- 1School of Bioengineering, Dalian University of Technology, Dalian, China
- 2Department of Chemistry, Middle East Technical University, Ankara, Turkey
The obligate biotrophic fungus Puccinia striiformis f. sp. tritici, which causes yellow (stripe) rust disease, is among the leading biological agents resulting in tremendous yield losses on global wheat productions per annum. The combatting strategies include, but are not limited to, fungicide applications and the development of resistant cultivars. However, evolutionary pressure drives rapid changes, especially in its “effectorome” repertoire, thus allowing pathogens to evade and breach resistance. The extracellular and intracellular effectors, predominantly secreted proteins, are tactical arsenals aiming for many defense processes of plants. Hence, the identity of the effectors and the molecular mechanisms of the interactions between the effectors and the plant immune system have long been targeted in research. The obligate biotrophic nature of P. striiformis f. sp. tritici and the challenging nature of its host, the wheat, impede research on this topic. Next-generation sequencing and novel prediction algorithms in bioinformatics, which are accompanied by in vitro and in vivo validation approaches, offer a speedy pace for the discovery of new effectors and investigations of their biological functions. Here, we briefly review recent findings exploring the roles of P. striiformis f. sp. tritici effectors together with their cellular/subcellular localizations, host responses, and interactors. The current status and the challenges will be discussed. We hope that the overall work will provide a broader view of where we stand and a reference point to compare and evaluate new findings.
Introduction
The underlying molecular mechanisms of the complex, dynamic, and multilayer nature of interactions between plants and pathogens are still elusive in many agriculturally crucial plant species and host-specific pathogens, such as Puccinia striiformis f. sp. tritici (Pst), causing yellow rust disease on wheat. Over the years, genetic studies using marker-assisted selection for producing yellow rust-resistant wheat cultivars not only laid the foundation to identify many resistance gene loci (Dracatos et al., 2016) but also allowed map-based cloning of some resistance genes. The advent of genome sequencing and other methods such as MutRenSeq (Steuernagel et al., 2016) and MutChromSeq (Sánchez-Martín et al., 2016) will facilitate the fast cloning of many new yellow rust R (YR) genes. Yellow rust disease is a devastating wheat disease; its destruction will be even more severe in a currently experienced and steadily elevated rapid climate change (Dudney et al., 2021), which further threatens food security. The ability to spread long distances and survival of over-seasoning cause widespread propagation and acceleration of the frequency of genetic variation over time (Jin et al., 2020). Genome-wide sequencing of Pst provides comprehensive data analysis as an eximious predictive tool and helps to understand the population characteristics that mirror genomic differences of races in different regions and allows diagnostics and surveillance in hot spot areas more concretely (Hubbard et al., 2015; Bueno-Sancho et al., 2017; Radhakrishnan et al., 2019). Additionally, the sequence information of many Pst races provides data to identify the pathogenic factors to excavate plant immunity and interactions between plants and pathogens.
The relationship between plants and pathogenic microorganisms is a co-evolutionary process. To cope with the invasion of complex pathogens, higher plants make use of a large number of cell surface and intracellular immune receptors to sense a variety of pathogenic signals and develop a complete immune system. On the other hand, pathogens need to overcome the host’s immune system for its differentiation and development to further their propagation. Plants have specific receptors to sense pathogens that are called pattern recognition receptors (PRRs), e.g., flagellin epitope (flg22) is a pathogen-associated molecular pattern (PAMP), which is recognized by a specific PRR named flagellin-sensing 2 (FLS2) (Boller and Felix, 2009; Zipfel, 2009). The detection of PAMPs by PRRs stimulates immunity, which is called PAMP-triggered immunity (PTI) (Jones and Dangl, 2006). Once PTI is activated, a series of signaling takes place to counter pathogen attacks, i.e., stomatal closure to prevent pathogen invasion, cell wall thickening and lignification or callose deposition, ion fluxes and oxidative burst; release of reactive oxygen species (ROS), synthesis and release of defense-related hormones; ethylene and salicylic acid (Zipfel and Robatzek, 2010). The PTI response is a massive, repelling, and intimidating shield against the pathogen nuisance. However, successful pathogens can evade, suppress, or manipulate the PTI phenomenon with the aid of specific and small proteinaceous compounds called effectors. Phytopathogen effectors could inhibit plant defense-related enzymes, block or seize recognition of PAMPs, and jam the signaling system. Plants developed another defense strategy to protect themselves against the effectors, which is called effector-triggered immunity (ETI) (Jones and Dangl, 2006). In ETI, effectors are sensed directly or indirectly by the cytoplasmic receptor proteins sharing common features such as N-terminal coiled-coil (CC) or Toll/interleukin-1 receptor (TIR) domains, nucleotide-binding (NB) domain, and leucine-rich region (LRR) (Kobe and Kajava, 2001; Bej et al., 2014) in the carboxyl-terminal. They are referred to as NB and oligomerization domain (NOD)-like receptors (NLRs), depending on the type of the domains, CC or TIR, then they are called CC NLRs (CNLs) and TIR-type sensor NLRs (TNLs), respectively. These two different domains also determine diverse paths of resistance responses (Feys et al., 2005; Day et al., 2006). The LRR of the receptor recognizes effectors secreted by a pathogen, whereas the NB domain is responsible for ATP/ADP binding (Jones and Dangl, 2006). Upon effector and ATP binding, NLR becomes activated (Wang et al., 2019a). Recently in a significant work, the three-dimensional (3D) structure of the active form of HOPZ-ACTIVATED RESISTANCE 1 (ZAR1) was reported (Wang et al., 2019a). It indicated that ZAR1 forms a pentameric complex through CC domains of the monomers on the plasma membrane (PM). The complex punctuates PM that works as a calcium channel. ZAR1 (HOPZ-ACTIVATED RESISTANCE 1) channel activity was shown to be required for triggering ROS prior to programmed cell death (PCD) (Wang et al., 2019b; Bi et al., 2021). Thus, terminating the pathogen invasion and achieving ETI and resistance. The discovery deepens our understanding of PCD at the molecular level for resistance that is activated by a host-specific effector; such effectors are specifically called avirulence factors (Avrs). Until the pathogen averts the recognition of its Avr effector by mutation, elimination, or evolving another effector that inhibits the inspection by NLRs, the cognate R protein maintains disease incompatibility of the plant or the resistance. Once Avr can no longer be sensed, the pathogen becomes virulent again, achieving compatibility.
Upon PCD, ETI-stimulated responses occur and lead to an array of secondary events similar to PTI. PTI can be considered as an extensive defense to a broad range of pathogens including non-host and non-adaptive pathogens due to PAMP perception, whereas ETI is an intensive defense against host-specific pathogens. Accumulating evidence suggests that PTI and ETI are indeed intertwined with each other. Some of the components in PTI and ETI are required for both types of immunity (Chang et al., 2022). PTI and ETI increase the effect of immunity synergistically. It is shown that PRRs are involved in PTI, recognizing apoplastic effectors, which are also required for intracellular ETI (Yuan et al., 2021; Ngou et al., 2021). On the other hand, ETI activates/enhances the expression of PTI signaling components (Pruitt et al., 2021; Tian et al., 2021).
Currently, we have a working model of CNL type of NLRs initiating ETI that is deciphered for ZAR1 (HOPZ-ACTIVATED RESISTANCE 1). A clear understanding of the mode of action will also be crucial for elucidating the mechanism of ETI in relation to PTI in wheat yellow rust resistance, provided that a PstAvr is discovered for a cloned cognate wheat YR gene.
Genomics, transcriptomics, and proteomics
Decline in the cost and advances in next-generation sequencing (NGS) enable generating genomic and transcriptomic data on much more complicated and challenging organisms, e.g., Pst. Thus, the ability to make comparisons between different Pst isolates and races and attempt to discover virulence and avirulence factors are made possible by sequencing. Similarly, transcript profiles of a pathogen could be monitored at different time intervals and under various environmental conditions during the infection processes—thanks to the accessibility of NGS. Despite a huge Pst genome and transcriptome sequence data availability in yellow rust disease, still, few effectors were investigated for their roles in detail.
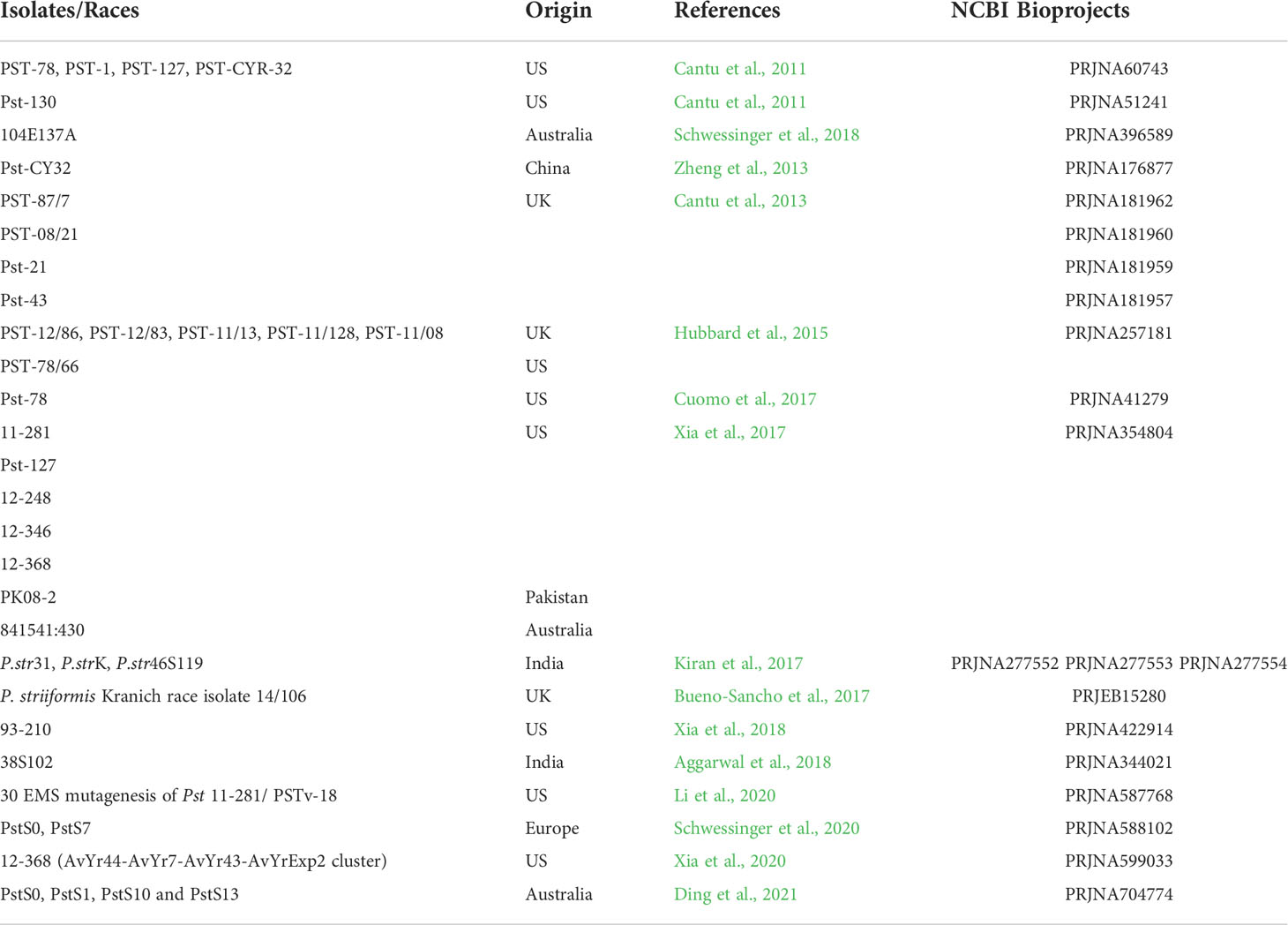
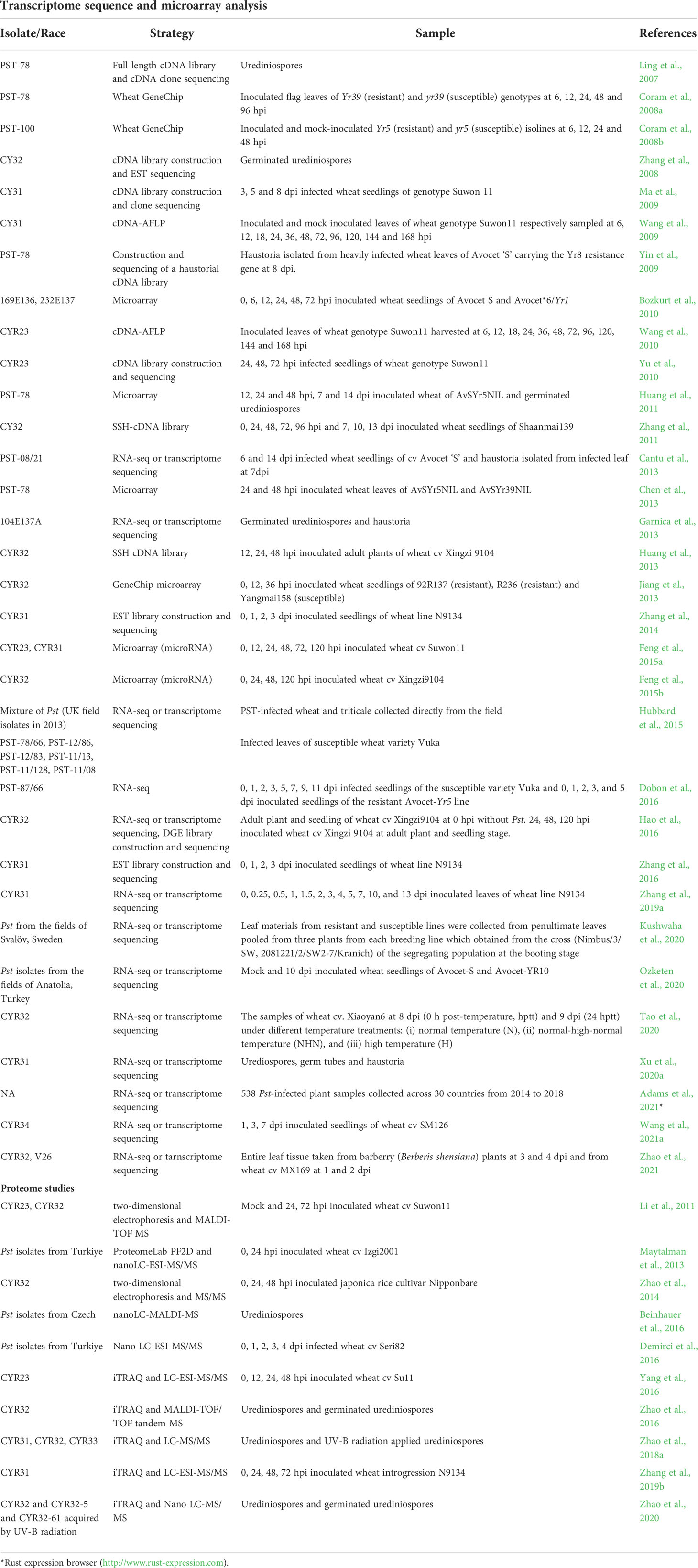
To present the current state of affairs, Tables 1 and 2 were organized using data reported in the literature. Pst can only be maintained as urediniospores on a living host, hence, the first identification of functional genes in disease was done on urediniospores of Pst-78 using a full-length cDNA library (Ling et al., 2007). Gene expression analysis was reported on germinated urediniospores of Pst-CYR32 using Expressed Sequence Tags (ESTs) (Zhang et al., 2008). Haustorium-specific genes were identified in the Pst-78 cDNA library, and authors defined proteins that are abundant in haustoria and secreted in various infection stages (Yin et al., 2009). A custom-made microarray chip was developed to reveal the expression profile of suspected genes obtained from past reports (Huang et al., 2011). The genome of Pst-130 was sequenced using NGS technology (Cantu et al., 2011). Chinese isolate Pst-CYR32 (09-001) was sequenced, and the origin of the isolate was analyzed by comparing four Pst isolates (Pst-CYR23, 104E137A, PK-CDRD, Hu09-2) from different geographical regions (Zheng et al., 2013). Pst-78 (2K-041) genome was published along with a detailed comparative analysis between Pgt and Ptt (Cuomo et al., 2017). Broad Institute released genome sequences of Pst-78 (2K-041), Pst-1 (3-5-79), Pst-127 (08-220), and PstCYR-32 (09-001) (Zheng et al., 2013; Cuomo et al., 2017; Xia et al., 2017). In addition, genome sequences of Puccinia graminis f. sp. tritici, Pgt (CRL 75-36-700-3), and Puccinia triticina, Ptt (BBDD), were published as a publicly available reference dataset, which is also useful for comparison studies (http://www.broadinstitute.org/) (Duplessis et al., 2011; Cuomo et al., 2017). The genome sequences of the four races, including Pst-87/7, Pst-08/21 (two UK races), Pst-21, and Pst-43 (US races), were reported in a publication by Cantu et al. Additionally, the gene expression data belonging to different time points [6 and 14 days post inoculation (dpi)] of the infection were described as well as haustorium-specific genes Cantu et al. (2013). Hubbard et al. (2015) surveyed Pst isolates collected from the UK fields of the United Kingdom in 2013. The authors investigated the evolutionary resemblance of harvested Pst isolates to the historical ones (14 UK and seven French isolates) and six additional isolates through whole-genome sequencing (WGS). It was discovered that the field isolates were not related to old isolates, but they possibly originated from foreign Pst populations. Garnica et al. (2013) reported the sequence data generated from both haustoria and germinated urediniospores of an Australian. In a later study, seven new races of Pst were sequenced using NGS and combined with seven older published genomes. A total of 14 races of Pst were subjected to correlation analyses in an attempt to predict Avr candidates (Xia et al., 2017). From the Indian subcontinent for the first time, Kiran et al. (2017) adopted NGS to sequence the genomes of P. striiformis pathotypes (46S 119, 31, and K). Eighty-one percent of the total annotated genes were successfully identified, and extracellularly secreted proteins were found to be very conserved in the three pathotypes.
Initial deep sequencing of the genomes of a Pst race is always laborious and costly but allows a valuable reference genome sequence for analyses of other races and transcriptome analyses at a selected state. Microarray profiling is still useful, but only a set of genes with known sequences could be monitored and investigated, not the novel ones. Although directly detecting proteins is valuable, proteome analyses generate a narrow range of information because of the low level and sometimes short duration of protein expressions; it is possible to miss key proteins. Nevertheless, a study about the proteome profile of compatible interactions between wheat and Pst revealed some of the proteins involved in pathogenesis (Demirci et al., 2016). Another proteome study listed proteins of Pst that are active in urediniospores and germ tubes using the isobaric tag for relative and absolute quantitation (iTRAQ) method and qRT-PCR for validation (Zhao et al., 2016). Alterations in the proteome content of urediniospores in response to the application of UV-B radiation were reported for three different Chinese races (CYR31, CYR32, CYR33) to elucidate deviations in virulence mechanisms (Zhao et al., 2018a).
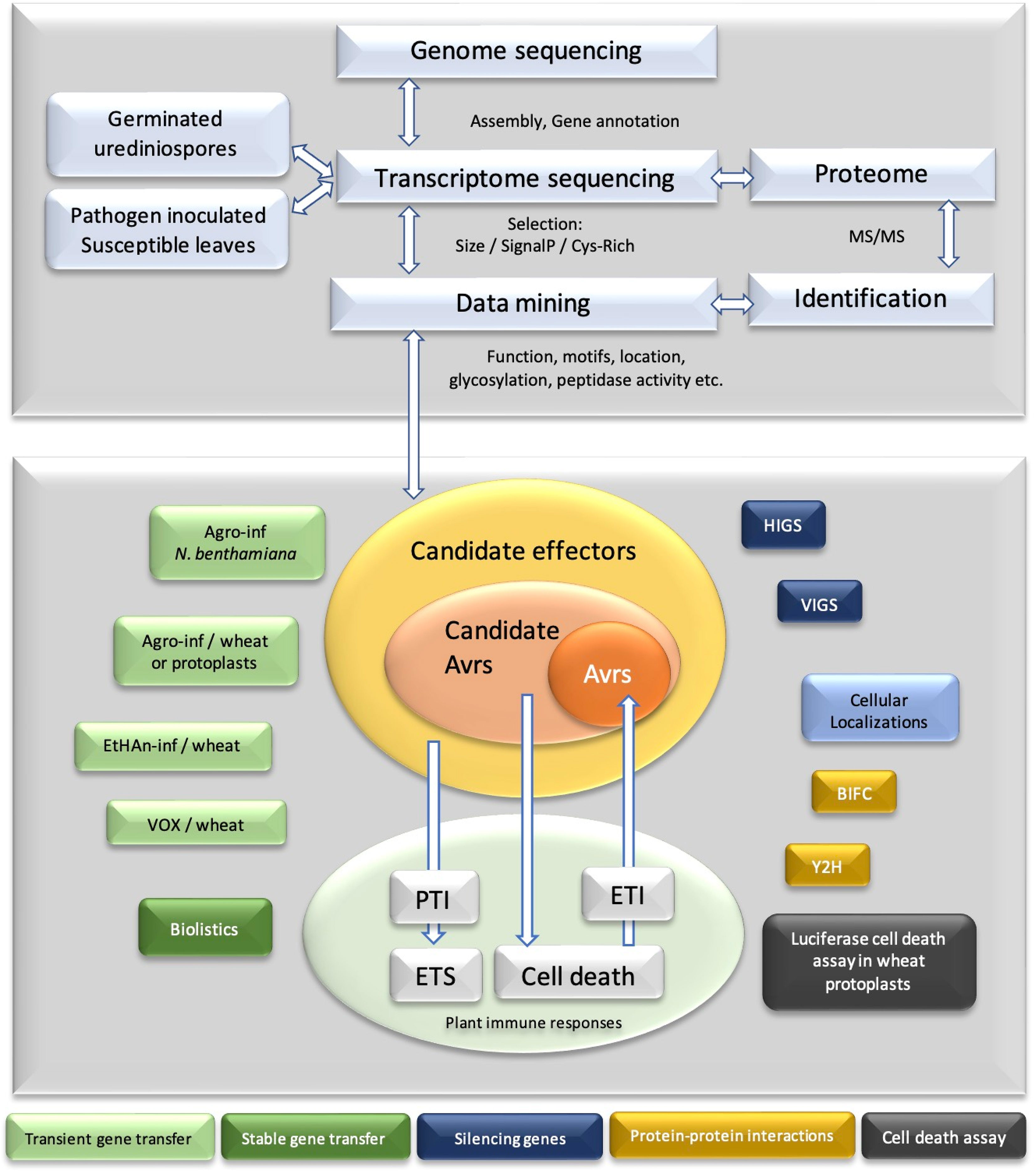
The effectiveness of omics technologies is obvious in providing bulk data on various races and different phases of the disease or the resistance. Hence, such data are the key resources for mining genes (Figure 1).
Data mining
The pace of effector evolution and the emergence of new races led to the amassed number of suspects in effector biology. The generated data of “omics” related to Pst help us to discover, compare, and pinpoint direct and indirect players in pathogenicity and plant resistance mechanisms. Data mining is a popular terminology to explain the studies conducted on big datasets using statistics, predictions, and deep machine learning to evaluate outcomes, to pinpoint crucial subsets of data from bulk collection, and to predict future patterns. Here, data mining is used as a terminology to cover all in silico strategies for handling and characterizing bulk data generated from various sequencing strategies to dissect the most relevant information.
Since the datasets obtained through sequencing are quite substantial and testing many numbers of uncovered genes is laborious, time-consuming, and costly, there is a need to pool the most probable sets of candidates so that they can be experimentally tested for function. Consequently, data mining is a useful strategy to narrow down candidate effectors. It uses our prior knowledge about effectors to predict new candidates. For instance, it is known that secreted proteins are important in achieving virulence. Hence, predicting the secreted proteome or “secretome” catalog of a pathogen is an initial step. Of course, the predicted subset may not be fully accurate or can be irrelevant to virulence, but it still concentrates to scan a shorter list of the genes. Each prediction and characterization will increase the success of trials. The first attempt to dissect the secretome was reported on the haustorial cDNA library of Pst-78 (Yin et al., 2009). Subsequently, the secretome and effectorome era has begun for many races. Abundant data generated with genomics, transcriptomics, and proteomics were subjected to secretome prediction and characterization by several studies (Cantu et al., 2011; Huang et al., 2011; Cantu et al., 2013; Garnica et al., 2013; Zheng et al., 2013; Demirci et al., 2016; Cuomo et al., 2017; Xia et al., 2017; Xia et al., 2018; Xia et al., 2020; Ozketen et al, 2020). Duplessis et al. (2011) published genome sequences of poplar leaf rust Melampsora larici-populina (Mlp) and wheat stem rust Pgt. Moreover, the group predicted the secretome of the pathogens and small secreted proteins. A pipeline to discover and characterize candidate effector proteins was defined in a hierarchical clustering study using the same data of pathogens, Mlp and Pgt (Saunders et al., 2012).
The core of secretome prediction is based on two rules: 1) the presence of secretion signal and 2) the absence of transmembrane helices. A protein could be secreted by either classical or non-classical pathways. In the classical pathway, the presence of an N-terminus secretion signal or signal peptide is required for translocation through the endoplasmic reticulum/Golgi-dependent secretory pathway (Nickel, 2003). The non-classical pathway lacks any secretion signal contradictory to a conventional path (Stein et al., 2014). However, secretome prediction is conducted frequently based on classical secretion even though some proteins follow non-classical pathways. The absence of any transmembrane helix is important to rule out any membrane-destined protein. After a secretome is defined, candidate effectors are predicted through certain parameters established on known effectors. Effector proteins are generally short in length. Some apoplastic effectors are rich in their cysteine content to provide stability in the hostile environment of the apoplast. Conserved motifs were also detected in the amino acid sequence of fungal effectors. Most notably, the [FYW]xC motif was identified in a number of candidate effectors of powdery mildew and rust (Godfrey et al., 2010). However, its significance is yet to be clarified. The haustoria provide a handy interface with enough proximity for the effector translocation in a pathogen-dependent or -independent manner. They generally show no homology to known domains except the ones associated with pathogenicity. Effectors could be encoded by genes with long intergenic regions, and they may contain internal repeats. Hence, it becomes possible to set an indefinite number of pipelines for effector mining using different filtering parameters based on known effector functions. A well-accepted pipeline was defined by Saunders et al. (2012) to pinpoint candidate-secreted effector proteins (CSEPs) of fungal pathogens (Duplessis et al., 2011; Saunders et al., 2012). The discovery of each novel effector offers new information for prediction. A list of generated software and databases for effector discovery and characterization is presented in Supplementary Table S1. Each one uses different strategies such as sequence similarity, biochemical nature of its composition, and presence of known signals and sequences for diverse sets of tasks including subcellular localization prediction, conserved domain discovery, structure, and function deduction. Among these strategies, machine learning is recently introduced to effector prediction. Algorithms compare and learn experimentally validated sets of positive and negative results in order to forecast a novel protein belonging to an appropriate group. For example, EffectorP is the first reported machine-learning program to predict effectors from other secreted proteins (Sperschneider et al., 2016). EffectorP 2.0, an upgrade for increased accuracy, is released (Sperschneider et al., 2018a). ApoplastP and Localizer are other programs to calculate the subcellular localization of an effector inside/outside the host plant (Sperschneider et al., 2017; Sperschneider et al., 2018b).
Data mining enables filtering amassed numbers of proteins for the probability of relevance, e.g., if the peptidase-like function in virulence is sought after, secretome repertoire can be monitored and sorted for peptidase domains. Undeniably, prediction does not mean that the sorted sets of proteins will always have peptidase function. Subsequently, they should be verified experimentally. Likewise, data mining offers candidate effector functional verifications. Each different pipeline yields a different list of candidate effectors. Hence, each catalog of candidates holds false positives and neglected false negatives. However, the advantages of mitigation of large datasets are greater than the disadvantages. Fungal effectors do not share conserved sequence motifs, sequence similarity, and common features in a broad spectrum. Hence, advances in data mining are essential for effector biology (Figure 1).
Functions and features of effectors
Sequencing of genomes and transcriptomes at various developmental stages following Pst infection and gene annotations have largely resolved the grouping of secreted proteins, which can be defined as candidate effectors. Nevertheless, for them to be identified as true pathogen effectors, experimental proof is required to assess their role in PTI and/or ETI, which necessitates labor-intensive experimental verifications of each effector candidate one by one to pinpoint the functions by elucidating the interactions with the host and even the other factors of a pathogen, determining targeted subcellular localizations, hence the mode of action.
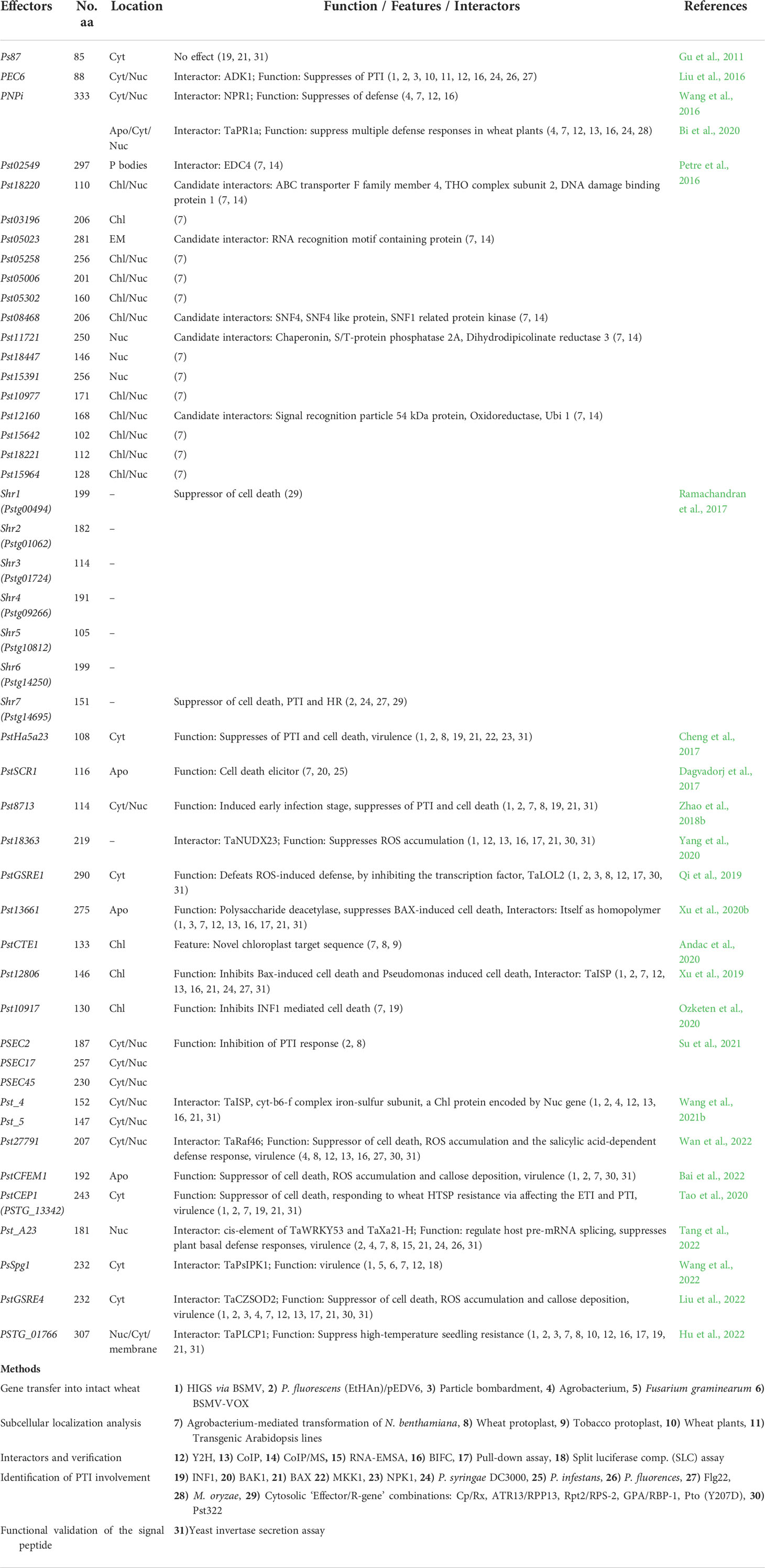
The following are the studied Pst effectors to date (Table 3). An effector candidate, Ps87, is discovered in the cDNA library of germinated urediniospores of Pst CYR32 (Zhang et al., 2008). Ps87 was reported to bear an RxLR-like motif (Gu et al., 2011). PEC6 (Pst effector candidate 6) was identified to interact with adenine kinase in host cells to suppress PTI by hindering ROS release and obstructing callose deposition (Liu et al., 2016). PNPi (Puccinia NPR1 interactor) has a DPBB-1 (Double-psi beta Barrel Domain-1) domain to interact with NPR1 (Non-expresser of PR genes 1), which is a central regulator of the defense response gene (Cao et al., 1994; Mou et al., 2003), in the nucleus to jam its interaction with the corresponding transcription factor of defense genes (Wang et al., 2016). Recently, it was shown that PNPi targets wheat pathogenesis-related protein TaPR1a in the apoplastic space and can suppress multiple defense responses in wheat plants by targeting different components (Bi et al., 2020). In another study, several Pst effector candidates were investigated to pinpoint their interactors inside host cells of Nicotiana benthamiana leaves (Petre et al., 2016). Ramachandran et al. (2017) studied nine Pst effectors, seven of which were reported to suppress cell death; Shr7 halted the PTI response stimulated by flagellin epitope (flg22) infiltration into N. benthamiana leaves (Ramachandran et al., 2017). These researchers assessed the ability of the effectors to suppress a hypersensitive response (HR) with known cytosolic Effector/R combinations, Cp/Rx, ATR13/RPP13, Rpt2/RPS2, and GPA/RBP1. PstHa5a23 is one of the candidate effectors that were identified in the haustorial cDNA library of Pst-78 (Yin et al., 2009). It was discovered that PstHa5a23 targets the cytoplasm and suppresses cell death triggered by INF1, BAX, MKK1, and NPK1. PstSCR1 (previously PstHa2a5) is shown to be induced during infection, and it enhances plant immunity, PTI. It elicits severe cell death upon translocation into the apoplastic fluid (Dagvadorj et al., 2017). An effector candidate (Pst8713) was shown for its ability to suppress the cell death triggered by INF1 and BAX hampering ROS and callose deposition (Zhao et al., 2018b). Candidate effector Pst_8713 is found to be highly expressed in early infection, is localized in the host cytoplasm and nucleus, and inhibits PTI-associated callose deposition. The effector PstGSRE1 acts as an important virulence factor targeting TaLOL2, which is a positive regulator of zinc finger protein transcription factor against stripe rust, proposed to block nuclear localization of TaLOL2 and inhibit host immunity (Qi et al., 2019). Pst18363 by Yang et al. (2020) was shown to interact with TaNUX23 and suppress ROS accumulation. Pst_12806, which has a predicted chloroplast transit peptide, is translocated into host plant chloroplasts, where it interacts with the Rieske domain of TaISP and attenuates photosynthesis rate, decreases ROS accumulation at the infection sites, and inhibits plant defenses (Xu et al., 2019). Pst13661 was identified as a polysaccharide deacetylase found to suppress BCL2 Associated X protein (BAX)-induced cell death (Xu et al., 2020a). PstCTE1 was shown to target chloroplast with an unknown targeting mechanism, since it lacks chloroplast-targeting transit peptide. Red Fluorescent Protein (RFP) blocking the N-terminal of PstCTE1 does not interfere with its destination to the chloroplast (Andac et al., 2020). PSEC2 and PSEC17 both appear in the cytoplasm and chloroplast inhibiting the PTI response of the host (Su et al., 2021). Two stripe rust effector proteins Pst_4 and Pst_5 weaken wheat resistance by inhibiting the entry of host ferritin into chloroplasts and interfere with chloroplast-mediated defense by binding to TaISP in the cytoplasm, by which they both inhibit the entry of TaISP into chloroplasts (Wang et al., 2021b). PSTG_10917 localizes in the chloroplast, and it can suppress cell death induced by INFESTIN 1 (IFN1) in an N. benthamiana heterologous expression system (Ozketen et al., 2020). The effector Pst27791 targets wheat Raf-like kinase TaRaf46 to interfere with host immunity including ROS accumulation, expression of Salicylic Acid (SA)-related defense genes TaPR1/2, and Mitogen-Activated Protein Kinase (MAPK) activation (Wan et al., 2022). A candidate effector protein PstCFEM1 facilitates Pst infection by suppressing ROS accumulation (Bai et al., 2022). Secreted protein PstCEP1 (PSTG_13342) has the function of suppressing PCD and responds to wheat high-temperature seedling-plant resistance via affecting PTI and ETI (Tao et al., 2020). Pst_A23 acts as a splicing regulator that directly binds the cis-elements of host genes, ultimately resulting in a reduction of the plant defense response (Tang et al., 2022). Recently found PstGSRE4 (Liu et al., 2022) is a glycine-serine-rich effector that interacts with wheat copper-zinc superoxide dismutase; TaCZSOD2 inhibits the dismutase activity. Host-induced gene silencing (HIGS) and the overexpression of the effector showed reduced virulence with increased H2O2 accumulation and increased virulence, respectively. One latest Pst effector, PsSpg1 (Wang et al., 2022), was shown to lack a typical fungal effector signal peptide, which comes as no surprise, since it was detected indirectly as an interactor while investigating the roles of a receptor-like cytoplasmic kinase (TaPsIPK1) that was induced by fungus inoculation, and it negatively regulates wheat resistance to yellow rust pathogen. TaPsIPK1 appears to be a susceptibility gene. PsSpg1 was shown to impede the virulence of multiple Pst races and promote parasitism via enhancing kinase activity and nuclear entry of TaPsIPK1. These effectors, except PsSpg1, regardless of their subcellular targets, are mostly found as PTI suppressors (Table 3, Figure 2). In susceptible plants, PTI is overcome by adapted pathogens in which virulence effectors are suppressed to defeat PTI. In the case of PsSpg1, it must act in effector-triggered susceptibility.
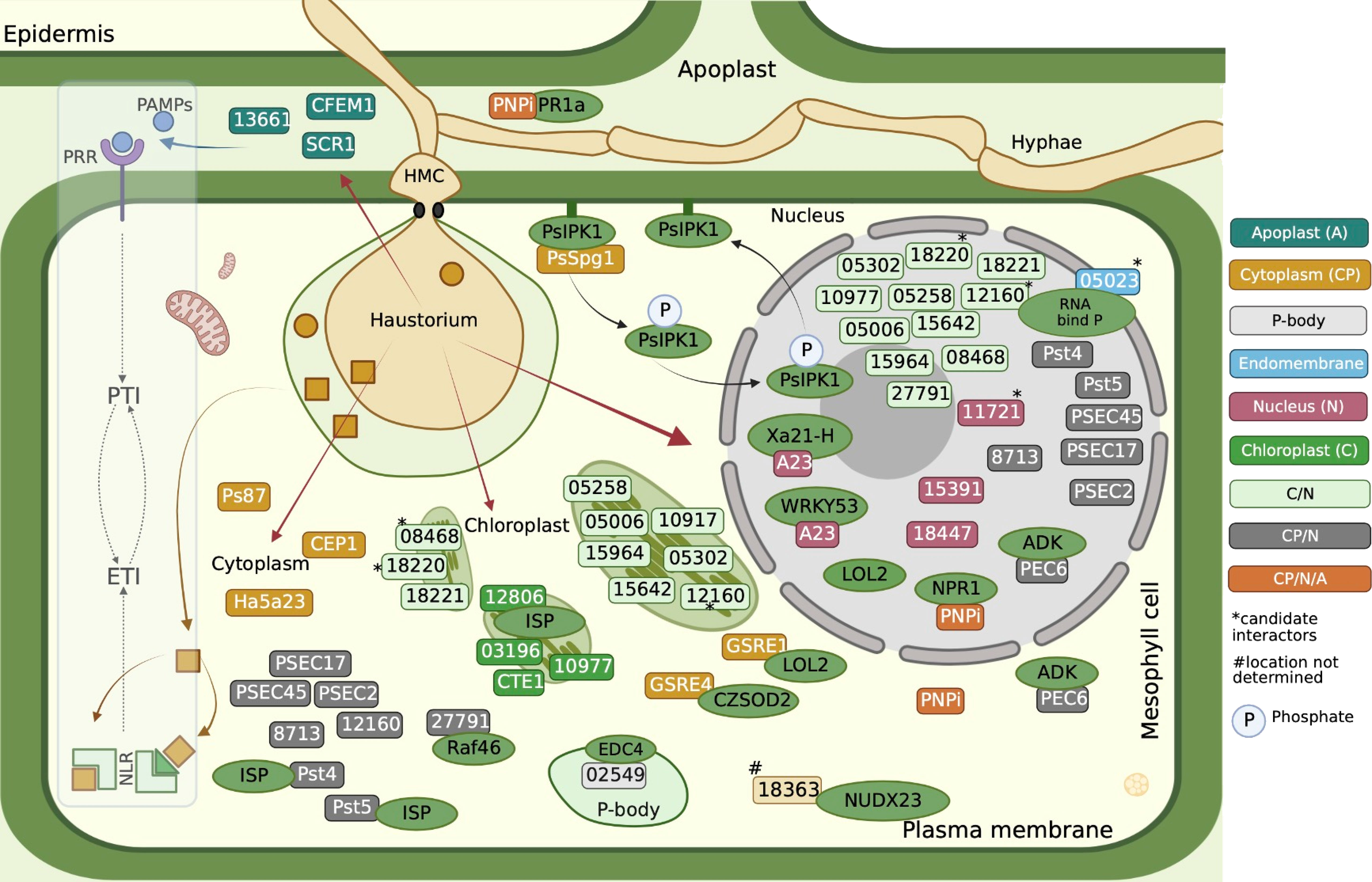
Figure 2 Cellular localization of Pst effectors and their known targets. The roles indicated in Table 3 are related to PTI. Any effector interacting directly or indirectly with NLR that generates ETI is not known. The red arrows point to the locations. Host interactors are shown in green oval boxes. *The candidate interactors were found but not confirmed (Table 3). ADK, Adenosine Kinase; CEP1, Candidate Effector Protein 1; CFEM1, Common in Fungal Extracellular Membrane Protein 1; CTE1, Chloroplast Targeting Effector 1; CZSOD2, Copper Zinc Superoxide Dismutase 2; EDC4, ENHANCER OF mRNA DECAPPING PROTEIN 4; ETI, Effector Triggered Immunity; GSRE1, Glycine-serine-rich Effector 1; GSRE4, Glycine-serine-rich Effector 4; ISP, Cytochrome b6–f complex iron–sulfur Subunit; LOL2, LSD1-One-Like-2, LSD1: Lysine Specific Demethylase 1; NLR, Nucleotide Binding Leucine-rich Receptors; NPR1, Non-expresser of PR genes 1; NUDX23, Nudix Hydrolase 23; PAMPs, Pathogen Associated Molecular Pattern; PNPi, Puccinia NPR1 interactor, NPR1: Non-expresser of PR genes 1; PR1a, Pathogenesis-related Protein 1a; PRR, Pattern Recognition Receptor; PsIPK1, Puccinia striiformis-Induced Protein Kinase 1; PsSpg1, Septum-promoting GTP-binding Protein 1; PTI, PAMP Triggered Immunity; Raf46, Raf-like Kinase; RNA bind P, RNA-binding Protein; SCR1, Small Cysteine-rich Protein 1; WRKY53, WRKY Transcription Factor 53; Xa21-H, Homologous to Xa21 in rice.
The current understanding of the research of Pst protein effector biology is still very limited; other unknown factors, both effectors of pathogen or host, may be facilitating the primary target interactors within the host cell. The activities of effectors most probably are transformed continuously in time and space.
The common methods used to determine the roles of effectors in PTI in the aforementioned studies were conducted in either whole plant (native host or model organism) or protoplasts. The assays contain detection of ROS generation, calcium production, activation of MAPK cascades, induction of defense-related genes, and callose deposition. These methods are well optimized in N. benthamiana (Chakravarthy et al., 2009; Nguyen et al., 2010). Also, the consequences of Pst effectors, which are overexpressed or silenced, were assessed using the determinants of PTI. For example, PstHa5a23 demonstrated the competence of the effector in suppressing cell death induced by the transient expression of INF1, BAX, MKK1, and NPK1 on N. benthamiana leaves (Cheng et al., 2017). Similarly, the influence of candidate effectors on non-host pathogens was studied using the heterologous system N. benthamiana.
In these studies, the experimental methods such as yeast two-hybrid and Co-Immunoprecipitation (Co-IP) and/or pulldown allowed detection of host protein interactors, which are the key determinants of understanding how an effector is mediating its influence. One of the strategies relying on chimeric effector-tagged protein fusions was established to capture interacting partners in vivo utilizing FLAG-tag and fluorescent tags (Win et al., 2011; Petre et al., 2016).
Another key determinant to elucidate the function of the effectors is the location of the effector both extracellularly and intracellularly. The location of a protein is meaningful for the evaluation of its biological function. It is expected for a protein to be present in the subcellular location of the interaction site. Similarly, the pathogen effector needs to travel to the location of its target. Exploiting this phenomenon, the biological function or pathogenicity attribution of an effector could be estimated. For instance, an apoplastic effector is more likely to establish favorable conditions by fighting host defensive measures such as defense enzymes. If an effector localizes in the nucleolus of the host cell, it should be expected to be involved in the regulation or interference of transcription. Thereby, investigating the localization site of an effector candidate illuminates its role. It is however imperative to compare and contrast the microscopic analyses with co-infiltration of known cellular markers fused to various fluorescent proteins, green fluorescent protein (GFP), Yellow Flurescent Protein (YFP), mCherry, etc. Agrobacterium tumefaciens-compatible plant destination vectors of “Gateway Cloning” methodology were designed previously (Karimi et al., 2002). The pK7FWG2 vector is one of the destination plasmids having a strong 35S promoter site of the cauliflower mosaic virus. Cloning the effector of choice into the plasmid results in effector-GFP fusion at the C-terminal end. The versatility of this system benefitted research related to candidate effector investigation (Liu et al., 2016; Petre et al., 2016; Dagvadorj et al., 2017; Evangelisti et al., 2017).
The studies in Table 3 uncovered the biological roles of numerous effectors of yellow rust fungi fused to fluorescent protein for their interacting partners and subcellular localizations. In the majority of these studies, N. benthamiana is used as a surrogate experimental plant system, which offers a great chance to scrutinize candidate effectors inside plant cells, despite being a non-homologous plant for Pst effectors.
Experimental approaches and limitations
There is a considerable number of methods in effectoromics research including in silico, in vitro, and in vivo approaches that aid in evaluating, dissecting, and filtering these gigantic datasets generated. The most laborious and time-consuming part is to validate each effector candidate for its functions using in planta and in vitro practices. Currently, a huge number of Pst candidate effectors are still awaiting full elucidation of functions in immune responses.
In the case of Pst parasite and wheat, to meet the following conditions is extremely challenging to study effectors with high-throughput experimental approaches. Not only is it preferred to study the effectors on their native host but also it is preferred that an effector in question can be overexpressed in the native parasitic organism with peptide tags and/or fused with fluorescent proteins, so recombinantly modified pathogen can be further investigated for functional analyses on the native host. Since Pst is an “obligate biotroph” and cannot be cultured in vitro, the pathogen cannot be genetically manipulated. The only form of Pst that can be obtained is a urediniospore, which can only be germinated on water-agar plates; the effective genetic manipulation of these entities is not currently available. Nevertheless, this limitation is overcome by expressing the Pst effectors or their interactors on wheat with various engineered biological entities, which are discussed below.
Agrobacterium tumefaciens mediated transient transformation of wheat by effectors
Inefficient transient transformation in wheat for the high-throughput screening of Pst candidate effectors directly on its host slows the research progress. A. tumefaciens-mediated transient gene transfer is ineffective in monocots due to compatibility issues (De Cleene and De Ley, 1976). Unlike N. benthamiana, simply put, Agrobacterium cannot transiently transform intact plant leaves of any wheat cultivar of interest as efficiently as needed. There are reports that when particular A. tumefaciens strains, LBA4404 and COR308, are used, the transient gene transfer on some wheat cultivars, e.g., Thatcher (Panwar et al., 2013; Cuomo et al., 2017), is successfully achieved; however, not every cultivar with a particular desired genetic background can be utilized. Thus, various Agrobacterium strains should be tested for each wheat variety of interest. It is reported that the recalcitrance of plant species to Agrobacterium is “primarily determined by the timing and the intensity at which host defense responses are activated” (Pitzschke, 2013). There are examples in the literature, e.g., for the closest yellow rust parasite, stem rust, where Agrobacterium strain AGL1 carrying AvrSr35 (Salcedo et al., 2017) and its cognate Sr35 co-infiltrated into barley successfully showed cell death due to ETI. Other examples are presented in Table 3.
Effector-to-host analyzer for transient transformation in wheat by effector
The other means of transient gene expression is the use of an engineered Pseudomonas fluorescens strain. A bacterial delivery system is engineered as Effector-to-Host Analyzer (EtHAn) by harnessing Type 3 Secretion System (T3SS) of Pseudomonas syringae pv. syringae-61 for effector delivery by stably integrating the hrp/hrc region into the genome of P. fluorescens Pf0-1 (Thomas et al., 2009). The pEDV6 gateway destination vector was constructed by manipulating the N-terminal amino acid sequence of AvrRPS4 for type 3 secretion of any effector of interest cloned into the vector (Sohn et al., 2007). There are concerns raised by the researchers about the lack of reproducibility of the observations with transient gene expression by EtHAn, including us. In our hands, for example, cell death once observed with a Pst effector was inconclusive in other trials (unpublished data). We suspect that very minor variations in plant growth conditions are the most probable cause for the observed irreproducibility. The list in Table 3 presents some of the successful applications.
Virus-mediated effector overexpression in wheat
A few other alternative means of gene transfer for overexpression in wheat is present, one of which is the utilization of engineered viruses, albeit with other limitations. Barley stripe mosaic virus (BSMV) is effectively used for gene silencing in wheat. However, its use for virus-mediated overexpression (VOX) is only possible for non-native or non-homologous genes; otherwise, instead of overexpression, silencing can occur. There is another limitation even if non-homologous genes are to be overexpressed, which is the requirement of a short insert. BSMV gamma-RNA genome cannot sustain large inserts of full non-homologous genes for overexpression. To overcome such limitations, there are efforts to split the gamma genome of BSMV to maintain stability (Lee et al., 2012). Recently, another virus, a monopartite foxtail mosaic virus (FoMV) was shown to stably overexpress longer proteins, with limitations of up to 600 amino acids (Bouton et al., 2018). There is only one example of VOX of Pst effector (232 amino acids) in wheat (Wang et al., 2022) (Table 3).
Effector gene silencing in wheat
Gene silencing strategy is an effective tool to investigate the biological significance of candidate genes by delivering the silencing constructs of antisense RNA. It is widely used as a gene validation tool for observing the change in phenotype. The virus-based elements are engineered for the expression of antisense RNA in host plants as in virus-induced gene silencing (VIGS). BSMV is the most commonly and effectively utilized virus for gene silencing in wheat. It is also used effectively for HIGS of the genes of Pst. VIGS is relatively straightforward and trouble-free in functional genomics of plants (Holzberg et al., 2002). In early applications of proviral DNA of BSMV, genomes were transcribed in vitro and the RNA generated was used for inoculations. Over a decade, viral RNA generated in vivo, N. benthamiana, sap containing the virus was used to rub-inoculate wheat seedlings (Yuan et al., 2011). The method reduced the cost and allowed its widespread application. HIGS exploits the ability of double-stranded RNA (dsRNA) or small interfering RNA (siRNA) translocation from a host into a pathogen (Nowara et al., 2010) upon expression of hairpin RNA, antisense, sense, or dsRNA. It is demonstrated that pathogen effector candidates could be subjected to HIGS for assessing the biological function of Pst effectors (Yin et al., 2011), since generated dsRNA and/or siRNAs can enter haustoria. Currently, host-induced silencing of the messages of Pst effectors is extensively conducted to assess the loss/reduced function of effectors (Table 3).
Effector delivery and expression in wheat protoplasts
The literature is full of use of protoplasts instead of intact plants for many purposes, especially if a robust transient transformation method of a plant is not available, or sometimes protoplasts allow better microscopic detections to assess not only the cellular targets of effectors but also cell death, ROS accumulation, etc. The optimized delivery of plasmids expressing the gene of interest is available for model plants and crops, many of which are referred to in the references of the studies listed in Table 3. The method takes advantage of polyethylene glycol (PEG)-mediated delivery of DNA and the use of enzymes for the removal of the cell wall. Nevertheless, the procedure must be optimized in each laboratory. A major shortcoming is the need to isolate protoplasts freshly. One of the best and most recent examples of protoplast microscopic analyses comes from the studies of ZAR1 (HOPZ-ACTIVATED RESISTANCE 1) (Bi et al., 2021), encouraging similar studies to be conducted with wheat protoplasts.
Conclusive remarks
The biological importance of effectors as virulence and avirulence determinants commenced the new era of effectoromics. Common features of the effectors such as secretion to apoplast or into the plant cell enabled the high-throughput discovery of effectors (Figure 1). Any identified unique feature compels further studies to elucidate their functions. Despite the available genome and transcriptome information and ongoing annotations of the genes of Pst, to entirely understand the biological and biochemical functions, interactions, cell entrance, organellar targeting, and the mechanism in induction or suppression of plant immunity of hundreds of Pst effectors, robust high-throughput functional analysis methods are needed. So far, a heterologous planta system, particularly the model organism N. benthamiana with immense examples and generated information, appears as the best system in understanding effector biology.
Most often, also in the case of Pst, the search and functional analyses of effectors aim to identify the Avrs. Despite the aforementioned limitations, it is still possible to test avirulence gene candidates by co-infiltrating N. benthamiana together with cognate wheat R-genes based on some criteria: 1) provided that the R-gene of interest is a singleton, or 2) if not, heterologous/non-native helper NLRs of N. benthamiana can execute an HR, or 3) if the cellular content and/or the genetic background of N. benthamiana is conducive for the avirulence factor activating its cognate R-protein. If these requirements are met, detecting cell death in N. benthamiana allows the verification of the effector being the avirulence factor of Pst. These requirements may limit the identification of many Avrs of Pst in a non-host system, N. benthamiana.
Currently, the use of wheat protoplasts might be the only means of testing Pst effectors for being avirulence factors for any cognate YR gene. A cell death detection assay is developed on wheat protoplasts. The protoplasts isolated can be used from a wheat line having a particular R gene by expressing effectors of a race with known virulence and avirulence pathogenicity possessing a cognate Avr to that of the R-gene. The method detects luciferase activity (Saur et al., 2019); it demonstrates the detection of significant loss of luciferase activity due to cell death by Avr sensed YR-protein, where the mesophyll protoplasts are transfected with the luciferase, a candidate Avr, and the YR-gene. However, the method can be very cumbersome for testing hundreds of effectors on many wheat lines with different YR genes. Thus, there is an urgent need for high-throughput transient gene expression methods in intact wheat plants. In our opinion, it appears that the best possible candidate approach could be virus-mediated overexpression of genes in wheat until the time a particular strain of A. tumefaciens is engineered for efficient transient gene expression. Most recently, BSMV-mediated VOX was successfully applied to express the predicted effector proteins from Pst to identify the AvrSr27 gene (Upadhyaya et al., 2021). In another study, the infiltration of purified AvrSr35, expressed with the intact signal peptide in Escherichia coli, into the wheat with Sr35 resistance gene resulted in an HR. The method is useful for determining Avr proteins in the true host (Salcedo et al., 2017). However, the applicability of this approach for high-throughput screening of effectors for seeking out Avrs can be cumbersome. In our opinion, the search for Avr genes of Pst requires genetic studies on isolates of the races, as it has been proven successful for finding Avr genes of wheat stem rust pathogen. The precisely identified virulence and avirulence of the Pst isolates of the same race can be utilized for comparing the genome and transcriptome sequences. Searching for natural mutations resulting in different virulence/avirulence by comparing the sequences leads to the identification of the candidate genes. For Pst, it is now possible to generate a segregating population of the isolates by fertilization, since its sexual reproduction is possible in the alternate host, which is determined to be barberry. On the segregating population, a high-density genetic map can be generated using single-nucleotide markers, determined by sequencing. Indeed, such a study produced candidate PstAvr genes by comparing the sequences of ethyl-methane sulfonate (EMS)-generated mutants with the progenitor isolate (Xia et al., 2020). Thus, the goal of identifying Avr genes of Pst is now within reach.
Thordal-Christensen proposed a model, known as the “iceberg model” as a view on ETI. In the model, it is pointed out that most of the NLRs, the effectors, and the effector targets keep one another in a silent state. The model helps explain the existence of many NLRs, effectors, and lesion mutants, also why many effectors appear to enhance virulence due to suppression of PTI (as in Table 3). It is argued that many of these effectors indeed cause effector-triggered susceptibility; when silenced, contributing to virulence indirectly, they may be misinterpreted as suppressors of PTI. In this model, it is claimed that what is observed most often is the tip of the iceberg (Thordal-Christensen, 2020). This view questions the presence of numerous NLRs and emphasizes that few are R-genes, and a few effectors are cognate Avrs. Finding the primary components of ETI (R/Avr) may be relatively easier than a full understanding of what is happening below the surface of the iceberg in plant immunity that requires time and huge effort.
Author contributions
MA, NW, and AO conceived the idea and wrote the manuscript. NW, YC, WJ, XZ, XRZ, YG and ZX gathered most of the literature regarding the topic. NW, YC, and WJ participated in the classification of the selected papers in informative tables. MA and YC prepared the figure. MA and NW revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from Dalian University of Technology (DUT18RC(3)062).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1012216/full#supplementary-material
References
Adams, T. M., Olsson, T. S. G., Ramírez-González, R. H., Bryant, R., Bryson, R., Campos, P. E., et al. (2021). Rust expression browser: An open-source database for simultaneous analysis of host and pathogen gene expression profiles with expVIP. BMC Genomics 22, 166. doi: 10.1186/s12864-021-07488-3
Aggarwal, R., Kulshreshtha, D., Sharma, S., Singh, V. K., Manjunatha, C., Bhardwaj, S. C., et al. (2018). Molecular characterization of Indian pathotypes of Puccinia striiformis f. sp. tritici and multigene phylogenetic analysis to establish inter- and intraspecific relationships. Genet. Mol. Biol. 41, 834–842. doi: 10.1590/1678-4685-GMB-2017-0171
Andac, A., Ozketen, A. C., Dagvadorj, B., Akkaya, M. S. (2020). An effector of Puccinia striiformis f. sp. tritici targets chloroplasts with a novel and robust targeting signal. Eur. J. Plant Pathol. 157, 751–765. doi: 10.1007/s10658-020-02033-6
Bai, X., Peng, H., Goher, F., Islam, M. A., Xu, S., Guo, J., et al. (2022). A candidate effector protein PSTCFEM1 contributes to virulence of stripe rust fungus and impairs wheat immunity. Stress Biol 2, 21. doi: 10.1007/s44154-022-00042-5
Beinhauer, J., Raus, M., Hanzalová, A., Horčička, P., Šebela, M. (2016). Intact spore MALDI-TOF mass spectrometry and proteomic analysis of Puccinia pathogenic fungi. Biochim. Biophys. Acta 1864, 1093–1103. doi: 10.1016/j.bbapap.2016.06.002
Bej, A., Sahoo, B. R., Swain, B., Basu, M., Jayasankar, P., Samanta, M. (2014). LRR search: An asynchronous server-based application for the prediction of leucine-rich repeat motifs and an integrative database of NOD-like receptors. Comput. Biol. Med. 53, 164–170. doi: 10.1016/j.compbiomed.2014.07.016
Bi, G., Su, M., Li, N., Liang, Y., Dang, S., Xu, J., et al. (2021). The ZAR1 (HOPZ-ACTIVATED RESISTANCE 1) resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528–3541.e12. doi: 10.1016/j.cell.2021.05.003
Bi, W., Zhao, S., Zhao, J., Su, J., Yu, X., Liu, D., et al. (2020). Rust effector PNPi interacting with wheat TaPR1a attenuates plant defense response. Phytopathol. Res. 2, 34. doi: 10.1186/s42483-020-00075-6
Boller, T., Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bouton, C., King, R. C., Chen, H., Azhakanandam, K., Bieri, S., Hammond-Kosack, K. E., et al. (2018). Foxtail mosaic virus: a viral vector for protein expression in cereals. Plant Physiol. 177, 1352–1367. doi: 10.1104/pp.17.01679
Bozkurt, T. O., Mcgrann, G. R. D., Maccormack, R., Boyd, L. A., Akkaya, M. S. (2010). Cellular and transcriptional responses of wheat during compatible and incompatible race-specific interactions with Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 11, 625–640. doi: 10.1111/j.1364-3703.2010.00633.x
Bueno-Sancho, V., Persoons, A., Hubbard, A., Cabrera-Quio, L. E., Lewis, C. M., Corredor-Moreno, P., et al. (2017). Pathogenomic analysis of wheat yellow rust lineages detects seasonal variation and host specificity. Genome Biol. Evol. 9, 3282–3296. doi: 10.1093/gbe/evx241
Cantu, D., Govindarajulu, M., Kozik, A., Wang, M., Chen, X., Kojima, K. K., et al. (2011). Next generation sequencing provides rapid access to the genome of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PloS One 6, e24230. doi: 10.1371/journal.pone.0024230
Cantu, D., Segovia, V., MacLean, D., Bayles, R., Chen, X., Kamoun, S., et al. (2013). Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics 14, 270. doi: 10.1186/1471-2164-14-270
Cao, H., Bowling, S. A., Gordon, A. S., Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. doi: 10.1105/tpc.6.11.1583
Chakravarthy, S., Velásquez, A. C., Martin, G. B. (2009). Assay for pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) in plants. J. Vis. Exp. 31, 1442. doi: 10.3791/1442
Chang, M., Chen, H., Liu, F., Fu, Z. Q. (2022). PTI and ETI: convergent pathways with diverse elicitors. Trends Plant Sci. 27, 113–115. doi: 10.1016/j.tplants.2021.11.013
Chen, X., Coram, T., Huang, X., Wang, M., Dolezal, A. (2013). Understanding molecular mechanisms of durable and non-durable resistance to stripe rust in wheat using a transcriptomics approach. Curr. Genomics 14, 111–126. doi: 10.2174/1389202911314020004
Cheng, Y., Wu, K., Yao, J., Li, S., Wang, X., Huang, L., et al. (2017). PSTha5a23, a candidate effector from the obligate biotrophic pathogen Puccinia striiformis f. sp. tritici, is involved in plant defense suppression and rust pathogenicity. Environ. Microbiol. 19, 1717–1729. doi: 10.1111/1462-2920.13610
Coram, T. E., Settles, M. L., Chen, X. (2008a). Transcriptome analysis of high-temperature adult-plant resistance conditioned by Yr39 during the wheat-Puccinia striiformis f. sp. tritici interaction. Mol. Plant Pathol. 9, 479–493. doi: 10.1111/J.1364-3703.2008.00476.X
Coram, T. E., Wang, M., Chen, X. (2008b). Transcriptome analysis of the wheat-Puccinia striiformis f. sp. tritici interaction. Mol. Plant Pathol. 9, 157–169. doi: 10.1111/J.1364-3703.2007.00453.X
Cuomo, C. A., Bakkeren, G., Khalil, H. B., Panwar, V., Joly, D., Linning, R., et al. (2017). Comparative analysis highlights variable genome content of wheat rusts and divergence of the mating loci. G3-Genes Genom. Genet. 7, 361–376. doi: 10.1534/g3.116.032797
Dagvadorj, B., Ozketen, A. C., Andac, A., Duggan, C., Bozkurt, T. O., Akkaya, ,. M. S. (2017). A Puccinia striiformis f. sp. tritici secreted protein activates plant immunity at the cell surface. Sci. Rep. 7, 1141. doi: 10.1038/s41598-017-01100-z
Day, B., Dahlbeck, D., Staskawicz, B. J. (2006). NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 18, 2782–2791. doi: 10.1105/tpc.106.044693
Demirci, Y. E., Inan, C., Günel, A., Maytalman, D., Mert, Z., Baykal, A. T., et al. (2016). Proteome profiling of the compatible interaction between wheat and stripe rust. Eur. J. Plant Pathol. 145, 941–962. doi: 10.1007/s10658-016-0882-1
Ding, Y., Cuddy, W. S., Wellings, C. R., Zhang, P., Thach, T., Hovmøller, M. S., et al. (2021). Incursions of divergent genotypes, evolution of virulence and host jumps shape a continental clonal population of the stripe rust pathogen Puccinia striiformis. Mol. Ecol. 30, 6566–6584. doi: 10.1111/mec.16182
Dobon, A., Bunting, D. C. E., Cabrera-Quio, L. E., Uauy, C., Saunders, D. G. O. (2016). The host-pathogen interaction between wheat and yellow rust induces temporally coordinated waves of gene expression. BMC Genomics 17, 380. doi: 10.1186/s12864-016-2684-4
Dracatos, P. M., Zhang, P., Park, R. F., McIntosh, R. A., Wellings, C. R. (2016). Complementary resistance genes in wheat selection ‘Avocet r’ confer resistance to stripe rust. Theor. Appl. Genet. 129, 65–76. doi: 10.1007/s00122-015-2609-7
Dudney, J., Willing, C. E., Das, A. J., Latimer, A. M., Nesmith, J. C. B., Battles, J. J. (2021). Nonlinear shifts in infectious rust disease due to climate change. Nat. Commun. 12, 5102. doi: 10.1038/s41467-021-25182-6
Duplessis, S., Cuomo, C. A., Lin, Y. C., Aerts, A., Tisserant, E., Veneault-Fourrey, C., et al. (2011). Obligate biotrophy features unraveled by the genomic analysis of rust fungi. PNAS 108, 9166–9171. doi: 10.1073/pnas.1019315108
Evangelisti, E., Gogleva, A., Hainaux, T., Doumane, M., Tulin, F., Quan, C., et al. (2017). Time-resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection-promoting Phytophthora palmivora effectors. BMC Biol. 15, 39. doi: 10.1186/s12915-017-0379-1
Feng, H., Sun, Y., Wang, B., Wang, X., Kang, Z. (2015a). Microarray-based identification of conserved microRNA from wheat and their expression profiles response to Puccinia striiformis f. sp. tritici. Can. J. Plant Pathol. 37, 82–91. doi: 10.1080/07060661.2014.999124
Feng, H., Wang, B., Zhang, Q., Fu, Y., Huang, L., Wang, X., et al. (2015b). Exploration of microRNAs and their targets engaging in the resistance interaction between wheat and stripe rust. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00469
Feys, B. J., Wiermer, M., Bhat, R. A., Moisan, L. J., Medina-Escobar, N., Neu, C., et al. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17, 2601–2613. doi: 10.1105/tpc.105.033910
Garnica, D. P., Upadhyaya, N. M., Dodds, P. N., Rathjen, J. P. (2013). Strategies for wheat stripe rust pathogenicity identified by transcriptome sequencing. PloS One 8, e67150. doi: 10.1371/journal.pone.0067150
Godfrey, D., Böhlenius, H., Pedersen, C., Zhang, Z., Emmersen, J., Thordal-Christensen, H. (2010). Powdery mildew fungal effector candidates share n-terminal Y/F/WxC-motif. BMC Genomics 11, 317. doi: 10.1186/1471-2164-11-317
Gu, B., Kale, S. D., Wang, Q., Wang, D., Pan, Q., Cao, H., et al. (2011). Rust secreted protein PS87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells. PloS One 6, e27217. doi: 10.1371/journal.pone.0027217
Hao, Y., Wang, T., Wang, K., Wang, X., Fu, Y., Huang, L., et al. (2016). Transcriptome analysis provides insights into the mechanisms underlying wheat plant resistance to stripe rust at the adult plant stage. PloS One 11, e0150717. doi: 10.1371/journal.pone.0150717
Holzberg, S., Brosio, P., Gross, C., Pogue, G. P. (2002). Barley stripe mosaic virus- induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313x.2002.01291.x
Huang, X., Chen, X., Coram, T., Wang, M., Kang, Z. (2011). Gene expression profiling of Puccinia striiformis f. sp. tritici during development reveals a highly dynamic transcriptome. J. Genet. Genomics 38, 357–371. doi: 10.1016/j.jgg.2011.07.004
Huang, X. L., Ma, J. B., Chen, X., Wang, X. J., Ding, K., Han, D. J., et al. (2013). Genes involved in adult plant resistance to stripe rust in wheat cultivar xingzi 9104. Physiol. Mol. Plant Pathol. 81, 26–32. doi: 10.1016/j.pmpp.2012.10.004
Hubbard, A., Lewis, C. M., Yoshida, K., Ramirez-Gonzalez, R. H., de Vallavieille-Pope, C., Thomas, J., et al. (2015). Field pathogenomics reveals the emergence of a diverse wheat yellow rust population. Genome Biol. 16, 23. doi: 10.1186/s13059-015-0590-8
Hu, Y., Su, C., Zhang, Y., Li, Y., Chen, X., Shang, H., et al. (2022). A puccinia striiformis f. sp. tritici effector inhibits high-temperature seedling-plant resistance in wheat. Plant J 112, 249–267. doi: 10.1111/tpj.15945
Jiang, Z., Ge, S., Xing, L., Han, D., Kang, Z., Zhang, G., et al. (2013). RLP1.1, a novel wheat receptor-like protein gene, is involved in the defence response against Puccinia striiformis f. sp. tritici. J. Exp. Bot. 64, 3735–3746. doi: 10.1093/jxb/ert206
Jin, Y., Kolmer, J., Szabo, L., Rouse, M. N., Hovmøller, M. S., Olivera, P. D., et al. (2020). “CHAPTER 4: Emergence and spread of new races of wheat rust fungi: continued threat to food security and prospects of genetic control,” in Emerging plant diseases and global food security. Eds. Ristaino, J. B., Records, A. (St. Paul, MN: American Phytopathological Society Press), 53–79. doi: 10.1094/9780890546383.004
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Karimi, M., Inzé, D., Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/s1360-1385(02)02251-3
Kiran, K., Rawal, H. C., Dubey, H., Jaswal, R., Bhardwaj, S. C., Prasad, P., et al. (2017). Dissection of genomic features and variations of three pathotypes of Puccinia striiformis through whole genome sequencing. Sci. Rep. 7, 42419. doi: 10.1038/srep42419
Kobe, B., Kajava, A. V. (2001). The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732. doi: 10.1016/s0959-440x(01)00266-4
Kushwaha, S. K., Vetukuri, R. R., Odilbekov, F., Pareek, N., Henriksson, T., Chawade, A. (2020). Differential gene expression analysis of wheat breeding lines reveal molecular insights in yellow rust resistance under field conditions. Agronomy 10, 1888. doi: 10.3390/agronomy10121888
Lee, W., Hammond-Kosack, K. E., Kanyuka, K. (2012). Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 160, 582–590. doi: 10.1104/pp.112.203489
Ling, P., Wang, M., Chen, X., Campbell, K. G. (2007). Construction and characterization of a full-length cDNA library for the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici). BMC Genomics 8, 145. doi: 10.1186/1471-2164-8-145
Liu, C., Pedersen, C., Schultz-Larsen, T., Aguilar, G. B., Madriz-Ordeñana, K., Hovmøller, M. S., et al. (2016). The stripe rust fungal effector PEC6 suppresses pattern-triggered immunity in a host species-independent manner and interacts with adenosine kinases. New Phytol 13 (3). doi: 10.1111/nph.14034
Liu, C., Wang, Y., Wang, Y., Du, Y., Song, C., Song, P., et al. (2022). Glycine-serine-rich effector PstGSRE4 in Puccinia striiformis f. sp. tritici inhibits the activity of copper zinc superoxide dismutase to modulate immunity in wheat. PloS Pathog. 18, e1010702. doi: 10.1371/journal.ppat.1010702
Li, H., Wei, G., Xu, J., Huang, L., Kang, Z. (2011). Identification of wheat proteins with altered expression levels in leaves infected by the stripe rust pathogen. Acta Physiol. Plant 33, 2423–2435. doi: 10.1007/s11738-011-0783-z
Li, Y., Xia, C., Wang, M., Yin, C., Chen, X. (2020). Whole-genome sequencing of Puccinia striiformis f. sp. tritici mutant isolates identifies avirulence gene candidates. BMC Genomics 21, 247. doi: 10.1186/s12864-020-6677-y
Ma, J., Huang, X., Wang, X., Chen, X., Qu, Z., Huang, L., et al. (2009). Identification of expressed genes during compatible interaction between stripe rust (Puccinia striiformis) and wheat using a cDNA library. BMC Genomics 10, 586. doi: 10.1186/1471-2164-10-586
Maytalman, D., Mert, Z., Baykal, A. T., Inan, C., Günel, A., Hasançebi, S. (2013). Proteomic analysis of early responsive resistance proteins of wheat (Triticum aestivum) to yellow rust (Puccinia striiformis f. sp. tritici) using ProteomeLab PF2D. Plant Omics J. 6, 24–35. doi: 10.5072/ZENODO.23126
Mou, Z., Fan, W., Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/s0092-8674(03)00429-x
Ngou, B. P. M., Ahn, H., Ding, P., Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Nguyen, H. P., Chakravarthy, S., Velásquez, A. C., McLane, H. L., Zeng, L., Nakayashiki, H., et al. (2010). Methods to study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol. Plant Microbe Interact. 23, 991–999. doi: 10.1094/MPMI-23-8-0991
Nickel, W. (2003). The mystery of nonclassical protein secretion. a current view on cargo proteins and potential export routes. Eur. J. Biochem. 270, 2109–2119. doi: 10.1046/j.1432-1033.2003.03577.x
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Ozketen, A. C., Andac-Ozketen, A., Dagvadorj, B., Demiralay, B., Akkaya, M. S. (2020). In-depth secretome analysis of Puccinia striiformis f. sp. tritici in infected wheat uncovers effector functions. Biosci. Rep. 40, BSR20201188. doi: 10.1042/BSR20201188
Panwar, V., McCallum, B., Bakkeren, G. (2013). Endogenous silencing of Puccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 73, 521–532. doi: 10.1111/tpj.12047
Petre, B., Saunders, D. G. O., Sklenar, J., Lorrain, C., Krasileva, K. V., Win, J., et al. (2016). Heterologous expression screens in Nicotiana benthamiana identify a candidate effector of the wheat yellow rust pathogen that associates with processing bodies. PloS One 11, e0149035. doi: 10.1371/journal.pone.0149035
Pitzschke, A. (2013). Agrobacterium infection and plant defense-transformation success hangs by a thread. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00519
Pruitt, R. N., Gust, A. A., Nürnberger, T. (2021). Plant immunity unified. Nat. Plants 7, 382–383. doi: 10.1038/s41477-021-00903-3
Qi, T., Guo, J., Liu, P., He, F., Wan, C., Islam, M. A., et al. (2019). Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol. Plant 12, 1624–1638. doi: 10.1016/j.molp.2019.09.010
Radhakrishnan, G. V., Cook, N. M., Bueno-Sancho, V., Lewis, C. M., Persoons, A., Mitiku, A. D., et al. (2019). MARPLE, a point-of-care, strain-level disease diagnostics and surveillance tool for complex fungal pathogens. BMC Biol. 17, 65. doi: 10.1186/s12915-019-0684-y
Ramachandran, S. R., Yin, C., Kud, J., Tanaka, K., Mahoney, A. K., Xiao, F., et al. (2017). Effectors from wheat rust fungi suppress multiple plant defense responses. Phytopathology 107, 75–83. doi: 10.1094/PHYTO-02-16-0083-R
Salcedo, A., Rutter, W., Wang, S., Akhunova, A., Bolus, S., Chao, S., et al. (2017). Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 358, 1604–1606. doi: 10.1126/science.aao7294
Sánchez-Martín, J., Steuernagel, B., Ghosh, S., Herren, G., Hurni, S., Adamski, N., et al. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17, 221. doi: 10.1186/s13059-016-1082-1
Saunders, D. G. O., Win, J., Cano, L. M., Szabo, L. J., Kamoun, S., Raffaele, S. (2012). Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PloS One 7, e29847. doi: 10.1371/journal.pone.0029847
Saur, I. M. L., Bauer, S., Lu, X., Schulze-Lefert, P. (2019). A cell death assay in barley and wheat protoplasts for identification and validation of matching pathogen AVR effector and plant NLR immune receptors. Plant Methods 15, 118. doi: 10.1186/s13007-019-0502-0
Schwessinger, B., Chen, Y., Tien, R., Vogt, J. K., Sperschneider, J., Nagar, R., et al. (2020). Distinct life histories impact dikaryotic genome evolution in the rust fungus Puccinia striiformis causing stripe rust in wheat. Genome Biol. Evol. 12, 597–617. doi: 10.1093/gbe/evaa071
Schwessinger, B., Sperschneider, J., Cuddy, W. S., Garnica, D. P., Miller, M. E., Taylor, J. M., et al. (2018). A near-complete haplotype-phased genome of the dikaryotic wheat stripe rust fungus Puccinia striiformis f. sp. tritici reveals high interhaplotype diversity. Mbio. 9, e02275-17. doi: 10.1128/mbio.02275-17
Sohn, K. H., Lei, R., Nemri, A., Jones, J. D. G. (2007). The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 19, 4077–4090. doi: 10.1105/tpc.107.054262
Sperschneider, J., Catanzariti, A. M., DeBoer, K., Petre, B., Gardiner, D. M., Singh, K. B., et al. (2017). LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 7, 44598. doi: 10.1038/srep44598
Sperschneider, J., Dodds, P. N., Gardiner, D. M., Singh, K. B., Taylor, J. M. (2018a). Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 19, 2094–2110. doi: 10.1111/mpp.12682
Sperschneider, J., Dodds, P. N., Singh, K. B., Taylor, J. M. (2018b). ApoplastP: prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol. 217, 1764–1778. doi: 10.1111/nph.14946
Sperschneider, J., Gardiner, D. M., Dodds, P. N., Tini, F., Covarelli, L., Singh, K. B., et al. (2016). EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol. 210, 743–761. doi: 10.1111/nph.13794
Stein, K. R., Giardina, B. J., Chiang, H. (2014). The non-classical pathway is the major pathway to secrete proteins in Saccharomyces cerevisiae. Clin. Exp. Pharmacol. 4, 155. doi: 10.4172/2161-1459.1000155
Steuernagel, B., Periyannan, S. K., Hernández-Pinzón, I., Witek, K., Rouse, M. N., Yu, G., et al. (2016). Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655. doi: 10.1038/nbt.3543
Su, Y., Chen, Y., Chen, J., Zhang, Z., Guo, J., Cai, Y., et al. (2021). Effectors of Puccinia striiformis f. sp. tritici suppressing the pathogenic-associated molecular pattern-triggered immune response were screened by transient expression of wheat protoplasts. Int. J. Mol. Sci. 22, 4985. doi: 10.3390/ijms22094985
Tang, C., Xu, Q., Zhao, J., Yue, M., Wang, J., Wang, X., et al. (2022). A rust fungus effector directly binds plant pre-mRNA splice site to reprogram alternative splicing and suppress host immunity. Plant Biotechnol. J. 20, 1167–1181. doi: 10.1111/pbi.13800
Tao, F., Hu, Y., Su, C., Li, J., Guo, L., Xu, X., et al. (2020). Revealing differentially expressed genes and identifying effector proteins of Puccinia striiformis f. sp. tritici in response to high-temperature seedling plant resistance of wheat based on transcriptome sequencing. mSphere 5, e00096–e00020. doi: 10.1128/msphere.00096-20
Thomas, W. J., Thireault, C. A., Kimbrel, J. A., Chang, J. H. (2009). Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J. 60, 919–928. doi: 10.1111/j.1365-313X.2009.03998.x
Thordal-Christensen, H.. (2020). A holistic view on plant effector-triggered immunity presented as an iceberg model. Cell Mol Life Sci 77, 3963–76. doi: 10.1007/s00018-020-03515-w
Tian, H., Wu, Z., Chen, S., Ao, K., Huang, W., Yaghmaiean, H., et al. (2021). Activation of TIR signalling boosts pattern-triggered immunity. Nature 598, 500–503. doi: 10.1038/s41586-021-03987-1
Upadhyaya, N. M., Mago, R., Panwar, V., Hewitt, T., Luo, M., Chen, J., et al. (2021). Genomics accelerated isolation of a new stem rust avirulence gene–wheat resistance gene pair. Nat. Plants 7, 1220–1228. doi: 10.1038/s41477-021-00971-5
Wang, Y., Huang, L., Luo, W., Jin, Y., Gong, F., He, J., et al. (2021a). Transcriptome analysis provides insights into the mechanisms underlying wheat cultivar Shumai126 responding to stripe rust. Gene 768, 145290. doi: 10.1016/j.gene.2020.145290
Wang, J., Hu, M., Wang, J., Qi, J., Han, Z., Wang, G., et al. (2019b). Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870. doi: 10.1126/science.aav5870
Wang, X., Liu, W., Chen, X., Tang, C., Dong, Y., Ma, J., et al. (2010). Differential gene expression in incompatible interaction between wheat and stripe rust fungus revealed by cDNA-AFLP and comparison to compatible interaction. BMC Plant Biol. 10, 9. doi: 10.1186/1471-2229-10-9
Wang, N., Tang, C., Fan, X., He, M., Gan, P., Zhang, S., et al. (2022). Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 185, 2961–2974.e19. doi: 10.1016/j.cell.2022.06.027
Wang, X., Tang, C., Zhang, G., Li, Y., Wang, C., Liu, B., et al. (2009). cDNA-AFLP analysis reveals differential gene expression in compatible interaction of wheat challenged with Puccinia striiformis f. sp. tritici. BMC Genomics 10, 289. doi: 10.1186/1471-2164-10-289
Wang, J., Wang, J., Hu, M., Wu, S., Qi, J., Wang, G. ,. H., et al. (2019a). Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364, eaav5868. doi: 10.1126/science.aav5868
Wang, X., Yang, B., Li, K., Kang, Z., Cantu, D., Dubcovsky, J. (2016). A conserved Puccinia striiformis protein interacts with wheat NPR1 and reduces induction of Pathogenesis-Related genes in response to pathogens. Mol. Plant Microbe Interact. 29, 977–989. doi: 10.1094/mpmi-10-16-0207-r
Wang, X., Zhai, T., Zhang, X., Tang, C., Zhuang, R., Zhao, H., et al. (2021b). Two stripe rust effectors impair wheat resistance by suppressing import of host fe–s protein into chloroplasts. Plant Physiol. 187, 2530–2543. doi: 10.1093/plphys/kiab434
Wan, C., Liu, Y., Tian, S., Guo, J., Bai, X., Zhu, H., et al. (2022). A serine-rich effector from the stripe rust pathogen targets a RAF-like kinase to suppress host immunity. Plant Physiol. 190, 762–778. doi: 10.1093/plphys/kiac218
Win, J., Kamoun, S., Jones, A. M. E. (2011). Purification of effector-target protein complexes via transient expression in Nicotiana benthamiana. Methods Mol. Biol. 712, 181–194. doi: 10.1007/978-1-61737-998-7_15
Xia, C., Lei, Y., Wang, M., Chen, W., Chen, X. (2020). An avirulence gene cluster in the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici) identified through genetic mapping and whole-genome sequencing of a sexual population. mSphere 5, e00128–e00120. doi: 10.1128/mSphere.00128-20
Xia, C., Wang, M., Cornejo, O. E., Jiwan, D. A., See, D. R., Chen, X. (2017). Secretome characterization and correlation analysis reveal putative pathogenicity mechanisms and identify candidate avirulence genes in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02394
Xia, C., Wang, M., Yin, C., Cornejo, O. E., Hulbert, S. H., Chen, X. (2018). Genome sequence resources for the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici) and the barley stripe rust pathogen (Puccinia striiformis f. sp. hordei). Mol. Plant Microbe Interact. 31, 1117–1120. doi: 10.1094/MPMI-04-18-0107-A
Xu, Q., Tang, C., Wang, X., Sun, S., Zhao, J., Kang, Z., et al. (2019). An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 10, 5571. doi: 10.1038/s41467-019-13487-6
Xu, Q., Tang, C., Wang, L., Zhao, C., Kang, Z., Wang, X. (2020a). Haustoria – arsenals during the interaction between wheat and Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 21, 83–94. doi: 10.1111/mpp.12882
Xu, Q., Wang, J., Zhao, J., Xu, J., Sun, S., Zhang, H., et al. (2020b). A polysaccharide deacetylase from Puccinia striiformis f. sp. tritici is an important pathogenicity gene that suppresses plant immunity. Plant Biotechnol. J. 18, 1830–1842. doi: 10.1111/pbi.13345
Yang, Q., Huai, B., Lu, Y., Cai, K., Guo, J., Zhu, X., et al. (2020). A stripe rust effector Pst18363 targets and stabilises TaNUDX23 that promotes stripe rust disease. New Phytol. 225, 880–895. doi: 10.1111/nph.16199
Yang, Y., Yu, Y., Bi, C., Kang, Z. (2016). Quantitative proteomics reveals the defense response of wheat against Puccinia striiformis f. sp. tritici. Sci. Rep. 6, 34261. doi: 10.1038/srep34261
Yin, C., Chen, X., Wang, X., Han, Q., Kang, Z., Hulbert, S. H. (2009). Generation and analysis of expression sequence tags from haustoria of the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. BMC Genomics 10, 626. doi: 10.1186/1471-2164-10-626
Yin, C., Jurgenson, J. E., Hulbert, S. H. (2011). Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 24, 554–561. doi: 10.1094/MPMI-10-10-0229
Yuan, C., Li, C., Yan, L., Jackson, A. O., Liu, Z., Han, C., et al. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PloS One 6, e26468. doi: 10.1371/journal.pone.0026468
Yuan, M., Ngou, B. P. M., Ding, P., Xin, X. F. (2021). PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62, 102030. doi: 10.1016/j.pbi.2021.102030
Yu, X., Wang, X., Wang, C., Chen, X., Qu, Z., Yu, X., et al. (2010). Wheat defense genes in fungal (Puccinia striiformis) infection. Funct. Integr. Genomics 10, 227–239. doi: 10.1007/s10142-010-0161-8
Zhang, H., Fu, Y., Guo, H., Zhang, L., Wang, C., Song, W., et al. (2019b). Transcriptome and proteome-based network analysis reveals a model of gene activation in wheat resistance to stripe rust. Int. J. Mol. Sci. 20, 1106. doi: 10.3390/ijms20051106
Zhang, H., Hu, W., Hao, J., Lv, S., Wang, C., Tong, W., et al. (2016). Genome-wide identification and functional prediction of novel and fungi-responsive lincRNAs in Triticum aestivum. BMC Genomics 17, 238. doi: 10.1186/s12864-016-2570-0
Zhang, H., Hu, Y., Wang, C., Ji, W. (2011). Gene expression in wheat induced by inoculation with Puccinia striiformis West. Plant Mol. Biol. Rep. 29, 458–465. doi: 10.1007/s11105-010-0245-6
Zhang, H., Mao, R., Wang, Y., Zhang, L., Wang, C., Lv, S., et al. (2019a). Transcriptome-wide alternative splicing modulation during plant-pathogen interactions in wheat. Plant Sci. 288, 110160. doi: 10.1016/j.plantsci.2019.05.023
Zhang, Y., Qu, Z., Zheng, W., Liu, B., Wang, X., Xue, X., et al. (2008). Stage-specific gene expression during urediniospore germination in Puccinia striiformis f. sp tritici. BMC Genomics 9, 203. doi: 10.1186/1471-2164-9-203
Zhang, H., Yang, Y., Wang, C., Liu, M., Li, H., Fu, Y., et al. (2014). Large-Scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genomics 15, 898. doi: 10.1186/1471-2164-15-898
Zhao, Y. Q., Cheng, P., Li, T. T., Wang, H. G. (2018a). Proteomic changes in urediniospores of puccinia striiformis f. sp. tritici after UV-b radiation. Int. J. Agric. Biol. 20, 1961–1969. doi: 10.17957/IJAB/15.0715
Zhao, Y., Cheng, P., Zhang, Y., Wang, H. (2020). Proteomic analysis of UV-b-induced virulence-mutant strains of Puccinia striiformis f. sp. tritici based on iTRAQ technology. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.542961
Zhao, J., Duan, W., Xu, Y., Zhang, C., Wang, L., Wang, J., et al. (2021). Distinct transcriptomic reprogramming in the wheat stripe rust fungus during the initial infection of wheat and barberry. Mol. Plant Microbe Interact. 34, 198–209. doi: 10.1094/MPMI-08-20-0244-R
Zhao, M., Wang, J., Ji, S., Chen, Z., Xu, J., Tang, C., et al. (2018b). Candidate effector Pst_8713 impairs the plant immunity and contributes to virulence of Puccinia striiformis f. sp. tritici. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01294
Zhao, J., Yang, Y., Kang, Z. (2014). Proteomic analysis of rice nonhost resistance to Puccinia striiformis f. sp. tritici using two-dimensional electrophoresis. Int. J. Mol. Sci. 15, 21644–21659. doi: 10.3390/ijms151221644
Zhao, J., Zhuang, H., Zhan, G., Huang, L., Kang, Z. (2016). Proteomic analysis of Puccinia striiformis f. sp. tritici (Pst) during uredospore germination. Eur. J. Plant Pathol. 144, 121–132. doi: 10.1007/s10658-015-0756-y
Zheng, W., Huang, L., Huang, J., Wang, X., Chen, X., Zhao, J., et al. (2013). High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat. Commun. 4, 2673. doi: 10.1038/ncomms3673
Zipfel, C. (2009). Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 12, 414–420. doi: 10.1016/j.pbi.2009.06.003
Keywords: Puccinia striiformis f. sp. tritici (Pst), effectors, secretome, wheat, yellow (stripe) rust, Nicotiana benthamiana, PTI, ETI
Citation: Wu N, Ozketen AC, Cheng Y, Jiang W, Zhou X, Zhao X, Guan Y, Xiang Z and Akkaya MS (2022) Puccinia striiformis f. sp. tritici effectors in wheat immune responses. Front. Plant Sci. 13:1012216. doi: 10.3389/fpls.2022.1012216
Received: 05 August 2022; Accepted: 10 October 2022;
Published: 07 November 2022.
Edited by:
Tesfaye Mengiste, Purdue University, United StatesReviewed by:
Vinay Panwar, Rothamsted Research, United KingdomDong Fang Ma, Yangtze University, China
Copyright © 2022 Wu, Ozketen, Cheng, Jiang, Zhou, Zhao, Guan, Xiang and Akkaya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Wu, d3VfbmFuQG1haWwuZGx1dC5lZHUuY24=; Ahmet Caglar Ozketen, YS5jYWdsYXJvemtldGVuQGdtYWlsLmNvbQ==; Mahinur S. Akkaya, bXNhQGRsdXQuZWR1LmNu
 Nan Wu1*
Nan Wu1* Ahmet Caglar Ozketen
Ahmet Caglar Ozketen Mahinur S. Akkaya
Mahinur S. Akkaya