- 1State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University, Guangzhou, China
- 2Guangdong Laboratory for Lingnan Modern Agriculture, South China Agricultural University, Guangzhou, China
- 3Guangdong Provincial Key Laboratory of Plant Molecular Breeding, College of Agriculture, South China Agricultural University, Guangzhou, China
- 4State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, China
Sheath blight, caused by Rhizoctonia solani, is a big threat to the global rice production. To characterize the early development of R. solani on rice leaf and leaf sheath, two genotypes, GD66 (a resistant genotype) and Lemont (a susceptible genotype), were observed using four cytological techniques: the whole-mount eosin B-staining confocal laser scanning microscopy (WE-CLSM), stereoscopy, fluorescence microscopy, and plastic semi-thin sectioning after in vitro inoculation. WE-CLSM observation showed that, at 12 h post-inoculation (hpi), the amount of hyphae increased dramatically on leaf and sheath surface, the infection cushions occurred and maintained at a huge number from about 18 to 36 hpi, and then the infection cushions disappeared gradually from about 42 to 72 hpi. Interestingly, R. solani could not only colonize on the abaxial surfaces of leaf sheath but also invade the paraxial side of the leaf sheath, which shows a different behavior from that of leaf. RNA sequencing detected 6,234 differentially expressed genes (DEGs) for Lemont and 7,784 DEGs for GD66 at 24 hpi, and 2,523 DEGs for Lemont and 2,719 DEGs for GD66 at 48 hpi, suggesting that GD66 is recruiting more genes in fighting against the pathogen. Among DEGs, resistant genes, such as OsRLCK5, Xa21, and Pid2, displayed higher expression in the resistant genotype than the susceptible genotype at both 24 and 48 hpi, which were validated by quantitative reverse transcription–PCR. Our results indicated that the resistance phenotype of GD66 was the consequence of recruiting a series of resistance genes involved in different regulatory pathways. WE-CLSM is a powerful technique for uncovering the mechanism of R. solani invading rice and for detecting rice sheath blight–resistant germplasm.
Introduction
Rice sheath blight, caused by Rhizoctonia solani, is a serious disease in many rice-producing countries (Zeng et al., 2017). The yield losses caused by sheath blight have exceeded other major diseases, making it the most serious rice disease in China (Zeng et al., 2011). Sheath blight caused about 20% yield losses in India, and it is also the most common rice disease in the Southern United States (Groth and Bond, 2007; Ghosh et al., 2016).
The soilborne fungal pathogen R. solani infects more than 250 plant species (Anderson, 1982). Plants immune to this pathogen have not been found. All the rice plants can be infected by this pathogen, but the serious extent varies among different cultivars. Large-scale screening in rice communities have found some rice varieties that showed partial resistance, such as Teqing (Li et al., 1995), Jasmine 85 (Pan et al., 1999; Zou et al., 2000; Park et al., 2008), Tetep (Channamallikarjuna et al., 2010), WSS2 (Sato et al., 2004), Pecos (Sharma et al., 2009), ARC10531 (Yadav et al., 2015), and GD66 (Zeng et al., 2017). Partial resistance means that they perform better resistance than other varieties, although none of which are immune to this pathogen.
Fungal pathogens usually produce some infection structures to invade and establish parasitic relationship with the host plant. The infection structures, such as infection cushion (Dodman et al., 1968; Dodman and Flentje, 1970), appressoria (Emmett and Parbery, 1975), infection peg, and haustorium (Riopel and Timko, 1995; Huang et al., 2003), are formed by specialized hyphae. Although there is still much debate about the details of the infection process, most researchers now agree that R. solani hyphae invade rice mainly in two ways: (1) Hyphae gather to form two specialized infection structures: infection cushion and appressoria on the surface of leaf sheath (Marshall, 1980a; Deising et al., 2000). The infection cushion may be conducive to the concentration of sheath blight pathogen energy and nutrition to infect the host and to destroy the plant defense system and obtain more nutrition and water from the host cells for the mass reproduction of the pathogen (Zhang et al., 2010). There are invasion nails and holes at the base of the contact surface between the infection cushion and the rice epidermis. After the invasive nail contacts the surface of leaf sheath, it secretes mucilaginous substances, which can poison plants (Matsuura, 1986). The infected nail became obviously thinner when invading epidermal cells, and it returned to its original diameter after entering cells (Tang, 2011). The mycelium expands and covers one side of the cell wall, it becomes very thin to pass through the cell wall, and then widens to restore the mycelium size (Zhang et al., 2010). (2) Hyphae can directly penetrate into rice epidermis without invasive structure. It penetrates rice epidermal cells and the wall between cells, invades rice leaf sheath from intercellular space, or directly invades host from natural orifice (mainly stomata) and wound (Marshall, 1980b; Tao, 1992; Zhang et al., 2010; Tang, 2011). Therefore, the infection structure is not a necessary device for penetration. Hyphae can form penetrating claws that exert great invasive pressure on rice epidermal cells to enter the host (Zhang et al., 2010; Tang, 2011). The primary hyphae form secondary hyphae after entering the leaf sheath, which grow in the host or expand in the intercellular space (Zhang et al., 2010). In addition, the mycelium is very thick and deformed in rice cells (Singh et al., 2003).
Genetic analysis revealed that sheath blight resistance in rice is controlled by quantitative trait loci (QTLs) with minor effects (Kump et al., 2011). In addition, the resistance is conferred by the additive effect of non–race-specific resistance QTLs (Pinson et al., 2005; Liu et al., 2009). Approximately 50 sheath blight–resistant QTLs have been detected on all the 12 rice chromosomes (Zuo et al., 2010; Xu et al., 2011), and QTL by environment interaction is frequently detected. More than 20 genes conferring sheath blight resistance have been cloned and characterized, including genes that positively regulate sheath blight resistance: OsWRKY4 (Wang et al., 2015a) OsWRKY13 (John and Subramanian, 2019), DOF11 (Kim et al., 2021), OsSWEET14 (Kim et al., 2021), OsPGIP1 (Wang et al., 2015b), OsRSR1 (Wang et al., 2021), OsGLP1 (Banerjee and Maiti, 2010), Os2H16 (Li et al., 2013), IDD13 (Sun et al., 2020), OsOSM1 (Xue et al., 2016), OsRLCK5 (Wang et al., 2021), OsWAK25 (Harkenrider et al., 2016), LPAl (Sun et al., 2019), OsPAL4 (Tonnessen et al., 2015), and OsCHI11 (Karmakar et al., 2016). Genes that negatively regulated sheath blight resistance were also characterized: OsWRKY45 (Shimono et al., 2012), OsWRKY53 (Yuan et al., 2020), OsSWEET11 (Gao et al., 2018), OsTrxm (Hu et al., 2021), IDD3 (Sun et al., 2020), OsNYC3 (Cao et al., 2022), OsGFl4e (Manosalva et al., 2011), OsMESL (Hu et al., 2021), OsBON1 (Yin et al., 2018), and OsAKT1 (Yuan et al., 2020). These sheath blight–resistant genes participated in different regulatory pathways, including reactive oxygen species (ROS), phytohormone, salicylic acid signaling, defense response to fungi and innate immune, sucrose transport, and other regulatory pathways.
Transcriptome analysis by RNA sequencing (RNA-seq) is a powerful tool to measure the spatial and temporal expression of various genes in response to outer stimulus and to demonstrate from the macroscopic view that different genes participated in different biological pathways (Słomnicka et al., 2021). The transcriptome profiles between the susceptible cultivar Lemont and the moderately resistant cultivar Teqing in response to sheath blight disease were studied, and it showed that regulation of photosynthesis, photorespiration, jasmonic acid, and phenylpropanoid pathways contribute to rice resistance to R. solani (Shu et al., 2019). Transcriptome analysis showed that the defense system in resistant rice cultivars involved the expression of PR genes, key transcription factors, PAL genes, and the enrichment of defense-related pathways (Yang et al., 2022). Comparative transcriptome analysis suggested that photosynthesis, photorespiration, and jasmonic acid and phenylpropanoid metabolism played important roles in disease resistance (Zhang et al., 2017).
Despite great efforts made in the rice sheath blight research communities, there are still many questions that need to be addressed: What is the detailed developmental progress of the R. solani hyphae on rice leaf and leaf sheath at different time spots under both fluorescence microscope and confocal microscope? Are there any differences between sheath blight–resistant and sheath blight–susceptible varieties during the hyphal growth? To breed a sheath blight–resistant cultivar, what can we learn from the transcriptome analysis results? To answer these questions, we first observed the hyphal growth on rice leaf and leaf sheath using a stereoscope, a fluorescence microscope, and the whole-mount eosin B-staining confocal laser scanning microscopy (WE-CLSM) to gain an overall picture of how the fungal hyphae were grown and spread on leaf and leaf sheath. We demonstrate that WE-CLSM can be used conveniently to observe the R. solani hyphae inside rice leaf or leaf sheath. This new method provides rapid observation of the cytological progress of R. solani hyphae invading rice that would benefit sheath blight studies in the future. Moreover, transcriptome analysis between a sheath blight–resistant genotype GD66 and a sheath blight–susceptible genotype Lemont was studied after pathogen inoculation to reveal the sheath blight–resistant mechanism and to discuss how to breed an ideal cultivar with good sheath blight resistance.
Materials and methods
Rice genotypes
Five rice genotypes (Lemont, Taichung65, Yinhesizhan, 8821, and GD66) were used in the present study. Lemont, Yinhesizhan, and GD66 were provided by the National Midterm Rice Genebank in China National Rice Research Institute (CNRRI). Taichung65 and 8821 were provided by the State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, South China Agricultural University (SCAU). Lemont is a japonica American cultivar. Taichung65 is a japonica cultivar originated from Taiwan, China. Yinhesizhan is an indica cultivar originated from Guangdong, China. 8821 is an indica cultivar developed by Zhaoqing Institute of Agricultural Sciences, Guangdong, China. GD66 is an indica restorer line developed by CNRRI. These genotypes were grown under a natural light in a greenhouse with the temperature ranged from 20°C to 35°C or in the field with common practices in South China Agricultural University (23.16N, 113.35E), Guangzhou, China.
R. solani isolate for inoculation
The R. solani isolate ZJ03, collected in Fuyang, Hangzhou, China, was used for inoculation in this study. The isolate ZJ03 has been used for testing rice sheath blight resistance and QTL mapping in some previous studies (Zeng et al., 2017; Chen et al., 2019; Zeng et al., 2021).
Inoculation and evaluation of sheath blight resistance in the field
The rice genotypes were sown on 25 February 2021 in the farm of SCAU, in Guangzhou, China. Fifty individual plants were planted in a plot for each genotype. The 50 plants were arranged in five rows with 10 plants in each row. The space between each row and different plants within each row were both 10 cm. At the late tillering stage, five plants in the middle of the third row were inoculated using ZJ03. A total of 10 tillers were inoculated for each genotype, i.e., two tillers for each individual plant. The inoculation method were described by Zou et al. (2000) and Zeng et al. (2017): toothpicks covering with mycelium were used to penetrate the third leaf sheath, counting from the top. Sheath blight resistance was evaluated 30 days after inoculation. The lesion length was measured vertically along the stem for each genotype. The visual rating system of 0 to 9 was used to evaluate each inoculated individual, where 0 indicated that the plant was immune to the pathogen and 9 indicated a dead or collapsed plant (Pinson et al., 2005).
In vitro inoculation
At the booting stage, the second leaf sheath counting from the top and the leaf that connected to the sheath were scissored off the plant and washed using tap water for surface cleaning. Then, the sheath and leaf were cut into uniform lengths of 10 cm from the middle and placed in the plastic Petri dish of 13 × 13 cm covered with two layers of sterile gauze according to the method described by Vijay et al. (2011) with some modifications: Three leaves or sheaths were placed in each Petri dish, and the bottom side of the leaf or sheath was covered with gauze. Three replications, i.e., nine leaves or sheaths in a total of three Petri dishes, were performed to ensure consistent and reliable results. ddH2O (20 ml) was added to each Petri dish to keep the gauze moisty. The R. solani isolate ZJ03 was cultured and maintained in the potato dextrose agar (PDA) medium at 28°C in a growth chamber. First, the ZJ03 sclerotia or mycelia were transferred into a new PDA medium. In about 3 days, the pathogen mycelia occupied the entire surface of the PDA medium. Then, a 5-mm-diameter PDA medium covered with mycelia was placed in the middle of the leaf or leaf sheath in the Petri dish (Figure S1). Finally, the Petri dishes were sealed with breathable sealing film and incubated in a growth chamber at 28 ± 1°C under a 16-h light/8-h dark condition. Sterile agar medium without pathogen mycelia was also placed on leaf or sheath surface as the control.
Fluorescence microscopy
At the booting stage, the second leaf sheath counting from the top and the leaf connected to the sheath were inoculated in vitro in Petri dishes as described above. About 1.5-cm2 leaf (or sheath) area at the inoculation site was observed at different time spots after inoculation under a microscope. At the time spot of every 4 hpi (Figures S2, S3), the PDA medium covered with fungal was removed from the leaf (or sheath) surface and observed using a stereoscope (Leica MZ16, Germany) with 2× lens. After stereoscopic observation, the materials were directly stained with 1% 4',6-diamidino-2-phenylindole (DAPI) solution for 2 min and observed under a fluorescence microscope (Leica DMRXA, Germany).
The whole-mount eosin B-staining confocal laser scanning microscopy
The WE-CLSM was applied to observe the mycelial growth of sheath blight pathogen following the previous protocol with minor modifications (Zeng et al., 2007): At the booting stage, rice leaf or sheath was collected every 6 h after in vitro inoculation and fixed in a formaldehyde–acetic acid–ethanol solution (70% ethanol:acetic acid:formaldehyde = 18:1:1, v/v) for at least 48 h. Then, the samples were washed with 50% ethanol and kept in 70% ethanol at 4°C. The samples were hydrated consecutively in 50%, 30%, and 10% ethanol and distilled water for 30 min at each concentration, respectively. Then, the samples were stained in eosin B solution (10 mg/L; dissolved in 4% sucrose solution) for 48 h. Finally, the samples were dehydrated sequentially in 10%, 30%, 50%, 70%, 90%, and 100% alcohol solution for 30 min at each concentration, respectively. The dehydrated samples were stored in 50% methyl salicylate (dissolved in pure alcohol) for 48 h. Finally, the samples were stored in pure methyl salicylate for 48 h and then observed under a Leica SPE laser scanning confocal microscope (Leica Microsystems, Heidelberg, Germany). Excitation wavelength was 543 nm, and emission wavelength was between 550 and 630 nm (Zeng et al., 2007). The leaf area about 0.5 cm2 at the inoculation position was observed under a confocal microscope.
Transcriptome analysis
Lemont (a sheath blight–susceptible cultivar) and GD66 (a sheath blight–resistant restorer line) were used for transcriptome analysis by RNA-seq. The young seedlings of these two genotypes were first grown in green house. At the late tillering stage, the plants were transferred into a growth chamber (DHP-9052, Shanghai, China, 28°C, 16-h light/8-h dark condition) and cultured for 10 days before inoculation. The toothpicks covering with mycelia were used to penetrate the sheath for inoculation using the method described by Zou et al. (2000) and Zeng et al. (2017).
Total RNAs were extracted from inoculated sheath at 24 and 48 h post-inoculation (hpi). Leaf sheath at the inoculation site was collected and stored in liquid nitrogen for RNA extraction. The total RNA of each sample was extracted using TRIzol reagent (Invitrogen, Waltham, MA, USA) following the manufacturer’s procedure. Transcriptome analyses were based on the results of three biological replicates.
RNA extraction, libraries construction, and sequencing were conducted on the basis of the manufacturer’s instructions of Illumina HiSeqTM2500 (LC Sciences, Hangzhou, China) by Biomarker Technologies (Beijing, China) (Yu et al., 2018). The differentially expressed genes (DEGs) were identified using DESeq software with the following criteria: fold change (FC) ≥ 2 and false discovery rate (FDR) ≤ 0.01. Genes with p-values < 0.05 were chosen for further analysis.
After selecting the DEGs, cluster analysis and Gene Ontology (GO) enrichment analysis were conducted using Cluster 3.0 software (Tokyo, Japan) and agriGO 2.0 (Beijing, China) (http://systemsbiology.cau.edu.cn/agriGOv2/SEAresult2.php), respectively. Venny 2.1 software (BioinfoGP, Madrid, Spanish) was used to identify the overlapped DEGs in different samples (http://bioinfogp.cnb.csic.es/tools/venny/). Heat map was drawn by TBtools (Chen et al., 2020). UpSet plot were delineated using an online OmicStudio tools (https://www.omicstudio.cn/tool). The MapMan software was downloaded from the Internet for MapMan analysis (https://mapman.gabipd.org).
qRT-PCR analysis
A total of seven candidate genes were selected for validation using quantitative reverse transcription (qRT)–PCR. The gene-specific primers (Table S1) for qRT-PCR were designed using a Primer Premier 5.0 software. The total RNA was extracted by AG RNAex Pro Reagent and was reverse-transcribed into cDNA with the Evo M-MLV RT Kit (Accurate Biotechnology Co., Ltd., Hunan, China). All operations were implemented according to the kit instructions. The amplification reaction was performed on the Lightcycler480 system (Roche, Basel, Switzerland) using the Advanced SYBR Green Supermix Kit (Bio-Rad, Hercules, CA, USA). The total volume of reaction system was 20 μl. The qRT-PCR cycles were performed using the following reaction procedure: 95°C for 30 s, 40 cycles of 95°C denaturation for 5 s, and 58°C annealing and extension for 20 s. The relative expression level of genes was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2013). The rice ubiquitin gene was used as an internal control to normalize the expression levels (Wu et al., 2014). All samples were conducted for three biological replications.
Results
Sheath blight resistance of different genotypes examined in the field
Five rice genotypes inoculated under the field condition showed different sheath blight resistance, with average lesion length ranging from 33.45 to 58.42 cm (Table 1). The sheath blight resistance of GD66 was significantly higher than that of Lemont in both lesion length and visual rating according to the Duncan’s multiple range test (Table 1, Figure S4).
Lesion on leaf surface observed after in vitro inoculation
Apparent lesions were not observed on leave surfaces of all genotypes from 0 to 12 hpi. Infection symptoms gradually arose at 16 hpi, and the leaf color started to turn yellow at the infection site. It can be easily found that the white hyphae covered the leaf surface at 16 hpi and formed a network structure. For most genotypes, the lesions stay yellow from 16 to 52 hpi and turned gray from 56 to 72 hpi. Typical sheath blight lesions usually accompanied with the gray color at 56 hpi for most rice genotypes. The GD66 was more sensitive in sheath blight reaction than the other four genotypes. Gray lesions were found in leaves of GD66 from 16 to 32 hpi, whereas the lesions of the other four genotypes were still yellow. Typical sheath blight lesions were found at 44 hpi for GD66, about 12 h much earlier than the other four genotypes (Figure 1).
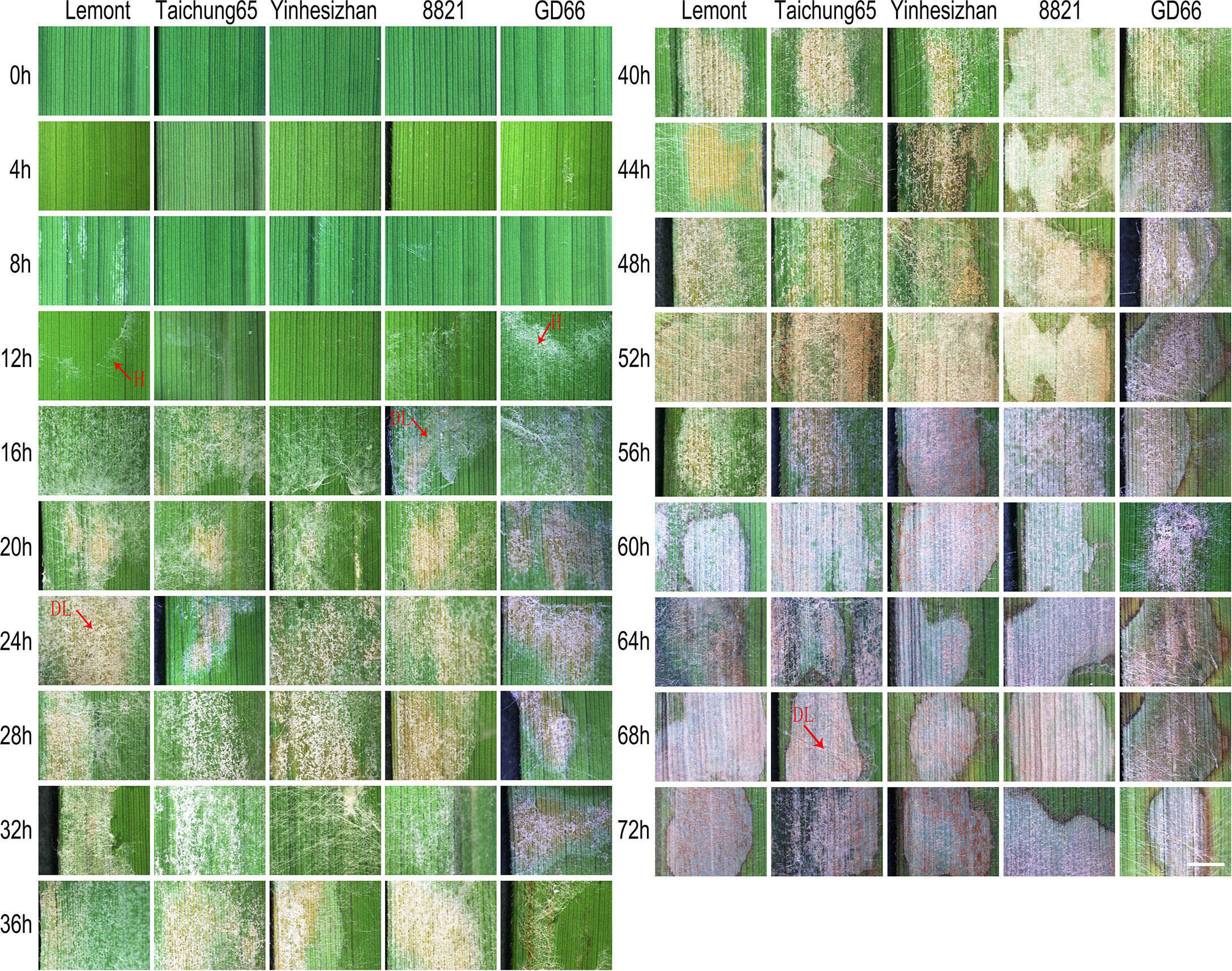
Figure 1 Sheath blight lesion development on rice leaf of five genotypes at every four h after in vitro inoculation observed under a stereoscopic microscope. Scale bar = 2 mm. H, hyphae; DL, disease lesion.
Lesion on sheath surface observed after in vitro inoculation
Although hyphae can be seen on sheath surface at 8 hpi, obvious symptoms were found at 24 hpi for Lemont, Taichung65, and Yinhesizhan and found at 32 hpi for GD66 and 8821. Typical sheath blight lesion appeared at 40 hpi for Taichung65, at 44 hpi for Lemont, at 48 hpi for Yinhesizhan, and at 60 hpi for GD66 and 8821, respectively (Figure 2).
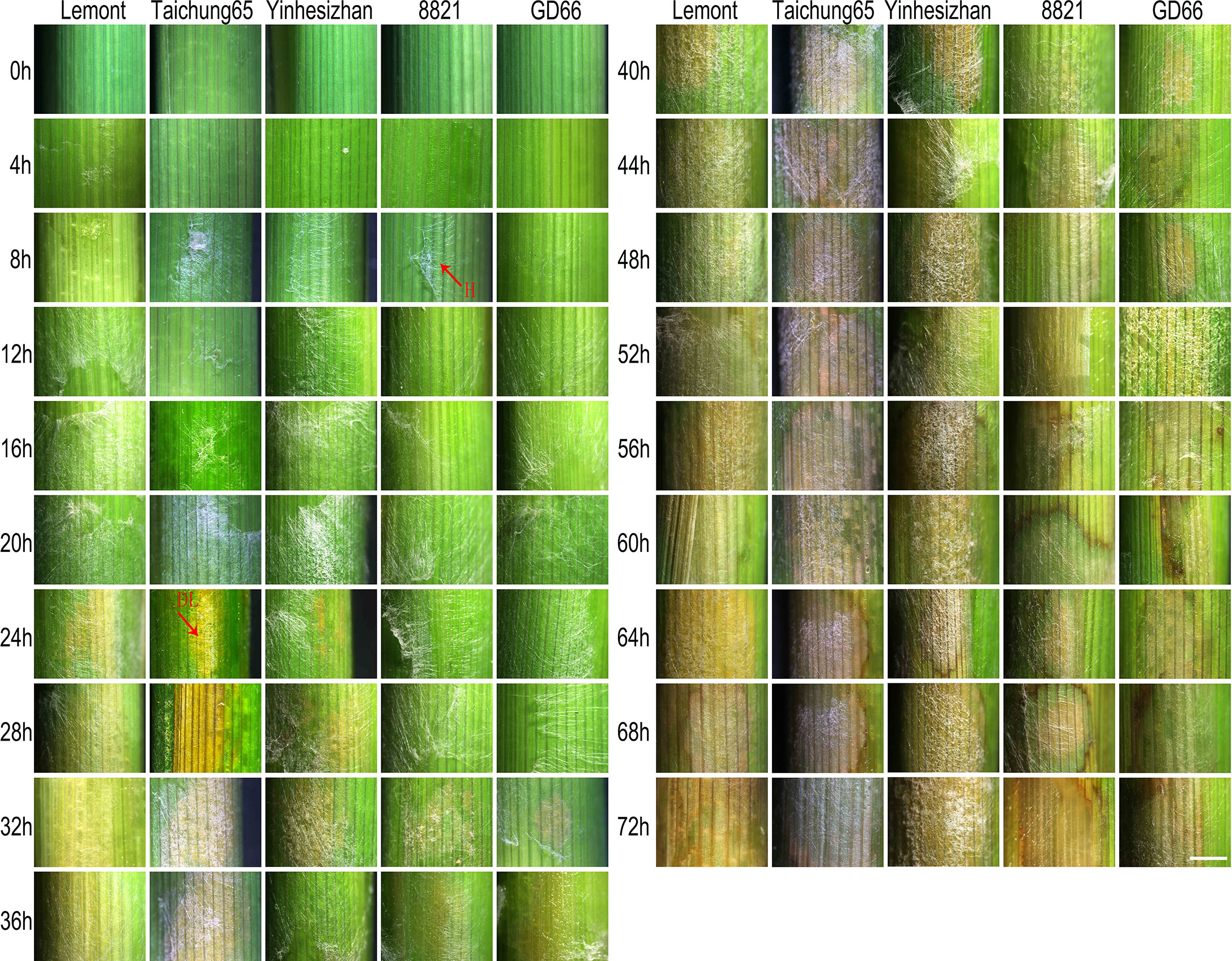
Figure 2 Sheath blight lesion development on rice sheath of five genotypes at every 4 h after in vitro inoculation observed under a stereoscopic microscope. Scale bar = 2 mm. H, hyphae; DL, disease lesion.
Developmental process of R. solani on rice leaf observed under a fluorescence microscope after in vitro inoculation
After stained with DAPI, the R. solani hyphae and infection cushions were clear. The bright color made them easy to be distinguished from rice leaf under a fluorescence microscope (Figure 3).
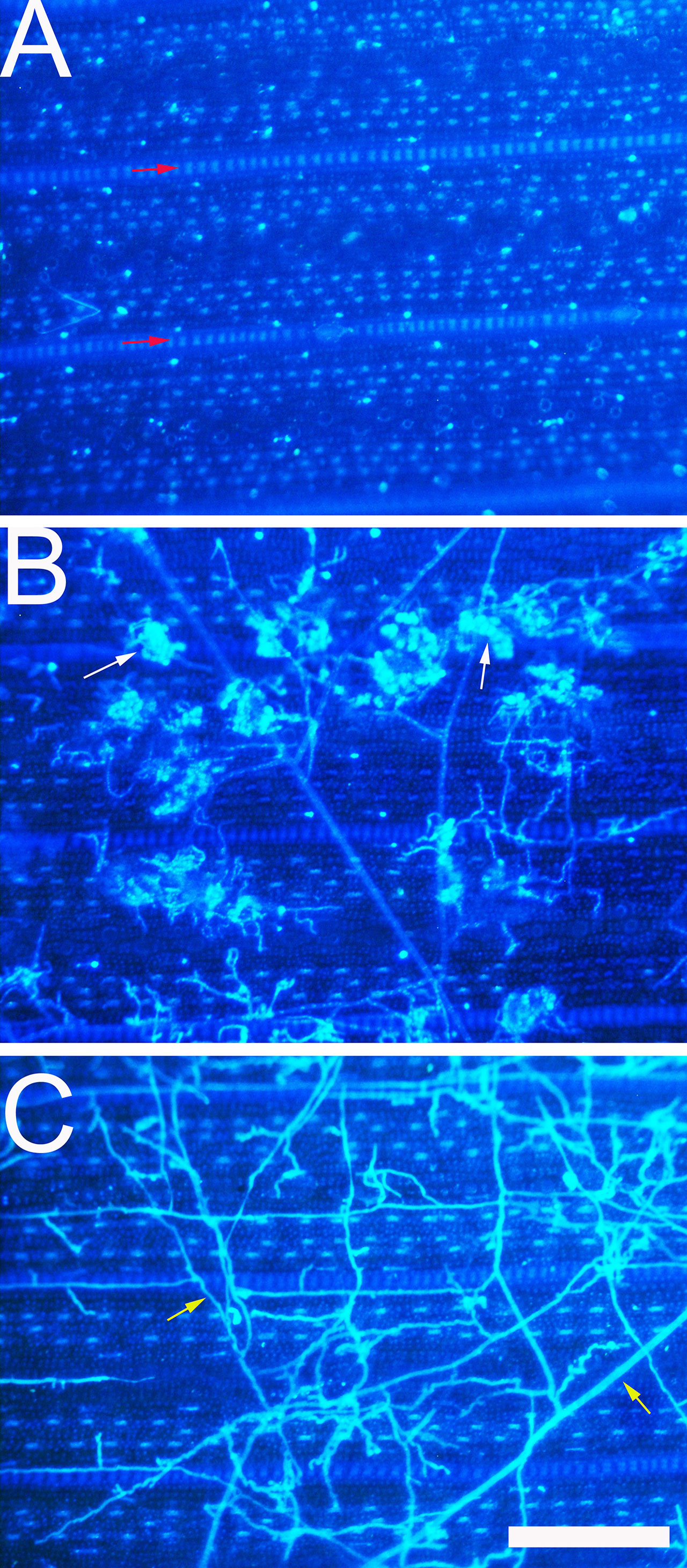
Figure 3 Development of Rhizoctonia solani hyphae and infection cushion on rice leaf observed under a fluorescence microscope. (A) The leaf surface of 8,821 at 0 hpi. Red arrow indicates vein. (B) The leaf surface of Lemont at 52 hpi. White arrow indicates the infection cushion of R. solani. (C) The leaf surface of Lemont at 72 hpi. Yellow arrow indicates hypha. Scale bar = 200 µm.
Hyphal growth was found on leaf surface at 4 hpi. From 4 to 8 hpi, the amount of hyphae was scarce. At 12 hpi, a large amount of hyphae covered the surface of rice leaves. A specialized penetration structure named infection cushion was clearly observed at 16 hpi. From 20 to 48 hpi, a large quantity of infection cushions occupied surface of leaves. The mount of infection cushions decreased gradually from 52 to 72 hpi (Figure 4).
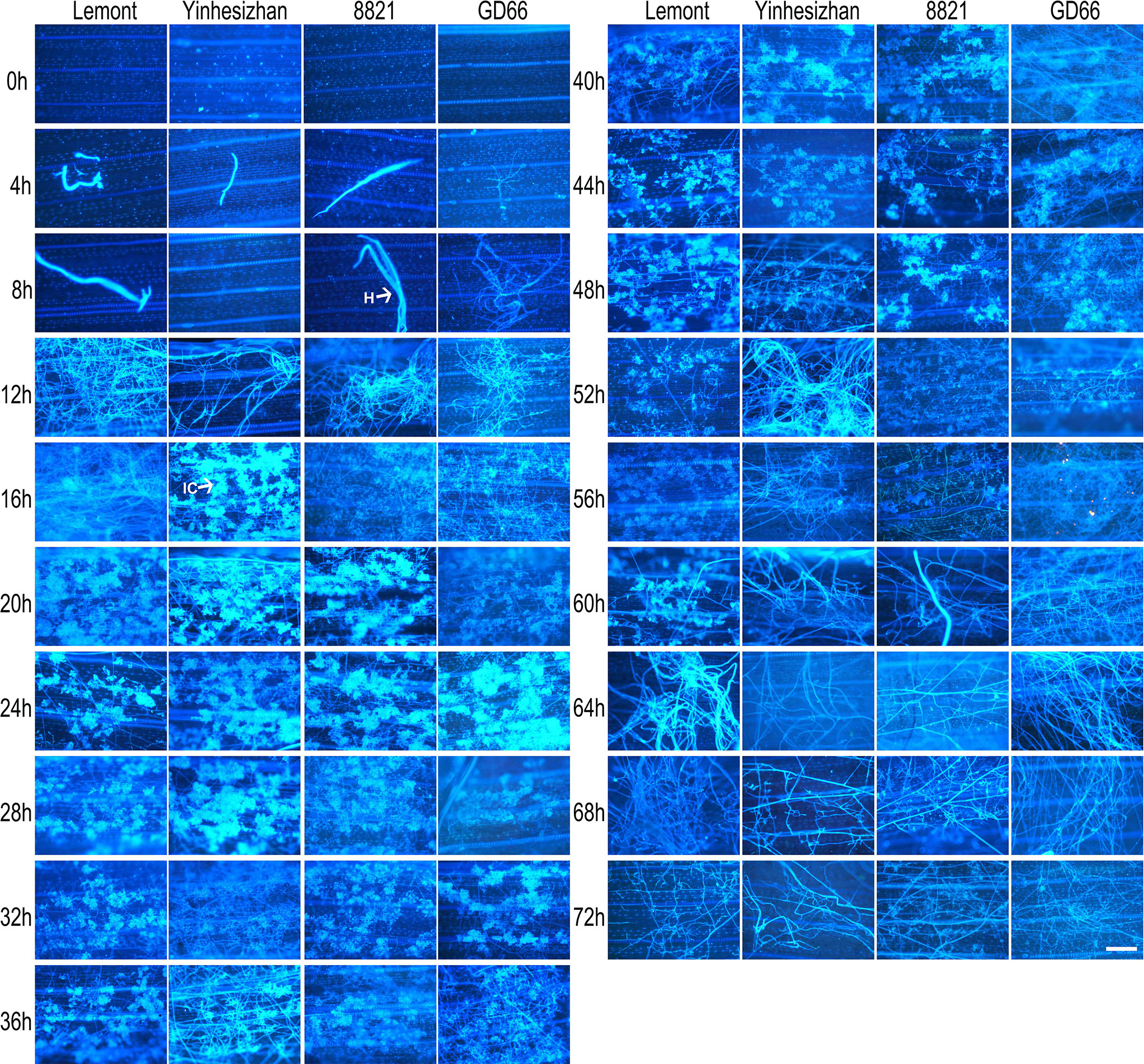
Figure 4 Development of Rhizoctonia solani hyphae and infection cushion on leaf of four rice genotypes at a serial time points after inoculation. The leaf samples were stained with DAPI and observed under a fluorescence microscope. Scale bar = 200 µm. H, hyphae; IC, infection cushions.
Developmental process of R. solani on rice sheath observed under a fluorescence microscope after in vitro inoculation
The development of hyphae and infection cushions on rice sheath was similar with those observed on rice leaf surface but with some differences. The infection cushions were first found at 12 hpi for Lemont, at 20 hpi for Yinhesizhan, at 24 hpi for 8821, and at 32 hpi for GD66, respectively. The infection cushion stayed a much shorter time on rice sheath surface compared with that on leaf surface (Figure 5). The infested structures spread along the edge of the lesion on the leaf, whereas there were only a few hyphae on the leaf sheath.
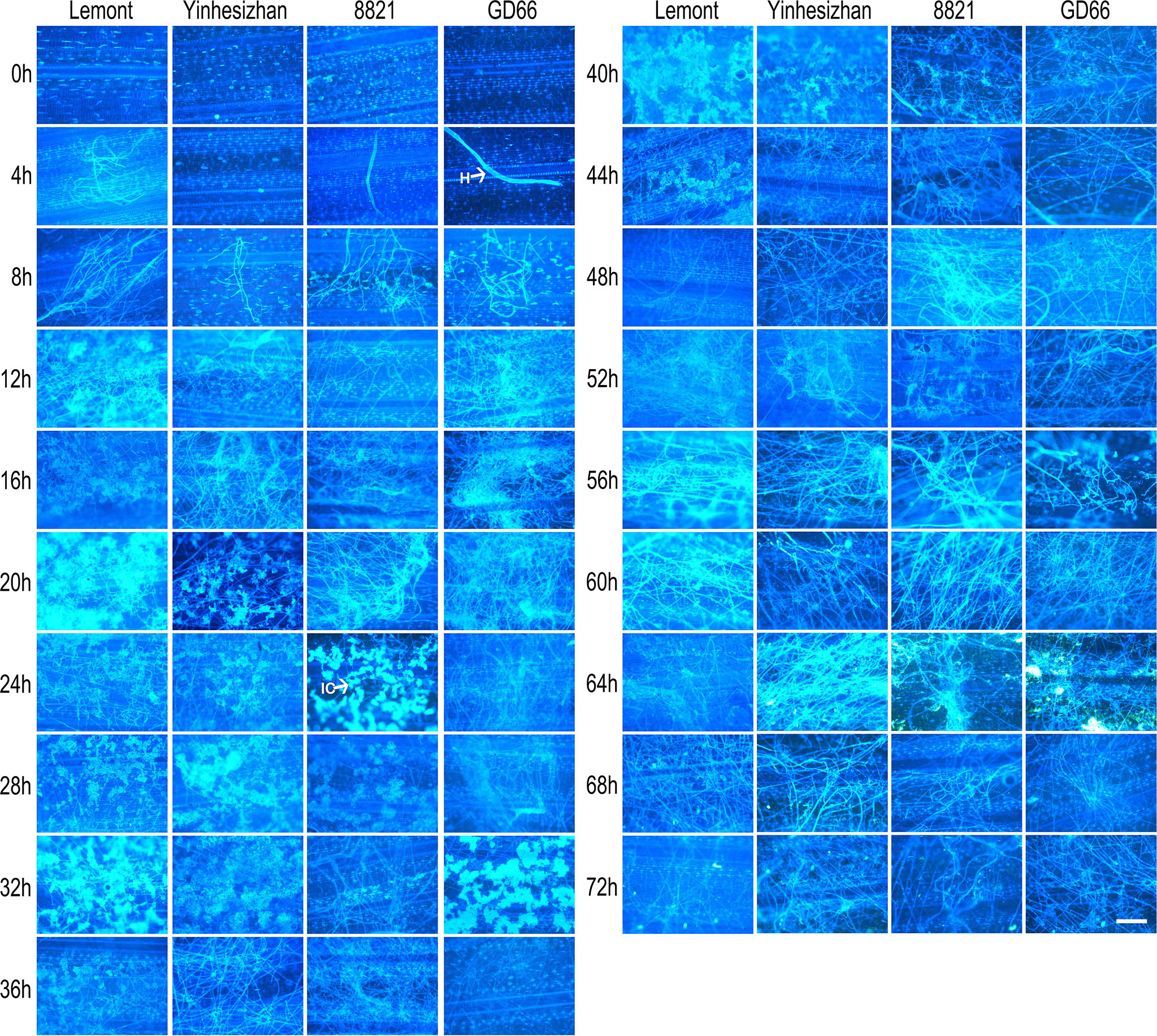
Figure 5 Development of Rhizoctonia solani hyphae and infection cushion on sheath of four rice genotypes at a serial time points after inoculation. The sheath samples were stained with DAPI and observed under a fluorescence microscope. Scale bar = 200 µm. H, hyphae; IC, infection cushions.
Developmental process of R. solani on rice leaf and sheath observed by WE-CLSM
WE-CLSM, which was developed by our research team (Zeng et al., 2007), was first employed to observe the developmental process of hyphae and infection cushions on rice leaf and sheath surface by using confocal microscopy. The hyphae or infection cushions of R. solani showed yellow or brighter color, whereas the leaf or sheath showed red color after stained with eosin B (Figure 6).
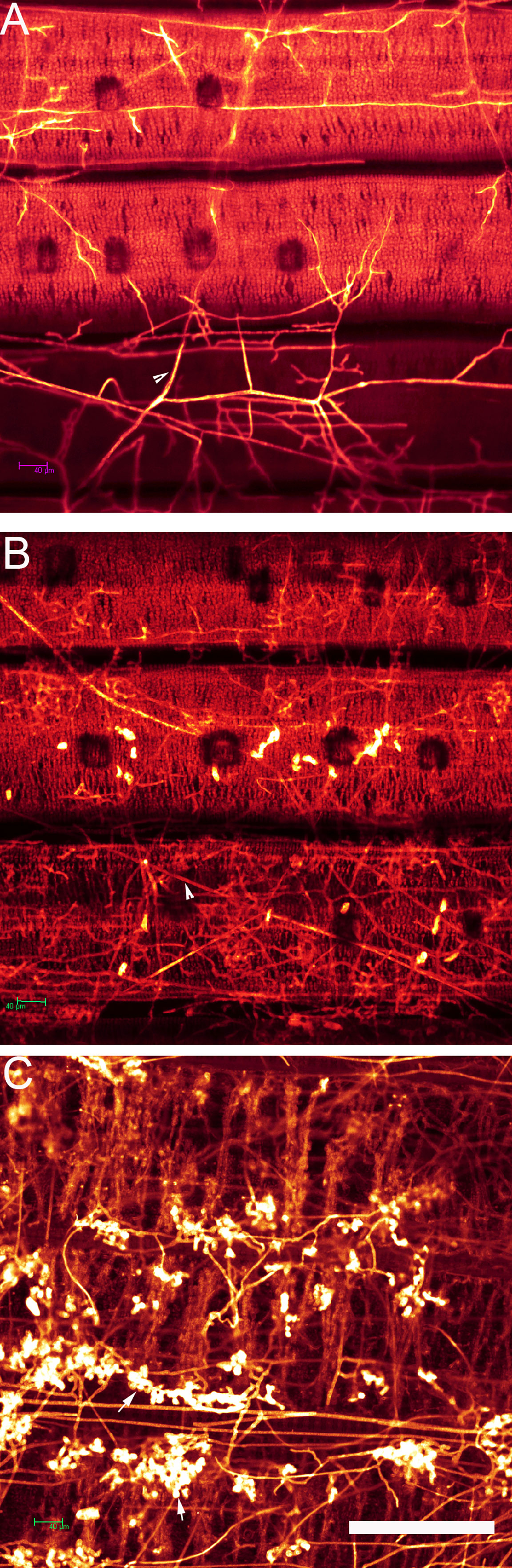
Figure 6 The whole-mount eosin B-staining confocal laser scanning microscopy developed in Xiangdong Liu’s laboratory was used for observing hyphal growth of Rhizoctonia solani on rice leaf. (A) Hyphae invading mesophyll cells on rice leaf at 12 h after in vitro inoculation. Arrowhead indicates the hypha. (B) Hyphae occupying a large number of spaces on rice leaf at 24 h after in vitro inoculation. Arrowhead indicates the hypha. (C) The mesophyll cells on rice leaf were almost destroyed and occupied by hyphae and infection cushions (arrows). Scale bar = 200 µm.
The hyphal growth was observed using WE-CLSM, and it was found that a large amount of hyphae covered leaf surface at 12 hpi. The infection cushions were clearly seen from 18 to 36 hpi, and they decreased gradually from 42 to 72 hpi (Figure 7). The situation was similar on sheath surface, but the infection cushions stayed a much shorter time on sheath surface than on leaf surface (Figure 8).
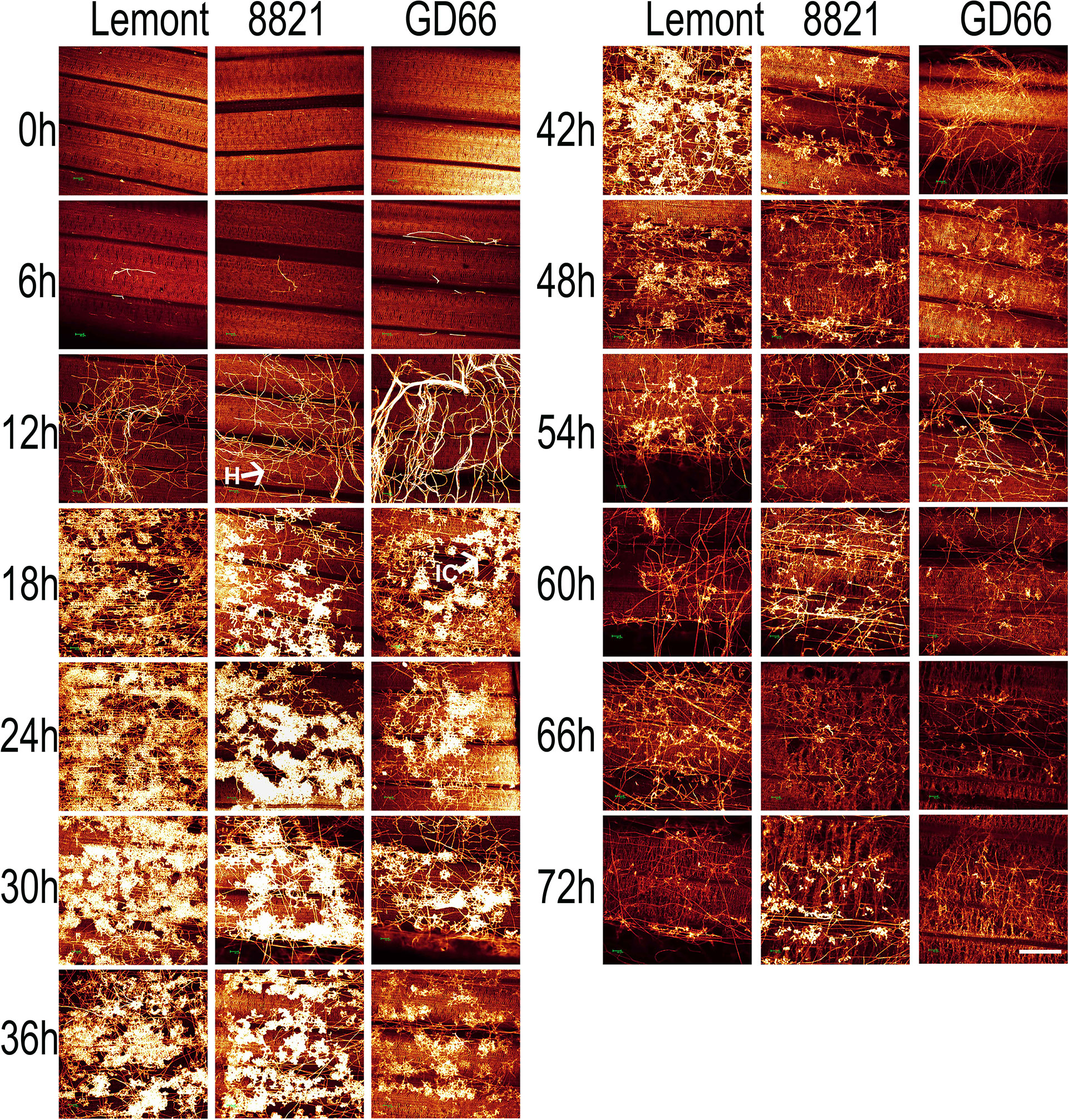
Figure 7 The whole-mount eosin B-staining confocal laser scanning microscopy showing developmental process of hyphae and infection cushions within rice leaf at a serial time point after inoculation. All images were obtained via the Leica SPE laser scanning confocal microscope. Excitation wavelength was 543 nm, and emission light was noticed between 550 and 630 nm. Scale bar = 200 µm. Three replications were applied for each sample. H, hyphae; IC, infection cushions.
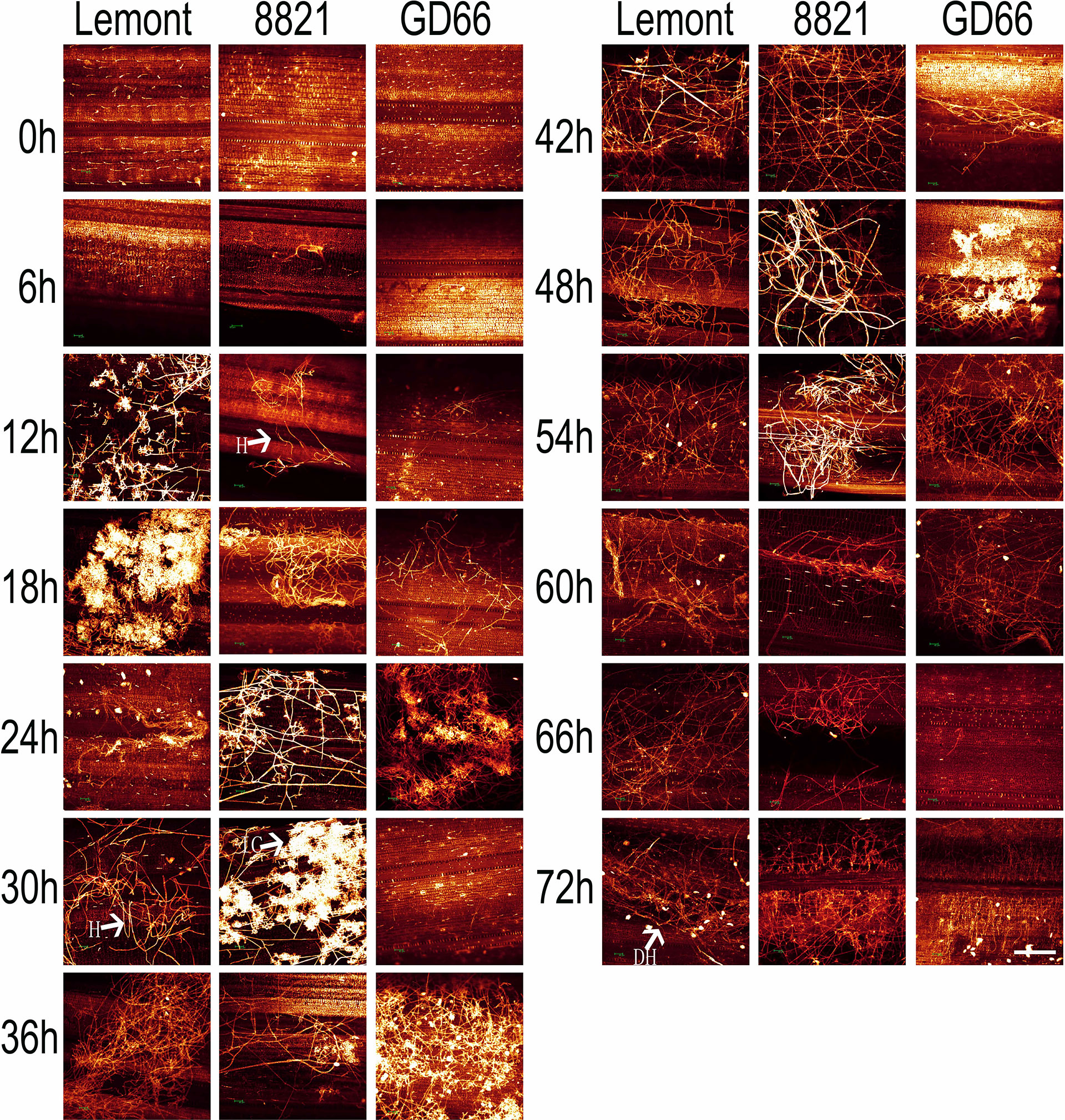
Figure 8 The whole-mount eosin B-staining confocal laser scanning microscopy showing developmental process of hyphae and infection cushions within rice sheath at a serial time point after inoculation. All images were obtained via the Leica SPE laser scanning confocal microscope. Excitation wavelength was 543 nm, and emission light was noticed between 550 and 630 nm. Scale bar = 200 µm. Three replications were applied for each sample. H, hyphae; IC, infection cushions. DH, decreased hyphae.
WE-CLSM can show not only the leaf or sheath surface but also different layers inside leaf or sheath. There were a large amount of hyphae found on the leaf surface of GD66 at 72 hpi (Figure 9). However, only a few hyphae were found invading GD66 mesophyll cells at 22 µm beneath leaf surface (Figure 9). At 21 µm beneath leaf surface, the amount of infection cushions decreased dramatically compared with that on the leaf surface (Figures 9C).
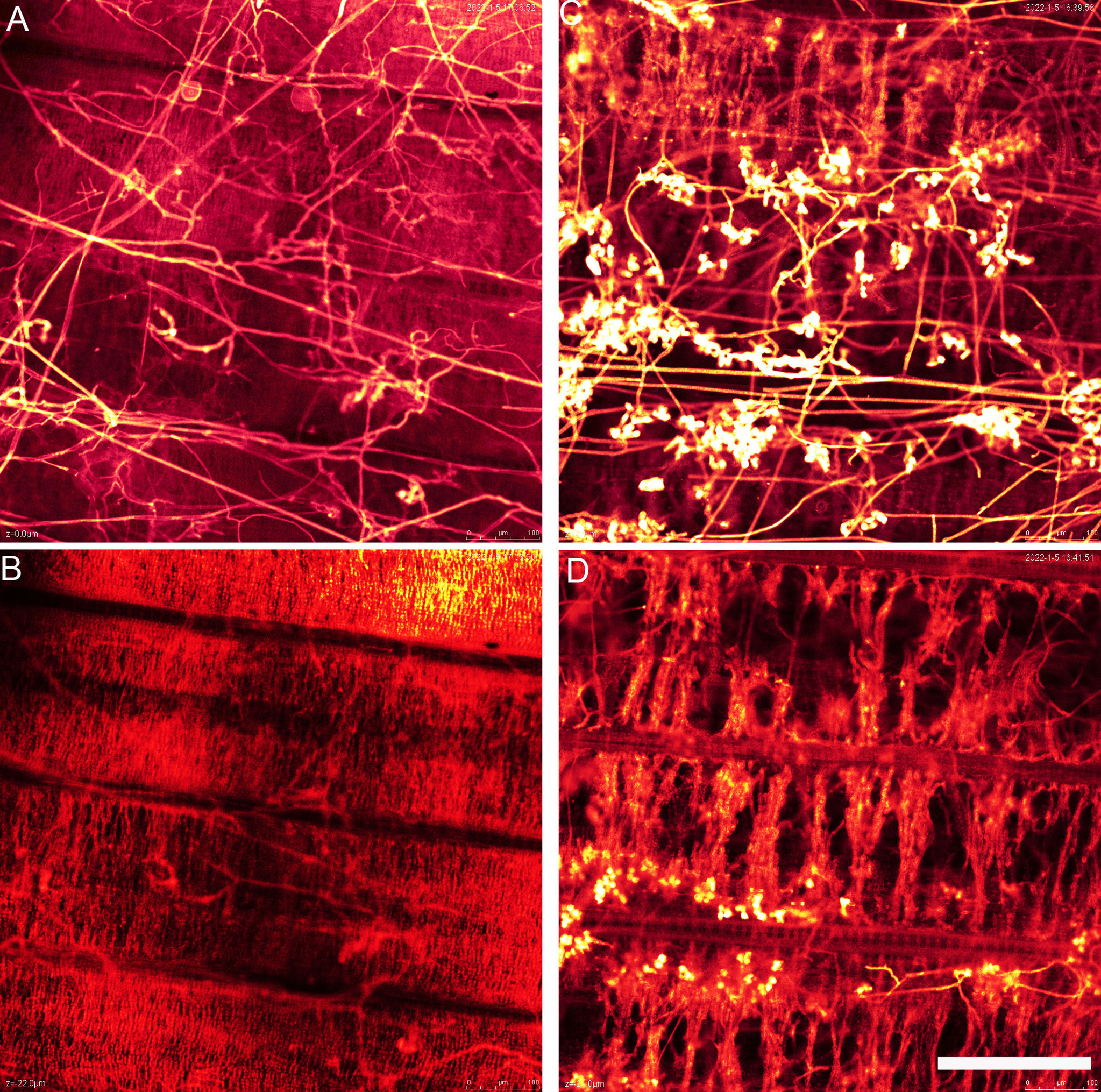
Figure 9 The hyphae above (A) and beneath (B) the GD66 leaf surface at 72 hpi as demonstrated by whole-mount eosin B-staining confocal laser scanning microscopy (WE-CLSM). (B) Twenty-two micrometers beneath GD66 leaf surface at 72 hpi. The infection cushions above (C) and beneath (D) the 8,821 leaf surface at 72 hpi as demonstrated by WE-CLSM. (D) Twenty-one micrometers beneath 8,821 leaf surface at 72 hpi. Scale bar = 200 µm.
Interestingly, R. solani could not only colonize on the abaxial surfaces of leaf sheath but also invade the paraxial side of the leaf sheath, suggesting that the hypha could pass through the surface of leaf sheath and move to the paraxial side and then colonize on both sides of leaf sheath. Cytological observation using semi-thin sections demonstrated this result (Figure S5). The observation in this study shows that the detailed positions where R. solani invading rice sheath were randomly distributed. WE-CLSM observation displayed that the R. solani could colonize on the paraxial side of leaf; however, it was not found on the abaxial side of the leaf, suggesting that the R. solani may have different infection mechanism between leaf sheath and leaf.
Statistical analysis of RNA-seq results at different time points after R. solani inoculation
To analyze the transcriptional response to the R. solani infection in rice, leaf sheaths were inoculated with ZJ03; RNA-seq analysis was performed on samples at 0, 24, and 48 hpi; and each time point was repeated thrice with a total of 18 samples. An overview of the sequencing and mapping results is shown in Supplementary Table S2. A total of 49.2-GB raw data were obtained from 18 samples. An average of 44,355,838 reads was obtained for each sample, with a Q30 quality score ≥ 93.55%. Filtered reads were aligned with the rice genome (Japanese Rice MSU_v7.0; 62,120 transcripts), which was obtained from the Genome databases for Plant Biology (https://plantbiology.aspb.org), resulting in 83.74% mapping percentage, indicating a high quality of the data.
Differential gene analysis in response to R. solani infection
A total of 6,234 and 2,523 DEGs were detected for Lemont at 24 and 48 hpi, respectively. In addition, 7,784 and 2,719 DEGs were detected for GD66 at 24 and 48 hpi, respectively (Figure 10). This indicated that the gene expression reached maximum extent at 24 hpi and then descended at that time for both genotypes.
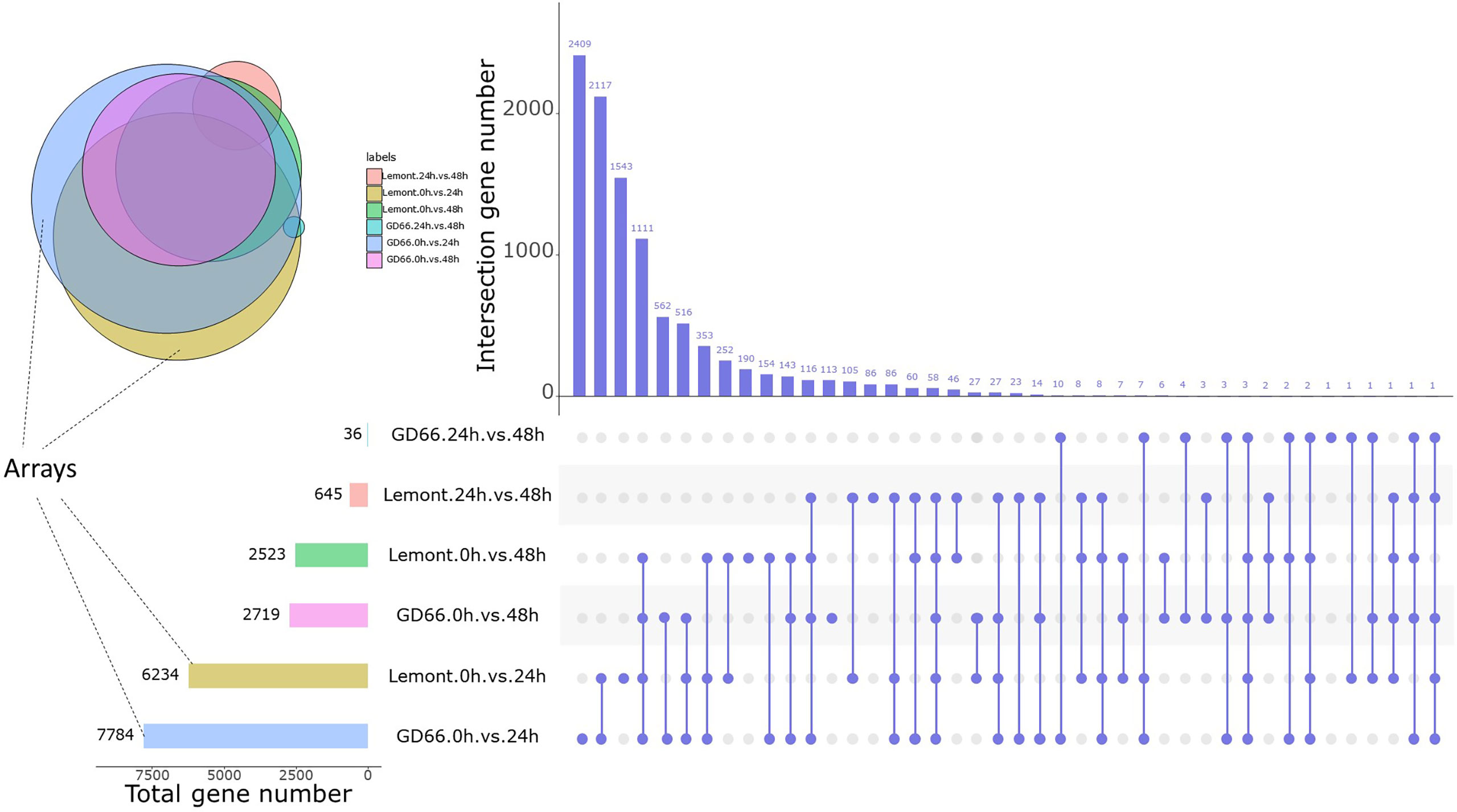
Figure 10 DEGs at three time points after inoculation between Lemont and GD66. The UpSet plot was used to visualize the total size and overlaps of DEGs in various data sets. The colorful bar charts indicate DEGs under single pairwise comparison; the left side of x-axis represents the number of DEGs. Column charts indicate DEGs under single or multiple comparisons. On the right side of x-axis, slate blue dots represent specific DEGs of a single comparison, slate blue lines connected by dots represent intersection DEGs of multiple comparisons, and the y-axis represents the number of DEGs corresponding to them. Colored circles indicate the Venn diagram of multiple comparisons of DEGs, with the same color corresponding to the colored bar chart on the left.
The number of DEGs was different between Lemont and GD66, and the upregulated genes exceeded those of the downregulated genes for both genotypes at different time points after inoculation: At 24 hpi, 6,234 DEGs were found in Lemont, with 3,244 upregulated genes and 2,990 downregulated genes. A total of 7,784 DEGs were identified in GD66, of which 4,177 were upregulated and 3607 were downregulated at 24 hpi. The same trend was found at 48 hpi (Table S3). It suggested that the resistance genotype GD66 recruited more genes than the susceptible genotype Lemont to fight against sheath blight pathogen.
We compared the sheath blight–resistant genotype GD66 and the susceptible genotype Lemont and found that there were 3,937, 4,021, and 1,848 DEGs at 0, 24, and 48 h, respectively (Figure 11). The results showed that there were great differences on gene expression before and after R. solani infection. The DEGs were also illustrated in a Venn diagram. It was clear that 1,293 DEGs were found to be continuously response in all time points (Figure 11).
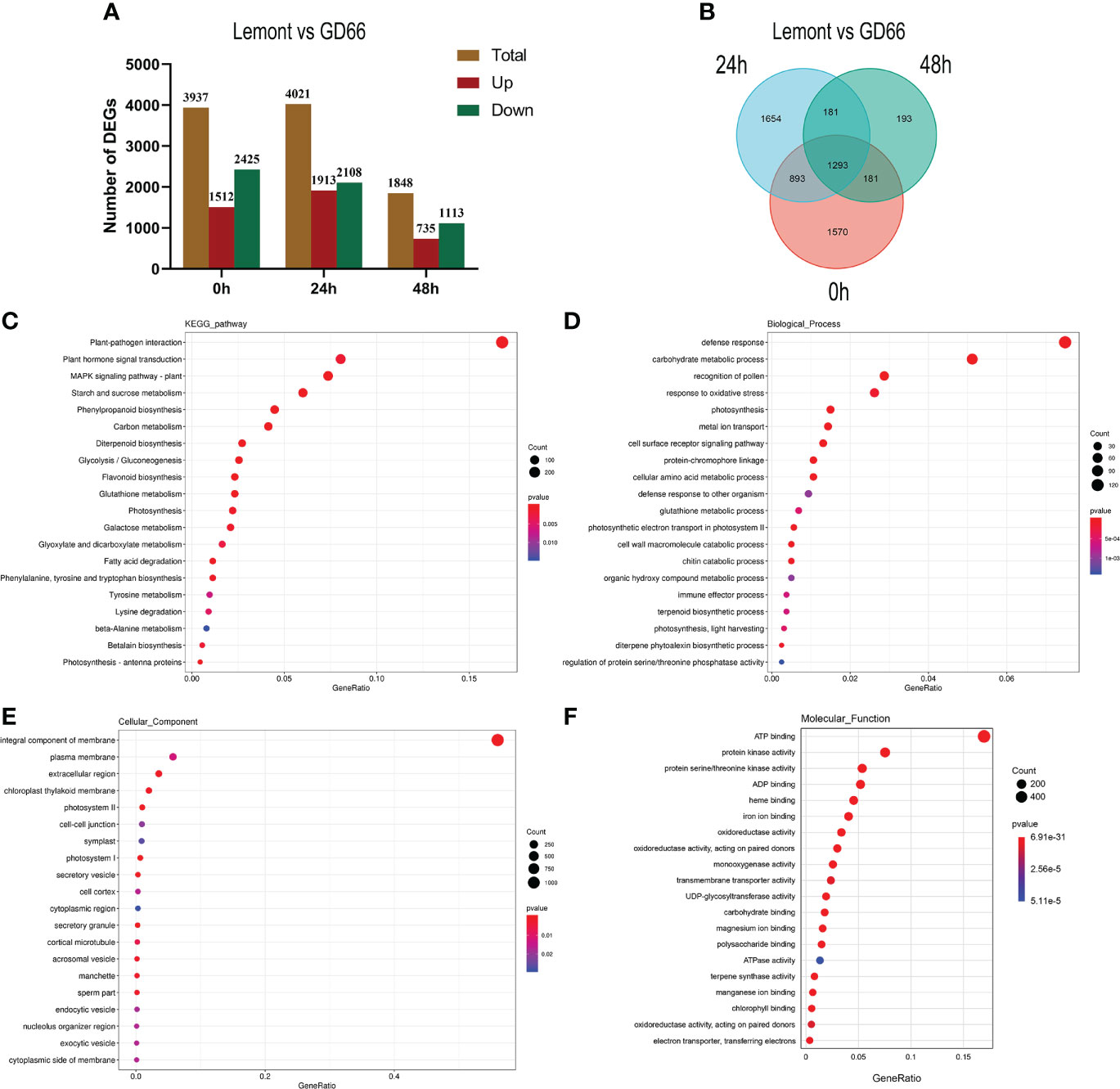
Figure 11 Differentially expressed genes (DEG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) and GO analysis. (A) Differentially expressed genes (DEG) between Lemont and GD66 detected at different time points after inoculation. Red color indicates upregulated genes, and green color indicates downregulated genes. (B) Venn diagram showing overlapping DEGs at three time points. (C) The KEGG pathway enrichment analysis of the DEGs between Lemont and GD66 at three time points. (D–F) Visualization of GO enrichment terms for total DEGs in Biological_Process, Cellular Component, and Molecular_Function Ontology, respectively.
Kyoto Encyclopedia of Genes and Genomes pathways and Gene Ontology annotation of the differentially expressed genes
A total of 5,965 DEGs were detected between Lemont and GD66 at different time points after inoculation (Figure 11). Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation revealed that the 5,965 DEGs were mapped to 132 KEGG pathways (p <0.05, Table S4). The highest enriched pathway was plant–pathogen interactions (KO04626), followed by plant hormone signal transduction (KO 04075), mitogen-activated protein kinase (MAPK) signaling pathway plant (KO04016), starch and sucrose metabolism (KO 00500), phenylpropanoid biosynthesis (KO 00940), and carbon metabolism (KO 01200) (Figure 11).
The 5,965 DEGs were annotated using GO database via the Blast2GO program (http://www.blast2go.com/). The most significantly enriched GO terms include the following: “defense response”, “carbohydrate metabolic process”, “recognition of pollen”, and “response to oxidative stress” in Biological Process Ontology (Figure 11, Table S5); “integral component of membrane”, “plasma membrane”, “extracellular region”, and “chloroplast thylakoid membrane” in Cellular Component Ontology (Figure 11, Table S6); “ATP binding”, “protein kinase activity”, “protein serine/threonine kinase activity”, and “ADP binding” in Molecular Function Ontology (Figure 11, Table S7). These results suggest that the complex molecular defense reaction was triggered in rice after R. solani invasion.
MapMan analysis
Because the DEGs reached maximum quantity at 24 hpi, we compared the DEGs between GD66 and Lemont at 24 hpi. The MapMan analysis showed that 392 DEGs were involved in the signaling regulation pathway. A total of 161, 147, 100, 62, 53, 37, 34, 28, 28, 24, and 23 DEGs were participated in the proteolysis, PR-protein, secondary metabolites, cell wall–related, abiotic stress, MYB transcription factor, ethylene, glutathione-S-transferase, redox state, peroxidases, and WRKY regulatory pathways, respectively. In addition, 19, 18, 16, 15, 14, 12, 12, 8, 5, 3, and 3 DEGs were involved in the glucanase, heat shock protein, auxin, Abscisic acid (ABA), Ethylene-responsive factor (ERF), salicylic acid, bZIP transcription factor, jasmonate, DNA binding with one finger (DOF), zinc finger protein, mitogen-activated protein (MAP) kinase, and brassinosteroid regulatory pathways, respectively (Figure 12, Table S8).
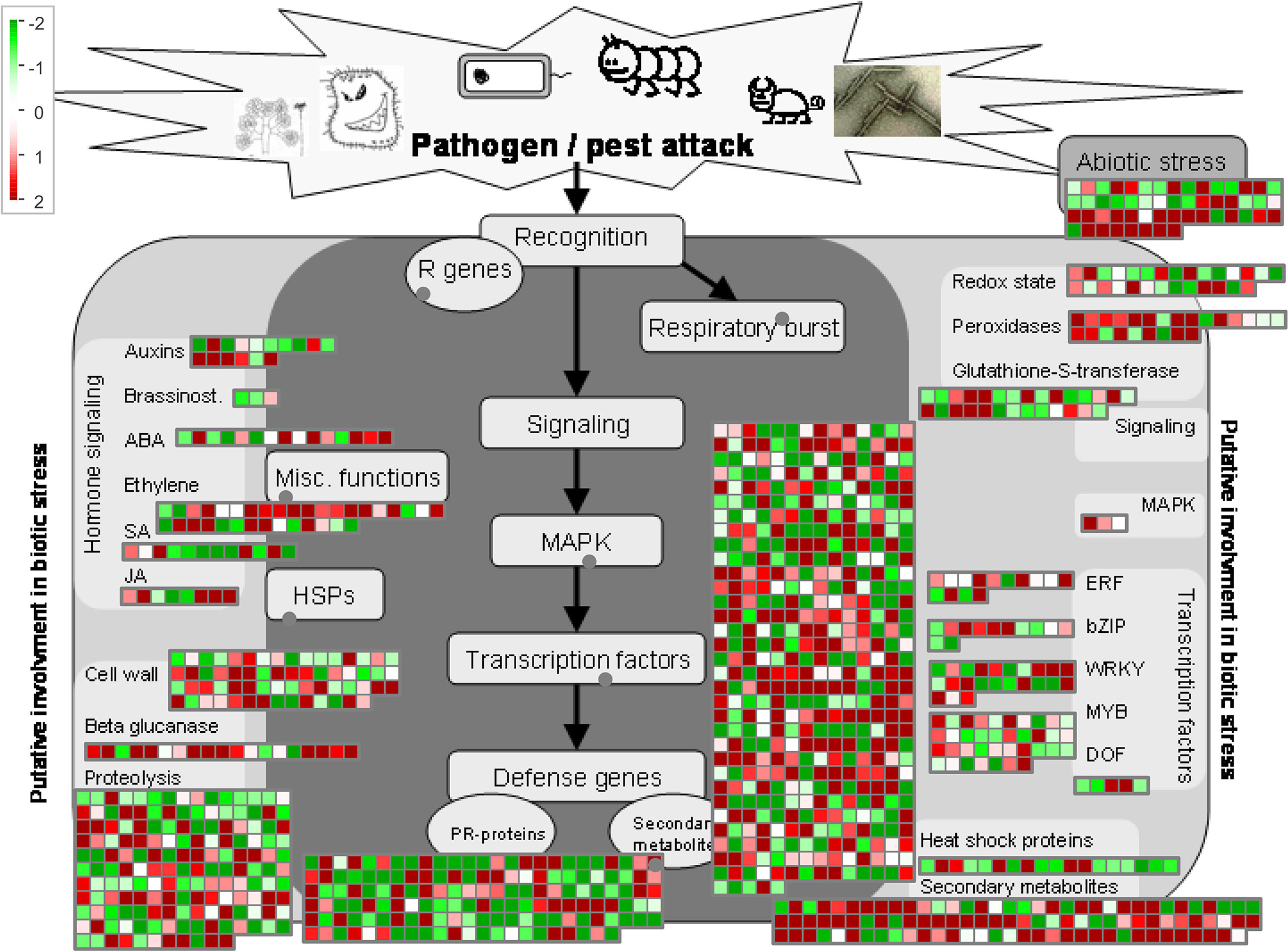
Figure 12 MapMan analysis of the DEGs between GD66 and Lemont at 24 hpi based on the log2FC value. Red, upregulate; green, downregulate.
Validation of RNA-seq results using qRT-PCR
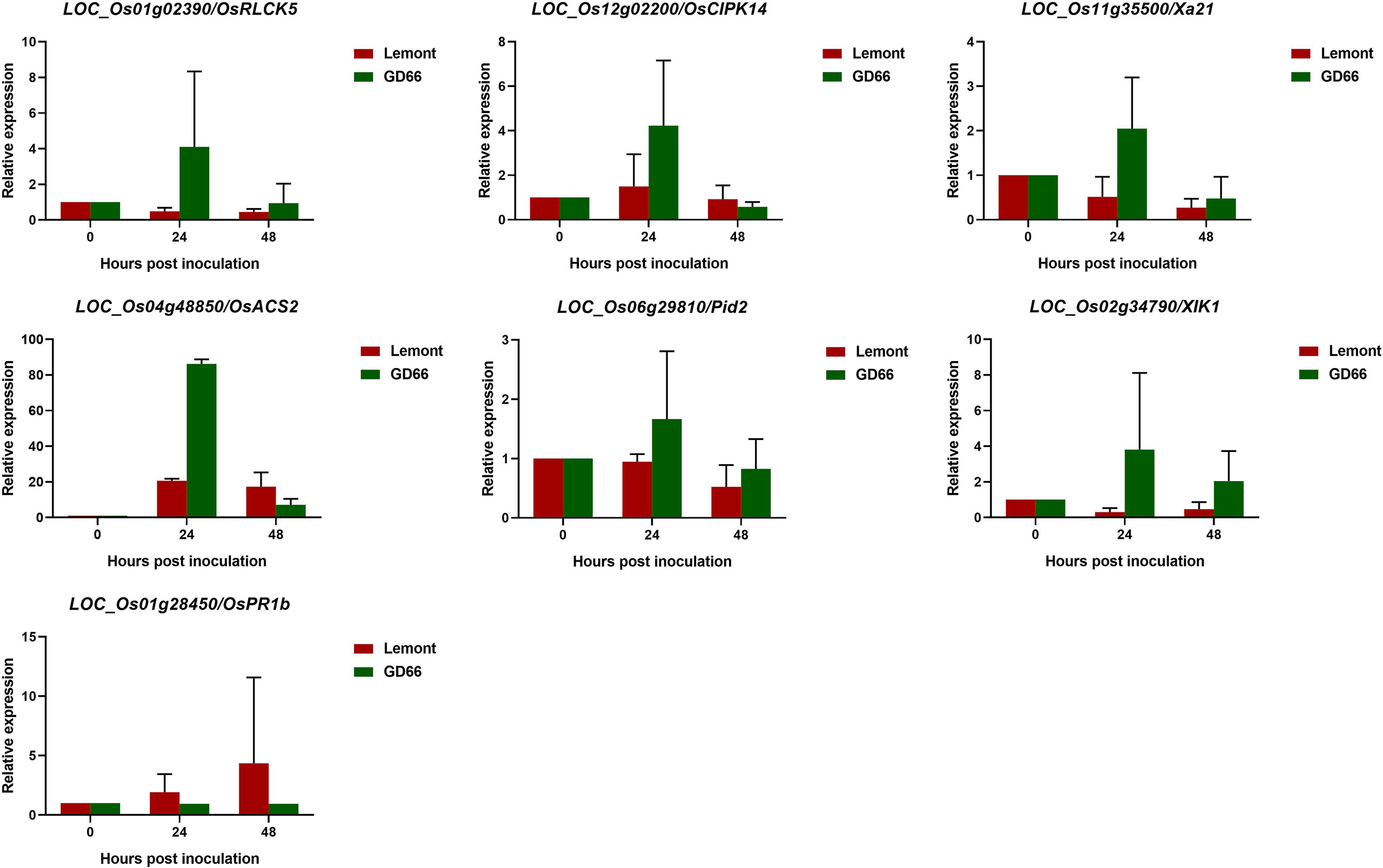
The RNA-seq data were further validated by qRT-PCR analysis. Seven rice genes that have been reported to be associated with disease resistance were chosen for qRT-PCR analysis. Melting curves of qPCR products showed a unique peak for all genes, suggesting a good specificity of the primers (Pictures not shown). Rice ubiquitin gene was used as the internal control, and the Ct values were normalized. The relative expression levels of the seven genes between Lemont and GD66 at different time points after inoculation were calculated. The qRT-PCR results showed the same trends with the RNA-seq data (Figure 13, Table S9), suggesting that the Illumina sequencing data were good and reliable. The qRT-PCR results confirmed that OsRLCK5, OsCIPK14, Xa21, OsACS2, Pid2, and XIK1 had higher expression in the resistant genotype than the susceptible genotype, whereas OsPR1b had lower expression.
Discussion
WE-CLSM is a wonderful method for observing hyphal development in rice leaf or leaf sheath after R. solani infection
The WE-CLSM was first developed in 2003 and named as “WE-CLSM” in 2007 in our research team laboratory for studying rice embryo sac and other rice tissues (Zhang et al., 2003; Zeng et al., 2007; Li et al., 2017). For the first time, we used this technique to study rice leaf or sheath after sheath blight infection in this study. The results showed that it was powerful for observing hyphal growth within rice leaf or sheath: The hyphae or infection cushions are in yellow or brighter color under the red mesophyll cell background, which makes them easy to be distinguished from the rice mesophyll cells under the confocal microscope (Figure 9). WE-CLSM has the advantage of displaying detailed information at any layers beneath the leaf epidermal cells without needing sections, which makes it more convenient than the conventional sectioning method. It was the first time to find that R. solani could not only colonize on the abaxial surfaces of leaf sheath but also invade the paraxial side of the leaf sheath. However, the R. solani could only colonize on the one side of leaf. The infested structures spread along the edge of the lesion on the leaf, whereas there were only a few hyphae found on the surface of leaf sheath. This could be explained by the results of plastic semi-thin sections and WE-CLSM observation: The hyphae and infested structures did not traverse the leaf, and infestation pads destroyed only the outer cell wall structure of the leaf. The hyphae formed clustered rod-like polymers that absorbed nutrients from the mesophyll cells inside the leaves. The hyphae or infested structures may not penetrate the tight tissue structure in the leaves, so it could only colonize on the paraxial side of leaf. There was a large amount of air in the cavity of the leaf sheath, and hyphae can spread and move freely. Moreover, the parenchyma cells in the inner layer of the leaf sheath were relatively easy to be destroyed. Therefore, the pathogen can penetrate and colonize on both sides of the leaf sheath. After entering the inner part and colonizing on the paraxial side of the leaf sheath, the infested structures decreased on the abaxial side of the leaf sheath. In addition, GD66 is a highly resistant genotype. In fact, GD66 is the most resistant genotype among 211 genotypes tested in six environments from 2012 to 2016 (Zeng et al., 2017). However, the lesion developed much earlier on leaves of GD66 than the other four genotypes as demonstrated in Figure 1. It suggested that the mechanism of R. solani–infecting rice sheath was different from that of leaf.
In addition to WE-CLSM, we also used a fluorescence microscope to observe the hyphal growth on rice leaf and leaf sheath. Both methods let the hyphae and infection cushions of R. solani clearer and easy to be distinguished from the leaf cells. However, the fluorescence microscopy can only display the situation on leaf surface; it cannot see through the leaf epidermis cells and show what is beneath the surface. However, WE-CLSM can demonstrate detailed information at any sections beneath leaf or sheath surface.
Using both fluorescence and WE-CLSM, it was found that the amount of hyphae increased from 4 to 12 hpi on leaf surface. In addition, the infection cushions formed at about 16 hpi and maintained large quantities from 20 to 48 hpi and then decreased gradually from 52 to 72 hpi on leaf surface. The situation was similar on sheath surface, but the infection cushions stayed shorter time on sheath surface than on leaf surface. We also found that the amount of hyphae or infection cushions decreased dramatically at 20 µm beneath leaf surface at 72 hpi by using confocal microscopy. In addition, we observed that sheath blight pathogen can penetrate the upper epidermis cells of sheath and then pass through the mesophyll cell and reach the lower epidermis cells (picture not shown). This suggested that the pathogen can harm rice sheath at two directions: (1) from the upper epidermis to the lower epidermis by entering the mesophyll cells at vertical direction and (2) occupying more area on sheath surfaces at horizontal direction.
DEGs between sheath blight–resistant and sheath blight–susceptible genotypes
In recent years, the advancement of next-generation sequencing technique has greatly promoted genome and transcriptome research in different organisms (Wang et al., 2009; Jain, 2012). RNA-seq, a sequencing technique, has been widely used in studying plant pathogenesis, host-defense responses, and mechanisms of plant–pathogen interactions (Kawahara et al., 2012; Lai and Mengiste, 2013; Okubara et al., 2014). To decipher the differential gene expression responses during the R. solani infection, a time-series RNA-seq analysis (i.e., 0, 24, and 48 h) was performed, and a total of 7,950 and 7,080 DEGs in GD66 and Lemont were identified, respectively. Our results are consistent with those by Samal et al. (2022), who detected more DEGs in the resistant genotype CR1014 than that in the susceptible genotype Swarna-Sub1. Similarly, stronger and earlier responses to pathogen invasion at the transcriptome level have been found in the resistant grapevine specie Vitis riparia infected with Plasmopara viticola (Polesani et al., 2010).
In the present study, a total of 3,937 DEGs were detected between Lemont and GD66 before pathogen inoculation, suggesting that these two genotypes had great differences in genetic background. The fact that there were more DEGs in GD66 than that in Lemont at both 24 and 48 hpi indicated that the resistant genotype GD66 recruited more genes than the susceptible genotype Lemont in fighting against the pathogen. Our results are different from some previous reports that more DEGs were found in sheath blight–susceptible cultivars than that in sheath blight–resistant cultivars (Zhang et al., 2017; Shi et al., 2020; Yang et al., 2022). It means that the GD66 may have different mechanism than other resistant cultivars during rice–R. solani interaction.
A comparison between the resistance variety Teqing and the susceptible variety Lemont showed that the DEGs mainly concentrated in the early stages after R. solani AG1 IA infection (Zhang et al., 2017). A similar result was also found in the present study. We found that the DEGs reached maximum extent at 24 hpi and then decreased at 48 hpi. It suggested that the changes in R. solani–induced gene expression were dynamic. This result is consistent with reports on different plants and emphasizes the importance of time series analyses in understanding R. solani–plant interaction (Moore et al., 2011; Windram et al., 2012; Zhu et al., 2016).
Genetic regulatory pathways involved in R. solani–rice interaction
Transcriptome analysis revealed complex gene regulatory network during R. solani infection in the present study. KEGG analysis showed that the DEGs were significantly enriched in pathways such as “plant–pathogen interactions”, ‘plant hormone signal transduction”, “MAPK signaling pathway”, “starch and sucrose metabolism”, and “phenylpropanoid biosynthesis”. Similar regulatory pathways have also been reported in some previous studies (Zhang et al., 2017; Yuan et al., 2018; Das et al., 2022). GO enrichment analysis showed that three important processes were involved during pathogen infection: (1) defense response, (2) carbohydrate metabolic process, and (3) oxidative stress response (Figure 11).
It has been reported that OsRLCK5 interacted with OsGRX20 and positively regulated rice resistance to R. solani (Wang et al., 2021). We found a higher expression of OsRLCK5 in the resistant genotype GD66 at both 24 and 48 hpi. The expression levels of OsRLCK5 are low in the susceptible genotype Lemont as shown by qRT-PCR analysis (Figure 13). We speculate that a higher expression of OsRLCK5 may contribute to the sheath blight resistance in GD66.
Rice blast is a serious rice disease. Our study showed that some blast resistance genes may participate in the sheath blight resistance. Pid2 encodes a receptor-like kinase protein, and it confers race-specific resistance to the Magnaporthe grisea strain, ZB15 (Chen et al., 2006). We found that the expression of Pid2 was higher in GD66 than that in Lemont at both 24 and 48 hpi (Figure 13). It suggested that the Pid2 may be related to sheath blight resistance. The Pi9, which encodes a nucleotide-binding site–leucine-rich repeat protein, is a broad-spectrum blast resistance gene in rice (Qu et al., 2006). We found that the expression of Pi9 (LOC_Os06g17900) was higher in the resistant genotype GD66 than that in the susceptible genotype Lemont before or after inoculation (Table S10). This suggested that the Pi9 may be related to the basic resistance of GD66. OsCPK12 encodes a rice calcium-dependent protein kinase, and it negatively regulates rice blast resistance (Asano et al., 2012). In the present study, a lower expression level of OsCPK12 in the resistance genotype GD66 and a higher expression in the susceptible genotype Lemont were detected. It suggested that the OsCPK12 may negatively regulate sheath blight resistance.
It has been reported that OslecRK, a rice lectin receptor-like kinase, is important for resistance to rice blast and bacterial blight diseases (Cheng et al., 2013). In the present study, the expression of OslecRK (LOC_Os04g12540) in GD66 was twice as that in Lemont at 24 hpi (Table S10). Two ethylene biosynthetic genes, OsACS1 and OsACS2, are induced by M. oryzae infection (Iwai et al., 2006). The expression of OsACS2 (LOC_Os04g48850) in GD66 was about five times as that in Lemont at 24 hpi based on the RNA-seq data (Table S9) or qRT-PCR result (Figure 13). This indicates that the biosynthetic gene that contributes to basal resistance against M. oryzae also responds during R. solani challenging and may be related to the resistance in GD66.
The WRKY transcription factor has been proven to be regulated in plant defense responses, including positive and negative regulation of disease resistance (Pandey and Somssich, 2009). OsWRKY24 encodes a protein that functions as a negative regulator of GA and ABA signaling (Zhang et al., 2009). We found that the OsWRKY24 (LOC_Os01g61080) was differentially expressed in GD66 and Lemont at 24 hpi. The expression of OsWRKY24 in GD66 was more than four times than that in Lemont (Table S10). It has been reported that overexpression of the OsWRKY89 gene enhanced resistance to rice blast fungus and white-backed planthopper (Wang et al., 2007). Our results showed that the expression of OsWRKY89 (LOC_Os11g02520) were lower in GD66 than that in Lemont before or after inoculation (Table S10). It seems that OsWRKY89 may not be involved in the sheath blight resistance of GD66. In addition, the expression of OsWRKY70 (LOC_Os05g39720) was significantly higher in GD66 than that in Lemont at 24 hpi (Table S10), suggesting that it may be related to sheath blight resistance in GD66.
OsERF922 is a rice transcription factor that negatively regulates rice resistance to M. oryzae (Liu et al., 2012). In our study, the expression of OsERF922 (LOC_Os01g54890) were lower in GD66 than that in Lemont before or after inoculation (Table S10), indicating that it may also negatively regulate sheath blight resistance in GD66.
It has been shown that OsCIPK14/15 play crucial role in the microbe-associated molecular pattern–induced defense signaling pathway in rice (Kurusu et al., 2010). In our study, the expression level of OsCIPK14 in GD66 was lower than that in Lemont before inoculation and at 48 hpi, but the expression of OsCIPK14 in GD66 was increased dramatically compared with that of Lemont at 24 hpi (Figure 13, Table S9). Whether OsCIPK14 participates in the regulation of sheath blight resistance needs to be clarified in the future.
XIK1 encodes leucine-rich repeat receptor-like kinase and positively regulates Xa21-mediated rice bacterial blight disease resistance (Hu et al., 2015). Our RNA-seq result showed that the expression of XIK1 (LOC_Os02g34790) was higher in GD66 than that in Lemont before or after R. solani inoculation (Table S9), suggesting that it may also involve in sheath blight resistance.
It has been reported that overexpression of OsPGIP1 enhances rice resistance to sheath blight (Chen et al., 2016). However, our RNA-seq data showed that the expression level of OsPGIP1 (LOC_Os05g01380) was lower in GD66 than that in Lemont before or after inoculation, suggesting that it may play an inessential role in the resistance genotype GD66.
A previous report showed that the levels of OsPR1b transcript was significantly upregulated in rice plant after infected by rice black-streaked dwarf virus (Lu et al., 2020). In the present study, we found that the expression of OsPR1b was significantly higher in Lemont than that in GD66 before or after inoculation (Figure 13, Table S9). How OsPR1b regulating sheath blight resistance needs to be characterized in the future.
Together, both RNA-seq and qRT-PCR results showed that genes participated in different regulatory pathways response to R. solani invasion: Genes relating to bacterial blight disease resistance (Xa21, XIK1), blast disease resistance (Pid2, OsACS2), rice black-streaked dwarf virus (OsPR1b), and other defense-related pathways altered significantly after R. solani infection.
We examined the DEGs between Lemont and GD66 and found that some of which contained typical NBS-LRR domain, such as LOC_Os01g52330, LOC_Os12g13550, and LOC_Os12g10710 (Table S8). These three genes have not been characterized in previous reports. Because they expressed differentially between GD66 and Lemont, they might be associated with the sheath blight resistance. These candidate genes can be tested further.
Insight in breeding a sheath blight–resistant rice cultivar
Genetic studies have shown that the rice sheath blight resistance is a typical quantitative trait controlled by many genes with minor effects. That means to breed a sheath blight–resistant cultivar, a lot of loci related to sheath blight resistance must be focused simultaneously. What genes should we focus? What is the target for selection in breeding? We recommend that the sheath blight–resistant genes characterized in previous studies should be the primary targets. We should also focus on genes that are participating in the sheath blight–resistant regulatory pathways identified by transcriptome analysis, although they may have not be cloned or characterized yet.
As we can learn from the present study, there were huge differences between the resistant genotype GD66 and the susceptible genotype Lemont at the transcriptome level. It is difficult to breed a sheath blight–resistant cultivar from a sheath blight–susceptible genetic background. In this study, we found that GD66 and 8821 are both superior genotypes with high resistance to sheath blight disease. If a sheath blight–susceptible cultivar was chosen as the parental parent in a breeding program, then we recommend using backcrossing to increase the sheath blight–resistant background of the maternal parent, such as 8821, to make sure that more sheath blight–resistant genes were selected in their descendants.
Conclusion
A systematic cytological observation of developmental characteristics of Rhizoctonia solani hyphae and infection cushions on rice leaf and sheath displayed that the amount of hyphae increased dramatically on leaf and sheath surface at 12 hpi and that the infection cushions occurred and maintained at a huge number from 18 to 36 hpi, and then, the infction cushions disappeared gradually from 42 to 72 hpi. The R. solani could colonize on the abaxial sheath first, then penetrate the epidermis cell to inner part of sheath, and finally invade the paraxial side of leaf sheath. A different behavior of R. solani was found in rice leaf. RNA-seq analysis revealed that the resistant genotype GD66 recruited more than 5,000 DEGs to fight against the pathogen, which associated with some important resistant pathways, including bacterial blight disease resistance (Xa21, XIK1), blast disease resistance (Pid2, OsACS2), rice black-streaked dwarf virus (OsPR1b), and other defense-related genes. These results suggest that the WE-CLSM is a powerful technique in observing development of R. solani hyphae and infection cushions on leaf or sheath surface, and this process was associated with many resistance gene expressions simultaneously.
Data availability statement
The data presented in the study are deposited in the NCBI SRA repository, accession number PRJNA886841.
Author contributions
SL: Conceptualization, methodology, investigation, and data curation, writing—original draft, and writing—review and editing. TW: Investigation, data curation, and supervision. GM: Investigation and supervision. JL: Investigation, data curation, and supervision. DL: Investigation and supervision. XL: Conceptualization, methodology, resources, funding acquisition, and writing—review and editing. YZ: Conceptualization, methodology, resources, writing—original draft, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Laboratory of Lingnan Modern Agriculture Project (NT2021001) and the Opening Foundation of State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources (202006).
Acknowledgments
The authors thank Ms. Shuhong Yu and other laboratory members for assistance in experiment. We thank Wang Ling and Huang Shiwen at CNRRI for kindly providing the R. solani ZJ03 isolate.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1055277/full#supplementary-material
References
Anderson, N. A. (1982). The genetics and pathology of Rhizoctonia solani. Annu. Rev. Phytopathol. 20, 329–347. doi: 10.1146/annurev.py.20.090182.001553
Asano, T., Hayashi, N., Kobayashi, M., Aoki, N., Miyao, A., Mitsuhara, I., et al. (2012). A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 69, 26–36. doi: 10.1111/j.1365-313X.2011.04766.x
Banerjee, J., Maiti, M. K. (2010). Functional role of rice germin-like protein1 in regulation of plant height and disease resistance. Biochem. Biophys. Res. Commun. 394, 178–183. doi: 10.1016/j.bbrc.2010.02.142
Cao, W. L., Zhang, H. M., Zhou, Y., Zhao, J. H., Lu, S. B., Wang, X. Q., et al. (2022). Suppressing chlorophyll degradation by silencing OsNYC3 improves rice resistance to Rhizoctonia solani, the causal agent of sheath blight. Plant Biotechnol. J. 20, 335–349. doi: 10.1111/pbi.13715
Channamallikarjuna, V., Sonah, H., Prasad, M., Rao, G. J. N., Chand, S., Upreti, H. C., et al. (2010). Identification of major quantitative trait loci qSBR11-1 for sheath blight resistance in rice. Mol. Breed. 25, 155–166. doi: 10.1007/s11032-009-9316-5
Chen, X. J., Chen, Y., Xu, B., Zhang, L. N., Chen, Z. X. (2016). Overexpression of OsPGIP1 enhances rice resistance to sheath blight. Plant Dis. 100, 388–395. doi: 10.1094/PDIS-03-15-0305-RE
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological bata. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, X. Y., Wu, Y., Guo, J. P., Du, B., Chen, R. Z., Zhu, L. L., et al. (2013). A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 76, 687–698. doi: 10.1111/tpj.12328
Chen, X. W., Shang, J. J., Chen, D. X. (2006). A b-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46, 794–804. doi: 10.1111/j.1365-313X.2006.02739.x
Chen, Y., Zeng, Y. X., Ji, Z. J., Liang, Y., Wen, Z. H., Yang, C. D. (2019). Identification of stable quantitative trait loci for sheath blight resistance using recombinant inbred line. Rice Sci. 26, 331–338. doi: 10.1016/j.rsci.2019.08.007
Das, A., Moin, M., Sahu, A., Kshattry, M., Kirti, P. B., Barah, P. (2022). Time-course transcriptome analysis identifies rewiring patterns of transcriptional regulatory networks in rice under Rhizoctonia solani infection. Gene 828, 146468–146484. doi: 10.1016/j.gene.2022.146468
Deising, H. B., Werner, S., Wernitz, M. (2000). The role of fungal appressoria in plant infection. Microbes Infect. 2, 1631–1641. doi: 10.1016/s1286-4579(00)01319-8
Dodman, R. L., Barker, K. R., Walker, J. C. (1968). A detailed study of the different modes of penetration by Rhizoctonia solani. Phytopathology California, USA: Berkeley 58, 1271–1276.
Dodman, R. L., Flentje, N. T. (1970). The mechanism and physiology of plant penetration by rhisoctonia solani (Berkeley, California, USA: University California Press).
Gao, Y., Chong, Z., Xiao, H., Zi, Y. W., Lai, M., Yuan, D. P., et al. (2018). Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol. Plant Pathol. 19, 2149–2161. doi: 10.1111/mpp.12689
Ghosh, P., Sen, S., Chakraborty, J., Das, S. (2016). Monitoring the efficacy of mutated Allium sativum leaf lectin in transgenic rice against Rhizoctonia solani. BMC Biotechnol. 16, 24–33. doi: 10.1186/s12896-016-0246-0
Groth, D. E., Bond, J. A. (2007). Effects of cultivars and fungicides on rice sheath blight, yield, and quality. Plant Dis. 91, 1647–1650. doi: 10.1094/PDIS-91-12-1647
Harkenrider, M., Sharma, R., Vleesschauwer, D. D., Tsao, L., Zhang, X. T., Chern, M., et al. (2016). Overexpression of rice wall-associated kinase 25 (OsWAK25) alters resistance to bacterial and fungal pathogens. PLoS One 11, e0147310. doi: 10.1371/journal.pone.0147310
Huang, G. H., Kang, Z. S., Zhu, Z. Y., Li, Z. Q. (2003). Histopathological and ultrastructural studies on development of puccinia recondita f.sp. tritici in a susceptible wheat cultivar. Acta Phytopathol. Sin. 33, 52–62.
Hu, H. T., Wang, J., Shi, C., Yuan, C., Peng, C. F., Yin, J. J., et al. (2015). A receptor like kinase gene with expressional responsiveness on Xanthomonas oryzae pv. oryzae is essential for Xa21-mediated disease resistance. Rice 8, 1–9. doi: 10.1186/s12284-014-0034-1
Hu, B., Zhou, Y., Zhou, Z. H., Sun, B., Lin, Y. J. (2021). Repressed OsMESL expression triggers reactive oxygen species mediated broad: pectrum disease resistance in rice. Plant Biotechnol. J. 19, 1511–1522. doi: 10.1111/pbi.13566
Iwai, T., Miyasaka, A., Seo, S., Ohashi, Y. (2006). Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 142, 1202–1215. doi: 10.1104/pp.106.085258
Jain, M. (2012). Next-generation sequencing technologies for gene expression profiling in plants. Brief. Funct. Genomics 11, 63–70. doi: 10.1093/bfgp/elr038
John, L. J., Subramanian, B. (2019). Gene network mediated by WRKY13 to regulate resistance against sheath infecting fungi in rice (Oryza sativa l.). Plant Sci. 280, 269–282. doi: 10.1016/j.plantsci.2018.12.017
Karmakar, S., Molla, K. A., Chanda, P. K., Sarkar, S. N., Datta, S. K., Datta, K. (2016). Green tissue-specific co-expression of chitinase and oxalate oxidase 4 genes in rice for enhanced resistance against sheath blight. Planta 243, 115–130. doi: 10.1007/s00425-015-2398-x
Kawahara, Y., Oono, Y., Kanamori, H., Matsumoto, T., Itoh, T., Minami, E. (2012). Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS One 7, e49423. doi: 10.1371/journal.pone.0049423
Kim, P., Xue, C. Y., Song, H. D., Gao, Y., Feng, L., Li, Y., et al. (2021). Tissue-specific activation of DOF11 promotes rice resistance to sheath blight disease and increases grain weight via activation of SWEET14. Plant Biotechnol. J. 19, 409–411. doi: 10.1111/pbi.13489
Kump, K. L., Bradbury, P. J., Wisser, R. J., Buckler, E. S., Belcher, A. R., Oropeza-Rosas, M. A., et al. (2011). Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 43, 163–168. doi: 10.1038/ng.747
Kurusu, T., Hamada, J., Nokajima, H., Kitagawa, Y., Kiyoduka, M., Takahashi, A., et al. (2010). Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin b-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells. Plant Physiol. 153, 678–692. doi: 10.4161/psb.5.8.12407
Lai, Z., Mengiste, T. (2013). Genetic and cellular mechanisms regulating plant responses to necrotrophic pathogens. Curr. Opin. Plant Biol. 16, 505–512. doi: 10.1016/j.pbi.2013.06.014
Li, N., Kong, L. G., Zhou, W. H., Zhang, X., Wei, S. T., Ding, X. H., et al. (2013). Overexpression of Os2H16 enhances resistance to phytopathogens and tolerance to drought stress in rice. Plant Cell Tissue Organ Cult. 115, 429–441. doi: 10.1007/s11240-013-0374-3
Li, Z., Pinson, S. R. M., Marchetti, M. A., Stansel, J. W., Park, W. D. (1995). Characterization of quantitative trait loci (QTLs) in cultivated rice contributing to field resistance to sheath blight (Rhizoctonia solani). Theor. Appl. Genet. 91, 382–388. doi: 10.1007/BF00220903
Li, X., Shahid, M. Q., Xia, J., Lu, Z. J., Fang, N., Wang, L., et al. (2017). Analysis of small RNAs revealed differential expressions during pollen and embryo sac development in autotetraploid rice. BMC Genomics 18, 129–146. doi: 10.1186/s12864-017-3526-8
Liu, D. F., Chen, X. J., Liu, J. Q., Ye, J. C., Guo, Z. J. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63, 3899–3911. doi: 10.1093/jxb/ers079
Liu, G., Jia, Y., Correa-Victoria, F. J., Prado, G. A., Yeater, K. M., McClung, A., et al. (2009). Mapping quantitative trait loci responsible for resistance to sheath blight in rice. Phytopathology 99, 1078–1084. doi: 10.1094/PHYTO-99-9-1078
Livak, K. J., Schmittgen, T. D. (2013). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, R. F., Liu, Z. Y., Shao, Y. D., Su, J. C., Li, X. J., Sun, F., et al. (2020). Nitric oxide enhances rice resistance to rice black-streaked dwarf virus infection. Rice 13, 24–36. doi: 10.1186/s12284-020-00382-8
Manosalva, P. M., Bruce, M., Leach, J. E. (2011). Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance. Plant J. 68, 777–787. doi: 10.1111/j.1365-313X.2011.04728.x
Marshall, D. S. (1980a). Infection cushion formation on rice sheaths by Rhizoctonia solani. Phytopathology 70, 947–950. doi: 10.1094/Phyto-70-947
Marshall, D. S. (1980b). Relation between infection by Rhizoctonia solani and R. oryzae and disease severity in rice. Phytopathology 70, 941–946. doi: 10.1094/Phyto-70-941
Matsuura, K. (1986). Scanning electron microscopy of the infection process of Rhizoctonia solani in leaf sheaths of rice plants. Phytopathology 76, 811–814. doi: 10.1094/Phyto-76-811
Moore, J. W., Loake, G. J., Spoel, S. H. (2011). Transcription dynamics in plant immunity. Plant Cell. 23, 2809–2820. doi: 10.1105/tpc.111.087346
Okubara, P. A., Dickman, M. B., Blechl, A., E. (2014). Molecular and genetic aspects of controlling the soilborne necrotrophic pathogens Rhizoctonia and Pythium. Plant Sci. 228, 61–70. doi: 10.1016/j.plantsci.2014.02.001
Pandey, S. P., Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Pan, X. B., Zou, J. H., Chen, Z. X., Lu, J. F., Yu, H. X., Li, H. T., et al. (1999). Tagging major quantitative trait loci for sheath blight resistance in a rice variety, jasmine 85. Chin. Sci. Bull. 44, 1783–1789. doi: 10.1007/BF02886159
Park, D. S., Sayler, R. J., Hong, Y. G., Nam, M. H., Yang, Y. (2008). A method for inoculation and evaluation of rice sheath blight disease. Plant Dis. 92, 25–29. doi: 10.1094/PDIS-92-1-0025
Pinson, S. R. M., Capdevielle, F. M., Oard, J. H. (2005). Confirming QTLs and finding additional loci conditioning sheath blight resistance in rice using recombinant inbred lines. Crop Sci. 45, 503–510. doi: 10.2135/cropsci2005.0503
Polesani, M., Bortesi, L., Ferrarini, A., Zamboni, A., Fasoli, M., Zadra, C., et al. (2010). General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species. BMC Genomics 11, 117–132. doi: 10.1186/1471-2164-11-117
Qu, S. H., Liu, G. F., Zhou, B., Bellizzi, M., Zeng, L. R., Dai, L. Y., et al. (2006). The broad-apectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172, 1901–1914. doi: 10.1534/genetics.105.044891
Riopel, J. L., Timko, M. P. (1995). “Haustorial initiation and differentiation” in Parasitic Plants, Eds. Press, M. C., Graves, J. D. (Boundary Row, London, UK: Chapman and Hall), 39–76.
Samal, P., Molla, K. A., Bal, A., Ray, S., Swain, H., Khandual, A., et al. (2022). Comparative transcriptome profiling reveals the basis of differential sheath blight disease response in tolerant and susceptible rice genotypes. Protoplasma 259, 61–73. doi: 10.1007/s00709-021-01637-x
Sato, H., Ideta, O., Ando, I., Kunihiro, Y., Hirabayashi, H., Iwano, M., et al. (2004). Mapping QTLs for sheath blight resistance in the rice line WSS2. Breed. Sci. 54, 265–271. doi: 10.1270/jsbbs.54.265
Sharma, A., McClung, A. M., Pinson, S. R. M., Kepiro, J. L., Shank, A. R., Tabien, R. E., et al. (2009). Genetic mapping of sheath blight resistance QTLs within tropical japonica rice cultivars. Crop Sci. 49, 256–264. doi: 10.2135/cropsci2008.03.0124
Shimono, M., Koga, H., Akagi, A., Hayashi, N., Goto, S., Sawada, M., et al. (2012). Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 13, 83–94. doi: 10.1111/j.1364-3703.2011.00732.x
Shi, W., Zhao, S. L., Liu, K., Sun, Y. B., Zhu, G. Y. (2020). Comparison of leaf transcriptome in response to Rhizoctonia solani infection between resistant and susceptible rice cultivars. BMC Genomics 21, 245–260. doi: 10.1186/s12864-020-6645-6
Shu, C. W., Zhao, M., Anderson, J. P., Garg, G., Singh, K. B., Zheng, W. B., et al. (2019). Transcriptome analysis reveals molecular mechanisms of sclerotial development in the rice sheath blight pathogen Rhizoctonia solani AG1-IA. Funct. Integr. Genomics 19, 743–758. doi: 10.1007/s10142-019-00677-0
Singh, A., Rohilla, R., Savary, S., Willocquet, L., Singh, U. S. (2003). Infection process in sheath blight of rice caused by Rhizoctonia solani. Indian Phytopathol. 56, 434–438. doi: 10.1016/S0021-8502(99)80406-6
Słomnicka, R., Olczak-Woltman, H., Sobczak, M., Bartoszewski, G. (2021). Transcriptome profiling of cucumber (Cucumis sativus l.) early response to Pseudomonas syringae pv. lachrymans. Int. J. Mol. Sci. 22, 4192–4209. doi: 10.3390/ijms22084192
Sun, Q., Li, D. D., Chu, J., Yuan, D. P., Li, S., Zhong, L. J., et al. (2020). Indeterminate domain proteins regulate rice defense to sheath blight disease. Rice 13, 15–26. doi: 10.1186/s12284-020-0371-1
Sun, Q., Li, T. Y., Li, D. D., Wang, Z. Y., Li, S., Li, D. P., et al. (2019). Overexpression of loose plant architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN-FORMED 1a in rice. Plant Biotechnol. J. 17, 855–857. doi: 10.1111/pbi.13072
Tang, C. Q. (2011). “Cytological observations of infection on rice leaves of rhizoctonia solani AG1-IA,” in Master's thesis (Sichuan: Sichuan Agriculture University).
Tao, J. F. (1992). Studies on the infection process of Rhizoctonia solani in rice. J. Sichuan Agric. Univ. 10, 471–477.
Tonnessen, B. W., Manosalva, P., Lang, J. M., Baraoidan, M., Bordeos, A., Mauleon, R., et al. (2015). Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 87, 273–286. doi: 10.1007/s11103-014-0275-9
Vijay, K., Reddy, M. S., Kloepper, J. W., Lawrence, K. S., Yellareddygari, S. K., Zhou, X. G., et al. (2011). Screening and selection of elite plant growth promoting rhizobacteria (PGPR) for suppression of Rhizoctonia solani and enhancement of rice seedling vigor. J. Pure Appl. Microbiol. 5, 641–651.
Wang, Z., Gerstein, M., Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wang, H. H., Hao, J. J., Chen, X. J., Hao, Z. N., Wang, X., Lou, Y. G., et al. (2007). Overexpression of rice WRKY89 enhances ultraviolet b tolerance and disease resistance in rice plants. Plant Mol. Biol. 65, 799–815. doi: 10.1007/s11103-007-9244-x
Wang, R., Lu, L. X., Pan, X. B., Hu, Z. L., Ling, F., Yan, Y., et al. (2015b). Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol. Biol. 87, 181–191. doi: 10.1007/s11103-014-0269-7
Wang, H. H., Meng, J., Peng, X. X., Tang, X. K., Zhou, P. L., Xiang, J. H., et al. (2015a). Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 89, 157–171. doi: 10.1007/s11103-015-0360-8
Wang, A. J., Shu, X. Y., Jing, X., Jiao, C. Z., Zheng, A. P. (2021). Identification of rice (Oryza sativa l.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnol. J. 19, 1553–1566. doi: 10.1111/pbi.13569
Windram, O. P., Madhou, P., Mchattie, S., Hill, C., Hickman, R. J., Cooke, E. J., et al. (2012). Arabidopsis defense against Botrytis cinerea: Chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Am. Soc Plant Biol. 24, 3530–3557. doi: 10.2307/41692823
Wu, J. W., Shahid, M. Q., Guo, H. B., Yin, W., Chen, Z. X., Wang, L., et al. (2014). Comparative cytological and transcriptomic analysis of pollen development in autotetraploid and diploid rice. Plant Reprod. 27, 181–196. doi: 10.1007/s00497-014-0250-2
Xue, X., Cao, Z. X., Zhang, X. T., Wang, Y., Zhang, Y. F., Chen, Z. X., et al. (2016). Overexpression of OsOSM1 enhances resistance to rice sheath blight. Plant Dis. 100, 1634–1642. doi: 10.1094/PDIS-11-15-1372-RE
Xu, Q., Yuan, X. P., Yu, H. Y., Wang, Y. P., Tang, S. X., Wei, X. H. (2011). Mapping quantitative trait loci for sheath blight resistance in rice using double haploid population. Plant Breed. 130, 404–406. doi: 10.1111/j.1439-0523.2010.01806.x
Yadav, S., Anuradha, G., Kumar, R. R., Vemireddy, L. R., Sudhakar, R., Donempudi, K., et al. (2015). Identification of QTLs and possible candidate genes conferring sheath blight resistance in rice (Oryza sativa l.). Springer plus. 4, 175–186. doi: 10.1186/s40064-015-0954-2
Yang, X. H., Gu, X., Ding, J. J., Yao, L. L., Gao, X. D., Zhang, M. M., et al. (2022). Gene expression analysis of resistant and susceptible rice cultivars to sheath blight after inoculation with Rhizoctonia solani. BMC Genomics 23, 278–293. doi: 10.1186/s12864-022-08524-6
Yin, X., Zou, B. H., Hong, X. X., Gao, M. J., Yang, W. B., Zhong, X. B., et al. (2018). Rice copine genes OsBON1 and OsBON3 function as suppressors of broad-spectrum disease resistance. Plant Biotechnol. J. 16, 1476–1487. doi: 10.1111/pbi.12890
Yuan, D. P., Xu, X. F., Hong, W. J., Wang, S. T., Jia, X. T., Liu, Y., et al. (2020). Transcriptome analysis of rice leaves in response to Rhizoctonia solani infection and reveals a novel regulatory mechanism. Plant Biotechnol. Rep. 14, 559–573. doi: 10.1007/s11816-020-00630-9
Yuan, Z. J., Zhang, Y., Xu, G. J., Bi, D. L., Qu, H. Y., Zou, X. W., et al. (2018). Comparative transcriptome analysis of Rhizoctonia solani-resistant and -susceptible rice cultivars reveals the importance of pathogen recognition and active immune responses in host resistance. J. Plant Biol. 61, 143–158. doi: 10.1007/s12374-017-0209-6
Yu, H., Shahid, M. Q., Li, R. B., Li, W., Liu, X. D. (2018). Genome-wide analysis of genetic variations and the detection of rich variants of NBS-LRR encoding genes in common wild rice lines. Plant Mol. Biol. Rep. 36, 618–630. doi: 10.1007/s11105-018-1103-1
Zeng, Y. X., Dong, J. J., Ji, Z. J., Yang, C. D., Liang, Y. (2021). A linear regression model for the prediction of rice sheath blight field resistance. Plant Dis. 105, 2964–2969. doi: 10.1094/PDIS-08-20-1681-RE
Zeng, Y. X., Hu, C. Y., Lu, Y. G., Li, J. Q., Liu, X. D. (2007). Diversity of abnormal embryo sacs in indica/japonica hybrids in rice demonstrated by confocal microscopy of ovaries. Plant Breed. 126, 574–580. doi: 10.1111/j.1439-0523.2007.01380.x
Zeng, Y. X., Ji, Z. J., Ma, L. Y., Li, X. M., Yang, C. D. (2011). Advances in mapping loci conferring resistance to rice sheath blight and mining Rhizoctonia solani resistant resources. Rice Sci. 18, 56–66. doi: 10.1016/S1672-6308(11)60008-5
Zeng, Y. X., Shi, J. S., Ji, Z. J., Wen, Z. H., Liang, Y., Yang, C. D. (2017). Genotype by environment interaction: The greatest obstacle in precise determination of rice sheath blight resistance in the field. Plant Dis. 101, 1795–1801. doi: 10.1094/PDIS-03-17-0435-RE
Zhang, J. F., Chen, L., Fu, C. L., Wang, L. X., Liu, H. N., Cheng, Y. Z., et al. (2017). Comparative transcriptome analyses of gene expression changes triggered by Rhizoctonia solani AG1 IA infection in resistant and susceptible rice varieties. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01422
Zhang, H. H., Feng, J. H., Lu, Y. G., Yang, B. Y., Liu, X. D. (2003). Observation on formation and development of autotetraploid rice embryo sac using laser scanning confocal microscope. J. Chin. Electron Microsc. Soc 22, 380–384. doi: 10.1023/A:1022289509702
Zhang, Z. L., Shin, M., Zou, X. L., Huang, J. Z., Ho, T. H. D., Shen, Q. J. (2009). A negative regulator encoded by a rice WRKY gene represses both abscisic acid and gibberellins signaling in aleurone cells. Plant Mol. Biol. 70, 139–151. doi: 10.1007/s11103-009-9463-4
Zhang, G. L., Yan, D. W., He, Z. H. (2010). Cytological characteristics of infection process by Rhizoctonia solani in rice. Chin. J. Cell Biol. 32, 451–455. doi: 10.1038/cr.2010.156
Zhu, C., Lin, A., Wang, L., Yin, P. P., Liu, C. L., Li, S. S., et al. (2016). De novo transcriptome analysis of Rhizoctonia solani AG1 IA strain early invasion in Zoysia japonica root. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00708
Zou, J. H., Pan, X. B., Chen, Z. X., Xu, J. Y., Lu, J. F., Zhai, W. X., et al. (2000). Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa l.). Theor. Appl. Genet. 101, 569–573. doi: 10.1007/s001220051517
Keywords: rice, Rhizoctonia solani, sheath blight, RNA-seq, confocal microscopy
Citation: Liu S, Wang T, Meng G, Liu J, Lu D, Liu X and Zeng Y (2022) Cytological observation and transcriptome analysis reveal dynamic changes of Rhizoctonia solani colonization on leaf sheath and different genes recruited between the resistant and susceptible genotypes in rice. Front. Plant Sci. 13:1055277. doi: 10.3389/fpls.2022.1055277
Received: 27 September 2022; Accepted: 11 October 2022;
Published: 03 November 2022.
Edited by:
Quan Xu, Shenyang Agricultural University, ChinaReviewed by:
Dezhi Wu, Hunan Agricultural University, ChinaMugui Wang, Center for Excellence in Molecular Plant Sciences (CAS), China
Yongxiang Huang, Guangdong Ocean University, China
Copyright © 2022 Liu, Wang, Meng, Liu, Lu, Liu and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangdong Liu, eGRsaXVAc2NhdS5lZHUuY24=; Yuxiang Zeng, enl4eWVzMUBzaW5hLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Sanglin Liu
Sanglin Liu Tianya Wang1,2,3†
Tianya Wang1,2,3† Xiangdong Liu
Xiangdong Liu Yuxiang Zeng
Yuxiang Zeng