- 1Biotechnology Division, CSIR-Institute of Himalayan Bioresource Technology, Palampur, Himachal Pradesh, India
- 2Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, India
The Myo-Inositol-1-phosphate synthase (MIPS) gene family is involved in the myo-inositol synthesis and plays a significant role in signal transduction, membrane biogenesis, oligosaccharides synthesis, auxin storage and transport, programmed cell death and abiotic stress tolerance in plants. This study comprehensively identified the MIPS genes in Rosaceae plant species, and 51 MIPS genes were identified from 26 Rosaceae species. The phylogenetic analysis divided the MIPSs into two clades (clade I; subfamily Amygdaloideae specific, and clade II; subfamily Rosoideae specific). MIPS genes of all 26 Rosaceae species consist of similar gene structure, motif and domain composition, which shows their conserved nature. The cis-regulatory elements (CREs) analysis revealed that most Rosaceae MIPS genes play a role in growth, development, and stress responses. Furthermore, the qRT-PCR analysis also revealed the involvement of RcMIPS gene in plant development and response to abiotic stresses, including drought and heat. The results of the present study contribute to the understanding of the biological function of Rosaceae MIPS genes, and that could be used in further functional validations.
Introduction
The Myo-inositol phosphate synthase (MIPS) is a key enzyme in the myo-inositol biosynthesis pathway, that produces myo-inositol (Loewus and Loewus, 1980). Myo-inositol is an important component that regulates plant growth and development processes (Martin, 1998), membrane biogenesis (Roth, 2004), oligosaccharides synthesis (Karner et al., 2004), auxin storage and transport (Abreu and Aragao, 2007), programmed cell death (Meng et al., 2009), phytic acid biosynthesis (Ali et al., 2013), etc.
MIPS enzymes have been ubiquitously found in a wide range of organisms, including higher plants and animals, parasites, fungi, green algae, bacteria, and archaea, and it is thought to be a primordial protein/gene (Majumder et al., 1997). Recent reports have also specified its functional diversity and shared evolutionary clues from archaea to higher plants and animals (Majumder et al., 2003). A genome-wide investigation of MIPS copies in plant kingdom was carried out to unveil the mystery of sequence conservation and divergence (Hazra et al., 2019). MIPS evolved from a single common algal ancestor to seed plants, and functional diversification is perhaps controlled by genomic variations. The differences in the number and properties of MIPS genes result from tandem, block, whole genome duplication and subsequent divergence. Different MIPS gene variants acquired distinct functions during duplication and speciation events (Hazra et al., 2019).
Additionally, MIPSs are also involved in abiotic stress regulation in plants (Taji et al., 2006; Kaur et al., 2013; Tan et al., 2013; Sharma et al., 2020; Alok et al., 2022). For instance, under NaCl treatment, salt-resistant rice varieties (Oryza sativa L.) show increased activity of the chloroplast version of MIPS either at the seedling stage or on fully grown plants (Raychaudhuri and Majumder, 1996). The expression of wheat TaMIPS2 was induced in various developmental seed stages during heat stress treatment. Unfertilized ovaries also showed greater TaMIPS2 transcript levels, with considerable quantities remaining during the recovery phase, indicating that MIPS is important for heat stress recovery and flower development (Khurana et al., 2012). The expression of the MfMIPS1 gene was induced by salt and drought stresses in Medicago falcata. The overexpressed MfMIPS1 tobacco lines have increased levels of Myo-inositol, leading to enhanced tolerance to multiple abiotic stress, i.e., cold, salt and drought stress (Tan et al., 2013). The IbMIPS1 overexpressed lines of sweet potato also exhibited increased salt and drought tolerance (Zhai et al., 2016). Recently, it was shown that MdMIPS1 overexpressing apple lines showed enhanced drought stress tolerance (Hu et al., 2022). The above studies suggested that the MIPS genes were involved in the regulation of various abiotic stresses in plants.
The MIPS genes have been investigated thoroughly in several plant species, including the model plant Arabidopsis (Johnson and Sussex, 1995), maize (Larson and Raboy, 1999), sesame (Chun et al., 2003), rice (Feng and Yoshida, 2004), soybean (Kumari et al., 2012), wild rice; Porteresia coarctata (Basak et al., 2018), cotton (Ma et al., 2019), and oat (Qin et al., 2021). Compared with other species, studies on the characterization of MIPS genes and their function in growth and abiotic stress regulation in the Rosaceae family are limited. Therefore, in this study, we comprehensively analysed the distribution of MIPS genes in 26 different species of the Rosaceae family. We also characterized the MIPS gene with respect to its exon-intron structure, motif analysis, conserved domain, phylogenetic relationship, cis-regulatory elements, and gene ontology. Furthermore, the expression pattern of the rose MIPS gene was also performed in different tissues and response to the drought and heat stresses using qRT-PCR analysis. Altogether, this comprehensive information on MIPS genes of Rosaceae could be used for further functional validation and also provide insight into their functional role during abiotic stresses in roses.
Materials and methods
Sequence retrieval and identification of MIPSs in Rosaceae family
The genome sequences of 26 Rosaceae species available at the Genome Database for Rosaceae (GDR) (https://www.rosaceae.org; Jung et al., 2019) were utilized for the annotation and identification of the MIPS genes (more details were provided in Table S1). The three Arabidopsis MIPS protein sequences (IDs: AT2G22240, AT4G39800 and AT5G10170) were retrieved from the TAIR database (https://www.arabidopsis.org/) used as a query in the BLASTp program (e-value < 1× 10-5) to obtain MIPSs of the Rosaceae. Following that, all MIPS proteins were further verified for the presence of MIPS signature domains (NAD_5 Binding; PF07994 and Inos-1-P Synth; PF01658) using the Pfam database (http://pfam.xfam.org; El-Gebali et al., 2019). The molecular characteristics (aa length, molecular weight and isoelectric point) were predicted using the online tool ExPASy (http://web.expasy.org/; Artimo et al., 2012).
Multiple sequence alignments and phylogenetic analysis of MIPSs
Using the MUSCLE package available in MEGA X software (Stecher et al., 2020), all 51 MIPS protein sequences representing 26 Rosaceae species were aligned. The phylogenetic tree was created using the maximum-likelihood (ML) method with Poisson model, pair-wise deletion and 1000 bootstraps in MEGA X software. The web programme iTOL (Letunic and Bork, 2007) has been used to visualize the phylogenetic tree.
Gene structure and motif analysis of MIPSs
The exon-intron structure of the MIPS gene was determined using the online server GSDS 2.0 (Gene Structure and Display server 2.0; http://gsds.cbi.pku.edu.cn/Gsds_abou.php). The conserved motif was predicted by using the Motif Elicitation (MEME) program with the parameter set to find 20 motifs (http://meme-suit.org; Bailey et al., 2015) and visualize the structure using the TBtool software (Chen et al., 2020).
Cis-regulatory elements analysis
To analyze the cis regulatory elements (CREs) of MIPSs, the upstream sequences (2,000 bp) of the start codon were fetched from the GDR database. The CREs were identified in MIPS genes using the PlantCARE online server (Lescot et al., 2002).
Nuclear localization signals, subcellular localization and GO analysis
The NLS in MIPS were predicted using the web tool cNLS Mapper (Kosugi et al., 2009). Using the WoLF PSOPT II online resource (https://www.genscript.com/wolf-psort.html?src=leftbar), the subcellular localization of Rosaceae MIPSs was predicted. The rose MIPS gene ID were subjected to the ShinyGO v0.75 database (Ge et al., 2020) to obtain gene ontology (GO) annotation.
Plant material and stress treatment
In this study, the rose genotype (Rosa chinensis) was used. The plants were grown in plastic pots containing soil, peat, and sand mixed in a ratio of 1:2:1 and under greenhouse conditions (photoperiod=14 h light/10 h dark and temperature=22 ± 2°C) for the expression analysis of the RcMIPS gene. For tissue-specific expression analysis, the following tissues, stem, leaf, bud and flower tissues were harvested at the flowering stage. To impose the heat stress (HS), plants were kept at 42°C for 24 h. For drought stress (DS) treatment, we irrigated the plants with 20% PEG 6000. Leaf samples were harvested at the following different times: 0h, 6h, 12h and 24h for both HS and DS. During the experiment, the control tissues were collected from the unstressed plants. In addition, we collected apple (Malus domestica) stem and leaf tissues for MdMIPS tissue-specific expression analyses. All of the tissues were collected in three biological replications and kept at -80°C till further use.
RNA isolation and quantitative real-time PCR
The PureLinkTMRNA Mini Kit (Invitrogen) was used to isolate total RNA according to the manufacturer’s instructions. Then total RNA was converted to cDNA using Thermo Scientific’s Verso cDNA Synthesis Kit. In the QuantStudio 3 applied biosystems Real-Time PCR System, quantitative RT-PCR (qRT-PCR) was performed using SYBR green master mix (Applied Biosystems, Foster City, USA). ApE–A plasmid Editor (https://jorgensen.biology.utah.edu/wayned/ape/) was used to generate primers for the specified RcMIPS; Forward primer–AGGAGGAACACTTTGATTGGTGG and Reverse primer–GGTTTGTGGAGCTGAAAGGTTC. As an internal control, the housekeeping gene RcActin (RchiOBHm_Chr1g0322051) was employed. For expression of MdMIPS, Forward primer– ACAACTACGAGACCACCGAG and Reverse primer– GTGAGGGTTGAGCCATTGTT, as well as an internal control nad5, was used (Menzel et al., 2002). Each sample received three biological replicates and three technical replicates for qRT-PCR. The 2−ΔΔCt technique was used to determine the relative expression level of RcMIPS gene (Livak and Schmittgen, 2001).
Results
Distribution of MIPS genes in Rosaceae family
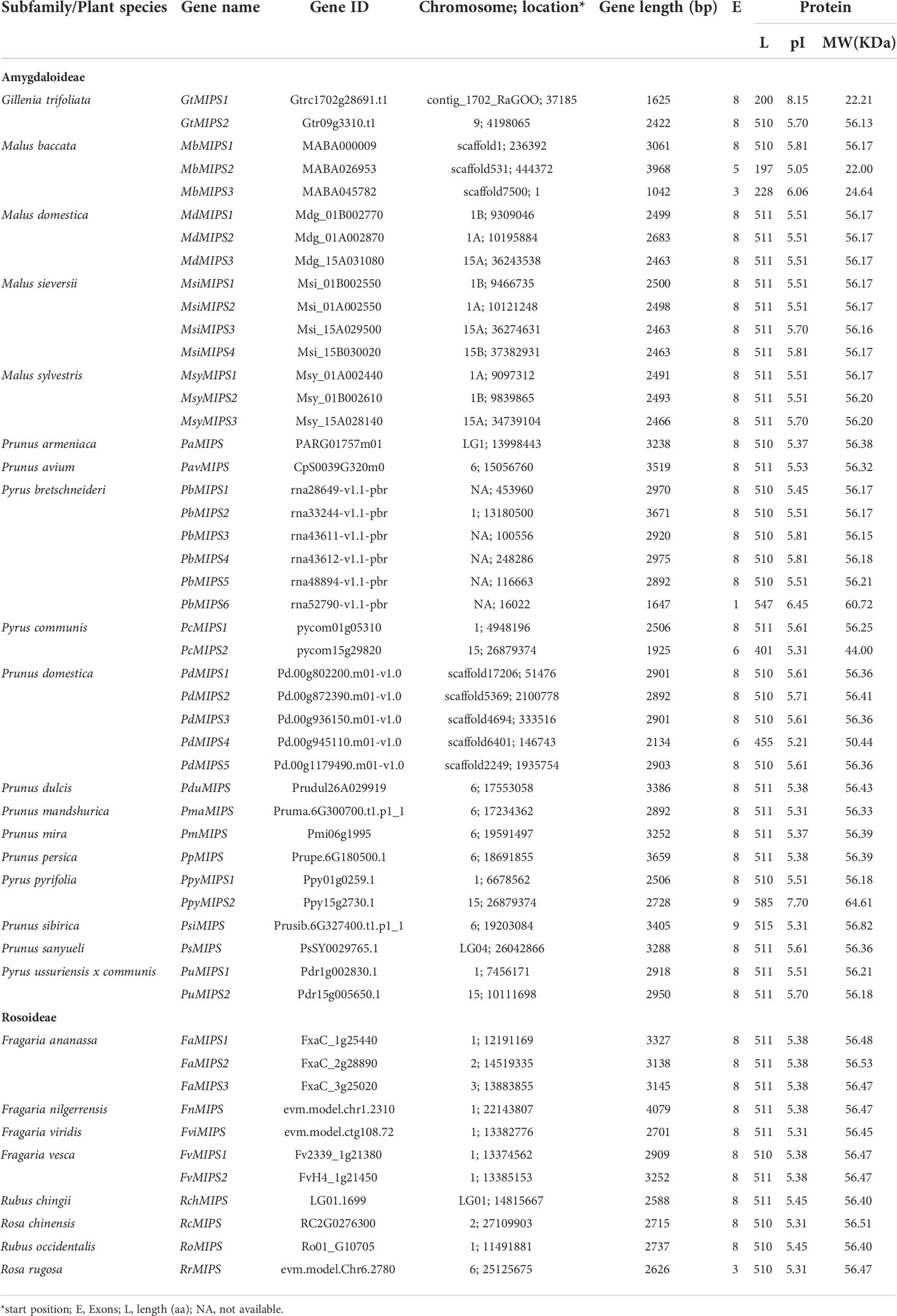
The genome-wide search through BLASTp searches and verification of the 26 Rosaceae species reference genomes identified a total of 51 MIPS genes (Table 1). These 26 species belong to two Rosaceae subfamilies (Amygdaloideae and Rosoideae). The genes were then named based on the reported genes in Arabidopsis and their chromosome location from top to bottom. The length of the Rosaceae MIPS gene ranged from 1042 (MbMIPS3) to 4079 bp (FnMIPS), and the average length was 2810 bp. The protein length ranged from 197aa (MbMIPS2) to 585 aa (PpyMIPS2), and the average length was 492 aa. The predicted molecular weight of Rosaceae MIPS protein varied from 22.0 kDa (MdMIPS2) to 64.6 kDa (PpyMIPS2), and the average molecular weight was 54.2 kDa. As shown in Table 1, there are variations in the number of MIPS genes among 26 Rosaceae species, it ranges from one (14 species see Table 1) to six (Pyrus bretschneideri). However, most of Rosaceae species (14 out of 26) were found to have single copy of MIPS gene and some have multiple copies like Gillenia trifoliata (2), Malus domestica (3), Malus sieversii (4), Prunus domestica (5), and Pyrus bretschneideri (6). To find out the reason behind the expansion of Rosaceae MIPS genes, we looked at gene duplication events in different Rosaceae genomes. A total of 43 pairs of duplications were found in the 26 Rosaceae species (for details see Table S2). We also calculated Ka/Ks value for duplicated gene pairs, the Ka/Ks values for all gene pairs were less than 1, with an average of 0.185 (Table S2). These results suggested that the MIPS genes in Rosaceae species are continuously evolving via purifying selection.
Phylogenetic analysis of Rosaceae MIPSs
To gain insights into the evolutionary relationship of the MIPSs in 26 Rosaceae species, a phylogenetic tree was made using ML-based multiple sequence alignments of 51 full-length protein sequences. The phylogenetic analysis divided the MIPSs into two clades (clade I and clade II) (Figure 1). Clade I had 40 MIPSs belonging to 18 species of Rosaceae subfamily Amygdaloideae and clade II had 11 MIPSs belonging to 8 species of Rosaceae subfamily Rosoideae (Figure 1).
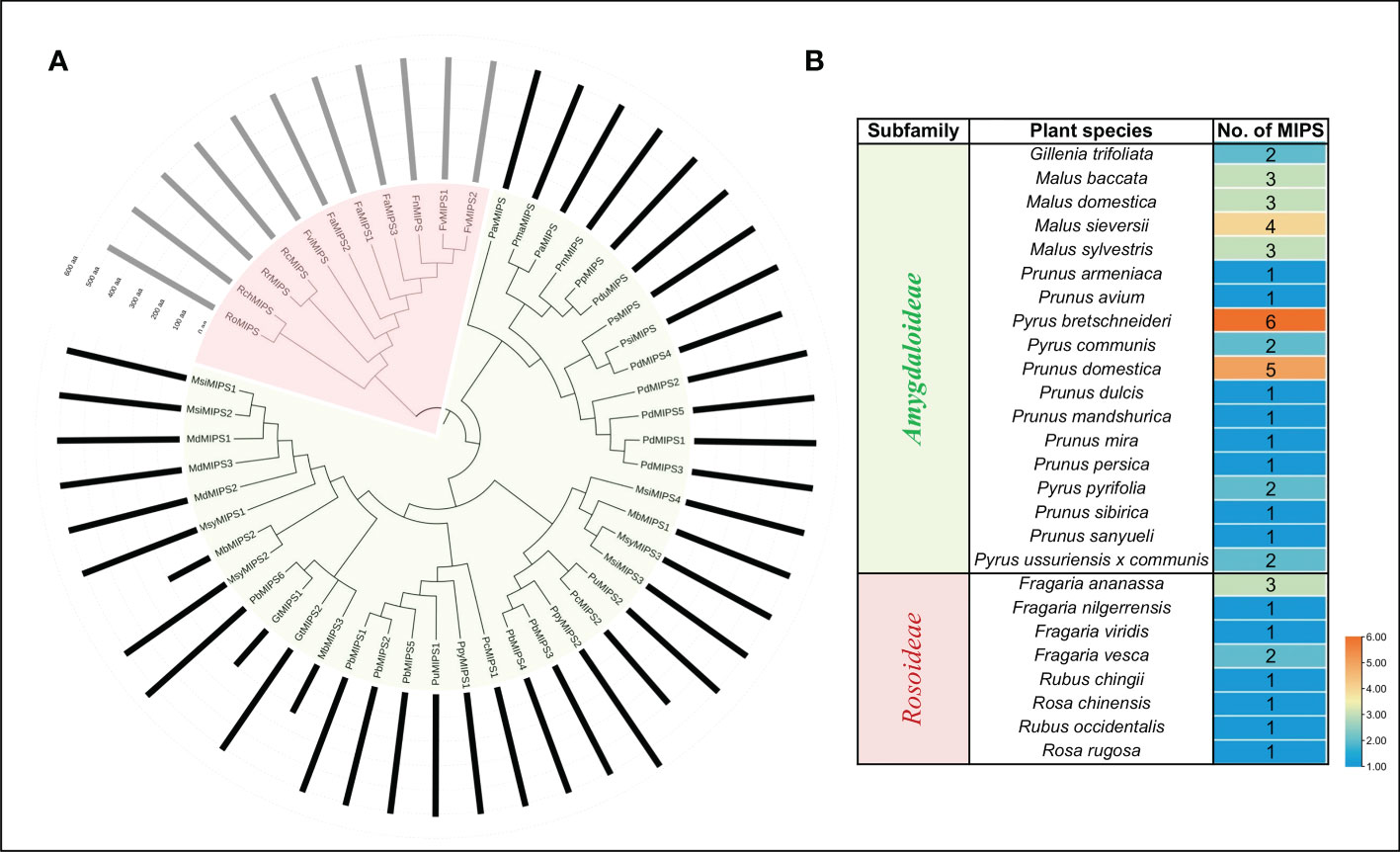
Figure 1 (A) Phylogenetic relationship among MIPSs belonging to the 26 Rosaceae plant species. (B) Table showing the number of MIPS genes present in each clade/sub-family.
Gene structure, motif and domain composition of MIPSs
The analysis of the exon-intron structures of the 51 MIPS genes in 26 Rosaceae species showed that the lengths of the MIPS genes were mainly distributed within the range of 1.5–4.0 kb. The number of exons ranges from one (PbMIPS6) to nine (PsiMIPS and PpyMIPS2). A total number of eight exons are most abundant and found in 43 MIPS genes (84%) (Figure 2). Motif distribution analysis predicted 20 different conserved motifs in 51 Rosaceae MIPS genes. The motif’s amino acid length was varied from 6 to 50. The 13 motifs (motifs 1 to 13) were present in all Rosaceae MIPS genes (except GtMIPS1, MbMIPs2, MbMIPS3, PbMIPS6 and PpyMIPS2), and the organization pattern of these 13 motifs in the MIPS protein sequence was similar in all 26 Rosaceae plant species (Figure 3). These data suggest that MIPS is a highly conserved Rosaceae species. The conservation of MIPS genes was further confirmed as all Rosaceae plant species MIPS protein possessed NAD binding domain (NAD_5_Binding; PF07994) and myo-inositol-1-phosphate synthase (Inos-1-P_synth; PF01658) domain (Figure 4A). Additionally, the stretches of conserved amino acid residues such as (1) GWGGNNG, (2) LWTANTERY, (3) INGSPQNTFVPG, and (3) GIKPLSIASYN were also present in Rosaceae plant species (Figure 4B).
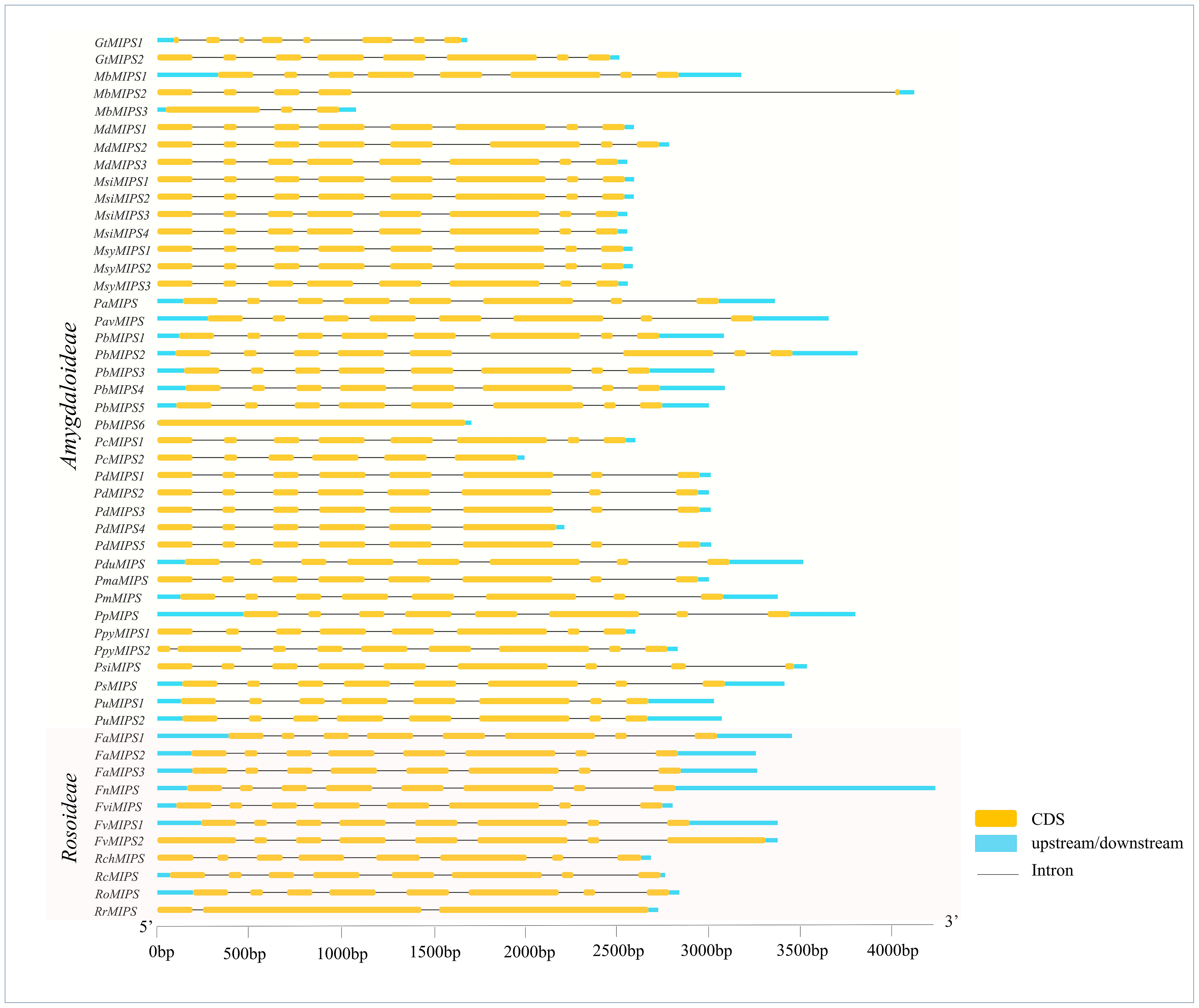
Figure 2 Exon–intron structures of Rosaceae MIPS genes. Exons are shown as yellow boxes, introns were denoted by thin dark grey lines and upstream/downstream regions are shown as sky-blue boxes. The lengths of exons and introns can be determined using the scale bar on the bottom.
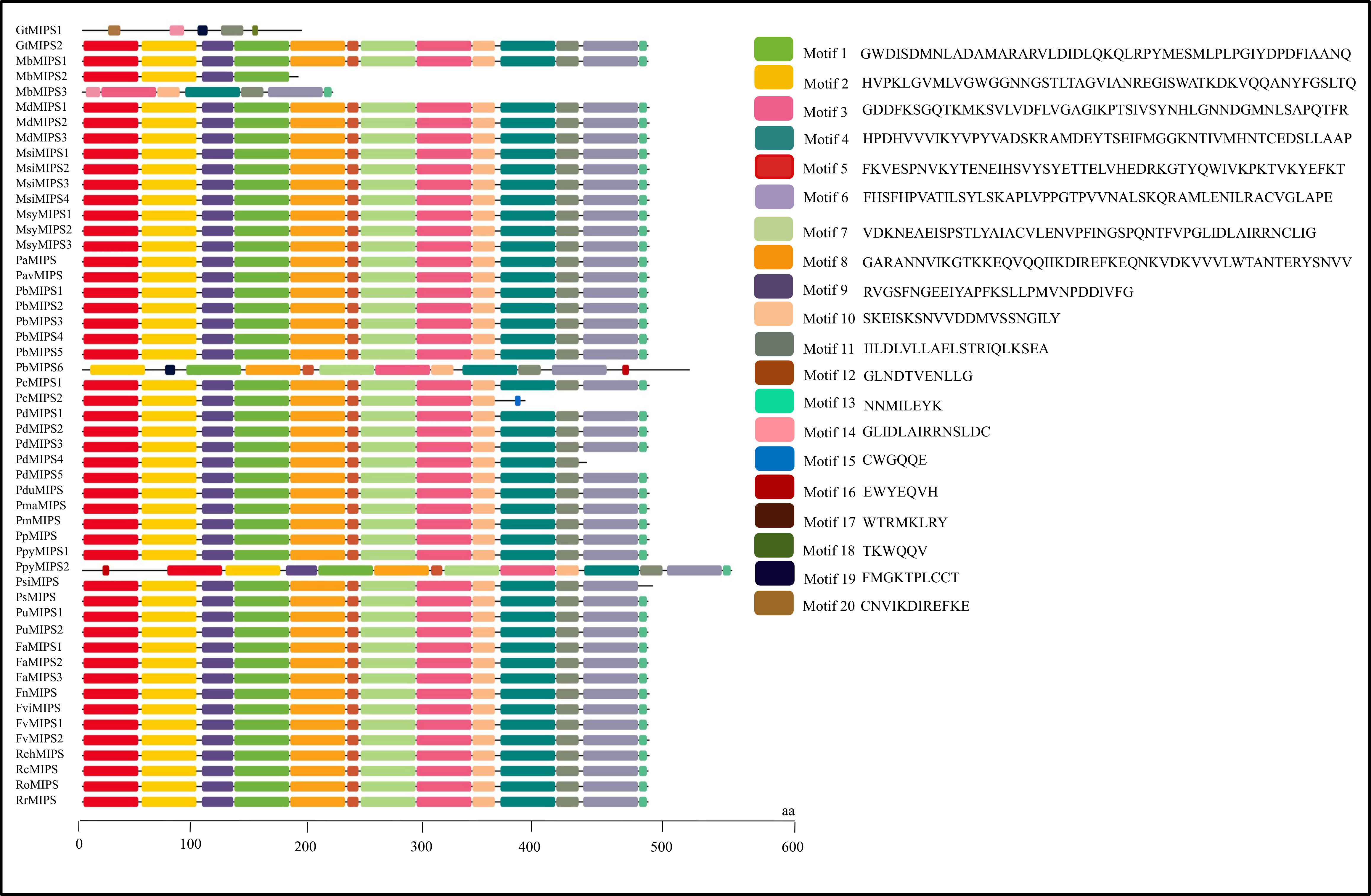
Figure 3 Motifs organization of Rosaceae MIPS proteins. Colour boxes represent the position of different motifs and box sizes show the length of motifs. Amino acid sequences for 20 different motifs (motif 1 to 20) are also provided on the right side.
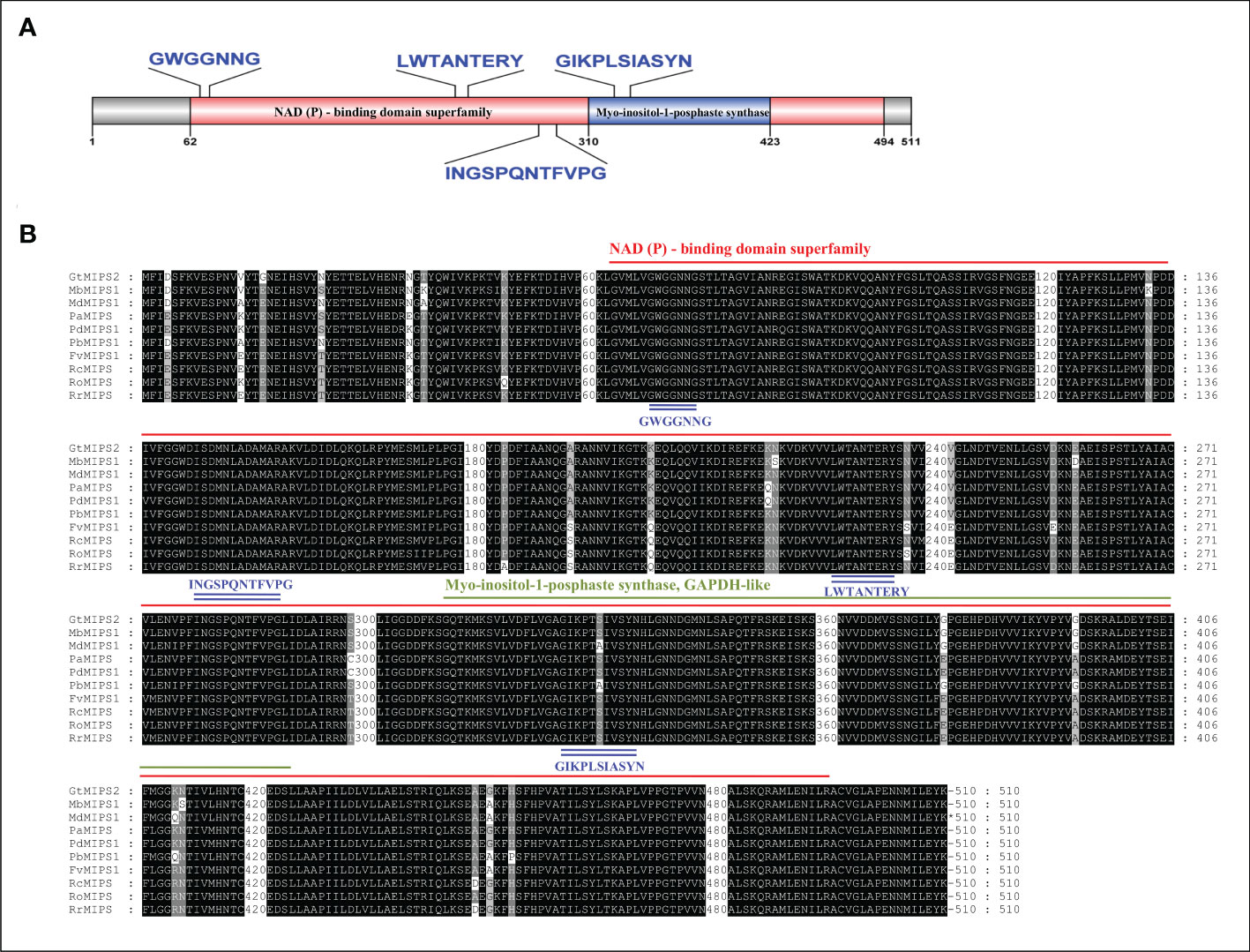
Figure 4 (A) Schematic representation of the conserved domain of Rosaceae MIPS protein. Two different conserved domains i.e., NAD binding domain (NAD_5_Binding) and myo-inositol-1-phosphate synthase (Inos-1-P_synth) are shown in different coloured boxes. (B) Multiple sequence alignment of ten representative Rosaceae MIPS proteins. Conserved domains are indicated with red and green underlines, the four stretches of conserved amino acid residues within domains are indicated by blue underlines.
Prediction of NLS and subcellular localization of MIPSs
The NLS prediction results showed that all the Rosaceae MIPSs had bipartite NLSs. The 51 Rosaceae MIPSs sequences have 41 NLSs sites (Supplementary Table S3). The PpyMIPS2 protein sequence also contains two monopartite sites in addition to bipartite NLSs. The results of subcellular localization prediction showed that all 51 Rosaceae MIPSs were localized in the cytoplasm (Supplementary Table S3).
Cis−regulatory elements analysis of MIPSs
To understand more about the possible role of MIPS genes in regulating growth and development and abiotic stress responses in Rosaceae species, we search for possible CREs in the two kb promoter regions of Rosaceae MIPSs genes. In total, 2243 representing 64 types of CREs were present in 51 Rosaceae MIPS genes (Figure 5). The preponderance of the cis-acting elements found were hormone response factors. The majority of the identified CREs were abiotic stress response elements (992; 44.22%), followed by plant growth-related elements (728; 32.45%), light-responsive elements (288; 12.83%), and hormone response factors (235; 10.47%).
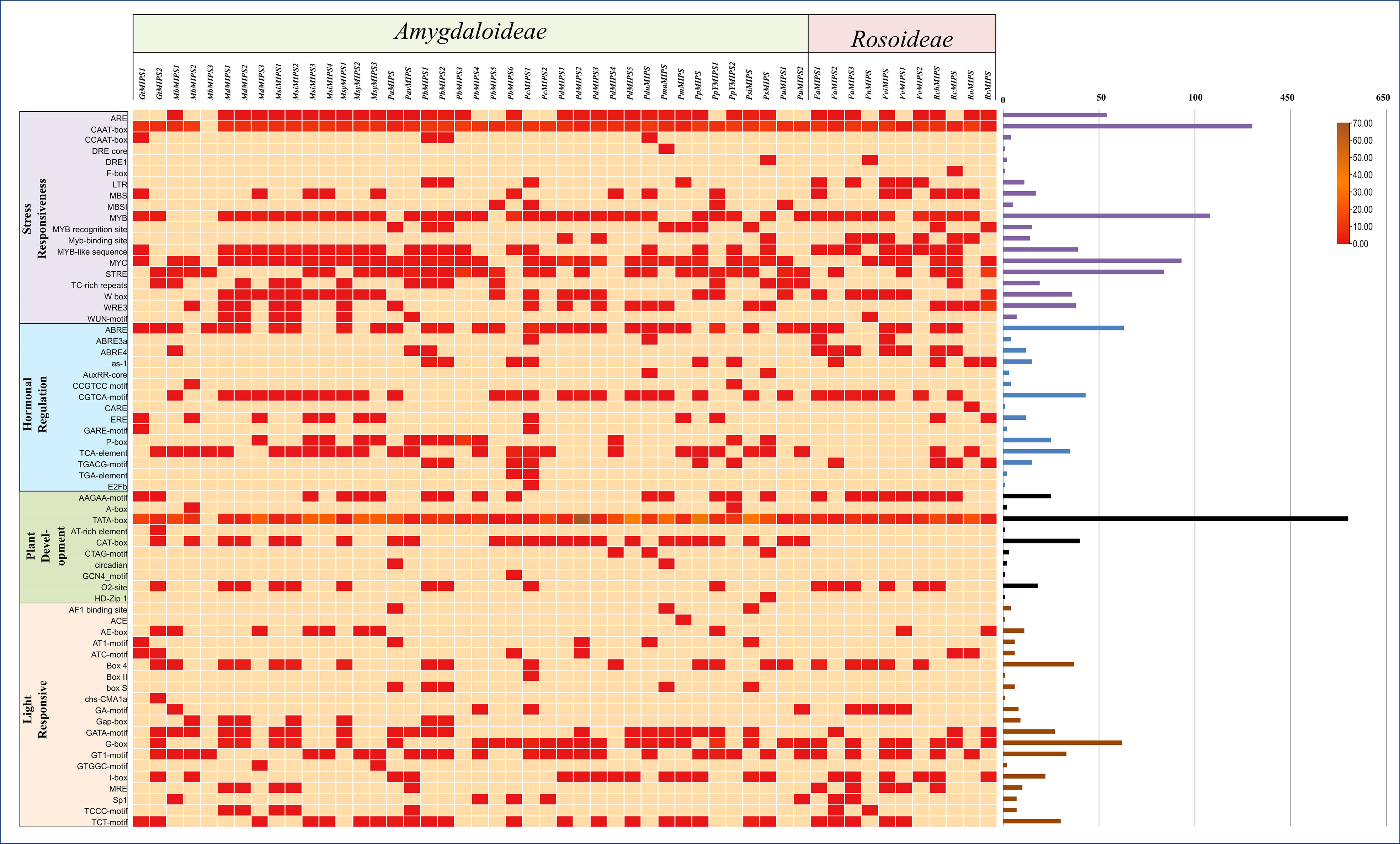
Figure 5 Showing the number of cis-regulatory elements (CREs) belonging to the following four categories (stress-responsive, hormonal regulation, plant development and light-responsive) per Rosaceae MIPS gene as a heatmap. Bar plots on the right represent the number of CREs for each MIPS gene.
Among the CREs of the stress response, drought-responsive CREs (ARE, DRE, MYB, MYC, MBS, F-box and ABRE) were predominantly present in the Rosaceae MIPS gene as compared to other stress-responsive elements. These findings suggest that Rosaceae MIPSs are involved in response to abiotic stresses, particularly drought. The 10 CREs involved in the growth and development of plants were identified in Rosaceae MIPS genes. These include AAGAA-motif, A-Box, TATA-box, AT-rich element, CAT-box, CTAG -motif, circadian, GCN4 motif, O2 site and HD-Zip1. The TATA-box and O2site CREs seemed to be the most prevalent and were detected in almost all MIPS genes. We also notice that three AT-rich element, CAT-box and CTAG -motif CREs found only in the Amygdaloideae subfamily of Rosaceae and absent in the Rosoideae subfamily. The 20 different types of light-responsive CREs were also predicted in Rosaceae MIPS genes. Among them, four CREs (Box 4, GATA-motif, GT1-motif and TCT-motif) seemed to be the most prevalent and appeared in almost all MIPS genes. Plant hormones-related CREs include the abscisic acid-responsive elements (ABRE), methyl jasmonate responsive elements (CGTCA-motif, TGACG-motif), salicylic acid-responsive elements (TCA-element), gibberellins responsive elements (P-box, GARE-motif), ethylene-responsive elements (ERE), and auxin-responsive elements (AuxRR-core, TGA-element). Altogether, these results indicate that MIPS genes may be involved in a variety of abiotic stress responses as well as plant growth regulation.
GO annotation of MIPS gene
To gain more insight into the function of the Rosaceae MIPS genes, we also performed a GO enrichment analysis for the RcMIPS gene. The results of GO enrichment analysis revealed that the RcMIPS gene was involved in the Inositol (phytic acid) biosynthesis, intramolecular lyase activity, Inositol (phytic acid) metabolic process and Polyol biosynthesis process (Figure 6). This GO annotation suggested that the MIPS genes might play an important role in phytic acid biosynthesis.
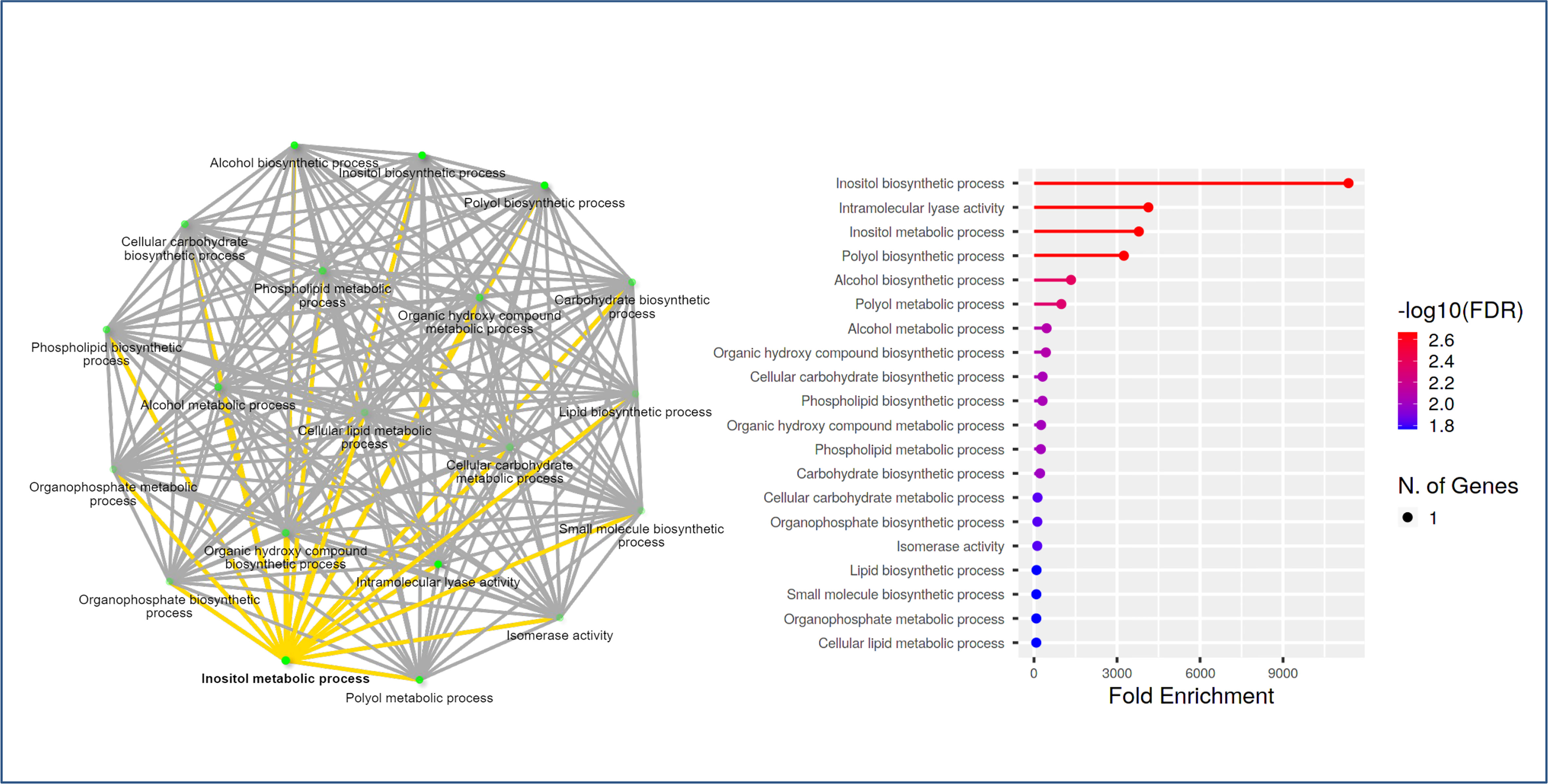
Figure 6 GO analysis using ShinyGo identified enriched biological processes of RcMIPS gene. The green dots represent the nodes for each GO biological process, while the lines (yellow and grey) represent the interaction between the nodes (minimum of 20% genes common between two connected) GO processes.
Expression profile of MIPS gene
To investigate the putative biological role of Rosaceae MIPS genes, we performed expression analysis in Malus domestica (subfamily, Amygdaloideae) and Rosa chinensis (subfamily, Rosoideae). In Malus domestica, the MIPS gene was expressed in the leaf and stem. The expression level of the MIPS gene in both tissues was almost similar; only a slightly higher expression (although non-significant) was observed in the leaf as compared to stem tissue (Figure 7A). In case of rose, a significant difference in the relative expression of RcMIPS gene was observed in different plant tissue (Figure 7A). Among the four tissues, least expression was observed in the stem, however, the flower bud showed maximum expression of RcMIPS (62.66 fold higher than the stem). In the leaf and flower tissues, RcMIPS expressed 54.39 fold and 12.58 fold higher than the stem, respectively.
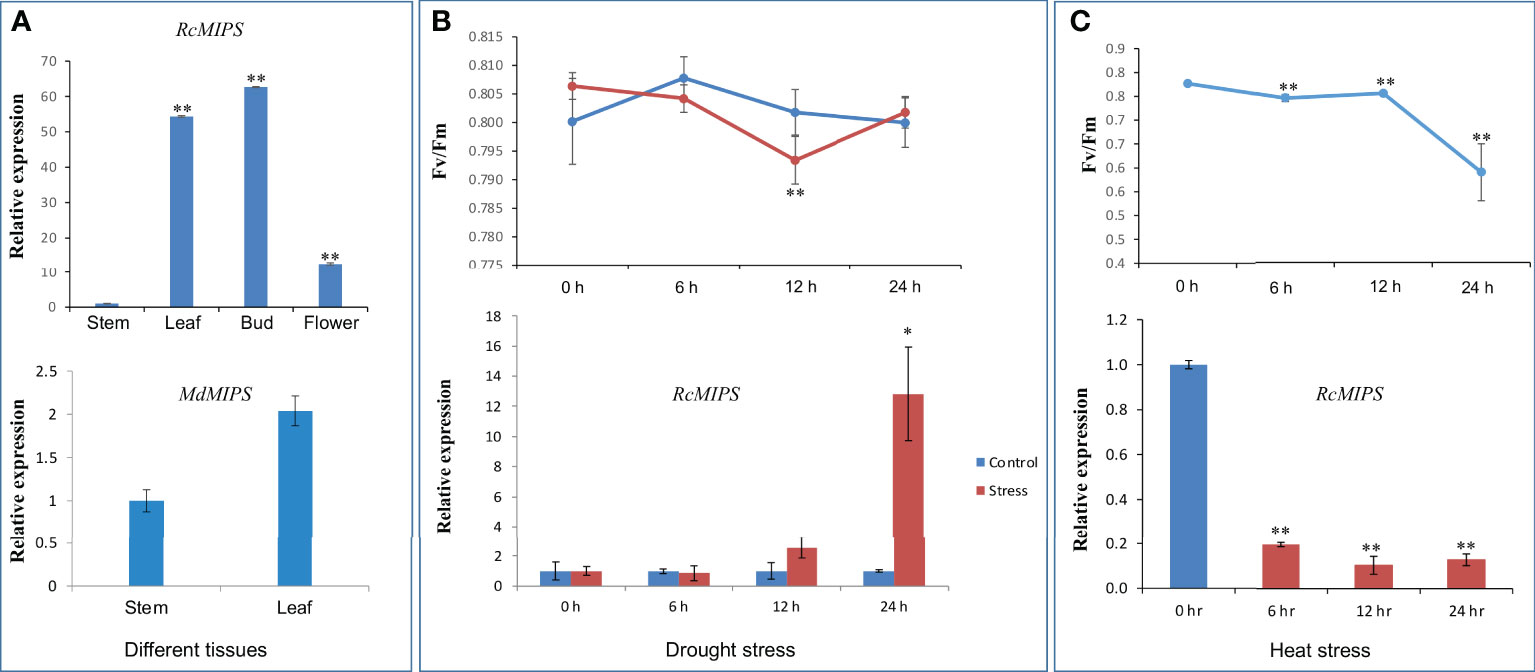
Figure 7 Relative expression level of MIPS in different plant tissue, during drought and heat stress, and effect of drought and heat stresses on quantum yield efficiency (Fv/Fm). (A) Relative expression level of RcMIPS and MdMIPS in different plant tissue. (B) Relative expression level of RcMIPS in leaf tissue at different times (0 h, 6 h, 12 h, and 24 h) treated with 20% PEG and effect of drought stress on quantum yield efficiency (Fv/Fm). (C) Relative expression level of RcMIPS in leaf tissue at different times (0 h, 6 h, 12 h, and 24 h) under heat stress at 42°C and effect of heat stress on quantum yield efficiency (Fv/Fm). * and ** represent the significant difference at p <0.05 and p <0.01, respectively using Student’s t test.
The significant difference of MIPS expression was observed in the case of rose; thus, further experiments were focused only in the rose. We performed drought stress treatment for six to 24 h on R. chinensis plants using 20% PEG-6000, and then we checked the expression of RcMIPS gene using qRT-PCR analysis. Expression analysis identified that under drought stress, RcMIPS gene was significantly upregulated (12.82-fold) as compared to control at only 24 h and before that its expression under drought stress was at par with control (Figure 7B). In the same tissues, we also calculated Fv/Fm to determine the physiological stress. As expected, at 0 h, no significant difference was observed in Fv/Fm values of control and drought stress tissue. Physiological stress was induced at 6 h and enhanced till 12 h, however, at 24 h physiological stress disappeared under drought stress. At 24 h, RcMIPS got upregulated and provided tolerance to plants to cope with stress conditions, and this might be one of the reasons that at 24 h no physiological stress was observed and Fv/Fm values of control tissue and drought stress tissue were almost same (Figure 7B). These results indicate that MIPS is possibly involved in drought stress tolerance in rose and a similar phenomenon might be operational in entire Rosaceae plant species.
We also explored the expression dynamics of RcMIPS under heat stress. For this purpose, plants were exposed to 42°C and leaf samples were collected at 0 h (control) and three-time points under heat stress i.e., 6 h, 12 h and 24 h. Under heat stress conditions, RcMIPS got significantly downregulated (Figure 7C). Further, physiological stress was also prominent in plants as Fv/Fm values significantly dropped under heat stress (Figure 7C).
Discussion
The MIPS is a key enzyme in the myo-inositol biosynthesis pathway and has been involved in multiple biological roles in plants i.e., signal transduction, membrane biogenesis, oligosaccharides synthesis, auxin storage and transport, programmed cell death, phytic acid biosynthesis (Martin, 1998; Roth, 2004; Karner et al., 2004; Abreu and Aragao, 2007; Meng et al., 2009; Ali et al., 2013; Hazra et al., 2019). MIPS genes have been reported in various organisms including, bacteria, fungi, animals, and plants (Abid et al., 2009). However, no systematic identification and characterization of MIPS genes in the Rosaceae family have been reported. Here, by utilizing the reference genome sequence data of 26 Rosaceae species, we have identified 51 MIPS genes in Rosaceae (Table 1). The gene structures, phylogenetic relationships, conserved domain and motifs, CREs, GO annotation and expression profiling during drought and heat stresses for RcMIPS were also analyzed.
The number of MIPS genes among 26 Rosaceae species ranges from one to six and more than 50% of Rosaceae species had a single copy of MIPS genes. In earlier studies conducted in plants, it was observed that algal members, bryophyte (Marchantia), Gymnosperm (Ginkgo), Amborella, and important dicots (Vitis vinifera, Eucalyptus grandis, and Daucus carota) had only one copy of MIPS gene (Hegeman et al., 2001; Abid et al., 2009; Hazra et al., 2019). More than one copy of MIPS (two to four) was also reported in several plant species including rice (two MIPS copies) (Khurana et al., 2012), Arabidopsis (three MIPS copies) (Mitsuhashi et al., 2008), cotton and soybean (four MIPS copies) (Chappell et al., 2006; Ma et al., 2019). These results suggested that MIPSs are highly conserved and expanded and diversified in some higher plants including some Rosaceae species (Gillenia trifoliata, Malus spp., Pyrus spp., Prunus spp., Fragaria spp.) via genome duplication and polyploidization events.
The phylogenetic analysis showed that 51 MIPSs belonging to 26 Rosaceae species were grouped into two distinct clades (Amygdaloideae and Rosoideae subfamily specific) (Figure 1). Interestingly, the MIPSs of 26 Rosaceae species were randomly dispersed in every clade, demonstrating that Rosaceae MIPSs were conserved and existed prior to Rosaceae plant species divergence. Further, most of the Rosaceae MIPS genes had similar exon-intron, motif and domain configurations (Figures 2 & 3). Earlier research has found that several key catalytic domains are highly conserved in MIPS amino acid sequence throughout evolutionary lines, indicating that they are catalytically active (Majumder et al., 2003; Hazra et al., 2019). The four highly conserved amino acid stretches i.e., GWGGNG, LWTANTERY, INGSPQNTFVPG and GIKPLSIASYN present in all 26 Rosaceae plant species (Figure 4) are also highly conserved in other plant species (Majumder et al., 2003; Hazra et al., 2019).
Most of the Rosaceae MIPS have NLS domains (Table S3), which are essential for these proteins’ precise targeting of the nucleus. The sub-cellular localization analysis predicts that Rosaceae MIPS were positioned in the cytoplasm (Table S3). As reported in earlier studies on different plant species MIPS is usually located in the cytoplasm but is also found in the nucleus, plasma membrane and endomembrane (Mitsuhashi et al., 2008; Donahue et al., 2010; Ray et al., 2010; Khurana et al., 2012; Ma et al., 2019). These results suggested that MIPS may be involved in signal transduction, membrane trafficking and regulation of gene expression.
We also predict 64 types CREs in the promoter of Rosaceae MIPS genes and these were associated with abiotic stress-response, plant development, light-responsive and hormonal regulation (Figure 5). In the promoter regions of MIPS genes in various plants, analogous CREs have been found (Khurana et al., 2012; Hazra et al., 2019; Ma et al., 2019). We also observed that drought-responsive CREs (ARE, DRE, MYB, MYC, MBS, F-box and ABRE) were predominantly present in the Rosaceae MIPS gene as compared to other stress-responsive elements. These findings suggest Rosaceae MIPS genes might play role in the growth and development, and stress response regulations in plants.
The MIPSs have been shown to be involved in the regulation of plant growth and development (Hazra et al., 2019). The significant difference (up to 62.66 fold) of RcMIPS in different plant parts like stem, leaf, bud, and flower suggested the important role of this gene in the growth and development of rose plants also (Figure 7A). MIPSs are also reported to play important role in abiotic stress tolerance including drought (Wei et al., 2010; Khurana et al., 2012; Kiranmai et al., 2018) and heat (Khurana et al., 2012). For instance, in groundnut, the MIPS gene was considered one of the drought-responsive genes and upregulated in transgenic lines which were more tolerant against drought as compared to wild type (Kiranmai et al., 2018). Similarly, in the case of other plant species like Jatropha curcas (Wang et al., 2011), Ricinus communis (Wei et al., 2010) Ipomea batata (Zhai et al., 2016) etc. MIPS genes were upregulated under drought stress. In the case of rice, splice variants of the MIPS gene (OsMIPS1) were also upregulated under drought stress (Khurana et al., 2012). Overexpression of chickpea CaMIPS2 gene in Arabidopsis transgenic plants shows improved tolerance to drought stress (Kaur et al., 2013). Ectopic expression of Medicago falcate MIPS (MfMIPS1) gene in tobacco enhanced resistance to drought stress (Tan et al., 2013). During the present study, significant enhancement in expression of MIPS gene was observed under drought stress in the case of rose and suggested the positive role of the MIPS gene in providing tolerance against drought case of rose (Figure 7B). Additionally, we also identified drought-responsive elements CREs (i.e., ABRE, DRE, MYB and MYC) in the promoter sequence of MIPS genes which further suggested the involvement of MIPS in providing tolerance to plants under limited water conditions. Perhaps a similar kind of function may also be expected in the entire Rosaceae family. Further, biochemical studies also found an increase in myo-inositol levels under drought stress in the case of Vigna umbellata (Wanek and Richter, 1997) and Cicer arietinum (Boominathan et al., 2004). Altogether, MIPS genes were found to be a positive regulator which got upregulated under drought stress and increase the synthesis of Myo-inositol and ultimately provide tolerance to plants under drought stress.
Under heat stress, the expression of MIPS was reported to be different in different tissues. In the case of wheat, TaMIPS got upregulated in root and shoot tissues, however, downregulated in flower tissues under heat stress (Khurana et al., 2012). During the present study, RcMIPS got downregulated under heat stress and consequently, physiological stress (in form of Fv/Fm) was also observed (Figure 7C). These results may be explained by the fact that the genotype used in the present study was R. chinensis, and R. chinensis is a heat-susceptible species (Qi et al., 2021). Further, in R. chinensis, RNA seq study also suggested that genes involved in carbohydrate biosynthesis and cell wall biosynthesis genes were downregulated and resulted into physiological stress in R. chinensis (Li et al., 2019). All these observations suggested that allele of the RcMIPS gene in R. chinensis might be a susceptible allele for heat stress which got downregulated under heat stress and resulted in physiological stress in plants. However, the situation might be reversible with heat tolerant rose accession where MIPS genes (with tolerant allele) perhaps provide tolerance to plants under heat stress.
In conclusion, we identified 51 MIPS genes in 26 Rosaceae species of plants which were grouped into two clades. Gene structure analysis suggested one to nine exons in MIPS and the total gene length of MIPS were found up to 4079 bp. In promoter sequence, the presence of different elements including abiotic stress-responsive elements suggested the important role of MIPSs in different plant developmental processes as well as abiotic stresses. Expression analysis of RcMIPS empathized the potential role of MIPS in plant development, drought and heat stress tolerance. Altogether, the present study provides compressive preliminary information on MIPS genes which can be a good foundation for functional characterization of this important gene family.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HG and PK: Performed experiments, analysed data and wrote the first draft. VG and VJ: Conceptualization, supervision, writing and editing. SK: Overall supervision and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the CSIR, India under Grant MLP201; the Department of Science and Technology (DST) for the INSPIRE faculty award; and Science and Engineering Research Board (SERB) for the Early Career Research Award.
Acknowledgments
This is a short text to acknowledge the contributions of specific colleagues, institutions, or agencies that aided the efforts of the authors. This study was supported by the Council of Scientific and Industrial Research (CSIR) for providing funds (MLP-201), VJ also thanks to the Science and Engineering Research Board (SERB) for the Early Career Research Award. VJ and VG thank the Department of Science and Technology (DST) for the INSPIRE faculty award. HG and PK thank CSIR for Junior Research Fellowship. This manuscript represents CSIR-IHBT communication number 5181.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1021297/full#supplementary-material
References
Abid, G., Silue, S., Muhovski, Y., Jacquemin, J. M., Toussaint, A., Baudoin, J. P. (2009). Role of myo-inositol phosphate synthase and sucrose synthase genes in plant seed development. Gene 439, 1–10. doi: 10.1016/j.gene.2009.03.007
Abreu, E. F. M., Aragao, F. J. L. (2007). Isolation and characterization of a myo-inositol-1-phosphate synthase gene from yellow passion fruit (Passifloraedulis f. flavicarpa) expressed during seed development and environmental stress. Ann. Bot. 99, 285–292. doi: 10.1093/aob/mcl256
Ali, N., Paul, S., Gayen, D., Sarkar, S. N., Datta, K., Datta, S. K. (2013). Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1, 3, 4, 5, 6-pentakisphosphate 2-kinase gene (IPK1). PLos One 8, e68161. doi: 10.1371/journal.pone.0068161
Alok, A., Singh, S., Kumar, P., Bhati, K. K. (2022). Potential of engineering the myo-inositol oxidation pathway to increase stress resilience in plants. Mol. Biol. Rep. 49, 8025–8035. doi: 10.1007/s11033-022-07333-0
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, 597–603. doi: 10.1093/nar/gks400
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, 39–49. doi: 10.1093/nar/gkv416
Basak, P., Sangma, S., Mukherjee, A., Agarwal, T., Sengupta, S., Ray, S., et al. (2018). Functional characterization of two myo-inositol-1-phosphate synthase (MIPS) gene promoters from the halophytic wild rice (Porteresia coarctata). Planta 248, 1121–1141. doi: 10.1007/s00425-018-2957-z
Boominathan, P., Shukla, R., Kumar, A., Manna, D., Negi, D., Verma, P. K., et al. (2004). Long term transcript accumulation during the development of dehydration adaptation in Cicer arietinum. Plant Physiol. 135, 1608–1620. doi: 10.1104/pp.104.043141
Chappell, A. S., Scaboo, A. M., Wu, X., Nguyen, H., Pantalone, V. R., Bilyeu, K. D. (2006). Characterization of the MIPS gene family in Glycine max. Plant Breed. 125, 493–500. doi: 10.1111/j.1439-0523.2006.01264.x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chun, J. A., Jin, U. H., Lee, J. W., Yi, Y. B., Hyung, N. I., Kang, M. H., et al. (2003). Isolation and characterization of a myo-inositol 1-phosphate synthase cDNA from developing sesame (Sesamum indicum l.) seeds: functional and differential expression, and salt-induced transcription during germination. Planta 216, 874–880. doi: 10.1007/s00425-002-0940-0
Donahue, J. L., Alford, S. R., Torabinejad, J., Kerwin, R. E., Nourbakhsh, A., Ray, W. K., et al. (2010). The arabidopsis thaliana myo-inositol 1-phosphate synthase1 gene is required for myo-inositol synthesis and suppression of cell death. Plant Cell 22, 888–903. doi: 10.1105/tpc.109.071779
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The pfam protein families database in 2019. Nucleic Acids Res. 47, 427–432. doi: 10.1093/nar/gky995
Feng, X., Yoshida, K. T. (2004). Molecular approaches for producing low-phytic-acid grains in rice. Plant Biotechnol. J. 21, 183–189. doi: 10.5511/plantbiotechnology.21.183
Ge, S. X., Jung, D., Yao, R. (2020). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. doi: 10.1093/bioinformatics/btz931
Hazra, A., Dasgupta, N., Sengupta, C., Das, S. (2019). MIPS: Functional dynamics in evolutionary pathways of plant kingdom. Genomics 111, 1929–1945. doi: 10.1016/j.ygeno.2019.01.004
Hegeman, C. E., Good, L. L., Grabau, E. A. (2001). Expression of d-myo-inositol-3-phosphate synthase in soybean. implications for phytic acid biosynthesis. Plant Physiol. 125, 1941–1948. doi: 10.1104/pp.125.4.1941
Hu, L. Y., Hong, Y. U. E., Zhang, J. Y., Yang-tian-su, L. I., Gong, X. Q., Kun, Z. H. O. U., et al. (2022). Overexpression of MdMIPS1 enhances drought tolerance and water-use efficiency in apple. J. Integr. Agric. 21, 1968–1981. doi: 10.1016/S2095-3119(21)63822-4
Johnson, M. D., Sussex, I. M. (1995). 1 l-myo-inositol 1-phosphate synthase from Arabidopsis thaliana. Plant Physiol. 107, 613–619. doi: 10.1104/pp.107.2.613
Jung, S., Lee, T., Cheng, C. H. (2019). 15 years of GDR: New data and functionality in the genome database for rosaceae. Nucleic Acids Res. 47, 1137–1145. doi: 10.1093/nar/gky1000
Karner, U., Peterbauer, T., Raboy, V., Jones, D. A., Hedley, C. L., Richter, A. (2004). myo-inositol and sucrose concentrations affect the accumulation of raffinose family oligosaccharides in seeds. J. Exp. Bot. 55, 1981–1987. doi: 10.1093/jxb/erh216
Kaur, H., Verma, P., Petla, B. P., Rao, V., Saxena, S. C., Majee, M. (2013). Ectopic expression of the ABA-inducible dehydration-responsive chickpea l-myo-inositol 1-phosphate synthase 2 (CaMIPS2) in arabidopsis enhances tolerance to salinity and dehydration stress. Planta 237, 321–335. doi: 10.1007/s00425-012-1781-0
Khurana, N., Chauhan, H., Khurana, P. (2012). Expression analysis of a heat-inducible, myo-inositol-1-phosphate synthase (MIPS) gene from wheat and the alternatively spliced variants of rice and Arabidopsis. Plant Cell Rep. 31, 237–251. doi: 10.1007/s00299-011-1160-5
Kiranmai, K., Rao, G. L., Pandurangaiah, M., Kumar, A. M., Reddy, V. A., Lokesh, U., et al. (2018). A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum lam. verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea l.) plants. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00346
Kosugi, S., Hasebe, M., Tomita, M., Yanagawa, H. (2009). Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. 106, 10171–10176. doi: 10.1073/pnas.090060410
Kumari, S., Jolly, M., Krishnan, V., Dahuja, A., Sachdev, A. (2012). Spatial and temporal expression analysis of d-myo-inositol 3-phosphate synthase (MIPS) gene family in Glycine max. Afr. J. Biotechnol. 11, 16443–16454. doi: 10.5897/AJB12.2811
Larson, S. R., Raboy, V. (1999). Linkage mapping of maize and barley myo-inositol 1-phosphate synthase DNA sequences: correspondence with a low phytic acid mutation. Theor. Appl. Genet. 99, 27–36. doi: 10.1007/s001220051205
Lescot, M., Patrice, D., Thijs, G., Marchal, K., Moreau, Y., de Peer, Y. V., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2007). Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128. doi: 10.1093/bioinformatics/btl529
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li, Z. Q., Xing, W., Luo, P., Zhang, F. J., Jin, X. L., Zhang, M. H. (2019). Comparative transcriptome analysis of Rosa chinensis ‘Slater's crimson china’ provides insights into the crucial factors and signaling pathways in heat stress response. Plant Physiol. Biochem. 142, 312–331. doi: 10.1016/j.plaphy.2019.07.002
Loewus, F. A., Loewus, M. W. (1980). “Myo-inositol: biosynthesis and metabolism,” in The biochemistry of plants: Carbohydrate structure and function. Ed. Preiss, J. (New York: Academic Press), 43–76.
Majumder, A. L., Chatterjee, A., Dastidar, K. G., Majee, M. (2003). Diversification and evolution of l-myo-inositol 1-phosphate synthase. FEBS Lett. 553, 3–10. doi: 10.1016/S0014-5793(03)00974-8
Majumder, A., Johnson, M. D., Henry, S. A. (1997). 1L-myo-inositol-1-phosphate synthase. Biochim. Biophys. Acta 1348, 245–256. doi: 10.1016/S0005-2760(97)00122-7
Martin, T. F. J. (1998). Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu. Rev. Cell Dev. Biol. 14, 231–264. doi: 10.1146/annurev.cellbio.14.1.231
Ma, R., Song, W., Wang, F., Cao, A., Xie, S., Chen, X., et al. (2019). A cotton (Gossypium hirsutum) myo-inositol-1-phosphate synthase (GhMIPS1D) gene promotes root cell elongation in arabidopsis. Int. J. Mol. Sci. 20, 1224. doi: 10.3390/ijms20051224
Meng, P. H., Raynaud, C., Tcherkez, G., Blanchet, S., Massoud, K., Domenichini, S., et al. (2009). Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in arabidopsis. PLos One 4, e7364. doi: 10.1371/journal.pone.0007364
Menzel, W., Jelkmann, W., Maiss, E. (2002). Detection of four apple viruses by multiplex RT-PCR assays with coamplification of plant mRNA as internal control. J. Virol. Methods 99, 81–92. doi: 10.1016/S0166-0934(01)00381-0
Mitsuhashi, N., Kondo, M., Nakaune, S., Ohnishi, M., Hayashi, M., Nishimura, I. H., et al. (2008). Localization of myo-inositol-1-phosphate synthase to the endosperm in developing seeds of arabidopsis. J. Exp. Bot. 59, 3069–3076. doi: 10.1093/jxb/ern161
Qin, D., Toyonaga, D., Saneoka, H. (2021). Characterization of myo-inositol-1-phosphate synthase (MIPS) gene expression and phytic acid accumulation in oat (Avena sativa) during seed development. Cereal Res. Commun 50, 379–384. doi: 10.1007/s42976-021-00186-6
Qi, W., Zhang, C., Wang, W., Cao, Z., Li, S., Li, H., et al. (2021). Comparative transcriptome analysis of different heat stress responses between self-root grafting line and heterogeneous grafting line in rose. Hortic. Plant J. 7, 243–255. doi: 10.1016/j.hpj.2021.03.004
Raychaudhuri, A., Majumder, A. L. (1996). Salinity-induced enhancement of l-myo-inositol 1-phosphate synthase in rice (Oryza sativa l.). Plant Cell Environ. 19, 1437–1442. doi: 10.1111/j.1365-3040.1996.tb00023.x
Ray, S., Patra, B., Das-Chatterjee, A., Ganguli, A., Majumder, A. (2010). Identification and organization of chloroplastic and cytosolic l-myo-inositol 1-phosphate synthase coding gene (s) in Oryza sativa: comparison with the wild halophytic rice, Porteresia coarctata. Planta 231, 1211–1227. doi: 10.1007/s00425-010-1127-8
Roth, M. G. (2004). Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 84, 699–730. doi: 10.1152/physrev.00033.2003
Sharma, N., Chaudhary, C., Khurana, P. (2020). Wheat myo-inositol phosphate synthase influences plant growth and stress responses via ethylene mediated signaling. Sci. Rep. 10, 1–14. doi: 10.1038/s41598-020-67627-w
Stecher, G., Tamura, K., Kumar, S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evo. 37, 1237–1239. doi: 10.1093/molbev/msz312
Taji, T., Takahashi, S., Shinozaki, K. (2006). Inositols and their metabolites in abiotic and biotic stress responses. Subcell. Biochem. 39, 239–264. doi: 10.1007/0-387-27600-9_10
Tan, J., Wang, C., Xiang, B., Han, R., Guo, Z. (2013). Hydrogen peroxide and nitric oxide mediated cold-and dehydration-induced myo-inositol phosphate synthase that confers multiple resistances to abiotic stresses. Plant Cell Environ. 36, 288–299. doi: 10.1111/j.1365-3040.2012.02573.x
Wanek, W., Richter, A. (1997). Biosynthesis and accumulation of d-ononitol in Vigna umbellata in response to drought stress. Physiol. Plant 101, 416–424. doi: 10.1111/j.1399-3054.1997.tb01016.x
Wang, Y., Huang, J., Gou, C. B., Dai, X., Chen, F., Wei, W. (2011). Cloning and characterization of a differentially expressed cDNA encoding myo-inositol-1-phosphate synthase involved in response to abiotic stress in Jatropha curcas. Plant Cell Tissue Organ Cult. 106, 269–277. doi: 10.1007/s11240-011-9917-7
Wei, W., Dai, X., Wang, Y., Gou, C. B., Chen, F. (2010). Cloning and expression analysis of 1 l-myo-inositol-1-phosphate synthase gene from Ricinus communis l. Z. Naturforsch. C. 65, 501–507. doi: 10.1515/znc-2010-7-814
Keywords: myo-inositol 1-phosphate synthase, phylogenetic analysis, gene structure, cis-elements, drought stress
Citation: Gangwar H, Kumari P, Gahlaut V, Kumar S and Jaiswal V (2022) Identification and comprehensive analysis of MIPSs in Rosaceae and their expression under abiotic stresses in rose (Rosa chinensis). Front. Plant Sci. 13:1021297. doi: 10.3389/fpls.2022.1021297
Received: 17 August 2022; Accepted: 20 October 2022;
Published: 03 November 2022.
Edited by:
Mohammad Irfan, Cornell University, United StatesCopyright © 2022 Gangwar, Kumari, Gahlaut, Kumar and Jaiswal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vijay Gahlaut, em9uZTR2aWpheUBnbWFpbC5jb20=; Vandana Jaiswal, dmFuZGFuYUBpaGJ0LnJlcy5pbg==
†These authors have contributed equally to this work and share first authorship
 Himanshi Gangwar
Himanshi Gangwar Priya Kumari
Priya Kumari Vijay Gahlaut
Vijay Gahlaut Sanjay Kumar
Sanjay Kumar Vandana Jaiswal
Vandana Jaiswal