
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 09 January 2023
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1040134
 Yadong Shao1†
Yadong Shao1† Shangtao Jiang1†
Shangtao Jiang1† Haiying Peng1
Haiying Peng1 Han Li1
Han Li1 Peigen Li1
Peigen Li1 Rou Jiang1
Rou Jiang1 Wenyi Fang1
Wenyi Fang1 Tingsu Chen2
Tingsu Chen2 Gaofei Jiang1
Gaofei Jiang1 Tianjie Yang1*
Tianjie Yang1* Savithri U. Nambeesan3*
Savithri U. Nambeesan3* Yangchun Xu1
Yangchun Xu1 Caixia Dong1*
Caixia Dong1*Background: Arbuscular mycorrhizal fungi (AMF) are beneficial soil fungi which can effectively help plants with acquisition of mineral nutrients and water and promote their growth and development. The effects of indigenous and commercial isolates of arbuscular mycorrhizal fungi on pear (Pyrus betulaefolia) trees, however, remains unclear.
Methods: Trifolium repens was used to propagate indigenous AMF to simulate spore propagation in natural soils in three ways: 1. the collected soil was mixed with fine roots (R), 2. fine roots were removed from the collected soil (S), and 3. the collected soil was sterilized with 50 kGy 60Co γ-radiation (CK). To study the effects of indigenous AMF on root growth and metabolism of pear trees, CK (sterilized soil from CK in T. repens mixed with sterilized standard soil), indigenous AMF (R, soil from R in T. repens mixed with sterilized standard soil; S, soil from S in T. repens mixed with sterilized standard soil), and two commercial AMF isolates (Rhizophagus intraradices(Ri) and Funneliformis mosseae (Fm)) inoculated in the media with pear roots. Effects on plant growth, root morphology, mineral nutrient accumulation, metabolite composition and abundance, and gene expression were analyzed.
Results: AMF treatment significantly increased growth performance, and altered root morphology and mineral nutrient accumulation in this study, with the S treatment displaying overall better performance. In addition, indigenous AMF and commercial AMF isolates displayed common and divergent responses on metabolite and gene expression in pear roots. Compared with CK, most types of flavones, isoflavones, and carbohydrates decreased in the AMF treatment, whereas most types of fatty acids, amino acids, glycerolipids, and glycerophospholipids increased in response to the AMF treatments. Further, the relative abundance of amino acids, flavonoids and carbohydrates displayed different trends between indigenous and commercial AMF isolates. The Fm and S treatments altered gene expression in relation to root metabolism resulting in enriched fructose and mannose metabolism (ko00051), fatty acid biosynthesis (ko00061) and flavonoid biosynthesis (ko00941).
Conclusions: This study demonstrates that indigenous AMF and commercial AMF isolates elicited different effects in pear plants through divergent responses from gene transcription to metabolite accumulation.
Pear(Pyrus sp.) is one of the most economically important fruit crops throughout the world. In pear cultivation, agricultural intensification, especially by increased use of fertilizers and pesticides has led to increased fruit yield (Foley et al., 2011). However, these practice reduce the abundance of keystone taxa and their network complexity in the root microbiome (Banerjee et al., 2019), which in-turn decreases microbial communities and impacts the soil ecological balance. Microbial communities play an indispensable role in nutrient cycling, organic matter decomposition, soil aggregate stabilization, and resistance to biotic and abiotic stress, demonstrating their importance in maintaining the multifunctionality of terrestrial ecosystems (Delgado-Baquerizo et al., 2016). AMF have been used as an alternative to chemical fertilizers to mitigate the impact of agricultural intensification on the soil microbial community, to restore soil ecological balance, and to improve plant growth and yield.
AMF form beneficial obligate symbiotic relationships with majority of plants, largely due to their prevalence in terrestrial ecosystems. In general, AMF hyphae exclusively colonize the root cortex and form an arbuscule, a highly branched structure, inside cortex cells (Berruti et al., 2013). AMF use their fine and extended extraradical hyphal network to explore parts of the soil volume that are not accessible by plant roots. This exploration by AMF allows for mineral and water uptake into plants in exchange for carbohydrates and lipids (Van't Padje et al., 2021), and release of photosynthates into the root zone by plants (Kaiser et al., 2015). Further, AMF can improve plant growth, root morphology, regulate host gene expression and cellular metabolism through various signaling molecules (Lopez-Raez et al., 2010; Rivero et al., 2015; Hill et al., 2018). Previous studies have shown that AMF can regulate the expression of genes associated with root development to promote plant root development, promote the expression of root transporter genes, and improve acquisition of mineral elements by plants (Liu et al., 2022; Wu et al., 2022). In a complimentary way, plant roots can regulate the colonization of AMF by changing the synthesis of its metabolites including that of fatty acids, flavonoids and signaling molecules (Larose et al., 2002; Jin et al., 2016; Kameoka et al., 2019).
There are limited studies on inoculations by commercial AMF isolates and their efficacy in orchard systems. Generally, studies indicate that commercial AMF isolates displayed growth promotion effects on a variety of plants. However, some studies also show that commercial AMF isolates inoculated into orchards can be inhibited by some strains of local bacterial communities, reducing their beneficial effects in the rhizosphere (Cruz-Paredes et al., 2020). Also, compared with multiple AMF, single strain inoculations are more likely affected by local microorganisms, limiting their colonization (Compant et al., 2010). The efficacy of mixed indigenous AMF composed of multiple AMF was more stable than that of single AMF isolates. A previous study showed that the largest and second strongest determinants of AMF community composition were the pool of available AMF and the functional traits of plant hosts, respectively (Smilauer et al., 2020). In long-term symbionts, host plants and AMF select one another to achieve optimal exchange strategy of carbohydrates and nutrients (Noe and Kiers, 2018). Different AMF taxa provide functional complementarity and differential nutritional benefits to the host plants. Thus it is expected that high community diversity is more beneficial for host plants than individual AMF isolates (Wagg et al., 2011; Huang et al., 2019). Application of indigenous AMF in pear orchards can not only promote the growth of pear seedlings, but also maintain the ecological balance of pear orchard soils, reduce the application of chemical fertilizer in agriculture, and maintain the diversity of soil microbial function. However, there are only a few studies on the effects of indigenous AMF on the AMF community composition in pear orchards (Yoshimura et al., 2013; Huang et al., 2019). And the effects of indigenous AMF on pear trees have hardly been studied. Previous study used Trifolium repens (It is often used to propagate AMF) as propagation material to propagate AMF spores in rhizosphere soil and root segments of citrus orchard in different ways and found that T. repens was an effective way to propagate indigenous AMF spores (Wu et al., 2019). In this study, T. repens was used as plant material to propagate AMF in rhizosphere soil and root of pear orchard to study the effects of indigenous AMF on pear trees.
Pyrus betulaefolia is a common pear rootstock widely used across the world and displays several disease resistance and nutrient efficiency characteristics (Yang et al., 2020). In this study, we sought to determine the effects of various AMF applications on P. betulaefolia plant growth, root morphology, mineral nutrient accumulation, metabolites and transcriptome responses. We hypothesized that: (1) the indigenous microorganisms or plant-specific metabolites in the orchard pear tree root affect the propagation and function of indigenous AMF; (2) the combined effects of indigenous AMF are more beneficial than commercial AMF isolates in enhancing pear growth; and (3) metabolism and gene expression responses of pear roots are specific to indigenous AMF and commercial AMF isolates.
Soil samples mixed with fine roots were collected from the rhizosphere at a depth between 0-20cm at a 15 year-old pear (P. betulaefolia) orchard which received combined application of chemical and organic fertilizers without AMF inoculum in Changzhou (31°61′N, 120°05′E), Jiangsu province, China. This orchard had been under active production for three years before collection of soil samples, thus the indigenous AMF community from the orchard soil was not likely disturbed by human factors. The collected soil mixed with fine roots were divided into three parts, the first part containing 20 g fine roots (R), the second part where fine roots were removed manually (S) and the third part (rhizosphere soil without fine roots) sterilized with 50 kGy 60Co γ-radiation (CK). The soil was air-dried and sieved (<2 mm) to achieve a high degree of homogeneity and to reduce variability among replicates.
Seeds of white clover (T. repens, purchased from farmers markets) were sterilized with 75% ethanol for 10 min and rinsed with distilled water. The white clover seeds (about 0.3 g) were sowed in plastic pots (about 0.25 gallons) containing 300 g soil (R, S and CK) mixed with 700 g sterilized sand and vermiculite (2:1, v:v; 0.11 MPa, 121 °C, 2 h). Each treatment was replicated three times, with a total of nine pots in this experiment. All the pots were grown in a greenhouse set at 70% relative humidity set on a light cycle of 15 h daytime (26 °C) and 9 h nighttime (24 °C) at the College of Resources and Environmental Sciences in Nanjing Agricultural University. Daytime illuminance, supplied by LED lighting was 1350 lx. After sowing, plants were irrigated with 50 mL of deionized water and fertilized every week with 50 mL half of low P (10% standard P concentration) Hoagland solution (pH 6.0). Plants were harvested at 80 d after sowing.
After harvest, the soil of the CK, R and S treatments were retained as mixed inoculum for treatment with P. betulaefolia. Seeds of pear plants (collected from wild pear trees, collected in November 2019 at Luoyang (34°32’N, 112°16’E), identified by the corresponding author and preserved in the Jiangsu Provincial Key Lab of Solid Organic Waste Utilization, Nanjing Agricultural University, ID Pb-2019-11) were sterilized with 2.5% of sodium hypochlorite solution for 15 min, rinsed with distilled water, and sown into autoclaved (121°C, 0.11 MPa, 0.5h) gauze for germination at 4 °C. After germination, seedlings were transferred to a growth chamber (16 h light/8 h darkness set at 25 ± 1 °C). After 18 d, pear seedlings with uniform size were selected. Standard soil from Baima National Agricultural Science and Technology Zone of Nanjing Agricultural University, sand and vermiculite were mixed in the volume ratio of 2:2:1, sterilized with gamma radiation (50 kGy 60Co γ-radiation). The seedlings were transferred to plastic pots (about 0.6 gallons) containing 3.1 kg standard soil and 300 g of R or S inoculum (main species include Claroideoglomus sp. and Acaulospora sp., unpublished data). To compare the effects of indigenous inoculum, 3.1 kg of standard soil was mixed with 300 g sterilized CK soil (CK), Rhizophagus intraradices (BGC BJ09) inoculum (Ri) and Funneliformis mosseae (BGC HUN03B) inoculum (Fm) (about 90 spores/g, approximately the same as the indigenous AMF spore density) separately. The commercial AM fungal strain R. intraradices and F. mosseae was provided by the Bank of Glomeromycota in China (BGC). The propagation methods of indigenous AMF and commercial AMF were the same, with the only difference being the original soil from the pear orchard and the commercial AMF substrate. Mycorrhizal inoculums included spores, infected root segments, and propagation substrates. However, after T. repens propagation, differences of mycorrhizal inoculums were lower among treatments, which are unlikely to result in differences in soil nutrition. In the actual treatments, the original pear tree soil accounted for a very small percentage (about 3%), further indicating that it had almost no impact on plant nutrition. Pear with all treatments were grown in a greenhouse at Baima National Agricultural Science and Technology Zone of Nanjing Agricultural University. One pear seedling of 18 d was used per pot and each treatment was replicated 4 times, for a total of 20 pots for the five treatments (R, S, CK, Ri, Fm).
Plants were harvested after 125 d and roots and shoots were separated. Roots were washed free of soil. Subsequently a portion of roots were frozen in liquid nitrogen and stored at -80 °C for root transcriptome sequencing and analysis. The residual roots were used for root architecture measurements using the LA1600 scanner (Canadian Regent Win RHizo2003b). Next, roots were dried in an oven set at 105°C for 30 min, followed by 75°C for 48 h and the dry biomass was measured. The leaves and stems were separated and the total leaf area was measured using a leaf area meter (SYE-YM02, China). The leaves were dried similar to the root tissue and measured for dry biomass.
A portion of residual root samples of T. repens and pear were stored in 50% ethanol for determination of mycorrhizal root colonization. Root segments were placed in 10% KOH (w/v) and incubated in a water bath set at 90 °C for approximately 15 min (white clover root) or 1 h (pear root), then soaked in an 10% hydrogen peroxide solution at room temperature, and stained with trypan blue for approximately 10 min. Spores in the rhizosphere soil of T. repens were isolated from approximately 30 g of air-dried soil by wet sieving, decanting, and centrifugation (3000 rpm, 2 min) through a 65%, (weight/volume) sucrose cushion. Spores were counted using a stereo microscope (40× magnification, BX43, Olympus, Japan). Soil hyphae were extracted from 5 g air dried rhizosphere soil using the membrane filter technique and stained with typan blue for approximately 10 min (Wu et al., 2019; Jiang et al., 2020). Calculations were performed as below:
The leaf, stem and other residual root samples were dried in an oven at 105 °C until constant weight, and then ground into a powder using a tissue lyser (TL2010S, DHS Life Science & Technology, China). The N in these samples was extracted by H2SO4-H2O2 method and determined by AutoAnalyzer (AA3, Seal, Germany). The P, K, Ca, Mg and Fe of samples was extracted by mixed acid method (concentrated nitric acid:perchloric acid = 4:1) and measured by the Inductively Coupled Plasma-Optical Emission spectroscopy (ICP-OES, Agilent 710, USA). The mineral accumulation (content) was calculated as the concentration of mineral nutrition × biomass.
After grinding with liquid nitrogen 25 mg of CK, R, S, Ri and Fm root samples were extracted in 500 μL solution containing methanol:water (3:1). Samples were mixed for 30 s using a vortex, homogenized at 35 hz for 4 min and sonicated for 5 min in an ice-water bath, these steps were repeated three times. Next, samples were incubated for 1 h at -40 °C and centrifuged at 12,000 rpm for 15 min at 4°C. The resulting supernatant was transferred to a fresh glass vial for analysis. The quality control (QC) sample was prepared by mixing an equal aliquot of the supernatants from all of the samples. Each treatment was repeated six times.
LC-MS/MS analyses were performed using an UHPLC system (Vanquish, Thermo Fisher Scientific) with a UPLC BEH Amide column (2.1 mm × 100 mm, 1.7 μm) coupled to Q Exactive HFX mass spectrometer (Orbitrap MS, Thermo). The mobile phase consisted of 25 mM ammonium acetate and 25 mM ammonia hydroxide in water (pH = 9.75) and acetonitrile. The column and auto-sampler temperature were set at 30 °C and 4 °C respectively, and the injection volume was 3 μL. The QE HFX mass spectrometer was used to acquire MS/MS spectra on information-dependent acquisition (IDA) mode in the acquisition software (Xcalibur, Thermo).
The relative abundance of metabolites was calculated using the following formula = 100 × (Peak area of class/Total peak area of metabolites).
Total RNA of root in S and Fm treatments was extracted using commercial kits by Guangdong Magigene Biotechnology Co. Ltd. (Guangzhou, China). RNA quantity was measured using Qubit 2.0 (Thermo Fisher Scientific, MA, USA) and Nanodrop One (Thermo Fisher Scientific, MA, USA). RNA integrity was determined using the Agilent 2100 system (Agilent Technologies, Waldbron, Germany). Whole mRNAseq libraries were generated using NEB Next® Ultra™ Nondirectional RNA Library Prep Kit for Illumina® (New England Biolabs, MA, USA) and sequenced on an Illumina Hiseq Xten platform and 150 bp paired-end reads were generated. Each treatment was repeated four times.
The raw data of fastq format were processed by Trimmomatic (v.0.36) to acquire the clean reads and mapped to NCBI Rfam databases, to remove the rRNA sequences by Bowtie2 (v2.33). Reference genome was Pyrus × bretschneideri (Chinese white pear) and downloaded in Genebank. The remaining mRNA sequences were mapped to the reference genome by Hisat2 (2.1.0). HTSeq-count (v0.9.1) was used to obtain the read count and gene function information.
To compare gene expression levels among different genes and different experiments, the RPKM (expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) of each gene was calculated. Read count of each gene obtained from HTSeq-count was used for differential expression analysis. Differentially expressed genes (DEGs) of two conditions/groups was performed using the edgeR (v3.16.5, http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) which accounts for the length and the number of genes. The resulting P-value was adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR). The cut off for differentially expressed genes was set at FPKM ≥ 2, FDR ≤ 0.05 and |log2(fold change)| ≥ 1. KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/) enrichment analysis of differentially expressed genes (FDR ≤ 0.05) were implemented by the clusterProfiler (v3.4.4, http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) (Kanehisa, 2019; Kanehisa et al., 2021). KEGG Pathways with FPKM ≥ 10 were selected to study the DEGs of fructose and mannose metabolism (ko00051), fatty acid biosynthesis (ko00061) and flavonoid biosynthesis (ko00941) in this study. The sequence raw data and the relevant data have been uploaded to the figshare database (https://figshare.com/s/b81b2c1bb72c49b2e) and Sequence Read Archive (SRA, BioProject: PRJNA761485).
Statistical analyses were carried out using ‘R’ statistical software (version 4.0.3.), implemented within the RStudio graphical user interface (RStudio Desktop, version 2022.07.0, Posit Software, PBC). The data were analyzed using one-way ANOVA (Table S2) to test for significance (p < 0.05) and Duncan’s test for multiple comparisons using the R package ‘agricolae’. Illustrations were made using Origin8.5 (Origin Inc., Chicago, USA) and ‘R’ statistical software (version 4.0.3.).
Before harvest, root mycorrhizal colonization was observed in the R and S treatments, but not found in CK treatment (Figure S1). Root mycorrhizal colonization and arbuscular abundance in S treatment was significantly higher than in the R treatment (Table 1). The spore density in the original rhizosphere soil of pear orchard was 21.5 spores/g, which increased significantly after propagation by 300% in R and by 347% in S treatments. The spore density and soil hyphal density after propagation was not significantly different between the R and S treatments (Table 1). There were no differences in N and P accumulation in the R and S treatments compared with CK treatment, and K accumulation was higher in the R treatment (Figure 1). Further, the dry biomass of the CK treatment was lower than that in the two other treatments (Figure 1).
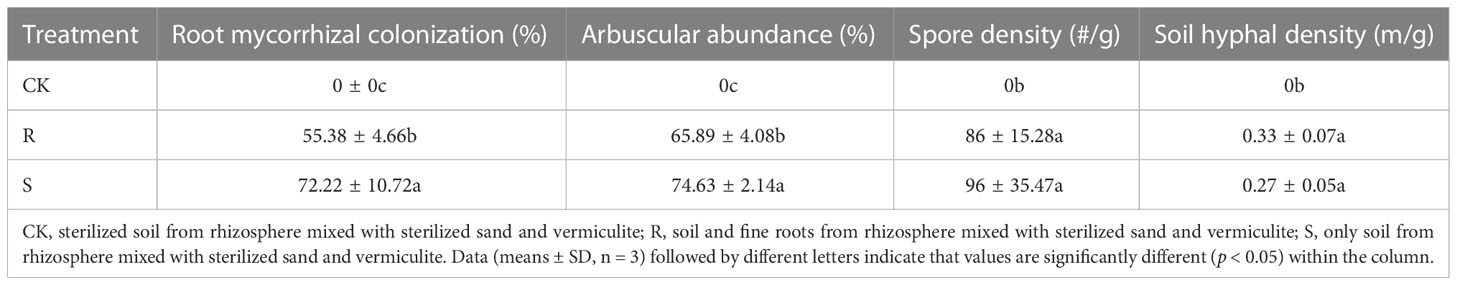
Table 1 The mycorrhizal propagation performance between R and S treatment in white clover root and rhizosphere.
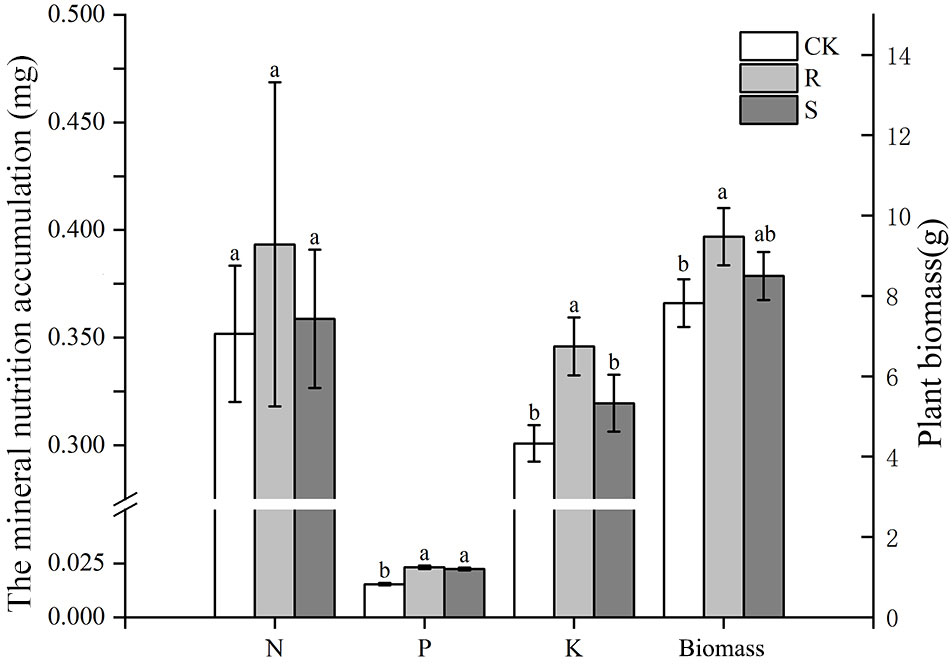
Figure 1 The mineral nutrition accumulation (N, P, and K) and biomass of white clover in different treatment. CK, sterilized soil from rhizosphere mixed with sterilized sand and vermiculite; R, soil and fine roots from rhizosphere mixed with sterilized sand and vermiculite; S, only soil from rhizosphere mixed with sterilized sand and vermiculite. Data (means ± SD, n = 3) followed by different letters indicate that values are significantly different (p < 0.05) within the treatments.
AMF treatments resulted in increased growth compared with the CK treatment. Shoot dry biomass, plant height, leaf number and leaf area were significantly increased in R, S, Fm and Ri treatments, compared to CK (Table 2). With the exception of shoot dry biomass, which was not significantly different among AMF treatments, the number of leaves, plant height and leaf area were greater in the S treatment.
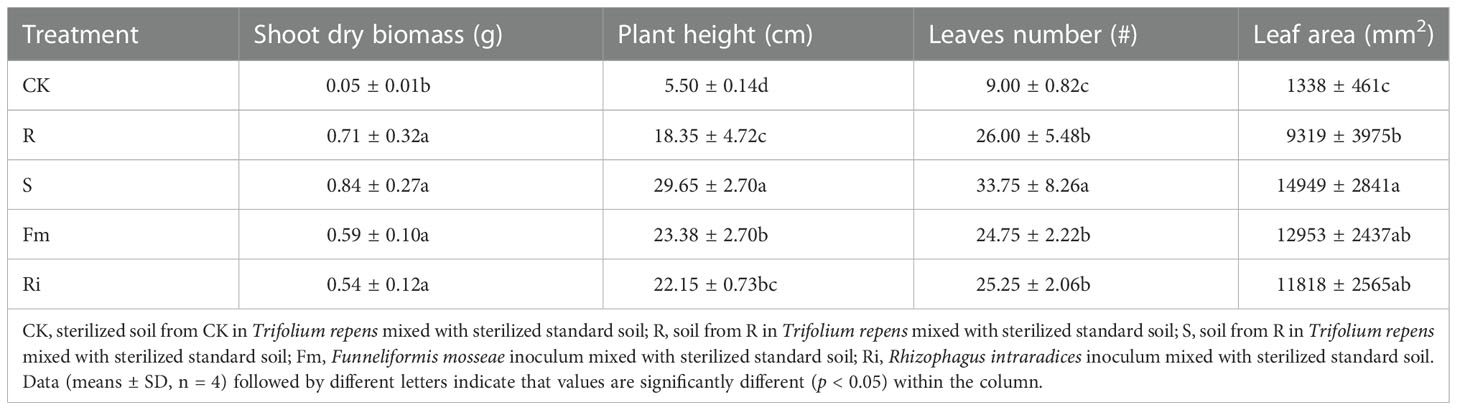
Table 2 The effects of indigenous AMF and commercial AMF isolates on growth performance of Pyrus betulaefolia.
The CK treatment did not display any AMF colonization in the root of pear plants, while mycorrhizal colonization was significantly affected by the diverse arbuscular mycorrhiza taxa (Table 3). Root mycorrhizal colonization among AMF treatments were ranked as S > R > Fm = Ri. However, the results also revealed that there were no significant differences among all treatments for the diversity of bacterial communities (Figures S2, S3). All of the root morphology attributes measured were higher in the treatments compared to CK (Table 3). Among the various treatments, root dry biomass was higher in Fm compared with the R treatment, root length was highest in the S treatment and, root volume and surface area were higher in the indigenous AMF treatments compared to the Ri isolate.
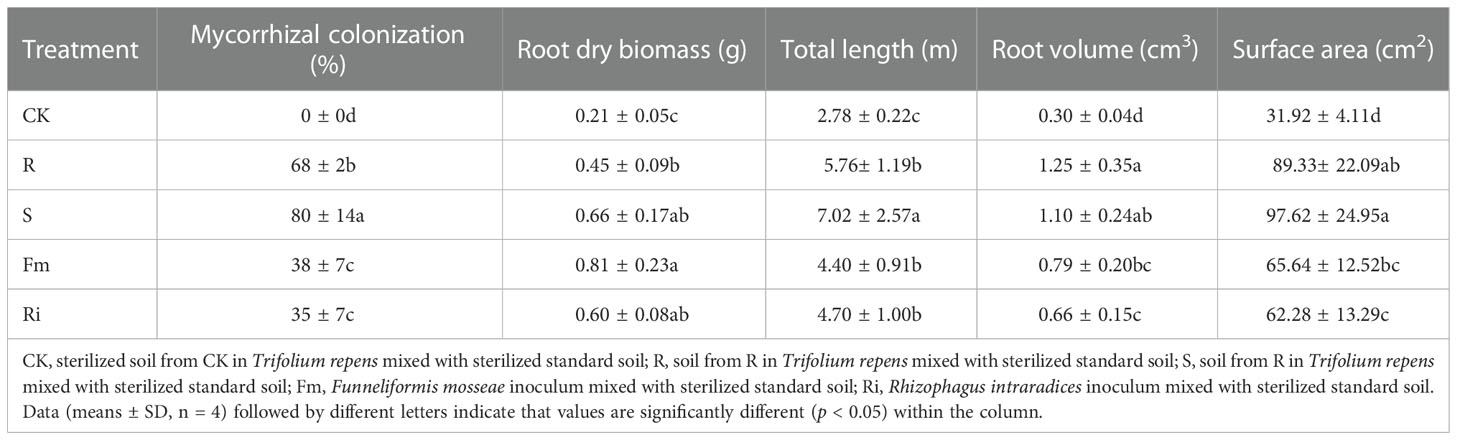
Table 3 The root mycorrhizal colonization of indigenous AMF and commercial AMF isolates and the effects on root morphology of Pyrus betulaefolia.
The effects of diverse AMF taxa on mineral nutrients in leaves, stems and roots of pear plants were determined. AMF inoculation treatments resulted in higher mineral nutrient accumulation than that in CK treatment in all tissues (Figure 2). Indigenous AMF treatment promoted mineral accumulation to a greater extent than the commercial AMF isolates in shoot, but the commercial AMF isolates resulted in better mineral accumulation in the root (Figure 2).
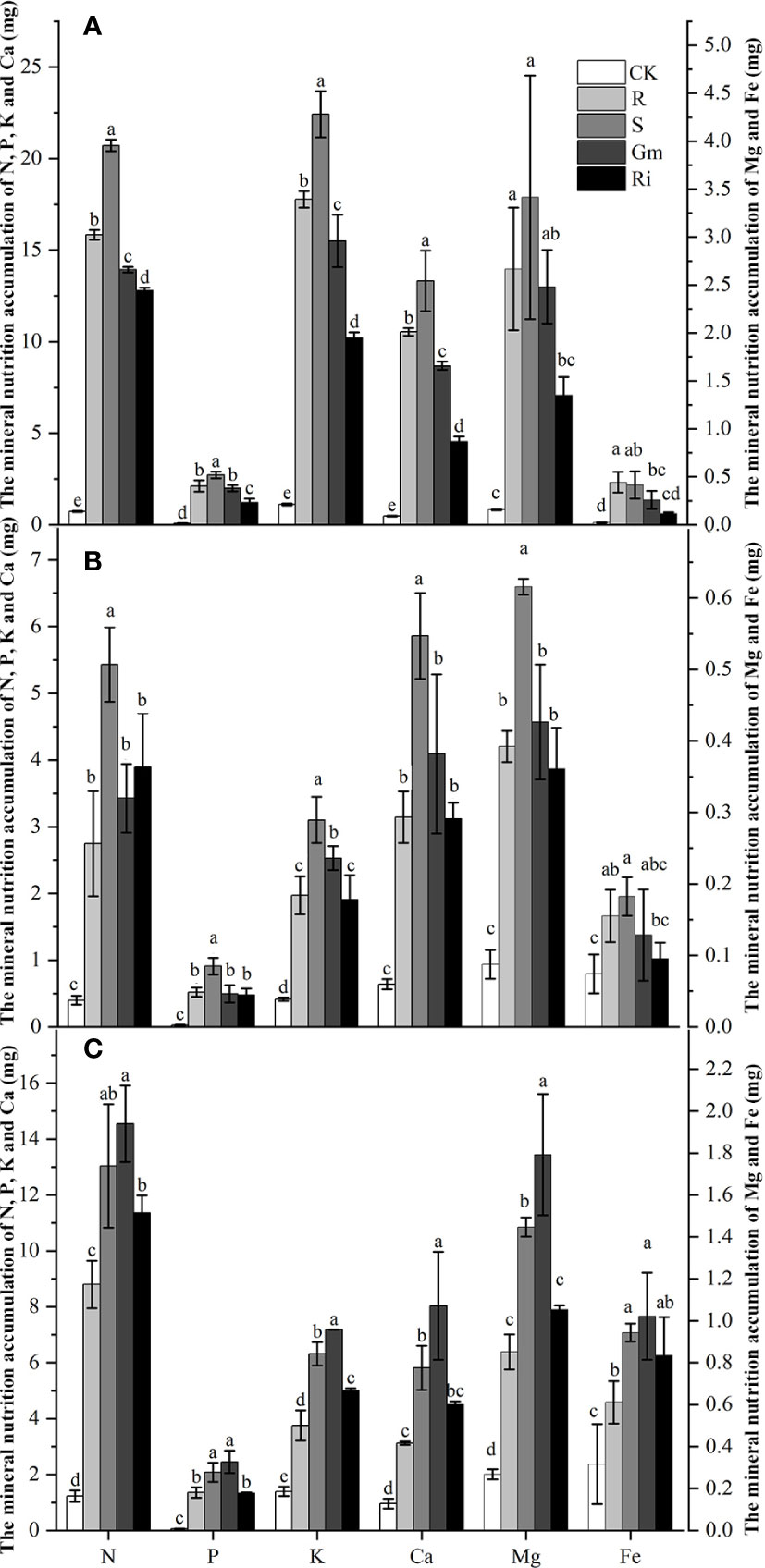
Figure 2 The mineral nutrition accumulation of Pyrus betulaefolia in different treatment. (A) Mineral nutrition accumulation of leaves (A), stem (B) and root (C). CK, sterilized soil from CK in Trifolium repens mixed with sterilized standard soil; R, soil from R in Trifolium repens mixed with sterilized standard soil; S, soil from S in Trifolium repens mixed with sterilized standard soil; Fm, Funneliformis mosseae inoculum mixed with sterilized standard soil; Ri, Rhizophagus intraradices inoculum mixed with sterilized standard soil. Data (means ± SD, n = 3) followed by different letters indicate that values are significantly different (p < 0.05) within the treatments.
In this study, 410 metabolites were detected in pear roots: mainly flavonoids, organo-oxygen compounds, prenol lipids, carboxylic acids and derivatives, glycerophospholipids, benzene and substituted derivatives, isoflavonoids, benzopyrans, coumarins and derivatives, organo-nitrogen compounds, steroids and steroid derivatives, glycerolipids and 2-arylbenzofuran flavonoids (Figure S4). In order to investigate differences in root metabolism, we focused on flavones, isoflavones, fatty acids, carbohydrates, amino acids, glycerolipids and glycerophospholipids that significantly differed across treatments.
In this study, compared with CK, multiple types of flavones, isoflavones, and carbohydrates decreased, whereas fatty acids, amino acids, glycerolipids, and glycerophospholipids increased in the AMF treatments (Figure 3).
Interestingly, the relative abundance of amino acids, glycerolipids, flavonoids, isoflavones, and fatty acids did not exhibit consistent changes in response to treatments. (Figure 3 and Table 4). Compared with CK, the relative abundance of flavonoids, carbohydrates, and fatty acids were significantly decreased in R, S, Fm and Ri treatments (Table 4). In addition, the relative abundance of amino acids, glycerophospholipids and glycerolipids in the AMF treatment were higher than CK (Table 4).
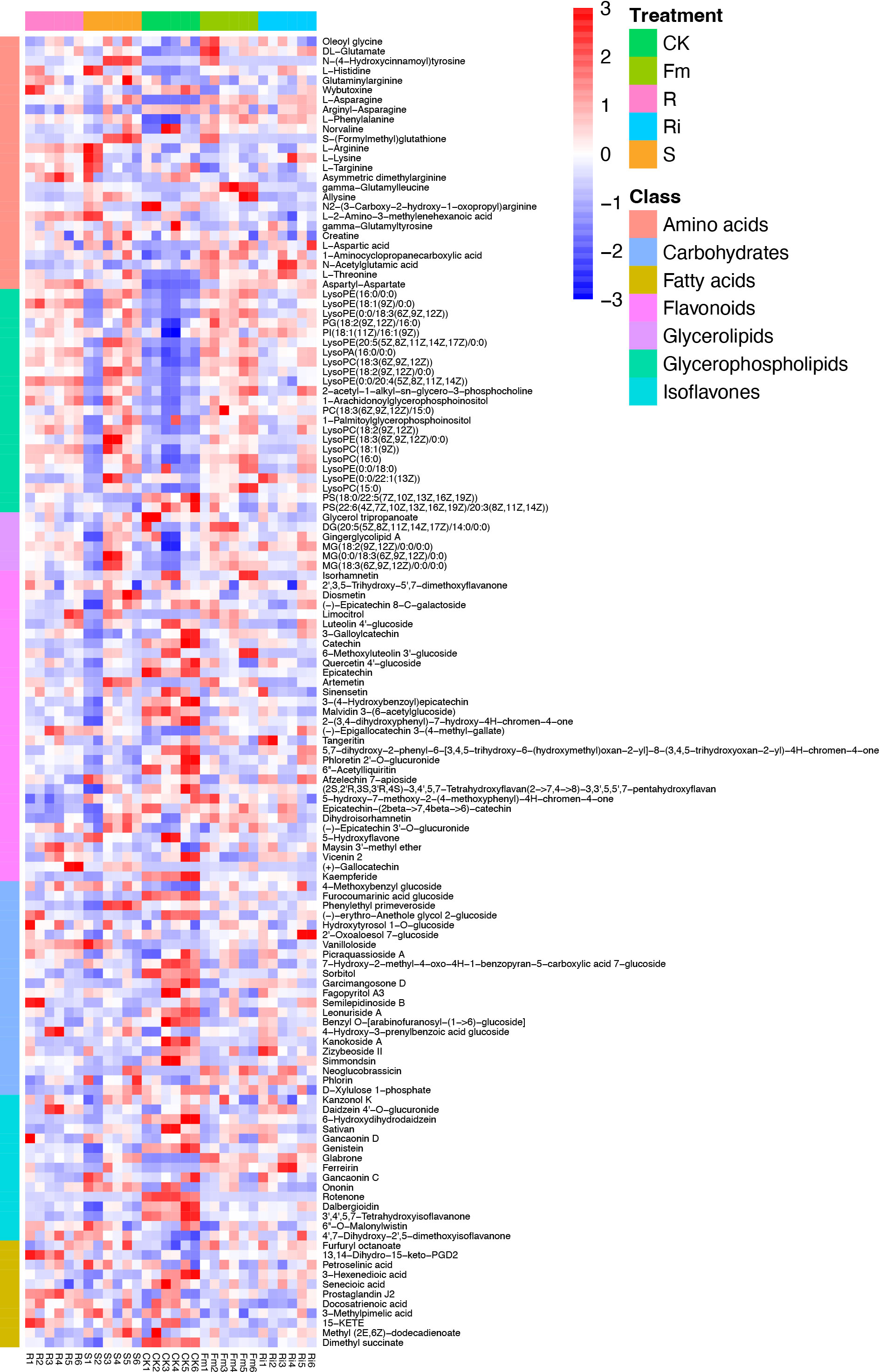
Figure 3 Heatmap of metabolites in Pyrus betulaefolia root. CK, sterilized soil from CK in Trifolium repens mixed with sterilized standard soil; R, soil from R in Trifolium repens mixed with sterilized standard soil; S, soil from S in Trifolium repens mixed with sterilized standard soil; Fm, Funneliformis mosseae inoculum mixed with sterilized standard soil; Ri, Rhizophagus intraradices inoculum mixed with sterilized standard soil.
The relative abundance of amino acids, flavonoids and carbohydrates showed different patterns between indigenous AMF and commercial AMF isolate treatments. The relative abundance of amino acids was higher in R and S, compared to Fm and CK treatments. The flavonoids and carbohydrates decreased significantly in the order, indigenous AMF, followed by specific isolates, and then by CK treatment.
To further compare the differences at the transcriptional levels between indigenous AMF and commercial AMF isolates, we sequenced the transcriptome of Fm and S, which resulted in better growth than other treatments, but also displayed significant differences in their metabolomes.
In the transcriptome 29,735,723 and 29,576,461 clean reads were obtained from the S and Fm root sample and accounted for 88.64% and 88.59% of the total sequences, respectively (Table S1). The reference genome for transcriptome analysis was the Chinese white pear (P. bretschneideri), and it resulted in matches of 76.30% and 76.56% of gene sequences for S and Fm, respectively (Table S1). We found a total of 1,217 differentially expressed genes (DEGs) for Fm/S, which included 682 up-regulated genes and 535 down-regulated genes (Figure S5).
KEGG analysis showed that the DEGs between Fm and S were primarily involved in metabolic pathways (Figure 4). The transcripts associated with metabolic pathways of fructose and mannose metabolism (ko00051), fatty acid biosynthesis (ko00061) and flavonoid biosynthesis (ko00941), were significantly different between Fm and S treatments. We found 7 DEGs in Fm compared with S related to the fructose and mannose metabolism; these genes encoded enzymes such as mannose-6-phosphate isomerase (MPI), diphosphate-dependent phosphofructokinase (PFP), fructose-bisphosphate aldolase, class I (ALDO), triosephosphate isomerase (TPI) and mannan endo-1,4-beta-mannosidase (MAN) (Table 5 and Figure S6). Compared to the S treatment, MAN (LOC103948539, LOC103960198), MPI (LOC103937460, LOC103948500), ALDO (LOC103934930) and TPI (LOC103965560) were significantly up-regulated by Fm treatment, while the PFP (LOC103955841) was significantly down-regulated. In the fatty acid biosynthesis pathway, the gene for the key enzyme of fatty acid biosynthesis, 3-oxoacyl-[acyl-carrier protein] reductase (FabG, LOC103960485), was significantly up-regulated and the acyl-[acyl-carrier-protein] desaturase (Fab2, LOC103939571) and long-chain acyl-CoA synthetase (FadD, LOC103964146) was down-regulated in Fm treatment. Four genes encoding fatty acyl-ACP thioesterase B (FATB) showed a different expression patterns: up-regulation of LOC103941054 and LOC103947075 and down-regulation of LOC103953493 and LOC103950938 (Table 5 and Figure S7). In the flavonoid biosynthesis pathway, compared to S, the genes encoding 5-O-(4-coumaroyl)-D-quinate 3’-monooxygenase (C3’H, LOC103944997), shikimate O-hydroxycinnamoyltransferase (HCT, LOC103960625), caffeoyl-CoA O-methyltransferase (CCoAOMT, LOC103965012) and flavonol synthase (FLS, LOC103930998 and LOC103961089) were up-regulated in Fm and the HCT(LOC103940185), bifunctional dihydroflavonol 4-reductase (DFR, LOC103954960) and FLS (LOC103938099) were down-regulated (Table 5 and Figure S8).
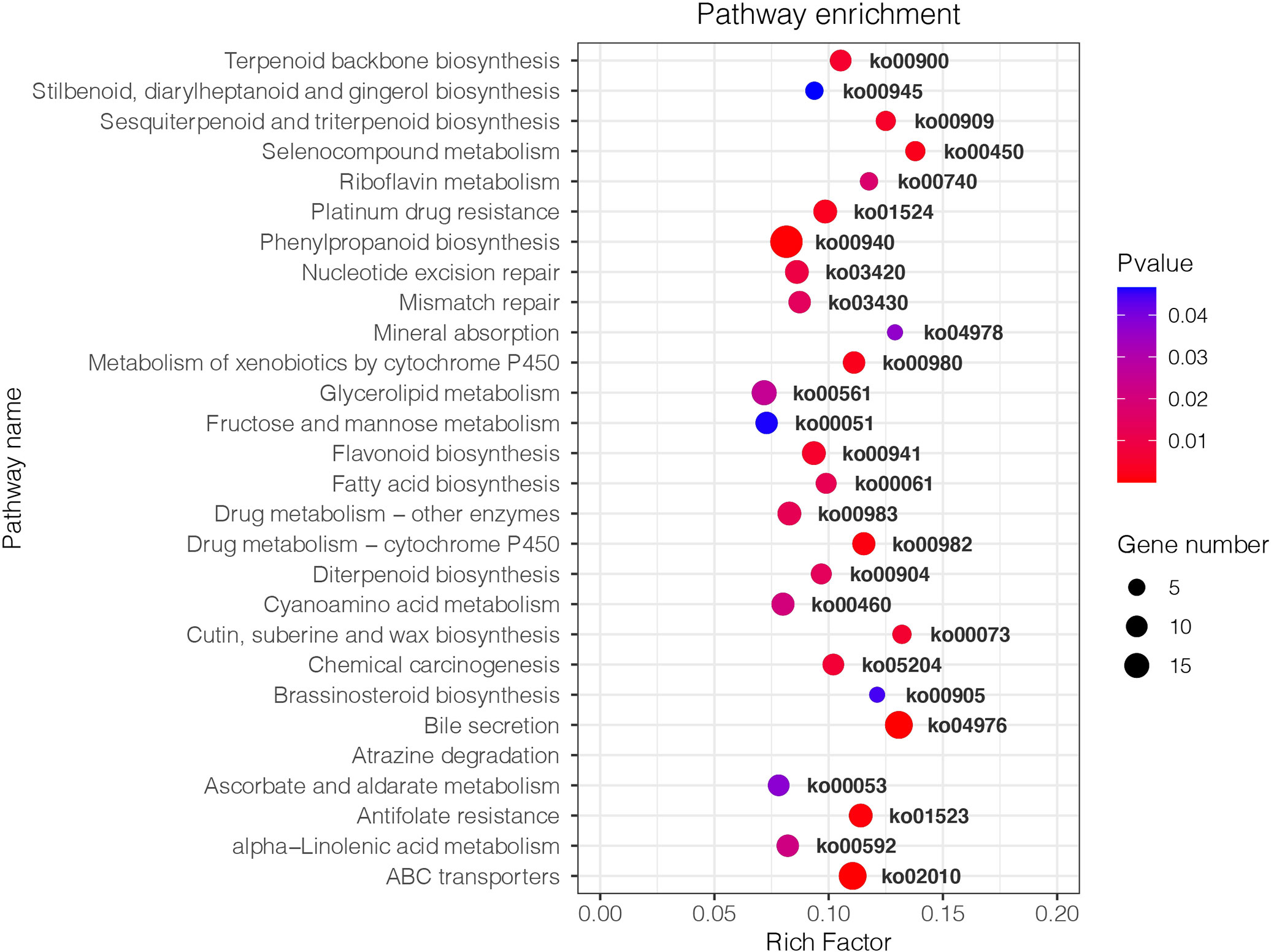
Figure 4 Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs in Pyrus betulaefolia roots between Fm and S. The size of the points represents the number of differentially expressed genes, and different colors represent different P-values. The larger the Rich Factor is the higher the degree of enrichment. S, soil from S in Trifolium repens mixed with sterilized standard soil; Fm, Funneliformis mosseae inoculum mixed with sterilized standard soil.
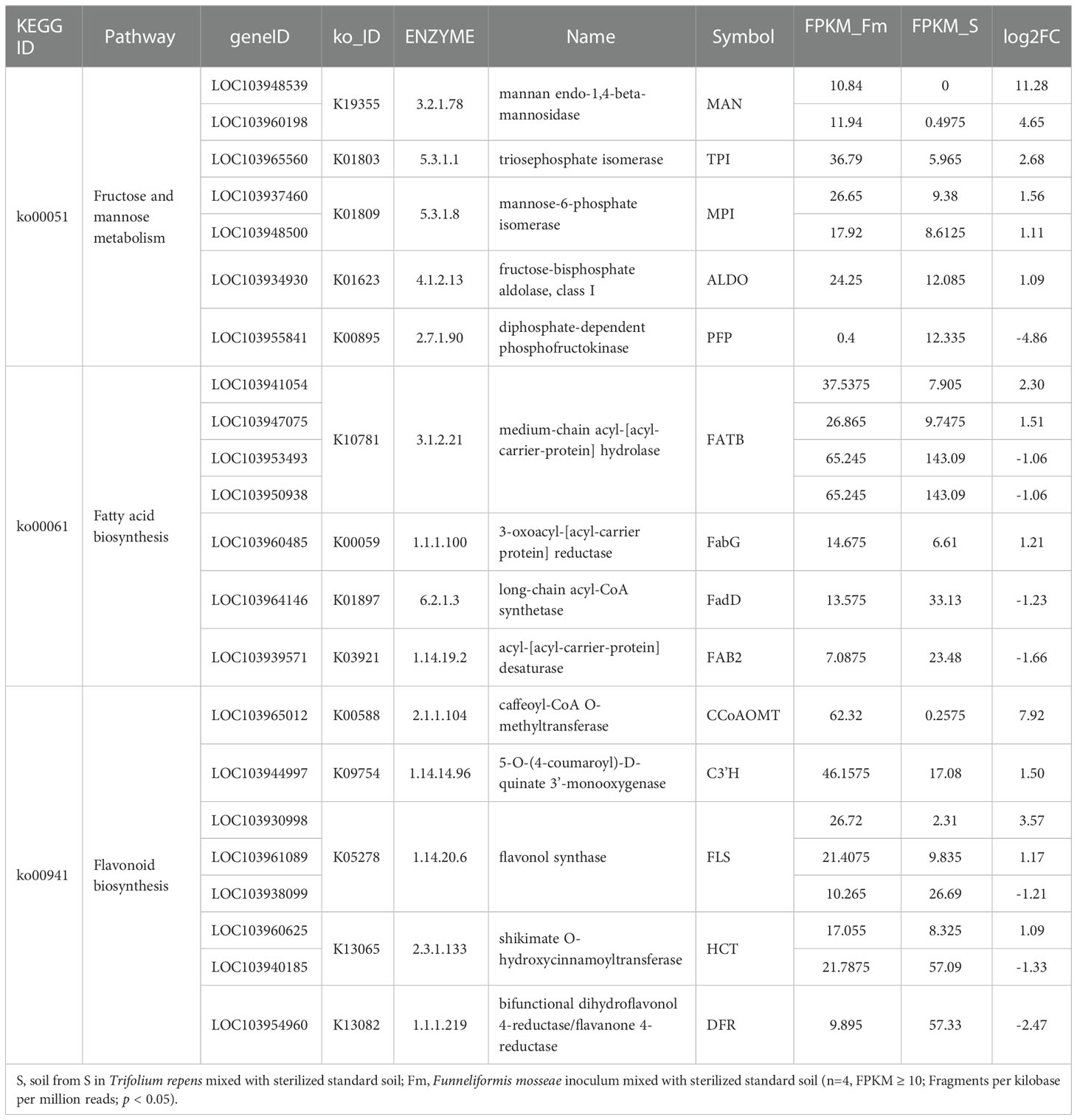
Table 5 KEGG enrichment analysis of fructose and mannose metabolism, fatty acid biosynthesis and flavonoid biosynthesis pathway in Pyrus betulaefolia roots between Fm and S.
In this study, we found that the mycorrhizal colonization in white clover root was significantly higher in S treatment, compared with the R treatment, and the accumulation of N, P, and K and biomass showed no differences, suggesting that the microorganisms in the root from pear plant could not assist the AMF proliferation. White clover may reshape the rhizosphere microorganism taxa through its root exudates to enrich its preferred microorganisms, resulting in microbial composition differences between propagation soil and original soil sample (Blažková et al., 2021). However, such changes may be transient, as the microorganism species ratios converged towards the spontaneously established composition within one plant generation (Blažková et al., 2021). In addition, the results also revealed there were no significant differences among all treatments for the diversity of bacterial communities (Figures S2, S3), indicating that bacterial communities tend to be identical through plant root selection and environmental factors in the process of culture (Fitzpatrick et al., 2018).
AMF form symbiotic associations with plant roots and enhance nutrient uptake, plant growth and tolerance to biotic and abiotic stresses (Berger and Gutjahr, 2021). The ability of plants to react to AMF with changes in morphology and/or performance in terms of yield is termed ‘AM responsiveness’ (Berger and Gutjahr, 2021). The effects of commercial AMF isolates on plants are often affected by a variety of factors (such as organic matter concentration, pH, total N, available P concentration, texture of soil, climate and climate) (Pellegrino et al., 2015; Islam et al., 2021). Previous studies have shown that commercial AMF does not always show growth-promoting effects, and sometimes requires long time for colonization due to various factors (Li et al., 2019; Islam et al., 2021). However, the commercial AMF can strongly affect the local AMF communities and change their original function; some commercial AMF may even inhibit the diversity of AMF community (depending on phylogenetic relatedness and life history strategies) (Mummey et al., 2009). In this study, plant performance in terms of growth parameters and mineral nutrition accumulation in the AMF treatment were greater than under no-AMF treatment, wherein indigenous AMF performed better than single isolates. Meanwhile, the effects on pear plant growth, root architecture and colonization of Fm and S were better than other treatments (Ri and R treatments), which can be attributed to compatibility. Such compatibility between AMF and plant has been previously noted among commercial AMF isolates belonging to different species, as well as among isolates of the same species (Jansa et al., 2008). The AM responsiveness of plants varied from negative to positive depending on plant and AMF species, mainly because of the mutual selection of the plant and the AMF owing to compatibility. Thus, in this study similar selection may allow for the pear plant to establish efficient symbionts with Fm rather than Ri.
A mixture of AMF species that are complementary in their functions provide greater benefit for the plant than any single species. High diversity of indigenous AMF provide more benefits for nutrient uptake, drought tolerance, and pathogen protection rather than commercial AMF isolates (Wagg et al., 2011; Berruti et al., 2015; Johnson and Gibson, 2020), because it allows for plants to select appropriate cooperative partners to minimize carbon consumption and maximize acquisition of soil resources (Whiteside et al., 2019). The benefits on pear growth of S treatment were higher than that for R in the present study, which is consistent with our first hypothesis. A possible explanation may be that the microorganisms enriched in the root of pear may also inhibit the formation of mycorrhiza. A previous study suggests that abiotic and biotic factors in soil can inhibit mycorrhizal activities (Cruz-Paredes et al., 2020). There was no difference in the alpha- and beta-diversity of bacterial communities among treatments (Figures S2, S3), indicating differences in bacterial communities may not be a primary reason for higher mycorrhizal colonization in both clover and pear trees in soil without roots. In addition, active compounds, such as soluble phenolic compounds from plant litter can also affect the colonization of AMF due to their toxicity effects. However there are only a few studies on soluble phenolic compounds from litter and their inhibitory effects on AMF abundance (Piotrowski et al., 2008). Such active compounds may stimulate antagonistic fungi that compete for space and nutrients, or result in a combination of both where AMF are suppressed and other fungi capable of phenolic detoxification proliferate (Piotrowski et al., 2008). In addition, niche competition between AMF and other fungi also affects AMF colonization. Such interactions warrant further studies (Xie et al., 2021). In general, indigenous AMF are more adapted to the local environment and thus show prominent effects in terms of growth improvement. However, in some ways, Fm appeared to work better, which may be related to differences in microbes in natural soils, which may inhibit the growth of pear trees (LÜ et al., 2018).
Interestingly, we found that the root mineral nutrient accumulation in Fm was higher than in S, but the shoot mineral nutrition accumulation, including leaves and stem, was lower. The discrepancies in the effects of mineral nutrients accumulation between the shoot and root may be attributed to the effects of the AMF community which directly regulate root metabolism or due to differences in root to shoot partitioning. This phenomenon indicates that some AMF taxa in indigenous AMF could promote translocation of nutrients in plants. A previous study reported that AMF could enhance the isotopic fractionation of Acacia caven xylem water, indicating that AMF can not only affect the plant xylem transport, but also result in selective transport of mineral nutrients (Poca et al., 2019). These findings support the second hypothesis that high AMF diversity is more effective than commercial AMF isolates in pear plant as a suite of complementary functions owing to greater diversity may allow for more comprehensive promotion of pear tree growth.
To further investigate the effects of indigenous AMF and commercial AMF isolates on plant root metabolism, we analyzed plant root metabolites across all treatments. In this study, AMF-inoculation mainly affected the accumulation of particular types of flavones, isoflavones, fatty acids, carbohydrates, amino acids, glycerolipids and glycerophospholipids. Results showed that AMF could decrease the flavonoids and carbohydrates and increase amino acids, glycerophospholipids and glycerolipids including their relative abundance. R. irregularis is a fatty acid auxotroph and fatty acids synthesized in the host plants are transferred to the fungus to sustain mycorrhizal colonization (Jiang et al., 2017). AMF uses lipids in roots as carbon sources, as well as sugars, which may promote the transformation of carbohydrates in roots into fatty acids. Subsequently, the lipid export pathway contributes a substantial amount of carbon to the AMF. This explains the reduced metabolite type and relative abundance of carbohydrates in the AMF treatment. Meanwhile, due to the consumption of fatty acids by AMF, the relative abundance of total fatty acids in roots is likely reduced.
The accumulation of certain flavonoids in mycorrhizal plants not only displayed time-specificity, but were also controlled by AMF species (Larose et al., 2002). Tian et al. (2021) found that higher concentrations of flavonoids (quercetin) in root exudates could enhance AMF colonization and biomass, indicating quercetin might be a key chemical signal affecting AM fungal associations (Tian et al., 2021). In our study, quercetin 4’-glucoside was higher in CK treatment than other AMF treatments (Figure 3), which could be attributed to the roots secreting specific flavonoids into the soil to induce mycorrhizal colonization, and reshaping of the AMF community. Moreover, flavonoids also have many other functions, including UV protection, as antioxidants, pigments, auxin transport regulation, defense against pathogens and signaling (Hassan and Mathesius, 2012). The flavonoids secretion of root enhanced by AMF is conducive to improve the adaptation of plants to the environment, and plays an important role in stabilizing the rhizosphere microorganisms and maintaining dynamic equilibrium.
The relative abundance of amino acids and carbohydrates showed substantially different trends between indigenous AMF treatment and commercial AMF isolates treatment. The higher relative abundance of amino acids in R and S and lower carbohydrates suggested that indigenous AMF enhanced the process of converting carbohydrates into amino acids, indicating indigenous AMF is more conducive to the accumulation of nitrogenous compounds and growth of the root system. Meanwhile, the relative abundance of flavonoids was higher in single isolates treatment, suggesting that single isolates required greater flavonoid biosynthesis in pear roots to enhance colonization than did the indigenous AMF. These results indicate that greater diversity in communities of indigenous AMF allow for functional complementarity within communities (Maherali and Klironomos, 2007).
Although indigenous AMF and commercial AMF isolates played a similar role in promoting pear tree growth, there were significant differences in metabolic pathways affected by them (Figure 3 and Table 4). The diversity of metabolome between indigenous AMF treatment and commercial AMF isolates treatment may be associated with differences in the root transcriptomes. To test our third hypothesis, we focused on the DEGs in the pear root in response to Fm and S treatments. The DEGs were enriched in fructose and mannose metabolism, fatty acid biosynthesis, and flavonoid biosynthesis pathways. The gene expression of MAN was up-regulated by around 11-and 5-fold and PFP was down-regulated by around 5-fold in the Fm compared to S suggesting that the encoded enzymes respond to different types of AMF. As a key modifying enzyme of mannans, MAN plays an important role in cell wall hydrolysis, endosperm weakening in seedling development, endosperm degradation after the completion of seed germination and anther and pollen development (Yuan et al., 2007). MAN up-regulation may enhance cell wall hydrolysis and contribute to modifying root morphology in the Fm treatment (Inukai et al., 2012). PFP catalyzes reversible interconversion of Fructose 6-phosphate to Fructose 1,6-bisphosphate and may function in glycolysis to facilitate the survival of plants in low Pi environments by recycling Pi from PPi and conserving limited ATP pools in roots of plants that do not form symbiotic associations with mycorrhizal fungi (Murley et al., 1998). The P accumulation of root was higher in Fm compared to S suggesting Fm has an advantage over S in Pi absorption (Figure 2). Potentially, increase Pi availability which may down-regulate PFP expression.
Several key genes associated with fatty acid biosynthesis were identified as DEGs in this study, including 3.1.2.14 (fatty acyl-ACP thioesterase B, FATB), 1.14.19.2 (acyl-[acyl-carrier-protein] desaturase, FAB2), 6.2.1.3 (long-chain acyl-CoA synthetase, FadD) and 1.1.1.100 (3-oxoacyl-[acyl-carrier protein] reductase, FabG). FAD2 in the endoplasmic reticulum catalyzes the desaturation of fatty acids attached to phospatidylcholine (PC) from PC-18: 1 to PC-18:2 (Okuley et al., 1994). It’s down-regulated expression likely resulted in inhibition of accumulation of hexadecenoic acid and octadecenoic acid in the Fm, compared to S treatment (Table 5 and Figure S5). Meanwhile, expression of the gene coding for FATB was up-regulated, likely enhancing the accumulation of octanoic acid, decanoic acid, dodecanoic acid, tetradecanoic acid and hexadecanoic acid. Sugiura et al. (2020) used eight saturated or unsaturated fatty acids (C12 to C18) and two β-monoacylglycerols to determine whether AM fungi can grow on medium supplied with fatty acids or lipids under asymbiotic conditions, demonstrating that myristate boosts the asymbiotic growth of AMF and can also serve as a carbon and energy source (Sugiura et al., 2020). These results suggest that Fm treatment can inhibit the biosynthesis of C16:0 and C18:0 and enhance the synthesis of tetradecanoic acid (myristic acid) in pear roots compared with S (Figure 5). The accumulation of myristic acid could stimulate spore germination and build-up of AMF biomass (Rillig et al., 2020), suggesting similar responses in pear roots.
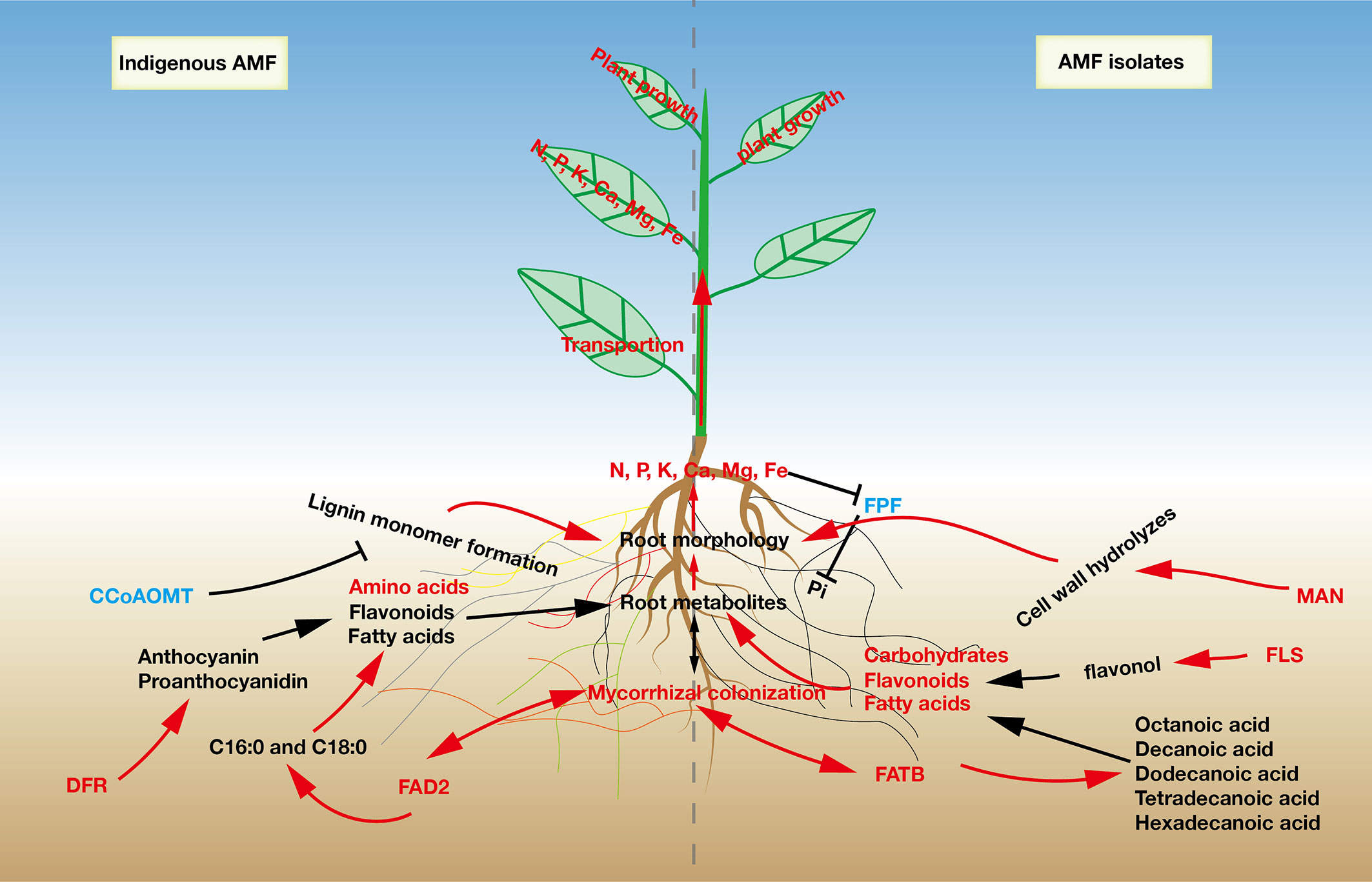
Figure 5 A proposed model to describe the effects of indigenous AMF and commercial AMF isolates on root responses in Pyrus betulaefolia.
In the flavonoid biosynthesis pathway, we focused on the genes encoding CCoAOMT, FLS and DFR. CCoAOMT has been suggested to be involved in a parallel pathway to lignin monomer formation, which promotes secondary cell wall formation (Marita et al., 2003). Down-regulation of CCoAOMT inhibited the synthesis of the secondary cell wall and allowed for root elongation (Toorchi et al., 2009), indicating that S may regulate lignin monomer formation pathway thereby affecting root morphology. The synthesis of flavonol aglycones has long been attributed to FLS, while DFR drives flux away from flavonols into anthocyanin and proanthocyanidin biosynthesis (Owens et al., 2008). The gene encoding FLS was up-regulated and that for DFR was down-regulated in the Fm treatment, indicating that Fm induced flavonoid synthesis toward flavonol synthesis, while S treatment resulted in diversion of flavonoid synthesis toward anthocyanin and proanthocyanidin biosynthesis (Figure 5).
To reduce the application of chemical fertilizers in pear production and to maintain the ecological health of the soil, we studied the effects of indigenous AMF and commercial AMF isolates on the differences in the growth of pear seedlings. In our study, we used two different AMF propagation methods (R and S) to simulate spore propagation in natural soils. After propagation, root mycorrhizal colonization and arbuscular abundance in S was significantly higher than in R, indicating that the microorganisms in the root from pear plant could limit AMF proliferation, which is consistent with our first hypothesis. These results showed that the inoculum with the plant root may contain plant-specific metabolites or indigenous microorganisms that inhibit AMF colonization of the next host plant. We also found that the combined effects of indigenous AMF on pear tree were better than that of commercial AMF isolates on plant growth, root morphology, and mineral nutrient accumulation. Analyses of transcriptomes and metabolomes demonstrated that pear roots displayed specific changes in metabolism and gene expression to respond to indigenous AMF and commercial AMF isolates, especially in the metabolism of flavones, carbohydrates and fatty acids. Taken together, our findings demonstrate the response of pear seedlings to indigenous AMF and commercial AMF isolates in aspects from the transcriptional level, metabolic capacity, mineral nutrient accumulation and plant growth, providing a new understanding of the relationship between AMF community and pear tree growth (Figure 5).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://figshare.com/s/b81244b2c1bb72c49b2e and https://www.ncbi.nlm.nih.gov/, PRJNA761485.
YX and CD conceived and designed the experiments; WF, HP and HL performed the experiments; YS, SJ, PL and RJ analyzed the data; YS and SJ wrote the manuscript; TC, SN, TY and GJ revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Science and Technology Support Program of Jiangsu Province (BE2018389), China Agriculture Research System (CARS-28-10), and the Guangxi science and technology research program (AD20159001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1040134/full#supplementary-material
Banerjee, S., Walder, F., Buchi, L., Meyer, M., Held, A. Y., Gattinger, A., et al. (2019). Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 13, 1722–1736. doi: 10.1038/s41396-019-0383-2
Berger, F., Gutjahr, C. (2021). Factors affecting plant responsiveness to arbuscular mycorrhiza. Curr. Opin. Plant Biol. 59, 101994. doi: 10.1016/j.pbi.2020.101994
Berruti, A., Borriello, R., Lumini, E., Scariot, V., Bianciotto, V., Balestrini, R. (2013). Application of laser microdissection to identify the mycorrhizal fungi that establish arbuscules inside root cells. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00135
Berruti, A., Lumini, E., Balestrini, R., Bianciotto, V. (2015). Arbuscular mycorrhizal fungi as natural biofertilizers: Let's benefit from past successes. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01559
Blažková, A., Jansa, J., Püschel, D., Vosátka, M., Janoušková, M. (2021). Is mycorrhiza functioning influenced by the quantitative composition of the mycorrhizal fungal community? Soil Biol. Bichem 157, 1082149. doi: 10.1016/j.soilbio.2021.108249
Compant, S., Clément, C., Sessitsc, A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Bichem 42, 669–678. doi: 10.1016/j.soilbio.20
Cruz-Paredes, C., Jakobsen, I., Nybroe, O. (2020). Different sensitivity of a panel of rhizophagus isolates to AMF-suppressive soils. Appl. Soil Ecol. 155, 103662. doi: 10.1016/j.apsoil.2020.103662
Delgado-Baquerizo, M., Maestre, F. T., Reich, P. B., Jeffries, T. C., Gaitan, J. J., Encinar, D., et al. (2016). Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7, 10541. doi: 10.1038/ncomms10541
Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., Johnson, M. T. J. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. P Natl. Acad. Sci. U.S.A. 115, E1157–E1165. doi: 10.1073/pnas.1717617115
Foley, J. A., Ramankutty, N., Brauman, K. A., Cassidy, E. S., Gerber, J. S., Johnston, M., et al. (2011). Solutions for a cultivated planet. Nature 478, 337–342. doi: 10.1038/nature10452
Hassan, S., Mathesius, U. (2012). The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 63, 3429–3444. doi: 10.1093/jxb/err430
Hill, E. M., Robinson, L. A., Abdul-Sada, A., Vanbergen, A. J., Hodge, A., Hartley, S. E. (2018). Arbuscular mycorrhizal fungi and plant chemical defence: Effects of colonisation on aboveground and belowground metabolomes. J. Chem. Ecol. 44, 198–208. doi: 10.1007/s10886-017-0921-1
Huang, Z., Zhao, F., Wang, M., Qi, K., Wu, J., Zhang, S. (2019). Soil chemical properties and geographical distance exerted effects on arbuscular mycorrhizal fungal community composition in pear orchards in jiangsu province, China. Appl. Soil Ecol. 142, 18–24. doi: 10.1016/j.apsoil.2019.05.017
Inukai, Y., Sakamoto, T., Morinaka, Y., Miwa, M., Kojima, M., Tanimoto, E., et al. (2012). ROOT GROWTH INHIBITING, a rice endo-1,4-β-d-Glucanase, regulates cell wall loosening and is essential for root elongation. J. Plant Growth Regul. 31, 373–381. doi: 10.1007/s00344-011-9247-3
Islam, M. N., Germida, J. J., Walley, F. L. (2021). Survival of a commercial AM fungal inoculant and its impact on indigenous AM fungal communities in field soils. Appl. Soil Ecol. 166, 103979. doi: 10.1016/j.apsoil.2021.103979
Jansa, J., Smith, F. A., Smith, S. E. (2008). Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi? New Phytol. 177, 779–789. doi: 10.1111/j.1469-8137.2007.02294.x
Jiang, S., Hu, X., Kang, Y., Xie, C., An, X., Dong, C., et al. (2020). Arbuscular mycorrhizal fungal communities in the rhizospheric soil of litchi and mango orchards as affected by geographic distance, soil properties and manure input. Appl. Soil Ecol. 152, 103593. doi: 10.1016/j.apsoil.2020.103593
Jiang, Y., Wang, W., Xie, Q., Liu, N., Liu, L., Wang, D., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1171–1175. doi: 10.1126/science.aam9970
Jin, Y., Liu, H., Luo, D., Yu, N., Dong, W., Wang, C., et al. (2016). DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 7, 12433. doi: 10.1038/ncomms12433
Johnson, N. C., Gibson, K. S. (2020). Understanding multilevel selection may facilitate management of arbuscular mycorrhizae in sustainable agroecosystems. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.627345
Kaiser, C., Kilburn, M. R., Clode, P. L., Fuchslueger, L., Koranda, M., Cliff, J. B., et al. (2015). Exploring the transfer of recent plant photosynthates to soil microbes: mycorrhizal pathway vs direct root exudation. New Phytol. 205, 1537–1551. doi: 10.1111/nph.13138
Kameoka, H., Tsutsui, I., Saito, K., Kikuchi, Y., Handa, Y., Ezawa, T., et al. (2019). Stimulation of asymbiotic sporulation in arbuscular mycorrhizal fungi by fatty acids. Nat. Microbiol. 4, 1654–1660. doi: 10.1038/s41564-019-0485-7
Kanehisa, M. (2019). Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. doi: 10.1002/pro.3715
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M., Tanabe, M. (2021). KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551. doi: 10.1093/nar/gkaa970
Larose, G., Chênevert, R., Moutoglis, P., Gagné, S., Piché, Y., Vierheilig, H. (2002). Flavonoid levels in roots ofMedicago sativa are modulated by the developmental stage of the symbiosis and the root colonizing arbuscular mycorrhizal fungus. J. Plant Physiol. 159, 1329–1339. doi: 10.1078/0176-1617-00896
Li, Y., Gan, Y., Lupwayi, N., Hamel, C. (2019). Influence of introduced arbuscular mycorrhizal fungi and phosphorus sources on plant traits, soil properties, and rhizosphere microbial communities in organic legume-flax rotation. Plant Soil 443, 87–106. doi: 10.1007/s11104-019-04213-8
Liu, C.-Y., Guo, X.-N., Wu, X.-L., Dai, F.-J., Wu, Q.-S. (2022). The comprehensive effects of rhizophagus intraradices and p on root system architecture and p transportation in citrus limon l. Agriculture 12, 317. doi: 10.3390/agriculture12030317
Lopez-Raez, J. A., Verhage, A., Fernandez, I., Garcia, J. M., Azcon-Aguilar, C., Flors, V., et al. (2010). Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway. J. Exp. Bot. 61, 2589–2601. doi: 10.1093/jxb/erq089
LÜ, L.-H., Srivastava, A. K., Shen, Y.-L., Wu, Q.-S. (2018). A negative feedback regulation of replanted soil microorganisms on plant growth and soil properties of peach. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 47, 255–261. doi: 10.15835/nbha47111344
Maherali, H., Klironomos, J. N. (2007). Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316, 1746–1748. doi: 10.1126/science.1143082
Marita, J. M., Ralph, J., Hatfield, R. D., Guo, D., Chen, F., Dixon, R. A. (2003). Structural and compositional modifications in lignin of transgenic alfalfa down-regulated in caffeic acid 3-o-methyltransferase and caffeoyl coenzyme a 3-o-methyltransferase. Phytochemistry 62, 53–65. doi: 10.1016/S0031-9422(02)00434-X
Mummey, D. L., Antunes, P. M., Rillig, M. C. (2009). Arbuscular mycorrhizal fungi pre-inoculant identity determines community composition in roots. Soil Biol. Biochem. 41, 1173–1179. doi: 10.1016/j.soilbio.2009.02.027
Murley, V. R., Theodorou, M. E., Plaxton, W. C. (1998). Phosphate starvation-inducible pyrophsphate-dependent phosphofructokinase occurs in plants whose roots do not form symbiotic associations with mycorrhizal fungi. Physiol. Plant 103, 405–414. doi: 10.1034/j.1399-3054.1998.1030314.x
Noe, R., Kiers, E. T. (2018). Mycorrhizal markets, firms, and Co-ops. Trends Ecol. Evol. 33, 777–789. doi: 10.1016/j.tree.2018.07.007
Okuley, J., Lightner, J., Feldmann, K., Yadav, N., Lark, E., Browsea, J. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6, 147–158.
Owens, D. K., Alerding, A. B., Crosby, K. C., Bandara, A. B., Westwood, J. H., Winkel, B. S. (2008). Functional analysis of a predicted flavonol synthase gene family in arabidopsis. Plant Physiol. 147, 1046–1061. doi: 10.1104/pp.108.117457
Pellegrino, E., Öpik, M., Bonari, E., Ercoli, L. (2015). Responses of wheat to arbuscular mycorrhizal fungi: A meta-analysis of field studies from 1975 to 2013. Soil Biol. Biochem. 84, 210–217. doi: 10.1016/j.soilbio.2015.02.020
Piotrowski, J. S., Morford, S. L., Rillig, M. C. (2008). Inhibition of colonization by a native arbuscular mycorrhizal fungal community via populus trichocarpa litter, litter extract, and soluble phenolic compounds. Soil Biol. Biochem. 40, 709–717. doi: 10.1016/j.soilbio.2007.10.005
Poca, M., Coomans, O., Urcelay, C., Zeballos, S. R., Bodé, S., Boeckx, P. (2019). Isotope fractionation during root water uptake by acacia caven is enhanced by arbuscular mycorrhizas. Plant Soil 441, 485–497. doi: 10.1007/s11104-019-04139-1
Rillig, M. C., Aguilar-Trigueros, C. A., Anderson, I. C., Antonovics, J., Ballhausen, M. B., Bergmann, J., et al. (2020). Myristate and the ecology of AM fungi: significance, opportunities, applications and challenges. New Phytol. 227, 1610–1614. doi: 10.1111/nph.16527
Rivero, J., Gamir, J., Aroca, R., Pozo, M. J., Flors, V. (2015). Metabolic transition in mycorrhizal tomato roots. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00598
Smilauer, P., Kosnar, J., Kotilinek, M., Smilauerova, M. (2020). Contrasting effects of host identity, plant community, and local species pool on the composition and colonization levels of arbuscular mycorrhizal fungal community in a temperate grassland. New Phytol. 225, 461–473. doi: 10.1111/nph.16112
Sugiura, Y., Akiyama, R., Tanaka, S., Yano, K., Kameoka, H., Marui, S., et al. (2020). Myristate can be used as a carbon and energy source for the asymbiotic growth of arbuscular mycorrhizal fungi. P Natl. Acad. Sci. U.S.A. 117, 25779–25788. doi: 10.1073/pnas.2006948117
Tian, B., Pei, Y., Huang, W., Ding, J., Siemann, E. (2021). Increasing flavonoid concentrations in root exudates enhance associations between arbuscular mycorrhizal fungi and an invasive plant. ISME J. 15, 1919–1930. doi: 10.1038/s41396-021-00894-1
Toorchi, M., Yukawa, K., Nouri, M. Z., Komatsu, S. (2009). Proteomics approach for identifying osmotic-stress-related proteins in soybean roots. Peptides 30, 2108–2117. doi: 10.1016/j.peptides.2009.09.006
Van't Padje, A., Oyarte Galvez, L., Klein, M., Hink, M. A., Postma, M., Shimizu, T., et al. (2021). Temporal tracking of quantum-dot apatite across in vitro mycorrhizal networks shows how host demand can influence fungal nutrient transfer strategies. ISME J. 15, 435–449. doi: 10.1038/s41396-020-00786-w
Wagg, C., Jansa, J., Schmid, B., van der Heijden, M. G. (2011). Belowground biodiversity effects of plant symbionts support aboveground productivity. Ecol. Lett. 14, 1001–1009. doi: 10.1111/j.1461-0248.2011.01666.x
Whiteside, M. D., Werner, G. D. A., Caldas, V. E. A., Van't Padje, A., Dupin, S. E., Elbers, B., et al. (2019). Mycorrhizal fungi respond to resource inequality by moving phosphorus from rich to poor patches across networks. Curr. Biol. 29, 2043–2050.e2048. doi: 10.1016/j.cub.2019.04.061
Wu, Q., He, J., SRIVASTAVA, A. K., Zhang, F., Zou, Y. N. (2019). Development of propagation technique of indigenous AMF and their inoculation response in citrus. Indian J. Agric. Sci. 89, 1190–11904. doi: 10.56093/ijas.v89i7.91696
Wu, T., Pan, L., Zipori, I., Mao, J., Li, R., Li, Y., et al. (2022). Arbuscular mycorrhizal fungi enhanced the growth, phosphorus uptake and pht expression of olive (Olea europaea l.) plantlets. PeerJ 10, e13813. doi: 10.7717/peerj.13813
Xie, L., Bi, Y., Ma, S., Shang, J., Hu, Q., Christie, P. (2021). Combined inoculation with dark septate endophytes and arbuscular mycorrhizal fungi: synergistic or competitive growth effects on maize? BMC Plant Biol. 21, 498. doi: 10.1186/s12870-021-03267-0
Yang, H., Li, Y., Jin, Y., Kan, L., Shen, C., Malladi, A., et al. (2020). Transcriptome analysis of pyrus betulaefolia seedling root responses to short-term potassium deficiency. Int. J. Mol. Sci. 21, 8857. doi: 10.3390/ijms21228857
Yoshimura, Y., Ido, A., Iwase, K., Matsumoto, T., Yamato, M. (2013). Communities of arbuscular mycorrhizal fungi in the roots of pyrus pyrifolia var. culta (Japanese pear) in orchards with variable amounts of soil-available phosphorus. Microbes Environ. 28, 105–111. doi: 10.1264/jsme2.me12118
Keywords: pear plant, arbuscular mycorrhizal fungi, plant growth, mineral nutrient accumulation, metabolomic, gene responses
Citation: Shao Y, Jiang S, Peng H, Li H, Li P, Jiang R, Fang W, Chen T, Jiang G, Yang T, Nambeesan SU, Xu Y and Dong C (2023) Indigenous and commercial isolates of arbuscular mycorrhizal fungi display differential effects in Pyrus betulaefolia roots and elicit divergent transcriptomic and metabolomic responses. Front. Plant Sci. 13:1040134. doi: 10.3389/fpls.2022.1040134
Received: 14 September 2022; Accepted: 12 December 2022;
Published: 09 January 2023.
Edited by:
Alessandra Turrini, University of Pisa, ItalyCopyright © 2023 Shao, Jiang, Peng, Li, Li, Jiang, Fang, Chen, Jiang, Yang, Nambeesan, Xu and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caixia Dong, Y3hkb25nQG5qYXUuZWR1LmNu; Savithri U. Nambeesan, Nambeesan, c3VuYW1iQHVnYS5lZHU=; Tianjie Yang, dGp5YW5nQG5qYXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.