
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 14 November 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1042889
This article is part of the Research TopicMolecular and Genetic Mechanisms of Chilling Tolerance in PlantsView all 6 articles
Dongnongdongmai No.1 (Dn1) is one of the few winter wheat varieties that can successfully overwinter at temperatures as low as -25°C or even lower. To date, few researches were carried to identify the freeze tolerance genes in Dn1 and applied them to improve plant resistance to extreme low temperatures. The basic helix-loop-helix (bHLH) transcription factor MYC2 is a master regulator in JA signaling, which has been reported to involve in responses to mild cold stress (2°C and 7°C). We hypothesized that MYC2 might be part of the regulatory network responsible for the tolerance of Dn1 to extreme freezing temperatures. In this study, we showed that wheat MYC2 (TaMYC2) was induced under both extreme low temperature (-10°C and-25°C) and JA treatments. The ICE-CBF-COR transcriptional cascade, an evolutionary conserved cold resistance pathway downstream of MYC2, was also activated in extreme low temperatures. We further showed that overexpression of any of the MYC2 genes from Dn1 TaMYC2A, B, D in Arabidopsis led to enhanced freeze tolerance. The TaMYC2 overexpression lines had less electrolyte leakage and lower malondialdehyde (MDA) content, and an increase in proline content, an increases antioxidant defences, and the enhanced expression of ICE-CBF-COR module under the freezing temperature. We further verified that TaMYC2 might function through physical interaction with TaICE41 and TaJAZ7, and that TaJAZ7 physically interacts with TaICE41. These results elucidate the molecular mechanism by which TaMYC2 regulates cold tolerance and lay the foundation for future studies to improve cold tolerance in plants.
Low temperature is an abiotic stress factor that seriously affect crop yield. When plants are exposed to a low-temperature environment, the cell membrane is affected and Reactive Oxygen Species (ROS) increase, which affects the normal growth of plants and reduces the crop yield (Liang et al., 2022). Wheat is one of the most important food crops in the world (Rizwan et al., 2019). The study of cold resistance in wheat is of great significance for improving wheat yield in low temperature areas. Dn1 is the first winter wheat variety that can safely overwinter at extreme temperatures of -30°C and has a recovery rate of up to 85% in the following spring. (Peng et al., 2021; Tian et al., 2021). Therefore, choosing Dn1 as the research object will be beneficial for exploring the molecular mechanism of freeze tolerance and for improving crop varieties.
Numerous genes have been identified as being involved in key mechanisms of cold stress (Castellani and Tipton, 2015; Albertos et al., 2019; Kidokoro et al., 2022). Among these, C-repeat/DREB binding factors (CBFs) are transcription factors that play significant roles in cold stress (Hwarari et al., 2022). When plants are exposed to cold stress, CBF genes are rapidly induced, and CBF proteins activate downstream target cold-responsive (COR) genes by binding to the C-repeat (CRT)/dehydration-responsive element (Kazan, 2015; Shi et al., 2018; Peng et al., 2022) to complete the self-protection of plants under cold stress. In Arabidopsis, three CBF family members (CBF1, CBF2, and CBF3) have been detected, that are tandemly distributed on chromosome 4. To date, researchers have detected CBFs in different plants and characterized their cold-tolerance functions (Wang et al., 2017a; Hwarari et al., 2022). In addition, as an MYC-type basic helix-loop-helix (bHLH) transcription factor, the inducer of CBF expression (ICE) plays a key role in cold resistance by activating the CBF gene (Chinnusamy et al., 2003). The ICE-CBF-COR transcriptional cascade has been established as a major regulatory mechanism for low-temperature signaling and freezing tolerance in Arabidopsis (Chinnusamy et al., 2003), rice (Xiang et al., 2007), and tomatoes (Juan et al., 2015). CBF-mediated transcriptional network is regulated at different levels in response to cold stress. Previous reports have shown that many pivotal protein kinases and TFs can interact with ICE-CBFs and affect plant cold tolerance by positively or negatively regulating them (Zhu, 2016; An et al., 2021b). However, little information is available for the molecular mechanism of ICE-CBF-COR module in winter wheat.
Plant hormones Jasmonic acid (JA) have important effects on the low-temperature stress response. In the JA signaling pathway, the F-box protein COI1 forms an SCF-type ubiquitin protein ligase E3 with SKP1 and CULLIN1. Jasmonate ZIM-domain (JAZ) acts as a repressor protein, and in the resting state, JAZs interact with downstream TFs and inhibit JA response. In the stimulated state, bioactive JA-ILE is generated, which interacts JAZ with SCFCOI1 and is then degraded by the 26S-proteasome to release transcription factors, and the genes regulated by transcription factors are expressed (Ruan et al., 2019; Wang et al., 2020). Low temperature stress increases JA levels in wheat (Soltész et al., 2013), rice (Du et al., 2013) and Arabidopsis (Hu et al., 2013), which are associated with increased expression of JA biosynthetic genes in rice (Du et al., 2013) and Arabidopsis (Hu et al., 2013), and decreased expression of JA catabolic genes in rice (Du et al., 2013). AtJAZ1 and AtJAZ4 can interact with AtICE1/2 and inhibit the stability of AtICE and its downstream CBF expression, thereby negatively regulating cold tolerance (Hu et al., 2013). In apple, JAZ1 and JAZ2 interact with BBX37, inhibit the interaction between BBX37 and ICE1, and negatively regulate CBF expression and cold tolerance (An et al., 2021a). These studies prove that there is a close relationship between the JA and CBF pathways. However, the relationship between JA and CBF in winter wheat under the extremely freezing temperature has not yet been reported.
MYC2 is the first described member of the MYC family (Boter et al., 2004), which is responsible for the activation of most JA-mediated responses (Schmiesing et al., 2016; Guo et al., 2022). Some studies have demonstrated that, in Arabidopsis, MYC2 can negatively regulate CAT2 to participate in plant responses to salt stress (Song et al., 2021).Simultaneously, MYC2 induced by water-spray stress promotes bHLH19 to directly activate the ORA47 promoter by interacting with bHLH19, thereby improving plant resistance (Chen et al., 2016; Van Moerkercke et al., 2019). MYC2 also interacts with ERD1 and LBD40 to improve drought tolerance in plants (Li et al., 2019; Wang et al., 2019). MYC2 plays a critical role in cold resistance. For instance, SlMYC2 regulates polyamine biosynthesis and thus participates in MeJA-induced cold tolerance in tomato fruits (Min et al., 2021). It has been shown that PtrMYC2 interacts with PtrBADH-I to improve glycine betaine and thus cold tolerance in Poncirus trifoliata. MaMYC2, a core transcription factor in the JA signaling pathway, positively regulates JA-mediated cold tolerance via interaction with MaICE1 (Peng et al., 2013; Zhao et al., 2013). Although MYC2 has been revealed to play an important role in plant abiotic stress response, the function of TaMYC2 in wheat under freeze stress remains unclear.
In this study, we investigated the molecular mechanism by which TaMYC2 regulates plant freeze tolerance under extreme freezing temperature. We found that TaMYC2 plays a positive regulatory role the in freezing stress response. Overexpression of TaMYC2 enhance the Arabidopsis freezing-resistance, including the accumulation of proline content and an increase of ROS-scavenging activity. TaMYC2 functions freezing-resistance possibly through the regulation of ICE-CBF-COR regulatory cascade. Our study demonstrated how TaMYC2 regulates JA-mediated freeze stress through TaICE41 in wheat and laid the foundation for improving crop tolerance to extreme freezing temperature stress in the future.
The winter wheat cultivar Dn1 was provided by the Northeast Agricultural University(NEAU). Dn1 was planted on 12 September 2021, in a test field on at the NEAU campus in Harbin, China. At the 2-3 leaf stage, the seedlings were divided into the following 2 groups: (1) seedlings sprayed with distilled water (control group) and (2) seedlings sprayed with 1.0 mM MeJA solution at a spray dosage of 44mL· m-2 (MeJA treatment group) (Zhao et al., 2019). Previous studies have shown that tillering nodes are the main components of overwintering plant (Bao et al., 2021). Tillering nodes were collected when the temperature naturally decreased to 5°C, -10°C and -25°C (average minimum temperature for 10 consecutive days in the field) (Supplementary Figure 1). To reduce the differences between the materials used and make the experimental results more representative, different wheat tiller nodes (approximately 1 cm) were combined into one sample (0.50 g), quickly frozen in liquid nitrogen, wrapped in tin foil, and stored in a refrigerator at -80°C (Tian et al., 2021; Liang et al., 2022).
The Arabidopsis thaliana ecotype Columbia (Col-0) and Arabidopsis thaliana mutant myc2 (atmyc2) were used in this study. Among them, atmyc2 (SALK_061267) was purchased from the Arabidopsis Information Resource (TAIR) website (http://www.arabidopsis.org/). The planting methods for Arabidopsis thaliana were based on previous studies (Tian et al., 2021). The incubation conditions were as follows: temperature 23°C, LED tube, photon flux density 120-150 μmol·m-2s-1, light 16h/dark 8h (Tian et al., 2021). The plates were incubated at 4°C for 3 d and then transferred to a growth chamber (temperature 23°C, LED tube, humidity 60%, photon flux density 120-150 μmol·m-2s-1, light 16h/dark 8h). For the cold acclimation treatment, 4 weeks old Arabidopsis were placed in an incubator at 4°C for 3 d and then in an incubator at -10°C for 5 h. The culture was then returned to room temperature for 7 d for observation.
Total RNA from the wheat tillering nodes was extracted using an Ultrapure RNA Kit (CWBIO, Jiangsu, China). The obtained RNA was reverse-transcribed into cDNA using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) was used for real-time fluorescence quantitative PCR, according to the manufacturer’s instructions. Quantitative primers were designed using WheatOmics (http://202.194.139.32/), and are described in detail in the Supplementary Table. Three replicates were established for each group of samples, and TaACTIN and AtACTIN were used as the internal reference gene for qRT-PCR. For Dn1, the tillering node is an important overwintering organ (Bao et al., 2021). Therefore, the tillering nodes of Dn1 under natural cooling conditions were selected for qRT-PCR. Relative expression levels were calculated using the 2 -ΔΔct method.
The sequences of the TaMYC2A, B, D genes were obtained from WheatOmics (http://202.194.139.32/) (Gene IDs: TraesCS1A02G193200, TraesCS1B02G208000 and TraesCS1D02G196900). PCR was used to amplify the complete DNA sequences (CDSs) of TaMYC2A, B, D. Primers used for PCR are listed in the Supplementary Table.
The nucleotide sequence of TaMYC2A, B, D and the amino acid sequences of TaMYC2A, B, D proteins were used to search for homologous genes and proteins in the NCBI database. The phylogenetic analysis was performed using MEGA 11. The phylogenetic trees were constructed according to the method of Liang et al. (Liang et al., 2022). The motif analysis was performed using the MEME (https://meme-suite.org/meme/tools/meme). The domain prediction was performed using NCBI Batch CD-Search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi).
Full-length CDSs of TaMYC2A, B, D were inserted into pCAMBIA1301 plant expression vectors to generate 35S:TaMYC2A, B, D constructs using User cloning technology (Clough and Bent, 1998). The constructed vector was then transferred into EHA105. Next, the 35S: TaMYC2A, B, D vectors were transformed into WT and atmyc2 mutant to generate overexpressed (OE TaMYC2A, B, D) and rescued (myc2/TaMYC2A, B, D) plants (Clough and Bent, 1998). The screening method for homozygous transgenic Arabidopsis was based on a previous study (Tian et al., 2021).
The full-length CDS of TaMYC2D, lacking its termination codon was inserted into the 35S: GFP vector (primers used are listed in the Supplementary Table). The recombinant vectors 35S: TaMYC2D -GFP and 35S: GFP (control) were instantaneously transformed into Nicotiana benthamiana using the leaf blade injection method (Liu et al., 2002). Tobacco plants were selected for each experiment, and the experiment was repeated thrice to eliminate interference from external factors. The transient expression of TaMYC2D was detected using a laser confocal microscope (TCS SP8; Leica, Germany).
The seeds of WT, atmyc2 and transgenic Arabidopsis were poured into EP tubes, 1 mL of 75% alcohol was added, and the seeds were sterilized by shaking for approximately 1 min. The alcohol was poured out and cleaned with distilled water, and the seeds were then sterilized with 2.5% (w/v) NaClO for 5 min and cleaned with distilled water. The washed WT, atmyc2, and transgenic Arabidopsis seeds were placed on 1/2 MS medium for the control group and 1/2 MS medium for the experimental group containing 50 µM MeJA. The culture dish was placed vertically in a light incubator (16/8h photoperiod) and root growth was observed. After approximately 7 d, photo graphs were taken. Finally, ImageJ software was used to measure the root length.
The relative electrolyte leakage (%), malondialdehyde (MDA) content and proline content in Arabidopsis leaves were estimated according to the methods described by Lu et al, Abrahám et al. and Ma et al. (Lu et al., 2007; Abrahám et al., 2010; Ma et al., 2015).
DAB and NBT staining were performed as previously described experimental methods (Tian et al., 2021). The H2O2 and contents were determined using an H2O2 and content determination kit (spectrophotometric method) (KeMing Biotechnology, Suzhou, China).
The activities of CAT and SOD were determined using a CAT and SOD determination kit (spectrophotometric method) (KeMing Biotechnology, Suzhou, China).
The coding regions of TaMYC2D and TaJAZ7 were cloned into a pGBKT7 (BD) vector. For TaMYC2D-pGBKT7, 200ng·ml-1 Aureobasidin (AbA) was used to inhibit auto activation. Similarly, the coding region of TaICE41 and TaJAZ7 were inserted into the vector pGADT7 (AD) (the primers used are listed in the Supplementary Table). The recombinant vector was then transferred into AH109. Next, positive clones were screened on SD/-Trp/-Leu synthetic defect medium and protein interactions were verified on SD/-Trp/-Leu/-His/-Ade synthetic defect medium (Peng et al., 2021; Liang et al., 2022).
For the BiFC assay, the full-length coding sequences of TaMYC2D and TaICE41, TaJAZ7 without their stop codons in fusion with GFPN and GFPC, respectively, were cloned into the pCAMBIA2300-BiFC vector. Negative control was set. The constructed vector was transferred into GV3101 strain and transiently expressed in tobacco. The experiment was performed as previously described (Peng et al., 2021). Tobacco plants were selected for each experiment, and the experiment was repeated thrice to eliminate interference from external factors. The GFP signals in the leaves were observed under a laser confocal microscope (TCS SP8, Leica, Germany).
In this study, error lines were obtained from three biological replicates. IBM SPSS Statistics 26 was used to analyze the mean values of the experimental data by one-way ANOVA analysis. The comparison method was the least significant difference (LSD), and the bar graphs with different letters indicated significant differences (p< 0.05). The bars were drawn using GraphPad prism 9.
To evaluate the effect of MeJA treatment on Dn1, we first examined the expression levels of TaPIEP1 (pathogen-induced ERF encoding gene) and TaPIMA1 (pathogen-induced MATE) genes, which were previously reported to be the marker genes in response to JA (Dong et al., 2010; Su et al., 2022). The results showed that at 5°C and -10°C, the expression of TaPIEP1 and TaPIMA1 in the MeJA treatment group was significantly higher than that in the control group (Figure 1A, B). This suggested that MeJA treatment can have an effect on Dn1 and has caused high expression of JA-induced genes.
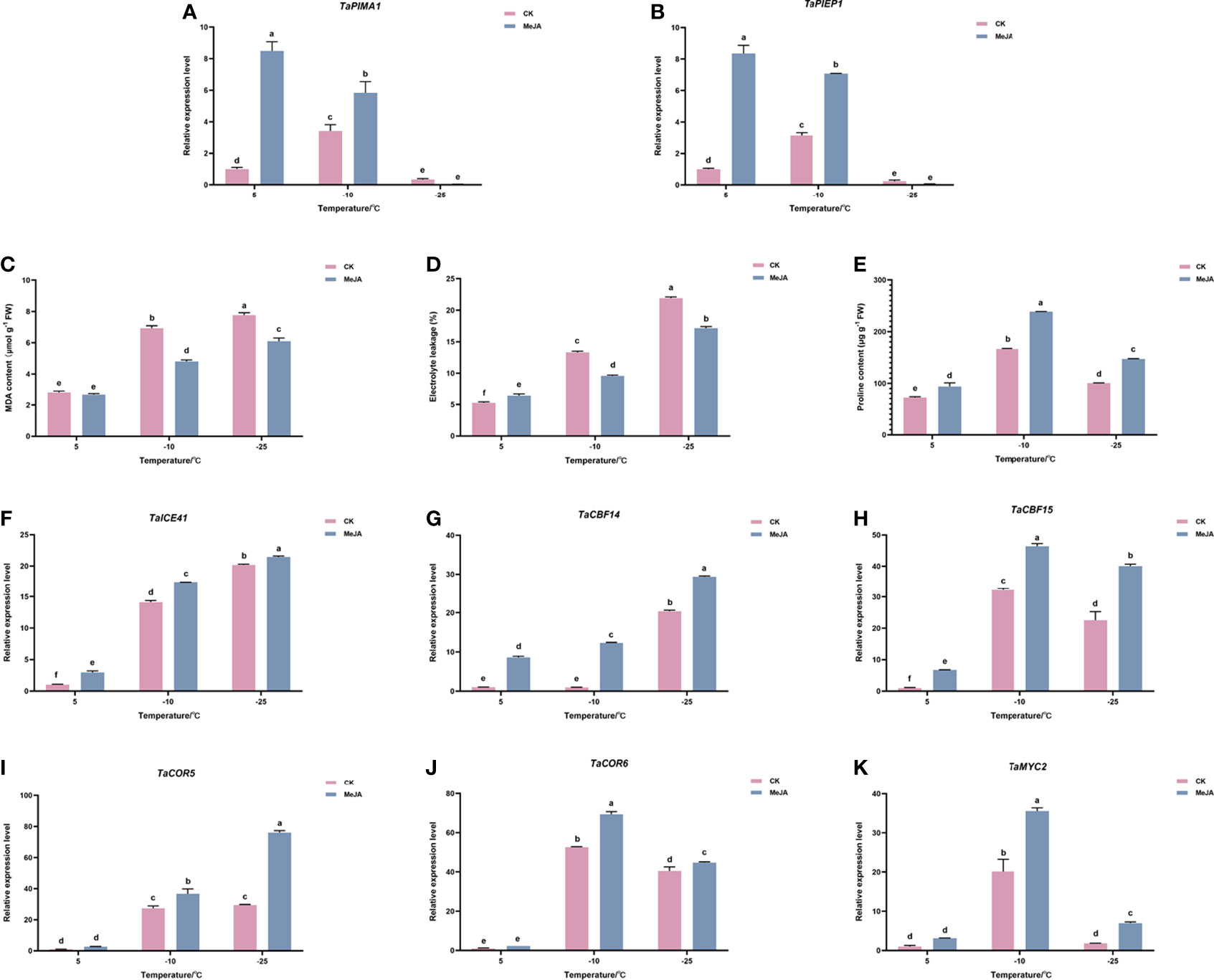
Figure 1 Effects of exogenous MeJA on (A) TaPIEP1 expression (B) TaPIMA1 expression (C) electronic leakage, (D) MDA content, (E) proline content, and (F–J) expression of ICE-CBF-COR cold resistance related genes. (K) TaMYC2 expression of Dn1 tillering node under cold stress. The error lines were obtained from three biological replicates. The mean values of the experimental data were analyzed by one-way ANOVA analysis. The comparison method was the least significant difference (LSD), and the bar graphs with different letters indicated significant differences (p < 0.05).
We then determined the effect of MeJA on physiologic parameters of freezing resistance in wheat. As shown in Figure 1C, D, the electrolyte leakage and MDA content of the control and MeJA treatment group did not change significantly at 5°C; however, when the temperature continued to decrease to -10°C or even -25°C, the electrolyte leakage and MDA content of the treatment group were significantly lower than those of the control group (Figure 1C, D). The proline content in the treatment group was always higher than that in the control group (Figure 1E).
Gene expression analysis of ICE-CBF-COR module showed that inducers of CBF expression (TaICE41), TaCBF (TaCBF14 and TaCBF15), and the cold-responsive genes TaCOR (TaCOR5 and TaCOR6) increased under the freezing temperature, reaching the highest levels at -25°C. Their expressions were upregulated after MeJA treatment, which indicated that MeJA treatment promoted freeze tolerance in plants by increasing the expression of ICE-CBF-COR genes. (Figure 1F–J).
Because MYC2 has been reported to act as an important regulator upstream of ICE-CBF-COR module, the expression of TaMYC2 in wheat freeze resistance was detected using quantitative RT-PCR (qRT-PCR). As shown in Figure 1K, TaMYC2 increased with a decrease in temperature, and the expression of TaMYC2 peaked at -10°C, but decreased to a certain extent at -25°C, which was still greater than that at 5°C. Under cold stress, the expression of TaMYC2 in the treatment group was greater than that in the control group, especially at -10°C, when the expression of TaMYC2 in the treatment group was 1.75 folds than that in the control group (Figure 1K). These results indicated that TaMYC2 expression is regulated by low temperature and MeJA.
Combined with the results that both freezing temperature and MeJA treatment up-regulated the expression of ICE-CBF-COR genes, we hypothesized that TaMYC2 may be involved in the regulation of wheat freezing resistance through the ICE-CBF-COR pathway.
Since wheat is a hexaploid plant with 3 genomes, the TaMYC2A, B, D sequences were obtained using WheatOmics 1.0. The TaMYC2A, B, D open reading frame (ORF) was isolated from the Dn1 tillering nodes based on putative sequence information available from the WheatOmics database. Sequence analysis showed that the TaMYC2A gene contained 2 exons and 1 intron, and TaMYC2B, D gene contained 1 exon. TaMYC2A, B encodes 693-amino-acids with an estimated molecular mass of 75.1KDa, and TaMYC2D encodes 695-amino-acids with an estimated molecular mass of 75.3KDa.
Phylogenetic analysis showed that TaMYC2A, B, D were most homologous to ZmMYC2 in Zea mays. In contrast, the relationship between Arabidopsis AtMYC2 and MaMYC2 was weak (Supplementary Figure 2). Motif analysis showed that the TaMYC2A, B, D proteins share 10 motifs with MYC2 proteins of tomato, tobacco, Arabidopsis, and maize (Supplementary Figure 2). Structural domain analysis revealed that TaMYC2A, B, D contained the structural domain features of the plant MYC-like bHLH proteins (Supplementary Figure 2).
To investigate the subcellular localization of TaMYC2, the coding sequence of TaMYB2D was expressed as a fusion of GFP in Nicotiana benthamiana. As is shown in Figure 2A, the fluorescence signal of 35S: GFP was detected in whole cells, whereas a strong fluorescence signal was only observed in the nucleus of 35S: TaMYC2D-GFP transformed cells, indicating that TaMYC2D is located in the nucleus.
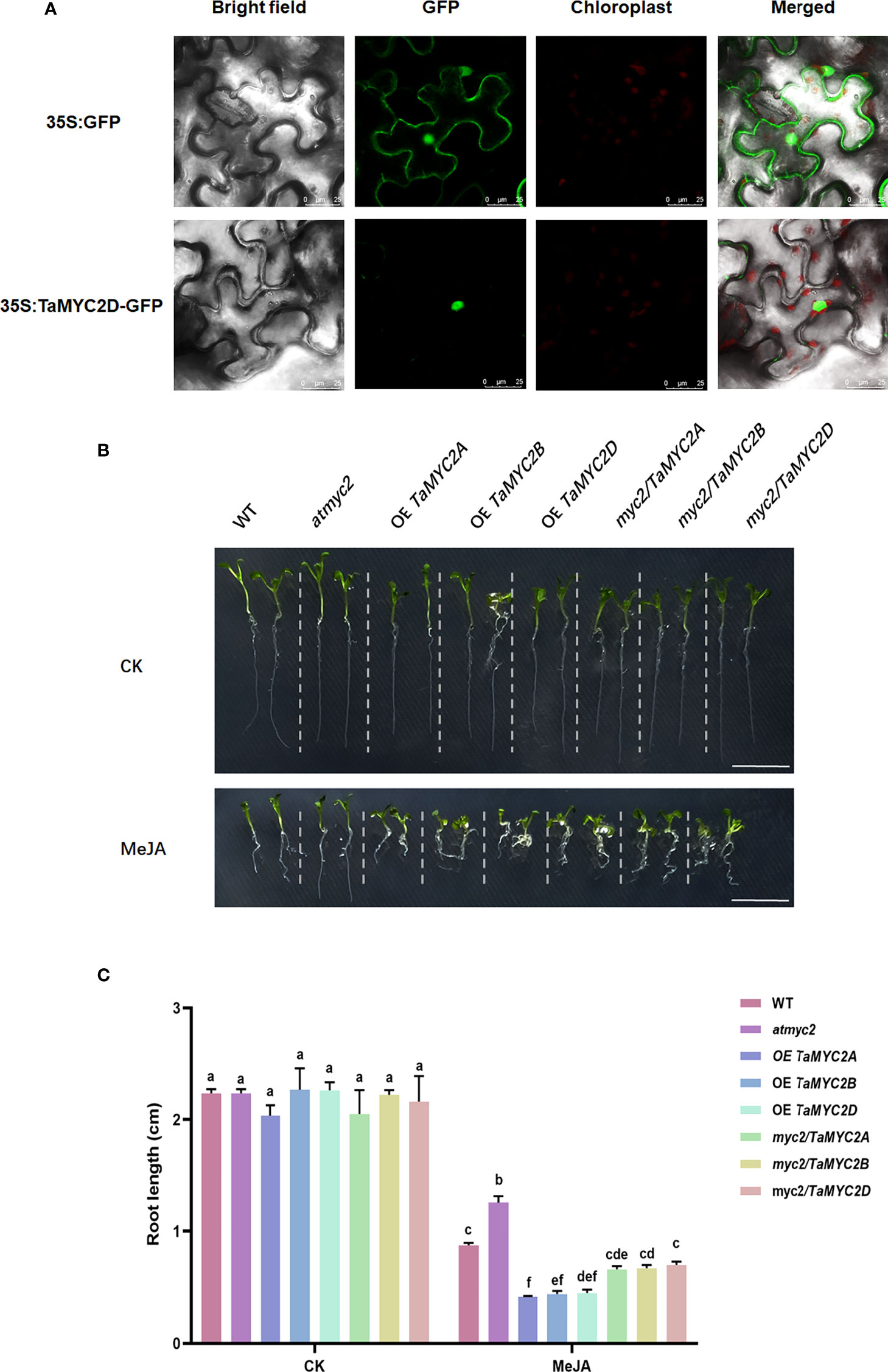
Figure 2 Subcellular localization of TaMYC2D in Nicotiana benthamiana and detection of JA sensitivity in TaMYC2A, B, D transgenic Arabidopsis root lengths. (A) Fluorescence detection of 35S: TaMYC2D-GFP fusion protein in tobacco leaves epidermal cells, and with 35S: GFP vector was used as a control (upper lane). (B) Root phenotype and (C) root length of Arabidopsis seedlings after 7 days of growth on medium with 50μM MeJA treated and control groups. The top to bottom on the right side of each bar chart corresponds to the left to right at the same treatment. The error lines were obtained from three biological replicates. The mean values of the experimental data were analyzed by one-way ANOVA analysis. The comparison method was the least significant difference (LSD), and the bar graphs with different letters indicated significant differences (p < 0.05).
Because JA signaling pathway can inhibit Arabidopsis root growth, and the myc2 mutant can affect this inhibition (Acosta et al., 2013), TaMYC2A, B, D were overexpressed in Arabidopsis myc2 mutant to assess their functions in JA signaling.
The results showed that there was no difference in the root growth phenotype among the lines on the control plates (Figure 2B). However, on the 1/2MS plate containing 50 µm MeJA, the root length of the WT was shortened by MeJA inhibition; atmyc2 had the longest root length, followed by WT and myc2/TaMYC2A, B, D, and OE TaMYC2A, B, D had the shortest root length (Figure 2B). Root length measurements were consistent with the phenotypic observations. The root length of OE TaMYC2A, B, D was approximately 2.6 folds shorter than that of atmyc2 (Figure 2C). This result demonstrates that TaMYC2 in transgenic plants could respond to the JA signaling pathway and thus inhibit root length.
As shown in Figure 3A, there were no clear differences in the phenotypes of these Arabidopsis lines at 23°C, including growth, leaf size, and color. As the temperature decreased, the leaves gradually became slightly transparent. After recovery, it was found that most of the leaves of WT Arabidopsis were yellow, the leaves of atmyc2 were the most yellow, and the leaves of OE TaMYC2A, B, D. Arabidopsis plants had almost no yellow leaves (Figure 3A).
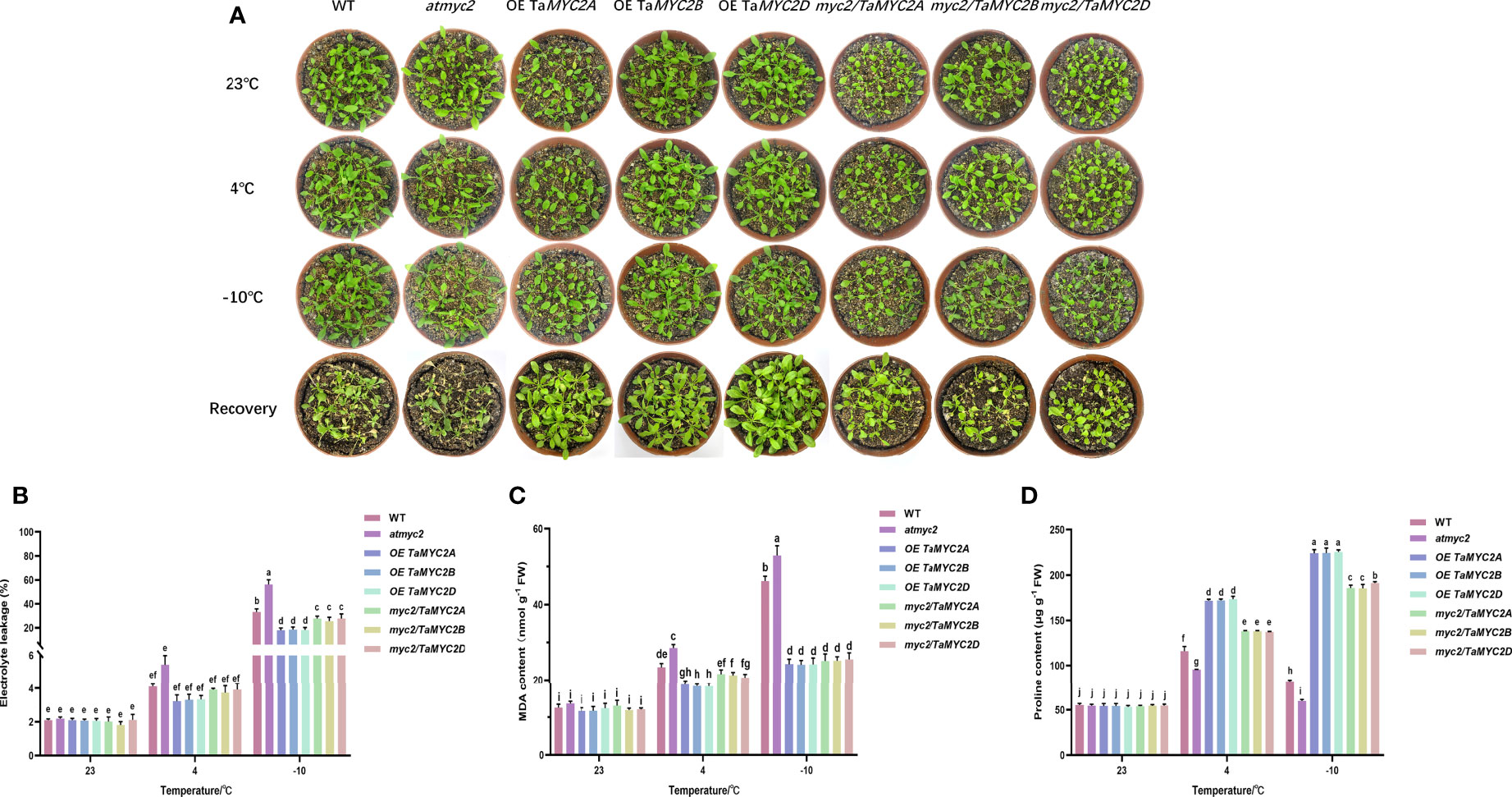
Figure 3 Phenotype investigation and physiological index measurement of WT, atmyc2 mutants and transgenic plants under cold stress. (A) Cold resistance phenotypes of WT, atmyc2 mutants and transgenic plants. (B) Electrolyte leakage, (C) MDA content, and (D) proline content of the WT, atmyc2, and transgenic plants. The top to bottom on the right side of each bar chart corresponds to the left to right of the same temperature. The error lines were obtained from three biological replicates. The mean values of the experimental data were analyzed by one-way ANOVA analysis. The comparison method was the least significant difference (LSD), and the bar graphs with different letters indicated significant differences (p < 0.05).
For the electrolyte leakage, MDA content, and proline content of the plants, there is no significant change between WT, atmyc2 and transgenic plants at 23°C, but when the temperature decreased to 4°C and -10°C, it was found that the electrolyte leakage and MDA of OE TaMYC2A, B, D Arabidopsis were lower than those of WT Arabidopsis and even lower than those of atmyc2 plants (Figure 3B, C). At the same time, the proline content in OE TaMYC2A, B, D Arabidopsis was the highest, and atmyc2 plants were the lowest under low-temperature stress (Figure 3D). This phenomenon was more obvious when the temperature was continuously reduced to -10°C.
Under cold stress, ROS are toxic to cells (Nadarajah, 2020). Analysis of ROS content and antioxidant enzyme activities showed that atmyc2 plants had the most colored leaves after DAB and NBT staining, followed by WT, myc2/TaMYC2A, B, D Arabidopsis, and OE TaMYC2A, B, D Arabidopsis (Figure 4A). H2O2 and contents in transgenic plants increased with a decrease in temperature (Figure 4B, C). Under the same low-temperature stress, the contents of H2O2 and in OE TaMYC2A, B, D Arabidopsis were the lowest, while the contents of H2O2 and in atmyc2 plants were the highest. CAT and SOD activities of OE TaMYC2A, B, D Arabidopsis were the strongest under low-temperature stress, followed by myc2/TaMYC2A, B, D Arabidopsis, and WT, whereas atmyc2 plants had the lowest activity (Figure 4D, E). The qRT-PCR results of antioxidase related genes (AtCAT1, AtCAT3, AtSOD1 and AtSOD2) showed that the expression levels of OE TaMYC2A, B, D and myc2/TaMYC2A, B, D were always higher than those of WT and atmyc2 under the same low temperature stress (Figure 4F–I).
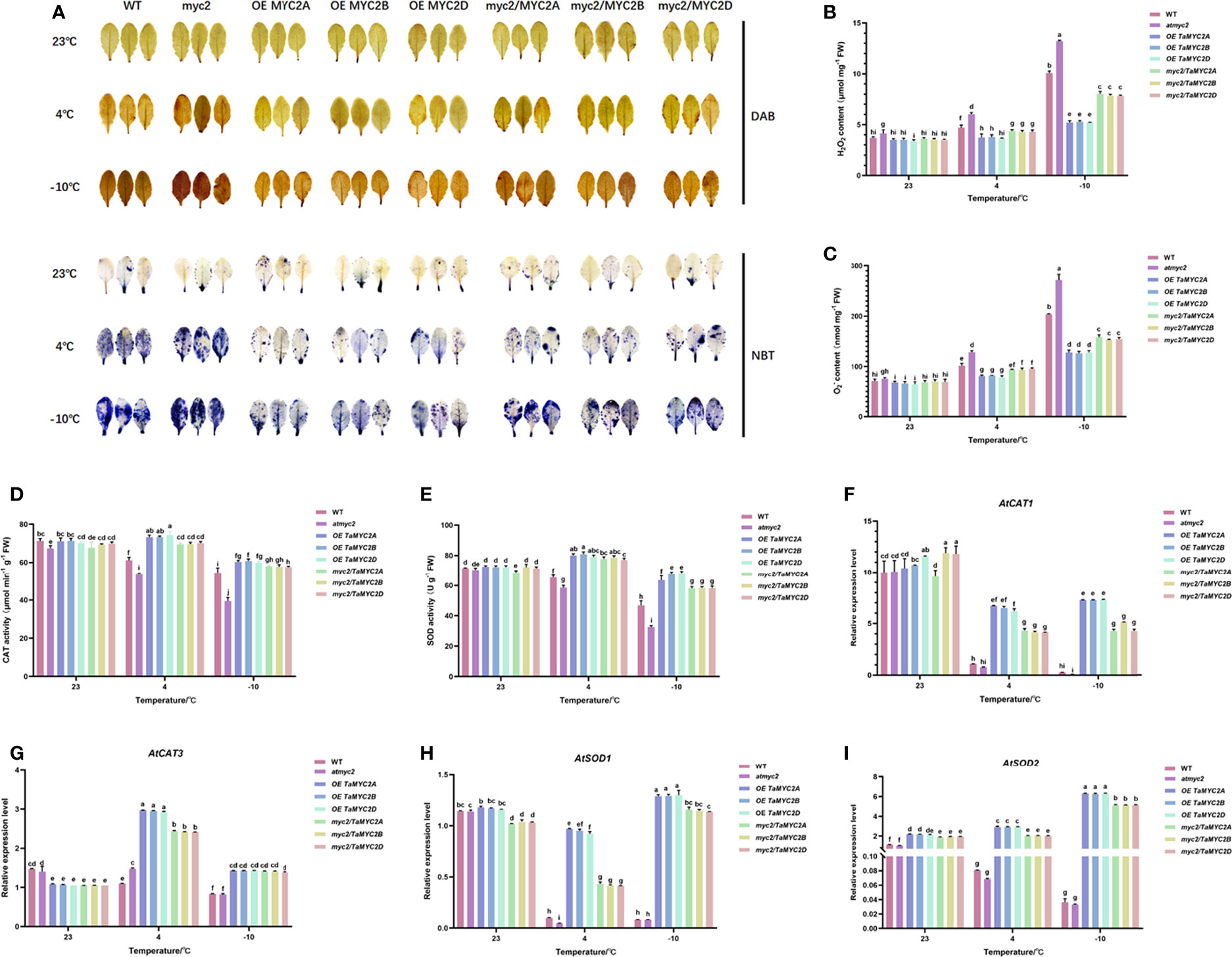
Figure 4 Histochemical staining and determination of ROS content and antioxidant enzymes in WT, atmyc2 mutants, and transgenic Arabidopsis under low temperature stress. (A) DAB (yellow) and NBT (blue) staining of the leaves under cold stress. (B, C) H2O2 and content determination. (D, E) Determination of CAT and SOD activity. Expression of (F) AtCAT1, (G) AtCAT3, (H) AtSOD1, (I) AtSOD2 in WT, atmyc2 mutants, and transgenic Arabidopsis under low temperature stress. The error lines were obtained from three biological replicates. The mean values of the experimental data were analyzed by one-way ANOVA analysis. The comparison method was the least significant difference (LSD), and the bar graphs with different letters indicated significant differences (p < 0.05).
To verify whether the improved freeze tolerance of TaMYC2A, B, D was related to ICE-CBF-COR pathway, we performed qRT-PCR for ICE-CBF-COR genes (AtICE1, AtCBF1, AtCBF2, AtCBF3, AtCOR15, AtCOR47, AtRD29A, and AtKIN1) in transgenic Arabidopsis under freezing stress. The results showed that the expression of ICE-CBF-COR genes in WT, atmyc2, OE TaMYC2A, B, D and myc2/TaMYC2A, B, D Arabidopsis plants did not change significantly at 23°C (Figure 5A–H). When the temperature was reduced to 4°C, most of the ICE-CBF-COR genes (AtICE1, AtCBF1, AtCBF3, AtCOR15, AtCOR47, AtKIN1) in OE TaMYC2A, B, D and myc2/TaMYC2A, B, D transgenic Arabidopsis were significantly greater than those in WT and atmyc2 Arabidopsis, except for AtCBF2 and AtRD29A genes (Figure 5A, B, D-F, H). When the temperature was lowered to -10°C, the expression of ICE-CBF-COR genes in OE TaMYC2A, B, D and myc2/TaMYC2A, B, D Arabidopsis were more significantly different from those in WT and atmyc2 Arabidopsis. At the same time, we found that there were no significant variations in the qRT-PCR results for TaMYC2A, B, D. This suggests that TaMYC2 enhances freeze tolerance in Arabidopsis, which is associated with the activation of the ICE-CBF-COR pathway.
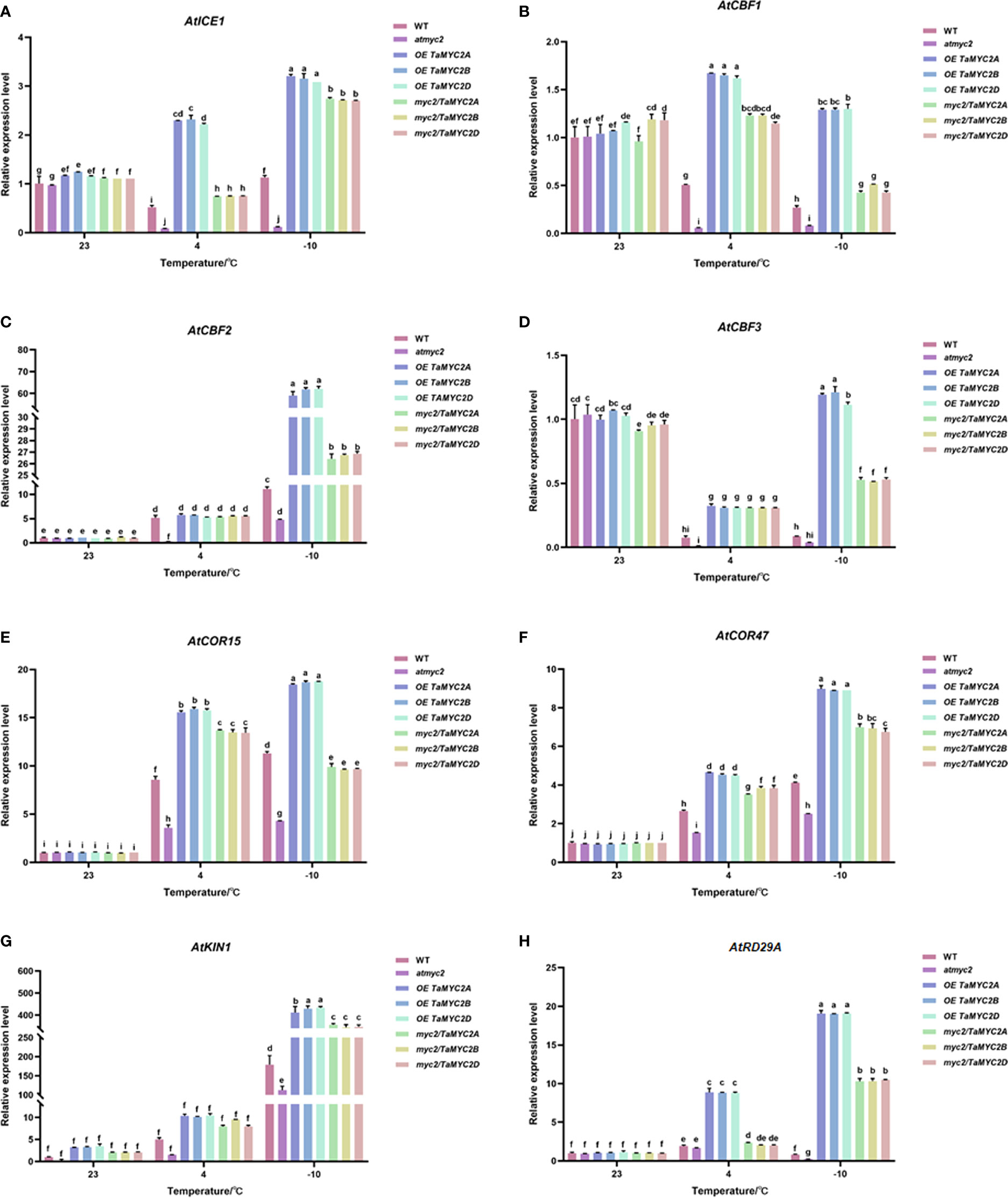
Figure 5 Effects of TaMYC2 on the ICE-CBF-COR module in Arabidopsis under cold stress. Expression of (A) AtICE1, (B) AtCBF1, (C) AtCBF2, (D) AtCBF3, (E) AtCOR15, (F) AtCOR47, (G) AtKIN1, (H) AtRD29A in WT, atmyc2 mutants, and transgenic Arabidopsis under low temperature stress. The top to bottom on the right side of each bar chart corresponds to the left to right sides of the same temperature. The error lines were obtained from three biological replicates. The mean values of the experimental data were analyzed by one-way ANOVA analysis. The comparison method was the least significant difference (LSD), and the bar graphs with different letters indicated significant differences (p < 0.05).
The interaction between TaMYC2D, TaICE41, and TaJAZ7 were assessed using the Y2H method. The results showed that TaMYC2D interacted with TaICE41 and TaJAZ7 (Figure 6A), and TaJAZ7 interacted with TaICE41 (Figure 6B)
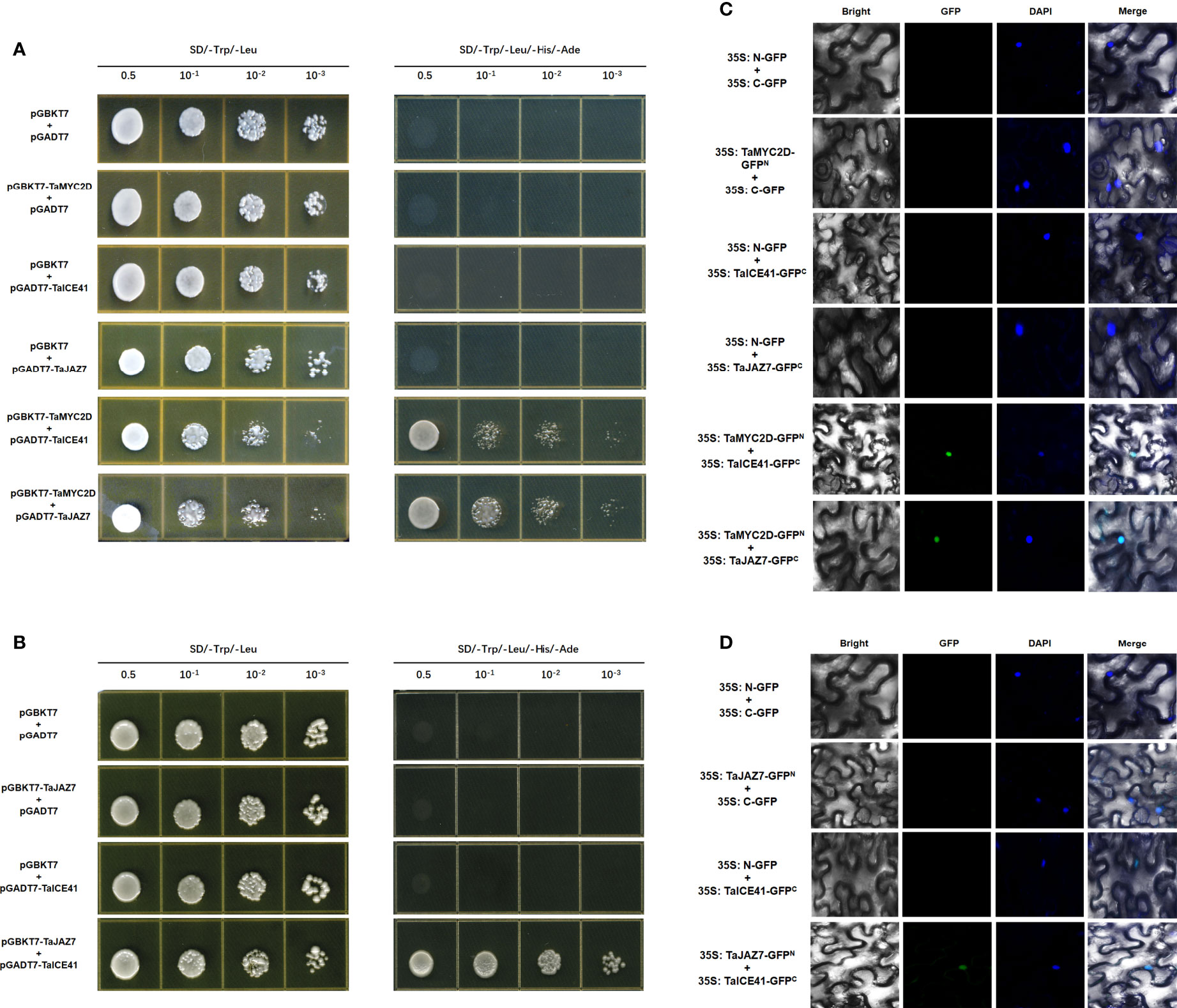
Figure 6 Protein interacts with protein. TaMYC2D protein physically interacts with TaICE41 and TaJAZ7 protein. (A) Y2H and (C) BiFC analysis. TaJAZ7 protein physically interacts with TaICE41 protein. (B) Y2H and (D) BiFC analysis.
To confirm the interactions between these proteins, BiFC test were performed using tobacco leaves. The results showed a GFP fluorescence signal was observed in the nucleus, when TaMYC2D co-expressed with TaICE41 and TaJAZ7, individually, and when TaICE41 was co-expressed with TaJAZ7 (Figure 6C, D).
The adaptation of plants to the environment is the result of long-term evolution, which is regulated by many internal and external factors, such as plant hormones, light, age. For example, day length and light quality significantly affect the frost tolerance of plants (Maibam et al., 2013). Blue light receptor CRY2 enhances plant low temperature tolerance through the COP1-HY5-BBX7/8 signaling pathway (Li and Hiltbrunner, 2021). The stability of red light receptor phyB protein enhances the freeze tolerance of plants at low temperature through the CBFs interact with PIF3 (Jiang et al., 2020). In addition, the freezing tolerance was increased with plant age at the vegetative stage (Zhao et al., 2022). In this study, we mainly focus on the influence of plant hormone JA on cold resistance of wheat under the same environmental conditions. We found that exogenous MeJA treatment significantly enhanced the freezing resistance of winter wheat Dn1.
Although the role of JA and JA-mediated CBF pathways in cold stress has also been confirmed in recent years (Hu et al., 2013; An et al., 2021a), a few researches were reported on the regulation of JA in freeze resistance of wheat under extremely low temperature. In the present study, we showed that application of MeJA to winter wheat Dn1 significantly enhanced the freeze resistance under the freezing temperature (-25°C) (Figure 1C–E). The expression of ICE-CBF-COR cascade, a well-known cold responsive gene module, and MYC2, a master transcription factor in JA signaling pathway were also increased in Dn1 after MeJA treatment under low-temperature stress (Figure 1F–J). These results suggest that JA signaling and ICE-CBF-COR cascade play a critical role in freezing stress responses in plants.
MYC2 plays a critical role in cold stress responses in many plants. Previous studies have shown that exogenous MeJA treatment increases the expression of MYC2 in banana (Zhao et al., 2013), tomato (Min et al., 2018) under mild-cold temperature (7°C and 2°C). In this study, we measured the changes in TaMYC2 expression under extremely low-temperature stress treated with MeJA, and the results showed that TaMYC2 expression increased with decreasing temperature, reaching a peak at -10°C (Figure 1K). Moreover, the expression of TaMYC2 was significantly increased by MeJA treatment (Figure 1K). These data indicated that TaMYC2 may be closely related to JA-mediated freeze tolerance.
TaMYC2 funtion as a key regulator in plant freezing resistance possibly through activating ICE-CBF-COR. Our result showed that TaMYC2A, B, D might activate the ICE-CBF-COR cold resistance pathway. This is different from previous study by Hu et al. (Hu et al., 2013), which detected the expression levels of CBF1, CBF2, and CBF3 in Arabidopsis thaliana WT and atmyc2 mutants at 4°C and found that there was no difference between WT and atmyc2 mutants (Hu et al., 2013). Regardless, our study demonstrated that the cold-resistance function of TaMYC2s in Arabidopsis and OE TaMYC2A, B, D-Arabidopsis can survive under the -10°C low temperature. Therefore, TaMYC2 can be potential genes for future genetic breeding to improve plant freezing resistance under extremely low temperature.
Most studies focused on the roles of MYC2 in activating the expression of ICE in ICE-CBF-COR module. To date, few studies was performed about the interaction between MYC2 and ICE in plants, except banana. Zhao et al. showed that MaMYC2 interacts with MaICE41 to participate in the cold resistance pathway of CBF at 7°C and JA pathway participates in the CBF signaling pathway through MaMYC2-MaICE1 (Zhao et al., 2013). In this study, we selected TaICE41, a sequence highly homologous to MaICE1 and AtICE1 in wheat. Considering the similarity in the domain, motif, and protein localization between TaMYC2 and MaMYC2, we hypothesized that TaMYC2 might also be linked to TaICE41. The TaMYC2A, B, D genes had the same effect on the freeze tolerance of transgenic Arabidopsis and the ICE-CBF-COR gene; therefore, we only selected TaMYC2D for the following interaction experiment. To test this hypothesis, we verified the interaction between TaMYC2 and TaICE41 by using Y2H and BiFC (Figure 6A, C). In addition, since MYC2 is thought to be repressed by JAZ7 in JA signaling, we detect the interaction between TaMYC2 and TaJAZ7, which has high homology with AtJAZ1 (Wang et al., 2017b). Our result showed that there is an interaction between TaMYC2D and TaJAZ7 (Figure 6A, C). These results suggest that TaJAZ7 may inhibit TaMYC2 and thus affect the effect of TaMYC2 on TaICE41, which in turn affects plant freezing tolerance. JA catalyzes the degradation of JAZ by SCFCOI1 ubiquitin ligase and 26S proteasome (Thines et al., 2007). This partly explains our previous finding that exogenous MeJA treatment increases TaMYC2 expression.
ROS are major regulatory molecules in plants and play a role in signaling events triggered by cellular metabolic disturbances and environmental stimuli (Waszczak et al., 2018). Under abiotic stress, ROS have dual functions; at high levels, they are toxic to cells, whereas appropriate levels of ROS can activate plant defense systems (Nadarajah, 2020). This study showed that the H2O2 and contents of OE TaMYC2A, B, D Arabidopsis and myc2/TaMYC2A, B, D Arabidopsis were lower than those of WT and atmyc2 under cold stress, both by chemical staining and quantitative determination. Simultaneously, the H2O2 and contents in myc2/TaMYC2A, B, D Arabidopsis were significantly lower than those in the atmyc2 mutant (Figure 4B, C). Furthermore, under cold stress, the CAT and SOD enzyme activities of the OE TaMYC2A, B, D Arabidopsis and myc2/TaMYC2A, B, D Arabidopsis were higher than those of the WT and atmyc2 mutant plants (Figure 4D, E). The antioxidase related genes (AtCAT1, AtCAT3, AtSOD1 and AtSOD2) showed that the expression levels of OE TaMYC2A, B, D and myc2/TaMYC2A, B, D were always higher than those of WT and atmyc2 under the same low temperature stress (Figure 4F–I).These results imply that OE TaMYC2A, B, D Arabidopsis plants could better maintain ROS homeostasis, followed by myc2/TaMYC2A, B, D Arabidopsis and then WT, and the ROS scavenging ability in atmyc2 mutants was the lowest. Thus, TaMYC2 may improve plant freezing tolerance by participating in ROS homeostasis.
In summary, our results demonstrate that TaMYC2 interacts with TaICE41 to activate the downstream CBF-COR pathway, thereby improving freeze tolerance in plants (Figure 7). Moreover, TaJAZ7 may interact with TaMYC2 to inhibit the transcriptional activity of TaICE41 or directly interfere with TaICE41 to inhibit its expression of TaICE41, thus negatively regulating freeze tolerance. These findings provided insights into the improvement of freeze tolerance in wheat, reveal the relationship between the JA and CBF pathways, and demonstrate the positive role of TaMYC2 in freeze tolerance.
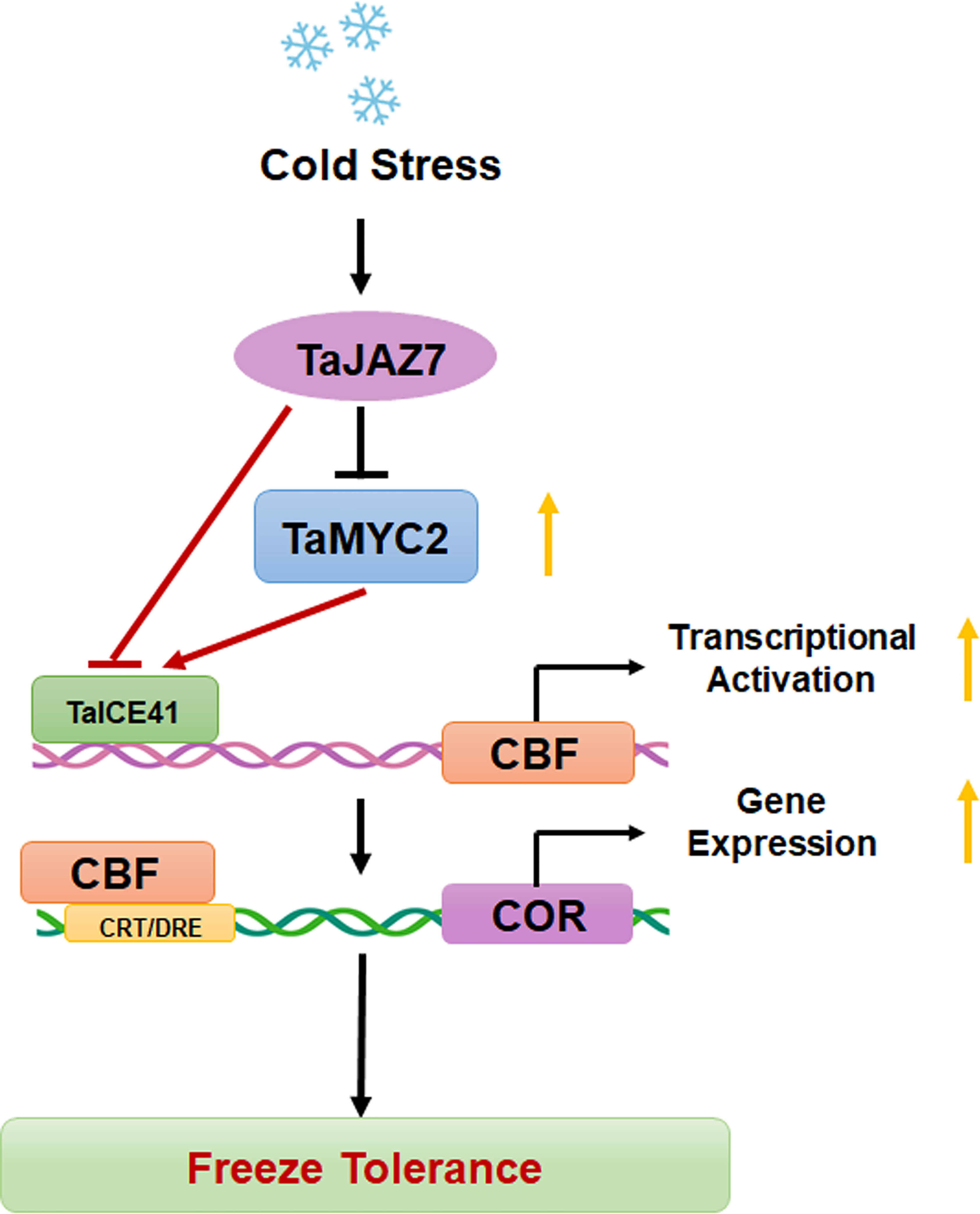
Figure 7 Working model diagram of the TaMYC2-TaICE41 module regulating cold stress in wheat. Yellow arrows represent the trends in gene expression.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
RW completed the experiments, data analysis and drafted the manuscript. MY, JqX helped in assisting with the experiments. JpX, XF, QX and JC participated in revising articles. DZ conceived and designed the study, obtained financial support, provided the study material and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Natural Science Foundation of China (31701348) and Natural Science Foundation of Heilongjiang Province (LH2021C022).
We sincerely appreciate the help of Professor Yunde Zhao (University of California, San Diego), Associate Researcher Yuying Wang (Institute of Botany, Chinese Academy of Sciences, China) and Associate Professor Libo Wang (Northeast Agricultural University, China) in the revision of the manuscript. We would like to thank KetengEdit (www.ketengedit.com) for its linguistic assistance during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1042889/full#supplementary-material
Abrahám, E., Hourton-Cabassa, C., Erdei, L., Szabados, L. (2010). Methods for determination of proline in plants. Methods Mol. Biol. 639, 317–331. doi: 10.1007/978-1-60761-702-0_20
Acosta, I. F., Gasperini, D., Chételat, A., Stolz, S., Santuari, L., Farmer, E. E. (2013). Role of NINJA in root jasmonate signaling. Proc. Natl. Acad. Sci. U.S.A. 110 (38), 15473–15478. doi: 10.1073/pnas.1307910110
Albertos, P., Wagner, K., Poppenberger, B. (2019). Cold stress signalling in female reproductive tissues. Plant Cell Environ. 42 (3), 846–853. doi: 10.1111/pce.13408
An, J. P., Wang, X. F., Zhang, X. W., You, C. X., Hao, Y. J. (2021a). Apple b-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 229 (5), 2707–2729. doi: 10.1111/nph.17050
An, J. P., Xu, R. R., Liu, X., Zhang, J. C., Wang, X. F., You, C. X., et al. (2021b). Jasmonate induces biosynthesis of anthocyanin and proanthocyanidin in apple by mediating the JAZ1-TRB1-MYB9 complex. Plant J. 106 (5), 1414–1430. doi: 10.1111/tpj.15245
Bao, Y., Yang, N., Meng, J., Wang, D., Fu, L., Wang, J., et al. (2021). Adaptability of winter wheat dongnongdongmai 1 (Triticum aestivum l.) to overwintering in alpine regions. Plant Biol. (Stuttg) 23 (3), 445–455. doi: 10.1111/plb.13200
Boter, M., Ruíz-Rivero, O., Abdeen, A., Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and arabidopsis. Genes Dev. 18 (13), 1577–1591. doi: 10.1101/gad.297704
Castellani, J. W., Tipton, M. J. (2015). Cold stress effects on exposure tolerance and exercise performance. Compr. Physiol. 6 (1), 443–469. doi: 10.1002/cphy.c140081
Chen, H. Y., Hsieh, E. J., Cheng, M. C., Chen, C. Y., Hwang, S. Y., Lin, T. P. (2016). ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol. 211 (2), 599–613. doi: 10.1111/nph.13914
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in arabidopsis. Genes Dev. 17 (8), 1043–1054. doi: 10.1101/gad.1077503
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 16 (6), 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dong, N., Liu, X., Lu, Y., Du, L., Xu, H., Liu, H., et al. (2010). Overexpression of TaPIEP1, a pathogen-induced ERF gene of wheat, confers host-enhanced resistance to fungal pathogen bipolaris sorokiniana. Funct. Integr. Genomics 10 (2), 215–226. doi: 10.1007/s10142-009-0157-4
Du, H., Liu, H., Xiong, L. (2013). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00397
Guo, Q., Major, I. T., Kapali, G., Howe, G. A. (2022). MYC transcription factors coordinate tryptophan-dependent defence responses and compromise seed yield in arabidopsis. New Phytol 236 (1), 132–145. doi: 10.1111/nph.18293
Hu, Y., Jiang, L., Wang, F., Yu, D. (2013). Jasmonate regulates the inducer of cbf expression-c-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in arabidopsis. Plant Cell 25 (8), 2907–2924. doi: 10.1105/tpc.113.112631
Hwarari, D., Guan, Y., Ahmad, B., Movahedi, A., Min, T., Hao, Z., et al. (2022). ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 23 (3), 1549. doi: 10.3390/ijms23031549
Jiang, B., Shi, Y., Peng, Y., Jia, Y., Yan, Y., Dong, X., et al. (2020). Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in arabidopsis. Mol. Plant 13 (6), 894–906. doi: 10.1016/j.molp.2020.04.006
Juan, J. X., Yu, X. H., Jiang, X. M., Gao, Z., Zhang, Y., Li, W., et al. (2015). Agrobacterium-mediated transformation of tomato with the ICE1 transcription factor gene. Genet. Mol. Res. 14 (1), 597–608. doi: 10.4238/2015.January.30.1
Kazan, K. (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20 (4), 219–229. doi: 10.1016/j.tplants.2015.02.001
Kidokoro, S., Shinozaki, K., Yamaguchi-Shinozaki, K. (2022). Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 27 (9), 922–935. doi: 10.1016/j.tplants.2022.01.008
Liang, Y., Xia, J., Jiang, Y., Bao, Y., Chen, H., Wang, D., et al. (2022). Genome-wide identification and analysis of bZIP gene family and resistance of TaABI5 (TabZIP96) under freezing stress in wheat (Triticum aestivum). Int. J. Mol. Sci. 23 (4), 2351. doi: 10.3390/ijms23042351
Li, J., Hiltbrunner, A. (2021). Is the Pr form of phytochrome biologically active in the nucleus? Mol. Plant 14 (4), 535–537. doi: 10.1016/j.molp.2021.03.002
Liu, Y., Schiff, M., Marathe, R., Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for n-mediated resistance to tobacco mosaic virus. Plant J. 30 (4), 415–429. doi: 10.1046/j.1365-313x.2002.01297.x
Li, Y., Yang, X., Li, X. (2019). Role of jasmonate signaling pathway in resistance to dehydration stress in arabidopsis. Acta Physiol. Plant. 41 (6), 100. doi: 10.1007/s11738-019-2897-7
Lu, P., Colombo, S. J., Sinclair, R. W. (2007). Cold hardiness of interspecific hybrids between pinus strobus and p. wallichiana measured by post-freezing needle electrolyte leakage. Tree Physiol. 27 (2), 243–250. doi: 10.1093/treephys/27.2.243
Ma, J., Du, G., Li, X., Zhang, C., Guo, J. (2015). A major locus controlling malondialdehyde content under water stress is associated with fusarium crown rot resistance in wheat. Mol. Genet. Genomics 290 (5), 1955–1962. doi: 10.1007/s00438-015-1053-3
Maibam, P., Nawkar, G. M., Park, J. H., Sahi, V. P., Lee, S. Y., Kang, C. H. (2013). The influence of light quality, circadian rhythm, and photoperiod on the CBF-mediated freezing tolerance. Int. J. Mol. Sci. 14, 11527–11543. doi: 10.3390/ijms140611527
Min, D., Zhou, J., Li, J., Ai, W., Li, Z., Zhang, X., et al. (2021). SlMYC2 targeted regulation of polyamines biosynthesis contributes to methyl jasmonate-induced chilling tolerance in tomato fruit. Postharvest Biol. Technol. 174, 111443. doi: 10.1016/j.postharvbio.2020.111443
Min, D., Li, F., Zhang, X., Cui, X., Shu, P., Dong, L., et al. (2018). SlMYC2 Involved in methyl jasmonate-induced tomato fruit chilling tolerance. J Agric Food Chem 66 (12), 3110–3117. doi: 10.1021/acs.jafc.8b00299.
Nadarajah, K. K. (2020). ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 21 (15), 5208. doi: 10.3390/ijms21155208
Peng, H. H., Shan, W., Kuang, J. F., Lu, W. J., Chen, J. Y. (2013). Molecular characterization of cold-responsive basic helix-loop-helix transcription factors MabHLHs that interact with MaICE1 in banana fruit. Planta 238 (5), 937–953. doi: 10.1007/s00425-013-1944-7
Peng, K., Tian, Y., Cang, J., Yu, J., Wang, D., He, F., et al. (2022). Overexpression of TaFBA-A10 from winter wheat enhances freezing tolerance in arabidopsis thaliana. J. Plant Growth Regul. 41 (1), 314–326. doi: 10.1007/s00344-021-10304-7
Peng, K., Tian, Y., Sun, X., Song, C., Ren, Z., Bao, Y., et al. (2021). Tae-miR399-UBC24 module enhances freezing tolerance in winter wheat via a CBF signaling pathway. J. Agric. Food Chem. 69 (45), 13398–13415. doi: 10.1021/acs.jafc.1c04316
Rizwan, M., Ali, S., Ali, B., Adrees, M., Arshad, M., Hussain, A., et al. (2019). Zinc and iron oxide nanoparticles improved the plant growth and reduced the oxidative stress and cadmium concentration in wheat. Chemosphere 214, 269–277. doi: 10.1016/j.chemosphere.2018.09.120
Ruan, J., Zhou, Y., Zhou, M., Yan, J., Khurshid, M., Weng, W., et al. (2019). Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 20 (10), 2479. doi: 10.3390/ijms20102479
Schmiesing, A., Emonet, A., Gouhier-Darimont, C., Reymond, P. (2016). Arabidopsis MYC transcription factors are the target of hormonal salicylic Acid/Jasmonic acid cross talk in response to pieris brassicae egg extract. Plant Physiol. 170 (4), 2432–2443. doi: 10.1104/pp.16.00031
Shi, Y., Ding, Y., Yang, S. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23 (7), 623–637. doi: 10.1016/j.tplants.2018.04.002
Soltész, A., Smedley, M., Vashegyi, I., Galiba, G., Harwood, W., Vágújfalvi, A. (2013). Transgenic barley lines prove the involvement of TaCBF14 and TaCBF15 in the cold acclimation process and in frost tolerance. J. Exp. Bot. 64 (7), 1849–1862. doi: 10.1093/jxb/ert050
Song, R. F., Li, T. T., Liu, W. C. (2021). Jasmonic acid impairs arabidopsis seedling salt stress tolerance through MYC2-mediated repression of CAT2 expression. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.730228
Su, Q., Rong, W., Zhang, Z. (2022). The pathogen-induced MATE gene TaPIMA1 is required for defense responses to rhizoctonia cerealis in wheat. Int. J. Mol. Sci. 23 (6), 3377. doi: 10.3390/ijms23063377
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448 (7154), 661–665. doi: 10.1038/nature05960
Tian, Y., Peng, K., Bao, Y., Zhang, D., Meng, J., Wang, D., et al. (2021). Glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase genes of winter wheat enhance the cold tolerance of transgenic arabidopsis. Plant Physiol. Biochem. 161, 86–97. doi: 10.1016/j.plaphy.2021.02.005
Van Moerkercke, A., Duncan, O., Zander, M., Šimura, J., Broda, M., Vanden Bossche, R., et al. (2019). A MYC2/MYC3/MYC4-dependent transcription factor network regulates water spray-responsive gene expression and jasmonate levels. Proc. Natl. Acad. Sci. U.S.A. 116 (46), 23345–23356. doi: 10.1073/pnas.1911758116
Wang, D. Z., Jin, Y. N., Ding, X. H., Wang, W. J., Zhai, S. S., Bai, L. P., et al. (2017a). Gene regulation and signal transduction in the ICE-CBF-COR signaling pathway during cold stress in plants. Biochem. (Mosc) 82 (10), 1103–1117. doi: 10.1134/s0006297917100030
Wang, Y., Qiao, L., Bai, J., Wang, P., Duan, W., Yuan, S., et al. (2017b). Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum l.). BMC Genomics 18 (1), 152. doi: 10.1186/s12864-017-3582-0
Wang, J., Song, L., Gong, X., Xu, J., Li, M. (2020). Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 21 (4), 1446. doi: 10.3390/ijms21041446
Wang, Y., Xu, H., Liu, W., Wang, N., Qu, C., Jiang, S., et al. (2019). Methyl jasmonate enhances apple’ cold tolerance through the JAZ–MYC2 pathway. Plant Cell Tissue Organ Cult. (PCTOC) 136 (1), 75–84. doi: 10.1007/s11240-018-1493-7
Waszczak, C., Carmody, M., Kangasjärvi, J. (2018). Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 69, 209–236. doi: 10.1146/annurev-arplant-042817-040322
Xiang, D., Zhang, Y., Yin, K. (2007). Transformation of ICE1 gene mediated by agrobacterium improves cold tolerance in transgenie rice. Chinese J Rice Sci. 21 (5), 482–486. doi: 10.16819/j.1001-7216.2007.05.007 (in Chinese)
Zhao, H., Fan, X., Luo, L., Zhang, R., Cang, J., Xu, Q., et al. (2019). Effects of MeJA on cold resistance physiology and expression of key genes in winter wheat under low temperature stress. J. Triticeae Crops 39 (4), 407–414. doi: 10.7606/j.issn.1009-1041.2019.04.05 (in Chinese)
Zhao, J., Shi, M., Yu, J., Guo, C. (2022). SPL9 mediates freezing tolerance by directly regulating the expression of CBF2 in arabidopsis thaliana. BMC Plant Biol. 22 (1), 59. doi: 10.1186/s12870-022-03445-8
Zhao, M.-L., Wang, J.-N., Shan, W. E. I., Fan, J.-G., Kuang, J.-F., Wu, K.-Q., et al. (2013). Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 36 (1), 30–51. doi: 10.1111/j.1365-3040.2012.02551.x
Keywords: wheat, TaMYC2, TaICE41, TaJAZ7, ICE-CBF-COR module, freeze resistance
Citation: Wang R, Yu M, Xia J, Xing J, Fan X, Xu Q, Cang J and Zhang D (2022) Overexpression of TaMYC2 confers freeze tolerance by ICE-CBF-COR module in Arabidopsis thaliana. Front. Plant Sci. 13:1042889. doi: 10.3389/fpls.2022.1042889
Received: 13 September 2022; Accepted: 28 October 2022;
Published: 14 November 2022.
Edited by:
Baohong Zou, Nanjing Agricultural University, ChinaReviewed by:
Jian Yan, South China Agricultural University, ChinaCopyright © 2022 Wang, Yu, Xia, Xing, Fan, Xu, Cang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Da Zhang, emhhbmdkYUBuZWF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.