- College of Resources, Sichuan Agricultural University, Chengdu, China
Nitrogen is one of the important nutrients required for plant growth and development. There is increasing evidences that almost all types of nitrogen metabolites affect, at least to some extent, auxin content and/or signaling in plants, which in turn affects seed germination, plant root elongation, gravitropism, leaf expansion and floral transition. This opinion focuses on the roles of nitrogen metabolites, , , tryptophan and NO and their synergistic effects with auxin on plant growth and development. Nitrate reductase (NR) converts nitrate into nitrite, and was roughly positive-correlated with the root auxin level, suggesting a crosstalk between nitrate signaling and auxin signaling. Abscisic Acid Responsive Element Binding Factor 3 (AFB3) and Tryptophan Aminotransferase of Arabidopsis 1 (TAA1) are also the key enzymes involved in nitrogen metabolite-regulated auxin biosynthesis. Recent advances in the crosstalk among , , tryptophan and NO in regulation to NR, AFB3 and TAA1 are also summarized.
Introduction
Nitrogen is one of the important nutrients required by plant growth and development. Plant roots can access nitrogen (N) in various forms which include organic compounds. The primary N forms root absorbs are ammonium (), nitrate (), and amino acids. Typically, the most plentiful source of N is nitrate (Miller et al., 2007; Kant et al., 2011).
The cytosolic enzyme nitrate reductase (NR) first converts nitrate inside the cell to nitrite, which is a rate-limiting step in the assimilation pathway. The nitrite is transported into the chloroplast (Fernandez and Galvan, 2007). Plastid nitrite reductase (NiR) catalyzes the conversion of nitrite to ammonium, which is then absorbed into carbon skeletons by producing glutamate via the glutamine synthetase/glutamine oxoglutarate aminotransferase (GS/GOGAT) cycle (Sanz-Luque et al., 2015). As a result, nitrate assimilation occurs via a rather straightforward linear process that includes two transport phases (nitrate and nitrite transport) and two reduction steps (involving NR and NiR) (Chamizo-Ampudia et al., 2017).
Roles of on plant growth and development
works as a signaling molecule to influence plant growth and development, as well as serving as a main N source for plants (Miller et al., 2007). This leads to a theory that plant cells needed an availability sensor for . Local availability regulates the expression of nitrate assimilation genes (Krapp et al., 2014), breaks seed dormancy (Alboresi et al., 2005; Liu et al., 2021), controls leaf morphogenesis (Yang et al., 2022), stimulates the formation and extension of lateral roots (Forde and Walch-Liu, 2009; Chen et al., 2018; Contreras-López et al., 2022) and postpones flowering (Yuan et al., 2016; Sanagi et al., 2021; Ye et al., 2021; Zhang S. et al., 2021). Lack of nitrate also affects tomato fruit yield and quality (Belfry et al., 2017) and maize stem internodes development (Peng et al., 2013).
The nitrate transporter NRT1.1 is a dual-affinity nitrate transceptor controlling the primary nitrate responses (nitrate signaling), in which expressions of nitrate assimilation genes and nitrate transporter genes are induced rapidly by nitrate treatments (Ho et al., 2009). NRT1.1 facilitates not only nitrate uptake but also auxin transport. Nitrate treatments repress NRT1.1-mediated auxin uptake, indicating that the nitrate signaling via NRT1.1 is correlated with a regulation of auxin transport (Krouk et al., 2010). Another report found that expression of Abscisic Acid Responsive Element Binding Factor 3 (AFB3) depends on the nitrate-transport function of NRT1.1 (Vidal et al., 2014).
Recently, the NIN-like protein (NLP) transcription factor NLP7 has also been suggested as a nitrate sensor (Liu et al., 2022). NLP7 is a crucial nitrate signaling regulator that binds directly to the TAR2 (Tryptophan Aminotransferase Related 2) promoter and activates its expression to sustain auxin signaling in the root primordia (Liu et al., 2017; Zhang T. T, et al., 2021).
Nitrate inhibits Ferredoxin-NADP+-Oxidoreductase (FNR1) expression, therefore causing declines in NADPH/NADP+ and ATP/AMP ratios, which in-turn promotes adenosine monophosphate-activated protein kinase (AMPK) activities and modulates their nuclear abundance (Yuan et al., 2016). KIN10 phosphorylates NLP7 to induce its cytoplasmic retention and the subsequent degradation, therefore repressing nitrate-regulated gene expression and inhibiting growth (Wang et al., 2021).
The nia1/nia2 (nitrate reductase) double mutant showed significantly low transcription levels of auxin biosynthesis/signaling genes and was insensitive to nitrogen changes. NR activity was roughly positive-correlated with the root auxin content, and there should be a crosstalk between nitrate signaling and auxin accumulation (Fu et al., 2020).
Another interesting study indicated that in the nia1/nia2 double mutant, the auxin signaling gene AFB3 expression was increased by the nitrate with the max level at 1 hour, as well as in the wild-type seedlings. Nevertheless contrasting to the wild-type seedlings, the AFB3 mRNA did not decrease in the nia1/nia2 double mutant after 1 hour (Vidal et al., 2010). The authors concluded that NR may regulate both auxin biosynthesis and auxin signaling, and some nitrate metabolite downstream of NR may control AFB3 expression indirectly (Vidal et al., 2010; Vidal et al., 2013).
Besides auxin, nitrate also regulates plant growth and development indirectly through interacting with cytokinin (Hu et al., 2020), ethylene (Zhou et al., 2022), abscisic acid (ABA; Sun et al., 2020), salicylic acid, gibberellins and brassinosteroids (Vega et al., 2019) (Figure 1).
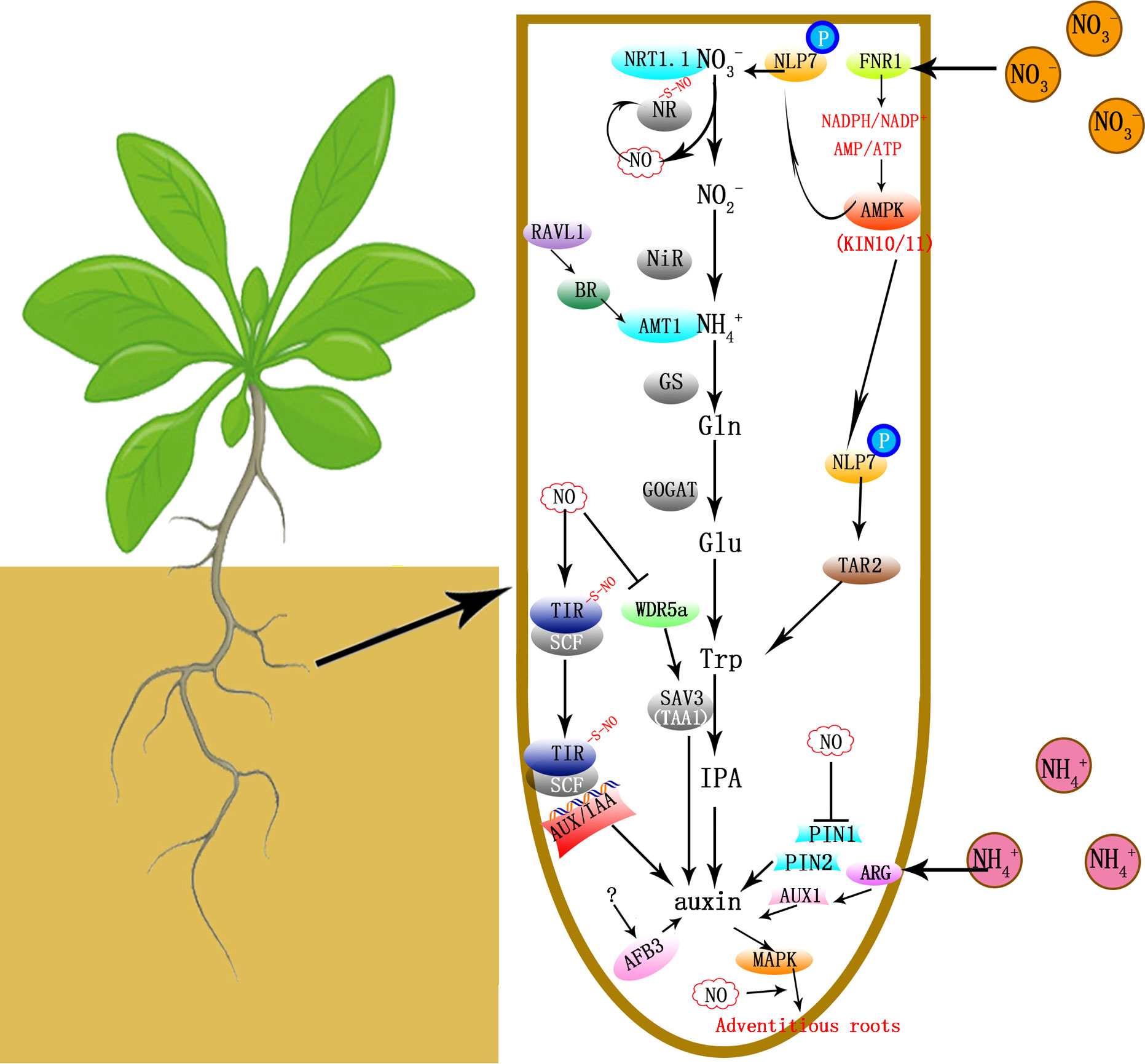
Figure 1 Metabolic pathways and signaling pathways that , , tryptophan and NO regulate auxin biosynthesis/signaling and plant growth/development. Nitrate reductase (NR) converts nitrate to nitrite. Then nitrite reductase (NiR) catalyzes the conversion of nitrite to ammonium, which is then converted to glutamate (Glu) via the glutamine (Gln) synthetase/glutamine oxoglutarate aminotransferase (GS/GOGAT) cycle and various amino acids, among which tryptophan is a key metabolite and the precursor of auxin. The nitrate transporter NRT1.1 is a nitrate transceptor controlling Abscisic Acid Responsive Element Binding Factor 3 (AFB3) expression. Nitrate inhibits Ferredoxin-NADP+-Oxidoreductase (FNR1) expression, therefore causing declines in NADPH/NADP+ and ATP/AMP ratios, which in-turn promotes adenosine monophosphate-activated protein kinase (AMPK; KIN10 and KIN11) activities and phosphorylates the NIN-like protein (NLP) transcription factor NLP7. NLP7 binds directly to the TAR2 (Tryptophan Aminotransferase Related 2) promoter and activates auxin signaling. Some nitrate metabolite downstream of NR might affect AFB3 transcription indirectly. NO is a by-product during NR functioning, however NO may deactivate NR by S-nitrosylation. When is applied to shoots, ARG1 (Altered Response to Gravity 1) induces the auxin influx carrier AUX1 expression and basipetal auxin transport via PIN-Formed2 (PIN2) in root apices. And mediated stresses were lessened when ABA signaling was activated (Li et al., 2012). And the brassinosteroid (BR) signaling transcription factor ABI3/VP1-Like 1 (RAVL1) controls BR-mediated activation of the ammonium transporter AMT1;2 and absorption. The Tryptophan Aminotransferase of Arabidopsis 1 (TAA1; SAV3) catalyzes the synthesis of indole-3-pyruvic acid (IPA) from Trp and controls hypocotyl elongation and leaf expansion responsive to N changes. NO inhibits WD40-REPEAT 5a (WDR5a), which induces TAA1 (SAV3) expression. NO also increases the auxin receptor Transport Inhibitor Response 1 (TIR1) - the transcriptional repressor Auxin/Indole-3-Acetic Acid (Aux/IAA) protein interaction, which facilities the E3-ubiquitin ligase complex SFC-mediated AUX/IAA degradation and enhances the expression of auxin-regulated genes. The mitogen-activated protein kinase (MAPK) signaling cascade is activated during the adventitious root formation induced by auxin in a NO-mediated pathway. NO also inhibites acropetal auxin transport by lowering the abundance of PIN1.
Roles of on plant growth and development
Numerous studies have shown that the early genomic responses of rice and Arabidopsis to exogenous result in a variety of distinct alterations in gene expression, metabolism, hormone signaling, redox state, and root system architecture (Lima et al., 2010; Patterson et al., 2010; Fernández-Crespo et al., 2015; Xuan et al., 2017; Xuan et al., 2019; Hachiya et al., 2021; Sun et al., 2021). Since many of these responses are not correlated with assimilation rate directly, has been suggested also as a signaling molecule; while the ammonium transporter AMT1 may serve as a sensor (Sonoda et al., 2003; Gaur et al., 2012). For example, AMT1;3 is necessary for -dependent lateral root branching in Arabidopsis (Lima et al., 2010).
The mediated suppression of root development is compromised in the auxin-resistant mutants aux1, axr1, and axr2 (Cao et al., 1993). And the auxin influx carrier AUX1 inhibits lateral root emergence when is applied to shoots (Li et al., 2011). The suppression of lateral root growth by is related with ethylene generation in shoots (Li et al., 2013). Zou et al. (2013) interestingly found that, under NH4+ stress, arg1 (Altered Response to Gravity 1) mutant displayed increased loss of root gravitropism. ARG1 is required for AUX1 protein expression and basipetal auxin transport via PIN-Formed2 (PIN2) in root apices. And mediated stresses were lessened when ABA signaling was activated (Li et al., 2012). Recently, it was discovered that an important brassinosteroid (BR) signaling transcription factor ABI3/VP1-Like 1 (RAVL1) controls BR-mediated activation of AMT1;2 and absorption in rice (Xuan et al., 2017) (Figure 1).
Roles of nitrogen metabolite tryptophan on plant growth and development
Nitrogen in plants first assimilates ammonia to glutamate, which is then converted to various amino acids, among which tryptophan is a key metabolite and the precursor of auxin. Given that auxin plays a key role in plant growth and development, we focused on tryptophan (Trp) and auxin metabolism and signaling in this review. There are two auxin biosynthesis pathways, the Trp-dependent and the Trp-independent pathways; while the tryptophan-dependent pathway is the main one (Korasick et al., 2013; Ljung, 2013). Auxin is a potential mediator of N signaling that auxin content is inversely associated with N status in various plants (Tian et al., 2008; Kiba et al., 2011; Ma et al., 2014). Furthermore, jasmonic acid (JA) locally produced in response to mechanical wounding triggers the de novo formation of auxin through the induction of Trp-dependent pathways (Zhang G. et al., 2019; Pérez-Alonso et al., 2021).
A research revealed that tryptophan’s role as an auxin precursor on root elongation is rather straightforward (Jing et al., 2009). Besides the promotion on auxin biosynthesis, exogenous tryptophan increased root length and plant height and improved plant resistance to stresses by enhancing C/N metabolism and related enzyme activities (Mustafa et al., 2018).
After N treatments, plant leaves become thicker and narrower, and the chlorophyll level increases. Our previous study found that the changes in leaf thickness and width were largely compromised in the shade avoidance 3 (sav3) mutant (Yang et al., 2022). The SAV3 protein catalyzes the synthesis of indole-3-pyruvic acid (IPA) from Trp, and is also named as Tryptophan Aminotransferase of Arabidopsis 1 (TAA1). SAV3 also controls hypocotyl elongation and leaf expansion under the shade condition (Tao et al., 2008), and regulates chlorophyll accumulation and nitrogen assimilation. Therefore SAV3 works as a master switch responsive to multiple environmental stimuli (Yang et al., 2022) (Figure 1).
NO and auxin synergistically regulate plant growth and development
Although direct nitric oxide synthase (NOS) has not been found in higher plants, it has been suggested that NR’s main role is to provide nitrite, which in turn can be further reduced to NO. In other words, NO is a by-product during nitrate assimilation (Chamizo-Ampudia et al., 2017).
Our previous study indicated that the NR protein can be S-nitrosated by NO. The S-nitrosylation status of NR is negatively correlated with its enzymatic activity. Thus NO generated through NR catalysis may deactivate the enzyme itself by this S-nitrosylation-dependent negative-feedback regulation (Fu et al., 2018).
Nitric oxide (NO) is a multi-purpose gaseous signaling molecule (Domingos et al., 2015; Simontacchi et al., 2015). NO and auxin interact with each other in controlling root development (Correa-Aragunde et al., 2004; Fernández-Marcos et al., 2011; Jin et al., 2011; Chen and Kao, 2012; Sun et al., 2017; Basu et al., 2021) and root hairs formation (Lombardo et al., 2006). The mitogen-activated protein kinase (MAPK) signaling cascade is activated during the adventitious root formation induced by auxin in a NO-mediated but cGMP-independent pathway. The stimulation of MAPK has been proposed in modulating mitotic processes in root cells (Pagnussat et al., 2004; Hu et al., 2005).
Some other studies suggested that NO may also function in signaling pathways upstream of auxin (Terrile et al., 2012; Liu et al., 2018). NO lowers the level of auxin in the root apex by inhibiting WD40-REPEAT 5a (WDR5a), which induces TAA1 (SAV3) and auxin accumulation (Liu et al., 2018). But NO increases the auxin receptor Transport Inhibitor Response 1 (TIR1) and the transcriptional repressor Auxin/Indole-3-Acetic Acid (Aux/IAA) protein interaction via S-nitrosylation on TIR1, which enhances the expression of auxin-regulated genes in the whole root (Terrile et al., 2012).
Additionally, NO-overproducing mutants and pharmacological treatments showed that, at high concentrations, NO inhibited acropetal auxin transport in Arabidopsis roots by lowering the abundance of the auxin efflux protein PIN1 through a proteasome-independent post-transcriptional mechanism (Fernández-Marcos et al., 2011).
Our previous study demonstrated that NO dramatically decreased monosaccharide catabolism by inhibiting sugar metabolic enzymes via S-nitrosylation. As a result, NO treatments reduced starch granule formation in root tips and compromised root gravitropism indirectly (Zhang et al., 2017).
Besides these putative mechanisms of NO on auxin transport and signaling, NO also regulates plant growth and development indirectly through interacting with ethylene (Du et al., 2014), cytokinin (Feng et al., 2013; Liu et al., 2013), ABA (Sang et al., 2008; Wang et al., 2015), gibberellin and light signaling (Bai et al., 2014).
Both nitrogen and NO treatments postpone plant flowering (He et al., 2004; Yuan et al., 2016). However, the high nitrogen condition reduced the amplitudes of transcripts of all circadian clock genes (Yuan et al., 2016). While NO enhanced the amplitudes of central oscillators, but reduced the amplitudes of circadian-clock output genes, GI (GIGANTEA) and CO (CONSTANS). NO induced S-nitrosation modification on GI and CO proteins, but not on the other circadian clock proteins (Zhang Z. W. et al., 2019). Thus nitrogen and NO rely on overlapping but different signaling pathways to regulate plant flowering (Figure 1).
Conclusions and perspectives
Nitrogen and its metabolites regulate plant growth and development through multiple and complex mechanisms. Nitrate assimilation metabolites, , , tryptophan and the by-product NO, as well as the key enzymes NR and TAA1 are all involved, with interacting with phytohormone signals. Interestingly, NO may deactivate NR by S-nitrosylation (Fu et al., 2018). Whether NO also generates a feedback regulation on TAA1 requires further studies.
It is interesting to note that the AFB3 transcript did not decrease in the NR-deficient mutant after 1 hour of nitrate treatment. Thus some nitrate metabolite downstream of NR might affect AFB3 transcription indirectly (Vidal et al., 2010; Vidal et al., 2013). Which metabolite plays the key role needs further investigations. Both nitrate and NO repress auxin accumulation by decreasing TAA1 expression (Liu et al., 2018; Yang et al., 2022). Whether TAA1 works upstream of AFB3 requires further explorations. And we also don’t know whether TAA1 activity is associated with the cellular tryptophan level.
Both ammonium poisoning and NO accumulation increase loss of root gravitropism and inhibit root elongation (Zou et al., 2013; Zhang et al., 2017). The crosstalk between signaling and NO metabolism in root morphogenesis would also be an interesting research direction.
Author contributions
SY conceived the project. Y-FF, X-YY, and Z-WZ. performed the literature search. Y-FF and SY wrote the manuscript with input from X-YY and Z-WZ. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Sichuan Province Youth Science and Technology Innovation Team (20CXTD0062) to SY and the Applied Basic Research Program of Sichuan Province (2020YJ0410) to Z-WZ.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alboresi, A., Gestin, C., Leydecker, M. T., Bedu, M., Meyer, C., Truong, H. N. (2005). Nitrate, a signal relieving seed dormancy in arabidopsis. Plant Cell Environ. 28, 500–512. doi: 10.1111/j.1365-3040.2005.01292.x
Bai, S., Yao, T., Li, M., Guo, X., Zhang, Y., Zhu, S., et al. (2014). PIF3 is involved in the primary root growth inhibition of arabidopsis induced by nitric oxide in the light. Mol. Plant 7, 616–625. doi: 10.1093/mp/sst142
Basu, S., Kumari, S., Kumar, A., Shahid, R., Kumar, S., Kumar, G. (2021). Nitro-oxidative stress induces the formation of roots' cortical aerenchyma in rice under osmotic stress. Physiol. Plant 172, 963–975. doi: 10.1111/ppl.13415
Belfry, K. D., Trueman, C., Vyn, R. J., Loewen, S. A., Van Eerd, L. L. (2017). Winter cover crops on processing tomato yield, quality, pest pressure, nitrogen availability, and profit margins. PloS One 12, e0180500. doi: 10.1371/journal.pone.0180500
Cao, Y., Glass, A. D. M., Crawford, N. M. (1993). Ammonium inhibition of arabidopsis root growth can be reversed by potassium and by auxin resistance mutations aux1, axr1, and axr2. Plant Physiol. 102, 983–989. doi: 10.1104/pp.102.3.983
Chamizo-Ampudia, A., Sanz-Luque, E., Llamas, A., Galvan, A., Fernandez, E. (2017). Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 22, 163–174. doi: 10.1016/j.tplants.2016.12.001
Chen, Y. H., Kao, C. H. (2012). Calcium is involved in nitric oxide- and auxin-induced lateral root formation in rice. Protoplasma 249, 187–195. doi: 10.1007/s00709-011-0277-2
Chen, H., Zhang, Q., Cai, H., Zhou, W., Xu, F. (2018). H2O2 mediates nitrate-induced iron chlorosis by regulating iron homeostasis in rice. Plant Cell Environ. 41, 767–781. doi: 10.1111/pce.13145
Contreras-López, O., Vidal, E. A., Riveras, E., Alvarez, J. M., Moyano, T. C., Sparks, E. E., et al. (2022). Spatiotemporal analysis identifies ABF2 and ABF3 as key hubs of endodermal response to nitrate. Proc. Natl. Acad. Sci. U.S.A. 119, e2107879119. doi: 10.1073/pnas.2107879119
Correa-Aragunde, N., Graziano, M., Lamattina, L. (2004). Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218, 900–905. doi: 10.1007/s00425-003-1172-7
Domingos, P., Prado, A. M., Wong, A., Gehring, C., Feijo, J. A. (2015). Nitric oxide: A multitasked signaling gas in plants. Mol. Plant 8, 506–520. doi: 10.1016/j.molp.2014.12.010
Du, J., Li, M., Kong, D., Wang, L., Lv, Q., Wang, J., et al. (2014). Nitric oxide induces cotyledon senescence involving co-operation of the NES1/MAD1 and EIN2-associated ORE1 signalling pathways in arabidopsis. J. Exp. Bot. 65, 4051–4063. doi: 10.1093/jxb/ert429
Feng, J., Wang, C., Chen, Q., Chen, H., Ren, B., Li, X., et al. (2013). S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat. Commun. 4, 1529. doi: 10.1038/ncomms2541
Fernández-Crespo, E., Scalschi, L., Llorens, E., García-Agustín, P., Camañes, G. (2015). NH4+ protects tomato plants against Pseudomonas syringae by activation of systemic acquired acclimation. J. Exp. Bot. 66, 6777–6790. doi: 10.1093/jxb/erv382
Fernandez, E., Galvan, A. (2007). Inorganic nitrogen assimilation in Chlamydomonas. J. Exp. Bot. 58, 2279–2287. doi: 10.1093/jxb/erm106
Fernández-Marcos, M., Sanz, L., Lewis, D. R., Muday, G. K., Lorenzo, O. (2011). Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN-FORMED 1 (PIN1)-dependent acropetal auxin transport. Proc. Natl. Acad. Sci. U.S.A. 108, 18506–18511. doi: 10.1073/pnas.1108644108
Forde, B. G., Walch-Liu, P. (2009). Nitrate and glutamate as environmental cues for behavioural responses in plant roots. Plant Cell Environ. 32, 682–693. doi: 10.1111/j.1365-3040.2008.01927.x
Fu, Y. F., Zhang, Z. W., Yang, X. Y., Wang, C. Q., Lan, T., Tang, X. Y., et al. (2020). Nitrate reductase is a key enzyme responsible for nitrogen-regulated auxin accumulation in arabidopsis roots. Biochem. Biophys. Res. Commun. 532 (4), 633–639. doi: 10.1016/j.bbrc.2020.08.057
Fu, Y. F., Zhang, Z. W., Yuan, S. (2018). Putative connections between nitrate reductase s-nitrosylation and NO synthesis under pathogen attacks and abiotic stresses. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00474
Gaur, V. S., Singh, U. S., Gupta, A. K., Kumar, A. (2012). Understanding the differential nitrogen sensing mechanism in rice genotypes through expression analysis of high and low affinity ammonium transporter genes. Mol. Biol. Rep. 39, 2233–2241. doi: 10.1007/s11033-011-0972-2
Hachiya, T., Inaba, J., Wakazaki, M., Sato, M., Toyooka, K., Miyagi, A., et al. (2021). Excessive ammonium assimilation by plastidic glutamine synthetase causes ammonium toxicity in Arabidopsis thaliana. Nat. Commun. 12, 4944. doi: 10.1038/s41467-021-25238-7
He, Y., Tang, R. H., Hao, Y., Stevens, R. D., Cook, C. W., Ahn, S. M., et al. (2004). Nitric oxide represses the arabidopsis floral transition. Science 305, 1968–1971. doi: 10.1126/science.1098837
Ho, C. H., Lin, S. H., Hu, H. C., Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. doi: 10.1016/j.cell.2009.07.004
Hu, X., Neill, S. J., Tang, Z., Cai, W. (2005). Nitric oxide mediates gravitropic bending in soybean roots. Plant Physiol. 137, 663–670. doi: 10.1104/pp.104.054494
Hu, S., Zhang, M., Yang, Y., Xuan, W., Zou, Z., Arkorful, E., et al. (2020). A novel insight into nitrogen and auxin signaling in lateral root formation in tea plant [Camellia sinensis (L.) o. kuntze]. BMC Plant Biol. 20, 232. doi: 10.1186/s12870-020-02448-7
Jin, C. W., Du, S. T., Shamsi, I. H., Luo, B. F., Lin, X. Y. (2011). NO synthase-generated NO acts downstream of auxin in regulating fe-deficiency-induced root branching that enhances fe-deficiency tolerance in tomato plants. J. Exp. Bot. 62, 3875–3884. doi: 10.1093/jxb/err078
Jing, Y., Cui, D., Bao, F., Hu, Z., Qin, Z., Hu, Y. (2009). Tryptophan deficiency affects organ growth by retarding cell expansion in arabidopsis. Plant J. 57, 511–521. doi: 10.1111/j.1365-313X.2008.03706.x
Kant, S., Bi, Y.-M., Rothstein, S. J. (2011). Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 62, 1499–1509. doi: 10.1093/jxb/erq297
Kiba, T., Kudo, T., Kojima, M., Sakakibara, H. (2011). Hormonal control of nitrogen acquisition: Roles of auxin, abscisic acid, and cytokinin. J. Exp. Bot. 62, 1399–1409. doi: 10.1093/jxb/erq410
Korasick, D. A., Enders, T. A., Strader, L. C. (2013). Auxin biosynthesis and storage forms. J. Exp. Bot. 64, 2541–2555. doi: 10.1093/jxb/ert080
Krapp, A., David, L. C., Chardin, C., Girin, T., Marmagne, A., Leprince, A. S., et al. (2014). Nitrate transport and signalling in arabidopsis. J. Exp. Bot. 65, 789–798. doi: 10.1093/jxb/eru001
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937. doi: 10.1016/j.devcel.2010.05.008
Li, G., Li, B., Dong, G., Feng, X., Kronzucker, H. J., Shi, W. (2013). Ammonium-induced shoot ethylene production is associated with the inhibition of lateral root formation in arabidopsis. J. Exp. Bot. 64, 1413–1425. doi: 10.1093/jxb/ert019
Li, B., Li, Q., Su, Y., Chen, H., Xiong, L., Mi, G., et al. (2011). Shoot-supplied ammonium targets the root auxin influx carrier AUX1 and inhibits lateral root emergence in arabidopsis. Plant Cell Environ. 34, 933–946. doi: 10.1111/j.1365-3040.2011.02295.x
Li, B., Li, Q., Xiong, L., Kronzucker, H. J., Krämer, U., Shi, W. (2012). Arabidopsis plastid AMOS1/EGY1 integrates abscisic acid signaling to regulate global gene expression response to ammonium stress. Plant Physiol. 160, 2040–2051. doi: 10.1104/pp.112.206508
Lima, J. E., Kojima, S., Takahashi, H., von Wirén, N. (2010). Ammonium triggers lateral root branching in arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 22, 3621–3633. doi: 10.1105/tpc.110.076216
Liu, W. Z., Kong, D. D., Gu, X. X., Gao, H. B., Wang, J. Z., Xia, M., et al. (2013). Cytokinins can act as suppressors of nitric oxide in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 1548–1553. doi: 10.1073/pnas.1213235110
Liu, K. H., Liu, M., Lin, Z., Wang, Z. F., Chen, B., Liu, C., et al. (2022). NIN-like protein 7 transcription factor is a plant nitrate sensor. Science 377, 1419–1425. doi: 10.1126/science.add1104
Liu, K. H., Niu, Y., Konishi, M., Wu, Y., Du, H., Sun Chung, H., et al. (2017). Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545, 311–316. doi: 10.1038/nature22077
Liu, M. Y., Tang, D., Shi, Y., Ma, L., Zhang, Q., Ruan, J. (2021). Foliar n application on tea plant at its dormancy stage increases the n concentration of mature leaves and improves the quality and yield of spring tea. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.753086
Liu, W.-C., Zheng, S.-Q., Yu, Z.-D., Gao, X., Shen, R., Lu, Y.-T. (2018). WD40-REPEAT 5a represses root meristem growth by suppressing auxin synthesis through changes of nitric oxide accumulation in arabidopsis. Plant J. 93, 883–893. doi: 10.1111/tpj.13816
Ljung, K. (2013). Auxin metabolism and homeostasis during plant development. Development 140, 943–950. doi: 10.1242/dev.086363
Lombardo, M. C., Graziano, M., Polacco, J. C., Lamattina, L. (2006). Nitric oxide functions as a positive regulator of root hair development. Plant Signal. Behav. 1, 28–33. doi: 10.4161/psb.1.1.2398
Ma, W., Li, J., Qu, B., He, X., Zhao, X., Li, B., et al. (2014). Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in arabidopsis. Plant J. 78, 70–79. doi: 10.1111/tpj.12448
Miller, A. J., Fan, X., Orsel, M., Smith, S. J., Wells, D. M. (2007). Nitrate transport and signalling. J. Exp. Bot. 58, 2297–2306. doi: 10.1093/jxb/erm066
Mustafa, A., Imran, M., Ashraf, M., Mahmood, K. (2018). Perspectives of using l-tryptophan for improving productivity of agricultural crops: A review. Pedosphere 28, 16–34. doi: 10.1016/S1002-0160(18)60002-5
Pagnussat, G. C., Lanteri, M. L., Lombardo, M. C., Lamattina, L. (2004). Nitric oxide mediates the indole acetic acid induction activation of a mitogen-activated protein kinase cascade involved in adventitious root development. Plant Physiol. 135, 279–286. doi: 10.1104/pp.103.038554
Patterson, K., Cakmak, T., Cooper, A., Lager, I., Rasmusson, A. G., Escobar, M. A. (2010). Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ. 33, 1486–1501. doi: 10.1111/j.1365-3040.2010.02158.x
Peng, Y., Li, C., Fritschi, F. B. (2013). Apoplastic infusion of sucrose into stem internodes during female flowering does not increase grain yield in maize plants grown under nitrogen-limiting conditions. Physiol. Plant 148, 470–480. doi: 10.1111/j.1399-3054.2012.01711.x
Pérez-Alonso, M. M., Sánchez-Parra, B., Ortiz-García, P., Santamaría, M. E., Díaz, I., Pollmann, S. (2021). Jasmonic acid-dependent MYC transcription factors bind to a tandem G-box motif in the YUCCA8 and YUCCA9 promoters to regulate biotic stress responses. Int. J. Mol. Sci. 22, 9768. doi: 10.3390/ijms22189768
Sanagi, M., Aoyama, S., Kubo, A., Lu, Y., Sato, Y., Ito, S., et al. (2021). Low nitrogen conditions accelerate flowering by modulating the phosphorylation state of FLOWERING BHLH 4 in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118, e2022942118. doi: 10.1073/pnas.2022942118
Sang, J., Zhang, A., Lin, F., Tan, M., Jiang, M. (2008). Cross-talk between calcium-calmodulin and nitric oxide in abscisic acid signaling in leaves of maize plants. Cell Res. 18, 577–588. doi: 10.1038/cr.2008.39
Sanz-Luque, E., Chamizo-Ampudia, A., Llamas, A., Galvan, A., Fernandez, E. (2015). Understanding nitrate assimilation and its regulation in microalgae. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00899
Simontacchi, M., Galatro, A., Ramos-Artuso, F., Santa-María, G. E. (2015). Plant survival in a changing environment: The role of nitric oxide in plant responses to abiotic stress. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00977
Sonoda, Y., Ikeda, A., Saiki, S., Wirén, N., Yamaya, T., Yamaguchi, J. (2003). Distinct expression and function of three ammonium transporter genes (OsAMT1;1 – 1;3) in rice. Plant Cell Physiol. 44, 726–734. doi: 10.1093/pcp/pcg083
Sun, X., Chen, F., Yuan, L., Mi, G. (2020). The physiological mechanism underlying root elongation in response to nitrogen deficiency in crop plants. Planta 251, 84. doi: 10.1007/s00425-020-03376-4
Sun, D., Fang, X., Xiao, C., Ma, Z., Huang, X., Su, J., et al. (2021). Kinase SnRK1.1 regulates nitrate channel SLAH3 engaged in nitrate-dependent alleviation of ammonium toxicity. Plant Physiol. 186, 731–749. doi: 10.1093/plphys/kiab057
Sun, H., Feng, F., Liu, J., Zhao, Q. (2017). The interaction between auxin and nitric oxide regulates root growth in response to iron deficiency in rice. Front. Plant Sci. 8, 2169. doi: 10.3389/fpls.2017.02169
Tao, Y., Ferrer, J., Ljung, K., Pojer, F., Hong, F., Long, J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049
Terrile, M. C., París, R., Calderón-Villalobos, L. I. A., Iglesias, M. J., Lamattina, L., Estelle, M., et al. (2012). Nitric oxide influences auxin signaling through s-nitrosylation of the arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 70, 492–500. doi: 10.1111/j.1365-313X.2011.04885.x
Tian, Q., Chen, F., Liu, J., Zhang, F., Mi, G. (2008). Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J. Plant Physiol. 165, 942–951. doi: 10.1016/j.jplph.2007.02.011
Vega, A., O'Brien, J. A., Gutiérrez, R. A. (2019). Nitrate and hormonal signaling crosstalk for plant growth and development. Curr. Opin. Plant Biol. 52, 155–163. doi: 10.1016/j.pbi.2019.10.001
Vidal, E. A., Álvarez, J. M., Gutiérrez, R. A. (2014). Nitrate regulation of AFB3 and NAC4 gene expression in arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Signal. Behav. 9, e28501. doi: 10.4161/psb.28501
Vidal, E. A., Araus, V., Lu, C., Parry, G., Green, P. J., Coruzzi, G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 4477–4482. doi: 10.1073/pnas.0909571107
Vidal, E. A., Moyano, T. C., Riveras, E., Contreras-López, O., Gutiérrez, R. A. (2013). Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Natl. Acad. Sci. U.S.A. 110, 12840–12845. doi: 10.1073/pnas.1310937110
Wang, P., Du, Y., Hou, Y. J., Zhao, Y., Hsu, C. C., Yuan, F., et al. (2015). Nitric oxide negatively regulates abscisic acid signaling in guard cells by s-nitrosylation of OST1. Proc. Natl. Acad. Sci. U.S.A. 112, 613–618. doi: 10.1073/pnas.1423481112
Wang, H., Han, C., Wang, J. G., Chu, X., Shi, W., Yao, L., et al. (2021). Regulatory functions of cellular energy sensor SnRK1 for nitrate signalling through NLP7 repression. Nat. Plants 8, 1094–1107. doi: 10.1038/s41477-022-01236-5
Xuan, Y. H., Duan, F. Y., Je, B. I., Kim, C. M., Li, T. Y., Liu, J. M., et al. (2017). Related to ABI3/VP1-like 1 (RAVL1) regulates brassinosteroid-mediated activation of AMT1;2 in rice (Oryza sativa). J. Exp. Bot. 68, 727–737. doi: 10.1093/jxb/erw442
Xuan, Y. H., Kumar, V., Han, X., Kim, S. H., Jeong, J. H., Kim, C. M., et al. (2019). CBL-INTERACTING PROTEIN KINASE 9 regulates ammonium-dependent root growth downstream of IDD10 in rice (Oryza sativa). Ann. Bot. 124, 947–960. doi: 10.1093/aob/mcy242
Yang, X. Y., Zhang, Z. W., Fu, Y. F., Feng, L. Y., Li, M. X., Kang, Q., et al. (2022). Shade avoidance 3 mediates crosstalk between shade and nitrogen in arabidopsis leaf development. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.800913
Ye, J. Y., Tian, W. H., Zhou, M., Zhu, Q. Y., Du, W. X., Jin, C. W. (2021). Improved plant nitrate status involves in flowering induction by extended photoperiod. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.629857
Yuan, S., Zhang, Z. W., Zheng, C., Zhao, Z. Y., Wang, Y., Feng, L. Y., et al. (2016). Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proc. Natl. Acad. Sci. U.S.A. 113, 7661–7666. doi: 10.1073/pnas.1602004113
Zhang, Z. W., Fu, Y. F., Zhou, Y. H., Wang, C. Q., Lan, T., Chen, G. D., et al. (2019). Nitrogen and nitric oxide regulate arabidopsis flowering differently. Plant Sci. 284, 177–184. doi: 10.1016/j.plantsci.2019.04.015
Zhang, T. T., Kang, H., Fu, L. L., Sun, W. J., Gao, W. S., You, C. X., et al. (2021). NIN-like protein 7 promotes nitrate-mediated lateral root development by activating transcription of TRYPTOPHAN AMINOTRANSFERASE RELATED 2. Plant Sci. 303, 110771. doi: 10.1016/j.plantsci.2020.110771
Zhang, Z. W., Luo, S., Zhang, G. C., Feng, L. Y., Zheng, C., Zhou, Y. H., et al. (2017). Nitric oxide induces monosaccharide accumulation through enzyme s-nitrosylation. Plant Cell Environ. 40, 1834–1848. doi: 10.1111/pce.12989
Zhang, S., Zhang, Y., Li, K., Yan, M., Zhang, J., Yu, M., et al. (2021). Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice. Curr. Biol. 31, 671–683.e5. doi: 10.1016/j.cub.2020.10.095
Zhang, G., Zhao, F., Chen, L., Pan, Y., Sun, L., Bao, N., et al. (2019). Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 5, 491–497. doi: 10.1038/s41477-019-0408-x
Zhou, Y., Ma, B., Tao, J. J., Yin, C. C., Hu, Y., Huang, Y. H., et al. (2022). Rice EIL1 interacts with OsIAAs to regulate auxin biosynthesis mediated by the tryptophan aminotransferase MHZ10/OsTAR2 during root ethylene responses. Plant Cell 34, 4366–4387. doi: 10.1093/plcell/koac250
Keywords: nitrogen metabolites, ammonium, nitrate reductase, nitric oxide, tryptophan aminotransferase, auxin signaling and transport
Citation: Fu Y-F, Yang X-Y, Zhang Z-W and Yuan S (2022) Synergistic effects of nitrogen metabolites on auxin regulating plant growth and development. Front. Plant Sci. 13:1098787. doi: 10.3389/fpls.2022.1098787
Received: 15 November 2022; Accepted: 07 December 2022;
Published: 20 December 2022.
Edited by:
Dong Meng, Cornell University, United StatesReviewed by:
Manrong Zha, Nanjing Agricultural University, ChinaMengxia Zhang, Cornell University, United States
Copyright © 2022 Fu, Yang, Zhang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu Yuan, cm91bmR0cmVlMzE4QGhvdG1haWwuY29t
 Yu-Fan Fu
Yu-Fan Fu Shu Yuan
Shu Yuan