
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 January 2023
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1044144
 Qiang Yang1†
Qiang Yang1† Yasir Sharif1†‡
Yasir Sharif1†‡ Yuhui Zhuang1
Yuhui Zhuang1 Hua Chen1
Hua Chen1 Chong Zhang1
Chong Zhang1 Huiwen Fu2
Huiwen Fu2 Shanshan Wang2
Shanshan Wang2 Tiecheng Cai1
Tiecheng Cai1 Kun Chen2
Kun Chen2 Ali Raza1‡
Ali Raza1‡ Lihui Wang2
Lihui Wang2 Weijian Zhuang1,2*‡
Weijian Zhuang1,2*‡Peanut is an important food and feed crop, providing oil and protein nutrients. Germins and germin-like proteins (GLPs) are ubiquitously present in plants playing numerous roles in defense, growth and development, and different signaling pathways. However, the GLP members have not been comprehensively studied in peanut at the genome-wide scale. We carried out a genome-wide identification of the GLP genes in peanut genome. GLP members were identified comprehensively, and gene structure, genomic positions, motifs/domains distribution patterns, and phylogenetic history were studied in detail. Promoter Cis-elements, gene duplication, collinearity, miRNAs, protein-protein interactions, and expression were determined. A total of 84 GLPs (AhGLPs ) were found in the genome of cultivated peanut. These GLP genes were clustered into six groups. Segmental duplication events played a key role in the evolution of AhGLPs, and purifying selection pressure was underlying the duplication process. Most AhGLPs possessed a well-maintained gene structure and motif organization within the same group. The promoter regions of AhGLPs contained several key cis-elements responsive to ‘phytohormones’, ‘growth and development’, defense, and ‘light induction’. Seven microRNAs (miRNAs) from six families were found targeting 25 AhGLPs. Gene Ontology (GO) enrichment analysis showed that AhGLPs are highly enriched in nutrient reservoir activity, aleurone grain, external encapsulating structure, multicellular organismal reproductive process, and response to acid chemicals, indicating their important biological roles. AhGLP14, AhGLP38, AhGLP54, and AhGLP76 were expressed in most tissues, while AhGLP26, AhGLP29, and AhGLP62 showed abundant expression in the pericarp. AhGLP7, AhGLP20, and AhGLP21, etc., showed specifically high expression in embryo, while AhGLP12, AhGLP18, AhGLP40, AhGLP78, and AhGLP82 were highly expressed under different hormones, water, and temperature stress. The qRT-PCR results were in accordance with the transcriptome expression data. In short, these findings provided a foundation for future functional investigations on the AhGLPs for peanut breeding programs.
Germin-like proteins (GLPs) are a group of pervasive water-soluble glycoproteins found in monocots, dicots, and gymnosperms (Bernier and Berna, 2001). GLP genes are widely distributed in the plant kingdom, and most of the genes are present in the form of multiple copies in plant genomes (Manosalva et al., 2009). Germins and germin-like proteins (GLPs) were initially discovered as germination-specific markers in germinating wheat seedlings (Lane et al., 1993). GLPs belong to the “Cupin superfamily” possessing β-sheet barrel (jellyroll beta-barrel structural domain) cupin domain (PF00190) with metal ion binding site at their C-terminus (Agarwal et al., 2009), and generally, these genes are encoded by two exons. Cupin superfamily is a functionally diverse family (Dunwell et al., 2004) with seed storage-related functions, such as vicilins (Gane et al., 1998). Members of this protein family contain two conserved motifs known as “germin box” (Lane et al., 1991; Yamahara et al., 1999). Both motifs of Germin-like proteins “G(x)5HxH(x)3,4E(x)6G” and “G(x)5PxG(x)2H(x)3N” are packed in a classic jellyroll beta-barrel structural domain (Woo et al., 2000). Crystallographic studies of barley germin proteins elaborated that six germin proteins form a stable hexamer structure, with each protein binding to a manganese ion (Woo et al., 2000). The monomer subunits of hexamer structure form “trimers of dimers,” in which ligands bound with the manganese ions similarly to that of manganese superoxide dismutase (Carter and Thornburg, 2000). Each monomer also contains an irregular extension at the N-terminus, and the domain shape of the N-terminus is generally conserved in many GLPs. Hexamer structure is composed of almost 1200 amino acid residues with a molecular weight of approximately 13 kDa (Lane, 2002), but on the contrary, a single copy of GLP has been discovered in rice which is capable of SOD activity even in its dimeric form (Banerjee and Maiti, 2010). Classification of germins and GLPs is challenging due to their structural and sequence similarities (Agarwal et al., 2009). Generally, the “true germins” are a group of well-conserved homogenous proteins uniquely found in cereals (Bernier and Berna, 2001; Davidson et al., 2009), while the germin-like proteins are heterogeneous proteins with wider distribution in the plant kingdom (Woo et al., 2000; Dunwell et al., 2008).
Germins and GLPs play a wide range of functions in plants, and mostly, their functions are associated with enzymatic reactions, stress responses, and cell wall synthesis (Dunwell et al., 2000; Sun et al., 2020). Members of this gene family are highly expressed under various biotic stresses, including bacterial pathogens responses such as bacterial rust resistance in peanut (Wang et al., 2013), fungal pathogens including powdery mildew (Erysiphe necator infection) responses in grapevine (Godfrey et al., 2007), Aspergillus flavus response in peanut (Knecht et al., 2010; Rietz et al., 2012), and viral pathogens responses in tobacco (Guevara-Olvera et al., 2012). GLPs have been involved in defense responses against fungal pathogens in cereals (Zimmermann et al., 2006). Transgenic tobacco and Arabidopsis plants overexpressing soybean and sunflower GLPs showed improved resistance to Sclerotinia sclerotiorum (Beracochea et al., 2015; Zhang et al., 2018). Transient overexpression of barley and wheat GLP4 (HvGLP4 and TaGLP4) increased resistance to B. graminis in transgenic Arabidopsis plants (Christensen et al., 2004). A newly discovered GLP member from upland cotton (Gossypium hirsutum) GhABP19 showed increased resistance against fungal pathogens, Fusarium oxysporum and Verticillium dahliae in transgenic Arabidopsis plants (Pei et al., 2019). GLPs are also highly expressed under abiotic stresses, including drought stress, salt stress, wound stress (Wang et al., 2013), high-temperature stress (Gangadhar et al., 2021), and heavy metals stress (Houde and Diallo, 2008). Recent studies have shown that OsGLP1 from rice helps plant acclimatize to UV radiations (He et al., 2021). GLPs also have regulatory effects in certain growth or development-related pathways, as it has been reported that a member of the GLP family in Gossypium barbadense (GbGLP2) is involved in secondary cell wall growth and ultimately controls fiber length (Sun et al., 2020).
Although some GLPs from various crop species have been studied in detail, the functions of most GLPs are unknown and uncharacterized. Mostly plant GLPs with unknown functions have been classified into various subfamilies (Carter and Thornburg, 1999; Carter and Thornburg, 2000). The true germins subfamily (cereals) contains the proteins with oxalate oxidase activity, while the members of subfamilies 1 and 2 are attributed with superoxide dismutase activity (SOD). Subfamily 3 includes the proteins with phosphodiesterase activity (3.1.4.1), while some studies have reported more subdivisions (Nakata et al., 2004). As in the case of barley, five subfamilies (HvGER1-HvGER5) have been reported (Lu et al., 2010).
Keeping in view the above reports, it is a fact that germins and GLPs are an important group of plant proteins playing a broad-spectrum role in plant growth regulation and defense responses. Despite the fact that GLPs have been comprehensively studied in some important plant species, including soybean (Lu et al., 2010), Arabidopsis (Rietz et al., 2012), rice (Li et al., 2016), and wheat (Yuan et al., 2021) and some of their members have been functionally characterized. Still, this gene family was yet to be comprehensively described in the peanut genome. Peanut (Arachis hypogea L.) is a popular food crop, and more than one hundred countries share its cultivation. It is an important source of vegetable oil, proteins, minerals, and dietary fibers (Toomer, 2018). Several biotic and abiotic stress agents affect the peanut plant throughout its lifecycle (Ali et al., 2020). Identification and application of novel genetic, genomics and gene editing resources can help to improve peanut growth and stress resistance (Yaqoob et al., 2023).
Peanut is now a resource-rich legume crop with massive transcriptome data (Clevenger et al., 2016; Sinha et al., 2020; Gangurde et al., 2021). Trait dissection and candidate gene discovery with linkage and association mapping attempted for several traits, for instance, yield traits (Gangurde et al., 2020; Pandey et al., 2020; Jadhav et al., 2021), fresh seed dormancy (Gangurde et al., 2020; Kumar et al., 2020; Bomireddy et al., 2022), disease resistance (Dodia et al., 2019; Shasidhar et al., 2020), and aflatoxin contamination (Pandey et al., 2019; Khan et al., 2020; Soni et al., 2020). The utilization of the available resources with new data is essential. In the present study, we used publicly available transcriptome datasets to investigate the expression levels of GLPs. The members of the GLP family in the genomes of cultivated peanut and its wild parents (Arachis duranensis and Arachis ipaensis) were identified by available genome annotations. The genomic distribution, conserved motifs organization, cis-regulatory elements, phylogenetic relationships, protein physicochemical properties, functional annotation (GO enrichment), prediction of miRNAs targeting the AhGLPs, and expression patterns in different tissues and under stress conditions were studied in detail. Expression patterns of selective genes were studied under different abiotic stresses by qRT-PCR analysis. This study will help to understand the role of GLPs in peanut against various biotic and abiotic stresses, growth and development and provide basic information for future studies.
Germin-like proteins were identified in a systematic way in peanut. First of all, Arabidopsis GLPs (Arabidopsis thaliana TAIR10-Thales cress) protein sequences were obtained from the Phytozome server (https://phytozome-next.jgi.doe.gov/) (Goodstein et al., 2012). The GLPs in diploid progenitors (A. duranensis and A. ipaensis) were searched from the PeanutBase database (https://www.peanutbase.org/home) (Bertioli et al., 2016) by BLASTp search by using AtGLPs protein sequences as queries. GLPs in diploid progenitors were also searched from the Legume Information System (https://legumeinfo.org/), using the keyword search “germin” and “PF00190” (Pfam accession number of Germin-like proteins). The protein sequences of identified GLPs from A. duranensis, A. ipaensis, and A. thaliana were used to search the GLPs in cultivated peanut from the Peanut Genome Resource (PGR) database (http://peanutgr.fafu.edu.cn/) (Zhuang et al., 2019). Cultivated peanut GLPs (AhGLPs) were also searched by keyword search “germin” in the PGR database. Finally, all identified proteins were checked for the presence of germin domain by Batch CD-search at NCBI (https://www.ncbi.nlm.nih.gov/). Duplicated IDs and different splicing variants of the same gene were removed to get the unique IDs.
The information about the chromosomal distribution of AhGLPs was obtained from the PGR database (http://peanutgr.fafu.edu.cn/). TBtools software (Chen et al., 2020) was used to map out the genes on chromosomes. The evolutionary relationships of AhGLPs with their homologs in A. duranensis (AdGLPs), A. ipaensis (AiGLPs), and model dicot plant A. thaliana (AtGLPs) were assessed by evolutionary phylogenetic analysis. Multiple sequence alignment analysis was performed by the ClustalW algorithm (Thompson et al., 2003), and a Neighbor-Joining phylogenetic tree (Saitou et al., 1987) with 1000 bootstrap replications was constructed in MEGA-X software (Kumar et al., 2018) with the Poisson model. The phylogenetic tree was beautified via the online program iTOL v6 (Letunic and Bork, 2021), available at (https://itol.embl.de/).
The gene structure (exon-intron distribution) analysis for AhGLPs was performed with the help of TBtools software by deploying the General Feature File (GFF3) downloaded from the PGR database (Zhuang et al., 2019). Conserved motifs of GLP genes were identified by MEME Suite (https://meme-suite.org/meme/) (Bailey et al., 2015) by employing the protein sequences. Parameters for motifs identification were set as; 6-200 optimum width range, the maximum number of Motifs: 10.
Subcellular localization of AhGLPs in different cell compartments was predicted by CELLO v2.5 (http://cello.life.nctu.edu.tw/) (Yu et al., 2006), and other physiochemical properties, including molecular weight (MW), theoretical Isoelectric point (PI), were predicted by Expasy server (https://web.expasy.org/protparam/) (Gasteiger et al., 2003; Gasteiger et al., 2005).
For the analysis of cis-regulatory elements of AhGLPs, the promoter sequences (2kb upstream of the start codon) were scanned at the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). Promoter sequences were accessed from the PGR database. Cis-elements were divided into four main categories based on their functions. These categories include light-responsive elements, hormones-responsive elements, growth and development-related elements, and stress-responsive elements.
The evolutionary genome conservations between three peanut species and Arabidopsis were analyzed by performing a comparative synteny analysis. The genome and GFF3 files of all these species were scanned for McScanX at TBtools software, and the resulting files were used for multiple synteny plots. To study the gene duplication in cultivated peanut, duplicated genes were identified by their phylogenetic relations and running an MCScanX for the whole peanut genome. The synonymous (Ks) and non-synonymous (Ka) substitution rates (Ka: No. of non-synonymous substitutions per non-synonymous site, Ks: No. of synonymous substitutions per synonymous site) were calculated by simple Ka/Ks calculator at TBtools software. Divergence time for duplicated gene pairs was calculated as ‘t=Ks/2r’ with the neutral substitution rate of r=8.12×10-9 (Bertioli et al., 2016). The orthologous GLP proteins in A. hypogea, A. duranensis, A. ipaensis, and A. thaliana were identified through orthovenn2 (https://orthovenn2.bioinfotoolkits.net/home) (Xu et al., 2019). Protein sequences of Arabidopsis and three peanut species were used to identify orthologous genes. The peanut species were assessed individually with each other and with Arabidopsis to identify orthologous gene clusters.
The coding sequences of AhGLPs were used to predict the putative miRNAs targeting the AhGLPs through the psRNATarget database (https://www.zhaolab.org/psRNATarget/) (Dai et al., 2018) following the default settings. The schematic diagram showing the interaction networks between miRNAs and AhGLPs was drawn by Cytoscape software version 3.8.2 (https://cytoscape.org/) (Shannon et al., 2003).
We performed the gene ontology (GO) analysis of AhGLPs to predict their functional annotation. For that purpose, AhGLP proteins were scanned at the EggNOG database (http://eggnog-mapper.embl.de/). GO enrichment analysis was performed in TBtools software from predicted GO annotations.
Protein-protein interactions were predicted based on studied Arabidopsis GLPs. STRING 11.5 tool (https://www.string-db.org/cgi/) was used to construct the interaction network between peanut and Arabidopsis GLPs. The top 10 interactions were predicted with a medium threshold level (0.4). MCL clustering with inflation parameter 10 was used, and dotted lines were used between cluster edges.
Transcriptome expression data were used to view the expression patterns of AhGLPs in different tissues, under phytohormones, water, and temperature treatments. Transcriptome expression data for different tissues (leaf, stem, stem tip, Inflorescence, root, root and stem, root tip, root nodule, gynophore/peg, pericarp, testa, cotyledons, and embryo), hormones (ABA, SA, Brassinolide, paclobutrazol, ethephon, and ddH2O), water (drought and normal irrigation) and temperature treatments (low temperature and room temperature) were accessed from the Peanut Genome Resource database (Zhuang et al., 2019). The tissue samples were taken at different growth stages, and their RNA was mixed for transcriptome analysis. For hormone treatment, samples were collected at different time points (3h, 6h, 12h, 24h, and 48h). For drought stress, peanut plants at the flowering stage (eight leaves) were exposed to stress by withholding the water, and samples were taken after 3d, 6d, 9d, and 12d after drought treatments. For temperature stress, young plants (four leaves) were kept at 4°C, and samples were taken at 3h, 6h, 12h, 24h, and 48h. RNA samples of different time points were mixed for transcriptome analysis. The log2 normalization Fragments Per Kilobase Million (FPKM) of AhGLPs were used for expression analysis, and log2 normalization values were used to construct the expression heatmaps.
For stress treatments, a widely cultivated peanut variety, Minhua-6 “M-6” seeds were grown in small plastic pots. At four-leaf stage, seedlings were treated with ABA (10 µg/mL) and low temperature (4°C). Samples were collected at 3, 6, 9, and 12 hours after stress treatments, while non-treated leaf samples (0 h) were taken as control (CK). RNA was extracted with the CTAB method with some modifications (Sharif et al., 2022) and cDNA was synthesized with the help of Evo M-MLV RT Kit (Hunan Aikerui Biological Engineering Co., Ltd. China) according to manufacturer guidelines. Peanut Actin gene was used as internal control, while qRT-PCR was performed as per our previous study (Sharif et al., 2022). Relative expression levels of selected genes were calculated by 2-ΔΔCT method (Livak and Schmittgen, 2001). Primers used for qRT-PCR are given in Supplementary Table 9. The graphs were drawn with GraphPad Prism 7.0 (Swift, 1997). The qRT-PCR results for the expression of selected genes under ABA and low temperature at different time points were subjected to analysis of variance (ANOVA) and Tuckey’s HSD test, to find the significant expression differences.
Thirty-seven GPLs in A. duranensis (AdGLPs) (V14167) (Supplementary Table 1) and 32 GLPs in A. ipaensis (AiGLPs) (K30076) (Supplementary Table 2) were identified as containing the germin domain PF00190. Then, 84 GLPs were found in A. hypogaea (AhGLPs) with the help of AtGLPs, AdGLPs, and AiGLPs. Previously, no systematic way to rename GLPs was available, and GLPs are mostly renamed according to their chromosomal/genomic locations. We renamed genes based on the genomic positions and chromosomal locations. Renaming based on genomic position as AhGLP1-AhGLP84 was further used for analysis. Similarly, 37 GLPs of A. duranensis were renamed as AdGLP1-AdGLP37, and 32 GLPs of A. ipaensis were renamed as AiGLP1-AiGLP32.
Germin-like proteins were unevenly distributed in the genome of cultivated peanut A. hypogea. The largest number of AhGLPs were present on chromosome Chr06 (16 out of 84), followed by 12 GLPs on the Chr16; 7 GLPs on Chr9 and Chr19 each; 5 GLPs on Chr01, Chr08, Chr13, and Chr18 each; 4 GLPs on Chr02, Chr03, Chr12 each; 2 GLPs on Chr15, Chr20 each. Chr04, Chr05, Chr10, Chr11, Chr14 possessed one GLP each. The chromosomal distribution patterns of AhGLPs are shown in Figure 1. One of the AhGLPs was present in the unassembled genome region. The genomic length of AhGLPs varied from 522 bp for AhGLP10 to 99291 bp for AhGLP29. The CDS length, protein length, and molecular weight of AhGLPs ranged from 522-2145 bp, 173-714 amino acids, and 18.74 to 81.40 KDa, respectively. Their theoretical isoelectric points varied from 5.01 (AhGLP73) to 9.42 (AhGLP29). Subcellular localization prediction of AhGLPs showed their distribution in different cell compartments, including the plasma membrane, nucleus, cytoplasm, chloroplast, mitochondria, and extracellular spaces. A large number of proteins are localized in more than one cell organelles. Detailed information on the physicochemical properties of AhGLPs is given in Table 1. The genomic distribution of GLPs in the diploid peanut species (A. duranensis and A. ipaensis) is shown in Supplementary Figures 1, 2. The genomic, CDS, protein lengths, and other physiochemical properties of GLPs in A. duranensis and A. ipaensis are given in Supplementary Tables 1, 2. Protein sequences of AhGLPs, AdGLPs, AiGLPs, and AtGLPs are given in Supplementary File 1.
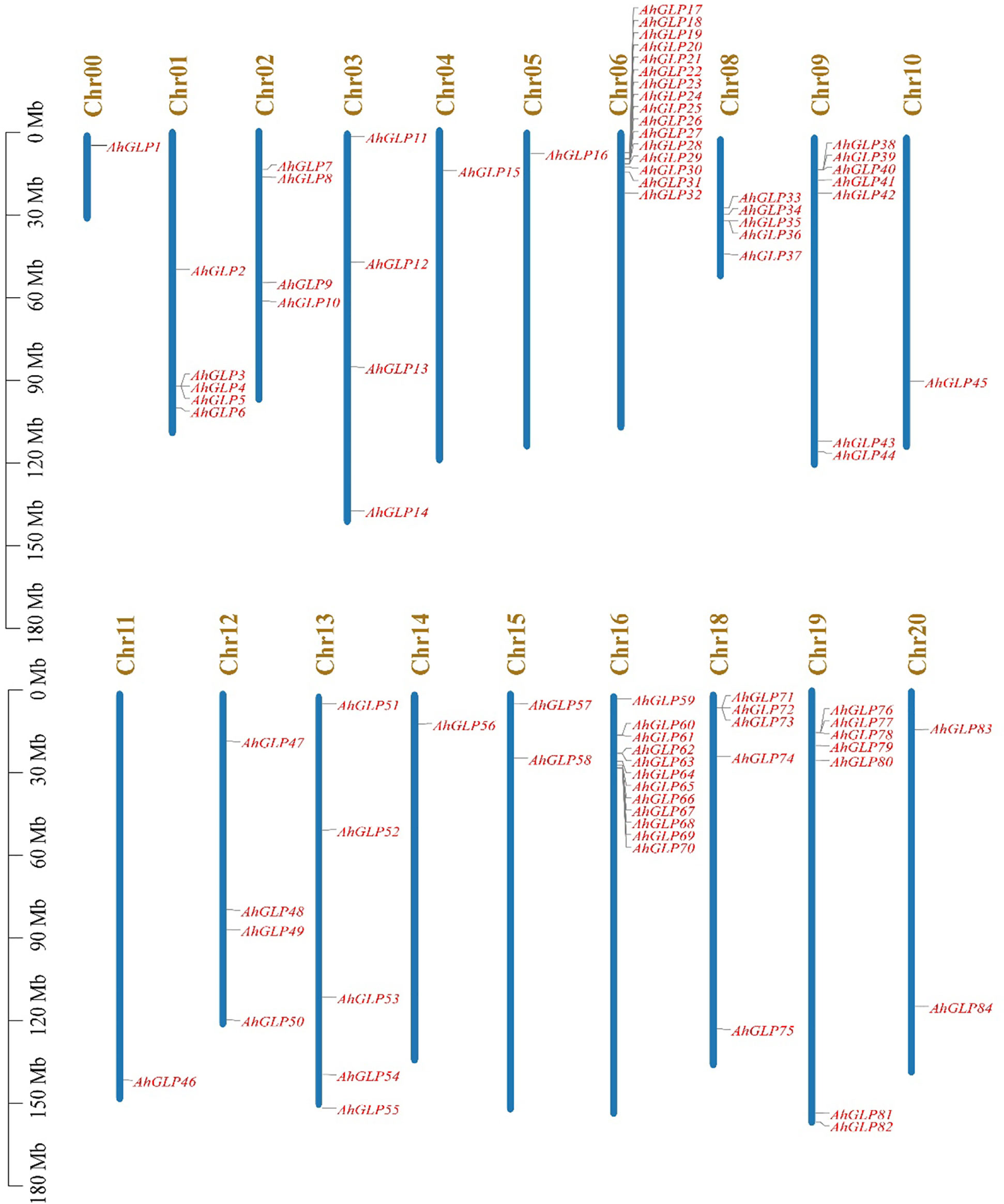
Figure 1 Chromosomal distribution of Arachis hypogaea Germin-like protein (AhGLPs) genes. The scale on left side indicates the chromosome length.
Several GLPs of cultivated peanut were composed of a single exon (29 out of 84), while the maximum number of exons was 10 (AhGLP64). Gene length variations of AhGLPs were in accordance with its diploid parents, as the smallest GLPs in diploid progenitors were AdGLP3 (552 bp), AdGLP34 (552 bp) (A. duranensis), and AiGLP32 (459 bp), AiGLP28 (552 bp) (A. ipaensis), similarly the smallest GLPs in cultivated peanut were AhGLP3, AhGLP10, AhGLP35, and AhGLP71 with the genomic length of 552 bp. But few genes in cultivated peanut possessed extraordinary long genomic sequences, for example, AhGLP62 with a genomic length of 71124 bp and AhGLP29 with a genomic length of 99291 bp (Figure 2). Scanning of protein sequences at the MEME server identified many common and unique motifs. Commonly shared motifs among genes tend to cluster in the same groups, referring to their similar functions. The length of motifs was also different, i.e., the maximum motif length was 50 amino acid residues (5th and 6th motifs), and the minimum motif length was 21 amino acid residues for the 2nd, 3rd, 7th, and 10th motifs (Supplementary Table 3). Conserved motifs distribution patterns of AhGLPs are shown in Figure 3A, and cupin domain locations are shown in Figure 3B. The gene structure and motif distribution patterns of AhGLPs are in agreement with A. duranensis and A. ipaensis GPLs (Supplementary Figures 3, 4, Supplementary Tables 4, 5).
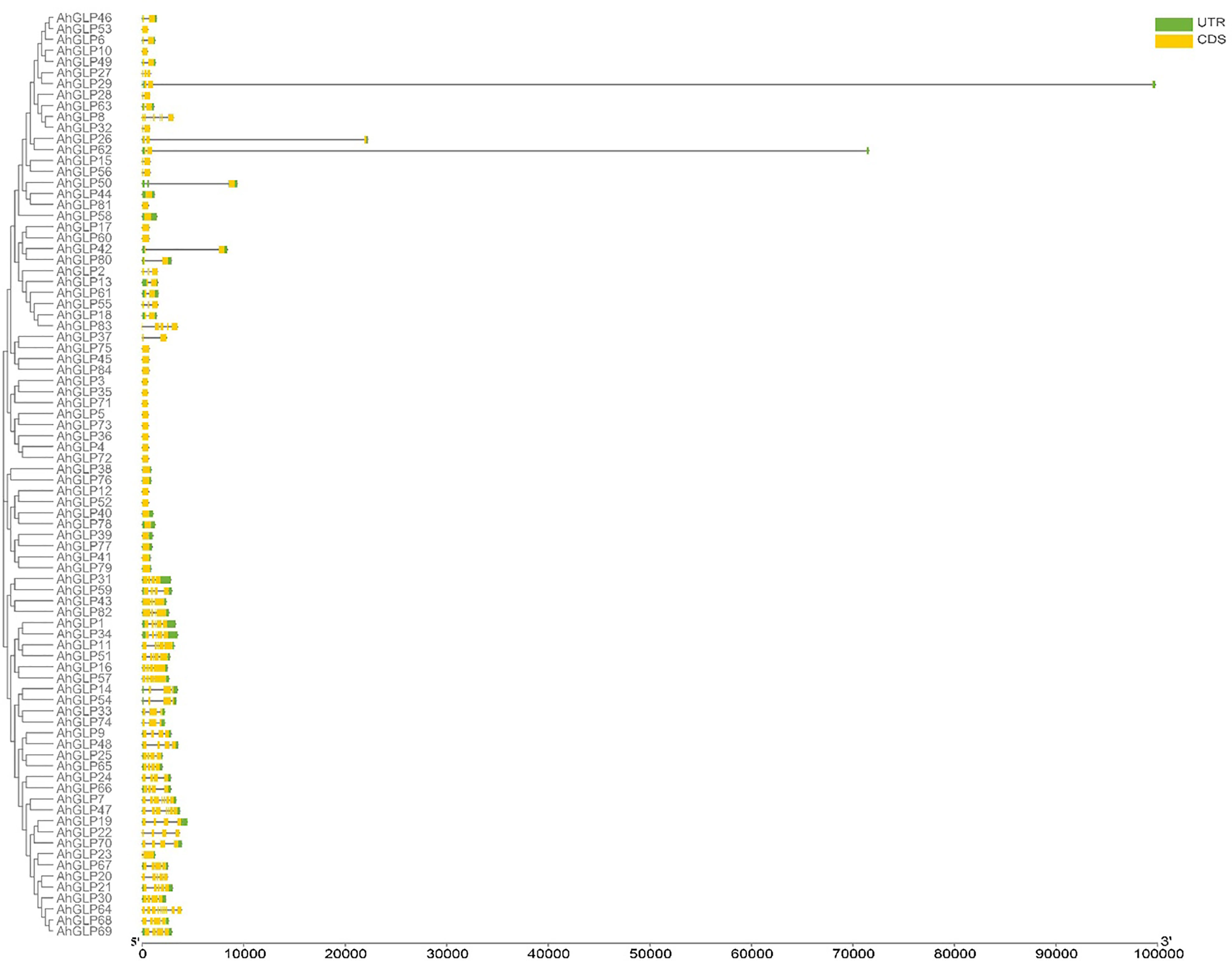
Figure 2 Gene structure (exons-introns distribution) of Arachis hypogaea Germin-like protein (AhGLPs) genes. Most AhGLPs are composed of a single exon, while the maximum number of exons is 10 (AhGLP64). AhAGLP26, AhGLP29, and AhGLP62 possessed very large intron sequences.
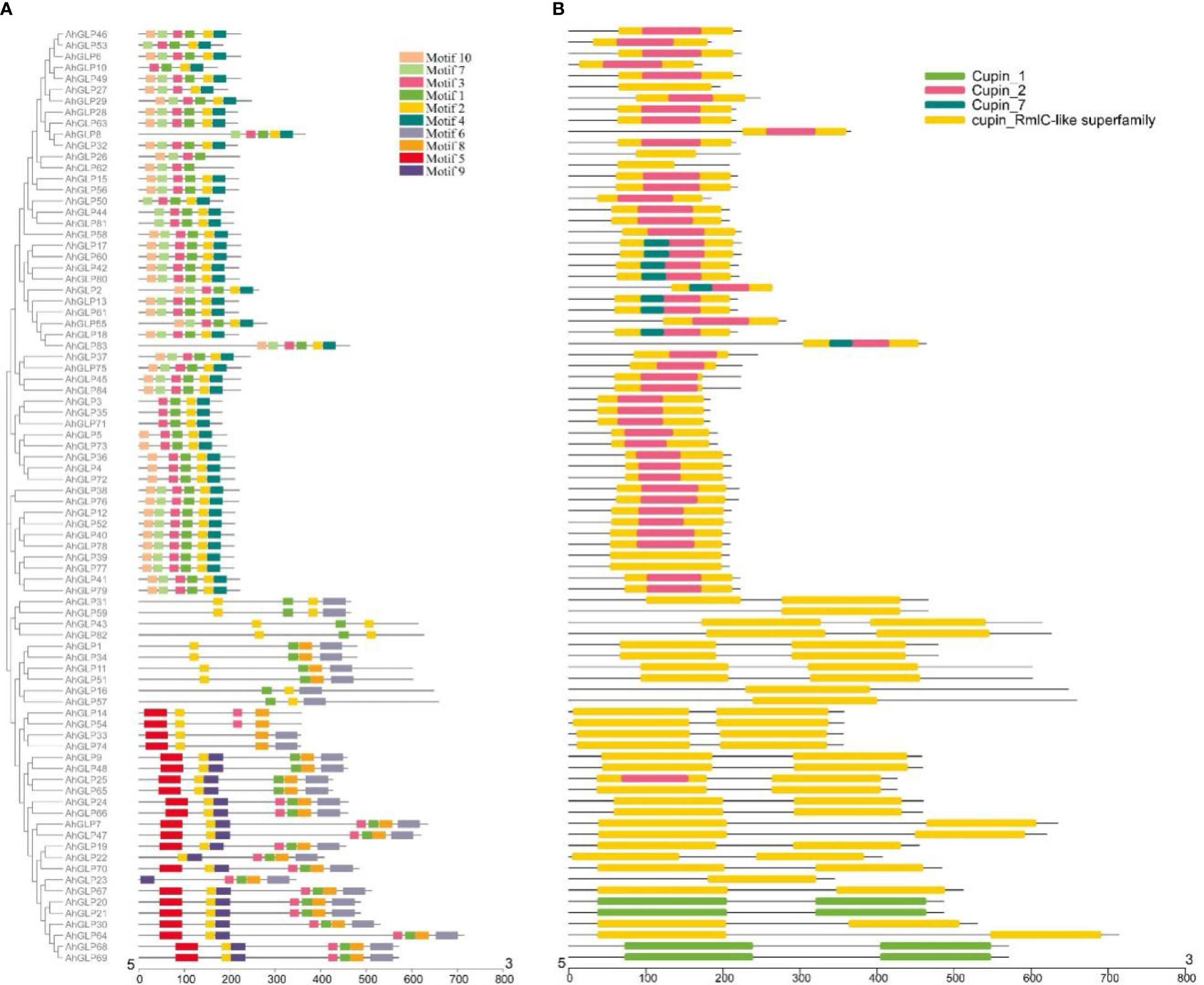
Figure 3 Conserved motifs distribution patterns (A) and presence of cupin domains (B) in Arachis hypogaea Germin-like proteins (AhGLPs). Commonly shared motifs among genes tend to cluster in the same groups, referring to their similar functions.
The evolutionary relationships of germin-like proteins of peanuts and Arabidopsis were elucidated by constructing the phylogenetic tree of their protein sequences. The higher number of GLPs present in cultivated peanut than in its diploid parents and Arabidopsis indicates a higher evolutionary rate of AhGLPs. Previous studies classified GLPs into six phylogenetic groups in barley, wheat, soybean, and rice (Lu et al., 2010; Barman and Banerjee, 2015; Li et al., 2016; Yuan et al., 2021), and the phylogenetic tree also divided peanut GLPs into six phylogenetic groups. All proteins of Arabidopsis were restricted to four groups such as groups 3, 4, 5, and 6 (Figure 4). GLPs in the first and second groups were only from peanut species, indicating that cultivated peanut was closer to its wild progenitors compared with Arabidopsis.
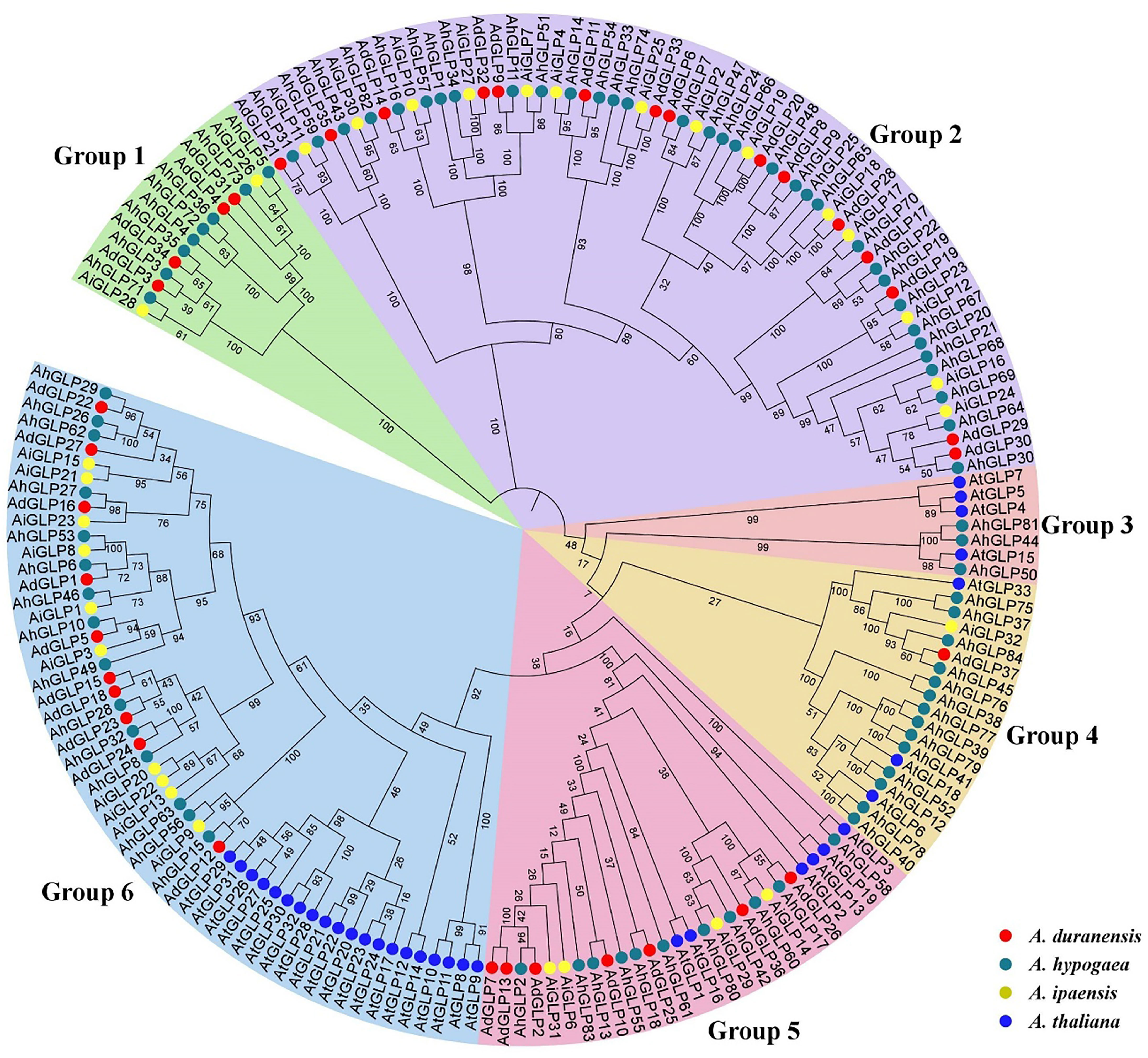
Figure 4 Phylogenetic relationships of Germin-like proteins of A. hypogea, A. duranensis, A. ipaensis, and A. thaliana. The phylogenetic tree classified peanut GLPs into six groups, while AtGLPs were restricted to only four groups. Germin-like proteins of cultivated peanut are phylogenetically closer to its wild parents as compared to Arabidopsis.
Identification of orthologous genes’ clusters is important to evaluate the polyploidization events during the evolutionary process of a gene family. The relative assessment was established to detect the orthologous gene clusters across A. hypogea, A. duranensis, A. ipaensis, and A. thaliana. Figure 5 elaborates the identified gene clusters and their overlapping regions. A. hypogea recorded maximum clusters followed by A. duranensis, A. ipaensis, and A. thaliana. Results showed that 18 gene clusters are solely composed of GLPs found in peanut species (diploid and tetraploid), which indicated that polyploidization had evolved new peanut-specific orthologous GLP gene clusters. We also constructed orthologous gene clusters among three peanut species (Supplementary Figure 5). Comparatively, 60, 52, 13, and 55 orthologous GLPs were found in A. hypogea, A. duranensis, A. ipaensis, and A. thaliana, respectively (Supplementary File 2). Thirty-six in-paralogous genes were identified in A. hypogea, eight and four in-paralogous genes were identified in A. duranensis and A. ipaensis, while 178 in-paralogous genes were found in A. thaliana. Surprisingly 12, 5, and 4 singletons were also found in A. hypogea, A. duranensis, and A. ipaensis, respectively. Results demonstrated that identified orthologous genes decrease with increased phylogenetic distances.
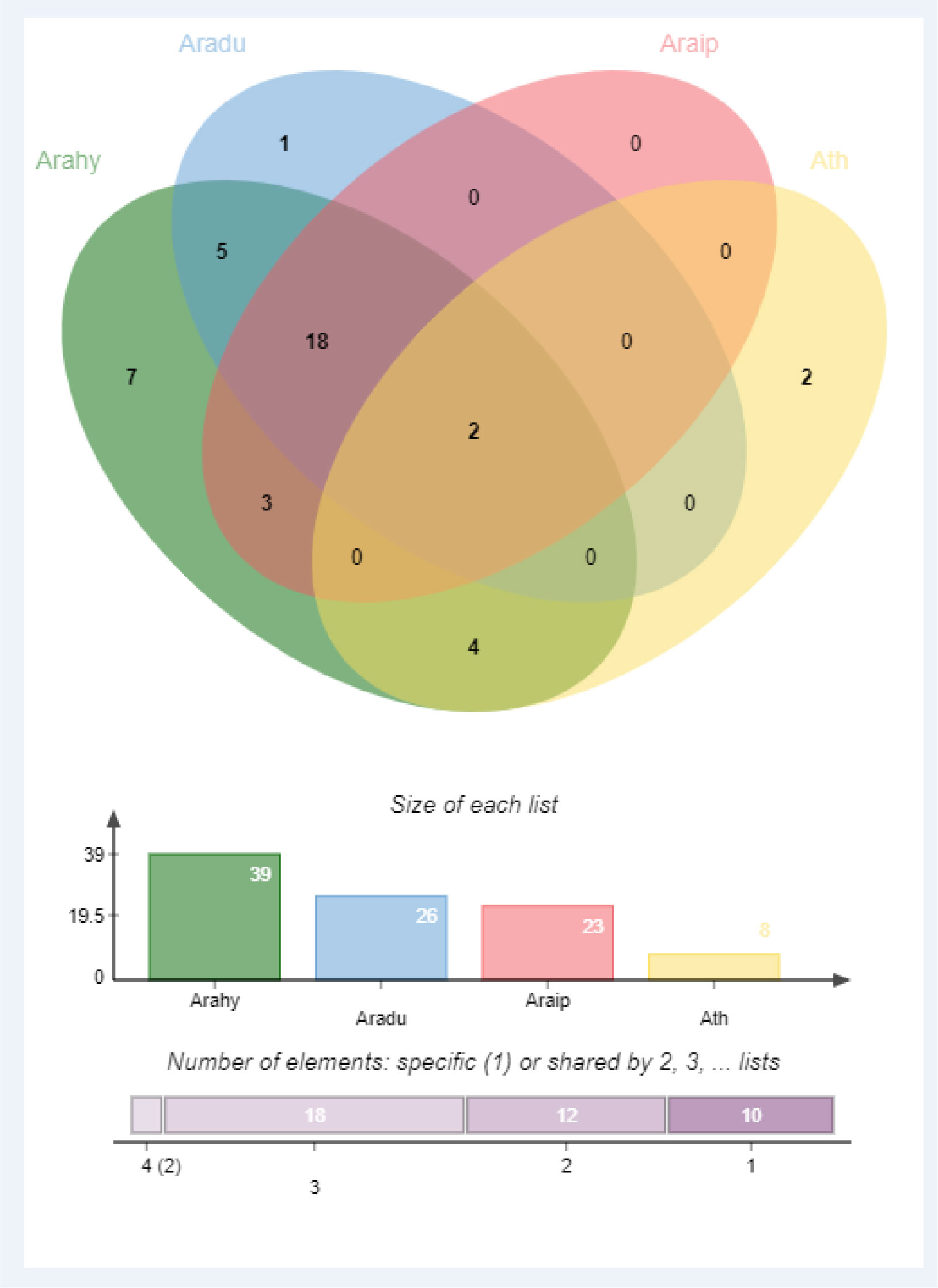
Figure 5 Orthologous genes’ clusters among A. hypogea, A. duranensis, A. ipaensis, and A. thaliana. A. hypogea recorded maximum clustered. Eighteen gene clusters are solely composed of GLPs found in peanut diploid and tetraploid species, indicating that polyploidization has evolved new peanut-specific orthologous GLP gene clusters.
For functional genomics studies, researchers must explore the genomic regions to identify the transcription factor binding sites or groups of sites constituting the cis-regulatory elements (Raza et al., 2021b). To predict the gene functions and their regulatory patterns, we searched the cis-regulatory elements of AhGLPs promoter regions. The cis-elements prediction results showed that in addition to core promoter elements (TATA-Box, CAAT-Box), a large number of important elements were also present (Figure 6). We classified these cis-regulatory elements into four groups according to their functions: ‘light-responsive’, ‘hormones responsive’, ‘growth and development-related’, and ‘stress-related’ elements (Figure 7). All 84 AhGLPs were enriched with hormones- and light-responsive elements, 77 genes were enriched with growth and development-related elements, and 71 genes were enriched with stress-responsive elements (Figure 7A). Thirteen genes were also enriched in RY-element, which is responsible for seed-specific regulation (Figure 7A) (Stålberg et al., 1993).

Figure 6 Cis-regulatory elements of AhGLP promoters. Cis-elements analysis revealed important elements responsive to light, hormones, growth and development, and stress responsiveness.
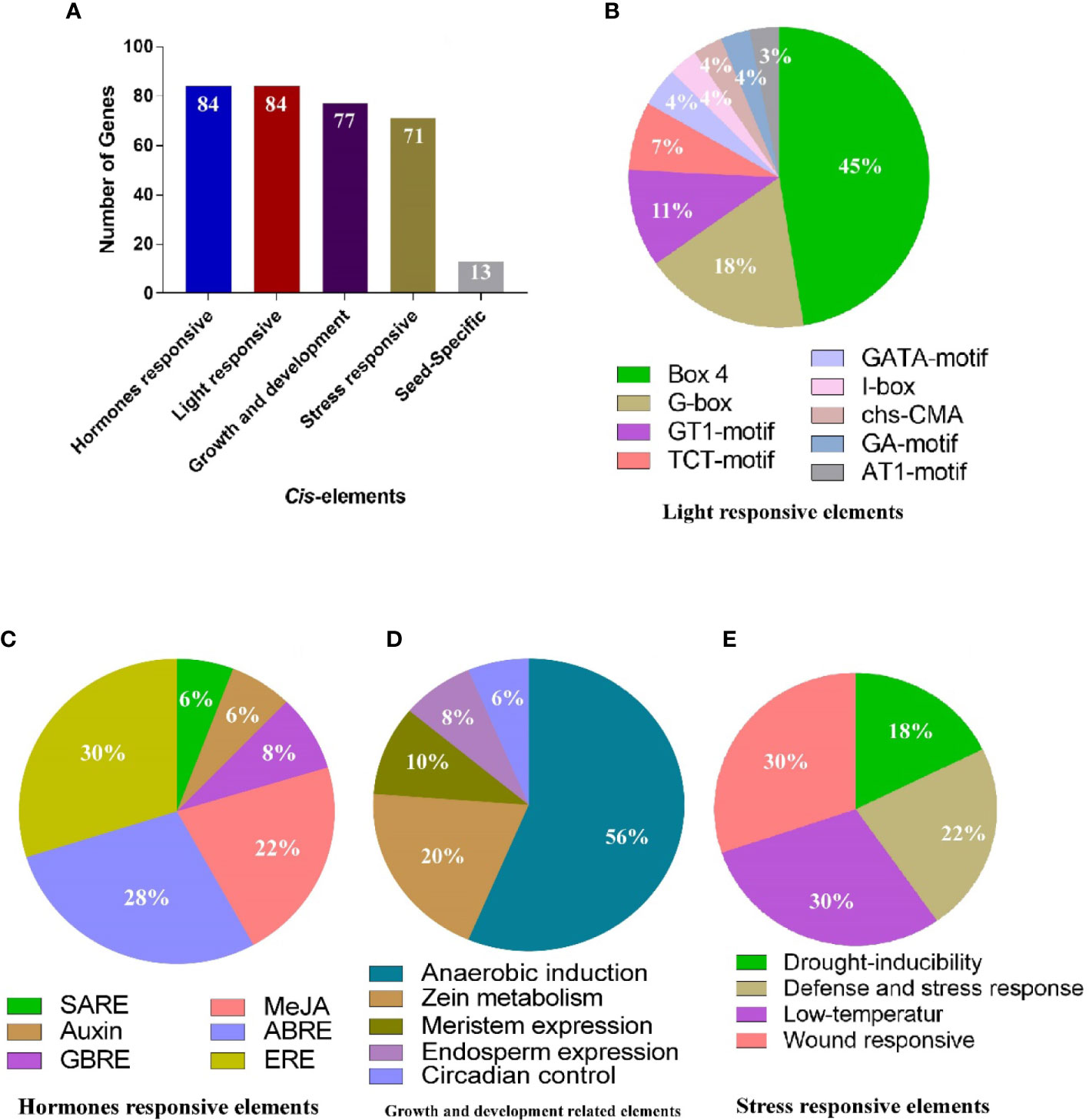
Figure 7 Cis-regulatory elements of AhGLP promoters. (A) the number of genes in different categories of elements. (B) composition of light-responsive, (C) hormone-responsive, (D) growth and development responsive, and (E) stress-responsive factors.
Light responsive elements mainly include Box-4, G-box, GT1-motif, GATA-motif, TCT-motif, GA-motif, chs-CMA element, I-box, and AT-1 motif (Figure 7B). Other light-responsive elements include 3-AF1 binding site, ATC-motif, AE-box, MRE element, Box II, CAG-motif, CGTCA-motif ATCT-motif, Gap-box, ACE element, TCCC-motif, GTGGC-motif, LAMP-element, LS7 element, and Sp1 element, were also present. Hormones responsive class includes abscisic acid-responsive (ABRE), auxin-responsive (AuxRE, AuxRR-core, CGTCA-motif, TGA-box), gibberellins responsive (P-box, GARE motif, TATC-box), methyl jasmonate responsive (CGTCA-motif, TGACG-motif), salicylic acid-responsive (SARE, TCA-element), and ethylene-responsive (ERE) elements (Figure 7C). Growth and development category contained anaerobic induction responsive (ARE), meristem expression responsive (CAT-box), endosperm expression related (GCN4-motif, AACA-motif), circadian control (CAAAGATATC), and zein metabolism-related (O2-site) elements (Figure 7D). The stress-responsive class further includes defense and stress response (TC-rich repeats), drought-responsive (MBS), low-temperature responsive (LTR), and wound-related (WUN-motif) elements (Figure 7E).
During the past decade, non-coding miRNAs have emerged as key regulators of post-transcriptional gene regulation (Chen et al., 2019; Raza et al., 2021a). Their roles have been worked out against many biotic and abiotic stresses (Sun et al., 2015; Ding et al., 2017). To better understand miRNAs modulating the post-transcriptional regulation of AhGLPs, the coding sequences of 84 AhGLPs were scanned at the psRNATarget database (https://www.zhaolab.org/psRNATarget/analysis?function=2) against the published A. hypogaea miRNAs. Scanning results predicted seven miRNAs from six different families targeting 25 peanut germin-like proteins (Supplementary Table 6). The miRNA ‘ahy-miR3514-3p’ targeted eight AhGLPs (the highest number of AhGLPs targeted by any miRNA). While ‘miRNA ahy-miR3511-5p’ and ‘ahy-miR3518’ targeted seven AhGLPs each. The miRNA ‘ahy-miR167-5p’ and ‘ahy-miR394’ targeted four AhGLPs each. The ‘ahy-miR3514-5p’ targeted three AhGLPs and ‘ahy-miR408-5p’ targeted only one peanut GLP (Figure 8A). The germin-like protein genes AhGLP1, AhGLP2, AhGLP13, AhLP18, AhGLP34, ahGLP43, AhGLP61, AhGLP82, and AhGLP83 each were targeted by more than one miRNAs (two) while the remaining genes were targeted by one miRNA. Some of the miRNAs target sites are shown in Figure 8B. Functional validation of the expression level of these miRNAs and their role in gene regulation in the peanut genome are potential future research dimensions.
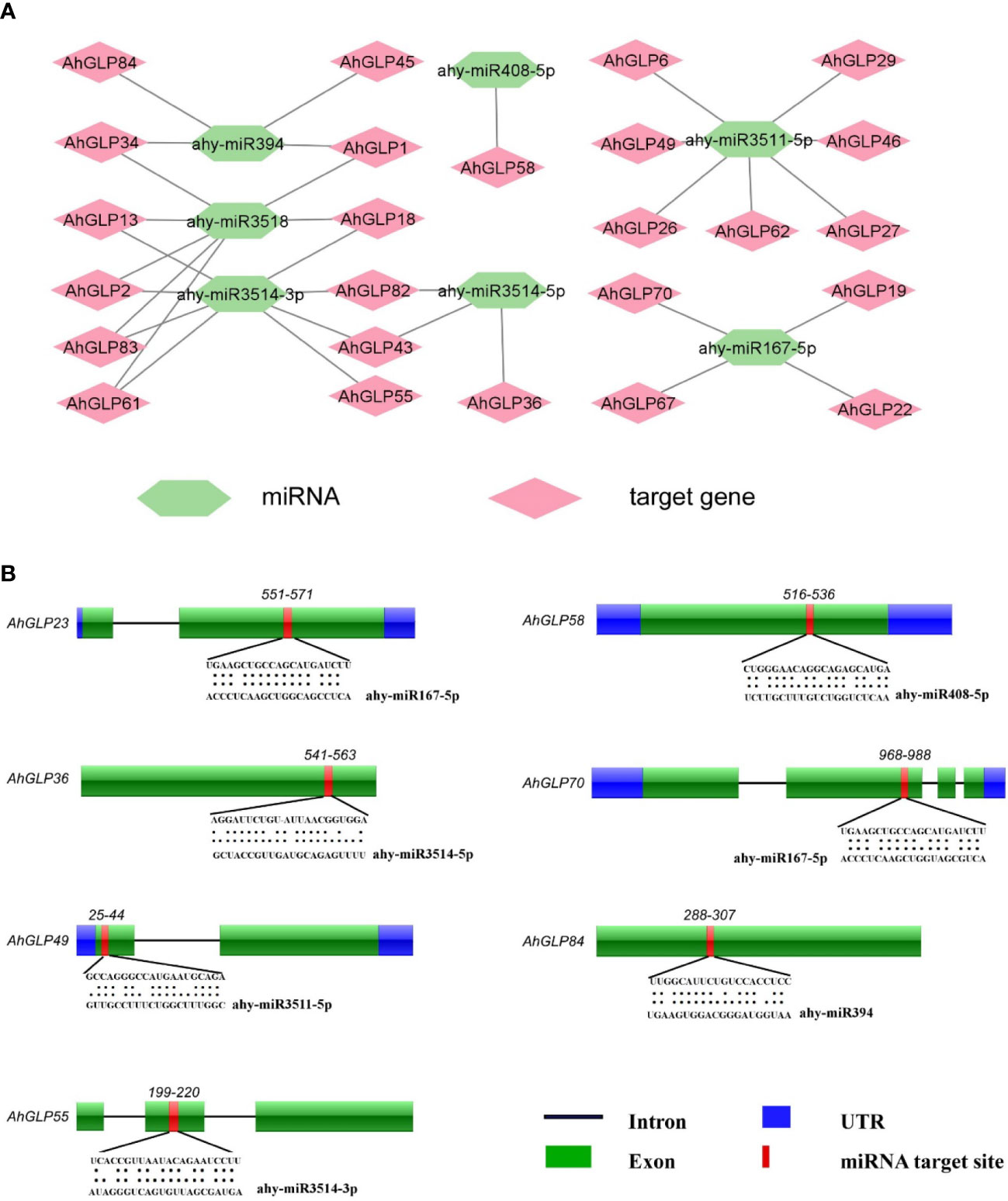
Figure 8 Predicted miRNAs targeting AhGLPs. (A) In total, 7 miRNAs were predicted targeting 25 Arachis hypogea Germin-like protein gens. (B) Schematic diagram of miRNAs target sites in some AhGLPs.
Gene duplication (segmental and tandem duplication) is a major force behind genome evolution (Su et al., 2021). So, gene duplication events for A. hypogea were evaluated. Mainly segmental gene duplication was found to be responsible for deriving the genome evolution (Figure 9A). Out of 84 AhGLPs, 38 were duplicated gene pairs (Table 2). The synonymous substitution rates Ks and non-synonymous substitution rates Ka were calculated by the simple Ka/Ks calculator. For each duplicated gene pair, evolutionary rates (Ka/Ks ratio) were calculated. The Ka/Ks=1 was considered neutral selection pressure, while Ka/Ks>1 was regarded as positive selection pressure, and Ka/Ks<1 was considered purifying selection pressure (Yang and Bielawski, 2000). Mainly purifying selection pressure was involved in gene duplication (Table 2). The expected divergence time ‘T’ of duplicated gene pairs was calculated as ‘T=Ks/2r’, where the exchange rate coefficient ‘r’ for the peanut is 8.12×10-9 (Bertioli et al., 2016). The divergence time varied from 0.43 MYA (million years ago) for gene pair AhGLP44:AhGLP81 to 166.24 MYA for AhGLP47:AhGLP66 (Table 2).
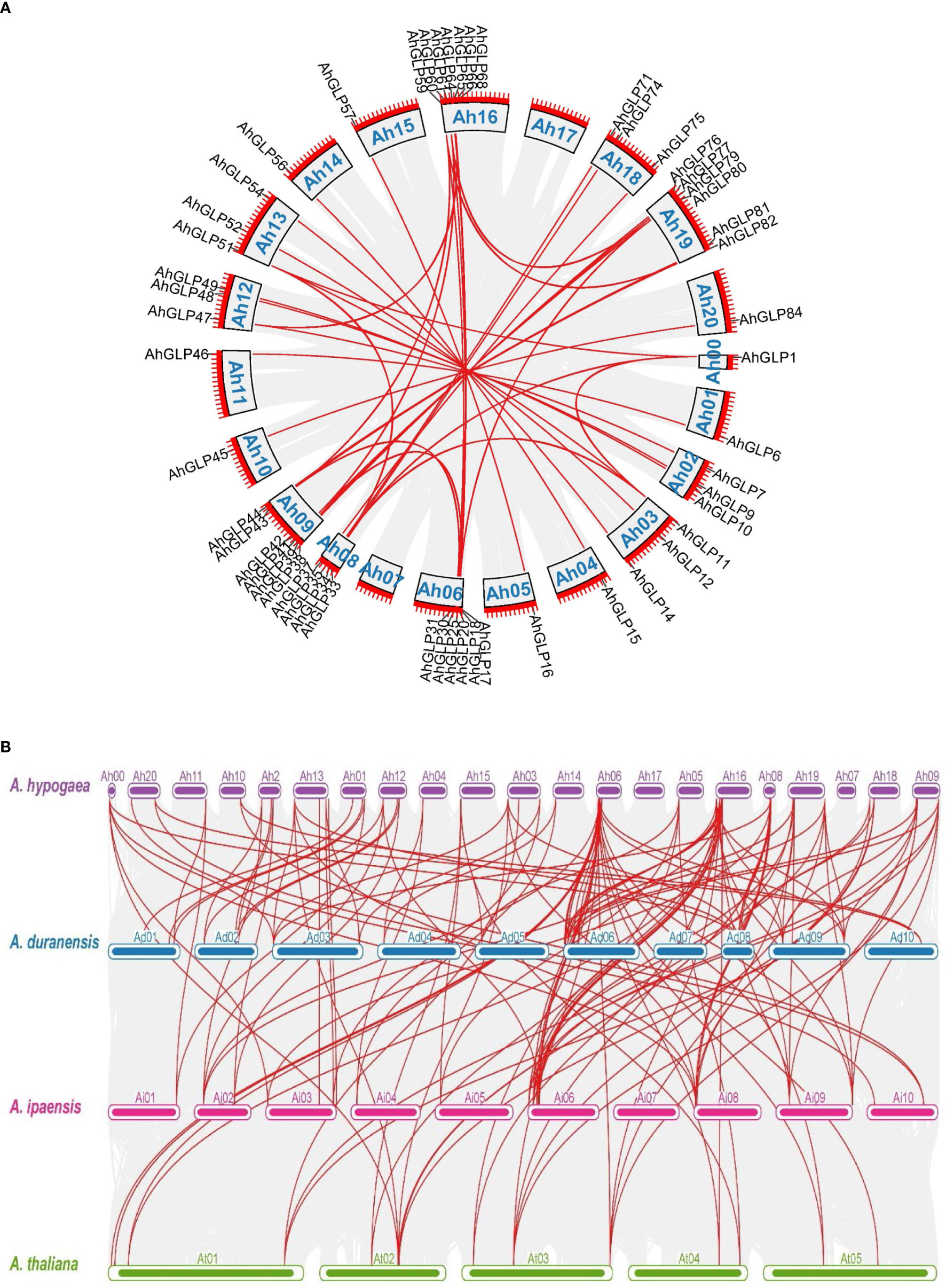
Figure 9 (A) Schematic diagram of gene duplication in AhGLPs, red lines indicate the duplicated gene pairs and grey lines in the background represent the duplicated regions among different chromosomes. (B) Synteny analysis among A hypogea, A duranensis, A ipaensis, and A thaliana. Synteny analysis showed key evolutionary relationships of GLPs in diploid and tetraploid peanut species. AhGLPs possessed highly conserved syntenic relationships with other peanut species as compared to Arabidopsis.
Comparative synteny analysis among A. hypogaea, A. duranensis, A. ipaensis, and A. thaliana represented remarkable evolutionary, duplication, expression, and functional relationships. Mainly AhGLPs showed significant syntenic relationships with its wild progenitors and Arabidopsis; however, the syntenic relationships of A. hypogaea were closer to its wild parents than Arabidopsis. Total 69 syntenic relationships of A. hypogaea were found in the genome of A. duranensis, and 57 syntenic relationships of AhGLPs were found in the genome of A. ipaensis. In contrast, 31 syntenic relationships were found among AhGLPs and AtGLPs (Supplementary File 3). The synteny analysis results showed that A. hypogaea is closer to its wild parents than Arabidopsis. The syntenic relations of A. hypogaea, A. duranensis, A. ipaensis, and A. thaliana are shown in Figure 9B.
The Functions of AhGLPs could be speculated on the basis of well-studied Arabidopsis GLPs. The protein-protein interaction network analysis of cultivated peanut GLPs were performed to understand the functions of GLP proteins on the basis of their orthologues in Arabidopsis. Protein interaction network prediction showed that AhGLPs have functions related to cysteine peroxidation 1 (PER1), contributing to inhibition of germination under stressed conditions (Figure 10). AhGLP82 has those related to oleosin2 (OLEO2) found in oil bodies and plays roles in freeze tolerance in seeds, and CUR3, a seed storage protein, mainly plays a role in response to ABA stress. AhGLP82 also possesses functions related to seed storage as of SESA2, SESA3, and SESA5. Other AhGLPs also had similar those and those related to proximal membrane proteins (AhGLP65 and AhGLP70). AhGLP83 was predicted to interact with Arabidopsis Chitinase-like protein (preventing the lignin accumulation in hypocotyls CTL2) and Laccase/diphenol oxidase family protein IRX12 (cell wall biosynthesis-related functions). Multiple sequence search method based on the scoring and integration of known and predicted associations results in comprehensive networks showed that some proteins, including AhGLP53, AhGLP58, AhGLP71, AhGLP73, AhGLP74, AhGLP71, and AhGLP84, did not show any interaction.
Gene ontology (GO) enrichment analysis of AhGLPs was performed to view their possible roles in biological processes (BP), molecular functions (MF), and cellular components (CC). GO enrichment results provided highly enriched terms related to BP, MF, and CC (Figure 10). AhGLPs were involved in several biological processes, including reproductive processes (GO:0048609 and GO:0032504), seed and fruit development (GO:0010431, GO:0048316, and GO:0010154), defense responses against various biotic and abiotic agents (GO:0050832, GO:0009620, GO:0098542), response to various phytohormones (GO:0009725, GO:0009735), interspecies interaction (GO:0044419), response to different chemicals (GO:0042221), responses to other organisms (GO:0098542), response to different endogenous and exogenous stimuli (GO:0043207, GO:0009607, GO:0050896), and postembryonic development (GO:0009791) etc. In the cellular component category (CC), AhGLPs are part of the cell wall (GO:0005618), cytoplasmic vesicle (GO:0031410), vesicle (GO:0031982), vacuole (GO:0005773), etc. For the molecular function category (MF), AhGLPs are involved in nutrient reservoir activity (GO:0045735) (the main molecular function of GLPs indeed). The detailed information about MF, BP, and CC categories, their associated GO IDs, and AhGLP members involved in these categories are given in Figure 11 and Supplementary File 4. Collectively it is evident from functional annotation analysis that AhGLPs play key roles in several biological, cellular, and molecular functions.
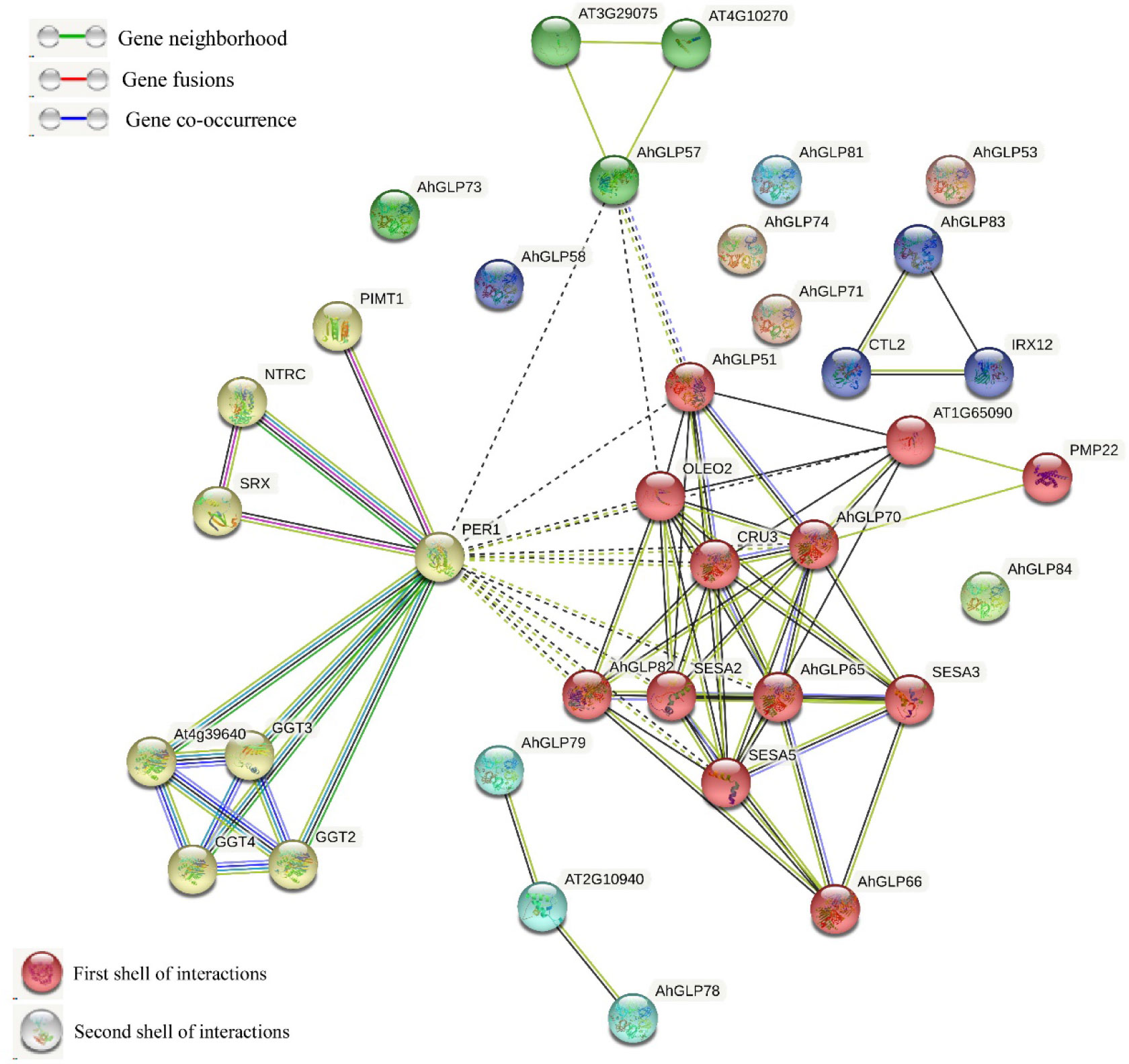
Figure 10 The predicted protein-protein interaction network of AhGLPs using the STRING database. Putative protein functions of AhGLPs are predicted on well-studied GLP orthologues in Arabidopsis.
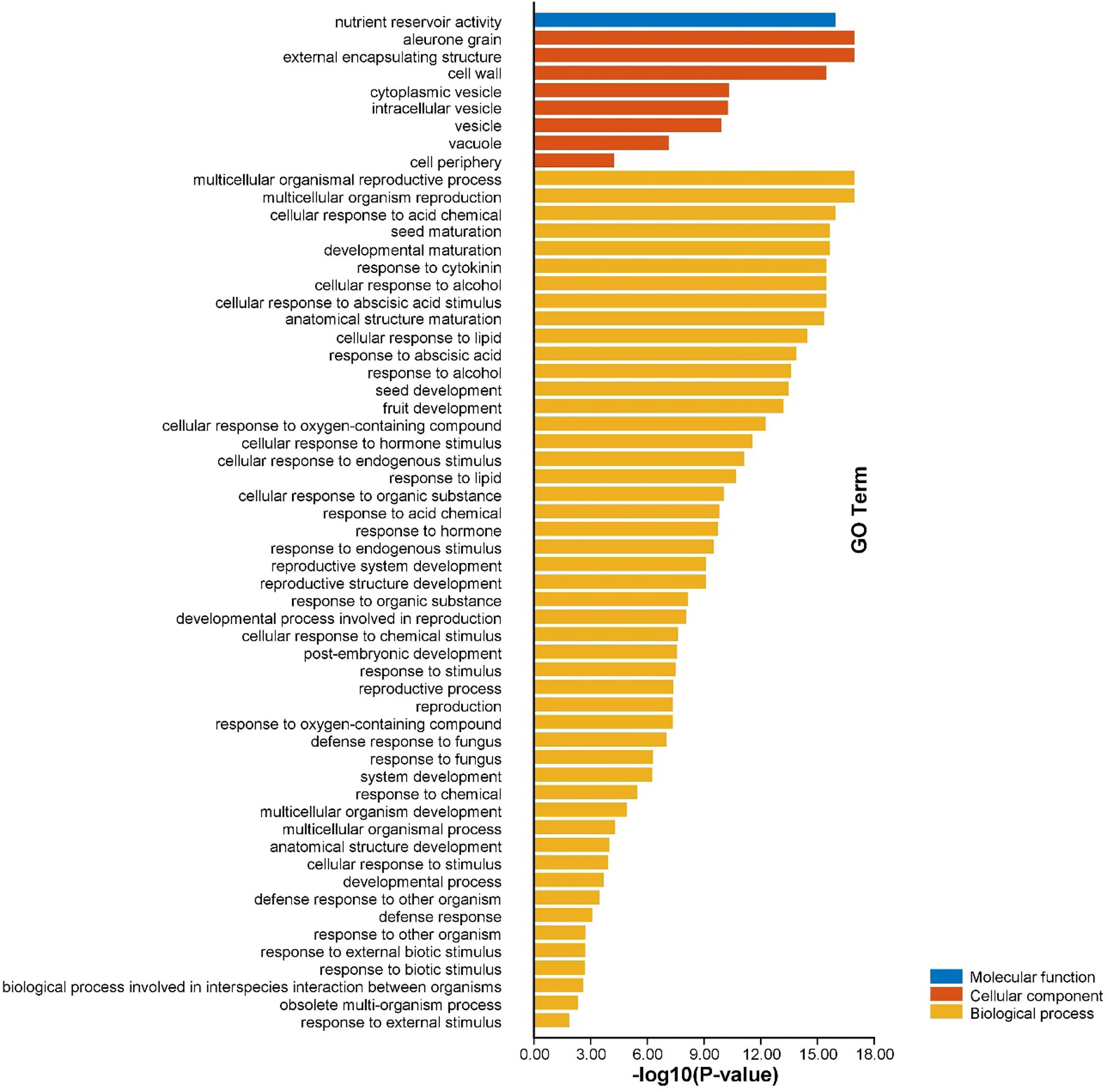
Figure 11 Functional annotation (GO) analysis of AhGLPs. The gene ontology enrichment analysis showed that AhGLPs are involved in all categories, including molecular function (MF), cellular component (CC), and biological processes (BP). AhGLPs are involved in a number of important biological processes, including growth and development related, biotic and abiotic stress response, and various stimulus responses.
Transcriptome expression data was used to view the expression levels of 84 AhGLPs in different tissues and under different abiotic stresses. AhGLPs possessed diverse expressions in different peanut tissues. Almost 25 genes showed expression responses in the embryo, including AhGLP7, AhGLP9, AhGLP11, AhGLP16, AhGLP19, AhGLP24, AhGLP47, AhGLP57, AhGLP66, AhGLP67, AhGLP68, AhGLP69, AhGLP70, AhGLP80 etc. Genes, including AhGLP14, AhGLP26, AhGLP29, and AhGLP62, were abundantly expressed in the pericarp. AhGLP3, AhGLP5, AhGLP32, AhGLP53, AhGL71, and AhGLP73 were uniquely down regulated in all tissues (Figure 12). Transcriptome expression data against different phytohormones including ABA, SA, Brassinolide, Paclobutrazol, and Ethephon treatment, water stress (drought and normal irrigation), temperature stress (low 4°C and room temperature 28°C) were also accessed to determine their stress responses. AhGLP12, AhGLP14, AhGLP18, AhGLP40, AhGLP52, AhGLP61, and AhGLP78 were upregulated under normal and hormonal treatment (Figure 13). AhGLP20, AhGLP21, AhGLP30, AhGLP43, AhGLP60, AhGLP64, AhGLP68, AhGLP69, and AhGLP82 were upregulated under paclobutrazol treatment as compared to control and other hormones. The expression responses of all genes to other hormones were similar to control. The expression responses of AhGLPs under different water treatments (drought and normal irrigation) were comparable to hormones. Some genes, including AhGLP12, AhGLP14, AhGLP24, AhGLP52, AhGLP54, and AhGLP74, were decreased under drought stress as compared to normal irrigation. But AhGLP31, AhGLP42, AhGLP50, AhGLP58, and AhGLP69 showed little increase under drought stress as compared to normal irrigation. A similar pattern was also observed under different temperature (low and normal) treatments. A few genes (AhGLP12, AhGLP14, AhGLP52, and AhGLP54) also showed decreased expression under low temperatures. The Fragments Per Kilobase Million (FPKM) values of transcriptome expression of AhGLPs in different tissues and under different stress conditions are given in Supplementary Tables 7, 8.
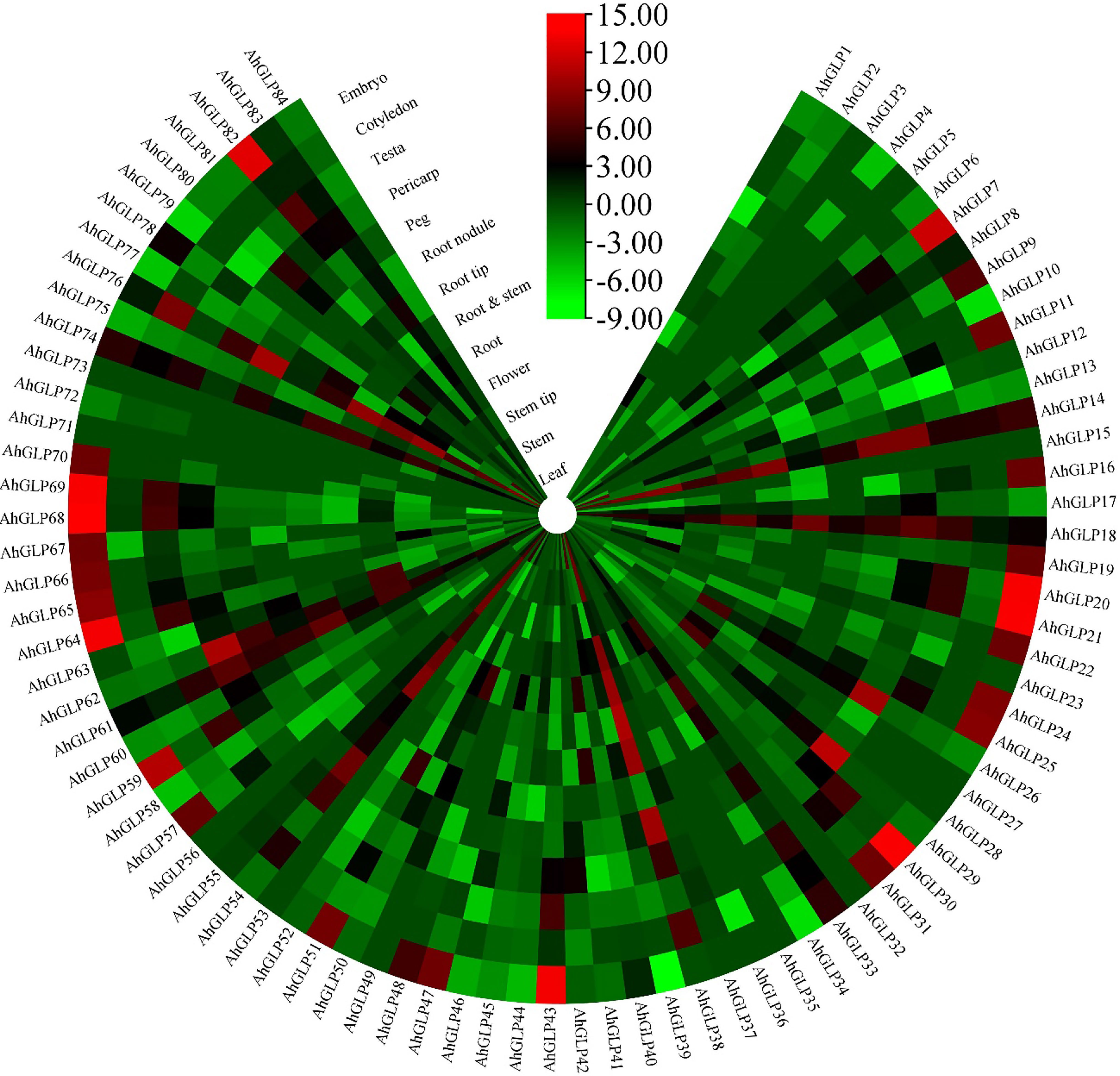
Figure 12 Transcriptome expression of AhGLPs in different tissues. AhGLPs possessed varying expression matrices in different tissues. Some genes are very lowly expressed in all tissues, while others are specifically expressed in leaf, pericarp, and embryo.
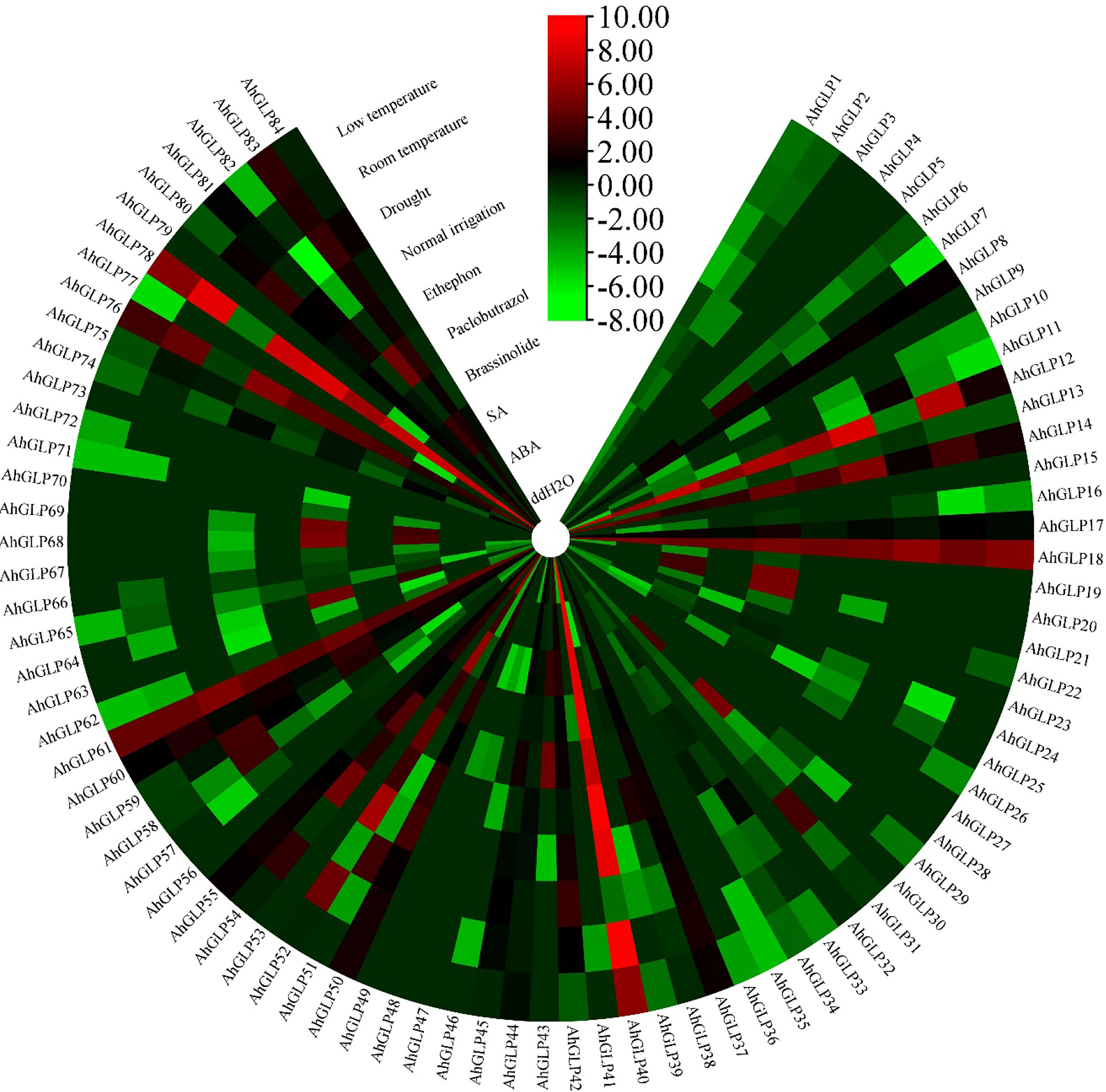
Figure 13 Transcriptome expression of AhGLPs under different hormones and stress conditions. Most of AhGLPs are not responsive to different stress treatments, while few genes AhGLP12, AhGLP18, AhGLP40, AhGLP61, and AhGLP78 showed good responses to all stress/normal environments.
For the validation of expression matrices, ten AhGLP genes with high transcriptome expression in response to ABA and low temperature (4 °C) were selected (Figure 13), and their expression was monitored with the help of quantitative real-time PCR. Most of the selected genes showed increased expression in response to ABA treatment except a few genes showed lower expression. Genes AhGLP14, AhGLP52 and AhGLP54 were downregulated in response to ABA treatment at all time points as compared to control. The expression of AhGLP14 and AhGLP54 was less downregulated at 9h, while the expression of AhGLP52 was less downregulated at 6h post-ABA treatment (Figure 14A). Under low-temperature stress, most of the genes were upregulated, except AhGLP40, AhGLP52, and AhGLP78, which were downregulated (Figure 14B). AhGLP40 showed a gradual increase in expression level but still its expression was lower under low temperature than control. The expression level of AhGLP54 at 9h was comparable to control, but it was downregulated at all other time points. In most cases, expression was highest at 6h after low-temperature treatment. Although there were some expression variations at different time points, overall, the qRT-PCR-based expression is in accordance with transcriptome expression. These results represent the reliability of the transcriptome expression data.
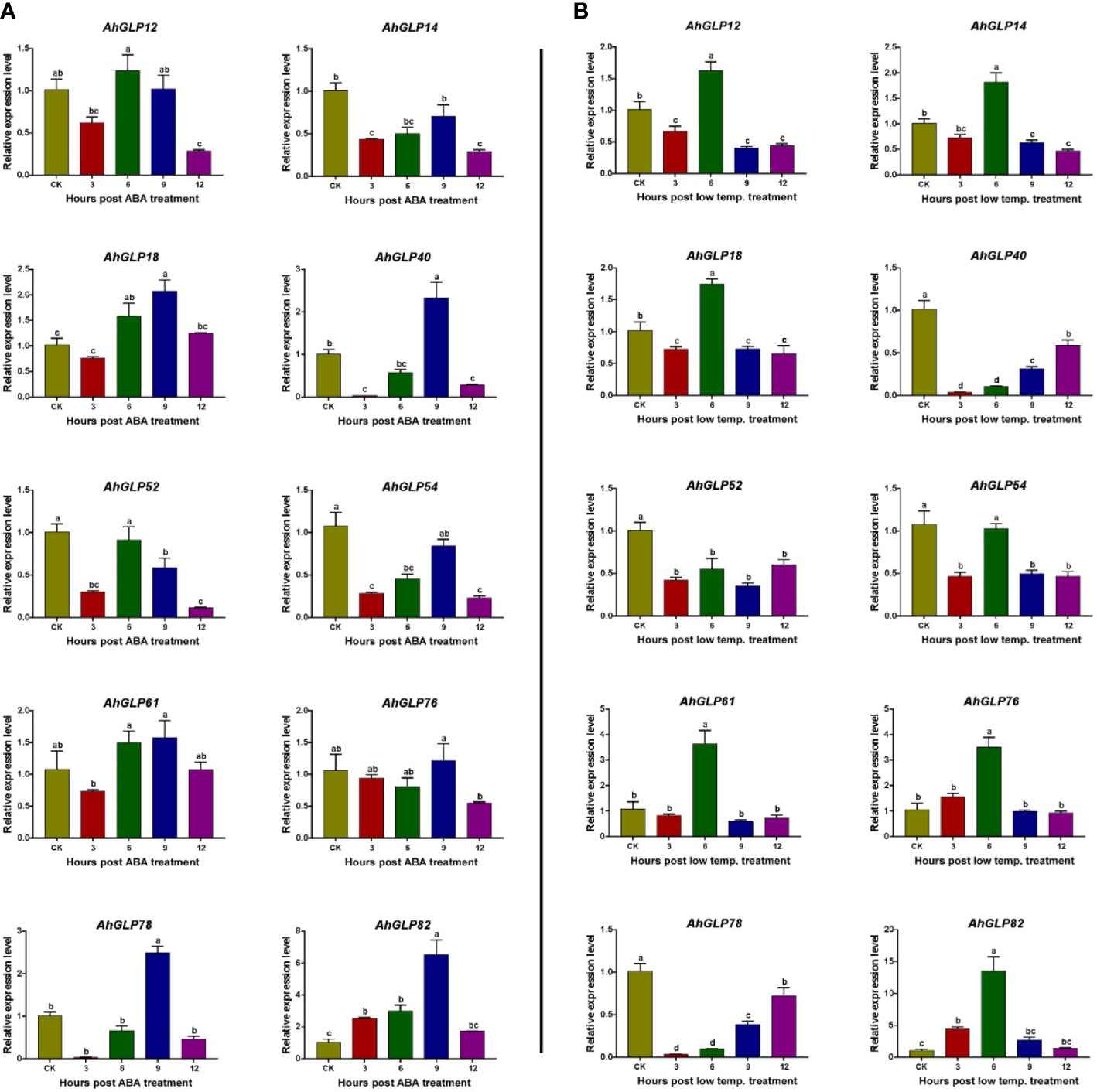
Figure 14 qRT-PCR-based expression profiling of 10 selected genes under (A) abscisic acid and (B) low-temperature stress. CK represents the control samples, while 3, 6, 9, and 12 represent the hours after stress (ABA/low temperature) treatment. Data were analyzed by 2-ΔΔCT method and statistical significance was determined by ANOVA. a, b, and c represents the significance levels among expression at different time points.
Peanut (Arachis hypogaea L.) is an important legume crop providing edible oil, proteins, food, and feed to humans and livestock. It is a staple grain in many Asian and African countries and constitutes a major part of their every meal. Like other crops, peanut has to face various bacterial, viral, and fungal pathogens and harsh environmental conditions (Zhang et al., 2017; Khan et al., 2020). Crops have evolved many defense mechanisms to combat unfavorable conditions. Genome duplications, gene family evolution, and seed dispersal mechanisms are among the strategies that plants adopt to survive. Different gene families have important roles in the plant kingdom. Thanks to the advances of functional genomics and high throughput sequencing technologies that have made it easy to identify the gene families and explore their roles in different metabolic and defense pathways. GLP family is among plant gene families playing key roles in plant defense and growth regulation (Wang et al., 2013). Previously GLP gene family has been studied at a genome-wide scale in some important crop species. Different numbers of GLP family members are found in different plants, mainly depending upon their ploidy level and genome size. To date, 32 GLP genes in Arabidopsis and 43 in rice (Li et al., 2016), 48 in barley (Zimmermann et al., 2006), 69 in soybean (Wang et al., 2014), and 258 in wheat (Yuan et al., 2021) have been reported. Based on the Arabidopsis GLPs and germin/Cupin domain (PF00190) search, we identified 37, 32, and 84 GLPs in A. duranensis, A. ipaensis, and A. hypogaea, respectively. The consensus sequences of cupin motifs in peant GLPs are given in Supplementary File 1. The number of genes belonging to a group/family in a species depends on genome size and complexity. The number of GLP homologs in Arabidopsis are lowest among all studied species, which is mainly due to smaller genome size and a smaller number of chromosomes. Previous studies reported 32 GLPs in Arabidopsis (Li et al., 2016), but we identified one more GLP member in Arabidopsis with careful analysis. Number of GLP homologs in peanut species show similar pattern as of other plant species. GLPs in diploid peanut species (A. duranensis and A. ipaensis) are very close to Arabidopsis. While the complex genome of A. hypogaea possessed a higher number of GLP homologs, as its genome is composed of two subgenomes that have undergone natural duplication, resulting in an allotetraploid species.
The large genome size and tetraploid nature of A. hypogaea genome have resulted in more GLPs than its diploid progenitors. Structural diversity of gene families has resulted due to the evolutionary process, and the number of exons/introns plays a key role in the expression of a gene (Hu et al., 2010; Guo and Qiu, 2013). Generally, the germin-like protein family is characterized by two exons (Li et al., 2016), but exon/intron number variations are very common. Many GLPs of wild peanut species were composed of two exons and a single intron (Supplementary Figures 3, 4). The maximum number of exons exceeded seven for A. duranensis and 15 for A. ipaensis. In contrast, most AhGLPs were composed of a single exon (Figure 2), and the maximum number of exons is 10 (Table 1). The presence of a single exon in most of AhGLPs might be due to intron deletion during the evolutionary process. Genome size variations and gene duplication events are key elements of genetic diversity. As genomic duplication is a key factor underlying gene families’ expression, diversification, and neofunctionalization, we also analyzed the duplication events in AhGLPs. Duplication analysis revealed 38 duplicated gene pairs in AhGLPs with varying divergence times (Table 2). Mostly segmental duplication events were found in the evolution of peanut germin-like proteins, and this type of duplication has been reported previously in rice and Arabidopsis (Li et al., 2016). The phylogenetic analysis previously divided the GLP family into six groups (Yuan et al., 2021); six groups were also found in peanut GLPs. In the phylogenetic classification, all GLPs of Arabidopsis were restricted to four groups, as described in previous studies (Li et al., 2016; Yuan et al., 2021). But the germin-like proteins of wild and cultivated peanut species were distributed among all six groups depicting a close relationship among peanut species (Figure 4).
Cis-elements of a genes’ promoter are significant as they determine the expression pattern, regulatory roles, and stress responses (Maruyama-Nakashita et al., 2005; Osakabe et al., 2014). We identified four types of cis-elements in AhGLPs promoters: light-responsive, hormones-responsive, growth and regulation-related and stress-related elements, and seed-specific expression-related elements (RY-repeat) were also found in some of AhGLPs (Figure 6). Micro-RNAs are critical regulatory elements and play important roles against different stresses (Su et al., 2021). For the past few years, miRNAs have been a hot research topic (Xie and Zhang, 2015; Ding et al., 2017; Chen et al., 2019). The present study predicted the putative miRNAs targeting AhGLPs from the published miRNAs database (Dai et al., 2018). We found seven miRNAs belonging to six families, targeting 25 AhGLPs (Supplementary Table 6 and Figure 7). Among these miRNAs, miRNA408 have been reported to be involved in responses to water deficit conditions, whereas it is upregulated in M. truncatula (Trindade et al., 2010) and downregulated in pea (Jovanović et al., 2014). It is also involved in drought tolerance in chickpea (Hajyzadeh et al., 2015). miR394 has been reported to improve drought and salt tolerance in Arabidopsis (Song et al., 2013), while miR167 has been reported to regulate the auxin response genes under blue light in Arabidopsis (Pashkovskiy et al., 2016). These reports suggest that predicted miRNAs targeting the AhGLPs might play key roles in response to different abiotic stresses.
Transcriptome-based expression profiling of AhGLPs revealed their expression under different abiotic stresses and in different tissues. A fraction of AhGLPs showed embryo-specific expression (Figure 12), while a few gens showed pericarp-specific expression. Interestingly pericarp specific genes (AhGLP26, AhGLP29, and AhGLP62) possessed the longest genomic sequences and longest introns among all AhGLPs (Figure 2 and Table 1), indicating some special role of long introns in pericarp-specific expression. Expression profiling under different stresses revealed their responses to drought and low temperature. These results are supported by previous studies on GLP genes (Lee and Lee, 2003; Komatsu et al., 2010). Hormones regulate a genes’ biochemical and physiological reactions through different signal transduction pathways (Fatima et al., 2021; Zaynab et al., 2021; Raza et al., 2022b). Drought is a major limiting factor for crop yields. And drought-smart breeding is a need of the day (Raza et al., 2022a). Transcriptome expression data revealed that some GLPs genes could be used for drought-smart breeding also. AhGLPs showed varying responses to different hormones treatment. Most of the genes were not affected by hormone treatment, some genes were downregulated, and a few were upregulated (Figure 12). Similar findings have also been reported for ABA, salt and drought stress (Wang et al., 2013; Liao et al., 2021). In response to different water and temperature treatments, a similar expression pattern was observed as in the case of hormones. AhGLP12, AhGLP18, AhGLP40, AhGLP61, and AhGLP78 showed higher expression under all hormones, water, and temperature treatments. Similar findings were also found by Huang et al. (2020). Similarly, drought is also a limiting factor and makes developing peanut seeds vulnerable to many pathogens. They performed RNA sequencing of peanut cultivar “Hua Yu 39” in response to Brassinolide treatment under drought and normal irrigation conditions (Huang et al., 2020). Their findings also support our transcriptome expression results. The qRT-PCR-based expression profiling of selected genes provided the validity of transcriptome expression. It provided some key genes responsive to ABA treatment, including, AhGLP18, AhGLP78 and AhGLP82. Similarly, real-time expression profiling of selected genes under low temperature provided that AhGLP12, AhGLP18, AhGLP61, AhGLP76, and AhGLP82 are key for low temperature responses. Overall, this study provides a deep insight into the molecular evolution and functioning of AhGLPs in cultivated peanut. And this study will be a base for further work on peanut germin-like proteins. Along with genomics, gene editing tools could be applied for orphan crop improvement, including cultivated peanut (Yaqoob et al., 2023).
This study identified 84 AhGLPs genes in the genome of cultivated peanut through a comprehensive genome-wide analysis. In the diploid progenitors of A. hypogaea (A. duranensis and A. ipaensis), 37 and 32 GLPs genes were identified, respectively. Phylogenetic and syntenic relations and gene structure analysis provided more profound insight into the evolutionary history of AhGLPs. Predicted miRNAs have shown their defensive roles against abiotic stress tolerance. Hence, they provide a future research dimension for stress-smart breeding. The transcriptome analysis showed that some genes responded significantly to hormones and abiotic stress stimuli. Some genes showed specific expression in pericarp and embryo, such as AhGLP7, AhGLP20, AhGLP21, AhGLP30, AhGLP43, AhGLP64, AhGLP68, AhGLP69, and AhGLP82. These genes and their promoters are valuable future candidates to express in embryo-specific manner or drive a gene in embryo-specific manner. Thus, additional investigations are required to confirm the purposeful role of GLPs in peanut growth, development, and response to numerous environmental stresses.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA480120.
WZ and HC conceived the idea and designed the study. QY, YS, YZ, HF, and SW analyzed the data and wrote the manuscript. CZ, TC, QY, KC, AR, and LW helped in literature search, and provided technical guidance. WZ, HC, and YZ supervised the work and edited the final version. QY and YS equally contributed to the manuscript. All authors have read and approved the final version of the manuscript.
This work was supported by grants from the National Natural Science Foundation (NSF) of China (U1705233, 31601337, and 32072103), the Science and Technology Foundation of Fujian Province of China (2017N0006), and Fujian Agriculture and Forestry University, Fuzhou, China.
The authors are thankful to the Center of Legume Plant Genetics and System Biology, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China, for providing the research facilities and technical guidance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1044144/full#supplementary-material
Agarwal, G., Rajavel, M., Gopal, B., Srinivasan, N. (2009). Structure-based phylogeny as a diagnostic for functional characterization of proteins with a cupin fold. PloS One 4, 5736. doi: 10.1371/journal.pone.0005736
Ali, N., Chen, H., Zhang, C., Khan, S. A., Gandeka, M., Xie, D., et al. (2020). Ectopic expression of AhGLK1b (GOLDEN2-like transcription factor) in arabidopsis confers dual resistance to fungal and bacterial pathogens. Genes 11, 343. doi: 10.3390/genes11030343
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, 39–49. doi: 10.1093/nar/gkv416
Banerjee, J., Maiti, M. K. (2010). Functional role of rice germin-like protein1 in regulation of plant height and disease resistance. Biochem. Biophys. Res. Commun. 394, 178–183. doi: 10.1016/j.bbrc.2010.02.142
Barman, A. R., Banerjee, J. (2015). Versatility of germin-like proteins in their sequences, expressions, and functions. Funct. Integr. Genomics 15, 533–548. doi: 10.1007/s10142-015-0454-z
Beracochea, V. C., Almasia, N. I., Peluffo, L., Nahirñak, V., Hopp, E., Paniego, N., et al. (2015). Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic arabidopsis thaliana. Plant Cell Rep. 34, 1717–1733. doi: 10.1007/s00299-015-1819-4
Bernier, F., Berna, A. (2001). Germins and germin-like proteins: Plant do-all proteins. but what do they do exactly? Plant Physiol. Biochem. 39, 545–554. doi: 10.1016/S0981-9428(01)01285-2
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K., et al. (2016). The genome sequences of arachis duranensis and arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446. doi: 10.1038/ng.3517
Bomireddy, D., Gangurde, S. S., Variath, M. T., Janila, P., Manohar, S. S., Sharma, V., et al. (2022). Discovery of major quantitative trait loci and candidate genes for fresh seed dormancy in groundnut. Agronomy 12, 404. doi: 10.3390/agronomy12020404
Carter, C., Thornburg, R. W. (1999). Germin-like proteins: Structure, phylogeny, and function. J. Plant Biol. 42, 97–108. doi: 10.1007/BF03031017
Carter, C., Thornburg, R. W. (2000). Tobacco nectarin I: purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. J. Biol. Chem. 275, 36726–36733. doi: 10.1074/jbc.M006461200
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Yang, Q., Chen, K., Zhao, S., Zhang, C., Pan, R., et al. (2019). Integrated microRNA and transcriptome profiling reveals a miRNA-mediated regulatory network of embryo abortion under calcium deficiency in peanut (Arachis hypogaea l.). BMC Genomics 20, 1–17. doi: 10.1186/s12864-019-5770-6
Christensen, A. B., Thordal-Christensen, H., Zimmermann, G., Gjetting, T., Lyngkjær, M. F., Dudler, R., et al. (2004). The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol. Plant-Microbe Interact. 17, 109–117. doi: 10.1094/MPMI.2004.17.1.109
Clevenger, J., Chu, Y., Scheffler, B., Ozias-Akins, P. (2016). A developmental transcriptome map for allotetraploid arachis hypogaea. Front. Plant Sci. 7 1446. doi: 10.3389/fpls.2016.01446
Dai, X., Zhuang, Z., Zhao, P. X. (2018). psRNATarget: A plant small RNA target analysis server, (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
Davidson, R. M., Reeves, P. A., Manosalva, P. M., Leach, J. E. (2009). Germins: A diverse protein family important for crop improvement. Plant Sci. 177, 499–510. doi: 10.1016/j.plantsci.2009.08.012
Ding, Y., Ma, Y., Liu, N., Xu, J., Hu, Q., Li, Y., et al. (2017). microRNAs involved in auxin signalling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 91, 977–994. doi: 10.1111/tpj.13620
Dodia, S. M., Joshi, B., Gangurde, S. S., Thirumalaisamy, P. P., Mishra, G. P., Narandrakumar, D., et al. (2019). Genotyping-by-sequencing based genetic mapping reveals large number of epistatic interactions for stem rot resistance in groundnut. Theor. Appl. Genet. 132, 1001–1016. doi: 10.1007/s00122-018-3255-7
Dunwell, J. M., Gibbings, J. G., Mahmood, T., Saqlan Naqvi, S. (2008). Germin and germin-like proteins: Evolution, structure, and function. Crit. Rev. Plant Sci. 27, 342–375. doi: 10.1080/07352680802333938
Dunwell, J. M., Khuri, S., Gane, P. (2000). Microbial relatives of the seed storage proteins of higher plants: Conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64, 153–179. doi: 10.1128/MMBR.64.1.153-179.2000
Dunwell, J. M., Purvis, A., Khuri, S. (2004). Cupins: The most functionally diverse protein superfamily? Phytochemistry 65, 7–17. doi: 10.1016/j.phytochem.2003.08.016
Fatima, M., Ma, X., Zhou, P., Zaynab, M., Ming, R. (2021). Auxin regulated metabolic changes underlying sepal retention and development after pollination in spinach. BMC Plant Biol. 21, 1–15. doi: 10.1186/s12870-021-02944-4
Gane, P. J., Dunwell, J. M., Warwickr, J. (1998). Modeling based on the structure of vicilins predicts a histidine cluster in the active site of oxalate oxidase. J. Mol. Evol. 46, 488–493. doi: 10.1007/PL00006329
Gangadhar, B. H., Mishra, R. K., Kappachery, S., Baskar, V., Venkatesh, J., Nookaraju, A., et al. (2021). Enhanced thermo-tolerance in transgenic potato (Solanum tuberosum l.) overexpressing hydrogen peroxide-producing germin-like protein (GLP). Genomics 113, 3224–3234. doi: 10.1016/j.ygeno.2021.07.013
Gangurde, S. S., Nayak, S. N., Joshi, P., Purohit, S., Sudini, H. K., Chitikineni, A., et al. (2021). Comparative transcriptome analysis identified candidate genes for late leaf spot resistance and cause of defoliation in groundnut. Int. J. Mol. Sci. 22, 4491. doi: 10.3390/ijms22094491
Gangurde, S. S., Wang, H., Yaduru, S., Pandey, M. K., Fountain, J. C., Chu, Y., et al. (2020). Nested-association mapping (NAM)-based genetic dissection uncovers candidate genes for seed and pod weights in peanut (Arachis hypogaea). Plant Biotechnol. J. 18, 1457–1471. doi: 10.1111/pbi.13311
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., Bairoch, A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., Bairoch, A. (2005). Protein identification and analysis tools on the ExPASy server. Proteomics Protoc. Handb., 571–607. doi: 10.1385/1-59259-890-0:571
Godfrey, D., Able, A. J., Dry, I. B. (2007). Induction of a grapevine germin-like protein (VvGLP3) gene is closely linked to the site of erysiphe necator infection: A possible role in defense? Mol. Plant-Microbe Interact. 20, 1112–1125. doi: 10.1094/MPMI-20-9-1112
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Guevara-Olvera, L., Ruíz-Nito, M., Rangel-Cano, R., Torres-Pacheco, I., Rivera-Bustamante, R., Muñoz-Sánchez, C., et al. (2012). Expression of a germin-like protein gene (CchGLP) from a geminivirus-resistant pepper (Capsicum chinense jacq.) enhances tolerance to geminivirus infection in transgenic tobacco. Physiol. Mol. Plant Pathol. 78, 45–50. doi: 10.1016/j.pmpp.2012.01.005
Guo, Y., Qiu, L.-J. (2013). Genome-wide analysis of the dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PloS One 8, e76809. doi: 10.1371/journal.pone.0076809
Hajyzadeh, M., Turktas, M., Khawar, K. M., Unver, T. (2015). miR408 overexpression causes increased drought tolerance in chickpea. Gene 555, 186–193. doi: 10.1016/j.gene.2014.11.002
He, Z.-D., Tao, M.-L., Leung, D. W. M., Yan, X.-Y., Chen, L., Peng, X.-X., et al. (2021). The rice germin-like protein OsGLP1 participates in acclimation to UV-b radiation. Plant Physiol. 186, 1254–1268. doi: 10.1093/plphys/kiab125
Houde, M., Diallo, A. O. (2008). Identification of genes and pathways associated with aluminum stress and tolerance using transcriptome profiling of wheat near-isogenic lines. BMC Genomics 9, 1–13. doi: 10.1186/1471-2164-9-400
Huang, L., Zhang, L., Zeng, R., Wang, X., Zhang, H., Wang, L., et al. (2020). Brassinosteroid priming improves peanut drought tolerance via eliminating inhibition on genes in photosynthesis and hormone signaling. Genes 11, 919. doi: 10.3390/genes11080919
Hu, R., Qi, G., Kong, Y., Kong, D., Gao, Q., Zhou, G. (2010). Comprehensive analysis of NAC domain transcription factor gene family in populus trichocarpa. BMC Plant Biol. 10, 1–23. doi: 10.1186/1471-2229-10-145
Jadhav, M. P., Gangurde, S. S., Hake, A. A., Yadawad, A., Mahadevaiah, S. S., Pattanashetti, S. K., et al. (2021). Genotyping-by-Sequencing based genetic mapping identified major and consistent genomic regions for productivity and quality traits in peanut. Front. Plant Sci. 2034. doi: 10.3389/fpls.2021.668020
Jovanović, Ž., Stanisavljević, N., Mikić, A., Radović, S., Maksimović, V. (2014). Water deficit down-regulates miR398 and miR408 in pea (Pisum sativum l.). Plant Physiol. Biochem. 83, 26–31. doi: 10.1016/j.plaphy.2014.07.008
Khan, S. A., Chen, H., Deng, Y., Chen, Y., Zhang, C., Cai, T., et al. (2020). High-density SNP map facilitates fine mapping of QTLs and candidate genes discovery for aspergillus flavus resistance in peanut (Arachis hypogaea). Theor. Appl. Genet. 133, 2239–2257. doi: 10.1007/s00122-020-03594-0
Knecht, K., Seyffarth, M., Desel, C., Thurau, T., Sherameti, I., Lou, B., et al. (2010). Expression of BvGLP-1 encoding a germin-like protein from sugar beet in arabidopsis thaliana leads to resistance against phytopathogenic fungi. Mol. Plant-Microbe Interact. 23, 446–457. doi: 10.1094/MPMI-23-4-0446
Komatsu, S., Kobayashi, Y., Nishizawa, K., Nanjo, Y., Furukawa, K. (2010). Comparative proteomics analysis of differentially expressed proteins in soybean cell wall during flooding stress. Amino Acids 39, 1435–1449. doi: 10.1007/s00726-010-0608-1
Kumar, R., Janila, P., Vishwakarma, M. K., Khan, A. W., Manohar, S. S., Gangurde, S. S., et al. (2020). Whole-genome resequencing-based QTL-seq identified candidate genes and molecular markers for fresh seed dormancy in groundnut. Plant Biotechnol. J. 18, 992–1003. doi: 10.1111/pbi.13266
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547. doi: 10.1093/molbev/msy096
Lane, B. G. (2002). Oxalate, germins, and higher-plant pathogens. IUBMB Life 53, 67–75. doi: 10.1080/15216540211474
Lane, B., Bernier, F., Dratewka-Kos, E., Shafai, R., Kennedy, T., Pyne, C., et al. (1991). Homologies between members of the germin gene family in hexaploid wheat and similarities between these wheat germins and certain physarum spherulins. J. Biol. Chem. 266, 10461–10469. doi: 10.1016/S0021-9258(18)99247-1
Lane, B., Dunwell, J. M., Ray, J., Schmitt, M., Cuming, A. (1993). Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 268, 12239–12242. doi: 10.1016/S0021-9258(18)31377-2
Lee, J.-Y., Lee, D.-H. (2003). Use of serial analysis of gene expression technology to reveal changes in gene expression in arabidopsis pollen undergoing cold stress. Plant Physiol. 132, 517–529. doi: 10.1104/pp.103.020511
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Liao, L., Hu, Z., Liu, S., Yang, Y., Zhou, Y. (2021). Characterization of germin-like proteins (GLPs) and their expression in response to abiotic and biotic stresses in cucumber. Horticulturae 7, 412. doi: 10.3390/horticulturae7100412
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li, L., Xu, X., Chen, C., Shen, Z. (2016). Genome-wide characterization and expression analysis of the germin-like protein family in rice and arabidopsis. Int. J. Mol. Sci. 17, 1622. doi: 10.3390/ijms17101622
Lu, M., Han, Y.-P., Gao, J.-G., Wang, X.-J., Li, W.-B. (2010). Identification and analysis of the germin-like gene family in soybean. BMC Genomics 11, 1–15. doi: 10.1186/1471-2164-11-620
Manosalva, P. M., Davidson, R. M., Liu, B., Zhu, X., Hulbert, S. H., Leung, H., et al. (2009). A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol. 149, 286–296. doi: 10.1104/pp.108.128348
Maruyama-Nakashita, A., Nakamura, Y., Watanabe-Takahashi, A., Inoue, E., Yamaya, T., Takahashi, H. (2005). Identification of a novel cis-acting element conferring sulfur deficiency response in arabidopsis roots. Plant J. 42, 305–314. doi: 10.1111/j.1365-313X.2005.02363.x
Nakata, M., Watanabe, Y., Sakurai, Y., Hashimoto, Y., Matsuzaki, M., Takahashi, Y., et al. (2004). Germin-like protein gene family of a moss, physcomitrella patens, phylogenetically falls into two characteristic new clades. Plant Mol. Biol. 56, 381–395. doi: 10.1007/s11103-004-3475-x
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., Tran, L. S. P. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49. doi: 10.1111/nph.12613
Pandey, M. K., Gangurde, S. S., Sharma, V., Pattanashetti, S. K., Naidu, G. K., Faye, I., et al. (2020). Improved genetic map identified major QTLs for drought tolerance-and iron deficiency tolerance-related traits in groundnut. Genes 12, 37. doi: 10.3390/genes12010037
Pandey, M. K., Kumar, R., Pandey, A. K., Soni, P., Gangurde, S. S., Sudini, H. K., et al. (2019). Mitigating aflatoxin contamination in groundnut through a combination of genetic resistance and post-harvest management practices. Toxins 11, 315. doi: 10.3390/toxins11060315
Pashkovskiy, P. P., Kartashov, A. V., Zlobin, I. E., Pogosyan, S. I., Kuznetsov, V. V. (2016). Blue light alters miR167 expression and microRNA-targeted auxin response factor genes in arabidopsis thaliana plants. Plant Physiol. Biochem. 104, 146–154. doi: 10.1016/j.plaphy.2016.03.018
Pei, Y., Li, X., Zhu, Y., Ge, X., Sun, Y., Liu, N., et al. (2019). GhABP19, a novel germin-like protein from gossypium hirsutum, plays an important role in the regulation of resistance to verticillium and fusarium wilt pathogens. Front. Plant Sci. 10, 583. doi: 10.3389/fpls.2019.00583
Raza, A., Mubarik, M. S., Sharif, R., Habib, M., Jabeen, W., Zhang, C., et al. (2022a). Developing drought-smart, ready-to-grow future crops. Plant Genome, e20279. doi: 10.1002/tpg2.20279
Raza, A., Salehi, H., Rahman, M. A., Zahid, Z., Haghjou, M. M., Najafi-Kakavand, S., et al. (2022b). Plant hormones and neurotransmitter interactions mediate antioxidant defenses under induced oxidative stress in plants. Front. Plant Sci. 13, 961872. doi: 10.3389/fpls.2022.961872
Raza, A., Su, W., Gao, A., Mehmood, S. S., Hussain, M. A., Nie, W., et al. (2021a). Catalase (CAT) gene family in rapeseed (Brassica napus l.): Genome-wide analysis, identification, and expression pattern in response to multiple hormones and abiotic stress conditions. Int. J. Mol. Sci. 22, 4281. doi: 10.3390/ijms22084281
Raza, A., Su, W., Hussain, M. A., Mehmood, S. S., Zhang, X., Cheng, Y., et al. (2021b). Integrated analysis of metabolome and transcriptome reveals insights for cold tolerance in rapeseed (Brassica napus l.). Front. Plant Sci. 1796. doi: 10.3389/fpls.2021.721681
Rietz, S., Bernsdorff, F. E., Cai, D. (2012). Members of the germin-like protein family in brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of sclerotinia sclerotiorum. J. Exp. Bot. 63, 5507–5519. doi: 10.1093/jxb/ers203
Saitou, N., Nei, M., Evolution (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharif, Y., Chen, H., Deng, Y., Ali, N., Khan, S., Zhang, C., et al. (2022). Cloning and functional characterization of a pericarp abundant expression promoter (AhGLP17-1P) from peanut (Arachis hypogaea l.). Front. Genet. 12. doi: 10.3389/fgene.2021.821281
Shasidhar, Y., Variath, M., Vishwakarma, M., Manohar, S., Gangurde, S., Sriswathi, M., et al. (2020). Improvement of three Indian popular groundnut varieties for foliar disease resistance and high oleic acid using SSR markers and SNP array in marker-assisted backcrossing. Crop J. 8, 1–15. doi: 10.1016/j.cj.2019.07.001
Sinha, P., Bajaj, P., Pazhamala, L. T., Nayak, S. N., Pandey, M. K., Chitikineni, A., et al. (2020). Arachis hypogaea gene expression atlas for fastigiata subspecies of cultivated groundnut to accelerate functional and translational genomics applications. Plant Biotechnol. J. 18, 2187–2200. doi: 10.1111/pbi.13374
Song, J. B., Gao, S., Sun, D., Li, H., Shu, X. X., Yang, Z. M. (2013). miR394 and LCR are involved in arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol. 13, 1–16. doi: 10.1186/1471-2229-13-210
Soni, P., Gangurde, S. S., Ortega-Beltran, A., Kumar, R., Parmar, S., Sudini, H. K., et al. (2020). Functional biology and molecular mechanisms of host-pathogen interactions for aflatoxin contamination in groundnut (Arachis hypogaea l.) and maize (Zea mays l.). Front. Microbiol. 11, 227. doi: 10.3389/fmicb.2020.00227
Stålberg, K., Ellerström, M., Josefsson, L.-G., Rask, L. (1993). Deletion analysis of a 2S seed storage protein promoter of brassica napus in transgenic tobacco. Plant Mol. Biol. 23, 671–683. doi: 10.1007/BF00021523
Sun, X., Xu, L., Wang, Y., Yu, R., Zhu, X., Luo, X., et al. (2015). Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus l.). BMC Genomics 16, 1–16. doi: 10.1186/s12864-015-1416-5
Sun, M., Ye, Z., Tan, J., Chen, S., Zhang, X., Tu, L. (2020). A cotton germin-like protein GbGLP2 controls fiber length via regulating genes involved in secondary cell wall synthesis. Mol. Breed. 40, 1–14. doi: 10.1007/s11032-020-01177-x
Su, W., Raza, A., Gao, A., Jia, Z., Zhang, Y., Hussain, M. A., et al. (2021). Genome-wide analysis and expression profile of superoxide dismutase (SOD) gene family in rapeseed (Brassica napus l.) under different hormones and abiotic stress conditions. Antioxidants 10, 1182. doi: 10.3390/antiox10081182
Swift, M. L. (1997). GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 37, 411–412. doi: 10.1021/ci960402j
Thompson, J. D., Gibson, T. J., Higgins, D. G. (2003). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinf. Chapter 2, Unit 2.3. doi: 10.1002/0471250953.bi0203s00
Toomer, O. T. (2018). Nutritional chemistry of the peanut (Arachis hypogaea). Crit. Rev. Food Sci. Nutr. 58, 3042–3053. doi: 10.1080/10408398.2017.1339015
Trindade, I., Capitão, C., Dalmay, T., Fevereiro, M. P., Dos Santos, D. M. (2010). miR398 and miR408 are up-regulated in response to water deficit in medicago truncatula. Planta 231, 705–716. doi: 10.1007/s00425-009-1078-0
Wang, T., Chen, X., Zhu, F., Li, H., Li, L., Yang, Q., et al. (2013). Characterization of peanut germin-like proteins, AhGLPs in plant development and defense. PloS One 8, e61722. doi: 10.1371/journal.pone.0061722
Wang, X., Zhang, H., Gao, Y., Sun, G., Zhang, W., Qiu, L. (2014). A comprehensive analysis of the cupin gene family in soybean (Glycine max). PloS One 9, e110092. doi: 10.1371/journal.pone.0110092
Woo, E.-J., Dunwell, J. M., Goodenough, P. W., Marvier, A. C., Pickersgill, R. W. (2000). Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7, 1036–1040. doi: 10.1038/80954
Xie, F., Zhang, B. (2015). Micro RNA evolution and expression analysis in polyploidized cotton genome. Plant Biotechnol. J. 13, 421–434. doi: 10.1111/pbi.12295
Xu, L., Dong, Z., Fang, L., Luo, Y., Wei, Z., Guo, H., et al. (2019). OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 47, W52–W58. doi: 10.1093/nar/gkz333
Yamahara, T., Shiono, T., Suzuki, T., Tanaka, K., Satoh, T., Takio, S., et al. (1999). Isolation of a germin-like protein with manganese superoxide dismutase activity from cells of a moss, barbula unguiculata. J. Biol. Chem. 274, 33274–33278. doi: 10.1074/jbc.274.47.33274
Yang, Z., Bielawski, J. P. (2000). Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15, 496–503. doi: 10.1016/S0169-5347(00)01994-7
Yuan, B., Yang, Y., Fan, P., Liu, J., Xing, H., Liu, Y., et al. (2021). Genome-wide identification and characterization of germin and germin-like proteins (GLPs) and their response under powdery mildew stress in wheat (Triticum aestivum l.). Plant Mol. Biol. Rep. 39, 1–12. doi: 10.1007/s11105-021-01291-w
Yaqoob, H., Tariq, A., Bhat, B. A., Bhat, K. A., Nehvi, I. B., Raza, A., Djalovic, I. , Prasad, P. V., Mir, R. A. (2023). Integrating genomics and genome editing for orphan crop improvement: A bridge between orphan crops and modern agriculture system. GM Crops & Food 14(1), 1–20. doi: 10.1080/21645698.2022.2146952
Yu, C. S., Chen, Y. C., Lu, C. H., Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins: Structure Function Bioinf. 64, 643–651. doi: 10.1002/prot.21018
Zaynab, M., Wang, Z., Hussain, A., Bahadar, K., Sajid, M., Sharif, Y., et al. (2021). ATP-binding cassette transporters expression profiling revealed its role in the development and regulating stress response in solanum tuberosum. Mol. Biol. Rep. 49, 1–14. doi: 10.1007/s11033-021-06697-z
Zhang, C., Chen, H., Cai, T., Deng, Y., Zhuang, R., Zhang, N., et al. (2017). Overexpression of a novel peanut NBS-LRR gene a h RRS 5 enhances disease resistance to r alstonia solanacearum in tobacco. Plant Biotechnol. J. 15, 39–55. doi: 10.1111/pbi.12589
Zhang, Y., Wang, X., Chang, X., Sun, M., Zhang, Y., Li, W., et al. (2018). Overexpression of germin-like protein GmGLP10 enhances resistance to sclerotinia sclerotiorum in transgenic tobacco. Biochem. Giophysical Res. Commun. 497, 160–166. doi: 10.1016/j.bbrc.2018.02.046
Zhuang, W., Chen, H., Yang, M., Wang, J., Pandey, M. K., Zhang, C., et al. (2019). The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 51, 865–876. doi: 10.1038/s41588-019-0402-2
Keywords: phylogenetic relations, gene evolution, environmental stress, functional annotation, micro-RNAs
Citation: Yang Q, Sharif Y, Zhuang Y, Chen H, Zhang C, Fu H, Wang S, Cai T, Chen K, Raza A, Wang L and Zhuang W (2023) Genome-wide identification of germin-like proteins in peanut (Arachis hypogea L.) and expression analysis under different abiotic stresses. Front. Plant Sci. 13:1044144. doi: 10.3389/fpls.2022.1044144
Received: 03 October 2022; Accepted: 20 December 2022;
Published: 23 January 2023.
Edited by:
Anil Kumar Singh, National Institute for Plant Biotechnology (ICAR), IndiaReviewed by:
Monika Bhuria, Panjab University, IndiaCopyright © 2023 Yang, Sharif, Zhuang, Chen, Zhang, Fu, Wang, Cai, Chen, Raza, Wang and Zhuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijian Zhuang, d2VpamlhbnpAZmFmdS5lZHUuY24=
†These authors have contributed equally to this work
‡ORCID: Yasir Sharif, orcid.org/0000-0001-6499-126X
Ali Raza, orcid.org/0000-0002-5120-2791
Weijian Zhuang, orcid.org/0000-0002-1340-5723
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.