- 1Department of Agronomy, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 2Key Laboratory for Tree Breeding and Germplasm Improvement, Southern Modern Forestry Collaborative Innovation Center, College of Forestry, Nanjing Forestry University, Nanjing, China
- 3College of Biotechnology, Jiangsu University of Science and Technology, Zhenjiang, China
- 4Hainan Institute of Zhejiang University, Sanya, China
A virus-induced gene silencing (VIGS) system was established to induce endogenous target gene silencing by post-transcriptional gene silencing (PTGS), which is a powerful tool for gene function analysis in plants. Compared with stable transgenic plant via Agrobacterium-mediated gene transformation, phenotypes after gene knockdown can be obtained rapidly, effectively, and high-throughput through VIGS system. This approach has been successfully applied to explore unknown gene functions involved in plant growth and development, physiological metabolism, and biotic and abiotic stresses in various plants. In this system, GhCLA1 was used as a general control, however, silencing of this gene leads to leaf albino, wilting, and plant death ultimately. As such, it cannot indicate the efficiency of target gene silencing throughout the whole plant growth period. To address this question, in this study, we developed a novel marker gene, Gossypium PIGMENT GLAND FORMATION GENE (GoPGF), as the control to trace the efficiency of gene silencing in the infected tissues. GoPGF has been proved a key gene in gland forming. Suppression of GoPGF does not affect the normal growth and development of cotton. The number of gland altered related to the expression level of GoPGF gene. So it is a good marker that be used to trace the whole growth stages of plant. Moreover, we further developed a method of friction inoculation to enhance and extend the efficiency of VIGS, which facilitates the analysis of gene function in both the vegetative stage and reproductive stage. This improved VIGS technology will be a powerful tool for the rapid functional identification of unknown genes in genomes.
Introduction
As a kind of hereditary immune mechanism prevalent in plants, virus-induced gene silencing (VIGS) belongs to post-transcriptional gene silencing (PTGS) (Lu et al., 2003). When carrying a vector with endogenous gene fragments of the host plant infection, VIGS can activate the plant’s own immune system. Plants recognize and degrade viral RNA, and also produce microRNAs that contain endogenous genes of interest. Through binding to the mRNAs of the target genes, these microRNAs can form a hairpin structure which is then degraded by the Dicer enzyme, leading to a decrease in the expression level of the gene of interest or loss of function (Senthil-Kumar and Mysore, 2011).
As one of the most widely used gene silencing systems, the tobacco rattle virus-induced gene silencing system (TRV-VIGS) has the advantages of high silencing efficiency, long duration, light symptoms of the host plant viruses, and no cover of the phenotype in various organizations that produce gene silencing. Above all, as it does not require the development of stable transformants, it is very suitable for characterizing gene function in plants with low transformation efficiency. Therefore, due to its validity and rapidity, TRV-VIGS has been successfully used in various plants, such as Nicotiana tabacum, Solanum lycopersicum, Gossypium hirsutum, Arabidopsis, Capsicum annuum, etc. (Tian et al., 2014).
The VIGS method utilizes the Cloroplastos alterados 1 gene (CLA1) as a common positive control, which results in photobleaching of the silenced regions (Gao et al., 2011a). Recently, VIGS technology has been used extensively to study the gene function of cotton, due to its discrete characters verification, such as Red plant (R1) (Cai et al., 2014), Okra leaf (L20) (Chang et al., 2016; Andres et al., 2017), Glandless (Gl2e) (Ma et al., 2016), Ligon-lintless (Li1) (Thyssen et al., 2017), Naked seed (N1) (Wan et al., 2016), Lint fiber development (Li3) (Wu et al., 2018), Axillary flowering (GbAF) (Si et al., 2018), Coiled-coil nucleotide-binding site leucine-rich repeat (CC-NBS-LRR) (Deng et al., 2019), APETALA 2/ethylene-responsive factor (AP2/ERF) (Pei et al., 2021), UBX-Domain Containing10 (Zang et al., 2021a) and novel stem pigment gland-forming gene (GoSPGF) (Zang et al., 2021b). There are many studies on the analysis of gene function in biotic and abiotic resistances including disease (Gao et al., 2011b), drought (Chen et al., 2015) and low temperature (Liu et al., 2015). However, after the whitening of the individual plants, the treated plants grow slowly and weakly compared to normal plants, due to their inability to complete the photosynthesis process. Finally, after the albino plant grows for about 2 months, it gradually withers (Figure 1). Therefore, this positive control is suitable only for identifying the phenotype at the seedling stage, and is not conducive to the study of the phenotype at the middle or late growth stage. Here, we report on a novel positive control gene GoPGF (Ma et al., 2016), which could constantly trace virus-induced gene silencing visually throughout the entire range of plant growth stages. Thus, it is suitable for studying traits such as buds, flowering, and fiber.
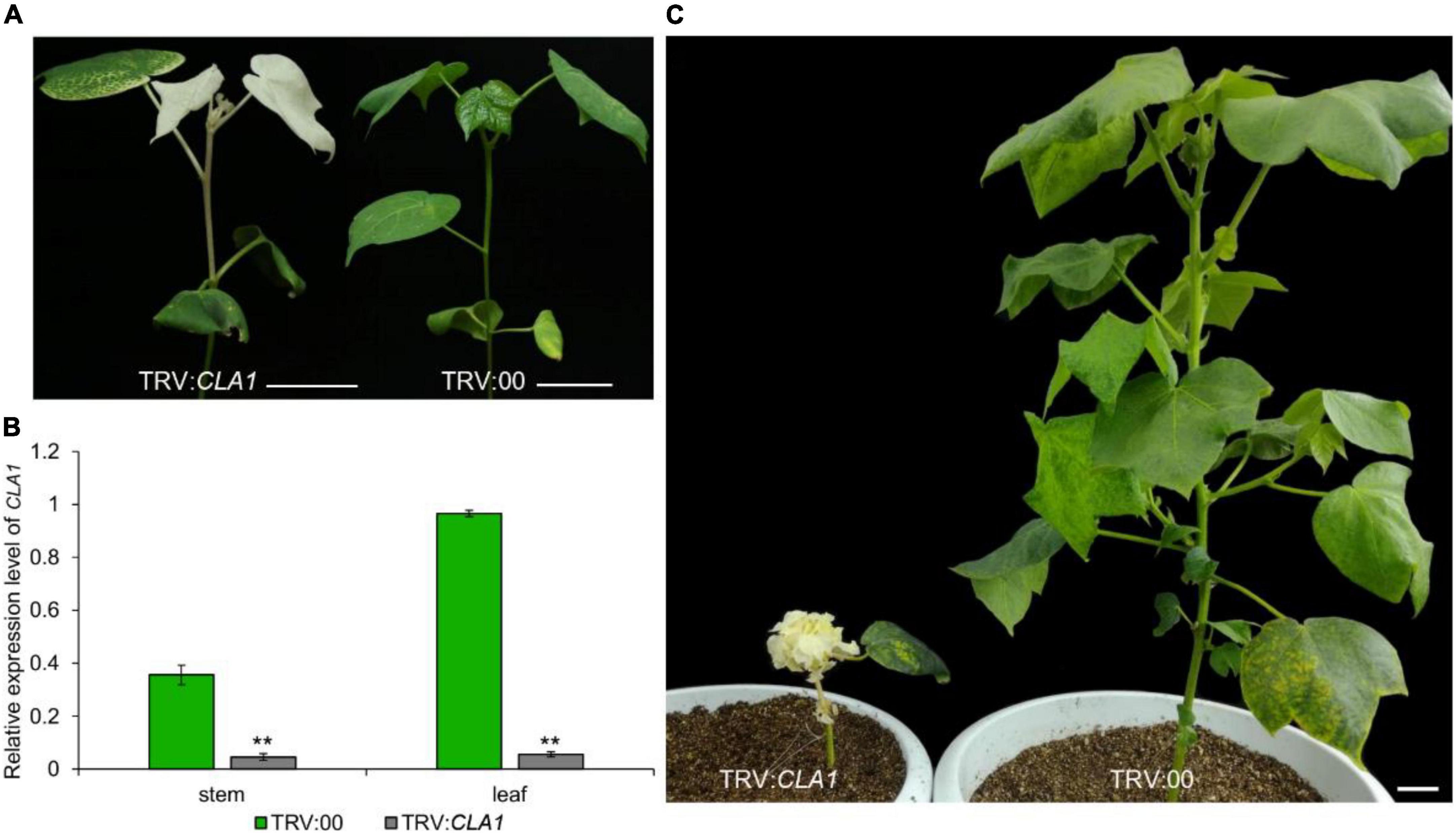
Figure 1. The gene expression and phenotype of the CLA1- silenced plants. (A) After 2 weeks of silencing the CLA1 gene, the leaves showed an albino phenotype. (B) After 2 weeks of silencing the CLA1 gene, the expression of genes in young leaves and young stems of albino and empty control plants were analyzed. (C) The phenotype of the plant was observed after silencing the CLA1 gene for 2 months. Error bars are s.d. of three biological repeats. **P < 0.01; Student’s t-test, n = 3. Bars = 5 cm.
Materials and methods
Plant materials
The materials used in the experiments were TM-1 with glands. TM-1 is a standard genetic line of Upland cotton and has broad leaves. The seeds were germinated and grown in a greenhouse. The seedlings from the cotyledon expansion to 4th true leaf emergence were used for the VIGS assays. Infiltrated plants were grown in a growth chamber at 23°C with a 16/8 h light/dark photoperiod. N. benthamiana was grown at 25°C in a semi-controlled walk-in chamber with a 16/8 h light to dark photoperiod for the generation of transgenic plants or agroinfiltration.
Recombinant virus-induced gene silencing vectors
The pTRV1, pTRV-CLA1, and pTRV-GoPGF vectors were stored in our laboratory. The three vectors were transformed into Agrobacterium tumefaciens strains GV3101.
Agrobacterium infiltration
pTRV1, pTRV-CLA1, and pTRV-GoPGF were grown overnight at 28°C in an antibiotic selection medium containing rifampicin and kanamycin at a concentration of 50 mg l–1. Agrobacterium cells were collected and resuspended in an infiltration medium (10 mM MgCl2, 10 mM MES and 200 μM acetosyringone), and adjusted OD600 to OD600 1.5. When silencing GoPGF gene separately Agrobacterium strains to contain the TRV1 and TRV-GoPGF vectors were mixed at a ratio of 1:1. While silencing the CLA1 and GoPGF genes at the same time, in the case of the Agrobacterium strains carrying pTRV-CLA1, pTRV- GoPGF, and pTRV1 vectors were mixed at a ratio of 1: 1: 2 and then injected into the cotton cotyledons. The treated cotton seedlings were grown in a growth chamber at 23°C with a 16/8 h light/dark photoperiod.
Friction inoculation in the virus-induced gene silencing system
The tobacco (N. benthamiana) grows to 6 leaves in a greenhouse at 25°C in a semi-controlled walk-in chamber with a 16/8 h light to dark photoperiod. TRV1 and TRV-CLA1 were grown overnight at 28°C in a LB medium containing antibiotics (50 μg/ml kanamycin, 25 μg/ml gentamicin), 10 mM MES and 20 μM acetosyringone. The cells were pelleted by centrifugation at 1,200 × g at room temperature for 8 min, and resuspended in an MMA (10 mM MES, 10 mM MgCl2, 200 μM acetosyringone) solution to the final OD600 of 1.5. Cell suspensions were incubated at room temperature for at least 3 h without shaking. The Agrobacterium strains containing the TRV1 and pTRV-CLA1 vectors were mixed at a ratio of 1:1 and infiltrated into tobacco leaves using a needleless syringe. The treated tobacco was grown in a greenhouse at 23°C with a 16/8 h light/dark photoperiod.
Tobacco was placed in the greenhouse after 2 weeks of growth, with the new leaves showing white spots. In the mortar, white-spotted tobacco leaves were added, along with a little 400 mesh quartz sand and 10 ml 0.01M phosphate buffer solution (PBS). These were then ground into a homogenate.
In the cotton plant, only the completely unfolded leaves were chosen, and a small quantity of 400 mesh quartz sand was sprinkled over the above mixture. A pestle was used to divert the prepared homogenate to gently rub over the selected cotton leaves. Friction control was implemented in order to penetrate the leaves, so that the leaves would be damaged to the epidermis. The purpose was intended to invade cotton leaves with the tobacco containing virus. The treated cotton was placed in a greenhouse at 23°C with a 16/8 h light/dark photoperiod. After 2–3 weeks, the new growing cotton leaves displayed whitening phenotypes.
RNA extraction and quantitative real-time PCR analysis
RNA was extracted from the stem, leaf, bract, and calyculus of TM-1 using the Biospin Plant Total RNA Extraction Kit (Hangzhou Bori Technology Co. Ltd, Hangzhou, China, cat: BSC65S1). First-strand cDNA was generated using a TransScript® One-Step gDNA Removal Kit and a cDNA Synthesis SuperMix (TransGen Biotech Co. Ltd, Beijing China, cat: AT311) according to the manufacturer’s instructions. The cotton Histone 3 gene (AF024716) was used as the housekeeping gene. The primers’ sequences were listed in Supplementary Table 1.
Results
The number of glands on the plants reflects the silencing efficiency of virus-induced gene silencing
The Gossypium species are characterized by the presence of small and darkly pigmented lysigenous glands on the surface of almost all the organs (Figure 2A and Supplementary Figure 1). Using a dominant glandless mutant Hai-1, we cloned the pigment gland formation regulator GoPGF which encodes a basic helix-loop-helix (bHLH) transcription factor (Ma et al., 2016). It is expressed in a constitutive manner in most organs and tissues including the stem, leaf, bract, etc. (Figure 2B), consistent with the presence of glands (Ma et al., 2016). GoPGF was proven to be specific to gland development and had no noticeable effect on plant growth and development (Gao et al., 2022). The silencing of GoPGF can lead to a complete glandless phenotype. However, we observed that there was a noticeable difference in the silencing efficiency in different VIGS-treated plants even though this applied to the same receptor material and was implemented with the same silencing vector injection. For example, a VIGS-silenced GoPGF plant displayed different gland phenotypes using the cotton genetic standard line TM-1 as the receptor. Some GoPGF-silenced plants did not develop glands at all, suggesting that GoPGF is completely silenced, while some plants showed reduced glands (Figure 2C). The relative expression of GoPGF gene is also down-regulated with a decrease in the number of glands (Figure 2D). Thus, the GoPGF gene can be utilized as a positive control and internal indicator to trace the silencing efficiency of the target gene through phenotype observation of the absence or presence of the gland and variation of gland numbers throughout the entire VIGS stage.
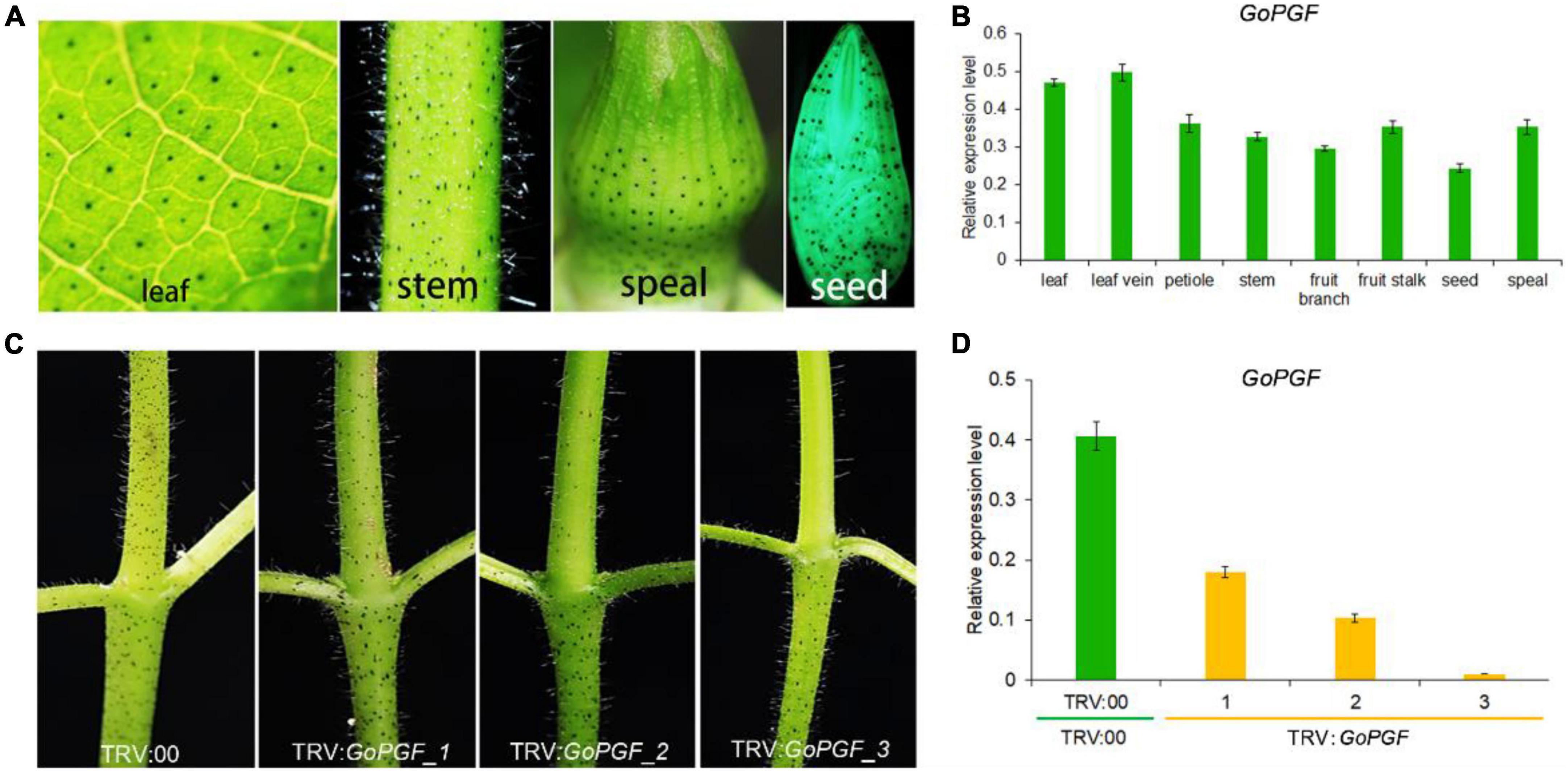
Figure 2. Gland phenotypes and gene expression level in different tissues of GoPGF-silenced plants. (A) Distribution of glandular phenotype in different tissues of cotton including leaf, stem, bud and seed. (B) Relative expression of the GoPGF gene in different tissues of cotton. Error bars are s.d. of three biological repeats. (C) Comparison of silencing efficiency in different VIGS plants. (D) The relative expression of the GoPGF gene was analyzed in the stems of different VIGS plants in panel (C). Error bars are s.d. of three biological repeats.
Assessment of degree and specificity of virus-induced endogenous GoPGF gene silencing in cotton
The pTRV-GoPGF silencing vector was constructed and injected into TM-1 to observe the glandular phenotype during the completed sequence of developmental stages. Silencing of the GoPGF gene resulted in the glandless phenotype in the new leaves and stems after 2–3 weeks of inoculation, while plants inoculated with empty vectors and the control plants displayed a glanded phenotype (Supplementary Figures 2A,B). qPCR analysis revealed that the transcripts of the GoPGF gene were significantly decreased in the glandless plants (Supplementary Figure 2). At the flowering stage, some GoPGF-silenced plants began to develop a few glands only in the 5-day-old young boll, but no glands were found in the top, middle and lower parts of the stems, the penultimate leaves and the sepals (Figure 3). The expression of the GoPGF gene in the six examined tissues was significantly lower than that of the empty vector control (Supplementary Figure 3). Therefore, the silencing efficiency of the endogenous gene can be maintained till the flowering stage. Moreover, when the GoPGF gene served as the internal control, the silencing efficiency of the target gene in the plant was directly reflected by the existence and the number variation of glands.

Figure 3. Comparison of the glandular phenotype of different tissues from GoPGF-VIGS plants and control at flowering stage. The tissues marked in the white box showed an enlargement of corresponding tissues including leaf, stem, bud and boll from the TRV:00 and TRV:PGF whole plants, respectively. The boll was collected 5 days after flowering.
GoPGF can be considered as an internal reference gene in a cotton virus-induced gene silencing system
According to previous reports, two target gene fragments can be constructed on one TRV vector to achieve the purpose of simultaneously silencing two genes (Pang et al., 2013). In this study, a mixed VIGS system was designed by equally mixing Agrobacterium containing TRV vectors of different target genes. We utilized the silencing of the CLA1 gene in order to produce leaf albino and silenced the GoPGF gene in order to create glandless traits to validate the above ideas. Agrobacterium strains carrying pTRV-CLA1, pTRV-GoPGF and pTRV1 vectors were mixed at the ratio of 1: 1: 2 and then injected into cotton cotyledons. At the same time, the Agrobacterium carrying pTRV-CLA1 and pTRV-GoPGF vectors were, respectively injected into the cotton cotyledons, and the TRV2 empty vector was adopted as the negative control. Two weeks later, the phenotype of the treated plants was observed. The RNA was extracted from the penultimate leaves of cotton plants, and the expression patterns of the CLA1 and GoPGF genes were analyzed. The plants injected with pTRV-CLA1 had an albino phenotype, while those plants injected with pTRV-GoPGF exhibited a glandless phenotype (Figure 4A). Meanwhile, the plants injected with pTRV-CLA1 mixed with pTRV-GoPGF exhibited the white and glandless phenotype simultaneously (Figure 4A). The phenotype of negative control did not change at all (Figure 4A). Expression pattern analysis confirmed that the expression of CLA1 and GoPGF was strongly inhibited when silencing these two genes independently (Figures 4B,C). By silencing pTRV-CLA1 and pTRV-GoPGF at the same time, the expression of both genes was also completely inhibited (Figures 4B,C). Moreover, it is completely feasible to determine the efficiency of the target gene silencing method using the GoPGF gene as an internal reference gene by alteration of the gland number.
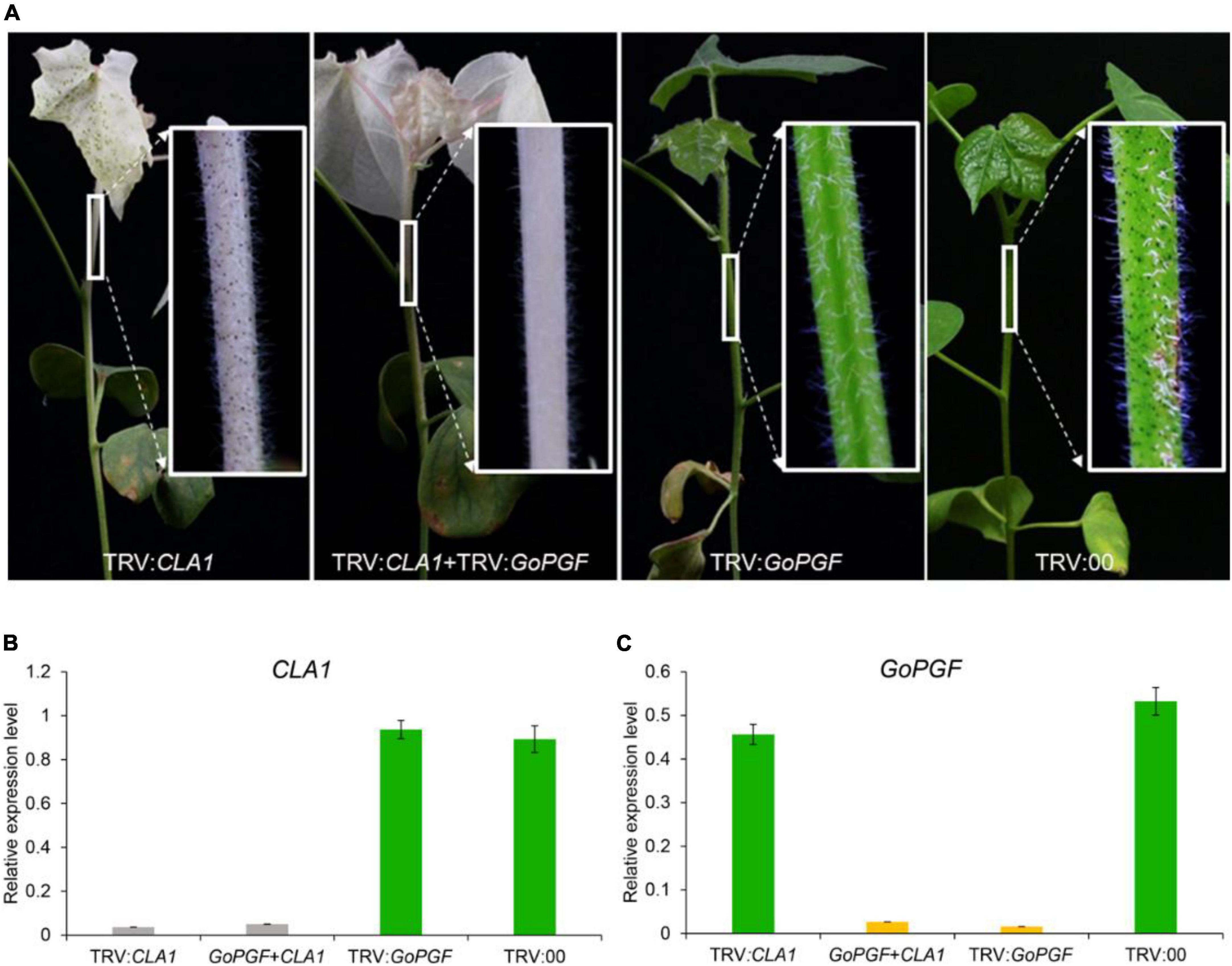
Figure 4. Phenotype and gene expression level of VIGS plants silenced by various combinations of the CLA1 and GoPGF gene. (A) Phenotype of VIGS plants silenced by CLA1 or GoPGF gene and both CLA1–silenced plants display a typical albino phenotype; the GoPGF-silenced plants show a glandless phenotype; VIGS plants showed albino and glandless phenotype when both were silenced simultaneously. (B) CLA1 gene relative expression in the stem marked white box in panel (A). Error bars are s.d. of three biological repeats. (C) GoPGF gene relative expression in the stem marked white box in panel (A). Error bars are s.d. of three biological repeats.
Friction inoculation can extend the duration of silencing of target genes
Though the VIGS method has a high silencing efficiency in cotton seedlings, the silencing efficiency after flowering is significantly reduced. Therefore, in order to render this method applicable to the study of gene function in flowering or the fiber elongation stage, we applied a friction inoculation method to extend the duration of silencing of target genes. In addition, the modified method can extend the duration of the VIGS in other plants (Becker and Lange, 2010; Senthil-Kumar and Mysore, 2011). With TM-1 as the receptor material, the CLA1 gene was used for validating the effectiveness of this approach (Gao et al., 2011a). First, the Agrobacterium strains carrying the CLA1 gene vector were injected into the Nicotiana tabacum, and then after 2 weeks, the newly grown tobacco leaf displayed a macular phenotype (Supplementary Figure 4A). Next, 1–3 g of tobacco leaves with the macular phenotype were collected and placed in a mortar. A small amount of PBS (0.01 M, pH = 7.2) was added and ground into juice. The cotton leaves were vaccinated and sprinkled with a trace of 500 mesh diamond. Upon dipping into the juice with a pestle and gently rubbing on the leaves, the leaves exhibited micro-wounds, but epidermal cells were not destroyed. Finally, the treated cotton plants were further grown in a greenhouse. After 2 weeks, the new cotton leaves grew albino (Supplementary Figure 4B). The relative expression of the CLA1 gene was analyzed by the whitening and normal leaves in the same position of the cotton plants. The expression of the CLA1 gene in the white leaves was significantly decreased (Supplementary Figure 4C). This result revealed that the friction inoculation method could prolong the silence duration of the target gene in the cotton plants.
Discussion and conclusions
The earliest application of VIGS in cotton was to construct a CLA1 gene fragment into a TRV vector and then transform it into Agrobacterium to inject cotton cotyledons. After 2–3 weeks, the whitening phenotype of the new leaves was observed, and the target gene was highly efficient (Gao et al., 2011a). Since then, many researchers have used this method for functional gene research and used the CLA1 gene as a positive control. For example, GhMLP28 is involved in resistance to Verticillium wilt (Yang et al., 2015), LMI1 regulates the development of the okra leaf (Chang et al., 2016; Andres et al., 2017) and GhSnRK2 participates in the regulation of drought resistance (Bello et al., 2014), etc. In Gossypium barbadense, VIGS systems were established using multiple marker genes, including GaPDS, GaCLA1, GaANS, and GaANR (Pang et al., 2013). In the above method, although the high silencing efficiency of the target gene in cotton VIGS system was established, the selection of the marker gene is not perfect. These marker genes have certain defects: CLA1 and PDS gene silencing plants have a whitening phenotype, but do not maintain long-term growth; in GaANS gene silencing plants, the leaves need to be treated before the phenotype can be observed. Compared with them, as a marker gene, GoPGF can well solve the above-mentioned limitations. When the expression of the GoPGF gene is silenced, it does not affect the normal growth of the cotton plant, and gene silencing efficiency can be visually observed through the number variation of glands.
In our study, after inoculation of two Agrobacterium strains (containing pTRV-CLA1 and pTRV-GoPGF plasmids, respectively) into the same plant, both whitening and glandless phenotypes appeared at the same time. It was demonstrated that mixing two gene-silencing vectors in VIGS can also silence two target genes simultaneously. This result was consistent with previous reports (Gao et al., 2011a). During the VIGS process, the growth environment of the plant determines the silencing efficiency of the target gene. Even though treated with the same material as inoculation receptor, silencing efficiency of target genes still exists individual difference. Therefore, GoPGF as an internal reference gene can reflect the silencing efficiency of the target gene in real time according to the number variation of glands in the newly grown tissue of the plant. Although the VIGS system can be applied to the study of the cotton growth period after flowering, the silencing efficiency of its target gene is much lower than that at the seedling stage (Wan et al., 2016). Thus, we developed an improved method of friction inoculation based on the principle that plant viruses can be transmitted between plants. We successfully verified the feasibility of cotton (Wu et al., 2018; Zang et al., 2021a). The method is to inoculate a TRV virus in the bud stage of cotton and to increase the silencing efficiency of the target gene in the newly grown fruit branch by spreading the virus in the cotton plant. Up to now, many genomes of many cotton species have been released and tens of thousands predicated coding genes have been annotated. However, the function of most genes is unknown, and it needs to be proved by experiments. As we all know, gain of function through transgenic expression of the candidate gene in the mutant varieties is the most direct and effective method for functional validation. But it is time consuming and laborious work and sometime difficult to implement because of genotype dependence of genetic transformation in cotton. Obviously, VIGS is a practicable method for quick gene function verification. Here, this improved VIGS method will facilitates the rapid validation of gene function especially during the development of cotton fiber initiation, elongation, lipid metabolism or under biotic and abiotic stresses.
In summary, we employed the GoPGF gene as a positive control and an internal reference gene during the VIGS process, so as to reflect the silencing efficiency of the target gene through existence and number variations of glands. The friction inoculation method is a necessary supplement and helps to investigate phenotype at later stages of development in cotton.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
ZS, HW, and YT conceived and designed the experiments. YH, ZS, and HW performed the experiments and wrote the manuscript. YT and ZZ participated in the experiments. ZS, HW, YT, TZ, and YH edited the manuscript. All authors read and approved the last version.
Funding
This work was financially supported in part by grants National Natural Science Foundation of China (31970320), the Fundamental Research Funds for the Central Universities (2021QN81011), the Xinjiang Production and Construction Corps (project of 2021AB008), the Hainan Yazhou Bay Seed Lab (project of B21HJ0223), and the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2019R01002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1020841/full#supplementary-material
References
Andres, R. J., Coneva, V., Frank, M. H., Tuttle, J. R., Samayoa, L. F., Han, S.-W., et al. (2017). Modifications to a Late Meristem Identity1 gene are responsible for the major leaf shapes of Upland cotton (Gossypium hirsutum L.). Proc. Natl. Acad. Sci. U S A. 114, E57–E66. doi: 10.1073/pnas.1613593114
Becker, A., and Lange, M. (2010). Vigs–genomics goes functional. Trends Plant Sci. 15, 1–4. doi: 10.1016/j.tplants.2009.09.002
Bello, B., Zhang, X., Liu, C., Yang, Z., Yang, Z., Wang, Q., et al. (2014). Cloning of Gossypium hirsutum sucrose non-fermenting 1-related protein kinase 2 gene (GhSnrk2) and its overexpression in transgenic Arabidopsis escalates drought and low temperature tolerance. PLoS One 9:e112269. doi: 10.1371/journal.pone.0112269
Cai, C., Zhang, X., Niu, E., Zhao, L., Li, N., Wang, L., et al. (2014). Ghpsy, a phytoene synthase gene, is related to the red plant phenotype in upland cotton (Gossypium hirsutum L.). Mol. Biol. Rep. 41, 4941–4952. doi: 10.1007/s11033-014-3360-x
Chang, L., Fang, L., Zhu, Y., Wu, H., Zhang, Z., Liu, C., et al. (2016). Insights into interspecific hybridization events in allotetraploid cotton formation from characterization of a gene-regulating leaf shape. Genetics 204, 799–806. doi: 10.1534/genetics.116.193086
Chen, T., Li, W., Hu, X., Guo, J., Liu, A., and Zhang, B. (2015). A cotton Myb transcription factor, Gbmyb5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 56, 917–929. doi: 10.1093/pcp/pcv019
Deng, J., Fang, L., Zhu, X., Zhou, B., and Zhang, T. (2019). A Cc-Nbs-Lrr gene induces hybrid lethality in cotton. J. Exp. Bot. 70, 5145–5156. doi: 10.1093/jxb/erz312
Gao, W., Zhu, X., Ding, L., Xu, B., Gao, Y., Cheng, Y., et al. (2022). Development of the engineered “glanded plant and glandless seed” cotton. Food Chem. Mol. Sci. 5:100130. doi: 10.1016/j.fochms.2022.100130
Gao, X., Britt, R. C. Jr., Shan, L., and He, P. (2011a). Agrobacterium-mediated virus-induced gene silencing assay in cotton. J. Vis. Exp. 54:e2938. doi: 10.3791/2938
Gao, X., Wheeler, T., Li, Z., Kenerley, C. M., He, P., and Shan, L. (2011b). Silencing Ghndr1 and Ghmkk2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305. doi: 10.1111/j.1365-313X.2011.04491.x
Liu, T., Guo, S., Lian, Z., Chen, F., Yang, Y., Chen, T., et al. (2015). A P4-Atpase gene Gbpatp of cotton confers chilling tolerance in plants. Plant Cell Physiol. 56, 549–557. doi: 10.1093/pcp/pcu200
Lu, R., Martin-Hernandez, A. M., Peart, J. R., Malcuit, I., and Baulcombe, D. C. (2003). Virus-induced gene silencing in plants. Methods 30, 296–303.
Ma, D., Hu, Y., Yang, C., Liu, B., Fang, L., Wan, Q., et al. (2016). Genetic basis for glandular trichome formation in cotton. Nat. Commun. 7:10456. doi: 10.1038/ncomms10456
Pang, J., Zhu, Y., Li, Q., Liu, J., Tian, Y., Liu, Y., et al. (2013). Development of Agrobacterium-mediated virus-induced gene silencing and performance evaluation of four marker genes in Gossypium barbadense. PLoS One 8:e73211. doi: 10.1371/journal.pone.0073211
Pei, Y., Zhang, J., Wu, P., Ye, L., Yang, D., Chen, J., et al. (2021). GoNe encoding a class Viiib Ap2/Erf is required for both extrafloral and floral nectary development in Gossypium. Plant J. 106, 1116–1127. doi: 10.1111/tpj.15223
Senthil-Kumar, M., and Mysore, K. S. (2011). New dimensions for Vigs in plant functional genomics. Trends Plant Sci. 16, 656–665.
Si, Z., Liu, H., Zhu, J., Chen, J., Wang, Q., Fang, L., et al. (2018). Mutation of Self-Pruning homologs in cotton promotes short-branching plant architecture. J. Exp. Bot. 69, 2543–2553. doi: 10.1093/jxb/ery093
Thyssen, G. N., Fang, D. D., Turley, R. B., Florane, C. B., Li, P., Mattison, C. P., et al. (2017). A Gly65Val substitution in an actin, Ghact_Li1, disrupts cell polarity and F−actin organization resulting in dwarf, lintless cotton plants. Plant J. 90, 111–121. doi: 10.1111/tpj.13477
Tian, J., Pei, H., Zhang, S., Chen, J., Chen, W., Yang, R., et al. (2014). Trv–Gfp: a modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J. Exp. Bot. 65, 311–322. doi: 10.1093/jxb/ert381
Wan, Q., Guan, X., Yang, N., Wu, H., Pan, M., Liu, B., et al. (2016). Small interfering Rna s from bidirectional transcripts of Gh Mml 3_A12 regulate cotton fiber development. New Phytol. 210, 1298–1310. doi: 10.1111/nph.13860
Wu, H., Tian, Y., Wan, Q., Fang, L., Guan, X., Chen, J., et al. (2018). Genetics and evolution of Mixta genes regulating cotton lint fiber development. New Phytol. 217, 883–895.
Yang, C.-L., Liang, S., Wang, H.-Y., Han, L.-B., Wang, F.-X., Cheng, H.-Q., et al. (2015). Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol. Plant 8, 399–411. doi: 10.1016/j.molp.2014.11.023
Zang, Y., Hu, Y., Xu, C., Wu, S., Wang, Y., Ning, Z., et al. (2021a). Ghubx controlling helical growth results in production of stronger cotton fiber. iscience 24:102930. doi: 10.1016/j.isci.2021.102930
Keywords: cotton, gene function analyses, VIGS (virus-induced gene silencing), marker gene, gland trait
Citation: Si Z, Wu H, Tian Y, Zhang Z, Zhang T and Hu Y (2022) Visible gland constantly traces virus-induced gene silencing in cotton. Front. Plant Sci. 13:1020841. doi: 10.3389/fpls.2022.1020841
Received: 16 August 2022; Accepted: 29 August 2022;
Published: 16 September 2022.
Edited by:
Xinyang Wu, China Jiliang University, ChinaReviewed by:
Jun Zhao, Zhenjiang Institute of Agricultural Sciences, ChinaLianguang Shang, Agricultural Genomics Institute at Shenzhen (CAAS), China
Copyright © 2022 Si, Wu, Tian, Zhang, Zhang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianzhen Zhang, Y290dG9uQHpqdS5lZHUuY24=; Yan Hu, MDAxNjIxMUB6anUuZWR1LmNu
†These authors have contributed equally to this work
 Zhanfeng Si
Zhanfeng Si Huaitong Wu
Huaitong Wu Yue Tian3†
Yue Tian3† Tianzhen Zhang
Tianzhen Zhang Yan Hu
Yan Hu