
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 October 2022
Sec. Plant Membrane Traffic and Transport
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1005020
This article is part of the Research Topic Metal Transport in Plants, Volume II View all 8 articles
Long non-coding RNAs (lncRNAs) are RNA molecules with functions independent of any protein-coding potential. A whole transcriptome (RNA-seq) study of Arabidopsis shoots under iron sufficient and deficient conditions was carried out to determine the genes that are iron-regulated in the shoots. We identified two previously unannotated transcripts on chromosome 1 that are significantly iron-regulated. We have called this iron-regulated lncRNA, CAN OF SPINACH (COS). cos mutants have altered iron levels in leaves and seeds. Despite the low iron levels in the leaves, cos mutants have higher chlorophyll levels than WT plants. Moreover, cos mutants have abnormal development during iron deficiency. Roots of cos mutants are longer than those of WT plants, when grown on iron deficient medium. In addition, cos mutant plants accumulate singlet oxygen during iron deficiency. The mechanism through which COS affects iron deficiency responses is unclear, but small regions of sequence similarity to several genes involved in iron deficiency responses occur in COS, and small RNAs from these regions have been detected. We hypothesize that COS is required for normal adaptation to iron deficiency conditions.
Iron is an essential micronutrient for plant development and is required in many enzymatic processes such as the electron transport chain of respiration and photosynthesis, and heme biosynthesis (Tanaka et al., 2011; Connorton et al., 2017). All organisms strictly regulate iron uptake to ensure that only the required amount of iron is present in cells and tissues (Ravet et al., 2009). For these reasons, plants have developed complex mechanisms to tightly regulate iron uptake, use and storage while adapting to changes in iron concentration in the environment.
As a transition metal, iron is redox active (Ward and Crichton, 2016). Iron deficiency decreases photochemical capacity and photosynthetic pigments (e.g., chlorophyll) by diminishing the number of photosynthetic units per unit leaf area (Spiller and Terry, 1980). Because iron deficiency decreases the concentration of photosynthetic pigments, it can promote photoinhibition (Godde and Hefer, 1994). This impairment of photosynthetic electron transport can result in accumulation of reactive oxygen species (ROS) (Yadavalli et al., 2012; Tewari et al., 2013). In order to maintain photosynthesis, plants must respond to changes in iron status without producing high levels of ROS (Kroh and Pilon, 2020). In the reaction center of photosystem II, the main ROS produced is singlet oxygen (Krieger-Liszkay, 2005). Aberrant production of ROS leads to photooxidative damage and eventually phytotoxicity in the organism (Halliwell and Gutteridge, 2015). Among all the different ROS, singlet oxygen is the major species causing photooxidative damage in plants (Triantaphylidès et al., 2008).
Plants respond to Fe-deficiency by enhancing their iron uptake capacity and iron use efficiency, but also by altering development. Iron is essential for chlorophyll biosynthesis and during iron-deficiency, the chlorophyll content of leaves decreases, a process known as iron chlorosis (Msilini et al., 2013). Under Fe-deficiency, plants display low levels of iron in leaves and later in development, in seeds (Waters et al., 2006). Moreover, during Fe-deficiency, roots are shorter compared to plants grown under Fe-sufficient conditions (Li et al., 2016; Zanin et al., 2017). Plant yield and quality is reduced during iron deficiency and this presumed to be the result of decreased photosynthesis associated with iron chlorosis (Zhang et al., 2019).
In response to Fe-deficiency, many changes occur at the transcriptional level, and this has been particularly well studied in roots, where the network of genes involved in regulating iron uptake has been defined. Expression of IRT1, FRO2 and a cascade of bHLH transcription factors are up-regulated in roots to activate the iron uptake machinery (Liang, 2022). While transcriptional response of the roots to Fe-deficiency is well studied, less is known about transcriptional changes occurring in leaves. Leaf responses to iron deficiency are rapid, and may occur even more quickly than responses in roots (Khan et al., 2018). Expression of HEMA1 and NYC1, which are genes involved in photosynthesis and tetrapyrrole biosynthesis, respectively, were downregulated in Fe-deficient leaves (Rodríguez-Celma et al., 2013). The role of transcription factor POPEYE (PYE) is well characterized in the roots as a negative regulator of iron uptake and homeostasis (Long et al., 2010). In a recent study, PYE was shown to be upregulated in leaves under Fe-deficiency, and to lessen the potential for photooxidative damage in leaves (Akmakjian et al., 2021) The Fe-S cluster transfer protein NEET has critical roles in iron homeostasis in leaves (Nechushtai et al., 2012). Disruption of the single Arabidopsis NEET gene triggers Fe-deficiency responses that result in iron over-accumulation in chloroplasts and enhanced accumulation in leaves (Zandalinas et al., 2020).
Under limited nutrient conditions plants adjust their metabolism to tolerate limited nutrient quantities. Nutrient economy strategies are used by plants in these situations to prioritize, recycle and remobilize limiting nutrients (Blaby-Haas and Merchant, 2013). Iron economy strategies are initiated when shoot demand exceeds supply by the roots (Urzica et al., 2012; Blaby-Haas and Merchant, 2013; Hantzis et al., 2018). Under iron limited conditions, limited amounts of iron are preferentially allocated to the most important functions. Specific changes occur at the transcript and protein level as part of iron economy strategy to help adjust the metabolism (Hantzis et al., 2018).
Long non-coding RNAs (lncRNAs) are a poorly understood, but common feature in eukaryotic genomes. The definition of a lncRNA comprises three criteria: the RNA molecule must have a function, to distinguish it from random products of transcriptional noise; this function must be independent of any protein-coding capacity of the RNA; and a lncRNA must be generated through mechanisms different from those that produce sRNAs (e.g., Dicer endonucleases) (Wierzbicki et al., 2021). lncRNAs vary in size and structure and can affect gene expression of their targets by modifying chromatin accessibility, modifying transcription itself; affecting splicing processes or affecting translation. lncRNAs can act locally (in cis) to regulate genes near the locus that encodes them, or at a distance (in trans) to affect expression of genes that are located in other parts of the genome (Chen et al., 2021; Wierzbicki et al., 2021). lncRNAs appear to be capable of producing sRNA through the action of Dicer (Ma et al., 2014). Such sRNA could also potentially account for biological activities of lncRNA in trans.
In this study, we have identified a previously unannotated lncRNA gene that we named CAN OF SPINACH (COS). COS expression is regulated by Fe-deficiency in both shoots and roots. Two cos mutant alleles were identified, and both had low levels of iron in shoots and seeds. Surprisingly, despite low iron levels, cos-2 shoots had increased levels of chlorophyll. Under Fe-deficiency conditions, cos mutants displayed abnormal growth. Also, cos mutants accumulate high levels of singlet oxygen in shoots, and appear to fail to make normal adaptations to low iron conditions. This work indicates that the previously unannotated lncRNA, COS, has a role iron homeostasis.
The wild type Arabidopsis thaliana accession Columbia-0 (Col-0) was used for all experiments; T-DNA insertion lines for COS (SAIL_445_G04; cos-1 and SALK_024945C; cos-2) were obtained from the Arabidopsis Biological Resource Center (ABRC). Homozygous mutant lines were confirmed by PCR. For soil growth, seeds were planted directly onto Pro-mix potting soil pre-treated with Gnatrol (Valent Bioscience Corporation) and stratified for three days at 4°C. Growth chamber conditions for soil-grown plants were 16 hours light (110 μmol·m–2·s–1), 8 hours dark at 22°C. For growth in sterile culture, sterilized seeds were plated directly on sterile petri plates containing 1/2X Murashige and Skoog medium (1/2X MS+Fe, prepared with 10X macronutrient solution, 100X micronutrient solution (without iron) and 100X Ferrous sulfate/chelate solution from PhytoTech Labs, Inc.) and placed at 4°C for three days. 1/2X MS-Fe was prepared using 10X macronutrient solution and 100X micronutrient solution (without iron). Growth chamber conditions for plate-grown plants were 16 hours light (110 μmol·m–2·s–1), 8 hours dark at 22°C.
Root and shoot tissues were disrupted in 1.5 mL Eppendorf tubes using a tissue homogenizer (Qiagen TissueLyser II) with 3.2 mm chrome steel beads (BioSpec Products). Total RNA was isolated using the Direct-zol™ RNA MiniPrep Kit (Zymo Research) with DNase I treatment according to the manufacturer’s instructions.
cDNA was synthesized from total RNA using SuperScript IV VILO Master Mix (Thermo Fisher Scientific) according to the manufacturer’s instructions. The RT-PCR and qRT-PCR reactions were carried out as described previously (Kumar et al., 2017). The primers used in this study are given in Supplementary Table 1.
Three biological replicates of three conditions were used for RNA-seq analysis. +Fe samples: plants were grown on 1/2X MS+ Fe for 14 days, then were transferred to fresh 1/2X MS+Fe for 3 days, and finally were transferred to 1/2X MS+Fe for 24 hours; -Fe samples: plants were grown on 1/2X MS+ Fe for 14 days, then were transferred to 1/2X MS-Fe for 3 days, and finally were transferred to fresh 1/2X MS-Fe for 24 hours; resupply samples: plants were grown on 1/2X MS+ Fe for 14 days, then were transferred to 1/2X MS-Fe for 3 days, and finally were transferred to 1/2X MS+Fe for 24 hours. Paired-end libraries were generated and sequenced by Novogene (Sacramento, CA).
Adapter sequences and low-quality reads were removed from raw reads with Trimmomatic v0.36.5 (Bolger et al., 2014). Mapping of reads to the Arabidopsis TAIR10 genome was carried out using HISAT2 v2.1.0 (Kim et al., 2015). Reads were assembled into transcripts and quantified using StringTie v1.3.3.2 (Pertea et al., 2015). Annotation was done using the TAIR10 genome GTF annotation file (www.arabidopsis.org). Differential gene expression analysis was performed using edgeR v3.20.7.2 through the Galaxy platform (Afgan et al., 2018). Minimum log2 fold change of 1 and Benjamini-Hochberg adjusted p-value of 0.05 were employed (Benjamini and Hochberg, 1995) as cutoff values.
Col-0, cos-1 and cos-2 seeds were grown on 1/2X MS+Fe plates for seven days and shifted to either 1/2X MS+Fe or 1/2X MS-Fe plates for three days. Plates were scanned using a flatbed scanner (Epson V700), and root length was measured using ImageJ software v1.52 (Schindelin et al., 2012).
Plants were grown directly on soil. Shoot tissues were collected when the first inflorescence became visible within the rosette, and fresh weight was determined. Chlorophyll extraction and quantification were performed as described previously (Waters et al., 2006).
Arabidopsis seeds for Col-0, cos-1 and cos-2 were grown directly on soil to determine iron content in shoots. Whole rosettes were collected when the first inflorescence became visible within the rosette and dried at 50°C for three days. To determine iron content in seeds, plants for Col-0, cos-1 and cos-2 were grown directly on soil until the seeds were mature. Seeds from individual plants were collected in Eppendorf tubes. ICP-MS analysis was carried out at the Ionomics Facility at the Donald Danforth Plant Science Center.
Seed weight was determined based on the weight of 10 batches of 100 seeds from three individual plants for each genotype. Weight per seed was calculated from the average of 10 seed batches. Seed number was determined by dividing the total weight of seeds by the individual seed weight.
Plants were grown on 1/2X MS+Fe plates for 14 days and shifted to either 1/2X MS+Fe or 1/2X MS-Fe plates for three days. The singlet oxygen sensor green (SOSG) staining procedure was performed as described by Akmakjian et al. (2021). Imaging of singlet oxygen detection was done on a Nikon MZ16 FA stereomicroscope equipped with a mercury lamp and a color camera.
Three biological replicates of three conditions were used for sRNA-seq analysis. +Fe samples: plants were grown on 1/2X MS+ Fe for 14 days, then were transferred to fresh 1/2X MS+Fe for 3 days, and finally were transferred to 1/2X MS+Fe for 24 hours; -Fe samples: plants were grown on 1/2X MS+ Fe for 14 days, then were transferred to 1/2X MS-Fe for 3 days, and finally were transferred to fresh 1/2X MS-Fe for 24 hours; resupply samples: plants were grown on 1/2X MS+ Fe for 14 days, then were transferred to 1/2X MS-Fe for 3 days, and finally were transferred to 1/2X MS+Fe for 24 hours.
Libraries were generated and sequenced by Novogene (Sacramento, CA). Cutadapt was used to remove adapter sequences (Martin, 2011). Shortstack was used to align, annotate, and quantify small RNAs (Axtell, 2013; Shahid and Axtell, 2014). The search size parameter for discovering sRNA loci with Shortstack was between 17 and 29 nt long. After processing the raw reads, raw read counts were used as input for differential expression analysis with DESeq2 (FDR < 10% and Log2FC ≥ |0.5|) (Love et al., 2014). Arabidopsis sRNA Target Prediction webserver at Donald Danforth Plant Science Center (wasabi.ddpsc.org/~apps/tp/) was used to predict the targets of the iron up-regulated sRNA candidates. The default scoring system was used for the analysis.
A whole transcriptome (RNAseq) study of Arabidopsis shoots under iron sufficient, deficient and resupplied conditions was carried out. The details of the growth conditions for this RNA-seq are given in the Materials and Methods section. Briefly, care was taken to match control and experimental growth as closely as possible, including all medium transfers. The -Fe samples were grown on iron depleted medium for a total of four days, while the ‘resupply’ samples experienced iron depleted medium for three days, followed by 24 hours on +Fe medium. The brief period of iron resupply is intended to capture those genes that respond very quickly to changes in iron status per se, as opposed to genes that may react slower in response to later changes in metabolism or growth after the plants have taken up sufficient iron during resupply.
In total 239 transcripts were significantly up- (154 transcripts) or down-regulated (85 transcripts) by iron deficiency in shoots. (Supplemental Data 1). We set a criterion for ‘rapid response to resupply’ if the comparison between the ‘resupply’ and +Fe samples was not statistically significant (i.e., p > 0.05). Expression of ~ 50% (73 out of 154) of the upregulated exhibit rapid response to resupply (Supplemental Table 1). Biological process GO term analysis was carried out against the PANTHER database (Mi et al., 2019; Thomas et al., 2022). No cut-offs were used for fold enrichment values, but only terms with adjusted p-value < 0.05 were included in the supplemental tables. Several genes with well defined roles in iron homeostasis were among the 73 ‘rapid response to resupply’ genes. In addition, we noted that the the GO term for cellular response to hypoxia was significantly enriched among ‘rapid response to resupply’ genes (Supplemental Table 2). There were also some transcripts that changed toward basal levels but were still elevated after 24 hours (e.g., one isoform of bHLH39), and there were a few rapid response to resupply genes with expression repressed below the basal (+Fe) levels (e.g., Histidine phosphotransmitter 4 (AHP4) and a gene encoding a DUF506 family protein) after 24 hours of iron resupply. The remaining 81 transcripts whose expression did not return to basal levels upon iron resupply were highly enriched for the GO term ‘removal of superoxide radicals’ but otherwise were not very different from the rapid response genes, in terms of biological process GO terms. (Supplemental Table 3).
Among the transcripts that were downregulated during iron deficiency, only about 25% (21 out of 85) of transcripts rapidly responded to iron resupply. The majority of these 21 transcripts were chloroplast localized and involved in functions such as metabolism and photosynthesis (e.g. NEET and NYC1 (Tanaka et al., 2011; Nechushtai et al., 2012), but no specific biological function GO term enrichment was present among these genes. Two chloroplast localized genes, FYD and CHLORIDE CHANNEL E (CLC-E) increased their expression beyond basal (+Fe) levels. Among the remaining 62 transcripts the two most enriched GO terms were for reactive oxygen species metabolic process and response to oxidative stress (Supplemental Table 4).
Among the differentially expressed genes, a particular locus containing three protein-coding genes (AT1G13607, AT1G13608, AT1G13609) was scored as having iron-deficiency-regulated gene expression. Two of the genes (AT1G13608, AT1G13609) appeared to be upregulated during iron deficiency, and these genes have been identified as iron-regulated in other analyses (Rodríguez-Celma et al., 2013; Naranjo-Arcos et al., 2017; Bastow et al., 2018; Nam et al., 2021; Peixoto et al., 2021). Upon closer inspection of the count data (FPKM values), we noticed that the iron-regulated reads were mostly not coming from the strand encoding AT1G13607, AT1G13608, and AT1G13609. Instead, we identified a previously unannotated transcript on the reverse strand that is significantly iron-regulated (Figures 1A, B). The AT1G13609 transcript on the top strand appears to be iron-regulated (Figure 1B) but the difference in FPKM was not significant. Moreover, the FPKM values (89.7 and 55.3) indicate that the unannotated bottom strand transcript is expressed at much higher levels than the top strand transcripts AT1G13607.1, AT1G13608.1, and AT1G13609.1 and AT1G13609.2 (0.00001, 0.000012, 1.68 and 34.1.) We note that the FPKM values for AT1G13607.1, AT1G13608.1, and AT1G13609.1 are lower than cutoff values that are often applied in plant RNAseq analyses, but we include them here to demonstrate the very low levels of expression of these transcripts (Golicz et al., 2018; Zheng et al., 2019; Zhang et al., 2020; Yu et al., 2022).
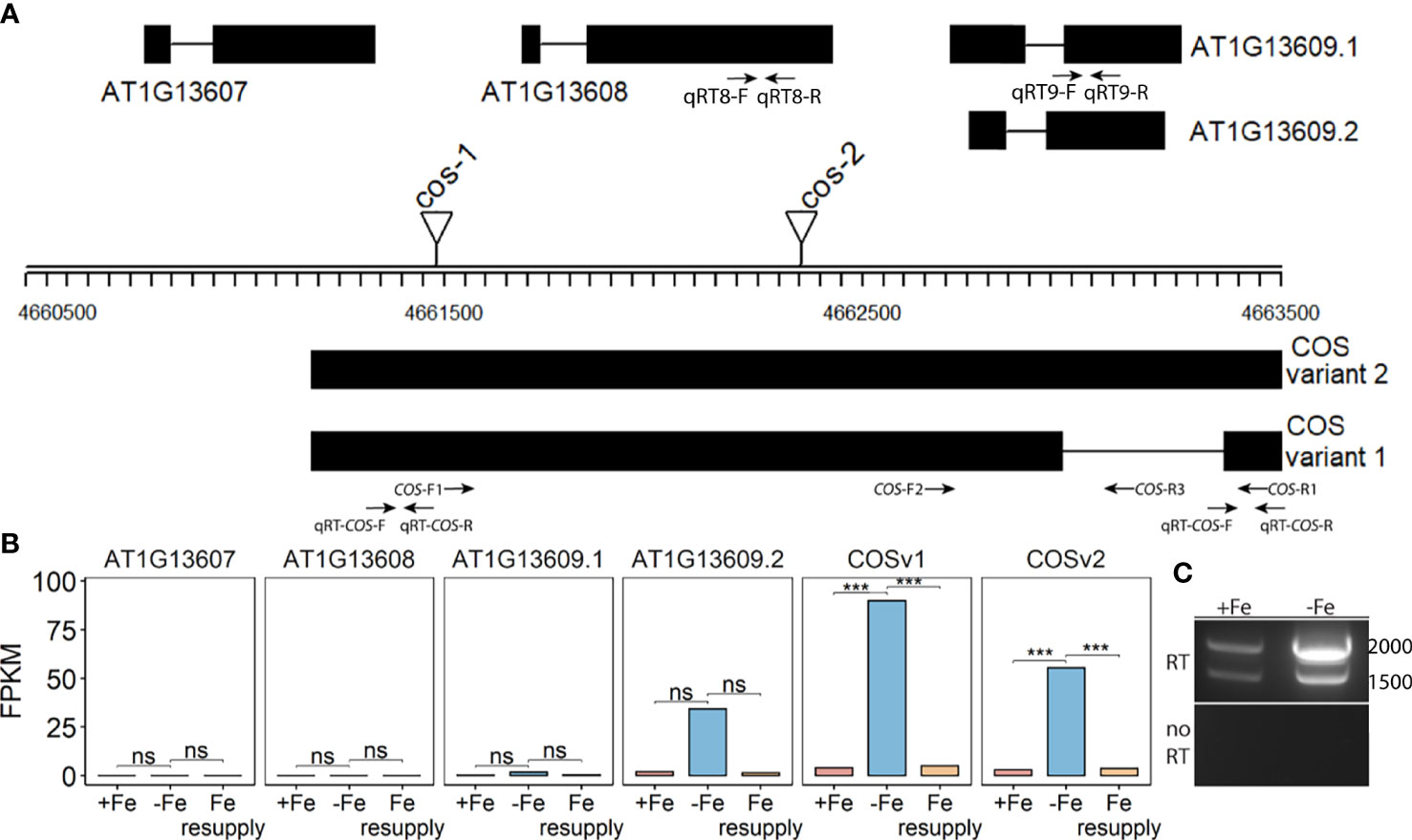
Figure 1 CAN OF SPINACH (COS), a novel long non-coding RNA (lncRNA). (A) Schematic representation of the locus. On the top strand, three genes (AT1G13607, AT1G13608 and AT1G13609) are annotated. The lncRNA COS is on the bottom strand. Thick boxes represent exons, lines represent introns. Positions of T-DNA insertion mutants used in this study are represented as triangles. Primers used in the study are represented as arrows at the bottom. (B) Bar plot representing the expression levels of various predicted transcripts under Fe sufficient, Fe deficient and Fe resupply conditions. Values from RNAseq are represented as fragments per kilobase of exon per million mapped fragments (FPKM). (C) RT-PCR using primers COS-F1 and COS-R1, using total RNA from shoots. Absence of genomic DNA contamination was confirmed by the lack of amplification of the no-RT control. cDNA was synthesized using oligodT(20). *** represents p-value ≤ 0.0001. ns, not significant.
Using primers located at the 5’ and 3’ ends of the putative transcript (COS-F1 and COS-R1), we were able to demonstrate the existence of the full length, spliced transcript using reverse transcription PCR (RT-PCR). These primers were specifically designed to only detect the expression of the putative transcript and not the coding genes on the top strand (Figure 1A). Both a spliced and an unspliced variant were amplified. Because cDNA synthesis was primed with oligodT, the transcript appears to be polyadenylated, as most plant lncRNAs are (Wang and Chekanova, 2017) (Figure 1C). We named the newly identified transcript, CAN OF SPINACH (COS). We carried out an ab initio prediction of any peptides COS might encode. We found that COS has the potential to encode only a small peptide of 57 aa long in a mRNA over 2 kb long. Furthermore, we carried out computational approaches to determine if COS is a long non-coding RNA (lncRNA). We used CPC (coding potential calculator) analysis, which is a tool to assess the protein coding potential of a transcript based on sequence intrinsic features (Kang et al., 2017). Both COS transcripts were classified as non-coding, with coding probabilities of 0.02 and 0.03, respectively. We also carried out PFAM domain analysis on all three reading frame translations of the COS RNA. (Mistry et al., 2021). No known protein domains were detected, further attesting that COS is a lncRNA.
Two different mutant alleles, cos-1 (SAIL_445_G04) and cos-2 (SALK_024945), in which T-DNAs are inserted in a COS exon (Figure 1A), were obtained, and homozygous mutant plants were established. We performed RT-PCR using two sets of primers, one upstream of the insertion sites and one downstream of the insertion sites. By sampling different parts of the COS lncRNA, we observed that the cos transcripts in the mutants are likely to be aberrant. Specifically, RT-PCR analysis of the COS transcript levels (Figure 2A) indicates that cos-1 has detectable RNA expression with both primer sets, but is detected at significantly lower levels with one of these sets. Thus, cos-1 may be a partial loss of function allele. The cos-2 allele has no detectable transcripts with one of the primer sets used and is therefore likely to be a total loss of function allele. It may produce an aberrant transcript that lacks a portion of the normal RNA. It is possible that cos-1 and cos-2 affect AT1G13608 on the top strand. We used quantitative reverse transcription PCR to investigate this, but it is important to understand that it was not possible to design a primer set for AT1G13608 that will not also amplify COS (Supplemental Figure 1). Analysis of AT1G13608 gene expression mirrors what was observed using primers specific to COS (Figure 2) Analysis of AT1G13609 expression (Supplemental Figure 1) indicates that expression of this gene is not significantly altered in either mutant. We cannot rule out that the mutations affect both COS and AT1G13608 expression, but note the very low expression levels of AT1G13608, which are below commonly used cutoffs for an expressed gene in an RNAseq dataset (Golicz et al., 2018; Zheng et al., 2019; Zhang et al., 2020; Yu et al., 2022) Although COS was first identified in Arabidopsis shoots, it is also expressed in the roots. The levels of COS transcripts in shoots and roots were similar when plants were grown with sufficient iron. During Fe-deficiency, the amount of COS transcript in roots trends lower, but this was not significant at our stringent P ≤ 0.01 criterion (Figure 2B).
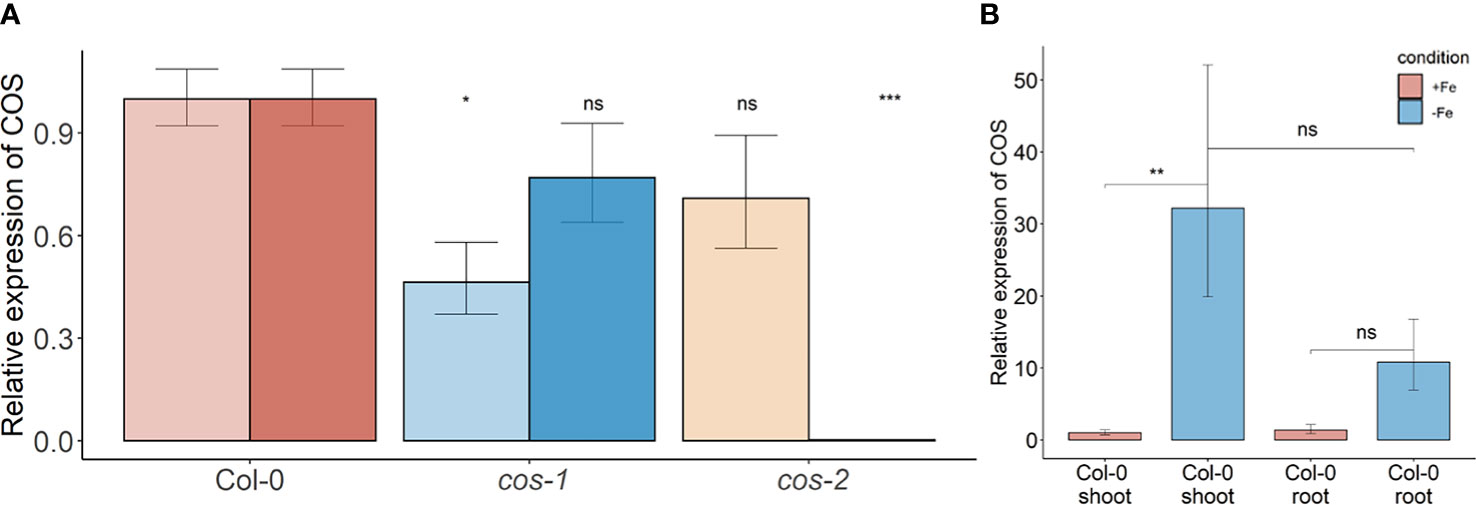
Figure 2 COS expression in WT and T-DNA mutants. (A) Two sets of primers were used for RT-PCR. The primer set upstream of the T-DNA insertion is designated with the lighter color, while the primer set downstream of the insertion is designated with darker color. Seedlings were grown on 1/2X MS agar with 50 μM Fe-EDTA for 5 d followed by a growth period of 3 d either on Fe-sufficient or Fe-deficient MS agar medium. Bars represent means ± SD of relative transcript levels normalized to ACT2 (n = 3). (B) Expression levels of COS transcript in WT shoots and roots. Seedlings were grown on 1/2X MS agar with 50 μM Fe-EDTA for 14 d followed by a growth period of 3 d either on Fe-sufficient or Fe-deficient MS agar medium. Bars represent means ± SD of relative transcript levels normalized to ACT2 (n = 3). * represents p-value ≤ 0.01. ** represents p-value ≤ 0.001, *** represents p-value ≤ 0.0001, ns, not significant.
Phenotypic characterization of T-DNA insertion mutants was carried out to further investigate the role of COS in iron homeostasis. We measured the accumulation of iron in the shoots of wild type (WT; Col-0) and cos mutant plants using ICP-MS (Figure 3A). The cos mutants have significantly less iron in their shoots than WT plants. The levels of Zn, Cu, and Mn in the mutants were not significantly different from WT (not shown). To complement and confirm these ICP-MS findings, we examined the expression of Ferritin 1 (FER1) in shoots. FER1 mRNA levels are commonly used as a marker for shoot iron status (Gaymard et al., 1996). In accord with shoot iron levels, FER1 expression was significantly lower in cos-2 mutants, while FER1 levels trended low, but were not significantly different in the partial loss of function allele cos-1 (Figure 3B). We postulated that the low iron levels in cos mutant plants might result in low iron levels in seeds. To examine this we analyzed iron accumulation in seeds. We found that seeds of cos mutants contain significantly less iron than WT seeds (Figure 3C). The levels of other metals in cos mutants were not significantly different from WT (not shown).
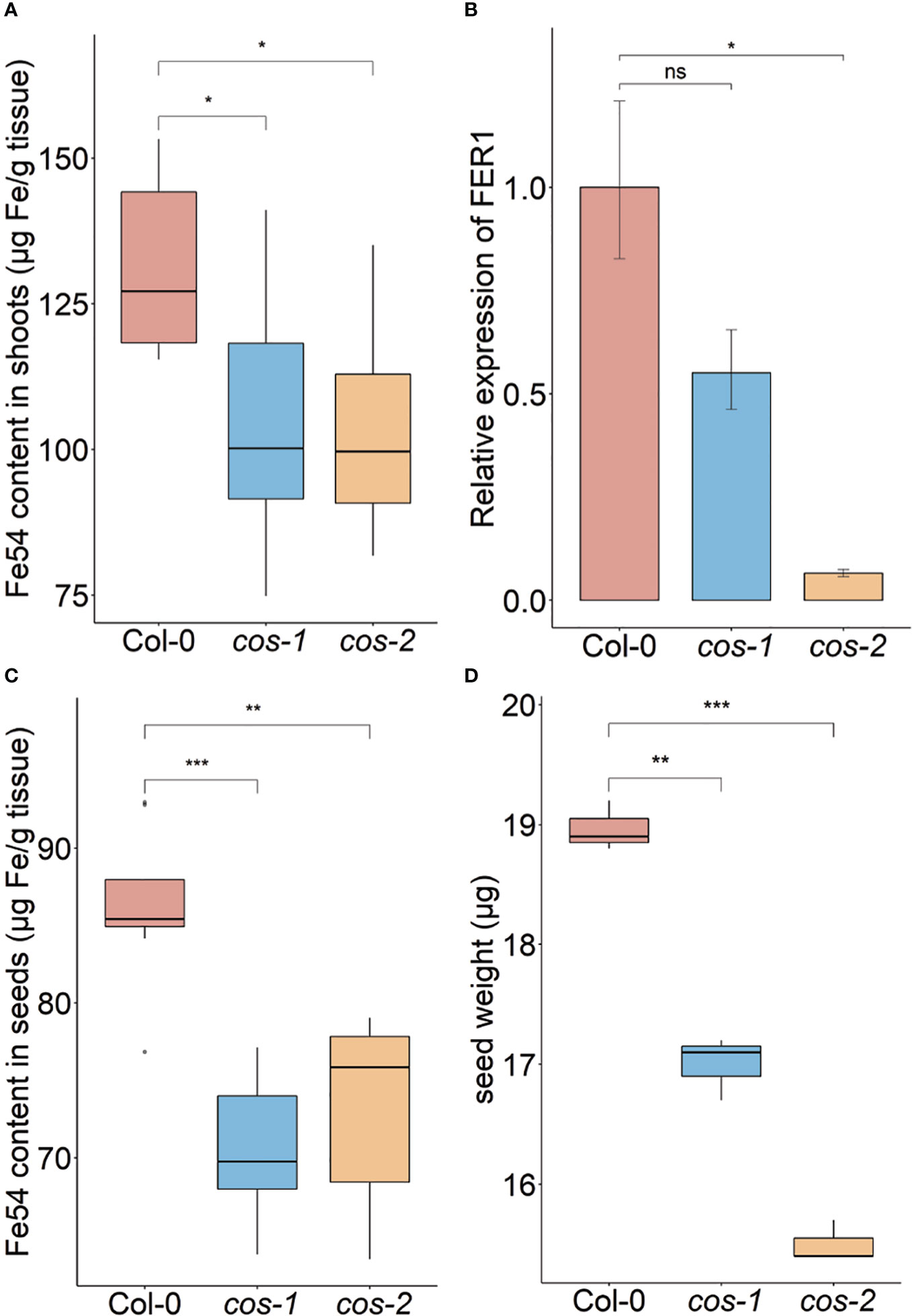
Figure 3 Phenotype of cos T-DNA mutants on soil. Iron concentration of shoots (A) and seeds (C) from plants grown on commercial potting mix. Results are given as μg Fe per gram of tissue (n = 8). (B) Expression levels of FER1 transcript in WT (Col-0) and cos mutants. Seedlings were grown on 1/2X MS agar with 50 μM Fe-EDTA for 14 d followed by a growth period of 3 d either on Fe-sufficient or Fe-deficient MS agar medium. Bars represent means ± SD of relative transcript levels normalized to ACT2 (n = 3). (D) Individual seed weight of WT and cos mutants was determined by sampling weights of 10 batches of 100 seeds. * represents p-value ≤ 0.01. ** represents p-value ≤ 0.001, *** represents p-value ≤ 0.0001, ns, not significant.
Since the iron levels in the cos seeds were low, we wondered whether this would affect seed development. We measured the seed weight and seed number in WT and cos mutants. The individual seed weight was decreased in cos mutants compared to WT seeds grown at the same time (Figure 3D). Both cos-1 and cos-2 displayed upward trends in seed number compared to WT, but these differences were not statistically significant (not shown). There is a minor (8-9%) defect in the rates of germination of cos mutant seeds compared to WT seeds (Supplemental Figure 2).
Generally, the cos mutants were not visibly different from WT plants, and the fresh weight of the shoots of cos mutant and WT plants at the time of bolting were indistinguishable (Supplemental Figure 3). One of the classic symptoms of iron deficiency is chlorosis (low chlorophyll), and many mutants that accumulate lower levels of iron in their tissues also exhibit iron deficiency chlorosis (Robinson et al., 1999; Curie et al., 2001; Vert et al., 2002; Colangelo and Guerinot, 2004; Waters et al., 2006). Because cos mutant shoots have low iron levels, we expected that they might have low chlorophyll levels. Surprisingly though, shoots of cos-2 knockout plants had significantly more chlorophyll than WT. The chlorophyll content of cos-1 mutants appeared to trend higher than WT but was not significantly higher (Figure 4A). Under iron deficient conditions, cos mutants displayed chlorosis similar to WT, however, due to the low levels of chlorophyll during iron deficiency we were not able measure the -Fe chlorophyll levels reliably.
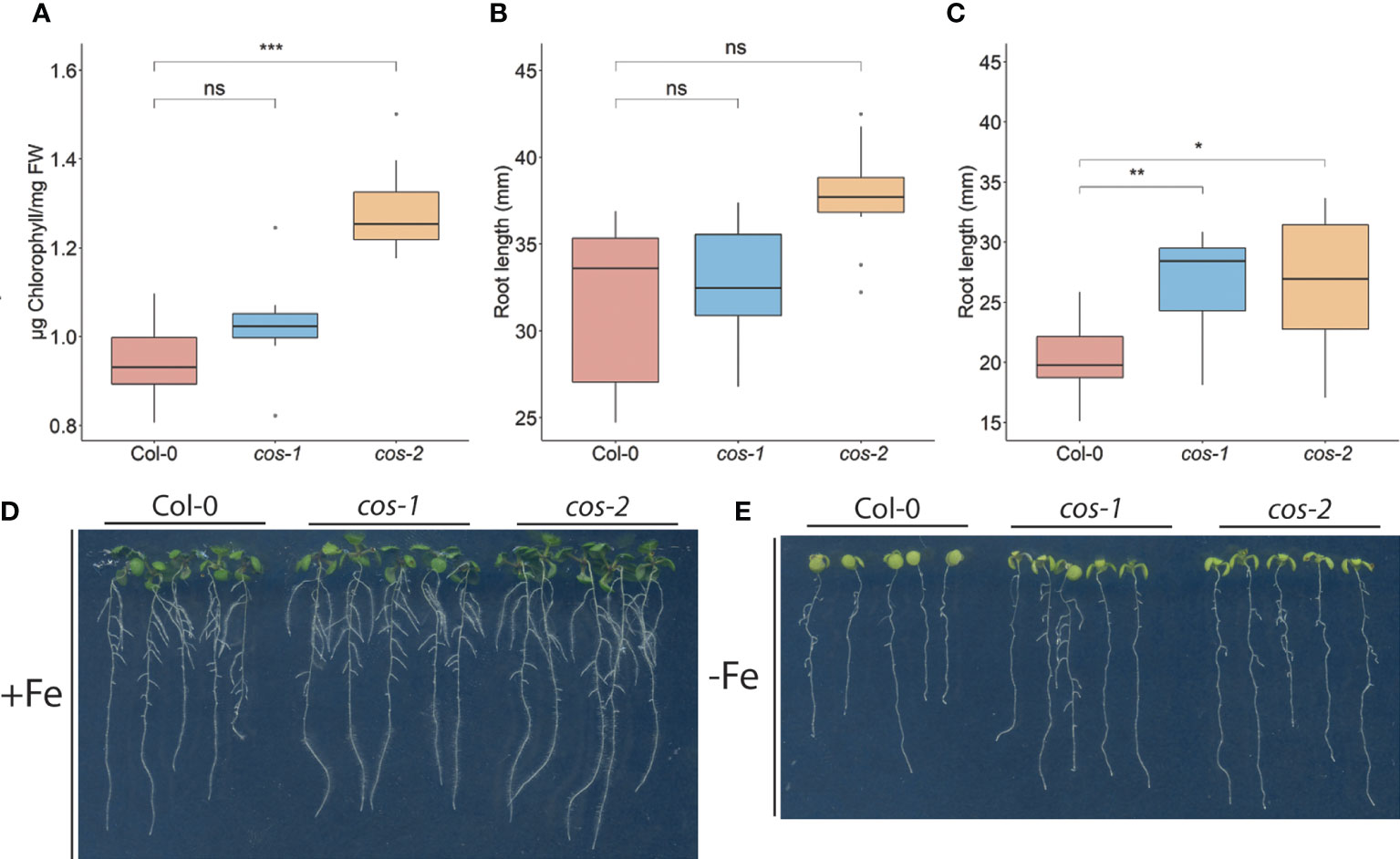
Figure 4 Phenotype of cos T-DNA mutants on 1/2X MS medium. (A) Total chlorophyll concentration of the shoot systems of plants grown on soil for 14 d. (B, C) Total root length of seedlings grown on 1/2X MS agar medium either with 50 μM Fe-EDTA (+Fe) or without Fe (-Fe) for 10 days. (D) Seedlings were grown on 1/2X MS agar medium either with 50 μM Fe-EDTA (+Fe) or without Fe (-Fe) for 10 days. * represents p-value ≤ 0.01, ** represents p-value ≤ 0.001, *** represents p-value ≤ 0.0001, ns, not significant.
We examined the root length of WT and cos mutants because we hypothesized that with low iron levels in shoots, cos mutants might show a root phenotype under Fe-deficiency. The root lengths of both cos mutants are similar to WT under iron sufficient conditions (Figure 4B). However, upon shifting plants to Fe-deficient environment for three days, both cos-1 and cos-2 had significantly longer root lengths than WT plants (Figure 4C, D).
Another symptom of iron deficiency is the up-regulation of transcripts for genes encoding the iron transporter, IRT1, and the iron-uptake regulatory gene FIT (Connolly et al., 2002; Colangelo and Guerinot, 2004). We examined IRT1 and FIT expression in WT and cos roots under Fe-sufficient and deficient conditions. Expression of IRT1 and FIT were similar to WT in cos roots under both conditions tested (Supplemental Figure 4), in spite of the low iron levels measured in cos mutant shoots and seeds.
Iron deficiency causes well documented negative effects on photosynthesis, and plants adapt to iron deficiency by decreasing the expression of particular iron containing proteins in leaves, while leaving the expression of presumably more essential iron proteins intact (Hantzis et al., 2018). During iron deficiency, plants produce reactive oxygen species, including singlet oxygen, and have specific responses that mitigate the damage that iron deficiency can cause in leaves (Akmakjian et al., 2021). We wondered whether disruption of COS affected these responses.
We investigated whether ROS production was affected in cos mutants. Expression of OXI1, an oxidative stress marker whose expression is induced by both singlet oxygen and hydrogen peroxide (Rentel et al., 2004), was significantly increased in cos mutants compared to WT under Fe-deficiency (Figure 5A). We then checked the expression of AT3G03810, a gene whose expression is induced specifically by singlet oxygen (Koh et al., 2016). During Fe-deficiency, AT3G03810 expression was significantly increased in cos mutants compared to WT (Figure 5B). The rise in expression of both of these genes during iron deficiency suggests that cos mutants might be accumulating singlet oxygen during Fe-deficiency.
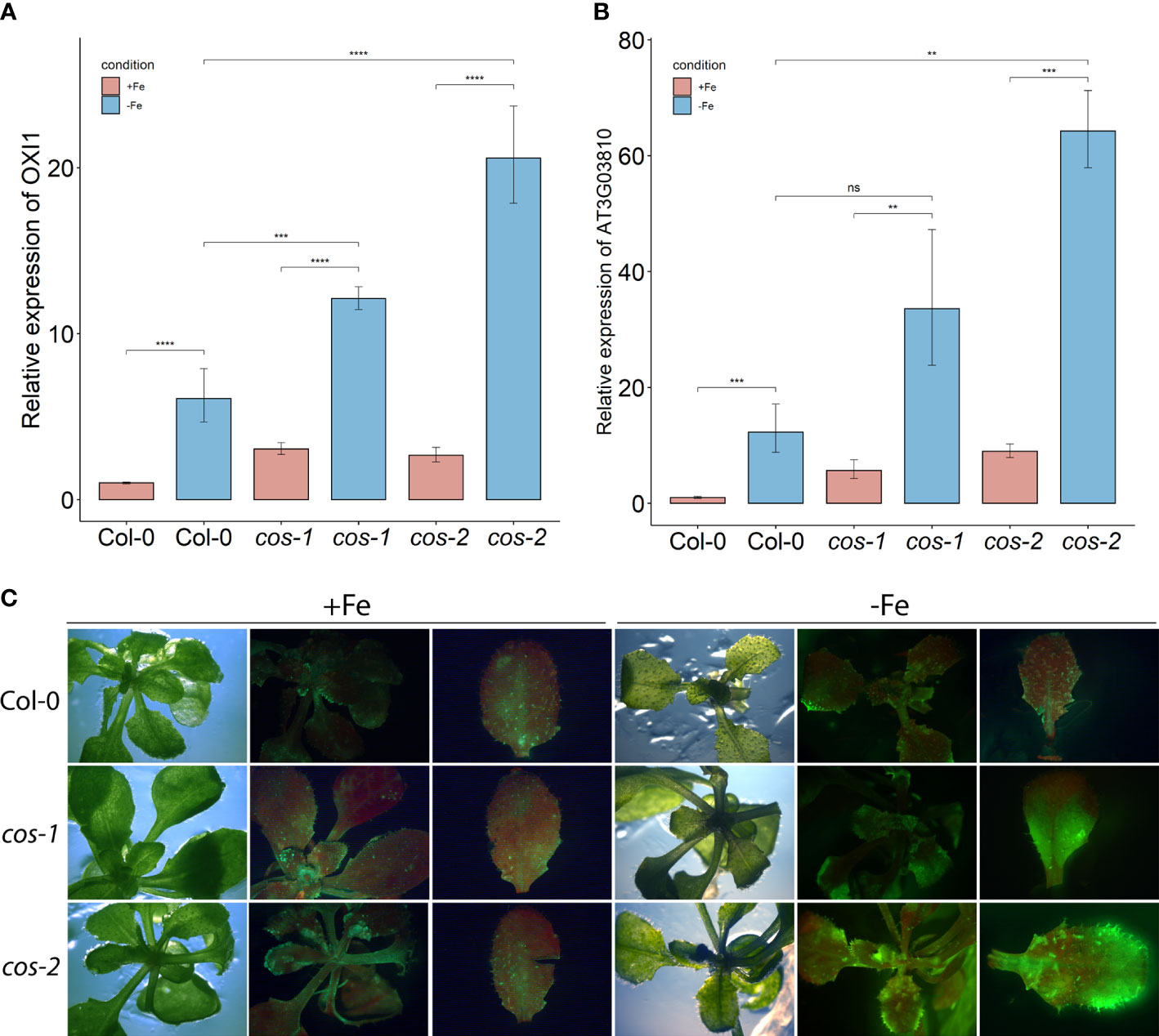
Figure 5 ROS in leaves of cos T-DNA mutants. RT-PCR of oxidative stress marker OXI1 (A) and singlet oxygen marker AT3G03810 (B) in WT and cos mutant shoots. Seedlings were grown on 1/2X MS agar with 50 μM Fe-EDTA for 14 d followed by a growth period of 3 d either on Fe-sufficient or Fe-deficient MS agar medium. Bars represent means ± SD of relative transcript levels normalized to ACT2 (n=3). (C) Singlet oxygen detection in cos mutants by SOSG. Seedlings were grown on 1/2X MS agar with 50 μM Fe-EDTA for 14 d followed by a growth period of 3 d either on Fe-sufficient or Fe-deficient MS agar medium. Seedlings were stained with SOSG and incubated under high light (500 µmol photons m−2 s−1) for 30 minutes. SOSG fluorescence is represented by green while chlorophyll autofluorescence is represented by red. * represents p-value ≤ 0.01, ** represents p-value ≤ 0.001, *** represents p-value ≤ 0.0001, **** represents p-value < 0.00001,, ns, not significant.
Next, we used the fluorescent stain Singlet Oxygen Sensor Green (SOSG; Invitrogen) to detect singlet oxygen production in cos mutants. Seedlings were vacuum infiltrated in the dark with SOSG, followed by a brief incubation under high light and subsequently imaged using a stereomicroscope. In WT plants, fluorescence from SOSG was barely detectable in either Fe-sufficient or deficient conditions. No fluorescence was detected in control plants that were infiltrated with buffer alone (Supplemental Figure 5). For the cos mutants, the fluorescence was similar to WT in Fe-sufficient conditions, but markedly brighter when plants were grown without iron, indicating that cos mutants accumulate high levels of singlet oxygen during Fe-deficiency (Figure 5C).
The transcription factors bHLH105/ILR3 and PYE were specifically shown to be required for photoprotection during Fe-deficiency and prevent accumulation of singlet oxygen (Akmakjian et al., 2021). PYE expression is altered in cos mutants (Figure 7A), where it is not strongly induced by iron deficiency. Expression of ILR3/bHLH105 is also altered in cos mutants, where basal expression levels are slightly, but significantly reduced (Figure 7C)
While analyzing the COS locus, we noticed an approximately 600 bp region within COS that had small (17-20 nt) sites of sequence complementarity to several well-known Fe-deficiency response genes (bHLH38, bHLH39, bHLH100, bHLH101, bHLH105/ILR3, bHLH115, BTSL1, FSD1, IMA1, PYE and VIT1; Figure 6; Liang, 2022). COS also contains two overlapping sites with sequence complementarity for the PHT4;4 gene, which is required for phosphate-deficiency induced inhibition of iron chlorosis (Nam et al., 2021; Figure 6). Since lncRNAs can produce sRNA, apparently through Dicer activity (Ma et al., 2014), we performed small RNA sequencing (sRNAseq) data from iron deficient and iron sufficient shoots, and discovered that there are small RNAs (sRNAs) generated, apparently from the larger COS transcript, that might target these Fe-deficiency genes, ultimately affecting their expression. The number of sRNAs emanating from COS is markedly increased during iron deficiency, and the sRNAs mapping to COS do not come from specific positions, and lack the distinctive, phased configuration characteristic of phasiRNAs (Liu et al., 2020; Figure 6) Instead, they map to a variety of apparently unphased and overlapping positions, many of which contain the small regions of sequence complementarity to Fe-deficiency response genes noted above (Figure 6).

Figure 6 Small RNA reads at the COS locus. Schematic representation of an ~600 bp region within COS that shares sequence complementarity to several Fe-deficiency genes, indicated with thick black lines labeled with putative target genes. At bottom, sRNAseq reads mapping to COS are shown from both +Fe and -Fe samples.
We checked the expression of some of these targets to test if their expression levels are affected in cos mutants. bHLH105/ILR3 expression, like the expression of other bHLH subgroup IVc transcription factors, is not affected by Fe-deficiency (Li et al., 2016), but as already noted, bHLH105/ILR3 expression is reduced in cos mutant plants (Figure 7C) Thus, COS does not seem to repress bHLH105/ILR3 expression. However, basal (+Fe) expression of the PYE gene is higher in cos mutants, as might be expected if COS repressed its expression. PYE mRNA levels normally increase during iron deficiency (Rodríguez-Celma et al., 2013), but cos mutant shoots failed to induce PYE expression under Fe-deficiency (Figure 7A). The pattern of expression of bHLH101 was similar to that of PYE—bHLH101 expression was higher in cos mutants grown on +Fe, but the normal iron deficiency induced increase in bHLH101 expression was attenuated (Figure 7B). Expression levels of IMA1 and BTSL1 were not different from WT under Fe-sufficient conditions, but, similar to PYE and bHLH101, their expression was attenuated during iron deficiency (Figures 7D, E). We did not see any difference in expression of bHLH38 in cos mutant shoots compared to WT under Fe-sufficient or deficient conditions (Figure 7F).
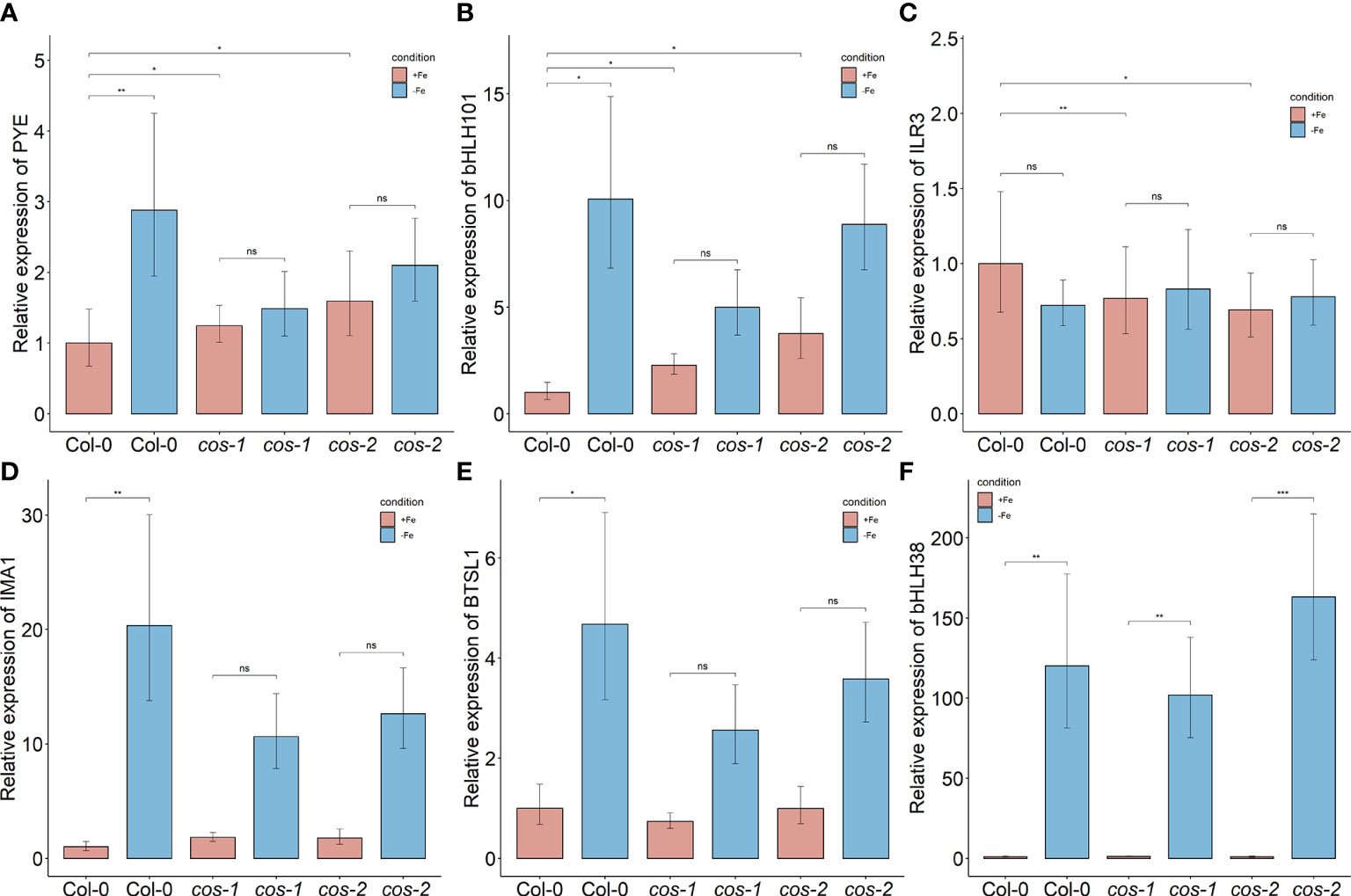
Figure 7 Expression of iron deficiency regulated genes in cos mutants. Expression levels of PYE (A), bHLH101 (B), ILR3/bHLH105 (C), IMA1 (D), BTSL1 (E), and bHLH38 (F) transcripts in WT shoots and roots. Seedlings were grown on 1/2X MS agar with 50 μM Fe-EDTA for 14 d followed by a growth period of 3 d either on Fe-sufficient or Fe-deficient MS agar medium. Bars represent means ± SD of relative transcript levels normalized to ACT2 (n = 3). * represents p-value ≤ 0.01. ** represents p-value ≤ 0.001, *** represents p-value ≤ 0.0001, ns, not significant.
In this study, we have identified a previously unannotated iron-regulated long non-coding RNA gene that we named CAN OF SPINACH (COS). It is possible that previous gene expression studies could have misidentified the genes on the opposite strand (AT1G13608 and/or AT1G13609) as iron-regulated, and that the truly iron-regulated mRNA from the locus is actually the COS lncRNA (Rodríguez-Celma et al., 2013; Naranjo-Arcos et al., 2017; Bastow et al., 2018; Nam et al., 2021; Peixoto et al., 2021). We detected a spliced and an unspliced variant of COS in whole shoots of Arabidopsis, both of which were polyadenylated (Figure 1C).
cos mutant plants have phenotypes that suggest abnormalities in iron homeostasis. They have reduced levels of iron in their shoots and seeds, which indicates that they are not taking up normal amounts of iron from the soil. In addition, and contrary to the expectation for a plant with lowered iron in tissues, the mutants have elevated chlorophyll levels (Figure 4A). This indicates that the plants are not responding to low shoot iron levels in a typical way. The roots of cos mutants also have abnormal growth, albeit only during iron deficiency. The cos roots are long compared to WT roots when they are grown without iron in the medium (Figures 4B, C). This phenotype can be interpreted in two ways. In one view, when iron is not available, root growth simply cannot continue owing to the lack of iron per se. However, it is possible that reduced root growth during iron deficiency is a low-iron-induced developmental program intended to protect the plant and reserve accumulated iron for more critical functions. The high chlorophyll/low iron phenotype of the mutant could be interpreted in a similar way: WT plants may reduce chlorophyll levels as an iron economy measure, when iron in the shoots is low. In cos mutants, that iron economy measure may have been disrupted, and the mutants produce high amounts of chlorophyll in spite of the low levels of iron in the shoots. If this view is correct, then the cos phenotypes could be interpreted as a failure of the plant to make normal adaptive growth alterations during iron deficiency. Since cos mutants seem to fail to adapt normally to iron, we considered whether cos mutants might have a reproductive penalty, and they do. The individual seed weight is lower in both cos mutants (Figure 3D), which could indicate minor problems with seed development. However, seed number was not strongly affected in the mutants.
Our singlet oxygen findings also support the idea that cos mutants do not adapt well to iron deficiency. One of the ways that Arabidopsis adapts to low iron is by changing gene expression to lessen the potential for photooxidative damage that occurs during iron deficiency (Akmakjian et al., 2021). Increased gene expression from the generic oxidative stress marker OXI1 and At3g01830, which is specifically induced by singlet oxygen, indicate that cos plants have increased singlet oxygen (Figures 5A, B), and direct histochemical staining for singlet oxygen indicates cos mutant plants are less able to prevent accumulation of singlet oxygen during iron deficiency. We speculate that this phenotype may reflect the inability of cos mutants to induce expression of PYE, and/or their lowered basal level of BHLH105/ILR3 expression (Figures 7A, C). Both PYE and BHLH105/ILR3 are required for this adaptation to low iron.
As part of this study, we performed a whole transcriptome analysis of Arabidopsis shoots under iron sufficient, deficient and resupplied conditions. We have compared our shoot iron transcriptome data to a publicly available root iron transcriptome (Rodríguez-Celma et al., 2013). When iron deficiency induced genes are compared between shoots and roots, we see that 20% of the transcripts are upregulated in both tissues. This indicates that shoots display a distinct response to Fe deficiency. Surprisingly, though, among the 20% that are up-regulated in both shoots and roots, the majority were key members of the root iron deficiency response. In roots, these ultimately affect expression of genes like IRT1 and FRO2 that have direct roles in iron uptake from the soil. These include bHLH38/39/100/101 (Wang et al., 2007; Yuan et al., 2008), and, PYE (Long et al., 2010). The function of these genes in shoots is largely unexplored, and their downstream targets are of considerable interest as components of the iron deficiency response of above ground tissues, which may include both adaptive responses (Akmakjian et al., 2021) and signaling of iron status through the plants (Grusak and Pezeshgi, 1996; Mendoza-Cózatl et al., 2014; Zhai et al., 2014; Kumar et al., 2017). Other known iron related genes included iron reductases (FRO2, FRO3) and transporters (OPT3, NRAMP4 and ZIF1), the NAS4 gene that produces the metal chelator nicotianamine, and the genes encoding IMA1/2/3 peptides that regulate BTS (Grillet et al., 2018; Li et al., 2021) and potentially, BTSL1 (Lichtblau et al., 2022).
About half of the iron deficiency induced transcripts in leaves decreased their expression after 24 hours of iron resupply. Among the 73 transcripts with ‘rapid response to resupply’, 20 transcripts are among the key iron deficiency response genes discussed above. Interestingly, for these 20 transcripts involved in iron deficiency response, their expression levels went down to basal levels after iron was resupplied. Only about 25% of the transcripts (21 out of 85) that were downregulated during iron deficiency responded rapidly to iron resupply. Interestingly, among these 21 transcripts chloroplast localized transcripts were highly abundant and these transcripts were involved in metabolism and photosynthesis. This might indicate how quickly photosynthesis is recovered after a period of iron deficiency.
In addition to mRNA sequencing of Arabidopsis shoots, we carried out sRNA sequencing of whole shoots under Fe-sufficient and deficient conditions. We noticed the presence of sRNAs that appear to emanate from the COS locus, and that are much more abundant in plants grown under iron deficiency. While searching for putative targets of these sRNAs, we found small regions of sequence complementarity to several important Fe-deficiency-induced genes (bHLH38, bHLH39, bHLH100, bHLH101, bHLH105/ILR3, bHLH115, BTSL1, FSD1, IMA1, PYE and VIT1) (Figure 6). This surprising finding led us to speculate that the sRNAs might affect the expression of these putative targets. However, analysis of the expression of the putative targets of some of these sRNAs indicated that most are not repressed by COS. Thus, while it is possible that the sRNAs have a regulatory function, it remains unclear at this time.
Alternatively, COS might affect expression of these putative targets through its secondary structure. lncRNAs can form secondary structures that bind genes in trans and affect their expression (Chen et al., 2021; Wierzbicki et al., 2021). Within these secondary structures, lncRNAs can have domains where base-pairing probabilities are increased, raising the possibility that the ~600 bp region of COS that contains complementary sequences for several iron homeostasis genes could be a biologically active region within the secondary structure of COS transcript. Interaction between the COS lncRNA and its targets could then affect the expression of these genes. Further investigation of the mechanism of COS lncRNA function should shed new light on the processes by which plants adapt to iron deficiency.
We particularly examined putative COS targets whose expression is associated with Fe-deficiency responses. Expression of some of these putative targets showed downward trends in expression in cos mutants, or failed to be fully induced during iron deficiency. We noted that putative target genes PYE, and bHLH101 display higher expression during iron sufficient growth of cos mutants. This is consistent with the possibility that COS negatively regulates their expression. However, PYE and bHLH101, which are both normally up-regulated during iron deficiency, both seem to have dampened iron deficiency responses in cos mutants. This may indicate a secondary effect in which gene expression is indirectly changed in cos mutants, perhaps owing to alterations in expression of upstream regulators like PYE and/or ILR3/bHLH115.
The original contributions presented in the study are publicly available. This data can be found here: GEO dataset, GSE209595.
EW conceived the original idea. AB carried out the experiments, analysed the data. AB and EW wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by grant from the National Science Foundation (NSF-IOS-1754966).
We thank Chris Phillips and Dan Jones at the UMass Morrill Greenhouses, for assistance with plant husbandry.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1005020/full#supplementary-material
Afgan, E., Baker, D., Batut, B., Van Den Beek, M., Bouvier, D., Ech, M., et al. (2018). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi: 10.1093/nar/gky379
Akmakjian, G. Z., Riaz, N., Guerinot, M.L. (2021). Photoprotection during iron deficiency is mediated by the bHLH transcription factors PYE and ILR3. Proc. Natl. Acad. Sci. U. S. A. 118, e2024918118. doi: 10.1073/pnas.2024918118
Axtell, M. J. (2013). ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA 19, 740–751. doi: 10.1261/rna.035279.112
Bastow, E. L., Garcia de la Torre, V. S., Maclean, A. E., Green, R. T., Merlot, S., Thomine, S., et al. (2018). Vacuolar iron stores gated by NRAMP3 and NRAMP4 are the primary source of iron in germinating seeds. Plant Physiol. 177, 1267–1276. doi: 10.1104/pp.18.00478
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Blaby-Haas, C. E., Merchant, S. S. (2013). Iron sparing and recycling in a compartmentalized cell. Curr. Opin. Microbiol. 16, 677–685. doi: 10.1016/j.mib.2013.07.019
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chen, Q., Liu, K., Yu, R., Zhou, B., Huang, P., Cao, Z., et al. (2021). From “Dark matter” to “Star”: Insight into the regulation mechanisms of plant functional long non-coding RNAs. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.650926
Colangelo, E. P., Guerinot, M. L. (2004). The essential basic helix-Loop-Helix protein FIT1 is required for the iron deficiency response. Plant Cell 16, 3400–3412. doi: 10.1105/tpc.104.024315
Connolly, E. L., Fett, J. P., Guerinot, M.L. (2002). Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14, 1347–1357. doi: 10.1105/tpc.001263
Connorton, J. M., Balk, J., Rodríguez-Celma, J. (2017). Iron homeostasis in plants-a brief overview. Metallomics 9, 813–823. doi: 10.1039/c7mt00136c
Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S. L., Briat, J. F., Walker, E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349. doi: 10.1038/35053080
Gaymard, F., Boucherez, J., Briat, J. F. (1996). Characterization of a ferritin mRNA from arabidopsis thaliana accumulated in response to iron through an oxidative pathway independent of abscisic acid. Biochem. J. 318, 67–73. doi: 10.1042/bj3180067
Godde, D., Hefer, M. (1994). Photoinhibition and light-dependent turnover of the D1 reaction-centre polypeptide of photosystem II are enhanced by mineral-stress conditions. Planta 193, 290–299. doi: 10.1007/BF00192543
Golicz, A. A., Singh, M. B., Bhalla, P. L. (2018). The long intergenic noncoding RNA (LincRNA) landscape of the soybean genome. Plant Physiol. 176, 2133–2147. doi: 10.1104/PP.17.01657
Grillet, L., Lan, P., Li, W., Mokkapati, G., Schmidt, W. (2018). IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat. Plants 4, 953–963. doi: 10.1038/s41477-018-0266-y
Grusak, M. A., Pezeshgi, S. (1996). Shoot-to-Root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol. 110, 329–334. doi: 10.1104/pp.110.1.329
Halliwell, B., Gutteridge, J. M. C. (2015). Reactive species can pose special problems needing special solutions: Some examples. Free Radicals Biol. Med., 354–410. doi: 10.1093/ACPROF:OSO/9780198717478.003.0007
Hantzis, L. J., Kroh, G. E., Jahn, C. E., Cantrell, M., Peers, G., Pilon, M., et al. (2018). A program for iron economy during deficiency targets specific fe proteins. Plant Physiol. 176, 596–610. doi: 10.1104/pp.17.01497
Kang, Y. J., Yang, D. C., Kong, L., Hou, M., Meng, Y. Q., Wei, L., et al. (2017). CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 45, W12–W16. doi: 10.1093/NAR/GKX428
Khan, M. A., Castro-Guerrero, N. A., McInturf, S. A., Nguyen, N. T., Dame, A. N., Wang, J., et al. (2018). Changes in iron availability in arabidopsis are rapidly sensed in the leaf vasculature and impaired sensing leads to opposite transcriptional programs in leaves and roots. Plant Cell Environ. 41, 2263–2276. doi: 10.1111/pce.13192
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Koh, E., Carmieli, R., Mor, A., Fluhr, R. (2016). Singlet oxygen-induced membrane disruption and serpin-protease balance in vacuolar-driven cell death. Plant Physiol. 171, 1616–1625. doi: 10.1104/PP.15.02026
Krieger-Liszkay, A. (2005). Singlet oxygen production in photosynthesis. J. Exp. Bot. (J Exp. Bot) 56, 337–346. doi: 10.1093/jxb/erh237
Kroh, G. E., Pilon, M. (2020). Iron deficiency and the loss of chloroplast iron-sulfur cluster assembly trigger distinct transcriptome changes in Arabidopsis rosettes. Metallomics 12, 1748–1764. doi: 10.1039/d0mt00175a
Kumar, R. K., Chu, H. H., Abundis, C., Vasques, K., Rodriguez, D. C., Chia, J. C., et al. (2017). Iron-nicotianamine transporters are required for proper long distance iron signaling. Plant Physiol. 175, 1254–1268. doi: 10.1104/pp.17.00821
Liang, G. (2022). Iron uptake, signaling, and sensing in plants. Plant Commun. 3, 100349. doi: 10.1016/j.xplc.2022.100349
Lichtblau, D. M., Schwarz, B., Baby, D., Endres, C., Sieberg, C., Bauer, P. (2022). The iron deficiency-regulated small protein effector FEP3/IRON MAN1 modulates interaction of BRUTUS-LIKE1 with bHLH subgroup IVc and POPEYE transcription factors. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.930049
Li, Y., Lu, C. K., Li, C. Y., Lei, R. H., Pu, M. N., Zhao, J. H., et al. (2021). IRON MAN interacts with BRUTUS to maintain iron homeostasis in arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 118, e2109063118. doi: 10.1073/pnas.2109063118
Liu, Y., Teng, C., Xia, R., Meyers, B. C. (2020). PhasiRNAs in plants: Their biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell 32, 3059–3080. doi: 10.1105/TPC.20.00335
Li, X., Zhang, H., Ai, Q., Liang, G., Yu, D. (2016). Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 170, 2478–2493. doi: 10.1104/pp.15.01827
Long, T. A., Tsukagoshi, H., Busch, W., Lahner, B., Salt, D. E., Benfey, P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in arabidopsis roots. Plant Cell Online 22, 2219–2236. doi: 10.1105/tpc.110.074096
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10. doi: 10.14806/ej.17.1.200
Ma, X., Shao, C., Jin, Y., Wang, H., Meng, Y. (2014). Long non-coding RNA: A novel endogenous source for the generation of dicer-like 1-dependent small RNAs in arabidopsis thaliana. RNA Biol. 11, 373–390. doi: 10.4161/rna.28725
Mendoza-Cózatl, D. G., Xie, Q., Akmakjian, G. Z., Jobe, T. O., Patel, A., Stacey, M. G., et al. (2014). OPT3 is a component of the iron-signaling network between leaves and roots and misregulation of OPT3 leads to an over-accumulation of cadmium in seeds. Mol. Plant 7, 1455–1469. doi: 10.1093/mp/ssu067
Mi, H., Muruganujan, A., Huang, X., Ebert, D., Mills, C., Guo, X., et al. (2019). Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703–721. doi: 10.1038/s41596-019-0128-8
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/NAR/GKAA913
Msilini, N., Essemine, J., Zaghdoudi, M., Harnois, J., Lachaâl, M., Ouerghi, Z., et al. (2013). How does iron deficiency disrupt the electron flow in photosystem I of lettuce leaves? J. Plant Physiol. 170, 1400–1406. doi: 10.1016/J.JPLPH.2013.05.004
Nam, H.-I., Shahzad, Z., Dorone, Y., Clowez, S., Zhao, K., Bouain, N., et al. (2021). Interdependent iron and phosphorus availability controls photosynthesis through retrograde signaling. Nat. Commun. 12, 1–13. doi: 10.1038/s41467-021-27548-2
Naranjo-Arcos, M. A., Maurer, F., Meiser, J., Pateyron, S., Fink-Straube, C., Bauer, P. (2017). Dissection of iron signaling and iron accumulation by overexpression of subgroup ib bHLH039 protein. Sci. Rep. 7, 10911. doi: 10.1038/s41598-017-11171-7
Nechushtai, R., Conlan, A. R., Harir, Y., Song, L., Yogev, O., Eisenberg-Domovich, Y., et al. (2012). Characterization of arabidopsis NEET reveals an ancient role for NEET proteins in iron metabolism. Plant Cell 24, 2139–2154. doi: 10.1105/tpc.112.097634
Peixoto, B., Moraes, T. A., Mengin, V., Margalha, L., Vicente, R., Feil, R., et al. (2021). Impact of the SnRK1 protein kinase on sucrose homeostasis and the transcriptome during the diel cycle. Plant Physiol. 187, 1357–1373. doi: 10.1093/plphys/kiab350
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Ravet, K., Touraine, B., Boucherez, J., Briat, J. F., Gaymard, F., Cellier, F. (2009). Ferritins control interaction between iron homeostasis and oxidative stress in arabidopsis. Plant J. 57, 400–412. doi: 10.1111/j.1365-313X.2008.03698.x
Rentel, M. C., Lecourieux, D., Ouaked, F., Usher, S. L., Petersen, L., Okamoto, H., et al. (2004). OXI1 kinase is necessary for oxidative burst-mediated signalling in arabidopsis. Nat 427 (6977), 858–861. doi: 10.1038/nature02353
Robinson, N. J., Procter, C. M., Connolly, E. L., Guerinot, M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. doi: 10.1038/17800
Rodríguez-Celma, J., Pan, I. C., Li, W., Lan, P., Buckhout, T. J., Schmidt, W. (2013). The transcriptional response of arabidopsis leaves to fe deficiency. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00276
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Shahid, S., Axtell, M. J. (2014). Identification and annotation of small RNA genes using ShortStack. Methods 67, 20–27. doi: 10.1016/j.ymeth.2013.10.004
Spiller, S., Terry, N. (1980). Limiting factors in photosynthesis: II. IRON STRESS DIMINISHES PHOTOCHEMICAL CAPACITY BY REDUCING THE NUMBER OF PHOTOSYNTHETIC UNITS 1, 2. Plant Physiol. 65, 121. doi: 10.1104/PP.65.1.121
Tanaka, R., Kobayashi, K., Masuda, T. (2011). Tetrapyrrole metabolism in arabidopsis thaliana. Arab. B. 9, e0145. doi: 10.1199/tab.0145
Tewari, R. K., Hadacek, F., Sassmann, S., Lang, I. (2013). Iron deprivation-induced reactive oxygen species generation leads to non-autolytic PCD in brassica napus leaves. Environ. Exp. Bot. 91, 74–83. doi: 10.1016/J.ENVEXPBOT.2013.03.006
Thomas, P. D., Ebert, D., Muruganujan, A., Mushayahama, T., Albou, L. P., Mi, H. (2022). PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 31, 8–22. doi: 10.1002/PRO.4218
Triantaphylidès, C., Krischke, M., Hoeberichts, F. A., Ksas, B., Gresser, G., Havaux, M., et al. (2008). Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 148, 960–968. doi: 10.1104/PP.108.125690
Urzica, E. I., Casero, D., Yamasaki, H., Hsieh, S. I., Adler, L. N., Karpowicz, S. J., et al. (2012). Systems and Trans -system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell 24, 3921–3948. doi: 10.1105/tpc.112.102491
Vert, G., Grotz, N., Dédaldéchamp, F., Gaymard, F., Guerinot, M., Briat, J.-F., et al. (2002). IRT1, an arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14, 1223–1233. doi: 10.1105/tpc.001388
Wang, H. L. V., Chekanova, J. A. (2017). “Long noncoding RNAs in plants,” in Advances in experimental medicine and biology (New York LLC: Springer), 133–154. doi: 10.1007/978-981-10-5203-3_5
Wang, H. Y., Klatte, M., Jakoby, M., Bäumlein, H., Weisshaar, B., Bauer, P. (2007). Iron deficiency-mediated stress regulation of four subgroup ib BHLH genes in arabidopsis thaliana. Planta 226, 897–908. doi: 10.1007/s00425-007-0535-x
Ward, R. J., Crichton, R. R. (2015). “Iron: Properties and Determination,” in Encyclopedia of Food and Health (Academic Press), 468–475. doi: 10.1016/B978-0-12-384947-2.00403
Waters, B. M., Chu, H.-H., Didonato, R. J., Roberts, L. A., Eisley, R. B., Lahner, B., et al. (2006). Mutations in arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141, 1446–1458. doi: 10.1104/pp.106.082586
Wierzbicki, A. T., Blevins, T., Swiezewski, S. (2021). Long noncoding RNAs in plants. Annu. Rev. Plant Biol. 72, 245–271. doi: 10.1146/annurev-arplant-093020-035446
Yadavalli, V., Neelam, S., Rao, A. S. V. C., Reddy, A. R., Subramanyam, R. (2012). Differential degradation of photosystem I subunits under iron deficiency in rice. J. Plant Physiol. 169, 753–759. doi: 10.1016/J.JPLPH.2012.02.008
Yuan, Y., Wu, H., Wang, N., Li, J., Zhao, W., Du, J., et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in arabidopsis. Cell Res. 18, 385–397. doi: 10.1038/cr.2008.26
Yu, N., Liang, Y., Wang, Q., Peng, X., He, Z., Hou, X. (2022). Transcriptomic analysis of OsRUS1 overexpression rice lines with rapid and dynamic leaf rolling morphology. Sci. Rep. 12, 6736. doi: 10.1038/s41598-022-10784-x
Zandalinas, S. I., Song, L., Sengupta, S., McInturf, S. A., Grant, D. A. G., Marjault, H. B., et al. (2020). Expression of a dominant-negative AtNEET-H89C protein disrupts iron–sulfur metabolism and iron homeostasis in arabidopsis. Plant J. 101, 1152–1169. doi: 10.1111/TPJ.14581
Zanin, L., Venuti, S., Zamboni, A., Varanini, Z., Tomasi, N., Pinton, R. (2017). Transcriptional and physiological analyses of fe deficiency response in maize reveal the presence of strategy I components and Fe/P interactions. BMC Genomics 18, 1–15. doi: 10.1186/S12864-016-3478-4/TABLES/3
Zhai, Z., Gayomba, S. R., Jung, H.-i., Vimalakumari, N. K., Pineros, M., Craft, E., et al. (2014). OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in arabidopsis. Plant Cell 26, 2249–2264. doi: 10.1105/tpc.114.123737
Zhang, X., Zhang, D., Sun, W., Wang, T. (2019). The adaptive mechanism of plants to iron deficiency via iron uptake, transport, and homeostasis. Int. J. Mol. Sci. 20, 2424. doi: 10.3390/ijms20102424
Zhang, H., Zhang, F., Yu, Y., Feng, L., Jia, J., Liu, B., et al. (2020). A comprehensive online database for exploring -20,000 public arabidopsis RNA-seq libraries Mol. Plant 13, 1231–1233. doi: 10.1016/j.molp.2020.08.001
Keywords: long non-coding RNA, iron deficiency, nutrient deficiency, Arabidopsis thaliana, oxidative stress, singlet oxygen
Citation: Bakirbas A and Walker EL (2022) CAN OF SPINACH, a novel long non-coding RNA, affects iron deficiency responses in Arabidopsis thaliana. Front. Plant Sci. 13:1005020. doi: 10.3389/fpls.2022.1005020
Received: 27 July 2022; Accepted: 13 September 2022;
Published: 05 October 2022.
Edited by:
Seçkin Eroğlu, Middle East Technical University, TurkeyReviewed by:
Alexandra Lešková, UMR5004 Biochimie et Physiologie Moléculaire des Plantes (BPMP), FranceCopyright © 2022 Bakirbas and Walker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elsbeth L. Walker, ZXdhbGtlckB1bWFzcy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.