
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 24 November 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1031679
Tomato is a drought-sensitive crop which has high susceptibility to adverse climatic changes. Dehydration-responsive element-binding (DREB) are significant plant transcription factors that have a vital role in regulating plant abiotic stress tolerance by networking with DRE/CRT cis-regulatory elements in response to stresses. In this study, bioinformatics analysis was performed to conduct the genome-wide identification and characterization of DREB genes and promoter elements in Solanum lycopersicum. In genome-wide coverage, 58 SlDREB genes were discovered on 12 chromosomes that justified the criteria of the presence of AP2 domain as conserved motifs. Intron–exon organization and motif analysis showed consistency with phylogenetic analysis and confirmed the absence of the A3 class, thus dividing the SlDREB genes into five categories. Gene expansion was observed through tandem duplication and segmental duplication gene events in SlDREB genes. Ka/Ks values were calculated in ortholog pairs that indicated divergence time and occurrence of purification selection during the evolutionary period. Synteny analysis demonstrated that 32 out of 58 and 47 out of 58 SlDREB genes were orthologs to Arabidopsis and Solanum tuberosum, respectively. Subcellular localization predicted that SlDREB genes were present in the nucleus and performed primary functions in DNA binding to regulate the transcriptional processes according to gene ontology. Cis-acting regulatory element analysis revealed the presence of 103 motifs in 2.5-kbp upstream promoter sequences of 58 SlDREB genes. Five representative SlDREB proteins were selected from the resultant DREB subgroups for 3D protein modeling through the Phyre2 server. All models confirmed about 90% residues in the favorable region through Ramachandran plot analysis. Moreover, active catalytic sites and occurrence in disorder regions indicated the structural and functional flexibility of SlDREB proteins. Protein association networks through STRING software suggested the potential interactors that belong to different gene families and are involved in regulating similar functional and biological processes. Transcriptome data analysis has revealed that the SlDREB gene family is engaged in defense response against drought and heat stress conditions in tomato. Overall, this comprehensive research reveals the identification and characterization of SlDREB genes that provide potential knowledge for improving abiotic stress tolerance in tomato.
Tomato (Solanum lycopersicum L.) is an economically valuable fruit worldwide. Its yield and development are acutely affected by harsh environmental conditions such as drought, salinity, and extreme temperatures. Salinity and drought stress conditions lead to harmful physiological effects—for instance, stunted plant growth, failure in seed germination, and reduction in fruit yield. Moreover, drought causes osmotic stress that results in the decrease of photosynthetic rate and plant development (Diouf et al., 2018; Zhou et al., 2019). In the FAO 2019 report, the data stated a substantial increase in tomato production to 182 million tons from 37 million tons that was in 2017.
The dehydration-responsive element-binding (DREB) gene family is one of the most prominent transcription regulators that bind with DRE/cis regulatory elements (CREs) to induce tolerance under biotic and abiotic stresses in plants (Du et al., 2018). It is also known as cis-binding factor, belongs to the ethylene-responsive element-binding factor (ERF) family, and contains conserved AP2 domain (Khan, 2011). The presence of valine (V) and glutamic acid (E) at the 14th and 19th positions in the AP2 domain adds more clarity to the DREB family from its other sub-family members. The AP2 domain contains 60 to 70 amino acid sequences that are responsible for plant defense mechanisms to combat stressful circumstances (Wu et al., 2015). YRG (19–22 aa long) and RAYDS are two conserved elements that are characteristic features of the AP2 domain. The core region of RAYD is supposed to be involved in the existence of an amphipathic α-helix (Chai et al., 2020). DREB is a sub-family of ERF transcription factors with conserved AP2 domain. The three-dimensional structure of AP2 domain was revealed in AtERF1 gene through nuclear magnetic resonance imaging that demonstrated the presence of an α-helix and three antiparallel β-sheets in its structural organization (Lata and Prasad, 2011; Li Z. et al., 2021). DREB genes attach to drought-responsive (DRE/CRT) elements with the assistance of a V residue, two W residues, and four R residues (Mushtaq et al., 2021). The phosphorylation of DREB genes is essential for their activation, supported by a threonine- or serine-rich sequence present next to the AP2 domain (Lata and Prasad, 2011). The DRE/CRT elements have a core motif consisting of ACCGAC/GCCGAC that directly interacts with DREB genes under abiotic stresses (Vazquez-Hernandez et al., 2017). The DREB gene family further splits into six classes, i.e. A1–A6, based on their fundamental structural and functional characteristics (Chai et al., 2020).
Subsequently, the DREB gene family has been identified in a number of plants including Arabidopsis (Hwang et al., 2012), pearl millet, strawberry, tobacco (Cheng et al., 2013), rice, lettuce (Park et al., 2020), bananas, wheat, bell pepper, maize (Liu et al., 2013), soybean (Hou et al., 2022), and potato (Mushtaq et al., 2021). Previous studies have identified DREB1, DREB2, and DREB3 in tomatoes. SlDREB1 has a remarkable role in response to drought stress. It enhances the concentration of soluble sugars and accumulates osmolytes that further balance the osmotic pressure and increase the tolerance response under water-deficient conditions (Jiang et al., 2017). SlDREB2 is involved in the regulation of stress signaling pathways. Studies have shown its contribution in the development of early vegetative and reproductive stages. It regulates the synthesis of proline that further assists plants in response to salt and hormonal changes (Hichri et al., 2016), whereas SlDREB3 is a negative regulator of the abscisic acid (ABA) pathway by altering ABA signaling. It is also known as the ripening gene by controlling the ethylene-dependent and ethylene-independent pathways (Gupta et al., 2022).
Despite this, limited studies have been conducted on the characterization of the DREB gene family in S. lycopersicum. The present research data extensively covers phylogenetic evaluation, gene structure analysis, conserved motifs, chromosomal localization, gene duplication events, synteny analysis for the detection of orthologues in interspecies, gene ontology, characterization of cis-regulatory elements, prediction of subcellular localization, protein modeling, prediction of pocket binding sites, disorder region analysis, and interactive networks with other proteins. Overall, our data has the potential information that serves as a strong infrastructure for unravelling the extensive structural and functional relationships of intricate genes for genetic engineering to enhance tolerance in tomato genotypes under environmental stresses.
To retrieve the amino acid sequences of DREB family members, the amino acid sequences of DREB genes of Arabidopsis thaliana were retrieved from the TAIR database (https://www.arabidopsis.org/) (Huala et al., 2001). These amino acid sequences were used as query sequences in BLASTp for the retrieval and identification of DREB family members in Solanum lycopersicum from Sol Genomics Networks (SGN) (https://solgenomics.net/). The criteria for BLASTp homology search was set to 1 × 10-5 against Solanum lycopersicum genome v4.0 (Mueller et al., 2005).
ScanProsite Server (https://prosite.expasy.org/scanprosite/) was used to identify a single AP2 domain, its sequence, and amino acid length throughout the DREB family (De Castro et al., 2006) NCBI CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (Yang et al., 2020) and Pfam (http://pfam.xfam.org/) (Mistry et al., 2021) were used for the verification of the complete AP2 domain. The ExPASY protparam tool (https://web.expasy.org/protparam/) (Gasteiger et al., 2005) was used to measure physicochemical properties such as the molecular weight (kDa), protein length, and isoelectric point (pI) of DREB protein sequences.
Verification of the conserved amino acid valine (V) at the 14th position and glutamic acid (E) at the 19th position alignment was conducted throughout the DREB protein sequences in S. lycopersicum which were retrieved through BLASTp using MEGA 11 software (www.megasoftware.net) using default settings (Tamura et al., 2021).
To classify SlDREB family members, retrieved amino acid sequences of Arabidopsis and S. lycopersicum were used. MEGA11 (www.megasoftware.net) software was used with default parameters for the multiple sequence alignment. Criteria for the construction of phylogenetic trees on MEGA 11 software (www.megasoftware.net) was set to maximum likelihood method with pairwise deletion, using the Poisson substitution model with bootstrap set to 1000 replicates (Tamura et al., 2021). The phylogenetic tree was used for the DREB family members classification in S. lycopersicum based on the classes in AtDREB proteins.
To generate the gene structure graph, Gene Structure Display Server (GSDS 2.0) (http://gsds.gao-lab.org0) was used which shows the number of introns and exons within a sequence (Hu et al., 2015). For this purpose, genomic and coding sequences (CDs) of all SlDREB genes were retrieved from the Phytozome. Comparison between these two sequences assist to generate intron-exon graphical illustration.
To detect motifs within the classes of SlDREB family members, MEME software version 5.3.2 (https://memesuite.org/meme/tools/meme) was selected and set parameters such as (i) an optimal width of motifs was 6-50 amino acids, (ii) zero or one occurrence per sequence of a single motif within the model, and (iii) number of motifs set to 20 (Bailey et al., 2015).
To map chromosomal positions of SlDREB genes, Toolkit Biologists Tools (TBtools) (https://github.com/CJ-Chen/TBtools) (Chen et al., 2018) software was used for the illustration of distances and physical location of SlDREBs.To evaluate respective time of divergence and selective pressure, Toolkit Biologists Tools (TBtools) software (https://github.com/CJ-Chen/TBtools) was used to measure the Ka and Ks values that denote non-synonymous and synonymous values. To calculate divergence time following formula was used T=Ks/2x*MYA, where x=6.56 x 10-9 and MYA= 10-6 (Yuan et al., 2015). Gene expansion follows two methods in gene duplication events. First method is tandem repeats if two genes present on the same chromosome are separated by fewer loci. Second method of gene expansion is segmental duplication (Laha et al., 2020).
For orthologues and synteny analysis of SlDREB genes with A. thaliana and S. tuberosum, gff3 annotation and genomic files were downloaded from Phytozome 13v database. To access the segmental duplication gene pairs Circos plot was constructed through Advanced Circos software and Dual synteny plotter was used for the visualization of orthologues of S. lycopersicum in the A. thaliana and S. tuberosum using TB Tools (https://github.com/CJ-Chen/TBtools) respectively.
To interpret Gene Ontology (GO), transcript sequences of SlDREBs were uploaded to Blast2Go software (https://www.blast2go.com/) (Conesa and Götz, 2008). Functions like functional annotation, BLASTx search, InterProScan, mapping, and annotation were executed with default parameters. Furthermore, these sequences were subjected to the investigation and graphical representation of biological processes (BP), cellular compartments, and molecular functions. The WoLF PSORT tool (https://wolfpsort.hgc.jp/) was used for the estimation of the subcellular localization of DREB genes (Horton et al., 2007).
To examine the promoter regions, Phytozomev13 database (https://phytozome.jgi.doe.gov/pz/portal.html) (Goodstein et al., 2012) was used for the retrieval of a 2.5-kbp upstream promoter sequence of the SlDREB genes of S. lycopersicum v4.0 genome. The promoter is an upstream region from the start site (ATG) which encodes for methionine (M) that signals for the translational start site. SGN (https://solgenomics.net/) was used to validate these promoter sequences. Plant CARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002) was used for the estimation of CREs. Toolkit Biologists Tools (TBtools) software was used to illustrate the graphical positions of CREs in sequences.
Five members were selected from five subgroups identified from the present study. The amino acid sequences of these members were used to perform secondary structure evaluation using the SOPMA server (https://npsaprabi.ibcp.fr/NPSA/npsa_sopma.html) (Geourjon and Deleage, 1995). To predict the 3D structure, the Phyre2 server (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) was employed on selected amino acid sequences. This software used modern methods for homology detection and assisted in the construction of reliable 3D structures (Kelley et al., 2015).
The predicted models were saved in PDB format. These models were subjected to Galaxy Refine server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) (Heo et al., 2013), PROCHECK server, and Ramachandran plot analysis (https://servicesn.mbi.ucla.edu/PROCHECK/) (Laskowski et al., 2006) that assist in model refinement and structural validation. To visualize the refined stable structures of SlDREB proteins, pdb-format files were utilized in ChimeraX (http://www.rbvi.ucsf.edu/chimerax/) software (Pettersen et al., 2021). To estimate the active catalytic sites, CASTp 3.0 (http://sts.bioe.uic.edu/) web server was used, whichs point out multiscale pockets on the structures of selected SlDREBs members (Tian et al., 2018). PrDOS (Protein DisOrder prediction System) server (http://prdos.hgc.jp/cgi-bin/top.cgi) (Ishida and Kinoshita, 2007) was utilized for the prediction of disorder regions within protein with default settings.
Proteins interact with many other proteins either belonging to the same class or different groups in response to regulating different signaling pathways. To evaluate the interaction of SlDREB proteins with other interactors, protein sequences of the selected SlDREB proteins were utilized in STRING Tool (Search Tool for the Retrieval of Interacting Genes; https://string-db.org/) (Szklarczyk et al., 2019) for the construction of interactive networks. The criterion for the confidence threshold was set to 0.40, and a maximum number of 10 interactors were selected in the first shell. Interactive networks showed the interconnection of query protein with other 10 interactors; some of them are interconnected.
The tomato transcriptome was retrieved from the Bio project NCBI (PRJNA635375) (https://www.ncbi.nlm.nih.gov/bioproject/635375). It encompasses gene expression data under drought and heat stresses through the Gene Expression Omnibus (GEO) NCBI (GSE151277) (https://www.ncbi.nlm.nih.gov/geo/) (Barrett et al., 2005). GEO datasets were refined through TBtools (https://github.com/CJ-Chen/TBtools) by using a table file manipulator to extract the gene expression of 58 SlDREB genes present in the tomato transcriptome. Subsequently, log2 base values were calculated for the normalization of the original dataset. An expression heat map was constructed through TBtools heat map illustrator to illustrate the upregulation and downregulation of SlDREB genes under drought and heat stress conditions.
Our results revealed 58 DREB protein sequences from tomato genome that show the maximum percentage identity and coverage with query sequences. The retrieved sequences, which were presented as SlDREB1 to SlDREB58, belong to DREB and ERF family according to Phytozome annotations (Supplementary Table S1). Scan prosite Server and CDD NCBI confirmed the occurrence of one conserved AP2 domain within each of the 58 SlDREB sequences. The chromosomal location, conserved domain length, protein length, and physiological characteristics such as isoelectric point and molecular weight (Mw) are indicated in Supplementary Table S1. The protein sequences of 58 SlDREB proteins are presented in Supplementary Table S2.
Multiple sequence alignment proved that the amino acids valine (V) and glutamic acid (E) at positions 14th and 19th within the AP2 domain were not conserved in the case of SlDREB sequences as demonstrated in Figure 1. Three SlDREB protein sequences (SlDREB6, SlDREB45, and SlDREB47) contain isoleucine (I) instead of valine (V) at position 14th. I can only replace V due to the same characteristics such as hydrophobicity, aliphatic, and nonpolar in physical as well as chemical features such as polarity. Among the 58 SlDREB sequences, 36 SlDREBs have (V) and (E) at the 14th and 19th positions. Glutamic acid (E) at 19th position is replaced by different amino acids like leucine (L) in seven SlDREBs, histidine (H) in six SlDREBs, valine (V) in two SlDREBs, glutamine in four SlDREBs, alanine (A) in one SlDREB, and aspartic acid (D) in two SlDREB sequences, respectively.

Figure 1 Multiple sequence alignment of S. lycopersicum dehydration-responsive element-binding (DREB) proteins. MEGA 11 software was employed using MUSCLE algorithm. The highlighted lines represent conserved amino acids and motifs, whereas motifs and amino acids enclosed in black lines represent variability and have active roles in protein structure and functionality.
Among these amino acids, E and D are hydrophilic, positively charged, and polar, Q is hydrophilic, uncharged, and polar, H is hydrophilic, positively charged, heterocycle, and polar, L and V are hydrophobic and non-polar, and A is hydrophobic, neutral, and non-polar in nature correspondingly (Supplementary Table S1). These sequences have a conserved WLG motif in its AP2 domain. All these amino acid modifications and features have a prominent effect on the binding capacity and interactions of SlDREB genes with DRE/GCC boxes and with other proteins in signaling pathways.
To identify DREB gene sub-classes, a phylogenetic analysis was assessed through MEGA 11 using MUSCLE algorithm at 1,000 bootstraps through the maximum likelihood method. Based on the classification in A. thaliana DREB family, the phylogenetic tree proved the absence of A3 sub-class in SlDREB genes because no SlDREB gene showed a homology with AT2G40220.1 (green color) which belongs to A3 sub-class as in Figure 2. SlDREB genes that show greater homology with each other are structurally more closely related than the AtDREB genes. A further investigation of the motifs and gene structures of the 58 SlDREB genes will prove their high homology.
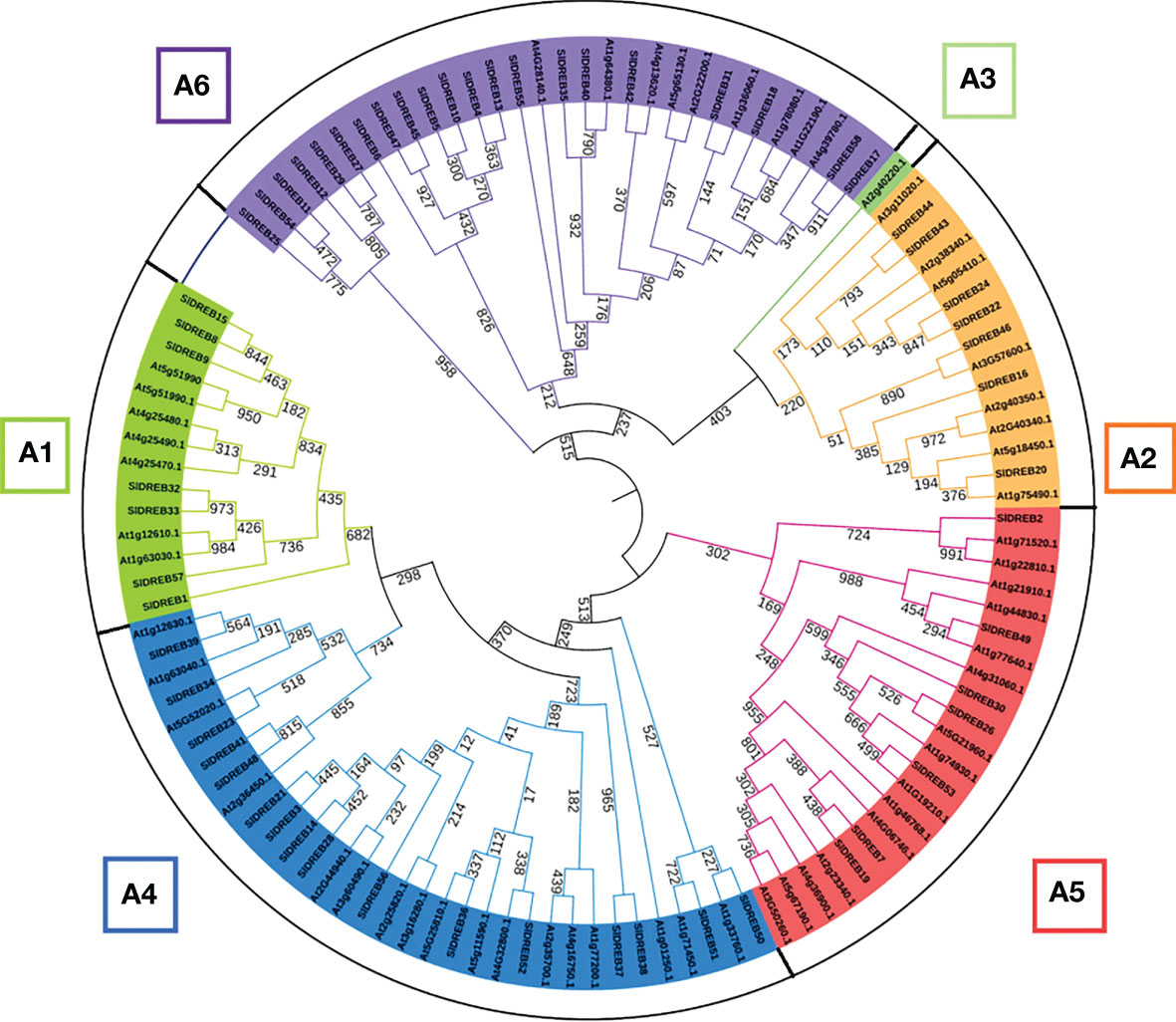
Figure 2 Phylogenetic tree of dehydration-responsive element-binding proteins of S. lycopersicum and A. thaliana. MEGA 11 software was utilized with 1,000 bootstrap and maximum likelihood method. Each color represents a specific class.
Gene structure is very crucial in the analysis of structural diversity of duplicated genes. Intron and exon organization reflects intuition about the structural diversity and similarity of DREB genes belonging to different classes in tomato. For the graphical illustration of gene structure organization, full-length cDNA was aligned and compared with genomic DNA of SlDREB sequences (Supplementary Table S2). The results indicated that only one intron and two exons were present in nine SlDREB genes (11, 12, 25, 27, 29, 34, 39, 42, and 58) and two introns and three exons were observed in the coding region of three SlDREB genes (17, 47, and 54). Except these 12 SlDREB genes, 46 other genes had only one exon in its coding region subsequent to the phylogenetic tree (Figure 3). A detailed motif analysis was conducted through MEME software as summarized in Supplementary Table S3). It assisted in the clarity of diversity among 58 SlDREB genes belonging to different classes. The results showed the variability in motifs of one class from another (Supplementary Table S4). A total of 20 motifs were discovered, from which motifs 1, 2, and 3 evaluated the presence of the AP2 domain which is a characteristic domain of DREB proteins. Motifs 1, 2, and 3 were present in all SlDREB proteins, with some degree of divergence except SlDREB7 (which belongs to A5 class) which showed the absence of motif 3 only. A1 class had motif 6 in all of its members and motif 17 in SlDREBs 32, 33, and 57. In A2 class, motif 7 was present in all of its members except SlDREB43, with motif 16 in SlDREBs 43 and 44 and motif 20 in SlDREBs 22 and 23. A4 class had motifs 10 and 12 in its members as follows: SlDREB 3, 21, 37, and 38 and SlDREB 37 and 38, respectively. A5 class shared only a few motifs with A1 and A4 classes. The following motifs corresponding to SlDREB genes were identified specifically in A6 class: motif 8 in 17, 18, 35, 40, and 58; motif 9 in 11, 25, and 54; motif 11 in 11, 12, 27, 25, 29, and 54; motif 14 in 5, 45, and 47; motif 15 in 5, 17, 18, 45, 47, and 55; and motifs 18 and 19 in 35 and 40 SlDREB genes, respectively. Motif 4 was observed in the entire A1 and A4 classes and in a member of A5 class (SlDREB49). Motif 5 was detected in a few members of A1 and A4 classes. Motif 13 was identified in A1, A4, and A5 groups. The same motif structure was observed among members of the same class. A motif analysis compared with a phylogenetic tree proved the evolutionary similarity of A1, A4, and A5 sub-classes that appeared in the same clade and their diversity with A2 and A6 classes which were associated in the second clade as presented in Figure 3.
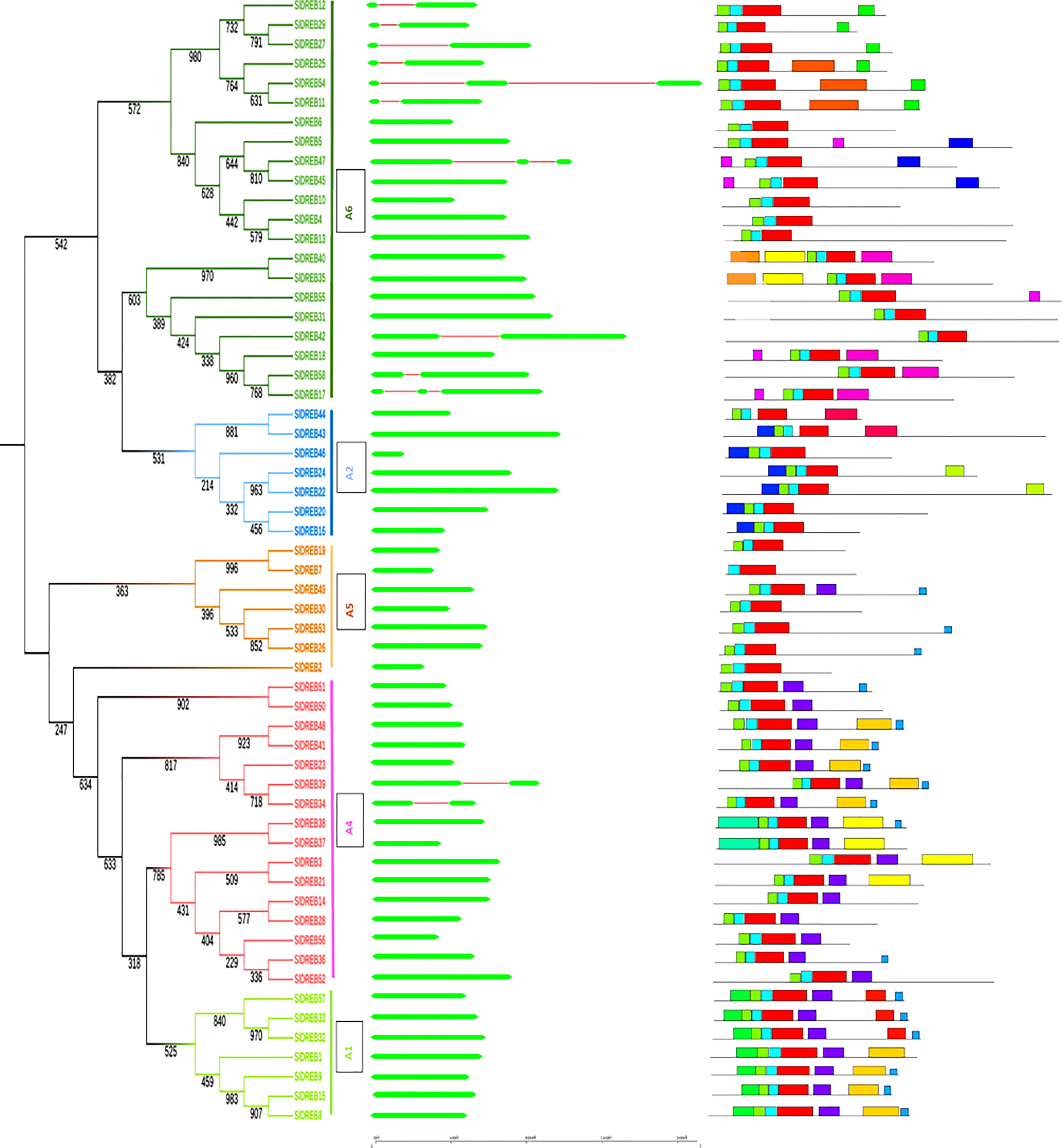
Figure 3 Intron–exon organizations of SlDREB genes. Gene structure was analyzed using Gene Structure Display Server. Here exons and introns are represented with green and red colors, respectively, whereas the scale shows the corresponding sizes of the genes. Twenty motifs predicted through MEME software within SlDREB protein sequences. The motifs were differentiated by distinctly colored boxes, whereas gray lines represented non-conserved regions.
To localize the 58 SlDREB genes, chromosomal localization was retrieved from Phytozome v.13 with its full length. TB Tools software was utilized for the physical mapping and distribution of all SlDREB genes across the tomato genome. All 58 SlDREB genes were represented in different colors that denoted their respective classes as indicated in the rectangular phylogenetic tree. Mapping demonstrated that genetic variation was observed in the gene distribution on 12 chromosomes. Chromosome 8 (chr08) had nine SlDREB genes positioned on it, which is the largest number compared with other chromosomes. However, three chromosomes (chr05, chr07, and chr09) comprised the least number of SlDREB genes which is 2. Chromosome 1 (chr01) had four SlDREB genes, chromosomes 2 and 11 (chr02 and 11) had three SlDREB genes, chromosome 3 (chr03) was comprised of eight SlDREB genes, chromosome 4 (chr04) consisted of five SlDREB genes, chromosomes 6 and 12 (chr06 and chr12) covered seven SlDREB genes, and chromosome 10 (chr10) positioned six SlDREB genes, respectively, as depicted in Figure 4. The plant genome undergoes genomic evolution due to gene expansion events that introduce new interactions within a genome and assist in the sustainability of unique functions. Two types of duplication events were observed in the tomato genome: tandem and segmental duplication. Tandem duplications were observed in eight gene pairs in which chromosome 8 (chr08) covered a large number of gene pairs which is three, namely, SlDREB 32–33, 34–39, and 35–40. Conversely, chromosomes 3, 6, and 11 (chr03, chr06, and chr11) was each comprised of only one gene pair (8–15), (27–29), and (50–51). Two pairs were positioned on chromosome 10 (chr10), which were SlDREB 43–44 and 45–47, represented by the red lines as given in Figure 4. However, segmental gene duplication event was observed in seven SlDREB gene pairs (36–52, 26–53, 11–54, 41–48, 17–58, 14–28, and 22–24) positioned on different chromosomes linked though the red lines in the Circos plot as illustrated in Figure 5.
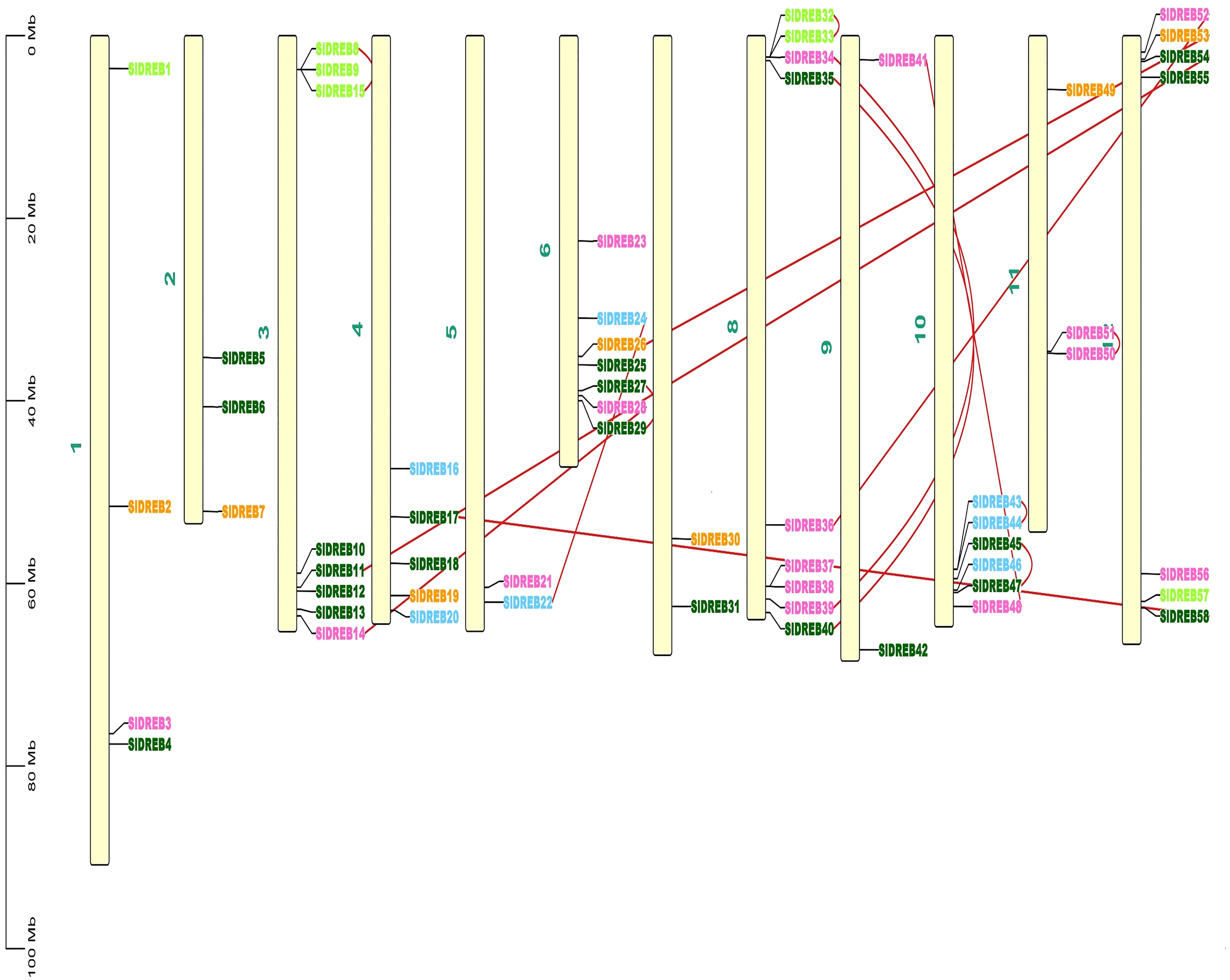
Figure 4 Chromosomal distribution of 58 SlDREB genes within the genome was illustrated by using TBtools. Each color of the SlDREB genes represents its class according to the phylogenetic tree. The scale shows the length of each chromosome. Red lines that linked the gene pairs present on the same chromosome denoted tandem duplication events.
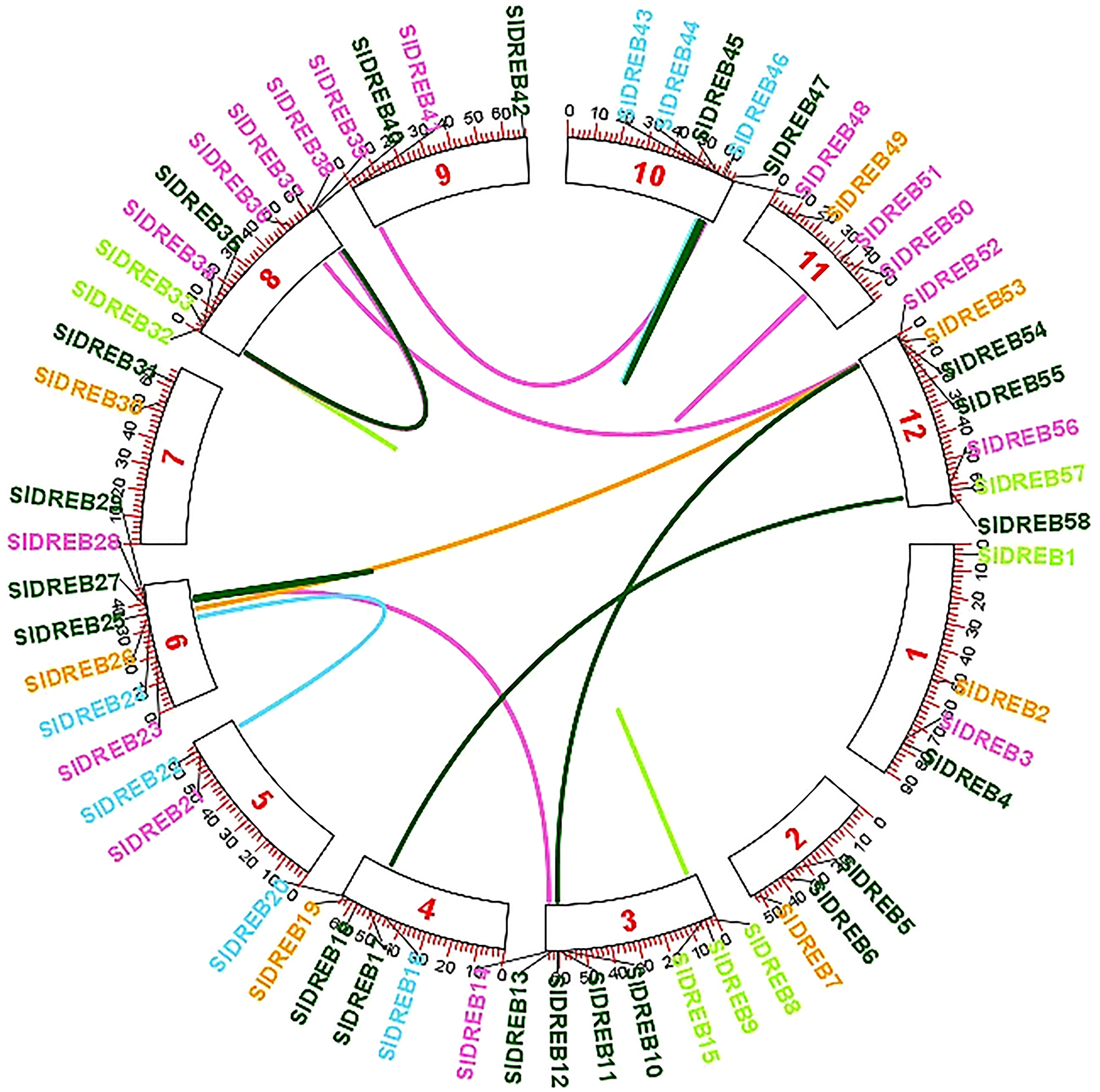
Figure 5 Segmental gene duplications were exhibited on the Circos plot of 58 SlDREB genes that was generated by the respective positions of each gene across 12 chromosomes. Colored lines indicated the segmental gene duplications among gene pairs situated on different chromosomes.
The Ka/Ks values of these 15 SlDREB gene pairs were measured for the estimation of divergence time and type of selection in each pair. If the Ka/Ks value is equal to 1, then the gene pair shows neutral selection. Although purification selection occurred in a gene pair if the Ka/Ks value was less than 1, on the contrary, Ka/Ks values less than 1 were denoted as a positive selection. In 15 pairs of SlDREB genes, duplication events occurred through purification selection and had a divergence time between 4.0 and 50 million years ago as represented in Supplementary Table S5. Synteny analysis was conducted for the evolutionary relationship that exhibited the collinearity of SlDREB genes in A. thaliana and in S. tuberosum by comparing their genomes with S. lycopersicum (Figure 6). The syntenic map revealed that 32 out of 58 SlDREB genes displayed a high-homology relationship in A. thaliana, whereas 47 out of 58 SlDREB genes revealed their orthologs in S. tuberosum as shown in Supplementary Table S5.
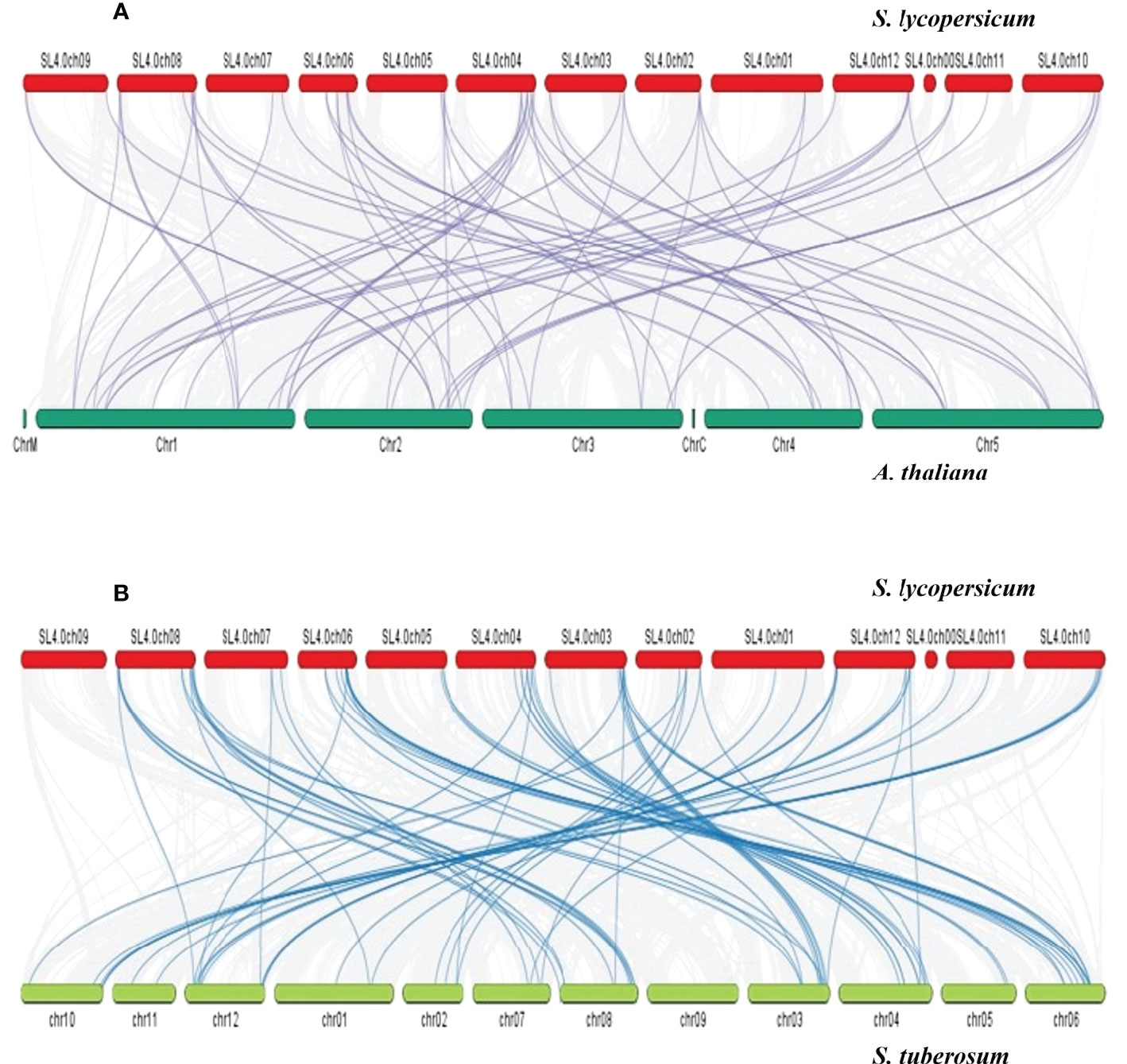
Figure 6 Synteny analysis was conducted among (A) S. lycopersicum and A thaliana and (B) S. lycopersicum and S. tuberosum for homology and evolutionary relationship. The red, dark green, and green bars denote the chromosomes of S. lycopersicum, A thaliana, and S. tuberosum. The gray lines indicate the collinear block within genomes. The purple and dark blue lines were designated to the respective homolog dehydration-responsive element-binding genes present between genomes.
GO annotations and Eggnog descriptions were performed on 58 SlDREB genes, followed by gene mapping, InterPro scan, and blast hits. Molecular functions, biological functions, and localization in cellular compartments of SlDREB genes were estimated. The results revealed, as shown in Figure 7A, that SlDREB genes were specifically involved in the transcriptional regulations, sequence-specific DNA binding that proved the involvement of these genes in stress-responsive gene regulations, growth, and development of tomato. Under molecular function analysis, SlDREB genes were involved in the following processes: 58 SlDREB genes play a role in defense mechanisms, 51 SlDREB genes regulate transcription factors, seven genes (SlDREB 16, 20, 22, 24, 43, 44, and 46) were involved in positive transcriptional regulation, two SlDREBs (11 and 54) respond to water deprivation, two SlDREBs (11 and 54) were linked with cutin biosynthesis, and two genes (SlDREB 11 and 54) were involved in the metabolic process as given in Figure 7B. The GO analysis showed that all 58 SlDREB genes were localized in the nucleus, as shown in Figure 7C and supported by Supplementary Table S6. The further subcellular localization of SlDREB genes was confirmed by WoLF PSORT software, and the results were depicted in the form of a heat map which exposed that SlDREB genes were highly concentrated in the nucleus following 13 other organelles, as shown in Figure 8 (Supplementary Table S6).
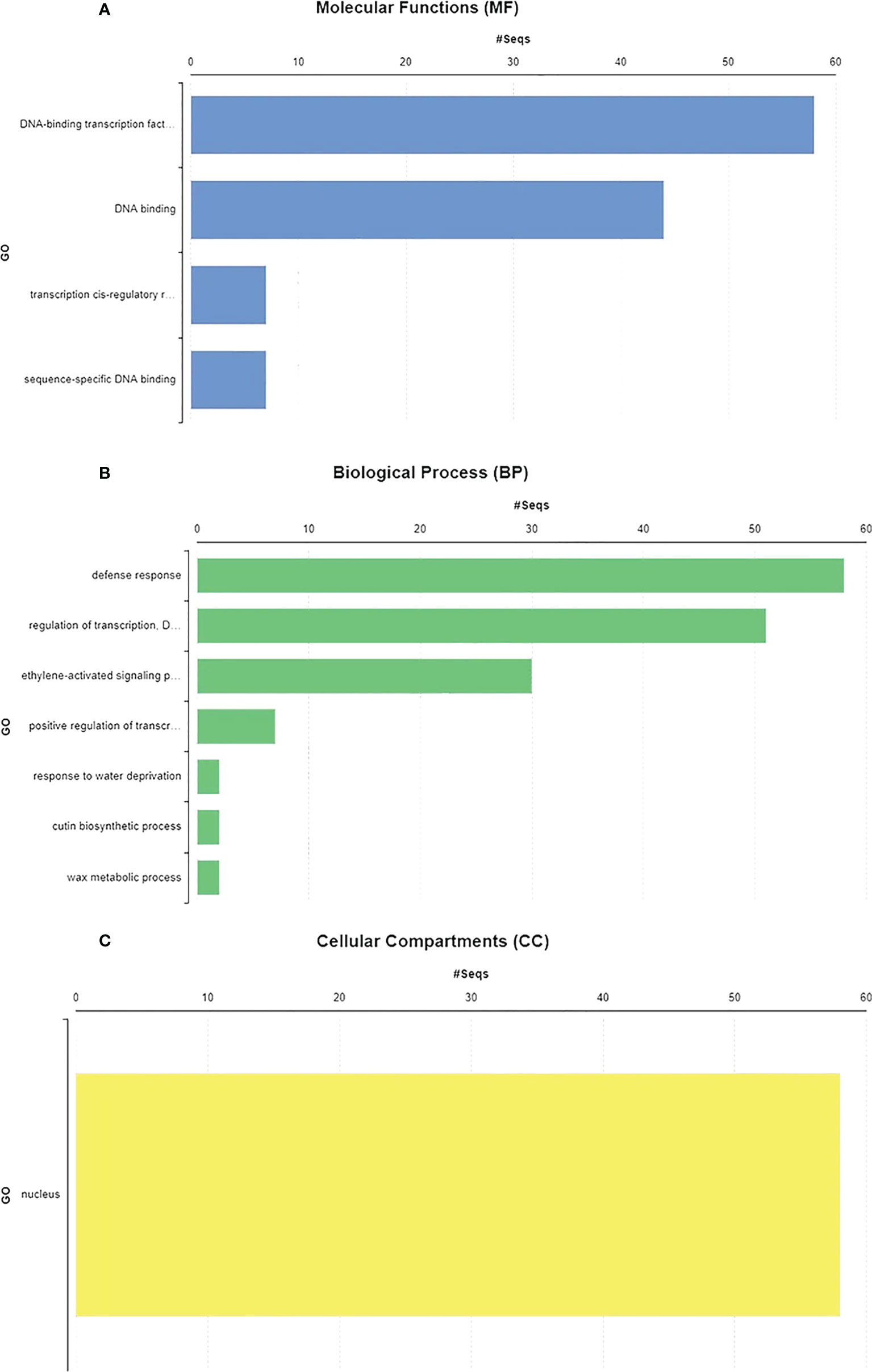
Figure 7 Gene ontology and functional analysis were performed by using Blast2GO software. (A) SlDREB genes involved in the respective molecular functions. (B) Biological processes conducted by these genes. (C) Cellular compartments comprised by SlDREB genes.
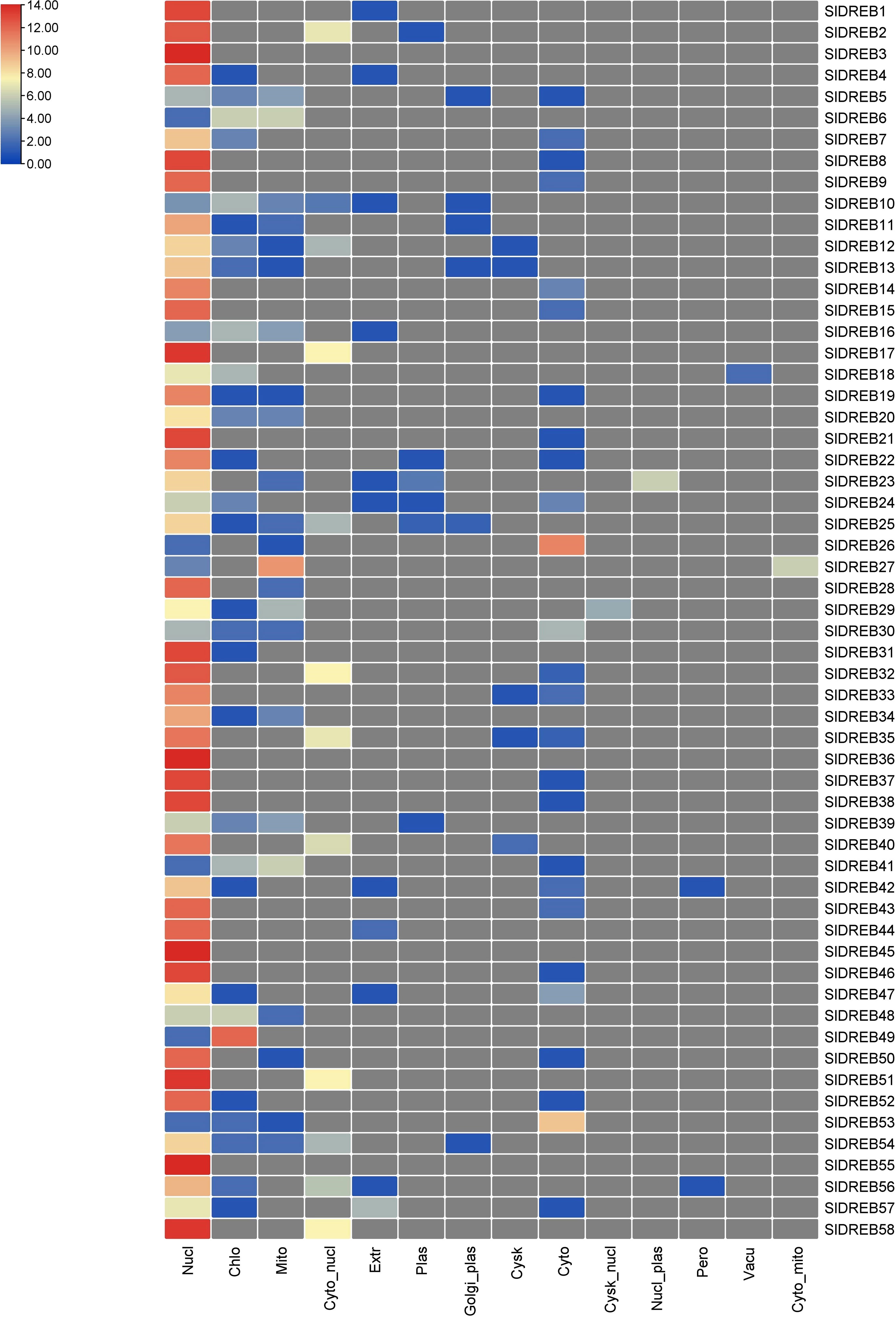
Figure 8 Illustration of cellular localization prediction of 58 SlDREB proteins. Each color indicates the level of abundance of SlDREB proteins in different cellular compartments.
Plant CARE program was employed on a 2,500-bp upstream region of 58 SlDREB genes for the analysis of potential cis regulatory elements that are involved in the transcriptional activity and response to biotic and abiotic stresses in tomato. According to the analysis, 103 motifs were found and classified into seven different categories depending on the major functions performed by them (Supplementary Table S7). These classes were (i) promoter-related, (ii) light-responsive, (iii) abiotic stress-responsive, (iv) hormone-related, (v) development-related, (vi) biotic stress-responsive, and (vii) unidentified cis regulatory elements that were demonstrated through a statistical analysis. Promoter-related cis elements covered the largest area (69%), followed by light-responsive (8%), abiotic stress-responsive (8%), hormone-related (6%), unidentified cis elements (5%), development-related (3%), and biotic stress-responsive (1%), as shown in Figure 9 (Supplementary Table S7).
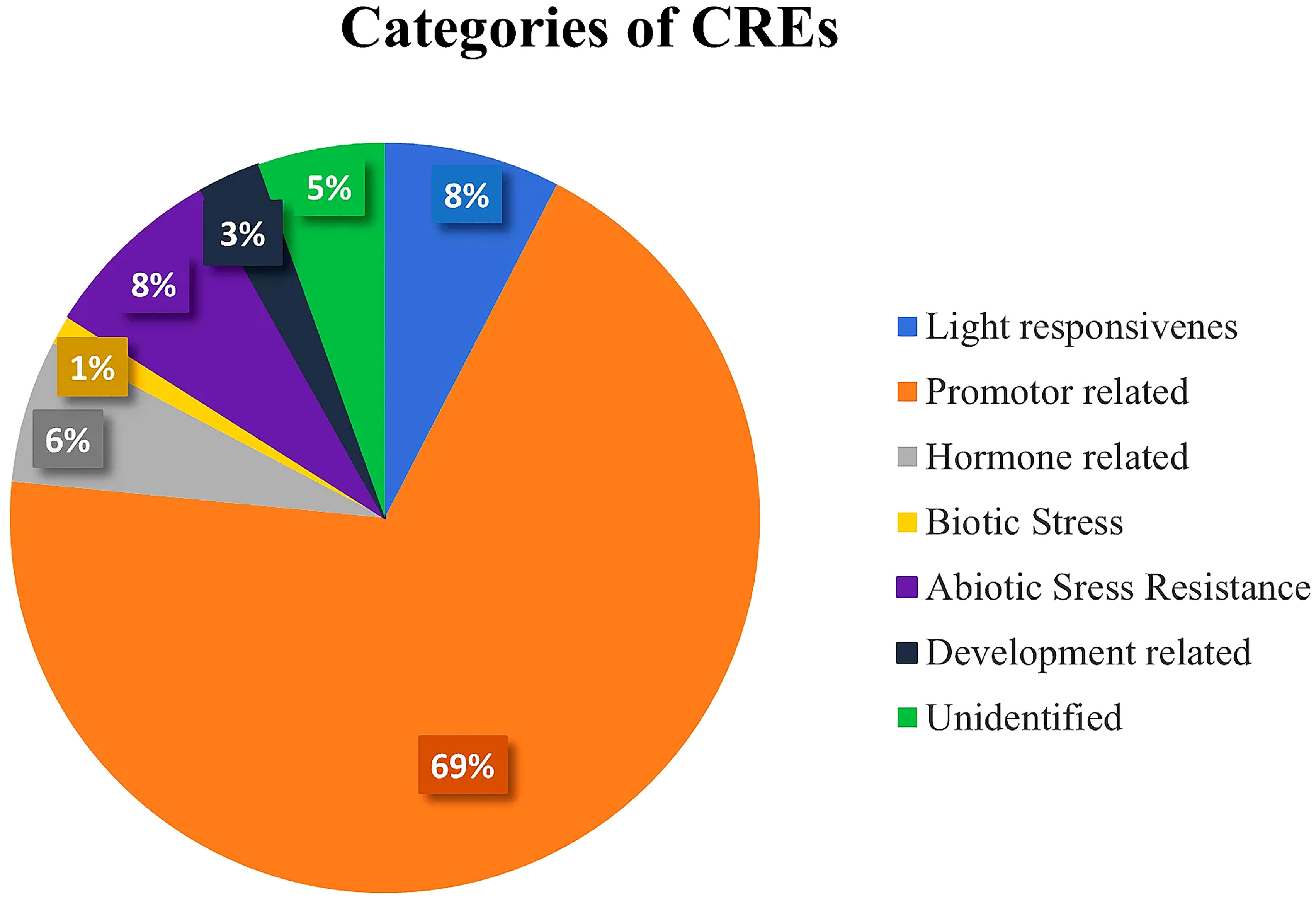
Figure 9 Pie chart representation of even categories of cis-regulatory elements with their respective percentages present in 58 SlDREB proteins.
Assessment in SlDREB proteins evaluated the presence of TATA-box, CAAT-box, AT~TATA box, A-box, and TATA elements that are involved in promoter binding site. TATA-box and CAAT-box were abundantly present in all 58 SlDREB genes that start the transcription by acting as a binding site for transcription factors, whereas TATA element assists in the binding of TATA-box and initiation site, and AT–TATA box and A-box are promoter binding sites as illustrated in Figure 10A. Light-responsive category was comprised of G-box or G-Box, TCT-motif, AE-box, GATA-motif, Sp1, GT1-motif, Box 4, ACE, LAMP-element, GA-motif, Gap-box, ATCT-motif, 3-AF1, chs-CMA1a, chs-CMA2a, AT1-motif, ATC-motif, TCCC-motif, I-box, chs-Unit 1 m1, AAAC-motif, Box II, CAG-motif, MRE, LS7, chs-CMA2b, ACA-motif, 3-AF3 binding site, and L-box. Box4 and G-box motifs contained the large area that maintainsed the transcription rate of light-controlled genes (Figure 10B).
In abiotic stress-responsive cis elements, the following motifs were revealed: AT-rich sequence, ARE, CCAAT-box, DRE core, DRE 1, GC-motif, LTR, MBS, MBS 1, STRE, TC rich repeats, MYB, MYC, MYB recognition site, MYB binding site, MYB like site, and AT-rich element as depicted in Figure 11A. Further results discovered ABRE, AuxRE, AuxRR-Core, CGTCA-Motif, ERE, GARE-motif, P-Box, TATC-box, TCA-element, TGACG-motif, TGA-element, ABRE4, ABRE2, ABRE3a, AT–ABRE, and TCA motifs in hormone-related cis element category. ABRE and ERE, also known as abscisic acid-responsive element and ethylene-responsive element, were greatly detected in SlDREB genes in response to hormone-related changes that were further linked to abiotic stress like drought. DREB is a sub-family of ethylene-responsive factor (ERF) gene family, so ERE elements also play an important role in its regulation. Recently, extensive studies on ERF family were conducted in Cucurbita moschata (Li et al., 2022). ABRE4, ABRE2, ABRE3a, and AT–ABRE are abscisic acid-responsive elements and act as binding sites. AuxRE, TGA-element, and AuxRR-Core were involved in response to auxin, whereas CGTCA-Motif and TGACG-motif act as methyl jasmonate-responsive elements. TCA-element involved in salicylic response, P-box, TATC-box, and GARE-motif act as gibberellin-responsive elements as provided in Figure 11B, respectively.
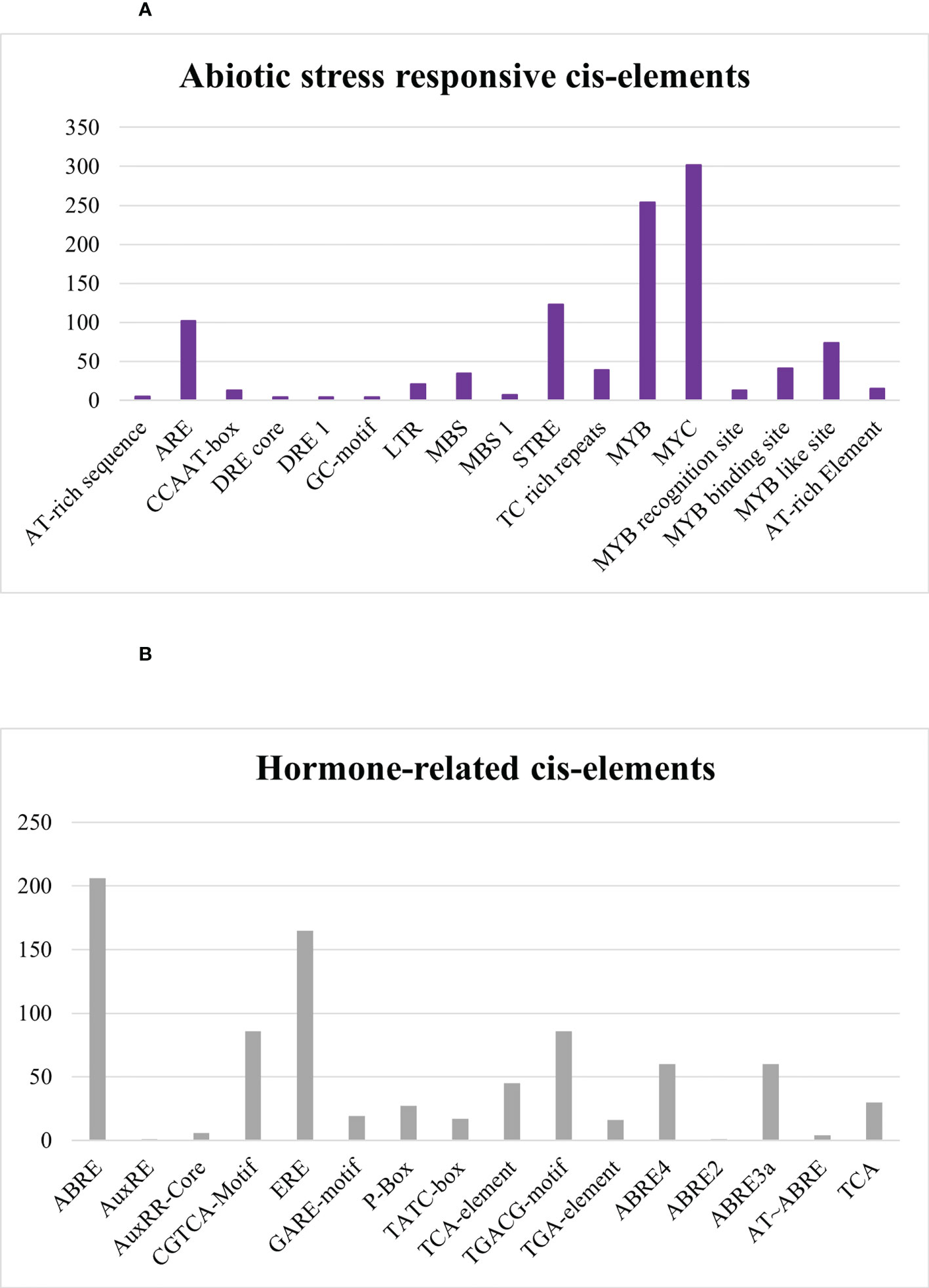
Figure 11 (A) Abiotic-stress-responsive cis regulatory elements. (B) Hormone-related cis regulatory elements.
In development-related cis elements, AAGAA-motif, CCGTCC-box, AP-1, AC-1, AC-II, as-I, CARE, circadian, CAT-box, dOCT, E2Fb, F-box, GCN4-motif, HD-zip 1, HD-zip 3, MSA like, NON, O2 Site, re2f1, RY element, AACA-motif, and CCGTCC motif were found. AAGAA-motif and as-1 element, available abundantly, are pathogenesis-related promoters. Other development-related cis elements have a role in meristem-specific stimulation, xylem-specific activation, palisade mesophyll cell differentiation, vascular development, biosynthesis and degradation of biological macromolecules, endosperm expression element, regulation of circadian rhythms, regulation of zein metabolism, seed development, cell cycle regulation, and mitosis-specific activator functioning as given in Figure 12A. In contrast, Box S, W-box, WRE 3, and WUN-motif cis regulatory elements were recognized in the biotic stress-responsive category, which interact with WRKY transcription factors against biotic stresses and act as a wound-responsive element as depicted in Figure 12B. However, Unnamed:2, Unnamed:4, Unnamed:6, CTAG, Unnamed:1, Unnamed:8, Unnamed:10, Unnamed:12, Unnamed:14, and Box III were considered as motifs with unknown functions. A graphical illustration of all cis regulatory elements equivalent to SlDREB genes is presented in Supplementary Figure S8. Identified cis elements might have an essential role in plant growth, development, and stress response.
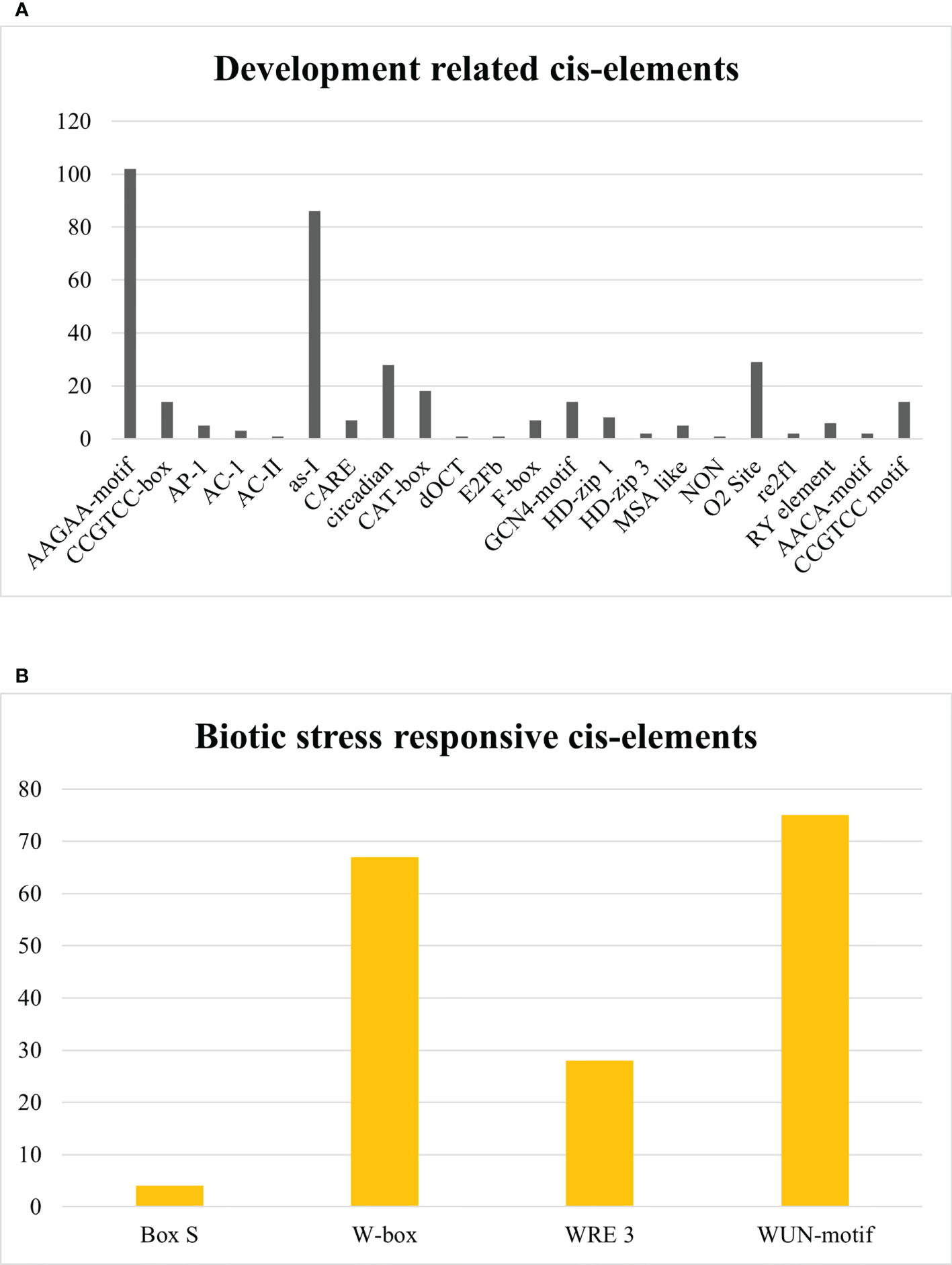
Figure 12 (A) Development-related cis regulatory elements. (B) Biotic-stress-responsive cis regulatory elements.
For protein modeling, five representatives (SlDREB15, SlDREB24, SlDREB51, SlDREB7, and SlDREB54) from each class (A1, A2, A4, A5, and A6) were selected and subjected to SOPMA online server. The analysis exhibited that each selected protein had different percentages of α-helices, β turn, random coils, and extended strands. These variations in different secondary factors contributed to the versatility of protein functions, stability, effects protein sequences, interactions with other molecules, and structure. The α-helices, β turn, random coils, and extended strands were detected to be flanked by 24.55–50.31%, 3.76–5.45%, 3.88–59.27%, and 4.91–12.73% as shown in Supplementary Table S9, respectively.
The prediction of 3D protein model assists in the analysis functions, cellular location, and their interaction with other proteins by filling the gap between structure and original protein sequence. The 3D structure of selected SlDREB proteins was predicted through Phyre2 server. Models with high percentage coverage (should be above 30%) and confidence were selected and subjected to further refinement for the construction of stable 3D structures.
For the structure refinement of SlDREB proteins, retrieved models from the Phyre2 server were subjected to the Galaxy refine server. These refined models were submitted to the PROCHECK server that was utilized for the evaluation and quality of these refined models. About 90% of residues in every SlDREB protein model were allowed in favorable regions when displayed in the Ramachandran plot that exhibited good stereo-chemical qualities of the generated model (Supplementary Table S10). Stable models were then visualized on ChimeraX software that exhibit a good quality of 3D structure with prominent α-helices and β turns represented in different colors (as depicted in Supplementary Table S11). To investigate the catalytic ligand binding sites in SlDREB proteins, CASTp 3.0 server was used. It assists in predicting the location and representation of concave regions of active pocket sites which were illustrated in the orange region on the 3D structure of proteins as given in Supplementary Table S12A.
Besides the location, the results also confirmed the presence of many important amino acid residues in active catalytic sites that involved the functional versatility of proteins, including LYS, PHE, GLN, ALA, ASN, THR, PRO, HIS, MET, ILE, GLY, ASP, VAL, and ARG. The detailed surface area and volumes of catalytic sites were also given in Supplementary Table S9. The disorder regions of SlDREB proteins were predicted using PrDOS software; it highlighted the disorder regions within the protein sequence as illustrated by the red regions (Supplementary Table S12B). The disorder regions had a significant role in the interactions and binding of proteins with other interactors, with high affinity depending on the cellular demand to cope with stresses or to regulate other metabolic pathways which was proved when the SlDREB proteins were subjected to STRING for the analysis of functional associations.
To depict the SlDREB protein associations with other proteins, STRING software, which provided 10 interactors associated with each SlDREB protein according to the criteria were employed. The full STRING network indicates both the functional and structural associations of proteins, whereas the colored line represents the type of interaction. It illustrated the interactive networks, associated in distinct colors, of SlDREB proteins with other proteins that belonged to different families, encoded by different genes, and contributed to the regulation of various biological and metabolic processes.
The interactive results of SlDREB1, member of the A1 subgroup, demonstrated the interactions with Solyc10g084370.1.1, Solyc02g077970.2.1, Solyc03g044300.2.1, Solyc02g082990.2.1, Solyc02g089570.2.1, Solyc02g089580.2.1, Solyc09g010820.2.1, Solyc02g077710.1.1, HSFA3, and Solyc02g065640.2.1 as given in Figure 13A. The results confirmed that Solyc10g084370.1.1 and Solyc09g010820.2.1 showed the presence of the SANT domain which belongs to the homeobox superfamily and a MYB-related protein that acts as DNA binding domain and regulates histone acetylation. The LEA protein family was observed in Solyc02g077970.2.1, also described as late-embryogenesis abundant 76-like protein that is involved in the defense of plant against environmental stresses. In Solyc03g044300.2.1, the presence of the AP2 domain showed its function as DNA binding domain and contributed to the regulation of transcription factor activity and defense response. The KIN1 and KIN 2 domains were detected in Solyc02g082990.2.1, Solyc02g089570.2.1, Solyc02g089580.2.1, and Solyc02g065640.2.1, which confirmed these interactors as stress-induced proteins in response to cold and abscisic acid (ABA). It is also known as cor6.6 in Arabidopsis. E-6 like protein (Solyc02g077710.1.1) plays a crucial role in early fiber development, and heat shock factor A3 (HSFA3) was detected in Solyc09g009100.2.1 interactor, which was involved in response to heat, DNA binding, and regulation of transcription.
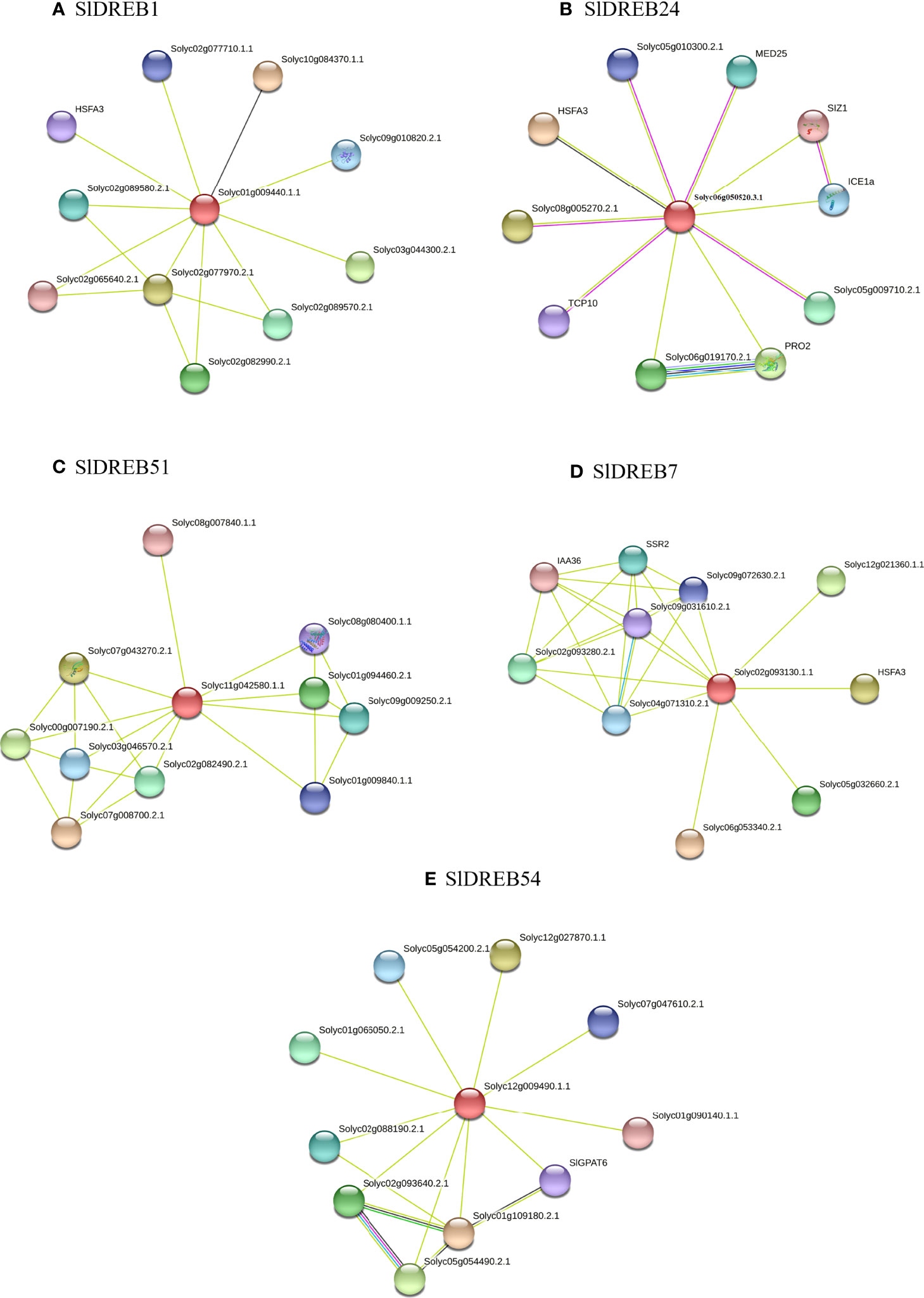
Figure 13 Interactive networks of SlDREB proteins through the STRING software. Protein interactions of SlDREB1 belong to A1 class denoted as (A), SlDREB24 member of A2 as (B), (C) SlDREB51, (D) SlDREB7, and (E) SlDREB 54 belong to A4, A5, and A6 classes, respectively. Colored networks depicted the associations depending on the information gathered from different databases, i.e., theoretical knowledge, experimental evidence, closely related genes, and protein model homology.
In SlDREB24, which belonged to A2 class, this was connected with HSFA3, Solyc08g005270.2.1, PRO2, Solyc06g019170.2.1, Solyc05g009710.2.1, MED25, ICE1a, Solyc05g010300.2.1, TCP10, and SIZ1 (Figure 13B). The HSFA3 (Solyc09g009100.2.1) protein activates in response to heat stress. Solyc08g005270.2.1 contains the RST domain, which interacts with transcription factors and acts as regulator under stress, hormonal, and developmental response, whereas PRO2 (Solyc08g043170.2.1) and Solyc06g019170.2.1 is known as delta-1-pyrroline-5-carboxylate synthase, which regulates the biosynthesis of amino acids, osmoregulation, phosphorylation in cytoplasm, metabolic process, and proline biosynthesis. In Solyc05g009710.2.1, MED25 (Solyc12g070100.1.1), the mediator complex subunit25_von Willebrand factor type A, is present and acts as a positive regulator and scaffold protein for basal transcription factors and RNA polymerase II. ICE1a (Solyc06g068870.2.1), also known as an inducer of CBI expression1, is a DNA-binding protein present in the nucleus and induces cold tolerance. The RPOLD domain in Solyc05g010300.2.1 contributes to protein dimerization, DNA binding, and regulation of transcription. Moreover, TCP10 (Solyc07053410.2.1) is responsible in protein–protein interactions and DNA binding transcription factor (TF) activity and SIZ1 (Solyc11g069160.1.1), also known as SP-RING-type zinc finger domain that plays a crucial role in zinc ion binding activity and protein SUMOylating during signaling pathways.
Similarly, the potential interactors for SlDREB51 (affiliated with A4 subgroup) were predicted and confirmed its associations with Solyc07g008700.2.1, Solyc07g043270.2.1, Solyc00g007190.2.1, Solyc01g094460.2.1, Solyc02g082490.2.1, Solyc09g009250.2.1, Solyc03g046570.2.1, Solyc01g009840.1.1, Solyc08g080400.1.1, and Solyc08g007840.1.1 as illustrated in Figure 13C. The FAR1-related sequence9 (Solyc07g008700.2.1) protein and Solyc07g043270.2.1 is a far-red elongated hypocotyl 3 isoform x involved in transcription regulation and circadian rhythms. The sigma factor PP2C-like phosphatases in Solyc00g007190.2.1 and Solyc02g082490.2.1 regulate protein dephosphorylation and serine/threonine phosphatase activity in the signaling pathway. Additionally, Solyc01g094460.2.1 contains AT-hook, and Solyc03g046570.2.1 (BES1-interacting MYC like protein) acts as a DNA-binding protein and controls protein dimerization activity. The Solyc01g009840.1.1 and Solyc08g080400.1.1 proteins belong to GRAS family that engaged in the gibberellin signaling pathway and are involved in plant growth and development. SlDREB51 also interacts with Solyc08g007840.1.1 (SlDREB34) to regulate the DNA binding transcription factor activity.
The STRING interactions predicted the association of the member of A5 subgroup (SlDREB7) with Solyc06g053340.2.1, HSFA3, Solyc12g021360.1.1, Solyc05g032660.2.1, Solyc02g093280.2.1, SSR2, Solyc04g071310.2.1, Solyc09g072630.2.1, Solyc09g031610.2.1, and IAA36, as depicted in Figure 13D. Solyc06g053340.2.1 belongs to the bZIP (basic-leucine zipper domain) family, also known as F box, which is linked with SKP1 protein that is involved in transcriptional regulations. HSFA3 (Solyc09g009100.2.1) is a heat shock transcription factor A3 type which responds to a feverish temperature, whereas the UVR domain containing protein Solyc06g053340.2.1, short-chain dehydrogenases/reductases (SDR) (Solyc05g032660.2.1), acts in chlorophyl(ide) b reductase activity. The basic helix–loop–helix (bHLH) protein (Solyc02g093280.2.1) regulates the circadian rhythm, response to cold and light stress, carpel development, and protein dimerization. Moreover, succinic semialdehyde reductase isofom2 SSR2 (Solyc03g121720) is an RNA recognition motif domain. The CDK-activating kinase assembly factor-related Solyc04g071310.2.1 is associated with kinase activity, nucleotide excision repair, and positive regulations. Furthermore, the ROP-interactive CRIB motif-containing protein 7 (Solyc09g072630.2.1) is a protein binding domain. Finally, the damaged DNA-binding 2 protein (Solyc09g031610.2.1) and auxin-responsive protein (IAA36) (Solyc06g066020.2.1) control DNA repair mechanism, UV damage excision repair, protein poly-ubiquitination, response to UV-A and UV-B, transcription, and auxin-activated signaling pathway.
Correspondingly, SlDREB54, a member of the A6 subgroup, displayed its connection with the Solyc01g109180.2.1 (long-chain acyl-CoA synthetase 2) which regulates beta-oxidation for the metabolism of lipid. Solyc12g027870.1.1 (cyclin-dependent kinase E1) acts as a catalytic domain, mediating phosphorylation and the kinase pathways. Solyc05g054490.2.1 is a member of 3-oxo-5-alpha-steroid 4-dehydrogenase family that is present in the membrane and involved in fatty acid biosynthesis. However, Solyc02g093640.2.1 is a SDR protein that is engaged in oxidoreductase activity. Solyc01g066050.2.1 is a transmembrane protein that belongs to tetratricopeptide repeat (TPR)-like superfamily protein present in the plasma membrane. Solyc02g088190.2.1 (SANT domain), Solyc05g054200.2.1, and Solyc07g047610.2.1 (transcriptional adapter ada2b isoform x1) respond to cold and are involved in histone acetylation and chromatin remodeling. SlGPAT6 (Solyc09g014350.2.1) is a glycerol-3-phosphate 2-O-acyltransferase 6 protein detected in cutin biosynthesis, and Solyc01g090140.1.1 protein [belonging to 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily] is involved in the formation of plant hormones like pigments, flavonoids, gibberellins, and oxidoreductase activity, as presented in Figure 13E.
Transcriptomic data of tomato seedlings under drought and heat stresses was retrieved from the GEO database and further refined to extract SlDREB expression levels (Supplementary Table S13). The mean values for similar sample sets were calculated to summarize the extensive datasets. Moreover, log2 base values were processed to quantify the expression levels of the respective genes (Supplementary Table S13). Furthermore, the expression levels were analyzed through an expression heat map (illustrated in Figure 14). The expression analysis of A1 sub-group demonstrates that SlDREB8 and SlDREB15 were upregulated under drought conditions and downregulated in response to heat stress. In addition, SlDREB9 showed an increase in expression profiles in response to drought and heat stresses. Moreover, SlDREB 32 and SlDREB33 were upregulated in response to drought stress. However, SlDREB1 indicated comparatively low expression profiles against both stresses, whereas SlDREB57 did not respond to drought and heat stress treatments.
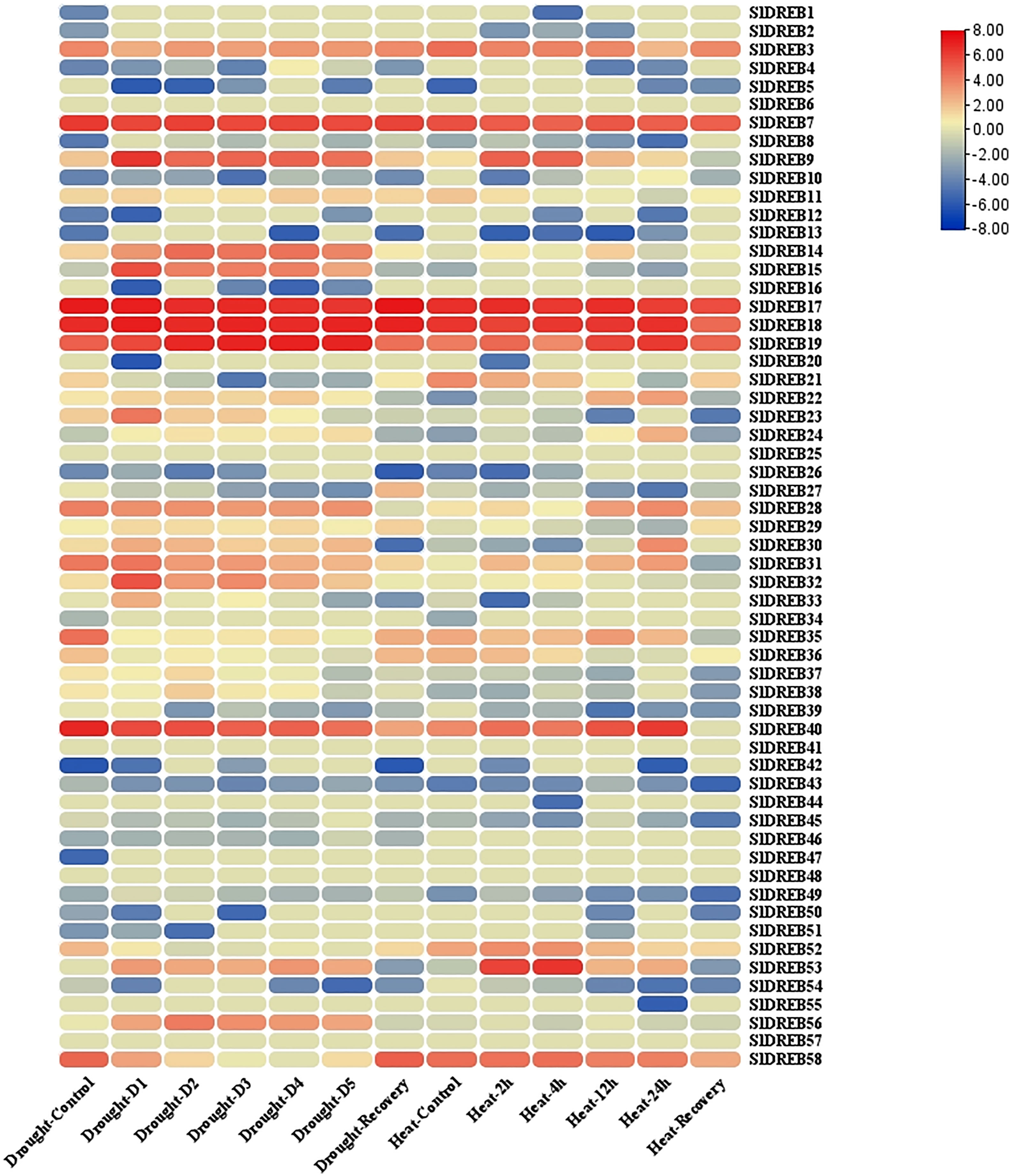
Figure 14 The expression analysis of 58 SlDREB has been illustrated in the form of an expression heat map. Almost 29 SlDREB genes were involved in response to drought and heat stress.
In A2 class, the transcriptomic expression of SlDREB22 and SlDREB24 was analyzed, which pointed towards a slight upregulation of both genes under drought stress; however, high expression profiles were reported in response to high temperature treatments. The results have shown that SlDREB43 was downregulated in response to heat and drought stresses, although SlDREB20 and SlDREB44 did not respond to both stresses. Moreover, SlDREB16 and SlDREB46 were downregulated against drought and not involved in response to heat stress.
The transcriptome analysis of the A4 class revealed that SlDREB36 highly responded towards both stresses. In addition, SlDREB37 and SlDREB838 were involved in defense signaling pathway and significantly expressed in response to water-deficient conditions, whereas SlDREB39 was downregulated under stressful conditions. The results have demonstrated that SlDREB3 was highly functional with significant expression profiles under water-deficient and high-temperature situations. Moreover, SlDREB14 showed substantial expression levels under both stresses. Similarly, SlDREB21 and SlDREB28 were upregulated against drought and heat stress to activate the defense signaling pathway. However, in the case of SlDREB34, SlDREB41, SlDREB48, SlDREB50, and SlDREB851, no significant expression profiles were indicated under stress conditions. However, high expression levels were reported for the SlDREB52 gene, whereas SlDREB56 was significantly upregulated in response to water-deficient conditions.
The transcriptome analysis data of A5 group members illustrated that SlDREB26 and SlDREB49 were downregulated whenever a plant undergoes heat and drought stresses. However, SlDREB2 did not respond to both situations. Subsequently, SlDREB7, SlDREB30, and SlDREB19 were highly expressive against drought stress. SlDREB7 and SlDREB9 were likewise upregulated in response to heat stress, whereas SlDREB30 showed an insignificant expression level against heat stress. Furthermore, SlDREB53 is involved in the drought and heat stress signaling pathway. The expression analysis of A6 class members revealed that SlDREB11 showed elevated expression profiles under drought and high temperature. However, SlDREB4, SlDREB5, SlDREB10, SlDREB27, SlDREB42, SlDREB45, and SlDREB54 demonstrate downregulation under both stresses, while SlDREB17, SlDREB18, and SlDREB40 showed high expression levels against heat and drought stress. Moreover, SlDREB25, SlDREB47, and SlDREB55 were not significantly responsive. On the other hand, SlDREB58 and SlDREB35 were highly expressed under heat stress and showed mild upregulation against drought stress. Lastly, SlDREB31 was significantly upregulated against drought stress as compared to high temperature stress. These results conclude that DREB genes have an important role against drought and heat stress signaling in tomato.
The present study covers extensive data on 58 SlDREB gene sequences. These 58 SlDREB genes were classified into A1–A6 classes based on the classification of AtDREB genes. The conserved amino acid analysis revealed that valine (V) is replaced by isoleucine (I) at the 14th position due to their same chemical structure, polarity, and characteristics in three SlDREB genes. Both are aliphatic, non-polar, and hydrophobic, whereas glutamic acid (E) is being replaced by V, Q, D, H, L, and A at the 19th position (Zhang et al., 2022). The genome-wide analysis discovered the presence of 58 SlDREB genes, which can vary in numbers in different plants due to the variance in genome sizes and genes present in their genome. The physiochemical properties, variation in amino acid structure, and protein binding affinity contribute multiplicity in protein function to withstand harsh environmental conditions (Salih et al., 2019; Panja et al., 2020). Thus, the variations in amino acids at conserved positions in SlDREB genes proved that these genes regulate many biological and molecular mechanisms associated with plant growth and interact with other stress-regulatory proteins. The total length of SlDREB proteins varied between 113 to 419 amino acids. Other properties like isoelectric point and molecular weight fluctuated between 4.42–9.41 and 12.35–46.94 kDa (Supplementary Table S1).
The phylogenetic analysis of SlDREB and AtDREB genes revealed that the variation in gene distribution patterns among A1–A6 classes, respectively, is due to genetic diversity. The study discovered the presence of seven members in A1 class and seven in A2 class, 16 members related to A4 class, seven members involved in A5, and 21 members present in A6 subgroup, respectively. It also demonstrated the absence of A3 class in the SlDREB gene family because no SlDREB member showed homology with AT2G40220.1 (member of A3 class) (Figure 2). Similarly, another related study reported the absence of A3 class in Ananas comosus (L.) Merr. (Chai et al., 2020). The research data illustrates that SlDREB24 (Solyc06g050520, accession no: AF500011 or DREB1) has a significant role under drought stress (Jiang et al., 2017). SlDREB22 (Solyc05g052410, GenBank accession: ADZ15315 or DREB2), known as SlDREB2, is likewise a potential regulator of stress-responsive genes and is expressed in early-seedling stage (Hichri et al., 2016). SlDREB18 (Solyc04g072900) (GenBank accession: AF506825.1), named LeDREB3, showed enhanced tolerance against many abiotic stresses like drought, salinity, and osmotic pressure (Islam and Wang, 2009, Wang G. et al., 2019), and SlDREB2 (Solyc06g066540) was recently characterized in response to heat stress (Mao et al., 2020). Furthermore, the gene structure analysis discovered the presence of introns in S. lycopersicum, unlike in Arabidopsis. The AtDREB genes showed the lack of intronic regions in its coding sequence, but studies revealed the occurrence of introns in various plants like S. tuberosum (Mushtaq et al., 2021), soybean (Zhou et al., 2020), Ananas comosus (L.) Merr., and Saccharum spontaneum (Zhen et al., 2021). Studies revealed that 12 SlDREB genes contain introns, out of which nine SlDREBs had one intron and the remaining three SlDREB genes entailed two introns. Thus, S. lycopersicum shows a similarity with Ananas comosus, Solanum tuberosum, Saccharum spontaneum, and soybean in terms of gene structure due to the presence of introns in the coding region. According to studies, gene structures with no or few introns are more active in response towards stress conditions (Jeffares et al., 2008). The difference in intron and exon organization clarifies the functional diversity of DREB genes that occurred due to variations in climatic conditions and stresses in a variety of plant species (Zhen et al., 2021).
To gain insight into the conserved motifs that have an important role in protein functionality and interactions, a motif analysis was accomplished. The results demonstrated the presence of conserved motifs in TFs on the same chromosome as in Ananas comosus (L.) Merr. and Solanum melongena L. (Li et al., 2021). In SlDREB proteins, 20 conserved motifs were discovered, out of which two motifs were present throughout all SlDREB sequences. It was observed that A1, A4, and A5 classes are more closely related, while A2 and A6 classes shared a combined clade due to the same conserved motifs. The AP2 domain consists of one alpha helix made up of 18 amino acids that assist in the interactions of DREB genes with other proteins, followed by three antiparallel beta sheets on the major groove of the DNA (Song et al., 2020). The identified 20 motifs in the SlDREB sequences have a variable composition that is the possible reason for the classification of DREB genes into different classes with high structural resemblance with the AP2 domain. Motifs 1, 2, and 3 are specifically present in the AP2 domain, which is the characteristic domain of DREB genes, whereas a difference in motif composition evaluated the formation of two clades in the phylogenetic tree. The difference in motif composition is responsible for the molecular interactions with different proteins and assists in DNA binding (Alberts et al., 2002; Defoort et al., 2018). This study proved that a similar pattern of intron–exon organization and conserved motifs was observed in the members belonging to same groups, thus elucidating the reliability of the phylogenetic relationship represented in Figure 3.
The random distribution of SlDREB genes provides an insight for their stability in a changing environment. Gene duplication events occurred in genes for adaptive evolution that is a major cause of plant diversity and adaptive specialization in genes (Flagel and Wendel, 2009). Polyploidy was observed in Arabidopsis, soybean, cotton, and potato (Nakano et al., 2006). Whole-genome duplication events occur in two ways: tandem duplication and segmental duplication. A tandem duplication event is responsible for the addition of a copied gene into the array, while segmental duplication is accountable for duplicating and spreading the fragments or the entire array. S. spontaneum, Z. mays, and P. trichocarpa go through both mechanisms of polyploidy. Whole-genome duplication was also examined in S. lycopersicum. Our results confirmed the occurrence of duplication events in 15 gene pairs, out of which eight gene pairs (32–33, 34–39, and 35–40) present on chromosome 08 (8–15, 27–29, and 50–51) exist on chromosomes 03, 06, and 11, and 43–44 and 45–47 belong to chromosome 10, undergoing tandem duplication events which were present on the same chromosomes (Figure 4).
In contrast, segmental duplication events were observed in seven gene pairs (36–52, 26–53, 11–54, 41–48, 17–58, 14–28, and 22–24) (Figure 5). In the tandem duplication method, gene pairs with the same cis regulatory elements, due to their presence on the same chromosome, were observed and involved in the regulation of similar functions (Flagel and Wendel, 2009). The present study’s data has revealed that the last gene duplication event occurred 4 million years ago in the 43–44 gene pair, which belongs to the A2 class and exhibits tandem duplication (Supplementary Table S5). Genes that are mostly involved in biological processes such as biotic and abiotic stress signaling undergo tandem duplications (Huang et al., 2020). To evaluate the presence of orthologous genes in interspecies, synteny analysis was employed. In total, 32 orthologs of SlDREB genes were found in Arabidopsis, whereas 47 orthologs were observed in S. tuberosum due to the close evolutionary relationship with S. lycopersicum, with both belonging to the same family (Figure 6). For the analysis of functional genomics and molecular processes conducted by each SlDREB gene, gene ontology annotation was employed. The results revealed that the DNA binding ability of SlDREB genes is uniform, based on studies conducted in P. vulgaris. The GO annotation analysis also discovered that the SlDREB genes primarily localized in the nucleus. Furthermore, the subcellular localization prediction showed the presence of SlDREBs in 13 other organelles as illustrated in the heat map (Figure 8).
In this study, 103 CREs were found in SlDREB sequences through promoter analysis. These 103 CREs have been classified into seven distinct categories depending on the functions and activation of the biological, cellular, and molecular pathways associated with them. Seven functional groups include light-responsive elements, hormone-related cis elements, development-related cis elements, biotic stress response elements, abiotic stress-responsive cis elements, promoter-related cis elements, and unidentified CREs (Ain-Ali et al., 2021). The promoter-related cis regulatory elements frequently occurred in all genes and secured about 69%, followed by light-responsive at 8%, abiotic stress-responsive at 8%, development-related at 6%, hormone-related at 3%, and biotic stress-responsive elements at 1% (Figure 9). The following cis elements are promoter-related: TATA-box, CAAT-box, AT~TATA box, A-box, and TATA elements. Among these elements, TATA-box and CAAT-box frequently occurred in every sequence and accommodated transcription factors by acting as their binding sites. The TATA element assists in the binding of TATA-box and activates the initiation site for transcription (Bae et al., 2015). The TATA box binds with RNA polymerase II for the initiation of transcription. AT~TATA box, and A-box are promoter binding sites. Moreover, A-box is an α-amylase promoter that is also identified in WRKY genes (Zhou et al., 2010). According to studies in rice, A-box assists in the regulation of the flowering stage (Mongkolsiriwatana et al., 2009). Thus, the analysis proved that SlDREB genes have a strong role in transcriptional regulation (Figure 10A).
The cis regulatory elements involved in light response are G-box or G-Box, TCT-motif, AE-box, GATA-motif, Sp1, GT1-motif, Box 4, ACE, LAMP-element, GA-motif, Gap-box, ATCT-motif, 3-AF1, chs-CMA1a, chs-CMA2a, AT1-motif, ATC-motif, TCCC-motif, I-box, chs-Unit 1 m1, AAAC-motif, Box II, CAG-motif, MRE, LS7, chs-CMA2b, ACA-motif, 3-AF3 binding site, and L-box. G-box and Box-4 abundantly appeared and regulated the transcription of light-responsive genes (Gilmartin et al., 1990). Moreover, G-box, ACE, Sp1, and GATA-motif also regulate the responses against abiotic stresses (Shariatipour and Heidari, 2020). The TCT motif controls photoperiodism, which is crucial for circadian rhythms for the proper development of plants and for the flowering stage (Zhao et al., 2018). Studies have evaluated the involvement of G-box in the phosphorylation of light-responsive genes through the calcium pathway upon receiving light signal and upregulated rd29 in response to drought as well (Kurt and Filiz, 2018). It is also involved in ethylene and ABA signaling pathway by acting as a binding site for bZIP proteins and responds to many abiotic stresses such as UV light and hormonal changes (Ma et al., 2013; Saidi and Hajibarat, 2018). Box-4 is a highly conserved DNA module in genes that are lightly sensitive. WRKY genes are also highly concentrated with box-4 element (Yin et al., 2013). The GT-1 motif is involved in the upregulation of light responses (Wang C. et al., 2019). The same pattern of cis elements was observed in S. tuberosum, with the exception of 3-AF1 binding site, 3-AF3 binding site, chs-CMA2b, and ACA motif elements that are only specified in S. lycopersicum (Figure 10B). The light-responsive elements are highly associated with photoperiods that regulate circadian rhythms and many other processes like flowering stage (Ain-Ali et al., 2021).
Abiotic stress conditions severely affect the growth and development of tomato crop. AT-rich sequence, ARE, CCAAT-box, DRE core, DRE 1, GC-motif, LTR, MBS, MBS 1, STRE, TC-rich repeats, MYB, MYC, MYB recognition site, MYB binding site, MYB like site, and AT-rich element are associated with abiotic stress-responsive elements (Figure 11A). MYB and MYC are dehydration-responsive motifs mostly observed in SlDREB gene promoters, whereas MBS and MBS1 act as binding sites for MYB that are engaged in drought response, biotic stress, and gene regulation of flavonoid biosynthesis (Li et al., 2015; Kaur et al., 2017). MYC is involved in the regulation of many hormone-related pathways. JA is one of the hormones mainly regulated through the MYC element in response to stresses (Yanfang et al., 2018). AT-rich elements act as binding site for AT-rich binding proteins and functions as activation mediator upon signal perception. Moreover, ARE is responsible for anaerobic induction and consists of two elements: GT and GC motif. GT motif has a high resemblance with MYB binding site and induces a response against anaerobic and water-deficient conditions (Dolferus et al., 2001). CCAAT-box acts as MYBHv1 binding site for heat shock factors against heat stress (Chaudhary et al., 2019). DRE core and DRE 1 are dehydration-responsive elements, whereas GC-motif is involved in anoxic-specific inducibility (Brown et al., 2003), LTR and STRE are low temperature stress-responsive elements, and TC-rich repeats are involved in defense signaling pathway (Lu et al., 2019; Niu et al., 2020). The cis elements associated with abiotic stress response appeared third in terms of occurrence and found to be consistent with prior studies conducted on S. tuberosum except the MYB like site.
Furthermore, among the results discovered were ABRE, AuxRE, AuxRR-Core, CGTCA-Motif, ERE, GARE-motif, P-Box, TATC-box, TCA-element, TGACG-motif, TGA-element, ABRE4, ABRE2, ABRE3a, AT~ABRE, and TCA motifs in the hormone-related cis elements category. Abscisic acid (ABA)-responsive element (ABRE) is associated with hormonal regulation and abiotic stress signaling. ABRE4, ABRE2, ABRE3a, and AT~ABRE act as binding sites (Verma et al., 2016). Another related study demonstrates that AuxRE, TGA, and AuxRR-Core cis elements are auxin-responsive and have a regulatory role in the expression of DREB genes under drought stress (Kolachevskaya et al., 2019). Moreover, auxin plays a significant role in plant development and defense signaling mechanisms against biotic and abiotic stresses, particularly in response to drought stress. Auxin, as a key growth hormone, assures the effective working of the plant physiology under stressful environments (Sharif et al., 2022), whereas CGTCA and TGACG-motif act as methyl jasmonate-responsive elements (Concha et al., 2013). Similar hormone-related elements were described in Saccharum spontaneum and Solanum tuberosum except ABRE4, ABRE2, ABRE3a, AT~ABRE, and TCA elements (Kaur et al., 2017). These hormone-related elements may have a direct or indirect interaction with signaling pathways in response to various stresses (Figure 11B).
In development-related cis elements, several motifs were discovered, such as AAGAA-motif, CCGTCC-box, AP-1, AC-1, AC-II, as-I, CARE, circadian, CAT-box, dOCT, E2Fb, F-box, GCN4-motif, HD-zip 1, HD-zip 3, MSA like, NON, O2 Site, re2f1, RY element, AACA-motif, and CCGTCC motif (Figure 12A). The AAGAA-motif and as-1 element, available abundantly, are pathogenesis-related promoters. The AAGAA-motif is responsible for the development of xylem and assists in water transportation. The As-1 element acts as a promoter binding site for many hormone-responsive genes, especially under auxin and methyl jasmonate stress signaling. The CCGTCC-box, CARE, and CAT-box act as cis elements in meristem-specific stimulation (Laloum et al., 2013). Moreover, AC-1 and AC-II are engaged in xylem-specific activation. HD-zip 1 and HD-zip 3 participate in palisade mesophyll cell differentiation and vascular development and regulate biological processes associated with macromolecules (Vulavala et al., 2017). AACA-motif and GCN4-motif act as endosperm expression elements and regulate the circadian rhythms (Kaur et al., 2017). O2 site is involved in the regulation of zein metabolism, and RY element assists in seed -specific regulation. However, E2Fb and F-box are engaged in cell cycle regulation, and MSA like is a mitosis-specific activator (Li L. et al., 2021). The present work demonstrates that S-box, W-box, WRE 3, and WUN-motif cis regulatory elements were recognized to function in response to biotic stresses and, furthermore, to interact with WRKY transcription factor which acts as a wound-responsive element as depicted in Figure 12B (Yin et al., 2013; Liu et al., 2016). According to another related study, W-box regulates seed dormancy and defense mechanisms against biotic stresses (Rinerson et al., 2015). These elements were also reported in Saccharum spontaneum and Solanum tuberosum. However, among 103 cis elements, 10 motifs which cover almost 5% occurrence were found with unidentified functions. Parallel studies revealed the existence of conserved motifs that might have a crucial role in many biological processes. Thus, the presence of these motifs in the promoter regions of SlDREB genes indicates their potentialities in plant defense signaling and processes associated with growth and development in tomato.
Moreover, structural variations were detected in five SlDREB representatives from each class through secondary and tertiary structure analysis, which gave insight to the development of stable protein structures. Secondary structures were created by SOPMA server that exhibited different percentages of α-helices, β turn, random coils, and extended strands. The spatial arrangement of amino acids present in the random coils of DREB proteins can vary depending on the intrinsic nature of protein. These factors contributed to the structural and functional versatility and molecular interactions of proteins (Vatansever et al., 2017). Furthermore, Phyre2 server assisted in the prediction of tertiary structures. Models that covered more than 30% coverage were further subjected for refinement. For structural refinement and validation, Galaxy Refine server and PROCHECK server were employed to complete the gaps between protein sequences and conformations to create stable protein structures and to check the stability through the generation of Ramachandran plots. Validation is considered as a core step in structural determination (Pražnikar et al., 2019). Studies reported that the residues lying from 85% to more than 90% in favored region is considered as a good model. Hence, this study indicated the presence of 91.2% for SlDREB1, 88.8% for SlDREB2, 89.4% for SlDREB51, 90.3% for SlDREB7, and 92.8% for SlDREB54 residues present in the favored region. The percentages were represented through a Ramachandran plot that further verified the models having good stereo-chemical properties (Supplementary Table S10). Stabilized models were visualized through ChimeraX that constructed high-quality models with their respective α-helix and β turns (Supplementary Table S11).
There are specific regions on proteins that are responsible to interact with other proteins and molecules in response to different cellular and biological processes, and these are known as ligand binding sites (Kawabata, 2010). The present study data has revealed the presence of active catalytic sites on five SlDREB protein structures that are crucial for binding with different proteins having variable binding capacities as indicated in orange region (Supplementary Table S12A). Each catalytic site is comprised of specific amino acids that contribute to the binding capacity, assortment in protein functions, interactions with other molecules, and structural versatility (Coleman and Sharp, 2010).
Furthermore, proteins consist of disorder regions that allow them to change their functions, expand their interactions, and induce structural versatility in response to the changing environment. These disorder regions are flexible parts that allow proteins to bind with greater affinity to different molecules and thus contribute to various defense signaling pathways and cell cycle processes. The disorder region analysis provides thorough understanding of the spatial regulations and interactions of signaling pathways associated with proteins. The presence of these flexible regions in transcription factors allow their binding with signaling cascades and other stress-responsive proteins (Pazos et al., 2013). Disorder regions were predicted in five representative SlDREB proteins (Supplementary Table S12B). The binding plasticity of disorder regions assists in the attachment to other gene families like NAC, GRAS, and dehydrins. These interactions generate a signaling cascade in developmental processes like seed development, response to hormonal changes, and drought stress (Hsiao, 2022).
Protein–protein interactions are necessary for the regulation of different developmental and adaptive processes for normal plant growth (Braun et al., 2013). Transcription factors stimulate other proteins and co-factors for the activation of distinct regulatory pathways that control transcription in response to various stresses (Francois et al., 2020). In response to both biotic and abiotic stresses, thousands of molecules like reactive oxygen species (ROS), hormones, and proteins interact at the cellular and molecular levels for the activation of kinases and other signaling cascades. For the complete integration of protein function, its cellular abundance, and biochemical activities, it is crucial to gain insight to interactive pathways (Struk et al., 2019). The protein–protein interactions of five SlDREB proteins were detected through STRING that revealed its physical and functional associations. In the present study (Figure 13), nodes indicate spliced isoforms and the interaction of query protein (red node) with different interactors due to post-translational modifications. Each node represents all the proteins produced by a single protein-coding gene locus. Moreover, colored edges represent the type of evidence in protein associations, blue and pink edges represent known interactions, and green, red, and dark blue colors are known for predicted interactions, whereas other colors indicate the evidence recovered from text mining or co-occurrence.
Moreover, our results demonstrate that SlDREB1 interacts with 10 interactors, namely: Solyc10g084370.1.1, Solyc02g077970.2.1, Solyc03g044300.2.1, Solyc02g082990.2.1, Solyc02g089570.2.1, Solyc02g089580.2.1, Solyc09g010820.2.1, Solyc02g077710.1.1, HSFA3, and Solyc02g065640.2.1. Out of 10 interactors, two members (Solyc10g084370.1.1 and Solyc09g010820.2.1) showed the presence of SANT domain which belongs to homeobox superfamily and a MYB-related protein that acts as a DNA binding domain and regulates histone acetylation (Boyer et al., 2004). Histone acetylation is essential in the cell cycle and in response to abiotic stress (light, heat, UVB, sugar, salt, and wound) and hormonal change (salicylic, ethylene, abscisic acid and auxin) (Jiang et al., 2020). Histone acetyltransferase genes previously discovered in rice (Liu et al., 2012), Triticum aestivum (Gao et al., 2021), and Arabidopsis thaliana (Pandey et al., 2002) (Solyc02g077970.2.1) belong to the LEA protein family which are connected with salinity and water stress signaling and have been reported in many plants like Vigna glabrescens (Singh et al., 2022), cotton (Magwanga et al., 2018), Salvia miltiorrhiza (Chen et al., 2021), and Triticum aestivum (Liu et al., 2019). The presence of AP2 domain was detected in one member (Solyc03g044300.2.1) that contributed to transcriptional regulation and responded to stresses as found in wheat (Magar et al., 2022) and castor bean (Xu et al., 2013). However, four members of SlDREB1 interactors—namely, Solyc02g082990.2.1, Solyc02g089570.2.1, Solyc02g089580.2.1, and Solyc02g065640.2.1—include the KIN1 and KIN2 domains, also known as cor6.6. in Arabidopsis, which is engaged in low temperature and responses to disturbance in abscisic acid levels, as abscisic acid is a vital hormone that activates in retort to stress stimuli (Wang et al., 1995). Solyc02g077710.1.1 is an E-6 like protein which is necessary for fiber development (John, 1996). Solyc09g009100.2.1 is the last interactor that is a heat shock transcription factor A3 (HSFA3) involved in response to heat stress, DNA binding, and regulation of transcription as illustrated in Figure 13A.
Several proteins were likewise associated with SlDREB24, which belongs to the A2 subgroup, i.e., HSFA3, Solyc08g005270.2.1, PRO2, Solyc06g019170.2.1, Solyc05g009710.2.1, MED25, ICE1a, Solyc05g010300.2.1, TCP10, and SIZ1 (Figure 13B). Solyc08g005270.2.1 contains the RST domain which is present in PARP and TAF4s factors and belongs to the SRO gene family. It is engaged in transcriptional regulation by interacting with transcription factors and maintains ribosyl transferase activity. Ribosyl transferase activity is linked with oxidative stress and responsible for ADP-ribosyl polymerases (Jaspers et al., 2010). Extensive analysis of RST domain has been carried out in wheat (Liu et al., 2014), tomato (Zhen et al., 2021), banana (Zhang et al., 2019), Sesamum indicum (Liu et al., 2021), and rice (You et al., 2014), whereas two members [Solyc08g043170.2.1 (PRO2) and Solyc06g019170.2.1) are known as delta-1-pyrroline-5-carboxylate synthase. These members are associated with proline biosynthesis and osmoregulation. Proline synthesis is important in plants as it works as an antioxidant and signaling molecule under stress conditions, which further activates the defense signaling cascades. Furthermore, it controls osmotic pressure and ROS levels, thus making the plant tolerant to stresses (Hayat et al., 2012). In Solyc05g009710.2.1, MED25 (Solyc12g070100.1.1) mediator complex subunit25_von Willebrand factor type A was detected, which performs as a scaffold protein for transcription factors and RNA polymerase II and regulates hormonal cascades under biotic and abiotic stresses (Kazan, 2017). MED25 has been reported in Arabidopsis, where it is discovered to deal with drought stress (Elfving et al., 2011). Additionally, ICE1a (Solyc06g068870.2.1), also known as inducer of CBI expression1, is a DNA binding protein present in the nucleus and confers tolerance in plants against cold stress (Chinnusamy et al., 2003). The RPOLD domain in Solyc05g010300.2.1 contributes to protein dimerization and cellular response to elevated temperatures. Moreover, TCP10 and SIZ1 (SP-RING-type zinc finger domain), detected in Solyc07053410.2.1 and Solyc11g069160.1.1, are responsible for protein–protein interactions and DNA binding TF activity and play a crucial role in zinc ion binding activity and protein SUMOylating. Studies in rice revealed that protein SUMOylation is a process in which local proteins act as signal transducers to activate signal cascades under stress (Raorane et al., 2013).
SlDREB51 likewise belongs to the A4 class. It shows interactions with Solyc07g008700.2.1, Solyc07g043270.2.1, Solyc00g007190.2.1, Solyc01g094460.2.1, Solyc02g082490.2.1, Solyc09g009250.2.1, Solyc03g046570.2.1, Solyc01g009840.1.1, Solyc08g080400.1.1, and Solyc08g007840.1.1 (Figure 13C). FAR1-related sequence9 (Solyc07g008700.2.1) protein and Solyc07g043270.2.1 are a far-red elongated hypocotyl 3 isoform x involved in transcription regulation, response to hormonal change, and light stress. Other functions that are revealed in other plants are the biosynthesis of chlorophyll and starch and circadian rhythms (Ma and Li, 2018). The sigma factor PP2C-like phosphatases in Solyc00g007190.2.1 and Solyc02g082490.2.1 regulate protein dephosphorylation and serine/threonine phosphatase activity in the signaling pathway. Additionally, Solyc01g094460.2.1 contains AT-hook and DUF 296 domain which is a domain of unknown function, and Solyc03g046570.2.1 (BES1-interacting MYC like protein) acts as a DNA binding protein and controls protein dimerization activity. SlDREB51 also interacts with Solyc08g007840.1.1 (SlDREB34), which demonstrated that SlDREB members are associated with each other for the regulation of DNA binding activity and response to biotic and abiotic stresses. The Solyc01g009840.1.1 and Solyc08g080400.1.1 proteins belong to the GRAS family, which is engaged in gibberellin signaling pathway and functions in plant growth and development. The GRAS family has been analyzed in various plants like rice, Populus and Arabidopsis (Liu and Widmer, 2014), Glycine max (Wang et al., 2020), Prunus mume (Lu et al., 2015), and tomato (Huang et al., 2015).
The present study has established that SlDREB7 is A5 class member and associated with the following proteins: Solyc06g053340.2.1, HSFA3, Solyc12g021360.1.1, Solyc05g032660.2.1, Solyc02g093280.2.1, SSR2, Solyc04g071310.2.1, Solyc09g072630.2.1, Solyc09g031610.2.1, and IAA36 (Figure 13D). One member (Solyc06g053340.2.1) belongs to the bZIP (basic-leucine zipper domain) family which is a prominent TF family associated with stress signaling pathways and developmental processes. In addition, studies on Arabidopsis and potato have revealed its functions in senescence, light and biotic stress response, and seed growth (Wang et al., 2021). Solyc06g053340.2.1 is a UVR domain-containing protein, and Solyc05g032660.2.1 is SDR, which shows oxidoreductase activity. SDR is of a heterogenous protein family which is involved in secondary metabolic and developmental pathways such as catabolism and flower growth (Moummou et al., 2012). However, Solyc02g093280.2.1 is recognized as bHLH DNA-binding protein that controls circadian rhythm, response to cold and light stress, carpel development, and protein dimerization. bHLH is a significant transcription factor family which is discovered in several plant species, i.e., Capsicum annuum L. (Zhang et al., 2020), tomato (Sun et al., 2015), and maize (Li et al., 2018). Moreover, succinic semialdehyde reductase isofom2 SSR2, recognized in Solyc03g121720, is an RNA recognition motif domain, and Solyc04g071310.2.1 is a CDK-activating kinase assembly factor-related protein that is associated with kinase activity and nucleotide excision repair, whereas ROP-interactive CRIB motif-containing protein 7 (Solyc09g072630.2.1) belongs to the Rho family that reacts to oxygen deficiency, cellular development, and hormone-mediated pathways (Yang and Fu, 2007). Finally, damaged DNA-binding 2 protein (Solyc09g031610.2.1) and auxin-responsive protein (IAA36) (Solyc06g066020.2.1) control the DNA repair mechanism, UV damage excision repair, protein poly-ubiquitination, response to UV-A and B, transcription, and auxin-activated signaling pathway (Reed, 2001).
STRING interactions revealed the network of SlDREB54 (member of the A6 subgroup) with Solyc01g109180.2.1 (long-chain acyl-CoA synthetase 2) which regulates and controls beta-oxidation for lipid metabolism. The second member is Solyc12g027870.1.1, a cyclin-dependent kinase E1 (CDKE1) that acts as a catalytic domain, mediating phosphorylation and the kinase pathways. In a study, CDKs revealed their functions in cell cycle regulations (Malumbres, 2014). Solyc05g054490.2.1 belongs to the 3-oxo-5-alpha-steroid 4-dehydrogenase family protein that functions in fatty acid biosynthesis. However, Solyc02g093640.2.1 is a fourth interactor (SDR protein) engaged in oxidoreductase activity. Solyc01g066050.2.1 (fifth interactor) is a transmembrane protein that belongs to the TPR-like superfamily present in the plasma membrane. The TPR family is involved in phytohormone signaling like abscisic acid, cytokinins, and gibberellins and also regulates ethylene signaling pathways. Furthermore, it regulates biological processes under osmotic stress conditions in plants (Schapire et al., 2006), and Solyc02g088190.2.1 consists of a SANT domain.
Moreover, Solyc05g054200.2.1 and Solyc07g047610.2.1 (transcriptional adapter ada2b isoform x1), associated with cold stress signaling, are involved in histone acetylation and chromatin remodeling. SlGPAT6 (Solyc09g014350.2.1) is a glycerol-3-phosphate 2-O-acyltransferase 6 protein detected in cutin biosynthesis in tomato (Petit et al., 2021). Additionally, SlDREB54 revealed its interaction with Solyc01g090140.1.1 protein [belongs to 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily] that functions in the biosynthesis of plant pigments, flavonoids, and gibberellins and regulates oxidoreductase activity (Kawai et al., 2014) (Figure 13E). The transcriptome analysis of tomato predicted that about 29 SlDREB genes are actively involved in regulating the defense pathways against abiotic stresses such as drought and heat stress. Only four DREB genes have already been characterized through an expression analysis that revealed their significant role against abiotic stresses. Recent studies conducted on SlDREB24 (Solyc06g050520, accession no: AF500011 or DREB1) showed its remarkable role in response to drought stress. It enhances the sugar concentration and accumulates osmolytes that further normalize the osmotic pressure and increase the response to drought stress (Jiang et al., 2017). SlDREB22 (Solyc05g052410, GenBank accession: ADZ15315 or DREB2), known as SlDREB2, is also a potential regulator of stress-responsive genes. According to spatiotemporal expression, this gene is also induced during early-seedling stage and is present in the vegetative parts of a plant. It is highly expressed in the leaves in defense against drought and salt stress (Hichri et al., 2016). Consequently, SlDREB18 (Solyc04g072900; GenBank accession: AF506825.1), named LeDREB3, has been reported to enhance the tolerance against many abiotic stresses like drought, salinity, and osmotic pressure. It is also known as the negative regulator of the ABA pathway, thus affecting many developmental processes like germination and senescence. It is also engaged in many physiological processes like root architecture development (Islam and Wang, 2009; Wang G. et al., 2019). Similar studies also revealed the expression of SlDREB28 (Solyc06g066540) under heat stress. It functions in response to many biotic and abiotic stresses such as salinity, drought by a fluctuating hormone level that contributes to increasing tolerance against stress. It assists in the biosynthesis of phytohormones (jasmonic acid, salicylic acid, and ethylene) and binds to DRE element under stress conditions (Mao et al., 2020). Other genes that were downregulated or not expressed under heat and drought stress might be involved in other abiotic stresses, developmental mechanisms, and the regulation of several stress-resistant genes involved in biotic responses. Previous studies on expression analysis exhibit the role of DREB genes in fruit development and ripening (Zhang et al., 2022). Overall, the present study data illustrates the comprehensive interactive networks of SlDREBs with various other stress-responsive proteins associated with the regulation of development, growth, and stress tolerance processes in tomato.
In conclusion, our results revealed a total of 58 SlDREB genes in tomato, with the presence of a conserved AP2 domain. Only three DREB genes indicated the replacement of valine with isoleucine due to the same polarity, structure, and chemical characteristics. Multiple sequence alignment and phylogenetic evaluation demonstrated the classification of SlDREB genes into five subgroups, excluding the A3 class, unlike Arabidopsis, which comprised A1–A6 classes. Gene duplication events proved the occurrence of polyploidy by tandem and segmental duplication methods. The evaluation of (Ka/Ks) values confirmed the existence of strong purification selection during the evolution of plants by calculating the divergence time. Moreover, synteny analysis revealed 32 and 47 orthologs in Arabidopsis and Solanum tuberosum, respectively. The cis-regulatory element analysis discovered 103 motifs that were further divided into seven categories. These elements have been implicated in light response, hormonal modifications, development, abiotic stress-responsive, biotic stress, and promoter binding sites. Gene ontology data illustrated that the SlDREB genes are involved in various biological and molecular processes. A subcellular localization assessment confirmed that the SlDREB genes primarily resided in the nucleus and were observed in 13 other organelles in variable concentrations. The secondary structure analysis assisted in determining essential points, whereas the tertiary structures of SlDREB proteins and validation of protein models provide a thorough understanding of homology modeling. The results of active ligand binding sites and disordered regions provide a deep insight into the protein’s structural and functional plasticity and versatility. Moreover, interactive network analysis demonstrated the connections of SlDREB proteins with other potential interactors. These interactions result in extensive signaling pathways and the functional diversity of SlDREB proteins. Transcriptome analysis revealed that 29 SlDREB genes regulate the defense signaling mechanisms against drought and heat stress. Overall, our data has potential information that serves as a robust infrastructure for unravelling the extensive structural and functional relationships of intricate genes requisite for genetic engineering, advance breeding, and biogenetics to develop stress tolerant/over-expressive tomato genotypes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
FM designed the project and scheme of work. HM and FM conducted the research, analyzed the data, and wrote the manuscript. FM, RA, and AG reviewed the manuscript. All authors contributed to the article and approved the submitted version.
We acknowledge the Higher Education Commission, Pakistan, and the National University of Sciences and Technology (NUST), Pakistan, for providing research facilities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1031679/full#supplementary-material
Ain-Ali, Q.-U., Mushtaq, N., Amir, R., Gul, A., Tahir, M., Munir, F. (2021). Genome-wide promoter analysis, homology modeling and protein interaction network of dehydration responsive element binding (DREB) gene family in solanum tuberosum. PloS One 16, e0261215. doi: 10.1371/journal.pone.0261215
Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., Walter, P. (2002). “DNA-Binding motifs in gene regulatory proteins,” in Molecular biology of the cell, 4th edition (New York: Garland Science).
Bae, S.-H., Han, H. W., Moon, J. (2015). Functional analysis of the molecular interactions of TATA box-containing genes and essential genes. PloS One 10, e0120848. doi: 10.1371/journal.pone.0120848
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Barrett, T., Suzek, T. O., Troup, D. B., Wilhite, S. E., Ngau, W.-C., Ledoux, P., et al. (2005). NCBI GEO: mining millions of expression profiles–database and tools. Nucleic Acids Res. 33, D562–D566. doi: 10.1093/nar/gki022
Boyer, L. A., Latek, R. R., Peterson, C. L. (2004). The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5, 158–163. doi: 10.1038/nrm1314
Braun, P., Aubourg, S., Van Leene, J., De Jaeger, G., Lurin, C. (2013). Plant protein interactomes. Annu. Rev. Plant Biol. 64, 161–187. doi: 10.1146/annurev-arplant-050312-120140
Brown, R. L., Kazan, K., Mcgrath, K. C., Maclean, D. J., Manners, J. M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1. 2 gene of arabidopsis. Plant Physiol. 132, 1020–1032. doi: 10.1104/pp.102.017814
Chai, M., Cheng, H., Yan, M., Priyadarshani, S., Zhang, M., He, Q., et al. (2020). Identification and expression analysis of the DREB transcription factor family in pineapple (Ananas comosus (L.) merr.). PeerJ 8, e9006. doi: 10.7717/peerj.9006
Chaudhary, R., Baranwal, V. K., Kumar, R., Sircar, D., Chauhan, H. (2019). Genome-wide identification and expression analysis of Hsp70, Hsp90, and Hsp100 heat shock protein genes in barley under stress conditions and reproductive development. Funct. Integr. Genomics 19, 1007–1022. doi: 10.1007/s10142-019-00695-y
Chen, C., Chen, H., He, Y., Xia, R. (2018). TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 289660, 289660. doi: 10.1101/289660
Cheng, H., Cai, H., An, Z., Huang, H. (2013). Comparative proteomics analysis revealed increased expression of photosynthetic proteins in transgenic tobacco by overexpression of'AtCBF1'gene. Plant Omics 6, 240–245. doi: 10.3316/informit.399083534423003
Chen, J., Li, N., Wang, X., Meng, X., Cui, X., Chen, Z., et al. (2021). Late embryogenesis abundant (LEA) gene family in salvia miltiorrhiza: identification, expression analysis, and response to drought stress. Plant Signaling Behav. 16, 1891769. doi: 10.1080/15592324.2021.1891769
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Coleman, R. G., Sharp, K. A. (2010). Protein pockets: inventory, shape, and comparison. J. Chem. Inf. Modeling. 50, 589–603. doi: 10.1021/ci900397t
Concha, C. M., Figueroa, N. E., Poblete, L. A., Oñate, F. A., Schwab, W., Figueroa, C. R. (2013). Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in fragaria chiloensis fruit. Plant Physiol. Biochem. 70, 433–444. doi: 10.1016/j.plaphy.2013.06.008
Conesa, A., Götz, S. (2008). Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832–619832. doi: 10.1155/2008/619832
De Castro, E., Sigrist, C. J., Gattiker, A., Bulliard, V., Langendijk-Genevaux, P. S., Gasteiger, E., et al. (2006). ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365. doi: 10.1093/nar/gkl124
Defoort, J., Van De Peer, Y., Vermeirssen, V. (2018). Function, dynamics and evolution of network motif modules in integrated gene regulatory networks of worm and plant. Nucleic Acids Res. 46, 6480–6503. doi: 10.1093/nar/gky468
Diouf, I. A., Derivot, L., Bitton, F., Pascual, L., Causse, M. (2018). Water deficit and salinity stress reveal many specific QTL for plant growth and fruit quality traits in tomato. Front. Plant Sci. 9, 279. doi: 10.3389/fpls.2018.00279
Dolferus, R., Klok, E. J., Ismond, K., Delessert, C., Wilson, S., Good, A., et al. (2001). Molecular basis of the anaerobic response in plants. IUBMB Life 51, 79–82. doi: 10.1080/15216540152122058
Du, X., Li, W., Sheng, L., Deng, Y., Wang, Y., Zhang, W., et al. (2018). Over-expression of chrysanthemum CmDREB6 enhanced tolerance of chrysanthemum to heat stress. BMC Plant Biol. 18, 1–10. doi: 10.1186/s12870-018-1400-8
Elfving, N., Davoine, C., Benlloch, R., Blomberg, J., Brännström, K., Müller, D., et al. (2011). The arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. 108, 8245–8250. doi: 10.1073/pnas.1002981108
Flagel, L. E., Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Francois, M., Donovan, P., Fontaine, F. (2020). Modulating transcription factor activity: Interfering with protein-protein interaction networks. Semin. Cell Dev. Biol. 99, 12–19. doi: 10.1016/j.semcdb.2018.07.019
Gao, S., Li, L., Han, X., Liu, T., Jin, P., Cai, L., et al. (2021). Genome-wide identification of the histone acetyltransferase gene family in triticum aestivum. BMC Genomics 22, 49. doi: 10.1186/s12864-020-07348-6
Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., Bairoch, A. (2005). Protein identification and analysis tools on the ExPASy server. Proteomics Protoc. Handb. 112, 571–607. doi: 10.1385/1-59259-890-0:571
Geourjon, C., Deleage, G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11, 681–684. doi: 10.1093/bioinformatics/11.6.681
Gilmartin, P. M., Sarokin, L., Memelink, J., Chua, N.-H. (1990). Molecular light switches for plant genes. Plant Cell 2, 369. doi: 10.1105/tpc.2.5.369
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Gupta, A., Upadhyay, R. K., Prabhakar, R., Tiwari, N., Garg, R., Sane, V. A., et al. (2022). SlDREB3, a negative regulator of ABA responses, controls seed germination, fruit size and the onset of ripening in tomato. Plant Sci. 319, 111249. doi: 10.1016/j.plantsci.2022.111249
Hayat, S., Hayat, Q., Alyemeni, M. N., Wani, A. S., Pichtel, J., Ahmad, A. (2012). Role of proline under changing environments: a review. Plant Signal Behav. 7, 1456–1466. doi: 10.4161/psb.21949
Heo, L., Park, H., Seok, C. (2013). GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 41, W384–W388. doi: 10.1093/nar/gkt458
Hichri, I., Muhovski, Y., Clippe, A., Žižková, E., Dobrev, P. I., Motyka, V., et al. (2016). SlDREB2, a tomato dehydration-responsive element-binding 2 transcription factor, mediates salt stress tolerance in tomato and a rabidopsis. Plant. Cell Environ. 39, 62–79. doi: 10.1111/pce.12591
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Hou, Z., Li, Y., Cheng, Y., Li, W., Li, T., Du, H., et al. (2022). Genome-wide analysis of DREB genes identifies a novel salt tolerance gene in wild soybean (Glycine soja). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.821647
Hsiao, A.-S. (2022). Plant protein disorder: spatial regulation, broad specificity, switch of signaling and physiological status. Frontiers in Plant Science 13, 904446. doi: 10.3389/fpls.2022.904446
Huala, E., Dickerman, A. W., Garcia-Hernandez, M., Weems, D., Reiser, L., Lafond, F., et al. (2001). The arabidopsis information resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 29, 102–105. doi: 10.1093/nar/29.1.102
Huang, Y., Liu, Y., Zhang, M., Chai, M., He, Q., Jakada, B. H., et al. (2020). Genome-wide identification and expression analysis of the ERF transcription factor family in pineapple (Ananas comosus (L.) merr.). PeerJ 8, e10014. doi: 10.7717/peerj.10014
Huang, W., Xian, Z., Kang, X., Tang, N., Li, Z. (2015). Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. BMC Plant Biol. 15, 1–18. doi: 10.1186/s12870-015-0590-6
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hwang, J. E., Lim, C. J., Chen, H., Je, J., Song, C., Lim, C. O. (2012). Overexpression of arabidopsis dehydration-responsive element-binding protein 2C confers tolerance to oxidative stress. Mol. Cells 33, 135–140. doi: 10.1007/s10059-012-2188-2
Ishida, T., Kinoshita, K. (2007). PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35, W460–W464. doi: 10.1093/nar/gkm363
Islam, M. S., Wang, M.-H. (2009). Expression of dehydration responsive element-binding protein-3 (DREB3) under different abiotic stresses in tomato. BMB. Rep. 42, 611–616. doi: 10.5483/BMBRep.2009.42.9.611
Jaspers, P., Overmyer, K., Wrzaczek, M., Vainonen, J. P., Blomster, T., Salojärvi, J., et al. (2010). The RST and PARP-like domain containing SRO protein family: analysis of protein structure, function and conservation in land plants. BMC Genomics 11, 170. doi: 10.1186/1471-2164-11-170
Jeffares, D. C., Penkett, C. J., Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Jiang, J., Ding, A. B., Liu, F., Zhong, X. (2020). Linking signaling pathways to histone acetylation dynamics in plants. J. Exp. Bot. 71, 5179–5190. doi: 10.1093/jxb/eraa202
Jiang, L., Wang, Y., Zhang, S., He, R., Li, W., Han, J., et al. (2017). Tomato SlDREB1 gene conferred the transcriptional activation of drought-induced gene and an enhanced tolerance of the transgenic arabidopsis to drought stress. Plant Growth Regul. 81, 131–145. doi: 10.1007/s10725-016-0195-6
John, M. E. (1996). Structural characterization of genes corresponding to cotton fiber mRNA, E6: reduced E6 protein in transgenic plants by antisense gene. Plant Mol. Biol. 30, 297–306. doi: 10.1007/BF00020115
Kaur, A., Pati, P. K., Pati, A. M., Nagpal, A. K. (2017). In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of arabidopsis thaliana and oryza sativa. PloS One 12, e0184523. doi: 10.1371/journal.pone.0184523
Kawabata, T. (2010). Detection of multiscale pockets on protein surfaces using mathematical morphology. Proteins.: Structure. Function. Bioinf. 78, 1195–1211. doi: 10.1002/prot.22639
Kawai, Y., Ono, E., Mizutani, M. (2014). Evolution and diversity of the 2–oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 78, 328–343. doi: 10.1111/tpj.12479
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Khan, M. S. (2011). The role of DREB transcription factors in abiotic stress tolerance of plants. Biotechnol. Biotechnol. Equip. 25, 2433–2442. doi: 10.5504/BBEQ.2011.0072
Kolachevskaya, O. O., Lomin, S. N., Arkhipov, D. V., Romanov, G. A. (2019). Auxins in potato: molecular aspects and emerging roles in tuber formation and stress resistance. Plant Cell Rep. 38, 681–698. doi: 10.1007/s00299-019-02395-0
Kurt, F., Filiz, E. (2018). Genome-wide and comparative analysis of bHLH38, bHLH39, bHLH100 and bHLH101 genes in arabidopsis, tomato, rice, soybean and maize: insights into iron (Fe) homeostasis. Biometals 31, 489–504. doi: 10.1007/s10534-018-0095-5
Laha, S. D., Dutta, S., Schäffner, A. R., Das, M. (2020). Gene duplication and stress genomics in brassicas: Current understanding and future prospects. J. Plant Physiol. 255, 153293. doi: 10.1016/j.jplph.2020.153293
Laloum, T., De Mita, S., Gamas, P., Baudin, M., Niebel, A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18, 157–166. doi: 10.1016/j.tplants.2012.07.004
Lata, C., Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, C., Ng, C. K.-Y., Fan, L.-M. (2015). MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 114, 80–91. doi: 10.1016/j.envexpbot.2014.06.014
Li, D., He, Y., Li, S., Shi, S., Li, L., Liu, Y., et al (2021). Genome-wide characterization and expression analysis of AP2/ERF genes in eggplant (Solanum melongena L.). Plant Physiol Biochem 167, 492–503.
Liu, S., Liu, S., Wang, M., Wei, T., Meng, C., Wang, M., et al. (2014). A wheat SIMILAR TO RCD-ONE gene enhances seedling growth and abiotic stress resistance by modulating redox homeostasis and maintaining genomic integrity. Plant Cell 26, 164–180. doi: 10.1105/tpc.113.118687
Liu, X., Luo, M., Zhang, W., Zhao, J., Zhang, J., Wu, K., et al. (2012). Histone acetyltransferases in rice (Oryza sativaL.): phylogenetic analysis, subcellular localization and expression. BMC Plant Biol. 12, 1–17. doi: 10.1186/1471-2229-12-145
Liu, S., Wang, X., Wang, H., Xin, H., Yang, X., Yan, J., et al. (2013). Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of zea mays l. PloS Genet. 9, e1003790. doi: 10.1371/journal.pgen.1003790
Liu, A., Wei, M., Zhou, Y., Li, D., Zhou, R., Zhang, Y., et al. (2021). Comprehensive analysis of SRO gene family in sesamum indicum (L.) reveals its association with abiotic stress responses. Int. J. Mol. Sci. 22, 13048. doi: 10.3390/ijms222313048
Liu, X., Widmer, A. (2014). Genome-wide comparative analysis of the GRAS gene family in populus, arabidopsis and rice. Plant Mol. Biol. Rep. 32, 1129–1145. doi: 10.1007/s11105-014-0721-5
Liu, H., Xing, M., Yang, W., Mu, X., Wang, X., Lu, F., et al. (2019). Genome-wide identification of and functional insights into the late embryogenesis abundant (LEA) gene family in bread wheat (Triticum aestivum). Sci. Rep. 9, 13375. doi: 10.1038/s41598-019-49759-w
Liu, L., Xu, W., Hu, X., Liu, H., Lin, Y. (2016). W-Box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 6, 1–9. doi: 10.1038/srep20881
Li, Z., Wang, G., Liu, X., Wang, Z., Zhang, M., Zhang, J. (2021). Genome-wide identification and expression profiling of DREB genes in saccharum spontaneum. BMC Genomics 22, 456. doi: 10.1186/s12864-021-07799-5
Li, Y., Wang, H., Zhang, Y., Martin, C. (2018). Can the world’s favorite fruit, tomato, provide an effective biosynthetic chassis for high-value metabolites? Plant Cell Rep. 37, 1443–1450. doi: 10.1007/s00299-018-2283-8
Li, Q., Zhang, L., Chen, P., Wu, C., Zhang, H., Yuan, J., et al. (2022). Genome-wide identification of APETALA2/ETHYLENE RESPONSIVE FACTOR transcription factors in cucurbita moschata and their involvement in ethylene response. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.847754
Lu, Y., Sun, J., Yang, Z., Zhao, C., Zhu, M., Ma, D., et al. (2019). Genome-wide identification and expression analysis of glycine-rich RNA-binding protein family in sweet potato wild relative ipomoea trifida. Gene 686, 177–186. doi: 10.1016/j.gene.2018.11.044
Lu, J., Wang, T., Xu, Z., Sun, L., Zhang, Q. (2015). Genome-wide analysis of the GRAS gene family in prunus mume. Mol. Genet. Genomics 290, 303–317. doi: 10.1007/s00438-014-0918-1
Magar, M. M., Liu, H., Yan, G. (2022). Genome-wide analysis of AP2/ERF superfamily genes in contrasting wheat genotypes reveals heat stress-related candidate genes. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.853086
Magwanga, R. O., Lu, P., Kirungu, J. N., Lu, H., Wang, X., Cai, X., et al. (2018). Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 19, 6. doi: 10.1186/s12863-017-0596-1
Ma, L., Li, G. (2018). FAR1-RELATED SEQUENCE (FRS) and FRS-RELATED FACTOR (FRF) family proteins in arabidopsis growth and development. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00692
Mao, L., Deng, M., Jiang, S., Zhu, H., Yang, Z., Yue, Y., et al. (2020). Characterization of the DREBA4-type transcription factor (SlDREBA4), which contributes to heat tolerance in tomatoes. Front. Plant Sci. 11, 554520. doi: 10.3389/fpls.2020.554520
Ma, S., Shah, S., Bohnert, H. J., Snyder, M., Dinesh-Kumar, S. P. (2013). Incorporating motif analysis into gene co-expression networks reveals novel modular expression pattern and new signaling pathways. PloS Genet. 9, e1003840. doi: 10.1371/journal.pgen.1003840
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Mongkolsiriwatana, C., Pongtongkam, P., Peyachoknagul, S. (2009). In silico promoter analysis of photoperiod-responsive genes identified by DNA microarray in rice (Oryza sativa l.). Agric. Natural Resour. 43, 164–177.
Moummou, H., Kallberg, Y., Tonfack, L. B., Persson, B., van der Rest, B. (2012). The plant short-chain dehydrogenase (SDR) superfamily: genome-wide inventory and diversification patterns. BMC Plant Biol. 12, 219. doi: 10.1186/1471-2229-12-219
Mueller, L. A., Solow, T. H., Taylor, N., Skwarecki, B., Buels, R., Binns, J., et al. (2005). The SOL genomics network. a comparative resource for solanaceae biology and beyond. Plant Physiol. 138, 1310–1317. doi: 10.1104/pp.105.060707
Mushtaq, N., Munir, F., Gul, A., Amir, R., Paracha, R. Z. (2021). Genome-wide analysis, identification, evolution and genomic organization of dehydration responsive element-binding (DREB) gene family in solanum tuberosum. PeerJ 9, e11647. doi: 10.7717/peerj.11647
Nakano, T., Suzuki, K., Fujimura, T., Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Niu, L., Chu, H. D., Tran, C. D., Nguyen, K. H., Pham, H. X., Le, D. T., et al. (2020). The GATA gene family in chickpea: structure analysis and transcriptional responses to abscisic acid and dehydration treatments revealed potential genes involved in drought adaptation. J. Plant Growth Regul. 39, 1647–1660. doi: 10.1007/s00344-020-10201-5
Pandey, R., Muèller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. doi: 10.1093/nar/gkf660
Panja, A. S., Maiti, S., Bandyopadhyay, B. (2020). Protein stability governed by its structural plasticity is inferred by physicochemical factors and salt bridges. Sci. Rep. 10, 1822. doi: 10.1038/s41598-020-58825-7
Park, S., Shi, A., Mou, B. (2020). Genome-wide identification and expression analysis of the CBF/DREB1 gene family in lettuce. Sci. Rep. 10, 1–14. doi: 10.1038/s41598-020-62458-1
Pazos, F., Pietrosemoli, N., García-Martín, J. A., Solano, R. (2013). Protein intrinsic disorder in plants. Front. Plant Sci. 4, 363. doi: 10.3389/fpls.2013.00363
Petit, J., Bres, C., Reynoud, N., Lahaye, M., Marion, D., Bakan, B., et al. (2021). Unraveling cuticle formation, structure, and properties by using tomato genetic diversity. Front. Plant Sci. 12, 778131. doi: 10.3389/fpls.2021.778131
Pettersen, E. F., Goddard, T. D., Huang, C. C., Meng, E. C., Couch, G. S., Croll, T. I., et al. (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82. doi: 10.1002/pro.3943
Pražnikar, J., Tomić, M., Turk, D. (2019). Validation and quality assessment of macromolecular structures using complex network analysis. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-38658-9
Raorane, M. L., Mutte, S. K., Varadarajan, A. R., Pabuayon, I. M., Kohli, A. (2013). Protein SUMOylation and plant abiotic stress signaling: in silico case study of rice RLKs, heat-shock and Ca(2+)-binding proteins. Plant Cell Rep. 32, 1053–1065. doi: 10.1007/s00299-013-1452-z
Reed, J. W. (2001). Roles and activities of Aux/IAA proteins in arabidopsis. Trends Plant Sci. 6, 420–425. doi: 10.1016/S1360-1385(01)02042-8
Rinerson, C. I., Scully, E. D., Palmer, N. A., Donze-Reiner, T., Rabara, R. C., Tripathi, P., et al. (2015). The WRKY transcription factor family and senescence in switchgrass. BMC Genomics 16, 1–17. doi: 10.1186/s12864-015-2057-4
Saidi, A., Hajibarat, Z. (2018). In silico analysis of floral MADS-BOX gene in brachypodium distachyon. Bionature 38(6), 366–375.
Salih, H., Odongo, M. R., Gong, W., He, S., Du, X. (2019). Genome-wide analysis of cotton C2H2-zinc finger transcription factor family and their expression analysis during fiber development. BMC Plant Biol. 19, 1–17. doi: 10.1186/s12870-019-2003-8
Schapire, A. L., Valpuesta, V., Botella, M. A. (2006). TPR proteins in plant hormone signaling. Plant Signaling Behav. 1, 229–230. doi: 10.4161/psb.1.5.3491
Shariatipour, N., Heidari, B. (2020). Meta-analysis of expression of the stress tolerance associated genes and uncover their-regulatory elements in rice (L.). Open Bioinf. J. 13, 39–49. doi: 10.2174/1875036202013010039
Sharif, R., Su, L., Chen, X., Qi, X. (2022). Involvement of auxin in growth and stress response of cucumber. Vegetable. Res. 2, 1–9. doi: 10.48130/VR-2022-0013
Singh, C. M., Kumar, M., Pratap, A., Tripathi, A., Singh, S., Mishra, A., et al. (2022). Genome-wide analysis of late embryogenesis abundant protein gene family in vigna species and expression of VrLEA encoding genes in vigna glabrescens reveal its role in heat tolerance. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.843107
Struk, S., Jacobs, A., Sánchez Martín-Fontecha, E., Gevaert, K., Cubas, P., Goormachtig, S. (2019). Exploring the protein–protein interaction landscape in plants. Plant. Cell Environ. 42, 387–409. doi: 10.1111/pce.13433
Sun, H., Fan, H.-J., Ling, H.-Q. (2015). Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16, 1–12. doi: 10.1186/s12864-014-1209-2
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Song, Q., Bari, A., Li, H., Chen, L. L. (2020). Identification and analysis of micro-exons in AP2/ERF and MADS gene families. FEBS Open Bio 10, 2564–2577.
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tian, W., Chen, C., Lei, X., Zhao, J., Liang, J. (2018). CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367.
Vatansever, R., Uras, M. E., Sen, U., Ozyigit, I. I., Filiz, E. (2017). Isolation of a transcription factor DREB1A gene from phaseolus vulgaris and computational insights into its characterization: protein modeling, docking and mutagenesis. J. Biomol. Structure. Dynamics. 35, 3107–3118. doi: 10.1080/07391102.2016.1243487
Vazquez-Hernandez, M., Romero, I., Escribano, M. I., Merodio, C., Sanchez-Ballesta, M. T. (2017). Deciphering the role of CBF/DREB transcription factors and dehydrins in maintaining the quality of table grapes cv. autumn royal treated with high CO2 levels and stored at 0 c. Front. Plant Sci. 8, 1591. doi: 10.3389/fpls.2017.01591
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 1–10. doi: 10.1186/s12870-016-0771-y
Vulavala, V. K., Fogelman, E., Rozental, L., Faigenboim, A., Tanami, Z., Shoseyov, O., et al. (2017). Identification of genes related to skin development in potato. Plant Mol. Biol. 94, 481–494. doi: 10.1007/s11103-017-0619-3
Wang, H., Datla, R., Georges, F., Loewen, M., Cutler, A. J. (1995). Promoters from kin1 and cor6.6, two homologous arabidopsis thaliana genes: transcriptional regulation and gene expression induced by low temperature, ABA, osmoticum and dehydration. Plant Mol. Biol. 28, 605–617. doi: 10.1007/BF00021187
Wang, L., Ding, X., Gao, Y., Yang, S. (2020). Genome-wide identification and characterization of GRAS genes in soybean (Glycine max). BMC Plant Biol. 20, 1–21. doi: 10.1186/s12870-020-02636-5
Wang, Q., Guo, C., Li, Z., Sun, J., Wang, D., Xu, L., et al. (2021). Identification and analysis of bZIP family genes in potato and their potential roles in stress responses. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.637343
Wang, C., Wang, Y., Pan, Q., Chen, S., Feng, C., Hai, J., et al. (2019). Comparison of trihelix transcription factors between wheat and brachypodium distachyon at genome-wide. BMC Genomics 20, 1–14. doi: 10.1186/s12864-019-5494-7
Wang, G., Xu, X., Wang, H., Liu, Q., Yang, X., Liao, L., et al. (2019). A tomato transcription factor, SlDREB3 enhances the tolerance to chilling in transgenic tomato. Plant Physiol. Biochem. 142, 254–262. doi: 10.1016/j.plaphy.2019.07.017
Wu, H., Lv, H., Li, L., Liu, J., Mu, S., Li, X., et al. (2015). Genome-wide analysis of the AP2/ERF transcription factors family and the expression patterns of DREB genes in moso bamboo (Phyllostachys edulis). PloS One 10, e0126657. doi: 10.1371/journal.pone.0126657
Xu, W., Li, F., Ling, L., Liu, A. (2013). Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis l.). BMC Genomics 14, 785. doi: 10.1186/1471-2164-14-785
Yanfang, Y., Kaikai, Z., Liying, Y., Xing, L., Ying, W., Hongwei, L., et al. (2018). Identification and characterization of MYC transcription factors in taxus sp. Gene 675, 1–8. doi: 10.1016/j.gene.2018.06.065
Yang, M., Derbyshire, M. K., Yamashita, R. A., Marchler-Bauer, A. (2020). NCBI's conserved domain database and tools for protein domain analysis. Curr. Protoc. Bioinf. 69, e90. doi: 10.1002/cpbi.90
Yang, Z., Fu, Y. (2007). ROP/RAC GTPase signaling. Curr. Opin. Plant Biol. 10, 490–494. doi: 10.1016/j.pbi.2007.07.005
Yin, G., Xu, H., Xiao, S., Qin, Y., Li, Y., Yan, Y., et al. (2013). The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 13, 1–19. doi: 10.1186/1471-2229-13-148
You, J., Zong, W., Du, H., Hu, H., Xiong, L. (2014). A special member of the rice SRO family, OsSRO1c, mediates responses to multiple abiotic stresses through interaction with various transcription factors. Plant Mol. Biol. 84, 693–705. doi: 10.1007/s11103-013-0163-8
Yuan, S., Xu, B., Zhang, J., Xie, Z., Cheng, Q., Yang, Z., et al. (2015). Comprehensive analysis of CCCH-type zinc finger family genes facilitates functional gene discovery and reflects recent allopolyploidization event in tetraploid switchgrass. BMC Genomics 16, 1–16. doi: 10.1186/s12864-015-1328-4
Zhang, Z., Chen, J., Liang, C., Liu, F., Hou, X., Zou, X. (2020). Genome-wide identification and characterization of the bHLH transcription factor family in pepper (Capsicum annuum l.). Front. Genet. 11, 570156. doi: 10.3389/fgene.2020.570156
Zhang, L., Zhou, D., Hu, H., Li, W., Hu, Y., Xie, J., et al. (2019). Genome-wide characterization of a SRO gene family involved in response to biotic and abiotic stresses in banana (Musa spp.). BMC Plant Biol. 19, 211. doi: 10.1186/s12870-019-1807-x
Zhang, L., Chen, L., Pang, S., Zheng, Q., Quan, S., Liu, Y., et al. (2022). Function analysis of the erf and dreb subfamilies in tomato fruit development and ripening. Front Plant Sci. 13.
Zhao, L., Li, M., Xu, C., Yang, X., Li, D., Zhao, X., et al. (2018). Natural variation in gm GBP 1 promoter affects photoperiod control of flowering time and maturity in soybean. Plant J. 96, 147–162. doi: 10.1111/tpj.14025
Zhen, L., Wang, G., Liu, X., Wang, Z., Zhang, M., Zhang, J. (2021). Genome-wide identification and expression profiling of DREB genes in saccharum spontaneum. BMC Genomics 22, 456. doi: 10.1186/s12864-021-07799-5
Zhou, R., Kong, L., Wu, Z., Rosenqvist, E., Wang, Y., Zhao, L., et al. (2019). Physiological response of tomatoes at drought, heat and their combination followed by recovery. Physiol. Plant. 165, 144–154. doi: 10.1111/ppl.12764
Zhou, L., Liu, Y., Liu, Z., Kong, D., Duan, M., Luo, L. (2010). Genome-wide identification and analysis of drought-responsive microRNAs in oryza sativa. J. Exp. Bot. 61, 4157–4168. doi: 10.1093/jxb/erq237
Keywords: Solanum lycopersicum, DREB gene family, cis-regulatory elements, protein modeling, interactive association networks, transcriptome analysis
Citation: Maqsood H, Munir F, Amir R and Gul A (2022) Genome-wide identification, comprehensive characterization of transcription factors, cis-regulatory elements, protein homology, and protein interaction network of DREB gene family in Solanum lycopersicum. Front. Plant Sci. 13:1031679. doi: 10.3389/fpls.2022.1031679
Received: 30 August 2022; Accepted: 25 October 2022;
Published: 24 November 2022.
Edited by:
George Popescu, Mississippi State University, United StatesReviewed by:
Christos Noutsos, State University of New York at Old Westbury, United StatesCopyright © 2022 Maqsood, Munir, Amir and Gul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Faiza Munir, ZmFpemEubXVuaXJAYXNhYi5udXN0LmVkdS5waw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.