
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 16 December 2022
Sec. Plant Systematics and Evolution
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1031087
 Xia Liu1,2†
Xia Liu1,2† Zhaoshan Wang2†
Zhaoshan Wang2† Wei Wang3†
Wei Wang3† Qinqin Huang1,4
Qinqin Huang1,4 Yanfei Zeng2
Yanfei Zeng2 Yu Jin5
Yu Jin5 Honglei Li1
Honglei Li1 Shuhui Du6
Shuhui Du6 Jianguo Zhang2*
Jianguo Zhang2*Introduction: Populus (Salicaceae) species harbour rich biodiversity and are widely distributed throughout the Northern Hemisphere. However, the origin and biogeography of Populus remain poorly understood.
Methods: We infer the divergence times and the historical biogeography of the genus Populus through phylogenetic analysis of 34 chloroplast fragments based on a large sample.
Results and Discussion: Eurasia is the likely location of the early divergences of Salicaceae after the Cretaceous-Paleogene (K-Pg) mass extinction, followed by recurrent spread to the remainder of the Old World and the New World beginning in the Eocene; the extant Populus species began to diversity during the early Oligocene (approximately 27.24 Ma), climate changes during the Oligocene may have facilitated the diversification of modern poplar species; three separate lineages of Populus from Eurasia colonized North America in the Cenozoic via the Bering Land Bridges (BLB); We hypothesize that the present day disjunction in Populus can be explained by two scenarios: (i) Populus likely originated in Eurasia and subsequently colonized other regions, including North America; and (ii) the fact that the ancestor of the genus Populus that was once widely distributed in the Northern Hemisphere and eventually wiped out due to the higher extinction rates in North America, similar to the African Rand flora. We hypothesize that disparities in extinction across the evolutionary history of Populus in different regions shape the modern biogeography of Populus. Further studies with dense sampling and more evidence are required to test these hypotheses. Our research underscores the significance of combining phylogenetic analyses with biogeographic interpretations to enhance our knowledge of the origin, divergence, and distribution of biodiversity in temperate plant floras.
A primary objective of study in the biological sciences is to achieve an understanding of the evolutionary history and biogeographic processes of species that are responsible for shaping their global patterns of variety. In the Northern Hemisphere, one of the best examples of a biogeographic pattern is known as the intercontinental disjunction. Some more traditional disjunct distribution patterns include northern temperate disjunctions (Milne, 2006). Commonly observed disjunctions are Madrean-Tethyan (Wen and Ickert-Bond, 2009) and the eastern Asian-Northeast American disjunctions (Wen, 1999; Wen, 2001; Hao et al., 2015). However, East Asian–Northwest American disjunctions (Wen, 1999; Nie et al., 2005; Zhou et al., 2012) have only infrequently been investigated utilizing current phylogenetic and biogeographic methods. Although the fact that numerous hypotheses regarding the causes of particular kinds of disjunctions have been put forward in research conducted by a variety of different groups, it has been determined that climate changes that occurred during the Cenozoic and two major migration routes, the Bering Land Bridge (BLB) and the North Atlantic Land Bridges (NALB) (Tiffney, 1985a; Tiffney, 1985b; Donoghue et al., 2001; Sanmartín et al., 2001; Zhang et al., 2013), are thought to be the main drivers for the biogeographic patterns across the different regions. Given the intricate biotic responses to these abiotic events that have been observed in the Northern Hemisphere, there is still much to learn about the historical development of disjunct distribution patterns and the underlying evolutionary mechanisms of species diversification. A deeper understanding of the evolution and geographic diversity of the Northern Hemisphere flora can be gained through large-scale biogeographic investigations of plant taxa that exhibit different intercontinental disjunctions (Hao et al., 2015).
Species of the genus Populus (Salicaceae), often known as poplars, cottonwoods and aspens, are widely distributed throughout the Northern Hemisphere from subtropical and boreal forests (Eckenwalder, 1996a; Cronk, 2005), and China has unique ecological conditions that make it an important natural distribution and diversity centre for poplars (Wan et al., 2013). A recent taxonomic study classified the 29 species in the genus Populus into six divisions (Abaso, Aigeiros, Leucoides, Populus, Tacamahaca, and Turanga) defined by the presence or absence of significant hybridization barriers (Eckenwalder, 1996a; Eckenwalder, 1996b). Most poplars exhibit a high level of ecological plasticity, with diverse adaptations and large population sizes. Section Abaso, with a single species P. mexicana Wesmael, and section Turanga, with three members, P. euphratica Olivier, P. pruinosa Schrenk and P. ilicifolia (Engl.) Rouleau, are restricted to warmer areas; species from sections Aigeros and Tacamahaca are found in riparian habitats; species from section Populus are found in drier areas; and most from section Leucoides live in swamps (Karrenberg et al., 2002). Additionally, poplars are known for their fast growth rates, extensive production of wood, along with their small genome sizes (Cronk, 2005; Hamzeh et al., 2006). Thus, poplars have been used as model organisms for many tree-related studies (Jansson and Douglas, 2007; Kuzovkina and Vietto, 2014). Populus is a genus that has a large number of species, has extensive records mainly across the Northern Hemisphere, and has unmistakable interspecific affinities (Liu et al., 2017). However, there are no phylogenetically based estimations of divergence times, and the biogeography of Populus is poorly understood. Currently, most research on the origin and evolution of Populus has been based exclusively on the fossil record (Manchester et al., 1986; Boucher et al., 2003; Manchester et al., 2006). Lineages and their biogeographic distributions have been mapped with the use of molecular phylogenies, and significant geological and ecological events have been more precisely dated using molecular clock, with fossil records serving as a calibrator for gene phylogenies (Berbee and Taylor, 2010).
To provide a timeline of a group’s evolutionary and biogeographic history, it is essential to have a solid understanding of that group’s fossil history (Helmstetter et al., 2019). The fossil records of Populus are extensive and common throughout the history of evolution (Supplementary Table S1). In China, the terrestrial strata of the Cretaceous and Tertiary are well developed and widely distributed, and abundant plant fossils have been preserved. Fossils that can be reliably attributed to P. latior Ai.Braum (Guo, 1975; Guo, 1983; Guo, 1985) have been dated to the late Cretaceous, corresponding to the Xigaze flora in Tibet, and another Cretaceous (Maestrichtian-Danian) fossil (72.1–66 million years ago, Ma) has been obtained from the Wuyun Formation in Jiayin, Heilongjiang (Guo, 1975; Guo, 1983; Guo, 1985; Tao and Xiong, 1986). According to fossil evidence, in the Cretaceous, Populus was widely dispersed in the Northern Hemisphere (Figure 1; Supplementary Table S1). In addition, all outgroups of Salicaceae and the greatest extant diversity of poplar are only found in Asia (Boucher et al., 2003; Cronk, 2005), but the basal group of Salicaceae and its fossils have not been found in North America. In the past few decades, robust phylogeny-based biogeography combined with molecular dating and ancestral area reconstruction methods has been frequently employed to trace the evolutionary and biogeographic history of species and factors driving the ranges of expansion of many plants (Riddle et al., 2008; Yu et al., 2010; Ali et al., 2012; Portelli and Quinteros, 2018). Nevertheless, the genesis and evolution of modern Populus biodiversity remain poorly understood. Here, we first evaluated divergence periods and recreated ancestral ranges to infer the historical processes that led to the modern global distribution of the genus Populus.
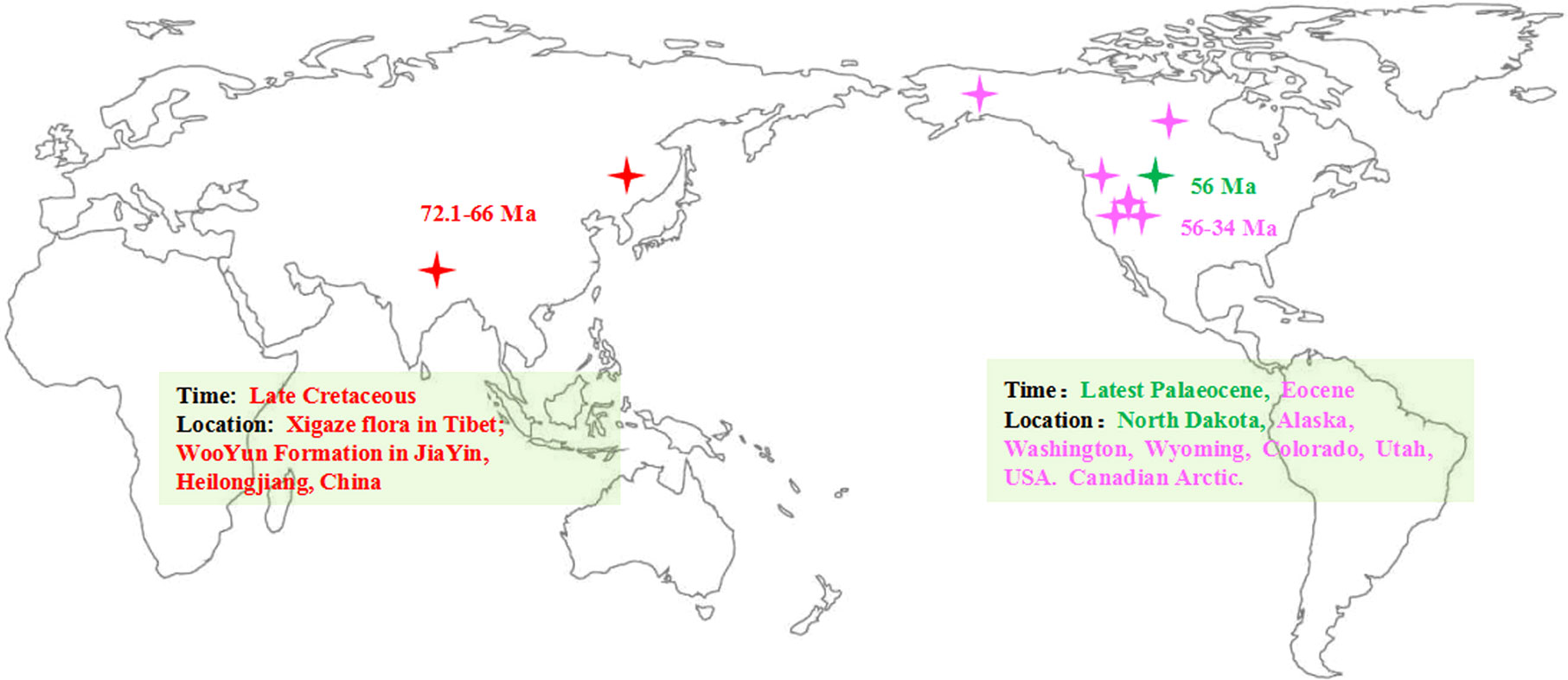
Figure 1 Map of the earlier fossil distribution of the genus Populus (the map adapt from Hao et al., 2015). The asterisk represented the location of the fossils.
Detailed procedures for DNA extraction, PCR, sequencing, and phylogenetic analysis, are provided (Liu et al., 2017). Briefly, we collected samples from all 34 species in the genus Populus, representing all six sections of the genus according to the taxonomy system of Eckenwalder (1996a) and Wu (1999); four species in the genus Salix, and three species of related genera of Salicaceae were sampled as outgroups for phylogenetic analyses. Details of voucher information and GenBank numbers for all samples (including the new species) are mentioned in Supplementary Table S2. In addition, the chloroplast (cp) fragment molecular dataset was used to reconstitute Populus species-level trees in this study, which were then utilized for divergence time calculation.
The sequences were aligned using ClustalX v1.83 (Thompson et al., 1997) and further adjusted manually in BioEdit (Hall, 1999). A number of loci or regions that were unable to be amplified were regarded as missing data. The 34 cpDNA fragments were combined into a single dataset for phylogenetic inference using maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI) with PAUP* v4.0b10 (Swofford, 2003), PhyML v2.4.4 (Guindon and Gascuel, 2003) and MrBayes v3.1.2 (Ronquist and Huelsenbeck, 2003), respectively.
The earliest fossil has been dated to 72.1–66 Ma (Guo, 1975, 1983, 1985; Tao and Xiong, 1986), this means that the most conservative minimum for the genus origin is 66 Ma, but it does not mean that 72.1 Ma is the conservative maximum, as Populus may have existed well before 72.1 Ma (though presently without such older fossils identified). A reasonable conservative maximum is 93 Ma, according to Davis et al. (2005) and the source (https://www.mobot.org/mobot/research/apweb/orders/malpighialeswebnew.htm ). Therefore, we utilized internal calibration to establish a divergence time of 66-93 Ma between Populus and Salix. As in prior research, a lognormal distribution with soft limits and confidence intervals was used by calibrating the means and standard deviations, allowing for the incorporation of error around the projected dates and stratigraphic assignment of the fossils themselves (Su et al., 2017). Divergence times were assessed utilizing BEAST v1.8.3 (Drummond et al., 2012) under a relaxed uncorrelated lognormal clock model utilizing the GTR+I+G substitution model evaluated by jModeltest v3.0.4 (Posada, 2008), birth-death tree prior, the relaxed lognormal clock model, and bootstrap rates using a calibrated rate ranging from 5.10e-4-9.19e-4 substitutions/site/myr (million years) based on Lanfear et al. (2013). Samples were taken every 5,000 generations from three independent runs of 500 million generations. After that, the results of the three separate runs were merged utilizing LogCombiner v2.0 (Drummond and Rambaut, 2007), and the first 10% of the samples were discarded as burn-in. Calculating the effective sample sizes of the parameters utilizing Tracer v1.7 (Rambaut et al., 2018) allowed us to evaluate convergence and ensure sufficient sampling. The values of the effective sample size (ESS) for all of the parameters exceeded 200. Trees were summarized as a maximum clade credibility (MCC) tree (maximum posterior probabilities) utilizing Tree Annotator v2.0 (Drummond and Rambaut, 2007), and the mean ages, 95% highest posterior density (HPD) intervals, posterior probabilities and substitution rates were calculated for each node. FigTree v1.4.2 (Rambaut, 2014) was utilized to depict the divergence tree.
Ancestral areas were reconstituted utilizing RASP v4.2 (Yu et al., 2020): (1) Statistical Dispersal-Violation Analysis (S-DIVA), which explicitly uses a whole posterior distribution of trees to account for both phylogenetic uncertainty and uncertainty in ancestral states, and (2) a Dispersal-Extinction-Cladogenesis (DEC) model in Lagrange analysis (Ree and Smith, 2008), which requires data on a single ultrametric-dated phylogeny and distributional information of extant species. Not only does the Lagrange analysis estimate the ancestral states, but it also calculates the probabilities of the areas at each node (Hao et al., 2015). These are helpful methodological instruments that eliminate the need for arbitrarily defined areas. The species ranges of the genus Populus were separated into 5 regions to accomplish biogeographic analysis. These regions were separated as follows: (A) Asia and surrounding areas, (B) Europe, (C) North America (including Mexico), (D) Central Asia, and (E) North Africa. Ancestral area reconstruction was performed using a condensed tree generated from the BEAST analysis. The biogeographical data were coded in accordance with the range of extant poplar species that were selected for this study.
We compiled the dataset by sampling the greatest number of Populus species to conduct phylogenetic analysis. The concatenated and aligned molecular datasets of 34 plastid fragments consisted of 36,031 base pairs (excluding outgroups). In the ML, the evolutionary model that provided the best match for the combined cpDNA datasets was the GTR+I+G model, while the TIM2+I+G model provided the best fit for the Bayesian analysis. Three methods, including MP, ML, and BI, utilized phylogenetic tree construction yielded similar topology. The chloroplast phylogenies also support Populus monophyly in the present research, confirming the conclusions of the vast majority of prior molecular phylogenetic investigations (Cervera et al., 2005; Liu et al., 2017; Wang et al., 2020). Populus species may be categorized into three largely supported clades that highly correspond to geographic distributions based on the combined cpDNA dataset analyses (Figure 2B). Clade I comprised all Eurasian species, with the exception of two species from North America (P. tremuloides and P. grandidentata) and one North African species (P. ilicifolia). Clade II comprised all the East Asian species that were well supported (96% MP, 98% BS, 1.00 BPP). These Asian species were all Tacamahaca species, but the extant Populus species in North America have differentiated into Aigeiros, Tacamahaca and Leucoides. Clade III consisted of the remaining seven North American species (Figure 2A). Our phylogenetic results obtained from the combined chloroplast dataset had high resolution.
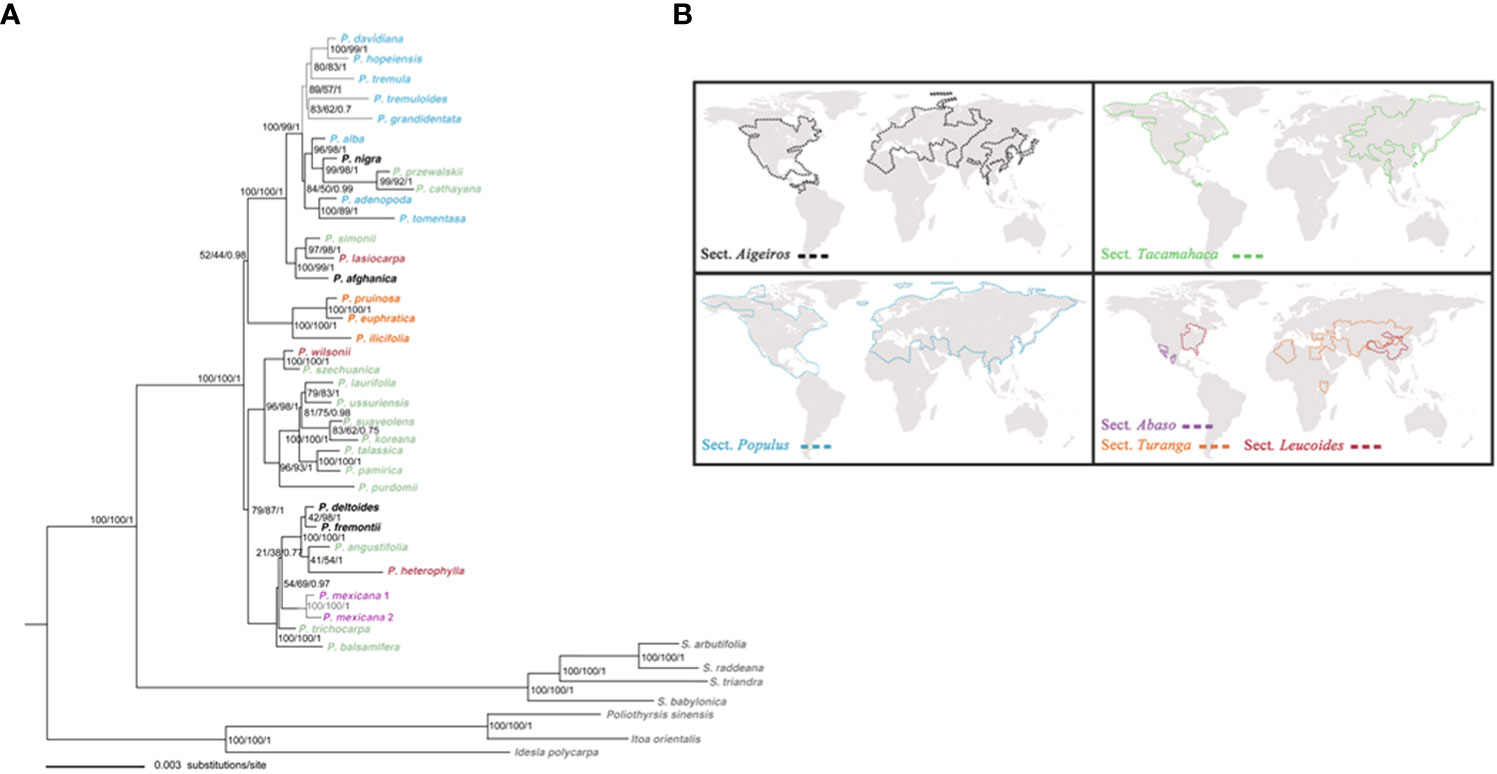
Figure 2 (A) The ML tree of Populus and outgroups based on combined 34 plastid fragments. Numbers above branches indicate bootstrap values for the MP and ML analyses, and Bayesian posterior probabilities, respectively. (B) The geographical distributions of six sections of genus Populus is modified from Wang et al. (2020).
Two hybrids, P. hopeiensis and P. tomentosa, were excluded in estimating the divergence time, because we mainly focused on divergence at the species level of Populus. A total of 39 sequences from the combined chloroplast dataset were used for divergence time estimation. In fact, excluding the two hybrids did not significantly affect the overall divergence-age estimates (Supplementary Figure S1), hence they were removed to improve the overall topological support values instead. Timetree revealed estimated divergence times between Salicaceae and Flacourtiaceae (91.75 Ma, 95% HPD: 81.74–115.79) and between Populus and Salix (68.67 Ma, 95% HPD: 63.73–75.05) (Figure 3A). The diversification of modern Populus species occurred in the late Oligocene at 27.24 Ma (95% HPD: 21.64–35.18). The divergence time estimation of Clade I taxa was 25.65 Ma (95% HPD: 19.87–31.96), which was slightly earlier than the divergence time of the Clade II and Clade III groups, which both diverged at 24.15 Ma (95% HPD: 19.03–29.12). Clade II comprises species from East Asia that first diverged at 17.56 Ma (95% HPD: 13.27–22.63). The divergence time corresponding to the rapid diversification of this clade was 11.96 Ma (95% HPD: 9.14–16.83). Clade III, a North American branch with a shared ancestor according to the plastid data, is the result of differentiation from a dispersal event from East Asian Tacamahaca species.
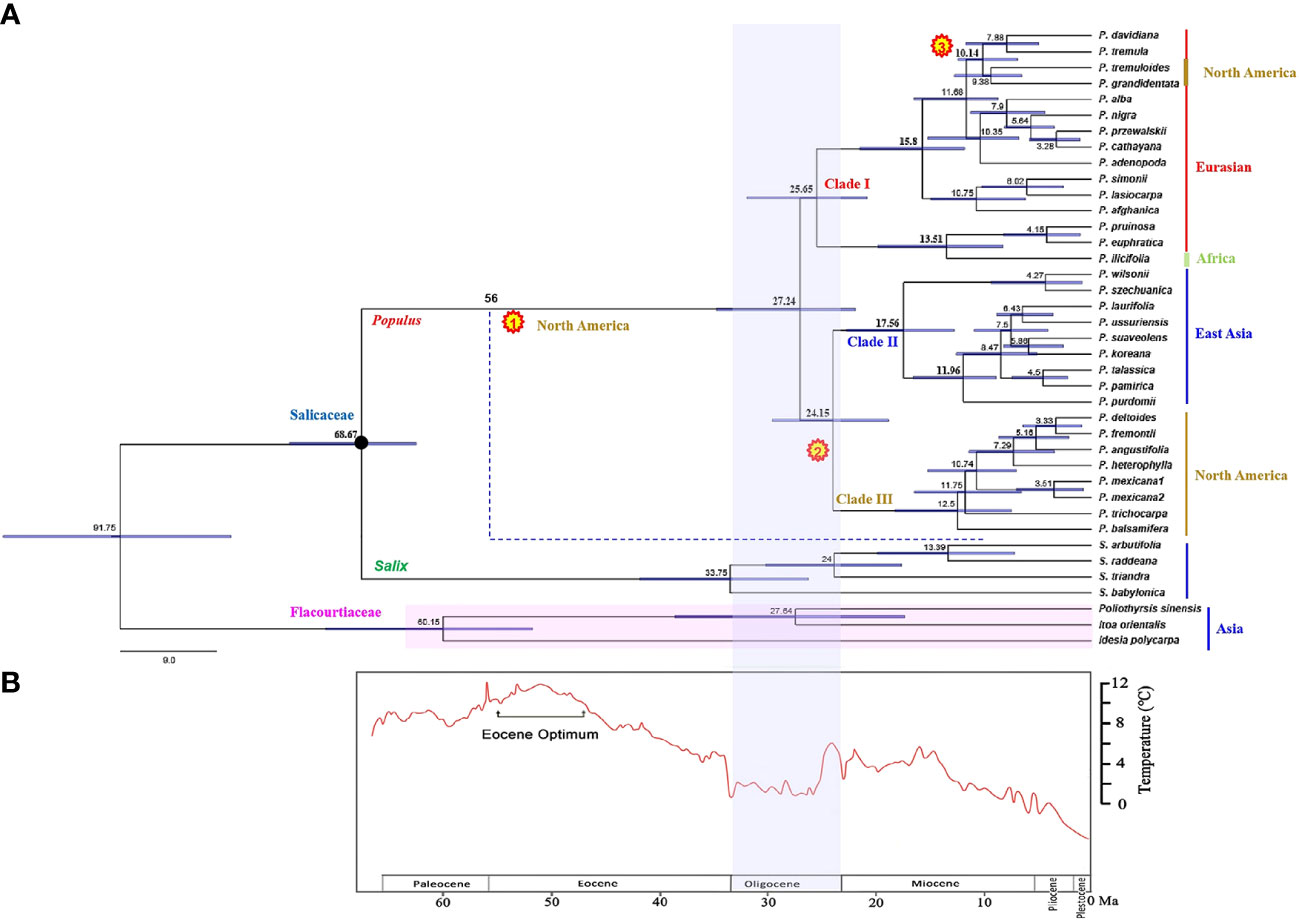
Figure 3 (A) Divergence time of Populus estimated from combined chloroplast DNA fragments. The black pie charts indicate the nodes calibrated by the fossils (66–93 Ma). The divergence time estimates shown on the nodes are median ages. Numbers in yellow pie charts show three times independently reached into North America events, respectively. The blue dotted line indicates an extinct branch that colonization of North America. The solid line of color next to the taxa names represent the different distribution of species. (B) The depiction of climate change past 60 million years before present is modified from Zachos et al. (2001).
The results from reconstructions of Populus’s ancestral area utilizing the DEC and S-DIVA models were very congruent (Figure 4). Multiple potential ancestral distribution patterns were predicted for each node; thus, we concentrated on nodes with the highest relative probabilities. The ancestral reconstruction at node 75 (ancestral node) indicates that the most likely ancestral region of the genus Populus is in Asia and adjacent areas (A) with later spread to other areas, and this node was strongly supported with a posterior probability value of 1.00. Throughout the evolution of the genus Populus, four dispersal episodes (D1–D4) and two vicariance events (V1–V2) may have occurred. A dispersal event was suggested at node 75 from A to C by ancestral reconstruction at nodes 74 and 60. At node 60, a vicariance event occurred (the most favoured ancestral area was AC at this node). One ancestral event persisted in area A, while the other diversified in area C; a second vicariance event was identified at node 67. The two model (DIVA and DEC) analyses all supported that Populus originated from Asia and neighbouring areas and subsequently expanded to Europe, North America and North Africa.
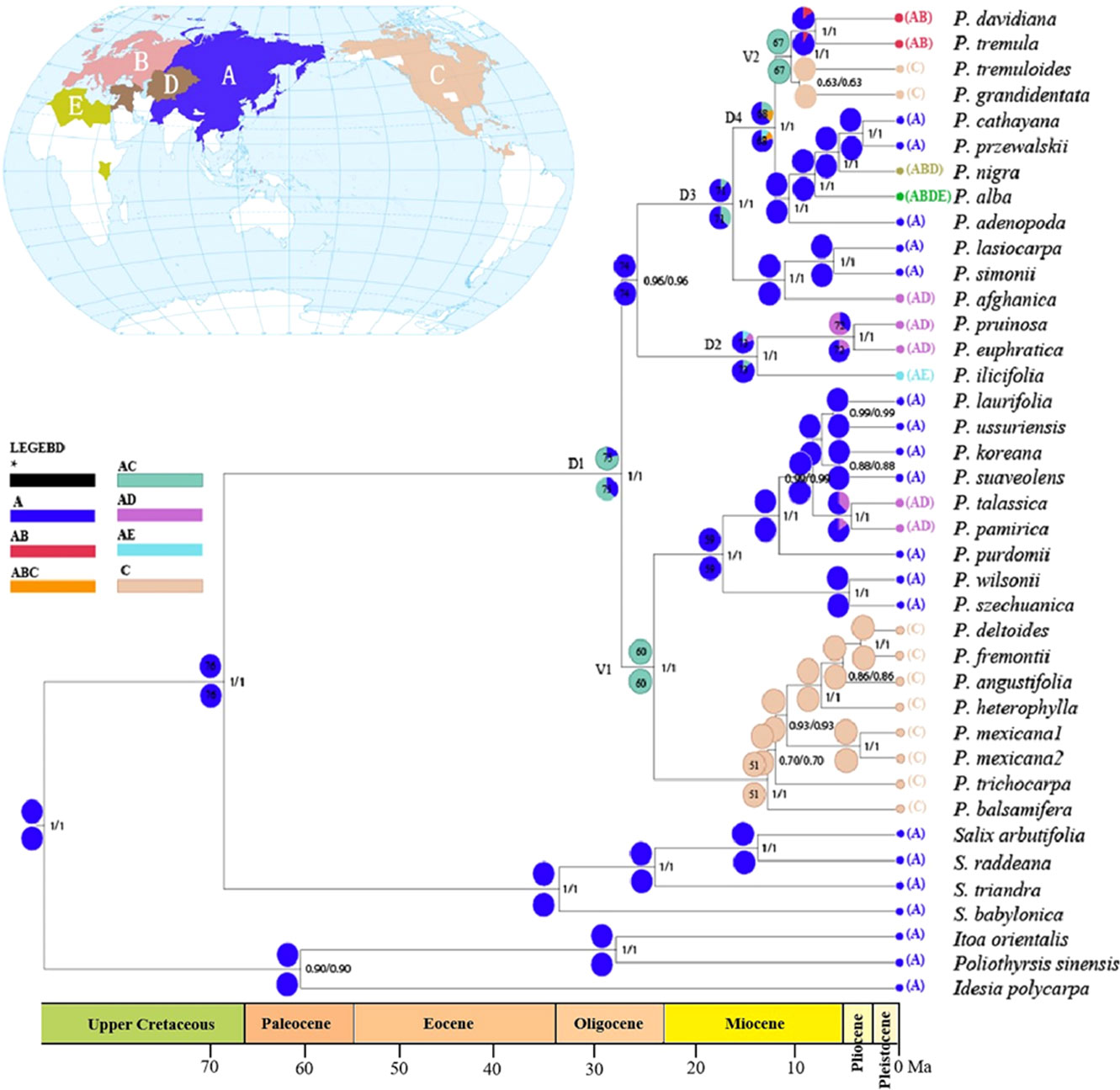
Figure 4 Ancestral area reconstruction for Populus based on statistical dispersal-vicariance analysis (S-DIVA) and dispersal-extinction-cladogenesis (DEC) overlaid onto the maximum clade credibility chronogram from BEAST. Current distributions are indicated before the species names. Five areas were defined: (A) Asia and neighboring areas, (B) Europe, (C) North America (including Mexico), (D) Center Asia, and (E) North Africa; black with an asterisk represents other ancestral ranges. Pie charts at nodes indicate probabilities of alternative ancestral ranges obtained by DIVA (below) and DEC (above). D1–4 and V1–2 indicate dispersal and vicariance events, respectively. Nodes of interests were marked as 1-11 in pie charts.
Based on our time estimates (Figure 3A) and ancestral area reconstruction (Figure 4), the stem age of Salicaceae was approximately 68.67 Ma (95% HPD: 63.73–75.05), which is in line with the oldest known Populus leaflet fossil record from East Asia (Guo, 1975; Guo, 1983; Guo, 1985; Tao and Xiong, 1986; Figure 1; Supplementary Table S1) and coincides with the K-Pg mass extinction boundary at approximately 66 Ma. The transition from the Eocene to the Oligocene was marked by a sudden cooling climate due to the establishment of permanent ice sheets in Antarctica (Figure 3B; Zachos et al., 2001). Global climatic change at the Eocene‐Oligocene boundary resulted in increased extinction and turnover of regional floras throughout the globe (Ivany et al., 2000; Retallack et al., 2004; Zanazzi et al., 2007; Sun et al., 2014; Nge et al., 2020). According to our results, the genus Populus forms a monophyletic group that separated into three distinct clades, which generally correspond to geographic distributions (Figure 2B). The three major branches of the lineages were very short in the phylogenetic tree (Figure 2A). Therefore, we speculated that the extant Populus species might originated from rapid radiation events in the Oligocene (Liu et al., 2017). During that period, the fluctuating global climates tended to be cooler and drier, the contemporary world’s general landform had already evolved, and severe climate changes were occurring (Tiffney and Manchester, 2001; Zachos et al., 2001). Until the Quaternary glaciations, the ensuing polar ice sheets expanded and retreated multiple times and covered the majority of North America and northern Europe (Zachos et al., 2001). Arid climatic belts also evolved in the core of Eurasia and North America during this time period (Milne and Abbott, 2002). Therefore, the cooling climate likely had a significant impact on the evolution and diversification of contemporary poplar species, which are primarily found in temperate and subtropical parts of the Northern Hemisphere. A previous study of organisms, such as Fagaceae (Manos and Stanford, 2001), Abies (Aguirre-Planter et al., 2012) and Populus (Du et al., 2015), demonstrated that the extreme climate changes that occurred throughout this time led to the dispersal of populations, the extinction of species, and the formation of new species. The estimated divergence of species in Clade I and Clade II occurred in the late Tertiary at 15.80 Ma (95% HPD: 12.16–22.04) and 17.56 Ma (95% HPD: 13.27–22.63), respectively (Figure 3A). During this period, the elevation of the Qinghai-Tibetan Plateau (QTP) drastically changed the terrain of eastern Asia and resulted in remarkable environmental variation, which greatly increased species diversity (Wei et al., 2010). The enormous plateau (Qinghai) and deserts from the north presumably acted as significant physical and ecological barriers that restricted migration and floristic exchange between East Asian species (Clade II) and those of Eurasia. Numerous plant genera, including Juniperus (Mao et al., 2010), Cedrus (Qiao et al., 2007), and Helleborus L. (Sun et al., 2001) have been documented to have a tight biogeographic relationship between the Mediterranean and the QTP.
As during the Palaeocene and Eocene (characterized by hot and humid climates) the boreotropics played a key role in facilitating the intercontinental exchange of tropical biota. However, the disruption that occurred during the late Eocene and early Oligocene, when the climate became cooler and drier, has been shown to have played an important role in forming the disjunct distribution patterns that are observed currently in several vascular plant lineages through vicariance (Baker and Couvreur, 2013; Thomas et al., 2015). The divergence time of current Populus species was about 27.24 Ma, which was consistent with the estimation of the destruction time of boreotropic flora. As for the species of Poulus species distributed in East Asia and North America, the former region often possesses a greater variety of species than the latter.
The relict branch formed the most recent common ancestor (MRCA) of extant Populus species was found in the early Oligocene (27.24 Ma) (Figure 3A). The climate during the Oligocene is characterized by climate fluctuations and a long-term cooling tendency (Figure 3B). Permanent Antarctic ice sheets lasted until the Late Oligocene, when a warming trend diminished their extent (Zachos et al., 2001). This warm phase reached its maximum extent during the late Mid-Miocene Climatic Optimum (MMCO, 15–17 Ma), which was then immediately followed by a progressive cooling of the temperature and the formation of a significant ice sheet in Antarctica by 10 Ma. (Flower and Kennett, 1995; Hao et al., 2015). These alterations in the Antarctic ice sheets are evidence of a globally significant climate oscillation that caused the BLB to become accessible for trade during a period of warmer temperatures (Tiffney, 1985a; Tiffney, 1985b). Since the Late Cretaceous period, the biological link that connects western North America and eastern Asia has remained mostly unchanged (Wen et al., 2016). Through the NALB and BLB, migration between Eurasia and North America was conceivable during the Cretaceous and Early Tertiary (Manchester et al., 2009). Throughout the majority of the Tertiary, the BLB was a key high-latitude link for plants between eastern Asia and North America (Tiffney, 1985a; Novacek, 1999).
Our ancestral reconstruction indicates that four dispersal (D1-D4) and two vicariance (V1-V2) events from Asia and neighbouring areas to North America have occurred (Figure 4). We found that Eurasian Populus species might have colonized the Americas three times through the BLB (Figure 5). Populus firstly reached North America via the BLB and/or NALB during the Paleo (approximately 56 Ma) or Upper Cretaceous (Figure 5A). The only remaining genetic traces of ancient species are from chloroplast capture (the chloroplast genome of one species was completely swapped out for that of another) by male plants of the ancestral species of P. mexicana, which also became extinct, such as P. cinnamomoides (Lesquereux) MacGinitie (MacGinitie, 1969; Eckenwalder, 1980), P. wilmattae Cockrell (Manchester et al., 1986), and P. tidwellii sp. n. (Manchester et al., 2006). Chloroplast capture is a common phenomenon in biological organisms (Petit et al., 2004). Well-noted examples of chloroplast capture have been reported, such as Eucalyptus (Rieseberg and Soltis, 1991), Helianthus (Ellis et al., 2008), Nothofagus (Acosta and Premoli, 2009), Lactuca (Wang et al., 2013) and Quercus (Deren et al., 2015). Among these convincing cases, both domestic and wild species are involved, and hybridization between intrageneric and intergeneric species occurs (Tsitrone et al., 2003). Thus, chloroplast capture is not rare and might be a common occurrence in Northern Hemisphere temperate plants. The second colonization was the dispersal of the ancestor species of the Tacamahaca section in east Asia during 24.15 Ma (95% HPD: 19.03–29.12; Figure 5B), which differentiated into today’s North American Populus species (P. balsamifera, P. trichocarpa, P. mexicana, P. heterophylla, P. angustifolia, P. fremontii, and P. deltoides). P. mexicana may be an ancient hybrid produced by the ancestral species that first dispersed to North America and retreated to Mexico to capture chloroplasts of the ancestors of P. trichocarpa and P. balsamifera that spread to North America for the second time, and the ancestor species of modern P. mexicana became extinct as the climate cooled. The third arrival in North America was the spread of the ancestor of Eurasian aspen at 10.14 Ma (95% HPD: 7.26–12.97), which and differentiated to form present-day P. tremuloides and P. grandidentata (Figure 5C). Both long-distance dispersals across the BLB and vicariance events probably contributed to the patterns of disjunction between eastern Asia and western North America in Populus. The resulting disjunctions have previously been inferred in plants (Donoghue and Smith, 2004; Wen et al., 2016) as well as animals (Sanmartín et al., 2001). These patterns of dispersal are mostly consistent with the hypothesis that the majority of the world’s surviving temperate forest flora began in east Asia and spread out over the course of the past 30 Ma (Donoghue and Smith, 2004; Wen et al., 2016). It is believed that the extensive pattern of divergent distributions of temperate forest species among Asia (mainly in East Asia) and North America is at least partially attributable to colonization across the BLB (Wen, 1999). However, another equally likely alternative scenario, given that fossils of the poplars are recorded in North America from the early Eocene, is that the ancestor of the genus Populus was once widespread across the Northern Hemisphere (including North America), but then due to higher extinction in Northern America, it was wiped out. Extinction and fragmentation of the original distributional range (vicariance) can occur if the changes caused by abiotic factors such as abrupt climatic or tectonic events are too rapid or large for species to adapt or migrate to more favourable areas (Wiens, 2004). For example, the famous ‘Rand Flora’, a continental-scale biogeographic disjunction pattern that suffered high extinction rates due to successive aridification waves, in northern and central Africa once had a widespread distribution, but now it is fragmented (Sanmartín et al., 2010; Pokorny et al., 2015). In addition, similar examples can be found in the Australian flora (Crisp and Cook, 2007; Nge et al., 2021). This uncertainty in our results may be due to the effects of extinction during Populus’s evolutionary history. Erroneous inferences about past geographical ranges are more likely when there are inconsistent extinction rates between different regions (Sanmartín and Meseguer, 2016). However, the Eurasian region has relatively higher Populus species diversity than North America (Boucher et al., 2003), a large number of Populus species living in Asia during the Eocene and Oligocene are well documented (Supplementary Table S1), and the less affected historical extinctions may have driven models towards an inference of Asia as an ancestral area. Biogeographic models and analyses based on current species distributions can be misleading due to the extinction of certain lineages (Sanmartín and Meseguer, 2016). Thus, the scenario of Eurasia as an ancestral area followed by long-distance dispersal should be treated with caution, and other alternative scenarios, such as an ancestral distribution that was once more widespread across the Northern Hemisphere (including North America) but destroyed by extinction, should also be considered in biogeographic studies.
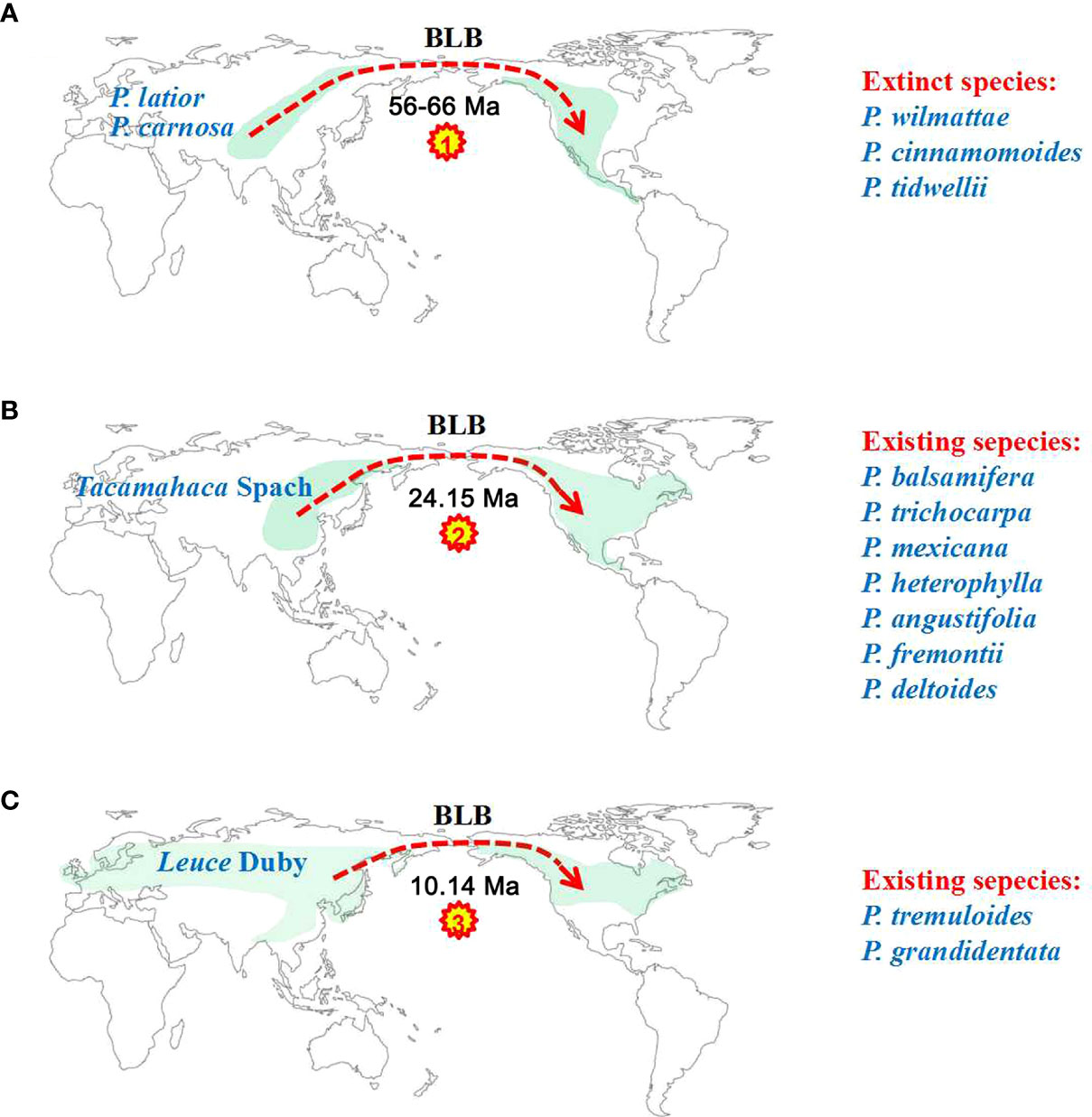
Figure 5 Three colonization events of North America by Eurasian poplar taxa via the BLB. (A) Descendants of the first colonization event of North America eventually became extinct. (B) The second colonization of Tacamahaca section species of North America from East Asia produced modern North American Populus species. (C) The third colonization event of North America formed P. tremuloides and P. grandidentata.
The diversification of the extant Populus is largely the result of in situ speciation with each of the defined areas during the early Oligocene and following a few independent dispersal and vicariance events at different time periods (Figures 3, 4). Our analyses indicate that the extant Populus may experienced three independent North American dispersal events via the BLB (Figure 5). An examination of the biogeography of a few further Eurasian species, such as Betulaceae (Forest et al., 2005), Rosaceae (Lo et al., 2009), and Sapindaceae (Forest et al., 2001; McClain and Manchester, 2001), indicates similar patterns. The ancestry of today’s poplars can be traced back to the early Oligocene (27.24 Ma), but their current diversity did not emerge until the early Miocene (17.56 Ma) (Figure 3A) based on our study. Several other important East Asian angiosperms also diverged about this time: Castanopsis (D. Don) Spach (approximately 18.5 Ma; Fagaceae; Xing et al., 2014), Michelia L. (approximately 21 Ma; Magnoliaceae; Nie et al., 2008), and Tribus Theeae DC. (approximately 19.4 Ma; Theaceae; Yu et al., 2017), which diverged in the early Miocene. The existence of several comparable fossil taxa on both sides of the Pacific implies that interactions between eastern Asia and North America were possible via the BLB from the Cretaceous to the late Neogene (Manchester et al., 2009). The MRCA of extant poplar species most likely survived because it was less affected by glaciers than North America and diverged in the Paratethys region of central Eurasia (mainly in East Asia) based on the Qaidam Basin leaf fossils of Populus (Song et al., 2020). Some potential sister groups or all Salicaceae outgroups exist only in Asia (Boucher et al., 2003), and similar patterns have been observed in Betulaceae (Forest et al., 2005), Rosaceae (Lo et al., 2009), and Sapindaceae (Forest et al., 2001; McClain and Manchester, 2001).
Within Clade I, there were two pairs of sister species (P. tremuloides and P. grandidentata), which were sister to Eurasian species, P. davidiana and P. tremula, exhibiting a Eurasia–North America temperate disjunct distribution (Figure 2). Their divergence time was estimated to be about 10.14 Ma (95% HPD: 7.26–12.97), which indicates that the long-distance dispersal across Eurasia and North America may have occurred through the BLB, resulting in P. tremuloides and P. grandidentata. This phenomenon is widespread in plants such as Fagaceae (Wen et al., 2016). The BLB broke down from 5.4 to 5.5 Ma (Gladenkov et al., 2002) and linked East Asia and North America before that period. The BLB has played a crucial role in the spread of numerous other species (Feng et al., 2012; Cai et al., 2014). The divergence of a pair of sister species, P. tremuloides and P. grandidentata (Figure 3A), may be related to the continuous decline in the climate on the North American continent 11.8 Ma (Zachos et al., 2001).
Our data suggest that most North American species (Clade III) and East Asian species (Clade II) were grouped together, Clade III split from Clade II at 24.15 Ma (95% HPD: 19.03–29.12), and its divergence time was approximately 12.50 Ma (95% HPD: 7.62–18.93) (Figure 2). Therefore, we speculated that dispersal events occurred among western North America and East Asia and that ancestral taxa likely spread to North America via the BLB. Clade III did not diversify immediately after it reached North America but only after the climate began to cool. Allopatric speciation and the breakage of biotic links between Tertiary floras of North America and East Asia may have been aided by the drastic climatic shifts that occurred between 15 Ma and the Last Glacial Maximum (Tiffney and Manchester, 2001; Zachos et al., 2001). North American Clade III is monophyletic, but it is divided into three sections (Aigeiros, Leucoides, Tacamahaca). Therefore, the North American lineages may well be a combination of both vicariance and dispersal events in different time periods.
Turanga is the basal section, as shown in Figure 2A, which accords with the findings of Wang et al. (2020). The Turanga section, with only three species, is mainly distributed in central Asia (P. euphratica and P. pruinosa) and northern Africa (P. ilicifolia). In addition, there were no differences in leaf morphology between P. ilicifolia and P. euphratica, but two sister species, P. euphratica and P. pruinosa were morphologically distinct. The majority of species in this division grow in tropical or subtropical conditions, showing that the genus Populus evolved in warmer environments. During the Oligocene or in the Middle Miocene, northern temperate mesic ancestors once included many Mediterranean species that can withstand drought, though just a few relics can be found in eastern Asia (Wen, 1999; Wen, 2001; Appelhans et al., 2016). These data, together with estimations of divergence time (Figure 3A), suggest that low-latitude Turanga species are likely relics of the Madrean-Tethyan vegetation zone. P. ilicifolia might be an earlier relict of Mediterranean species following its origin from subtropical central Asia and subsequent spread to North Africa, where P. ilicifolia and P. euphratica arose (13.51 Ma, 95% HPD: 8.53–20.02). P. pruinosa diverged from P. euphratica at approximately 4.15 Ma (95% HPD: 2.37–8.53). The divergence time of these two species coincides with that of the most recent increase in the QTP (An et al., 2001), suggesting that the uplift of the QTP might have been a major factor driving the origin and diversification of Populus. Although the chloroplast datasets analyzed here provide the species-level framework of Populus, more evidence for relationships within the genus is still needed. Particularly, nuclear genes evidence are required to explain complex evolutionary history of the genus at future studies, because chloroplast datasets usually show a strong geographic pattern not reflecting of true species relationships. In addition, chloroplast datasets and nuclear datasets, as in most cases, are in conflict which each other.
Our study applied divergence-time estimations and biogeographic models to investigate the origin and ancestral area of Populus, providing a robust framework for further research on the genus. Our study indicates that the present-day disjunction in Populus can be explained by Populus likely originating in Eurasia and subsequently colonizing other regions, including North America. The climatic change in the Oligocene-Miocene likely drove the diversification of present-day poplar species, the modern North American lineages that may well be a combination of both vicariance and dispersal events from different time periods. An alternative scenario of a once widespread ancestral distribution now erased by mass extinction, similar to the African Rand flora, should also be considered in the biogeographic history of Populus. Further studies with dense sampling and more evidence are required to test these hypotheses.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JZ and ZW designed the study. XL, SD and QH prepared materials and performed most of the experiments. XL, WW, HL, YZ and YJ performed the data analysis. XL and ZW wrote the manuscript. All authors have read and commented on the paper. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (31470665).
We sincerely thank Xingguang Li from Hubei Engineering Technology Research Center, Hubei, China, for bioinformatics software help; and we would like to thank Peng Guo and Guoyun Zhang for their assistance in the lab. We also thank two reviewers and the Associate Editor for their comments that helped to improve the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1031087/full#supplementary-material
Acosta, M. C., Premoli, A. C. (2009). Evidence of chloroplast capture in south American Nothofagus (subgenus Nothofagus, nothofagaceae). Mol. Phylogenet. Evol. 54, 235–242. doi: 10.1016/j.ympev.2009.08.008
Aguirre-Planter, É, Jaramillo-Correa, J. P., Gómez-Acevedo, S., Khasa, D. P., Bousquet, J., Eguiarte, L. E. (2012). Phylogeny, diversification rates and species boundaries of Mesoamerican firs (Abies, pinaceae) in a genuswide context. Mol. Phylogenet. Evol. 62, 263–274. doi: 10.1016/j.ympev.2011.09.021
Ali, S. S., Yu, Y., Pfosser, M., Wetschnig, W. (2012). Inferences of biogeographical histories within subfamily hyacinthoideae using s-DIVA and Bayesian binary MCMC analysis implemented in RASP (Reconstruct ancestral state in phylogenies). Ann. Bot. 109, 95–107. doi: 10.1093/aob/mcr274
An, Z. S., Kutzbach, J. E., Prell, W. L., Porter, S. C. (2001). Evolution of Asian monsoons and phased uplift of the himalaya-Tibetan plateau since late Miocene times. Nature. 411, 62–66. doi: 10.1038/35075035
Appelhans, M. S., Krohm, S., Manafzadeh, S., Wen, J. (2016). Phylogenetic placement of psilopeganum, a rare monotypic genus of rutaceae (the citrus family) endemic to China. J. Syst. Evol. 54, 535–544. doi: 10.1111/jse.12208
Baker, W. J., Couvreur, T. L. P. (2013). Global biogeography and diversification of palms sheds light on the evolution of tropical lineages. I. historical biogeography. J. Biogeogr. 40, 274–285. doi: 10.1111/j.1365-2699.2012.02795.x
Berbee, M. L., Taylor, J. W. (2010). Dating the molecular clock in fungi-how close are we? Fungal. Biol. Rev. 24, 1–16. doi: 10.1016/j.fbr.2010.03.001
Boucher, L. D., Manchester, S. R., Judd, W. S. (2003). An extinct genus of salicaceae based on twigs with attached flowers, fruits, and foliage from the Eocene green river formation of Utah and Colorado, USA. Am. J. Bot. 90, 1389–1399. doi: 10.3732/ajb.90.9.1389
Cai, Q., Tulloss, R. E., Tang, L. P., Tolgor, B., Zhang, P., Chen, Z. H., et al. (2014). Multi-locus phylogeny of lethal amanitas: Implications for species diversity and historical biogeography. BMC Evol. Biol. 14, 143. doi: 10.1186/1471-2148-14-143
Cervera, M. T., Storme, V., Soto, A., Ivens, B., Van, Montagu, M., Rajora, O. P., et al. (2005). Intraspecific and interspecific genetic and phylogenetic relationships in the genus Populus based on AFLP markers. Theor. Appl. Genet. 111, 1440–1456. doi: 10.1007/s00122-005-0076-2
Crisp, M. D., Cook, L. G. (2007). A congruent molecular signature of vicariance across multiple plant lineages. Mol. Phylogenet. Evol. 43, 1106–1117. doi: 10.1016/j.ympev.2007.02.030
Cronk, Q. C. B. (2005). Plant eco-devo: The potential of poplar as a model organism. New Phytol. 166, 39–48. doi: 10.1111/j.1469-8137.2005.01369.x
Davis, C. C., Webb, C. O., Wurdack, K. J., Jaramillo, C. A., Donoghue, M. J. (2005). Explosive radiation of malpighiales supports a mid-Cretaceous origin of modern tropical rain forests. Am. Naturalist. 165, E36–E65. doi: 10.1086/428296
Deren, A. R. E., Andrew, L. H., Antonio, G. R., Jeannine, C. B. (2015). Historical introgression among the American live oaks and the comparative nature of tests for introgression. Evolution. 69, 2857–2601. doi: 10.1111/evo.12758
Donoghue, M. J., Bell, C. D., Li, J. H. (2001). Phylogenetic patterns in northern hemisphere plant geography. Int. J. Plant Sci. 162, S41–S52. doi: 10.1086/323278
Donoghue, M. J., Smith, S. A. (2004). Patterns in the assembly of the temperate forest around the northern hemisphere. Philos. T. R. Soc B. 359, 1633–1644. doi: 10.1098/rstb.2004.1538
Drummond, A. J., Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. doi: 10.1186/1471-2148-7-214
Drummond, A. J., Suchard, M. A., Xie, D., Rambaut, A. (2012). Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Phylogenet. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Du, S. H., Wang, Z. S., Ingvarsson, P. K., Wang, D. S., Wang, J. H., Wu, Z. Q., et al. (2015). Multilocus analysis of nucleotide variation and speciation in three closely related Populus (Salicaceae) species. Mol. Ecol. 24, 4994–5005. doi: 10.1111/mec.13368
Eckenwalder, J. E. (1980). Foliar heteromorphism in Populus (Salicaceae), a source of confusion in the taxonomy of tertiary leaf remains. Syst. Bot. 5, 366–383. doi: 10.2307/2418518
Eckenwalder, J. E. (1996a). “Systematics and evolution of populus,” in Biology of populus. Eds. Stettler, R. F., Heilman, J. P. E., Hinckley, T. M. (Ottawa, Canada: National Research Council of Canada Research Press), 7–32.
Eckenwalder, J. E. (1996b). Taxonomic signal and noise in multivariate interpopulational relationships in Populus mexicana (Salicaceae). Syst. Bot. 21, 261–271. doi: 10.2307/2419658
Ellis, J. R., Bentley, K. E., McCauley, D. E. (2008). Detection of rare paternal chloroplast inheritance in controlled crosses of the endangered sunflower helianthus verticillatus. Heredity 100, 574–580. doi: 10.1038/hdy.2008.11
Feng, B., Xu, J. P., Wu, G., Hosen, M. I., Zeng, N. K., Li, Y. C., et al. (2012). DNA Sequences analyses reveal abundant diversity, endemism and evidence for Asian origin of the Porcini mushroom. PloS One 7, e37567. doi: 10.1371/journal.pone.0037567
Flower, B. P., Kennett, J. P. (1995). Middle Miocene deepwater paleoceanography in the southwest pacific: Relations with east Antarctic ice sheet development. Paleoceanogr. Paleocl. 10, 1095–1112. doi: 10.1029/95PA02022
Forest, F., Drouin, J. N., Charest, R., Brouillet, L., Bruneau, A. (2001). A morphological phylogenetic analysis of Aesculus l. and Billia peyr. (Sapindaceae). Can. J. Bot. 79, 154–169. doi: 10.1139/b00-146
Forest, F., Savolainen, V., Chase, M. W., Lupia, R., Bruneau, A., Crane, P. R. (2005). Teasing apart molecular-versus fossil-based error estimates when dating phylogenetic trees: A case study in the birch family (Betulaceae). Syst. Bot. 30, 118–133. doi: 10.1600/0363644053661850
Gladenkov, A. Y., Oleinik, A. E., Marincovich, L., Barinov, K. B. (2002). A refined age for the earliest opening of Bering strait. Palaeogeogr. Palaeoclimatol. Palaeoecol. 183, 321–328. doi: 10.1016/S0031-0182(02)00249-3
Guindon, S., Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Bot. 52, 696–704. doi: 10.1080/10635150390235520
Guo, S. X. (1975). “The plant of the xigaze group from mount jolmo lungma region,” in Report of scientific expedition to mt. jolmo lungma region (Beijing, China: Science Press), 411–425. Tibet Scientific Expedition Team, Academia Sinica.
Guo, S. X. (1983). “Discussion on the ecological environment of late Cretaceous and tertiary plant geography in China,” in Palaeogeographic fauna of China (Beijing, China: Science Press), 164–184. Basic Theories of Paleontology Series Editorial Committee.
Guo, S. X. (1985). Preliminary interpretation of tertiary climate by using megafossil floras in China. Palae. Catha. 2, 169–175.
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment program for windows 95/98/NT. Nucleic Acids Symposium Series. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hamzeh, M., Périnet, P., Dayanandan, S. (2006). Genetic relationships among species of Populus (Salicaceae) based on nuclear genomic data. J. Torrey Bot. Soc 133, 519–527. doi: 10.3159/1095-5674(2006)133[519:GRASOP]2.0.CO;2
Hao, Z. Z., Liu, Y. Y., Nazaire, M., Wei, X. X., Wang, X. Q. (2015). Molecular phylogenetics and evolutionary history of sect. quinquefoliae (Pinus): Implications for northern hemisphere biogeography. Mol. Phylogenet. Evol. 87, 65–79. doi: 10.1016/j.ympev.2015.03.013
Helmstetter, A. J., Buggs, R. J. A., Lucas, S. J. (2019). Repeated long-distance dispersal and convergent evolution in hazel. Sci. Rep-UK. 9, 16016. doi: 10.1038/s41598-019-52403-2
Ivany, L., Patterson, W., Lohmann, K. (2000). Cooler winters as a possible cause of mass extinctions at the Eocene/Oligocene boundary. Nature. 407, 887–890. doi: 10.1038/35038044
Jansson, S., Douglas, C. J. (2007). Populus: a model system for plant biology. Ann. Rev. Plant Biol. 58, 435–458. doi: 10.1146/annurev.arplant.58.032806.103956
Karrenberg, S., Edwards, P. J., Kollmann, J. (2002). The life history of Salicaceae living in the active zone of floodplains. Freshw. Biol. 47, 733–748. doi: 10.1046/j.1365-2427.2002.00894.x
Kuzovkina, Y. A., Vietto, L. (2014). An update on the cultivar registration of Populus and Salix (Salicaceae). Skvortsovia. 1, 133–148.
Lanfear, R., Ho, S. Y. W., Davies, T. J., Moles, A. T., Aarssen, L., Swenson, N. G., et al. (2013). Taller plants have lower rates of molecular evolution. Nat. Communication. 4, 1879. doi: 10.1038/ncomms2836
Liu, X., Wang, Z. S., Shao, W. H., Ye, Z. Y., Zhang, J. G. (2017). Phylogenetic and taxonomic status analyses of the abaso section from multiple nuclear genes and plastid fragments reveal new insights into the north America origin of Populus (Salicaceae). Front. Plant Sci. 7, 2022. doi: 10.3389/fpls.2016.02022
Lo, E. Y. Y., Stefanović, S., Christensen, K. I., Dickinson, T. A. (2009). Evidence for genetic association between East Asian and western north American Crataegus l. (Rosaceae) and rapid divergence of the eastern north American lineages based on multiple DNA sequences. Mol. Phylogenet. Evol. 51, 157–168. doi: 10.1016/j.ympev.2009.01.018
MacGinitie, H. D. (1969). The Eocene green river flora of northwestern Colorado and northeastern Utah Vol. 83 (University of California: Publ. Geol. Sci), 1–140.
Manchester, S. R., Chen, Z. D., Lu, A. M., Uemura, K. (2009). Eastern Asian Endemic seed plant genera and their paleogeographic history throughout the northern hemisphere. J. Syst. Evol. 47, 1–42. doi: 10.1111/j.1759-6831.2009.00001.x
Manchester, S. R., Dilcher, D. L., Tidwell, W. D. (1986). Interconnected reproductive and vegetative remains of Populus (Salicaceae) from the middle Eocene green river formation, northeastern Utah. Am. J. Bot. 73, 156–160. doi: 10.1002/j.1537-2197.1986.tb09691.x
Manchester, S. R., Judd, W. S., Handley, B. (2006). Foliage and fruits of early poplars (Salicaceae: Populus) from the Eocene of Utah, Colorado, and Wyoming. Int. J. Plant Sci. 167, 897–908. doi: 10.1086/503918
Manos, P. S., Stanford, A. M. (2001). The historical biogeography of fagaceae: Tracking the tertiary history of temperate and subtropical forests of the northern hemisphere. Int. J. Plant Sci. 162, S77–S93. doi: 10.1086/323280
Mao, K. S., Hao, G., Liu, J. Q., Adams, R. P., Milne, R. I. (2010). Diversification and biogeography of Juniperus (Cupressaceae): variable diversification rates and multiple intercontinental dispersals. New Phytol. 188, 254–272. doi: 10.1111/j.1469-8137.2010.03351.x
McClain, A. M., Manchester, S. R. (2001). Dipteronia (Sapindaceae) from the tertiary of north America and implications for the phytogeographic history of the aceroideae. Am. J. Bot. 88, 1316–1334. doi: 10.2307/3558343
Milne, R. I. (2006). Northern hemisphere plant disjunctions: A window on tertiary land bridges and climate change? Ann. Bot. 98, 465–472. doi: 10.1093/aob/mcl148
Milne, R. I., Abbott, R. J. (2002). The origin and evolution of tertiary relict floras. Adv. Bot. Res. 38, 281–314. doi: 10.1016/S0065-2296(02)38033-9
Nge, F. J., Biffin, E., Thiele, K. R., Waycott, M. (2020). Extinction pulse at Eocene–oligocene boundary drives diversification dynamics of two Australian temperate floras. Proc. R. Soc B. 287, 20192546. doi: 10.1098/rspb.2019.2546
Nge, F. J., Biffin, E., Waycott, M., Thiele, K. R. (2021). Phylogenomics and continental biogeographic disjunctions: insight from the Australian starflowers (Calytrix). Am. J. Bot. 109, 291–308. doi: 10.1002/ajb2.1790
Nie, Z. L., Wen, J., Azuma, H., Qiu, Y. L., Sun, H., Meng, Y., et al. (2008). Phylogenetic and biogeographic complexity of magnoliaceae in the northern hemisphere inferred from three nuclear data sets. Mol. Phylogenet. Evol. 48, 1027–1040. doi: 10.1016/j.ympev.2008.06.004
Nie, Z. L., Wen, J., Sun, H., Bartholomew, B. (2005). Monophyly of Kelloggia torrey ex benth. (Rubiaceae) and evolution of its intercontinental disjunction between western north America and eastern Asia. Am. J. Bot. 92, 642–652. doi: 10.3732/ajb.92.4.642
Novacek, M. J. (1999). 100 million years of land vertebrate evolution: the cretaceousearly tertiary transition. Ann. Mo. Bot. Gard. 86, 230–258. doi: 10.2307/2666178
Petit, R. J., Bodenes, C., Ducousso, A., Roussel, G., Kremer., A. (2004). Hybridization as a mechanism of invasion in oaks. New Phytol. 161, 151–164. doi: 10.1046/j.1469-8137.2003.00944.x
Pokorny, L., Riina, R., Mairal, M., Meseguer, A. S., Culshaw, V., Cendoya, J., et al. (2015). Living on the edge: timing of rand flora disjunctions congruent with ongoing aridification in Africa. Front. Genet. 6, 154. doi: 10.3389/fgene.2015.00154
Portelli, S. N., Quinteros, A. S. (2018). Phylogeny, time divergence, and historical biogeography of the south American Liolaemus alticolor-bibronii group (Iguania: Liolaemidae). PeerJ. 6, e4404. doi: 10.7717/peerj.4404
Posada, D. (2008). jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Qiao, C. Y., Ran, J. H., Li, Y., Wang, X. Q. (2007). Phylogeny and biogeography of Cedrus (Pinaceae) inferred from sequences of seven paternal chloroplast and maternal mitochondrial DNA regions. Ann. Bot-London. 100, 573–580. doi: 10.1093/aob/mcm134
Rambaut, A. (2014) FigTree v1.4.2. Available at: http://tree.bio.ed.ac.uk/software/figtree/.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ree, R. H., Smith, S. A. (2008). Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14. doi: 10.1080/10635150701883881
Retallack, G. J., Orr, W. N., Prothero, D. R., Duncan, R. A., Kester, P. R., Ambers, C. P. (2004). Eocene-Oligocene extinction and paleoclimatic change near Eugene, Oregon. GSA Bulletin. 116, 817–839. doi: 10.1130/B25281.1
Riddle, B. R., Dawson, M. N., Hadly, E. A., Hafner, D. J., Hickerson, M. J., Mantooth, S. J., et al. (2008). The role of molecular genetics in sculpting the future of integrative biogeography. Prog. Phys. Geog. 32, 173–202. doi: 10.1177/0309133308093822
Rieseberg, L. H., Soltis, D. E. (1991). Phylogenetic consequences of cytoplasmic gene flow in plants. Evol. Trends Plants 5, 65–84. doi: 10.1007/BF00021248
Ronquist, F., Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sanmartín, I., Enghoff, H., Ronquist, F. (2001). Patterns of animal dispersal, vicariance and diversification in the Holarctic. Biol. J. Linn. Soc. 73, 345–390. doi: 10.1006/bijl.2001.0542
Sanmartín, I., Anderson, C. L., Alarcon, M., Ronquist, F., Aldasoro, J. J. (2010). Bayesian Island biogeography in a continental setting: the rand flora case. Biol.Lett. 6, 703–707. doi: 10.1098/rsbl.2010.0095
Sanmartín, I., Meseguer, A. S. (2016). Extinction in phylogenetics and biogeography: from timetrees to patterns of biotic assemblage. Front. Genet. 7, 35. doi: 10.3389/fgene.2016.00035
Song, B. W., Robert, A. S., Zhang, K. X., Ji, J. L., Alexander, F., Alice, C. H., et al. (2020). Qaidam basin leaf fossils show northeastern Tibet was high, wet and cool in the early oligocene. Earth Planet. Sc. Lett. 537, 116175. doi: 10.1016/j.epsl.2020.116175
Sun, H., Mclewin, W., Michael, F. F. (2001). Molecular phylogeny of Helleborus (Ranunculaceae), with an emphasis on the East Asian-Mediterranean disjunction. Taxon 50, 1001–1018. doi: 10.2307/1224717
Sun, J. M., Ni, X. J., Bi, S. D., Wu, W. Y., Ye, J., Meng, J., et al. (2014). Synchronous turnover of flora, fauna and climate at the Eocene–oligocene boundary in Asia. Sci. Rep-UK. 4, 7463. doi: 10.1038/srep07463
Su, C. Y., Shi, Q. H., Sun, X. Y., Ma, J. Y., Li, C. X., Hao, J. S., et al. (2017). Dated phylogeny and dispersal history of the butterfly subfamily nymphalinae (Lepidoptera: Nymphalidae). Sci. Rep-UK. 7, 8799. doi: 10.1038/s41598-017-08993-w
Swofford, D. L. (2003). PAUP*: Phylogenetic analysis using parsimony (* and other methods), version 4.0. b10 (Sunderland, MA: Sinauer Associates).
Tao, J. R., Xiong, X. Z. (1986). The latest Cretaceous flora of heilongjiang province and the floristic relationship between east Asia and north America. Acta Phytotaxon. Sin. 24, 1–15.
Thomas, D. C., Chatrou, L., Stull, G., Johnson, D. M., Harris, D. J., Thongpairoj, U., et al. (2015). The historical origins of palaeotropical intercontinental disjunctions in the pantropical flowering plant family annonaceae. Perspect. Plant Ecol. 17, 1–16. doi: 10.1016/j.ppees.2014.11.001
Thompson, J., Higgins, D., Gibson, T. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic. Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tiffney, B. H. (1985a). The Eocene north Atlantic land bridge and its importance in tertiary and modern phytogeography of the northern hemisphere. J. Arnold. Arbor. 66, 243–273. doi: 10.5962/bhl.part.13183
Tiffney, B. H. (1985b). Perspectives on the origin of the floristic similarity between eastern Asia and eastern north America. J. Arnold. Arbor. 66, 73–94. doi: 10.5962/bhl.part.13179
Tiffney, B. H., Manchester, S. R. (2001). The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the northern hemisphere tertiary. Int. J. Plant Sci. 162, S3–S17. doi: 10.1086/323880
Tsitrone, A., Kirkpatrick, M., Levin, D. A. (2003). A model for chloroplast capture. Evolution 57, 1776–1782. doi: 10.1554/02-746
Wang, Z. H., Peng, H., Kilian, N. (2013). Molecular phylogeny of the Lactuca alliance (Cichorieae subtribe lactucinae, asteraceae) with focus on their Chinese centre of diversity detects potential events of reticulation and chloroplast capture. PloS One 8, e82692. doi: 10.1371/journal.pone.0082692
Wang, M. C., Zhang, L., Zhang, Z. Y., Li, M. M., Wang, D. Y., Zhang, X., et al. (2020). Phylogenomics of the genus Populus reveals extensive interspecific gene flow and balancing selection. New. Phytol. 225, 1370–1382. doi: 10.1111/nph.16215
Wan, X. Q., Zhang, F., Zhong, Y., Ding, Y. H., Wang, L. W., Hu, T. X. (2013). Study of genetic relationships and phylogeny of the native Populus in southwest China based on nucleotide sequences of chloroplast trnT-trnF and nuclear DNA. Plant Syst. Evol. 299, 57–65. doi: 10.1007/s00606-012-0702-9
Wei, X. X., Yang, Z. Y., Li, Y., Wang, X. Q. (2010). Molecular phylogeny and biogeography of pseudotsuga (Pinaceae): Insights into the floristic relationship between Taiwan and its adjacent areas. Mol. Phylogenet. Evol. 55, 77–785. doi: 10.1016/j.ympev.2010.03.007
Wen, J. (1999). Evolution of eastern Asian and eastern north American disjunct distributions in flowering plants. Ann. Rev. Ecol. Syst. 30, 421–455. doi: 10.1146/annurev.ecolsys.30.1.421
Wen, J. (2001). Evolution of eastern Asian-eastern north American biogeographic disjunctions: A few additional issues. Int. J. Plant Sci. 162, S117–S122. doi: 10.1086/322940
Wen, J., Ickert-Bond, S. M. (2009). Evolution of the madrean-tethyan disjunctions and the north and south American amphitropical disjunctions in plants. J. Syst. Evol. 47, 331–348. doi: 10.1111/j.1759-6831.2009.00054.x
Wen, J., Nie, Z. L., Ickert-Bond, S. M. (2016). Intercontinental disjunctions between eastern Asia and western north America in vascular plants highlight the biogeographic importance of the Bering land bridge from late Cretaceous to neogene. J. Syst. Evol. 54, 469–490. doi: 10.1111/jse.12222
Wiens, J. J. (2004). Speciation and ecology revisited: phylogenetic niche conservatism and the origin of species. Evolution. 58, 193–197. doi: 10.1111/j.0014-3820.2004.tb01586.x
Wu, Z. Y. (1999). “Salicaceae,” in Flora of China, vol. 20 . Eds. Wu, Z. Y., Raven, P. H., Hong, D. Y. (Beijing: Science Press).
Xing, Y. W., Onstein, R. E., Carter, R. J., Stadler, T., Linder, H. P. (2014). Fossils and a large molecular phylogeny show that the evolution of species richness, generic diversity, and turnover rates are disconnected. Evolution 68, 2821–2832. doi: 10.1111/evo.12489
Yu, Y., Blair, C., He, X. (2020). RASP4: Ancestral state reconstruction tool for multiple genes and characters. Mol. Biol. Evol. 87, 46–49. doi: 10.1093/MOLBEV/MSZ257
Yu, X. Q., Gao, L. M., Soltis, D. E., Soltis, P. S., Yang, J. B., Fang, L., et al. (2017). Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol. 215, 1235–1248. doi: 10.1111/nph.14683
Yu, Y., Harris, A., He, X. (2010). S-DIVA (Statistical dispersal-vicariance analysis): a tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 56, 848–850. doi: 10.1016/j.ympev.2010.04.011
Zachos, J., Pagani, M., Sloan, L., Thomas, E., Billups, K. (2001). Trends, rhythms, and aberrations in global climate 65 ma to present. Science 292, 686–693. doi: 10.1126/science.1059412
Zanazzi, A., Kohn, M., MacFadden, B. J., Terry, D. O. (2007). Large Temperature drop across the Eocene-oligocene transition in central north America. Nature. 445, 639–642. doi: 10.1038/nature05551
Zhang, J. B., Li, R. Q., Xiang, X. G., Manchester, S. R., Lin, L., Wang, W., et al (2013). Integrated fossil and molecular data reveal the biogeographic diversification of the Eastern Asian-Eastern North American disjunct hickory genus (Carya Nutt.). PLoS One, 8, e70449. doi: 10.1371/journal.pone.0070449
Keywords: biogeography, divergence time, molecular phylogeny, Populus, bering land bridge
Citation: Liu X, Wang Z, Wang W, Huang Q, Zeng Y, Jin Y, Li H, Du S and Zhang J (2022) Origin and evolutionary history of Populus (Salicaceae): Further insights based on time divergence and biogeographic analysis. Front. Plant Sci. 13:1031087. doi: 10.3389/fpls.2022.1031087
Received: 29 August 2022; Accepted: 23 November 2022;
Published: 16 December 2022.
Edited by:
Daniel Potter, University of California, Davis, United StatesReviewed by:
Francis J. Nge, Institut de Recherche Pour le Développement (IRD), FranceCopyright © 2022 Liu, Wang, Wang, Huang, Zeng, Jin, Li, Du and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianguo Zhang, emhhbmdqZ0BjYWYuYWMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.