
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 15 December 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1087388
This article is part of the Research TopicFormation, Regulation and Affecting Factors of Fruit Quality of Horticultural CropsView all 7 articles
 Xiaoshan Jin1†
Xiaoshan Jin1† Chenxia Cheng1†
Chenxia Cheng1† Qi Qi1
Qi Qi1 Suping Zhou2
Suping Zhou2 Caihong Wang1
Caihong Wang1 Yong Zhang1
Yong Zhang1 Chao Sun1
Chao Sun1 Yuling Wang1
Yuling Wang1 Ruihong Dang3*
Ruihong Dang3* Shaolan Yang1*
Shaolan Yang1*The hard-end is a disorder of pear fruit, however, the mechanisms underlying its development remain unknown. In this study, we found that the hard-end fruit contained a higher transcript abundance level of ethylene-response factor 1b-like (PpERF1b-like) and released more ethylene compared to normal pear. In the ethephon treated normal fruit, flesh tissues accumulated more lignin together with elevated expression of PpERF1b-like. Overexpressing PpERF1b-like transiently in fruit and stably in callus increased lignin accumulation and the expression of lignin biosynthesis genes; the opposite results were observed in fruit showing repressed expression of PpERF1b-like. These results confirmed the role of PpERF1b-like in promoting hard-end formation through promoting lignin synthesis. This study provided valuable information for further clarifying the regulation of hard-end formation in pear.
Hard-end fruit is a physiological disorder affecting the marketable quality of pear (Pyrus pyrifolia) (Raese, 1993). In recent years, it has been occurring very frequently on pear ‘Whangkeumbae’ (P. pyrifolia). The hard-end fruits have protruding calyx with copper-green tough apex (Raese and Drake, 2006). Lu et al. (2015) reported that the hard-end pears contain higher lignin content and more stony cells compared to normal pears, especially a large amount of lignin is deposited in the peel tissue. Lignin deposition on cell wall is critical for plant growth and development (Dixon et al., 2001; Rogers and Campbell, 2004). The secondary cell wall of stony cell is mainly composed of lignin and cellulose (Zhang et al., 2021), and the biosynthesis and deposition of lignin and cellulose in pear flesh are closely related to the generation of stony cells (Cai et al., 2010; Brahem et al., 2017).
The development of hard-end fruit is usually accompanied by lignification (Feng et al., 2021), but the correlation between the two processes remains to be clarified. Ethylene plays a regulatory role in many processes during plant growth and development (Abeles et al., 1992). Studies have shown that ethylene promotes lignification in bamboo shoot (Luo et al., 2021), mung bean (Huang et al., 2013), and loquat (Shan et al., 2008). Ethylene-response factor (ERF) belongs to the AP2/ERF (APETALA2/Ethylene-Responsive Factor) transcription factor (TF) superfamily, it plays some roles in providing resistance to biotic and abiotic stresses (Xu et al., 2011). In ethylene treated banana fruit, the expression levels of MaERF9 and MaERF11 were increased, which concurs with the involvement of AP2/ERF TFs in ethylene biosynthesis and signal transduction (Xiao et al., 2013).
The first ERF TF was isolated from tobacco (Nicotiana tabacum) as a critical gene in ethylene signal transduction pathway of plants (Ohme-Takagi and Shinshi, 1995; Bleecker and Kende, 2000). In recent years, roles of ERF TFs in regulating responses to stress-induced ethylene accumulation have been studied on tomato (Liu et al., 2014; Nakano et al., 2014), kiwifruit (Yin et al., 2012), grape (Licausi et al., 2010), apple (Wang et al., 2007), sweet orange (Xie et al., 2014), and melon (Ma et al., 2015). Additionally, the ERF TFs can also affect the lignification process of plants by influencing lignin biosynthesis. For instance, the EjAP2-1 which was isolated from loquat (Eriobotrya japonica), was reported to inhibit the activity of promoter for the lignin biosynthesis gene Ej4CL1, and enhance the activation effect of EjMYB2 on lignin biosynthesis by interacting with EJMYB1/2, thus negatively regulating the lignification of loquat fruit (Zeng et al., 2015). Guo et al. (2016) also reported that in island cotton, GbERF1-like up-regulates the expression of genes in lignin biosynthesis pathway, and the increased lignin accumulation helps to improve the resistance of cotton to verticillium wilt.
In our studies, the expression of PpERF1b-like TF was found to be significantly up-regulated in hard-end pears by using RNA-Seq analysis (Li et al., 2019) and RT-qPCR verification. The function and mechanism of PpERF1b-like TF were investigated using transient overexpression of the gene in normal fruit and transgenic calli derived from flesh tissues of pear ‘Whangkeumbae’. The results presented here will not only help understanding the hand-end formation in pear fruit, but also the process of lignin biosynthesis and its interaction with ethylene release.
Ten-year-old pear ‘Whangkeumbae’ trees were grown in two orchards located in Laiyang City, Shandong Province, China. Normal fruit were picked in the orchard with low incidence rate (Orchard 1), and the hard-end fruit in the orchard with high incidence rate (Orchard 2). Pears were harvested at 90 and 120 days after anthesis, respectively. Samples were transported to the laboratory within two hours after picking from the trees. Normal fruit of uniform size with no defects stored at 20 °C were used in fruit injection experiments for studying transient overexpression of genes. After removal of seeds or peel, fruit flesh tissues were cut into small pieces (approximately 1 cm3), immediately frozen in liquid nitrogen, and stored at -80 °C until analysis.
The ethylene production rate was measured using fruit harvested at 90 and 120 days after anthesis by using a GC-2010 Plus Gas Chromatograph (Shimadzu, Kyoto, Japan). The hard-end and normal fruit with no visible defects (including damages from wound, disease or insects) were selected and then placed in a closed container for 1 hour. Gas was extracted, and ethylene content was determined according to Yang et al. (2015). Five fruits per sampling point and 3 biological replicates per treatment group were used in ethylene assay.
The total RNA was extracted from flesh tissues of pear ‘Whangkeumbae’ using RNA Plant Reagent (TianGen, Shanghai, China) according to the manufacturer’s instructions. The first strand cDNA was synthesized with 1 μg RNA using the HiScript II Q RT SuperMix for qPCR (+ gDNA wiper) (Vazyme, Nanjing, China). The RT-qPCR was performed on a Light Cycler® 480 instrument (Roche, Basel, Switzerland), using a program comprising an initial denaturation at 94 °C for 5 min, followed by 42 cycles of 94 °C for 15 s and 60 °C for 1 min. A negative control without template cDNA for each primer pair was included in each run. For each group of samples, three biological replicates and three technical replicates/sample were included. A pear Actin gene (LOC125473976, NCBI database) was used as the house keeping gene (internal reference) to normalize the relative gene transcript abundance levels. The relative gene expression was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2000). The primers used for the RT-qPCR analysis were designed using NCBI Primer designing tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and are listed in Table 1.
Isolation of total RNA and synthesis of cDNA followed the same protocol used in RT-qPCR analysis. The PCR reaction was conducted using a program comprising of initial denaturation at 94°C for 5 minutes followed by 35 cycles of 94°C for 30 seconds, 58°C for 30 seconds and 72°C for 1 minute; with a final extension at 72°C for 10 minutes. The PCR primers used to clone PpERF1b-like are listed in Table 2. The open reading frame (ORF) of PpERF1b-like was amplified using the Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China). The PCR products were cloned onto pMD-19-T vector (Takara, Dalian, China).
The full-length cDNA and the antisense fragment of PpERF1b-like that were isolated by digestion with restriction enzymes Xba I and Sma I were ligated onto the expression vector, Super1300, driven a 35S promoter. The fusion vectors, Super1300-sense-PpERF1b-like and Super1300-antisense-PpERF1b-like were introduced into Agrobacterium tumefaciens GV3101 using the freeze-thaw method (Weigel and Glazebrook, 2006). The transient overexpression assay of sense-PpERF1b-like, antisense-PpERF1b-like and empty vector in ‘Whangkeumbae’ normal pear fruit was conducted according to a method described previously (Li et al., 2019). The primers used to construct the expression vectors are listed in Table 2.
Subcellular localization of the PpERF1b-like TF in onion (Allium cepa) cells was conducted using A. tumefaciens harboring the pSuper1300-PpERF1b-like following the method of Sun et al. (2007) with some modifications. Onion scales were incubated at 28 °C for 24 h in dark. For infection, the onion peels were submerged in A. tumefaciens suspension (OD600 = 0.6-0.8) and supplemented with acetosyringone (20 mg/L) for 15-20 min. The onion peels were then transferred to 1/2 MS solid medium supplemented with acetosyringone (20 mg/L) followed by incubation at 28 °C for 24 hours in dark. The infected cells producing green fluorescent protein (GFP) were observed and imaged using EVOS™ FL Auto 2 Imaging System (EVOS FL AUTO 2.0, Thermo Fisher, Waltham, America). Positive signals indicate the subcellular localization of PpERF1b-like TF.
During harvest season of pear ‘Whangkeumbae’, normal and hard-end fruit were treated with ethephon, and water was used as non-treated control. One half of the fruit was soaked in 400 mg/L ethephon solution and the other half in water, each for 5 minutes. The treated fruits were stored for 3 days before stained with phloroglucinol.
The Wiesner reaction assay using phloroglucinol-HCl staining was conducted using tissue sections in order to visualize lignified structures (Blanco-Portales et al., 2002). Sections of 2 mm in thickness were cut along the equator of the fruit, and stained with Wiesner reagent for 10 min. Lignified tissues appeared pink or fuchsia in color. Images were captured using a camera (ZXUS 230 HS; Canon, Tokyo, Japan). After staining in the Wiesner reagent for 5 minutes, the thin-sections were observed under a Leica Fluorescence Microscope and imaged using the CCD image acquisition system on the same microscope. The lignin content was measured as previously described by Dyckmans et al. (2002). The stone cell area was counted by Adobe Photoshop 2020.
The pear calli were induced from the flesh of ‘Nanshui’ (P. pyrifolia) of fruitlets in our laboratory, and the pear calli were cultured following the method of Wang et al. (2021). The calli were transformed with pSuper1300-PpERF1b-like-GFP and pSuper1300-GFP as the empty vector control, using the Agrobacterium-mediated genetic transformation method described by Wang et al. (2021) with some modifications. Pear calli were inoculated with A. tumefaciens GV3101 harboring the vector constructs after incubation for 20-30 min at 24 °C. After co-culture on plates containing acetosyringone (100 µg/mL) in dark for 3 days, calli were sub-cultured onto selection plates containing cephalosporin (300 mg/L) and hygromycin (10 mg/L) for 25 days. Newly-formed calli were subcultured to fresh selection media monthly until tissues with no sign of brown and died cells were obtained. Tissue culture was conducted at 24 °C in a growth chamber.
Data were analyzed using Microsoft Excel software. Significant differences between sample groups were calculated using GraphPad Prism 8 and data were graphed in the same program.
Ethylene production rate was measured for normal and hard-end fruit with obvious phenotype at 90 and 120 days after anthesis (Figure 1A), respectively. The ethylene production rate of hard-end fruit was higher than the normal fruit on both sampling dates (Figure 1B). The results showed that ethylene production was positively correlated with the occurrence of hard-end pear fruit.

Figure 1 Ethylene production rate of ‘Whangkeumbae’ at 90 and 120 days after anthesis. (A) Phenotypes of normal and hard-end fruit of pear ‘Whangkeumbae’ at 90 and 120 days after anthesis. Scale bars = 150μm. (B) Ethylene production rate of normal and hard-end fruit at 90 and 120 days after anthesis. Data are given as mean ± SD (n = 4). The asterisk represents a significant difference (***p <0.001; Student’s t-test).
PpERF1b-like was identified as one of the different expressed genes (DEGs) between hard-end and normal fruit harvested on 90 and 120 days after anthesis. The raw RNA seq sequences were deposited in NCBI database (NCBI SRA Accession: SRP063385). In this study, the transcript level of PpERF1b-like in pear ‘Whangkeumbae’ was higher in hard-end fruit harvested on the 90 days after anthesis (Figure 2A). Moreover, results from RT-qPCR analysis showed that the relative transcript abundance level of PpERF1b-like in hard-end fruit was significantly higher than that in normal fruit from the two harvests (Figure 2B). These data indicate a positive correlation between the expression of PpERF1b-like and the occurrence of hard-end fruit. A phylogenetic tree was constructed using the amino acid sequence of PpERF1b-like and other ERFs from a variety of plant species. The results showed that PpERF1b-like protein has the closest relationship with PbrERF1b protein from P. bretschneideri Rehd (Figure 2C). The analysis for subcellular localization confirmed that the PpERF1b-like-GFP fusion protein was localized in the nuclei of onion epidermal cells, indicating that the PpERF1b-like is a nucleus protein (Figure 2D).
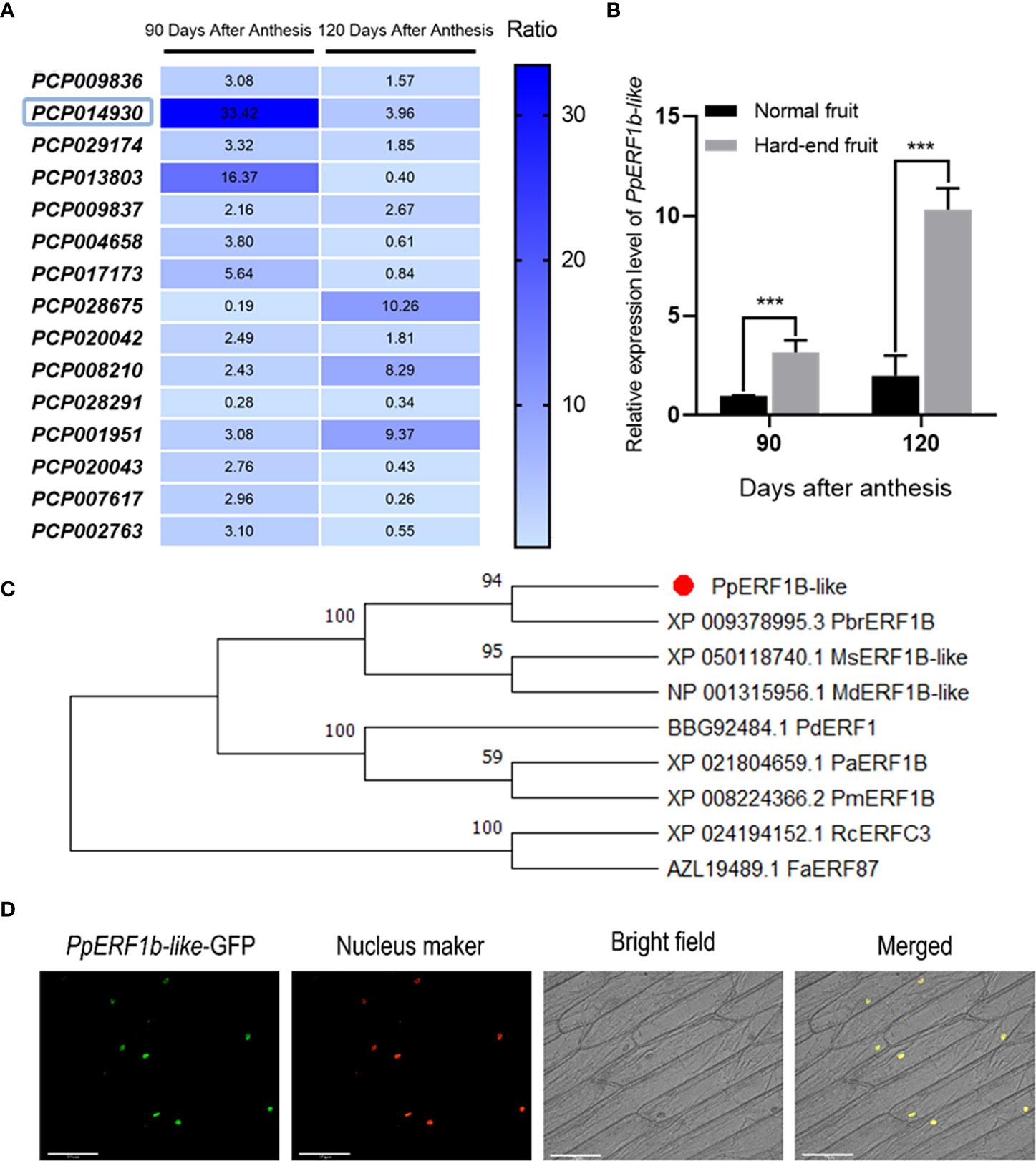
Figure 2 Transcriptome sequencing and subcellular localization of PpERF1b-like. (A) Screening of PpERF1b-like by transcriptome sequencing. (B) The relative expression level of PpERF1b-like in normal and hard-end fruit of pear ‘Whangkeumbae’ at 90 and 120 days after anthesis. Data are given as mean ± SD (n = 4). The asterisk represents a significant difference (***p <0.001; Student’s t-test). (C) Phylogenetic analysis of PpERF1b-like. PpERF1b-like protein sequences (indicated by red figure) were aligned with ERFs from P. bretschneideri Rehd (Pbr), Malus sylvestris (Ms), M. domestica (Md), P. dulcis (Pd), P. avium (Pa), P. mume (Pm) and Rosa chinensis (Rc). The phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replications in MEGA 11 software. Sequences were obtained from the National Center for Biotechnology Information nucleotide database (http://www.ncbi.nlm.nih.gov/nucleotide/). Bootstrap values are indicated above the branches. (D) Subcellular localization of PpERF1b-like-green fluorescent protein (GFP) fusion protein was determined by infecting onion epidermal cells and imaged under a EVOS FL Auto 2 imaging system. Scale bars = 125μm.
The effect of ethephon treatment on fruit lignification was further analyzed using normal and hard-end fruits treated with 400 mg/L ethephon. Based on the intensity of lignin staining, both normal and hard-end fruit accumulated more lignin after ethephon treatments compared to the control group (Figure 3A). The lignin content of ethephon treated fruits was significantly higher than that of the water-treated control samples (Figure 3B). The relative expression level of PpERF1b-like showed the same pattern with higher abundance of transcripts in ethephon-treated compared to the non-treated groups (Figure 3C). These results indicate that accumulation of lignin and the expression of PpERF1b-like were enhanced by ethephon treatment of pears.
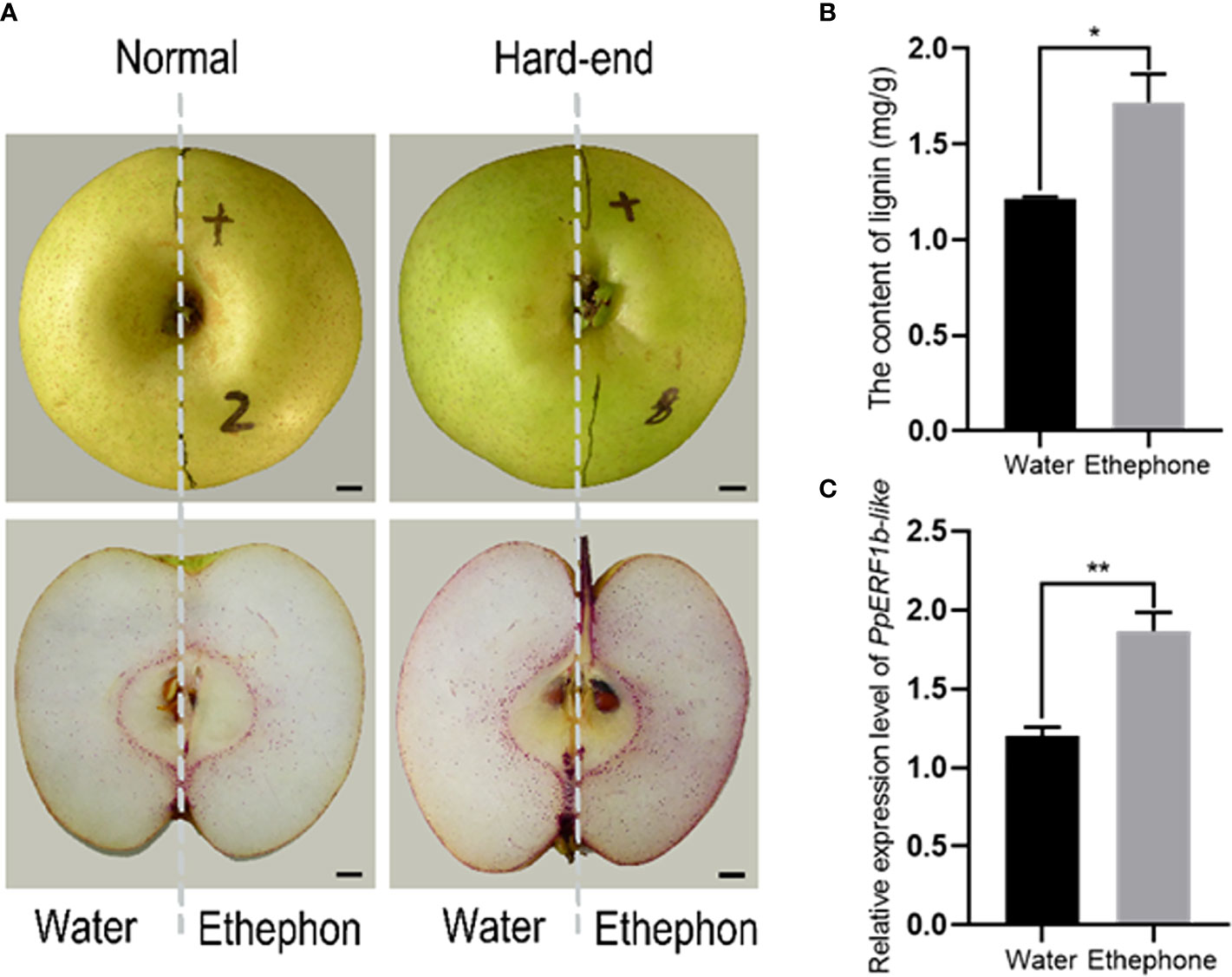
Figure 3 Ethephon promoted lignin accumulation and induced the expression of PpERF1b-like. (A) Half of the fruit was treated with water and the other half with ethephon. Scale bars = 100μm. (B), The content of lignin. (C) The relative expression level of PpERF1b-like. Data are given as mean ± SD (n = 3). The asterisk represents a significant difference (*p <0.05; **p <0.01) using Student’s t-test.
On the fruit, the sites inoculated with the PpERF1b-like overexpression vector appeared to be more lignified (Figure 4A). The expression of PpERF1b-like was analyzed in fruit tissues surrounding the injection site. The relative expression level of PpERF1b-like in fruit injected with the sense-PpERF1b-like vector was significantly higher than the control (inoculated with empty vector), whereas it was significantly repressed by inoculation of the antisense-PpERF1b-like vector (Figure 4B).

Figure 4 The relative expression level of PpERF1b-like in pear fruit surrounding the site of infiltration.in transient overexpression fruit. (A) Phenotype of transient overexpression of fruit. Scale bars = 100μm. (B) The relative expression level of PpERF1b-like in pear fruit surrounding the site of infiltration. Data are given as mean ± SD (n = 4). a-c, the different letters indicate significant differences between groups (p <0.05; Duncan’s multiple-range test). Ev: Empty vector, Sense: Sense-PpERF1b-like, Antisense: Antisense-PpERF1b-like.
In order to clarify the function of PpERF1b-like in regulating lignin biosynthesis in pear, the thin-sections of fruit inoculated with the gene constructs were stained in Wiesner reagent. As shown in Figure 5A, the lignin stain of fruit with transient overexpression of PpERF1b-like gene was much deeper than the transient overexpression assay of antisense-PpERF1b-like and empty vector. The frozen tissue section prepared using the inoculated fruit also showed that the number of stony cells in the injection site of PpERF1b-like gene increased significantly, whereas the injection site with antisense-PpERF1b-like contained a smaller number of stony cells (Figure 5A). Furthermore, the fruit injection site with PpERF1b-like gene has the largest area occupied with stony cells (Figure 5B). These results demonstrated that PpERF1b-like has a positive regulatory effect on fruit lignification of ‘Whangkeumbae’ pear. The expression of lignin biosynthesis related genes was analyzed in tissues collected around the injection site. As shown in Figures 5C–F, the relative expression levels of four genes related to lignin biosynthesis, Pp4CL3 (Figure 5C), PpCAD1 (Figure 5D), PpCAD2 (Figure 5E) and PpCCR (Figure 5F), were significantly higher in fruit showing transient overexpression of PpERF1b-like, compared to the control group. Conversely, the expression of these genes was repressed in fruit with overexpression of antisense-PpERF1b-like. When combining all the data from lignin staining and changes in the expression of genes affecting lignin biosynthesis, it can be concluded that the PpERF1b-like gene has a positive regulatory function during lignin synthesis which affects the occurrence of hard-end fruit.
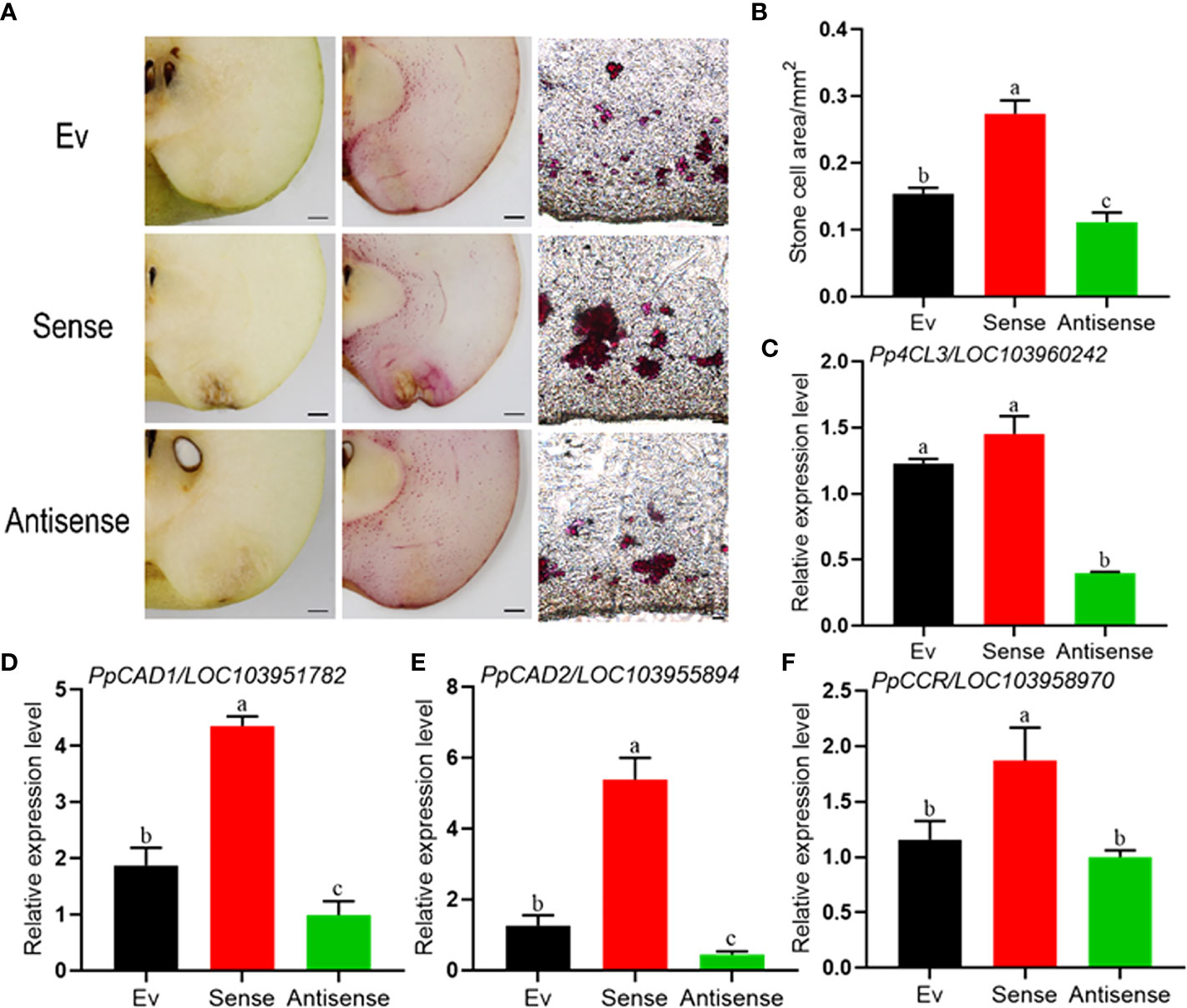
Figure 5 Transient overexpression of sense-PpERF1b-like promotes lignin synthesis and the relative expression levels of lignin synthesis related genes in pear fruit surrounding the site of infiltration. (A) Lignin staining and frozen section staining in the injection site of transient overexpression fruit. Ev: Empty vector, Sense: sense-PpERF1b-like, Antisense: antisense-PpERF1b-like. Scale bars = 100μm. (B) The area of stone cells in the injection site. Data are given as mean ± SD (n=4). Bars with different letters indicate significant differences between groups (p <0.05; Student’s t-test). (C–F) The relative expression levels of lignin synthesis related genes (Pp4CL3, PpCAD1, PpCAD2, PpCCR) in transient overexpression fruit. Data are given as mean ± SD (n=4). a-c, the different letters indicate significant differences between groups (p <0.05; Duncan’s multiple-range test). Ev: Empty vector, Sense: sense-PpERF1b-like, Antisense: antisense-PpERF1b-like.
Transgenic calli with stable overexpression of PpERF1b-like were stained with Wiesner reagents (Figures 6A, B). Calli infected with PpERF1b-like turned red which corresponds to high lignin content, whereas no such changes were shown in calli inoculated with the control (empty vector). The relative expression level of PpERF1b-like in PpERF1b-like-overexpressing pear calli was significantly higher than the control transformed with the empty vector (Figure 6C). The relative expression levels of lignin biosynthesis related genes, Pp4CL3 (Figure 6D), PpCAD1 (Figure 6E), PpCAD2 (Figure 6F) and PpCCR (Figure 6G), were also significantly higher in PpERF1b-like-overexpressing calli than those from the control group (Figures 6D–G). These results further validated the role of PpERF1b-like TF in enhancing lignin biosynthesis.
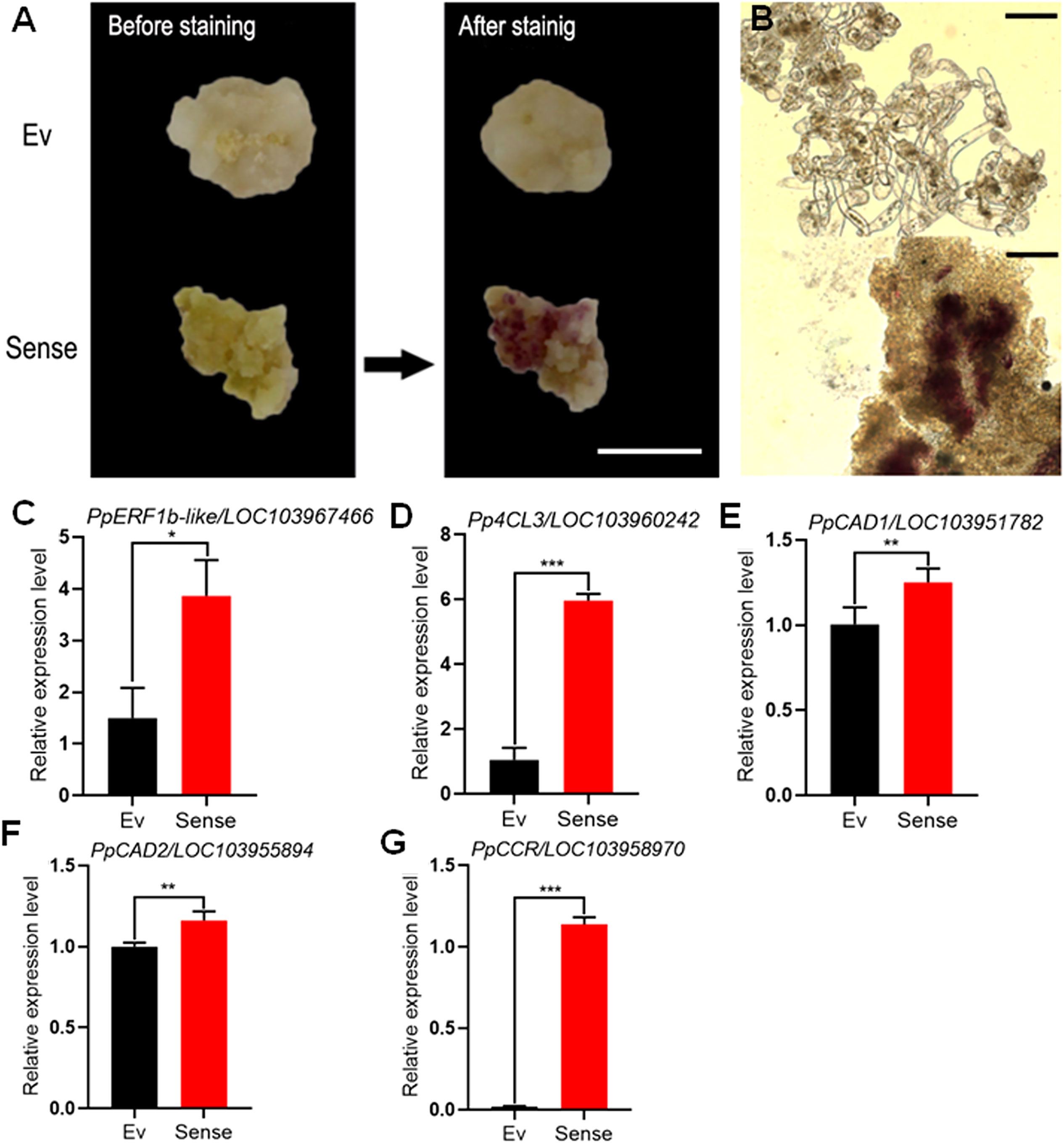
Figure 6 Genetic transformation of callus. (A) Whole tissue lignin staining. Scale bar = 100μm. Ev: Empty vector, Sense: sense-PpERF1b-like. (B) Lignin staining of thin-sections of pear. Scale bars = 50μm. (C) The relative expression level of PpERF1b-like in transgenic callus. Data are given as mean ± SD (n = 4). The asterisk represents a significant difference (*p < 0.05; **p < 0.01; Student’s t-test). (D–G) The relative expression levels of lignin biosynthesis related genes (Pp4CL3, PpCAD1, PpCAD2, PpCCR) in transgenic calli. Data are given as mean ± SD (n = 4). The asterisk represents a significant difference (*p < 0.05; **p < 0.01; ***p < 0.001; Student’s t-test). Ev: Empty vector, Sense: sense-PpERF1b-like.
APETALA2/Ethylene Response Factor (AP2/ERF) TFs comprise one of the largest TF families in plants, and they are involved in a variety of signal transduction and growth regulation (Gu et al., 2017). The ERF subfamily is primarily located downstream of ethylene signal transduction and is involved in many biochemical pathways, such as metabolic regulation (Yu et al., 2012), responses to biotic and abiotic stress factors (Yao et al., 2017), hormone signal transduction pathways in plants (Müller and Munné-Bosch, 2015) and plant development (Banno et al., 2001). This study provided experimental evidences for the function of an ethylene response factor, PpERF1b-like, isolated from pear ‘Whangkeumbae’. In fruit harvested at 90 and 120 days after anthesis, the ethylene production rate and the relative expression level of PpERF1b-like in hard-end fruit were both higher than those in normal fruit. It can thus postulate that the expression level of PpERF1b-like is positively related with the physiological disorder in pear.
Lignin is a group of aromatic polymers formed by oxidative coupling of 4-hydroxyphenylpropane through phenylpropane metabolic pathway (Boerjan et al., 2003; Ralph et al., 2004). When plants are challenged with stress conditions, lignin deposition on the secondary cell wall leads to wall thickening which increases the hardness of fruit (Tao et al., 2009; Vanholme et al., 2010). This study shows that ethephon treatment promoted the accumulation of lignin, and the number of stone cells in the treated fruits was also increased as well as the expression of ethylene response factor PpERF1b-like. In a study using ethephon treated mung bean roots, it induced expression of peroxidase (POD) which is a key gene catalyzing lignin formation, and more extensive lignification of stems (Huang et al., 2013). As a response factor of ethylene, PpERF1b-like may also participate in the regulation of lignin biosynthesis similarly, and eventually affects the occurrence of hard-end fruit in pear.
Based on results from lignin staining, pear flesh tissues became more lignified when they were injected with the sense-PpERF1b-like-overexpressing vector, and concurrently, the number of stony cells was also increased, compared to the tissues injected with empty vector. However, there was no significant change in the degree of lignification in tissues injected with antisense-PpERF1b-like in spite of fewer stony cells. In Wiesner staining, pear calli infected with PpERF1b-like turned red, but this reaction was not obvious in pear calli infected with empty vector. Our results are consistent with a previous study on pear ‘Whangkeumbae’ where lignin staining, lignin content and the number of stony cells in hard-end fruit were all significantly higher than normal fruit (Lu et al., 2015). Taken together, these results indicate that PpERF1b-like should positively regulate the synthesis of lignin.
According to a study on Isatis indigotica, the AP2/ERF TFs positively regulate lignin biosynthesis by activating salicylic acid signal transduction and lignin pathway genes (Ma et al., 2017). Lasserre et al. (2008) also found that the expression level of AtERF38 gene was positively correlated with the thickening of secondary wall in seeds and this process is intertwined with lignin biosynthesis. In this study, overexpressing sense-PpERF1b-like transiently in pear ‘Whangkeumbae’ fruit or stably in calli both led to an elevated levels of expression of PpERF1b-like and lignin biosynthesis related genes. On the contrary, expression of these genes were repressed in the fruit injected with antisense-PpERF1b-like. Future studies will investigate if the PpERF1b-like gene directly or indirectly regulate lignin biosynthesis genes, as well as its connection to the formation of hard-end pears.
This study reports that the relative expression level of PpERF1b-like was positively correlated with higher lignin content in hard-end pears, indicating that PpERF1b-like is a key gene affecting fruit lignification. Results from transient expression of PpERF1b-like in normal fruit of pear ‘Whangkeumbae’ and in callus tissue provided experimental evidences supporting the role of PpERF1b-like in regulating the expression of lignin-related genes, and affecting the occurrence of fruit calyx end hardening.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
Publicly available datasets were analyzed in this study. This data can be found here: NCBI SRA Accession: SRP063385.
Study design: CC, RD, and SY. Data collection: CC, YZ, CS, and QQ. Data analysis: XJ, CC, CW, and SY. Data interpretation: CC. Software: XJ, RD, and YZ. Supervision: CW, RD, and SY. Visualization: XJ, QQ, and RD. Writing—original draft: XJ and SZ. Writing—review and editing: XJ, CC, SZ, RD and SY. All authors contributed to the article and approved the submitted version.
This work was supported by the Project of Shandong Natural Science Foundation (ZR2021MC005, ZR2017MC006), Project of National Natural Science Foundation of China (31201608), the Breeding Plan of Shandong Provincial Qingchuang Research Team (2019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abeles, F. B., Morgan, P. W., Saltveit, M. E., Jr. (1992). Ethylene in plant biology (San Diego: Academic Press).
Banno, H., Ikeda, Y., Niu, Q. W., Chua, N. H. (2001). Overexpression of arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell. 13, 2609–2618. doi: 10.1105/tpc.010234
Blanco-Portales, R., Medina-Escobar, N., López-Ráez, J. A., González-Reyes, J. A., Villalba, J. M., Moyano, E., et al. (2002). Cloning, expression and immunolocalization pattern of a cinnamyl alcohol dehydrogenase gene from strawberry (Fragaria x ananassa cv. chandler). J. Exp. Bot. 53 (375), 1723–1734. doi: 10.1093/jxb/erf029
Bleecker, A. B., Kende, H. (2000). Ethylene: A gaseous signal molecule in plants. Ann. Rev. Cell Dev. Biol. 16 (1), 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Boerjan, W., Ralph, J., Baucher, M. (2003). Lignin biosynthesis. Ann. Rev. Plant Biol. 54 (1), 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Brahem, M., Renard, C. M. G. C., Gouble, B., Bureau, S., Le Bourvellec, C. (2017). Characterization of tissue specific differences in cell wall polysaccharides of ripe and overripe pear fruit. Carbohyd Polym. 156, 152–164. doi: 10.1016/j.carbpol.2016.09.019
Cai, Y., Li, G., Nie, J., Lin, Y., Fan, N., Zhang, J., et al. (2010). Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 125 (3), 374–379. doi: 10.1016/j.scienta.2010.04.029
Dixon, R. A., Chen, F., Guo, D., Parvathi, K. (2001). The biosynthesis of monolignols: a “metabolic grid” or independent pathways to guaiacyl and syringyl units. Phytochemistry 57 (7), 1069–1084. doi: 10.1016/S0031-9422(01)00092-9
Dyckmans, J., Flessa, H., Brinkmann, K., Mai, C., Polle, A. (2002). Carbon and nitrogen dynamics in acid detergent fibre lignins of beech (Fagus sylvatica l.) during the growth phase. Plant Cell Environ. 25 (4), 469–478. doi: 10.1046/J.1365-3040.2002.00826.x
Feng, X. X., Liu, X. F., Jia, Y. X., Liu, H. F., Li, L. L. (2021). Transcriptomic analysis of hardened ‘Suli’ pear (Pyrus bretschneideri rehd) for identification of key genes for lignin biosynthesis and accumulation. Horticulturae 7 (11), 467. doi: 10.3390/horticulturae7110467
Gu, C., Guo, Z. H., Hao, P. P., Wang, G. M., Jin, Z. M., Zhang, S. L. (2017). Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 58, 6. doi: 10.1186/s40529-016-0159-1
Guo, W., Jin, L., Miao, Y., He, X., Hu, Q., Guo, K., et al. (2016). An ethylene response-related factor, GbERF1-like, from gossypium barbadense improves resistance to verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91 (3), 305–318. doi: 10.1007/s11103-016-0467-6
Huang, W. N., Liu, H. K., Zhang, H. H., Chen, Z., Guo, Y. D., Kang, Y. F. (2013). Ethylene-induced changes in lignification and cell wall-degrading enzymes in the roots of mung bean(Vigna radiata)sprouts. Plant Physiol. Bioch. 73, 412–419. doi: 10.1016/j.plaphy.2013.10.020
Lasserre, E., Jobet, E., Llauro, C., Delseny, M. (2008). AtERF38 (At2g35700), an AP2/ERF family transcription factor gene from arabidopsis thaliana, is expressed in specific cell types of roots, stemsand seeds that undergo suberization. Plant Physiol. Bioch. 46 (12), 1051–1061. doi: 10.1016/j.plaphy.2008.07.003
Licausi, F., Giorgi, F. M., Zenoni, S., Osti, F., Pezzotti, M., Perata, P. (2010). Genomic and transcriptomic analysis of the AP2/ERF superfamily in vitis vinifera. BMC Genomics 11 (1), 719. doi: 10.1186/1471-2164-11-719
Li, M. T., Cheng, C. X., Zhang, X. F., Zhou, S. P., Wang, C. H., Ma, C. H., et al. (2019). PpNAC187 enhances lignin synthesis in ‘Whangkeumbae’ pear (Pyrus pyrifolia) ‘Hard-end’ fruit. Molecules 24 (23), 4338. doi: 10.3390/molecules24234338
Liu, M., Diretto, G., Pirrello, J., Roustan, J. P., Li, Z., Giuliano, G., et al. (2014). The chimeric repressor version of an ethylene response factor (ERF) family member,Sl-ERF. B3, shows contrasting effects on tomato fruit ripening. New Phytol. 203 (1), 206–218. doi: 10.1111/nph.12771.
Livak, K., Schmittgen, T. (2000). Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Ct method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, G. L., Li, Z. J., Zhang, X. F., Wang, R., Yang, S. L. (2015). Expression analysis of lignin associated genes in hard end pear (Pyrus pyrifolia whangkeumbae) and its response to calcium chloride treatment conditions. J. Plant Growth Regul. 34 (2), 251–262. doi: 10.1007/s00344-014-9461-x
Luo, Z. S., Feng, S. M., Pang, J., Mao, L. C., Shou, H. L., Xie, J. W. (2012). Effect of heat treatment on lignification of postharvest bamboo shoots (Phyllostachys praecox f. prevernalis). Food Chem. 135 (4), 2182–2187. doi: 10.1016/j.foodchem.2012.07.087
Ma, R., Xiao, Y., Lv, Z., Tan, H. X., Chen, R., Li, Q., et al. (2017). AP2/ERF transcription factor, li049, positively regulates lignan biosynthesis in isatis indigotica through activating salicylic acid signaling and lignan/lignin pathway genes. Front. Plant Sci. 8, 361–1377. doi: 10.3389/fpls.2017.01361
Ma, Y., Zhang, F., Bade, R., Daxibater, A., Men, Z., Hasi, A. (2015). Genome-wide identification and phylogenetic analysis of the ERF gene family in melon. J. Plant Growth Regul. 34 (1), 66–77. doi: 10.1007/s00344-014-9443-z
Müller, M., Munné-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Nakano, T., Fujisawa, M., Shima, Y., Ito, Y. (2014). The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 65 (12), 3111–3119. doi: 10.1093/jxb/eru154
Ohme-Takagi, M., Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 7 (2), 173–182. doi: 10.1105/tpc.7.2.173
Raese, J. (1993). Effect of calcium sprays on control of black end, fruit quality, yield and mineral composition of ‘Bartlett’ pears. VI Int. Symp Pear Grow. 367, 314–322. doi: 10.17660/ActaHortic.1994.367.47
Raese, J., Drake, S. (2006). Calcium foliar sprays for control of alfalfa greening, cork spot, and hard end in a’njou’ pears. J. Plant Nutr. 29, 543–552. doi: 10.1080/01904160500526683
Ralph, J., Lundquist, K., Brunow, G., Lu, F., Boerjan, W. (2004). Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem. Rev. 3 (1), 29–60. doi: 10.1023/B:PHYT.0000047809.65444.a4
Rogers, L. A., Campbell, M. M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 164 (1), 17–30. doi: 10.1111/j.1469-8137.2004.01143.x
Shan, L. L., Li, X., Wang, P., Cai, C., Zhang, B., Sun, C. D., et al. (2008). Characterization of cDNAs associated with lignification and their expression profiles in loquat fruit with different lignin accumulation. Planta 227 (6), 1243–1254. doi: 10.1007/s00425-008-0696-2
Sun, W., Cao, Z., Li, Y., Zhao, Y., Zhang, H. (2007). A simple and effective method for protein subcellular localization using Agrobacterium-mediated transformation of onion epidermal cells. Biologia 62 (5), 529–532. doi: 10.2478/s11756-007-0104-6
Tao, S., Khanizadeh, S., Zhang, H., Zhang, S. (2009). Anatomy, ultrastructure and lignin distribution of stone cells in two pyrus species. Plant Sci. 176 (3), 410–419. doi: 10.1016/j.plantsci.2008.12.011
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153 (3), 895–905. doi: 10.1104/pp.110.155119
Wang, A., Tan, D., Takahashi, A., Li, T. Z., Harada, T. (2007). MdERFs, two ethylene-response factors involved in apple fruit ripening. J. Exp. Bot. 58 (13), 3743–3748. doi: 10.1093/jxb/erm224
Wang, X., Zhou, F., Liu, J., Liu, W., Zhang, S., Li, D., et al. (2021). Establishment of efficient callus genetic transformation system for Pyrus armeniacaefolia. Sci. Hortic. 289 (17), 110429. doi: 10.1016/j.scienta.2021.110429
Weigel, D., Glazebrook, J. (2006). Transformation of agrobacterium using the freeze-thaw method. CSH Protoc. 7, 1031–1036. doi: 10.1101/pdb.prot4666
Xiao, Y. Y., Chen, J. Y., Kuang, J. F., Wei, S., Hui, X., Jiang, Y. M., et al. (2013). Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J. Exp. Bot. 64 (8), 2499–2510. doi: 10.1093/jxb/ert108
Xie, X. L., Shen, S. L., Yin, X. R., Xu, Q., Grierson, D., Ferguson, I., et al. (2014). Isolation, classification and transcription profiles of the AP2/ERF transcription factor superfamily in citrus. Mol. Biol. Rep. 41 (7), 4261–4271. doi: 10.1007/s11033-014-3297-0
Xu, Z. S., Chen, M., Li, L. C., Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53 (7), 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Yang, S. L., Zhang, X. N., Lu, G. L., Wang, C. R., Wang, R. (2015). Regulation of gibberellin on gene expressions related with the lignin biosynthesis in ‘Wangkumbae’ pear (Pyrus pyrifolia nakai) fruit. Plant Growth Regul. 76 (2), 127–134. doi: 10.1007/s10725-014-9982-0
Yao, Y., He, R. J., Xie, Q. L., Zhao, X. H., Deng, X. M., He, J. B., et al. (2017). ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog d (RbohD)-dependent mechanism in response to different stresses in arabidopsis. New Phytol. 213, 1667–1681. doi: 10.1111/nph.14278
Yin, X. R., Allan, A. C., Xu, Q., Burdon, J., Dejnoprat, S., Chen, K. S., et al. (2012). Differential expression of kiwifruit ERF genes in response to postharvest abiotic stress. Postharvest Biol. Tec. 66, 1–7. doi: 10.1016/j.postharvbio.2011.11.009
Yu, Z. X., Li, J. X., Yang, C. Q., Hu, W. L., Wang, L. J., Chen, X. Y. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in artemisia annua l. Mol. Plant 5, 353–365. doi: 10.1093/mp/ssr087
Zeng, J. K., Li, X., Xu, Q., Chen, J. Y., Yin, X. R., Ferguson, I. B., et al. (2015). EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factor. Plant Biotechnol. J. 13 (9), 1325–1334. doi: 10.1111/pbi.12351
Keywords: pear, hard-end, PpERF1b-like, ethephon, lignin
Citation: Jin X, Cheng C, Qi Q, Zhou S, Wang C, Zhang Y, Sun C, Wang Y, Dang R and Yang S (2022) PpERF1b-like enhances lignin synthesis in pear (Pyrus pyrifolia) ‘hard-end’ fruit. Front. Plant Sci. 13:1087388. doi: 10.3389/fpls.2022.1087388
Received: 02 November 2022; Accepted: 28 November 2022;
Published: 15 December 2022.
Edited by:
Alfred (Heqiang) Huo, University of Florida, United StatesReviewed by:
Zhang Shaoli, Ludong University, ChinaCopyright © 2022 Jin, Cheng, Qi, Zhou, Wang, Zhang, Sun, Wang, Dang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaolan Yang, c2hhb2xhbnlhbmdAMTI2LmNvbQ==; Ruihong Dang, ZHJoMzRAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.