- 1Key Laboratory of Non-coding RNA Transformation Research of Anhui Higher Education Institution, Yijishan Hospital of Wannan Medical College, Wuhu, China
- 2Anhui Provincial Clinical Research Center for Critical Respiratory Disease, Wuhu, China
- 3Forestry College, Central South University of Forestry and Technology, Changsha, China
- 4School of Landscape and Ecological Engineering, Hebei University of Engineering, Handan, China
- 5College of Basic Medical Sciences, Hubei University of Chinese Medicine, Wuhan, China
- 6Hunan Institute of Microbiology, Changsha, China
Reproductive growth and vegetative growth are a pair of main contradictions in the process of plant growth. Flowering, as part of reproductive growth, is a key switch in the life cycle of higher plants, which affects the yield and economic benefits of plants to a certain extent. The Euphorbiaceae species, including castor bean (Ricinus communis), physic nut (Jatropha curcas), tung tree (Vernicia fordii), cassava (Manihot esculenta), and rubber tree (Hevea brasiliensis), have important economic values because they are raw materials for the production of biodiesel, rubber, etc. The flowering mechanisms are still excluded in the Euphorbiaceae species. The flowering-related genes of Arabidopsis thaliana (Arabidopsis) were used as a reference to determine the orthologs of these genes in Euphorbiaceae genomes. The result showed that 146, 144, 114, 114, and 149 of 207 A. thaliana genes were respectively matched to R. communis, V. fordii, J. curcas, H. brasiliensis, and M. esculenta. These identified genes were clustered into seven pathways including gibberellins, floral meristem identity (FMI), vernalization, photoperiod, floral pathway integrators (FPIs), and autonomous pathways. Then, some key numbers of flowering-related genes are widely conserved in the Euphorbiaceae genomes including but not limited to FPI genes LFY, SOC1, FT, and FMI genes AG, CAL, and FUL. However, some genes, including FRI, FLC, and GO, were missing in several or all five Euphorbiaceae species. In this study, we proposed the putative mechanisms of flowering-related genes to control flowering and provided new candidate flowering genes for using marker-assisted breeding to improve variety quality.
Introduction
Flowering is a key switch in the high plant life cycle. The developmental transition from vegetative growth to reproductive growth is regulated by multiple signaling pathways (Simpson and Dean, 2002). No matter when seeds and fruits are harvested, flowering is a premise for crop production in agriculture or forestry (Bluemel et al., 2015). The regulation of flowering time plays a very important role in the adaptation of crops to specific growth regions, so flowering time is a key topic of primary importance in agriculture or forestry. The identification and understanding of the function and structure of flowering-related genes may lay the foundation for further use of molecular-assisted breeding to cultivate new crop varieties with altered flowering times (Kim A. M. et al., 2013; Liu Y. et al., 2020). The introduction of early flowering-related genes may allow multiple rounds of cropping in single seasons or short growing seasons (Jung et al., 2012; Peng et al., 2015; Liu Y. et al., 2020). Additionally, transfer of genes that participated in late flowering may help increase the yield of crops by extending the time of vegetative growth (Putterill et al., 2004; Liu Y. et al., 2020).
For understanding the mechanism of plant flowering, many researchers have made some important progress on the molecular basis of flowering (Hecht et al., 2005; Mouhu et al., 2009; Jung et al., 2012; Peng et al., 2015; Liu Y. et al., 2020). In Arabidopsis thaliana (Arabidopsis), Fornara have isolated some mutants with loss of function and then identified more than 180 genes involved in regulating flowering time (Fornara et al., 2010). A result of the genetic analysis of A. thaliana mutants controlling flowering time suggested that the process of flowering involved a complex cross talk between different pathways responding to endogenous factors and environmental signals. The flowering transition process is mainly controlled by environmental signals (such as inter temperature (vernalization) and day length (photoperiod)) to ensure timely flowering (Bernier and Périlleux, 2005). Additionally, flowering time may be affected by ambient temperature, but the molecular mechanism of this pathway is still in the preliminary research stage (Lee et al., 2008; Cho et al., 2017). In addition to these external factors, the researchers also found that four floral pathways are closely related to flowering time in A. thaliana: GA (gibberellin) pathway, including autonomous pathway, vernalization response pathway, and photoperiod response pathway (Sheldon et al., 2000; Searle and Coupland, 2004; Simpson, 2004; Trevaskis et al., 2007; Jackson, 2009; Mutasa-Göttgens and Hedden, 2009). Compared to the wild type, mutations in genes such as FPA, FVE, LD, and FCA that participated in the autonomous pathway led to flowering under both short days and long days (Koornneef et al., 1991; Marquardt et al., 2006). On the contrary, mutations in genes participated in the long-day pathway, such as FT, GI, and CO, resulting in later flowering in long days but no delay in flowering under short days in comparison to the wild type (Koornneef et al., 1991; Cheng and Wang, 2005). Recently, in the control of vernalization and the circadian clock, the flowering mechanism has made great progress. The FLC containing a MADS-box domain seems to act as a flowering suppressor in A. thaliana, and its level is reduced after vernalization (Michaels and Amasino, 1999). The change of DNA methylation status was caused by the vernalization, which led to the repression of FLC expression (Bastow et al., 2004; Jean Finnegan et al., 2005). ZTL and FKF1, belonging to circadian clock-related genes, act as a bridge between circadian clock control and photoperiodic light signaling (Hoecker, 2005; Baudry et al., 2010). Researchers have confirmed that many flowering-related genes are widely conserved in the plant genomes. Putterill reported that about 85% of A. thaliana genes exist in other plant genomes; flowering-related genes have been identified and isolated from Acacia mangium, Lotus corniculatus, Medicago truncatula, Glycine max, and carnation (Putterill et al., 2004). Using genes related to flowering time from A. thaliana to detect the flowering-related genes in other plant genomes is an effective way (Hecht et al., 2005; Mouhu et al., 2009; Jung et al., 2012; Peng et al., 2015; Liu Y. et al., 2020), and these data provide resources for us to further understand the flowering time control beyond A. thaliana.
Bolting and flowering are the most important key life-history traits in the plant life cycle, which exercise far-reaching influence on evolution, gene flow, reproductive suitability, mating opportunities, and patterns (Post et al., 2008; Jung et al., 2016). The flowering strategies of plants show great diversity under different habitats and environmental conditions. Drought stress affects flowering, and this process was reported to promote flowering in Sapium sebiferum, Citrus latifolia, and A. thaliana (Southwick and Davenport, 1986; Riboni et al., 2013; Yang et al., 2015). The GIGANTEA (GI) gene, which is a repressor of FT from the photoperiod pathway, can accelerate flowering by suppressing CYCLING DOF FACTOR (CDF) or by binding to the FT promoter (Sawa and Kay, 2011). In addition, GI genes respond to conditions under salt, drought, and cold stresses, thereby helping plants adapt to unfavorable environments (Kim W.-Y. et al., 2013; Riboni et al., 2013; Fornara et al., 2015). The life cycles of plants can be adjusted systematically to adapt to different climates and latitudes. The model specie A. thaliana, which is an annual plant, can respond to vernalization and long day lengths. The Euphorbiaceae species castor bean (Ricinus communis) is an annual plant while tung tree (Vernicia fordii), physic nut (Jatropha curcas), cassava (Manihot esculenta), and rubber tree (Hevea brasiliensis) are perennials. To further elucidate the molecular mechanism of flowering in Euphorbiaceae, a large number of researchers have conducted limited studies. In contrast, most of the studies were carried out in the model plant A. thaliana. In J. curcas, RNA-seq analysis and molecular biology experiments have been carried out to identify and confirm genes controlling floral organ development and flowering time (Brasileiro et al., 2012; Li et al., 2014). In V. fordii, phenological, morphological, and histological experiments of tung flowers were conducted to give a comprehensive study of the flower biology and ontogeny (Li et al., 2020). Recently, the drafts of the R. communis, V. fordii, J. curcas, M. esculenta, and H. brasiliensis genome sequences were sequenced and reported (Chan et al., 2010; Wang W. et al., 2014; Ha et al., 2019; Zhang et al., 2019b; Liu J. et al., 2020). This genomic information lays a strong foundation for the genome-wide comparison of flowering-related genes between these Euphorbiaceae species and A. thaliana. The purpose of our study is to detect gene homologs associated with flowering among five Euphorbiaceae species by searching these whole-genome sequences. The divergence of Euphorbiaceae species R. communis, V. fordii, J. curcas, M. esculenta, and H. brasiliensis in genome duplication causes them to have different genome complexities and sizes (Chan et al., 2010; Wang W. et al., 2014; Ha et al., 2019; Zhang et al., 2019b; Liu J. et al., 2020). From this perspective, both of the differences among homologous genes and the distribution of the homologs were highlighted among these five Euphorbiaceae genomes. The results of this study obtained a large amount of gene resources, which provided a solid material basis for understanding the flowering mechanism of Euphorbiaceae and also provided a certain reference of other species.
Materials and methods
Data retrieval
Firstly, we obtained the genomes of J. curcas, M. esculenta, and R. communi from Phytozome and downloaded the genomes of V. fordii and H. brasiliensis from NCBI. Subsequently, a total of 207 A. thaliana genes involved in the flowering pathway as query sequences were obtained from the published paper and downloaded from TAIR. According to the HMM models and BlastP software, the flowering-related genes were identified in five Euphorbiaceae species. The obtained flowering-related genes were further confirmed by searching against NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with non-redundant protein sequences.
Domain analysis
We further analyzed all obtained flowering-related proteins with InterProScan using default parameters (Jones et al., 2014). In our study, we only chose the longest sequence from alternatively spliced transcripts for further analysis.
Orthologous analysis and evolutionary tree constructe
To identify the orthologs between five Euphorbiaceae and A. thaliana, we carried out a collinear analysis using MCScanX (Wang et al., 2012). Firstly, we generated two files: proteins file and GFF file. Then, the proteins file was used to carry out a BlastP analysis with E-value 10-5. Finally, we plotted a collinear diagram by loading the BlastP file and GFF file. An evolutionary tree was constructed between five Euphorbiaceae and A. thaliana according to the methods of previous studies (Cao et al., 2020).
Results and discussion
Identification of flowering-related genes in Euphorbiaceae
To predict the orthologs of corresponding flowering-related genes of A. thaliana in the five Euphorbiaceae genomes, this review used 207 A. thaliana genes involved in the flowering pathway as query sequences. Among these genes, Jung (Jung et al., 2012) used an ortholog-based method to detect 24 genes with regulating flowering time, and Fornara (Fornara et al., 2010) have confirmed that 183 genes are involved in flowering regulatory pathways in a previous study. These genes for flowering pathways are mainly involved in the photoperiod pathway, the vernalization pathway, the gibberellin (GA) pathway, and the autonomous pathway, along with genes for floral meristem identity and floral pathway integrators (FPIs). In general, a gene can contain multiple functions involving different pathways; each gene was assigned to a single pathway according to its main function. Based on the HMM models, MCScanX, and BlastP software, the orthologs of A. thaliana flowering-related genes were identified in five Euphorbiaceae species (Figure 1), as described in several published papers (Cao et al., 2019a; Cao et al., 2019b; Cao et al., 2020). Finally, we found that 146, 144, 114, 114, and 149 of 207 A. thaliana genes were respectively matched to R. communis, V. fordii, J. curcas, H. brasiliensis, and M. esculenta genes and the numbers of these orthologous genes in each Euphorbiaceae were 176, 169, 130, 171, and 257, respectively (Table 1, Table S1, and Table S2).
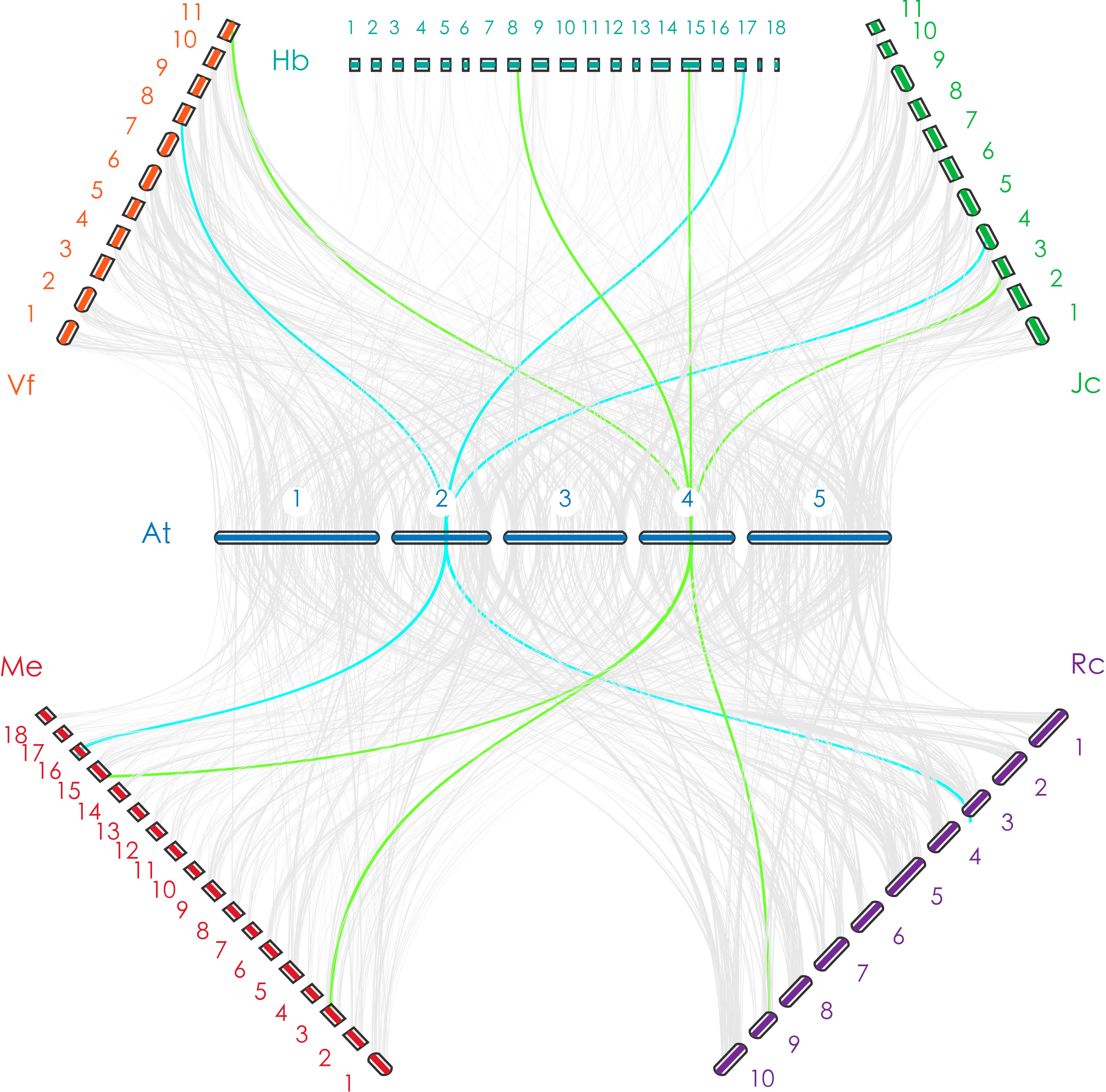
Figure 1 The orthologous relationship between Arabidopsis thaliana and Euphorbiaceae. The blue line indicates a one-to-one orthologous relationship (take ELF3 as an example); the green line indicates a one-to-one or one-to-two orthologous relationship (take ESD4 as an example).
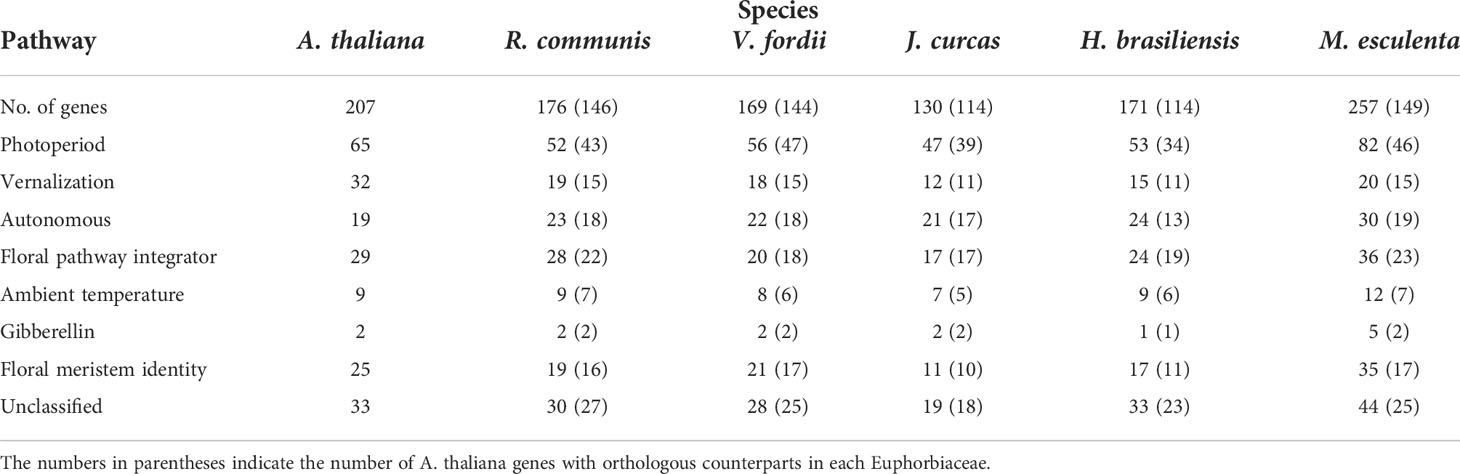
Table 1 The number of orthologous genes of Arabidopsis thaliana flowering-related genes in the Ricinus communis, Jatropha curcas, Vernicia fordii, Manihot esculenta, and Hevea brasiliensis genomes.
Photoperiod pathway
Light is one of the most important environmental regulators that affect the flowering in plants (Li et al., 2016). The photoperiod pathway is the main way for plants to monitor the light environment to perceive the time of day and season (Searle and Coupland, 2004). The number of identified A. thaliana flowering-related genes participating in the photoperiod pathway is 65, only 47 of which contain orthologs in the Euphorbiaceae genomes. R. communis, V. fordii, and J. curcas contain 52, 56, and 47 genes, respectively, which are part of the photoperiod pathway. We also identified 53 genes in H. brasiliensis and 82 genes in M. esculenta as putative orthologs of these A. thaliana flowering genes (Table 1). A recent whole-genome duplication episode that occurred in both M. esculenta and H. brasiliensis (Figure 2), but not in R. communis, V. fordii, and J. curcas, probably played key roles in the expansion of flowering-related genes in M. esculenta (Chan et al., 2010; Wang W. et al., 2014; Ha et al., 2019; Zhang et al., 2019b; Liu J. et al., 2020). However, these genes of H. brasiliensis have experienced a gene loss event after a recent whole-genome duplication (WGD) event, resulting in its number less than M. esculenta. Among 65 genes involved in the photoperiod pathway in A. thaliana, 18 genes were not found to have orthologs in the five Euphorbiaceae genomes.
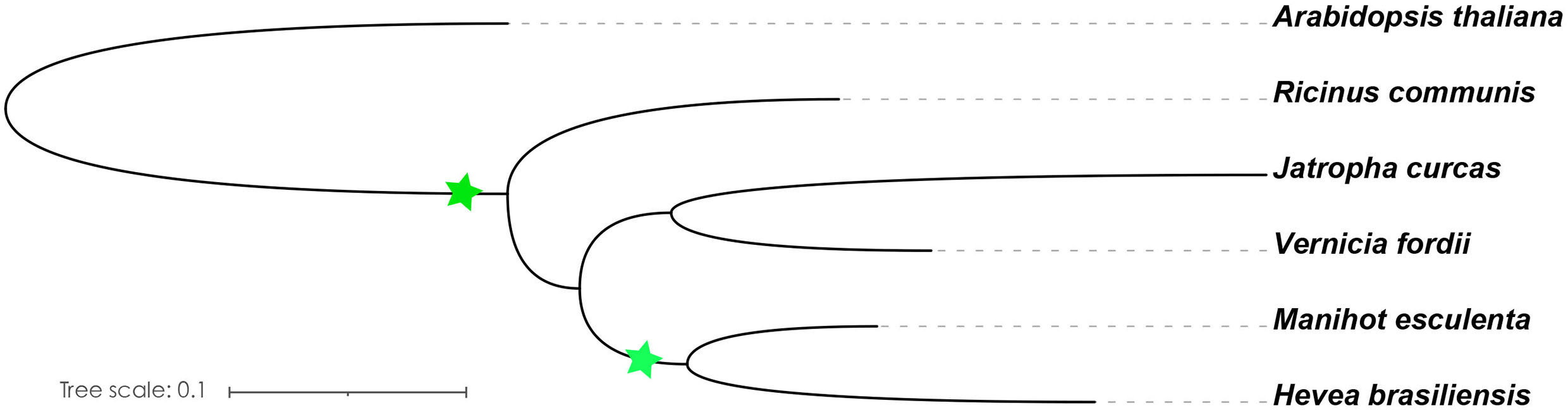
Figure 2 Species evolution tree of A. thaliana and Euphorbiaceae species. The OrthoFinder was used to investigate the single-copy orthologs (Emms and Kelly, 2019), and IQ-TREE was used to construct a species evolution tree (Nguyen et al., 2015). The green star represents the WGD event.
In A. thaliana, the PHYTOCHROME(PHY)A, PHYB, PHYC, PHYD, and PHYE genes encoded phytochrome proteins that perceived the red and far-red light (Clack et al., 1994). Additionally, PHYA is unique because it can be activated by light and can be degraded specifically (Weller et al., 2004; Debrieux and Fankhauser, 2010). Three, five, three, three, and three orthologous genes of A. thaliana PHY genes were detected in R. communis, V. fordii, J. curcas, M. esculenta, and H. brasiliensis, respectively. CRYPTOCHROME 1 (CRY1) and CRY2 encoded cryptochrome proteins which are sensitive to blue light (Ahmad et al., 1998; Holtkotte et al., 2017). The A. thaliana CRY genes contain two, two, two, two, and three homolog genes in R. communis, V. fordii, J. curcas, H. brasiliensis, and M. esculenta, respectively, while no homolog of CRY2 was detected in H. brasiliensis. Light is a key signal not only for plant growth and development but also for photosynthetic energy production. The light induction of circadian clock associated 1 (CCA1) and late elongated hypocotyl (LHY) genes was affected by the multiple events; the complex of PHYB–PIF3 binds promoter regions of these two genes (Imaizumi, 2010). In the study, regardless of the CCA1, LHY, or PIF3 gene, we have identified a single-copy orthologous gene in R. communis, V. fordii, and J. curcas, M. esculenta, and H. brasiliensis.
The central oscillator of the circadian clock was produced by CCA1 and LHY together with TIMING OF CAB EXPRESSION 1 (TOC1) (Alabadıí et al., 2001). TOC1 encodes a nuclear protein containing a CONSTANS, CO-like, and TOC1 (CCT) motif, and the expression of these genes is regulated by the antagonism of CCA1 or LHY (Alabadıí et al., 2001). R. communis, V. fordii, J. curcas, H. brasiliensis, and M. esculenta have one, one, one, one, and two orthologous genes of TOC1, respectively. Three, two, one, six, and six orthologous genes belonging to the PRR family were identified in R. communis, V. fordii, J. curcas, H. brasiliensis, and M. esculenta, respectively. LOV KELCH PROTEIN 2 (LKP2), FLAVIN-BINDING KELCH REPEAT F-BOX 1 (FKF1), and ZEITLUPE (ZTL) blue-light photoreceptors contribute to regulate the photoperiodic flowering and circadian clock pathway (Imaizumi et al., 2005; Baudry et al., 2010). These proteins have a LOV (light-oxygen voltage-sensing) domain, repeated Kelch motifs, an F-box, and a PAS (Per-Arnt-Sim) domain (Boss et al., 2004). GIGANTEA (GI) is co‐expressed with an F‐box gene FKF1 and binds both to CYCLING DOF FACTOR 1 (CDF1)–CDF5 and to GI under light conditions (Sawa et al., 2007; Song et al., 2012). CONSTANS (CO) encodes crucial regulators of floral transition, day-length perception, and photoperiodic gene expression, and its expression is directly suppressed by CYCLING DOF FACTORS (CDFs) (Fornara et al., 2009; Goralogia et al., 2017). The upregulation of CO during development marks the timing of flowering, as it causes the accumulation of high FT transcripts to the threshold level required to trigger flowering (Valverde et al., 2004; Wang C.-Q. et al., 2014). A schematic of this GI–CO–FT regulatory module in the photoperiod pathway has also been found in other plants and is highly conserved, such as soybean, maize, and rice (Hayama et al., 2003; Zhang et al., 2019a). The LKP2, FKF1, and/or ZTL genes contain 11 orthologous genes among these five Euphorbiaceae genomes. Remarkably, we also noted that the GI gene was conserved because R. communis, J. curcas, H. brasiliensis, and M. esculenta have a single-copy gene, while this gene contains two orthologous genes in V. fordii.
Vernalization pathway
Cold winter reduces the reproductive success rate of plants growing in temperate regions; therefore, these plants have produced the vernalization pathway during evolution, requiring a period of low temperature before the transition of flora (Sung and Amasino, 2004; Jung and Müller, 2009; Bouché et al., 2017). The vernalization pathway is involved in the transduction process and the signal perception that flowering occurs after winter (Luo and He, 2020; Matar et al., 2020). The previously studies revealed that 32 flowering-related genes of A. thaliana played key roles in the vernalization pathway. Among these genes, 18 genes have not been detected in the five Euphorbiaceae genomes. V. fordii, R. communis, J. curcas, and H. brasiliensis contain 14, 15, 9, and 11 genes, respectively, which are likely related to the vernalization pathway. In M. esculenta, we identified 17 genes to be orthologous genes of these A. thaliana genes.
FLOWERING LOCUS C (FLC) is a key gene for vernalization, which can effectively inhibit flowering (Putterill et al., 2004; Deng et al., 2011; Madrid et al., 2020). The repressor of FLC is the REDUCED VERNALIZATION RESPONSE 1 (VRN1) and VRN2 (Putterill et al., 2004). Among them, VRN1 encodes a transcription factor that contains a B3 DNA-binding domain, and VRN2 encodes a protein with a zinc-finger motif belonging to polycomb group (PcG) proteins (Gendall et al., 2001; Zhou et al., 2019). Both VRN1 and VRN2 can affect the expression of FLC, indicating that epigenetic changes in chromatin structure at the locus of FLC are a molecular machinery basis for this cellular vernalization memory (Jean Finnegan et al., 2005; Whittaker and Dean, 2017). A. thaliana VRN1 was conserved in R. communis, V. fordii, J. curcas, and M. esculenta containing a single-copy gene. V. fordii and M. esculenta each contain a single-copy VRN2 gene, and two and two orthologous genes were found in R. communis and H. brasiliensis, respectively. Recently, some researchers have found some equivalents of the A. thaliana FLC regulators, such as VERNALIZATION INDEPENDENCE 3 and 4 (VIP3 and VIP4), PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 (PIE1), PHOTOPERIOD-EARLY FLOWERING IN SHORT DAYS (EFS), and EARLY IN SHORT DAYS 4 (ESD4). PIE1, an imitation switch (ISWI) family member, played important roles in floral repression and FLC activation (Noh and Amasino, 2003; Ojolo et al., 2018). ESD4, a SUMO-specific protease, is required for FLC expression (Jean Finnegan et al., 2005). The FLC chromatin structure depends on EFS containing histone-lysine N-methyltransferase activity. VIP3 contains multiple WD repeats, and VIP4 is a novel protein playing key roles in the PAF1 transcriptional complex (Takagi and Ueguchi, 2012). R. communis, V. fordii, J. curcas, and M. esculenta each have a single-copy PIE1 gene, and two orthologous genes were found in H. brasiliensis. Both EFS and ESD4 have one orthologous gene in R. communis, V. fordii, and J. curca and two orthologous genes in M. esculenta and H. brasiliensis. A. thaliana VIP3 and VIP4 were conserved in these five Euphorbiaceae genomes containing a single-copy gene, except for H. brasiliensis which possesses two orthologous VIP4 genes. A. thaliana PAF1 and PAF2 were conserved in these five Euphorbiaceae genomes containing a single-copy gene, except for J. curca which does not contain any orthologous PAF2 gene. Also, some researchers have found that there is a vernalization response FLC-independent because flc null mutants contain functions with a vernalization-sensitive phenotype (Michaels and Amasino, 1999; Michaels and Amasino, 2001). After vernalization of the FLC null mutant, the expression of FT and SOC1 was upregulated, suggesting that FLC-independent and -dependent vernalization branches share a common target (Moon et al., 2003). Alexandre and Hennig found that the MADS transcription factor AGAMOUS-LIKE 24 (AGL24) may be a target because vernalization upregulates the expression of AGL24, which provides an FLC-independent pathway for regulating flowering time (Alexandre and Hennig, 2008). There are three M. esculenta genes, two R. communis genes, two V. fordii genes, one J. curcas gene, and one H. brasiliensis gene identified to be orthologous to AGL24.
Autonomous pathways
Autonomous pathways include posttranscriptional genes and epigenetic regulation, which can control the flowering time in plants (Simpson and Dean, 2002; Simpson, 2004). There are 19 genes involved in the autonomous pathway in A. thaliana. V. fordii, R. communis, J. curcas, M. esculenta, and H. brasiliensis have 19, 20, 18, 26, and 22 genes, respectively, which are part of the autonomous pathway. The functions of the autonomous pathway repress flowering by promoting the accumulation of an mRNA that is a MADS-domain transcription factor, FLOWERING LOCUS C (FLC) (Michaels and Amasino, 2001). FCA is an RNA-binding protein that can interact with FY to downregulate the expression of FLC (Quesada et al., 2003), thereby promoting flowering. A. thaliana FCA was conserved in these five Euphorbiaceae genomes containing a single-copy gene, except for H. brasiliensis which does not possess any orthologous FCA. FY contains one or two orthologous genes in these five Euphorbiaceae genomes. FLOWERING LATE KH MOTIF (FLK), an RNA-binding protein, can regulate the autonomous pathway via FLC (Lim et al., 2004). FPA encodes a protein involved in floral induction having RNA-recognition motifs (Schomburg et al., 2001). FVE, a retinoblastoma-related protein with a WD-repeat domain, can bind to chromatin and regulate flowering time (Ausín et al., 2004). FPA has one orthologous gene in R. communis, J. curcas, and M. esculenta. FLK contains one or two homologs in these five Euphorbiaceae genomes. The MSI family contains four members, such as FVE, which have five, six, five, seven, and five orthologs in V. fordii, J. curcas, R. communis, M. esculenta, and H. brasiliensis, respectively. LUMINIDEPENDENS (LD) encodes a nuclear protein with a homeodomain, which plays an important role in RNA processing, such as FPA and FCA. FLOWERING LOCUS D (FLD) is another autonomous gene that can inhibit the expression of FLC to control flowering time. A. thaliana FLD and LD were conserved in these five Euphorbiaceae genomes with a single-copy gene, except for H. brasiliensis which does not contain any orthologous FLD gene and contains two orthologous LD genes.
Floral pathway integrator
The signaling pathways that transmit and receive input signals include the autonomous pathway, the ambient temperature, the vernalization, and the photoperiod pathways (Andrés and Coupland, 2012). Floral pathway integrator genes can integrate the input from these pathways (Simpson and Dean, 2002; Van Dijk and Molenaar, 2017). Although there are multiple pathways with associated genes involved in the regulation of flowering, the expression level of FT largely determines the flowering time. FT, SOC1, and LFY integrate multiple pathways and then make a single decision of developmental (Moon et al., 2005). In A. thaliana, 29 genes were identified to classify as FPIs. V. fordii, J. curcas, R. communis, M. esculenta, and H. brasiliensis contain 19, 16, 24, 34, and 23 genes, respectively, which are putatively associated with FPIs.
The transcriptional activation of FT, an activator of flowering, can be induced by the activation of the photoperiod flowering pathway (Steinbach and Hennig, 2014). The function of FT is mainly as a mobile flowering signal, which is generated in the leaves and then transferred to the shoot apical meristem (SAM) (Turck et al., 2008). In SAM, FT interacts with FD to produce an FT–FD complex and then activates other FPI genes, such as LFY and SOC1 (Kaneko-Suzuki et al., 2018; Li et al., 2019). In the study, we identified one, one, five, three, and one gene in V. fordii, J. curcas, R. communis, M. esculenta, and H. brasiliensis to be orthologous to FT and its homolog TWIN SISTER OF FT (TSF), respectively. FT and floral repressor terminal flower 1 (TFL1), belonging to the same Raf family, contain antagonistic functions. Each V. fordii, J. curcas, R. communis, and M. esculenta possess one ortholog of TFL1. SOC1 and LFY genes play vital roles in the regulation of the flowering network. SOC1 can link floral development and floral induction by regulating the expression of LFY. Each R. communis, H. brasiliensis, and M. esculenta contain two orthologs of SOC1, and the remaining two species contain one SOC1 ortholog. LFY contains one orthologous gene in R. communis, J. curcas, and V. fordii, while LFY contains two orthologs in M. esculenta and H. brasiliensis. In this study, we also considered GENERAL REGULATORY FACTOR (GRF) and NUCLEAR FACTOR Y (NF-Y) due to these two transcription factors that have been confirmed to be involved in flower development. For NF-Y genes encoding the basic helix–loop–helix ID factors, V. fordii, J. curcas, R. communis, H. brasiliensis, and M. esculenta have three, seven, nine, six, and 11 orthologs, respectively. There are 11 genes of the GRF family containing 12, 6, 10, 12, and 17 homologs of V. fordii, J. curcas, R. communis, H. brasiliensis, and M. esculenta, respectively.
Ambient temperature pathway
The biomass and architecture of plants can be dramatically affected by changes in ambient temperature (Wigge, 2013). The global temperatures seem to be rising, so understanding how plants respond to changes in ambient temperatures can help plants adapt to different climatic conditions. In response to changes in ambient temperatures, plants can make corresponding measures to control flowering time. The floral integrator FT can be activated independently of CO expression at high temperatures and seems to partially mediate the ambient temperature pathway (Wigge, 2011). SHORT VEGETATIVE PHASE (SVP), a MADS-box gene, can negatively regulate the expression of FT by directly binding to the FT sequence (Lee et al., 2013). FLC interacts with SVP to generate a complex to control the flowering time (Li et al., 2008). We also found genes homologous to SVP in these five Euphorbiaceae genomes. The FVE and FCA in the autonomous pathway have been confirmed to take part in the perception of ambient temperatures affecting flowering time. Different ambient temperatures can activate different photoreceptors. For example, PHYE is a contributor to the main phytochrome at 16°C. The PSEUDO-RESPONSE REGULATOR 7 (PRR7) and PRR9 genes expressed in the morning contain dual functions in the circadian rhythm, participating in the regulation of the central oscillator and the transmission of light signals to the clock (Farré et al., 2005). The MADS-box gene, FLOWERING LOCUS M (FLM), can inhibit flowering in response to temperature. At ambient temperature, EARLY FLOWERING 3 (ELF3) and TFL1 have complementary effects on regulating flowering time (Strasser et al., 2009). Actually, Arabidopsis thermosensitive flowering may require genes from many different pathways, which are involved in the control of FT expression.
Gibberellin pathway
Among various phytohormones, gibberellin can induce flower formation and promote flowering in the model plant A. thaliana (Fornara et al., 2010). The GA pathway acts by inducing LFY, SOC1, FT, etc. The O-linked N-acetylglucosamine transferase SPINDLY (SPY) acts as a negative regulator to regulate the gibberellin (GA) signaling pathway (Jacobsen and Olszewski, 1993). This gene contains two orthologs in M. esculenta and one in each of the remaining four Euphorbiaceae genomes.
Floral meristem identity genes
The meristem identity genes can be classified as the shoot meristem identity genes and the floral meristem identity genes, and the latter genes are necessary for the developing floral primordia (Grandi et al., 2012). In the initial stage of flower development, flower meristems will not form organs, but their size will increase to a certain extent. Among these genes, the functions of CAULIFLOWER (CAL), FRUITFUL (FUL), APETALA 1 (AP1), UNUSUAL FLORAL ORGANS (UFO), and LFY can promote floral meristem identity. LFY, a meristem identity gene, is a key player in flower development (Silva et al., 2016). Three genes, namely, SHOOTMERISTEMLESS (STM), WUSCHEL (WUS), and TFL1, play key roles in maintaining the identity of inflorescence shoot meristems (Sablowski, 2007; Kim M. Y. et al., 2013). Some genes, such as PISTILLATA (PI), AGAMOUS (AG), APETALA 3 (AP3), and AP2, have also been examined because these genes may be involved in mediating floral organ identity and meristem function (Riechmann et al., 1996; Fornara et al., 2010). In A. thaliana, 25 genes were found to contribute to the development of floral organs and the establishment of floral meristems. Remarkably, five genes, namely, SEPALLATA3 (SEP3), APETALA 3 (AP3), SEEDSTICK (STK), ENHANCER OF AG-4 2 (HUA2), and PISTILLATA, were not examined in any of these five Euphorbiaceae genomes. V. fordii, J. curcas, R. communis, H. brasiliensis, and M. esculenta contain 20, 20, 18, 15, and 33 orthologs of A. thaliana genes, respectively. APETALA 1 (AP1), CAL, and FUL were paralogs, each encoding a MADS-box domain (Alvarez-Buylla et al., 2006). AP2 belongs to the ethylene-responsive element-binding protein (EREBP) family encoding the AP2 domain that is involved in the control of flower. These data indicate that the transcriptional regulatory network plays an important role in the specification of floral organs and meristems. UFO is an F-box protein that is required for bract suppression and floral-meristem identity (Hepworth et al., 2006). V. fordii contains three orthologs of AP1, and one ortholog of AP2. R. communis has two orthologs of AP1 and one ortholog of AP2. One ortholog of AP1 and two orthologs of AP2 were found in H. brasiliensis. There are four and two orthologs of AP1 and AP2 identified in M. esculenta separately, while only two orthologs of AP1 were detected in J. curcas. The number of homologs of CAL in J. curcas, V. fordii, and R. communis is two, while M. esculenta contains three CAL orthologs. The function of FUL mainly affects many biological processes including but not limited to controlling cauline leaf morphology, meristem identity, and flowering time. AG is required to specify the identity of floral organs in A. thaliana. By contributing to meristem and floral organ identity, members of the SEP family are required to specify the “floral state”. UFO has been reported to be involved in both floral organ and meristem development (Hepworth et al., 2006). There are multiple FUL copies in R. communis and M. esculenta while FUL is conserved having a single-copy gene in H. brasiliensis, J. curcas, and V. fordii. J. curcas lack AG orthologs, but this gene is conserved in four other genomes containing a single-copy gene. J. curcas, R. communis, and H. brasiliensis have one, two, and two members of the SEP family, respectively, while V. fordii contains six and M. esculenta has five members of the SEP family. UFO contains one homolog gene in J. curcas, while the other four genomes lack UFO homologs.
Comparison of flowering-related genes in A. thaliana and Euphorbiaceae genomes
In A. thaliana, 207 flowering-related genes were identified to play important roles in the control of flowering time. In this study, we found that 39 genes did not contain orthologous counterparts among these five Euphorbiaceae genomes, such as FLC, CO-LIKE (COL), FRI, and MAF (Tables S1 and Table S2). The lack of some flowering-related genes orthologs has been confirmed by previously published papers (Kim M. Y. et al., 2013). For example, Kim found that Lotus corniculatus, Glycine max, and Medicago truncatula lack 56 orthologous of A. thaliana flowering-related genes, such as CO and ELF4, indicating that the photoperiod pathway is regulated in a CO-independent manner in legume species (Kim M. Y. et al., 2013). In our study, R. communis, V. fordii, and H. brasiliensis lack CO orthologs, and J. curcas and M. esculenta contain two CO orthologs, suggesting that different plants have evolved different ways for activating the photoperiod pathway. The transcriptional activation of FT is achieved by CO directly binding its promoter, which may recruit different microRNAs or DNA-binding proteins to medicate FT expression (Fujiwara et al., 2008). This way has been mentioned for the photoperiod but the CO-independent pathway to control flowering time by regulating the expression of FT. Indeed, some researchers have found a CO-independent pathway to regulate the expression of FT which is directly activated by miR172 or GI (Jung et al., 2007; Sawa and Kay, 2011).
As central players in the autonomous pathway and vernalization pathway, both FLC and FRI genes were involved in regulating the flowering time in A. thaliana (Fornara et al., 2010). In our study, we did not detect the homologs of FRI and FLC, as well as the MAFs (FLC homologs). This phenomenon has been found in several previous studies. The FLC-FRI module mechanistically interacts to prevent A. thaliana from flowering before vernalization (Townsend et al., 2018). Silencing FLC genes to regulate flowering time is one of the most typical examples of the role of chromatin remodeling and non-coding RNA in epigenetic control (Wigge, 2011). In germplasms that require long-term vernalization, FLC expression is reactivated after unsaturated vernalization, but this reactivation gradually weakens with increasing cold exposure (Nishio et al., 2020). In our study, the FLC locus was not found in vernalization-non-responsive R. communis, V. fordii, H. brasiliensis, J. curcas, and M. esculenta, which were consistent with their characteristics. The deletion of some key genes in the flowering pathway may help to solve other problems related to molecular circuits in the flowering pathway.
Distribution of flowering-related genes and putative gene regulatory network in the Euphorbiaceae genomes
In this study, we distributed all orthologs of flowering-related genes in A. thaliana throughout the genomes of V. fordii, J. curcas, R. communis, H. brasiliensis, and M. esculenta. There are similar numbers of flowering-related orthologs in V. fordii, J. curcas, R. communis, and H. brasiliensis. Comparisons among paralogous orthologous genes of these five Euphorbiaceae genomes suggest that an ancient WGD was shared by these five genomes, and a recent WGD occurred prior to the split of the H. brasiliensis and M. esculenta (Chan et al., 2010; Wang W. et al., 2014; Ha et al., 2019; Zhang et al., 2019b; Liu J. et al., 2020). We noted that H. brasiliensis has also experienced two WGDs, but there are similar numbers of flowering-related orthologs in V. fordii, J. curcas, R. communis, and H. brasiliensis. However, the number of flowering-related genes contained in M. esculenta is much greater than the other four Euphorbiaceae genomes. These data may reflect a recent WGD which appears to have produced about two times more homologs genes than are found in V. fordii, J. curcas, and R. communis (Figure 2). However, the number of flowering-related genes in H. brasiliensis may have experienced a gene loss event after two WGDs, which ultimately resulted in the number of genes being basically the same as the other three Euphorbiae species. In the study, although we found that some number of flowering-related genes have multiple copies in these Euphorbiaceae genomes, several flowering-related genes still exist and remain in the form of single copies (Figure 1). In V. fordii, J. curcas, and R. communis, 123, 100, and 120 genes of A. thaliana are conserved in the form of single copies, respectively, while both of H. brasiliensis and M. esculenta contain 61 single copies. Totally, nine common genes were identified as conserved single-copy genes among these five Euphorbiaceae species (Figure 1).
The probability of blooming costs is related to the success rate of reproduction, which may have led to plants evolving to develop a set of metabolic and genetic mechanisms to sense and respond to changes in their own environment (Liu Y. et al., 2020). The regulatory network of the SAM destiny is complex, which depends on multiple exogenous and endogenous factors (Liu Y. et al., 2020). In recent decades, both complex networks and mechanisms of flowering have been well studied in the model plant A. thaliana (Fornara et al., 2010). However, few studies of flowering have been performed in the Euphorbiaceae species. Based on the above results, the regulatory network of flowering was inferred in Euphorbiaceae plants (Figure 3). FT controls the flowering time by converging environmental signals sensed by leaves, such as temperature and photoperiod (Corbesier et al., 2007; Wigge, 2011; Liu Y. et al., 2020). The circadian clock in leaves of the plant can receive day-length information. The circadian clock which included GI and PRR7 can regulate the expression of FT and positively control flowering (Farré et al., 2005; Mizoguchi et al., 2005). In the phloem of the leaf, FT will be transcribed and translated, and then its proteins move to the shoot apex (Corbesier et al., 2007). The transcription level of the circadian clock gene GI is regulated by abiotic stresses such as cold, drought, and salt stress, which were related to floral induction (Riboni et al., 2013; Fornara et al., 2015). Many physiological processes including fruit ripening, seed germination, flowering, and growth require GA to participate. DELLA protein, a negative regulator of GA signaling, belongs to the GRAS family (Yoshida et al., 2014). In the SAM, the expression of the FMI identity gene AP1 can be directly regulated by FT, while the control of LFY requires FT to activate SOC1 (Lee and Lee, 2010). In plants, there is an FT-SOC1-LFY model to activate LFY to induce flowering. In our study, the homologous genes of these FT, SOC1, and LFY were identified in these Euphorbiaceae genomes, indicating that this model played a major role in these species.
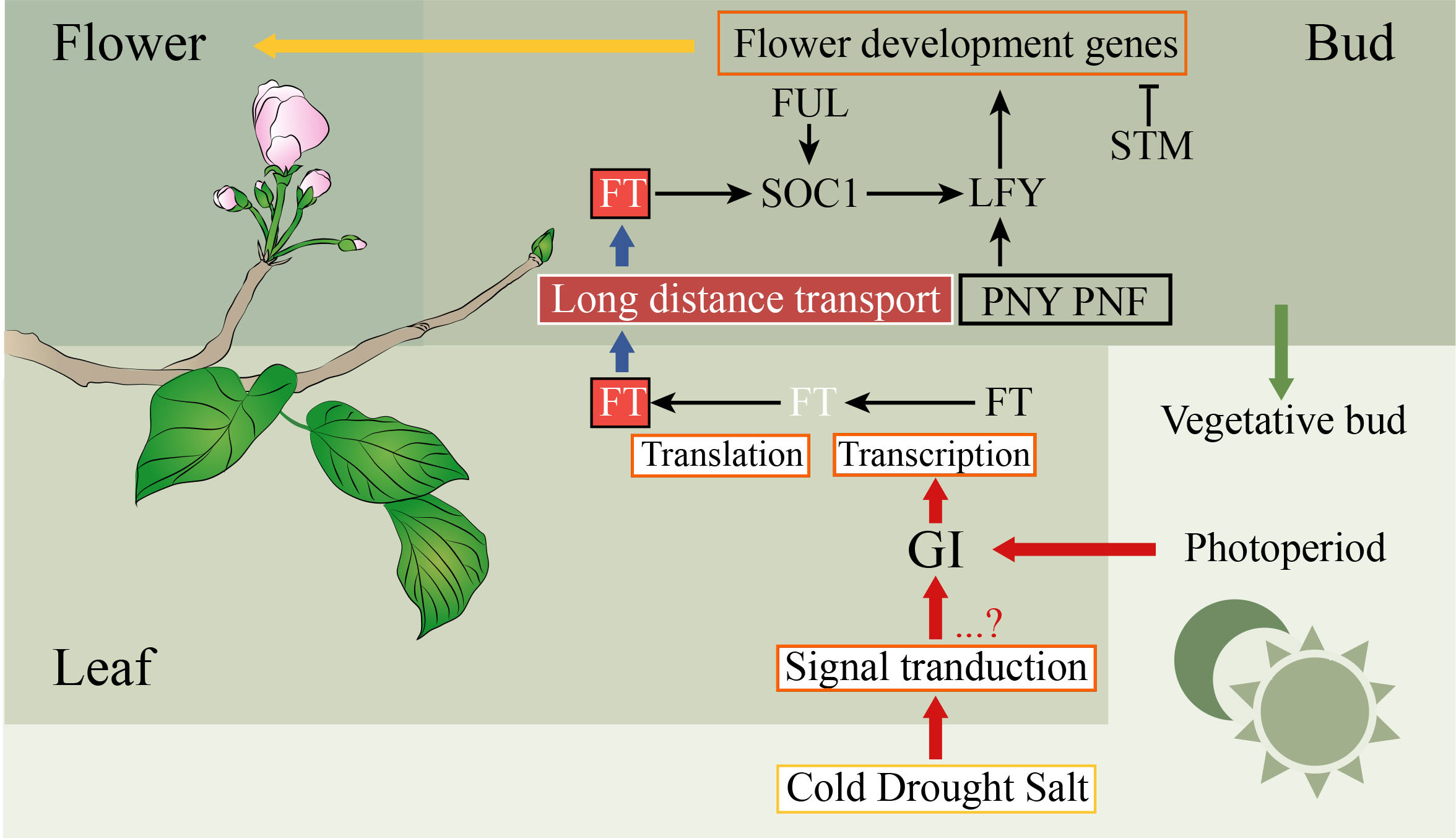
Figure 3 Simplified pathways to participate in the regulation of flowering in plants. The positive and negative controls are represented by arrows and perpendicular lines, respectively. The physiological processes, mRNA, genes, and proteins are represented by orange frames, white letters without frames, black letters without frames, and other frames, respectively.
Conclusion
In the study, the orthologous counterparts of A. thaliana flowering-related genes were identified in five Euphorbiaceae species, V. fordii, J. curcas, R. communis, M. esculenta, and H. brasiliensis. Most A. thaliana flowering-related genes have been detected in Euphorbiaceae genomes, suggesting that basic flowering pathways may be relatively conservative in different plants during evolution. These data will provide new perspectives and potential candidate genes to regulate the timing of flowering on molecular processes in Euphorbiaceae, which have important economic values.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
LJ designed this research and then wrote the manuscript. LJ, TF and JX participated in the evaluation of the manuscript revision. LW and JX prepared the figures and tables. LW, LZ and JX contributed to the provided guidance of the whole study. TF and JX reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Fund Project of Hunan Province (Grant No. 2021JJ41068 and 2020JJ4049) and the Outstanding Youth of the Education Department of Hunan Province (Grant No. 20B617).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1015114/full#supplementary-material
Supplementary Table 1 | The list of homologys of the A. thaliana flowering‐related genes in Euphorbiaceae species, including castor bean (Ricinus communis), physic nut (Jatropha curcas), tung tree (Vernicia fordii), cassava (Manihot esculenta), and rubber tree (Hevea brasiliensis).
Supplementary Table 2 | Lack of homology of the A. thaliana flowering‐related genes in Euphorbiaceae species, including castor bean (Ricinus communis), physic nut (Jatropha curcas), tung tree (Vernicia fordii), cassava (Manihot esculenta), and rubber tree (Hevea brasiliensis).
References
Ahmad, M., Jarillo, J. A., Smirnova, O., Cashmore, A. R. (1998). Cryptochrome blue-light photoreceptors of arabidopsis implicated in phototropism. Nature 392, 720–723. doi: 10.1038/33701
Alabadıí, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Más, P., Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Alexandre, C. M., Hennig, L. (2008). FLC or not FLC: the other side of vernalization. J. Exp. Bot. 59, 1127–1135. doi: 10.1093/jxb/ern070
Alvarez-Buylla, E. R., García-Ponce, B., Garay-Arroyo, A. (2006). Unique and redundant functional domains of APETALA1 and CAULIFLOWER, two recently duplicated arabidopsis thaliana floral MADS-box genes. J. Exp. Bot. 57, 3099–3107. doi: 10.1093/jxb/erl081
Andrés, F., Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Ausín, I., Alonso-Blanco, C., Jarillo, J. A., Ruiz-García, L., Martínez-Zapater, J. M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36, 162–166. doi: 10.1038/ng1295
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Baudry, A., Ito, S., Song, Y. H., Strait, A. A., Kiba, T., Lu, S., et al. (2010). F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control arabidopsis clock progression. Plant Cell 22, 606–622. doi: 10.1105/tpc.109.072843
Bernier, G., Périlleux, C. (2005). A physiological overview of the genetics of flowering time control. Plant Biotechnol. J. 3, 3–16. doi: 10.1111/j.1467-7652.2004.00114.x
Bluemel, M., Dally, N., Jung, C. (2015). Flowering time regulation in crops–what did we learn from arabidopsis? Curr. Opin. Biotechnol. 32, 121–129. doi: 10.1016/j.copbio.2014.11.023
Boss, P. K., Bastow, R. M., Mylne, J. S., Dean, C. (2004). Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16, S18–S31. doi: 10.1105/tpc.015958
Bouché, F., Woods, D. P., Amasino, R. M. (2017). Winter memory throughout the plant kingdom: different paths to flowering. Plant Physiol. 173, 27–35. doi: 10.1104/pp.16.01322
Brasileiro, B. G., Dias, D.C.F.D.S., Bhering, M. C., Dias, L. (2012). Floral biology and characterization of seed germination in physic nut (Jatropha curcas l.). Rev. Bras. Sementes. 34, 556–560. doi: 10.1590/S0101-31222012000400005
Cao, Y., Liu, M., Long, H., Zhao, Q., Jiang, L., Zhang, L. (2020). Hidden in plain sight: Systematic investigation of leucine-rich repeat containing genes unveil the their regulatory network in response to fusarium wilt in tung tree. Int. J. Biol. Macromol. 163, 1759–1767. doi: 10.1016/j.ijbiomac.2020.09.106
Cao, Y., Liu, W., Zhao, Q., Long, H., Li, Z., Liu, M., et al. (2019a). Integrative analysis reveals evolutionary patterns and potential functions of SWEET transporters in euphorbiaceae. Int. J. Biol. macromol. 139, 1–11. doi: 10.1016/j.ijbiomac.2019.07.102
Cao, Y., Meng, D., Han, Y., Chen, T., Jiao, C., Chen, Y., et al. (2019b). Comparative analysis of b-BOX genes and their expression pattern analysis under various treatments in dendrobium officinale. BMC Plant Biol. 19, 245. doi: 10.1186/s12870-019-1851-6
Chan, A. P., Crabtree, J., Zhao, Q., Lorenzi, H., Orvis, J., Puiu, D., et al. (2010). Draft genome sequence of the oilseed species ricinus communis. Nat. Biotechnol. 28, 951–956. doi: 10.1038/nbt.1674
Cheng, X. F., Wang, Z. Y. (2005). Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in arabidopsis thaliana. Plant J. 43, 758–768. doi: 10.1111/j.1365-313X.2005.02491.x
Cho, L. H., Yoon, J., An, G. (2017). The control of flowering time by environmental factors. Plant J. 90, 708–719. doi: 10.1111/tpj.13461
Clack, T., Mathews, S., Sharrock, R. A. (1994). The phytochrome apoprotein family inArabidopsis is encoded by five genes: the sequences and expression ofPHYD andPHYE. Plant Mol. Biol. 25, 413–427. doi: 10.1007/BF00043870
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of arabidopsis. science 316, 1030–1033. doi: 10.1126/science.1141752
Debrieux, D., Fankhauser, C. (2010). Light-induced degradation of phyA is promoted by transfer of the photoreceptor into the nucleus. Plant Mol. Biol. 73, 687–695. doi: 10.1007/s11103-010-9649-9
Deng, W., Ying, H., Helliwell, C. A., Taylor, J. M., Peacock, W. J., Dennis, E. S. (2011). FLOWERING LOCUS c (FLC) regulates development pathways throughout the life cycle of arabidopsis. Proc. Natl. Acad. Sci. 108, 6680–6685. doi: 10.1073/pnas.1103175108
Emms, D. M., Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14. doi: 10.1186/s13059-019-1832-y
Farré, E. M., Harmer, S. L., Harmon, F. G., Yanovsky, M. J., Kay, S. A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the arabidopsis circadian clock. Curr. Biol. 15, 47–54. doi: 10.1016/j.cub.2004.12.067
Fornara, F., De Montaigu, A., Coupland, G. (2010). SnapShot: control of flowering in arabidopsis. Cell 141, 550–550. doi: 10.1016/j.cell.2010.04.024
Fornara, F., De Montaigu, A., Sánchez-Villarreal, A., Takahashi, Y., Ver Loren Van Themaat, E., Huettel, B., et al. (2015). The GI–CDF module of arabidopsis affects freezing tolerance and growth as well as flowering. Plant J. 81, 695–706. doi: 10.1111/tpj.12759
Fornara, F., Panigrahi, K. C. S., Gissot, L., Sauerbrunn, N., Rühl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Fujiwara, S., Oda, A., Yoshida, R., Niinuma, K., Miyata, K., Tomozoe, Y., et al. (2008). Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in arabidopsis. Plant Cell 20, 2960–2971. doi: 10.1105/tpc.108.061531
Gendall, A. R., Levy, Y. Y., Wilson, A., Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in arabidopsis. Cell 107, 525–535. doi: 10.1016/S0092-8674(01)00573-6
Goralogia, G. S., Liu, T. K., Zhao, L., Panipinto, P. M., Groover, E. D., Bains, Y. S., et al. (2017). CYCLING DOF FACTOR 1 represses transcription through the TOPLESS co-repressor to control photoperiodic flowering in arabidopsis. Plant J. 92, 244–262. doi: 10.1111/tpj.13649
Grandi, V., Gregis, V., Kater, M. M. (2012). Uncovering genetic and molecular interactions among floral meristem identity genes in arabidopsis thaliana. Plant J. 69 (5), 881–893. doi: 10.1111/j.1365-313X.2011.04840.x
Ha, J., Shim, S., Lee, T., Kang, Y. J., Hwang, W. J., Jeong, H., et al. (2019). Genome sequence of jatropha curcas l., a non-edible biodiesel plant, provides a resource to improve seed-related traits. Plant Biotechnol. J. 17, 517–530. doi: 10.1111/pbi.12995
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Hecht, V., Foucher, F., Ferrándiz, C., Macknight, R., Navarro, C., Morin, J., et al. (2005). Conservation of arabidopsis flowering genes in model legumes. Plant Physiol. 137, 1420–1434. doi: 10.1104/pp.104.057018
Hepworth, S. R., Klenz, J. E., Haughn, G. W. (2006). UFO In the arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression. Planta 223, 769–778. doi: 10.1007/s00425-005-0138-3
Hoecker, U. (2005). Regulated proteolysis in light signaling. Curr. Opin. Plant Biol. 8, 469–476. doi: 10.1016/j.pbi.2005.07.002
Holtkotte, X., Ponnu, J., Ahmad, M., Hoecker, U. (2017). The blue light-induced interaction of cryptochrome 1 with COP1 requires SPA proteins during arabidopsis light signaling. PloS Genet. 13, e1007044. doi: 10.1371/journal.pgen.1007044
Imaizumi, T. (2010). Arabidopsis circadian clock and photoperiodism: time to think about location. Curr. Opin. Plant Biol. 13, 83–89. doi: 10.1016/j.pbi.2009.09.007
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., Kay, S. A. (2005). FKF1 f-box protein mediates cyclic degradation of a repressor of CONSTANS in arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Jackson, S. D. (2009). Plant responses to photoperiod. New Phytol. 181, 517–531. doi: 10.1111/j.1469-8137.2008.02681.x
Jacobsen, S. E., Olszewski, N. E. (1993). Mutations at the SPINDLY locus of arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896. doi: 10.1105/tpc.5.8.887
Jean Finnegan, E., Kovac, K. A., Jaligot, E., Sheldon, C. C., James Peacock, W., Dennis, E. S. (2005). The downregulation of FLOWERING LOCUS c (FLC) expression in plants with low levels of DNA methylation and by vernalization occurs by distinct mechanisms. Plant J. 44, 420–432. doi: 10.1111/j.1365-313X.2005.02541.x
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., Mcanulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Jung, C., Müller, A. E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573. doi: 10.1016/j.tplants.2009.07.005
Jung, W. Y., Park, H. J., Lee, A., Lee, S. S., Kim, Y.-S., Cho, H. S. (2016). Identification of flowering-related genes responsible for differences in bolting time between two radish inbred lines. Front. Plant Sci. 7, 1844. doi: 10.3389/fpls.2016.01844
Jung, J.-H., Seo, Y.-H., Seo, P. J., Reyes, J. L., Yun, J., Chua, N.-H., et al. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in arabidopsis. Plant Cell 19, 2736–2748. doi: 10.1105/tpc.107.054528
Jung, C.-H., Wong, C. E., Singh, M. B., Bhalla, P. L. (2012). Comparative genomic analysis of soybean flowering genes. PloS One 7, e38250. doi: 10.1371/journal.pone.0038250
Kaneko-Suzuki, M., Kurihara-Ishikawa, R., Okushita-Terakawa, C., Kojima, C., Nagano-Fujiwara, M., Ohki, I., et al. (2018). TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD. Plant Cell Physiol. 59, 458–468. doi: 10.1093/pcp/pcy021
Kim, W.-Y., Ali, Z., Park, H. J., Park, S. J., Cha, J.-Y., Perez-Hormaeche, J., et al. (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in arabidopsis. Nat. Commun. 4, 1–13. doi: 10.1038/ncomms2846
Kim, M. Y., Kang, Y. J., Lee, T., Lee, S. H. (2013). Divergence of flowering-related genes in three legume species. Plant Genome 6, 1–12. doi: 10.3835/plantgenome2013.03.0008
Koornneef, M., Hanhart, C. J., van der Veen, J. H. (1991). A genetic and physiological analysis of late flowering mutants in arabidopsis thaliana. Mol. Gen. Genet. MGG. 229, 57–66. doi: 10.1007/BF00264213
Lee, J., Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Lee, J. H., Lee, J. S., Ahn, J. H. (2008). Ambient temperature signaling in plants: an emerging field in the regulation of flowering time. J. Plant Biol. 51, 321–326. doi: 10.1007/BF03036133
Lee, J. H., Ryu, H.-S., Chung, K. S., Posé, D., Kim, S., Schmid, M., et al. (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628–632. doi: 10.1126/science.1241097
Li, L., Li, X., Liu, Y., Liu, H. (2016). Flowering responses to light and temperature. Sci. China Life Sci. 59, 403–408. doi: 10.1007/s11427-015-4910-8
Li, W., Liu, M., Dong, X., Cao, H., Zhang, L. (2020). Flower biology and ontogeny of the tung tree (Vernicia fordii hemsl.). Trees 34, 1363–1381. doi: 10.1007/s00468-020-02041-3
Li, D., Liu, C., Shen, L., Wu, Y., Chen, H., Robertson, M., et al. (2008). A repressor complex governs the integration of flowering signals in arabidopsis. Dev. Cell 15, 110–120. doi: 10.1016/j.devcel.2008.05.002
Li, C., Luo, L., Fu, Q., Niu, L., Xu, Z.-F. (2014). Isolation and functional characterization of JcFT, a FLOWERING LOCUS T (FT) homologous gene from the biofuel plant jatropha curcas. BMC Plant Biol. 14, 125. doi: 10.1186/1471-2229-14-125
Lim, M.-H., Kim, J., Kim, Y.-S., Chung, K.-S., Seo, Y.-H., Lee, I., et al. (2004). A new arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS c. Plant Cell 16, 731–740. doi: 10.1105/tpc.019331
Liu, Y., Hao, X., Lu, Q., Zhang, W., Zhang, H., Wang, L., et al. (2020). Genome-wide identification and expression analysis of flowering-related genes reveal putative floral induction and differentiation mechanisms in tea plant (Camellia sinensis). Genomics. 112 (3), 2318–2326. doi: 10.1016/j.ygeno.2020.01.003
Liu, J., Shi, C., Shi, C.-C., Li, W., Zhang, Q.-J., Zhang, Y., et al. (2020). The chromosome-based rubber tree genome provides new insights into spurge genome evolution and rubber biosynthesis. Mol. Plant 13, 336–350. doi: 10.1016/j.molp.2019.10.017
Li, D., Zhang, H., Mou, M., Chen, Y., Xiang, S., Chen, L., et al. (2019). Arabidopsis class II TCP transcription factors integrate with the FT–FD module to control flowering. Plant Physiol. 181, 97–111. doi: 10.1104/pp.19.00252
Luo, X., He, Y. (2020). Experiencing winter for spring flowering: A molecular epigenetic perspective on vernalization. J. Integr. Plant Biol. 62, 104–117. doi: 10.1111/jipb.12896
Madrid, E., Chandler, J. W., Coupland, G. (2020). Gene regulatory networks controlled by FLOWERING LOCUS c that confer variation in seasonal flowering and life history. J. Exp. Bot 72 (1), 4–14. doi: 10.1093/jxb/eraa216
Marquardt, S., Boss, P. K., Hadfield, J., Dean, C. (2006). Additional targets of the arabidopsis autonomous pathway members, FCA and FY. J. Exp. Bot. 57, 3379–3386. doi: 10.1093/jxb/erl073
Matar, S., Kumar, A., Holtgräwe, D., Weisshaar, B., Melzer, S. (2020). The transition to flowering in winter rapeseed during vernalization. Plant. Cell Environment. 44 (2), 506–518. doi: 10.1111/pce.13946
Michaels, S. D., Amasino, R. M. (1999). FLOWERING LOCUS c encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Michaels, S. D., Amasino, R. M. (2001). Loss of FLOWERING LOCUS c activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941. doi: 10.1105/tpc.13.4.935
Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., et al. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in arabidopsis. Plant Cell 17, 2255–2270. doi: 10.1105/tpc.105.033464
Moon, J., Lee, H., Kim, M., Lee, I. (2005). Analysis of flowering pathway integrators in arabidopsis. Plant Cell Physiol. 46, 292–299. doi: 10.1093/pcp/pci024
Moon, J., Suh, S. S., Lee, H., Choi, K. R., Hong, C. B., Paek, N. C., et al. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in arabidopsis. Plant J. 35, 613–623. doi: 10.1046/j.1365-313X.2003.01833.x
Mouhu, K., Hytönen, T., Folta, K., Rantanen, M., Paulin, L., Auvinen, P., et al. (2009). Identification of flowering genes in strawberry, a perennial SD plant. BMC Plant Biol. 9, 122. doi: 10.1186/1471-2229-9-122
Mutasa-Göttgens, E., Hedden, P. (2009). Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 60, 1979–1989. doi: 10.1093/jxb/erp040
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nishio, H., Iwayama, K., Kudoh, H. (2020). Duration of cold exposure defines the rate of reactivation of a perennial FLC orthologue via H3K27me3 accumulation. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-020-72566-7
Noh, Y.-S., Amasino, R. M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in arabidopsis. Plant Cell 15, 1671–1682. doi: 10.1105/tpc.012161
Ojolo, S. P., Cao, S., Priyadarshani, S., Li, W., Yan, M., Aslam, M., et al. (2018). Regulation of plant growth and development: a review from a chromatin remodeling perspective. Front. Plant Sci. 9, 1232. doi: 10.3389/fpls.2018.01232
Peng, F. Y., Hu, Z., Yang, R.-C. (2015). Genome-wide comparative analysis of flowering-related genes in arabidopsis, wheat, and barley. Int. J. Plant Genomics 2015, 874361. doi: 10.1155/2015/874361
Post, E. S., Pedersen, C., Wilmers, C. C., Forchhammer, M. C. (2008). Phenological sequences reveal aggregate life history response to climatic warming. Ecology 89, 363–370. doi: 10.1890/06-2138.1
Putterill, J., Laurie, R., Macknight, R. (2004). It's time to flower: the genetic control of flowering time. Bioessays 26, 363–373. doi: 10.1002/bies.20021
Quesada, V., Macknight, R., Dean, C., Simpson, G. G. (2003). Autoregulation of FCA pre-mRNA processing controls arabidopsis flowering time. EMBO J. 22, 3142–3152. doi: 10.1093/emboj/cdg305
Riboni, M., Galbiati, M., Tonelli, C., Conti, L. (2013). GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1. Plant Physiol. 162, 1706–1719. doi: 10.1104/pp.113.217729
Riechmann, J. L., Krizek, B. A., Meyerowitz, E. M. (1996). Dimerization specificity of arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. 93, 4793–4798. doi: 10.1073/pnas.93.10.4793
Sablowski, R. (2007). Flowering and determinacy in arabidopsis. J. Exp. Bot. 58, 899–907. doi: 10.1093/jxb/erm002
Sawa, M., Kay, S. A. (2011). GIGANTEA directly activates flowering locus T in arabidopsis thaliana. Proc. Natl. Acad. Sci. 108, 11698–11703. doi: 10.1073/pnas.1106771108
Sawa, M., Nusinow, D. A., Kay, S. A., Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Schomburg, F. M., Patton, D. A., Meinke, D. W., Amasino, R. M. (2001). FPA, a gene involved in floral induction in arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13, 1427–1436. doi: 10.1105/TPC.010017
Searle, I., Coupland, G. (2004). Induction of flowering by seasonal changes in photoperiod. EMBO J. 23, 1217–1222. doi: 10.1038/sj.emboj.7600117
Sheldon, C. C., Finnegan, E. J., Rouse, D. T., Tadege, M., Bagnall, D. J., Helliwell, C. A., et al. (2000). The control of flowering by vernalization. Curr. Opin. Plant Biol. 3, 418–422. doi: 10.1016/S1369-5266(00)00106-0
Silva, C. S., Puranik, S., Round, A., Brennich, M., Jourdain, A., Parcy, F., et al. (2016). Evolution of the plant reproduction master regulators LFY and the MADS transcription factors: the role of protein structure in the evolutionary development of the flower. Front. Plant Sci. 6, 1193. doi: 10.3389/fpls.2015.01193
Simpson, G. G. (2004). The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of arabidopsis flowering time. Curr. Opin. Plant Biol. 7, 570–574. doi: 10.1016/j.pbi.2004.07.002
Simpson, G. G., Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. doi: 10.1126/science.296.5566.285
Song, Y. H., Smith, R. W., To, B. J., Millar, A. J., Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049. doi: 10.1126/science.1219644
Southwick, S. M., Davenport, T. L. (1986). Characterization of water stress and low temperature effects on flower induction in citrus. Plant Physiol. 81, 26–29. doi: 10.1104/pp.81.1.26
Steinbach, Y., Hennig, L. (2014). Arabidopsis MSI1 functions in photoperiodic flowering time control. Front. Plant Sci. 5, 77. doi: 10.3389/fpls.2014.00077
Strasser, B., Alvarez, M. J., Califano, A., Cerdán, P. D. (2009). A complementary role for ELF3 and TFL1 in the regulation of flowering time by ambient temperature. Plant J. 58, 629–640. doi: 10.1111/j.1365-313X.2009.03811.x
Sung, S., Amasino, R. M. (2004). Vernalization and epigenetics: how plants remember winter. Curr. Opin. Plant Biol. 7, 4–10. doi: 10.1016/j.pbi.2003.11.010
Takagi, N., Ueguchi, C. (2012). Enhancement of meristem formation by bouquet-1, a mis-sense allele of the VERNALIZATION INDEPENDENCE 3 gene encoding a WD 40 repeat protein in arabidopsis thaliana. Genes to Cells 17, 982–993. doi: 10.1111/gtc.12014
Townsend, T., Albani, M., Wilkinson, M., Coupland, G., Battey, N. (2018). The diversity and significance of flowering in perennials. Annu. Plant Rev. Online 181-197. doi: 10.1002/9781119312994.apr0202
Trevaskis, B., Hemming, M. N., Dennis, E. S., Peacock, W. J. (2007). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12, 352–357. doi: 10.1016/j.tplants.2007.06.010
Turck, F., Fornara, F., Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. doi: 10.1126/science.1091761
Van Dijk, A. D. J., Molenaar, J. (2017). Floral pathway integrator gene expression mediates gradual transmission of environmental and endogenous cues to flowering time. PeerJ 5, e3197. doi: 10.7717/peerj.3197
Wang, W., Feng, B., Xiao, J., Xia, Z., Zhou, X., Li, P., et al. (2014). Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 5, 1–9. doi: 10.1038/ncomms6110
Wang, C.-Q., Guthrie, C., Sarmast, M. K., Dehesh, K. (2014). BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription, defining a flowering time checkpoint in arabidopsis. Plant Cell 26, 3589–3602. doi: 10.1105/tpc.114.130252
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49. doi: 10.1093/nar/gkr1293
Weller, J. L., Batge, S. L., Smith, J. J., Kerckhoffs, L. H. J., Sineshchekov, V. A., Murfet, I. C., et al. (2004). A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome a. Plant Physiol. 135, 2186–2195. doi: 10.1104/pp.103.036103
Whittaker, C., Dean, C. (2017). The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33, 555–575. doi: 10.1146/annurev-cellbio-100616-060546
Wigge, P. A. (2011). FT, a mobile developmental signal in plants. Curr. Biol. 21, R374–R378. doi: 10.1016/j.cub.2011.03.038
Wigge, P. A. (2013). Ambient temperature signalling in plants. Curr. Opin. Plant Biol. 16, 661–666. doi: 10.1016/j.pbi.2013.08.004
Yang, M., Wu, Y., Jin, S., Hou, J., Mao, Y., Liu, W., et al. (2015). Flower bud transcriptome analysis of sapium sebiferum (Linn.) roxb. and primary investigation of drought induced flowering: pathway construction and G-quadruplex prediction based on transcriptome. PloS One 10, e0118479. doi: 10.1371/journal.pone.0118479
Yoshida, H., Hirano, K., Sato, T., Mitsuda, N., Nomoto, M., Maeo, K., et al. (2014). DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc. Natl. Acad. Sci. 111, 7861–7866. doi: 10.1073/pnas.1321669111
Zhang, L., Jiang, Y., Zhu, Y., Su, W., Long, T., Huang, T., et al. (2019a). Functional characterization of GI and CO homologs from eriobotrya deflexa nakai forma koshunensis. Plant Cell Rep. 38, 533–543. doi: 10.1007/s00299-019-02384-3
Zhang, L., Liu, M., Long, H., Dong, W., Pasha, A., Esteban, E., et al. (2019b). Tung tree (Vernicia fordii) genome provides a resource for understanding genome evolution and improved oil production. Genom. Proteomics Bioinf. 17, 558–575. doi: 10.1016/j.gpb.2019.03.006
Keywords: Euphorbiaceae, flowering-related genes, pathway, mechanisms, family
Citation: Jiang L, Fan T, Wang L, Zhang L and Xu J (2022) Divergence of flowering-related genes to control flowering in five Euphorbiaceae genomes. Front. Plant Sci. 13:1015114. doi: 10.3389/fpls.2022.1015114
Received: 09 August 2022; Accepted: 20 September 2022;
Published: 19 October 2022.
Edited by:
Hui Song, Qingdao Agricultural University, ChinaReviewed by:
Chongchong Yan, Anhui Academy of Agricultural Sciences (CAAS), ChinaLiwei Zheng, Zhengzhou University, China
Copyright © 2022 Jiang, Fan, Wang, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihu Wang, d2FuZ2xpaHVAaGViZXUuZWR1LmNu; Lin Zhang, bHpoYW5nc3NAbXNuLmNvbQ==; Jun Xu, eHVqdW5zZWFAMTYzLmNvbQ==
 Lan Jiang
Lan Jiang Tingting Fan
Tingting Fan Lihu Wang
Lihu Wang Lin Zhang
Lin Zhang Jun Xu6*
Jun Xu6*