- 1Center for Agricultural Synthetic Biology, University of Tennessee Institute of Agriculture, Knoxville, TN, United States
- 2Department of Plant Sciences, University of Tennessee Institute of Agriculture, Knoxville, TN, United States
- 3Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, United States
- 4Environmental Molecular Sciences Laboratory (EMSL), Pacific Northwest National Laboratory (PNNL), Richland, WA, United States
- 5Department of Plant Sciences, University of California, Davis, Davis, CA, United States
Abiotic stresses can cause significant damage to plants. For sustainable bioenergy crop production, it is critical to generate resistant crops to such stress. Engineering promoters to control the precise expression of stress resistance genes is a very effective way to address the problem. Here we developed stably transformed Populus tremula × Populus alba hybrid poplar (INRA 717-1B4) containing one-of-six synthetic drought stress-inducible promoters (SDs; SD9-1, SD9-2, SD9-3, SD13-1, SD18-1, and SD18-3) identified previously by transient transformation assays. We screened green fluorescent protein (GFP) induction in poplar under osmotic stress conditions. Of six transgenic lines containing synthetic promoter, three lines (SD18-1, 9-2, and 9-3) had significant GFP expression in both salt and osmotic stress treatments. Each synthetic promoter employed heptamerized repeats of specific and short cis-regulatory elements (7 repeats of 7-8 bases). To verify whether the repeats of longer sequences can improve osmotic stress responsiveness, a transgenic poplar containing the synthetic promoter of the heptamerized entire SD9 motif (20 bases, containing all partial SD9 motifs) was generated and measured for GFP induction under osmotic stress. The heptamerized entire SD9 motif did not result in higher GFP expression than the shorter promoters consisting of heptamerized SD9-1, 9-2, and 9-3 (partial SD9) motifs. This result indicates that shorter synthetic promoters (~50 bp) can be used for versatile control of gene expression in transgenic poplar. These synthetic promoters will be useful tools to engineer stress-resilient bioenergy tree crops in the future.
Introduction
Abiotic stress causes various damage to agricultural crops and ecosystems, resulting in severe crop yield loss and extra cost for ecosystem recovery (He et al., 2018; El-Sappah et al., 2022). Synthetic biology has tremendous potential to improve trait delivery in food and biofuel crops to resist abiotic stress. For example, recent advances in synthetic biology enabled the installation of genetic circuits and assembly of multigene constructs in transgenic plants for agricultural and pharmaceutical purposes (Lomonossoff and D'Aoust, 2016; Vazquez-Vilar et al., 2018; Andres et al., 2019; Sainsbury, 2020; Yang et al., 2022). While synthetic gene constructs can provide significant versatility, they give rise to a major challenge in the control of gene expression, which is necessary for regulating targeted proteins and the final production of the metabolites of interest (Liu and Stewart, 2015). To address this challenge, many researchers have attempted to control gene expression by generating antisense transcripts against the target transcript or by regulation at the post-transcriptional or post-translational levels (Swiezewski et al., 2009; Martinez de Alba et al., 2013; Van Lijsebettens and Grasser, 2014; Merchante et al., 2017). However, these methods all require sufficient transcription initiation, which is the most efficient target for gene expression controllers. Therefore, the selection of proper promoters is an essential component in both approaches.
In the past, Arabidopsis has been engineered to tolerate abiotic stresses by ectopic expression of the gene encoding dehydration-responsive element binding protein 1A (DREB1A) driven by the 35S Cauliflower mosaic virus (CaMV) promoter (Kasuga et al., 1999). However, the constitutive overexpression of DREB1A was shown to impair plant growth. In contrast to constitutive promoters, specific inducible native promoters or synthetic promoters are useful to express genes of interest in a spatiotemporal fashion. For example, native stress-responsive promoters of Rab16 A and COR14A were used to induce the synthesis of proteins responding to abscisic acid (ABA), salt, and cold to improve plant growth under stress (RoyChoudhury et al., 2007; Li et al., 2013; Singhal et al., 2016). However, native promoters, because of their plethora of recognizable transcription factor binding sequences (cis-elements) associated with stress response and other factors, have complex transcription cues. In order to express stress-responsive genes under a specific stress condition, synthetic promoters made with optimal cis-elements can prove useful. One such application of stress-responsive synthetic promoters was in Arabidopsis, in which three synthetic promoters (Ap, Dp, ANDp) were developed from Arabidopsis stress responsive promoters of responsive to desiccation 29A (RD29A), RD 29B, dehydration-responsive elements (DREs), and ABA responsive elements (ABREs) (Zhang et al., 2022). These promoters were used to induce the gene expressions of cytosolic ABA receptor kinase 1 (CARK1) and regulatory components of ABA receptor 11 (RCAR11), resulting in tolerance against drought stress in Arabidopsis (Ge et al., 2018).
Synthetic promoters have been engineered using cis-elements and other transcriptional motifs have been designed and produced for various applications (Ali and Kim, 2019). The positioning and ordering of cis-elements in synthetic promoters are important to optimize their function. Recently, Cai et al. (2020) demonstrated an effective methodology for rationally designing short synthetic promoters (Cai et al., 2020). The rational design of a synthetic promoter was conducted with 3-4 copies of a short cis-element identified from constitutive promoters relating to pathogen recognition promoter sequences. This short synthetic promoter functioned consistently in transient expression tests using protoplasts of three different plant species, suggesting that short synthetic promoters containing homo- or hetero-repeats of key sequences within active cis-elements can enhance targeted gene expression in an orthogonal manner. Another recent study showed the orthogonal regulatory effect of synthetic activators and repressors and their interaction with synthetic promoters using the adopted orthogonal regulatory system of yeast (Belcher et al., 2020). Transient expression assays in Nicotiana benthamiana and stable transgenic Arabidopsis were validated sequentially by showing synthetic promoter activation and repression relative to the addition of synthetic promoters.
Owing to the accessibility of genetic information, a reliable transformation system, and abundant experimental work, hybrid poplar species are apt woody biofuel feedstock models (Tuskan et al., 2006; Ma et al., 2019). Populus also provides economically valuable bioproducts such as paper, cellulose, wood, and fiber. Poplar has been engineered for improved biofuel production by reducing recalcitrance of lignocellulosic biomass or increasing biomass stock polymers such as cellulose and hemicellulose for conversion to biofuel (Bryant et al., 2020). The overexpression of selected genes or the production of RNA interference fragments of target genes has mainly been controlled by constitutive promoters such as the 35S CaMV promoter or ubiquitin promoter. Additionally, native promoters such as those from the vascular-related NAD transcription factor (VND (SND/NST)) or MYB genes, which serve as regulatory transcription factors for secondary cell wall synthesis and biomass biosynthesis-related enzymes, such as cellulase, have been studied broadly and recently used to develop synthetic promoters in Arabidopsis (Endo et al., 2015; Nakano et al., 2015). Based on native promoter sequence information, small fragments of a secondary cell wall NAC binding element (SNBE) functioned as a synthetic promoter constructed for CRISPR/Cas9 gene editing knock-out of CINNAMOTYL-CoA REDUCTASE 1 (CCR1) or stem tissue-specific expression of CCR1 in ccr1 mutant, resulting in tissue-specific lignin content modification in Arabidopsis (De Meester et al., 2018; Yu et al., 2021). In these studies, the synthetic SNBE promoters were shown to be tissue-specific, showing promise for synthetic promoter motif utility for biomass optimization. Currently however, applicable promoters are limited to biosynthesis processes, not in other areas such as stress response in poplar. Also, most of this work has been performed in herbaceous species, and yet to be confirmed in a woody plant such as poplar.
Our lab recently reported on synthetic drought (SD) inducible promoters designed by computational DNA motif analysis software from the promoter sequences of stress-responsive co-expressed genes in the poplar genome (Yang et al., 2021). The heptamerized 7-8 base repeats of conserved DNA motifs from drought stress-inducible promoters were screened in transient transformation assays using poplar leaf mesophyll protoplasts, agroinfiltration in N. benthamiana leaves, and in stably transgenic Arabidopsis. The objective of the present study was to assess the osmotic stress responsiveness of six SD inducible promoters in stably transformed poplar. We also scrutinized the effect of different sequence lengths in a synthetic promoter from a DNA motif with heptamerized repeats of the full-length SD9 motif (20 bases) compared with heptamerized partial sequences (7-8 bases) from the SD9.
Materials and methods
Plants
Populus tremula × Populus alba female INRA 717 1B4 clones (717-1B4) were kindly donated by Dr. Steven Strauss (Oregon State University). Small plants were propagated in solidified rooting media (Yang et al., 2021) in Magenta GA7 boxes (Bioworld, Dublin, OH, USA), and grown in a growth chamber (Percival Scientific, Fontana, WI, USA) with 16/8 light/dark conditions at 23 °C with 150 μmol/m2s irradiance. The rooted plants were transferred to ProMix BK25 (Premier Tech, Quakertown, PA, USA) in 1 L pots, and then acclimated for 2 weeks in the same chamber. The propagated plants were grown in a greenhouse at the University of Tennessee, Knoxville, and watered every two days. The potted plants were fertilized with a 14-14-14 nutrient solution (Seed World, Odessa, FL, USA) every three weeks.
Nicotiana benthamiana seeds were germinated and grown in ProMix BK25 after 4 days of stratification using sterile water at 4°C in the dark. Plants were grown under the same growth chamber conditions as poplar.
Generation of binary gene constructs containing inducible synthetic promoters
The binary backbone vectors including a kanamycin resistance gene as a transgenic plant selective marker were adopted from our lab’s published work and used for agroinfiltration testing and transgenic poplar generation (Yang et al., 2021). We screened the same binary vector constructs carrying SD9-1 (a total of 56 bases), 9-2 (49 bases), 9-3 (49 bases), 13-1 (49 bases), 18-1 (56 bases), and 18-3 (49 bases) synthetic promoters in this study. A new synthetic promoter, denoted SD9 (140 bases), was generated from the overlapped core sequences of SD9-1, 9-2, and 9-3, was inserted into the same backbone vector using Golden Gate cloning (Engler and Marillonnet, 2014). The synthesized fragment sequences are listed in Table S1. The oligonucleotides for double-strand synthetic promoter fragments were synthesized by Integrated DNA Technologies (IDT, Coralville, IA, USA). All DNA sequences were verified by commercial Sanger sequencing (Psomagen, Rockville, MA, USA).
Transient agroinfiltration assays in N. benthamiana leaves
The binary gene constructs incorporating our synthetic promoters were used for transient agrobacterium infiltration assays. The processes for Agrobacterium transformation, agroinfiltration of 1-month-old N. benthamiana leaves, osmotic stress treatment, and subsequent fluorescence measurements were performed as previously described (Sparkes et al., 2006; Yang et al., 2021). The GFP fluorescence intensity was measured using a Fluorolog®-3 (HORIBA, Kyoto, JAPAN) fluorescence spectrophotometer using direct scans on intact leaves.
Agrobacterium-mediated poplar transformation and genotyping
Agrobacterium-mediated poplar leaf disk transformation was performed with the method established by Song et al. (Song et al., 2006). Leaf disks were collected from two-month-old 717-1B4 poplar plant leaves grown in the Magenta GA7 boxes. Following kanamycin (50 μg/ml) selection regenerated shoots were rooted in rooting medium including activated charcoal (5 g/L) (Kang et al., 2009). The rooted shoots were transplanted into 1 L pots following the protocol described above. After transplanting to soil and growing in the growth chamber for three months, all plants were transplanted into 8 L pots to be grown in the greenhouse.
For genotyping of transgenic poplar, the genomic DNA was extracted from the leaves of each genetic event with a DNeasy® Plant Mini kit (Qiagen, Germantown, MD, USA). PCR was performed by DreamTaq DNA polymerase Master Mixture (Thermo Fisher Scientific) with the reaction sequence as follows: 1 cycle of 95°C for 2 min, 30 cycles repeating 95°C for 30 sec, 57°C for 30 sec, and 72°C for 1 min/1 kb, and 1 cycle of 72°C for 7 min. Two separate reactions were performed, using primers targeting GFP and primers targeting PtUBCc as an internal control for genomic DNA qualification. Bands were separated on 1.2% agarose gels (1 × TAE).
Ramet propagation of transgenic poplar and osmotic stress treatment
The poplar ramets were propagated from cuttings from the original transgenic plant that grew for at least 6 months in potting mix, and then the ramets were grown for another six to nine months. Fully expanded leaves of propagated ramets were used for stress-responsive testing under high salinity and polyethylene glycol (PEG) treatments. The solutions of 250 mM NaCl (Fisher scientific, Hampton, NH, USA) and 20% PEG 6000 (EMD Millipore Burlington, MA, USA) were prepared with deionized (DI) and autoclaved water. The solutions were directly sprayed onto fully expanded leaves positioned from the fifth to seventh leaf from the main stem apex for 3 days (once daily at 11:00 a.m.). The sprayed leaves were sealed to prevent drying. Leaves in the same position in other ramets were sprayed with DI water as mock control. All stress treatments were performed between March and April of 2021. The stress response was determined by assessing GFP fluorescence using a Fluorolog®-3 (HORIBA, Kyoto, Japan) spectrofluorometer. The corresponding image of multi-channel fluorescence detection was generated with the same leaf using the fluorescence-inducing laser projector (FILP) (Rigoulot et al., 2021). The laser power, filter conditions, and exposure time are described in each figure legend. The image conversion was performed by macro function operated in Image J software (Schneider et al., 2012).
Statistical analysis
Technical and biological repeat numbers are described in individual figure legends. Statistical analysis of all measurements was performed by the Student’s t-test protocol integrated in R (R core team, 2014).
Results
Stably transformed poplar with osmotic stress-induced synthetic promoters
To assess our six synthetic promoters (SD9-1, 9-2, 9-3, 13-1, 18-1, and 18-3) in planta, we first generated stably transformed poplar each carrying one of the six SD promoters. These six short synthetic promoters were produced from poplar A-domain promoter sequences, which activated downstream GFP expression in transient agroinfiltration assays on N. benthamiana leaves under water deficit or high salinity conditions (Yang et al., 2021). Among these synthetic promoters, SD9-2 and SD18-1 reliably induced GFP in transgenic Arabidopsis. However, four other synthetic promoters remained to be tested in planta, as well as testing all six promoters in the poplar system.
At least 10 different transgenic events were obtained for each promoter construct via Agrobacterium-mediated leaf disk transformation followed by kanamycin selection (data not shown). Figure S1 summarizes representative images of transformation steps. We confirmed the presence of the GFP gene in genomic DNA isolated from seven randomly selected transgenic poplar plants, and three different transgenic events were propagated by cuttings to produce more ramets for the experiments described in the following sections (Figure S2).
Determination of optimal concentrations for salinity and PEG treatments
To determine the optimal conditions for inducing osmotic stress in transgenic poplar, we measured GFP fluorescence on leaves that were sprayed with various concentrations of NaCl and PEG for 3 days. When we first pre-screened by spraying 250 mM NaCl and 30% PEG on several randomly selected transgenic lines, GFP was detected most readily in transgenic poplar lines containing the SD9-2 (SD9-2::GFP) and SD18-1 (SD18-1::GFP), synthetic promoters, respectively. Therefore, we used two different transgenic events of SD9-2::GFP for salt stress experiments and two events of SD18-1::GFP for PEG-induced osmotic stress. A transgenic line containing the empty vector was used as a comparator.
In the NaCl treatment test, GFP fluorescence correlated with salt concentrations up to 250 mM (Figure 1A). Concentrations of 275 mM and 300 mM resulted in leaf wilting and yellowing, which interfered with the fluorescence measurements (data not shown). Therefore, we selected 250 mM NaCl concentration (~20 ml per spray) to apply to plants every day for 3 days for further salinity treatment tests.
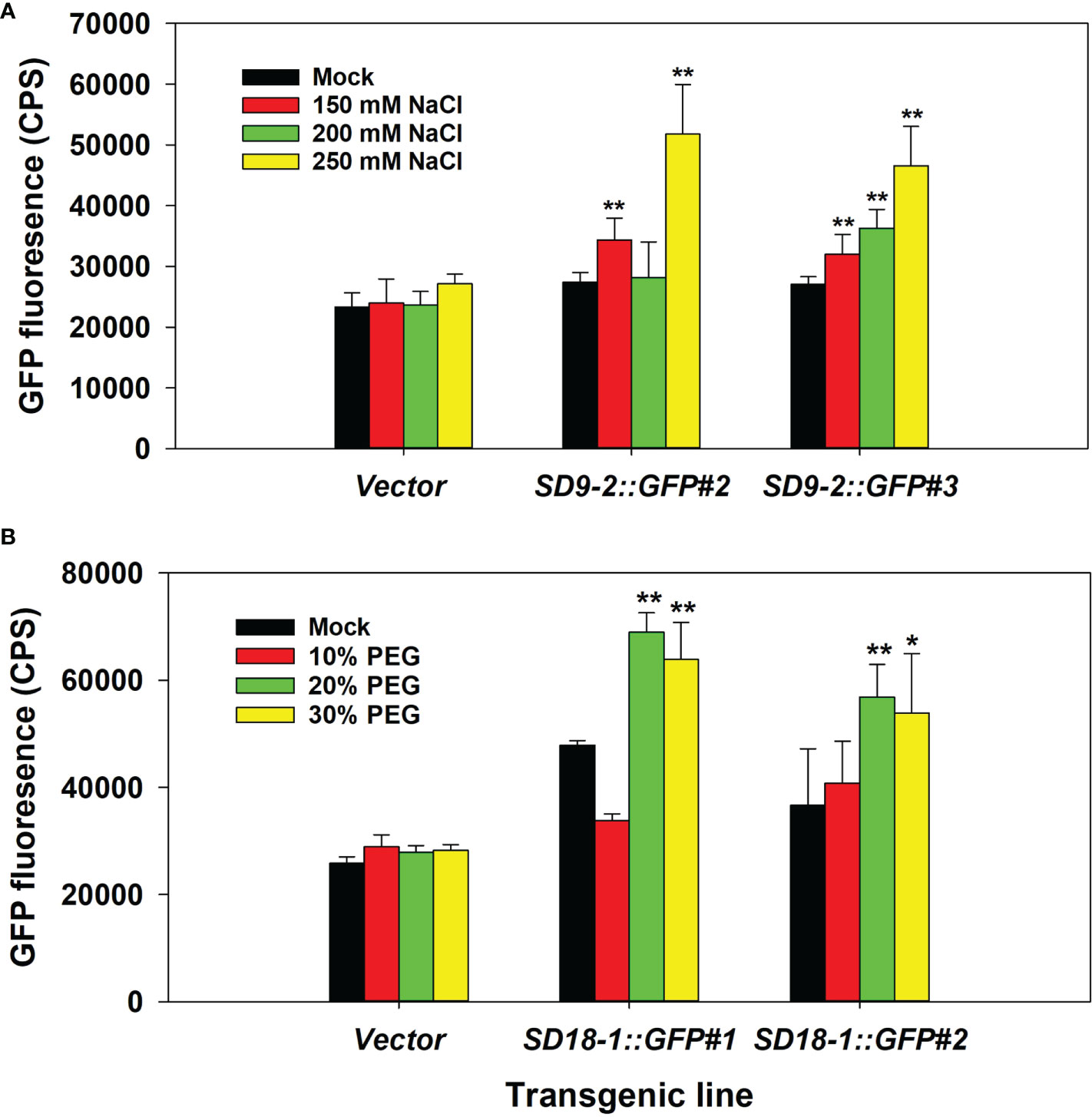
Figure 1 Determination of optimal NaCl and PEG treatment concentrations. The fifth to seventh positioned leaves of six-month-old ramets were used. (A) GFP fluorescence measurement after 150, 200, and 250 mM NaCl spray treatment for 3 days in counts per second (CPS). Vector control and two transgenic events of SD9-2:GFP as representative transgenic lines were tested. (B) GFP fluorescence measurement after 10, 20, and 30% PEG spray treatment for 3 days. Vector control and two transgenic events of SD18-1:GFP were used as representative transgenic lines. The bar shows the average of GFP intensity of the abaxial side of treated leaves (n=3). Asterisks denote significant difference of GFP expression between mock control and stress treated leaf via t-test (*P < 0.05, **P < 0.01).
For testing water-deficit conditions, we initially tested withholding water for up to 10 days before measurements. However, leaves were severely shrunken, which affected GFP measurements. Thus, we used PEG chemical treatments to simulate water-deficit conditions that allow proper GFP fluorescence measurement. When leaves were treated with 10, 20, and 30% PEG, the maximum GFP fluorescence was reached in 20% PEG treatments. Therefore, 20% of PEG solutions were used for water-deficit stress measurements in further testing (Figure 1B).
Synthetic promoter activity using the SD9 motif with heptamerized partial cis-regulatory sequences (7-8 bases) versus a heptamerized full SD9 motif (20 bases)
To identify if the synthetic promoter activity is dependent on core sequence length, we assessed whether a synthetic promoter formation with repeats of the full SD9 motif sequence (20 bases) functions more effectively than the heptamerized partial sequences (7-8 bases; SD9-1, SD9-2, and SD9-3). For this purpose, we generated a synthetic promoter of the entire SD9 motif (20 bases) and cloned it into a binary vector (Figures 2A, B).
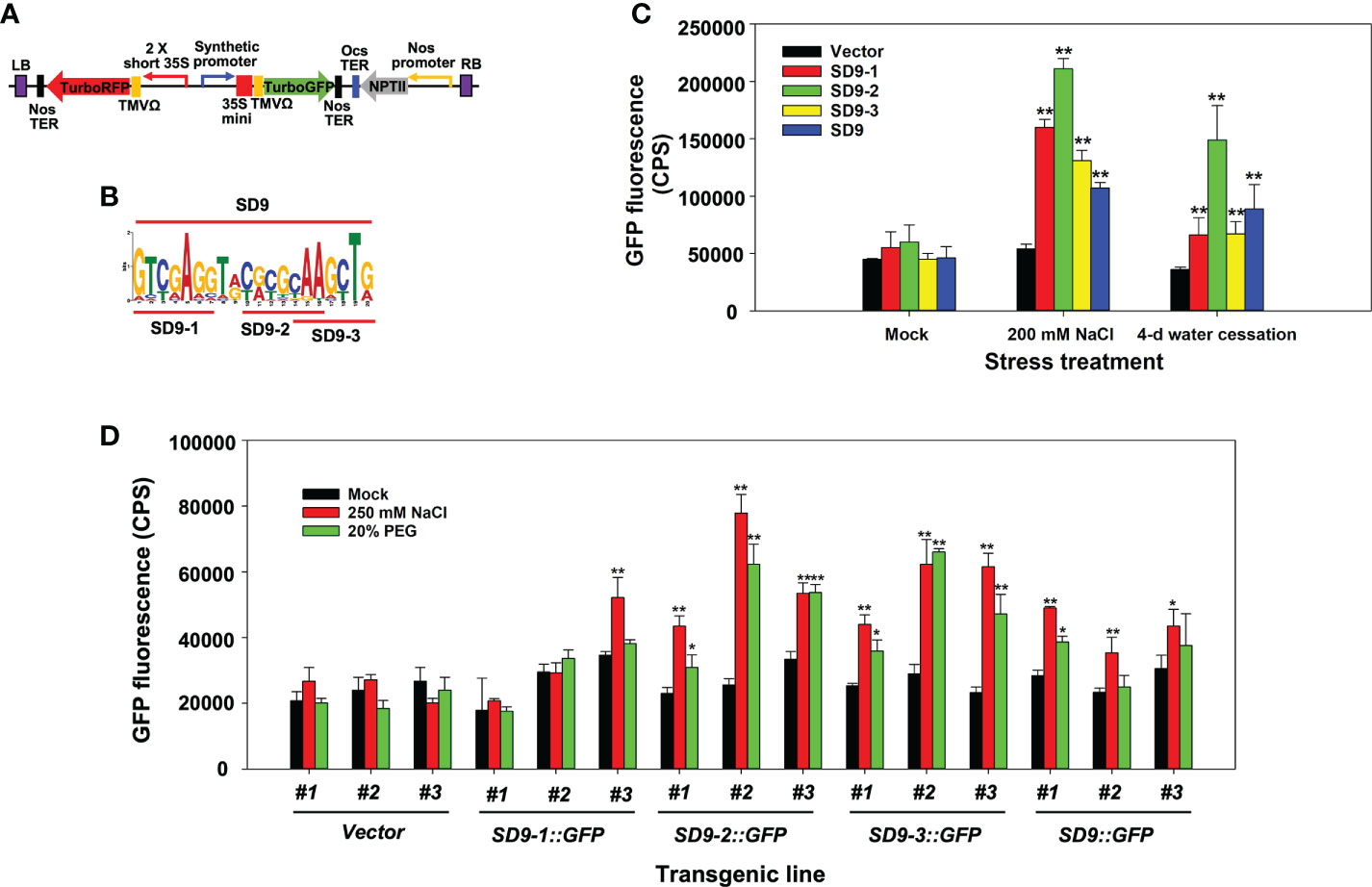
Figure 2 Stress induction of synthetic promoters consisting of heptamerized partial or full sequences from the SD9 motif. (A) Binary vector construct containing the synthetic promoter fused to TurboGFP. TurboRFP was driven by a 2 × short 35S constitutive promoter to monitor transformation. Heptamerized partial or full sequences from the SD9 motif (panel (B) were inserted into this backbone plasmid. (B) LOGO plot of full and partial DNA sequences of SD9. This sequence was heptamerized in working plasmids. (C) Transient expression analysis by agroinfiltration on N. benthamiana leaves. Each bar represents the average of GFP fluorescence measured by Fluorolog fluorescence spectrophotometry (n=3) in CPS. (D) Stress-inducible analysis of synthetic promoters consisting of heptamerized partial or full sequences from the SD9 motif in stable transgenic poplar. The GFP expression in the transgenic poplar leaves each carrying synthetic promoters in high salinity or drought-like conditions. For both tests, the leaves of six-month-old propagated clones were sprayed with 250 mM NaCl or 20% PEG for 3 days. Mock control was sprayed with DI water for the same period. The bar shows the average of GFP intensity of the abaxial side of treated leaves (n=3). In panel c and d, error bar represents standard deviation. The statistical significance of each value against vector control was determined by t-test (*P < 0.05, **P < 0.01).
Transient Agrobacterium infiltration experiments on leaves of N. benthamiana were performed to visualize the synthetic promoter activity in leaves under high salinity and severe water-deficit stress. Separate treatments of watering with 200 mM NaCl and water cessation were used to generate osmotic stress conditions after agroinfiltration (Yang et al., 2021). SD9::GFP had consistently lower GFP expression compared to the partial sequence repeats while SD9-2::GFP induced higher promoter activity and GFP gene expression than the others under both high salt and water-deficit conditions (Figure 2C). Interestingly, all synthetic promoters were more induced by salt than water deficit stress.
These results were confirmed in stably transformed poplar plants with the same gene constructs used for transient agroinfiltration assays. Three different transgenic events were established for each gene construct transformation (Figure 2D). Leaves of six-month-old propagated transgenic poplar ramets were sprayed separately with 250 mM NaCl or 20% PEG for 3 days. Leaves of SD9::GFP, SD9-2::GFP, and SD9-3::GFP transgenic poplars showed up to 2-fold higher GFP fluorescence than mock treatments after exposure to 250 mM NaCl (Figure 2D). These synthetic promoters also induced GFP expression in 20% PEG treatment, but the fluorescence intensity was relatively lower than that of salt treated leaves (Figure 2D). In line with our observations in the transient agroinfiltration tests, the heptamerized SD9-2 and 9-3 synthetic promoters induced around 1.5-fold higher than that of the complete SD9 in the 250 mM NaCl treatment. In contrast with the agroinfiltration tests, the heptamerized SD9-1 did not induce GFP expression for either stress condition.
To summarize, three transgenic poplar lines containing the heptamerized SD9-2, 9-3, and the entire SD9 synthetic promoter showed responses to NaCl and PEG. However, the synthetic promoters constructed with heptamerized partial sequences of SD9 (SD9-2 and 9-3) showed higher promoter activity than the heptamerized SD9 full sequence (SD9) in high salinity and PEG treatments.
Osmotic stress responses of stably transformed poplar carrying synthetic promoters derived from the SD9 motif
To verify the osmotic stress responsiveness of SD9-1, 9-2, and 9-3 synthetic promoters in stable transgenic plants, we measured GFP fluorescence in transformed poplar after 250 mM or 20% PEG treatments for 3 days. Among three SD9-derived synthetic promoters, the transgenic poplar containing SD9-1 (SD9-1::GFP) did not display GFP induction despite previous transient transformation tests that showed GFP induction under osmotic stress (Figure S3; Yang et al., 2021). However, all three transgenic event lines carrying the SD9-2 synthetic promoter (SD9-2::GFP) had significantly induced higher GFP expression (1.5 to 2.6-fold) in 250 mM NaCl treatment than mock control (Figure 3B). SD9-2::GFP displayed 1.2 ~ 1.8-fold GFP fluorescence increase in response to the 20% PEG treatments (Figure 3B). Multi-channel fluorescence scanning images of salt or PEG treated leaves using FILP showed clear and consistent activation of GFP fluorescence (Figure 3C). RFP fluorescence, an internal control for monitoring correct transformation, was detected in all mock control and treated leaves, confirming that the gene constructs were transformed stably into transgenic poplar (Figure 3C). The binary vector construct for poplar transformation is shown in Figure 3A.
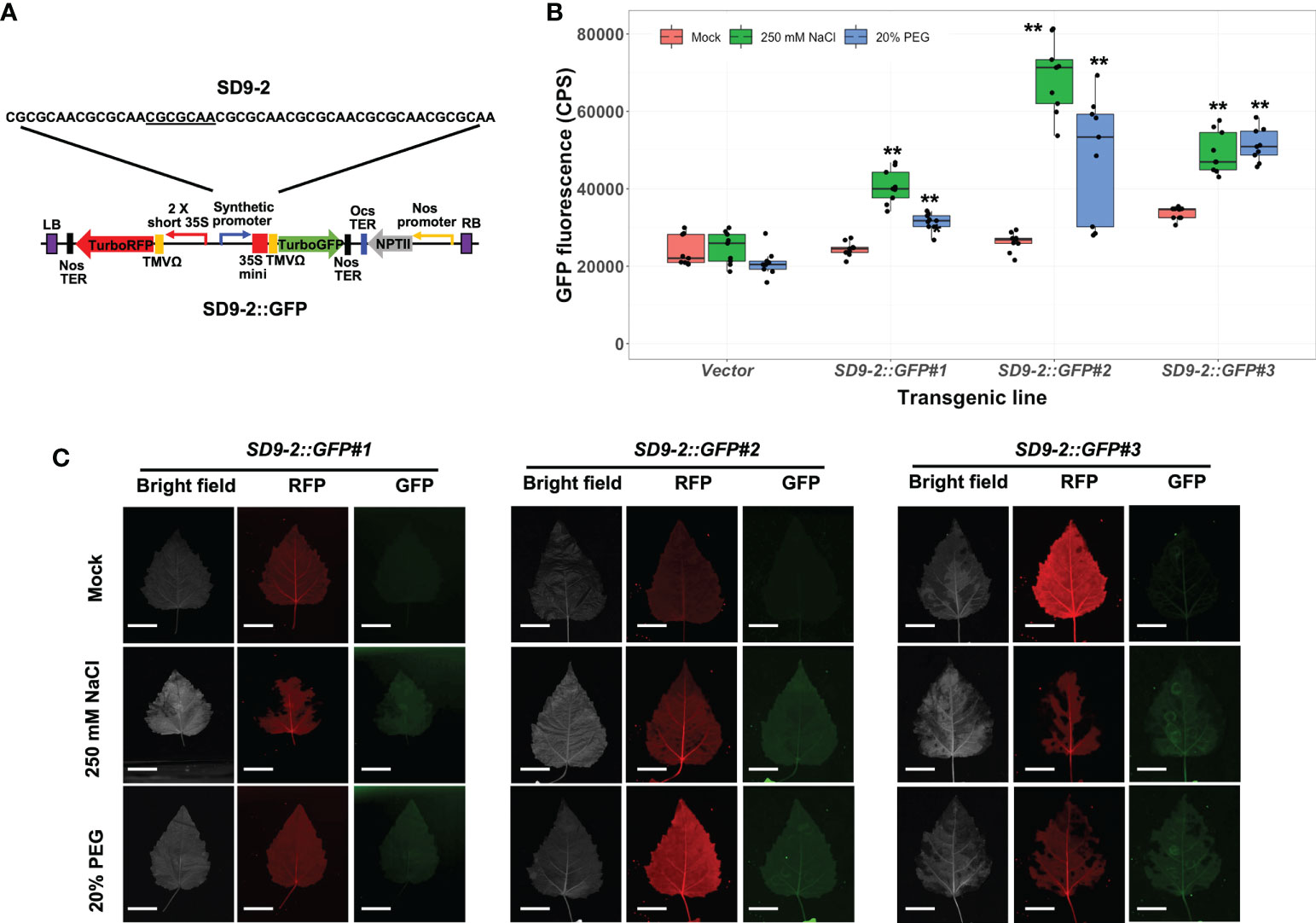
Figure 3 Osmotic stress induction of GFP fluorescence in transgenic poplar containing SD9-2 synthetic promoter (SD9-2::GFP). Three different transgenic poplar lines were propagated by cutting and grown in the greenhouse. (A) SD9-2::GFP gene construct for generating transgenic poplar. Heptamerized 7 base sequence (basic sequence of CGCGCAA; SD9-2) was inserted into this binary vector. (B) GFP fluorescence in transgenic poplar carrying SD9-2 synthetic promoter under osmotic stress treatments. Six- to nine-month-old transgenic poplar lines were treated with 250 mM NaCl or 20% PEG. NaCl and PEG solutions were sprayed for 3 days on the leaves selected between fifth and seventh positions from the apex. Water was sprayed for mock treatment (mock) following the same guidelines. CPS values of GFP fluorescence were measured by Fluorolog fluorescence spectrophotometry at emission wavelength of 502 nm under a fixed excitation wavelength of 482 nm. Triplicate experiments were performed with three different clones (n=9). Asterisks denote significant difference of GFP expression between mock control and stress treated leaf via t-test (*P < 0.05, **P < 0.01). (C) Visualizations of induced RFP and GFP fluorescence detected by FILP, using the same stress-treated leaves. RFP was used as an internal control to monitor for transformation. Exposure time for images was 100 ms. Scale bars represent 2 cm of length at a detection distance of 3 m.
The transgenic poplar lines carrying the SD9-3 synthetic promoter (SD9-3::GFP) had GFP fluorescence ranging from 1.5 to 2.0-fold higher in 250 mM NaCl-treated leaves compared to the mock control (Figure 4B). In the 20% PEG treatments, the GFP fluorescence ranged from 1.4- to 1.8-fold higher (Figure 4B). The fluorescence images taken by FILP were consistent with the fluorolog-measured GFP fluorescence (Figure 4C). The GFP fluorescence intensity for SD9-3 was similar to the range recorded for SD9-2. In summary, of the three SD9-derived synthetic promoters, SD9-2 and SD9-3 demonstrated similarly high responses to high salinity and PEG, while SD9-1 did not respond in transgenic poplar lines.
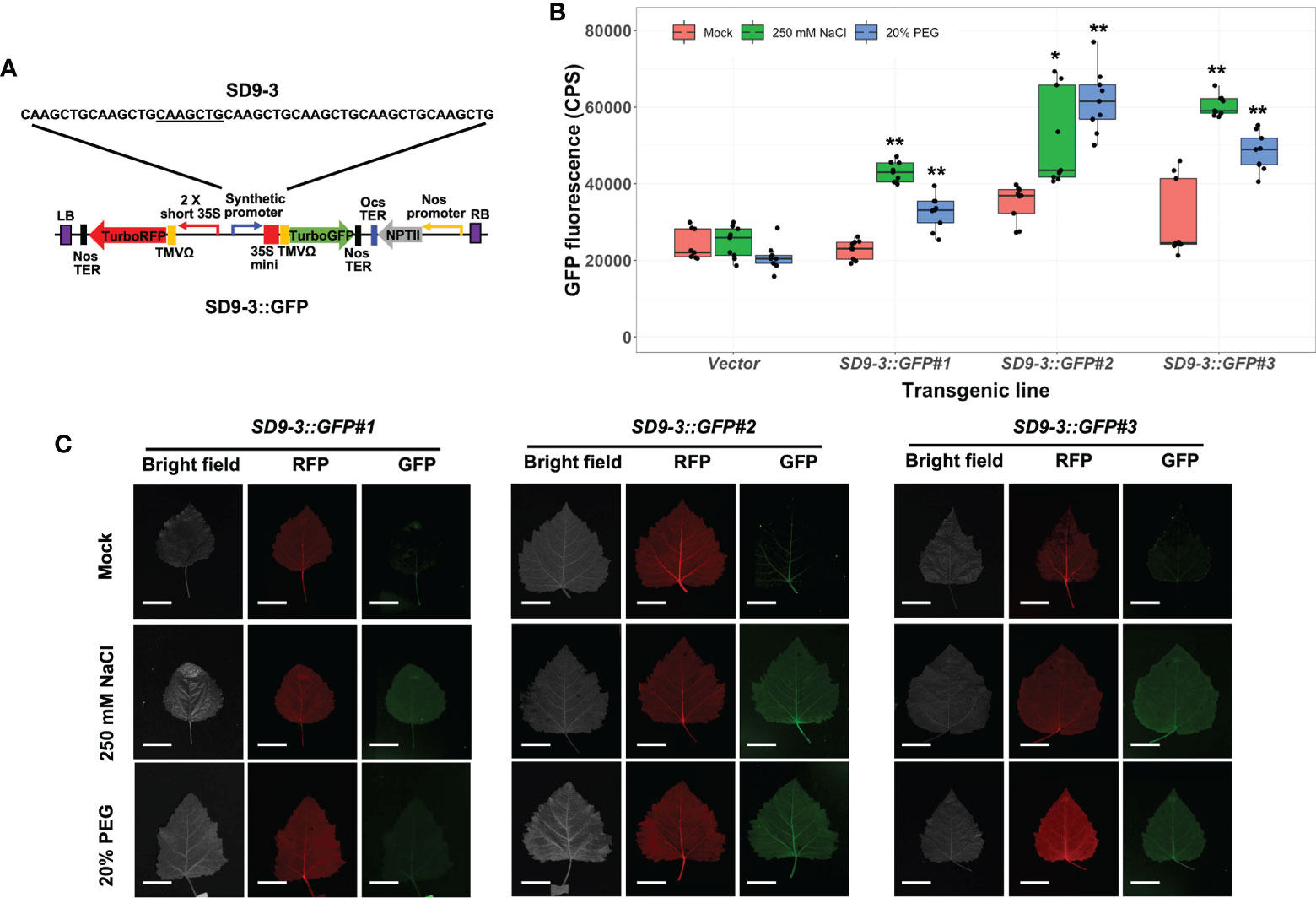
Figure 4 Osmotic stress induction of GFP fluorescence in transgenic poplar carrying SD9-3 synthetic promoter (SD9-3::GFP). Three different transgenic poplar lines were propagated by cutting and grown in the greenhouse. (A) SD9-3::GPF gene construct for generating transgenic poplar. Heptamerized 7 bases (CAAGCTG; SD9-3) was inserted into this binary vector fused to GFP. (B) GFP fluorescence in transgenic poplars containing SD9-3 synthetic promoter under osmotic stress treatments. Six- to nine-month-old transgenic poplar lines were treated with 250 mM NaCl, or 20% PEG. NaCl and PEG solutions were sprayed for 3 days on the leaves selected between fifth and seventh positioned from apex. Water was sprayed for mock treatment (mock). CPS value of GFP fluorescence was measured at emission wavelength of 502 nm under a fixed excitation wavelength of 482 nm measured by Fluorolog fluorescence spectrophotometry in CPS. Triplicate experiments were performed with three clones (n=9). Asterisks denote significant difference of GFP expression between mock control and stress treated leaf via t-test (*P < 0.05, **P < 0.01). (C) Visualization of induced RFP and GFP fluorescence detected by FILP, using the same stress-treated leaves. RFP was used as an internal control to confirm gene construct transformation. Exposure time for images was 100 ms. Scale bars represent 2 cm of length at a detection distance of 3 m.
Osmotic stress responses of stably transformed poplar carrying synthetic promoters derived from the SD13 and SD18 motifs
We verified osmotic stress sensitivity of other SD promoters in stably transformed poplar plants after 250 mM NaCl and 20% PEG treatments for 3 days. Two synthetic promoters from the SD13 motif (SD13-1 and 13-2) and three synthetic promoters with the SD18 motif (SD18-1, 18-2, and 18-3), which presented together with SD9 in the poplar A-domain, were tested (Yang et al., 2021). Of these GFP fused binary vectors, the SD13-1, 18-1, and 18-3 synthetic promoters were significantly activated by high salinity and water-deficit stresses. However, present transgenic poplar plants containing SD13-1 (SD13-1::GFP) and SD18-3 synthetic promoters (SD18-3::GFP) did not have increased GFP fluorescence when treated in the 250 mM NaCl or 20% PEG treatment (Figure S3). Of these three synthetic promoters, only transgenic poplar with the SD18-1 synthetic promoter (SD18-1::GFP) had significantly induced GFP expression in the range from 1.4- to 1.5- fold and 1.5- to 1.7-fold higher GFP fluorescence in NaCl and PEG treatments, respectively (Figure 5B). As we observed in SD9-2::GFP and SD9-3::GFP, the FILP images were consistent with measured GFP fluorescence (Figure 5C). However, the GFP fluorescence value was 0.7-fold lower (average of all GFP fluorescence in three transgenic events) than that of SD9-2::GFP and SD9-3::GFP.
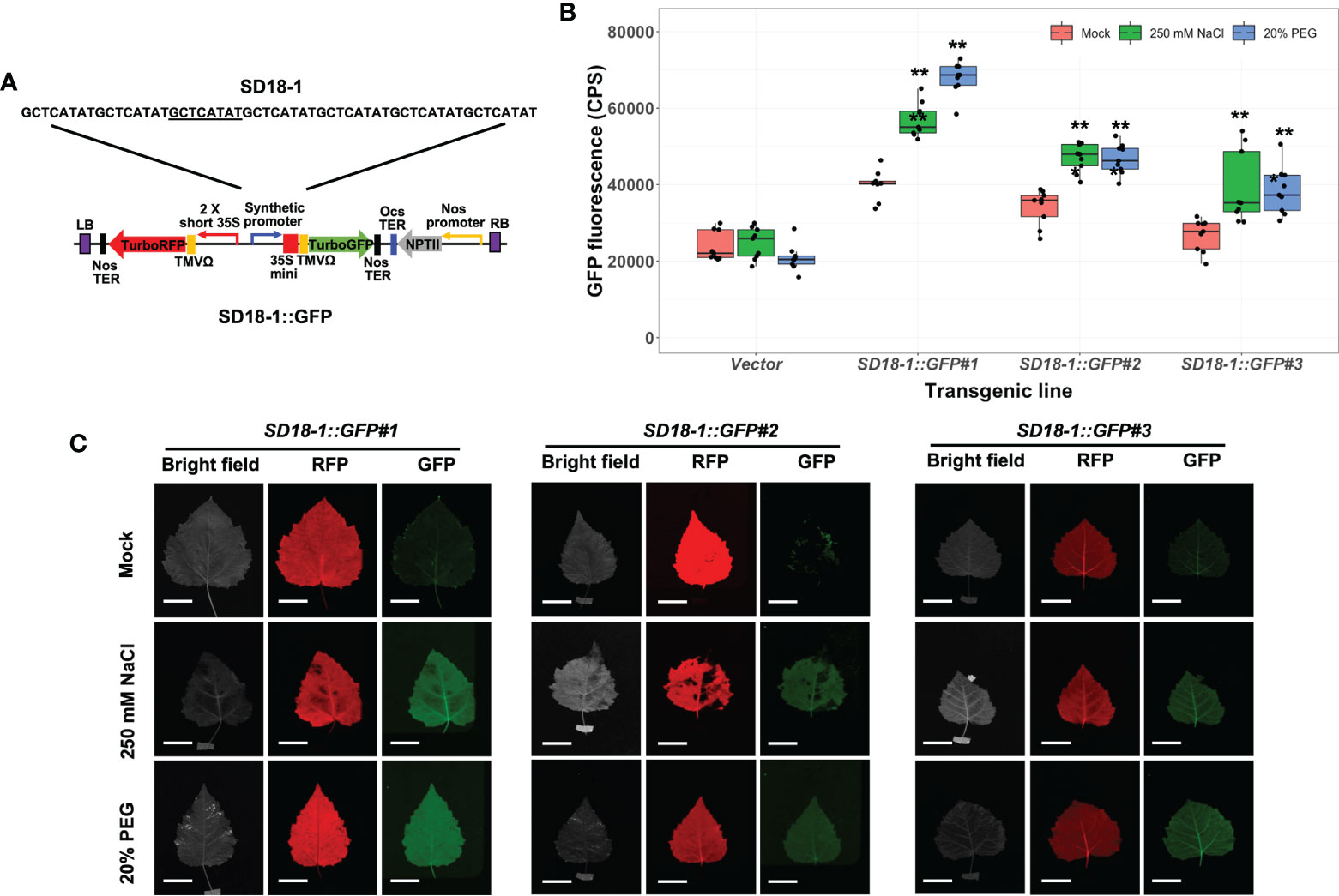
Figure 5 Stress induction of GFP fluorescence in transgenic poplar harboring SD18-1 synthetic promoter (SD18-1::GFP) by osmotic stress. Three different transgenic poplar lines were propagated by cutting and grown in a greenhouse. (A) SD18-1::GFP gene construct for generating transgenic poplar. Heptamerized 8-base sequence (GCTCATAT; SD18-1) was inserted into binary vector fused to GFP. (B) GFP fluorescence in transgenic poplars harboring SD18-1 synthetic promoter under osmotic stress treatments. Six- to nine-month-old transgenic poplar lines were treated with 250 mM NaCl or 20% PEG. NaCl and PEG solutions were sprayed for 3 days on the leaves selected between fifth and seventh positions from apex. Water was sprayed for mock treatment following the same guidelines (mock). CPS of GFP fluorescence was measured at emission wavelength of 502 nm under a fixed excitation wavelength of 482 nm measured by Fluorolog fluorescence spectrophotometry. Triplicate experiments were performed with three different clones (n=9). Asterisks denote significant difference of GFP induction between mock control and stress treated leaf via t-test (*P < 0.05, **P < 0.01). (C) The image of induced RFP and GFP fluorescence detected by FILP, using the same stress-treated leaves. RFP was used as an internal control to confirm gene construct transformation. Exposure time for images was 100 ms. Scale bars represent 2 cm of length at a detection distance of 3 m.
Summary of transient and stable transformation tests of SD synthetic promoters
In summary, we performed transient transformation using poplar leaf mesophyll protoplasts and agroinfiltration of N. benthamiana leaves, as well as stable transformation of poplar using the same binary vectors each carrying one of our synthetic promoters. Through GFP fluorescence assays, we identified the stress responsiveness of poplar osmotic stress-induced synthetic promoters. The results for these assays are summarized in Table 1. Out of eight SD-series synthetic promoters, six synthetic promoters including SD9-1, 9-2, 9-3, 13-1, 18-1, and 18-3 activated downstream GFP expression in transient transformation tests under osmotic stress conditions (Yang et al., 2021). Three SD synthetic promoters including SD9-2, 9-3, and 18-1 also consistently induced GFP expression in stable transgenic poplars (Figures 3–5). These three synthetic promoters responded to both high salinity and PEG treatments. The osmotic stress responsiveness of synthetic promoters was reliably screened in both transiently and stably transformed plant cells although a small number of biases still existed between both transformations. Three SDs were confirmed as osmotic stress-responsible synthetic promoters through all tests.
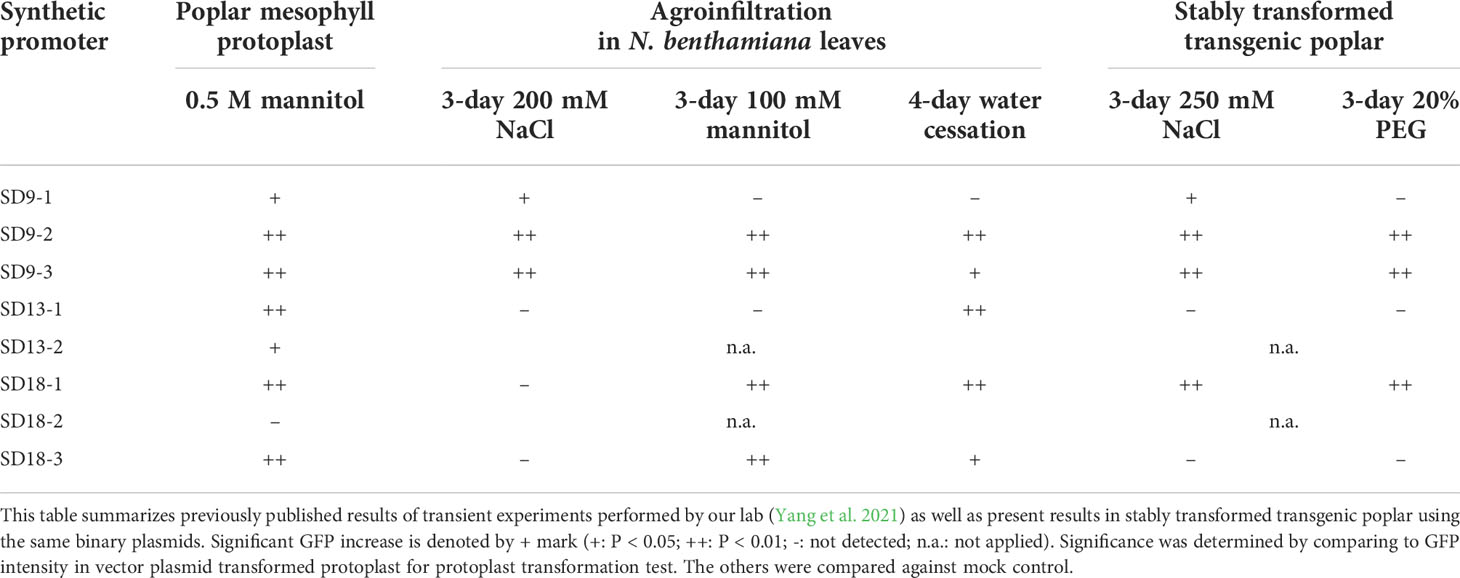
Table 1 Summary of GFP induction of SD synthetic promoters in transient and stably transformed plants under osmotic stress conditions. .
Discussion
Modern synthetic biology techniques for transgenic plant generation have been regarded as powerful tools for revolutionizing agriculture. Promoter modification with native or synthetic promoters is a powerful approach for tuning targeted gene expression in transgenic plants. To control gene expression at the optimal timing and with the appropriate transcript abundance, it is necessary to design versatile synthetic promoters, which considers optimal sequence selection, copy number and spacing optimization, and proper orientation and order of components within the synthetic promoter. Most of all, it is important that researchers select constituent components that maximize the activity of promoter constructs including three important elements: a) the core promoter, b) appropriate TF binding site(s) in the proximal promoter, and c) other additive elements such as enhancers or insulators (repressors) as needed for the application of interest (Dey et al., 2015). To date, native and synthetic promoters for this purpose have been mostly reported in a few species (Dey et al., 2015; Liu and Stewart, 2016; Ali and Kim, 2019).
We previously reported poplar osmotic stress-responsive synthetic promoters that were based on native promoter sequences from Populus. The CaMV 35S core and translation leader 5’ untranslated region (UTR) sequences were fused to a GFP reporter gene, and GFP activity was assayed by transient screening using poplar protoplasts and agroinfiltration (Yang et al., 2021). The present study confirmed the positive activity of these synthetic promoters in stably transformed poplar plants under osmotic stress (Figures 3–5, and Table 1). The synthetic promoters are composed of synthetic heptamer repeats fused with the -46 35S minimal core promoter, which includes a TATA boxn, and a TMV Ω 5’ UTR leader sequence to maximize translational efficiency. Recent studies using STARR-seq analysis showed that the most robust core promoters have the TATA box at the -30 to -40 bp location from the transcriptional starting site (TSS) (Jores et al., 2020; Jores et al., 2021). They found that core promoters, which include the TATA box, activated downstream gene expression in a species-specific manner. In addition, they also identified peak promoter activity when the core promoter worked in tandem with other transcription initiation factors such as an initiator and pyrimidine-rich promoter (TC motif or Y-patch) (Jores et al., 2021). Several DNA sequence studies have used high throughput techniques to study core promoter sequences from yeast, Drosophila melanogaster, and human cells (Lubliner et al., 2015; Arnold et al., 2017; van Arensbergen et al., 2017; Weingarten-Gabbay et al., 2019; de Boer et al., 2020; Kotopka and Smolke, 2020). These studies revealed that the core promoter elements and TF-binding sites in the region close to the TSS cooperate to mediate the gene transcription. Other cis-elements such as enhancers and insulators can be added distal to the promoter region for the regulation of synthetic promoter activity (Lenhard et al., 2012; Beagrie and Pombo, 2016; Haberle and Stark, 2018). However, these findings had limited applications in plant systems because core promoter elements including the TATA box and the initiator (Ins) elements were identified by computational analysis without reliable confirmation in planta (Yamamoto et al., 2007; Morton et al., 2014; Srivastava et al., 2014; Jores et al., 2020). It was recently reported, using massively parallel reporter assays in tobacco leaves and maize protoplasts, that the core promoter elements including the TATA box as well as promoter GC content and promoter proximal transcription factor binding sites affect the strength of most promoters in the genes of Arabidopsis, maize, and sorghum (Jores et al., 2021). The study demonstrated that promoter-proximal cis-regulatory elements work in tandem with the core promoter sequence to determine expression strength in tobacco leaves and maize protoplasts (Jores et al., 2021). In addition, another study screened a 37-promoter library comprised of fourteen common plant active promoters, seven 5’ UTRs, and eleven 3’ UTRs in transfected N. benthamiana leaves, canopies, and protoplasts. This study showed that 3’ UTR components are also important for maximizing promoter activity, in addition to upstream regulatory elements (Pfotenhauer et al., 2022). Therefore, understanding the cooperation of entire promoter compositions including the core elements, proximal cis-elements close to the TSS, other factors such as enhancers and insulators (or repressors), and 3’ UTR of transcription terminator is crucial for the design of versatile synthetic promoters. Although our present gene construct demonstrated coordination between each component, it is yet to be determined if further optimization is possible for gene expression in poplar. In order to generate the optimized synthetic promoter, it will be necessary to test other features such as replacement of the current virus core promoter with an intact poplar core promoter or linkage with other candidate cis-elements.
Another important factor for consideration is the ability of proximal cis-elements of the core promoter fragment to modify and boost promoter activity (Cai et al., 2020; Yang et al., 2021). In the present study as well as our previously published results, we tested many unknown cis-elements from the poplar A-domain as well as salt stress-responsive promoters found in the poplar genome (Yang et al., 2021). Using heptamerized 7-20 bp core sequences, we successfully showed GFP induction in poplar leaf protoplasts, transiently agroinfiltrated N. benthamiana leaves, and stably-transformed transgenic Arabidopsis and poplar plants after osmotic stress treatments (Figures 2–5 and Yang et al., 2021). Our results showed that the promoter activity of the heptamerized entire SD9 motif including SD9-1, 9-2, and 9-3, resulted in lower GFP induction, compared to that in separate SD9-2 and 9-3 short DNA motifs, despite those sequences being contained within SD9 (Figure 2). The higher synthetic promoter activity using 7 copies of basic cis-elements supports the observation that a 3-4 repeat combination of different cis-elements had stronger promoter activity than a single copy of a short cis-element (Cai et al., 2020). Interestingly, our results showed that the synthetic promoter containing 7 repeats of a short sequence induced stronger downstream gene expression than that of the same repeats of a longer sequence, indicating that promoter activity is not associated with the length of its sequence per se. These results demonstrate the importance of understanding individual cis-element features and their interactions to rationally design functional synthetic promoters.
Cooperation or repression effects via trans factors, such as the transcription factor protein (TF) or its complex, may also be important considerations for the optimization of synthetic promoter activity. There are many crucial transcription regulators that maintain typical cellular events for cell development and determination, tissue homeostasis, response to environmental stimuli, and disease processes (Maurano et al., 2012; Andersson et al., 2014; Liu and Stewart, 2015).
Promoters have been classically defined as an activating regulatory element together with an enhancer that interacts with the main promoter to amplify transcription dose (Shlyueva et al., 2014; Beagrie and Pombo, 2016; Haberle and Stark, 2018). With the advancement of modern genomic techniques, it has been discovered that the locations of the enhancer and promoter relative to the TSS, as well as the interaction between these factors, are important factors for transcription regulation (Andersson and Sandelin, 2020). Promoters serve a critical role in establishing baseline transcriptional capacity through the recruitment of proteins, including transcription factors. Because of the complexity of the TF complex and its simultaneous role as an activator and repressor, it is still difficult to explain how and where TFs and their cognizant binding of cis-elements interact for synthetic promoter activity. As high-throughput sequencing continues to advance, it will become easier to elucidate the tissue-specific or cell-specific expression of the TF complex through techniques using single-cell analysis (Dorrity et al., 2021; Marand et al., 2021; Zhang et al., 2021), allowing the use of TF-modulation of synthetic promoters in plant systems (Belcher et al., 2020). The synthetic neurospora-based ‘Q-system’ has previously been applied to enhance synthetic promoters to tune signal specificity and sensitivity in Arabidopsis and potato (Persad et al., 2020; Persad-Russell et al., 2022). Modifying or identifying a similar orthogonal system could help to improve synthetic promoter activity in other species such as poplar.
To date, the functional identification of distal regulators such as enhancers and silencers in the poplar genome has been scarce. Further studies are needed for the identification of enhancers to maximize synthetic promoter activity. In addition, assessing tissue- or cell-specific factors from various promoters can contribute to the generation of multiple functional synthetic promoters based on our current design. To our knowledge, the present study is the first to confirm osmotic stress-responsive synthetic promoter designs in a transgenic woody plant. We expect to expand upon this success and develop more useful techniques for industrial applications, contributing to the use of poplar and other woody plant species as a sustainable feedstock for biofuel applications in marginal areas with salty soil and subject to drought conditions. Utilizing new promoter sequences as outlined in this study is the first step in developing stronger abiotic stress-responsive synthetic promoters for long-lived trees.
Conclusion
To construct valuable stress-responsive synthetic promoters for use in woody plants, we characterized transgenic poplar lines containing the SD promoters consisting of 7 copies of short core sequences. Three synthetic promoters responded positively against osmotic stresses in transgenic poplar. In addition, higher GFP induction by short SDs (~50 bases) containing 7 copies of shorter core sequences than the one with longer (~140 bases) sequences implies that longer sequences are not necessarily required for optimal gene expression. The present study confirmed the stress responsiveness of SDs in transgenic poplar, activities of which were predicted by transient transformation assay following the promoter synthesis from computational analysis.
The present results show a good process from core sequence selection to promoter verification of developing synthetic promoters in woody plants. However, further work is needed to develop optimized synthetic promoters for activation under specific abiotic stresses. Rational design using optimal positioning and ordering of core sequences is necessary. Furthermore, effective components such as enhancers and orthogonal factors are required for the amplification of current synthetic promoters’ activities. In the future, the identified synthetic promoters will be fused with stress-responsive genes and stress-related hormone genes, which can enhance stress tolerance and rapid recovery of cell damage in transgenic wood.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
YY contributed to designing and performing the experiments, analyzing the data, and writing the manuscript. YS, TC, JL, and MP generated transgenic poplar and cared for plants. YS and TC additionally helped with collecting fluorescence measurement data. AA, EB, and CNS conceived of the study and its design and coordination and assisted with interpretation of results and versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by funding from the Biological and Environmental Research in the U.S. Department of Energy Office of Science (DE-SC0018347).
Acknowledgments
We are especially appreciative of our collaboration with Steve DiFazio and his ideas and discussions with him. We also appreciate the support of technical staff and students that contributed to the work in our laboratories.
Conflict of interest
CNS is an inventor on synthetic promoter and biotechnology intellectual property that is assigned to the University of Tennessee Research Foundation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1011939/full#supplementary-material
References
Ali, S., Kim, W. C. (2019). A fruitful decade using synthetic promoters in the improvement of transgenic plants. Front. Plant Sci. 10, 1433. doi: 10.3389/fpls.2019.01433
Andersson, R., Gebhard, C., Miguel-Escalada, I., Hoof, I., Bornholdt, J., Boyd, M., et al. (2014). An atlas of active enhancers across human cell types and tissues. Nature 507 (7493), 455–461. doi: 10.1038/nature12787
Andersson, R., Sandelin, A. (2020). Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 21 (2), 71–87. doi: 10.1038/s41576-019-0173-8
Andres, J., Blomeier, T., Zurbriggen, M. D. (2019). Synthetic switches and regulatory circuits in plants. Plant Physiol. 179 (3), 862–884. doi: 10.1104/pp.18.01362
Arnold, C. D., Zabidi, M. A., Pagani, M., Rath, M., Schernhuber, K., Kazmar, T., et al. (2017). Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat. Biotechnol. 35 (2), 136–144. doi: 10.1038/nbt.3739
Beagrie, R. A., Pombo, A. (2016). Gene activation by metazoan enhancers: Diverse mechanisms stimulate distinct steps of transcription. Bioessays 38 (9), 881–893. doi: 10.1002/bies.201600032
Belcher, M. S., Vuu, K. M., Zhou, A., Mansoori, N., Agosto Ramos, A., Thompson, M. G., et al. (2020). Design of orthogonal regulatory systems for modulating gene expression in plants. Nat. Chem. Biol. 16 (8), 857–865. doi: 10.1038/s41589-020-0547-4
Bryant, N. D., Pu, Y., Tschaplinski, T. J., Tuskan, G. A., Muchero, W., Kalluri, U. C., et al. (2020). Transgenic poplar designed for biofuels. Trends Plant Sci. 25 (9), 881–896. doi: 10.1016/j.tplants.2020.03.008
Cai, Y. M., Kallam, K., Tidd, H., Gendarini, G., Salzman, A., Patron, N. J. (2020). Rational design of minimal synthetic promoters for plants. Nucleic Acids Res. 48 (21), 11845–11856. doi: 10.1093/nar/gkaa682
de Boer, C. G., Vaishnav, E. D., Sadeh, R., Abeyta, E. L., Friedman, N., Regev, A. (2020). Deciphering eukaryotic gene-regulatory logic with 100 million random promoters. Nat. Biotechnol. 38 (1), 56–65. doi: 10.1038/s41587-019-0315-8
De Meester, B., de Vries, L., Ozparpucu, M., Gierlinger, N., Corneillie, S., Pallidis, A., et al. (2018). Vessel-specific reintroduction of CINNAMOYL-COA REDUCTASE1 (CCR1) in dwarfed ccr1 mutants restores vessel and xylary fiber integrity and increases biomass. Plant Physiol. 176 (1), 611–633. doi: 10.1104/pp.17.01462
Dey, N., Sarkar, S., Acharya, S., Maiti, I. B. (2015). Synthetic promoters in planta. Planta 242 (5), 1077–1094. doi: 10.1007/s00425-015-2377-2
Dorrity, M. W., Alexandre, C. M., Hamm, M. O., Vigil, A. L., Fields, S., Queitsch, C., et al. (2021). The regulatory landscape of Arabidopsis thaliana roots at single-cell resolution. Nat. Commun. 12 (1), 3334. doi: 10.1038/s41467-021-23675-y
El-Sappah, A. H., Rather, S. A., Wani, S. H., Elrys, A. S., Bilal, M., Huang, Q., et al. (2022). Heat stress-mediated construaints in maize (Zea mays) production: Challenges and solutions. Front. Plant Sci. 13, 879366. doi: 10.3389/fpls.2022.879366
Endo, H., Yamaguchi, M., Tamura, T., Nakano, Y., Nishikubo, N., Yoneda, A., et al. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56 (2), 242–254. doi: 10.1093/pcp/pcu134
Engler, C., Marillonnet, S. (2014). Golden gate cloning. Methods Mol. Biol. 1116, 119–131. doi: 10.1007/978-1-62703-764-8_9
Ge, H., Li, X., Chen, S., Zhang, M., Liu, Z., Wang, J., et al. (2018). The expression of CARK1 or RCAR11 driven by synthetic promoters increases drought tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 19 (7), 1945. doi: 10.3390/ijms19071945
Haberle, V., Stark, A. (2018). Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 19 (10), 621–637. doi: 10.1038/s41580-018-0028-8
He, M., He, C. Q., Ding, N. Z. (2018). Abiotic stresses: general defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 9, 1771. doi: 10.3389/fpls.2018.01771
Jores, T., Tonnies, J., Dorrity, M. W., Cuperus, J. T., Fields, S., Queitsch, C. (2020). Identification of plant enhancers and their constituent elements by STARR-seq in tobacco leaves. Plant Cell 32 (7), 2120–2131. doi: 10.1105/tpc.20.00155
Jores, T., Tonnies, J., Wrightsman, T., Buckler, E. S., Cuperus, J. T., Fields, S., et al. (2021). Synthetic promoter designs enabled by a comprehensive analysis of plant core promoters. Nat. Plants 7 (6), 842–855. doi: 10.1038/s41477-021-00932-y
Kang, B., Osburn, L., Kopsell, D., Tuskan, G. A., Cheng, Z. (2009). Micropropagation of Populus trichocarpa ‘Nisqually-1’: The genotype deriving the Populus reference genome. Plant Cell Tiss Organ Cult 99, 251–257. doi: 10.0007/s11240-009-9596-9
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17 (3), 287–291. doi: 10.1038/7036
Kotopka, B. J., Smolke, C. D. (2020). Model-driven generation of artificial yeast promoters. Nat. Commun. 11 (1), 2113. doi: 10.1038/s41467-020-15977-4
Lenhard, B., Sandelin, A., Carninci, P. (2012). Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 13 (4), 233–245. doi: 10.1038/nrg3163
Liu, W., Stewart, C. N., Jr. (2015). Plant synthetic biology. Trends Plant Sci. 20 (5), 309–317. doi: 10.1016/j.tplants.2015.02.004
Liu, W., Stewart, C. N., Jr. (2016). Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 37, 36–44. doi: 10.1016/j.copbio.2015.10.001
Li, M., Wang, X., Cao, Y., Liu, X., Lin, Y., Ou, Y., et al. (2013). Strength comparison between cold-inducible promoters of arabidopsis cor15a and cor15b genes in potato and tobacco. Plant Physiol. Biochem. 71, 77–86. doi: 10.1016/j.plaphy.2013.06.021
Lomonossoff, G. P., D'Aoust, M. A. (2016). Plant-produced biopharmaceuticals: A case of technical developments driving clinical deployment. Science 353 (6305), 1237–1240. doi: 10.1126/science.aaf6638
Lubliner, S., Regev, I., Lotan-Pompan, M., Edelheit, S., Weinberger, A., Segal, E. (2015). Core promoter sequence in yeast is a major determinant of expression level. Genome Res. 25 (7), 1008–1017. doi: 10.1101/gr.188193.114
Marand, A. P., Chen, Z., Gallavotti, A., Schmitz, R. J. (2021). A cis-regulatory atlas in maize at single-cell resolution. Cell 184 (11), 3041-3055.e3021. doi: 10.1016/j.cell.2021.04.014
Martinez de Alba, A. E., Elvira-Matelot, E., Vaucheret, H. (2013). Gene silencing in plants: a diversity of pathways. Biochim. Biophys. Acta 1829 (12), 1300–1308. doi: 10.1016/j.bbagrm.2013.10.005
Maurano, M. T., Humbert, R., Rynes, E., Thurman, R. E., Haugen, E., Wang, H., et al. (2012). Systematic localization of common disease-associated variation in regulatory DNA. Science 337 (6099), 1190–1195. doi: 10.1126/science.1222794
Ma, J., Wan, D., Duan, B., Bai, X., Bai, Q., Chen, N., et al. (2019). Genome sequence and genetic transformation of a widely distributed and cultivated poplar. Plant Biotechnol. J. 17 (2), 451–460. doi: 10.1111/pbi.12989
Merchante, C., Stepanova, A. N., Alonso, J. M. (2017). Translation regulation in plants: an interesting past, an exciting present and a promising future. Plant J. 90 (4), 628–653. doi: 10.1111/tpj.13520
Morton, T., Petricka, J., Corcoran, D. L., Li, S., Winter, C. M., Carda, A., et al. (2014). Paired-end analysis of transcription start sites in Arabidopsis reveals plant-specific promoter signatures. Plant Cell 26 (7), 2746–2760. doi: 10.1105/tpc.114.125617
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6, 288. doi: 10.3389/fpls.2015.00288
Persad, R., Reuter, D. N., Dice, L. T., Nguyen, M. A., Rigoulot, S. B., Layton, J. S., et al. (2020). The q-system as a synthetic transcriptional regulator in plants. Front. Plant Sci. 11, 245. doi: 10.3389/fpls.2020.00245
Persad-Russell, R., Mazarei, M., Schimel, T. M., Howe, L., Schmid, M. J., Kakeshpour, T., et al. (2022). Specific bacterial pathogen phytosensing is enabled by a synthetic promoter-transcription factor system in potato. Front. Plant Sci. 13, 873480. doi: 10.3389/fpls.2022.873480
Pfotenhauer, A. C., Occhialini., A., Nguyen, M. A., Scott, H., Dice, L. T., Harbison, S. A., et al. (2022). Building the plant synbio toolbox through combinatorial analysis of DNA regulatory elements. ACS Synthetic Biol. 11, 2741–2755. doi: 10.1021/acssynbio.2c00147
R core team (2014). R: A language and environment for statistical computing. r foundation for statistical computing (Vienna, Austria). Available at: http://www.R-project.org/.
Rigoulot, S. B., Schimel, T. M., Lee, J. H., Sears, R. G., Brabazon, H., Layton, J. S., et al. (2021). Imaging of multiple fluorescent proteins in canopies enables synthetic biology in plants. Plant Biotechnol. J. 19 (4), 830–843. doi: 10.1111/pbi.13510
RoyChoudhury, A., Roy, C., Sengupta, D. N. (2007). Transgenic tobacco plants overexpressing the heterologous lea gene Rab16A from rice during high salt and water deficit display enhanced tolerance to salinity stress. Plant Cell Rep. 26 (10), 1839–1859. doi: 10.1007/s00299-007-0371-2
Sainsbury, F. (2020). Innovation in plant-based transient protein expression for infectious disease prevention and preparedness. Curr. Opin. Biotechnol. 61, 110–115. doi: 10.1016/j.copbio.2019.11.002
Schneider, C. A., Rasband, W. S., Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 (7), 671–675. doi: 10.1038/nmeth.2089
Shlyueva, D., Stampfel, G., Stark, A. (2014). Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15 (4), 272–286. doi: 10.1038/nrg3682
Singhal, P., Jan, A. T., Azam, M., Haq, Q. M. R. (2016). Plant abiotic stress: a prospective strategy of exploiting promoters as alternative to overcome the escalating burden. Front. Life Sci. 9 (1), 52–63. doi: 10.1080/21553769.2015.1077478
Song, J., Lu, S., Chen, Z. Z., Lourenco, R., Chiang, V. L. (2006). Genetic transformation of Populus trichocarpa genotype nisqually-1: a functional genomic tool for woody plants. Plant Cell Physiol. 47 (11), 1582–1589. doi: 10.1093/pcp/pcl018
Sparkes, I. A., Runions, J., Kearns, A., Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1 (4), 2019–2025. doi: 10.1038/nprot.2006.286
Srivastava, R., Rai, K. M., Srivastava, M., Kumar, V., Pandey, B., Singh, S. P., et al. (2014). Distinct role of core promoter architecture in regulation of light-mediated responses in plant genes. Mol. Plant 7 (4), 626–641. doi: 10.1093/mp/sst146
Swiezewski, S., Liu, F., Magusin, A., Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an arabidopsis polycomb target. Nature 462 (7274), 799–802. doi: 10.1038/nature08618
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, populus trichocarpa (Torr. & Gray). Science 313 (5793), 1596–1604. doi: 10.1126/science.1128691
van Arensbergen, J., FitzPatrick, V. D., de Haas, M., Pagie, L., Sluimer, J., Bussemaker, H. J., et al. (2017). Genome-wide mapping of autonomous promoter activity in human cells. Nat. Biotechnol. 35 (2), 145–153. doi: 10.1038/nbt.3754
Van Lijsebettens, M., Grasser, K. D. (2014). Transcript elongation factors: shaping transcriptomes after transcript initiation. Trends Plant Sci. 19 (11), 717–726. doi: 10.1016/j.tplants.2014.07.002
Vazquez-Vilar, M., Orzaez, D., Patron, N. (2018). DNA Assembly standards: Setting the low-level programming code for plant biotechnology. Plant Sci. 273, 33–41. doi: 10.1016/j.plantsci.2018.02.024
Weingarten-Gabbay, S., Nir, R., Lubliner, S., Sharon, E., Kalma, Y., Weinberger, A., et al. (2019). Systematic interrogation of human promoters. Genome Res. 29 (2), 171–183. doi: 10.1101/gr.236075.118
Yamamoto, Y. Y., Ichida, H., Abe, T., Suzuki, Y., Sugano, S., Obokata, J. (2007). Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res. 35 (18), 6219–6226. doi: 10.1093/nar/gkm685
Yang, Y., Chaffin, T. A., Ahkami, A. H., Blumwald, E., Stewart, C. N., Jr. (2022). Plant synthetic biology innovations for biofuels and bioproducts. Trends Biotechnol. (in press). doi: 10.1016/j.tibtech.2022.09.007
Yang, Y., Lee, J. H., Poindexter, M. R., Shao, Y., Liu, W., Lenaghan, S. C., et al. (2021). Rational design and testing of abiotic stress-inducible synthetic promoters from poplar cis-regulatory elements. Plant Biotechnol. J. 19 (7), 1354–1369. doi: 10.1111/pbi.13550
Yu, H., Liu, C., Dixon, R. A. (2021). A gene-editing/complementation strategy for tissue-specific lignin reduction while preserving biomass yield. Biotechnol. Biofuels 14 (1), 175. doi: 10.1186/s13068-021-02026-5
Zhang, T. Q., Chen, Y., Liu, Y., Lin, W. H., Wang, J. W. (2021). Single-cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root. Nat. Commun. 12 (1), 2053. doi: 10.1038/s41467-021-22352-4
Keywords: Synthetic biology, synthetic promoter, osmotic stress, poplar, cis-regulatory elements, bioenergy
Citation: Yang Y, Shao Y, Chaffin TA, Lee JH, Poindexter MR, Ahkami AH, Blumwald E and Stewart CN Jr (2022) Performance of abiotic stress-inducible synthetic promoters in genetically engineered hybrid poplar (Populus tremula × Populus alba). Front. Plant Sci. 13:1011939. doi: 10.3389/fpls.2022.1011939
Received: 04 August 2022; Accepted: 28 September 2022;
Published: 18 October 2022.
Edited by:
Ahmad Arzani, Isfahan University of Technology, IranReviewed by:
Ahmed H. El-Sappah, Zagazig University, EgyptHan Xiao, Center for Excellence in Molecular Plant Sciences (CAS), China
Copyright © 2022 Yang, Shao, Chaffin, Lee, Poindexter, Ahkami, Blumwald and Stewart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: C. Neal Stewart Jr, bmVhbHN0ZXdhcnRAdXRrLmVkdQ==
 Yongil Yang
Yongil Yang Yuanhua Shao
Yuanhua Shao Timothy A. Chaffin1,2
Timothy A. Chaffin1,2 Jun Hyung Lee
Jun Hyung Lee Magen R. Poindexter
Magen R. Poindexter Amir H. Ahkami
Amir H. Ahkami Eduardo Blumwald
Eduardo Blumwald