
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 September 2022
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1001427
This article is part of the Research Topic Interactive Effects of Plant Growth-Promoting Microbes and Nanoparticles on the Physiology, Growth, and Yield of Crops View all 11 articles
 Sagheer Ahmad1
Sagheer Ahmad1 Guizhen Chen1
Guizhen Chen1 Jie Huang1
Jie Huang1 Kang Yang1
Kang Yang1 Yang Hao1
Yang Hao1 Yuzhen Zhou1
Yuzhen Zhou1 Kai Zhao1,2
Kai Zhao1,2 Siren Lan1
Siren Lan1 Zhongjian Liu1*
Zhongjian Liu1* Donghui Peng1*
Donghui Peng1*Biological control is a safe way of combating plant diseases using the living organisms. For the precise use of microbial biological control agents, the genetic information on the hypersensitive response (HR), and defense-related gene induction pathways of plants are necessary. Orchids are the most prominent stakeholders of floriculture industry, and owing to their long-awaited flowering pattern, disease control is imperative to allow healthy vegetative growth that spans more than 2 years in most of the orchids. We observed leaf-less flowering in three orchid species (Cymbidium ensifolium, C. goeringii and C. sinense). Using these materials as reference, we performed transcriptome profiling for healthy leaves from non-infected plants to identify genes specifically involved in plant-pathogen interaction pathway. For this pathway, a total of 253 differentially expressed genes (DEGs) were identified in C. ensifolium, 189 DEGs were identified in C. goeringii and 119 DEGs were found in C. sinense. These DEGs were mainly related to bacterial secretion systems, FLS2, CNGCs and EFR, regulating HR, stomatal closure and defense-related gene induction. FLS2 (LRR receptor-like serine/threonine kinase) contained the highest number of DEGs among three orchid species, followed by calmodulin. Highly upregulated gene sets were found in C. sinense as compared to other species. The great deal of DEGs, mainly the FLS2 and EFR families, related to defense and immunity responses can effectively direct the future of biological control of diseases for orchids.
Plants have developed a multi-layered defense system to resist the invasion of pathogens (Kaur et al., 2022). In the primary response, plants recognize pathogens by using PTI (PAMP-triggered immunity) which uses cell-surface pattern-recognition receptors (PRRs). Defense-related genes are activated to produce antimicrobial compounds through MAPK signaling pathway triggered by FLS2 and EFR (Nishad et al., 2020). The increase in the concentration of Ca2+ in the cytosol regulates the production of ROS (reactive oxygen species) and HR (hypersensitive response)/programmed cell death. The secondary response is called ETI (effector-triggered immunity). In this response, pathogens use secretion systems to inject effector proteins into the plant cells to suppress PTI. Pathogens also invade the immunity of the host plants by manipulating their hormone signaling pathways. In response to pathogen effectors, plants use intracellular surveillance proteins called R proteins to monitor the presence of the virulence proteins from pathogens. The ETI arrests pathogen growth by localized programmed cell death, which causes cultivar-specific disease resistance (Nishad et al., 2020).
FLAGELLIN SENSING 2 (FLS2) is an important regulator of plant immunity against pathogenic bacteria (Zipfel et al., 2004). It acts as a PRR (pattern recognition receptor) for flagellin, which is the building block of bacterial flagellum and is perceived as PAMP (pathogen-associated molecular pattern) protein in animals and plants (Gómez-Gómez and Boller, 2000; Hayashi et al., 2001). PRRs proteins contain single transmembrane bearing N-terminal receptor-like domain that recognizes ligands. FLS2 is the first PRR identified in plants and belongs to a large family of receptor kinases (Robatzek and Wirthmueller, 2013). It can recognize a conserved 22-amino acid peptide called flg22 in the N-terminus of flagellin (Felix et al., 1999; Bauer et al., 2001; Chinchilla et al., 2006). In Arabidopsis, other PRRs include EFR (EF-Tu receptor) that identifies the elf18 peptide corresponding to the N-terminus of the bacterial EF-Tu, and CERK1 (chitin elicitor receptor kinase 1) that recognizes the chitin of fungi and the bacterial peptidoglycans (Zipfel et al., 2006; Miya et al., 2007; Petutschnig et al., 2010; Shafique et al., 2011; Willmann et al., 2011). In rice, chitin-elicitor-binding protein, a receptor protein, functions together with OsCERK1 for chitin perception (Shimizu et al., 2010). The receptor kinase XA21 uses the XA21 peptide sensing to create resistance against Xanthomonas oryzae bacteria (Lee et al., 2009). Tomato receptor proteins LeEix1/2 confer the perception of xylanase from fungi in the ethylene-induction pathway (Ron and Avni, 2004). Moreover, PRRs can perform heterologous functions (Monaghan and Zipfel, 2012). For example, Ve1 is a PRR in tomato and mediates immunity against Verticillium fungus, while it can also provide resistance against Verticillium in Arabidopsis (Fradin et al., 2011; De Jonge et al., 2012).
FLS2 regulates the ROS (reactive oxygen species) production, the defense-related hormones (salicylic acid and ethylene), deposition of secondary compounds (e.g., callose) and the transcriptional reprogramming involving WRKY TFs (Boller and Felix, 2009), thereby establishing plant immunity. FLS2 makes a stable complex with BRI-associated kinase 1 (BAK1) in a flg22-dependent manner (Robatzek and Wirthmueller, 2013). BAK1 is a regulatory transmembrane receptor kinase and a member of SERK (somatic embryo receptor kinase) family; it involves brassinosteroid signaling and flg22-activated multiple immune responses (Li et al., 2002; Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011; Schwessinger et al., 2011). BAK1 and FLS2 interact with BIK1 (botrytis-induced kinase 1) and its homologs (PBL1, PBL2, and PBS1); while BIK1 is receptor-like cytoplasmic kinase (Robatzek and Wirthmueller, 2013). MAPK (mitogen-activated protein kinase) cascades work downstream of FLS2-BAK1 complex in response to flg22 (Asai et al., 2002; Ichimura et al., 2006). Moreover, FLS2-BAK1 dimerization activates CDPK (calcium dependent protein kinase) signaling pathways (Boudsocq et al., 2010).
Calcium signaling plays important roles in symbiotic and pathogenic plant-microbe interactions (Zipfel and Oldroyd, 2017; Mubeen et al., 2022; Rhodes et al., 2022). PRRs trigger a transiently rapid influx in the cytoplasmic calcium, depending upon the PRR complex and downstream RLCKs (receptor-like cytoplasmic kinases; Ranf et al., 2012, 2014; Li et al., 2014; Monaghan et al., 2015; Ahmad et al., 2018). Recently, a number of calcium channels have been implicated, such as CNGCs (cyclic nucleotide-gated channels; Rhodes et al., 2022). The NRL-triggered cell death, known as HR (hypersensitive response), depends on calcium (Bashir et al., 2013). Treatment with calcium channel blockers and calcium chelators can inhibit cell death (Levine et al., 1996; Grant et al., 2000; Ali et al., 2007).
Perception of PAMP by PRRs instigates a defense response called PTI (pattern-triggered immunity) to inhibit pathogenic infections (Macho and Zipfel, 2014). Against it, pathogens use effector molecules to suppress PTI (Wang et al., 2022) and plants have evolved intracellular receptors to counter effector molecules. These receptors are called NLR (nucleotide-binding, leucine-rich repeats) proteins which activate ETI (effector-triggered immunity; Ngou et al., 2022). PTI and ETI have been considered as two independent pathway layers regulating plant immune system (Rhodes et al., 2022). PRR activation also triggers the phosphorylation of MAPK cascades. MAP kinase kinase kinase (MAPKKK/MEKK) activation triggers the downstream MAP kinase kinase (MAPKK/MKK), which stimulates the downstream MAPK/MPK (Asai et al., 2002; Ichimura et al., 2002; Akram et al., 2014; He et al., 2018; Komis et al., 2018).
The Orchidaceae falls among the largest and the highly evolved monocot plant families (Roberts and Dixon, 2008). Approximately, 70,000 orchid species have been cultivated worldwide as medicinal and ornamental plants (Wong, 2002). The Cymbidium orchids are best known for their ideal characteristics and aesthetic appeal around the world (Cribb, 2014). However, there are a number of biotic (viral, bacterial and fungal diseases) and abiotic (salinity and drought stress) factors that seriously affect the quality and production of orchids (Tuhid et al., 2012; Akram et al., 2019; Ren et al., 2020). For example, Tobamovirus and CymMV (Cymbidium mosaic virus) are the lethal viral pathogens causing necrosis, chlorosis and dwarfism in orchids, which causes huge ornamental and economic losses to orchids (Koh et al., 2014). A little information is available on the immunity mechanism of orchids, although their long vegetative phase requires a strong defense strategy to cope with pathogens. Such studies indicate the orchid immunity and defense responses for some specific diseases. A transcriptome-wide analysis for multiple orchid species has not been described, especially the Cymbidium orchids. Moreover, a leaf-less control has not been discussed before, that can produce much more genetic information on immunity regulation in the healthy leaves. For an effective biological control, the understanding of orchids defense machinery is a demanding area of research. Therefore, this study identifies an ideal leaf-less control to study the genetic makeup of immunity responses for Cymbidium orchids. Three ideal Cymbidium orchids, C. ensifolium, C. goeringii and C. sinense were used to perform reference-based transcriptome analysis and then genes related to plant-pathogen interaction pathway and MAPK-signaling pathways were ascertained. This study provides a broader image of orchid defense mechanism and maximizes the immunity information than ever before. The outcomes, thus, suggest the effective immunity mechanism to devise biological control strategy for orchids and other floriculture crops.
Three orchid species (C. ensifolium, C. goeringii, and C. sinense) were grown through protocorms in long jars with special media containing 6-BA (8.0 mg L−1), NAA (0.5 mg L−1), sugar (35 g L−1), activated carbon (1.5 g L−1) and agar (7.0 g L−1). The chamber temperature was set to 26 ± 2 oC with a photoperiod of 12 h/day and light intensity of 2,500–3,000 Lx.
Two types of protocorms were obtained for each species; at an age of 6 months, the leaf-less plants which produced flowers without vegetative growth and the normal plants with leaf and root growth. Three replicates were obtained for each pattern of each species for RNA Sequencing.
A total of 18 (6 tissues in 3 replicates) tissues were obtained from 6 month old plants to extract RNA suing the TaKaRa kit. The total RNA was used to prepare cDNA libraries. The mRNA was obtained with Oligotex Midi Kit for mRNA (Qiagen, Germany) and the quality assessment was done with Nano-Drop spectrophotometer (Thermo Fisher Scientific, United States). The cDNA libraries were prepared by using Illumina protocol. The library product evaluation was made through Agilent 2,200 TapeStation and Qubit®2.0 (Life Technologies, United States), followed by product dilution to 10 pM to generate in situ clusters on HiSeq2500 pair-end flow cells and pair-end sequencing (2 × 100). Finally, the reference-based transcriptome sequencing was performed using the reference genomes of each species. Gene expression was measures as FPKM (fragments per kilobase per transcript per million mapped reads).
The assembled genes were mapped to publically available databases, such as NR (non-redundant), GO (Gene Ontology), KEGG (Kyoto Encyclopedia of Genes and Genomes) and KO (KEGG ortholog) databases using the BLASTX program with a threshold E-value ≤10-5. The GO and KEGG annotations were further classified into functional categories and pathways using the phyper function in R software. The corrected p-value was obtained by FDR and the terms with functional Q-value ≤0.05 were considered as significantly enriched (Ahmad et al., 2021).
The Bowtie2 program (v2.4.4, Johns Hopkins University) was used to align clean reads to genomic sequences and the expression level of each sample was calculated using RSEM (v1.2.8) with default parameters. Then the DEGseq package (1.10.1) was used in R software to obtain DEGs. The DEGs were sorted at a threshold value of p < 0.001 and the log2FC > 1 (Ahmad et al., 2022).
From the DEGs, we filtered the genes related to two key pathways that play important roles in the regulation of defense responses and plant immunity against pathogens. These pathways included Plant-pathogen interaction pathway (ko04626) and MAPK signaling pathway (ko04016). The genes for each pathway were divided into different categories according to their up- and down-regulated trends.
The common gene names were obtained for defense-related genes for three species and their protein sequences were run on online string facility1 to generate a network. The network annotation was shown as GO biological processes for all the genes.
The top five highly upregulated and highly downregulated genes were identified for each species in plant-pathogen interaction pathway. Their expression intensities were drawn as heatmap using TBtools.2 Finally, the pathway integrators were shown for pathogen stress in a combined form for three species.
The expression patterns for leaf and leaf-less flowers were observed for each species using the empirical cutoff values of genes with positive expressions. The boxplots in Figure 1 shows the expression distribution of the FPKM values of genes. The boxplots curtail the uniform distribution of median and quartile values of DEGs among samples of each species.
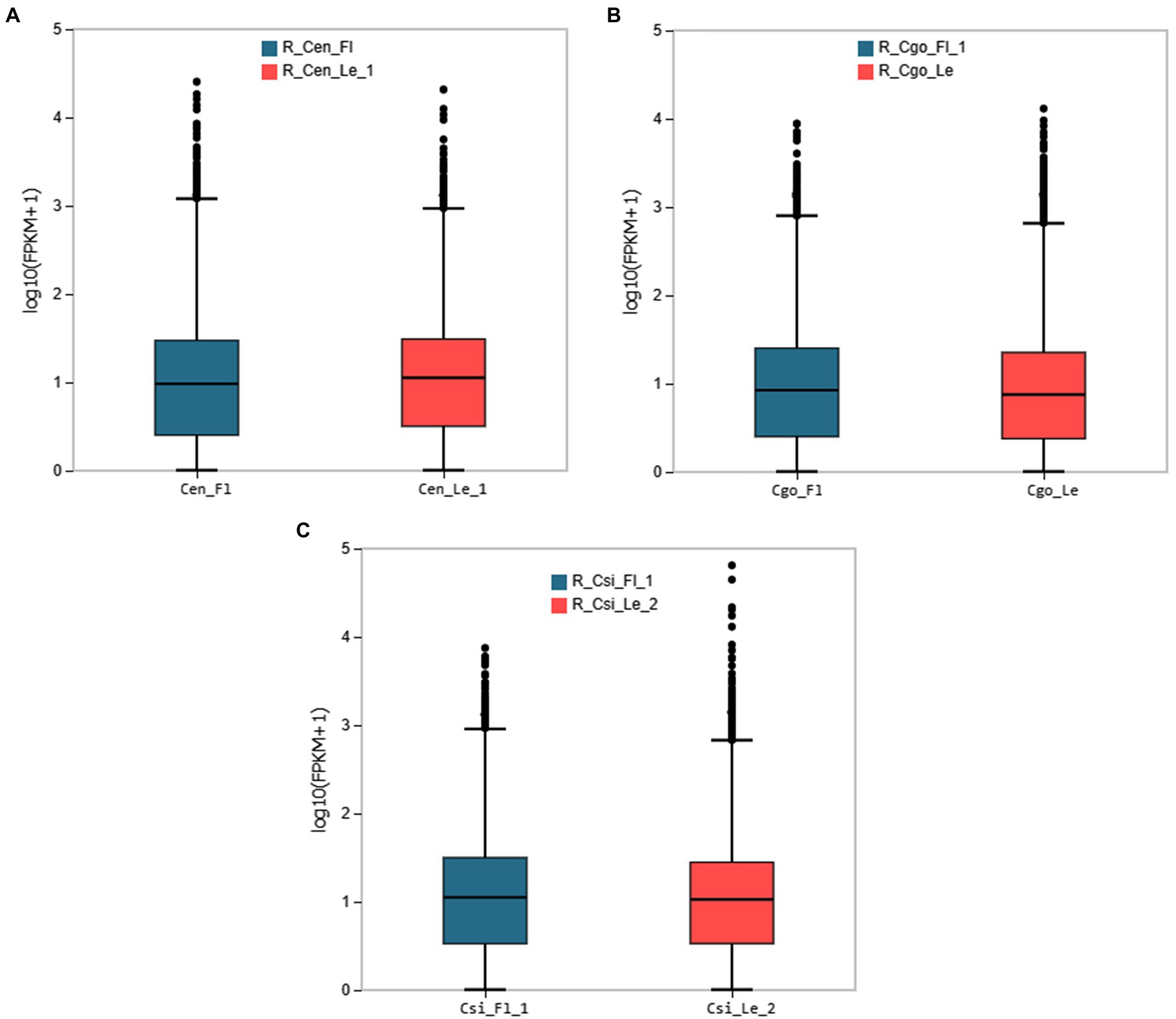
Figure 1. The boxplot distribution of gene expression from the two tissues of each species; (A) C. ensifolium, (B) C. goeringii, (C) C. sinense. The X-axis shows the sample name; the Y-axis represents log10(FPKM+1). The boxplot of each area shows five statistics (from top to bottom are the upper limit, upper quartile, median, and lower quartile, respectively).
The DEGs were compared between leaf-less flowers and leaf samples within each species separately (Figure 2). The highest number of up- and down-regulated genes (4089) can be seen for C. goeringii (Figure 2B), followed by C. ensifolium (3414; Figure 2A) and C. sinense (2807; Figure 2C). C. ensifolium showed the highest number of leaf-specific upregulated genes (2167) as compared to C. goeringii (1827) and C. sinense (1442). Interestingly, the highest numbers of downregulated genes were observed in the leaves of C. goeringii (2262), which was significantly different from C. ensifolium (1247) and C. sinense (1365).
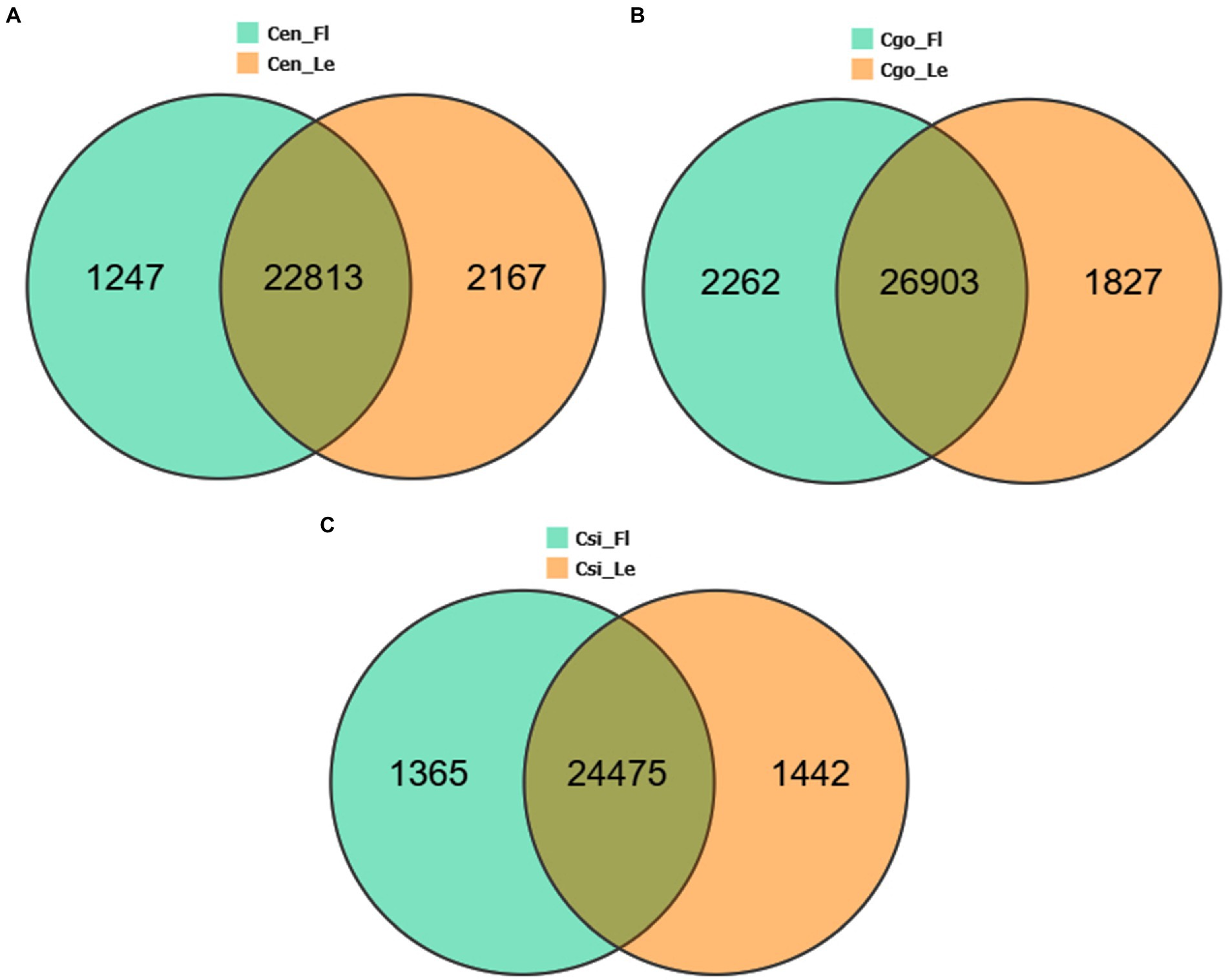
Figure 2. Individual species tissue-specific and common DEGs among the leaf-less control and leaf samples of C. ensifolium (A), C. goeringii (B), and C. sinense (C).
The GO annotation was obtained for individual species (Figure 3). For C. ensifolium the maximum number of genes was related to cellular process and metabolic process in the GO biological process category (Figure 3A). For the cellular component category, the highest number of genes was enriched for cellular anatomical entities followed by intracellular components. The major molecular functions were related to catalytic activity and binding (Figure 3A). In the case of C. goeringii (Figure 3B), the GO annotations were similar to C. ensifolium. Although similar GO enrichments were observed for C. sinense (Figure 3C), however, the number of genes were less than other two species.
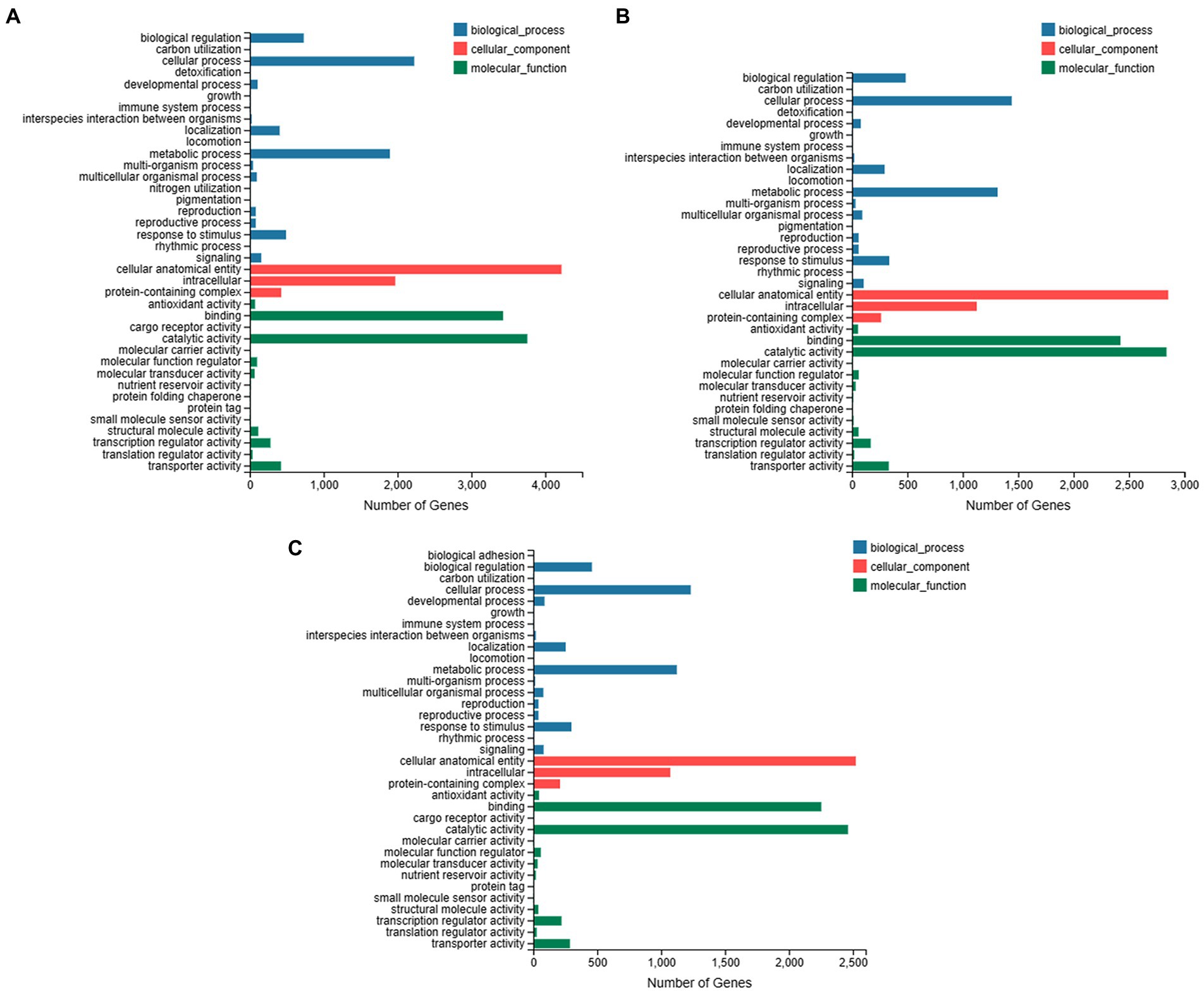
Figure 3. Overview of GO enrichments for C. ensifolium (A), C. goeringii (B), and C. sinense (C). The DEGs enrichment is shown in three GO categories, including biological process, cellular component and molecular function. The y-axis shows the GO category and the x-axis shows the number of genes enriched to each category.
The plant-pathogen interaction pathway (ko04626) was counted among the most enriched KEGG pathways in the three orchid species (Figure 4). Highly enriched number of genes for this pathway were found in C. ensifolium (Figure 4A), followed by C. goeringii (Figure 4B) and C. sinense (Figure 4C). MAPK signaling pathway was prominent among the other pathways in C. ensifolium and C. goeringii, however, it was not prominent in C. sinense. The other major pathways with significant gene enrichment included plant hormone signal transduction, phenylpropanoid biosynthesis, and starch and sucrose metabolism, which may indirectly play roles in plant defense and immunity responses.
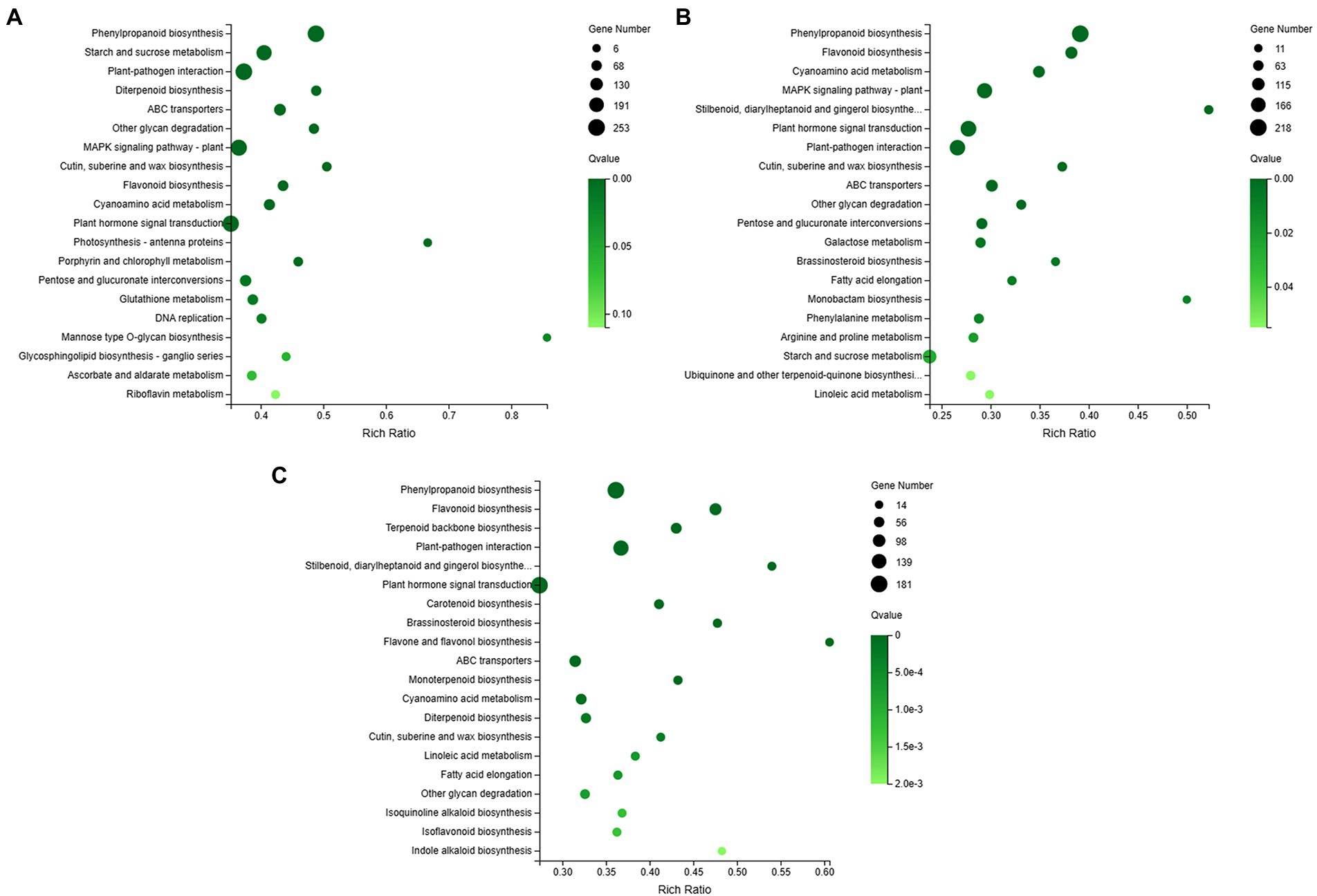
Figure 4. Overview of KEGG enrichments for C. ensifolium (A), C. goeringii (B), and C. sinense (C). The y-axis shows the KEGG pathway names and the x-axis shows the pathway-richness ratio. The size of the black circles shows the number of genes enriched to each pathway; bigger the circle size more is the number of genes.
A number of plant-pathogen interaction response pathways were observed in the three orchid species (Figure 5). All the routes were converged to the three key responses, i.e., defense related gene induction, stomatal closure and hypersensitive response. The stimulation originates from the cell membrane and the final responses are shown in the cytoplasm of plant cells. Four key response generators were observed in the three species, including CNGCs, FLS2, EFR and bacterial secretion system (Supplementary Tables 1–3). In C. ensifolium, the genes related to CNGCs were upregulated in leaves as compared to leaf-less flowers (Figure 5A). Contrarily, the EFR related genes were downregulated in the leaves. The FLS2 contained both upregulated and downregulated genes. The other solely upregulated genes were related to Rboh in the CNGCs routes, MEKK1 in the FLS2 route and Pti1 in the bacterial secretion route. All other regulators contained both upregulated and downregulated genes (Figure 5B). Higher number of downregulated genes were observed in C. ensifolium leaves than upregulated genes among the 27 expressed pathway stimulators (Figure 5B). Moreover, NHO1 was the only defense-related gene downregulated in the leaves of three species.
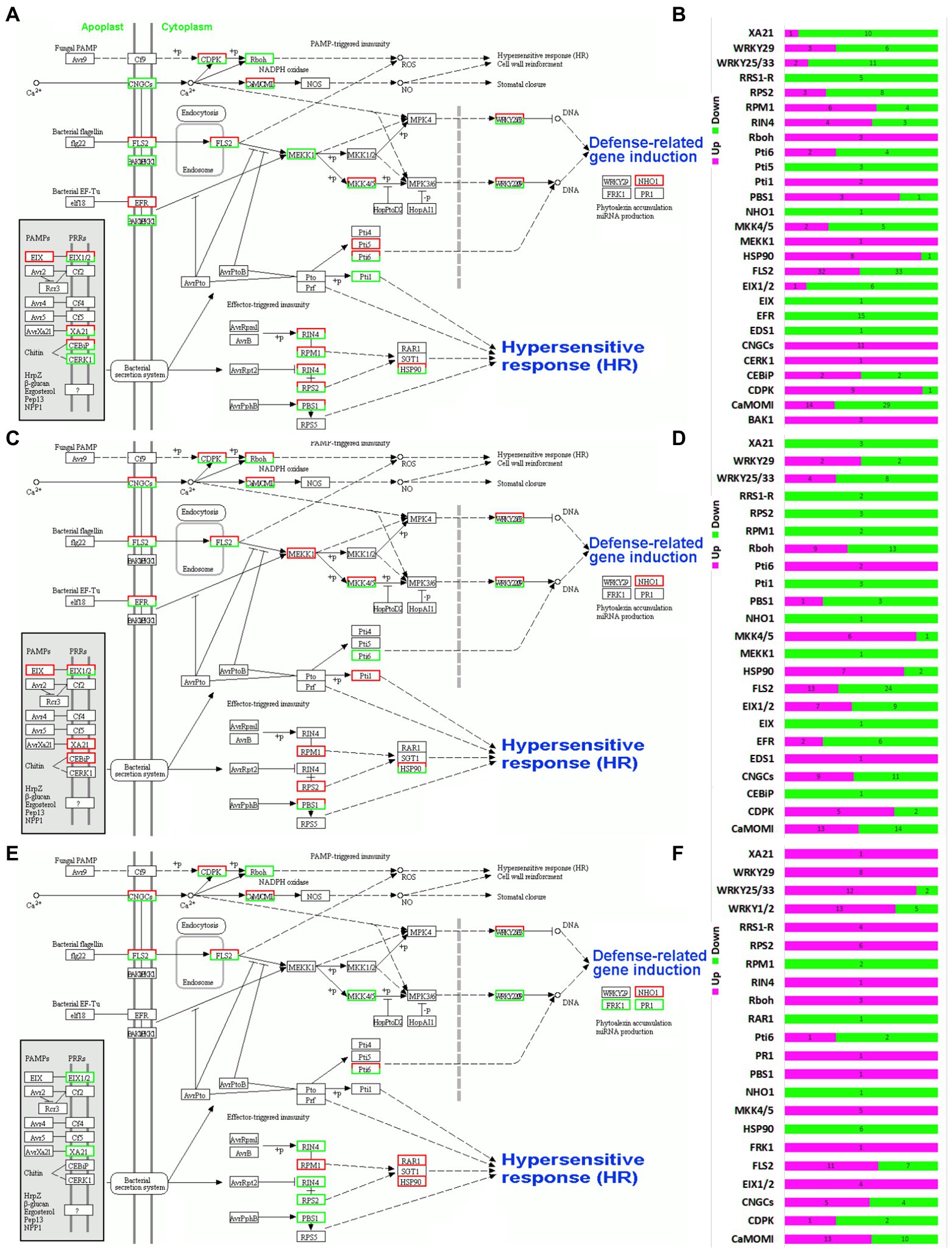
Figure 5. Overview of defense response pathway and the distribution of up- and downregulated pathway integrators for C. ensifolium (A,B), C. goeringii (C,D), and C. sinense (E,F).
Contrary to C. ensifolium the EFR contained both upregulated and downregulated genes in C. goeringii (Figure 5C). Moreover, the MEKK1 and Pti1 were completely downregulated, while Pti6 was upregulated as compared to C. ensifolium. In the bacterial secretion route, the RPM1 and RPS2 were completed downregulated in the C. goeringii leaves as compared to C. ensifolium (Figure 5C). Similar to C. ensifolium, NHO1 was the only defense related gene downregulated in the leaves of C. goeringii. Moreover, the number of downregulated genes was higher than the upregulated genes (Figure 5D).
The defense-related genetic map of C. sinense was much different than other two species (Figures 5E,F). In the CNGCs rout, Rboh genes were completely upregulated in the leaves. In the FLS2 route, MKK4/5 and WRKY22 and WRKY29 were also upregulated. In the bacterial secretion route, the RIN4, RPS2 and PBS1 were upregulated, while, the RAR1 and HSP90 were completely downregulated. Three defense-related genes were induced in C. sinense as compared to other two species. Here, the FRK1 and PR1 were upregulate and NHO1 was downregulated. Most of the genes were upregulated in C. sinense contrary to other two species, while a few were downregulated (Figure 5F).
The protein sequences of plant-pathogen interaction and MAPK signaling pathway genes were run on the online string database to see their interaction patterns (Figure 6A). All the important regulators of plant defense and immunity were interconnected. The interconnected proteins were mainly involved in calcium mediated signaling (GO:0019722), cellular macromolecule metabolic process (GO:0044260), defense responses (GO:0006952), defense response to bacteria (GO:0042742), hormone-mediated signaling (GO:0009755), immune system process (GO:0002376), and JA-mediated signaling (GO:2000022; Figure 6C).
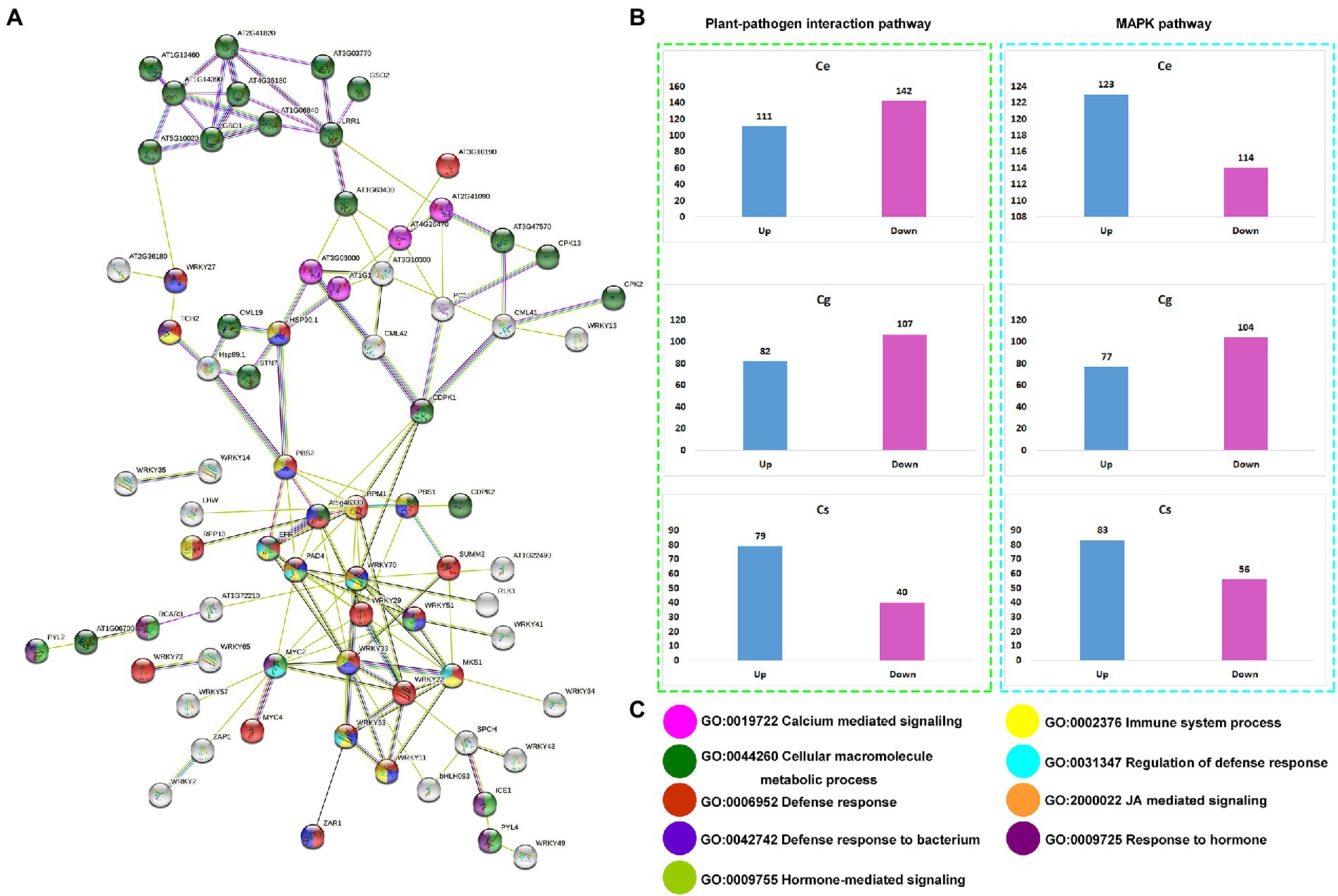
Figure 6. Identification string-based interaction network for all the genes expressed in three orchid species (A), up- and downregulated genes for plant-pathogen interaction pathway and MAPK signaling pathway for three orchid species (B), and the representation of major biological process enriched in the interaction network (C).
Up-and down-regulation profiles were compared for both the pathways (Figure 6B). For plant-pathogen interaction pathway the number of downregulated genes was higher than the number of upregulated genes in C. ensifolium and C. goeringii as compared to C. sinense, where the number of downregulated genes (40) was significantly lower than the number of upregulated genes (79; Figure 6B). For MAPK signaling pathway, the number of upregulated genes was higher than that of downregulated genes in C. ensifolium and C. sinense. However, the number of upregulated genes (77) was significantly lower than the number of downregulated genes (104) in C. goeringii.
The leaf-less controls provides the best control to study the defense and immunity mechanism in the leaf (Figure 7A). Using this control, we found the top 5 highly upregulated and highly downregulated genes in the leaves of three orchid species (Figure 7B). In C. ensifolium, the downregulated genes included calmodulin, three EFRs and one FLS2, while the upregulated genes included three FLS2s, CDPK and WRKY2. In C. goeringii, the downregulated genes included FLS2, MAPK4, rhamnogalacturonan endolyase, Pti1-like and HtpG, while the upregulated genes included three disulfide isomerases, MAPK4/5 and ULK4. In the case of C. sinense, the downregulated genes included two WRKY2, FLS2, calmodulin and rhamnogalacturonan endolyase, while the upregulated genes were CML41, FLS2, CML30, WRKY72, and RPM1 (Figure 7B).
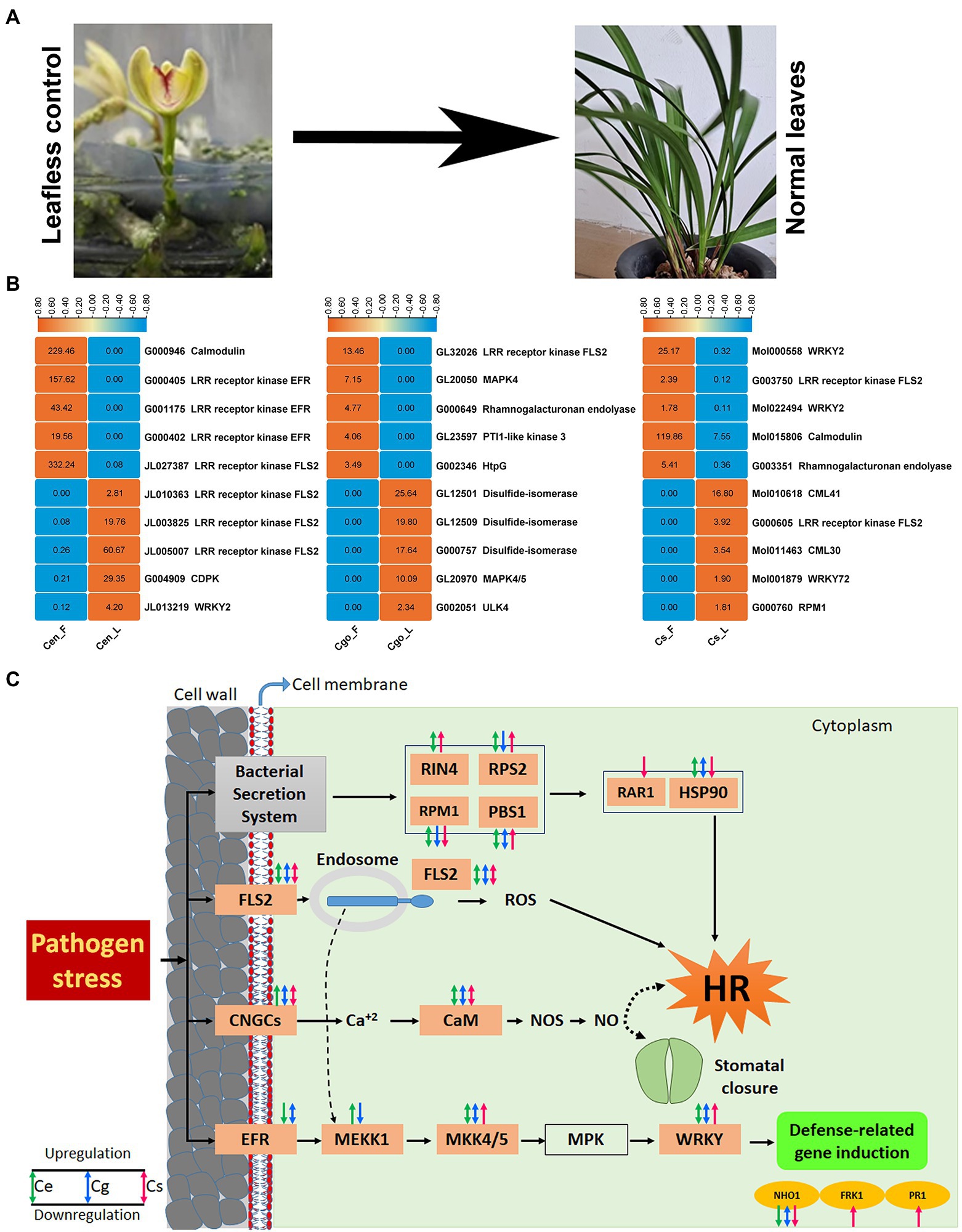
Figure 7. Leaf-less control and the healthy leaf samples used for sequencing analysis (A), top 5 highly upregulated and highly downregulated defense-related DEGs (B), and the final pathway of pathogen stress showed by three orchid species (C). The orange boxes show the genes expressed in either of the species.
In the orchid species, the defense-related responses are induced by four key routes, including bacterial secretion system, FLS2, CNCGs, and EFR (Figure 7C). The bacterial secretion system stimulates four proteins in orchids, such as RIN4, RPS2, RPM1 and PBS1. RIN4 was not expressed in C. goeringii, while the remaining three were expressed in all the three species. These proteins ultimately instigate hypersensitive response (HR) through the stimulation of RAR1 and HSP90. RAR1 was only downregulated in C. sinense, while it was not expressed in other two species. HSP90 was downregulated in C. sinense and showed both up- and down-regulation in C. ensifolium and C. goeringii. FLS2 also triggers HR through reactive oxygen species (ROS). Both up- and downregulation profiles were found for FLS2 in the three orchid species. The CNGCs use calcium signaling to induce CaM, which then cause HR and stomatal closure through NOS and NO signals. Both up- and downregulation profiles were found for CNGCs and CaM in the three orchid species.
The EFR routes works differently than the previous three routes and cause the induction of defense-related genes. It involves multiple regulatory before the final response. These regulators include MEKK1, MKK4/5, MPK and WRKY, which finally induce defense-related genes, such as NHO1, FRK1 and PR1. EFR and MEKK1 genes were not expressed in C. sinense, while MPK genes were not expressed in any of the species. The MKK4/5 and WRKY were expressed in all the species. Among the defense-related genes, the NHO1 was expressed in all the species, while the FRK1 and PR1 were expressed only in C. sinense (Figure 7C).
Orchids share a huge portion of floriculture industry. Obtaining virus free orchids through meristem culturing has been used to control the spread of diseases (Panattoni et al., 2013). However, tissue culturing of orchids is very costly and time-consuming and their long vegetative phase (2–3 years) requires a permanent solution against pathogens. The mechanisms of plant defense and immunity against pathogens have been documented in numerous model plants. However, limited genomic information has been presented on the orchids, especially the Cymbidium orchids. A detailed and genome-wide searching of defense-related genes would facilitate the future breeding programs for disease-resistance in Orchidaceae.
Reference-based transcriptome sequencing produced significantly upregulated and downregulated gene profiles for leaf-less control and leaf samples for each of three orchid species, C. ensifolium, C. goeringii and C. sinense (Figure 2). Cellular and metabolic process were the most enriched GO biological processes, mainly involving catalytic and binding activities in the cellular anatomical entities (Figure 3). Plant-pathogen interaction and MAPK signaling pathways were highly enriched KEGG terms in the three orchid species (Figure 4) among the other pathways including, mainly, plant hormone signal transduction and phenylpropanoid biosynthesis pathways. The data enrichment suggests the significance of plant defense and immunity responses of orchids.
Highly upregulated and downregulated transcripts associated with plant-pathogen interaction were induced for the leaves of three orchid species. For example, LRR receptor kinases encoding transcripts were highly expressed in the leaves as compared to leaf-less controls. EFR and FLS2, LRR receptor-like kinases, are the primary response elements and act to recognize bacterial epitopes elf18 and flg22 (Figures 5A,C,E; Saijo et al., 2009; Ahmad et al., 2014). A total of 65 FLS2-related transcripts (32 upregulated and 33 downregulated) were found in the leaves of C. ensifolium (Figure 5D), while 15 EFRs were found, which were all downregulated. In C. goeringii, 37 FLS2s were found (13 upregulated and 24 downregulated; Figure 5F), while there were 8 EFRs (2 upregulated and 6 downregulated). C. sinense contained 18 FLS2s (11 upregulated and 7 downregulated) and no EFR expressed here (Figure 5F).
CML and CaM are calcium sensors and play pivotal roles in calmodulin signaling pathways involving oxidative burst and cell death regulation in the infected plants enduring HR (Harding et al., 1997; Harding and Roberts, 1998; Poovaiah et al., 2013). A total of 43 CaM transcripts were identified in C. ensifolium (14 upregulated and 29 downregulated; Figure 5B), 27 in C. goeringii (13 upregulated and 14 downregulated; Figure 5D), and 23 CaMs were identified in C. sinense (13 upregulated and 10 downregulated; Figure 5F).
Plants use specific pattern recognizing receptors to identify microbes. These receptors are activated by MAMPs (microbe associated molecular patterns), which results into MTI (MAMP-triggered immunity). However, clever pathogens bypass MTI using virulence effectors, thereby causing pathogen proliferation. In the resistant genotypes, intracellular immune receptors detect these effectors and start defense responses called ETI (effector triggered immunity), including HR and transcriptional reprogramming in order to halt the growth of pathogens. Pseudomonas syringae secretes AvrRpm1 effector into the host cell to induce virulence. RIN4 negatively regulates MTI and its modification in the presence of effectors like ArvRpm1. The RPM1 and RPS2 are a nucleotide binding leucine-rich repeat sensors. RPM1 perceives the perturbation of RIN4 in disease resistance, which also activates the RPS2, causing an enhanced immune response (Cherkis et al., 2012). Heat shock proteins (HSP90) involve detoxification and stress responses (Azaiez et al., 2009). The AvrB effector of P. syringae suppresses PTI by using RAR1, an HSP90 cochaperone required for ETI, indicating that RAR1 is a negative regulator of PTI (Shang et al., 2006). Our data included all the components responding to ETI, including RIN4, RPS2, RPM1, PBS1, HSP90, which were expressed in all the species, except RAR1 which was only expressed but downregulated in the leaves of C. sinense (Figure 7C), suggesting the immunity level of Cymbidium orchids against RAR-mediated pathogenic attack.
Among the highly expressed genes, EFRs were downregulated in the leaves of C. ensifolium, while FLS2s were upregulated. A number of WRKY transcription factors were differentially expressed, such as WRKY25, WRKY29, and WRKY33 (Figures 5B,D,F). WRKY TFs have admitted role in the regulation of plant defense responses (Eulgem, 2006; Knoth et al., 2007; Azaiez et al., 2009; Faiz et al., 2022). WRKY2 represses the basal immunity of barley by directly targeting PAMP recognition receptor genes (Yu et al., 2022). It was upregulated in the leaves of C. ensifolium, while downregulated in the leaves of C. sinense (Figure 7), suggesting that orchids may differ in their immunity responses even within the same genus. Similarly FLS2 has also dual roles with both upregulated and downregulated genes in the three species.
Previous studies show that flg22 triggers MAP kinases, such as MEKK1, MKK4/5 and MPK (Nuhse et al., 2000; Asai et al., 2002). Three flg22-inducible genes, including NHO1, FRK1, and WRKY29 (An and Mou, 2012), are induced by this cascade of kinases in the EFR pathway. We found up- and downregulated transcripts for EFR, MEKK1, MKK4/5, and defense-related genes, such as NHO1, FRK1, and PR1 (Figure 7C). NHO1 was expressed in all the species, while FRK1 and PR1 were upregulated only in C. sinense. Our study, thus, presents a general comparison among three orchid species for a transcriptome-wide mining of defense related genes. This information can be used to plant biological control strategies for orchids and floriculture crops.
This is the first study that uses leaf-less control to mine the defense and immunity related gene profiles in three Cymbidium orchids. A number of upregulated and downregulated genes were identified in four major routes of pathogen stress, including bacterial secretion system, FLS2, CNGCs and EFR. These routes mainly regulate three processes, namely, hypersensitive response (HR), stomata closure and defense-related gene induction. The regulation of immunity response was a little different in C. sinense as compared to C. ensifolium and C. goeringii. For example, two of the defense related genes (FRK1 and PR1) were upregulated only in C. sinense, while they did not express in other two species. Overall, the results present a broad picture of defense machinery of Cymbidium orchids, which can be used to ameliorate pathogen affliction through biological control.
The transcriptome sequences described in this article have been submitted to The National Genomics Data Center (NGDC, https://ngdc.cncb.ac.cn) under accession number PRJCA009885.
SA: conceptualization and writing—original draft. GC: data curation and software. JH: investigation. KY: data curation. YH: software. YZ: visualization, investigation, and editing. KZ: data curation and editing. SL: software and editing. ZL: supervision, conceptualization, and funding acquisition. DP: supervision, conceptualization, funding acquisition, and writing—reviewing and editing. All authors contributed to the article and approved the submitted version.
This work was supported by The National Natural Science Foundation of China (no. 32071815); The National Key Research and Development Program of China (2019YFD1001000); The National Key Research and Development Program of China (2018YFD1000401); The Innovation and Application Engineering Technology Research Center of Ornamental Plant Germplasm Resources in Fujian Province (115-PTJH16005); and National Natural Science Foundation of China (no. 32101583).
We are thankful to funding agencies for funding support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1001427/full#supplementary-material
1. ^https://string-db.org/cgi/input?sessionId=HofWDb4VJD8K (Accessed August 21, 2022).
2. ^https://github.com/CJ-Chen/TBtools/releases (Accessed August 21, 2022).
Ahmad, A., Shafique, S., and Shafique, S. (2014). Intracellular interactions involved in induced systemic resistance in tomato. Sci. Hortic. 176, 127–133. doi: 10.1016/j.scienta.2014.07.004
Ahmad, A., Yasin, N. A., Ibrahim, A., Shahzadi, I., Gohar, M., Bashir, Z., et al. (2018). Modelling of cotton leaf curl viral infection in Pakistan and its correlation with meteorological factors up to 2015. Clim. Dev. 10, 520–525. doi: 10.1080/17565529.2017.1318738
Ahmad, S., Chen, J., Chen, G., Huang, J., Hao, Y., Shi, X., et al. (2022). Transcriptional proposition for uniquely developed protocorm flowering in three orchid species: resources for innovative breeding. Front. Plant Sci. 13:942591. doi: 10.3389/fpls.2022.942591
Ahmad, S., Lu, C., Gao, J., Ren, R., Wei, Y., Wu, J., et al. (2021). Genetic insights into the regulatory pathways for continuous flowering in a unique orchid Arundina graminifolia. BMC Plant Biol. 21:587. doi: 10.1186/s12870-021-03350-6
Akram, W., Anjum, T., Ahmad, A., and Moeen, R. (2014). First report of Curvularia lunata causing leaf spots on Sorghum bicolor from Pakistan. Plant Dis. 98:1007. doi: 10.1094/PDIS-12-13-1291-PDN
Akram, W., Aslam, H., Ahmad, S. R., Anjum, T., Yasin, N. A., Khan, W. U., et al. (2019). Bacillus megaterium strain A12 ameliorates salinity stress in tomato plants through multiple mechanisms. J. Plant Interact. 14, 506–518. doi: 10.1080/17429145.2019.1662497
Ali, R., Ma, W., Lemtiri-Chlieh, F., Tsaltas, D., Leng, Q., Von Bodman, S., et al. (2007). Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19, 1081–1095. doi: 10.1105/tpc.106.045096
An, C., and Mou, Z. (2012). Non-host defense response in a novel Arabidopsis-Xanthomonas citri subsp. citri pathosystem. PLoS One 7:e31130. doi: 10.1371/journal.pone.0031130
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W.-L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Azaiez, A., Boyle, B., Levée, V., and Séguin, A. (2009). Transcriptome profiling in hybrid poplar following interactions with Melampsora rust fungi. Mol. Plant-Microbe Interact. 22, 190–200. doi: 10.1094/MPMI-22-2-0190
Bashir, Z., Ahmad, A., Shafique, S., Anjum, T., Shafique, S., and Akram, W. (2013). Hypersensitive response—a biophysical phenomenon of producers. Eur. J. Microbiol. Immunol. 3, 105–110. doi: 10.1556/EuJMI.3.2013.2.3
Bauer, Z., Gómez-Gómez, L., Boller, T., and Felix, G. (2001). Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. J. Biol. Chem. 276, 45669–45676. doi: 10.1074/jbc.M102390200
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe–associated molecular patterns and danger signals by pattern-recognition. Ann. Rev. Plant Biol 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Boudsocq, M., Willmann, M. R., Mccormack, M., Lee, H., Shan, L., He, P., et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422. doi: 10.1038/nature08794
Cherkis, K. A., Temple, B. R., Chung, E.-H., Sondek, J., and Dangl, J. L. (2012). AvrRpm1 missense mutations weakly activate RPS2-mediated immune response in Arabidopsis thaliana. PLoS One 7:e42633. doi: 10.1371/journal.pone.0042633
Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476. doi: 10.1105/tpc.105.036574
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
De Jonge, R., Peter Van Esse, H., Maruthachalam, K., Bolton, M. D., Santhanam, P., Saber, M. K., et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. 109, 5110–5115. doi: 10.1073/pnas.1119623109
Eulgem, T. (2006). Dissecting the WRKY web of plant defense regulators. PLoS Pathog. 2:e126. doi: 10.1371/journal.ppat.0020126
Faiz, S., Yasin, N. A., Khan, W. U., Shah, A. A., Akram, W., Ahmad, A., et al. (2022). Role of magnesium oxide nanoparticles in the mitigation of lead-induced stress in Daucus carota: modulation in polyamines and antioxidant enzymes. Int. J. Phytoremediation 24, 364–372. doi: 10.1080/15226514.2021.1949263
Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. doi: 10.1046/j.1365-313X.1999.00265.x
Fradin, E. F., Abd-El-Haliem, A., Masini, L., Van Den Berg, G. C., Joosten, M. H., and Thomma, B. P. (2011). Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. doi: 10.1104/pp.111.180067
Gómez-Gómez, L., and Boller, T. (2000). FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A., and Mansfield, J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23, 441–450. doi: 10.1046/j.1365-313x.2000.00804.x
Harding, S. A., Oh, S.-H., and Roberts, D. M. (1997). Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 16, 1137–1144. doi: 10.1093/emboj/16.6.1137
Harding, S. A., and Roberts, D. M. (1998). Incompatible pathogen infection results in enhanced reactive oxygen and cell death responses in transgenic tobacco expressing a hyperactive mutant calmodulin. Planta 206, 253–258. doi: 10.1007/s004250050397
Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., et al. (2001). The innate immune response to bacterial flagellin is mediated by toll-like receptor 5. Nature 410, 1099–1103. doi: 10.1038/35074106
He, Y., Zhou, J., Shan, L., and Meng, X. (2018). Plant cell surface receptor-mediated signaling: a common theme amid diversity. J. Cell Sci. 131:jcs209353. doi: 10.1242/jcs.209353
Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M., He, K., Li, J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. 104, 12217–12222. doi: 10.1073/pnas.0705306104
Ichimura, K., Casais, C., Peck, S. C., Shinozaki, K., and Shirasu, K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281, 36969–36976. doi: 10.1074/jbc.M605319200
Ichimura, K., Shinozaki, K., Tena, G., Sheen, J., Henry, Y., Champion, A., et al. (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308. doi: 10.1016/S1360-1385(02)02302-6
Kaur, S., Samota, M. K., Choudhary, M., Choudhary, M., Pandey, A. K., Sharma, A., et al. (2022). How do plants defend themselves against pathogens-biochemical mechanisms and genetic interventions. Physiol. Mol. Biol. Plants 28, 485–504. doi: 10.1007/s12298-022-01146-y
Knoth, C., Ringler, J., Dangl, J. L., and Eulgem, T. (2007). Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant-Microbe Interact. 20, 120–128. doi: 10.1094/MPMI-20-2-0120
Koh, K. W., Lu, H.-C., and Chan, M.-T. (2014). Virus resistance in orchids. Plant Sci. 228, 26–38. doi: 10.1016/j.plantsci.2014.04.015
Komis, G., Šamajová, O., Ovečka, M., and Šamaj, J. (2018). Cell and developmental biology of plant mitogen-activated protein kinases. Annu. Rev. Plant Biol. 69, 237–265. doi: 10.1146/annurev-arplant-042817-040314
Lee, S.-W., Han, S.-W., Sririyanum, M., Park, C.-J., Seo, Y.-S., and Ronald, P. C. (2009). A type I–secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326, 850–853. doi: 10.1126/science.1173438
Levine, A., Pennell, R. I., Alvarez, M. E., Palmer, R., and Lamb, C. (1996). Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr. Biol. 6, 427–437. doi: 10.1016/S0960-9822(02)00510-9
Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E., and Walker, J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. doi: 10.1016/S0092-8674(02)00812-7
Li, L., Li, M., Yu, L., Zhou, Z., Liang, X., Liu, Z., et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338. doi: 10.1016/j.chom.2014.02.009
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Monaghan, J., Matschi, S., Romeis, T., and Zipfel, C. (2015). The calcium-dependent protein kinase CPK28 negatively regulates the BIK1-mediated PAMP-induced calcium burst. Plant Signal. Behav. 10:e1018497. doi: 10.1080/15592324.2015.1018497
Monaghan, J., and Zipfel, C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. doi: 10.1016/j.pbi.2012.05.006
Mubeen, S., Shahzadi, I., Akram, W., Saeed, W., Yasin, N. A., Ahmad, A., et al. (2022). Calcium nanoparticles impregnated With Benzenedicarboxylic acid: a new approach to alleviate combined stress of DDT and cadmium in Brassica alboglabra by modulating bioacummulation, antioxidative machinery and osmoregulators. Front. Plant Sci. 13, –825829. doi: 10.3389/fpls.2022.825829
Ngou, B. P. M., Ding, P., and Jones, J. D. (2022). Thirty years of resistance: zig-zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/plcell/koac041
Nishad, R., Ahmed, T., Rahman, V. J., and Kareem, A. (2020). Modulation of plant defense system in response to microbial interactions. Front. Microbiol. 11:1298. doi: 10.3389/fmicb.2020.01298
Nuhse, T. S., Peck, S. C., Hirt, H., and Boller, T. (2000). Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 275, 7521–7526. doi: 10.1074/jbc.275.11.7521
Panattoni, A., Luvisi, A., and Triolo, E. (2013). Elimination of viruses in plants: twenty years of progress. Span. J. Agric. Res. 11, 173–188. doi: 10.5424/sjar/2013111-3201
Petutschnig, E. K., Jones, A. M., Serazetdinova, L., Lipka, U., and Lipka, V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285, 28902–28911. doi: 10.1074/jbc.M110.116657
Poovaiah, B., Du, L., Wang, H., and Yang, T. (2013). Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol. 163, 531–542. doi: 10.1104/pp.113.220780
Ranf, S., Eschen-Lippold, L., Fröhlich, K., Westphal, L., Scheel, D., and Lee, J. (2014). Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol. 14, 1–15. doi: 10.1186/s12870-014-0374-4
Ranf, S., Grimmer, J., Pöschl, Y., Pecher, P., Chinchilla, D., Scheel, D., et al. (2012). Defense-related calcium signaling mutants uncovered via a quantitative high-throughput screen in Arabidopsis thaliana. Mol. Plant 5, 115–130. doi: 10.1093/mp/ssr064
Ren, R., Wei, Y., Ahmad, S., Jin, J., Gao, J., Lu, C., et al. (2020). Identification and characterization of NPR1 and PR1 homologs in Cymbidium orchids in response to multiple hormones, salinity and viral stresses. Int. J. Mol. Sci. 21:1977. doi: 10.3390/ijms21061977
Rhodes, J., Zipfel, C., Jones, J. D., and Ngou, B. P. M. (2022). Concerted actions of PRR-and NLR-mediated immunity. Essays Biochem. doi: 10.1042/EBC20220067
Robatzek, S., and Wirthmueller, L. (2013). Mapping FLS2 function to structure: LRRs, kinase and its working bits. Protoplasma 250, 671–681. doi: 10.1007/s00709-012-0459-6
Roberts, D. L., and Dixon, K. W. (2008). Orchids. Curr. Biol. 18, R325–R329. doi: 10.1016/j.cub.2008.02.026
Ron, M., and Avni, A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16, 1604–1615. doi: 10.1105/tpc.022475
Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A., Holton, N., et al. (2011). The Arabidopsis leucine-rich repeat receptor–like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455. doi: 10.1105/tpc.111.084301
Saijo, Y., Tintor, N., Lu, X., Rauf, P., Pajerowska-Mukhtar, K., Häweker, H., et al. (2009). Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 28, 3439–3449. doi: 10.1038/emboj.2009.263
Schwessinger, B., Roux, M., Kadota, Y., Ntoukakis, V., Sklenar, J., Jones, A., et al. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7:e1002046. doi: 10.1371/journal.pgen.1002046
Shafique, S., Anjum, T., Shafique, S., Ahmad, A., Akram, W., and Bashir, Z. (2011). Induction of systemic defenses in plants under the activity of dynamic inducers. Mycopathologia 9, 95–104.
Shang, Y., Li, X., Cui, H., He, P., Thilmony, R., Chintamanani, S., et al. (2006). RAR1, a central player in plant immunity, is targeted by pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. 103, 19200–19205. doi: 10.1073/pnas.0607279103
Shimizu, T., Nakano, T., Takamizawa, D., Desaki, Y., Ishii-Minami, N., Nishizawa, Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. doi: 10.1111/j.1365-313X.2010.04324.x
Tuhid, N.H., Abdullah, N.E., Khairi, N., Saaid, M., Shahrizam, M., and Hashim, H. (2012). “A statistical approach for orchid disease identification using RGB color,” in 2012 IEEE control and system graduate research colloquium: IEEE, 382–385.
Wang, Y., Pruitt, R. N., Nürnberger, T., and Wang, Y. (2022). Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 20, 449–464. doi: 10.1038/s41579-022-00710-3
Willmann, R., Lajunen, H. M., Erbs, G., Newman, M.-A., Kolb, D., Tsuda, K., et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. 108, 19824–19829. doi: 10.1073/pnas.1112862108
Wong, S. (2002). Orchid viruses: a compendium. Orchid Biology: Reviews and Perspectives. 8, 505–546.
Yu, D., Fan, R., Zhang, L., Xue, P., Liao, L., Hu, M., et al. (2022). HvWRKY2 acts as an immunity suppressor and targets HvCEBiP to regulate powdery mildew resistance in barley. Crop J., in press. doi: 10.1016/j.cj.2022.05.010
Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J. D., Boller, T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts agrobacterium-mediated transformation. Cell 125, 749–760. doi: 10.1016/j.cell.2006.03.037
Zipfel, C., and Oldroyd, G. E. (2017). Plant signalling in symbiosis and immunity. Nature 543, 328–336. doi: 10.1038/nature22009
Keywords: leaf-less control, orchids, pathogens, biological control, immunity, defense
Citation: Ahmad S, Chen G, Huang J, Yang K, Hao Y, Zhou Y, Zhao K, Lan S, Liu Z and Peng D (2022) Beauty and the pathogens: A leaf-less control presents a better image of Cymbidium orchids defense strategy. Front. Plant Sci. 13:1001427. doi: 10.3389/fpls.2022.1001427
Received: 23 July 2022; Accepted: 16 August 2022;
Published: 13 September 2022.
Edited by:
Waheed Akram, University of the Punjab, PakistanReviewed by:
Muhammad Furqan Ashraf, UiT The Arctic University of Norway, NorwayCopyright © 2022 Ahmad, Chen, Huang, Yang, Hao, Zhou, Zhao, Lan, Liu and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongjian Liu, empsaXVAZmFmdS5lZHUuY24=; Donghui Peng, ZmpwZGhAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.