- 1Research Institute of Subtropical Forestry, Chinese Academy of Forestry, Fuyang, Hangzhou, Zhejiang, China
- 2Henan Institute of Science and Technology Department of Resources & Environment, Xinxiang, Henan, China
Pecan (Carya illinoinensis Wang. K. Koch) is a deciduous tree of the Juglandaceae family with important economic value worldwide. Anthracnose of the pecan leaves and shuck is a devastating disease faced by pecan-growing areas in China. However, the causal species occurring on pecan remain largely unidentified. we collected samples of diseased pecan from the provinces of China, Leaves and fruits affected by anthracnose were sampled and subjected to fungus isolation, The morphological characters of all strains were observed and compared; Multi-locus phylogenetic analyses [Internally transcribed spacer (ITS), Actin (ACT), Calmodulin (CAL), Chitin synthase (CHS1), Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and b-tubulin (TUB2)] were performed on selected representative strains; examine their pathogenicity on leaves of pecan.The results showed that: (1) resulting in a total of 11 Colletotrichum isolates, Two Colletotrichum species were identifified to be C. fioriniae and C. fructicola; (2) Pathogenicity tests revealed that both species caused black spots on pecan leaves and fruit, The virulence of the different isolates varied substantially, with C. fioriniae PCJD179 being the most virulent; (3) The susceptibility levels of pecan tree varieties, ‘Mahan’ and ‘Kanza’, were determined, No significant differences were observed in the lesion sizes produced by the various isolates in ‘Kanza’, while there were signifificant differences in ‘Mahan’. This study is thefifirst to determine that C. fructicola and C. fioriniaecan cause anthracnose in pecan in China. It improves the understanding of the species that cause anthracnose in pecan and provides useful information for the effective control of this disease in China.
Introduction
Pecan (Carya illinoinensis Wang. K. Koch) is a deciduous tree of the Juglandaceae family. It is an important economic nut-producing tree worldwide. In China, pecan is planted mostly from 24 to 40° N in 14 provinces, mainly in Anhui, Zhejiang, Jiangsu, and Yunnan (Yao et al., 2014). In 2014, commercial pecan orchards covered ~8,500 ha in China (Zhang et al., 2015). By the end of 2020, the total planting area in the country had grown to approximately 68,000 ha. The national output in 2020 was 2,000 tons, while it was 4,500 tons in 2021 (personal investigation).
Colletotrichum is an important pathogenic fungus worldwide. It has numerous species and a wide plant host range. However, studies on its pathogenicity against pecan are limited. Pecan anthracnose caused by Glomerella cingulata (stonem.) was first reported in Georgia as early as 1914 (Rand, 1914). Subsequently, three Colletotrichum spp. were established to infect pecan: C. gloeosporioides, C. siamense, and C. nymphaeae (Mantz et al., 2010; Poletto et al., 2019; Zhang et al., 2019; Oh et al., 2021).
Anthracnose of the leaves and shuck in pecan is among the most devastating diseases that inflict the pecan-growing areas of China. On leaves, the lesions are irregular, necrotic, and usually surrounded by chlorotic rings. They can merge to form large necrotic areas. On the pecan shucks, the lesions are black, irregular, and slightly sunken (Figures 1A–C). Light yellow spots appear first, which enlarge and finally develop into irregularly shaped lesions with a chlorotic halo (Zhang et al., 2019). Visible symptoms of shuck include darkening, indentations, and irregularly shaped lesions (Oh et al., 2021). Other symptoms consist of circular lesions of up to 5 mm with clearly delineated or cracked centers, dark brown margins, and a yellowish halo on the leaves (Poletto et al., 2019). Serious infections by such fungi can thus result in autumn defoliation, fruit drop, and a decline in nut quality (Worley, 1979). It is crucial to accurately grasp the control time and select efficient fungicides, which requires the accurate identification of Colletotrichum spp. Therefore, the objective of the present study was to characterize and identify Colletotrichum isolates obtained from pecan tissues using morphological and molecular tools and to determine their pathogenicity in pecan.

Figure 1 Visible symptoms caused by Colletotrichum spp. on pecan. (A) Irregular necrotic spots on leaves. (B) Irregular grey lesions on the pericarp. (C) Internal symptoms in the fruit.
Materials and methods
Sampling and fungal isolation
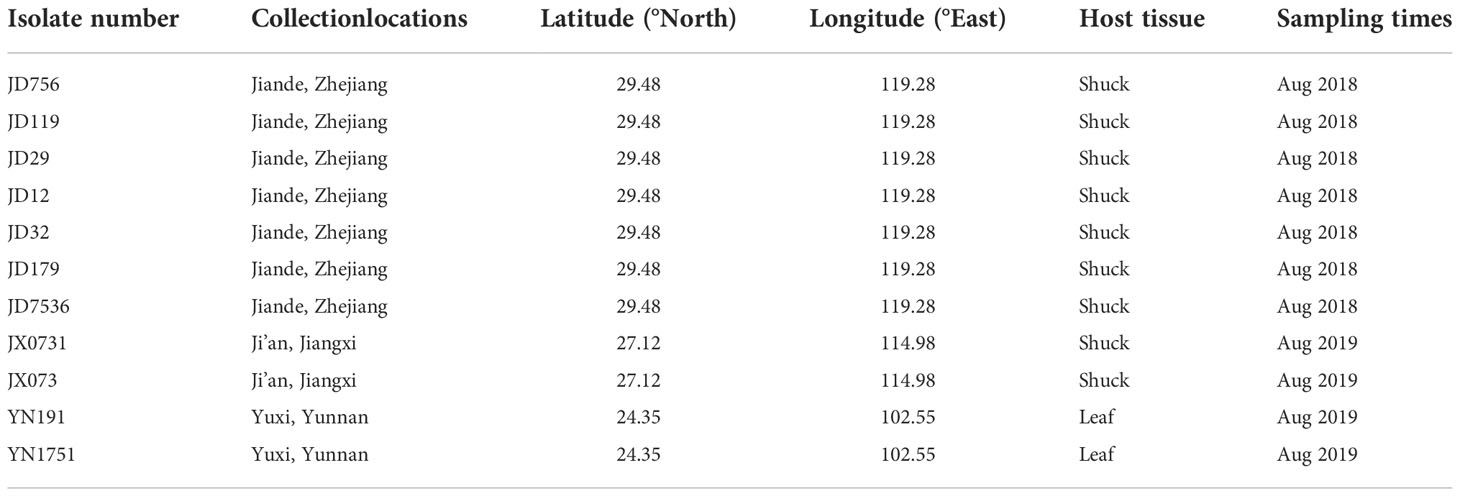
Samples of pecan leaves and shuck displaying typical anthracnose symptoms were collected from pecan trees in Jiande, Zhejiang Province; Ji’an, Jiangxi Province; and Yuxi, Yunnan Province in August 2018 and 2019 (Table 1). This period generally covers the early nut maturation and filling stages. Sampling followed the procedures described by Cai et al. (2009) and Prihastuti et al. (2009). Briefly, leaf and fruit surfaces were rinsed with distilled water, disinfected with 75% alcohol for 30 s, disinfected with 3% sodium hypochlorite for 2–3 min, rinsed with sterile water thrice, and dried with sterilized paper. Pieces were sliced from the junctions of diseased and healthy tissues, which were cut into squares with a side length of 3–4 mm. The excised tissues were placed onto potato dextrose agar (PDA, 20% diced potato, 2% glucose, and 1.5% agar in distilled water) plates and incubated at 28°C.Conidia were also collected, suspended in sterilized water, diluted to a concentration of 1 × 104 conidia per mL, and spread onto the surface of water agar (WA, 2% agar in distilled water) to generate discrete colonies (Choi et al., 1999). Six single colonies of each isolate were picked up with a sterilized needle (insect pin, 0.5 mm diameter) and transferred onto PDA plates.
Morphological characterization
The isolates were purified using single spore isolation and stored at 4°C on PDA slants for further use. They were stored on filter paper at -80°C for long-term preservation. Isolates were transferred from PDA slants to PDA plates and cultivated at 28°C under a 12-h photoperiod for 14 days. The following morphological characteristics were recorded: conidia, appressoria, conidiomata, and conidiophores. The mean lengths and widths of 100 randomly selected conidia from each isolate were measured using 100× magnification in a microscope (Nikon Ti-S inverted microscope, Japan). Among the 36 isolates thus obtained, 11 representative isolates were selected for further multi-locus phylogenetic analyses based on geographical location, morphology (e.g., colony shape and color and other physical characteristics of aerial mycelia and conidia), and ITS sequences.
DNA extraction
The genomic DNA of each isolate was isolated from 0.5 g of fresh hyphae using a DNA extraction kit (TaKaRa Bioengineering Co., Ltd, Dalian, China) and stored at −20°C. The ITS, GAPDH, ACT, TUB2, CHS-1, and CAL sequences were amplified and sequenced as previously described in Weir et al. (2012). The primers used, along with their respective sequences, are presented in Table 2.
PCR amplification
The PCR was performed according to the methods of Weir et al. (2012). All the PCR reactions were conducted in 25-μl volumes containing 12.5 μl PCR MasterMix (TIANGEN BIOTECH (BEIJING) CO., LTD., Beijing, China), 10 μM primers (both), 1 μl of the template DNA (20 ng μl-1), and 9.5 μl of double-distilled H2O. The PCR program to amplify ACT and GAPDH included a denaturation step at 94°C for 5 min, followed by 35 cycles of 94°C for 45 s, 59°C for 30 s, and 72°C for 2 min. The final cycle comprised 72°C for 10 min. The amplifications of ITS, TUB2, CHS-1, and CAL were also performed similarly with annealing temperatures of 56, 58, 58, and 57°C, respectively. The amplification products were analyzed on a 1.0% agarose gel in tris/borate/EDTA buffer. The PCR products were then purified and sequenced at Hangzhou Shangya Biotechnology Co., Ltd (Zhejiang, China).
Phylogenetic analysis
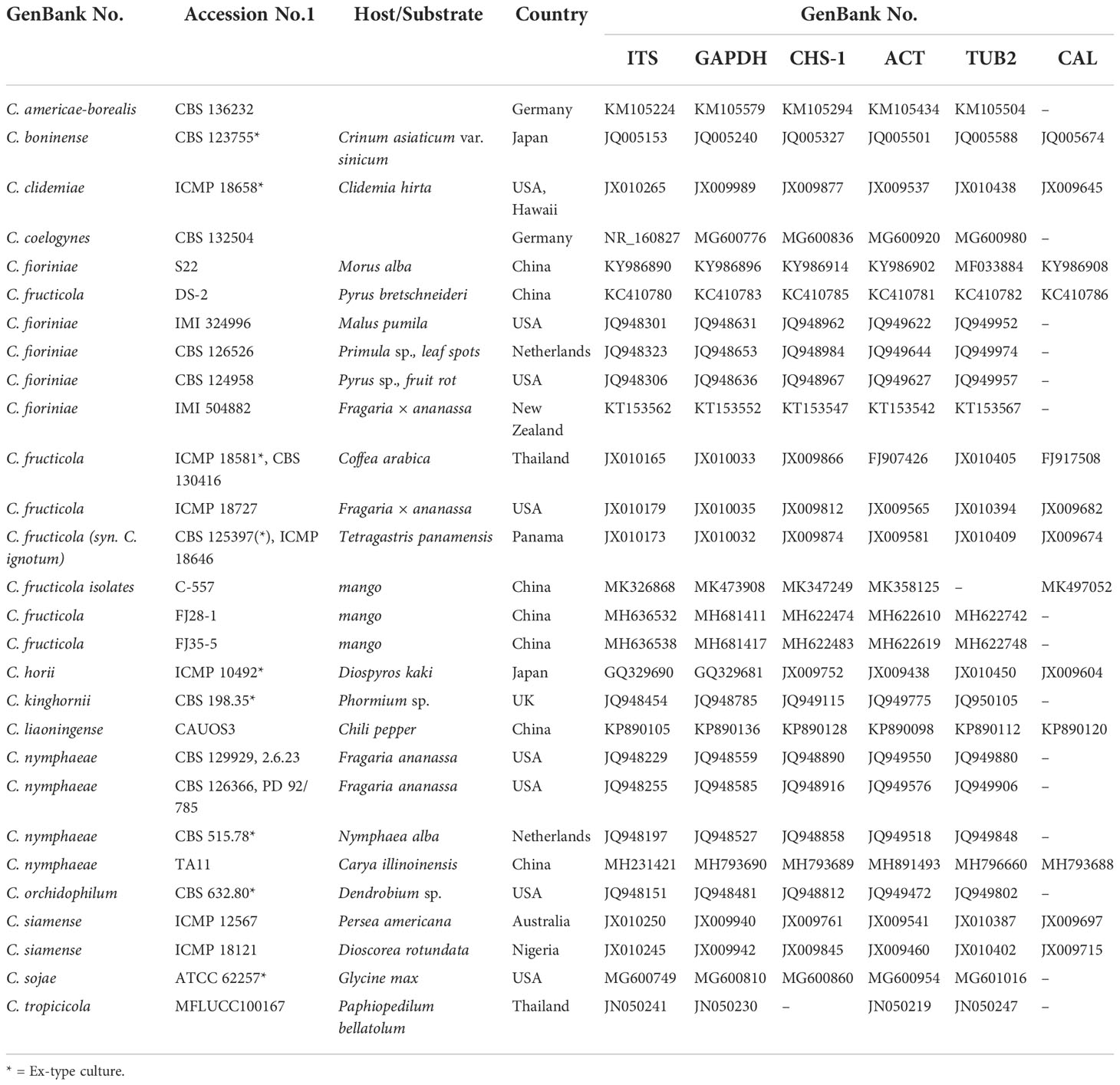
The reference sequences for ITS, GAPDH, CHS-1, ACT, TUB2, and CAL were downloaded from NCBI (Table 3). The forward and reverse sequences were edited and assembled using DNAMAN v. 8.0 (Lynnon Biosoft). According to the sequence of ITS-ACT-CAL-CHS-1-GAPDH-TUB2, six loci were combined and aligned using BioEdit v. 7.2.5 (Hall, 1999). Bayesian analyses were performed on concatenated alignments using MrBayes v. 3.2.1 (Ronquist et al., 2012). Maximum likelihood (ML) analyses were performed on the multilocus alignment using IQ-TREE (Nguyen et al., 2015), and the nucleotide substitution models were selected by Model Finder (Kalyaanamoorthy et al., 2017).
Pathogenicity assay
Same-sized healthy lesion-free leaves were collected from ‘Mahan’ subjects growing in a disease-free orchard located in Jiande, Zhejiang Province. Fruits were collected in the same manner as the leaves a month before harvest. Pathogenicity tests were conducted on pecan following the methods described by Kanchana-udomkan et al. (2004); Than et al. (2008), and Fu et al. (2019). Healthy pecan leaves and fruits were separately surface sterilized in 1% NaClO for 5 min, washed twice with sterile-distilled water, and air dried on sterile filter paper. Each fruit was inoculated with 20 μl of a conidial suspension (1 × 107 conidia/ml). The suspension was injected into the surface of non-wounded fruit using a microsyringe (Eppendorf, Shanghai, China). Control leaves and fruits were treated with 20 μl of distilled water. Each isolate was inoculated into five replicate fruits. The inoculated fruits were incubated in a moist chamber at 28°C and were examined daily for symptoms for 9 d.
Virulence assay
Virulence assay was performed in vivo on seven Colletotrichum spp. identified as representative isolates. These were selected according to the species, orchard province, and morphological characteristics. The virulence assay was carried out in a greenhouse of the Institute of Subtropical Forestry, Chinese Academy at Forestry Sciences (Zhejiang, China) from late June to August 2020. During the 5-year black spot resistance investigation on different cultivars preserved in the Pecan Resource Garden in Jiande City, Zhejiang, China, we found highly resistant cultivars, such as ‘Kanza’, and susceptible cultivars, such as ‘Mahan’. In this experiment, annually grafted container seedlings of the highly resistant variety, ‘Kanza’, and the susceptible variety, ‘Mahan’, served as plant materials. The fourth, fifth, and sixth pairs (from bottom to top) of pinnate compound leaves were selected, surface sterilized with 75% ethanol, and rinsed with sterile water. Each leaf was impaled at its middle near the midvein and wounded five times in a 5 mm region with a sterilized needle. The 1 × 107 ml-1 spore suspension was dropped on the wounds using a pipette. Some wounded leaves also served as controls and were inoculated with the same amount of sterile water instead of the spore suspension. Sterile damp absorbent cotton was placed on the pinnate compound leaves after air-drying, and sealed bags were used to retain the moisture. The experiment was set up with three biological replicates per isolate. The bags and cotton were removed after two days. The pathogenicity of each isolate was determined by evaluating the diameters of the disease lesions after 18 days. The one-way analysis of variance was performed with SPSS v. 16.0 software; means were compared using Duncan’s test at the significance level of 0.05.
Results
Fungal isolates
From 2018 to 2019, pecan leaves and shuck displaying anthracnose symptoms were collected from three pecan orchards in three provinces of China. Colletotrichum isolates associated with pecan anthracnose were collected from 60 affected pecan samples. Leaves were collected for fungal isolation, resulting in a total of 36 Colletotrichum isolates identified based on morphology and ITS sequences. A total of 11 representative isolates were chosen for further analyses based on their morphology (colony shape, color, and conidial morphology), ITS sequences, types of symptoms, and origin (Table 1). At least two isolates from each field were chosen for further analysis.
Phylogenetic analyses
In total, 36 Colletotrichum isolates were obtained from three pecan orchards. Eleven single-spore representative isolates were used for the following morphological characterization and phylogenetic analyses (Table 4). These comprised two C. fructicola isolates from leaves and six C. fioriniae and three C. fructicola isolates from the shuck.
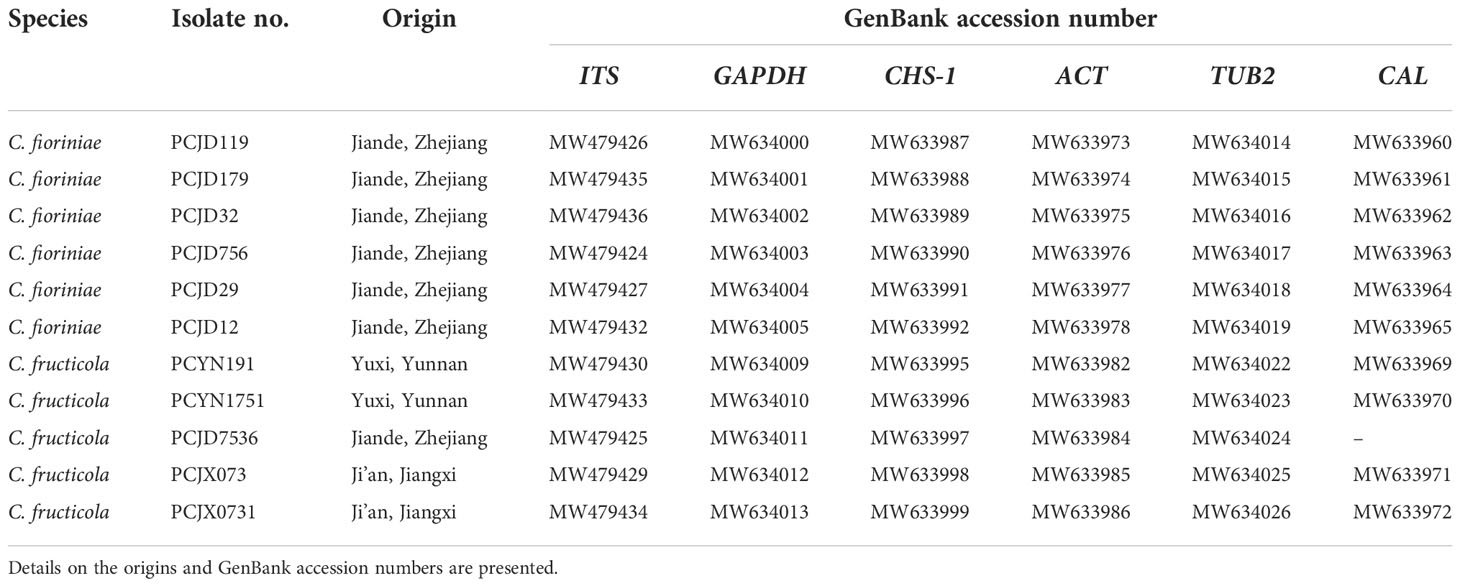
Table 4 List of 11 representative isolates of two Colletotrichum spp. collected from pecan in China.
Phylogenetic analyses on the six loci (ITS, ACT, CAL, GADPH, CHS-1, and TUB2) of the Colletotrichum spp. included 11 isolates. The sequences of ITS, GAPDH, ACT, TUB2, CHS-1, and CAL genes of Colletotrichum spp. isolates from pecan were deposited to Genbank (Table 4). They were compared with reference sequences of Colletotrichum isolated from other countries and plant hosts available on GenBank (Table 3). The C. boninense (CBS 123755*) was used as the outgroup. For the maximum likelihood inference, the best fit model for the six loci was set at HKY+I+G4+F with UFbootstrap 20000. For the Bayesian inference, the best fit model was HKY+F+G4 with a gamma rate for six loci. The posterior probabilities of the Bayesian tree were consulted to confirm the topology of the maximum likelihood tree. Phylogenetic analysis provided enough information to distinguish two Colletotrichum species; six and five isolates belonged to C. fioriniae and C. fructicola, respectively (Figure 2). Among the seven isolates collected from the orchards in Jiande, six were C. fioriniae and one was C. fructicola. The four isolates from the orchards in Yunnan and Jiangxi were all C. fructicola (Figure 2).
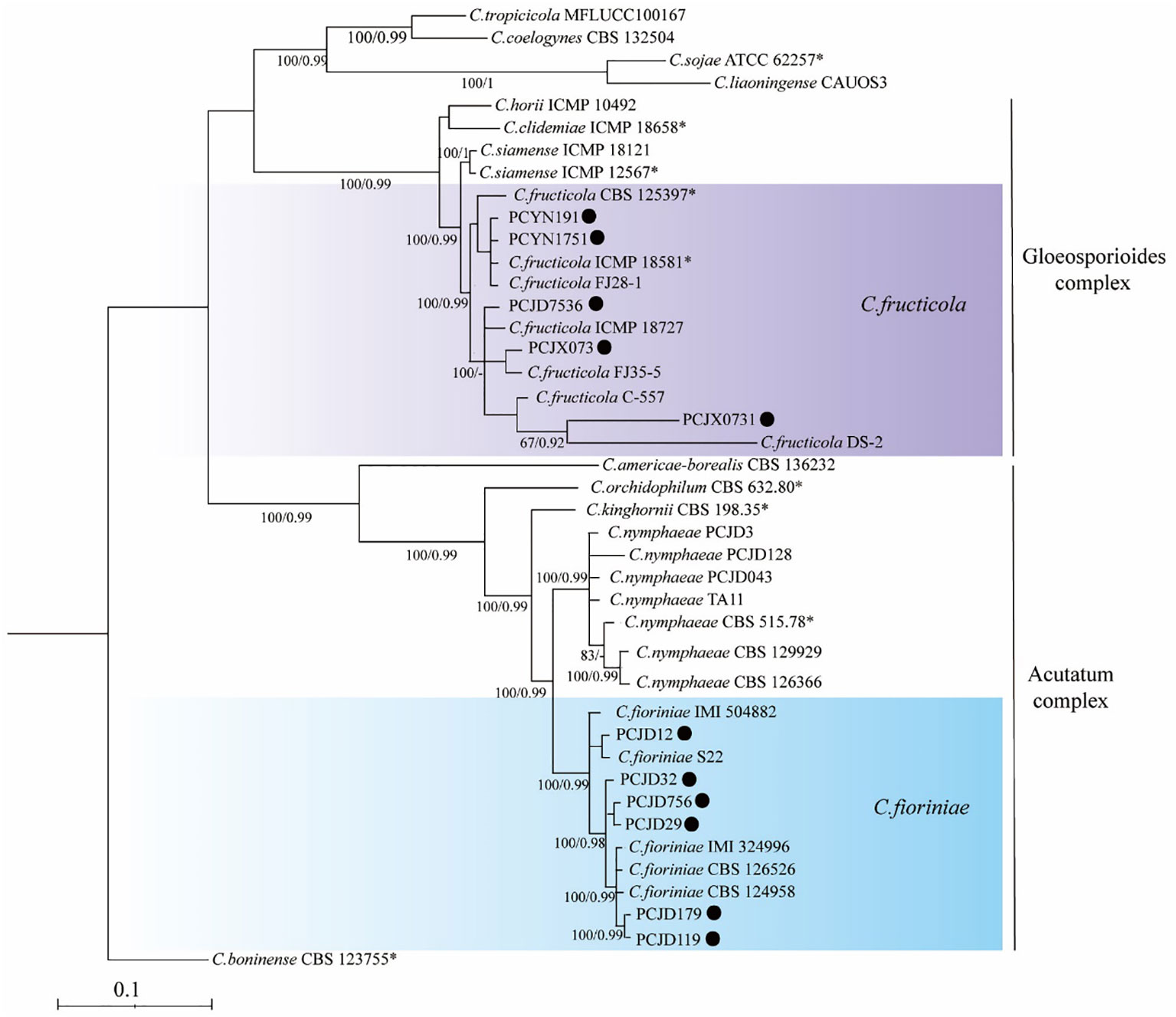
Figure 2 A maximum likelihood phylogenetic tree based on concatenated sequences of internal transcribed spacer (ITS), actin (ACT), calmodulin (CAL), glyceraldehyde-3phosphate dehydrogenase (GAPDH), β-tubulin (TUB2), and chitin synthase 1 (CHS-1). The tree illustrates the relationships between Colletotrichum fioriniae and C. fructicola. C. boninense (CBS 123755*) was used as an outgroup. Bayesian posterior probability values ≥ 0.90 and UFbootstrap support values ≥ 50% are shown at the nodes.
Morphological characteristics
The diameter of the colonies was measured daily for five days to calculate their mycelial growth rate (mm/d). The shape, color, and density of colonies were recorded after 14 days. Differences in colony morphology were observed between the two species identified when grown on PDA. Colonies generally showed dense, white to greyish or red growth (Figure 3). The mycelia of C. fructicola appeared dark grey on PDA plates after 14 days; their orange conidial masses were yellow and cylindrical or oval with an area of 16.8 ± 1.5 × 5.4 ± 0.4 μm2. Colletotrichum fioriniae was pinkish in color after 14 days of growth on PDA, and the conidial masses were orange. Its conidia were spindle-shaped with an area of 15.5 ± 1.5 × 5.2 ± 0.3 μm2. The mycelium growth rate varied from 12.3 to 15.6 mm/day (average = 14.6 mm/day) for C. fructicola and 10.3 to 13.2 mm/day (average = 11.24 mm/day) for C. fioriniae.
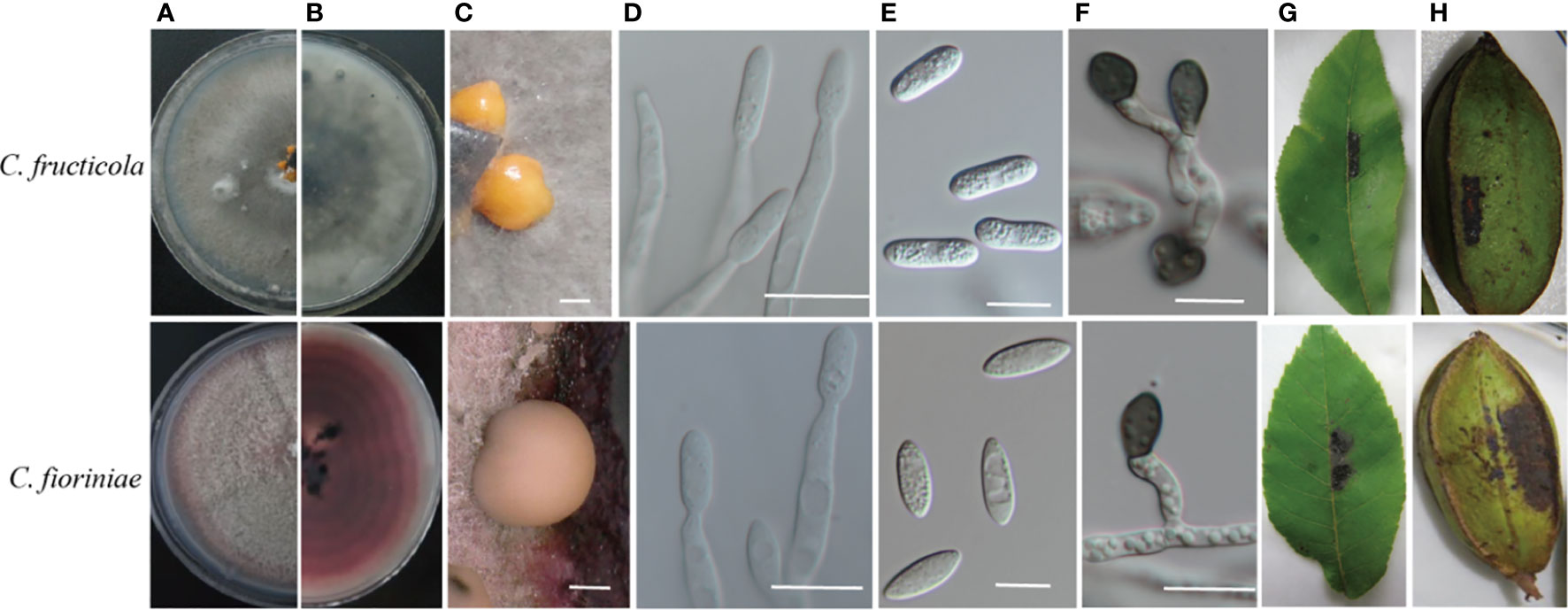
Figure 3 Cultural characteristics and pathogenicity of two Colletotrichum species isolated from the pecan tree. (A, B) front and back views of 14-days-old PDA culture; (C)conidiomata; (D) conidiophores; (E) conidia; (F) appressoria. The symptoms were caused by (G, H) appearing on the leaves 6 days after inoculation and on the shuck of the fruit 20 days after inoculation with conidia. (C, E, F): Bar = 10 µm. (D): Bar = 50 µm.
Pathogenicity and virulence assays
All the Colletotrichum isolates were pathogenic to the detached pecan leaves and shucks. The inoculated leaves, pericarps, and nuts showed necrotic spots, while these tissues remained healthy in the controls. No lesions were induced in the control tissues that had been inoculated with sterile water. The morphology of fungal colonies re-isolated from the symptomatic shuck was the same as those produced by the original isolate used for inoculation, satisfying Koch’s postulate (Figure 3).
In the virulence assay, the seven isolates caused symptoms on the leaves of 1-year-old pecan seedlings and showed different levels of virulence. The lesion diameters on the attached leaves start to differ 18 days after inoculation with various isolates (Figures 4A, B). No significant differences were observed in the lesion sizes produced on the highly resistant variety, ‘Kanza’, by the various isolates (p = 0.29), while there were significant differences on the susceptible variety, ‘Mahan’ (p = 0.001), indicating different virulence levels among the seven isolates. PCJD179 was noted as the strongest.
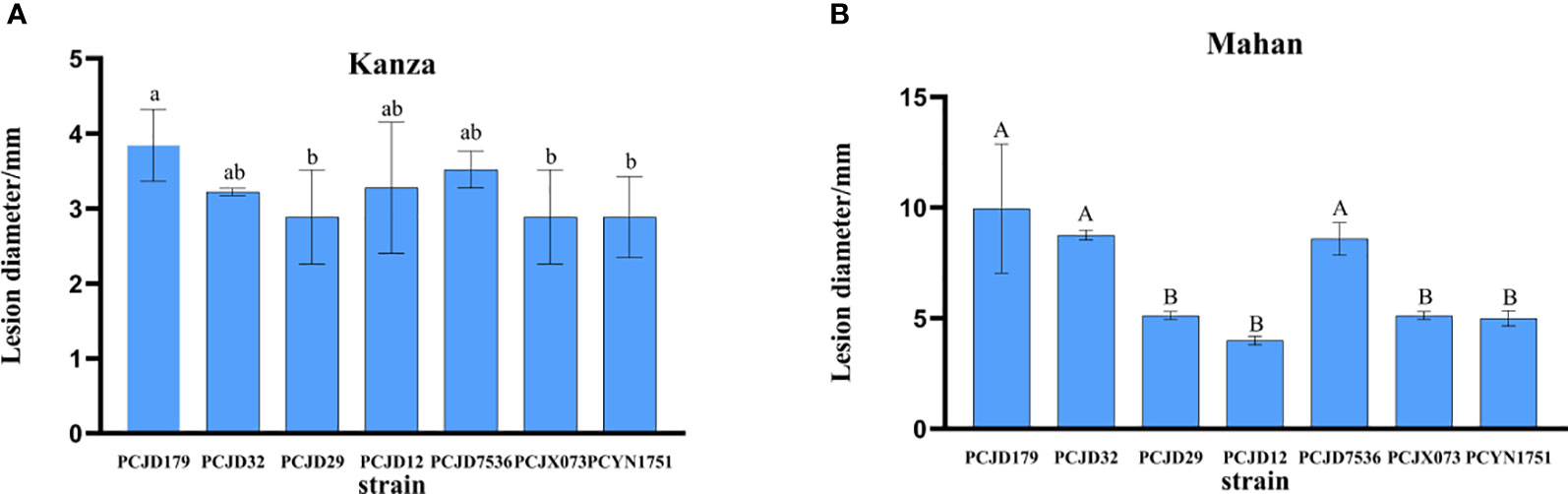
Figure 4 Lesion lengths on wounded pecan leaves at 18 dpi with spore suspensions of seven Colletotrichum spp. Different lowercase letters above the error bars indicate significant differences at p = 0.05. Different capital letters above the error bars indicate significant differences at p = 0.01. (A) ‘Kanza’; (B) ‘Mahna’.
The virulence assay showed that the lesions caused by Colletotrichum spp. Were mainly restricted to the inoculated areas of the pecan variety, ‘Kanza’, and their diameters did not extend beyond 4 mm (Figure 4A). There were significant differences in the infection lesions on the ‘Mahan’ variety. The lesion diameters caused by PCJD179, PCJD32, and PCJD29 isolates of C. fioriniae and the PCJD7536 isolate of C. fructicola all exceeded 5.1 mm on ‘Mahan’. Among them, the diameter of the lesion caused by the isolate PCJD179 reached up to 10 mm, indicating that the isolate was relatively the higher virulent (Figure 4B).
Additionally, different isolates of Colletotrichum species displayed pathogenic differences. For example, the C. fioriniae isolates PCJD179, PCJD32, and PCJD29 produced large necrotic spots (5.1–10.0 mm). However, the spot diameters caused by PCJD12 in this species were 4.0 mm, and they were concentrated near the inoculation point. The spots produced by C. fructicola isolate, PCJD7536, were also significantly larger than those produced by isolates, PCJX073 and PCYN1751.
Discussion
Colletotrichum spp. are important plant pathogenic fungi that cause a variety of plant diseases (Cannon et al., 2012; Dean et al., 2012; Diao et al., 2017; Guarnaccia et al., 2017). Previously, the taxonomy of Colletotrichum spp. has mainly been based on the host range and morphological characteristics (Von Arx, 1957; Sutton, 1980). Traditional classification methods do not effectively distinguish between the relatively complex Colletotrichum species because of their relatively high levels of genetic variability (Cai et al., 2009; Hyde et al., 2009; Rojas et al., 2010; Cannon et al., 2012; Damm et al., 2012a; Damm et al., 2012b). ITS sequence primers were designed to amplify the ribosomal genes of fungi. This method is a powerful means to identify and phylogenetically analyze many species at once (White et al., 1990). However, some complex species cannot be effectively identified using the ITS region alone because their support rates are low. In recent years, the identification of Colletotrichum species has significantly improved by combinations of multi-gene sequence and morphological analyses. Genes that produce good differentiation include glyceraldehyde 3-phosphate dehydrogenase (GAPDH), calmodulin (CAL), actin (ACT), β-tubulin (TUB2), and chitin synthase (CHS-1) (Damm et al., 2012a; Damm et al., 2012b; Weir et al., 2012; Damm et al., 2013; Damm et al., 2014; Yan et al., 2015; Guarnaccia et al., 2017). As a result, complexes with 15 and 14 species have been identified in the genus Colletotrichum (Marin-Felix et al., 2017; Damm et al., 2019). To better characterize the pathogen, multiple traits of the Colletotrichum spp. causing anthracnose in pecan in China are worth studying using phylogenetic analyses. Here, we detected 11 isolates from three orchards and identified them as C. fioriniae and C. fructicola. This is the first report of the two species being pathogens of pecan in China. This is also the first report of C. fioriniae being a pathogen of pecan worldwide. Thus, our results increase the understanding of the pathogenicity of Colletotrichum species against pecan.
C. fructicola was the first species isolated from shuck and leaves of pecan in Zhejiang, Yunnan, and Jiangxi provinces. The five isolates of C. fructicola mainly infected the shuck and leaves and were clustered on two branches of the phylogenetic tree. The pathogenic bacteria isolated from the diseased leaves of pecan trees from Yunnan were C. fructicola. The dominant species in the anthracnose of Chinese Camellia oleifera leaves was C. fructicola, which is in agreement with the observations of Li et al. (2016) and Wang et al. (2020). The six isolates of C. fioriniae were all isolated from the shuck and were clustered on four branches of the phylogenetic tree. This indicated that the pathogenicity of some fungi in the Colletotrichum genus may be tissue-specific (Fu et al., 2019; Wang et al., 2020). Moreover, anthracnose-causing Colletotrichum in Camellia oleifera Abel has previously been reported to differ in the composition, structure, and dominant phyla on leaves and fruit, where dominant phyla presented genetic differentiation among the various geographically separated populations (Li et al., 2016; Wang et al., 2020). Due to the small sample size, our study failed to reflect the distribution of Colletotrichum species and genetic variation in the mechanisms by which they cause anthracnose in pecan trees. This information could provide a theoretical basis for formulating targeted disease prevention and control strategies.
C. fructicola can cause anthracnose in the plants of several genera, including Citrus reticulata, Capsicum annuum, Camellia sinensis, and Mangifera indica (Huang et al., 2013; Lima et al., 2013; Liu et al., 2015; Diao et al., 2017). This Colletotrichum species has been associated with a certain geographical preference and is mainly distributed in the Yangtze River Basin in China (Fu et al., 2019). We found that C. fructicola was isolated from Zhejiang, Jiangxi, and Yunnan provinces. Additionally, the isolates from these three locations were clustered on two branches of the phylogenetic tree. The PCJD7536, PCJX073, and PCJX0731 isolates isolated from the shuck were clustered on one branch, and the PCYN1751 and PCYN191 isolates from the leaves were clustered on one branch. The pecans in Jiangxi Province were introduced from Jiande, Zhejiang Province. PCJD7536, PCJX073, and PCJX0731 were clustered in one branch, which might be related to the plants’ origin or the host tissues (Wang et al., 2020; Fu et al., 2019). In China, the cultivation of pecan was initiated in Yunnan and Zhejiang in the 1960s. Because of the recent initiation of large cultivation of pecan in Jiangxi, most of the pecan here was cultivated from Zhejiang-primed cultivars. Thus, the introduced species were genetically similar. In Yunnan, pecan grows as a native variety. The local climate and ecological environment differ from Zhejiang and are geographically distant. A larger number of samples are thus needed for further in-depth investigations.
Differences were revealed in the pathogenicity of the species or isolates of Colletotrichum. C. fioriniae and C. fructicola cause disease on the leaves and shuck of pecan trees, while the seven isolates of both species presented varying degrees of pathogenicity. Pathogenicity differentiation between and within the Colletotrichum spp. has been previously reported in chili (Capsicum spp.), citrus (Citrus spp.), and pear (Pyrus spp). Pathogenicity associated with various regions, varieties, and tissues of the host is also established (Diao et al., 2017; Guarnaccia et al., 2017; Fu et al., 2019). The present study preliminarily determined the pathogenicity of Colletotrichum which inflicts pecan trees with anthracnose. The symptoms of anthracnose are affected by various factors, including temperature, relative humidity, variety, and the number of pathogens (Han et al., 2016; Mo et al., 2018). There may be some differences between the results of virulence assays performed in the laboratory and those on trees growing in natural environmental conditions in the orchards.
Data availability statement
The dataets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JC conceived this project. JC and YZ designed experiments and interpreted the results. JC wrote the manuscript. FZ and JS provided technical guidance for the experiment. JC, DW, and YZ performed the experiments and analyzed the data. XY provided experimental materials and funds. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the Fundamental Research Funds for the Central Non-Profit Research Institution of CAF (CAFYBB2020SY014).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Cai, L., Hyde, K. D., Taylor, P. W. J., Weir, B. S., Waller, J. M., Abang, M. M., et al. (2009). A polyphasic approach for studying Colletotrichum. Fungal Divers. 39, 183–204. doi: 10.1016/j.riam.2009.11.001
Cannon, P. F., Damm, U., Johnston, P. R., Weir, B. S. (2012). Colletotrichum-current status and future directions. Stud. Mycol. 73, 181–213. doi: 10.3114/sim0014
Carbone, I., Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 91, 553–556. doi: 10.2307/3761358
Choi, Y. W., Hyde, K. D., Ho, W. H. (1999). Single spore isolation of fungi. Fungal Diversity 3, 29–38.
Damm, U., Cannon, P. F., Liu, F., Barreto, R. W., Guatimosim, E., Crous, P. W. (2013). The Colletotrichum orbiculare species complex: important pathogens of field crops and weeds. Fungal Divers. 61, 29–59. doi: 10.1007/s13225-013-0255-4
Damm, U., Cannon, P. F., Woudenberg, J. H. C., Crous, P. W. (2012b). The Colletotrichum acutatum species complex. Stud. Mycol. 73, 37–113. doi: 10.3114/sim0010
Damm, U., Cannon, P. F., Woudenberg, J. H. C., Johnston, P. R., Weir, B. S., Tan, Y. P., et al. (2012a). The Colletotrichum boninense species complex. Stud. Mycol. 73, 1–36. doi: 10.3114/sim0002
Damm, U., O’Connell, R. J., Groenewald, J. Z., Crous, P. W. (2014). The Colletotrichum destructivum species complex hemibiotrophic pathogens of forage and field crops. Stud. Mycol. 79, 49–84. doi: 10.1016/j.simyco.2014.09.003
Damm, U., Sato, T., Alizadeh, A., Groenewald, J. Z., Crous, P. W. (2019). The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Stud. Mycol. 92, 1–46. doi: 10.1016/j.simyco.2018.04.001
Dean, R., Kan, J. L. V., Pretorius, Z. A., Hammond-kosack, K. H., Pietro, A. D., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Diao, Y. Z., Zhang, C., Liu, F., Wang, W. Z., Liu, L., Cai, L., et al. (2017). Colletotrichum species causing anthracnose disease of chili in China. Persoonia 38, 20–37. doi: 10.3767/003158517X692788
Fu, M., Crous, P. W., Bai, Q., Zhang, P. F., Xiang, J., Guo, Y. S., et al. (2019). Colletotrichum species associated with anthracnose of Pyrus spp. in China. Persoonia 42, 1–35. doi: 10.3767/persoonia.2019.42.01
Glass, N. L., Donaldson, G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330. doi: 10.1002/bit.260460112
Guarnaccia, V., Groenewald, J. Z., Polizzi, G., Crous, P. W. (2017). High species diversity in Colletotrichum associated with citrus diseases in Europe. Persoonia 39, 32–50. doi: 10.3767/persoonia.2017.39.02
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Han, Y. C., Zeng, X. G., Xiang, F. Y., Ren, L., Chen, F. Y., Gu, Y. C. (2016). Distribution and characteristics of Colletotrichum spp. associated with anthracnose of strawberry in hubei, China. Plant Dis. 100, 996–1006. doi: 10.1094/PDIS-09-15-1016-RE
Huang, F., Chen, G. Q., Hou, X., Fu, Y. S., Cai, L., Hyde, K. D., et al. (2013). Colletotrichum species associated with cultivated citrus in China. Fungal Divers. 61, 61–74. doi: 10.1007/s13225-013-0232-y
Hyde, K. D., Cai, L., Cannon, P. F., Crouch, J. A., Crous, P. W., Damm, U., et al. (2009). Colletotrichum-names in current use. Fungal Divers. 39, 147–182. doi: 10.1016/j.riam.2009.11.001
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kanchana-udomkan, C., Taylor, P. W. J., Mongkolporn, O. (2004). Development of a bioassay to study anthracnose infection of Capsicum chinense jacq. fruit caused by Colletotrichum capsici. Thai J. Agric. Sci. 37, 293–297.
Lima, N. B., De, A., Batista, M. V., Morais, J. M. A., Barbosa, M. A. G., Michereff, S. J., et al. (2013). Five Colletotrichum species are responsible for mango anthracnose in northeastern Brazil. Fungal Divers. 61, 75–88. doi: 10.1007/s13225-013-0237-6
Liu, F., Weir, B. S., Damm, U., Crous, P. W., Wang, Y., Liu, B., et al. (2015). Unravelling Colletotrichum species associated with camellia: employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia 35, 63–86. doi: 10.3767/003158515X687597
Li, H., Zhou, G. Y., Liu, J. Y., Xu, J. P. (2016). Population genetic analyses of the fungal pathogen Colletotrichum fructicola on tea-oil trees in China. PloS One 11, e0156841. doi: 10.1371/journal.pone.0156841
Mantz, G., Minhot, R., Morrelli, G., Maiale, S. (2010). First report of Colletotrichum gloeosporioides causing pecan anthracnose in Argentina. J. Plant Pathol. 92, 544. doi: 10.2307/41998837
Marin-Felix, Y., Groenewald, J. Z., Cai, L., Chen, Q., Marincowitz, S., Barnes, I., et al. (2017). Genera of phytopathogenic fungi: GOPHY 1. Stud. Mycol. 86, 99–216. doi: 10.1016/j.simyco.2017.04.002
Mo, J., Zhao, G., Li, Q., Soalngi, G. S., Tang, L., Guo, T. X., et al. (2018). Identification and characterization of Colletotrichum species associated with mango anthracnose in guangxi, China. Plant Dis. 102, 1283–1289. doi: 10.1094/PDIS-09-17-1516-RE
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
O’Donnell, K., Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7, 103–116. doi: 10.1006/mpev.1996.0376
O’Donnell, K., Nirenberg, H. I., Aoki, T., Cigelnik, E. (2000). A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycosci. 41, 61–78. doi: 10.1007/BF02464387
Oh, J. Y., Heo, J. I., Lee, D. H. (2021). First report of anthracnose on pecan (Carya illinoinensis) caused by colletotrichum siamense in Korea. Plant Dis. 105, 3296. doi: 10.1094/PDIS-12-20-2709-PDN
Poletto, T., Muniz, M. F. B., Blume, E., Fantinel, V. S., Reiniger, L. R. S., Brioso, P. S. T., et al. (2019). First report of Colletotrichum nymphaeae causing anthracnose on Carya illinoinensis in Brazil. Plant Dis. 103, 3277. doi: 10.1094/PDIS-04-19-0896-PDN
Prihastuti, H., Cai, L., Chen, H., Mckenzie, E. H. C., Hyde, K. D. (2009). Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 39, 89–109. doi: 10.1016/j.riam.2009.11.001
Rojas, E. I., Rehner, S. A., Samuels, G. J., Van Bael, S. A., Herre, E. A., Cannon, P., et al. (2010). Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panama: multilocus phylogenies distinguish pathogen and endophytes. Mycologia 102, 1318–1338. doi: 10.3852/09-244
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sutton, B. C. (1980). “Fungi imperfecti with pycnidia, acervuli and stromata,” in The coelomycetes (Kew, UK: C. M. I.), 523–537. https://doi.org/Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panamá: multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes RID C-7783-2011.
Templeton, M. D., Rikkerink, E. H., Solon, S. L., Crowhurst, R. N. (1992). Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene 122, 225–230. doi: 10.1016/0378-1119(92)90055-T
Than, P. P., Jeewon, R., Hyde, K. D., Pongsupasamit, S., Mongkolporm, O., Taylor, P. W. J. (2008). Characterization and pathogenicity of Colletotrichum species associated with anthracnose on chilli (Capsicum spp.) in Thailand. Plant Pathol. 57, 562–572. doi: 10.1111/j.1365-3059.2007.01782.x
Wang, Y. X., Chen, J. Y., Cheng, J. Y., Zheng, L., Huang, J. B., Li, D. W. (2020). Identification and characterization of Colletotrichum species associated with anthracnose disease of camellia oleifera in China. Plant Dis. 104, 474–482. doi: 10.1094/PDIS-11-18-1955-RE
Weir, B. S., Johnston, P. R., Damm, U. (2012). The Colletotrichum gloeosporioides species complex. Stud. Mycol. 73, 115–180. doi: 10.3114/sim0011
White, T. J., Bruns, T., Lee, S., Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. pages 315-322 in: PCR protocols: A guide to methods and applications. Eds. Innis, M., Gelfand, D., Sninsky, J., White, T. (San Diego, CA: Academic Press). doi: 10.0000/PMID1793
Worley, R. E. (1979). Pecan yield, quality, nutlet set, and spring growth as a response to time of fall defoliation. J. Am. Soc Hortic. Sci. 104, 192–194. doi: 10.1007/BF00038955
Yan, J. Y., Jayawardena, M. M. R. S., Goonasekara, I. D., Wang, Y., Zhang, W., Liu, M., et al. (2015). Diverse species of Colletotrichum associated with grapevine anthracnose in China. Fungal Divers. 71, 233–246. doi: 10.1007/s13225-014-0310-9
Yao, X. H., Chang, J., Wang, K. L. (2014). The research proceeding of pecan in China (Beijing: Science Press), 109–111.
Zhang, Y. B., Meng, K., Shu, J. P., Zhang, W., Wang, H. J. (2019). First report of anthracnose on pecan (Carya illinoinensis) caused by Colletotrichum nymphaeae in China. Plant Dis. 103, 6, 1432. doi: 10.1094/PDIS-11-18-1968-PDN
Keywords: pecan, black-spot diseases, colletotrichum spp., pathogen identification, disease resistance
Citation: Chang J, Zhai F, Zhang Y, Wang D, Shu J and Yao X (2022) Identification and characterization of Colletotrichum fioriniae and C. fructicola that cause anthracnose in pecan. Front. Plant Sci. 13:1043750. doi: 10.3389/fpls.2022.1043750
Received: 14 September 2022; Accepted: 07 November 2022;
Published: 23 November 2022.
Edited by:
Youxiong Que, Fujian Agriculture and Forestry University, ChinaReviewed by:
Andreia Loureiro, University of Lisbon, PortugalNimal Adikaram, National Institute of Fundamental Studies (NIFS), Sri Lanka
Copyright © 2022 Chang, Zhai, Zhang, Wang, Shu and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yabo Zhang, eWJ6aGFuZ0BjYWYuYWMuY24=
 Jun Chang
Jun Chang Fengyan Zhai2
Fengyan Zhai2 Yabo Zhang
Yabo Zhang