
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 October 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1019512
This article is part of the Research Topic Cereal Leaf Blights, Volume II View all 10 articles
 Zhibin Feng1†
Zhibin Feng1† Mingzhi Xu2,3†
Mingzhi Xu2,3† Jin Yang2,3†
Jin Yang2,3† Renhong Zhang2,3
Renhong Zhang2,3 Zigui Geng2,3
Zigui Geng2,3 Tingting Mao2,3,4
Tingting Mao2,3,4 Yuting Sheng2,3,4
Yuting Sheng2,3,4 Limin Wang2,3,4
Limin Wang2,3,4 Juan Zhang2,3,4*
Juan Zhang2,3,4* Hongxia Zhang2,4,5*
Hongxia Zhang2,4,5*Rhizoctonia solani Kühn naturally infects and causes Sheath blight disease in cereal crops such as wheat, rice and maize, leading to severe reduction in grain yield and quality. In this work, a new bacterial strain Bacillus halotolerans LDFZ001 showing efficient antagonistic activity against the pathogenic strain Rhizoctonia solani Kühn sh-1 was isolated. Antagonistic, phylogenetic and whole genome sequencing analyses demonstrate that Bacillus halotolerans LDFZ001 strongly suppressed the growth of Rhizoctonia solani Kühn sh-1, showed a close evolutionary relationship with B. halotolerans F41-3, and possessed a 3,965,118 bp circular chromosome. Bioinformatic analysis demonstrated that the genome of Bacillus halotolerans LDFZ001 contained ten secondary metabolite biosynthetic gene clusters (BGCs) encoding five non-ribosomal peptide synthases, two polyketide synthase, two terpene synthases and one bacteriocin synthase, and a new kijanimicin biosynthetic gene cluster which might be responsible for the biosynthesis of novel compounds. Gene-editing experiments revealed that functional expression of phosphopantetheinyl transferase (SFP) and major facilitator superfamily (MFS) transporter genes in Bacillus halotolerans LDFZ001 was essential for its antifungal activity against R. solani Kühn sh-1. Moreover, the existence of two identical chitosanases may also make contribution to the antipathogen activity of Bacillus halotolerans LDFZ001. Our findings will provide fundamental information for the identification and isolation of new sheath blight resistant genes and bacterial strains which have a great potential to be used for the production of bacterial control agents.
Importance: A new Bacillus halotolerans strain Bacillus halotolerans LDFZ001 resistant to sheath blight in wheat is isolated. Bacillus halotolerans LDFZ001 harbors a new kijanimicin biosynthetic gene cluster, and the functional expression of SFP and MFS contribute to its antipathogen ability.
Wheat sheath blight has become one of worldwide disease causing severe yield loss crop plants (Ghosh et al., 2017). Necrotrophic fungus Rhizoctonia solani Kühn, which could produce host specific phytotoxins as pathogenicity or virulence factors, resulting in its widely spread and difficult to control, has been identified as the causal agent of sheath blight in many crops (Srivastava et al., 2016). Although Bacillus species have been widely used to control sheath blight, the antagonistic activity is still not high enough and needs to be improved (Abbas et al., 2019).
To date, various Bacillus species have been commercially used as pesticides, surfactants, and biological agents for flavor enhancing and nutrition supplementation, and about half of the commercially available bacterial control agents were originated from Bacillus species (Stein, 2005; Ongena and Jacques, 2008; Tareq et al., 2012; Tareq et al., 2014; Kim et al., 2017b; Pereira et al., 2019). The secondary metabolites, hydrolases and peptides, such as ribosomally synthesized and post-translationally modified peptides (RiPPs), nonribosomally synthesized peptides (NRPs), antitumor polyketides (PKs) and terpenes, which constitute a rich assortment of biologically active small molecules in the bio-control process, play a crucial role in plant pathogen inhibition (Tosato et al., 1997; Duitman et al., 1999; Tsuge et al., 2001; Luo et al., 2015b; Torres et al., 2015; Nair et al., 2016; Saggese et al., 2018). Based on the sequences of their genomes, gene clusters corresponding to different biological active molecules have been cloned, and their biological functions have been identified in different bacterial species (Gao et al., 2017; Jin et al., 2017).
To understand the functions of biological active molecules produced by microorganism for plant pathogen protection, complete genome sequencing has been taken as an efficient strategy (Chen et al., 2009b; He et al., 2013; Guo et al., 2015; Shaligram et al., 2016; Jin et al., 2017; Jadeja et al., 2019; Lu et al., 2019; Pereira et al., 2019). Compared with the genome sequence of B. subtilis 168, the first sequenced model organism of Bacillus species, different sequences responsible for cell wall and antibiotic synthesis have been identified, implying the functional difference of different genome sequences in Bacillus species (Guo et al., 2013; Sabaté and Audisio, 2013; Bóka et al., 2019). In the genome of B. subtilis 168, almost 4% of the whole genome was predicted to be responsible for the encoding of multifunctional enzymes involved in antibiotic biosynthesis (Kunst et al., 1997). By comparing the genome sequences between different microorganisms, some new gene clusters synthesizing novel antimicrobial products have been predicted (Chen et al., 2009a; Luo et al., 2015a; Mobegi et al., 2017).
Surfactins, the biosurfactant molecules widely identified in Bacillus species, have showed multiple bio-activities (Kim et al., 2017a; Li et al., 2021; Han et al., 2022). As a member of lipopeptide family, surfactins synergistically affected the bio-control effectivity of Bacillus species (Li et al., 2016; Kim et al., 2017b). Full genome sequence annotation analysis indicated that a surfactin operon, including srfA, srfB, srfC, and srfD, was responsible for the biosynthesis of surfactin (Nakano et al., 1991). Meanwhile, a genetic locus sfp, encoding a phosphopantetheine transferase, was also required for surfactin production (Wu et al., 2019). Although B. subtilis strain 168 possessed a complete srf operon, it was unable to produce surfactin due to a frameshift in the SFP gene, which resulted in the production of an inactive phosphopantetheine transferase. Integration of a functional SFP gene restored the ability of B. subtilis 168 to synthesize surfactin (Reuter et al., 1999; Wu et al., 2019).
In addition to SFP gene, other genes in srf operon, such as major facilitator superfamily (MFS) proteins, were also identified via genome sequence analysis. To date, MFS transporter was the largest transporter superfamily, including over 10,000 members divided into 74 families (Marger and Saier, 1993; Saier et al., 1999; Wang et al., 2020; Saier et al., 2021). MFS transporters could facilitate the transport of a variety of substrates, including ions, sugar phosphates, drugs, nucleosides, amino acids and peptides, across cytoplasmic and internal membranes (Saier and Paulsen, 2001; Lorca et al., 2007; Chen et al., 2008; Yen et al., 2010). Crystallographic structures of MFS members consisted of a typical 12 transmembrane segments and a unique intracellular four-helix domain (Madej and Kaback, 2013). Most MFS transporters in some bacteria transported specific substrates and were closely related with the immunological issues such as virus invasion and drug resistance (Manel et al., 2005).
Chitosanase could specifically catalyse the hydrolysis of the β-1,4-glycosidic linkage in chitosan, to produce chitosan oligosaccharides, the only natural alkaline amino oligosaccharides widely used in pharmaceutical, food and cosmetic industry (Park et al., 1999; Kurakake et al., 2000; Omumasaba et al., 2000; Yoon et al., 2002; Wang et al., 2008; Johnsen et al., 2010; Kang et al., 2012). Chitosanases also have a function in plant pathogen suppression (Zhao et al., 2011). In prokaryotes, chitosanase, whose target chitosan is not a constituent of the cells, is alleged to work as extracellular enzymes (de Araújo et al., 2016; Park et al., 1999; Liang et al., 2014). A gene cluster encoding chitosanase, which was able to prevent plant from the infection by Plasmodiophora brassicae, a common pathogen that causes clubroot disease, has been identified (Guo et al., 2013).
In the past years, a number of Bacillus strains have been isolated and the possible functions of some gene clusters in their genomes have been examined. However, Bacillus strain showing significantly protection of wheat from sheath blight disease caused by Rhizoctonia solani Kühn is still not identified. In the present study, a new Bacillus species with high antifungal activity and great potential for sheath blight protection in cereal crops was isolated, and its genome sequence and the possible genes responsible for the antifungal activity were investigated.
Bacillus halotolerans LDFZ001 (B. halotolerans LDFZ001) was isolated from the sandy soil collected from the coastal zone of Yantai city, Shandong province, China, using serial dilution plating methods. Single colony was cultured on LB medium and stored at -80°C. The control strains Bacillus subtillis 168 (B. subtillis 168) and Bacillus halotolerans F41-3 (B. halotolerans F4103) were purchased from BioSciBio (Hangzhou, China). The pathogenic strain Rhizoctonia solani Kühn sh-1 is a collection in our lab. All the bacterial and fungus strains used in this study were listed in Table S1.
For the cultivation of B. halotolerans LDFZ001, B. subtillis 168 and B. halotolerans F41-3, nutrient broth (NB) liquid medium consisting of 3 g beef extract, 10 g peptone, 5 g NaCl, 2 g MgCl2 per liter was used. For the growth and preservation of the pathogenic strain Rhizoctonia solani Kühn sh-1, potato dextrose agar (PDA) medium consisting of 200 g Patato Destrose Agar (Coolaber, Beijing, China), 20 g sucrose, 20 g agar, was used.
The cloning plasmid pEASY-T1 was purchased from TransGen Biotech (Beijing, China). The expression plasmid pET28a (+) and the host strain E. coli BL21 (DE3) were purchased from Novagen (Shanghai, China). The plasmid pJOE8999 for gene editing with CRISPR-Cas 9 system is a collection in our lab. All the materials for gene cloning and protein purification were purchased from TaKaRa Biotechnology (Dalian, China). HisTrap HP and HiTrap Desalting were purchased from GE Healthcare (München, Germany). Chitosan for enzyme assay was obtained from Sigma-Aldrich (St. Louis, USA). Other chemicals were purchased from Sangon Biotech (Shanghai, China).
The morphology of purified strain B. halotolerans LDFZ001 was observed with light microscope (Olympus BX41, Japan) at a magnification of × 1000. Genomic DNA, isolated from the purified bacterial strain was used as template to amplify the 16S rRNA gene. The corresponding sequence was applied to phylogenetic analysis using MEGA 7.0 (Kumar et al., 2018).
For antifungal activity analysis, pathogenic microbe strain R. solani Kühn sh-1 was inoculated at the center of PDA medium plate and cultured at 28°C alone or with B. halotolerans LDFZ001, Bacillus subtillis 168 or B. halotolerans F41-3 for three days, which was inoculated as a scratch line under the R. solani Kühn sh-1 inoculation spot on each plate. Then, the inhibition rate (IR) of B. halotolerans LDFZ001, B. subtillis 168 and B. halotolerans F41-3 against R. solani Kühn sh-1 was evaluated as described previously (Chen et al., 2019).
For the inhibition efficiency analysis of B. halotolerans LDFZ001, B. subtillis 168 and B. halotolerans F41-3 on Rhizoctonia solani Kühn sh-1 caused wheat sheath blight, Rhizoctonia solani Kühn sh-1 cultured on NB solid medium at 28°C for 24 were collected by centrifugation, washed two times with 0.05 M sodium phosphate buffer (pH7.2), and re-suspended with it to a final concentration of 2×108 CFU/mL. One-week-old wheat seedlings germinated on filter paper soaked with sterile water were sprayed with 10 mL of the prepared bacterial solution. After 24 h, a 5 mm agar block of Rhizoctonia solani Kühn sh-1 grown on NB solid medium were inoculated to the middle of hypocotyls. After incubated at 25°C for 5 days, the phenotypes of seedlings were observed. The disease severity was graded according to the previous reported standard (Chen et al., 2019). The disease incidence rate (DIR) was generated using the following formula:
N represents the total number of investigated plants and n is the number of infected plants (Li et al., 2019). In our study, 30 plants were selected to conserve and investigate the DIR for each treatment.
Cells of B. halotolerans LDFZ001, B. subtillis 168 and B. halotolerans F41-3 were cultivated in NB liquid medium at 28°C for 48 h. The concentrations of the fermentation broths were calibrated to OD600 = 1.0. Then 100mL of the calibrated fermentation broths from these three bacteria were separately centrifuged at 8000 rpm/min for 10min to remove the bacteria. The supernatants were modified to pH=2 by 6mol/L hydrochloric acid and placed in 4°C refrigerator for more than 12h. After centrifugation at 8000 rpm/min for 10 min, the precipitate was collected. The crude extract was washed twice and dissolved in 100μ of methanol for HPLC analysis and antifungal activity assay. For HPLC analysis, crude extracts were filtered with a 0.22 μm membrane filter. Mobile phase was a mixture of acetonitrile and H2O (85:15, v/v). The flow rate was 1.00 mL/min. The injection volume was 10μL. The temperature was set at 28°C. Agilent C18 (250×4.6mm, 5µm) column was use and the detection wavelength was 210nm. Lipopeptides were analyzed using an Ultra high liquid chromatography system with a high resolution mass spectrometer (MS) as described previously (Chen et al., 2018).
The Oxford cup method was performed to analyze the antifungal activity of crude lipopeptides. Rhizoctonia solani Kühn sh-1 cultured on NB liquid medium at 28°C for 24 h were spread evenly on PDA solid medium. Two days later, four agar blocks (3mm×3mm) with the Rhizoctonia solani Kühn sh-1 were cut and place on a PDA solid medium plate, in the center of which a sterilized Oxford cup was placed. Using ddH2O as a control, 200μL of crude lipopeptide was dripped into the Oxford cup, which was taken away after 12h. The plates containing crude lipopeptide and agar blocks of the Rhizoctonia solani Kühn sh-1 were placed in 28°C for 72h. The inhibition zones were observed, photographed and measured.
High-quality genomic DNA isolated from B. halotolerans LDFZ001 with Wizard® Genomic DNA Purification Kit (Promega) according to manufacturer’s protocol was quantified with TBS-380 fluorometer (Turner BioSystems Inc., Sunnyvale, CA), and applied to a combination of PacBio RS II Single MoleculeReal Time (SMRT) and Illumina sequencing platforms for sequencing. Then, data generated were analyzed using I-Sanger Cloud Platform (www.i-sanger.com) from Shanghai Majorbio (Shanghai, China).
CDS, tRNA and rRNA were respectively predicted with Glimmer (version 3.02, http://cbcb.umd.edu/software/glimmer/), tRNA-scan-SE (version 1.23, http://lowelab.ucsc.edu/tRNAs.can-SE) and Barrnap (version 1.2, http://www.cbs.dtu.dk/services/RNAmmer/). The predicted CDSs were annotated from the non-redundant (NR) NCBI database, Swiss-Prot (http://uniprot.org), Pfam, GO, COG (http://www.ncbi.nlm.nih.gov/COG) and KEGG (http://www.genome.jp/kegg/) database using sequence alignment tools BLAST, Diamond and HMMER. Briefly, each set of query proteins was aligned with the databases, and annotations of best-matched subjects (e-value< 10-5) were obtained for gene annotation.
Secondary metabolite gene clusters were predicted with the online tools NP searcher (http://dna.sherman.lsi.umich.edu/) and antiSMASH (http://antismash.secondarymetabolites.org/). The genome of B. halotolerans LDFZ001 in a circular format was obtained using Circos.
Deletion of the gene’s chromosomal region was performed as described previously (Altenbuchner, 2016). PCR fragments from the upward and downward regions of the gene were inserted into the downstream of T7 promoter in plasmid pJOE8999. To generate plasmids pJOEsfp and pJOEmfs for B. halotolerans LDZF001 transformation, the sgDNAs of the relative genes to be deleted based on the database from the internet (http://crispor.tefor.net/crispor.py) were separately inserted into these resultant plasmids via homologous recombination (Anagnostopoulos and Spizizen, 1961; Ge et al., 2015; Altenbuchner, 2016). Using conventional two-step procedure, the strain was cultured in a minimal medium (Peptone 10g/L, Yeast extract 5g/L, NaCl 10g/L, Sorbitol 91g/L, pH7.0) to an of OD600 value of 0.8 at 30°C. After harvested via centrifugation at 4 °C, the cell pellet was re-suspended in ETM buffer (Sorbitol 91g/L, Mannitol 91g/L, 10% Glycerol (v/v), pH 7.0) for electroporation (1800v, 200Ω, 25μF). Competent cells containing pJOEsfp or pJOEmfs plasmid were recovered in RM medium (Peptone 10g/L, Yeast extract 5g/L, NaCl 10g/L, Sorbitol 91g/L, Mannitol 69g/L, pH7.0) and spread on the RM solid plates with supplemented with 40 mg/L kanamycin. The transformant colonies were tested with PCR after the plasmid was cured by being cultured at 37°C for 12-16h.
For heterologous expression of Csn-gene1288 and Csn-gene2656, the two chitosanase genes from B. halotolerans LDFZ001 were cloned into pET28a(+) to generate the recombinant plasmids pET28a-Csn1288 and pET28a-Csn2656 via homologous recombination (Ge et al., 2015). The resultant constructs were subsequently transformed into E. coli BL21 (DE3). After induced with IPTG, the recombinant proteins were purified via immobilized metal affinity chromatography. Chitosanase activity was measured as describe previously (Kurakake et al., 2000; Kang et al., 2012). The reaction was performed at 50°C for 15 min in sodium acetate buffer (pH5.4), followed by the determination of reducing sugar using DNS methods. The optimal condition assays were performed by measuring the activity in sodium acetate buffer (pH 3.6-7.0) at specific temperatures ranging from 30°C to 75°C. All the primer sequences used in this study were shown in Table S2.
For the antifungal assay, three replicates were performed. Student’s t-test with IBM SPSS Statistics 21 was performed to generate every P value (*P< 0.05).
To identify new antagonistic strain against fungal pathogen, we isolated a total number of 26 bacterial clones from the sandy soil in the coastal zone of Yantai, Shandong Province of China, and assessed their antagonistic activities against the pathogenic strain Rhizoctonia solani Kühn sh-1 which caused sheath blight disease in most crop plants. We found that one clone, numbered as LDFZ001, displayed very strong in vitro antagonistic activity against Rhizoctonia solani Kühn sh-1. Therefore, LDFZ001 was chosen for further studies. It is well known that Bacillus species possessed antagonistic activity against various pathogenic fungi. Therefore, two Bacillus strains, B. subtillis 168 and B. halotolerans F41-3, along with LDFZ001, were respectively co-cultivated with Rhizoctonia solani Kühn sh-1 on PDA medium. LDFZ001 effectively suppressed the radical growth of Rhizoctonia solani Kühn Sh-1, whereas B. subtillis 168 and B. halotolerans F41-3 did not. LDFZ001 exhibited a 98.8%, whereas B. subtillis 168 and B. halotolerans F41-3 only showed 2.3% and 1.4%, inhibition rates on the radical growth of Rhizoctonia solani Kühn Sh-1, respectively (Figures 1A, C).
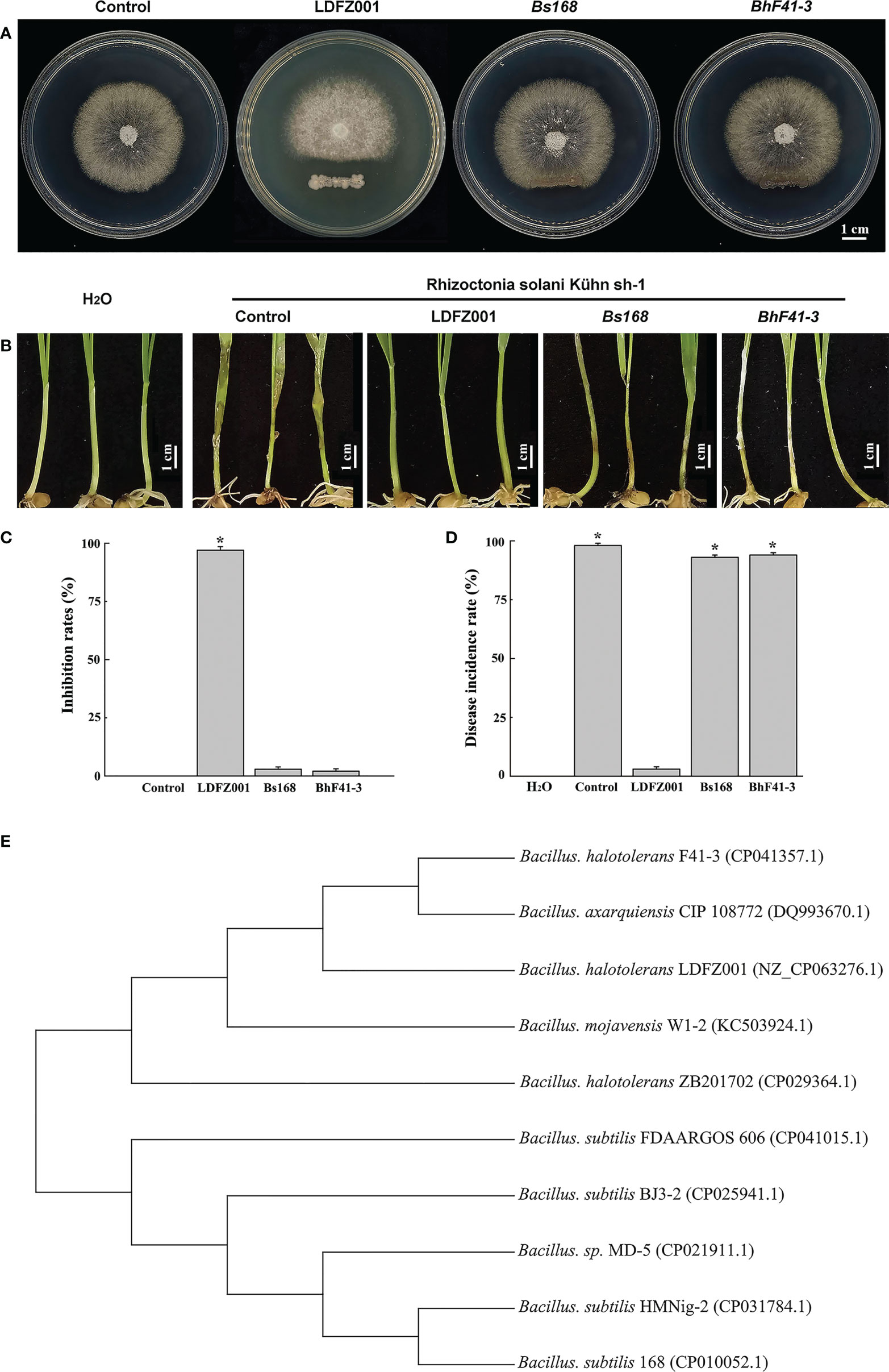
Figure 1 Antipathogen activity and phylogenetic relationship analyses. (A) Comparison of the antipathogen activity of B halotolerans LDFZ001 (LDFZ001) with its relative strains Bacillus subtillis 168 (Bs168) and Bacillus halotolerans F41-3 (BhF41-3) against R. solani Kühn sh-1, a pathogenic fungus strain cased sheath blight disease in crop plants. R. solani Kühn sh-1 inoculated at the center of PDA medium plate was cultured three days at 28°C alone or co-cultured with LDFZ001, Bs168 or BhF41-3, which was inoculated as a scratch line under the R. solani Kühn sh-1 inoculation spot on each plate. (B) Susceptibility analysis of wheat seedlings pretreated with LDFZ001, Bs168 and BhF41-3 to R. solani Kühn Sh-1.The sodium phosphate buffer was used as negative control (control). Wheat seedlings without any treatment were shown in the left as a control, too. (C) Inhibition rates (IRs) of B halotolerans LDFZ001, B subtillis 168 and B halotolerans F41-3 against R. solani Kühn sh-1 in image (A). (D) Disease incidence rates (DIRs) in image (B). Values are the mean ± SD from three independent experiments (n = 30). *P< 0.05. (E) Phylogenetic tree of B halotolerans LDFZ001 and other related taxa was generated based on 16S rRNA sequence. MEGA 7 was used to align the sequences.
Since Rhizoctonia solani Kühn Sh-1 can produce basidiospores, which cause the damping off and stem rot in wheat seedlings. We further compared the protective ability of B. subtillis 168, B. halotolerans F41-3 and LDFZ001 against wheat sheath blight on wheat seedlings caused by R. solani Kühn sh-1. We observed that one-week-old wheat seedlings pretreated with LDFZ001 successfully protected the occurrence of wheat sheath blight, but seedlings pretreated with B. subtillis 168 or B. halotolerans F41-3 did not. The seedlings were susceptible to R. solani Kühn Sh-1 and showed almost the same wheat sheath blight phenotype as did the negative control seedlings treated with sodium phosphate buffer, accompanied with a very high disease incidence rate (Figures 1B, D).
To determine the properties of LDFZ001, we performed morphological observation. LDFZ001 colonies cultured on LB medium plate formed approximate circles with creamy smooth surface and regular edge. Morphological analysis with light microscopy demonstrated that LDFZ001 cells were gram-positive and in rod shape (data not shown). To further determine the phylogenetic relationship of LDFZ001 with other bacterial strains, a neighbor-joining tree based on 16S rRNA sequence was constructed with MEGA 7.0. Compared with other Bacillus family members, LDFZ001 showed close evolutionary relationship with B. halotolerans F41-3 and B. mojavensis W1-2 (Figure 1E). Therefore, LDFZ001 was named as Bacillus halotolerans LDFZ001, and was deposited in the China General Microbiological Culture Collection Center with an accession number of CGMCC 7187.
Lipopeptides play a crucial role in the protection of plants from fungal pathogen attack. To understand whether lipopeptides also make a contribution to the antagonistic activity of B. halotolerans LDFZ001 against Rhizoctonia solani Kühn Sh-1, lipopeptide extracts from B. subtillis 168, B. halotolerans F41-3 and B. halotolerans LDFZ001 were separately isolated. Similarly, lipopeptide extract from B. halotolerans LDFZ001 significantly prohibited the growth of Rhizoctonia solani Kühn Sh-1. A clear inhibition zone of about 6.11 cm2 against Rhizoctonia solani Kühn sh-1 was observed. However, lipopeptide extracts from B. subtillis 168 and B. halotolerans F41-3 did not. The growth of Rhizoctonia solani Kühn Sh-1 was about the same as that on the control plate (Figure 2A). We then carried out HPLC assays with the lipopeptide extracts from B. subtillis 168, B. halotolerans F41-3 and B. halotolerans LDFZ001, a serial of distinct peaks in the profile of B. halotolerans LDFZ001, but not in the profiles of B. subtillis 168 and B. halotolerans F41-3, were observed (Figures 2B, C). Using UPLC-ESI-MS, these distinct peaks were determined as antifungal lipopeptide surfactin A (Figures 2B, C). Ions of m/z values 1022.67, 1036.69 and 1058.67 were also detected in previous reports (Roongsawang et al., 2002; Chen et al., 2019).
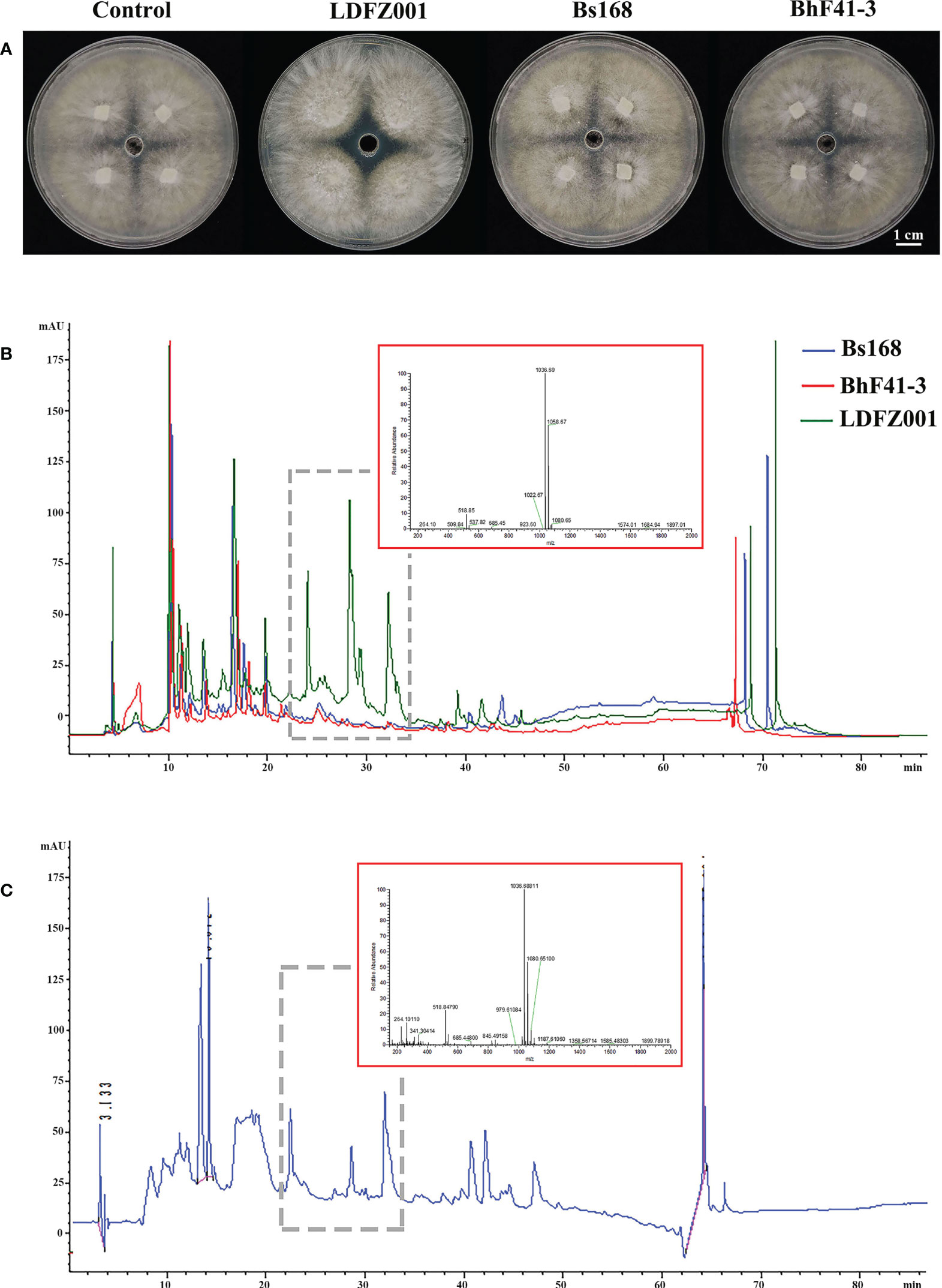
Figure 2 Lipopeptide extracts from B halotolerans LDFZ001 (LDFZ001), Bacillus subtillis 168 (Bs168) and Bacillus halotolerans F41-3 (BhF41-3) were used for antifungal activity and HPLC analyses. (A) Antifungal activity assays of putative lipopeptides against R. solani Kühn sh-1 (B, C) Profiling comparisons of HPLC and the extracted ion flow spectra of m/z 200–2000 from different strains.
To identify the responsible genes for the antifungal activity of B. halotolerans LDFZ001, we performed whole genome sequencing of B. halotolerans LDFZ001. The whole genome was assembled into a 3,965,118 bp circular chromosome with an average genome coverage depth of 475.27-fold and a G+C content of 43.92 (Figure 3). B. halotolerans LDFZ001 shares very high sequence identity (97.98%) with B. halotolerans F41-3. A lower sequence identity, from 82.30% to 77.80%, was also observed between B. halotolerans LDFZ001 and B. subtilis 168, B. intestinalis T30, B. subtilis ATCC 6633, B. subtilis FDAARGOS 606 and B. subtilis BJ3-2. Pairwise comparisons for the average nucleotide identity (ANI) and in silico DNA-DNA hybridization (DDH) between B. halotolerans LDFZ001 and these Bacillus family members revealed that the ANI values ranged from 87.11 to 97.98% and the DDH values ranged from 32.7 to 81.92% (Table 1). Based on the high ANI and DDH values between B. halotolerans LDFZ001 and B. halotolerans F41-3, both of them should belong to the same Bacillus species.
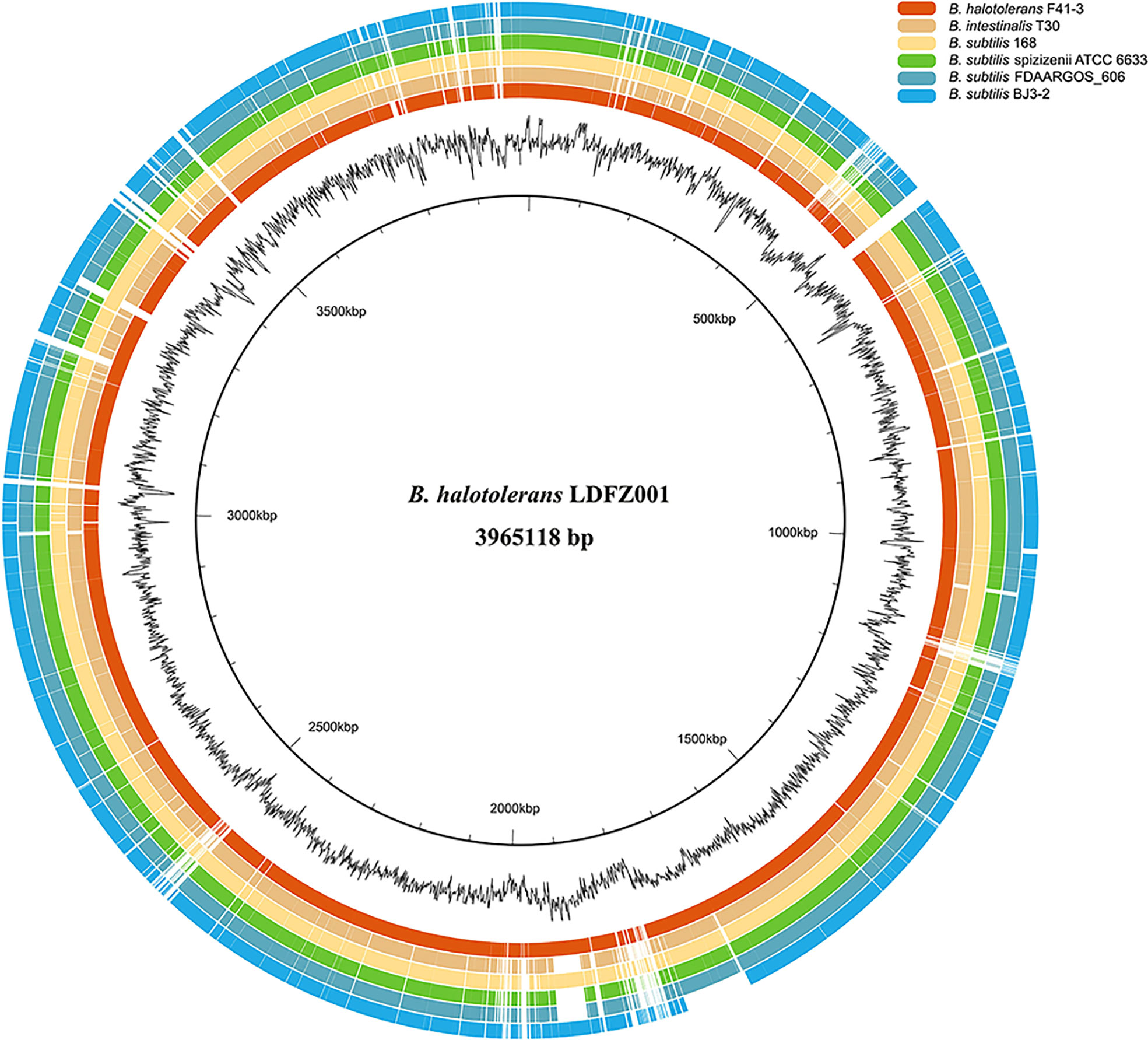
Figure 3 Genome comparison of B halotolerans LDFZ001 with six of its close Bacillus species. The innermost black circle represents the genome of B halotolerans LDFZ001 followed by its GC content and the Bacillus genomes.
We further performed genome sequence analysis with Glimmer 3.02 and GeneMarks. The whole genome of B. halotolerans LDFZ001 was composed of 4,126 coding sequences (CDSs), 73 tRNAs and 24 rRNAs. The 3,500,484 bp CDSs, with an average gene length of 848 bp, accounted for 88.28%, whereas the 22 tandem repeats, with a total DNA length of 6773 bp, accounted for 0.19%, of the whole chromosome DNA (Figure S1). Functional classification of clusters of orthologous gene (COG) showed that the predicted genes in B. halotolerans LDFZ001 genome were distributed into 4 COG categories (information storage and processing, metabolism, cellular processes and signaling, and poorly characterized), including 21 COG types and 1237 metabolism, 586 cellular processes and signaling, 498 information storage and processing, and 836 functionally poorly characterized genes (Figure 4). The complete genome sequence of B. halotolerans LDFZ001, with an accession number of NZ_CP063276.1, has been deposited in the NCBI GenBank.
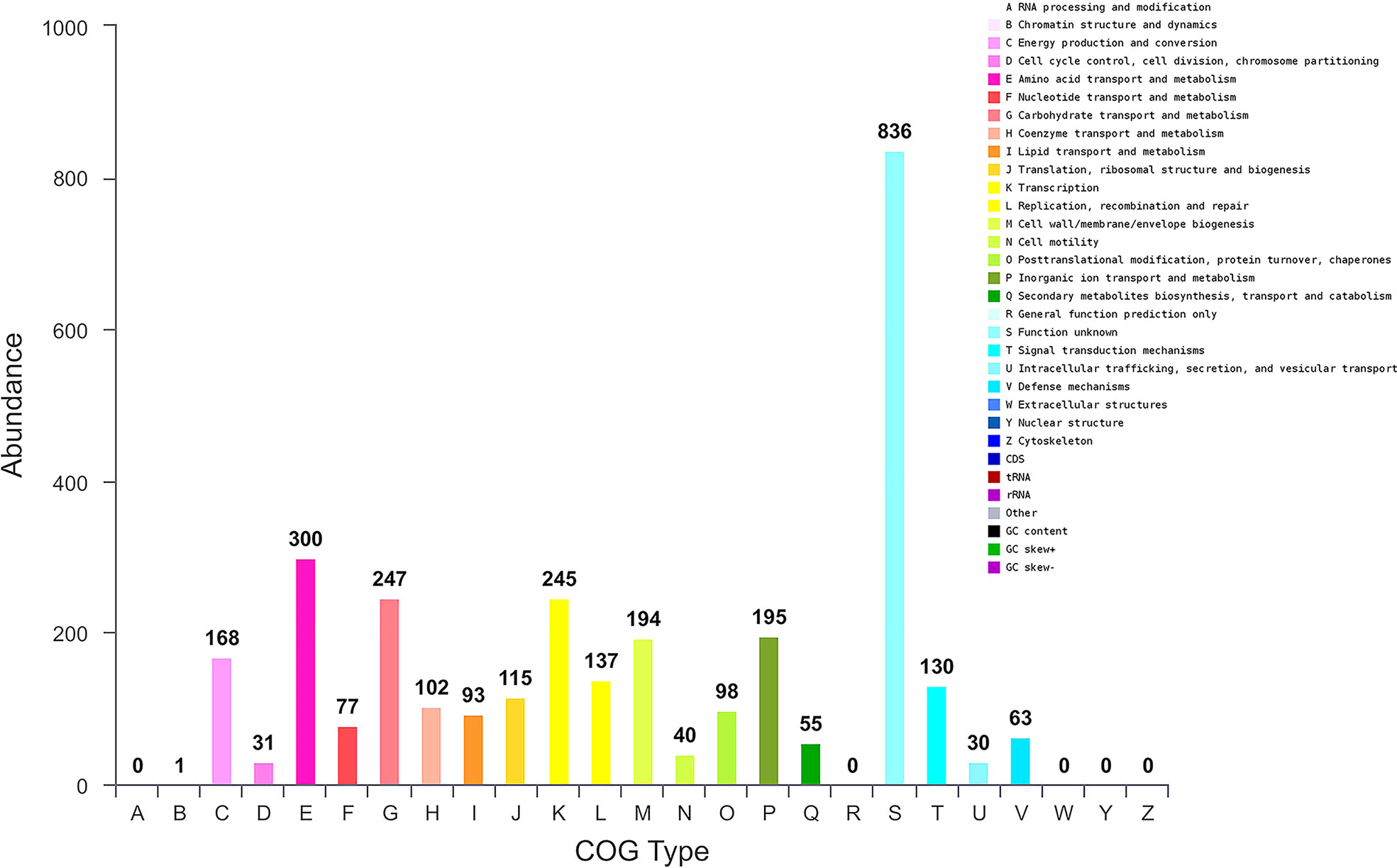
Figure 4 Genome sequence analysis of B halotolerans LDFZ001. Gene numbers involved in different pathways.
As one of the well commercialized biological control strains, Bacillus family can produce a diverse variety of secondary metabolites. Based on antiSMASH, a total number of 10 secondary metabolite biosynthetic gene clusters (BGCs), encoding 5 non-ribosomal peptide synthases, 2 polyketide synthase, 2 terpene synthases and 1 bacteriocin synthase, were predicted in the B. halotolerans LDFZ001 genome (Table 2). These enzymes are involved in the biosynthesis of various secondary metabolites, such as lipopeptides (surfactin and fengycin), lantipeptides (Kijanimicin and subtilosin A), dipeptide antibiotic (bacilysin), polyketides (Bacillaene), siderophores (bacillibactin) and unknown terpenes.
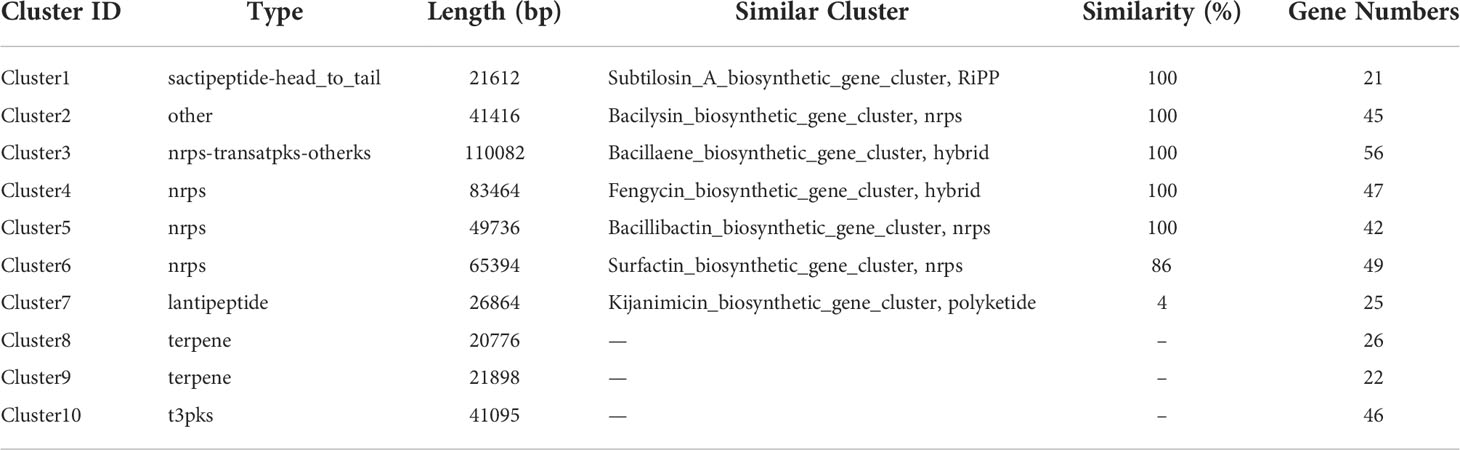
Table 2 Biosynthetic gene clusters for secondary metabolites in the genome of B. halotolerans LDFZ001.
Based on the annotation of secondary metabolite biosynthesis gene clusters, B. halotolerans LDFZ001 has a great potential to produce novel antibiotics. Therefore, we compared the BGCs of B. halotolerans LDFZ001 with other previously reported bacterial strains. Different from the bacilysin, bacillaene, fengycin, bacillibactin and subtilosin A biosynthesis clusters, which shared 100% similarity, and the surfactin biosynthesis cluster, which shared 86% similarity, a new kijanimicin biosynthesis cluster, which shared only 4% similarity, with the reported genome sequences of other bacterial strains, was identified. The schematic representation of the entire gene cluster exhibited that this novel gene cluster contained two lanthionine synthtesase C-like proteins, followed by seven ORFs encoding ATP-binding cassette (ABC) transporter proteins responsible for the translocation of a variety of metabolite compounds across membranes. Although surfactin biosynthesis cluster mainly consists of four genes, srfAA, srfAB, srfAC and srfAD, surfactin biosynthesis depends on the phosphopantetheinyl transferase and its adjacent genes. Sequence analysis showed that B. halotolerans LDFZ001, and the reference strains B. subtilis 168 and B. halotolerans F41-3, all possessed a complete surfactin biosynthesis cluster. However, a frameshift in sfp gene, downstream the surfactin biosynthesis cluster in B. subtilis 168, and a frameshift in the open reading frame of MFS transporter encoding gene, adjacent to srfAD in B. halotolerans F41-3, were observed. But no mutation in these two genes was observed in B. halotolerans LDFZ001 (Figure 5A).
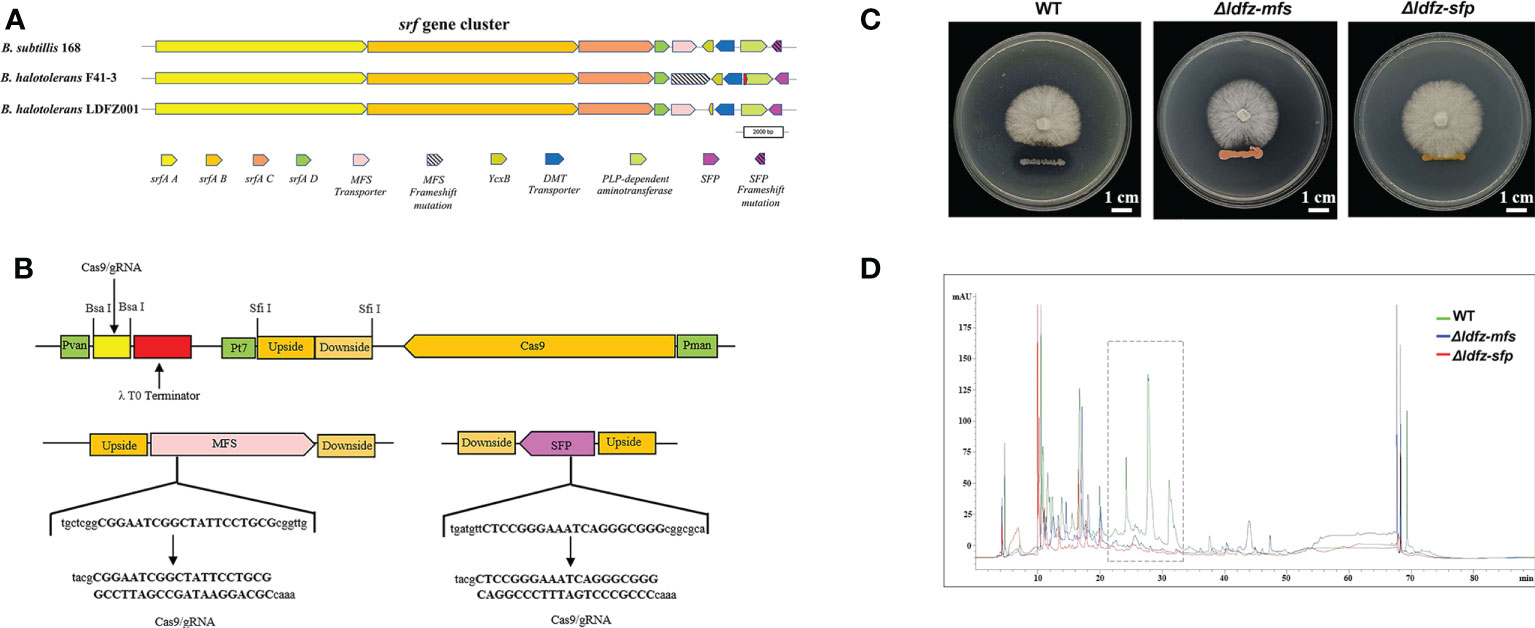
Figure 5 Functional analysis of SFP and MFS transporter genes. (A) Comparison of core genes in the srf gene clusters from B halotolerans LDFZ001, B subtillis 168 and B halotolerans F41-3. (B) A schematic map to show the gRNA sequence positions of SFPe and MFS genes. (C) Δldfz-sfp and Δldfz-mfs generated by respectively editing the SFP and MFS in B halotolerans LDFZ001 led to the loss of antifungal activity. (D) HPLC profile of lipopeptide extracts from WT and mutant Δldfz-sfp and Δldfz-mf.
Using CRISPR-Cas9 system, genes encoding SFP and MFS in B. halotolerans LDFZ001 were edited separately by a deletion of part of the gene sequences (Figure 5B). Two B. halotolerans LDFZ001 mutants, Δldfz-sfp and Δldfz-mfs were generated. Confrontation experiments showed that, unlike the wild type B. halotolerans LDFZ001, both Δldfz-sfp and Δldfz-mfs lost their antifungal activity against Rhizoctonia solani Kühn sh-1 (Figure 5C). Further HPLC analysis showed that the content of lipopeptide surfactin A was significantly decreased in these two mutants (Figure 5D).
Carbohydrate-active enzymes (CAZymes) can break down cell wall polysaccharides to trigger the death of fungal cells. We found that B. halotolerans LDZF001 harbored a large group of potential glycoside hydrolases, including 62 predicted CAZymes such as glycoside hydrolases, glycosyl transferases, carbohydrate esterases and carbohydrate-binding modules (Figure 6A). Interestingly, two members of the glycoside hydrolase GH46 family, Csn-gene1288 and Csn-gene2656, were predicted to be putative chitosanases, with a very high (90.6%) amino acid identity. Sequence alignment analysis of the deduced Csn-gene1288 and Csn-gene2656 with those of other bacterial chitosanases revealed that they were separated into two different branches (Figure 6B). Further enzyme activity analysis with thin-layer chromatography (TLC) showed that the hydrolysates of chitosan hydrolysed by both purified Csn-gene1288 and Csn-gene2656 were (GlcN)2, (GlcN)3, (GlcN)4, (GlcN)5 and (GlcN)6, with (GlcN) 3, (GlcN)4, and (GlcN)5 as the major products (Figure 6C). The catalytic properties of Csn-gene1288 and Csn-gene2656, with the optimal catalytic condition of pH5.4 at 50°C, were coincidentally similar to those of chitosanases from other Bacillus species (Figures 6D, E).
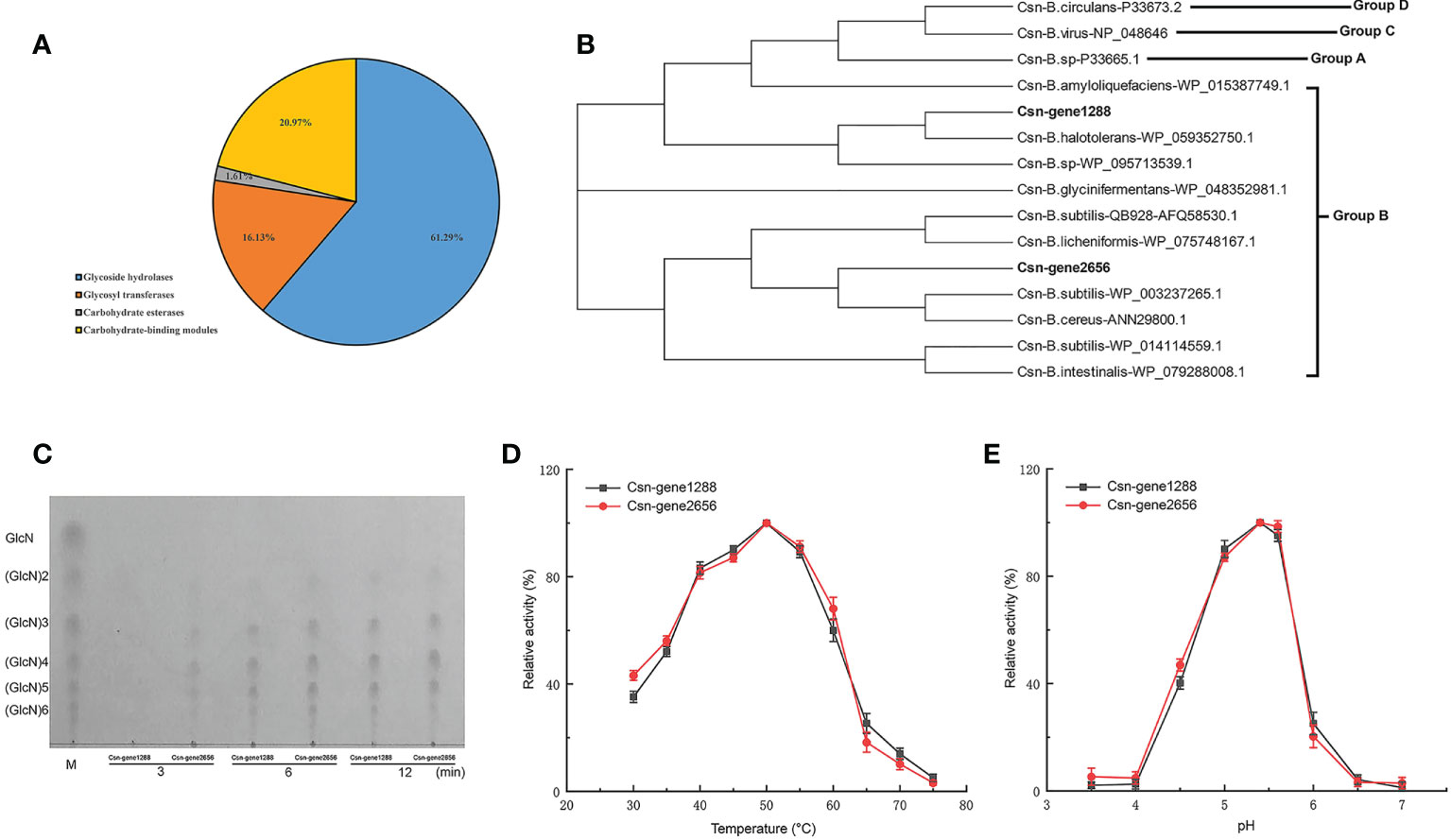
Figure 6 Carbohydrate-active enzyme activity analysis of the two chitosanase csn-gene1288 and csn-gene2656. (A) Statistical chart of annotated carbohydrate-active enzyme in B halotolerans LDFZ001. (B) Sequence alignment of csn-gene1288 and csn-gene2656 with that of other GH46 chitosanases. (C) Time course profiles of hydrolysis products of csn-gene1288 and csn-gene2656 enzymes on chitosan. (D) Effects of temperature on the enzyme activities of csn-gene1288 and csn-gene2656. (E) Effects of pH on the enzyme activities of csn-gene1288 and csn-gene2656.
Biocontrol microorganisms have been well commercialized as the source of microbial pesticides. They can either be directly used or processed into pesticides. The biocontrol activities of microorganisms were largely determined by the active metabolites and hydrolases they produced (Pal et al., 2000; Nguyen et al., 2017; Chen et al., 2018). During our screening of antifungal pathogen bacterial strains, a new clone, B. halotolerans LDFZ001, wich could effectively inhibit the growth of fungal pathogen Rhizoctonia solani Kühn sh-1, was isolate (Figures 1A, B). Phylogenetic and whole genome sequencing analyses implied that B. halotolerans LDFZ001 and B. halotolerans F41-3 fell into the same bacterial strain category (Figure 1C). Although the whole genomic sequence of B. halotolerans LDFZ001 was smaller than that of B. halotolerans F41-3, they shared as high as 97.98% amino acid sequence identity (Figure 3; Table 1).
To assess the antifungal pathogen activity of B. halotolerans LDFZ001, we subsequently performed antagonistic activity analysis. B. halotolerans LDFZ001 exhibited very strong suppression, whereas the control strain B. subtillis 168 and B. halotolerans F41-3 showed nearly no inhibition, against the sheath blight pathogen strain R. solani Kühn sh-1 (Figure 2A). Further genome sequence analysis revealed that B. halotolerans LDFZ001 genome contained ten gene clusters related to lipopeptide and hydrolase biosynthesis (Table 2). Numerous studies have showed that lipopeptides and hydrolases play crucial roles in antifungal protection. We observed that in contrast to other biocontrol strains, B. halotolerans LDFZ001 contained two novel terpene gene clusters, one novel Type III PKS, and a new kijanimicin biosynthesis cluster. The new gene cluster, involved in the biosynthesis of kijanimicin, only shared 4% amino acid similarity with other reported genome sequences in Bacillus species (Table 1). With its specific functional mechanism, kijanimicin has provided a vital view for antibiotics research (Castiglione et al., 2008; Crowther et al., 2013). The disclosure of gene cluster for putative kijanimicin biosynthesis in the B. halotolerans LDFZ001 genome will provide an important gene source for the future study.
In Bacillus species, surfactin is a strong biological surfactant essential for the formation of mycelium (Mukherjee et al., 2006; Shen et al., 2010). Although it does not directly inhibit the growth of plant pathogenic fungi, surfactin can effectively enhance the anti-fungal activity of other lipopeptides (Kobayashi et al., 2002; Kim et al., 2017b). To date, the biosynthetic mechanism of surfactin has been deeply investigated. The srf gene cluster has been reported to be sfp-dependent. During the domestication of B. subtilis 168, the mutation in sfp gene caused the loss of its ability to produce NRPs (Wu et al., 2019). Consistently, our genome sequence analysis revealed that B. halotolerans LDFZ001 contained complete sfp gene reading frame in its srf gene cluster, indicating that it had ability to produce surfactin to suppress the growth of pathogenic fungi (Figures 5A–C). MFS transporters also play a vital role in many substances transport in both eukaryotes and prokaryotes (Saier and Paulsen, 2001; Lorca et al., 2007; Chen et al., 2008; Yen et al., 2010). We observed that, although B. halotolerans F41-3 has a complete sfp gene, a frame shift mutation was found in the open reading frame of the MFS transporter gene adjacent to srfAD. Therefore, the reduced antipathogen activity against R. solani Kühn sh-1 in B. subtillis 168 and B. halotolerans F41-3 could be due to the mutations in the SFP and MFS genes, as confirmed by the SFP and MFS gene-editing analysis with CRISPR-Cas9 system in B. halotolerans LDFZ001 (Figures 5A–C).
In prokaryotes, gene duplication only occurred among gene products in high demand, such as rRNA and histones, due to the limited sequence size (Zhang, 2003). However, to adapt the environmental changes, specific genes could be generated, giving a great contribution to the divergence of microbes (He and Zhang, 2005; Conant and Wolfe, 2008; Serres et al., 2009; Innan and Kondrashov, 2010). Although chitosanases were found to be widely distributed in both eukaryotes and prokaryotes, chitosanase gene duplication has rarely occurred (Hurst and Smith, 1998; Zhang, 2003). Two chitosanase genes in the genome of B. halotolerans LDFZ001 were observed, for the firstly time, in Bacillus species (Figures 6B–E). The presence of two redundant chitosanases implied that B. halotolerans LDFZ001 has adjusted itself to survival the variable conditions. The high enzyme activity ascribed to the duplication of chitosanase gene will also expand its utilization potential for biocontrol, chitosan production and environmental improvement of B. halotolerans LDFZ001.
Taken together, a new bacterial strain B. halotolerans LDFZ001 against sheath blight disease caused by R. solani Kühn sh-1 was isolated. The growth suppression ability of B. halotolerans LDFZ001 on R. solani Kühn sh-1 could be ascribed to the functional expression of SFP and MFS genes. Two redundant chitosanases, which implied the evolutionary adaption to the environment via gene duplication, were also verified in B. halotolerans LDFZ001. Our findings in this study will provide fundamental information for new candidate gene identification and bacterial strain commercialization in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, NZ_CP063276.1.
This article does not contain any studies with human participants or animals performed by any of the authors.
ZF, MX, RZ, JY, and ZG conducted experiments. TM and YS analyzed data. JZ, ZF, and HZ wrote the manuscript. HZ polished the manuscript. All authors contributed to the article and approved the submitted version.
This work has been jointly supported by the following grants: The National Mega Project of GMO Crops of China (2016ZX08004-002-006); The National Natural Science Foundation of China (31870576, 32071733, 31901572); The Natural Science Foundation of Shandong Province, China(ZR2019PC015, ZR2021QC140); The Cooperation Project of University and Local Enterprise in Yantai of Shandong Province (2021XDRHXMPT09); The Modern Agricultural Industry Technology System Innovation Team of Shandong Province of China (SDAIT-02-05).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1019512/full#supplementary-material
Supplementary Figure 1 | A genome map of B. halotolerans LDFZ001.
Supplementary Table 1 | Strains and plasmids used in this study.
Supplementary Table 2 | Oligonucleotides used in this study.
Abbas, A., Khan, S. U., Khan, W. U., Saleh, T. A., Khan, M. H. U., Ullah, S., et al. (2019). Antagonist effects of strains of bacillus spp. against rhizoctonia solani for their protection against several plant diseases: Alternatives to chemical pesticides. C R Biol. 342, 124–135. doi: 10.1016/j.crvi.2019.05.002
Altenbuchner, J. (2016). Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl. Environ. Microbiol. 82, 5421–5427. doi: 10.1128/AEM.01453-16
Anagnostopoulos, C., Spizizen, J. (1961). Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81, 741–746. doi: 10.1128/jb.81.5.741-746.1961
Bóka, B., Manczinger, L., Kocsubé, S., Shine, K., Alharbi, N. S., Khaled, J. M., et al. (2019). Genome analysis of a Bacillus subtilis strain reveals genetic mutations determining biocontrol properties. World J. Microbiol. Biotechnol. 35, 52. doi: 10.1007/s11274-019-2625-x
Castiglione, F., Lazzarini, A., Carrano, L., Corti, E., Ciciliato, I., Gastaldo, L., et al. (2008). Determining the structure and mode of action of microbisporicin a potent lantibiotic active against multiresistant pathogens. Chem. Biol. 15, 22–31. doi: 10.1016/j.chembiol.2007.11.009
Chen, L., Heng, J., Qin, S., Bian, K. (2018). A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against fusarium head blight. PLoS. One 13, e0198560. doi: 10.1371/journal.pone.0198560
Chen, X. H., Koumoutsi, A., Scholz, R., Borriss, R. (2009a). More than anticipated-production of antibiotics and other secondary metabolites by Bacillus amyloliquefaciens FZB42. J. Mol. Microbiol. Biotechnol. 16, 14–24. doi: 10.1159/000142891
Chen, X. H., Koumoutsi, A., Scholz, R., Schneider, K., Vater, J., Süssmuth, R., et al. (2009b). Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J. Biotechnol. 140, 27–37. doi: 10.1016/j.jbiotec.2008.10.011
Chen, D. E., Podell, S., Sauer, J. D., Swanson, M. S., Saier, M. H. (2008). The phagosomal nutrient transporter (Pht) family. Microbiol. (Reading) 154, 42–53. doi: 10.1099/mic.0.2007/010611-0
Chen, L., Wu, Y. D., Chong, X. Y., Xin, Q. H., Wang, D. X., Bian, K. (2019). Seed-borne endophytic Bacillus velezensis LHSB1 mediate the biocontrol of peanut stedm rot caused by scleotium rolfsii. J. Appl. Microbiol. 128, 803–813. doi: 10.1111/jam.14508
Conant, G. C., Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Crowther, G. S., Baines, S. D., Todhunter, S. L., Freeman, J., Chilton, C. H., Wilcox, M. H. (2013). Evaluation of NVB302 versus vancomycin activity in an in vitro human gut model of Clostridium difficile infection. J. Antimicrob. Chemother. 68, 168–176. doi: 10.1093/jac/dks359
de Araújo, N. K., Pimentel, V. C., da Silva, N. M.P., de Araújo Padilha, C. E., de Macedo, G. R., Dos Santos, E. S. (2016). Recovery and purification of chitosanase produced by Bacillus cereus using expanded bed adsorption and central composite design. J. Sep. 39, 709–716. doi: 10.1002/jssc.201500900a
Duitman, E. H., Hamoen, L. W., Rembold, M., Venema, G., Seitz, H., Saenger, W., et al. (1999). The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. U S A. 96, 13294–13299. doi: 10.1073/pnas.96.23.13294
Gao, W., Liu, F., Zhang, W., Quan, Y., Dang, Y., Feng, J., et al. (2017). Mutations in genes encoding antibiotic substances increase the synthesis of poly-γ-glutamic acid in Bacillus amyloliquefaciens LL3. MicrobiologyOpen 6, e00398. doi: 10.1002/mbo3.398
Ge, J., Li, W., Zhao, Q., Li, N., Chen, M., Zhi, P., et al. (2015). Architecture of the mammalian mechanosensitive Piezo1 channel. Nature 527, 64–69. doi: 10.1038/nature15247
Ghosh, S., Kanwar, P., Jha, G. (2017). Alterations in rice chloroplast integrity, photosynthesis and metabolome associated with pathogenesis of rhizoctonia solani. Sci. Rep. 7, 41610. doi: 10.1038/srep41610
Guo, S., Li, X., He, P., Ho, H., Wu, Y., He, Y. (2015). Whole-genome sequencing of Bacillus subtilis XF-1 reveals mechanisms for biological control and multiple beneficial properties in plants. J. Ind. Microbiol. Biotechnol. 42, 925–937. doi: 10.1007/s10295-015-1612-y
Guo, S., Mao, Z., Wu, Y., Hao, K., He, P., He, Y. (2013). Genome sequencing of Bacillus subtilis strain XF-1 with high efficiency in the suppression of plasmodiophora brassicae. Genome Announc. 1, e0006613. doi: 10.1128/genomeA.00066-13
Han, V. C., Yu, N. H., Yoon, H., Ahn, N. H., Son, Y. K., Lee, B. H., et al. (2022). Identification, characterization, and efficacy evaluation of Bacillus velezensis for shot-hole disease biocontrol in flowering cherry. Plant Pathol. J. 38, 115–130. doi: 10.5423/PPJ.OA.01.2022.0004
He, P., Hao, K., Blom, J., Rückert, C., Vater, J., Mao, Z., et al. (2013). Genome sequence of the plant growth promoting strain Bacillus amyloliquefaciens subsp. plantarum B9601-Y2 and expression of mersacidin and other secondary metabolites. J. Biotechnol. 164, 281–291. doi: 10.1016/j.jbiotec.2012.12.014
He, X., Zhang, J. (2005). Gene complexity and gene duplicability. Curr. Biol. 15, 1016–1021. doi: 10.1016/j.cub.2005.04.035
Hurst, L. D., Smith, N. G. C. (1998). The evolution of concerted evolution. Proc. R Soc. Lond Ser. B. 265, 121–127. doi: 10.1098/rspb.1998.0272
Innan, H., Kondrashov, F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108. doi: 10.1038/nrg2689
Jadeja, N. B., Moharir, P., Kapley, A. (2019). Genome sequencing and analysis of strains bacillus sp. AKBS9 and Acinetobacter sp. AKBS16 for biosurfactant production and bioremediation. Appl. Biochem. Biotechnol. 187, 518–530. doi: 10.1007/s12010-018-2828-x
Jin, Q., Jiang, Q., Zhao, L., Su, C., Li, S., Si, F., et al. (2017). Complete genome sequence of Bacillus velezensis S3-1, a potential biological pesticide with plant pathogen inhibiting and plant promoting capabilities. J. Biotechnol. 259, 199–203. doi: 10.1016/j.jbiotec.2017.07.011
Johnsen, M. D., Hansen, O. C., Stougaard, P. (2010). Isolation, characterization and heterologous expression of a novel chitosanase from janthinobacterium sp. strain 4239. Microb. Cell Fact 9, 5. doi: 10.1186/1475-2859-9-5
Kang, L. X., Chen, X. M., Fu, L., Ma, L. X. (2012). Recombinant expression of chitosanase from Bacillus subtilis HD145 in pichia pastoris. Carbohydr. Res. 352, 37–43. doi: 10.1016/j.carres.2012.01.025
Kim, K., Lee, Y., Ha, A., Kim, J. I., Park, A. R., Yu, N. H., et al. (2017a). Chemosensitization of fusarium graminearum to chemical fungicides using cyclic lipopeptides produced by Bacillus amyloliquefaciens strain JCK-12. Front. Plant Sci. 8, 2010. doi: 10.3389/fpls.2017.02010
Kim, Y. T., Park, B. K., Kim, S. E., Lee, W. J., Moon, J. S., Cho, M. S., et al. (2017b). Organization and characterization of genetic regions in Bacillus subtilis subsp. krictiensis ATCC55079 associated with the biosynthesis of iturin and surfactin compounds. PloS One 12, e0188179. doi: 10.1371/journal.pone.0188179
Kobayashi, D., Kondo, K., Uehara, N., Otokozawa, S., Tsuji, N., Yagihashi, A., et al. (2002). Endogenous reactive oxygen species is an important mediator of miconazole antifungal effect. Antimicrob. Agents Chemother. 46, 3113–3117. doi: 10.1128/aac.46.10.3113-3117.2002
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G., Azevedo, V., et al. (1997). The complete genome sequence of the gram-positive bacterium bacillus subtilis. Nature 390, 249–256. doi: 10.1038/36786
Kurakake, M., Yo-u, S., Nakagawa, K., Sugihara, M., Komaki, T. (2000). Properties of chitosanase from bacillus cereus S1. Curr. Microbiol. 40, 6–9. doi: 10.1007/s002849910002
Liang, T. W., Chen, Y. Y., Pan, P. S., Wang, S. L. (2014). Purification of chitinase/chitosanase from Bacillus cereus and discovery of an enzyme inhibitor. Int. J. Biol. Macromol. 63, 8–14. doi: 10.1016/j.ijbiomac.2013.10.027
Li, Y., Guo, Q., Wei, X., Xue, Q., Lai, H. (2019). Biocontrol effects of penicillium griseofulvum against monkshood (Aconitum carmichaelii debx.) root diseases caused by sclerotium rolfsiii and fusarium spp. J. Appl. Microbiol. 127, 1532–1545. doi: 10.1111/jam.14382.l
Li, X., Munir, S., Xu, Y., Wang, Y., He, Y. (2021). Combined mass spectrometry-guided genome mining and virtual screening for acaricidal activity in secondary metabolites of Bacillus velezensis W1. RSC Adv. 11, 25441–25449. doi: 10.1039/d1ra01326b
Li, X., Zhang, Y., Wei, Z., Guan, Z., Cai, Y., Liao, X. (2016). Antifungal activity of isolated Bacillus amyloliquefaciens SYBC H47 for the biocontrol of peach gummosis. PloS One 11, e0162125. doi: 10.1371/journal.pone.0162125
Lorca, G. L., Barabote, R. D., Zlotopolski, V., Tran, C., Winnen, B., Hvorup, R. N., et al. (2007). Transport capabilities of eleven gram-positive bacteria: comparative genomic analyses. Biochim. Biophys. Acta 1768, 1342–1366. doi: 10.1016/j.bbamem.2007.02.007
Luo, C., Liu, X., Zhou, X., Guo, J., Truong, J., Wang, X., et al. (2015b). Unusual biosynthesis and structure of locillomycins from Bacillus subtilis 916. Appl. Environ. Microbiol. 81, 6601–6609. doi: 10.1128/AEM.01639-15
Luo, C., Liu, X., Zhou, H., Wang, X., Chen, Z. (2015a). Nonribosomal peptide synthase gene clusters for lipopeptide biosynthesis in Bacillus subtilis 916 and their phenotypic functions. Appl. Environ. Microbiol. 81, 422–431. doi: 10.1128/AEM.02921-14
Lu, J. Y., Zhou, K., Huang, W. T., Zhou, P., Yang, S., Zhao, X., et al. (2019). A comprehensive genomic and growth proteomic analysis of antitumor lipopeptide bacillomycin lb biosynthesis in Bacillus amyloliquefaciens X030. Appl. Microbiol. Biotechnol. 103, 7647–7662. doi: 10.1007/s00253-019-10019-6
Madej, M. G., Kaback, H. R. (2013). Evolutionary mix-and-match with MFS transporters II. Proc. Natl. Acad. Sci. U.S.A. 110, E4831–E4838. doi: 10.1073/pnas.1319754110
Manel, N., Battini, J. L., Taylor, N., Sitbon, M. (2005). HTLV-1 tropism and envelope receptor. Oncogene 24, 6016–6025. doi: 10.1038/sj.onc.1208972
Marger, M. D., Saier, M. H., Jr. (1993). A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 18, 13–20. doi: 10.1016/0968-0004(93)90081-w
Mobegi, F. M., Zomer, A., de Jonge, M. I., van Hijum, S. A. F. T. (2017). Advances and perspectives in computational prediction of microbial gene essentiality. Brief Funct. Genomics 16, 70–79. doi: 10.1093/bfgp/elv063
Mukherjee, S., Das, P., Sen, R. (2006). Towards commercial production of microbial surfactants. Trends Biotechnol. 24, 509–515. doi: 10.1016/j.tibtech.2006.09.005
Nair, D., Vanuopadath, M., Nair, B. G., Pai, J. G., Nair, S. S. (2016). Identification and characterization of a library of surfactins and fengycins from a marine endophytic bacillus sp. J. Basic Microbiol. 56 (11), 1159–1172. doi: 10.1002/jobm.201600029
Nakano, M. M., Magnuson, R., Myers, A., Curry, J., Grossman, A. D., Zuber, P. (1991). srfA is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J. Bacteriol. 173 (5), 1770–1778. doi: 10.1128/jb.173.5.1770-1778.1991
Nguyen, P. A., Strub, C., Fontana, A., Schorr-Galindo, S. (2017). Crop molds and mycotoxins: alternative management using biocontrol. Biol. Control 104, 10–27. doi: 10.1016/j.biocontrol.2016.10.004
Omumasaba, C. A., Yoshida, N., Sekiguchi, Y., Kariya, K., Ogawa, K. (2000). Purification and some properties of a novel chitosanase from Bacillus subtilis KH1. J. Gen. Appl. Microbiol. 46 (1), 19–27. doi: 10.2323/jgam.46.19
Ongena, M., Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16 (3), 115–125. doi: 10.1016/j.tim.2007.12.009
Pal, K. K., Tilak, K. V., Saxena, A. K., Dey, R., Singh, C. S. (2000). Antifungal characteristics of a fluorescent Pseudomonas strain involved in the biological control of Rhizoctonia solani. Microbiol. Res. 155 (3), 233–242. doi: 10.1016/S0944-5013(00)80038-5
Park, J. K., Shimono, K., Ochiai, N., Shigeru, K., Kurita, M., Ohta, Y., et al. (1999). Purification, characterization, and gene analysis of a chitosanase (ChoA) from matsuebacter chitosanotabidus 3001. J. Bacteriol. 181 (21), 6642–6649. doi: 10.1128/JB.181.21.6642-6649.1999
Pereira, J. Q., Ritter, A. C., Cibulski, S., Brandelli, A. (2019). Functional genome annotation depicts probiotic properties of Bacillus velezensis FTC01. Gene 713, 143971. doi: 10.1016/j.gene.2019.143971
Reuter, K., Mofid, M. R., Marahiel, M. A., Ficner, R. (1999). Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4’-phosphopantetheinyl transferase superfamily. EMBO J. 18 (23), 6823–6831. doi: 10.1093/emboj/18.23.6823
Roongsawang, N., Thaniyavarn, J., Thaniyavarn, S., Kameyama, T., Haruki, M., Imanaka, T., et al. (2002). Isolation and characterization of a halotolerant bacillus subtilis BBK-1 which produces three kinds of lipopeptides: bacillomycin l, plipastatin, and surfactin. Extremophiles 6, 499–506. doi: 10.1007/s00792-002-0287-2
Sabaté, D. C., Audisio, M. C. (2013). Inhibitory activity of surfactin, produced by different Bacillus subtilis subsp. subtilis strains, against listeria monocytogenes sensitive and bacteriocin-resistant strains. Microbiol. Res. 168 (3), 125–129. doi: 10.1016/j.micres.2012.11.004
Saggese, A., Culurciello, R., Casillo, A., Corsaro, M. M., Ricca, E., Baccigalupi, L. (2018). A marine isolate of Bacillus pumilus secretes a pumilacidin active against staphylococcus aureus. Mar. Drugs 16 (6), 180. doi: 10.3390/md16060180
Saier, M. H., Beatty, J. T., Goffeau, A., Harley, K. T., Heijne, W. H., Huang, S. C., et al. (1999). The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1 (2), 257–279.
Saier, M. H., Jr., Paulsen, I. T. (2001). Phylogeny of multidrug transporters. Semin. Cell Dev. Biol. 12 (3), 205–213. doi: 10.1006/scdb.2000.0246
Saier, M. H., Reddy, V. S., Moreno-Hagelsieb, G., Hendargo, K. J., Zhang, Y., Iddamsetty, V., et al. (2021). The transporter classification database (TCDB): 2021 update. Nucleic Acids Res. 49 (D1), D461–D467. doi: 10.1093/nar/gkaa1004
Serres, M. H., Kerr, A. R., McCormack, T. J., Riley, M. (2009). Evolution by leaps: gene duplication in bacteria. Biol. Direct 4, 46. doi: 10.1186/1745-6150-4-46
Shaligram, S., Kumbhare, S. V., Dhotre, D. P., Muddeshwar, M. G., Kapley, A., Joseph, N., et al. (2016). Genomic and functional features of the biosurfactant producing bacillus sp. AM13. Funct. Integr. Genomics 16 (5), 557–566. doi: 10.1007/s10142-016-0506-z
Shen, H. H., Thomas, R. K., Penfold, J., Fragneto, G. (2010). Destruction and solubilization of supported phospholipid bilayers on silica by the biosurfactant surfactin. Langmuir 26, 7334–7342. doi: 10.1021/la904212x
Srivastava, S., Bist, V., Srivastava, S., Singh, P. C., Trivedi, P. K., Asif, M. H., et al. (2016). Unraveling aspects of bacillus amyloliquefaciens mediated enhanced production of rice under biotic stress of rhizoctonia solani. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00587
Stein, T. (2005). Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56 (4), 845–857. doi: 10.1111/j.1365-2958.2005.04587.x
Tareq, F. S., Kim, J. H., Lee, M. A., Lee, H. S., Lee, Y. J., Lee, J., et al. (2012). Ieodoglucomides a and b from a marine-derived bacterium. Bacillus licheniformis. Org. Lett. 14 (6), 1464–1467. doi: 10.1021/ol300202z
Tareq, F. S., Lee, M. A., Lee, H. S., Lee, Y. J., Lee, J. S., Hasan, C. M., et al. (2014). Noncytotoxic antimicrobial linear lipopeptides from a marine bacterium. Bacillus subtilis. Org. Lett. 16 (3), 928–931. doi: 10.1021/ol403657r
Torres, M. J., Petroselli, G., Daz, M., Erra-Balsells, R., Audisio, M. C. (2015). Bacillus subtilis subsp. subtilis CBMDC3f with antimicrobial activity against gram-positive foodborne pathogenic bacteria: UV-MALDI-TOF MS analysis of its bioactive compounds. World J. Microbiol. Biotechnol. 31 (6), 929–940. doi: 10.1007/s11274-015-1847-9
Tosato, V., Albertini, A. M., Zotti, M., Sonda, S., Bruschi, C. V. (1997). Sequence completion, identification and definition of the fengycin operon in bacillus subtilis 168. Microbiol. (Reading) 143 (Pt11), 3443–3450. doi: 10.1099/00221287-143-11-3443
Tsuge, K., Akiyama, T., Shoda, M. (2001). Cloning, sequencing, and characterization of the iturin a operon. J. Bacteriol. 183 (21), 6265–6273. doi: 10.1128/JB.183.21.6265-6273.2001
Wang, S. L., Chen, S. J., Wang, C. L. (2008). Purification and characterization of chitinases and chitosanases from a new species strain pseudomonas sp. TKU015 using shrimp shells as a substrate. Carbohydr. Res. 343 (7), 1171–1179. doi: 10.1016/j.carres.2008.03.018
Wang, S. C., Davejan, P., Hendargo, K. J., Javadi-Razaz, I., Chou, A., Yee, D. C., et al. (2020). Expansion of the major facilitator superfamily (MFS) to include novel transporters as well as transmembrane-acting enzymes. Biochim. Biophys. Acta Biomembr. 1862 (9), 183277. doi: 10.1016/j.bbamem.2020.183277
Wu, Q., Zhi, Y., Xu, Y. (2019). Systematically engineering the biosynthesis of a green biosurfactant surfactin by bacillus subtilis 168. Metab. Eng. 52, 87–97. doi: 10.1016/j.ymben.2018.11.004
Yen, M. R., Chen, J. S., Marquez, J. L., Sun, E. I., Saier, M. H. (2010). Multidrug resistance: phylogenetic characterization of superfamilies of secondary carriers that include drug exporters. Methods Mol. Biol. 637, 47–64. doi: 10.1007/978-1-60761-700-6_3
Yoon, H. G., Lee, K. H., Kim, H. Y., Kim, H. K., Shin, D. H., Hong, B. S., et al. (2002). Gene cloning and biochemical analysis of thermostable chitosanase (TCH-2) from Bacillus coagulans CK108. Biosci. Biotechnol. Biochem. 66 (5), 986–995. doi: 10.1271/bbb.66.986
Zhang, J. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18 (6), 292–298. doi: 10.1016/S0169-5347(03)00033-8
Keywords: Bacillus halotolerans LDFZ001, Rhizoctonia solani Kühn, antagonistic activity, sheath blight, wheat
Citation: Feng Z, Xu M, Yang J, Zhang R, Geng Z, Mao T, Sheng Y, Wang L, Zhang J and Zhang H (2022) Molecular characterization of a novel strain of Bacillus halotolerans protecting wheat from sheath blight disease caused by Rhizoctonia solani Kühn. Front. Plant Sci. 13:1019512. doi: 10.3389/fpls.2022.1019512
Received: 18 August 2022; Accepted: 03 October 2022;
Published: 17 October 2022.
Edited by:
Kostya Kanyuka, National Institute of Agricultural Botany (NIAB, United KingdomReviewed by:
Wei Wang, Chinese Academy of Tropical Agricultural Sciences, ChinaCopyright © 2022 Feng, Xu, Yang, Zhang, Geng, Mao, Sheng, Wang, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Zhang, anVhbnpoNzRAbGR1LmVkdS5jbg==; Hongxia Zhang, aHh6aGFuZ0BzaWJzLmFjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.