- 1College of Horticulture, Qingdao Agricultural University, Qingdao, China
- 2Qingdao Key Laboratory of Genetic Improvement and Breeding in Horticulture Plants, Qingdao, China
- 3College of Life Science, Qingdao Agricultural University, Qingdao, China
Δ1-Pyrroline-5-carboxylate synthetase (P5CS) acts as the rate-limiting enzyme in the biosynthesis of proline in plants. Although P5CS plays an essential role in plant responses to environmental stresses, its biological functions remain largely unclear in pear (Pyrus betulifolia). In the present study, 11 putative pear P5CSs (PbP5CSs) were identified by comprehensive bioinformatics analysis and classified into five subfamilies. Segmental and tandem duplications contributed to the expansion and evolution of the PbP5CS gene family. Various cis-acting elements associated with plant development, hormone responses, and/or stress responses were identified in the promoters of PbP5CS genes. To investigate the regulatory roles of PbP5CS genes in response to abiotic and biotic stresses, gene expression patterns in publicly available data were explored. The tissue-specific expressional dynamics of PbP5CS genes indicate potentially important roles in pear growth and development. Their spatiotemporal expression patterns suggest key functions in multiple environmental stress responses. Transcriptome and real-time quantitative PCR analyses revealed that most PbP5CS genes exhibited distinct expression patterns in response to drought, waterlogging, salinity-alkalinity, heat, cold, and infection by Alternaria alternate and Gymnosporangium haraeanum. The results provide insight into the versatile functions of the PbP5CS gene family in stress responses. The findings may assist further exploration of the physiological functions of PbP5CS genes for the development and enhancement of stress tolerance in pear and other fruits.
Introduction
It is well known that abiotic stresses impose severe restrictions on plant growth. These environmental constraints restrict cultivation sites, and diminish agricultural productivity worldwide (Zhu, 2001). To cope with different stresses, plants have evolved multiple mechanisms from physiological, morphological, and molecular perspectives (Tan et al., 2013). Proline plays an essential role in plant growth, development, and stress responses (Anton et al., 2020). It is a compatible solute and a scavenger of reactive oxygen species that provides protection against oxidative damage in plants (Szabados and Savouré, 2010). Free proline is rapidly accumulated in plant cells for adaptation to drought, cold and salinity (Hayat et al., 2012).
Proline biosynthesis involves evolutionarily conserved metabolic pathways in bacteria and higher organisms (Rai and Penna, 2013). Proline biosynthesis uses ornithine or glutamic acid as the substrate, with glutamic acid being preferred under stress conditions (Delauney and Verma, 1993). Δ1-Pyrroline-5-carboxylate synthetase (P5CS) is a key enzyme in the biosynthesis of proline in plants, and it regulates proline content by catalyzing the rate-limiting step in the glutamate pathway (Chen et al., 2013). An increase in P5CS activity can stimulate accumulation of proline to improve osmotic adjustment in plants under environmental stresses (Guan et al., 2014; Anton et al., 2020). P5CS can control proline biosynthesis by transcriptional regulation (Yoshiba et al., 1995). There are two isoforms of P5CS in Arabidopsis thaliana, P5CS1 and P5CS2 (Turchetto-Zolet et al., 2009; Funck et al., 2020). Both P5CS isoforms are mainly present in the cytosol, but may be localized in plastids under stress conditions (Székely et al., 2008). However, other P5CS homologues should exist in the plant genome, some of which may be highly stress-induced and essential for proline accumulation.
Since P5CS plays a crucial role in proline biosynthesis, many attempts have been made to enhance proline accumulation by manipulating the P5CS gene in order to improve plant stress tolerance. Studies have shown that an increase in P5CS enzyme can induce the accumulation of proline in plants, favoring osmotic adjustment under environmental stresses (Peng et al., 1996; Anton et al., 2020). Over-expression of P5CS increased proline content and oxidative stress tolerance in several plants, such as rice (Oryza sativa), A. thaliana, switchgrass (Panicum virgatum), and Stipa purpurea under salt and drought stresses (Kumar et al., 2010; Chen et al., 2013; Guan et al., 2020; Yang D. et al., 2021). Moreover, in oriental hybrid lily (Lilium spp.), LhSorP5CS expression was up-regulated by mannitol and abscisic acid treatments, accompanied by increased proline accumulation (Wang et al., 2017). In addition to abiotic stress, biotic stress also leads to alterations in proline metabolism. For example, P5CS2 expression was up-regulated in response to infection by Pseudomonas syringae in A. thaliana (Fabro et al., 2004).
Pear (Pyrus spp.) fruits have high nutritional value and are popular among consumers. Pears are reproduced primarily through grafting, with Pyrus betulifolia as one of the major rootstocks used in China. Although P5CS genes have been identified in Eugenia uniflora, S. purpurea, rice, and lily (Kumar et al., 2010; Wang et al., 2017; Anton et al., 2020; Yang D. et al., 2021), no comprehensive study of the pear P5CS (PbP5CS) gene family has been reported. Following the release of the pear genome (Dong et al., 2020), we can now systematically analyze the putative functions of PbP5CS genes. In the present study, 11 members of the PbP5CS gene family were identified. The PbP5CS genes were characterized in terms of gene structures and phylogenetic relationships. Their tissue expression profiles and the expression patterns under different stress conditions were analyzed. The results of PbP5CS gene analysis provide insight into the functional role of this gene family in pear.
Materials and methods
Identification of P5CS genes in the pear genome
Amino acid sequences of the model plant A. thaliana P5CS were obtained from the Arabidopsis Information Resource database (https://www.arabidopsis.org/). Using these sequences as queries, the pear genome database was screened by BLASTp (E-value <1e-5). The complete genome assembly of pear (Pyrus betulifolia Bunge.) and the complete proteome sequence file were obtained from the Genome Database for Rosaceae (https://www.rosaceae.org/). Putative P5CS genes were confirmed by BLASTp searches of the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov/). Other information obtained from this database included chromosome number, gene accession numbers, predicted masses of proteins encoded by P5CS genes, and genomic information. The isoelectric point, grand average of hydropathicity (GRAVY), and molecular weight of P5CS proteins were calculated via the ExPasy website (Duvaud et al., 2021). Subcellular locations of pear P5CS proteins were predicted using WoLF PSORT II (Horton et al., 2007).
Construction of P5CS phylogenetic trees
P5CS amino acid sequences from apple (Malus domestica), peach (Prunus persica), poplar (Populus trichocarpa), and A. thaliana were downloaded from Ensembl (https://plants.ensembl.org/index.html). ClustalW v1.83 (Hung et al., 2015) was used for multiple sequence alignments of P5CS proteins. The Muscle module within MEGA 7.0 (Kumar et al., 2016) was used to align the sequences of full-length proteins. Construction of phylogenetic trees based on PbP5CS protein sequences was performed using the neighbor-joining approach with Poisson model, pairwise deletion, and 1000 bootstrap replicates.
Analysis of conserved motifs, conserved domains, and gene structure
Conserved motifs of all P5CS proteins were identified using the online MEME analysis tool (Bailey et al., 2015) with the maximum number of motifs set at 10, and default values for all other parameters. The NCBI CDD database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi/) was used to analyze the conserved domains of 11 PbP5CS protein sequences, and P5CS genes’ domain information data were retained. The information of exon (coding sequence), intron, and untranslated region for 11 PbP5CS genes was determined according to the alignments of their sequences in the P. betulifolia genome database (https://www.rosaceae.org/species/pyrus_betulifolia/genome_v1.0/). To compare conserved motifs, conserved domains, and gene structures of different groups, TBtools software (Chen et al., 2020) was used for clustering, drawing phylogenetic trees, and mapping conserved motifs of PbP5CS. Exon-intron structures were visualized using Gene Structure Display Server 2.0 (Hu et al., 2014).
Chromosomal localization and duplication analysis
The chromosomal localization of each PbP5CS gene was determined based on physical location information obtained from the pear genome database (https://www.rosaceae.org/species/pyrus_betulifolia/genome_v1.0/). Then, a gene localization and distribution map was drawn using TBtools. Pear genome proteins data were self-compared by BLASTp, and fragment replication type and tandem repeat replication type of PbP5CSs were analyzed by MCScanX (Wang et al., 2012). Tandem duplicated genes were identified by analyzing physical locations on specific chromosomes. MCScanX was used to assess syntenic blocks for PbP5CS genes, as well as those between pear and A. thaliana, between pear and apple, between pear and peach, and between pear and poplar
Cis-element analysis of PbP5CS gene promoters
The promoter sequences of 2000 bp regions upstream of each PbP5CS gene-coding region were retrieved from the pear genome database. PlantCARE (Lescot et al., 2002) was then used to annotate elements, and elements related to hormones, stress, growth, and development were selected for location distribution mapping.
Expression profiles of PbP5CSs in various tissues
Expression patterns of PbP5CS genes in various tissues were acquired from RNA sequencing (RNA-seq) data available in the NCBI database (https://www.ncbi.nlm.nih.gov/sra/?term=SRP230672/; SRA accession no.: SRP230672). Fragments per kilobase of exon per million mapped reads (FPKM) values were used to estimate gene expression levels. Multi Experiment Viewer (Saeed et al., 2006) was used to evaluate and graphically characterize means of expression values for each gene in all tissues. A heatmap of PbP5CS genes was generated using the OmicShare Tool (https://www.omicshare.com/tools/).
Plant growth conditions and different stress treatments
Pear seeds after vernalization were sown in nutritive soil (65% fertile garden soil, 25% burning soil, 10% fine sand, 0.4% calcium-magnesium-phosphate fertilizer). All materials were kept in a plant incubator. When seedlings grew to the five-leaf stage, they were transplanted into wet vermiculite in pots (7 cm × 7 cm × 10 cm) and kept in an artificial climate room. Pear seedlings received Hoagland solution every 3 days and were grown at 23 ± 2°C with a light intensity of 800 µmol m-2·s-1. Two-month-old seedlings were used to measure mRNA expression levels of PbP5CSs under biotic and abiotic stress conditions. Drought stress of potted pear plants was inflicted by withholding water for 20 days (Yang S. et al., 2021); waterlogging stress was inflicted by submerging plants in water (Yu et al., 2019); salinity-alkalinity stress was performed at a ratio of 1:1.4 NaCl and NaHCO3 (Zhang et al., 2020); cold stress was simulated at 4°C (Xi et al., 2011); heat stress was simulated at 40°C (Liu et al., 2013). Pear rust was applied by infecting leaves with Gymnosporangium haraeanum Syd. (Li et al., 2006). Pear leaves were taken at 0, 1, 3, and 6 days after abiotic stresses, and at 0, 6, 12, and 24 hours after biotic stress. The samples (with three independent biological replications) were immediately frozen in liquid nitrogen and stored at -80°C until analysis. To explore the gene expression profiles of PbP5CSs in response to salt, cold, drought, and Alternaria alternate infection, pear RNA-seq datasets were retrieved from published supplemental datasets (SRA accession nos.: SRP077703, SRP287704, SRP148620, and SRP276846).
Measurements of P5CS enzyme activities
P5CS enzyme activities were measured using a commercial kit (Suzhou Geruisi Biotechnology, Suzhou, China) following the manufacturer’s instructions. Each experiment was independently repeated three times.
Real-time quantitative PCR analysis
Total RNA extraction from leaf samples was performed using the method of Ma et al. (2022). First-strand cDNA was prepared using PrimeScript RTase (TaKaRa Biotechnology, Dalian, China). Primers for qPCR were designed using Primer Premier 5.0 (Premier Biosoft International, Silicon Valley, CA, USA). Primer sequences are detailed in Supplementary Table S1. A LightCycler R 480 SYBR Green Master (Roche, Mannheim, Germany) was used for qPCR assays with a LightCycler R 480 II system (Roche, Rotkreuz, Switzerland). Relative expression levels of the target genes were calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001) and normalized against the Actin gene (GenBank: AB190176).
Results
Identification of P5CS genes in pear
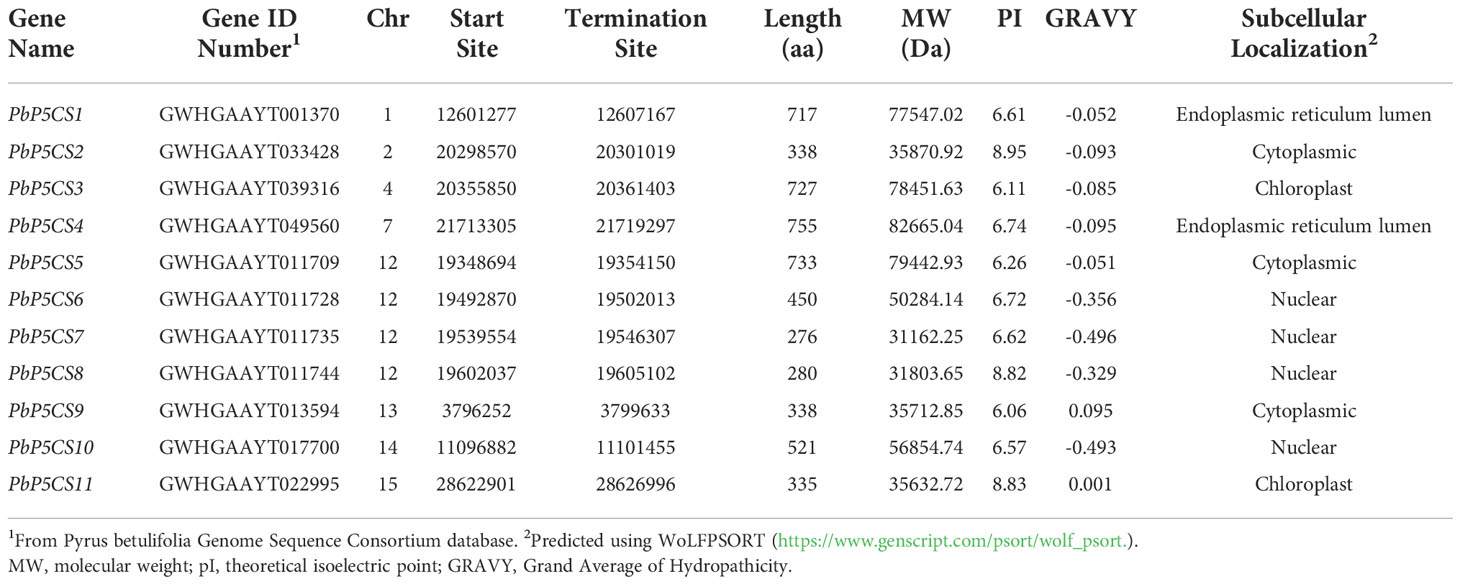
Based on the conserved domains of protein sequences, 11 P5CS protein sequences were screened and named PbP5CS1−PbP5CS11 according to their chromosomal sequences and positions. Detailed physical and chemical characterizations of PbP5CS proteins are listed in Table 1. The 11 PbP5CS proteins have different numbers of amino acids; PbP5CS7 is the shortest (276 amino acids), while PbP5CS4 is the longest (755 amino acids). The molecular weight of PbP5CSs ranged from 31.16 kDa (PbP5CS7) to 82.67 kDa (PbP5CS4). The isoelectric point values of PbP5CS proteins ranged from 6.06 (PbP5CS9) to 8.95 (PbP5CS2). Except for PbP5CS9 and PbP5CS11, the GRAVY values of other PbP5CSs were less than zero. The predicted subcellular localizations were cytoplasm for PbP5CS2, PbP5CS5, and PbP5CS9, endoplasmic reticulum for PbP5CS1 and PbP5CS4, chloroplast for PbP5CS3 and PbP5CS11, and nucleus for the other PbP5CSs.
Phylogenetic relationships of PbP5CS members
To explore the evolutionary relationships of PbP5CS members, a phylogenetic tree was built using 50 conserved domain sequences of P5CS proteins from pear (11), apple (8), peach (14), poplar (13), and A. thaliana (4; Figure 1). In the tree, PbP5CS, MdP5CS, PpP5CS, PtP5CS, and AtP5CS were classified into five groups (Groups I, II, III, IV, and V). Notably, P5CS genes of woody plants (pear, apple, peach, and poplar) clustered together. Most of the pear P5CSs also clustered together with proteins from A. thaliana, consistent with the closer relationship of pear to eudicots. The distribution of pear P5CSs was uneven in these groups. Group I was the largest with 24 members, including nearly half of all pear P5CSs (PbP5CS1, PbP5CS3, PbP5CS4, PbP5CS5, and PbP5CS8). PbP5CS9 and PbP5CS11 were in both Groups II and IV, PbP5CS6 and PbP5CS7 were in Group III, and PbP5CS2 and PbP5CS10 were in Group V.
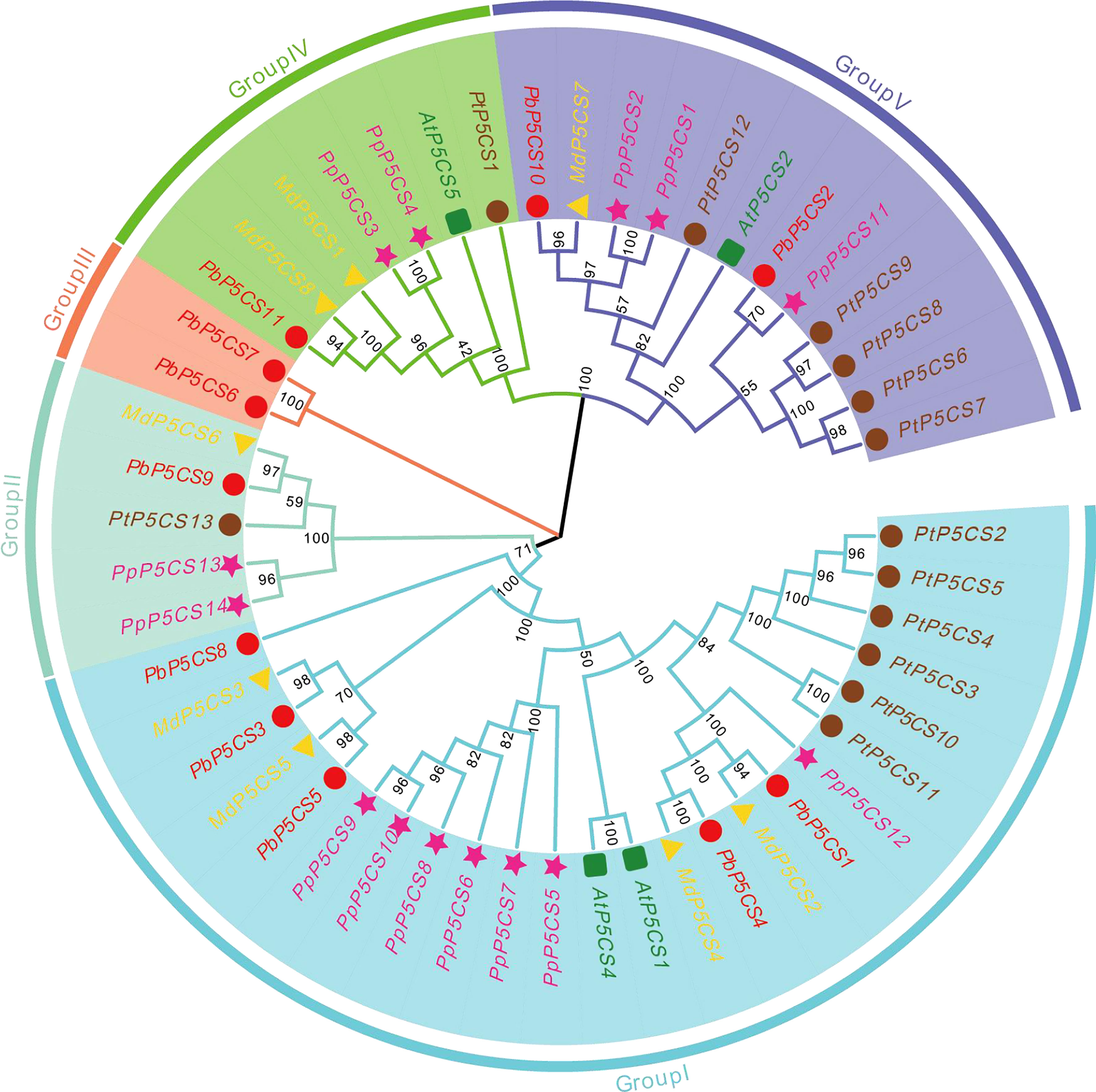
Figure 1 Phylogenetic tree of P5CS proteins constructed using the neighbor-joining method with P5CS domains from pear Pyrus betulifolia (red circles), Malus domestica (yellow triangles), Populus trichocarpa (pink pentagons), Prunus persica (brown circles), and Arabidopsis thaliana (green squares). Members are divided into Groups I, II, III, IV, and V.
Conserved protein motifs and exon-intron structures of PbP5CS genes
A total of 10 conserved motifs were predicted in PbP5CSs (Figure 2B; Supplementary Table S2), ranging from 21 to 100 amino acids in length. Interestingly, we observed that Motifs 1, 2, 3, 6, 7, 9, and 10 were present only in Group I members, which might contribute to the functional divergence of P5CS genes. Motif 5 was found not only in all members of Group I, but also in PbP5CS6 and PbP5CS7 in Group III, this suggests that PbP5CS6 and PbP5CS7 may have evolved from Group I. Group II only comprised Motif 4, Group IV only contained Motif 8. As Motifs 4 and 8 were found in PbP5CS2 and PbP5CS10 in Group V, PbP5CS2 and PbP5CS10 may have evolved from Groups II and IV (Figures 2A, B). We also found that four genes in Group I (PbP5CS1, PbP5CS3, PbP5CS4, and PbP5CS5) had more than 20 exons, while all others carried between six and 11 exons (Figure 2C).
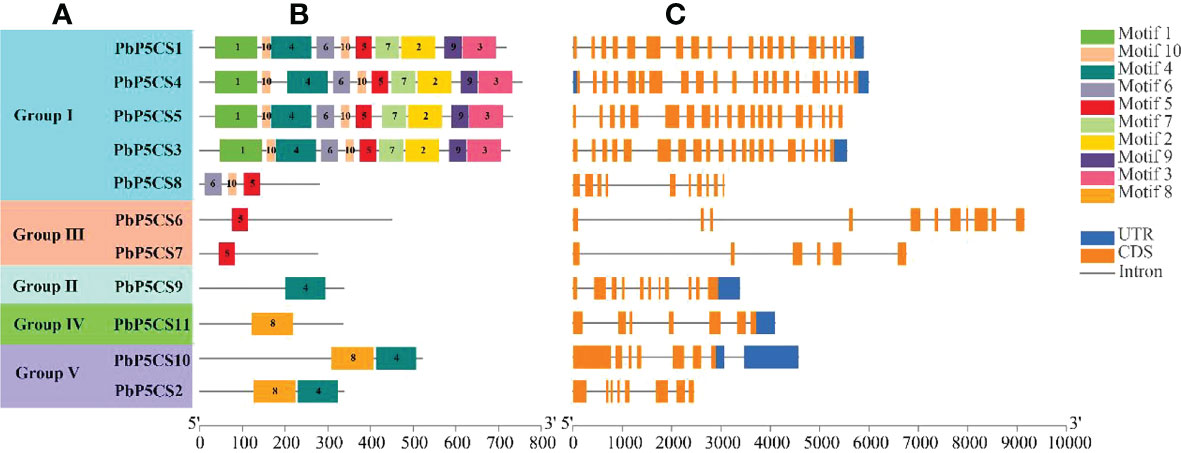
Figure 2 Schematic representation of protein and gene structures of pear P5CS (PbP5Cs) genes. (A) Phylogenetic relationships. (B) Motif composition. (C) Gene structure. Motifs 1−8 identified using the MEME search tool are marked on protein sequences in each clade (I−V). The length and order of each motif corresponds to the actual length and position in the protein sequences. Coding sequence and untranslated regions are represented by filled orange and dark blue boxes, respectively.
Chromosomal locations and homologous genotypes of PbP5CS genes
According to their annotated genomic locations, we found that the 11 PbP5CSs were widely distributed among the pear chromosomes. Chromosome 12 contained four PbP5CS genes, whereas Chromosomes 1, 2, 4, 7, 13, 14, and 15 had only one gene (Figure 3). PbP5CS1, PbP5CS3, PbP5CS4, and PbP5CS5 (six pairs) were segmental (Figure 4A). In order to further explore the homologous gene relationships of PbP5CSs, we compared the physical locations of P5CS genes among the genomes of pear, apple, peach, poplar, and A. thaliana (Figure 4B). P5CS genes showed intimate collinear relationships in these five species. P5CSs in pear and apple showed the closest collinear relationship, with seven P5CS genes in apple sharing a close evolutionary relationship with PbP5CS genes. Moreover, six P5CS genes in each of peach and poplar, as well as two P5CS genes in A. thaliana, shared close evolutionary relationships with PbP5CS genes (Supplementary Table S3).
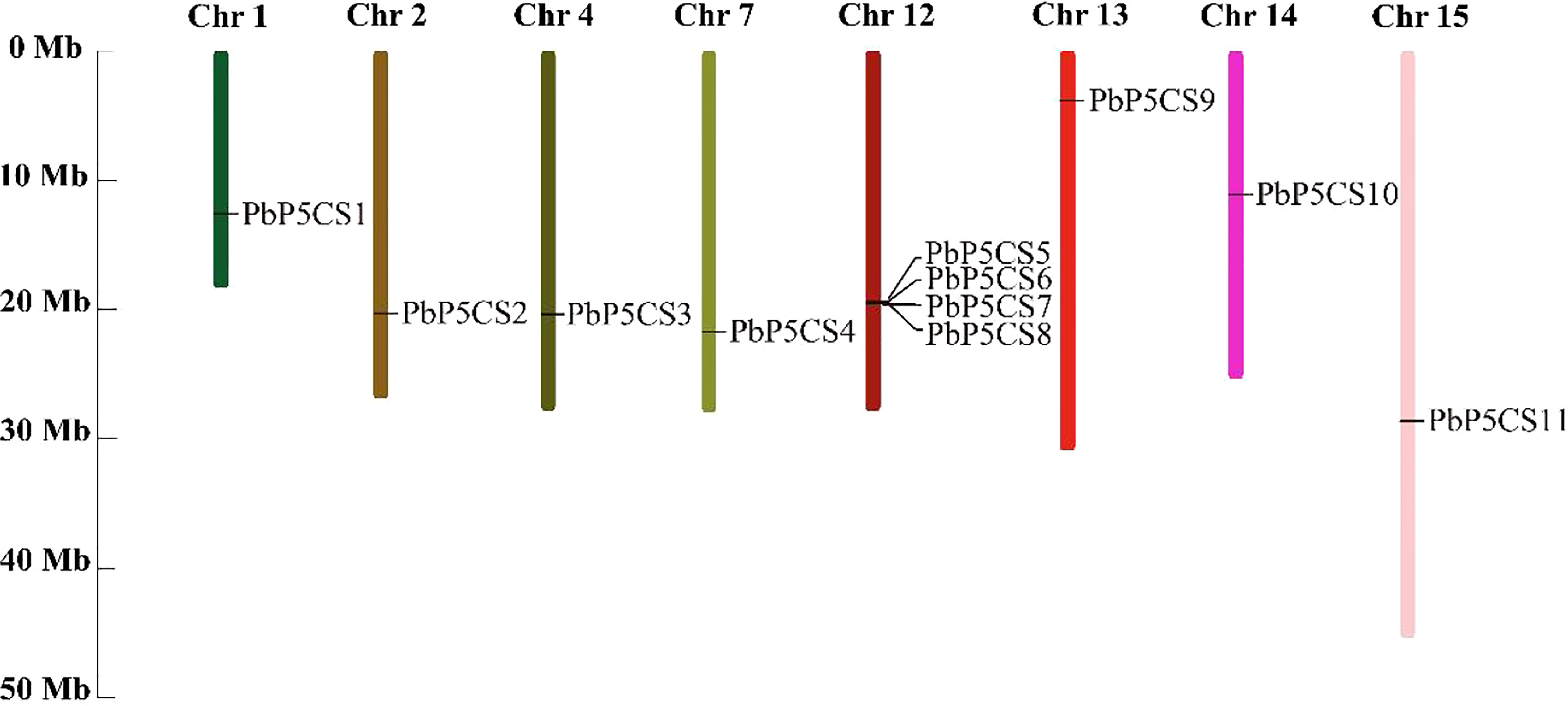
Figure 3 Chromosomal distribution of PbP5CS genes. Chromosomal mapping was based on the physical position (Mb) in 17 pear chromosomes. The scale on the left is in megabases (Mb). Chromosome numbers are indicated at the top of each bar.
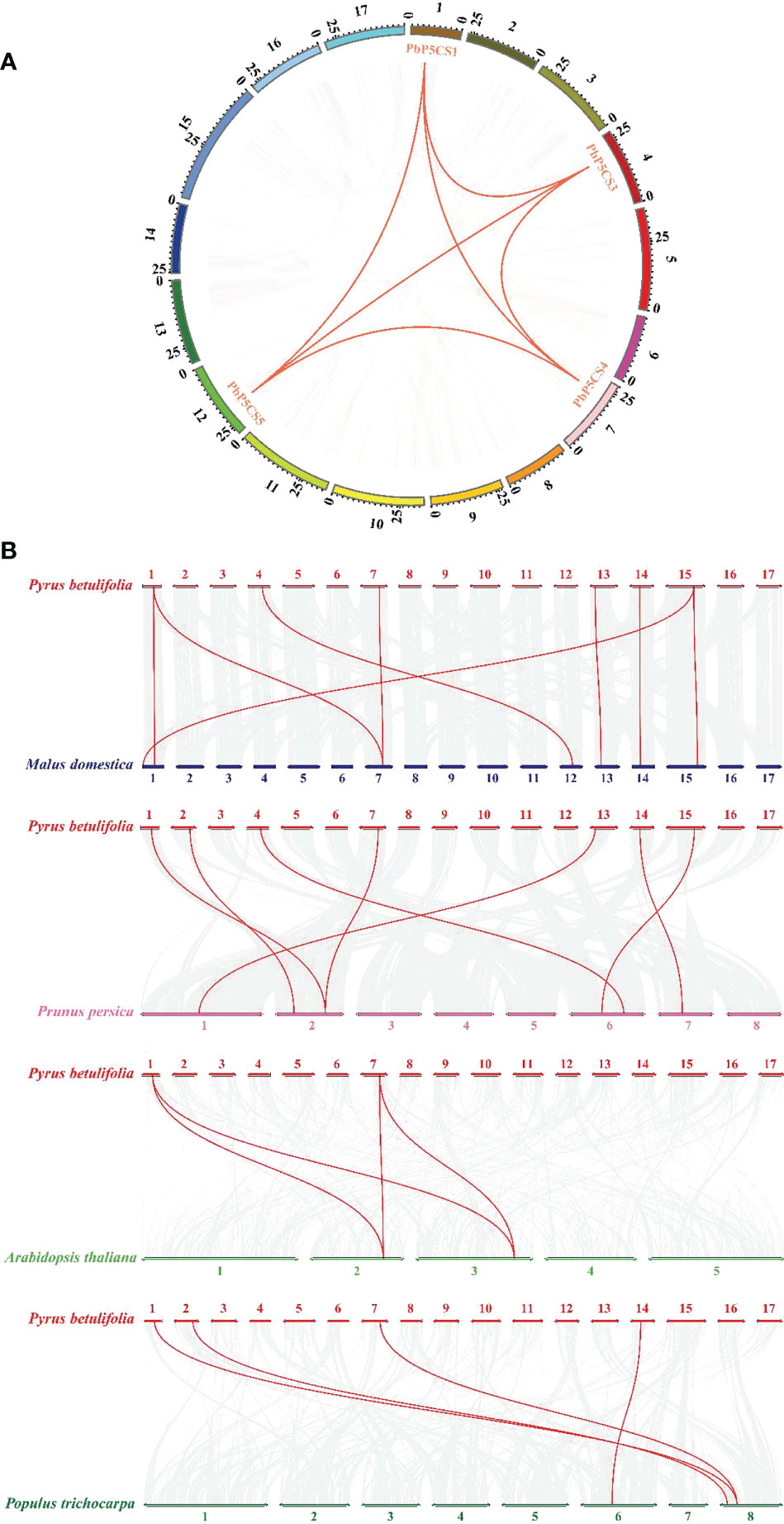
Figure 4 Synteny analysis of P5CS genes. (A) Synteny analysis of PbP5CS genes in pear. Gray lines indicate collinear blocks in the whole Pyrus betulifolia genome, and red lines indicate duplicated PbP5CS gene pairs. (B) Synteny analysis of P5CS genes in the P. betulifolia, Populus trichocarpa, Arabidopsis thaliana, Malus domestica, and Prunus persica genomes. Red lines highlight syntenic P5CS gene pairs.
Promoter cis-regulatory elements of PbP5CS genes
To better understand the gene functions and transcriptional regulation of PbP5CSs, we analyzed cis-elements in the promoter regions of PbP5CSs (Figure 5). The conventional promoter element CAAT-box was found in all PbP5CS promoters. Various cis-elements related to plant growth, development, and responses to stresses and phytohormones were also identified (Supplementary Table S4). Additionally, the conventional promoter element GC-motif was present in PbP5CS2, the seed-specific regulation element (RY-element) was found in the promoter of PbP5CS7, and the endosperm expression regulation element (GCN4_motif) was observed in the promoters of PbP5CS1 and PbP5CS8. The zein metabolism regulation element (O2-site) was identified in the promoters of seven PbP5CS genes.
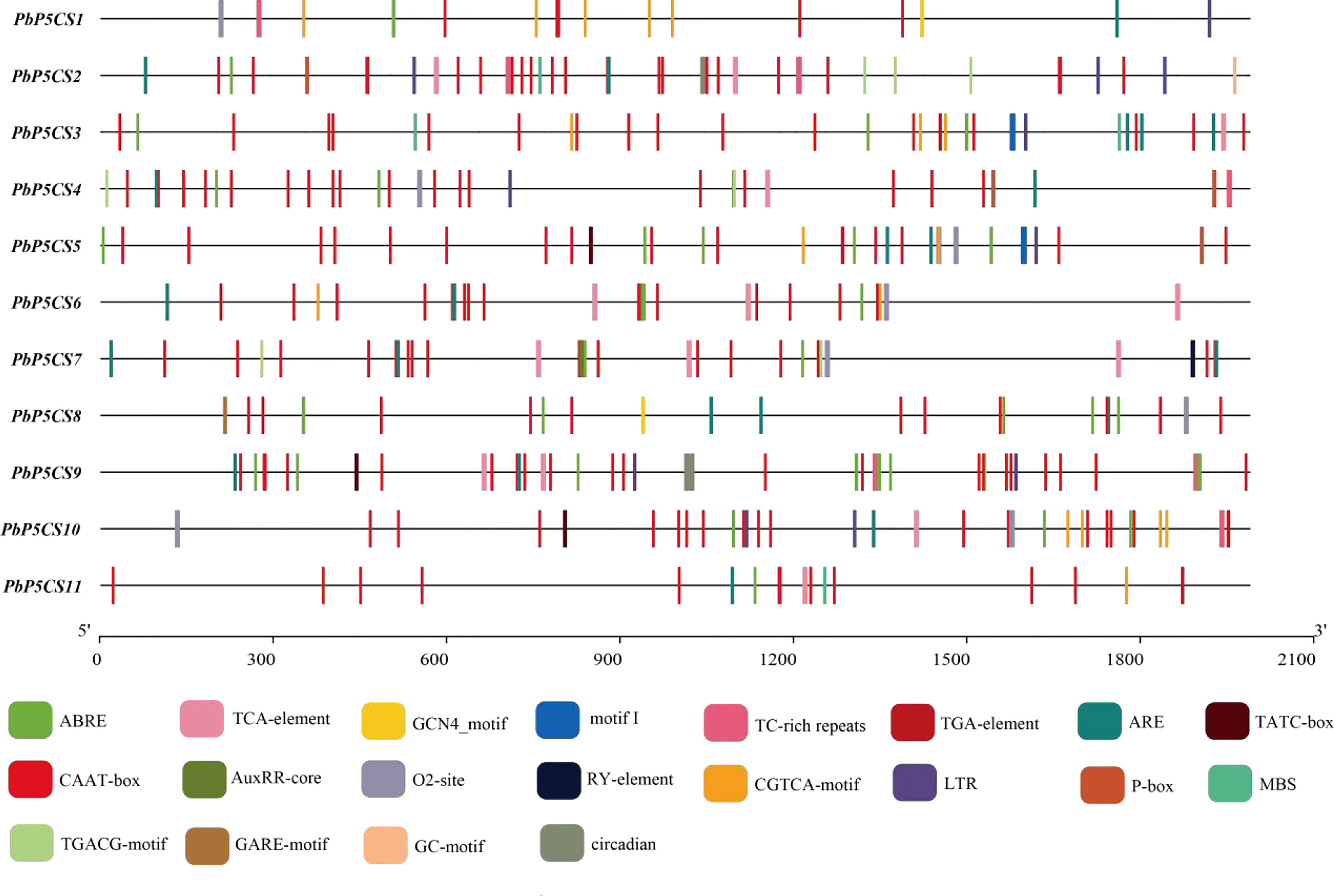
Figure 5 Promoter cis-regulatory element analysis of PbP5CS genes. Different colors indicate various promoter elements in PbP5CS genes.
Among the cis-elements that respond to plant hormones, the abscisic acid responsive element (ABRE) was present in the promoter of all PbP5CS genes, methyl jasmonate responsive elements (TGACG-motif and CGTCA-motif) were observed in the promoters of all PbP5CS genes excluding PbP5CS8, and salicylic acid-responsive element (TCA-element) was found in eight PbP5CS genes. Gibberellin responsive elements (GARE-motif, P-box, and TATC-box) and auxin responsive elements (AUXRR-core and TGA) were identified in six PbP5CS genes. We also found the stress-related cis-acting element (ARE) involved in anaerobic induction in all PbP5CS genes. Furthermore, defense and stress response element (TC-rich repeat), drought-inducible response element (MBS), low temperature responsiveness (LTR), and circadian features were identified in the promoters of PbP5CS genes (Figure 5; Supplementary Table S4).
Tissue-specific expression patterns of PbP5CS genes
To investigate the putative roles of the PbP5CS genes in pear development, we analyzed organic-specific expression patterns of PbP5CSs. Expression patterns of PbP5CS genes in five different tissues (petal, stigma, leaf, ovary, and shoot) were analyzed using publicly available gene expression data (SRP230672). Some PbP5CSs were expressed at considerably high levels in specific tissues (Figure 6). For example, three PbP5CSs (PbP5CS1, PbP5CS5, and PbP5CS8) displayed higher expression levels in the petal than other tissues, implying that they play important roles in pear petal growth and development.
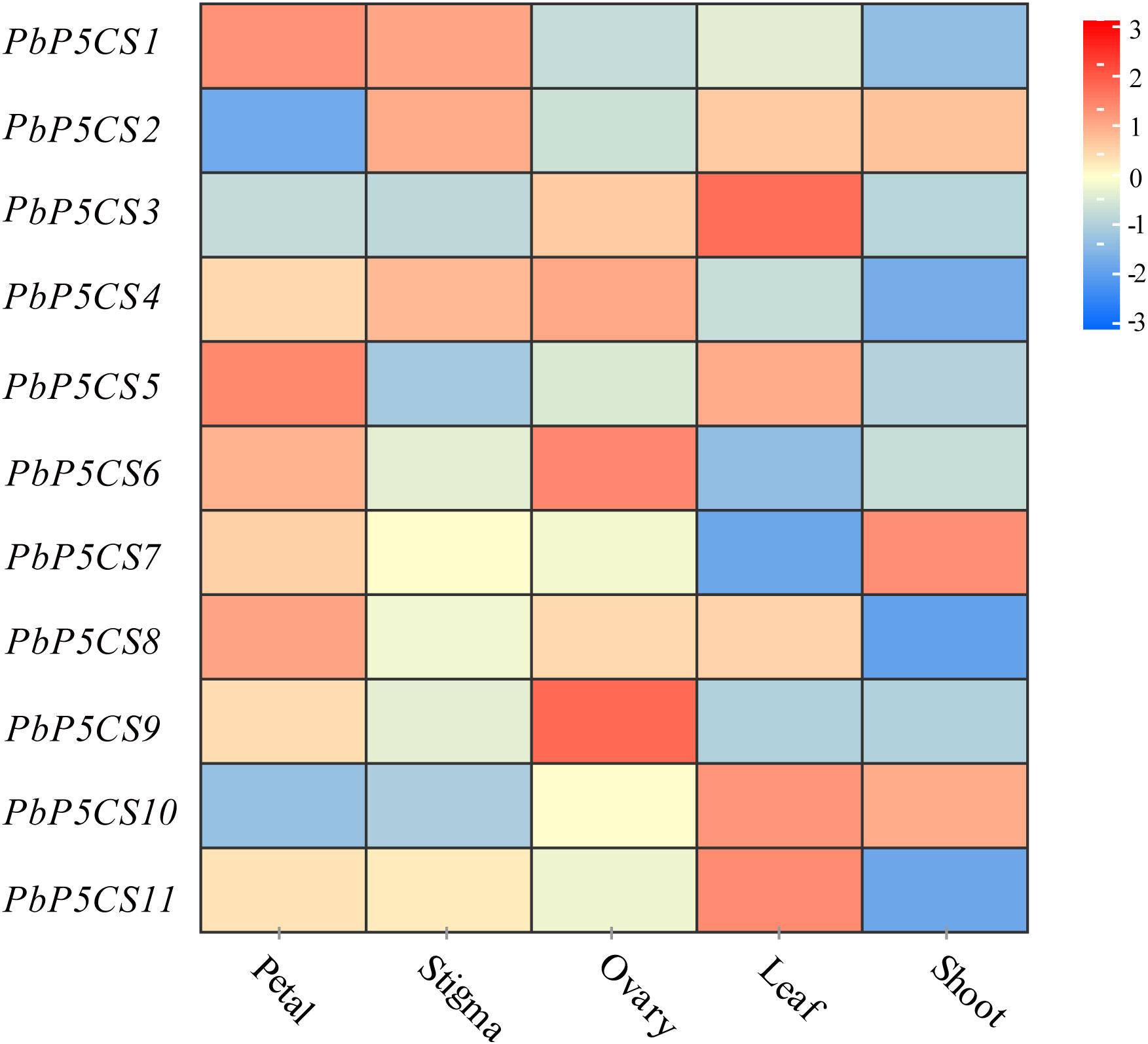
Figure 6 Expression patterns of PbP5CS genes in different tissues of pear. Microarray analysis results of PbP5CS genes in Pyrus bretschneideri ‘Yali’ were downloaded from NCBI GEO DataSets (SRP230672). Red and blue boxes indicate high and low expression levels, respectively, for each gene.
PbP5CS1, PbP5CS2, and PbP5CS4 exhibited high levels of expression in the stigma, which suggests that they may play roles in stigma growth and development. PbP5CS4, PbP5CS6, and PbP5CS9 were preferentially expressed in the ovary, which suggests that they may take part in in ovary growth and development. PbP5CS3, PbP5CS5, PbP5CS10, and PbP5CS11 showed relatively high expression levels in the leaf, which means that they could participate in leaf development. PbP5CS7 showed higher expression in the shoot than other tissues, which reflects its possible role in shoot growth (Supplementary Table S5). These results suggest that PbP5CS genes have different expression patterns, and may play diverse roles in pear during growth and development of different tissues.
Expression patterns of PbP5CS genes under abiotic and biotic stresses
To explore whether the P5CS enzyme plays an essential role under biotic and abiotic stresses, we measured the enzyme activities of P5CS under drought, waterlogging, salinity-alkalinity, cold, heat, and G. haraeanum infection (Supplementary Figure S1). The enzyme activities of P5CS gradually increased under the different stresses. We then investigated expression patterns of PbP5CS genes in response to different stresses. With regard to abiotic stresses, RNA-seq datasets for pear subjected to drought (SRP148620), salt (SRP077703), and cold (SRP287704) were explored. In general, expression levels of PbP5CS1, PbP5CS3, PbP5CS4, PbP5CS5, PbP5CS6, and PbP5CS11 were up-regulated by all abiotic stress treatments, indicating potential roles of these PbP5CSs in abiotic stress responses (Figure 7; Supplementary Table S6). Under drought treatment, PbP5CS1, PbP5CS2, PbP5CS3, PbP5CS4, PbP5CS5, PbP5CS6, PbP5CS7, PbP5CS9, and PbP5CS11 were up-regulated, but PbP5CS8 and PbP5CS10 were repressed in response to short-term drought stress (Figure 7A). Furthermore, PbP5CS2, PbP5CS3, PbP5CS5, and PbP5CS8 were significantly up-regulated at 72 hours of NaCl treatment (Figure 7B). PbP5CS6 and PbP5CS9 were significantly induced at 50 days of cold treatment (Figure 7C). These results indicate that the responsive PbP5CSs may be involved in plant defense mechanisms under both short- and long-term abiotic stress.
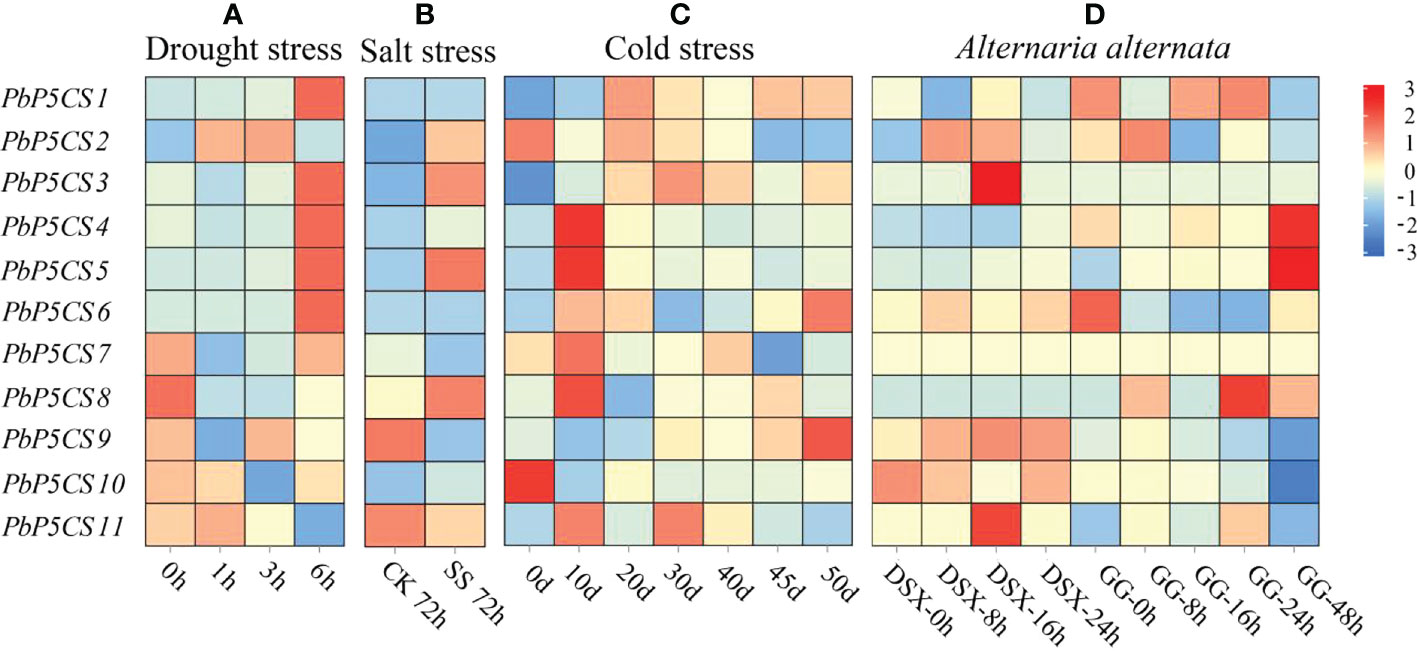
Figure 7 Expression patterns of PbP5CS genes in response to abiotic and biotic stresses. (A) Gene expression patterns in Pyrus bretschneideri at 1, 3, and 6 hours of drought treatment (GEO series SRP148620). (B) Gene expression patterns in Pyrus betulifolia at 72 hours of salt stress treatment (GEO series SRP077703). CK, control; SS, salt stress treatment. (C) Gene expression patterns in Pyrus bretschneideri ‘Suli’ at 0, 10, 20, 30, 40, 45, and 50 days of cold treatment (GEO series SRP287704). (D) Gene expression patterns in Pyrus pyrifolia ‘Deshengxiang’ (DSX) and Pyrus pyrifolia ‘Guiguan’ (GG) at 0, 8, 12,16, 24, and 48 hours post-infection by Alternaria alternate (GEO series SRP276846). Data are expressed as fragments per kilobase of exon per million mapped reads. Blue and red blocks indicate decreased and increased transcription levels, respectively.
To explore the potential roles of PbP5CS genes in responses to biotic stresses, we investigated RNA-seq datasets from the infection experiment of pear with A. alternate (SRP276846). PbP5CS1 and PbP5CS2 were significantly up-regulated at 24 and 8 hours post-infection, respectively. PbP5CS3, PbP5CS9, and PbP5CS11 showed significant up-regulation at 16 hours post-infection. PbP5CS4, PbP5CS5, and PbP5CS8 were markedly induced at 48 hours post-infection. However, PbP5CS6 and PbP5CS10 were distinctively repressed following A. alternate infection, whereas PbP5CS7 did not respond to infection (Figure 7D).
To validate previous RNA-seq data and reveal more details of PbP5CSs in stress responses, we investigated the transcription levels of PbP5CS genes under different environmental stresses, including drought, waterlogging, salinity-alkalinity, cold, heat, and G. haraeanum infection. Under drought stress, nine (PbP5CS1, PbP5CS2, PbP5CS3, PbP5CS4, PbP5CS5, PbP5CS6, PbP5CS7, PbP5CS9, and PbP5CS11) and two (PbP5CS8 and PbP5CS10) PbP5CSs were up-regulated and down-regulated, respectively (Figure 8A). Moreover, seven (PbP5CS2, PbP5CS3, PbP5CS4, PbP5CS5, PbP5CS7, PbP5CS9, and PbP5CS11) and four (PbP5CS3, PbP5CS8, PbP5CS9, and PbP5CS10) PbP5CS genes were respectively up-regulated and down-regulated under waterlogging treatment (Figure 8B). Similarly, eight PbP5CS genes (PbP5CS1, PbP5CS2, PbP5CS3, PbP5CS4, PbP5CS7, PbP5CS9, PbP5CS10, and PbP5CS11) showed increased expression levels and three PbP5CSs (PbP5CS5, PbP5CS6, and PbP5CS8) were down-regulated by salinity-alkalinity treatment (Figure 8C). Additionally, the transcription levels of seven PbP5CS genes (PbP5CS1, PbP5CS3, PbP5CS4, PbP5CS5, PbP5CS9, PbP5CS10, and PbP5CS11) were increased under cold stress (Figure 8D). Under heat stress, seven PbP5CS genes (PbP5CS1, PbP5CS2, PbP5CS4, PbP5CS5, PbP5CS6, PbP5CS7, and PbP5CS11) were up-regulated, and the remaining four PbP5CSs (PbP5CS3, PbP5CS8, PbP5CS9, and PbP5CS10) were down-regulated (Figure 8E).
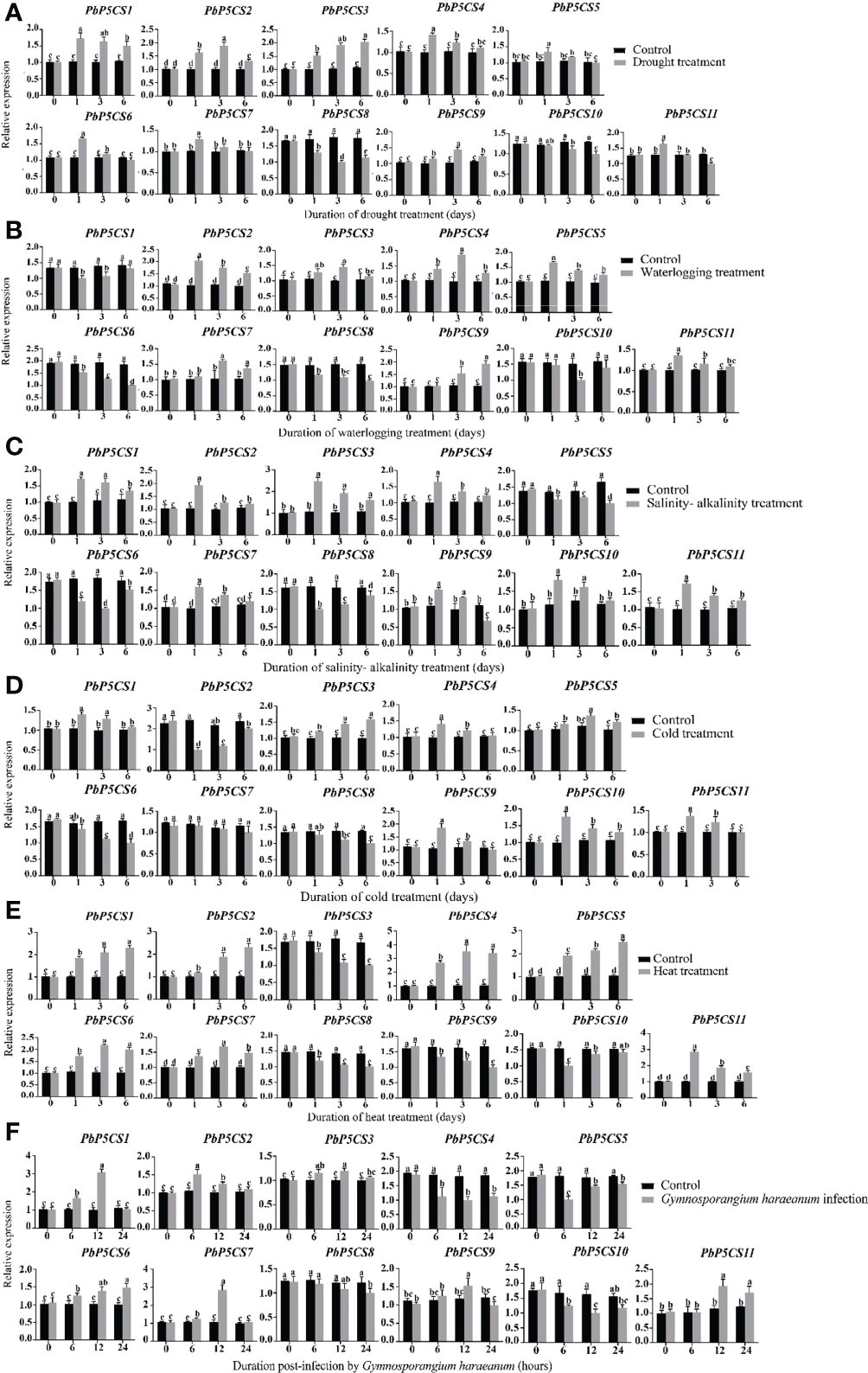
Figure 8 Real-time quantitative PCR (qPCR) analysis of pear P5CS genes in response to (A) drought, (B) waterlogging, (C) salinity-alkalinity, (D) cold, (E) heat, and (F) Gymnosporangium haraeanum infection stresses. Data were normalized against expression levels of the Actin gene. Mean values were calculated from three independent replicates. Vertical bars indicate standard error of the mean. Different lowercase letters indicate significant differences between treatments according to Fisher’s least significant difference (P < 0.05).
Pear rust caused by G. haraeanum is one of the main diseases affecting pear production. To understand the potential functions of PbP5CS genes in response to biotic stress, transcript levels of 11 PbP5CSs were measured by qPCR in pear subjected to G. haraeanum infection. Seven (PbP5CS1, PbP5CS2, PbP5CS3, PbP5CS6, PbP5CS7, PbP5CS9, and PbP5CS11) and four (PbP5CS4, PbP5CS5, PbP5CS8, and PbP5CS10) PbP5CS genes were up-regulated and down-regulated, respectively (Figure 8F). Interestingly, PbP5CS4 and PbP5CS11 were also strongly induced by drought, waterlogging, salinity-alkalinity, cold, and heat, suggesting that these two genes might be candidate genes for mitigating abiotic stresses. In particular, PbP5CS11 was induced dramatically by various abiotic and biotic stresses (Figure 8).
Discussion
P5CS genes play key roles in biochemical and physiological processes in response to various stressors in plants (Chen et al., 2013; Yang D. et al., 2021). Therefore, studying the functions of P5CS gene families in inhospitable environments can provide valuable information on the mechanisms underlying plant adaptation. In the current work, we performed a genome-wide analysis of P5CSs in pear by considering their gene structures, phylogenetic relationships, cis-acting elements, linkage group organization, and duplication events. We also explored their possible roles in plant tissues and responses to stress. The genome-wide results for PbP5CS genes not only provides novel insights into their physiological functions, but also a foundation for functional research on these genes during pear growth and development.
The expansion of gene family members is driven by gene duplication events, either segmental or tandem duplications, during plant evolution (Guo et al., 2014; Wang et al., 2022). Most plants have two P5CS isoforms, as demonstrated for Phaseolus vulgaris, Lotus japonicus, and Brassica napus (Xue et al., 2009; Chen et al., 2013; Signorelli and Monza, 2017). In the present work, 11 P5CS genes were identified in the pear genome (Table 1; Figure 1), more than apple (8) and A. thaliana (4), but fewer than poplar (13) and peach (14). The variation might be due to gene duplication differences, considered a fundamental driving force in the evolution of genomes (Kong et al., 2007). Gene duplications can provide raw materials for new genes, leading to the emergence of new functions.
Our phylogenetic analysis of P5CS proteins among pear, apple, peach, poplar, and A. thaliana showed that the proteins formed species-specific clusters (Figure 1). This result indicates that P5CS proteins have been highly conserved during evolution. Furthermore, there were six segmental duplication pairs (PbP5CS1/PbP5CS3, PbP5CS1/PbP5CS4, PbP5CS1/PbP5CS5, PbP5CS3/PbP5CS4, PbP5CS3/PbP5CS5, and PbP5CS4/PbP5CS5) in pear (Figure 4A). These results demonstrate that segmental duplication plays a vital role in driving the expansion of the pear PbP5CS gene family. The synteny analysis of P5CS genes in pear, apple, peach, poplar, and A. thaliana showed that PbP5CS genes shared higher homology with P5CS genes in apple than in peach, poplar, and A. thaliana (Figure 4B; Supplementary Table S3). However, PbP5CS1 and PbP5CS4 were collinear with the P5CS genes of the other four species (Supplementary Table S3), indicating that PbP5CS1 and PbP5CS4 in different plants may have evolved from a common ancestor.
Variation in introns and exons plays is essential for the evolution of different genes (Mustafin and Khusnutdinova, 2015; Rogers, 1990). Introns play major roles in gene evolution (Rose, 2008). Our analysis of P5CS gene structure revealed that all PbP5CS genes contained different numbers of exons and introns (Figure 2C), indicating functional diversity among PbP5CS genes. In addition, cis-regulatory element analysis revealed the presence of a series of abiotic/biotic stress responsive cis-acting elements, such as ARE, ABRE, MBS, LTR, and AuxRR-core, in the promoter regions of PbP5CS genes (Figure 5; Supplementary Table S4). This implies that the PbP5CS genes perform potential functions in response to abiotic and biotic stresses. The different cis-regulatory elements in P5CS genes presumably allows them to exert diverse effects on plant growth and development, including under different stress conditions.
Proline has been shown to be critically involved in a number of plant developmental processes, such as pollen fertility, root elongation, embryo development, and floral transition (Trovato et al., 2018). P5CS1 predominantly contributes to stress-induced proline accumulation, and P5CS2 is mainly involved in plant growth and development (Funck et al., 2020). For example, an increase in proline content was accompanied by markedly high expression of BnP5CS in flowers of B. napus, suggesting possible contribution of proline to flower development (Xue et al., 2009). In pear, eight PbP5CS genes (PbP5CS1, PbP5CS4, PbP5CS5, PbP5CS6, PbP5CS7, PbP5CS8, PbP5CS9, and PbP5CS11) displayed high expression levels in the petal (Figure 6), which further indicates that proline is a key factor in floral development (Trovato et al., 2018). Furthermore, PbP5CS1, PbP5CS2, and PbP5CS4 were highly expressed in the stigma, PbP5CS4, PbP5CS6, and PbP5CS9 showed relatively high expression levels in the ovary, and PbP5CS3, PbP5CS10, and PbP5CS11 were predominantly expressed in the leaf (Figure 6). All these PbP5CS members likely function in the particular pear tissues.
Proline accumulation is mainly regulated by the P5CS enzyme in plant cells under stress conditions (Yang D. et al., 2021). Increasing evidence demonstrates that P5CS genes participate in plant development, biological regulation, and stress responses, and they play an essential role in plant resistance to different abiotic stresses (Zhang et al., 1995; Parida et al., 2008; Bandurska et al., 2017). P5CS is a key enzyme enhancing oxidative stress tolerance in plants under salt and drought stresses (Kumar et al., 2010; Rai and Penna, 2013). P5CS activity and expression levels were up-regulated in barley (Hordeum vulgare), cotton (Gossypium hirsutum), and S. purpurea under drought stress conditions (Parida et al., 2008; Bandurska et al., 2017; Yang D. et al., 2021). In pear, P5CS enzyme activity was induced under six different stresses (Supplementary Figure S1). The expression of PbP5CS1, PbP5CS2, PbP5CS3, PbP5CS4, PbP5CS5, PbP5CS6, and PbP5CS11 was significantly induced in response to drought stress according to RNA-seq and qPCR data (Figures 7, 8), indicating roles for these genes in drought stress tolerance by regulating P5CS enzyme activities. Moreover, most pear P5CSs (PbP5CS1, PbP5CS2, PbP5CS3, PbP5CS4, PbP5CS5, PbP5CS6, PbP5CS8, PbP5CS10, and PbP5CS11) clustered together with proteins from A. thaliana (Figure 1). The AtP5CS gene was induced by high salt treatment in A. thaliana (Yoshiba et al., 1995). Similarly, PbP5CS2, PbP5CS4, and PbP5CS10 expression was induced in response to salt and salinity-alkalinity stresses, according to RNA-seq and qPCR data (Figures 7, 8C), indicating putative functions for these genes in pear salt or salinity-alkalinity stress tolerance. Furthermore, AmP5CS was rapidly initiated by heat stress in grey mangrove (Avicennia marina) (Liu and Wang, 2020). PvP5CS was prominently up-regulated in common bean (Phaseolus vulgaris), which enhanced tolerance under cold stress (Atienza et al., 2004; Chen et al., 2009; Chen et al., 2013). CpP5CS can be induced by both heat and cold stress in papaya (Carica papaya) (Zhu et al., 2012). In the current work, P5CS enzyme activity was gradually increased, and the expression of PbP5CS1, PbP5CS4, PbP5CS5, and PbP5CS11 was significantly induced by heat and cold. Additionally, the enzyme activity of P5CS was induced, and PbP5CS2, PbP5CS3, PbP5CS4, PbP5CS5, PbP5CS7, PbP5CS9, and PbP5CS11 were up-regulated by waterlogging stress (Supplementary Figure S1; Figure 8). The collective results indicate that these PbP5CS genes also regulate P5CS enzyme activities to mitigate various abiotic stresses.
Regarding biotic stresses, AtP5CS2 participates in the A. thaliana hypersensitive response induced by avirulent Pseudomonas spp. (Fabro et al., 2004). Herein, PbP5CS1, PbP5CS2, PbP5CS3, PbP5CS6, PbP5CS7, PbP5CS9, and PbP5CS11 were up-regulated in response to G. haraeanum infection (Figure 8). Accordingly, these PbP5CSs might participate in the pathogen response pathway. However, PbP5CS4, PbP5CS5, PbP5CS8, and PbP5CS10 were down-regulated in response to G. haraeanum infection, suggesting that the four genes may function through different mechanisms to protect against biotic stimuli. All identified PbP5CS members were differentially regulated by both biotic and abiotic stresses, indicating that these genes are likely to mediate plant defense mechanisms in pear. Currently, the biological functions of most PbP5CS genes in plant developmental and defense processes remain unknown. The present bioinformatic and expression analyses of PbP5CS genes provide valuable information for screening candidate genes, and the results are helpful to further investigate the functions of this gene family in pear.
Conclusions
Eleven PbP5CS genes were identified in pear, and a systematic study of the PbP5CS gene family was carried out. The comprehensive analyses encompassed conserved domains, gene structures, and phylogenetic relationships, in addition to gene duplications, chromosome locations, cis-acting elements, and expression patterns. There were various cis-acting elements in the PbP5CS promoter sequences, suggesting that PbP5CSs act in complex networks regulating plant development and responses to stresses. Transcriptome and qPCR analyses revealed that PbP5CS genes are likely to take part in plant response to biotic and abiotic stresses. Our genome-wide analysis of PbP5CSs provides evidence for the functions of this gene family in pear. Further studies on PbP5CS genes are underway to verify their functions in stressed environments.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
CM and CW conceived and designed the research. MW, MZ, MY, XZ, YT, ZS, and XL performed the experiments, conducted the field work, and analyzed the data. MC and MW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Funds for Modern Agricultural Industry Technology System in Shandong Province (SDAIT-06-06), the Shandong Provincial Natural Science Foundation (ZR2019BC038), and the High-level Scientific Research Foundation of Qingdao Agricultural University (Grant 663/1121043), China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1066765/full#supplementary-material
References
Anton, D. B., Guzman, F. L., Vetö, N. M., Krause, F. A., Kulcheski, F. R., Coelho, A. P. D., et al. (2020). Characterization and expression analysis of P5CS (Δ1-pyrroline-5-carboxylate synthase) gene in two distinct populations of the Atlantic forest native species Eugenia uniflora l. Mol. Biol. Rep. 47, 1033–1043. doi: 10.1007/s11033-019-05195-7
Atienza, S. G., Faccioli, P., Perrotta, G., Dalfino, G., Zschiesche, W., Humbeck, K., et al. (2004). Large Scale analysis of transcripts abundance in barley subjected to several single and combined abiotic stress conditions. Plant Sci. 167, 1359–1365. doi: 10.1016/j.plantsci.2004.07.006
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bandurska, H., Niedziela, J., Pietrowska-Borek, M., Nuc, K., Chadzinikolau, T., Radzikowska, D. (2017). Regulation of proline biosynthesis and resistance to drought stress in two barley (Hordeum vulgare l.) genotypes of different origin. Plant Physiol. Biochem. 118, 427–437. doi: 10.1016/j.plaphy.2017.07.006
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Xia, R. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J. B., Wang, S. M., Jing, R. L., Mao, X. G. (2009). Cloning the PvP5CS gene from common bean (Phaseolus vulgaris) and its expression patterns under abiotic stresses. J. Plant Physiol. 166 (1), 12–19. doi: 10.1016/j.jplph.2008.02.010
Chen, J. B., Yang, J. W., Zhang, Z. Y., Feng, X. F., Wang, S. M. (2013). Two P5CS genes from common bean exhibiting different tolerance to salt stress in transgenic Arabidopsis. J. Genet. 92 (3), 461–469. doi: 10.1007/s12041-013-0292-5
Delauney, A. J., Verma, D. P. S. (1993). Proline biosynthesis and osmoregulation in plants. Plant J. 4, 215–223. doi: 10.1046/j.1365-313X.1993.04020215.x
Dong, X., Wang, Z., Tian, L., Zhang, Y., Qi, D., Huo, H., et al. (2020). De novo assembly of a wild pear (Pyrus betuleafolia) genome. Plant Biotechnol. J. 18 (2), 581–595. doi: 10.1111/pbi.13226
Duvaud, S., Gabella, C., Lisacek, F., Stockinger, H., Ioannidis, V., Durinx, C. (2021). Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 49, 216–227. doi: 10.1093/nar/gkab225
Fabro, G., Kovács, I., Pavet, V., Szabados, L., Alvarez, M. E. (2004). Proline accumulation and AtP5CS2 gene activation are induced by plant-pathogen incompatible interactions in Arabidopsis. Mol. Plant Microbe Interact. 17 (4), 343–350. doi: 10.1094/MPMI.2004.17.4.343
Funck, D., Baumgarten, L., Stift, M., von Wirén, N., Schönemann, L. (2020). Differential contribution of P5CS isoforms to stress tolerance in Arabidopsis. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.565134
Guan, C., Cui, X., Liu, H. Y., Li, X., Li, M. Q., Zhang, Y. W. (2020). Proline biosynthesis enzyme genes confer salt tolerance to switchgrass (Panicum virgatum l.) in cooperation with polyamines metabolism. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00046
Guan, C., Ji, J., Guan, W., Feng, Y., Li, X., Jin, C., et al. (2014). A Lycium chinense-derived P5CS-like gene is regulated by water defcit-induced endogenous abscisic acid and overexpression of this gene enhances tolerance to water defcit stress in Arabidopsis. Mol. Breed. 34, 1109–1124. doi: 10.1007/s11032-014-0103-6
Guo, Z., Jiang, W., Lages, N., Borcherds, W., Wang, D. (2014). Relationship between gene duplicability and diversifiability in the topology of biochemical networks. BMC Genomics 15 (1), 577. doi: 10.1186/1471-2164-15-577
Hayat, S., Hayat, Q., Alyemeni, M. N., Wani, A. S., Pichtel, J., Ahmad, A. (2012). Role of proline under changing environments: a review. Plant Signal Behav. 7 (11), 1456–1466. doi: 10.4161/psb.21949
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, 585–587. doi: 10.1093/nar/gkm259
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2014). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Hung, C. L., Lin, Y. S., Lin, C. Y., Chung, Y. C., Chung, Y. F. (2015). CUDA ClustalW: An efficient parallel algorithm for progressive multiple sequence alignment on multi-GPUs. Comput. Biol. Chem. 58, 62–68. doi: 10.1016/j.compbiolchem.2015.05.004
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Ma, H., DePamphilis, C. W. (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885. doi: 10.1111/j.1365-313X.2007.03097.x
Kumar, V., Shriram, V., Kishor, P., Jawali, N., Shitole, M. (2010). Enhanced proline accumulation and salt stress tolerance of transgenic indica rice by over-expressing P5CSF129A gene. Plant Biotechnol. Rep. 4, 37–48. doi: 10.1007/s11816-009-0118-3
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, B., Dong, X., Zhang, Z., Zhao, H., Lian, S., Li, B. (2006) Optimum time for control of pear rust (Gymnosporangium haraeanum syd.) with fungicides in laiyang plant protection. Available at: https://en.cnki.com.cn/Article_en/CJFDTOTAL-ZWBH200601020.htm.
Liu, J., Wang, Y. S. (2020). Proline metabolism and molecular cloning of AmP5CS in the mangrove Avicennia marina under heat stress. Ecotoxicol. 29 (6), 698–706. doi: 10.1007/s10646-020-02198-0
Liu, D. F., Zhang, D., Liu, G. Q., Hussain, S., Teng, Y. W. (2013). Influence of heat stress on leaf ultrastructure, photosynthetic performance, and ascorbate peroxidase gene expression of two pear cultivars (Pyrus pyrifolia). J. Zhejiang Univ. Sci. B. 14 (12), 1070–1083. doi: 10.1631/jzus.B1300094
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Ma, C., Bian, C., Liu, W., Sun, Z., Xi, X., Guo, D., et al. (2022). Strigolactone alleviates the salinity-alkalinity stress of Malus hupehensis seedlings. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.901782
Parida, A. K., Dagaonkar, V. S., Phalak, M. S., Aurangabadkar, L. P. (2008). Differential responses of the enzymes involved in proline biosynthesis and degradation in drought tolerant and sensitive cotton genotypes during drought stress and recovery. Acta Physiol. Plant 30, 619–627. doi: 10.1007/s11738-008-0157-3
Peng, Z., Lu, Q., Verma, D. P. (1996). Reciprocal regulation of delta 1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol. Gen. Genet. 253 (3), 334–341. doi: 10.1007/pl00008600
Rai, A. N., Penna, S. (2013). Molecular evolution of plant P5CS gene involved in proline biosynthesis. Mol. Biol. Rep. 40 (11), 6429–6435. doi: 10.1007/s11033-013-2757-2
Rogers, J. H. (1990). The role of introns in evolution. FEBS Lett. 268(2):339–43. doi: 10.1016/0014-5793(90)81282-s
Rose, A. B.. (2008). Intron-mediated regulation of gene expression. Curr. Top. Microbiol. Immunol. 326 (2), 277–290. doi: 10.1007/978-3-540-76776-3_15
Saeed, A. I., Bhagabati, N. K., Braisted, J. C., Sharov, V., Howe, E. A., Li, J. W., et al. (2006) TM4 microarray software suite. Methods Enzymol. 411, 134–193. doi: 10.1016/S0076-6879(06)11009-5
Signorelli, S., Monza, J. (2017). Identification of Δ1-pyrroline 5-carboxylate synthase (P5CS) genes involved in the synthesis of proline in Lotus japonicus. Plant Signal Behav. 12 (11), e1367464. doi: 10.1080/15592324.2017.1367464
Szabados, L., Savouré, A. (2010). Proline: A multifunctional amino acid. Trends Plant Sci. 15, 89–97. doi: 10.1016/j.tplants.2009.11.009
Székely, G., Abrahám, E., Cséplo, A., Rigó, G., Zsigmond, L., Csiszár, J., et al. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53 (1), 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Tan, C. M., Chen, R. J., Zhang, J. H., Gao, X. L., Li, L. H., Wang, P. R., et al. (2013). OsPOP5, a prolyl oligopeptidase family gene from rice confers abiotic stress tolerance in Escherichia coli. Int. J. Mol. Sci. 14, 20204–20219. doi: 10.3390/ijms141020204
Trovato, M., Mattioli, R., Costantino, P. (2018). From a. rhizogenes RolD to plant P5CS: Exploiting proline to control plant development. Plants (Basel). 7 (4), 108. doi: 10.3390/plants7040108
Turchetto-Zolet, A. C., Margis-Pinheiro, M., Margis, R. (2009). The evolution of pyrroline-5-carboxylate synthase in plants: a key enzyme in proline synthesis. Mol. Genet. Genomics 281 (1), 87–97. doi: 10.1007/s00438-008-0396-4
Wang, L., Guo, Z., Zhang, Y., Wang, Y., Yang, G., Yang, L., et al. (2017). Characterization of LhSorP5CS, a gene catalyzing proline synthesis in oriental hybrid lily Sorbonne: molecular modelling and expression analysis. Bot. Stud. 58 (1), 10. doi: 10.1186/s40529-017-0163-0
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi: 10.1093/nar/gkr1293
Wang, P., Wang, Z., Guan, L., Muhammad, S. H., Maazullah, N., Yuan, Y., et al. (2022). Versatile physiological functions of the nudix hydrolase family in berry development and stress response in grapevine. J. Integr. Agriculture. 21, 91–112. doi: 10.1016/S2095-3119(20)63490-6
Xi, L., Xu, K., Qiao, Y., Qu, S., Zhang, Z., Dai, W. (2011). Differential expression of ferritin genes in response to abiotic stresses and hormones in pear (Pyrus pyrifolia). Mol. Biol. Rep. 38, 4405–4413. doi: 10.1007/s11033-010-0568-2
Xue, X., Liu, A., Hua, X. (2009). Proline accumulation and transcriptional regulation of proline biosynthesis and degradation in Brassica napus. BMB Rep. 42, 28–34. doi: 10.5483/bmbrep.2009.42.1.028
Yang, S., Bai, M., Hao, G., Zhang, X., Guo, H., Fu, B. (2021b). Transcriptome survey and expression analysis reveals the adaptive mechanism of ‘Yulu xiang’ pear in response to long-term drought stress. PloS One 16, e0246070. doi: 10.1371/journal.pone.0246070
Yang, D., Ni, R., Yang, S., Pu, Y., Qian, M., Yang, Y., et al. (2021a). Functional characterization of the Stipa purpurea P5CS gene under drought stress conditions. Int. J. Mol. Sci. 22, 9599. doi: 10.3390/ijms22179599
Yoshiba, Y., Kiyosue, T., Katagiri, T., Ueda, H., Mizoguchi, T., Yamaguchi-Shinozaki, K., et al. (1995). Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J. 7, 751–760. doi: 10.1046/j.1365-313x.1995.07050751.x
Yu, F., Liang, K., Fang, T., Zhao, H., Han, X., Cai, M., et al. (2019). A group VII ethylene response factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotechnol. J. 17 (12), 2286–2298. doi: 10.1111/pbi.13140
Zhang, H., Huo, Y., Xu, Z., Guo, K., Wang, Y., Zhang, X., et al. (2020). Physiological and proteomics responses of nitrogen assimilation and glutamine/glutamine family of amino acids metabolism in mulberry (Morus alba l.) leaves to NaCl and NaHCO3 stress. Plant Signal Behav. 15 (10), 1798108. doi: 10.1080/15592324.2020.1798108
Zhang, C. S., Lu, Q., Verma, D. P. S. (1995). Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase, a bifunctional enzyme catalyzing the first two steps of proline biosynthesis in plants. J. Biol. Chem. 270, 20491–20496. doi: 10.1074/jbc.270.35.20491
Zhu, J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–71. doi: 10.1016/S1360-1385(00)01838-0
Zhu, X. Y., Li, X. P., Zou, Y., Chen, W. X., Lu, W. J. (2012). Cloning, characterization and expression analysis of Δ1-pyrroline-5-carboxylate synthetase (P5CS) gene in harvested papaya (Carica papaya) fruit under temperature stress. Food Res. Int. 49 (1), 272–279. doi: 10.1016/j.foodres.2012.08.003
Keywords: P5CS, Pyrus betulifolia, transcriptome, stress response, biotic stress
Citation: Ma C, Wang M, Zhao M, Yu M, Zheng X, Tian Y, Sun Z, Liu X and Wang C (2022) The Δ1-pyrroline-5-carboxylate synthetase family performs diverse physiological functions in stress responses in pear (Pyrus betulifolia). Front. Plant Sci. 13:1066765. doi: 10.3389/fpls.2022.1066765
Received: 11 October 2022; Accepted: 07 November 2022;
Published: 24 November 2022.
Edited by:
Tuanhui Bai, Henan Agricultural University, ChinaReviewed by:
Changhai Liu, Northwest A&F University, ChinaQinglong Dong, Agricultural University of Hebei, China
Copyright © 2022 Ma, Wang, Zhao, Yu, Zheng, Tian, Sun, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caihong Wang, Y2h3NjA2OEAxMjYuY29t
†These authors have contributed equally to this work
 Changqing Ma
Changqing Ma Mengqi Wang1,2†
Mengqi Wang1,2† Xiaodong Zheng
Xiaodong Zheng Caihong Wang
Caihong Wang