
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 December 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1073856
Tobacco black shank caused by Phytophthora nicotianae is a devastating disease that causes huge losses to tobacco production across the world. Investigating the regulatory mechanism of tobacco resistance to P. nicotianae is of great importance for tobacco resistance breeding. The jasmonate (JA) signaling pathway plays a pivotal role in modulating plant pathogen resistance, but the mechanism underlying JA-mediated tobacco resistance to P. nicotianae remains largely unclear. This work explored the P. nicotianae responses of common tobacco cultivar TN90 using plants with RNAi-mediated silencing of NtCOI1 (encoding the perception protein of JA signal), and identified genes involved in this process by comparative transcriptome analyses. Interestingly, the majority of the differentially expressed bHLH transcription factor genes, whose homologs are correlated with JA-signaling, encode AtBPE-like regulators and were up-regulated in NtCOI1-RI plants, implying a negative role in regulating tobacco response to P. nicotianae. A subsequent study on NtbHLH49, a member of this group, showed that it’s negatively regulated by JA treatment or P. nicotianae infection, and its protein was localized to the nucleus. Furthermore, overexpression of NtbHLH49 decreased tobacco resistance to P. nicotianae, while knockdown of its expression increased the resistance. Manipulation of NtbHLH49 expression also altered the expression of a set of pathogen resistance genes. This study identified a set of genes correlated with JA-mediated tobacco response to P. nicotianae, and revealed the function of AtBPE-like regulator NtbHLH49 in regulating tobacco resistance to this pathogen, providing insights into the JA-mediated tobacco responses to P. nicotianae.
Tobacco production is hampered by diverse diseases. Tobacco black shank, caused by the hemibiotrophic pathogen Phytophthora nicotianae, is one of the most devastating diseases to tobacco cultivation (Meng et al., 2014; Wang and Jiao, 2019). P. nicotianae is a species of Phytophthora that belongs to the class Oomycetes in the kingdom Chromista and has over 100 species causing destructive damage to a range of plants (Kroon et al., 2012). P. nicotianae is a typical soil-borne disease that can infect tobacco plants at any growth stage and affects a broad variety of plants, including Solanaceae crops, horticultural plants, and fruit trees (Kroon et al., 2012). Infection by P. nicotianae could cause a sudden withering of the entire plant, as well as root rot and the formation of black patches on the stem near the ground level (Meng et al., 2014). In recent decades, many chemicals have been employed to control tobacco black shank, but their efficacy is rather limited (Meng et al., 2014). Moreover, the majority of chemical pesticides may cause significant environmental pollution (Alexandrino et al., 2022). Development of P. nicotianae resistant tobacco cultivars is still one of the most economical and effective approaches to reduce yield loss caused by this pathogen, therefore, identification of genes associated with this pathogen is of great importance.
Studies have shown that plant resistance to Phytophthora pathogens is influenced by environmental variables and phytohormones. For example, abiotic stresses and the induced phytohormone abscisic acid (ABA) may attenuate the severity of disease caused by the Phytophthora pathogen in plants (Roubtsova and Bostock, 2009; Dileo et al., 2010). Reducing reactive oxygen species (ROS) and ethylene (ET) in susceptible tobacco plants may increase pathogen resistance to P. nicotianae (Wi et al., 2012; Cho et al., 2013). Salicylic acid (SA) plays a positive role in regulating the resistance of potato (Solanum tuberosum) plants to P. infestans (Halim et al., 2007; Zhou et al., 2018). Furthermore, studies on Arabidopsis AtRTP5 showed that this factor could regulate the resistance to Phytophthora pathogen by modulating the biosynthesis of both SA and jasmonate (JA), implying that JA is involved in regulating plant resistance to Phytophthora (Li et al., 2020). The involvement of JA in tobacco resistance to Phytophthora pathogen was also indicated (Long et al., 2021). However, the mechanism underlying JA-mediated tobacco resistance to P. nicotianae is still largely unclear.
The JA-signaling pathway plays essential role in modulating plant responses to biotic and abiotic stresses (Howe et al., 2018). Extensive researches suggest that JA-signaling pathway is indispensable for plant resistance to both fungal pathogens (e.g., Alternaria brassicicola, Botrytis cinerea) and bacterial pathogens (e.g., Pseudomonas syringae spp.) (Thomma et al., 1998; Kloek et al., 2001). JA-signaling pathway functions in a complicated crosstalking network with SA, ET, and other phytohormones during the regulation of plant resistance to pathogenic microbes (Pieterse et al., 2012). JA signal is perceived by a multi-component complex consisting of COIl (CORONATINE INSENSITIVE1), JAZ (jasmonate ZIM-domain) protein and an inositol pentyl phosphate molecule (Yan et al., 2009). Perception of JA-Ile (the bioactive derivative of JA) by the receptor complex leads to the degradation of JAZ repressor protein via the 26S proteasome pathway, resulting in the release of transcription activators that tune up the downstream JA-responses (Yan et al., 2009). The F-box protein COI1 is a key factor in JA signal transduction. COI1 dysfunction could result in a loss of JA responses (Xie et al., 1998). Studies showed that mutations in COI1 could alter plant resistance to bacterial and fungal pathogens (Kloek et al., 2001; Zhang et al., 2017), and that the JAZ repressor protein is also involved in the regulation of plant resistance to pathogens (Thatcher et al., 2016). Downstream regulators, such as bHLH transcription factors, MYB transcription factors and ERFs (ethylene responsive factors), were found to play important roles in plant resistance to pathogen attack (Müller and Munné-Bosch, 2015; Kazan, 2015; Thatcher et al., 2016; Ren et al., 2020; Ren et al., 2022). Identification the downstream regulators that control tobacco responses to Phytophthora pathogen is of great importance for dissecting the function of JA-signaling in Phytophthora resistance (Thaler et al., 2004; Li et al., 2020).
In order to discover regulators conducting the JA-mediated P. nicotianae resistance, we compared P. nicotianae resistance between NtCOI1-silenced plants (NtCOI1-RI) and control plants, and identified genes involved in JA-mediated responses to this pathogen using comparative transcriptome analysis. A set of genes encoding AtBPE-like bHLH transcription factor were found to be up-regulated in NtCOI1-RI plants. Further study on NtbHLH49, one of these regulators, shows that overexpression of NtbHLH49 decreased tobacco resistance to P. nicotianae, while knockdown of this gene increased this resistance. The expression of a set of pathogen resistance genes was regulated by NtbHLH49 during tobacco response to P. nicotianae. These findings extend our understanding of plant responses to Phytophthora pathogens and provide fundamental molecular information for tobacco resistance breeding against P. nicotianae.
Wild type plants of Nicotiana tabacum cv. TN90, previously developed NtCOI1-silenced plants (NtCOI1-RI) and empty-vector-transformed control plants (Wang et al., 2014) were employed for the JA-mediated pathogen resistance assay and gene expression assay of this study. For developing transgenic plants overexpressing NtbHLH49, the full-length coding sequence of NtbHLH49 amplified with primer 5’-CACCATGGATATGGATAGCAAGAATG-3’ and 5’-TATGATCGCCTTACCTGCAACC-3’ was cloned into pENTR-D-TOPO vector (Invitrogen, USA) and then integrated into a binary vector pMDC83-Flag, which was modified from pMDC83-GFP (Curtis and Grossniklaus, 2003), to generate the gene overexpression vector. In order to develop transgenic plants for RNAi-mediated gene silencing of NtbHLH49, the 5’-end region of NtbHLH49 amplified with primer 5’-CACCATGGATATGGATAGCAAGAATG-3’ and 5’-GGGTTCATCATATCACTGAGGT-3’ was cloned into pENTR-D-TOPO vector and integrated into the binary vector pHZPRi-Hyg (Wang et al., 2014) to generate the gene silencing vector. The obtained binary vectors were introduced into Agrobacterium tumefaciens LBA4404 to develop transgenic plants as described previously (Wang et al., 2014). Transgenic plants verified to overexpress NtbHLH49 (NtbHLH49-OE) or have NtbHLH49 silenced (NtbHLH49-RI) were utilized for further assays.
For tobacco cultivation, surface-sterilized seeds of tobacco plants were sown on 1/2 MS medium supplemented with 25 mg/L hygromycin, and the resistant seedlings were placed into soil pots and validated by PCR tests. Tobacco plants were cultivated in an indoor growth room at 25°C with a photoperiod of 14 h light/10 h dark. 5-week-old seedlings were used for pathogen challenges, phytohormone treatment and transcriptional assays.
For phytohormone treatment, 5-week-old tobacco seedlings were sprayed with solutions containing 100 μM MeJA (methyl jasmonate) and 0.005% Tween 20. Control plants were sprayed with water containing 0.005% Tween 20. Then, tobacco leaves were detached from plants at the indicated time point for further assays.
The oomycete pathogen P. nicotianae (kept in our laboratory) was streaked on oat medium (30 g oatmeal and 20 g agar in 1 L ddH2O, pH 7.5) and cultured at 28°C for ten days before inoculating tobacco leaves. Tobacco leaves of each experimental group were divided into two groups, and inoculated with P. nicotianae mycelium (4 mm blocks from the growth medium) or mock (medium blocks only), respectively. Three independent replicates were set up for each treatment group. The inoculated leaves were incubated in dark at 28°C. After 72 hours of infection, the disease symptom was inspected. No visible symptom or formation of necrosis spots in leaves indicates resistance to P. nicotianae, and the development of water-soaking patches indicates susceptibility to P. nicotianae. The number of leaves with necrosis spots, water-soaking patches, or invisible symptoms was surveyed to characterize the P. nicotianae resistance of tobacco plants. In addition, a set of these leaves were collected at the indicated time points for transcriptional analyses.
Total RNAs were extracted from tobacco leaf samples using a RN38-EASY SPIN Plus Plant Kit (BioTeke, China) following the manufacturer’s instruction. The RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Transcriptome sequencing, i.e. next-generation RNA sequencing (RNA-seq), was conducted on an Illumina HiSeq 4000TM platform (Illumina Inc., USA) by Allwegene Technology (Beijing, China) with cDNA libraries constructed for each of the samples. Adapter-related reads, low-quality reads (>50% with quality scores ≤20) and reads containing more than 10% unknown nucleotides were removed from the data to generate reliable clean reads for read assembly. The alignment analysis was performed using TopHat 2.1.1 software. The RNA seq data presented in the study are deposited in the NCBI Sequence Read Archive (SRA) repository, accession number PRJNA901343.
The raw sequencing reads were pre-processed by FastQC (http://cufflinks.cb.umd.edu/). The processed reads were mapped to Nicotiana tabacum cv. TN90 genome (https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/715/135/GCF_000715135.1_Ntab-TN90/) and assembled using TopHat 2.1.1 software with the reference annotation. The FPKM (fragments per kilobase of exon model per million mapped reads) values for each gene and the DEGs (differentially expressed genes) were calculated with Cufflinks V2.2.l. The DEGs between NtCOI1-RI plants and control plants were determined according to the fold change and q-value.
Total RNAs of tobacco leaf samples were prepared as described above. First-strand cDNAs were synthesized using a PrimeScript RT reagent Kit (Takara Bio, Japan). The qRT-PCR amplification was performed in a 25μl reaction. The reaction conditions were as followings: an initial denaturation at 95°C for 1 min; 45 cycles of 95°C for 20 s, 59°C for 20 s and 72°C for 30 s; melting curve analysis between 50-95°C. Three biological replicates were performed for each gene. The relative transcription level of each gene was determined using the 2-ΔΔCt method (Livak and Schmittgen, 2001), with Actin gene serving as an internal control. Gene-specific primers used for validation of DEGs, identification of NtbHLH49-OE and NtbHLH49-RI plants, and determination of the expression of pathogen resistance genes in transgenic plants are listed in Supplementary Table 1.
For determining the subcellular localization of NtbHLH49, its coding sequence was cloned into pENTR-D-TOPO vector as described above. Then, NtbHLH49 was integrated into pMDC83-GFP vector (Curtis and Grossniklaus, 2003) to generate pMDC83-NtbHLH49-GFP vector for expressing the NtbHLH49-GFP fusion protein, and a modified pMDC83-GFP vector for expressing GFP alone was used as control. The obtained vectors were later introduced into A. tumefaciens LBA4404, and introduced into Nicotiana tabacum BY-2 cells as described by (Goossens et al., 2003). The subcellular localization of NtbHLH49-GFP and GFP in BY-2 cells was observed under a Leica TCSSP8 confocal microscope.
Plants with RNAi-mediated gene silencing of NtCOI1, encoding the JA-Ile receptor protein, were employed to investigate the changes in tobacco responding to P. nicotianae upon dysfunction of the JA-signaling. When the leaves of NtCOI1-RI plants were infected with P. nicotianae, a substantially more severe water-soaking disease symptom was observed in the majority of them after 72 hours incubation at 28°C (Figure 1A). On the contrary, the majority of leaves from control plants showed a hypersensitive reaction, with necrosis lesions forming around the P. nicotianae infection sites (Figure 1A). A quantitative assessment of the severity of the diseases revealed that less than 30% of the leaves of the control plants developed water-soaking patches, but nearly 70% of the leaves of NtCOI1-RI plants formed water-soaking spots (Figure 1B). Around 60% of the leaves of control plants had necrosis lesions, but only about 20% of the leaves of NtCOI1-RI plants had necrosis lesions (Figure 1B). The number of leaves with no visible symptom was slightly greater in control plant than in NtCOI1-RI plants (Figure 1B). These findings support a positive role for NtCOI1 and the JA-signaling pathway in controlling tobacco resistance to this pathogen.
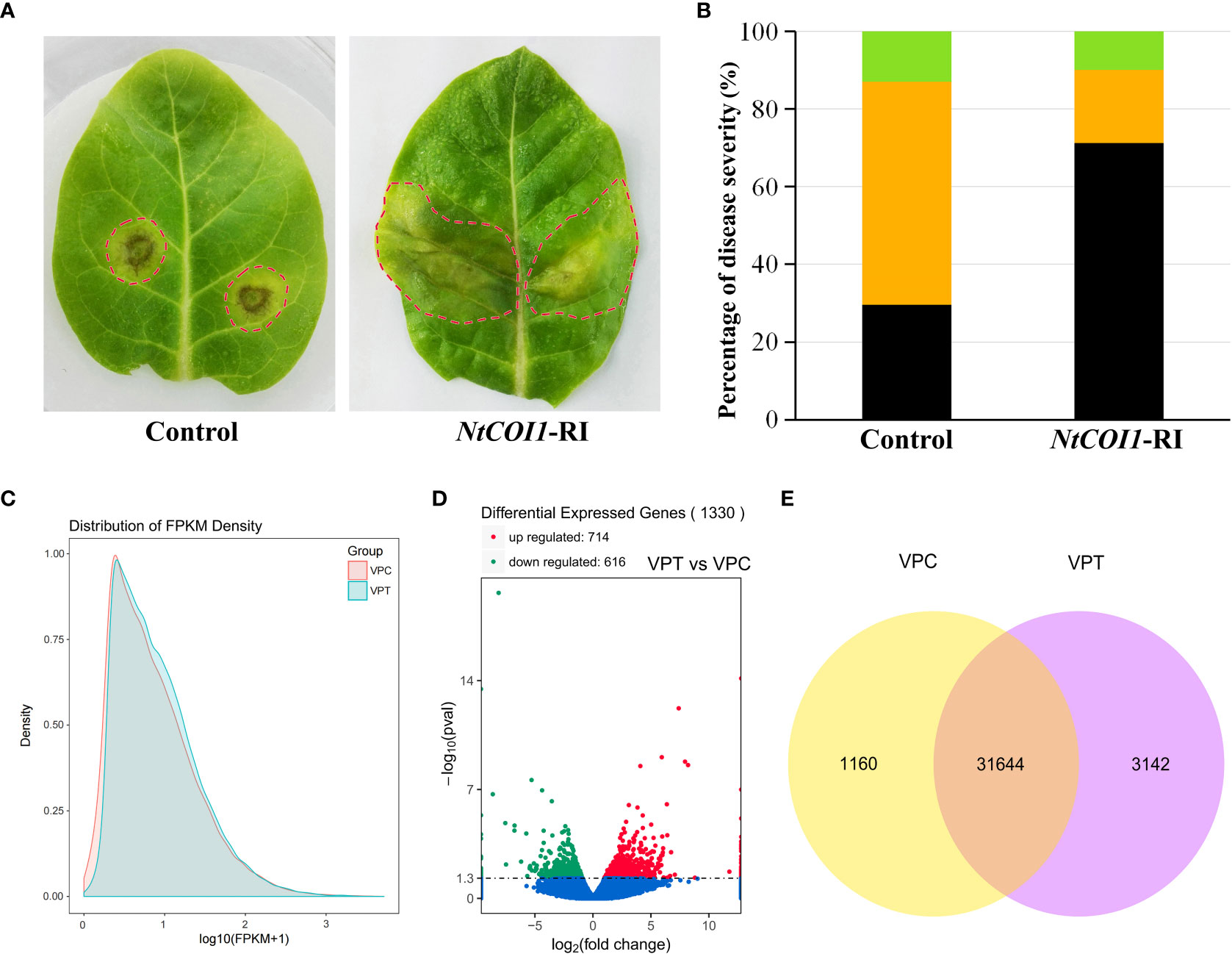
Figure 1 Responses of tobacco plants to P. nicotianae infection. (A) Representative symptoms of control and NtCOI1-RI tobacco leaves infected by P. nicotianae. The leaf area with visible symptom was indicated with dashed-circle. (B) Percentiles of tobacco leaves with differential symptom severity. The black color indicates water-soaking symptom, the orange color indicates necrosis symptom, and the green color indicates no visible symptom. (C, D) The density distribution (C), volcano plot (D) and Venn diagram (E) of the differentially expressed genes (DEGs) between control and NtCOI1-RI plants after P. nicotianae infection. VPT indicates NtCOI1-RI plants, and VPC indicates control plants.
In order to elucidate the mechanism underlying JA/NtCOI1-mediated tobacco resistance to P. nicotianae, the infected leaf samples from NtCOI1-RI plants and control plants were subjected to a high-throughout RNA-seq assay on an Illumina second-generation high-throughput sequencing platform using the PE150 sequencing strategy. The overall number of mapped reads for control plants is 160,863,066, including 3,934,116 multiple mapped reads and 156,928,950 uniquely mapped reads, and the total number of mapped reads for NtCOI1-RI plants is 141,680,162 including 3,141,296 multiple mapped reads and 138,538,866 uniquely mapped reads (Table 1; Supplementary Table 2). The expression levels of DEGs (differentially expressed genes) were calculated based on the number of uniquely mapped reads using the FPKM (fragments per kilobase of exon model per million mapped reads) method. Among these DEGs, 31,644 genes were expressed in both NtCOI1-RI plants and control plants, whereas 1,160 genes and 3,142 genes were specifically expressed in NtCOI1-RI plants and control plants, respectively (mean FPKM value>1.0). In the NtCOI1-RI plants, 616 genes were found to be down-regulated and 714 genes to be up-regulated. The density distribution, volcano plot and Venn diagram of the FPKM were shown in Figures 1C–E to visually exhibit the gene expression profiles between NtCOI1-RI plants and control plants. In order to verify the reliability of the gene expression data obtained from the RNA-seq, 9 genes with differential expression levels between NtCOI1-RI plants and control plants were chosen and subjected to a qRT-PCR validation assay, which suggested that the gene expression data obtained by RNA-seq are valid for further analysis (Supplementary Table 3; Supplementary Figure 1).
A total of 1082 DEGs were annotated in the GO (gene ontology) database using GOseq software (Young et al., 2010), and the GO terms were classified into three ontologies, including biological process, molecular function and cellular component. There were 877 up-regulated and 768 down-regulated DEGs (NtCOI1-RI plants vs. Control plants) in the enrichment analysis by GO terms. The GO terms enriched from down-regulated DEGs fell into the categories of systemic acquired resistance, defense response, incompatible interaction, immune response, innate immune response, immune system process, response to other organism, response to biotic stimulus, defense response, defense/immunity protein activity, signaling receptor activity, hormone activity, pathogenesis (Figure 2A; Table 2; Supplementary Table 4). These pieces of information are consistent with the decreased P. nicotianae resistance of NtCOI1-RI plants. On the other hand, the GO terms enriched from up-regulated DEGs comprise only a few abiotic-responsive DEGs, and the majority of them are engaged in mitotic cell cycle, transportation and other biological processes that have little relevance with pathogen responses (Figure 2A).
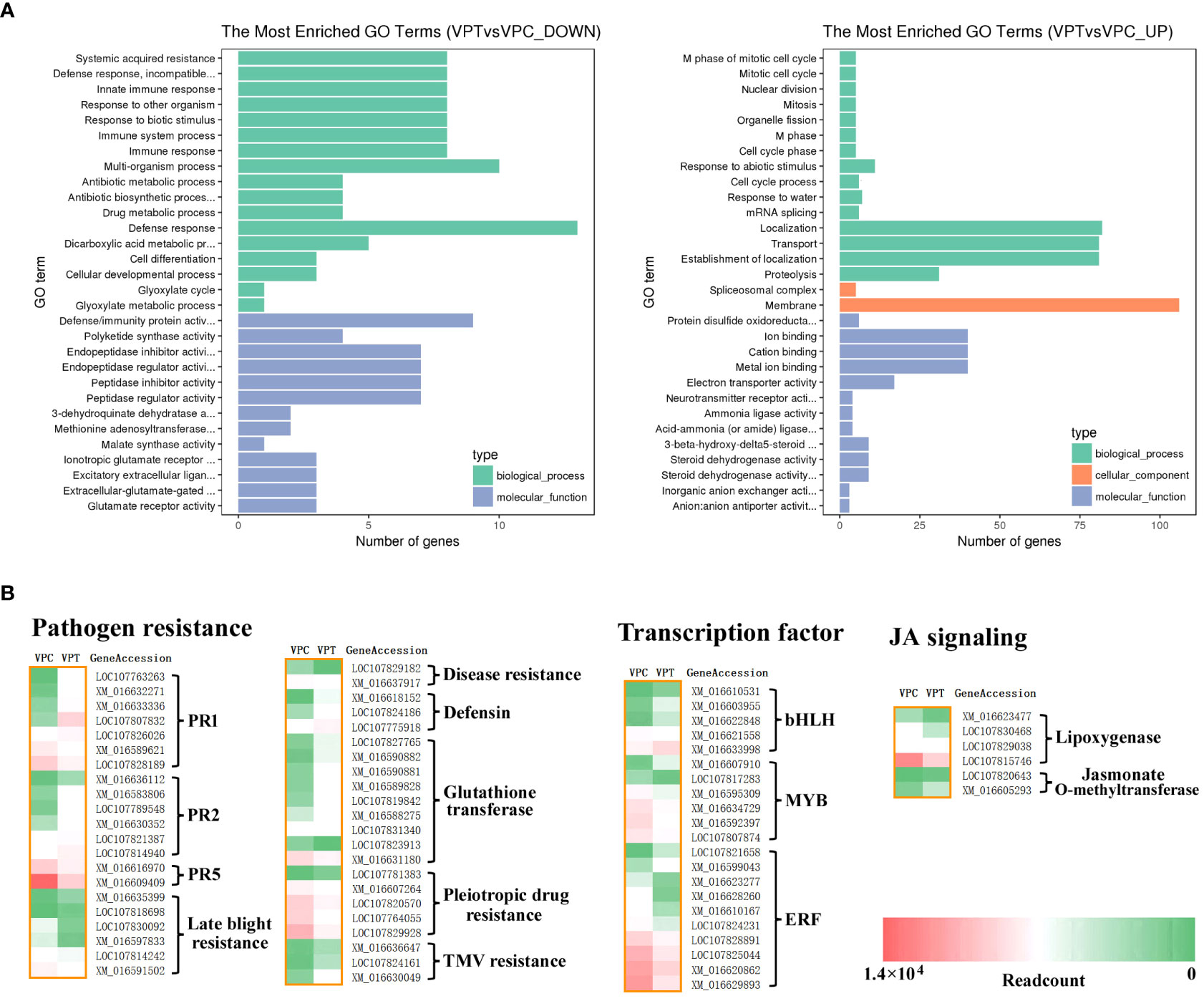
Figure 2 Analysis of DEGs between control and NtCOI1-RI plants. (A) The enrichment of down-regulated (left) and up-regulated (right) DEGs in NtCOI1-RI plants compared with control by GO terms. (B) Diagrammatic profile of the differentially expressed pathogen resistance gens, transcription factor genes, and JA-signaling genes in control and NtCOI1-RI plants. Color-bar at bottom-right corner indicates the number of readcounts. VPT indicates NtCOI1-RI plants and VPC indicates control plants.
Further analysis was carried out to mine DEGs corresponding to pathogen resistance genes, transcription regulatory genes, and JA-signaling related genes relevant to pathogen resistance. The pathogen resistance genes included PR (pathogenesis-related) genes, glutathione transferase genes, late blight resistance genes, pleiotropic drug resistance genes, and TMV resistance genes (Figure 2B). The groups that have the majority of genes up-regulated in NtCOI1-RI plants comprise PR1 genes, PR2 genes (encoding glucanase that cleavages β-1,3-glucans present in fungal cell wall), TMV (tobacco mosaic virus)-resistance genes, defensin genes, and glutathione transferase genes (Figure 2B). The groups that have most genes down-regulated in NtCOI1-RI plants include PR5 (osmotin) genes, late blight resistance genes, disease resistance genes, and pleiotropic drug resistance genes (Figure 2B). Among the differentially expressed transcription regulatory genes, 5 are bHLH transcription factor genes, 6 are MYB transcription factor genes, and 10 are ERF genes. Furthermore, the identified DEGs also contain a couple of genes engaged in the production or perception of JA (Figure 2B), such as genes encoding lipoxygenase (α-linolenic acid metabolism) and jasmonate O-methyltransferase. These findings indicated a sophisticated gene regulation during the JA-mediated P. nicotianae responses.
As described above, the expression of 5 bHLH genes was found to be altered by the silencing of NtCOI1. Interestingly, 4 of them are AtBPE-like regulators belonging to the C-group bHLHs of tobacco (Rushton et al., 2008; Figures 2C, 3A). In order to analyze the phylogenetic relationship of these regulators (i.e., NtbHLH49/130/74/128) to other C-group bHLH transcription factors, the bHLH domains of a set of representative bHLHs were retrieved for the genomic data (Rushton et al., 2008) and subjected to a phylogenetic analysis. As shown in Figure 3B, tobacco AtBPE-like regulators could be classified into four subgroups including CI, CII, CIII, and CIV. While AtBPE is close to the CI subgroup members, NtbHLH49 belongs to CII subgroup, NtbHLH74 belongs to CIII subgroup, and NtbHLH130/128 belong to CIV subgroup. The sequence alignment suggested that NtbHLH130/74/128 had a complete bHLH domain that was highly similar to that of AtBPE, however, NtbHLH49 had only a partial bHLH domain (Figure 3B). These findings cued us to perform further studies to explore the roles of NtbHLH49 in regulating tobacco responses to P. nicotianae attack.
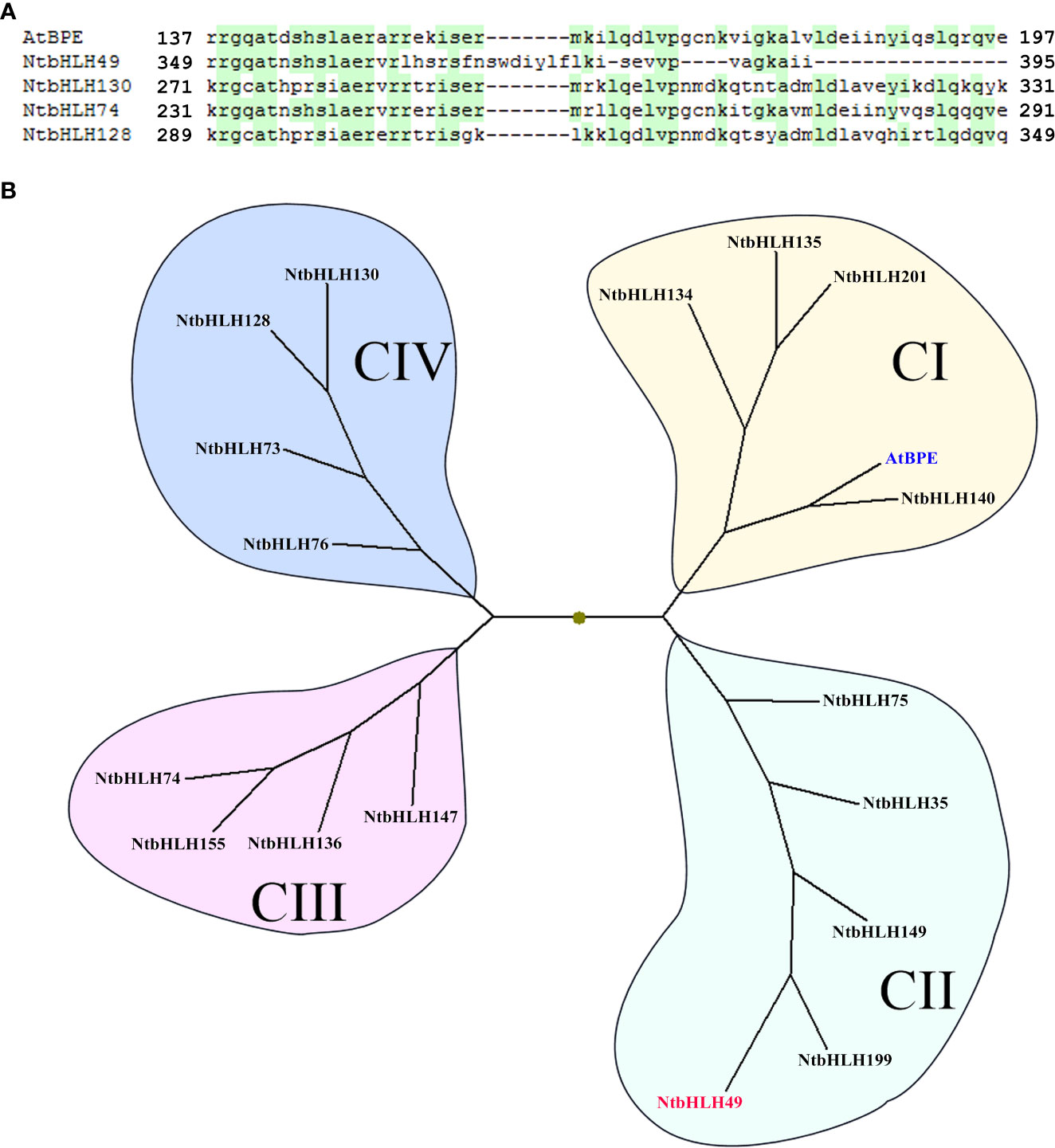
Figure 3 Sequence alignment (A) and phylogenetic analyses (B) of tobacco AtBPE-like transcription factors. GenBank accession for AtBPE (in blue) is NP_564749, and those for tobacco bHLHs are as followings: NtbHLH49 (in red; XM_016610531), NtbHLH130 (XM_016622848), NtbHLH74 (XM_016633998), NtbHLH128 (XM_016621558), NtbHLH201 (XP_016471633), NtbHLH199 (XP_016500166), NtbHLH134 (XP_016477426), NtbHLH135 (XP_016443360), NtbHLH136 (XP_016462422), NtbHLH140 (XP_016494316), NtbHLH147 (XP_016441121), NtbHLH149 (XP_016478982), NtbHLH155 (XP_016462240), NtbHLH35 (NP_001312938), NtbHLH73 (XP_016497820), NtbHLH75 (XP_016438666), NtbHLH76 (XP_016447869).
In order to determine the expression pattern of NtbHLH49 in tobacco, we first analyzed its expression in the leaves of NtCOI1-RI plants and control plants. The results showed that the expression level of NtbHLH49 in NtCOI1-RI plants was about 6 folds of that in control plants (Figure 4A), confirming that its expression is negatively regulated by the JA signal perception protein NtCOI1. Then, the expression of NtbHLH49 responding to P. nicotianae attack and MeJA treatment was determined. Two days after the inoculation of P. nicotianae, the expression of NtbHLH49 was suppressed and it was suppressed by over 50% after 3 d of infection (Figure 4B). Upon MeJA treatment, the expression of NtbHLH49 began to decrease after 1 h of treatment, attenuated by nearly 50% after 3 h of treatment, and decreased by about 70% after 12 h of treatment (Figure 4C). These data suggest that the expression of NtbHLH49 is negatively regulated by P. nicotianae attack or MeJA treatment, which indicate that NtbHLH49 may be a negative regulator of tobacco in responding to P. nicotianae.
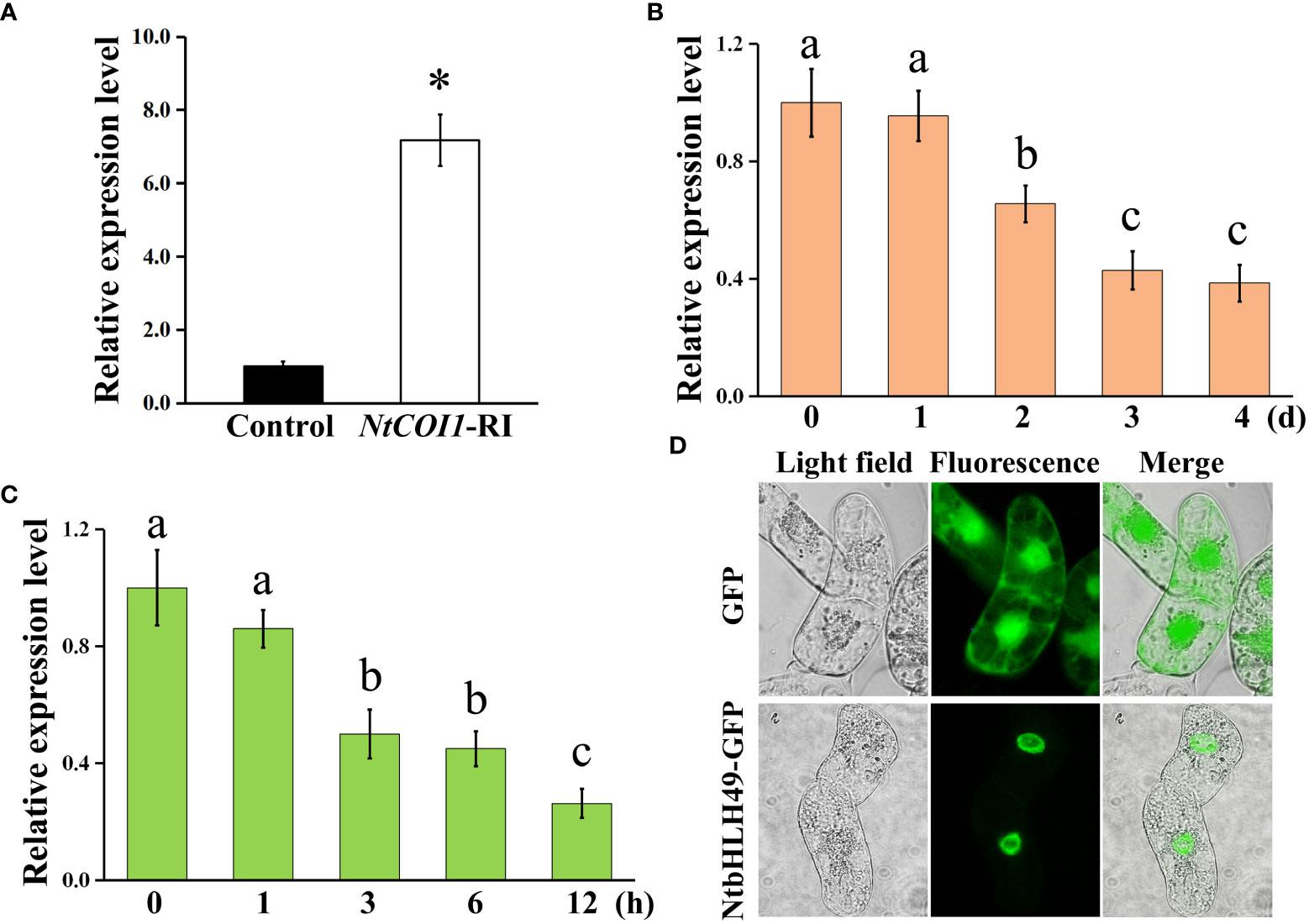
Figure 4 Gene expression pattern and subcellular localization of NtbHLH49. (A) Relative expression of NtbHLH49 in control and NtCOI1-RI plants. (B) Relative expression of NtbHLH49 in tobacco leaves after different time of P. nicotianae infection. (C) Relative expression of NtbHLH49 in tobacco leaves after different time of MeJA treatment. Actin gene was used as an internal control. The expression level of NtbHLH49 in control plants (A) or the time point ‘0’ (B, C) was set as “1”. Data are shown as means ± SD (n = 3). Asterisk in (A) indicates significant difference from control (Student's t-test; P<0.05). Lower-case letters (i.e., a, b, and c) in (B–C) indicate significant difference among the values (p<0.05). (D) Subcellular localization of NtbHLH49-GFP fusion protein and GFP alone (control) in BY2 cells.
Subsequently, the subcellular localization of NtbHLH49 was investigated by developing transgenic BY-2 cell lines that expressing GFP (yellow fluorescent protein; as control) and the C-terminal GFP fusion of NtbHLH49. As shown in Figure 4D, the fluorescence of GFP protein was observed in the cytoplasm and nucleus, while that of the fusion protein NtbHLH49-GFP was only detected in the nucleus. These results showed that NtbHLH49 is a nucleus-localized transcription factor.
Transgenic tobacco plants overexpressing NtbHLH49 (NtbHLH49-OE) or having NtbHLH49 knocked down by RNAi-mediated gene silencing (NtbHLH49-RI) were developed to further investigate the roles of NtbHLH49 in regulating tobacco resistance to P. nicotianae. Three lines that had NtbHLH49 overexpressed more than 10 folds and another three lines that had the expression of NtbHLH49 suppressed by near 80% percent were employed for further gene function analyses (Figures 5A, B). After infection with P. nicotianae, most leaves from control plants formed small necrosis lesions around the inoculation sites, but the leaves from NtbHLH49-OE plants developed much severe water-soaking disease symptom around the inoculation sites (Figure 5C). In contrast, most leaves from the NtbHLH49-RI plants had near invisible necrosis lesions around the inoculation sites, showing an increased resistance to P. nicotianae. Quantification of the severity of the diseases showed that control plants had less than 20% of leaves developed water-soaking patches, about 60% of leaves formed necrosis lesions and near 20% of leaves without visible symptom, while NtbHLH49-OE plants had over 50% of leaves developed water-soaking spots, near 40% of leaves formed necrosis lesions and less than 10% of leaves without visible symptom (Figure 5D). The NtbHLH49-RI plants had around 70% of leaves without visible symptom, around 20% of leaves formed necrosis lesions and less than 5% of leaves had water-soaking patches (Figure 5D). These findings suggest that NtbHLH49 plays a negative role in controlling tobacco resistance to P. nicotianae.
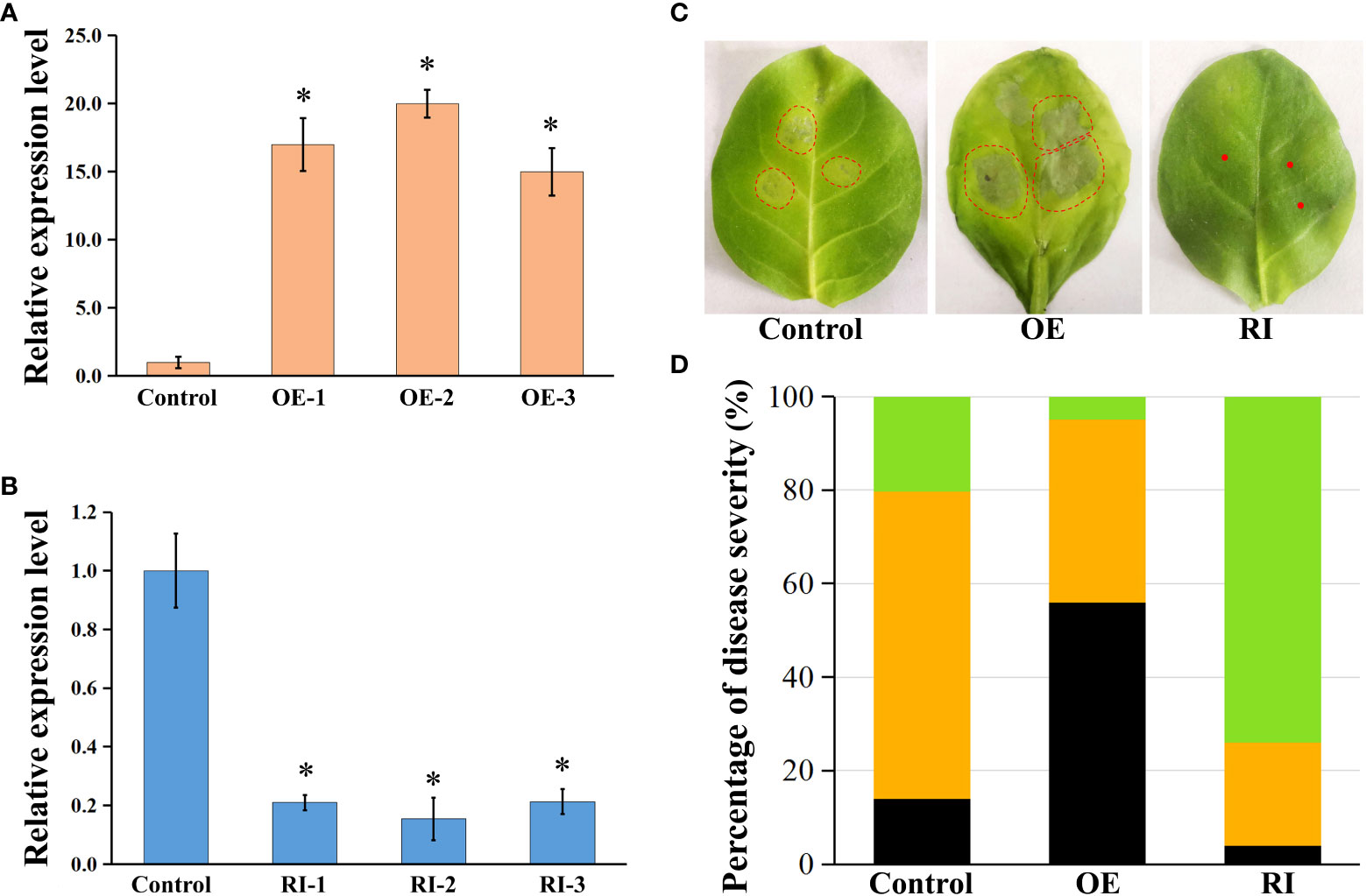
Figure 5 Roles of NtbHLH49 in regulating tobacco resistance to P. nicotianae. (A, B) Relative expression of NtbHLH49 in NtbHLH49-OE and NtbHLH49-RI plants. Tobacco Actin gene was used as an internal control. The expression level of NtbHLH49 in control plant was set as “1”. OE-1/2/3 and RI-1/2/3 indicate different transgenic lines. Data are shown as means ± SD (n = 3). Asterisks indicate significant difference from control (Student's t-test; P<0.05). (C) Representative symptoms of control, NtbHLH49-OE and NtbHLH49-RI tobacco leaves infected by P. nicotianae. The leaf area with visible symptom was indicated with dashed-circle. (D) Percentiles of control, NtbHLH49-OE and NtbHLH49-RI tobacco leaves with differential symptom severity. The black color indicates water-soaking symptom, the orange color indicates necrosis symptom, and the green color indicates no visible symptom.
The expression of pathogen resistance genes were inspected to reveal the mechanism underlying NtbHLH49-mediated tobacco resistance to P. nicotianae. These tested genes included two PR1 genes, two PR2 genes, two PR5 genes, two late blight resistance genes, and two disease resistance genes. The results showed that one PR1 gene (GenBank accession: XM_016632271) was increased 5 fold in the NtbHLH49-OE plants but was suppressed in the NtbHLH49-RI lines, and that the other PR1 gene (GenBank accession: XM_016589621) was suppressed in the NtbHLH49-OE plants but was accentuated over 5 folds in the NtbHLH49-RI lines (Figure 6), suggesting that NtbHLH49 displayed differential regulation on the expression of specific PR1 genes. Both of the PR2 genes (GenBank accession: XM_016583806, LOC107789548) were increased in the NtbHLH49-OE plants and were suppressed in the NtbHLH49-RI plants (Figure 6), showing a positive regulatory of NtbHLH49 on their expression. On the contrary, both the PR5 genes (GenBank accession: XM_016616970, XM_016609409) were decreased in the NtbHLH49-OE plants and were increased in the NtbHLH49-RI plants (Figure 6), indicating a negative regulatory role on the tested PR5 genes. Furthermore, NtbHLH49 showed a negative regulatory role on the expression of the two tested late blight resistance genes (GenBank accession: XM_016597833, XM_016591502), and also negatively regulated the expression of the two disease resistance genes (GenBank accession: LOC107829182, XM_016637917) as shown in Figure 6. The differential regulation of the tested pathogen resistance genes indicated a complicated role of NtbHLH49 in mediating tobacco resistance to P. nicotianae.
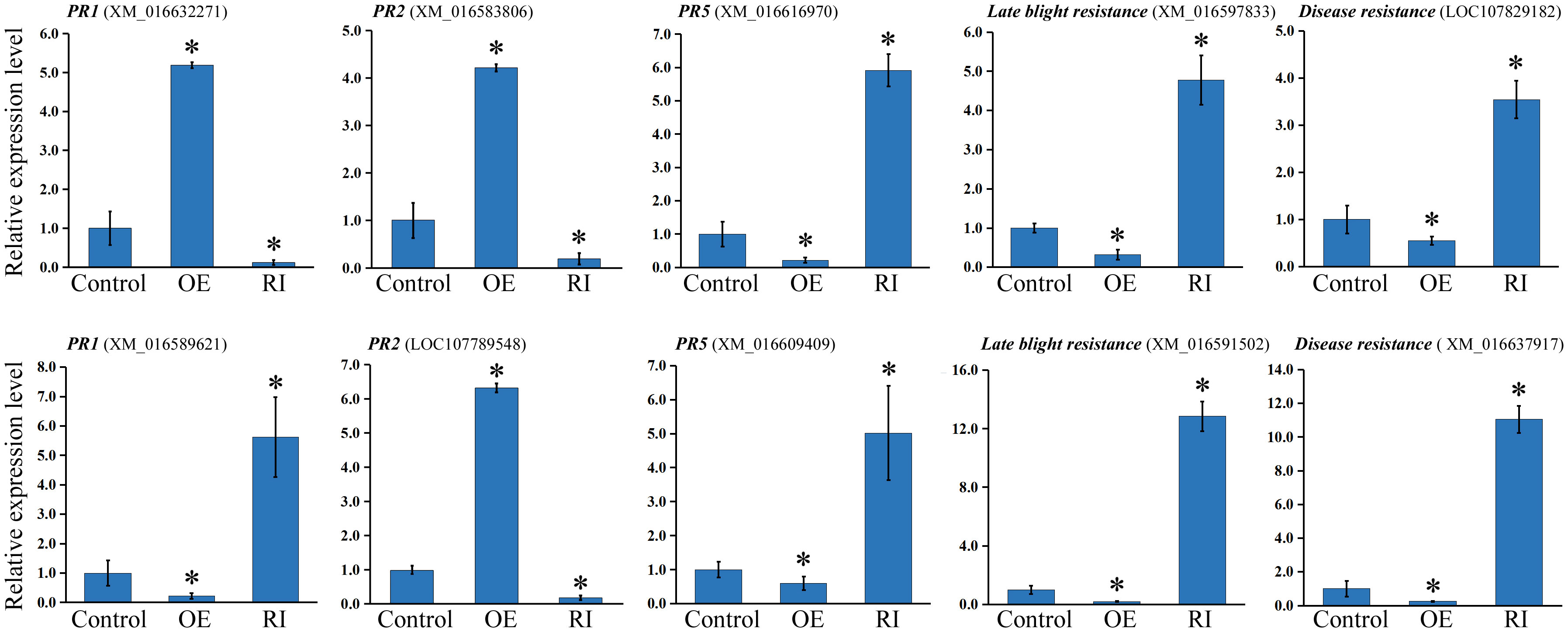
Figure 6 Relative expression of pathogen resistance genes in control, NtbHLH49-OE (OE) and NtbHLH49-RI (RI) plants. Tobacco Actin gene was used as an internal control. The expression level of each gene in control plant was set as “1”. Each value is the average (±SD) of three independent transgenic lines. Asterisks indicate significant difference from control (Student’s t-test; P < 0.05).
Plant resistance to pathogen attack is regulated by multiple phytohormones, such as JA, ET, SA, ABA etc. (Robert-Seilaniantz et al., 2011; Mishra et al., 2020). The JA-/ET-signaling pathway mediates defense against necrotrophic or hemibiotrophic infections (Robert-Seilaniantz et al., 2011). Available studies suggest that plant resistance to Phytophthora pathogens is regulated by environmental stresses and phytohormones (Dong et al., 2015; Shibata et al., 2016; Yang et al., 2019). For instance, P. sojae RXLR effector PsAvh238 suppresses plant defenses against P. sojae by targeting GmACSs, an enzyme that catalyzes ACC formation during ET biosynthesis (Yang et al., 2019). The expression of GmCYP82A3, a soybean CYP82 gene functioning in JA/ET-meidated defensive responses, enhanced N. benthamiana resistance to P. parasitica (Yan et al., 2016). Additionally, Arabidopsis resistance to Phytophthora pathogen is modulated by both the SA and JA signaling pathways (Li et al., 2020). These pieces of evidence also indicated the importance of JA-signaling in plant responses to Phytophthora pathogen. Utilizing previously developed NtCOI1-silenced plants, this work investigated the roles of JA-signaling pathway in mediating tobacco responses to P. nicotianae and analyzed the correlated transcriptional regulations using a transcriptome-sequencing approach.
The transcriptome analysis identified a total of 1330 DEGs between NtCOI1-RI plants and control plants infected with P. nicotiane. 1082 of these DEGs were annotated in the GO database, with 877 being up-regulated and 768 being down-regulated in NtCOI1-RI plants. The GO annotation revealed that a large number of pathogen resistance genes, such as those involved in systemic acquired resistance, immune/defense response, defense/immunity protein, biotic stimulus response, pathogenesis, and phytohormone-signaling related, were down-regulated in NtCOI1-RI plants. The analysis identified some pathogen resistance genes, transcription regulator genes and phytohormone-signaling related genes. These findings showed that the JA-signaling pathway could manipulate both functional genes and transcription regulatory genes during tobacco responding to P. nicotiane attack.
Extensive studies showed that dysfunction of JA-signal perception protein COI1 could alter plant resistance to both bacterial and fungal pathogens (Kloek et al., 2001; Zhang et al., 2017), and that the downstream regulators, such as bHLH transcription factors and MYB transcription factors, played important roles in the JA-mediated plant resistance to pathogen attack (Thatcher et al., 2016; Ren et al., 2020; Ren et al., 2022). The differentially expressed transcription factor genes included a set of bHLH transcription factor genes, MYB transcription factor genes, and ERF genes that may play roles in transducing JA signals and regulating JA-mediated pathogen responses (Solano et al., 1998; Lorenzo et al., 2003). bHLH transcription factors could regulate plant resistance to multiple pathogens, such as Botrytis cinerea and Sclerotinia sclerotiorum in Arabidopsis and so on (Kazan and Manners, 2013; Li et al., 2018; Teng et al., 2021). They have also been indicated in the regulation of plant Phytophthora pathogen resistance. In soybean, the bHLH transcription factor GmPIB1 enhances resistance to P. sojae and reduces reactive oxygen species accumulation (Cheng et al., 2018). Potato (Solanum tuberosum) bHLH transcription factor StCHL1 and its N. benthamiana homolog could enhance the leaf colonization by P. infestans and suppress cell immunity (Turnbull et al., 2017). Genome-wide transcriptome of N. benthamiana also suggested that bHLH genes may play important roles in Phytophthora pathogen resistance (Yu et al., 2019). Employing common tobacco cultivar TN90, this study revealed that 5 bHLHs transcription factor genes were differentially expressed between NtCOI1-silenced plants and control. Intriguingly, the majority of these bHLH transcription factor genes were AtBPE-like regulators and were up-regulated in NtCOI1-RI plants, which suggested a negative role in regulating tobacco resistance to P. nicotianae. Further study on NtbHLH49, one of these bHLH transcription factor genes, found that its expression was negatively regulated by JA treatment or P. nicotianae infection, and that its overexpression decreased and its gene knockdown increased tobacco resistance to P. nicotianae. Therefore, these findings suggested that NtbHLH49 is a negative regulator during tobacco responding to P. nicotianae. Subsequent transcription assays demonstrated that NtbHLH49 regulated the expression of a set of pathogen resistance genes including PR1, PR2, PR5, and late blight resistance genes etc., and that it exhibited differential regulation on the specific homolog of these pathogen resistance genes, showing a complicated regulatory function of NtbHLH49 in tobacco. Taken together, these assays revealed the function of AtBPE-like regulator NtbHLH49 in regulating tobacco resistance to P. nicotianae, and extended the knowledge of bHLH transcription factors in mediating tobacco responses to P. nicotianae.
Using physiological experiments and a high-throughout sequencing approach, this study identified the functional and regulatory genes that may be involved in the JA-mediated P. nicotianae responses of tobacco. The results revealed that a set of pathogen resistance genes, transcription factor genes, and JA-signaling genes took part in tobacco resistance to this pathogen, which also indicated the involvement of a group of AtBPE-like regulators in this process. Subsequent studies on NtbHLH49 showed that its gene expression was negatively regulated by JA treatment or P. nicotianae infection, and it played a negative role in regulating tobacco resistance to P. nicotianae. Findings of this study provide insights into the regulatory roles of AtBPE-like bHLH transcription factors in tobacco responses to P. nicotianae and are helpful to unravel the underlying mechanism for tobacco resistance against this pathogen.
The names of the repository/repositories and accession number(s) can be found at NCBI, PRJNA901343.
WW and JZ conceived the study and designed the experiments. WW, JZ, YC, and XY performed the experiments, and produced the figures. All authors analyzed the data. WW and JZ wrote the manuscript, and FW, JY, and XW helped in the manuscript revision. All authors contributed to the article and approved the submitted version.
This work is supported by the Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (ASTIP-TRIC04), China National Tobacco Corporation [110202101055(LS-15)] and Sichuan Tobacco Company (SCYC202014). This study received funding from China National Tobacco Corporation and Sichuan Tobacco Company. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1073856/full#supplementary-material
Alexandrino, D. A. M., Almeida, C. M. R., Mucha, A. P., Carvalho, M. F. (2022). Revisiting pesticide pollution: The case of fluorinated pesticides. Environ. pollut. 292, 118315. doi: 10.1016/j.envpol.2021.118315
Cheng, Q., Dong, L., Gao, T., Liu, T., Li, N., Wang, L., et al. (2018). The bHLH transcription factor GmPIB1 facilitates resistance to phytophthora sojae in glycine max. J. Exp. Bot. 69, 2527–2541. doi: 10.1093/jxb/ery103
Cho, K., Kim, Y., Wi, S. J, Seo, J. B., Kwon, J., Chung, J. H., et al. (2013). Metabolic survey of defense responses to a compatible hemibiotroph, phytophthora parasitica var. nicotianae, in ethylene signaling-impaired tobacco. J. Agric. Food Chem. 4, 8477–8489. doi: 10.1021/jf401785w
Curtis, M. D., Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Dileo, M. V., Pye, M. F., Roubtsova, T. V., Duniway, J. M., Macdonald, J. D., Rizzo, D. M., et al. (2010). Abscisic acid in salt stress predisposition to phytophthora root and crown rot in tomato and chrysanthemum. Phytopathology 100, 871–879. doi: 10.1094/PHYTO-100-9-0871
Dong, L. D., Cheng, Y. X., Wu, J. J., Cheng, Q., Li, W. B., Fan, S. J., et al. (2015). Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to phytophthora sojae in soybean. J. Exp. Bot. 66, 2635–2647. doi: 10.1093/jxb/erv078
Goossens, A., Häkkinen, S. T., Laakso, I., Seppänen-Laakso, T., Biondi, S., De Sutter, V., et al. (2003). A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. U.S.A. 8, 8595–8600. doi: 10.1073/pnas.1032967100
Halim, V. A., Eschen-Lippold, L., Altmann, S., Birschwilks, M., Scheel, D., Rosahl, S. (2007). Salicylic acid is important for basal defense of solanum tuberosum against phytophthora infestans. Mol. Plant Microbe Interact. 20, 1346–1352. doi: 10.1094/MPMI-20-11-1346
Howe, G. A., Major, I. T., Koo, A. J. K. (2018). Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 69, 387–415. doi: 10.1146/annurev-arplant-042817-040047
Kazan, K. (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20, 219–229. doi: 10.1016/j.tplants.2015.02.001
Kazan, K., Manners, J. M. (2013). MYC2: the master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Kloek, A. P., Verbsky, M. L., Sharma, S. B., Schoelz, J. E., Vogel, J., Klessig, D. F., et al. (2001). Resistance to pseudomonas syringae conferred by an arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26, 509–522. doi: 10.1046/j.1365-313x.2001.01050.x
Kroon, L. P., Brouwer, H., de Cock, A. W., Govers, F. (2012). The genus phytophthora anno 2012. Phytopathology 102, 348–364. doi: 10.1094/PHYTO-01-11-0025
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li, X., Yang, R., Chen, H. (2018). The arabidopsis thaliana mediator subunit MED8 regulates plant immunity to botrytis cinerea through interacting with the basic helix-loop-helix (bHLH) transcription factor FAMA. PloS One 13, e0193458. doi: 10.1371/journal.pone.0193458
Li, W., Zhao, D., Dong, J., Kong, X., Zhang, Q., Li, T., et al. (2020). AtRTP5 negatively regulates plant resistance to phytophthora pathogens by modulating the biosynthesis of endogenous jasmonic acid and salicylic acid. Mol. Plant Pathol. 21, 95–108. doi: 10.1111/mpp.12883
Long, J., Yang, M., Zuo, C., Song, N., He, J. M., Zeng, J., et al. (2021). Requirement of jasmonate signaling for defense responses against alternaria alternata and phytophthora nicotiane in tobacco. Crop Sci. 61, 4273–4283. doi: 10.1002/csc2.20625
Lorenzo, O., Piqueras, R., Sanchez-Serrano, J. J., Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Meng, Y. L., Zhang, Q., Ding, W., Shan, W. X. (2014). Phytophthora parasitica: a model oomycete plant pathogen. Mycology 5, 43–51. doi: 10.1080/21501203.2014.917734
Mishra, B., Bose, S. K., Sangwan, N. S. (2020). Comparative investigation of therapeutic plant withania somnifera for yield, productivity, withanolide content, and expression of pathway genes during contrasting seasons. Ind. Crop Prod. 154, 112508. doi: 10.1016/j.indcrop.2020.112508
Müller, M., Munné-Bosch, S. (2015). Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Pieterse, C. M., van der Does, D., Zamioudis, C., Leon-Reyes, A., Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Ren, H., Bai, M., Sun, J., Liu, J., Ren, M., Dong, Y., et al. (2020). RcMYB84 and RcMYB123 mediate jasmonate-induced defense responses against botrytis cinerea in rose (Rosa chinensis). Plant J. 103, 1839–1849. doi: 10.1111/tpj.14871
Ren, Y., Zou, W., Feng, J., Zhang, C., Su, W., Zhao, Z., et al. (2022). Characterization of the sugarcane MYC gene family and the negative regulatory role of ShMYC4 in response to pathogen stress. Ind. Crop Prod 176, 114292. doi: 10.1016/j.indcrop.2021.114292
Robert-Seilaniantz, A., Grant, M., Jones, J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicyate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Roubtsova, T. V., Bostock, R. M. (2009). Episodic abiotic stress as a potential contributing factor to onset and severity of disease caused by phytophthora ramorum in rhododendron and viburnum. Plant Dis. 93, 912–918. doi: 10.1094/PDIS-93-9-0912
Rushton, P. J., Bokowiec, M. T., Han, S., Zhang, H., Brannock, J. F., Chen, X., et al. (2008). Tobacco transcription factors: novel insights into transcriptional regulation in the solanaceae. Plant Physiol. 147, 280–295. doi: 10.1104/pp.107.114041
Shibata, Y., Ojika, M., Sugiyama, A., Yazaki, K., Jones, D. A., Kawakita, K., et al. (2016). The full-size ABCG transporters Nb-ABCG1 and Nb-ABCG2 function in pre- and post-invasion defense against phytophthora infestans in nicotiana benthamiana. Plant Cell 28, 1163–1181. doi: 10.1105/tpc.15.00721
Solano, R., Stepanova, A., Chao, Q., Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Teng, Z., Zheng, W., Yu, Y., Hong, S. B., Zhu, Z., Zang, Y. (2021). Effects of BrMYC2/3/4 on Plant Development, glucosinolate metabolism, and Sclerotinia sclerotiorum Resistance in Transgenic Arabidopsis thaliana. Front. Plant Sci. 12, 707054. doi: 10.3389/fpls.2021.707054
Thaler, J. S., Owen, B., Higgins, V. J. (2004). The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 135, 530–538. doi: 10.1104/pp.104.041566
Thatcher, L. F., Cevik, V., Grant, M., Zhai, B., Jones, J. D., Manners, J. M., et al. (2016). Characterization of a JAZ7 activation-tagged arabidopsis mutant with increased susceptibility to the fungal pathogen fusarium oxysporum. J. Exp. Bot. 67, 2367–2386. doi: 10.1093/jxb/erw040
Thomma, B. P., Eggermont, K., Penninckx, J. A., Mauch-Mani, B., Vogelsang, R., Cammue, B. P. A., et al. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in arabidopsid are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 95, 15107–15111. doi: 10.1073/pnas.95.25.15107
Turnbull, D., Yang, L., Naqvi, S., Breen, S., Welsh, L., Stephens, J., et al. (2017). RXLR effector AVR2 up-regulates a brassinosteroid-responsive bHLH transcription factor to suppress immunity. Plant Physiol. 174, 356–369. doi: 10.1104/pp.16.01804
Wang, W. J., Jiao, F. C. (2019). Effectors of phytophthora pathogens are powerful weapons for manipulating host immunity. Planta 250, 413–425. doi: 10.1007/s00425-019-03219-x
Wang, W. J., Liu, G. S., Niu, H. X., Timko, M. P., Zhang, H. B. (2014). The f-box protein COI1 functions upstream of MYB305 to regulate primary carbohydrate metabolism in tobacco. J. Exp. Bot. 65, 2147–2160. doi: 10.1093/jxb/eru084
Wi, S. J., Ji, N. R., Park, K. Y. (2012). Synergistic biosynthesis of biphasic ethylene and reactive oxygen species in response to hemibiotrophic phytophthora parasitica in tobacco plants. Plant Physiol. 159, 251–265. doi: 10.1104/pp.112.194654
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., Turner, J. G. (1998). COI1: an arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Yan, Q., Cui, X., Lin, S., Gan, S., Xing, H., Dou, D. (2016). GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway enhances plant resistance to biotic and abiotic stresses. PloS One 11, e0162253. doi: 10.1371/journal.pone.0162253
Yan, J., Zhang, C., Gu, M., Bai, Z., Zhang, W., Qi, T., et al. (2009). The arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236. doi: 10.1105/tpc.109.065730
Yang, B., Wang, Y. Y., Guo, B. D., Jing, M. F., Zhou, H., Li, Y. F., et al. (2019). The phytophthora sojae RXLR effector Avh238 destabilizes soybean Type2 GmACSs to suppress ethylene biosynthesis and promote infection. New Phytol. 222, 425–437. doi: 10.1111/nph.15581
Young, M. D., Wakefield, M. J., Smyth, G. K., Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. doi: 10.1186/gb-2010-11-2-r14
Yu, J., Ai, G., Shen, D., Chai, C., Jia, Y., Liu, W., et al. (2019). Bioinformatical analysis and prediction of nicotiana benthamiana bHLH transcription factors in phytophthora parasitica resistance. Genomics 111, 473–482. doi: 10.1016/j.ygeno.2018.03.005
Zhang, L., Zhang, F., Melotto, M., Yao, J., He, S. Y. (2017). Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 68, 1371–1385. doi: 10.1093/jxb/erw478
Keywords: Phytophthora nicotianae, tobacco, jasmonate, AtBPE-like, NtbHLH49
Citation: Wang W, Zhang J, Cao Y, Yang X, Wang F, Yang J and Wang X (2022) NtbHLH49, a jasmonate-regulated transcription factor, negatively regulates tobacco responses to Phytophthora nicotianae. Front. Plant Sci. 13:1073856. doi: 10.3389/fpls.2022.1073856
Received: 19 October 2022; Accepted: 16 November 2022;
Published: 06 December 2022.
Edited by:
Miguel Gonzalez-Guzman, University of Jaume I, SpainReviewed by:
Yan Li, Sichuan Agricultural University, ChinaCopyright © 2022 Wang, Zhang, Cao, Yang, Wang, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjing Wang, d2FuZ3dlbmppbmdAY2Fhcy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.