
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 09 December 2022
Sec. Functional and Applied Plant Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1064847
This article is part of the Research TopicGenomics in Plant Sciences: Understanding and Development of Stress-Tolerant PlantsView all 12 articles
 Pradeep K. Papolu1†
Pradeep K. Papolu1† Muthusamy Ramakrishnan1,2*†
Muthusamy Ramakrishnan1,2*† Sileesh Mullasseri3
Sileesh Mullasseri3 Ruslan Kalendar4,5
Ruslan Kalendar4,5 Qiang Wei2
Qiang Wei2 Long−Hai Zou1
Long−Hai Zou1 Zishan Ahmad2
Zishan Ahmad2 Kunnummal Kurungara Vinod6
Kunnummal Kurungara Vinod6 Ping Yang1,7*
Ping Yang1,7* Mingbing Zhou1,7*
Mingbing Zhou1,7*Long terminal repeat retrotransposons (LTR retrotransposons) are the most abundant group of mobile genetic elements in eukaryotic genomes and are essential in organizing genomic architecture and phenotypic variations. The diverse families of retrotransposons are related to retroviruses. As retrotransposable elements are dispersed and ubiquitous, their “copy-out and paste-in” life cycle of replicative transposition leads to new genome insertions without the excision of the original element. The overall structure of retrotransposons and the domains responsible for the various phases of their replication is highly conserved in all eukaryotes. The two major superfamilies of LTR retrotransposons, Ty1/Copia and Ty3/Gypsy, are distinguished and dispersed across the chromosomes of higher plants. Members of these superfamilies can increase in copy number and are often activated by various biotic and abiotic stresses due to retrotransposition bursts. LTR retrotransposons are important drivers of species diversity and exhibit great variety in structure, size, and mechanisms of transposition, making them important putative actors in genome evolution. Additionally, LTR retrotransposons influence the gene expression patterns of adjacent genes by modulating potential small interfering RNA (siRNA) and RNA-directed DNA methylation (RdDM) pathways. Furthermore, comparative and evolutionary analysis of the most important crop genome sequences and advanced technologies have elucidated the epigenetics and structural and functional modifications driven by LTR retrotransposon during speciation. However, mechanistic insights into LTR retrotransposons remain obscure in plant development due to a lack of advancement in high throughput technologies. In this review, we focus on the key role of LTR retrotransposons response in plants during heat stress, the role of centromeric LTR retrotransposons, and the role of LTR retrotransposon markers in genome expression and evolution.
Eukaryotic genomes contain repetitive elements, such as transposable elements (TEs), that are present in multiple copies throughout the genome. TEs are tandemly arrayed, interspersed throughout the genome, and can be processed as pseudogenes. TEs are major components of eukaryotic genomes and can change their position within genomes (Lisch, 2013; Bourque et al., 2018). TEs were first described in maize by Barbara McClintock in the middle of the twentieth century and she named them jumping genes (Ravindran, 2012; Goodier, 2016). Although TEs are a source of spontaneous mutations, their expression and activity can also increase the stress response to different biotic and abiotic stresses (Ramakrishnan et al., 2021). Moreover, TE specificity has now been associated with the adaptation of plants to a range of these stresses. TEs have deep evolutionary origins and continuous diversification and come in a bewildering variety of forms and shapes (Bourque et al., 2018; Klein and O’Neill, 2018) in most eukaryotic genomes (Wicker et al., 2007; Muñoz-López and García-Pérez, 2010; Gorbunova et al., 2021). TEs are primarily classified into DNA transposons (Class II) and retrotransposons (Class I) based on their mechanism of transposition (Boeke et al., 1985). Both classes are further divided into subclasses based on the mechanism of chromosomal integration. Class I has two major classes, Long Terminal Repeats (LTR) retrotransposons (LTR retrotransposons) and non-LTR retrotransposons (Wessler et al., 1995). LTR retrotransposons and related elements are abundant in plant genomes and include functional genes encoding structural and enzymatic proteins (Galindo-González et al., 2017). LTR retrotransposon mobility is ensured through an RNA intermediate, allowing a copy-and-paste approach for their transposition. Their encoded RNA is reverse transcribed using their own (or not) encoded enzymes that reform a double-stranded DNA from the single-stranded RNA matrix at a new location. LTR retrotransposon integration occurs by cleavage and strand-transfer reaction catalyzed by an integration, similar to retroviruses (Brown et al., 1987). On the other hand, non-LTR retrotransposons include both long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs)s (Luan et al., 1993).
The two major superfamilies of LTR retrotransposons are Ty1/Copia and Ty3/Gypsy, which are classified based on the retroviral structural homology and domain order organization of the pol gene (Capy, 2005). These LTR retrotransposons exist universally in plant and animal genomes (Malik and Eickbush, 2001; Mangiavacchi et al., 2021). LTR-RTs are more active in plants and their functions are fine-tuned by epigenetic modifications. Although LTR retrotransposons in plants have attracted great attention in recent years, a more comprehensive understanding of the diverse functions of LTR retrotransposons can be gained from further studies. This review provides an overview of the processes associated with LTR retrotransposons involved in precise gene regulation in the plant genome. We also focus on the key role of LTR retrotransposon in plant heat response. Further, we discuss the LTR-derived small interfering RNA (siRNAs), LTR retrotransposon delivery system, centromeric LTR retrotransposons, the application of LTR-based molecular markers, and their contributions towards genome expression and evolution.
LTR retrotransposons comprise about 10% to 90% of the total eukaryote genome in most plants. The distribution of LTRs differs among the major families of Ty3/Gypsy and Ty1/Copia elements in all plant genomes (Supplementary Table 1). Ty3/Gypsy elements are enriched in euchromatic sub-telomeric regions, whereas, Ty1/Copia elements are more frequent in heterochromatic pericentromeric regions (Jedlicka et al., 2019). Moreover, Ty3/Gypsy elements play crucial roles in host epigenetic response and are more heterogenous than Ty1/Copia elements. Although both families are found in a large number of copies in higher plants, these families were first identified in Drosophila (Sant et al., 2000). Members of these superfamilies differ primarily in the arrangement of the gene coding for polymerase function within the polyprotein (POL) region. Ty1/Copia elements have a pol gene organized as the domains protease (PR), integrase (INT), reverse transcriptase (RT), and ribonuclease H (RNase H) (PR-INT-RT-RNase H). Ty3/Gypsy elements are organized as PR-RT-RNase H-INT domains (Sant et al., 2000).
LTR retrotransposons from genomes of about 300 plant species have been identified and are associated with diverse structural, functional annotation, and classification information (Zhou et al., 2021). Thus, this information may provide useful resources for investigating the evolutionary dynamics and functional implications of LTR retrotransposons in plant genomes (Kalendar et al., 2004; Moisy et al., 2014; Kalendar et al., 2020). Moreover, understanding the evolutionary forces governing TE polymorphism is crucial to understanding phenotypic variation in plants (Catlin and Josephs, 2022). Therefore, exploring the role of TEs leading to phenotypic variation and its regulation in plants has significant economic importance in the development of more efficient crops (Kalendar et al., 2008; Malaviya et al., 2021).
The impact of TEs on the structure, function and evolution of multiple plant genes have paved the way for epigenetic techniques that address diverse stresses in various crop species. TEs can be highly sensitive to different abiotic and biotic stresses, including salt, cold, heat, wounds, and infections (Mhiri et al., 1997; Ivashuta et al., 2002; Grandbastien et al., 2005; Buchmann et al., 2009; Naito et al., 2009; Ito et al., 2011; Lanciano and Mirouze, 2018). Several studies (Table 1) revealed that LTR retrotransposons become activated under certain epigenetic processes, such as siRNA regulation, DNA methylation, LTR retrotransposon integration, and chromatin modification (Grandbastien, 2015; Schorn et al., 2017). Moreover, LTR retrotransposons play a crucial role in the regulation of gene activity at the transcriptional and post-transcriptional level and in genome epigenetic regulation of stress resistance in a wide range of organisms (Galindo-González et al., 2017).
The transcriptional gene silencing of several LTR retrotransposons of Arabidopsis is accomplished by the loss of nucleosome and heterochromatin decondensation, which was restored upon recovery from heat stress (Pecinka et al., 2010). This indicates the role of environmental stress leading to epigenetic regulation. Moreover, heat-activated LTR retrotransposons play a crucial role in shaping a genome over an evolutionary period (Wessler, 1996; Masuta et al., 2018). Recently, we reported that the role of two LTR retrotransposons, PHRE1 and PHRE2 (Ty3/Gypsy), in Moso bamboo (Phyllostachys edulis) indicated that the 5’ LTR acts as a promoter and can increase transposition activity during heat stress (Papolu et al., 2021).
A heat-responsive ONSEN retrotransposon is conserved among the Brassica species, and Adzuki bean exhibited upregulated transcript levels, and full-length extrachromosomal DNA accumulated in the stress-treated plants (Boonjing et al., 2020). The ONSEN family in most species of Brassicaceae showed integration into active chromatin, which was promoted by heat stress (Ito et al., 2013). Furthermore, there is a correlation between the heat-responsive elements (HREs) of Copia families and putative high-affinity heat shock factor binding HREs within the LTRs in seven Brassicaceae species. Moreover, the strong HRE of ONSEN is conserved over millions of years (Pietzenuk et al., 2016).
The active full-length Ty1/Copia, GBRE-1, showed increased expression under heat stress in Gossypium hirsutum, and its expression was similar to that of the ONSEN retrotransposon (Cao et al., 2015). The heat stress response and heat accumulation of Ty3/Gypsy retrotransposon in Cryptomeria japonica exhibited differential expression due to preheating treatment with heat shock factors, indicating the impact of LTR retrotransposons in the regulation of heat response systems in plants (Ujino-Ihara, 2020).
Several studies revealed the active role of ONSEN in regulating heat stress (Cavrak et al., 2014; Nozawa et al., 2021), including the regulatory role of siRNA. In Arabidopsis, ONSEN is activated by protracted exposure to heat stress (Ito et al., 2011; Matsunaga et al., 2012; Matsunaga et al., 2015; Ito et al., 2016). The genetic consequences of transposition bursts of the Arabidopsis LTR retrotransposon Copia78 family generated a novel progeny of chromosomally integrated LTRs consisting of a high frequency of intrafamily recombination and significant sequence diversity of LTR retrotransposons under heat stress (Sanchez et al., 2017). However, the role of LTRs, especially the Ty1/Copia and the Ty3/Gypsy superfamilies, requires further investigations to reveal their role in heat stress regulation. Such investigations will further the possibilities of developing crops to increase resistance to heat stresses due to global warming.
Small noncoding RNAs (sRNAs) are the sequence-specific modulators of gene expression and precisely involved in the regulation of plant immunity (Borges and Martienssen, 2015). sRNAs interfere with the expression of particular genes with complementary nucleotide sequences by degrading mRNA after transcription, thus preventing translation (Laganà et al., 2015). Based on differences in biogenesis and function, sRNAs can be classified into several major classes, including: microRNAs (miRNAs), hairpin-derived siRNAs (hp-siRNAs), natural antisense siRNAs (natsiRNAs), heterochromatic siRNAs (hetsiRNAs) and secondary siRNAs. miRNAs and siRNAs are the two major classes of plant sRNAs. The role of miRNAs in plant development, immunity, and intracellular immune receptors is well documented (Song et al., 2019; Wang S. et al., 2021; Dong et al., 2022). siRNAs are best known for their role in silencing viral RNAs, replication, and genome reprogramming (Kong et al., 2022).
siRNAs are specifically generated from double-stranded RNA (dsRNA) precursors derived from noncoding transcripts, inverted repeat sequences, sense and anti-sense transcripts, and exogenous RNAs (Xie et al., 2004). The dsRNAs are primarily processed into mature 21-24-nt siRNAs by various Dicer-like enzymes (DCL 1-4) and loaded into AGOs to form RISCs. DCL1 processes primary miRNAs into 21-nt-long mature miRNAs. DCL2 is involved in antiviral strategies and cleaves viral dsRNA into 21-22 nt long siRNAs, which target viral transcripts. DCL3 is involved in silencing processes targeting TEs and produces siRNAs approximately 24 nt in length. Finally, DCL4 generates 21-nt transacting siRNAs (tasiRNAs), which silence specific genes. siRNAs can be divided into two main classes: RDR6-dependent secondary siRNAs and RNA polymerase IV-dependent siRNAs (P4-siRNAs) (Kong et al., 2022). Secondary siRNAs are generated by transcripts from noncoding genes, e.g., tasiRNA loci, and protein-coding genes within large gene families, e.g., the nucleotide-binding leucine-rich repeats (NB -LLRs) (Sanan-Mishra et al., 2021). P4-siRNAs, especially 24-nt long, are mainly produced by heterochromatic regions, and TEs are linked to RdDM to induce transcriptional gene silencing (Ito, 2012; Lopez-Gomollon and Baulcombe, 2022).
siRNA pathways are significantly involved in retrotransposon silencing and may mediate different forms of epigenetic regulation in plants (Figure 1) (Lippman et al., 2003). In addition, siRNAs derived from TEs act as a trigger for host silencing mechanisms (Table 2). For example, siRNA silencing of a different class of LTR-retrotransposon mutants was shown to impact retrotransposon methylation, chromatin remodeling, and histone modification in Arabidopsis (Lippman et al., 2003). The mutagenic activity of LTR retrotransposons, especially in the pollen vegetative nucleus of Arabidopsis, is suppressed by siRNA silencing that may transmit the TEs to next-generation offspring (Slotkin et al., 2009). Remarkably, siRNAs suppress transposons by RNA-directed DNA methylation (RdDM), thus in turn leading to TEs becoming epigenetically silenced (Nosaka et al., 2012). In maize, loss of RNA-dependent RNA polymerase 2 (RDR2) function in the mediator of paramutation1 (mop1) results in the reactivation of transcriptionally silenced mutator retrotransposon and a substantial reduction in the accumulation of siRNAs. This suggests that the RDR2 pathway is an independent mechanism for silencing LTR retrotransposons in complex genomes like maize (Jia et al., 2019).
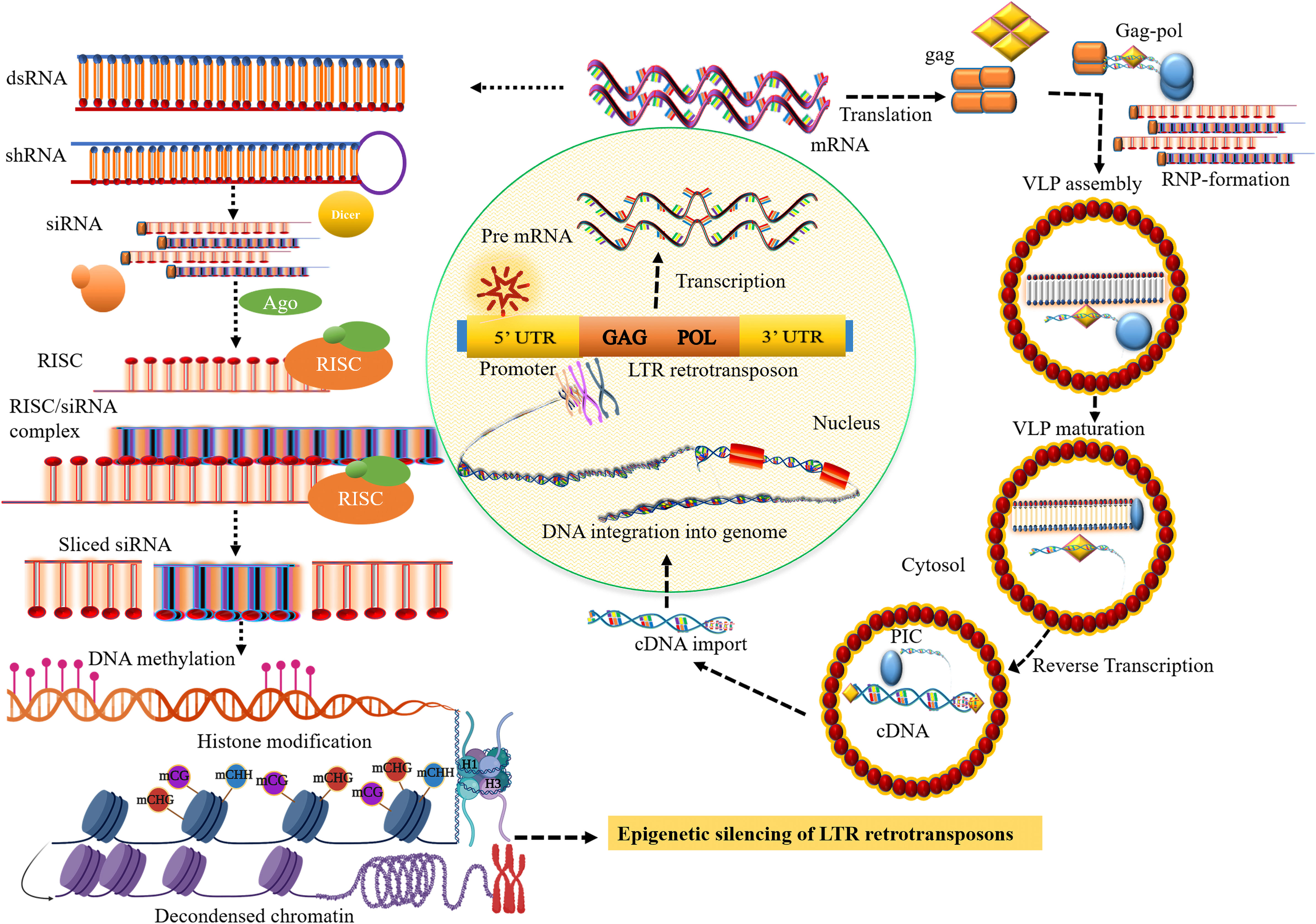
Figure 1 Small RNA biosynthesis and the transposition mechanism of long terminal repeat (LTR) retrotransposons (LTR retrotransposons). The transposition cycle initiates with the transcription of LTR retrotransposon that produces RNA template strand and major structural proteins for reverse transcription. After cleavage of the major structural gag-pol polyprotein by the viral protease (PR) activity, the gag protein-containing capsid and nucleic acid-binding domain involve the formation of cytoplasmic virus-like particles (VLPs). The polyprotein, pol comprising catalytic domain for replication encodes pepsin-like aspartate proteases, integrase, reverse transcriptase, and ribonuclease H proteins, which are crucial for reverse transcription and transposition of retrotransposons. Collectively with the RNA template, reverse transcription most likely takes place within VLPs that produce the cDNA which is then imported into the nucleus. The integrase that involves the formation of DNA nicks at the target sites is inserted into a new chromosomal locus to generate a new copy of retrotransposons and their insertion into the genome. The core siRNA silencing pathway: dsRNA or shRNA is processed into siRNA duplexes by Dicer RNase III. Subsequently, the siRNA or RNA-induced silencing complex (RISC) then binds to the complementary sequence of the target mRNA resulting in the degradation of the target transcript, establishing methylation of DNA through RdDM, and inducing histone modification, and heterochromatin formation.
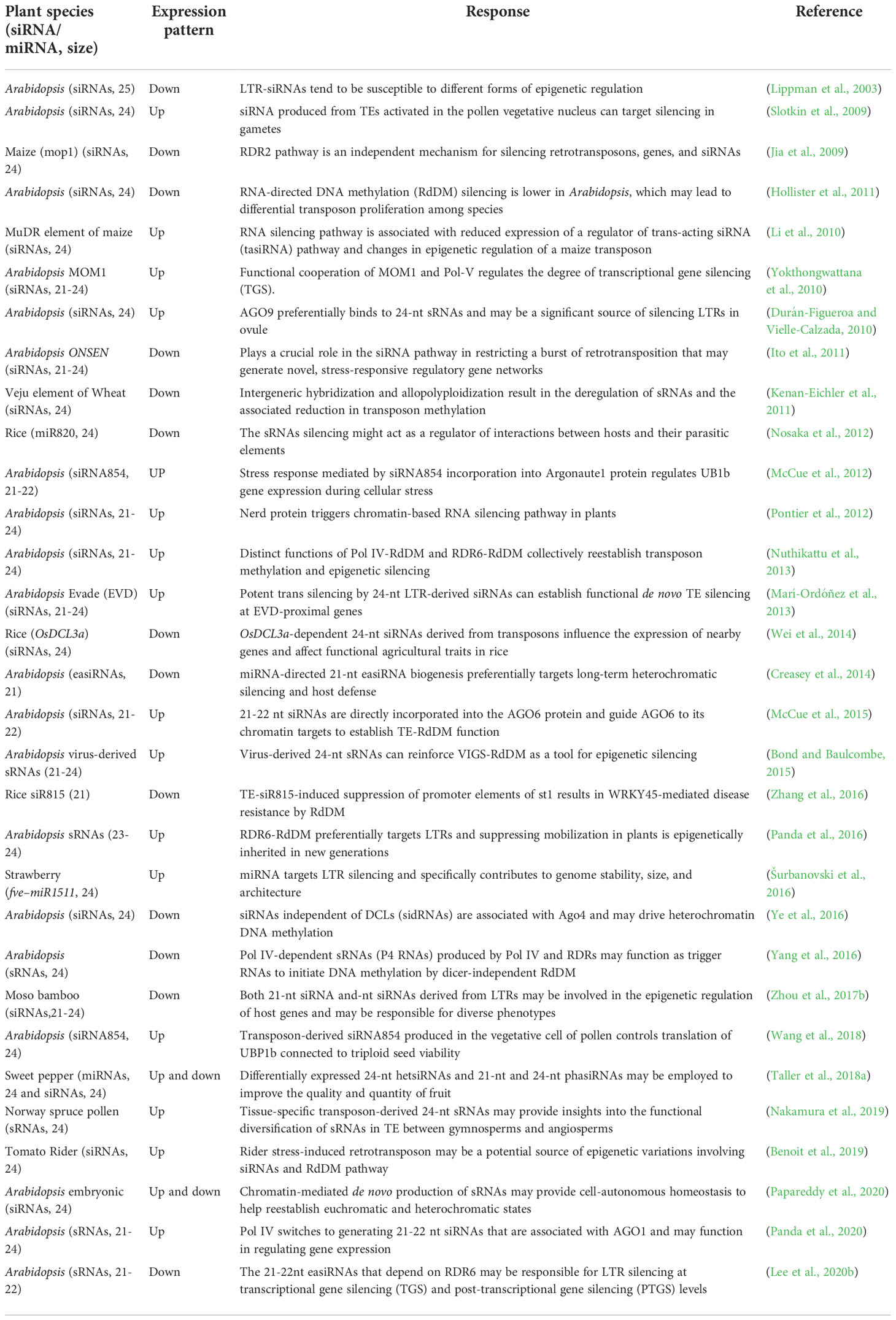
Table 2 List of small interfering RNA (siRNA), micro RNAs (miRNAs), and small RNAs (sRNAs) derived from LTR retrotransposons and their functions.
In Arabidopsis, siRNA targeted LTR retrotransposons are associated with reduced gene expression due to RdDM silencing. However, the effect of RdDM silencing was lower in A. lyrata, and thus showed differential transposon proliferation among species (Hollister et al., 2011). In addition, the transcriptionally active LTR retrotransposons in Arabidopsis produced RdDM-dependent siRNAs, indicating the function of RNA-dependent RNA Polymerase 6 (RDR6) and RNA Polymerase IV (Pol IV). These are independent in the silencing of TEs, in which Pol IV-RdDM functions to initiate TE silencing in an RNA Polymerase II expression-independent manner. In contrast, RDR6-RdDM functions to recognize active Polymerase II-derived TE mRNA transcripts to reestablish DNA methylation and TE silencing (Nuthikattu et al., 2013). Moreover, the targeting specificity of RDR6-RdDM function for full-length LTR retrotransposons in Arabidopsis have full-length transposon mRNA to be cleaved by primary 21-22-nt siRNAs and thus the RNA cleavage specificity drives the initiation of epigenetic transcriptional silencing targeted to LTR retrotransposons and transgenes (Panda et al., 2016). The function of DNA methylation to transcriptionally active LTR retrotransposons has demonstrated that mRNA-derived 21-22-nt siRNAs are directly incorporated into the ARGONAUTE 6 (AGO6) protein and in turn guide the AGO6 to its chromatin targets to establish epigenetic transcriptional silencing of TEs in RdDM (McCue et al., 2015).
Recently, Nerd, a plant-specific GW repeat protein triggered by siRNA-dependent DNA methylation in Arabidopsis, was found to play a central role in integrating chromatin-based RNA silencing supported by binding both histone H3 and Ago2 proteins and to contribute to siRNA accumulation at a Nerd-targeted locus of LTR retrotransposons. This suggests that RdDM might preferentially target LTR retrotransposons and other repeat sequences (Pontier et al., 2012). The establishment of virus-induced gene silencing (VIGS) mediated RdDM function in Arabidopsis requires RNA Polymerase V (Pol V) and de novo methyltransferase 2 (DRM2). However, dicer-like-3 and Pol IV pathway components are not required for such functions. Perhaps the DNA methylation in VIGS is guided by virus-derived 21-22-nt siRNAs, thus suggesting VIGS-RdDM is a tool for retrotransposon silencing in Arabidopsis (Bond and Baulcombe, 2015). Later, the retrotransposon virus-like particles in Arabidopsis are activated by DDM1 mutations, giving rise to 21-22-nt siRNA through RNA-dependent RNA polymerase 6 (RDR6). This suggests that virus-like particle (VLP) DNA could also provide a powerful tool for identifying active LTR retrotransposons from the complex genome and their control at the transcriptional and post-transcriptional levels (Lee et al., 2020a). However, TE-derived siR815 drives RdDM of ST1 promoter and leads to transcriptional suppression of ST1, which abolished the WRKY45 transcription factor in rice resistance to Xanthomonas oryzae (Zhang et al., 2016).
The stress-induced full-length Rider LTR retrotransposons in the tomato genome indicate that RdDM controls Rider activity through siRNA production and DNA methylation, which may contribute to phenotypic variation through epigenetic alteration induced during environmental stress (Benoit et al., 2019). Furthermore, Arabidopsis mutations in the Argonaute9 protein (AGO9) indicated that AG09 can interact with 24-nt small RNAs (sRNA) corresponding to LTR retrotransposons expression in the ovule. AGO9 is also necessary for silencing repetitive genomic regions involved in heterochromatin formation. Thus, the AGO9-dependent pathway may be responsible for the epigenetic control of gametogenesis in plants (Durán-Figueroa and Vielle-Calzada, 2010). In a recent report on pepper, pepper-specific heterochromatin-associated 24-nt siRNAs (hetsiRNAs) and 21-24-nt phased siRNAs (phasiRNAs) produced from transposons were preferentially expressed in seeds and placenta, indicating that pepper fruit quality and quantity is associated with changes in sRNA abundance (Taller et al., 2018b). The dynamics of TE-derived embryonic siRNAs in Arabidopsis could promote re-methylation of euchromatic and heterochromatic TEs in a new generation, therefore the decondensed chromatin-mediated 24-nt siRNA transcription may provide cell-autonomous silencing of transposons (Papareddy et al., 2020). The TE-siRNAs generated by plant-specific Pol IV can participate in RdDM, whereas other siRNAs and microRNAs (21-22-nt) are associated with Argonaute1 (AGO1), suggesting that Pol I-dependent 21-22-nt siRNAs may participate in post-transcriptional regulation (Panda et al., 2016; Panda et al., 2020).
In maize, a link between the vegetative phase and the initiation of epigenetic silencing of MuDR retrotransposon is associated with a reduction of mutant expression during plant development. This is associated with an increase in trans-acting siRNA (LRRs) levels, which in turn is responsible for silencing epigenetic regulation of the MuDR element (Li et al., 2010). The regulatory interplay between MOM1 mutants of LTR-retrotransposon in Arabidopsis and RNA polymerase-V may regulate the intensity and siRNA accumulation at the transgenic locus and the transcriptional gene silencing at the locus is accompanied by DNA methylation (Yokthongwattana et al., 2010). The heat-induced ONSEN retrotransposon in Arabidopsis showed its accumulation was stimulated in mutants deficient in the biogenesis of siRNAs, suggesting a considerable role of the siRNA pathway triggered by environmental stress during retrotransposition (Ito et al., 2011).
In wheat, high-throughput sRNA sequencing in parental, hybrid, and allopolyploid plants showed that miRNAs and the TE-derived siRNAs respond differently to changes at the ploidy level, and the siRNA pools were significantly reduced upon allopolyploidization. This, in turn, causes siRNA deregulation and the associated reduction in CpG methylation of LTR retrotransposons, which may contribute to genome instability at the initial stage of speciation (Kenan-Eichler et al., 2011). The Fatima family LTR retrotransposons of polyploid wheat are highly specific to B-genome and proliferated before allopolyploid wheat formation (Salina et al., 2011). Likewise, in hexaploidy wheat TriRe-1, LTR retrotransposons have a specific amplification history of B-genome progenitors, implying that genome-specific TriRe-1 may be utilized for the development of wheat molecular markers (Monden et al., 2014b).
In rice, a transposon produces microRNA820 (miR820) to suppress host silencing. The miR820 negatively regulates the expression of de novo DNA methyltransferase gene OsDRM2, indicating that transposon-derived siRNA silencing might act as a regulator of interactions between the host and their TEs (Nosaka et al., 2012). The Dicer-like 3 homolog OsDCL3a produces 24-nt siRNAs that target gibberellin (GA) and brassinosteroid (BR) homeostasis-related genes by association with TEs, which suppress the expression of nearby genes and may control important agricultural traits in rice (Wei et al., 2014). Whereas Dicer-like (DCL) proteins and 24-nt siRNAs are not required for DNA methylation at RdDM target loci, P4 sRNA transcripts generated by Pol IV and RNA-dependent RNA polymerases (RDRs) may function as RNA-triggered gene silencing of retrotransposons to initiate DNA methylation through the RdDM pathway (Yang et al., 2016). However, the biogenesis of TE heterochromatin-associated siRNAs in Arabidopsis is mechanically distinct from gene-regulating microRNAs (miRNA) or tasiRNAs. This suggests that the TE-derived siRNA854 regulates UBP1b mutant gene expression during the stress response, and the accumulation of siRNA854 is under the same trans-generational epigenetic regulation and inheritance pattern as the Arabidopsis LTR retrotransposons (McCue et al., 2012). Evd LTR-derived 24-nt siRNAs can silence transactive Evd copies in Arabidopsis. Reciprocal crossing between F11 and F14 plants resulted in the silencing of all F11-derived Evd copies. In addition, an Evd RNA and 3′ gag-derived siRNAs of 21-22 nt were below detection in F1 plants, indicating effective trans silencing by LTR-triggered 24-nt siRNAs (Marí-Ordóñez et al., 2013).
In Arabidopsis, Post-transcriptional gene silencing (PTGS) mediated by miRNA-directed siRNA biogenesis specifically targets retrotransposon transcripts, whereas transcriptional gene silencing (TGS) of LTR retrotransposons is mediated by 24-nt heterochromatic (het) siRNA. Together, LTR retrotransposons give rise to the most abundant 21-nt epigenetically activated siRNAs (easiRNAs) in ddm1 and methyltransferase1 (met1) mutants, and in the nucleus of pollen grains and callus cultures. Consequently, this supports an antagonistic relationship between PTGS and TGS in plants (Creasey et al., 2014).
In moso bamboo, both 21-nt siRNA and 24-nt siRNA have targets within LTR regions of retrotransposons. The high number of siRNAs derived from LTR retrotransposons may be responsible for diverse phenotypes of moso bamboo (Zhou et al., 2017a). The silencing mechanism of LTR retrotransposons is mediated by the most abundantly expressed miRNA, fve-miR1511. This fve-miR1511 is generated from a single locus that specifically targets LTR transcripts at the PBS site for methionyl initiator tRNA, which is essential for reverse transcription. This may contribute to features such as genome stability size and architecture in strawberries (Šurbanovski et al., 2016). The distinct class of 24-nt siRNAs independent of Dicer-like 3 (DCL3) is associated with effector AGO4 and is capable of driving DNA methylation and is subsequently subjected to 3’-5’ exonucleolytic activity for maturation. Therefore, this class may be the initial trigger of de novo DNA methylation (Ye et al., 2016).
In addition, the transposon-associated sRNAs in pollen and cell culture of Norway spruce are responsible for tissue-specific and environmentally induced gene repression. This may provide insights into the diversification process of sRNA in transposon silencing between angiosperms and gymnosperms (Nakamura et al., 2019). The enhanced retrotransposon expression in Botrytis cinerea leads to the suppression of plant defense-related genes during infection. Retrotransposons are pathogenicity factors that manipulate host gene expression by encoding trans-species sRNAs (BcsRNAs) and therefore have a broad impact on host-microbe interactions and pathology (Porquier et al., 2021).
Previously, understanding of the sRNA activity in plants generally came from their prominent functions in plant development. Now, there is a greater understanding of the complex molecular mechanisms involved in sRNA biogenesis and function in plants. sRNAs play a significant role in the diversification and specialization of gene silencing. This is because there are several pathways for sRNA biogenesis and function, which are related to evolution. However, most sRNA classes contribute to biotic and abiotic stress and transgenerational inheritance, and the stability of acquired sRNA-based responses has not been characterized.
However, unless sRNAs in isolated cell types and single cells can be profiled, understanding of the specificities and interplay between the different gene-silencing mechanisms operating in plant cells will remain limited. Therefore, focused research on the aspects described above is necessary to manage stress-induced agricultural losses and the development of stress-resistant crops.
Nanomaterial-mediated delivery of biomolecules and therapeutics has been extensively studied in animals, but its potential for plant-based systems lags behind (Demirer et al., 2019a). Several previous studies have used nanoparticles to deliver plasmid DNAs (Cunningham et al., 2018; Wang J. W. et al., 2019; Lv et al., 2020), proteins (Wang J. W. et al., 2019; Wang J. W. et al., 2021), small interfering RNAs (Demirer et al., 2020), and intact plant cells (Serag et al., 2011; Demirer et al., 2019a). Carbon nanotubes have been used to perform stable genetic transformation in bacterial (Castillo et al., 2021; Weise et al., 2022) and mammalian (Golestanipour et al., 2018) cells. In our recent study, we used for the first time an efficient polyethylenimine (PEI)-walled carbon nanotube (SWNT) diffusion method to introduce the LTR retrotransposon plasmid DNA into Moso bamboo plants without transgene integration (Papolu et al., 2021) (Figure 2). We found that internalization of nanoparticles in the intact plant cells resulted in increased GFP expression in the leaves after 72 hours. The carbon nanotubes enable the transport of plasmids without integration of transgenes into crop plants (Kwak et al., 2019). GFP were expressed in various tissues such as roots, leaves, protoplasts, and immature tissues (Ali et al., 2022). Enhanced GFP expression in leaf protoplasts by the use of carbon nanomaterials has been demonstrated in arugula (Eruca sativa), Gossypium hirsutum (cotton), and wheat (Triticum aestivum) (Demirer et al., 2019b; Kwak et al., 2019). The use of nanoparticle-mediated transformation has also been demonstrated for siRNA gene silencing production (Demirer et al., 2020; Zhang et al., 2021). Further focused studies on LTR retrotransposon delivery system are required to explore the molecular mechanisms of LTR retrotransposons in the plant genome.
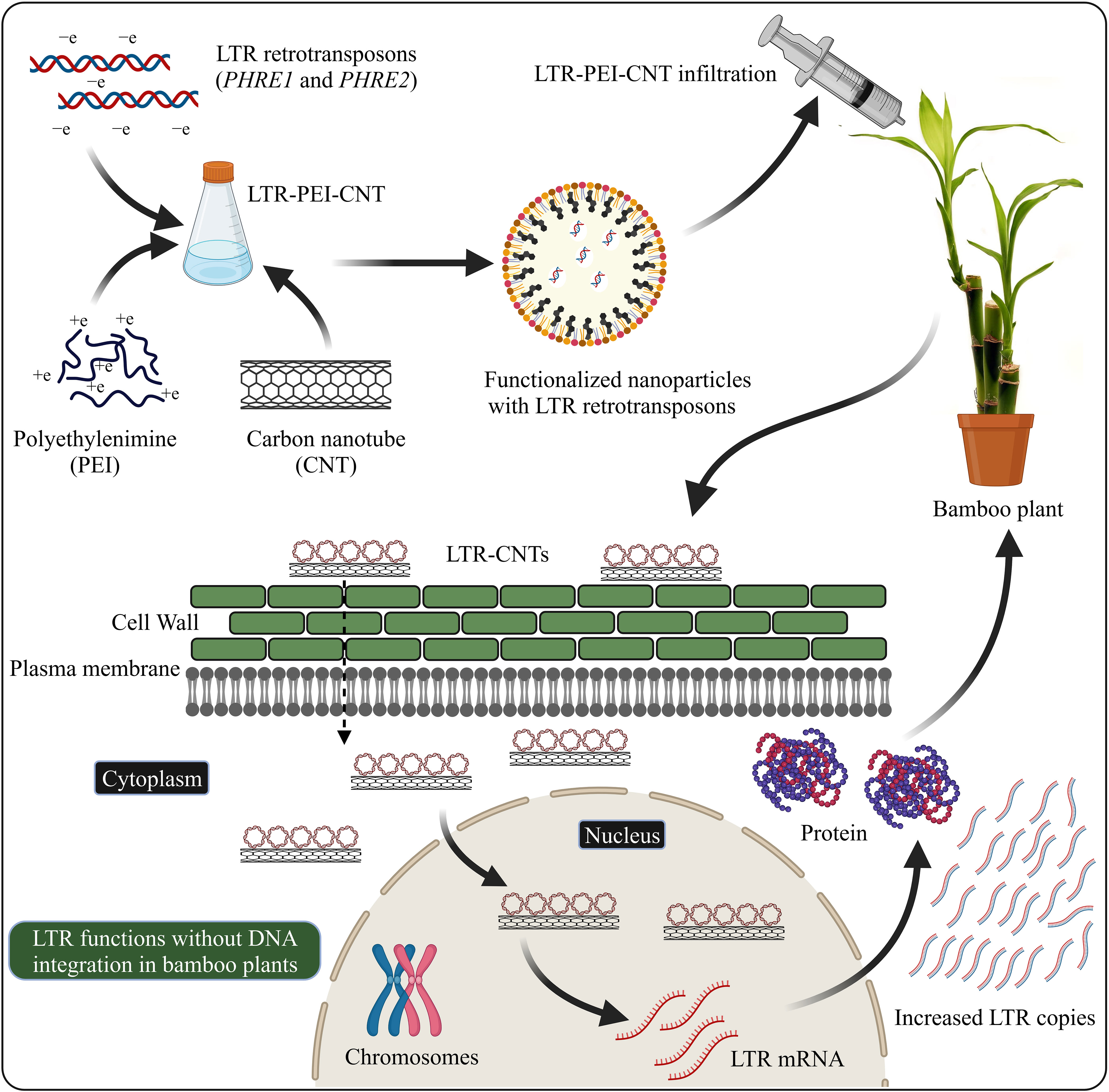
Figure 2 LTR retrotransposons with carboxylated carbon nanotubes (COOH-CNTs) and polyethylenimine (PEI) delivery into bamboo leaves. Covalently modified COOH-CNTs with PEI carrying a net positive charge are incubated with negatively charged LTR vectors. Bamboo leaves infiltrated with LTR–CNTs produce LTR transcripts and proteins and increased LTR copies, without the genome integration. The schematic representation is based on Demirer et al., (2019a) and Papolu et al. (2021) and created with BioRender.com.
LTR retrotransposons are greatly responsible for plant genome evolution and are enriched in the pericentromeric region of host genomes. Active retrotransposable elements are also highly mutagenic and often target protein-coding genes for insertion. In addition, these elements cause chromosome breakage, illegitimate recombination, and genome rearrangement. Therefore, active retrotransposable elements are recognized to play a central role in maintaining chromatin structures, centromeric functions, and regulation of gene expression in their hosts (Shapiro, 2014). Moreover, they are largely responsible for plant genome size variation (Girard and Freeling, 1999; Slotkin and Martienssen, 2007). Centromeric sequences play a central role in chromosome distribution during the mitotic and meiotic cell lifecycle (de Castro Nunes et al., 2018). Centromeric retrotransposons (CR) were first discovered in the grass as centromere-specific sequences (Miller et al., 1998; Presting et al., 1998). Remarkably, in plants, they are usually surrounded and dispersed by LTR retrotransposon sequences (Neumann et al., 2011). The centromere-targeting retrotransposable elements can replace centromeric tandem repeats that bind centromere-specific proteins and may act as a substrate for the efficient repair of frequent double-stranded breaks (Presting, 2018). The centromere-specific histone H3 (CENH3)-associated sequences of centromeric retrotransposons and satellite DNAs are the important structural elements in epigenetic centromere function (Keçeli et al., 2020). Retrotransposable elements can be used to deduce centromere positions, as some elements target active centromeres during integration (Presting, 2018). However, the roles of retrotransposable elements in centromere functions remain unclear. Centromere-targeting elements may be able to replace centromeric tandem repeats. Therefore, centromeric retrotransposons of several plant species have been investigated during the last two decades.
In wheat, the FISH analysis revealed that the sequence of pHind258 was homologous to integrase and the LTRs of centromeric Ty3-gypsy retrotransposons of cereal species (Ito et al., 2004). A 67-kb clone (R11H) containing Ty3/gypsy retrotransposon-related sequences was also identified, which showed strong hybridization signals on the centromeres (Fukui et al., 2001). The expansion of centromeric retrotransposon sequences on dicentric chromosomes to chromosome arms and the formation of multiple centromeres in wheat-rye hybrids may be responsible for chromosome breakage in the next-generation offspring and may be associated with chromosomal rearrangement, stability, and novel chromosome formations (Guo et al., 2016).
In cotton species, centromere-associated sequences are composed of A and D genomes, and the location of the functional centromere co-localizes with centromere retrotransposon hybridization on metaphase mitotic chromosomes. Additionally, FISH and dot-blot hybridization revealed that centromere retrotransposons are present only in D-genome diploid species, indicating that retrotransposons may have invaded the A-genome centromere from the D genome during allopolyploidization (Luo et al., 2012). In addition, LTRs generated from a sequenced bacterial artificial chromosome (BAC) were located in the D progenitor in Gossypium raimondii but not in the A progenitor G. herbaceum, indicating that the centromeric regions of triploid cotton may be derived from D progenitor (Zhang et al., 2014). Moreover, centromeric retrotransposable elements from the different progenitor genomes may become activated during genomic instability following allopolyploidization (Divashuk et al., 2016). Thus, allopolyploid offers an opportunity to understand the evolution of centromeric sequences from resident TEs (Hartley and O’Neill, 2019).
In maize, centromeric retrotransposons represent a transcriptionally active component of centromeres from a wide range of angiosperm species and play a central role in plant centromere evolution (Neumann et al., 2011). However, a recent study revealed that the centromeric retrotransposons can give rise to CRM1 and CRM4 tandem repeats in maize. Nevertheless, maize centromeres are fluid genomic regions whose borders are heavily influenced by the interplay of retrotransposons and epigenetic marks. Distinct CRM1TR sequence variation may lead to gene conversion, which is the main cause of sequence variation and may increase the size of the satellite repeat locus (Sharma et al., 2013). Furthermore, FISH and chromatin immunoprecipitation (ChIP) with anti-CENH3 antibodies in maize and soybean revealed that centromeres differ in size and contain a higher density of CENH3 chip reads, indicating that the tandem satellite repeats and interspersed centromeric retrotransposons may be shaped primarily by retrotransposons (Wolfgruber et al., 2009; Tek et al., 2010). Additionally, various repetitive elements in maize, including centromeric retrotransposon, CentC, and CentA, are found preferentially near the centromeres of the A chromosome hybridized to distinct sites from centromere on the B chromosome, revealing a high concentration of centromeric repeats at the major location on the B chromosome (Lamb et al., 2005). A comparative genomic analysis of centromeric retrotransposons in maize revealed that the maize B chromosome co-existed with the A chromosome during retrotransposition, suggesting that the B chromosome had its origins from A chromosome elements (Theuri et al., 2005). The cores of maize centromeres contain primarily CentC arrays and a cluster of centromere-specific retrotransposons of maize. The structural relationship between CentC, centromeric retrotransposons, and CENH3 was visualized by sequential detection procedure on stretched centromeres, demonstrating that the maize centromeres constantly incorporate oat CENH3 nucleosomes (Jin et al., 2005).
Tobacco cell lines have been identified with an expression of a HaloTag7-fused CENH3 centromeric-tandem repetitive DNA sequences located with CENH3 by a HaloTag7-based chromatin affinity purification system. Further, FISH and ChIP analysis indicated that repeats were chromosome-specific centromeric retrotransposons (Nagaki et al., 2012). Moreover, the centromeric retrotransposons derived from BAC clones act as centromeric DNA sequences in tobacco and the estimated amplification timings of centromeric retrotransposons were different in the two ancestral diploid species of tobacco, indicating that retrotransposons accumulate especially in CENH3-binding regions of tobacco species (Nagaki et al., 2011).
In Brassica species, centromere retrotransposons are the major repeats in centromeric and pericentromeric heterochromatin, and the distribution of the species in allotetraploid relatives indicates that repetitive elements are A-genome specific (Lim et al., 2007). In addition, ChIP and immunostaining analysis with anti-CENH3 antibodies showed that both centromere-specific retrotransposons and centromeric tandem repeats represent a dominant component of the diploid and allotetraploid Brassica species and are directly associated with CENH3 proteins (Wang et al., 2011). Recently, the centromeric-specific retrotransposon in Brassica species showed that the centromeric repeats spread and proliferated between the diploid species possessing A, B, or C genomes after polyploidization, implying that centromeric retrotransposons are particularly important in the evolution and polyploidization of the Brassica genome (Wang G.-X. et al., 2019). Furthermore, the repetitive elements in Brassica species that are conserved in pericentromeres, sub-telomeres, and telomeres rapidly diverged during the evolution of A/C and B genome lineages. Furthermore, these repeats may be associated with genomic stability and may provide insights into genome evolution during Brassica polyploidization (Koo et al., 2011). BACs derived from the rapid proliferation of nested LTR retrotransposons in Brassica species may play an evolutionarily important role in the formation of centromere regions (Wei et al., 2013).
In rice, the contribution of LTR retrotransposons to the evolution of gene structure and function indicates that Ty3/Gypsy elements are more abundant than Ty1/Copia elements, and the intrachromosomal distribution of retrotransposons across chromosome 10 is non-random with the highest density being present in the pericentromeric region (Gao et al., 2004). Moreover, the structural features of LTR retrotransposons in rice indicated that centromeric retrotransposons and CentO satellite repeats are harbored in the core region of the rice chromosome 4-specific centromere, indicating the fragmental duplication of arrays of satellite repeats is mainly responsible for the amplification of centromere satellite DNA and rapid reshuffling of CentO satellites (Ma and Bennetzen, 2006). Although the centers of rice centromeres are occupied by a CentO satellite repeat and a centromere-specific retrotransposon, the CentO satellite is quantitatively variable among 12 rice chromosomes and is interrupted by centromeric retrotransposons, therefore suggesting that CentO satellite and centromere-specific retrotransposons may be the key DNA components for centromere function in rice (Cheng et al., 2002).
The position of CENH3 nucleosomes in rice centromeres is regularly spaced with 155-bp periodicity on CentO satellite repeats but not on non-CentO sequences, suggesting that centromeric repeats evolve for the stabilization of CENH3 nucleosomes (Zhang et al., 2013). Evidence also suggests that suppression of LTR retrotransposon proliferation through the formation of heterochromatin may be an advantage in large genomes in eukaryotes that have a high content of LTR retrotransposons (Cossu et al., 2017). The centromeric retrotransposons of rice are enriched with heterochromatin and its constitutive sequences are transcribed in all the tested rice organs. The centromeric transcripts are differentially processed into sRNAs, indicating a crucial role in the RNAi-mediated pathway for heterochromatin formation and centromere function (Neumann et al., 2007).
Recently, the phylogenetic relationships of centromeric retrotransposons in grasses show that horizontal transfer of centromeric retrotransposon between oryzoid (rice) and panicoid (maize, sorghum, Setaria, Panicum, and Coix) lineages and interelement recombination are important factors in the evolution of centromeric retrotransposons (Sharma and Presting, 2014).
In sugarcane, the characterization of centromere-associated DNA sequences indicated that centromeric retrotransposable elements and centromeric tandem repeats may directly interact with CENH3 in sugarcane centromeres (Nagaki and Murata, 2005). Moreover, the centromeric satellites had the formation and evolutionary stability for 7 million years and exhibited different ploidy levels and unusually longer monomeric repeats that lacked translation phasing on the CENH3 nucleosomes. This indicates that they originated from a retrotransposon and may form extrachromosomal circular DNAs (eccDNAs) (Huang et al., 2021).
In the grass family, centromere-specific retrotransposons discovered in BAC clones revealed that both centromere-specific and non-centromere-specific repeats are the primary DNA elements of maize centromeres and may play a significant role in grass family evolution (Nagaki et al., 2003). Similarly, a centromeric LTR retrotransposon of Brachypodium distachyon derived from centromeric BAC sequences was found in high copy number and is enriched in B. distachyon centromeric regions, indicating that Brachypodium centromeric retrotransposons are highly divergent among other grass species (Qi et al., 2013).
In the potato genome, retrotransposon-related sequences and satellite repeat-based centromeres can rapidly proliferate from neocentromeres by de novo amplification and can associate with the CENH3 nucleosome (Gong et al., 2012). The LTR retrotransposons identified using BAC inserts in Beta species have a chromodomain that is highly similar to centromeric retrotransposons in rice, maize, and barley. Based on sequence diversity, LTRs may have been transposed within the last 60 000 years, indicating that their large-scale genomic organization and transcriptional activity may play an important structural role in centromeres of chromosomes (Weber and Schmidt, 2009).
The annotations and comparison of the centromeric region of Coffea, which is rich in several centromeric retrotransposon family elements, showed that the role of LTR retrotransposons may be more diverse in plants and may extend beyond the chromodomains (de Castro Nunes et al., 2018). The centromeric region of tomato chromosome 12 is composed of nested repeat sequences, including LTR retrotransposons and chloroplast DNA insertions. A block of CAA trinucleotide microsatellite repeats was found in the centromere and pericentromeric region of chromosome 12, suggesting that microsatellite arrays like CAA blocks may be a component of tomato centromeres (Yang et al., 2005). A high copy number of tandem repeats in Allium species is located in all chromosomes and differs in sequence, structure, chromosome level, and genome organization. These repeats are transcribed and associated with the insertions of retrotransposons and organelle DNA, which can be used for future applications of its association with kinetochore protein CENH3 (Kirov et al., 2020). Likewise, the chromosomal organization of centromeric retrotransposons in the genomes of Allium cepa and A. fistulosum are localized in centromeric regions and the chromosomes of A. fistulosum are expressed less in centromeric regions and were abundant in other chromosomal regions (Kiseleva et al., 2014). Holocentromeres in Rhynchospora pubera is composed of centromeric units interspersing the gene containing chromatin. A cell-dependent shuffling of multiple centromeric units results in the formation of functional centromeres during mitosis; genome-wide analysis indicated that different types of holocentromeres may exist in different species, with and without repetitive elements among eukaryotes (Marques et al., 2015).
In Arabidopsis, the centromere-enriched retrotransposons are significantly diverged between two different species and can target their integration preferentially into the centromere region on each of the different chromosomes in the karyotype (Birchler and Presting, 2012). Furthermore, the structure and organization of centromere-specific retrotransposons and CentO-F satellites in Oryza brachyantha indicate that CentO-F satellites are located within the chromosomal regions and are characterized by tandemly repeated satellite DNA flanked by centromeric retrotransposons. This may explain its potential impact on functional centromeres in Oryza species (Yi et al., 2013). FRetro3 centromeric retrotransposons are located in the functional domains of O. brachyantha centromeres and have replaced centromeric retrotransposons of rice as dominant centromeric retroelements in Oryza species (Gao et al., 2009). The retrotransposon of A. lyrata Tal1 was introduced into Arabidopsis by tissue culture-mediated transformation and showed that the highest retrotransposed copies were found in centromeric repeats of Arabidopsis, which suggests dynamic controls for the evolution of the retrotransposon-rich heterochromatin regions (Tsukahara et al., 2012). Furthermore, the structural heterozygosity and chromosomal rearrangements of tissue-specific retrotransposons and tandem repeat copy number in Aegilops speltoides indicate that the tissue-specific pattern of retrotransposons emerges during cell proliferation and this may reflect the reorganization of individual genomes under rapid environmental changes (Shams and Raskina, 2018). However, significant advancements in epigenetics and different types of plant centromeres may be essential to increase the number of sequenced genomes (Oliveira and Torres, 2018).
Over the last two decades, several varied approaches have been used to study the genomes of many plant species. Studies on agriculturally important plant species are particularly important. Following genome sequencing of crop plants, genome sequencing within the genus should be the next targeted research for genomic analysis. Further research should be conducted on genome organization and comparisons at the chromosome, sequence, functional, and evolutionary levels (Voronova et al., 2020).
Several studies demonstrated that LTR retrotransposons participate in centromere-specific transposition and may be a driving force in plant centromere evolution. However, there are many mechanisms involved in the organization of genome functions and in maintaining complex programs of genome organization. Therefore, studies resolving the questions above require novel technologies in molecular biological, cytogenetic, biochemical, and genetic methods. Such studies may provide a clearer understanding of the relationship between plant evolution and LTR retrotransposons.
Retrotransposable and related elements are highly abundant in eukaryotic genomes and insert into new genomic locations by a mechanism that involves the reverse transcription of an RNA intermediate. Changes in the copy number of repeat elements and internal rearrangements on both homologous chromosomes occur after the induction of recombinational processes during the meiotic prophase. The insertion of LTR retrotransposons is random and occurs in the transposition process in the continuous evolution of a species. This can provide a wealth of information for the study of evolution, species, and genome differentiation.
Retrotransposon-based DNA marker applications have become a key element of research on genetic variability and diversity (Vuorinen et al., 2018; Ghonaim et al., 2020; Kalendar et al., 2021b). The scope of their usage includes creating genetic maps and the identification of individuals or lines carrying certain genetic polymorphic variations (Khapilina et al., 2021a). LTR retrotransposon-derived molecular genetic marker systems have been employed in deciphering the genetic diversity of crop plants (Kalendar et al., 1999; Kalendar and Schulman, 2006; Kalendar et al., 2011; Kalendar et al., 2018; Kalendar et al., 2021a). The retrotransposon-based marker systems are highly effective in detecting the effects of environmental stress on retrotransposon activation (Kalendar et al., 2008; Belyayev et al., 2010). Moreover, the detection of TE expression, including polymorphisms and the diversity of the transposon transcriptional landscape, may provide new insight into host-TE interactions (Lanciano and Cristofari, 2020). In addition, LTR retrotransposons are associated with key genes involved in potential applications of genome assembly, genome variation, gene tagging, and functional analysis of genes, indicating their crucial role as markers in molecular breeding (Potter, 2005).
In pepper (C. annuum), LTR retrotransposons were inserted 6 million years ago and exhibit chromosomal insertional preferences, which may be a useful tool to design species-specific retrotransposon-based markers (Yañez-Santos et al., 2021). The combination of active LTR retrotransposons and Inter-Retrotransposon Amplified Polymorphism (IRAP) markers (Kalendar et al., 1999; Hosid et al., 2012) may be a suitable system for genetic fidelity assessment of tissue-culture-generated plants in sugarcane (Shingote et al., 2019) and better germplasm management in Xanthosoma and Colocasia (Doungous et al., 2015). The IRAP marker system in LTR retrotransposon insertions of flax genome appeared to be suitable for the identification of retrotransposon polymorphisms and showed a high level of plant adaptation in a radioactive environment (Smýkal et al., 2011; Lancíková and Žiarovská, 2020). The IRAP and REMAP markers of the cassava genome produced high polymorphism and may be suitable for the investigation of genetic diversity and relationships among cassava cultivars (Kuhn et al., 2016). A comparative analysis of two LTR retrotransposons, BARE-1 and Jeli, may provide a potential source of polymorphic Sequence-Specific Amplification Polymorphism (SSAP) markers for genetic diversity in diploid wheat (Konovalov et al., 2010). The LTR retrotransposon based SSAP markers in cashew and myrtle genomes exhibited a significantly higher proportion of polymorphic markers than those of AFLP (Waugh et al., 1997; Syed et al., 2005; Woodrow et al., 2012).
The genetic maps generated with several retrotransposon-based markers such as iPBS (inter-Priming Binding Site) (Kalendar et al., 2010; Doungous et al., 2020; Khapilina et al., 2021b) and REMAP (REtrotransposon-Microsatellite Amplified Polymorphism) (Kalendar et al., 1999) exhibited regions of different marker densities, indicating that the distribution of retrotransposons in lentil is non-random and widespread throughout the lentil genome. This may be useful in lentil breeding by marker-assisted selection (Rey-Banos et al., 2017). The development of Retrotransposon-Based Insertion Polymorphism (RBIP) markers (Flavell et al., 1998) derived from sweet potatoes can determine intraspecific variability. These markers can also be used as core primer pairs for evaluating genetic diversity and constructing linkage maps of various plant species, guiding breeding and germplasm research (Meng et al., 2021). The RBIP marker was shown to be duplicated several times during the development of Asian pear cultivars and may provide a comprehensive picture of the complex relationship and evolution of Pyrus species (Jiang et al., 2015). Likewise, genome-wide analysis of RBIP markers in the Melilotus genome revealed considerable polymorphism information content (PIC), indicating that these markers are highly informative and may be used for implementing genetic improvement in the Melilotus genus (Ouyang et al., 2021). Furthermore, RBIP markers used for DNA profiling of Japanese, Chinese, and European pear cultivars revealed that retrotransposons have transposed during Asian pear evolution or reflect the genetic relationship between Asian and European pears. Thus, suitable combinations of retrotransposon insertions may be useful for cultivar-specific DNA markers (Kim et al., 2012). The polymorphism markers generated from several retrotransposon families and the effectiveness of the dominant (IRAP) and codominant (RBIP) marker systems for assessing the genetic diversity among different potato varieties were compared. Distinct DNA profiles for Ty1/Copia and Ty3/Gypsy retrotransposons are active in the genome and may contribute to potato genome organization (Sharma and Nandineni, 2014). High-throughput RBIP data analysis indicated that may strongly support the model of independent domestications for Pisum sativum species, which in turn provides a broad understanding of the diversity and evolution of Pisum (Jing et al., 2010). Likewise, a wide variety of LTR retrotransposon-based markers generated from peas, broad beans, and Norway spruce may be useful in revealing polymorphisms associated with the corresponding retrotransposons within the Pisum genus (Pearce et al., 1999). The non-random distribution of abundant LTR retrotransposons within the lentil genome indicates that defective and non-autonomous retrotransposons are highly frequent and maybe a suitable source of genetic markers for further genetic analysis (Rey-Banos et al., 2017). The novel Ty1/Copia and Ty3/Gypsy LTR retrotransposons derived from Lilium species indicate that they were non-autonomous retrotransposons. IRAP analysis using the LTR sequence of these retrotransposons may provide a new approach to analyzing the species relationship among Lilium species (Lee et al., 2015). In Cleistogenes songorica and strawberry genomes, various LTR retrotransposon-based molecular markers were developed and exhibited a high level of polymorphism frequency and high transferability of polymorphic primer pairs. This suggests that RBIP markers may be useful in future studies on genetic diversity, QTL mapping, population structure, and the evolution of germplasm accessions in C. songorica and related grasses (Monden et al., 2014a; Ma et al., 2022). Several LTR retrotransposon markers derived from chokecherry genome sequences indicated that retrotransposon markers in map construction and genetic mapping may facilitate genetic research in Rosaceous species (Liang et al., 2016).
Evolution is primarily a change in physiological and genetic composition; therefore, variation is a significant process in evolution. Like in most eukaryotes, TEs are the most variable parts of the plant genome (Lisch, 2013). TEs can make dramatic differences in the overall architecture of the genomes of even closely related plant species. Moreover, TEs make up most of all plant DNA (Bennetzen et al., 2005). Gene inactivation is one of the most common TE-induced phenotypic changes. Therefore, the propensity of some TEs to insert into or near genes has been successfully utilized for generating new null mutations (Hsing et al., 2007; Settles et al., 2007; Candela and Hake, 2008), and this is also a major driver of genome size evolution (Hawkins et al., 2006; Neumann et al., 2006; Piegu et al., 2006) Therefore, the evolutionary potential of TEs, especially LTR retrotransposons, should be thoroughly explored to gain a better understanding on the evolutionary characteristics of plants. Retroviruses and LTR retrotransposons share similar gene architecture, but LTR retrotransposons lack the envelop gene and an extracellular stage in their lifecycle. It has been proposed that these retroviruses emerged from the LTR retrotransposon family Ty3/Gypsy by acquiring the envelope gene (Malik and Eickbush, 2001), but this evolutionary relationship is not confirmed.
Genome relationships and LTR retrotransposon diversity can be used to understand the genomic relationship among the members of a genus or family in plants. Recently, genome relationships and LTR retrotransposon diversity in three cultivated Capsicum strains were analyzed and a close relationship among the species was revealed (de Assis et al., 2020). Moreover, genome-wide analysis of LTR retrotransposons and their impact on evolution has been explored in several plants (Roulin et al., 2009; Beulé et al., 2015; Giordani et al., 2016; Mascagni et al., 2017; Keidar et al., 2018; Liu et al., 2018; Akakpo et al., 2020; Mascagni et al., 2020; Ouyang et al., 2021). TE amplification is the main mechanism behind plant genome size increase and evolution (Gantuz et al., 2022). The proliferation of LTR retrotransposons is related to genome reorganization caused by hybridization or polyploidization (Vicient and Casacuberta, 2017). Moreover, allopolyploidization is associated with rapid structural and functional alterations of genomes (Leitch and Leitch, 2008) and this is recognized as the major mechanism behind adaptation and speciation in the plant kingdom (Ramsey and Schemske, 1998). In addition, polyploidy increases genome size and activates TE amplification, and the resultant genome rearrangement may alter their balance in epigenetic silencing (O'Neill et al., 1998; Ozkan et al., 2001; Madlung et al., 2005; Petit et al., 2007; Parisod et al., 2009). TEs are known to associate with recombination-driven sequence loss that leads to major structural changes (Parisod et al., 2010). In plants, TE abundance is correlated with the recombination rate of some TE families (Daron et al., 2014). In maize, LTR retrotransposons are enriched in regions of low recombination (Stitzer et al., 2021). Moreover, a negative correlation between LTR retrotransposons and recombination was also reported in many other plant species, such as soya bean, rice, and bread wheat (Tian et al., 2009; Tian et al., 2012; Daron et al., 2014). Angiosperm genomes are unstable at the level of chromosome number, genome size, and repetitive DNA content; most genes are found as single-gene groups surrounded by nested TEs (Sanmiguel and Bennetzen, 1998). Furthermore, in maize, any two alleles of the same gene diverged >2 million years ago (Wang and Dooner, 2006). Although gene content and organization are mostly similar, variation in copy number and gene order has been observed in grass plants (Bennetzen, 2007; Springer et al., 2009). However, variation in copy number and its influence on genome order and evolution should be explored to gain a better understanding of the influence of LTR retrotransposons in plant evolution.
In general, LTR retrotransposons are one of the key elements that drive evolution by mechanisms of recombination and gene duplications. Moreover, TEs affect the genome when the mobile elements are closer to the genome or even from a considerable distance. This is because TEs can move. Therefore, TEs, especially LTR retrotransposons, have a significant role in the evolution of the plant kingdom because of their wide occurrence. Further focused studies are required to explore the role and the exact process of LTR retrotransposons in plant evolution, which may provide further insight into the molecular mechanisms of evolution in the plant kingdom.
1. Retrotransposable elements represent up to 90% of the total genome in most plants. Several studies describe the role of LTR retrotransposons in epigenetic regulation. Exploring the role of retrotransposable elements leading to phenotypic variation and its regulation in plants may have significant economic importance in the field of plant breeding and agriculture.
2. Investigations on the role of LTR retrotransposons, especially the Ty1/Copia and the Ty3/Gypsy superfamilies, may reveal their roles in heat stress regulation, which will provide a further understanding of the possibilities of developing smart crops that are resistant to heat stresses due to global warming.
3. Genetic engineering methods and epigenetic modifications using LTR retrotransposons may have future scope in the field of smart agriculture by developing smarter crops.
4. Further research should focus on profiling sRNAs in isolated cell types and single cells. This may further understand the specificities and interplay between the different gene-silencing mechanisms in plant cells.
5. There is currently a limited understanding of most sRNA classes that contribute to biotic and abiotic stress and the transgenerational inheritance and stability of acquired sRNA-based responses. This should be a focus of further research in the development of stress-resilient crops and plant breeding in general.
6. LTR retrotransposons participate in centromere-specific transposition and may be a driving force in plant centromere evolution. Further studies should focus on genome organization and comparisons at the chromosome, sequence, functional, and evolutionary levels.
7. Genome-sequencing studies on agriculturally important plant species are important; genome sequencing within the genus should be targeted for subsequent research.
8. Significant advancements in epigenetics and different types of plant centromeres are required to increase the number of sequenced genomes. This increase should further understand the relationship between plant evolution and LTR retrotransposons.
9. Further investigations are necessary to gain a better understanding of the variation in copy number of LTR retrotransposons and its influence on evolution and genetic variation.
10. TEs, especially LTR retrotransposons, contribute significantly to intraspecific phenotypic variation in plants. Therefore, understanding the dynamics governing LTR retrotransposons is a crucial research focus for evolutionary biologists.
Retrotransposons are a class of mobile genetic elements that are universally distributed in plant genomes. Their distribution and transposition activities are significantly associated with plant evolution. Several studies of LTR retrotransposons have provided valuable insights into the mechanism of the genome evolution of plants. The genomes of most plant species exhibit dynamic variations in size and other structural features of LTRs. In chromatin modification, reduced DNA methylation often promotes the expression of retrotransposons. A wide variety of genetic factors are responsible for retrotransposon expressions, such as miRNAs, ncRNAs, piRNAs, RdRPs, risiRNAs, siRNAs, ta-siRNAs, ra-siRNAs, nat-siRNAs, dsRNAs, endo-siRNAs, viRNAs, heterochromatin, DNA methylation, histone post-translational modifications, and gene silencing pathways. Moreover, the potential biological functions of plant sRNAs to acquire information from different tissues and shift it across generations may improve future plant research. The development of RNA biogenesis mechanisms leads to the regulation of biological processes coupled with plant development and environmental responses. Retrotransposable elements, considered a kind of genetic pool, have tremendous potential in genome analysis, biodiversity research, gene mapping, gene cloning, and functional analysis.
A high proportion of LTR retrotransposons are involved in multiple epigenetic mechanisms, including stress tolerance, transpositional activity, regulation of gene expression, DNA methylation, histone modification, and chromatin remodeling and their interconnected networks in the plant genome. Increasing research interest in such epigenetic mechanisms may contribute to a greater understanding of their central role in genome organization and evolution. Therefore, an integrated TE database with epigenetic information will be a valuable resource for future research focused on assessing the possible contribution of LTR retrotransposons to develop single-molecule real-time sequencing and transcriptome variations resulting from advancements of genome annotation and investigations of plant genetic diversity. Moreover, advancement in the forthcoming reference genomes in association with novel sequence technologies may lead to the implementation of long-read sequencing. This will further enhance understanding of various aspects of genome disruption of LTR retrotransposons.
Environmental stresses affecting crops grown under field conditions are a major threat in the global warming era, and the activities of retrotransposons show a close relationship with such stresses. During environmental stress, LTR retrotransposons are more active and induce mutational and insertional polymorphisms. LTR retrotransposon-mediated molecular genetic markers are a highly polymorphic and efficient system. Moreover, as this does not influence genetic structure across species, DNA marker investigations will be a promising tool for exploring crop diversity and germplasm. Furthermore, several studies revealed that centromere-specific retrotransposons are conserved in pericentromeres, sub-telomeres, and telomeres and have rapidly diverged during the evolution of A, B, and C diploid genome lineages. Moreover, recent developments in genomics-based on whole-genome sequencing and 3D nuclear organization, allele-specific histone modification, and RNA Pol II binding profiles may facilitate the understanding of epigenetic regulation of differential gene expression between homologous chromosomes.
Another consideration is nanoparticle-based biomolecule delivery systems. In these systems, biomolecules such as DNA, RNA, and protein can be efficiently delivered and incorporated into the plant genome. This method can be utilized to make desirable epigenetic modifications in crop plants. In addition, high-throughput sequencing technology combined with artificial intelligence approaches for big data analysis may be beneficial in providing a more comprehensive picture of the interplay between LTR retrotransposon-induced epigenetic changes. Further collaborative studies are required to understand the complexity of LTR retrotransposons in evolutionary and organismal biology.
PKP and MR planned, designed, and wrote the review. PKP, MR, SM, QW, RK, PY, and MZ outlined and edited the review. PKP, MR, RK, QW, SM, LHZ, ZA, KKV, PY, and MZ edited and revised the review.
This work was funded by a grant from the National Natural Science Foundation of China (Grant Nos 31870656). This review was also supported by grants from the Zhejiang Provincial Natural Science Foundation of China (Grant No. LZ19C160001). The authors are grateful for the support of Metasequoia Faculty Research Start-up Funding (grant number 163100028) at the Bamboo Research Institute, Nanjing Forestry University for the co-first author MR.
The authors wish to thank the University of Helsinki Language Centre, Finland for the outstanding editing and proofreading of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1064847/full#supplementary-material
Akakpo, R., Carpentier, M. C., Ie Hsing, Y., Panaud, O. (2020). The impact of transposable elements on the structure, evolution and function of the rice genome. New Phytol. 226, 44–49. doi: 10.1111/nph.16356
Ali, Z., Serag, M. F., Demirer, G. S., Torre, B., di Fabrizio, E., Landry, M. P., et al. (2022). DNA–Carbon nanotube binding mode determines the efficiency of carbon nanotube-mediated DNA delivery to intact plants. ACS Appl. Nano Mater. 5, 4663–4676. doi: 10.1021/acsanm.1c03482
Belyayev, A., Kalendar, R., Brodsky, L., Nevo, E., Schulman, A. H., Raskina, O. (2010). Transposable elements in a marginal plant population: Temporal fluctuations provide new insights into genome evolution of wild diploid wheat. Mobile DNA 1, 1–16. doi: 10.1186/1759-8753-1-6
Bennetzen, J. L. (2007). Patterns in grass genome evolution. Curr. Opin. Plant Biol. 10, 176–181. doi: 10.1016/j.pbi.2007.01.010
Bennetzen, J. L., Ma, J., Devos, K. M. (2005). Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 95, 127–132. doi: 10.1093/aob/mci008
Benoit, M., Drost, H.-G., Catoni, M., Gouil, Q., Lopez-Gomollon, S., Baulcombe, D., et al. (2019). Environmental and epigenetic regulation of rider retrotransposons in tomato. PloS Genet. 15, e1008370. doi: 10.1371/journal.pgen.1008370
Beulé, T., Agbessi, M. D., Dussert, S., Jaligot, E., Guyot, R. (2015). Genome-wide analysis of LTR-retrotransposons in oil palm. BMC Genomics 16, 1–14. doi: 10.1186/s12864-015-2023-1
Birchler, J. A., Presting, G. G. (2012). Retrotransposon insertion targeting: A mechanism for homogenization of centromere sequences on nonhomologous chromosomes. Genes Dev. 26, 638–640. doi: 10.1101/gad.191049.112
Boeke, J. D., Garfinkel, D. J., Styles, C. A., Fink, G. R. (1985). Ty Elements transpose through an RNA intermediate. Cell 40, 491–500. doi: 10.1016/0092-8674(85)90197-7
Bond, D. M., Baulcombe, D. C. (2015). Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in arabidopsis thaliana. Proc. Natl. Acad. Sci. 112, 917–922. doi: 10.1073/pnas.1413053112
Boonjing, P., Masuta, Y., Nozawa, K., Kato, A., Ito, H. (2020). The effect of zebularine on the heat-activated retrotransposon ONSEN in arabidopsis thaliana and vigna angularis. Genes Genet. Syst. 95 (4), 165–172. doi: 10.1266/ggs.19-00046
Borges, F., Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Bourque, G., Burns, K. H., Gehring, M., Gorbunova, V., Seluanov, A., Hammell, M., et al. (2018). Ten things you should know about transposable elements. Genome Biol. 19, 1–12. doi: 10.1186/s13059-018-1577-z
Brown, P. O., Bowerman, B., Varmus, H. E., Bishop, J. M. (1987). Correct integration of retroviral DNA in vitro. Cell 49, 347–356. doi: 10.1016/0092-8674(87)90287-X
Buchmann, R. C., Asad, S., Wolf, J. N., Mohannath, G., Bisaro, D. M. (2009). Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83, 5005–5013. doi: 10.1128/JVI.01771-08
Candela, H., Hake, S. (2008). The art and design of genetic screens: Maize. Nat. Rev. Genet. 9, 192–203. doi: 10.1038/nrg2291
Cao, Y., Jiang, Y., Ding, M., He, S., Zhang, H., Lin, L., et al. (2015). Molecular characterization of a transcriptionally active Ty1/copia-like retrotransposon in gossypium. Plant Cell Rep. 34, 1037–1047. doi: 10.1007/s00299-015-1763-3
Capy, P. (2005). Classification and nomenclature of retrotransposable elements. Cytogenetic Genome Res. 110, 457–461. doi: 10.1159/000084978
Castillo, A. E. D. R., De León-Rodriguez, A., Terrones, M., de la Rosa, A. P. B. (2021). Multi-walled carbon nanotubes enhance the genetic transformation of bifidobacterium longum. Carbon 184, 902–909. doi: 10.1016/j.carbon.2021.08.052
Catlin, N. S., Josephs, E. B. (2022). The important contribution of transposable elements to phenotypic variation and evolution. Curr. Opin. Plant Biol. 65, 102140. doi: 10.1016/j.pbi.2021.102140
Cavrak, V. V., Lettner, N., Jamge, S., Kosarewicz, A., Bayer, L. M., Mittelsten Scheid, O. (2014). How a retrotransposon exploits the plant's heat stress response for its activation. PloS Genet. 10, e1004115. doi: 10.1371/journal.pgen.1004115
Cheng, Z., Dong, F., Langdon, T., Ouyang, S., Buell, C. R., Gu, M., et al. (2002). Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14, 1691–1704. doi: 10.1105/tpc.003079
Cossu, R. M., Casola, C., Giacomello, S., Vidalis, A., Scofield, D. G., Zuccolo, A. (2017). LTR Retrotransposons show low levels of unequal recombination and high rates of intraelement gene conversion in large plant genomes. Genome Biol. Evol. 9, 3449–3462. doi: 10.1093/gbe/evx260
Creasey, K. M., Zhai, J., Borges, F., Van Ex, F., Regulski, M., Meyers, B. C., et al. (2014). miRNAs trigger widespread epigenetically activated siRNAs from transposons in arabidopsis. Nature 508, 411–415. doi: 10.1038/nature13069
Cunningham, F. J., Goh, N. S., Demirer, G. S., Matos, J. L., Landry, M. P. (2018). Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 36, 882–897. doi: 10.1016/j.tibtech.2018.03.009
Daron, J., Glover, N., Pingault, L., Theil, S., Jamilloux, V., Paux, E., et al. (2014). Organization and evolution of transposable elements along the bread wheat chromosome 3B. Genome Biol. 15, 1–15. doi: 10.1186/s13059-014-0546-4
de Assis, R., Baba, V. Y., Cintra, L. A., Gonçalves, L. S. A., Rodrigues, R., Vanzela, A. L. L. (2020). Genome relationships and LTR-retrotransposon diversity in three cultivated capsicum L.(Solanaceae) species. BMC Genomics 21, 1–14. doi: 10.1186/s12864-020-6618-9
de Castro Nunes, R., Orozco-Arias, S., Crouzillat, D., Mueller, L. A., Strickler, S. R., Descombes, P., et al. (2018). Structure and distribution of centromeric retrotransposons at diploid and allotetraploid coffea centromeric and pericentromeric regions. Front. Plant Sci. 9, 175. doi: 10.3389/fpls.2018.00175
Demirer, G. S., Zhang, H., Goh, N. S., González-Grandío, E., Landry, M. P. (2019a). Carbon nanotube–mediated DNA delivery without transgene integration in intact plants. Nat. Protoc. 14, 2954–2971. doi: 10.1038/s41596-019-0208-9
Demirer, G. S., Zhang, H., Goh, N. S., Pinals, R. L., Chang, R., Landry, M. P. (2020). Carbon nanocarriers deliver siRNA to intact plant cells for efficient gene knockdown. Sci. Adv. 6, eaaz0495. doi: 10.1126/sciadv.aaz0495
Demirer, G. S., Zhang, H., Matos, J. L., Goh, N. S., Cunningham, F. J., Sung, Y., et al. (2019b). High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 14, 456–464. doi: 10.1038/s41565-019-0382-5
Divashuk, M. G., Khuat, T. M. L., Kroupin, P. Y., Kirov, I. V., Romanov, D. V., Kiseleva, A. V., et al. (2016). Variation in copy number of Ty3/Gypsy centromeric retrotransposons in the genomes of thinopyrum intermedium and its diploid progenitors. PloS One 11, e0154241. doi: 10.1371/journal.pone.0154241
Dong, Q., Hu, B., Zhang, C. (2022). microRNAs and their roles in plant development. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.824240
Doungous, O., Kalendar, R., Adiobo, A., Schulman, A. H. (2015). Retrotransposon molecular markers resolve cocoyam (Xanthosoma sagittifolium) and taro (Colocasia esculenta) by type and variety. Euphytica 206, 541–554. doi: 10.1007/s10681-015-1537-6
Doungous, O., Kalendar, R., Filippova, N., Ngane, B. K. (2020). Utility of iPBS retrotransposons markers for molecular characterization of African gnetum species. Plant Biosystems 154 (5), 587–592. doi: 10.1080/11263504.2019.1651782
Durán-Figueroa, N., Vielle-Calzada, J.-P. (2010). ARGONAUTE9-dependent silencing of transposable elements in pericentromeric regions of arabidopsis. Plant Signaling Behav. 5, 1476–1479. doi: 10.4161/psb.5.11.13548
Flavell, A. J., Knox, M. R., Pearce, S. R., Ellis, T. N. (1998). Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J. 16, 643–650. doi: 10.1046/j.1365-313x.1998.00334.x
Fukui, K.-N., Suzuki, G., Lagudah, E. S., Rahman, S., Appels, R., Yamamoto, M., et al. (2001). Physical arrangement of retrotransposon-related repeats in centromeric regions of wheat. Plant Cell Physiol. 42, 189–196. doi: 10.1093/pcp/pce026
Galindo-González, L., Mhiri, C., Deyholos, M. K., Grandbastien, M.-A. (2017). LTR-Retrotransposons in plants: Engines of evolution. Gene 626, 14–25. doi: 10.1016/j.gene.2017.04.051
Gantuz, M., Morales, A., Bertoldi, M. V., Ibañez, V. N., Duarte, P. F., Marfil, C. F., et al. (2022). Hybridization and polyploidization effects on LTR-retrotransposon activation in potato genome. J. Plant Res. 135, 81–92. doi: 10.1007/s10265-021-01354-9
Gao, D., Gill, N., Kim, H. R., Walling, J. G., Zhang, W., Fan, C., et al. (2009). A lineage-specific centromere retrotransposon in oryza brachyantha. Plant J. 60, 820–831. doi: 10.1111/j.1365-313X.2009.04005.x
Gao, L., McCarthy, E. M., Ganko, E. W., McDonald, J. F. (2004). Evolutionary history of oryza sativa LTR retrotransposons: A preliminary survey of the rice genome sequences. BMC Genomics 5, 1–18. doi: 10.1186/1471-2164-5-18
Gaubert, H., Sanchez, D. H., Drost, H.-G., Paszkowski, J. (2017). Developmental restriction of retrotransposition activated in arabidopsis by environmental stress. Genetics 207, 813–821. doi: 10.1534/genetics.117.300103
Ghonaim, M., Kalendar, R., Barakat, H., Elsherif, N., Ashry, N., Schulman, A. H. (2020). High-throughput retrotransposon-based genetic diversity of maize germplasm assessment and analysis. Mol. Biol. Rep. 47, 1589–1603. doi: 10.1007/s11033-020-05246-4
Giordani, T., Cossu, R. M., Mascagni, F., Marroni, F., Morgante, M., Cavallini, A., et al. (2016). Genome-wide analysis of LTR-retrotransposon expression in leaves of populus× canadensis water-deprived plants. Tree Genet. Genomes 12, 1–14. doi: 10.1007/s11295-016-1036-5
Girard, L., Freeling, M. (1999). Regulatory changes as a consequence of transposon insertion. Dev. Genet. 25, 291–296. doi: 10.1002/(SICI)1520-6408(1999)25:4<291::AID-DVG2>3.0.CO;2-5
Golestanipour, A., Nikkhah, M., Aalami, A., Hosseinkhani, S. (2018). Gene delivery to tobacco root cells with single-walled carbon nanotubes and cell-penetrating fusogenic peptides. Mol. Biotechnol. 60, 863–878. doi: 10.1007/s12033-018-0120-5
Gong, Z., Wu, Y., Koblížková, A., Torres, G. A., Wang, K., Iovene, M., et al. (2012). Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24, 3559–3574. doi: 10.1105/tpc.112.100511
Goodier, J. L. (2016). Restricting retrotransposons: a review. Mobile DNA 7, 1–30. doi: 10.1186/s13100-016-0070-z
Gorbunova, V., Seluanov, A., Mita, P., McKerrow, W., Fenyö, D., Boeke, J. D., et al. (2021). The role of retrotransposable elements in ageing and age-associated diseases. Nature 596, 43–53. doi: 10.1038/s41586-021-03542-y
Grandbastien, M.-A. (2015). LTR Retrotransposons, handy hitchhikers of plant regulation and stress response. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 1849, 403–416. doi: 10.1016/j.bbagrm.2014.07.017
Grandbastien, M.-A., Audeon, C., Bonnivard, E., Casacuberta, J., Chalhoub, B., Costa, A.-P., et al. (2005). Stress activation and genomic impact of Tnt1 retrotransposons in solanaceae. Cytogenetic Genome Res. 110, 229–241. doi: 10.1159/000084957
Guo, X., Su, H., Shi, Q., Fu, S., Wang, J., Zhang, X., et al. (2016). De novo centromere formation and centromeric sequence expansion in wheat and its wide hybrids. PloS Genet. 12, e1005997. doi: 10.1371/journal.pgen.1005997
Hartley, G., O’Neill, R. J. (2019). Centromere repeats: Hidden gems of the genome. Genes 10, 223. doi: 10.3390/genes10030223
Hawkins, J. S., Kim, H., Nason, J. D., Wing, R. A., Wendel, J. F. (2006). Differential lineage-specific amplification of transposable elements is responsible for genome size variation in gossypium. Genome Res. 16, 1252–1261. doi: 10.1101/gr.5282906
He, P., Ma, Y., Dai, H., Li, L., Liu, Y., Li, H., et al. (2012). Characterization of the hormone and stress-induced expression of FaRE1 retrotransposon promoter in strawberry. J. Plant Biol. 55, 1–7. doi: 10.1007/s12374-011-9180-9
Hollister, J. D., Smith, L. M., Guo, Y.-L., Ott, F., Weigel, D., Gaut, B. S. (2011). Transposable elements and small RNAs contribute to gene expression divergence between arabidopsis thaliana and arabidopsis lyrata. Proc. Natl. Acad. Sci. 108, 2322–2327. doi: 10.1073/pnas.1018222108
Hosid, E., Brodsky, L., Kalendar, R., Raskina, O., Belyayev, A. (2012). Diversity of long terminal repeat retrotransposon genome distribution in natural populations of the wild diploid wheat Aegilops speltoides. Genetics 190, 263–274. doi: 10.1534/genetics.111.134643
Hsing, Y.-I., Chern, C.-G., Fan, M.-J., Lu, P.-C., Chen, K.-T., Lo, S.-F., et al. (2007). A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 63, 351–364. doi: 10.1007/s11103-006-9093-z
Huang, Y., Ding, W., Zhang, M., Han, J., Jing, Y., Yao, W., et al. (2021). The formation and evolution of centromeric satellite repeats in saccharum species. Plant J. 106, 616–629. doi: 10.1111/tpj.15186
Ito, H. (2012). Small RNAs and transposon silencing in plants. Development Growth Differ. 54, 100–107. doi: 10.1111/j.1440-169X.2011.01309.x
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., Paszkowski, J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119. doi: 10.1038/nature09861
Ito, H., Kim, J.-M., Matsunaga, W., Saze, H., Matsui, A., Endo, T. A., et al. (2016). A stress-activated transposon in arabidopsis induces transgenerational abscisic acid insensitivity. Sci. Rep. 6, 1–12. doi: 10.1038/srep23181
Ito, H., Nasuda, S., Endo, T. R. (2004). Erratum: A direct repeat sequence associated with the centromeric retrotransposons in wheat. Genome 47, 998–998. doi: 10.1139/g04-901
Ito, H., Yoshida, T., Tsukahara, S., Kawabe, A. (2013). Evolution of the ONSEN retrotransposon family activated upon heat stress in brassicaceae. Gene 518, 256–261. doi: 10.1016/j.gene.2013.01.034
Ivashuta, S., Naumkina, M., Gau, M., Uchiyama, K., Isobe, S., Mizukami, Y., et al. (2002). Genotype-dependent transcriptional activation of novel repetitive elements during cold acclimation of alfalfa (Medicago sativa). Plant J. 31, 615–627. doi: 10.1046/j.1365-313X.2002.01383.x
Jedlicka, P., Lexa, M., Vanat, I., Hobza, R., Kejnovsky, E. (2019). Nested plant LTR retrotransposons target specific regions of other elements, while all LTR retrotransposons often target palindromes and nucleosome-occupied regions: In silico study. Mobile DNA 10, 1–14. doi: 10.1186/s13100-019-0186-z
Jia, H. M., Jia, H. J., Cai, Q. L., Wang, Y., Zhao, H. B., Yang, W. F., et al. (2019). The red bayberry genome and genetic basis of sex determination. Plant Biotechnol. J. 17, 397–409. doi: 10.1111/pbi.12985
Jia, Y., Lisch, D. R., Ohtsu, K., Scanlon, M. J., Nettleton, D., Schnable, P. S. (2009). Loss of RNA–dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PloS Genet. 5, e1000737. doi: 10.1371/journal.pgen.1000737
Jiang, S., Zong, Y., Yue, X., Postman, J., Teng, Y., Cai, D. (2015). Prediction of retrotransposons and assessment of genetic variability based on developed retrotransposon-based insertion polymorphism (RBIP) markers in pyrus l. Mol. Genet. Genomics 290, 225–237. doi: 10.1007/s00438-014-0914-5
Jing, R., Vershinin, A., Grzebyta, J., Shaw, P., Smýkal, P., Marshall, D., et al. (2010). The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evol. Biol. 10, 1–20. doi: 10.1186/1471-2148-10-44
Jin, W., Lamb, J. C., Vega, J. M., Dawe, R. K., Birchler, J. A., Jiang, J. (2005). Molecular and functional dissection of the maize b chromosome centromere. Plant Cell 17, 1412–1423. doi: 10.1105/tpc.104.030643
Kalendar, R., Amenov, A., Daniyarov, A. (2018). Use of retrotransposon-derived genetic markers to analyse genomic variability in plants. Funct. Plant Biol. 46, 15–29. doi: 10.1071/FP18098
Kalendar, R., Antonius, K., Smýkal, P., Schulman, A. H. (2010). iPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 121, 1419–1430. doi: 10.1007/s00122-010-1398-2
Kalendar, R., Flavell, A., Ellis, T., Sjakste, T., Moisy, C., Schulman, A. H. (2011). Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 106, 520–530. doi: 10.1038/hdy.2010.93
Kalendar, R., Grob, T., Regina, M., Suoniemi, A., Schulman, A. (1999). IRAP and REMAP: Two new retrotransposon-based DNA fingerprinting techniques. Theor. Appl. Genet. 98, 704–711. doi: 10.1007/s001220051124
Kalendar, R., Raskina, O., Belyayev, A., Schulman, A. H. (2020). Long tandem arrays of Cassandra retroelements and their role in genome dynamics in plants. Int. J. Mol. Sci. 21, 2931. doi: 10.3390/ijms21082931
Kalendar, R., Muterko, A., Boronnikova, S. (2021a). Retrotransposable elements: DNA fingerprinting and the assessment of genetic diversity. Methods in molecular biology 2222, 263–286. doi: 10.1007/978-1-0716-0997-2_15
Kalendar, R., Sabot, F., Rodriguez, F., Karlov, G. I., Natali, L., Alix, K. (2021b). Editorial: Mobile elements and plant genome evolution, comparative analyzes and computational tools. Front. Plant Sci. 12, 735134. doi: 10.3389/fpls.2021.735134
Kalendar, R., Schulman, A. H. (2006). IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nat. Protoc. 1, 2478–2484. doi: 10.1038/nprot.2006.377
Kalendar, R., Tanskanen, J., Chang, W., Antonius, K., Sela, H., Peleg, O., et al. (2008). Cassandra Retrotransposons carry independently transcribed 5S RNA. Proc. Natl. Acad. Sci. U.S.A. 105, 5833–5838. doi: 10.1073/pnas.0709698105
Kalendar, R., Vicient, C. M., Peleg, O., Anamthawat-Jonsson, K., Bolshoy, A., Schulman, A. H. (2004). Large Retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166, 1437–1450. doi: 10.1534/genetics.166.3.1437
Keçeli, B. N., Jin, C., Van Damme, D., Geelen, D. (2020). Conservation of centromeric histone 3 interaction partners in plants. J. Exp. Bot. 71, 5237–5246. doi: 10.1093/jxb/eraa214
Keidar, D., Doron, C., Kashkush, K. (2018). Genome-wide analysis of a recently active retrotransposon, au SINE, in wheat: content, distribution within subgenomes and chromosomes, and gene associations. Plant Cell Rep. 37, 193–208. doi: 10.1007/s00299-017-2213-1
Kenan-Eichler, M., Leshkowitz, D., Tal, L., Noor, E., Melamed-Bessudo, C., Feldman, M., et al. (2011). Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188, 263–272. doi: 10.1534/genetics.111.128348
Khapilina, O., Raiser, O., Danilova, A., Shevtsov, V., Turzhanova, A., Kalendar, R. (2021a). DNA Profiling and assessment of genetic diversity of relict species allium altaicum pall. on the territory of Altai. PeerJ 9, e10674. doi: 10.7717/peerj.10674
Khapilina, O., Turzhanova, A., Danilova, A., Tumenbayeva, A., Shevtsov, V., Kotukhov, Y., et al. (2021b). Primer binding site (PBS) profiling of genetic diversity of natural populations of endemic species Allium ledebourianum schult. BioTech 10, 23. doi: 10.3390/biotech10040023
Kim, H., Terakami, S., Nishitani, C., Kurita, K., Kanamori, H., Katayose, Y., et al. (2012). Development of cultivar-specific DNA markers based on retrotransposon-based insertional polymorphism in Japanese pear. Breed. Sci. 62, 53–62. doi: 10.1270/jsbbs.62.53
Kirov, I., Odintsov, S., Omarov, M., Gvaramiya, S., Merkulov, P., Dudnikov, M., et al. (2020). Functional allium fistulosum centromeres comprise arrays of a long satellite repeat, insertions of retrotransposons and chloroplast DNA. Front. Plant Sci. 11, 1668. doi: 10.3389/fpls.2020.562001
Kiseleva, A., Kirov, I., Khrustaleva, L. (2014). Chromosomal organization of centromeric Ty3/gypsy retrotransposons in allium cepa l. and allium fistulosum l. Russian J. Genet. 50, 586–592. doi: 10.1134/S102279541404005X
Klein, S. J., O’Neill, R. J. (2018). Transposable elements: genome innovation, chromosome diversity, and centromere conflict. Chromosome Res. 26, 5–23. doi: 10.1007/s10577-017-9569-5
Kong, X., Yang, M., Le, B. H., He, W., Hou, Y. (2022). The master role of siRNAs in plant immunity. Mol. Plant Pathol. 23, 1565–1574. doi: 10.1111/mpp.13250
Konovalov, F. A., Goncharov, N. P., Goryunova, S., Shaturova, A., Proshlyakova, T., Kudryavtsev, A. (2010). Molecular markers based on LTR retrotransposons BARE-1 and jeli uncover different strata of evolutionary relationships in diploid wheats. Mol. Genet. Genomics 283, 551–563. doi: 10.1007/s00438-010-0539-2
Koo, D.-H., Hong, C. P., Batley, J., Chung, Y. S., Edwards, D., Bang, J.-W., et al. (2011). Rapid divergence of repetitive DNAs in brassica relatives. Genomics 97, 173–185. doi: 10.1016/j.ygeno.2010.12.002
Kuhn, B., Mangolin, C. A., Souto, E. R., Vicient, C. M., Machado, M. F. (2016). Development of retrotransposon-based markers IRAP and REMAP for cassava (Manihot esculenta). Gen. Mol. Res. 15 (2), gmr.15027149. doi: 10.4238/gmr.15027149
Kwak, S.-Y., Lew, T. T. S., Sweeney, C. J., Koman, V. B., Wong, M. H., Bohmert-Tatarev, K., et al. (2019). Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 14, 447–455. doi: 10.1038/s41565-019-0375-4
Laganà, A., Veneziano, D., Russo, F., Pulvirenti, A., Giugno, R., Croce, C. M., et al. (2015). Computational design of artificial RNA molecules for gene regulation. RNA Bioinformatics 1269, 393–412. doi: 10.1007/978-1-4939-2291-8_25
Lamb, J. C., Kato, A., Birchler, J. A. (2005). Sequences associated with a chromosome centromeres are present throughout the maize b chromosome. Chromosoma 113, 337–349. doi: 10.1007/s00412-004-0319-z
Lanciano, S., Cristofari, G. (2020). Measuring and interpreting transposable element expression. Nat. Rev. Genet. 21, 721–736. doi: 10.1038/s41576-020-0251-y
Lanciano, S., Mirouze, M. (2018). Transposable elements: All mobile, all different, some stress responsive, some adaptive? Curr. Opin. Genet. Dev. 49, 106–114. doi: 10.1016/j.gde.2018.04.002
Lancíková, V., Žiarovská, J. (2020). Inter-retrotransposon amplified polymorphism markers revealed long terminal repeat retrotransposon insertion polymorphism in flax cultivated on the experimental fields around Chernobyl. J. Environ. Sci. Health Part A 55, 957–963. doi: 10.1080/10934529.2020.1760016
Lee, S. C., Ernst, E., Berube, B., Borges, F., Parent, J.-S., Ledon, P., et al. (2020a). Arabidopsis LTR retrotransposons and their regulation by epigenetically activated small RNA. bioRxiv 2020.01.24.919167. doi: 10.1101/2020.01.24.919167
Lee, S. C., Ernst, E., Berube, B., Borges, F., Parent, J.-S., Ledon, P., et al. (2020b). Arabidopsis retrotransposon virus-like particles and their regulation by epigenetically activated small RNA. Genome Res. 30, 576–588. doi: 10.1101/gr.259044.119
Lee, S.-I., Kim, J.-H., Park, K.-C., Kim, N.-S. (2015). LTR-Retrotransposons and inter-retrotransposon amplified polymorphism (IRAP) analysis in Lilium species. Genetica 143, 343–352. doi: 10.1007/s10709-015-9833-6
Lee, Y.-S., Maple, R., Dürr, J., Dawson, A., Tamim, S., Del Genio, C., et al. (2021). A transposon surveillance mechanism that safeguards plant male fertility during stress. Nat. Plants 7, 34–41. doi: 10.1038/s41477-020-00818-5
Leitch, A., Leitch, I. (2008). Genomic plasticity and the diversity of polyploid plants. science 320, 481–483. doi: 10.1126/science.1153585
Liang, Y., Lenz, R. R., Dai, W. (2016). Development of retrotransposon-based molecular markers and their application in genetic mapping in chokecherry (Prunus virginiana l.). Mol. Breed. 36, 1–10. doi: 10.1007/s11032-016-0535-2
Li, H., Freeling, M., Lisch, D. (2010). Epigenetic reprogramming during vegetative phase change in maize. Proc. Natl. Acad. Sci. 107, 22184–22189. doi: 10.1073/pnas.1016884108
Lim, K. B., Yang, T. J., Hwang, Y. J., Kim, J. S., Park, J. Y., Kwon, S. J., et al. (2007). Characterization of the centromere and peri-centromere retrotransposons in brassica rapa and their distribution in related brassica species. Plant J. 49, 173–183. doi: 10.1111/j.1365-313X.2006.02952.x
Lippman, Z., May, B., Yordan, C., Singer, T., Martienssen, R., Becker, P. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PloS Biol. 1, e67. doi: 10.1371/journal.pbio.0000067
Lisch, D. (2013). How important are transposons for plant evolution? Nat. Rev. Genet. 14, 49–61. doi: 10.1038/nrg3374
Liu, S., De Jonge, J., Trejo-Arellano, M. S., Santos-González, J., Köhler, C., Hennig, L. (2021). Role of H1 and DNA methylation in selective regulation of transposable elements during heat stress. New Phytol. 229, 2238–2250. doi: 10.1111/nph.17018
Liu, Z., Liu, Y., Liu, F., Zhang, S., Wang, X., Lu, Q., et al. (2018). Genome-wide survey and comparative analysis of long terminal repeat (LTR) retrotransposon families in four gossypium species. Sci. Rep. 8, 1–10. doi: 10.1038/s41598-018-27589-6
Lopez-Gomollon, S., Baulcombe, D. C. (2022). Roles of RNA silencing in viral and non-viral plant immunity and in the crosstalk between disease resistance systems. Nat. Rev. Mol. Cell Biol. 23, 645–662. doi: 10.1038/s41580-022-00496-5
Luan, D. D., Korman, M. H., Jakubczak, J. L., Eickbush, T. H. (1993). Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72, 595–605. doi: 10.1016/0092-8674(93)90078-5
Luo, S., Mach, J., Abramson, B., Ramirez, R., Schurr, R., Barone, P., et al. (2012). The cotton centromere contains a Ty3-gypsy-like LTR retroelement. PloS One 7, e35261. doi: 10.1371/journal.pone.0035261
Lv, Z., Jiang, R., Chen, J., Chen, W. (2020). Nanoparticle-mediated gene transformation strategies for plant genetic engineering. Plant J. 104, 880–891. doi: 10.1111/tpj.14973
Ma, J., Bennetzen, J. L. (2006). Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc. Natl. Acad. Sci. 103, 383–388. doi: 10.1073/pnas.0509810102
Madlung, A., Tyagi, A. P., Watson, B., Jiang, H., Kagochi, T., Doerge, R. W., et al. (2005). Genomic changes in synthetic arabidopsis polyploids. Plant J. 41, 221–230. doi: 10.1111/j.1365-313X.2004.02297.x
Malaviya, D. R., Roy, A. K., Kaushal, P., Pathak, S., Kalendar, R. (2021). Phenotype study of multifoliolate leaf formation in trifolium alexandrinum l. PeerJ. 9, e10874. doi: 10.7717/peerj.10874
Malik, H. S., Eickbush, T. H. (2001). Phylogenetic analysis of ribonuclease h domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 11, 1187–1197. doi: 10.1101/gr.185101
Mangiavacchi, A., Liu, P., Della Valle, F., Orlando, V. (2021). New insights into the functional role of retrotransposon dynamics in mammalian somatic cells. Cell. Mol. Life Sci. 78, 5245–5256. doi: 10.1007/s00018-021-03851-5
Marí-Ordóñez, A., Marchais, A., Etcheverry, M., Martin, A., Colot, V., Voinnet, O. (2013). Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45, 1029–1039. doi: 10.1038/ng.2703
Marques, A., Ribeiro, T., Neumann, P., Macas, J., Novák, P., Schubert, V., et al. (2015). Holocentromeres in rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proc. Natl. Acad. Sci. 112, 13633–13638. doi: 10.1073/pnas.1512255112
Mascagni, F., Giordani, T., Ceccarelli, M., Cavallini, A., Natali, L. (2017). Genome-wide analysis of LTR-retrotransposon diversity and its impact on the evolution of the genus helianthus (L.). BMC Genomics 18, 1–16. doi: 10.1186/s12864-017-4050-6
Mascagni, F., Vangelisti, A., Usai, G., Giordani, T., Cavallini, A., Natali, L. (2020). A computational genome-wide analysis of long terminal repeats retrotransposon expression in sunflower roots (Helianthus annuus l.). Genetica 148, 13–23. doi: 10.1007/s10709-020-00085-4
Masuda, S., Nozawa, K., Matsunaga, W., Masuta, Y., Kawabe, A., Kato, A., et al. (2016). Characterization of a heat-activated retrotransposon in natural accessions of arabidopsis thaliana. Genes Genet. Syst. 91 (6), 293–299. doi: 10.1266/ggs.16-00045
Masuta, Y., Kawabe, A., Nozawa, K., Naito, K., Kato, A., Ito, H. (2018). Characterization of a heat-activated retrotransposon in vigna angularis. Breed. Sci. 68, 168–176. doi: 10.1270/jsbbs.17085
Masuta, Y., Nozawa, K., Takagi, H., Yaegashi, H., Tanaka, K., Ito, T., et al. (2017). Inducible transposition of a heat-activated retrotransposon in tissue culture. Plant Cell Physiol. 58, 375–384. doi: 10.1093/pcp/pcw202
Matsunaga, W., Kobayashi, A., Kato, A., Ito, H. (2012). The effects of heat induction and the siRNA biogenesis pathway on the transgenerational transposition of ONSEN, a copia-like retrotransposon in arabidopsis thaliana. Plant Cell Physiol. 53, 824–833. doi: 10.1093/pcp/pcr179
Matsunaga, W., Ohama, N., Tanabe, N., Masuta, Y., Masuda, S., Mitani, N., et al. (2015). A small RNA mediated regulation of a stress-activated retrotransposon and the tissue specific transposition during the reproductive period in arabidopsis. Front. Plant Sci. 6, 48. doi: 10.3389/fpls.2015.00048
Ma, T., Wei, X., Zhang, Y., Li, J., Wu, F., Yan, Q., et al. (2022). Development of molecular markers based on LTR retrotransposon in the cleistogenes songorica genome. J. Appl. Genet. 63, 61–72. doi: 10.1007/s13353-021-00658-9
McCue, A. D., Nuthikattu, S., Reeder, S. H., Slotkin, R. K. (2012). Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PloS Genet. 8, e1002474. doi: 10.1371/journal.pgen.1002474
McCue, A. D., Panda, K., Nuthikattu, S., Choudury, S. G., Thomas, E. N., Slotkin, R. K. (2015). ARGONAUTE 6 bridges transposable element m RNA-derived si RNA s to the establishment of DNA methylation. EMBO J. 34, 20–35. doi: 10.15252/embj.201489499
Meng, Y., Su, W., Ma, Y., Liu, L., Gu, X., Wu, D., et al. (2021). Assessment of genetic diversity and variety identification based on developed retrotransposon-based insertion polymorphism (RBIP) markers in sweet potato (Ipomoea batatas (L.) lam.). Sci. Rep. 11, 1–12. doi: 10.1038/s41598-021-95876-w
Mhiri, C., Morel, J.-B., Vernhettes, S., Casacuberta, J. M., Lucas, H., Grandbastien, M.-A. (1997). The promoter of the tobacco Tnt1 retrotransposon is induced by wounding and by abiotic stress. Plant Mol. Biol. 33, 257–266. doi: 10.1023/A:1005727132202
Miller, J. T., Dong, F., Jackson, S. A., Song, J., Jiang, J. (1998). Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics 150, 1615–1623. doi: 10.1093/genetics/150.4.1615
Moisy, C., Schulman, A. H., Kalendar, R., Buchmann, J. P., Pelsy, F. (2014). The Tvv1 retrotransposon family is conserved between plant genomes separated by over 100 million years. Theor. Appl. Genet. 127, 1223–1235. doi: 10.1007/s00122-014-2293-z
Monden, Y., Fujii, N., Yamaguchi, K., Ikeo, K., Nakazawa, Y., Waki, T., et al. (2014a). Efficient screening of long terminal repeat retrotransposons that show high insertion polymorphism via high-throughput sequencing of the primer binding site. Genome 57, 245–252. doi: 10.1139/gen-2014-0031
Monden, Y., Takai, T., Tahara, M. (2014b). Characterization of a novel retrotransposon TriRe-1 using nullisomic-tetrasomic lines of hexaploid wheat (Faculty of Agriculture, Okayama University) 103, 21–30.
Muñoz-López, M., García-Pérez, J. L. (2010). DNA Transposons: Nature and applications in genomics. Curr. Genomics 11, 115–128. doi: 10.2174/138920210790886871
Nagaki, K., Murata, M. (2005). Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosome Res. 13, 195–203. doi: 10.1007/s10577-005-0847-2
Nagaki, K., Shibata, F., Kanatani, A., Kashihara, K., Murata, M. (2012). Isolation of centromeric-tandem repetitive DNA sequences by chromatin affinity purification using a HaloTag7-fused centromere-specific histone H3 in tobacco. Plant Cell Rep. 31, 771–779. doi: 10.1007/s00299-011-1198-4
Nagaki, K., Shibata, F., Murata, M. (2011). A mosaic structure of centromeric DNA in tobacco. Genes Genet. Syst. 86, 434–434. GENETICS SOC JAPAN NATIONAL INST GENETICS YATA 1111, MISHIMA, SHIZUOKA-KEN.
Nagaki, K., Song, J., Stupar, R. M., Parokonny, A. S., Yuan, Q., Ouyang, S., et al. (2003). Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163, 759–770. doi: 10.1093/genetics/163.2.759
Naito, K., Zhang, F., Tsukiyama, T., Saito, H., Hancock, C. N., Richardson, A. O., et al. (2009). Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461, 1130–1134. doi: 10.1038/nature08479
Nakamura, M., Köhler, C., Hennig, L. (2019). Tissue-specific transposon-associated small RNAs in the gymnosperm tree, Norway spruce. BMC Genomics 20, 1–10. doi: 10.1186/s12864-019-6385-7
Neumann, P., Koblízková, A., Navrátilová, A., Macas, J. (2006). Significant expansion of vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 173, 1047–1056. doi: 10.1534/genetics.106.056259
Neumann, P., Navrátilová, A., Koblížková, A., Kejnovský, E., Hřibová, E., Hobza, R., et al. (2011). Plant centromeric retrotransposons: a structural and cytogenetic perspective. Mobile DNA 2, 1–16. doi: 10.1186/1759-8753-2-4
Neumann, P., Yan, H., Jiang, J. (2007). The centromeric retrotransposons of rice are transcribed and differentially processed by RNA interference. Genetics 176, 749–761. doi: 10.1534/genetics.107.071902
Nie, Q., Qiao, G., Peng, L., Wen, X. (2019). Transcriptional activation of long terminal repeat retrotransposon sequences in the genome of pitaya under abiotic stress. Plant Physiol. Biochem. 135, 460–468. doi: 10.1016/j.plaphy.2018.11.014
Nosaka, M., Itoh, J.-I., Nagato, Y., Ono, A., Ishiwata, A., Sato, Y. (2012). Role of transposon-derived small RNAs in the interplay between genomes and parasitic DNA in rice. PLoS Genet. 8 (9), e1002953. doi: 10.1371/journal.pgen.1002953
Nozawa, K., Chen, J., Jiang, J., Leichter, S. M., Yamada, M., Suzuki, T., et al. (2021). DNA Methyltransferase CHROMOMETHYLASE3 prevents ONSEN transposon silencing under heat stress. PloS Genet. 17, e1009710. doi: 10.1371/journal.pgen.1009710
Nuthikattu, S., McCue, A. D., Panda, K., Fultz, D., DeFraia, C., Thomas, E. N., et al. (2013). The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162, 116–131. doi: 10.1104/pp.113.216481
O'Neill, R. J. W., O'Neill, M. J., Graves, J. A. M. (1998). Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393, 68–72. doi: 10.1038/29985
Oliveira, L. C., Torres, G. A. (2018). Plant centromeres: genetics, epigenetics and evolution. Mol. Biol. Rep. 45, 1491–1497. doi: 10.1007/s11033-018-4284-7
Ouyang, Z., Wang, Y., Ma, T., Kanzana, G., Wu, F., Zhang, J. (2021). Genome-wide identification and development of LTR retrotransposon-based molecular markers for the melilotus genus. Plants 10, 890. doi: 10.3390/plants10050890
Ozkan, H., Levy, A. A., Feldman, M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–triticum) group. Plant Cell 13, 1735–1747. doi: 10.1105/tpc.010082
Panda, K., Ji, L., Neumann, D. A., Daron, J., Schmitz, R. J., Slotkin, R. K. (2016). Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 17, 1–19. doi: 10.1186/s13059-016-1032-y
Panda, K., McCue, A. D., Slotkin, R. K. (2020). Arabidopsis RNA polymerase IV generates 21–22 nucleotide small RNAs that can participate in RNA-directed DNA methylation and may regulate genes. Philos. Trans. R. Soc. B 375, 20190417. doi: 10.1098/rstb.2019.0417
Papareddy, R. K., Páldi, K., Paulraj, S., Kao, P., Lutzmayer, S., Nodine, M. D. (2020). Chromatin regulates expression of small RNAs to help maintain transposon methylome homeostasis in arabidopsis. Genome Biol. 21, 1–24. doi: 10.1186/s13059-020-02163-4
Papolu, P. K., Ramakrishnan, M., Wei, Q., Vinod, K. K., Zou, L.-H., Yrjala, K., et al. (2021). Long terminal repeats (LTR) and transcription factors regulate PHRE1 and PHRE2 activity in moso bamboo under heat stress. BMC Plant Biol. 21, 1–19. doi: 10.1186/s12870-021-03339-1
Parisod, C., Alix, K., Just, J., Petit, M., Sarilar, V., Mhiri, C., et al. (2010). Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 186, 37–45. doi: 10.1111/j.1469-8137.2009.03096.x
Parisod, C., Salmon, A., Zerjal, T., Tenaillon, M., Grandbastien, M. A., Ainouche, M. (2009). Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in spartina. New Phytol. 184, 1003–1015. doi: 10.1111/j.1469-8137.2009.03029.x
Pearce, S. R., Stuart-Rogers, C., Knox, M. R., Kumar, A., Ellis, T. N., Flavell, A. J. (1999). Rapid isolation of plant Ty1-copia group retrotransposon LTR sequences for molecular marker studies. Plant J. 19, 711–717. doi: 10.1046/j.1365-313x.1999.00556.x
Pecinka, A., Dinh, H. Q., Baubec, T., Rosa, M., Lettner, N., Scheid, O. M. (2010). Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in arabidopsis. Plant Cell 22, 3118–3129. doi: 10.1105/tpc.110.078493
Peng, Y., Zhang, Y., Gui, Y., An, D., Liu, J., Xu, X., et al. (2019). Elimination of a retrotransposon for quenching genome instability in modern rice. Mol. Plant 12, 1395–1407. doi: 10.1016/j.molp.2019.06.004
Petit, M., Lim, K. Y., Julio, E., Poncet, C., De Borne, F. D., Kovarik, A., et al. (2007). Differential impact of retrotransposon populations on the genome of allotetraploid tobacco (Nicotiana tabacum). Mol. Genet. Genomics 278, 1–15. doi: 10.1007/s00438-007-0226-0
Piegu, B., Guyot, R., Picault, N., Roulin, A., Saniyal, A., Kim, H., et al. (2006). Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in oryza australiensis, a wild relative of rice. Genome Res. 16, 1262–1269. doi: 10.1101/gr.5290206
Pietzenuk, B., Markus, C., Gaubert, H., Bagwan, N., Merotto, A., Bucher, E., et al. (2016). Recurrent evolution of heat-responsiveness in brassicaceae COPIA elements. Genome Biol. 17, 1–15. doi: 10.1186/s13059-016-1072-3
Pontier, D., Picart, C., Roudier, F., Garcia, D., Lahmy, S., Azevedo, J., et al. (2012). NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in arabidopsis. Mol. Cell 48, 121–132. doi: 10.1016/j.molcel.2012.07.027
Porquier, A., Tisserant, C., Salinas, F., Glassl, C., Wange, L. E., Enard, W., et al. (2021). Retrotransposons as pathogenicity factors of the plant pathogenic fungus botrytis cinerea. Genome Biol. 22, 225. doi: 10.1186/s13059-021-02446-4
Potter, S. (2005). Long terminal repeat (Ltr) type retrotransposons in populus species: A uniquely abundant and informative class of molecular markers for forest biotechnology. For. Genet. 12 (1), 35–44.
Presting, G. G. (2018). Centromeric retrotransposons and centromere function. Curr. Opin. Genet. Dev. 49, 79–84. doi: 10.1016/j.gde.2018.03.004
Presting, G. G., Malysheva, L., Fuchs, J., Schubert, I. (1998). A TY3/GYPSY retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 16, 721–728. doi: 10.1046/j.1365-313x.1998.00341.x
Qi, L., Wu, J., Friebe, B., Qian, C., Gu, Y., Fu, D., et al. (2013). Sequence organization and evolutionary dynamics of brachypodium-specific centromere retrotransposons. Chromosome Res. 21, 507–521. doi: 10.1007/s10577-013-9378-4
Ramakrishnan, M., Satish, L., Kalendar, R., Narayanan, M., Kandasamy, S., Sharma, A., et al. (2021). The dynamism of transposon methylation for plant development and stress adaptation. Int. J. Mol. Sci. 22, 11387. doi: 10.3390/ijms222111387
Ramsey, J., Schemske, D. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics 29 (1), 467–501.
Ravindran, S. (2012). Barbara McClintock and the discovery of jumping genes. Proc. Natl. Acad. Sci. 109, 20198–20199. doi: 10.1073/pnas.1219372109
Rey-Banos, R., Saenz de Miera, L. E., García, P., Pérez de la Vega, M. (2017). Obtaining retrotransposon sequences, analysis of their genomic distribution and use of retrotransposon-derived genetic markers in lentil (Lens culinaris medik.). PloS One 12, e0176728. doi: 10.1371/journal.pone.0176728
Roulin, A., Piegu, B., Fortune, P. M., Sabot, F., d'Hont, A., Manicacci, D., et al. (2009). Whole genome surveys of rice, maize and sorghum reveal multiple horizontal transfers of the LTR-retrotransposon Route66 in poaceae. BMC Evol. Biol. 9, 1–10. doi: 10.1186/1471-2148-9-58
Salina, E. A., Sergeeva, E. M., Adonina, I. G., Shcherban, A. B., Belcram, H., Huneau, C., et al. (2011). The impact of Ty3-gypsy group LTR retrotransposons Fatima on b-genome specificity of polyploid wheats. BMC Plant Biol. 11, 1–14. doi: 10.1186/1471-2229-11-99
Sanan-Mishra, N., Abdul Kader Jailani, A., Mandal, B., Mukherjee, S. K. (2021). Secondary siRNAs in plants: Biosynthesis, various functions, and applications in virology. Front. Plant Sci. 12, 610283. doi: 10.3389/fpls.2021.610283
Sanchez, D. H., Gaubert, H., Drost, H.-G., Zabet, N. R., Paszkowski, J. (2017). High-frequency recombination between members of an LTR retrotransposon family during transposition bursts. Nat. Commun. 8, 1–7. doi: 10.1038/s41467-017-01374-x
Sanmiguel, P., Bennetzen, J. L. (1998). Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 82, 37–44. doi: 10.1006/anbo.1998.0746
Sant, V., Sainani, M., Sami-Subbu, R., Ranjekar, P., Gupta, V. (2000). Ty1-copia retrotransposon-like elements in chickpea genome: their identification, distribution and use for diversity analysis. Gene 257, 157–166. doi: 10.1016/S0378-1119(00)00405-4
Schorn, A. J., Gutbrod, M. J., LeBlanc, C., Martienssen, R. (2017). LTR-Retrotransposon control by tRNA-derived small RNAs. Cell 170, 61–71. e11. doi: 10.1016/j.cell.2017.06.013
Serag, M. F., Kaji, N., Gaillard, C., Okamoto, Y., Terasaka, K., Jabasini, M., et al. (2011). Trafficking and subcellular localization of multiwalled carbon nanotubes in plant cells. ACS Nano 5, 493–499. doi: 10.1021/nn102344t
Settles, A. M., Holding, D. R., Tan, B. C., Latshaw, S. P., Liu, J., Suzuki, M., et al. (2007). Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8, 1–12. doi: 10.1186/1471-2164-8-116
Shams, I., Raskina, O. (2018). Intraspecific and intraorganismal copy number dynamics of retrotransposons and tandem repeat in aegilops speltoides tausch (Poaceae, triticeae). Protoplasma 255, 1023–1038. doi: 10.1007/s00709-018-1212-6
Shapiro, J. A. (2014). Epigenetic control of mobile DNA as an interface between experience and genome change. Front. Genet. 5. doi: 10.3389/fgene.2014.00087
Sharma, V., Nandineni, M. R. (2014). Assessment of genetic diversity among Indian potato (Solanum tuberosum l.) collection using microsatellite and retrotransposon based marker systems. Mol. Phylogenet. Evol. 73, 10–17. doi: 10.1016/j.ympev.2014.01.003
Sharma, A., Presting, G. G. (2014). Evolution of centromeric retrotransposons in grasses. Genome Biol. Evol. 6, 1335–1352. doi: 10.1093/gbe/evu096
Sharma, A., Wolfgruber, T. K., Presting, G. G. (2013). Tandem repeats derived from centromeric retrotransposons. BMC Genomics 14, 1–11. doi: 10.1186/1471-2164-14-142
Shingote, P. R., Mithra, S. A., Sharma, P., Devanna, N. B., Arora, K., Holkar, S. K., et al. (2019). LTR Retrotransposons and highly informative ISSRs in combination are potential markers for genetic fidelity testing of tissue culture-raised plants in sugarcane. Mol. Breed. 39, 25. doi: 10.1007/s11032-019-0931-5
Slotkin, R. K., Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. doi: 10.1038/nrg2072
Slotkin, R. K., Vaughn, M., Borges, F., Tanurdžić, M., Becker, J. D., Feijó, J. A., et al. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472. doi: 10.1016/j.cell.2008.12.038
Smýkal, P., Bačová-Kerteszová, N., Kalendar, R., Corander, J., Schulman, A. H., Pavelek, M. (2011). Genetic diversity of cultivated flax (Linum usitatissimum l.) germplasm assessed by retrotransposon-based markers. Theor. Appl. Genet. 122, 1385–1397. doi: 10.1007/s00122-011-1539-2
Song, X., Li, Y., Cao, X., Qi, Y. (2019). MicroRNAs and their regulatory roles in plant–environment interactions. Annu. Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Springer, N. M., Ying, K., Fu, Y., Ji, T., Yeh, C.-T., Jia, Y., et al. (2009). Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PloS Genet. 5, e1000734. doi: 10.1371/journal.pgen.1000734
Stitzer, M. C., Anderson, S. N., Springer, N. M., Ross-Ibarra, J. (2021). The genomic ecosystem of transposable elements in maize. PloS Genet. 17, e1009768. doi: 10.1371/journal.pgen.1009768
Sun, L., Jing, Y., Liu, X., Li, Q., Xue, Z., Cheng, Z., et al. (2020). Heat stress-induced transposon activation correlates with 3D chromatin organization rearrangement in arabidopsis. Nat. Commun. 11, 1–13. doi: 10.1038/s41467-020-15809-5
Šurbanovski, N., Brilli, M., Moser, M., Si-Ammour, A. (2016). A highly specific micro RNA-mediated mechanism silences LTR retrotransposons of strawberry. Plant J. 85, 70–82. doi: 10.1111/tpj.13090
Syed, N., Sureshsundar, S., Wilkinson, M., Bhau, B., Cavalcanti, J., Flavell, A. (2005). Ty1-copia retrotransposon-based SSAP marker development in cashew (Anacardium occidentale l.). Theor. Appl. Genet. 110, 1195–1202. doi: 10.1007/s00122-005-1948-1
Takehira, K., Hayashi, Y., Nozawa, K., Chen, L., Suzuki, T., Masuta, Y., et al. (2021). DRD1, a SWI/SNF-like chromatin remodeling protein, regulates a heat-activated transposon in arabidopsis thaliana. Genes Genet. Syst. 96 (3), 151–158. doi: 10.1266/ggs.21-00005
Taller, D., Balint, J., Gyula, P., Nagy, T., Barta, E., Baksa, I., et al. (2018a). Expansion of capsicum annum fruit is linked to dynamic tissue-specific differential expression of miRNA and siRNA profiles. PloS One 13, e0200207. doi: 10.1371/journal.pone.0200207
Taller, D., Bálint, J., Gyula, P., Nagy, T., Barta, E., Baksa, I., et al. (2018b). Correction: Expansion of capsicum annum fruit is linked to dynamic tissue-specific differential expression of miRNA and siRNA profiles. PloS One 13, e0203582. doi: 10.1371/journal.pone.0203582
Tek, A. L., Kashihara, K., Murata, M., Nagaki, K. (2010). Functional centromeres in soybean include two distinct tandem repeats and a retrotransposon. Chromosome Res. 18, 337–347. doi: 10.1007/s10577-010-9119-x
Theuri, J., Phelps-Durr, T., Mathews, S., Birchler, J. (2005). A comparative study of retrotransposons in the centromeric regions of a and b chromosomes of maize. Cytogenetic Genome Res. 110, 203–208. doi: 10.1159/000084953
Tian, Z., Rizzon, C., Du, J., Zhu, L., Bennetzen, J. L., Jackson, S. A., et al. (2009). Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res. 19, 2221–2230. doi: 10.1101/gr.083899.108
Tian, Z., Zhao, M., She, M., Du, J., Cannon, S. B., Liu, X., et al. (2012). Genome-wide characterization of nonreference transposons reveals evolutionary propensities of transposons in soybean. Plant Cell 24, 4422–4436. doi: 10.1105/tpc.112.103630
Tittel-Elmer, M., Bucher, E., Broger, L., Mathieu, O., Paszkowski, J., Vaillant, I. (2010). Stress-induced activation of heterochromatic transcription. PloS Genet. 6, e1001175. doi: 10.1371/journal.pgen.1001175
Tsukahara, S., Kawabe, A., Kobayashi, A., Ito, T., Aizu, T., Shin-i, T., et al. (2012). Centromere-targeted de novo integrations of an LTR retrotransposon of arabidopsis lyrata. Genes Dev. 26, 705–713. doi: 10.1101/gad.183871.111
Ujino-Ihara, T. (2020). Transcriptome analysis of heat stressed seedlings with or without pre-heat treatment in cryptomeria japonica. Mol. Genet. Genomics 295, 1163–1172. doi: 10.1007/s00438-020-01689-3
Vicient, C. M., Casacuberta, J. M. (2017). Impact of transposable elements on polyploid plant genomes. Ann. Bot. 20 (2), 195–207. doi: 10.1093/aob/mcx078
Voronova, A., Belevich, V., Jansons, A., Rungis, D. (2014). Stress-induced transcriptional activation of retrotransposon-like sequences in the scots pine (Pinus sylvestris l.) genome. Tree Genet. Genomes 10, 937–951. doi: 10.1007/s11295-014-0733-1
Voronova, A., Jansons, Ā, Ruņǵis, D. (2011). Expression of retrotransposon-like sequences in scots pine (Pinus sylvestris) in response to heat stress. Environ. Exp. Biol. 9, 121–127.
Voronova, A., Rendón-Anaya, M., Ingvarsson, P., Kalendar, R., Ruņǵis, D. (2020). Comparative study of pine reference genomes reveals transposable element interconnected gene networks. Genes 11, 1216. doi: 10.3390/genes11101216
Vuorinen, A. L., Kalendar, R., Fahima, T., Korpelainen, H., Nevo, E., Schulman, A. H. (2018). Retrotransposon-based genetic diversity assessment in wild emmer wheat (Triticum turgidum ssp. dicoccoides). Agronomy 8, 107. doi: 10.3390/agronomy8070107
Wang, J. W., Cunningham, F. J., Goh, N. S., Boozarpour, N. N., Pham, M., Landry, M. P. (2021). Nanoparticles for protein delivery in planta. Curr. Opin. Plant Biol. 60, 102052. doi: 10.1016/j.pbi.2021.102052
Wang, Q., Dooner, H. K. (2006). Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc. Natl. Acad. Sci. 103, 17644–17649. doi: 10.1073/pnas.0603080103
Wang, J. W., Grandio, E. G., Newkirk, G. M., Demirer, G. S., Butrus, S., Giraldo, J. P., et al. (2019). Nanoparticle-mediated genetic engineering of plants. Mol. Plant 12, 1037–1040. doi: 10.1016/j.molp.2019.06.010
Wang, G., He, Q., Liu, F., Cheng, Z., Talbert, P. B., Jin, W. (2011). Characterization of CENH3 proteins and centromere-associated DNA sequences in diploid and allotetraploid brassica species. Chromosoma 120, 353–365. doi: 10.1007/s00412-011-0315-z
Wang, G.-X., He, Q.-Y., Zhao, H., Cai, Z.-X., Guo, N., Zong, M., et al. (2019). ChIP-cloning analysis uncovers centromere-specific retrotransposons in brassica nigra and reveals their rapid diversification in brassica allotetraploids. Chromosoma 128, 119–131. doi: 10.1007/s00412-019-00701-z
Wang, Y., Liang, W., Tang, T. (2018). Constant conflict between gypsy LTR retrotransposons and CHH methylation within a stress-adapted mangrove genome. New Phytol. 220, 922–935. doi: 10.1111/nph.15209
Wang, S., Liang, H., Xu, Y., Li, L., Wang, H., Sahu, D. N., et al. (2021). Genome-wide analyses across viridiplantae reveal the origin and diversification of small RNA pathway-related genes. Commun. Biol. 4, 412. doi: 10.1038/s42003-021-01933-5
Waugh, R., McLean, K., Flavell, A., Pearce, S., Kumar, A., Thomas, B., et al. (1997). Genetic distribution of bare–1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol. Gen. Genet. MGG 253, 687–694. doi: 10.1007/s004380050372
Weber, B., Schmidt, T. (2009). Nested Ty3-gypsy retrotransposons of a single beta procumbens centromere contain a putative chromodomain. Chromosome Res. 17, 379–396. doi: 10.1007/s10577-009-9029-y
Wei, L., Gu, L., Song, X., Cui, X., Lu, Z., Zhou, M., et al. (2014). Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc. Natl. Acad. Sci. 111, 3877–3882. doi: 10.1073/pnas.1318131111
Weise, K., Winter, L., Fischer, E., Kneis, D., de la Cruz Barron, M., Kunze, S., et al. (2022). Multiwalled carbon nanotubes promote bacterial conjugative plasmid transfer. Microbiol. Spectr. 10, e00410–e00422. doi: 10.1128/spectrum.00410-22
Wei, L., Xiao, M., An, Z., Ma, B., Mason, A. S., Qian, W., et al. (2013). New insights into nested long terminal repeat retrotransposons in brassica species. Mol. Plant 6, 470–482. doi: 10.1093/mp/sss081
Wessler, S. R. (1996). Plant retrotransposons: turned on by stress. Curr. Biol. 6, 959–961. doi: 10.1016/S0960-9822(02)00638-3
Wessler, S. R., Bureau, T. E., White, S. E. (1995). LTR-Retrotransposons and MITEs: important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5, 814–821. doi: 10.1016/0959-437X(95)80016-X
Wicker, T., Sabot, F., Hua-Van, A., Bennetzen, J. L., Capy, P., Chalhoub, B., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982. doi: 10.1038/nrg2165
Wolfgruber, T. K., Sharma, A., Schneider, K. L., Albert, P. S., Koo, D.-H., Shi, J., et al. (2009). Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic loci shaped primarily by retrotransposons. PloS Genet. 5, e1000743. doi: 10.1371/journal.pgen.1000743
Woodrow, P., Pontecorvo, G., Ciarmiello, L. F. (2012). Isolation of Ty1-copia retrotransposon in myrtle genome and development of s-SAP molecular marker. Mol. Biol. Rep. 39, 3409–3418. doi: 10.1007/s11033-011-1112-8
Xie, Z., Johansen, L. K., Gustafson, A. M., Kasschau, K. D., Lellis, A. D., Zilberman, D., et al. (2004). Genetic and functional diversification of small RNA pathways in plants. PloS Biol. 2, e104. doi: 10.1371/journal.pbio.0020104
Yañez-Santos, A. M., Paz, R. C., Paz-Sepúlveda, P. B., Urdampilleta, J. D. (2021). Full-length LTR retroelements in capsicum annuum revealed a few species-specific family bursts with insertional preferences. Chromosome Res. 29, 261–284. doi: 10.1007/s10577-021-09663-4
Yang, T.-J., Lee, S., Chang, S.-B., Yu, Y., de Jong, H., Wing, R. A. (2005). In-depth sequence analysis of the tomato chromosome 12 centromeric region: Identification of a large CAA block and characterization of pericentromere retrotranposons. Chromosoma 114, 103–117. doi: 10.1007/s00412-005-0342-8
Yang, D.-L., Zhang, G., Tang, K., Li, J., Yang, L., Huang, H., et al. (2016). Dicer-independent RNA-directed DNA methylation in arabidopsis. Cell Res. 26, 66–82. doi: 10.1038/cr.2015.145
Ye, R., Chen, Z., Lian, B., Rowley, M. J., Xia, N., Chai, J., et al. (2016). A dicer-independent route for biogenesis of siRNAs that direct DNA methylation in arabidopsis. Mol. Cell 61, 222–235. doi: 10.1016/j.molcel.2015.11.015
Yi, C., Zhang, W., Dai, X., Li, X., Gong, Z., Zhou, Y., et al. (2013). Identification and diversity of functional centromere satellites in the wild rice species oryza brachyantha. Chromosome Res. 21, 725–737. doi: 10.1007/s10577-013-9374-8
Yokthongwattana, C., Bucher, E., Čaikovski, M., Vaillant, I., Nicolet, J., Scheid, O. M., et al. (2010). MOM1 and pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J. 29, 340–351. doi: 10.1038/emboj.2009.328
Zhang, W., Cao, Y., Wang, K., Zhao, T., Chen, J., Pan, M., et al. (2014). Identification of centromeric regions on the linkage map of cotton using centromere-related repeats. Genomics 104, 587–593. doi: 10.1016/j.ygeno.2014.09.002
Zhang, H., Cao, Y., Xu, D., Goh, N. S., Demirer, G. S., Cestellos-Blanco, S., et al. (2021). Gold-nanocluster-mediated delivery of siRNA to intact plant cells for efficient gene knockdown. Nano Lett. 21, 5859–5866. doi: 10.1021/acs.nanolett.1c01792
Zhang, T., Talbert, P. B., Zhang, W., Wu, Y., Yang, Z., Henikoff, J. G., et al. (2013). The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres. Proc. Natl. Acad. Sci. 110, E4875–E4883. doi: 10.1073/pnas.1319548110
Zhang, H., Tao, Z., Hong, H., Chen, Z., Wu, C., Li, X., et al. (2016). Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants 2, 1–8. doi: 10.1038/nplants.2016.16
Zhou, M., Hu, B., Zhu, Y. (2017a). Genome-wide characterization and evolution analysis of long terminal repeat retroelements in moso bamboo (Phyllostachys edulis). Tree Genet. Genomes 13, 43. doi: 10.1007/s11295-017-1114-3
Zhou, S.-S., Yan, X.-M., Zhang, K.-F., Liu, H., Xu, J., Nie, S., et al. (2021). A comprehensive annotation dataset of intact LTR retrotransposons of 300 plant genomes. Sci. Data 8, 1–9. doi: 10.1038/s41597-021-00968-x
Keywords: transposable element, retrotransposons, LTR, genetic diversity, siRNAs, RdDM pathways, Ty1/copia, Ty3/gypsy
Citation: Papolu PK, Ramakrishnan M, Mullasseri S, Kalendar R, Wei Q, Zou L−H, Ahmad Z, Vinod KK, Yang P and Zhou M (2022) Retrotransposons: How the continuous evolutionary front shapes plant genomes for response to heat stress. Front. Plant Sci. 13:1064847. doi: 10.3389/fpls.2022.1064847
Received: 08 October 2022; Accepted: 21 November 2022;
Published: 09 December 2022.
Edited by:
Hussain Touseef, Matimate Agromart Pvt. Ltd. (Sevama AgriClinic Laboratory), IndiaReviewed by:
Karthikeyan Adhimoolam, Jeju National University, South KoreaCopyright © 2022 Papolu, Ramakrishnan, Mullasseri, Kalendar, Wei, Zou, Ahmad, Vinod, Yang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muthusamy Ramakrishnan, cmFta3lAbmpmdS5lZHUuY24=; Ping Yang, eWFuZ3BpbmdAemFmdS5lZHUuY24=; Mingbing Zhou, emhvdW1pbmdiaW5nQHphZnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.