
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 01 December 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1014176
This article is part of the Research TopicThe Evolution, Diversity and Functions of Alternative Splicing (AS) in Plant Responses to Abiotic StressView all 5 articles
In wheat production, appropriate flowering time and ideal plant architecture are the prerequisites for high grain yield. Alternative splicing (AS) is a vital process that regulates gene expression at the post-transcriptional level, and AS events in wheat have been found to be closely related to grain-related traits and abiotic stress tolerance. However, AS events and their biological roles in regulating flowering time and plant architecture in wheat remain unclear. In this study, we report that TaNAK1 undergoes AS, producing three splicing variants. Molecular characterization of TaNAK1 and its splicing variants demonstrated that all three protein isoforms have a conserved NB-ARC domain and a protein kinase domain, but the positions of these two domains and the length of the protein kinase domains are different among them, implying that they may have different three-dimensional structures and therefore have different functions. Further investigations showed that the two splicing variants of TaNAK1, TaNAK1.1 and TaNAK1.2, exhibited different expression patterns during wheat growth and development, while the other one, TaNAK1.3, was not detected. Subcellular localization demonstrated that TaNAK1.1 was mainly localized in the cytoplasm, while TaNAK1.2 was localized in the nucleus and cytoplasm. Both TaNAK1.1 and TaNAK1.2 exhibit protein kinase activity in vitro. Ectopic expression of TaNAK1.1 and TaNAK1.2 in Arabidopsis demonstrated that these two splicing variants play opposite roles in regulating flowering time and plant architecture, resulting in different seed yields. TaNAK1.2 positive regulates the transition from vegetative to reproductive growth, plant height, branching number, seed size, and seed yield of Arabidopsis, while TaNAK1.1 negatively regulates these traits. Our findings provide new gene resource for regulating flowering time and plant architecture in crop breeding for high grain yield.
Wheat (Triticum aestivum L.), one of the major staple crops worldwide, provides more than one fifth calories and protein for mankind. Improving wheat production is critical to world food and nutrition security (FAO, http://faostat.fao.org). In crop production, appropriate flowering and ideal plant architecture are the prerequisites for high yield (Wang et al., 2018). The appropriate flowering time is crucial for wheat to deal with adverse environmental factors, such as cold, freezing, high temperature, rainstorm, and various diseases, to complete life generation and to maintain species reproduction (Worland, 1996). Meanwhile, it is also conducive to get high yield and better quality through the coordination of vegetative and reproductive growth (Kamran et al., 2014). Therefore, in the long-term practice of wheat cultivation, domestication and production, flowering time has been regarded as one of the important agronomic traits. Plant architecture refers to the structure and three-dimensional organization of all organs of a plant, which includes aspects such as stem height, tillering/branching pattern and number, leaf shape and arrangement, the number of inflorescences/flowers and their distribution, as well as root morphology and distribution. The significance of plant architecture in agriculture is that it is regulated by both genetic and environmental factors. This confers extensive phenotypic plasticity that enables plants to adapt to different environment, facilitating selection for favorable traits such as grain yield and harvest indices (Reinhardt and Kuhlemeier, 2002). Crop plant architecture is regarded as one of the most crucial factors that affect grain yield. Wheat breeders have been focused on its architecture, including plant height, tiller number and angle, leaf shape, size, and angle, and spike morphology since the Green Revolution, the development and cultivation of semi-dwarf wheat and rice varieties that have greatly increased crop production since 1960 (Wang et al., 2018; Li et al., 2019). Therefore, it is of great significance to explore and identify the genes regulating flowering time and plant architecture in wheat, understand their molecular mechanism, and realize the fine regulation of flowering time and plant architecture, so as to improve grain yield and ensure food security.
Alternative splicing (AS) is a vital process for gene expression regulation at the post-transcriptional level, in which a pre-mRNA can produce multiple transcripts through different splicing modes to improve transcriptome plasticity and proteome diversity, to increase functional complexity (Yu et al., 2016; Ananda et al., 2022), and even to engender trait diversity (Gao et al., 2021). Therefore, AS plays an important role in plant adaptation and evolution. AS mainly includes exon skipping (ES), intron retention (IR), alternative 5’ splice site (alt 5’SS), alternative 3’ splice site (alt 3’SS), variable first exon (VFE), variable last exon (VLE) and mutually exclusive exons (MEE) (Ule and Blencowe, 2019). AS can generate truncated proteins, disrupting main domains to form non-functional proteins (Kalyna et al., 2012) or annulling interaction with other proteins and thereby impeding the formation of functional protein complexes (Qin et al., 2017). Moreover, some splicing variants can antagonistically compete with other variant and interfere with its function in a negative manner (Posé et al., 2013; Ren et al., 2021). Increasing evidences have demonstrated that AS participates in the regulation of plant growth and development. In Arabidopsis, the MADS-box transcription factor genes FLOWERING LOCUS M (FLM) and SHORT VEGETATIVE PHASE (SVP) have key functions in regulation of flowering. FLM undergoes temperature-dependent alternative splicing, and antagonistic FLM variants, FLM-β and FLM-δ, compete for interaction with the floral repressor SVP to regulate flowering (Posé et al., 2013). AS of Delay of Germination1 (DOG1) produces two mRNA variants, lgDOG1 and shDOG1. In contrast to lgDOG1, shDOG1 is translated and functions to promote seed dormancy in Arabidopsis (Cyrek et al., 2016). AS is involved in the regulation of some grain-related traits. In rice, AS of OsLG3b, a gene encoding MADS-box transcription factor1 (OsMADS1), produces the truncated proteins that are positively associated with grain length and contributes to grain yield (Yu et al., 2018). In Oryza officinalis Wall. ex Watt, MKK3 encoding mitogen-activated protein kinase 3 is subject to alternative splicing and produces splicing variants, and overexpressing four of the five OsMKK3 splicing variants leads to reduced grain length and width (Pan et al., 2021). Wheat has very huge and complex genome, and substantial wheat genes have complicated roles (Singh et al., 2018). The genome-wide AS profiling uncovered a very complex AS landscape in wheat, exhibiting that almost 22% of genes occur AS (Yu et al., 2020). Increasing evidences have uncovered that AS events in wheat are enormously related to grain-related traits such as polyphenol oxidase activity (Sun et al., 2010), starch synthase activity (Zhou et al., 2018), and grain weight and size (Ren et al., 2021). AS is also related to abiotic stresses tolerance (Egawa et al., 2006; Liu et al., 2018) and disease resistance (Wang et al., 2020; Sánchez-Martín et al., 2021). However, AS events and their biological roles in regulating wheat flowering time and plant architecture are poorly understood.
Our previous study identified a wheat specific-miRNA tae-miR5048 from wheat small RNA (sRNA) library, which is highly expressed in seedlings and flag leaves (Han et al., 2014). Recently, a RLK gene (ID TraeCS4B01G313900) in IWGSC RefSeq v1.0) (https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Annotations/v1.0/), was identified to be one of tae-miR5048 target genes by using psRNA Target software (http://plantgrn.noble.org/psRNATarget/) and Degradome sequencing (unpublished). However, its biological function remains elusive. In this study, we found that this RLK gene is subject to AS and produces three splicing variants encoding the NB-ARC (Nucleotide-binding adaptor shared by APAF-1, R proteins and CED-4) domain and protein kinase domain proteins, thereby named as TaNAK1. Molecular characterization of TaNAK1 and its three splice variants demonstrated that all three protein isoforms have a conserved NB-ARC domain and a protein kinase domain, but the positions of these two domains and the length of the protein kinase domains are different among them, implying that they may have different three-dimensional structures and therefore have different biological functions. Further investigations showed that the two splicing variants of TaNAK1, TaNAK1.1 and TaNAK1.2, exhibited different expression patterns during wheat growth and development, while the other splice variant, TaNAK1.3, was not detected. Subcellular localization showed that TaNAK1.1 was mainly localized in the cytoplasm, while TaNAK1.2 was localized in the nucleus and cytoplasm. Both TaNAK1.1 and TaNAK1.2 exhibit the activity of protein kinase in vitro. Ectopic expression of TaNAK1.1 and TaNAK1.2 in Arabidopsis demonstrated that these two splicing variants play opposing roles in regulating flowering time and plant architecture, resulting in different seed yields. Our findings provide new gene resource for regulating flowering time and plant architecture in crop breeding for high yield.
Wheat cultivar cv. Chinese Spring was used to clone the cDNA of TaNAK1, and winter wheat cv. Xiaoyan 6 was used to determine the expression patterns of the splicing variants of TaNAK1 during wheat growth and development. After imbibition, the seeds of Chinese Spring were placed on wet filter paper and germinated at the condition of 22°C, 16 h light/8 h dark, and 75% relative humidity. Leaf and root samples were collected from wheat seedlings at the two-leaf stage, rapidly frozen in liquid nitrogen, and then stored at -80°C for RNA extraction and cDNA cloning. Xiaoyan 6 was planted in the experimental plots at Northwest A & F University, Yangling, Shaanxi, China (longitude 108°4′E, latitude 34°17′N) during the natural growing seasons in 2018 and 2020. Ten wheat organs/tissues, including root (R), stem (S), leaf (L) of five-leaf-stage seedlings, flag leaf (FL) from wheat plants at heading stages, the 5 cm long young ear (YS5) from wheat plants at booting stage, and the 10 cm long spike (YS10) from wheat plants at heading stage, the grains 5, 10, 15, 20, and 25 days post-anthesis (GR5, GR10, GR15, and GR20, respectively), were collected and rapidly frozen in liquid nitrogen and stored at -80°C for further usage, three independent biological replicates being included for individual tissues/organs.
Arabidopsis (ecotype Columbia-0) was used to conducted genetic transformation in this study. The seeds of Arabidopsis were sterilized and sowed after 2 days imbibition at 4°C, and the plants were cultured in a phytotron with 22°C, the photoperiod of 16 h light/8 h dark, the relative humidity of 70% and light intensity of 130-150 μmol/m2/s.
A wheat gene (ID TraeCS4B01G313900), which was identified as one of the tae-miR5048 targets (unpublished data), is annotated as a receptor kinase gene (RK) and is subject to AS, generating three splicing variants (TraeCS4B01G313900.1, TraeCS4B01G313900.2, and TraeCS4B01G313900.3), based on the annotation in IWGSC RefSeq v1.0 (https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Annotations/v1.0/). Here, the gene is tentatively named as TaRK4B-1, and the three splicing variants are named as TaRK4B-1.1, TaRK4B-1.2, and TaRK4B-1.3, respectively. The genome DNA, cDNA, and the CDS sequences of TaRK4B-1 were downloaded from IWGSC RefSeq v1.0, and used to analyze the gene structure. SMART (http://smart.embl.de/) was used to identify conserved domains contained in target proteins. DNAMAN v9 (https://www.lynnon.com/qa.html) was used for multi-sequences alignment.
Total RNA of different organs/tissues from wheat or Arabidopsis was extracted using Polysaccharide and Polyphenol total RNA isolation Kit (BioTeke Corporation, Beijing, China) according to the manufacturer’s instruction. The quality of RNA samples obtained was assessed by 2.0% agarose gel electrophoresis. First strand cDNA synthesis was performed using Takara’s reverse transcription kit PrimeScript ™ IV 1st strand cDNA Synthesis Mix (Takara, Japan).
Because of the very high sequence similarity of TaNAK1 splicing variants, it was not possible to design specific primers for quantitative analysis of individual splicing variants through real-time quantitative RT-PCR (qRT-PCR). Therefore, the expression levels of alternative splicing variants of TaNAK1 in different developmental organs/tissues of wheat cv. Xiaoyan 6 were detected by semi-qRT-PCR with gene-specific primer pair (Supplementary Table S1) and the cDNA from different wheat tissues as templates, TaACTIN (TraesCS1A01G020500) being used as a reference gene.
Due to the very high sequence similarity of TaNAK1 splicing variants, a universal primer pair (TaNAK1-F/R) that can amplify all the alternative splicing variants of TaNAK1 was designed (Supplementary Table S1) according to the CDS sequences of TaNAK1 downloaded from IWGSC RefSeq v1.0. The alternative splicing variants of TaNAK1 were amplified by PCR with the primer pair and wheat leaf and grain cDNA mixture as template. PCR products were purified and ligated to a pMD™19-T vector (Takara Biotechnology (Dalian) Co. Ltd., Japan) for sequencing. At least five clones of each PCR product were selected for sequencing.
In order to reveal the subcellular localization of TaNAK1.1 and TaNAK1.2, the coding sequences of TaNAK1.1 and TaNAK1.2 were separately amplified using the primer pair containing restriction enzyme sites TaNAK1-SalI-F/TaNAK1-BamHI-R (Supplementary Table S1) and pMD19-T-TaNAK1.1 and pMD19-T-TaNAK1.2 as a template, respectively. The coding sequences of the amino terminus of two protein isoforms (1-257aa and 1-353 aa in TaNAK1.1, 1-306aa and 1-421 aa in TaNAK1.2) that include only their kinase domains or both the kinase domains and the NA-ARC domains were also separately amplified with primer pairs TaNAK1-HindIII-F/TaNAK1.1-N1-SalI-R, TaNAK1-HindIII-F/TaNAK1.1-N1-SalI-R, TaNAK1-HindIII-F/TaNAK1.2-N1-SalI-R, and TaNAK1-HindIII-F/TaNAK1.2-N2-SalI-R. The coding sequences of the carboxyl terminal that contain NB-ARC domains and all subsequent sequences) of the two protein isoforms were amplified with primer pair TaNAK1-C-SalI-F/TaNAK1-BamHI-R. These PCR products were separately digested by their restriction enzymes and subcloned into the p16318-GFP expression vector between the CaMV35S promoter and the GFP gene and fused with GFP in frame, generating transient expression vectors p16318-TaNAK1.1-GFP, p16318-TaNAK1.2-GFP, p16318-TaNAK1.11-257-GFP, p16318-TaNAK1.11-353-GFP, p16318-TaNAK1.21-306-GFP, p16318-TaNAK1.21-421-GFP and p16318-TaNAK1.1/1.2c-terminal-GFP, respectively. The resulted constructs and the control vector p16318-GFP were separately introduced into strain DH5α, and the endotoxin free plasmids of these vectors were extracted with GoldHi EndoFree Plasmid Maxi Kit (BOYAO Biotechnology, Shanghai, China). The p16318-TaNAK1.1-GFP, p16318-TaNAK1.2-GFP, p16318-TaNAK1.11-257-GFP, p16318-TaNAK1.11-353-GFP, p16318-TaNAK1.21-306-GFP p16318-TaNAK1.21-421-GFP, and p16318-TaNAK1.1/1.2c-terminal-GFP as well as the control vector p16318-GFP were separately transformed or co-transformed with nuclear localization marker vector NLS-mCherry into wheat mesophyll protoplasts that were released from leaves of 6-10 days wheat seedlings by polyethylene glycol (PEG)-mediated transient expression system, followed by incubation in the dark at 25°C for 15h (Yoo et al., 2007). Moreover, the recombinant vectors were separately bombarded into onion epidermal cells via a gene gun system (Bio-Rad, United States), with p35S::GFP as a control, and the transformed cells were incubated on 1/2 Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) in light or darkness for 36–48 h at 28°C. GFP, the subcellular localization of fusion proteins and mCherry in the transformed cell was observed with Leica inverted microscope DMi8 (Leica, Germany).
In order to detect the kinase activity of TANAK.1 and TaNAK1.2, we first constructed prokaryotic expression vectors of the kinase domains of TaNAK1.1 and TaNAK1.2 using vector PGEX-6P-1 as a backbone. In detail, the coding sequences of the protein kinase domains in TaNAK1.1 and TaNAK1.2 were separately amplified with pMD19-T-TaNAK1.1 or pMD19-T-TaNAK1.2 as a template and primer pair containing restriction site, namely TaNAK1-kinase-EcoRI-F/TaNAK1.1-kinase-XhoI-R or TaNAK1-kinase-EcoRI-F/TaNAK1.2-kinase-SalI-R (Supplementary Figure S1). The PCR products were double digested with restriction enzymes, and subcloned into the prokaryotic expression vector pGEX-6P-1, respectively, and fused with the GST tag, generating prokaryotic expression vectors pGEX-6P-1-TaNAK1.144-257 and pGEX-6P-1-TaNAK1.244-306. These recombinant expression vectors were first confirmed by sequencing, and then the correct constructs were separately transferred into prokaryotic expression strain Rosetta (DE3) (Novagen, Malaysia). These transformants were used to express fusion protein GST-TaNAK1.144-257 or GST-TaNAK1.244-306 in large quantities at the condition of 0.5 mM IPTG, 28°C, 180 rpm, and the target proteins were purified with GST-Resin (Shang Hai Yue-ke Biotechnology CO.,LTD).
The kinase activity of the purified target proteins was determined by in vitro autophosphorylation activity. The buffer of kinase reaction in vitro includes 100 mM Tris, pH7.5, 10 mM MgCl2, 2 mM DTT (Dithiothreitol), and 0.5 mM ATP (Enders et al., 2017). CIAP (Takara Biotechnology (Dalian) Co. Ltd) was used for dephosphorylation reaction of target protein. The mixture of the purified protein and the kinase reaction buffer with or without CIAP was incubated at 25°C for 30 min, and 5×SDS PAGE buffer was used to stop the reactions. After the reactions being stopped, the target proteins were separated by 12.5% SDS–polyacrylamide gel electrophoresis (PAGE) at 15 mA for 1.5 h and transferred to nitrocellulose membranes. Anti-GST (Absin, Shanghai) was used to detect the target proteins with GST Tags. Phosphorylated target protein was detected using Phos-tag™ BTL-111 1mM (Lumiprobe, USA) according to a previous report (Kinoshita et al., 2012).
The coding sequences of TaNAK1.1 and TaNAK1.2 were amplified using primer pair TaNAK1-XbaI-F/TANAK1-BglII-R (Supplementary Table S1) and the pMD19-T-TaNAK1.1 or the pMD19-T-TaNAK1.2 as templates, respectively. The PCR products were double digested with restriction enzymes XbaI and BglII, and separately subcloned to pCAMBIA1304 (p1304) vector, between the CaMV35S promoter and the NOS terminator, generating plant expression vectors p1304-TaNAK1.1 and p1304-TaNAK1.2 (named as p35S::TaNAK1.1 and p35S::TaNAK1.2, respectively, for simplicity). The resulted expression vectors were first confirmed by sequencing and then separately transformed into Agrobacterium tumefaciens EHA105. The wild type plants of Arabidopsis (ecotype Columbia-0) were transformed with Agrobacterium tumefaciens EHA105 containing p35S::TaNAK1.1 or p35S::TaNAK1.2, respectively, by floral-dip method (Zhang et al., 2006). The mature seeds of the transformed Arabidopsis were cultured in the soil, and the plants were screened by spraying with 100 mg/L glufosinate (Basta) into the leaves of seedlings, and the positive transgenic plants that have resistance to Basta were transplanted and grown in phytotron as described above. The independent transgenic lines were continuously self-pollinated until homozygous lines were obtained.
Arabidopsis leaf samples of different genotypes were used to extract total RNA to determine the expression levels of the transgene TaNAK1.1 or TaNAK1.2, and the flowering-related hub genes AtFLC, AtFT, AtLFY, and AtAP1 by qRT-PCR. The cDNA products of transgenic lines and wild-type plants were normalised using AtACTIN (AT3G18780) as an internal reference gene, and three independent biological replicates were included. The PCR was conducted in triplicate for each RNA sample/primer combination. The primer sequences used were shown in Supplementary Table S1. The program of qRT-PCR was as follows: denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. The qRT-PCR was performed on CFX96 Touch Real-Time PCR Detection System (Bio-Rad, USA) using TB Green Premix Ex TaqII (TaKaRa Biotechnology (Dalian) Co. Ltd, Japan). For each PCR, the specificity of the amplification was validated and CT (the threshold cycle above background) values were calculated using Bio-Rad Cycler software, and PCR efficiency close to 100%.
The relative expression levels of the tested genes were analyzed according to the comparative 2−ΔΔCT method reported previously (Livak and Schmittgen, 2001).
The transition from vegetative to reproductive growth in Arabidopsis was expressed as bolting time (the time from sowing to the primary inflorescence reaching 1.0 cm in length). The number of rosette leaves was counted one week after bolting. Plant height and total number of branches per plant, including primary and secondary branches, were measured at maturity stage. After harvest, the total seed weight per plant and shoot biomass per plant were weighed using an electronic balance, and Harvest Index is calculated by dividing seed yield per plant by biomass per plant. More than 12 plants were tested for each independent transgenic line and wild-type. The seeds of different genotypic Arabidopsis were photographed with a stereomicroscope (SMZ25, Nikon) and Image J (https://imagej.nih.gov/ij/) was used for seed size statistics. More than 20 seeds were detected for each of the independent transgenic lines.
Statistical analysis of the expression levels of target genes or phenotypic data was conducted using SPSS Statistics 26 (https://www.ibm.com/products/spss-statistics) and Duncan one-way ANOVA was performed. A significant difference was considered when P<0.05, and multiple comparisons or pairwise comparison (Student’s t test) were performed.
In order to uncover the differences among the three splicing variants of TaRK4B-1, we first analyzed the genomic structure of TaRK4B-1 based on the IWGSC RefSeq v1.0 (Singh et al., 2018), and found that the full-length of TaRK4B-1 locus encompasses a genomic DNA fragment of 5799 bp, including exons, introns, and 5’- and 3’- UTR (Figure 1A). We then compared the gene sequence with the three splicing variants and discovered that TaRK4B-1.2 is a constitutive variant containing an ORF of 2646 bp, a 5’-UTR of 212 bp, and a 3’-UTR of 404 bp. TaRK4B-1.1 arises from ES, leading to a deletion of 204 bp (772-975 bp) in the CDS, compared with TaRK4B-1.2. While TaRK4B-1.3 is produced by VFE and Alt 3’ ss, resulting in deletion of 204 bp (1-165 and 772-810 bp) in the CDS, compared to TaRK4B-1.2 (Figure 1A).
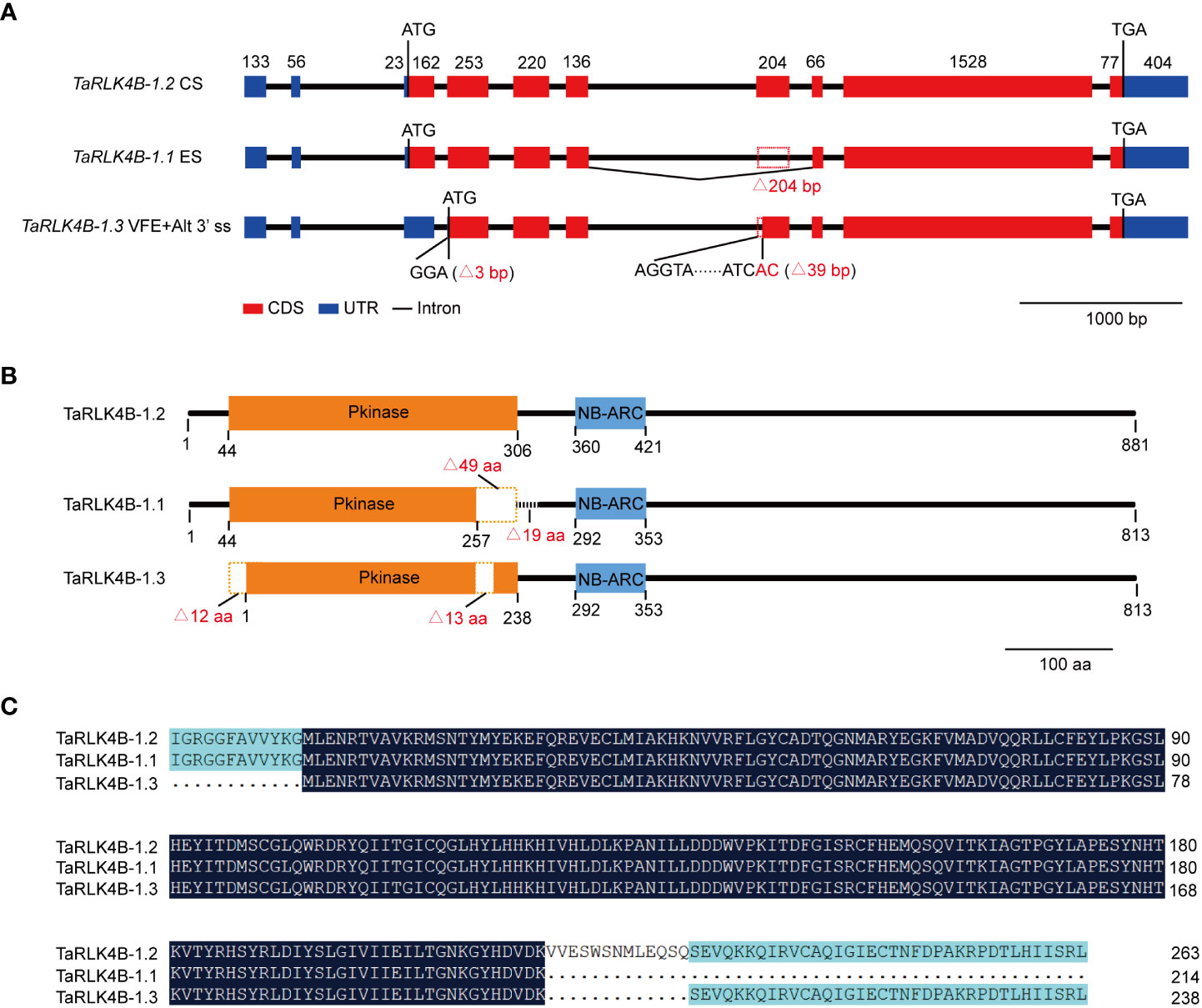
Figure 1 TaRK4B-1 is subject to alternative splicing. (A) Gene structure of three alternative splicing variants TaRK4B-1.1, TaRK4B-1.2 and TaRK4B-1.3. CS, constitutive splicing; ES, exon skipping; VFE, variable first exon; Alt 3’ ss, alternative 3’ splice site. (B) Schematic representation of the three TaRK4B-1 isoforms; Pkinase, protein kinase domain; NB-ARC, NB-ARC domain. (C) Amino acid sequence alignment of the protein kinase domains in the three TaRK4B-1 isoforms.
We further analyzed these splicing variants and their encoded proteins and found that TaRK4B-1.2 encodes a protein composed of 881 amino acids (aa), that possesses two domains, namely the protein kinase domain (44-306 aa) and the NB-ARC domain (360-421 aa) (Figure 1B, Supplementary Figure S1). Compared with TaRK4B-1.2, TaRK4B-1.1 misses a 204 bp fragment and the deletion does not result in frameshift mutations, thereby encoding a truncated protein isoform with a deletion of 49 aa (257-306 aa) in the protein kinase domain and subsequent 19 aa (307-325 aa), compared to the TaRK4B-1.2 (Figures 1B, C). Thus, the TaRK4B-1.1 contains a truncated protein kinase domain (44-257 aa) and the conserved NB-ARC domain (292-353 aa) (Figures 1B, C, Supplementary Figure S1). While TaRK4B-1.3 produces a truncated protein isoform with a deletion of 56 aa (1-56 aa) in the amino terminal and a deletion of 13 aa (257-270 aa) within the protein kinase domain, compared with the TaRK4B-1.2; thus the TaRK4B-1.3 also possesses a truncated protein kinase domain (1-238 aa) and the conserved NB-ARC domain (292-353 aa) (Figures 1B, C, Supplementary Figure S1). Taken together, the three protein isoforms, TaRK4B-1.1, TaRK4B-1.2, and TaRK4B-1.3, all have the conserved NB-ARC domain and the protein kinase domain. However, the positions of the two domains and the length of the protein kinase domains are different among them, implying that they may have different three-dimensional structures and therefore have different biological functions. The three-dimensional structures of these three protein isoforms were predicated and the result exhibited their different three-dimensional structures (Supplementary Figure S2).
Considering the fact that TaRK4B-1 gene encodes the proteins containing both the NB-ARC domain and the protein kinase domain in wheat, it was renamed as NB-ARC domain containing protein kinase 1 (TaNAK1), correspondingly, the splicing variants TaRK4B-1.1, TaRK4B-1.2, and TaRK4B-1.3 were renamed as TaNAK1.1, TaNAK1.2, and TaNAK1.3, respectively, in this study.
Knowledge about the spatiotemporal expression of a gene might provide clues on where the gene functions. In order to explore the biological role of the three splicing variants of TaNAK1 in wheat, we first analyzed the expression patterns of TaNAK1 in different developmental stages and different tissues/organs of wheat cv. Xiaoyan 6 by conducting semi-qRT-PCR. The result showed that only two splicing variants TaNAK1.1 and TaNAK1.2 were detected, and they exhibit different expression patterns across ten tissues/organs, including R, S, L, FL, YS5, YS10, GR5, GR10, GR15, and GR20 (Figure 2A). TaNAK1.1 is expressed in GR5, GR10, and GR15 to different degrees, with the highest expression level in GR15, while TaNAK1.2 is mainly expressed in L and FL, with much higher expression level in FL than in L (Figure 2A). The splicing variant TaNAK1.3 was not detected in any of the ten tissues tested (Figure 2A), implying its abundance too low to be measured. These results suggested that the two splicing variants TaNAK1.1 and TaNAK1.2 might play crucial and different biological roles in wheat growth and development. Therefore, TaNAK1.1 and TaNAK1.2 will be further investigated in subsequent studies.
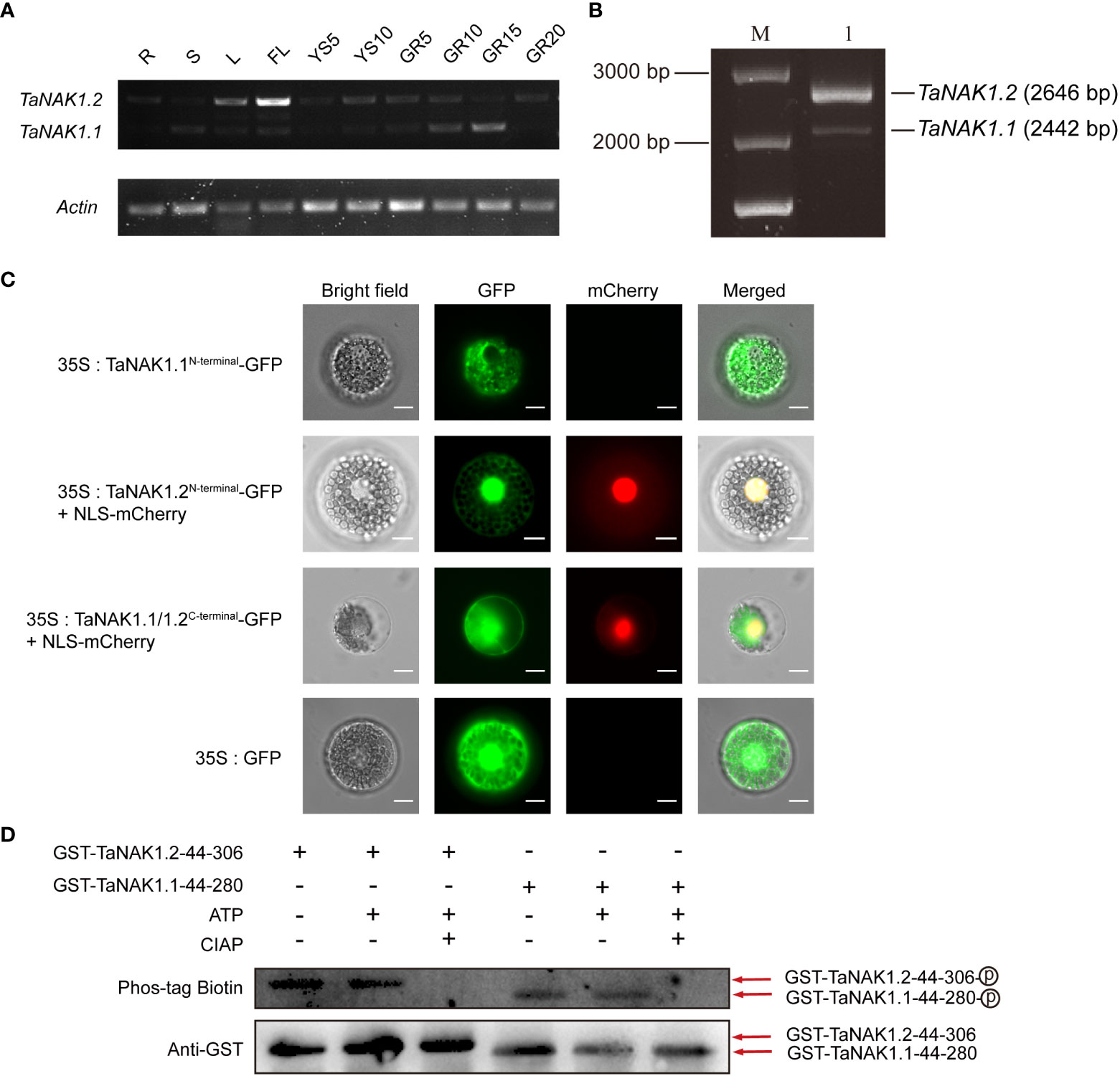
Figure 2 Expression patterns of TaNAK1 splicing variants and subcellular localization and the autophosphorylation activities of two protein isoforms TaNAK1.1 and TaNAK1.2. (A) The spatiotemporal expression profiles of the splicing variants of TaNAK1 in wheat detected by semi-quantitative reverse transcriptase-polymerase chain reaction with Actin (TraesCS1A01G020500) as a reference gene. TaNAK1.1 and TaNAK1.2, two splicing variants of TaNAK1; Actin, a wheat housekeeping gene (TraesCS1A01G020500). R, root; S, stem; L, leaf; FL, flag leaf; YS5, the 5 cm long young earfrom wheat plants at booting stage; YS10, the 10 cm long spike from wheat plants at heading stage; GR5, GR10, GR15, and GR20, the grains 5, 10, 15, 20, and 25 days post-anthesis, respectively. (B) Amplification products of the TaNAK1 transcripts obtained by RT-PCR; M, 250 bp DNA marker; 1, the products amplified with the gene-specific primer pair and the cDNA of wheat leaf as a template. (C) Subcellular localization of two protein isoforms TaNAK1.1 and TaNAK1.2 in wheat protoplasts. TaNAK1.1N-terminal, the N-terminals containing both the kinase domain and the NB-ARC domain (1-353 aa) of TaNAK1.1; TaNAK1.2N-terminal, the N-terminals containing both the kinase domain and the NB-ARC domain (1-421 aa) of TaNAK1.2; TaNAK1.1/1.2C-terminal, the C-terminal containing NB-ARC domain and all subsequent sequences (521aa at the carboxyl end) of TaNAK1.1/TaNAK1.2; NLS-mCherry, the Nuclear Localization Sequence short peptide fused with mCherry (red fluorescent protein). The vector control (35S:GFP) and fusion protein vectors (35S: TaNAK1.1N-terminal-GFP, 35S:TaNAK1.2N-terminal-GFP, and TaNAK1.1/1.2C-terminal-GFP) were each introduced into wheat protoplasts. GFP, the fusion proteins, and mCherry (red fluorescence) were observed with laser scanning confocal microscope; and Co-localization analysis of 35S:TaNAK1.2N-terminal-GFP or TaNAK1.1/1.2C-terminal-GFP (green) and NLS-mCherry (red) was observed by overlay. Bar = 10 μm. (D) The autophosphorylation activities of two protein isoforms TaNAK1.1 and TaNAK1.2 in vitro. Purified fusion proteins GST-TaNAK1.144-257 and GST-TaNAK1.244-306 were incubated independently with ATP and with or without CIAP in kinase reaction buffer, and their autophosphorylation activities were detected using Phos-tag Biotin (Wako, Japan). TaNAK1.144-257and TaNAK1.244-306 represents the kinase domains of TaNAK1.1 and TaNAK1.2, respectively. GST-TaNAK1.144-257 and GST-TaNAK1.244-306 indicate the protein kinase domains of TaNAK1.1 and TaNAK1.2 fused with GST, respectively. CIAP: Calf intestine alkaline phosphatase; Phos-tag Biotin: Biotinylated Phos-tag(Lumiprobe, USA); Anti-GST, GST antibody (Absin, Shanghai); : Phosphorylation.
To better understand the biological functions of TaNAK1, we first cloned and sequenced the complete coding sequences of TaNAK1.1 and TaNAK1.2 (Figure 2B). The results showed that the obtained sequence was identical to those described in IWGSC RefSeq v1.0. We then conducted subcellular localization of TaNAK1.1 and TaNAK1.2 by transient expression of fusion proteins TaNAK1.1-GFP and TaNAK1.2-GFP in wheat protoplasts and onion epidermal cells under the control of the CaMV35S promoter. Unfortunately, no green fluorescent signal was detected in transformed wheat protoplasts and onion epidermal cells, and we guessed that the fusion proteins were too large to be expressed transiently in wheat protoplast and onion epidermal cells. We next transiently expressed GFP fused with the N-terminals only containing the kinase domain (TaNAK1.11-257 and TaNAK1.21-306), the N-terminals containing both the kinase domain and the NB-ARC domain (TaNAK1.11-353 and TaNAK1.21-421), and the C-terminal containing NB-ARC domain and all subsequent sequences (521aa at the carboxyl end) of TaNAK1.1/TaNAK1.2 (named as TaNAK1.1/1.2C-terminal for simplicity), respectively, in wheat protoplasts and onion epidermal cells, with the vector 35S::GFP being used as a positive control. The results exhibited that the green fluorescence of fusion proteins TaNAK1.11-257-GFP (GFP fused with the N-terminal containing only the kinase domain of TaNAK1.1) and TaNAK1.11-353-GFP (GFP fused with the N-terminal containing both the kinase domain and the NB-ARC domain of TaNAK1.1) was mainly observed in the cytoplasm of the transformed wheat protoplasts and onion epidermal cells (Figure 2C and Supplementary Figure S3). While the green fluorescence of fusion proteins TaNAK1.21-306-GFP (GFP fused with the N-terminal containing only the kinase domain of TaNAK1.2) and TaNAK1.21-421-GFP (GFP fused with the N-terminal containing both the kinase domain and the NB-ARC domain of TaNAK1.2) was observed in both the cytoplasm and the nucleus (Figure 2C and Supplementary Figure S3); And the green fluorescence of fusion protein TaNAK1.1/1.2C-terminal-GFP was observed in the cytoplasm and nucleus, which is similar to that of the control GFP (Figure 2C and Supplementary Figure S3). These suggested that TaNAK1.1 and TaNAK1.2 have different subcellular localization in wheat, and their subcellular localization was mainly determined by their respective N-terminal kinase domain sequences; TaNAK1.1 was located in cytoplasm, while TaNAK1.2 was located in both cytoplasm and nucleus.
Since the predicted kinase domains of the two protein isoforms TaNAK1.1 and TaNAK1.2 are different, we wondered whether they both have kinase activity. The kinase domains of TaNAK1.1 (44-257 aa) and TaNAK1.2 (44-306 aa) were expressed in vitro, and their autophosphorylation activities were detected using Phos-tag Biotin (Wako, Japan). The results showed that both protein isoforms could be autophosphorylated in vitro (Figure 2D), indicating that they both have kinase activity. However, whether there is a difference in kinase activity between these two protein isoforms needs further investigate.
To explore the effects of the TaNAK1 splicing variants on plant growth and development, expression vectors p35S::TaNAK1.1 and p35S::TaNAK1.2 as well as empty vector p35S::Null as a control were constructed and transferred into Arabidopsis ecotype Col-0, respectively, through Agrobacterium-mediated inflorescence dipping method (Zhang et al., 2006). More than 10, 10, and 5 independent transgenic Arabidopsis lines were developed for p35S::TaNAK1.1, p35S::TaNAK1.2, and p35S::Null construct, respectively. And finally, at least 5 independent transgenic homozygous lines for each genotype were obtained for further analysis. The mRNA levels of the TaNAK1 splicing variants were first quantified in the transgenic lines carrying p35S::TaNAK1.1 (TaNAK1.1-OEs) or p35S::TaNAK1.2 (TaNAK1.2-OEs), and the transgenic lines carrying empty vector p35S::Null (EV) as well as wild-type Arabidopsis ecotype Col-0 (Col-0) by qRT-PCR. The results showed that TaNAK1.1-OEs and TaNAK1.2-OEs had significantly high abundance of the target genes, compared with Col-0 and EV, which had similar levels (Figure 3A), TaNAK1.1-OE6, 7, 9 and TaNAK1.2-OE5, 9, 10 being shown as representatives of TaNAK1.1-OEs and TaNAK1.2-OEs, respectively.
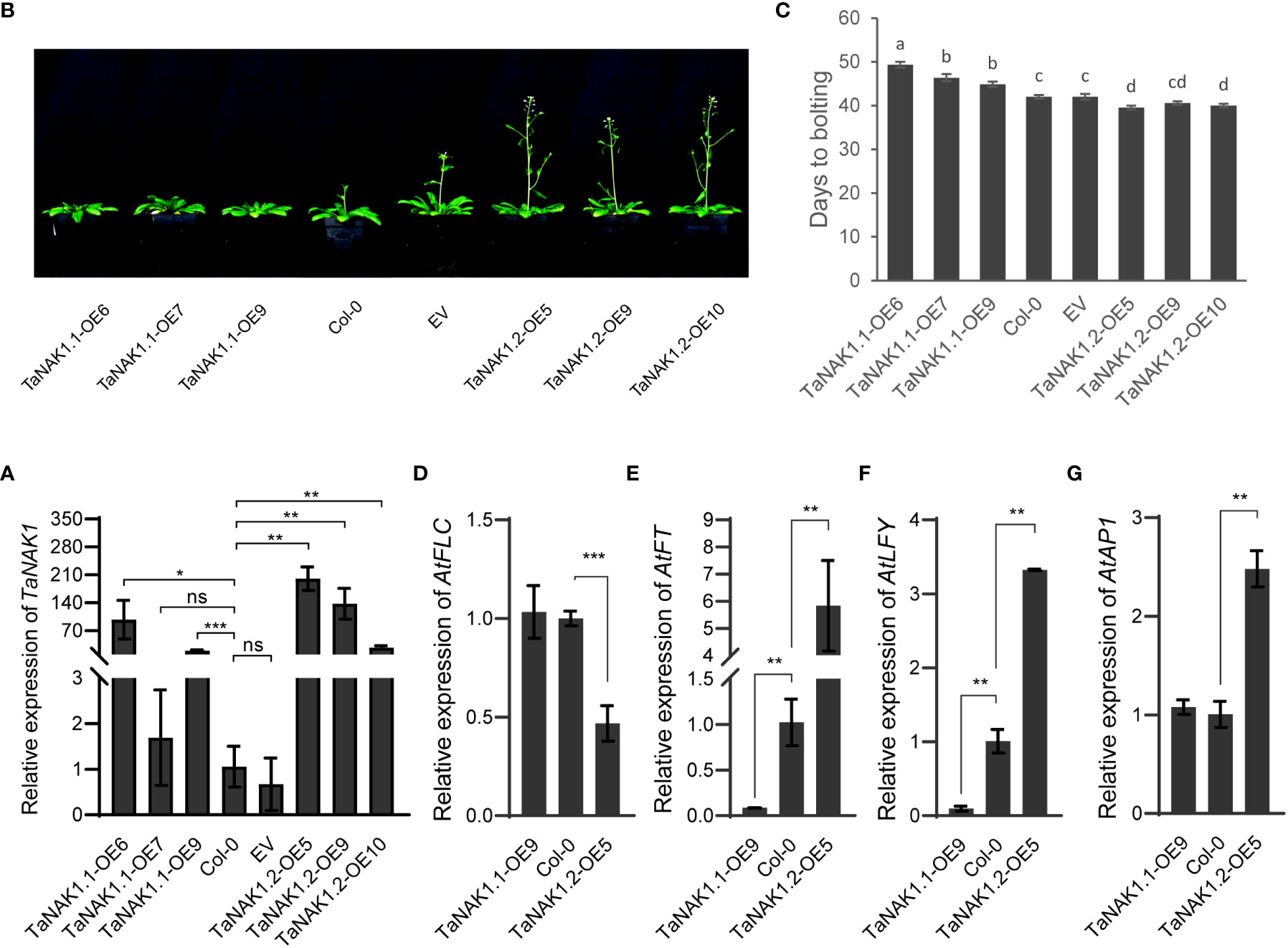
Figure 3 The effects of two TaNAK1 splicing variants on the transition time from vegetative to reproductive growth of Arabidopsis. (A) The relative mRNA abundance of the TaNAK1 splicing variants in transgenic lines and wild-type Arabidopsis plants determined by real-time quantitative RT-PCR (qRT–PCR). Values are given as the mean ± SE; three independent biological replicates included. * and ** indicate significant differences at P < 0.05 and P < 0.01 levels, respectively, compared with the Col-0 using Student’s t test; ns represent no significant differences. (B) The phenotypes of bolting in different genotypes of Arabidopsis. (C) Bolting time in different genotypes of Arabidopsis. Bolting time (the time from sowing to the primary inflorescence reaching 1.0 cm in length) represents the transition from vegetative to reproductive growth in Arabidopsis. (D-G) The relative expression levels of flowering-related genes AtFLC, AtFT, AtLFY, and AtAP1 in different genotypes of Arabidopsis at 40 days post germination. Col-0: wild type Arabidopsis; EV: transgenic lines carrying empty vector p35S::Null; TaNAK1.1-OEs and TaNAK1.2-OEs: transgenic lines carrying expression vector p35S::TaNAK1.1 and p35S::TaNAK1.2, respectively. The data indicate means ± SE (n ≥ 12), and significance analysis was performed using Duncan one-way Anova; The different lowercase letters above the error bars indicate the different significance level at P < 0.05; ** and *** indicate significant differences at P < 0.01 and P < 0.001 levels, respectively, compared with the Col-0 using Student’s t test. ns, no significant difference.
The phenotypes of TaNAK1.1-OEs, TaNAK1.2-OEs, EVs, and wild-type plants (Col-0) were then observed throughout the growth period of Arabidopsis. Firstly, very different bolting times were observed among these different genotypes of Arabidopsis (Figure 3B). Compared with Col-0, TaNAK1.1-OEs exhibited a delay in bolting time (3-7 days late), TaNAK1.1-OE6, TaNAK1.1-OE7, and TaNAK1.1-OE9 being shown as representatives (Figures 3B, C). In contrast, TaNAK1.2-OEs showed an advanced bolting time (2-3 days early), TaNAK1.2-OE5, TaNAK1.2-OE9, and TaNAK1.2-OE10 being shown as representatives (Figures 3B, C). While EV showed a similar bolting time with Col-0. Secondly, the very different number of rosette leaves was discovered among these different genotypes of Arabidopsis (Supplementary Figure S3). Compared with Col-0, TaNAK1.1-OEs had significantly increased number of rosette leaves at 7 days after bolting, but TaNAK1.2-OEs showed significantly decreased number of rosette leaves (Supplementary Figures S4A, B), which may be attributed to the length of their different vegetative growth periods. TaNAK1.1-OEs had an extended vegetative growth time due to late bolting, however TaNAK1.2-OEs had shortened vegetative growth time owing to early bolting. Taken together, TaNAK1 splicing variants TaNAK1.1 and TaNAK1.2 have opposite effects on regulating flowering time in transgenic Arabidopsis, TaNAK1.1 delays, while TaNAK1.2 promotes the transition of Arabidopsis from vegetative growth to productive.
In Arabidopsis, flowering integration factor FLOWERING LOCUS T (FT) is involved in regulating the expression of transcription factors FLEAY (LFY) and APETALA1 (AP1), the accumulation of appropriate amount of LFY and AP1 is the key to flower organ formation (Corbesier et al., 2007; Mathieu et al., 2007). But FLOWERING LOCUS C (FLC) as a negative regulator of flowering inhibits the transcription of AtFT (Helliwell et al., 2006; Krzymuski et al., 2015; Kinmonth-Schultz, 2021). To further elucidate the mechanism by which TaNAK1.1 and TaNAK1.2 affect flowering time, we investigated their effects on the above four hub genes regulating flowering, using TaNAK1.1-OE9 and TaNAK1.2-OE5 as representatives of TaNAK1.1-OEs and TaNAK1.2-OEs, respectively. The results exhibited that compared with Col-0, the expression levels of AtFLC and AtAP1 were not significantly changed in TaNAK1.1-OE9, and the expression levels of AtFT and AtLFY were significantly decreased. However, the expression levels of AtFLC were significantly decreased in TaNAK1.2-OE5, and the expression levels of AtFT, AtLFY, and AtAP1 were significantly increased, compared with Col-0 (Figures 3D-G). These indicated that overexpression of TaNAK1.1 downregulated the expression of AtFT and AtLFY in transgenic Arabidopsis, while overexpression of TaNAK1.2 downregulated the expression of AtFLC and thereby enhanced the expression of AtFT, AtLFY and AtAP1 in transgenic Arabidopsis. These results are consistent with the phenotypes of Arabidopsis transition from vegetative growth to reproductive growth.
The plant height and branch number of Arabidopsis at maturity were also investigated. The results showed that compared with the wild-type plants Col-0, TaNAK1.1-OEs exhibited decreased plant heights, while TaNAK1.2-OEs increased plant heights (Figures 4A, B), suggesting that TaNAK1.1 and TaNAK1.2 have opposite effects on plant height, TaNAK1.1 and TaNAK1.2 positively and negatively regulating plant height of transgenic Arabidopsis, respectively. Moreover, TaNAK1.1-OEs had the significantly reduced number of branches (11-13 branches), but TaNAK1.2-OEs showed the significantly increased number of branches (27-34 branches), compared with the wild-type plants Col-0 (20 branches) (Figure 4C); indicating that TaNAK1.1 and TaNAK1.2 have opposite effects on the number of branches of Arabidopsis.

Figure 4 The effects of two TaNAK1 splicing variants on plant architecture and seed yield related traits of Arabidopsis. (A) Images of different genotypes of Arabidopsis at maturity. Bar = 10 cm. (B, C) Plant height, the number of branches per plant of different genotypes of Arabidopsis. (D) Images of mature seeds from different genotypes of Arabidopsis. Bar = 0.5 mm. (E-J) Seed length, seed width, seed size, seed weight per plant, biomass per plant and harvest index of different genotypes of Arabidopsis. Col-0: wild type Arabidopsis; EV: transgenic lines carrying empty vector p35S::Null; TaNAK1.1-OE and TaNAK1.2-OE: transgenic lines carrying expression vector p35S::TaNAK1.1 and p35S::TaNAK1.2, respectively. The data indicate means ± SE (n ≥ 12), and significance analysis was performed using Duncan one-way Anova; the different lowercase letters above the error bars indicate the different significance at P < 0.05 level.
We further examined the seed yield–related traits of Arabidopsis, including seed size, seed yield per plant, biomass per plant, and harvest coefficient. The results indicated that the seed length (0.4760-0.5543 mm) and width (0.2805 mm-0.3059 mm) of TaNAK1.2-OEs were significantly higher than those of the wild type (seed length: 0.4339 mm, seed width: 0.2722 mm) (Figures 4D-F). The detection of the seed sizes showed that compared with Col-0 (0.0963 mm2), TaNAK1.1-OEs had significantly decreased seed size (0.0938-0.0962 mm2), but TaNAK1.2-OEs increased seed size (0.1002-0.1174 mm2) (Figures 4D, G). Moreover, TaNAK1.1-OEs had lower biomass per plant (0.40-0.45 g) and seed yield per plant (0.098-0.100 g) than Col-0 (0.52 g, 0.15 g), however, TaNAK1.2-OEs possessed higher biomass per plant (0.69-0.77 g) and seed yield per plant (0.22-0.27 g) (Figures 4H, I). These suggest that TaNAK1.1 and TaNAK1.2 have opposite effects on the biomass and seed yield of transgenic Arabidopsis. Furthermore, TaNAK1.1-OEs had increased harvest index, but TaNAK1.2-OEs reduced harvest index, compared with the wild-type plants Col-0 (Figure 4J).
Taken together, ectopic expression of the two splicing variants, TaNAK1.1 and TaNAK1.2, confers opposite effects on flowering time, plant architecture, and seed-yield related traits of Arabidopsis, TaNAK1.1 negatively regulating the transition from vegetative growth to reproductive growth, plant height, branch number, biomass and seed yield per plant, while TaNAK1.2 positively regulating these traits.
The existence of AS in plants indicates the specialization of gene function in biological evolution, which is a universal phenomenon in organisms. The expression patterns of different transcripts produced by the same gene, the functional domain positions of the proteins encoded by these transcripts, and the biological roles of these proteins may be different (Swarup et al., 2016; Qin et al., 2017; Huang et al., 2019). For examples, in rice, 18 nitrate and di/tripeptide transporter (NPF) genes undergo alternative splicing, producing 36 different transcripts. Of these, OsNPF7.1 and OsNPF7.4 exhibited opposite expression patterns in axillary buds, the expression levels of the two alternative splicing variants determine the growth of axillary buds, especially for the second bud, and subsequently affect the tillering number of rice (Huang et al., 2019). In Musa nana Lour., MYB16 gene generates two alternative splicing variants, MaMYB16L and MaMYB16S, which play opposite functions in the regulation of fruit ripening. MaMYB16L is a transcription inhibitor that directly downregulates the expression of MaDREB2 to inhibit fruit ripening; MaMYB16S loses its ability to bind to DNA but can competitively bind with MaMYB16L, forming inactive dimers that can promote fruit ripening (Jiang et al., 2021). In wheat, the Gγ subunit gene TaGS3 undergoes AS and produces five splicing variants, namely, the constitutive TaGS3.1, the truncated TaGS3.2–3.4 with the C-terminal Cys-rich region missing, and the truncated TaGS3.5 that contains disrupted OSR domain; and the five splicing variants exhibited expression divergence in wheat polyploidization, and differential function in controlling grain weight and size (Ren et al., 2021).
In this study, the two transcripts of TaNAK1, TaNAK1.1 and TaNAK1.2, were experimentally identified. Constitutive TaNAK1.2 encodes a protein composed of 881 amino acids that contains a protein kinase domain (44-306 aa) and a NB-ARC domain (360-421 aa), while TaNAK1.1 misses an exon with a length of 204 bp, compared with TaRK4B-1.2, thereby encoding a truncated protein isoform with a deletion of 49 aa (257-306 aa) in the kinase domain and subsequent 19 aa (307-325 aa), compared with TaRK4B-1.2 (Figures 1B, C); The positions of the two functional domains and the length of the protein kinase domains between the two protein isoforms are different, which makes them to have different three-dimensional structures (Supplementary Figure S2) and therefore have different biological functions. In vitro autophosphorylation activity analysis of using Phos-tag Biotin (Wako, Japan) showed that both TaNAK1.1 and TaNAK1.2 have protein kinase activity (Figure 2D). Furthermore, the subcellular localization of the protein isoforms was conducted in this stuby. Because the full-length of the two protein isoforms fused with GFP was too large to be expressed in wheat protoplasts and in onion epidermal cells, we tried to construct different length of TaNAK1.1 and TaNAK1.2 into expression vector to solve this problem. The result exhibited that the fusion proteins TaNAK1.11-257-GFP and TaNAK1.11-353-GFP was mainly observed in the cytoplasm of the transformed wheat protoplasts and onion epidermal cells, while the proteins TaNAK1.21-306-GFP and TaNAK1.21-421-GFP was observed in the cytoplasm and nucleus; And the fusion protein TaNAK1.1/1.2 C-terminal-GFP was observed in the cytoplasm and nucleus, which is similar as that of the control GFP (Figure 2C and Supplementary Figure S3). These suggested that TaNAK1.1 and TaNAK1.2 have different subcellular localization in wheat, TaNAK1.1 mainly in the cytoplasm, while TaNAK1.2 in both the nucleus and cytoplasm; And their subcellular localization was mainly determined by their respective N-terminal kinase domain sequences. These data suggested that these two protein isoforms might have different biological roles. Ectopic expression of the two splicing variants, TaNAK1.1 and TaNAK1.2, has opposite effects on flowering time, plant architecture, and seed-yield related traits of Arabidopsis, TaNAK1.1 negatively regulates the transition time from vegetative growth to reproductive growth, plant height, branching number, biomass and seed yield per plant, while TaNAK1.2 positively regulates these traits (Figures 3 and Figure 4). Based on previous relevant reports, we preliminarily suppose that there may be two main reasons for this difference. One is that the deletion of a 204-bp exon leads to a 49-aa difference in the kinase domains and a subsequent 19-aa difference between TaNAK1.1 and TaNAK1.2, which results in a difference in subcellular localization of the two protein isoforms. This may be the key to the phenotypic differences. Another is that the differences in the positions of two functional domains and the length of the protein kinase domains of TaNAK1.1 and TaNAK1.2 may result in their different three-dimensional structures and therefore binding different substrates. This might be one of the possible reasons for the phenotypic differences. However, this needs further demonstrate.
Flowering at the right moment is crucial for the reproductive success of flowering plants. The regulation of flowering time in model plant species Arabidopsis has been extensively studied. A great deal of information is available about the factors involved and how they interact genetically. For examples, in Arabidopsis leaves, SHORT VEGETATIVE PHASE (SVP) and FLC repress FT transcription (Helliwell et al., 2006; Jang et al., 2010). FT is produced in leaves and moves to shoot apical meristem (SAM) to interact with FD to form complex that activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) and AP1 expression on SAM (Abe et al., 2005; Yoo et al., 2005; Corbesier et al., 2007; Mathieu et al., 2007). In addition, LFY plays a key role in the integration of flowering signals parallel to FT to activate floral meristem identity genes such as AP1 triggering flowering (William et al., 2004; Abe et al., 2005). And FT can activate LFY expression through the transcription factor SOC1 (Yoo et al., 2005; Lee et al., 2008). A core regulatory network of eight integrated flowering genes has been established (Jaeger et al., 2013; Leal Valentim et al., 2015). In general, the floral pathway integrator genes FT and LFY play key roles in higher plants. Environmental and endogenous floral promoting signals stimulate the expression of floral pathway integrator genes FT and LFY. Elevated levels of FT and LFY proteins stimulate the expression of floral meristem identity genes, such as AP1, leading to flowering.
In the current study, in order to elucidate the mechanisms by which TaNAK1.1 and TaNAK1.2 differentially affect the transition from vegetative growth to reproductive growth in Arabidopsis, we investigated their effects on the four hub genes regulating flowering, including AtFLC, AtFT, AtLFY, and AtAP1, using TaNAK1.1-OE9 and TaNAK1.2-OE5 as the representatives of TaNAK1.1-OEs and TaNAK1.2-OEs, respectively. Our results demonstrated that ectopic overexpression of TaNAK1.1 and TaNAK1.2 had opposite effects on the expression of AtFT and AtLFY in transgenic lines, TaNAK1.2 upregulating while TaNAK1.1 downregulating the expression of AtFT and AtLFY (Figures 3E, E). These are consistent with the phenotypes of advanced flowering in TaNAK1.2-OEs and delayed flowering in TaNAK1.1-OEs, indicating that the main reason for TaNAK1.1 and TaNAK1.2 differentially affecting Arabidopsis transition from vegetative growth to reproductive growth may be that they differentially regulate the expression of AtFT and AtLFY genes in Arabidopsis.
Plant architecture is defined as the three-dimensional organization of the whole plant. Tillering/branching and plant height are crucial traits that determine plant architecture and grain yield of cereal crops (Khush, 2003; Wang and Li, 2008; Alqudah et al., 2016; Jian et al., 2022). Plant architecture is one of the most important factors affecting grain yield. Rice researchers have proposed a model of ideal plant architecture (IPA) that should have a low tiller number; a high number of productive tillers (9-10); 200–250 grains per panicle; erect leaves; and vigorous and deep root systems (Wang and Li, 2008). A striking example is the Green Revolution, the development of semi-dwarf wheat and rice varieties that have greatly increased crop production since 1960. Great advances have been made in understanding the molecular mechanisms underlying plant structure formation over the past decade (Lee et al., 2008; Wang et al., 2018). Numerous studies have revealed the genetic basis of rice tillering using natural or artificially induced mutants, transgenic plants as well as specific loss-of-function mutants generated by genome editing, uncovering the functions of vital genes such as MOC1 (MONOCULM1) that encodes a transcriptional regulator of axillary meristem formation and can be used to enhance the number of tillers (Li et al., 2003), and CCA1 (CIRCADIAN CLOCK ASSOCIATED 1) and PRR1 (PSEUDORESPONSE REGULATOR 1) that have opposite effects on tiller numbers: overexpressing CCA1 or knocking-out PRR1 reduces tiller number, whereas knocking-out CCA1 or overexpressing PRR1 exhibits the opposite effect (Wang et al., 2020). In addition, CCA1 has also been demonstrated to regulate IPA1/SPL14, a transcription factor of the SQUAMOSA promoter binding protein family, which has been implicated in the ideal plant architecture of rice (Jiao et al., 2010; Miura et al., 2010). In barley, alternative splicing of a gene HORVU2Hr1G098820, which encodes a trypsin family protein, produces a non-functional protein that results in excessive tillering and semi-dwarfing, leading to increased yield (Hua et al., 2020). In rice, two splicing variants of OsNPF7, OsNPF7.1 and OsNPF7.4, exhibited opposite expression patterns in axillary buds under different nitrogen concentrations, the expression levels of OsNPF7.1 and OsNPF7.4 determining the axillary bud outgrowth and subsequently influencing the tiller number, therefore, differentially regulating tiller and grain yield (Huang et al., 2019). In the present study, the two splicing variants of TaNAK1, TaNAK1.1 and TaNAK1.2, exhibit differential expression patterns during wheat growth and development, TaNAK1.1 mainly is expressed in developmental grains, whereas TaNAK1.2 in L and FL (Figure 2A), this suggesting that TaNAK1.1 and TaNAK1.2 might play crucial but different biological roles in wheat growth and development. The two protein isoforms, TaNAK1.1 and TaNAK1.2, exhibit different subcellular localization in wheat, TaNAK1.1 mainly in the cytoplasm, while TaNAK1.2 in the cytoplasm and nucleus (Figure 2C). Further analysis of the kinase activity of TaNAK1.1 and TaNAK1.2 demonstrated that they both have kinase activity in vitro (Figure 2D). However, whether they may bind different substrates needs to be further studied. In order to rapidly explore the effects of TaNAK1.1 and TaNAK1.2 on plant architecture and seed yield, transgenic Arabidopsis lines overexpressing the two splicing variants were developed and investigated. Our research data indicated that the two splicing variants have opposite effects on plant height and branch number of Arabidopsis, TaNAK1.2 positively regulates plant height and the number of branching, while TaNAK1.1 negatively regulates these traits (Figures 4A-C). In addition, TaNAK1.1 and TaNAK1.2 have opposite effects on Arabidopsis seed-size, biomass and seed yield per plant as well as harvest index, TaNAK1.2 increases seed size, biomass and seed yield per plant, and harvest index, while TaNAK1.1 decreases these traits (Figures 4D-J). In short, TaNAK1.1 and TaNAK1.2 have different effects on plant architecture and seed-yield related traits of Arabidopsis. Combining with differential expression patterns and subcellular localization of TaNAK1.1 and TaNAK1.2 in wheat, we speculate that TaNAK1.1 and TaNAK1.2 may have different functions in wheat growth and development. However, whether TaNAK1.1 and TaNAK1.2 play different roles in regulating wheat plant architecture and grain yield remains to be further verified by using transgenic wheat and loss-of-function mutants of TaNAK1 generated by CRISPR/Cas9 genome editing, and this research work is in progress.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
HXZ and MM conceived and designed the experiments. BW and XZ conducted the experiments and did data analysis. KH, HYZ and SZ did investigation on the phenotypic traits of Arabidopsis. XL provided with helpful discussion. BW and XZ wrote the draft. HXZ and MM revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was financially supported by the Natural Science Foundation of China (32072003 and 32072059), and the Key Research and Development Program of Shaanxi Province (2021NY-079).
We give thanks to Prof. Rudi Appels, Honorary Professor, University of Melbourne, for reading and improving the draft of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer HM declared a shared affiliation with the authors to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1014176/full#supplementary-material
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Alqudah, A. M., Koppolu, R., Wolde, G. M., Graner, A., Schnurbusch, T. (2016). The genetic architecture of barley plant stature. Front. Genet. 7. doi: 10.3389/fgene.2016.00117
Ananda, G. K. S., Norton, S. L., Blomstedt, C., Furtado, A., Møller, B. L., Gleadow, R., et al. (2022). Transcript profiles of wild and domesticated sorghum under water-stressed conditions and the differential impact on dhurrin metabolism. Planta 255, 1–19. doi: 10.1007/s00425-022-03831-4
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Cyrek, M., Fedak, H., Ciesielski, A., Guo, Y., Sliwa, A., Brzezniak, L., et al. (2016). Seed dormancy in Arabidopsis is controlled by alternative polyadenylation of DOG1. Plant Physiol. 170, 947–955. doi: 10.1104/pp.15.01483
Egawa, C., Kobayashi, F., Ishibashi, M., Nakamura, T., Nakamura, C., Takumi, S. (2006). Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet. Systems 81, 77–91. doi: 10.1266/ggs.81.77
Enders, T. A., Frick, E. M., Strader, L. C. (2017). An Arabidopsis kinase cascade influences auxin-responsive cell expansion. Plant J. 92, 68–81. doi: 10.1111/tpj.13635
Gao, P., Quilichini, T. D., Zhai, C., Qin, L., Nilsen, K. T., Li, Q., et al. (2021). Alternative splicing dynamics and evolutionary divergence during embryogenesis in wheat species. Plant Biotechnol. J. 19, 1624–1643. doi: 10.1111/pbi.13579
Han, R., Jian, C., Lv, J. Y., Yan, Y., Chi, Q., Li, Z., et al. (2014). Identification and characterization of microRNAs in the flag leaf and developing seed of wheat (Triticum aestivum l.). BMC Genomics 15, 289. doi: 10.1186/1471-2164-15-289
Helliwell, C. A., Wood, C. C., Robertson, M., James Peacock, W., Dennis, E. S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46, 183–192. doi: 10.1111/j.1365-313X.2006.02686.x
Huang, W., Nie, H., Feng, F., Wang, J., Lu, K., Fang, Z. (2019). Altered expression of OsNPF7.1 and OsNPF7.4 differentially regulates tillering and grain yield in rice. Plant Sci. 283, 23–31. doi: 10.1016/j.plantsci.2019.01.019
Hua, W., Tan, C., Xie, J., Zhu, J., Shang, Y., Yang, J., et al. (2020). Alternative splicing of a barley gene results in an excess-tillering and semi-dwarf mutant. Theor. Appl. Genet. 133, 163–177. doi: 10.1007/s00122-019-03448-4
Jaeger, K. E., Pullen, N., Lamzin, S., Morris, R. J., Wigge, P. A. (2013). Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell. 25, 820–833. doi: 10.1105/tpc.113.109355
Jang, S., Torti, S., Coupland, G. (2010). Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. Cell Mol. Biol. 60, 614–625. doi: 10.1111/j.1365-313X.2009.03986.x
Jiang, G., Zhang, D., Li, Z., Liang, H., Deng, R., Su, X., et al. (2021). Alternative splicing of MaMYB16L regulates starch degradation in banana fruit during ripening. J. Integr. Plant Biol. 63, 1341–1352. doi: 10.1111/jipb.13088
Jian, C., Hao, P., Hao, C., Liu, S., Mao, H., Song, Q., et al. (2022). The miR319/TaGAMYB3 module regulates plant architecture and improves grain yield in common wheat (Triticum aestivum). New Phytol. 235, 1515–1530. doi: 10.1111/nph.18216
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Kalyna, M., Simpson, C. G., Syed, N. H., Lewandowska, D., Marquez, Y., Kusenda, B., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40, 2454–2469. doi: 10.1093/nar/gkr932
Kamran, A., Iqbal, M., Spaner, D. (2014). Flowering time in wheat (Triticum aestivum l.): a key factor for global adaptability. Euphytica 197, 1–26. doi: 10.1007/s10681-014-1075-7
Khush, G. (2003). Productivity improvements in rice. Nutr. Rev. 61, S114–S116. doi: 10.1301/nr.2003.jun.S114-S116
Kinmonth-Schultz, H., Lewandowska-Sabat, A., Imaizumi, T., Ward, JK., Rognoli, OA., Fjellheim, S. (2021). Flowering Times of Wild Arabidopsis Accessions From Across Norway Correlate With Expression Levels of FT, CO, and FLC Genes. Plant Sci. 12, 747740. doi: 10.3389/fpls.2021.747740
Kinoshita, E., Kinoshita-Kikuta, E., Sugiyama, Y., Fukada, Y., Ozeki, T., Koike, T. (2012). Highly sensitive detection of protein phosphorylation by using improved phos-tag biotin. Proteomics 12, 932–937. doi: 10.1002/pmic.201100639
Krzymusk, M., Andrés, F., Cagnola, J. I, Seonghoe, J., Yanovsky, M., Coupland, G., et al (2015). The dynamics of FLOWERING LOCUS T expression encodes long-day information. Plant J. 83, 952–961. doi: 10.1111/tpj.12938
Leal Valentim, F., Mourik, S.v., Pose, D., Kim, M. C., Schmid, M., van Ham, R. C., et al. (2015). A quantitative and dynamic model of the Arabidopsis flowering time gene regulatory network. PLoS One 10, e0116973. doi: 10.1371/journal.pone.0116973
Lee, J., Oh, M., Park, H., Lee, I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J. 55, 832–843. doi: 10.1111/j.1365-313x.2008.03552.x
Li, H., Li, J., Song, J., Zhao, B., Guo, C., Wang, B., et al. (2019). An auxin signaling gene BnaA3.IAA7 contributes to improved plant architecture and yield heterosis in rapeseed. New Phytol. 222, 837–851. doi: 10.1111/nph.15632
Li, X., Qian, Q., Fu, Z., Wang, Y., Xiong, G., Zeng, D., et al. (2003). Control of tillering in rice. Nature 422, 618–621. doi: 10.1038/nature01518
Liu, Z., Qin, J., Tian, X., Xu, S., Wang, Y., Li, H., et al. (2018). Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum l.). Plant Biotechnol. J. 16, 714–726. doi: 10.1111/pbi.12822
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mathieu, J., Warthmann, N., Küttner, F., Schmid, M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17, 1055–1060. doi: 10.1016/j.cub.2007.05.009
Miura, K., Ikeda, M., Matsubara, A., Song, X.-J., Ito, M., Asano, K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Pan, Y. H., Gao, L. J., Liang, Y. T., Zhao, Y., Liang, H. F., Chen, W. W., et al. (2021). OrMKK3 influences morphology and grain size in rice. J. Plant Biol. 64, 1–14. doi: 10.1007/s12374-020-09290-2
Posé, D., Verhage, L., Ott, F., Yant, L., Mathieu, J., Angenent, G. C., et al. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. doi: 10.1038/nature12633
Qin, Z., Wu, J., Geng, S., Feng, N., Chen, F., Kong, X., et al. (2017). Regulation of FT splicing by an endogenous cue in temperate grasses. Nat. Commun. 8, 14230. doi: 10.1038/ncomms14320
Reinhardt, D., Kuhlemeier, C. (2002). Plant architecture. EMBO Rep. 3, 846–851. doi: 10.1093/embo-reports/kvf177
Ren, X., Zhi, L., Liu, L., Meng, D., Su, Q., Batool, A., et al. (2021). Alternative splicing of TaGS3 differentially regulates grain weight and size in bread wheat. Int. J. Mol. Sci. 22, 11692. doi: 10.3390/ijms222111692
Sánchez-Martín, J., Widrig, V., Herren, G., Wicker, T., Zbinden, H., Gronnier, J., et al. (2021). Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants 7, 327–341. doi: 10.1038/s41477-021-00869-2
Singh, K., Chhuneja, P., Gupta, O. P., Jindal, S., Yadav, B. (2018). Shifting the limits in wheat research and breeding using a fully annotated refrence genome. Science 361, 1–13. doi: 10.112/science.aar7191
Sun, Y., He, Z., Ma, W., Xia, X. (2010). Alternative splicing in the coding region of ppo-A1 directly influences the polyphenol oxidase activity in common wheat (Triticum aestivum l.). Funct. Integr. Genomics 11, 85–93. doi: 10.1007/s10142-010-0201-4
Swarup, R., Crespi, M., Bennett, M. J. (2016). One gene, many proteins: Mapping cell-specific alternative splicing in plants. Dev. Cell. 39, 383–385. doi: 10.1016/j.devcel.2016.11.002
Ule, J., Blencowe, B. J. (2019). Alternative splicing regulatory networks: Functions, mechanisms, and evolution. Mol. Cell. 76, 329–345. doi: 10.1016/j.molcel.2019.09.017
Wang, F., Han, T., Song, Q., Ye, W., Song, X., Chu, J., et al. (2020). The rice circadian clock regulates tiller growth and panicle development through strigolactone signaling and sugar sensing. Plant Cell. 32, 3124–3138. doi: 10.1105/tpc.20.00289
Wang, Y., Li, J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279. doi: 10.1146/annurev.arplant.59.032607.092902
Wang, B., Smith, S. M., Li, J. (2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468. doi: 10.1146/annurev-arplant-042817040422
Wang, X., Zhang, H., Nyamesorto, B., Luo, Y., Mu, X., Wang, F., et al. (2020). A new mode of NPR1 action via an NB-ARC–NPR1 fusion protein negatively regulates the defence response in wheat to stem rust pathogen. New Phytol. 228, 959–972. doi: 10.1111/nph.16748
William, D. A., Su, Y., Smith, M. R., Lu, M., Baldwin, D. A., Wagner, D. (2004). Genomic identification of direct target genes of LEAFY. Proc. Natl. Acad. Sci. 101, 1775–1780. doi: 10.1073/pnas.0307842100
Worland, A. J. (1996). The influence of flowering time genes on environmental adaptability in European wheats. Euphytica 89, 49–57. doi: 10.1007/BF00015718
Yoo, S.-D., Cho, Y.-H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yoo, S. K., Chung, K. S., Kim, J., Lee, J. H., Hong, S. M., Yoo, S. J., et al. (2005). Constans activates suppressor of overexpression of constans 1 through flowering locus T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778. doi: 10.1104/pp.105.066928
Yu, K., Feng, M., Yang, G., Sun, L., Qin, Z., Cao, J., et al. (2020). Changes in alternative splicing in response to domestication and polyploidization in wheat. Plant Physiol. 184, 1955–1968. doi: 10.1104/pp.20.00773
Yu, J., Miao, J., Zhang, Z., Xiong, H., Zhu, X., Sun, X., et al. (2018). Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 16, 1667–1678. doi: 10.1111/pbi.12903
Yu, H., Tian, C., Yu, Y., Jiao, Y. (2016). Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana. Mol. Plant 9, 749–752. doi: 10.1016/j.molp.2015.12.018
Zhang, X. R., Henriques, R., Lin, S. S., Niu, Q. W., Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Keywords: wheat, TaNAK1, alternative splicing, flowering time, plant architecture, seed yield
Citation: Wu B, Zhang X, Hu K, Zheng H, Zhang S, Liu X, Ma M and Zhao H (2022) Two alternative splicing variants of a wheat gene TaNAK1, TaNAK1.1 and TaNAK1.2, differentially regulate flowering time and plant architecture leading to differences in seed yield of transgenic Arabidopsis. Front. Plant Sci. 13:1014176. doi: 10.3389/fpls.2022.1014176
Received: 08 August 2022; Accepted: 14 November 2022;
Published: 01 December 2022.
Edited by:
Enamul Huq, The University of Texas at Austin, United StatesReviewed by:
Bin Zhang, Beijing Vegetable Research Center, ChinaCopyright © 2022 Wu, Zhang, Hu, Zheng, Zhang, Liu, Ma and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huixian Zhao, aHh6aGFvMjEyQG53YWZ1LmVkdS5jbg==; Meng Ma, ZGlrNzI1QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.