind to hydroxyl groups of proteins strategically involved in signal transduction or interfere with cationic co-factors of enzymes regulating pathogenesis-related genes, acting as a modulator affecting the timing and degree of plant defense responses, the same way secondary messengers induce systemic resistance. As a result, Si may induce resistance by interacting with several factors within plant stress signaling systems (Fauteux et al., 2005).
A transcriptomic study revealed minimal indication of regulation of a particular metabolic activity, and the response to the supply of Si on control (uninfected) plants was restricted to 47 genes of various functions (Chain et al., 2009). Bgt inoculation led to an upregulation of a plethora of stress- and metabolism-related genes while simultaneously downregulating the expression of genes involved in photosynthesis. Si-treatment to Bgt-infected plants halted the spread of disease and resulted in an almost complete reversal of the genes controlled by only Bgt effects (Chain et al., 2009). These findings imply that Si has a little effect on a plant’s transcriptome in the absence of stress, even in the case of a monocot that accumulates a lot of Si, like wheat. Conversely, the transcriptome alterations generated by Bgt were counteracted by the addition of Si, which had the added advantage of reducing the impact of biotic stress (Chain et al., 2009). About 900 genes reacting to pathogen infection were changed in control leaves of wheat-Bgt infected plants, whereas the pathogen altered very few genes in Si-treated plants, indicating that the stress caused by Bgt invasion was almost eliminated by Si (Chain et al., 2009). As a result, Si appears to minimize the effect of pathogen infection on the transcriptome of host plants, most probably by limiting the exploitation of pathogen virulence factors, rather than promoting resistance via transcriptional reprogramming of defense-related genes (Van Bockhaven et al., 2015). Si can induce the formation of a thicker cellulose membrane, whereas the severity of the disease in wheat in the field can be reduced by the double cuticle layer, density of long and short silicified plant epidermal cells, thick silica layer below the cuticle thereby curbing yield losses (Pozza et al., 2015). It was also established that, as the Si content in the culture medium increased, the pathogen index decreased, the grain mass and number together with the dry weight of shoots increased significantly (Moldes et al., 2016). Multivariant analysis revealed that an increase in Si treatments significantly reduced the antioxidant activities of some biochemical parameters such as superoxide dismutase, catalase, and ascorbate peroxidase. Furthermore, increase in Si doses resulted in a decrease in Bgt proliferation in foliar surfaces. It was concluded that an increase in Si concentration and reduced activity of antioxidant enzymes is closely associated with a reduction of powdery mildew in wheat (Moldes et al., 2016). Incorporating the Si functions in reduction of plant stresses into development of molecular methods can now be facilitated by recent advances in genomics and metabolomics. It has been shown that Si-application reduces biotic stress over time, which may be useful for ecologically integrated strategies that aim to improve resistance of crops to biotic stresses and boost yield without resorting to pesticides.
Undoubtedly, wheat powdery mildew is among the most destructive diseases that are threatening global wheat production. To combat this disease, breeders can utilize genetically resistant cultivars, which is both cost-effective and chemical-free. As genomics of agricultural plants and their linked wild species improves, the gene pool available for enhancing powdery mildew resistance may be increased and widely utilized. Future breeding programs should aim to find exploited and unexploited Pm genes, and the introgression of these genes, to develop a large number of pathogen-resistant cultivars. Characterizing wheat germplasm at the pathological level, followed by molecular characterization utilizing linked markers for known Pm genes, would be one of the most efficient approaches for combating the expanding problem of biotic stresses in wheat. Classical breeding has been successful in amassing resistance, and cultivars with reasonable levels of resistance have been identified by germplasm screenings. Resistance characterization and efficient application in wheat breeding have been enhanced by the adoption of modern phenotyping methodologies, the extraordinary growth of genetic resources, and the introduction of speed breeding tools.
Despite the recent technological and scientific advancements in the management of wheat disease, plant pathogens continue to threaten global wheat production. Climate change may directly or indirectly affect fungal pathogens and their respective diseases they cause. Factors such as rainfall, temperature, relative humidity, wind speed and direction, affect the pathogenesis, invasive growth, proliferation, spread and the ultimate survival of the pathogen. The most significant factors in determining the success or failure of a specific host-pathogen interaction, as well as the spread and survival of the pathogen are temperature, relative humidity and precipitation. Therefore, before investigating the effects of climate on powdery mildew development, it is of great importance to predict the possible repercussions of the climate change on the host, pathogen, their interaction, population dynamics, micro-evolutionary developments and the structure of the agro-system community. Powdery mildew can therefore be effectively managed by harnessing genetic resistance and integrated powdery mildew management methods to develop more durable wheat resistant cultivars.
Conceptualization, JM; literature search, JM; writing—original draft preparation, JM; writing—review and editing, JM, JC, and WY; funding acquisition, WY. All authors contributed to the article and approved the submitted version.
This work was funded by the Natural Science Foundation of China (No. 301871915, 32172367), Natural Science Foundation of Hebei Province (C2020204071), Modern Agricultural Industry System of Wheat Industry in Hebei Province (No. HBCT2018010204).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acevedo-Garcia, J., Spencer, D., Thieron, H., Reinstädler, A., Hammond-Kosack, K., Phillips, A. L., et al. (2017). Mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 15 (3), 367–378. doi: 10.1111/pbi.12631
Adachi, H., Tsuda, K. (2019). Convergence of cell-surface and intracellular immune receptor signalling. New Phytol. 221 (4), 1676–1678. doi: 10.1111/nph.15634
Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., Rogers, J., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 361 (6403), eaar7191. doi: 10.1126/science.aar7191
Arya, G. C., Sarkar, S., Manasherova, E., Aharoni, A., Cohen, H. (2021). The plant cuticle: An ancient guardian barrier set against long-standing rivals. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.663165
Ashapkin, V. V., Kutueva, L. I., Aleksandrushkina, N. I., Vanyushin, B. F. (2020). Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses. Int. J. Mol. Sci. 21 (20), 7457. doi: 10.3390/ijms21207457
Avni, R., Nave, M., Barad, O., Baruch, K., Twardziok, S. O., Gundlach, H., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science. 357 (6346), 93–97. doi: 10.1126/science.aan0032
Azevedo, C., Betsuyaku, S., Peart, J., Takahashi, A., Noël, L., Sadanandom, A., et al. (2006). Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25 (9), 2007–2016. doi: 10.1038/sj.emboj.7601084
Baggerman, G., Vierstraete, E., De Loof, A., Schoofs, L. (2005). Gel-based versus gel-free proteomics: a review. Comb. Chem. High Throughput Screen. 8 (8), 669–677. doi: 10.2174/138620705774962490
Bélanger, R. R., Benhamou, N., Menzies, J. G. (2003). Cytological evidence of an active role of silicon in wheat resistance to powdery mildew (Blumeria graminis f. sp. tritici). Phytopathol. 93 (4), 402–412. doi: 10.1094/phyto.2003.93.4.402
Bhaskar, P. B., Raasch, J. A., Kramer, L. C., Neumann, P., Wielgus, S. M., Austin-Phillips, S., et al. (2008). Sgt1, but not Rar1, is essential for the RB-mediated broad-spectrum resistance to potato late blight. BMC Plant Biol. 8 (1), 1–9. doi: 10.1186/1471-2229-8-8
Bhuiyan, N. H., Liu, W., Liu, G., Selvaraj, G., Wei, Y., King, J. (2007). Transcriptional regulation of genes involved in the pathways of biosynthesis and supply of methyl units in response to powdery mildew attack and abiotic stresses in wheat. Plant Mol. Biol. 64 (3), 305–318. doi: 10.1007/s11103-007-9155-x
Bhuiyan, N. H., Selvaraj, G., Wei, Y., King, J. (2009). Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60 (2), 509–521. doi: 10.1093/jxb/ern290
Bhullar, N. K., Zhang, Z., Wicker, T., Keller, B. (2010). Wheat gene bank accessions as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project'. BMC Plant Biol. 10 (1), 88. doi: 10.1186/1471-2229-10-88
Bianchi, M. W., Roux, C., Vartanian, N. (2002). Drought regulation of GST8, encoding the arabidopsis homologue of ParC/Nt107 glutathione transferase/peroxidase. Physiol. Plant 116 (1), 96–105. doi: 10.1034/j.1399-3054.2002.1160112.x
Bindschedler, L. V., Panstruga, R., Spanu, P. D. (2016). Mildew-omics: how global analyses aid the understanding of life and evolution of powdery mildews. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00123
Bolouri Moghaddam, M. R., Van den Ende, W. (2012). Sugars and plant innate immunity. J. Exp. Bot. 63 (11), 3989–3998. doi: 10.1093/jxb/ers129
Bourras, S., Kunz, L., Xue, M., Praz, C. R., Müller, M. C., Kälin, C., et al. (2019). The AvrPm3-Pm3 effector-NLR interactions control both race-specific resistance and host-specificity of cereal mildews on wheat. Nat. Commun. 10 (1), 2292. doi: 10.1038/s41467-019-10274-1
Bourras, S., McNally, K. E., Ben-David, R., Parlange, F., Roffler, S., Praz, C. R., et al. (2015). Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell. 27 (10), 2991–3012. doi: 10.1105/tpc.15.00171
Bourras, S., McNally, K. E., Müller, M. C., Wicker, T., Keller, B. (2016). Avirulence genes in cereal powdery mildews: The gene-for-gene hypothesis 2.0. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00241
Bourras, S., Praz, C. R., Spanu, P. D., Keller, B. (2018). Cereal powdery mildew effectors: a complex toolbox for an obligate pathogen. Curr. Opin. Microbiol. 46, 26–33. doi: 10.1016/j.mib.2018.01.018
Brunner, S., Hurni, S., Streckeisen, P., Mayr, G., Albrecht, M., Yahiaoui, N., et al. (2010). Intragenic allele pyramiding combines different specificities of wheat Pm3 resistance alleles. Plant J. 64 (3), 433–445. doi: 10.1111/j.1365-313X.2010.04342.x
Burdon, J. J., Barrett, L. G., Rebetzke, G., Thrall, P. H. (2014). Guiding deployment of resistance in cereals using evolutionary principles. Evol. Appl. 7 (6), 609–624. doi: 10.1111/eva.12175
Buscaill, P., Rivas, S. (2014). Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 20, 35–46. doi: 10.1016/j.pbi.2014.04.004
Cao, H. H., Wang, S. H., Liu, T. X. (2014). Jasmonate-and salicylate-induced defenses in wheat affect host preference and probing behavior but not performance of the grain aphid, Sitobion avenae. Insect Sci. 21 (1), 47–55. doi: 10.1111/1744-7917.12023
Cao, A., Xing, L., Wang, X., Yang, X., Wang, W., Sun, Y., et al. (2011). Serine/threonine kinase gene Stpk-v, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. U S A. 108 (19), 7727–7732. doi: 10.1073/pnas.1016981108
Chadick, J. Z., Asturias, F. J. (2005). Structure of eukaryotic mediator complexes. Trends Biochem. Sci. 30 (5), 264–271. doi: 10.1016/j.tibs.2005.03.001
Chain, F., Côté-Beaulieu, C., Belzile, F., Menzies, J. G., Bélanger, R. R. (2009). A comprehensive transcriptomic analysis of the effect of silicon on wheat plants under control and pathogen stress conditions. Mol. Plant Microbe Interact. 22 (11), 1323–1330. doi: 10.1094/mpmi-22-11-1323
Chaudhari, P., Ahmed, B., Joly, D. L., Germain, H. (2014). Effector biology during biotrophic invasion of plant cells. Virulence. 5 (7), 703–709. doi: 10.4161/viru.29652
Chaudhary, J., Khatri, P., Singla, P., Kumawat, S., Kumari, A., R, V., Vikram, A., et al. (2019). Advances in omics approaches for abiotic stress tolerance in tomato. Biol. (Basel). 8, 4. doi: 10.3390/biology8040090
Chen, R., Jiang, H., Li, L., Zhai, Q., Qi, L., Zhou, W., et al. (2012). The arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell. 24 (7), 2898–2916. doi: 10.1105/tpc.112.098277
Chen, F., Jia, H., Zhang, X., Qiao, L., Li, X., Zheng, J., et al. (2019a). Positional cloning of PmCH1357 reveals the origin and allelic variation of the Pm2 gene for powdery mildew resistance in wheat. Crop J. 7 (6), 771–783. doi: 10.1016/j.cj.2019.08.004
Chen, R., Li, M., Zhang, H., Duan, L., Sun, X., Jiang, Q., et al. (2019b). Continuous salt stress-induced long non-coding RNAs and DNA methylation patterns in soybean roots. BMC Genomics 20 (1), 730. doi: 10.1186/s12864-019-6101-7
Chen, T., Xiao, J., Xu, J., Wan, W., Qin, B., Cao, A., et al. (2016). Two members of TaRLK family confer powdery mildew resistance in common wheat. BMC Plant Biol. 16 (1), 27. doi: 10.1186/s12870-016-0713-8
Chen, W., Zhu, Q., Liu, Y., Zhang, Q. (2017). Chromatin remodeling and plant immunity. Adv. Protein Chem. Struct. Biol. 106, 243–260. doi: 10.1016/bs.apcsb.2016.08.006
Chethana, K. W. T., Jayawardena, R. S., Chen, Y.-J., Konta, S., Tibpromma, S., Abeywickrama, P. D., et al. (2021). Diversity and function of appressoria. Pathog. 10 (6), 746. doi: 10.3390/pathogens10060746
Creutz, C. E., Tomsig, J. L., Snyder, S. L., Gautier, M. C., Skouri, F., Beisson, J., et al. (1998). The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J. Biol. Chem. 273 (3), 1393–1402. doi: 10.1074/jbc.273.3.1393
Dangl, J. L., Horvath, D. M., Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science. 341 (6147), 746–751. doi: 10.1126/science.1236011
Debona, D., Rodrigues, F. A., Datnoff, L. E. (2017). Silicon's role in abiotic and biotic plant stresses. Annu. Rev. Phytopathol. 55 (1), 85–107. doi: 10.1146/annurev-phyto-080516-035312
Deleris, A., Halter, T., Navarro, L. (2016). DNA Methylation and demethylation in plant immunity. Annu. Rev. Phytopathol. 54, 579–603. doi: 10.1146/annurev-phyto-080615-100308
Deng, Y., Zhai, K., Xie, Z., Yang, D., Zhu, X., Liu, J., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science. 355 (6328), 962–965. doi: 10.1126/science.aai8898
Ding, B., Wang, G. L. (2015). Chromatin versus pathogens: the function of epigenetics in plant immunity. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00675
Dodds, P. N., Lawrence, G. J., Catanzariti, A. M., Teh, T., Wang, C. I., Ayliffe, M. A., et al. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. U S A. 103 (23), 8888–8893. doi: 10.1073/pnas.0602577103
Dowen, R. H., Pelizzola, M., Schmitz, R. J., Lister, R., Dowen, J. M., Nery, J. R., et al. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. U S A. 109 (32), E2183–E2191. doi: 10.1073/pnas.1209329109
Epstein, E. (1972). “Mineral nutrition of plants,” in Principles and perspectives 2nd Ed (Sunderland: Sinauer Associates Inc).
Espinas, N. A., Saze, H., Saijo, Y. (2016). Epigenetic control of defense signaling and priming in plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01201
Exley, C. (1998). Silicon in life: a bioinorganic solution to bioorganic essentiality. J. Inorg. Biochem. 69 (3), 139–144. doi: 10.1016/S0162-0134(97)10010-1
Fauteux, F., Rémus-Borel, W., Menzies, J. G., Bélanger, R. R. (2005). Silicon and plant disease resistance against pathogenic fungi. FEMS Microbiol. Letters. 249 (1), 1–6. doi: 10.1016/j.femsle.2005.06.034
Filha, M. S. X., Rodrigues, F. A., Domiciano, G. P., Oliveira, H. V., Silveira, P. R., Moreira, W. R. (2011). Wheat resistance to leaf blast mediated by silicon. Aust. Plant Pathol. 40 (1), 28–38. doi: 10.1007/s13313-010-0010-1
Fu, Y., Zhang, H., Mandal, S. N., Wang, C., Chen, C., Ji, W. (2016). Quantitative proteomics reveals the central changes of wheat in response to powdery mildew. J. Proteomics. 130, 108–119. doi: 10.1016/j.jprot.2015.09.006
Gawroński, P., Witoń, D., Vashutina, K., Bederska, M., Betliński, B., Rusaczonek, A., et al. (2014). Mitogen-activated protein kinase 4 is a salicylic acid-independent regulator of growth but not of photosynthesis in Arabidopsis. Mol. Plant 7 (7), 1151–1166. doi: 10.1093/mp/ssu060
Geng, S., Kong, X., Song, G., Jia, M., Guan, J., Wang, F., et al. (2019). DNA Methylation dynamics during the interaction of wheat progenitor Aegilops tauschii with the obligate biotrophic fungus Blumeria graminis f. sp. tritici. New Phytol. 221 (2), 1023–1035. doi: 10.1111/nph.15432
Gevaert, K., Vandekerckhove, J., Vandekerckhove, J. (2011). Gel-free proteomics: Methods and protocols. 1st Edn (Cham: Springer). doi: 10.1007/978-1-61779-148-2
Glawe, D. A. (2008). The powdery mildews: a review of the world's most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol. 46, 27–51. doi: 10.1146/annurev.phyto.46.081407.104740
Godfray, H. C., Beddington, J. R., Crute, I. R., Haddad, L., Lawrence, D., Muir, J. F., et al. (2010). Food security: the challenge of feeding 9 billion people. Science. 327 (5967), 812–818. doi: 10.1126/science.1185383
Gong, H., Jiao, Y., Hu, W. W., Pua, E. C. (2005). “Expression of glutathione-s-transferase and its role in plant growth and development in vivo and shoot morphogenesis in vitro.,” in Plant mol. biol, (Springer) 57, 53–66. doi: 10.1007/s11103-004-4516-1
Guerriero, G., Hausman, J. F., Ezcurra, I. (2015). WD40-repeat proteins in plant cell wall formation: Current evidence and research prospects. Front. Plant Sci. 6, 1112doi: 10.3389/fpls.2015.01112
Hao, Y., Xu, S., lyu, Z., Wang, H., Kong, L., Sun, S. (2021). Comparative analysis of the glutathione s-transferase gene family of four triticeae species and transcriptome analysis of GST genes in common wheat responding to salt stress. Int. J. Genom. 2021, 6289174. doi: 10.1155/2021/6289174
He, Y., Li, Z. (2018). Epigenetic environmental memories in plants: establishment, maintenance, and reprogramming. Trends Genet. 34 (11), 856–866. doi: 10.1016/j.tig.2018.07.006
Henderson, I. R., Jacobsen, S. E. (2007). Epigenetic inheritance in plants. Nature. 447 (7143), 418–424. doi: 10.1038/nature05917
Herrera-Foessel, S. A., Singh, R. P., Lillemo, M., Huerta-Espino, J., Bhavani, S., Singh, S., et al. (2014). Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 127 (4), 781–789. doi: 10.1007/s00122-013-2256-9
Hewitt, T., Müller, M. C., Molnár, I., Mascher, M., Holušová, K., Šimková, H., et al. (2021). A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 229 (5), 2812–2826. doi: 10.1111/nph.17075
He, H., Zhu, S., Zhao, R., Jiang, Z., Ji, Y., Ji, J., et al. (2018). Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 11 (6), 879–882. doi: 10.1016/j.molp.2018.03.004
Hofius, D., Schultz-Larsen, T., Joensen, J., Tsitsigiannis, D. I., Petersen, N. H., Mattsson, O., et al. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 137 (4), 773–783. doi: 10.1016/j.cell.2009.02.036
Hu, P., Liu, J., Xu, J., Zhou, C., Cao, S., Zhou, W., et al. (2018). A malectin-like/leucine-rich repeat receptor protein kinase gene, RLK-v, regulates powdery mildew resistance in wheat. Mol. Plant Pathol. 19 (12), 2561–2574. doi: 10.1111/mpp.12729
Hurni, S., Brunner, S., Buchmann, G., Herren, G., Jordan, T., Krukowski, P., et al. (2013). Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 76 (6), 957–969. doi: 10.1111/tpj.12345
Islam, W., Tayyab, M., Khalil, F., Hua, Z., Huang, Z., Chen, H. Y. H. (2020). Silicon-mediated plant defense against pathogens and insect pests.Pestic. Biochem. Physiol. 168, 104641. doi: 10.1016/j.pestbp.2020.104641
Jankovics, T., Komáromi, J., Fábián, A., Jäger, K., Vida, G., Kiss, L. (2015). New insights into the life cycle of the wheat powdery mildew: Direct observation of ascosporic infection in Blumeria graminis f. sp. tritici. Phytopathol 105 (6), 797–804. doi: 10.1094/phyto-10-14-0268-r
Jaswal, R., Kiran, K., Rajarammohan, S., Dubey, H., Singh, P. K., Sharma, Y., et al. (2020). Effector biology of biotrophic plant fungal pathogens: Current advances and future prospects. Microbiol. Res. 241, 126567. doi: 10.1016/j.micres.2020.126567
Jenuwein, T., Allis, C. D. (2001). Translating the histone code. Science. 293 (5532), 1074–1080. doi: 10.1126/science.1063127
Jing, Y., Liu, J., Liu, P., Ming, D., Sun, J. (2019). Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci. Rep. 9 (1), 5691. doi: 10.1038/s41598-019-42177-y
Jing, Y., Zhang, D., Wang, X., Tang, W., Wang, W., Huai, J., et al. (2013). Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell. 25 (1), 242–256. doi: 10.1105/tpc.112.105742
Jin, Y., Liu, H., Tiantian, G., Xing, L., Han, G., Ma, P., et al. (2022). PM2b, a CC-NBS-LRR protein, interacts with TaWRKY76-d to regulate powdery mildew resistance in common wheat. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.973065
Jin, Y., Xu, H., Ma, P., Fu, X., Song, L., Xu, Y., et al. (2018). Characterization of a new Pm2 allele associated with broad-spectrum powdery mildew resistance in wheat line subtil. Sci. Rep. 8 (1), 475. doi: 10.1038/s41598-017-18827-4
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature. 444 (7117), 323–329. doi: 10.1038/nature05286
Jones, J. D., Vance, R. E., Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science. 354 (6316), aaf6395. doi: 10.1126/science.aaf6395
Jones, H., Whipps, J. M., Gurr, S. J. (2001). The tomato powdery mildew fungus Oidium neolycopersici. Mol. Plant Pathol. 2 (6), 303–309. doi: 10.1046/j.1464-6722.2001.00084.x
Kamakura, T., Yamaguchi, S., Saitoh, K., Teraoka, T., Yamaguchi, I. (2002). A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol. Plant Microbe Interact. 15 (5), 437–444. doi: 10.1094/mpmi.2002.15.5.437
Kang, Y., Zhou, M., Merry, A., Barry, K. (2020). Mechanisms of powdery mildew resistance of wheat – a review of molecular breeding. Plant Pathol. 69 (4), 601–617. doi: 10.1111/ppa.13166
Kanyuka, K., Rudd, J. J. (2019). Cell surface immune receptors: the guardians of the plant’s extracellular spaces. Curr. Opin. Plant Biol. 50, 1–8. doi: 10.1016/j.pbi.2019.02.005
Kanzaki, H., Yoshida, K., Saitoh, H., Fujisaki, K., Hirabuchi, A., Alaux, L., et al. (2012). Arms race co-evolution of Magnaporthe oryzae AVR-pik and rice pik genes driven by their physical interactions. Plant J. 72 (6), 894–907. doi: 10.1111/j.1365-313X.2012.05110.x
Karpenko, V., Pavlyshyn, S., Prytulіak, R., Naherniuk, D. (2019). Content of malondialdehyde and activity of enzyme glutathione-s-transferase in the leaves of emmer wheat under the action of herbicide and plant growth regulator. Agron. Res. 17, 144–154. doi: 10.15159/ar.19.014
Kidd, B. N., Edgar, C. I., Kumar, K. K., Aitken, E. A., Schenk, P. M., Manners, J. M., et al. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in arabidopsis. Plant Cell. 21 (8), 2237–2252. doi: 10.1105/tpc.109.066910
Kim, J. M., To, T. K., Matsui, A., Tanoi, K., Kobayashi, N. I., Matsuda, F., et al. (2017). Acetate-mediated novel survival strategy against drought in plants. Nat. Plants. 3, 17097. doi: 10.1038/nplants.2017.97
Kong, L., Chang, C. (2018). Suppression of wheat TaCDK8/TaWIN1 interaction negatively affects germination of Blumeria graminis f.sp. tritici by interfering with very-long-chain aldehyde biosynthesis. Plant Mol. Biol. 96 (1-2), 165–178. doi: 10.1007/s11103-017-0687-4
Kong, L., Liu, Y., Wang, X., Chang, C. (2020a). Insight into the role of epigenetic processes in abiotic and biotic stress response in wheat and barley. Int. J. Mol. Sci. 21 (4), 1480. doi: 10.3390/ijms21041480
Kong, L., Zhi, P., Liu, J., Li, H., Zhang, X., Xu, J., et al. (2020b). Epigenetic activation of enoyl-CoA reductase by an acetyltransferase complex triggers wheat wax biosynthesis. Plant Physiol. 183 (3), 1250–1267. doi: 10.1104/pp.20.00603
Krattinger, S. G., Kang, J., Bräunlich, S., Boni, R., Chauhan, H., Selter, L. L., et al. (2019). Abscisic acid is a substrate of the ABC transporter encoded by the durable wheat disease resistance gene Lr34. New Phytol. 223 (2), 853–866. doi: 10.1111/nph.15815
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 323 (5919), 1360–1363. doi: 10.1126/science.1166453
Kurdistani, S. K., Grunstein, M. (2003). Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4 (4), 276–284. doi: 10.1038/nrm1075
Law, J. A., Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11 (3), 204–220. doi: 10.1038/nrg2719
Lei, Y., Wang, M., Wan, A., Xia, C., See, D. R., Zhang, M., et al. (2017). Virulence and molecular characterization of experimental isolates of the stripe rust pathogen (Puccinia striiformis) indicate somatic recombination. Phytopathol. 107 (3), 329–344. doi: 10.1094/phyto-07-16-0261-r
Le, T. N., Schumann, U., Smith, N. A., Tiwari, S., Au, P. C., Zhu, Q. H., et al. (2014). DNA Demethylases target promoter transposable elements to positively regulate stress responsive genes in arabidopsis. Genome Biol. 15 (9), 458. doi: 10.1186/s13059-014-0458-3
Liang, P., Liu, S., Xu, F., Jiang, S., Yan, J., He, Q., et al. (2018). Powdery mildews are characterized by contracted carbohydrate metabolism and diverse effectors to adapt to obligate biotrophic lifestyle. Front. Microbiol. 9 (3160). doi: 10.3389/fmicb.2018.03160
Liang, Y., Xia, Y., Chang, X., Gong, G., Yang, J., Hu, Y., et al. (2019). Comparative proteomic analysis of wheat carrying Pm40 response to Blumeria graminis f. sp. tritici using two-dimensional electrophoresis. Int. J. Mol. Sci. 20 (4), 933. doi: 10.3390/ijms20040933
Li, M., Dong, L., Li, B., Wang, Z., Xie, J., Qiu, D., et al. (2020). A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol. 228 (3), 1027–1037. doi: 10.1111/nph.16761
Li, H., Dong, Z., Ma, C., Tian, X., Xiang, Z., Xia, Q., et al. (2019). Discovery of powdery mildew resistance gene candidates from Aegilops biuncialis chromosome 2Mb based on transcriptome sequencing. PloS One 14 (11), e0220089. doi: 10.1371/journal.pone.0220089
Li, C. X., Hu, L., Xu, W. G., Zhang, L., Dong, H. B., Wang, H. W. (2013). Differentially expressed wheat genes in response to powdery mildew infection. Ann. Appl. Biol. 163 (2), 209–217. doi: 10.1111/aab.12047
Li, Z., Lan, C., He, Z., Singh, R. P., Rosewarne, G. M., Chen, X., et al. (2014). Overview and application of QTL for adult plant resistance to leaf rust and powdery mildew in wheat. Crop Sci. 54 (5), 1907–1925. doi: 10.2135/cropsci2014.02.0162
Liu, Y., Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. doi: 10.1146/annurev-arplant-042811-105441
Liu, S., Griffey, C. A., Maroof, M. A. S. (2001). Identification of molecular markers associated with adult plant resistance to powdery mildew in common wheat cultivar Massey. Crop Sci. 41 (4), 1268–1275. doi: 10.2135/cropsci2001.4141268x
Liu, J., Sun, L., Chen, Y., Wei, L., Hao, Y., Yu, Z., et al. (2020). The regulatory network of CMPG1-V in wheat-Blumeria graminis f. sp. tritici interaction revealed by temporal profiling using RNA-seq. Int. J. Mol. Sci. 21 (17), 5967. doi: 10.3390/ijms21175967
Liu, X., Wang, J., Fan, B., Shang, Y., Sun, Y., Dang, C., et al. (2018). A COI1 gene in wheat contributes to the early defence response against wheat powdery mildew. J. Phytopathol. 166 (2), 116–122. doi: 10.1111/jph.12667
Liu, J., Zhang, T., Jia, J., Sun, J. (2016). The wheat mediator subunit TaMED25 interacts with the transcription factor TaEIL1 to negatively regulate disease resistance against powdery mildew. Plant Physiol. 170 (3), 1799–1816. doi: 10.1104/pp.15.01784
Liu, J., Zhi, P., Wang, X., Fan, Q., Chang, C. (2019). Wheat WD40-repeat protein TaHOS15 functions in a histone deacetylase complex to fine-tune defense responses to Blumeria graminis f. sp. tritici. J. Exp. Bot. 70 (1), 255–268. doi: 10.1093/jxb/ery330
Li, F., Vierstra, R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17 (9), 526–537. doi: 10.1016/j.tplants.2012.05.006
Li, G., Wu, Y., Liu, G., Xiao, X., Wang, P., Gao, T., et al. (2017a). Large-Scale proteomics combined with transgenic experiments demonstrates an important role of jasmonic acid in potassium deficiency response in wheat and rice. Mol. Cell. Proteom. 16 (11), 1889–1905. doi: 10.1074/mcp.RA117.000032
Li, J., Yang, X., Liu, X., Yu, H., Du, C., Li, M., et al. (2017b). Proteomic analysis of the compatible interaction of wheat and powdery mildew (Blumeria graminis f. sp. tritici). Plant Physiol. Biochem. 111, 234–243. doi: 10.1016/j.plaphy.2016.12.006
Li, A., Zhang, R., Pan, L., Tang, L., Zhao, G., Zhu, M., et al. (2011). Transcriptome analysis of H2O2-treated wheat seedlings reveals a H2O2-responsive fatty acid desaturase gene participating in powdery mildew resistance. PloS One 6 (12), e28810. doi: 10.1371/journal.pone.0028810
Li, A. L., Zhu, Y. F., Tan, X. M., Wang, X., Wei, B., Guo, H. Z., et al. (2008). Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum l.). Plant Mol. Biol. 66 (4), 429–443. doi: 10.1007/s11103-007-9281-5
Lu, P., Guo, L., Wang, Z., Li, B., Li, J., Li, Y., et al. (2020). A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 11 (1), 680. doi: 10.1038/s41467-020-14294-0
Luo, M., Yu, C.-W., Chen, F.-F., Zhao, L., Tian, G., Liu, X., et al. (2012). Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in arabidopsis. PloS Genet. 8 (12), e1003114. doi: 10.1371/journal.pgen.1003114
Luyckx, M., Hausman, J.-F., Lutts, S., Guerriero, G. (2017). Silicon and plants: current knowledge and technological perspectives. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00411
Maccaferri, M., Harris, N. S., Twardziok, S. O., Pasam, R. K., Gundlach, H., Spannagl, M., et al. (2019). Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51 (5), 885–895. doi: 10.1038/s41588-019-0381-3
Mandal, M. S., Fu, Y., Zhang, S., Ji, W. (2014). Proteomic analysis of the defense response of wheat to the powdery mildew fungus, Blumeria graminis f. sp. tritici. Protein J. 33 (6), 513–524. doi: 10.1007/s10930-014-9583-9
Manser, B., Koller, T., Praz, C. R., Roulin, A. C., Zbinden, H., Arora, S., et al. (2021). Identification of specificity-defining amino acids of the wheat immune receptor Pm2 and powdery mildew effector AvrPm2. Plant J. 106 (4), 993–1007. doi: 10.1111/tpj.15214
Mapuranga, J., Zhang, N., Zhang, L., Chang, J., Yang, W. (2022b). Infection strategies and pathogenicity of biotrophic plant fungal pathogens. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.799396
Mapuranga, J., Zhang, N., Zhang, L., Liu, W., Chang, J., Yang, W. (2022c). Harnessing genetic resistance to rusts in wheat and integrated rust management methods to develop more durable resistant cultivars. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.951095
Mapuranga, J., Zhang, L., Zhang, N., Yang, W. (2022a). The haustorium: The root of biotrophic fungal pathogens. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.963705
Matzke, M. A., Mosher, R. A. (2014). RNA-Directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15 (6), 394–408. doi: 10.1038/nrg3683
Maurya, S., Singh, M., Kumar, S., Lakhran, L., Kumar, N., Prajapati, S. (2021). Breeding approaches for disease resistance in crop plants: A review. Ann. Clin. Lab. Sci. 4 (2), 1022 Available at: http://meddocsonline.org.
Ma, P., Wu, L., Xu, Y., Xu, H., Zhang, X., Wang, W., et al. (2021). Bulked segregant RNA-seq provides distinctive expression profile against powdery mildew in the wheat genotype YD588. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.764978
McDonald, B. A., Linde, C. (2002). The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica. 124 (2), 163–180. doi: 10.1023/A:1015678432355
McIntosh, R. A., Dubcovsky, J., Rogers, J. W., Xia, X. C., Raupp, W. (2019). Catalogue of 505 gene symbols for wheat: 2019 supplement: In annual wheat 506. ed. raupp, w. j. (Manhattan, USA: Newsletter). Ann. Wheat Newslett. 506, 98–113.
Meldau, S., Baldwin, I. T., Wu, J. (2011). SGT1 regulates wounding- and herbivory-induced jasmonic acid accumulation and Nicotiana attenuata's resistance to the specialist lepidopteran herbivore Manduca sexta. New Phytol. 189 (4), 1143–1156. doi: 10.1111/j.1469-8137.2010.03558.x
Minina, E. A., Bozhkov, P. V., Hofius, D. (2014). Autophagy as initiator or executioner of cell death. Trends Plant Sci. 19 (11), 692–697. doi: 10.1016/j.tplants.2014.07.007
Moldes, C. A., de Lima Filho, O. F., Merini, L. J., Tsai, S. M., Camiña, J. M. (2016). Occurrence of powdery mildew disease in wheat fertilized with increasing silicon doses: a chemometric analysis of antioxidant response. Acta Physiol. Plant 38 (8), 206. doi: 10.1007/s11738-016-2217-4
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47 (12), 1494–1498. doi: 10.1038/ng.3439
Mueller, L. A., Goodman, C. D., Silady, R. A., Walbot, V. (2000). AN9, a petunia glutathione s-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 123 (4), 1561–1570. doi: 10.1104/pp.123.4.1561
Mundt, C. C. (2018). Pyramiding for resistance durability: Theory and practice. Phytopathol. 108 (7), 792–802. doi: 10.1094/phyto-12-17-0426-rvw
Muskett, P., Parker, J. (2003). Role of SGT1 in the regulation of plant R gene signalling. Microbes Infect. 5 (11), 969–976. doi: 10.1016/s1286-4579(03)00183-7
Narlikar, G. J., Sundaramoorthy, R., Owen-Hughes, T. (2013). Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154 (3), 490–503. doi: 10.1016/j.cell.2013.07.011
Parlange, F., Roffler, S., Menardo, F., Ben-David, R., Bourras, S., McNally, K. E., et al. (2015). Genetic and molecular characterization of a locus involved in avirulence of Blumeria graminis f. sp. tritici on wheat Pm3 resistance alleles. Fungal Genet. Biol. 82, 181–192. doi: 10.1016/j.fgb.2015.06.009
Pazdiora, P. C., da Rosa Dorneles, K., Forcelini, C. A., Del Ponte, E. M., Dallagnol, L. J. (2018). Silicon suppresses tan spot development on wheat infected by Pyrenophora tritici-repentis. Europ. J. Plant Pathol. 150 (1), 49–56. doi: 10.1007/s10658-017-1251-4
Pazdiora, P. C., da Rosa Dorneles, K., Morello, T. N., Nicholson, P., Dallagnol, L. J. (2021). Silicon soil amendment as a complement to manage tan spot and fusarium head blight in wheat. Agron. Sust. Dev. 41 (2), 21. doi: 10.1007/s13593-021-00677-0
Pei, D., Zhang, W., Sun, H., Wei, X., Yue, J., Wang, H. (2014). Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum l.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 33 (10), 1697–1710. doi: 10.1007/s00299-014-1648-x
Pozza, E. A., Pozza, A. A. A., Botelho, D. M., d., S. (2015). Silicon in plant disease control. Rev. Ceres. 62, 323–331. doi: 10.1590/0034-737X201562030013
Praz, C. R., Bourras, S., Zeng, F., Sánchez-Martín, J., Menardo, F., Xue, M., et al. (2017). AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus. New Phytol. 213 (3), 1301–1314. doi: 10.1111/nph.14372
Rahman, A., Wallis, C. M., Uddin, W. (2015). Silicon-induced systemic defense responses in perennial ryegrass against infection by. Magnaporthe oryzae. Phytopathol. 105 (6), 748–757. doi: 10.1094/PHYTO-12-14-0378-R
Ramirez-Prado, J. S., Piquerez, S. J. M., Bendahmane, A., Hirt, H., Raynaud, C., Benhamed, M. (2018). Modify the histone to win the battle: chromatin dynamics in plant–pathogen interactions. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00355
Ramos-Cruz, D., Troyee, A. N., Becker, C. (2021). Epigenetics in plant organismic interactions. Curr. Opin. Plant Biol. 61, 102060. doi: 10.1016/j.pbi.2021.102060
Rémus-Borel, W., Menzies, J. G., Bélanger, R. R. (2005). Silicon induces antifungal compounds in powdery mildew-infected wheat. Physiol. Mol. Plant Pathol. 66 (3), 108–115. doi: 10.1016/j.pmpp.2005.05.006
Sánchez-Martín, J., Steuernagel, B., Ghosh, S., Herren, G., Hurni, S., Adamski, N., et al. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17 (1), 1–7. doi: 10.1186/s13059-016-1082-1
Sánchez-Martín, J., Widrig, V., Herren, G., Wicker, T., Zbinden, H., Gronnier, J., et al. (2021). Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants. 7 (3), 327–341. doi: 10.1038/s41477-021-00869-2
Sharma, N., Gautam, A. (2019). Early pathogenicity events in plant pathogenic fungi: A comprehensive review. Biol. Forum Int. J. 11, 24–34.
Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Bariana, H., Bansal, U., McCallum, B., et al. (2012). “Lr34/Yr18/Sr57/Pm38/Bdv1/Ltn1 confers slow rusting, adult plant resistance to puccinia graminis tritici,” in Proceedings of the 13th international cereal rusts and powdery mildews conference, Beijing, China. 173.
Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Lan, C. X., Basnet, B. R., Bhavani, S., et al. (2013). “Pleiotropic gene Lr46/Yr29/Pm39/Ltn2 confers slow rusting, adult plant resistance to wheat stem rust fungus,” in Proceedings BGRI 2013 Technical Workshop, New Delhi, India. 19–22.
Singh, S. P., Hurni, S., Ruinelli, M., Brunner, S., Sanchez-Martin, J., Krukowski, P., et al. (2018). Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol. Biol. 98 (3), 249–260. doi: 10.1007/s11103-018-0780-3
Srichumpa, P., Brunner, S., Keller, B., Yahiaoui, N. (2005). Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 139 (2), 885–895. doi: 10.1104/pp.105.062406
Stael, S., Kmiecik, P., Willems, P., van der Kelen, K., Coll, N. S., Teige, M., et al. (2015). Plant innate immunity–sunny side up? Trends Plant Sci. 20 (1), 3–11. doi: 10.1016/j.tplants.2014.10.002
Te Beest, D. E., Paveley, N. D., Shaw, M. W., van den Bosch, F. (2008). Disease-weather relationships for powdery mildew and yellow rust on winter wheat. Phytopathol. 98 (5), 609–617. doi: 10.1094/phyto-98-5-0609
Turrà, D., Segorbe, D., Di Pietro, A. (2014). Protein kinases in plant-pathogenic fungi: conserved regulators of infection. Annu. Rev. Phytopathol. 52, 267–288. doi: 10.1146/annurev-phyto-102313-050143
Valluru, R., Van den Ende, W. (2011). Myo-inositol and beyond–emerging networks under stress. Plant Sci. 181 (4), 387–400. doi: 10.1016/j.plantsci.2011.07.009
Van Bockhaven, J., Steppe, K., Bauweraerts, I., Kikuchi, S., Asano, T., Höfte, M., et al. (2015). Primary metabolism plays a central role in moulding silicon-inducible brown spot resistance in rice. Mol. Plant Pathol. 16 (8), 811–824. doi: 10.1111/mpp.12236
Voigt, C. A., Schäfer, W., Salomon, S. (2006). A comprehensive view on organ-specific callose synthesis in wheat (Triticum aestivum l.): glucan synthase-like gene expression, callose synthase activity, callose quantification and deposition. Plant Physiol. Biochem. 44 (4), 242–247. doi: 10.1016/j.plaphy.2006.05.001
Wang, Z., Cheng, J., Fan, A., Zhao, J., Yu, Z., Li, Y., et al. (2018). LecRK-V, an l-type lectin receptor kinase in Haynaldia villosa, plays positive role in resistance to wheat powdery mildew. Plant Biotechnol. J. 16 (1), 50–62. doi: 10.1111/pbi.12748
Wang, Q., Guo, J., Jin, P., Guo, M., Guo, J., Cheng, P., et al. (2022a). Glutathione s-transferase interactions enhance wheat resistance to powdery mildew but not wheat stripe rust'. Plant Physiol. 190 (2), 1418–1439. doi: 10.1093/plphys/kiac326
Wang, X., Kong, L., Zhi, P., Chang, C. (2020). Update on cuticular wax biosynthesis and its roles in plant disease resistance. Int. J. Mol. Sci. 21 (15), 5514 doi: 10.3390/ijms21155514
Wang, Y., Qiao, L., Bai, J., Wang, P., Duan, W., Yuan, S., et al. (2017). Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum l.). BMC Genomics 18 (1), 1–19. doi: 10.1186/s12864-017-3582-0
Wang, X., Wang, X., Chen, X.-R., Wu, J., Gou, J.-Y., Zhang, M., et al. (2022b). Editorial: Molecular interactions between crops and phytopathogens, volume I: Wheat and maize. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.979855
Wang, B., Xie, C., Xin, M., Song, N., Ni, Z., Sun, Q. (2012). Comparative proteomic analysis of wheat response to powdery mildew infection in wheat Pm30 near-isogenic lines. J. Phytopathol. 160 (5), 229–236. doi: 10.1111/j.1439-0434.2012.01889.x
Wang, X., Zhi, P., Fan, Q., Zhang, M., Chang, C. (2019). Wheat CHD3 protein TaCHR729 regulates the cuticular wax biosynthesis required for stimulating germination of Blumeria graminis f.sp. tritici. J. Exp. Bot. 70 (2), 701–713. doi: 10.1093/jxb/ery377
Wu, Q., Chen, Y., Li, B., Li, J., Zhang, P., Xie, J., et al. (2022). Functional characterization of powdery mildew resistance gene MlIW172, a new Pm60 allele and its allelic variation in wild emmer wheat. J. Genet. Genom. 49 (8), 787–795. doi: 10.1016/j.jgg.2022.01.010
Wu, X. X., Xu, X. F., Ma, X., Chen, R. Z., Li, T. Y., Cao, Y. Y. (2019). Virulence structure and its genetic diversity analyses of Blumeria graminis f. sp. tritici isolates in China. BMC Evol. Biol. 19 (1), 183. doi: 10.1186/s12862-019-1511-3
Xie, J., Guo, G., Wang, Y., Hu, T., Wang, L., Li, J., et al. (2020). A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol. 228 (3), 1011–1026. doi: 10.1111/nph.16762
Xing, L., Hu, P., Liu, J., Witek, K., Zhou, S., Xu, J., et al. (2018). Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 11 (6), 874–878. doi: 10.1016/j.molp.2018.02.013
Xing, L., Qian, C., Cao, A., Li, Y., Jiang, Z., Li, M., et al. (2013). The Hv-SGT1 gene from Haynaldia villosa contributes to resistances towards both biotrophic and hemi-biotrophic pathogens in common wheat (Triticum aestivum l.). PloS One 8 (9), e72571. doi: 10.1371/journal.pone.0072571
Xin, M., Wang, X., Peng, H., Yao, Y., Xie, C., Han, Y., et al. (2012). Transcriptome comparison of susceptible and resistant wheat in response to powdery mildew infection. Genom. Proteom. Bioinform. 10 (2), 94–106. doi: 10.1016/j.gpb.2012.05.002
Xu, Q., Truong, T. T., Barrero, J. M., Jacobsen, J. V., Hocart, C. H., Gubler, F. (2016). A role for jasmonates in the release of dormancy by cold stratification in wheat. J. Exp. Bot. 67 (11), 3497–3508. doi: 10.1093/jxb/erw172
Yadav, B., Kaur, V., Narayan, O. P., Yadav, S. K., Kumar, A., Wankhede, D. P. (2022). Integrated omics approaches for flax improvement under abiotic and biotic stress: Current status and future prospects. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.931275
Yahiaoui, N., Brunner, S., Keller, B. (2006). Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J. 47 (1), 85–98. doi: 10.1111/j.1365-313X.2006.02772.x
Yahiaoui, N., Kaur, N., Keller, B. (2009). Independent evolution of functional Pm3 resistance genes in wild tetraploid wheat and domesticated bread wheat. Plant J. 57 (5), 846–856. doi: 10.1111/j.1365-313X.2008.03731.x
Yahiaoui, N., Srichumpa, P., Dudler, R., Keller, B. (2004). Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 37 (4), 528–538. doi: 10.1046/j.1365-313x.2003.01977.x
Yue, J., Sun, H., Zhang, W., Pei, D., He, Y., Wang, H. (2015). Wheat homologs of yeast ATG6 function in autophagy and are implicated in powdery mildew immunity. BMC Plant Biol. 15, 95. doi: 10.1186/s12870-015-0472-y
Yu, A., Lepère, G., Jay, F., Wang, J., Bapaume, L., Wang, Y., et al. (2013). Dynamics and biological relevance of DNA demethylation in arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. U S A. 110 (6), 2389–2394. doi: 10.1073/pnas.1211757110
Zeller, F. J. (1973). 1B/1R wheat-rye chromosome substitutions and translocations. Columbia, USA. Univ. Missouri., 209–221.
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91 (4), 714–724. doi: 10.1111/tpj.13599
Zhang, X., Wang, G., Qu, X., Wang, M., Guo, H., Zhang, L., et al. (2022). A truncated CC-NB-ARC gene TaRPP13L1-3D positively regulates powdery mildew resistance in wheat via the RanGAP-WPP complex-mediated nucleocytoplasmic shuttle. Planta. 255 (3), 60. doi: 10.1007/s00425-022-03843-0
Zhang, H., Yang, Y., Wang, C., Liu, M., Li, H., Fu, Y., et al. (2014). Large-Scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genomics 15 (1), 898. doi: 10.1186/1471-2164-15-898
Zhang, J., Zhang, P., Dodds, P., Lagudah, E. (2020). How target-sequence enrichment and sequencing (TEnSeq) pipelines have catalyzed resistance gene cloning in the wheat-rust pathosystem. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00678
Zhang, D., Zhu, K., Dong, L., Liang, Y., Li, G., Fang, T., et al. (2019). Wheat powdery mildew resistance gene Pm64 derived from wild emmer (Triticum turgidum var. dicoccoides) is tightly linked in repulsion with stripe rust resistance gene Yr5. Crop J. 7 (6), 761–770. doi: 10.1016/j.cj.2019.03.003
Zhao, Y., Dong, W., Zhang, N., Ai, X., Wang, M., Huang, Z., et al. (2014). A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 164 (2), 1068–1076. doi: 10.1104/pp.113.227595
Zhi, P., Kong, L., Liu, J., Zhang, X., Wang, X., Li, H., et al. (2020). Histone deacetylase TaHDT701 functions in TaHDA6-TaHOS15 complex to regulate wheat defense responses to Blumeria graminis f.sp. tritici. Int. J. Mol. Sci. 21 (7), 2640 doi: 10.3390/ijms21072640
Zhou, W., Qian, C., Li, R., Zhou, S., Zhang, R., Xiao, J., et al. (2018). TaNAC6s are involved in the basal and broad-spectrum resistance to powdery mildew in wheat. Plant Sci. 277, 218–228. doi: 10.1016/j.plantsci.2018.09.014
Zhu, S., Liu, C., Gong, S., Chen, Z., Chen, R., Liu, T., et al. (2022). Orthologous genes Pm12 and Pm21 from two wild relatives of wheat show evolutionary conservation but divergent powdery mildew resistance. Plant Commun. 8, 100472. doi: 10.1016/j.xplc.2022.100472
Zhu, X., Yang, K., Wei, X., Zhang, Q., Rong, W., Du, L., et al. (2015). The wheat AGC kinase TaAGC1 is a positive contributor to host resistance to the necrotrophic pathogen Rhizoctonia cerealis. J. Exp. Bot. 66 (21), 6591–6603. doi: 10.1093/jxb/erv367
Zou, B., Ding, Y., Liu, H., Hua, J. (2018a). Silencing of copine genes confers common wheat enhanced resistance to powdery mildew. Mol. Plant Pathol. 19 (6), 1343–1352. doi: 10.1111/mpp.12617
Zou, S., Shi, W., Ji, J., Wang, H., Tang, Y., Yu, D., et al. (2022). Diversity and similarity of wheat powdery mildew resistance among three allelic functional genes at the Pm60 locus. Plant J. 110 (6), 1781–1790. doi: 10.1111/tpj.15771
Keywords: wheat, powdery mildew, Blumeria graminis f. sp. tritici, disease resistance, resistance genes
Citation: Mapuranga J, Chang J and Yang W (2022) Combating powdery mildew: Advances in molecular interactions between Blumeria graminis f. sp. tritici and wheat. Front. Plant Sci. 13:1102908. doi: 10.3389/fpls.2022.1102908
Received: 19 November 2022; Accepted: 07 December 2022;
Published: 16 December 2022.
Edited by:
Hongjie Feng, National Key Laboratory of Cotton Biology (CAAS), ChinaReviewed by:
Hong Zhang, Northwest A&F University, ChinaCopyright © 2022 Mapuranga, Chang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxiang Yang, d2VueGlhbmd5YW5nMjAwM0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.