- 1Center for Genomics and Biotechnology, Haixia Institute of Science and Technology, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Fujian Breeding Engineering Technology Research Center for Longan & Loquat, Fruit Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 3College of Life Sciences, Fujian Normal University, Fuzhou, China
- 4College of Mechanical and Electrical Engineering, Fujian Agriculture and Forestry University, Fuzhou, China
- 5Department of Plant Biology, University of Illinois at Urbana-Champaign, Urbana, IL, United States
Longan (Dimocarpus longan Lour.) is an important economic crop widely planted in tropical and subtropical regions, and flower and fruit development play decisive effects on the longan yield and fruit quality formation. MCM1, AGAMOUS, DEFICIENS, Serum Response Factor (MADS)-box transcription factor family plays important roles for the flowering time, floral organ identity, and fruit development in plants. However, there is no systematic information of MADS-box family in longan. In this study, 114 MADS-box genes were identified from the longan genome, phylogenetic analysis divided them into type I (Mα, Mβ, Mγ) and type II (MIKC*, MIKCC) groups, and MIKCC genes were further clustered into 12 subfamilies. Comparative genomic analysis of 12 representative plant species revealed the conservation of type II in Sapindaceae and analysis of cis-elements revealed that Dof transcription factors might directly regulate the MIKCC genes. An ABCDE model was proposed for longan based on the phylogenetic analysis and expression patterns of MADS-box genes. Transcriptome analysis revealed that MIKCC genes showed wide expression spectrums, particularly in reproductive organs. From 35 days after KClO3 treatment, 11 MIKC genes were up-regulated, suggesting a crucial role in off-season flower induction, while DlFLC, DlSOC1, DlSVP, and DlSVP-LIKE may act as the inhibitors. The gene expression patterns of longan fruit development indicated that DlSTK, DlSEP1/2, and DlMADS53 could be involved in fruit growth and ripening. This paper carried out the whole genome identification and analysis of the longan MADS-box family for the first time, which provides new insights for further understanding its function in flowers and fruit.
Introduction
The MADS-box genes, widely distributed in fungi, plants, and animals, encode a large transcription factor family. These genes with diverse functions play important roles in plant development, signal transduction, and stress responses (Riechmann and Meyerowitz, 1997; Messenguy and Dubois, 2003). The family name “MADS” consists of the initials of its earliest genes, MCM1 in yeast, AGAMOUS (AG) in Arabidopsis thaliana (Arabidopsis), DEFICIENS in Antirrhinum majus, and Serum Response Factor (SRF) in humans (Passmore et al., 1988; Sommer et al., 1990; Yanofsky et al., 1990). Each MADS protein possesses a conserved domain of about 60 amino acids named MADS-box at the N-terminus, which recognizes and binds to CArG boxes (CC[A/T]6GG) (De Bodt et al., 2003). At an evolutionary level, all the members can be divided into two lineages, type I and type II, according to their relationships to animal SRF-like and MEF2-like genes, respectively (Alvarez-Buylla et al., 2000). In general, type I MADS genes have simple structures with 1–2 exons and there are few reports on their functions. The type II genes were also named MIKC genes due to their four domain structures: the highly conserved MADS-box (M) domain, the less-conserved intervening (I) domain, the moderately conserved keratin-like (K) domain, and the variable C-terminal (C) region (Theissen et al., 1996; Kaufmann et al., 2005). In Arabidopsis MADS-box genes were separated into five groups including Mα, Mβ, Mγ, Mδ, and MIKC based on the phylogenetic analysis (Parenicova et al., 2003). These MIKC type transcription factors, only found in the plant, can be subdivided into MIKCC and MIKC* groups based on differences in I and K domains and have been characterized functionally in the regulation of growth and development (Theissen et al., 2000; Becker and Theissen, 2003). These type II genes may be separated from the ancestor of extant land plants (Becker and Theissen, 2003).
In seed plants, extensive research has been performed since the MIKCC type genes were demonstrated to be involved in floral organ identity, the control of flowering time, and seed development. They can be further classified into 12 clades (Becker and Theissen, 2003), for example, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and SHORT VEGETATIVE PHASE (SVP) participate in the regulation of flowering time; APETALA 1 (AP1) is not only a floral meristem identity gene, but also the floral organ identity gene; FRUITFULL (FUL) regulates cell differentiation during fruit development (Coen and Meyerowitz, 1991; Gu et al., 1998; Lee and Lee, 2010). With extensive researches, the well-known ABCDE model was built from studies in Arabidopsis and Antirrhinum majus, which explains floral organ identity (Coen and Meyerowitz, 1991; Theissen, 2001; Theissen and Saedler, 2001). Within the floral meristem, A + E genes specify sepals, A + B + E genes specify petals, B + C + E genes determine stamens, and C + E genes direct carpels, in addition, D + E genes are involved in ovule development. In the model plant Arabidopsis, A-class genes are represented by AP1, APETALA2 (AP2), FUL, CAULIFLOWER (CAL), and AGAMOUS-LIKE 79 (AGL79) (Jofuku et al., 1994), B class correspond to genes from APETALA3 (AP3) and PISTILLATA (PI) (Goto and Meyerowitz, 1994; Yang et al., 2003), C class includes AG (Yanofsky et al., 1990), D class includes SHATTERPROOF (SHP) and SEEDSTICK (STK) and E class include 4 SEPALLATA genes (SEP1, SEP2, SEP3, and SEP4) (Pelaz et al., 2000; Pinyopich et al., 2003; Ditta et al., 2004; Zahn et al., 2006).
Longan (Dimocarpus longan Lour., 2n = 2x = 30), a member of the Sapindaceae family, was derived and widely cultivated in Southeast Asia (Lai et al., 2000). As a famous subtropical fruit tree, longan commonly called “dragon eye” in China is of high nutritional and medicinal value (Mei et al., 2014). Floral induction (FI) is the biological basis for the development of floral organs and fruit. However, in longan, FI is sensitive to temperature and thus floral reversion occurs frequently (Chen et al., 2009). Began in the late 1990s, potassium chlorate (KClO3) has been heavily input into longan thanks to the discovery of flowering induction by KClO3 at almost any time of the year (Subhadrabandhu and Yapwattanaphun, 2001; Manochai et al., 2005). Interestingly, off-season FI by KClO3 was only found in longan, even its close species litchi (Litchi chinensis Sonn.) has never been affected by KClO3. Several studies have investigated the physiological mechanisms of FI by KClO3, indicating that starch, sucrose contents, and cytokinin may play an important role (Sringarm et al., 2009; Chang, 2010). In addition, “Sijimi” (“SJ”) longan, performs a unique trait of perpetual flowering (PF), flower and bears fruit throughout the year without external environment conditions (Zhang H.N. et al., 2016). Longan likely has a special FI mechanism among the perennial fruit species. In this decade, the molecular genetics of flowering in model plants has a great advanced development (Fornara et al., 2010), but the research progress of flowering regulation in perennial fruit trees is very limited due to their long juvenile period and complex genetic backgrounds. Several flowering genes have been cloned and analyzed in longans such as FLOWERING LOCUS T (FT), AP1, and LEAFY (LFY) (Winterhagen et al., 2013), however, no research has been carried out on the MADS-box family in longan.
Recently, we sequenced and assembled the “Shixia” (“SX”) longan genome, this chromosome level genome can assist us with the analysis of MADS-box genes from the entire genome. Here, we identified 114 MADS-box genes from the longan genome for the first time, with the phylogenetics, gene evolution, conserved motif, and cis-element analysis was performed. We also determined the expression profiles of longan MADS-box genes particularly the ABCDE model genes during off-season FI under KClO3 and fruit development. This work provides an overview and information useful for future functional analysis of longan MADS-box genes in the reproductive process.
Materials and Methods
Data Retrieval and Plant Materials
The Arabidopsis MADS-box genes were retrieved from The Arabidopsis Information Resource (TAIR)1. The MADS-box genes of Amborella trichopoda, Citrus clementina, Citrus sinensis, Vitis vinifera, Oryza sativa, Sorghum bicolor, Zea mays, Musa acuminate, Nymphaea colorata, and Physcomitrella patens were described in previous reports (Aono and Hasebe, 2006; Arora et al., 2007; Zhao et al., 2011; Hou et al., 2013; Project, 2013; Wang et al., 2015; Liu et al., 2017; Zhang et al., 2020). The genomic data of Selaginella moellendorffii and Chlamydomonas reinhardtii were obtained from Phytozome2 and Xanthoceras sorbifolium was downloaded from GigaDB3. The evolutionary relationships of the above species were obtained from National Center for Biotechnology Information (NCBI)4. The RNA-seq data from ten tissues (root, stem, leaf, dormant bud, flower bud, flower, young fruit, pulp, pericarp, and seed) of “Sijimi” (“SJ”) longan were downloaded from the NCBI under accession number: PRJNA329283 and PRJNA387674 from previous studies (Lin et al., 2017; Jue et al., 2019).
The collection of Dimocarpus longan and the performance of experimental research on such plants complied with the national guidelines of China. We collected apical buds under CK and KClO3 (99% active ingredient) treatment from 4∼6 years old “Shixia” longan in Maoming, Guangdong, China at ten-time points (from day 0 to day 54 including November 18, 2016, November 23, 2016, November 28, 2016; December 3, 2016, December 8, 2016, December 13, 2016, December18, 2016, December 23, 2016, December 29, 2016, and January 11, 2017). Four whorls of floral organs including sepal, petal, stamen, carpel (pistil without ovary) and ovary of “Baoshi No. 1” (“BS-1”) and fruit of n were collected in Germplasm Repository of Longan (Dimocarpus longan), Fuzhou City, Ministry of Agriculture. All samples were collected and stored at −80°C until RNA-Seq. A pair-end library was made using the Illumina® TruSeq™ RNA Sample Preparation Kit [RS-122-2001(2), Illumina] and sequencing using Illumina X ten.
Identification and Features of MADS-Box Family in Longan
Two strategies were used to identify the MADS-box transcription factor family: the hidden Markov model (HMM) profile of SRF-TF domains (PF00319) from the Pfam database5 was used as a query to identify MADS-box sequences with HMMER version 3 (Eddy, 2011) against the longan genome with a threshold of e-value ≤ 1 × 10–5. In addition, MADS protein sequences of Arabidopsis were used as queries to search against the longan genome using the BLASTP program (Altschul et al., 1990) with an e-value cutoff of 1e−5 and identity > 40%. Subsequently, these proteins were submitted to the Pfam database and NCBI Conserved Domain Search6 to confirm the presence and completeness of the MADS domain. Candidate genes without the MADS domain were re-annotated manually with the assistance of FGENESH (Solovyev et al., 2006). The physical-chemical properties of longan MADS-box genes were predicted with the online tool ProtParam from ExPASy (see text footnote 6).
Phylogenetic Analysis
Sequence alignments of MADS-box genes were performed using the MUSCLE program in MEGA X (Kumar et al., 2018) with default parameters. The NJ (neighbor-joining) phylogenetic tree was constructed using MEGA X with the following parameters: Poisson model and pairwise deletion, bootstrap for 1000 replicates. Maximum-likelihood (ML) phylogenetic trees were constructed using FastTree software (Price et al., 2009) with the LG model. Arabidopsis MADS-box genes (AtMADS) were used to assist classification (Theissen et al., 2000).
Chromosome Locations and Synteny Analysis
To analyze the synteny for longan MADS-box genes (DlMADS), the BLASTP program (e-value < 1e−5), and MCScanX (Wang et al., 2012) were used. The tool ‘duplicate gene classifier’ was used to classify the origins of duplicate genes for longan.
Gene Structure, Conserved Motif, and Cis-Element Analysis
Gene structures of MADS-box genes were extracted from the General Feature Format (GFF) file and the diagram was drawn with the online program Gene Structure Display Server (GSDS7). Conserved motifs were identified using Multiple EM for Motif Elicitation (MEME, version 5.38) with the following parameters: 10 different motifs, Minimum Motif Width 10, Maximum Motif Width 100. The identified motifs were sent to Interpro9 for annotation.
The 1,500-bp sequences upstream of the start codon of each longan MADS-box gene were extracted to predict cis-acting elements through PlantCARE10. We further detected the conserved motifs from promoters of MIKCC genes using MEME with p-values < 0.05 and compared the motifs with known transcription factor binding sites (TFBS) from JASPAR Core plants 2018 database11 by performing Motif Comparison Tool TOMTOM (TOMTOM version 5.312) with e-value < 0.05.
RNA-Seq Data Analysis
Raw reads from RNA-seq were trimmed with Trimmomatic version 0.39 (Bolger et al., 2014) to remove adaptor sequences and low-quality reads. The high-quality reads were mapped to the “SX” longan genome using HISAT2 version 2.1 (Kim et al., 2015). The expression levels of MADS-box genes were normalized to fragments per kilobase of exons per million fragments mapped (FPKM) using StringTie version 2.1.2 (Kovaka et al., 2019). We defined genes with an FPKM value < 1 as not expressed, genes with FPKM value > 1 as lowly expressed, and values > 10 as highly expressed. Genes with FPKM value > 100 were extremely highly expressed. The differentially expressed genes (DEGs) were identified using the R package DESeq2 version 1.3 (Love et al., 2014). The gene co-expression networks were constructed using the WGCNA (Langfelder and Horvath, 2008) package with the filtered genes (FPKM > 1 at least one sample). Software Cytoscape version 3.7.1 (Shannon et al., 2003) was used to visualize the gene interaction networks of MADS-box genes.
Expression Validation of DlMADS by qRT-PCR
The cDNA for qRT-PCR was synthesized using the StarScript II First-strand cDNA Synthesis Mix with gDNA Remover (GenStar, A224-10). Gene-specific primers were designed using the online tool PrimerQuest13 (Supplementary Table 1). Two longan actin genes (Dil.10g021740.1 and Dil.06g016430.1) were used as the internal control and three replicates were performed (Jue et al., 2019). The qRT-PCR amplification was carried out using 2× RealStar Green Fast Mixture (GenStar, A301-10) on a Multicolor Real-Time PCR Detection System (Bio-Rad) using the protocol for this kit: 95°C for 2 min, 40 cycles of 95°C for 15 s and 60°C for 30 s. The relative expression levels of the candidate genes were calculated using the 2−ΔΔCt method.
Results
Identification of MADS-Box Genes in Longan
To identify the MADS-box gene family, both the hidden Markov model (HMM) profile (PF00319) and 107 Arabidopsis MADS-box protein sequences were used as queries to perform HMMER and BLASTP against the “SX” longan genome. A total of 117 candidate MADS-box genes were identified. Among them, nine genes that did not have MADS domain-coding sequences were manually re-annotated with the assistance of the online tool FGENESH. Three re-annotated sequences without MADS domain were excluded from further analysis. Finally, 114 complete MADS-box genes were confirmed and named as DlMADS1-DlMADS114 based on their genomic location in the chromosome (Supplementary Table 2). The length and molecular weight of 114 MADS-box proteins ranged from 64 AA and 7358.57 Da (DlMADS85) to 643 AA and 72378.92 Da (DlMADS101), with isoelectric points in the range of 5.06 to 10.49 (Supplementary Table 2). This result showed divergences in physicochemical properties among MADS-box family members in longan.
Phylogenetic and Evolutionary Analysis
To classify and examine the evolutionary relationship among MADS-box genes, we constructed a neighbor-joining (NJ) phylogenetic tree with alignments of longan and Arabidopsis MADS-box protein sequences. The result showed that DlMADS were divided into 5 groups, as in Arabidopsis (Parenicova et al., 2003; Supplementary Figure 1). Of these, 63 longan MADS-box genes were assigned to type I including 39 genes in Mα, 10 genes in Mβ, 14 genes in Mγ. 51 DlMADS were classified as type II including 36 MIKCC and 15 MIKC* type genes.
The MADS-box family is widely distributed in plants, and to understand the gene evolution in longan, we searched for MADS-box genes in the genomes of 15 representative plant species for comparative genomic analysis. Nine of them have been previously reported (see Methods), while MADS-box genes of the other six plant species were identified and classified from their genomes. As shown in Figure 1, both type I and type II have single genes in C. reinhardtii (Algae), supporting the view that these two types of MADS-box genes are very ancient and originated before the origin of Embryophyte. The MADS-box genes were expanded in Angiospermae due to the ε-whole-genome duplication (WGD). Compared to type I, the number of type II genes is relatively conserved in monocots and eudicots. In addition, longan has the largest number (114) of MADS-box genes, similar to litchi.
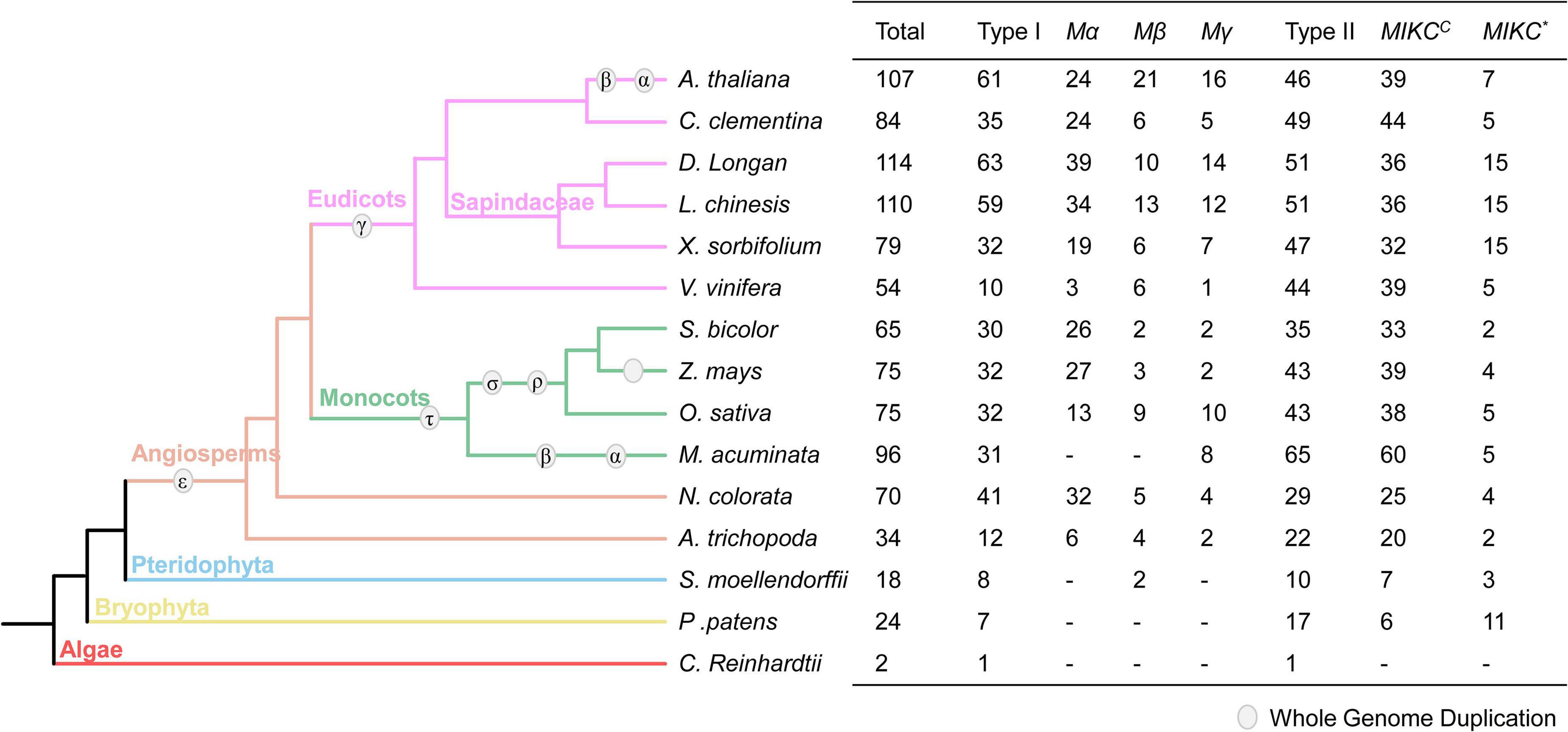
Figure 1. The MADS-box gene family in 15 species. The evolutionary relationship of 15 species on the left of the figure was obtained from NCBI (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi). The right of the figure shows the number detail of the MADS-box family in each species. Type I MADS-box genes contain three groups: Mα, Mβ, Mγ, and type II genes are subdivided into MIKCC and MIKC* groups.
In order to further classify the MIKCC genes, two phylogenetic trees were constructed using MADS-box proteins from Arabidopsis, longan, litchi, and Xanthoceras sorbifolium (yellowhorn) using the maximum-likelihood (ML) and neighbor-joining (NJ) methods (Figure 2 and Supplementary Figure 2). The structures of the two trees based on NJ and ML methods were similar, indicating a reliable subfamily division. The longan MIKCC genes could be divided into 12 subgroups with Arabidopsis genes as a reference, and no novel subfamily was found, indicating the conservation of the gene evolution in dicot species. The Flowering Locus C (FLC) clade of Arabidopsis contains one FLC gene and five homologs, MADS-AFFECTING FLOWERING 1-5 (MAF1-5), which was attributed to the two rounds of Arabidopsis specific WGD. Only one gene was found in each Sapindaceae FLC clade, which was similar to the result in Citrus (Hou et al., 2013). Similar to FLC, subgroups TT16 and AGAMOUS-like 6 (AGL6) contained the minimum number (one) of longan type II genes. For the SVP clade, there are two genes (SVP and AGL24) in Arabidopsis, with the SVP subgroup consisting of the largest number (11) of MIKCC type genes in longan, indicating that some of the loss and duplication events probably occurred after the divergence of two the species. As dormancy-associated MADS-box (DAM) genes, which were highly homologous to SVP and AGL24, have been proven to affect dormancy in peach (Bielenberg et al., 2008), we collected reported DAM gene sequence data from Prunus persica (peach), Prunus mume (plum), Pyrus pyrifolia (pear), and Malus domestica (apple) to generate a phylogenetic tree with longan and litchi (Supplementary Figure 3). Except for the main SVP and AGL24 clade, all the Rosaceae DAM genes cluster in one clade, with nine longan MADS genes (named as DlSVP-LIKE) and seven litchi MADS genes located in an individual cluster.
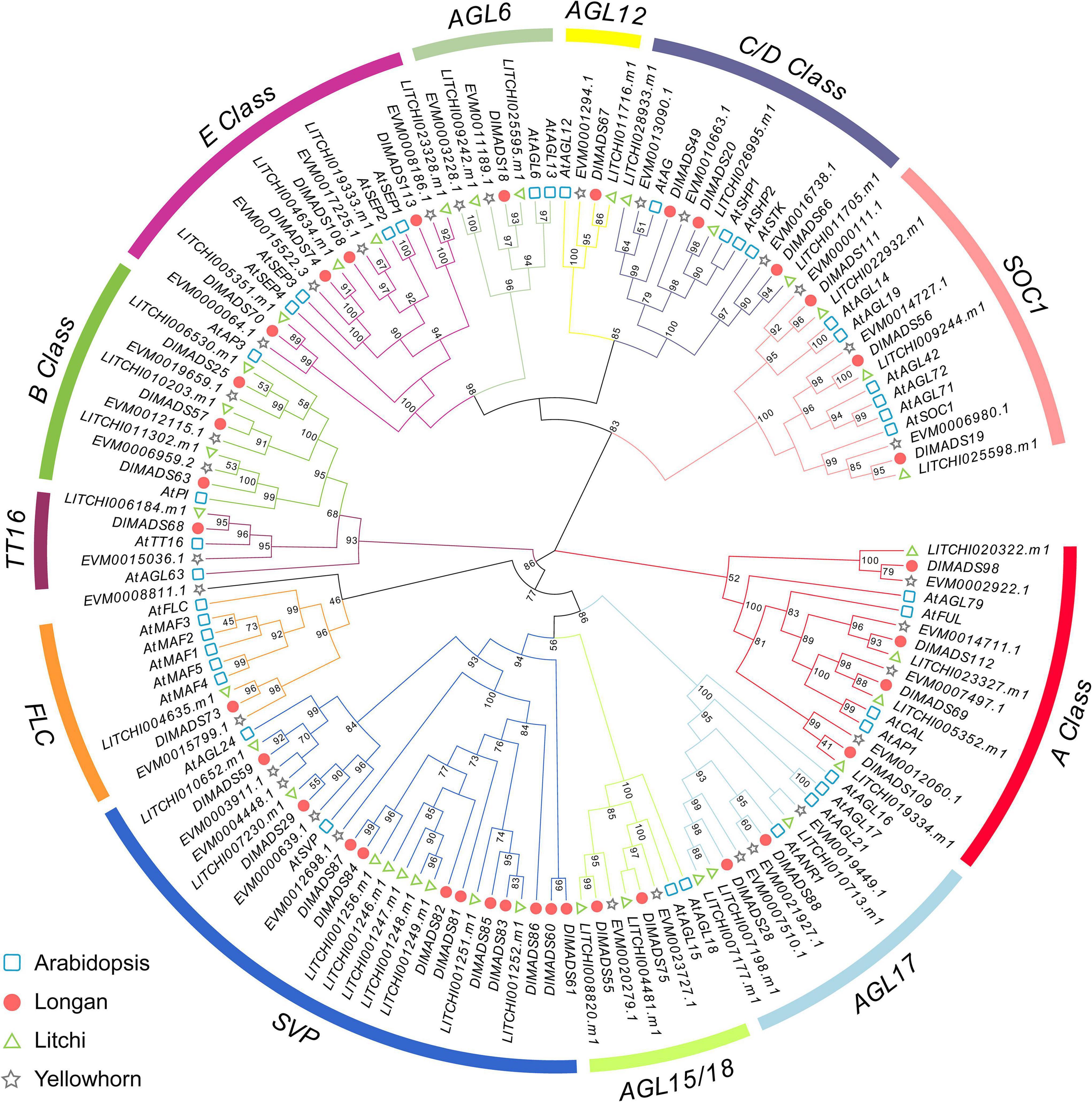
Figure 2. Phylogenetic analysis of type II MADS-box transcription factors. The maximum-likelihood (ML) tree was constructed with MIKCC proteins sequences from Arabidopsis (At), longan (Dl), litchi (LITCHI), and yellowhorn (EVM).
Genomic Distribution and Gene Duplication
The 114 MADS-box genes were unevenly distributed on 15 chromosomes of the longan genome (Figure 3A). At least 3 DlMADS were found in each chromosome, chromosome 11 had the largest number of family members at 20, and only three genes were located on chromosomes 4 and 12. Gene duplication has been considered the driving force for species evolution, and WGD events have probably occurred in many eukaryotes, sometimes more than once (Van de Peer, 2004). Previous research has established that over 90% of the increase in the number of regulatory genes was caused by the three WGD events in Arabidopsis (Maere et al., 2005). In this study, we investigated gene duplication events in Arabidopsis, longan, litchi, and yellowhorn genome using MCScan X. Of the genes of the longan MADS-box family, 44 (38.6%) originated from WGD or segmental duplication, 20 (17.5%) appeared to have been created through tandem duplication, 22 (19.3%) were proximal duplicated genes and 28 (24.6%) were dispersed duplicated genes (Table 1). In Arabidopsis, nearly half of the MADS-box genes (46.7%) originated from dispersed duplication, while tandem duplication events were more widespread in the MADS-box genes which indicates they made a valuable contribution to the evolution of the longan MADS-box family. Of significance, at least 26 genes of the Mα group were clustered on special regions of chromosomes 2, 7, and 13, indicating that tandem duplications were the main force driving the expansion of this MADS-box group.
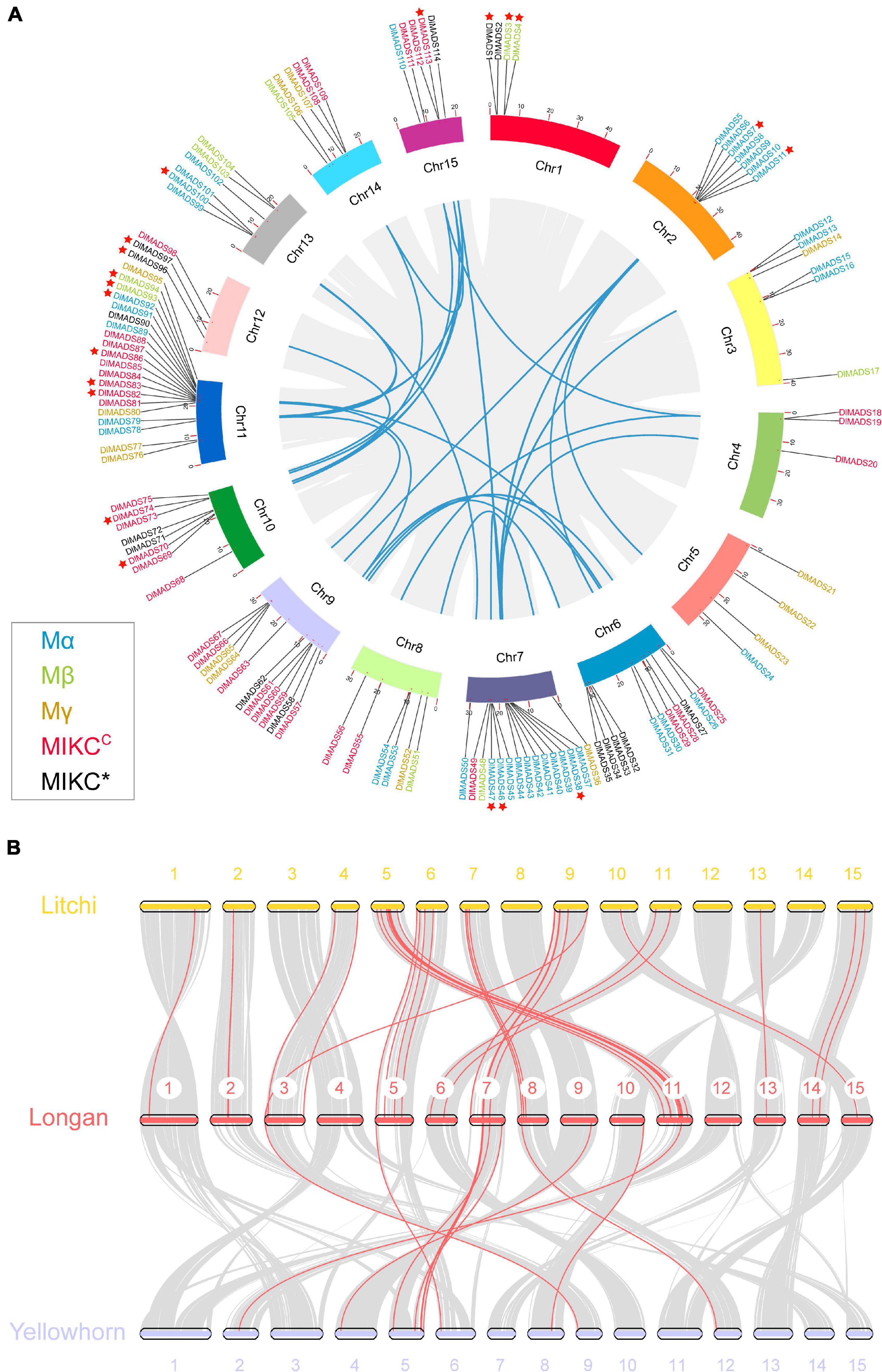
Figure 3. Chromosomal location and synteny analysis of the MADS-box genes. (A) A total of 114 MADS-box genes are located in 15 chromosomes with different colors. Gene pairs of WGD or segmental duplication are linked using blue lines. Tandem duplication genes are marked by red stars. Gene labels of five types: Mα, Mβ, Mγ, MIKC*, and MIKCC are denoted by blue, green, yellow, red, and black, respectively. (B) Synteny relationship of type I MADS-box genes from longan, litchi, and yellowhorn genomes. Gray lines in the background indicate the collinear blocks within two genomes. Red lines highlight the syntenic type I MADS-box gene pairs.
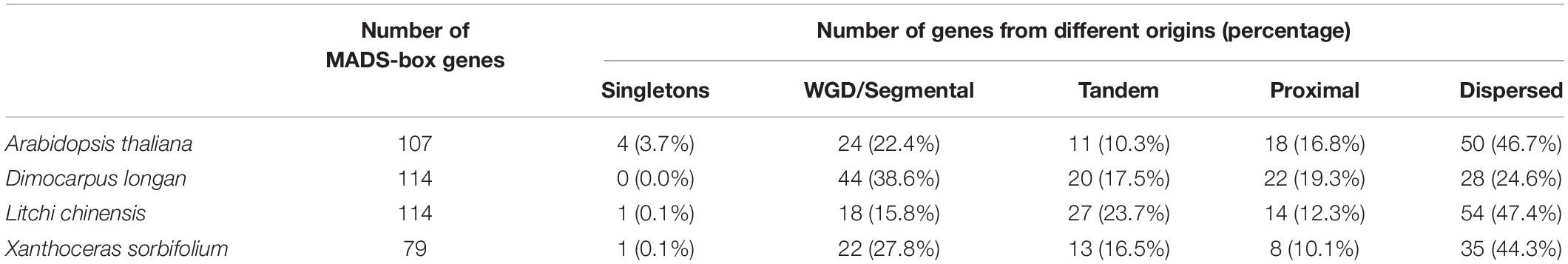
Table 1. Numbers of MADS-box genes from different origins in Arabidopsis, longan, litchi, and yellowhorn genomes.
In Sapindaceae, type II gene numbers were generally conserved, while type I gene numbers varied. Longan and litchi have consistent numbers of MADS-box genes in all the subgroups with the exception of Mβ and Mγ (Figure 1). Yellowhorn had much fewer type I MADS-box genes (32) than both longan (63) and litchi (59). Therefore, we constructed the comparative syntenic maps among longan, litchi, and yellowhorn (Figure 3B). For type I, 44 orthologous gene pairs were identified between longan and litchi, indicating close relationships. In comparison to longan and litchi genomes, gene concentration was observed in which was mainly caused by the loss of type I genes in the corresponding longan chromosomes 1, 2, 3, 6, 11, 13, 14, and 15. Moreover, the synonymous substitution rates (Ka/Ks) of the gene pairs were calculated to identify the evolutionary forces. All of the 58 orthologous gene pairs had Ka/Ks < 1 (Supplementary Table 3), suggesting that purifying selection may be the dominant force driving the evolution of Sapindaceae type I MADS-box genes.
Gene Structure and Conserved Motifs of Longan MADS-Box Family
To assess the structural diversity of longan MADS-box genes, the GSDS program was used to display intron-exon organization. The results showed that the structures of type I genes were short and simple with the presence of 0–2 introns except for DlMASD101 with 9 introns (Supplementary Figure 4). The full-genome lengths of DlMADS40 and DlMADS42 were significantly longer at 6503 and 9040 bp. Through the survey of transposable element (TE) sequences, we found that 9 and 5 TEs were located in DlMADS40 and DlMADS42, respectively (Supplementary Table 4). Compared with type I, type II genes contain more exons in the range of 0–12, which is in contrast to the previous report that MIKC genes have a common structure of 1–6 exons (Johansen et al., 2002). In addition, the average length of MIKC* genes (3,900 bp) was shorter than MIKCC genes (8,800 bp).
Since MIKC type genes with complex structures play functional roles in developmental processes in plants (Becker and Theissen, 2003), we analyzed the conserved motifs of longan MIKC type proteins using the MEME program. A total of 10 conserved motifs were identified among 51 MIKC protein sequences (Figure 4 and Supplementary Table 5) and the number of motifs in DlMADS ranged from 1 to 7 and each subfamily had similar motif compositions. Motif 1 which was annotated as MADS domain was found in nearly all protein sequences except for DlMADS86, 84, 87 in the SVP class and DlMADS1 in the MIKC* class, with the distribution of the unknown motif 4 being similar to motif 1, suggesting that these two motifs play crucial functions. Motif 2, 3, and 7 may be fragments of the K-box and are distributed in most MIKCC members, but motif 5 annotated as K-box was only found in MIKC* class. The result reveals that the primary differences between MIKCC and MIKC* proteins are variations in the K domain.
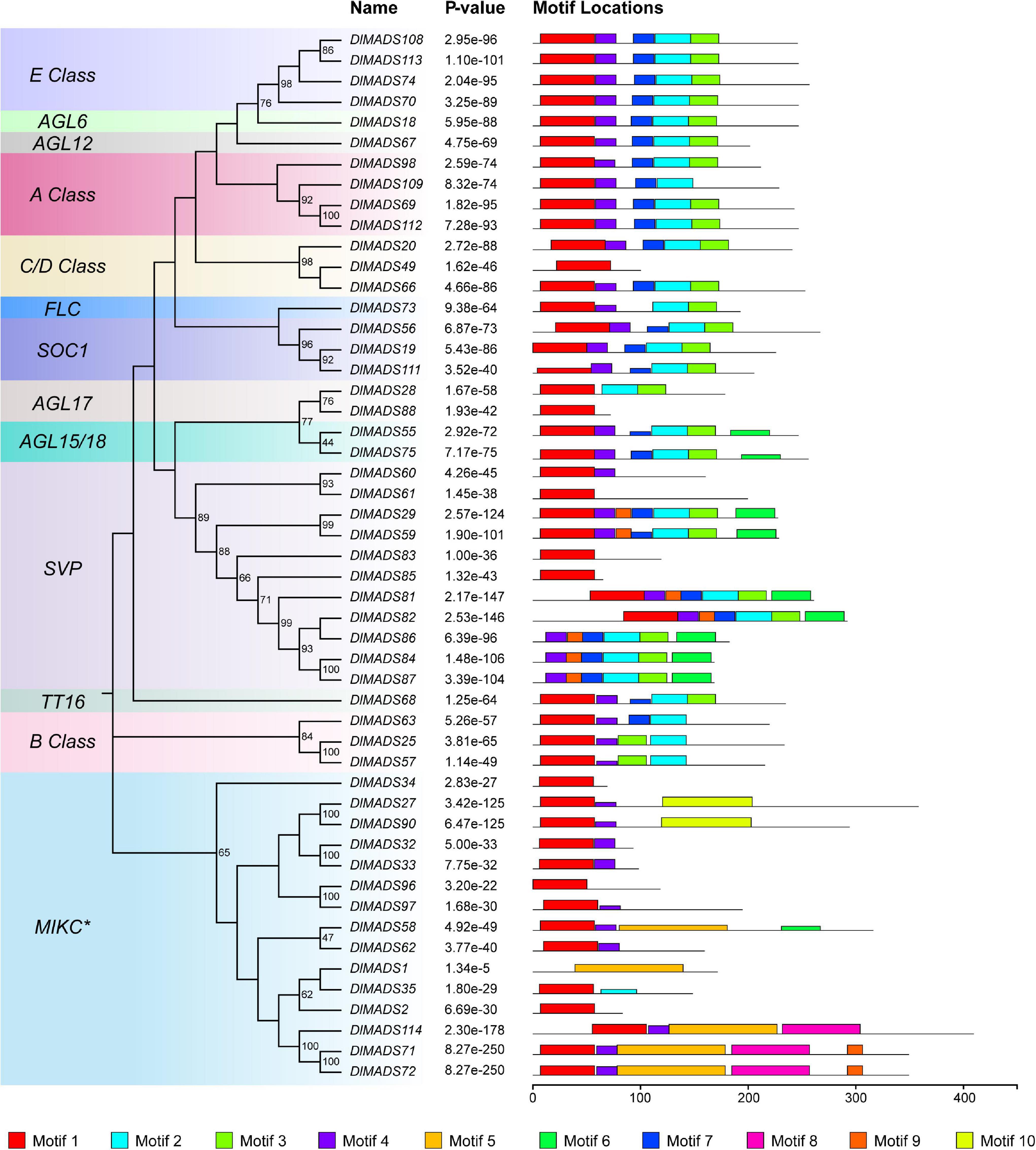
Figure 4. Conserved motif compositions of longan type II MADS-box proteins. The neighbor-joining tree was constructed with the aligned protein sequences of longan type II MADS-box genes. 10 motifs were identified and displayed in different colors.
Cis-Elements and Potential Transcription Factor Binding Sites
Transcription factors (TFs) control and regulate gene expression through binding cis-elements in the promoters of target genes. To investigate the regulatory gene networks of the MADS-box family, we analyzed the cis-elements of the upstream 1,500-bp sequences of DlMADS based on the PlantCARE database. Cis-elements associated with 11 biological processes such as light-responsiveness and the circadian clock were annotated in DlMADS (Supplementary Table 6). Of note, the number of functional elements in MIKCC type genes is significantly less than that of other genes (Figure 5A). Light-responsive boxes existed in all MADS-box genes. Hormone-related (ABRE, TGA-element, AuxRR-core, GARE-motif, P-box, etc.) and defense and stress-responsive (WUN-motif, LTR, TC-rich repeats, ARE) cis-elements were found in most family members. Additionally, DlMADS1 belonging to the MIKC* group had the most cis-elements amongst the gene families. In the MIKCC subfamily, the hormone-related elements were more frequently located in SEP, FLC, and SVP clades.
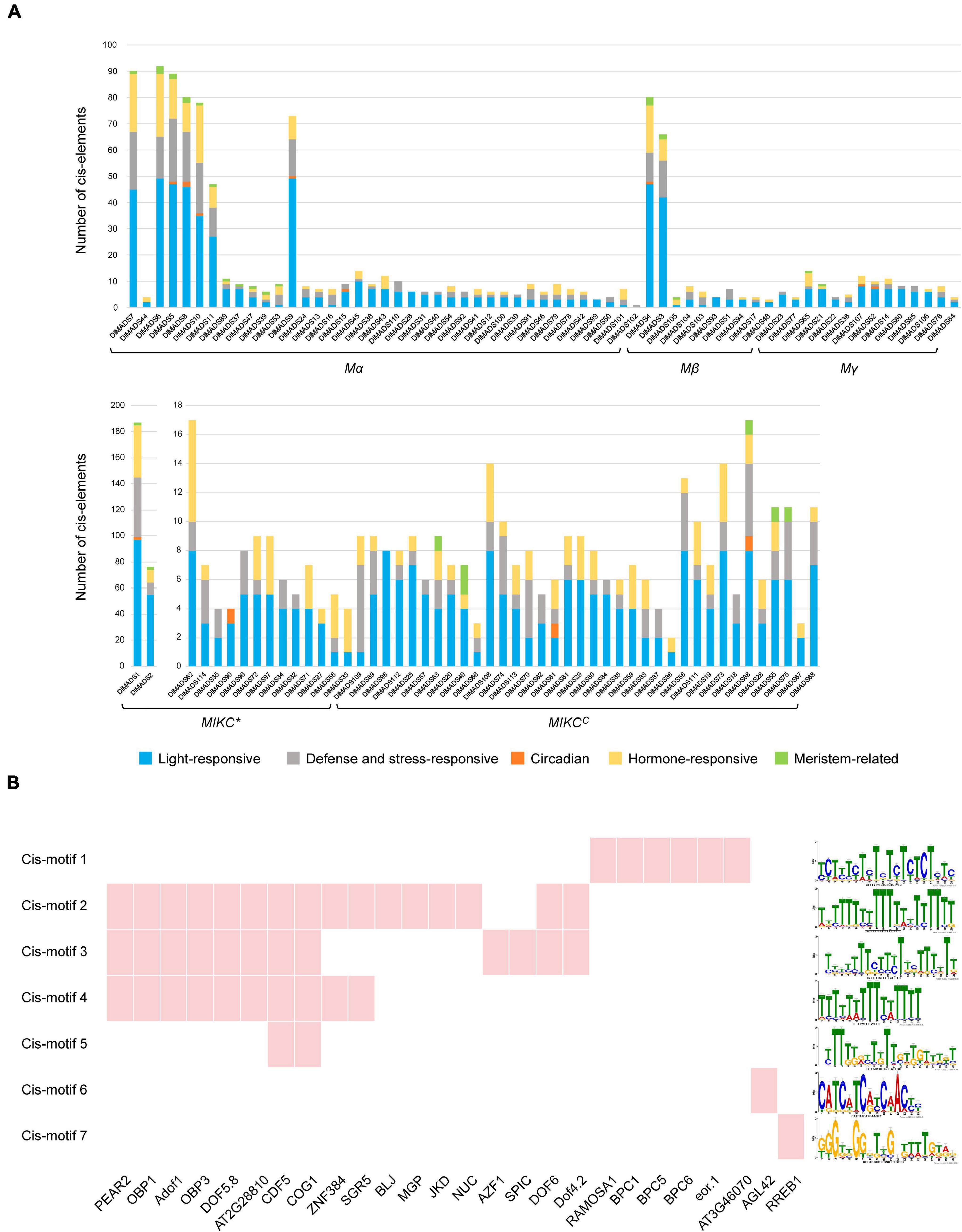
Figure 5. Prediction of cis-elements and transcription factors of DlMADS. (A) The number of cis-elements related to light-responsive, defense and stress-responsive, circadian, hormone-responses and meristem. The upstream 1.5 kb sequences of all longan MADS-box genes were analyzed through PlantCARE. (B) The cis-motifs and transcription factors potentially target 36 MIKCC genes. Red indicates the transcription factor binding sites were found in promoter motifs of MIKCC genes.
Transcription factors can directly bind to specific sequences in promoters of target genes called binding sites to affect gene expression. In order to further identify the transcription factors which potentially control the MIKCC type genes, the promoters of 36 MIKCC genes were processed using MEME software to locate cis-motifs. As a result, 20 enriched motifs were found and a total of 26 TFs along with their DNA bind sites were identified in 7 motifs by comparison with the JASPAR database (Figure 5B and Supplementary Table 7). 21 TFs were C2H2 zinc finger proteins particularly the DNA-binding One Zinc Finger (Dof) transcription factor family. For example, CYCLING DOF FACTOR 5 (CDF5) and COGWHEEL1 (COG1) binding sites were found in 4 motifs. Other TFs belong to BARLEY B RECOMBINANT/BASIC PENTACYSTEINE (BBR/BPC), MADS-box family and, ETS-related genes.
A Proposed ABC(D)E Model in Longan
Extensive research showed that five classes of homeotic genes called the ABCDE model, control floral development at the molecular level. Phylogenetics and classification based on the model plant as reference allowed the identification of the putative functions of longan MADS-box genes. As shown in Figure 6A, only one copy of an A-class functional MADS-box gene was found in Amborella and four homologs were confirmed in rice, Arabidopsis, and the three Sapindaceae species. Phylogeny indicated that A-class genes were present in a one-to-one orthologous pattern in the Sapindaceae species. AP1 and CAL originated from a recent duplication event and have both partially redundant and unique functions (Alvarez-Buylla et al., 2000), and only one gene of longan (DlMADS69) was found in AP1/CAL clade, named as DlAP1. In addition, the number of AP2 genes in the AP2/EREBP family was highly conserved in tested Angiosperm (Figure 6A). Both AP3 and PI which diverged in the ancestors of angiosperms experienced loss events in eudicots, and C and D class genes formed a very close sister group, with each Sapindaceae species having one AG, one STK, and one SHP gene. SEP genes have roles throughout five whorls of floral organs and were duplicated during angiosperm evolution. Four SEP homologs were found in longan, litchi, and yellowhorn. Overall, these results demonstrated that the number of ABCDE genes in Sapindaceae was similar to the model plant Arabidopsis.
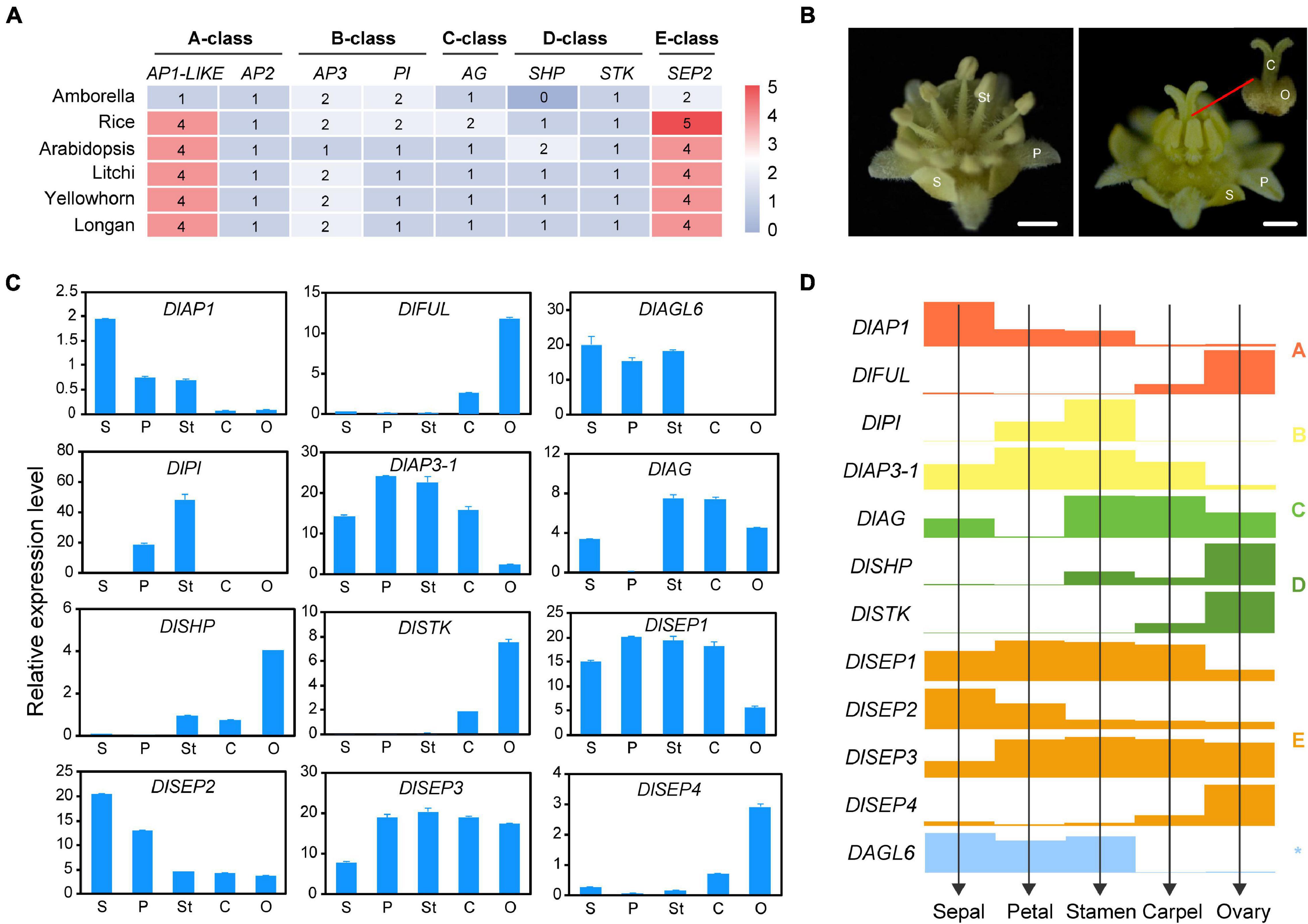
Figure 6. The deduced ABCDE model in longan. (A) The heat map shows the number of ABCDE classes genes in Amborella, Arabidopsis, rice, and longan. (B) Mature male (left) and female (right) flowers of longan. Five types of floral organs including sepal (S), petal (P), stamen (St), carpel (C), ovary (O) were collected for gene expression analysis. (C) Expression analysis of ABCDE classes genes in floral organs in (B) by using qRT-PCR. (D) The ABCDE model in longan is proposed based on the gene expression levels (bar heights) from (C).
In order to investigate the correlation between ABCDE genes and longan floral organs, we analyzed the expression patterns of certain genes in mature four whorls of floral organs (sepals, petals, stamens, carpel) and ovary by performing qRT-PCR (Figures 6B,C). Although twelve selected genes could be detected in most organs, they showed high specificity in different organs. For A-class, DlAP1 was mainly expressed in the first three whorls and DlMADS112 (DlFUL) was mainly expressed in carpel and ovary. DlMADS63 (DlPI) of B class was only detected in petal and stamen, DlMADS25 (DlAP3-1) also showed relatively high expression in these two organs, even though it could be found in all five organs. The only C class gene DlMADS49 (DlAG) was detected in sepal, stamen, carpel, and ovary, especially in stamen and carpel. Two D-class genes, DlMADS20 (DlSHP) and DlMADS66 (DlSTK) were expressed in reproductive organs (stamen and pistil) and showed relatively high expression levels in the ovary which contains ovules. Interestingly, four members of the E class showed different expression patterns in floral organs suggestive of diversified gene function. The expression levels of DlMADS74 and 113 (DlSEP1 and 3) were well-proportioned in five orangs. However, DlMADS108 (DlSEP2) showed a similar expression pattern with DlAP1, and DlMADS70 (DlSEP4) was preferentially expressed in the ovary like DlSTK.
Besides the ABCDE genes, AGL6 has been demonstrated to control floral development. In Arabidopsis flowers, AGL6 could be detected in all floral organs and in developing ovules (Koo et al., 2010). However, the transcripts of DlMADS18 (DlAGL6) were only detected in the first three whorls of organs (sepals, petals, and stamens), which suggested that this gene may not control longan ovule development. These results allowed us to predict the ABC(D)E model in longan (Figure 6D).
Tissue-Specific Expression Patterns of DlMADS
To investigate the expression pattern of longan MASD-box genes, previously published transcriptome data from multiple tissues including root, stem, leaf, dormant bud (before the emergence of floral primordia), flower bud, flower, young fruit, pericarp, pulp, and the seed of “SJ” longan were used for analysis (Lin et al., 2017; Jue et al., 2019). In line with previous studies (Kofuji et al., 2003; Duan et al., 2015), almost all genes from type I displayed low expression levels (FPKM value < 1) in nine tissues (Supplementary Table 8). In addition, DlMADS21 was highly expressed specifically in roots and DlMADS53 showed extremely high expression in pulp (Supplementary Figure 5), which suggests that these two genes play roles in root and fruit development, respectively. Moreover, DlMADS45 was primarily expressed in root, stem, and seeds. DlMADS65 was lowly expressed in multiple tissues. In the MIKC* group, DlMADS71, 72, 114 with more protein motifs in the K-box region than others showed higher expression in nearly all tissues, which provides evidence to the fundamental role of the K-box domain. The expression level of DlMADS27 and DlMADS58 in flower buds and flowers were higher than other organs, indicating crucial roles in the flowering process.
MIKCC type genes have important functions in the productive organs in plants (Becker and Theissen, 2003). In longan, genes of MIKCC displayed a wide range of expression levels in different tissues, among which 11 (30.6%), 18 (50.0%), 17 (47.2%), 17 (47.2%), 24 (66.7%), 20 (55.6%), 16 (44.5%), 16 (44.5%), 8 (22.2%), 14 (38.9%), and 16 (44.5%) were expressed (FPKM value > 1) in root, stem, leaf, dormant bud, flower bud, flower, young fruit, pulp, pericarp and seed, respectively (Supplementary Figure 6). Although expression patterns of DlMADS were conserved within each subfamily, several genes showed different expression levels in tested tissues such as DlAP1, DlFUL, and DlMADS109 in the A-class group. Our analysis showed that expression patterns of MIKCC genes in longan tissues were classified into four clusters (Figure 7A). In cluster 1, most genes show little expression but several genes such as SVP class members were expressed in leaf, stem, and dormant bud. DlMADS67 and DlMADS88 were specifically expressed in roots. In cluster 2, DlMADS was mainly expressed in vegetative tissues. DlMADS73 (DlFLC), DlFUL, and DlMADS29 (DlSVP) of cluster 3 displayed extensive-expression levels in nearly all tested tissues. In cluster 4, DlAGL6 and 9 genes of the ABCDE classes were mainly expressed in the reproductive organs including flower bud, flower, fruit, pericarp, and seed. From this finding, it is clear that MIKCC type DlMADS might perform significant roles in flowering and fruit development as opposed to other organs.
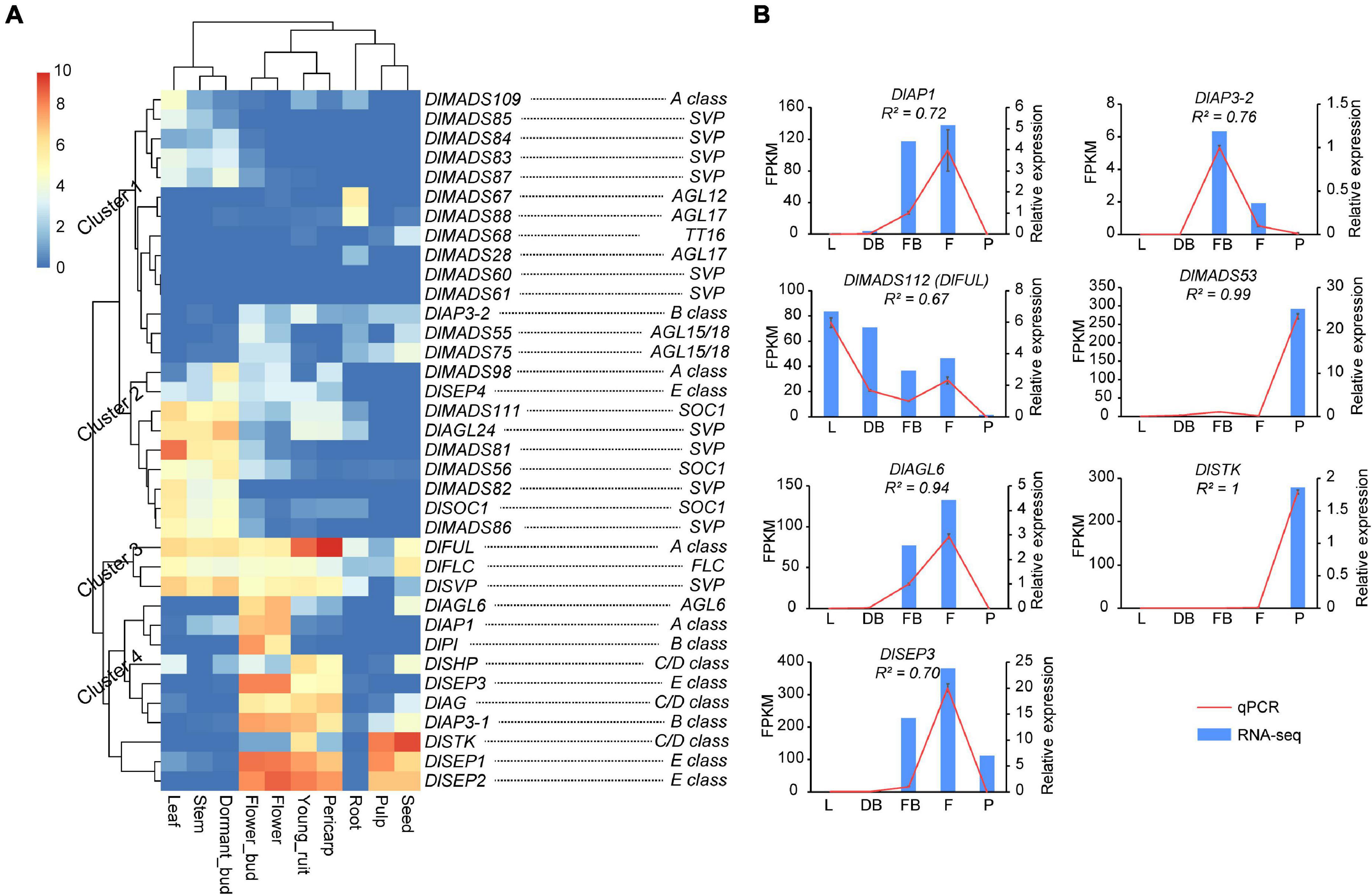
Figure 7. The expressional pattern of MIKCC genes in longan. (A) Expression heat maps of MIKCC genes in nine tissues (root, stem, leaf, dormant bud, flower bud, flower, fruit, pericarp, and seed) of “SJ” longan Expression values were normalized by log2(FPKM+1). (B) Detection of MADS-box genes in longan based on qRT-PCR. Six MADS-box genes were selected to detect the expression level in leaf (L), dormant bud (DB), floral bud (FB), flower (F), and pulp (F) of “XC” longan. Data were normalized to Actin1 and Actin2 genes, and the vertical bars indicate standard deviation.
To further survey the expression patterns of key DlMADS having reliable transcriptional support, quantitative RT-PCR was performed using total RNA isolated from leaf, dormant bud (before the emergence of floral primordia), floral bud (floral primordia), flower, and pulp (Figure 7B). Among seven selected genes, DlAP1, DlMADS57 (DlAP3-2), and DlAGL6 were only detected in floral or flower tissues, suggesting a unique function in flowering. DlSEP3 showed high expression in the floral bud, flower, and pulp tissues. Moreover, DlMADS53 showed specific high expression in pulp, consistent with the result of RNA-seq.
Differential Expression of DlMADS in Off-Season Flower Induction
The compound potassium chlorate (KClO3) has been widely used to induce flowering in longan since the last century (Subhadrabandhu and Yapwattanaphun, 2001). In this study, we detected the expression of MADS-box genes in apical buds at ten-time points using untreated controls (CK) and KClO3 treatments (see Methods). A total of 34 genes were expressed in more than one stage (FPKM > 1), including 26 MIKCC type genes, 3 MIKC* genes, and 5 genes of type I (Supplementary Table 9).
Among these, we found 14 genes showed significant differential expression levels (| logFC| > 2 and P-value < 0.05) after KClO3 treatment, including the ABCE model genes and other flowering time integrators (Figure 8A). DlAP1, DlMADS98, and DlSEP4 were up-regulated from day 41 to day 54, especially DlAP1 with the highest FPKM value. It is interesting to note that DlAP3-1, DlAP3-2, DlPI, DlAG, DlSEP1/2/3, and DlAGL6 showed the same patterns with upregulation on day 54. Therefore, these up-regulated genes could be considered as promoting factors for off-season flowering induction. In addition, DlMADS111, DlFLC, and DlMADS82 displayed a decreased pattern from day 35 to day 54, suggesting that these may inhibit longan flowering in this time period.
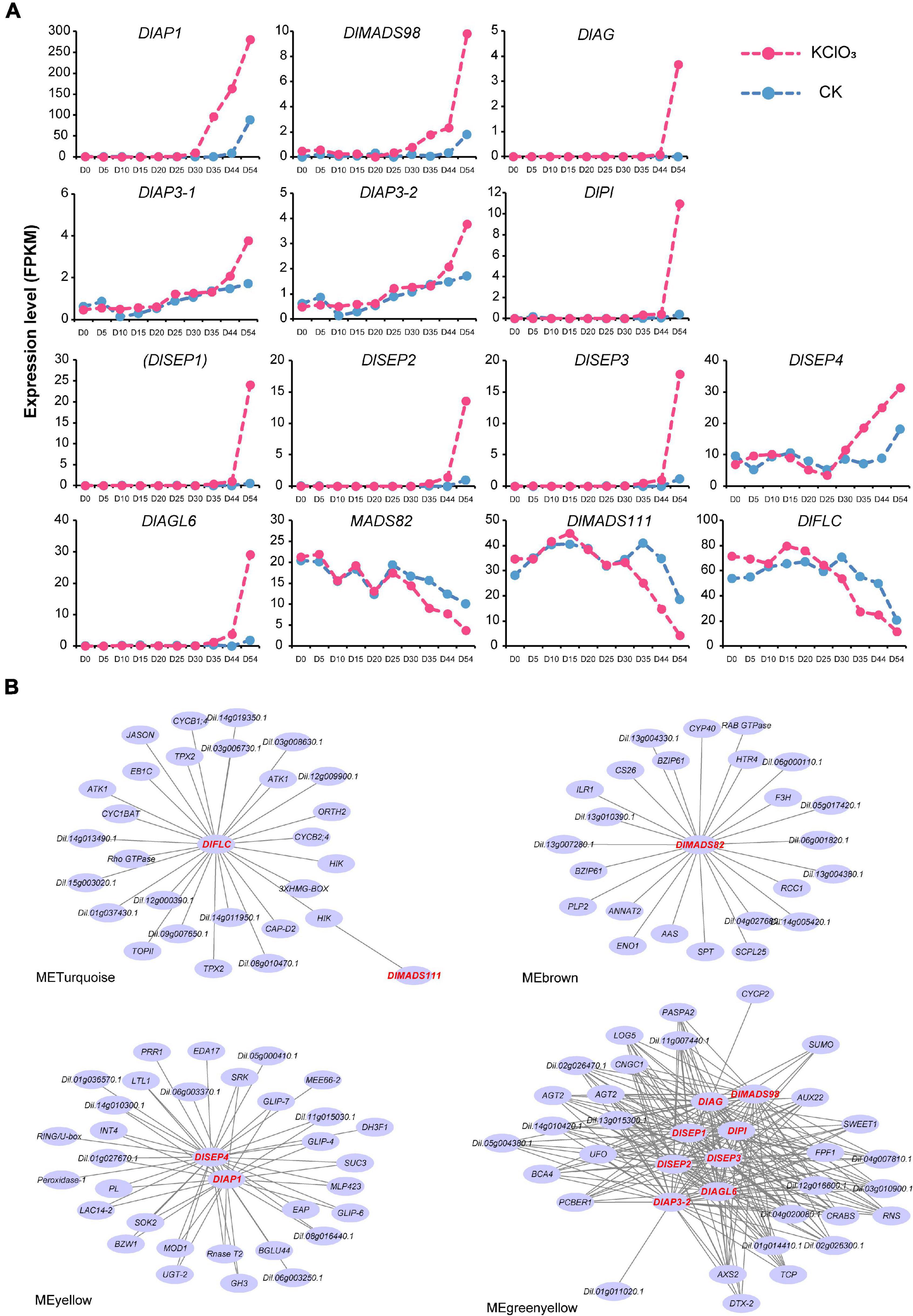
Figure 8. Transcript analysis of longan MADS-box genes in off-season flower induction. (A) Expression profile of differentially expressed MADS-box genes during 10 stages (day 0∼54) under CK and KClO3 treatment. (B) Co-expression networks of differential expression MADS-box genes during off-season flower induction. Each node represents a gene and the lines between nodes represent co-expression correlations.
To assess the relationships between DlMADS and other genes throughout the genome that may affect off-season flowering, the expressed genes were classified into 16 modules by weighted gene co-expression network analysis (WGCNA) (Supplementary Figure 7). The result showed that 14 differentially expressed MADS-box genes were distributed in four modules (MEyellow, MEbrown, MEturquoise, and MEgreenyellow). Based on the differentially expressed MADS-box genes, we constructed the co-expression networks and found that 113 genes were presumed to interact with them (Figure 8B). Based on the Arabidopsis thaliana flowering interactive database14, we found five flower-related genes interacted with MADS-box genes in the networks including UNUSUAL FLORAL ORGANS (UFO), TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP), CRABS, FLOWERING PROMOTING FACTOR1 (FPF1), and HOTHEAD (HTH) (Kania et al., 1997; Krolikowski et al., 2003; Hepworth et al., 2006; Gross et al., 2018; Zhao et al., 2020). Besides these flower-related genes, (UDP-Glycosyltransferase-2) UGT-2, (SPATULA) SPT, and SWEET1 were predicted to co-expressed with the MADS-box genes although have not yet been reported for the function related to flower. Whether these genes play functions in longan flower induction and development needs more experiments to confirm. In addition, 8 DlMADS (DlMADS98, DlAG, DlSEP1/2/3, DlPI, DlAP3-2, and DlAGL6) were found in the green-yellow module and DlAGL6 was the top hub gene with the highest kME value (0.99) indicating the central role in flower development (Figure 8B).
Moreover, we also compared the expression levels of DlMADS between “SX” and “SJ” during three floral transition stages (T1: dormant bud; T2: floral primordia; T3: floral organ formation). As a result, seven MADS-box genes were significantly differentially expressed in two accessions (Supplementary Figure 8). The expression of DlMADS111, DlAGL24, DlFLC, and DlSOC1 was higher in “SX_T1” than that in “SJ_T1,” and they were all downregulated in the following two stages except for DlSOC1. DlAP1 was upregulated in T1 and T2 in both accessions but showed higher expression levels in “SX” than that in “SJ.” On the contrary, the DlMADS98 displayed a higher expression in “SJ” than that in “SX” in the entire stages.
Expression Profiling of DlMADS During Fruit Development
It was reported that the MADS-box gene family have functions in fruit development and ripening (Moore et al., 2002), and our present study analyzed the expression pattern of DlMADS in six fruit developmental stages at 80, 100, 110, 120, 130, and 140 DAF (days after flowering of female flower) of 10 years “XC” tree by RNA-seq. 21 MADS-box genes were expressed in at least one stage of fruit development (Figure 9A and Supplementary Table 10). Ten genes (DlMADS75, 70, 59, 55, 72, 18, 114, 29, 49, 68) were only expressed at low levels in the first two stages of fruit development (DAF 80 and DAF 100). DlAP3-1, DlAP3-2, and DlMADS71 were down-regulated gradually during developmental stages and DlFLC showed the reverse trend. DlSTK and DlSHP exhibited the highest expression levels in DAF 80, while the expression of DlSTK rose in later stages and DlSHP decreased until it was not expressed at all. Increasing expression from DAF 80 to DAF 130 was found in DlSEP1 and DlSEP2 indicating that they may be involved in the fruit ripening process. It is significant that DlMADS53 (Mα) showed an extremely high expression level suggesting important functions in longan fruit development. The RNA-seq results of DlAP3-2, DlSTK, DlFLC, DlFUL, and DlMADS53 were corroborated by qRT-PCR. The relative expression levels were in agreement with the FPKM, confirming the accuracy of our transcriptomic analysis (R2 > 0.9; Figure 8B).
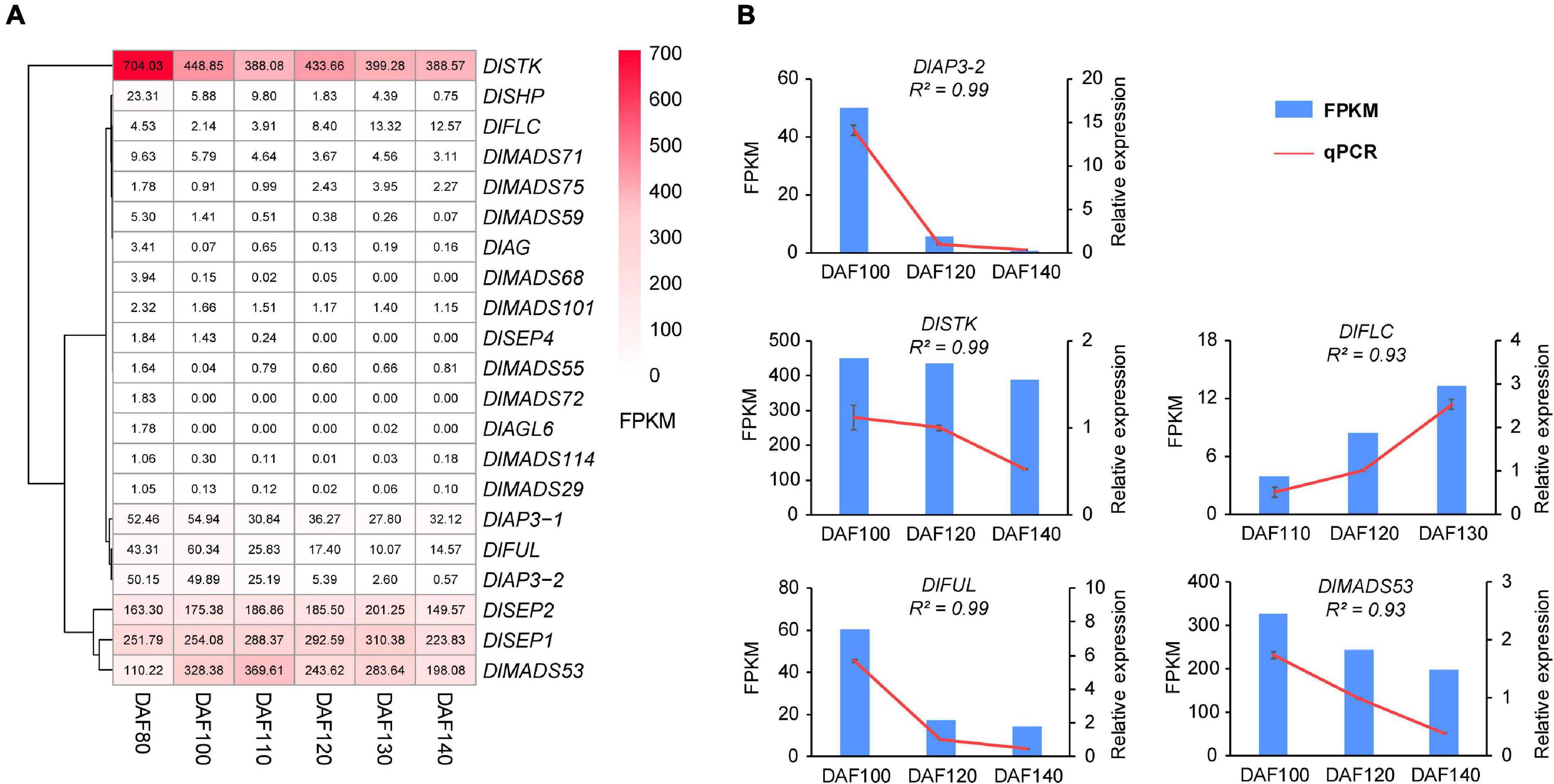
Figure 9. The expression pattern of differentially expressed DlMADS during the fruit development stage. (A) Heat maps based on FPKM for differentially expressed DlMADS at three fruit developmental stages (80∼140 days after flowering). (B) Detection of five differentially expressed DlMADS at three selected stages by using qRT-PCR. The bar and line graphs are derived from RNA-seq and qRT-PCR data, respectively.
Discussion
Given the developments in genome sequencing, there are numerous reports on the important MADS-box family in various plants such as Arabidopsis (Parenicova et al., 2003), rice (Arora et al., 2007), soybean (Fan et al., 2013), and banana (Liu et al., 2017). In this study, we completed the identification of the MADS-box gene in “SX” longan genome for the first time, and 114 MADS-box members were identified and classified into type I: Mα (39), Mβ (10), Mγ (14), and type II: MIKCC (36), MIKC* (14) (Supplementary Table 2). Combined with the relative representative plant species, it is possible to understand the evolution of MADS-box genes.
Evolution of MADS-Box Family
In this study, we identified the MADS-box genes from 15 representative species (Figure 1). The primitive lower plant, Chlamydomonas reinhardtii, only contained two genes belonging to both type I and type II, in accordance with the view that these two types of genes appeared by a duplication event before the divergence of plants and animals (Alvarez-Buylla et al., 2000). Overall, the number of MADS-box genes in Angiosperms is larger than that in Pteridophyta, Bryophyta, and Alga. A large expansion of MIKCC genes was found in basal angiosperms, given that flowering is a typical feature of high plants, we predict MIKCC genes may be endowed with new functions as plants evolved especially in flowering and development. In monocots and eudicots, type II genes showed relative conservation and the number of type I genes varied greatly in eudicots. It was found that longan contained the largest number of family genes, and we speculate that members of the MADS-box family were likely more active for tandem duplication after the γ-WGD event in longan than other eudicot species (Table 1). Furthermore, the degree of retention of type I genes seems to be higher than in other angiosperms. MIKC* genes expanded much more and they mainly originated from WGD/Segmental and dispersed duplication.
In Sapindaceae, the three examined genomes have no additional WGD after the γ-WGD, while, both longan and lichee have large numbers of MADS-box genes as the tandem duplication events frequently occurred in the longan genome. This phenomenon indicated that longan and lichee probably have complex molecular mechanisms for flower development due to the interaction of large sets of MADS-box genes for the regulation of gene networks. Sapindaceae could be classified into two subfamilies including Sapindoideae and Dodonaeoideae (Harrington et al., 2005). The divergence of the two subfamilies resulted in a great variation in the number of the type I genes particularly for Mβ and Mγ (Figure 1), which was probably caused by the functional redundancy of the two subgroups of MADS-box genes since these genes have few reports detailing their functions (Masiero et al., 2011). MIKCC genes were demonstrated to be involved in floral organ identity, the control of flowering time, and seed development. The relatively consistent number of MIKCC genes in Sapindaceae is likely attributed to the functional conservation of these genes in floral organ development.
Dof Family May Regulate MIKCC Genes in the Flowering Process
The potential transcription factors which control MIKCC genes were identified, and 26 TF binding sites were found in promoters of most MIKCC genes (Supplementary Table 7). The majority of TFs contain C2H2 zinc finger domain such as the Dof family, which is involved in photoperiod inducement of flowering and flowering development (Yanagisawa, 2002). In Arabidopsis, CYCLING DOF FACTOR (CDF) is the well-known inhibitory factor of flowering which can repress the expression of FT and CONSTANS (CO), it can be degraded by ubiquitin-protein KELCH REPEAT, F-BOX 1 (FKF1) (Imaizumi et al., 2005). OBF4 Binding Protein 3 (OBP3) and Cogwheel 1 (COG1) modulate phytochrome to further influence flowering (Park et al., 2003; Ward et al., 2005). Other research showed that maize transcription factor Zmdof1 regulates pollen formation and development (Chen et al., 2012). In rice, overexpressing OsDof12 led to early flowering and up regulation of OsMADS14 (Li et al., 2009). The previous study has demonstrated that Dof proteins were able to recognize (A/T)AAAG sequence as the core motif (Chen et al., 1996; Yanagisawa and Schmidt, 1999). In this study, similar binding sites of CDF5, COG1, FOXP1, OBP3, and other Dof genes were found in cis-motif 2, 3, 4, and 5 (Figure 5B). It could be speculated that Dof TFs may directly regulate MIKCC genes in longan, however, this speculation needs further investigation.
BBR-BASIC PENTACYSTEINE (BPC) identified in Arabidopsis is a small transcription factor family of seven members (Meister et al., 2004). BPC1 binds to the STK promoter at GA consensus sequences and regulates the expression of STK (Kooiker et al., 2005). In addition, chromatin immunoprecipitation analysis showed that BPCs also bind to the GA boxes in vivo, and their mutation can result in suppressed expression of the STK promoter (Simonini et al., 2012). Our data showed that three genes of the BBR-BPC family probably bind sequences in cis-motif 1, which provides evidence that BPC binding sites are important for STK and other MIKCC genes (Figure 5B). Moreover, AGAMOUS-LIKE 42 (AGL42) is a member of the SOC1 subfamily and controls the floral transition in the axillary meristem, and SOC1 directly binds to CArG-boxes from other SOC1 class genes (Dorca-Fornell et al., 2011). SOC1 plays a central role in the flowering pathway (Moon et al., 2003; Yoo et al., 2005), and a recent study showed that SOC1 also binds to its own locus and several flowering genes including the MADS-box members (Immink et al., 2012). Cis-motif 6 annotated as SOC1 binding sites was found in the promoter of 15 MIKCC genes (Supplementary Table 7), perhaps SOC1 class members regulate other MADS-box genes during the longan flowering process.
Functional Conservation of ABCDE Model Genes in Longan
Longan is monecious and very little is known on longan floral biology. We collected four whorls of organs and ovaries from male and female flowers to predict the ABCDE model of longan (Figure 6). The expression of A-class DlAP1 was notably higher in sepal than in other floral organs indicating the sepal identity of A function. Its close paralogs DlFUL was mainly expressed in the ovary. In Arabidopsis, FUL is required for valve differentiation and expansion in the ovary after fertilization (Zhang Y. et al., 2016). Generally, B class genes were restricted to expressed in the second and third whorls in model plants (Jack et al., 1992; Guo et al., 2015). In our results, DlPI was only detected in petals and stamens, which is in line with its expected function in petal and stamen identity specification. DlAP3-1 was also expressed in the other two organs (sepal and carpel), indicating neo-functionalization in longan after the divergence of PI and AP3. Regarding the C class, AG was the first floral organs identity gene found to determine the stamen and carpel in Arabidopsis (Yanofsky et al., 1990). DlAG displayed a higher expression level in the stamen and carpel than the other organs, suggesting that DlAG maintains the C function of specifying both male and female reproductive organs in longan. A recent study revealed that Arabidopsis AG regulated sepal senescence (Jibran et al., 2017). In this study, the transcripts of DlAG were also found in sepals, supporting a potential role for sepal senescence in longan. Both DlSHP and DlSTK were relatively highly expressed in the ovary, we, therefore, hypothesized that DlSHP and DlSTK perform class D functions in longan ovule development.
All four members (SEP1/2/3/4) play redundant functions in determining floral organ identity in Arabidopsis (Zahn et al., 2006). A similar result was found in longan with four DlSEP having different expression profiles in tested organs. DlSEP1 and 3 seem to account for a major position of E function due to their wider and higher expression level. DlSEP4 might regulate the ovule development together with DlFUL, DlSHP, and DlSTK in the early stage. AGL6 class was closely related to SEP in the phylogeny (Figure 2; Becker and Theissen, 2003), and orthologs of AGL6 in rice, maize, and petunia have been regarded as regulators for multiple floral organ identity like Class E function (Ohmori et al., 2009; Rijpkema et al., 2009). Our analysis showed that DlAGL6 possibly controls sepal, petal, and stamen of longan, not like TaAGL6 which is required for all four whorls of wheat floral organs (Kong et al., 2021). Overall, the expression patterns of ABCDE model genes in longan are similar to typical models in Eudicots (Figure 6D).
The MADS Genes Are Involved in Flowering and Respond to Potassium Chlorate
MADS-box genes play significant roles in flowering processes in plants. Expression analysis allowed us to predict the function of longan MADS-box genes. Our results revealed a diverse expression pattern in different longan tissues. Compared to previous studies (Kofuji et al., 2003), the expression of most genes in type I and MIKC* was not detected, with their function remaining unclear and it is possible they are only be expressed in specific cells or under specific conditions. Nevertheless, we found several genes such as DlMADS21 and DlMADS53 in root and pulp, suggesting these subfamilies with few studies never play a negligible role (Supplementary Figure 5). These type I and MIKC* genes should be investigated in future work.
Flowering is the key factor in yield determination (Bangerth, 2009), but the problem of poor production persisted for a long time owing to low flowering rates in longan. Normally, flower induction in longan needs a period of low temperature (Suttitanawat et al., 2012), and longan can respond to KClO3 for floral bud formation. The molecular mechanism associated with off-season flowering induction is of great value in scientific research and is of practical significance.
MIKC genes are of vital importance in reproductive development, especially the well know ABCDE model (Coen and Meyerowitz, 1991). In the present study, many MIKC genes seem to involve in the longan flowering process, as highly expressed genes usually play important roles in plant development. For example, DlAP1, DlPI, DlAGL6 were highly expressed specifically in flower buds and flowers (Figure 7), which highlights their key roles in flowering. DlAG, DlSHP, DlFUL, DlSEP1/2/3 showed high expression levels in multiple tissues, inferring their function in regions beyond just flowers. More importantly, 11 DlMADS were up-regulated after KClO3 treatment (Figure 8A), highlighting the involvement of MADS-box genes in off-season FI of longan.
Normally, LFY was considered as a determinant of flower initiation that plays a central role in flower pathways (Fornara et al., 2010). Recently, LFY was confirmed as a pioneer transcription factor to promote floral transition via upregulating AP1 (Jin et al., 2021). However, the ortholog of LFY in longan showed no apparent changes during flower induction in “SJ” (Jue et al., 2019). On further investigation, we found the expression of DlAP1 in “SJ” was significantly lower than in “SX” especially in the floral primordia stage, even though it was induced during floral transition in both accessions (Supplementary Figure 8). This result implied that the molecular pathways of flowering in “SJ” may differ from those in the other longan accessions. One A-class gene, DlMADS98, showed a higher expression level in “SJ.” It is likely that the DlMADS98 displayed function complementation with DlAP1 in off-season FI.
In Arabidopsis, FLC and its five homologs, MAF1-5 co-regulates flowering time by temperature-dependent alternative splicing (Scortecci et al., 2001; Bastow et al., 2004). Flowering induction in longan also requires low temperature (vernalization), thus, FLC might be functionally constrained in longan. DlFLC showed extensive-expression especially in longan vegetative and reproductive tissues and slightly decreased during off-season flowering. Significantly, the expression of DlFLC was lower in “SJ” than that in “SX” during the whole stages (Supplementary Figure 8). Given that FLC is regarded as a repressor in different flowering pathways (Rouse et al., 2002), we could speculate that DlFLC may be an inhibitor of off-season FI in longan. SVP is another important gene in response to environmental temperature to regulate plant flowering. The SVP class showed obvious expansion and the collinearity analysis implied it experienced tandem gene duplications like the DAM genes in Rosaceae (Supplementary Figure 3). RNA-seq and qRT-PCR analysis showed that SVP class genes such as DlMADS82 tend to be expressed in leaf tissue and dormant buds, and this gene was also suppressed by KClO3 (Figures 7, 8). This leads us to speculate that SVP class genes could regulate vegetative development and be responsible for apical bud formation in response to dormancy.
Moreover, SOC1 responds to multiple floral induction pathways including photoperiod, vernalization, and autonomous (Lee et al., 2000). It acts downstream of FLOWERING LOCUS T (FT) and regulates the expression of LFY which promotes transcription of AP1 (Immink et al., 2012). On the other hand, it was also identified as a FLC suppressor (Lee et al., 2000). Contrary to the previous study, DlSOC1 exhibited little expression in flowers or floral buds and was down-regulated after KClO3 treatment. It is also notable that DlMADS111 (SOC1 clade) and DlSOC1 showed lower expression levels in “SJ.” Whether SOC1 clade genes have altered functions in longan remains to be elucidated. From these results, it seems that genes in DlFLC, DlSVP, and DlSOC1 have similar functions in maintaining vegetative growth and inhibiting floral transition, with further studies being required to verify the function of these genes.
Potential Roles of Longan MADS Genes in Fruit Development
Longan fruit, which contains many nutrients such as vitamins, carbohydrates, mineral elements, and amino acids, plays an important role in human anti-cancer, anti-aging, and brain development (Rangkadilok et al., 2005). The growth and development of longan fruit usually take 100–150 days. During this period, there are obvious changes in fruit size, nutritional composition, sweetness, and flavor, which are regulated by external environmental conditions, plant hormones, and many genes. In the present study, 9 of the MADS-box genes were highly expressed in fruit (Figure 9). FRUITFULL FUL is known for its role in controlling flowering time, carpel identity, fruit development, and leaf morphology (Coen and Meyerowitz, 1991; Gu et al., 1998; Torti et al., 2012). Functional studies in multiple species indicated that FUL orthologs have a conserved function for regulating fruit development even in fruits with diverse morphologies and structures (Jaakola et al., 2010; Bemer et al., 2012; Pabón-Mora et al., 2012; Fujisawa et al., 2014). Not surprisingly, in longan, DlFUL showed extremely high expression levels in young fruit, pericarp, and flowers, suggesting that this gene also has a conserved function for regulating the flowering time and fruit development (Figures 7, 9).
Recent research showed that ectopic expression of MdPI controls apple fruit tissue growth and shape, indicating that the functions of B class genes are not limited to the development of petals and stamen (Yao et al., 2018). Analysis of Sl-AGL11 overexpression in tomatoes revealed that the D class gene could regulate early fleshy fruit development (Huang et al., 2017). In this study, two DlAP3 genes were not only expressed in flowers, but also expressed in fruit (seed, pericarp, and pulp) with different patterns during fruit development. The transcript of DlSTK has detected all the examined fruit organs with high levels in the pulp. It is likely that longan B and D class genes experienced different evolutionary fates from Arabidopsis and played functions for fruit development. In addition, a preview study has reported that LeMADS-RIN is necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus (Vrebalov et al., 2002). Arabidopsis SEP genes were the homologs of LeMADS-RIN. We determined that DlSEP1 and 2 were extremely highly expressed with expression tending to increase during fruit development, which suggests DlSEP1 and 2 may act as regulators of longan fruit ripening, with further studies being required to verify their function. It is noteworthy that a type I gene DlMADS53 only expressed in the pulp of longan and remained at high expression levels during fruit ripening. To the best of our knowledge, there is no study about the orthologs of DlMADS53 in plants. This result provides a new clue for the functional study of the MADS-box in the plants.
Conclusion
This study identified 114 MADS-box genes including 63 types I and 51 types II genes in the longan genome. Thirteen subfamilies of MIKC genes were identified through phylogenetic analysis and the main difference between MIKCC and MIKC* proteins were the K-box domain. Analysis of cis-elements revealed that Dof transcription factors might directly regulate the MIKCC genes. Phylogeny and gene expressional analysis showed that the composition and expression of the ABCDE genes were conserved in longan. We also provided expression information for DlMADS in vegetative and reproductive tissues and this data led us to conclude that MIKCC genes play crucial roles in flowering. Several genes such as DlSTK, DlSEP1/2, and DlMADS53 could be involved in fruit growth and ripening. In summary, this comprehensive analysis provided the basic resources to examine the molecular regulation of MADS-box genes in the longan reproduction process.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA741049.
Author Contributions
JZ conceived the study and designed the experiments. BW, WH, YF, XF, JF, TZ, SZ, RM, and JZ carried out the experiments and analyzed the data. BW and JZ wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Key Research and Developmental Program of China (2018YFD1000104), the Project of Fujian Academy of Agricultural Sciences (AB2017-4), and the National Natural Science Foundation of China (31701884).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.813798/full#supplementary-material
Footnotes
- ^ http://www.arabidopsis.org/
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://gigadb.org/
- ^ https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi
- ^ http://Pfam.sanger.ac.uk/
- ^ https://web.expasy.org/protparam/
- ^ http://gsds.gao-lab.org/
- ^ http://meme-suite.org/tools/meme
- ^ https://www.ebi.ac.uk/interpro/search/sequence/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://jaspar.genereg.net
- ^ https://meme-suite.org/meme/tools/tomtom
- ^ https://sg.idtdna.com/pages/tools/primerquest
- ^ http://www.phytosystems.ulg.ac.be/florid/
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Alvarez-Buylla, E. R., Pelaz, S., Liljegren, S. J., Gold, S. E., Burgeff, C., Ditta, G. S., et al. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. U.S.A. 97, 5328–5333. doi: 10.1073/pnas.97.10.5328
Aono, N., and Hasebe, M. (2006). Functional analysis of MADS-box genes in Physcomitrella patens. Plant Cell Physiol. Suppl. 2006, 498–498. doi: 10.14841/jspp.2006.0.498.0
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 8:242. doi: 10.1186/1471-2164-8-242
Bangerth, K. F. (2009). Floral induction in mature, perennial angiosperm fruit trees: similarities and discrepancies with annual/biennial plants and the involvement of plant hormones. Sci. Hortic. 122, 153–163. doi: 10.1016/j.scienta.2009.06.014
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Becker, A., and Theissen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489. doi: 10.1016/s1055-7903(03)00207-0
Bemer, M., Karlova, R., Ballester, A. R., Tikunov, Y. M., Bovy, A. G., Wolters-Arts, M., et al. (2012). The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24, 4437–4451. doi: 10.1105/tpc.112.103283
Bielenberg, D. G., Wang, Y., Li, Z., Zhebentyayeva, T., Fan, S., Reighard, G. L., et al. (2008). Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genom. 4, 495–507. doi: 10.1007/s11295-007-0126-9
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chang, Q. (2010). Study on Relationship Between Off-Season Floral Induction in Longan and Nutrition of the Carbon and Nitrogen. Master’s thesis. Fuzhou: Fujian Agriculture & Forestry University.
Chen, S., Liu, H., Chen, W., Xie, D., and Zheng, S. (2009). Proteomic analysis of differentially expressed proteins in longan flowering reversion buds. Sci. Hortic. 122, 275–280. doi: 10.1016/j.scienta.2009.05.015
Chen, W., Chao, G., and Singh, K. B. (1996). The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 10, 955–966. doi: 10.1046/j.1365-313x.1996.10060955.x
Chen, X., Wang, D., Liu, C., Wang, M., Wang, T., Zhao, Q., et al. (2012). Maize transcription factor Zmdof1 involves in the regulation of Zm401 gene. Plant Growth Regul. 66, 271–284. doi: 10.1007/s10725-011-9651-5
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
De Bodt, S., Raes, J., Van de Peer, Y., and Theissen, G. (2003). And then there were many: MADS goes genomic. Trends Plant Sci. 8, 475–483. doi: 10.1016/j.tplants.2003.09.006
Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., and Yanofsky, M. F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. doi: 10.1016/j.cub.2004.10.028
Dorca-Fornell, C., Gregis, V., Grandi, V., Coupland, G., Colombo, L., and Kater, M. M. (2011). The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 67, 1006–1017. doi: 10.1111/j.1365-313X.2011.04653.x
Duan, W., Song, X., Liu, T., Huang, Z., Ren, J., Hou, X., et al. (2015). Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol. Genet. Genom. 290, 239–255. doi: 10.1007/s00438-014-0912-7
Eddy, S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Fan, C. M., Wang, X., Wang, Y. W., Hu, R. B., Zhang, X. M., Chen, J. X., et al. (2013). Genome-wide expression analysis of soybean MADS genes showing potential function in the seed development. PLoS One 8:e62288. doi: 10.1371/journal.pone.0062288
Fornara, F., de Montaigu, A., and Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550–550.e552. doi: 10.1016/j.cell.2010.04.024
Fujisawa, M., Shima, Y., Nakagawa, H., Kitagawa, M., Kimbara, J., Nakano, T., et al. (2014). Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26, 89–101. doi: 10.1105/tpc.113.119453
Goto, K., and Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Gross, T., Broholm, S., and Becker, A. (2018). CRABS CLAW acts as a bifunctional transcription factor in flower development. Front. Plant Sci. 9:835. doi: 10.3389/fpls.2018.00835
Gu, Q., Ferrándiz, C., Yanofsky, M. F., and Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517.
Guo, S., Sun, B., Looi, L. S., Xu, Y., Gan, E. S., Huang, J., et al. (2015). Co-ordination of flower development through epigenetic regulation in two model species: rice and Arabidopsis. Plant Cell Physiol. 56, 830–842. doi: 10.1093/pcp/pcv037
Harrington, M., Edwards, K., Johnson, S., Chase, M., and Gadek, P. (2005). Phylogenetic inference in Sapindaceae sensu lato using plastid matK and rbcL DNA sequences. Syst. Bot. 30, 366–382. doi: 10.1600/0363644054223549
Hepworth, S. R., Klenz, J. E., and Haughn, G. W. (2006). UFO in the Arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression. Planta 223, 769–778. doi: 10.1007/s00425-005-0138-3
Hou, X.-J., Liu, S.-R., Khan, M. R. G., Hu, C.-G., and Zhang, J.-Z. (2013). Genome-wide identification, classification, expression profiling, and SSR marker development of the MADS-Box gene family in citrus. Plant Mol. Biol. Report. 32, 28–41. doi: 10.1007/s11105-013-0597-9
Huang, B., Routaboul, J. M., Liu, M., Deng, W., Maza, E., Mila, I., et al. (2017). Overexpression of the class D MADS-box gene Sl-AGL11 impacts fleshy tissue differentiation and structure in tomato fruits. J. Exp. Bot. 68, 4869–4884. doi: 10.1093/jxb/erx303
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., and Kay, S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Immink, R. G. H., Posé, D., Ferrario, S., Ott, F., Kaufmann, K., Valentim, F. L., et al. (2012). Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 160, 433–449. doi: 10.1104/pp.112.202614
Jaakola, L., Poole, M., Jones, M. O., Kämäräinen-Karppinen, T., Koskimäki, J. J., Hohtola, A., et al. (2010). A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 153, 1619–1629. doi: 10.1104/pp.110.158279
Jack, T., Brockman, L. L., and Meyerowitz, E. M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. doi: 10.1016/0092-8674(92)90144-2
Jibran, R., Tahir, J., Cooney, J., Hunter, D. A., and Dijkwel, P. P. (2017). Arabidopsis AGAMOUS regulates sepal senescence by driving jasmonate production. Front. Plant Sci. 8:2101. doi: 10.3389/fpls.2017.02101
Jin, R., Klasfeld, S., Zhu, Y., Fernandez Garcia, M., Xiao, J., Han, S.-K., et al. (2021). LEAFY is a pioneer transcription factor and licenses cell reprogramming to floral fate. Nat. Commun. 12:626. doi: 10.1038/s41467-020-20883-w
Jofuku, K. D., den Boer, B. G., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Johansen, B., Pedersen, L. B., Skipper, M., and Frederiksen, S. (2002). MADS-box gene evolution-structure and transcription patterns. Mol. Phylogenet. Evol. 23, 458–480. doi: 10.1016/s1055-7903(02)00032-5
Jue, D., Sang, X., Liu, L., Shu, B., Wang, Y., Liu, C., et al. (2019). Comprehensive analysis of the longan transcriptome reveals distinct regulatory programs during the floral transition. BMC Genom. 20:126. doi: 10.1186/s12864-019-5461-3
Kania, T., Russenberger, D., Peng, S., Apel, K., and Melzer, S. (1997). FPF1 promotes flowering in Arabidopsis. Plant Cell 9, 1327–1338. doi: 10.1105/tpc.9.8.1327
Kaufmann, K., Melzer, R., and Theissen, G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. doi: 10.1016/j.gene.2004.12.014
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kofuji, R., Sumikawa, N., Yamasaki, M., Kondo, K., Ueda, K., Ito, M., et al. (2003). Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 20, 1963–1977. doi: 10.1093/molbev/msg216
Kong, X., Wang, F., Geng, S., Guan, J., Tao, S., Jia, M., et al. (2021). The wheat AGL6-like MADS-box gene is a master regulator for floral organ identity and a target for spikelet meristem development manipulation. Plant Biotechnol. J. 20, 75–88. doi: 10.1111/pbi.13696
Koo, S. C., Bracko, O., Park, M. S., Schwab, R., Chun, H. J., Park, K. M., et al. (2010). Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J. 62, 807–816. doi: 10.1111/j.1365-313X.2010.04192.x
Kooiker, M., Airoldi, C. A., Losa, A., Manzotti, P. S., Finzi, L., Kater, M. M., et al. (2005). BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 17, 722–729. doi: 10.1105/tpc.104.030130
Kovaka, S., Zimin, A. V., Pertea, G. M., Razaghi, R., Salzberg, S. L., and Pertea, M. (2019). Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20:278. doi: 10.1186/s13059-019-1910-1
Krolikowski, K. A., Victor, J. L., Wagler, T. N., Lolle, S. J., and Pruitt, R. E. (2003). Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J. 35, 501–511. doi: 10.1046/j.1365-313X.2003.01824.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lai, Z.-X., Chen, C. L., Zeng, L. H., and Chen, Z. G. (2000). Somatic embryogenesis in longan (Dimocarpus longan Lour.). Somat. Embryogen. Woody Plants 6, 415–431. doi: 10.1007/978-94-017-3030-3_13
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 9:559. doi: 10.1186/1471-2105-9-559
Lee, H., Suh, S. S., Park, E., Cho, E., Ahn, J. H., Kim, S. G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. doi: 10.1101/gad.813600
Lee, J., and Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Li, D., Yang, C., Li, X., Gan, Q., Zhao, X., and Zhu, L. (2009). Functional characterization of rice OsDof12. Planta 229, 1159–1169. doi: 10.1007/s00425-009-0893-7
Lin, Y., Min, J., Lai, R., Wu, Z., Chen, Y., Yu, L., et al. (2017). Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. Gigascience 6, 1–14. doi: 10.1093/gigascience/gix023
Liu, J., Zhang, J., Zhang, J., Miao, H., Wang, J., Gao, P., et al. (2017). Genome-wide analysis of banana MADS-box family closely related to fruit development and ripening. Sci. Rep. 7:3467. doi: 10.1038/s41598-017-03897-1
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
Manochai, P., Sruamsiri, P., Wiriya-alongkorn, W., Naphrom, D., Hegele, M., and Bangerth, F. (2005). Year around off season flower induction in longan (Dimocarpus longan, Lour.) trees by KClO3 applications: potentials and problems. Sci. Hortic. 104, 379–390. doi: 10.1016/j.scienta.2005.01.004
Masiero, S., Colombo, L., Grini, P. E., Schnittger, A., and Kater, M. M. (2011). The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 23, 865–872. doi: 10.1105/tpc.110.081737
Mei, Z. Q., Fu, S. Y., Yu, H. Q., Yang, L. Q., Duan, C. G., Liu, X. Y., et al. (2014). Genetic characterization and authentication of Dimocarpus longan Lour. using an improved RAPD technique. Genet. Mol. Res. 13, 1447–1455. doi: 10.4238/2014.March.6.3
Meister, R. J., Williams, L. A., Monfared, M. M., Gallagher, T. L., Kraft, E. A., Nelson, C. G., et al. (2004). Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 37, 426–438. doi: 10.1046/j.1365-313X.2003.01971.x
Messenguy, F., and Dubois, E. (2003). Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316, 1–21. doi: 10.1016/s0378-1119(03)00747-9
Moon, J., Suh, S. S., Lee, H., Choi, K. R., Hong, C. B., Paek, N. C., et al. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623. doi: 10.1046/j.1365-313x.2003.01833.x
Moore, S., Vrebalov, J., Payton, P., and Giovannoni, J. (2002). Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato. J. Exper. Bot. 53, 2023–2030. doi: 10.1093/jxb/erf057
Ohmori, S., Kimizu, M., Sugita, M., Miyao, A., Hirochika, H., Uchida, E., et al. (2009). MOSAIC FLORAL ORGANS1, an AGL6-like mads box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21, 3008–3025. doi: 10.1105/tpc.109.068742
Pabón-Mora, N., Ambrose, B. A., and Litt, A. (2012). Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 158, 1685–1704. doi: 10.1104/pp.111.192104
Parenicova, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Park, D. H., Lim, P. O., Kim, J. S., Cho, D. S., Hong, S. H., and Nam, H. G. (2003). The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 34, 161–171. doi: 10.1046/j.1365-313x.2003.01710.x
Passmore, S., Maine, G. T., Elble, R., Christ, C., and Tye, B. K. (1988). Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. J. Mol. Biol. 204, 593–606. doi: 10.1016/0022-2836(88)90358-0
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Project, A. G. (2013). The Amborella genome and the evolution of flowering plants. Science 342:1241089. doi: 10.1126/science.1241089
Rangkadilok, N., Worasuttayangkurn, L., Bennett, R. N., and Satayavivad, J. (2005). Identification and quantification of polyphenolic compounds in longan (Euphoria longana Lam.) fruit. J. Agric. Food Chem. 53, 1387–1392. doi: 10.1021/jf0403484
Riechmann, J. L., and Meyerowitz, E. M. (1997). MADS domain proteins in plant development. Biol. Chem. 378, 1079–1101.
Rijpkema, A. S., Zethof, J., Gerats, T., and Vandenbussche, M. (2009). The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J. 60, 1–9. doi: 10.1111/j.1365-313X.2009.03917.x
Rouse, D. T., Sheldon, C. C., Bagnall, D. J., Peacock, W. J., and Dennis, E. S. (2002). FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J. 29, 183–191. doi: 10.1046/j.0960-7412.2001.01210.x
Scortecci, K. C., Michaels, S. D., and Amasino, R. M. (2001). Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26, 229–236. doi: 10.1046/j.1365-313x.2001.01024.x
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Simonini, S., Roig-Villanova, I., Gregis, V., Colombo, B., Colombo, L., and Kater, M. M. (2012). Basic pentacysteine proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis. Plant Cell 24, 4163–4172. doi: 10.1105/tpc.112.103952
Solovyev, V., Kosarev, P., Seledsov, I., and Vorobyev, D. (2006). Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7:S10. doi: 10.1186/gb-2006-7-s1-s10
Sommer, H., Beltrán, J. P., Huijser, P., Pape, H., Lönnig, W. E., Saedler, H., et al. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 9, 605–613.
Sringarm, K., Potchanasin, P., Sruamsiri, P., and Bangerth, K. F. (2009). Floral induction (FI) in longan (Dimocarpus longan, Lour.) trees—The possible participation of endogenous hormones: II. Low temperature and potassium chlorate effects on hormone concentrations in and their export out of leaves. Sci. Hortic. 122, 295–300. doi: 10.1016/j.scienta.2008.11.031
Subhadrabandhu, S., and Yapwattanaphun, C. (2001). Regulation off-season flowering of longan in Thailand. Acta Hortic. 558, 193–198. doi: 10.17660/ActaHortic.2001.558.26
Suttitanawat, P., Sruamsiri, P., and Sringarm, K. (2012). Changes in cytokinins concentrations during induction period of longan cv. Daw in sand culture. J. Agric. Technol. 8, 2353–2362.
Theissen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. doi: 10.1016/s1369-5266(00)00139-4
Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Münster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149.
Theissen, G., Kim, J. T., and Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. doi: 10.1007/bf02337521
Theissen, G., and Saedler, H. (2001). Plant biology. Floral quartets. Nature 409, 469–471. doi: 10.1038/35054172
Torti, S., Fornara, F., Vincent, C., Andrés, F., Nordström, K., Göbel, U., et al. (2012). Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a Leucine-rich repeat protein that promotes flowering. Plant Cell 24, 444–462. doi: 10.1105/tpc.111.092791
Van de Peer, Y. (2004). Computational approaches to unveiling ancient genome duplications. Nat. Rev. Genet. 5, 752–763. doi: 10.1038/nrg1449
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. doi: 10.1126/science.1068181
Wang, L., Yin, X., Cheng, C., Wang, H., Guo, R., Xu, X., et al. (2015). Evolutionary and expression analysis of a MADS-box gene superfamily involved in ovule development of seeded and seedless grapevines. Mol. Genet. Genom. 290, 825–846. doi: 10.1007/s00438-014-0961-y
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Ward, J. M., Cufr, C. A., Denzel, M. A., and Neff, M. M. (2005). The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17, 475–485. doi: 10.1105/tpc.104.027722
Winterhagen, P., Tiyayon, P., Samach, A., Hegele, M., and Wünsche, J. N. (2013). Isolation and characterization of FLOWERING LOCUS T subforms and APETALA1 of the subtropical fruit tree Dimocarpus longan. Plant Physiol. Biochem. 71, 184–190. doi: 10.1016/j.plaphy.2013.07.013
Yanagisawa, S. (2002). The Dof family of plant transcription factors. Trends Plant Sci. 7, 555–560. doi: 10.1016/S1360-1385(02)02362-2
Yanagisawa, S., and Schmidt, R. J. (1999). Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17, 209–214. doi: 10.1046/j.1365-313X.1999.00363.x
Yang, Y., Fanning, L., and Jack, T. (2003). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33, 47–59. doi: 10.1046/j.0960-7412.2003.01473.x
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Yao, J.-L., Xu, J., Tomes, S., Cui, W., Luo, Z., Deng, C., et al. (2018). Ectopic expression of the PISTILLATA homologous MdPI inhibits fruit tissue growth and changes fruit shape in apple. Plant J. 2:e00051. doi: 10.1002/pld3.51
Yoo, S. K., Chung, K. S., Kim, J., Lee, J. H., Hong, S. M., Yoo, S. J., et al. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778. doi: 10.1104/pp.105.066928
Zahn, L. M., Feng, B., and Ma, H. (2006). Beyond the ABC-model: regulation of floral homeotic genes. Adv. Bot. Res. 44, 163–207.
Zhang, H. N., Shi, S. Y., Li, W. C., Shu, B., Liu, L. Q., Xie, J. H., et al. (2016). Transcriptome analysis of ‘Sijihua’ longan (Dimocarpus longan L.) based on next-generation sequencing technology. J. Hortic. Sci. Biotechnol. 91, 180–188. doi: 10.1080/14620316.2015.1133539
Zhang, Y., Shen, Y. Y., Wu, X. M., and Wang, J. B. (2016). The basis of pod dehiscence: anatomical traits of the dehiscence zone and expression of eight pod shatter-related genes in four species of Brassicaceae. Biol. Plant. 60, 343–354. doi: 10.1007/s10535-016-0599-1
Zhang, L., Chen, F., Zhang, X., Li, Z., Zhao, Y., Lohaus, R., et al. (2020). The water lily genome and the early evolution of flowering plants. Nature 577, 79–84. doi: 10.1038/s41586-019-1852-5
Zhao, Y., Broholm, S. K., Wang, F., Rijpkema, A. S., Lan, T., Albert, V. A., et al. (2020). TCP and MADS-Box transcription factor networks regulate heteromorphic flower type identity in Gerbera hybrida. Plant Physiol. 184, 1455–1468. doi: 10.1104/pp.20.00702
Keywords: longan, MADS-box, ABCDE model, KClO3, flower, fruit development
Citation: Wang B, Hu W, Fang Y, Feng X, Fang J, Zou T, Zheng S, Ming R and Zhang J (2022) Comparative Analysis of the MADS-Box Genes Revealed Their Potential Functions for Flower and Fruit Development in Longan (Dimocarpus longan). Front. Plant Sci. 12:813798. doi: 10.3389/fpls.2021.813798
Received: 12 November 2021; Accepted: 16 December 2021;
Published: 27 January 2022.
Edited by:
Shunquan Lin, South China Agricultural University, ChinaReviewed by:
Hui Song, Qingdao Agricultural University, ChinaQilin Tian, Minnan Normal University, China
Lisha Shen, Temasek Life Sciences Laboratory, Singapore
Copyright © 2022 Wang, Hu, Fang, Feng, Fang, Zou, Zheng, Ming and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jisen Zhang, emppc2VuQGZhZnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Baiyu Wang
Baiyu Wang Wenshun Hu
Wenshun Hu Yaxue Fang
Yaxue Fang Xiaoxi Feng
Xiaoxi Feng Jingping Fang
Jingping Fang Tengyue Zou4
Tengyue Zou4 Ray Ming
Ray Ming Jisen Zhang
Jisen Zhang