
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 09 September 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1009575
This article is part of the Research TopicApplication of Multi-Omics to Important Traits of Ornamental and Beverage PlantsView all 20 articles
 Man Zhang1†
Man Zhang1† Jianshuang Shen2†
Jianshuang Shen2† Yutong Wu1†
Yutong Wu1† Xiaolu Zhang1
Xiaolu Zhang1 Zhengtian Zhao1
Zhengtian Zhao1 Jia Wang1
Jia Wang1 Tangren Cheng1
Tangren Cheng1 Qixiang Zhang1
Qixiang Zhang1 Huitang Pan1*
Huitang Pan1*Leaf color is one of the most important features for plants used for landscape and ornamental purposes. However, the regulatory mechanism of yellow leaf coloration still remains elusive in many plant species. To understand the complex genetic mechanism of yellow-leaf Forsythia, we first compared the pigment content and leaf anatomical structure of yellow-leaf and green-leaf accessions derived from a hybrid population. The physiological and cytological analyses demonstrated that yellow-leaf progenies were chlorophyll deficient with defected chloroplast structure. With comparative transcriptome analysis, we identified a number of candidate genes differentially expressed between yellow-leaf and green-leaf Forsythia plants. Among these genes, we further screened out two candidates, ChlH (magnesium chelatase Subunit H) and POLGAMMA2 (POLYMERASE GAMMA 2), with consistent relative-expression pattern between different colored plants. To verify the gene function, we performed virus-induced gene silencing assays and observed yellow-leaf phenotype with total chlorophyll content reduced by approximately 66 and 83% in ChlH-silenced and POLGAMMA2-silenced plants, respectively. We also observed defected chloroplast structure in both ChlH-silenced and POLGAMMA2-silenced Forsythia. Transient over-expression of ChlH and POLGAMMA2 led to increased chlorophyll content and restored thylakoid architecture in yellow-leaf Forsythia. With transcriptome sequencing, we detected a number of genes related to chlorophyll biosynthesis and chloroplast development that were responsive to the silencing of ChlH and POLGAMMA2. To summarize, ChlH and POLGAMMA2 are two key genes that possibly related to yellow-leaf coloration in Forsythia through modulating chlorophyll synthesis and chloroplast ultrastructure. Our study provided insights into the molecular aspects of yellow-leaf Forsythia and expanded the knowledge of foliage color regulation in woody ornamental plants.
The genus of Forsythia belongs to the family of Oleaceae, and is consisted of many deciduous species that are commonly used ornamental plants (Shen et al., 2019). Forsythia species are prized for their arching branches, golden blossoms, and early flowering features and are widely cultivated worldwide. The fruits of Forsythia were often used for medical treatment and herbal drug industries (Guo et al., 2007; Piao et al., 2008). During the cultivation history of Forsythia, many morphologically diverged cultivars were evolved and selected. Forsythia koreana “Suwon Gold” is a yellow-leaf cultivar, forming magnificent golden canopy throughout growing seasons (Wang et al., 2017). Comparing with green-leaf cultivars, F. koreana “Suwon Gold” is less vigorous and its leaves are more susceptible to sun burn during summer (Wang et al., 2017). Though great effort has been made to breed for colored leaf cultivars, the mechanism controlling yellow-leaf coloration in Forsythia remains unknown.
Leaf coloration is an important ornamental aspect for plants used in landscape applications. Various leaf color can enhance the ornamental effect of plant species and compensate for the monotonous color during growing period (Dong et al., 2020). Different leaf color often reflects the composition and content of major pigments, including chlorophyll, carotenoids, and flavonoids in the mesophyll cells (Sinkkonen et al., 2012; Yang et al., 2015). For example, the redness of red maple leaves is a result of low chlorophyll content and accumulated cyanidin (Chen et al., 2019). Chlorophyll and carotenoids are two major pigments that determine the leaf yellowing in plants (Luo et al., 2019; Mo-zhen et al., 2020). Previous studies have characterized more than twenty enzymatic steps required for chlorophyll biosynthesis (Sobotka, 2013). The first step is the insertion of Mg2+ into protoporphyrin IX (PP IX) to form Mg-PP IX, which is catalyzed by Mg-chelatase (Terry et al., 2002). Mg-chelatase is a protein complex that comprised of subunits encoded by ChlI (magnesium chelatase Subunit I), ChlD (magnesium chelatase Subunit D), and ChlH (magnesium chelatase Subunit H) (Wang and Grimm, 2015). Apart from catalyzing the chlorophyll branch, ChlH also participates in plastid-nuclear signaling pathway that regulate the expression of photosynthesis-related nuclear genes in Arabidopsis (Mochizuki et al., 2001). Mutations in ChlH gene led to underdeveloped thylakoid membrane and low chlorophyll content in rice (Jung et al., 2003). DVR encodes the 3,8-divinyl protochlorophyllide a 8-vinyl reductase that catalyzing another indispensable step for chlorophyll biosynthesis (Pogson et al., 2015). DVR mutants exhibited yellow-green phenotype with reduced chlorophyll content, arrested chloroplast development, and attenuated photosynthesis activity in rice (Rodríguez et al., 2016). Mutation in genes involved in chlorophyll degradation can also produce leaf color mutants (Zhao et al., 2020). For example, a yellow-leaf mutant of Cymbidium sinense is caused by the up-regulation of two enzymes, CLH2 (CHLOROPHYLLASE 2) and RCCR (red chlorophyll catabolite reductase), which promote the breakdown of chlorophyll a (Amancio et al., 2015).
On the other hand, abnormal chloroplast development can also affect the chlorophyll accumulation and cause yellow leaf phenotype (Zhao et al., 2020). For example, the Arabidopsis “cue” mutants, exhibiting yellow-green and pale-green leaves, have delayed chloroplast differentiation and impaired chloroplast structure (Loìpez-Juez et al., 1998). Chloroplast development is regulated by the coordination of nuclear and chloroplast genes that mediate transcription and translation, thylakoid formation, pigment synthesis, plastid-nuclear signaling transduction, and chloroplast division (Pogson and Albrecht, 2011; Zhao et al., 2020). Any mutation in these genes may impair the chloroplast development and functioning (Kessler and Schnell, 2009). A genetic screen in yellow-leaf soybean detected a point mutation in YL (YELLOW LEAF), a chloroplast localized gene homologous to the Arabidopsis ORRM1 (ORGANELLE RRM PROTEIN 1), which regulates chloroplast RNA-editing and photosynthesis (Zhu et al., 2020). In Arabidopsis, the mutation in SCO1 (SNOWY COTYLEDON 1), which encodes the chloroplast elongation factor G, led to cotyledon albinism due to improperly developed chloroplasts (Ruppel and Hangarter, 2007). Despite the research advancements in leaf color regulation, the mechanism underlying leaf yellowing may differ across different plant species.
To understand the physiological basis of yellow leaf-color in Forsythia, we compared the pigment and anatomical characteristics of yellow-leaf and green-leaf accessions from a F1 hybrid population generated in a previous study (Wang et al., 2017). Subsequently, we performed comparative transcriptome analysis and obtained a number of differentially expressed genes that possibly related to the leaf color difference between yellow-leaf and green-leaf Forsythia. Among the candidate genes, we screened out two genes, ChlH and POLGAMMA2, with consistent expression patterns among different leaf-color progeny lines regardless of light-intensities. We further verified their functions with virus-induced gene silencing and transient over-expression technique in Forsythia and other model plant species. By analyzing the transcriptome change after gene silencing, we explored the possible genetic mechanism of ChlH and POLGAMMA2 in regulating leaf-color in Forsythia. Our study revealed the physiological and transcriptional characteristics of yellow-leaf Forsythia, and provided valuable information for future breeding of colorful foliage plants.
To reduce the genetical difference between two Forsythia species, we hybridized F. koreana “Suwon Gold” (♂) with F. “Courtaneur” (♀) and generated progenies with clear leaf-color variations (from green to yellow) in a previous study (Wang et al., 2017). All hybrids were planted in the nursery at National Engineering Research Center for Floriculture (40°02′ N, 115°50′ E) in Beijing, China. We randomly selected thirty progeny strains of yellow-leaf and green-leaf plants and propagated ten clones for each strain from stem cuttings. To study the effect of light intensity on leaf-coloration, we selected ten green-leaf (L1 group) and yellow-leaf (L2 group) lines with three biological replicates for each group, and cultivated them under the natural light conditions (daily light intensity range from 1,300 to 1,500 μmol⋅m–2⋅s–1). Similarly, we selected ten green-leaf (S1 group) and ten yellow-leaf accessions (S2 group) for shade treatment (daily light intensity 390–450 μmol⋅m–2⋅s–1). These two light-intensity treatments were performed from May 3, 2017 to June 3, 2017 with the same environmental temperatures. Mature leaves (the third and fourth leaves from the top) of new branches in each Forsythia plant were sampled for further analysis.
To characterize the four treatment groups, we measured the color of mature leaves for plants in L1, L2, S1, and S2 group with the fifth edition Royal Horticultural Society Color Chart, respectively. Approximately 150 mg fresh leaves were collected for each sample and immersed into 20 ml of 80% acetone (v/v) at room temperature in the dark for 24 h. The absorbance of supernatant was measured for each group at 470 nm, 646, and 663 nm using BioMate 3S UV-visible spectrophotometer (Thermo Fisher Scientific, Shanghai, China) with three technical replicates. The chlorophyll (Chl a, Chl b, and total Chl) and carotenoid (Car) contents were determined according to Lichtenthaler (1987). The Chl/Car ratio was calculated as the total chlorophyll content to total carotenoids content.
Mature leaves were selected and fixed in FAA solution (acetic acid: 70% alcohol = 1:3). Cross sections (12 mm thick) were sliced using a microtome, stained with 50% water-soluble safranin and fast green, and mounted in gelatin-glycerine. For each leaf sample, five to ten sections were randomly selected for observation under microscope ZEISS Scope A1. The leaf thickness, thickness of upper epidermis, lower epidermis, palisade, and spongy parenchyma were measured using Image-Pro Plus 6.0. The CTR (cell tense ratio) and SR (spongy ratio) values were also calculated for three sections of three progeny lines randomly selected for each color group (He et al., 2017).
For transmission electron microscopy (TEM) analyses, mature leaves of each group were sampled and cut into 2 mm × 4 mm pieces fixed with 3% glutaraldehyde solution (glutaraldehyde solution is prepared with 0.1 mol/L phosphate buffer, pH 7.2) at 4°C for at least 2 hours. The samples were then rinsed with phosphate buffer for 3–5 times and fixed in 1% osmium tetroxide solution at 4°C overnight, followed by dehydration using gradient ethanol solutions (50, 70, 95, and 100% alcohol). Samples were embedded in epoxy resin and sliced with LEICA DC61 ultra-thin microtome (Leica Microsystems, Germany). The ultrathin sections were examined under JEM-1010 electron microscope (JEOL, Tokyo, Japan). For statistical analysis, we conducted at least three independent measurements for each parameter in ten mesophyll cells of three randomly selected leaves for each group. The leaf anatomical features and chloroplast structure were also examined and compared for the newly sprouted leaves of plants before and after transient silencing and over-expression of candidate genes.
Leaves were collected from ten randomly selected progeny lines of each color group (L1, S1, L2, S2) with three biological replicates. Total RNA was extracted and the RNA integrity was assessed using Agilent 2100 bioanalyzer. A total of twelve sequencing libraries were prepared and sequenced on Illumina Hiseq 2500 platform (Novogene Bioinformatics Technology Co., Ltd., Beijing, China). Raw reads were deposited in NCBI SRA archive as BioProject PRJNA437859 (SRA accession: SRS3039871-SRS3039882).
Raw reads were cleaned by filtering adaptor and low quality sequences with fastp v0.19.7 (Chen et al., 2018). We mapped all clean reads to the reference genome of F. suspensa (Li et al., 2020) using HISAT (Kim et al., 2015) and sorted the alignment with Samtools (Li et al., 2009). Transcript abundance level was estimated with Stringtie (Li and Dewey, 2011) and was normalized to FPKM value (Fragment Per kilo bases per Million reads). Differential expression analysis was performed on four groups using DEGseq R package (Wang et al., 2010). We defined genes with adjusted p-value ≤ 0.05 and fold change ≥2.0 as differentially expressed genes (DEGs). GO and KEGG enrichment analyses were performed on the DEGs using the “phyper” function in R. GO terms and KEGG pathways with FDR < 0.05 were considered as significantly enriched.
To determine the transcriptional profile established by the silencing of FsChlH and FsPOLGAMMA2, we performed RNA-seq on leaves of green-leaf Forsythia before and after ChlH and POLGAMMA2 were transiently silenced, respectively. The procedure of sampling, RNA extraction, library construction, and sequencing were the same as described above. The raw sequence data generated correspondingly were deposited in NCBI SRA archive under BioProject PRJNA756703 (SRA accession: SRR15559813-SRR15559824). Differential expression analysis was conducted by comparing the FsChlH-silenced (CY) and FsPOLGAMMA2-silenced (PY) plants with their green-leaf vector controls (CG and PG). The DEGs were defined as genes with adjusted p-value ≤ 0.05 and fold change ≥2.0 in the differential expression analysis.
To validate the results of RNA-seq and expression pattern of candidate genes, sample RNA was extracted with Plant RNA Kit (Omega Bio-Tek, United States) and was reverse transcribed using the Prime Script RT reagent Kit with gDNA Eraser (Takara Bio, Japan) following the manufacturer’s protocols. Quantitative real-time RT-PCR was performed using TB Green® Premix Ex Taq II (Takara Bio, Japan) on CFX Connect Real-Time System (Bio-Rad Laboratories, Inc., United States) following the temperature setting: 30 s at 94°C, 40 cycles of 94°C for 5 s, 60°C for 30 s, and 72°C for 30 s. Relative gene expression was performed using the 2–ΔΔCt method with CYCLOPHILIN gene used as reference gene (Shen et al., 2019). The primers of selected DEGs for qRT-PCR assays were designed with Primer Premier 5 software (Premier, United States) and were provided (Supplementary Table 1). Each qRT-PCR experiment was performed with three biological replicates and three technical replicates. qRT-PCR experiments were also performed to evaluate the candidate gene expression in plants with target genes transiently silenced or over-expressed. For model plant systems, ACTIN genes were used as internal reference for tomato (Chang et al., 2018) and tobacco (Aziz et al., 2019) in the qRT-PCR analysis.
Full length sequence of FsChlH and FsPOLGAMMA2 were cloned and were inserted into the pSuper1300 vector to generate the pSuper1300-FsChlH and pSuper1300-FsPOLGAMMA2 for gene over-expression. Gene-specific fragments of FsChlH and FsPOLGAMMA2 between 300 and 500 bp were cloned and transferred into the pTRV2 vector, which encodes the RNA2 genome of TRV (Tobacco Rattle Virus) (Fu et al., 2005). Primers used for preparing the expression vectors above were provided (Supplementary Table 2). All constructed vectors were sequencing confirmed by Sangon Biotech Inc. (Shanghai, China) and were transformed into the Agrobacterium GV3101 strain cells.
For transient over-expression assay, the transformed agrobacteria with pSuper1300-FsChlH and pSuper1300-FsPOLGAMMA2 were used to infect yellow-leaf plants following previously reported protocols (Yamamoto et al., 2018). With leaf veins as the boundary, we infected only one side of the leaf, leaving the other side as the control. For VIGS assay, the green-leaf Forsythia plants were infiltrated with Agrobacterium strains containing the mixture of pTRV2 and pTRV2-FsChlH, pTRV2, and pTRV2-FsPOLGAMMA2, with empty-loaded pTRV2 as the mock based on previously established protocol (Shen et al., 2021). For each treatment, six individual plants were infected, kept in dark for 24 h, and were maintained in climatic chamber [light intensity 300 μmol⋅m–2⋅s–1; day length: 16 h (22°C)/8 h (18°C); relative humidity 60%]. Similarly, VIGS studies were performed in tomato (Solanum lycopersicum) and tobacco (Nicotiana benthamiana) following the standard protocols (Senthil-Kumar and Mysore, 2011). The plants with transient silencing or over-expression of target genes were examined for leaf pigment content and chloroplast structure following same protocols described above.
All data were analyzed using SPSS software v.17.0 and the statistical significances of leaf pigment contents, anatomical features, and gene expression levels were assessed using one-way ANOVA followed by multiple comparisons using Duncan’s Multiple Range Test (DMRT).
The yellow-leaf progenies with yellowish leaf (YG11A-YGGN144A) under natural light conditions convert to yellow-green (YGG144A-C) when transferred to low light-intensity condition (Figure 1A). While the virescent leaves of green-leaf lines (YGG146A-B) converted to green (GG137A-GGN137C) after shade treatment (Figure 1A). The pigment content analysis revealed that Chl a, Chl b, total Chl content, and carotenoid of yellow-leaf lines under natural light conditions (L2) were 0.12, 0.26, 0.38, and 0.29 mg/g, respectively (Table 1). In contrast, the Chl a, b, and carotenoid content in green-leaf lines (L1) were 0.77, 0.29, and 0.61 mg/g, respectively (Table 1). After the shading treatment, chlorophyll and carotenoid contents were both significantly increased in green-leaf and yellow-leaf plants (Table 1). The Chl/Car ratio in yellow-leaf plants after shading treatment is significantly higher than that of the yellow-leaf plants under normal daylight condition (Table 1). However, there is no significant differences in Chl/Car ratio between yellow-leaf and green-leaf plants under natural light conditions (Table 1). We observed significantly higher level of Chl a in green-leaf lines than that of yellow-leaf lines in both natural light and shade conditions. Moreover, no significant difference was observed in Chl b content among the four treatment groups (Table 1). Therefore, the accumulation of Chl a content is likely one major cause for the leaf-color difference between green-leaf and yellow-leaf Forsythia lines.
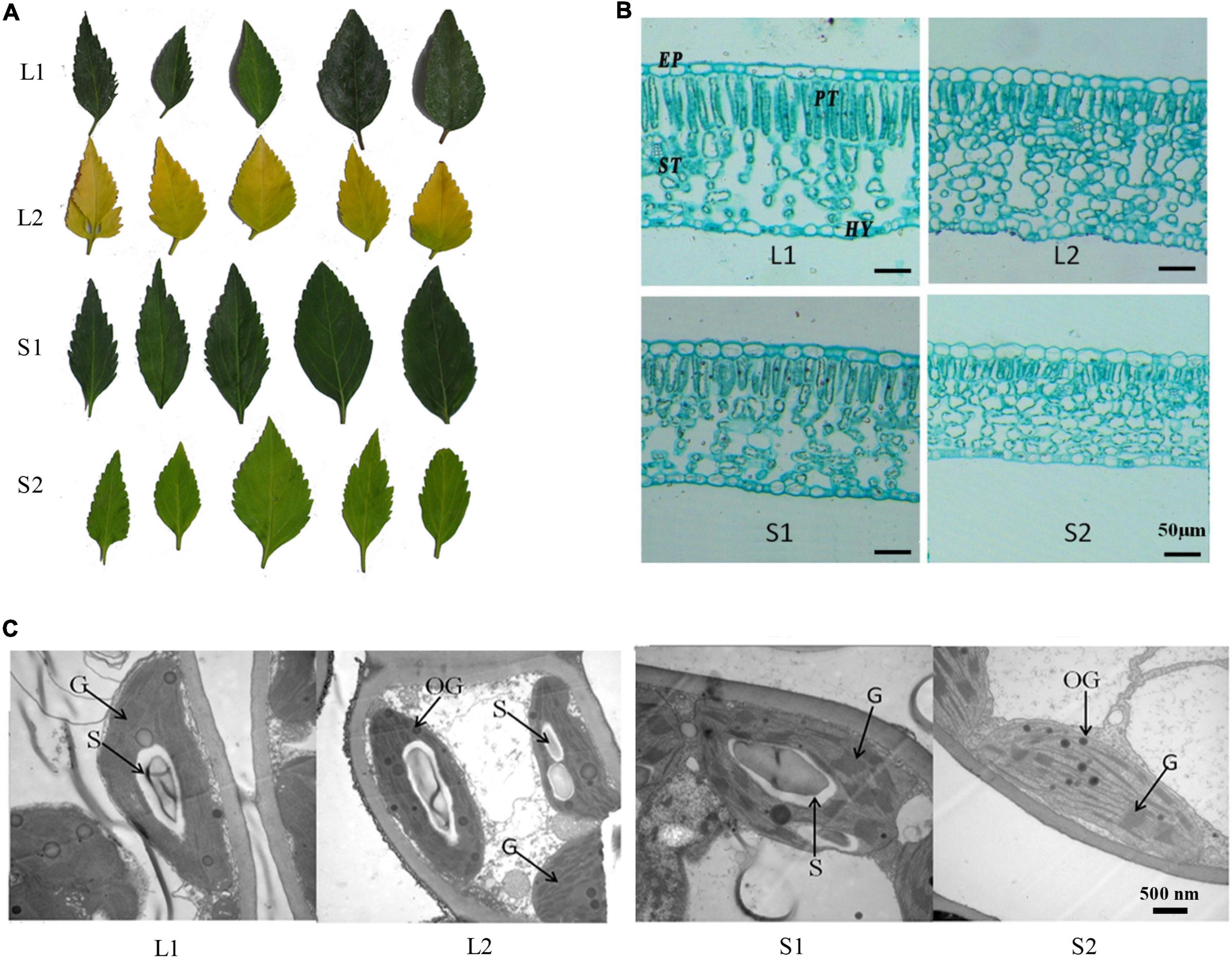
Figure 1. Phenotypic analysis of yellow-leaf and green-leaf Forsythia plants under different light-intensity treatments. (A) Leaf color of four groups (L1: green-leaf Forsythia under natural light condition; L2: yellow-leaf Forsythia under natural light condition; S1: green-leaf Forsythia under shade condition; S2: yellow-leaf Forsythia under shade condition). (B) Leaf anatomy of plants in four groups. EP refers to epidermis; PT refers to palisade tissue; ST refers to spongy tissue; HY refers to Hypodermis. (C) Chloroplast ultrastructure of plants in four groups. G refers to granum; S refers to starch grain; OG refers to osmiophilic globule.
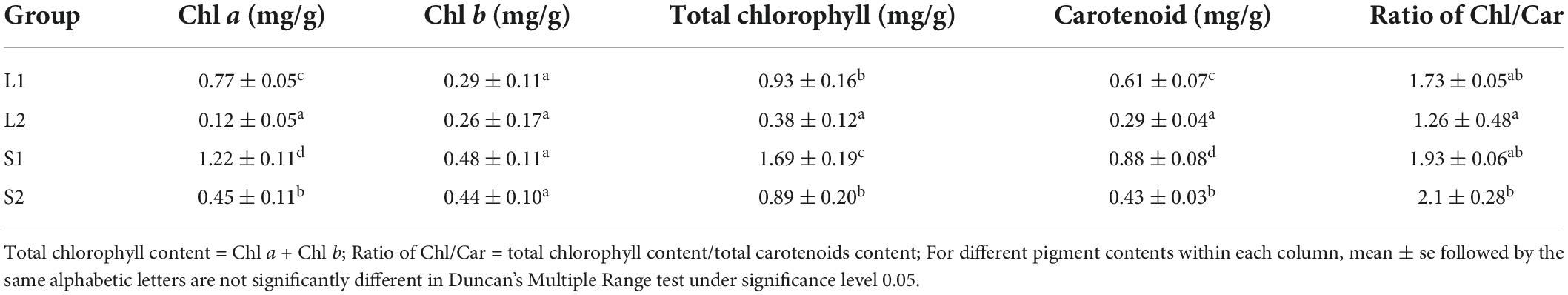
Table 1. The photosynthetic pigment content in green-leaf and yellow-leaf plants under different light intensities.
To compare the leaf anatomical features in four treatment groups, we measured the thickness of four tissue types: epidermis (EP), palisade tissue (PT), sponge tissue (ST), hypoderm (HY). The thickness of EP, PT, and CTR (Palisade thickness/Leaf thickness) of yellow-leaf Forsythia plants (L2 and S2) were significantly lower than those of green-leaf Forsythia plants (L1 and S1) (Supplementary Table 3 and Figure 1B). After shade treatment, the sponge tissue thickness in both yellow-leaf (L2) and green-leaf Forsythia plants (L1) decreased dramatically (Supplementary Table 3 and Figure 1B). By examining the chloroplast structure with TEM, we observed abnormal chloroplast structure in yellow-leaf plants (Figure 1C). Under natural light conditions, the chloroplasts of the green-leaf Forsythia plants (L1) were well-developed with normally stacked grana (Figure 1C). Each chloroplast contains 1–2 huge starch grains and a few plastoglobules. However, yellow-leaf Forsythia plants (L2) contained chloroplasts lacking grana with only abnormally arranged lamellar structures (Figure 1C and Supplementary Table 4). After shading treatment, the chloroplasts of green-leaf plants (S1) remain normal, while a few loosely packed thylakoids with few layers were visible in the chloroplasts of yellow-leaf (S2) Forsythia plants (Figure 1C and Supplementary Table 4). Moreover, the number of osmiophilic globules was significantly increased in yellow-leaf Forsythia plants than that of green-leaf lines in shade condition (Figure 1C and Supplementary Table 4). These results suggested that the chloroplast development and chlorophyll biosynthesis were disrupted in the yellow-leaf Forsythia plants and these defects cannot be fully recovered in low irradiance condition.
To understand the molecular mechanism of yellow leaf coloration in Forsythia, we performed RNA-seq on leaves of yellow-leaf and green-leaf plants. After trimming adapters and filtering out low-quality reads, we obtained a total of 120.22 Gb clean reads generated from twelve sequencing libraries (Supplementary Table 5). With differential expression analysis, we identified a total of 1820 DEGs between yellow-leaf and green-leaf Forsythia lines under natural light conditions and 346 DEGs after shade treatment (Supplementary Figure 1). By comparing the DEG sets from high and low light-intensity treatments, we obtained 157 common DEGs. To validate the accuracy of RNA-seq results, we performed qRT-PCR analysis and observed that the relative gene expression pattern of the selected DEGs is in agreement with that in transcriptome data (Supplementary Figure 2). Among the DEGs identified in normal light condition, 1011 DEGs were up-regulated and 809 DEGs were down-regulated. Functional enrichment analysis revealed that these DEGs were significantly enriched in oxidation-reduction process (GO:0055114), carbohydrate metabolic process (GO:0005975), and DNA repair (GO:0006281) (Supplementary Table 6). Among the DEGs identified between yellow-leaf and green-leaf lines after shade treatment, we identified 127 up-regulated DEGs and 219 down-regulated DEGs. These DEGs were found mainly involved in biological processes, such as nucleic acid-templated transcription (GO:0097659), galactose metabolic process (GO:0006012), and oxidation-reduction process (GO:0055114) (Supplementary Figure 1 and Supplementary Table 6).
To screen for candidate genes associated with yellow-leaf phenotype, we mainly focused on analyzing the expression profiles of DEGs involved in pigment biosynthesis and chloroplast development and only considered those with consistent relative expression pattern among different leaf-color plants for downstream functional verification. In total, we identified 19 DEGs related to chlorophyll biosynthesis and three DEGs related to carotenoid biosynthesis (Supplementary Table 7). We observed the down-regulation of ChlH, CRDs (COPPER RESPONSE DEFECT 1), DVR, and HEMA1 (glutamyl-tRNA reductase) in the yellow-leaf plants comparing to green-leaf lines, which is consistent with the low chlorophyll content in yellow-leaf plants under natural light conditions (Figure 2A). In contrast, a few genes promoting the biosynthesis of Chl a, such as ChlM (magnesium protoporphyrin IX methyltransferase), CHlI (magnesium chelatase subunit I), GSA1 (GLUTAMATE-1-SEMIALDEHYDE-2,1-AMINOMUTASE), and HEMBs-HEMGs displayed higher expression level in yellow-leaf plants (Figure 2). CAO2 (CHLOROPHYLL A OXYGENASE), the key enzyme involved in Chl b biosynthesis, also showed lower expression level in yellow-leaf plants (Figure 2). We also observed a number of DEGs displaying contrasting expression patterns between yellow-leaf and green-leaf plants after shade treatment (Figure 2A). For example, the transcription levels of CAO1-2, CRD1-2, DVR, HEMA1, and PORA1-3 (PROTOCHLOROPHYLLIDE OXIDOREDUCTASE A) were low in yellow-leaf plants, but were strongly up-regulated under low light-intensity condition (Figure 2A). Among all the DEGs related chlorophyll biosynthesis, ChlH were consistently down-regulated in yellow-leaf lines regardless of the light-intensity condition (Figure 2A). Thus, we considered ChlH as one of the key candidate genes for the following functional analysis.
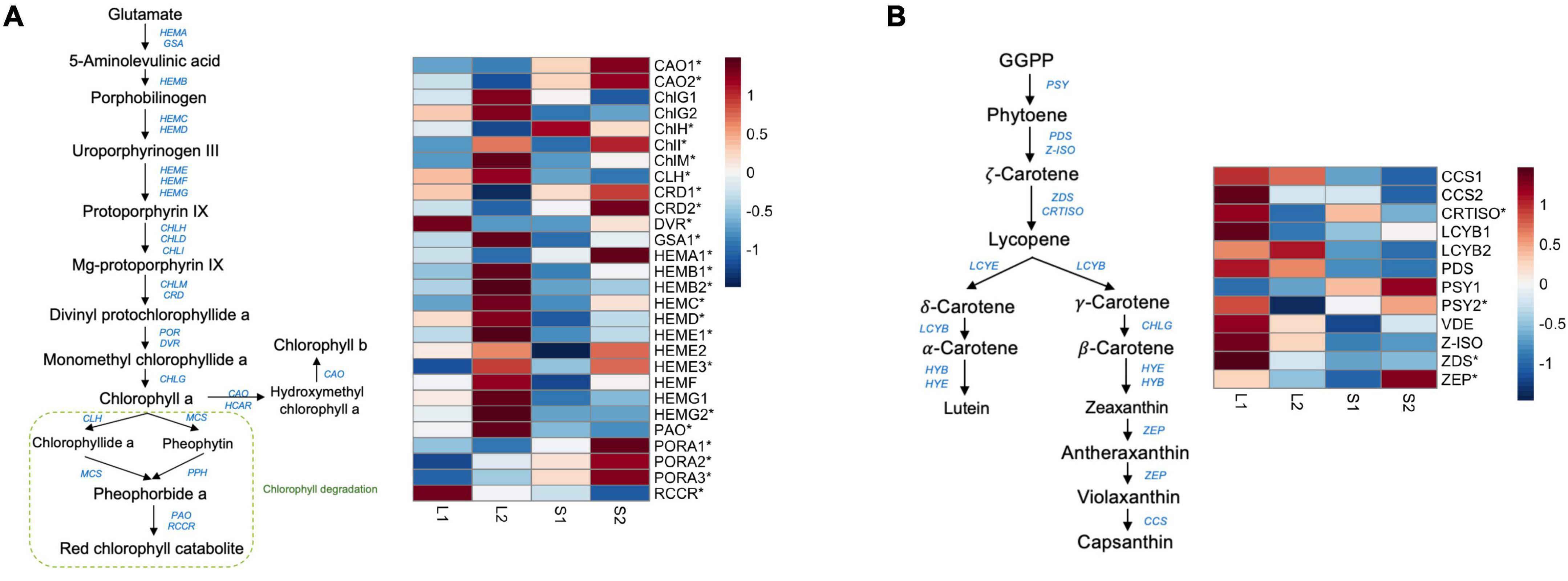
Figure 2. The expression profile of genes involved in chlorophyll and carotenoid biosynthesis. (A) The candidate genes in the chlorophyll biosynthetic pathway and their expression profiles across four treatment groups; (B) the candidate genes in the carotenoid biosynthetic pathway and their expression profiles across four treatment groups. The star symbols highlight the DEGs in either group comparisons.
We also detected four DEGs that were involved in carotenoid synthesis (Supplementary Table 7). These include PSY2 (PHYTOENE SYNTHASE), CRTISO (CAROTENOID ISOMERASE), ZEP (ZEAXANTHIN EPOXIDASE), and ZDS (ZETA-CAROTENE DESATURASE). All four genes were relatively down-regulated in yellow-leaf lines under natural light conditions (Figure 2B). However, the expression of PSY2 and ZEP was significantly induced in shade condition (Figure 2B). ZDS, on the other hand, remained lowly expressed in both green-leaf and yellow-leaf Forsythia plants after shade treatment (Figure 2B). CRTISO was consistently down-regulated in yellow-leaf plants under both natural-light and shade conditions (Figure 2B). Since the content level of carotenoids is not the major reason for yellow leaf-color, we excluded the candidate genes related to carotenoid biosynthesis from further functional investigations.
Furthermore, we examined the expression pattern of DEGs that are related to photosynthesis, chlorophyll binding and chloroplast development. Among them, three genes were annotated to lipid metabolism, including GLTP3 (GLYCOLIPID TRANSFER PROTEIN 3), LTPL101 (lipid-transfer family protein), and GDPD3 (GLYCEROPHOSPHODIESTER PHOSPHODIESTERASE 3). The expression levels of GLTP3 and GDPD3 were significantly higher in yellow-leaf plants, but were then dropped in shading condition (Supplementary Figure 3). The expression of LTPL101 was low in natural light conditions, and were induced in both green and yellow plants after shading treatment (Supplementary Figure 3). We observed the transcriptional pattern change of photosynthesis-related DEGs as the irradiance change. For example, PsaAs (PHOTOSYSTEM I SUBUNIT A), PsbAs (PHOTOSYSTEM II REACTION CENTER PROTEIN A), and PsbBs (PHOTOSYSTEM II REACTION CENTER PROTEIN B) were up-regulated in yellow-leaf plants (Supplementary Figure 3). PsaHs (PHOTOSYSTEM I SUBUNIT H2), PsbYs (PHOTOSYSTEM II BY), and PsbP2 (PHOTOSYSTEM II SUBUNIT P) were relatively up-regulated in green-leaf plants, but their expression levels were higher in yellow-leaf plants after transferred to shade condition (Supplementary Figure 3). Additionally, we characterized a few candidate genes, such as PTAC3 (PLASTID TRANSCRIPTIONALLY ACTIVE 3) and POLGAMMA2, that putatively regulate chloroplast development. PTAC3 is an essential component of PEP complex (plastid encoded RNA polymerase) that regulate the photosynthetic gene expression, chloroplast development and functioning (Kindgren and Strand, 2015). The expression of PTAC3 was high in yellow-leaf plants under natural light, but it significantly decreased in shading condition (Supplementary Figure 3). POLGAMMA2 encodes a nuclear-encoded organelle DNA polymerase targeting to mitochondria and plastids (Baruch-Torres and Brieba, 2017). The expression of POLGAMMA2 was relatively low in yellow-leaf Forsythia lines under both natural light and shading conditions, showing relative expression pattern consistent with leaf color change among samples. Therefore, we also tested the function of POLGAMMA2 with the following functional assays.
To further elucidate the function of ChlH and POLGAMMA2, we performed VIGS assays in green-leaf Forsythia plants (Figure 3). We observed normal green colored leaves and shoots regenerated from the Forsythia injected with empty-loaded TRV2 (vector control). However, leaves of plants expressing TRV:FsChlH and TRV:FsPOLGAMMA2 showed bright yellow-leaf phenotype (Figure 3A). The younger leaves displayed bright yellow-color in the silenced plants, while the mature leaves showed variegated leaves (Figure 3A). We measured the relative expression of ChlH and POLGAMMA2 in the newly generated shoots from FsChlH-silenced and FsPOLGAMMA2-silenced plants at 15 days post the inoculation. qRT-PCR analysis showed that ChlH and POLGAMMA2 were significantly down-regulated in the FsChlH- and FsPOLGAMMA2-silenced plants, respectively (Figure 3B). We also observed significantly reduced chlorophyll and carotenoid content in the silenced plants (Figure 3C and Supplementary Table 8). The total chlorophyll decreased about 66.2 and 83.1% in FsChlH-silenced and FsPOLGAMMA2-silenced Forsythias, respectively (Supplementary Table 8). Moreover, we observed slightly increased number of chloroplasts with fewer grana stacks in FsChlH-silenced Forsythia plants (Figure 3D and Supplementary Table 9). In FsPOLGAMMA2-silenced plants, the thylakoid membrane system was defective with improperly organized granum structures (Figure 3D and Supplementary Table 9). The number of starch grains showed no difference between FsChlH-silenced and the control, but was significantly dropped in FsPOLGAMMA2-silenced plants (Supplementary Table 9).
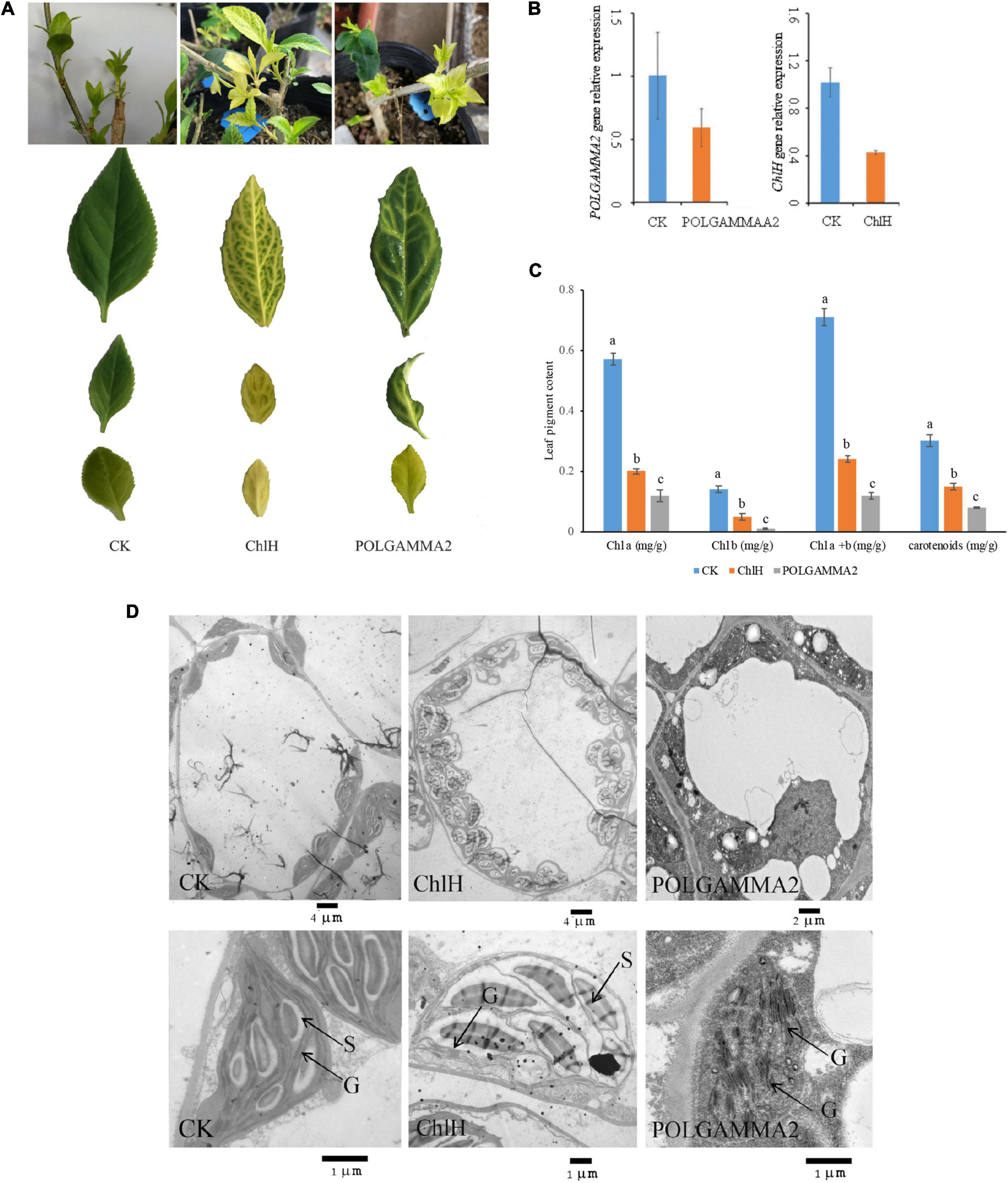
Figure 3. The leaf color and anatomy change in green-leaf Forsythia plants with candidate genes transiently silenced. (A) The leaf color of CK (vector control), FsChlH-silenced, and FsPOLGAMMA2-silenced plants; (B) the relative gene expression of POLGAMMA2 and ChlH in FsPOLGAMMA2-silenced and FsChlH-silenced plants, respectively; (C) the content of Chl a, Chl b, total Chl, and carotenoid in CK, FsChlH-silenced, and FsPOLGAMMA2-silenced Forsythia plants; (D) the chloroplast ultrastructure of CK, FsChlH-silenced, and FsPOLGAMMA2-silenced plants.
To test if the functional role of ChlH and POLGAMMA2 are conserved in other plant species, we used VIGS technique to reduce the transcription level of ChlH and POLGAMMA2 gene in tobacco (Nicotiana benthamiana) and tomato (Solanum lycopersicum). We observed similar leaf color change in ChlH- and POLGAMMA2-silenced plants (Figure 4A). The newly sprouted leaves from ChlH-silenced tobacco (N-ChlH) and tomato (S-ChlH) showed yellow-color at 20 days after inoculation. The leaf color of POLGAMMA2-silenced tobacco (N-POLGAMMA2) and tomato (S-POLGAMMA2) were faded into white color (Figure 4A). The expression of ChlH and POLGAMMA2 was barely detected in tomato and tobacco leaves expressing TRV:ChlH and TRV:POLGAMMA2, respectively (Figure 4B). Additionally, both chlorophyll and carotenoid content were significantly reduced with the down-regulation of ChlH and POLGAMMA2 in tomato and tobacco leaves (Figure 4C and Supplementary Tables 10, 11). The total chlorophyll decreased by 53 and 54% in ChlH-silenced tobacco and tomato, and by 76 and 86% in POLGAMMA2-silenced tobacco and tomato, respectively (Supplementary Tables 10, 11). In ChlH-silenced tomato and tobacco plants, the width of chloroplasts was reduced by approximately 30% and the number of chloroplasts was reduced by 20% (Supplementary Tables 12, 13). We observed loosely packed grana with fewer layers of thylakoids in ChlH-silenced tomato and tobacco plants (Supplementary Figure 4). In FsPOLGAMMA2-silenced plants, the thylakoid membrane system was severely disrupted with no obvious granum structures observed (Supplementary Figure 4 and Supplementary Tables 12, 13).
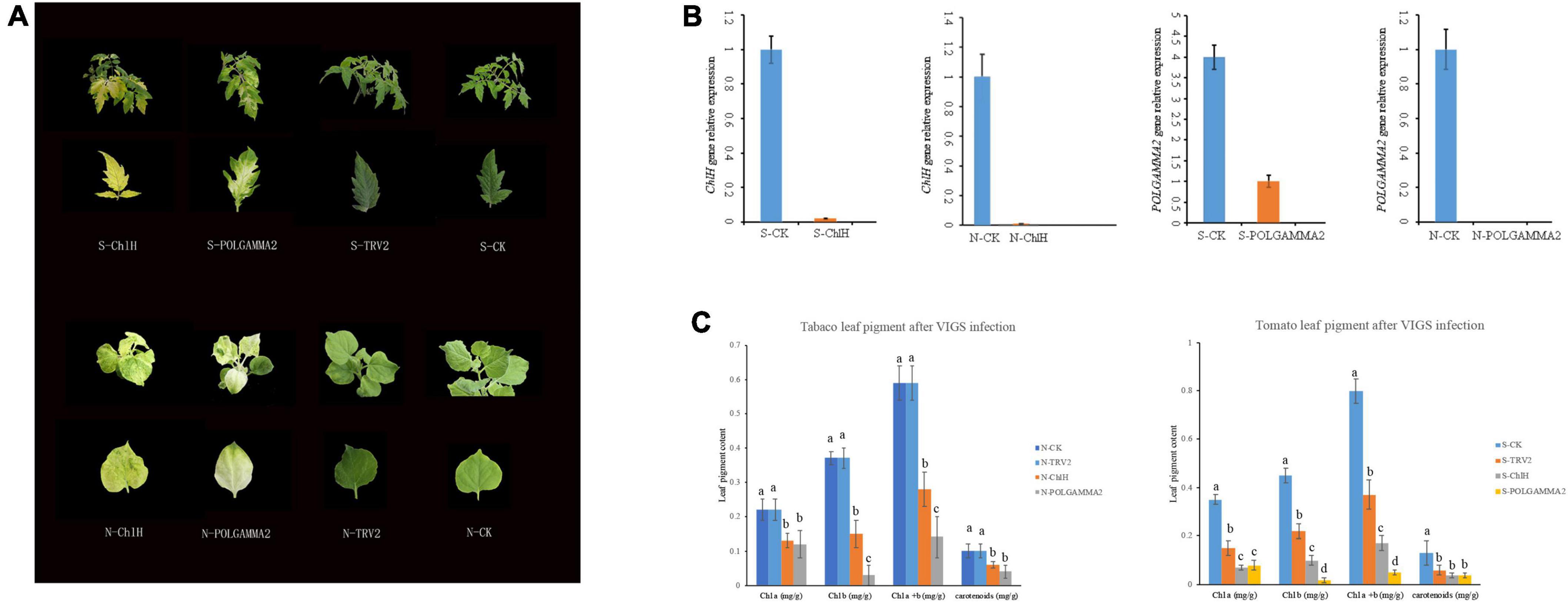
Figure 4. The leaf color change in tobacco and tomato plants after transiently silencing candidate genes. (A) The leaf color of ChlH-silenced (S/N-ChlH), POLGAMMA2-silenced (S/N-POLGAMMA2), TRV2 (vector control; S/N-TRV2), and non-infected (S/N-CK) tomato (S-) and tobacco (N-) plants. (B) The relative gene expression of ChlH and POLGAMMA2 in ChlH-silenced and POLGAMMA2-silenced tomato (S-) and tobacco (N-) plants, respectively. (C) The content of Chl a, Chl b, total Chl, and carotenoid in CK, TRV2, ChlH-silenced, and POLGAMMA2-silenced tomato (S-) and tobacco (N-) plants.
To further examine if the expression of ChlH and POLGAMMA2 can rescue the leaf-color, we performed transient over-expression of these two genes in yellow-leaf Forsythia plants. With qRT-PCR assays, we found that the expression of ChlH and POLGAMMA2 in the over-expression plants were twice as high as that of the control (Figure 5C). However, no obvious leaf color change was observed after the transient over-expression of FsChlH and FsPOLGAMMA2 at 20 days post inoculation (Figure 5A). We also detected significantly increased content of Chl a and Chl b in plants with ChlH and POLGAMMA2 transiently over-expressed (Figure 5B and Supplementary Table 14). The total chlorophyll content of yellow-leaf plants increased about two-fold after transient over-expressing ChlH and POLGAMMA2, while carotenoids remained unchanged in ChlH and POLGAMMA2 over-expressed leaves (Figure 5B and Supplementary Table 14). Though the grana structures remained defected in plants over-expressing ChlH or POLGAMMA2, we observed some lamellae structures of thylakoids in plants over-expressing POLGAMMA2 (Figure 5D and Supplementary Table 15).
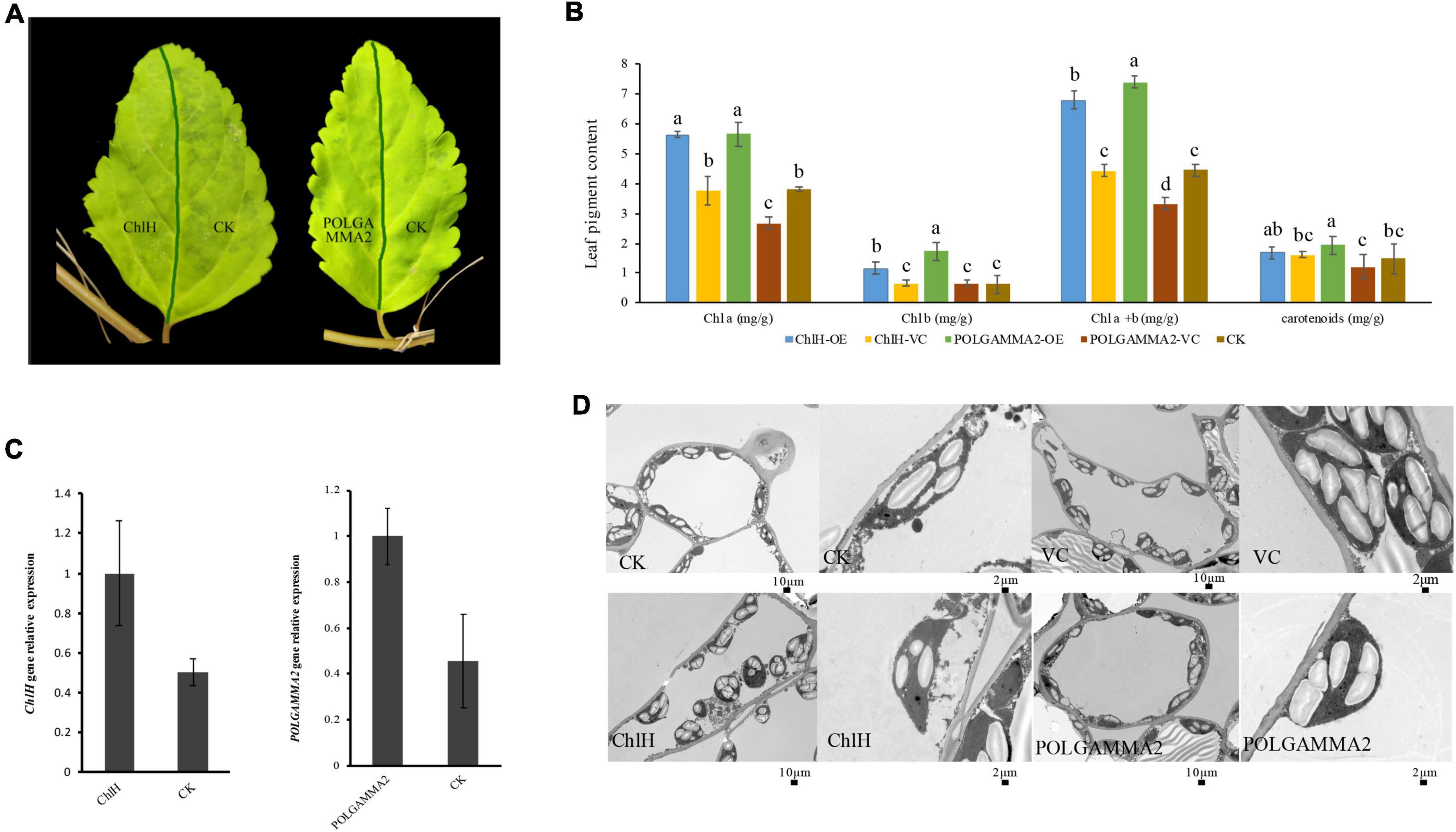
Figure 5. The leaf color and anatomy change in yellow-leaf Forsythia plants with candidate genes transiently over-expressed. (A) The leaf color of ChlH-OE, POLGAMMA2-OE, and the non-infected (CK) leaves of Forsythia. (B) The content of Chl a, Chl b, total Chl, and carotenoid in ChlH-OE, POLGAMMA2-OE, vector control, and the non-infected plants. (C) The relative gene expression of ChlH and POLGAMMA2 in ChlH-OE and POLGAMMA2-OE plants, respectively. (D) The chloroplast structure of CK, VC (vector control), ChlH-OE, and POLGAMMA2-OE plants.
To understand the mechanism of yellow-leaf phenotype in FsChlH- and FsPOLGAMMA2-silenced plants, we performed transcriptome sequencing on leaves collected from the agroinfiltrated plants with TRV:ChlH (CY), TRV:POLGAMMA2 (PY), and the vector control plants (CG and PG). In total, we identified 2040 DEGs between CY and CG, and 77 DEGs between PY and PG. The DEGs that are responsive to the decreasing level of ChlH are enriched in biological processes including photosynthesis (GO:0015979), chloroplast organization (GO: 0009658), response to light stimulus (GO:0009416), chlorophyll biosynthetic process (GO:0015995), and carotenoid biosynthetic process (GO:0016117) (Supplementary Table 16). In the ChlH-silenced plants, we observed decreased level of ChlI and ChlD, which encode two subunits of magnesium chelatase that catalyzing the insertion of Mg2+ into protoporphyrin IX in the first step of chlorophyll biosynthesis (Figure 6A). The transcription level of other chlorophyll biosynthesis genes (HEMC, HEMAs, DVR, and PORAs) were also reduced significantly with the decreasing level of ChlH (Figure 6A). Additionally, a few chloroplast development related genes were found significantly repressed in ChlH-silenced plants, including PTACs (plastid encoded RNA polymerases), CPN60A (CHAPERONIN-60ALPHA) that involved in Rubisco folding, CRB (CHLOROPLAST RNA BINDING), CLPP4 (CLP PROTEASE P4), and CLPP6 (CLP PROTEASE P6) that involved in chloroplast organization, TIC110 (TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 110) that regulate chloroplast biogenesis (Chen and Li, 2017), and ALB3 (ALBINO 3) that regulate thylakoid membrane organization (Figure 6B). Moreover, we observed reduced transcription of carotenoid biosynthesis genes, such as PSY2, LYC (LYCOPENE CYCLASE), and ZDS, and a few carotenoid degradation genes including CYP97A3 (CYTOCHROME P450-TYPE MONOOXYGENASE 97A3) and BETA-OHASE 1 (BETA-HYDROXYLASE 1) in the ChlH-silenced plants (Supplementary Figure 5A). The expression of genes that involved in the photosynthesis were also found decreased, including the PHOTOSYSTEM I (PsaAs, PsaBs, PsaG, PsaL, PsaO) and PHOTOSYSTEM II subunits (PsbB2, PsbP2, PsbE, PsbCs, PsbRs), LHCA1s (PHOTOSYSTEM I LIGHT HARVESTING COMPLEX GENE 1), and LHB1B1s (LIGHT-HARVESTING CHLOROPHYLL-PROTEIN COMPLEX II SUBUNIT B1) (Supplementary Figure 5B).
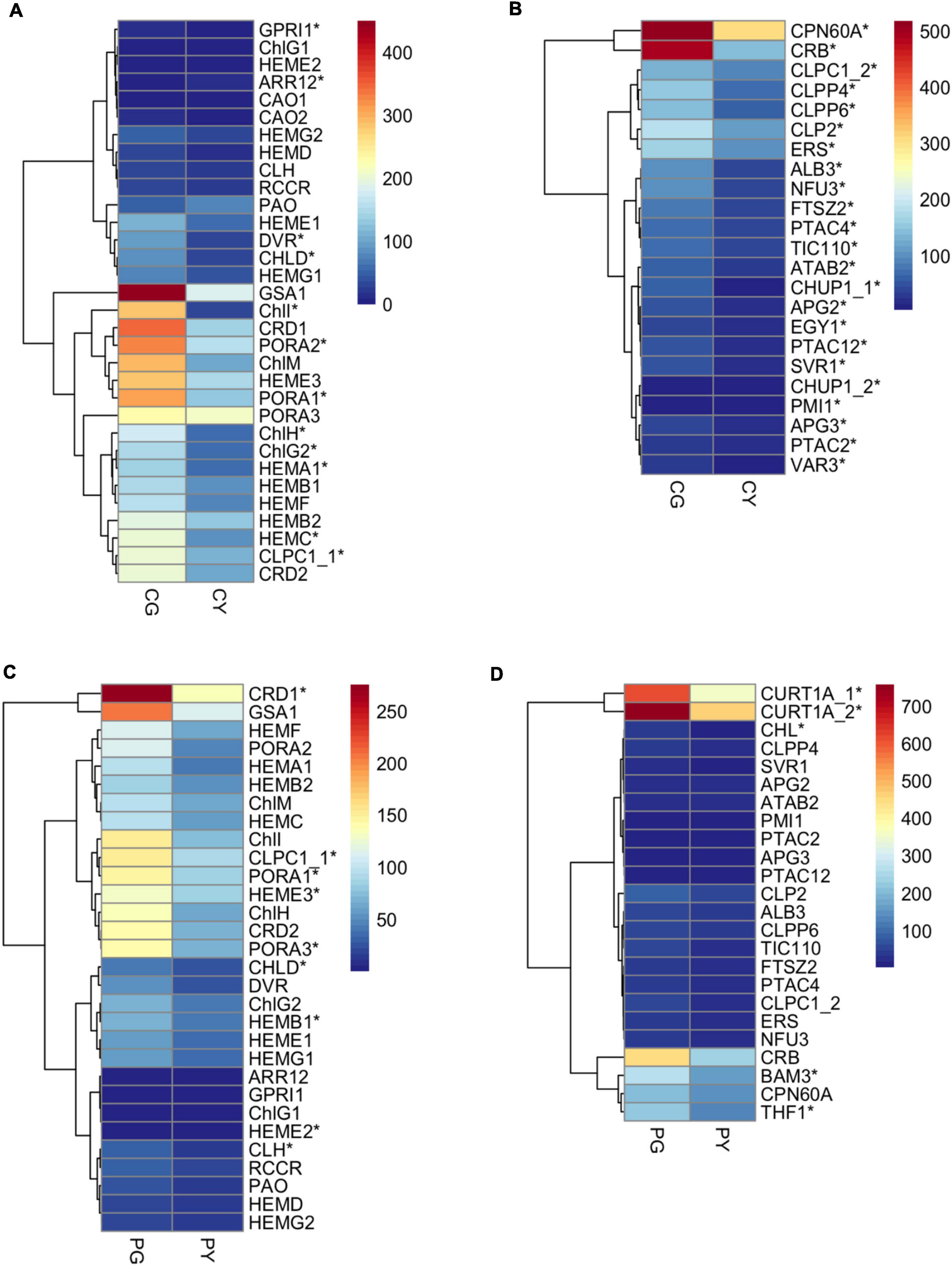
Figure 6. The expression pattern of key genes related to chlorophyll biosynthesis and chloroplast development in the transiently silenced Forsythia plants. (A) DEGs related to chlorophyll biosynthesis in the comparison of CG vs. CY; (B) DEGs related to chloroplast development in the comparison of CG vs. CY; (C) DEGs related to chlorophyll biosynthesis in the comparison of PG vs. PY; (D) DEGs related to chloroplast development in the comparison of PG vs. PY.
Genes with drastic expression change after silencing of FsPOLGAMMA2 are mostly involved in biological processes including phenylpropanoid metabolic process (GO:0009698), response to oxidative stress (GO: 0006979), and cellular lipid metabolic process (GO: 0044255) (Supplementary Table 17). The silencing of POLGAMMA2 led to reduced transcript levels of chlorophyll synthetic genes, such as ChlI, ChlH, PORA1, PORA3, HEME3, CRD1, and carotenoid biosynthetic gene such as PSY2, CCS2 (plastid encoded RNA polymerase), and ZEP (Figure 6C and Supplementary Figure 5C). Additionally, the genes related to chlorophyll binding and photosynthesis, including PsaG, PsaH, PsaL, PsbO2, PsbP2, PsbR1, PsbYs, LHB1B1, and LHCAs, were also significantly reduced (Supplementary Figure 5D). Moreover, we observed strong suppression of thylakoid development genes, including the CHL (CHLOROPLASTIC LIPOCALIN) that protects thylakoidal membrane lipids (Levesque-Tremblay et al., 2009), THF1 (THYLAKOID FORMATION1) that involved in the vesicle-mediated formation of thylakoid membrane (Ma et al., 2015), and CURT1A (CURVATURE THYLAKOID 1A) that is essential for thylakoid granum assembly (Armbruster et al., 2013; Figure 6D). The reduced transcription of genes related to pigment biosynthesis and chloroplast development may lead to the yellow-leaf phenotype in plants with FsChlH and FsPOLGAMMA2 gene silenced.
Leaf coloration is an important attribute for ornamental plants. The diversity of leaf color can enhance the ornamental effect of landscape and extend the duration (Dong et al., 2020). Chlorophyll, carotenoid, and flavanols are three major types of pigments that determines the foliage color in plants (Zhao et al., 2020). So far, leaf color mutants have been characterized in many plant species, such as Arabidopsis (Jarvis et al., 1998), rice (Khan et al., 2021), wheat (Zhang et al., 2020), poplar (Ramamoorthy et al., 2019), and tomato (Mo-zhen et al., 2020). Yellow-leaf phenotype is often the direct consequence of changed contents in chlorophyll and carotenoids (Dong et al., 2020). For example, previous study reported reduced chlorophyll and increased lutein in golden-leaf gingko mutant (Li et al., 2018). Another study in Populus deltoids Marsh revealed that the disruption in chlorophyll synthesis or catabolism lead to lower level of chlorophyll and carotenoids in the yellow-leaf mutants (Ramamoorthy et al., 2019). In addition to pigment abundance, defective chloroplast development and ultrastructure can also impact leaf color (Dong et al., 2020). A spontaneous yellow-green leaf color mutant in Triticum aestivum L. exhibited reduced chlorophyll content and abnormal chloroplast development (Wu et al., 2018). In the leaf-color mutants of Anthurium andraeanum “Sonate,” the number of chloroplasts was significant reduced with ruptured chloroplasts in leaf mesophyll cells (Yang et al., 2015).
In our study, we have examined the physiological and anatomic characteristics of yellow-leaf Forsythia lines. We found that the yellow-leaf is determined by the combined effects of chlorophyll deficiency and disrupted chloroplast structure. Previous studies have demonstrated that different light intensities can affect the synthesis/degradation of photosynthetic pigments, the chloroplast development, and organelle structure (Kong et al., 2016). Upon light exposure, chlorophylls are synthesized, photo-oxidized and degraded rapidly (Zhu et al., 2017). Excessive light usually leads to plants with decreased chlorophyll content due to inhibited chloroplast formation and reduced thylakoid membrane appression (Fu et al., 2012). On the contrary, plants grown in shady environment acclimate and develop larger, thinner foliage with increased chlorophyll production and more appressed thylakoids to optimize the energy usage and conservation (Middleton, 2001; Anderson et al., 2008). Long-term deficient light can lead to degradation of chlorophyll, resulting in yellow leaves (Lee et al., 2014). Thus, we also evaluated the leaf anatomy and chloroplast ultrastructure of progeny lines grown in low light-intensity conditions. Both yellow-leaf and green-leaf lines displayed thinner leaves and palisade tissues after shade treatment, a phenomenon commonly observed in other plant species (Kong et al., 2016). Despite severely distorted grana, a few thylakoid structures were observed in the yellow-leaf plants grown under shady environment with chlorophyll content significantly increased, suggesting the thylakoid membrane appression and chlorophyll synthesis may be slightly enhanced when exposed to low irradiance condition. However, the low light intensity is not sufficient to restore the defected thylakoid membrane system and recover leaf-greening in the yellow-leaf Forsythia plants. Additionally, the photosynthetic capacity of the hybrid population was assessed previously by Wang et al. (2017). The photosynthetic performance of yellow-leafed plants is generally weaker than that of green-leafed plants. The reduced photochemical efficiency may be due to incomplete photosynthetic system on the underdeveloped chloroplast thylakoid development in yellow-leaf Forsythia plants (Wang et al., 2017). These results suggested that the deficiency in chlorophyll content and distorted chloroplast structure are likely two major causes of yellow-leaf coloration in Forsythia independent of light intensity.
The formation of yellow-leaf is controlled by complex genetic network and environmental factors. Any mutations in genes related to pigment synthesis and metabolism, chloroplast transcription, and plastid-nuclear signal transduction can directly or indirectly disrupt pigment biosynthesis and stability, resulting in leaf discoloration (Zhao et al., 2020). In previous studies, RNA-seq approach has been widely applied to investigate candidate genes related to yellow-leaf coloration in plants (Zhao et al., 2020). For instance, the transcriptome study on golden leaf ginkgo mutants revealed that the down-regulated chlorophyll biosynthetic gene as well as up-regulated genes related to chlorophyll degradation and carotenoid biosynthesis likely caused lower chlorophyll and higher carotenoid detected in yellow-leaf mutants (Li et al., 2018). Similar findings were reported in yellow-leaf Shumard oak (Dong et al., 2020). A comprehensive study of yellow-leaf mutant of Lagerstroemia indica identified eleven candidates involved in chlorophyll metabolism, photosynthesis, and chloroplast development that may be responsible for altered chlorophyll content level and hindered chloroplast development in the mutants (Li et al., 2015).
In order to understand the molecular basis of yellow-leaf phenotype, we performed transcriptome sequencing on leaves of yellow-leaf and green-leaf Forsythia hybrids grown under different light-intensity conditions. Based on the comparative transcriptome analysis, we identified a large number of DEGs that were annotated to different biological processes including carbohydrate metabolic process, oxidation-reduction process, pigment biosynthesis, and lipid metabolism. These DEGs from various genetic pathways suggested a complex regulatory network mediating the yellow-leaf phenotype in Forsythia. Since reduced chlorophyll content and abnormal chloroplast structure is observed in yellow-leaf plants, we mainly focused on the DEGs that associated with chlorophyll biosynthesis and chloroplast development. Meanwhile, the physiological experiments showed that the leaf color as well as chlorophyll content were consistently lower in yellow-leaf plants comparing to green-leaf lines given different light conditions. Therefore, we further searched for candidate gene displaying consistent expression patterns between yellow-leaf and green-leaf plants under different light intensities to narrow down the candidate gene set.
Among the chlorophyll biosynthetic genes, we observed significant down-regulation of ChlH in the chlorophyll-deficient yellow-leaf lines in both natural and shade conditions and considered ChlH for functional validation. On the other hand, we screened out POLGAMMA2 as another candidate gene as result of its consistently down-regulation in yellow-leaf Forsythia lines matching the chlorophyll content and leaf color change among different groups. POLGAMMA2 encodes a nuclear-encoded organelle DNA polymerase responsible for the replication of mitochondria and plastid genome (Parent et al., 2011). POLGAMMA2, also known as POLYMERASE I A (Pol1A), is one of the two Plant Organellar DNA polymerases (POPs) in flowering plants (García-Medel et al., 2019; Ji and Day, 2020). POLGAMMA2 is mainly involved in DNA replication, while its paralog POLGAMMA1 serves an additional role in DNA repair (Parent et al., 2011; Morley et al., 2019). In Arabidopsis, plants with mutation in either POLGAMMA2 or POLGAMMA1 can survive with no visible growth defects despite an 30% decrease in organelle DNA copy number, while deletions of both genes are lethal (Parent et al., 2011; Morley and Nielsen, 2016; Morley et al., 2019). Whether POLGAMMA2 affect chloroplast development is worthy testing in Forsythia species.
To investigate the functions of these two candidate genes, we transiently silenced ChlH and POLGAMMA2 in green-leaf Forsythia plants and observed yellow-leaf phenotype with significant reduction of chlorophyll and carotenoid content in both ChlH-silenced and POLGAMMA2-silenced plants. We observed severely disintegrated thylakoid membranes in FsPOLGAMMA2-silenced plants, similar as the chloroplast structure observed in yellow-leaf Forsythia. While the defects of granum structures in FsChlH-silenced plants were less severe than that of yellow leaf Forsythia lines. In pea, the virus-induced gene silencing of ChlH also led to lower chlorophyll accumulation, undeveloped thylakoid membranes, and reduced photosynthesis in yellow-leaf mutants (Luo et al., 2013). The less disrupted chloroplasts in FsChlH-silenced Forsythia is likely dependent on the degree of ChlH gene down-regulation. We also validated the function of ChlH and POLGAMMA2 in model plant species. The FsChlH-silenced tomato and tobacco plants both exhibited yellow leaves with altered chloroplast ultrastructure, which is consistent with tobacco plants with endogenous ChlH gene silenced (Hiriart et al., 2002). On the other hand, paled colored leaves with disassembled thylakoid layers were observed in POLGAMMA2-silenced plants. Previous works showed that POLYMERASE I A (Pol1A) and POLYMERASE I B (Pol1B), designated as POLGAMMA2 and POLGAMMA1, respectively, were organellar DNA polymerases known as Plant Organellar DNA Polymerases (POPs). Most flowering plants harbor two POP genes, while the dicot Solanaceae species, such as Solanum lycopersicum and Nicotiana tomentosiformis, contain only one POP gene (Ji and Day, 2020). It likely that the virus-induced silencing of FsPOLGAMMA2 severely disrupted the DNA replication within chloroplasts, leading to severely disrupted chloroplast development in tobacco and tomato.
Furthermore, we detected increased chlorophyll content in the yellow-leaf plants with ChlH or POLGAMMA2 transiently over-expressed. The expression of ChlH failed to restore the chloroplast structure, while the expression of POLGAMMA2 partially restored the thylakoid membranes, suggesting that POLGAMMA2 may have an additional role in chloroplast development though regulating chloroplast DNA replication. Though the transient over-expression of ChlH and POLGAMMA2 caused the increment of chlorophyll content, however, the induced expression of these two genes are not sufficient for complete restoration of thylakoid membranes in yellow-leaf plants. It is likely due to the limited efficacy of transient assays that may remain active within only a few days (Wroblewski et al., 2005). Another putative explanation is that there are other casual genes participating in the formation of yellow-leaf Forsythia. In addition to genes directly involved in pigment synthesis and chloroplast structure, there are other genetic factors influencing the leaf color through mediating processes such as chloroplast gene transcription and translation, plastid-nuclear signaling, and ROS scavenging system (Pogson and Albrecht, 2011; Cheng et al., 2022). With comparative transcriptome sequencing approach, we are only able to detect functional candidate genes with significant fold-change between yellow-leaf and green-leaf lines. The underlying genetic mechanism of yellow-leaf coloration may still remain incomplete. In the future studies, genetic mapping approaches can be applied to further narrow down the candidate gene list and elucidate the casual mechanism of yellow-leaf coloration in Forsythia.
To further understand the functional mechanism of ChlH and POLGAMMA2, we performed RNA-seq on the green-leaf Forsythia plants with ChlH and POLGAMMA2 silenced, respectively. The silencing of ChlH gene leads to decreased abundance of a large number of genes that involved in chlorophyll and carotenoid biosynthesis, photosynthesis, and chloroplast development. ChlH is a multifunctional protein coordinating the chlorophyll biosynthesis and plastid-to-nucleus retrograde signaling (Ibata et al., 2016). In Arabidopsis, the ChlH mutant displayed repressed plastid signaling with decreased level of photosynthesis-associated nuclear genes, such as LHCB1 (Schlicke et al., 2014). Since chlorophyll biosynthesis is tightly coupled with thylakoid membrane development, the chlorophyll deficiency triggered by ChlH silencing may indirectly impair the transcription of genes regulating chloroplast biogenesis and thylakoid membranes assembly, therefore leading to undeveloped chloroplast structure in Forsythia (Hiriart et al., 2002).
POLGAMMA2 is an organelle localized DNA polymerase associated with DNA replication in chloroplast (Morley and Nielsen, 2016). The modified transcription level of POLGAMMA2 can directly affect the chloroplast DNA copy number and influence chloroplast development related genes indirectly. When POLGAMMA2 is transiently silenced in Forsythia, we observed strong reduction of chlorophyll and carotenoid synthetic genes, which correlates with the deficiency in chlorophyll and carotenoid. We also observed similar yellow-leaf phenotype in tomato and tobacco with POLGAMMA2 transiently silenced, indicating the functional conservation of POLGAMMA2. Moreover, a few genes that possibly regulating thylakoidal membrane formation and architecture were also found repressed after the silencing of POLGAMMA2. CURT1 is an essential gene localized to grana margin and is required for thylakoid membrane bending and the granum stacking (Armbruster et al., 2013). In Arabidopsis, the thylakoids are disorganized with unstacked membranes in the CURT1 mutants (Armbruster et al., 2013). THF1 encodes an imported chloroplast protein that regulate the thylakoid membrane biogenesis and organization during early stage of chloroplast development (Wang et al., 2004). The loss function in THF1 lead to reduced chlorophyll level, less efficient photosynthesis, and defected thylakoid stacking, resulting in variegated leaves in Arabidopsis (Wang et al., 2004). In Forsythia, the silencing of POLGAMMA2 may somehow disrupt the photosynthetic pigment accumulation and thylakoid membrane organization through repressing the transcription of chlorophyll synthesis genes, such as ChlH and PORA, and chloroplast development related genes, such as THF1 and CURT1. Taken together, the inhibition of ChlH and POLGAMMA2 is likely required for the yellow-leaf Forsythia through influencing the transcription of genes related to chlorophyll biosynthesis and chloroplast structure intactness. In the current study, we only characterized the functional role of candidate DEGs with consistent expression pattern regardless of light-intensity change. Future research can focus on the candidate genes responsive to light-intensity change and test their functional roles in regulating pigment synthesis and thylakoid structure in Forsythia.
To conclude, our study characterized the yellow-leaf Forsythia with reduced chlorophyll and carotenoid content, as well as defected chloroplast structure. The transcriptome sequencing and expression pattern analysis screened out two candidate genes, ChlH and POLGAMMA2, with expression profiles strongly correlated with chlorophyll content and leaf color variation among different leaf-colored Forsythia lines. The transient silencing of ChlH and POLGAMMA2 lead to reduced chlorophyll level and disrupted chloroplast structure, while the transient over-expression of these two genes promoted the restoration of thylakoid architecture in yellow-leaf Forsythia. By examining genes with transcriptional change responsive to the silencing of these two genes, we conclude that ChlH and POLGAMMA2 may be related to the yellow-leaf phenotype in Forsythia through directly or indirectly mediating genes involved in chlorophyll biosynthesis and chloroplast development. In general, our study provides a deep understanding in the physiological as well as molecular mechanism underlying the yellow-leaf coloration in Forsythia.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
HP conceived the experiments. MZ, JS, and YW performed the experiments and wrote the manuscript. ZZ, XZ, TC, JW, and QZ assisted in analyzing the data and finalizing the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Beijing Municipal Science and Technology Project (Z181100002418006), the Fundamental Research Fund for the Central University (2015ZCQ-YL-03), and the World-Class Discipline Construction and Characteristic Development Guidance Funds for Beijing Forestry University (2019XKJS0323).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1009575/full#supplementary-material
Amancio, S., Zhu, G., Yang, F., Shi, S., Li, D., Wang, Z., et al. (2015). Transcriptome characterization of Cymbidium sinense ‘Dharma’ using 454 pyrosequencing and its application in the identification of genes associated with leaf color variation. PLoS One 10:e0128592. doi: 10.1371/journal.pone.0128592
Anderson, J. M., Chow, W. S., and De Las Rivas, J. (2008). Dynamic flexibility in the structure and function of photosystem II in higher plant thylakoid membranes: The grana enigma. Photosynth. Res. 98, 575–587. doi: 10.1007/s11120-008-9381-3
Armbruster, U., Labs, M., Pribil, M., Viola, S., Xu, W., Scharfenberg, M., et al. (2013). Arabidopsis curvature thylakoid1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell 25, 2661–2678. doi: 10.1105/tpc.113.113118
Aziz, E., Batool, R., Akhtar, W., Rehman, S., Gregersen, P. L., and Mahmood, T. (2019). Expression analysis of the polyphenol oxidase gene in response to signaling molecules, herbivory and wounding in antisense transgenic tobacco plants. 3 Biotech 9:55. doi: 10.1007/s13205-019-1587-x
Baruch-Torres, N., and Brieba, L. G. (2017). Plant organellar DNA polymerases are replicative and translesion DNA synthesis polymerases. Nucleic Acids Res. 45, 10751–10763. doi: 10.1093/nar/gkx744
Chang, T., Zhang, Y., Xu, H., Shao, X., Xu, Q., Li, F., et al. (2018). Osmotic adjustment and up-regulation expression of stress-responsive genes in tomato induced by soil salinity resulted from nitrate fertilization. Int. J. Agric. Biol. Eng. 11, 126–136. doi: 10.25165/j.ijabe.20181103.2952
Chen, L.-J., and Li, H.-M. (2017). Stable megadalton TOC-TIC supercomplexes as major mediators of protein import into chloroplasts. Plant J. 92, 178–188. doi: 10.1111/tpj.13643
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, Z., Lu, X., Xuan, Y., Tang, F., Wang, J., Shi, D., et al. (2019). Transcriptome analysis based on a combination of sequencing platforms provides insights into leaf pigmentation in Acer rubrum. BMC Plant Biol. 19:240. doi: 10.1186/s12870-019-1850-7
Cheng, M., Meng, F., Mo, F., Chen, X., Zhang, H., and Wang, A. (2022). Insights into the molecular basis of a yellow leaf color mutant (ym) in tomato (Solanum lycopersicum). Sci. Hortic. 293:110743. doi: 10.1016/j.scienta.2021.110743
Dong, X., Huang, L., Chen, Q., Lv, Y., Sun, H., and Liang, Z. (2020). Physiological and anatomical differences and differentially expressed genes reveal yellow leaf coloration in Shumard Oak. Plants 9:169. doi: 10.3390/plants9020169
Fu, D.-Q., Zhu, B.-Z., Zhu, H.-L., Jiang, W.-B., and Luo, Y.-B. (2005). Virus-induced gene silencing in tomato fruit. Plant J. 43, 299–308. doi: 10.1111/j.1365-313X.2005.02441.x
Fu, W., Li, P., and Wu, Y. (2012). Effects of different light intensities on chlorophyll fluorescence characteristics and yield in lettuce. Sci. Hortic. 135, 45–51. doi: 10.1016/j.scienta.2011.12.004
García-Medel, P. L., Baruch-Torres, N., Peralta-Castro, A., Trasviña-Arenas, C. H., Torres-Larios, A., and Brieba, L. G. (2019). Plant organellar DNA polymerases repair double-stranded breaks by microhomology-mediated end-joining. Nucleic Acids Res. 47, 3028–3044. doi: 10.1093/nar/gkz039
Guo, H., Liu, A.-H., Ye, M., Yang, M., and Guo, D.-A. (2007). Characterization of phenolic compounds in the fruits of Forsythia suspensa by high-performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21, 715–729. doi: 10.1002/rcm.2875
He, N., Liu, C., Tian, M., Li, M., Yang, H., Yu, G., et al. (2017). Variation in leaf anatomical traits from tropical to cold-temperate forests and linkage to ecosystem functions. Funct. Ecol. 32, 10–19. doi: 10.1111/1365-2435.12934
Hiriart, J.-B., Lehto, K., Tyystjärvi, E., Junttila, T., and Aro, E.-M. (2002). Suppression of a key gene involved in chlorophyll biosynthesis by means of virus-inducing gene silencing. Plant Mol. Biol. 50, 213–224. doi: 10.1023/A:1016000627231
Ibata, H., Nagatani, A., and Mochizuki, N. (2016). CHLH/GUN5 function in tetrapyrrole metabolism is correlated with plastid signaling but not ABA responses in guard cells. Front. Plant Sci. 7:1650. doi: 10.3389/fpls.2016.01650
Jarvis, P., Chen, L.-J., Li, H.-M., Peto, C. A., Fankhauser, C., and Chory, J. (1998). An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103. doi: 10.1126/science.282.5386.100
Ji, J., and Day, A. (2020). Construction of a highly error-prone DNA polymerase for developing organelle mutation systems. Nucleic Acids Res. 48, 11868–11879. doi: 10.1093/nar/gkaa929
Jung, K.-H., Hur, J., Ryu, C.-H., Choi, Y., Chung, Y.-Y., Miyao, A., et al. (2003). Characterization of a rice chlorophyll-deficient mutant using the T-DNA Gene-Trap system. Plant Cell Physiol. 44, 463–472. doi: 10.1093/pcp/pcg064
Kessler, F., and Schnell, D. (2009). Chloroplast biogenesis: Diversity and regulation of the protein import apparatus. Curr. Opin. Cell Biol. 21, 494–500. doi: 10.1016/j.ceb.2009.03.004
Khan, A., Jalil, S., Cao, H., Sunusi, M., Tsago, Y., Chen, J., et al. (2021). Identification and fine mapping of candidate gene for yellow leaf mutant (ygl54) exhibiting yellow leaf colour in rice. Russ. J. Plant Physiol. 68, 1069–1078. doi: 10.1134/S1021443721060078
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kindgren, P., and Strand, Å (2015). Chloroplast transcription, untangling the Gordian Knot. New Phytol. 206, 889–891. doi: 10.1111/nph.13388
Kong, D.-X., Li, Y.-Q., Wang, M.-L., Bai, M., Zou, R., Tang, H., et al. (2016). Effects of light intensity on leaf photosynthetic characteristics, chloroplast structure, and alkaloid content of Mahonia bodinieri (Gagnep.) Laferr. Acta Physiol. Plant. 38:120. doi: 10.1007/s11738-016-2147-1
Lee, E., Ahn, H., and Choe, E. (2014). Effects of light and lipids on chlorophyll degradation. Food Sci. Biotechnol. 23, 1061–1065. doi: 10.1007/s10068-014-0145-x
Levesque-Tremblay, G., Havaux, M., and Ouellet, F. (2009). The chloroplastic lipocalin AtCHL prevents lipid peroxidation and protects Arabidopsis against oxidative stress. Plant J. 60, 691–702. doi: 10.1111/j.1365-313X.2009.03991.x
Li, B., and Dewey, C. N. (2011). RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, L.-F., Cushman, S. A., He, Y.-X., and Li, Y. (2020). Genome sequencing and population genomics modeling provide insights into the local adaptation of weeping Forsythia. Hortic. Res. 7:130. doi: 10.1038/s41438-020-00352-7
Li, W.-X., Yang, S.-B., Lu, Z., He, Z.-C., Ye, Y.-L., Zhao, B.-B., et al. (2018). Cytological, physiological, and transcriptomic analyses of golden leaf coloration in Ginkgo biloba L. Hortic. Res. 5:12. doi: 10.1038/s41438-018-0015-4
Li, Y., Zhang, Z., Wang, P., Wang, S. A., Ma, L., Li, L., et al. (2015). Comprehensive transcriptome analysis discovers novel candidate genes related to leaf color in a Lagerstroemia indica yellow leaf mutant. Genes Genomics 37, 851–863. doi: 10.1007/s13258-015-0317-y
Lichtenthaler, H. K. (1987). “Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes,” in Methods in enzymology, eds L. Packer and R. Douce (New York, NY: Academic Press), 50–382. doi: 10.1515/znc-2001-11-1225
Loìpez-Juez, E., Paul Jarvis, R., Takeuchi, A., Page, A. M., and Chory, J. (1998). New Arabidopsis cue mutants suggest a close connection between plastid- and phytochrome regulation of nuclear gene expression. Plant Physiol. 118, 803–815. doi: 10.1104/pp.118.3.803
Luo, F., Cheng, S.-C., Cai, J.-H., Wei, B.-D., Zhou, X., Zhou, Q., et al. (2019). Chlorophyll degradation and carotenoid biosynthetic pathways: Gene expression and pigment content in broccoli during yellowing. Food Chem. 297:124964. doi: 10.1016/j.foodchem.2019.124964
Luo, T., Luo, S., Araújo, W. L., Schlicke, H., Rothbart, M., Yu, J., et al. (2013). Virus-induced gene silencing of pea CHLI and CHLD affects tetrapyrrole biosynthesis, chloroplast development and the primary metabolic network. Plant Physiol. Biochem. 65, 17–26. doi: 10.1016/j.plaphy.2013.01.006
Ma, Z., Wu, W., Huang, W., and Huang, J. (2015). Down-regulation of specific plastid ribosomal proteins suppresses thf1 leaf variegation, implying a role of THF1 in plastid gene expression. Photosynth. Res. 126, 301–310. doi: 10.1007/s11120-015-0101-5
Middleton, L. (2001). Shade-tolerant flowering plants: Adaptations and horti-cultural implications. Leuven: International Society for Horticultural Science (ISHS), 95–102.
Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. U.S.A. 98, 2053–2058. doi: 10.1073/pnas.98.4.2053
Morley, S. A., and Nielsen, B. L. (2016). Chloroplast DNA Copy number changes during plant development in organelle DNA polymerase mutants. Front. Plant Sci. 7:57. doi: 10.3389/fpls.2016.00057
Morley, S. A., Peralta-Castro, A., Brieba, L. G., Miller, J., Ong, K. L., Ridge, P. G., et al. (2019). Arabidopsis thaliana organelles mimic the T7 phage DNA replisome with specific interactions between Twinkle protein and DNA polymerases Pol1A and Pol1B. BMC Plant Biol. 19:241. doi: 10.1186/s12870-019-1854-3
Mo-zhen, C., Hao-nan, Q., Fu-lei, M., Jiangang, Y., Lei, Z., and He, Z. (2020). The chlorophyll biosynthesis and degradation and chloroplast structure in Tomato yellow mutant. J. Hortic. Sci. Res. 3, 113–124. doi: 10.36959/745/406
Parent, J.-S., Lepage, E., and Brisson, N. (2011). Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol. 156, 254–262. doi: 10.1104/pp.111.173849
Piao, X.-L., Jang, M. H., Cui, J., and Piao, X. (2008). Lignans from the fruits of Forsythia suspensa. Bioorganic Med. Chem. Lett. 18, 1980–1984. doi: 10.1016/j.bmcl.2008.01.115
Pogson, B. J., and Albrecht, V. (2011). Genetic dissection of chloroplast biogenesis and development: An overview. Plant Physiol. 155, 1545–1551. doi: 10.1104/pp.110.170365
Pogson, B. J., Ganguly, D., and Albrecht-Borth, V. (2015). Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 1847, 1017–1024. doi: 10.1016/j.bbabio.2015.02.003
Ramamoorthy, S., Zhang, S., Wu, X., Cui, J., Zhang, F., Wan, X., et al. (2019). Physiological and transcriptomic analysis of yellow leaf coloration in Populus deltoides Marsh. PLoS One 14:e0216879. doi: 10.1371/journal.pone.0216879
Rodríguez, F., Garrido, J. L., Sobrino, C., Johnsen, G., Riobó, P., Franco, J., et al. (2016). Divinyl chlorophyllain a in the marine eukaryotic protist Alexandrium ostenfeldii (Dinophyceae). Environ. Microbiol. 18, 627–643. doi: 10.1111/1462-2920.13042
Ruppel, N. J., and Hangarter, R. P. (2007). Mutations in a plastid-localized elongation factor G alter early stages of plastid development in Arabidopsis thaliana. BMC Plant Biol. 7:37. doi: 10.1186/1471-2229-7-37
Schlicke, H., Hartwig, A. S., Firtzlaff, V., Richter, A. S., Glässer, C., Maier, K., et al. (2014). Induced deactivation of genes encoding chlorophyll biosynthesis enzymes disentangles tetrapyrrole-mediated retrograde signaling. Mol. Plant 7, 1211–1227. doi: 10.1093/mp/ssu034
Senthil-Kumar, M., and Mysore, K. S. (2011). Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato. Plant Biotechnol. J. 9, 797–806. doi: 10.1111/j.1467-7652.2011.00589.x
Shen, J., Si, W., Wu, Y., Xu, Y., Wang, J., Cheng, T., et al. (2021). Establishment and Verification of an efficient virus-induced gene silencing system in Forsythia. Hortic. Plant J. 7, 81–88. doi: 10.1016/j.hpj.2020.09.001
Shen, J., Wu, Y., Jiang, Z., Xu, Y., Zheng, T., Wang, J., et al. (2019). Selection and validation of appropriate reference genes for gene expression studies in Forsythia. Physiol. Mol. Biol. Plants 26, 173–188. doi: 10.1007/s12298-019-00731-y
Sinkkonen, A., Somerkoski, E., Paaso, U., Holopainen, J. K., Rousi, M., and Mikola, J. (2012). Genotypic variation in yellow autumn leaf colours explains aphid load in silver birch. New Phytol. 195, 461–469. doi: 10.1111/j.1469-8137.2012.04156.x
Sobotka, R. (2013). Making proteins green; biosynthesis of chlorophyll-binding proteins in cyanobacteria. Photosynth. Res. 119, 223–232. doi: 10.1007/s11120-013-9797-2
Terry, M. J., Linley, P. J., and Kohchi, T. (2002). Making light of it: The role of plant haem oxygenases in phytochrome chromophore synthesis. Biochem. Soc. Trans. 30, 604–609. doi: 10.1042/bst0300604
Wang, J.-Y., Shen, J.-S., Gu, M., Wang, J., Cheng, T.-R., Pan, H.-T., et al. (2017). Leaf Coloration and Photosynthetic Characteristics of Hybrids between Forsythia ‘Courtaneur’ and Forsythia koreana ‘Suwon Gold’. HortScience 52, 1661–1667. doi: 10.21273/hortsci12177-17
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, P., and Grimm, B. (2015). Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth. Res. 126, 189–202. doi: 10.1007/s11120-015-0154-5
Wang, Q., Sullivan, R. W., Kight, A., Henry, R. L., Huang, J., Jones, A. M., et al. (2004). Deletion of the chloroplast-localized thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 136, 3594–3604. doi: 10.1104/pp.104.049841
Wroblewski, T., Tomczak, A., and Michelmore, R. (2005). Optimization of agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3, 259–273. doi: 10.1111/j.1467-7652.2005.00123.x
Wu, H., Shi, N., An, X., Liu, C., Fu, H., Cao, L., et al. (2018). Candidate Genes for yellow leaf color in common wheat (Triticum aestivum L.) and major related metabolic pathways according to transcriptome profiling. Int. J. Mol. Sci. 19:1594. doi: 10.3390/ijms19061594
Yamamoto, T., Hoshikawa, K., Ezura, K., Okazawa, R., Fujita, S., Takaoka, M., et al. (2018). Improvement of the transient expression system for production of recombinant proteins in plants. Sci. Rep. 8, 1–10. doi: 10.1038/s41598-018-23024-y
Yang, Y., Chen, X., Xu, B., Li, Y., Ma, Y., and Wang, G. (2015). Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum ‘Sonate’. Front. Plant Sci. 6:139. doi: 10.3389/fpls.2015.00139
Zhang, H., Chen, Y., Niu, Y., Zhang, X., Zhao, J., Sun, L., et al. (2020). Characterization and fine mapping of a leaf yellowing mutant in common wheat. Plant Growth Regul. 92, 233–247. doi: 10.1007/s10725-020-00633-0
Zhao, M.-H., Li, X., Zhang, X.-X., Zhang, H., and Zhao, X.-Y. (2020). Mutation mechanism of leaf color in plants: A review. Forests 11:851. doi: 10.3390/f11080851
Zhu, X., Chen, J., Qiu, K., and Kuai, B. (2017). Phytohormone and light regulation of chlorophyll degradation. Front. Plant Sci. 8:1911. doi: 10.3389/fpls.2017.01911
Keywords: Forsythia, yellow-leaf, ChlH, POLGAMMA2, transcriptome sequencing, chlorophyll biosynthesis, chloroplast development
Citation: Zhang M, Shen J, Wu Y, Zhang X, Zhao Z, Wang J, Cheng T, Zhang Q and Pan H (2022) Comparative transcriptome analysis identified ChlH and POLGAMMA2 in regulating yellow-leaf coloration in Forsythia. Front. Plant Sci. 13:1009575. doi: 10.3389/fpls.2022.1009575
Received: 02 August 2022; Accepted: 24 August 2022;
Published: 09 September 2022.
Edited by:
Penghui Li, Anhui Agricultural University, ChinaReviewed by:
Yaping Kou, Insititute of Vegetables and Flowers (CAAS), ChinaCopyright © 2022 Zhang, Shen, Wu, Zhang, Zhao, Wang, Cheng, Zhang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huitang Pan, aHRwYW5AYmpmdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.