- 1Guangdong Provincial Key Laboratory of Plant Adaptation and Molecular Design, Guangzhou Key Laboratory of Crop Gene Editing, Innovative Center of Molecular Genetics and Evolution, School of Life Sciences, Guangzhou University, Guangzhou Higher Education Mega Center, Guangzhou, China
- 2Suihua Branch Institute, Heilongjiang Academy of Agricultural Sciences, Suihua, Heilongjiang, China
IQM, a plant-specific calmodulin-binding protein, plays multiple roles in plant growth and development. Although a comprehensive analysis has been carried out on the IQM family genes in Arabidopsis and rice, the number and functions of IQM genes in other species have not been explored. In this study, we identified 15 members of the soybean (Glycine max) IQM gene family using BLASTP tools. These members were distributed on 12 soybean chromosomes and constitute six pairs caused by fragment duplication events. According to phylogeny, the 15 genes were divided into three subfamilies (I, II, and III), and members of the same subfamily had similar gene and protein structures. Yeast two-hybrid experiments revealed that the IQ motif is critical for the binding of GmIQM proteins to GmCaM, and its function is conserved in soybean, Arabidopsis, and rice. Based on real-time PCR, the soybean IQM genes were strongly induced by PEG and NaCl, suggesting their important biological functions in abiotic stress responses. Overall, this genome-wide analysis of the soybean IQM gene family lays a solid theoretical foundation for further research on the functions of GmIQM genes and could serve as a reference for the improvement and breeding of soybean stress resistance traits.
Introduction
Many signaling pathways and complex networks are involved in the response of plant to biotic and abiotic stresses (Saijo and Loo, 2020; Zhang et al., 2022). Ca2+ is an important second messenger that plays an important role in plant adaptation to external stimuli, thereby regulating multiple physiological processes and signaling transduction through Ca2+ sensors and their target proteins (Reddy et al., 2011; Carafoli and Krebs, 2016; Edel et al., 2017). The calcium sensors in higher plants include calmodulin (CaM), CaM-like proteins (CML), calcium-dependent protein kinase (CDPK), and calcineurin B-like proteins (CBL). Among these calcium sensors, CaMs are highly conserved acidic heat-stable proteins. There are seven CaMs in the model plant, Arabidopsis (Arabidopsis thaliana), and their amino acid sequences are highly similar, with only one to five amino acid differences (Williams, 1992; Chin and Means, 2000; DeFalco et al., 2009; Ranty et al., 2016; Aldon et al., 2018). In most cases, a single CaM has no biochemical or enzymatic activity, but acts by binding to Ca2+ and various downstream target proteins called CaM-binding proteins (CaMBPs), including kinases, cytoskeletal proteins, transcription factors, and metabolic enzymes that are involved in plant development, metabolic regulation, defense, and stress responses (Yang and Poovaiah, 2003; Bouché et al, 2005; Truman et al., 2013; Ali et al., 2020).
IQM has been identified as a calcium-independent CaMBP family with an IQ motif in Arabidopsis (Zhou et al., 2010). The IQ motif is the first recognized calcium-independent CaM-binding domain, and its complete amino acid sequence is IQxxxRGxxxR (Rhoads and Friedberg, 1997). In addition to the IQM family, there are four other classes of the IQ motif-containing protein family in plants: the myosin protein family (Reddy and Day, 2001), the calmodulin-binding transcription activator (CAMTA) family (Bouché et al., 2002), the cyclic nucleotide-gated channel (CNGC) family (Talke et al., 2003), and the IQ67-domain containing protein (IQD) family (Abel et al, 2005). Members of these five families differ in the number and distribution of IQ motifs. Compared to other families, only few studies have been performed on the function of the IQM family.
To date, the biological functions of IQM genes have only been investigated in Arabidopsis. There are six IQM members, IQM1 to IQM6, in Arabidopsis. The N-terminal of the typical IQMs has a sequence homologous to the pea heavy-metal induced protein 6A (PHMIP 6A), in which the IQ motif is located, while their C-terminal has a fragment homologous to the ribosome inactivating protein, trichosanthin (Zhou et al., 2010). IQM1 plays an important role in the modulation of stomatal movement by affecting the ROS content (Zhou et al., 2012) and is a key regulator in plant disease defense mediated by JA signaling (Lv et al., 2019). AtIQM5 regulates flowering by modulating juvenile-to-adult transition (Gong et al., 2016) and promotes lateral root and callus formation by interacting with IAAs (Zhang et al., 2022). IQM4 affects seed dormancy and germination by regulating ABA biosynthesis and signaling in seeds (Zhou et al., 2018). Although the partial roles and internal mechanisms of IQMs in Arabidopsis have been revealed in these studies, research on the role of IQM family members in other species is still lacking. Besides Arabidopsis, whole genome analysis of the IQM family genes has only been performed for rice (Fan et al., 2021).
In this study, 15 non-redundant soybean (Glycine max) IQM genes were identified using bioinformatics analysis on a genome-wide scale and molecular biology techniques. The phylogenetic relationships, gene structure, conserved motifs, chromosome location, evolutionary pattern analysis, yeast two-hybrid analysis, and expression profile in response to abiotic stress and hormones of the soybean IQM family were discussed. Our results provide a theoretical basis for subsequent functional analysis of soybean IQM genes and insights for improving soybean resistance to biotic and abiotic stresses.
Materials and methods
Identification of soybean IQM proteins
Six Arabidopsis IQM protein sequences were used as reference, and their homologous sequences in soybean were retrieved from Phytozome v13 database (http://phytozome-next.jgi.doe.gov ) through Protein Basic Local Alignment Search Tool (BLASTP) program. The Pfamscan (https://www.ebi.ac.uk/Tools/pfa/pfamscan/ ) and SMART (http://smart.embl-heidelberg.de/ ) databases were used to annotate the conserved domains of candidate sequences. Finally, proteins containing the complete IQ motif were identified as members of the soybean IQM family. Information on soybean IQM genes, including CDSs, genome sequences, location coordinates, lengths of open reading frames (ORF), number of amino acids, and molecular weight, were acquired from Phytozome v13. The physicochemical characteristics of the GmIQMs were generated using ExPASy (http://web.expasy.org/protparam/ ). Subcellular localization was predicted using the WoLF PSORT program (http://wolfpsort.org ).
Phylogenetic analysis
To examine the intraspecific and interspecific evolutionary relationships of IQM genes, the predicted IQM protein sequences of multiple species were obtained from the corresponding databases. Arabidopsis and rice IQM protein sequences were downloaded from TAIR (http://www.arabidopsis.org) and RAP-DB (http://rapdb.dna.affrc.go.jp/ ), respectively, whereas soybean, alfalfa, tomato, maize, sorghum, and Brachypodium distachyon IQM protein sequences were retrieved from Glycine max Wm82.a2.v1, Medicago truncatula Mt4.0v1, Solanum lycopersicum ITAG4.0, Zea mays B84 v1.2, Sorghum bicolor v3.1.1, and Brachypodium distachyon v3.1 of Phytozome v13. ClustalW software was used for multisequence alignment. The parameters were set to the system default values [Pairwise Alignment—Gap Opening Penalty:10.00; Gap Extension Penalty: 0.10. Multiple Alignment—Gap Opening Penalty:10.00; Gap Extension Penalty: 0.20. Use Negative Matrix: off; Delay Divergent Cutoff(30)]. MEGA11 software was used to construct an unrooted phylogenetic tree using the neighbor-joining (NJ) method, and bootstrap analysis was conducted using 1,000 replicates (Wu et al., 2016). The generated phylogenetic tree was identified using iTOL (https://itol.embl.de/ ).
Analysis of the structure of soybean IQM gene and protein
To compare the gene structures of GmIQMs, the distribution of exons and introns was analyzed using GSDS2.0 (http://gsds.cbi.pku.edu.cn) (Mei et al., 2021). The gff3 format files of soybean IQM genes downloaded from Phytozome v13 database were imported, then the system will automatically generate the structure chart contained CDS, UTR and Intro. To understand the similarities and differences in protein structures, the conserved motifs of encoded GmIQM proteins were identified using the MEME database (https://meme-suite.org/meme/ ), the maximum number of motifs was set to 15.
Chromosomal localization and gene duplication
The distribution image of GmIQM genes on the soybean chromosome was generated by MG2C (http://mg2c.iask.in/mg2c_v2.1/ ) according to the gene information retrieved from the Phytozome v13 database. Gene duplication analysis was performed as previously described (Feng et al., 2014). The SoyBase browser (http://soybase.org/gb2/gbrowse/gmax1.01) was used to search for duplicate gene pairs. Coparalogs were considered to be duplicated in tandem if they were on the same chromosome and separated by five or fewer genes in a 100-kb region (Wang et al., 2010); otherwise, they were deemed to be fragment duplications. To further analyze the divergence of duplicated genes, the synonymous substitution rate (Ks) and non-synonymous substitution rate (Ka) were calculated using TBtools software. According to a rate of 6.1×10-9 substitutions per site per year, the divergence time (T) was calculated using the Ks value and the formula: T = Ks/(2×6.1×10-9)10-6 Mya (Lynch and Conery, 2000).
Plant materials and growth conditions
The Arabidopsis culture conditions were obtained from Lv et al, 2019. Arabidopsis (Arabidopsis thaliana) Col-0 seeds were sown in nutrient soil after 3 days of vernalization at 4°C and then cultured at 22°C under 16 h light/8 h dark conditions for approximately 4 weeks. Rosettes were cut before flowering to prepare protoplasts for subcellular localization and BiFC assays.
Soybean (Glycine max) Williams82 seeds were placed on absorbent paper and placed in the dark at room temperature for three days to germinate. Seedlings that grew consistently in a sponge hole tray were cultured in 1/4 strength Hoagland’s solution for 14 days under 16 h light/8 h dark conditions at 25°C and 6000 lx with 80% relative humidity. Subsequently, 50 mM NaHCO3, 150 mM NaCl, 20% (w/v) PEG6000, 100 µM ABA, 100 µM MeJA, and 2 mM SA were added to Hoagland’s solution to simulate various abiotic stress conditions. Equal amounts of leaves and roots were collected for RNA isolation at 0, 3, 6, 12, and 24 h after treatment.
Yeast two-hybrid assays
Yeast two-hybrid assays were performed according to the manufacturer’s protocol. Briefly, the full-length CDS of GmCaM (Glyma.19G121900) and 15 GmIQMs were cloned separately into pGADT7 and pGBKT7. The plasmids, AD-GmCaM and BD-GmIQMs, were co-transformed into AH109 yeast cells using the PEG/LiAC method. Interactions in yeast were tested on SD/-Trp/-Leu/-His and SD/-His-Trp-Leu-Ade plates containing 20 μg/mL x-α-gal. Cotransforming with the AD empty vector and BD-empty vector was used as a negative control, while AD-AtCAT2 and BD-AtIQM1 was used as a positive control (Lv et al., 2019).
For construction BD-GmIQMLQ/VQ yeast expression vectors, LQ or VQ motif in CDS of five representative GmIQMs (GmIQM1a, 112-113aa LQ; GmIQM2a, 135-136aa LQ; GmIQM3a, 63-64aa VQ; GmIQM1e, 127-128aa LQ; GmIQM6a, 66-67aa LQ) was deleted using the PCR product fragment splicing method in vitro. Mutant protein sequences were cloned into the pGBKT7 vector. Thereafter, GmCaM binding to the mutant GmIQMs was analyzed using the yeast two-hybrid assay described above.
Subcellular localization and BiFC assays
The protoplasmic transformation for both experiments was performed as previously described (Lv et al., 2019). For subcellular localization, the full-length CDS of 15 IQM genes in soybean Williams82 was cloned into the pGreenII-35S-GFP plant expression vector, respectively. Thereafter, pGreenII-35S-GFP-GmIQMs plasmids were transformed into protoplasts isolated from the leaves of 4-week-old Arabidopsis wild-type Col-0. After 16 h of incubation at 22°C, the GFP signal was observed using a Zeiss LSM800 confocal microscope at 488 nm absorption and 507 nm emission wavelengths. The pGreenII-35S-GFP empty vector was used as a negative control.
For the BiFC assays, the CDSs of GmIQM1d/GmIQM2c and GmCaM were cloned into binary pSAT1-nEYFP and pSAT1-cEYFP vectors, respectively. Thereafter, the pSAT1-nEYFP-GmIQM1d/GmIQM2c and pSAT1-cEYFP-GmCaM plasmids were transformed into protoplasts isolated from the leaves of 4-week-old Col-0. After 16 h of incubation at 22°C, the EYFP signal was observed using a Zeiss LSM800 confocal microscope at 488 nm absorption and 530 nm emission wavelengths. Co-transformation with nEYFP empty vector and cEYFP-GmCaM served as negative controls.
RNA extraction and RT-qPCR analyses
RNA was isolated using the Eastep Super Total RNA Extraction Kit (Promega, LS1040) according to the manufacturer’s protocol. For the tissue-specific expression analysis, total RNA was extracted from roots (no nodule), cotyledons, stems, leaves, flowers, pods and nodules of Williams82. For the gene expression analysis under abiotic and biotic stress, total RNA was extracted from 14-days-old leaves and roots of Williams82, with or without treatment. RT-qPCR was performed as previously described (Bu et al., 2014; Lv et al., 2019), and first-strand complementary DNA (cDNA) synthesis was performed using the PrimeScript RT Reagent Kit (Takara, RR047A), according to the manufacturer’s instructions. cDNAs were used as templates for RT-qPCR with gene-specific primers. Primer sequences are listed in Supplemental Table S2. RT-qPCR was performed using 384-well plates with SYBR Premix Ex Taq II (Takara, RR820A) and a Roche Light Cycler 480 Real-Time PCR system. The soybean tubulin gene (Glyma.05G207500) was used as an internal control for mRNA (Li et al., 2017) and the relative expression levels of the genes were calculated using the 2−CT method. Each sample was collected from five independent plants, independent biological assays were repeated three times, and similar results were obtained. Data from one representative replicate are shown. Values are means average from three technical measurements.
Results
Identification of IQM gene family in soybean genome
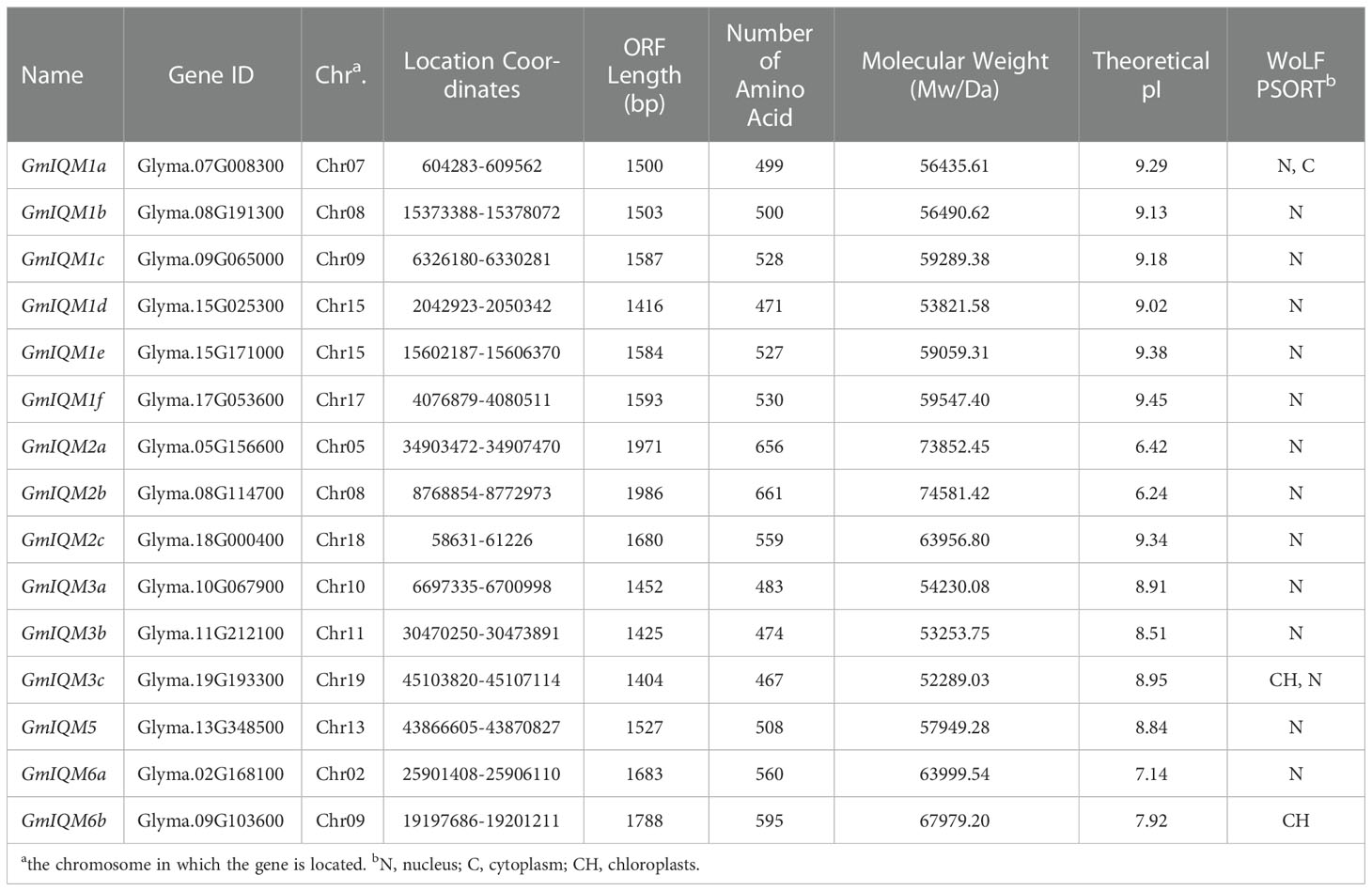
As revealed in previous studies, IQM proteins are plant-specific calcium-independent calmodulin-binding proteins that contain one IQ motif (Zhou et al., 2010). To identify the IQM gene family in soybean, we performed BLASTP analysis of the Glycine max genome via alignment with reported IQM proteins in Arabidopsis. Finally, 15 candidate GmIQM genes containing the IQ calmodulin-binding motifs were confirmed using conserved domain analysis. According to the order on chromosomes of GmIQMs and their homology relationship with AtIQMs, these 15 GmIQM genes were named GmIQM1a-1f, GmIQM2a-2c, GmIQM3a-3c, GmIQM5, and GmIQM6a/6b. The characteristics of each IQM protein, including location on chromosome, location coordinates, open reading frame (ORF) length, number of amino acids, molecular weight, theoretical pI (isoelectric points), and predicted subcellular localization, are listed in Table 1. According to these data, the protein sequences of the 15 soybean IQM genes did not significantly differ in length (467-661 aa) and molecular weight (52.29-74.58 kDa). Further, most proteins had high isoelectric points (pI 7.14-9.34), except GmIQM2a (pI 6.42) and GmIQM2b (pI 6.24). Almost all GmIQM proteins were predicted to localize in the nucleus, while only few were predicted to localize in the chloroplast or cytoplasm (Table 1).
Protein interaction analysis of GmIQMs with GmCaM
Based on our previous study, IQMs can interact with CaMs in Arabidopsis and rice (Zhou et al., 2012; Fan et al, 2021 suggesting that binding to CaM is a common characteristic of IQM proteins. To further verify IQM in soybean, yeast two-hybrid experiments were carried out to determine whether IQM could also be combined with CaM in soybean. The results showed that only two vector combinations of AD-GmCaM and BD-GmIQM1d/2c could not grow on SD/-Trp/-Leu/-His triple dropout and SD/-Trp/-Leu/-Ade/-His quadruple dropout culture media (Figure 1A), indicating that most GmIQMs could interact with GmCaM in yeast cells, except GmIQM1d and GmIQM2c.
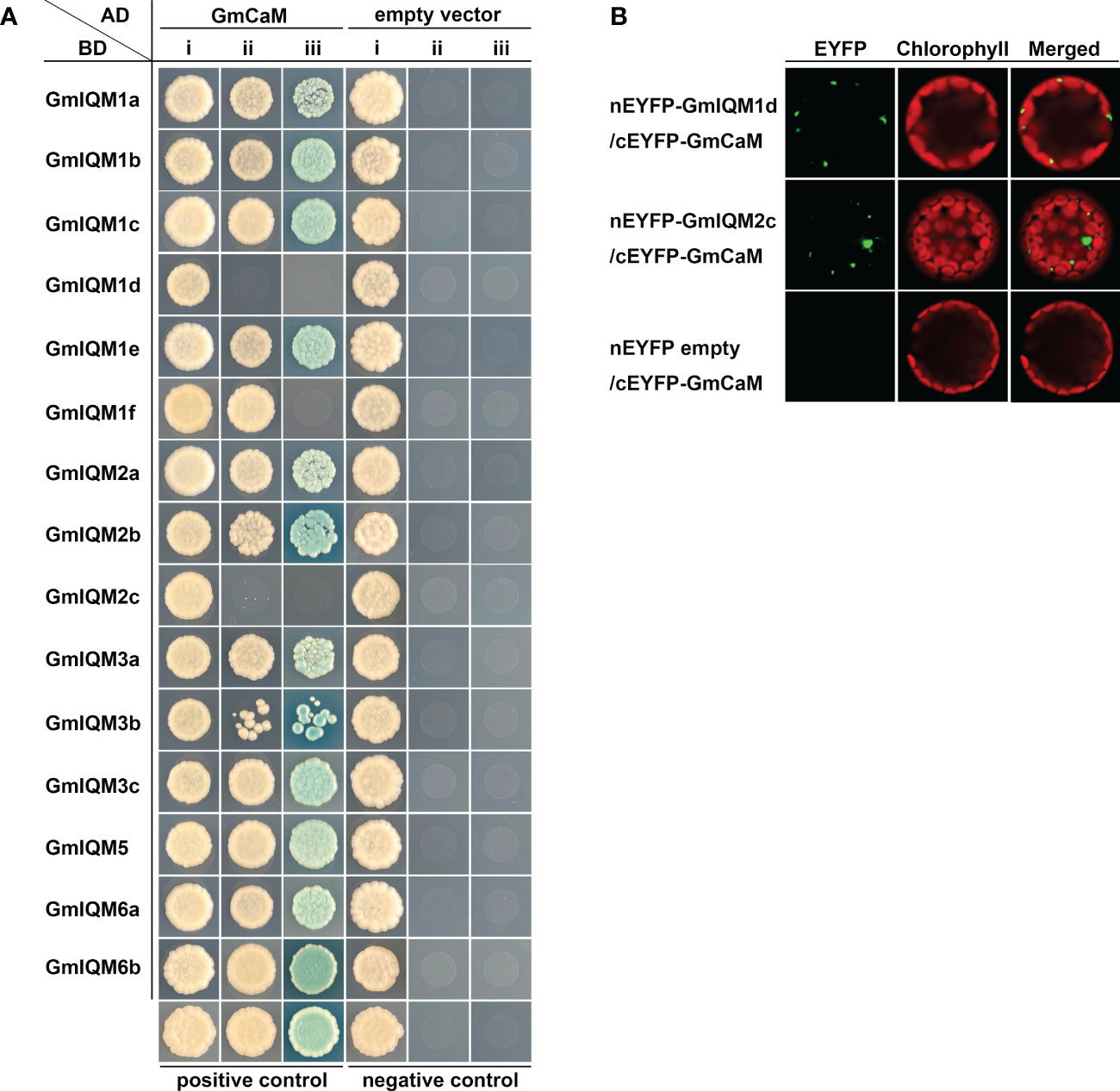
Figure 1 Interaction Between Gmiqms And Gmcam. (A) GmIQMs interact with GmCaM in yeast cells. pGADT7-GmCaM and pGBKT7-GmIQMs were co-transformed into AH109 yeast cells, which were grown on SD/-Trp-Leu (i), SD/-Trp-Leu-His (ii) and SD/-Trp-Leu-His-Ade containing 20 μg/ml x-α-gal (iii) for 3 days. Co-transformation with the AD empty vector and BD-empty vector served as negative controls. AD-AtCAT2 and BD-AtIQM1 were used as positive controls. (B) BiFC assays to detect the interaction between GmIQM1d/GmIQM2c and GmCaM. nEYFP-GmIQM1d/GmIQM2c and cEYFP-GmCaM plasmids were co-transformed into protoplasts isolated from the leaves of 4-week-old Col-0 plants. After 16 h of incubation, the EYFP signals were observed using a laser confocal microscope. nEYFP-empty vector and cEYFP-GmCaM were employed as negative controls.
To further assess the interaction between GmIQM1d/GmIQM2c and GmCaM in vivo, an enhanced yellow fluorescent protein (EYFP)-based bimolecular fluorescence complementation (BiFC) assay was performed. Plasmids carrying nEYFP-GmIQM1d/GmIQM2c and cEYFP-GmCaM fused expression vectors were co-transformed into protoplasts of Arabidopsis Col-0. The co-transformation with the nEYFP empty vector and cEYFP-GmCaM was used as negative controls. Fluorescence signals were observed in the test group using a laser confocal microscope (Figure 1B).These results revealed that GmIQM1d/GmIQM2c interacts with GmCaM in vivo and a particular component of plant cells, which is nonexistent in yeast cells, and may be indispensable for their interactions. Overall, the reliability of soybean IQM proteins was confirmed using protein interaction assays.
Phylogenetic and structural analyses of soybean IQM genes
To understand the similarity and evolutionary relationship between GmIQM proteins, we constructed an unrooted phylogenetic tree of 15 soybean IQM protein sequences. Based on the results, all members of the soybean IQM gene family could be divided into three major subfamilies (I, II, and III; Figure 2A). Similar to the analysis results for Arabidopsis and rice, the IQM members in soybeans mainly belonged to subfamilies I and II. Notably, subfamily I had the most members, with seven genes, while subfamily III had the fewest IQM members, with three genes. Further, the most closely related members within each subfamily formed sister-gene pairs.
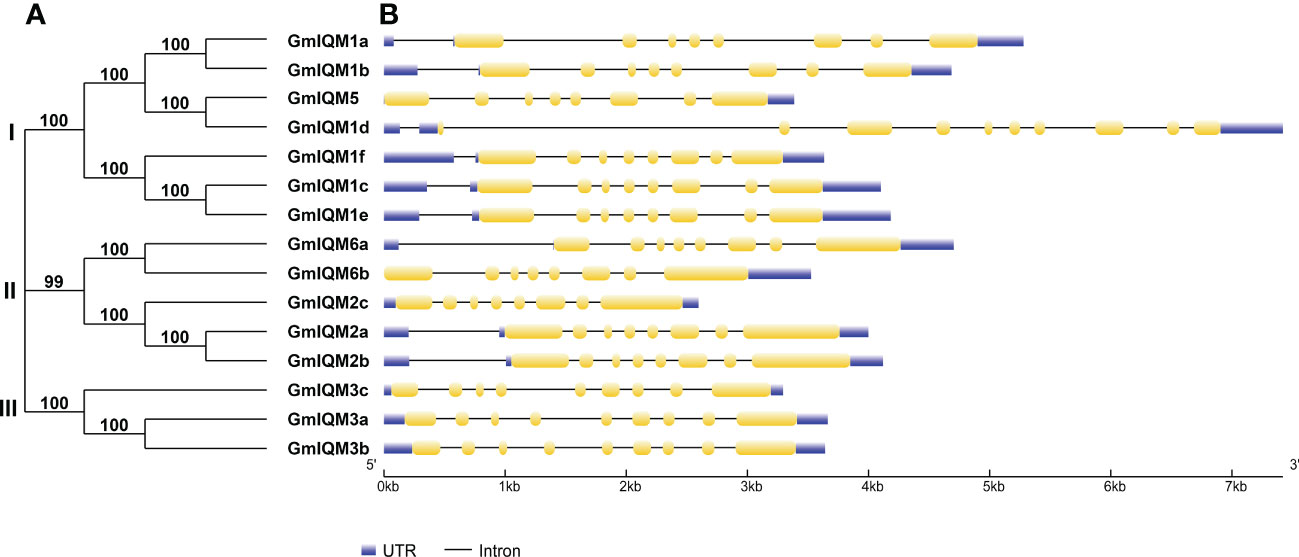
Figure 2 Phylogenetic relationships and gene structures of the GmIQM genes. (A) Unrooted phylogenetic tree of GmIQMs. The full-length amino acid sequences of 15 GmIQMs were aligned by ClustalW, and the unrooted phylogenetic tree was constructed using MEGA11 by the neighbor-joining method. The number of bootstrap values was 1000 replicates. (B) Exon/intron organization of soybean IQM genes. Yellow boxes represent exons, black lines represent introns, blue boxes represent Untranslated regions (UTRs). The sizes of exons and introns can be estimated using the scale at the bottom.
The diversity of gene structure is well known as an important foundation for gene family classification (Zhao et al., 2017; Mengarelli and Zanor, 2021; Zhou et al., 2021). To further detect the features of each IQM subfamily in the soybean genome, the exon/intron organization of GmIQM genes was generated (Figure 2B). The structure chart revealed that most soybean IQM genes (eleven/fifteen) contained eight exons, except GmIQM1d, which contained ten exons, and three members of subfamily III (GmIQM3a, 3b, and 3c), which had nine exons. The relatively uniform exon numbers indicate the structural conservation of GmIQM.
The most closely related GmIQM gene pairs in the same subfamily shared similar gene structures, both in exon distribution and intron length. Nevertheless, one pair (GmIQM1d/GmIQM5) displayed an obvious discrepancy (Figure 2B); compared with GmIQM5, GmIQM1d had two extra short coding sequences in front of the first exon, which may be due to intron gain events during the long evolutionary period.
The conserved domains or motifs of 15 GmIQM genes were examined using the MEME database. Fifteen potentially conserved motifs were identified (Figure 3A; Figure S1, and S2). Motif 5 was annotated to encode the IQ motif based on a data search of Pfamscan and SMART. The distribution of motifs in 15 GmIQMs is consistent with the gene clustering that
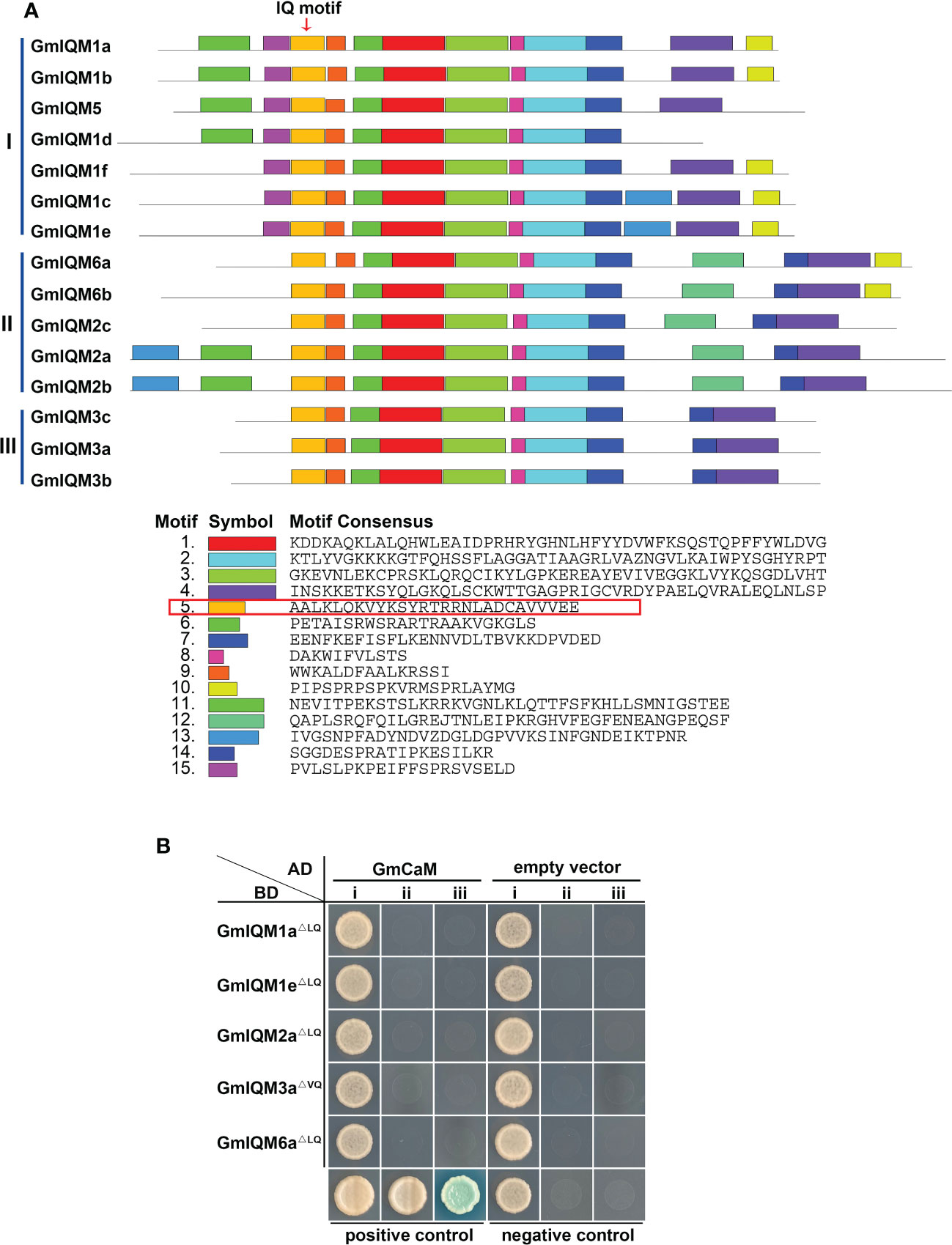
Figure 3 Conserved motif in GmIQM proteins. (A) Motif distribution in the IQM proteins of soybean. Motifs of the GmIQM proteins were identified using the MEME online program. The maximum number of motifs was set as 15. Fifteen motifs were represented by different colors; each motif consensus sequence is listed below, red box represents the amino acid sequence of the IQ motif, arrow above the diagram represents the location of the IQ motif. (B) Functional validation of the IQ motif in GmIQM proteins. AD-GmCaM and BD-GmIQMLQ/VQ were co-transformed into yeast cell AH109 grown on SD/-Trp-Leu (i), SD/-Trp-Leu-His (ii), and SD/-Trp-Leu-His-Ade containing 20 μg/ml x-α-gal (iii) for 3 days. Co-transformation with the AD empty vector and BD-empty vector was performed for use as the negative controls. AD-AtCAT2 and BD-AtIQM1 served as the positive controls.
Most closely gene pairs in the same subfamilies had identical motifs, except GmIQM1d/GmIQM5 (Figure 3A), suggesting functional similarities or redundancies among these GmIQM proteins. Further, the 15 GmIQM proteins had common motifs (motifs 1, 2, 3, 5, 6, 7, 8, and 9); however, some motifs were recognized to be specific to certain subfamilies. For example, motifs 12 and 15 are unique to subfamilies II and I, respectively. Thus, variations in these specific motifs may cause functional differentiation of the IQM proteins in soybean.
The IQ motif is key to the interaction between CaM and CaMBP (Rhoads and Friedberg, 1997). To determine whether the combination of GmCaM and GmIQMs is dependent on the IQ motif, we constructed a BD vector with the soybean IQM protein sequence deleted 6Q (LQ/VQ) motif. Thereafter, AD-GmCaM and BD-LQ/VQ were co-transformed into yeast cell AH109. As all transformants did not grow on the SD/-Trp/-Leu/-Ade/-His quadruple dropout culture medium (Figure 3B), the IQ motif was identified to be necessary for functional GmIQM and its conservation in different species.
Chromosomal locations and gene duplication
To accurately understand the orientation of the 15 soybean IQM genes on each chromosome, a chromosomal map we constructed based on the location information retrieved from the soybean database. The distributions of these genes on chromosomes appeared to be wide but unbalanced. Fifteen GmIQM genes were mapped on 12 of the 20 soybean chromosomes. Nine chromosomes, including chromosomes 2, 5, 7, 10, 11, 13, 17, 18, and 19, contained only one IQM gene. Three chromosomes, including chromosomes 8, 9, and 15, had two IQM genes, while the remaining eight soybean chromosomes did not contain any IQM gene (Figure 4). This biased distribution pattern of IQM genes has been observed in Arabidopsis and rice genomes (Zhou et al, 2010; Fan et al, 2021). Although two IQM genes were present on chromosomes 8, 9, and 15, however, there was no clustering from GmIQM genes on the same chromosomes, suggesting that the expansion of IQM family genes in soybean may not have been produced from tandem duplications.
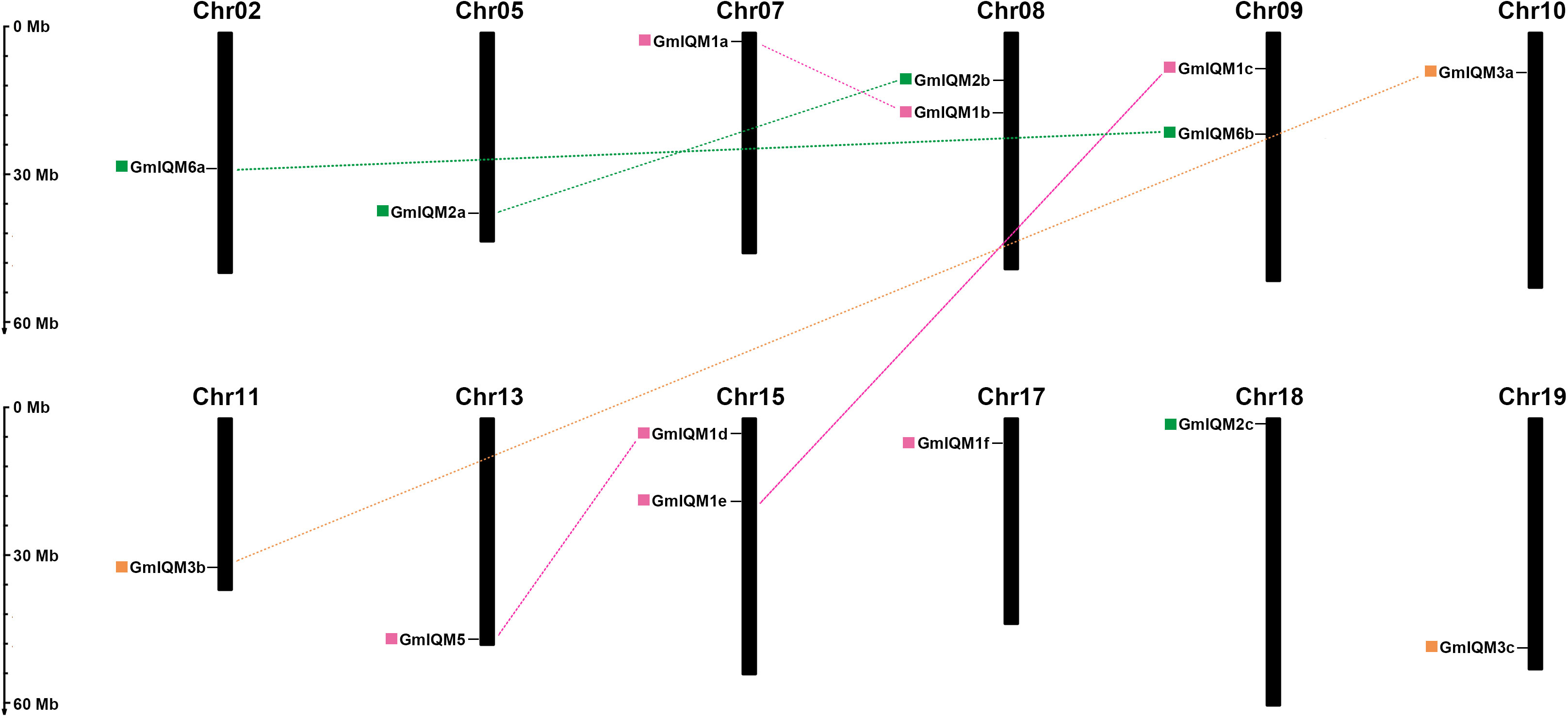
Figure 4 Chromosomal locations and segmental duplication of soybean IQM genes.The fifteen IQM genes are widely mapped to 12 of the 20 chromosomes in soybean. Chromosome numbers are located at the top of each vertical bar. The duplicated paralogous pairs of GmIQM gene are connected with dotted lines of the same color. The colored boxes in front of the gene names represent different subfamilies (pink, subfamily I; green, subfamily II; orange, subfamily III). Scale on the left represents the chromosome length.
Gene duplication is considered an important source of biological evolution. There are three main types of gene duplication: segmental duplication, tandem duplication, and transposition (Sankoff, 2001; Flagel and Wendel, 2009; Panchy et al., 2016). Among these duplication types, segmental duplication is a major contributor to the amplification of many gene families (Cannon et al., 2004; Zhu et al., 2020; Li et al., 2021; Wang M, et al., 2021; Zhao et al., 2021);. Herein, gene duplication events were examined to further understand the expansion mechanism of the IQM family in soybean. According to the data of synteny (Glycine recent duplication) from SoyBase browser, we found six pairs of
GmIQM genes located in duplicated blocks, indicating that these genes were generated by segmental duplication. The remaining three genes (GmIQM1f, 2c, and 3c) lacked duplicated pairs in their corresponding synteny blocks (Figure 4 and Table 2), which aligns with the results of clustering analysis. We determined whether tandem duplication also played a role in adjacent GmIQM genes on the same chromosome. It has been reported that a pair of genes is separated by three or fewer genes within a 100-kb region on a chromosome, this may be due to tandem duplication (Wang et al., 2010; Feng et al., 2014; Wang X, et al., 2021). According to this criterion, no pair was generated by tandem duplication of the soybean IQM genes. Therefore, segmental duplication was identified to contribute significantly to the expansion of the IQM gene family in soybean.
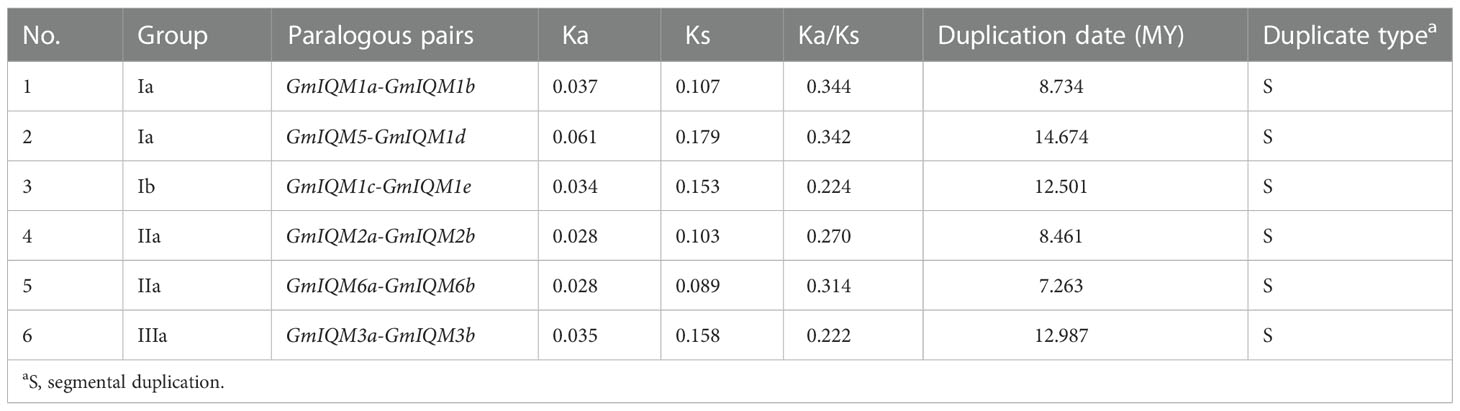
To understand the evolutionary selection of duplicated GmIQM genes, we calculated the Ka/Ks ratio (substitution ratio of non-synonymous/synonymous) for each pair of duplicated GmIQM genes. In general, Ka/Ks = 1 indicates that both genes drift neutrally, Ka/Ks>1 indicates accelerated evolution with positive selection, and Ka/Ks<1 indicates a functional constraint with negative or purifying selection of the genes (Hurst, 2002; Nekrutenko et al., 2002; Feng et al., 2014; Ma et al., 2014). The Ka/Ks ratios from all duplicated gene pairs were found to be less than 0.4 (Table 2), suggesting that the evolution of the soybean IQM gene family was mainly influenced by negative or purifying selection, thereby limiting the functional differentiation of duplicated genes. Based on the divergence rate of 6.1×10-9 synonymous mutations per site per year (Lynch and Conery, 2000; Zhang et al., 2011; Chen et al., 2014; Hyun et al., 2014; Meng et al., 2015; Zhang et al., 2019), the duplication of these paralogous pairs was estimated to occur between 7.263 and 14.674 million years ago (Mya) (Table 2).
Phylogenetic comparison of the IQM genes in various species
Seventy-four putative IQM protein sequences from eight species, including four dicotyledons, Glycine max (15), Arabidopsis thaliana (6), Solanum lycopersicum (9), Medicago sativa (8); and four monocotyledons, Oryza sativa (8), Zea mays (11), Sorghum bicolor (10), and Brachypodium distachyon (7), were analyzed using the neighbor-
joining (NJ) method to further assess the phylogenetic relationship of IQMs in plants. All IQMs were found to cluster into three distinct subfamilies (I, II, and III) (Figure 5). This result aligns with the classification of GmIQM genes; subfamilies I and II had the most members (77%) in the combined phylogenetic tree, while subfamily III had the fewest genes (23%). Each subfamily was further divided into two subgroups (a and b) of dicots and monocots (Figure 5), indicating that significant differences in the structure and function of IQM genes between dicots and monocots.
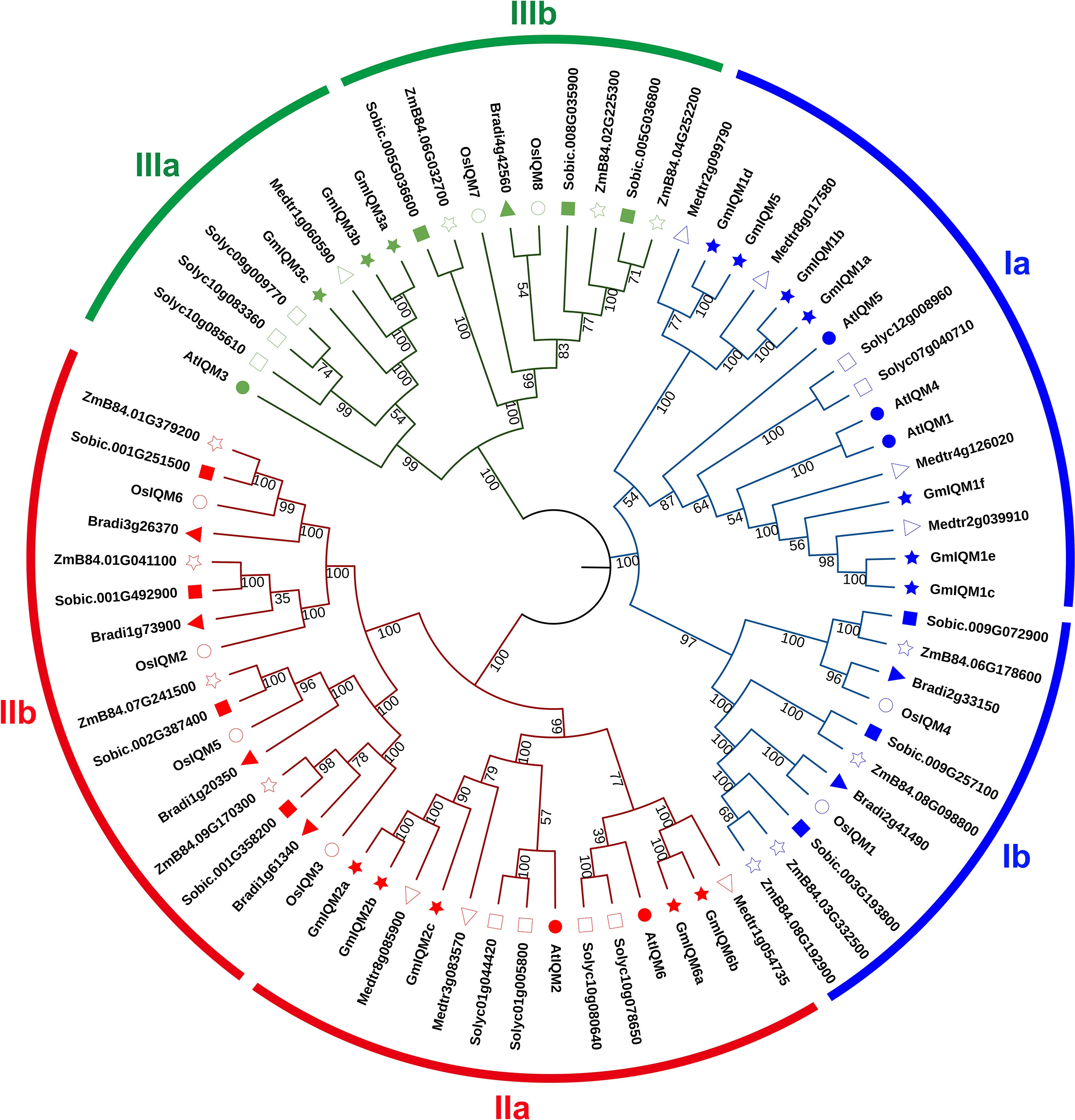
Figure 5 Phylogenetic relationships of IQMs in eight species.The full-length amino acid sequences of 74 IQMs were aligned using ClustalW, and the unrooted phylogenetic tree was constructed using MEGA11 by the neighbor-joining method. The number of bootstrap values was 1000 replicates. At, Arabidopsis; Gm, soybean; Medtr, alfalfa; Solyc, tomato; Os, rice; ZmB84, maize; Sobic, sorghum; Bradi, Brachypodium distachyon. Blue, subfamily I; red, subfamily II; green, subfamily III. The shapes preceding the gene numbers represent different species.
Subcellular localization of GmIQMs
The subcellular localization of 15 soybean IQM proteins was determined to further confirm their functional sites in cells. The full-length CDS sequence of each GmIQM was cloned and constructed into a pGreenII-35S-GFP vector, and different plasmids were transformed into protoplasts of Arabidopsis wild-type Col-0. After incubation, the GFP signal was observed under a confocal microscope (Figure 6). The results showed that GmIQM1a and GmIQM1b were only located in the nucleus; GmIQM3a, 3b, and 3c were located in the nucleus and cytoplasm; and the remaining IQMs were mainly localized in the cytoplasm. These location patterns matched the clustering results, members of the closest relative clade had the same localization in the cell, indicating that these genes may have the same or similar functions. Further, the practical localization of these genes did not completely match the prediction by the Wolf PSORT programs.
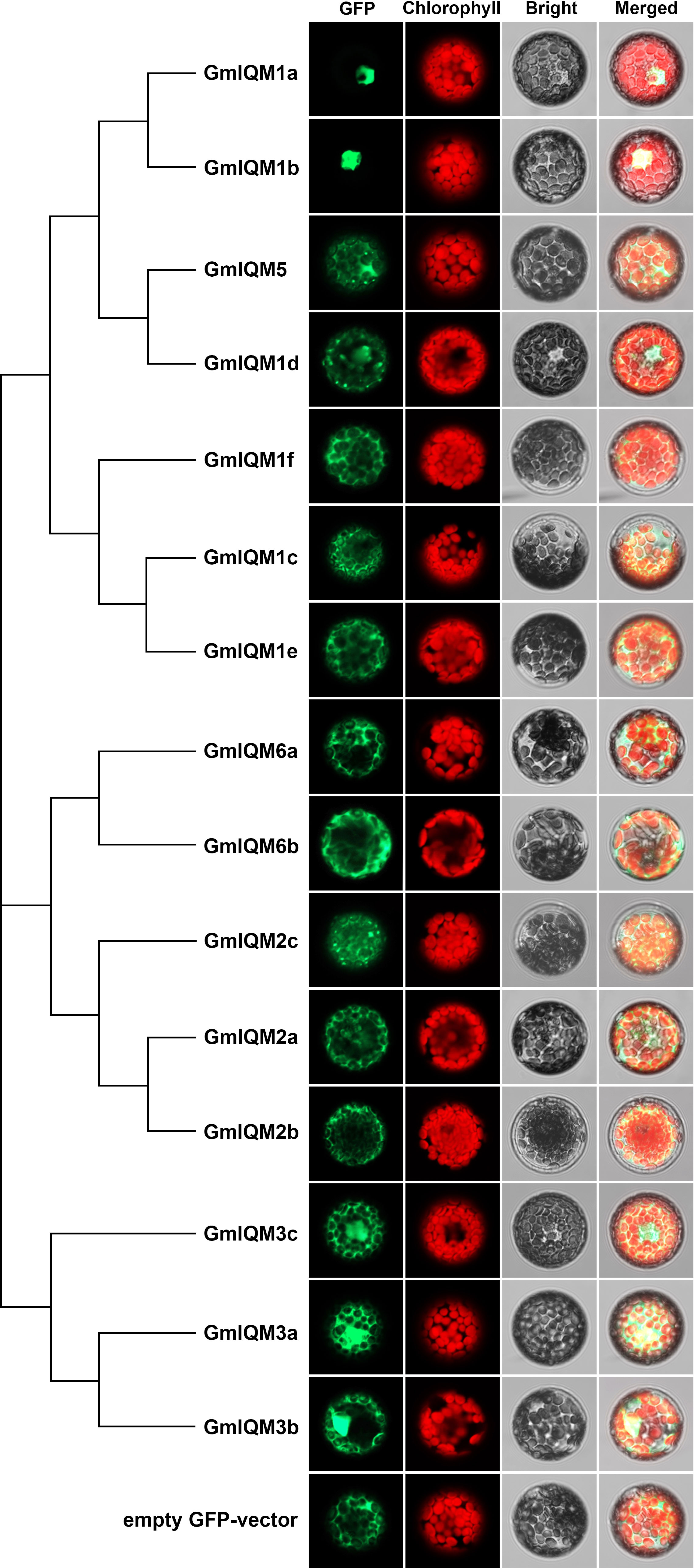
Figure 6 Subcellular localization of fifteen GmIQM proteins in Arabidopsis protoplasts.The plasmids containing pGreenII-35S-GFP-GmIQMs vector were transformed into protoplasts isolated from the leaves of 4-week-old Col-0 plants. After 16 h of incubation, the GFP signals were observed using a laser confocal microscope. Transformed pGreenII-35S-GFP empty vector was employed as the negative control. GFP, green fluorescent protein; Chlorophyll, chlorophyll auto-fluorescence; Bright, bright field; Merged, merged GFP, chlorophyll and bright.
Tissue-specific expression of GmIQM genes
Every stage of plant growth and development is closely regulated by a large number of genes. To understand the potential role of soybean IQM genes in the plant life cycle, quantitative polymerase chain reaction (qRT-PCR) was performed to determine the expression levels of IQM genes in soybean roots (no nodule), cotyledons, stems, leaves, flowers, seeds, pods, and nodules respectively. As depicted in the heat map, soybean IQM genes from the same subfamily or the most closely related paralogous gene pairs had similar gene expression patterns. For example,
Three members of subfamily III (GmIQM3a, 3b, and 3c) were significantly expressed in eight tissues. However, in subfamily I, only two genes, GmIQM1c and GmIQM1e, which are sister pairs in the closest clade, had universal expression, and the remaining genes (GmIQM1a, GmIQM1b, GmIQM5, GmIQM1d, and GmIQM1f) were not expressed in most tissues (Figure 7). Notably, some GmIQM genes have tissue-specific expression characteristics, such as GmIQM5 and GmIQM1d, which had markedly high transcript abundance in nodules but were not expressed in other tissues, and IQM6a, which only had a slight expression in flowers (Figure 7), suggesting that these genes might have vital functions at specific stages of soybean development.
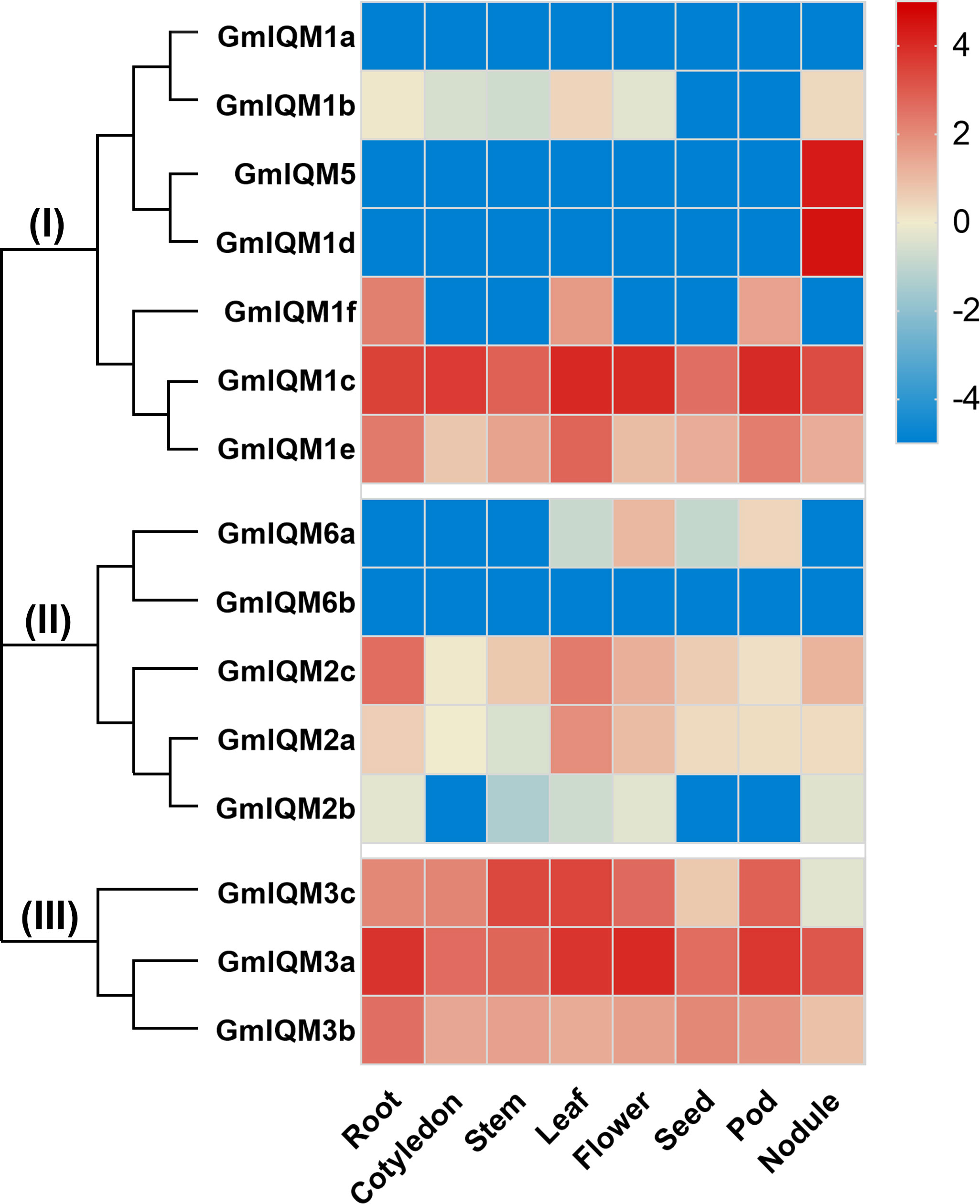
Figure 7 Heat map of real-time quantitative PCR analysis results of GmIQM in eight tissues.Total RNA was extracted from roots, cotyledons, stems, leaves, flowers, pods, and nodules. Each sample was collected from five independent plants, independent biological assays were repeated three times, and similar results were obtained. Data from one representative replicate are shown. Values are means average from three technical measurements. Color scale at the right side of the image represents log10 of the gene expression values. Red indicates high expression level and blue indicates low expression level.
Analysis of cis-acting elements of GmIQM gene promoters
The type and number of cis-acting elements in gene promoters are well known to largely determine gene function. To explore the potential role of soybean IQM genes, we analyzed the GmIQM gene promoter sequences within 2 kb upstream of the start codon using the PlantCARE database. In addition to the core elements, TATA-box and CAAT-box, various stress and hormone response elements were found in the promoter of each soybean IQM gene, such as MBS, GT1, ERE, MYB, MYC (drought, high salt, low temperature, etc.), ABRE (ABA), CGTCA-motif, TGACG-motif (MeJA), and TCA-element (SA) (Figure 8). Therefore, GmIQMs could play a role in the response of soybean to biotic and abiotic stresses. However, the number and distribution range of abiotic response elements were significantly higher than those of MeJA and SA stress-related elements, especially MYB and MYC, which existed in almost all soybean IQM promoters. Thus, the soybean IQM gene family may be more important for abiotic stress response than biotic defense.
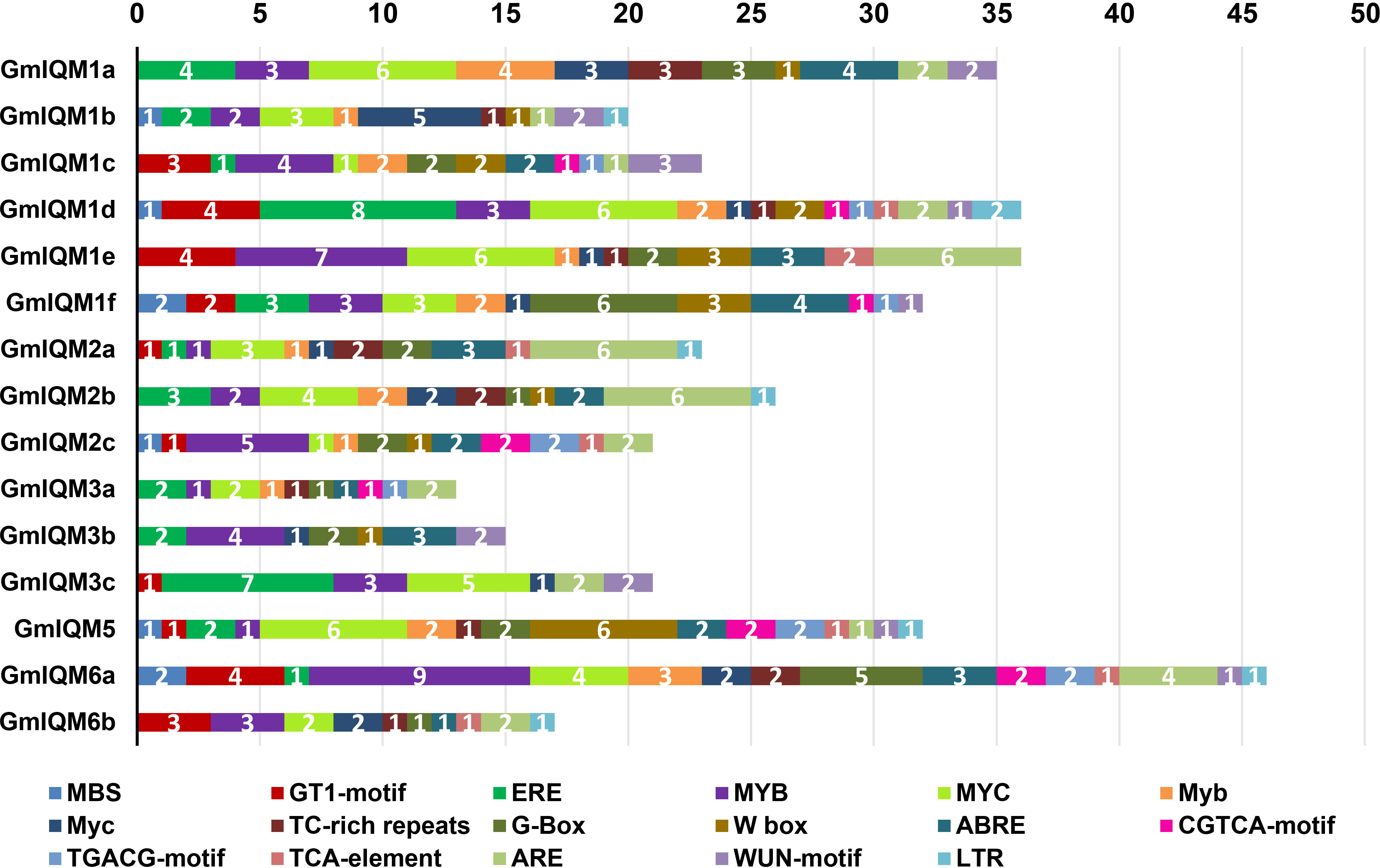
Figure 8 Putative cis-acting elements of the IQM promoters predicted by PlantCARE.The different cis-elements are indicated by different colors. The numbers on the box represent the count of cis-elements in the promoter.
Effects of abiotic and biotic stress on GmIQM gene expression
According to the analysis of cis-acting elements in promoters, IQM genes are most likely regulated by abiotic stresses and hormonal stimuli in soybean. To further confirm the function of the GmIQM genes, soybean wild-type Willims82 was treated with different stressors, including NaCl, NaHCO3, PEG, SA, ABA, and MeJA, to simulate saline, alkaline, drought, and biological stress conditions. After treatment, the expression levels of eight representative IQM genes in soybean leaves and roots were determined using qRT-PCR. Gene expression profiling revealed that these IQM genes were strongly induced by NaCl and PEG, both in the leaves and roots (Figure 9). Thus, these IQM genes play an important role in the response to high salinity and drought conditions in soybean. These genes were also upregulated by NaHCO3, SA, ABA, and MeJA; however, compared with NaCl and PEG, the induction degree of their expression levels was relatively low under these stresses, and the transcriptional change mainly occurred in the roots, and not in the leaves (Figure 9). These results indicated that IQM family genes also function in alkali and biological stress responses to some extent.
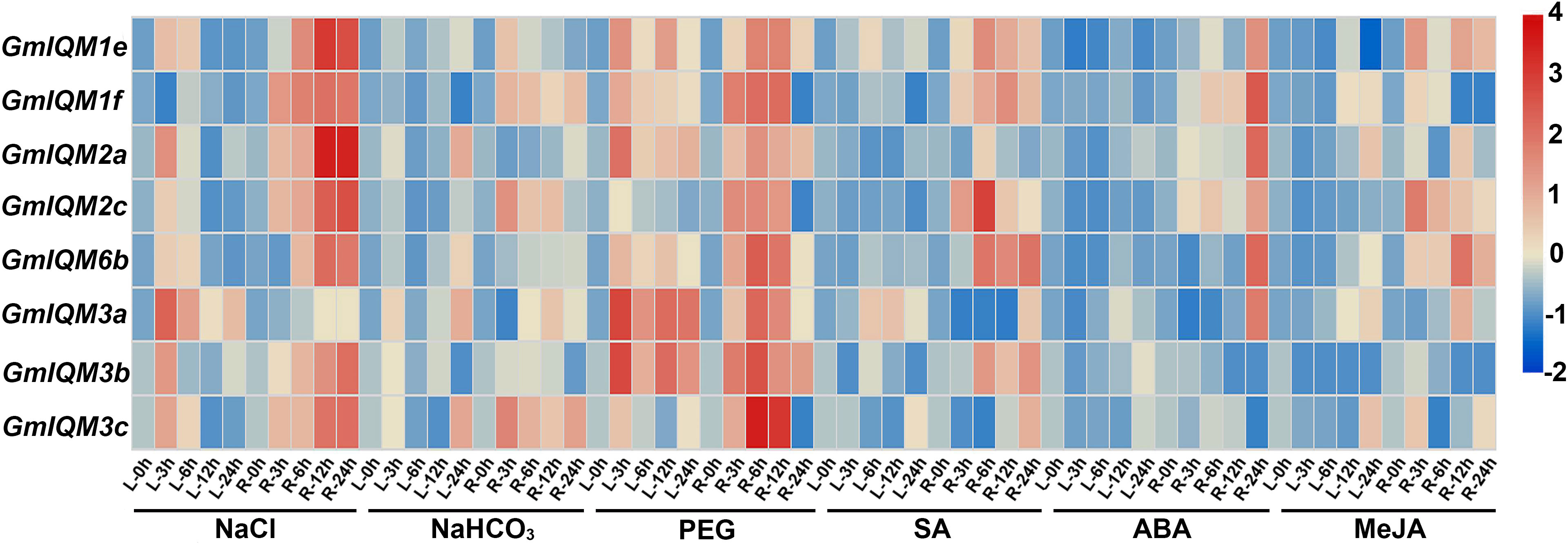
Figure 9 Heat map of the expression of GmIQMs in leaves and roots under different treatments.NaCl, NaHCO3, PEG, SA, ABA, and MeJA were used to treat soybean. Total RNA was then extracted from the roots and leaves at indicated times. Each sample was collected from five independent plants, independent biological assays were repeated three times, and similar results were obtained. Data from one representative replicate are shown. Values are means average from three technical measurements. Color scale at the right side of the image represents log10 of the gene expression values. Red indicates high expression level and blue indicates low expression level. L, leaves; and R, roots.
Discussion
As plant-specific calcium signaling components, IQM genes have been postulated to play a critical role in the crosstalk between multiple signaling pathways in the context of plant growth and development (Lv et al., 2019; Zhang et al., 2022). In our previous studies, we conducted a systematic and comprehensive analysis of IQM family genes in Arabidopsis and rice (Zhou et al., 2010; Fan et al., 2021). However, the number and functions of IQM genes in other species remain unclear. Herein, we identified 15 IQM genes in soybean, and employed bioinformatics to analyze the soybean IQM family genes at the whole-genome level.
Compared to Arabidopsis (six members) and rice (eight members), the IQM gene family in soybean is by far the largest. In fact, the number of IQM genes in soybean is 2.5-fold that of Arabidopsis. This doubling of the gene number has been observed in many other soybean gene families. For example, IQD, another IQ-containing calmodulin binding protein family, has 67 members in soybean, which is more than twice that in Arabidopsis (33 members) (Abel et al., 2005; Feng et al., 2014). The basic leucine zipper (b-ZIP) transcription factor family contains 78 members in Arabidopsis and 160 members in soybean (Jakoby et al., 2002; Dröge-Laser et al., 2018; Zhang et al., 2018). The genome-wide duplication (WGD) events in soybean evolutionary history may explain this phenomenon. WGD is very common in plants, leading to double gene copies in the genome. The functional divergence of duplicate gene pairs is the source of new genes. Soybean has been reported to experience at least two WGD events, approximately 59 and 13 million years ago, resulting in a highly duplicated genome with nearly 75% of the genes present in multiple copies (Schmutz et al., 2010). Therefore, a greater expansion of IQM genes may have occurred in the soybean genome than in other species. According to our analysis, six paralogous pairs were present among the 15 GmIQM genes, and all six pairs were caused by segmental duplication, but not tandem duplication (Table 2). Such findings indicate that segmental duplication played a more important role in the long evolution of soybean IQM genes. The Ka/Ks ratio was less than 1 for all 6 pairs of duplicated genes, indicating that these gene pairs were subjected to purifying selection (Hurst, 2002). The calculations also revealed that the duplication events of the 6 paralogous pairs in the soybean IQM family occurred between 7.26 and 14.27Mya, which was during the recent WGD of soybean. This date predates the duplication of the IQM gene pairs in rice (11.9-19.8 Mya; Fan et al., 2021).
By comparing the predicted IQM sequences from multiple species, the IQM genes in three different subfamilies were further divided into two groups: dicots(Arabidopsis, soybean, alfalfa, and tomato) and monocots (rice, maize, sorghum, and Brachypodium distachyon), indicating that the IQM gene sequences in monocots and dicots are conserved and significantly different. In general, two genes from ortholog pairs in phylogenetic trees are often considered to have a common ancestor and may have similar functions (Fan et al., 2021). In our study, most of the closest ortholog gene pairs in monocotyledons comprised maize/sorghum and rice/Brachypodium distachyon, indicating that IQM genes from the four monocots species are more closely related to each other than to those from the same species in the three subfamilies. However, for dicotyledons, the closest gene pair were paralogous gene from the same species. Thus, the IQM family genes of dicots are more closely conserved than those of monocots. Nevertheless, the IQM genes of the same species were not clustered in the closest proximity within the dicot sub-branch, but were dispersed among different dicot species, indicating that there was a common IQM ancestor in different dicots. Further, the closestgene pairs were not found between monocotyledons and dicotyledons, indicating that the ancestral IQM genes did not exist before the dicot-monocot divergence.
According to a previous study, IQM is a calcium-independent calmodulin-binding protein family that contains one IQ motif with a sequence composition of IQxxxRGxxxR or its more relaxed version, [ILV]QxxxRxxxx [R, K] (Feng et al., 2014). The IQ motifs of 15 soybean IQM genes are listed in Table S1. The IQ motif in soybean IQM genes was [ILV]Qxxx[K/R]xxxxR, which is more inclined to its relaxed version; however, the amino acid sequence at position 6 of most IQ motifs was K instead of R, except GmIQM3a, GmIQM 3b, and GmIQM 3c. IQM can bind to CaMs through the IQ motif in Arabidopsis and rice, indicating that the IQ motif is key for the IQM gene to play a role in calcium signaling. To demonstrate the reliability of the predicted soybean IQ motif, we validated the role of the IQ motif using yeast two-hybrid and bimolecular fluorescence complementation (BiFC) experiments. Based on our results, the 15 soybean IQMs could bind to GmCaM protein in yeast or plant cells. However, the GmIQM protein-deleted IQ motif completely lost its ability to bind GmCaM, suggesting that the role of the IQ motif is conserved and crucial in Arabidopsis, rice and soybean. These results also indicate that the soybean IQM genes identified in this study were reliable.
Soybean IQM genes were divided into three subfamilies according to their sequences. This clustering is supported by the results of structural analysis of IQM genes and proteins. Members in the same groups were found to be highly conserved in terms of intron length, exon number, and motif distribution. For example, all five members of subfamily II contained eight exons, while three members of subfamily III contained nine exons (Figure 2),and the motifs 15 and 12 were unique to subfamilies I and II, respectively (Figure 3). The differentiation of gene sequences leads to diversity in gene function. This was confirmed by the isoelectric point (pI) values, subcellular localization, and gene expression analysis. The pI values of 15 IQM genes ranged from 6.24 to 9.45; however, the paralogous pairs or genes in the same subfamily share very similar parameters (Table 1). For example, the pI values of GmIQM1a/1b and GmIQM2a/2b were 9.29/9.13 and 6.42/6.24, respectively. The location patterns of 15 GmIQM genes matched the clustering results, with the closest related clade members having the same intracellular localization, such as GmIQM1a and GmIQM1b were localized only in the nucleus, whereas GmIQM3a and GmIQM3b in both the nucleus and cytoplasm, indicating that genes adjacent to each other on the phylogenetic tree may have the same or similar functions (Figure 6). In addition, the real-time PCR also revealed that the paralogous pairs had similar tissue expression patterns (Figure 7). These results provide strong evidence to support the clustering of soybean IQM family genes and imply the complex functions of GmIQM genes.
Members of a single-gene family may have different biological functions. In Arabidopsis, AtIQM1 modulates stomatal movement by affecting ROS levels (Zhou et al., 2012) and participates in plant disease response signaling by promoting the synthesis of JA (Lv et al., 2019); AtIQM4 is positively involved in seed dormancy and germination by regulating ABA content (Zhou et al., 2018); AtIQM5 regulates flowering by modulating the juvenile-to-adult transition (Gong et al., 2017) and interacts with IAAs, a key repressor of auxin signaling, to promote lateral root and callus formation (Zhang et al., 2022). These studies revealed multiple roles of AtIQM genes throughout the plant life cycle. However, the functions of the IQM genes in soybean have not been reported. To preliminarily explore the potential function of the GmIQMs, we conducted an expression analysis of the 15 GmIQM genes in different tissues or organs using qRT-PCR. The absence of GmIQM1a and GmIQM6b in eight different tissues indicates that these genes may be pseudogenes or may be expressed at a specific time point in the course of soybean life or in response to an external stimulus. GmIQM1c, GmIQM1e, GmIQM3a, GmIQM3b, and GmIQM3c were highly abundant in various tissues, suggesting that these genes may play multiple roles in soybean development. Finally, GmIQM1d and GmIQM5 were specifically and markedly expressed only in nodules, suggesting their important functions in nitrogen fixation or communication between plants and rhizosphere microorganisms. GmIQM6a was hardly expressed in other tissues, except flowers, indicating that this gene may only play a specific role during soybean flowering. These results imply that widely expressed genes may play a role in multiple aspects of the entire plant life course, and other IQM genes with little or no expression may be activated in response to certain environmental stimuli with irritability.
After treatment with NaCl, NaHCO3, PGE, SA, ABA, and MeJA, the expression levels of representative GmIQM genes (GmIQM1e, 1f, 2a, 2c, 3a, 3b, 3c, 6b) were significantly induced, especially with PEG and NaCl, indicating that GmIQMs may play an important role in salt and drought resistance. However, our results do not exclude the role of soybean IQM genes in the response to alkali and other abiotic stresses, and disease and insect invasion in soybean. The promoter analysis also showed that most soybean IQM genes contained multiple cis-acting elements related to abiotic and biotic stresses, but there were fewer related to biotic stresses. The results of the two parts confirmed each other, which further proved the credibility of the GmIQMs function in abiotic and biotic stress responses. There are six IQM genes, including GmIQM1a, 1b, 1d, 2b, GmIQM5 and 6a that are not expressed or expressed at very low levels in soybean leaves and roots, and whose transcript levels remain unchanged under various treatments (results are not shown), indicating that these genes may function only transiently at a particular time point.
In conclusion, fifteen IQM genes were identified in soybean. These genes have a variety of functions based on their structures, especially in response to abiotic stress (salt and drought) reactions in soybean, in which they may play an important role. Our systematic analysis of IQM family genes provides a theoretical foundation and a clear direction for subsequent in-depth research on the biological functions of each soybean IQM genes.
Data availability statement
The datasets presented in this article are not readily available because. Requests to access the datasets should be directed toY2hhbmdlbnRpYW5AYWxpeXVuLmNvbQ==.
Author contributions
CT, and TL designed the experiments, supervised the study, and managed the projects. TL performed most of the research and drafted manuscript. QL performed bioinformatics analysis and charting. HX participated in some of the experiments. TF, YZ, JW and CT analyzed and discussed the results. All authors contributed to the article and approved the submitted the version.
Funding
This work was funded by the National Natural Science Foundation of China (311770342, 32201800). This work was also supported by Natural Science Foundation of Guangdong Province (2020A1515011423), Major Program of Guangdong Basic and Applied Research (2021A1515110522).
Acknowledgments
We thank XIALAB of South China Agricultural University for help and communication on bioinformatics related issues.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1093589/full#supplementary-material
References
Abel, S., Savchenko, T., Levy, M. (2005). Genome-wide comparative analysis of the IQD gene families in arabidopsis thaliana and oryza sativa. BMC evolutionary Biol. 5, 72. doi: 10.1186/1471-2148-5-72
Aldon, D., Mbengue, M., Mazars, C., Galaud, J. P. (2018). Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 19 (3), 665. doi: 10.3390/ijms19030665
Ali, E., Raza, M. A., Cai, M., Hussain, N., Shahzad, A. N., Hussain, M., et al. (2020). Calmodulin-binding transcription activator (CAMTA) genes family: Genome-wide survey and phylogenetic analysis in flax (Linum usitatissimum). PloS One 15 (7), e0236454. doi: 10.1371/journal.pone.0236454
Bouché, N., Scharlat, A., Snedden, W., Bouchez, D., Fromm, H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277 (24), 21851–21861. doi: 10.1074/jbc.M200268200
Bouché, N., Yellin, A., Snedden, W. A., Fromm, H. (2005). Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 56, 435–466. doi: 10.1146/annurev.arplant.56.032604.144224
Bu, Q., Lv, T., Shen, H., Luong, P., Wang, J., Wang, Z., et al. (2014). Regulation of drought tolerance by the f-box protein MAX2 in arabidopsis. Plant Physiol. 164 (1), 424–439. doi: 10.1104/pp.113.226837
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Carafoli, E., Krebs, J. (2016). Why calcium? how calcium became the best communicator. J. Biol. Chem. 291 (40), 20849–20857. doi: 10.1074/jbc.R116.735894
Chen, X., Chen, Z., Zhao, H., Zhao, Y., Cheng, B., Xiang, Y. (2014). Genome-wide analysis of soybean HD-zip gene family and expression profiling under salinity and drought treatments. PloS One 9 (2), e87156. doi: 10.1371/journal.pone.0087156
Chin, D., Means, A. R. (2000). Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 10 (8), 322–328. doi: 10.1016/S0962-8924(00)01800-6
DeFalco, T. A., Bender, K. W., Snedden, W. A. (2009). Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 425 (1), 27–40. doi: 10.1042/BJ20091147
Dröge-Laser, W., Snoek, B. L., Snel, B., Weiste, C. (2018). The arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 45 (Pt A), 36–49. doi: 10.1016/j.pbi.2018.05.001
Edel, K. H., Marchadier, E., Brownlee, C., Kudla, J., Hetherington, A. M. (2017). The evolution of calcium-based signalling in plants. Curr. biology: CB 27 (13), R667–R679. doi: 10.1016/j.cub.2017.05.020
Fan, T., Lv, T., Xie, C., Zhou, Y., Tian, C. (2021). Genome-wide analysis of the IQM gene family in rice (Oryza sativa l.). Plants (Basel Switzerland) 10 (9), 1949. doi: 10.3390/plants10091949
Feng, L., Chen, Z., Ma, H., Chen, X., Li, Y., Wang, Y., et al. (2014). The IQD gene family in soybean: structure, phylogeny, evolution and expression. PloS One 9 (10), e110896. doi: 10.1371/journal.pone.0110896
Flagel, L. E., Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183 (3), 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Gong, L., Chen, J., Zhou, Y., Huang, X., Tian, C. E. (2016). Disruption of IQM5 delays flowering possibly through modulating the juvenile-to-adult transition. Acta Physiologiae Plantarum 39, 21. doi: 10.1007/s11738-016-2314-4
Hurst, L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends genetics: TIG 18 (9), 486. doi: 10.1016/s0168-9525(02)02722-1
Hyun, T. K., Eom, S. H., Han, X., Kim, J. S. (2014). Evolution and expression analysis of the soybean glutamate decarboxylase gene family. J. Biosci. 39 (5), 899–907. doi: 10.1007/s12038-014-9484-2
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in arabidopsis. Trends Plant Sci. 7 (3), 106–111. doi: 10.1016/S1360-1385(01)02223-3
Li, Y., Chen, Q., Nan, H., Li, X., Lu, S., Zhao, X., et al. (2017). Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PloS One 12 (6), e0179554. doi: 10.1371/journal.pone.0179554
Li, K., Ma, B., Shen, J., Zhao, S., Ma, X., Wang, Z., et al. (2021). The evolution of the expansin gene family in brassica species. Plant Physiol. biochemistry: PPB 167, 630–638. doi: 10.1016/j.plaphy.2021.08.033
Lv, T., Li, X., Fan, T., Luo, H., Xie, C., Zhou, Y., et al. (2019). The calmodulin-binding protein IQM1 interacts with CATALASE2 to affect pathogen defense. Plant Physiol. 181 (3), 1314–1327. doi: 10.1104/pp.19.01060
Lynch, M., Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Sci. (New York N.Y.) 290 (5494), 1151–1155. doi: 10.1126/science.290.5494.1151
Ma, H., Feng, L., Chen, Z., Chen, X., Zhao, H., Xiang, Y. (2014). Genome-wide identification and expression analysis of the IQD gene family in populus trichocarpa. Plant science: an Int. J. Exp. Plant Biol. 229, 96–110. doi: 10.1016/j.plantsci.2014.08.017
Mei, C., Liu, Y., Dong, X., Song, Q., Wang, H., Shi, H., et al. (2021). Genome-wide identification and characterization of the potato IQD family during development and stress. Front. Genet. 12, 693936. doi: 10.3389/fgene.2021.693936
Mengarelli, D. A., Zanor, M. I. (2021). Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 253 (1), 15. doi: 10.1007/s00425-020-03537-5
Meng, X., Wang, C., Rahman, S. U., Wang, Y., Wang, A., Tao, S. (2015). Genome-wide identification and evolution of HECT genes in soybean. Int. J. Mol. Sci. 16 (4), 8517–8535. doi: 10.3390/ijms16048517
Nekrutenko, A., Makova, K. D., Li, W. H. (2002). The K(A)/K(S) ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res. 12 (1), 198–202. doi: 10.1101/gr.200901
Panchy, N., Lehti-Shiu, M., Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171 (4), 2294–2316. doi: 10.1104/pp.16.00523
Ranty, B., Aldon, D., Cotelle, V., Galaud, J. P., Thuleau, P., Mazars, C. (2016). Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 7, 327. doi: 10.3389/fpls.2016.00327
Reddy, A. S., Ali, G. S., Celesnik, H., Day, I. S. (2011). Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23 (6), 2010–2032. doi: 10.1105/tpc.111.084988
Reddy, A. S., Day, I. S. (2001). Analysis of the myosins encoded in the recently completed arabidopsis thaliana genome sequence. Genome Biol. 2 (7), RESEARCH0024. doi: 10.1186/gb-2001-2-7-research0024
Rhoads, A. R., Friedberg, F. (1997). Sequence motifs for calmodulin recognition. FASEB journal: Off. Publ. Fed. Am. Societies Exp. Biol. 11 (5), 331–340. doi: 10.1096/fasebj.11.5.9141499
Saijo, Y., Loo, E. P. (2020). Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 225 (1), 87–104. doi: 10.1111/nph.15989
Sankoff, D. (2001). Gene and genome duplication. Curr. Opin. Genet. Dev. 11 (6), 681–684. doi: 10.1016/S0959-437X(00)00253-7
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463 (7278), 178–183. doi: 10.1038/nature08670
Talke, I. N., Blaudez, D., Maathuis, F. J., Sanders, D. (2003). CNGCs: prime targets of plant cyclic nucleotide signalling? Trends Plant Sci. 8 (6), 286–293. doi: 10.1016/S1360-1385(03)00099-2
Truman, W., Sreekanta, S., Lu, Y., Bethke, G., Tsuda, K., Katagiri, F., et al. (2013). The CALMODULIN-BINDING PROTEIN 60 family includes both negative and positive regulators of plant immunity. Plant Physiol. 163 (4), 1741–1751. doi: 10.1104/pp.113.227108
Wang, M., Chen, B., Zhou, W., Xie, L., Wang, L., Zhang, Y., et al. (2021). Genome-wide identification and expression analysis of the AT-hook motif nuclear localized gene family in soybean. BMC Genomics 22 (1), 361. doi: 10.1186/s12864-021-07687-y
Wang, L., Guo, K., Li, Y., Tu, Y., Hu, H., Wang, B., et al. (2010). Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10, 282. doi: 10.1186/1471-2229-10-282
Wang, X., Wu, M. H., Xiao, D., Huang, R. L., Zhan, J., Wang, A. Q., et al. (2021). Genome-wide identification and evolutionary analysis of RLKs involved in the response to aluminium stress in peanut. BMC Plant Biol. 21 (1), 281. doi: 10.1186/s12870-021-03031-4
Williams, R. J. (1992). Calcium and calmodulin. Cell calcium 13 (6-7), 355–362. doi: 10.1016/0143-4160(92)90049-X
Wu, M., Li, Y., Chen, D., Liu, H., Zhu, D., Xiang, Y. (2016). Genome-wide identification and expression analysis of the IQD gene family in moso bamboo (Phyllostachys edulis). Sci. Rep. 6, 24520. doi: 10.1038/srep24520
Yang, T., Poovaiah, B. W. (2003). Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 8 (10), 505–512. doi: 10.1016/j.tplants.2003.09.004
Zhang, X., Feng, Y., Cheng, H., Tian, D., Yang, S., Chen, J. Q. (2011). Relative evolutionary rates of NBS-encoding genes revealed by soybean segmental duplication. Mol. Genet. genomics: MGG 285 (1), 79–90. doi: 10.1007/s00438-010-0587-7
Zhang, M., Liu, Y., Shi, H., Guo, M., Chai, M., He, Q., et al. (2018). Evolutionary and expression analyses of soybean basic leucine zipper transcription factor family. BMC Genomics 19 (1), 159. doi: 10.1186/s12864-018-4511-6
Zhang, Z., Zhao, Y., Feng, X., Luo, Z., Kong, S., Zhang, C., et al. (2019). Genomic, molecular evolution, and expression analysis of NOX genes in soybean (Glycine max). Genomics 111 (4), 619–628. doi: 10.1016/j.ygeno.2018.03.018
Zhang, H., Zhu, J., Gong, Z., Zhu, J. K. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23 (2), 104–119. doi: 10.1038/s41576-021-00413-0
Zhao, W., Cheng, Y. H., Zhang, C., Shen, X. J., You, Q. B., Guo, W., et al. (2017). Genome-wide identification and characterization of the GmSnRK2 family in soybean. Int. J. Mol. Sci. 18 (9), 1834. doi: 10.3390/ijms18091834
Zhao, K., Chen, S., Yao, W., Cheng, Z., Zhou, B., Jiang, T. (2021). Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 21 (1), 122. doi: 10.1186/s12870-021-02879-w
Zhou, Y., Chen, Y., Yamamoto, K. T., Duan, J., Tian, C. E. (2010). Sequence and expression analysis of the arabidopsis IQM family. Acta Physiologiae Plantarum 32 (1), 191–198. doi: 10.1007/s11738-009-0398-9
Zhou, Y., Duan, J., Fujibe, T., Yamamoto, K. T., Tian, C. E. (2012). AtIQM1, a novel calmodulin-binding protein, is involved in stomatal movement in arabidopsis. Plant Mol. Biol. 79 (4-5), 333–346. doi: 10.1007/s11103-012-9915-0
Zhou, Z., Lakhssassi, N., Knizia, D., Cullen, M. A., El Baz, A., Embaby, M. G., et al. (2021). Genome-wide identification and analysis of soybean acyl-ACP thioesterase gene family reveals the role of GmFAT to improve fatty acid composition in soybean seed. Theor. Appl. Genet. 134 (11), 3611–3623. doi: 10.1007/s00122-021-03917-9
Zhou, Y., Wu, J., Xiao, W., Chen, W., Chen, Q., Fan, T., et al. (2018). Arabidopsis IQM4, a novel calmodulin-binding protein, is involved with seed dormancy and germination in arabidopsis. Front. Plant Sci. 9, 721. doi: 10.3389/fpls.2018.00721
Keywords: IQM, identification, family analysis, soybean, CaMBP
Citation: Lv T, Liu Q, Xiao H, Fan T, Zhou Y, Wang J and Tian C-e (2023) Genome-wide identification and analysis of the IQM gene family in soybean. Front. Plant Sci. 13:1093589. doi: 10.3389/fpls.2022.1093589
Received: 09 November 2022; Accepted: 13 December 2022;
Published: 06 January 2023.
Edited by:
Humira Sonah, Laval University, CanadaReviewed by:
Shumayla., Panjab University, IndiaSantosh Rajput, Dryland Genetics Inc., United States
Giriraj Kumawat, ICAR Indian Institute of Soybean Research, India
Copyright © 2023 Lv, Liu, Xiao, Fan, Zhou, Wang and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianxiao Lv, bGpsX3Z2QGd6aHUuZWR1LmNu; Chang-en Tian, Y2hhbmdlbnRpYW5AYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work
 Tianxiao Lv
Tianxiao Lv Qiongrui Liu
Qiongrui Liu Hong Xiao
Hong Xiao Tian Fan
Tian Fan Yuping Zhou
Yuping Zhou Jinxing Wang2
Jinxing Wang2 Chang-en Tian
Chang-en Tian