- Liaoning Key Laboratory for Biological Invasions and Global Changes, College of Bioscience and Biotechnology, Shenyang Agricultural University, Shenyang, Liaoning, China
Soil nitrogen forms are important for exotic plant invasions. However, little effort has been made to study the molecular mechanisms underlying the utilization of different N forms in co-occurring invasive and native plants. The invasive plant Xanthium strumarium prefers nitrate relative to ammonium, and mainly invades nitrate-dominated environments, while it co-occurring native congener X. sibiricum prefers ammonium. Here, we addressed the genetic bases for the interspecific difference in ammonium use and the effects of gibberellin (GA). Twenty-six transcripts related with GA biosynthesis and ammonium utilization were induced by ammonium in X. sibiricum, while only ten in X. strumarium and none for ammonium uptake. XsiAMT1.1a, XsiGLN1.1 and XsiGLT1b may be crucial for the strong ability to absorb and assimilate ammonium in X. sibiricum. All tested transcripts were significantly up-regulated by GA1 and GA4 in X. sibiricum. XsiGA3OX1a, which was also induced by ammonium, may be involved in this regulation. Consistently, glutamine synthetase activity increased significantly with increasing ammonium-N/nitrate-N ratio for X. sibiricum, while decreased for X. strumarium. Our study is the first to determine the molecular mechanisms with which invasive and native plants use ammonium differently, contributing to understanding the invasion mechanisms of X. strumarium and its invasion habitat selection.
Introduction
Nitrogen (N) is an essential component of proteins and nucleic acids, and thus plays critical roles in plant growth and development (Feng et al., 2009; Xu et al., 2012). In many ecosystems, N is an important limiting environmental resource for plants (Thomas et al., 2015). The two forms of inorganic N, i.e., nitrate and ammonium, are the main N source for plants in most ecosystems, and different plants prefer to use different N forms (Zhang et al., 2018; Sun et al., 2021). Rice (Oryza sativa L.), potato (Solanum tuberosum L.), tea trees (Camellia sinensis L.) and invasive plant Flaveria bidentis prefer ammonium, while wheat (Triticum aestivum L.), maize (Zea mays L.), cotton (Gossypium hirsutum L.) and invasive plant Ipomoea cairica prefer nitrate (Li et al., 2013; Huangfu et al., 2016; Chen and Chen, 2019). Our previous study showed that Xanthium sibiricum grows better in ammonium relative to nitrate, while its invasive congener X. strumarium grows better in nitrate treatment (Luo et al., 2022). However, the molecular mechanism underlying the better growth of X. sibiricum in ammonium is still unknown.
Plants absorb ammonium by ammonium transporter (AMT). AtAMT1.1, 1.2, 1.3 and 1.5 are highly expressed in roots and all involved in ammonium uptake (Gazzarrini et al., 1999; Yuan et al., 2007). Ammonium uptakes were significantly reduced in the mutants of atamt1.1, 1.2 and 1.3 (Kaiser et al., 2002; Loque et al., 2006; Yuan et al., 2007). Before being used to synthesize organics, ammonium is firstly assimilated into glutamine and glutamate by glutamine synthetase (GS) and glutamate synthase (GOGAT), respectively. AtGS1 and AtGS2 were highly expressed in roots and shoots, respectively (Ishiyama et al., 2004). AtGLN1.1, one of GS1 subfamily members, was more highly expressed under N starvation than other members of GS1 subfamily. AtGLN1.2 was induced by ammonium and more highly expressed under 10 mmol L-1 ammonium treatment than other members of GS1 subfamily. The GS activity of AtGLN1.3 was stronger than that of other members of GS1 subfamily (Ishiyama et al., 2004). The GS activity was reduced in the mutant of AtGLN1.2 which had low ammonium assimilation ability (Lothier et al., 2011). For GOGAT in Arabidopsis, all family members are involved in ammonium assimilation (Li et al., 2017). AtGLU1 was highly expressed in leaves, while AtGLU2 and AtGLT1 were highly expressed in roots (Coschigano et al., 1998; Lancien et al., 2002).
Plant hormones can regulate utilization of different N forms (O'Brien et al., 2016). It has been demonstrated that nitrate can induce expressions of some cytokinin biosynthesis related genes, including IPT3 (isopentenyl transferase 3) and CYP735A (encoding a cytochrome P450 monooxygenase), promoting cytokinin synthesis (Takei et al., 2004; Wang et al., 2004; Sakakibara, 2006). In turn, cytokinin can induce expressions of some nitrate utilization related genes, including NRT1.3 (low-affinity nitrate transporter) and NIA1 (nitrate reductase) (Brenner et al., 2005; Kiba et al., 2005; Sakakibara et al., 2006). However, few studies determined the interaction between hormones and ammonium. Recently, Li et al. (2019) found that shoot ethylene content increased in A. thaliana under high level of ammonium. Compared with the wild type, the mutant of EIN3, which acts as a key regulator in ethylene signal pathway, can increase ammonium tolerance. Gibberellin GA3 was also found to improve ammonium uptake and utilization in rice by up-regulating expressions of OsAMT1.1 and OsGS1.2 (Li et al., 2018).
Xanthium strumarium, native to north America, is a noxious invasive plant in north China (Feng, 2020). It prefers to absorb nitrate relative to ammonium, and grows better in nitrate (Luo et al., 2022). Consistently, it often invades disturbed habitats with nitrate as dominant soil N form. In contrast, its co-occurring native congener X. sibiricum prefers to use ammonium, and grows better in ammonium (Luo et al., 2022). The difference in N form utilization between these Xanthium species may be associated with invasion success of the invader, and its invasiveness evolution under the background of global change such as N deposition. The ratio of nitrate-N to ammonium-N in the atmospheric deposition N increases gradually in recent years in China (Yu et al., 2019). Thus, it is important to elucidate the mechanisms underlying this difference. In this study, we explored the molecular mechanisms underlying the strong ability to absorb and assimilate ammonium for X. sibiricum. We hypothesize that (1) more transcripts related with ammonium uptake and assimilation such as AMT, GS and GOGAT are induced by ammonium, and/or more highly expressed in X. sibiricum than in X. strumarium; (2) the same for the transcripts related with syntheses of some hormones, and these hormones up-regulate expressions of some transcripts related with ammonium uptake and assimilation. Our study is the first one that explored the molecular mechanisms underlying the difference in response to different N forms between invasive and native plants.
Materials and methods
Materials
The invasive plant Xanthium strumarium and its native congener X. sibiricum were used in this study. The seeds of these species were collected in Gaomabao village, Faku County, Shenyang City, Liaoning Province (42° 27′ N, 123° 0′ E; asl. 73 m), where no herbicides were applied to control the invader. Seed germination and seedling preparation were the same as in our previous study (Luo et al., 2022). Three similar-sized seedlings of each of the two species were grown separately in 1/2 Hoagland nutrient solution without N for 2 d, and then treated with 5 mmol L-1 KNO3 and NH4Cl, respectively. Four hour later, the fine roots of three seedlings of each species were collected separately for RNA sequencing (three biological replicates).
RNA extraction and sequencing
The roots of the above-mentioned seedlings were collected for total RNA extraction using the MiniBEST universal RNA extraction kit (Takara, Dalian, China). The quality and quantity of the RNA extracts were assessed using 1% agarose gel electrophoresis, NanoDrop (Thermo Fisher Scientific, Inc., Waltham, USA.) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, USA), respectively. In order to acquire the full-length sequences of transcriptomes, PacBio sequencing (third-generation sequencing) was carried out firstly, as there was no genome data for the two Xanthium species. Then, HiSeq sequencing (second-generation sequencing) was performed in order to analyze the expression levels of the transcripts responding to different N forms. The sequencing methods were the same as in our previous study (Luo et al., 2022).
Bioinformatics analysis
High-quality clean data were acquired using Cutadapt (version 1.9.1). The data were then assembled according to a method described by Grabherr et al. (2011). Transcript annotation was carried out based on non-redundant protein (NR), KEGG, GO and Swiss-Prot. Relative expression level (FPKM value) of each transcript was calculated using RSEM (v1.2.6). The expression difference in each transcript between ammonium and nitrate treatments was conducted for each species using the Bioconductor package DESeq (Li and Dewey, 2011). The transcripts were considered as differentially expressed transcripts (DETs) if their Benjamini and Hochberg (BH)-adjusted P-values (FDR) ≤ 0.05 and the magnitudes of the difference ≥ 2-fold (Luo et al., 2022). GO terms and KEGG pathway enrichment analyses were conducted for the DETs using GO-TermFinder and Kyoto Encyclopedia of Genes and Genomes (KEGG) database, respectively. Significant enrichments were also determined by BH-adjusted P-values (FDR) ≤ 0.05.
To determine the transcripts that may be responsible for efficient use of ammonium in the two Xanthium species, the expressions of the DETs belonging to AMT, GLN, GLU and GLT members were presented using heat maps, which were constructed using the pheatmap package in R (Costa et al., 2020). The differences in expressions of the DETs related with gebberalin (GA) metabolism were also analyzed, as GA may regulate expressions of AMT, GLN, GLU and GLT members. The methods had been described in detail in our previous study (Luo et al., 2022).
Effects of gibberellin (GA) on expressions of related transcripts
Based on our results, GA was the main hormone significantly affected by N forms, and thus we determined its effects on expressions of the transcripts related with ammonium uptake and assimilation for X. sibiricum. The transcripts (to be measured) were determined based on our RNA sequencing data: expressed the most highly among the transcripts of each related gene family and induced by ammonium. In this study, the transcripts included an AMT member (XsiAMT1.1a), a GLN member (XsiGLN1.1) and a GLT member (XsGLT1b). The seedlings after N starvation (2 d) were treated for 10 h in the nutrient solution including 5 mmol L-1 NH4Cl with and without addition of 1 mg L-1 GA1 and GA4, respectively. Then, the fine roots were separately collected for measuring the relative expression levels of the transcripts using quantitative real-time PCR (three biological replicates). Quantitative real-time PCR was performed as described in reference (Luo et al., 2022). Primers were displayed in Table S1. The differences for these transcripts between GA treatments were tested using one-way ANOVA (SPSS Inc., Chicago, IL, USA).
Effects of nitrogen forms on growth and glutamine synthetase
The seedlings of X. sibiricum and X. strumarium were treated separately with two ratios (nitrate-N: ammonium-N = 1: 9 and 9: 1) and two N levels (50% and 100% N concentrations of Hoagland nutrient) for 45 days. In this experiment, mixed N forms were used as the species, especially X. strumarium, could not grow well for long time in pure ammonium condition based on our preliminary observations. After treatment, recently matured leaves and fine roots were collected separately for each of six seedlings per species (six replicates), quickly frozen in liquid nitrogen, and stored in -80 °C for measuring glutamine synthetase activity, which was determined according to Li (2000). Six seedlings per species per treatment were harvested separately, oven dried at 60 °C for 3 d, and then weighed using an analytical balance. One-way ANOVA was used to test the differences among different N treatments for the same species, and independent sample t-test was used to analyze the differences between the two species under the same N treatment. The analyses were carried out using SPSS (Inc., Chicago, IL, USA).
Results
Overview of ammonium-responsive transcripts
The reads and measured bases ranged from 3.99 × 107 to 6.38 × 107 and from 5.99 × 109 to 9.57 × 109, respectively, and the Q30 values were all above 90% (Table S2), indicating that our mRNA sequencing results are valid. There were 4166 differentially expressed transcripts (DETs) between ammonium and nitrate treatments in the native plant Xanthium sibiricum, which were much more than those in the invasive plant X. strumarium (2907; Figure 1). In addition, much more DETs were induced by ammonium (expressed more highly under ammonium relative to nitrate treatment) in X. sibiricum than in the invader (3050 vs. 1737). In contrast, the proportion of nitrate-induced transcripts in total DETs was much higher in the invader than in X. sibiricum (40% vs. 27%), while the absolute numbers of the nitrate-induced DETs were similar for these two species (1170 vs 1116). GO terms analysis showed that the number of DETs enriched in molecular function was the largest, followed successively by those enriched in biological process and cellular component (Figure S1), suggesting that these two species responded to the conversion of different N forms mainly by changing molecular function. Most of the DETs of the two Xanthium species were enriched in catalytic activity, binding, metabolic process, and cellular process.
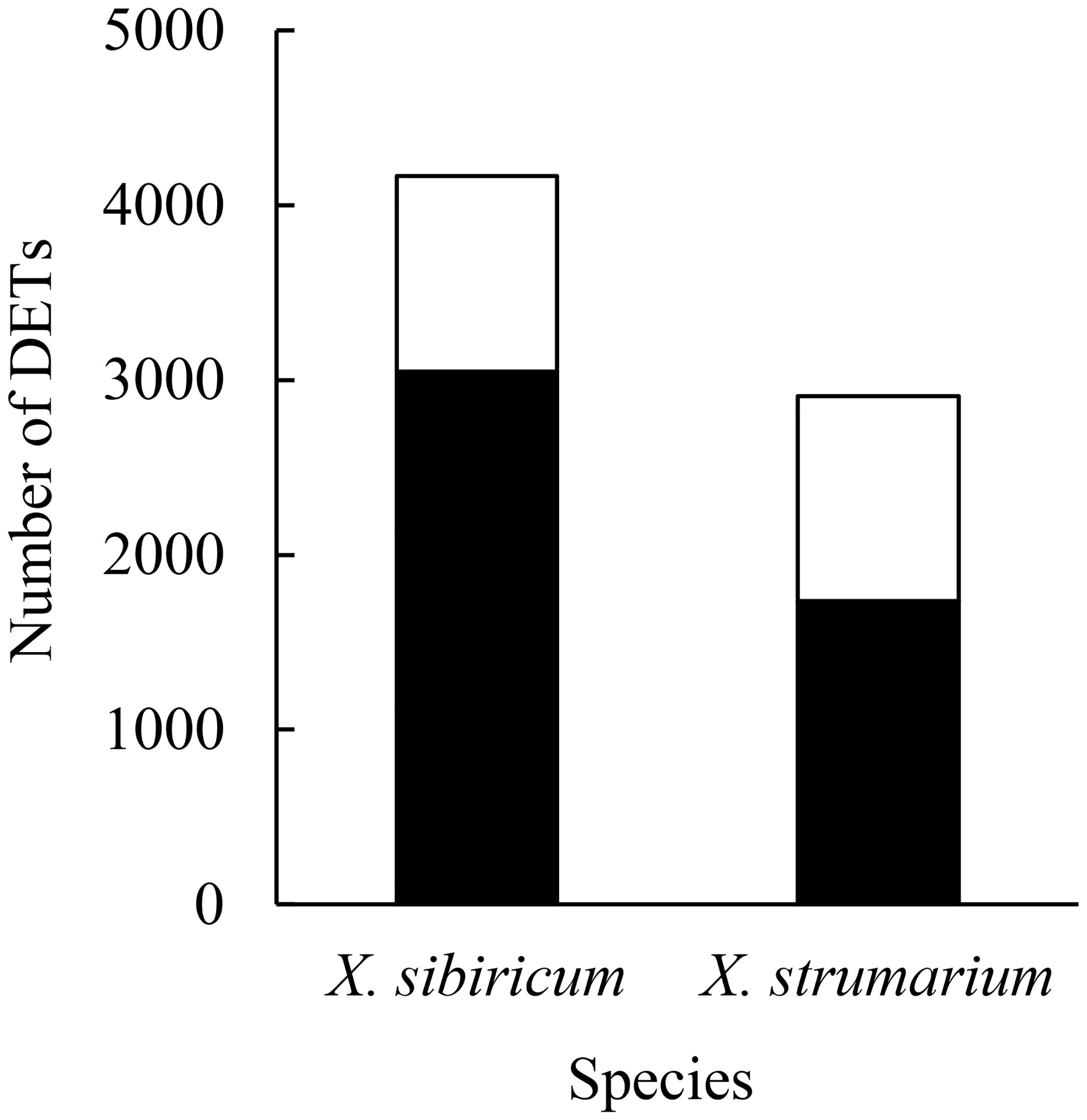
Figure 1 The numbers of differently expressed transcripts (DETs) between ammonium and nitrate treatments in Xanthium sibiricum and X. strumarium, respectively. Closed columns indicate the DETs induced by ammonium, i.e., expressed more highly under amonium relative to nitrate. Open columns indicate the DETs induced by nitrate.
KEGG pathway enrichment analysis of ammonium-responsive transcripts
Among the top 30 pathways in which the DETs were significantly enriched for each of the Xanthium species, there were 18 common pathways for X. sibiricum and X. strumarium, and 12 unique pathways for each species (Figure 2). Among the unique pathways for each species, the numbers of the DETs enriched in biosynthesis of amino acids (followed by that in nitrogen metabolism), and starch and sucrose metabolism (followed by that in inositol phosphate metabolism) were the highest for X. sibiricum and X. strumarium, respectively. The ratio of the enriched DETs to total transcripts (rich factors) in diterpenoid biosynthesis (followed by taurine and hypotaurine metablism) and riboflavin metabolism (followed by glucosinolate biosynthesis) were the highest for X. sibiricum and X. strumarium, respectively. Interestingly, the DETs enriched in diterpenoid biosynthesis were involved in the biosynthesis and metabolism of gibberellin (GA) for X. sibiricum (Figure 3).
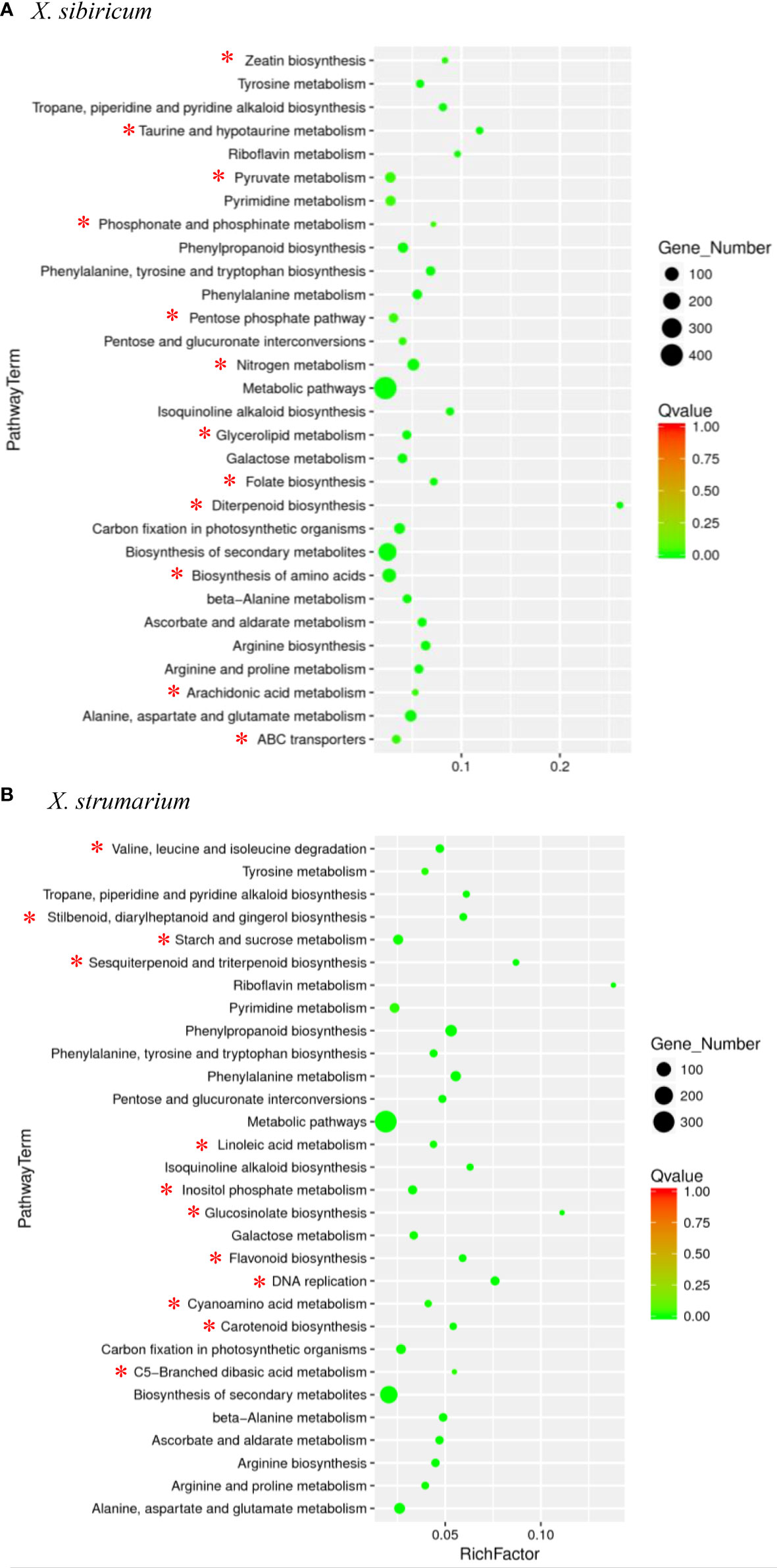
Figure 2 Pathways enrichment analysis for differently expressed transcripts (DETs) in Xanthium sibiricum (A) and X. strumarium (B). Circle size indicates the number of DETs enriched in each pathway. Q value indicates significance of the enrichment, increasing from red to green. Rich factor represents the ratio of the enriched DETs to total transcripts in this pathway. The 12 unique pathways for each species are marked with *, and the 18 common pathways are not.
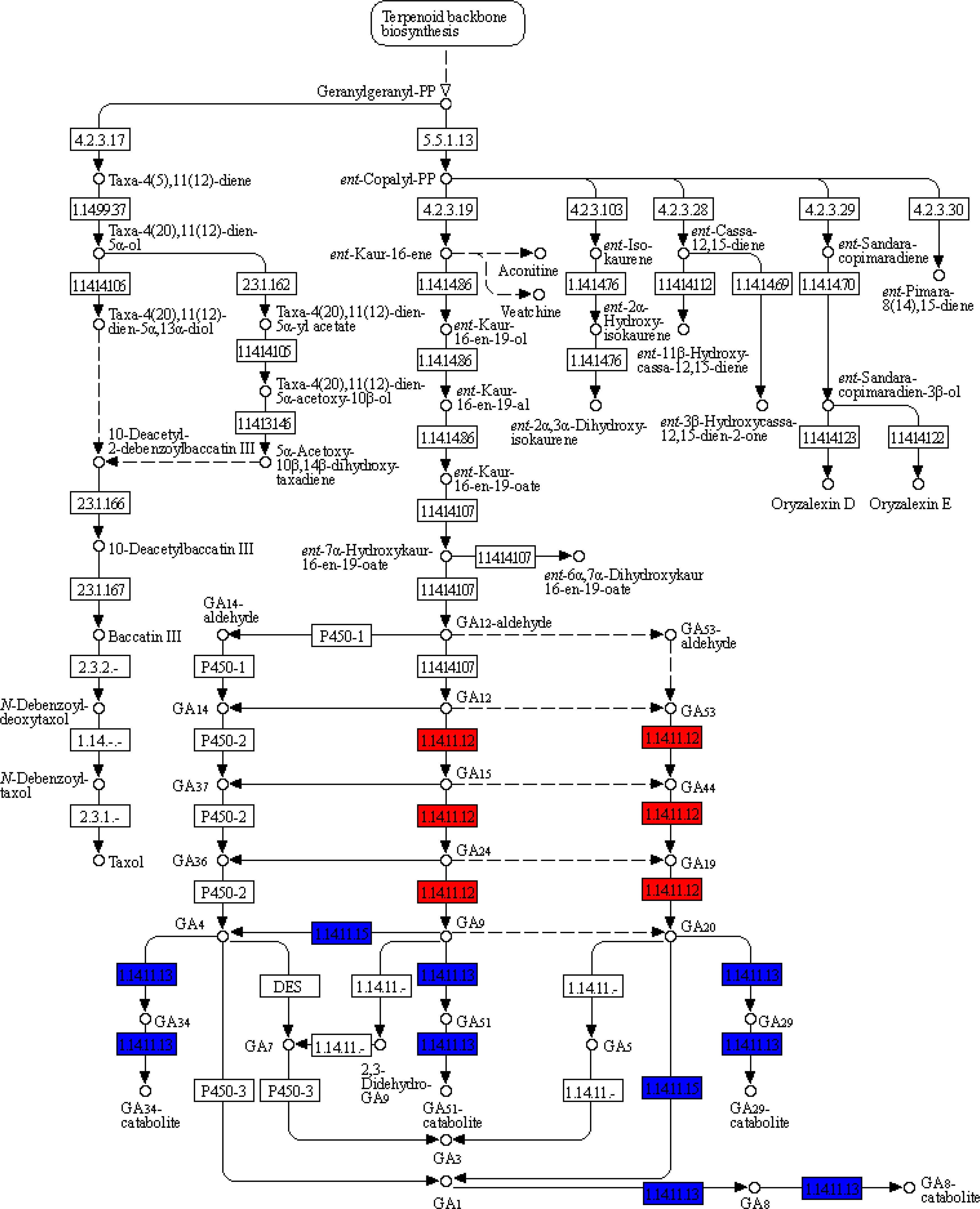
Figure 3 Differently expressed transcripts (DETs) of Xanthium sibiricum involved in GA biosynthesis and metabolism. Blue and red words represent that this kind of DETs were induced by ammonium and nitrate, respectively.
Expression analysis of ammonium-use related transcripts
In X. sibiricum, the transcripts of gibberellin 2beta-dioxygenase (GA2OX) [EC:1.14.11.13] and gibberellin 3beta-dioxygenase (GA3OX) [EC:1.14.11.15] were induced by ammonium, while the transcripts of gibberellin-44 dioxygenase (GA20OX) [EC:1.14.11.12] were induced by nitrate (Figure 3). In total, there were six DETs encoding these three enzymes in X. sibiricum: three for GA2OX, two for GA3OX and one for GA20OX (Figures 4A, S2). All these DETs were induced by ammonium except XsiGA20OX1 which was induced by nitrate. Among the six DETs, XsiGA3OX1a was the most highly expressed (Figure 4A). For X. strumarium, however, only one DET (XstGA2OX1) related with GA metabolism were identified, which was induced by ammonium.

Figure 4 Expression patterns of the differently expressed transcripts (DETs) related with gibberellin synthesis and metabolism (A), ammonium transporter (B), glutamine synthetase (C) and glutamate synthase (D) in Xanthium sibiricum and X. strumarium. For X. strumarium, there were no DETs related with ammonium transporter. Different colors represent different expression levels (increasing from green to red). A and N indicate ammonium and nitrate treatments, respectively. The names of the transcripts were gased on our phylogenetic analyses (Figures S2-5).
Seven ammonium transporter (AMT) transcripts, four glutamine synthetase (GS) transcripts and 12 glutamate synthase (GOGAT) transcripts were differently expressed in X. sibiricum between ammonium and nitrate treatments (Figures 4B–D, S3-5). Except XsiAMT2.2 and XsiGLT2 (induced by nitrate), all remaining 21 DETs of X. sibiricum were induced by ammonium. XsiAMT1.1a, XsiGLN1.1 and XsiGLT1b were the most highly expressed among these differently expressed AMT, GS and GOGAT transcripts, respectively. For X. strumarium, however, no ammonium uptake related transcript and only 10 ammonium assimilation related transcripts (2 GS and 8 GOGAT) were differently expressed in ammonium and nitrate. All DETs of X. strumarium were induced by ammonium except XstGLT2a, which was induced by nitrate. The sequences of XstGLT1a and XstGLT1b were similar with that of XsiGLT1b (Figure S5), but their expression levels were lower than that of XsiGLT1b (Figure 4D).
Our quantitative real-time PCR also showed that XsiGA3OX1a, XsiAMT1.1a, XsiGLN1.1 and XsiGLT1b were expressed much more highly in ammonium relative to nitrate treatment (Figure 5). Moreover, XsiAMT1.1a, XsiGLN1.1 and XsiGLT1b were induced by GA1 and GA4 in ammonium, and the effects of GA4 were stronger than those of GA1 (Figure 6).
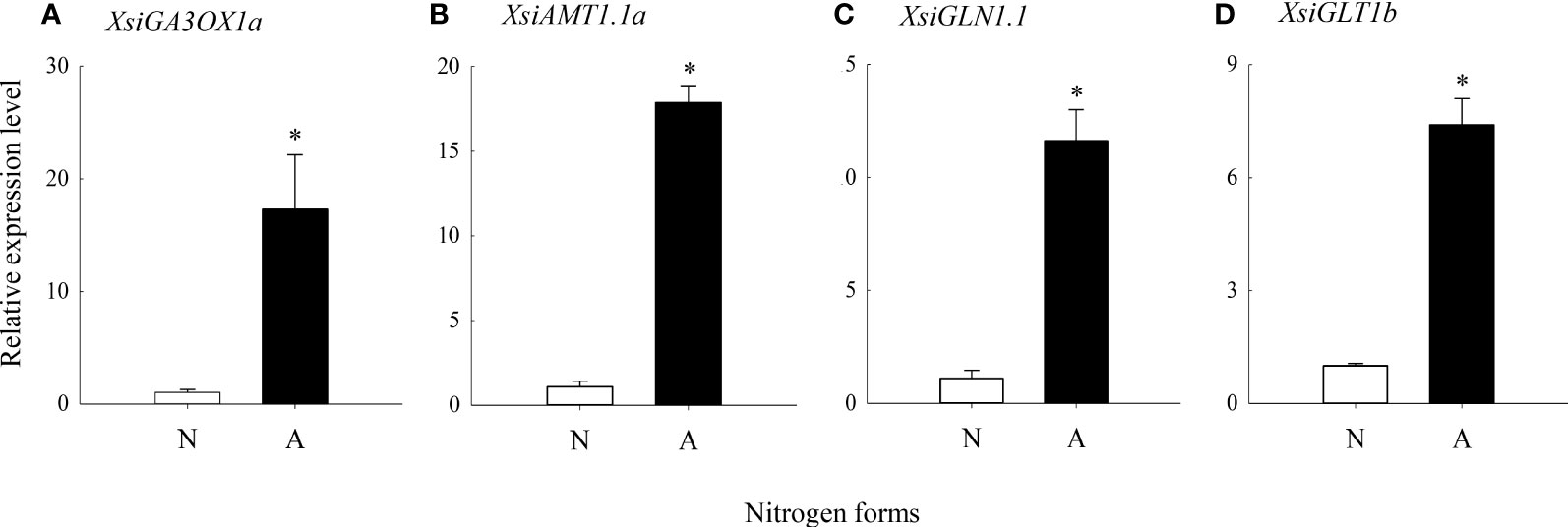
Figure 5 Differences in expression levels of XsiGA3OX1a (A), XsiAMT1.1a (B), XsiGLN1.1 (C) and XsiGLT1b (D) between nitrate (N; open bars) and ammonium (A; closed bars) treatments based on our quantitative real-time PCR. XsiActin1 was used as internal controls. Means + SD (n = 3). Asterisk indicates significant difference between ammonium and nitrate treatments (P< 0.05; independent sample t-test). The names of the transcripts were based on our phylogenetic analyses (Figures S2-5).
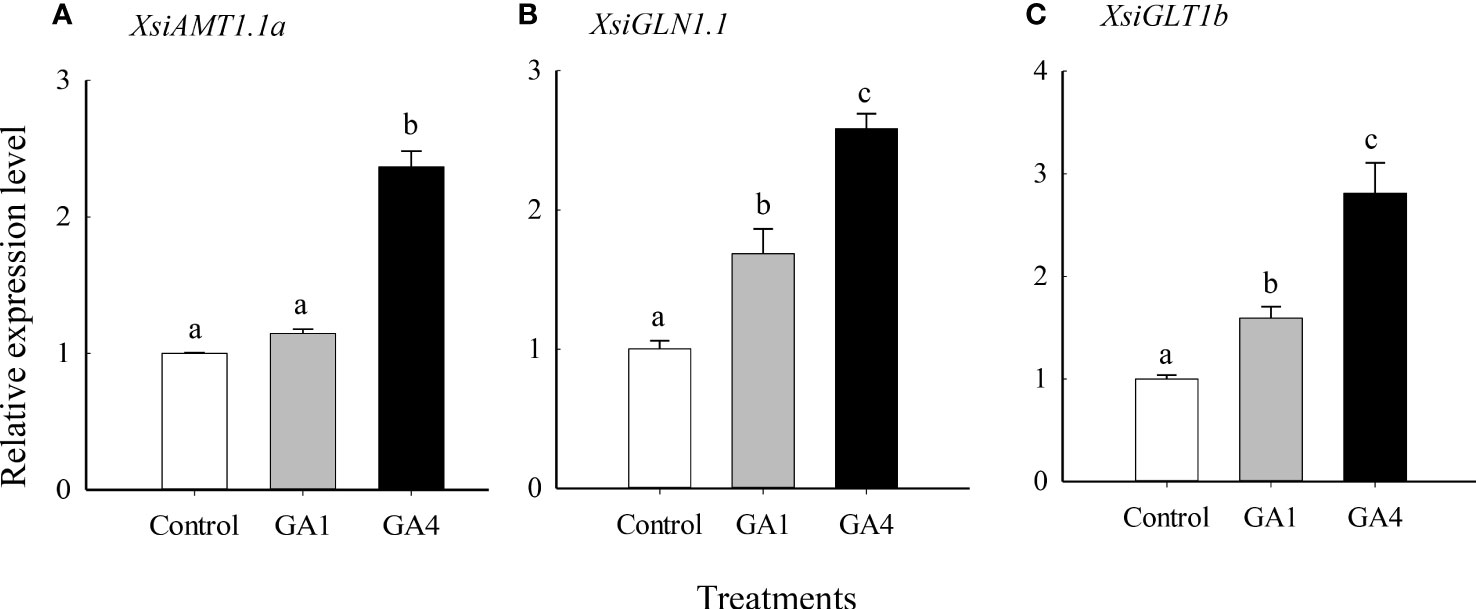
Figure 6 Effects of gibberellin (GA1 and GA4) on expression levels of XsiAMT1.1a (A), XsiGLN1.1 (B) and XsiGLT1b (C). White bars, grown in nutrient solution including 5 mmol L-1 ammonium and without GA1 and GA4 addition; gray bars, grown in nutrient solution including 5 mmol L-1 ammonium and with 1 mmol L-1 GA1 addition; black bars, grown in nutrient solution including 5 mmol L-1 ammonium and with 1 mmol L-1 GA4 addition. XsiActin1 was used as internal controls. Means + SD (n = 3). Different letters indicate significant difference between treatments (P< 0.05; one-way ANOVA). The names of the transcripts were based on our phylogenetic analyses (Figures S3-5).
Growth and glutamine synthetase activity
Under the low ratio (1/9), total biomass (not significant in 0.5 N), leaf and root GS activities were significantly higher for X. sibiricum than for X. strumarium, regardless of N levels (Figures 7, 8). Under the high ratio (9/1), however, the results were reversed. With decreasing ratio, X. sibiricum increased total biomass, leaf and root GS activities significantly, while X. strumarium decreased. Under the same ratio, total biomass (except X. strumarium under 1/9 of ratio), GS activities in leaves (except X. strumarium under 1/9 of ratio) and roots (except X. sibiricum under 9/1 of ratio) increased significantly with increasing N level.
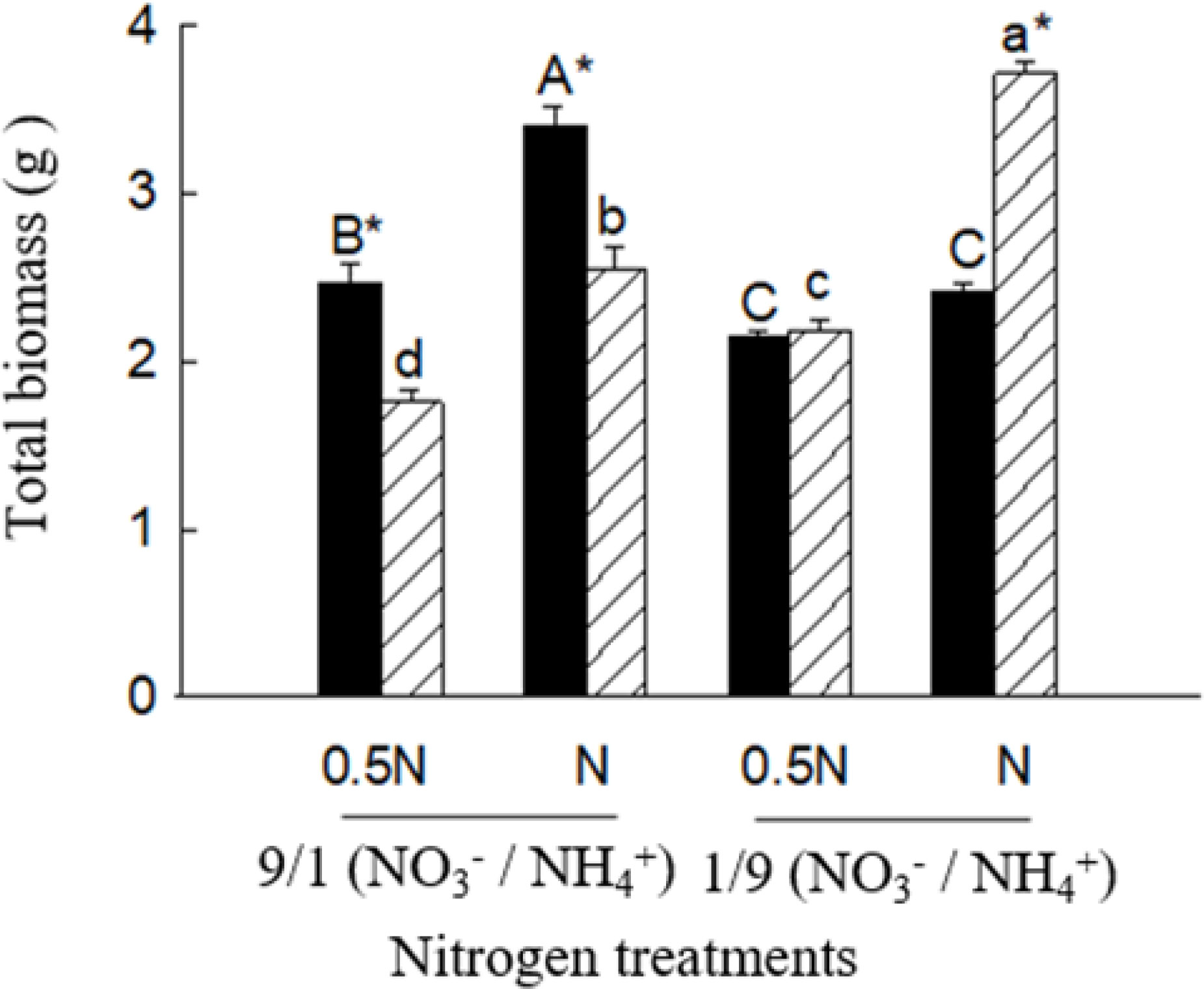
Figure 7 Total biomass of Xanthium sibiricum (striped bars) and X. strumarium (closed bars) in different nitrogen treatments. 0.5 N and 1 N indicate 50% and 100% nitrogen concentration of the Hoagland nutrient solution, respectively. Means + SE (n = 6). Different upper- and lowercase letters indicate significant dfferences among nitrogen treatments for X. strumarium and X. sibiricum, respectively (P< 0.05; one-way ANOVAs); * indicates significant difference between the invasive and native species in the same treatment (P< 0.05; independent sample t-test).
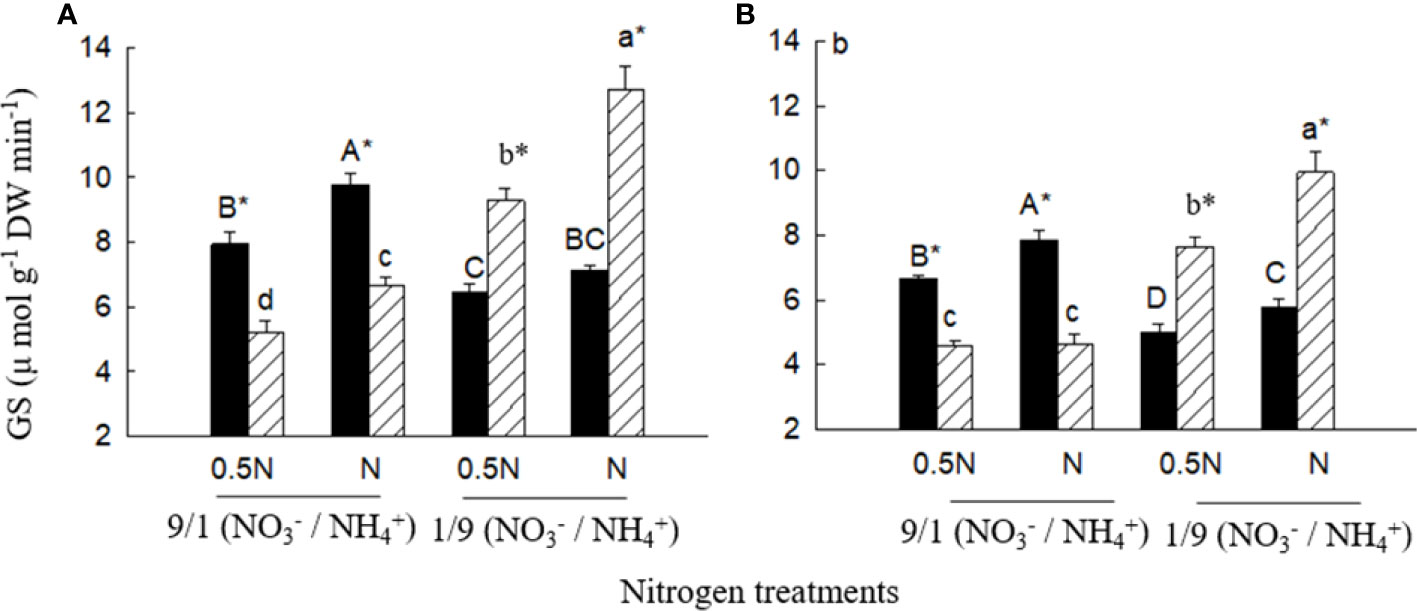
Figure 8 Glutamine synthetase (GS) activities in leaves (A) and roots (B) of Xanthium sibiricum (striped bars) and X. strumarium (closed bars) in different nitrogen treatments. 0.5 N and 1 N indicate 50% and 100% nitrogen concentration of the Hoagland nutrient solution, respectively. Means + SE (n = 6). Different upper- and lowercase letters indicate significant dfferences among nitrogen treatments for X. strumarium and X. sibiricum, respectively (P< 0.05; one-way ANOVAs); * indicates significant difference between the invasive and native species in the same treatment (P< 0.05; independent sample t-test).
Discussion
Our results showed that many transcripts related with GA biosynthesis and metabolism, N uptake and assimilation were induced by ammonium in the native plant Xanthium sibiricum, and many of the transcripts related with N uptake and assimilation were up regulated by GA (Figures 4–6). These results indicate that in response to ammonium X. sibiricum may increase GA biosynthesis firstly, then increase its abilities of N uptake and assimilation, and thus growth (Figure 8). For the invasive plant X. strumarium, however, only nine N assimilation related transcripts were induced by ammonium, and no AMT transcripts were induced by ammonium (Figures 4B–D). In contrast, the relative quantity of nitrate-induced transcripts was much more in the invader than in X. sibiricum (Figure 1). All DETs enriched in DNA replication pathway are induced by nitrate for the invader (Figure S6), which indicates that nitrate may increase DNA replication and thus growth in the invader, providing molecular evidence for its strong ability to use nitrate. These results explain the reasons that X. sibiricum responded more strongly to ammonium, while the invader more strongly to nitrate (Figure 7; Luo et al., 2022). Exotic plants may be more like to invade the habitats where the dominant soil N form is what they prefer (Feng, 2020; Sun et al., 2021). Flaveria bidentis and Ipomoea cairica prefer ammonium and nitrate, respectively, and grow better in the habitats with related N forms as main soil N sources (Huangfu et al., 2016; Chen and Chen, 2019). Our study is the first one that determines the molecular mechanisms for co-occurring invasive and native plants to use different N forms, contributing to understanding the selection of invasion habitats of alien plants.
Nitrogen uptake and assimilation
As our hypothesis, more transcripts related with ammonium uptake and assimilation (AMT, GS and GOGAT) were induced by ammonium in X. sibiricum than in X. strumarium (21 vs. 9; Figures 4B–D). Among the six ammonium-induced AMT transcripts, XsiAMT1.1a was the most highly expressed (Figure 4B). XsiAMT1.1a, XsiAMT1.1b, XsiAMT1.1c, XsiAMT1.2a and XsiAMT1.2b are clustered together with AtAMT 1.1, 1.2, 1.3 and 1.5 in evolutionary tree (Figure S3), which all have ammonium uptake activities (Li et al., 2017). These transcripts (especially XsiAMT1.1a and XsiAMT1.2a) may be important for X. sibiricum’s higher uptake rate of ammonium relative to nitrate N, and thus faster growth in ammonium (Luo et al., 2022). Rice prefers ammonium relative to nitrate, which may be associated with its origin habitat (flooded wetland) where ammonium is the dominant N form (Xu et al., 2012), and many AMT members such as OsAMT1.1 and OsAMT1.2 were highly expressed and induced by ammonium (Sonoda et al., 2003; Li et al., 2016). Overexpression of OsAMT1.1 improves ammonium uptake in rice, while the reverse is true for disruption of OsAMT1.1 (Kumar et al., 2006; Li et al., 2016). However, XsiAMT2.2 may not be important for X. sibiricum to absorb ammonium, as it was induced by nitrate (Figure 4B), and clustered together with AtAMT2.1 in the evolutionary tree (Figure S3), which has not been found to have ammonium uptake activity (Li et al., 2017).
For X. sibiricum, XsiGLN1.1, XsiGLN1.3 and XsiGLN1.4 may all contribute to its fast ammonium assimilation and thus fast growth in ammonium, as they were highly expressed (especially XsiGLN1.1), induced by ammonium (Figure 4C), and clustered together with AtGLN1.1 and AtGLN1.2 in the evolutionary tree (Figure S4), which have been proved to have ammonium assimilation activity (Li et al., 2017). Coding genes of glutamine synthase were identified in many other species such as barley, rice, maize, Arabidopsis thaliana and so on (Martin et al., 2006; Brauer et al., 2011; Goodall et al., 2013; Konishi et al., 2014). AtGLN1.2, AtGLT1 and HvGS1.3 were highly expressed and induced by ammonium (Goodall et al., 2013; Konishi et al., 2014). Overexpression of OsGS1 improves spikelet yield and N-use efficiency in rice (Brauer et al., 2011). Similarly, overexpression of ZmGS1 increases maize grain yield by 45% (Martin et al., 2006). Consistent with the ammonium-induced expressions of XsiGLN1.1, XsiGLN1.3 and XsiGLN1.4, leaf and root GS activities increased for X. sibiricum with increasing environmental ammonium to nitrate ratio, and were always higher than those of X. strumarium under high ammonium to nitrate ratio (Figure 8).
Among the eleven ammonium-induced GOGAT transcripts in X. sibiricum, XsiGLT1b (followed by XsiGLU1 and XsiGLT1h) were the most highly expressed (Figure 4D), which may contribute to ammonium assimilation, and thus to growth of X. sibiricum. Chichkova et al. (2001) found that high expression levels of GOGAT genes improve N assimilation in tobacco. Overexpression of OsGOGAT improves grain filling in rice (Yamaya et al., 2002). In contrast, knockout of OsNADH-GOGAT1 gene reduces total amino acid content and yield in rice (Tamura et al., 2010).
For X. sibiricum, much more DETs were induced by ammonium than by nitrate (Figure 1). Yang et al. (2018) also found that ammonium-induced DETs were 1.5 times higher than nitrate-induced DETs in tea tree, which also prefers ammonium compared with nitrate. In addition, the number of the DETs enriched in biosynthesis of amino acids pathways was much greater than that enriched in any other 11 unique pathways for X. sibiricum, and most of the DETs enriched in this pathway were induced by ammonium (Figures 2, S7). The biosynthesis of histidine, phenylalanine, glutamate, glutamine, proline, and arginine may be enhanced by ammonium (Figure S7). For X. strumarium, however, DETs were not significantly enriched in this pathway (Figure 2). It has been reported that the concentrations of glutamate, glutamine, and asparagine in rice and Myriophyllum aquaticum increase with the increase of ammonium treatment concentration and time (Yang et al., 2017; Zhang et al., 2021). However, N storage in glutamate (C/N = 5/1), glutamine (C/N = 2.5/1) and asparagine (C/N = 2/1) may cause carbohydrate deficiency and inhibit plant growth (Bittsanszky et al., 2015). In order to reduce carbon consumption, N can also be stored in arginine (C/N = 1.5/1), which had the lowest C/N ratio among all amino acids. Thus, the fast amino acid synthesis and conversion may enhance the ability to use ammonium for X. sibiricum.
Our results also indicate that X. sibiricum and X. strumarium may share some similar strategies to acclimate different N forms. Sixty percent of the top 30 pathways with DETs significantly enriched were common in these two species, including ascorbate and aldarate metabolism pathways (Figure 2). In this pathway, ascorbate peroxidase and ascorbate are involved in reactive oxygen species (ROS) quenching, and therefore decrease oxidative damage to plants in ammonium. It has been found that plants can accumulate ammonium in different tissues, which are seriously harmful to plants (Apel and Hirt, 2004).
Gibberellin and its functions
Our research found five ammonium-induced GA biosynthesis and metabolism related DETs in X. sibiricum. Among them, XsiGA3OX1a was the most highly expressed, followed by XsiGA2OX2 and XsiGA2OX1b (Figure 4A). It was reported that GA3OX was participated in biosynthesis of GA1 and GA4, which are two main active forms of GA (Yamaguchi et al., 1998; Ueguchi-Tanaka et al., 2007). The induction of XsiGA3OX1a may enhance the contents of GA1 and GA4 in ammonium for X. sibiricum. Supplement of exogenous GA1 or GA4 up regulated expressions of all ammonium-utilization related transcripts tested in X. sibiricum, including XsiAMT1.1a, XsiGLN1.1 and XsiGLT1b (Figure 6), and the effects of GA4 were higher than those of GA1, indicating that GA4 play more important roles in ammonium-utilization. GA4, as well as other bioactive forms of GA, play critical roles in many developmental processes, including seed germination, root elongation, flowering, and fruit development (Hedden and Sponsel, 2015; Binenbaum et al., 2018). Dai et al. (2016) also found that GA influenced invasion success of the exotic plant Wedelia trilobata. However, few studies explore the roles of GA in utilization of ammonium, and the regulations of N forms on expression of GA biosynthesis and metabolism related transcripts is still unknown. Recently, it was reported that growth-regulating factor 4 (GRF4), which is involved in GA signal pathway, can improve ammonium uptake and rice growth (Li et al., 2018). ChIP–PCR showed that GRF4 can bind to the promoter regions of OsAMT1.1 and OsGS1.2 (Li et al., 2018). Our results provided direct evidence that GA, especially GA4, is involved in regulations of ammonium uptake and assimilation.
In addition, some DETs were also significantly enriched in the pathway of zeatin biosynthesis for X. sibiricum but not for X. strumarium (Figure 2). tRNA dimethylallyl transferase [EC:2.5.1.75] and cis-zeatin O-glucosyl transferase [EC:2.4.1.215] were induced by ammonium (Figure S8). These results indicate that different forms of zeatin may also be involved in the stronger ability to use ammonium for X. sibiricum relative to X.strumarium.
Conclusion
Xanthium sibiricum increases expressions of many transcripts related with ammonium uptake and assimilation in response to ammonium, improving ammonium uptake, assimilation and thus growth. XsiAMT1.1a, XsiGLN1.1, XsiGLT1b, XsiGA3OX1a, XsiGA3OX1b, XsiGA2OX2 as well as others may be important for the strong ability to use ammonium in X. sibiricum. Gibberellin, especially GA4, is involved in the regulation of ammonium utilization. Ammonium regulates expressions of some transcripts related with GA biosynthesis and metabolism, which in turn induces expressions of transcripts related with ammonium uptake and assimilation. Zeatins may also be involved in the regulation of ammonium utilization in X. sibiricum. Our study elucidates the molecular mechanisms with which the Xanthium species acclimate to different soil nitrogen forms, contributing to understanding the invasion mechanisms of X. strumarium and its invasion habitat selection.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA751368 https://www.ncbi.nlm.nih.gov/, PRJNA784152.
Author contributions
CZ, Y-LF: Conceived the ideas and designed methodology. CZ, J-JL, J-BZ, ZZ, S-TW: Conducted the experiment. CZ, J-JL, X-JZ, T-SF: Analyzed the data. CZ, Y-LF: Wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (31971557, 31670545 and 32001235).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1035137/full#supplementary-material
References
Apel, K., Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Binenbaum, J., Weinstain, R., Shani, E. (2018). Gibberellin localization and transport in plants. Trends Plant Sci. 23, 410–421. doi: 10.1016/j.tplants.2018.02.005
Bittsanszky, A., Pilinszky, K., Gyulai, G., Komives, T. (2015). Overcoming ammonium toxicity. Plant Sci. 231, 184–190. doi: 10.1016/j.plantsci.2014.12.005
Brauer, E. K., Rochon, A., Bi, Y.-M., Bozzo, G. G., Rothstein, S. J., Shelp, B. J. (2011). Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase1. Physiol. Plant 141, 361–372. doi: 10.1111/j.1399-3054.2011.01443.x
Brenner, W. G., Romanov, G. A., Kollmer, I., Burkle, L., Schmulling, T. (2005). Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 44, 314–333. doi: 10.1111/j.1365-313X.2005.02530.x
Chen, W.-B., Chen, B.-M. (2019). Considering the preferences for nitrogen forms by invasive plants: a case study from a hydroponic culture experiment. Weed Res. 59, 49–57. doi: 10.1111/wre.12344
Chichkova, S., Arellano, J., Vance, C. P., Hernandez, G. (2001). Transgenic tobacco plants that overexpress alfalfa NADH-glutamate synthase have higher carbon and nitrogen content. J. Exp. Bot. 52, 2079–2087. doi: 10.1093/jexbot/52.364.2079
Coschigano, K. T., Melo-Oliveira, R., Lim, J., Coruzzi, G. M. (1998). Arabidopsis gls mutants and distinct Fd-GOGAT genes: implications for photorespiration and primary nitrogen assimilation. Plant Cell 10, 741–752. doi: 10.1105/tpc.10.5.741
Costa, C. P., Duennes, M. A., Fisher, K., Der, J. P., Watrous, K. M., Okamoto, N., et al. (2020). Transcriptome analysis reveals nutrition- and age-related patterns of gene expression in the fat body of pre-overwintering bumble bee queens. Mol. Ecol. 29, 720–737. doi: 10.1111/mec.15361
Dai, Z. C., Fu, W., Qi, S. S., Zhai, D. L., Chen, S. C., Wan, L. Y., et al. (2016). Different responses of an invasive clonal plant Wedelia trilobata and its native congener to gibberellin: implications for biological invasion. J. Chem. Ecol. 42, 85–94. doi: 10.1007/s10886-016-0670-6
Feng, Y.-L., Lei, Y.-B., Wang, R.-F., Callaway, R. M., Valiente-Banuet, A., Inderjit, et al. (2009). Evolutionary tradeoffs for nitrogen allocation to photosynthesis versus cell walls in an invasive plant. Proc. Natl. Acad. Sci. U S A. 106, 1853–1856. doi: 10.1073/pnas.0808434106
Gazzarrini, S., Lejay, L., Gojon, A., Ninnemann, O., Frommer, W. B., von Wiren, N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into arabidopsis roots. Plant Cell 11, 937–948. doi: 10.1105/tpc.11.5.937
Goodall, A. J., Kumar, P., Tobin, A. K. (2013). Identification and expression analyses of cytosolic glutamine synthetase genes in barley (Hordeum vulgare l.). Plant Cell Physiol. 54, 492–505. doi: 10.1093/pcp/pct006
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hedden, P., Sponsel, V. (2015). A century of gibberellin research. J. Plant Growth Regul. 34, 740–760. doi: 10.1007/s00344-015-9546-1
Huangfu, C., Li, H., Chen, X., Liu, H., Wang, H., Yang, D. (2016). Response of an invasive plant, Flaveria bidentis, to nitrogen addition: a test of form-preference uptake. Biol. Invasions .18, 3365–3380. doi: 10.1007/s10530-016-1231-1
Ishiyama, K., Inoue, E., Watanabe-Takahashi, A., Obara, M., Yamaya, T., Takahashi, H. (2004). Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. J. Biol. Chem. 279, 16598–16605. doi: 10.1074/jbc.M313710200
Kaiser, B. N., Rawat, S. R., Siddiqi, M. Y., Masle, J., Glass, A. D. M. (2002). Functional analysis of an Arabidopsis T-DNA "knockout" of the high-affinity NH4+ transporter AtAMT1;1. Plant Physiol. 130, 1263–1275. doi: 10.1104/pp.102.010843
Kiba, T., Naitou, T., Koizumi, N., Yamashino, T., Sakakibara, H., Mizuno, T. (2005). Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the his->Asp phosphorelay circuitry. Plant Cell Physiol. 46, 339–355. doi: 10.1093/pcp/pci033
Konishi, N., Ishiyama, K., Matsuoka, K., Maru, I., Hayakawa, T., Yamaya, T., et al. (2014). NADH-dependent glutamate synthase plays a crucial role in assimilating ammonium in the Arabidopsis root. Physiol. Plant 152, 138–151. doi: 10.1111/ppl.12177
Kumar, A., Kaiser, B. N., Siddiqi, M. Y., Glass, A. D. M. (2006). Functional characterisation of OsAMT1.1 overexpression lines of rice, Oryza sativa. Funct. Plant Biol. 33, 339–346. doi: 10.1071/FP05268
Lancien, M., Martin, M., Hsieh, M.-H., Leustek, T., Goodman, H., Coruzzi, G. M. (2002). Arabidopsis glt1-t mutant defines a role for NADH-GOGAT in the non-photorespiratory ammonium assimilatory pathway. Plant J. 29, 347–358. doi: 10.1046/j.1365-313x.2002.01218.x
Li, H.-S. (2000). Principles and techniques of plant physiological and biochemical experiment (Beijing, China:Higher Education Press).
Li, B., Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li, H., Hu, B., Chu, C. (2017). Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J. Exp. Bot. 68, 2477–2488. doi: 10.1093/jxb/erx101
Li, C., Tang, Z., Wei, J., Qu, H., Xie, Y., Xu, G. (2016). The OsAMT1.1 gene functions in ammonium uptake and ammonium-potassium homeostasis over low and high ammonium concentration ranges. J. Genet. Genomics 43, 639–649. doi: 10.1016/j.jgg.2016.11.001
Li, S., Tian, Y., Wu, K., Ye, Y., Yu, J., Zhang, J., et al. (2018). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560, 595–600. doi: 10.1038/s41586-018-0415-5
Li, S. X., Wang, Z. H., Stewart, B. A. (2013). Responses of crop plants to ammonium and nitrate n. Adv. Agron. 118, 205–397. doi: 10.1016/B978-0-12-405942-9.00005-0
Li, G., Zhang, L., Wang, M., Di, D., Kronzucker, H. J., Shi, W. (2019). The Arabidopsis AMOT1/EIN3 gene plays an important role in the amelioration of ammonium toxicity. J. Exp. Bot. 70, 1375–1388. doi: 10.1093/jxb/ery457
Loque, D., Yuan, L., Kojima, S., Gojon, A., Wirth, J., Gazzarrini, S., et al. (2006). Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 48, 522–534. doi: 10.1111/j.1365-313X.2006.02887.x
Lothier, J., Gaufichon, L., Sormani, R., Lemaitre, T., Azzopardi, M., Morin, H., et al. (2011). The cytosolic glutamine synthetase GLN1;2 plays a role in the control of plant growth and ammonium homeostasis in Arabidopsis rosettes when nitrate supply is not limiting. J. Exp. Bot. 62, 1375–1390. doi: 10.1093/jxb/erq299
Luo, J.-J., Gao, Y.-M., Feng, W.-W., Liu, M.-C., Qu, B., Zhang, C., et al. (2022). Stronger ability to absorb nitrate and associated transporters in the invasive plant Xanthium strumarium compared with its native congener. Environ. Exp. Bot. 198, 104851. doi: 10.1016/j.envexpbot.2022.104851
Martin, A., Lee, J., Kichey, T., Gerentes, D., Zivy, M., Tatout, C., et al. (2006). Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18, 3252–3274. doi: 10.1105/tpc.106.042689
O'Brien, J. A., Vega, A., Bouguyon, E., Krouk, G., Gojon, A., Coruzzi, G., et al. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9, 837–856. doi: 10.1016/j.molp.2016.05.004
Sakakibara, H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. doi: 10.1146/annurev.arplant.57.032905.105231
Sakakibara, H., Takei, K., Hirose, N. (2006). Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 11, 440–448. doi: 10.1016/j.tplants.2006.07.004
Sonoda, Y., Ikeda, A., Saiki, S., von Wiren, N., Yamaya, T., Yamaguchi, J. (2003). Distinct expression and function of three ammonium transporter genes (OsAMT1;1-1;3) in rice. Plant Cell Physiol. 44, 726–734. doi: 10.1093/pcp/pcg083
Sun, S.-M., Chen, J.-X., Feng, W.-W., Zhang, C., Huang, K., Guan, M., et al. (2021). Plant strategies for nitrogen acquisition and their effects on exotic plant invasions. Biodiversity Sci. 29, 72–80. doi: 10.17520/BIODS.2020072
Takei, K., Ueda, N., Aoki, K., Kuromori, T., Hirayama, T., Shinozaki, K., et al. (2004). AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 45, 1053–1062. doi: 10.1093/pcp/pch119
Tamura, W., Hidaka, Y., Tabuchi, M., Kojima, S., Hayakawa, T., Sato, T., et al. (2010). Reverse genetics approach to characterize a function of NADH-glutamate synthase1 in rice plants. Amino Acids 39, 1003–1012. doi: 10.1007/s00726-010-0531-5
Thomas, R. Q., Brookshire, E. N., Gerber, S. (2015). Nitrogen limitation on land: how can it occur in earth system models? Glob Chang Biol. 21, 1777–1793. doi: 10.1111/gcb.12813
Ueguchi-Tanaka, M., Nakajima, M., Motoyuki, A., Matsuoka, M. (2007). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58, 183–198. doi: 10.1146/annurev.arplant.58.032806.103830
Wang, R., Tischner, R., Gutierrez, R. A., Hoffman, M., Xing, X., Chen, M., et al. (2004). Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136, 2512–2522. doi: 10.1104/pp.104.044610
Xu, G., Fan, X., Miller, A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi: 10.1146/annurev-arplant-042811-105532
Yamaguchi, S., Smith, M. W., Brown, R. G., Kamiya, Y., Sun, T. (1998). Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10, 2115–2126. doi: 10.1105/tpc.10.12.2115
Yamaya, T., Obara, M., Nakajima, H., Sasaki, S., Hayakawa, T., Sato, T. (2002). Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. J. Exp. Bot. 53, 917–925. doi: 10.1093/jexbot/53.370.917
Yang, H.-C., Kan, C.-C., Hung, T.-H., Hsieh, P.-H., Wang, S.-Y., Hsieh, W.-Y., et al. (2017). Identification of early ammonium nitrate-responsive genes in rice roots. Sci. Rep. 7, 16885. doi: 10.1038/s41598-017-17173-9
Yang, Y., Wang, F., Wan, Q., Ruan, J. (2018). Transcriptome analysis using RNA-seq revealed the effects of nitrogen form on major secondary metabolite biosynthesis in tea (Camellia sinensis) plants. Acta Physiol. Plant 40, 127. doi: 10.1007/s11738-018-2701-0
Yuan, L., Loque, D., Kojima, S., Rauch, S., Ishiyama, K., Inoue, E., et al. (2007). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19, 2636–2652. doi: 10.1105/tpc.107.052134
Yu, G. R., Jia, Y. L., He, N. P., Zhu, J. X., Chen, Z., Wang, Q. F. (2019). Stabilization of atmospheric nitrogen deposition in China over the past decade. Nat. Geosci. 12, 424–429. doi: 10.1038/s41561-019-0352-4
Zhang, J., Cai, Z., Mueller, C. (2018). Terrestrial n cycling associated with climate and plant-specific n preferences: a review. Eur. J. Soil Sci. 69, 488–501. doi: 10.1111/ejss.12533
Keywords: ammonium, assimilation, gibberellin, transcriptome analysis, uptake
Citation: Zhang C, Luo J-J, Zuo J-B, Zhang Z, Wang S-T, Zhang X-J, Fu T-S and Feng Y-L (2022) Transcripts related with ammonium use and effects of gibberellin on expressions of the transcripts responding to ammonium in two invasive and native Xanthium species. Front. Plant Sci. 13:1035137. doi: 10.3389/fpls.2022.1035137
Received: 02 September 2022; Accepted: 11 October 2022;
Published: 26 October 2022.
Edited by:
Mirjana Ljubojević, University of Novi Sad, SerbiaReviewed by:
Velimir Mladenov, University of Novi Sad, SerbiaAdalbert Balog, Sapientia Hungarian University of Transylvania, Romania
Copyright © 2022 Zhang, Luo, Zuo, Zhang, Wang, Zhang, Fu and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Long Feng, ZnlsQHN5YXUuZWR1LmNu
†These authors have contributed equally to this work
 Chang Zhang
Chang Zhang Jia-Jun Luo†
Jia-Jun Luo† Zheng Zhang
Zheng Zhang Xiao-Jia Zhang
Xiao-Jia Zhang Yu-Long Feng
Yu-Long Feng