
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 03 February 2023
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1086007
This article is part of the Research TopicEverything Edamame: Biology, Production, Nutrition, Sensory and Economics, Volume IIView all 8 articles
 Zhibo Wang1
Zhibo Wang1 Dajun Yu2
Dajun Yu2 Gota Morota3
Gota Morota3 Kshitiz Dhakal1
Kshitiz Dhakal1 William Singer1
William Singer1 Nilanka Lord1
Nilanka Lord1 Haibo Huang2
Haibo Huang2 Pengyin Chen4
Pengyin Chen4 Leandro Mozzoni5
Leandro Mozzoni5 Song Li1
Song Li1 Bo Zhang1*
Bo Zhang1*The sucrose and Alanine (Ala) content in edamame beans significantly impacts the sweetness flavor of edamame-derived products as an important attribute to consumers’ acceptance. Unlike grain-type soybeans, edamame beans are harvested as fresh beans at the R6 to R7 growth stages when beans are filled 80-90% of the pod capacity. The genetic basis of sucrose and Ala contents in fresh edamame beans may differ from those in dry seeds. To date, there is no report on the genetic basis of sucrose and Ala contents in the edamame beans. In this study, a genome-wide association study was conducted to identify single nucleotide polymorphisms (SNPs) related to sucrose and Ala levels in edamame beans using an association mapping panel of 189 edamame accessions genotyped with a SoySNP50K BeadChip. A total of 43 and 25 SNPs was associated with sucrose content and Ala content in the edamame beans, respectively. Four genes (Glyma.10g270800, Glyma.08g137500, Glyma.10g268500, and Glyma.18g193600) with known effects on the process of sucrose biosynthesis and 37 novel sucrose-related genes were characterized. Three genes (Gm17g070500, Glyma.14g201100 and Glyma.18g269600) with likely relevant effects in regulating Ala content and 22 novel Ala-related genes were identified. In addition, by summarizing the phenotypic data of edamame beans from three locations in two years, three PI accessions (PI 532469, PI 243551, and PI 407748) were selected as the high sucrose and high Ala parental lines for the perspective breeding of sweet edamame varieties. Thus, the beneficial alleles, candidate genes, and selected PI accessions identified in this study will be fundamental to develop edamame varieties with improved consumers’ acceptance, and eventually promote edamame production as a specialty crop in the United States.
Edamame, vegetable or edible soybean, is a nutritious food source of protein, isoflavones, dietary fibers, and vitamins (Mahoussi et al., 2020; Carneiro et al., 2021; Dhakal et al., 2021; Yu et al., 2022). Edamame has been cultivated and consumed in east Asian countries for centuries and documented edamame varieties are mainly originated from this area (Carneiro et al., 2021; Dhakal et al., 2021; Yu et al., 2022). In the past two decades, production and breeding of locally adapted edamame varieties have been reported in North America, and its consumption has increased (Wolfe et al., 2018). More attention has been paid to edamame as a potential alternative crop among growers and processors in the mid-Atlantic and Southeast United States (Carson et al., 2011; Ogles et al., 2016).
Typically, edamame is eaten as a snack or added to salads, stews and soups (Kumar et al., 2011; Carneiro et al., 2021). Sensory surveys of edamame have shown that the sweetness of edamame bean is positively correlated with consumer acceptance (Yu et al., 2022). Two components, sucrose and Ala, are the main contributors to the sweetness of edamame products (Hou et al., 2009; Yu et al., 2022). To promote domestic production and consumption of edamame, the varieties with improved seed sweetness are needed to match American consumer expectations for edamame products. Therefore, in addition to improving the nutritional content and agronomic properties of edamame, breeders must also consider edamame sensory attributes, such as appearance, flavor and texture, especially sweetness, to increase domestic market (Carneiro et al., 2020). Newly developed edamame variety with improved natural seed sweetness also helps reduce supplementation of added artificial sweeteners.
Soybean seeds typically contain about 35% carbohydrates on a dry matter basis, of which about 40% and 60% are soluble sugars and insoluble sugars, respectively (Sui et al., 2020). Soluble sugars usually consist of 5% sucrose, 1.5% raffinose, and 3% stachyose (Wang et al., 2014; Sui et al., 2020). Sucrose content varies widely among soybean accessions. The sucrose content in cultivated and wild soybean seeds is usually 4-6% and 3-4%, respectively (Sui et al., 2020). Numerous genetic studies on the inheritance of sucrose content in soybean seeds have been reported. Based on the recombinant inbred line (RIL) population crossed between V71-370 and PI 407162, 17 QTL associated with sucrose content were identified on chromosomes (Chr) 5, 7, 8, 13, 15, 19 and 20 (Maughan et al., 2000). Using RIL populations from the ‘Keunolkong’ × ‘Shinpaldalkong’ cross, six QTL associated with sucrose content were characterized on Chr 2, 11, 12, 16 and 19 (Kim et al., 2005). A major QTL on Chr 6 in the Sat_213-Satt643 interval was identified from two F2 populations, which could explain 76% of the observed genetic variation of sucrose content (Skoneczka et al., 2009). A sucrose QTL was reported on Chr 11 from an F2 population derived from V97-3000 × V99-5089 (Wang et al., 2014) while four QTL were identified on Chr 6, 8, 16 and 20 from a cross between G. max (Williams 82 (WM82)) and G. soja (PI 483460B) (Patil et al., 2018). Five SNPs located on Chr 4, 6, 7, 11, and 12 were detected to be associated with sucrose content in a genome-wide association study (GWAS) analysis of 178 diverse soybean accessions (Sui et al., 2020). In summary, there was a total of 362 SNPs and 31 bi-parental QTL (Quantitative Trait Locus) being identified to associate with soybean sucrose content. As another major sweetness contributor, the content of Ala also varies widely in soybean seeds, 15 - 23 mg g-1 dry seed (Qin et al., 2019). A GWAS reported 11 SNPs being related with Ala content in soybean seeds (Qin et al., 2019).
As the cost of genotyping has decreased and statistical methods have been improved, GWAS has become a popular method for understanding the genetic basis of complex traits, enabling more concise breeding of various crop species including soybean, cotton, wheat, rice, maize, and sorghum (Khan et al., 2018; Liu et al., 2018; Jiang et al., 2020; Sui et al., 2020; Gahlaut et al., 2021; Verma et al., 2022; Singer et al., 2022). GWAS is also complementary to QTL mapping and linkage analysis, where QTL analysis deals with the contribution of a locus to variation in continuous trait while GWAS studies the association between alleles and traits (Liu et al., 2018). In soybean, GWAS has been used to characterize the molecular basis of complex traits, as it significantly increases the range of phenotypic variation based on the accumulation of historical recombination events. Specifically, GWAS has been used for agronomic traits (Chang et al., 2018), seed quality traits including protein, oil, and fatty acid content (Zhang et al., 2019; Zhao et al., 2019), disease resistance (Hanson et al., 2018), and abiotic stress adaptation (Do et al., 2019). Advances in genome sequencing have accelerated the resolution and accuracy of the application of GWAS in soybean research (Li et al., 2015).
Because edamame beans are harvested when the seeds are immature, the chemical properties of edamame, especially the sucrose and Ala levels are different from those in dry soybean seeds. As a result, the genetic basis responsible for the sucrose and Ala contents between edamame and dry seeds might be different. The objective of this study was to investigate the genes associated with sucrose and Ala contents in edamame through a GWAS approach in order to better assist edamame breeding on sensory and flavor.
A total of 189 collected edamame accessions (Table S1) were planted at Blacksburg (Virginia), Portageville (Missouri), and Fayetteville (Arkansas) in the years of 2018 and 2019. The seeds were sown in 3 m row and 0.75 m row spacing (with a seeding rate of approximately 70,000 plants per hectare). Field trials were performed with three to four replications in a randomized complete block design (RCBD). The edamame beans were harvested at R6-R7 stages when the bean was filled 80-90% of the pod capacity to quantify sucrose and Ala contents.
Sucrose and Ala contents in edamame beans were measured using the procedures described by Yu et al. (2021) (Yu et al., 2021; Yu et al., 2022). Briefly, the freeze-dried beans were ground to powder to pass through a 500-μm sieve. Subsequently, 0.15 g of the powder was mixed with 1.5 mL of deionized (DI) water and then shaken for 2 hours at room temperature. The mixture was centrifuged at 13,500 g for 10 minutes, and 750 μL of supernatant was collected and shaken with an equal volume of acetonitrile for 10 minutes. With another centrifuge at 13,500 g for 10 minutes, 750 μL of supernatant was collected and filtered by a 0.20-μm membrane filter. The pass-through was measured for its sucrose and Ala contents by high performance liquid chromatography (HPLC).
Sucrose content was determined by HPLC with a refractive index detector (RID) (Agilent Technologies, Santa Clara, CA, USA). A Luna Omega 3 μm SUGAR column (150 × 4.6 mm, Phenomenex, Torrance, CA, USA) was applied for sugar separation at 40°C in which acetonitrile/water (75:25 v:v) was used as the mobile phase. The flow rate was 1 mL/min and the injection volume was 5 μL.
To determine the Ala content, HPLC was performed using online derivatization with o-Phthalaldehyde (OPA), and the Agilent AdvanceBio Amino Acid Analysis (AAA) C18 column with a 1200 diode array detector at λ = 338 nm. Mobile phase A contained 10 mM Na2HPO4, and 10 mM Na2B4O7 with pH 8.2 while mobile phase B mixed acetonitrile, methanol, and DI water (45:45:10, v: v: v). The flow rate was set at 1.5 mL/min with a gradient program for separation with details described in Yu et al. (2021). Each sample for sucrose and Ala measurements had two technical replicates which were averaged to account for biological and equipment variation.
A discriminant analysis of principal components (DAPC) was conducted to assess the population structure of the panel by using the adegenet package in R (Jombart et al., 2010). A total of three principal components were retained to maximize cumulative variance and Bayesian information criterion (BIC) was used to identify an optimal number of clusters.
Best linear unbiased estimates (BLUE) of genotypes were inferred while accounting for systematic effects. The BLUE values were obtained using the equation below,
Where yijk is a vector of the of sucrose or Ala content for the kth genotype in the ith replication and the jth environment, μis the grand mean, ri is the fixed effect of replication, bj is the fixed effect of environment (combined analysis only), gk is the fixed effect of genotype, and eijk is the model residual.
The genetic signals associated with the sucrose content and Ala content in edamame beans were identified using a SoySNP50K iSelect BeadChip (Song et al., 2013). The association between 41,985 SNPs and traits was determined using a mixed linear model (MLM) implemented in the R package rrBLUP (Endelman, 2011). In total, we generated 10 phenotypic datasets for exploring the SNPs that associated with the sucrose and Ala contents. There are six datasets for six individual environments, Blacksburg in 2018 or 2019, Portageville in 2018 or 2019, and Fayetteville in 2018 or and 2019. The two years’ data for the same location were combined and displayed as: Blacksburg combined dataset, Portageville combined dataset, and Fayetteville combined dataset. At last, BLUE (Haslett and Puntanen, 2009) was introduced to adjust the phenotypic data scored across all locations and years.
A modified Šidák correction (αsid = 1 − (1 − α)1/m) for multiple testing was used to derive a statistical significance threshold. The effective number of markers (Meff) was calculated to replace m, and thus, the adjusted significance threshold at α = 5% and the suggestive threshold at α = 10% were (P) > 3.01 and (P) > 2.70, respectively (Li and Ji, 2005). The R packages ggplot2 and dplyr were used to generate Manhattan and quantile-quantile (QQ) plots.
The mean, standard deviation, and distribution of sucrose content were calculated and analyzed (Figure 1 and Table S2). Sucrose contents across all environments displayed normal, continuous distributions with a grand mean of 47.84 mg g -1 fresh bean and an average standard deviation (SD) of 16.06 mg g -1 fresh bean. Edamame sucrose distributions were displayed for all environments (Figure 1A), Blacksburg 2018 and 2019 (Figure 1B), Portageville 2018 and 2019 (Figure 1C), and Fayetteville 2018 and 2019 (Figure 1D). Specifically, Blacksburg 2018, 2019; Portageville 2018, 2019; and Fayetteville 2018, 2019 environments had means and SDs of 42.29, 65.77, 39.98, 44.47, 37.38, and 48.10 mg g -1 fresh bean and 8.39, 18.34, 8.29, 14.07, 15.17, and 11.98 mg g -1 g fresh bean, respectively.
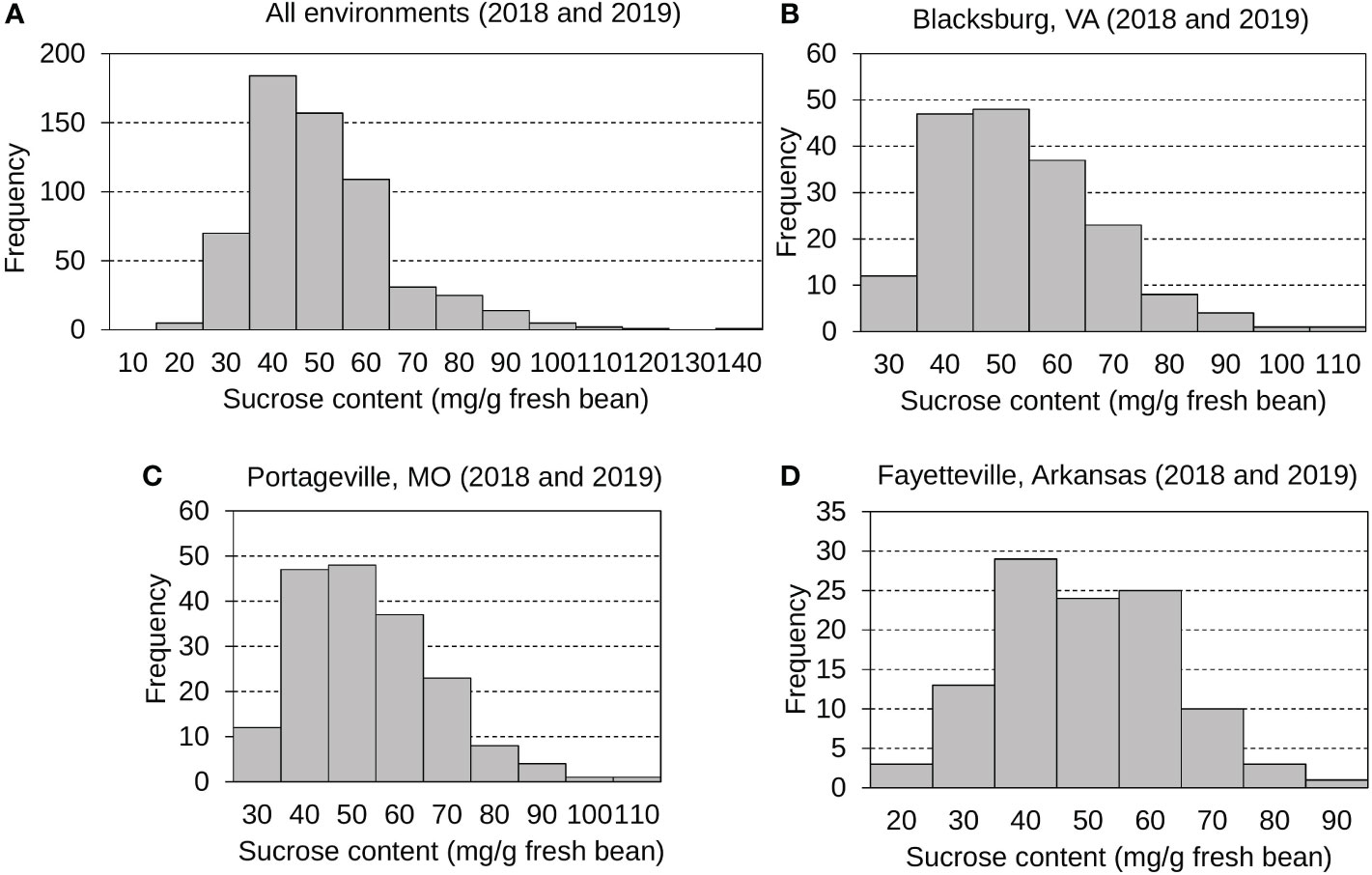
Figure 1 Frequency distributions displaying edamame bean sucrose contents collected from all environments (A), Blacksburg, VA (B), Portageville, MO (C), and Fayetteville, AR (D).
As another significant contributor to edamame sweetness for customers (Yu et al., 2021), the mean, standard deviation, and distribution of Ala content in fresh edamame beans were also examined and investigated (Figure 2 and Table S2). Generally, the Ala content varies more greatly than the sucrose content. Edamame Ala distributions were displayed for all environments (Figure 2A), Blacksburg 2018 and 2019 (Figure 2B), Portageville 2018 and 2019 (Figure 2C), and Fayetteville 2018 and 2019 (Figure 2D). Ala contents for Fayetteville displayed a normal, continuous distribution while it across other environments displayed skewed distributions. It had a grand mean of 1.51 mg g -1 fresh bean and an average SD of 1.20 mg g -1 fresh bean. Blacksburg 2018, 2019; Portageville 2018, 2019; and Fayetteville 2018, 2019 environments had Ala content means and SDs of 0.91, 1.62, 1.83, 1.60, 1.53, and 1.72 mg g -1 fresh bean and 0.68, 1.54, 0.96, 1.38, 0.96, and 1.07 mg g -1 g fresh bean, respectively. Samples grown in Blacksburg 2019 had significantly higher Ala content than that in 2018 while the Ala content in edamame beans from other two locations did not show notable difference across years of 2018 and 2019.
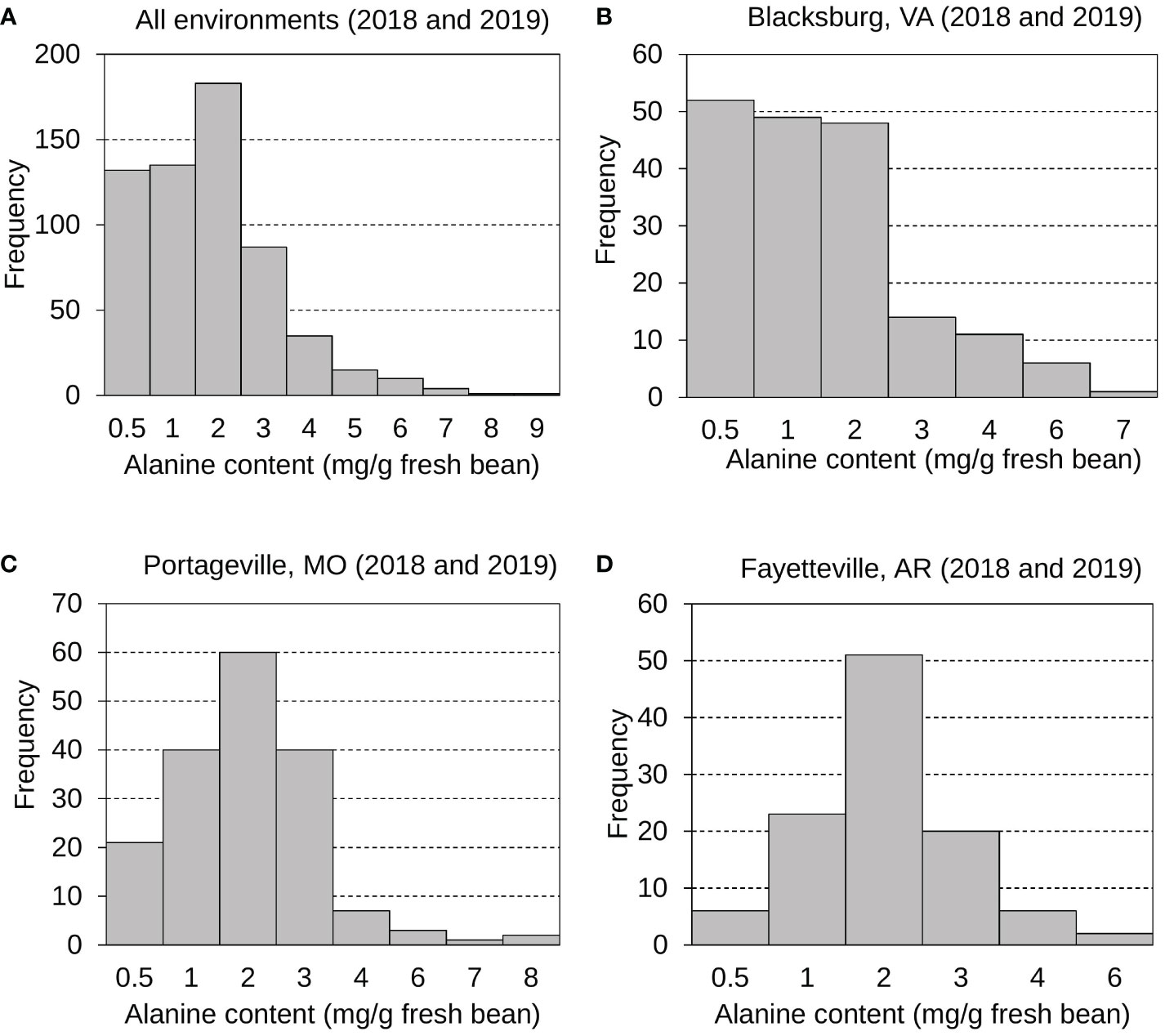
Figure 2 Frequency distributions displaying edamame bean proteinogenic Ala contents collected from all environments (A), Blacksburg, VA (B), Portageville, MO (C), and Fayetteville, AR (D).
Although the consumption of edamame has increased in the United States in the past 20 years, most edamame is still imported from Asia (Wolfe et al., 2018). Thus, in present study, all accessions are original from East Asia, including South Korea, North Korea, Japan, mainland China, and Taiwan, China except for PI 548624, which was from Delaware, United States.
Using the R package DAPC, commonly used in GWAS (Jombart et al., 2010), 189 edamame accessions were classified into three genetic clusters (Figure 3). Cluster I has 27 accessions that are all from South Korea classified at mature group IV (Table S1). Cluster II has 54 accessions including 49 from South Korea, two from North Korea, two from northern China and one from Japan. With the maturity group information from ars-grin.org, the majority of the germplasm in this cluster still belongs to group IV (Table S1). The remaining edamame accessions were classified into cluster III with relatively high diversity in both geographic origin and maturity (Table S1). Specifically, this cluster includes 75 from Japan, 21 from South Korea, two from North Korea, one from the United States, 6 from mainland China, and three from Taiwan, China with maturity groups from III to VI.
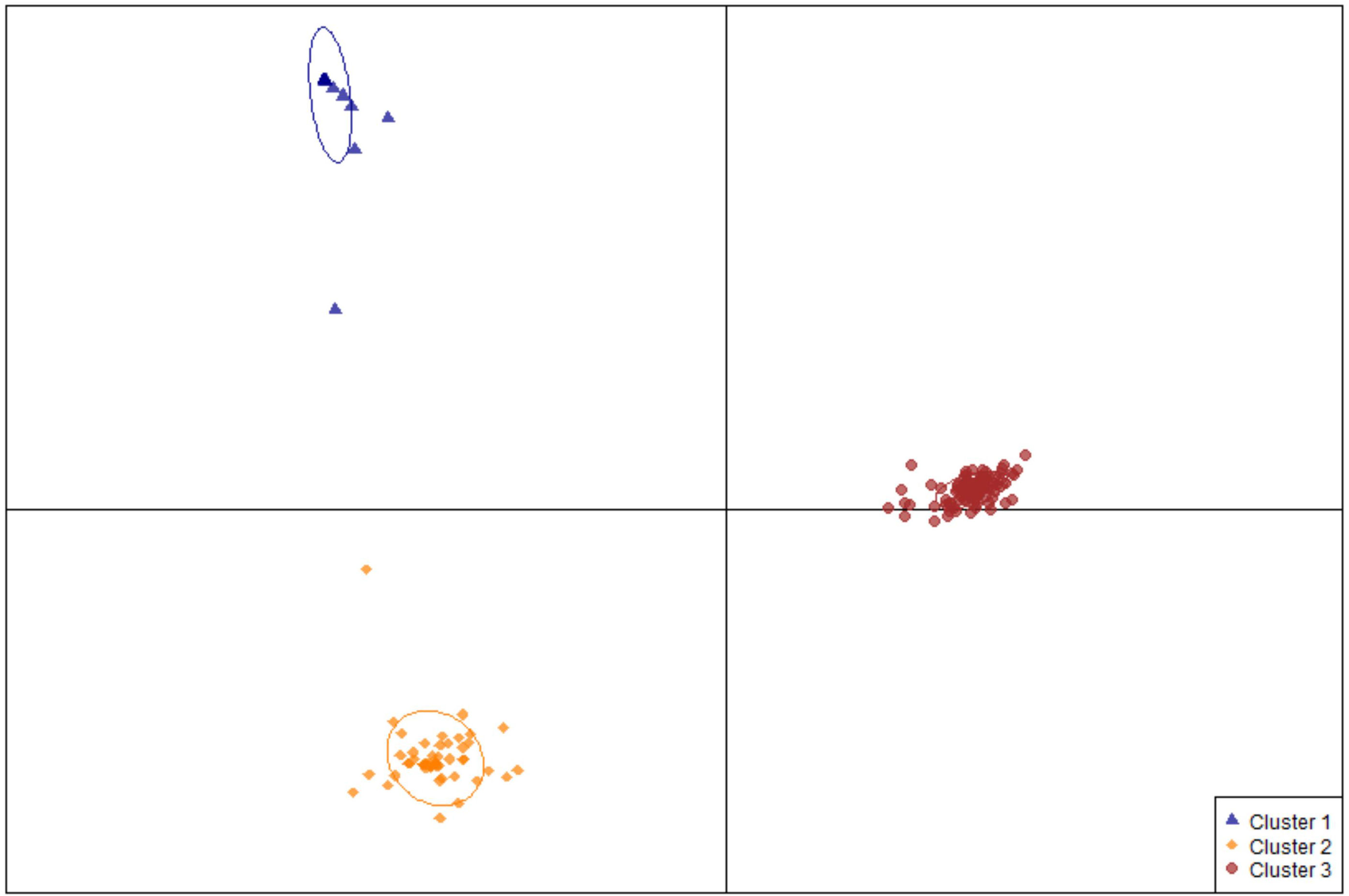
Figure 3 Population structure of edamame germplasms used in this study. A scatter plot depicting the three clusters identified as likely subpopulations within the 189 accessions: cluster I (blue triangle, n = 27), cluster II (gold diamonds, n = 54), cluster III (red circles, n = 107).
SNPs associated with sucrose content were identified using the MLM. The SNPs associated with edamame sucrose and Ala contents were determined by significant and suggestive thresholds of -log10(P) > 3.01 (α = 0.05, the blue line in Manhattan plot in Figures 4, 5, S1, S2) and -log10(P) > 2.70 (α = 0.10, the red line in Manhattan plot in Figures 4, 5, S1, S2), using a modified Šidák (Li and Ji, 2005) adjustment.
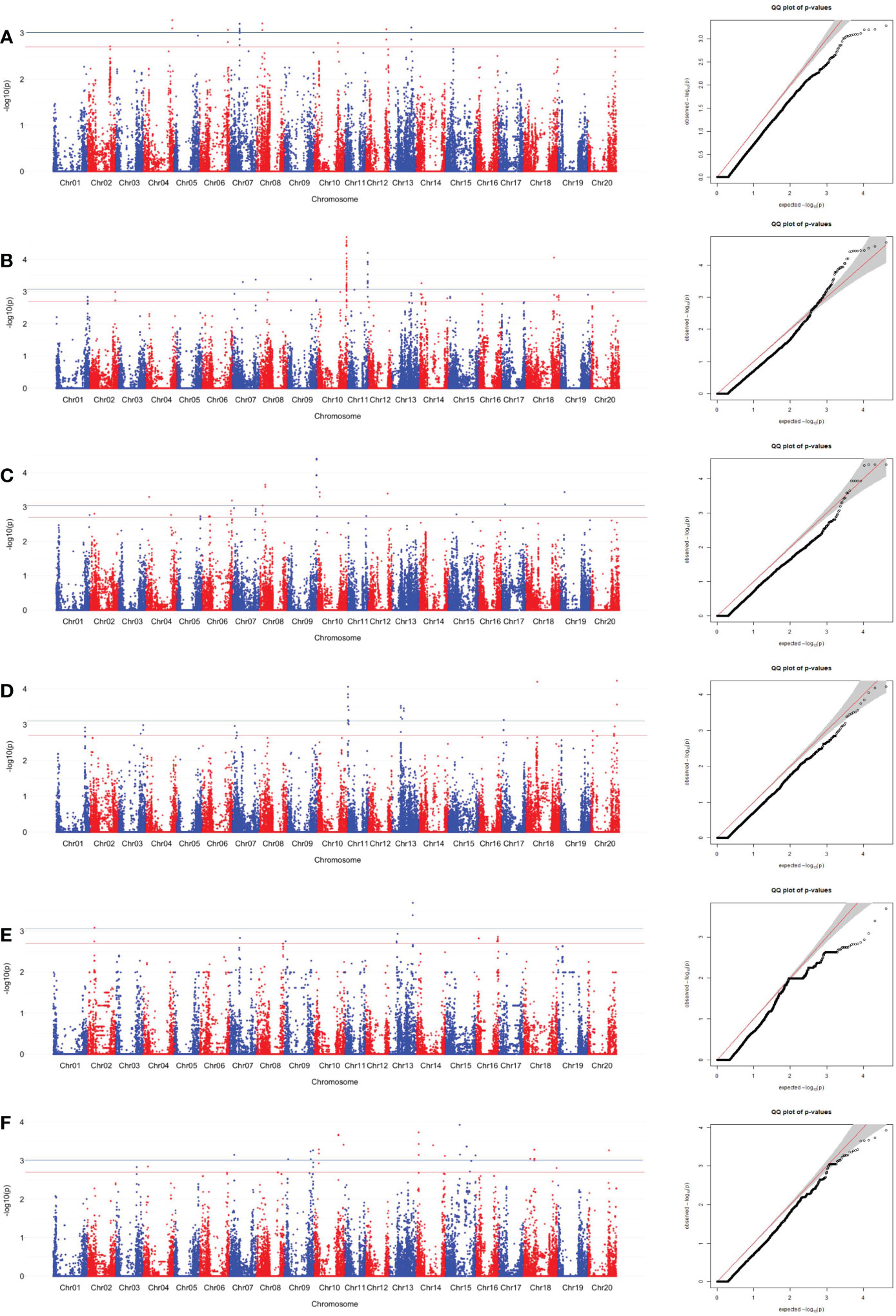
Figure 4 SNP associations for sucrose content in edamame beans harvest from (A) Blacksburg, VA, 2018, (B) Blacksburg, VA, 2019, (C) Portageville, MO, 2018, (D) Portageville, MO, 2019, (E) Fayetteville, AR, 2018, (F) Fayetteville, AR, 2019, are displayed in Manhattan plots with chromosomes in alternating colors, significance thresholds-log10(P) > 3.01 (the blue line) and suggestive threshold-log10(P) > 2.70 (the red line). Each respective QQ plot displays observed-log10(P) against expected-log10(P).
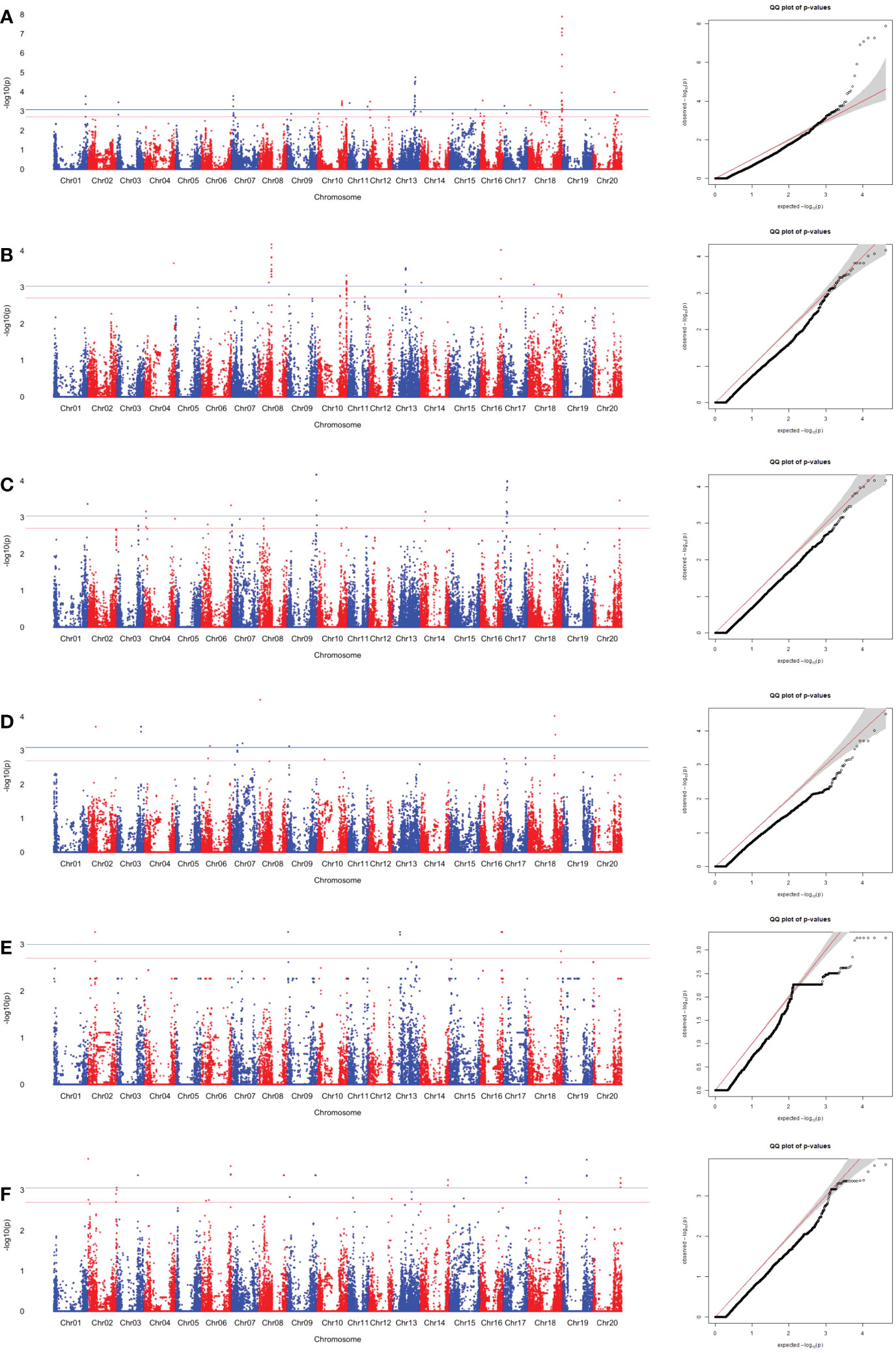
Figure 5 SNP associations for Ala content in edamame beans harvest from (A) Blacksburg, VA, 2018, (B) Blacksburg, VA, 2019, (C) Portageville, MO, 2018, (D) Portageville, MO, 2019, (E) Fayetteville, AR, 2018, (F) Fayetteville, AR, 2019, are displayed in Manhattan plots with chromosomes in alternating colors, significance thresholds-log10(P) > 3.01 (the blue line) and suggestive threshold-log10(P) > 2.70 (the red line). Each respective QQ plot displays observed-log10(P) against expected-log10(P).
A total of 43 SNPs was identified as being associated with sucrose contents in at least two phenotypic datasets (Table S3). Figure 3 and Figure S1 visualized the associations between the SNPs and sucrose content in the individual dataset and combined datasets, respectively.
Fayetteville 2018 dataset, Fayetteville 2019 dataset, and Fayetteville combined dataset had four, 16, and 22 overlapped associated SNPs, respectively; Portageville 2018 dataset, Portageville 2019 dataset, and Portageville combined dataset had three, eight, and 13 overlapped associated SNPs, respectively; and Blacksburg 2018 dataset, Blacksburg 2019 dataset, and Blacksburg combined dataset had two, five, and seven overlapped associated SNPs, respectively. In addition, BLUE combined dataset had nine overlapped associated SNPs. It was noticed that 40 SNPs associated with two phenotypic datasets while 3 SNPs associated with three phenotypic datasets. They were: ss715599697 (Chr 8) being associated with Blacksburg 2019, Portageville combined, and BLUE combined; ss715602625 (Chr 8) being associated with Portageville 2018, Fayetteville combined, and BLUE combined; ss715618513 being associated with Blacksburg combined, Fayetteville 2018 and 2019. The SNP associations are distributed on 11 chromosomes in the soybean genome: Chromosome 3, 6, 8, 9, 10, 13, 14, 16, 17, 18, and 20. Chr 18 contained 12 SNP associations, which was the most, followed by seven SNP associations in Chr 13, five SNP associations in both Chr 14 and Chr 8, three SNP associations in both Chr 6 and Chr 20, two SNP associations in three chromosomes including Chr 9, Chr 16 and Chr 17, one SNP association in both Chr 3 and Chr 10.
A total of 25 SNPs was identified as being associated with Ala contents in edamame beans in at least two phenotypic datasets (Table S4). Figure 5 and Figure S2 visualized the associations between the SNPs with the sucrose content in the individual dataset, and combined datasets, respectively. BLUE combined dataset had the 19 overlapped associated SNPs, what was the most among all the phenotypic datasets. Fayetteville 2018 dataset, Fayetteville 2019 dataset, and Fayetteville combined dataset had one, three, and four overlapped associated SNPs, respectively; Portageville 2018 dataset, Portageville 2019 dataset, and Portageville combined dataset had five, one, and eight overlapped associated SNPs, respectively; and Blacksburg 2018 dataset and Blacksburg combined dataset had seven and six overlapped associated SNPs, respectively.
Although there were 35 (above the significant threshold) and 20 (above the suggestive threshold) SNPs being identified to associate with Alaine content in the Blacksburg 2019 dataset (Figure 4), none of them overlapped with the SNPs derived by other phenotypic datasets. Twenty-one SNPs associated with two phenotypic datasets while four SNPs associated with three phenotypic datasets. Interestingly, the four SNPs were adjacent in the Chr 8: ss715628063, ss715628063, ss715628067 all being associated with Portageville 2018, Portageville combined, and BLUE combined while ss715628064 being associated with Blacksburg 2018, Portageville 2018, and BLUE combined. The associations distributed in nine chromosomes in soybean genome: Chr 2, 6, 8, 10, 11, 13, 14, 17, 18. Chr 17 contained the 10 SNP associations, which was the most, followed by five SNP associations in Chr 14, four SNP associations in Chr 13, and one SNP association in Chr 2, Chr6, Chr 8, Chr 10, Chr 11, and Chr 18.
In total, 39 sucrose-related candidate genes from WM82 were found within 10 kb flanking regions of individual significant SNP (Table S5). A number of gene models were found on eleven chromosomes: one on Chr 3, three on Chr 6, four on Chr 8, two on Chr 9, one on Chr 10, seven on Chr 13, five on Chr 14, two on Chr 16, two on Chr 17, nine on Chr 18, three on Chr 20 (Table S5). These candidate genes have various functions in the numerous biological and biochemical processes. Interestingly, there were two genes (Glyma.08g137500 and Glyma.10g270800) being annotated to function as the trehalose-6-phosphate synthase (TPS). TPS is able to catalyze the production of trehalose-6-phosphate (T6P), which can convert to trehalose after consecutive dephosphorylation by trehalose-6-phosphate phosphatase (TPP) (Ponnu et al., 2011; Yadav et al., 2014). T6P has been shown to regulate sucrose utilization in plants since the precursors of T6P are derived from the sucrose metabolism (Yadav et al., 2014). Thus, the two genes might serve as a key component regulating the sucrose content in edamame beans.
In addition, there were two SNPs (ss715607940 and ss715631133) being identified only associated with the sucrose content in the edamame beans harvested by Blacksburg 2019. Within the flanking regions of these two significant SNPs, two genes were found to have the potential to control the sucrose content. Glyma.18g193600 and Glyma.10g268500 located near the ss715631133 and ss715607940 on Chr 18 and Chr 10, respectively. Glyma.18g193600 encoded a Fructose-1,6-bisphosphatase (FBPase, EC:3.1.3.11) protein, which was characterized as a key enzyme in the plant sucrose synthesis pathway, in the Calvin cycle (Ge et al., 2020). Glyma.10g268500 encoded a Fructose-1, 6-bisphosphate aldolase (FBA, EC 4.1.2.13) protein enzyme that is a key plant enzyme being involved in glycolysis, gluconeogenesis, and the Calvin cycle (Lv et al., 2017). It is worthy to investigate the potential function of the fructose-metabolism-related genes on regulating the edamame bean sucrose content for the basic edamame genetics research and applied edamame breeding.
In total, 25 Ala-related candidate genes from WM82 were found within 10 kb flanking regions of each significant SNP (Table S6). A number of gene models were found on nine chromosomes: one on Chr 2, one on Chr 6, one on Chr 8, one on Chr 10, one on Chr 11, four on Chr 13, five on Chr 14, 10 on Chr 17, and one on Chr 18 (Table S6). Based on the annotated information in the soybase, these candidate gene models belong to several protein families with various metabolic and biosynthesis implications including DNA binding, development and programmed cell death, protein kinase, etc. It is noteworthy there were two genes (Glyma.14g201100 andGlyma.18g269600) being annotated to function as the O-methyltransferase (OMT) while the gene of Glyma.17g070500 was annotated to encode a S-adenosylmethionine (SAM) decarboxylase. Both OMT and SAM are involved in the biochemical pathway from methionine to cysteine, which is located at the upstream pathway from cysteine to Ala (Morishige et al., 2010; Hildebrandt et al., 2015). It implies that the three candidate genes above might have the potential in controlling metabolism of Ala in edamame beans that is worthy for further validation and investigation.
The sucrose and Ala in edamame beans are two desirable traits for sweet taste and flavor of edamame and its derived food product to promote edamame as a nutritional specialty crop in the world. Therefore, varieties with high sucrose and Ala levels are urgently needed in the edamame industry. In the current study, 189 potential edamame accessions with large seed size (> 20 g/100 seeds) (Table S1) were collected. The sucrose and Ala contents were determined in the seeds obtained from three states for two years. A GWAS study was employed to explore the genetic controls for these two traits using SoySNP50K iSelect BeadChip (Song et al., 2013).
Major seed composition characteristics, such as amino acid, oil, and sucrose content, are significantly affected by environments (Bandillo et al., 2015; Chaudhary et al., 2015). The sucrose and Ala levels in the edamame beans were different across three locations (Figures 1, 2 and Table S2). It might be due to the diversity of temperatures and precipitation rates among three locations, affecting plant growth and/or seed development (Zeng et al., 2014). It was reported that decreased soil moisture content increased the sucrose level (Wang et al., 2021). Blacksburg (VA), 2019 had a much higher precipitation (42.15 inches) than 2018 (59.35 inches), which may explain why the average sucrose content in edamame harvested by Blacksburg 2019 was higher than that in 2018. The similar negative relationship was also observed between the edamame bean sucrose content and the precipitation level at the location of Portageville, MO. It had 48.57 inches precipitation in the year of 2018 and an average sucrose content was 39.98 mg g-1 fresh bean, while it had 61.84 inches precipitation in the year of 2019 and an average sucrose content was 44.47 mg g-1 fresh bean. However, the similar trend was not found between the edamame bean sucrose content and the precipitation level at the location of Fayetteville, AR. It implies that the sucrose content in fresh edamame bean is a complex trait affected by multiple environmental factors and more research efforts need to be invested to decouple their relationship. In contrast to sucrose content, Ala content was distributed more divergently. There were few reports characterizing the genetic basis of Ala synthesis and signaling in the soybean/edamame beans. Our study can lay a foundation of understanding the impact of environmental factors on the edamame Ala content in order to boost future breeding edamame varieties with sweeter flavor.
The long-term goal of this study is to serve as the foundation for breeding edamame varieties with improved sensory adapted to distinctive environments under the global climate change challenge. Therefore, in addition to investigating the genetic causes regulating sucrose and Ala contents in edamame beans by GWAS, there is an urgent need to select a collection of sweet edamame germplasms with moderate diversity.
By summarizing the sucrose and Ala contents in the edamame beans, three PI accessions were selected for their high sucrose and high Ala content. PI 532469, PI 243551, and PI 407748 had sucrose content means and SDs of 63.69, 71.67, 65.01 mg g-1 fresh bean and 6.53, 29.3, 16.91 mg g-1 fresh bean while they had Ala content means and SDs of 4.60, 3.73, 3.62 mg g-1 fresh bean and 1.42, 2.75, 1.90 mg g-1 fresh bean. They had high content of sucrose and Ala in more than one tested environment, displaying consistent sweetness. The three “sweet” accessions characterized in present study had the comparable sucrose content but significantly high Ala content. By contrast, VT Sweet, a recent released edamame cultivar (Zhang et al., 2021), had 69.22 ± 21.65 mg sucrose g-1 fresh bean and 0.59 ± 0.07 mg Ala g-1 fresh bean in the Blacksburg extension trial, 2019. PI532469 and PI243551 both originated from Japan and PI407748 originated from China. PI243551 and PI407748 belong to maturity group III while the information of maturity information remains unclear for PI532469. These three accessions can serve as genetic resources in edamame breeding to provide sweet trait.
As shown in Figure 3 and Table S1, 188 out of 189 edamame accessions used in this study originated from East Asia whereas only PI548624 is from the United States, mainly because soybean is original from China, and the communities in East Asia started consuming edamame centuries ago. The current edamame germplasm from East Asia provides the materials for other global regions, including the United States for edamame breeding.
Although the geographical origins of 188 germplasms is East Asia, the R package DAPC was still able to distinguish them (Jombart et al., 2010). All 27 accessions in cluster I and 49 out of 54 accessions in cluster II originated from South Korea (Table S1). In addition, most accessions in these two clusters belong to maturity group IV (Table S1), implying their potential adaptability to the south-central United States. The edamame germplasms in cluster III display a higher divergency in both geographic origin and maturity (Table S1). They were documented originally in Japan, South Korea, North Korea, mainland China, and Taiwan, China with maturity groups of III, IV, V or even later (Table S1). The diverse germplasms in cluster III have the potential to promote the edamame as a specialty crop adapting to all states of America.
In present study, the sucrose and Ala contents were measured in the edamame beans harvested from six environmental combinations (location × year). However, because of the environmental variation and genotype by environment interaction, the SNPs associated with sucrose and Ala were different at each environment. To diminish the variations caused by local environments and fluctuation of annual weather conditions, adjusted phenotype based on BLUE was used to characterize sucrose and Ala related genes in the edamame beans (Haslett and Puntanen, 2009). The sucrose and Ala contents of six combinations were combined for the phenotyping input. With the combined data, two SNPs coupling with Ala and one SNP associated with sucrose were identified, which were different from the SNPs derived from the phenotyping data of combinations of certain location × certain year. The functions of these SNPs in the sucrose and Ala signaling require to be validated by further experimental evidence.
Soluble sugar content in soybean seeds, especially sucrose content, significantly affects the taste and flavor of soybean foods such as soy milk and tofu (Kim et al., 2005). In addition to its important nutritional value in seeds, sucrose is the main photosynthesis product of higher plants. It is not only a carbon base for plant physiological metabolism, but also a signal molecule that coordinates the relationship between plant sources and sinks. Sucrose is involved in various metabolic and biosynthetic processes and plays a crucial role in plant growth and seed development (Ruan, 2014). Furthermore, sucrose has important functions in responses to various abiotic stresses (Rosa et al., 2009; Du et al., 2020).
In this study, a total of 41 candidate genes were screened in genomic regions of 45 SNPs (Table S5). Further investigations of their biological functions can assist selection for edamame germplasms with higher sucrose content. Of the candidate genes, four leucine-rich repeat-containing genes were identified (Table S5). Leucine-rich repeats (LRR) containing genes represent a large and complex gene family in plants, and are functional in various biological and biochemical processes including development, plant immunity, and responses to environmental stimuli (Dufayard et al., 2017). Essential for plant growth and development, sucrose is engaged in plant defense by activating plant immune responses against pathogens (Tauzin and Giardina, 2014). During infection, pathogens reallocate the plant sugars for their own needs forcing the plants to modify their sugar content and triggering their defense responses (Tauzin and Giardina, 2014). Thus, we assume that the LRR domain contains genes allied with sucrose content that might be involved in the plant defense behavior mediated by sucrose signaling.
In addition, 18 enzyme-type genes with catalytic function and two kinase-type genes were also characterized to associate with sucrose content in the edamame beans (Table S5). Due to the importance of sucrose for plants, its synthesis, accumulation and signaling in plants are regulated with a subtle manner. A large number of enzymes along with kinases have been reported in charge of sucrose homeostasis. For instance, the members in two kinase families including SNRK and CDPK, are broadly demonstrated to participate in the synthesis, transportation, and signaling of sucrose (Hulsmans et al., 2016; Crizel et al., 2020). The functions of candidate enzyme and kinase genes derived from GWAS analysis of sucrose content in this study will be noteworthy to investigate to reveal the sucrose related mechanism in soybean seeds and edamame beans.
To date, 37 QTL and 362 genes were identified that are related with sucrose content in dried soybean seeds (https://www.soybase.org/search/index.php?searchterm=sucrose). By comparison with these reported genetic sources responsible for soybean sucrose content, two genes (Gm10g268500 and Gm18g193600) obtained from this study were confirmed to have effects in regulating sucrose content and encode two enzyme proteins that locate adjacently in the sucrose synthesis pathway. The first one is FBA, encoded by Gm10g268500. It catalyzes the reversible aldol cleavage of fructose-1,6-bisphosphate (FBP) into dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP), either in glycolysis or gluconeogenesis and in the Calvin-Benson cycle (Lv et al., 2017). The second gene is FBPase, encoded by Gm18g193600. It catalyzes the conversion of FBP to fructose-6-phosphate in the Calvin cycle and eventually to starch in chloroplast, while it catalyzes the conversion of triose phosphates to sucrose in the sucrose-biosynthesis pathway in cytosol (Serrato et al., 2009). Both FBA and FBPase are the key enzymes in the plant sucrose synthesis pathway, in the Calvin cycle, and also play important roles in photosynthesis regulation in green plants (Serrato et al., 2009; Lv et al., 2017). These reactions are essential for carbon fixation and sucrose metabolism. The results confirmed once again that the two genes have solid effects in regulating sucrose content. Hence, the value of these two genes for increasing the content of sucrose in edamame was worth exploring in the future.
In addition to FBPase and FBA genes, two TPS genes, Gm10g270800 and Glyma.08g137500), were unveiled to couple with sucrose content in the edamame beans. TPS catalyzes the synthesis of T6P from UDP-glucose (UDPG) and glucose-6-P (G6P), which are derived from the sucrose metabolism (Ponnu et al., 2011; Yadav et al., 2014). A regulatory loop consisting of T6P, SnRK1, and bZIP11 is reported controlling sucrose availability and utilization (Delatte et al., 2011). In addition, T6P has recently emerged as an important signaling metabolite, regulating carbon assimilation, plant sugar status, and plant development (Ponnu et al., 2011; Zhang et al., 2017). Our results suggested that TPS and T6P-related metabolism might be involved in manipulating edamame bean sucrose content, which has never been reported in grain type soybean basic genetics. It implies the genetic base for sucrose content in fresh edamame bean might vary from that in dried soybean seed and provide a novel insight for edamame bean sucrose genetics as well as a promising foundation for edamame breeding with improved flavor.
Catabolic pathways for Ala in plants is very short, where cysteine desulfurase (EC 2.8.1.7) catalyzes the conversion from cysteine to Ala, and Ala is directly converted to pyruvate by Ala aminotransferase (EC 2.6.1.2) (Hildebrandt et al., 2015). Labeling experiments using 13CO2 revealed that de novo biosynthesis rate of Ala was much rapidly than several other amino acids including phenylAla, glycine, and serine, glutamate, threonine, lysine, leucine, isoleucine, proline, and asparagine (Hildebrandt et al., 2015). This result is consistent with the relatively simple and short synthesis pathway of Ala. To date, only 30 candidate genes and six QTL were recorded to be associated with Ala content in soybean plants (https://www.soybase.org/search/index.php?searchterm=Ala). The quantity of genetic sources impacting the Ala content is much less than the essential amino acids. Compared with grain-type soybean, Ala content in edamame beans is extremely important because it is one of the two major contributors for the sweetness of edamame bean and partially determines consumer acceptance. Additionally, Ala was reported as a generic stress response molecule involved in protecting plants from extreme temperature, hypoxia, drought, and heavy metal shock (Parthasarathy et al., 2019). There was also evidence that Ala participated in lignin biosynthesis and ethylene production. The present study can assist the genetic understanding of Ala metabolism in the soybean/edamame beans and guide the edamame breeding with a goal of more sweetness.
A total of 25 candidate genes were screened in the respective genomic regions of 25 SNPs (Table S6). In the future, the SNPs and/or loci with validated function will be capable to boost selection for edamame with higher Ala content germplasms. Three genes encoding DNA-binding proteins and two kinase genes were demonstrated to be associated with Ala content (Table S6). None of the above genes was strongly overlapped with the current known Ala-allied QTL and genes obtained from the soybase, probably because they are genes responsible for Ala content in edamame beans other than the dried soybean seeds. Their functions would be explored in the future with the intention of uncovering the mechanism of Ala synthesis, accumulation, transportation and signaling.
Additionally, nine enzyme-type genes with catalytic functions were found to associate with edamame Ala content (Table S6). Uniquely, a guanosine monophosphate (GMP) synthetase (EC 6.3.5.2) gene was revealed to associate with Ala content in the edamame beans (Table S6). GMP synthetase is an amidotransferase that catalyzes the amination of xanthosine 5’-monophosphate to form GMP in the presence of glutamine and ATP. Glutamine hydrolysis produces the necessary amino group while ATP hydrolysis drives the reaction (Nakamura et al., 1995). Moreover, GMP synthetase is involved in amino acid metabolism because it generates L-glutamate from L-glutamine (Yoo et al., 2020). Hence, we suspected that GMP synthetase genes might be also involved in the Ala generation and homeostasis in edamame beans.
A SAM decarboxylase gene (Gm17g070500) was identified to ally with Ala content (Table S6). SAM is a key intermediate component linking the conversion from methionine and cysteine, which locates at the upstream of the Ala synthesis pathway (Hildebrandt et al., 2015). Additionally, two OMT genes (Glyma.14g201100 and Glyma.18g269600) were found to link with Ala content. OMT is a type of methyltransferase enzyme transferring a methyl group on a molecule (Morishige et al., 2010). Similarly, OMT was reported being engaged in the biochemical pathway from methionine to cysteine, followed by the Ala synthesis pathway. Nevertheless, the direct evidence for the impact of these gene on the Ala signaling pathway and synthesis remains unclear yet. In the future, we will employ in vitro enzyme assay trial to explore its substrate and biochemical function, and might apply the gene editing tool to characterize this gene’s in vivo function related to Ala.
By analysis of sucrose and Ala levels in 189 edamame accessions at three locations in two years, three PI accessions were selected as the high sucrose and high Ala accessions that are able to serve the breeding of sweeter edamame varieties. A total of 45 and 25 SNPs was identified to associate sucrose content and Ala content, respectively, based on phenotypic data derived 189 edamame accessions and SoySNP50K iSelect BeadChip by GWAS. The PI accessions, beneficial alleles, and candidate genes identified in this study will be fundamental for breeding sweeter edamame varieties with improved seed levels of sucrose and Ala, and eventually promote the consumers’ acceptance of edamame as a specialty crop in the United States.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
ZW and BZ designed the experiment. BZ selected the edamame panel and provided plant samples. BZ, NL, LM, PC led the field trials at VA, MO and AR. DY led the measurements of sucrose and Ala contents in the edamame beans. KD and GM combined the phenotypic data using BLUE. ZW, KD, GM and SL did the GWAS analysis. ZW, DY, and GM wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the USDA-NIFA, Grant No. 2018-51181-28384, Accession No. 1016465.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1086007/full#supplementary-material
Supplementary Figure 1 | SNP associations for edamame sucrose content from (A) BLUE combined dataset, (B) Blacksburg, VA, combined, (C) Portageville, MO, combined, (D) Fayetteville, AR, combined, are displayed in Manhattan plots with chromosomes in alternating colors, significance thresholds-log10(P) > 3.01 (the blue line) and suggestive threshold-log10(P) > 2.70 (the red line). Each respective QQ plot displays observed-log10(P) against expected-log10(P).
Supplementary Figure 2 | SNP associations for edamame Ala content from (A) BLUE combined dataset, (B) Blacksburg, VA, combined, (C) Portageville, MO, combined, (D) Fayetteville, AR, combined, are displayed in Manhattan plots with chromosomes in alternating colors, significance thresholds-log10(P) > 3.01 (the blue line) and suggestive threshold-log10(P) > 2.70 (the red line). Each respective QQ plot displays observed-log10(P) against expected-log10(P).
Supplementary Table 1 | Germplasms utilized in present study and their populational structure
Supplementary Table 2 | Sucrose and Ala contents in the edamame beans harvested from Blacksburg (VA), Portageville (MO), and Fayetteville (AR) in the years of 2018 and 2019.
Bandillo, N., Jarquin, D., Song, Q., Nelson, R., Cregan, P., Specht, J., et al. (2015). A population structure and genome-wide association analysis on the USDA soybean germplasm collection. Plant Genome 8 eplantgenome2015 04, 0024. doi: 10.3835/plantgenome2015.04.0024
Carneiro, R. C. V., Duncan, S. E., O'keefe, S. F., Yin, Y., Neill, C. L., Zhang, B. (2020). Sensory and consumer studies in plant breeding: A guidance for edamame development in the U.S. Front. Sustain. Food Syst. 4. doi: 10.3389/fsufs.2020.00124
Carneiro, R., Duncan, S., O'keefe, S., Yu, D., Huang, H., Yin, Y., et al. (2021). Utilizing consumer perception of edamame to guide new variety development. Front. Sustain. Food Syst. 4. doi: 10.3389/fsufs.2020.556580
Carson, L. C., Freeman, J. H., Zhou, K., Welbaum, G., Reiter, M. (2011). Cultivar evaluation and lipid and protein contents of Virginia-grown edamame. HortTechnology 21, 131–135. doi: 10.21273/HORTTECH.21.1.131
Chang, F., Guo, C., Sun, F., Zhang, J., Wang, Z., Kong, J., et al. (2018). Genome-wide association studies for dynamic plant height and number of nodes on the main stem in summer sowing soybeans. Front. Plant Sci. 9, 1184. doi: 10.3389/fpls.2018.01184
Chaudhary, J., Patil, G. B., Sonah, H., Deshmukh, R. K., Vuong, T. D., Valliyodan, B., et al. (2015). Expanding omics resources for improvement of soybean seed composition traits. Front. Plant Sci. 6, 1021. doi: 10.3389/fpls.2015.01021
Crizel, R. L., Perin, E. C., Vighi, I. L., Woloski, R., Seixas, A., Da Silva Pinto, L., et al. (2020). Genome-wide identification, and characterization of the CDPK gene family reveal their involvement in abiotic stress response in fragaria x ananassa. Sci. Rep. 10, 11040. doi: 10.1038/s41598-020-67957-9
Delatte, T. L., Sedijani, P., Kondou, Y., Matsui, M., De Jong, G. J., Somsen, G. W., et al. (2011). Growth arrest by trehalose-6-phosphate: an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiol. 157, 160–174. doi: 10.1104/pp.111.180422
Dhakal, K., Zhu, Q., Zhang, B., Li, M., Li, S. (2021). Analysis of shoot architecture traits in edamame reveals potential strategies to improve harvest efficiency. Front. Plant Sci. 12, 614926. doi: 10.3389/fpls.2021.614926
Do, T. D., Vuong, T. D., Dunn, D., Clubb, M., Valliyodan, B., Patil, G., et al. (2019). Identification of new loci for salt tolerance in soybean by high-resolution genome-wide association mapping. BMC Genomics 20, 318. doi: 10.1186/s12864-019-5662-9
Dufayard, J. F., Bettembourg, M., Fischer, I., Droc, G., Guiderdoni, E., Perin, C., et al. (2017). New insights on leucine-rich repeats receptor-like kinase orthologous relationships in angiosperms. Front. Plant Sci. 8, 381. doi: 10.3389/fpls.2017.00381
Du, Y., Zhao, Q., Chen, L., Yao, X., Zhang, W., Zhang, B., et al. (2020). Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol. Biochem. 146, 1–12. doi: 10.1016/j.plaphy.2019.11.003
Endelman, J. B. (2011). Ridge regression and other kernels for genomic selection with r package rrBLUP. Plant Genome 4, 250–255. doi: 10.3835/plantgenome2011.08.0024
Gahlaut, V., Jaiswal, V., Balyan, H. S., Joshi, A. K., Gupta, P. K. (2021). Multi-locus GWAS for grain weight-related traits under rain-fed conditions in common wheat (Triticum aestivum l.). Front. Plant Sci. 12, 758631. doi: 10.3389/fpls.2021.758631
Ge, Q., Cui, Y., Li, J., Gong, J., Lu, Q., Li, P., et al. (2020). Disequilibrium evolution of the fructose-1,6-bisphosphatase gene family leads to their functional biodiversity in gossypium species. BMC Genomics 21, 379. doi: 10.1186/s12864-020-6773-z
Hanson, A. A., Lorenz, A. J., Hesler, L. S., Bhusal, S. J., Bansal, R., Michel, A. P., et al. (2018). Genome-wide association mapping of host-plant resistance to soybean aphid. Plant Genome 11(3), 1–12. doi: 10.3835/plantgenome2018.02.0011
Haslett, S. J., Puntanen, S. (2009). Equality of BLUEs or BLUPs under two linear models using stochastic restrictions. Stat. Papers 51, 465–475. doi: 10.1007/s00362-009-0219-7
Hildebrandt, T. M., Nesi, N., A., A., Braun, H. P. (2015). Amino acid catabolism in plants. Mol. Plant 8, 1563–1579. doi: 10.1016/j.molp.2015.09.005
Hou, A., Chen, P., Alloatti, J., Li, D., Mozzoni, L., Zhang, B., et al. (2009). Genetic variability of seed sugar content in worldwide soybean germplasm collections. Crop Sci. 49, 903–912. doi: 10.2135/cropsci2008.05.0256
Hulsmans, S., Rodriguez, M., De Coninck, B., Rolland, F. (2016). The SnRK1 energy sensor in plant biotic interactions. Trends Plant Sci. 21, 648–661. doi: 10.1016/j.tplants.2016.04.008
Jiang, S., Zhang, H., Ni, P., Yu, S., Dong, H., Zhang, A., et al. (2020). Genome-wide association study dissects the genetic architecture of maize husk tightness. Front. Plant Sci. 11, 861. doi: 10.3389/fpls.2020.00861
Jombart, T., Devillard, S., Balloux, F. (2010). Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11, 94. doi: 10.1186/1471-2156-11-94
Khan, M. A., Tong, F., Wang, W., He, J., Zhao, T., Gai, J. (2018). Analysis of QTL-allele system conferring drought tolerance at seedling stage in a nested association mapping population of soybean [Glycine max (L.) merr.] using a novel GWAS procedure. Planta 248, 947–962. doi: 10.1007/s00425-018-2952-4
Kim, H.-K., Kang, S.-T., Cho, J.-H., Choung, M.-G., Suh, D.-Y. (2005). Quantitative trait loci associated with oligosaccharide and sucrose contents in soybean (Glycine max l.). J. Plant Biol. 48, 106–112. doi: 10.1007/BF03030569
Kumar, V., Rani, A., Goyal, L., Pratap, D., Billore, S. D., Chauhan, G. S. (2011). Evaluation of vegetable-type soybean for sucrose, taste-related amino acids, and isoflavones contents. Int. J. Food Properties 14, 1142–1151. doi: 10.1080/10942911003592761
Li, J., Ji, L. (2005). Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 95, 221–227. doi: 10.1038/sj.hdy.6800717
Li, Y. H., Reif, J. C., Ma, Y. S., Hong, H. L., Liu, Z. X., Chang, R. Z., et al. (2015). Targeted association mapping demonstrating the complex molecular genetics of fatty acid formation in soybean. BMC Genomics 16, 841. doi: 10.1186/s12864-015-2049-4
Liu, R., Gong, J., Xiao, X., Zhang, Z., Li, J., Liu, A., et al. (2018). GWAS analysis and QTL identification of fiber quality traits and yield components in upland cotton using enriched high-density SNP markers. Front. Plant Sci. 9, 1067. doi: 10.3389/fpls.2018.01067
Lv, G. Y., Guo, X. G., Xie, L. P., Xie, C. G., Zhang, X. H., Yang, Y., et al. (2017). Molecular characterization, gene evolution, and expression analysis of the fructose-1, 6-bisphosphate aldolase (FBA) gene family in wheat (Triticum aestivum l.). Front. Plant Sci. 8, 1030. doi: 10.3389/fpls.2017.01030
Mahoussi, K. A. D., Eric, E. A., Symphorien, A., Florent, J.-B. Q., Flora, J. C., Achille, E. A., et al. (2020). Vegetable soybean, edamame: Research, production, utilization and analysis of its adoption in Sub-Saharan Africa. J. Horticulture Forestry 12, 1–12. doi: 10.5897/JHF2019.0604
Maughan, P. J., Maroof, M. A. S., Buss, G. R. (2000). Identification of quantitative trait loci controlling sucrose content in soybean (Glycine max). Mol. Breeding 6(1), 105–111.
Morishige, T., Tamakoshi, M., Takemura, T., Sato, F. (2010). “Molecular characterization of O-methyltransferases involved in isoquinoline alkaloid biosynthesis in coptis japonica,” in Proceedings of the Japan Academy. Series B, Physical and Biological Sciences, Vol. 86. 757–768.
Nakamura, J., Straub, K., Wu, J., Lou, L. (1995). The glutamine hydrolysis function of human GMP synthetase. identification of an essential active site cysteine. J. Biol. Chem. 270, 23450–23455. doi: 10.1074/jbc.270.40.23450
Ogles, C. Z., Guertal, E. A., Weaver, D. B. (2016). Edamame cultivar evaluation in central Alabama. Agron. J. 108, 2371–2378. doi: 10.2134/agronj2016.04.0218
Parthasarathy, A., Savka, M. A., Hudson, A. O. (2019). The synthesis and role of beta-alanine in plants. Front. Plant Sci. 10, 921. doi: 10.3389/fpls.2019.00921
Patil, G., Vuong, T. D., Kale, S., Valliyodan, B., Deshmukh, R., Zhu, C., et al. (2018). Dissecting genomic hotspots underlying seed protein, oil, and sucrose content in an interspecific mapping population of soybean using high-density linkage mapping. Plant Biotechnol. J. 16, 1939–1953. doi: 10.1111/pbi.12929
Ponnu, J., Wahl, V., Schmid, M. (2011). Trehalose-6-phosphate: connecting plant metabolism and development. Front. Plant Sci. 2, 70. doi: 10.3389/fpls.2011.00070
Qin, J., Shi, A., Song, Q., Li, S., Wang, F., Cao, Y., et al. (2019). Genome wide association study and genomic selection of amino acid concentrations in soybean seeds. Front. Plant Sci. 10, 1445. doi: 10.3389/fpls.2019.01445
Rosa, M., Prado, C., Podazza, G., Interdonato, R., Gonzalez, J. A., Hilal, M., et al. (2009). Soluble sugars–metabolism, sensing and abiotic stress: a complex network in the life of plants. Plant Signaling Behav. 4, 388–393. doi: 10.4161/psb.4.5.8294
Ruan, Y. L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. doi: 10.1146/annurev-arplant-050213-040251
Serrato, A. J., Dios Barajas-Lopez, De, J., C., Sahrawy, M. (2009). Changing sugar partitioning in FBPase-manipulated plants. J. Exp. Bot. 60, 2923–2931. doi: 10.1093/jxb/erp066
Singer, W. M., Shea, Z., Yu, D., Huang, H., Mian, M. A. R., Shang, C., et al. (2022). Genome-wide association study and genomic selection for proteinogenic methionine in soybean seeds. Front. Plant Sci. 13, 859109. doi: 10.3389/fpls.2022.859109
Skoneczka, J. A., Maroof, M. A. S., Shang, C., Buss, G. R. (2009). Identification of candidate gene mutation associated with low stachyose phenotype in soybean line PI200508. Crop Sci. 49, 247–255. doi: 10.2135/cropsci2008.07.0403
Song, Q., Hyten, D. L., Jia, G., Quigley, C. V., Fickus, E. W., Nelson, R. L., et al. (2013). Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PloS One 8, e54985. doi: 10.1371/journal.pone.0054985
Sui, M., Wang, Y., Bao, Y., Wang, X., Li, R., Lv, Y., et al. (2020). Genome-wide association analysis of sucrose concentration in soybean (Glycine max l.) seed based on high-throughput sequencing. Plant Genome 13, e20059. doi: 10.1002/tpg2.20059
Tauzin, A. S., Giardina, T. (2014). Sucrose and invertases, a part of the plant defense response to the biotic stresses. Front. Plant Sci. 5, 293. doi: 10.3389/fpls.2014.00293
Verma, R. K., Chetia, S. K., Sharma, V., Baishya, S., Sharma, H., Modi, M. K. (2022). GWAS to spot candidate genes associated with grain quality traits in diverse rice accessions of north East India. Mol. Biol. Rep 49, 5365–5377. doi: 10.1007/s11033-021-07113-2
Wang, Y., Chen, P., Zhang, B., Singh, R. (2014). Quantitative trait loci analysis of soluble sugar contents in soybean. Plant Breed. 133, 493–498. doi: 10.1111/pbr.12178
Wang, X., Wu, Z., Zhou, Q., Wang, X., Song, S., Dong, S. (2021). Physiological response of soybean plants to water deficit. Front. Plant Sci. 12, 809692. doi: 10.3389/fpls.2021.809692
Wolfe, E., Popp, M., Bazzani, C., Nayga, R. M., Danforth, D., Popp, J., et al. (2018). Consumers’ willingness to pay for edamame with a genetically modified label. Agribusiness 34, 283–299. doi: 10.1002/agr.21505
Yadav, U. P., Ivakov, A., Feil, R., Duan, G. Y., Walther, D., Giavalisco, P., et al. (2014). The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 65, 1051–1068. doi: 10.1093/jxb/ert457
Yoo, H. C., Yu, Y. C., Sung, Y., Han, J. M. (2020). Glutamine reliance in cell metabolism. Exp. Mol. Med. 52, 1496–1516. doi: 10.1038/s12276-020-00504-8
Yu, D., Lin, T., Sutton, K., Lord, N., Carneiro, R., Jin, Q., et al. (2021). Chemical compositions of edamame genotypes grown in different locations in the US. Front. Sustain. Food Syst. 5. doi: 10.3389/fsufs.2021.620426
Yu, D., Lord, N., Polk, J., Dhakal, K., Li, S., Yin, Y., et al. (2022). Physical and chemical properties of edamame during bean development and application of spectroscopy-based machine learning methods to predict optimal harvest time. Food Chem. 368, 130799. doi: 10.1016/j.foodchem.2021.130799
Zeng, A., Chen, P., Shi, A., Wang, D., Zhang, B., Orazaly, M., et al. (2014). Identification of quantitative trait loci for sucrose content in soybean seed. Crop Sci. 54, 554–564. doi: 10.2135/cropsci2013.01.0036
Zhang, B., Lord, N., Kuhar, T., Duncan, S., Huang, H., Ross, J., et al. (2021). ‘VT sweet’: A vegetable soybean cultivar for commercial edamame production in the mid-Atlantic USA. J. Plant Registrations 16, 29–33. doi: 10.1002/plr2.20140
Zhang, T., Wu, T., Wang, L., Jiang, B., Zhen, C., Yuan, S., et al. (2019). A combined linkage and GWAS analysis identifies QTLs linked to soybean seed protein and oil content. Int. J. Mol. Sci. 20(23), 5915. doi: 10.3390/ijms20235915
Zhang, W., Zhang, N. S., Zhao, J. J., Guo, Y. P., Zhao, Z. Y., Mei, L. X. (2017). Potassium fertilization improves apple fruit (Malus domestica borkh. cv. Fuji) development by regulating trehalose metabolism. J. Hortic. Sci. Biotechnol. 92, 539–549. doi: 10.1080/14620316.2017.1304165
Keywords: GWAS, edamame, sucrose, alanine, sensory
Citation: Wang Z, Yu D, Morota G, Dhakal K, Singer W, Lord N, Huang H, Chen P, Mozzoni L, Li S and Zhang B (2023) Genome-wide association analysis of sucrose and alanine contents in edamame beans. Front. Plant Sci. 13:1086007. doi: 10.3389/fpls.2022.1086007
Received: 31 October 2022; Accepted: 29 December 2022;
Published: 03 February 2023.
Edited by:
Bourlaye Fofana, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Milad Eskandari, University of Guelph, CanadaCopyright © 2023 Wang, Yu, Morota, Dhakal, Singer, Lord, Huang, Chen, Mozzoni, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Zhang, emhhbmc3NkB2dC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.