
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 09 January 2023
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1058421
This article is part of the Research TopicWomen in Plant Development and EvoDevo: 2022View all 10 articles
Reprogramming of microspores development towards embryogenesis mediated by stress treatment constitutes the basis of doubled haploid production. Recently, compounds that alter histone post-translational modifications (PTMs) have been reported to enhance microspore embryogenesis (ME), by altering histones acetylation or methylation. However, epigenetic mechanisms underlying ME induction efficiency are poorly understood. In this study, the epigenetic dynamics and the expression of genes associated with histone PTMs and ME induction were studied in two bread wheat cultivars with different ME response. Microspores isolated at 0, 3 and 5 days, treated with 0.7M mannitol (MAN) and 0.7M mannitol plus 0.4µM trichostatin A (TSA), which induced ME more efficiently, were analyzed. An additional control of gametophytic development was included. Microspores epigenetic state at the onset of ME induction was distinctive between cultivars by the ratio of H3 variants and their acetylated forms, the localization and percentage of labeled microspores with H3K9ac, H4K5ac, H4K16ac, H3K9me2 and H3K27me3, and the expression of genes related to pollen development. These results indicated that microspores of the high responding cultivar could be at a less advanced stage in pollen development. MAN and TSA resulted in a hyperacetylation of H3.2, with a greater effect of TSA. Histone PTMs were differentially affected by both treatments, with acetylation being most concerned. The effect of TSA was observed in the H4K5ac localization pattern at 3dT in the mid-low responding cultivar. Three gene networks linked to ME response were identified. TaHDT1, TaHAG2, TaYAO, TaNFD6-A, TabZIPF1 and TaAGO802-B, associated with pollen development, were down-regulated. TaHDA15, TaHAG3, TaHAM, TaYUC11D, Ta-2B-LBD16 TaMS1 and TaDRM3 constituted a network implicated in morphological changes by auxin signaling and cell wall modification up-regulated at 3dT. The last network included TaHDA18, TaHAC1, TaHAC4, TaABI5, TaATG18fD, TaSDG1a-7A and was related to ABA and ethylene hormone signaling pathways, DNA methylation and autophagy processes, reaching the highest expression at 5dT. The results indicated that TSA mainly modified the regulation of genes related to pollen and auxin signaling. This study represents a breakthrough in identifying the epigenetic dynamics and the molecular mechanisms governing ME induction efficiency, with relevance to recalcitrant wheat genotypes and other crops.
Microspore embryogenesis (ME) constitutes a process in which the microspore changes its developmental pathway from a gametophytic towards a sporophytic. This change is triggered by a stress treatment, followed by an in vitro culture phase in which embryos and haploid plants are finally formed (Soriano et al., 2013). Doubled haploid (DH) plants can be obtained spontaneously by nuclear fusion or endoreduplication, during the first microspore divisions, or can be induced by the action of chromosome doubling agents (González-Melendi et al., 2005; Griggs and Zheng, 2016). DH plants have a great value to produce new varieties or inbred lines because complete homozygosis is achieved in one generation.
In recent years, increasing evidence has been accumulated on how the dynamics of epigenetic processes regulates gene expression and establishes genomic elements in adaptive responses to developmental transitions. Epigenetic modifications have been reported to play an important role in reproductive development, abiotic stress response and plant regeneration, processes which are involved in ME (Begcy and Dresselhaus, 2018; Lee and Seo, 2018; Mozgova et al., 2019; Ono and Kinoshita, 2021).
Changes in histone post-translational modifications (PTMs), DNA methylation as well as regulation of small RNA and long noncoding RNA pathways have been described as the major epigenetic mechanisms in plants (Pikaard and Scheid, 2014). PTMs of histone-tails can alter the interaction between histones and DNA directly or by recruiting specific effectors such as transcriptional regulators or chromatin remodelers (for review see Tessarz and Kouzarides, 2014). Acetylation and methylation are the two most common histone PTMs involved in activation and repression of transcription, respectively. Histone deacetylases (HDAC) and acetyltransferases (HAT) regulate the balance of histone acetylation by modifying lysine residues, leading to a chromatin compaction/decompaction that facilitates the repression/activation of gene transcription (for review see Bannister and Kouzarides, 2011). Histone lysine residues can be mono-, di- or tri-methylated in a dynamic way by the action of histone lysine methyltransferases (HKMT) and histone demethylases (for review see Bannister and Kouzarides, 2011).
Histone hyperacetylation and DNA hypomethylation have been directly associated to ME induction by the application of a heat stress treatment in the model species Brassica napus (Solis et al., 2012; Li et al., 2014a). Furthermore, the application of inhibitors of enzymes involved in the modification of epigenetic marks has been shown to be a successful strategy to increase the efficiency of ME (for review see Testillano, 2019). Thus, trichostatin A (TSA), a HDAC inhibitor acting over acetylation of histones and other proteins related to cytoskeleton, increased the efficiency of microspore reprogramming in Brassica (Li et al., 2014a; Zhang et al., 2016) and barley (Pandey et al., 2017). Studies performed in barley indicated that there was a very intricate interaction between the histone acetylation and methylation after TSA application (Pandey et al., 2017). Other histone deacetylases inhibitors such as suberoylanilide hydroxamic acid (SAHA) and sodium butyrate have also been used to enhance ME in Brassica (Zhang et al., 2016). Also, the application of 5-azacytidine, which causes a global DNA demethylation, or BIX, a histone lysine methyltransferase inhibitor, promotes ME in Brassica, barley and triticale (Solis et al., 2015; Berenguer et al., 2017; Nowicka et al., 2019).
Different approaches have been described for the application of TSA to increase ME efficiency in wheat. Thus, green plant regeneration was enhanced in different cultivars applying TSA in the microspore isolation procedure or in the culture medium, after a cold stress treatment (Jiang et al., 2017; Wang et al., 2019). However, a comparison of different strategies showed that the application of 0.4 µM TSA simultaneously with 0.7 M mannitol for 5 days was the most efficient for DH production, increasing twice and four times the number of green plants compared to mannitol in a high and a mid-low responding bread wheat cultivars, respectively (Castillo et al., 2020).
Although different protocols are available for ME in wheat (Castillo et al., 2021; Jensen et al., 2022), their efficiency depends largely on the genotype (Lantos et al., 2013; Weigt et al., 2019; Castillo et al., 2019). The genetic control of ME is quite complex (Muñoz-Amatriaín et al., 2008; Nielsen et al., 2015; Abd ElFath et al., 2020). Besides the genetic control, the epigenetic state of the microspores before ME induction could also play and important role. Thus, microspores from a high ME responding line of triticale had a higher level of DNA methylation than a low responding one (Nowicka et al., 2019). Different levels of endogenous ABA as well as gene transcripts related to ME have been also described in genotypes with different ME response in Brassica, wheat and triticale (Dubas et al., 2013; Sánchez-Díaz et al., 2013; Nowicka et al., 2019).
Little is known about the dynamics of histone PTMs and their impact on transcriptional regulation of wheat ME. A recent study applying TSA after a cold treatment shows that TSA up-regulates genes involved in cell division, DNA organization, signaling cascades, hormone mediated signaling, and cell wall and cytoskeleton modifications (Jensen, 2021). To provide new insights into wheat ME induction mechanisms, the dynamics of acetylation and methylation histone PTMs was analyzed with a mannitol and a mannitol plus TSA treatment, which induced ME more efficiently, in two bread wheat cultivars with different ME response. The expression of genes involved in PTMs and key genes for ME and their correlations were also studied. For the first time, the epigenetic dynamics and gene regulation are discussed in wheat ME induction depending on cultivars and treatments.
The spring cultivars (cvs) of bread wheat Pavon and Caramba, which have a high and a mid-low ME response, respectively, were used in this study. Growth conditions of the mother plants and harvest and sterilization of spikes were performed as described by Castillo et al. (2021). Anthers containing most microspores at the mid to late uninucleated stage, determined by DAPI (4´, 6-Diamidine-2’-phenylindole dihydrochloride) staining were excised and brought under stress treatments. For selection of the spikes at this stage of development, tillers with 3-5 cm length from the distal part of the spike to the basal part of the flag leaf were harvested. Some spikes were harvested two days later to allow microspores to progress into gametophytic development (GD).
Comparison of different TSA application strategies showed that the incorporation of TSA in a mannitol treatment was the best to enhance wheat ME (Castillo et al., 2020). Therefore, the effect of 0.4 µM TSA (Sigma T8852) in combination with 0.7 M mannitol stress treatment was assayed. TSA was dissolved in 1% DMSO (final concentration in the medium). Freshly excised anthers were randomly distributed in 6 cm Ø Petri dishes containing solidified 0.7 M mannitol stress medium (SM medium, Castillo et al., 2021) (MAN) or in mannitol plus 0.4 µM TSA (TSA) (Castillo et al., 2020) and placed in the dark at 25°C. Microspores from anthers after 3 and 5 days of MAN (3dT-MAN, 5dT-MAN) and TSA (3dT-TSA, 5dT-TSA) treatments from Pavon and Caramba were isolated following the protocol described by Castillo et al. (2000). Isolated microspores from freshly harvested anthers (0dT) were used as control. In addition, isolated microspores at GD were used as a second control to discriminate the gametophytic and the sporophytic pathways.
Isolated microspores of Pavon and Caramba from 0dT, GD, 3dT-MAN, 5dT-MAN, 3dT-TSA, and 5dT-TSA were used. Leaves from 30 day-old plants, young embryos (14 days after pollination) and pollen grains were also included in this analysis. Proteins were extracted using the buffer containing 150 mM NaCl, 2 mM EDTA and 50 mM NaH2PO4, 2% SDS, pH 6. After homogenization, samples were boiled (95°C 10 min) and centrifuged twice at 15000 g, 10 min at 4°C. Protein extracts were quantified using Qubit Protein BR Assay kit following manufacturer’s instructions in fluorimeter Qubit4. Ten µg of total proteins from all samples, except from embryos (5 µg), were electrophoresed on a Mini-PROTEAN TGX 12% SDS-PAGE gel under reducing conditions. Proteins were transferred to a polyvinylidene difluoride membrane and blocked with 5% BSA in TBS-T (20 mM Tris, 150 mM NaCl, pH 7.5, and 0.1% Tween 20). The blots were incubated for 2 h with anti-histone H3 (clone A3S, Millipore), anti-histone H4 (clone 62-141-13, Millipore), anti-pan-acetyl-histone H3 (K9, K14, K18, K23, K27) (ab47915, Abcam) or anti-pan-acetyl-histone H4 (K5, K8, K12, K16) (PA1-84526 Invitrogen), all of them at 1:1000 dilution. Secondary goat anti-rabbit IgG peroxidase conjugated antibody (AP132P, Millipore) was used at a 1:1000 dilution and signals were detected using enhanced chemiluminescence (Clarity Western ECL substrate, BIORAD). Western blot was performed in two independent biological replicates.
Isolated microspores of both cultivars from 0dT, GD, 3dT-MAN, 5dT-MAN, 3dT-TSA, and 5dT-TSA were fixed in 4% paraformaldehyde and 2% sucrose in PBS, pH 7.3, overnight at 4°C. After three washes in PBS for 10 min, microspores were pre-embedded in 2% agarose and post-fixed in the same fixative solution during 2 h at 4°C. The samples were washed twice in PBS before being dehydrated in an ethanol series (30, 50 and 70%) at 4°C, processed in an automatic Vacuum Infiltration Processor (Sakura 4893-Floor E150, BAYER, Barcelona, Spain), and embedded in paraffin blocks. Four µm-thick sections were cut using a rotary microtome (Microm HM 340E, Bio Optica, Milan, Italy) with steel blades, and were collected on special slides (Menzel-Glaser, J1800AMNN, Braunschweig, Germany) to facilitate their adherence and avoid their loss during the performance of the immunohistochemical techniques.
The sections were deparaffinized with xylene and rehydrated with graded ethanol to water. Before the incubation with the primary antibodies, samples were treated with 0.05 M NH4Cl and 0.05 M NaBH4 in TBS (0.05 M TRIS, 0.5 M NaCl, pH 7.36) to partially reduce the autofluorescence of aldehydes (highly abundant in the exine of microspores) and were blocked with a 2% BSA solution at RT. The following primary antibodies, raised in rabbit, were used: anti-histone H3K9ac (EMD Millipore, Cat. No. 06-942; dilution 1:100), anti-histone H4K5ac (EMD Millipore, Cat. No. 07-327; dilution 1:100), anti-histone H4K16ac (EMD Millipore, Cat. No. 07-329; dilution 1:100), anti-histone H3K9me2 (EMD Millipore, Cat. No. 07-441; dilution 1:100) and anti-histone H3K27me3 (EMD Millipore, Cat. No. 07-449; dilution 1:50). The samples were incubated with primary antibodies overnight at 4°C. Goat anti-rabbit Alexa Fluor 488 (Sigma, Cat. No. SAB4600044; dilution 1:100) was applied as secondary antibody during 30 min at RT. Fluoromount G with DAPI (Invitrogen, Cat. No. 00-4959-52) was used as an aqueous mounting medium.
To study the localization pattern of epigenetic marks and to quantify the number of labeled microspores and nuclei, an inverted microscope Nikon Eclipse-T300 was used and fluorochrome Alexa Fluor 488 was excited at 460 nm, and emission was detected with a 505-520 nm long-pass filter. Microspores nuclei were counterstained with DAPI, excited at 350 nm in combination with a 400-420 nm long-pass filter. Images were recorded by the Digital sight DS-5MC camera with NIS-D software. Immunohistochemical analysis was performed in two independent biological replicates, containing an average of five technical replicates for each mark (H3K9ac, H4K5ac, H4K16ac, H3K9me2 and H3K27me3). An average of 1500 and 1000 microspores and 400 and 250 nuclei from Pavon and Caramba were analyzed, respectively.
Genes associated with ME induction response were selected based on a previous study performed in barley (Sánchez-Díaz, 2014). In this study, an Affymetrix Barley1 GeneChip microarray analysis was carried out on anthers before stress treatment, after 4 days of 0.7 M mannitol treatment, and after 4 and 8 days of culture, following the procedures described in Muñoz-Amatriaín et al. (2006). Identification and clustering of the differentially expressed genes allowed the selection of candidate genes that characterized barley ME induction. Among them, 20 barley probe set related to HDACs, HATs, and major processes involved in ME induction were selected (Supplementary Table 1).
Wheat genes with homology to barley probe set were identified based on sequence homology by BLASTN of barley sequences exported from HarvEST : Barley version 2.26 (http://www.harvest.ucr.edu), against Triticum aestivum IWGSC (Genomic sequence) on Ensembl Plants, which hosts the RefSeq v1.0 assembly from the IWGSC (http://plants.ensembl.org/Triticum_aestivum/Tools/Blast) or against WheatExp: An Expression Database for Polyploid Wheat (https://wheat.pw.usda.gov/WheatExp/). Overlapping genes with higher scores were selected, and homologues genes in bread wheat and orthologues in Arabidopsis thaliana and Oryza sativa Japonica Group were identified on Ensembl Plants. Only TaMS1 gene was directly selected from literature (Supplementary Table 1).
If primers were not available in the literature, the cDNA sequence of genes based on Ensembl Plants/Triticum_aestivum was used for designing specific primer pairs by Primer-BLAST (NCBI), which uses Primer3 (Ye et al., 2012) (Supplementary Table 2).
Selected genes ontology analyses were performed based on RefSeq v1.0 assembly by AgriGO v2.0, a database (Tian et al., 2017) (Supplementary Table 3).
Total RNA of three independent biological replicates from 0dT, GD, 3dT-MAN, 5dT-MAN, 3dT-TSA, and 5dT-TSA from Pavon and Caramba was isolated using TRIzol Reagent (Ambion, Life technologies) and passed through RNeasy columns (Qiagen) for further clean up. Double-stranded cDNA was synthesized using the M-MLV Reverse Transcriptase kit from Promega. qRT-PCR reactions were performed with PowerUp SYBR Green Master Mix (Applied Biosystems) and ROX (ROX Reference Dye, Invitrogen). The reaction conditions were optimized at: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, in the QuantStudio3 System (Applied Biosystems), using the Ta.27771 and Ta.2291 as reference genes (Paolacci et al., 2009). Two technical replicates were performed for each biological replicate in each gene.
The expVIP Wheat Expression Browser (http://www.wheat-expression.com) was used to study the expression of selected genes in different studies (Ramírez-González et al., 2018). The relative transcript abundance (per 10 million reads) from each gene obtained from the databases was presented as a heat map (Graphad Prism 7).
A densitometry analysis was performed on images of western blotting membranes, obtained in the ChemiDoc Imaging System (BIO-RAD), using the gel analyzer tool from Image-J software. Densitometry data allowed the estimation of the H3.1/H3.2, H3.1ac/H3.1, H3.2ac/H3.2 and H4ac/H4 ratios. In the immunofluorescence analysis, the following variables were calculated: the percentage of labeled microspores, that is, the number of microspores with each epigenetic marks over the total number of microspores, and the percentage of labeled nuclei, that is, the number of microspores with each epigenetic marks in the nuclei over the total number of microspores stained with DAPI. Normality and homogeneity of variance were tested using the Shapiro-Wilk and Levene´s tests, respectively. Data from the qRT-PCR reactions were analyzed using the Livak (2-∆∆CT) calculation method (Livak and Schmittgen, 2001). For the calculation of the standard errors, the values of the relative expression level (2-∆∆CT) were used. For statistical analysis, the values of fold change (2 -∆∆CT) were transformed with log2 to correct the heterogeneity of variance. A one-way analysis of variance (ANOVA) was used, and significant differences among treatments were determined by the Duncan test (p ≤0.05). Statistical analysis of these variables was performed using IBM SPSS statistics version 27.0.1. (IBM Corp, 2020).
All possible pairwise correlations of the expression of the studied genes over samples were calculated using Pearson´s coefficient. A principal component analysis (PCA) was performed to analyze the association between relative gene expression and the course of ME inducing treatments, using the program R 4.1.3 (R Core Team, 2021).
To determine if MAN and TSA treatments modified microspores global histone acetylation, a western blot assay was performed using a pan-acetylated Lys antibody for histones H3 and H4 (Figures 1A, B). Antibodies against unmodified histone H3 and H4 were used as controls. In the text, only statistically significant differences were described. However, in some cases a tendency to statistical significance has been described as slight.
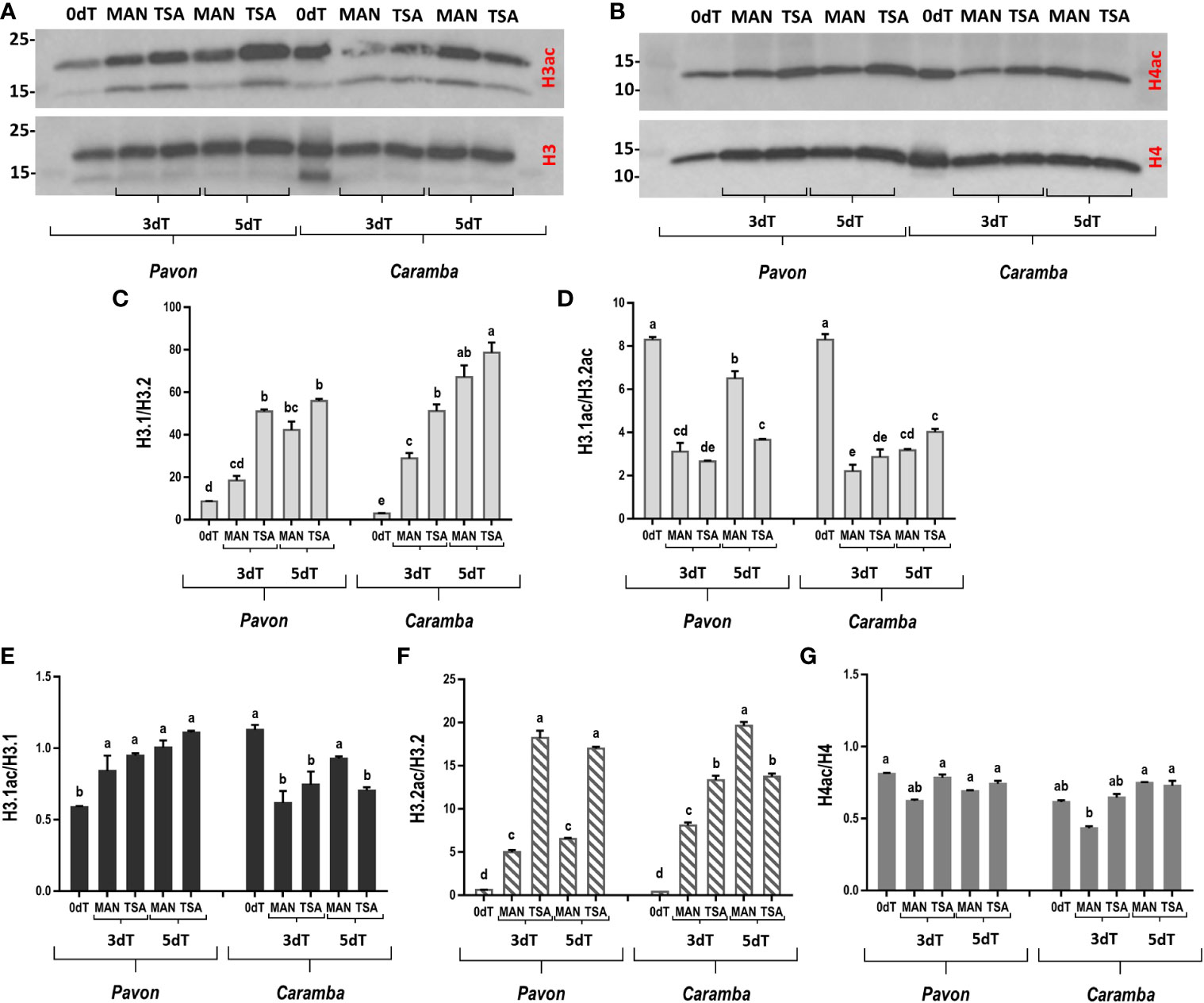
Figure 1 Effect of MAN and TSA treatments on histones H3 and H4 acetylation. (A, B) Immunoblot of histones H3 and H4 and pan-acetylated H3 and H4 (H3ac and H4ac, respectively). (C, D) Quantitative analysis of the H3.1/H3.2 and H3.1ac/H3.2ac ratios, respectively. (E, F) Quantitative analysis of the H3.1 and H3.2 acetylation ratio (G) Quantitative analysis of the H4 acetylation ratio. Microspores before stress treatment (0dT), after 3 and 5 days with mannitol treatment (3dT-MAN, 5dT-MAN), and after 3 and 5 days with mannitol plus TSA treatment (3dT-TSA, 5dT-TSA) from Pavon (on the left) and Caramba (on the right). Bars represent the standard errors of the means. Histone H3.1/H3.2, H3.1ac/H3.2ac, H3.1ac/H3.1, H3.2ac/H3.2 and H4ac/H4 ratios with the same letter within each ratio are not significantly different (P˂0.05) according to the Duncan test.
Two histone H3 variants with different molecular weight (17 KDa and 14 KDa, approximately) were detected in both cultivars (Figure 1A). These proteins corresponded to the major H3.1 variant and the minor H3.2 variant (H3.3-type, Waterborg, 1991). Quantitative analysis of the H3.1/H3.2 ratio showed a low value at 0dT, which increased greatly with MAN and TSA treatments, reaching the highest value at 5dT (Figure 1C). This ratio was higher with TSA than with MAN in both cultivars at 3dT. Given the change in the H3.1/H3.2 ratio during ME induction, the study was extended to young zygotic embryos, leaves, and GD, including mature pollen grains (Supplementary Figure 1A). The two variants were observed in all samples, although there were differences between cultivars, particularly in leaves (Supplementary Figures 1A, B). It was noteworthy the increase of H3.1/H3.2 in mature pollen (P) in both cultivars (Supplementary Figure 1B). Embryos showed the highest and the lowest amount of H3.1 and H3.2 variants, respectively, but quantification of H3.1/H3.2 was not possible.
Acetylation of both H3.1 and H3.2 variants was observed in all microspore samples (Figure 1A). The highest H3.1ac/H3.2ac ratio was showed at 0dT, and treatments reduced this ratio in both cultivars, especially at 3dT (Figure 1D). However, it increased greatly at 5dT-MAN in Pavon. Acetylation ratio of each variant was also studied (Figures 1E, F). At 0dT, H3.1ac/H3.1 was twice higher in Caramba than in Pavon. MAN and TSA treatments increased this ratio in Pavon but decreased it in Caramba (Figure 1E). H3.2ac/H3.2 had low and similar values at 0dT, but MAN and TSA induced H3.2 acetylation in both cultivars (Figure 1F). However, TSA enhanced acetylation up to approximately 4-fold over MAN at 3dT and 5dT in Pavon, but 1.5-fold only at 3dT in Caramba.
Analysis with H4 antibody showed that it was acetylated in all samples at a similar ratio to those of H3.1 variant (Figures 1B, E, G). It should be noted that MAN caused a slight decrease in H4 acetylation at 3dT in both cultivars, followed by an increase at 5dT in Caramba, reaching similar values in both cultivars (Figure 1G). The acetylation ratio at 3dT-TSA was slightly higher than at 3dT-MAN, but similar values were observed at 5dT.
Epigenetic dynamics were characterized by the pattern of intracellular localization and the percentage of labeled microspores or nuclei with specific antibodies for histone acetylation (H3K9ac, H4K5ac, H4K16ac) and methylation (H3K9me2 and H3K27me3). The percentage of labeled nuclei was only noted if it was different from that of the labeled microspores.
Uninucleate microspores from the two cultivars at 0dT showed large differences in epigenetic state, although they were at a similar morphological stage of development according to microscopic visualization. The sum of the percentages of microspores with acetylation marks was 35.85% in Pavon but only 9.29% in Caramba, with the values for each mark being lower in Caramba (Figure 2A). In contrast to acetylation, the sum of the percentages of microspores with methylation marks was lower in Pavon (12.89%) than in Caramba (33.14%) (Figure 2C). It should be noted the low number of microspores with H3K27me3 and the absence of H3K9me2 mark in Pavon. Furthermore, all acetylation marks were exclusively located in the nucleus in both cultivars (Figures 3A, B). H3K27me3 was observed in the cytoplasm (including the vacuole) in both cultivars and only in the nucleus in Pavon (Figures 3A, B).
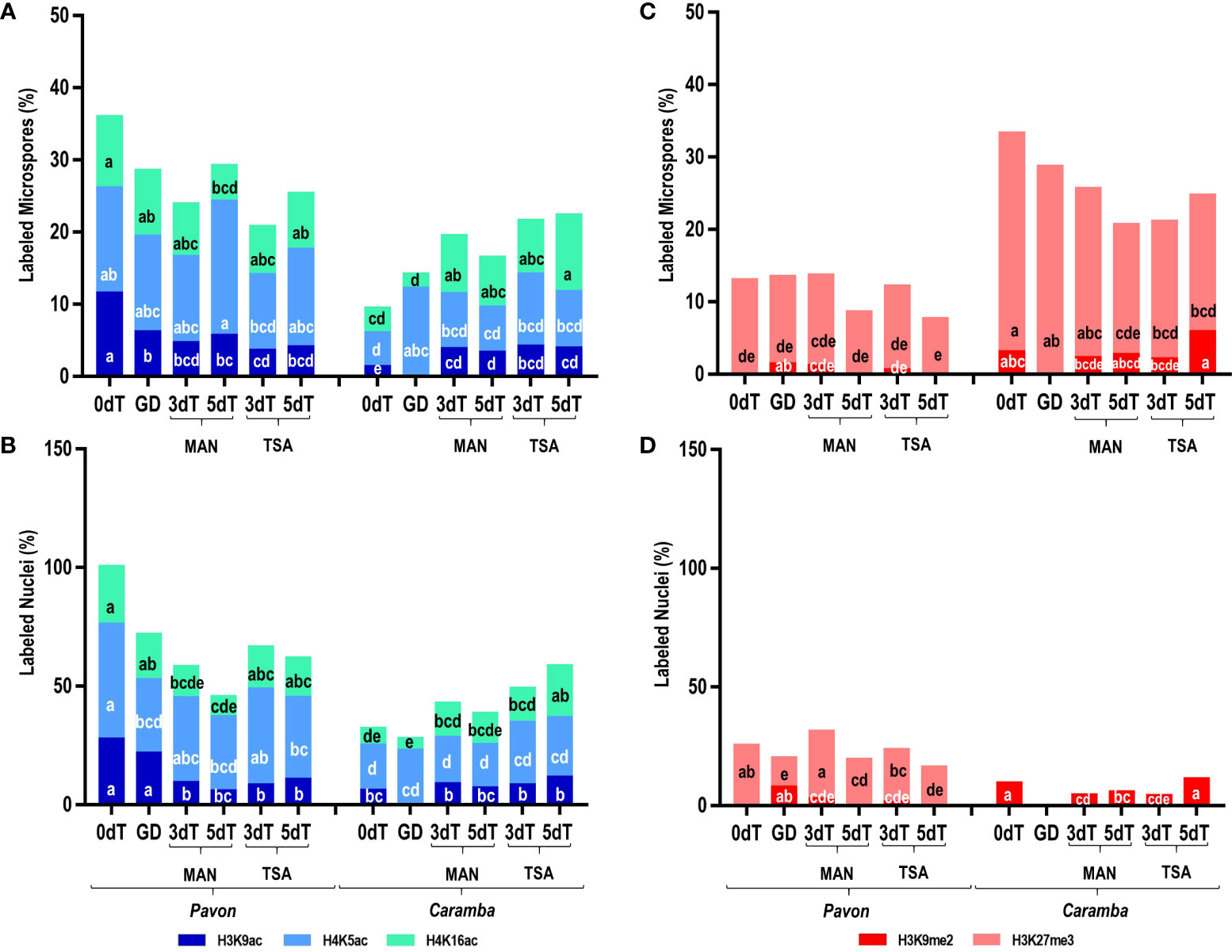
Figure 2 Effect of MAN and TSA on histone PTMs. (A, B) Percentage of labeled microspores and nuclei, respectively, with the acetylation marks H3K9ac (dark blue), H4K5ac (light blue), H4K16ac (green). (C, D) Percentage of labeled microspores and nuclei, respectively, with the methylation marks H3K9me2 (red), H3K27me3 (pink). Microspores before stress treatment (0dT), microspores under gametophyte development (GD), microspores after 3 and 5 days with mannitol treatment (3dT-MAN, 5dT-MAN) and after 3 and 5 days with mannitol plus TSA treatment (3dT-TSA, 5dT-TSA) from Pavon (on the left) and Caramba (on the right). Percentages of labeled microspore and nuclei with the same letter within each epigenetic mark are not significantly different (P˂0.05) according to the Duncan test.
Differences in the dynamics of PTMs at GD were also observed between cultivars. The sum of percentages of acetylation decreased in Pavon as compared with 0dT (28.39% vs 35.85%), mainly due to the lower value of H3K9ac (6.03% vs 11.40%) (Figure 2A). There was a global increase of acetylation in Caramba (14.01%) due to H4K5ac (12.07%), which reached similar percentages to Pavon (13.25%). H4K5ac mark was observed in the nucleus and the cytoplasm (including the vacuole) in both cultivars and H3K9ac was absent in Caramba (Figures 3A, B). Concerning methylation, H3K9me2 was detected in a low percentage of nuclei (7.31%) in Pavon and was absent in Caramba (Figure 2D), and H3K27me3 was localized in the cytoplasm (including the vacuole) in both cultivars (Figure 3B).
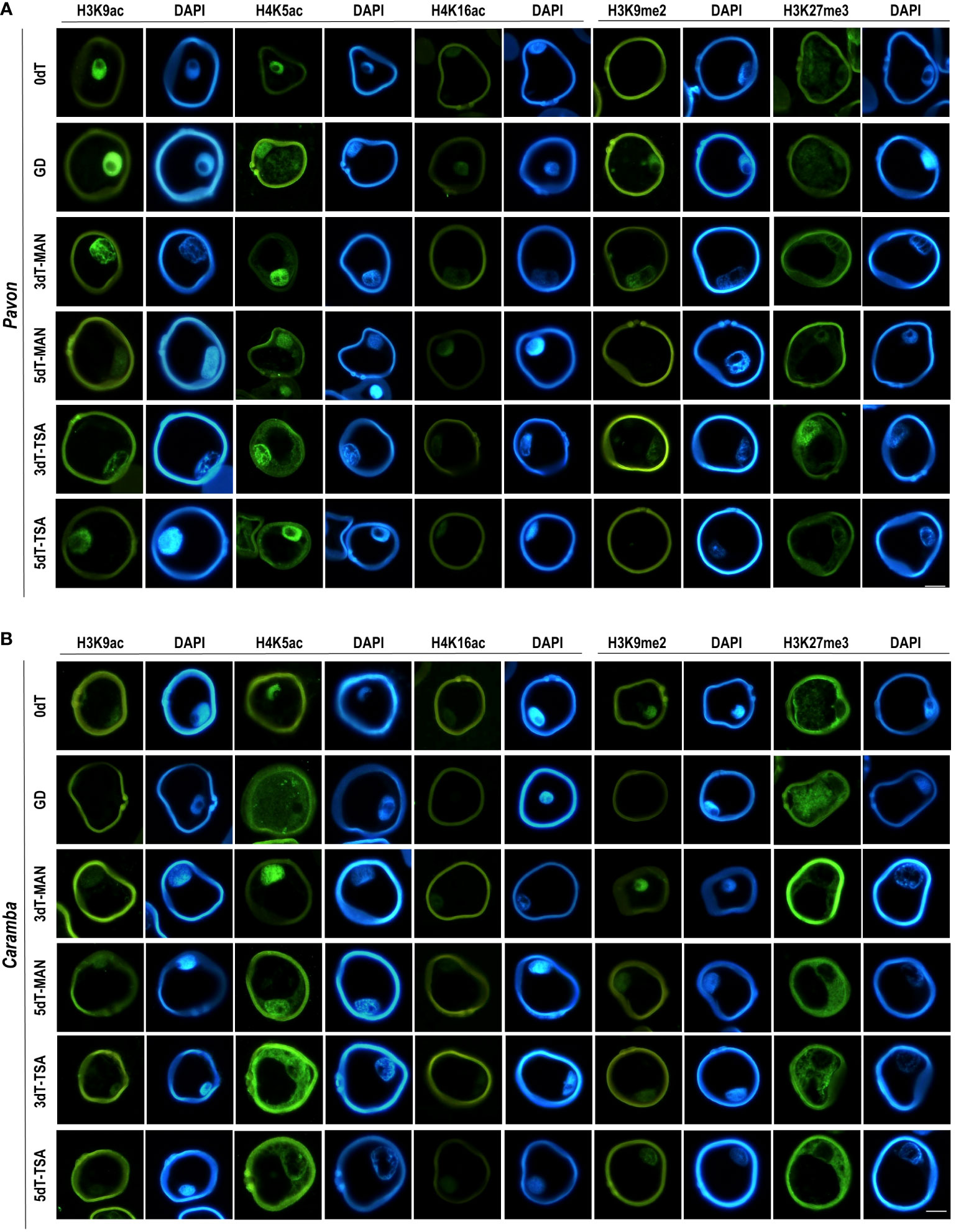
Figure 3 Immunolocalization of histone PTMs in microspores before stress treatment (0dT), microspores under gametophytic development (GD), microspores after 3 and 5 days with mannitol treatment (3dT-MAN, 5dT-MAN) and after 3 and 5 days with mannitol plus TSA treatment (3dT-TSA, 5dT-TSA) from Pavon (A) and Caramba (B). H3K9ac, H4K5ac, H4K16ac, H3K9me2 and H3K27me3 antibodies labeling in green, DAPI in blue. Scale bar = 20 µm.
Induction treatment with MAN modified the epigenetic state of microspores compared to 0dT and GD, and determined differences between cultivars. In Pavon, the sum of the percentages of acetylated microspores at 3dT (23.76%) decreased as compared to 0dT (35.85%) or GD (28.39%), mainly due to lower value of H3K9ac (4.51% vs 11.40%) (Figure 2A). A decrease was also observed in the percentage of labeled nuclei with H4K16ac (Figure 2B). However, the percentage of acetylated microspores rose at 5dT-MAN (29.07%), mainly due to the H4K5ac mark, although this percentage decreased in the nuclei (Figure 2B). In Caramba, the dynamics of epigenetic marks was different from that of Pavon. An increase in the percentage of acetylated microspores was observed at 3dT with respect to 0dT (19.36% vs 9.29%), being significant for H3K9ac (3.67% vs 1.19%) and H4K16ac (8.06% vs 3.42%) (Figure 2A). Nevertheless, the differences were not significant in the nuclei (Figure 2B). A slight decrease in all acetylation marks was noticed at 5dT (Figure 2A).
Regarding methylation, the percentage of labeled microspores with H3K9me2 was very low at 3dT-MAN (1.08%), and it was absent at 5dT-MAN in Pavon (Figure 2C). In Caramba, this percentage was not modified with MAN, but the percentage of labeled nuclei decreased (Figure 2D). Furthermore, the percentage of microspores with H3K27me3 decreased at 5dT (17.98%) compared to 0dT (30.18%).
The localization of H4K5ac and H3K27me3 with MAN also revealed differences between the two cultivars. H4K5ac was detected in the nucleus and cytoplasm at both 3dT and 5dT in Pavon (Figure 3A). In Caramba, it was only observed in the nucleus at 3dT, but also in the cytoplasm at 5dT (Figure 3B). H3K27me3 was present in the nucleus and cytoplasm in Pavon (Figure 3A), but only in the cytoplasm in Caramba (Figure 3B).
TSA treatment produced modifications in the epigenetic state of the microspores compared to MAN. TSA decreased the percentages of microspores with acetylation marks in Pavon (20.61% at 3dT-TSA and 25.25% at 5dT-TSA vs 23.76% at 3dT-MAN and 29.07% at 5dT-MAN) (Figure 2A). However, the sum of the percentages of labeled nuclei was higher at 5dT-TSA than at 5dT-MAN, mainly due to the H4K16ac mark (Figure 2B). In Caramba, higher percentages of acetylated microspores were observed with TSA than with MAN, especially at 5dT (22.24% vs 16.37%), highlighting the increase of the H4K16ac mark. The most remarkable change produced by TSA was the localization of H4K5ac in both the nucleus and cytoplasm at 3dT-TSA in Caramba (Figure 3B). Finally, TSA resulted in an increase in the percentage of labeled nuclei with H3K9me2 at 5dT-TSA compared to 5dT-MAN in Caramba, and a decrease in the percentage of nuclei with H3K27me3 at 3dT-TSA in Pavon (Figure 2D).
The immunohistochemical analysis revealed that MAN and TSA modified the epigenetic state of microspores. Therefore, the dynamics of transcriptional changes in genes controlling histone acetylation state and genes involved in key processes of ME induction produced by treatments was studied. The candidate genes were selected based on a previous study on barley ME induction (Sánchez-Díaz, 2014). Genes were associated with GO terms as a source of information about gene functions (Supplementary Table 3).
Histone acetylation depends on the balance between HDAC and HAT. In this study, six wheat HDAC genes (TaHDA8, TaHDA15, TaHDA18, TaHDT1, TaHDT2 and TaTAF14) and five HAT genes (TaHAG2, TaHAG3, TaHAM, TaHAC1 and TaHAC4) were studied. Three HDAC genes were not considered for further analysis as TaHDA8 was not amplified, and TaHDT2 and TaTAF14 showed no differences between the samples.
In microspores at 0dT, only two HATs genes (TaHAC4 and TaHAG2) showed differential expression between the two cultivars. Both genes were expressed at a lower level in Pavon, although the differences were only significant in TaHAC4 (Figure 4). Also noteworthy was the high expression level of TaHDT1 in both cultivars. In microspores at GD, TaHDT1 was down-regulated in both cultivars, TaHAG2 in Caramba, and TaHAC4 was up-regulated in Pavon (Figure 4).
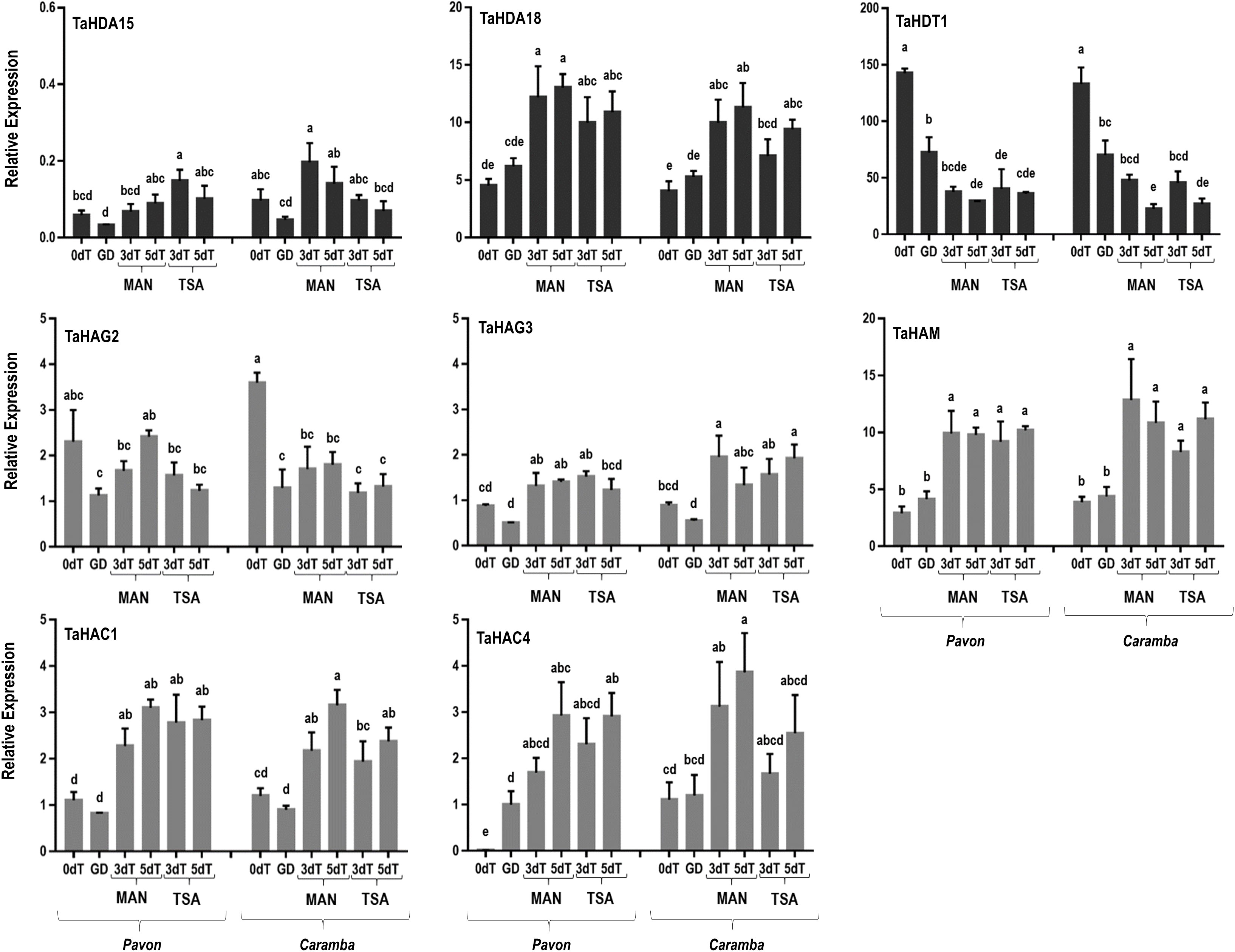
Figure 4 qRT-PCR analysis of histone deacetylases genes (TaHDA15, TaHDA18, TaHDT1, TaHDT2, TaTAF14) and histone acetyltransferases genes (TaHAG2, TaHAG3, TaHAM, TaHAC1, TaHAC4) in microspore before stress treatment (0dT), microspores under gametophytic development (GD), microspores after 3 and 5 days with mannitol treatment (3dT-MAN, 5dT-MAN) and after 3 and 5 days with mannitol plus TSA treatment (3dT-TSA, 5dT-TSA) from Pavon (on the left) and Caramba (on the right). Data were normalized using Ta.27771 and Ta.2291 as endogenous control genes. Bars represent the standard errors of the means. Relative expression values with the same letter are not significantly different (P<0.05) according to the Duncan test.
MAN treatment modified the expression of most HDAC and HAT genes (Figure 4). The decrease in TaHDT1, compared to 0dT, was even higher than at GD, reaching the lowest values at 5dT in both cultivars. A similar decrease in TaHAG2 throughout MAN treatment was observed in Caramba, whereas this gene was only slightly down-regulated at 3dT in Pavon. The TaHDA18 and four HAT genes (TaHAG3, TaHAM, TaHAC1 and TaHAC4) were up-regulated at 3dT in both cultivars. The expression at 3dT and 5dT in most of the above mentioned genes were similar in both cultivars. TaHDA15 was not induced in any cultivar, but Pavon showed a lower expression level than Caramba at 3dT (Figure 4).
TSA showed a tendency to up-regulate most genes in Pavon and to down-regulate them in Caramba, compared to MAN (Figure 4). Notably, TSA treatment caused an increase in TaHDA15 expression at 3dT in Pavon. In addition, Pavon showed a lower TaHAG3 expression than Caramba at 5dT-TSA.
The transcriptome and RNA-Seq data set available on the expVIP Wheat Expression Browser was used to analyze the expression of the selected genes under different stresses and in different wheat tissues (Supplementary Figure 2). TaHDA15 and TaHAG2 were specifically expressed in the spike, whereas a low expression of TaHAC1 and TaHAC4 was observed in all tissues. In the RNA-Seq data of bread wheat cv Svilena microspores induced to ME by cold, only TaHAG3 and TaHAM were up-regulated after stress (Supplementary Figure 3).
Candidate genes involved in key processes associated with ME induction, such as histone methylation, DNA silencing, nuclear fusion, pollen wall development, autophagy, ABA signaling, auxin synthesis, and stress tolerance were also examined.
In microspores at 0dT, four genes showed differential expression between cultivars: TaDRM3 (DOMAINS REARRANGED METHYLTRANSFERASE 3), encoding a DNA methyltransferase; TaMS1 (MALE STERILITY 1), that encodes a protein required for exine development; TaATG18fD (AUTOPHAGY-ASSOCIATED 18fD); and Ta-2B-LBD16 (LATERAL ORGAN BOUNDARIES DOMAIN 16), encoding a plant-specific transcription factor (Figure 5). Pavon showed higher expression of TaMS1 and Ta-2B-LBD16 and lower expression of TaDRM3 and TaATG18fD than Caramba (Figure 5). Two genes TaABI5 (ABCISIC ACID-INSENSITIVE 5) and TaYUC11D (INDOLE-3-PYRUVATE MONOOXYGENASE YUCCA 11D), that encode a basic leucine zipper transcription factor and an auxin synthesis limiting enzyme, respectively, were also differentially expressed between cultivars at a very low level. TaNFD6-A (NUCLEAR FUSION DEFECTIVE 6-A), encoding a mitochondrial ribosomal protein involved in nuclear membrane fusion, showed a slightly down-regulation in Pavon.
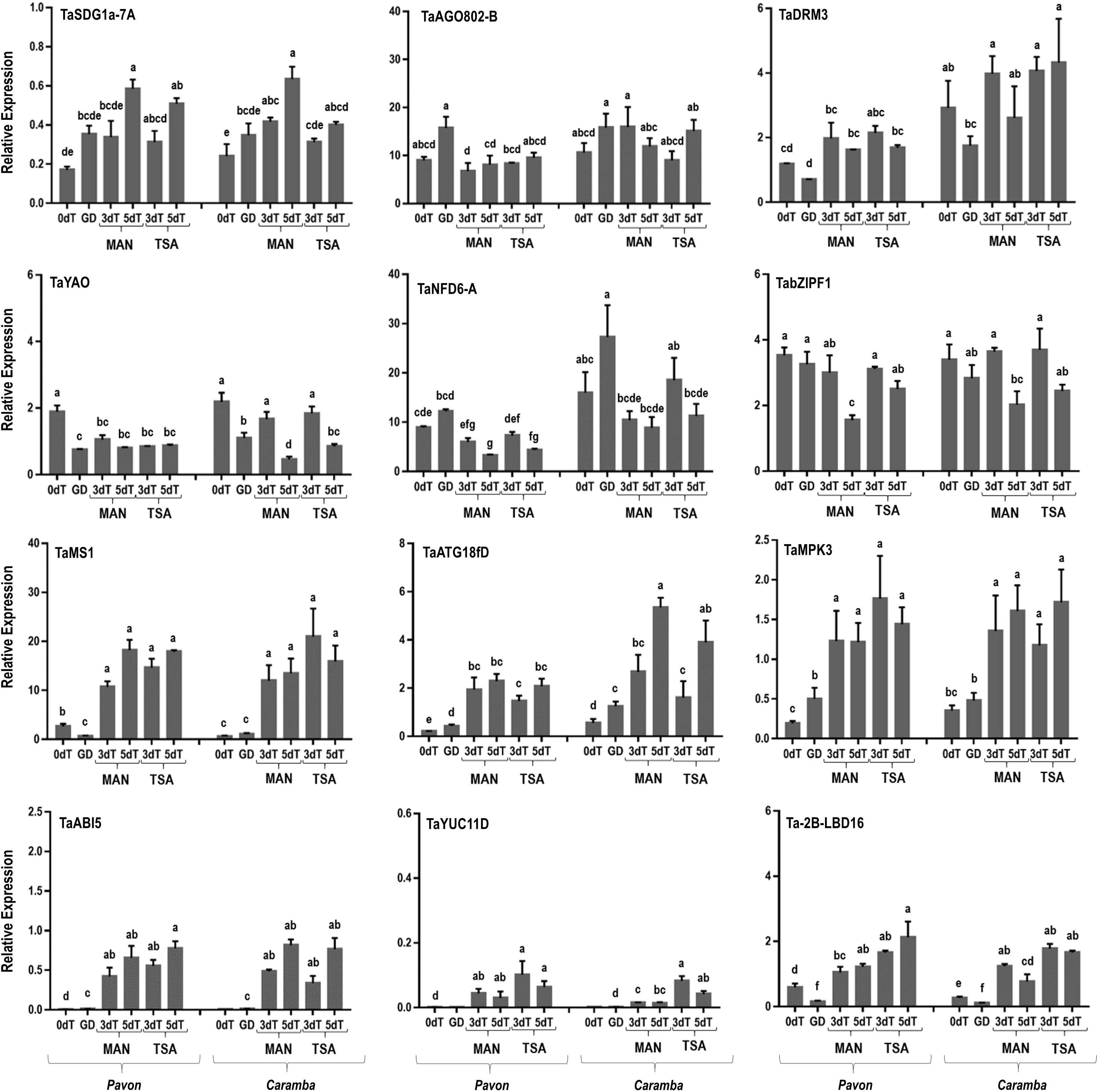
Figure 5 qRT-PCR analysis of key genes involved in microspore embryogenesis induction. Microspores before stress treatment (0dT), microspores under gametophytic development (GD), microspores after 3 and 5 days with mannitol treatment (3dT-MAN, 5dT-MAN) and after 3 and 5 days with mannitol plus TSA treatment (3dT-TSA, 5dT-TSA) from Pavon (on the left) and Caramba (on the right). Data were normalized using Ta.27771 and Ta.2291 as endogenous genes control. Bars represent the standard errors of the means. Relative expression values with the same letter are not significantly different (P˂0.05) according to the Duncan test.
Changes in the expression of several genes were observed when comparing GD with 0dT (Figure 5). A reduction in TaMS1 expression was shown in Pavon. TaYAO (YAOZHE), that encodes a protein involved in rRNA processing, and Ta-2B-LBD16 were down-regulated in both cultivars. In contrast, up-regulation of TaMPK3 (MITOGEN-ACTIVATED PROTEIN KINASE 3) in Pavon and TaATG18fD in both cultivars was observed. Genes TaDRM3, TaYAO, TaNFD6-A, TaATG18fD and TaYUC11D were differentially expressed between cultivars, with Pavon showing a lower level.
MAN treatment at 3dT increased the expression of most genes (TaMS1, TaATG18fD, TaMPK3, TaABI5, TaYUC11D, and Ta-2B-LBD16) compared to 0dT in both cultivars (Figure 5). Interestingly, TaSDG1a-7A (SET DOMAIN GENE 1a-7A), encoding a histone methyltransferase, was up-regulated only in Caramba and TaYAO was the only gene down-regulated in Pavon. Differences between cultivars were also observed in TaAGO802-B, TaDRM3, TaYAO, and TaYUC11D, being TaYUC11D the only one with a higher expression in Pavon. When MAN treatment was extended from 3 to 5 days, some genes changed their expression. TaSDG1a-7A increased in Pavon, and TaATG18fD in Caramba. Also, TaYAO, TabZIPF1 (BASIC LEUCINE ZIPPER F1), described as an E2F-RELATED (E2F) transcription factor, and Ta-2B-LBD16 were down-regulated in Caramba. Differences between cultivars were also noticed at 5dT-MAN, with Ta-2B-LBD16 showing higher expression, and TaNFD6-A and TaATG18fD lower in Pavon.
TSA only resulted in small changes at 3dT compared to MAN in Caramba. A higher expression of TaYUC11D was accompanied by a slightly lower expression of TaAGO802-B. Furthermore, TabZIPF1 in Pavon and TaYAO and Ta-2B-LBD16 in Caramba were up-regulated at 5dT-TSA (Figure 5).
The analysis of the transcriptome and RNA-Seq data set available showed that all genes had a low expression level, except TaAGO802-B, TabZIPF1, TaMPK3 and Ta-2B-LBD16 (Supplementary Figure 2). Some of these genes were associated with stress response, such as TaMPK3 and Ta-2B-LBD16, while TaABI5 and TaNFD6-A were specifically expressed in grains and roots, respectively, and TaAGO802-B and TaMS1 in spikes. Differences in the expression between homologues of TaMS1, TabZIPF1 and TaMPK3 were also detected. In the RNA-Seq data of ME induction, few of the studied genes were expressed. TaAGO802-B was shown as a constitutive gene, whereas TaYAO was mainly expressed in embryogenic microspores, TaATG18fD in cold treated and embryogenic microspores, and TabZIPF1 in fresh and cold treated microspores (Supplementary Figure 3).
Pearson correlation coefficients were calculated to examine the association between the relative expression of genes determining histone acetylation status and genes involved in processes related to ME induction. The data from all samples (0dT, GD, 3dT-MAN, 5dT-MAN, 3dT-TSA, and 5dT-TSA) in the two cultivars were used.
Two groups could be clearly distinguished in the correlation matrix (Figure 6A). The first revealed a strong positive correlation between the histone deacetylase TaHDT1 gene and the set of genes TaYAO, TaNFD6-A and TabZIPF1, and a negative correlation with most of the other genes. TaHAG2 acetyltransferase was positively correlated with TaHDT1 and TaYAO but was independent of TaDRM3, TaNFD6-A and TabZIPF1. The majority of HDACs and HATs (TaHDA15 and TaHDA18, TaHAG3, TaHAM, TaHAC1 and TaHAC4) were in the second group that was positively associated with TaSDG1a-7A, TaMS1, TaATG18fD, TaMPK3, TaABI5, TaYUC11D and Ta-2B-LBD16 (Figure 6A).
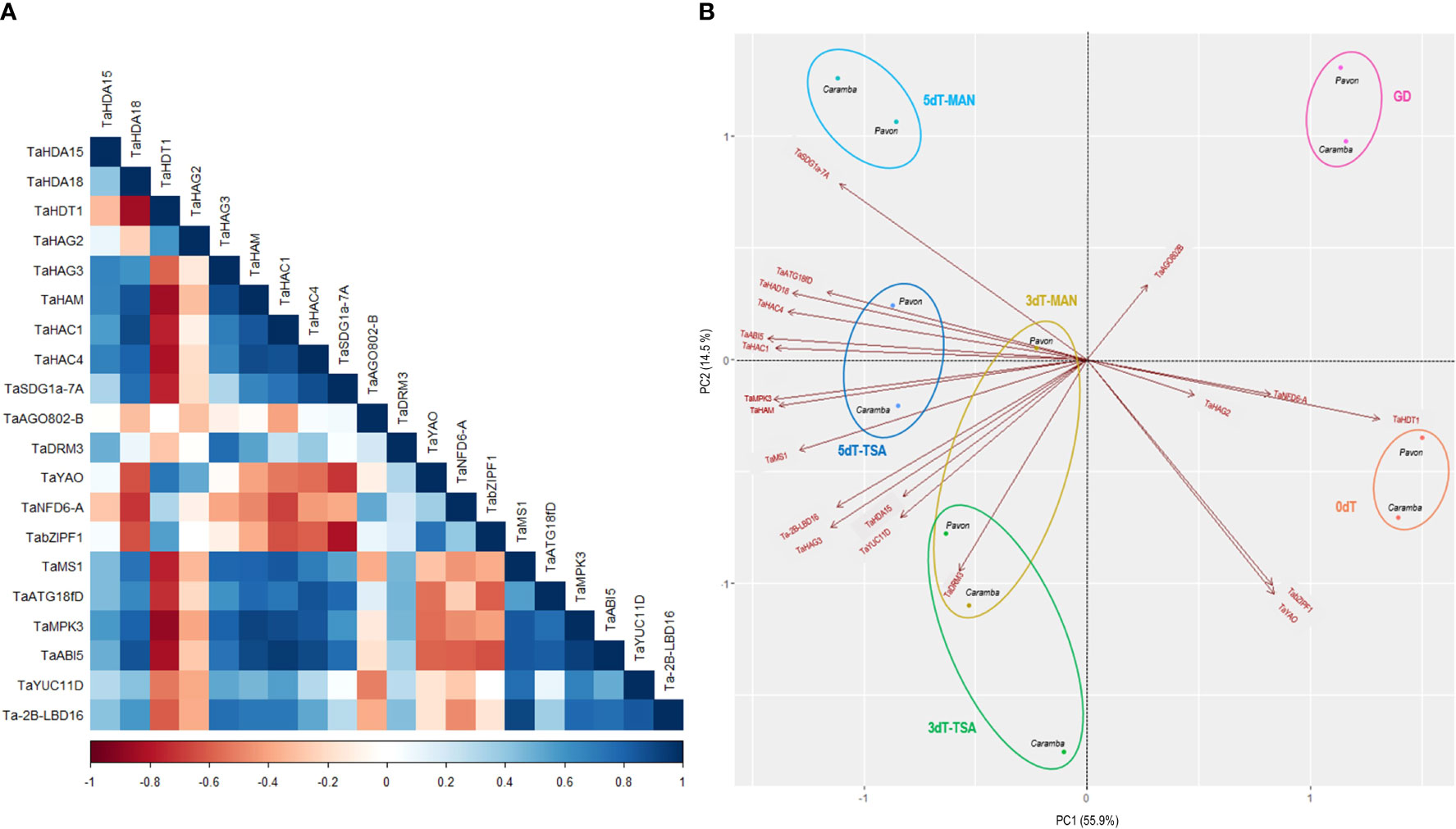
Figure 6 (A) Pearson correlation coefficients of histone deacetylases, histone acetyltransferases and key genes involved in ME induction, considering all samples of the two cultivars. (B) Principal component analysis (PCA) plot of the 20 genes in Pavon and Caramba. Microspores before stress treatment (0dT), microspores under gametophytic development (GD), microspores after3 and 5 days with mannitol treatment (3dT-MAN, 5dT-MAN) and after 3 and 5 days with mannitol plus TSA treatment (3dT-TSA, 5dT-TSA).
Principal component analysis (PCA) was performed to identify the genes associated with wheat ME induction. The data set included the relative expression of the genes in all samples from Pavon and Caramba (Figure 6B). The two components analysis accounted for 70.4% of the total variance, 55.9% explained by PC1 and 14.5% by PC2. PC1 was mainly determined by TaHDA18, TaHDT1, TaHAM, TaHAC1, TaHAC4, TaMS1, TaMPK3 and TaABI5 genes, and PC2 by TaHDA15, TaHAG3, TaSDG1a-7A, TaDRM3, TaYAO, TabZIPF1, TaYUC11D and Ta-2B-LBD16 genes.
PC1 differentiated untreated (0dT) and GD from MAN and TSA microspores of both cultivars, on the right and left respectively (Figure 6B). PC1 showed that TaHDT1 was strongly associated with microspores at 0dT and GD. TaYAO and TabZIPF1 contributed to the differentiation between 0dT and GD microspores. On the opposite position in PC1, TaHDA18, TaHAM, TaHAC1, and TaHAC4 were positively associated with stress treated microspores, accompanied by TaMS1, TaMPK3, TaABI5. The differences between 3dT and 5dT were determined by a strong contribution of TaSDG1a-7A, TaHDA15, TaHAG3, TaDRM3, TaYUC11D and Ta-2B-LBD16.
Increasing evidence indicates that epigenetic changes are involved in the regulation of biological processes including stress response and developmental reprogramming, both implicated in ME induction (for review see Lloyd and Lister, 2022). Histone PTMs, mainly acetylation and methylation, and DNA methylation have been directly associated with ME induction. Our previous report showed that the simultaneous application of mannitol and TSA increased ME efficiency compared to mannitol in two bread wheat cultivars with different ME response (Castillo et al., 2020). However, it remains a challenge to ascertain the mechanisms by which chromatin organization responds to transcriptional requirements and how TSA modifies them during ME induction.
Pollen development is characterized by specific chromatin changes, including the dynamics of H3 variants, the progressive loss of active histone marks and the accumulation of repressive marks (Borg and Berger, 2015). Furthermore, transcriptome complexity is significantly reduced after pollen mitosis I (PMI) (Liu et al., 2018a). However, uninucleate microspores (0dT) become transcriptionally active just before PMI, initiating the transition from sporophyte to gametophyte (Nelms and Walbot, 2022).
Histone variants contribute to define different chromatin states (for a review see Probst et al., 2020). The replicative histone H3.1 variant is associated with gene silencing in heterochromatin and the replacement H3.3 variant with active transcription and euchromatin regions (for a review see Probst et al., 2020). The H3.2 identified in wheat belongs to the histone H3.3 class (Waterborg, 1991). According to Otero et al. (2016), the ratio between H3.1 and H3.3 could be related to a proliferative state of the cell in Arabidopsis. Interestingly, the H3.1/H3.2 ratio was higher in Pavon than in Caramba at 0dT, that could indicate an elevated cell division potential of Pavon (Otero et al., 2016). Differences in H3.1 acetylation have been associated with diverse heterochromatin domains and compaction levels (Jasencakova et al., 2001). The lower H3.1 acetylation suggests a higher heterochromatin compaction in Pavon (Figure 1).
One of the major epigenetic differences between cultivars at 0dT was the higher percentages of acetylated microspores in Pavon, highlighting the percentage of H3K9ac (Figure 3A). At GD, the percentage of labeled microspores with H3K9ac decreased in Pavon whereas this mark was absent in Caramba (Figure 3A). It is known that H3K9ac mark is present in microspores but is removed during pollen development (Pandey et al., 2013; Liu et al., 2018a). In accordance, Pavon microspores could be at a less advanced stage in pollen development. Curiously, H4K5ac was localized in the cytoplasm, including the vacuole, in both cultivars at GD (Figure 2), the increase of labeled microspores with this mark in Caramba being noteworthy (Figure 3A). The presence of H4K5ac in the cytoplasm has been previously described in barley and Brachypodium (Braszewska-Zalewska et al., 2013; Wolny et al., 2014). These authors propose that newly synthetized acetylated H4, prior to DNA replication, is present in the cytoplasm while the recycling of acetylated H4 takes place in the vacuole. Differences in H4K5ac dynamics suggests that Pavon is less primed to enter in DNA replication. Histone acetylation depends on the balance between HDAC and HAT. No major differences in HDAC or HAT expression were observed between cultivars at 0dT, except for TaHAC4 that showed a lower level in Pavon (Figure 4). TaHAC4 has been associated with the enrichment of H3K9ac in Arabidopsis (Li et al., 2022). In contrast, the expression level of this gene was negatively correlated with the percentage of microspores with H3K9ac in both cultivars.
In Arabidopsis, the diploid to haploid transition is governed by the loss of H3K9me2 mark, and the transition back to diploid by the loss of H3K27me3 (Borg et al., 2021). However, H3K9me2 mark persisted throughout pollen development in rye and barley (Houben et al., 2011; Pandey et al., 2013). In this study, H3K9me2 was absent in Pavon and present in Caramba at 0dT, and the opposite was observed at GD (Figure 2). This could be associated with small adjustments to control transposon reactivation in the two cultivars during pollen development (Pecinka et al., 2010). Interestingly, in Caramba, H3K27me3 was exclusively in the cytoplasm that could also be related to an accumulation of histones prior to DNA replication as indicated in barley (Pandey et al., 2017). However, the progressive decrease of the percentage of labeled nuclei and the enrichment of H3K27me3 in the cytoplasm reinforce the less advanced stage in pollen development of Pavon microspores.
Differences in the epigenetic state of the cultivars at 0dT were barely reflected at the transcriptional level of the genes involved in ME induction, with the exception of TaDRM3 and TaNFD6-A. DRM3 participates in de novo DNA methylation via RNA (RdDM) and is crucial to maintain CHH methylation, although the levels in microspores are low (Henderson et al., 2010; Gehring, 2019). TaNFD6-A encodes a protein related to nuclear membrane fusion, playing an important role in female and male gametogenesis (Portereiko et al., 2006). At 0dT, Caramba microspores had a higher expression of both genes, but TaDRM3 decreased and TaNFD6-A increased in both cultivars at GD. Other genes related to pollen development, as TaMS1 and Ta-2B-LBD16 (Li et al., 2021; Xu et al., 2021), had a higher expression at 0dT in Pavon and were down-regulated at GD in both cultivars. The expression pattern of these genes confirmed that Pavon microspores were more delayed in pollen development.
The relationship between epigenetic modifications, transcriptional changes and ME induction is largely unknown. The analysis of epigenetic dynamics in coordination with gene regulation offers new opportunities to study the mechanisms underlying microspore reprogramming towards embryogenesis.
H3 variants are involved in transcriptional regulation in response to stress (Yung et al., 2021). However, no previous information about the role of histone variants in ME induction is available. In this study, MAN and TSA raised the H3.1/H3.2 ratio in both cultivars, highlighting the TSA effect at 3dT (Figure 1). These results agreed with a high cell division potential (Otero et al., 2016), and with the higher number of pro-embryos produced by TSA (Castillo et al., 2020). Histone hyperacetylation processes were observed in MAN and TSA (Figure 1), accordingly with previous results reported in Brassica ME induction by heat and TSA (Li et al., 2014a). The results obtained in this study confirm the higher hyperacetylation capacity of the H3.2 variant described by Waterborg (1991). The effect of TSA on H3.2 hyperacetylation enhancement at 3dT (Figure 1) could be an important factor for ME induction in both cultivars. MAN and TSA caused only minor H4 deacetylation/acetylation processes, although H4 acetylation was slightly higher with TSA in both cultivars at 3dT.
Long-duration and medium-intensity stress treatments have epigenetic characteristics that allow for a greater adaptation and survival (Pecinka et al., 2010). Thus, acetylation is positively associated with the activation of stress response genes in plants (Kong et al, 2020), but deacetylation is also essential to cause growth responses to warm temperature (Tasset et al., 2018).
In this study, the effect of the MAN and TSA on histone acetylation depended on the epigenetic state of the microspores at 0dT. A strong decrease in the percentage of microspores with H3K9ac in Pavon and a large increase in H3K9ac and H4K16ac in Caramba were observed at 3dT (Figure 3A). Interestingly, both PTMs were detected in the nuclei after treatments in this study, whereas H3K9ac was mainly located in the cytoplasm in barley (Pandey et al., 2017). The cytoplasmic H4K5ac did prove to be distinctive between treatments in Caramba, emerging at 3dT with TSA but at 5dT with MAN (Figure 2B). As mentioned above, the presence of H4K5ac in the cytoplasm could point out to a nascent acetylated H4 for DNA synthesis. Therefore, TSA could accelerate the replication processes in the mid-low responding cultivar. Curiously, one effect of TSA in maize seedlings is the hyperacetylation of H4K5, which is associated with cell cycle arrest in prophase (Wang et al., 2016). Furthermore, DNA replication arrested in G1 is related to the accumulation of the cytoplasmic H4K5ac signals in dry seeds of Brachypodium (Wolny et al., 2014).
The percentages of labeled microspores with H3K9me2 and H3K27me3 silencing marks during treatments seemed to be determined by the cultivar and the epigenetic state of the microspore at 0dT, similarly to histone acetylation. In MAN and TSA, H3K9me2 emerged at 3dT in Pavon but it was present in Caramba at 0dT and throughout treatments. A striking increase in the percentage of labeled microspores at 5dT-TSA was observed in Caramba (Figure 3B). However, the presence of H3K9me2 was noticed during ME induction in a high responding barley cultivar (Pandey et al., 2017). The loss of H3K27me3 accompanies callus induction and plant regeneration in Arabidopsis (He et al., 2012; Yan et al., 2020). In this study, the presence of this mark in Pavon nuclei could indicate some level of silencing, which is not contradictory to an active response to ME induction. In Caramba, H3K27me3 is restrictive to the cytoplasm as in a barley high responding cultivar to ME (Pandey et al., 2017). Even if there were differences between cultivars, it was not possible to establish a direct correlation between methylation marks and ME induced by MAN and TSA.
The gene networks regulated by histone PTMs are still poorly characterized (Kumar et al., 2021). In addition, few reports are focused on transcriptional changes accompanying wheat ME induction (Seifert et al., 2016; Jensen, 2021), and the direct impact of PTMs dynamics on key genes related to ME is unknown.
After 3 days of MAN or TSA treatments, most genes studied were up-regulated compared to 0dT and GD. Only a few genes associated with 0dT or GD in PCA analysis, TaHDT1, TaHAG2, TaYAO, TaNFD6-A, TabZIPF1 and TaAGO802-B were down-regulated (Figures 4, 5, 6). TaYAO, TaNFD6-A, TabZIPF1 and TaAGO802 were directly related to different pollen development processes as 18S pre-rRNA processing, zinc homeostasis, DNA silencing, or other unknown functions (Portereiko et al., 2006; Li et al., 2010; Evens et al., 2017; Akond et al., 2020). Disruption of the gametophytic developmental pathway has been reported as a key process in barley ME induction with mannitol (Muñoz-Amatriaín et al., 2006; Muñoz-Amatriaín et al., 2009). In this study, a progressive down-regulation of most genes associated with the disruption of the gametophytic development was observed, with a tendency to be faster in Pavon. Surprisingly, TSA caused a slower down-regulation of TaYAO and TaNFD6-A in Caramba, and TabZIPF1 in Pavon. TaAGO802-B deserves an especial mention, since it exhibited a remarkable difference between MAN and TSA, with opposite expression in Caramba at 3dT (Figure 5).
The epigenetic control of the transcriptional regulation of these genes could be associated with TaHDT1 and TaHAG2. TaHDT1 sharply decreased at 3dT in both treatments and cultivars as compared to 0dT. This gene is a negative regulator of biotic defense response in wheat (Zhi et al., 2020). Accordingly, the activation of pathogen defense genes has been previously described in barley and wheat ME induction (Muñoz-Amatriaín et al., 2006; Soriano, 2008). Moreover, HDT1 binds to the YAO promoter during rRNA processing (Luo et al., 2022). Therefore, the down-regulation of TaHDT1 could lead to a reduction on rRNA processing, allowing the elimination of gametophytic development by prioritizing the synthesis of stress-response RNAs. TaHAG2 is activated in response to drought and medium temperature stresses (Gao et al., 2021; Li et al., 2022). In this study, TaHAG2 was down-regulated with the stress, showing only slight differences between treatments.
TaHDA15, TaHAG3, TaHAM, TaYUC11D, Ta-2B-LBD16, TaMS1 and TaDRM3 constitute a network according to a PCA analysis (Figure 6), characterized by activation a 3dT and few expression changes at 5dT (Figures 4, 5). TaYUC11D is a limiting enzyme in auxin synthesis (Yang et al., 2021). In ME, auxin accumulation is localized in uninucleate microspores after induction in Brassica (Dubas et al., 2014; Soriano et al., 2014). De novo auxin biosynthesis is noticed at early multicellular embryo in barley and Brassica ME, increasing during embryo development. (Rodríguez-Sanz et al., 2014; Pérez-Pérez et al., 2019). In addition, it has been suggested that YUC11 is responsible of auxin biosynthesis require for zygotic embryo polarization in Arabidopsis (Larsson et al., 2017). Furthermore, YUCCA genes also play a crucial role in morphology changes associated with survival in a medium-long temperature stress (Casal and Balasubramanian, 2019). Interestingly, TSA increased the expression of TaYUC11D, particularly in the mid-low responding cultivar Caramba (Figure 5). These results agree with the up-regulation of auxin-related genes produced by TSA in Brassica ME (Li et al., 2014a). Moreover, TSA also caused an up-regulation of the auxin related gene Ta-2B-LBD16, in Caramba at 5dT. Ta-2B-LBD16 is an orthologist of AtLBD40 that promotes auxin signaling-dependent lateral organ development (Zentella et al., 2007; Okushima et al., 2007). TSA has also been associated with an up-regulation of genes encoding cell wall mobilization enzymes in Brassica ME (Li et al., 2014a). In this study, TaMS1, which is required for pollen exine development (Liu et al., 2018b), was up-regulated not only by TSA but also by MAN. A special mention of TaDRM3, a DNA silencing component, should also be made since it was up-regulated by treatments, maintaining the differences between cultivars previously observed at 0dT.
Diverse histone acetylation regulatory genes are included in this network, but only TaHAG3 and TaHAM showed slight differences between treatments in Caramba, highlighting HAG3 expression at 5dT-TSA (Figure 4). Interestingly, orthologists of TaHDA15, TaHAG3 and TaHAM have been described to promote hypocotyl elongation by up-regulation of auxin response and cell wall modifications genes in darkness in Arabidopsis (Tang et al., 2017; Woloszynska et al., 2018; Peng et al., 2018). Therefore, histone acetylation could be directly involved in the up-regulation of auxin related genes in this network.
The last network was characterized by the highest expression levels at 5dT, including TaABI5, TaATG18fD, TaSDG1a-7A, TaHDA18, TaHAC1 and TaHAC4 (Figure 6). The role of ABA in ME induction is unclear (for review see Żur et al., 2015). It has been proposed that each genotype requires a basal level of ABA to initiate a signaling cascade for ME (Żur et al., 2012). However, TaABI5 was expressed at low levels at 0dT, and slightly increased throughout the treatments at a similar rate in the two cultivars. This could be in line with the requirement of the novo ABA synthesis to induce barley ME reported by Hoekstra et al. (1996). The up-regulation of BnATG8, accompanied by an accumulation of autophagosomes, has been previously described in Brassica ME (Berenguer et al., 2020). In this study, the mid-low responding cultivar had a higher expression of TaATG18fD, suggesting a greater cellular component recycling in stress response. Finally, the gene TaSDG1a-7A, an orthologist of AtCLF (CURLY LEAF) (Strejčková et al., 2020) that is a PRC2 component (polycomb repressive complex 2), was up-regulated particularly at 5dT-MAN. The CLF/H3K27me3 complex could play a role in controlling the transcription of developmental genes in stress-response (Avramova, 2015). The up-regulation of CLF has been previously associated with ME induction by mannitol in barley (Muñoz-Amatriaín et al., 2009), and by cold in perennial ryegrass (Begheyn et al., 2018).
The TaHDA18, TaHAC1 and TaHAC4 genes could be implicated in the epigenetic control of the network. It has been reported that they are related to the regulation of developmental processes mediated by hormone signaling, including ABA, ethylene or auxins (Xu et al., 2005; Li et al., 2014b; Li et al., 2022). These genes showed a slightly higher expression with MAN than TSA, especially in Caramba.
In this report, for the first time, a comprehensive analysis of the epigenetic dynamics involved in ME induction has been performed applying two treatments of different efficiency, in two bread wheat cultivars with different response. The epigenetic state of the microspores before induction was distinctive between cultivars. Evidences obtained in this study allow us to conclude that microspores of the high responding cultivar at the onset of ME induction were at a less advanced stage in pollen development and had a higher cell division potential. The initial differences determined the dynamics of histone PTMs to MAN and TSA. A response pattern could be established for the percentages of acetylation but not for methylation, during treatments. MAN and TSA increased the H3.1/H3.2 ratio and the hyperacetylation of H3. Moreover, there was a down-regulation of pollen developmental genes and an up-regulation of genes related to stress and hormonal response and morphological changes. The effect of TSA pointed out the most significant processes for wheat ME induction. In the mid-low responding cultivar, TSA enhanced histone hyperacetylation and cell division potential, and promoted an early initiation of DNA replication mechanisms. In addition, the down-regulation of pollen development genes was slowed down whereas the expression of genes associated with morphological changes, mainly mediated by auxin signaling, was increased. This study answers key questions about the role of epigenetic modifications and their relationship with transcriptional requirements in wheat ME induction, and it is a starting point for new studies and approaches to improve ME efficiency in a wide range of wheat genotypes and other crops.
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Conceptualization AMC and MPV; methodology IV-R, AMC, MAB, and MPV; microspore and RNA sampling IV-R, AMC, and MPV; immunofluorescence analysis IV-R, AMC, and MAB; gene expression analysis IV-R and MPV; writing-original draft preparation IV-R, AMC, and MPV; writing-review and editing IV-R, AMC, MAB, and MPV; supervisor, project administration and funding acquisition AMC and MPV. All authors contributed to the article and approved the submitted version.
This research was supported by Project AGL2016-77211-R and PID2020-119540RB-100 from State R&D Program Oriented to the Challenges of the Spanish Society and funds for Reference Research Groups recognized by Diputación General de Aragón, Spain (Group A08-20R). IV-R was funded by the Spanish Ministry of Science and Innovation grant no. BES-2017-080970 (linked to the project AGL2016-77211-R).
The authors would like to thank Asunción Costar for her assistance growing the plant material and Begoña Echávarri and Patricia Fustero for their assistance collecting microspores samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1058421/full#supplementary-material
Abd ElFatah, B. E. S., Sayed, M. A., ElSanusy, S. A. (2020). Genetic analysis of anther culture response and identification of QTLs associated with response traits in wheat (Triticum aestivum l.). Mol. Biol. Rep. 47, 9289–9300. doi: 10.1007/s11033-020-06007-z
Akond, Z., Rahman, H., Ahsan, M. A., Mosharaf, M. P., Alam, M., Mollah, M. N. H. (2020). Bioinformatic analysis based genome-wide identification, characterization, diversification and regulatory transcription components of RNA silencing machinery genes in wheat (Triticum aestivum l.). bioRxiv. doi: 10.1101/2020.05.21.108100
Avramova, Z. (2015). Transcriptional ‘memory’ of a stress: transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 83, 149–159. doi: 10.1111/tpj.12832
Bannister, A., Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi: 10.1038/cr.2011.22
Batra, R., Gautam, T., Pal, S., Chaturvedi, D., Jan, I., Balyan, H. S., et al. (2020). Identification and characterization of SET domain family genes in bread wheat (Triticum aestivum l.). Sci. Rep. 10, 1–14. doi: 10.1038/s41598-020-71526-5
Begcy, K., Dresselhaus, T. (2018). Epigenetic responses to abiotic stresses during reproductive development in cereals. Plant Reprod. 31, 343–355. doi: 10.1007/s00497-018-0343-4
Begheyn, R. F., Yates, S. A., Sykes, T., Studer, B. (2018). Genetic loci governing androgenic capacity in perennial ryegrass (Lolium perenne l.). G3: Genes Genomes Genet. 8, 1897–1908. doi: 10.1534/g3.117.300550
Berenguer, E., Bárány, I., Solís, M. T., Pérez-Pérez, Y., Risueño, M. C., Testillano, P. S. (2017). Inhibition of histone H3K9 methylation by BIX-01294 promotes stress-induced microspore totipotency and enhances embryogenesis initiation. Front. Plant Sci. 8, 1161. doi: 10.3389/fpls.2017.01161
Berenguer, E., Minina, E. A., Carneros, E., Bárány, I., Bozhkov, P. V., Testillano, P. S. (2020). Suppression of metacaspase-and autophagy-dependent cell death improves stress-induced microspore embryogenesis in Brassica napus. Plant Cell Physiol. 61, 2097–2110. doi: 10.1093/pcp/pcaa128
Borg, M., Berger, F. (2015). Chromatin remodelling during male gametophyte development. Plant J. 83, 177–188. doi: 10.1111/tpj.12856
Borg, M., Jiang, D., Berger, F. (2021). Histone variants take center stage in shaping the epigenome. Curr. Opin. Plant Biol. 61, 101991. doi: 10.1016/j.pbi.2020.101991
Braszewska-Zalewska, A. J., Wolny, E. A., Smialek, L., Hasterok, R. (2013). Tissue-specific epigenetic modifications in root apical meristem cells of hordeum vulgare. PloS One 8, e69204. doi: 10.1371/journal.pone.0069204
Casal, J. J., Balasubramanian, S. (2019). Thermomorphogenesis. Annu. Rev. Plant Biol. 70, 321–346. doi: 10.1146/annurev-arplant-050718-095919
Castillo, A. M., Allué, S., Costar, A., Álvaro, F., Vallés, M. P. (2019). Doubled haploid production from Spanish and central European spelt by anther culture. J. Agric. Sci. Technol. 21, 1313–1324.
Castillo, A. M., Valero-Rubira, I., Allué, S., Costar, A., Vallés, M. P. (2021). “Bread wheat doubled haploid production by anther culture”. Doubled haploid Technology: Methods Protocols ed Seguí-Simarro, J. M. (Humana New York NY) 227–244.
Castillo, A. M., Valero-Rubira, I., Burrell, M. A., Allué, S., Costar, A., Vallés, M. P. (2020). Trichostatin a affects developmental reprogrammning of bread wheat microspores towarsd an embryogenic route. Plants 9, 1442.
Castillo, A. M., Valles, M. P., Cistué, L. (2000). Comparison of anther and isolated microspore cultures in barley. effects of culture density and regeneration medium. Euphytica 113, 1–8. doi: 10.1023/A:1003937530907
Dubas, E., Janowiak, F., Krzewska, M., Hura, T., Żur, I. (2013). Endogenous ABA concentration and cytoplasmic membrane fluidity in microspores of oilseed rape (Brassica napus l.) genotypes differing in responsiveness to androgenesis induction. Plant Cell Rep. 32, 1465–1475. doi: 10.1007/s00299-013-1458-6
Dubas, E., Moravčíková, J., Libantová, J., Matušíková, I., Benková, E. (2014). Żur, I The influence of heat stress on auxin distribution in transgenic B. napus microspores and microspore-derived embryos. al Protoplasma 251, 1077–1087. doi: 10.1007/s00709-014-0616-1
Evens, N. P., Buchner, P., Williams, L. E., Hawkesford, M. J. (2017). The role of ZIP transporters and group f bZIP transcription factors in the zn-deficiency response of wheat (Triticum aestivum). Plant J. 92, 291–304. doi: 10.1111/tpj.13655
Gao, S., Li, L., Han, X., Liu, T., Jin, P., Cai, L., et al. (2021). Genome-wide identification of the histone acetyltransferase gene family in Triticum aestivum. BMC Genomics 22, 1–17. doi: 10.1186/s12864-020-07348-6
Gehring, M. (2019). Epigenetic dynamics during flowering plant reproduction: evidence for reprogramming? New Phytol. 224, 91–96. doi: 10.1111/nph.15856
González-Melendi, P., Ramirez, C., Testillano, P. S., Kumlehn, J., Risueño, M. C. (2005). Three dimensional confocal and electron microscopy imaging define the dynamics and mechanisms of diploidisation at early stages of barley microspore-derived embryogenesis. Planta 222, 47–57. doi: 10.1007/s00425-005-1515-7
Griggs, R., Zheng, M. Y. (2016). Nuclear fusion during early stage of microspore embryogenesis indicates chromosome doubling in wheat (Triticum aestivum). Am. J. Plant Sci. 7, 489–499. doi: 10.4236/ajps.2016.73043
He, C., Chen, X., Huang, H., Xu, L. (2012). Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured arabidopsis tissues. PloS Genet. 8, e1002911. doi: 10.1371/journal.pgen.1002911
Henderson, I. R., Deleris, A., Wong, W., Zhong, X., Chin, H. G., Horwitz, G. A., et al. (2010). The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA–directed DNA methylation in arabidopsis thaliana. PloS Genet. 6, e1001182. doi: 10.1371/journal.pgen.1001182
Hoekstra, S., Hoekstra, I. R., Hoekstra, R. A., Hoekstra, E. (1996). The interaction of 2, 4-d application and mannitol pretreatment in anther and microspore culture of hordeum vulgareL. cv. igri. J. Plant Physiol. 148, 696–700. doi: 10.1016/S0176-1617(96)80370-1
Houben, A., Kumke, K., Nagaki, K., Hause, G. (2011). CENH3 distribution and differential chromatin modifications during pollen development in rye (Secale cereale l.). Chromosome Res. 19, 471–480. doi: 10.1007/s10577-011-9207-6
Jasencakova, Z., Meister, A., Schubert, I. (2001). Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma 110, 83–92. doi: 10.1007/s004120100132
Jensen, T. (2021). The impact of cold and trichostatin a treatment on the transcriptome of wheat microspores. university of lethbridge (Canada). ProQuest Dissertations Theses 137.
Jensen, T., Bodell, K., Jiang, F., Laurie, J. D. (2022). “Doubled haploid production through microspore culture,” in Accelerated breeding of cereal crops. Eds. Bilichak, A., Laurie, J. D.(Humana, New York, NY), 151–166.
Jiang, F., Ryabova, D., Diedhiou, J., Hucl, P., Randhawa, H., Marillia, E. F., et al. (2017). Trichostatin a increases embryo and green plant regeneration in wheat. Plant Cell Rep. 36, 1701–1706. doi: 10.1007/s00299-017-2183-3
Jin, P., Gao, S., He, L., Xu, M., Zhang, T., Zhang, F., et al. (2020). Genome-wide identification and expression analysis of the histone deacetylase gene family in wheat (Triticum aestivum l.). Plants 10, 19. doi: 10.3390/plants10010019
Kong, L., Liu, Y., Wang, X., Chang, C. (2020). Insight into the role of epigenetic processes in abiotic and biotic stress response in wheat and barley. Int. J. Mol. Sci. 21, 1480. doi: 10.3390/ijms21041480
Kumar, V., Thakur, J. K., Prasad, M. (2021). Histone acetylation dynamics regulating plant development and stress responses. Cell. Mol. Life Sci. 78, 4467–4486. doi: 10.1007/s00018-021-03794-x
Lantos, C., Weyen, J., Orsini, J. M., Gnad, H., Schlieter, B., Lein, V., et al. (2013). Efficient application of in vitro anther culture for different European winter wheat (Triticum aestivum l.) breeding programmes. Plant Breed. 132, 149–154. doi: 10.1111/pbr.12032
Larsson, E., Vivian-Smith, A., Offringa, R., Sundberg, E. (2017). Auxin homeostasis in arabidopsis ovules is anther-dependent at maturation and changes dynamically upon fertilization. Front. Plant Sci. 8, 1735. doi: 10.3389/fpls.2017.01735
Lee, K., Seo, P. J. (2018). Dynamic epigenetic changes during plant regeneration. Trends Plant Sci. 23, 235–247. doi: 10.1016/j.tplants.2017.11.009
Li, H., Liu, H., Pei, X., Chen, H., Li, X., Wang, J., et al. (2022). Comparative genome-wide analysis and expression profiling of histone acetyltransferases and histone deacetylases involved in the response to drought in wheat. J. Plant Growth Regul. 41, 1065–1078. doi: 10.1007/s00344-021-10364-9
Li, H. J., Liu, N. Y., Shi, D. Q., Liu, J., Yang, W. C. (2010). YAO is a nucleolar WD40-repeat protein critical for embryogenesis and gametogenesis in arabidopsis. BMC Plant Biol. 10, 1–12. doi: 10.1186/1471-2229-10-169
Li, H., Soriano, M., Cordewener, J., Muiño, J. M., Riksen, T., Fukuoka, H., et al. (2014a). The histone deacetylase inhibitor trichostatin a promotes totipotency in the Male gametophyte. Plant Cell 26, 195–209. doi: 10.1105/tpc.113.116491
Liu, L., Lu, Y., Wei, L., Yu, H., Cao, Y., Li, Y., et al. (2018a). Transcriptomics analyses reveal the molecular roadmap and long non-coding RNA landscape of sperm cell lineage development. Plant J. 96, 421–437. doi: 10.1111/tpj.14041
Liu, H., Wang, J., Li, C., Qiao, L., Wang, X., Li, J., et al. (2018b). Phenotype characterisation and analysis of expression patterns of genes related mainly to carbohydrate metabolism and sporopollenin in male-sterile anthers induced by high temperature in wheat (Triticum aestivum). Crop Pasture Sci. 69, 469–478. doi: 10.1071/CP18034
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li, J., Wang, Z., Chang, Z., He, H., Tang, X., Ma, L., et al. (2021). A functional characterization of TaMs1 orthologs in poaceae plants. Crop J. 9, 1291–1300. doi: 10.1016/j.cj.2020.12.002
Li, C., Xu, J., Li, J., Li, Q., Yang, H. (2014b). Involvement of arabidopsis HAC family genes in pleiotropic developmental processes. Plant Signaling Behav. 9, 426–435. doi: 10.1093/pcp/pct180
Lloyd, J. P., Lister, R. (2022). Epigenome plasticity in plants. Nat. Rev. Genet. 23, 55–68. doi: 10.1038/s41576-021-00407-y
Luo, Y., Shi, D. Q., Jia, P. F., Bao, Y., Li, H. J., Yang, W. C. (2022). Nucleolar histone deacetylases HDT1, HDT2, and HDT3 regulate plant reproductive development. J. Genet. Genomics 49, 30–39. doi: 10.1016/j.jgg.2021.10.002
Mozgova, I., Mikulski, P., Pecinka, A., Farrona, S. (2019). Epigenetic mechanisms of abiotic stress response and memory in plants. Epigenet. Plants Agronomic Importance: Fundamentals Appl. Eds. Alvarez-Venegas, R., De-la-Peña, C., Casas-Mollano, J. (Springer, Cham), 1–64. doi: 10.1007/978-3-030-14760-0_1
Muñoz-Amatriaín, M., Castillo, A. M., Cistué, L., Vallés, M. P. (2008). Identification and validation of QTLs for green plant percentage in barley (Hordeum vulgare l.) anther culture. Mol. Breed. 22, 119–129. doi: 10.1007/s11032-008-9161-y
Muñoz-Amatriaín, M., Svensson, J. T., Castillo, A. M., Cistué, L., Close, T. J., Vallés, M. P. (2006). Transcriptome analysis of barley anthers: effect of mannitol treatment on microspore embryogenesis. Physiologia Plantarum 127, 551–560. doi: 10.1111/j.1399-3054.2006.00729.x
Muñoz-Amatriaín, M., Svesson, J. T., Castillo, A. M., Cistué, L., Close, T. J., Vallés, M. P. (2009). “Expression profiles in barley microspore embryogenesis,” in Advanced in haploid production in higher plants. Eds. Touraev, A., Forster, B. P., Jain, S. M. (Dordrecht: Springer), 127–134.
Nelms, B., Walbot, V. (2022). Gametophyte genome activation occurs at pollen mitosis I in maize. Science 375, 424–429. doi: 10.1126/science.abl7392
Nielsen, N. H., Andersen, S. U., Stougaard, J., Jensen, A., Backes, G., Jahoor, A. (2015). Chromosomal regions associated with the in vitro culture response of wheat (Triticum aestivum l.) microspores. Plant Breed. 134, 255–263. doi: 10.1111/pbr.12257
Nowicka, A., Juzoń, K., Krzewska, M., Dziurka, M., Dubas, E., Kopeć, P., et al. (2019). Chemically-induced DNA de-methylation alters the effectiveness of microspore embryogenesis in triticale. Plant Sci. 287, 110189. doi: 10.1016/j.plantsci.2019.110189
Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., Tasaka, M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in arabidopsis. Plant Cell 19, 118–130. doi: 10.1105/tpc.106.047761
Ono, A., Kinoshita, T. (2021). Epigenetics and plant reproduction: Multiple steps for responsibly handling succession. Curr. Opin. Plant Biol. 61, 102032. doi: 10.1016/j.pbi.2021.102032
Otero, S., Desvoyes, B., Peiró, R., Gutierrez, C. (2016). Histone H3 dynamics reveal domains with distinct proliferation potential in the arabidopsis root. Plant Cell 28, 1361–1371. doi: 10.1105/tpc.15.01003
Pandey, P., Daghma, D. S., Houben, A., Kumlehn, J., Melzer, M., Rutten, T. (2017). Dynamics of post-translationally modified histones during barley pollen embryogenesis in the presence or absence of the epi-drug trichostatin a. Plant Reprod. 30, 95–105. doi: 10.1007/s00497-017-0302-5
Pandey, P., Houben, A., Kumlehn, J., Melzer, M., Rutten, T. (2013). Chromatin alterations during pollen development in hordeum vulgare. Cytogenetic Genome Res. 141, 50–57. doi: 10.1159/000351211
Paolacci, A. R., Tanzarella, O. A., Porceddu, E., Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10, 1–27. doi: 10.1186/1471-2199-10-11
Pecinka, A., Dinh, H. Q., Baubec, T., Rosa, M., Lettner, N., Scheid, O. M. (2010). Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in arabidopsis. Plant Cell 22, 3118–3129. doi: 10.1105/tpc.110.078493
Peng, M., Li, Z., Zhou, N., Ma, M., Jiang, Y., Dong, A., et al. (2018). Linking PHYTOCHROME-INTERACTING FACTOR to histone modification in plant shade avoidance. Plant Physiol. 176, 1341–1351. doi: 10.1104/pp.17.01189
Pérez-Pérez, Y., El-Tantawy, A. A., Solís, M. T., Risueño, M. C., Testillano, P. S. (20191200). Stress-induced microspore embryogenesis requires endogenous auxin synthesis and polar transport in barley. Front. Plant Sci. 10.
Pikaard, C. S., Scheid, O. M. (2014). Epigenetic regulation in plants. Cold Spring Harbor Perspect. Biol. 6, a019315. doi: 10.1101/cshperspect.a019315
Placido, D. F., Campbell, M. T., Folsom, J. J., Cui, X., Kruger, G. R., Baenziger, P. S., et al. (2013). Introgression of novel traits from a wild wheat relative improves drought adaptation in wheat. Plant Physiol. 161, 1806–1819. doi: 10.1104/pp.113.214262
Portereiko, M. F., Sandaklie-Nikolova, L., Lloyd, A., Dever, C. A., Otsuga, D., Drews, G. N. (2006). NUCLEAR FUSION DEFECTIVE1 encodes the arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol. 141, 957–965. doi: 10.1104/pp.106.079319
Probst, A. V., Desvoyes, B., Gutierrez, C. (2020). Similar yet critically different: the distribution, dynamics and function of histone variants. J. Exp. Bot. 71, 5191–5204. doi: 10.1093/jxb/eraa230
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361, eaar6089.
R Core Team (2021). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Rodríguez-Sanz, H., Solís, M. T., López, M. F., Gómez-Cadenas, A., Risueño, M. C., Testillano, P. S. (2014). Auxin biosynthesis, accumulation, action and transport are involved in stress-induced microspore embryogenesis initiation and progression in Brassica napus. Plant Cell Physiol. 56, 1401–1417. doi: 10.1093/pcp/pcv058
Sánchez-Díaz, R. A. (2014). Estudio de los mecanismos moleculares implicados en la embriogénesis de la microspora en cebad y trigo (University of Zaragoza – Estación Experimental Aula Dei (Spain). http://hdl.handle.net/10261/94534
Sánchez-Díaz, R. A., Castillo, A. M., Vallés, M. P. (2013). Microspore embryogenesis in wheat: New marker genes for early, middle and late stages of embryo development. Plant Reprod. 26, 287–296. doi: 10.1007/s00497-013-0225-8
Seifert, F., Bössow, S., Kumlehn, J., Gnad, H., Scholten, S. (2016). Analysis of wheat microspore embryogenesis induction by transcriptome and small RNA sequencing using the highly responsive cultivar “Svilena. BMC Plant Biol. 16, 1–16. doi: 10.1186/s12870-016-0782-8
Singh, M., Singh, S., Randhawa, H., Singh, J. (2013). Polymorphic homoeolog of key gene of RdDM pathway, ARGONAUTE4_9 class is associated with pre-harvest sprouting in wheat (Triticum aestivum l.). PloS One 8, e77009. doi: 10.1371/journal.pone.0077009
Solís, M. T., El-Tantawy, A. A., Cano, V., Risueño, M. C., Testillano, P. S. (2015). 5-azacytidine promotes microspore embryogenesis initiation by decreasing global DNA methylation, but prevents subsequent embryo development in rapeseed and barley. Front. Plant Sci. 6, 472. doi: 10.3389/fpls.2015.00472
Solís, M. T., Rodríguez-Serrano, M., Meijón, M., Cañal, M. J., Cifuentes, A., Risueño, M. C., et al. (2012). DNA Methylation dynamics and MET1a-like gene expression changes during stress-induced pollen reprogramming to embryogenesis. J. Exp. Bot. 63, 6431–6444. doi: 10.1093/jxb/ers298
Soriano, M. (2008). Embriogénesis de la microspora en trigo panadero: optimización de la producción de plantas doblehaploides y estudio de genes de estrés durante el pretratamiento (University of Zaragoza – Estación Experimental Aula Dei (Spain).
Soriano, M., Li, H., Boutilier, K. (2013). Microspore embryogenesis: Establishment of embryo identity and pattern in culture. Plant Reprod. 26, 181–196. doi: 10.1007/s00497-013-0226-7
Soriano, M., Li, H., Jacquard, C., Angenent, G. C., Krochko, J., Offringa, R., et al. (2014). Plasticity in cell division patterns and auxin transport dependency during in vitro embryogenesis in brassica napus. Plant Cell 26 (6), 2568–2581. doi: 10.1105/tpc.114.126300
Strejčková, B., Čegan, R., Pecinka, A., Milec, Z., Šafář, J. (2020). Identification of polycomb repressive complex 1 and 2 core components in hexaploid bread wheat. BMC Plant Biol. 20, 1–13.
Tang, Y., Liu, X., Liu, X., Li, Y., Wu, K., Hou, X. (2017). Arabidopsis NF-YCs mediate the light-controlled hypocotyl elongation via modulating histone acetylation. Mol. Plant 10, 260–273. doi: 10.1016/j.molp.2016.11.007
Tasset, C., Singh Yadav, A., Sureshkumar, S., Singh, R., van der Woude, L., Nekrasov, M., et al. (2018). POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in arabidopsis thaliana. PloS Genet. 14, e1007280. doi: 10.1371/journal.pgen.1007280
Tessarz, P., Kouzarides, T. (2014). Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 15, 703–708. doi: 10.1038/nrm3890
Testillano, P. S. (2019). Microspore embryogenesis: targeting the determinant factors of stress-induced cell reprogramming for crop improvement. J. Exp. Bot. 70, 2965–2978. doi: 10.1093/jxb/ery464
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 45, 122–129. doi: 10.1093/nar/gkx382
Wang, H. M., Enns, J. L., Nelson, K. L., Brost, J. M., Orr, T. D., Ferrie, A. M. R. (2019). Improving the efficiency of wheat microspore culture methodology: Evaluation of pretreatments, gradients, and epigenetic chemicals. Plant Cell Tissue Organ Culture 139, 589–599. doi: 10.1007/s11240-019-01704-5
Wang, Z., Li, J., Chen, S., Heng, Y., Chen, Z., Yang, J., et al. (2017). Poaceae-specific MS1 encodes a phospholipid-binding protein for male fertility in bread wheat. Proc. Natl. Acad. Sci. 114, 12614–12619. doi: 10.1073/pnas.1715570114
Wang, Z., Zhang, R., Cheng, Y., Lei, P., Song, W., Zheng, W., et al. (2021). Genome-wide identification, evolution, and expression analysis of LBD transcription factor family in bread wheat (Triticum aestivum l.). Front. Plant Sci. 12, 1870. doi: 10.3389/fpls.2021.721253
Wang, P., Zhang, H., Hou, H., Wang, Q., Li, Y., Huang, Y., et al. (2016). Cell cycle arrest induced by inhibitors of epigenetic modifications in maize (Zea mays) seedling leaves: characterization of the process and possible mechanisms involved. New Phytol. 211, 646–657. doi: 10.1111/nph.13942
Waterborg, J. H. (1991). Multiplicity of histone h3 variants in wheat, barley, rice, and maize. Plant Physiol. 96, 453–458. doi: 10.1104/pp.96.2.453
Weigt, D., Kiel, A., Siatkowski, I., Zyprych-Walczak, J., Tomkowiak, A., Kwiatek, M. (2019). Comparison of the androgenic response of spring and winter wheat (Triticum aestivum l.). Plants 9, 49. doi: 10.3390/plants9010049
Wolny, E., Braszewska-Zalewska, A., Hasterok, R. (2014). Spatial distribution of epigenetic modifications in brachypodium distachyon embryos during seed maturation and germination. PloS One 9, e101246. doi: 10.1371/journal.pone.0101246
Woloszynska, M., Gagliardi, O., Vandenbussche, F., De Groeve, S., Alonso Baez, L., Neyt, P., et al. (2018). The elongator complex regulates hypocotyl growth in darkness and during photomorphogenesis. J. Cell Sci. 131, jcs203927.
Xu, J., Hu, P., Tao, Y., Song, P., Gao, H., Guan, Y. (2021). Genome-wide identification and characterization of the lateral organ boundaries domain (LBD) gene family in polyploid wheat and related species. Peer J. 9, e11811. doi: 10.7717/peerj.11811
Xu, C. R., Liu, C., Wang, Y. L., Li, L. C., Chen, W. Q., Xu, Z. H., et al. (2005). Histone acetylation affects expression of cellular patterning genes in the arabidopsis root epidermis. Proc. Natl. Acad. Sci. 102, 14469–14474. doi: 10.1073/pnas.0503143102
Yan, A., Borg, M., Berger, F., Chen, Z. (2020). The atypical histone variant H3. 15 promotes callus formation in arabidopsis thaliana. Development 147, dev184895.
Yang, Y., Xu, T., Wang, H., Feng, D. (2021). Genome-wide identification and expression analysis of the TaYUCCA gene family in wheat. Mol. Biol. Rep. 48, 1269–1279. doi: 10.1007/s11033-021-06197-0
Yan, J., Su, P., Wei, Z., Nevo, E., Kong, L. (2017). Genome-wide identification, classification, evolutionary analysis and gene expression patterns of the protein kinase gene family in wheat and Aegilops tauschii. Plant Mol. Biol. 95, 227–242. doi: 10.1007/s11103-017-0637-1
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., Madden, T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 13, 1–11. doi: 10.1186/1471-2105-13-S6-S1
Yung, W. S., Li, M. W., Sze, C. C., Wang, Q., Lam, H. M. (2021). Histone modifications and chromatin remodelling in plants in response to salt stress. Physiologia Plantarum 173, 1495–1513. doi: 10.1111/ppl.13467
Zentella, R., Zhang, Z. L., Park, M., Thomas, S. G., Endo, A., Murase, K., et al. (2007). Global analysis of DELLA direct targets in early gibberellin signaling in arabidopsis. Plant Cell 19, 3037–3057. doi: 10.1105/tpc.107.054999
Zhang, T., Liu, P., Zhong, K., Zhang, F., Xu, M.He, l.et al). wheat yellow mosaic virus NIb interacting with host light induced protein (LIP) facilitates its infection through perturbing the abscisic acid pathway in wheat. Biology 8, 80. doi: 10.3390/biology8040080
Zhang, L., Zhang, Y., Gao, Y., Jiang, X., Zhang, M., Wu, H., et al. (2016). Effects of histone deacetylase inhibitors on microspore embryogenesis and plant regeneration in pakchoi (Brassica rapa ssp. chinensis L.) Sci. Hortic. 209, 61–66. doi: 10.1016/j.scienta.2016.05.001
Zhi, P., Kong, L., Liu, J., Zhang, X., Wang, X., Li, H., et al. (2020). Histone deacetylase TaHDT701 functions in TaHDA6-TaHOS15 complex to regulate wheat defense responses to Blumeria graminis f. sp tritici Int. J. Mol. Sci. 21, 2640. doi: 10.3390/ijms21072640
Zur, I., Dubas, E., Krzewska, M., Janowiak, F. (2015). Current insights into hormonal regulation of microspore embryogenesis. Front. Plant Sci. 6, 424.
Keywords: bread wheat, microspore embryogenesis, mannitol, trichostatin A, histones, epigenetics, gene expression
Citation: Valero-Rubira I, Castillo AM, Burrell MÁ and Vallés MP (2023) Microspore embryogenesis induction by mannitol and TSA results in a complex regulation of epigenetic dynamics and gene expression in bread wheat. Front. Plant Sci. 13:1058421. doi: 10.3389/fpls.2022.1058421
Received: 30 September 2022; Accepted: 07 December 2022;
Published: 09 January 2023.
Edited by:
Jana Krajnakova, New Zealand Forest Research Institute Limited (Scion), New ZealandReviewed by:
Giorgio Perrella, University of Milan, ItalyCopyright © 2023 Valero-Rubira, Castillo, Burrell and Vallés. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Pilar Vallés, dmFsbGVzQGVlYWQuY3NpYy5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.