- Department of Systems Biotechnology, Konkuk University, Seoul, South Korea
SHORT-ROOT (SHR) is a mobile transcription factor that plays important roles in ground tissue patterning, stem cell niche specification and maintenance, and vascular development in Arabidopsis roots. Although mRNA and protein of SHR are also found in hypocotyls, inflorescence stems, and leaves, its role in the above-ground organs has been less explored. In most developmental cases, SHR, together with its partner SCARECROW (SCR), regulates the expression of downstream target genes in controlling formative and proliferative cell divisions. Accumulating evidence on the regulatory role of SHR in shoots suggests that SHR may also play key roles in the above-ground organs. Interestingly, recent work has provided new evidence that SHR is also required for cell elongation in the hypocotyl of the etiolated seedling. This suggests that the novel roles of SHR and SHR-mediated regulatory networks can be found in shoots. Furthermore, comparative research on SHR function in roots and shoots will broaden and deepen our understanding of plant growth and development.
Introduction
Roots of an individual plant play crucial roles in i) acquiring water and nutrients, ii) supporting the plant, iii) synthesizing plant hormones, iv) storing nutrients and metabolites, and v) interacting with soil microbiome (Schiefelbein and Benfey, 1991; Benfey et al., 2010; Petricka et al., 2012). Therefore, understanding the molecular mechanisms controlling root growth and development is of prime importance. Due to the simple cellular organization and a plethora of molecular, genetic, and genomic resources, the model plant Arabidopsis thaliana (Arabidopsis) has enormously contributed to broadening and deepening our understanding of root growth and development (Schiefelbein and Benfey, 1991; Dolan et al., 1993; Benfey et al., 2010; Petricka et al., 2012).
Three decades ago, in an attempt to isolate mutants with abnormal root structures in Arabidopsis, the Philip Benfey lab, then at New York University, identified a mutant that exhibited a short-root growth phenotype (Benfey et al., 1993). Since the recessive mutation resulted in determinate root growth, the mutant was named “short-root (shr)” (Benfey et al., 1993). In addition to abnormal root growth, detailed phenotypic analyses revealed that shr possessed no endodermis, the innermost ground tissue (GT) with the Casparian strip (Benfey et al., 1993). Therefore, shr had only a single GT layer between the epidermis and the stele instead of the two layers found in the wild-type (WT) root (Benfey et al., 1993). The root radial pattern defect in shr was traced back to the heart-stage embryo (Scheres et al., 1995). Furthermore, when crossed with the fass mutant with the multiple GT layers, the endodermis was not restored in the shr fass double mutant, indicating that the specification and differentiation of the endodermis depended on SHR function (Scheres et al., 1995).
Similarly, another recessive mutant, scarecrow (scr), also had one GT layer in roots (Scheres et al., 1995). Unlike shr, the remaining GT layer in scr showed both endodermis and cortex characteristics, indicating that the periclinal (parallel to the growth axis) formative division to separate the two layers was flawed (Scheres et al., 1995; Di Laurenzio et al., 1996). The SCR gene was identified, and its expression was detected in the quiescent center (QC), cortex/endodermis initial (CEI), cortex/endodermis initial daughter (CEID), and endodermis (Di Laurenzio et al., 1996). Four years after SCR cloning, the SHR gene was also identified and shown to encode a similar transcription factor to SCR, belonging to the GRAS family (Pysh et al, 1999; Helariutta et al., 2000). Interestingly, SHR mRNA was observed in the stele. However, the protein moved outward to the tissues (QC, CEI, and CEID) where SCR was expressed, indicating that SHR acted as a mobile transcription factor (Helariutta et al., 2000; Nakajima et al., 2001; Gallagher et al., 2004; Gallagher and Benfey, 2009). Moreover, SHR interacted with SCR in the nuclei of the endodermis to control the SCR expression for proper radial patterning (Cui et al., 2007; Koizumi et al., 2012a; Koizumi et al., 2012b). In addition to SCR, SCARECROW-LIKE23 (SCL23), the closest SCR homolog, was also shown to play a role in the specification of endodermis cell fate (Long et al., 2015a). Furthermore, JACKDAW (JKD) and its related BIRD transcription factors [also known as INDETERMINATE DOMAIN (IDD)] interacted with SHR to restrict SHR from moving beyond the endodermis (Welch et al., 2007; Long et al., 2015b; Moreno-Risueno et al., 2015; Long et al., 2017; Figure 1, left). Other factors, such as RETINOBLASTOMA-RELATED (RBR) and CYCLIN D6;1 (CYCD6;1), also played a role in controlling the formative division to generate the cortex and endodermis (Sozzani et al., 2010; Cruz-Ramírez et al., 2012; Figure 1, left). Recently, SHR homologs were identified in the roots with multiple GT layers such as date palms, legumes, maize, and Setaria (Setaria viridis) (Xiao et al., 2019; Dong et al., 2021; Ortiz-Ramírez et al., 2021; Xu et al., 2021; Wang et al., 2022). Indeed, the SHR-mediated regulatory networks also controlled GT formation across species, resulting in generation of a multilayered cortex (Hernández-Coronado and Ortiz-Ramírez, 2021). Unlike SHR in the Arabidopsis root, all three maize SHR homologs (ZmSHR1, ZmSHR2, and ZmSHR2-h) were predominantly expressed in the endodermis, revealed by single-cell RNA sequencing and in situ RNA hybridization (Ortiz-Ramírez et al., 2021). Moreover, the ZmSHR1 protein was hypermobile, moving from the endodermis to the cortex layers. Interestingly, the Zmshr2 Zmshr2-h double mutant had reduced cortex numbers instead of missing the endodermis, indicating that SHR in maize played a critical role in expansion of the cortex tissue (Ortiz-Ramírez et al., 2021). In addition, its role in cortex multiplication was validated in another monocot Setaria, monitored by phenotypic analyses of the loss-of-function mutants of the two Setaria SHR homologs (SvSHR1 and SvSHR2). Indeed, the Svshr1 Svshr2 double mutant showed substantially reduced cortex layers (Ortiz-Ramírez et al., 2021). Therefore, it was suggested that hypermobility of the SHR proteins was common in monocots, which played an important role in multilayered cortex development (Wu et al., 2014; Hernández-Coronado and Ortiz-Ramírez, 2021; Ortiz-Ramírez et al., 2021). Nonetheless, elucidating the role of SHR and its regulatory networks in root radial patterning is still an active subject of research (Zhang et al., 2018; Tian et al., 2022; Yang et al., 2022).
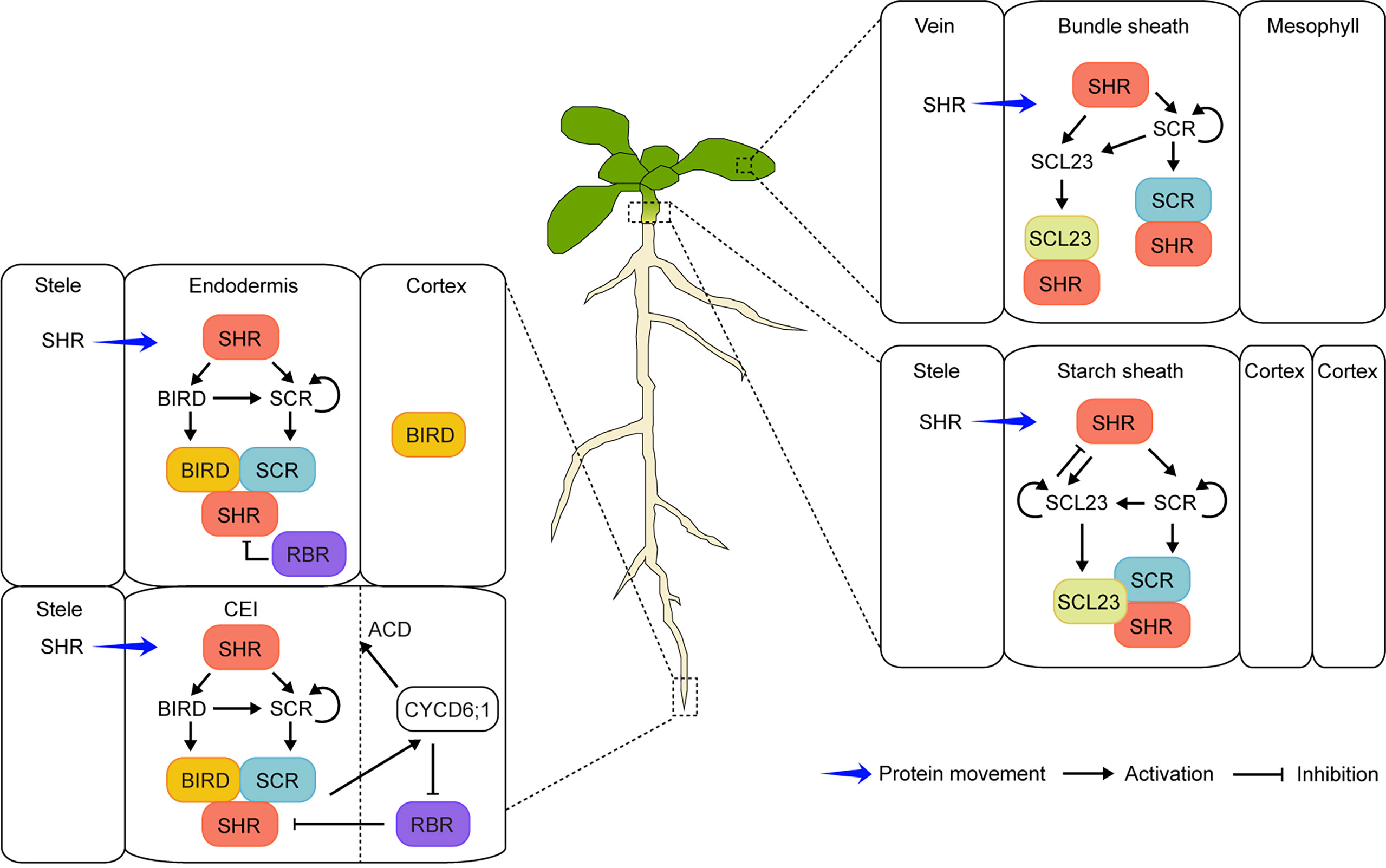
Figure 1 Schematic model of the SHR-mediated regulatory networks in the endodermis development of Arabidopsis roots and shoots. In roots (left), SHR protein moves from the stele into the endodermis and CEI (cortex/endodermis initial) where it activates the expression of the downstream target SCR and BIRD genes. SHR forms protein complexes with SCR and BIRD, resulting in the confinement of SHR in the endodermis and CEI. The protein complexes induce the CYCD6;1 expression, which subsequently inhibits the negative regulator RBR from interacting with the SHR/SCR complex and promotes the asymmetric cell division (ACD) of CEI. In leaves and hypocotyls (right), SHR as a mobile regulator activates the expression of both SCR and SCL23 in the endodermis and its equivalents (bundle sheath in leaves and starch sheath in hypocotyls). Protein complexes of SHR-SCR, SHR-SCL23, or SHR-SCR-SCL23 can be formed, which prevents SHR from moving beyond the endodermis. Moreover, SCL23 negatively regulates SHR function in hypocotyls.
In addition to radial pattern formation, SHR is involved in the specification and maintenance of the root stem cell niche (Sabatini et al., 2003; Qi et al., 2019). The shr mutant displayed a loss of functional QC and a reduction of the meristem size, thereby resulting in determinate root growth (Benfey et al., 1993; Helariutta et al., 2000; Sabatini et al., 2003). PLETHORA (PLT) transcription factors were shown to specify and maintain the QC and stem cell niche (Aida et al., 2004; Galinha et al., 2007). However, it was suggested that PLTs and SHR acted in parallel pathways in QC and stem cell niche specification and maintenance (Aida et al., 2004; Galinha et al., 2007; Santuari et al., 2016; Pardal and Heidstra, 2021).
Due to its localization in the root stele (Helariutta et al., 2000; Nakajima et al., 2001), it was reasonable to speculate that SHR might play a role in root vascular development. Indeed, mutations in SHR caused reduced cell numbers in the root vasculature (Levesque et al., 2006; Yu et al., 2010) and ectopic metaxylem differentiation in place of protoxylem (Carlsbecker et al., 2010; Yu et al., 2010; Miyashima et al., 2011). For example, SHR and SCR activated the expression of two microRNA (miRNA165 and 166) genes in the endodermis. The resulting miRNA165/166 with gradients restricted their target mRNAs, class III HOMEODOMAIN LEUCINE ZIPPER (HD-ZIP III) mRNAs at post-transcriptional levels for xylem patterning (Carlsbecker et al., 2010; Miyashima et al., 2011). In addition to xylem patterning, shr exhibited severe developmental defects in phloem development (Kim et al., 2020). These studies indicated that SHR non-cell-autonomously exerted its decisive role on the formative cell division for xylem and phloem development. Interestingly, it was demonstrated that SHR controlled cytokinin homeostasis by directly activating the expression of CYTOKININ OXIDASE3 (CKX3) (Cui et al., 2011; Yang et al., 2021). These findings suggested that spatiotemporal regulation of cytokinin levels might be achieved by SHR in the periphery of the root xylem axis (Cui et al., 2011; Yang et al., 2021).
In addition to its role in cell division, recent work revealed that shr displayed a drastic reduction in cell elongation in the root maturation zone, suggesting that SHR also played a role in root cell elongation by regulating redox homeostasis (Fu et al., 2021).
Since the first characterization of shr, detailed studies have provided insights into its regulatory role in plant roots. Nevertheless, much is still to be learned by unveiling SHR-mediated plant developmental networks.
Discussion
The very first report of SHR’s involvement in the above-ground organs came from the serendipitous finding that both hypocotyl and inflorescence stem of shoot gravitropism7 (sgr7) displayed no response to a change of gravity vector (Fukaki et al., 1998). The sgr7 mutant turned out to be allelic to shr and had no endodermis/starch sheath in hypocotyls and stems, similar to shr roots (Fukaki et al., 1998). In addition, SHR formed protein complexes with SCL23; therefore, the SHR-SCR-SCL23 module played a role in the formation of the functional bundle sheath (also known as endodermis equivalent) in Arabidopsis hypocotyls (Figure 1, right). These studies indicated that a common molecular mechanism exerted decisive control on the specification and differentiation of the endodermis and its equivalents in shoots and roots (Fukaki et al., 1998; Yoon et al., 2016; Kim et al., 2017).
Besides the phenotypic perturbations in hypocotyl and stem radial patterning, the shoot growth of shr was evidently retarded, thereby resulting in a stunted plant at maturity (Figures 2A,B). In addition, shr exhibited substantial reductions in fresh and dry weights, which were comparable to approximately one-tenth of the WT levels (Figures 2C,D). The identification and characterization of the SHR gene and its expression patterns indicated that SHR might play a role in shoot development (Helariutta et al., 2000). Nonetheless, compared to what we have learned about SHR and its regulatory networks in roots, its role in shoots has been less explored.
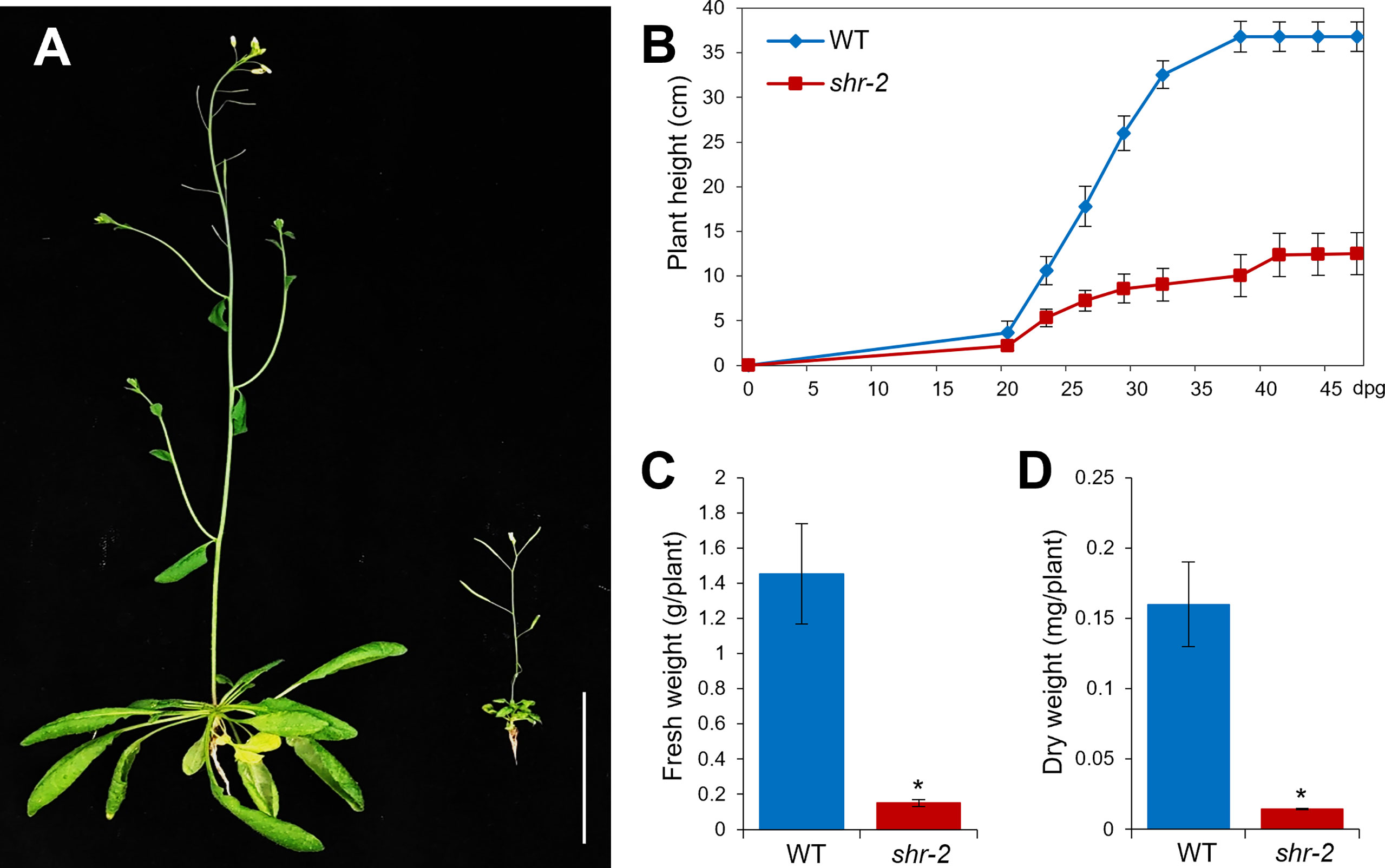
Figure 2 Shoot growth phenotypes of Arabidopsis wild-type (WT) and shr adult plants. (A) Approximately 6-week-old WT and shr-2 plants. Scale bar: 5 cm. (B) Lengths of inflorescence stems of WT and shr-2 plants at different time points (dpg: days postgermination). (C) Fresh weights of ~6-week-old WT and shr-2 plants. (D) Dry weights of ~6-week-old WT and shr-2 plants. The data are shown as mean ± SEM (n > 30). Statistical significance was determined by Student′s t-test compared with WT (*P < 0.05).
In leaves, both mRNA and protein of SHR were also detected in the vascular bundle and the surrounding bundle sheath (Dhondt et al., 2010; Gardiner et al., 2011; Cui et al., 2014). The size of rosette leaves was severely reduced in shr compared to that in WT, suggesting that SHR was involved in proliferative cell division in developing leaves (Dhondt et al., 2010). Moreover, in shr leaves, cells in the bundle sheath were rather irregular in shape and became larger than those observed in WT (Cui et al., 2014). Therefore, cells surrounding the vascular core appeared to become mesophyll-like in leaves. As in roots and hypocotyls (Long et al., 2015a; Yoon et al., 2016), the SHR-SCR-SCL23 regulatory module was critically involved in the specification and maintenance of the endodermis equivalent in leaves (Cui et al., 2014; Figure 1, right). These studies provided new insights into the role of SHR as a critical regulator in formative and proliferative cell divisions in Arabidopsis leaves.
In hypocotyls and stems, shr had reduced xylem and phloem areas, resulting in hypocotyls and stems with smaller diameters (Ko et al., 2022). Because the post-transcriptional interaction between miRNA165/166 and HD-ZIP IIIs was known to play a crucial role in radial patterning of the shoot vasculature (Emery et al., 2003; Kim et al., 2005), it will be interesting to investigate whether SHR regulates the expression of miRNA165/166 in these organs, as in the root vascular development (Carlsbecker et al., 2010; Miyashima et al., 2011; Kim et al., 2020).
In the etiolated seedling, SHR was also critically involved in controlling hypocotyl cell elongation (Dhar et al., 2022). Hypocotyl cell length in the etiolated shr seedling was discernibly reduced, compared to that in WT (Dhar et al., 2022). Indeed, SHR controlled the cell elongation process via transcriptional regulation of a group of xyloglucan endotransglucosylase/hydrolase (XTH) genes encoding cell wall remodeling enzymes (Dhar et al., 2022). In most cases, SHR acted together with SCR to regulate the expression of downstream target genes (Helariutta et al., 2000; Cui et al., 2007; Carlsbecker et al., 2010; Sozzani et al., 2010; Cruz-Ramírez et al., 2012; Hirano et al., 2017; Long et al., 2017). Unlike the known mode of action, SHR activated the expression of the three XTH genes (XTH18, XTH22 and XTH24) in a SCR-independent manner (Dhar et al., 2022). SHR is well known to play key roles in regulating formative and proliferative cell divisions. In the etiolated seedling, however, SHR was required for cell elongation. Therefore, this finding indicated that SHR might play previously uncharacterized roles in Arabidopsis shoots.
Since the identification and characterization of shr were first reported (Benfey et al., 1993), there have been tremendous efforts to isolate homologous genes of SHR and elucidate their function in diverse species. Research in monocots, such as rice and maize, provided new insights into SHR’s role in shoots. For example, Kamiya et al. (2003) identified two rice SHR homologs (OsSHR1 and OsSHR2) and reported that OsSHR1 was expressed during stomata development. Likewise, the maize SHR homologs (ZmSHR1 and ZmSHR2) were shown to be involved in the development of Kranz anatomy and C4 physiology in leaves (Slewinski, 2013; Fouracre et al., 2014; Slewinski et al., 2014; Schuler et al., 2018). In particular, the Zmshr1 mutant exhibited alterations in patterning and spacing of vascular, bundle sheath and mesophyll cells in maize leaves (Slewinski et al., 2014). Therefore, these reports indicated that the SHR-mediated networks distinctly controlled both vascular and stomata patterning in monocot leaves (Schuler et al., 2018). Thus, it will be interesting to investigate whether SHR also regulates stomata development in Arabidopsis leaves.
Although recent studies have identified new regulatory aspects of SHR in shoots, more research is still required (e.g., the interplay between SHR and plant hormones that modulates the growth and development of the above-ground organs at all developmental phases). Together with what we have learned about SHR and its regulatory networks in roots and shoots so far, the time is coming closer to appreciate the whole picture of what role the master regulator SHR plays in plant growth and development. So, it is time to look up!
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
EKY, JO, and JL conceived and designed the research plans. EKY, JO, and JL wrote the manuscript. EKY and JO designed the figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Konkuk University Research Fund 2017.
Acknowledgments
We thank members of the Lim laboratory for comments on the manuscript. We also would like to thank Editage (www.editage.co.kr) for English language editing. We apologize to all colleagues whose work has not been mentioned due to space limitations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. doi: 10.1016/j.cell.2004.09.018
Benfey, P. N., Bennett, M., Schiefelbein, J. (2010). Getting to the root of plant biology: impact of the Arabidopsis genome sequence on root research. Plant J. 61, 992–1000. doi: 10.1111/j.1365-313X.2010.04129.x
Benfey, P. N., Linstead, P. J., Roberts, K., Schiefelbein, J. W., Hauser, M. T., Aeschbacher, R. A. (1993). Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119, 57–70. doi: 10.1242/dev.119.1.57
Carlsbecker, A., Lee, J. Y., Roberts, C. J., Dettmer, J., Lehesranta, S., Zhou, J., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321. doi: 10.1038/nature08977
Cruz-Ramírez, A., Diaz-Trivino, S., Blilou, I., Grieneisen, V. A., Sozzani, R., Zamioudis, C., et al. (2012). A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150, 1002–1015. doi: 10.1016/j.cell.2012.07.017
Cui, H., Hao, Y., Kovtun, M., Stolc, V., Deng, X. W., Sakakibara, H., et al. (2011). Genome-wide direct target analysis reveals a role for SHORT-ROOT in root vascular patterning through cytokinin homeostasis. Plant Physiol. 157, 1221–1231. doi: 10.1104/pp.111.183178
Cui, H., Kong, D., Liu, X., Hao, Y. (2014). SCARECROW, SCR-LIKE 23 and SHORT-ROOT control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J. 78, 319–327. doi: 10.1111/tpj.12470
Cui, H., Levesque, M. P., Vernoux, T., Jung, J. W., Paquette, A. J., Gallagher, K. L., et al. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316, 421–425. doi: 10.1126/science.1139531
Dhar, S., Kim, J., Yoon, E. K., Jang, S., Ko, K., Lim, J. (2022). SHORT-ROOT controls cell elongation in the etiolated arabidopsis hypocotyl. Mol. Cells 45, 243–256. doi: 10.14348/molcells.2021.5008
Dhondt, S., Coppens, F., De Winter, F., Swarup, K., Merks, R. M., Inzé, D., et al. (2010). SHORT-ROOT and SCARECROW regulate leaf growth in Arabidopsis by stimulating s-phase progression of the cell cycle. Plant Physiol. 154, 1183–1195. doi: 10.1104/pp.110.158857
Di Laurenzio, L., Wysocka-Diller, J., Malamy, J. E., Pysh, L., Helariutta, Y., Freshour, G., et al. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433. doi: 10.1016/S0092-8674(00)80115-4
Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., et al. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84. doi: 10.1242/dev.119.1.71
Dong, W., Zhu, Y., Chang, H., Wang, C., Yang, J., Shi, J., et al. (2021). An SHR-SCR module specifies legume cortical cell fate to enable nodulation. Nature 589, 586–590. doi: 10.1038/s41586-020-3016-z
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. doi: 10.1016/j.cub.2003.09.035
Fouracre, J. P., Ando, S., Langdale, J. A. (2014). Cracking the kranz enigma with systems biology. J. Exp. Bot. 65, 3327–3339. doi: 10.1093/jxb/eru015
Fukaki, H., Wysocka-Diller, J., Kato, T., Fujisawa, H., Benfey, P. N., Tasaka, M. (1998). Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 14, 425–430. doi: 10.1046/j.1365-313X.1998.00137.x
Fu, J., Zhang, X., Liu, J., Gao, X., Bai, J., Hao, Y., et al. (2021). A mechanism coordinating root elongation, endodermal differentiation, redox homeostasis and stress response. Plant J. 107, 1029–1039. doi: 10.1111/tpj.15361
Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., et al. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057. doi: 10.1038/nature06206
Gallagher, K. L., Benfey, P. N. (2009). Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J. 57, 785–797. doi: 10.1111/j.1365-313X.2008.03735.x
Gallagher, K. L., Paquette, A. J., Nakajima, K., Benfey, P. N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14, 1847–1851. doi: 10.1016/j.cub.2004.09.081
Gardiner, J., Donner, T. J., Scarpella, E. (2011). Simultaneous activation of SHR and ATHB8 expression defines switch to preprocambial cell state in Arabidopsis leaf development. Dev. Dyn. 240, 261–270. doi: 10.1002/dvdy.22516
Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., et al. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567. doi: 10.1016/S0092-8674(00)80865-X
Hernández-Coronado, M., Ortiz-Ramírez, C. (2021). Root patterning: tuning SHORT ROOT function creates diversity in form. Front. Plant Sci. 12, 745861. doi: 10.3389/fpls.2021.745861
Hirano, Y., Nakagawa, M., Suyama, T., Murase, K., Shirakawa, M., Takayama, S., et al. (2017). Structure of the SHR–SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD. Nat. Plants 3, 17010. doi: 10.1038/nplants.2017.10
Kamiya, N., Itoh, J., Morikami, A., Nagato, Y., Matsuoka, M. (2003). The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J. 36, 45–54. doi: 10.1046/j.1365-313X.2003.01856.x
Kim, G., Dhar, S., Lim, J. (2017). The SHORT-ROOT regulatory network in the endodermis development of Arabidopsis roots and shoots. J. Plant Biol. 60, 306–313. doi: 10.1007/s12374-017-0134-8
Kim, J., Jung, J.-H., Reyes, J. L., Kim, Y. S., Kim, S. Y., Chung, K. S., et al. (2005). microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 42, 84–94. doi: 10.1111/j.1365-313X.2005.02354.x
Kim, H., Zhou, J., Kumar, D., Jang, G., Ryu, K. H., Sebastian, J., et al. (2020). SHORTROOT-mediated intercellular signals coordinate phloem development in Arabidopsis roots. Plant Cell 32, 1519–1535. doi: 10.1105/tpc.19.00455
Koizumi, K., Hayashi, T., Gallagher, K. L. (2012a). SCARECROW reinforces SHORT-ROOT signaling and inhibits periclinal cell divisions in the ground tissue by maintaining SHR at high levels in the endodermis. Plant Signal Behav. 7, 1573–1577. doi: 10.4161/psb.22437
Koizumi, K., Hayashi, T., Wu, S., Gallagher, K. L. (2012b). The SHORT-ROOT protein acts as a mobile, dose-dependent signal in patterning the ground tissue. Proc. Natl. Acad. Sci. U.S.A. 109, 13010–13015. doi: 10.1073/pnas.1205579109
Ko, K., Yoon, E. K., Dhar, S., Oh, J., Lim, J. (2022). A SHORTROOT-mediated transcriptional regulatory network for vascular development in the Arabidopsis shoot. J. Plant Biol. 65, 341–355. doi: 10.1007/s12374-022-09355-4
Levesque, M. P., Vernoux, T., Busch, W., Cui, H., Wang, J. Y., Blilou, I., et al. (2006). Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 4, e143. doi: 10.1371/journal.pbio.0040143
Long, Y., Goedhart, J., Schneijderberg, M., Terpstra, I., Shimotohno, A., Bouchet, B. P., et al. (2015a). SCARECROW-LIKE23 and SCARECROW jointly specify endodermal cell fate but distinctly control SHORT-ROOT movement. Plant J. 84, 773–784. doi: 10.1111/tpj.13038
Long, Y., Smet, W., Cruz-Ramírez, A., Castelijns, B., de Jonge, W., Mähönen, A. P., et al. (2015b). Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell 27, 1185–1199. doi: 10.1105/tpc.114.132407
Long, Y., Stahl, Y., Weidtkamp-Peters, S., Postma, M., Zhou, W., Goedhart, J., et al. (2017). In vivo FRET-FLIM reveals cell-type-specific protein interactions in Arabidopsis roots. Nature 548, 97–102. doi: 10.1038/nature23317
Miyashima, S., Koi, S., Hashimoto, T., Nakajima, K. (2011). Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138, 2303–2313. doi: 10.1242/dev.060491
Moreno-Risueno, M. A., Sozzani, R., Yardimci, G. G., Petricka, J. J., Vernoux, T., Blilou, I., et al. (2015). Transcriptional control of tissue formation throughout root development. Science 350, 426–430. doi: 10.1126/science.aad1171
Nakajima, K., Sena, G., Nawy, T., Benfey, P. N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311. doi: 10.1038/35095061
Ortiz-Ramírez, C., Guillotin, B., Xu, X., Rahni, R., Zhang, S., Yan, Z., et al. (2021). Ground tissue circuitry regulates organ complexity in maize and Setaria. Science 374, 1247–1252. doi: 10.1126/science.abj2327
Pardal, R., Heidstra, R. (2021). Root stem cell niche networks: it’s complexed! insights from Arabidopsis. J. Exp. Bot. 72, 6727–6738. doi: 10.1093/jxb/erab272
Petricka, J. J., Winter, C. M., Benfey, P. N. (2012). Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63, 24.1–24.28. doi: 10.1146/annurev-arplant-042811-105501
Pysh, L. D., Wysocka-Diller, J. W., Camilleri, C., Bouchez, D., Benfey, P. N. (1999). The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119. doi: 10.1046/j.1365-313X.1999.00431.x
Qi, L., Zhang, X., Zhai, H., Liu, J., Wu, F., Li, C., et al. (2019). Elongator is required for root stem cell maintenance by regulating SHORTROOT transcription. Plant Physiol. 179, 220–232. doi: 10.1104/pp.18.00534
Sabatini, S., Heidstra, R., Wildwater, M., Scheres, B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17, 354–358. doi: 10.1101/gad.252503
Santuari, L., Sanchez-Perez, G. F., Luijten, M., Rutjens, B., Terpstra, I., Berke, L., et al. (2016). The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell 28, 2937–2951. doi: 10.1105/tpc.16.00656
Scheres, B., Laurenzio, L. D., Willemsen, V., Hauser, M.-T., Janmaat, K., Weisbeek, P., et al. (1995). Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121, 53–62. doi: 10.1242/dev.121.1.53
Schiefelbein, J. W., Benfey, P. N. (1991). The development of plant roots: new approaches to underground problems. Plant Cell 3, 1147–1154. doi: 10.1105/tpc.3.11.1147
Schuler, M. L., Sedelnikova, O. V., Walker, B. J., Westhoff, P. (2018). And langdale, J SHORTROOT-mediated increase in stomatal density has no impact on photosynthetic efficiency. A.Plant Physiol. 176, 757–772. doi: 10.1104/pp.17.01005
Slewinski, T. L. (2013). Using evolution as a guide to engineer kranz-type C4 photosynthesis. Front. Plant Sci. 4, 212. doi: 10.3389/fpls.2013.00212
Slewinski, T. L., Anderson, A. A., Price, S., Withee, J. R., Gallagher, K., Turgeon, R. (2014). Short-root1 plays a role in the development of vascular tissue and kranz anatomy in maize leaves. Mol. Plant 7, 1388–1392. doi: 10.1093/mp/ssu036
Sozzani, R., Cui, H., Moreno-Risueno, M., Busch, W., Van Norman, J., Vernoux, T., et al. (2010). Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466, 128–132. doi: 10.1038/nature09143
Tian, Y., Zhao, N., Wang, M., Zhou, W., Guo, J., Han, C., et al. (2022). Integrated regulation of periclinal cell division by transcriptional module of BZR1-SHR in Arabidopsis roots. New Phytol. 233, 795–808. doi: 10.1111/nph.17824
Wang, C., Li, M., Zhao, Y., Liang, N., Li, H., Li, P., et al. (2022). SHORT-ROOT paralogs mediate feedforward regulation of d-type cyclin to promote nodule formation in soybean. Proc. Natl. Acad. Sci. U.S.A. 119, e2108641119. doi: 10.1073/pnas.2108641119
Welch, D., Hassan, H., Blilou, I., Immink, R., Heidstra, R., Scheres, B. (2007). Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 21, 2196–2204. doi: 10.1101/gad.440307
Wu, S., Lee, C.-M., Hayashi, T., Price, S., Divol, F., Henry, S., et al. (2014). A plausible mechanism, based upon SHORT-ROOT movement, for regulating the number of cortex cell layers in roots. Proc. Natl. Acad. Sci. U.S.A. 111, 16184–16189. doi: 10.1073/pnas.1407371111
Xiao, T. T., Raygoza, A. A., Pérez, J. C., Kirschner, G., Deng, Y., Atkinson, B., et al. (2019). Emergent protective organogenesis in date palms: a morpho-devo-dynamic adaptive strategy during early development. Plant Cell 31, 1751–1766. doi: 10.1105/tpc.19.00008
Xu, X., Crow, M., Rice, B. R., Li, F., Harris, B., Liu, L., et al. (2021). Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev. Cell 56, 557–568. doi: 10.1016/j.devcel.2020.12.015
Yang, B. J., Minne, M., Brunoni, F., Plačková, L., Petřík, I., Sun, Y., et al. (2021). Non-cell autonomous and spatiotemporal signalling from a tissue organizer orchestrates root vascular development. Nat. Plants 7, 1485–1494. doi: 10.1038/s41477-021-01017-6
Yang, L., Zhu, M., Yang, Y., Wang, K., Che, Y., Yang, S., et al. (2022). CDC48B facilitates the intercellular trafficking of SHORT-ROOT during radial patterning in roots. J. Integr. Plant Biol. 64, 843–858. doi: 10.1111/jipb.13231
Yoon, E. K., Dhar, S., Lee, M.-H., Song, J. H., Lee, S. A., Kim, G., et al. (2016). Conservation and diversification of the SHR-SCR-SCL23 regulatory network in the development of the functional endodermis in Arabidopsis shoots. Mol. Plant 9, 1197–1209. doi: 10.1016/j.molp.2016.06.007
Yu, N. I., Lee, S. A., Lee, M. H., Heo, J. O., Chang, K. S., Lim, J. (2010). Characterization of SHORT-ROOT function in the Arabidopsis root vascular system. Mol. Cells 30, 113–119. doi: 10.1007/s10059-010-0095-y
Keywords: Arabidopsis, GRAS transcription factor, shoot development, root development, SCARECROW (SCR), SHORT-ROOT (SHR)
Citation: Yoon EK, Oh J and Lim J (2022) (Don’t) Look Up!: Is short-root just a short‐root plant? Front. Plant Sci. 13:1069996. doi: 10.3389/fpls.2022.1069996
Received: 14 October 2022; Accepted: 31 October 2022;
Published: 17 November 2022.
Edited by:
Yuling Jiao, Peking University, ChinaReviewed by:
Wenkun Zhou, China Agricultural University, ChinaCopyright © 2022 Yoon, Oh and Lim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Lim, amxpbUBrb25rdWsuYWMua3I=
†Present address: Eun Kyung Yoon, Plant Biotechnology Research Center, Ghent University Global Campus, Incheon, South Korea
‡These authors have contributed equally to this work and share first authorship
 Eun Kyung Yoon
Eun Kyung Yoon Jiyeong Oh
Jiyeong Oh Jun Lim
Jun Lim