
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 November 2022
Sec. Plant Nutrition
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1039041
This article is part of the Research Topic Roles of Nitrogen in Plant Adaptation to Nutrient Deficiency and Ionic Stress View all 5 articles
 Alessandra Koltun1†
Alessandra Koltun1† Rodolfo A. Maniero1†
Rodolfo A. Maniero1† Marielle Vitti1
Marielle Vitti1 Nathalia de Setta2,3
Nathalia de Setta2,3 Ricardo F. H. Giehl4
Ricardo F. H. Giehl4 Joni E. Lima1,5*
Joni E. Lima1,5* Antonio Figueira1*
Antonio Figueira1*AMMONIUM TRANSPORTER/METHYLAMMONIUM PERMEASE/RHESUS (AMT) family members transport ammonium across membranes in all life domains. Plant AMTs can be categorized into AMT1 and AMT2 subfamilies. Functional studies of AMTs, particularly AMT1-type, have been conducted using model plants but little is known about the function of AMTs from crops. Sugarcane (Saccharum spp.) is a major bioenergy crop that requires heavy nitrogen fertilization but depends on a low carbon-footprint for competitive sustainability. Here, we identified and functionally characterized sugarcane ScAMT2;1 by complementing ammonium uptake-defective mutants of Saccharomyces cerevisiae and Arabidopsis thaliana. Reporter gene driven by the ScAMT2;1 promoter in A. thaliana revealed preferential expression in the shoot vasculature and root endodermis/pericycle according to nitrogen availability and source. Arabidopsis quadruple mutant plants expressing ScAMT2;1 driven by the CaMV35S promoter or by a sugarcane endogenous promoter produced significantly more biomass than mutant plants when grown in NH4+ and showed more 15N-ammonium uptake by roots and nitrogen translocation to shoots. In A. thaliana, ScAMT2;1 displayed a Km of 90.17 µM and Vmax of 338.99 µmoles h-1 g-1 root DW. Altogether, our results suggest that ScAMT2;1 is a functional high-affinity ammonium transporter that might contribute to ammonium uptake and presumably to root-to-shoot translocation under high NH4+ conditions.
Nitrogen (N) is the most abundant mineral element present in plant tissues, and nitrate () and ammonium (NH4+) are the primary inorganic sources absorbed by roots of higher plants. NH4+ is transported across cell membranes by proteins of the AMMONIUM TRANSPORTER/METHYLAMMONIUM PERMEASE/RHESUS (AMT/MEP/Rh) family (Gazzarrini et al., 1999; Loqué and von Wirén, 2004), which are present in all living organisms (Li et al., 2009; McDonald et al., 2012). Plant AMTs can be further categorized into AMT1 and AMT2 (Loqué and von Wirén, 2004; McDonald et al., 2012). AMT1-type proteins share an evolutionary history related to prokaryotic NH4+ transporters, while AMT2-type proteins are homologues of the methylammonium permease (MEP) family (von Wittgenstein et al., 2014). The number of AMT family members varies considerably among plant species, displaying a variety of expression patterns, spatial regulations, substrate affinities, and presumed functions (Yuan et al., 2007; Guether et al., 2009; Yuan et al., 2009; McDonald et al., 2012; Li et al., 2016; Giehl et al., 2017; Song et al., 2017).
In Arabidopsis thaliana, four root-expressed AMT1 proteins are responsible for high-affinity NH4+ uptake, namely, AtAMT1;1, AtAMT1;2, AtAMT1;3, and AtAMT1;5 (Loqué et al., 2006; Yuan et al., 2007), while AtAMT1;4 plays a major role in NH4+ uptake in pollen grains (Yuan et al., 2009). Functional studies revealed that the membrane proteins AtAMT1;1 and AtAMT1;3 are active in the rhizodermis, cortex, and root hairs and are responsible for approximately two-thirds of the NH4+ uptake capacity by the symplastic route, further supported by AtAMT1;5 in radial transport (Loqué et al., 2006; Yuan et al., 2007). In addition, the apoplastic NH4+ pool can enter the root symplast by AtAMT1;2 activity in the plasma membrane of endodermal and cortical cells (Yuan et al., 2007). Functional and regulatory characterization of AMT1 proteins in other plant species, including crops (von Wirén et al., 2000; Suenaga et al., 2003; D’Apuzzo et al., 2004; Couturier et al., 2007; Gu et al., 2013; Koegel et al., 2013), have confirmed their dominant role in high-affinity NH4+ uptake in roots (Loqué et al., 2006; Yuan et al., 2007; Gu et al., 2013).
In contrast to AMT1-type proteins, there is less information about the physiological functions of AMT2 proteins. The sole AMT2 member present in the A. thaliana genome, AtAMT2;1, was shown to have a minor role in ammonium uptake under N-deficient conditions (Giehl et al., 2017). However, at elevated N levels, AtAMT2;1 mediates ammonium accumulation in xylem sap and contributes to long-distance ammonium translocation from roots to shoots (Giehl et al., 2017). AMT2 members have been investigated in a few crops, such as wheat (Li et al., 2017b; Jiang et al., 2019), sorghum (Koegel et al., 2013) and maize (Dechorgnat et al., 2019), but various aspects of AMT2 function and regulation remain to be addressed.
Sugarcane (Saccharum spp.) is a robust feedstock for bioenergy production due to its remarkably high aboveground biomass, including culms with high sucrose content (Tilman et al., 2009; Waclawovsky et al., 2010; Hoang et al., 2015). High N fertilizer rates are applied to boost sugarcane production; however, the crop responds poorly to N fertilization, and N losses can reach up to 50% (Franco et al., 2008; Robinson et al., 2011). The reason behind the high N loss in sugarcane remains elusive (Thorburn et al., 2017; de Castro et al., 2018; Lima et al., 2022). The low nitrogen use efficiency (NUE) of sugarcane represents high economic and environmental costs (Thorburn et al., 2011; Skocaj et al., 2013), reducing the long-term sustainability of this bioenergy crop (Erisman et al., 2010). Various forms of N losses from the soil (volatilization, N2O emission, nitrate leaching, and run-off) negatively impact ecosystems (McAllister et al., 2012). Therefore, it is essential to improve sugarcane NUE to help maintain its competitiveness and sustainability as a bioenergy crop (Thorburn et al., 2017).
NUE is a complex trait involving N uptake, assimilation, and remobilization during plant development (Dobermann, 2005; Li et al., 2017a; Sharma and Bali, 2018). Prospecting genes involved in NUE is challenging, particularly in crops with a highly polyploid and complex genome, such as modern sugarcane cultivars (Thirugnanasambandam et al., 2018). Sugarcane achieves optimal growth and yield under the supply of mixed ammonium and nitrate sources (Otto et al., 2016), yet sugarcane roots have a physiological preference for ammonium over nitrate uptake under N-sufficient (Robinson et al., 2011) or N-limited conditions (Lima et al., 2022). Therefore, the sugarcane preference for NH4+ may also mean that transporters that are potentially involved in radial NH4+ transport in roots and/or root-to-shoot NH4+ translocation may affect the overall NUE of sugarcane, which led us to investigate AMTs to determine the potential role of AMTs in improving NUE in sugarcane. We started by functionally characterizing AMT1-type ammonium transporter members of sugarcane (unpublished results).
In the present work, we searched for AMT2-type ammonium transporters in the sugarcane genome by screening clones from a bacterial artificial chromosome (BAC) library (Tomkins et al., 1999). We then functionally characterized ScAMT2;1 by complementing ammonium transport-defective mutants of Saccharomyces cerevisiae (triple mepΔ) and A. thaliana (qko). The analysis of the ScAMT2;1 expression profile in response to various N conditions in sugarcane, together with promoter analysis driving a reporter gene, allowed some insight on the regulation of AMT2;1 in NH4+ transport in response to changes in external N availability and source. Altogether, the evidence suggests a role for AMT2;1 in ammonium uptake and a presumed contribution to root-to-shoot translocation.
AMT2 sequences were searched in a sugarcane BAC library from the commercial cultivar ‘R570’ (Tomkins et al., 1999). Analyses of ScAMT2;1 expression in sugarcane organs upon various N sources and levels were performed using the commercial cultivar SP80-3280. The S. cerevisiae mutant 31019b (triple mepΔ: mep1, mep2::LEU2, mep3::KanMX2, ura3) (Marini et al., 1997) defective for ammonium uptake was complemented with AtAMT1;1 or ScAMT2;1. The Arabidopsis genotype Columbia-0 (Col-0), the quadruple AMT-knockout mutant qko (amt1;1, amt1;2, amt1;3, and amt2;1) (Yuan et al., 2007), and the respective complemented lines were used in complementation assays.
Sugarcane AMTs were sought in a BAC library that consists of 269 plates with 384 clones each in a total of 103,296 clones representing a 4.5X coverage of the sugarcane genome (Tomkins et al., 1999). The search was performed by real-time PCR amplification of the three-dimensional pool of clones (de Setta et al., 2014). For that, A. thaliana and Oryza sativa AMT2;1 were used to find orthologue sequences in the sugarcane expressed sequence tag (SUCEST) database (https://sucest-fun.org/) to design the primers (Supplementary Table S1). First, superpools were screened for positive blocks, and positive blocks were further screened for the specific coordinates of positive clones, which were then isolated for confirmation and sequenced using the 454/Roche sequencing platform, assembled, and automated annotated as previously described (de Setta et al., 2014).
AMT gene automated annotation was curated using Artemis Genome Browser and Annotation Tool (v. 16.0.11) (Rutherford et al., 2000), and sorghum AMT2 was used as a reference. ScAMTs were aligned with AMTs from maize, rice, sorghum, and S. spontaneum by ClustalW (Thompson et al., 2003), including a sugarcane (‘SP80-3280’) AMT2;1 root-expressed sequence, identified here as ‘comp105883’ (NCBI id# OM966894). The evolutionary history was inferred by using the Maximum Likelihood method and JTT matrix-based model using MEGA11 (Tamura et al., 2021). A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+G, parameter = 1,1177)]. This analysis involved a total of 537 positions in the final amino acid alignment. A physical map of genomic sequences (~100 kb) containing AMT2;1 from sugarcane (Saccharum spp. ‘R570’) BAC clones (032_A12, 038_G02, 118_C18, 216_D16, and 235_F05) and S. bicolor (chromosome 9; NC_012878) was manually generated.
ScAMT2;1 identified in the BAC clones were analyzed to select the sequence to be functionally characterized. Regulatory upstream (~ 3 kb from the start codon) and coding regions were aligned and compared by ClustalW using BioEdit (Hall, 1999). Conceptually translated amino acid sequences were analyzed for specific elements/domains of the MEP/AMT/Rh transporter superfamily using Prosite (Hulo et al., 2006), TMHMM (Krogh et al., 2001), and WebLogo (Crooks et al., 2004). The presence of transposable elements (TEs) in the ScAMT2;1 regulatory region was predicted by Censor (Kohany et al., 2006) using the Viridiplantae database, and the co-occurrence of transcription factor-binding sites (TFBSs) was analyzed by PlantPAN 2.0 (Chow et al., 2016).
‘SP80-3280’ plantlets derived from in vitro meristem culture were grown hydroponically in 5 L plastic pots with full-strength nutrient solution (Hoagland and Arnon, 1950) containing 1 mM NH4NO3 (pH adjusted to 5.8) under greenhouse conditions for three months. The nutrient solution was aerated and renewed weekly. Prior to treatment, plants received a nutrient solution containing 2 mM NH4NO3 for 2 d. Subsequently, the plants were subjected to either an N-free nutrient solution (-N), 2 mM NH4NO3 (+N), 4 mM KNO3 (), 4 mM NH4Cl (), or 5 mM NH4NO3 (high N) for 14 d. Roots, culms, and young (+1) and mature (+3) leaves were collected, frozen in liquid N and stored at -80°C. Three plants per treatment were used for ScAMT2;1 tissue-specific expression.
Arabidopsis seeds were surface sterilized and grown for 30 d in substrate and vermiculite (1:1) in a growth chamber at 22°C, 80% humidity, and a 16/8 h light/dark phase at 200 µmol m-2 s-1. For the selection of transgenic events and experiments in agar plates, seeds were sown onto modified half-strength MS with 1 mM NH4NO3 as the sole N source, with the pH adjusted to 5.8. After a 4 d vernalization at 4°C in the dark, plates were placed in a growth cabinet at 24°C, 16/8 h light/dark phases, and 100 μmol m-2 s-1. For experiments in agar plates, Arabidopsis seeds were kept for 3 d in half-strength MS medium with 5 mM KNO3, with plates positioned vertically. Seedlings were then transferred onto media supplemented with various N sources at the indicated concentrations under the same environmental conditions. Treatments included either 0.5 mM KNO3 or 2 mM NH4Cl for experiments with plants bearing p35S::ScAMT2;1 and 2 mM KNO3 or 0.2, 2, and 4 mM NH4Cl for experiments with qko+p2ScAMT2;1::ScAMT2;1 plants (sugarcane endogenous promoter). After 14 d of treatment, seedlings were harvested, and the dry or fresh weight was measured.
Total RNA was isolated from sugarcane leaves as described (Leal et al., 2007) or from Arabidopsis using TRIzol (Thermo Fisher Scientific; Waltham, MS, USA). cDNA was synthesized using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific). Primers were designed based on the ScAMT2;1 sequence from clone BAC 118_C18 (Supplementary Table S1). RT–qPCR was performed with 5 µL of KAPA SYBR FAST (Kapa Biosystems, Wilmington, MA, USA), 0.2 µM of each primer (Supplementary Table S1), and 1 µL of diluted cDNA (1:10) in a final volume of 10 µL. Reactions were run in a RotorGene-6000 (Qiagen) with the following settings: 50°C for 10 min and 95°C for 2 min, followed by 40 cycles of 95°C for 20 s, 62°C for 25 s, and 72°C for 25 s. UBIQUITIN2 was used as a reference gene in sugarcane (ScUBQ2) and Arabidopsis (AtUBQ2) (Supplementary Table S1). All reactions were performed in triplicate with three biological replicates. Relative expression levels were calculated as described (Livak and Schmittgen, 2001). Normalization is indicated for each experiment.
The full-length ScAMT2;1 coding sequence from clone BAC 118_C18 was synthesized (Biomatik; Cambridge, Ontario, Canada); AtAMT1;1 was used as a positive control because of its well-established function, and the empty vector was used as a negative control (final constructs in Supplementary Table S2). AtAMT1;1 and ScAMT2;1 sequences were amplified from Arabidopsis cDNA or synthetic vector, respectively, with primers containing restriction enzyme sites (Supplementary Table S1), cloned into pGEM-T Easy (Promega, Madison, WI, USA), sequenced, and subcloned into the expression vector pDR196 (Rentsch et al., 1995). Triple mepΔ (strain 31019b) yeast cells were transformed by the lithium acetate method (Gietz and Schiestl, 2007). Confirmed positive clones were inoculated in liquid YNB-AA/AS (0.17% yeast nitrogen base without amino acids or ammonium sulfate) containing 1 mM arginine (positive control) and 50 mg L-1 ampicillin for 36 h at 30°C at 200 rpm. A growth test was performed with a serial dilution (DO600nm= 1, and subsequent dilution of 10-1, 10-2, and 10-3) plated onto YNB-AA/AS supplemented with 3% glucose and one source of N [0.5, 2, 3, or 5 mM NH4Cl (NH4+), 100 mM methylammonium (MeA), or 1 mM arginine (Arg)]. MES-Tris was added at 20 mM to maintain the pH at 5.0, 6.0, or 7.5. The plates were incubated at 30°C for 6 d.
The ScAMT2;1 promoter region fragment from clones BAC 118_C18 (2,936 bp; p1ScAMT2;1) and BAC 235_F05 (2,962 bp; p2ScAMT2;1), hereafter called endogenous promoters, as well as the coding region from the synthetic ScAMT2;1 gene, were amplified (see above). The amplified products were cloned into pDONR or pCR8 (Thermo Fisher Scientific) and then subcloned into the final pMDC vectors (primers in Supplementary Table S1; vectors and final constructs in Supplementary Table S2) using the Gateway system (Thermo Fisher Scientific). Arabidopsis plants (Col-0 or qko) were transformed by floral dipping (Narusaka et al., 2010) using Agrobacterium tumefaciens GV3101 bearing the constructs indicated in Supplementary Table S2. Transformed lines were selected for hygromycin resistance. Homozygous lines were confirmed by PCR and RT–qPCR.
Seedlings of Arabidopsis transgenic lines containing the GUS reporter gene (uidA, pMDC164) driven by ScAMT2;1 endogenous promoter were cultivated on half-strength MS media without N (-N) or supplied with 2 mM NH4Cl or 1 mM NH4NO3 as the sole N source for up to 10 d. For GUS staining, plants were transferred to buffer containing 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X-Gluc; Jersey Lab and Glove Supply, Livingston, NJ, USA) at 37°C for 4 h 30 min and then washed in 70% ethanol (Jefferson et al., 1987). Plant tissues were analyzed and photographed under a Nikon SMZ18 stereo microscope.
Wild-type Arabidopsis and homozygous (T3) ScAMT2;1-complemented qko plants were hydroponically grown in N-sufficient conditions (2 mM NH4NO3) with pH adjusted to 5.8 with 2-(N-morpholino)ethanesulfonic acid (MES) for 40 d. For qko+p35S::ScAMT2;1 lines, plants were subjected to N deficiency (-N, no N) or N sufficiency (+N, 1 mM NH4NO3), whereas qko+p2ScAMT2;1::ScAMT2;1 plants were transferred to -N, 2 mM KNO3 or NH4Cl as sole N sources. After 3 d under treatment, plants were exposed to a short-term 15N-ammonium influx assay with a 10 min incubation in a full-strength nutrient solution containing 0.2 mM (15NH4)2SO4 (60% of 15N-ammonium). To assess 15N accumulation in roots and shoots, plants were subjected to -N for 3 d and then transferred to a 15N-labelled nutrient solution with 2 mM (15NH4)2SO4 (60% of 15N-ammonium) for 1 h. For concentration-dependent influx of NH4+ into roots of qko and qko+p35S::ScAMT2;1 lines, 40-d-old plants grown hydroponically under the same conditions mentioned above were transferred to -N for 3 d. Roots were then incubated for 10 min in full nutrient solution containing increasing concentrations 0, 25, 50 100, 150, 200, 300, and 500 mM of (15NH4)2SO4 (60% of 15N-ammonium). To further assess the contribution of AMT2;1 to 15N root-to-shoot translocation, 40-d-old hydroponically grown qko and qko+ p2ScAMT2::ScAMT2;1 plants were subjected to -N for 3 d followed by one h-root exposure to a nutrient solution containing 0.2 mM or 4 mM (15NH4)2SO4 (60% of 15N-ammonium). In all 15N experiments, roots were first rinsed with 1 mM CaSO4 for 1 min before exposure to 15N, followed by washing with 1 mM CaSO4 prior to sample collection. Roots and shoots were collected separately, dried, ground, and analyzed for total 15N content using continuous-flow isotope ratio mass spectrometry (ANCA SL, Sercon, Cheshire, UK).
A completely randomized design was used in all experiments. The number of biological replicates is indicated for each experiment. Analysis of variance (ANOVA) was performed, and means were compared using Tukey’s test at 5% significance or t test (p ≤ 0.10 and p ≤ 0.05), as indicated for each experiment, using SAS software (SAS Institute Inc., Cary, NC, USA).
Through real-time amplification of the three-dimensional pool of BAC clones using AMT2;1-specific primers followed by screening for the specific coordinates of positive clones, we identified five clones containing sequences closely related to ScAMT2;1 transcript, namely, BAC 032_A12, BAC 038_G02, BAC 118_C18, BAC 216_D16, and BAC 235_F05 (Supplementary Figure S1). Each BAC clone contained a unique locus, except for BAC 216_D16 and BAC 235_F05, which shared the same protein sequence. The BAC 032_A12, BAC 038_G02, and BAC 118_C18 AMT2.1 loci were phylogenetically closer to the root-expressed assembled transcript (comp105883). On the other hand, BAC 216_D16 and BAC 235_F05 did not group closely with the transcript sequence (Supplementary Figure S1).
As sugarcane cultivars are polyploids derived from interspecific crosses between S. officinarum and S. spontaneum (Nogueira et al., 2005; Graff et al., 2011), the identified BAC clones containing distinct ScAMT2;1 loci are expected to differ for the surrounding topology (Figure 1). Of the five clones, BAC 216_D16 and BAC 235_F05 were highly similar, with 62.0% identity.
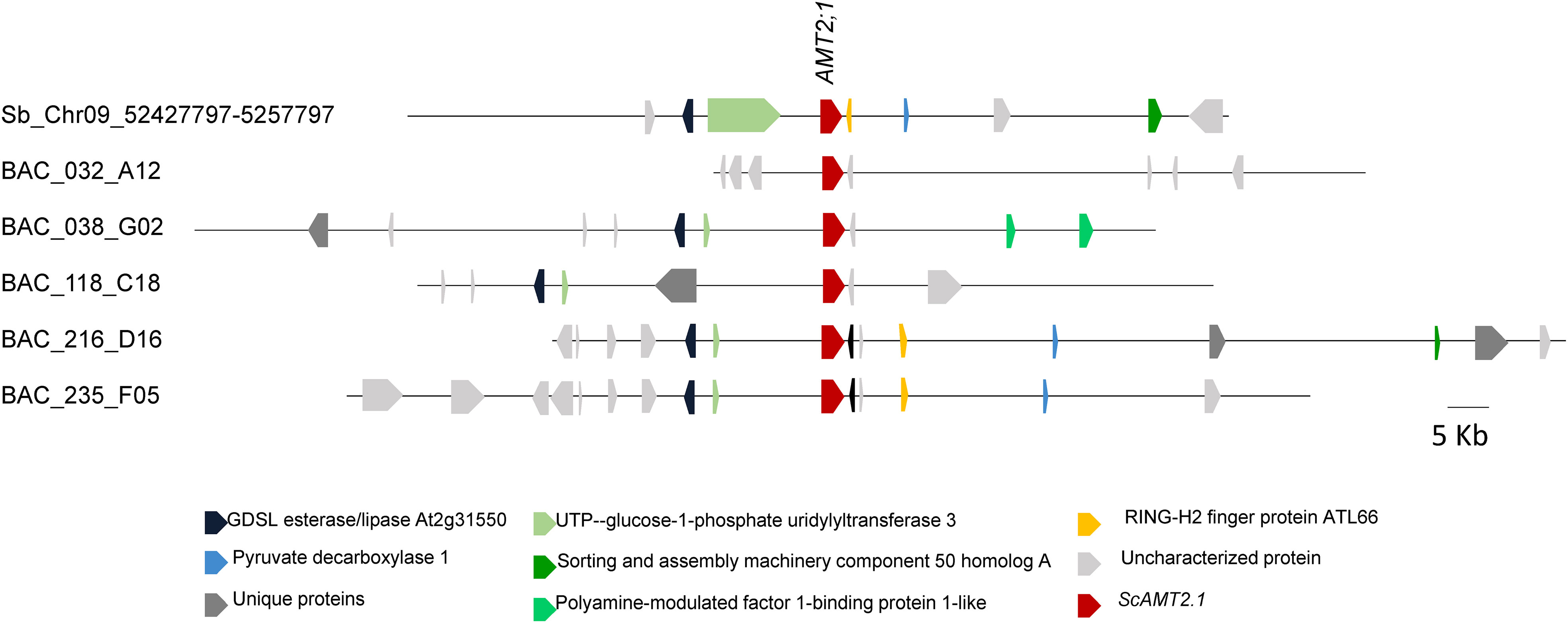
Figure 1 Physical map of genomic sequences (100 Kb) containing AMT2;1 from sugarcane (Saccharum spp. ‘R570’) BAC clones (032_A12, 038_G02, 118_C18, 216_D16, and 235_F05) and S. bicolor (chromosome 9; NC_012878).
The alignment of the deduced amino acid of AMT2;1 sequences from the BAC clones, comp105883, and one S. spontaneum AMT2;1 (Wu et al., 2021) indicated that BAC 118_C18 had an identical protein sequence as the transcribed assembled sequence (comp105883) (Supplementary Figure S3A). The complete ScAMT2;1 gene from BAC 118_C18 (NCBI# OM471796) is 1,473 bp long with three exons encoding 490 amino acids, conceptually estimated to be a protein of 52 kDa (Supplementary Figure S2B). Similar gene structure and protein features were observed for S. spontaneum SsAMT2;1 from the chromosome 7 (Sspon.03G0003380-4D; Wu et al., 2021) (Supplementary Figure S2B; Supplementary Figure S3A). In addition, the presumed ScAMT2;1 amino acid sequence from BAC 118_C18 was identical to the SsAMT2;1 protein Sspon.03G0003380-4D (Supplementary Figure S3A) and contained the expected 11 transmembrane domains predicted by TMHMM (Supplementary Figure S3B), along with the conserved signature motif for the MEP/AMT/Rh superfamily (Supplementary Figure S3C).
Various plant AMT1 and AMT2 sequences, including the identified sugarcane members, were compared to verify the conservation of C- and N-terminal regions concerning amino acids essential for transport function (Supplementary Figure S4). In the N-terminus of the tomato protein LeAMT1;1, two cysteines (C3 and C27) have been proven to be fundamental for AMT1 oligomer stability (Graff et al., 2011). While AMT1 proteins except for SlAMT1;3 contained these two conserved Cys residues, these residues were absent in all AMT2 homologues (Supplementary Figures S4A, C). In the C-terminus, some residues have been associated with transport regulation, including glycine-456 (G456; SlAMT1;1) and threonine-460 (T460) (Ludewig et al., 2003). G456 was found in all AMTs evaluated to date, whereas T460 was absent in all AMT2 subfamily members, including ScATM2;1 (Supplementary Figures S4B, D).
Multiple alignment of the various ScAMT2;1 promoter sequences identified in the BAC clones (approximately 3 kb upstream of the predicted translation start codon) allowed the arbitrary separation of the clones into two groups, in which sequences from BAC 032_A12, BAC 038_G02, and BAC 118_C18 were more similar between each other, differing from BAC 216_D16 and BAC 235_F05 (Supplementary Figure S5A). We analyzed whether this separation could be due to transposable element (TE) insertions, which are commonly found in promoter regions of sugarcane sequences (de Setta et al., 2014). TE insertion was assessed by Censor, which identified repetitive elements by comparison with known repeats and assigned a score for probability. The results reinforced the similarity of BAC 216_D16 and BAC 235_F05, showing a similar TE insertion profile (Supplementary Figure S5C). To further investigate the presence of regulatory elements and presumed synteny of regulatory motifs, we chose clones from each group, BAC 118_C18 and BAC 235_F05. Only a few conserved regions exist between the selected regulatory regions, indicating significant variation between the two ScAMT2;1 promoters (Supplementary Figure S5C). As loci from BAC 118_C18 were not functional in driving the expression of uidA in the GUS assay (see below), the sequence from the BAC 235_F05 clone was chosen to be further analyzed as a functional ScAMT2;1 endogenous promoter. Concerning the gene sequence, ScAMT2;1 from BAC 118_C18 was selected for functional validation due to greater similarity with the root-expressed ScAMT2;1 sequence comp105883 and S. spontaneum Sspon.03G0003380-4D (Supplementary Figure S2B).
The transcriptional profile of ScAMT2;1 was examined in the organs of sugarcane plants grown under various N conditions. At the stage of generative growth (90-d-old plants) under N-sufficient conditions, ScAMT2;1 was expressed in all organs analyzed, with more transcript accumulation in roots, but it was also largely expressed in mature leaves, followed by young leaves, and less abundant in culms (Figure 2A). To assess how ScAMT2;1 expression is regulated by N supply, transcript levels were determined in various organs in plants grown in nutrient solution containing distinct N sources or without N for 14 d and compared with the +N treatment (Figure 2B). In the presence of nitrate as the sole N source, ScAMT2;1 transcripts accumulated approximately 2- to 3-fold more in roots, mature leaves, and culms but not in young leaves compared with plants grown in ammonium nitrate (+N). Thus, exposing the plants at the same high N level (4 mM) but changing the source from 2 mM NH4NO3 to 4 mM KNO3 (4 mM N with no ammonium) was sufficient to induce ScAMT2;1 expression in roots and shoots more than the change from 2 mM NH4NO3 to 4 mM NH4Cl.
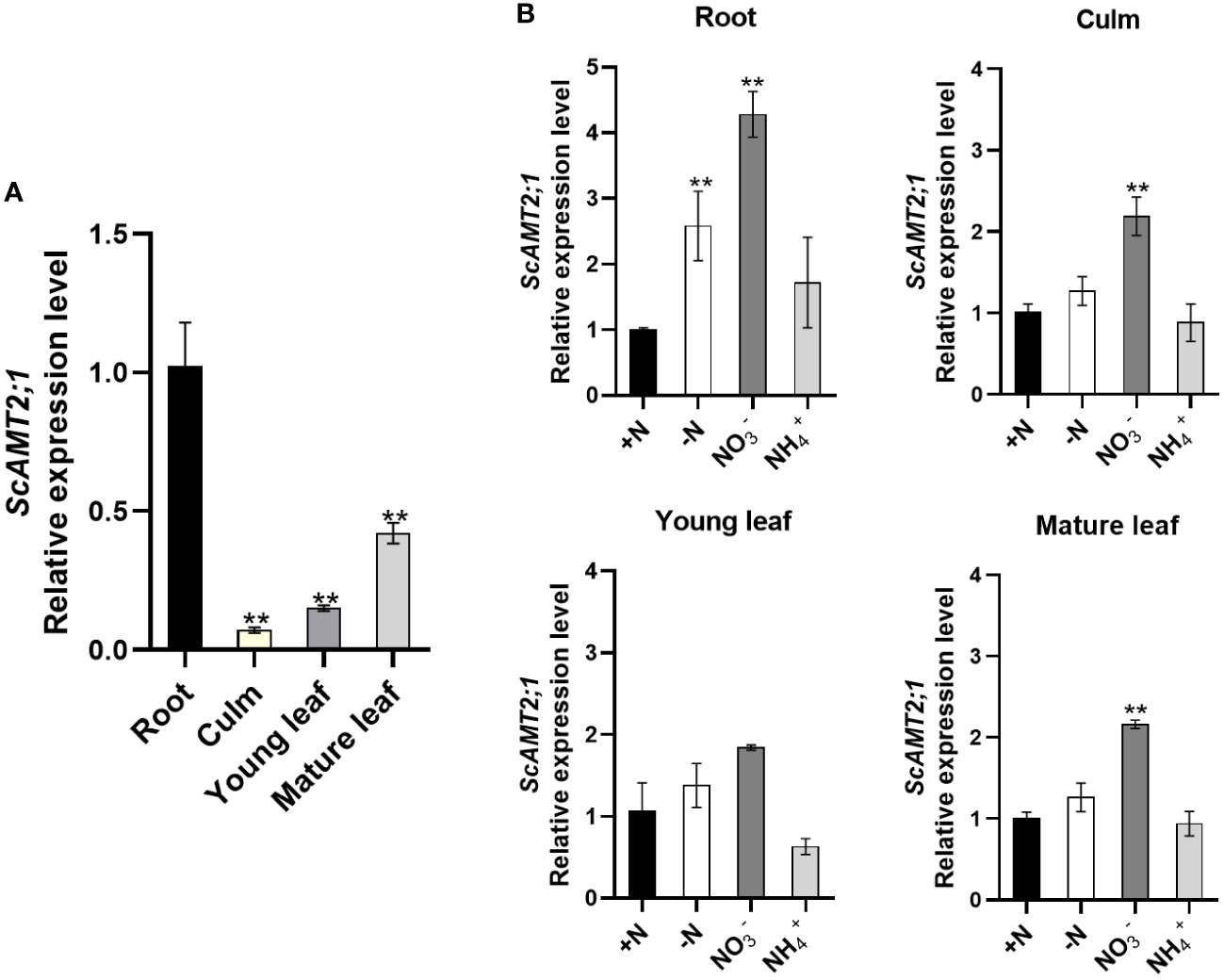
Figure 2 Expression of ScAMT2;1 in sugarcane organs after subjecting plants to distinct inorganic N sources or no N. (A) RT–qPCR analysis of ScAMT2;1 expression in roots, culms, and young (+1) or mature (+3) leaves of sugarcane grown under 2 mM NH4NO3 for 2 d. Bars indicate ± SE (n = 3). ScUBQ2 was adopted as a reference gene. Gene expression levels were normalized to expression levels in culms. Asterisks represent significant differences in relation to culms according to Student’s t test (p < 0.01). (B) ScAMT2;1 relative expression levels in sugarcane roots, culm, young, and mature leaves of plants under +N: 2 mM NH4NO3, -N: no N; : 4 mM KNO3, or NH4+: 4 mM NH4Cl for 14 d. ScUBQ2 was used as a reference gene. The gene expression level was normalized to the +N treatment. Bars indicate ± SE (n = 3). Asterisks represent significant differences between treatments and +N according to Student’s t test (p < 0.01).
To further investigate whether ScAMT2;1 is transcriptionally modulated by N availability, the expression profile was evaluated in sugarcane plants grown under 5 mM NH4NO3 (high N) or no N (-N) for 10 d (Supplementary Figure S6). Transient and temporal transcript accumulation was detected in N-deficient mature leaves relative to high N supply. In culms, ScAMT2;1 transcripts showed some accumulation in both treatments; however, this transcriptional response was not observed in roots or young leaves. Altogether, these observations indicate that the N source and the plant N status modulate the expression of ScAMT2;1 in sugarcane.
To investigate whether the selected ScAMT2;1 gene (BAC118_C18) encodes a functional ammonium transporter, we complemented the S. cerevisiae triple mep mutant (31019b) (Marini et al., 1997). The positive control (triple mep complemented with AtAMT1;1) completely restored growth under all N conditions tested (Figure 3). By increasing the external NH4+ concentration, triple mep cells complemented with ScAMT2;1 showed slightly more growth than the negative control suggesting that ScAMT2;1 is a functional protein that mediates ammonium transport. At 5 mM ammonium, the growth of the triple mep complemented with ScAMT2;1 was strongly pH dependent. The ScAMT2;1-expressing triple mep grew slightly better than the negative control (empty pDR196) at a pH of 5.0 and 6.0. Raising the pH further to 7.5 may have increased the concentration of ammonia (NH3), resulting in similar growth between triple mep expressing ScAMT2;1 and the negative control.
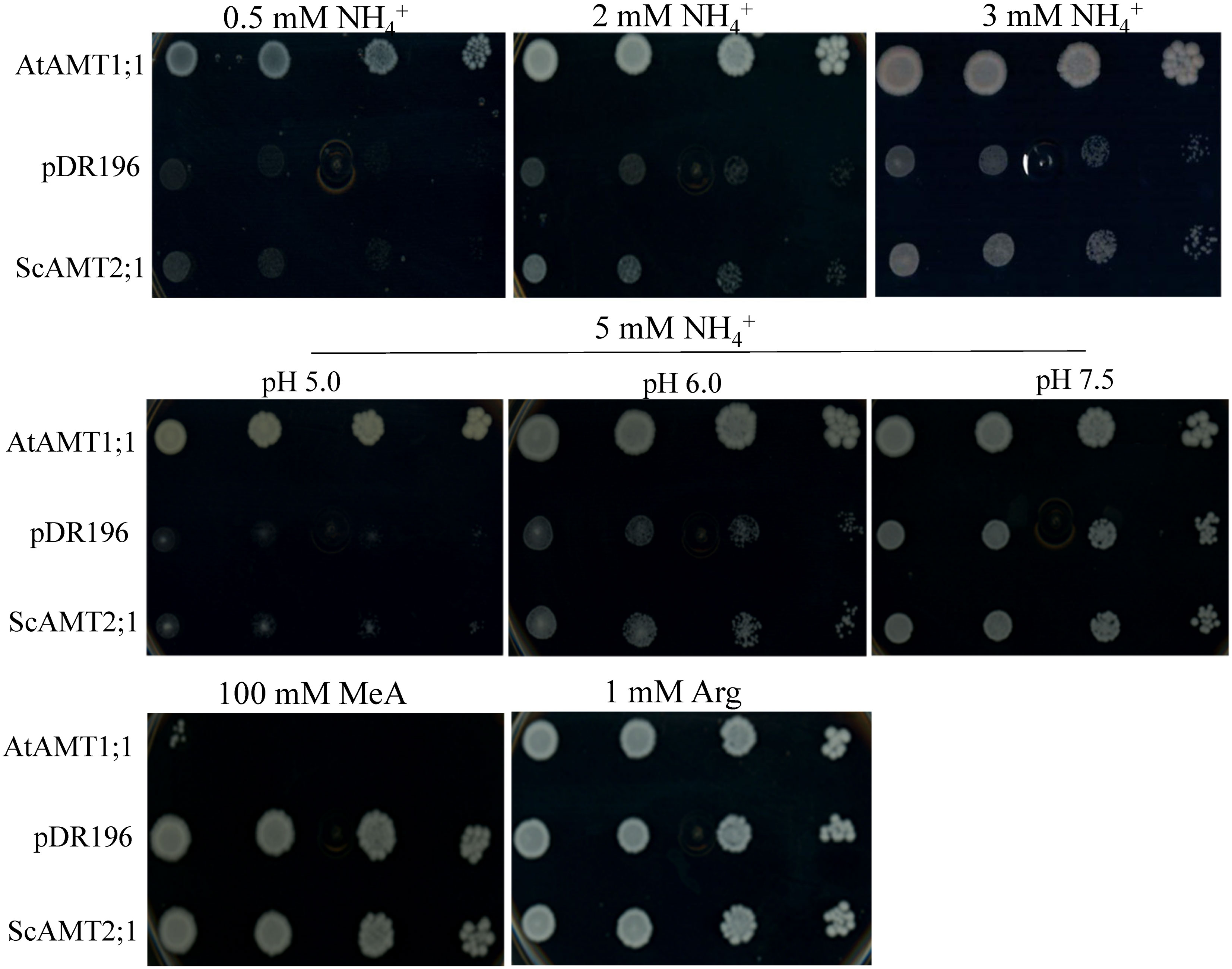
Figure 3 Functional complementation of the yeast mutant defective for ammonium uptake. Growth of the ammonium transporter-deficient yeast strain triple mepΔ (31019b) expressing AtAMT1;1 (positive control), the empty pDR196 vector (negative control), or ScAMT2;1 on media supplied with 0.5 to 5 mM ammonium chloride (NH4+), 100 mM methylammonium (MeA), or 1 mM arginine (Arg; positive control) as the sole N source. Culture media pH was adjusted to 6.0 when not indicated otherwise. Each transformant line was grown to OD600nm = 1 and plated in concentrated and sequential four 10-fold dilutions.
In contrast to type 1 AMT proteins, AMT2 has been proposed to be impermeable to the transport of the ammonium toxic analogue methylammonium (MeA) (Sohlenkamp et al., 2000; Sohlenkamp et al., 2002). The growth of triple mep complemented with ScAMT2;1 was evaluated on media supplemented with 100 mM MeA (Figure 3). The toxic effect of MeA drastically reduced the growth of triple mep cells expressing AtAMT1;1, whereas those complemented with ScAMT2;1 or the empty vector displayed no visible sensitivity towards MeA.
ScAMT2;1 driven by the CaMV35S promoter (p35S) was expressed in the Arabidopsis quadruple AMT mutant line (qko) (Yuan et al., 2007). Three independent T3 homozygous lines were characterized for ScAMT2;1 expression in relation to Col-0 plants (Supplementary Figure S7) and then used for phenotypic evaluation. ScAMT2;1-complemented events grown in the presence of ammonium as the only N source accumulated significantly more total dry biomass than qko, with values approximately 65% (event #1) and 51% (event #2) higher under 2 mM NH4+ (Figures 4A, B). In contrast, no significant difference between qko and the complemented lines was observed when only nitrate was supplied (Figures 4A, B), suggesting that the ectopic expression of ScAMT2;1 restored the qko mutant growth phenotype only under ammonium nutrition, likely by mediating ammonium uptake into roots.
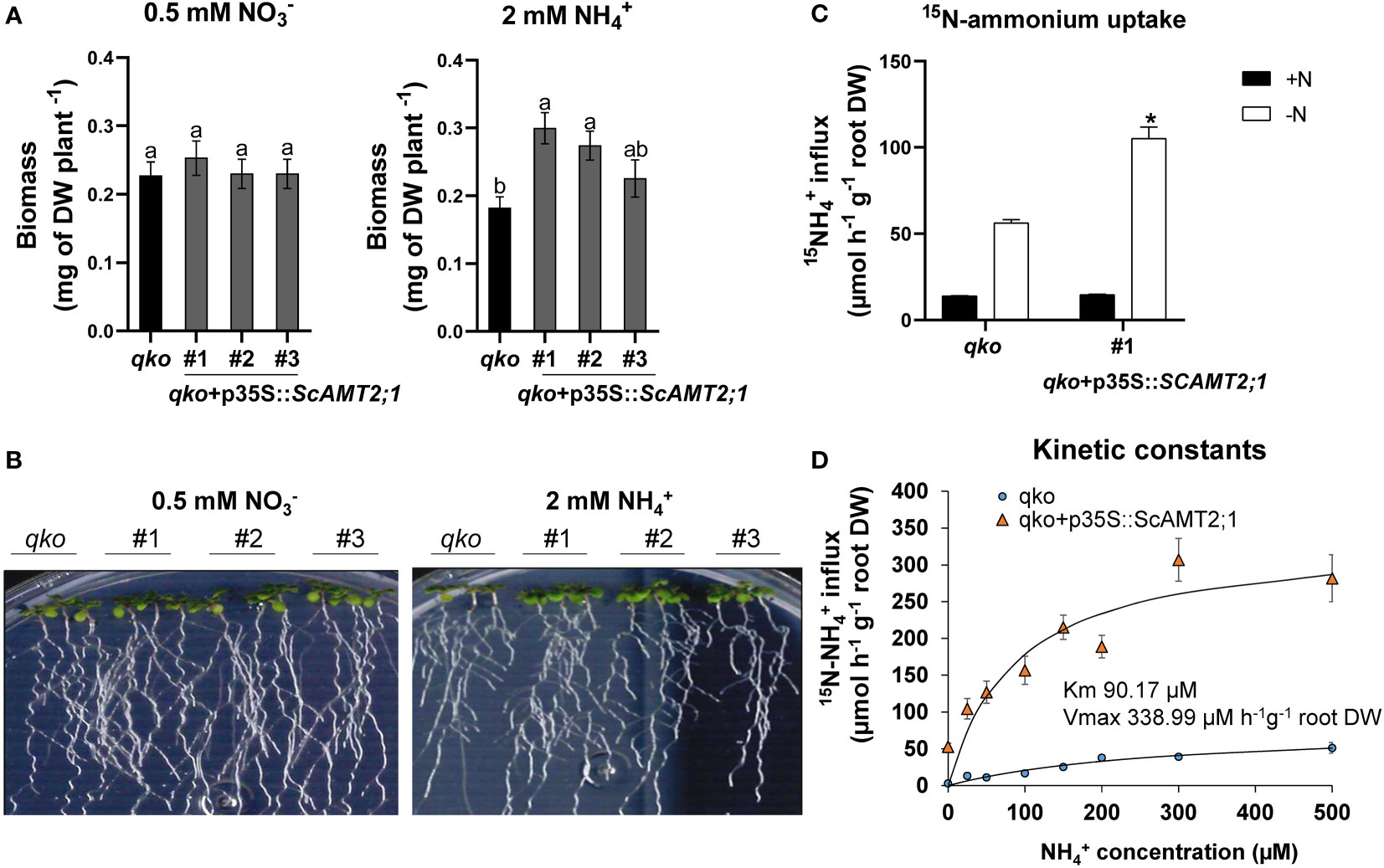
Figure 4 Functional evaluation of complemented Arabidopsis qko mutant. (A) Total biomass (DW, dry weight) of homozygous lines of Arabidopsis overexpressing ScAMT2;1 (qko+p35S::ScAMT2;1) subjected to either 0.5 mM KNO3 () or 2 mM NH4Cl (NH4+) for 14 d. Bars indicate means ± SE (n = 6). Different letters indicate significant differences among means according to Tukey’s test (p ≤ 0.05). (B) Phenotype of qko and transgenic events grown in vitro under different N sources. (C) Influx of 15N-labeled ammonium into the roots of qko and a transgenic line (#1) overexpressing ScAMT2;1 subjected to either N-free (-N) or 0.2 mM NH4+ (+N) nutrient solution for 3 d. Bars indicate means ± SE (n = 4). Asterisks indicate significant differences between qko and the transgenic line according to Student’s t test (p < 0.05). (D) Concentration-dependent influx of 15NH4+ into roots of qko or qko+p35S::ScAMT2;1 (#1). Symbols indicate six biological replicates (n = 6).
We then evaluated the short-term influx of 15N-NH4+ in the qko ScAMT2;1-complemented lines. Under -N, the root ammonium uptake capacity of qko+p35S::ScAMT2;1 increased by 87% compared with qko (Figure 4C), corroborating the function of ScAMT2;1 in NH4+ uptake in roots. To estimate the substrate affinity of ScAMT2;1, six-week-old ScAMT2;1-overexpressing (p35S) qko plants were grown under -N for 3 d, followed by concentration-dependent 15N-NH4+ influx analyses. In this experiment, ScAMT2;1 function was saturated above 90 µM (Figure 4D). The estimated net ammonium influx fitted the Michaelis–Menten equation well, resulting in a Km = 90.17 µM and a Vmax of 338.99 µmoles h-1 g-1 root DW, determined by subtracting the values of qko (Figure 4D). These results demonstrate that the ScAMT2;1 protein can contribute to high-affinity ammonium transport in planta.
To help determining the ScAMT2;1 function, we conducted localization experiments in Arabidopsis by expressing the GUS reporter gene driven by the ScAMT2;1 regulatory region from BAC 118_C18 (p1ScAMT2;1) and BAC 235_F05 (p2ScAMT2;1). No reporter expression was detected with the promoter p1ScAMT2;1, which was apparently nonfunctional (Supplementary Figure S8). Arabidopsis lines expressing p2ScAMT2;1::GUS allowed tracing promoter activity in vascular bundles and outermost cells in leaves, either under ammonium or ammonium nitrate (Figure 5A). In contrast, leaves from N-deficient plants displayed no p2ScAMT2;1 activity in outer cells, and activity appeared to predominate at vascular bundles (Figure 5A). In roots, GUS was mainly detected in the innermost tissues (Figure 5B). Altogether, these results suggest that the ScAMT2;1 regulatory region is associated with root and leaf vascular tissues, but tissue-specific expression depends particularly on the N status in leaves rather than roots.
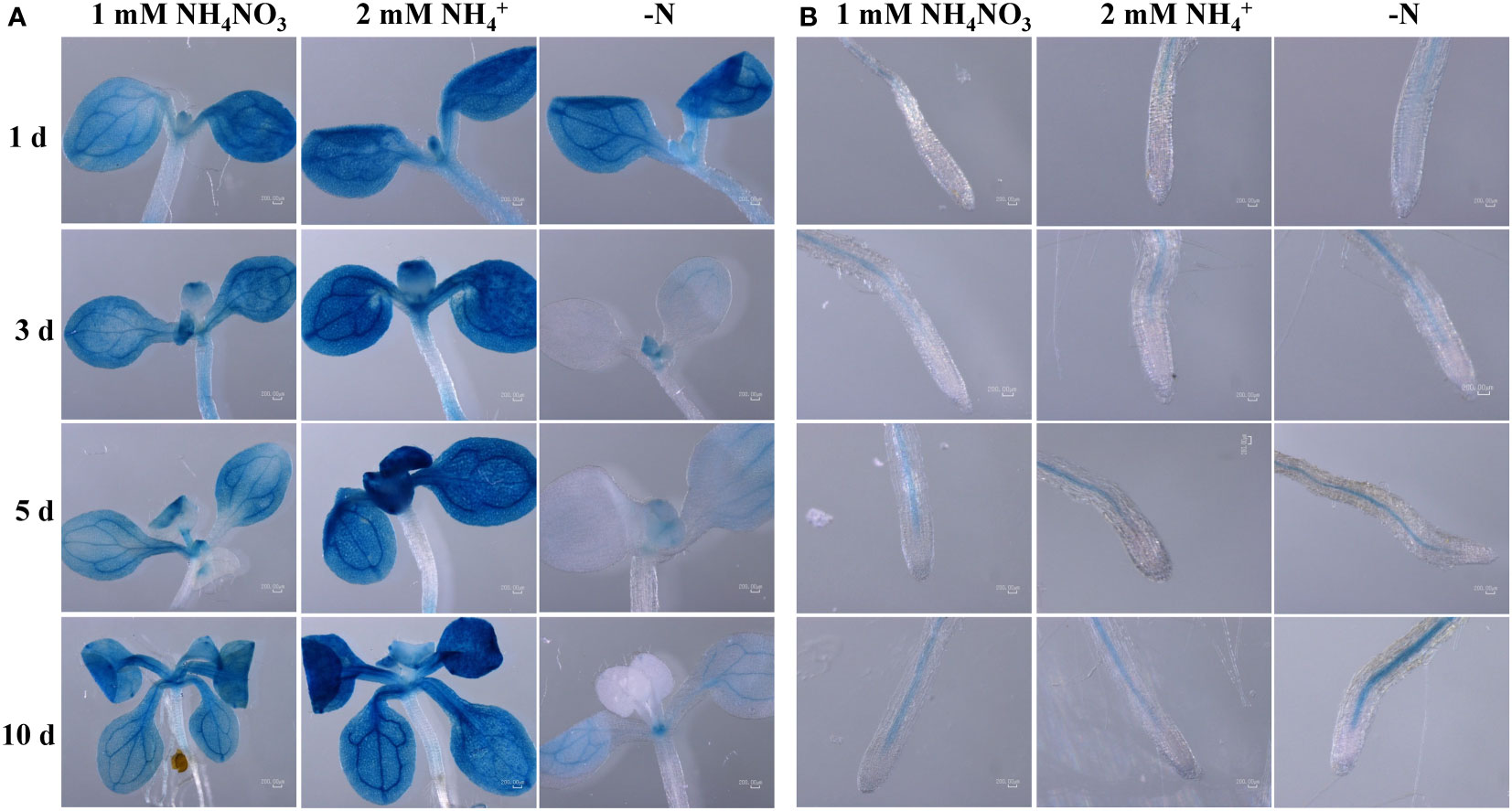
Figure 5 Endogenous ScAMT2;1 promoter (p2) driving GUS expression in Col-0 Arabidopsis plants subjected to 1 mM NH4NO3, 2 mM NH4+, or no N for 1, 3, 5, and 10 d in (A) shoots; and (B) roots. The blue color shows GUS activity. Bars = 200 μm.
We then assessed the contribution of ScAMT2;1 to ammonium uptake by generating qko lines complemented with ScAMT2;1 driven by the endogenous regulatory region p2ScAMT2;1. While all p2ScAMT2;1::ScAMT2;1-complemented lines and qko grew similarly on agar medium supplemented with either 2 mM nitrate or 0.2 mM ammonium (Figure 6A), the total biomass of qko+p2ScAMT2;1::ScAMT2;1 plants was clearly superior to that of qko under higher external NH4+ concentrations. At 2 mM NH4+, qko+p2ScAMT2;1::ScAMT2;1 accumulated approximately 83% (#1), 103% (#2), or 28% (#3) more shoot biomass than qko (Figure 6A). The biomass accumulation for plants grown at 4 mM NH4+ was 102% (event #1), 75% (event #2), or 107% (event #3) higher than the control plants (Figure 6A). These results suggest that ScAMT2;1 under the control of the sugarcane endogenous promoter significantly increased biomass at elevated external ammonium levels and confirm the functionality of ScAMT2;1 in facilitating NH4+ uptake.
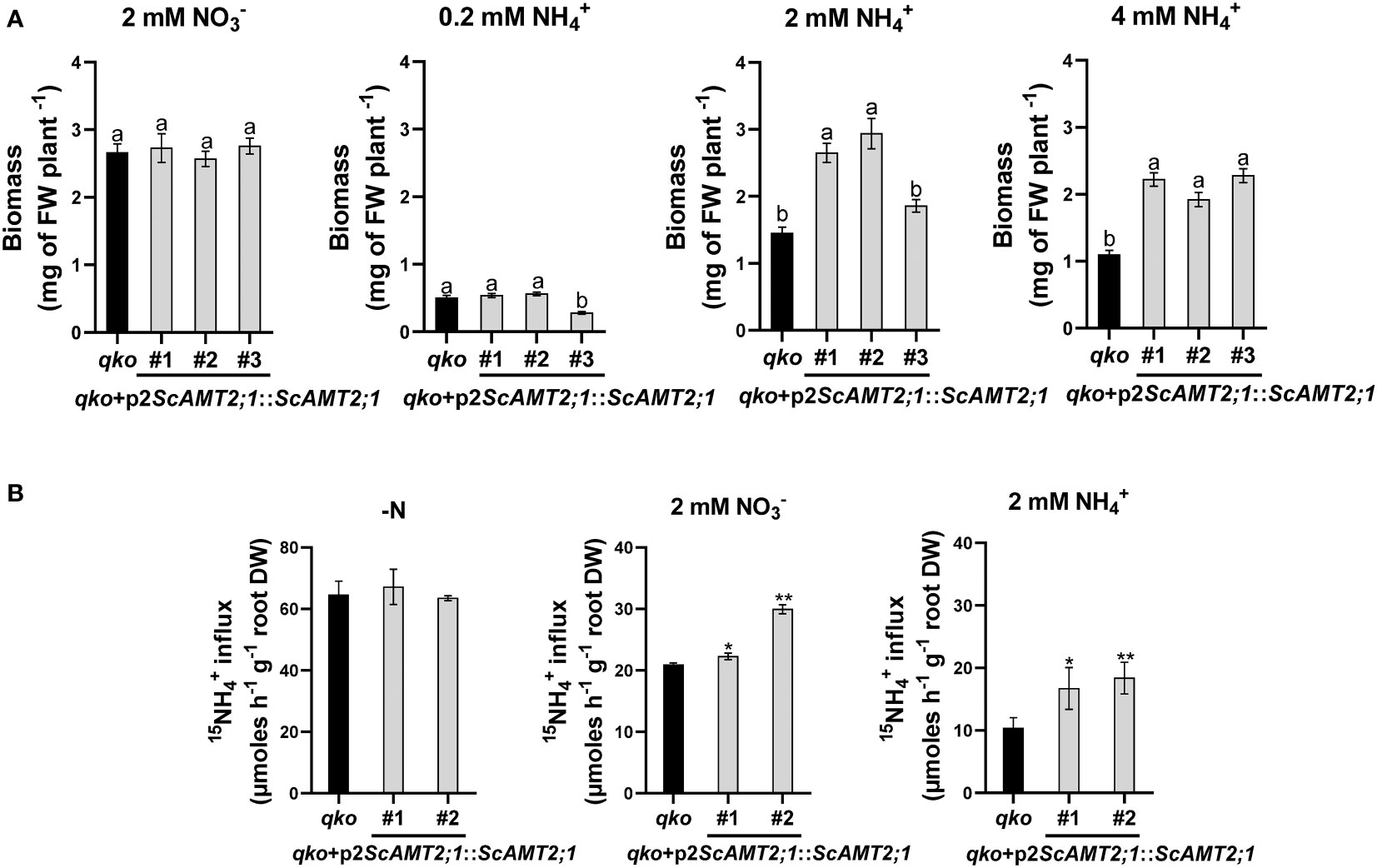
Figure 6 Biomass accumulation in Arabidopsis mutant plants and lines complemented with ScAMT2;1 driven by the sugarcane endogenous promoter. (A) Fresh weight of homozygous lines of qko+p2ScAMT2;1::ScAMT2;1 subjected to 2 mM KNO3 () or 0.2, 2, and 4 mM NH4Cl (NH4+) for 14 d. Bars indicate means ± SE (n = 30). Different letters indicate significant differences among means according to Tukey’s test (p ≤ 0.05). (B) Influx of 15N-labeled ammonium (NH4+) into roots of qko and qko+p2ScAMT2;1::ScAMT2;1 lines upon 3-d exposure to N-free (-N), 2 mM , or 2 mM NH4+ nutrient solution. Bars indicate means ± SE (n = 4). Asterisks indicate significant differences between qko and transgenic plants according to Student’s t test (*p < 0.10 and **p < 0.05).
To evaluate the regulatory level of the response of the ScAMT2;1 promoter to high external N supply, short-term 15N-ammonium influx analysis was performed with qko+p2ScAMT2;1::ScAMT2;1 plants in the presence of 2 mM of either ammonium or nitrate or no N (-N). The influx of 15N-NH4+ into the roots of qko and complemented lines subjected to -N was similar and nonsignificant (Figure 6B). Complementation with ScAMT2;1 significantly increased uptake levels by 6% (#1) and 43% (#2) compared with qko when subjected to 2 mM nitrate and by 61% (#1) and 78% (#2) in complemented plants subjected to 2 mM ammonium (Figure 6B). Altogether, these results indicate that the regulation of ScAMT2;1 in ammonium uptake depends strictly on the preconditioning of plants to an externally high N form but not to -N.
Our experiments indicated that ScAMT2;1 contributes to root ammonium uptake mainly in NH4+-supplied plants (Figure 6) and that the ScAMT2;1 promoter (p2) drives gene expression in the inner vascular root cells (Figure 5). These results prompted us to evaluate whether ScAMT2;1 mediates root-to-shoot ammonium transport under ammonium supply. To this end, we evaluated ScAMT2;1-specific functions by estimating 15N accumulation in roots and shoots of qko and qko+p2ScAMT2;1::ScAMT2;1 lines subjected to either 0.2 mM or 4 mM 15N-NH4+ for 1 h to allow time for root-to-shoot translocation (Figure 7A). At 0.2 mM 15N-NH4+ supply, no significant 15N was accumulated in shoots compared with qko, whereas some 15N accumulation in roots occurred for one transgenic line (event #2). When plants were grown in the presence of 4 mM 15N-NH4+, roots of qko and complemented lines accumulated 15N in a similar pattern. In contrast, significantly more 15N accumulated in the shoots of both qko+p2ScAMT2;1::ScAMT2;1 lines, approximately 35% and 25% more than in qko shoots (Figure 7A). The rate of 15N accumulated in the shoot in relation to the whole plant was 7 and 6.3% for the qko+p2ScAMT2;1::ScAMT2;1 lines, significantly superior to the 4.8% observed in qko when plants were subjected to 4 mM 15N-NH4+ (Figure 7B). These results suggest that ScAMT2;1 activity in roots might contribute to ammonium translocation to shoots under high external ammonium conditions.

Figure 7 15N accumulation in roots and shoots of qko and complemented qko transgenic lines expressing ScAMT2;1 under the regulation of its sugarcane endogenous promoter (p2). The assay was performed in plants after 3 d of exposure to an N-free nutrient solution, followed by 1 h of treatment in either a 0.2 mM or 4.0 mM 15NH4+ solution. Bars indicate means ± SE (n = 5). Asterisks indicate significant differences in root and shoot 15NH4+ accumulation between qko and transgenic plants according to Student’s t test (*p < 0.1 and **p < 0.05). (B) Root-to-shoot translocation of 15N in percent of 15N accumulation in shoots in relation to the whole plant. Asterisks indicate significant differences between qko and transgenic plants according to Student’s t test (** p < 0.05).
AMT2 proteins have been demonstrated to exhibit transport properties distinct from those of the AMT1 subfamily (Sohlenkamp et al., 2002; Mayer and Ludewig, 2006; Neuhäuser et al., 2009). However, little is known about the physiological roles of AMT2-type ammonium transporters. In Arabidopsis thaliana, the only AMT2 member was shown to play a critical role in root-to-shoot partitioning of ammonium (Giehl et al., 2017). Here, we provide evidence that ScAMT2;1 might be involved in sugarcane root ammonium uptake at elevated external substrate levels.
We started by identifying AMT2s in the sugarcane genome by searching a BAC library from the commercial cultivar R570 (Tomkins et al., 1999). The screening allowed the identification of five ScAMT2;1 sequences. Modern sugarcane cultivars are derived from interspecific crosses between S. officinarum and S. spontaneum, followed by backcrossing to S. officinarum, producing a highly complex genome (Thirugnanasambandam et al., 2018). Therefore, considering the high polyploidy and redundant character of the sugarcane genome (Garsmeur et al., 2018; Zhang et al., 2018), a series of in silico conceptual analyses was performed with the various upstream regulatory regions and gene sequences of the ScAMT2;1 loci found in the five BAC clones to define the target for functional characterization. Based on the phylogenetic analysis, it was possible to infer that BAC 216_D16 and BAC 235_F05 clones contained ScAMT2;1 alleles distinct from BAC 038_G02, BAC 032_A12, and BAC 118_C18, which was corroborated by analyzing their regulatory regions. The ScAMT2;1 coding sequence from BAC 118_C18 was chosen because it demonstrated the highest similarity to a root-expressed sequence detected by us in sugarcane (NCBI# OM966894). In addition, ScAMT2;1 from BAC 118_C18 was structurally identical to an AMT2;1 from S. spontaneum (Sspon.03G0003380-4D; Supplementary Figure S2), with an identical deduced protein (Supplementary Figure S3A). The conceptually translated ScAMT2;1 protein from clone BAC 118_C18 displays the conserved 11 presumed transmembrane domains, amino and carboxy-terminal, facing the inner and outer parts of the plasma membrane, respectively, and contains the superfamily signature motif, strongly indicating that this gene is an ammonium transporter (Supplementary Figure S2) (Marini et al., 1997; Marini and André, 2000; Schwacke et al., 2003; Loqué and von Wirén, 2004; Ellerbeck et al., 2013). It will be necessary to evaluate whether the other ScAMT2;1 alleles are functional and present the same expression and regulation patterns and transport characteristics as the one evaluated here.
ScAMT2;1 expression in sugarcane was more pronounced in roots and, to a minor extent, in aboveground organs, especially mature leaves (Figure 2A). Similar patterns were found for AMT2;1 expression in A. thaliana (Sohlenkamp et al., 2002), O. sativa (Suenaga et al., 2003), Lotus japonicus (Simon-Rosin et al., 2003), and S. bicolor (Koegel et al., 2013), in all cases showing expression in distinct plant organs. For instance, the homologue PbAMT2 from Pyrus betulaefolia was shown to be expressed in stems, petioles, and leaves but primarily in roots (Li et al., 2016), similar to the ScAMT2;1 expression pattern described here for sugarcane. Conversely, PtAMT2;1 from P. trichocarpa was shown to be exclusively expressed in roots (Couturier et al., 2007). Analysis of the expression profile in sugarcane roots showed that ScAMT2;1 transcription was slightly but significantly induced by the N status and significantly induced by (possibly sensing the lack of NH4+) in roots, culms, and mature leaves (Figure 2B). In Arabidopsis roots, AtAMT2;1 is induced by N starvation and weakly repressed by nitrate (Giehl et al., 2017), whereas the poplar homologue PtrAMT2;1 is not regulated by N (Couturier et al., 2007), suggesting distinct regulation according to species.
The ScAMT2;1 endogenous promoter drove the expression of GUS in Arabidopsis mainly at endodermal and pericycle cells in the innermost root tissue, with apparently more expression in roots subjected to ammonium compared with N deficiency. Previously, the activity of the Arabidopsis AMT2;1 promoter was shown to become more confined to root endodermal and particularly pericycle cells when plants were exposed to high ammonium concentrations (Giehl et al., 2017). N deficiency, in turn, shifted the expression of AtAMT2;1 towards the outer cells (Giehl et al., 2017). Our results with the heterologous expression of the ScAMT2;1 promoter in Arabidopsis provide initial evidence that ScAMT2;1 expression is concentrated on vascular and immediately surrounding tissues in roots and shoots. To confirm the predicted tissue-specific localization of ScAMT2;1 in sugarcane, future studies based on in situ hybridization or transient or stable expression of AMT2;1::GUS/GFP directly in sugarcane will be necessary. Nevertheless, functional evaluation of sugarcane sequences, such as genes and regulatory regions, in a model plant provides initial inference before narrowing down to the target organism, such as described for sugarcane gene functional analysis (Wang et al., 2021; Chai et al., 2022).
Complementation of the ammonium uptake-defective yeast mutant (triple mepΔ) suggested that ScAMT2;1 is a functional NH4+ transporter, despite its lower substrate affinity than AtAMT1;1, similar to what had been previously described for AtAMT2;1 (Sohlenkamp et al., 2000). The expression of OsAMT2;1 in the same yeast mutant supported cell growth on media containing 5 mM NH4+ but not on 1 mM NH4+ (Suenaga et al., 2003). The poplar homologue PtrAMT2;1 complemented the triple mepΔ cells on 1 mM NH4+ (Couturier et al., 2007), while the homologues AtAMT2;1 (Sohlenkamp et al., 2000), LjAMT2;1 (Simon-Rosin et al., 2003), PbAMT2, and PbAMT3 (Li et al., 2016) restored the growth of the same yeast mutant strain cells on 0.5 mM NH4+ or even lower N concentrations, indicating a diverse biochemical transport capacity of the various AMT2 homologues.
Sugarcane ScAMT2;1 restored the growth of the triple mepΔ yeast in a pH-dependent manner, increasing activity consistently as the pH was raised from 5.0 to 6.0 while achieving the same growth of the negative control triple mepΔ+pDR196 at pH 7.5 (Figure 3), suggesting NH3 diffusion (Martinelle et al., 1996; Sohlenkamp et al., 2002). The apparent Vmax of AtAMT2;1 determined in yeast also increased at higher pH values (Sohlenkamp et al., 2002). Ammonia is a weak base (pKa 9.25), with more than 99% protonated at neutral external pH. Thus, elevating the pH from 5.0 to 7.5 increases the concentration of NH3 by 30-fold, while that of NH4+ remains almost constant (Sohlenkamp et al., 2002). The complementation of triple mepΔ by ScAMT2;1 at lower pH suggests that NH4+ rather than NH3 is the substrate for ScAMT2;1. Notably, as a common transport mechanism performed by AMT2 proteins, NH4+ appears to be deprotonated before transport, and NH3 permeates through the transporter pore (Khademi et al., 2004; Guether et al., 2009; Neuhäuser et al., 2009; Akgun and Khademi, 2011; Ariz et al., 2018). In yeast, ScAMT2;1 was unable to transport methylammonium (MeA), as triple mepΔ cells expressing ScAMT2;1 exhibited similar growth as cells expressing the empty vector (Figure 3). Previously, LjAMT2;1 and AtAMT2;1 were shown to be impermeable to MeA and to perform electroneutral transport of uncharged ammonia with a low transport capacity (Simon-Rosin et al., 2003; Neuhäuser et al., 2009). Thus, these three plant AMT2 homologues exhibit pH-dependent activity, being less active in acidic extracellular environments and displaying similar biochemical properties for NH4+ uptake and possible cotransport of NH3/H+ through the protein lumen (Neuhäuser et al., 2009). Certainly, expression analysis in Xenopus oocyte cells would be needed to validate this hypothesis.
The function of ScAMT2;1 was further supported by ectopic expression in the Arabidopsis qko mutant. Arabidopsis qko lines overexpressing ScAMT2;1 accumulated more biomass under NH4+ nutrition as the only N source (Figure 4). Short-term influx analysis in qko complemented lines expressing ScAMT2;1 allowed us to estimate a Km equal to 90.17 µM and a Vmax of 338.99 µmol h-1 g-1 root DW, suggesting that ScAMT2;1 contributes to high-affinity ammonium transport in planta. The estimated Km value of ScAMT2;1 is higher than those determined for AtAMT1;1 and AtAMT2;1 via 13N-ammonium in yeast (22 and 21 µM at pH 6.1, respectively) (Sohlenkamp et al., 2002). However, despite the similar Km values, the ammonium transport capacity of AtAMT2;1 was at least 10 times lower than that of AtAMT1;1 at a pH of 5.0 and 6.1. Nevertheless, at a pH of 7.5, the transport capacity of the two transporters appeared to be similar (Sohlenkamp et al., 2002).
Initial evidence for the possible function of ScAMT2;1 was obtained by heterologous expression of ScAMT2;1 in Arabidopsis driven by one of its endogenous promoters (p2ScAMT2;1). Expression of p2ScAMT2;1::ScAMT2;1 complemented the growth of qko plants under ammonium supply and significantly increased net ammonium influx only at high external N concentrations (Figure 6). These results and the localization of p2ScAMT2;1 promoter activity (Figure 5) suggest that ScAMT2;1 mediates ammonium import from the apoplast, which is in agreement with previous studies with AtAMT2;1 (Sohlenkamp et al., 2002; Yuan et al., 2007; Neuhäuser et al., 2009). In Arabidopsis roots, AtAMT2;1 contributes to 10%–25% of the overall ammonium uptake rate at high external ammonium concentrations, whereas under N deficiency, AtAMT2;1 activity occurs in outer cell layers and supports root ammonium uptake capacity in the millimolar concentration range (Giehl et al., 2017).
Based on the localization of p2ScAMT2;1 promoter activity, we hypothesized that ScAMT2;1 might be involved in root-to-shoot NH4+ translocation. Our findings indicated a contribution of ScAMT2;1 to shoot ammonium translocation only in fully ammonium- or nitrate-supplied plants (Figure 7). The increased root-to-shoot NH4+ translocation may have resulted from an increased ScAMT2;1-facilitated radial transport of ammonium towards the root vascular tissue, altering N partitioning between roots and shoots and impacting the N nutrition of the shoot. Although glutamine is the predominant organic N form translocated in the xylem of ammonium-supplied oilseed rape (Finnemann and Schjoerring, 1999) and A. thaliana plants (Lam et al., 1995), ammonium can represent 11% of the total N translocated in the xylem sap, reaching up to 18 mM in Arabidopsis vasculature (Giehl et al., 2017). Hence, increased root-to-shoot translocation of ammonium provides a stable supply of N to the shoots in response to high N availability. In Arabidopsis, a concerted function of AtAMT2;1 and GLN1;2 in roots is proposed to determine ammonium translocation and assimilation in response to high N supply (Giehl et al., 2017). Likewise, our results showing ScAMT2;1 activity in the innermost root cell suggest that this protein might also provide ammonium for the N assimilation pathway in sugarcane roots upon high ammonium supply. While GS1 activity has not been linked to yield gain or improved NUE in sugarcane genotypes (Robinson et al., 2007), ammonium is preferentially acquired by sugarcane roots (Robinson et al., 2011; Lima et al., 2022). Therefore, the critical role of ammonium translocation to shoots in response to plant nutritional status might significantly impact vegetative biomass in sugarcane plants. The mechanisms involved in root-to-shoot N transport, recycling, and remobilization are paramount for improving plant performance and NUE and can certainly decrease the need for fertilizers and strengthen sustainable sugarcane crop production. Our preliminary findings suggest that ScAMT2;1 might contribute to ammonium uptake in sugarcane roots in response to high external N availability in addition to presumably contributing to root-to-shoot ammonium translocation by facilitating its radial transport towards the vascular system, which may finally contribute to enhanced shoot growth under abundant N supply.
We conclude that ScAMT2;1 is a functional ammonium transporter as it was able to complement the defective Arabidopsis mutant and partially complement yeast. Estimation of Km and Vmax indicated ScAMT2;1 to be a high-affinity ammonium transporter. In sugarcane, ScAMT2;1 is expressed in different organs, with the highest expression in roots induced by external nitrate (possibly lack of ammonium). When expressed in A. thaliana, ScAMT2;1 promoter activity can be detected in the innermost cell layers of roots and the vasculature of leaves, and it can increase ammonium translocation from root to shoot. Our findings suggest that ScAMT2;1 might contribute to ammonium uptake in sugarcane roots in response to high external N availability and to probably contribute to root-to-shoot ammonium translocation by facilitating its radial transport towards the vascular system.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
AF and JL designed the research project. AK and RM performed experiments. JL, RM, and AK analyzed and interpreted the data. AK, RM, and NS performed bioinformatic analysis. NS helped to analyze/interpret the genomic data from BAC clones. MV contributed with analyses and protocols. AK and JL wrote the manuscript. RG critically reviewed the manuscript and helped with data interpretation. AF and JL supervised the project and experiments. All authors contributed to the article and approved the submitted version.
This work was supported by FAPESP (The Sao Paulo Research Foundation) through Regular Research Grants (16/14669-8; 2013/15989-8), and fellowships to JL (2010/11313-1), and RM (2017/00460-2). Additional support came from CNPq (Brazilian National Council for Scientific and Technological Development) and CAPES (Coordination for the Improvement of Higher Education Personnel). AF is a recipient of a CNPq research fellowship (310645/2021-2).
Seeds of qko were gently provided by Prof. Dr. Nicolaus von Wirén from the Leibniz Institute of Plant Genetics and Crop Plant Research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1039041/full#supplementary-material
Akgun, U., Khademi, S. (2011). Periplasmic vestibule plays an important role for solute recruitment, selectivity, and gating in the Rh/Amt/MEP superfamily. Proc. Natl. Acad. Sci. 108, 3970–3975. doi: 10.1073/pnas.1007240108
Ariz, I., Boeckstaens, M., Gouveia, C., Martins, A. P., Sanz-Luque, E., Fernández, E., et al. (2018). Nitrogen isotope signature evidences ammonium deprotonation as a common transport mechanism for the AMT-Mep-Rh protein superfamily. Sci. Adv. 4, eaar3599. doi: 10.1126/sciadv.aar3599
Chai, Z., Fang, J., Yao, W., Zhao, Y., Cheng, G., Akbar, S., et al. (2022). ScGAIL, a sugarcane n-terminal truncated DELLA-like protein, participates in GA signaling in arabidopsis. J. Exp. Bot 73, 3462–3476. doi: 10.1093/jxb/erac056
Chow, C.-N., Zheng, H.-Q., Wu, N.-Y., Chien, C.-H., Huang, H.-D., Lee, T.-Y., et al. (2016). PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44, D1154–D1160. doi: 10.1093/nar/gkv1035
Couturier, J., Montanini, B., Martin, F., Brun, A., Blaudez, D., Chalot, M. (2007). The expanded family of ammonium transporters in the perennial poplar plant. New Phytol. 174, 137–150. doi: 10.1111/j.1469-8137.2007.01992.x
Crooks, G. E., Hon, G., Chandonia, J.-M., Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
D’Apuzzo, E., Rogato, A., Simon-Rosin, U., el Alaoui, H., Barbulova, A., Betti, M., et al. (2004). Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol. 134, 1763–1774. doi: 10.1104/pp.103.034322
de Castro, S. G. Q., Magalhães, P. S. G., Franco, H. C. J., Mutton, M.Â. (2018). Harvesting systems, soil cultivation, and nitrogen rate associated with sugarcane yield. Bioenergy Res. 11, 583–591. doi: 10.1007/s12155-018-9917-0
Dechorgnat, J., Francis, K. L., Dhugga, K. S., Rafalski, J. A., Tyerman, S. D., Kaiser, B. N. (2019). Tissue and nitrogen-linked expression profiles of ammonium and nitrate transporters in maize. BMC Plant Biol. 19, 1–13. doi: 10.1186/s12870-019-1768-0
de Setta, N., Monteiro-Vitorello, C. B., Metcalfe, C. J., Cruz, G. M. Q., del Bem, L. E., Vicentini, R., et al. (2014). Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics 15, 1–18. doi: 10.1186/1471-2164-15-540
Dobermann, A. R. (2005). Nitrogen use efficiency-state of the art. Agron–Fac. Publ. 316. Available at: https://digitalcommons.unl.edu/agronomyfacpub/316
Ellerbeck, M., Schüßler, A., Brucker, D., Dafinger, C., Loos, F., Brachmann, A. (2013). Characterization of three ammonium transporters of the glomeromycotan fungus Geosiphon pyriformis. Eukaryot Cell 12, 1554–1562. doi: 10.1128/EC.00139-13
Erisman, J. W., van Grinsven, H., Leip, A., Mosier, A., Bleeker, A. (2010). Nitrogen and biofuels; an overview of the current state of knowledge. Nutr. Cycl Agroecosyst 86, 211–223. doi: 10.1007/s10705-009-9285-4
Finnemann, J., Schjoerring, J. K. (1999). Translocation of NH4+ in oilseed rape plants in relation to glutamine synthetase isogene expression and activity. Physiol. Plant 105, 469–477. doi: 10.1034/j.1399-3054.1999.105311.x
Franco, H. C. J., Trivelin, P. C. O., Faroni, C. E., Vitti, A. C., Otto, R. (2008). Aproveitamento pela cana-de-açúcar da adubação nitrogenada de plantio. Rev. Bras. Cienc Solo 32, 2763–2770. doi: 10.1590/S0100-06832008000700021
Garsmeur, O., Droc, G., Antonise, R., Grimwood, J., Potier, B., Aitken, K., et al. (2018). A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 9, 1–10. doi: 10.1038/s41467-018-05051-5
Gazzarrini, S., Lejay, L., Gojon, A., Ninnemann, O., Frommer, W. B., von Wirén, N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into arabidopsis roots. Plant Cell 11, 937–947. doi: 10.1105/tpc.11.5.937
Giehl, R. F. H., Laginha, A. M., Duan, F., Rentsch, D., Yuan, L., von Wirén, N. (2017). A critical role of AMT2; 1 in root-to-shoot translocation of ammonium in arabidopsis. Mol. Plant 10, 1449–1460. doi: 10.1016/j.molp.2017.10.001
Gietz, R. D., Schiestl, R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34. doi: 10.1038/nprot.2007.13
Graff, L., Obrdlik, P., Yuan, L., Loqué, D., Frommer, W. B., von Wirén, N. (2011). N-terminal cysteines affect oligomer stability of the allosterically regulated ammonium transporter LeAMT1; 1. J. Exp. Bot. 62, 1361–1373. doi: 10.1093/jxb/erq379
Gu, R., Duan, F., An, X., Zhang, F., von Wirén, N., Yuan, L. (2013). Characterization of AMT-mediated high-affinity ammonium uptake in roots of maize (Zea mays l.). Plant Cell Physiol. 54, 1515–1524. doi: 10.1093/pcp/pct099
Guether, M., Neuhauser, B., Balestrini, R., Dynowski, M., Ludewig, U., Bonfante, P. (2009). A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol. 150, 73–83. doi: 10.1104/pp.109.136390
Hall, T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hoagland, D. R., Arnon, D. I. (1950). The water-culture method for growing plants without soil. California Agricultural Experiment Station, Circular 347.
Hoang, N., Furtado, A., Botha, F. C., Simmons, B. A., Henry, R. J. (2015). Potential for genetic improvement of sugarcane as a source of biomass for biofuels. Front. Bioeng Biotechnol. 3, 182. doi: 10.3389/fbioe.2015.00182
Hulo, N., Bairoch, A., Bulliard, V., Cerutti, L., de Castro, E., Langendijk-Genevaux, P. S., et al. (2006). The PROSITE database. Nucleic Acids Res. 34, D227–D230. doi: 10.1093/nar/gkj063
Jefferson, R. A., Kavanagh, T. A., Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Jiang, J., Zhao, J., Duan, W., Tian, S., Wang, X., Zhuang, H., et al. (2019). TaAMT2; 3a, a wheat AMT2-type ammonium transporter, facilitates the infection of stripe rust fungus on wheat. BMC Plant Biol. 19, 1–11. doi: 10.1186/s12870-019-1841-8
Khademi, S., O’Connell, J., Remis, J., Robles-Colmenares, Y., Miercke, L. J. W., Stroud, R. M. (2004). Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 1979) 305, 1587–1594. doi: 10.1126/science.1101952
Koegel, S., Ait Lahmidi, N., Arnould, C., Chatagnier, O., Walder, F., Ineichen, K., et al. (2013). The family of ammonium transporters (AMT) in Sorghum bicolor: two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol. 198, 853–865. doi: 10.1111/nph.12199
Kohany, O., Gentles, A. J., Hankus, L., Jurka, J. (2006). Annotation, submission and screening of repetitive elements in repbase: RepbaseSubmitter and censor. BMC Bioinf. 7, 1–7. doi: 10.1186/1471-2105-7-474
Krogh, A., Larsson, B., von Heijne, G., Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Lam, H.-M., Coschigano, K., Schultz, C., Melo-Oliveira, R., Tjaden, G., Oliveira, I., et al. (1995). Use of arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7, 887. doi: 10.1105/tpc.7.7.887
Leal, G. A., Jr., Albuquerque, P. S. B., Figueira, A. (2007). Genes differentially expressed in Theobroma cacao associated with resistance to witches’ broom disease caused by Crinipellis perniciosa. Mol. Plant Pathol. 8, 279–292. doi: 10.1111/j.1364-3703.2007.00393.x
Li, H., Cong, Y., Chang, Y., Lin, J. (2016). Two AMT2-type ammonium transporters from Pyrus betulaefolia demonstrate distinct expression characteristics. Plant Mol. Biol. Rep. 34, 707–719. doi: 10.1007/s11105-015-0957-8
Li, H., Hu, B., Chu, C. (2017a). Nitrogen use efficiency in crops: lessons from arabidopsis and rice. J. Exp. Bot. 68, 2477–2488. doi: 10.1093/jxb/erx101
Li, T., Liao, K., Xu, X., Gao, Y., Wang, Z., Zhu, X., et al. (2017b). Wheat ammonium transporter (AMT) gene family: diversity and possible role in host–pathogen interaction with stem rust. Front. Plant Sci. 8, 1637. doi: 10.3389/fpls.2017.01637
Lima, J. E., Serezino, L. H. D., Alves, M. K., Tagliaferro, A. L., Vitti, M., Creste, S., et al. (2022). Root nitrate uptake in sugarcane (Saccharum spp.) is modulated by transcriptional and presumably posttranscriptional regulation of the NRT2. 1/NRT3. 1 transport system. Mol. Genet. Genomics 297, 1403–1421. doi: 10.1007/s00438-022-01929-8
Li, B.-Z., Merrick, M., Li, S.-M., Li, H.-Y., Zhu, S.-W., Shi, W.-M., et al. (2009). Molecular basis and regulation of ammonium transporter in rice. Rice Sci. 16, 314–322. doi: 10.1016/S1672-6308(08)60096-7
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loqué, D., von Wirén, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55, 1293–1305. doi: 10.1093/jxb/erh147
Loqué, D., Yuan, L., Kojima, S., Gojon, A., Wirth, J., Gazzarrini, S., et al. (2006). Additive contribution of AMT1; 1 and AMT1; 3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient arabidopsis roots. Plant J. 48, 522–534. doi: 10.1111/j.1365-313X.2006.02887.x
Ludewig, U., Wilken, S., Wu, B., Jost, W., Obrdlik, P., el Bakkoury, M., et al. (2003). Homo-and hetero-oligomerization of ammonium transporter-1 uniporters. J. Biol. Chem. 278, 45603–45610. doi: 10.1074/jbc.M307424200
Marini, A.-M., André, B. (2000). In vivo n-glycosylation of the Mep2 high-affinity ammonium transporter of saccharomyces cerevisiae reveals an extracytosolic n-terminus. Mol. Microbiol. 38, 552–564. doi: 10.1046/j.1365-2958.2000.02151.x
Marini, A.-M., Soussi-Boudekou, S., Vissers, S., Andre, B. (1997). A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell Biol. 17, 4282–4293. doi: 10.1128/MCB.17.8.4282
Martinelle, K., Westlund, A., Häggström, L. (1996). Ammonium ion transport–a cause of cell death. Cytotechnology 22, 251–254. doi: 10.1007/BF00353945
Mayer, M., Ludewig, U. (2006). Role of AMT1; 1 in acquisition in Arabidopsis thaliana. Plant Biol. 8, 522–528. doi: 10.1055/s-2006-923877
McAllister, C. H., Beatty, P. H., Good, A. G. (2012). Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnol. J. 10, 1011–1025. doi: 10.1111/j.1467-7652.2012.00700.x
McDonald, T. R., Dietrich, F. S., Lutzoni, F. (2012). Multiple horizontal gene transfers of ammonium transporters/ammonia permeases from prokaryotes to eukaryotes: toward a new functional and evolutionary classification. Mol. Biol. Evol. 29, 51–60. doi: 10.1093/molbev/msr123
Narusaka, M., Shiraishi, T., Iwabuchi, M., Narusaka, Y. (2010). The floral inoculating protocol: a simplified Arabidopsis thaliana transformation method modified from floral dipping. Plant Biotechnol. 27, 349–351. doi: 10.5511/plantbiotechnology.27.349
Neuhäuser, B., Dynowski, M., Ludewig, U. (2009). Channel-like NH3 flux by ammonium transporter AtAMT2. FEBS Lett. 583, 2833–2838. doi: 10.1016/j.febslet.2009.07.039
Nogueira, E. M., Olivares, F. L., Japiassu, J. C., Vilar, C., Vinagre, F., Baldani, J. I., et al. (2005). Characterization of glutamine synthetase genes in sugarcane genotypes with different rates of biological nitrogen fixation. Plant Sci. 169, 819–832. doi: 10.1016/j.plantsci.2005.05.031
Otto, R., Castro, S. A. Q., Mariano, E., Castro, S. G. Q., Franco, H. C. J., Trivelin, P. C. O. (2016). Nitrogen use efficiency for sugarcane-biofuel production: what is next? Bioenergy Res. 9, 1272–1289. doi: 10.1007/s12155-016-9763-x
Rentsch, D., Laloi, M., Rouhara, I., Schmelzer, E., Delrot, S., Frommer, W. B. (1995). NTR1 encodes a high affinity oligopeptide transporter in arabidopsis. FEBS Lett. 370, 264–268. doi: 10.1016/0014-5793(95)00853-2
Robinson, N., Brackin, R., Vinall, K., Soper, F., Holst, J., Gamage, H., et al. (2011). Nitrate paradigm does not hold up for sugarcane. PloS One 6, e19045. doi: 10.1371/journal.pone.0019045
Robinson, N., Fletcher, A., Whan, A., Critchley, C., von Wirén, N., Lakshmanan, P., et al. (2007). Sugarcane genotypes differ in internal nitrogen use efficiency. Funct. Plant Biol. 34, 1122–1129. doi: 10.1071/FP07183
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M.-A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. doi: 10.1093/bioinformatics/16.10.944
Schwacke, R., Schneider, A., van der Graaff, E., Fischer, K., Catoni, E., Desimone, M., et al. (2003). ARAMEMNON, a novel database for arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26. doi: 10.1104/pp.011577
Sharma, L. K., Bali, S. K. (2018). A review of methods to improve nitrogen use efficiency in agriculture. Sustainability 10, 51. doi: 10.3390/su10010051
Simon-Rosin, U., Wood, C., Udvardi, M. K. (2003). Molecular and cellular characterisation of LjAMT2; 1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol. Biol. 51, 99–108. doi: 10.1023/A:1020710222298
Skocaj, D. M., Everingham, Y. L., Schroeder, B. L. (2013). Nitrogen management guidelines for sugarcane production in Australia: can these be modified for wet tropical conditions using seasonal climate forecasting? Springer Sci. Rev. 1, 51–71. doi: 10.1007/s40362-013-0004-9
Sohlenkamp, C., Shelden, M., Howitt, S., Udvardi, M. (2000). Characterization of arabidopsis AtAMT2, a novel ammonium transporter in plants. FEBS Lett. 467, 273–278. doi: 10.1016/S0014-5793(00)01153-4
Sohlenkamp, C., Wood, C. C., Roeb, G. W., Udvardi, M. K. (2002). Characterization of arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol. 130, 1788–1796. doi: 10.1104/pp.008599
Song, S., He, Z., Huang, X., Zhong, L., Liu, H., Sun, G., et al. (2017). Cloning and characterization of the ammonium transporter genes BaAMT1; 1 and BaAMT1; 3 from Chinese kale. Hortic. Environ. Biotechnol. 58, 178–186. doi: 10.1007/s13580-017-0168-3
Suenaga, A., Moriya, K., Sonoda, Y., Ikeda, A., von Wirén, N., Hayakawa, T., et al. (2003). Constitutive expression of a novel-type ammonium transporter OsAMT2 in rice plants. Plant Cell Physiol. 44, 206–211. doi: 10.1093/pcp/pcg017
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thirugnanasambandam, P. P., Hoang, N. V, Henry, R. J. (2018). The challenge of analyzing the sugarcane genome. Front. Plant Sci. 9, 616. doi: 10.3389/fpls.2018.00616
Thompson, J. D., Gibson, T. J., Higgins, D. G. (2003). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinf. 2–3. doi: 10.1002/0471250953.bi0203s00
Thorburn, P. J., Biggs, J. S., Palmer, J., Meier, E. A., Verburg, K., Skocaj, D. M. (2017). Prioritizing crop management to increase nitrogen use efficiency in Australian sugarcane crops. Front. Plant Sci. 8, 1504. doi: 10.3389/fpls.2017.01504
Thorburn, P. J., Jakku, E., Webster, A. J., Everingham, Y. L. (2011). Agricultural decision support systems facilitating co-learning: a case study on environmental impacts of sugarcane production. Int. J. Agric. Sustain 9, 322–333. doi: 10.1080/14735903.2011.582359
Tilman, D., Socolow, R., Foley, J. A., Hill, J., Larson, E., Lynd, L., et al. (2009). Beneficial biofuels–the food, energy, and environment trilemma. Science 1979(325), 270–271. doi: 10.1126/science.1177970
Tomkins, J. P., Yu, Y., Miller-Smith, H., Frisch, D. A., Woo, S. S., Wing, R. A. (1999). A bacterial artificial chromosome library for sugarcane. Theor. Appl. Genet. 99, 419–424. doi: 10.1007/s001220051252
von Wirén, N., Lauter, F.-R., Ninnemann, O., Gillissen, B., Walch-Liu, P., Engels, C., et al. (2000). Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J. 21, 167–175. doi: 10.1046/j.1365-313x.2000.00665.x
von Wittgenstein, N. J. J. B., Le, C. H., Hawkins, B. J., Ehlting, J. (2014). Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol. Biol. 14, 1–17. doi: 10.1186/1471-2148-14-11
Waclawovsky, A. J., Sato, P. M., Lembke, C. G., Moore, P. H., Souza, G. M. (2010). Sugarcane for bioenergy production: an assessment of yield and regulation of sucrose content. Plant Biotechnol. J. 8, 263–276. doi: 10.1111/j.1467-7652.2009.00491.x
Wang, J., Li, Y., Ching, M. W., Beuchat, G., Chen, L.-Q. (2021). Identification and analysis of stem-specific promoters from sugarcane and energy cane for oil accumulation in their stems. Glob Change Biol. Bioenergy 13, 1515–1527. doi: 10.1111/gcbb.12872
Wu, Z, Gao, X, Zhang, N, Feng, X, Huang, Y, Zeng, Q, et al. (2021). Genome-wide identification and transcriptional analysis of ammonium transporters in Saccharum. Genomics 113, 1671–1680. doi: 10.1111/gcbb.12872
Yuan, L., Graff, L., Loqué, D., Kojima, S., Tsuchiya, Y. N., Takahashi, H., et al. (2009). AtAMT1; 4, a pollen-specific high-affinity ammonium transporter of the plasma membrane in arabidopsis. Plant Cell Physiol. 50, 13–25. doi: 10.1093/pcp/pcn186
Yuan, L., Loqué, D., Kojima, S., Rauch, S., Ishiyama, K., Inoue, E., et al. (2007). The organization of high-affinity ammonium uptake in arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19, 2636–2652. doi: 10.1105/tpc.107.052134
Keywords: ammonium uptake, AMT2 subfamily, nitrogen use efficiency, quadruple mutant, transport kinetics, xylem loading
Citation: Koltun A, Maniero RA, Vitti M, de Setta N, Giehl RFH, Lima JE and Figueira A (2022) Functional characterization of the sugarcane (Saccharum spp.) ammonium transporter AMT2;1 suggests a role in ammonium root-to-shoot translocation. Front. Plant Sci. 13:1039041. doi: 10.3389/fpls.2022.1039041
Received: 07 September 2022; Accepted: 31 October 2022;
Published: 18 November 2022.
Edited by:
Jing Che, Institute of Soil Science (CAS), ChinaReviewed by:
Jiang Wang, University of Illinois at Urbana-Champaign, United StatesCopyright © 2022 Koltun, Maniero, Vitti, de Setta, Giehl, Lima and Figueira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joni E. Lima, bGltYWpvbmlAZ29vZ2xlbWFpbC5jb20=; Antonio Figueira, ZmlndWVpcmFAY2VuYS51c3AuYnI=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.