
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 30 September 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1001206
This article is part of the Research Topic Adaptation Mechanisms of Grass and Forage Plants to Stressful Environments View all 42 articles
 Andong Yu1,2
Andong Yu1,2 Xueqian Jiang1
Xueqian Jiang1 Yan Sun2
Yan Sun2 Qiannan Hu2
Qiannan Hu2 Xiaoxi Zhu1
Xiaoxi Zhu1 Junmei Kang1
Junmei Kang1 Lin Chen1
Lin Chen1 Lin Liu3
Lin Liu3 Linfeng Hao3
Linfeng Hao3 Qingchuan Yang1
Qingchuan Yang1 Ruicai Long1*
Ruicai Long1* Mingna Li1*
Mingna Li1*Uridine diphosphate glycosyltransferases (UGTs) are enzymes that catalyze glycosylation modifications and play an essential role in regulating plant metabolism. Alfalfa (Medicago sativa L.) is the most important legume in the world due to its high yields and protein content; however, the UGT genes in alfalfa have not yet been studied. Identifying UGT genes with metabolic roles in alfalfa is essential for identifying and modifying genetic traits that are relevant to yield and quality. In this study, 90 of the 239 UGT genes identified from the alfalfa “Zhongmu No. 1” genome database were found to be related to secondary metabolism, and a series of gene family characterization analyses were conducted on each. The results demonstrated that all 90 UGT genes were unevenly distributed on eight chromosomes with few introns and that tandem duplications were the crucial driving force expanding the UGT family in alfalfa. Notably, the 90 UGT genes can be clustered into ten evolutionary groups which contain specific PSPG motifs, and genes in these ten groups have specific tissue expressions. This suggests that the UGT genes in each group could have similar glycosylation roles corresponding to analogous secondary metabolites in alfalfa. Additionally, multiple cis-acting elements found in MsUGT promoter regions, such as phytohormone and flavonoids, indicate that 90 UGT members could be induced by these features, which are also related to secondary metabolism. Therefore, our study identified 90 UGT members inten evolutionary groups that are likely related to glycosylation modifications with secondary metabolites in alfalfa. These findings help uncover pivotal regulatory mechanisms associated with secondary metabolism in plant yield and quality and contribute to genetic modification and breeding in alfalfa and other plant species.
Glycosyltransferases (GTs) are enzymes that utilize activated glycosyl donors as substrates to regulate biochemical properties and subcellular localization by catalyzing the glycosylation reactions of molecules such as proteins, lipids, plant hormones, and phenylpropane compounds (Song et al., 2018; Speeckaert et al., 2020). In plants, glycosylation can preserve the diversity of small molecular compounds and plays an important role in regulating biological processes such as seed germination, growth, flowering, fruiting, and senescence (Li H. et al., 2020; Wu et al., 2020). The GT family 1 (GT1), which is referred to as UDP glycosyltransferase (UGT), mainly catalyzes UDP sugar transfer to specific receptors, such as proteins, nucleic acids, antibiotics, alkaloids, and plant hormones, and is the largest glycosyltransferase gene family in the plant kingdom (Zhang et al., 2020).
UGT is ubiquitous existed in both unicellular algae, where it has a simple structure, and higher plants, where it has a complicated structure (Rehman et al., 2018; Song et al., 2018). The first plant UGT family gene, Bronze1, which encodes a UGT enzyme that can synthesize flavonoid glycosides and regulate the accumulation of melanin in maize (Zea mays) seeds, was identified by Barbara McClintock in 1977 when she was studying the genetic traits of transposons in maize (Dooner and Nelson, 1977). Currently, many UGT gene families have been identified in several crops and plant species at the whole genome-wide level, such as wheat (Triticum aestivum) (He et al., 2018), soybean (Glycine max) (Yano et al., 2018), rice (Oryza sativa) (Dong et al., 2020), and Arabidopsis thaliana (Chen T. T. et al., 2020). Meanwhile, the sophisticated roles of plant hormone regulation, biotic and abiotic stress adaptation, yield and quality enhancement, and secondary metabolism adjustment, that UGTs played, have also been highlighted (Mohnike et al., 2021; Kurze et al., 2022; Wang et al., 2022; Zhang et al., 2022).
For instance, UGTs such as UGT84B1 and UGT76F1 in Arabidopsis (Aoi et al., 2020; Chen L. et al., 2020), CsUGT85A53 in Camellia sinensis (Jing et al., 2020), UGT76C2 in Arabidopsis (Li Y. et al., 2020), and the promoter of SbUGT in poplar (Scutellaria barbata) (Li et al., 2019), reportedly participate in regulating the glycosylation modifications of auxin (IAA), abscisic acid (ABA), and cytokinin (CK), respectively, controlling the growth and development of the roots, flowering, and root buds, respectively. Furthermore, UGTs such as AtUGT71C3 can compromise the excessive defense response of Arabidopsis by regulating the steady state of MeSA and SA (Chen et al., 2019), while the down-regulation of CsUGT91Q2 decreased the scavenging capacity of reactive oxygen species and caused sensitivity to low-temperature stress (Zhao et al., 2020). Moreover, UGTs such as GhUGT103, GhUGT105 (Xiao et al., 2019), and OsUGT83A1 (Dong et al., 2020) were involved in regulating fiber development in upland cotton (Gossypium hirsutum L.) and grain size in rice, and have been considered important traits of grain yield.
In particular, UGTs such as UGT79B2 and UGT79B3 in Arabidopsis (Li et al., 2017), ZmUFGT2 (UGT78D2) in maize (Li et al., 2018), and UGT87A1 in Carex rigescens (Zhang K. et al., 2021), are involved in secondary metabolism via the glycosylation of flavonoids, which are a kind of natural polyphenolic secondary metabolite with antioxidant and free radical scavenging functions in different tissues (Rojas Rodas et al., 2014; Huo et al., 2021). Additionally, these UGTs can catalyze glycosylation reactions from anthocyanins, kaemferol, and quercetin to the corresponding glycosides in vivo. Decreasing the expression of CsUGT91Q2 resulted in the decrease of neroli glycoside and caused sensitivity to low-temperature stress (Zhao et al., 2020), indicating that UGT acts on glycosylation terpenoids, which are the largest category of secondary metabolites (with more than 40,000 structures) and have several biological functions such as anti-tumor, antibacterial, hypoglycemic and economic value due to their flavor, tastants, and colorants (Chan et al., 2016; Kurze et al., 2022). Moreover, studies have demonstrated that the soyasaponins of soybean (Yano et al., 2018) and rebaudioside A (Reb A) of Stevia rebaudiana (Kim et al., 2019) can be converted by UGT91H9 and UGT76G1 respectively, leading to better taste and higher quality. Generally, the glycosylation modification functions that plant UGTs have on various secondary metabolites can generate active molecules that are involved in several processes related to biological regulation (Rehman et al., 2018; Yu et al., 2022). Therefore, studying the functions of UGTs involved in secondary metabolism is essential for identifying and modifying genetic traits highly relevant to crop yield and quality production.
There is a highly conserved plant secondary product glycosyltransferase box (PSPG) composed of 44 amino acids at the C-terminal of the UGT gene family protein (Cheng et al., 2019). A previous study proposed that the whole folding and core region of UGT proteins in plants are conserved since the C-terminal domain of UGT proteins mostly recognizes the same or similar glycosyl donors, and the N-terminal domain recognizes specific receptors (Nair et al., 2020). This indicates that the PSPG motif is an important binding region that can recognize the UDP sugar donor of uridine, which contributes to glycosylating specific secondary metabolites in plants (Cheng et al., 2019). While the evolutionary relationships of the UGT gene family members are related and have similar exon and intron numbers and sequence characteristics (Xiao et al., 2019), 122, 148, 184, and 128 UGT genes have been identified in Arabidopsis, maize, grape (Vitis vinifera L.) and soybean, respectively, using the PSPG motif (Rehman et al., 2018). This suggests that UGTs have certain specificity in different plant species. For example, after comparing the amino acid sequences encoded by five UGT genes related to terpenoid glycosylation in Panax notoginseng, the positions of 17 amino acids were found to be highly conservative, while the other positions vary (Wang et al., 2020). Therefore, it is necessary to identify and characterize the specific UGT gene family in certain plant species before investigating the UGTs involved in secondary metabolic regulation.
Alfalfa (Medicago sativa L.) is the most important and widespread legume plant worldwide and is prized for its high protein content, nutritional quality, yield, and environmental adaptability (Barros et al., 2019; Shen et al., 2020). Studying the molecular regulatory role of UGT genes in alfalfa is important for improving its yield and quality through genetic engineering. However, few studies have researched UGT genes in alfalfa, and the biological role of UGTs associated with secondary metabolism has not been understood until now. Recently, the genome sequence data of the autotetraploid plant alfalfa has been gradually completed (Chen H. et al., 2020; Shen et al., 2020; Long et al., 2022), providing valuable genomic information and making it possible to identify and research the function of the UGT gene family in alfalfa at the genome-wide level. In this study, we performed a systematic genome-wide analysis to identify UGT family genes in alfalfa and screened 90 of them for their association with secondary metabolism. These 90 UGT genes were then clustered into ten phylogenetic groups, and their evolutionary relationship, conserved motif, chromosomal distribution, gene structure, cis-acting elements, intra- and inter-species collinearity, and tissue expression patterns were analyzed. This study aimed to identify the UGT gene family in alfalfa involved in secondary metabolism at the genome-wide level and clarify their gene and protein molecular characteristics, evolutionary relationship, and major induced factors. The results provide comprehensive genetic information and outline the molecular features of UGT genes related to secondary metabolism in alfalfa.
Alfalfa (Medicago sativa L. cv. Zhongmu-1) seeds preserved in our lab were used as the plant material. Alfalfa seeds were germinated on filter paper immersed with ultrapure water in a petri dish in a growth chamber (GXZ-500, Jiangnan, China) 16 h light (1,200–1,250 μmol m–2 s–1) at 25°C and 8 h darkness at 20°C. After 7 days of germination, the seedlings were transferred to a plastic cuboid container (25 cm × 20 cm × 7.5 cm) with 1/2 Hoagland nutrient solution under controlled conditions. For the transcriptional expression assay, the 4-week-old Hoagland solution was used to culture alfalfa seedlings under identical growth conditions. Samples were obtained from seven sections, including the new leaves (NL), the mature leaves (ML), the first real leaves (FL), and the stems of each 1/3 position from top to bottom: upper stems (US), middle stems (MS) and bottom stems (BS), and root, with three biological replicates for each (Ma et al., 2021).
The M. sativa “Zhongmu No. 1” genome information (Shen et al., 2020) was downloaded from the figshare website1. UGT protein sequences of Arabidopsis were retrieved from the P450 database2. Alfalfa UGT genes were confirmed using the local BLAST with a cutoff E-value of 1.0 × 10–10 and TBtools software (Huazhong Agricultural University, Wuhan, China) (Chen C. et al., 2020). The UGT protein sequences of alfalfa were aligned using the Hidden Markov Model (HMM) model on the Pfam online website3 (Finn et al., 2014). The UDPGT domain (PF00201) was used to identify UGT members in the Pfam database4 (Finn et al., 2014).
UGT proteins identified in alfalfa were annotated in the eggNOG-Mapper database5 functional annotation enrichment (Huerta-Cepas et al., 2019). UGT proteins involved in secondary metabolism were considered by the Pathway Name and Description in the KEGG Pathway Database6 (Antonov et al., 2010), including metabolic pathways (K01115), glucosinolate biosynthesis (K11280), biosynthesis of various plant secondary metabolites (K21371), monoterpenoid biosynthesis (K21373, K21374), anthocyanin biosynthesis (K12930, K12938, K17193), zeatin biosynthesis (K13493, K13494, K13495, K13496), and phenylpropanoid biosynthesis (K12356).
Basic physical and chemical characteristics, including the sequence start position, sequence end position, length, molecular weight, isoelectric point, and Grand average of hydropathicity (GRAVY) of UGT genes in alfalfa, which were screened by KEGG functional enrichment, were obtained from the ExPASy website7 (Gasteiger et al., 2003). Subcellular localization prediction was performed by WoLF PSORT8 (Horton et al., 2007).
Divergent UGT protein sequences were removed from the identified sequences after alignment with MEGA 11 software. The phylogenetic tree was built using MEGA11 software (Tamura et al., 2021) with UGT protein sequences from alfalfa, Arabidopsis, and maize by the neighbor-joining (NJ) method (bootstrap values for 1,000 replicates) and was visualized using the Evolview website9. UGT proteins in alfalfa were classified according to the 14 reported evolutionary groups in Arabidopsis (A-N) and the other three new groups found in maize (O-Q). The conserved PSPG motif, composed of 44 amino acids in each UGT phylogenetic group, was analyzed using MEME software10 (Bailey et al., 2009).
Information on the chromosomal location of UGT genes was retrieved and generated by TBtools. Ten motifs with lengths of 15–50 amino acids in UGT proteins from alfalfa were identified using the online MEME Suite (see text footnote 10) (Bailey et al., 2009). The MAST file was obtained from the MEME website, and the Newick tree string was visualized in TBtools. The exon and intron structures of MsUGT genes were analyzed using TBtools software, inputting gene annotation GFF files and Newick tree string.
The promoter regions of MsUGT genes (2,000 bp sequence upstream of the DNA genome sequences) were obtained from the M. sativa genome database. PlantCARE database11 was used to predict the cis-acting elements (Lescot et al., 2002). The results containing promoter information and the phylogenetic evolution of the UGT gene family in alfalfa were visualized using TBtools software.
The identification of gene duplication patterns in MsUGT genes was performed using multiple collinear scanning toolkits (MCScanX) with an E-value set to 10–5. The syntenic relationship between MsUGT genes and UGT genes from Medicago truncatula, Arabidopsis thaliana, Oryza sativa, and Glycine max were determined using the Dual Synteny Plotter function in the TBtools software. The necessary genome sequence was obtained from the BioMart Ensemble plants database12.
Total plant RNA was extracted using the total RNA Extraction kit (Shanghai Promega Biological Products, China), according to the manufacturer’s instructions. Complementary DNA was generated with reverse transcriptase (TaKaRa, Beijing, China) at 42°C for 2 min, then at 37°C for 15 min, and finally at 85°C for 5 s. Quantitative real-time PCR (qRT-PCR) was conducted with 2 × EasyTaq® PCR SuperMix (+ dye) (TransGen Biotech, Beijing, China), according to the manufacturer’s instructions, on the CFX96 Touch™ RT-PCR system (BioRad, Los Angeles, CA, United States). Three technical repeats were performed for each sample. The specific primer sequences of MsUGT genes for qRT-PCR determination are provided in Supplementary Table 1. The Ms-ACTIN gene was used as an internal control (Supplementary Table 1). The relative transcriptional expressions of each gene were analyzed based on the 2–ΔΔCt method (Livak and Schmittgen, 2001). SPSS 20 software was used to process the experimental data by the analysis of variance (ANOVA). The least significant difference method (LSD, P < 0.05 level) was used for statistical testing (Ma et al., 2016). The results were completed and presented using Graphpad Prism 8.
To identify the UGT gene family in alfalfa, a local blast analysis was conducted on “Zhongmu No. 1” protein sequences using 122 UGT protein sequences from Arabidopsis. In total, 239 putative UGTs containing the UDPGT domain (PF00201) in alfalfa were obtained and were named according to their chromosomal location. In addition, these 239 UGT genes were enriched to KEGG pathways corresponding to the eggNOG-Mapper database (Figure 1). The results demonstrated that 90 genes were enriched in the functional pathways of monoterpenoid biosynthesis (54), zeatin biosynthesis (17), anthocyanin biosynthesis (14), glucosinolate biosynthesis (9), biosynthesis of various plant secondary metabolites (5), benzoxazinoid biosynthesis (3), and phenylpropanoid biosynthesis (3) (Figure 1), which are considered the pathways associated with secondary metabolism. This indicates that monoterpenoid biosynthesis is the dominant pathway associated with UGT genes in alfalfa, while zeatin biosynthesis, anthocyanin biosynthesis, and glucosinolate biosynthesis are the main functional pathways of enriched UGT genes. Therefore, based on these screening results, we analyzed the molecular characteristics, phylogenetic relationships, chromosomal localization, structural characterization, conserved motifs, cis-acting elements, intra- and interspecies collinearity, and expression pattern of these 90 UGT members to represent UGT genes related to secondary metabolism in alfalfa.
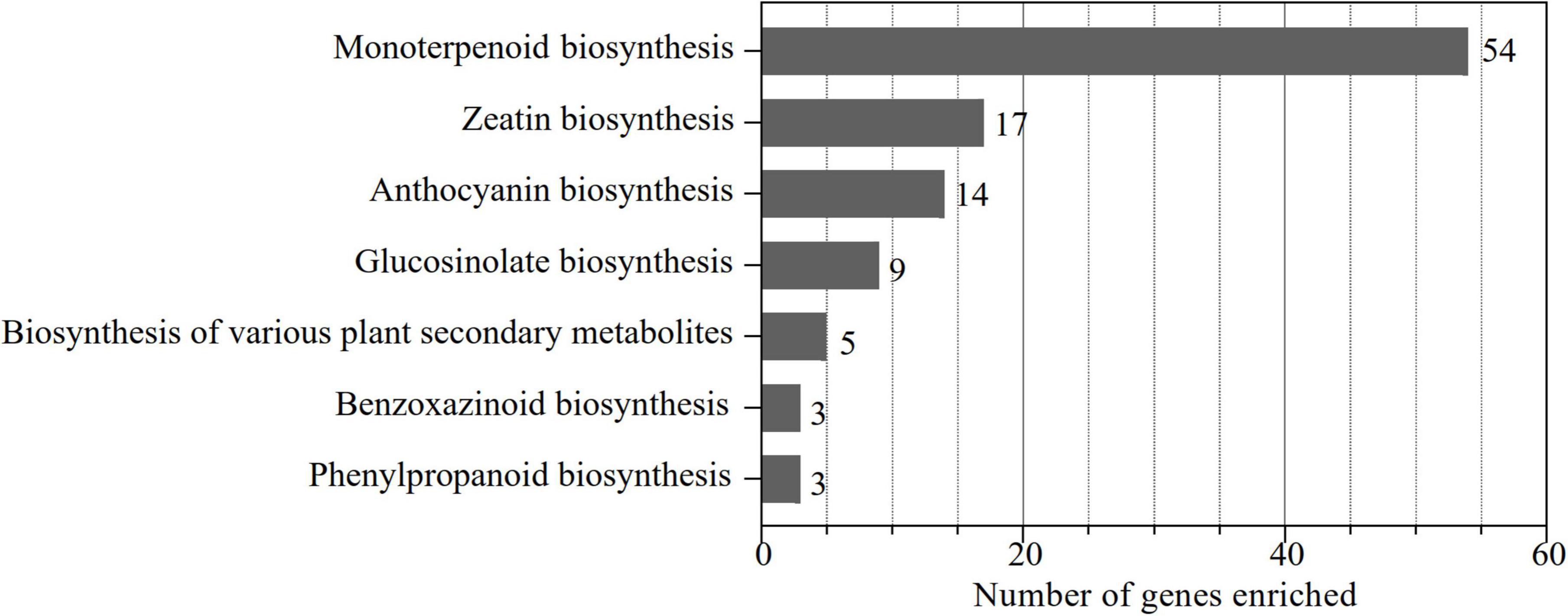
Figure 1. Number of UGT genes involved in secondary metabolism-related pathways. The KO identifiers (called K numbers) we considered associated with the secondary metabolism network are displayed: monoterpenoid biosynthesis (K21373, K21374), zeatin biosynthesis (K13493, K13494, K13495, K13496), anthocyanin biosynthesis (K12930, K12938, K17193), glucosinolate biosynthesis (K11280), biosynthesis of various plant secondary metabolites (K21371), phenylpropanoid biosynthesis (K12356), and benzoxazinoid biosynthesis (K13227).
To characterize the 90 UGTs in alfalfa screened above, the basic molecular characteristics of the proteins were analyzed. The gene name, gene ID based on the “Zhongmu No. 1” database, the start and end point, the number of amino acids, molecular weight (Mw), isoelectric point (pI), and GRAVY of each gene are displayed in Supplementary Table 2. The amino acid length of MsUGTs varied among the 90 proteins, from 82 to 1,683 amino acids. The theoretical pI and Mw ranged from 4.94 to 8.86 and from 9,097.77 Da to 188,543.37 Da, respectively. Analysis of predicting subcellular localization found that 47 MsUGT members were localized in the chloroplast; 27 and 9 MsUGTs were distributed in the cytoplasm and nucleus, respectively; while a small number of MsUGT proteins were widely located in the mitochondria (2), extracell (1), peroxisome (1) and plasma membrane (1). These results provide an integrative view of the molecular characteristics of these 90 UGT members.
To better understand the evolutionary relationship of the 90 UGT genes associated with secondary metabolism, phylogenetic analysis was conducted based on the amino acid sequence similarity and topological structure (Figure 2). The results demonstrated that the 90 UGT proteins were classified into ten different evolutionary groups (Figure 2A). The four groups with the largest numbers were G, L, A, and D, with 33, 14, 10, and 10 UGT members, respectively; the three groups with the fewest UGT proteins were groups F, H, and P, with 2, 3 and 3, respectively; and none were found in groups B, C, I, K, M, N, and Q in alfalfa. Moreover, ten phylogenetic groups of the MsUGT genes were further distinguished by the symbolic PSPG motif (Figure 2B). While the PSPG boxes were presented in each phylogenetic group, the PSPG sequences in different groups were not completely identical (Figure 2B). Twenty-two and 32 amino acid residues of PSPG motifs were fully conserved in groups D, J, and L and in groups H and P, respectively. Additionally, 16 amino acid residues were the same in these sequences, and groups A, E, F, G, and O had low sequence similarity with other groups, though they were conservative within the group.
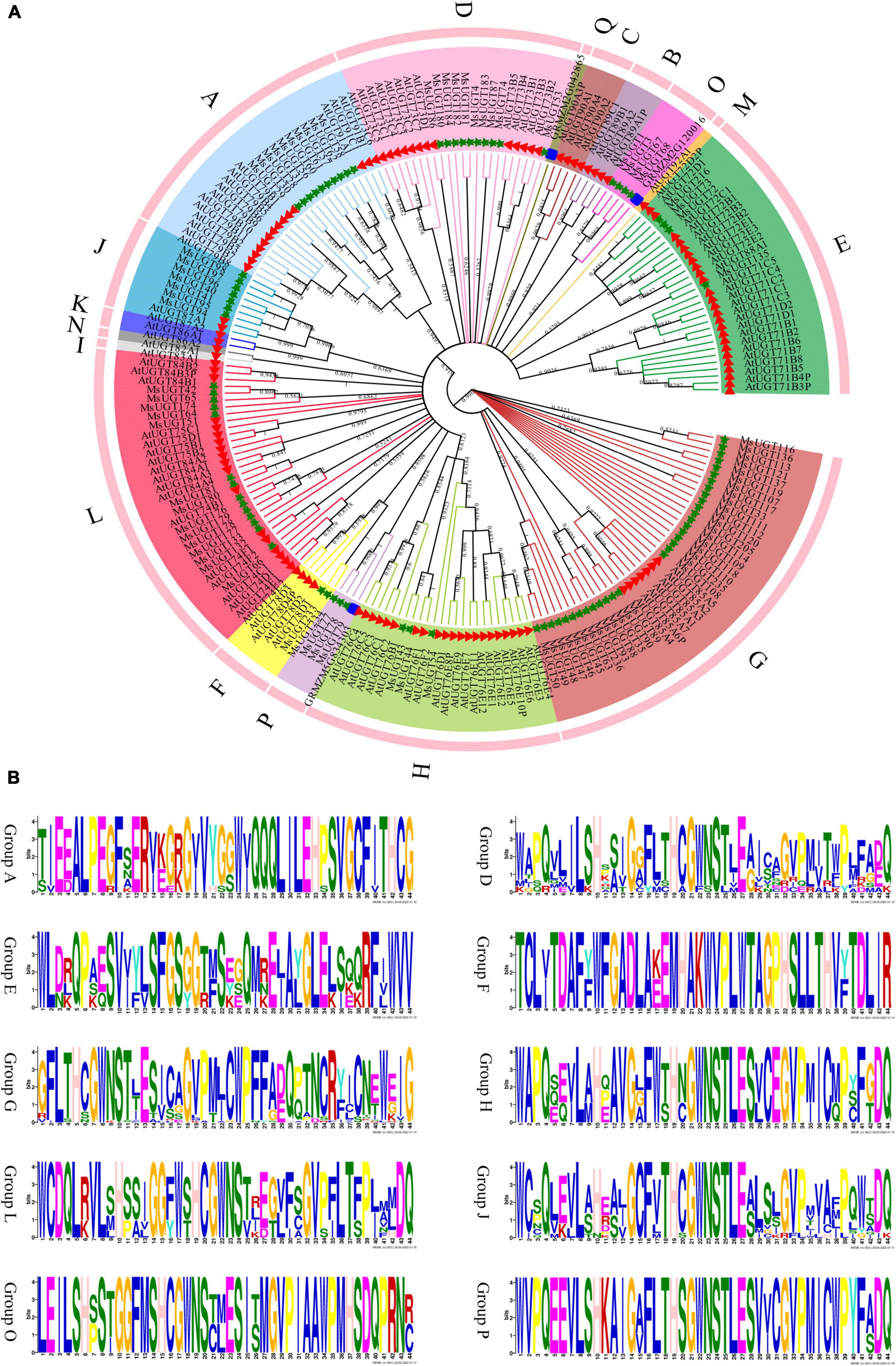
Figure 2. Phylogenetic and PSPG box analyses of MsUGT proteins associated with secondary metabolism in alfalfa. (A) The phylogenetic tree is generated by the NJ method with 1,000 repeats from the sequences of Arabidopsis thaliana, Zea mays, and M. sativa. (B) The PSPG motif of each phylogenetic group is shown.
To explore the genomic distribution of alfalfa UGT genes, the chromosomal location of MsUGT genes was investigated based on the genome annotation database (Figure 3 and Supplementary Table 3). As shown in Figure 3, 88 UGTs were unevenly distributed across eight chromosomes, and two UGTs were identified in the scaffolds. Twenty UGTs were distributed on chromosome 6, and 19, 14, and 14 UGTs were distributed on chromosomes 5, 4, and 3. Notably, 23 gene pairs were involved in tandem duplications covering 39 UGT genes, which were clustered into seven alfalfa chromosomes (Figure 3).
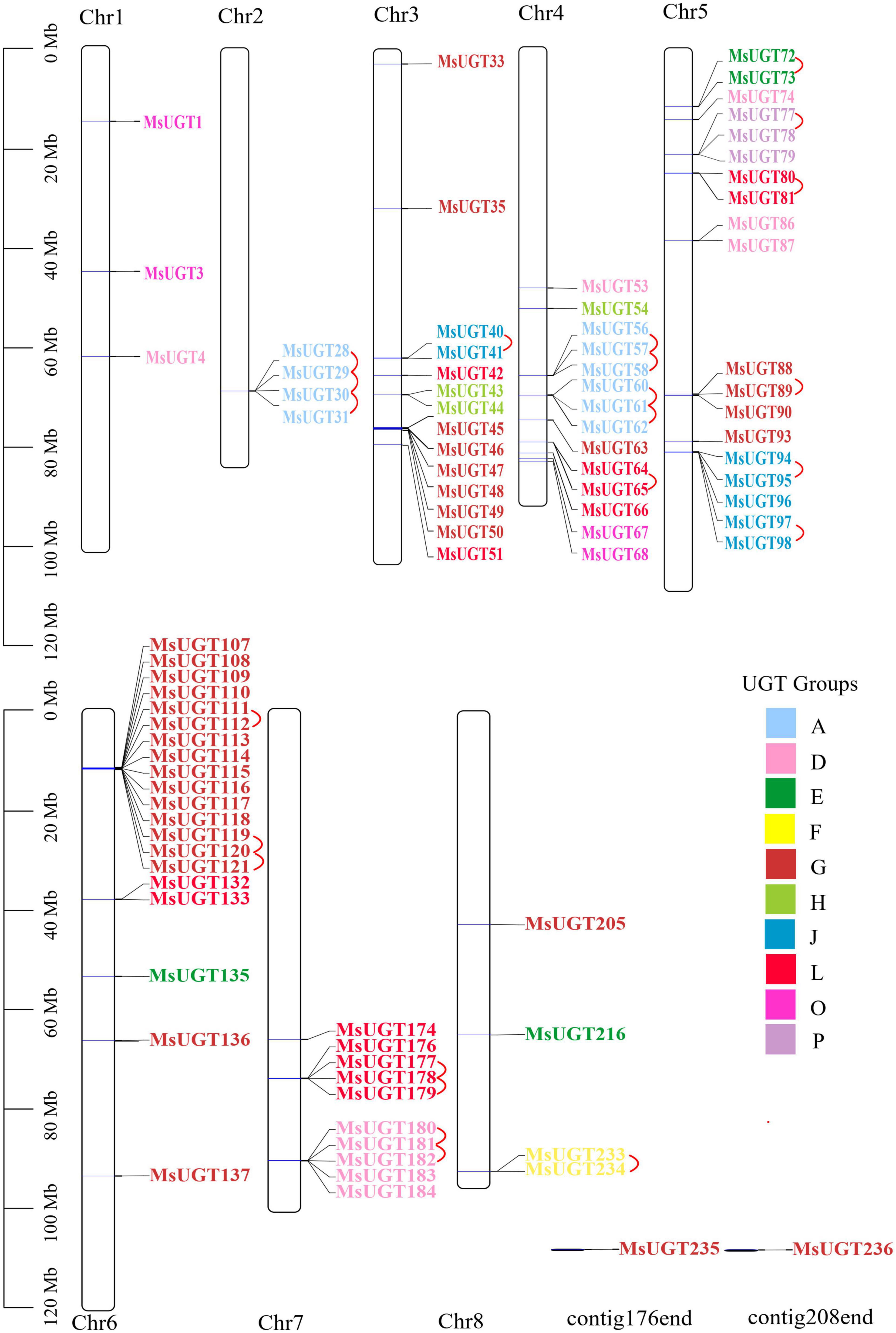
Figure 3. Chromosomal location and tandem duplication event analyses of MsUGT genes associated with secondary metabolism in alfalfa. The tandem duplicated gene pairs are connected by red lines. Each phylogenetic group is indicated by the same color used in the evolutionary tree in Figure 2A.
Analysis of the gene structure and motifs was constructed with the phylogenetic tree (Figure 4A). Gene structure analysis demonstrated that the intron numbers of UGT genes associated with secondary metabolism ranged from 0 to 23 (Figure 4B). Most UGT genes contained 0–2 introns, MsUGT74 had 23 (the most), and 26 genes (MsUGT29, 30, 31, 44, 56, 57, 60, 61, 62, 64, 67, 68, 72, 73, 79, 86, 93, 107, 110, 132, 180, 181, 182, 183, 216, and 235) lacked introns (Table 1). We also identified ten motifs with 15–50 amino acids (Figure 4C and Supplementary Table 4). Most UGT proteins contained 6–10 conserved motifs, and UGT proteins of the same branch had similar conserved motif composition and sorting order, suggesting that MsUGT proteins in the same group could share similar functions.
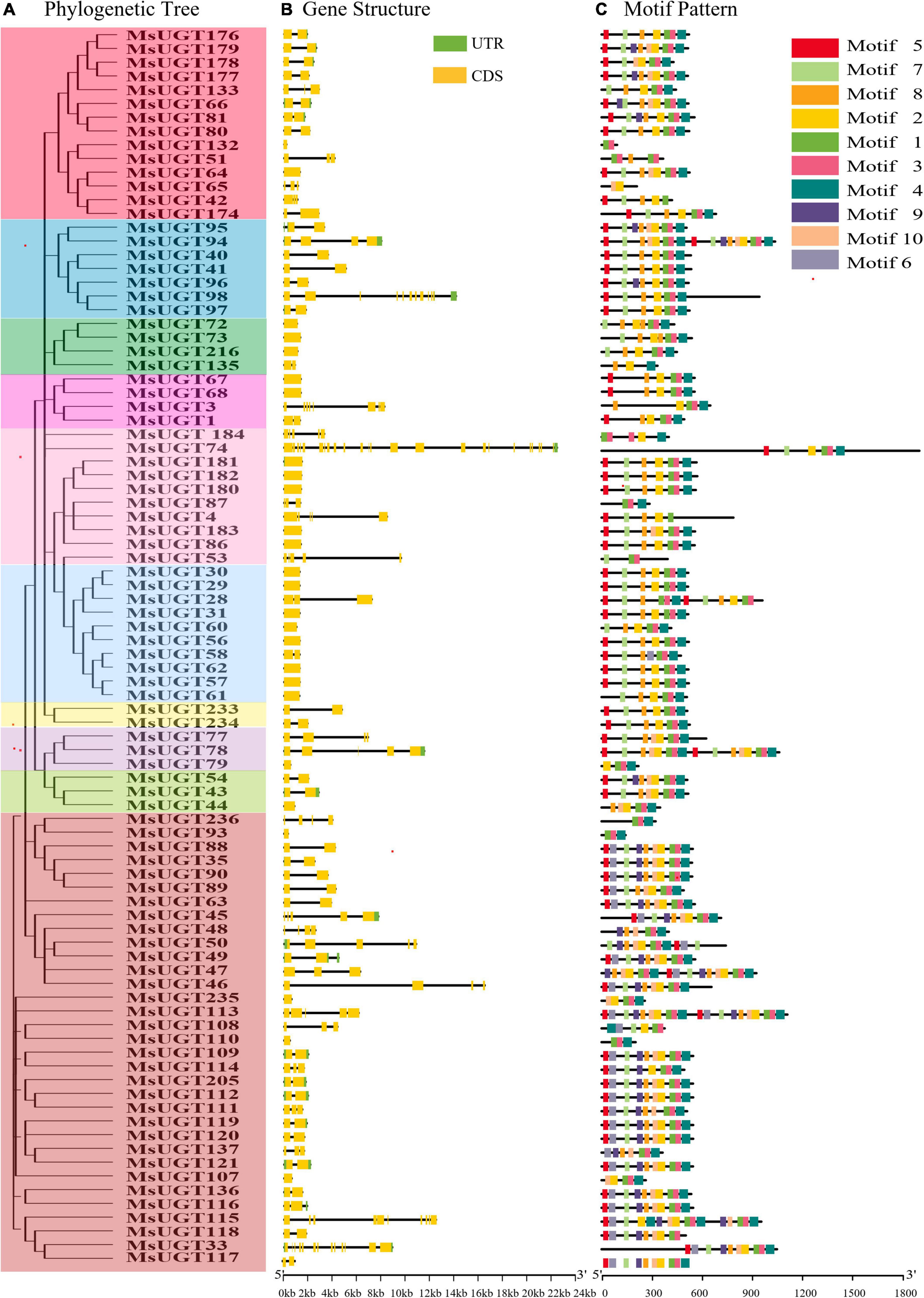
Figure 4. The phylogenetic relationship, exon-intron structure, and conservative motif analysis of 90 UGT genes associated with secondary metabolism in alfalfa. (A) The phylogenetic tree of 90 MsUGTs was constructed by the NJ method with 1,000 repeats. (B) The exon-intron structure analysis of 90 UGT genes was represented in detail. (C) The conservative motif analysis of 90 MsUGTs was predicted. Each phylogenetic group is indicated by the same color used in the evolutionary tree in Figure 2A.
To better investigate the characteristics of UGT promoters, the cis-acting elements were analyzed (Figure 5 and Supplementary Table 5). The results demonstrated that the cis-acting elements of promoter regions of the UGT gene family were classified into five categories, including light responsiveness, plant hormone response, resistant response, promoter core elements (TATA-box), and common cis-acting element (CAAT-box). Except for the promoter core elements and common cis-acting elements, the number of light-responsive elements was the largest, with 2,676. Phytohormone-responsive elements were primarily related to GA, MeJA, IAA, ABA, and SA, and data for three abiotic stress response elements such as drought, low temperature, and salt stress, are displayed in detail (Figure 5). MsUGTs contained 11 flavonoid biosynthesis-related elements due to the potential involvement of MsUGT genes in secondary metabolism. Overall, the cis-element analysis indicated that UGT genes could respond to different kinds of metabolic processes.
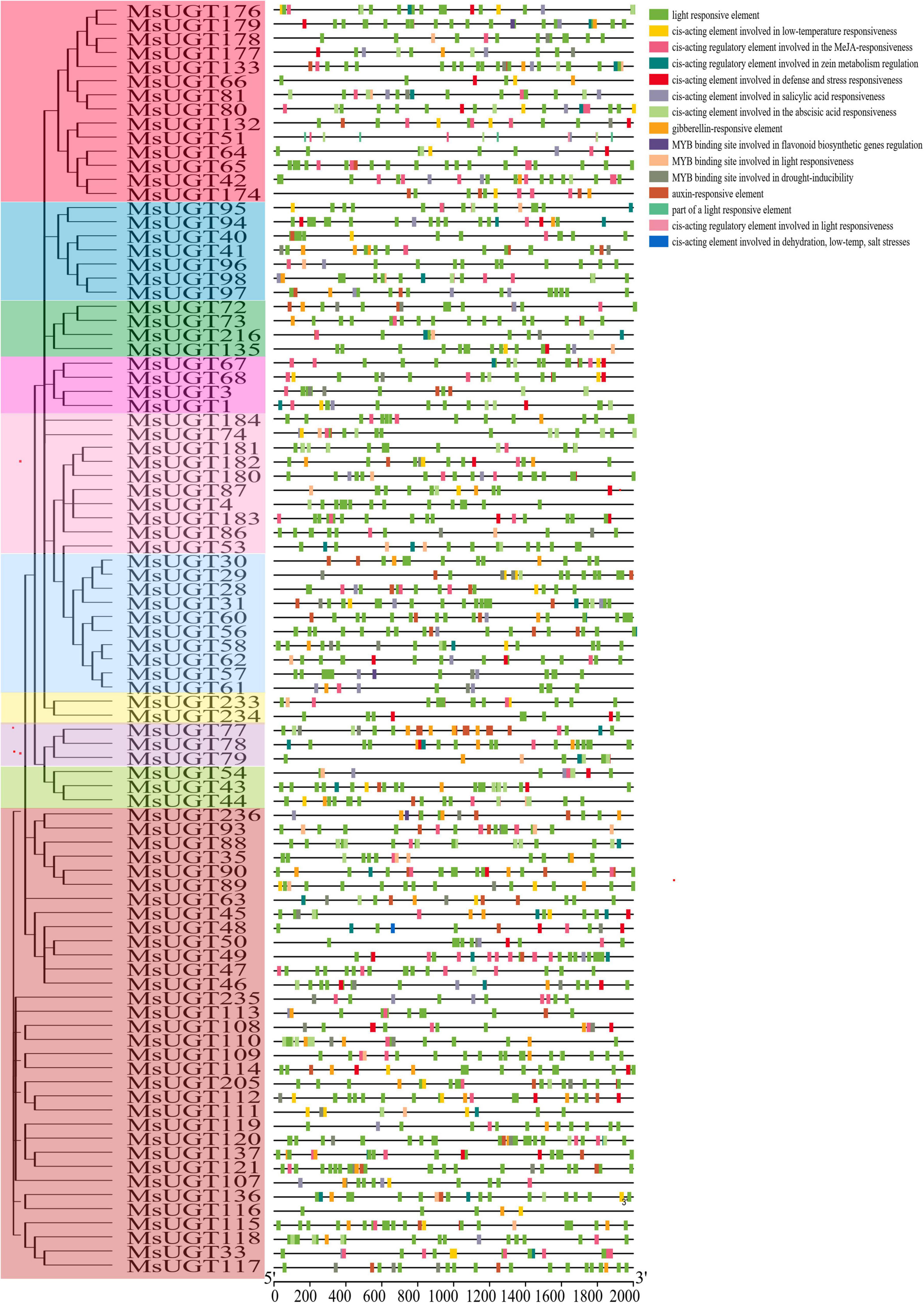
Figure 5. Cis-acting elements analysis in the promoter region of UGT genes associated with secondary metabolism in alfalfa. Each phylogenetic group is indicated by the same color used in the evolutionary tree in Figure 2A.
To explore the collinearity of the MsUGT gene family between and across species, we constructed syntenic maps of MsUGT genes with those in M. truncatula and three other model plants of Arabidopsis thaliana, Glycine max, and Oryza sativa (Figure 6 and Supplementary Table 6). A segmental duplication event was identified from MsUGTs (MsUGT4/MsUGT182), which was distributed on chromosomes 1 and 7 (Figure 6A). A total of 118 corresponding orthologs were identified, including 55, 16, 45, and 2 genes from M. truncatula, Arabidopsis, soybean, and rice, respectively (Figure 6B). These genes were unevenly distributed on eight chromosomes in alfalfa, while chr3 and chr5 identified more collinear genes than the others. These results indicate that MsUGT genes displayed different numbers of syntenic lines with four species, while most paralogous MsUGTs pairs could have variable expression patterns.
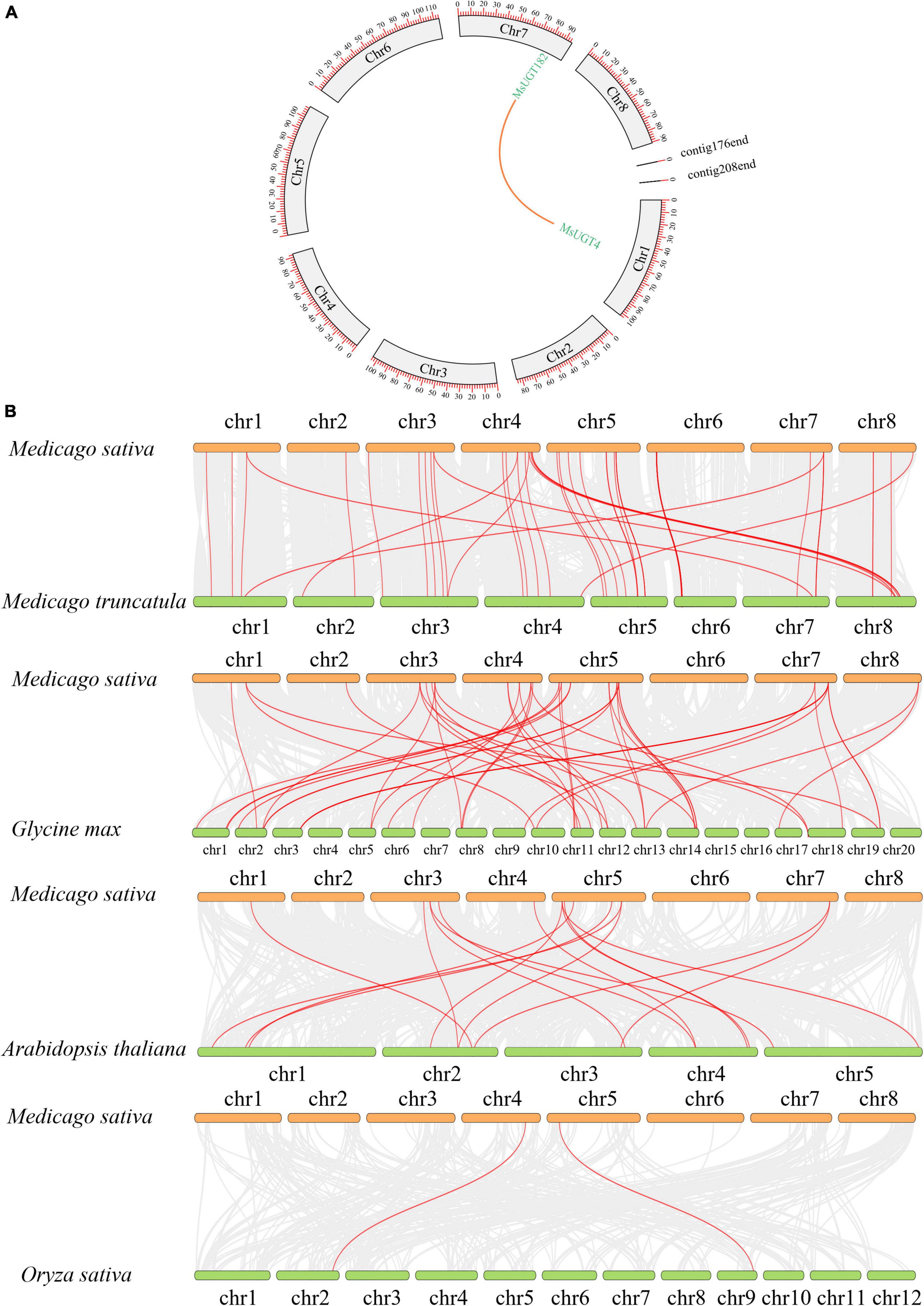
Figure 6. MsUGT gene duplications in the genomes between and within species. (A) The duplicated event is a collinearity block in the alfalfa genome, and the segmental duplication between MsUGT4 and MsUGT182 is drawn with an orange line. (B) The collinear blocks of UGT genes between M. sativa and M. truncatula, Glycine max, Arabidopsis thaliana, and Oryza sativa, respectively, are indicated by red lines.
To investigate the tissue expression profiles of the 90 UGT gene members associated with secondary metabolism in alfalfa, we performed a transcriptional expression analysis of one representative gene from each evolutionary group (Figure 7 and Supplementary Table 7). The results demonstrated that the expression of MsUGT28, MsUGT54, and MsUGT79 from groups A, H, and P have similar tissue expression profiles and were highest in the leaves and lowest in the roots (Figure 7A and Supplementary Table 7). MsUGT28 expression in leaves was significantly higher than in the roots and stems, and the highest expression occurred in NL, reaching 43.52; the expression of MsUGT79 was similar to that of MsUGT28, while its highest expression level appeared in NL, reaching 86.02, and its expression in three stem tissues also reached 17.21, 27.74 and 31.71; the expression level of MsUGT54 reached 27.03, 5.64, and 4.80 in FL, NL, and ML, respectively, which was significantly higher than in the stems and roots.
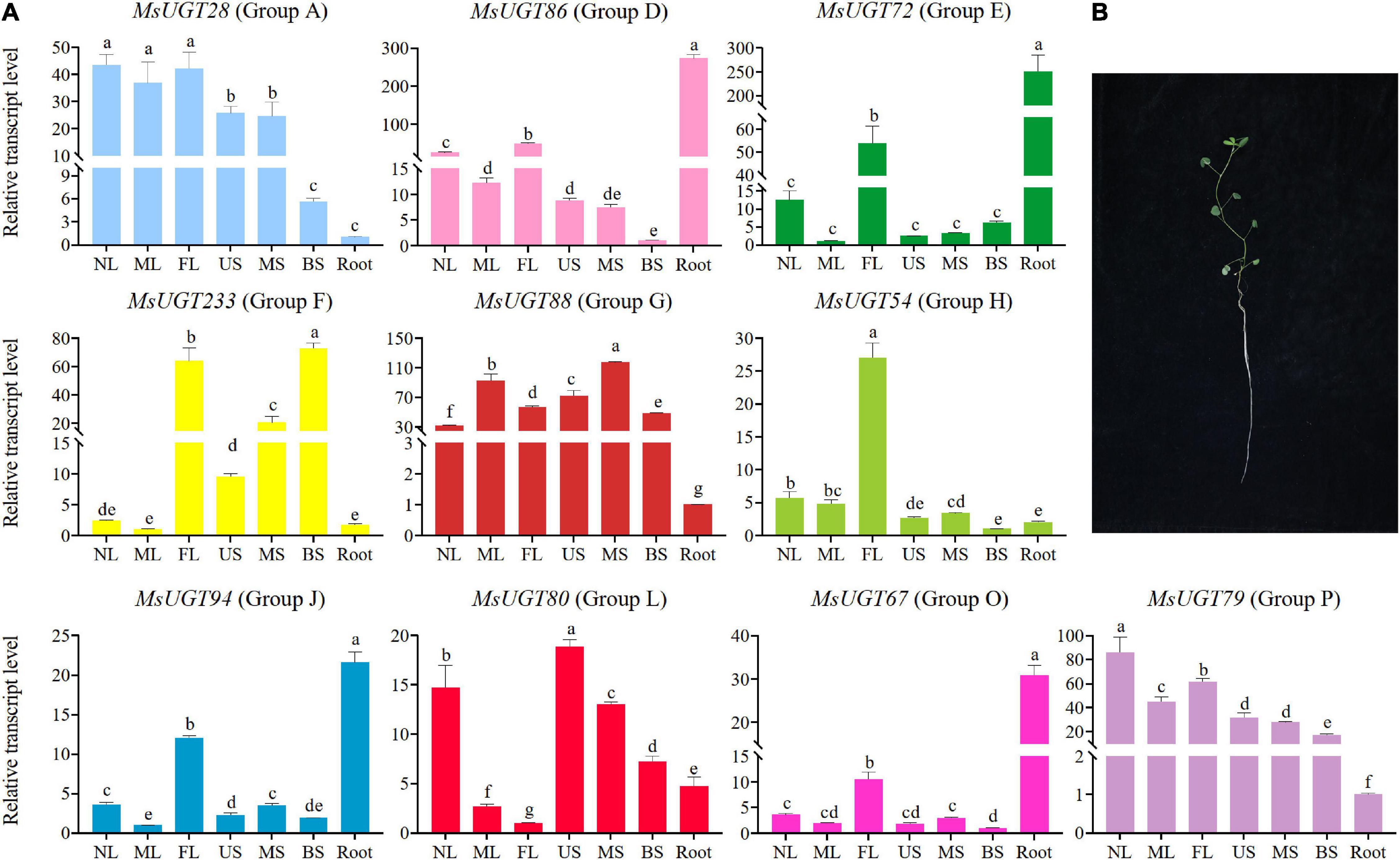
Figure 7. Transcriptional expression analysis of MsUGT genes of 10 groups in seven different tissues. (A) Genes of MsUGT28, 86, 72, 233, 88, 54, 94, 80, 67, and 79 were selected from groups A, D, E, F, G, H, J, L, O, and P, respectively. The gene expression represented in each evolutionary group is shown with the same color as the phylogenetic tree in Figure 2A. (B) Tissues are harvested for transcriptional expression. NL, new leaves; ML, mature leaves; FL, the first real leaves; US, Upper stems; MS, middle stems; BS, bottom stems; Root.
The expression of MsUGT86, MsUGT72, MsUGT94, and MsUGT67 from groups D, E, J, and O in the roots reached 273.23, 250.60, 21.58, and 30.89, respectively, which was higher than in other tissues. Of these, the expression of MsUGT86 in three stem tissues from the bottom to the top was the lowest, and the expression in the leaves exceeded that in the stems, reaching 24.59, 12.27, and 48.53, respectively. The tissue expression patterns of MsUGT72, MsUGT94, and MsUGT67 in FL were 53.79, 12.02, and 10.52, which was significantly higher than in other leaves and stems.
However, some UGT genes in each evolutionary group showed specific tissue expression profiles, such as UGT88, UGT233, and UGT80 from groups G, F, and L, respectively. The expression of MsUGT88 in group G exceeded 30 in both the stems and leaves, which was much higher than in the roots. The expression of MsUGT233 in group F showed the highest expression in FL and BS, with 63.86 and 72.9, respectively, while MS was 20.48, which was significantly higher than in NL, ML, US, and root tissues. The expression of MsUGT80 in group L was the highest in US, 18.86, and decreased from NL and MS, BS, root, ML, and FL. In summary, MsUGT28 in group A, MsUGT54 in group H, and MsUGT79 in group P had the highest expression in leaves; MsUGT233 in group F, MsUGT88 in group E, and MsUGT80 in group L had the highest expression in stem tissues; MsUGT86 in group D, MsUGT72 in group E, MsUGT94 in group J, and MsUGT67 in group O had the highest expression in the roots. These results demonstrate that MsUGT genes related to secondary metabolism could function in the full plant, and that MsUGT genes in each evolutionary group had tissue-specific expression profiles.
Alfalfa is the most important legume in the world due to its high yield, high quality, and environmental adaptability, while the glycosylation reactions catalyzed by UGTs are necessary for maintaining and enhancing metabolic homeostasis by regulating flavonoids, terpenoids, lignin, and other metabolites. Accordingly, identifying and characterizing the UGT genes involved in secondary metabolism is important for improving yield, quality, and stress tolerance traits in alfalfa. In this study, 90 UGT genes associated with secondary metabolism screened by KEGG pathway functional enrichment were identified in alfalfa. They were classified into ten evolutionary groups, and their basic molecular characteristics, evolutionary genetic information, gene structure, motif conservation, promoter region function prediction, intra- and inter-species collinearity, and expression patterns were analyzed. Based on these results, their basic molecular features, genetic evolution properties related to secondary metabolism, and cis-element analysis were assessed.
In our study, MsUGT genes associated with secondary metabolism had similar conservative motifs and gene structures, demonstrating that the number of genes containing 0–2 introns adds up to 74 and accounts for 82.2% to the 90 members. The gene structures were similar to the results of previous studies of UGT genes in upland cotton (Xiao et al., 2019), Arabidopsis (Wu et al., 2020), and pomelo (Citrus grandis) (Wu et al., 2020). Of the 36 UGT members of upland cotton (Xiao et al., 2019), 17 and 19 genes contained one and two introns, respectively, while approximately 50% of the genes encoding glycosyltransferase in Arabidopsis and pomelo had no introns, most remaining UGTs contained one or two introns, and few genes contained multiple introns (Wu et al., 2020). The motif sequences of UGT members in groups with evolutionary relationships were similar, indicating that similarities in the motif location and gene structure of MsUGTs in the same subfamily sustained the phylogenetic classification of MsUGTs.
Our data also demonstrated that of the identified 90 UGT genes, 23 tandem duplications primarily occurred on chromosomes 2, 4, 5, 6, and 7, and one segmental duplication occurred on chromosomes 1 and 7. It is speculated that the variation in the number of UGT family members can be influenced by gene duplication events (Zhang H. et al., 2021). The tandem and segmental duplications of the genome provided more gene copies for generating novel gene functions and expanding gene families in alfalfa. However, compared with the seven segmental duplications in sacred lotus (Nelumbo nucifera) (Li H. et al., 2020), 24 gene segmental duplication events in cassava (Manihot esculenta Crantz) (Wu et al., 2021), and 28 segmental duplications in Broussonetia papyrifera (Wang et al., 2021), alfalfa has fewer segmental duplication genes and only one pair of highly homologous genes involving MsUGT4 and MsUGT182. This indicates that tandem duplication events are more crucial than segmental duplications for MsUGT expansion. Furthermore, compared with the 16 and 2 corresponding orthologs identified in Arabidopsis and rice, the number of corresponding orthologs in M. truncatula and soybean was higher (36 and 40), demonstrating that UGT families could be related to the evolution of dicots but not monocots, and that whole genome duplication events could have contributed more to the evolution of species within the same family (Zhang H. et al., 2021).
The 90 UGT genes were clustered into ten evolutionary groups after confirming the UGT gene families in Arabidopsis and maize. Of them, ten genes (MsUGT4, 53, 74, 86, 87, 180, 181, 182, 183, and 184) were identified in group D in alfalfa, which has been confirmed to catalyze terpenes and flavonoids; four UGTs (MsUGT72, 73, 135, and 216) were identified in group E, which catalyzes phenylpropanoids (Song et al., 2019). For example, the UGT 72 family in group E has been shown to glycosylate two classes of phenylpropanoids (monolignols and flavonoids) (Speeckaert et al., 2022). Moreover, PSPG has been highlighted to play a critical role in regulating secondary plant metabolites (Zhang et al., 2018; Wang et al., 2020), and the PSPG analysis performed on the 90 MsUGT proteins in all ten evolutionary groups demonstrated that each evolutionary group had PSPG box in alfalfa. Accordingly, positions 1 (W), 4 (Q), 10 (H), 14 (G), 16 (F), 19–24 (HCGWNS), 27 (E), 32 (G), 34 (P), 39 (P), 43 (E/D) and 44 (Q) were highly conserved in MsUGT members in group D, J, and L, which indicate that crucial amino acids act on triterpenoids in Panax notoginseng (Wang et al., 2020). Therefore, differences in the glycosylation reactions of secondary metabolites among the evolutionary groups and the specificities in PSPG motif analyses further verified the involvement of these 90 UGT genes in secondary metabolic biosynthesis.
Our analysis demonstrated that ten UGT genes from each group showed different expression patterns in alfalfa tissues. Some MsUGT members were preferentially expressed in tissues as the stem, root, or leaves, which could be related to the higher accumulation of flavonoids and terpenoids. For instance, MsUGTs in groups A, H, and P, and UGT genes in groups D, E, J, and O have higher expression in leaves and root tissues, respectively, which is where flavone glucuronides primarily accumulate (Adiji et al., 2021). The highest expression levels of MsUGT233, MsUGT88, and MsUGT80 were in BS, MS, and US, respectively, which are responsible for the glycosylation of the secondary metabolite lignin (Lin et al., 2016). This indicates that the expression of UGTs associated with secondary metabolism could differ widely from the specific active sites of glycosylation actions.
A remarkable amount of phytohormone response elements were obtained in the promoter regions of the 90 MsUGT genes, suggesting that glycosylation reactions could be induced by these phytohormones. Plant hormones play an important role in responding to various stresses and normal growth and developmental progress (Tian et al., 2017; Radojicic et al., 2018; Shu et al., 2018). UGTs, such as UGT84A2 (Zhang et al., 2017), UGT75B1 (Chen T. T. et al., 2020) and UGT76D1 (Huang et al., 2018) in Arabidopsis could participate in the glycosylation of IAA, ABA, and SA, respectively, and regulate flowering, stress resistance, and systemic acquired immunity. However, recent studies have proposed that plant hormones could complete their functional role connected with some secondary metabolites by the UGTs. For example, GSA1 in rice had glycosyltransferase activity on flavonoids and lignitol monomers, indirectly affecting flavonoid-mediated auxin polar transport and the expression of related genes (Dong et al., 2020). Therefore, this might be an important line of UGT research in future studies.
In this study, eleven flavonoid response elements were identified in the promoter element, indicating that some UGTs in alfalfa could also be induced by flavonoids. Flavonoids are multifunctional secondary metabolites that are important for plants and human health, which can effectively delay antioxidation and anti-inflammatory effects in the human body (Yin et al., 2017a). A previous study demonstrated that CsUGT75L12 in tea plants can transfer sugar molecules to the C7 hydroxyl group of flavonoids, catalyzing the generation of flavonoid 7-O-glycosides (Dai et al., 2017); Plant secondary metabolites can be coupled with hormones to play an important role in responding to growth and developmental processes (Dong et al., 2020). The final step in the biosynthesis of particular flavonoids was glycosylation modifications by UGTs, while changes in the bioactivity, stability, and solubility of flavonoid compounds could ultimately affect the quality, yield, and adaptation of legumes to environmental changes (Yin et al., 2017a,b).
In this study, we identified 239 MsUGT genes from the alfalfa genome and further screened 90 of them that are involved in secondary metabolic pathways. The basic molecular characteristics, motifs, gene structure, and duplication events of the 90 MsUGT genes were assayed and demonstrated. The genetic evolution properties, including group classifications and PSPG motifs related to secondary metabolism, were analyzed. Tissue differential expression analysis revealed the tissue-specific expression patterns of MsUGT genes in each evolutionary group. Cis-elements analysis of the promoter regions predicted that UGTs could be induced by various phytohormones and flavonoids. Therefore, our study provides comprehensive genetic information and molecular features of the UGT genes associated with secondary metabolism in alfalfa, which helps reveal some of the regulatory mechanisms associated with secondary metabolites that can improve yield and quality in alfalfa and other plant species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
AY and XZ collected plant materials. AY and XJ performed the experiment. AY, XJ, LL, and LH analyzed the data. AY, XJ, and QH processed the figures and tables. YS, RL, and ML conceived and designed the experiment. AY, RL, and ML wrote the manuscript. AY, JK, LC, QY, RL, and ML revised and finalized the manuscript. All authors have read and approved the final manuscript.
This work was supported by Key project of Science and technology vitalize Mongolia action (grant no. NMKJXM202110-5), China Agriculture Research System of MOF and MARA, China (grant no. CARS-34), and Agricultural Science and Technology Innovation Program, China (grant no. ASTIP-IAS14).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1001206/full#supplementary-material
Adiji, O. A., Docampo-Palacios, M. L., Alvarez-Hernandez, A., Pasinetti, G. M., Wang, X., and Dixon, R. A. (2021). UGT84F9 is the major flavonoid UDP-glucuronosyltransferase in Medicago truncatula. Plant Physiol. 185, 1617–1637. doi: 10.1093/plphys/kiab016
Antonov, A. V., Schmidt, E. E., Dietmann, S., Krestyaninova, M., and Hermjakob, H. (2010). R spider: a network-based analysis of gene lists by combining signaling and metabolic pathways from Reactome and KEGG databases. Nucleic Acids Res. 38, W78–W83. doi: 10.1093/nar/gkq482
Aoi, Y., Hira, H., Hayakawa, Y., Liu, H., Fukui, K., Dai, X., et al. (2020). UDP-glucosyltransferase UGT84B1 regulates the levels of indole-3-acetic acid and phenylacetic acid in Arabidopsis. Biochem. Biophys. Res. Commun. 532, 244–250. doi: 10.1016/j.bbrc.2020.08.026
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208.
Barros, J., Temple, S., and Dixon, R. A. (2019). Development and commercialization of reduced lignin alfalfa. Curr. Opin. Biotechnol. 56, 48–54.
Chan, W. K., Tan, L. T., Chan, K. G., Lee, L. H., and Goh, B. H. (2016). Nerolidol: A sesquiterpene alcohol with multi-faceted pharmacological and biological activities. Molecules 21:529
Chen, L., Wang, W. S., Wang, T., Meng, X. F., Chen, T. T., Huang, X. X., et al. (2019). Methyl salicylate glucosylation regulates plant defense signaling and systemic acquired resistance. Plant Physiol. 180, 2167–2181.
Chen, T. T., Liu, F. F., Xiao, D. W., Jiang, X. Y., Li, P., Zhao, S. M., et al. (2020). The Arabidopsis UDP-glycosyltransferase75B1, conjugates abscisic acid and affects plant response to abiotic stresses. Plant Mol. Biol. 102, 389–401. doi: 10.1007/s11103-019-00953-4
Chen, L., Huang, X. X., Li, Y. J., and Hou, B. K. (2020). Glycosyltransferase UGT76F1 is involved in the temperature-mediated petiole elongation and the BR-mediated hypocotyl growth in Arabidopsis. Plant Signal. Behav. 15:1777377. doi: 10.1080/15592324.2020.1777377
Chen, H., Zeng, Y., Yang, Y., Huang, L., Tang, B., Zhang, H., et al. (2020). Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 11:2494. doi: 10.1038/s41467-020-16338-x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, Y., Zhang, J., Shao, Y., Xu, Y., Ge, H., Yu, B., et al. (2019). Enzyme-catalyzed glycosylation of curcumin and its analogues by glycosyltransferases from Bacillus subtilis ATCC 6633. Catalysts 9:734.
Dai, X., Zhuang, J., Wu, Y., Wang, P., Zhao, G., Liu, Y., et al. (2017). Identification of a Flavonoid Glucosyltransferase Involved in 7-OH Site Glycosylation in Tea plants (Camellia sinensis). Sci. Rep. 7:5926. doi: 10.1038/s41598-017-06453-z
Dong, N. Q., Sun, Y., Guo, T., Shi, C. L., Zhang, Y. M., Kan, Y., et al. (2020). UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 11:2629. doi: 10.1038/s41467-020-16403-5
Dooner, H. K., and Nelson, O. E. (1977). Controlling element-induced alterations in UDPglucose: flavonoid glucosyltransferase, the enzyme specified by the bronze locus in maize. Proc. Natl. Acad. Sci. U.S.A. 74, 5623–5627. doi: 10.1073/pnas.74.12.5623
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: The protein families database. Nucleic Acids Res. 42, D222–D230.
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
He, Y., Ahmad, D., Zhang, X., Zhang, Y., Wu, L., Jiang, P., et al. (2018). Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 18:67. doi: 10.1186/s12870-018-1286-5
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 35, W585–W587.
Huang, X. X., Zhu, G. Q., Liu, Q., Chen, L., Li, Y. J., and Hou, B. K. (2018). Modulation of Plant Salicylic Acid-Associated Immune Responses via Glycosylation of Dihydroxybenzoic Acids. Plant Physiol. 176, 3103–3119. doi: 10.1104/pp.17.01530
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernandez-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Huo, K., Chen, Y., Sui, L., Wang, Y., Fu, Y., Pei, X., et al. (2021). Transient expression and enzymatic assay identified uridine-diphosphate glucosyltransferases related to flavonoid glycosylation in Vernonia amygdalina leaves. Ind. Crops Prod. 172:114005.
Jing, T., Zhang, N., Gao, T., Wu, Y., Zhao, M., Jin, J., et al. (2020). UGT85A53 promotes flowering via mediating abscisic acid glucosylation and FLC transcription in Camellia sinensis. J. Exp. Bot. 71, 7018–7029. doi: 10.1093/jxb/eraa373
Kim, M. J., Zheng, J., Liao, M. H., and Jang, I. C. (2019). Overexpression of SrUGT76G1 in Stevia alters major steviol glycosides composition towards improved quality. Plant Biotechnol. J. 17, 1037–1047. doi: 10.1111/pbi.13035
Kurze, E., Wust, M., Liao, J., Mcgraphery, K., Hoffmann, T., Song, C., et al. (2022). Structure-function relationship of terpenoid glycosyltransferases from plants. Nat. Prod. Rep. 39, 389–409. doi: 10.1039/d1np00038a
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van, P. Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, H., Yang, X., Lu, M., Chen, J., and Shi, T. (2020). Gene expression and evolution of Family-1 UDP-glycosyltransferases—insights from an aquatic flowering plant (sacred lotus). Aquat. Bot. 166:103270.
Li, Y., Liu, F., Li, P., Wang, T., Zheng, C., and Hou, B. (2020). An Arabidopsis Cytokinin-modifying glycosyltransferase UGT76C2 improves drought and salt tolerance in rice. Front Plant Sci 11:560696. doi: 10.3389/fpls.2020.560696
Li, P., Li, Y. J., Zhang, F. J., Zhang, G. Z., Jiang, X. Y., Yu, H. M., et al. (2017). The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 89, 85–103. doi: 10.1111/tpj.13324
Li, W., Zhai, L., Strauss, S. H., Yer, H., Merewitz, E., Chen, J., et al. (2019). transgenic reduction of cytokinin levels in roots inhibits root-sprouting in Populus. Plant Physiol. 180, 1788–1792.
Li, Y. J., Li, P., Wang, T., Zhang, F. J., Huang, X. X., and Hou, B. K. (2018). The maize secondary metabolism glycosyltransferase UFGT2 modifies flavonols and contributes to plant acclimation to abiotic stresses. Ann. Bot. 122, 1203–1217. doi: 10.1093/aob/mcy123
Lin, J. S., Huang, X. X., Li, Q., Cao, Y., Bao, Y., Meng, X. F., et al. (2016). UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J. 88, 26–42. doi: 10.1111/tpj.13229
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408.
Long, R., Zhang, F., Zhang, Z., Li, M., Chen, L., Wang, X., et al. (2022). Genome assembly of alfalfa cultivar zhongmu-4 and identification of SNPs associated with agronomic traits. Genomics Proteomics Bioinformatics 13, S1672–S1229. doi: 10.1016/j.gpb.2022.01.002
Ma, D., Liu, B., Ge, L., Weng, Y., Cao, X., Liu, F., et al. (2021). Identification and characterization of regulatory pathways involved in early flowering in the new leaves of alfalfa (Medicago sativa L.) by transcriptome analysis. BMC Plant Biol. 21:8. doi: 10.1186/s12870-020-02775-9
Ma, X., Xu, Q., Meyer, W. A., and Huang, B. (2016). Hormone regulation of rhizome development in tall fescue (Festuca arundinacea) associated with proteomic changes controlling respiratory and amino acid metabolism. Ann. Bot. 118, 481–494. doi: 10.1093/aob/mcw120
Mohnike, L., Rekhter, D., Huang, W., Feussner, K., Tian, H., Herrfurth, C., et al. (2021). The glycosyltransferase UGT76B1 modulates N-hydroxy-pipecolic acid homeostasis and plant immunity. Plant Cell 33, 735–749.
Nair, P. C., Chau, N., Mckinnon, R. A., and Miners, J. O. (2020). Arginine-259 of UGT2B7 Confers UDP-Sugar Selectivity. Mol. Pharmacol. 98, 710–718. doi: 10.1124/molpharm.120.000104
Radojicic, A., Li, X., and Zhang, Y. (2018). Salicylic Acid: A double-edged sword for programed cell death in plants. Front. Plant Sci. 9:1133. doi: 10.3389/fpls.2018.01133
Rehman, H. M., Nawaz, M. A., Shah, Z. H., Ludwig-Muller, J., Chung, G., Ahmad, M. Q., et al. (2018). Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 8:1875. doi: 10.1038/s41598-018-19535-3
Rojas Rodas, F., Rodriguez, T. O., Murai, Y., Iwashina, T., Sugawara, S., Suzuki, M., et al. (2014). Linkage mapping, molecular cloning and functional analysis of soybean gene Fg2 encoding flavonol 3-O-glucoside (1 –> 6) rhamnosyltransferase. Plant Mol. Biol. 84, 287–300. doi: 10.1007/s11103-013-0133-1
Shen, C., Du, H., Chen, Z., Lu, H., Zhu, F., Chen, H., et al. (2020). The chromosome-level genome sequence of the autotetraploid alfalfa and resequencing of core germplasms provide genomic resources for alfalfa research. Mol. Plant 13, 1250–1261. doi: 10.1016/j.molp.2020.07.003
Shu, K., Zhou, W., Chen, F., Luo, X., and Yang, W. (2018). Abscisic acid and gibberellins antagonistically mediate plant development and abiotic stress responses. Front. Plant Sci. 9:416. doi: 10.3389/fpls.2018.00416
Song, C., Hartl, K., Mcgraphery, K., Hoffmann, T., and Schwab, W. (2018). Attractive but Toxic: Emerging roles of glycosidically bound volatiles and glycosyltransferases involved in their formation. Mol. Plant 11, 1225–1236. doi: 10.1016/j.molp.2018.09.001
Song, Z., Niu, L., Yang, Q., Dong, B., Wang, L., Dong, M., et al. (2019). Genome-wide identification and characterization of UGT family in pigeonpea (Cajanus cajan) and expression analysis in abiotic stress. Trees 33, 987–1002.
Speeckaert, N., Adamou, N. M., Hassane, H. A., Baldacci-Cresp, F., Mol, A., Goeminne, G., et al. (2020). Characterization of the UDP-glycosyltransferase UGT72 Family in poplar and identification of genes involved in the glycosylation of monolignols. Int. J. Mol. Sci. 21:5018. doi: 10.3390/ijms21145018
Speeckaert, N., El Jaziri, M., Baucher, M., and Behr, M. (2022). UGT72, a Major glycosyltransferase family for flavonoid and monolignol homeostasis in plants. Biology (Basel) 11:441. doi: 10.3390/biology11030441
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis Version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tian, H., Lv, B., Ding, T., Bai, M., and Ding, Z. (2017). Auxin-BR interaction regulates plant growth and development. Front. Plant Sci. 8:2256. doi: 10.3389/fpls.2017.02256
Wang, D., Wang, J., Shi, Y., Li, R., Fan, F., Huang, Y., et al. (2020). Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab. Eng. 61, 131–140. doi: 10.1016/j.ymben.2020.05.007
Wang, F., Su, Y., Chen, N., and Shen, S. (2021). Genome-wide analysis of the UGT gene family and identification of flavonoids in Broussonetia papyrifera. Molecules 26:3449. doi: 10.3390/molecules26113449
Wang, X., Wang, J., Cui, H., Yang, W., Yu, B., Zhang, C., et al. (2022). The UDP-glycosyltransferase MtUGT84A1 regulates anthocyanin accumulation and plant growth via JA signaling in Medicago truncatula. Environ. Exp. Bot. 201:104972.
Wu, B., Liu, X., Xu, K., and Zhang, B. (2020). Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (Citrus grandis) fruit. BMC Plant Biol. 20:459. doi: 10.1186/s12870-020-02655-2
Wu, C., Dai, J., Chen, Z., Tie, W., Yan, Y., Yang, H., et al. (2021). Comprehensive analysis and expression profiles of cassava UDP-glycosyltransferases (UGT) family reveal their involvement in development and stress responses in cassava. Genomics 113, 3415–3429. doi: 10.1016/j.ygeno.2021.08.004
Xiao, X., Lu, Q., Liu, R., Gong, J., Gong, W., Liu, A., et al. (2019). Genome-wide characterization of the UDP-glycosyltransferase gene family in upland cotton. 3 Biotech 9:453. doi: 10.1007/s13205-019-1984-1
Yano, R., Takagi, K., Tochigi, S., Fujisawa, Y., Nomura, Y., Tsuchinaga, H., et al. (2018). Isolation and characterization of the Soybean Sg-3 gene that is involved in genetic variation in sugar chain composition at the C-3 position in soyasaponins. Plant Cell Physiol. 59, 792–805. doi: 10.1093/pcp/pcy019
Yin, Q., Shen, G., Chang, Z., Tang, Y., Gao, H., and Pang, Y. (2017a). Involvement of three putative glucosyltransferases from the UGT72 family in flavonol glucoside/rhamnoside biosynthesis in Lotus japonicus seeds. J. Exp. Bot. 68, 597–612.
Yin, Q., Shen, G., Di, S., Fan, C., Chang, Z., and Pang, Y. (2017b). Genome-wide identification and functional characterization of UDP-glucosyltransferase genes involved in flavonoid biosynthesis in Glycine max. Plant Cell Physiol. 58, 1558–1572.
Yu, A. D., Liu, L., Long, R. C., Kang, J. M., Chen, C., Yang, Q. Q., et al. (2022). Function and application prospect of plant UDP-glycosyltransferase(UGT). Plant Physiol. J. 58, 631–642.
Zhang, G. Z., Jin, S. H., Li, P., Jiang, X. Y., Li, Y. J., and Hou, B. K. (2017). Ectopic expression of UGT84A2 delayed flowering by indole-3-butyric acid-mediated transcriptional repression of ARF6 and ARF8 genes in Arabidopsis. Plant Cell Rep. 36, 1995–2006. doi: 10.1007/s00299-017-2225-x
Zhang, H., Liu, X., Wang, X., Sun, M., Song, R., Mao, P., et al. (2021). Genome-wide identification of GRAS gene family and Their Responses to Abiotic Stress in Medicago sativa. Int. J. Mol. Sci. 22:7729. doi: 10.3390/ijms22147729
Zhang, K., Sun, Y., Li, M., and Long, R. (2021). CrUGT87A1, a UDP-sugar glycosyltransferases (UGTs) gene from Carex rigescens, increases salt tolerance by accumulating flavonoids for antioxidation in Arabidopsis thaliana. Plant Physiol. Biochem. 159, 28–36. doi: 10.1016/j.plaphy.2020.12.006
Zhang, P., Zhang, Z., Zhang, L., Wang, J., and Wu, C. (2020). Glycosyltransferase GT1 family: Phylogenetic distribution, substrates coverage, and representative structural features. Comput. Struct. Biotechnol. J. 18, 1383–1390. doi: 10.1016/j.csbj.2020.06.003
Zhang, Y., Guo, W., Chen, L., Shen, X., Yang, H., Fang, Y., et al. (2022). CRISPR/Cas9-Mediated targeted mutagenesis of GmUGT enhanced soybean resistance against leaf-chewing insects through flavonoids biosynthesis. Front. Plant Sci. 13:802716. doi: 10.3389/fpls.2022.802716
Zhang, Z., Zhuo, X., Yan, X., and Zhang, Q. (2018). Comparative genomic and transcriptomic analyses of family-1 UDP glycosyltransferase in Prunus Mume. Int. J. Mol. Sci. 19:3382. doi: 10.3390/ijms19113382
Keywords: Medicago sativa, UDP glycosyltransferase, secondary metabolism, flavonoids, terpenoids, phylogenetic analysis
Citation: Yu A, Jiang X, Sun Y, Hu Q, Zhu X, Kang J, Chen L, Liu L, Hao L, Yang Q, Long R and Li M (2022) Genome-wide identification, characterization, and expression analysis of UDP-glycosyltransferase genes associated with secondary metabolism in alfalfa (Medicago sativa L.). Front. Plant Sci. 13:1001206. doi: 10.3389/fpls.2022.1001206
Received: 23 July 2022; Accepted: 23 August 2022;
Published: 30 September 2022.
Edited by:
Jing Zhang, Nanjing Agricultural University, ChinaReviewed by:
Ke Teng, Beijing Academy of Agricultural and Forestry Sciences, ChinaCopyright © 2022 Yu, Jiang, Sun, Hu, Zhu, Kang, Chen, Liu, Hao, Yang, Long and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruicai Long, bG9uZ3J1aWNhaUBjYWFzLmNu; Mingna Li, bGltaW5nbmFAY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.