
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 31 October 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1032838
This article is part of the Research TopicPlant Genomics and Pathogenomics: From Technology to Application in Improving Crop Disease ResistanceView all 6 articles
In plants, a family of terpene synthases (TPSs) is responsible for the biosynthesis of terpenes and contributes to species-specific diversity of volatile organic compounds, which play essential roles in fitness of plants. However, little is known about the TPS gene family in peach and/or nectarine (Prunus persica L.). In this study, we identified 40 PpTPS genes in peach genome v2.0. Although these PpTPSs could be clustered into five classes, they distribute in several gene clusters of three chromosomes, share conserved exon-intron organizations, and code similar protein motifs. Thirty-five PpTPSs, especially PpTPS2, PpTPS23, PpTPS17, PpTPS18, and PpTPS19, altered their transcript levels after inoculation with Botryosphaeria dothidea, a cause of peach gummosis, compared to the mock treatments, which might further affect the contents of 133 terpenoids at 48 hours and/or 84 hours post inoculations in the current-year shoots of ‘Huyou018’, a highly susceptible nectarine cultivar. Moreover, about fifteen PpTPSs, such as PpTPS1, PpTPS2, PpTPS3, and PpTPS5, showed distinct expression patterns during fruit development and ripening in two peach cultivars, yellow-fleshed ‘Jinchun’ and white-fleshed ‘Hikawa Hakuho’. Among them, the transcription level of chloroplast-localized PpTPS3 was obviously related to the content of linalool in fruit pulps. In addition, elevated concentrations (0.1 g/L to 1.0 g/L) of linalool showed antifungal activities in PDA medium. These results improve our understanding of peach PpTPS genes and their potential roles in defense responses against pathogens.
Volatile organic compounds (VOCs) are essential for the interaction of plants with pollinators, seed-spreaders, herbivores, and microorganisms (Dudareva et al., 2013; Bouwmeester et al., 2019). Among the four classes of plant VOCs (terpenoids, phenylpropanoids/benzenoids, fatty acid derivatives, and amino acid derivatives), terpenoids constitute the largest and most diverse class of secondary metabolites with many volatile constituents (Dudareva et al., 2006; Dudareva et al., 2013). Terpenoids are derived from two common C5-isoprene building units, isopentenyl diphosphate (IPP) and its allylic isomer, dimethylallyl diphosphate (DMAPP) (McGarvey and Croteau, 1995). Both IPP and DMAPP are substrates for short-chain prenyltransferases, which produce prenyl diphosphate precursors, farnesyl diphosphate, geranyl diphosphate, and geranylgeranyl diphosphate (Vranova et al., 2013). A family of terpene synthases/cyclases (TPSs) is responsible for the synthesis of volatile terpenoids from prenyl diphosphate precursors in cytosol and/or plastids (Chen et al., 2011). Although two compartmentally separated pathways, mevalonic acid (MVA) and methylerythritol phosphate (MEP), are responsible for the formation of IPP and DMAPP in plants, cross-flows between MVA and MEP pathways have been identified (Vranova et al., 2013). Therefore, the divergent function and subcellular localization of TPSs determine the diversity of volatile terpenoids. In general, through specific activities of TPSs, the MVA pathway generates volatile sesquiterpenes (C15) in cytosol and peroxisome, whereas the MEP pathway generates volatile hemiterpenes (C5), monoterpenes (C10), and diterpenes (C20) in plastids (McGarvey and Croteau, 1995; Sapir-Mir et al., 2008; Vranova et al., 2013).
TPS genes have been widely found in bacteria, fungi, and plants, but their proteins show high structural similarity and share a common evolutionary origin. Notably, the size of the TPS gene family shows divergence in different lineages (Chen et al., 2011). For example, only a single functional TPS gene, coding a copalyl synthase/kaurene synthase (CPS/KS), was found in the bryophyte Physcomitrella patens, but over 100 TPS gene models were found in Vitis vinifera (Hayashi et al., 2006; Martin et al., 2010). Usually, a typical/complete TPS protein consists of two independent domains at the N-terminal and C-terminal. The N-terminal domain of the protein has features of the active site of a CPS, whereas the KS activity is located in the C-terminal of the protein (Hayashi et al., 2006). In most cases, functional TPS genes lose activity in either the CPS- or KS-type domains in both gymnosperms and angiosperms, that is, these TPSs only act as monofunctional TPSs, whereas the unique TPS in P. patens is a bifunctional enzyme (Keeling et al., 2010). This gene family is divided into seven subfamilies (TPS-a through TPS-g) based on sequence relatedness, functional assessment, and gene architecture (Aubourg et al., 2002; Alquezar et al., 2017; Chen et al., 2017; Yu et al., 2020; Zhou et al., 2020). TPSs from related plant species tend to cluster more than enzymes with similar functions, thus challenging the substrate/product predictions based on sequence similarities (Bohlmann et al., 1998; Aubourg et al., 2002). These results suggest that the expansion of TPS gene family can be species- and/or lineage-specific. As a result, the majority of TPS genes are generated through tandem duplications rather than whole-genome duplications or segmental duplications (Yu et al., 2020; Zhou et al., 2020). Moreover, the roles of some TPS proteins can be variable and associated with the availability of substrates and the microenvironment of distinct compartments (Aharoni et al., 2004). For example, two sesquiterpene synthases, TPS21 and TPS11, account for the biosynthesis of nearly all 20 sesquiterpenes found in the floral volatile blend in Arabidopsis (Tholl et al., 2005).
Since terpenoid-derived VOCs are pivotal to the survival and fitness of plants, the importance and potential roles of TPSs in the biosynthesis of VOCs and highly complicated interactions/conversations (ecological correlations) between plants and other living beings, including humans, animals, plants, and microbes have attracted attention (Dudareva et al., 2013; Song et al., 2018; Huang et al., 2019). Conversely, TPSs predominantly affect the overall flower and fruit aroma, which determines the multiplication capacity and economic value of plants. For example, Cstps1, a sesquiterpene synthase encoding-gene, is involved in the production of valencene, the key aroma compound in Citrus fruits (Sharon-Asa et al., 2003). The evolution of TPS20-related terpene synthases influences chemical diversity in the glandular trichomes of the wild tomato relative Solanum habrochaites (Gonzales-Vigil et al., 2012). The sequence variation of Fragaria ananassa Nerolidol Synthase1 (FaNES1) gene in cultivated strawberry might be the genetic basis of fruit flavor selections, which contributes to the higher levels of linalool and nerolidol than wild F. vesca (Aharoni et al., 2004). Meanwhile, TPSs provide basal products for the biosynthesis of toxic metabolites against undesirable invaders. For example, rice OsTPS24 encodes a jasmonate-responsive monoterpene synthase that produces an antibacterial ɤ-terpinene against Xanthomonas oryzae pv. oryzae (Xoo), but it does not show significant antifungal activity against the blast fungus Magnaporthe oryzae (Yoshitomi et al., 2016). However, rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in plants and its overexpression leads to enhanced resistance to Magnaporthe oryzae (Chen et al., 2018). In addition, OsTPS46, a rice terpene synthase, confers natural resistance to bird cherry-oat aphid (Sun et al., 2017). In soybean, GmAFS encodes an (E, E)-α-farnesene synthase and has defensive roles in both below-ground and above-ground organs of soybean against nematodes and insects, respectively (Lin et al., 2017).
Peach (Prunus persica L.), which has flavorful aroma, is a popular and economically important fruit crop worldwide. However, because of the absence of efficient genetic resistance, most peach cultivars suffer from the destructive gummosis disease in high temperature and humidity areas, such as Japan, south of China and USA. Fungal peach gummosis is mainly caused by Botryosphaericeae pathogens (Lasiodiplodia theobromae, Botryosphaeria dothidea, Diplodia seriata, Neofusicoccum parvum) (Wang et al., 2011; Ye et al., 2020). Although there were several studies elucidated the roles of TPSs in flavour formation of peach fruit, little is known about their function in defense responses (Liu et al., 2017; Wei et al., 2021; Wei et al., 2022). In this study, the genome-wide identification and preliminary bioinformatics analyses of PpTPS genes were performed. An ‘omics’ approach combining non-targeted metabolomics based on ultra-high performance liquid chromatography-mass spectroscopy (UHPLC-MS), gas chromatography-mass spectroscopy (GC-MS) and transcriptome was performed, the aim is to investigate the potential roles of PpTPS genes and terpenes in defense responses of peach plants to pathogens.
Genome sequences of Prunus persica were downloaded from Genome Database for Rosaceae (https://www.rosaceae.org/species/prunus_persica/genome_v2.0.a1). The hidden Markov model profile of the metal binding domain (PF01397), C-terminal metal binding domain (PF03936), and N-terminal domain (PF19086) was downloaded from the Pfam database (http://pfam.xfam.org). Redundant sequences were deleted using Blastp, and only the longest sequences among alternative splicing variants were retained.
The amino acid sequences of TPSs with defined functions in rice (Oryza sativa), apple (Malus domestica), grapevine (Vitis vinifera), strawberry (Fragaria ananassa and F. vesca), kiwifruit (Actinidia chinensis and A. arguta), pineapple (Ananas comosus), poplar (Populus trichocarpa), cotton (Gossypium hirsutum), sweet orange (Citrus sinensis), and Arabidopsis were downloaded from the National Center for Biotechnology Information (NCBI) according to the accession number of each gene (Aharoni et al., 2004; Tholl et al., 2005; Martin et al., 2010; Nieuwenhuizen et al., 2013; Nieuwenhuizen et al., 2015; Yoshitomi et al., 2016; Alquezar et al., 2017; Chen et al., 2017; Sun et al., 2017; Chen et al., 2018; Huang et al., 2018). These sequences were used to generate a phylogenetic tree based on Multiple Sequence Comparison by Log-Expectation (MUSCLE) alignment, then phylogenetic and molecular evolutionary analyses were conducted using MEGA version 11 (Tamura et al., 2021). Neighbor-joining analysis with pairwise deletion was performed using the Jones-Taylor-Thornton model. Bootstrap analysis was performed with 1000 replicates to assess the level of statistical support for each tree node. The putative protein domains of PpTPSs were identified using the MEME Suite (http://meme-suite.org/meme), while the gene structures of PpTPSs were analyzed using the Gene Structure Display Server (http://gsds.gao-lab.org). The upstream 1.5 kb region of the translation start site of the PpTPS genes was used for putative promoter cis-acting element analysis in PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html). The motifs putatively involved in different types were summarized. For synteny analysis, synteny blocks between peach and Citrus sinensis and Arabidopsis thaliana genomes were obtained from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/index/downloads).
Two early-maturity cultivars with different fruit flesh colors, ‘Jinchun’ (yellow-fleshed) and ‘Hikawa Hakuho’ (white-fleshed), were selected to investigate the accumulation of linalool and transcription change of PpPTS genes during developmental stages. Three trees for each cultivar were grown under consistent and standard agronomic practices for fertilization, irrigation, fruit thinning, pruning, and pest and disease control in the Zhuanghang experimental field of Shanghai Academy of Agricultural Sciences (SAAS), Shanghai, China (121.45°E, 30.92°N). Fruits were collected at five sampling times (day 35 (S1), 50 (S2), 65 (S3), 80 (S4), 90 (S5), 95(S6)) after full blossom. For each sampling stage, 20 fruits were selected and taken to the laboratory immediately. The mesocarp of each fruit were cut into small pieces and frozen instantly in liquid nitrogen and stored at -80°C for linalool quantification and RNA sequencing. Linalool was quantified using a modified method based on the description of Liu and Wei et al. (Liu et al., 2017; Wei et al., 2021).
Three pathogens isolations, designated as Botryosphaeria dothidea SHTLJ001 (OP340986), Botrytis cinerea SHTLK001 (OP340985), and Monilinia fruticola SHTHF001 (OP340987) were collected from the peach experimental trial fields of Shanghai Academy of Agricultural Sciences, which were purified through multiple subculture of hyphae and identified by sequencing of their ITSs with common primers ITS1 and ITS4. The current-year shoots of the susceptible cultivar ‘Huyou018’ were inoculated with B. dothidea SHTLJ001 using the method described in Gao et al. (Gao et al., 2016). In brief, the surface-sterilized peach shoots were cut into 15 cm-long segments and then wounded with a sterilized needle. A single mycelial plug (5 mm in diameter) of B. dothidea SHTLJ001 was placed onto the wound point. Shoot segments inoculated with sterile PDA medium without B. dothidea SHTLJ001 were treated as controls. The inoculated and control shoots were placed in square plastic boxes laying multiple layers of wet filter papers. The boxes were covered with clear plastic wrap to keep 90% relative humidity and then placed in a light incubator at 28/25°C, with a photoperiod of 12/12 h light (5000 lux)/dark.
To measure the relative contents of terpenoids in peach shoots post B. dothidea inoculations, the tissue measuring 0.5 cm in diameter was cut from the lesion area and frozen at -80°C at 0 h, 48 h, and 84 h after inoculation. The extraction and relative quantification of terpenoids was performed according to the method described in Sawada et al. (Sawada et al., 2009). Briefly, 50 mg of freeze-dried and ground sample was transferred to a 2 mL tube, and 700 μL of extract solution (methanol/water=3:1, precooled at -40°C, containing internal standard) was added. After being vortexed for 30 s, the samples were homogenized at 35 Hz for 4 min and sonicated for 5 min in an ice-water bath. Homogenization and sonication were performed three times. After being extracted overnight at 4°C on a shaker, the samples were centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant was carefully filtered through a 0.22 μm microporous membrane, and then the resulting supernatants were diluted 20 times with methanol/water mixture (v:v=3:1, containing an internal standard), vortexed for 30 s, and transferred to 2 mL glass vials. Next, 40 μL of each sample was pooled as quality control samples. The samples were used to perform UHPLC-MS analyses with previous methods (Sawada et al., 2009).
Total RNA extraction and first-strand complementary DNA (cDNA) synthesis for both shoots and fruit pulp were carried out using the method described in Li et al. (2015). The construction of RNA sequencing libraries and the quantification of gene expression level were generated according to previous study (Pertea et al., 2015). The FPKM of each gene was calculated based on the length of the gene and the read count mapped to that gene. The change of gene transcription level in each stage was compared with the first sampling stage.
The CDS of PpTPS1, PpTPS3, and PpTPS4 were cloned from the cDNA library of ‘Huyou018’ with gene-specific primers (Supplementary Table 1). Then, the comfirmed CDS sequences were subcloned into pCambia1302-GFP vector. The fusion proteins were transiently expressed in the fully expanded young leaves of six-week-old Nicotiana benthamiana via Agrobacterium-midiated infiltration (Wydro et al., 2006). Images were obtained with a LEICA laser scaning confocal microscope at 3 days post infiltrations.
To test the antifungal activities of linalool, the growth speed assays of three fungi species were performed on PDA solid medium containing different concentrations of linalool. The pure linalool (ANPEL Laboratory Technologies, Shanghai) were dissolved in the 1 mL ethanol (Sinopharm Chemical Reagent Co., Ltd, Shanghai) to prepare a mixture liquid. Then, different volumes of mixture liquid were added into 200 mL sterilized PDA medium (prior to solidification). The equal amount of fresh hyphal mass were inoculated at the center of new PDA plates with a needle and the diameter of colony were measured at 3 days post inoculations.
The experiment was arranged in a completely randomized design with three replications, and collected data were statistically analyzed using multivariate analysis methods in SAS computer software (SAS Version 9.2, Institute). Analysis of variance (ANOVA) was used to determine the overall statistical significance of the data at the significance level of P <0.05, and the data were represented as average ± STDEV (n = 3). The heatmaps were visualized using TBtools software (Chen et al., 2020).
Based on the genome sequences of peach (Lovell v2.0.a1), a total of 45 candidate TPS genes were preliminarily identified, and their deduced proteins contain at least one of the three core Pfam domains (PF01397, PF03936, and PF19086) that are essential to the catalytic activities of TPS. Five of them (Prupe.4G193900, Prupe.4G194000, Prupe.4G194700, Prupe.4G238800, and Prupe.4G265600) were too short (the length of amino acid sequences is shorter than 250) to code a single functional domain in length; therefore, they were discarded. There were three PpTPS genes (Prupe.4G030400, Prupe.4G029900, and Prupe.4G030300) have been named as PpTPS1, PpTPS2 and PpTPS3 in previous studies (Liu et al., 2017; Wei et al., 2021). According to the chromosome localization of the genes, the remaining 37 peach TPS genes were named as Prunus persica Terpene Synthases 4 to 40 (PpTPS4-PpTPS40) (Table 1). These PpTPS genes are only distributed in three chromosomes of peach (chromosome 3, 4, and 8). Among them, chromosome 4 contains 34 PpTPSs, which intensively occur in several gene clusters, whereas chromosome 8 only has 1 PpTPS (PpTPS40) (Table 1). The phylogenetic analysis grouped these 40 PpTPSs into 5 clades corresponding to the TPS classes a (22), b (8), c (1), e/f (7), and g (2) (Figure 1A). The synteny analysis of TPS genes among P. persica, A. thaliana, and C. sinensis suggested that only few PpTPS genes showed synteny relations and the Ka/Ks ratio of these synteny genes are far less than 1.0 (Figure 1B, Supplementary Table 2), which means that most of these PpTPS genes were generated after lineage separation and those synteny genes have undergone purifying selections. Detailed information concerning each putative PpTPS gene is presented in Table 1.
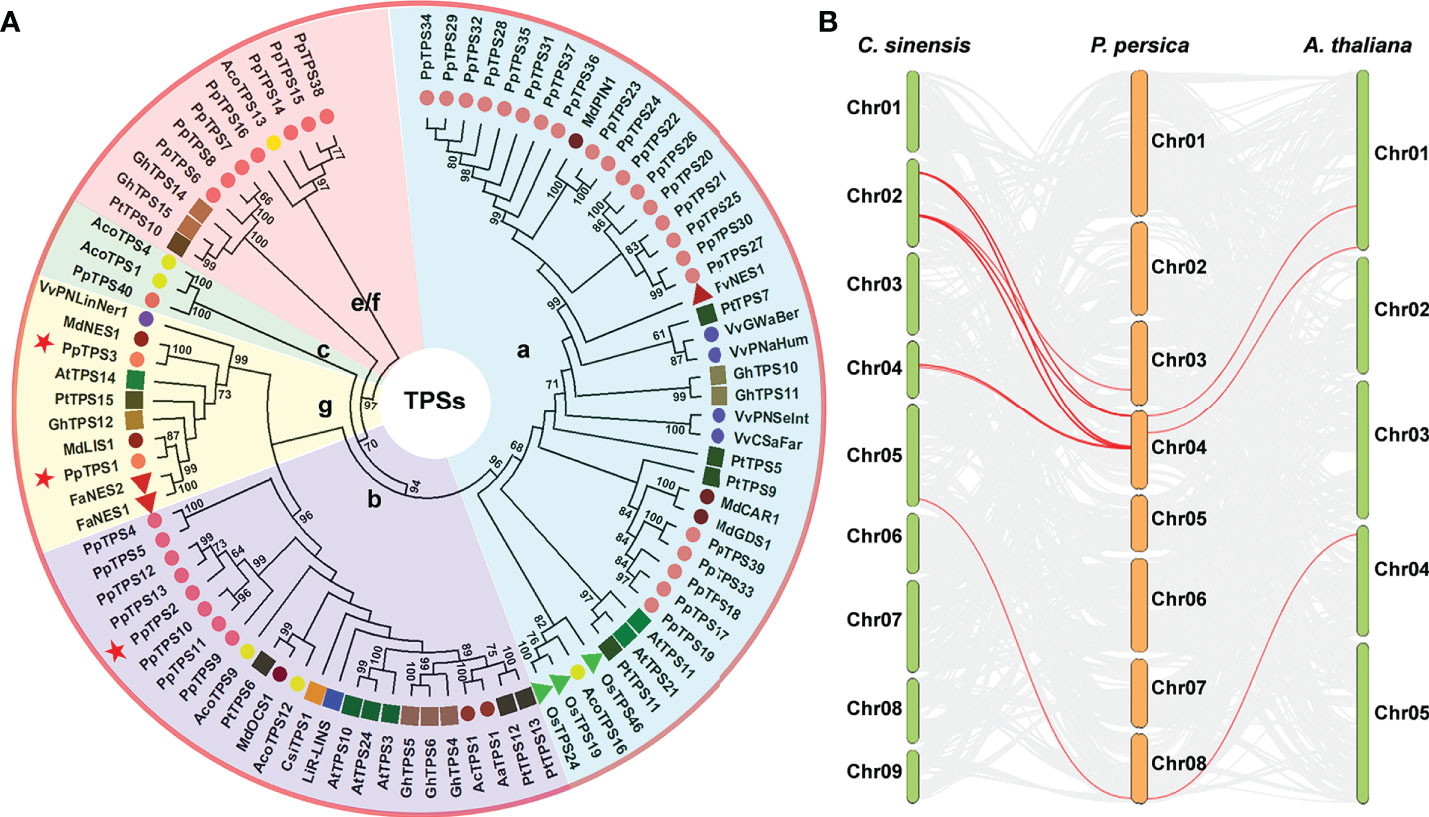
Figure 1 Evolutionary analysis of PpTPS genes. (A) A phylogenetic tree that was constructed using the full-length amino acid sequences of 40 TPSs from Prunus persica and 50 function-defined TPSs from Arabidopsis thaliana, kiwifruit (Actinidia arguta, Actinidia chinensis), pineapple (Ananas comosus), sweet orange (Citrus sinensis), strawberry (Fragaria ananassa, Fragaria vesca), cotton (Gossypium hirsutum), apple (Malus domestica), rice (Oryza sativa), poplar (Populus trichocarpa), and grapevine (Vitis vinifera). The phylogenetic tree was constructed with MEGA 11 using the Neighbor-joining method and the amino acid sequences were obtained from the previous studies (Aharoni et al., 2004; Tholl et al., 2005; Martin et al., 2010; Nieuwenhuizen et al., 2013; Nieuwenhuizen et al., 2015; Yoshitomi et al., 2016; Alquezar et al., 2017; Chen et al., 2017; Sun et al., 2017; Chen et al., 2018; Huang et al., 2018). The detailed accession numbers of all the TPS proteins were presented in Supplementary Table 3. Red stars denote three function-defined PpTPS proteins in previous studies. (B) The synteny analysis of TPS genes from Prunus persica, Arabidopsis thaliana, and Citrus sinensis.
To better understand the evolutionary relationships and structural characteristics of PpTPSs, the exon-intron organizations of these genes and the motifs of the deduced proteins were predicted using online tools. The PpTPSs that belong to class a, b, and g share similar gene structure with a smaller number of exons (4 to 8) and shorter coding DNA sequences (CDSs) (260 to 629 bp), whereas the members classified into class c and class e/f show a similar gene structure with a greater number of exons (6 to 14) and longer CDSs (275 to 837 bp) (Table 1, Figures 2A, B). The gene structure of PpTPSs directly determines the number and organization of protein domains/motifs, when the parameter is set as five conserved motifs to be predicted using the motif-based sequence analysis tools in the MEME Suite (http://meme-suite.org/meme/) (Supplementary Figure 1). Thirty PpTPSs contain the conserved DDxxD motif (Motif 2 in Figure 2C), four PpTPSs have mutant DDxxD motif, and six PpTPSs have thoroughly lost this motif (Table 1, Figure 2C). Even among 30 PpTPSs with DDxxD motif, 5 PpTPSs have lost the NSE/DTE motif, which means that only 25 PpTPSs might have kaurene synthase activities, because both the DDxxD and NSE/DTE motifs are essential to the metal dependent ionization of the prenyl diphosphate substrate. In addition, the unique class c TPS, PpTPS40, has a DxDD motif that is important for copalyl synthase activities, which suggests that it might play a role in the protonation-initiated cyclization of geranylgeranyl diphosphate to CPP (Table 1).
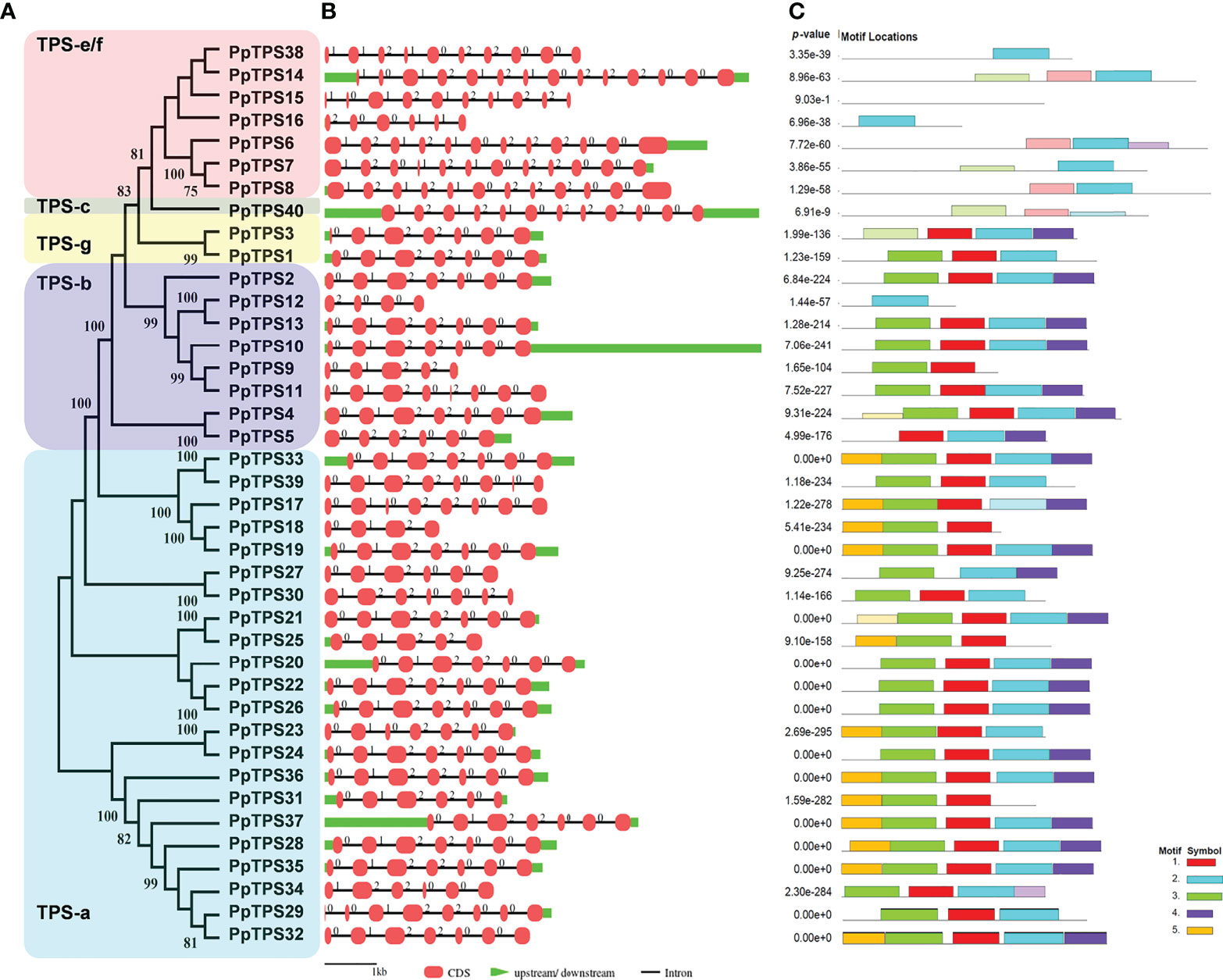
Figure 2 Structural analysis of PpTPS genes. (A) A phylogenetic tree of forty full-length PpTPS proteins. (B) A schematic graph shows the exon-intron organizations of PpTPS genes. (C) The organization of five protein motifs in PpTPSs predicted by MEME online tools. The detailed sequences of protein domains in PpTPSs were presented in Supplementary Figure 1.
To explore the potential roles of PpTPSs in the defense response of peach against fungus that induces peach gummosis, we analyzed the transcript levels (FPKM values) of 40 PpTPSs in the shoot of ‘Huyou018’ after inoculation with B. dothidea via RNA sequencing (Supplementary Table 4). The majority of PpTPSs showed down-regulation at the transcription level, and some PpTPSs showing elevated transcription levels in mock treatment (potato dextrose agar, PDA, medium inoculations) were down-regulated at the transcription level after inoculation with pathogens (Figure 3A). Several PpTPSs including PpTPS2, PpTPS17, PpTPS18, PpTPS19, and PpTPS23 were up-regulated at the transcription level post inoculation, but they showed differences (Figure 3A). For example, PpTPS2 and PpTPS23 were gradually up-regulated at the transcription level, whereas PpTPS17, PpTPS18, and PpTPS19 were up-regulated at 48 h post inoculation (hpi), and then they reduced their expression until 84 hpi (Figures 3B, C).
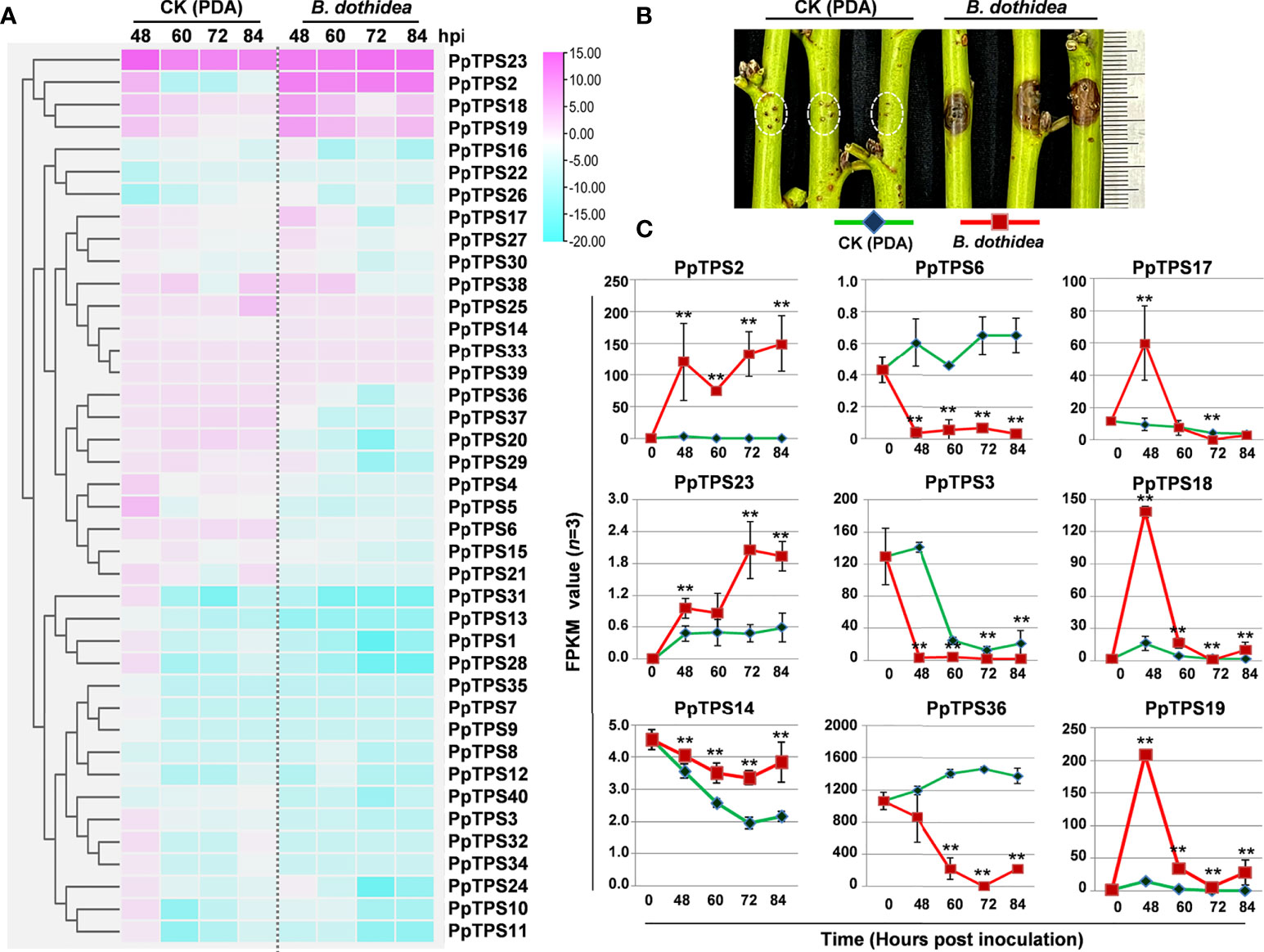
Figure 3 Expression profiles of PpTPS genes in peach shoots post B. dothidea inoculations determined by RNA sequencing. (A) A heatmap shows the relative transcript levels (FPKM value) of forty PpTPS genes in the shoots of ‘Huyou018’ after PDA medium (Mock) and B. dothidea inoculations. The color scale represents log(fold change, 2) values of PpTPSs transcript levels at each time point (48 h, 60 h, 72 h, and 84) compared with 0 h after each treatment, with pink denoting increased transcript abundance and blue denoting decreased transcript abundance. (B) The representative photograph shows the phenotype of peach shoots at 48 h after B. dothidea inoculations. (C) Line graphs show the detailed FPKM value of nine selected PpTPS genes after PDA medium (Mock) and B. dothidea inoculations. The asterisks denote that the FPKM values of a gene in B. dothidea inoculations were significantly different with that in Mock (p<0.05). The detailed FPKM values of PpTPSs in pathogen inoculation assays were presented in Supplementary Table 4.
To further verify the relationship between the transcriptions of PpTPSs with the accumulation of terpenoid metabolites in the shoot post inoculation with B. dothidea, we measured the content of multiple metabolites in the same batches of samples used for RNA sequencing. A total of 133 terpenoid metabolites were detected, including 18 monoterpenes, 23 diterpenes, 26 sesquiterpenes, 51 triterpenes, 14 iridoids, and one capsanthin (Figure 4, Supplementary Table 5). Based on the peak areas of each metabolite obtained from UHPLC-MS assays, we found that the relative contents of many detected terpenes (22% of monoterpenes, 52% of diterpenes, 19% of sesquiterpenes, 33% of triterpenes, and 21% of iridoids) were elevated in response to the inoculation with B. dothidea, especially at 84 hpi. The relative contents of some terpenoids also changed after mock treatments.

Figure 4 The relative contents of 133 terpenoids in peach shoots post B. dothidea inoculations determined by UHPLC-MS. (A–D) Heatmaps show the relative content of eighteen monoterpenes (A), twenty-three diterpenes (B), twenty-six sesquiterpenes (C), and 65 triterpenes and iridoids (D) in the shoot of ‘Huyou018’ after PDA medium (Mock) and B. dothidea inoculations, respectively. The color scale represents relative contents of terpenoids at each time point (0 h, 48 h, 84 h) with pink denoting relatively higher terpenoids contents and blue denoting relatively lower terpenoids contents. A red asterisk in (A) points out the linalool oxide. The detailed relative contents of 133 terpenoids were presented in Supplementary Table 5.
To explore the potential function of PpTPSs in the development of peach fruits, we analyzed the transcript levels (FPKM values) of 40 PpTPSs during fruit development based on RNA sequencing data (Supplementary Table 6, Figure 5A). Approximately half of the PpTPSs showed down-regulated transcript levels at different sampling times compared to the first sampling time (S1); thus, these PpTPS genes could reduce their expression during peach fruit development (Figure 5B). Fifteen PpTPSs showed consistent transcript levels during the entire process of peach fruit development, suggesting that they are not directly associated with fruit development (Figure 5B). However, five PpTPSs exhibited elevated transcript levels at the middle or late stages of peach fruit development either in yellow-fleshed cultivar ‘Jinchun’ or in white-fleshed cultivar ‘Hikawa Hakuho’; thus, these PpTPSs can contribute to the biosynthesis of aroma-related terpenoids (Figure 5B). Most PpTPSs showed similar expression profiles in these two peach cultivars, whereas several PpTPSs exhibited distinct expression characteristics. For example, the transcript levels of PpTPS2 and PpTPS18 were up-regulated in ‘Jinchun’ but reduced at the same sampling time points in ‘Hikawa Hakuho’ (Figure 5B). In contrast, the transcript levels of PpTPS5 and PpTPS40 were up-regulated in ‘Hikawa Hakuho’ but down-regulated in ‘Jinchun’. The transcript levels of PpTPS1 and PpTPS38 only showed up-regulation in ‘Jinchun’, but those of PpTPS15 and PpTPS20 only showed up-regulation in ‘Hikawa Hakuho’ (Figure 5B). These results suggest that the expression of PpTPSs is cultivar-specific.
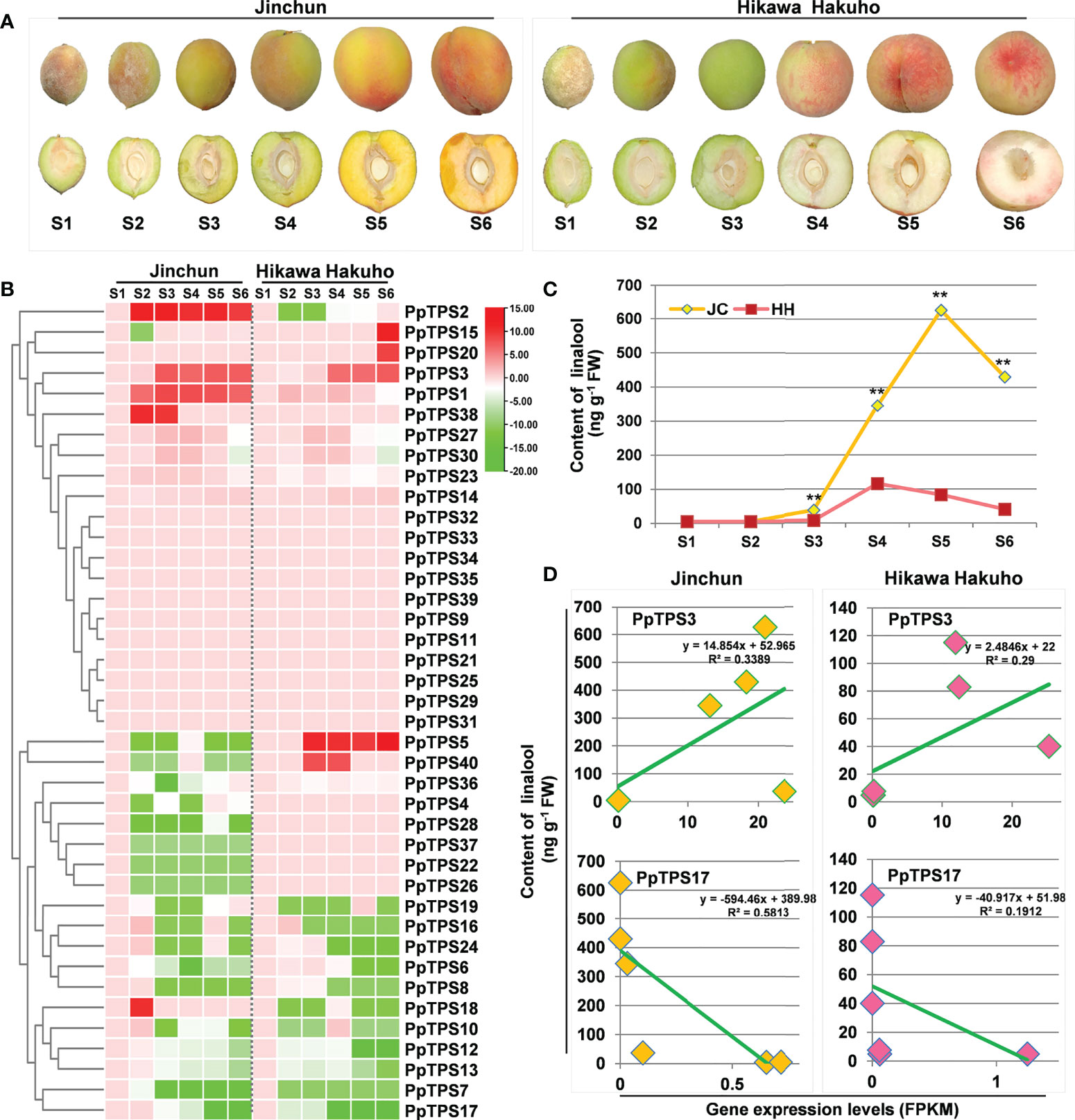
Figure 5 Expression profiles of PpTPS genes during fruit development determined by RNA sequencing. (A) Representative photographs show the peach fruits at each sampling time point. (B) A heatmap shows the relative transcript levels (FPKM value) of forty PpTPSs in fruit pulps of ‘Jinchun’ (JC) and ‘Hikawa Hakuho’ (HH) at different stages of fruit development and ripening stages (35 d (S1), 50 d (S2), 65 d (S3), 80 d (S4), 90 d (S5), 95 d (S6)). The color scale represents log (fold change, 2) values of PpTPSs transcript levels at each development stages compared with S1, with red denoting increased transcript abundance and green denoting decreased transcript abundance. The detailed FPKM values of PpTPSs during fruit development were presented in Supplementary Table 6. (C) A line graph shows the content of linalool in fruit pulps of ‘Jinchun’ and ‘Hikawa Hakuho’ at different stages of fruit development. The asterisks denote that the linalool contents in ‘Jinchun’ were significantly higher than that in ‘Hikawa Hakuho’ at each sampling time site (p<0.05). (D) Line graphs show the linear correlations between the relative transcript levels of PpTPSs (PpTPS3 and PpTPS17) and the contents of linalool in fruit pulps of ‘Jinchun’ and ‘Hikawa Hakuho’ during fruit development.
To further verify the relationship between the transcriptions of PpTPSs and the accumulation of terpenoid metabolites in peach fruits, we measured the content of linalool, the predominant terpene accumulated in peach fruits, in the same batches of samples used for RNA sequencing. The linalool content gradually elevated during the early stages of fruit development both in ‘Jinchun’ and ‘Hikawa Hakuho’, and peaked at S5 and/or S4, respectively, then decreased during fruit ripening stages (Figure 5C). Obviously, the fruit pulp of ‘Jinchun’ contains higher linalool content than that of ‘Hikawa Hakuho’ at all sampling times (Figure 5C). Correlation analysis demonstrated that the expression profiles of PpTPS3 exhibited positive linear correlations with the linalool content both in ‘Jinchun’ and ‘Hikawa Hakuho’ (Figure 5D). However, the expression profiles of PpTPS17 exhibited negative linear correlations with the linalool content in both cultivars (Figure 5D).
For TPS proteins, the subcellular localization determine their substrates availabilities, therefore, it should be an essential character of each TPS individual. To preliminarily investigate the subcellular localization of PpTPSs, we selected three representative PpTPSs, including PpTPS1, PpTPS3, and PpTPS4, that showed elevated transcription levels post pathogen inoculations or during the fruit development process, fused their CDS with GFP gene, and then transiently expressed these fusion genes in the leaves of tobacco. As shown in Figure 6, PpTPS3-GFP showed specific chloroplast localizations in the leaves of tobacco, while PpTPS1-GFP and PpTPS4-GFP not only exhibited co-localization with chloroplasts, but also partially distributed in nucleus and cytoplasm.
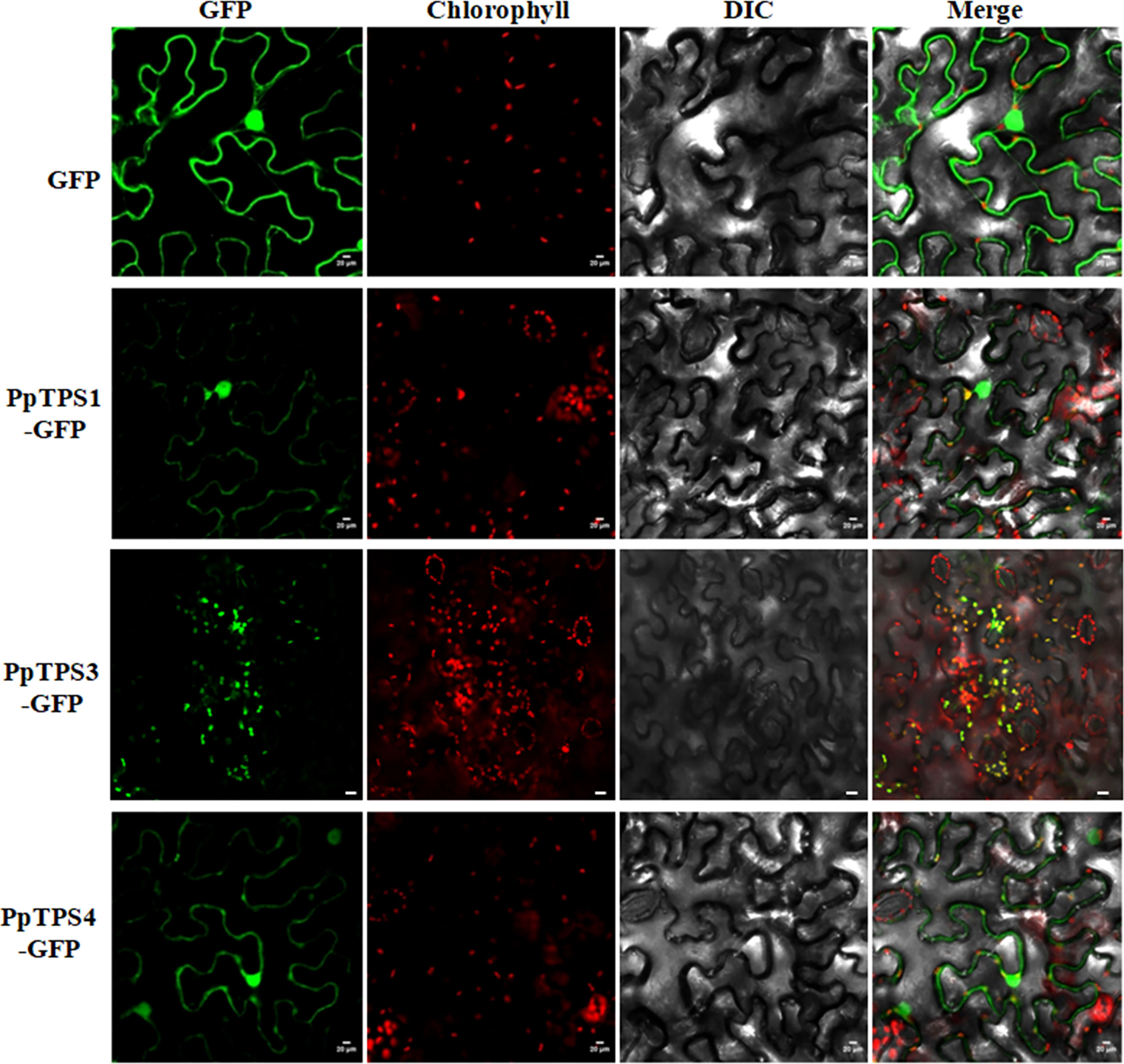
Figure 6 Subcellular localizations of three PpTPS-GFP fusion proteins in the leaves of Nicotiana benthamiana. The primers used for gene cloning were listed in Supplementary Table 1.
To examine whether the metabolites of PpTPSs coding enzymes are involved in peach defense responses against fungi or not, we analyzed the antifungal activities of linalool in PDA medium. As shown in Figure 7A, though lower levels of linalool did not suppress the hyphal growth of fruit parasitic pathogens (B. cinerea and M. fruticola), but the elevated linalool contents (>0.1 g/L) could significantly limit the hyphal growth of all the three main peach pathogens in PDA medium (Figure 7B). This result suggested that linalool might have broad-spectrum antifungal activities.
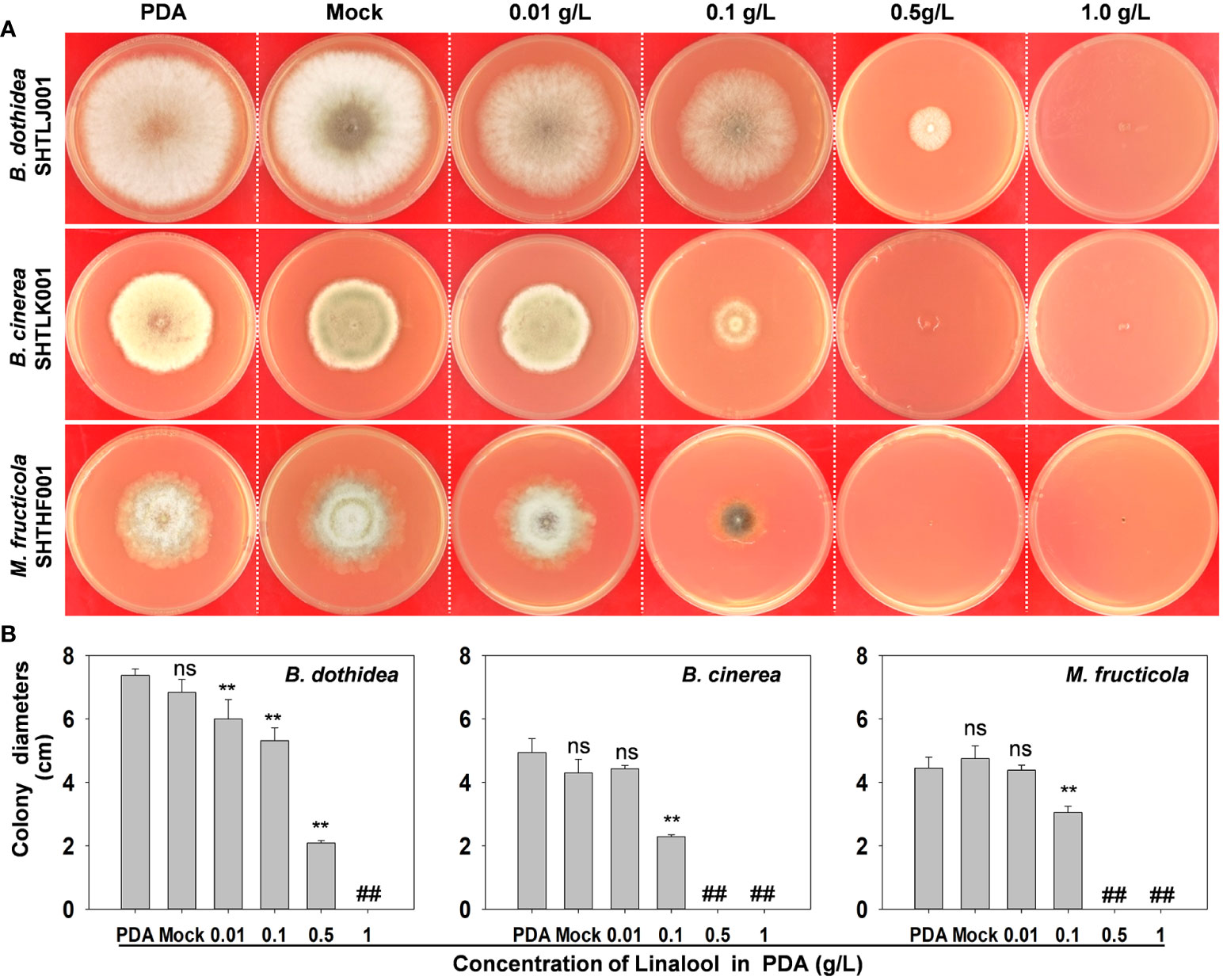
Figure 7 Antifungal activities of linalool in PDA medium. (A) Representative photographs show the growth of three pathogens on the PDA mediums containing different concentrations of linalool at 3 days post inoculation.(B) Statistic of the colony diameters of three pathogens on PDA mediums at 3 days post inoculation. The asterisks denote that the colony diameters of each pathogen in those PDA mediums containing linalool were significantly lower than that in PDA mediums without ethanol (Mock) and linalool (p<0.05), ‘##’ indicates no data were obtained and ‘ns’ means the difference was not significant.
To explain the likely mechanisms of the distinct expression patterns of different PpTPS members either in defense response or in fruit floral-note aroma formation, we analyzed the distributions of cis-acting regulatory elements in the promoter regions of 40 PpTPS genes. Over 15 cis-acting regulatory elements were identified, and they were widely distributed in these promoters. The top six identified elements are MYC (-CATTTG-), G-box (-CACGTT/C-), ABRE (-ACGTG-), CGTCA-box (-CGTCA-/-TGACG-), ARE (-AAACCA-), and as-1 (-TGACG-) (Figures 8A, B). All of them were distributed in the promoters of at least 30 PpTPSs. In addition, multiple types of MYB transcription factor binding sites, including ARE, Myb (-TAACTG-), MYB-binding site (-CAACAG-), MYB-like sequence (-TAACCA-), and MBS (-CAACTG-) were identified (Figure 8B). The comparison of the promoter sequences of PpTPS1, PpTPS2, PpTPS3, PpTPS17, PpTPS18, and PpTPS19 showed that the presence or absence of ARE, TC-rich repeats (-GTTTTCTTAC-), and TCA-element (-TCAGAAGAGG-) might impact the expression profiles of PpTPSs in the shoot after inoculation with B. dothidea (Figure 8C).
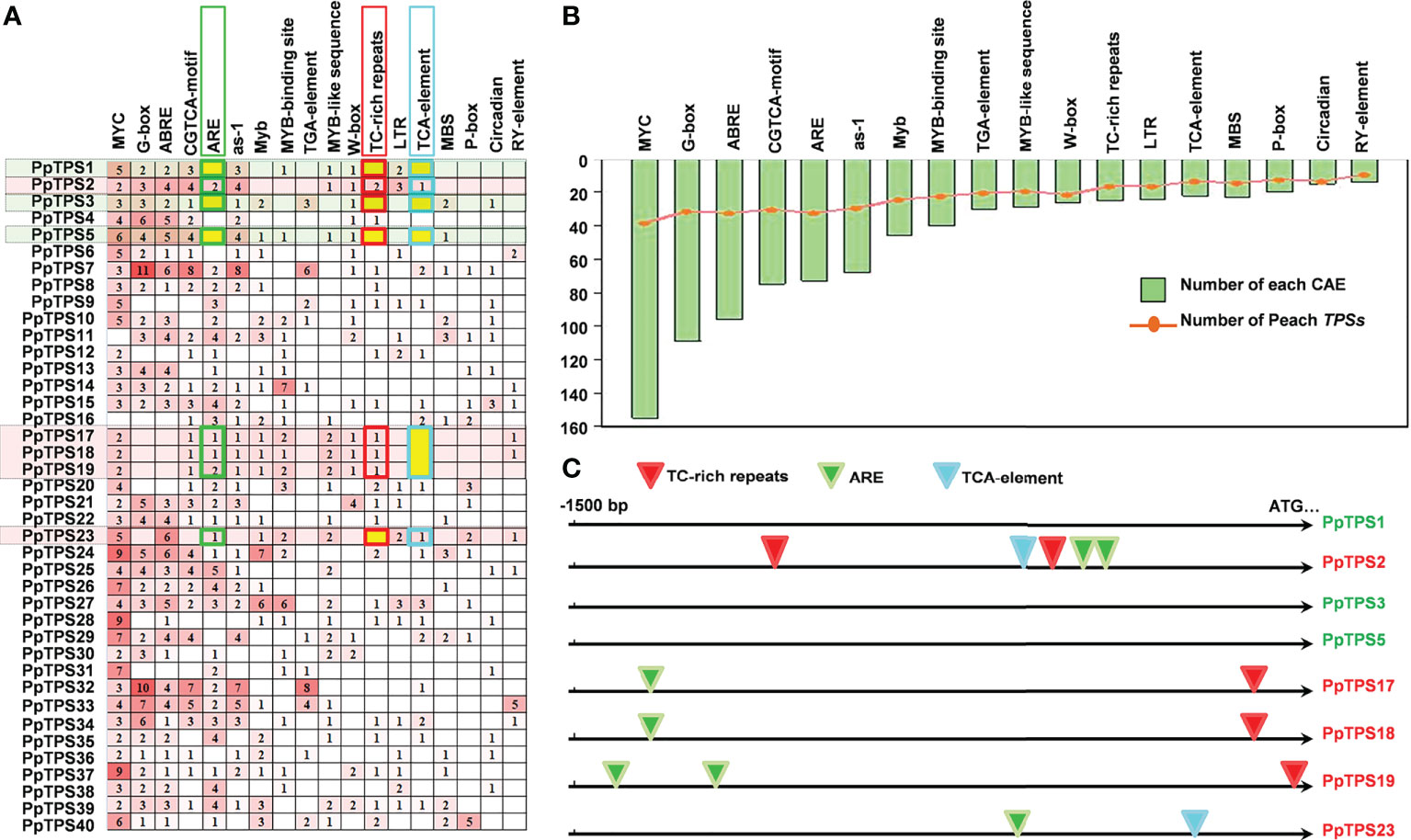
Figure 8 Distribution of cis-acting regulatory elements in the promoter of PpTPS genes. (A) A heatmap shows the number of different cis-acting regulatory elements in the promoter of forty PpTPSs. (B) A bar graph shows the total number of each cis-acting regulatory element identified in the promoter of forty PpTPSs. The curve point denotes the number of PpTPSs having each cis-acting regulatory element. (C) A schematic shows the distribution of three SA-associated cis-acting regulatory elements in the promoters of eight selected PpTPSs.
Plant VOCs can affect the disease resistance and economical value of agricultural plants, especially horticultural crops (Aharoni et al., 2004; Lin et al., 2017; Liu et al., 2017; Chen et al., 2018; Huang et al., 2018; Wei et al., 2021). Owing to the unique function of TPSs in the biosynthesis of terpenoids, which is the largest group of plant-released VOCs, studies similar to the present analysis in Arabidopsis, tomato (Solanum lycopersicum), carrot (Daucus carota), cotton (Gossypium hirsutum), pineapple (Ananas comosus), apple (Malus domestica), grapevine (Vitis vinifera), tea (Camellia sinensis), holy basil (Ocimum sanctum), and Cannabis sativa have identified numerous TPS genes (Aubourg et al., 2002; Martin et al., 2010; Falara et al., 2011; Nieuwenhuizen et al., 2013; Alquezar et al., 2017; Chen et al., 2017; Keilwagen et al., 2017; Huang et al., 2018; Kumar et al., 2018; Allen et al., 2019; Yu et al., 2020; Zhou et al., 2020; Zhang et al., 2021). However, except for the preliminary identification and evolution analysis of TPS genes in Rosaceae and few studies reporting the roles of three PpTPSs (PpTPS1, PpTPS2 and PpTPS3) in the biosynthesis of flavor-related linalool and (E,E)-α-farnesene, respectively, limited information is available about this important gene family in peach (Liu et al., 2017; Wei et al., 2021; Zhang et al., 2021; Wei et al., 2022; Zhang et al., 2022). In this study, we investigated expression profiles of peach TPS genes in current-year shoots post B. dothidea inoculations and/or in fruit pulps during fruit development, and tried to explore the potential roles of PpTPSs and/or their metabolites in disease resistance.
TPS genes widely found in bacteria, fungi, and plants are likely derived from a common ancestor gene and then undergo species-specific expansion (Chen et al., 2011). Compared to other higher plants, peach has a smaller quantity of TPS genes (Martin et al., 2010; Nieuwenhuizen et al., 2013; Zhou et al., 2020; Zhang et al., 2021) (Table 1, Figure 1A) because Prunoideae has not experienced recent whole-genome duplication events (Zhang et al., 2021). In addition, the TPS genes intensively distribute in several gene clusters of three chromosomes in peach genome (Table 1, Figure 1A), indicating that these chromosome regions harboring PpTPS genes have not undergone large-scale segment duplications or recombination in the form of crossing over between nonhomologous chromosomes. Alternatively, tandem duplications may be the cause of TPS gene family expansion (copy number variations) in peach. We showed that the gene structures of PpTPSs were highly similar but the loss of exons in some duplicated PpTPSs led to variations in key protein motifs or missing protein motifs (Figures 2A, B). Being similar to other angiosperm plants, though 25 deduced PpTPSs contain the intact KS activity center, only the unique class c member PpTPS40 still harbors a CPS activity center (Table 1), which means that the bifunctional diterpene synthase could be absent in peach.
The distinctive and rich flavor of peach fruit, which is a key trait of peach attracting consumers, is attributed to the mixture of volatiles, sugars, and acids. To date, more than 100 volatile chemicals have been identified in peach fruit, and linalool is the main odorant that affects fruit aroma and consumer preference (Eduardo et al., 2010). In this study, we observed that linalool is a dominant terpenoid detected in fruit pulps of two peach cultivars (‘Jinchun’ and ‘Hikawa Hakuho’) using GC-MS (Figure 5C). Among 40 PpTPSs, the transcript level (FPKM) of PpTPS3 showed positive linear correlations with the linalool content, suggesting that PpTPS3 can directly contribute to the biosynthesis of linalool (Figure 5D). In a recent study, this gene has been identified to encode a linalool synthase in peach fruit (Wei et al., 2021). PpTPS2 showed distinct expression profiles in ‘Jinchun’ and ‘Hikawa Hakuho’ during fruit development and has been identified to encode an UV-B-induced sesquiterpene synthase that promotes the accumulation of (E,E)-α-farnesene in peach fruits (Liu et al., 2017) (Figure 5D).
Fungi-induced perennial gummosis on trunks and branches, caused by multiple fungi classified as Botryosphaericeae (Lasiodiplodia theobromae, Botryosphaeria dothidea, Diplodia seriata, Neofusicoccum parvum), is a major problem in peach orchards worldwide (Wang et al., 2011). Several studies reported that the overexpression of TPS genes could improve the resistance of plants against bacteria, fungi, and nematodes, owing to the ectopic accumulations of different terpenoids, including linalool, (S)-limonene, and gamma-terpinene (Shimada et al., 2014; Yoshitomi et al., 2016; Lin et al., 2017; Shimada et al., 2017; Chen et al., 2018; Huang et al., 2018). In this study, most of the 40 PpTPS genes showed changed transcript levels and over 100 terpene-related metabolites exhibited altered contents in the shoots post B. dothidea inoculation, indicating the terpenoid-related transcription and metabolism were strongly induced during fungal gummosis (Figures 3 and 4). Although over 30 PpTPS genes were down-regulated at the transcription levels after inoculation with pathogens, several PpTPS genes (PpTPS2, PpTPS17, PpTPS18, PpTPS19, and PpTPS23) were gradually up-regulated and/or were predominantly up-regulated at the early stages (48 hpi) of pathogen infections, suggesting their potential importance in this bioprocess (Figure 3C). While most of the detected metabolites showed reduced relative contents at 48 hpi compared to 0 hpi, over half of these diterpenes and triterpenes displayed substantial accumulations at 84 hpi (Figure 4), suggesting that the metabolism of terpenoids were finely regulated in peach shoots during pathogen infections.
To support the above speculations, we also examined the antifungal activities of linalool in PDA medium, and found that high concentrations (0.1 g/L to 1.0 g/L) of linalool could fully limit the hyphal growth of B. dothidea, B. cinerea, and M. fruticola, which cause devastating diseases in peach (Figures 7A, B). This result suggests that linalool and/or it-related terpenes could be effective chemical weapons of peach against necrotrophic pathogens. However, the transcription of PpTPS3, the most important contributor of linalool biosynthesis, was down-regulated during B. dothidea inoculations compared to PDA treatments in the shoot of ‘Huyou018’ (Figure 3C), indicating the defense-related metabolisms of peach could be hijacked by pathogens via transcriptional regulations.
TPSs are vital terminal enzymes catalyzing the formation of plant terpenoids by using various prenyl diphosphate precursors as substrates, but the transcription of TPS genes can be controlled by many transcription factors. For example, in flowers of Arabidopsis, AtMYC2 interacts with DELLA proteins to regulate sesquiterpene synthase gene expression (Hong et al., 2012). In flowers of monocotyledonous plant Freesia hybrida, two interacted transcription factors, FhMYB21L2 and FhMYC2, aggressively regulate the transcription of linalool synthesis gene FhTPS1 (Yang et al., 2020). In the leaves of sweet orange, CiMYB42 regulates the expression of CiOSC by binding to the -TTGTTG- sequence (type II MYB core) (Zhang et al., 2020). In peach fruit, transcription factor PpbHLH1 and PpERF61 activate PpTPS1 and PpTPS3 expression by directly binding to E-box and DRE/CRT cis-element in their promoters, respectively (Wei et al., 2021; Wei et al., 2022). These results suggest that bHLH, ERF and MYB transcription factors can be the major upstream regulators of TPS genes. In this study, we also observed that MYC2, G-box, ABRE, CGTCA-motif, and multiple types of MYB-binding sites are widely distributed in the promoters of almost all the PpTPSs (Figures 8A, B). ABRE is not directly associated with bHLH and/or MYB transcription factors, but ABA signaling is highly associated with these transcription factors, which indirectly affects the biosynthesis of terpenoids via ABRE (Abe et al., 2003). In addition, SA-related cis-acting regulatory elements, including as-1, ARE, TCA-element, and TC-rich repeats, might also affect the distinct expression profiles of PpTPSs. For example, the presence or absence of ARE, TC-rich repeats, and TCA-elements is associated with the pathogen-induced up-regulations after inoculation with B. dothidea (Figure 8C). Although the experimental evidences uncovering the relationships between phytohormone signalings and terpenes biosynthesis is lacking, we speculated that the cross-talks between different phytohormone signaling pathways could fine tune the terpenes biosynthesis via various transcription factors. Therefore, more efforts are required to elucidate the roles of divergent phytohormone and transcription factor in regulating terpene biosynthesis in the further studies.
In this study, we identified, classified, and named TPS family genes in P. persica and assessed the expression profiles of them in current-year shoots post B. dothidea inoculations and/or in fruit pulps during fruit development via RNA sequencing. Being highly similar with previous studies, PpTPS1, PpTPS2, and PpTPS3 exhibited dominant transcript levels and significantly elevated their transcript levels during fruit development and ripening in two peach cultivars, suggesting their essential roles in the formation of terpene-derived peach flavors and fruit quality. Moreover, we also observed that over 20 PpTPSs showed reduced transcript levels in defense responses against B. dothidea, and 10 PpTPSs exhibited elevated transcript levels during the same process. Meanwhile, the results of UHPLC-MS and GC-MS analyses revealed that the contents of different terpenoids were altered in defense responses of peach. These results suggest the potential importance of PpTPSs in peach defense responses. Notably, the transcription levels of PpTPS3 exhibited positive linear correlations with the linalool contents in fruit pulps, suggesting its important roles in linalool biosynthesis. In addition, we also provide evidences to support the effective antifungal activities of linalool in PDA medium. Taken together, our results provide a foundation for further studies to explore the function of PpTPS genes in response to biotic stress stimuli; however, additional studies are required to elucidate the detailed function of each TPS protein in this economically important fruit crop.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject accession number: PRJNA828349.
XL and ZY initiated the project, designed the experiment. MS, JD, HZ and XZ collected the samples. YH, XL and MZ analyzed and processed the raw data. YH and XL drafted the manuscript. XL and ZY reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by funds from Shanghai Science and Technology committee Rising-Star Program (19QB1404600), the National Key Research and Development Program of China (2019YFD1000801), the Outstanding Team Program of Shanghai Academy of Agricultural Science (Grant No. 2022–004).
We are grateful for the help of Shanghai BIOTREE Biological Technology Co., Ltd. (Shanghai, China) in linalool and terpenoids measurement.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1032838/full#supplementary-material
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 (1), 63–78. doi: 10.1105/tpc.006130
Aharoni, A., Giri, A. P., Verstappen, F. W. A., Bertea, C. M., Sevenier, R., Sun, Z. K., et al. (2004). Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16 (11), 3110–3131. doi: 10.1105/tpc.104.023895
Allen, K. D., McKernan, K., Pauli, C., Roe, J., Torres, A., Gaudino, R. (2019). Genomic characterization of the complete terpene synthase gene family from Cannabis sativa. PloS One 14 (9), e0222363. doi: 10.1371/journal.pone.0222363
Alquezar, B., Rodriguez, A., de la Pena, M., Pena, L. (2017). Genomic analysis of terpene synthase family and functional characterization of seven sesquiterpene synthases from Citrus sinensis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01481
Aubourg, S., Lecharny, A., Bohlmann, J. (2002). Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. molecular genetics and genomics 267, 6, 730–745. doi: 10.1007/s00438-002-0709-y
Bohlmann, J., Meyer-Gauen, G., Croteau, R. (1998). Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proceedings of the national academy of sciences of the united states of America. 95, 4126–4133. doi: 10.1073/pnas.95.8.4126
Bouwmeester, H., Schuurink, R. C., Bleeker, P. M., Schiestl, F. (2019). The role of volatiles in plant communication. Plant J. 100 (5), 892–907. doi: 10.1111/tpj.14496
Chen, X. J., Chen, H., Yuan, J. S., Kollner, T. G., Chen, Y. Y., Guo, Y. F., et al. (2018). The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol. J. 16 (10), 1778–1787. doi: 10.1111/pbi.12914
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Tholl, D., Bohlmann, J., Pichersky, E. (2011). The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66 (1), 212–229. doi: 10.1111/j.1365-313X.2011.04520.x
Chen, X. E., Yang, W., Zhang, L. Q., Wu, X. M., Cheng, T., Li, G. L. (2017). Genome-wide identification, functional and evolutionary analysis of terpene synthases in pineapple. Computional Biol. Chem. 70, 40–48. doi: 10.1016/j.compbiolchem.2017.05.010
Dudareva, N., Klempien, A., Muhlemann, J. K., Kaplan, I. (2013). Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 198 (1), 16–32. doi: 10.1111/nph.12145
Dudareva, N., Negre, F., Nagegowda, D. A., Orlova, I. (2006). Plant volatiles: Recent advances and future perspectives. Crit. Rev. Plant Sci. 25 (5), 417–440. doi: 10.1080/07352680600899973
Eduardo, I., Chietera, G., Bassi, D., Rossini, L., Vecchietti, A. (2010). Identification of key odor volatile compounds in the essential oil of nine peach accessions. J. Sci. Food Agric. 90 (7), 1146–1154. doi: 10.1002/jsfa.3932
Falara, V., Akhtar, T. A., Nguyen, T. T., Spyropoulou, E. A., Bleeker, P. M., Schauvinhold, I., et al. (2011). The tomato terpene synthase gene family. Plant Physiol. 157 (2), 770–789. doi: 10.1104/pp.111.179648
Gao, L., Wang, Y., Li, Z., Zhang, H., Ye, J., Li, G. (2016). Gene expression changes during the gummosis development of peach shoots in response to Lasiodiplodia theobromae infection using RNA-seq. Front. Physiol. 7. doi: 10.3389/fphys.2016.00170
Gonzales-Vigil, E., Hufnagel, D. E., Kim, J., Last, R. L., Barry, C. S. (2012). Evolution of TPS20-related terpene synthases influences chemical diversity in the glandular trichomes of the wild tomato relative Solanum habrochaites. Plant J. 71 (6), 921–935. doi: 10.1111/j.1365-313X.2012.05040.x
Hayashi, K., Kawaide, H., Notomi, M., Sakigi, Y., Matsuo, A., Nozaki, H. (2006). Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 580 (26), 6175–6181. doi: 10.1016/j.febslet.2006.10.018
Hong, G. J., Xue, X. Y., Mao, Y. B., Wang, L. J., Chen, X. Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24 (6), 2635–2648. doi: 10.1105/tpc.112.098749
Huang, A. C. C., Jiang, T., Liu, Y. X., Bai, Y. C., Reed, J., Qu, B. Y., et al. (2019). A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364 (6440), eaau6389. doi: 10.1126/science.aau6389
Huang, X. Z., Xiao, Y. T., Kollner, T. G., Jing, W. X., Kou, J. F., Chen, J. Y., et al. (2018). The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell Environ. 41 (1), 261–274. doi: 10.1111/pce.13088
Keeling, C. I., Dullat, H. K., Yuen, M., Ralph, S. G., Jancsik, S., Bohlmann, J. (2010). Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiology 152 3, 1197–1208. doi: 10.1104/pp.109.151456
Keilwagen, J., Lehnert, H., Berner, T., Budahn, H., Nothnagel, T., Ulrich, D., et al. (2017). The terpene synthase gene family of carrot (Daucus carota l.): Identification of QTLs and candidate genes associated with terpenoid volatile compounds. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01930
Kumar, Y., Khan, F., Rastogi, S., Shasany, A. K. (2018). Genome-wide detection of terpene synthase genes in holy basil (Ocimum sanctum l.). PloS One 13 (11), e0207097. doi: 10.1371/journal.pone.0207097
Li, X. W., Jiang, J., Zhang, L. P., Yu, Y., Ye, Z. W., Wang, X. M., et al. (2015). Identification of volatile and softening-related genes using digital gene expression profiles in melting peach. Tree Genet. Genomes 11, 71. doi: 10.1007/s11295-015-0891-9
Lin, J. Y., Wang, D., Chen, X. L., Kollner, T. G., Mazarei, M., Guo, H., et al. (2017). An (E,E)-alpha-farnesene synthase gene of soybean has a role in defence against nematodes and is involved in synthesizing insect-induced volatiles. Plant Biotechnol. J. 15 (4), 510–519. doi: 10.1111/pbi.12649
Liu, H. R., Cao, X. M., Liu, X. H., Xin, R., Wang, J. J., Gao, J., et al. (2017). UV-B irradiation differentially regulates terpene synthases and terpene content of peach. Plant Cell Environ. 40 (10), 2261–2275. doi: 10.1111/pce.13029
Martin, D. M., Aubourg, S., Schouwey, M. B., Daviet, L., Schalk, M., Toub, O., et al. (2010). Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 10, 226. doi: 10.1186/1471-2229-10-226
McGarvey, D. J., Croteau, R. (1995). Terpenoid metabolism. Plant Cell 7 (7), 1015–1026. doi: 10.1105/tpc.7.7.1015
Nieuwenhuizen, N. J., Chen, X. Y., Wang, M. Y., Matich, A. J., Perez, R. L., Allan, A. C., et al. (2015). Natural variation in monoterpene synthesis in kiwifruit: Transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-Like transcription factors. Plant Physiol. 167 (4), 1243–1258. doi: 10.1104/pp.114.254367
Nieuwenhuizen, N. J., Green, S. A., Chen, X. Y., Bailleul, E. J. D., Matich, A. J., Wang, M. Y., et al. (2013). Functional genomics reveals that a compact terpene synthase gene family can account for terpene volatile production in apple. Plant Physiol. 161 (2), 787–804. doi: 10.1104/pp.112.208249
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 (3), 290–295. doi: 10.1038/nbt.3122
Sapir-Mir, M., Mett, A., Belausov, E., Tal-Meshulam, S., Frydman, A., Gidoni, D., et al. (2008). Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiol. 148 (3), 1219–1228. doi: 10.1104/pp.108.127951
Sawada, Y., Akiyama, K., Sakata, A., Kuwahara, A., Otsuki, H., Sakurai, T., et al. (2009). Widely targeted metabolomics based on Large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol. 50, 37–47. doi: 10.1093/pcp/pcn183
Sharon-Asa, L., Shalit, M., Frydman, A., Bar, E., Holland, D., Or, E., et al. (2003). Citrus fruit flavor and aroma biosynthesis: Isolation, functional characterization, and developmental regulation of Cstps1, a key gene in the production of the sesquiterpene aroma compound valencene. Plant J. 36 (5), 664–674. doi: 10.1046/j.1365-313x.2003.01910.x
Shimada, T., Endo, T., Fujii, H., Rodriguez, A., Pena, L., Omura, M. (2014). Characterization of three linalool synthase genes from Citrus unshiu marc. and analysis of linalool-mediated resistance against Xanthomonas citri subsp citri and Penicilium italicum in citrus leaves and fruits. Plant Sci. 229, 154–166. doi: 10.1016/j.plantsci.2014.09.008
Shimada, T., Endo, T., Rodriguez, A., Fujii, H., Goto, S., Matsuura, T., et al. (2017). Ectopic accumulation of linalool confers resistance to Xanthomonas citri subsp citri in transgenic sweet orange plants. Tree Physiol. 37 (5), 654–664. doi: 10.1093/treephys/tpw134
Song, C. K., Hartl, K., McGraphery, K., Hoffmann, T., Schwab, W. (2018). Attractive but toxic: Emerging roles of glycosidically bound volatiles and glycosyltransferases involved in their formation. Mol. Plant 11 (10), 1225–1236. doi: 10.1016/j.molp.2018.09.001
Sun, Y., Huang, X. Z., Ning, Y. S., Jing, W. X., Bruce, T. J. A., Qi, F. J., et al. (2017). TPS46, a rice terpene synthase conferring natural resistance to bird cherry-oat aphid, Rhopalosiphum padi (Linnaeus). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00110
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11 molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tholl, D., Chen, F., Petri, J., Gershenzon, J., Pichersky, E. (2005). Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 42 (5), 757–771. doi: 10.1111/j.1365-313X.2005.02417.x
Vranova, E., Coman, D., Gruissem, W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700. doi: 10.1146/annurev-arplant-050312-120116
Wang, F., Zhao, L. N., Li, G. H., Huang, J. B., Hsiang, T. (2011). Identification and characterization of Botryosphaeria spp. causing gummosis of peach trees in hubei province, central China. Plant Dis. 95 (11), 1378–1384. doi: 10.1094/PDIS-12-10-0893
Wei, C. Y., Li, M. T., Cao, X. M., Jin, Z. N., Zhang, C., Xu, M., et al. (2022). Linalool synthesis related PpTPS1 and PpTPS3 are activated by transcription factor PpERF61 whose expression is associated with DNA methylation during peach fruit ripening. Plant Sci. 317, 111200. doi: 10.1016/j.plantsci.2022.111200
Wei, C. Y., Liu, H. R., Cao, X. M., Zhang, M. L., Li, X., Chen, K. S., et al. (2021). Synthesis of flavour-related linalool is regulated by PpbHLH1 and associated with changes in DNA methylation during peach fruit ripening. Plant Biotechnol. J. 19 (10), 2082–2096. doi: 10.1111/pbi.13638
Wydro, M., Kozubek, E., Lehmann, P. (2006). Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim. Polonica. 53 (2), 289–298. doi: 10.18388/abp.2006_3341
Yang, Z. Z., Li, Y. Q., Gao, F. Z., Jin, W., Li, S. Y., Kimani, S., et al. (2020). MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 71 (14), 4140–4158. doi: 10.1093/jxb/eraa184
Ye, Z. W., Li, X. W., Zhou, J. Y., Su, M. S., Ma, Y. P., Wang, N. N., et al. (2020). Advances in peach gummosis disease. Acta Agriculturae Shanghai 36 (2), 146–150. doi: 10.15955/j.issn1000-3924.2020.02.24
Yoshitomi, K., Taniguchi, S., Tanaka, K., Uji, Y., Akimitsu, K., Gomi, K. (2016). Rice terpene synthase 24 (OsTPS24) encodes a jasmonate-responsive monoterpene synthase that produces an antibacterial gamma-terpinene against rice pathogen. J. Plant Physiol. 191, 120–126. doi: 10.1016/j.jplph.2015.12.008
Yu, Z., Zhao, C., Zhang, G., Teixeira da Silva, J. A., Duan, J. (2020). Genome-wide identification and expression profile of TPS gene family in Dendrobium officinale and the role of DoTPS10 in linalool biosynthesis. Int. J. Mol. Sci. 21 (15), 5419. doi: 10.3390/ijms21155419
Zhang, P., Liu, X. F., Xu, X., Wang, F. S., Long, J. H., Shen, W. X., et al. (2020). The MYB transcription factor CiMYB42 regulates limonoids biosynthesis in citrus. BMC Plant Biol. 20, 254. doi: 10.1186/s12870-020-02475-4
Zhang, A. D., Xiong, Y. H., Fang, J., Jiang, X. H., Wang, T. F., Liu, K. C., et al. (2022). Diversity and functional evolution of terpene synthases in rosaceae. Plants (Basel) 11 (6), 736. doi: 10.3390/plants11060736
Zhang, A. D., Zhou, H., Jiang, X. H., Han, Y. P., Zhang, X. J. (2021). The draft genome of a flat peach (Prunus persica l. cv. '124 pan') provides insights into its good fruit flavor traits. Plants (Basel) 10 (3), 538. doi: 10.3390/plants10030538
Keywords: peach (Prunus persica L.), terpene synthase genes, gummosis disease, terpenoids, linalool
Citation: Li X, Hu Y, Su M, Zhang M, Du J, Zhou H, Zhang X and Ye Z (2022) Genome-wide analysis of terpene synthase gene family to explore candidate genes related to disease resistance in Prunus persica. Front. Plant Sci. 13:1032838. doi: 10.3389/fpls.2022.1032838
Received: 31 August 2022; Accepted: 28 September 2022;
Published: 31 October 2022.
Edited by:
Victoria Pastor, University of Jaume I, SpainReviewed by:
Jian Guo, Shandong Agricultural University, ChinaCopyright © 2022 Li, Hu, Su, Zhang, Du, Zhou, Zhang and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengwen Ye, eWV6aGVuZ3dlbjEzMDBAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.