
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 September 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1016551
This article is part of the Research Topic Dissection of Physiological and Genetics Essences of Stress Resistance and High Yield in Triticeae Crops View all 8 articles
 Naicui Wei1†
Naicui Wei1† ShengQuan Zhang2†
ShengQuan Zhang2† Ye Liu1
Ye Liu1 Jie Wang1
Jie Wang1 Bangbang Wu3
Bangbang Wu3 Jiajia Zhao3
Jiajia Zhao3 Ling Qiao3
Ling Qiao3 Xingwei Zheng3
Xingwei Zheng3 Juanling Wang1,3*
Juanling Wang1,3* Jun Zheng1,3*
Jun Zheng1,3*In arid and semi-arid regions, coleoptile length is a vital agronomic trait for wheat breeding. The coleoptile length determines the maximum depth that seeds can be sown, and it is critical for establishment of the crop. Therefore, identifying loci associated with coleoptile length in wheat is essential. In the present study, 282 accessions from Shanxi Province representing wheat breeding for the Loess Plateau were grown under three experimental conditions to study coleoptile length. The results of phenotypic variation indicated that drought stress and light stress could lead to shortening of coleoptile length. Under drought stress the growth rate of environmentally sensitive cultivars decreased more than insensitive cultivars. The broad-sense heritability (H2) of BLUP (best linear unbiased prediction) under various conditions showed G × E interaction for coleoptile length but was mainly influenced by heredity. Correlation analysis showed that correlation between plant height-related traits and coleoptile length was significant in modern cultivars whereas it was not significant in landraces. A total of 45 significant marker-trait associations (MTAs) for coleoptile length in the three conditions were identified using the 3VmrMLM (3 Variance-component multi-locus random-SNP-effect Mixed Linear Model) and MLM (mixed linear model). In total, nine stable genetic loci were identified via 3VmrMLM under the three conditions, explaining 2.94–7.79% of phenotypic variation. Five loci on chromosome 2B, 3A, 3B, and 5B have not been reported previously. Six loci had additive effects toward increasing coleoptile length, three of which are novel. Molecular markers for the loci with additive effects on coleoptile length can be used to breed cultivars with long coleoptiles.
The coleoptile is a conical sheath-like structure that protects the young cotyledons and apical meristem of monocotyledonous plants during germination and emergence (Pucciariello, 2020). The wheat coleoptile is composed of inner and outer epidermis with vascular bundles and long and narrow cells, mostly parenchyma, covered by stratum corneum (Hohl and Schopfer, 1992; Liao et al., 2011). In arid and semi-arid regions, coleoptile length is a vital agronomic trait in wheat breeding. It is critical for wheat establishment because its length determines the maximum depth that seeds can be sown (Li et al., 2017). Wheat seedling emergence and survival in arid areas is dependent on sufficient soil moisture. Seedlings emergence will be uneven or even fail when the soil moisture content is lower than 16–18% (Li et al., 2014). Especially in years with less rainfall, wheat tends to be sown deeper (Mohan et al., 2013; Francki et al., 2021). Wheat cultivars with long coleoptiles can be sown deep to ensure that the seeds can absorb enough moisture for germination and stand establishment, thereby enabling normal yield (Anzooman et al., 2018). Therefore, dissecting the genetic basis of coleoptile length is important for the improvement of wheat in arid and semi-arid regions.
Differences in moisture and temperature leads to the differences in coleoptile length in various regions. The wheat growing areas of the north central United States have a continental climate, with severe winters, hot summers, and a short rainy season. There is no direct and indirect interference in breeding, so the average length of wheat coleoptiles was about 7 cm (Sidhu et al., 2020). However, the rainfall in southeast and southwest of Australia was evenly distributed, with a long rainy season. Wheat cultivars with long coleoptiles were preferred in breeding, resulting in cultivars with an average coleoptile length of 8–11 cm (Rebetzke et al., 2001, 2014). The coleoptile length of wheat is a quantitative trait under polygenic control. It is influenced by both genotype and environment and can be adjusted by genetic improvement (Yang et al., 2007). With the development of molecular biology and genomics in the field of wheat breeding, some progress had been made in QTL mapping of coleoptile length. Quantitative trait loci (QTL) for traits related to coleoptile length have been reported on all chromosomes by using bi-parental and multi-parental mapping populations. The pleiotropic genes Rht1 and Rht2, located, respectively, on chromosomes 4B and 4D, reduce both coleoptile length and plant height (Rebetzke et al., 2004, 2007a). Using recombinant inbred lines (RILs), main effect QTL related to coleoptile length, such as qCL.3B.1 and qCL.4B.1, were found to account for 6.6–19.9% of the phenotypic variation (Singh et al., 2015). Also using RILs, a major QTL on chromosomes 4DL was found to explain 33% of phenotypic variation, and Rht1 and Rht2 were also detected (Yu and Bai, 2010). Genome-wide association study (GWAS) is another widely used approach that can reduce the limitations of bi-parental or multi-parental QTL identification. The dwarf genes Rht1 and Rht2 were located via association analysis using a global wheat collection of 893 accessions grown in various environments (Li et al., 2016). Seventeen marker-trait associations (MTAs) for coleoptile length were identified in 707 Chinese landraces, explaining 2.73–8.17% of the phenotypic variation (Ma et al., 2020). A study in the United States using association analysis of a diverse population of 473 wheat cultivars and a population of 279 wheat breeding lines identified three major markers of environmental stability for coleoptile length, which explained 4–7% of the phenotypic variation (Merrick et al., 2021). However, there are few studies reported using GWAS to identify novel loci for coleoptile length. Few of these GWAS literatures address the relationship between coleoptile length and plant height-related agronomic traits. Although association analysis has many advantages, such as being less time-consuming with large allele coverage and high mapping accuracy, the false positive rate of the loci identified is high (Zhang et al., 2006; Yang et al., 2007). In addition, most models of association analysis do not decompose additive and dominant effects, resulting in loci that cannot be stably identified across environments (Wang et al., 2016; Wen et al., 2018). The 3VmrMLM (3 Variance-component multi-locus random-SNP-effect Mixed Linear Model) distinguishes various confounding effects by pre-estimating the genotype effect, and is more conducive to the detection of loci (Li et al., 2022). Only a few studies have used GWAS to identify novel loci for coleoptile length compared with other major agronomic traits such as plant height, thousand-grain weight, and grain number per spike. Furthermore, some of the markers used were SSR, which are few in number and low in density, and there is no report on the application of markers to breeding for coleoptile length (Spielmeyer et al., 2007). Therefore, using more effective methods to identify QTL for coleoptile length is of great significance for wheat improvement.
The Loess Plateau is famous for the world’s largest and deepest loess deposits (Fu et al., 2017). Shanxi Province in China is in the Loess Plateau, accounting for more than a quarter of the Loess Plateau’s total area (Wang et al., 2022). This region has a long history of wheat cultivation, but water resources are scarce. Shanxi province has annual rainfall between 400 and 650 mm, which is typical for semi-arid regions (Zheng et al., 2021). Therefore, Shanxi wheat cultivars are suitable for studying QTL for wheat drought resistance-related agronomic traits. The present study is needed because (1) the number of loci and markers for wheat coleoptile length is low compared to other agronomic traits, (2) the relationship between coleoptile length and plant height-related agronomic traits is unclear, and (3) it is unclear whether light and dark treatments affect coleoptile length differences among genotypes. With a GWAS approach, 241 modern cultivars, 41 landraces, and 3VmrMLM analysis were used to identify significant loci of coleoptile length, and provide a basis for molecular marker-assisted selection for coleoptile length in wheat.
A diverse hexaploid wheat collection of 282 accessions from Shanxi Province was used (Supplementary Table 1). These genotypes have significant phenotypic differences and include 126 irrigated and 115 dryland modern wheat cultivars. The remaining 41 experimental materials were landraces from Shanxi Province selected from the Chinese wheat core collection (Zheng et al., 2021).
Seeds of each line were carefully cleaned with 3% H2O2 and distilled water and evaluated in three treatments (C1–C3; Sidhu et al., 2020). C1 condition represents normal control with three replicates (E1–E3), C2 condition represents drought treatment with two replicates (E5–E6), and C3 condition represents light treatment with two replicates (E8–E9). E4, E7, and E10 are the mean values of each treatment, respectively. C1:E1–E4 20°C in darkness with 150 ml distilled water; C2:E5–E7 20°C in darkness with 150 ml, 20% PEG-6000 hypertonic solution to simulate drought stress; C3:E8–E10 20°C, 12 h darkness + 12 h light at 500μmol⋅m–2⋅s–1 with 150 ml distilled water. In each replicate, twenty seeds per genotype were placed and germinated on a filter paper measuring 40 cm × 60 cm. After seven days, the coleoptile lengths of five individual plants for each cultivar were measured. Coleoptile length was recorded as the distance from the seed to where the first leaf broke through the coleoptile sheath (Bai et al., 2004). For all 282 accessions, data were collected for plant height, uppermost internode length, and internode length of basal sections I–IV from 2020 to 2021 field plots grown in Yaodu District (P1:36°48′N, 111°30′E) and Yanhu District (P2:35°20′N, 110°59′E) in Shanxi Province.
Genomic DNA was extracted using the cetyl trimethyl ammonium bromide (CTAB) method (Zheng et al., 2019). DNA was sent to the MOL-BREEDING Company (Shijiazhuang, China) for high-throughput genotyping using wheat GenoBaits technology (14868 mSNP, 37669 SNP). After eliminating markers with an allele frequency (MAF) < 0.05, > 10% missing data, or > 20% heterozygosity (Arora et al., 2017; Jung et al., 2021), a total of 9,793 high-quality SNPs were identified for GWAS analyses (Zheng et al., 2021).
Two models were used for association analysis between coleoptile length and SNP markers. Marker-trait associations were tested using the 3VmrMLM in the RStudio software and markers with a threshold of LOD score ≥3.99 were considered as significant associations (Wang et al., 2016; Li et al., 2022). Similarly, marker-trait associations were tested using the mixed linear model (MLM) in TASSEL 5.0 software and markers with a threshold of -log10(p-value) ≥ 3.99 were used as the screening criterion (Zhang et al., 2010; Li et al., 2016). GWAS was conducted with ten datasets, including E1 to E10. Manhattan was generated via RStudio software to visualize the GWAS results. The linkage disequilibrium (LD) of each SNP marker was extended on each chromosome (Zheng et al., 2021). The extended region where the LD between nearby SNPs and the peak SNP decayed to R2 = 0.2 was defined as the local LD-based QTL interval (Zheng et al., 2019). For each associated locus, the p-value and QTL intervals of the peak SNP defined the significance and the interval of the locus (Zhang et al., 2016).
To facilitate the identification of genes in functional regions, the chromosomal region was delimited at 6.11 Mb (Yao et al., 2021). Genes within the target region were identified using the genome browser (JBrowse) on the WheatOmics-bata website1 (Ma et al., 2021). The sequences of common wheat genes were retrieved based on the intervals of major QTL identified from https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Annotations/v1.0/ (Zheng et al., 2019). Functional annotation of genes in segments were carried out in the Gene Ontology (GO) database using the R package cluster Profiler. The RNA-seq of genes within the target region were retrieved from Wheat Expression Browser.2 Statistical calculations of phenotypic data were performed by SAS V8.03 (Smith et al., 1998). BLUP calculation, phenotypic variance analysis, and correlation analysis were performed by JMP Pro164 and IBM SPSS Statistics 26 software.5 Data was plotted via RStudio6 and Origin 2018.7
Coleoptile length within the 282 wheat accessions was normally distributed across in all ten datasets (Supplementary Figure 1). Under the C1 condition, coleoptile lengths ranged from 2.80 to 7.03 cm and the coefficient of variation was 17.78%, indicating that there were significant differences in coleoptile length among the test materials. Compared with C1, coleoptile lengths in C2 and C3 decreased by 19.21 and 20.83%, respectively (Figure 1 and Supplementary Table 2).
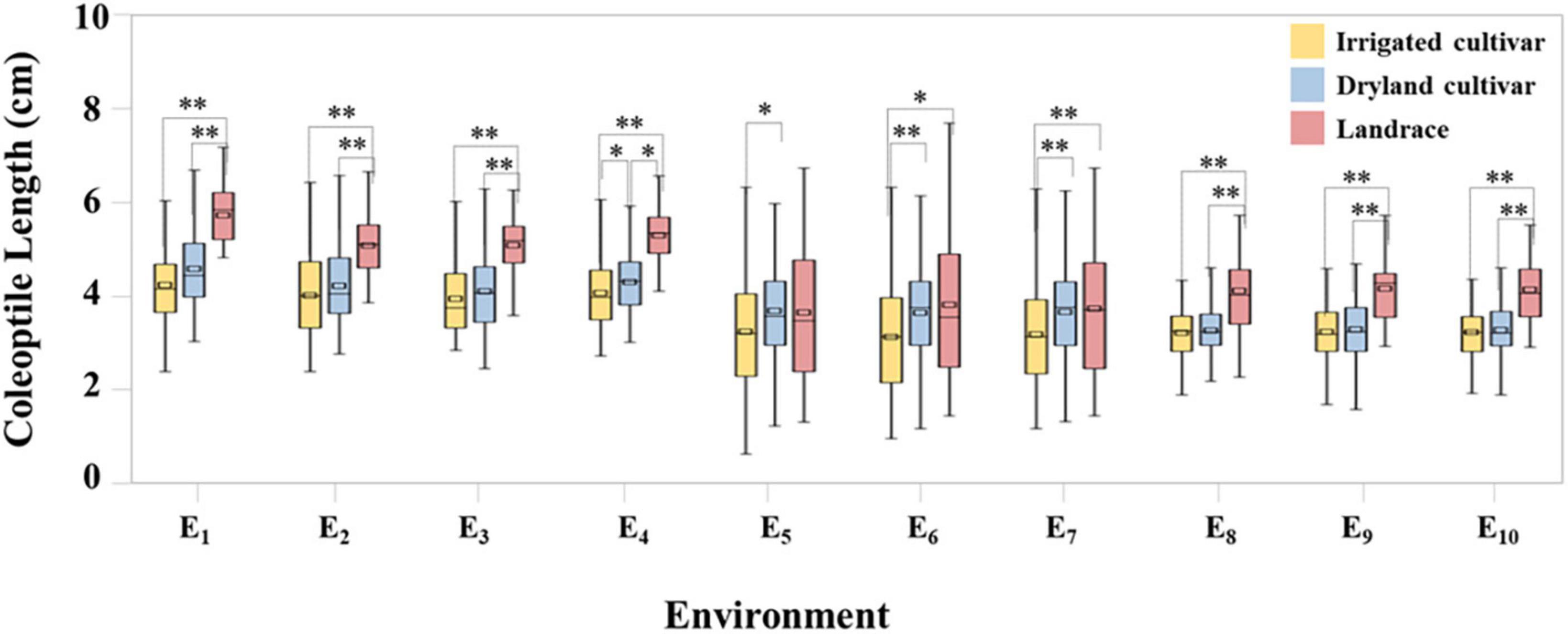
Figure 1. Coleoptile length of irrigated cultivars, dryland cultivars, and landraces. * and ** represent significance level of P < 0.05 and P < 0.01, respectively.
In C1 the coleoptile length of landraces was the greatest followed by dryland cultivars with the coleoptile lengths of the irrigated cultivars being the shortest. Among the test materials, the coleoptile of Yulanmai was the longest and that of Shengmai 104 was the shortest. In C2 the greatest decrease in coleoptile length occurred in landraces (28.73%) versus dryland cultivars (14.12%) or irrigated cultivars (20.78%), indicating that landraces were more sensitive to drought stress and dryland cultivars were more adapted to stress conditions (Table 1). Linfeng 3 and Chang 5222 were the most sensitive to drought stress, with a decrease in coleoptile length by 77.31 and 67.41%, respectively. In C3, the coleoptile length of most cultivars was reduced indicating that light had a significant effect on the coleoptile length (Table 1). In the field, the wheat coleoptile is generated in a darkness within the soil, therefore darkness must be used experimentally to simulate the natural environment. Seven cultivars, including Jinmai 47, Zeyou 2 (Jinmai 76), Changmai 6686, Jintai 9923, Jinmai 102, Chang 5804 and Chang 6154, showed no significant differences in coleoptile length under the three conditions and were thus insensitive to the treatment conditions.
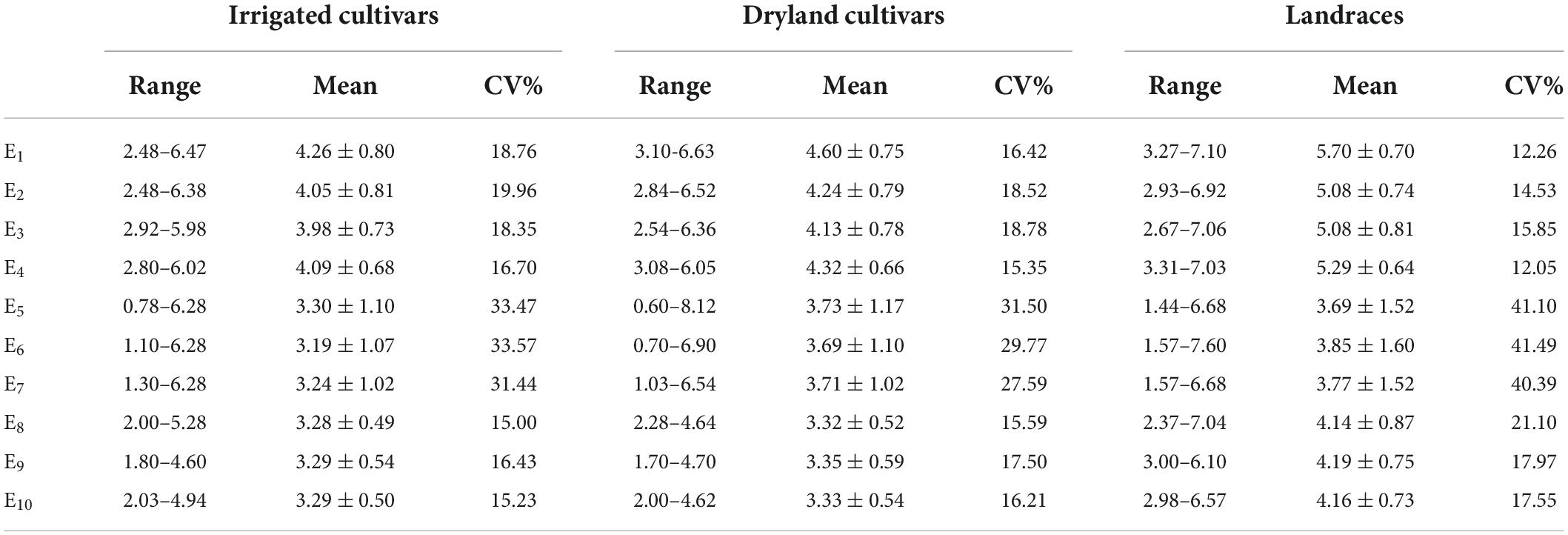
Table 1. Coleoptile length of irrigated cultivars, dryland cultivars, and landraces in various environments.
Broad-sense heritability (H2) is defined in each environment and BLUP value. The estimated H2 for the coleoptile length was greater than 85% (H2(C1) = 87.62%; H2(C2) = 87.17%;H2(C3) = 91.84%) under the three conditions, indicating the trait was mainly influenced by heredity. H2 of BLUP value was 66.43%, indicating that phenotypic variation was affected by environmental factors. The different H2 values showed that there was G × E interaction for coleoptile length, but the effect of genotype was greater than that of environment. Both C2 and C3 produced a significant reduction in coleoptile length compared with C1, indicating that environmental differences caused overall phenotypic variation (Supplementary Table 2). Pearson’s coefficient of correlation in the three conditions, C3 was the highest, C2 was the second, and C1 was the lowest (Supplementary Table 3). The correlation coefficients of coleoptile length among environments were highly significant and ranged from 0.23 to 0.96 (P < 0.01), which also indicated that coleoptile length was affected by the environment.
The accessions were divided into two types according to the DTC (drought tolerance coefficient) values. The coleoptile length of the insensitive cultivars such as Jinmai 47, Jinmai 919 and Linyou 2069 did not change significantly across environments. The sensitive cultivars such as Chang 6388, Shengmai 20 and Yulanmai showed clear changes in the coleoptile length across environments. Jinmai 47 has been used as a trial reference in the dryland wheat regional trial in the northern part of Huanghuai wheat region since it was released in the 1990s (Zheng et al., 2018). Yulanmai is an excellent landrace that was widely grown in the 1950s and 1960s. By investigating the dynamic changes of coleoptile length of Jinmai 47 and Yulanmai over seven days (Figure 2), the overall trend of the two cultivars was the same with the growth rate highest from day 2 to day 3, and the coleoptile length reaching a maximum from day 5 to day 6. However, the growth rate changes differed. The growth rate of Jinmai 47 under C1 was higher than that under C2 from day 1 to day 3 and lower than that under C2 from day 3 to day 6. Specifically, the maximum coleoptile length under stress conditions decreased by 3.99% compared with C1. However, the growth rate of Yulanmai under C2 from day 1 to day 5 was lower than that under C1 and the maximum coleoptile length decreased by 10.67% compared with C1. Thus, the differences in the growth rate of the cultivars under drought stress led to different coleoptile lengths.
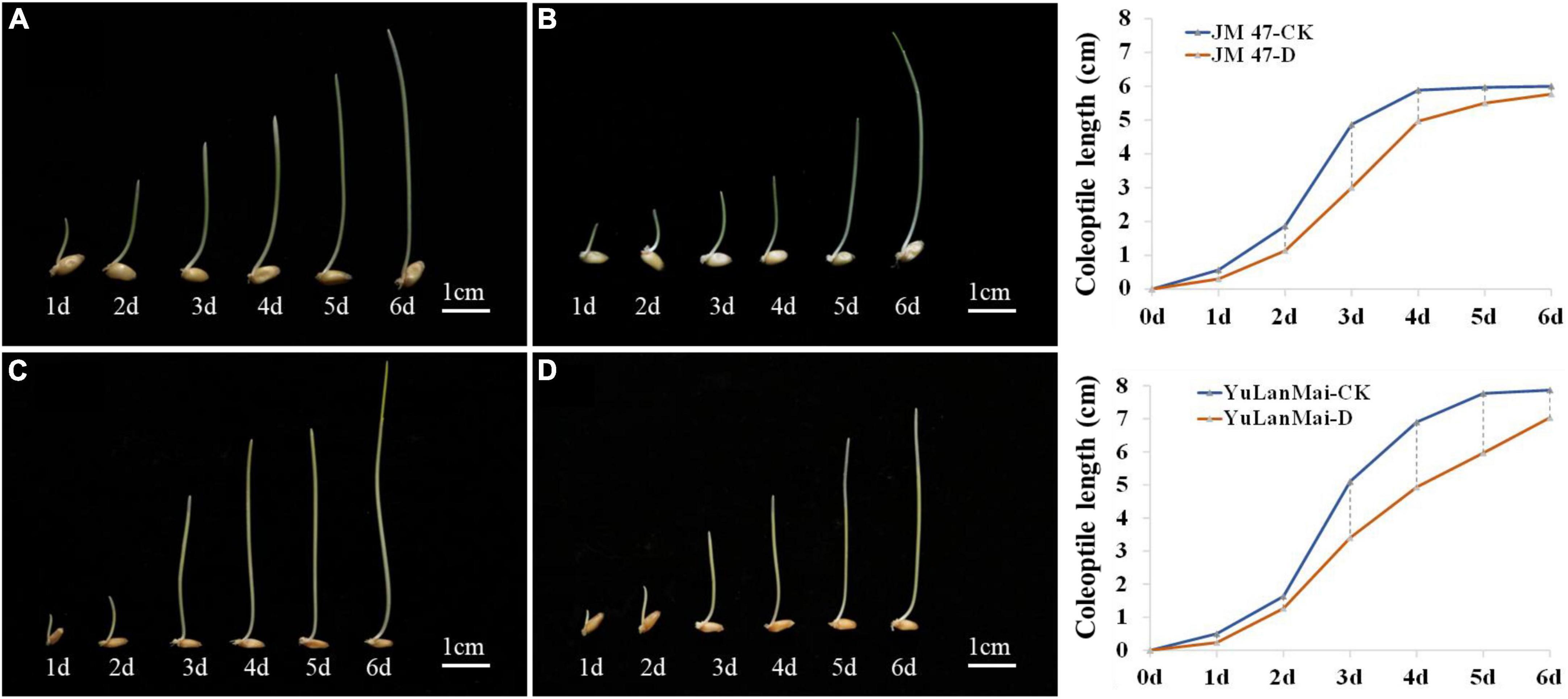
Figure 2. Dynamic changes of coleoptile length in a growth cycle. (A) Growth dynamics of Jinmai 47 under C1; (B) Growth dynamics of Jinmai 47 under C2; (C) Growth dynamics of Yulanmai wheat under C1; (D) Growth dynamics of Yulanmai under C2; The graph on the right is the growth curve of Jinmai 47 and Yulanmai respectively (X is the growth days, Y is the coleoptile length).
To determine whether the coleoptile length of the 282 wheat accessions was associated with plant height-related traits, correlation analyses was performed between coleoptile length and each of the assessed agronomic traits. The coleoptile length was significantly positively correlated (P < 0.05, P < 0.01) with plant height, internode length below the spike, and the internode length of basal sections I–IV (Supplementary Table 4). The correlation coefficients between coleoptile length and the plant height-related traits were significant in modern cultivars, but not significant in landraces (Supplementary Tables 5–7). The highest correlation coefficient was between coleoptile length and plant height. The Pearson’s coefficient of correlation between coleoptile length and the length of each stem section was higher in basal sections II and III, followed by that for the lower internode below the spike and basal section IV with the lowest correlation for basal section I (Supplementary Table 4). Except for basal sections I and IV, the correlation in the irrigated cultivars was significantly higher than in dryland cultivars. These results indicate a greater coleoptile length at the seedling stage of wheat may result in an increase of the plant height and the length of each stem section at the adult stage.
A total of seven MTAs were detected using MLM, but only 1A_298114344 was stably associated (Supplementary Table 8). A total of 53 MTAs distributed on all chromosomes except 3D, 4B, 4D, and 7D were identified via 3VmrMLM. Among these, 13, 34, and 6 were on genomes A, B, and D, respectively, and the phenotypic variation explanation rate (R2) ranged from 2.69 to 14.23% (Supplementary Table 9). 3VmrMLM detected more stable loci and previously reported loci, possibly because all types of effects can be estimated via 3VmrMLM (Li et al., 2022). 3VmrMLM first scanned all markers at the genome-wide level to obtain potential loci, put them into the multi-locus model for selection, and obtained significant loci (Li et al., 2022). Therefore, 3VmrMLM is better at detecting stable loci compared with MLM. Five MTAs were detected by both models.
In total, nine stable genetic loci were identified via 3VmrMLM under three conditions (Table 2 and Figure 3). Three loci with R2 from 3.47 to 5.72% were identified under C1, three loci with R2 from 4.23 to 7.79% were identified under C2, and two loci with R2 from 3.80 to 5.33% were identified under C3. 3B_810370914 with an R2 from 2.94 to 4.30% was detected in both C1 and C2. According to the Zheng et al. (2021) analysis of LD attenuation distance, the MTAs 4A_597823528 and 4A_604382166 were the same locus and explained 3.47–4.00% of the phenotypic variation. The loci significantly associated with 2B_60980523, 3A_650566192, 3B_519815931, 5B_68179721, and 5B_610798888 have not been reported previously.
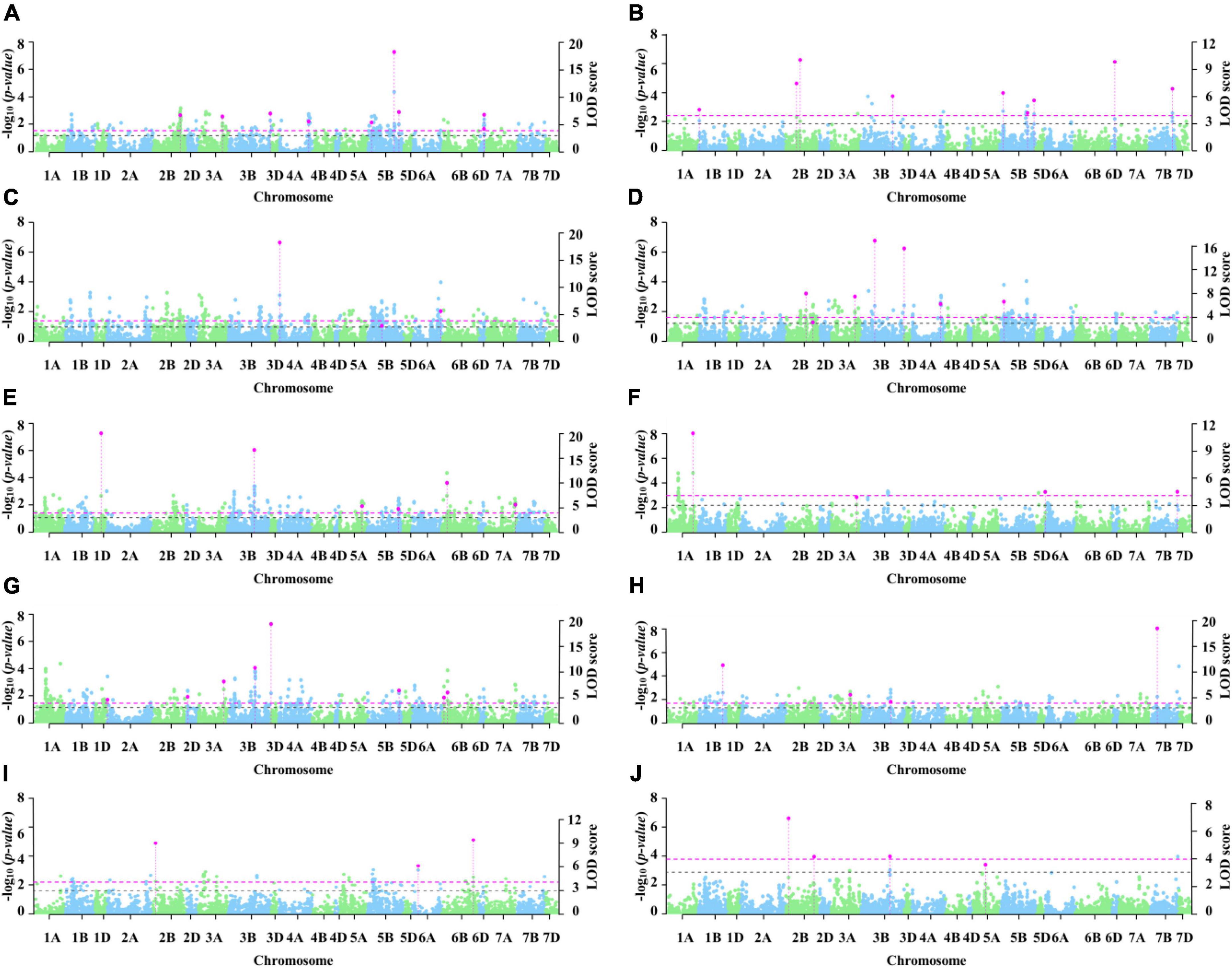
Figure 3. Manhattan plot of significant loci for coleoptile length. The left y axis reports -log10(p-values), which are obtained from single-marker genome-wide scanning for all the markers in the first step of 3VmrMLM, and the right y axis reports LOD scores, which are obtained from a likelihood ratio test for significant and suggested QTL in the second step of 3VmrMLM. In the Manhattan plots, the dashed pink line represents the threshold LOD = 3.99 and the dashed black line represents the threshold LOD = 3.0 (A–J represent E1, E2, E3, E4, E5, E6, E7, E8, E9, and E10, respectively).
To understand the biological significance among genes, these genes were annotated by Gene Ontology (GO) analysis. Overall, 514 high confidence genes from the nine functional intervals were annotated. We found that the GO domains were mainly enriched in the molecular function, including binding and catalytic activity. In the region harboring 2B_60980523, five of these genes were involved in response to oxidative stress according to gene functional annotations in the GO public database. In the region harboring 3B_559967593, three genes encoding expansin, including TraesCS3B02G347800, TraesCS3B02G347900, and TraesCS3B02G348000. In the region harboring 3B_810370914, we identified three genes encoding peroxidase, including TraesCS3B02G577900, TraesCS3B02G578000, and TraesCS3B02G578600. TraesCS4A02G298900 on chromosome 4B was involved in the regulation of seed germination. In addition, we identified seven genes that respond to defense conditions, including TraesCS3B02G579300, TraesCS3B02G579400, TraesCS3B02G584700, TraesCS3B02G349500, TraesCS 3B02G349600, TraesCS3B02G349600, TraesCS3B02G349800. TraesCS5B02G439000 was associated with a light intensity stimulus.
The expression levels of genes in functional interval in various tissues were analyzed. The results revealed high variation among the expression patterns of different tissues. Some genes in functional interval had higher expression levels in coleoptiles, and these genes had also higher expression in first leaf, stems axis and shoots. However, some genes in functional interval had relatively high expression levels in the coleoptiles, while these genes were expressed at relatively low levels in roots, endosperm, and radicle (Figure 4).
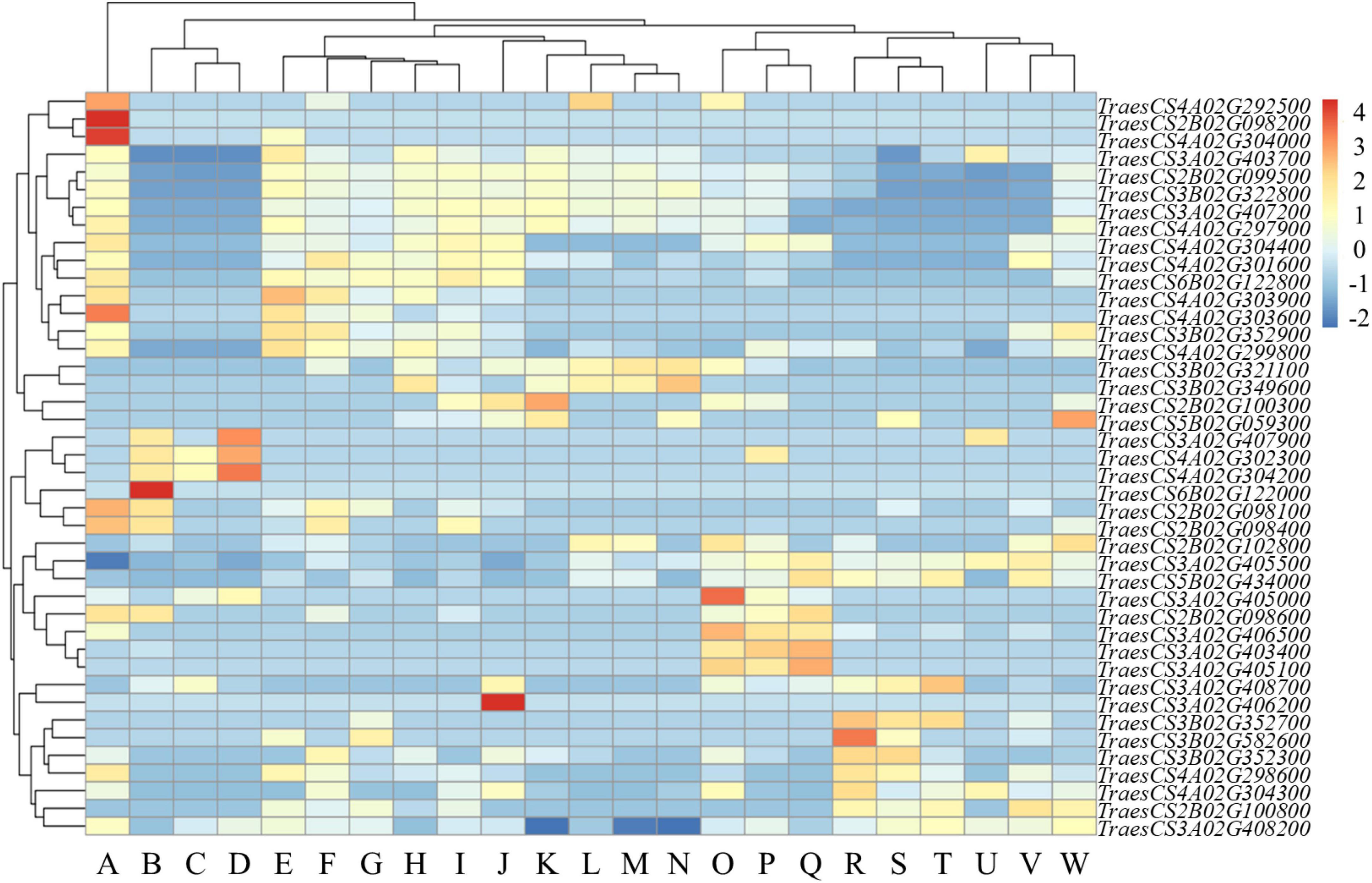
Figure 4. Expression profile of functional interval genes. Based on the Z-score of the TPM plotting Heatmap, – 2 to 4 represent expression levels from low to high. (A: coleoptile; B: grain; C: endosperm; D: aleurone layer; E: stem axis; F: first leaf sheath; G: seedling; H: first leaf blade; I: shoots; J: leaf; K: flag leaf; L: glumes; M: awns; N: flag leaf blade; O: lemma and rachis; P: spikelets; Q: stigma or ovary; R: radicle; S: roots; T: root apical meristem; U: embryo; V: shoot apical meristem; W: stem).
We found that TraesCS2B02G098200 was only expressed on coleoptiles (Figure 4). TraesCS2B02G098200 is homologous to OsPrx116 (Os07g49360) in rice. The functional annotation of TraesCS2B02G098200 was for the peroxidase activity. Peroxidase is an important antioxidant in plants, which can alleviate oxidative damage under abiotic stress (Gao et al., 2010; Qayyum et al., 2021). Therefore, the specific expression of TraesCS2B02G098200 gene in the coleoptile could improve the tolerance of wheat to stress, thereby maintaining early vigor.
There was a significant difference in the trend of DTC values between irrigated cultivars and dryland cultivars with year of release. DTC values were higher in dryland cultivars (0.83) than in irrigated cultivars (0.79). Figure 5A, indicating the effect of artificial selection by dryland cultivars was higher than in modern cultivars. The overall DTC value trend over time for dryland cultivars was stable at 0.85 before the 2010s. The DTC value of dryland cultivars increased to 1.04 after 2010s, indicating that breeders further increased the effect of artificial selection. The DTC values of irrigated cultivars decreased over time, which mainly due to the improvement of soil irrigation level and the decrease of attention by breeders to coleoptile length.
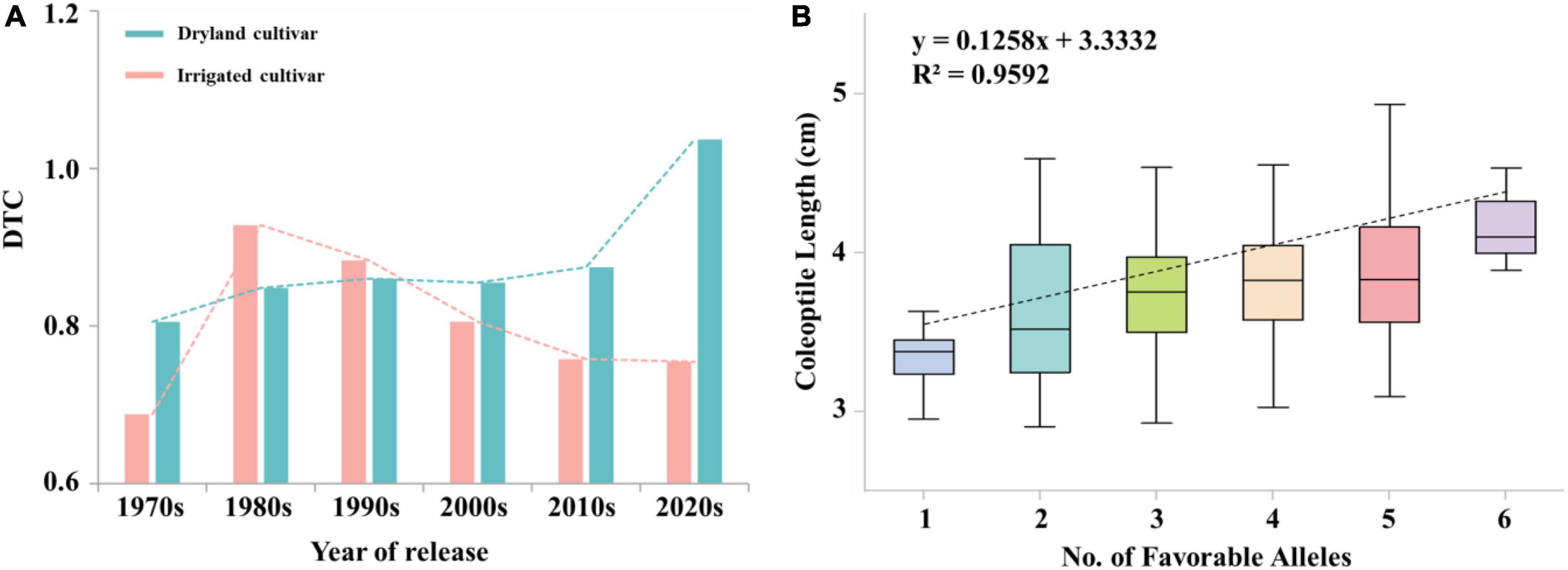
Figure 5. (A) The trend of DTC values for irrigated cultivars and dryland cultivars over time. (B) Linear regression between the number of favorable alleles in the population and coleoptile length. X and Y in the equation represent the number of favorable alleles and the coleoptile length, respectively.
The frequency of favored alleles within the 282 materials was the highest in landraces (58.81%) and lowest in irrigated cultivars (40.44%). Huoshaotou had the highest frequency of favored alleles (88.89%) and exhibited a greater coleoptile length. In seven cultivars, including Jinmai 47, Zeyou 2 (Jinmai 76), Changmai 6686 and Jintai 9923, the favorable alleles had a significant positive effect at 3B_810370914, 4A_597823528, 5B_681790721. In addition, the proportion of favored allele for each locus was different, which indicated that these important loci had experienced different degrees of selection during wheat breeding. For example, the frequencies of favored alleles for loci 3B_810370914 and 4A_597823528 were 93.26 and 86.17%, respectively, whereas the frequency for locus 3B_519815931 was only 8.87%.
Drought is the main limiting factor for increasing yield, especially at the seedling stage of wheat (Sadok, 2017; Lin et al., 2019). In arid areas, longer wheat coleoptiles help overcome water limitation be enabling deeper planting for better stand establishment. This trait has been widely recognized by breeders (Ma et al., 2020). The coleoptile length of landraces in Shanxi Province is longer, making them more suitable for local semi-arid climate (Figure 1). Wheat with long coleoptiles emerged more frequently than wheat with short coleoptiles, especially when sown deep (Rebetzke et al., 2005, 2007a,2007b). Deeper sowing also assists in reducing removal of seeds by birds and rodents and in avoiding phytotoxicity associated with some pre-emergent herbicides (Brown et al., 2003). Selection of wheat cultivars with long coleoptile is an important component of improving emergence, weed suppression and grain yield in low rainfall regions (Farhad et al., 2014). Thus, the coleoptile length is essential for successful emergence and early plant vigor in arid and semi-arid regions. Nutrients in seeds are consumed during the growth of coleoptile. The main factor affecting coleoptile elongation is moisture (Rebetzke et al., 2007a; Sun et al., 2022). In the present study, the growth states of coleoptiles under various conditions were consistent in general, but the growth rates were quite different. Coleoptile elongation is mainly the extension of coleoptile cells. The turgor pressure distributed in the epidermis and internal tissues provides the impetus for cell elongation (Li and Wang, 1996; Chen and Chen, 2002). The expansion of coleoptiles is closely related to cell wall elongation, keeping the cell wall in a relaxed state (Gao et al., 2008). Under water stress, cell elongation is inhibited due to decreased sensitivity of cell wall (Wang et al., 1999). For example, the longitudinal growth of the coleoptiles of Jinmai 47 and Yulanmai under normal water conditions (C1) were maintained by continuous turgor pressure, and the cell wall did not significantly restrict elongation. Under water limited conditions (C2), the elongation growth of Jinmai 47 and Yulanmai was restricted by the cell wall and the maximum elongation rate was significantly reduced. Yulanmai was sensitive to water deficit, whereas Jinmai 47 was not significantly affected. Compared with Jinmai 47, the growth rate of Yulanmai decreased significantly during the growth of coleoptiles, resulting in a significant shortening of coleoptile length. Thus, the coleoptile length is one of the indicators of drought resistance ability and it can be measured at the seedling stage for selection purposes.
No consistent relationship between coleoptile length and plant height has been established. For 124 European modern winter wheat cultivars, plant height and coleoptile length showed a significant positive correlation, but the correlation between coleoptile length and the length of each section in the stem was not analyzed (Liatukas and Ruzgas, 2011). There was no correlation between coleoptile length and plant height for 707 Chinese landraces in China (Ma et al., 2020). Landraces were used in the present study as well as modern cultivars, which are more easily used in breeding. Consistent with previous studies, the correlation between plant height-related traits and coleoptile length was significant in the modern cultivars but not significant in the landraces. The coleoptile length of the modern cultivars was positively correlated with the plant height- related traits (P < 0.05 or P < 0.01). Thus, the increase of the coleoptile length of the modern cultivars could possibly promote the increase of the length of each stem section at the adult stage, and eventually lead to the increase of the plant height.
A total of nine stable loci were identified via 3VmrMLM in all ten datasets. The B genome may have more genes controlling coleoptile length (Sidhu et al., 2020). In the present study, 58.14% of the MTAs were located on the B genome and only 27.91% were located on the A genome, which supports the important role of the B genome for the coleoptile length trait. Three loci identified in the present study, 3B_810370914, 4A_597823528, and 6B_118976998, likely have been reported previously. Rebetzke et al. (2007a,2014) reported the DArT marker gwm637 and SNP marker wsnp_Ex_c13615_21393511 for coleoptile length on chromosome 4A. These markers coincide with the physical location of 4A_597823528. Ma et al. (2020) reported the locus QCl. sicau-3B.2 for coleoptile length on chromosome 3B. This marker coincides with the physical location of 3B_810370914, indicating that these two markers likely represent the same locus. Similarly, QCI.sicau-6B.1 coincided with the physical location of 6B_118976998, which indicated that two markers likely represent the same locus. In addition, 3B_559967593 and 6B_118976998 were reported to be significantly associated with plant height (Zanke et al., 2014). In the present study, 3B_559967593 was also identified as associated with coleoptile length and was located 6.31 Mb distant from the marker BobWhite_c19725_1329 of plant height on chromosome 3B (Zanke et al., 2014). Similarly, we also identified 6B_118976998, which was approximately 3.03Mb distant from the marker BS00084314_51 of plant height on chromosome 6B (Zanke et al., 2014). The results indicate that 3B_559967593 and 6B_118976998 are pleiotropic, affecting both plant height and coleoptile length (Zanke et al., 2014). The loci associated with coleoptile length located on 2B (60.98 Mb), 3A (650.57 Mb), 3B (519.82 Mb), and 5B (68.18 Mb and 610.80 Mb) have not been reported previously and are likely novel. These results not only show the accuracy of the association analysis in the present work, but also confirm the high efficiency of 3VmrMLM for QTL mapping. The phenotypic variation explanation rate of these loci ranged from 2.94 to 7.79%, which is consistent with results from a previous study (Merrick et al., 2021). Among the markers identified, 6B_118976998 had the highest effect, which could explain up to 7.79% of the phenotypic variation, followed by 3B_519815931, which explained 6.51%. There is a significant correlation between plant height and coleoptile length in wheat with dwarfing genes compared to wheat without dwarfing genes (Worland and Sayers, 1995; Rebetzke et al., 2007a). Five MTAs were detected by both 3VmrMLM and MLM. Among these MTAs, we identified 7B_702845873, which was about 8.39 Mb distant from the Rht13 gene (Ellis et al., 2005). Li et al. (2015) and Tian et al. (2017) reported the markers Xbarc103 and Xwmc256 for the dwarf gene Rht24 (QPH.caas-6A) on chromosome 6A. These markers coincided with the physical location of 6A_585345446 in the present study, indicating that all three markers are linked to the Rht24 locus. These results indicated that the dwarfing genes Rht13 and Rht24 are associated with reductions in both plant height and coleoptile length. Therefore, the positive correlation between coleoptile length and plant height parameters among Chinese modern cultivars and its absence in Chinese landraces could be due to the Green Revolution in China in the 1960s, which was the result of breeding (Peng, 1999; Li and Shan, 2022).
Breeders seek to maintain long coleoptile lengths in semi-dwarf genotypes (Li et al., 2017). However, it is a challenge to breed semi-dwarf long-coleoptile wheat cultivars, which due to the complex relationship between coleoptile lengths and plant height. Landraces have wide adaptability to local environments, which had the highest numbers of favorable alleles. The number of favored alleles in dryland cultivars was slightly higher than that in irrigated cultivars among the modern cultivars, which was consistent with the shorter coleoptile length and lower favorable allelic variation frequency for the trait in irrigated cultivars. This result is likely because dryland cultivars are dependent on rainfall, whereas irrigated cultivars are dependent on soil irrigation levels. Since the advent of the Green Revolution, the negative selection of coleoptile favorable alleles has been excessive during positive selection for reduced plant height (Spielmeyer et al., 2007). Therefore, the breeding of long coleoptiles in wheat should focus on dryland cultivars and landraces in the future, which could enable combining longer coleoptiles with semi-dwarf plant height. Among seven cultivars that were insensitive to the treatment conditions in the present study the number of favorable alleles in Jinmai 47, Zeyou 2 (Jinmai 76), Changmai 6686, and Chang6154 was higher than in other three cultivars, and loci 3B_810370914, 4A_597823528, and 5B_68179721 all had favorable alleles. This result may explain the high seedling emergence rate and stress resistance of Jinmai 47, which has been used as a trial reference in the dryland wheat regional trials. Six loci had additive effects toward increasing coleoptile length, including 3A_650566192, 3B_559967593, 3B_810370914, 4A_597823528, 5B_68179721, and 5B_610798888. The average coleoptile length increased as the number of favored alleles increased (Figure 5B). Among the six loci, 3A_650566192, 5B_68179721, and 5B_610798888 are novel loci related to coleoptile length and these markers can be used to develop molecular markers for selecting long coleoptile cultivars.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
NW and SZ performed the phenotypic evaluation, conducted data analysis, and drafted the manuscript. YL, JiW, BW, JiZ, and LQ performed the phenotypice valuation and helped with data analysis. JiZ, LQ, XZ, and JuZ helped to draft the manuscript. JuZ and JuW designed and coordinated the study and revised the manuscript. All authors have read and approved the final manuscript for publication.
This work was supported by the research program sponsored by the State Key Laboratory of Sustainable Dryland Agriculture (in preparation), Shanxi Agricultural University (No. 202002-1), and the Shanxi Province Research Program (20210302124505).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1016551/full#supplementary-material
Anzooman, M., Dang, Y. P., Christopher, J., Mumford, M. H., Menzies, N. W., and Kopittke, P. M. (2018). Greater emergence force and hypocotyl cross sectional area may improve wheat seedling emergence in sodic conditions. Plant Sci. 277, 188–195. doi: 10.1016/j.plantsci.2018.09.007
Arora, S., Singh, N., Kaur, S., Bains, N. S., Uauy, C., Poland, J., et al. (2017). Genome-wide association study of grain architecture in wild wheat Aegilops tauschii. Front. Plant Sci. 8:886. doi: 10.3389/fpls.2017.00886
Bai, G., Das, M. K., Carver, B. F., Xu, X., and Krenzer, E. G. (2004). Covariation for microsatellite marker alleles associated with Rht8 and coleoptile length in winter wheat. Crop Sci. 44, 1187–1194. doi: 10.2135/cropsci2004.1187
Brown, P. R., Singleton, G. R., Tann, C. R., and Mock, I. (2003). Increasing sowing depth to reduce mouse damage to winter crops. Crop Prot. 22, 653–660.
Chen, A. G., and Chen, J. H. (2002). The mechanism of coleoptiles elongation and its physiological and ecological responses. J. Shandong Agric. Univ (Natural Science). 33, 438–441.
Ellis, M. H., Rebetzke, G. J., Azanza, F., Richards, R. A., and Spielmeyer, W. (2005). Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theor. Appl. Genet. 111, 423–430. doi: 10.1007/s00122-005-2008-6
Farhad, M., Hakim, M. A., Alam, M. A., and Barma, N. C. D. (2014). Screening wheat genotypes for coleoptile length: A trait for drought tolerance. Am. J. Agric. For. 2237. doi: 10.11648/j.ajaf.20140206.11
Francki, M. G., Stainer, G. S., Walker, E., Rebetzke, G. J., Stefanova, K. T., and French, R. J. (2021). Phenotypic evaluation and genetic analysis of seedling emergence in a global collection of wheat genotypes (Triticum aestivum L.) under limited water availability. Front. Plant Sci. 12:796176. doi: 10.3389/fpls.2021.796176
Fu, B., Wang, S., Liu, Y., Liu, J., Liang, W., and Miao, C. (2017). Hydrogeomorphic ecosystem responses to natural and anthropogenic changes in the Loess Plateau of China. Annu. Rev. Earth Planet. Sci. 45, 223–243. doi: 10.1146/annurev-earth-063016-020552
Gao, C., Wang, Y., Liu, G., Wang, C., Jiang, J., and Yang, C. (2010). Cloning of ten peroxidase (POD) genes from Tamarix hispida and characterization of their responses to abiotic stress. Plant Mol. Biol. Rep. 28, 77–89. doi: 10.1007/s11105-009-0129-9
Gao, Q., Zhao, M. R., Li, F., Guo, Q. F., Xing, S. C., and Wang, W. (2008). Expansins and coleoptile elongation in wheat. Protoplasma 233, 73–81. doi: 10.1007/s00709-008-0303-1
Hohl, M., and Schopfer, P. (1992). Physical extensibility of maize coleoptile cell walls: Apparent plastic extensibility is due to elastic hysteresis. Planta 187, 498–504. doi: 10.1007/BF00199968
Jung, W. J., Lee, Y. J., Kang, C. S., and Seo, Y. W. (2021). Identification of genetic loci associated with major agronomic traits of wheat (Triticum aestivum L.) based on genome-wide association analysis. BMC Plant Biol. 21:418. doi: 10.1186/s12870-021-03180-6
Li, G., Bai, G., Carver, B. F., Elliott, N. C., Bennett, R. S., Wu, Y., et al. (2017). Genome-wide association study reveals genetic architecture of coleoptile length in wheat. Theor. Appl. Genet. 130, 391–401. doi: 10.1007/s00122-016-2820-1
Li, G., Xu, X., Bai, G., Carver, B. F., Hunger, R., Bonman, J. M., et al. (2016). Genome-wide association mapping reveals novel QTL for seedling leaf rust resistance in a worldwide collection of winter wheat. Plant Genome 9. doi: 10.3835/plantgenome2016.06.0051
Li, L. C., and Wang, X. C. (1996). Effects of water deficit on plant cell wall and its relation to cell elongation. Plant Physiol. J. 32, 321–327. doi: 10.13592/j.cnki.ppj.1996.05.001
Li, M., Zhang, Y. W., Zhang, Z. C., Xiang, Y., Liu, M. H., Zhou, Y. H., et al. (2022). A compressed variance component mixed model for detecting QTNs and QTN-by-environment and QTN-by-QTN interactions in genome-wide association studies. Mol. Plant 15, 630–650. doi: 10.1016/j.molp.2022.02.012
Li, X., Xia, X., Xiao, Y., He, Z., Wang, D., Trethowan, R., et al. (2015). QTL mapping for plant height and yield components in common wheat under water-limited and full irrigation environments. Crop Pasture Sci. 66, 660–670. doi: 10.1071/CP14236
Li, Y. C., Yuan, S. J., Guo, X. M., Liu, J. M., Liang, H., Bai, Y. M., et al. (2014). Effect of soil moisture on growth and yield of winter wheat. J. Meteorol. Environ. 30, 90–97.
Li, Y. D., and Shan, X. H. (2022). Gibberellin metabolism regulation and green revolution. Biotechnol. Bull. 38, 195–204. doi: 10.13560/j.cnki.biotech.bull.1985.2021-0389
Liao, M. L., Peng, Z. S., Yang, Z. J., Wei, S. H., and Ouyang, Z. M. (2011). Effect of shortstalk gene on coleoptile growth of tetroploid wheat. Seed 30, 49–52. doi: 10.16590/j.cnki.1001-4705
Liatukas, Z., and Ruzgas, V. (2011). Coleoptile length and plant height of modern tall and semi-dwarf European winter wheat varieties. Acta Soc. Bot. Pol. 80, 197–203. doi: 10.5586/asbp.2011.018
Lin, Y., Yi, X., Tang, S., Chen, W., Wu, F., Yang, X., et al. (2019). Dissection of phenotypic and genetic variation of drought-related traits in diverse Chinese wheat landraces. Plant Genome 12, 1–14. doi: 10.3835/plantgenome2019.03.0025
Ma, J., Lin, Y., Tang, S., Duan, S., Wang, Q., Wu, F., et al. (2020). A genome-wide association study of coleoptile length in different Chinese wheat landraces. Front. Plant Sci. 11:677. doi: 10.3389/fpls.2020.00677
Ma, S. W., Wang, M., Wu, J. H., Guo, W. L., Chen, Y. M., Li, G. W., et al. (2021). WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 14, 1965–1968. doi: 10.1016/j.molp.2021.10.006
Merrick, L. F., Burke, A. B., Zhang, Z. W., and Carter, A. H. (2021). Comparison of single-trait and multi-trait genome-wide association models and inclusion of correlated traits in the dissection of the genetic architecture of a complex trait in a breeding program. Front. Plant Sci. 12(undefined):772907. doi: 10.3389/fpls.2021.772907
Mohan, A., Schillinger, W. F., and Gill, K. S. (2013). Wheat seedling emergence from deep planting depths and its relationship with coleoptile length. PLoS One 8:e73314. doi: 10.1371/journal.pone.0073314
Peng, J. R. (1999). Rht gene for green revolution in wheat and gibberellin signal conduction. J. Agric. Sci. Technol. 1, 60–61.
Pucciariello, C. (2020). Molecular mechanisms supporting rice germination and coleoptile elongation under low oxygen. Plants (Basel) 9:1037. doi: 10.3390/plants9081037
Qayyum, A., Al Ayoubi, S., Sher, A., Bibi, Y., Ahmad, S., Shen, Z., et al. (2021). Improvement in drought tolerance in bread wheat is related to an improvement in osmolyte production, antioxidant enzyme activities, and gaseous exchange. Saudi J. Biol. Sci. 28, 5238–5249. doi: 10.1016/j.sjbs.2021.05.040
Rebetzke, G. J., Appels, R., Morrison, A. D., Richards, R. A., McDonald, G., Ellis, M. H., et al. (2001). Quantitative trait loci on chromosome 4B for coleoptile length and early vigour in wheat (Triticum aestivum L.). Austr. J. Agric. Res. 52, 1221–1234. doi: 10.1071/ar01042
Rebetzke, G. J., Bruce, S. E., and Kirkegarrd, J. A. (2005). Longer coleoptiles improve emergence through crop residues to increase seedling number and biomass in wheat. Plant Soil 272, 87–100.
Rebetzke, G. J., Ellis, M. H., Bonnett, D. G., and Richards, R. A. (2007a). Molecular mapping of genes for coleoptile growth in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 114, 1173–1183. doi: 10.1007/s00122-007-0509-1
Rebetzke, G. J., Richards, R. A., Fettell, N. A., Long, M., Condon, A. G., Forrester, R. I., et al. (2007b). Genotypic increases in coleoptile length improves stand establishment, vigour and grain yield of deep-sown wheat. Field Crops Res. 1, 10–23. doi: 10.1016/j.fcr.2006.05.001
Rebetzke, G. J., Richards, R. A., Sirault, X. R. R., and Morrison, A. D. (2004). Genetic analysis of coleoptile length and diameter in wheat. Austr. J. Agric. Res. 55, 733–743.
Rebetzke, G. J., Verbyla, A. P., Verbyla, K. L., Morell, M. K., and Cavanagh, C. R. (2014). Use of a large multiparent wheat mapping population in genomic dissection of coleoptile and seedling growth. Plant Biotechnol. J. 12, 219–230. doi: 10.1111/pbi.12130
Sadok, W. (2017). “Wheat,” in Water-conservation traits to increase crop yields in water-deficit environments: Case studies, ed. T. R. Sinclair (Cham: Springer International Publishing), 85–92.
Sidhu, J. S., Singh, D., Gill, H. S., Brar, N. K., Qiu, Y., Halder, J., et al. (2020). Genome-wide association study uncovers novel genomic regions associated with coleoptile length in hard winter wheat. Front. Genet. 10:1345. doi: 10.3389/fgene.2019.01345
Singh, K., Shukla, S., Kadam, S., Semwal, V. K., Singh, N. K., and Khanna-Chopra, R. (2015). Genomic regions and underlying candidate genes associated with coleoptile length under deep sowing conditions in a wheat RIL population. J. Plant Biochem. Biotechnol. 24, 324–330. doi: 10.1007/s13562-014-0277-3
Smith, S. E., Kuehl, R. O., Ray, I. M., Hui, R., and Soleri, D. (1998). Evaluation of simple methods for estimating broad-sense heritability in stands of randomly planted genotypes. Crop Sci. 38, 1125–1129. doi: 10.2135/cropsci1998.0011183X003800050003x
Spielmeyer, W., Hyles, J., Joaquim, P., Azanza, F., Bonnett, D., Ellis, M. E., et al. (2007). A QTL on chromosome 6A in bread wheat (Triticum aestivum) is associated with longer coleoptiles, greater seedling vigour and final plant height. Theor. Appl. Genet. 115, 59–66. doi: 10.1007/s00122-007-0540-2
Sun, N. N., Zhou, Q., Zhi, L., Qiao, P. F., Mu, L. M., Ni, S. L., et al. (2022). Identification of drought resistance at seedling stage and correlation analysis between drought resistance indexes and important agronomic characters of 240 wheat materials. Acta Agronomica Sinica. 31, 137–143. doi: 10.7606/ji.ssn.1004-1389.2022.02.003
Tian, X., Wen, W., Xie, L., Fu, L., Xu, D., Fu, C., et al. (2017). Molecular mapping of reduced plant height gene Rht24 in bread wheat. Front. Plant Sci. 8:1379. doi: 10.3389/fpls.2017.01379
Wang, J., Yang, C., Zhao, W., Wang, Y., Qiao, L., Wu, B., et al. (2022). Genome wide association study of grain hardness and novel Puroindoline alleles in common wheat. Mol. Breed. 42:40. doi: 10.1007/s11032-022-01303-x
Wang, S. B., Feng, J. Y., Ren, W. L., Huang, B., Zhou, L., Wen, Y. J., et al. (2016). Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 6:19444. doi: 10.1038/srep19444
Wang, W., Zou, Q., Yang, J., and Zhou, X. (1999). The dynamic characteristics of coleoptile growth under water stress in different drought-resistant wheats. Plant Physiol. Commun. 35, 359–362. doi: 10.13592/j.cnki.ppj.1999.05.002
Wen, Y. J., Zhang, H., Ni, Y. L., Huang, B., Zhang, J., Feng, J. Y., et al. (2018). Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Brief. Bioinform. 19, 700–712. doi: 10.1093/bib/bbw145
Worland, A. J., and Sayers, E. J. (1995). Rht1 (B. dw), an alternative allelic variant for breeding semi-dwarf wheat varieties. Plant Breed. 114, 397–400.
Yang, X. H., Yan, J. B., Zheng, Y. P., Yu, J. M., and Li, J. S. (2007). Reviews of association analysis for quantitative traits in plants. Acta Agronomica Sinica. 33, 523–530.
Yao, F., Guan, F., Duan, L., Long, L., Tang, H., Jiang, Y., et al. (2021). Genome-wide association analysis of stable stripe rust resistance loci in a chinese wheat landrace panel using the 660K SNP Array. Front. Plant Sci. 12:783830. doi: 10.3389/fpls.2021.783830
Yu, J. B., and Bai, G. H. (2010). Mapping quantitative trait loci for long coleoptile in Chinese wheat landrace Wangshuibai. Crop Sci. 50, 43–50. doi: 10.2135/cropsci2009.02.0065
Zanke, C. D., Ling, J., Plieske, J., Kollers, S., Ebmeyer, E., Korzun, V., et al. (2014). Whole genome association mapping of plant height in winter wheat (Triticum aestivum L.). PLoS One 9:e113287. doi: 10.1371/journal.pone.0113287
Zhang, X. H., Warburton, M. L., Setter, T., Liu, H. J., Xue, Y. D., Yang, N., et al. (2016). Genome-wide association studies of drought-related metabolic changesin maize using an enlarged SNP panel. Theor. Appl. Genet. 129, 1449–1463. doi: 10.1007/s00122-016-2716-0
Zhang, X. Y., Tong, Y. P., You, G. X., Hao, C. Y., Ge, H. M., Wang, L. F., et al. (2006). Hitchhiking effect mapping: A new approach for discovering agronomic important genes. Sci. Agric. Sin. 39, 1526–1535.
Zhang, Z., Ersoz, E., Lai, C. Q., Todhunter, R. J., Tiwari, H. K., Gore, M. A., et al. (2010). Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42, 355–360. doi: 10.1038/ng.546
Zheng, J., Li, X. H., Zhao, J. J., Shang, B. H., Cao, Y., Ma, X. F., et al. (2018). Genetic diversity analysis of wheat cultivars in shanxi province. J. Plant Genet. Resour. 19, 699–706. doi: 10.13430/j.cnki.jpgr.20171105001
Zheng, X. W., Qiao, L., Liu, Y., Wei, N. C., Zhao, J. J., Wu, B. B., et al. (2021). Genome-wide association study of grain number in common wheat from shanxi under different water regimes. Front. Plant Sci. 12:806295. doi: 10.3389/fpls.2021.806295
Keywords: Shanxi wheat, coleoptile length, drought stress, GWAS, 3VmrMLM
Citation: Wei N, Zhang S, Liu Y, Wang J, Wu B, Zhao J, Qiao L, Zheng X, Wang J and Zheng J (2022) Genome-wide association study of coleoptile length with Shanxi wheat. Front. Plant Sci. 13:1016551. doi: 10.3389/fpls.2022.1016551
Received: 11 August 2022; Accepted: 29 August 2022;
Published: 21 September 2022.
Edited by:
Shuaifeng Geng, Institute of Crop Sciences (CAAS), ChinaReviewed by:
Chenyang Hao, Institute of Crop Sciences (CAAS), ChinaCopyright © 2022 Wei, Zhang, Liu, Wang, Wu, Zhao, Qiao, Zheng, Wang and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juanling Wang, MTM5OTQyNjc1MDhAMTYzLmNvbQ==; Jun Zheng, c3hua3l6akAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.