- 1Jilin Provincial Engineering Laboratory of Plant Genetic Improvement, College of Plant Science, Jilin University, Changchun, China
- 2Institute of Agricultural Biotechnology, Jilin Academy of Agricultural Sciences, Changchun, China
The AP2/ERF family is a large group of plant-specific transcription factors that play an important role in many biological processes, such as growth, development, and abiotic stress responses. OsDREB2B, a dehydration responsive factor (DRE/CRT) in the DREB subgroup of the AP2/ERF family, is associated with abiotic stress responses, such as cold, drought, salt, and heat stress, in Arabidopsis or rice. However, its role in regulating plant growth and development in rice is unclear. In this study, we reported a new function of OsDREB2B, which negatively regulates plant height in rice. Compared with wild type (WT), OsDREB2B-overexpressing (OE) rice exhibited dwarf phenotypes, such as reduction in plant height, internode length, and seed length, as well as grain yield, while the knockout mutants developed by CRISPR/Cas9 technology exhibited similar phenotypes. Spatial expression analysis revealed that OsDREB2B was highly expressed in the leaf sheaths. Under exogenous GA3 application, OsDREB2B expression was induced, and the length of the second leaf sheath of the OsDREB2B-OE lines recovered to that of the WT. OsDREB2B localized to the nucleus of the rice protoplast acted as a transcription activator and upregulated OsAP2-39 by directly binding to its promoter. OsDREB2B-OE lines reduced endogenous bioactive GA levels by downregulating seven GA biosynthesis genes and upregulating eight GA deactivation genes but not GA signaling genes. The yeast two-hybrid assay and bimolecular fluorescence complementation assay showed that OsDREB2B interacted with OsWRKY21. In summary, our study suggests that OsDREB2B plays a negative role in rice growth and development by regulating GA metabolic gene expression, which is mediated by OsAP2-39 and OsWRKY21, thereby reducing GA content and rice plant height.
Introduction
DREB is a major subgroup of AP2/ERF transcription factor family, which plays an important role in many biological processes such as growth, development and abiotic stress responses in plant. It has been reported that the DREB genes in rice affect abscisic acid (ABA) biosynthesis and signaling, as well as crosstalk between signal transduction pathways for biotic and abiotic stress responses, such as OsDREB2A (Zhang et al., 2013), OsDREB6 (Ke et al., 2014), and OsARAG1 (Zhao et al., 2010). A total of 170 AP2/ERF genes have been identified in rice genome, which are divided into five major groups, namely AP2 (APETALA2), DREB (dehydration-responsive element binding protein), ERF (ethylene responsive factor), RAV (related to ABI3/VP1) and Soloist 5 (Rashid et al., 2012; Gu et al., 2017). Some AP2/ERF transcription factors affect GA metabolism by regulating the expression of GA metabolic genes, thus regulating plant growth and development, such as OsEATB (Qi et al., 2011), OsRPH1 (Ma et al., 2020), and OsAP2-39 (Yaish et al., 2010).
Rice is one of the most important food crops in the world, and it plays an important role in ensuring food security. Plant height is one of the most important factors determining rice plant morphology. An appropriate plant height can reduce the density of leaves, increase yield, and prevent lodging (Sasaki et al., 2002; Spielmeyer et al., 2002). Plant height is regulated by various hormones (Iwamoto et al., 2011), such as gibberellin (GA) (Hedden, 2003; Ueguchi-Tanaka et al., 2005; Liu et al., 2018; Wu et al., 2021), brassinosteroid (BR) (Hong et al., 2002; Tong et al., 2014), indole-3-acetic acid (Aux/IAA) (Huang et al., 2016; Zhang et al., 2016), and strigolactones (SLs) (Zhou et al., 2013). GA can significantly promote elongation of the second leaf sheath in GA-deficient OsRPH1-overexpressing rice (Ma et al., 2020). BR can greatly increase the leaf angles of the flag leaves in rice, and BR-insensitive OsOFP22-overexpressing rice has a significantly reduced lamina joint angle in flag leaves compared to the WT (Chen et al., 2021). Aux/IAA regulates phototropism/gravitropism, root formation, apical dominance, stem/hypocotyl elongation, and leaf expansion. The overexpression of OsIAA1 changes the leaf angle (Song et al., 2009), and the overexpression of OsIAA4 changes the tiller angle (Song and Xu, 2013).
Among these, GA is one of the most important hormones affecting plant height. A total of 136 GA species have been identified; among them, GA1, GA3, GA4, and GA7 have been proven to be endogenous bioactive species in plants, and some have at least some intrinsic bio-activities, but they do not exist in vegetative tissues and therefore may have no regulatory function (Hedden, 2002). Numerous rice GA mutants are deficient in the biosynthesis of GA or in the perception of GA signals (Hedden, 2003; Ueguchi-Tanaka et al., 2005; Liu et al., 2018), including GA-deficient and GA-insensitive mutants. GA-deficient mutants are caused by mutations of enzymes involved in GA biosynthesis or GA deactivation, and not in GA perception, as external GA application allows normal height to be recovered (Ashikari et al., 2002; Hedden, 2003; Sakamoto et al., 2004). GA-insensitive mutants are caused by gene mutations involved in GA signal transduction, and the content of endogenous bioactive GAs is usually much higher than that of the WT (Ueguchi-Tanaka et al., 2000; Sasaki et al., 2003; Ueguchi-Tanaka et al., 2005).
In rice, GA metabolic genes include catalytic enzymes in the early steps of GA biosynthesis, such as CPS, KS, KO, and KAO, which are encoded by single genes (Yamaguchi, 2008). In the late stages of GA biosynthesis, GA20ox, GA3ox, and GA2ox are encoded by multi-gene families. Among them, GA20ox and GA3ox convert GA precursors into bioactive GAs through a cascade of catalytic oxidation (Hedden and Thomas, 2012), while GA2ox is critical for GA deactivation (Thomas et al., 1999). In the GA signaling pathway in rice, GID1 encodes a soluble GA receptor (Ueguchi-Tanaka et al., 2005), and GID2 is an F-box subunit of Skp1-Cullin-F box protein (SCF) E3 ubiquitin ligase, facilitating SLENDER RICE 1 (SLR1) degradation by the 26S proteasome in the presence of GA (Sasaki et al., 2003; Itoh et al., 2005).
OsDREB2B gene belongs to the DREB subgroup and responds to abiotic stress, such as drought, heat stress, and cold, in Arabidopsis or rice (Chen et al., 2008; Matsukura et al., 2010; Sakata et al., 2014). OsDREB2B overexpression effectively improved tolerance to drought stress in rice (Chen et al., 2008) and heat shock and drought stress in Arabidopsis (Matsukura et al., 2010). Interestingly, there is a relationship between cold stress and GA metabolism. For example, under low-temperature conditions, the mRNA expression of OsDREB2B is upregulated, while that of the GA biosynthesis genes GA20ox3 and GA3ox1 is downregulated in rice (Zhang et al., 2013; Sakata et al., 2014). SlDREB, a DREB subgroup gene in tomato (Solanum lycopersicum), downregulates the expression of GA-related genes, and its overexpression in transgenic tomato results in dwarfing, including restriction of internode elongation (Li et al., 2012). Overexpression of OsAP2-39, an ERF subgroup of AP2/ERF family transcription factor, can reduce plant height in overexpressing transgenic rice by upregulating the GA catabolic gene OsEUI (Elongated Uppermost Internode) (Yaish et al., 2010), which encodes for a cytochrome P450 monooxygenase, an enzyme that deactivates GA through an epoxidation reaction (Ma et al., 2006; Zhu et al., 2006; Zhang et al., 2008).
Many AP2/ERF transcription factors regulate plant growth and development and stress response by interacting with other transcription factors, such as BR regulated (Xie et al., 2019), WRKY (Tripathi et al., 2015), MYB (Zeng et al., 2015), and zinc finger transcription factors (Hawku et al., 2021). One of the WRKY transcription factor, OsWRKY21 (LOC_Os01g60640) has been reported to be involved in rice growth and development by regulating the expression of GA metabolism and cell wall biosynthesis-related genes. Overexpressing of OsWRKY21 in rice exhibited a semi-dwarf phenotype, earlier heading dates, and shorter stem internodes (Wei et al., 2021).
In contrast to the wealth of knowledge obtained on stress response, information about the regulation of growth and development by OsDREB2B remains limited. In the present study, we focused on OsDREB2B regulating plant growth and development. OsDREB2B-overexpressing rice possessed a dwarf phenotype of plant height, internode length and seed length in rice. We examined the GA-mediated physiological process of second leaf sheath elongation, the contents of various forms of endogenous GA levels, and the expression alteration of GA-related genes in OsDREB2B-overexpressing and knockout mutant. In addition, OsDREB2B downstream target genes and interaction proteins were also elucidated. Our results indicate that OsDREB2B plays a negative role in rice growth and development by regulating GA metabolic gene expression, which is mediated by OsAP2-39 and OsWRKY21, thereby reducing the GA content and plant height in rice.
Materials and methods
Plant materials
The full-length CDS of OsDREB2B was cloned and fused to the overexpression vector pCUbi1390, which was driven by the maize Ubiquitin promoter by PstI and HindIII sites to generate the pUbi::OsDREB2B construct. pUbi::OsDREB2B was transformed into Japonica rice variety Kitaake via Agrobacterium (EHA105-) mediated transformation (Nishimura et al., 2006). The T3 homozygous lines were screened by molecular detection and hygromycin resistance selection. The primers used in the plant expression vector construction are listed in Table S1.
Construction and identification of OsDREB2B mutants
CRISPR/Cas9 genome editing technology was used to obtain osdreb2b mutants. Selection of Cas9/gRNA target site and vector construction were conducted according to previous reports (Li et al., 2017). The OsDREB2B-pBGK032 vector was introduced into Agrobacterium EHA105 and transformed into Kitaake (Nishimura et al., 2006). Homozygous osdreb2b mutants were screened by testing the specific site of the Cas9 label using PCR analysis and DNA sequencing of the OsDREB2B‐specific editing site. The primer sequences used for plasmid construction and mutant identification are listed in Table S1.
Phenotype analysis, statistical analysis, and growth conditions
All plants were grown in the field at Jilin University in Changchun, Jilin province, China, under normal cultivation condition. The phenotypes of OE and mt lines at seedling stage were evaluated after growing hydroponically for 14 days under 12 h light at 30°C/12 h dark at 24°C. Then, the seedlings were transplanted into the field during the rice-growing season. Seed length, width, and thickness were measured after air drying of fully developed grains. Student’s t-tests were used to determine the statistical significance of phenotypic differences between 10 seedlings of the mt and OE lines.
GA3 treatments
The WT seeds were germinated in distilled water and grown in kimura B culture solution. After culturing for 14 days, the seedlings were treated with 10, 50, and 100 µM GA3 (Sigma-Aldrich, Shanghai, China). Leaf sheaths and leaves were collected at 0, 1, 6, and 12 h after treatment. The experiment was performed with three biological replicates.
The exogenous GA3 rescue assay was performed as previously described (Ma et al., 2020). WT and transgenic rice seedlings at the three-leaf stage were incubated in kimura B culture solution for 10 days and then treated with 10, 50, and 100 µM GA3. The length of the second leaf sheath was measured every 6 h in three trials, with 20 individuals per trial. These experiments were repeated three times. Values are means ± SD.
Measurement of GA content
The endogenous GA content of the WT, OE, and mt lines was measured by high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS). One gram of shoot tissue from 10d-old rice seedlings was collected for endogenous GA measurements. The quantification of endogenous GAs was performed as described by Li et al. (2017). Three biological replicates and three technical replicates were performed for each sample, and a Student’s t-test was used to determine statistical significance.
RNA isolation and quantitative real-time PCR analysis
Total RNA was extracted from the leaves of 14d-old rice seedlings using an RNA Prep Pure Plant Kit (Tiangen Co., Beijing, China) and was reverse transcribed using a SuperScript II Kit (TaKaRa, Tokyo, Japan). Real-time PCR was performed using one SGExcel FastSYBR qPCR Mixture (Sangon Biotech, Shanghai, China) on an ABI One Step Plus PCR System (Applied Biosystems, Foster City, CA, USA). Each reaction contained 10 µL SGExcel FastSYBR qPCR Mixture, 0.2 µM primer, and 1 µL template cDNA. The PCR reaction parameters were 95°C for 2 min (1 cycle), 95°C for 15 s, and 60°C for 20 s (40 cycles), followed by melting curve analysis at 95°C for 60 s, 55°C for 30 s, and 95°C for 30 s. The rice OsUbiquitin and OsActin genes were used as internal controls (Zheng et al., 2015; Mao et al., 2019). Three biological replicates and three technical replicates were performed for each qRT-PCR analysis. The primer sequences used for qRT-PCR are listed in Table S1.
Transactivation activity assay in yeast
The full-length open reading frames (ORF) of OsDREB2B were inserted into the pBridge vector to form pBridge-OsDREB2B. pBridge-OsRPH1 was used as a positive control, and the empty pBridge vector was used as a negative control (Ma et al., 2020). The resultant constructs were transformed into yeast strain AH109 (Clontech). The positive transformants were verified on triple dropout media (SD/-Trp-Ade-His) and dropped on quatuor dropout media (SD/-Trp-Leu-Ade-His). All protocols were performed according to the manufacturer’s user manual (Clontech, Mountain View, CA, USA).
Subcellular localization of OsDREB2B in rice protoplasts
For the subcellular localization of OsDREB2B, the coding region of OsDREB2B was inserted into the pAN580-GFP vector driven by the CaMV35S promoter to form the OsDREB2B-GFP construct. The D53-RFP fusion protein was used as a nuclear marker (Zhou et al., 2013). Rice protoplast preparation and transformation were performed as previously described by Wang et al. (2016). The transient expression constructs were transformed into rice protoplast cells and incubated in the dark at 28°C for 12–14 h. Fluorescent images were observed using a confocal laser microscope (Carl Zeiss, aJena, Germany).
cDNA library
A rice cDNA library was constructed using total RNA extracted from 2-week-old rice seedlings. The first-strand cDNA was synthesized by Oligo (dT) primers using the SMARTM cDNA Library Construction Kit (Clontech) with 1 μg of total RNA as template and then double-stranded cDNA was synthesized by long-distance PCR (LD PCR). After a quality check, the dscDNA was inserted into the pGADT7-SfiI vector after SfiI digestion and then transformed into an E. coli electro-cell host strain. Six single positive colonies grown on LB medium (Amp 100 µg/mL) were randomly selected to perform PCR amplification with pGADT7 primers: forward, 5’-TAATACGACTCACTATAGG-3’; reverse, 5’-GGCAAAACGATGTATAAATGA-3’. Yeast co-transformed with pGBKT7-53 (+) and pGADT7 served as a positive control, and yeast co-transformed with pGBKT7-Lam (−) and pGADT7 served as a negative control (Chen et al., 2017). The resultant constructs were transformed into yeast strain AH109 (Clontech), and the positive transformants were verified on quatuor dropout media (SD/-Trp-Leu-Ade-His) and dropped on quatuor dropout media (SD/-Trp-Leu-Ade-His+X-gal).
Yeast-two hybrid assay
The cDNA fragment encoding OsDREB2B was amplified and cloned into a pGBKT7 vector to form pGBKT7-OsDREB2B as the ‘bait’ using a gateway recombination system. A cDNA library prepared from rice seedlings was used to perform the Y2H screening, and positive clones were identified by DNA sequencing. The coding region of OsWRKY21 was inserted into the pGADT7 vector as the ‘prey’. The ‘bait’ and ‘prey’ constructs were co-transformed into yeast strain AH109 (Clontech). These positive transformants were verified on triple dropout media (SD/-Trp-Leu-His) and then dropped on quatuor dropout media (SD/-Trp-Leu-Ade-His). All protocols were performed according to the manufacturer’s user manual (Clontech Laboratories).
Bimolecular fluorescence complementation assays
The cDNA fragment encoding OsWRKY21 was amplified and cloned into a pXY105 vector carrying the N-terminal half of YFP (nYFP) under the control of a CaMV35S promoter to form nYFP-OsWRKY21, and cDNA-encoding OsDREB2B was cloned into the pxy 103 vector carrying the C-terminal half of YFP (cYFP) under the control of a CaMV35S promoter to form cYFP-OsDREB2B. nYFP-OsWRKY21 and cYFP-OsDREB2B were co-transformed into the rice protoplasts. The D53-RFP fusion protein was used as a nuclear marker. The transformed rice protoplasts were incubated at 23°C in the dark for 12–14 h. Fluorescent images were observed using Axio Observer D1 (Carl Zeiss, Jena, Germany).
Yeast one-hybrid assays
The full-length CDS of OsDREB2B was amplified and cloned into pJG4-5 vector to form pJG4-5-OsDREB2B, and the OsAP2-39 promoter (~1500 bp) was cloned into reporter vector pLacZi-2µ. pJG4-5-OsDREB2B and OsAP2-39pro:LacZ were then co-transformed with the fusion protein into yeast strain EGY48 (Clontech). The transformants were cultured on SD/Trp-Ura-plates at 30°C for 3–4 days. DNA sequencing was used to identify putative positive clones. A single positive colony was streaked on the SD/Trp-Ura chromogenic plate and incubated at 30°C for 3 days (Lin et al., 2007).
Dual luciferase reporter assay
The OsAP2-39 promoter (~1500 bp) was inserted into vector pGreen II 0800 containing the luciferase reporter gene (LUC) to generate a reporter vector, and the CDS of OsDREB2B was inserted into the Flag vector to generate the effector vector. Subsequently, the reporter vector was co-transformed via PEG into rice protoplasts together with an empty Flag vector (negative control) or the effector vector (Flag-OsDREB2B) (Yoo et al., 2007). After culturing at 28°C for 12–14 hours, LUC and reniral siferase (REN) activity levels of rice protoplasts were measured using the dual luciferase reporter assay system (E1910, Promega). The transient expression of LUC driven by the OsAP2-39 promoter was normalized to the expression of the internal control reporter (REN).
Results
Tissue-specific and GA-responsive expression profiles of OsDREB2B
In order to elucidate whether the biological function of OsDREB2B (LOC_Os05g27930) is related to GA, we examined the tissue-specific and GA-responsive expression of OsDREB2B by using qRT-PCR. As shown in Figure 1A, the transcript of OsDREB2B was expressed in many organs, such as the leaf sheath, young leaf, stem, flag leaf, unopened spike and opened spike. Notably, it was significantly highly expressed in the leaf sheaths, reaching at least eight times the unopened spike, while weak expression was observed in other organs (Figure 1A). GA is a hormone that promotes the growth and development of rice, mainly regulating the elongation of leaf sheaths and internodes. To analyze the GA responsive expression of OsDREB2B, WT rice treated with different concentrations of exogenous GA3, and then its leaf sheaths and leaves were collected at different time points for qRT-PCR. With the increase of GA3 concentration, the transcription level of OsDREB2B increased: at 100 μM, it rapidly changed 2.9-fold within 1 h (Figures 1B–D). Therefore, we suggest that OsDREB2B may be involved in GA-regulated biological processes, probably in plant growth and development.
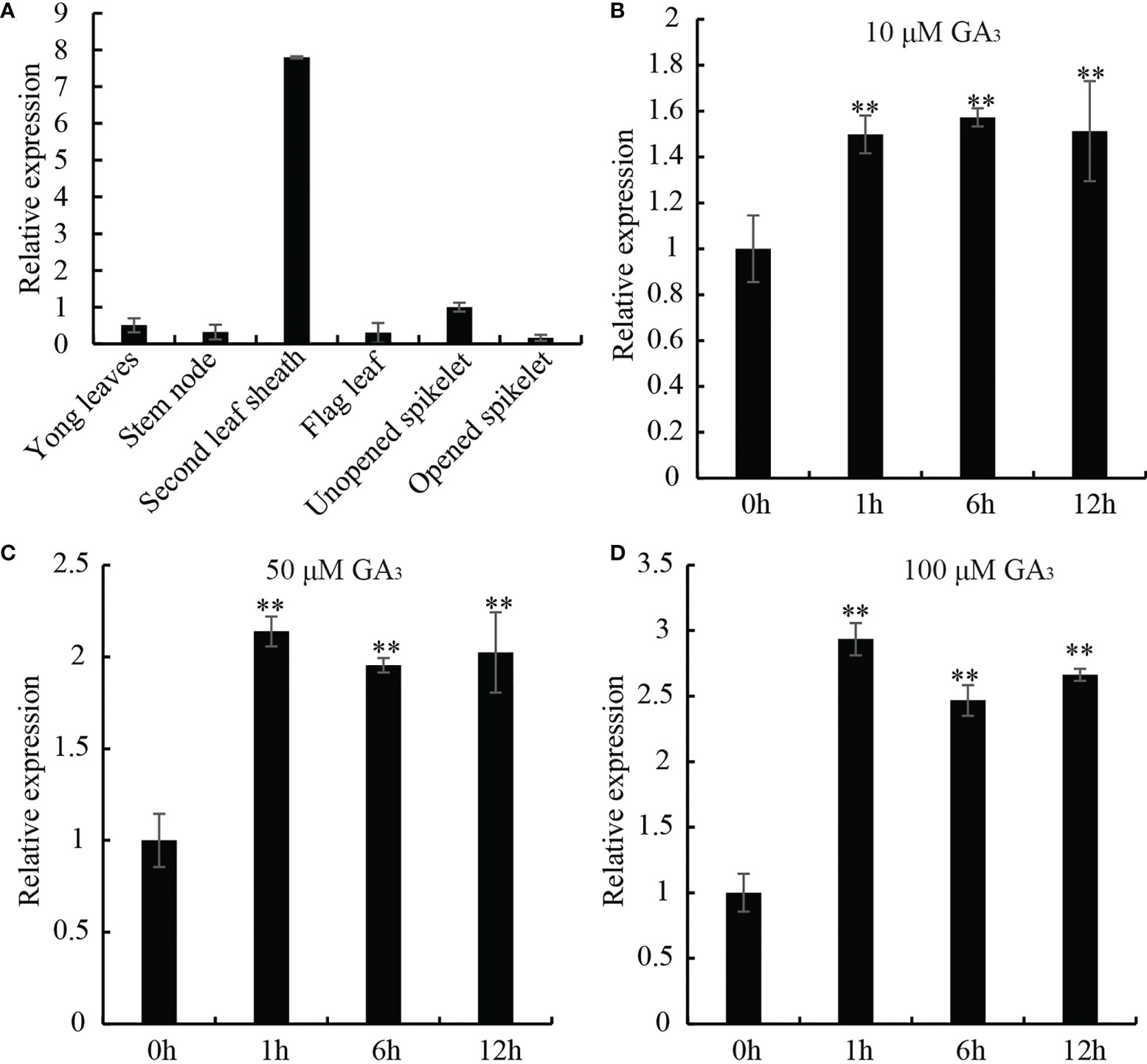
Figure 1 Expression pattern of OsDREB2B in rice. (A) Transcript levels of OsDREB2B in various tissues and organs tested by qRT-PCR. Transcript level of unopened spikelet was set to 1. Data are the mean ± SD (n = 3). (B–D) Expression profiles of OsDREB2B in response to different concentrations of GA3 application analyzed by qRT-PCR. Student’s t-test was used to generate the P values. **Significant differences between the WT and OE plants at P = 0.01.
Overexpressing of OsDREB2B in rice has a dwarf phenotype
OsDREB2B overexpressing and knock-out rice were constructed to elucidate its role in plant growth and development. Plant overexpression vector pUbi::OsDREB2B was constructed and transformed into the recipient rice variety ‘Kitaake’ (Figure 2A). Through phenotype evaluation, we screened OE lines that exhibited a dwarf phenotype compared to the WT (Kitaake). We obtained 14 independent T0 generations of OsDREB2B-OE lines, and the expression levels of the transgene were assessed (Figure 2B). We selected two lines showing high expression levels of the transgene (OE-1 and OE-2), and their T3 homozygous lines were selected for further study (Figure 2C). Both OE lines showed dwarf phenotype, with significantly lower plant height, internode length and seed length than WT (Figures 2D–K). In addition, many yield component factors, such as panicle number per plant, number of seeds per panicle and seed setting rate, were also significantly reduced (Table S2). At the seedling stage, the average lengths of the second sheath of OE-1 (3.07 cm) and OE-2 (2.95 cm) were only 70.9% and 68.1% of the WT (4.33 cm), respectively (Figures 2D, E). At maturity, the average plant heights of OE-1 (68.76 cm) and OE-2 (64.16 cm) were only 81.3% and 75.9% of the WT (84.56 cm), respectively (Figures 2F, G). A comparison of internode length between the OE lines and the WT showed that four internodes (first, second, third, and fourth) were shorter in the OE lines than in the WT (Figures 2H, I). These results suggest that overexpression of OsDREB2B has a negative regulatory effect on rice plant height.
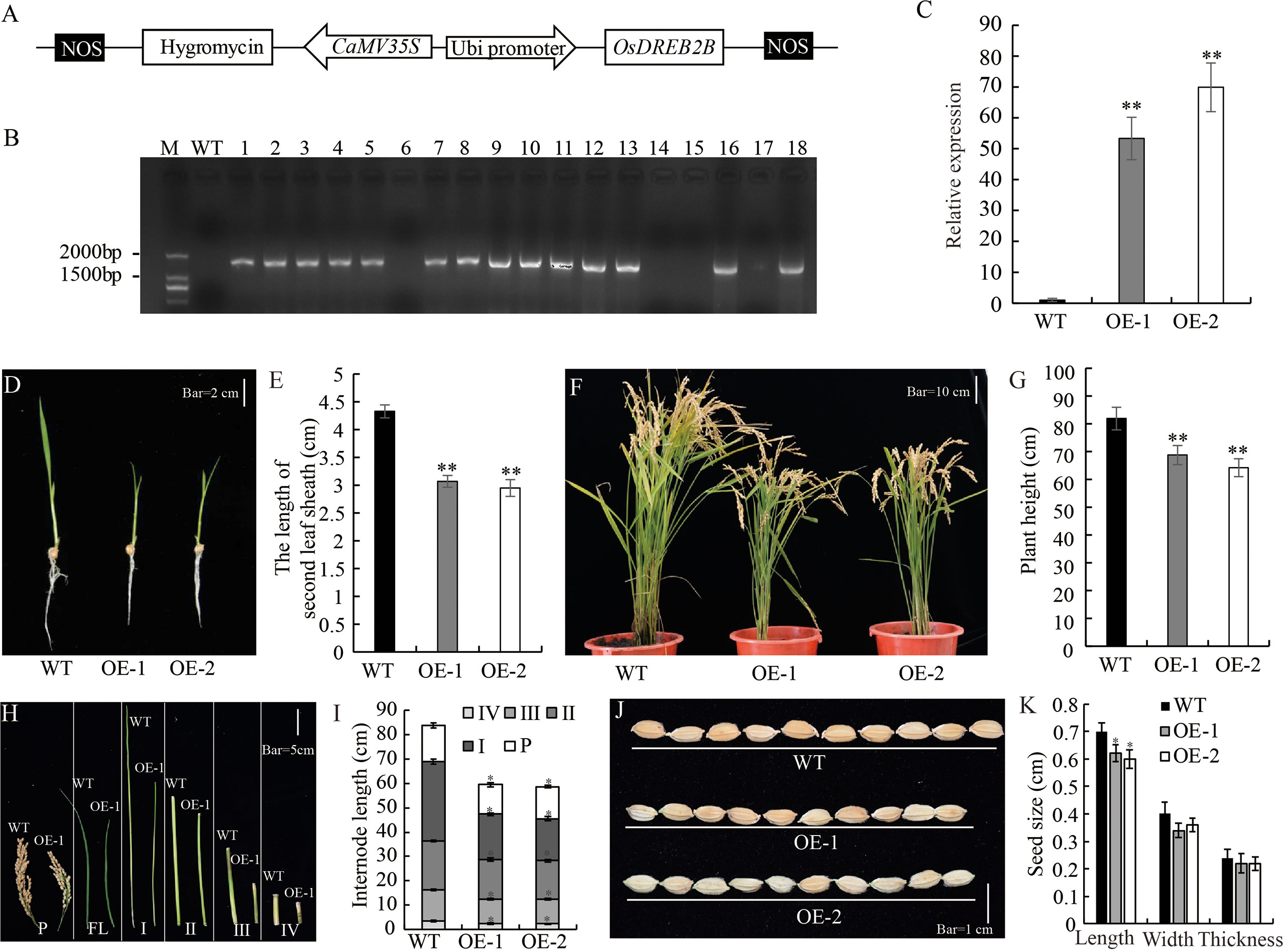
Figure 2 Development and morphology of OsDREB2B-OE rice. (A) Schematic representation of OsDREB2B plant expression vector. OsDREB2B was expressed under the control of maize ubiquitin (Ubi) promoter, and the hygromycin (hgy) gene was under the control of 35S promoter as a selection marker in rice. (B) PCR analyses for T0 transgenic rice using construct-specific primers. M, marker; WT, Kitaake; 1~18, independent transformants of T0 generation. (C) The expression of OsDREB2B were analyzed by qRT-PCR in WT and DE. The data was presented as the mean ± SE values (n = 3) from two independent experiments. (D, E) Whole-plant morphology of the OE lines and the WT at the seedling stage. Bar = 2 cm. (F, G) Whole plant morphology of OsDREB2B-OE plants and the WT at maturity. Bar = 10 cm. (H) Panicles and internodes of the main culms of OE-1 (right) and the WT (left). Bar = 5 cm. (I) Internode lengths of OE lines and the WT. (J) Seed morphology. Bar = 1 cm. (K) Seed size of the OE lines and the WT. Single asterisk indicates significant differences at P ≤ 0.05 compared with the WT and double asterisks indicate significant differences at P ≤ 0.01 compared with the WT in Student’s t-test.
To further test the role of OsDREB2B in plant growth regulation, we generated OsDREB2B-knockout mutants (osdreb2b) using the CRISPR/Cas9 system by OsDREB2B genome editing in the WT. By DNA sequencing, two independent knockout lines, mt-1 and mt-2, were selected which were deleted with one or two nucleotides in the second exon of OsDREB2B, respectively (Figures 3A, B). The length of the second sheath of mt-1 and mt-2 seedlings was similar to that of the WT in 10 d-old seedlings (Figures 3C, D). In addition, the length, width, and thickness of the seeds of mt-1 and mt-2 were similar to those of the WT (Figures 3E, F). These results suggest that repression of OsDREB2B has no effect on rice growth and development.
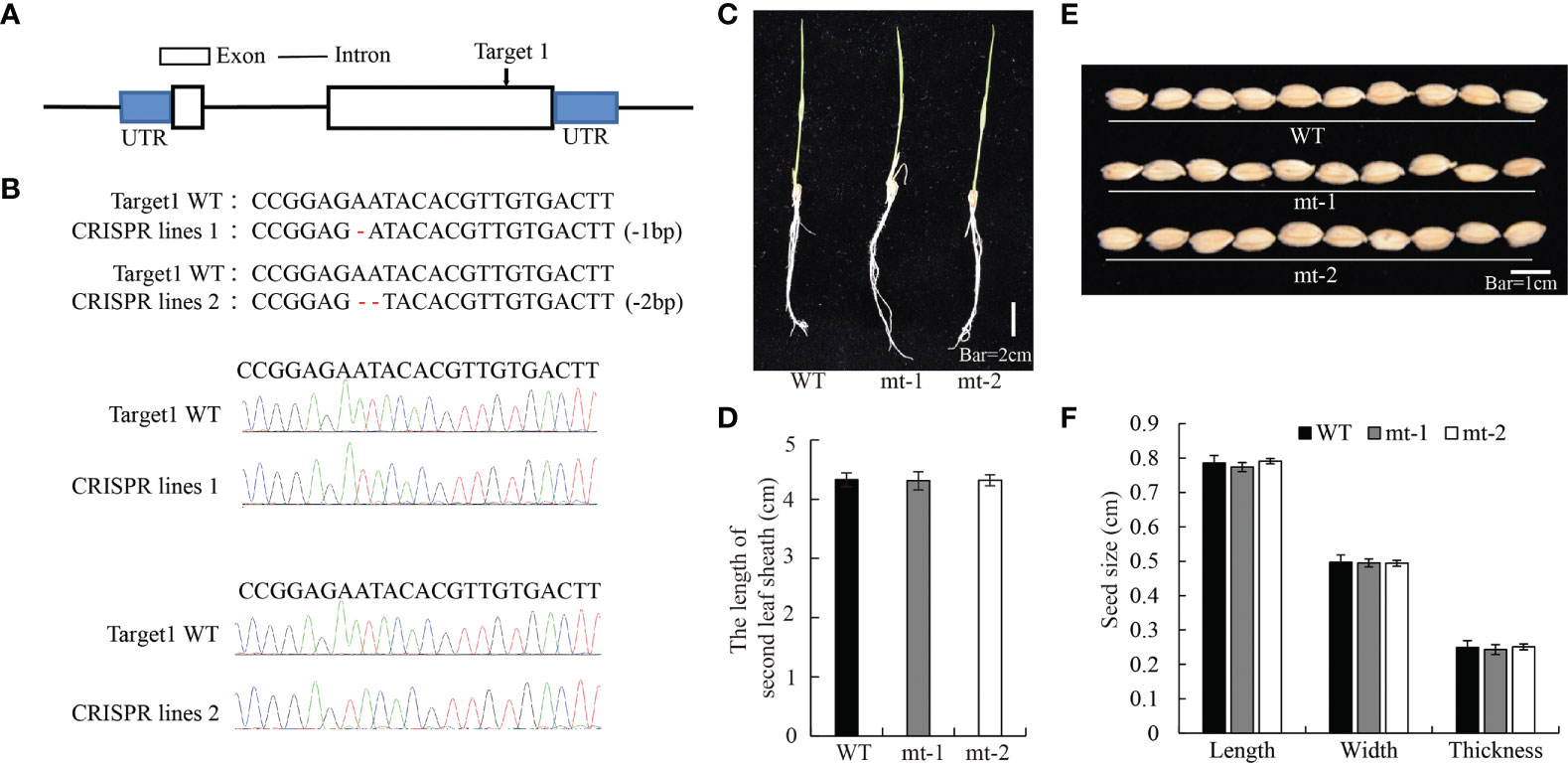
Figure 3 Development and morphology of OsDREB2B-mt rice. (A, B) Osdreb2b mutants were produced in rice using a cluster of regularly spaced short palindrome repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system. (C) Whole-plant morphology of the mt lines and the WT at the seedling stage. Bar = 2 cm. (D) The second leaf sheath length of OE lines and the WT. (E) Seed morphology. Bar = 1 cm. (F) Seed size of the OE lines and the WT.
Dwarf phenotype of OsDREB2B-OE seedlings is rescued by exogenous GA3 application in rice
GA-deficient rice was dwarfed, and the short length of the second leaf sheath was rescued by exogenous GA3 application (Spielmeyer et al., 2002; Ma et al., 2020). To elucidate whether the dwarf phenotype of the OsDREB2B-OE lines was due to reduced endogenous GA content, the seedling growth of OE lines and WT rice was investigated under exogenous GA3 treatment. 10 d-old seedlings were treated with different concentrations of GA3 (0, 10, 50, and 100 μM) at different time points (Figure 4). The elongation rate of the second leaf sheath increased with the increase of GA3 concentration, and it was faster in the OE lines than in the WT. At the concentration of 100 μM, the length of the second leaf sheath of the OE lines was almost as long as that of the WT plants within 48 h after treatment (Figure 4D). In addition, exogenous GA3 was also applied in the mt lines, but there was no significant difference with the WT (data not shown). Therefore, exogenous GA3 application could completely restore the shortened second leaf sheath of the OE lines, supporting the idea that dwarfism in OE lines was not caused by blocking the GA signaling pathway but might be caused by a deficiency in endogenous GA content.
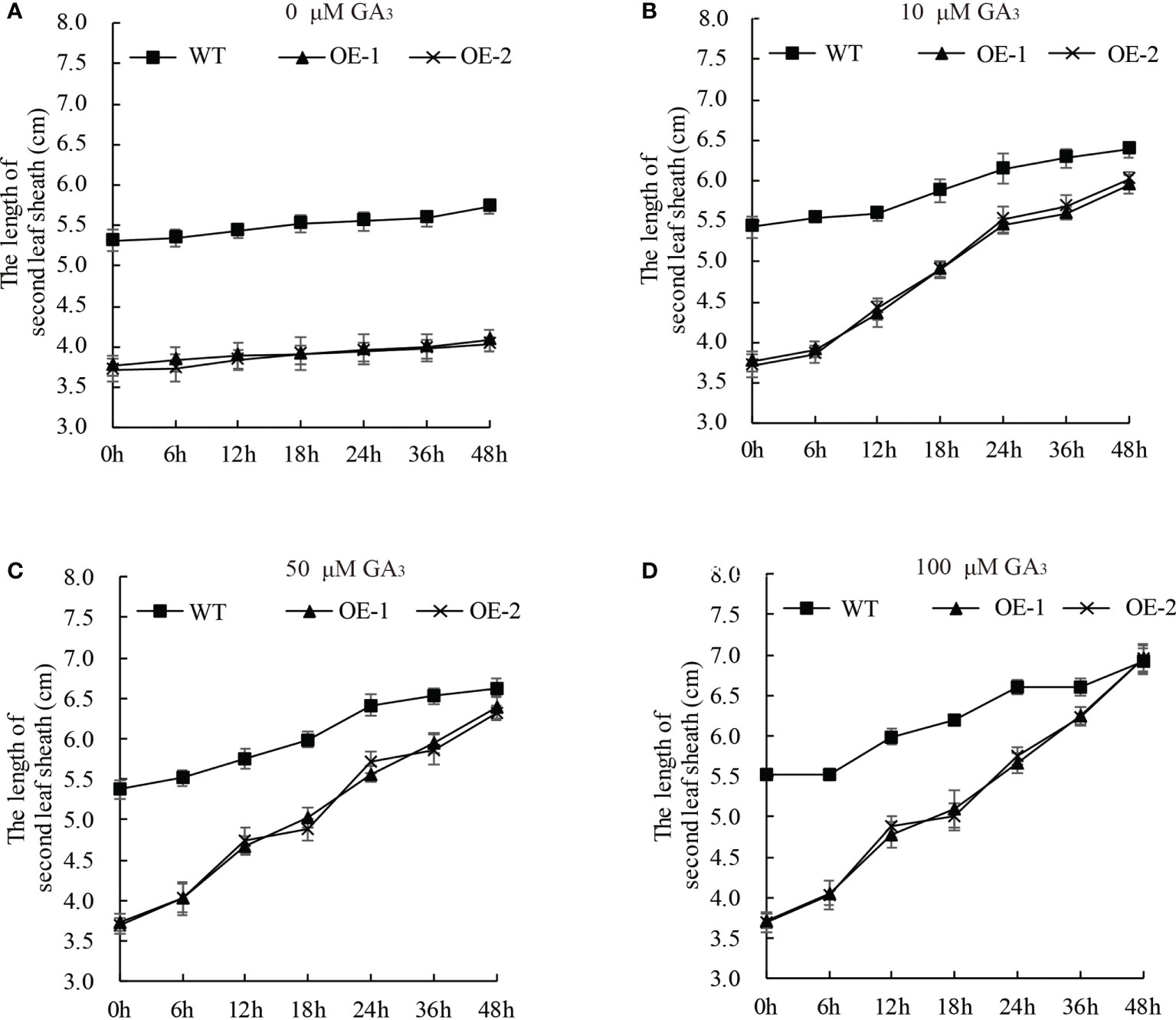
Figure 4 The dwarf phenotype of the OE plants can be rescued by GA3. Ten-day-old OE and WT seedlings were treated with 0 (A), 10 (B), 50 (C), and 100 (D) μM GA3. At least 10 rice seedlings from each individual line were measured. Data are presented as the average of 10 samples per genotype ( ± SD).
Bioactive GA content is reduced in OsDREB2B-OE rice
To confirm our hypothesis, we quantified the endogenous GA content in 10 d-old seedlings of OE lines and WT by high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) analysis. The content and distribution patterns of 15 GA species were analyzed, including active forms, as well as their metabolites and precursors (Figure 5A). Comparing the GA content between OE and WT: in the GA12-pathway, four GA species, including bioactive GA4 and its precursors (GA15, GA24, and GA9) decreased, while GA51 (the catabolite of GA9) increased, and GA12 (the first precursor of bioactive GA4) and GA34 (the catabolite of GA4) were not detected; in the GA53-pathway, five GA species, including bioactive GA1 and GA3, as well as their precursors (GA44, GA19, and GA20), decreased; however, GA53 (the first precursor of bioactive GA1) and two catabolites, GA29 and GA8, increased (Figures 5A, B).
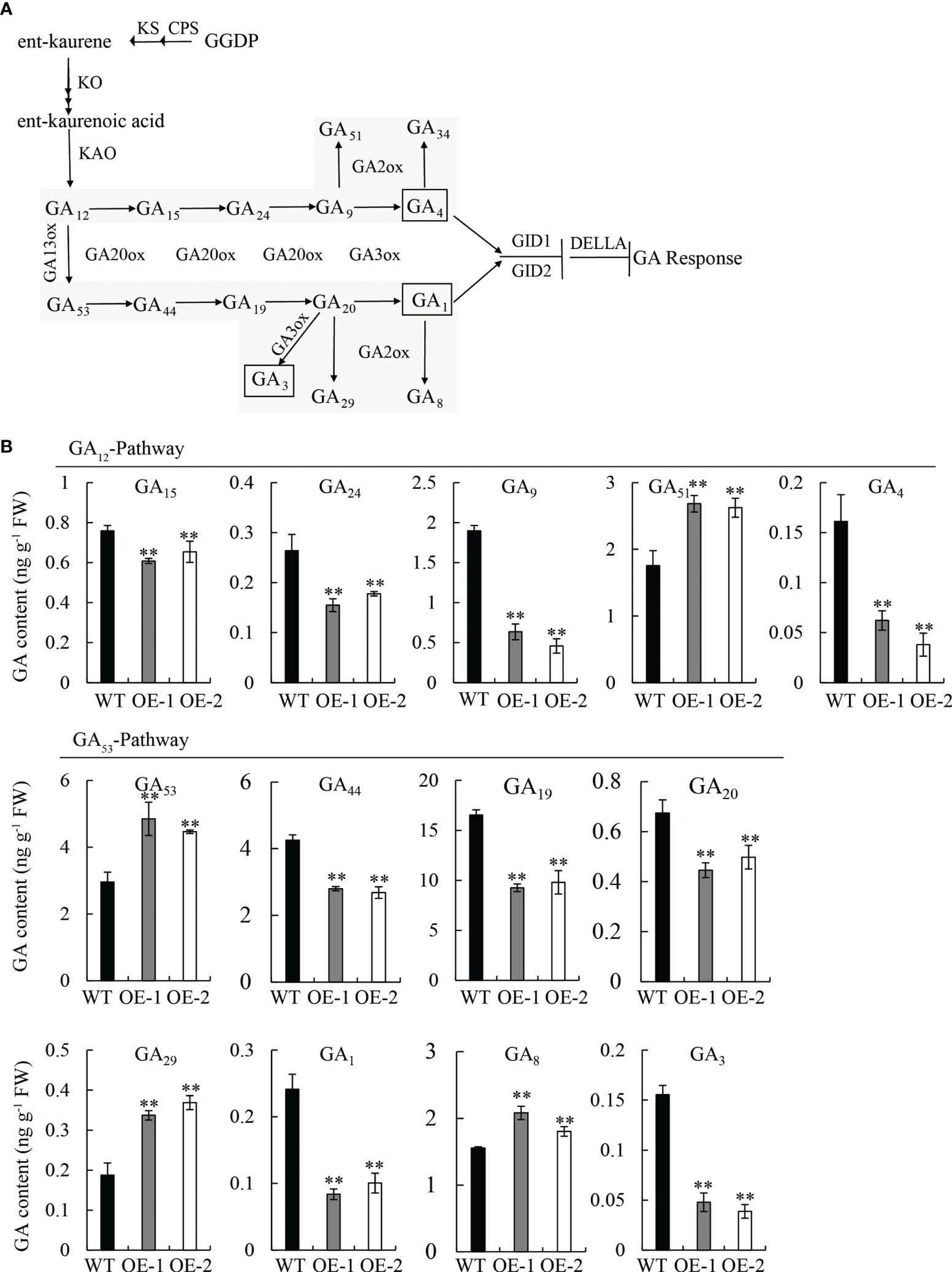
Figure 5 Measurement of endogenous GA in seedlings of the WT and OE lines. (A) Comparison of major gibberellin (GA) biosynthetic pathways in plants. (B) Content of GA species in the GA12-pathway and GA53-pathway. FW, fresh weight. Student’s t-tests were used to determine statistical significance. FW, fresh weight; error bars indicate ± SE (n = 3). Double asterisks indicate significant differences at P ≤ 0.01 compared with the WT in Student’s t-test.
Strikingly, the content of the three bioactive GAs (GA1, GA3, and GA4) was significantly reduced (P ≤ 0.01) in the OE lines compared to the WT, exhibiting a reduction of 62.5, 75, and 68.7%, respectively (Figure 5B); however, no significant difference was observed between mutant lines (mt-1 and mt-2) and the WT (Figure S2). The overexpression of OsDREB2B led to a decrease in endogenous bioactive GA content, but its repression had no effect on GA content, indicating that OsDREB2B negatively regulates GA content.
OsDREB2B downregulates GA biosynthesis genes but upregulates GA deactivation genes
To confirm the involvement of OsDREB2B as a negative regulator in GA content, the transcription levels of most enzymes that catalyze the early steps and the late steps of GA biosynthesis, as well as GA signaling factors, were examined via qRT-PCR in 10 d-old seedlings of OE lines and WT. Interestingly, the expression level of several genes in the OE lines, such as OsCPS1, OsKS, OsKO2, and OsKAO in the early GA biosynthesis step, was significantly (P ≤ 0.01) downregulated, which decreased by 44.4, 51.8, 55.5, and 83.7%, respectively, compared to the WT (Figures 5A, 6A). Similarly, the expression level of the late-step GA biosynthesis genes, including OsGA20ox1–OsGA20ox2 and OsGA3ox1–OsGA3ox2, were significantly (P ≤ 0.01) downregulated in OE lines compared to the WT (Figures 5A, 6B). In contrast, the expression levels of GA deactivation-related genes, such as OsEUI (Figure 6A), OsGA2ox1, OsGA2ox3-6, OsGA2ox8, and OsGA2ox9, were significantly (P ≤ 0.01) upregulated in OE compared to the WT (Figures 5A, 6C).
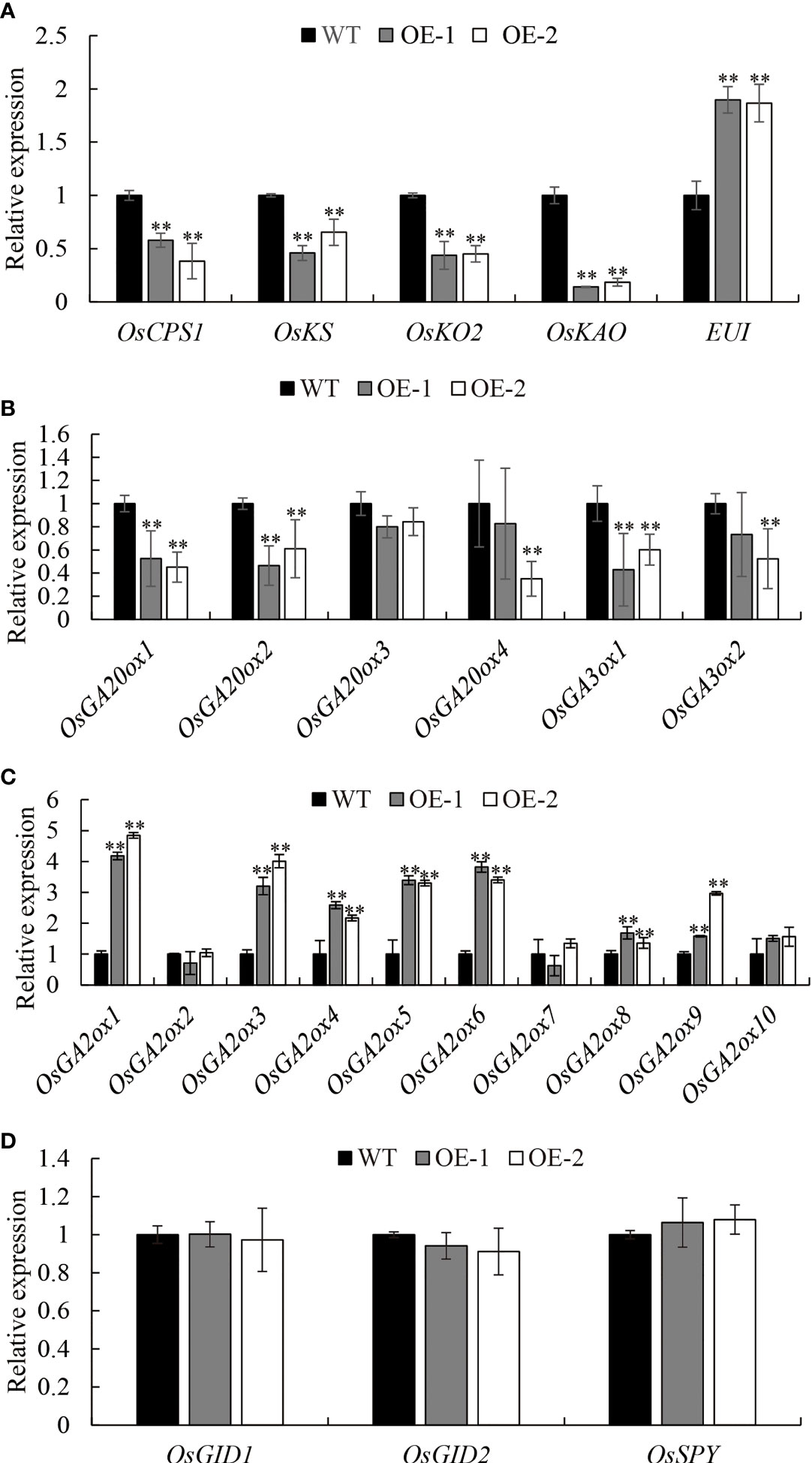
Figure 6 Expression analysis of GA metabolic (A–C) and signal genes (D). Transcript levels from the WT were set to 1. Data are the mean ± SD (n = 3). Student’s t-tests were used to generate the P values. **Significant differences between the WT and OE lines at P = 0.01.
In addition, the transcript of GA signaling factors OsGID1, OsGID2, and OsSPY in the OE lines did not show significant differential expression compared to the WT (Figure 6D), which means that the GA signaling pathway was not blocked by OsDREB2B. This is consistent with the phenomenon of short leaf sheath rescue after exogenous GA3 treatment (Figure 4). Taken together, OsDREB2B negatively regulates bioactive GA content not only by downregulating GA biosynthetic genes, but also by upregulating GA deactivation genes, instead of blocking GA signaling factors.
OsDREB2B functions as a transcriptional activator
Transcription factors regulate downstream gene expression by binding to cis-elements on genomic DNA sequences in the nucleus. To determine whether the OsDREB2B protein has trans-activation activity, it was fused to the GAL4-DNA binding domain in the pBridge vector and then expressed in yeast. The transformants of pBridge-OsDREB2B and the positive control pBridge-OsRPH1 grew well on quatuor dropout media (SD/-Trp-Leu-Ade-His), while the negative control pBridge failed to grow (Figure 7A). OsDREB2B fusion with the GAL4 DNA-binding domain of pBridge effectively activated transcription in yeast cells. Compared to the negative control, pBridge-OsDREB2B still grew normally on 75 mM of 3-AT after being transferred to the defective media (SD/-Trp-Leu-Ade-His) containing different concentrations of 3-amino-1, 2, 4-triazole (3-AT) and incubated at 30°C for 3 days (Figure S3A). These results indicate that OsDREB2B acts as a transcriptional activator in yeast.
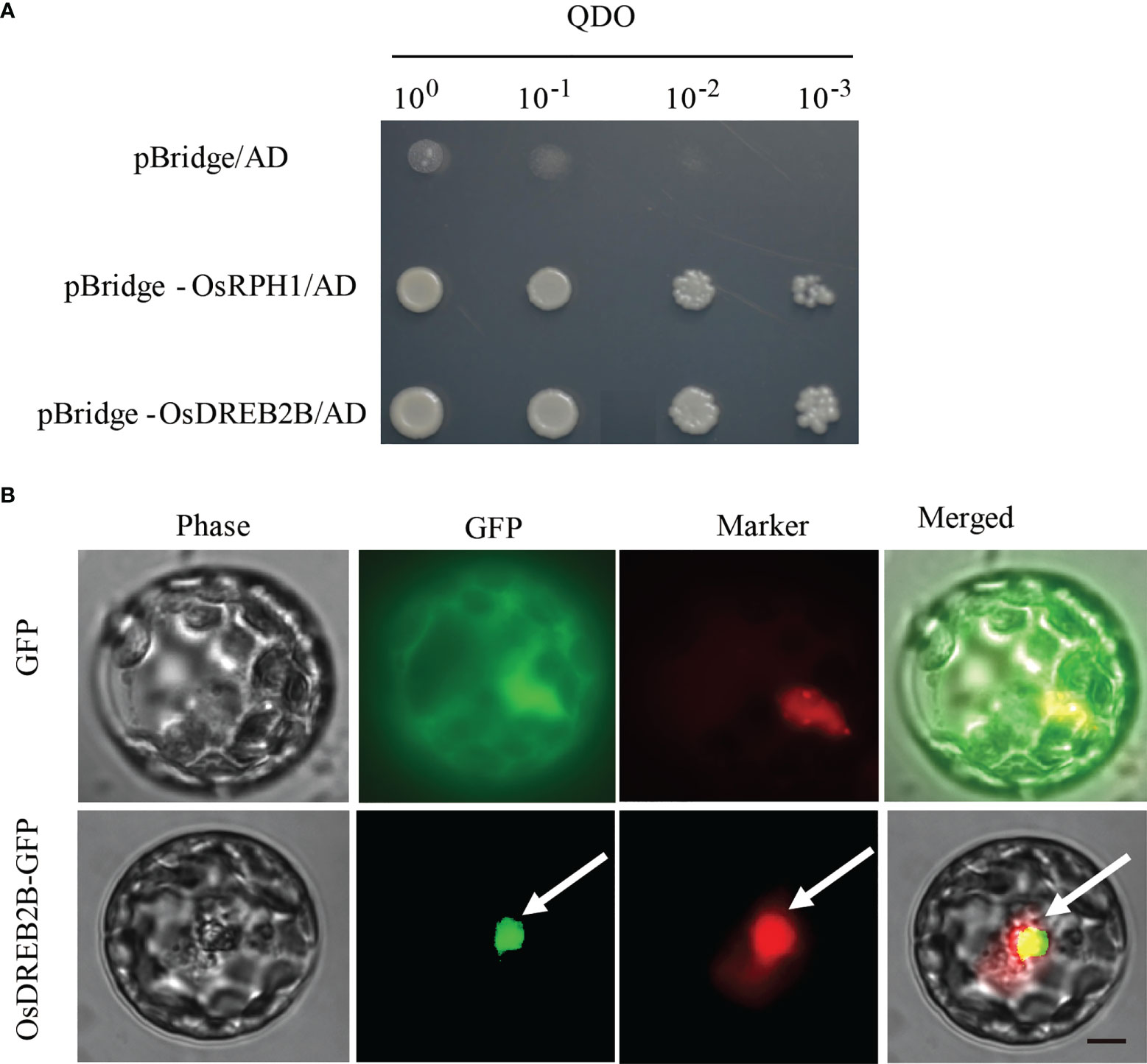
Figure 7 Trans-activation activity and subcellular localization of OsDREB2B. (A) Trans-activation analysis of the OsDREB2B protein in yeast. BD, GAL4-DNA binding domain. The OsRPH1 protein was used as a positive control. (B) Subcellular localization of OsDREB2B-GFP fusion proteins in rice protoplast cells. D53-RFP was used as a nuclear marker. Bar = 10 µm.
To verify the subcellular localization pattern of OsDREB2B, an OsDREB2B-GFP fusion protein under the control of the CAMV35S promoter (p35S::OsDREB2B-GFP) was constructed and then transiently expressed in rice protoplasts. As shown in Figure 7B, the control smGFP was uniformly distributed throughout the cell, while the OsDREB2B-GFP fusion protein was exclusively localized to the nucleus. Therefore, we suggest that OsDREB2B functions as a transcriptional activator in rice.
OsDREB2B upregulates the expression of OsAP2-39 by directly binding to its promoter
The transcript expression level of OsAP2-39 was significantly upregulated in the OsDREB2B-OE lines compared to the WT (Figure 8A). In a promoter assay using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), OsAP2-39 has a DRE/CRT cis-element in the promoter region, however, OsEUI does not have any DRE/CRT or GCC-box in the promoter region (Table S4). Therefore, we speculated that OsDREB2B directly binds to OsAP2-39 promoter region to activate its expression, excluding the possibility that OsEUI may be a direct downstream target of OsDREB2b.
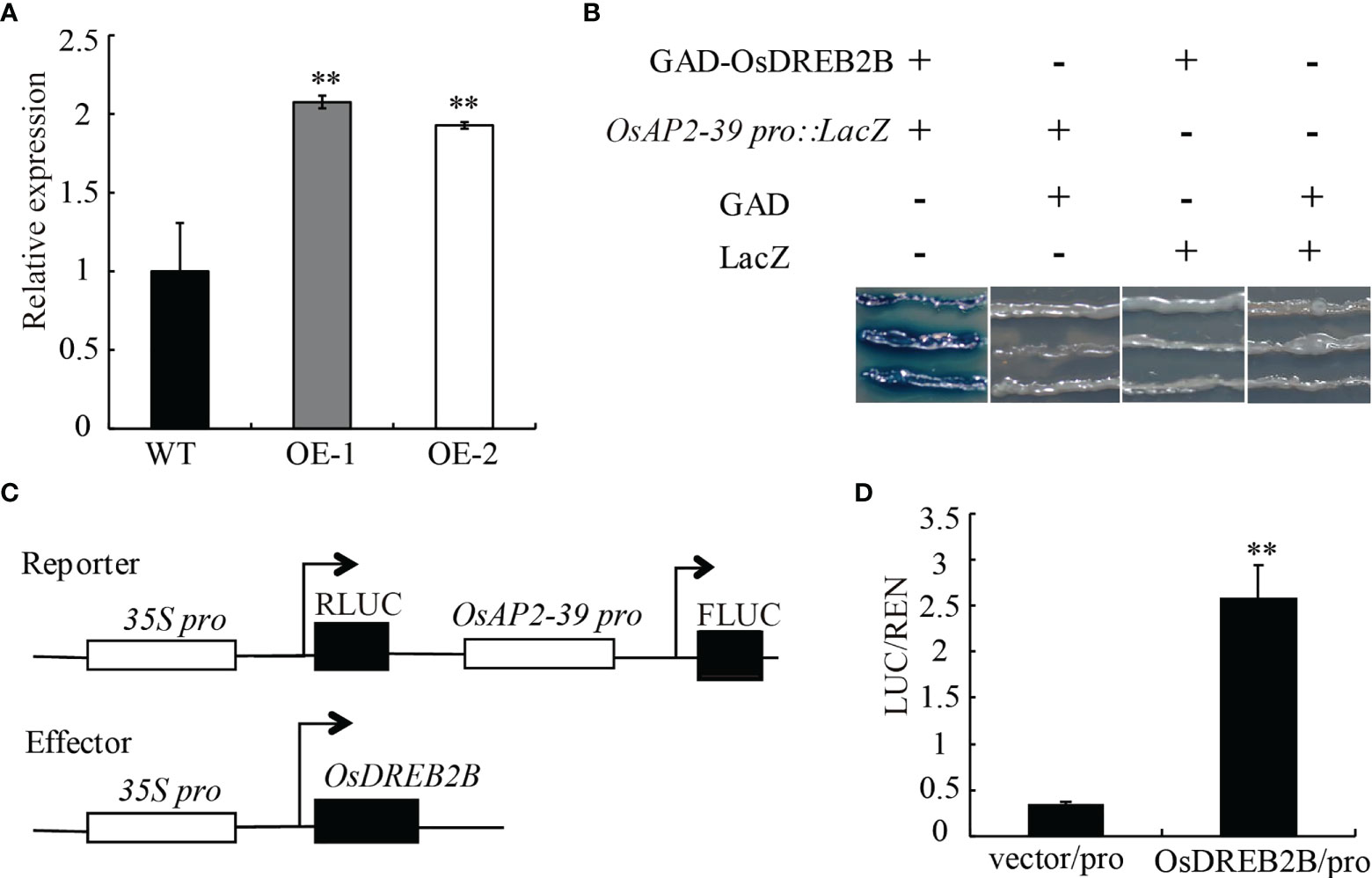
Figure 8 Direct regulation of OsAP2-39 by OsDREB2B. (A) Relative expression of the OsAP2-39 gene in the WT and OE lines. (B) Combination of OsDREB2B and OsAP2-39 in yeast. GAD-OsDREB2B activates the expression of LacZ reporter genes driven by OsAP2-39 promoters in yeast. GAD, GAL4 transcriptional activation domain. (C) Schematic diagram of the carrier in the dual luciferase reporter system. (D) In vivo dual luciferase reporter assay. Flag-OsDREB2B and OsAP2-39pro:FLUC, Flag, and OsAP2-39pro:FLUC were co-transformed into rice protoplasts and incubated at 28°C for 12–14 (h) FLuc and RLuc activities were measured, and Renilla luciferase (RLuc) was used as an internal control. Student’s t-tests were used to generate the P values. **Significant differences between the WT and OE plants at P = 0.01.
To confirm the in vivo binding of OsDREB2B to the OsAP2-39 promoter, yeast one-hybrid (Y1H) and dual-luciferase assays were performed. In the Y1H assays, the full-length DREB2B protein was fused to the GAL4 transcriptional activation domain (GAD) and the OsAP2-39 promoter was fused to the LacZ reporter genes. A blue color reaction occurred when GAD-OsDREB2B and OsAP2-39Pro : LacZ were co-transformed into yeast cells (Figure 8B). In dual-luciferase assays, the OsAP2-39 promoter was fused to the luciferase (LUC) reporter gene, and OsDREB2B was driven by the 35S as the effector (Figure 8C). When co-transfected with OsAP2-39pro:FLUC in rice protoplasts, the relative FLuc/RLuc activity ratios harboring Flag-OsDREB2B were significantly higher than that when co-transfected with an empty vector (Figure 8D). These results revealed that OsDREB2B directly binds to the promoter of OsAP2-39 and activates its expression.
OsDREB2B interacts with OsWRKY21 in vivo
Some AP2/ERF transcription factors interact with various proteins to exert redundant functions. To identify the potential interactive protein of OsDREB2B, yeast two-hybrid (Y2H) screening was performed using a rice cDNA library. Autoactivation activity verification of three truncated bait vectors (full length, N-terminal, and C-terminal of OsDREB2B) showed that the C-terminus was required for the autoactivation activity of OsDREB2B. Therefore, the truncated version of OsDREB2B-N without the C-terminus was used for screening (Figure S3).
In total, 49 proteins were screened as potential proteins that interacted with OsDREB2B-N (Table S3). Most proteins grew normally on the four-deficiency media (SD/-Trp-Leu-Ade-His) and 7 of them had deep blue color reactions (Figure S4). Among these, a protein of interest, OsWRKY21 (Serial No. was N-7 in Table S3 and No.7 in Figure S4), which is involved in rice growth and development, was identified.
To further verify the physical interaction between OsDREB2B and OsWRKY21, a one-on-one Y2H assay was performed. As shown in Figure 9A, OsDREB2B-pGBKT7 and AD-OsWRKY21 interacted when co-transformed into yeast. In addition, the interaction between OsDREB2B and OsWRKY21 was verified by the bimolecular fluorescence complementation (BiFC) assay in rice protoplast. Interestingly, the fluorescence signal of YFP was evidently detected in the nucleus of the protoplast when OsDREB2B-nYFP and OsWRKY21-cYFP were co-transfected, whereas no signal was detected in the co-transfection of OsDREB2B-nYFP and cYFP or OsWRKY21-cYFP and nYFP (Figure 9B). Therefore, OsDREB2B physically interacted with OsWRKY21 in yeast and in the nucleus of the rice protoplast.
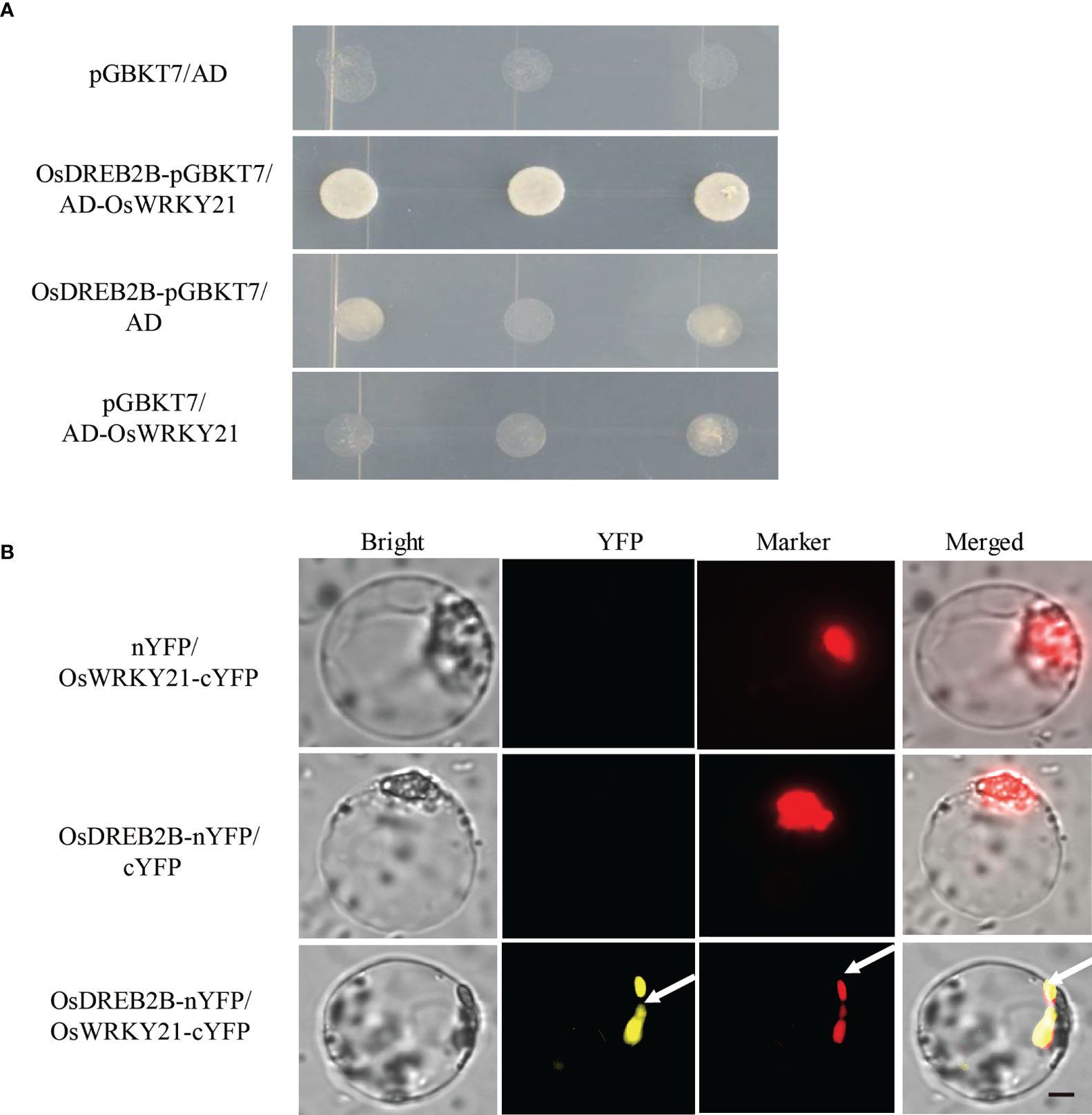
Figure 9 OsDREB2B physically interacts with OsWRKY21 in yeast and plant cells. (A) The interaction of OsDREB2B and OsWRKY21 was detected in yeast cells. AD, activation domain; BD, GAL4-DNA binding domain. (B) BiFC was used to detect the interaction of OsDREB2B and OsWRKY21 in rice protoplasts. Bright, bright field; YFP, YFP fluorescence; Marker, marker fluorescence. Bar = 10 µm.
Discussion
AP2/ERF transcription factor OsDREB2B belongs to the DREB subgroup, which is reported to respond to abiotic stress, such as drought, heat stress, and cold in Arabidopsis or rice (Chen et al., 2008; Matsukura et al., 2010; Sakata et al., 2014). However, little is known about the molecular mechanism of OsDREB2B in plant growth and development in rice. In this study, OsDREB2B acted as a dual regulator of GA metabolic gene expression mediated by OsAP2-39 and OsWRKY21, which reduced GA content and led to the reduction of plant height in rice.
GA is a hormone that promotes the growth and development of rice, mainly regulating the elongation of leaf sheaths and internodes. In our study, the OE lines exhibited dwarf phenotypes, which were mainly manifested in the shortening of leaf sheaths and internode length (Figures 2, S1), while the phenotypes of the mt lines were similar to the WT (Figure 3). These observations revealed that overexpression of OsDREB2B disturbs many aspects of plant growth and development, while its inhibition has no effect on the growth of rice.
In addition, exogenous GA3 application significantly promoted rice leaf sheath elongation and affected the transcription of OsDREB2B (Figures 1, 4). This phenomenon was similar to that of GA-deficient mutants (Ashikari et al., 2002; Gomi et al., 2004; Sakamoto et al., 2004). Furthermore, the expression of many GA metabolic genes was changed, while that of GA signaling genes showed no difference between the OE lines and the WT (Figures 5A, 6). These results revealed that OsDREB2B is involved in GA metabolism related to plant growth retardation and not the GA signaling pathway.
Among GA metabolic genes, GA20ox and GA3ox convert GA precursors into bioactive GAs through a cascade of catalytic oxidation reactions (Hedden and Thomas, 2012), while GA2ox is recognized as critical for GA deactivation (Thomas et al., 1999). The rice dwarf mutant d18 and the green-revolution variety, semi-dwarf 1 (sd1), are resulted from mutation in GA biosynthesis genes, OsGA3ox2 and GA20ox2, respectively (Itoh et al., 2001; Spielmeyer et al., 2002). Mutations in GA20ox and GA3ox in maize (dwarf1) and barley (sdw1/denso) (Spray et al., 1996; Jia et al., 2015) and overexpression of deactivation genes (OsGA2ox1, OsGA2ox6, and OsGA2ox9) in rice (Sakamoto et al., 2001; Sasaki et al., 2003; Lo et al., 2008; Huang et al., 2010) caused moderate plant height reductions by decreasing the GA content. These circumstances occurred simultaneously in our OsDREB2B-OE rice, which is similar to our previously identified AP2/ERF transcription factor, OsRPH1 (Ma et al., 2020). The bioactive GA content in the OE lines was significantly (P ≤ 0.01) lower than that of the WT (Figure 5), while that in the mt lines was not (Figure S2). Moreover, compared to the WT, most of the bioactive GA precursors were also decreased, while GA53 and some inactive catabolites were increased in the OE lines (Figure 5). The downregulation of most GA20ox and GA3ox may lead to the accumulation of GA53, the first precursor of GA1, while the upregulation of GA2ox may lead to the increase of three catabolites, such as GA29, GA8 and GA51 (Figures 5, 6). The altered expression of GA metabolic genes in OE lines agreed with the results of a decrease in bioactive GA content, showing that the GA biosynthesis genes were downregulated and GA deactivation genes were upregulated. These results indicate that OsDREB2B, as a dual regulator of GA metabolic genes, negatively regulates the expression of GA synthetic genes, while it positively regulates GA deactivation genes, resulting in a decrease in bioactive GAs in the rice.
AP2/ERF proteins have been shown to be integrators of biotic and abiotic stress responses by combining with cis-acting elements in the promoter region of the target genes, such as ethylene-response elements (GCC-box, core motif:A/GCCGCC) and dehydration-responsive element/C-repeats (DRE/CRT, core motif: G/ACCGAC) (Allen et al., 1998). DREB subgroup proteins contain an AP2-conserved domain that specifically binds to the DRE/CRT cis-acting elements of its regulatory gene (Shi et al., 2018). Overexpression of a DREB gene in tomato, SlDREB, affects GA biosynthesis by downregulating the key genes involved in GA biosynthesis, thus limiting internode elongation and leading to dwarfing. SlDREB likely acts as a direct repressor of SlCPS in plants by binding to the DRE/CRT elements of SlCPS (Li et al., 2012). Therefore, we hypothesized that OsDREB2B may have functions similar to those of SlDREB. OsAP2-39 is an AP2/ERF protein belonging to the ERF subgroup that could strongly bind to the GCC box in the promoter of its target GA deactivation gene OsEUI, an enzyme that catalyzes 16α, 17-epoxidation of non-13-hydroxylated GAs, which has been shown to deactivate GAs in rice (Yaish et al., 2010). The OsDREB2B protein has transactivation activity and is localized to the nucleus (Figure 7). Transcriptional expression of OsAP2-39 was upregulated in our OE lines (Figure 8A), and OsDREB2B was directly combined with the OsAP2-39 promoter (Figures 8B–D). OsDREB2B could be specifically bound to DRE/CRT elements (Chen et al., 2008), and a DRE/CRT cis-element was observed in the promoter region of OsAP2-39 (Table S4), indicating that OsDREB2B directly regulates the expression of OsAP2-39 by binding to a DRE/CRT element in its promoter. In this study, among the genes whose transcriptional expression was altered (Figure 6), four GA metabolic genes had at least one GCC box, and five genes had DRE/CRT cis-elements in their promoter region (Table S4), indicating that OsDREB2B may directly regulate the genes harboring DRE/CRT elements and indirectly regulate genes harboring a GCC-box by binding to the OsAP2-39 promoter.
The OsWRKY21-overexpressing rice showed dwarf phenotypes and regulated the expression of GA metabolism and cell wall biosynthesis-related genes (Wei et al., 2021). In our study, OsDREB2B physically interacted with OsWRKY21, which was verified by the Y2H assay and BiFC assay (Figure 9). Interestingly, among the 15 genes whose expression changed, 10 genes contained at least one repeat of a W-box in their promoter (Table S4). The OsDREB2B and OsWRKY21 interactions may regulate the expression of the GA metabolic genes that harbor a W-box and/or DRE/CRT cis-elements in their promoters. The detailed molecular mechanism of protein–protein interaction on plant growth and development will be elucidated by the generation of OsDREB2B overexpression plants in oswrky21 knockout mutants or/and OsWRKY21 overexpression plants in osdreb2 knockout mutants in future studies.
We proposed a model of OsDREB2B controlling plant height in rice through GA metabolism (Figure 10). OsDREB2B interacts with OsWRKY21 and directly binds to the promoters, which contain DRE/CRT-box or W-box elements, and indirectly binds to the GCC-box by activating OsAP2-39 to upregulate GA deactivation genes. In contrast, unidentified transcription repressors inhibit OsAP2-39, and the OsWRKY21/OsDREB2B complex binds to the promoter region of GA biosynthesis genes, resulting in transcription inhibition. The positive and negative regulation of GA metabolic genes leads to a decrease in bioactive GAs, thus reducing plant height in rice.
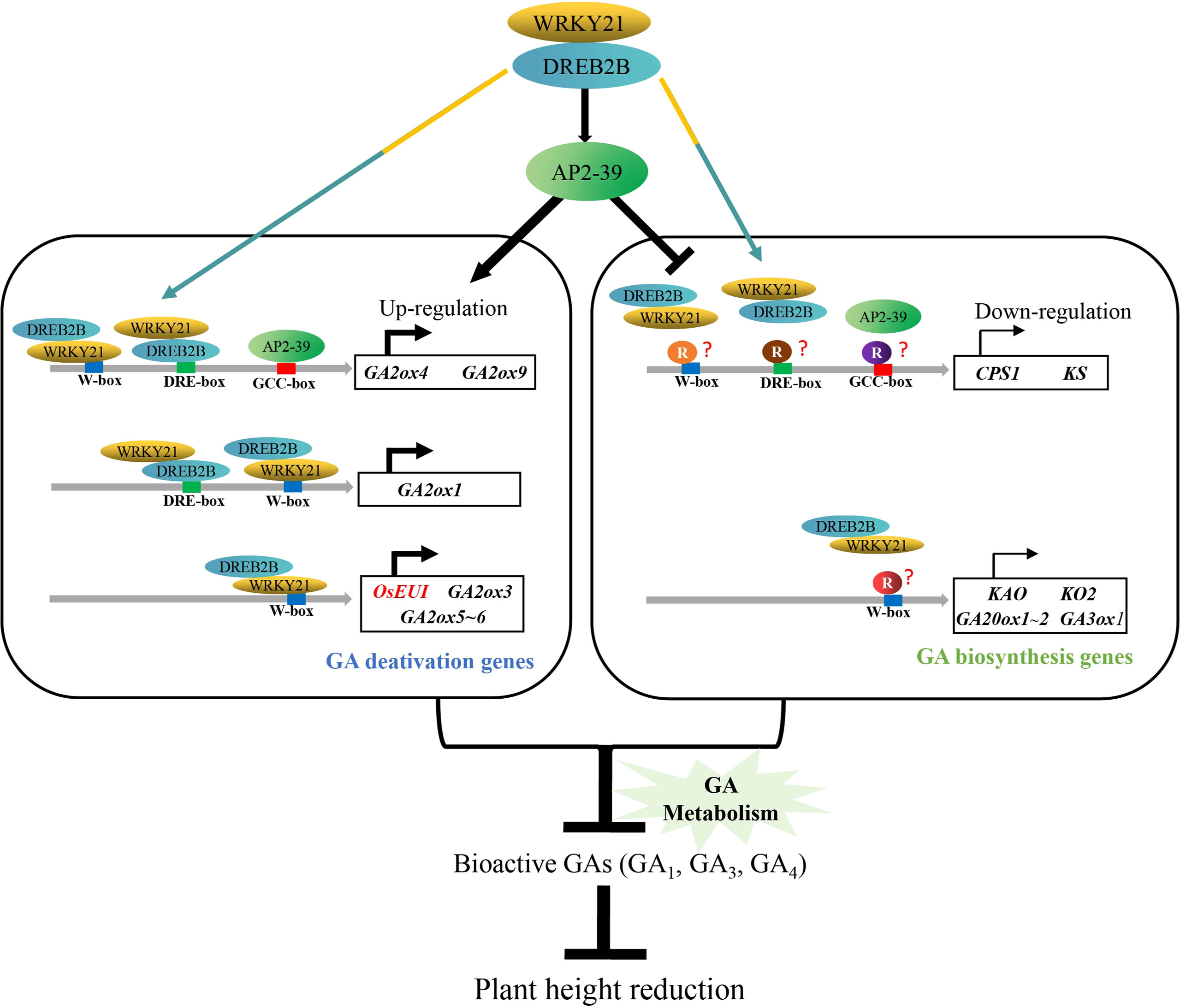
Figure 10 Proposed model of OsDREB2B regulatory mechanism in plant height reduction in rice. Colored spindles indicate different transcription factors, and colored circles indicate different repressors. Colored rectangles indicate the cis-element binding sequence. Colored arrows indicate that OsDREB2B/WRKY21 complex can bind directly to the cis-element (s) on the promoters of GA metabolic genes. The black arrows represent the activation effect, and the hammer lines indicate the inhibition effect. Curved arrows in the dark indicate the transcriptional expression of genes, and the thickness of the line represents the relative transcription level. Genes in red letters are based on experimental evidence, genes in black letters are based on bioinformatic prediction.
In summary, OsDREB2B is an AP2/ERF transcription factor that negatively regulates plant height by modulating bioactive GA content via downregulating GA biosynthesis genes and upregulating GA deactivation genes by binding to the OsAP2-39 promoter and interacting with OsWRKY21. The crosstalk mechanism of OsDREB2B on plant growth and stress responses will be further clarified in future studies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
ZM, Y-MJ, XD, and WJ designed the experiments. ZM, LH, TW, Y-MJ, and YZ performed the experiments. LH and TW analyzed the data. ZM, Y-MJ, TW, XD and WJ participated in the manuscript writing and amending. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Agricultural Major Project of Jilin Provincial Science and Technology Development Plan of China (20210302008NC), Jilin Provincial Agricultural Science and Technology Innovation Project of China (CXGC2021ZY110), and Scientific Research Project of Education Department of Jilin Province of China (JJKH20211130KJ).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1007811/full#supplementary-material
References
Allen, M. D., Yamasaki, K., Ohme, T. M., Tateno, M., Suzuki, M. (1998). A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5484–5496. doi: 10.1093/emboj/17.18.5484
Ashikari, M., Sasaki, A., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Datta, S., et al. (2002). Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice ‘Green revolution’. Breed. Sci. 52, 143–150. doi: 10.1270/jsbbs.52.143
Chen, Z. S., Liu, X. F., Wang, D. H., Chen, R., Zhang, X. L., Xu, Z. H., et al. (2017). Transcription factor OsTGA10 is a target of the MADS protein OsMADS8 and is required for tapetum development. Plant Physiol. 176, 01419. doi: 10.1104/pp.17.01419
Chen, J. Q., Meng, X. P., Zhang, Y., Xia, M., Wang, X. P. (2008). Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 30, 2191–2198. doi: 10.1007/s10529-008-9811-5
Chen, H. Y., Yu, H., Jiang, W. Z., Li, H. Y., Wu, T., Chu, J. F., et al. (2021). Overexpression of ovate family protein 22 confers multiple morphological changes and represses gibberellin and brassinosteroid signaling in transgenic rice. Plant Sci. 304, 110734. doi: 10.1016/j.plantsci.2020.110734
Gomi, K., Sasaki, A., Itoh, H., Ueguchi-Tanaka, M., Ashikari, M., Kitano, H., et al. (2004). GID2, an f-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37, 626–634. doi: 10.1111/j.1365-313X.2003.01990.x
Gu, C., Guo, Z. H., Hao, P. P., Wang, G. M., Jin, Z. M., Zhang, S. L. (2017). Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Botanical Stud. 58, 6. doi: 10.1186/s40529-016-0159-1
Hawku, M. D., Goher, F., Islam, M. A., Guo, J., He, F., Bai, X., et al. (2021). TaAP2-15, an AP2/ERF transcription factor, is positively involved in wheat resistance to puccinia striiformis f. sp. tritici. Int. J. Mol. Sci. 22, 2080. doi: 10.3390/ijms22042080
Hedden, P. (2002). Gibberellin metabolism and its regulation. J. Plant Growth Regul. 20, 317–318. doi: 10.1007/s003440010039
Hedden, P. (2003). The genes of the green revolution. Trends Genet. 19, 5–9. doi: 10.1016/S0168-9525(02)00009-4
Hedden, P., Thomas, S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25. doi: 10.1042/BJ20120245
Hong, Z., Ueguchi-Tanaka, M., Shimizu-Sato, S., Inukai, Y., Fujioka, S., Shimada., Y., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, c-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. doi: 10.1046/j.1365-313X.2002.01438.x
Huang, J., Li, Z. Y., Zhao, D. Z. (2016). Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci. Rep. 6, 29938. doi: 10.1038/srep29938
Huang, J., Tang, D., Shen, Y., Qin, B., Hong, L., You, A., et al. (2010). Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa l.). J. Genet. Genomics 37, 23–36. doi: 10.1016/S1673-8527(09)60022-9
Itoh, H., Sasaki, A., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Hasegawa, Y., et al. (2005). Dissection of the phosphorylation of rice DELLA protein, slender rice1. Plant Cell Physiol. 46, 1392–1399. doi: 10.1093/pcp/pci152
Itoh, H., Ueguchi-Tanaka, M., Sentoku, N., Kitano, H., Matsuoka, M., Kobayashi, M., et al. (2001). Cloning and functional analysis of two gibberellin 3beta-hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. United States America 98, 8909–8914. doi: 10.1073/pnas.14123 9398
Iwamoto, M., Kiyota, S., Hanada, A., Yamaguchi, S., Takano, M. (2011). The multiple contributions of phytochromes to the control of internode elongation in rice. Plant Physiol. 157, 1187–1195. doi: 10.1104/pp.111.184861
Jia, Q. J., Li, C. D., Shang, Y., Zhu, J. H., Hua, W., Wang, J. M., et al. (2015). Molecular characterization and functional analysis of barley semi-dwarf mutant riso no. 9265. BMC Genomics 16, 927.
Ke, Y. G., Yang, Z. J., Yu, S. W., Li, T. F., Luo, L. J. (2014). Characterization of OsDREB6 responsive to osmotic and cold stresses in rice. J. Plant Biol. 42, 9264–9269. doi: 10.1007/s12374-013-0480-0
Li, D., Guo, Z., Liu, C., Li, J., Chen, Y. (2017). Quantification of near-attomole gibberellins in floral organs dissected from a single arabidopsis thaliana flower. Plant J. 91, 547–557. doi: 10.1111/tpj.13580
Lin, R. C., Ding, L., Casola, C., Ripoll, D. R., Feschotte, C., Wang, H. Y. (2007). Transposase-derived transcription factors regulate light signaling in arabidopsis. Science 318, 1302–1305. doi: 10.1126/science.1146281
Li, J., Sima, W., Ouyang, B., Wang, T., Ziaf, K., Luo, Z., et al. (2012). Tomato SlDREB gene restricts leaf expansion and internode elongation by downregulating key genes for gibberellin biosynthesis. J. Exp. Bot. 63, 6407–6420. doi: 10.1093/jxb/ers295
Liu, C., Zheng, S., Gui, J. S., Fu, C. J., Yu, H., Song, D. L., et al. (2018). Shortened basal internodes encodes a gibberellin 2-oxidase and contributes to lodging resistance in rice. Mol. Plant 11, 288–299. doi: 10.1016/j.molp.2017.12.004
Lo, S. F., Yang, S. Y., Chen, K. T., Hsing, Y. I., Zeevaart, J. A. D., Chen, L. J., et al. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20, 2603–2618. doi: 10.1105/tpc.108.060913
Mao, D., Xin, Y., Tan, Y., Hu, X., Bai, J., Liu, Z.-y., et al. (2019). Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl. Acad. Sci. United States America 116, 3494–3501.
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283, 185–196. doi: 10.1007/s00438-009-0506-y
Ma, Z. M., Wu, T., Huang, K., Jin, Y. M., Zhao, L., et al. (2020). A novel AP2/ERF transcription factor, OsRPH1, negatively regulates plant height in rice. Front. Plant Sci. 11, 1–15. doi: 10.3389/fpls.2020.00709
Ma, H. L., Zhang, S. B., Ji, L., Zhu, H. B., Yang, S. L., Fang, X. J., et al. (2006). Fine mapping and in silico isolation of the EUI1 gene controlling upper internode elongation in rice. Plant Mol. Biol. 60, 87–94. doi: 10.1007/s11103-005-2762-5
Nishimura, A., Aichi, I., Matsuoka, M. (2006). A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 1, 2796–2802. doi: 10.1038/nprot.2006.469
Qi, W. W., Sun, F., Wang, Q. J., Chen, M. L., Huang, Y. Q., Feng, Y. Q., et al. (2011). Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 157, 216–228. doi: 10.1104/pp.111.179945
Rashid, M., Guangyuan, H., Guangxiao, Y., Hussain, J., Xu, Y. (2012). AP2/ERF transcription factor in rice: genome-wide canvas and syntenic relationships between monocots and eudicots. Evolutionary Bioinf. 8, 321–355. doi: 10.4137/EBO.S9369
Sakamoto, T., Kobayashi, M., Itoh, H., Tagiri, A., Kayano, T., Tanaka, H., et al. (2001). Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 125, 1508–1516. doi: 10.1104/pp.125.3.1508
Sakamoto, T., Miura, K., Itoh, H., Tatsumi, T., Ueguchi-Tanaka, M., Ishiyama, K., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134, 1642–1653. doi: 10.1104/pp.103.033696
Sakata, T., Oda, S., Tsunaga, Y., Shomura, H., Kawagishi-Kobayashi, M., Aya, K., et al. (2014). Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol. 164, 2011–2019. doi: 10.1104/pp.113.234401
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. doi: 10.1038/416701a
Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., et al. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an f-box mutant. Science 299, 1896–1898. doi: 10.1126/science.1081077
Shi, Y. T., Ding, Y. L., Yang, S. H. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23, 623–637. doi: 10.1016/j.tplants.2018.04.002
Song, Y. L., Xu, Z. F. (2013). Ectopic overexpression of an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) gene OsIAA4 in rice induces morphological changes and reduces responsiveness to auxin. Int. J. Mol. Sci. 14 (7), 13645–13656. doi: 10.3390/ijms140713645
Song, Y., You, J., Xiong, L. (2009). Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol. Biol. 70, 297–309. doi: 10.1007/s11103-009-9474-1
Spielmeyer, W., Ellis, M. H., Chandler, P. M. (2002). Semidwarf (sd-1), "green revolution" rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. United States America 99, 9043–9043.
Spray, C. R., Kobayashi, M., Suzuki, Y., Phinney, B. O., Macmillan, G. J. (1996). The dwarf-1 (dt) mutant of zea mays blocks three steps in the gibberellinbiosynthetic pathway. Proc. Natl. Acad. Sci. United States America 93, 10515–10518. doi: 10. 1073/pnas.93.19.10515
Thomas, S. G., Phillips, A. L., Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellins deactivation. Proc. Natl. Acad. Sci. United States America 96, 4698–4703. doi: 10.1073/pnas.96.8.4698
Tong, H. N., Xiao, Y. H., Liu, D. P., Gao, S. P., Liu, L. C., Yin, Y. H., et al. (2014). Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26, 4376–4393. doi: 10.1105/tpc.114.132092
Tripathi, P., Rabara, R. C., Choudhary, M. K., Miller, M. A., Huang, Y. S., Shen, Q. X. J., et al. (2015). The interactome of soybean GmWRKY53 using yeast 2-hybrid library screening to saturation. Plant Signaling Behav. 10, e1028705. doi: 10.1080/15592324.2015.1028705
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). Gibberellin insensitive dwarf1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
Ueguchi-Tanaka, M., Fujisawa, Y., Kobayashi, M., Ashikari, M., Iwasaki, Y., Kitano, H., et al. (2000). Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. United States America 97, 11638–11643. doi: 10.1073/pnas.97.21.11638
Wang, Y., Wang, C., Zheng, M., Lyu, J., Xu, Y., Li, X., et al. (2016). WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiol. 170, 2110–2123. doi: 10.1104/pp.15.01949
Wei, X. S., Zhou, H. L., Xie, D. Y., Li, J. G., Yang, M. C., Chang, T. L., et al. (2021). Genome-wide association study in rice revealed a novel gene in determining plant height and stem development, by encoding a WRKY transcription factor. Int. J. Mol. Sci. 22, 8192. doi: 10.3390/ijms22158192
Wu, K., Xu, H., Gao, X. H., Fu, X. D. (2021). New insights into gibberellin signaling in regulating plant growth-metabolic coordination. Curr. Opin. Plant Biol. 63, 102074. doi: 10.1016/j.pbi.2021.102074
Xie, Z. L., Nolan, T., Jiang, H., Tang, B. Y., Zhang, M. C., Li, Z. H., et al. (2019). The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in arabidopsis. Plant Cell 31, 1788–1806. doi: 10.1105/tpc.18.00918
Yaish, M. W., El-Kereamy, A., Zhu, T., Beatty, P. H., Good, A. G., Bi, Y. M., et al. (2010). The APETALA-2-Like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PloS Genet. 6, e1001098. doi: 10.1371/journal.pgen.1001098
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zeng, J. K., Li, X., Xu, Q., Chen, J. Y., Yin, X. R., Ferguson, I. B., et al. (2015). EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 13, 1325–1334. doi: 10.1111/pbi.12351
Zhang, X. X., Tang, Y. J., Ma, Q. B., Yang, C. Y., Mu, Y. H., Suo, H. C., et al. (2013). OsDREB2A, a rice transcription factor, significantly affects salt tolerance in transgenic soybean. PloS One 8, e83011. doi: 10.1371/journal.pone.0083011
Zhang, S. Z., Wu, T., Liu, S. J., Liu, X., Jiang, L., Wan, J. M. (2016). Disruption of OsARF19 is critical for floral organ development and plant architecture in rice (Oryza sativa l.). Plant Mol. Biol. Rep. 34, 748–760. doi: 10.1007/s11105-015-0962-y
Zhang, Y. Y., Zhu, Y. Y., Peng, Y., Yan, D. W., Li, Q., Wang, J. J., et al. (2008). Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice. Cell Res. 18, 412–421. doi: 10.1038/cr.2008.28
Zhao, L. F., Hu, Y. B., Chong, R., Wang, T. (2010). ARAG1, an ABA-responsive DREB gene, plays a role in seed germination and drought tolerance of rice. Ann. Bot. 105, 401–409. doi: 10.1093/aob/mcp303
Zheng, M., Wang, Y. H., Wang, Y. L., Wang, C. M., Ren, Y. L., Lv, J., et al. (2015). DEFORMED FLORAL ORGAN1 (DFO1) regulates floral organ identity by epigenetically repressing the expression of OsMADS58 in rice (Oryza sativa). New Phytol. 206, 1476–1490. doi: 10.1111/nph.13318
Zhou, F., Lin, Q. B., Zhu, L. H., Ren, Y. L., Zhou, K. N., Shabek, N., et al. (2013). D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. doi: 10.1038/nature12878
Keywords: AP2/ERF transcription factor, OsDREB2B, plant height, leaf sheath, gibberellin, rice
Citation: Ma Z, Jin Y-M, Wu T, Hu L, Zhang Y, Jiang W and Du X (2022) OsDREB2B, an AP2/ERF transcription factor, negatively regulates plant height by conferring GA metabolism in rice. Front. Plant Sci. 13:1007811. doi: 10.3389/fpls.2022.1007811
Received: 31 July 2022; Accepted: 05 October 2022;
Published: 28 October 2022.
Edited by:
Jiangqi Wen, Oklahoma State University, United StatesReviewed by:
Xia Yang, Institute of Botany, Chinese Academy of Sciences (CAS), ChinaYunqing Yu, Donald Danforth Plant Science Center, United States
Wenjing Yao, Nanjing Forestry University, China
Copyright © 2022 Ma, Jin, Wu, Hu, Zhang, Jiang and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinglin Du, ZHV4aW5nbGluMjAwNEAxNjMuY29t; Wenzhu Jiang, and6MTk3NUBqbHUuZWR1LmNu
†These authors have contributed equally to this work
 Ziming Ma
Ziming Ma Yong-Mei Jin
Yong-Mei Jin Tao Wu
Tao Wu Lanjuan Hu
Lanjuan Hu Ying Zhang2
Ying Zhang2 Wenzhu Jiang
Wenzhu Jiang