
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 February 2022
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.831140
This article is part of the Research Topic Mechanisms of Abiotic Stress Responses and Tolerance in Plants: Physiological, Biochemical and Molecular Interventions, volume II View all 37 articles
A correction has been applied to this article in:
Corrigendum: Genome-Wide Identification and Expression Profiling of Germin-Like Proteins Reveal Their Role in Regulating Abiotic Stress Response in Potato
 Madiha Zaynab1†
Madiha Zaynab1† Jiaofeng Peng2†
Jiaofeng Peng2† Yasir Sharif3
Yasir Sharif3 Mahpara Fatima4
Mahpara Fatima4 Mohammed Albaqami5
Mohammed Albaqami5 Rashid Al-Yahyai6
Rashid Al-Yahyai6 Khalid Ali Khan7,8,9
Khalid Ali Khan7,8,9 Saqer S. Alotaibi10
Saqer S. Alotaibi10 Ibrahim A. Alaraidh11
Ibrahim A. Alaraidh11 Hassan O. Shaikhaldein11
Hassan O. Shaikhaldein11 Shuangfei Li1*
Shuangfei Li1*Germin and germin-like proteins (GLPs) perform a significant role in plants against biotic and abiotic stress. To understand the role of GLPs in potato, a comprehensive genome-wide analysis was performed in the potato genome. This study identified a total of 70 StGLPs genes in the potato genome, distributed among 11 chromosomes. Phylogenetic analysis exhibited that StGLPs were categorized into six groups with high bootstrap values. StGLPs gene structure and motifs analysis showed a relatively well-maintained intron–exon and motif formation within the cognate group. Additionally, several cis-elements in the promoter regions of GLPs were hormones, and stress-responsive and different families of miRNAs target StGLPs. Gene duplication under selection pressure also exhibited positive and purifying selections in StGLPs. In our results, the StGLP5 gene showed the highest expression in response to salt stress among all expressed StGLPs. Totally 19 StGLPs genes were expressed in response to heat stress. Moreover, three genes, StGLP30, StGLP17, and StGLP14, exhibited a relatively higher expression level in the potato after heat treatment. In total, 22 genes expressed in response to abscisic acid (ABA) treatment indicated that ABA performed an essential role in the plant defense or tolerance mechanism to environmental stress. RNA-Seq data validated by RT-qPCR also confirm that the StGLP5 gene showed maximum expression among selected genes under salt stress. Concisely, our results provide a platform for further functional exploration of the StGLPs against salt and heat stress conditions.
Germin-like proteins (GLPs) are pervasive proteins regarded as water-soluble glycoproteins present in either cell apoplast or its extracellular matrix (Lane et al., 1992; Dunwell et al., 2008). These proteins were first identified in wheat germinating embryos (Lu et al., 2010). At present, several proteins have been identified in different plant groups that exhibit 30–70% sequence homogeneity with wheat germin proteins (Finn et al., 2007).
Germin-like genes are expressed in embryos, leaves, stems, roots, flowers, and seeds of plants responding to various environmental stimuli based upon the species or genes under consideration (Wang et al., 2013). Abiotic and biotic stresses significantly affect plant growth and production worldwide (Raza et al., 2020; Qasim et al., 2021, 2018a; Raza, 2021). Various evidence has been given as an indication of GLPs involvement in defense and tolerance mechanisms of plants (Breen and Bellgard, 2010). Incidents like pathogen infections, insect infestation, or exposure to various chemicals such as salicylic acid, hydrogen peroxide, or ethylene may trigger the expression level of GLPs (Godfrey et al., 2007). Many internal elements have influenced the expression of germin, as in wheat, where the germin gene gf-2.8 is induced by hormone auxin (Schweizer et al., 1999). Transient overexpression and silencing of some Hordeum vulgare GLP genes have imparted enhanced resistance against fungal infection of powdery mildew (Schweizer et al., 1999). The promoter alternative of rice oxalate oxidase genes envisaged in the resistance against Magnaporthe oryzae. For some subfamilies, transitory and stable expression depicted the superoxide dismutase activity of the encoded protein (EC1.15.1.1) (Zimmermann et al., 2006). The silencing of GLPs in Nicotiana attenuate has proved fruitful in enhancing performance against herbivores (Lou and Baldwin, 2006). Fluctuation in the transcription of germin genes in Brassica napus and nearly associated Arabidopsis is found during the circadian cycle (Heintzen et al., 1994). These GLPs are associated with the protective mechanisms of cereals against the infection of fungal pathogens (Lane, 2000). Studies have revealed that the transfer of GLPs from soybean and sunflower to Nicotiana tabacum can increase its resistance against Sclerotinia sclerotiorum (Beracochea et al., 2015; Zhang et al., 2018). Transient-enhanced expression of TaGLP4 and HvGLP4 makes wheat and barley plants more resistant to Blumeria graminis. At the same time, the ephemeral gene that is silenced by utilizing RNA interference minimizes the basal resistance in cereals (Christensen et al., 2004, p. 4). A study is conducted to explore that the regulatory factors in various GLP gene promoters and several regulatory factors are associated with many essential roles with variable copy numbers and dispersed in locations of various positive or negative strands. Cotton GLP as GhABP19 is involved in various essential tasks in the exhibition of resistance to Verticillium and Fusarium wilt in crops (Pei et al., 2019, p. 19). A few GLP subfamily members are intricated in a vast range of fungal disease resistance in the tissues of leaves (Davidson et al., 2009).
Germin-like protein genes have variable expression and multiple tasks in several tissues and at different phases of plant growth processes (Barman and Banerjee, 2015). This gene family is one of those gene families that are crucial and indispensable for the defense and stress tolerance mechanisms of plants. Expression of these genes varies against various abiotic stress such as salt (Wang et al., 2013), heavy metal (Berna and Bernier, 1999), and drought stress. Moreover, some purposes of GLP genes have been characterized in Hordeum vulgare (Zimmermann et al., 2006), Arabidopsis thaliana (Rietz et al., 2012), Oryza sativa (Li et al., 2016), and Glycine max (Wang et al., 2014). The expression levels of many rice GLP genes were altered under salt stress (Das et al., 2019). OsGLP8-14 was found to be involved in salt stress response (Banerjee et al., 2017). Liao et al. (2021) reported the differential expression of CsGLP genes in response to salt, drought, and ABA treatments (Liao et al., 2021).
Potato (Solanum tuberosum L.) is an economically essential field crop and is widely grown as a basic food commodity worldwide. About 368,168,914 tons of potatoes were harvested in 2018 on 17,578,672 ha of land, and over 1 billion people consumed them (Rashid et al., 2021). Like other plant species, potato yield is also in jeopardy due to many biotic and abiotic environmental fluctuations (Zaynab et al., 2021d). To date, the expression analysis of GLP genes against abiotic stress has not been reported in potatoes. Therefore, this study used genome-wide analysis to find GLP genes in the potato genome. Furthermore, their evolutionary relationships, synteny analysis, gene structures, conserved motifs, cis-elements, and miRNA predictions were investigated. Expression profiling in many tissues/organs and response to various hormones and abiotic stresses have been substantially explored, enabling us to explore and understand these GLPs in extensive detail in the potato genome.
In this study, we used two methods to identify GLP genes from the genome of the Solanum tuberosum. The first method was the Hidden Markov Model (HMM), and the second one was the BLASTP search (Raza et al., 2021; Zaynab et al., 2021d). Potato genome sequences were retrieved from JGI Phytozome 12.01 database (Goodstein et al., 2012). GLP amino acids sequences of A. thaliana were taken from TAIR2 (Rhee, 2003) database and used as a query to run BLASTP search. The HMM file with Pfam ID PF00190 containing GLP genes was downloaded from Pfam3 (El-Gebali et al., 2019) database. Moreover, a local HMMER 3.14 webserver was utilized to identify the GLP genes with default parameters (Finn et al., 2015). Eventually, a total of 70 GLP genes were identified from the genome of the potato. Non-redundant genes with conserved domains were serially named through StGLP1–StGLP70 and used for further analysis.
Physiochemical characteristics, including molecular weight and isoelectric points, were identified through ProtParam5 (Gasteiger et al., 2005).
In order to investigate the evolutionary history of the StGLP family genes, a phylogenetic tree was constructed among the amino acid sequences of Solanum lycopersicum, S. tuberosum, and A. thaliana. We used MEGA 76 (Kumar et al., 2018) for the construction of phylogenetic tree. The neighbor-joining method was applied to make a phylogenetic tree with 1,000 bootstrap values. TBtools software7 was deployed to construct the gene structure of StGLPs (Chen et al., 2020). Conserved motifs present in StGLPs sequences were recognized through MEME8 (Bailey et al., 2009) server. For comparative synteny analysis, Circoletto Tool9 was used.
Data related to the chromosomal location was taken from general feature format (GFF) files of S. tuberosum genome database (PGSC Ref seq v4.03) (The Potato Genome Sequencing Consortium, 2011). The localization of 70 identified genes on chromosomes was performed by utilizing TBtools (Chen et al., 2020). To explore the putative cis-elements in the promoter region of StGLPs, we extracted a sequence of 2 Kb upstream of start codons in the genome of S. tuberosum. Afterward, to analyze the promoter region of each gene, we deployed them to PlantCARE10 (Lescot, 2002) and figures made by TBtools (Chen et al., 2020). Besides, the synonymous and non-synonymous substitution rate was computed by employing KaKs Calculator 2.0 software using the MYN procedure. Ks and Ka refer to the number of synonymous and non-synonymous substitutions concerning the synonymous non-synonymous site (Wang et al., 2010). Divergence time (t = Ks/2r) was estimated through exchange rate as (r = 2.6 × 10–9) (Li et al., 2019).
For identifying miRNA targeting StGLPs, psRNAtarget11 was used with default parameters (Dai et al., 2018). For illustrating the protein--protein interaction network, the STRING 11.012 server was employed, and for reference, the genome of S. tuberosum was used. The exhibit specifications were propped as the confidence value is 0.9 for network; and 10 for edges evidence and maximum number of interaction.
In order to investigate the expression of StGLP genes in various tissues and under various stress conditions, the transcriptome data were obtained from the available public database BioProject ID: Project: PRJEB2430. For the expression analysis of StGLP genes in different tissues and under different stresses, fragments per kilobase million (FPKM) were observed (Zaynab et al., 2021d). The obtained data were arranged according to their expression in different tissues. The observed FPKM values were utilized to construct a heatmap through TBtools after the normalization of log2 values.
The rooster variety of S. tuberosum was selected as the sample for these experiments, which were conducted at the experimental station of the National Institute for Genomics and Advanced Biotechnology (NIGAB), Islamabad, Pakistan. After 6 weeks of growth, the sample was treated with 150 mM NaCl for 0 and 24 h, respectively, for salt treatment. The first time point (0 h) served as a control. After NaCl treatment, leaves were taken, and samples were immediately frozen in liquid nitrogen and stored at −80°C for total RNA extraction.
Total RNA was extracted using TransZol Up Plus RNA Kit (TransGen Biotech, Beijing, China). For cDNA synthesis, cDNA Synthesis SuperMix was purchased from the above-mentioned Chinese company. Instructions given by manufacturers were followed during the whole process. The comprehensive knowledge about reactions of RT-qPCR has been reported in our earlier published work (Zaynab et al., 2021a). In our study, the expression data were evaluated by using the 2–ΔΔCT methods (Zaynab et al., 2021c), and the elongation factor 1-alpha was chosen as a housekeeping gene (Zaynab et al., 2021b). The primers used in this study are given in Supplementary Table 1.
A total of 70 GLP genes were identified in the potato genome by employing sequences of A. thaliana as a query (Table 1). In this study, Batch-CD and SMART were utilized to confirm the domain with Pfam ID (PF00190). All these 70 genes contain a domain with Pfam ID (PF00190). The protein sequences of A. thaliana, S. lycopersicum, and identified StGLPs are given in Supplementary Table 2. Amino acids length varied from 94 to 567, the molecular weight from 10.650 to 65.780 kDa, and the isoelectric point values from 4.46 to 9.74 (Table 1). A phylogenetic tree was constructed using the neighbor-joining method through MEGA7 to illustrate the phylogenetic relationship among A. thaliana, S. lycoperiscum, and S. tuberosum. This study results showed that the phylogenetic tree was divided into six groups. Group-6 is the largest group, and group-3 is the smallest among all groups (Figure 1). Exons and introns are two main determinants in the gene family evolution. The detailed analysis of the phylogenetic relationship and gene composition illustration corroborated our understanding of the structures of StGLPs genes (Figure 2). The results revealed that numerical attributes of exon and intron in StGLP genes possessed high inconsistency in numbers of exons and introns (Figure 2). In this study, the StGLP46 gene possessed the most extended sequence. Some genes such as StGLP41 and StGLP38 exhibited similarities in exon numbers. Additionally, StGLP21 and StGLP57 genes were equal in exons numbers but differed in their sequence lengths. For determining structural diversification and functional characterization of these StGLPs, we analyzed full-length sequences of 70 StGLPs through MEME software to localize their conserved motifs. The motif results revealed that a total of ten motifs were identified (Figure 2). The sequences of predicted motifs are given in Supplementary Table 3. All of the StGLPs genes contained motif 1 except genes StGLP StGLP18, StGLP26, StGLP29, StGLP31, StGLP51, StGLP52, StGLP63, StGLP65, StGLP21, StGLP6, StGLP20, StGLP12, StGLP22, StGLP8, and StGLP27. The length of the motif was varied from 21 to 50 amino acids. The highest 50 amino acids were possessed by motif 3,7, and 10, while motif 2 and 5 contained the lowest 21 amino acids. In our results, motif 4 had 39 amino acids, motif 8 had 35 amino acids, while motifs 1 and 9 had 41 amino acids in their sequence. In short, the reliability of group organization was exceedingly encouraged by analyzing conserved motif conformation, genetic structures, and phylogenetic relationship, indicating that StGLP proteins possess enormously well-sustained amino acid members and remain inside a group. Thus, it can be inferred that proteins with identical motifs and structures may possess functional similarities.
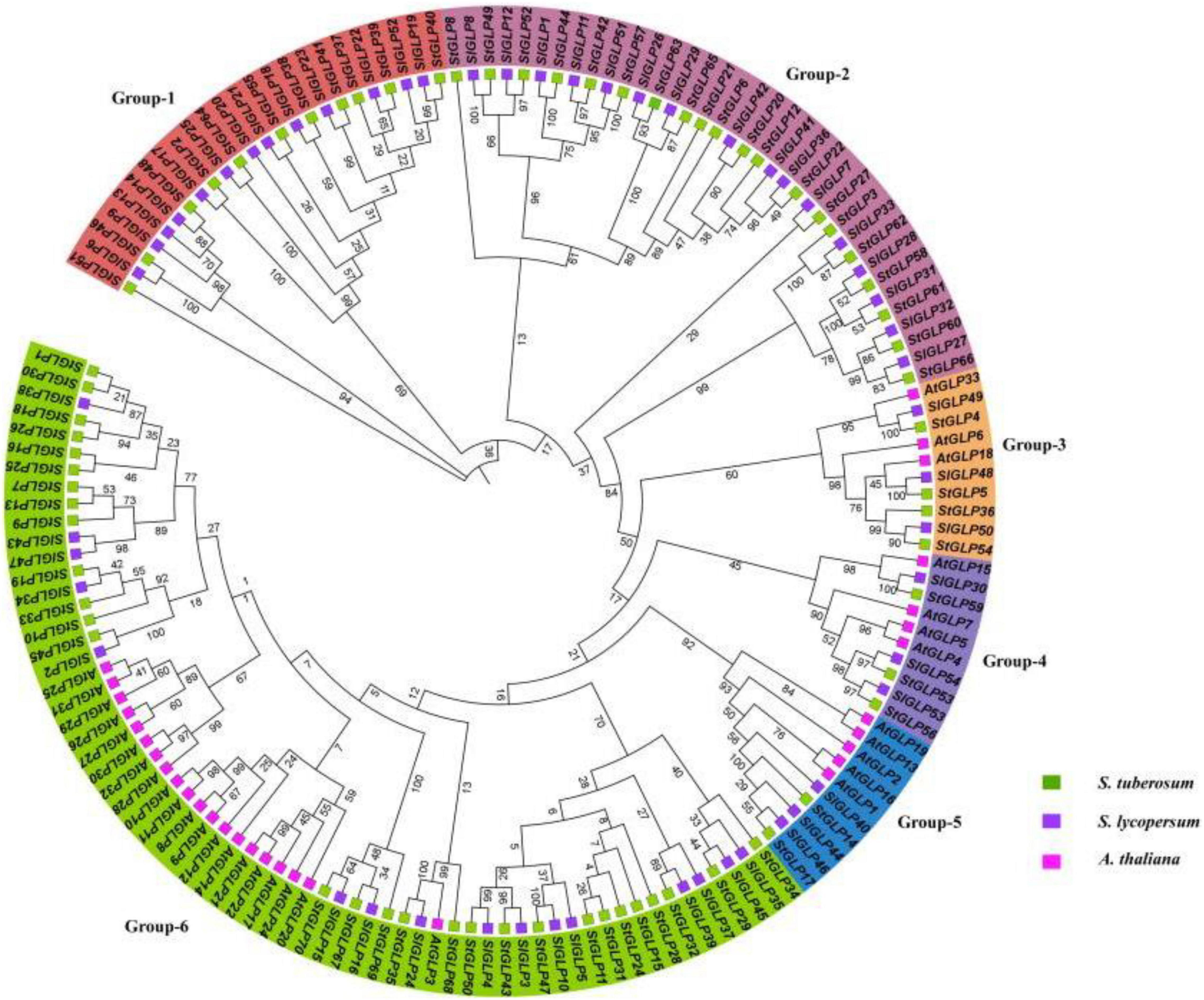
Figure 1. Phylogenetic tree of GLP proteins from Arabidopsis, S. lycoperiscum, and S. tuberosum. The colored arcs indicate different groups. The stars colors represent proteins of Arabidopsis, S. lycoperiscum, and S. tuberosum.
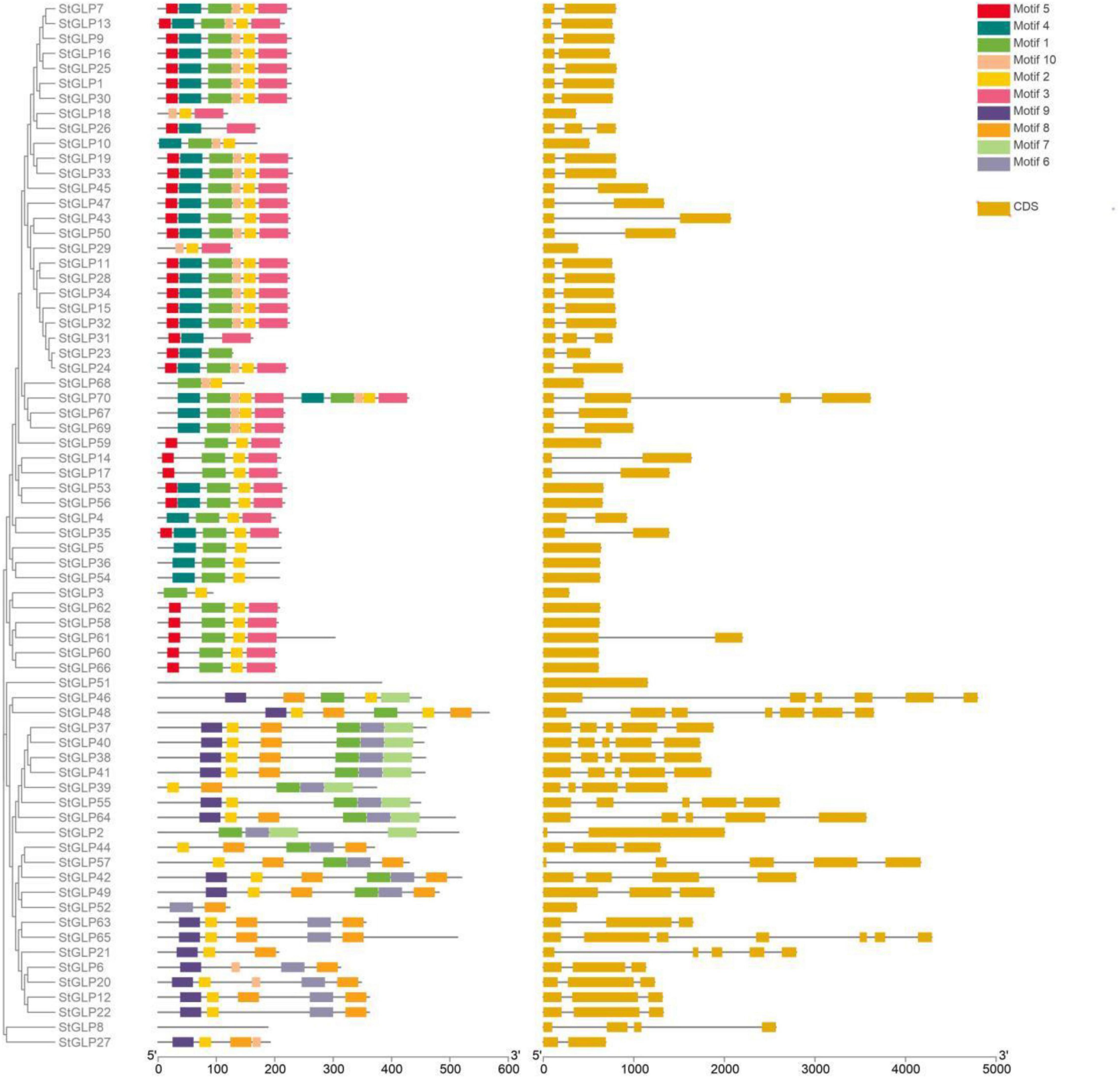
Figure 2. Distribution of exons, introns, and untranslated regions (UTR) in StGLP gene sequences and distributions of de novo MEME motifs in the StGLP proteins.
Chromosomal distribution analysis of StGLPs depicted their unequal localization on all chromosomes (Figure 3). A maximum number of 29 genes were present on the 1st chromosome, while the 9th chromosome had 11 genes. There are 9 genes present on chromosome number 11. Only 2 genes are located on chromosome number 02 (Figure 3) in our results. Furthermore, there was no gene located on chromosome number 08. To determine the rate of molecular evolution of each gene pair that has been duplicated, the Ka to Ks (synonymous/non-synonymous substitution) ratios were calculated. When the value of the ratio is more than 1, then the selection effect will be positive; if its value is less than 1, then purifying selection will be exhibited, while when this ratio’s value is equal to 1, then there will be the presence of neutral selection in gene pairs (Yang and Bielawski, 2000). The cosmic majority of StGLPs genes exhibited a value of Ks more than 0.52, and the corresponding deviation time may be more than 100 million years ago (MYA). In this study, duplicated genes (StGLP4/StGLP35) showed the Ks value 3.54 while its corresponding time may be 682.30 MYA (Table 2). Comparative synteny analysis was analyzed among A. thaliana, S. tuberosum, and S. lycoperiscum which exhibited a phenomenal relationship in the evolution expression, duplication, triplication, and function of genes. Genes sequence of SlGLP17 exhibited synteny with the sequence of StGLP2. At the same time, two potato genes, StGLP20 and StGLP22, showed synteny with the SlGLP41 gene of tomato. The comparative synteny analysis revealed the conservation and duplication of inward and outward tangling events in ribbons, respectively (Figure 4).
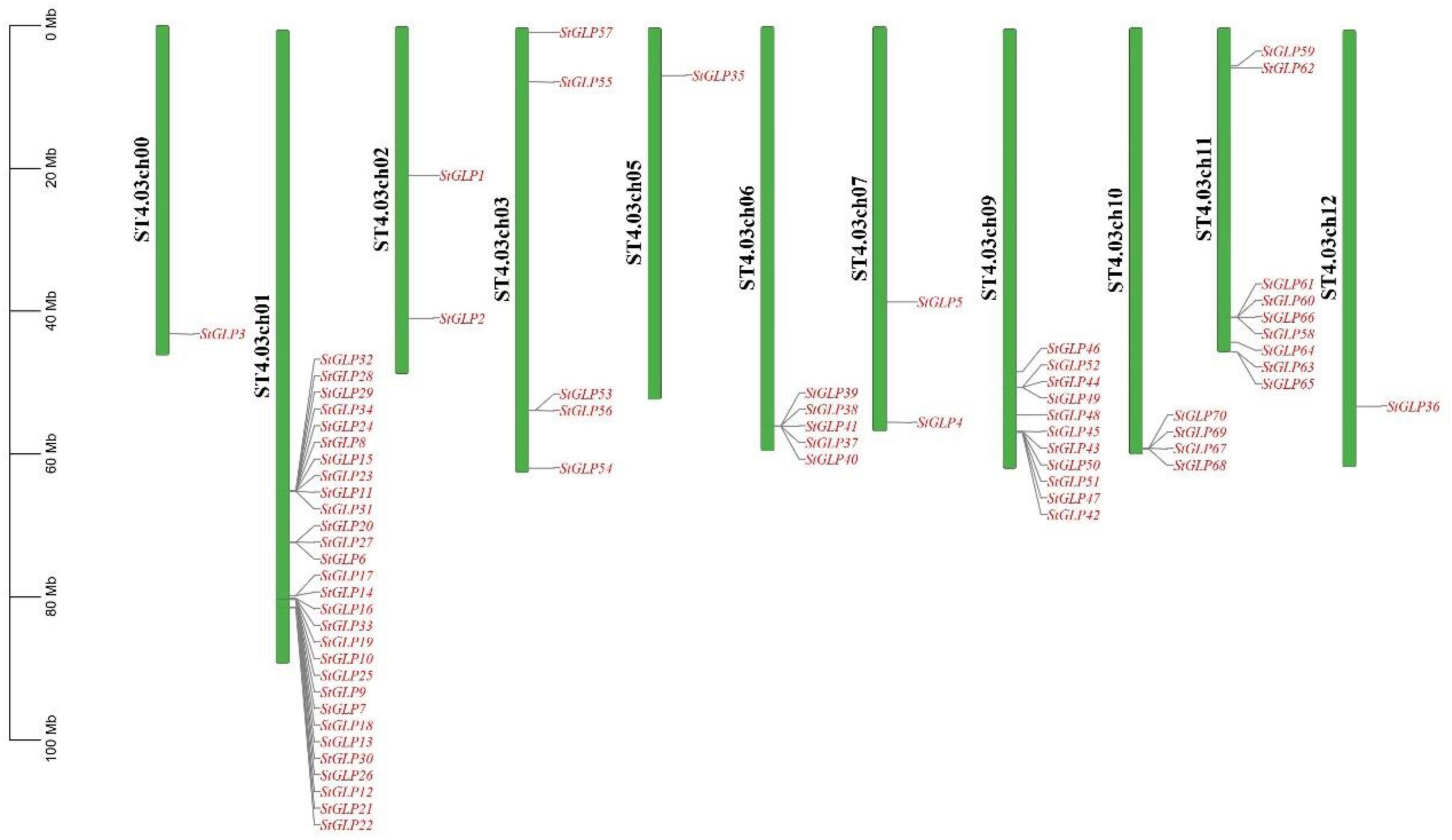
Figure 3. Chromosomal distribution of StGLP genes. Green color shows chromosomes, red color shows StGLP genes, and black color shows the number of chromosomes.
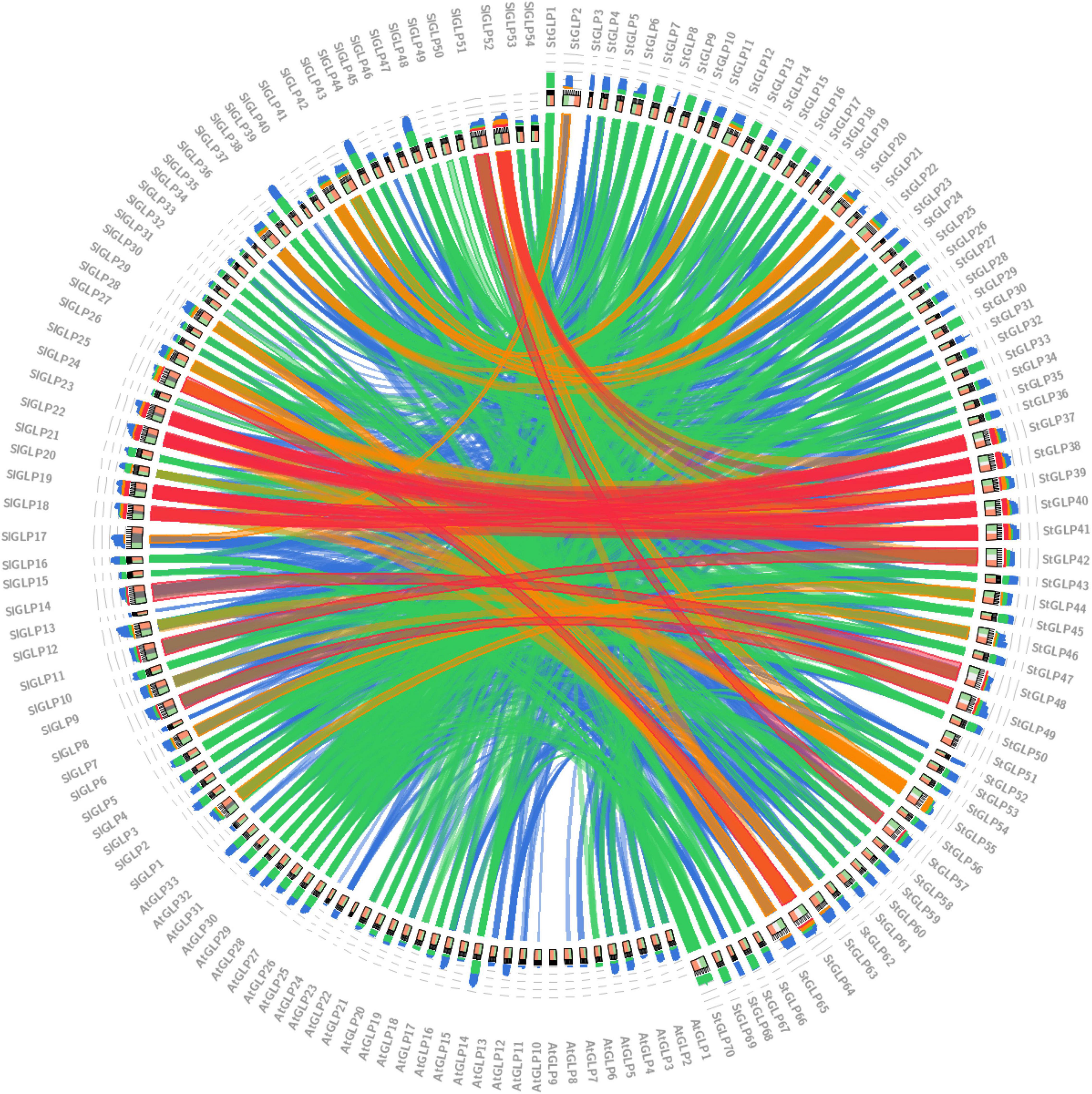
Figure 4. Synteny analysis of GLPs among potato, tomato, and Arabidopsis genomes. In comparative synteny analysis, the color and intensity of inward and outward tangling of ribbons showed conservation and duplication events, respectively.
To observe the StGLPs gene function and their regulatory role, promoter cis-elements were analyzed using the PlantCARE database. The cis-elements that are associated with plant hormones were identified. As shown in Figure 5, the number of genes associated with phytohormones indicates the importance of these genes in phytohormone stress. Furthermore, the role of anaerobic, light, and low-temperature responsive factors in this study was discovered (Figure 5). Mainly several elements that are responsive to light were categorized to be extensively distributed in all genes, indicating the significant role of StGLPs in response to light stress. Generally, findings envisaged that diversification in the expression levels of various StGLPs might be attributed to different plant hormones and environmental stress. The Hormones- and stress-related cis-elements found in the promoter regions of StGLPs are given in Supplementary Table 4.
Many researchers have reported that microRNA mediated a significant role in enhancing stress response in plants (Su et al., 2021; Xu et al., 2021). Thus, for an in-depth comprehension of miRNA-triggered post-transcriptional modulations of StGLPs, we identified miRNAs targeting StGLPs genes. Several sites that are targeted by miRNA are illustrated in Supplementary Table 1. Our findings revealed that seven members of the miR166 family target six genes, including StGLP8, StGLP54, StGLP23, StGLP24, StGLP38, and StGLP28. A gene, StGLP2, was targeted by a single member of the miR169 family. Targets StGLP51, StGLP48, and StGLP47 (Figure 6 and Supplementary Table 5) are all three members of the miRNA gene family. The findings of our study revealed that two members of the miR8040 aimed at two genes, StGLP51 and StGLP62. A gene StGLP55 was the target of two members of the miR1886 family. Our results mainly estimated two genes, StGLP40 and StGLP8, to target numerous miRNAs.
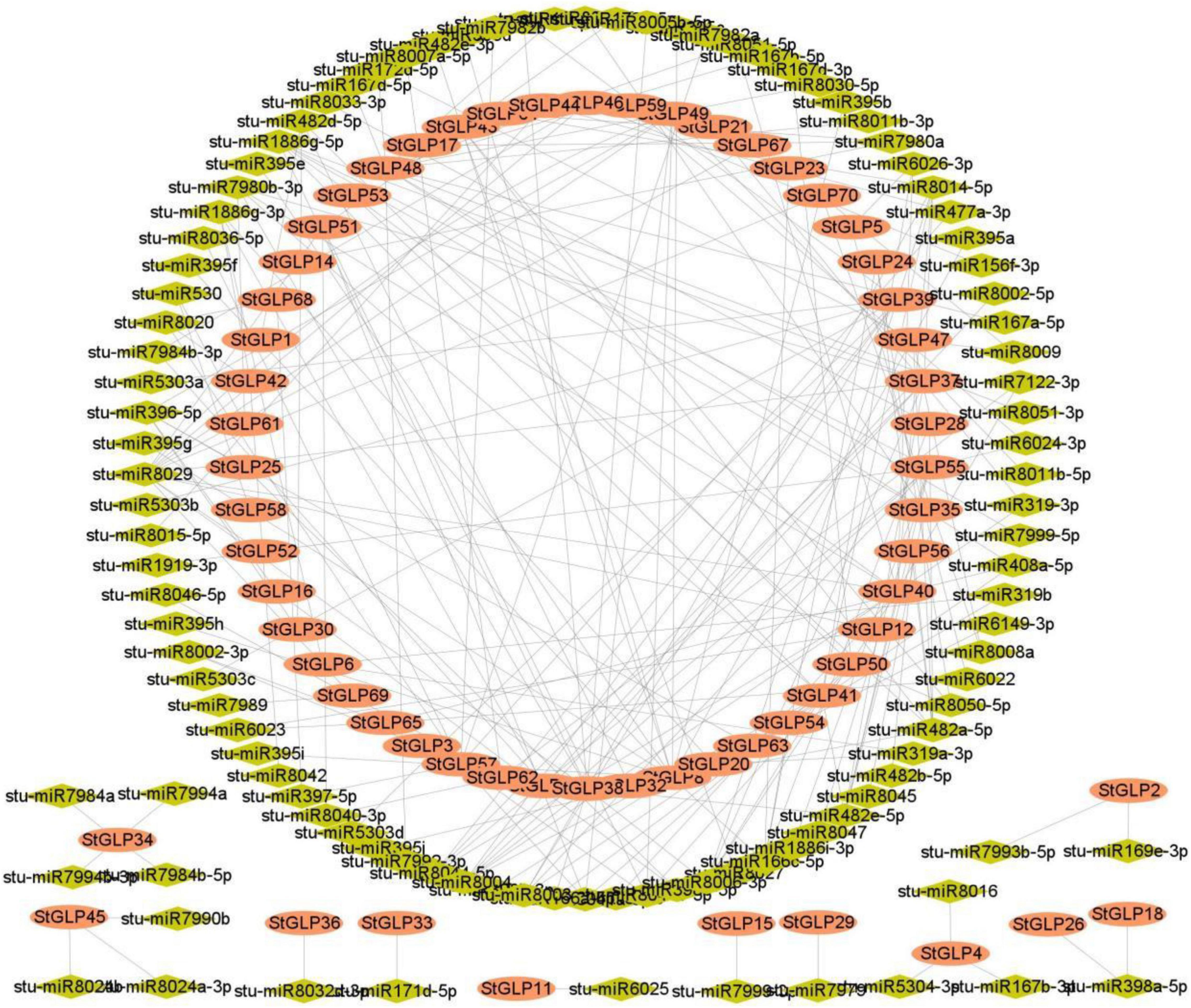
Figure 6. miRNAs targeting StGLP genes. Network figure of anticipated miRNAs targeting the StGLP genes. Green color represents miRNAs, while orange color corresponds to the target StGLP gene.
The protein–protein interaction network provided details about StGLPs in the genome of potatoes. The interaction network was observed among several proteins, including StGLP57, StGLP44, StGLP46, StGLP48, and StGLP2. These proteins depicted the concurrence, co-expression, fusion, and homology of genes. Likewise, another framework was also identified among StGLP53, StGLP63, and StGLP4 protein. Among all StGLPs, two proteins, StGLP48 and StGLP46, are responsible for framing a protein–protein interaction network, so these can be reputed as hub proteins of StGLPs (Figure 7).
Expression profiling of StGLPs revealed that many genes exhibited high expression in the tissues of roots than that in the tissues of stems and leaves. Some genes showed expression in some tissues, while some genes have no expression in others. For example, the StGLP17 gene showed a higher expression level in the tissues of roots, leaves, and stems, while the StGLP26 and StGLP45 genes showed higher expression levels in the tissues of roots and stems. Furthermore, in the tissues of leaves, StGLP5 genes showed the maximum expression in our results (Figure 8).
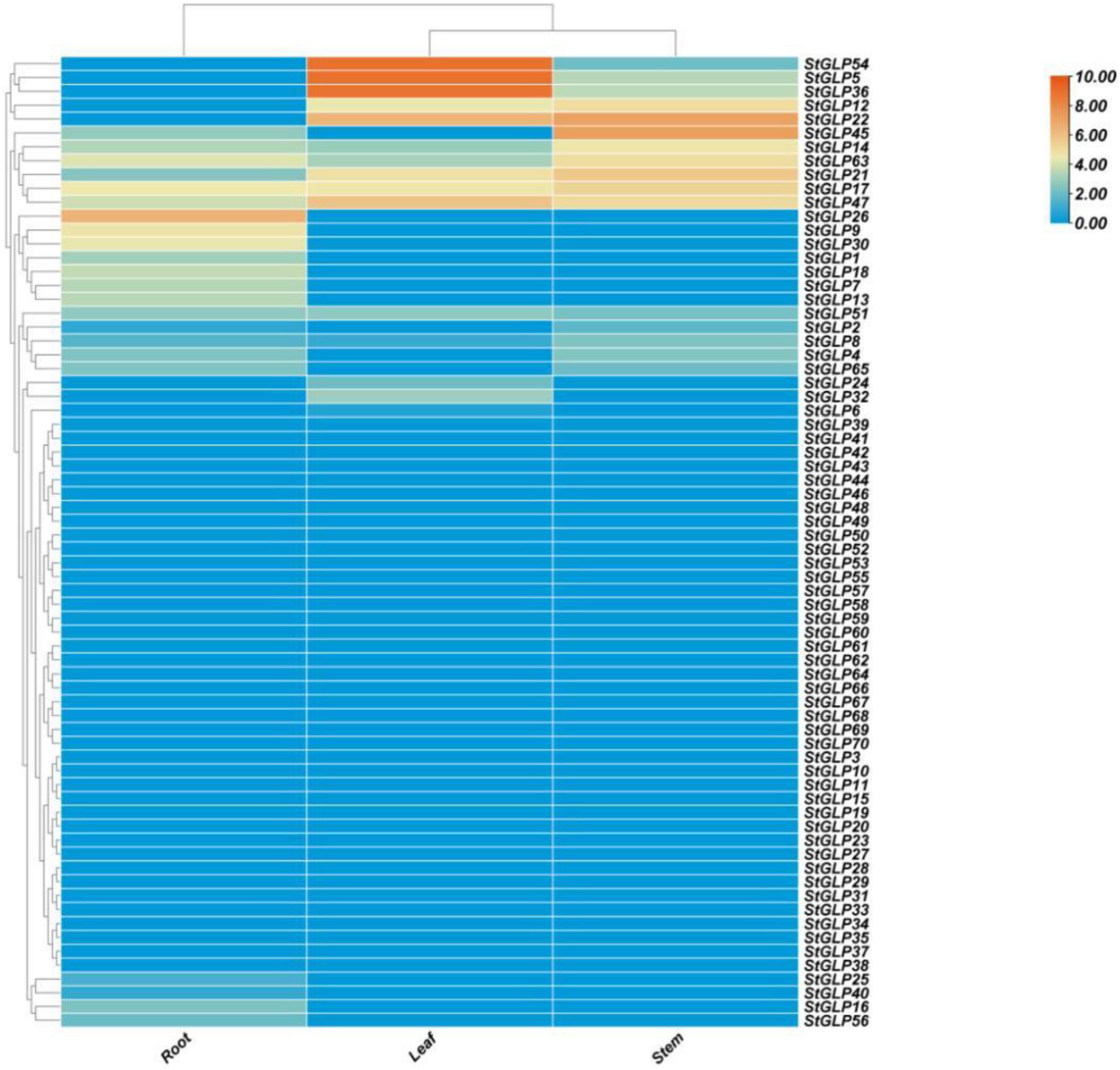
Figure 8. Expression profiling of StGLP genes in leaves, roots, and stems. The orange and blue colors display high- to low-expression levels. The expression heat map was created by taking log10 normalized values of fragments per kilobase million (FPKM).
Transcriptional profiling of GLPs was performed when potato plants were subjected to heat stress. StGLPs may help potato plants cope better with high temperatures, according to some research. A total of 19 StGLPs genes were expressed in response to heat stress. Moreover, three genes, StGLP30, StGLP17, and StGLP14, exhibited a relatively higher expression level in potatoes after heat treatment (Figure 9). In our results, StGLP30 gene expression was higher than StGLP17 and StGLP14.
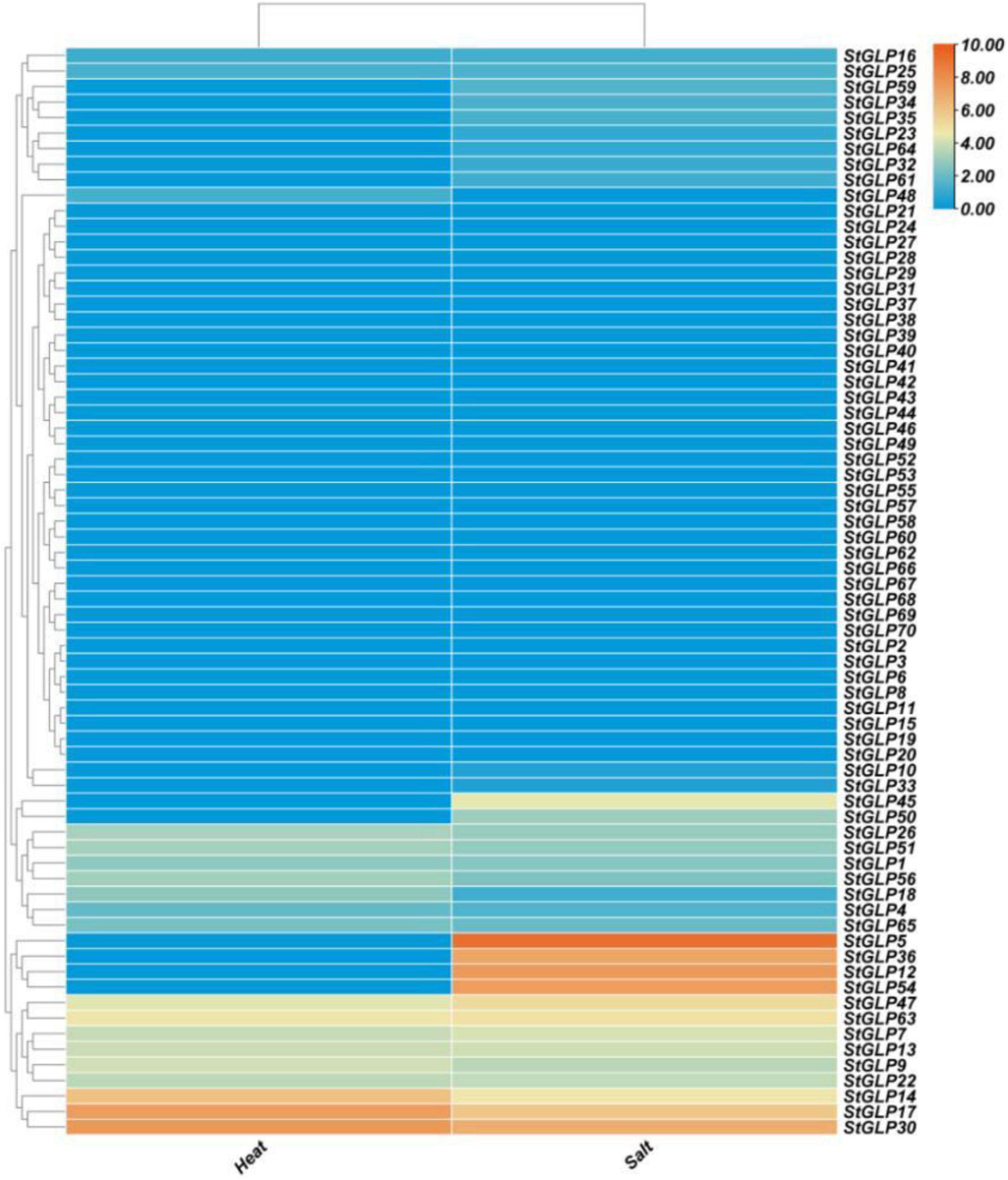
Figure 9. Expression profiling of StGLP genes in response to salt and heat treatments. The orange and blue colors display high- to low-expression levels. The expression heat map was created by taking log10 of fragments per kilobase million (FPKM).
Furthermore, StGLP16 genes showed the lowest expression compared with other expressed StGLPs after heat treatment. Likewise, 29 GLPs were expressed in response to salt treatment. This study showed that StGLP54, StGLP12, and StGLP5 genes were highly expressed in potatoes after salt treatment (Figure 9), but the expression level of the StGLP5 gene was relatively higher as compared to StGLP12 and StGLP54 genes in the potato genome.
Three hormones, abscisic acid (ABA), gibberellic acid (GA3), and indole acetic acid (IAA), were selected to evaluate the hormonal response of StGLPs.
This study results showed that 22 StGLP genes were expressed after the treatment of ABA. Similarly, in the case of IAA treatment, 24 StGLP genes were expressed. Furthermore, the StGLP12 gene was highly expressed compared with others after the treatment of ABA, while StGLP5 showed the highest expression after IAA treatment (Figure 10). Additionally, 27 StGLPs were expressed when treated with GA3, and the expression of gene StGLP14 was highest among all 27 expressed genes (Figure 10). The upregulation of StGLPs after ABA, IAA, and GA3 treatment revealed the significant role of StGLP genes in mitigating the lethal effects of different hormonal and environmental stress.

Figure 10. Expression profiling of StGLP genes in response to ABA, IAA, and GA3. The orange and blue colors display high- to low-expression levels. The expression heat map was created by taking log10 of fragments per kilobase million (FPKM).
Real-time-qPCR was performed to confirm the expression of five genes, namely, StGLP63, StGLP17, StGLP51, StGLP14, and StGLP47, in the tissues of roots, stems, and leaves (Figure 11). Two genes, namely, StGLP17 and StGLP14, exhibited a comparatively higher expression level in the tissues of stems than that in the tissues of roots and leaves. In contrast, the expression of StGLP47 was relatively more in the tissues of leaves than that in the tissues of roots and stems. Generally, findings depicted that StGLP genes may perform particular functions in potato plant growth and developmental process. Similarly, RT-qPCR was performed to confirm the expression of five genes, namely, StGLP5, StGLP12, StGLP30, StGLP 36, and StGLP54, in leaves treated with 150 mM NaCl. The results showed that the StGLP5 genes showed the maximum expression after 24 h of NaCl stress. In addition, genes such as StGLP12, StGLP 36, and StGLP54 were highly expressed with the extension of stress time. Moreover, StGLP30 showed expression but low expression compared with the other four genes (Figure 12).
Potato is an integral part of the food chain worldwide (Zaynab et al., 2017b). Potato plants are subjected to different biotic and abiotic stresses (Zaynab et al., 2021b). GLPs have a significant role in plant signaling and response to various environmental stresses (Yuan et al., 2021). Members of GLPs show variations in their expression and possess considerable functions in several tissues at different phases of growth cycle of the plant (Barman and Banerjee, 2015). This crucial gene family carried out vital defense mechanisms and is an essential part of plant defense processes. The expression of the essential genes increases exponentially when plants encounter stress like salinity (Wang et al., 2013; Zaynab et al., 2021d), water shortage (Komatsu et al., 2010), heavy metals (Berna and Bernier, 1999), and heat stress (Qasim et al., 2018b). Various functions of GLPs have also been determined in different crops such as Hordeum vulgare (Zimmermann et al., 2006), A. thaliana (Rietz et al., 2012), Oryza sativa (Li et al., 2016), and Glycine max (Wang et al., 2014). But till now, there has been no considerable in-depth study of GLPs in the potato. These days, the whole-genomic sequence of potato is comfortably accessible, allowing us to conduct genome-wide analysis of the GLP genes family that may prove worthy for further research.
Structural analysis of StGLPs will prove beneficial for their functional characterization. The utilization of the evolutionary relic of GLPs has established that the evolution of genes has been affected by the arrangement of their exons and introns (Moore and Purugganan, 2005; Flagel and Wendel, 2009). This is in correlation with previous scientists finding that genes with short introns or no introns are retained in the genomic systems of plants during the evolutionary process. At the same time, the expression levels of such genes are also low in plants (Mattick and Gagen, 2001). Moreover, the compact arrangement of genes favors the expression response of the GLPs against different exo or endogenous stimuli (Jeffares et al., 2008). Our findings through structural analysis of these genes inferred that those sequences of StGLP depicted a similar number of exons–introns with identical functional properties because they may have arisen through duplication events in the course of evolution (Waqas et al., 2019).
Studying the defense mechanisms employed by plants against various types of environmental stress, cis-elements were investigated in the promoter region of these genes. According to our findings, different cis-elements were identified that show their response against light and hormone stress. Several identified cis-elements were associated with ABA, SA, GA, MeJA, low temperature, anaerobic induction, and light. Earlier reports have attributed the stress response of plants to these cis-acting elements (Osakabe et al., 2014). Cis-factors are integral parts of plant stress response machinery as they regulate stress-responsive genes (Wu et al., 2014). So, these crucial Cis-acting sites of StGLPs indicate their behavior when different stresses are imposed.
All identified StGLP genes were classified into six clades. Similarly, phylogenetic grouping was also observed in other plants. The comparative phylogenetic analysis revealed that the organization of S. lycoperiscum, S. tuberosum, and A. thaliana proteins was relatively similar in six clades, indicating that all StGLP genes in these groups may have descended from a common ancestor. Previous studies (Li et al., 2016) reported the classification of GLPs into six clades in rice and Arabidopsis, supporting our results.
The size of the genome, duplication events in the genome, and the distribution of genes are major determinants of genetic variability in land plants. Genetic duplication property has long been recognized throughout the origins of evolutionary novelty, expression, and complexity of the gene families. We also noticed some duplications in StGLPs that may carry out vital roles in their amplification. As gene duplication is an imperative character in neofunctionalization, expansion, and variability among gene families (Lavin et al., 2005), similarly, the arrangement and localization of StGLP genes at chromosomes will enable breeders of potato to breed better varieties with desirable features.
Several types of research have depicted the response of plants to salinity and other abiotic stresses, and it is now well understood that miRNAs and their objects have direct impacts on the stress tolerance processes of plants (Li, 2015; Villanueva et al., 2016). This study recognized micro RNAs from various families targeting StGLP genes. Likewise, the unique role of miR156 has been widely documented during various abiotic stress in several plants (Cui et al., 2014; Kohli et al., 2014). Similarly, a study on cotton established a critical role of miR827 in response to saline stress (Covarrubias and Reyes, 2010); miR167 has also been recognized as one of the significant role players in response to various stressed environmental conditions (Khraiwesh et al., 2012). Concisely, these results are in concordance with our findings that revealed that miR156, miR167, and miR827 are significant determinants of tolerance in plants against several stresses as they alter the expression of StGLP genes in the genome of potatoes.
Germin-like gene miRNAs are generally present in embryos, cotyledons, leaves, stems, flowers, roots, and seeds. Some are expressed in response to different environmental conditions; this depends upon genes under analysis or species in which they are expressed. Many authentications have indicated that various GLPs may have a critical role in the defense mechanisms of plants (Breen and Bellgard, 2010). So, the expression profiling of GLPs in the potato is helpful for their in-depth comprehension. Many genes exhibited a high level of expression in tissues, suggesting the putative role of these genes in potato growth and developmental processes. Our RT-qPCR findings revealed that the upregulation of genes in the tissues of roots and stems indicated the role of GLP in potato plant growth.
Furthermore, GLPs expression was observed in response to hormones and abiotic stress. These findings are following earlier reports that GLPs are an integral part of the defense system in saline tolerance (Wang et al., 2013), drought stress (Komatsu et al., 2010), cold tolerance, and heavy metals tolerance (Berna and Bernier, 1999). Up till now, numerous researches have depicted that GLPs are significant players of plant stress response mechanisms. In our results, StGLP14, StGLP17, and StGLP30 are upregulated in response to heat stress, and StGLP5, StGLP12, and StGLP54 are highly expressed in response to salt stress supported by the findings of Wang et al. (2013). Hormones may alter the physio-biochemical pathways or metabolisms of plants by different pathways of signal transduction (Zaynab et al., 2017a; Fatima et al., 2021). ABA and IAA are major hormones of the immune system in plants. Several studies have established that besides being part of plant response to different stresses, GLPs also are involved in developing systems and hormonal signaling (Wang et al., 2020). To study how hormone signaling affects the production of StGLPs, leaves of potato were treated with ABA, GA3, and IAA, and the expression of genes was investigated. There was the induction of 22 genes on the treatment of ABA, 24 genes on the treatment of IAA, and 27 genes on the treatment of GA3, suggesting that various members of StGLPs carry out crucial roles in the plant defense system induced by these hormones. When the treatment of ABA and IAA was made, increasing the expression level of potato GLPs exhibited that these hormones possess an essential part in this defense mechanism, similar to the results obtained by Wang et al. (2020). The strong correlation between clusters of genes and their expression was noted under various stresses and tissues.
In this study, we identified 70 GLPs in the genome of the potato by genome-wide analysis. The gene structure, synteny, phylogenetic motifs, promotor, and miRNA analysis were performed to enhance our comprehension. The expression profiling after various stresses has been analyzed. The findings exhibited that GLPs genes responded significantly against hormonal and abiotic stresses, strengthening our comprehension. In our results, the StGLP5 gene showed the highest expression in response to salt stress. So, further analysis is needed to corroborate the persistent role of GLPs genes in response to salt stress.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
MZ, JP, and YS gave the idea. MF, MA, and RA-Y performed the experiments. KK and SA wrote the manuscript. SL revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Shenzhen Special Project for Sustainable Development (KCXFZ20201221173211033). The authors appreciate the support of the Research Center for Advanced Materials Science (RCAMS) at King Khalid University Abha, Saudi Arabia, through a grant KKU/RCAMS/G002-21. Taif University Researchers Supporting Project number (TURSP-2020/38), Taif University, Taif, Saudi Arabia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.831140/full#supplementary-material
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Banerjee, J., Gantait, S., and Maiti, M. K. (2017). Physiological role of rice germin-like protein 1 (OsGLP1) at early stages of growth and development in indica rice cultivar under salt stress condition. Plant Cell Tissue Organ. Cult. 131, 127–137. doi: 10.1007/s11240-017-1270-z
Barman, A. R., and Banerjee, J. (2015). Versatility of germin-like proteins in their sequences, expressions, and functions. Funct. Integr. Genomics 15, 533–548. doi: 10.1007/s10142-015-0454-z
Beracochea, V. C., Almasia, N. I., Peluffo, L., Nahirñak, V., Hopp, E. H., Paniego, N., et al. (2015). Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic Arabidopsis thaliana. Plant Cell Rep. 34, 1717–1733. doi: 10.1007/s00299-015-1819-4
Berna, A., and Bernier, F. (1999). Regulation by biotic and abiotic stress of a wheat germin gene encoding oxalate oxidase, a H2O2-producing enzyme. Plant Mol. Biol. 39, 539–549. doi: 10.1023/A:1006123432157
Breen, J., and Bellgard, M. (2010). Germin-like proteins (GLPs) in cereal genomes: gene clustering and dynamic roles in plant defence. Funct. Integr. Genomics 10, 463–476. doi: 10.1007/s10142-010-0184-1
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Christensen, A. B., Thordal-Christensen, H., Zimmermann, G., Gjetting, T., Lyngkjær, M. F., Dudler, R., et al. (2004). The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. MPMI 17, 109–117. doi: 10.1094/MPMI.2004.17.1.109
Covarrubias, A. A., and Reyes, J. L. (2010). Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ. 33, 481–489. doi: 10.1111/j.1365-3040.2009.02048.x
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., and Lin, H. X. (2014). The miR156-SPL 9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
Dai, X., Zhuang, Z., and Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
Das, A., Pramanik, K., Sharma, R., Gantait, S., and Banerjee, J. (2019). In-silico study of biotic and abiotic stress-related transcription factor binding sites in the promoter regions of rice germin-like protein genes. PLoS One 14:e0211887. doi: 10.1371/journal.pone.0211887
Davidson, R. M., Reeves, P. A., Manosalva, P. M., and Leach, J. E. (2009). Germins: a diverse protein family important for crop improvement. Plant Sci. 177, 499–510. doi: 10.1016/j.plantsci.2009.08.012
Dunwell, J. M., Gibbings, J. G., Mahmood, T., and Saqlan Naqvi, S. M. (2008). Germin and germin-like proteins: evolution, structure, and function. Crit. Rev. Plant Sci. 27, 342–375. doi: 10.1080/07352680802333938
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Fatima, M., Ma, X., Zhou, P., Zaynab, M., and Ming, R. (2021). Auxin regulated metabolic changes underlying sepal retention and development after pollination in spinach. BMC Plant Biol. 21:166. doi: 10.1186/s12870-021-02944-4
Finn, R. D., Clements, J., Arndt, W., Miller, B. L., Wheeler, T. J., Schreiber, F., et al. (2015). HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38. doi: 10.1093/nar/gkv397
Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J., Hotz, H.-R., et al. (2007). The Pfam protein families database. Nucleic Acids Res. 36, D281–D288. doi: 10.1093/nar/gkm960
Flagel, L. E., and Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook, ed. J. M. Walker (Totowa, NJ: Humana Press), 571–607.
Godfrey, D., Able, A. J., and Dry, I. B. (2007). Induction of a grapevine germin-like protein (VvGLP3) gene is closely linked to the site of erysiphe necator infection: a possible role in defense? MPMI 20, 1112–1125. doi: 10.1094/MPMI-20-9-1112
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Heintzen, C., Fischer, R., Melzer, S., Kappeler, S., Apel, K., and Staiger, D. (1994). Circadian oscillations of a transcript encoding a germin-like protein that is associated with cell walls in young leaves of the long-day plant Sinapis alba L. Plant Physiol. 106, 905–915. doi: 10.1104/pp.106.3.905
Jeffares, D. C., Penkett, C. J., and Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Khraiwesh, B., and Zhu, J.-K., and Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta Gene Regul. Mech. 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Kohli, D., Joshi, G., Deokar, A. A., Bhardwaj, A. R., Agarwal, M. Katiyar-Agarwal, S., et al. (2014). Identification and characterization of wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing. PLoS One 9:e108851. doi: 10.1371/journal.pone.0108851
Komatsu, S., Kobayashi, Y., Nishizawa, K., Nanjo, Y., and Furukawa, K. (2010). Comparative proteomics analysis of differentially expressed proteins in soybean cell wall during flooding stress. Amino Acids 39, 1435–1449. doi: 10.1007/s00726-010-0608-1
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lane, B. G. (2000). Oxalate oxidases and differentiating surface structure in wheat: germins. Biochem. J. 349, 309–321. doi: 10.1042/bj3490309
Lane, B. G., Cuming, A. C., Fregeau, J., Carpita, N. C., Hurkman, W. J., Bernier, F., et al. (1992). Germin isoforms are discrete temporal markers of wheat development. Pseudogermin is a uniquely thermostable water-soluble oligomeric protein in ungerminated embryos and like germin in germinated embryos, it is incorporated into cell walls. Eur. J. Biochem. 209, 961–969. doi: 10.1111/j.1432-1033.1992.tb17369.x
Lavin, M., Herendeen, P. S., and Wojciechowski, M. F. (2005). Evolutionary rates analysis of leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 54, 575–594. doi: 10.1080/10635150590947131
Lescot, M. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, G., Hou, M., Liu, Y., Pei, Y., Ye, M., Zhou, Y., et al. (2019). Genome-wide identification, characterization and expression analysis of the non-specific lipid transfer proteins in potato. BMC Genomics 20:375. doi: 10.1186/s12864-019-5698-x
Li, L., Xu, X., Chen, C., and Shen, Z. (2016). Genome-wide characterization and expression analysis of the germin-like protein family in rice and Arabidopsis. IJMS 17:1622. doi: 10.3390/ijms17101622
Li, S. (2015). The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signal. Behav. 10:e1044192. doi: 10.1080/15592324.2015.1044192
Liao, L., Hu, Z., Liu, S., Yang, Y., and Zhou, Y. (2021). Characterization of germin-like proteins (GLPs) and their expression in response to abiotic and biotic stresses in cucumber. Horticulturae 7:412. doi: 10.3390/horticulturae7100412
Lou, Y., and Baldwin, I. T. (2006). Silencing of a germin-like gene in Nicotiana attenuata improves performance of native herbivores. Plant Physiol. 140, 1126–1136. doi: 10.1104/pp.105.073700
Lu, M., Han, Y.-P., Gao, J.-G., Wang, X.-J., and Li, W.-B. (2010). Identification and analysis of the germin-like gene family in soybean. BMC Genomics 11:620. doi: 10.1186/1471-2164-11-620
Mattick, J. S., and Gagen, M. J. (2001). The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol. Biol. Evol. 18, 1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951
Moore, R. C., and Purugganan, M. D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8, 122–128. doi: 10.1016/j.pbi.2004.12.001
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. P. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49. doi: 10.1111/nph.12613
Pei, Y., Li, X., Zhu, Y., Ge, X., Sun, Y., Liu, N., et al. (2019). GhABP19, a novel germin-like protein from Gossypium hirsutum, plays an important role in the regulation of resistance to Verticillium and Fusarium wilt pathogens. Front. Plant Sci. 10:583. doi: 10.3389/fpls.2019.00583
Qasim, M., Baohua, W., Zou, H., Lin, Y., Dash, C. K., Bamisile, B. S., et al. (2018a). Phylogenetic relationship and genetic diversity of citrus psyllid populations from China and Pakistan and their associated Candidatus bacterium. Mol. Phylogenet. Evol. 126, 173–180. doi: 10.1016/j.ympev.2018.04.028
Qasim, M., Lin, Y., Dash, C. K., Bamisile, B. S., Ravindran, K., Islam, S. U., et al. (2018b). Temperature-dependent development of Asian citrus psyllid on various hosts, and mortality by two strains of Isaria. Microb. Pathog. 119, 109–118. doi: 10.1016/j.micpath.2018.04.019
Qasim, M., Xiao, H., He, K., Omar, M. A. A., Hussain, D., Noman, A., et al. (2021). Host-pathogen interaction between Asian citrus psyllid and entomopathogenic fungus (Cordyceps fumosorosea) is regulated by modulations in gene expression, enzymatic activity and HLB-bacterial population of the host. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 248:109112. doi: 10.1016/j.cbpc.2021.109112
Rashid, M.-O., Li, J.-H., Liu, Q., Wang, Y., and Han, C.-G. (2021). Molecular detection and identification of eight potato viruses in Gansu province of China. Curr. Plant Biol. 25:100184. doi: 10.1016/j.cpb.2020.100184
Raza, A. (2021). Eco-physiological and biochemical responses of rapeseed (Brassica napus L.) to abiotic stresses: consequences and mitigation strategies. J. Plant Growth Regul. 40, 1368–1388. doi: 10.1007/s00344-020-10231-z
Raza, A., Ashraf, F., Zou, X., Zhang, X., and Tosif, H. (2020). “Plant adaptation and tolerance to environmental stresses: mechanisms and perspectives,” in Plant Ecophysiology and Adaptation under Climate Change: Mechanisms and Perspectives I, ed. M. Hasanuzzaman (Singapore: Springer Singapore), 117–145.
Raza, A., Su, W., Gao, A., Mehmood, S. S., Hussain, M. A., Nie, W., et al. (2021). Catalase (CAT) gene family in rapeseed (Brassica napus L.): genome-wide analysis, identification, and expression pattern in response to multiple hormones and abiotic stress conditions. IJMS 22:4281. doi: 10.3390/ijms22084281
Rhee, S. Y. (2003). The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31, 224–228. doi: 10.1093/nar/gkg076
Rietz, S., Bernsdorff, F. E. M., and Cai, D. (2012). Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J. Exp. Bot. 63, 5507–5519. doi: 10.1093/jxb/ers203
Schweizer, P., Christoffel, A., and Dudler, R. (1999). Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J. 20, 541–552. doi: 10.1046/j.1365-313X.1999.00624.x
Su, W., Raza, A., Gao, A., Jia, Z., Zhang, Y., Hussain, M. A., et al. (2021). Genome-wide analysis and expression profile of superoxide dismutase (SOD) gene family in rapeseed (Brassica napus L.) under different hormones and abiotic stress conditions. Antioxidants 10:1182. doi: 10.3390/antiox10081182
The Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi: 10.1038/nature10158
Villanueva, M. A. Islas-Flores, T., and Ullah, H. (2016). Signaling through WD-repeat proteins in plants. Front. Plant Sci. 7:1157. doi: 10.3389/fpls.2016.01157
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinform. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, H., Zhang, Y., Xiao, N., Zhang, G., Wang, F., Chen, X., et al. (2020). Rice GERMIN-LIKE PROTEIN 2-1 functions in seed dormancy under the control of abscisic acid and gibberellic acid signaling pathways. Plant Physiol. 183, 1157–1170. doi: 10.1104/pp.20.00253
Wang, T., Chen, X., Zhu, F., Li, H., Li, L., Yang, Q., et al. (2013). Characterization of peanut germin-like proteins, AhGLPs in plant development and defense. PLoS One 8:e61722. doi: 10.1371/journal.pone.0061722
Wang, X., Zhang, H., Gao, Y., Sun, G., Zhang, W., and Qiu, L. (2014). A comprehensive analysis of the cupin gene family in soybean (Glycine max). PLoS One 9:e110092. doi: 10.1371/journal.pone.0110092
Waqas, M., Azhar, M. T., Rana, I. A., Azeem, F., Ali, M. A., Nawaz, M. A., et al. (2019). Genome-wide identification and expression analyses of WRKY transcription factor family members from chickpea (Cicer arietinum L.) reveal their role in abiotic stress-responses. Genes Genom. 41, 467–481. doi: 10.1007/s13258-018-00780-9
Wu, J., Wang, J., Pan, C., Guan, X., Wang, Y., Liu, S., et al. (2014). Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS One 9:e103032. doi: 10.1371/journal.pone.0103032
Xu, T., Zhang, L., Yang, Z., Wei, Y., and Dong, T. (2021). Identification and functional characterization of plant MiRNA under salt stress shed light on salinity resistance improvement through MiRNA manipulation in crops. Front. Plant Sci. 12:665439. doi: 10.3389/fpls.2021.665439
Yang, Z., and Bielawski, J. P. (2000). Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15, 496–503. doi: 10.1016/s0169-5347(00)01994-7
Yuan, B., Yang, Y., Fan, P., Liu, J., Xing, H., Liu, Y., et al. (2021). Genome-wide identification and characterization of germin and germin-like proteins (GLPs) and their response under powdery mildew stress in wheat (Triticum aestivum L.). Plant Mol. Biol. Rep. [Epub ahead of print]. doi: 10.1007/s11105-021-01291-w
Zaynab, M., Wang, Z., Hussain, A., Bahadar, K., Sajid, M., Sharif, Y., et al. (2021d). ATP-binding cassette transporters expression profiling revealed its role in the development and regulating stress response in Solanum tuberosum. Mol. Biol. Rep. [Epub ahead of print]. doi: 10.1007/s11033-021-06697-z
Zaynab, M., Chen, H., Chen, Y., Ouyang, L., Yang, X., Xu, W., et al. (2021a). Transcriptome analysis revealed transporter proteins role in the growth of Labrenzia sp. PO1 and SY1. J. King Saud Univ. Sci. 33:101433. doi: 10.1016/j.jksus.2021.101433
Zaynab, M., Hussain, A., Sharif, Y., Fatima, M., Sajid, M., Rehman, N., et al. (2021b). Mitogen-activated protein kinase expression profiling revealed its role in regulating stress responses in potato (Solanum tuberosum). Plants 10:1371. doi: 10.3390/plants10071371
Zaynab, M., Pan, D., Fatima, M., Sharif, Y., Chen, S., and Chen, W. (2021c). Proteomics analysis of Cyclobalanopsis gilva provides new insights of low seed germination. Biochimie 180, 68–78. doi: 10.1016/j.biochi.2020.10.008
Zaynab, M., Kanwal, S., Hussain, I., Qasim, M., Noman, A., Iqbal, U., et al. (2017b). Rice chitinase gene expression in genetically engineered potato confers resistance against Fusarium solani and Rhizictonia solani. PSM Microbiol. 2, 63–73.
Zaynab, M., Kanwal, S., Furqan, M., Islam, W., Noman, A., Ali, G. M., et al. (2017a). Proteomic approach to address low seed germination in Cyclobalnopsis gilva. Biotechnol. Lett. 39, 1441–1451. doi: 10.1007/s10529-017-2393-3
Zhang, Y., Wang, X., Chang, X., Sun, M., Zhang, Y., Li, W., et al. (2018). Overexpression of germin-like protein GmGLP10 enhances resistance to Sclerotinia sclerotiorum in transgenic tobacco. Biochem. Biophys. Res. Commun. 497, 160–166. doi: 10.1016/j.bbrc.2018.02.046
Keywords: abiotic stress, gene expression, gene structure, miRNA, RNA-seq, stress responses
Citation: Zaynab M, Peng J, Sharif Y, Fatima M, Albaqami M, Al-Yahyai R, Khan KA, Alotaibi SS, Alaraidh IA, Shaikhaldein HO and Li S (2022) Genome-Wide Identification and Expression Profiling of Germin-Like Proteins Reveal Their Role in Regulating Abiotic Stress Response in Potato. Front. Plant Sci. 12:831140. doi: 10.3389/fpls.2021.831140
Received: 08 December 2021; Accepted: 31 December 2021;
Published: 17 February 2022.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Naeem Iqbal, Muhammad Nawaz Shareef University of Agriculture, PakistanCopyright © 2022 Zaynab, Peng, Sharif, Fatima, Albaqami, Al-Yahyai, Khan, Alotaibi, Alaraidh, Shaikhaldein and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuangfei Li, c2ZsaUBzenUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.