- 1Shanghai Key Laboratory of Plant Molecular Sciences, Development Center of Plant Germplasm Resources, College of Life Sciences, Shanghai Normal University, Shanghai, China
- 2Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
Cryptic splice sites in eukaryotic genome are generally dormant unless activated by mutation of authentic splice sites or related splicing factors. How cryptic splice sites are used remains unclear in plants. Here, we identified two cryptic splicing regulators, RBP45d and PRP39a that are homologs of yeast U1 auxiliary protein Nam8 and Prp39, respectively, via genetic screening for suppressors of the virescent sot5 mutant, which results from a point mutation at the 5’ splice site (5’ ss) of SOT5 intron 7. Loss-of-function mutations in RBP45d and PRP39a significantly increase the level of a cryptically spliced variant that encodes a mutated but functional sot5 protein, rescuing sot5 to the WT phenotype. We furtherly demonstrated that RBP45d and PRP39a interact with each other and also with the U1C, a core subunit of U1 snRNP. We found that RBP45d directly binds to the uridine (U)-rich RNA sequence downstream the 5’ ss of SOT5 intron 7. However, other RBP45/47 members do not function redundantly with RBP45d, at least in regulation of cryptic splicing. Taken together, RBP45d promotes U1 snRNP to recognize the specific 5’ ss via binding to intronic U-rich elements in plants.
Introduction
Removal of introns from precursor mRNA (pre-mRNA), namely RNA splicing, plays an important role in regulation of gene expression at the posttranscriptional level in eukaryotes. It has been estimated that about 95% of human genes with multiple exons undergo alternative splicing and produce more than one mRNA variants, which significantly expands the human proteome (Chen and Manley, 2009). If pre-mRNA splicing is not accurately processed, it will lead to generation of various altered splicing events and subsequent nonfunctional proteins. In human genetic diseases, 10% are caused by abnormal splicing (Montes et al., 2019). Likewise, regulation of alternative splicing is also one of the important mechanisms regulating plant growth and development in responses to various environmental and developmental cues (Reddy et al., 2013; Staiger and Brown, 2013).
Pre-mRNA splicing is catalyzed by a macromolecular complex called spliceosome, which consists of five core small nuclear ribonucleoprotein particles (U snRNPs) and a number of auxiliary proteins (Chen and Manley, 2009). Coordination of U snRNPs with their auxiliary proteins and many non-snRNP proteins plays an important role in the recognition of splice sites and splicing efficiency. The intron usually contains a number of essential degenerate consensus motifs interacting with U snRNPs, such as at the 5’ and 3’ splice sites (ss), where the most conserved dinucleotides are GU and AG, respectively, and at the branch point site (BP), where A is present 20 to 50 nucleotides (nt) upstream of the 3’ss. In addition, exons and introns may contain cis-elements that can enhance or silence splicing and regulate constitutive splicing or alternative splicing via interaction with splicing regulatory factors (Chen and Manley, 2009; Lee and Rio, 2015). Interestingly, plant introns have evolved a number of distinct features from those in yeast and animals (Brown et al., 1996). For example, in plants, an intron is usually smaller in size and richer in A and U, the branch point sequence is not obvious, and the pyrimidine tract located between the BPS and 3’ ss is mostly U (Brown et al., 1996). It has been demonstrated that the intronic AU-richness is essential for efficient splicing and 5’ ss selection in dicots (McCullough et al., 1993; Gniadkowski et al., 1996).
RNA splicing is initiated by base-pairing of the 5’ end of the U1 snRNA to the 5’ ss. In yeast and mammals, the 5’ ss consensus sequence for the major U2-type (GT-AG) introns can complement perfectly to that of the U1 snRNA (Roca et al., 2008), which provides a mechanistic explanation for the important role of U1 snRNP in selection of 5’ ss. Besides U1 snRNA, U1 snRNP has seven core subunits called Sm proteins common in all the U snRNPs and three U1-specific proteins (U1-70K, U1A, and U1C) (Will and Luhrmann, 2011). It has been implicated that U1C is essential for U1 snRNP function by interacting with the Sm protein and recognizing the sequence of the 5’ ss (Nelissen et al., 1994; Du and Rosbash, 2002). In contrast, yeast Saccharomyces cerevisiae U1 snRNA (568 nts) is significantly larger than mammalian U1 snRNA (De Bortoli et al., 2019), and its U1 snRNP contains seven additional U1 auxiliary proteins including Prp39, Prp40, Snu71, Snu56, Luc7, Prp42, and Nam8 (Li et al., 2017). Among these U1 auxiliary proteins, the yeast Nam8 was reported to directly bind to the uridine (U)-rich sequence downstream of the 5’ ss and U1 snRNP at the same time, so as to effectively help splicing of introns with non-classical 5’ ss sequences (Puig et al., 1999). The human homologous protein of Nam8, TIA-1 (T-cell intracellular antigen-1) has been implicated in U1 snRNP recognition of the 5’ ss and alternative splicing regulation of various pre-mRNA via binding to U-rich motifs located downstream of 5’ ss and interaction with the U1C (Le Guiner et al., 2001; Forch et al., 2002). These data indicate that selection of 5’ ss is finely regulated not only by the complementary degree between the 5’ ends of the U1 snRNA and introns but also by many other factors, such as intronic U-rich elements and auxiliary splicing factors. Interestingly, there are eight Nam8 homologs, called the RNA binding protein (RBP) 45/47 family in Arabidopsis (Lorkovic et al., 2000; Peal et al., 2011). Recently, one member of the RBP45/47 family, RBP45d, was reported to associate with U1 snRNP via interacting with PRP39a and regulate alternative splicing (AS) for a specific set of genes (Chang et al., 2022). In addition, the Arabidopsis genome encodes many homologs of the seven yeast U1 auxiliary proteins (Wang and Brendel, 2004; Reddy et al., 2013). Thus, plant U1 snRNP has been suggested to be similar to or more complex than that in yeast. Although increasing genetic evidence supports that these U1 auxiliary proteins play an important role in plant growth, development and stress response (Wang et al., 2007; Zhan et al., 2015; de Francisco Amorim et al., 2018; Hernando et al., 2019), molecular mechanisms by which they regulate U1 snRNP function remain largely unknown.
There are large numbers of splice sites, known as cryptic splice sites in Eukaryotic genomes, which are generally dormant or used only at low levels unless activated by mutations of nearby authentic splice sites (Roca et al., 2003; Kapustin et al., 2011). Notably, about 9% of all mutations reported in the human gene mutation database are splicing mutations (Anna and Monika, 2018). It is important to be able to predict cryptic splice sites that might be activated in genetic disease so that effective therapeutic strategies can be designed. On the other hand, mutations leading to activation of cryptic splice sites in crops have generated new genetic germplasm resources, such as waxy rice, low phytic acid soybean, and pale green Chinese cabbage (Isshiki et al., 1998; Yuan et al., 2012; Zhao et al., 2022). It is generally accepted that cryptic splice sites are suppressed by nearby stronger splice sites and that splice site selection can be viewed as a competition between the various potential splice sites in a pre-mRNA (Kapustin et al., 2011). And it is also proposed that cryptic splice sites are regulated by various RNA binding proteins including heterogeneous ribonucleoproteins (hnRNP) and RNA recognition motif (RRM)-containing SR (serine and arginine-rich) proteins, which are also the alternative splicing regulators (Anna and Monika, 2018). Little is known about splicing factors involved in regulating cryptic splicing in plants. In this study, we showed that Arabidopsis RBP45d facilitates U1 snRNP to preferentially select a cryptic splice site that is activated by mutations of the intronic 5’ ss via directly binding to U-rich sequences downstream of the 5’ ss. Our results suggest that RBP45d regulates cryptic splice site selection in plants.
Results
An intron mutation leads to activation of two cryptic splice sites in sot5
We previously reported that the Arabidopsis PPR protein SOT5 (At1g30610), also named EMB2279, is required for intron splicing of the two chloroplast housekeeping genes, rpl2 and trnk (Huang et al., 2018). The sot5 (emb2279-3) mutant contains a G to A mutation at the first base (+1) of the seventh intron and displays a leaf virescent phenotype (Figures 1A, C). This point mutation leads to accumulation of the three alternative splicing variants in sot5, compared to the wild type (WT) (Figure 1B). DNA sequencing of these cloned PCR products indicated that the longest one retained the whole seventh intron with 84 nts (Figure 1B and Figure S1), the middle transcript retained 9 nts more at the 3’ ss of exon 6 (Figure 1B and Figure S1), and the shortest one lacked 22 nts at the 3’ end of exon 6 (Figure 1B and Figure S1). These results indicate that the mutation of the first invariant G at the 5’ ss of intron 7 results in intron retention and activation of two cryptic splice sites named -22 ss and +10 ss (Figure 1C). Since the level of band d was much higher than that of band c, the cryptic splicing mainly occurred at -22 ss in sot5 (Figure 1B). Polypeptide prediction indicated that the -22 ss product encodes a truncated protein with only 6 PPR motifs that should be loss of function (Figures 1D, E); the intron-retained transcript encodes a probably partially functional protein with 10 PPR motifs (Chen et al., 2020); the +10 ss product encodes almost the same protein as the WT one except the seventh PPR motif is slightly changed (Figures 1D, E, Figure S2). Thus, these results interpret well why sot5 displays a virescent phenotype, but the emb2279-1 knockout mutant is embryo lethal.
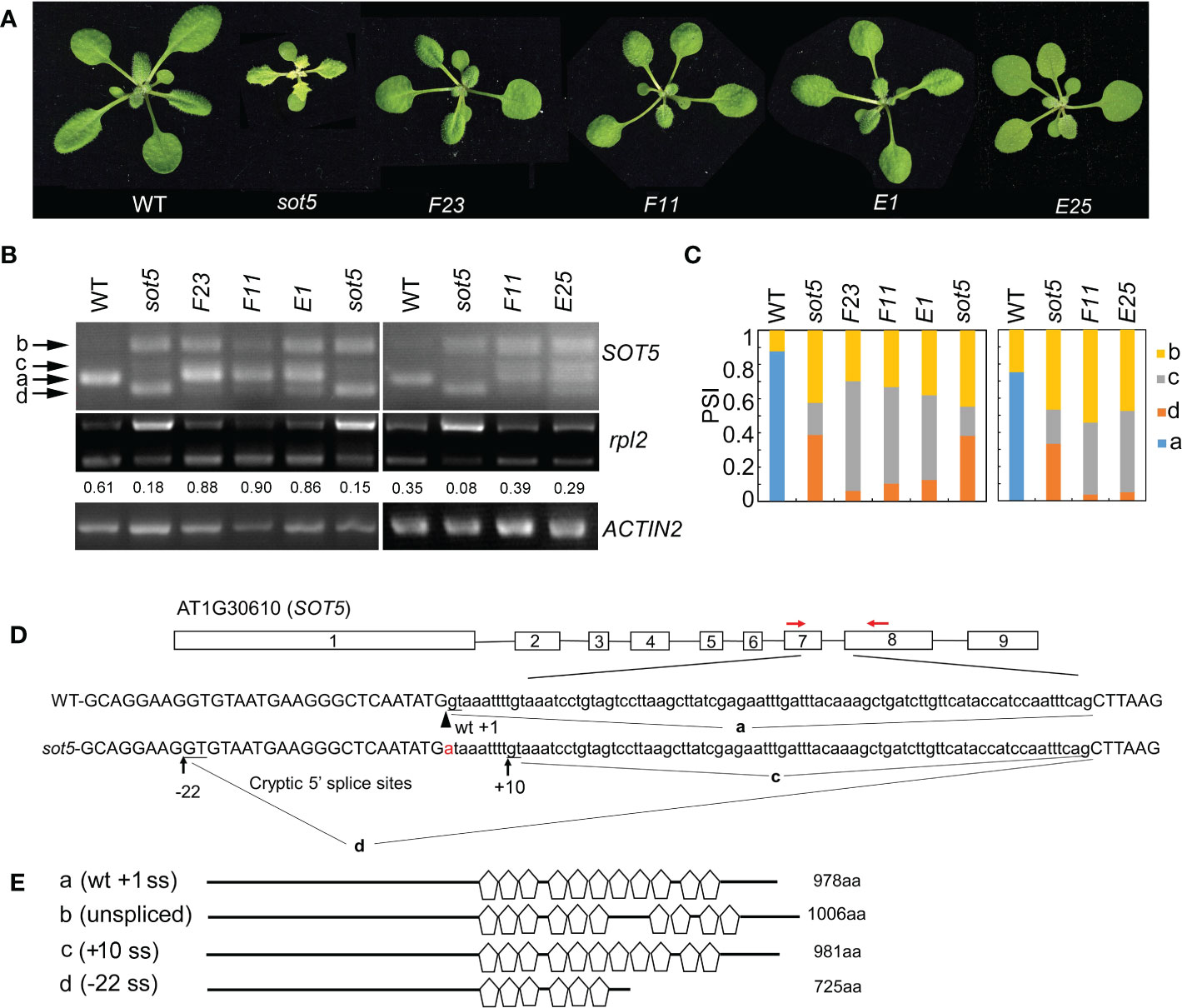
Figure 1 Genetic screening for suppressors of the sot5 viresent phenotype reveals altered cryptic 5’ splice sites by second mutations. (A) Four suppressor lines of sot5 display WT-like phenotype. The representative 30-d-old plants were shown. (B) Semi-quantitative RT-PCR of SOT5 intron 7 and plastid rpl2 intron in WT, sot5, and the suppressor lines. ACTIN2 was used as an internal control. Band a shows the WT splicing product. Band b, c, and d correspond to the splicing variants containing intron 7, derived from splice site +10 ss and -22 ss, respectively. The number under the rpl2 gel band represent the percentage of mature rpl2 transcripts which quantified by Image J software. (C) Quantification of the four bands (a–d) of SOT5 intron7 RT-PCR products in each genotype showed in (B) by Image J software. PSI (percent spliced in index) indicates the percentage of each variant in total transcripts. Three biological replicates (independent pools of aerial parts of plants) were analyzed, and one representative result is shown. (D) The schematic diagram of SOT5 gene structure and intron 7 splicing variants. The black boxes indicate exons and the black lines indicate introns. The authentic 5’ ss in WT is shown by the arrowhead. The G to A point mutation in sot5 is shown by the red letter. Black arrows indicate two cryptic splice sites, +10 ss and -22 ss. a, c and d indicated the splicing variants corresponding to the bands in (B). Red arrows indicate the primers used for RT-PCR. (E) Schematic diagrams of the mutated SOT5 proteins encoded by the variants corresponding to the bands in (B). Pentagons indicate PPR motifs. The numbers indicate the residue number of each protein.
Genetic screening for suppressors of sot5 reveals altered cryptic splice sites usage by second mutations
The weak allele sot5, in which an intron mutation leads to generation of three abnormal transcripts, provides a good material to study regulation of the cryptic splice site usage. To dissect the genetic pathway for the cryptic splice site selection of SOT5 intron 7, we screened for sot5 suppressors which exhibit a WT-like phenotype using the ethyl methanesulfonate (EMS)-mutagenized sot5 seeds. We obtained a number of WT-like suppressor lines for the sot5 mutant (Figure S3). In this study, four suppressor lines, namely F23, F11, E1 and E25, were selected for further analysis based on their similar phenotype and alternative splicing pattern of SOT5 intron7 (Figures 1A–C and Figure S3). Compared to sot5, all these suppressor lines produced a lower level of -22 ss transcripts (band d in Figure 1B), but a higher level of +10 ss transcripts (band c in Figure 1B), suggesting a positive correlation between the green leaf and the level of the +10 ss transcripts. In sot5 mutant, the splicing efficiency of plastid rpl2 and trnK intron is significantly decreased (Huang et al., 2018). Indeed, the suppressors of sot5 were able to recover the splicing defect of plastid rpl2 intron, which is consistent with their WT-like phenotype (Figure 1B). In addition, our genetic analyses showed that the suppressor phenotype was caused by a single gene mutation, and interestingly F23, F11 and E1 were allelic while E25 was mutated in another gene. Taken together, our data suggest that the sot5 phenotype is suppressed by second mutations that can modulate cryptic splice site selection and splicing efficiency.
Loss-of-function mutations in RBP45d suppress the sot5 phenotype
To isolate the suppressor gene in F23, we produced a mapping population from the cross between F23 and Landsberg erecta (Ler). Through Mapping-By-Sequencing (MBS) technique, the gene responsible for phenotypic suppression of sot5 was mapped to the region with the length ~8 M bp on chromosome 5, which contains 7 point mutations (Figures S4A, B). Among these 7 mutations, one stop codon-gain mutation from C to T occurred at 187 bp of the CDS in the locus At5g19350, which encodes RBP45d, a member of the RBP45/47 family with triple RNA recognition motifs (RRM) (Figures 2A and S4B). Considering that sot5-activated cryptic splicing efficiency is altered in F23, we speculated that RBP45d was the most likely gene to suppress sot5. Thus, we sequenced the RBP45d gene in F11 and E1, which were genetically testified as F23 alleles. Indeed, F11 has a point mutation from G to A at the 481 bp position from the start codon of the RBP45d CDS, resulting in the substitution of glycine (G) by arginine (R) at the 161 amino acid residue in the conserved RRM2 domain, whereas E1 contains a premature stop codon mutation from G to A at the 80 bp position from the start codon of the RBP45d CDS (Figure 2A).
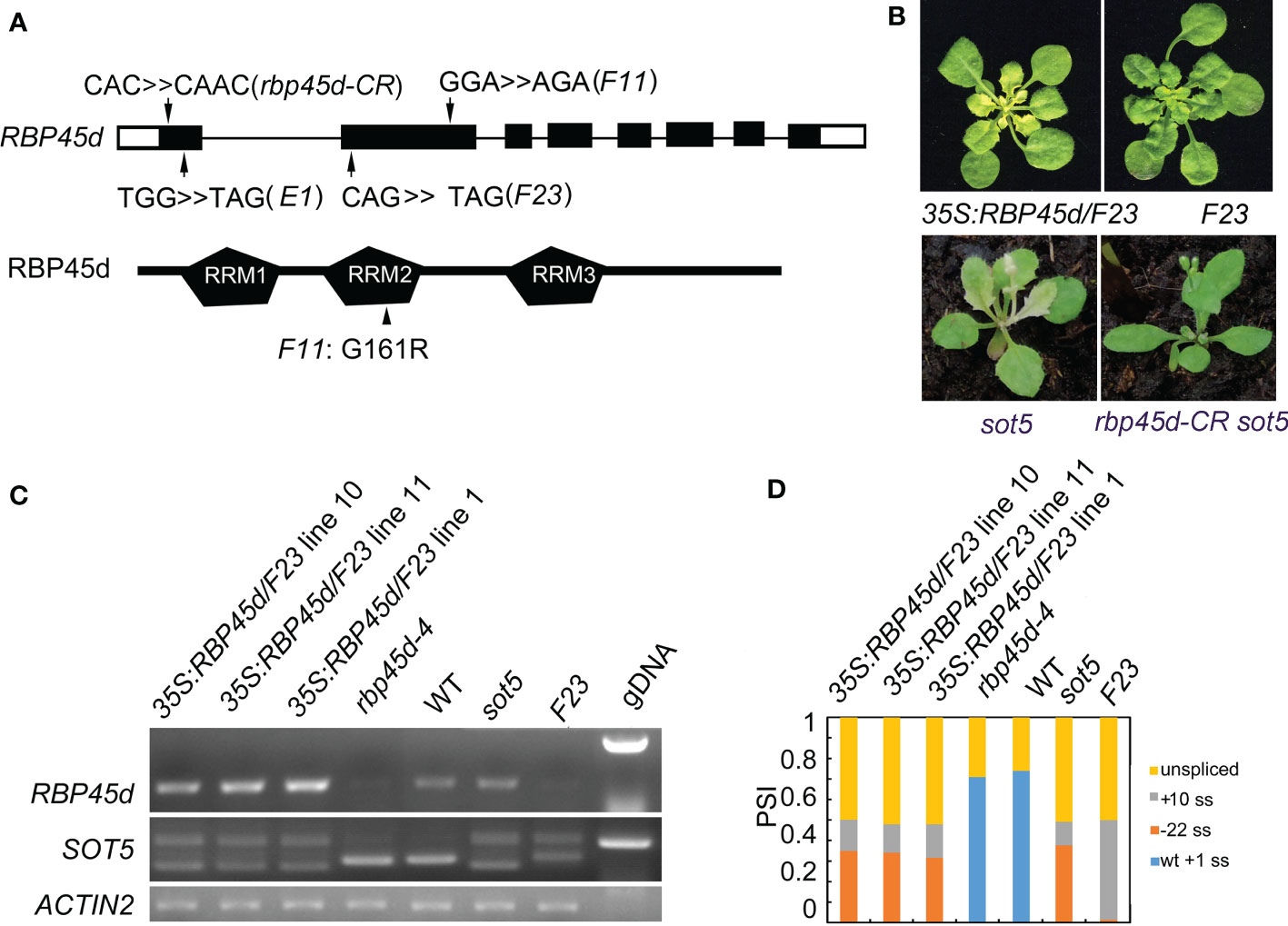
Figure 2 Mutations in RBP45d suppress the sot5 phenotype. (A) Schematic diagrams of RBP45d gene structure and RBP45d protein with the three RRM domains. Arrows indicate the mutation sites in E1, F23 and F11 lines. The upper case letters indicate the mutated genetic codon(s) in E1 and F23 which become the stop codons, and that in F11 which leads to replacement of the amino acid residue from G to R in the conservative region of the RRM2 domain (arrowhead). The black boxes indicate exons in UTR and the black rectangles indicate exons in CDS and the black lines indicate introns. (B) Overexpression of RBP45d CDS in F23 restores the phenotype. And the rbp45d-CR sot5 double mutant showed the WT-like phenotype. One of representative 35S:RBP45d/F23 line was shown. (C) RT-PCR analysis of RBP45d expression and the splicing products of SOT5 intron 7 in three 35S:RBP45d/F23 transgenic lines, rbp45d-4, WT, sot5, and F23 plants. ACTIN2 was used as an internal control and gDNA is used as a control. Two biological replicates (independent pools of aerial parts of plants) were analyzed, and one representative result is shown. (D) Quantification of the splicing variants from SOT5 intron 7 shown in (C). PSI (percent spliced in index) indicates the percentage of each variant in total transcripts.
To verify the mapping result, we overexpressed the native RBP45d CDS in F23. Our data showed that transgenic lines 35S:RBP45d/F23 not only displayed the sot5 phenotype (Figure 2B), but also had the same alternative splicing pattern as the sot5 mutant (Figures 2C, D). In addition, we generated a knockout allele of RBP45d (rbp45d-CR, with one base insertion at the first exon) in sot5 mutant background by CRISPR/CAS9 technique, which presented WT-like phenotype (Figures 2A, B). These results indicated that the phenotypic recovery of sot5 is attributed to the loss-of-function mutation in RBP45d. We then isolated the single mutant, named rbp45d-4, from the F2 population of the cross between F23 and Col-0 and rbp45d-5 from the F2 population of the cross between F11 and Col-0. RT-PCR analysis showed that RBP45d transcripts were almost undetectable in rbp45d-4 and F23 (Figure 2C), indicating that the point mutation in rbp45d-4 triggers the nonsense-mediated mRNA decay (NMD) pathway. Taken together, our results imply that RBP45d mutations are responsible for the recovery of sot5 leaf virescence.
Mutations of other RBP45/47 members cannot suppress the phenotype of sot5
It has been reported that the Arabidopsis genome has eight members in the RBP45/47 family (Wang and Brendel, 2004). RT-PCR analysis and eFP Browser (http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi) showed that they are ubiquitously expressed in all organs including roots, inflorescences and leaves (Figure S5). Subcellular localization experiment showed that RBP45a, RBP45b, RBP45c, RBP45d and RBP47b were localized in the both nucleus and cytoplasm, while RBP47c’ mainly localized in the cytoplasm (Figure S6). The mutant phenotype and physiological function of RBP45/47 family members were not well characterized yet. To know whether other members of the RBP45/47 family can function in the same manner as RBP45d to suppress the sot5 phenotype, we identified four T-DNA insertion mutants rbp45a, rbp45c, rbp47a and rbp47b from the ABRC stock (Figures 3A, B), and made their double mutants with sot5 by genetic crossing. All the four single mutants displayed the WT-like phenotype under normal growth conditions, while the double mutants with sot5 showed the sot5-like phenotype (Figure 3C). Furtherly, RT-PCR analysis showed that the SOT5 intron7 splicing pattern was not altered in these mutants when compared with that in sot5 and F23 (Figure 3D). Therefore, our data indicate that rbp45a, rbp45c, rbp47a and rbp47b cannot suppress the phenotype of sot5.
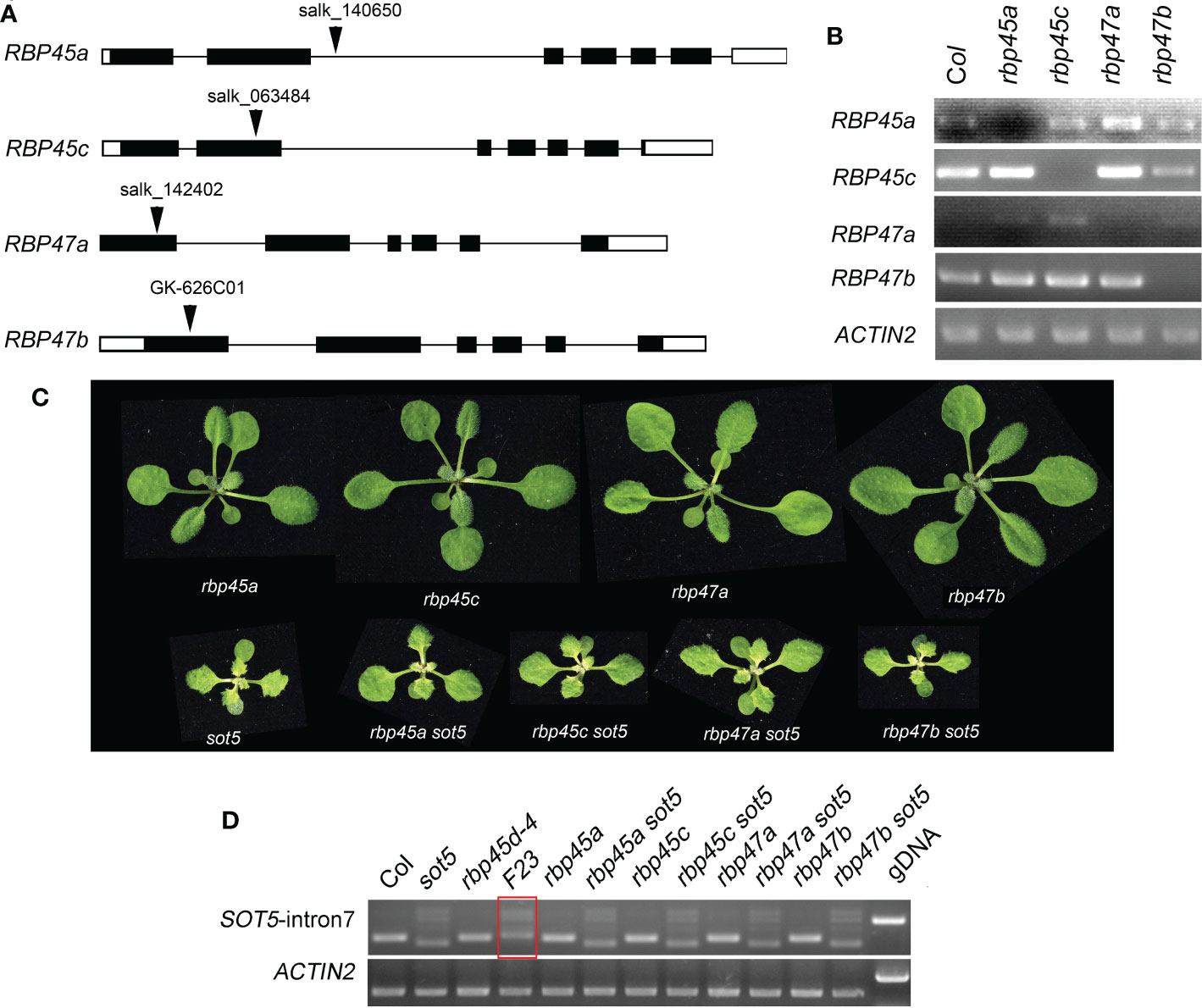
Figure 3 Mutations in other RBP45/47 paralogs cannot suppress the viresent phenotype of sot5. (A) Identification of T-DNA insertions in RBP45a, RBP45c, RBP47a and RBP47b genes. The black boxes indicate exons in UTR and the black rectangles indicate exons in CDS and the black lines indicate introns. Arrowheads indicate the insertional position of T-DNA for each gene. (B) RT-PCR analysis of gene expression in the T-DNA knockout mutants. ACTIN2 was used as an internal control. Two biological replicates (independent pools of aerial parts of plants) were analyzed, and one representative result is shown. (C) Phenotypes of single and double mutants as indicated. (D) Semi-quantitative RT-PCR showed the splicing pattern of SOT5 intron 7 in sot5 RBP45a, sot5 RBP45c, sot5 RBP47a and sot5 RBP47b double mutant when compared with that in F23 (sot5 rbp45d-4). The red box highlighted the altered splicing pattern of SOT5 intron 7 in F23..
RBP45d promote flowering and root growth that different from other RBP45/47 members
Recently, Chang et al. (2022) and Wang et al. (2021) reported that RBP45d regulates temperature-responsive flowering in Arabidopsis. In this study, we also observed the later flowering phenotype of rbp45d mutants including rbp45d-4, rbp45d-5 and rbp45d-CR (a knockout rbp45d mutant created by CRISPR/CAS9 technique) under 16h light/8h dark growth conditions (Figure 4A upper panel and Figure S7). The number of rosette leaves for rbp45d mutants to flower were significantly bigger than that for WT (Figure 4B the left panel and Figures S7B and S7C). However, there were no significant difference in flowering time between WT and other rbp45/47 plants (Figures 4A, B). In addition, we found that loss-of-function mutations in RBP45d but not in other RBP45/47 genes led to the significant shorter primary root phenotype, compared with that of WT, when seedlings were vertically grown on 1/2 MS plates (Figure 4A lower panel, Figure S7). For 5-day-old seedlings, primary root length of rbp45d-4 and rbp45d-5 was 64% and 80% of WT, respectively (Figure 4A lower panel and Figure 4B). Such a difference in primary root length became much larger between 10-day-old rbp45d and WT or other rbp45/47 seedlings. These results suggest that the biological function of RBP45d is different from that of other RBP45/47 paralogs.
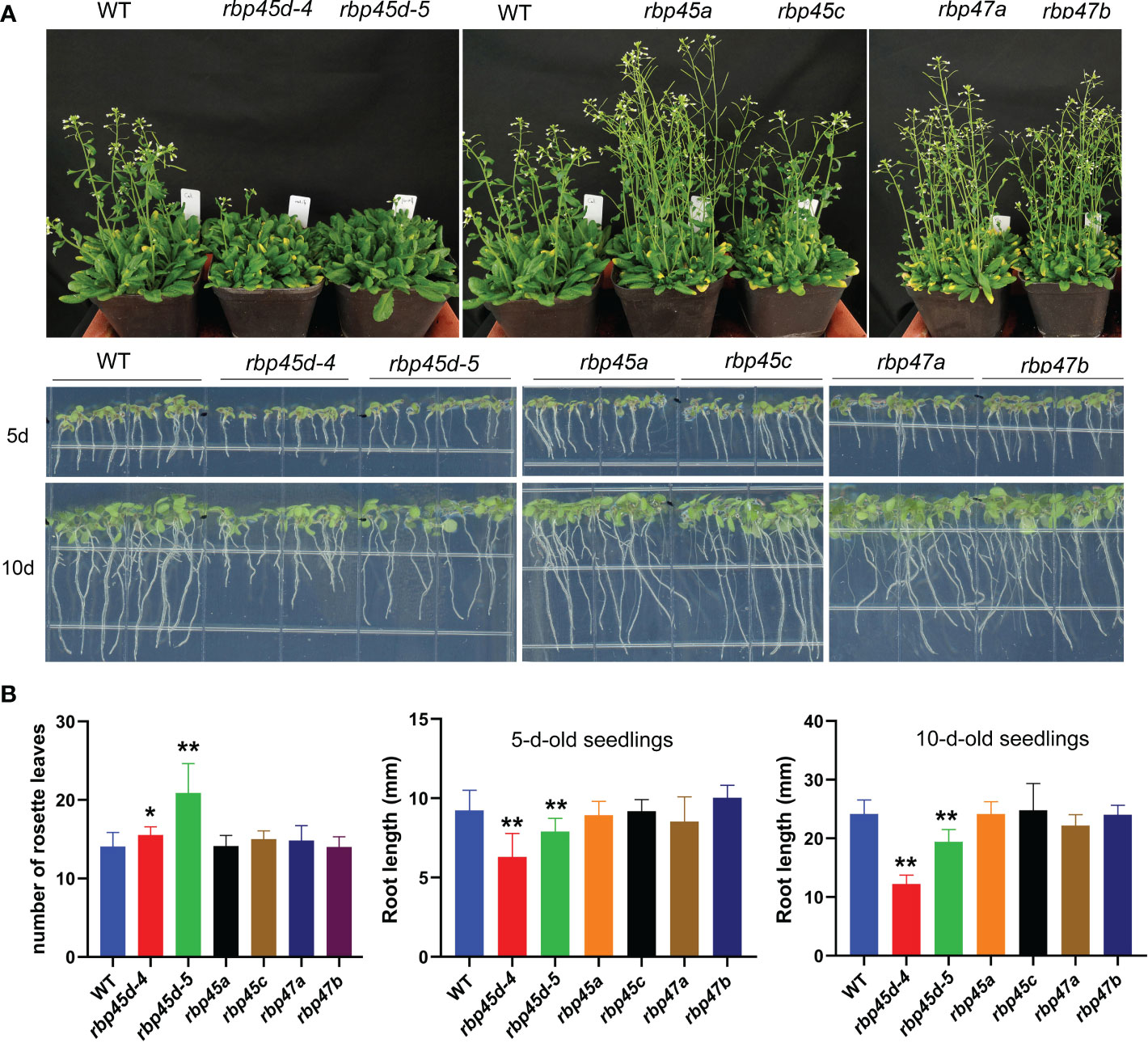
Figure 4 RBP45d promote flowering and root growth that different from other RBP45/47 members. (A) Phenotypic comparison among rbp45/47 mutants. The upper panel shows the phenotype of 45d-old rbp45/47 mutants grown under LD conditions (16h light/8h dark photoperiod). The lower panel shows 5-d- and 10-d-old rbp45/47 mutants vertically grown on 1/2 MS plates. (B) The left panel shows the rosette leaf number of each genotype at bolting in LD conditions. The middle and right panels show root length of 5-d and 10-d-old seedlings, respectively. The data are shown means ± SD (n = 20). Student’s t-test, * P < 0.05, ** P < 0.01, compared to WT. The experiments were performed at least three times.
RBP45d physically interacts with the U1 snRNP core subunit U1C
RBP45/47 proteins are homologous with yeast Nam8p protein and human TIA-1 protein. It was reported that TIA-1 can directly interact with U1C, the core protein of U1 snRNP (Forch et al., 2002). And the cryo-electron microscopy structure of the yeast Saccharomyces cerevisiae prespliceosome revealed that yeast Nam8 protein physically interacts with the U1C. Thus, we proposed that RBP45d may function in a similar manner with TIA-1 and Nam8. We then used yeast two hybrid (Y2H) and BiFC techniques to detect whether RBP45d interacts directly with U1C, U1 70K and U1A of U1 snRNP in Arabidopsis. Y2H results showed that RBP45d directly interacted with U1C, but not with U1A and U1 70K (Figure 5A). Consistently, BiFC analysis also revealed that RBP45d was bound to U1C in both the nucleus and cytoplasm in Arabidopsis protoplasts (Figure 5B), which is consistent with their subcellular localization (Figure S6). Taken together, these results indicate that RBP45d interacts with U1C in vitro and in vivo.
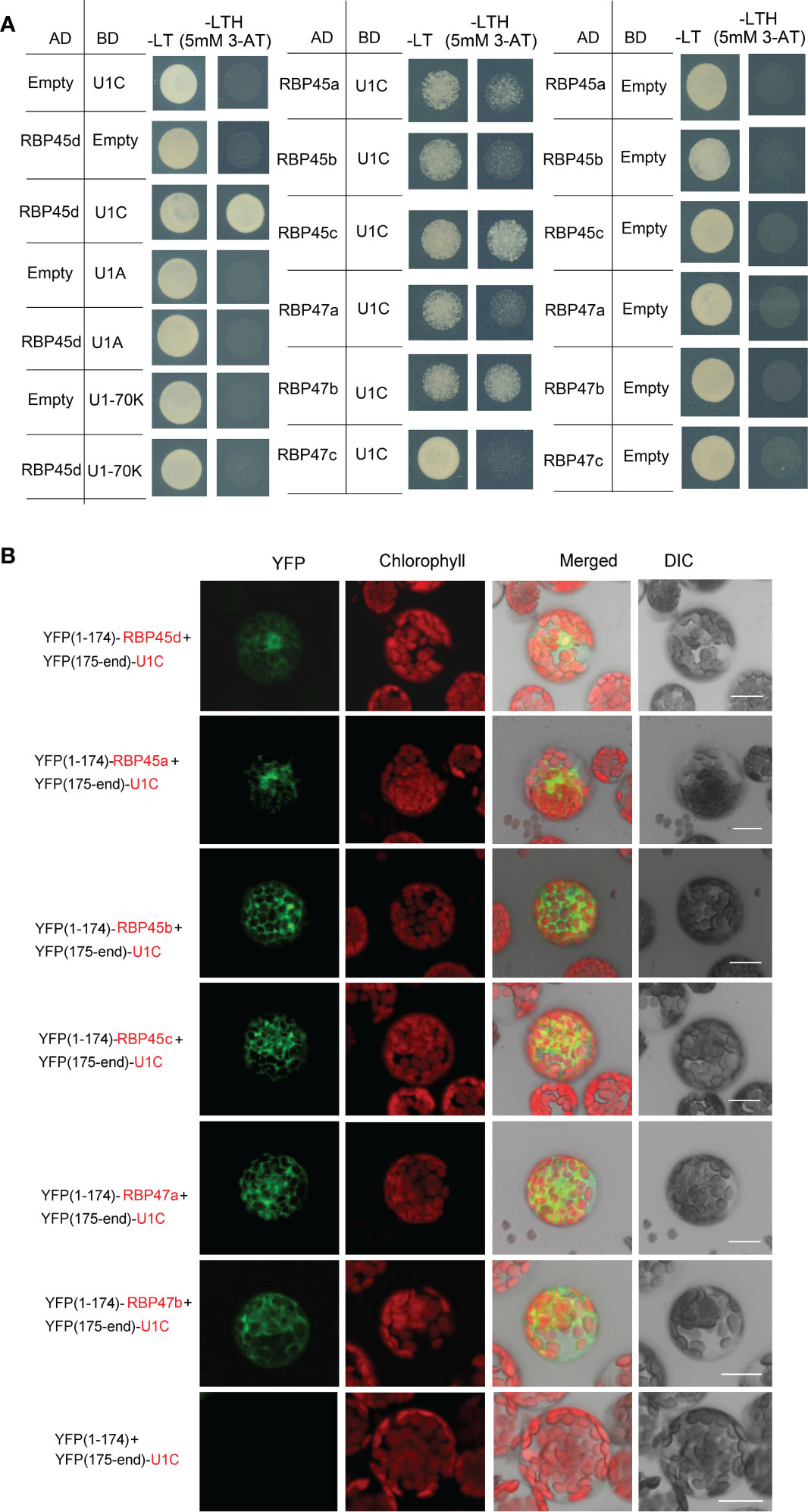
Figure 5 RBP45d and other RBP45/47 members interact with U1C. (A) Yeast two-hybrid assays show the interaction between RBP45d and U1C, U1A or U1 70K and the interaction between U1C and RBP45a, RBP45b, RBP45c, RBP47a, RBP47b or RBP47c. The transformed yeast cells were grown on -LT (SD-Leu/-Trp) mediums and -LTH+3-AT (SD- Leu/-Trp/-His with 5 mM 3-AT) mediums. (B) BiFC assay of the interaction between U1C and RBP45d, RBP45a, RBP45b, RBP45c, RBP47a or RBP47b. YFP, fluorescence of yellow fluorescent protein; DIC, Differential interference contrast microscopy; Merge, merge of YFP, DIC and chlorophyll. Bar = 15 μm.
Meanwhile, Y2H analysis showed that except for RBP47C, the RBP45/47 members interacted with U1C (Figure 5A), whereas BiFC assays indicated that all the tested RBP45/47 paralogs interacted with U1C mainly in the cytoplasm (Figure 5). Thus, our results indicate that the biological significance of U1C interaction with RBP45d may differs from that with other RBP45/47 members.
RBP45d is an intronic U-rich element binding factor
The above results suggest that RBP45d is recruited to the U1 snRNP via interacting with U1C, which is consistent with the working model of yeast Nam8 and human TIA1. It indicates the molecular mechanism of RBP45d regulating 5’ss selection is also conserved in plants. Then we asked whether RBP45d can interact with U-rich sequences. In the sot5 mutant, the G to A mutation at the 5’ ss of intron 7 results in activation of the two cryptic splice sites (-22 ss and +10 ss) and preferential utilization of the -22 ss (Figures 1B, C). In contrast, the absence of RBP45d in the suppressor lines F23, F11 and E1 leads to preferentially utilize +10 ss (Figure 1B), implying that RBP45d plays an important role in controlling the the cryptic splice sites usage. Search for U-rich elements within sot5 intron 7 indeed revealed the presence of the AUAAAUUUU sequence downstream of the -22 ss (Figure 6A). We proposed that RBP45d can bind to this U-rich sequence and subsequently promote the upstream -22 ss selection.
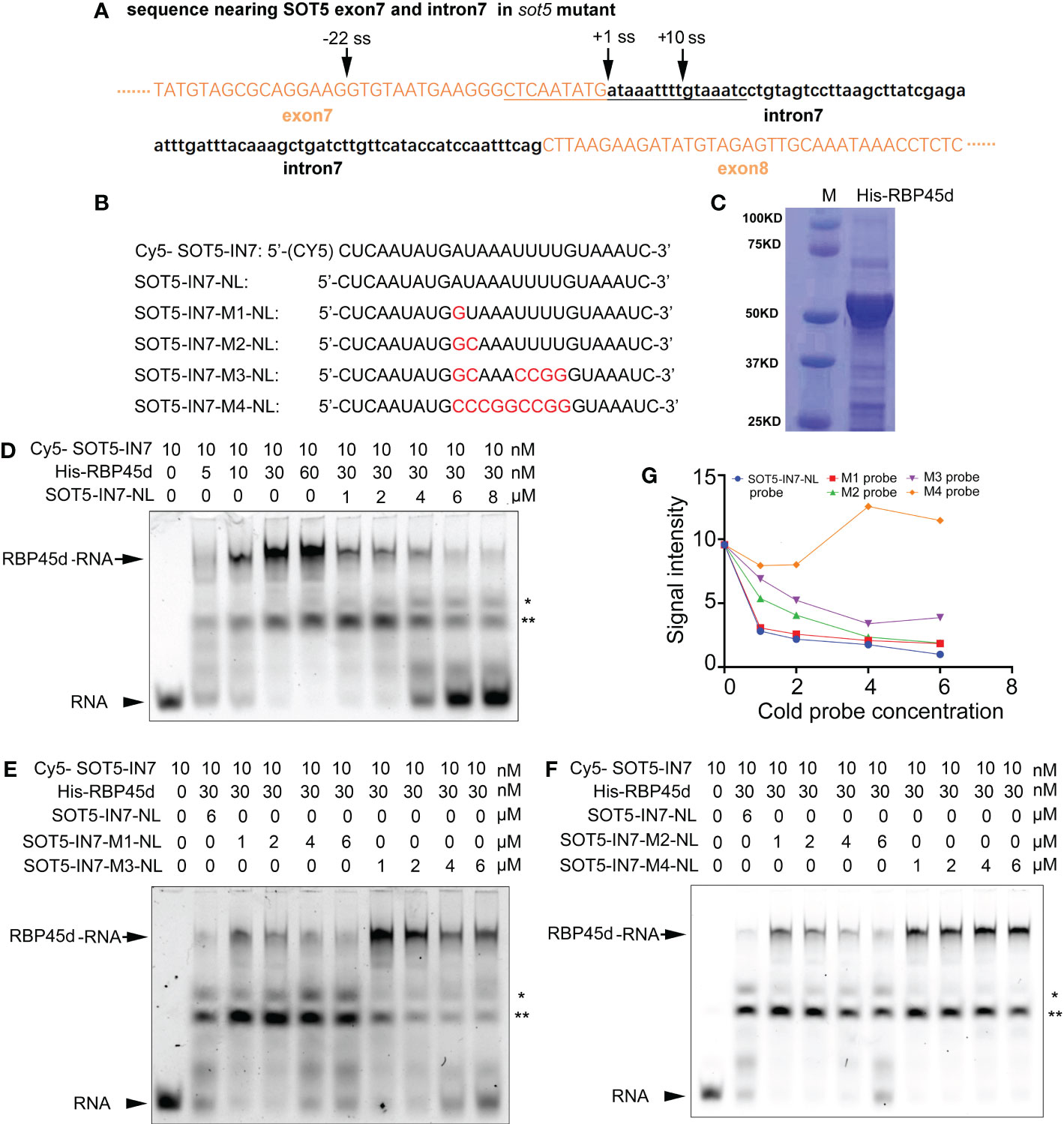
Figure 6 REMSA shows direct binding of RBP45d to the U-rich sequence downstream of the -22 ss of SOT5 intron 7. (A) DNA sequence nearby the cryptic 5’ splice sites in sot5. Upper and lower case letters indicate exonic and intronic sequences in SOT5 gene, respectively. The underlined sequence is used as a template for RNA probe synthesis. Arrows, splice sites. (B) Probe names and sequences used for REMSA. The mutation sites in the probes were highlighted with red color. (C) Coomassie brilliant-blue staining of the purified His-RBP45d protein separated by SDS-PAGE. (D) RBP45d protein can directly bind to the Cy5-SOT5-IN7 RNA probe. (E) Competitive binding assay of unlabeled probes SOT5-IN7-M1-NL and SOT5-IN7-M3-NL with the labeled Cy5-SOT5-IN7 probe. (F) Competitive binding assay of unlabeled probes SOT5-IN7-M2-NL and SOT5-IN7-M4-NL with the labeled Cy5-SOT5-IN7 probe. Arrows in (D–F) show the protein-RNA complex. Arrowheads in (D–F) show the free RNA probe. * and ** in (D–F) indicate non-specific bands. (G) Relative signal intensity of the protein-RNA complex in (D–F) was quantified by ImageJ software. The signal intensity of the protein-RNA band in the lane using 6 μM unlabeled SOT5-IN7-NL to compete with the labeled Cy5-SOT5-IN7 probe is artificially set to 1 in (D–F). For REMSA assays, three technical replicates were analyzed, and the results have the same trend. So one representative result is shown.
To test this hypothesis, we carried out RNA electrophoretic mobility shift assay (REMSA) with a series of synthesized 26 nt RNA oligo probes including the U-rich element (SOT5-IN7) and the mutated variants (U-poor or U-less, SOT5-IN7-M1~4), and purified His-tagged RBP45d protein from E. coli (Figures 6B, C). REMSA results showed that RBP45d bound to the Cy5-SOT5-IN7 probe in a dose-dependent manner, and the unlabeled SOT5-IN7-NL probe was able to well compete with the labeled probe in RBP45d binding (Figure 6D). Further analysis showed the unlabeled SOT5-IN7-M1-NL and SOT-IN7-M2-NL probes, in which the first A and AU of intron 7 were mutated into G and GC, respectively, still efficiently blocked RBP45d binding to the labeled Cy5-SOT5-IN7 probe (Figures 6E, F), whereas the unlabeled SOT5-IN7-M3-NL probe with mutations at the first AU and 4 continuous U of intron 7 significantly reduced the binding affinity to RBP45d (Figure 6E). Meanwhile, the unlabeled SOT5-IN7-M4-NL probe completely lost the binding affinity to RBP45d when all the AU-rich element was mutated into the CG-rich one (Figure 6F). Quantification of the signal intensity of these protein-RNA bands revealed that the affinity of RBP45d binding to RNA increased with the increase in U and/or A levels of the probes (Figure 6G). Thus, our results suggest that RBP45d can bind to the U-rich sequence element, and binding affinity between RBP45d and RNA is dependent on the content of U and/or A.
RBP45d also modulates cryptic splice site selection in the clpR4-3 mutant
Previously, we reported the G to A mutation at the 5’ ss (+1G-to-A) of ClpR4 intron 2 in the clpR4-3 mutant, which displays virescent leaves (Wu et al., 2013). Similarly, the mutation of the authentic splice site leads to retention of intron 2 and activation of the two cryptic splice sites (+16 ss and +49 ss) (Figure 7A), producing three different transcripts, named band a, b, and c that are not detected in WT (Figure 7C). To examine whether RBP45d is involved in regulation of alternative splicing of ClpR4 intron 2 in clpR4-3, we constructed the rbp45d-4 clpR4-3 double mutant by genetic crossing. Although rbp45d-4 was unable to suppress the virescent phenotype of clpR4-3 (Figure 7B), the splice sites usage and splicing efficiency of ClpR4 mRNA were clearly altered in the double mutant, where band b products significantly decreased while band d products appeared and band c products increased slightly, compared with those in the single clpR4-3 mutant (Figures 7C, D). Sequencing of the cloned PCR products showed that band b, c and d products were derived from the cryptic splice sites +49 ss, +16 ss, and -96 ss, respectively (Figures 7A, C and Figure S8). These results suggest that the absence of RBP45d in clpR4-3 allows the U1 snRNP complex to preferentially recognize the cryptic splice sites -96 ss and +16 ss, instead of the cryptic splice site +49 ss. We found a five consecutive U (U-rich) element downstream of the cryptic splice site +49 ss (the underlined letters in Figure 7A). It is possible that RBP45d binds to this U-rich element in ClpR4 intron 2, and subsequently promotes the cryptic +49 ss selection in clpR4-3.
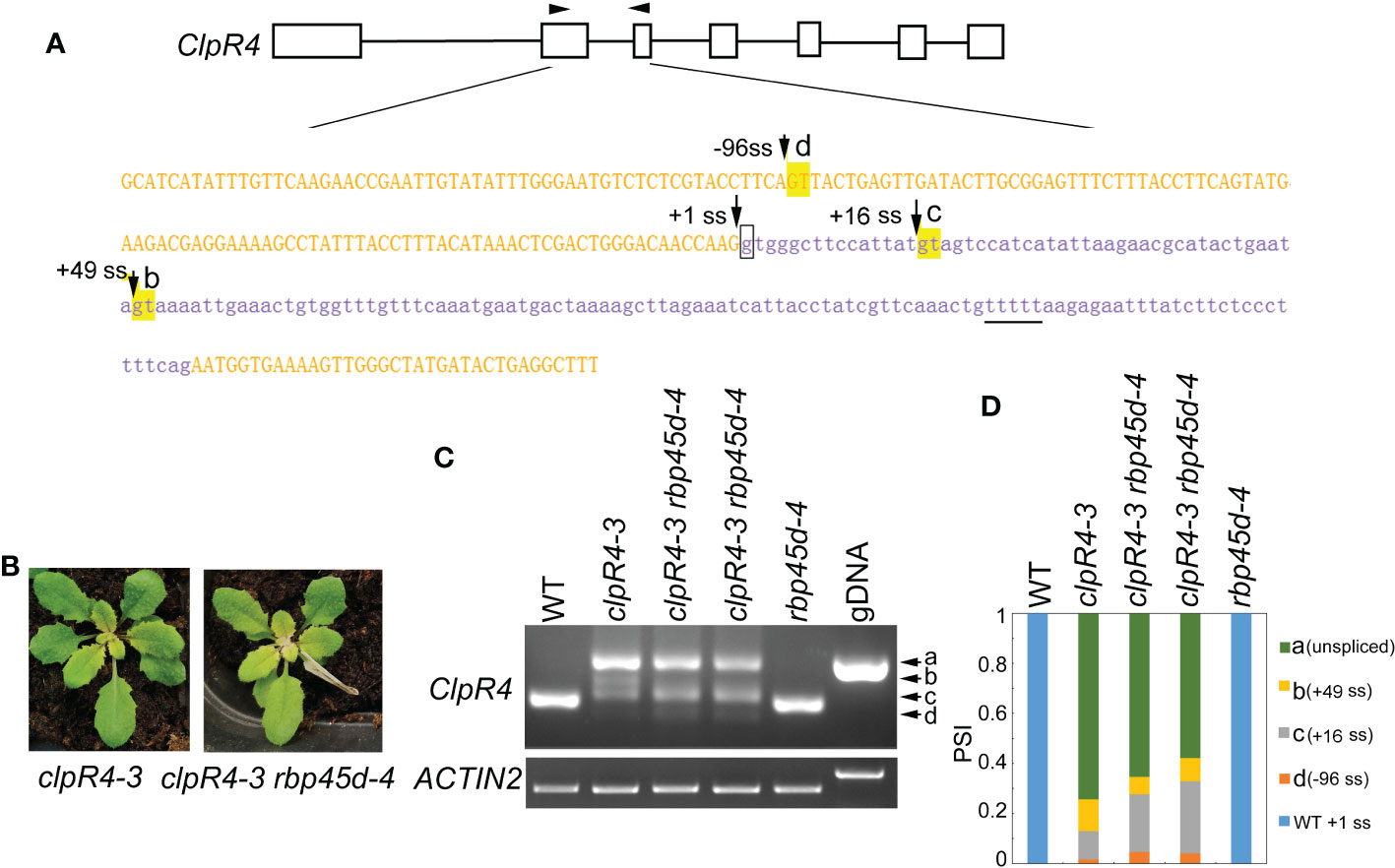
Figure 7 RBP45d regulates the cryptic 5’ splice site selection and splicing efficiency of ClpR4 intron 2 in clpR4-3. (A) Schematic diagrams of the ClpR4 gene and sequences of ClpR4 exon 2 and intron 2. The black boxes indicate exons and the black lines indicate introns. Upper and lower case letters indicate exon and intron sequences, respectively. The arrows labelled with d, c and b indicate the activated cryptic 5’ splice sites at the -96 ss, +16 ss, and +49 ss, respectively. The arrow labelled with +1 ss indicates the authentic splice site, and boxed g indicates the point mutation in clpR4-3. The arrowheads indicate the primers used for RT-PCR. The T-rich sequence downstream the splice site +49ss is underlined. (B) Phenotype of clpR4-3 and clpR4-3 rbp45d-4 mutants. (C) Splicing patterns of ClpR4 intron 2 in WT, clpR4-3 and clpR4-3 rbp45d-4 plants. Band a corresponds to the transcript retained intron 2, whereas band b, c and d correspond to the transcripts spliced at +49 ss, +16 ss and -96 ss, respectively. ACTIN2 is used as an internal control. Two biological replicates (independent pools of aerial parts of plants) were analyzed, and one representative result is shown. (D) Relative levels of the PCR product bands in (C) quantified by ImageJ software. PSI (percent spliced in index) indicates the percentage of each variant in total transcripts.
Mutations in PRP39a suppress the sot5 phenotype
To clone the suppressor gene in the E25 line, which displays a similar phenotype to F23, F11 and E1 but was not allelic with them, we used the backcrossed F2 population between E25 and sot5 to clone the suppressor gene via the MutMap approach. Our data showed that the suppressor gene was located in a region (~1.3 M) containing 15 candidate genes on Chromosome 1 (Figure S9). Interestingly, we found a G to A mutation at the 5’ ss of the eleventh intron in the PRP39a gene, which encodes a U1 snRNP auxiliary protein PRP39a (Figure 8A). To verify the result, we identified a T-DNA insertion mutant prp39a (prp39a-1, salk_133733) from the ABRC stock, and made the sot5 prp39a-1 double mutant by genetic crossing. The double mutant exhibited the same phenotype as prp39a-1 (Figure 8B). Likewise, PCR analysis showed that the pattern of splicing products in sot5 prp39a was similar to that in E25 (Figures 8C, D). In addition, we overexpressed the PRP39a genomic DNA in E25, and found that the phenotype and the splicing pattern of the transgenic lines were the same as those of sot5 (Figures 8C, D). Taken together, our results suggest that PRP39a mutations contribute to the suppression of sot5 in E25.
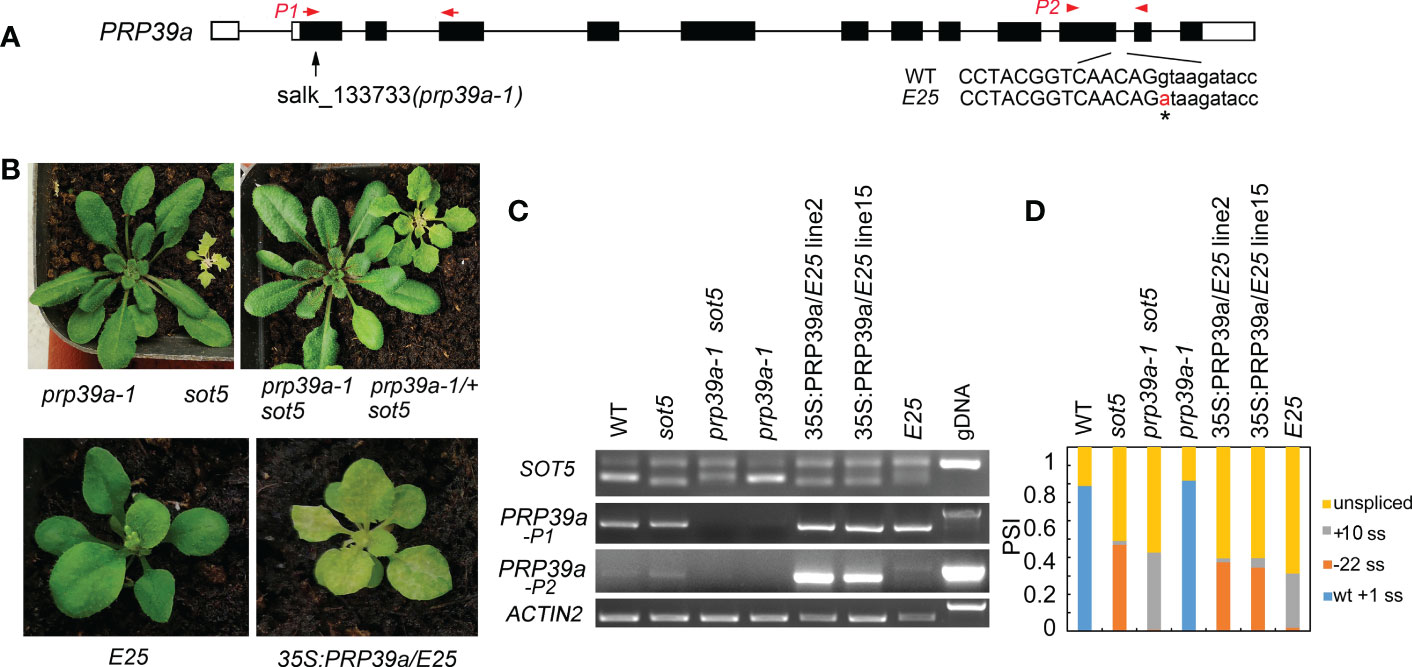
Figure 8 PRP39a is a new suppressor gene isolated from E25. (A) The schematic presentation of the PRP39a gene. The black boxes indicate exons and the black lines indicate introns. Black arrow showed the site of T-DNA insertion. The G to A point mutation of E25 was indicated by *. And red arrow and arrowhead showed the primer pair P1 and P2 used for RT-PCR, respectively. (B) Upper panel, phenotypes of prp39a-1, sot5, sot5 prp39a-1 and PRP39a-1/+ sot5. Lower panel, phenotypes of E25 and 35S:PRP39a-gDNA/E25 transgenic plants. (C) RT-PCR analysis of the PRP39a-1, sot5 prp39a-1 and 35S:PRP39a-gDNA/E25 transgenic plants. ACTIN2 is used as an internal control. Two biological replicates (independent pools of aerial parts of plants) were analyzed, and one representative result is shown. (D) Quantification of the SOT5 PCR product bands in (C). PSI (percent spliced in index) indicates the percentage of each variant in total transcripts.
RBP45d specifically interacts with PRP39a
The yeast Prp39 and it paralog Prp42 form the scaffold connecting auxiliary proteins associated with U1 snRNP to the core (Li et al., 2017). RT-PCR analysis and eFP browser showed that Arabidopsis PRP39a is ubiquitously expressed (Figure S10A). However, PRP39a protein specifically localized in nucleus, which is different from that of RBP45d (Figure S10B). Our genetic screening of sot5 suppressors indicated that RBP45d and PRP39a act in the same genetic pathway with regard to SOT5 intron 7 alternative splicing. In addition, the prp39a-1 mutant exhibited the same shorter primary root phenotype as rbp45d, compared with that of WT (Figure S11). We then examined whether PRP39a and RBP45d physically interact each other in vitro and in vivo. Y2H results showed that PRP39a directly interacted with RBP45d, but not with other RBP45/47 members (Figure 9A). Consistently, BiFC analysis showed that PRP39a and RBP45d interacted specifically in the nucleus (Figure 9B), which is consistent with the nucleus localization of PRP39a (Figure S10). However, we did not detect the fluorescence signal between PRP39a and RBP47b (Figure 9B), indicating that they are not associated each other in vivo. These results are consistent with previously reported that the role of RBP45d in alternative splicing relies on its specific interaction with PRP39a (Chang et al., 2022). Meanwhile both Y2H and BiFC results showed that PRP39a was able to interact with U1C in the nucleus (Figure 9B). Taken together, our results suggest that Arabidopsis RBP45d, PRP39a and U1C function in a similar way to that in yeast with regard to alternative splicing regulation.
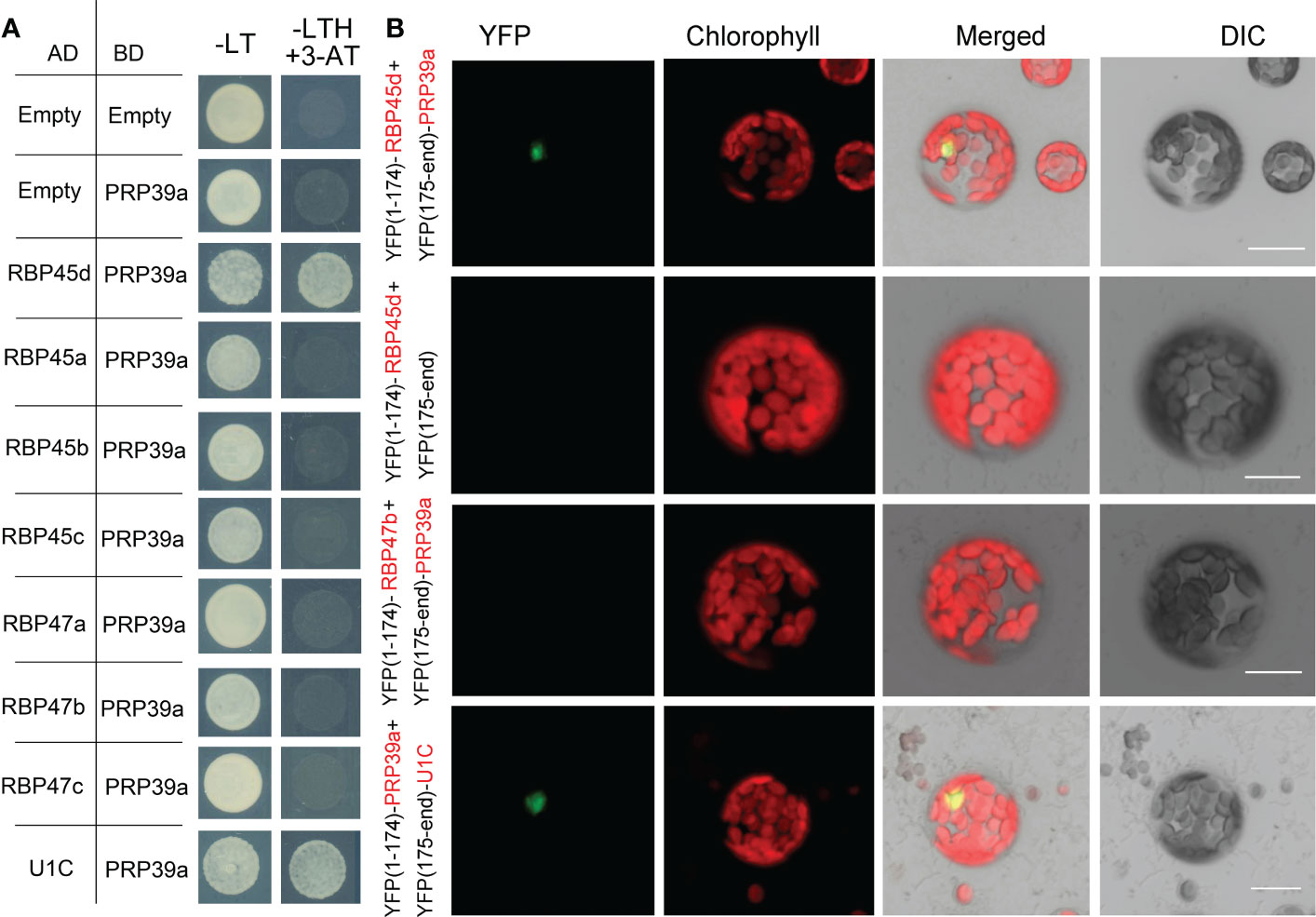
Figure 9 PRP39a interacts with RBP45d and U1C. (A) Yeast two-hybrid assays of the interaction between PRP39a and RBP45/47 members or U1C. The transformed yeast cells were grown on -LT (SD-Leu/-Trp) mediums and -LTH+3-AT (SD- Leu/-Trp/-His with 5 mM 3-AT) mediums. (B) BiFC assay of the interaction between PRP39a and RBP45d, RBP47b or U1C. YFP, fluorescence of yellow fluorescent protein; DIC, Differential interference contrast microscopy; Merge, merge of YFP, DIC and chlorophyll. Bar = 15 μm.
Discussion
In this study, we used a powerful genetic approach to screen phenotypic suppressors of the sot5 mutant, which contains a G to A mutation at the first nucleotide of intron 7 (Huang et al., 2018), and isolated two suppressor genes, RBP45d and PRP39a. RBP45d and PRP39a are yeast Nam8 and Prp39 homologs, respectively, which are auxiliary components of the U1 snRNP complex. Although they have been demonstrated to play important roles in alternative splicing in yeast, human and Arabidopsis (Puig et al., 1999; Le Guiner et al., 2001; Forch et al., 2002; Chang et al., 2022), their function in cryptic splice site selection was not reported yet. Here we collected several lines of evidence substantially supporting that RBP45d and PRP39a are two trans-acting splicing factors that facilitate U1 snRNP to select specific 5’ ss. First, loss-of-function mutations in RBP45d and PRP39a significantly alter 5’ cryptic splice site usage in the sot5 mutant. Second, RBP45d physically interacts with the core subunit U1C of U1 snRNP and the U1 auxiliary factor PRP39a. Third, RBP45d can bind to U-rich sequences and its binding affinity is positively correlated to the U content of the sequence. Taken together, our data suggest that RBP45d promotes 5’ ss selection by directly binding to its downstream U-rich cis-element and then recruits U1 snRNP to the splice site by interacting with both PRP39a and U1C.
The working model of RBP45d and PRP39a in facilitating U1 snRNP to select specific 5’ ss
It is well known that U1 snRNP plays a critical role in initiation of 5’ ss recognition by base pairing of U1 snRNA and 5’ ss consensus sequence. And several U1 snRNP auxiliary proteins such as Nam8, Luc7 and RBM25 can also influence 5’ ss selection, especially when the 5’ss sequence are not conserved (Fortes et al., 1999; Puig et al., 1999; Forch et al., 2002; Puig et al., 2007; Zhou et al., 2008). Based on the data reported previously, the Arabidopsis U1 snRNP component RBP45d is required for alternative splicing for a set of genes (Kanno et al., 2020; Chang et al., 2022). Consistently, the two RBP45d homologs, yeast Nam8 and human TIA-1, are implicated in alternative splicing via binding to the U-rich region downstream of a weak 5’ ss (Puig et al., 1999; Le Guiner et al., 2001; Aznarez et al., 2008; Qiu et al., 2011). The similar molecular function of Nam8, TIA-1, and RBP45d is attributed to their highly conserved domains including the three RRM domains and the C-terminal Gln-rich tail. Cryo electron microscopy (cryo-EM) analysis of prespliceosome structure revealed that Nam8 or TIA-1 binds to U-rich sequences through the RRM2 domain to define the use of weak 5’ ss, and interacts with the U1C-terminal region near the U1-5’ ss helix via the RRM3 domain (Plaschka et al., 2018). Recently, Cryo-EM structure of the yeast U1 snRNP showed that the alternative splicing factors Prp39 and Prp42 form heterodimer that acts as a central scaffold connecting all auxiliary proteins associated with U1 snRNP to the core (Li et al., 2017). In human, there is no Prp42 homolog, but the Prp39 homolog PrpF39 forms a homodimer that acts as the Prp39/Prp42 heterodimer in U1 snRNP. In addition, Nam8/TIA-1 interacts with the Prp39/Prp42 heterodimer by its RRM3 domain and C-terminal tail (Plaschka et al., 2018). The structural and biochemical data provide insight into alternative splicing mediated by Nam8/TIA-1 and Prp39/prp42. Although the U1 snRNP complex and structure remain to be dissected in plants, most of the counterparts of yeast U1 snRNP core subunits and auxiliary proteins are present in Arabidopsis and encoded by multiple genes (Wang and Brendel, 2004). Considering that the basic mechanism of pre-mRNA splicing is highly conserved in all eukaryotic organisms, we propose that RBP45d functions as a U1 snRNP auxiliary factor in the same way as Nam8/TIA-1. As expectedly, our data showed that RBP45d can not only bind to U1C (the core protein of U1 snRNP) and PRP39a (the scaffold protein connecting the auxiliary proteins to the core), but also to the downstream U-rich RNA sequence near 5’ ss of SOT5 intron 7. Accordingly, we suggest a working model that RBP45d can recruit spliceosome to specific 5’ ss and enhance the stability of spliceosome and pre-mRNA complex by directly binding to U-rich RNA sequences downstream of 5’ ss (Figure 10).
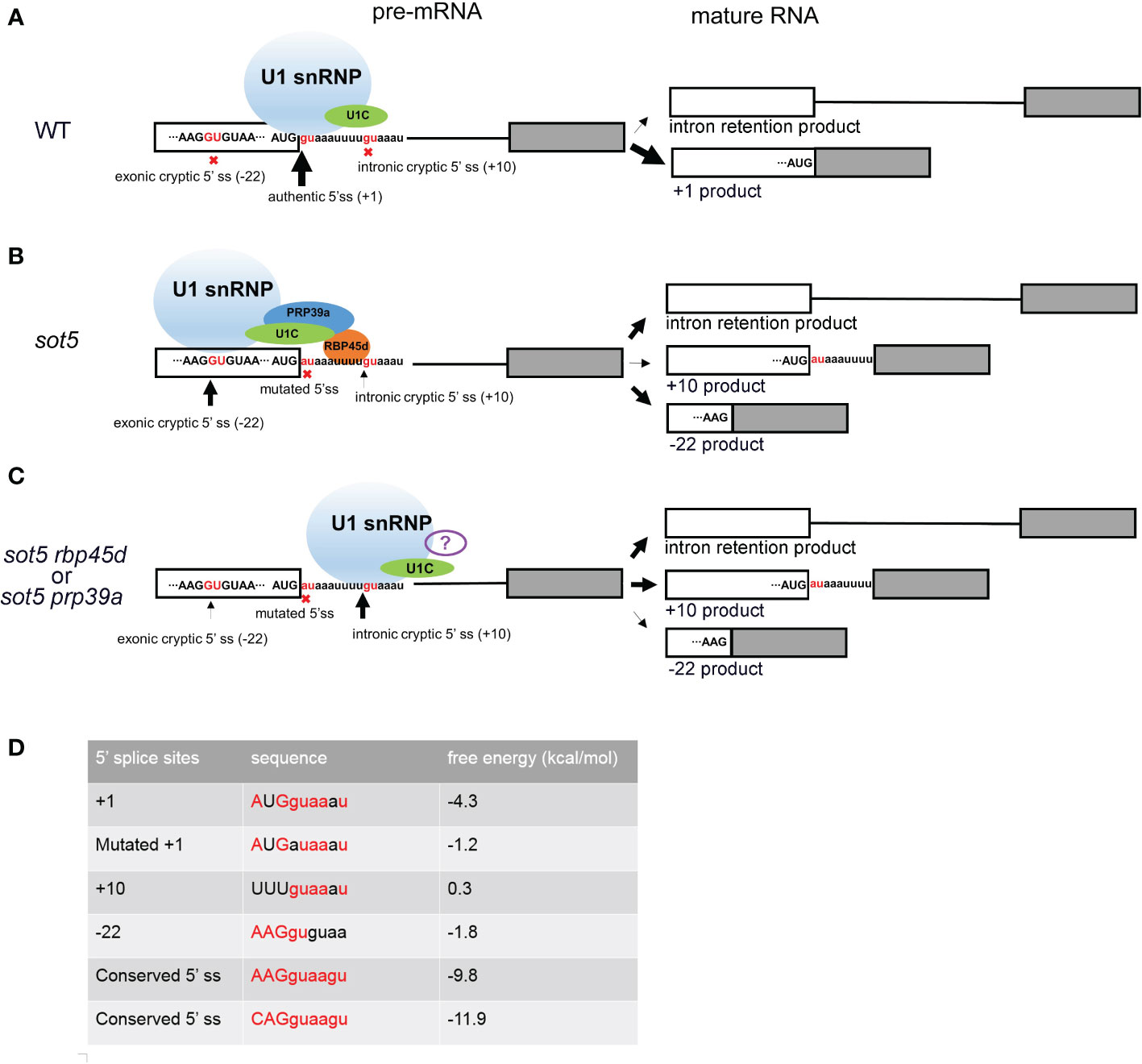
Figure 10 Working model of RBP45d and PRP39a in 5’ cryptic splice site selection. (A) In WT plant, most SOT5 transcripts are excised intron 7 and only a very small proportion of SOT5 transcripts retained intron 7. Since the authentic 5’ ss is strong enough to well base pair with U1 snRNA and recruit U1 snRNP, no cryptic 5’ ss nearby were used. (B) In sot5, the mutation of the authentic 5’ ss of SOT5 intron7 led to the reduced base pairing with U1 snRNA, so that the exonic and intronic cryptic 5’ ss nearby are activated. Beside, nearly half transcripts retained intron 7. In this scenario, RBP45d binds to the U-rich element located downstream of the exonic cryptic 5’ ss and recruits U1 snRNP to select the exonic cryptic 5’ ss by physically interacting with both U1C and PRP39a. It is also likely that the RBP45d bound to the U-rich element which is proximal to the intronic cryptic 5’ ss masks this 5’ ss. (C) When RBP45d or PRP39a is absent or disrupted, the masked intronic cryptic 5’ ss is exposed and is preferred by the U1 snRNP complex (lacking PRP39a and RBP45d or with some other unknown splicing factors). The white and gray boxes indicate the exon 7 and exon 8 of SOT5 transcripts, respectively. The black lines indicate the intron 7. The sequence nearby the 5’ ss of intron 7 was shown and the invariant GU dinucleotides were colored by red. The nine nucleotides AUGguaaau indicate the authentic 5’ ss in WT. The nine nucleotides AUGauaaau indicate the mutated 5’ ss in sot5. The nine nucleotides AAGGUGUAA indicate the exonic cryptic 5’ ss in SOT5 exon 7. The nine nucleotides uuuguaaau indicate the intronic cryptic 5’ ss in SOT5 intron7. The red crosses indicate the splice sites were not used. The black arrows indicate the usage of the 5’ ss and the thickness of the arrows indicate the frequency of the splice site usage. (D) Comparison of the conserved degree of these 5’ ss and the free energy of the RNA duplex formed by the 5’ ss based pairing with the 5’ end of U1 snRNA. The conserved 5’ ss sequence well based paired with the 5’ end of U1 snRNA, the free energy of the RNA duplex is low. On the contrast, if the 5’ ss sequence did not based paired with the 5’ end of U1 snRNA, the free energy of the RNA duplex is relatively high. The conserved nucleotides are highlighted by red color. The free energy was calculated by The RNAeval web server (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAeval.cgi).
RBP45d and PRP39a regulate cryptic splice site selection in 5’ ss mutants
Cryptic splice sites are repressed by complicated mechanisms in eukaryotes. Study of many model organisms revealed that when the authentic splice site is mutated, the cryptic splice sites close to the authentic splice site will be activated (Roca et al., 2003). Usually, there are multiple cryptic splice sites nearby. The mechanism of which cryptic splice site is preferentially selected is not very clear. It is reported that according to the conservation degree of the sequence near 5’ ss, the strong or weak 5’ ss can be predicted. The more conservative the motif is (i.e. it can be perfectly base paired with U1 snRNA), the stronger the splice site is. And the strong splice site will be preferentially and efficiently spliced. On the contrary, the less conservative the motif, the weaker the splice site is (Roca et al., 2003; Dawes et al., 2022). However, when there are AU-rich or U-rich sequences in the intron, this prediction is not accurate, because the U-rich sequence downstream of the weak splice site will be bound by specific splicing factors, such as Nam8p or TIA-1, and the weak splice site will be activated in this scenario. It is reported the AU-rich sequences in plant introns are more abundant and the sequence of 5’ ss is more variable. Therefore, the selection of plant splice sites may be more dependent on the location and abundance of AU- rich or U-rich sequence of introns (McCullough et al., 1993). But what trans-acting factor that acting on the AU- rich or U-rich sequence in plants remains unclear. Our study showed that RBP45d is a such splicing factor for U-rich introns splicing in plants. When the 5’ ss of SOT5 intron7 is mutated, two cryptic splice sites at position - 22 and + 10 are activated, but the + 10 ss (splice site) is much weaker than the - 22 ss. RBP45d or PRP39a mutation reverse the usage of two cryptic splice sites (Figure 10). By carefully analyzing the sequences downstream of these two cryptic splicing sites, we found that there is a U- rich sequence that can be bound by RBP45d downstream of - 22 ss. In addition, we demonstrated that RBP45d physically interacted with PRP39a, consistent with the results that they act on the same genetic pathway.
The possible different roles of U1 snRNP auxiliary proteins RBP45d, PRP39a, LUC7 paralogs and RBM25 in regulating 5’ ss selection
CryoEM revealed that yeast U1 snRNP contains seven additional U1 auxiliary proteins including Prp39, Prp40, Snu71, Snu56, Luc7, Prp42, and Nam8 (Li et al., 2017). Yeast Luc7, a Zinic-finger RNA binding protein, is also involved in 5 ‘ss recognition (Fortes et al., 1999). It is suggested that interaction of yeast Luc7 with the upstream exon stabilizes the pre-mRNA–U1 snRNP interaction. Similarly, human hLuc7A affects 5’ss selection possibly via the formation of a network of protein-RNA interactions that stabilize pre-mRNA–U1 snRNP interaction and thus formation of the commitment complex. (Puig et al., 2007). There are three LUC7 paralogs (LUC7A, LUC7B and LUC7RL) in Arabidopsis genome and they specifically promote splicing of a subset of terminal introns and affects plant development and adaptation to stress (de Francisco Amorim et al., 2018). Whether the LUC7 family members acted at the same pathway with RBP45d and PRP39a remains unclear. Arabidopsis RBM25 (homolog of yeast Snu71) was aslo an U1 auxiliary protein, but it acts differently with RBP45d to regulate alternative splicing (Kanno et al., 2017). Human RBM25 was reported to specifically bind to Bcl-x RNA through a CGGGCA sequence located within exon 2 and enhance Bcl-xs 5’ ss selection and thus regulate apoptotic cell death (Zhou et al., 2008). In addition, human RBM25 was found to associate with a U1 snRNP associated factor, hLuc7A by Co-IP (Zhou et al., 2008). These results indicate that the different U1 snRNP auxiliary protein may bind different cis-element in exons or introns to stabilize U1 snRNP and pre-mRNA complex and differentially regulate the alternative splicing of the pre-mRNA. The relationship among RBP45d, RBM25, and LUC7 family members remains to be elucidated.
It is noted that in Arabidopsis, although some of the U1 snRNP auxiliary proteins are encoded by two or more genes, they generally do not function redundantly in pre-mRNA splicing. RBP45d is a good example. Besides, Kanno et al., 2017 reported that Arabidopsis PRP39a mutation changes the alternative splicing of GFP reporter gene, while PRP39b cannot, indicating that they have different functions in splicing regulation. Probably, LUC7A, LUC7B and LUC7RL may also preserved their unique function in regulating pre-mRNA splicing. How these Arabidopsis U1 snRNP auxiliary proteins regulate 5’ss selection remains to be explored.
The distinct function of RBP45d from other members
There are eight paralogs in the Arabidopsis RBP45/47 family. Whether they function redundantly or not remains unclear. Here we showed that RBP45d functions differently with other RBP45/47 members. The specificity of RBP45d in pre-mRNA splicing is supported by the following evidence. The sot5 phenotype can be rescued by mutations in RBP45d but not in other RBP45/47 members. Only RBP45d is interacted with PRP39a, since other RBP45/47 members lack the long C-terminal tail that was demonstrated to play an important role in protein-protein interaction between RBP45d and PRP39a (Chang et al., 2022). Furthermore, we observed that rbp45d mutants have short primary root and late flowering phenotypes, which are not present in other rbp45/47 mutants (Figure 4 and Figure S7). However, we found some common features in all the RBP45/47 members. For example, they are localized in both the nucleus and cytoplasm (Figure S5), and they all interact with the U1 snRNP core subunit U1C (Figure S6). Thus, it is likely that other RBP45/47 members are also involved in splicing regulation through the mechanisms different from those of RBP45d. It is noted that most of the RBP45/47 members including RBP45d are localized both in nucleus and cytoplasm, indicating that they may also play roles in mRNA regulation in cytoplasm. It will be interesting to investigate biological functions of other RBP45/47 members as well as the molecular mechanisms by which they act in plants.
It is interestingly that the Tabaco NtRBP45 was recently reported to facilitate post-transcriptional gene silencing (PTGS) by promoting siRNA production (Zhang et al., 2020). While the Aarabidopsis RBP45d was reported to promote RNA-directed DNA methylation (RdDM), similar to the known components of RdDM (NRPD2A, RDR2, AGO4 and AGO6). It is suggested that RBP45d facilitates siRNA production by stabilizing either the precursor RNA or the slicer protein (Wang et al., 2021). Several research groups have also found mutations of some mRNA splicing factors reduced the level of siRNA dependent on POL IV, but it is unclear how these splicing factors affect the level of siRNA (Ausin et al., 2012; Dou et al., 2013; Huang et al., 2013; Zhang et al., 2013). It will be interesting to explore how the splicing factors function in RNA-directed DNA methylation in the future.
Materials and methods
Plant materials and growth conditions
The Arabidopsis ecotype Columbia-0 (Col-0) was used as the wild type (WT) in this study. The mutants sot5 and clpR4-3 were previously described (Huang et al., 2018, Wu et al., 2013). The prp39a-1 (salk_133733), rbp45a (salk_140650), rbp45c (salk_063484), rbp47a (salk_142402), and rbp47b (GK-626C01) mutants were obtained from the ABRC stock center (https://abrc.osu.edu/) and were genotyped by PCR using gene specific primers (Table S1). Double mutants prp39a-1 sot5, rbp45a sot5, rbp45c sot5 rbp47a sot5 rbp47b sot5, and clpR4-3 rbp45d-4 were identified from F2 generations derived from crosses between single mutants by the PCR-based genotyping procedure. Seeds were surface-sterilized by 75% ethanol and stratified at 4°C for 3 days, and then sown onto half-strength Murashige and Skoog (MS) medium with 0.5% sucrose. Seeds were also directly sown in soil and grown in a phytotron with long-day conditions (16 h light/8 h dark) and light intensity (100 μmol photons m-2 s-1) at 22°C.
EMS mutagenesis, gene cloning, plasmid construction and transformation
In order to obtain sot5 suppressors, sot5 seeds were ethyl methanesulfonate (EMS) mutagenized (0.1%). About 30000 M2 plants from 5000 M1 lines were screened. Several suppressor lines that displayed WT-like green leaves were obtained from the M2 generation. To clone F23, we produced an F2 population from the cross between F23 and Landsberg erecta (Ler). In the F2 population, about eighty F23 plants were pooled for Whole Genome Resequencing and Mapping-By-Sequencing (MBS) analysis (OE Biotech Co.,Ltd). To clone the E25 gene, we crossed the E25 with sot5 and generated the BC1 F2 population. About thirty E25 plants were pooled for Whole Genome Resequencing and Mutmap analysis (Biomarker Technologies).
The RBP45d coding DNA sequence (CDS) and PRP39a genomic DNA were cloned into the pENTR SD/D-TOPO entry vector (Invitrogen), and then recombined into the pGWB2 destination vector, respectively. The destination vector containing RBP45d was transformed into F23 mutant and the destination vector containing PRP39a was transformed into E25 mutant according to the method as our previously described (Huang et al., 2018). To obtain recombinant RBP45d protein, the RBP45d CDS was transferred into the pet 51b vector and then was fused with His tag at its N-terminal.
Generation of rbp45d-CR mutant lines by CRISPR/Cas9-mediated genome editing
The rbp45d-CR mutant line was generated by CRISPR/Cas9-mediated genome editing. The sgRNA sequences were designed by the online software CRISPR-P v2.0 (Liu et al., 2017) and CRISPR-GE (Xie et al., 2017) to target exon1 of the RBP45d gene. The binary vector pYAO-Cas9-1300 (Feng et al., 2018) was linearized using BsaI (ThermoFisher Scientific, UK) endonuclease. Two complementary sgRNA nucleotides were synthesized and annealed to generate double-strand DNA with appropriate overhangs on both ends and then inserted into the pYAO-Cas9-1300 vector. The vector was transformed into the Agrobacterium tumefaciens strain GV3101 competence cells. And the GV3101 with the vector was transformed into the Col-0 plants by the floral dip method. The transgenic plants of T1 generation were screened by Hygromycin B resistance, DNA from positive lines was extracted and PCR and sequencing were performed to identify mutations in sgRNA target sequences. Finally, we screened the Cas9-free background and homozygous RBP45d gene editing lines in T2 generation plants to obtain rbp45d-CR mutant lines.
Analysis of RT-PCR and sequencing of the cloned PCR products
For RT-PCR, total RNAs were extracted from seedlings or different plant tissues by RNA Easy Fast Plant Tissue Kit (TIANGEN, DP452) according to the manufacturer’s instructions. DNase I treatment and RT-PCR analysis were conducted as previously described (Huang et al., 2018). Semi-quantitative RT-PCR was carried out using the gene-specific primers (Table S1). To identify the splicing variants, the PCR products amplified by the primers spanning the introns were purified and cloned into pMD19-T vector (TaKaRa, D102A). About 10~20 clones of each PCR products were sequenced and aligned with the corresponding genes by SnapGene software.
Subcellular localization of RBP45/47 members and PRP39a fused with YFP and BiFC assay
To express YFP-fused protein, the full length CDS of the RBP45a, RBP45b, RBP45c, RBP45d, RBP47a, RBP47b, RBP47c, RBP47c’, PRP39a and U1C genes were amplified from the cDNA of WT seedlings using the primers (Table S1). The sequences were cloned into the pENTR SD/D-TOPO entry vector (Invitrogen) and then recombined into the p2YWG7 vector (http://gateway.psb.ugent.be/vector/show/p2GWY7/search/index/) or BiFC vctors pE3130 and pE3136 (http://www.bio.purdue.edu/people/faculty/gelvin/nsf/protocols_vectors.htm). The purified plasmid was transformed into the Arabidopsis protoplasts according to the method (Wu et al., 2009). The BiFC vector combinations are indicated in the Figures 5 and 10.
Yeast two hybrid
The CDS of RBP45a, RBP45b, RBP45c, RBP45d, RBP47a, RBP47b, RBP47c, PRP39a U1A, U1C and U1-70K were clone into the pENTR vector and then recombined into pDEST22 and pDEST32 destination vectors, respectively. For yeast two-hybrid assays, plasmids were transformed into yeast strain AH109 by the lithium chloride–polyethylene glycol method according to the manufacturer’s manual (Clontech). The transformants were selected on SD-Leu-Trp plates. The protein–protein interactions were tested on SD-Trp-Leu-His plates with or without 5mM 3-amino-1,2,4-triazole (3AT).
Protein expression, purification and RNA electrophoretic mobility shift assay
The RBP45d CDS was cloned into the expression vector pet51b, and protein expression was induced using 0.5 mM isopropyl-b–D–thiogalactopyranoside at 16°C for 12 h in E. coli strain Rosetta. The protein was purified as described by Zhu et al. (2020). The RNA probes were chemically synthesized, and their 5’-end were labeled by Cy5 (Sangon Biotech (Shanghai) Co., Ltd). The sequences of the probes were as follows:
Cy5-SOT5-IN7: 5’-(CY5) CUCAAUAUGAUAAAUUUUGUAAAUC-3’;
The corresponding non-labeled and mutated probes were also chemically synthesized for competition assays.
SOT5-IN7-NL: 5’-CUCAAUAUGAUAAAUUUUGUAAAUC-3’
SOT5-IN7-M1-NL: 5’-CUCAAUAUGGUAAAUUUUGUAAAUC-3’
SOT5-IN7-M2-NL: 5’-CUCAAUAUGGCAAAUUUUGUAAAUC-3’
SOT5-IN7-M3-NL: 5’-CUCAAUAUGGCAAACCGGGUAAAUC-3’
SOT5-IN7-M4-NL: 5’-CUCAAUAUGCCCGGCCGGGUAAAUC-3’
For EMSA assay, RBP45d protein and RNAs were incubated at room temperature for 30 min in the reaction solution (10 mM HEPES pH 7.3, 20 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol, 5% glycerol (v/v) and 0.1 μg tRNA). The reactants were analyzed using a native polyacrylamide gel, and signals were detected by Azure Biosystems C600.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author. The accession numbers of the gene described in the study are listed in Table S2.
Author contributions
WH and JH conceived and designed the research. WH, LZ, YajZ, JC, YawZ, FL and XC performed the experiments. WH and JH supervised the experiments and wrote the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32171293), The Fund of Innovation Program of Shanghai Municipal Education Commission (2021‐01‐07‐00‐02‐E00117), and Shanghai Engineering Research Center of Plant Germplasm Resources (17DZ2252700).
Acknowledgments
We thank the ABRC for providing the prp39a-1 (salk_133733), rbp45a (salk_140650), rbp45c (salk_063484), rbp47a (salk_142402), and rbp47b (GK-626C01) seeds. We thank Yu Fang, Xiong Renxue and Wang Yanzhu for purifying the RBP45d protein. We thank Zhang hui and Wu Wenjuan for designing the sgRNA targeted RBP45d.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1086506/full#supplementary-material
References
Anna, A., Monika, G. (2018). Splicing mutations in human genetic disorders: examples, detection, and confirmation. J. Appl. Genet. 59, 253–268. doi: 10.1007/s13353-018-0444-7
Ausin, I., Greenberg, M. V., Li, C. F., Jacobsen, S. E. (2012). The splicing factor SR45 affects the RNA-directed DNA methylation pathway in arabidopsis. Epigenetics 7, 29–33. doi: 10.4161/epi.7.1.18782
Aznarez, I., Barash, Y., Shai, O., He, D., Zielenski, J., Tsui, L. C., et al. (2008). A systematic analysis of intronic sequences downstream of 5’ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 18, 1247–1258. doi: 10.1101/gr.073155.107
Brown, J. W., Smith, P., Simpson, C. G. (1996). Arabidopsis consensus intron sequences. Plant Mol. Biol. 32, 531–535. doi: 10.1007/BF00019105
Chang, P., Hsieh, H. Y., Tu, S. L. (2022). The U1 snRNP component RBP45d regulates temperature-responsive flowering in arabidopsis. Plant Cell 34, 834–851. doi: 10.1093/plcell/koab273
Chen, M., Manley, J. L. (2009). Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754. doi: 10.1038/nrm2777
Chen, J., Zhu, H., Huang, J., Huang, W. (2020). A new method for functional analysis of plastid EMBRYO-DEFECTIVE PPR genes by efficiently constructing cosuppression lines in arabidopsis. Plant Methods 16, 154. doi: 10.1186/s13007-020-00696-0
Dawes, R., Joshi, H., Cooper, S. T. (2022). Empirical prediction of variant-activated cryptic splice donors using population-based RNA-seq data. Nat. Commun. 13, 1655. doi: 10.1038/s41467-022-29271-y
De Bortoli, F., Neumann, A., Kotte, A., Timmermann, B., Schuler, T., Wahl, M. C., et al. (2019). Increased versatility despite reduced molecular complexity: evolution, structure and function of metazoan splicing factor PRPF39. Nucleic Acids Res. 47, 5867–5879. doi: 10.1093/nar/gkz243
de Francisco Amorim, M., Willing, E. M., Szabo, E. X., Francisco-Mangilet, A. G., Droste-Borel, I., Macek, B., et al. (2018). The U1 snRNP subunit LUC7 modulates plant development and stress responses via regulation of alternative splicing. Plant Cell. 30, 2838–2854. doi: 10.1105/tpc.18.00244
Dou, K., Huang, C. F., Ma, Z. Y., Zhang, C. J., Zhou, J. X., Huang, H. W., et al. (2013). The PRP6-like splicing factor STA1 is involved in RNA-directed DNA methylation by facilitating the production of pol V-dependent scaffold RNAs. Nucleic Acids Res. 41, 8489–8502. doi: 10.1093/nar/gkt639
Du, H., Rosbash, M. (2002). The U1 snRNP protein U1C recognizes the 5’ splice site in the absence of base pairing. Nature 419, 86–90. doi: 10.1038/nature00947
Feng, Z., Zhang, Z., Hua, K., Gao, X., Mao, Y., Botella, J. R., et al. (2018). A highly efficient cell division-specific CRISPR/Cas9 system generates homozygous mutants for multiple genes in arabidopsis. Int. J. Mol. Sci. 19. doi: 10.3390/ijms19123925
Forch, P., Puig, O., Martinez, C., Seraphin, B., Valcarcel, J. (2002). The splicing regulator TIA-1 interacts with U1-c to promote U1 snRNP recruitment to 5’ splice sites. EMBO J. 21, 6882–6892. doi: 10.1093/emboj/cdf668
Fortes, P., Bilbao-Cortes, D., Fornerod, M., Rigaut, G., Raymond, W., Seraphin, B., et al. (1999). Luc7p, a novel yeast U1 snRNP protein with a role in 5’ splice site recognition. Genes Dev. 13, 2425–2438. doi: 10.1101/gad.13.18.2425
Gniadkowski, M., Hemmings-Mieszczak, M., Klahre, U., Liu, H. X., Filipowicz, W. (1996). Characterization of intronic uridine-rich sequence elements acting as possible targets for nuclear proteins during pre-mRNA splicing in nicotiana plumbaginifolia. Nucleic Acids Res. 24, 619–627. doi: 10.1093/nar/24.4.619
Hernando, C. E., Garcia Hourquet, M., de Leone, M. J., Careno, D., Iserte, J., Mora Garcia, S., et al. (2019). A role for pre-mRNA-PROCESSING PROTEIN 40C in the control of growth, development, and stress tolerance in arabidopsis thaliana. Front. Plant Sci. 10, 1019. doi: 10.3389/fpls.2019.01019
Huang, C. F., Miki, D., Tang, K., Zhou, H. R., Zheng, Z., Chen, W., et al. (2013). A pre-mRNA-splicing factor is required for RNA-directed DNA methylation in arabidopsis. PloS Genet. 9, e1003779. doi: 10.1371/journal.pgen.1003779
Huang, W., Zhu, Y., Wu, W., Li, X., Zhang, D., Yin, P., et al. (2018). The pentatricopeptide repeat protein SOT5/EMB2279 is required for plastid rpl2 and trnK intron splicing. Plant Physiol. 177, 684–697. doi: 10.1104/pp.18.00406
Isshiki, M., Morino, K., Nakajima, M., Okagaki, R. J., Wessler, S. R., Izawa, T., et al. (1998). A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5’ splice site of the first intron. Plant J. 15, 133–138. doi: 10.1046/j.1365-313X.1998.00189.x
Kanno, T., Lin, W. D., Fu, J. L., Chang, C. L., Matzke, A. J. M., Matzke, M. (2017). A genetic screen for pre-mRNA splicing mutants of arabidopsis thaliana identifies putative U1 snRNP components RBM25 and PRP39a. Genetics 207, 1347–1359. doi: 10.1534/genetics.117.300149
Kanno, T., Venhuizen, P., Wu, M. T., Chiou, P., Chang, C. L., Kalyna, M., et al. (2020). A collection of pre-mRNA splicing mutants in arabidopsis thaliana. G3 (Bethesda) 10, 1983–1996. doi: 10.1534/g3.119.400998
Kapustin, Y., Chan, E., Sarkar, R., Wong, F., Vorechovsky, I., Winston, R. M., et al. (2011). Cryptic splice sites and split genes. Nucleic Acids Res. 39, 5837–5844. doi: 10.1093/nar/gkr203
Lee, Y., Rio, D. C. (2015). Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323. doi: 10.1146/annurev-biochem-060614-034316
Le Guiner, C., Lejeune, F., Galiana, D., Kister, L., Breathnach, R., Stevenin, J., et al. (2001). TIA-1 and TIAR activate splicing of alternative exons with weak 5’ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 276, 40638–40646. doi: 10.1074/jbc.M105642200
Li, X., Liu, S., Jiang, J., Zhang, L., Espinosa, S., Hill, R. C., et al. (2017). CryoEM structure of saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing. Nat. Commun. 8, 1035. doi: 10.1038/s41467-017-01241-9
Liu, H., Ding, Y., Zhou, Y., Jin, W., Xie, K., Chen, L. L. (2017). CRISPR-p 2.0: An improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. doi: 10.1016/j.molp.2017.01.003
Lorkovic, Z. J., Wieczorek Kirk, D. A., Klahre, U., Hemmings-Mieszczak, M., Filipowicz, W. (2000). RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA 6, 1610–1624. doi: 10.1017/S1355838200001163
McCullough, A. J., Lou, H., Schuler, M. A. (1993). Factors affecting authentic 5’ splice site selection in plant nuclei. Mol. Cell Biol. 13, 1323–1331. doi: 10.1128/mcb.13.3.1323-1331.1993
Montes, M., Sanford, B. L., Comiskey, D. F., Chandler, D. S. (2019). RNA Splicing and disease: Animal models to therapies. Trends Genet. 35, 68–87. doi: 10.1016/j.tig.2018.10.002
Nelissen, R. L., Will, C. L., van Venrooij, W. J., Luhrmann, R. (1994). The association of the U1-specific 70K and c proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 13, 4113–4125. doi: 10.1002/j.1460-2075.1994.tb06729.x
Peal, L., Jambunathan, N., Mahalingam, R. (2011). Phylogenetic and expression analysis of RNA-binding proteins with triple RNA recognition motifs in plants. Mol. Cells 31, 55–64. doi: 10.1007/s10059-011-0001-2
Plaschka, C., Lin, P. C., Charenton, C., Nagai, K. (2018). Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature 559, 419–422. doi: 10.1038/s41586-018-0323-8
Puig, O., Bragado-Nilsson, E., Koski, T., Seraphin, B. (2007). The U1 snRNP-associated factor Luc7p affects 5’ splice site selection in yeast and human. Nucleic Acids Res. 35, 5874–5885. doi: 10.1093/nar/gkm505
Puig, O., Gottschalk, A., Fabrizio, P., Seraphin, B. (1999). Interaction of the U1 snRNP with nonconserved intronic sequences affects 5’ splice site selection. Genes Dev. 13, 569–580. doi: 10.1101/gad.13.5.569
Qiu, Z. R., Schwer, B., Shuman, S. (2011). Determinants of Nam8-dependent splicing of meiotic pre-mRNAs. Nucleic Acids Res. 39, 3427–3445. doi: 10.1093/nar/gkq1328
Reddy, A. S., Marquez, Y., Kalyna, M., Barta, A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25, 3657–3683. doi: 10.1105/tpc.113.117523
Roca, X., Olson, A. J., Rao, A. R., Enerly, E., Kristensen, V. N., Borresen-Dale, A. L., et al. (2008). Features of 5’-splice-site efficiency derived from disease-causing mutations and comparative genomics. Genome Res. 18, 77–87. doi: 10.1101/gr.6859308
Roca, X., Sachidanandam, R., Krainer, A. R. (2003). Intrinsic differences between authentic and cryptic 5’ splice sites. Nucleic Acids Res. 31, 6321–6333. doi: 10.1093/nar/gkg830
Staiger, D., Brown, J. W. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25, 3640–3656. doi: 10.1105/tpc.113.113803
Wang, B. B., Brendel, V. (2004). The ASRG database: identification and survey of arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 5, R102. doi: 10.1186/gb-2004-5-12-r102
Wang, C., Tian, Q., Hou, Z., Mucha, M., Aukerman, M., Olsen, O. A. (2007). The arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep. 26, 1357–1366. doi: 10.1007/s00299-007-0336-5
Wang, L., Xu, D., Scharf, K., Frank, W., Leister, D., Kleine, T. (2021). The RNA-binding protein RBP45D of arabidopsis promotes transgene silencing and flowering time. Plant J 109, 1397–1415. doi: 10.1111/tpj.15637
Will, C. L., Luhrmann, R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3. doi: 10.1101/cshperspect.a003707
Wu, F.-H, Shen, S.-C., Lee, L.-Y., Lee, S.-H., Chan, M.-T., Lin, C.-S., et al. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5.
Wu, W., Zhu, Y., Ma, Z., Sun, Y., Quan, Q., Li, P., et al. (2013). Proteomic evidence for genetic epistasis: ClpR4 mutations switch leaf variegation to virescence in arabidopsis. Plant J. 76, 943–956. doi: 10.1111/tpj.12344
Xie, X., Ma, X., Zhu, Q., Zeng, D., Li, G., Liu, Y. G. (2017). CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 10, 1246–1249. doi: 10.1016/j.molp.2017.06.004
Yuan, F. J., Zhu, D. H., Tan, Y. Y., Dong, D. K., Fu, X. J., Zhu, S. L., et al. (2012). Identification and characterization of the soybean IPK1 ortholog of a low phytic acid mutant reveals an exon-excluding splice-site mutation. Theor. Appl. Genet. 125, 1413–1423. doi: 10.1007/s00122-012-1922-7
Zhang, C. J., Zhou, J. X., Liu, J., Ma, Z. Y., Zhang, S. W., Dou, K., et al. (2013). The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in arabidopsis. EMBO J. 32, 1128–1140. doi: 10.1038/emboj.2013.49
Zhang, W., Zhu, Z., Du, P., Zhang, C., Yan, H., Wang, W., et al. (2020). NtRBP45, a nuclear RNA-binding protein of nicotiana tabacum, facilitates post-transcriptional gene silencing. Plant Direct 4, e00294. doi: 10.1002/pld3.294
Zhan, X., Qian, B., Cao, F., Wu, W., Yang, L., Guan, Q., et al. (2015). An arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 6, 8139. doi: 10.1038/ncomms9139
Zhao, Y., Huang, S., Wang, N., Zhang, Y., Ren, J., Zhao, Y., et al. (2022). Identification of a biomass unaffected pale green mutant gene in Chinese cabbage (Brassica rapa l. ssp. pekinensis). Sci. Rep. 12, 7731. doi: 10.1038/s41598-022-11825-1
Zhou, A., Ou, A. C., Cho, A., Benz, E. J., Jr., Huang, S. C. (2008). Novel splicing factor RBM25 modulates bcl-x pre-mRNA 5’ splice site selection. Mol. Cell Biol. 28, 5924–5936. doi: 10.1128/MCB.00560-08
Keywords: Arabidopsis, 5’ splice site selection, cryptic splice site, U1 snRNP, RBP45d, PRP39a, U-rich sequence
Citation: Huang W, Zhang L, Zhu Y, Chen J, Zhu Y, Lin F, Chen X and Huang J (2022) A genetic screen in Arabidopsis reveals the identical roles for RBP45d and PRP39a in 5’ cryptic splice site selection. Front. Plant Sci. 13:1086506. doi: 10.3389/fpls.2022.1086506
Received: 01 November 2022; Accepted: 02 December 2022;
Published: 22 December 2022.
Edited by:
Jin-Gui Chen, Oak Ridge National Laboratory (DOE), United StatesReviewed by:
Sureshkumar Balasubramanian, Monash University, AustraliaAlan Rose, University of California, Davis, United States
Copyright © 2022 Huang, Zhang, Zhu, Chen, Zhu, Lin, Chen and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jirong Huang, aHVhbmdqckBzaG51LmVkdS5jbg==
 Weihua Huang
Weihua Huang Liqun Zhang
Liqun Zhang Yajuan Zhu2
Yajuan Zhu2 Jirong Huang
Jirong Huang