- 1Department of Agronomy, Iowa State University, Ames, IA, United States
- 2Department of Plant Science, Penn State University, University Park, PA, United States
- 3Ecosystem Science and Management, Penn State University, University Park, PA, United States
- 4Department of Agricultural Biotechnology and Molecular Biology, Dr. Rajendra Prasad Central Agricultural University, Samastipur, India
- 5Centre for Crop Health, University of Southern Queensland (USQ), Toowoomba, QLD, Australia
- 6Genetics Gains Research Program, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Hyderabad, India
- 7Division of Crop Improvement, Indian Council of Agricultural Research (ICAR)-Sugarcane Breeding Institute, Coimbatore, India
- 8Saskatoon Research and Development Centre, Agriculture and Agri-Food Canada, Saskatoon, SK, Canada
- 9State Agricultural Biotechnology Centre, Crop Research Innovation Centre, Food Futures Institute, Murdoch University, Murdoch, WA, Australia
Chickpea (Cicer arietinum L.) is one of the major pulse crops, rich in protein, and widely consumed all over the world. Most legumes, including chickpeas, possess noticeable amounts of raffinose family oligosaccharides (RFOs) in their seeds. RFOs are seed oligosaccharides abundant in nature, which are non-digestible by humans and animals and cause flatulence and severe abdominal discomforts. So, this study aims to identify genetic factors associated with seed oligosaccharides in chickpea using the mini-core panel. We have quantified the RFOs (raffinose and stachyose), ciceritol, and sucrose contents in chickpea using high-performance liquid chromatography. A wide range of variations for the seed oligosaccharides was observed between the accessions: 0.16 to 15.13 mg g-1 raffinose, 2.77 to 59.43 mg g-1 stachyose, 4.36 to 90.65 mg g-1 ciceritol, and 3.57 to 54.12 mg g-1 for sucrose. Kabuli types showed desirable sugar profiles with high sucrose, whereas desi types had high concentrations RFOs. In total, 48 single nucleotide polymorphisms (SNPs) were identified for all the targeted sugar types, and nine genes (Ca_06204, Ca_04353, and Ca_20828: Phosphatidylinositol N-acetylglucosaminyltransferase; Ca_17399 and Ca_22050: Remorin proteins; Ca_11152: Protein-serine/threonine phosphatase; Ca_10185, Ca_14209, and Ca_27229: UDP-glucose dehydrogenase) were identified as potential candidate genes for sugar metabolism and transport in chickpea. The accessions with low RFOs and high sucrose contents may be utilized in breeding specialty chickpeas. The identified candidate genes could be exploited in marker-assisted breeding, genomic selection, and genetic engineering to improve the sugar profiles in legumes and other crop species.
Introduction
Chickpea (Cicer arietinum L.) is one of the founder crops domesticated between 9,000–11,000 years ago and is an important ancient legume grown and consumed all over the globe (Lev-Yadun et al., 2000). As a legume crop, it is often grown as rotational crops with cereals to enhance yield because of their ability to fixing atmospheric nitrogen (Graham and Vance, 2003). Chickpea is rich in carbohydrates (60-65%), protein (20-22%), fat (6%), and rich in dietary fiber, as well as minerals (phosphorus, calcium, magnesium, iron, and zinc) and vitamins (β-carotene, thiamin, riboflavin, and niacin) (Jukanti et al., 2012). The major pulse grain constituents are carbohydrates, based on their polymeric structure that can be classified as monosaccharides (ribose, fructose, and glucose), disaccharides (sucrose, maltose, melibiose), oligosaccharides (raffinose, stachyose, verbascose, ajugose, and ciceritol) and polysaccharides (Chibbar et al., 2010). Among the oligosaccharides, α-galacto-oligosaccharides (α-GOS) are known as raffinose family oligosaccharides (RFOs) (Sosulski et al., 1982). The major RFOs found in chickpea include raffinose, stachyose, and verbascose. However, ciceritol does not belong to the RFOs since its structure is different from α-GOS and can rapidly undergo a hydrolysis process, so unlike raffinose and stachyose, ciceritol does not cause flatulence in humans and animals (Quemener and Brillouet, 1983).
RFOs are the single most deterrent factor for the rapid adoption of legumes in mainstream food usage in humans and animals (Delumen, 1992; Elango et al., 2022a). Humans and animals lack the enzyme α-galactosidase to degrade α-galactosides (RFOs), which results in the accumulation of undigested RFOs in the large intestine of the digestive system, which ultimately causes flatulence and abdominal discomforts due to the production of flatulent gases by colonic bacteria through fermenting the un-digested RFOs present in the guts (Calloway and Murphy, 1968; Sosulski et al., 1982; Singh, 1985; Han and Baik, 2006). Though, RFOs have been reported to have a beneficial effect on gut microflora (Van den Ende, 2013; Su et al., 2019), and play a role in seed germinability and biotic and abiotic stress tolerance in crop plants (Gulewicz et al., 2002; Taji et al., 2002; Nishizawa-Yokoi et al., 2008; Dobrenel et al., 2013; Van den Ende, 2013; Gangl and Tenhaken, 2016; Yan et al., 2022). However, we do not know what the right concentration is needed to benefit humans, animals, and plants concerning RFOs.
Food processing can eliminate RFOs at varying degrees and significantly increase dietary fraction availability in food (Jood et al., 1985; Egbe and Akinyele, 1990; Aguilera et al., 2009). However, these techniques often come with trade-offs; most techniques are time-consuming, lead to loss of nutrients, and sometimes have consumer acceptability issues. Therefore, identifying sources of variation for developing desirable sugar-type cultivars through crop breeding is very important. Screening and identification of low RFOs have been carried out in many economically important legume crops such as lentil (Tahir et al., 2011), chickpea (Raja et al., 2015; Gangola et al., 2016), pea (Peterbauer et al., 2003), soybean (Blackman et al., 1992; Dierking and Bilyeu, 2008; Obendorf and Górecki, 2012), mung bean and urd bean (Souframanien et al., 2014), whereas, very limited efforts have been taken toward the identification of genomic regions associated with RFOs in crop plants. In this context, our study aims to identify the genetic factors responsible for seed oligosaccharides in chickpea through genome-wide association mapping using the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) mini-core collection.
Materials and methods
Plant materials
The chickpea mini-core collection consisting of 211 accessions from 24 countries (Asia, Africa, Europe, North, and South American regions) was obtained from the genetic resources division of International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), India (Upadhyaya and Ortiz, 2001). Field experiments were performed in a randomized complete block design (RCBD) with three replications in the 2010 winter season at the Department of Pulses (11.0232° N latitude, 76.9293° E longitude, 426.72 m altitude), Tamil Nadu Agricultural University (TNAU), Coimbatore, India. Each accession was grown in a single row in a 3 m long plot. Seeds from each replicate of individual accessions were harvested at physiological maturity and stored at 4°C until analysis was performed. A standard agronomic package of practices was followed to achieve the best crop establishment.
Sugar extraction and quantification
The seeds of each chickpea accession were grounded, and the flours were used to extract soluble sugars. One gram of flour samples was taken into a screw cap vial and mixed with 10 ml of 50% ethanol, and vortexed briefly. After adding ethanol, samples were shaken horizontally using a water bath shaker maintained at 50° C for 30 min at 100 rpm. The incubated vial was centrifuged at 4000 rpm for 5 min, then 5 ml of supernatant was taken and mixed with 7 ml of acetonitrile (high-performance liquid chromatography (HPLC)-grade) to precipitate the soluble proteins. The mixture (5 ml supernatant + 7 ml acetonitrile) was incubated at room temperature for two hours. After incubation, the mixture was centrifuged at 3670 g for 5 min, and one ml aliquot of the supernatant was collected. The collected supernatant was dried at 50°C and resuspended with 500 μl 65% HPLC-grade acetonitrile and filtered through a 0.2 μm membrane filter and transferred to HPLC vials. Standards of sucrose, raffinose, and stachyose were purchased from Sigma-Aldrich, Bengaluru, India and ciceritol was purchased from Clearsynth, Hyderabad, India. Three different concentrations, 1.25 mg ml-1, 2.5 mg ml-1, and 5.0 mg ml-1, were prepared and included in each batch of samples to obtain the standard curve. The concentration of different sugars (sucrose, raffinose, stachyose, and ciceritol) was determined using the HPLC (Shimadzu, Kyoto, Japan), which consisted of an LC20AD pump and a RID-10A refraction index detector. Sugar concentrations were determined using the peak area of the sample in comparison with standards.
Marker-trait association analyses
We performed genome-wide association mapping analysis using 673,115 single nucleotide polymorphisms (SNPs), where the SNP calls for 211 genotypes were obtained from Varshney et al. (2019). As reported in Varshney et al. (2019), for calling SNPs, the clean reads were mapped on to the reference genome of chickpea genotype CDC Frontier using SOAP2. We then used SOAPsnp3 to calculate the likelihood of all possible genotypes for each sample. In order to filter out low-quality variants, the loci with sequencing depth higher than 10,000 and lower than 400, mapping times higher than 1.5, or quality scores lower than 20, were filtered out. The loci with estimated allele frequency not equal to 0 or 1 were determined as SNPs. After obtaining the SNPs, we also determined the genotype of each individual at the SNP locus by assigning the most likely genotype from the SOAPsnp3 result of each sample. We have used the Fixed and random model Circulating Probability Unification (FarmCPU) model (Liu et al., 2016) in the Genome Association and Prediction Integrated Tool (GAPIT3) package (Wang and Zhang, 2021) to identify significant marker-trait associations (MTAs). GAPIT estimated the allelic effect for the significant SNPs identified. Sign (+/-) of the allelic effect estimate is relative to the alphabetical order of the nucleotides. MTAs were selected for p-value <10-5. Gene annotations were determined from the reference genome of chickpeas released in 2013 (Varshney et al., 2013). Genes within the flanking regions of 50kb upstream and downstream of the significantly called SNPs were collected first, and among them, only the genes already annotated in the chickpea genome with predicted function or found orthologs in other model plants were selected as candidate genes. Distance from the SNP (bp) is calculated as the distance from the SNP location to the start site of the upstream or downstream genes. If the SNP is located within a gene, the distance is calculated as the distance to the start site of the gene.
Results
Phenotypic variation and correlations among seed oligosaccharides
We have observed wide variations for all sugars measured in the ICRISAT chickpea mini-core collection. Among the morphotypes, Kabuli-type chickpeas exhibited higher sucrose and total sugar contents. In contrast, the desi-type chickpeas showed higher RFOs (raffinose and stachyose) and ciceritol contents in seeds (Figure 1). Mini-core collection showed wide seed oligosaccharide variations: 0.16 to 15.13 mg g-1, 2.77 to 59.43 mg g-1, 4.36 to 90.65 mg g-1, 3.57 to 54.12 mg g-1 for raffinose, stachyose, ciceritol, and sucrose with an average of 4.61, 28.02, 34.48, and 23.11 mg g-1 flour sample, respectively (Table 1). We have observed significant positive correlations among seed oligosaccharides measured: sucrose, ciceritol, and stachyose have strong positive correlations with total sugars; moderate correlations were observed between sucrose and ciceritol, and ciceritol and stachyose (Figure 2). Whereas raffinose had a low level of positive correlations with all other seed oligosaccharides (Figure 2).
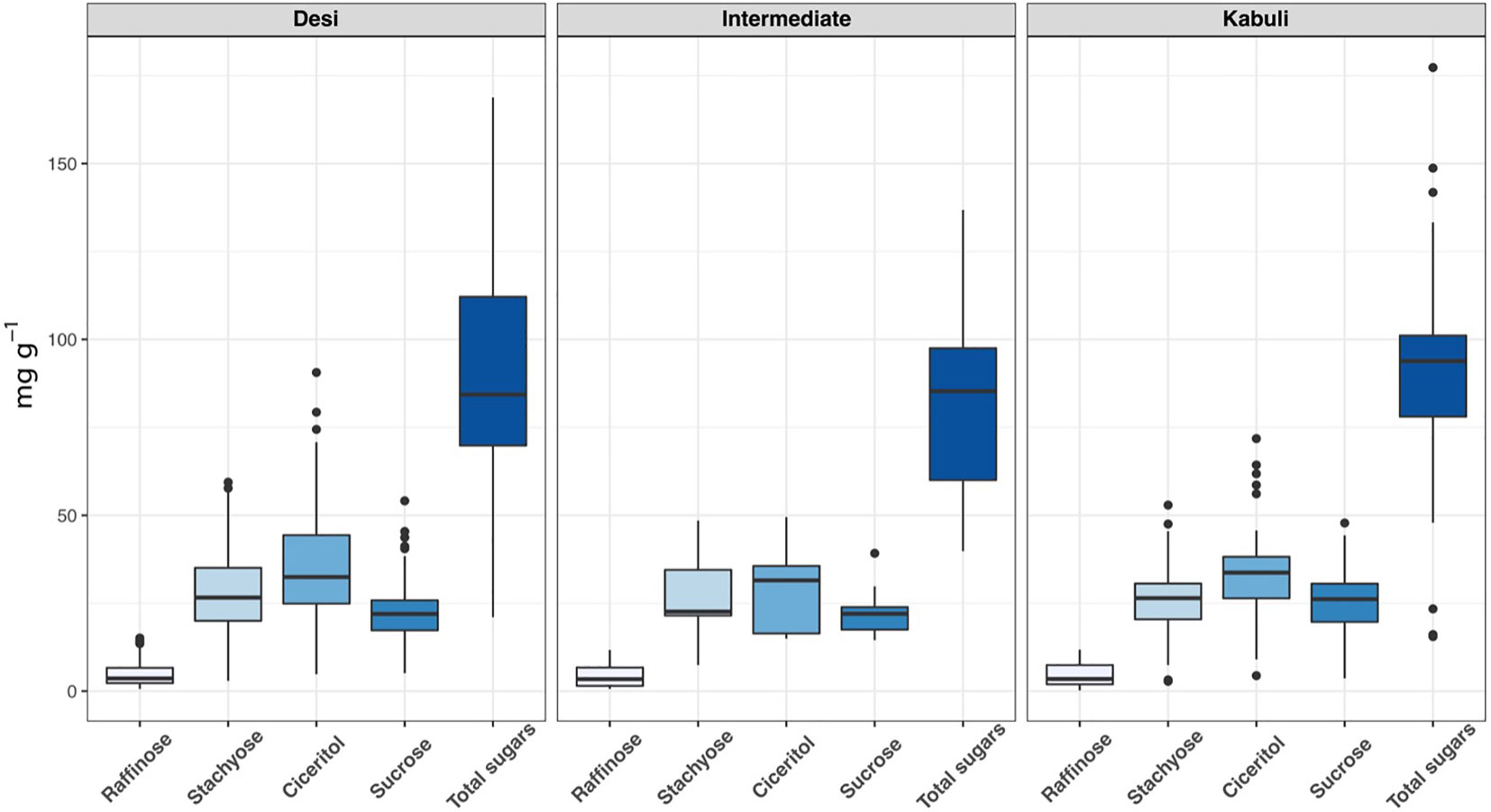
Figure 1 Seed oligisaccharides variations among desi, kabuli, and intermediate types in the ICRISAT chickpea mini-core panel.
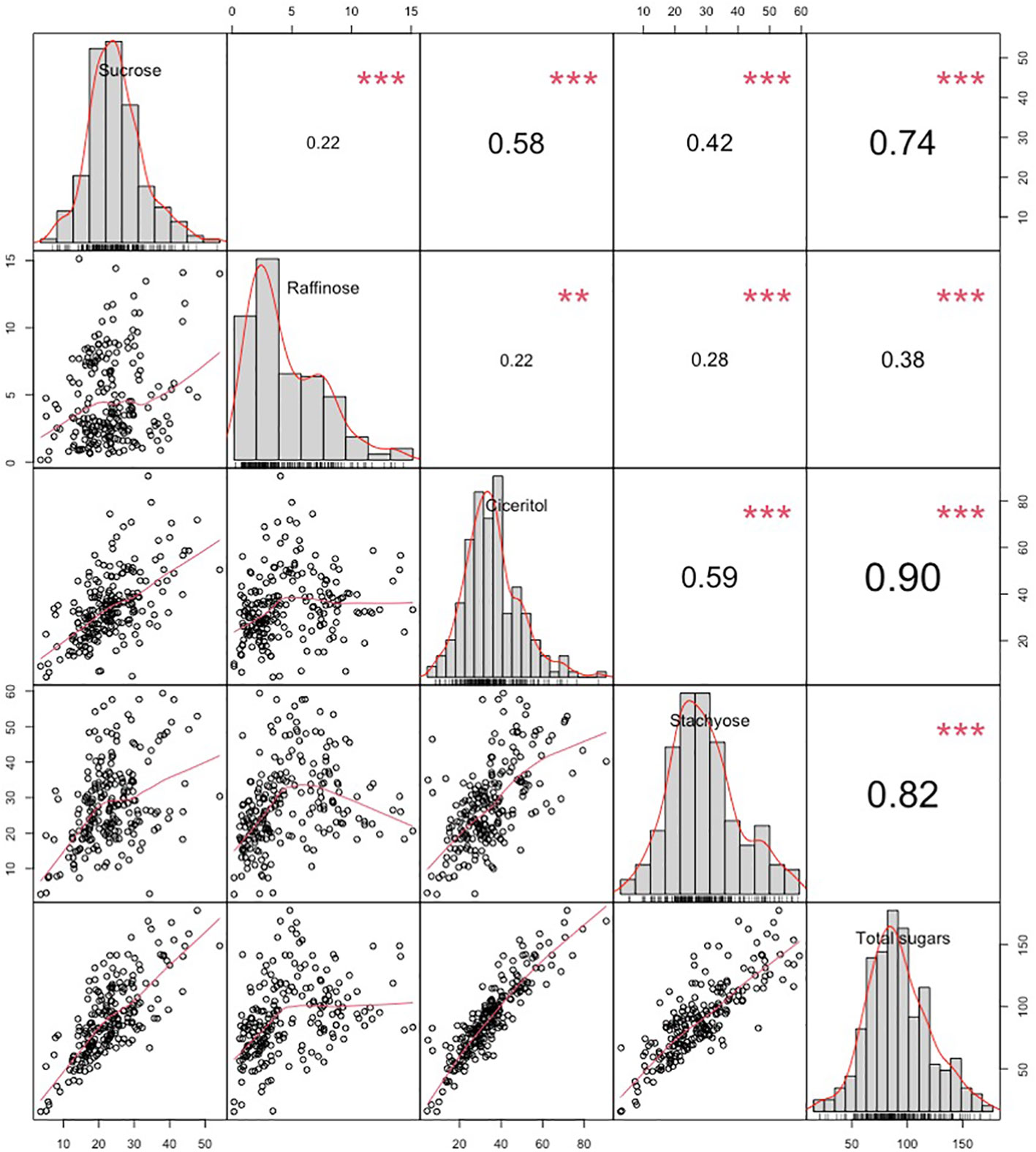
Figure 2 Variation and Pearson pairwise correlations of sucrose, raffinose, ciceritol, stachyose, and total sugars in chickpea. Upper diagonal: Pearson correlation coefficients between every two traits. Mid-diagonal: Histograms of sucrose, raffinose, ciceritol, stachyose, and total sugars. Lower diagonal: Bivariate scatter plots of correlations between every two traits with a fitted line. **Significant at the .01 probability level. ***Significant at the .001 probability level.
Marker trait associations
Genome-wide association mapping identified 48 SNPs associated with the seed oligosaccharide contents in chickpeas (Table 2). The largest number of associated markers (12) were detected on chromosome 4 (Table 2), and there were 16 SNPs found to be significantly associated with raffinose content in chickpea, which is the most associated markers compared with other three sugars: 7 SNPs for ciceritol and stachyose respectively, and 9 SNPs for sucrose, and 8 SNPs for total sugar content in chickpea (Table 2; Figure 3).
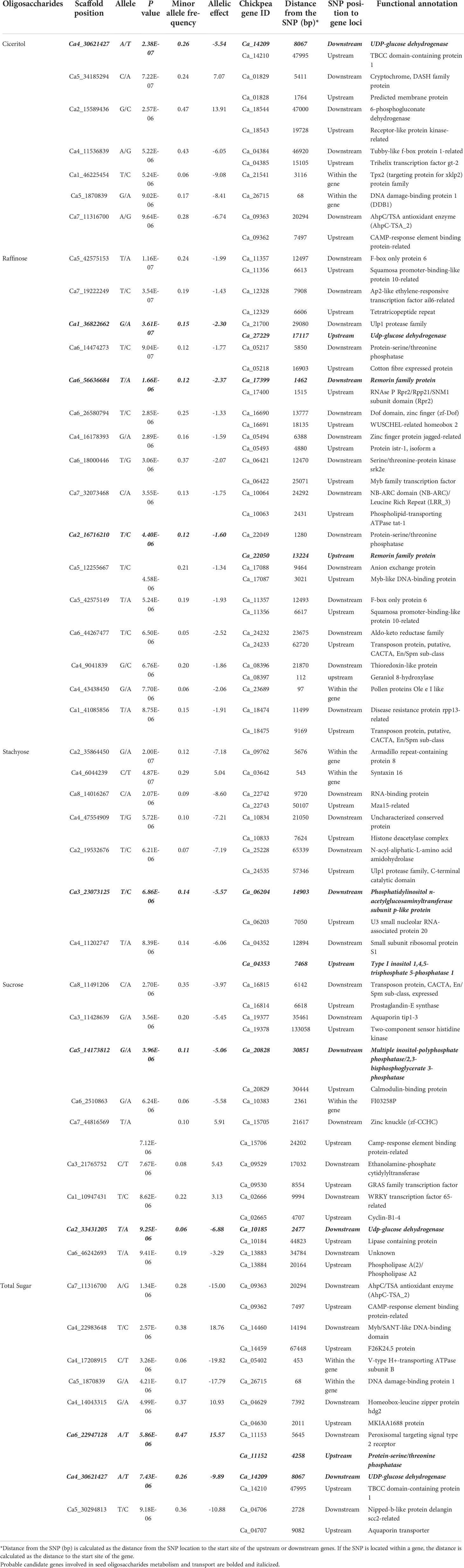
Table 2 List of significant single nucleotide polymorphic associations, the genes tagged by significant single nucleotide polymorphic markers, and candidate genes identified based on proximity to the significant markers and their description for ciceritol, raffinose, stachyose, sucrose, and total sugars in chickpea.
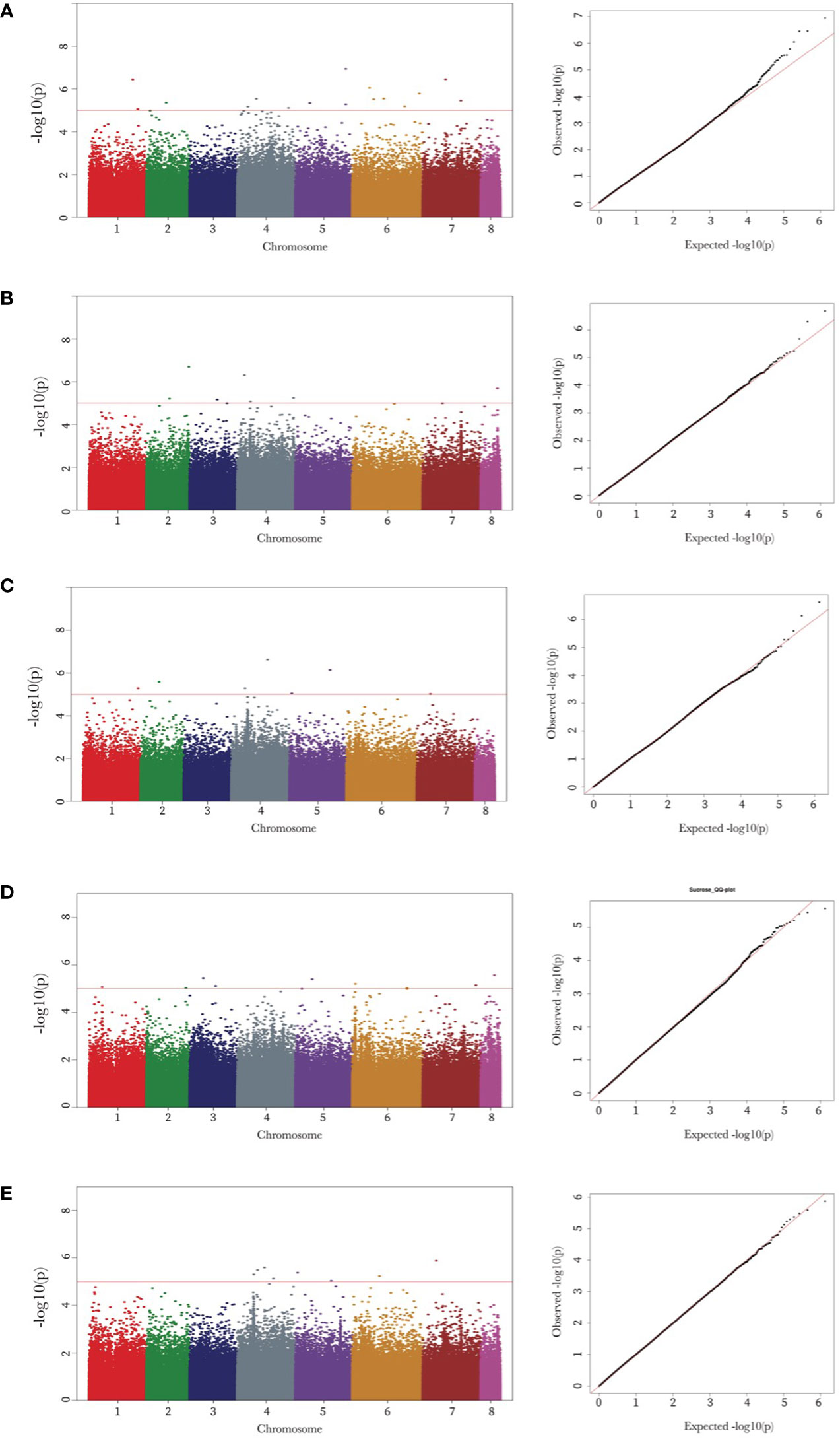
Figure 3 Manhattan and quantile-quantile (Q-Q) plots of raffinose, stachyose, ciceritol, sucrose, and total sugars in chickpea. Manhattan and Q-Q plots of the seed oligosaccharides from a to e are as follows: raffinose (A), stachyose (B), ciceritol (C), sucrose (D), and total sugars (E). Negative log10 transformed P values (y-axis) are plotted against the physical single nucleotide polymorphism (SNP) position on each chromosome (x-axis). Each circle represents a SNP, and the corresponding SNPs were mentioned trait wise. The dotted red line represents the genome-wide significance threshold as determined by Bonferroni correction at.05. Regions with negative log10 P values above the threshold contain quantitative trait loci candidates.
Probable candidate genes
We identified 80 probable candidate genes for all the seed oligosaccharides measured in this study (Table 2). Among them, nine genes were recognized with annotated functions highly associated with sugar biosynthesis and transportation. Four genes that are important components in inositol biosynthesis: two associated with stachyose (Ca_06204: phosphatidylinositol N-acetylglucosaminyltransferase; Ca_04353: Type I inositol 1,4,5-trisphosphate 5-phosphatase), and two associated with sucrose (Ca_20828: inositol-polyphosphate phosphatase; Ca_10185: UDP-glucose dehydrogenase) have been identified as probable candidates for seed oligosaccharide biosynthesis in chickpeas (Table 2). There were also three candidate genes identified playing an important role in RFOs transportation: two of the genes (Ca_17399 and Ca_22050) are associated with raffinose content, and the other gene (Ca_11152) is linked with total sugar content in chickpea seeds, and all of them encodes remorin protein, which regulates the carbohydrate translocation in plants. The other group of important genes identified in our study includes Ca_14209 (associated with two traits: ciceritol and total sugar contents) and Ca_27229 (associated with raffinose content), and both genes encode UDP-glucose dehydrogenase (UGD) and likely interfere with RFOs biosynthesis as a competitor for the upstream precursor compound (Joët et al., 2009).
A candidate gene Ca_23689 harboring SNP Ca4_43438450 was associated with raffinose content, and GO annotation indicates gene Ca_23689 encodes structural constituent of the cell wall (Table 2). A precedent study has discovered that over-expression of raffinose synthase (rfs) resulted in increased biomass and total cellulose content in the cell wall (Unda et al., 2017). Gene Ca_03642 was associated with chickpea stachyose content in seed (Table 2). The GO term functional annotation suggests that gene Ca_03642 involves intracellular protein transport, vesicle-mediated transport, and membrane fusion. Studies have demonstrated that stachyose is the primary photoassimilate and transport sugar in legumes (Peterbauer et al., 2001; Qiu et al., 2015). Another gene, Ca_09762, which is predicted as involved in calcium ion transmembrane transport, was also associated with stachyose content (Table 2). Meanwhile, we identified another SNP locus (Ca6_2510863) in the gene Ca_10383 associated with sucrose content in chickpea seed, and the functional prediction of gene Ca_10383 is ATP hydrolysis coupled proton transport. A previous study suggested that stachyose and sucrose may also be accumulated in the vacuole by stachyose and Sucrose/H+ antiporter mechanisms, which is an ATPase energized vacuolar uptake process (Keller, 1992).
Two SNP loci (Ca_46225454 in gene Ca_21541 and Ca5_1870839 in gene Ca_26715) were associated with ciceritol content in chickpea seeds (Table 2). Gene Ca_21541 is predicted as a TPX2 (targeting protein for Xklp2) family protein. SNP Ca_46225454 is also adjacent to a sequence fragment ortholog of AT5G40690, which encodes for methyltransferase activity. Ciceritol is the end product of the inositol methylation process, which explains the role of the identified SNP Ca_46225454 in ciceritol biosynthesis in chickpeas. Gene Ca_26715 is an endochitinase A-like protein. Chitinase is known to protect plants against abiotic and biotic stresses (de Las Mercedes Dana et al., 2006; Karlsson and Stenlid, 2008; Xin et al., 2021; Zhang et al., 2022). The association between endochitinase and ciceritol content in chickpea suggests a metabolic link between the ciceritol pathway and the pathway leading to biotic and abiotic resistance. Two SNPs were identified for total sugar content: SNP Ca4_17208915 in gene Ca_05402 and Ca5_1870839 in gene Ca_26715. Gene Ca_05402 involves ATP hydrolysis coupled proton transport. Gene Ca_26715 is predicted as an A-like endochitinase (Table 2).
Discussion
Breeding specialty chickpeas
Identification of germplasm lines with good nutritional quality parameters is key for developing cultivars for different end users. In this study, we have identified varying sugar profile accessions that could be exploited as a base in breeding specialty chickpeas. Especially the accessions with high sucrose (ICC 12564 and ICC 9137) and low RFOs (ICC 6263 and ICC 13816) are desirable for making delicacies like hummus, besan laddoo, Mysore Pak, besan barfi, Puran Poli, and other confectionaries without refined sugars and or artificial sweetening agents. The low RFO lines could be exploited in human and animal food and feed industries. Stachyose and raffinose are considered the most undesirable oligosaccharides present in chickpea. The low stachyose and raffinose accessions ICC 13816 and ICC 6263 can be used in breeding programs as a unique germplasm resource to develop chickpea varieties with improved nutrient utilization and digestibility. The chickpea mini-core collection also shows a considerable amount of variation for ciceritol among accessions. It is believed that these compounds play an important role in protecting plants and seeds against drought stress (Keller and Ludlow, 1993). Ciceritol, a new trisaccharide do not correlate with flatulence, was found high in chickpea accessions reported by Quemener and Brillouet (1983) and Xiaoli et al. (2008). Further research indicated that ciceritol also plays an important role in improving gut health by enhancing the growth of Lactobacillus, Enterococcus, and Bifidobacterium spp in addition to the production of short-chain fatty acids and is used as a potential source of prebiotics (Zhang et al., 2017). Therefore, increasing ciceritol relatively decreases the flatus potential of chickpea.
Seed oligosaccharide candidates
There are four genes - two associated with stachyose (Ca_06204: phosphatidylinositol N-acetylglucosaminyltransferase; Ca_04353: Type I inositol 1,4,5-trisphosphate 5-phosphatase) and two associated with sucrose (Ca_20828: inositol-polyphosphate phosphatase; Ca_10185:UDP-glucose dehydrogenase) – have been recognized as indispensably involved in various signaling pathways in plants via mediating the phospholipidation and phosphatization of Myo-inositol and its derivatives like sucrose and RFOs. Studies reported the crosstalk linkage between inositol signaling and sugar metabolism in plants (Saddhe et al., 2021; Lou et al., 2007; Yang et al., 2007). In 2008, Ananieva et al. (2008) discovered the in-vitro interactions between the myo-inositol polyphosphate 5-phosphatase (5PTase13) and the sucrose nonfermenting-1-related kinase (SnRK1.1) and identified 5PTase13 regulated SnRK1 activity under different sugar conditions. Plant SnRK1 is known for its key role at the interface among sugar metabolism, stress signaling, and other physiological developmental processes like seed germination and seedling growth (Radchuk et al., 2006; Baena-González et al., 2007; Jossier et al., 2009; Hulsmans et al., 2016; Elango et al., 2022b). SnRK1 has been identified in higher plants and two other subfamilies – SnRK2 and SnRK3 (Halford and Hey, 2009). Purcell et al. (1998) demonstrated that SnRK1 plays an essential role in the regulation of Suc synthase expression in potatoes (Solanum tuberosum), and later Tiessen et al. (2003) recognized that SnRK1 also regulates starch biosynthesis. The same finding about SnRK1’s role involved in starch synthesis was also identified in pollen grains of barley (Hordeum vulgare) (Zhang et al., 2001). Our findings indicate that in chickpea, the stachyose and sucrose biosynthesis is mediated by various kinds of inositol-polyphosphate phosphatase, potentially through their regulation of SnRK families. Additionally, the genes involved in the RFO biosynthetic pathway have been identified in soybean (Glycine max) and common bean (Phaseolus vulgaris). de Koning et al. (2021) identified three galactinol synthase (GolS) genes in common bean, named PvGolS1, PvGolS2, and PvGolS3. GolS crosslinks between inositol and RFO biosynthesis, and GolS are the primary checkpoint for RFO biosynthesis via inositols (Sengupta et al., 2015).
We also found that three candidate genes encode for remorin protein – two genes (Ca_17399 and Ca_22050) are associated with raffinose content, and the other gene (Ca_11152) is linked with total sugar content in chickpea seeds. RFOs serve as a major transport form of carbohydrates in the vascular system in plants (Ayre et al., 2003; Johnson et al., 2020; Ren et al., 2021). Remorin is a kind of plant-specific membrane-bound protein and has been identified in Arabidopsis thaliana, Nicotiana tabacum, Medicago truncatula, and Lycopersicon esculentum (Watson et al., 2003; Bariola et al., 2004; Marmagne et al., 2004; Mongrand et al., 2004; Sazuka et al., 2004; Nelson et al., 2006; Valot et al., 2006). In Arabidopsis, 16 genes have been identified in the REM family, and among them, REM1.2, REM1.3, and REM1.4 from the REM1 subfamily were found to exist ubiquitously in the majority of the tissues (Huang et al., 2019). Previous studies demonstrated that remorin proteins are localized in the plasma membrane and plasmodesmata of phloem companion cells and regulate photoassimilate translocation via reducing plasmodesmata permeability in the symplastic system in rice (Oryza sativa) (Gui et al., 2014). As an example, in rice, over-expressed remorin gene gsd1-D in the dominant mutant (grain setting defect1-Dominant) showed a grain setting-deficient phenotype of reduced grain setting rate, reversible accumulation of carbohydrate in leaves, and reduced synthesis of soluble sugar concentration in phloem exudates (Gui et al., 2014).
Three candidate genes are also identified as UDP-glucose dehydrogenase (UGD) – gene Ca_14209 is simultaneously associated with two traits (ciceritol and total sugar contents). The other two genes are Ca_27229 (associated with raffinose content) and Ca_10185 (associated with sucrose content). UGD is a key enzyme in carbohydrate metabolism and has been identified in soybean (Glycine max), maize (Zea mays L.), cotton (Gossypium hirsutum), and Arabidopsis (Arabidopsis thaliana) (Kärkönen et al., 2005; Kohlberger et al., 2018; Jia et al., 2021). The overexpression of UDG-coding gene PeUGDH4 in Arabidopsis leads to a significant increase in hemicellulose synthesis (Yang et al., 2020), indicating its critical role in plant cell wall synthesis. UGD converts UDP-glucose to UDP-glucuronic acid, providing the precursor for hemicellulose and pectin biosynthesis - the two confound components in the primary cell wall matrix (Oka and Jigami, 2006). And later, UGD has also been identified to be highly involved in the secondary cell wall construction in Moso bamboo (Phyllostachys edulis) (Yang et al., 2020). Besides being incorporated into the cell wall, the remainder forms of carbohydrates can be small molecule oligosaccharides such as RFOs. The RFOs biosynthesis pathways started with UDP-galactose being converted to UDP-galacturonic acid (UDP-GalA) by UG4E UDP-glucose 4′-epimerase; or alternatively UDP-glucose being converted to myo-inositol (Karner et al., 2004; Joët et al., 2009). Then both UDP-GAL and myo-inositol can be the precursors of the galactinol biosynthesis by galactinol synthase GolS (Keller and Pharr, 1996). Galactinol is believed to be the only known galactosyl donor to RFOs (Sprenger and Keller, 2000). In summary, the cell wall polysaccharide (CWP) and RFOs biosynthesis pathways are interconnected but also competitive for the upstream precursor UDP-glucose. The increase in UGD activity could lead to the increased production of CWP; however, at the same time, it could diminish RFOs and other carbohydrates production.
Conclusion
The present study identified potential candidate genes regulating the biosynthesis and transport of seed oligosaccharides in chickpea. We have identified 48 SNPs associated with five sugar types. Nine genes (Ca_06204, Ca_04353, and Ca_20828: Phosphatidylinositol N-acetylglucosaminyltransferase; Ca_17399 and Ca_22050: Remorin proteins; Ca_11152: Protein-serine/threonine phosphatase; Ca_10185, Ca_14209, and Ca_27229: UDP-glucose dehydrogenase) were identified as potential candidate genes for sugar metabolism and transport in chickpea. The accessions with low RFOs and high sucrose contents may be utilized in breeding specialty chickpeas. The identified candidates could be exploited in marker-assisted breeding, genomic selection, and genetic engineering to improve the sugar profiles in legumes and other crop species.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Author contributions
DE- designed experiments, collected and analyzed data, and wrote the manuscript. WW- analyzed data and wrote the manuscript. MT- analyzed the data. SS- designed experiments, collected and analyzed data. BR- analyzed and wrote the manuscript. RV- edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The open access publication fees for this article were covered by the Iowa State University Library.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1024543/full#supplementary-material
References
Aguilera, Y., Martín-Cabrejas, M. A., Benítez, V., Mollá, E., López-Andréu, F. J., Esteban, R. M. (2009). Changes in carbohydrate fraction during dehydration process of common legumes. J. Food Composition Anal. 22, 678–683. doi: 10.1016/j.jfca.2009.02.012
Ananieva, E. A., Gillaspy, G. E., Ely, A., Burnette, R. N., Erickson, F.l. (2008). Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiol. 148, 1868–1882. doi: 10.1104/PP.108.130575
Ayre, B. G., Keller, F., Turgeon, R. (2003). Symplastic continuity between companion cells and the translocation stream: Long-distance transport is controlled by retention and retrieval mechanisms in the phloem. Plant Physiol. 131, 1518–1528. doi: 10.1104/PP.012054
Baena-González, E., Rolland, F., Thevelein, J. M., Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. doi: 10.1038/NATURE06069
Bariola, P. A., Retelska, D., Stasiak, A., Kammerer, R. A., Fleming, A., Hijri, M., et al. (2004). Remorins form a novel family of coiled coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Mol. Biol. 55, 579–594. doi: 10.1007/S11103-004-1520-4
Blackman, S. A., Obendorf, R. L., Leopold, A. C. (1992). Maturation proteins and sugars in desiccation tolerance of developing soybean seeds. Plant Physiol. 100, 225–230. doi: 10.1104/pp.100.1.225
Calloway, D. H., Murphy, E. L. (1968). The use of expired air to measure intestinal gas formation. Ann. N Y Acad. Sci. 150, 82–95. doi: 10.1111/J.1749-6632.1968.TB19034.X
Chibbar, R. N., Ambigaipalan, P., Hoover, R. (2010). REVIEW: Molecular diversity in pulse seed starch and complex carbohydrates and its role in human nutrition and health. Cereal Chem. 87, 342–352. doi: 10.1094/CCHEM-87-4-0342
de Koning, R., Kiekens, R., Toili, M. E. M., Angenon, G. (2021). Identification and expression analysis of the genes involved in the raffinose family oligosaccharides pathway of phaseolus vulgaris and glycine max. Plants 10, 1–21. doi: 10.3390/plants10071465
de Las Mercedes Dana, M., Pintor-Toro, J. A., Cubero, B. (2006). Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol. 142, 722. doi: 10.1104/PP.106.086140
Delumen, B. O. (1992). Molecular strategies to improve protein-quality and reduce flatulence in legumes - a review. Food Structure 11, 33–46.
Dierking, E. C., Bilyeu, K. D. (2008). Association of a soybean raffinose synthase gene with low raffinose and stachyose seed phenotype. Plant Genome 1, 135–145. doi: 10.3835/plantgenome2008.06.0321
Dobrenel, T., Marchive, C., Azzopardi, M., Clément, G., Moreau, M., Sormani, R., et al. (2013). Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR? Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00093
Egbe, I. A., Akinyele, I. O. (1990). Effect of cooking on the antinutritional factors of lima beans (Phaseolus lunatus). Food Chem. 35, 81–87. doi: 10.1016/0308-8146(90)90022-V
Elango, D., Rajendran, K., van der Laan, L., Sebastiar, S., Raigne, J., Thaiparambil, N. A., et al. (2022a). Raffinose family oligosaccharides: Friend or foe for human and plant health? Front. Plant Sci. 13. doi: 10.3389/FPLS.2022.829118/BIBTEX
Elango, D., Wang, X., Bhatnagar, R. S., Tan, Q., Gaffoor, I., Hu, Z., et al. (2022b). Association genetics of early season cold and late season frost tolerance in sorghum bicolor. Crop Sci. 62, 1844–1865. doi: 10.1002/csc2.20810
Gangl, R., Tenhaken, R. (2016). Raffinose Family Oligosaccharides Act As Galactose Stores in Seeds and Are Required for Rapid Germination of Arabidopsis in the Dark. Front Plant Sci 7, 1–15. doi: 10.3389/fpls.2016.01115
Gangola, M. P., Jaiswal, S., Kannan, U., Gaur, P. M., Båga, M., Chibbar, R. N. (2016). Galactinol synthase enzyme activity influences raffinose family oligosaccharides (RFO) accumulation in developing chickpea (Cicer arietinum l.) seeds. Phytochemistry 125, 88–98. doi: 10.1016/j.phytochem.2016.02.009
Graham, P. H., Vance, C. P. (2003). Legumes: Importance and constraints to greater use. Plant Physiol. 131, 872–877. doi: 10.1104/PP.017004
Gui, J., Liu, C., Shen, J., Li, L. (2014). Grain setting defect1, encoding a remorin protein, affects the grain setting in rice through regulating plasmodesmatal conductance. Plant Physiol. 166, 1463–1478. doi: 10.1104/PP.114.246769
Gulewicz, P., Szymaniec, S., Bubak, B., Frias, J., Vidal-Valverde, C., Trojanowska, K., et al. (2002). Biological activity of α-galactoside preparations from lupinus angustifolius l. and pisum sativum l. seeds. J. Agric. Food Chem. 50, 384–389. doi: 10.1021/jf010973y
Halford, N. G., Hey, S. J. (2009). Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem. J. 419, 247–259. doi: 10.1042/BJ20082408
Han, I. H., Baik, B. K. (2006). Oligosaccharide content and composition of legumes and their reduction by soaking, cooking, ultrasound, and high hydrostatic pressure. Cereal Chem. 83, 428–433. doi: 10.1094/CC-83-0428
Huang, D., Sun, Y., Ma, Z., Ke, M., Cui, Y., Chen, Z., et al. (2019). Salicylic acid-mediated plasmodesmal closure via remorin-dependent lipid organization. Proc. Natl. Acad. Sci. U.S.A. 116, 21274–21284. doi: 10.1073/PNAS.1911892116
Hulsmans, S., Rodriguez, M., de Coninck, B., Rolland, F. (2016). The SnRK1 energy sensor in plant biotic interactions. Trends Plant Sci. 21, 648–661. doi: 10.1016/J.TPLANTS.2016.04.008/ATTACHMENT/3B97F173-DE7E-48F8-83F5-FF0FBC3B99C5/MMC1.MP4
Jia, T., Ge, Q., Zhang, S., Zhang, Z., Liu, A., Fan, S., et al. (2021). UDP-Glucose dehydrogenases: Identification, expression, and function analyses in upland cotton (Gossypium hirsutum). Front. Genet. 111648. doi: 10.3389/FGENE.2020.597890/BIBTEX
Joët, T., Laffargue, A., Salmona, J., Doulbeau, S., Descroix, F., Bertrand, B., et al. (2009). Metabolic pathways in tropical dicotyledonous albuminous seeds: Coffea arabica as a case study. New Phytol. 182, 146–162. doi: 10.1111/J.1469-8137.2008.02742.X
Johnson, N., Johnson, C. R., Thavarajah, P., Kumar, S., Thavarajah, D. (2020). The roles and potential of lentil prebiotic carbohydrates in human and plant health. Plants People Planet 2, 310–319. doi: 10.1002/PPP3.10103
Jood, S., Mehta, U., Bhat, C. M., Singh, R. (1985). Effect of processing on flatus-producing factors in legumes. J. Agric. Food Chem. 33, 268–271. doi: 10.1021/jf00062a028
Jossier, M., Bouly, J. P., Meimoun, P., Arjmand, A., Lessard, P., Hawley, S., et al. (2009). SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in arabidopsis thaliana. Plant J. 59, 316–328. doi: 10.1111/J.1365-313X.2009.03871.X
Jukanti, A. K., Gaur, P. M., Gowda, C. L. L., Chibbar, R. N. (2012). Nutritional quality and health benefits of chickpea (Cicer arietinum l.): a review. Br. J. Nutr. 108, S11–S26. doi: 10.1017/S0007114512000797
Kärkönen, A., Murigneux, A., Martinant, J. P., Pepey, E., Tatout, C., Dudley, B. J., et al. (2005). UDP-Glucose dehydrogenases of maize: a role in cell wall pentose biosynthesis. Biochem. J. 391, 409–415. doi: 10.1042/BJ20050800
Karlsson, M., Stenlid, J. (2008). Comparative evolutionary histories of the fungal chitinase gene family reveal non-random size expansions and contractions due to adaptive natural selection. Evol. Bioinf. 2008, 47–60. doi: 10.4137/ebo.s604
Karner, U., Peterbauer, T., Raboy, V., Jones, D. A., Hedley, C. L., Richter, A. (2004). Myo-inositol and sucrose concentrations affect the accumulation of raffinose family oligosaccharides in seeds. J. Exp. Bot. 55, 1981–1987. doi: 10.1093/JXB/ERH216
Keller, F. (1992). Transport of stachyose and sucrose by vacuoles of Japanese artichoke (Stachys sieboldii) tubers. Plant Physiol. 98, 442–445. doi: 10.1104/PP.98.2.442
Keller, F., Ludlow, M. M. (1993). Carbohydrate metabolism in drought-stressed leaves of pigeonpea (Cajanus cajan). J. Exp. Bot. 44, 1351–1359. doi: 10.1093/jxb/44.8.1351
Keller, F., Pharr, D. M. (1996). “Metabolism of carbohydrates in sinks and sources: galactosyl-sucrose oligosaccharides,” in Photoassimilate distribution in plants and crops. Eds. Zamski, E., Schaffer, A. A. (New York: Taylor & Francis), 157–183.
Kohlberger, M., Thalhamer, T., Weiss, R., Tenhaken, R. (2018). Arabidopsis MAP-kinase 3 phosphorylates UDP-glucose dehydrogenase: a key enzyme providing UDP-sugar for cell wall biosynthesis. Plant Mol. Biol. Rep. 36, 870. doi: 10.1007/S11105-018-1130-Y
Lev-Yadun, S., Gopher, A., Abbo, S. (2000). The cradle of agriculture. Sci. (1979) 288, 1602–1603. doi: 10.1126/science.288.5471.1602
Liu, X., Huang, M., Fan, B., Buckler, E. S., Zhang, Z. (2016). Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PloS Genet. 12, e1005767. doi: 10.1371/journal.pgen.1005767
Lou, Y., Gou, J. Y., Xue, H. W. (2007). PIP5K9, an arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19, 163–181. doi: 10.1105/TPC.106.045658
Marmagne, A., Roue, M. A., Ferro, M., Rolland, N., Alcon, C., Joyard, J., et al. (2004). Identification of new intrinsic proteins in arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 3, 675–691. doi: 10.1074/MCP.M400001-MCP200
Mongrand, S., Morel, J., Laroche, J., Claverol, S., Carde, J. P., Hartmann, M. A., et al. (2004). Lipid rafts in higher plant cells: purification and characterization of triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279, 36277–36286. doi: 10.1074/JBC.M403440200
Nelson, C. J., Hegeman, A. D., Harms, A. C., Sussman, M. R. (2006). A quantitative analysis of arabidopsis plasma membrane using trypsin-catalyzed 18O labeling. Mol. Cell. Proteomics 5, 1382–1395. doi: 10.1074/MCP.M500414-MCP200/ATTACHMENT/F5709B47-DBEB-49EE-9EE7-676C9E281C9C/MMC1.ZIP
Nishizawa-Yokoi, A., Yabuta, Y., Shigeoka, S. (2008). The contribution of carbohydrates including raffinose family oligosaccharides and sugar alcohols to protection of plant cells from oxidative damage. Plant Signal Behav. 3, 1016–1018. doi: 10.4161/psb.6738
Obendorf, R. L., Górecki, R. J. (2012). Soluble carbohydrates in legume seeds. Seed Sci. Res. 22, 219–242. doi: 10.1017/S0960258512000104
Oka, T., Jigami, Y. (2006). Reconstruction of de novo pathway for synthesis of UDP-glucuronic acid and UDP-xylose from intrinsic UDP-glucose in saccharomyces cerevisiae. FEBS J. 273, 2645–2657. doi: 10.1111/J.1742-4658.2006.05281.X
Peterbauer, T., Karner, U., Mucha, J., Mach, L., Jones, D. A., Hedley, C. L., et al. (2003). Enzymatic control of the accumulation of verbascose in pea seeds. Plant Cell Environ. 26, 1385–1391. doi: 10.1046/j.0016-8025.2003.01063.x
Peterbauer, T., Lahuta, L. B., Blöchl, A., Mucha, J., Jones, D. A., Hedley, C. L., et al. (2001). Analysis of the raffinose family oligosaccharide pathway in pea seeds with contrasting carbohydrate composition. Plant Physiol. 127, 1764–1772. doi: 10.1104/pp.010534
Purcell, P. C., Smith, A. M., Halford, N. G. (1998). Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J. 14, 195–202. doi: 10.1046/J.1365-313X.1998.00108.X
Qiu, D., Vuong, T., Valliyodan, B., Shi, H., Guo, B., Shannon, J. G., et al. (2015). Identification and characterization of a stachyose synthase gene controlling reduced stachyose content in soybean. Theor. Appl. Genet. 128, 2167–2176. doi: 10.1007/S00122-015-2575-0
Quemener, B., Brillouet, J. M. (1983). Ciceritol, a pinitol digalactoside form seeds of chickpea, lentil and white lupin. Phytochemistry 22, 1745–1751. doi: 10.1016/S0031-9422(00)80263-0
Radchuk, R., Radchuk, V., Weschke, W., Borisjuk, L., Weber, H. (2006). Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 140, 263. doi: 10.1104/PP.105.071167
Raja, R. B., Balraj, R., Agasimani, S., Dinakaran, E., Thiruvengadam, V., Kannan Bapu, J. R., et al. (2015). Determination of oligosaccharide fraction in a worldwide germplasm collection of chickpea ('Cicer arietinum’L.) using high performance liquid chromatography. Aust. J. Crop Sci. 9, 605–613.
Ren, Y., Li, M., Guo, S., Sun, H., Zhao, J., Zhang, J., et al. (2021). Evolutionary gain of oligosaccharide hydrolysis and sugar transport enhanced carbohydrate partitioning in sweet watermelon fruits. Plant Cell 33, 1554–1573. doi: 10.1093/PLCELL/KOAB055
Saddhe, A. A., Manuka, R., Penna, S. (2021). Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiol Plant 171, 739–755. doi: 10.1111/PPL.13283
Sazuka, T., Keta, S., Shiratake, K., Yamaki, S., Shibata, D. (2004). A proteomic approach to identification of transmembrane proteins and membrane-anchored proteins of arabidopsis thaliana by peptide sequencing. DNA Res. 11, 101–113. doi: 10.1093/DNARES/11.2.101
Sengupta, S., Mukherjee, S., Basak, P., Majumder, A. L. (2015). Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00656
Singh, U. (1985). Nutritional quality of chickpea (Cicer arietinum l.): current status and future research needs. Plant Foods Hum. Nutr. 35, 339–351. doi: 10.1007/BF01091779
Sosulski, F. W., Elkowicz, L., Reichert, R. D. (1982). Oligosaccharides in eleven legumes and their air-classified protein and starch fractions. J. Food Sci. 47, 498–502. doi: 10.1111/j.1365-2621.1982.tb10111.x
Souframanien, J., Roja, G., Gopalakrishna, T. (2014). Genetic variation in raffinose family oligosaccharides and sucrose content in blackgram [Vigna mungo l. (Hepper)]. J. Food Legumes 27, 37–41.
Sprenger, N., Keller, F. (2000). Allocation of raffinose family oligosaccharides to transport and storage pools in ajuga reptans: the roles of two distinct galactinol synthases. Plant J. 21, 249–258. doi: 10.1046/J.1365-313X.2000.00671.X
Su, H., Chen, J., Miao, S., Deng, K., Liu, J., Zeng, S., et al. (2019). Lotus seed oligosaccharides at various dosages with prebiotic activity regulate gut microbiota and relieve constipation in mice. Food and Chemical Toxicology 134, 1–12. doi: 10.1016/j.fct.2019.110838
Tahir, M., Lindeboom, N., Båga, M., Vandenberg, A., Chibbar, R. (2011). Composition and correlation between major seed constituents in selected lentil (Lens culinaris medik) genotypes. Can. J. Plant Sci. 91, 825–835. doi: 10.4141/cjps2011-010
Taji, T., Ohsumi, C., Iuchi, S., Seki, M., Kasuga, M., Kobayashi, M., et al. (2002). Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in arabidopsis thaliana. Plant J. 29, 417–426. doi: 10.1046/j.0960-7412.2001.01227.x
Tiessen, A., Prescha, K., Branscheid, A., Palacios, N., McKibbin, R., Halford, N. G., et al. (2003). Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J. 35, 490–500. doi: 10.1046/J.1365-313X.2003.01823.X
Unda, F., Kim, H., Hefer, C., Ralph, J., Mansfield, S. D. (2017). Altering carbon allocation in hybrid poplar (Populus alba × grandidentata) impacts cell wall growth and development. Plant Biotechnol. J. 15, 865–878. doi: 10.1111/pbi.12682
Upadhyaya, H. D., Ortiz, R. (2001). A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theor. Appl. Genet. 102, 1292–1298. doi: 10.1007/s00122-001-0556-y
Valot, B., Negroni, L., Zivy, M., Gianinazzi, S., Dumas-Gaudot, E. (2006). A mass spectrometric approach to identify arbuscular mycorrhiza-related proteins in root plasma membrane fractions. Proteomics 6 (Suppl 1), S145–S155. doi: 10.1002/PMIC.200500403
Van den Ende, W. (2013). Multifunctional fructans and raffinose family oligosaccharides. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00247
Varshney, R. K., Song, C., Saxena, R. K., Azam, S., Yu, S., Sharpe, A. G., et al. (2013). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246. doi: 10.1038/nbt.2491
Varshney, R. K., Thudi, M., Roorkiwal, M., He, W., Upadhyaya, H. D., Yang, W., et al. (2019). Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 51, 857–864. doi: 10.1038/s41588-019-0401-3
Wang, J., Zhang, Z. (2021). GAPIT version 3: Boosting power and accuracy for genomic association and prediction. Genomics Proteomics Bioinf. 19, 629–640. doi: 10.1016/j.gpb.2021.08.005
Watson, B. S., Asirvatham, V. S., Wang, L., Sumner, L. W. (2003). Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol. 131, 1104–1123. doi: 10.1104/PP.102.019034
Xiaoli, X., Liyi, Y., Shuang, H., Wei, L., Yi, S., Hao, M., et al. (2008). Determination of oligosaccharide contents in 19 cultivars of chickpea (Cicer arietinum l) seeds by high performance liquid chromatography. Food Chem. 111, 215–219. doi: 10.1016/J.FOODCHEM.2008.03.039
Xin, Y., Wang, D., Han, S., Li, S., Gong, N., Fan, Y., et al. (2021). Characterization of the chitinase gene family in mulberry (Morus notabilis) and MnChi18 involved in resistance to botrytis cinerea. Genes (Basel) 13, 1–12. doi: 10.3390/GENES13010098/S1
Yan, S., Liu, Q., Li, W., Yan, J., Fernie, A. R. (2022). Raffinose Family Oligosaccharides: Crucial Regulators of Plant Development and Stress Responses. CRC Crit Rev Plant Sci 41, 286–303. doi: 10.1080/07352689.2022.2111756
Yang, Y., Kang, L., Wu, R., Chen, Y., Lu, C. (2020). Genome-wide identification and characterization of UDP-glucose dehydrogenase family genes in moso bamboo and functional analysis of PeUGDH4 in hemicellulose synthesis. Sci. Rep. 2020 10:1 10, 1–13. doi: 10.1038/s41598-020-67227-8
Yang, J. I., Perera, I. Y., Brglez, I., Davis, A. J., Stevenson-Paulik, J., Phillippy, B. Q., et al. (2007). Increasing plasma membrane Phosphatidylinositol(4,5)Bisphosphate biosynthesis increases phosphoinositide metabolism in nicotiana tabacum. Plant Cell 19, 1603–1616. doi: 10.1105/TPC.107.051367
Zhang, Y. J., Ren, L. L., Lin, X. Y., Han, X. M., Wang, W., Yang, Z. L. (2022). Molecular evolution and functional characterization of chitinase gene family in populus trichocarpa. Gene 822, 1–10. doi: 10.1016/J.GENE.2022.146329
Zhang, Y., Shewry, P. R., Jones, H., Barcelo, P., Lazzeri, P. A., Halford, N. G. (2001). Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J. 28, 431–441. doi: 10.1046/J.1365-313X.2001.01167.X
Keywords: anti-nutritional factors (ANF), flatus potential, marker trait associations, prebiotics, raffinose family oligosaccharides (RFOs), specialty chickpeas
Citation: Elango D, Wang W, Thudi M, Sebastiar S, Ramadoss BR and Varshney RK (2022) Genome-wide association mapping of seed oligosaccharides in chickpea. Front. Plant Sci. 13:1024543. doi: 10.3389/fpls.2022.1024543
Received: 02 September 2022; Accepted: 10 October 2022;
Published: 24 October 2022.
Edited by:
Jianghua Chen, Key Laboratory of Tropical Plant Resource and Sustainable Use (CAS), ChinaReviewed by:
Eleonora Cominelli, National Research Council (CNR), ItalyUsman Aziz, Northwestern Polytechnical University, China
Copyright © 2022 Elango, Wang, Thudi, Sebastiar, Ramadoss and Varshney. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dinakaran Elango, ZGVsYW5nb0BpYXN0YXRlLmVkdQ==
†These authors have contributed equally to this work
‡ORCID: Dinakaran Elango, orcid.org/0000-0003-2226-486X
 Dinakaran Elango
Dinakaran Elango Wanyan Wang
Wanyan Wang Mahender Thudi
Mahender Thudi Sheelamary Sebastiar
Sheelamary Sebastiar Bharathi Raja Ramadoss
Bharathi Raja Ramadoss Rajeev K. Varshney
Rajeev K. Varshney