
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 September 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1005895
This article is part of the Research Topic Forage Crop Improvement for Improved Livestock Production and Nutrition View all 16 articles
Phosphorus (P) is an indispensable mineral nutrient for plant growth and agricultural production. Plants acquire and redistribute inorganic phosphate (Pi) via Pi transporters (PHT1s/PTs). However, apart from MtPT4, functions of the M. truncatula (Medicago truncatula) PHT1s remain unclear. In this study, we evaluated the function of the PHT1 family transporter MtPT5 in M. truncatula. MtPT5 was closely related to AtPHT1; 1 in Arabidopsis (Arabidopsis thaliana) and GmPT7 in soybean (Glycine max). MtPT5 was highly expressed in leaves in addition to roots and nodules. Ectopic expression of MtPT5 complemented the Pi-uptake deficiency of Arabidopsis pht1;1Δ4Δ double mutant, demonstrating the Pi-transport activity of MtPT5 in plants. When overexpressing MtPT5 in M. truncatula, the transgenic plants showed larger leaves, accompanying with higher biomass and Pi enrichment compared with wild type. All these data demonstrate that MtPT5 is important for leaf growth and Pi accumulation of M. truncatula and provides a target for molecular breeding to improve forage productivity.
Phosphorus (P) is an essential mineral nutrient for plant growth. It plays various biological functions and is a major determinant of crop production (Raghothama, 1999). Inorganic phosphate (Pi) is the main form of P that can be absorbed by plant roots (Chiou and Lin, 2011; Vincent et al., 2012; López-Arredondo et al., 2014). The total P level in soil is high, but the soluble Pi is always limited due to its low mobility, as well as precipitation and fixation (Marschner and Rimmington, 1988; Yan et al., 2021). It has been reported that about 70% of cultivated land in the world is deficient in plant-available Pi. P is one of the limiting factors for cultivated plants (Smith and Schindler, 2009; Péret et al., 2011; López-Arredondo et al., 2013). To maintain crop yield, the usage of P fertilizer is increased annually (Dobre et al., 2014; Heuer et al., 2017). However, excessive fertilizer is not only a waste, but also leads to environmental issues (Zhang et al., 2013; Zak et al., 2018; Che et al., 2020).
Plants absorb and translocate Pi via Pi transporters (PHT1s/PTs; Versaw and Garcia, 2017; Dai et al., 2022). Hence, PHT1 genes are potential targets for improving plants Pi efficiency and benefiting yields (Veneklaas et al., 2012). Most of the PHT1 genes are root-specific, while some are highly expressed in the aerial part or nodules and involved in Pi redistribution (Chen et al., 2019; Wang et al., 2020). The firstly identified PHT1 gene was Pho84 cloned from Saccharomyces cerevisiae (Bun-ya et al., 1991). Then, numerous PHT1s have been identified in plants including Arabidopsis (Arabidopsis thaliana; Muchhal et al., 1996; Shin et al., 2004), M. truncatula (Medicago truncatula; Liu et al., 2008), rice (Oryza sativa L.; Liu et al., 2011), maize (Zea mays L.; Wang et al., 2020) and soybean (Glycine max; Chen et al., 2019). Nine PHT1 members were identified in Arabidopsis (Mudge et al., 2002). Among them, AtPHT1;1 and AtPHT1;4 play predominant roles in Pi uptake (Shin et al., 2004). GmPT7 was reported to be responsible for Pi uptake from soil into nodules and distribution to the fixation zones. Overexpression of GmPT7 promotes plant growth and soybean yield (Chen et al., 2019). When overexpressing OsPT1 in rice, transgenic plants accumulated more Pi in shoots and displayed increased tiller numbers compared with wild-type plants (Seo et al., 2008). Thus, investigation of the functions of PHT1s provides an efficient route for improving plants nutrient efficiency.
Currently, 11 PHT1s were identified in M. truncatula (Liu et al., 2008). Yeast kinetics assays showed that MtPT1, MtPT2, MtPT3, and MtPT4 are low-affinity Pi transporters. MtPT1, MtPT2, MtPT3, and MtPT5 share 84% sequence identity, but only MtPT5 displayed high affinity for Pi (Liu et al., 2008). MtPT4 is highly expressed in mycorrhizal roots, responsible for Pi acquisition from arbuscules (Harrison et al., 2002). It is also expressed in plant root tip in the absence of the arbuscular mycorrhizal (AM) fungus and modulates root branching, whereas it does not significantly affect Pi accumulation in plants without AM symbiosis (Cao et al., 2020). Recently, MtPT6 was reported to be involved in Pi uptake by heterologous expression of MtPT6 in Arabidopsis pht1;1 or pht1;4 mutant. However, the role of MtPT6 in M. truncatula is unknown (Volpe et al., 2016). Information on the functions of PHT1s in Medicago is still limited.
In this study, we identified the role of MtPT5 in leaf growth and Pi accumulation of M. truncatula. MtPT5 is highly expressed in roots, leaves, and nodules and is low-Pi inducible. MtPT5 can rescue the Pi-uptake deficiency of Arabidopsis pht1;1Δ4Δ double mutant, indicating the Pi transport activity of MtPT5 in plants. When overexpressing MtPT5 in M. truncatula, the transgenic plants displayed larger leaf size and higher Pi content. These data demonstrate that MtPT5 plays important roles in M. truncatula vegetable growth and Pi nutrition.
Medicago truncatula ecotype R108, Arabidopsis thaliana ecotype Wassilewskija ecotype (Ws) and pht1;1Δ4Δ mutant were used in this study. For germination of M. truncatula, seeds were placed on wet filter paper at 4°C for 2 days. Then, the imbibed seeds were transferred to chamber with illumination of 120 μmol m−2 s−1, temperature 24°C, and 16 h light/8 h dark photoperiod for 4 days. The seedlings were grown in 1/2 Hoagland or in soil for experiments. For nodulation, four-day-old seedlings were incubated with Sm1021 resuspended in 1/2 Hoagland, then transferred to soil and injected with Sm1021 every 2 days for 1 month. Different organs were harvested separately for RNA extraction. For Arabidopsis germination, the seeds were kept at 4°C for 2 days for imbibition, then transferred to medium with 200 μM arsenate or 1/2 MS under normal conditions (120 μmol m−2 s−1, 22°C, 16 h light/8 h dark).
For Arabidopsis, the 20-day-old seedings grown on 1/2 MS medium were harvested for Pi content measurement. For M. truncatula, the top leaflets of 3-month-old plants grown in soil and 20-day-old seedlings grown in 1/2 Hoagland were collected. The measurement was assayed as described in the previous report (Ames, 1996). Briefly, different samples were collected and frozen in liquid nitrogen immediately. Pi was extracted in the buffer containing acetic acid at 42°C for 30 min. Pi concentration was measured at 820 nm wavelength using universal microplate spectrophotometer (BioTek Power Wave XS2). The Pi content was calculated based on the concentration and fresh weight of different samples.
The full-length coding sequence (CDS) of MtPT5 was cloned into pTOPO-TA Simple vector (Science Tool) for sequencing. Then the sequence-verified MtPT5 CDS was constructed into BamH I-linearized pCAMBIA1302 vector to generate 35S:MtPT5 plasmid via homologous recombination. The recombinant vector was used for plant transformation. For Arabidopsis (pht1;1Δ4Δ mutant), floral dip method was used as described (Clough and Bent, 1998) using Agrobacterium tumefaciens strain GV3101. The transformants were obtained on MS medium containing 50 mg/l hygromycin. For M. truncatula, the construct was introduced into R108 leaves via Agrobacterium EHA105-mediated transformation as described previously (Cosson et al., 2006). The transgenic M. truncatula plants were identified by PCR using vector-specific primers. T2 and T3 transgenic lines were used for Arabidopsis and M. truncatula separately in this study.
For quantification of gene expression, total RNA was isolated using Eastep Super Total RNA Extraction KIT (Promega) and quantified by nanodrop. 1 μg RNA was used for reverse-transcription using the PrimeScript II 1st Strand cDNA Synthesis Kit (Takara). qRT-PCR was performed using 2 × EasyTaq® PCR SuperMix (TransGen Biotech) on CFX96 system (Bio-Rad). MtActin11 was used to calculate the relative quantitative results for M. truncatula. The transcripts of MtPT5 in R108, pht1;1Δ4Δ mutant and pht1;1Δ4Δ/MtPT5 were tested by RT-PCR using cDNAs as templates. EF1a was amplified as a quantitative control.
PHT1 amino acid sequences were obtained from NCBI1 and EnsemblPlants.2 Amino acid sequences were firstly aligned using CluxtalX. The neighbor-joining tree was conducted in MEGA5 using bootstrap method (900 replicates) on poisson model.
Significant differences were determined by One-way ANOVA with Tukey test or Student’s t-test using SigmaPlot 12.5 software.
It has been reported that there are 11 PHT1 transporters in M. truncatula (Cao et al., 2020). We identified another two members (Mt4g083960 and Mt5g068140) by searching Ensemblplants.3 All members shared the common secondary structures with 11 predicted transmembrane domains (TM) separated by a large hydrophilic loop between TM6 and TM7 (Supplementary Figure S1). The signature GGDYPLSATIxSE (Karandashov and Bucher, 2005; Loth-Pereda et al., 2011) was identified and conserved among all MtPHT1s, except two of them. The signature of Mt1g069930 was modified with a Thr (T) replaced by a Val (V), and Mt1g074940 was modified with an Ala (A) replaced by a Ser (S; Supplementary Figure S1). The amino acid sequences of PHT1 proteins from M. truncatula, Arabidopsis, soybean maize and rice were used for constructing the neighbor-joining tree (Figure 1). The analysis showed that Mt1g074930 (MtPT5) was clustered phylogenetically to AtPHT1;1 and GmPT7, showing 80% and 86% amino acid sequence identities, respectively.
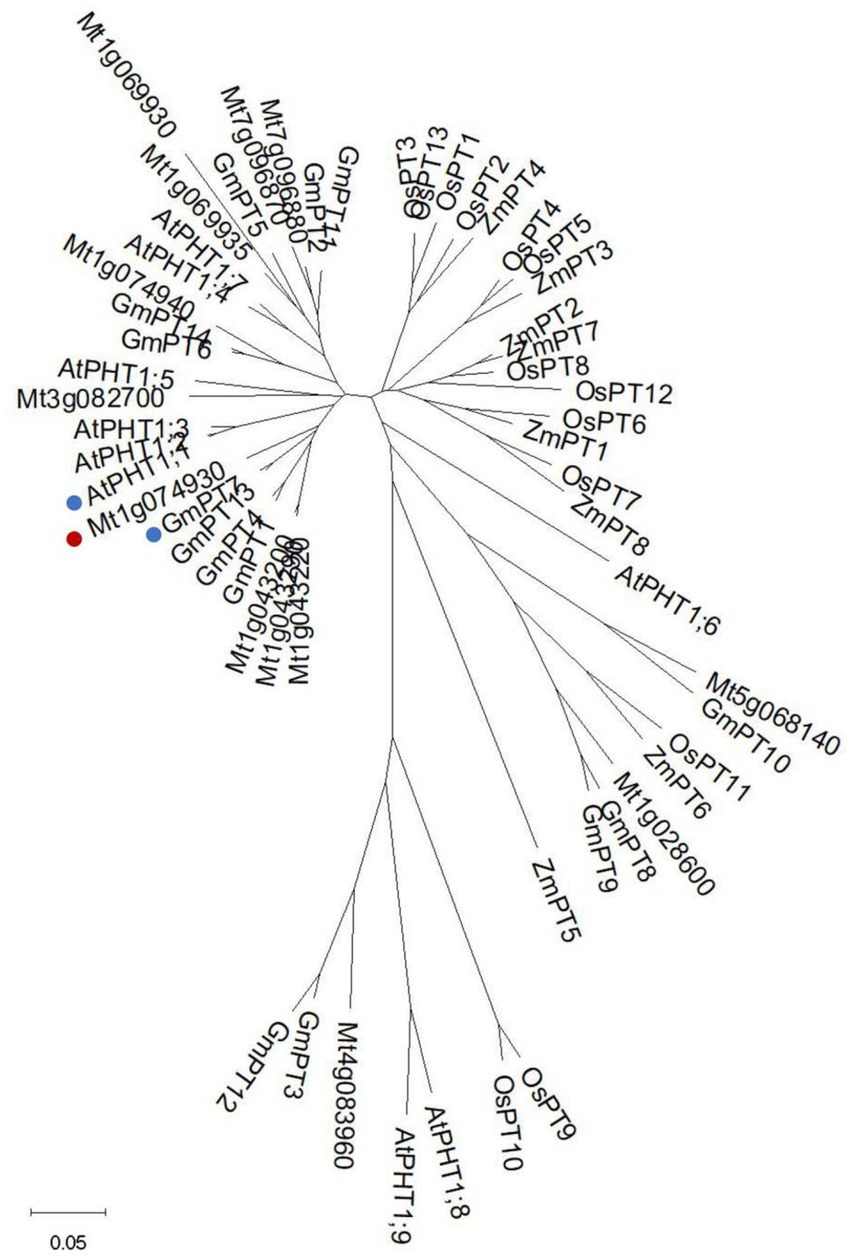
Figure 1. Phylogenetic analysis of PHT1s from different species. Phylogenetic tree of PHT1s from Medicago truncatula, Arabidopsis, soybean, maize, and rice. The tree was generated as described in materials and methods. Mt1g074930 (MtPT5) was labeled with a red spot. AtPHT1;1 and GmPT7 were labeled with blue spots. The bar shows 0.05 amino acid substitutions per site.
MtPT1, MtPT2, and MtPT3 are paralogues of MtPT5 in M. truncatula (Liu et al., 2008). The coding sequences of MtPT1, MtPT2, and MtPT3 share 97% identity. A single pair of primers were used to test the expression of these three genes. Quantitative RT-PCR (qRT-PCR) analysis showed that MtPT1/2/3 was predominantly expressed in roots and nodules and nearly undetectable in shoots (Figure 2A). The transcription abundance of MtPT5 was around four-to six-fold higher than that of MtPT1/2/3 in the underground tissues. In addition, MtPT5 was also highly expressed in shoots (Figure 2A). The expression pattern of MtPT5 in the aerial part was further tested. qRT-PCR results showed that MtPT5 was mainly expressed in leaves (Supplementary Figure S2). Pi starvation analysis showed that MtPT5 was induced under Pi-deficient condition (Figure 2B), in accordance with the previous report (Liu et al., 2008). The expression profile suggests that MtPT5 probably have multiple functions in plants and crucial roles in leaves. Leaf is important for plant yield. Hence, MtPT5 was further analyzed in Pi nutrition and development of M. truncatula.
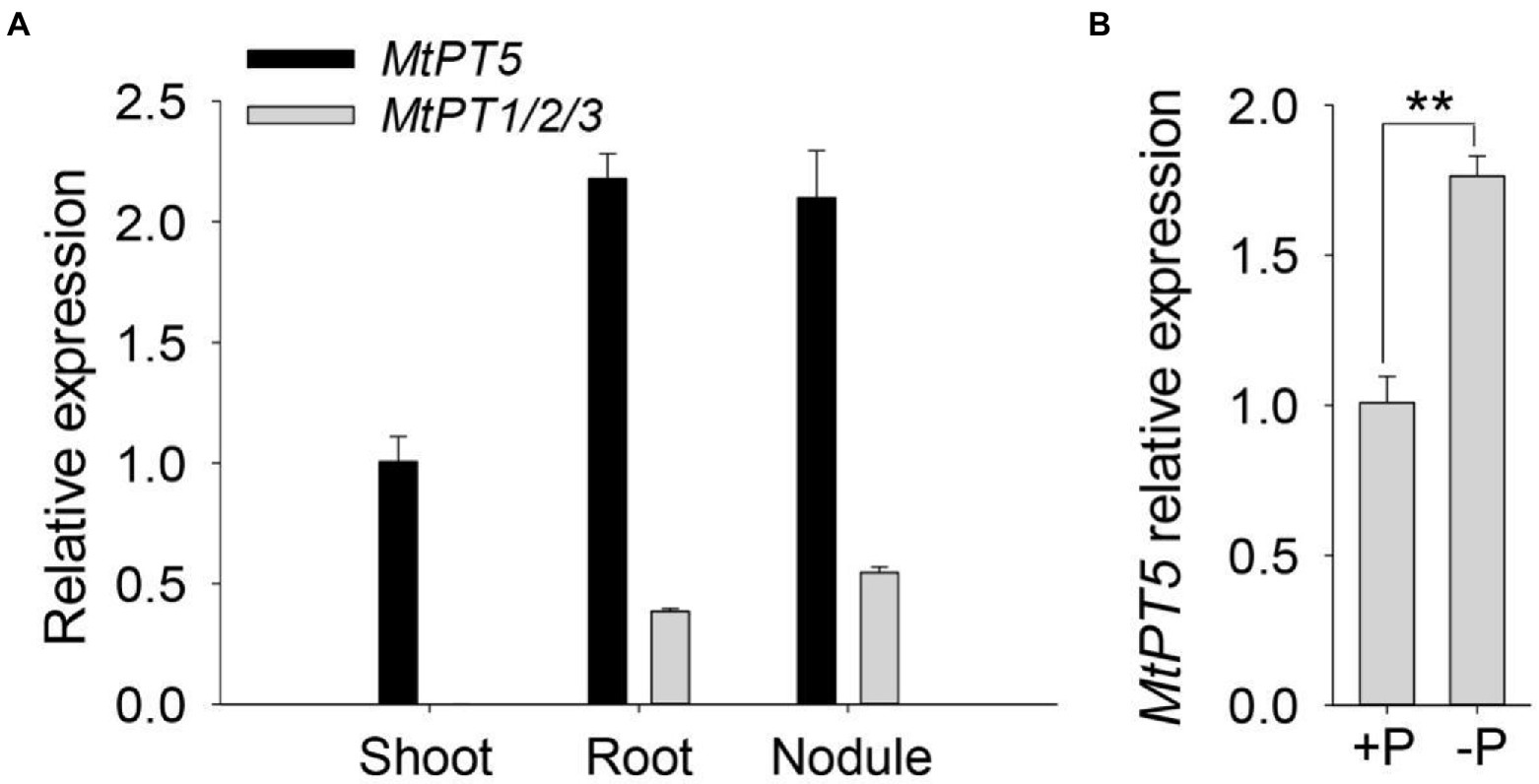
Figure 2. Expression profiles of MtPTs in M. truncatula. (A) qRT-PCR analysis of MtPT1/2/3 and MtPT5 in different tissues of M. truncatula. Four-day-old wild-type seedlings (R108) were incubated with Sm1021 resuspended in 1/2 Hoagland and then transferred to soil and injected with Sm1021 every 2 days for 1 month. Shoots, roots, and nodules were harvested, respectively, for RNA extraction. Data represent mean ± SE (n = 3). (B) qRT-PCR analysis of MtPT5 in wild-type seedlings (R108) during phosphate starvation. Four-day-old M. truncatula seedlings were transferred to hydroponic solution with Pi (+P) or solution without Pi (−P) for 5 days. The whole seedlings were used for RNA extraction. Data represent mean ± SE (n = 3). ** indicates significant difference at p < 0.01 (Student’s t-test).
AtPHT1;1 and AtPHT1;4 are two major Pi transporters in Arabidopsis. The double mutant pht1;1Δ4Δ displayed dramatic reduction in Pi uptake capacity and Pi content compared with wild type (Shin et al., 2004). To examine the Pi uptake activity of MtPT5 in plants, the coding sequence of MtPT5 driven by 35S promoter (35S:MtPT5) was introduced into pht1;1Δ4Δ. Two independent transgenic lines, 35S:MtPT5/pht1;1Δ4Δ-1 and 35S:MtPT5/pht1;1Δ4Δ-2, were used in this study. RT-PCR analysis showed that the MtPT5 transcripts were present in the two transgenic lines and not detectable in wild type (Ws) and pht1;1Δ4Δ mutant (Figure 3A). The fresh weight (FW) measurement showed that loss of PHT1;1 and PHT1;4 led to about 27% reduction in pht1;1Δ4Δ mutant biomass compared with wild type, similar to the previous report (Shin et al., 2004). Meanwhile, the biomasses of 35S:MtPT5/pht1;1Δ4Δ transgenic lines could be rescued to the level of wild type (Supplementary Figure S3). This indicates that MtPT5 can rescue the morphological defects of pht1;1Δ4Δ mutant. Next, we tested the Pi contents in different genotypic Arabidopsis seedlings grown under Pi-sufficient condition (1/2 MS). Pi content in pht1;1Δ4Δ mutant was significantly reduced compared with wild type, while the two overexpression lines exhibited similar Pi contents with wild type (Figure 3B). These data suggest that MtPT5 can complement the Pi-uptake deficiency of pht1;1Δ4Δ mutant.
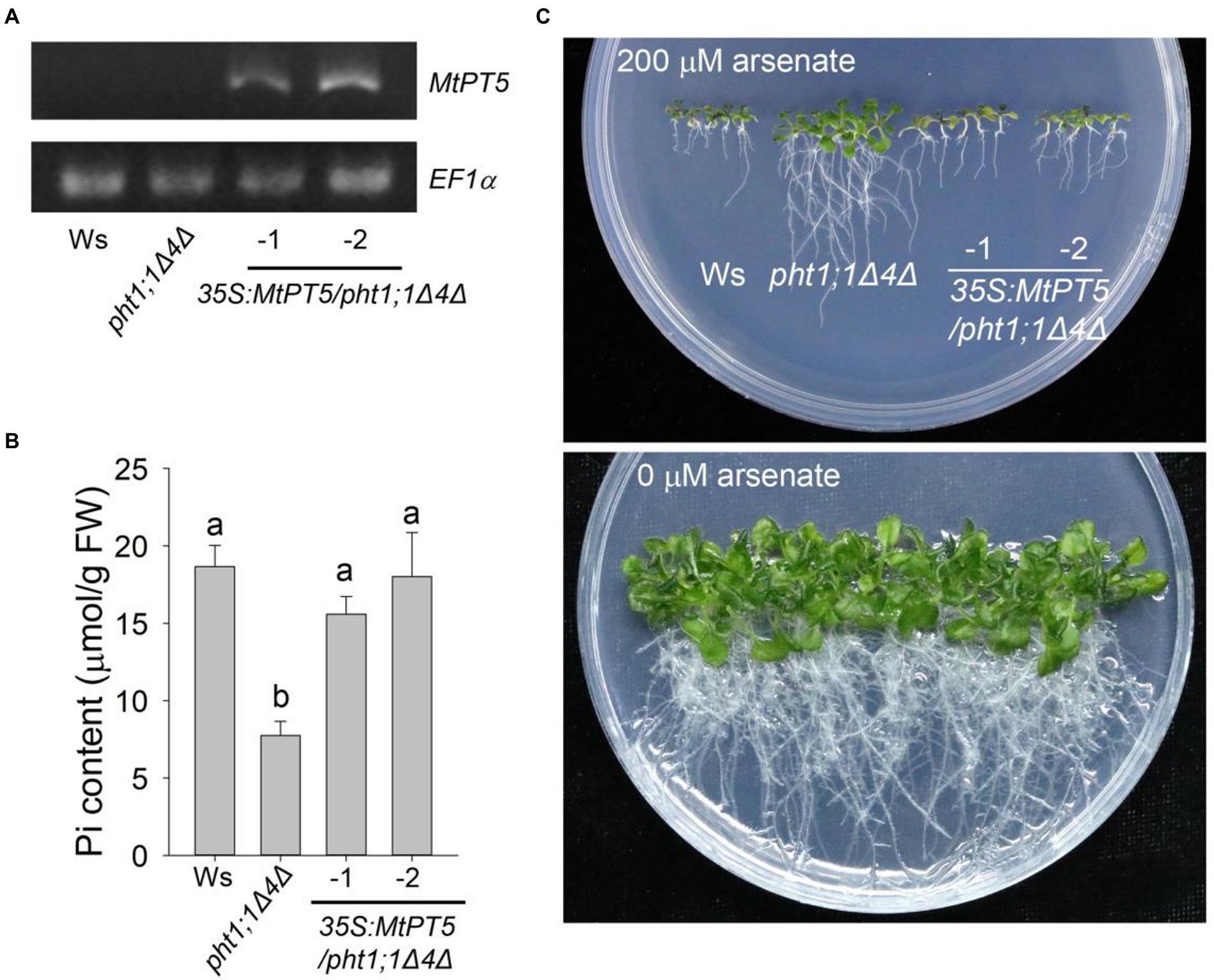
Figure 3. Ectopic expression of MtPT5 enhanced Pi acquisition in Arabidopsis. (A) RT-PCR analysis of MtPT5 in wild-type Arabidopsis (Ws), pht1;1Δ4Δ double mutant and 35S:MtPT5/pht1;1Δ4Δ transgenic plants using MtPT5-specific primers. The housekeeping gene EF1α was used as an internal control. (B) Pi content measurement of wild-type Arabidopsis (Ws), pht1;1Δ4Δ double mutant and 35S:MtPT5/pht1;1Δ4Δ transgenic plants. The whole seedlings grown on 1/2 MS for 20 days were collected for Pi extraction. Data represent mean ± SE (n = 3). Different letters indicate significant difference at p < 0.05 (One-way ANOVA, Tukey test). (C) Image of wild-type Arabidopsis (Ws), pht1;1Δ4Δ double mutant and 35S:MtPT5/pht1;1Δ4Δ transgenic plants grown on 1/2 MS containing 200 μM arsenate or without arsenate for 20 days.
Arsenate is a toxic metalloid structurally analogous of Pi and is transported into root cells mainly via PHT1 transporters (Catarecha et al., 2007; Castrillo et al., 2013; Wang et al., 2014). Phenotypes of wild type, pht1;1Δ4Δ mutant and 35S:MtPT5/pht1;1Δ4Δ transgenic plants were compared on the medium with or without arsenate. When grown on the medium with 200 μM arsenate, the pht1;1Δ4Δ mutant showed an arsenate-tolerant phenotype as previously reported (Shin et al., 2004), while the wild type and 35S:MtPT5/pht1;1Δ4Δ seedlings were hypersensitive to arsenate with dramatically shorter roots and smaller shoots (Figure 3C). Taken together, these data indicate that MtPT5 has Pi transport capacity and positively modulates Pi uptake in plants.
Given that MtPT5 was induced by low-Pi stress, two independent MtPT5-overexpressing lines, 35S:MtPT5-1 and 35S:MtPT5-2, were generated to examine the physiological role of MtPT5 in M. truncatula. qRT-PCR analysis showed that both MtPT5-overexpressing lines had significantly increased MtPT5 transcripts compared with wild-type M. truncatula (Figure 4A). We performed phenotypic tests on wild type and MtPT5-overexpressing plants. In both hydroponic culture and soil pots, the MtPT5-overexpressing lines displayed larger leaves compared with wild type (Figures 4B–D). Quantifications of leaf area confirmed this phenotype (Figure 4E). Meanwhile, leaf biomasses of the MtPT5-overexpressing plants were significantly higher than that of wild type (Figure 4F). These morphological traits indicate that overexpression of MtPT5 promotes leaves growth in M. truncatula.
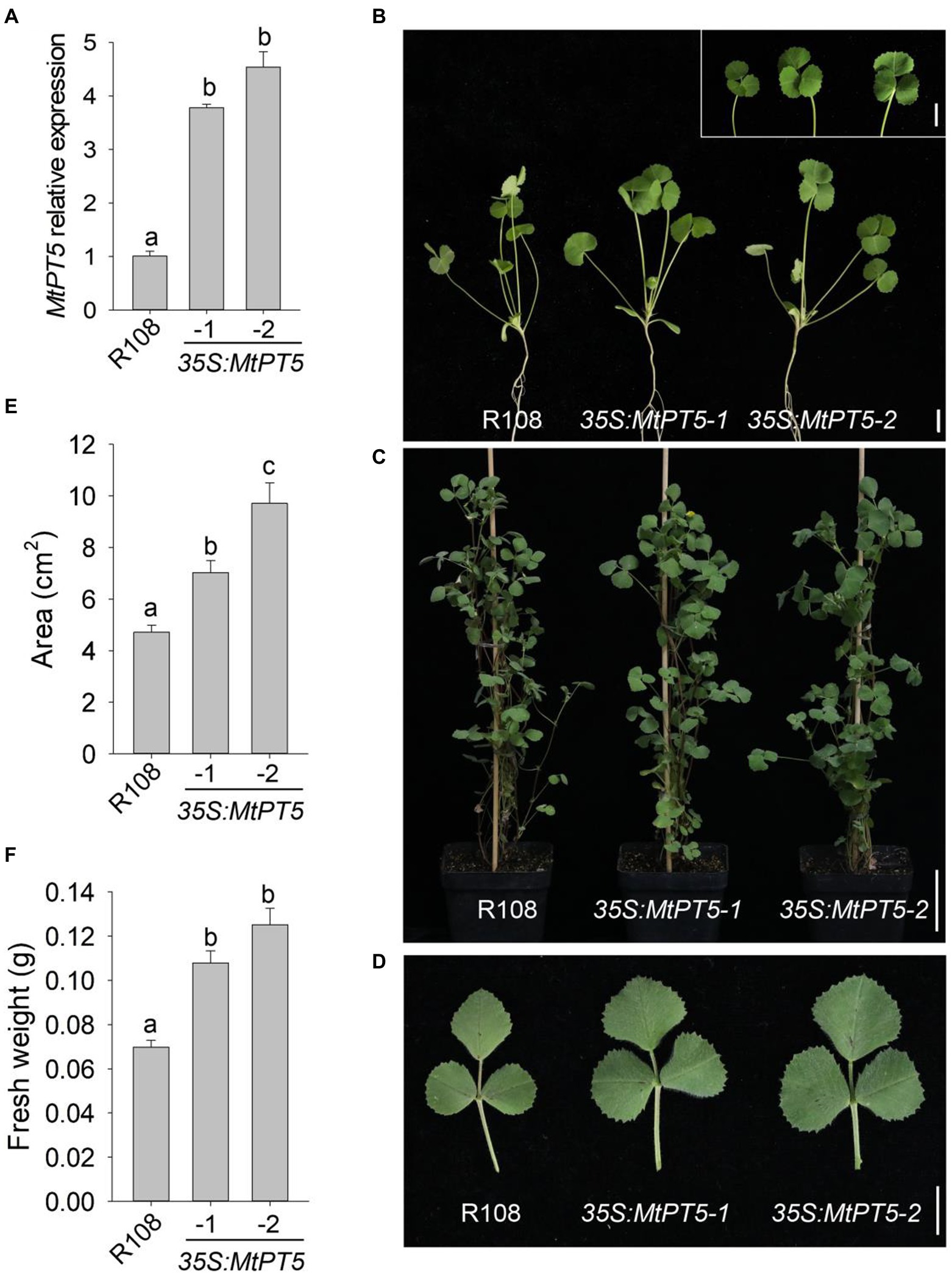
Figure 4. Overexpression of MtPT5 promotes leaves growth of M. truncatula. (A) qRT-PCR analysis of MtPT5 in wild-type M. truncatula (R108) and MtPT5-overexpressing plants (35S:MtPT5-1 and 35S:MtPT5-2). Data represent mean ± SE (n = 3). Different letters indicate significant difference at p < 0.05 (One-way ANOVA, Tukey test). (B–D) Phenotypic comparation of wild type (R108) and MtPT5-overexpressing plants. (B) Four-day-old seedlings were transferred to 1/2 Hoagland and grown for 20 days. Inset is leaves detached from the indicated plants. Bars = 1 cm. (C) Plants grown in soil for 3 months. Bar = 5 cm. (D) Top leaflets detached from (C). Bar = 1 cm. (E) Quantification of leaf areas. The fourth leaf of wild type (R108) and MtPT5-overexpressing plants shown in (B) were taken for quantification. Data represent mean ± SE (n = 8). Different letters indicate significant difference at p < 0.05 (One-way ANOVA, Tukey test). (F) Fresh weight of four expanded leaves detached from (B). Data represent mean ± SE (n = 8). Different letters indicate significant difference at p < 0.05 (One-way ANOVA, Tukey test).
To explore the function of MtPT5 in M. truncatula Pi nutrition, we measured the Pi content in leaves of wild type and MtPT5-overexpressing lines. The top leaflets of plants grown in soil for 3 months were collected for Pi extraction. The measurement showed that relative to wild type, the Pi content in MtPT5-overexpressing plants increased dramatically, especially in 35S:MtPT5-2 line (Figure 5A). The Pi contents of whole plants were also measured. Various plants grown in hydroponic culture for 20 days were harvested. The MtPT5-overexpressing plants showed significantly higher Pi contents than wild type (Figure 5B). Taken together, these data indicate that overexpression of MtPT5 enhances Pi accumulation in M. truancatula.
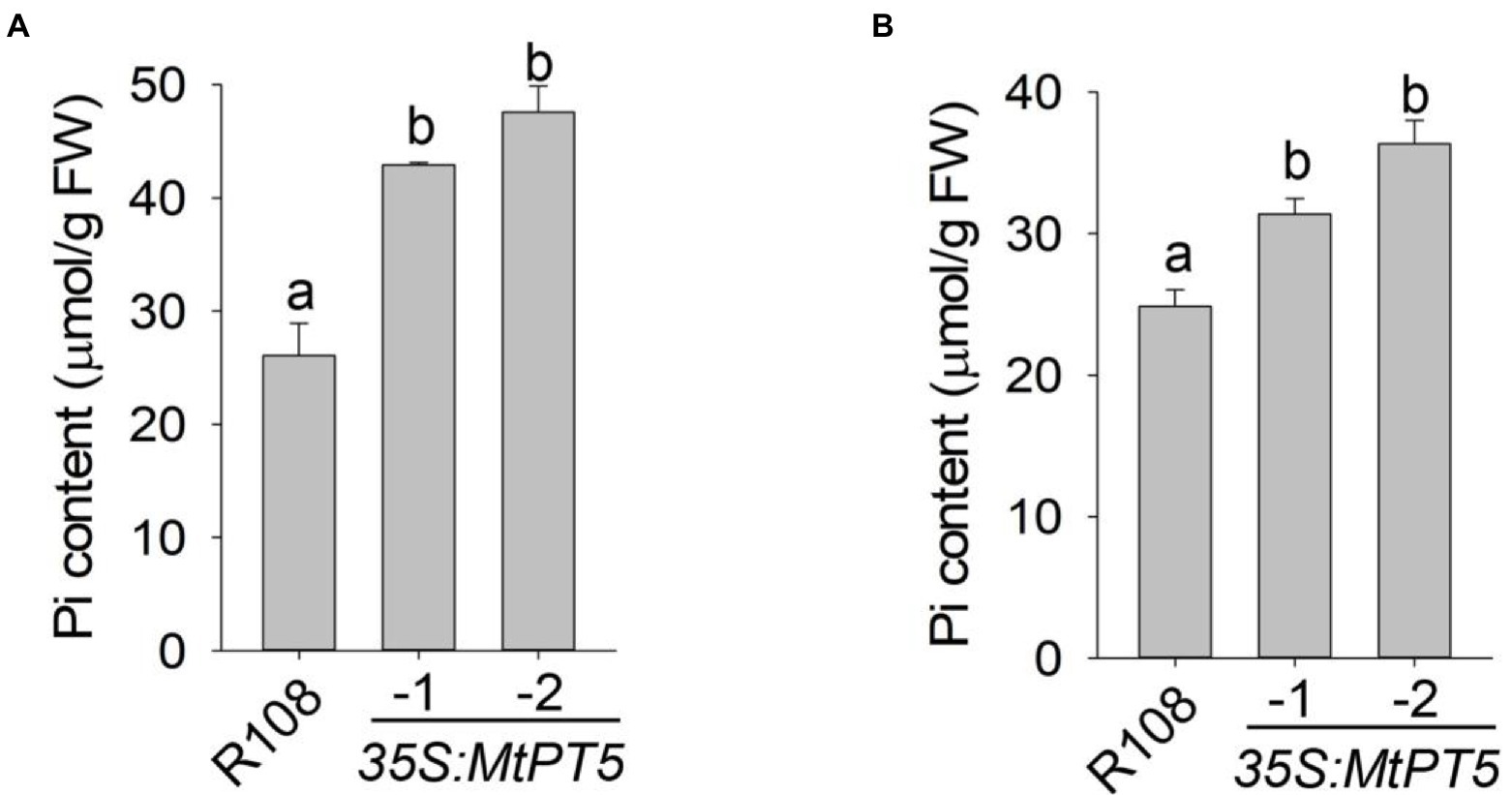
Figure 5. Overexpression of MtPT5 enhances Pi accumulation of M. truncatula. (A) Pi content in top leaflets of wild type (R108) and MtPT5-overexpressing plants grown in soil for 3 months. Data represent mean ± SE (n = 3). Different letters indicate significant difference at p < 0.05 (One-way ANOVA, Tukey test). (B) Pi content of wild type (R108) and 35S:MtPT5-overexpressing plants. Four-day-old seedlings were transferred to 1/2 Hoagland and grown for 20 days, then the whole plants were taken for Pi extraction. Data represent mean ± SE (n = 5). Different letters indicate significant difference at p < 0.05 (One-way ANOVA, Tukey test).
Phosphorus (P) is a major determinant of agriculture production. Plants absorb Pi via PHT1 transporters (Harrison et al., 2002), while some of them participate in Pi translocation and remobilization among different organs and tissues (Chang et al., 2019; Wang et al., 2020). It provides opportunities for improving crop performance by studying the functions of PHT1s (Chen and Liao, 2017; Han et al., 2022). Currently, 11 PHT1s have been found in M. truncatula. MtPT4 is responsible for Pi acquisition from mycorrhiza and plant root branching (Harrison et al., 2002; Volpe et al., 2016). MtPT6 was reported to promote Pi acquisition in Arabidopsis (Cao et al., 2020). Except for MtPT4, the functions of other PHT1s in M. truncatula are still unclear. In this study, we uncovered that Pi transporter MtPT5 plays an important role in leaf growth and Pi accumulation in M. truncatula.
We found two more PHT1s (Mt4g083960 and Mt5g068140) in M. truncatula by searching Ensemblplants.4 Alignment analysis showed that the 13 PHT1s all contained 12 predicted transmembrane domains, in accordance with the previous report (Pedersen et al., 2013). To choose one member of PHT1 family for further study in M. truncatula, phylogenetic tree was firstly constructed using PHT1s from M. truncatula, Arabidopsis, soybean, maize, and rice. The analysis showed that MtPT5 was closely related to AtPHT1;1 and GmPT7. AtPHT1;1 is an essential Pi transporter in Arabidopsis. Under Pi sufficient condition, the mutation of PHT1;1 leads to about 50% reduction of Pi uptake compared with wild-type plants. The Pi uptake of pht1;1Δ4Δ mutant reduces about 75% compared with wild type (Shin et al., 2004). GmPT7 is a nodule-located Pi transporter and responsible for the direct Pi acquisition from soil and Pi translocation from nodules to plant. Overexpression of GmPT7 improves shoot P content, nitrogen (N) content and soybean yield (Chen et al., 2019). The phylogenetic analysis indicates that MtPT5 probably have essential roles in M. truncatula Pi nutrition. The amino acid sequence of MtPT5 shared 84% identity with MtPT1, MtPT2 and MtPT3, whereas MtPT5 displayed an opposite affinity for Pi (Liu et al., 2008). This indicates the multiple functions of different PHT1s in Pi utilization even though PHT1s share high amino acid identities.
MtPT5 was reported to be a membrane-located high-affinity Pi transporter (Liu et al., 2008). To examine the Pi uptake activity of MtPT5 in plants, the coding sequence of MtPT5 driven by 35S promoter (35S:MtPT5) was constructed and introduced into Arabidopsis double mutant pht1;1Δ4Δ. Phenotypic analysis showed that pht1;1Δ4Δ mutant displayed an arsenate-tolerant phenotype, while the wild type and 35S:MtPT5/pht1;1Δ4Δ plants displayed arsenate-toxic phenotype. Pi content measurement showed that the Pi content in pht1;1Δ4Δ was significantly reduced compared with wild type, in accordance with previously reported (Shin et al., 2004). The Pi contents in 35S:MtPT5/pht1;1Δ4Δ transgenic lines were rescued to the level of wild type. Taken together, these data demonstrate that MtPT5 has the Pi-transporter activity in plants.
To identify the function of MtPT5 in M. truncatula, two independent MtPT5-overexpressing lines (35S:MtPT5-1 and 35S:MtPT5-2) were generated with significantly higher MtPT5 transcript levels. The MtPT5-overexpressing lines displayed larger leaves compared with wild type, and the leaf biomasses of the transgenic plants were increased dramatically. The Pi contents of top leaflets and whole plant in MtPT5-overexpressing lines were much higher than that in wild-type plants. These data demonstrate that overexpression of MtPT5 enhances M. truncatula leaf growth and Pi accumulation.
Expression analysis showed that MtPT5 was highly accumulated in shoots, roots and nodules. Previous reports demonstrated that ZmPT7, which is expressed in both roots and leaves, participates in Pi acquisition and redistribution in maize (Wang et al., 2020). GmPT7, located to nodules, is responsible for the direct Pi uptake from soil and translocation to fixation zones (Chen et al., 2019). The expression profile of MtPT5 suggests that it probably have multiple functions in Pi nutrition. In this study, we demonstrate that MtPT5 plays a vital role in Pi accumulation, and overexpression of MtPT5 promotes the leaf growth of M. truncatula dramatically. Leaf size is a vital trait to improve the yield and quality of forage, such as legume alfalfa (Medicago sativa L.) (Warman et al., 2011; Zhang et al., 2019). It was reported that about 70% protein of alfalfa is stored in leaves, while the cellulose content in leaves is only 1/3 of that in stems (Yang et al., 2016). Hence, our study provides a clue for elevating alfalfa Pi efficiency and genetic breeding.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
XW and QY planned and designed the research. XW and CW performed the experiments. XW wrote the manuscript. QY supervised this work and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by China Postdoctoral Science Foundation (2021M693441).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1005895/full#supplementary-material
1. ^https://www.ncbi.nlm.nih.gov/
2. ^http://plants.ensembl.org/Medicago_truncatula/Info/Index
3. ^http://plants.ensembl.org/Medicago_truncatula/Info/Index
4. ^http://plants.ensembl.org/Medicago_truncatula/Info/Index
Ames, B. N. (1996). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8, 115–118.
Bun-ya, M., Nishimura, M., Harashima, S., and Oshima, Y. (1991). The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 11, 3229–3238.
Cao, Y., Liu, J., Li, Y., Zhang, J., Li, S., An, Y., et al. (2020). Functional analysis of the phosphate transporter gene MtPT6 from Medicago truncatula. Front. Plant Sci. 11:620377. doi: 10.3389/fpls.2020.620377
Castrillo, G., Sánchez-Bermejo, E., de Lorenzo, L., Crevillén, P., Fraile-Escanciano, A., Tc, M., et al. (2013). WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell 25, 2944–2957. doi: 10.1105/tpc.113.114009
Catarecha, P., Segura, M. D., Franco-Zorrilla, J. M., García-Ponce, B., Lanza, M., Solano, R., et al. (2007). A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19, 1123–1133. doi: 10.1105/tpc.106.041871
Chang, M. X., Gu, M., Xia, Y. W., Dai, X. L., Dai, C. R., Zhang, J., et al. (2019). OsPHT1;3 mediates uptake, translocation, and remobilization of phosphate under extremely low phosphate regimes. Plant Physiol. 179, 656–670. doi: 10.1104/pp.18.01097
Che, J., Yamaji, N., Miyaji, T., Mitani-Ueno, N., Kato, Y., Shen, R. F., et al. (2020). Node-localized transporters of phosphorus essential for seed development in rice. Plant Cell Physiol. 61, 1387–1398. doi: 10.1093/pcp/pcaa074
Chen, L., and Liao, H. (2017). Engineering crop nutrient efficiency for sustainable agriculture. J. Integr. Plant Biol. 59, 710–735. doi: 10.1111/jipb.12559
Chen, L., Qin, L., Zhou, L., Li, X., Chen, Z., Sun, L., et al. (2019). A nodule-localized phosphate transporter GmPT7 plays an important role in enhancing symbiotic N2 fixation and yield in soybean. New Phytol. 221, 2013–2025. doi: 10.1111/nph.15541
Chiou, T. J., and Lin, S. I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Clough, S. J., and Bent, A. F. (1998). Floraldip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cosson, V., Durand, P., d’Erfurth, I., Kondorosi, A., and Ratet, P. (2006). Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 343, 115–127. doi: 10.1007/978-1-4939-1695-5_4
Dai, C., Dai, X., Qu, H., Men, Q., Liu, J., Yu, L., et al. (2022). The rice phosphate transporter OsPHT1;7 plays a dual role in phosphorus redistribution and anther development. Plant Physiol. 188, 2272–2288. doi: 10.1093/plphys/kiac030
Dobre, P., Jurcoane, S., Cristea, S., Matei, F., Moraru, A. C., and Dincă, L. (2014). Influence of N, P chemical fertilizers, row distance and seeding rate on camelina crop. Agrolife Sci. J. 3:1
Han, Y., White, P. J., and Cheng, L. (2022). Mechanisms for improving phosphorus utilization efficiency in plants. Ann. Bot. 129, 247–258. doi: 10.1093/aob/mcab145
Harrison, M. J., Dewbre, G. R., and Liu, J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14, 2413–2429. doi: 10.1105/tpc.004861
Heuer, S., Gaxiola, R., Schilling, R., Herrera-Estrella, L., Lopez-Arredondo, D., Wissuwa, M., et al. (2017). Improving phosphorus use efficiency: a complex trait with emerging opportunities. Plant J. 90, 868–885. doi: 10.1111/tpj.13423
Karandashov, V., and Bucher, M. (2005). Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 10, 22–29. doi: 10.1016/j.tplants.2004.12.003
Liu, F., Chang, X. J., Ye, Y., Xie, W. B., Wu, P., and Lian, X. M. (2011). Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant 4, 1105–1122. doi: 10.1093/mp/ssr058
Liu, J., Versaw, W. K., Pumplin, N., Gomez, S. K., Blaylock, L. A., and Harrison, M. J. (2008). Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J. Biol. Chem. 283, 24673–24681. doi: 10.1074/jbc.M802695200
López-Arredondo, D. L., Leyva-González, M. A., Alatorre-Cobos, F., and Herrera-Estrella, L. (2013). Biotechnology of nutrient uptake and assimilation in plants. Int. J. Dev. Biol. 57, 595–610. doi: 10.1387/ijdb.130268lh
López-Arredondo, D. L., Leyva-González, M. A., González-Morales, S. I., LópezBucio, J., and Herrera-Estrella, L. (2014). Phosphate nutrition: improving low-phosphate tolerance in crops. Annu. Rev. Plant Physiol. 65, 95–123. doi: 10.1146/annurev-arplant-050213-035949
Loth-Pereda, V., Orsini, E., Courty, P. E., Lota, F., Kohler, A., Diss, L., et al. (2011). Structure and expression profile of the phosphate Pht1 transporter gene family in mycorrhizal Populus trichocarpa. Plant Physiol. 156, 2141–2154. doi: 10.1104/pp.111.180646
Marschner, H., and Rimmington, G. (1988). Mineral nutrition of higher plants. Plant Cell Environ. 11, 147–148.
Muchhal, U. S., Pardo, J. M., and Raghothama, K. G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 93, 10519–10523. doi: 10.1073/pnas.93.19.10519
Mudge, S. R., Rae, A. L., Diatloff, E., and Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31, 341–353. doi: 10.1046/j.1365-313X.2002.01356.x
Pedersen, B. P., Kumar, H., Waight, A. B., Risenmay, A. J., Roe-Zurz, Z., Chau, B. H., et al. (2013). Crystal structure of a eukaryotic phosphate transporter. Nature 496, 533–536. doi: 10.1038/nature12042
Péret, B., Clément, M., Nussaume, L., and Desnos, T. (2011). Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci. 16, 442–450. doi: 10.1016/j.tplants.2011.05.006
Raghothama, K. G. (1999). Phosphate acquisition. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 50, 665–693. doi: 10.1146/annurev.arplant.50.1.665
Seo, H. M., Jung, Y., Song, S., Kim, Y., Kwon, T., Kim, D. H., et al. (2008). Increased expression of OsPT1, a high-affinity phosphate transporter, enhances phosphate acquisition in rice. Biotechnol. Lett. 30, 1833–1838. doi: 10.1007/s10529-008-9757-7
Shin, H., Shin, H. S., Dewbre, G. R., and Harrison, M. J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low-and high-phosphate environments. Plant J. 39, 629–642. doi: 10.1111/j.1365-313X.2004.02161.x
Smith, V. H., and Schindler, D. W. (2009). Eutrophication science: where do we go from here? Trends Ecol. Evol. 24, 201–207. doi: 10.1016/j.tree.2008.11.009
Veneklaas, E. J., Lambers, H., Bragg, J., Finnegan, P. M., Lovelock, C. E., Plaxton, W. C., et al. (2012). Opportunities for improving phosphorususe efficiency in crop plants. New Phytol. 195, 306–320. doi: 10.1111/j.1469-8137.2012.04190.x
Versaw, W. K., and Garcia, L. R. (2017). Intracellular transport and compartmentation of phosphate in plants. Curr. Opin. Plant Biol. 39, 25–30. doi: 10.1016/j.pbi.2017.04.015
Vincent, A. G., Schleucher, J., Grobner, G., Vestergren, J., Persson, P., Jansson, M., et al. (2012). Changes in organic phosphorus composition in boreal forest humus soils: The role of iron and aluminium. Biogeochemistry 108, 485–499. doi: 10.1007/s10533-011-9612-0
Volpe, V., Giovannetti, M., Sun, X. G., Fiorilli, V., and Bonfante, P. (2016). The phosphate transporters LjPT4 and MtPT4 mediate early root responses to phosphate status in non-mycorrhizal roots. Plant Cell Environ. 39, 660–671. doi: 10.1111/pce.12659
Wang, F., Cui, P. J., Tian, Y., Huang, Y., Wang, H. F., Liu, F., et al. (2020). Maize ZmPT7 regulates pi uptake and redistribution which is modulated by phosphorylation. Plant Biotechnol. J. 18, 2406–2419. doi: 10.1111/pbi.13414
Wang, H., Xu, Q., Kong, Y. H., Chen, Y., Duan, J. Y., Wu, W. H., et al. (2014). Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 164, 2020–2029. doi: 10.1104/pp.113.235077
Warman, L., Moles, A. T., and Edwards, W. (2011). Not so simple after all: searching for ecological advantages of compound leaves. Oikos 120, 813–821. doi: 10.1111/j.1600-0706.2010.19344.x
Yan, X., Chen, X., Ma, C., Cai, Y., and Zhang, F. (2021). What are the key factors affecting maize yield response to and agronomic efficiency of phosphorus fertilizer in China? Field Crop Res 270:108221. doi: 10.1016/j.fcr.2021.108221
Yang, Q. C., Kang, J. M., Zhang, T. J., Liu, F. Q., Long, R. C., and Sun, Y. (2016). Distribution, breeding and utilization of alfalfa germplasm resources. Chin. Sci. Bull. 61, 261–270.
Zak, D., Kronvang, B., Carstensen, M. V., Hoffmann, C. C., Kjeldgaard, A., Larsen, S. E., et al. (2018). Nitrogen and phosphorus removal from agricultural runoff in integrated buffer zones. Environ. Sci. Technol. 52, 6508–6517. doi: 10.1021/acs.est.8b01036
Zhang, F., Chen, X., and Vitousek, P. (2013). An experiment for the world. Nature 497, 33–35. doi: 10.1038/497033a
Keywords: phosphate, PHT1 transporter, MtPT5, Medicago truncatula, leaf growth, Pi accumulation
Citation: Wang X, Wei C, He F and Yang Q (2022) MtPT5 phosphate transporter is involved in leaf growth and phosphate accumulation of Medicago truncatula. Front. Plant Sci. 13:1005895. doi: 10.3389/fpls.2022.1005895
Received: 28 July 2022; Accepted: 19 August 2022;
Published: 06 September 2022.
Edited by:
Jiyu Zhang, Lanzhou University, ChinaReviewed by:
Hao Lin, Biotechnology Research Institute (CAAS), ChinaCopyright © 2022 Wang, Wei, He and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingchuan Yang, cWNoeWFuZzY2QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.